Ras
Ras belongs to a class of small GTPase and is involved in transmitting signals within cells.
Products for Ras
- Cat.No. Product Name Information
-
GC64657
(E/Z)-ZINC09659342
(E/Z)-ZINC09659342 is an inhibitor of Lbc-RhoA interaction.
-
GC19541
(rac)-Antineoplaston A10
(rac)-Antineoplaston A10 is the racemate of Antineoplaston A10

-
GC69904
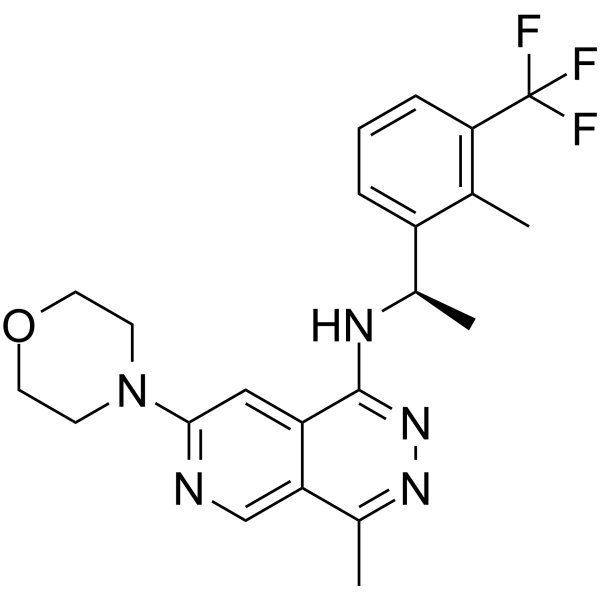
(S)-JDQ-443
(S)-JDQ-443 is an isomer of JDQ-443. JDQ-443 is an orally effective and selective covalent KRAS G12C inhibitor. JDQ-443 has anti-tumor activity.
-
GC32770
1A-116
1A-116, a potent Rac1 inhibitor, is specific for W56 residues, can prevent EGF-induced Rac1 activation and block Rac1-P-Rex1 interaction. 1A-116 can induce apoptosis and inhibit cell proliferation, migration and cycle progression in a concentration-dependent manner. 1A-116 also demonstrates a high antimetastatic activity in vivo.
-
GC15478
6H05
K-Ras inhibitor
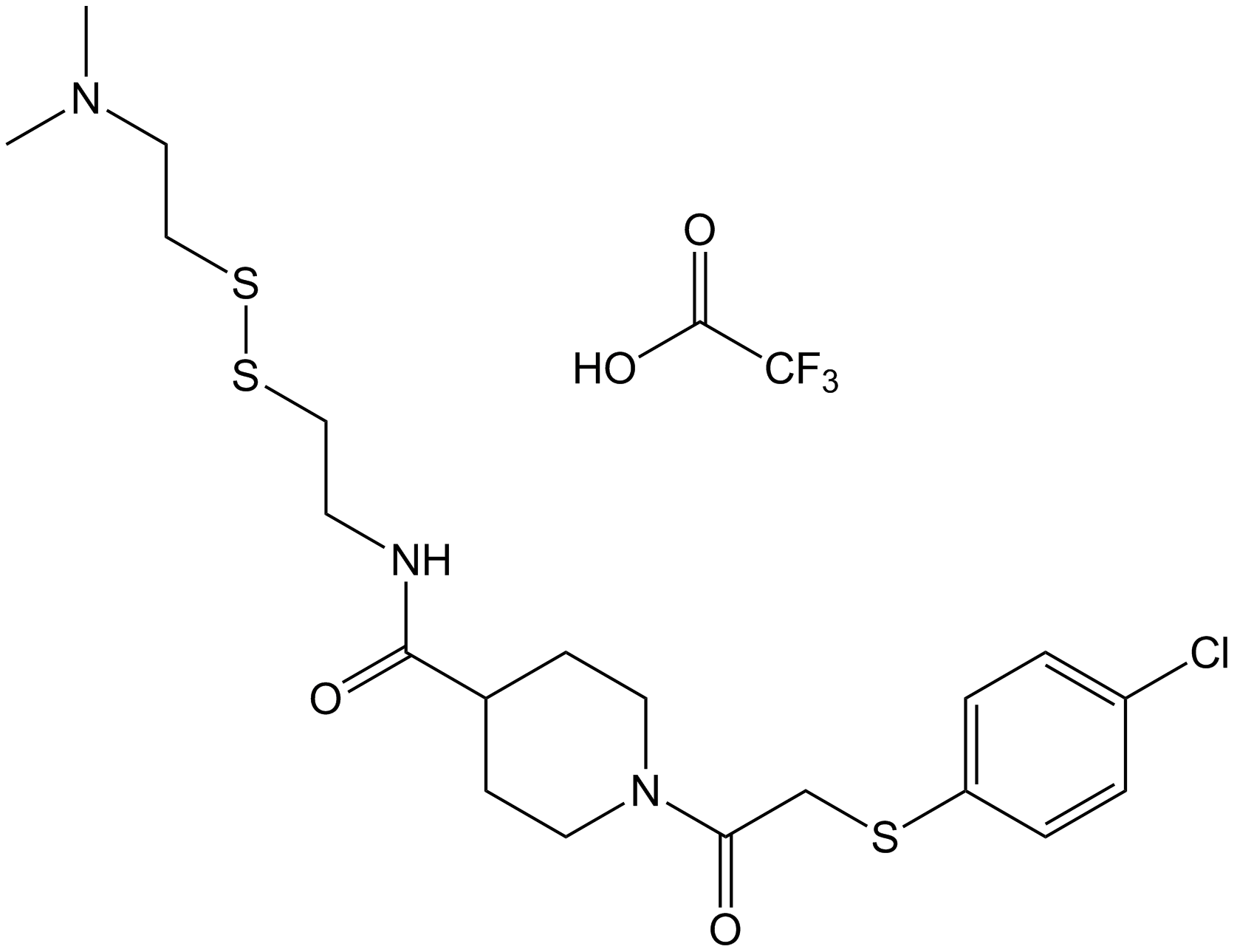
-
GC60533
6H05 (TFA)
An allosteric inhibitor of oncogenic K-Ras(G12C)
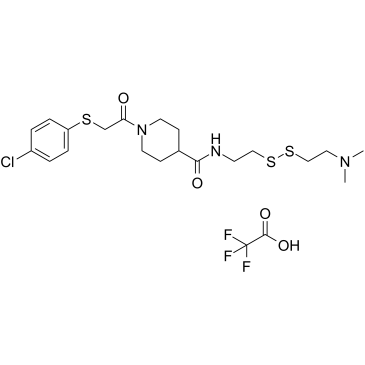
-
GC34427
6H05 trifluoroacetate (K-Ras inhibitor)
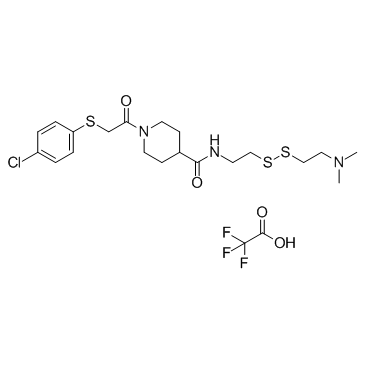
-
GC19546
AMG-510 racemate
Sotorasib (AMG-510) racemate is the racemate of Sotorasib (AMG-510). AMG-510 is a potent, orally bioavailable, and selective KRAS G12C covalent inhibitor with anti-tumor activity.
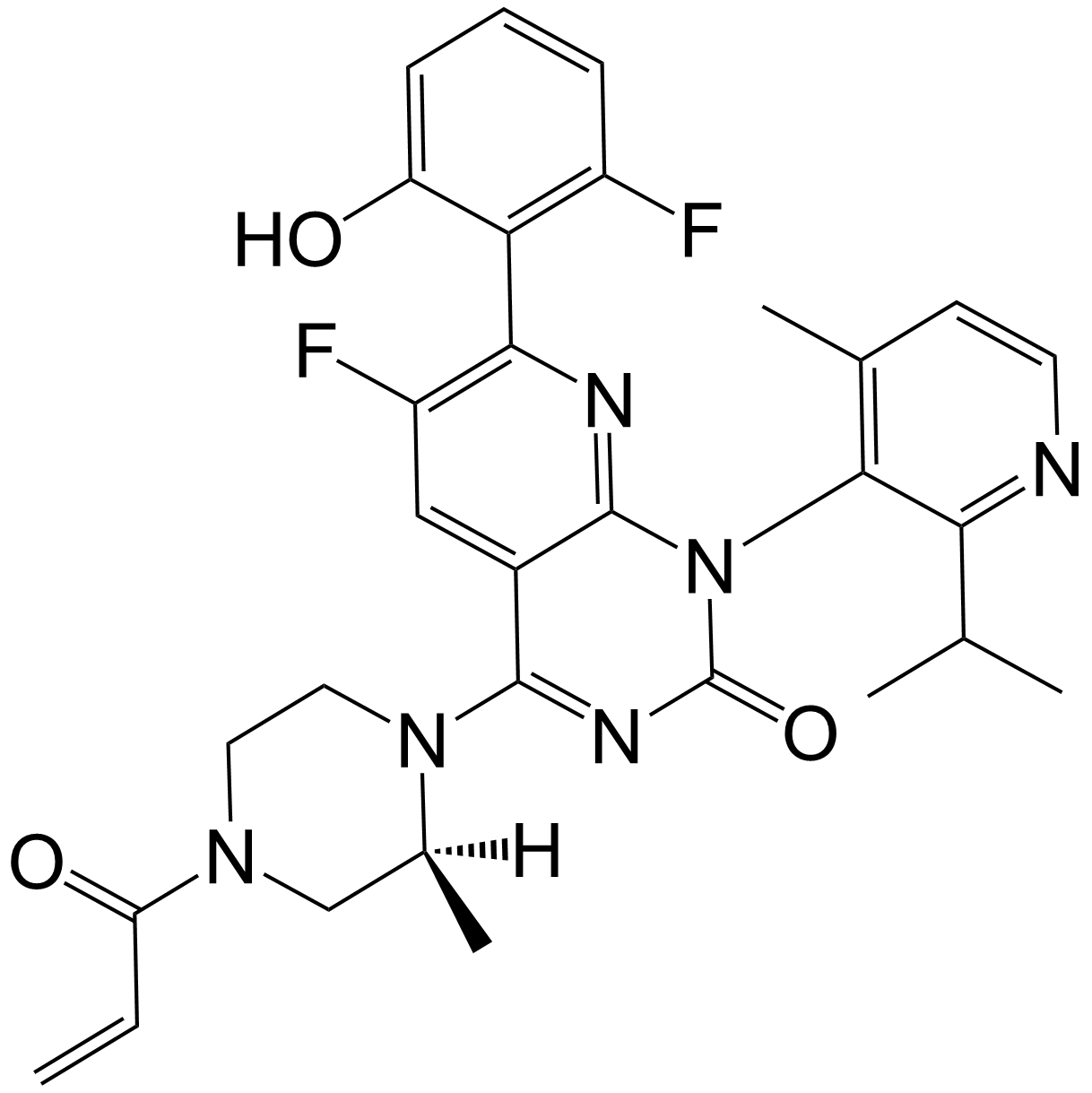
-
GC35361
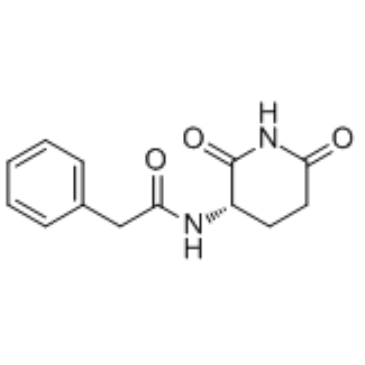
Antineoplaston A10
Antineoplaston A10, a naturally occurring substance in human body, is a Ras inhibitor potentially for the treatment of glioma, lymphoma, astrocytoma and breast cancer.
-
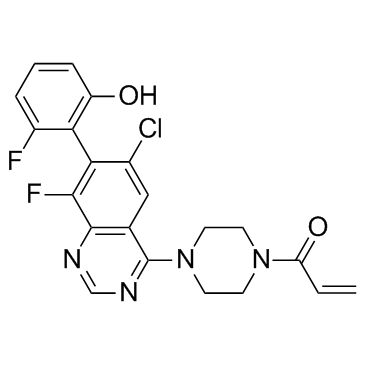
GC34114
ARS-1323
ARS-1323, the racemate of ARS-1620, is a novel inhibitor of mutant K-ras G12C extracted from patent WO 2015054572 A1.
-
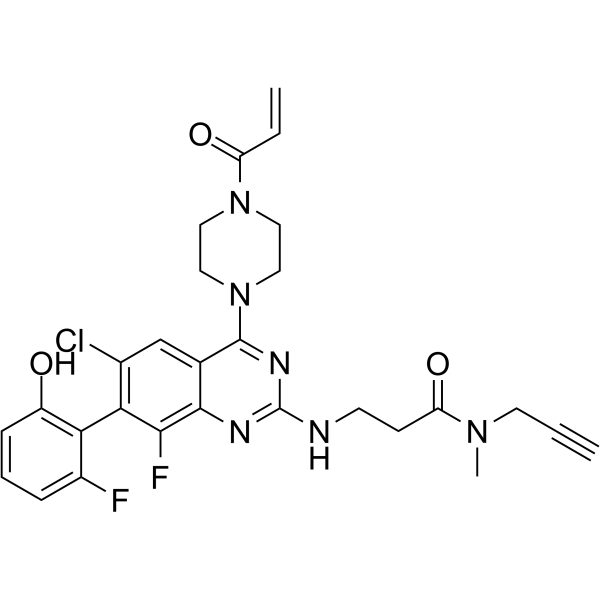
GC65480
ARS-1323-alkyne
ARS-1323-alkyne, a switch-II pocket (S-IIP) inhibitor, is a conformational specific chemical reporter of KRASG12C nucleotide state in living cells.
-
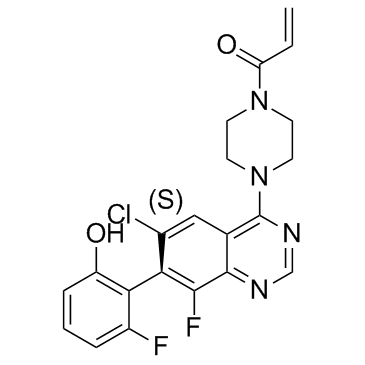
GC34055
ARS-1620
ARS-1620 is an atropisomeric selective KRASG12C inhibitor with desirable pharmacokinetics.
-
GC34091
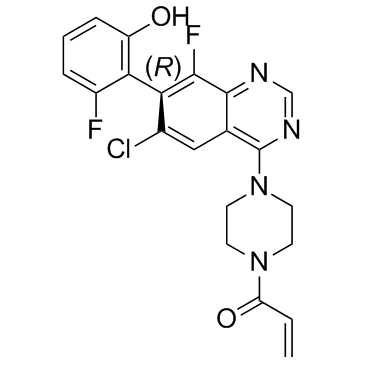
ARS-1630
ARS-1630, a less active enantiomer of ARS-1620, is a novel inhibitor of mutant K-ras G12C extracted from patent WO 2015054572 A1.
-
GC19037
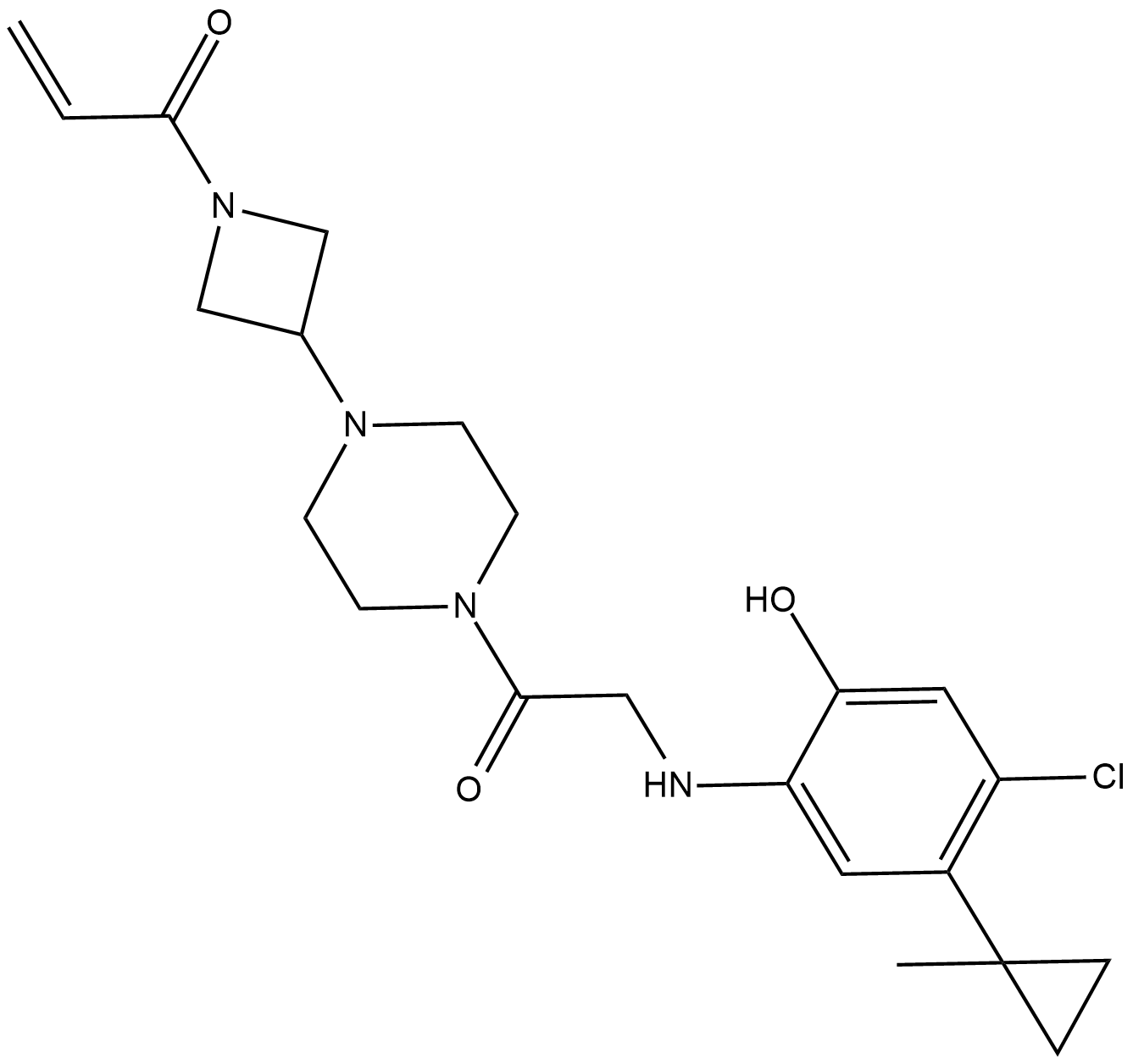
ARS-853
ARS-853 is a selective, covalent KRASG12C inhibitor with an IC50 of 2.5 uM.
-
GC64341
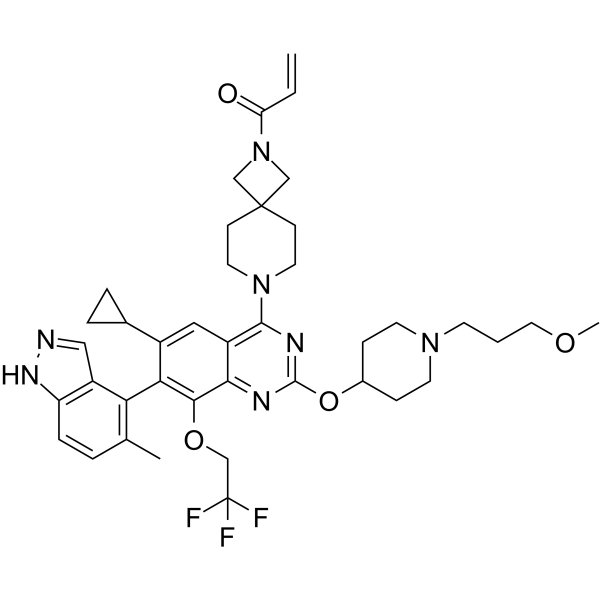
ASP2453
ASP2453 is a potent, selective and covalent KRAS G12C inhibitor. ASP2453 inhibits the Son of Sevenless (SOS)-mediated interaction between KRAS G12C and Raf with an IC50 value of 40 nM.
-
GC46893
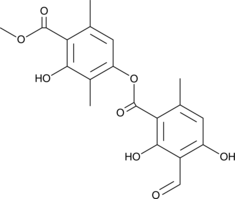
Atranorin
A depside lichen metabolite with diverse biological activities
-
GC33186
BAY-293
An inhibitor of the K-Ras-SOS1 protein-protein interaction
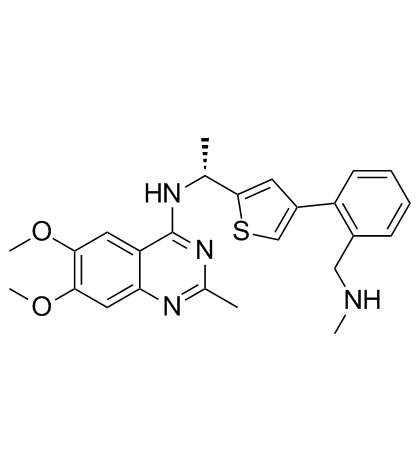
-
GC33340
BDP9066
BDP9066 is a potent and selective myotonic dystrophy-related Cdc42-binding kinase MRCK inhibitor with an IC50 of 64 nM for MRCKβ in SCC12 cells, Ki values of 0.0136 nM and 0.0233 nM for MRCKα/β in house determinations, respectively. BDP9066 has therapeutic effect on skin cancer by reducing substrate phosphorylation.
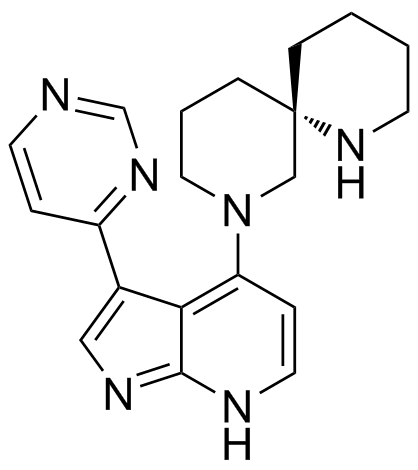
-
GC35513
BI-2852
BI-2852 is a KRAS inhibitor for the switch I/II pocket (SI/II-pocket) by structure-based drug design with nanomolar affinity. BI-2852 is mechanistically distinct from covalent KRASG12C inhibitor (binds to switch II pocket) and binds ten-fold more strongly to active KRASG12D?versus KRASwt?(740?nM vs 7.5?μM). BI-2852 blocks GEF, GAP, and effector interactions with KRAS, leading to inhibition of downstream signaling and an antiproliferative effect in KRAS mutant cells.
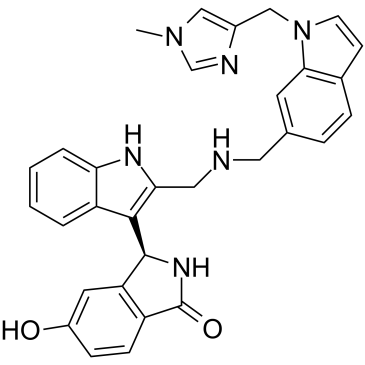
-
GC68760
BI-2865
BI-2865 is a non-covalent pan-KRAS inhibitor. BI-2865 binds to KRAS WT, G12C, G12D, G12V and G13D mutants with KD values of 6.9, 4.5, 32, 26 and 4.3 nM respectively. BI-2865 inhibits the proliferation of BaF3 cells expressing KRAS G12C, G12D or G12V mutations (average IC50: approximately 140 nM).
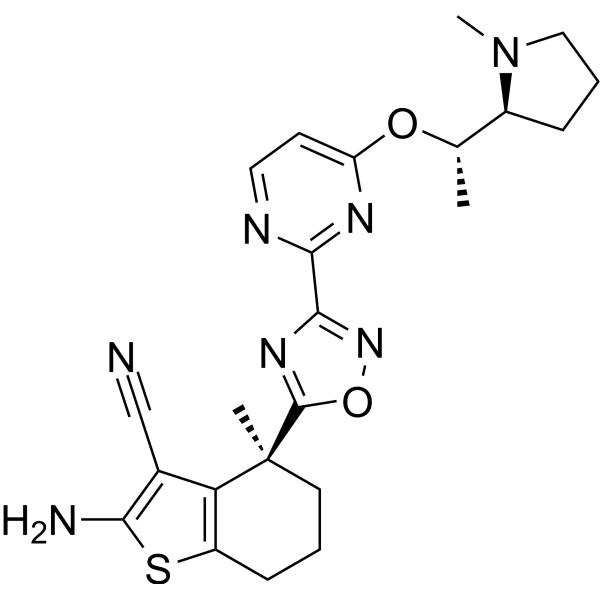
-
GC10612
BQU57
Derivative of RBC8
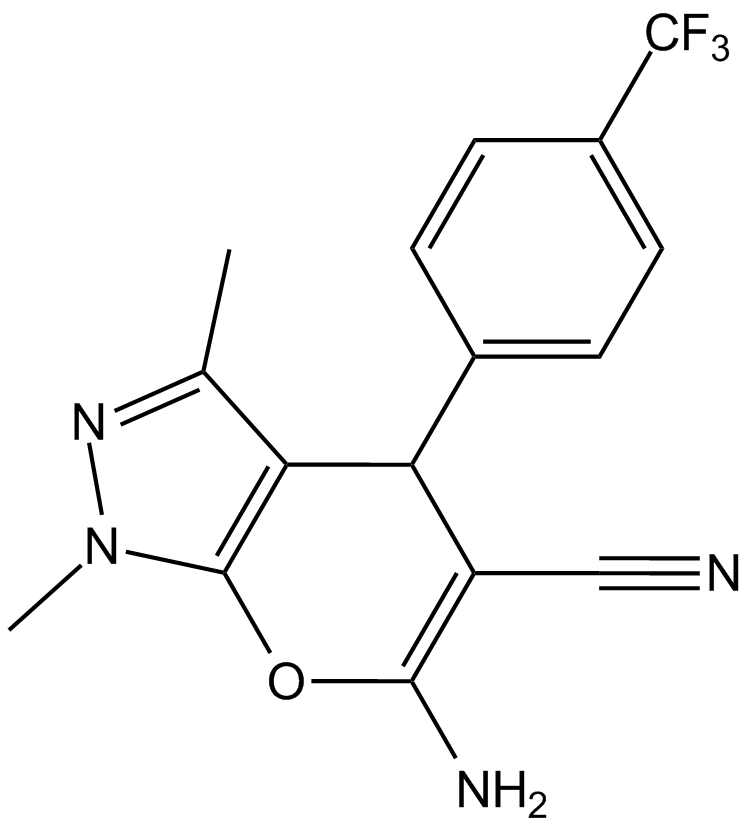
-
GC16692
Casin
GTPase Cdc42 inhibitor
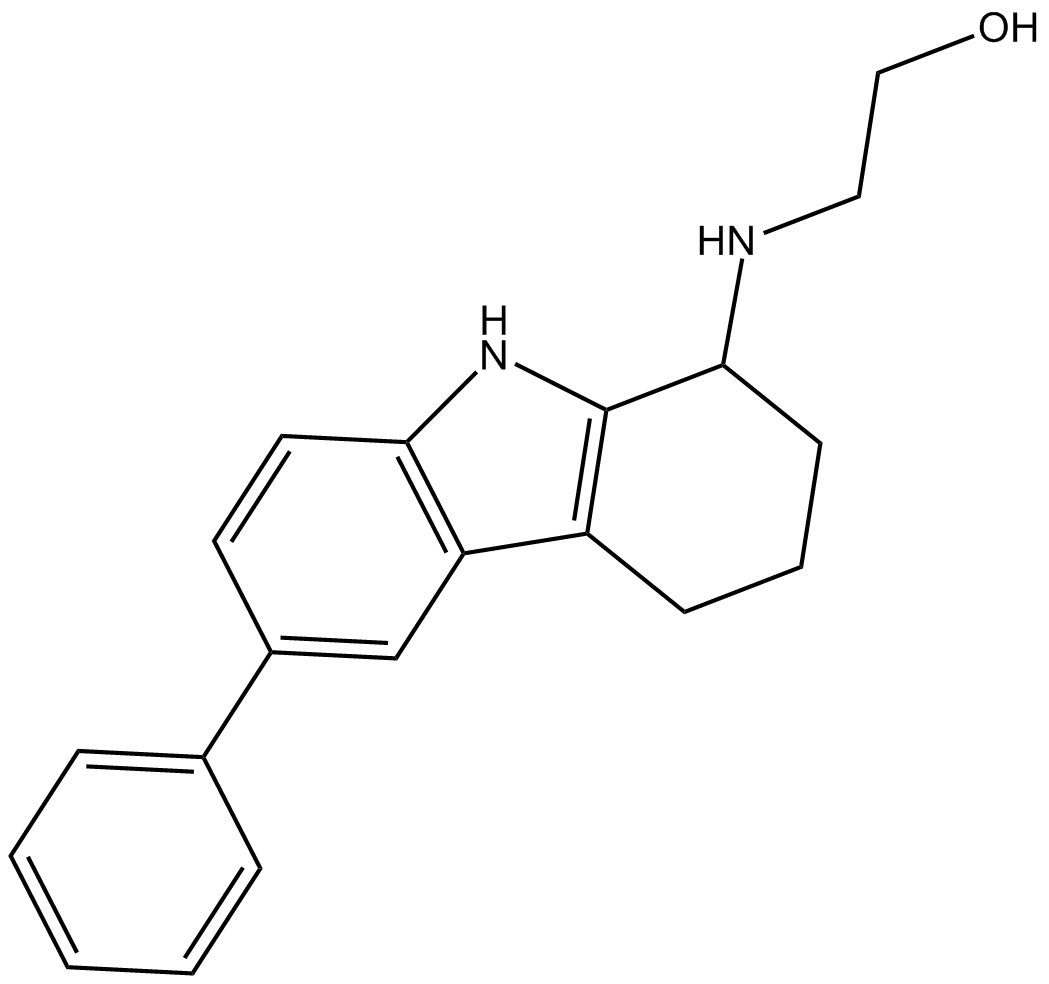
-
GC14266
CCG 203971
Antifibrotic agent
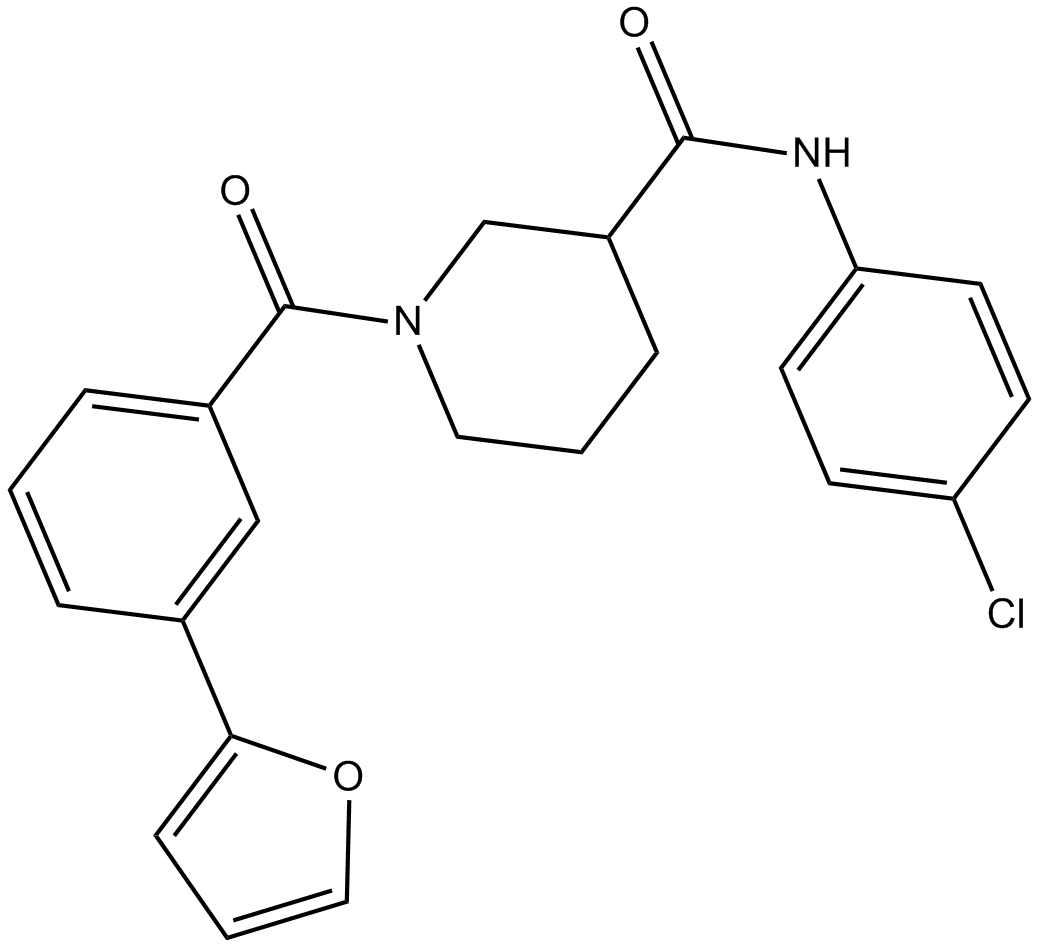
-
GC12795
CCG-1423
RhoA inhibitor
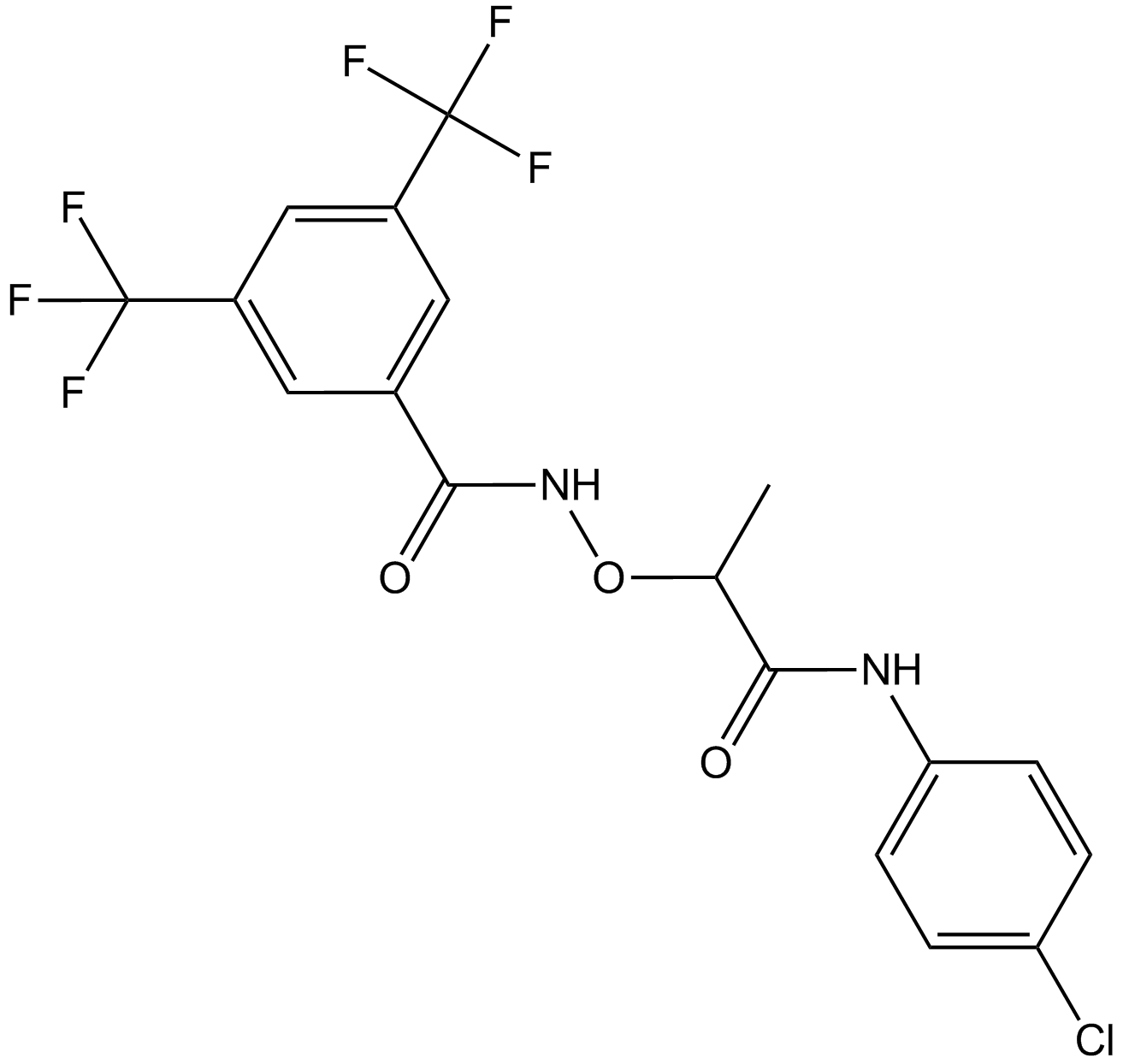
-
GC38898
CCG-222740
CCG-222740 is an orally active and selective Rho/myocardin-related transcription factor (MRTF) pathway inhibitor. CCG-222740 is also a potent inhibitor of alpha-smooth muscle actin protein expression. CCG-222740 effectively reduces fibrosis in skin and blocks melanoma metastasis.
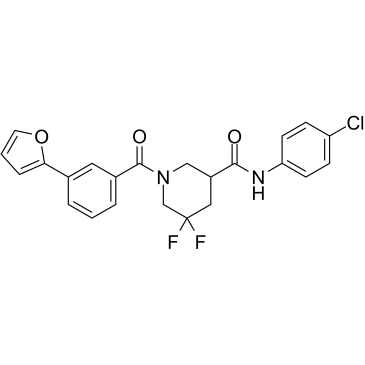
-
GC12948
CID-1067700
competitive inhibitor of nucleotide binding by Ras-related GTPases
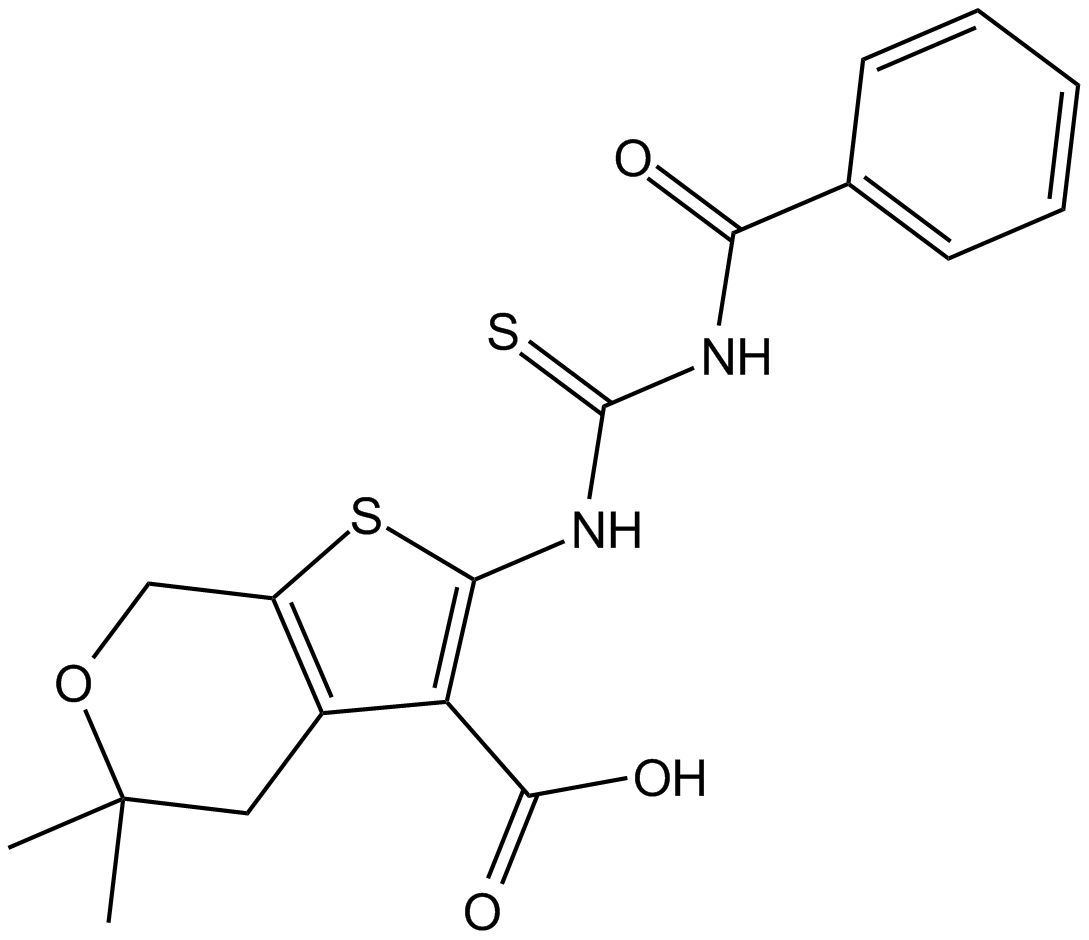
-
GC62347
CMC2.24
CMC2.24 (TRB-N0224), an orally active tricarbonylmethane agent, is effective against pancreatic tumor in mice by inhibiting Ras activation and its downstream effector ERK1/2 pathway.
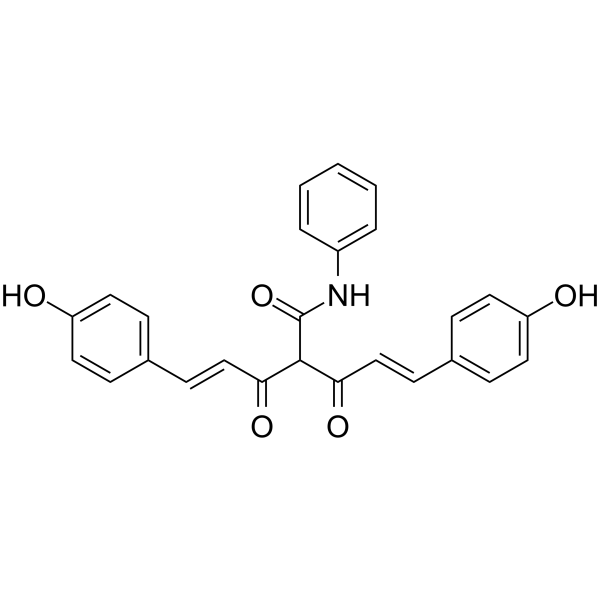
-
GC13076
Deltarasin
A KRAS inhibitor
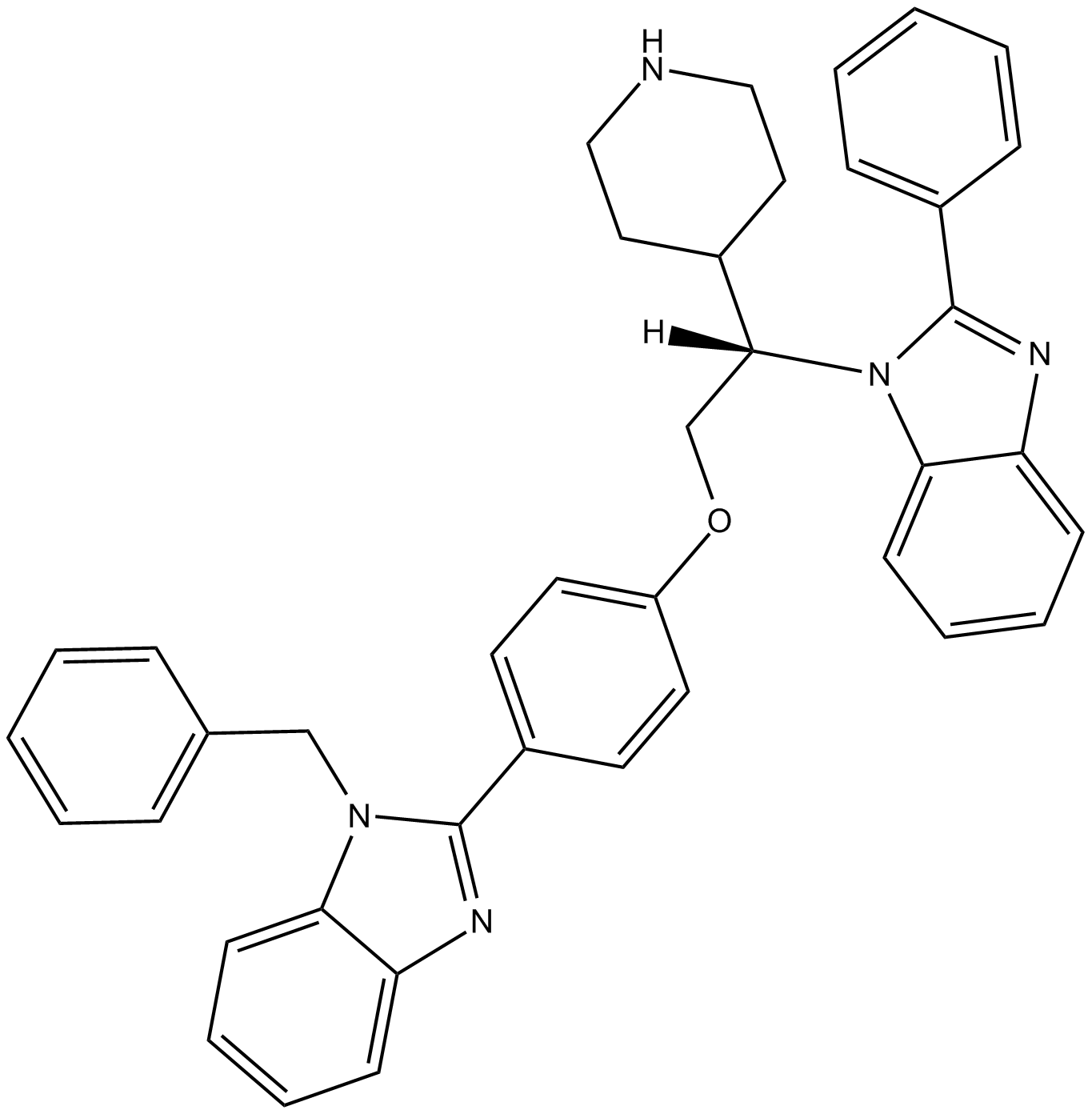
-
GC12060
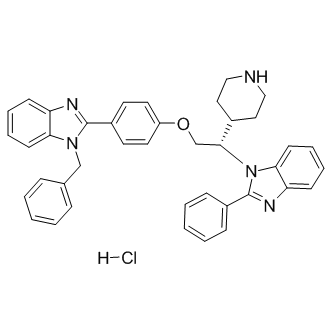
Deltarasin hydrochloride
inhibitor of KRAS-PDEδ interaction, potent and selective
-
GC35859
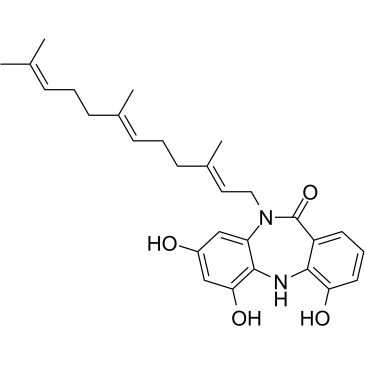
Diazepinomicin
Diazepinomicin (TLN-4601) is a secondary metabolite produced by Micromonospora sp. Diazepinomicin (TLN-4601) inhibits the EGF-induced Ras-ERK MAPK signaling pathway and induces apoptosis. An anti-tumor agent for K-Ras mutant models.
-
GC65270
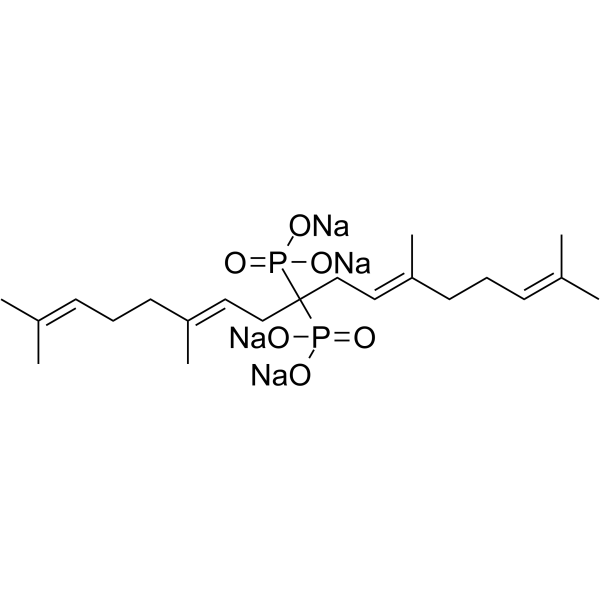
Digeranyl bisphosphonate
Digeranyl bisphosphonate (DGBP) is a potent geranylgeranylpyrophosphate (GGPP) synthase inhibitor, which inhibits geranylgeranylation of Rac1.
-
GC10030
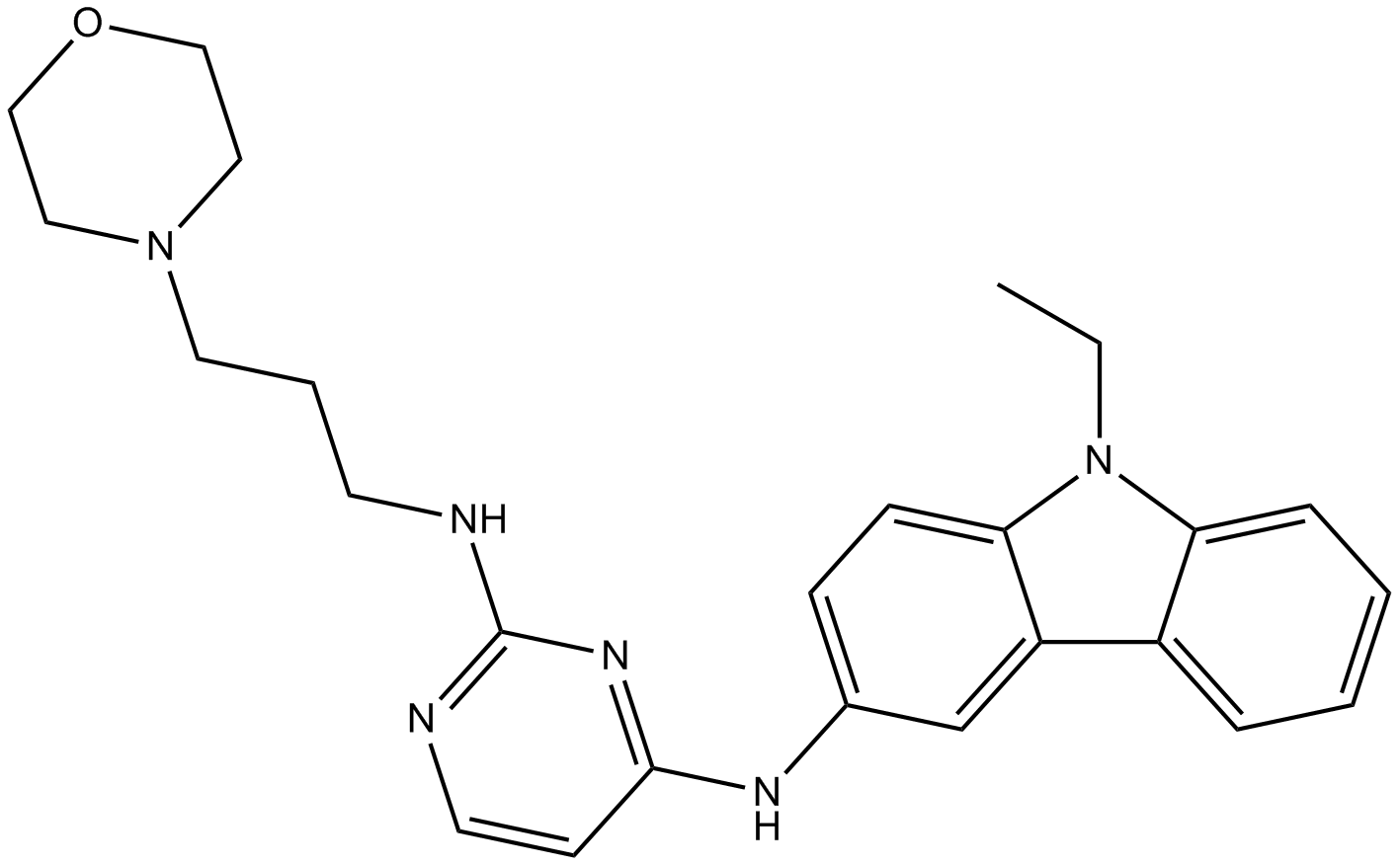
EHop-016
Rac1/Rac3 GTPase inhibitor,potent and specific
-
GC10660
EHT 1864
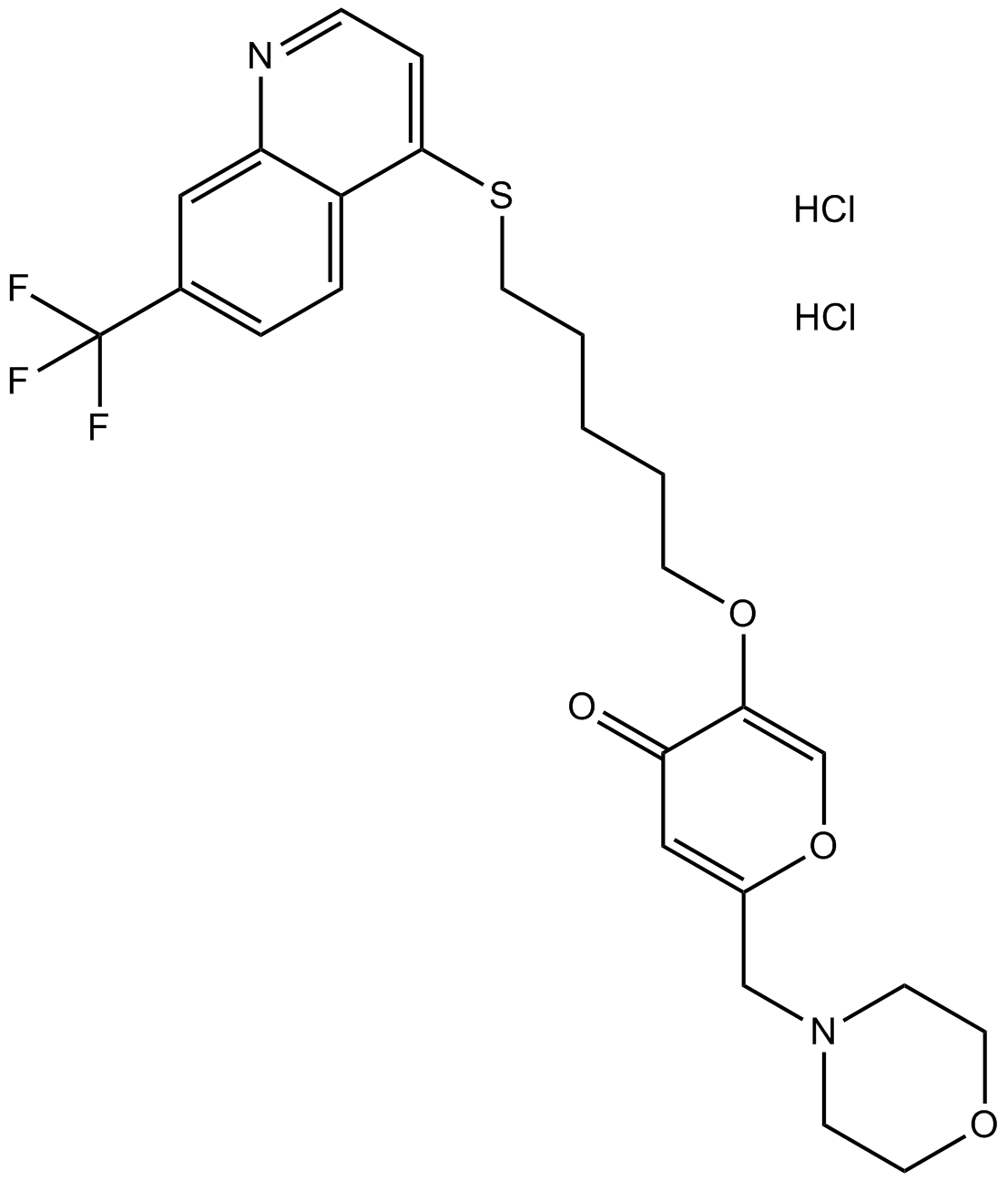
Rac family small GTPases inhibitor
-
GC66473
ESI-08
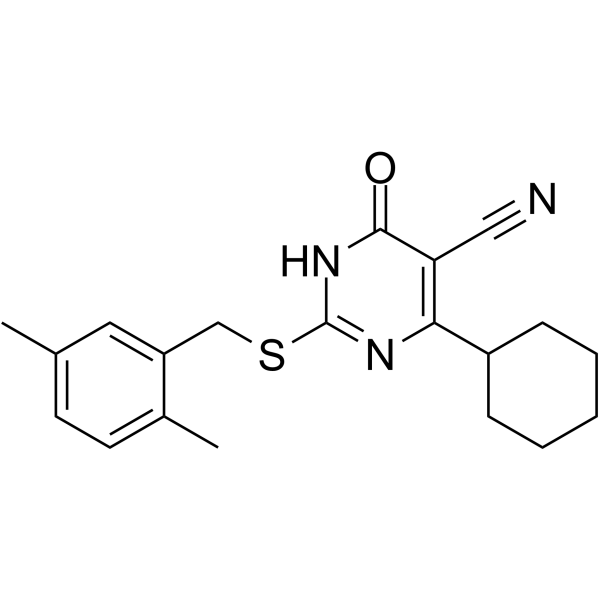
ESI-08 is a potent and selective EPAC antagonist, which can completely inhibit both EPAC1 and EPAC2 (IC50 of 8.4 μM) activity. ESI-08 selectively blocks cAMP-induced EPAC activation, but does not inhibit cAMP-mediated PKA activation.
-
GC64723
Garsorasib
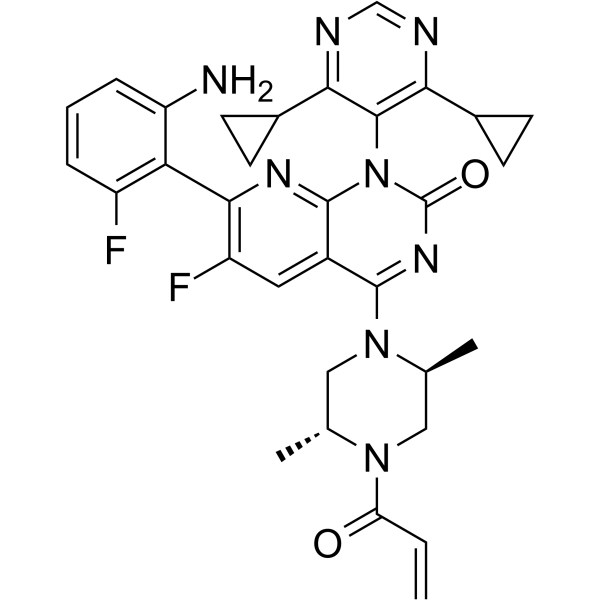
Garsorasib is a potent inhibitor of KRAS G12C with an IC50 of 10 nM. Garsorasib has the potential for the research of various cancer such as pancreatic cancer, endometrial cancer, colorectal cancer, or lung cancer (non-small cell lung cancer) (extracted from patent WO2020233592A1, compound 2).
-
GC64510
GDC-6036
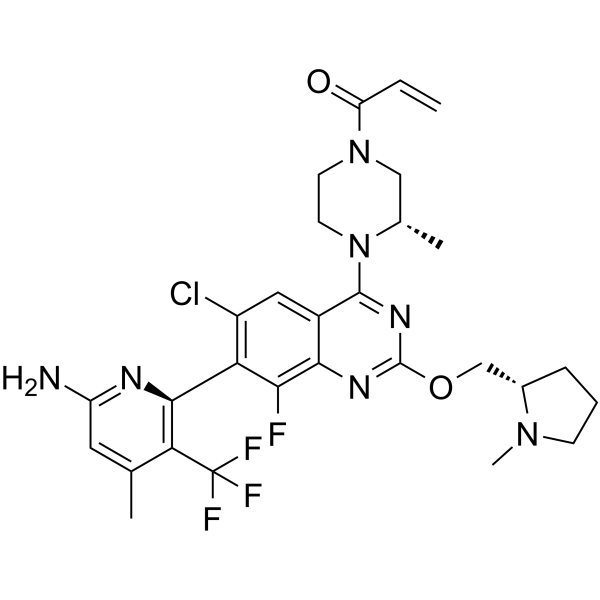
GDC-6036 (compound 17a) is a potent K-Ras G12C inhibitor with an IC50 of <0.01 μM. GDC-6036 has an EC50 of 2 nM in K-Ras G12C-alkylation HCC1171 cells.
-
GC68004
GDC-6036-NH
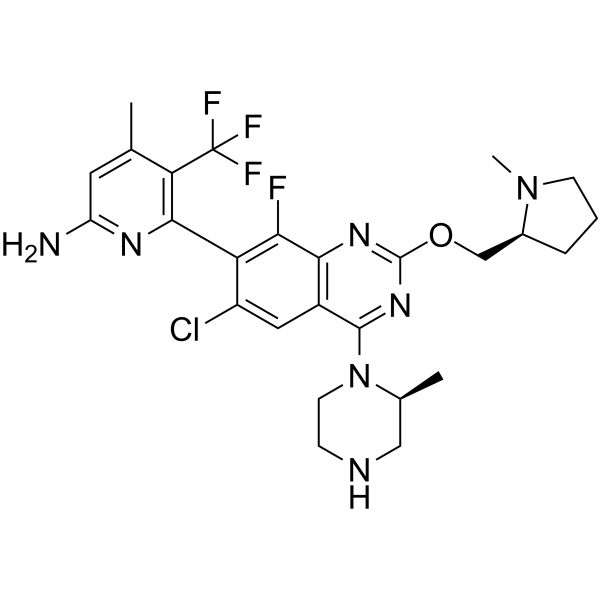
-
GC64216
GGTI-286
GGTI-286, a potent and cell-permeable GGTase I inhibitor, is 25-fold more potent (IC50=2 μM) than the corresponding methyl ester of FTI-276. GGTI-286 selectively inhibits geranylgeranylation of Rap1A over farnesylation of H-Ras in NIH3T3 cells (IC50s =2 and >30 μM, respectively). GGTI-286 also potently inhibits oncogenic K-Ras4B stimulation with an IC50 of 1 μM.
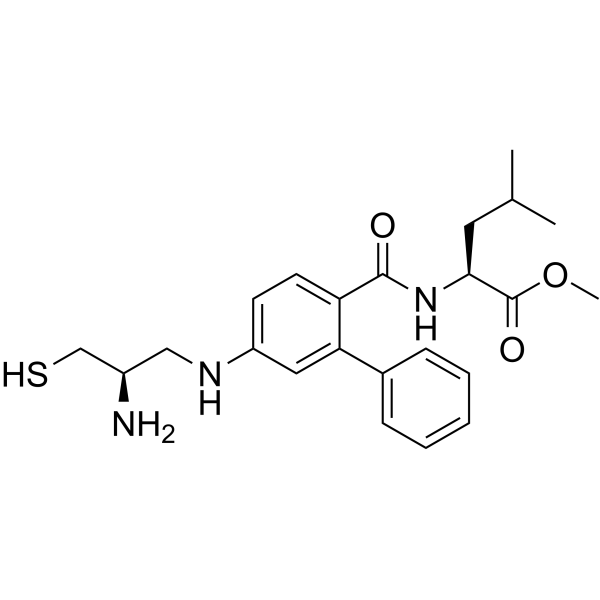
-
GC64784
GGTI-286 hydrochloride
GGTI-286 hydrochloride, a potent GGTase I inhibitor, is 25-fold more potent (IC50=2 μM) than the corresponding methyl ester of FTI-276. GGTI-286 hydrochloride selectively inhibits geranylgeranylation of Rap1A over farnesylation of H-Ras in NIH3T3 cells (IC50s =2 and >30 μM, respectively). GGTI-286 hydrochloride also potently inhibits oncogenic K-Ras4B stimulation with an IC50 of 1 μM.
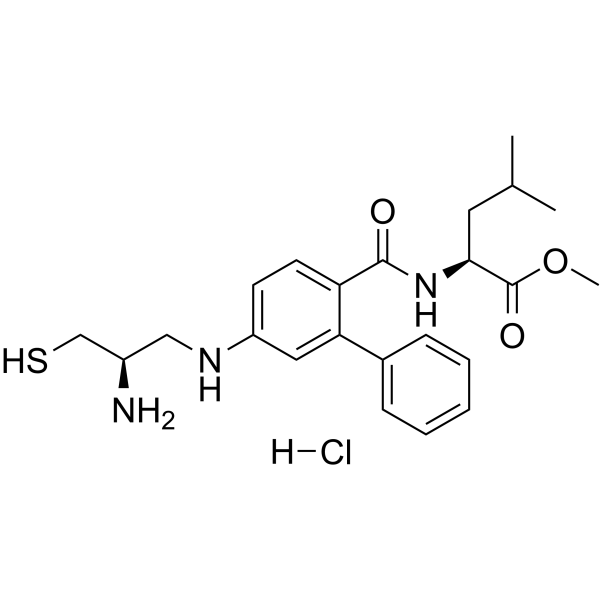
-
GC36134
GGTI298
GGTI298 is a CAAZ peptidomimetic geranylgeranyltransferase I (GGTase I) inhibitor, strongly inhibiting the processing of geranylgeranylated Rap1A with little effect on processing of farnesylated Ha-Ras, with IC50 values of 3 and > 20 μM in vivo, respectively.
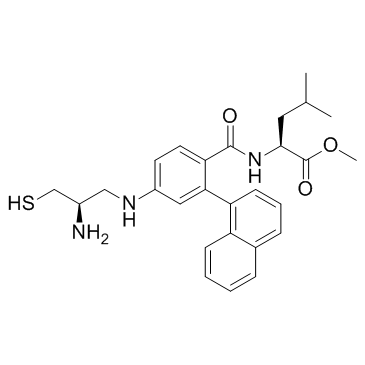
-
GC19164
GGTI298 Trifluoroacetate
GGTI298 Trifluoroacetate is a CAAZ peptidomimetic geranylgeranyltransferase I (GGTase I) inhibitor, which can inhibit Rap1A with IC50 of 3 uM; little effect on Ha-Ras with IC50 of >20 uM.
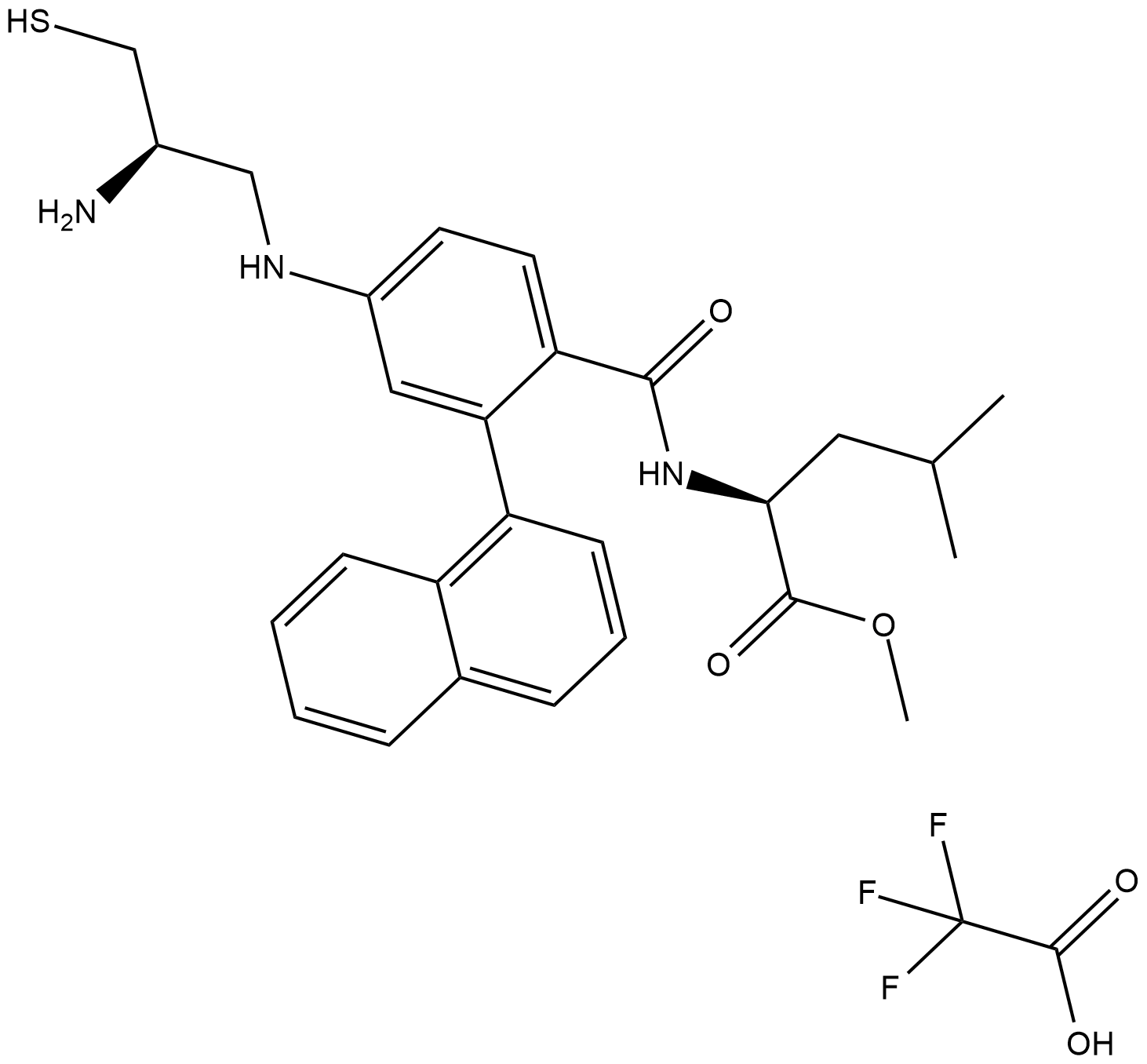
-
GC63564
JDQ-443
JDQ-443 is an orally active, potent, selective, and covalent KRAS G12C inhibitor (extracted from patent WO2021120890A1). JDQ-443 shows antitumor activity.
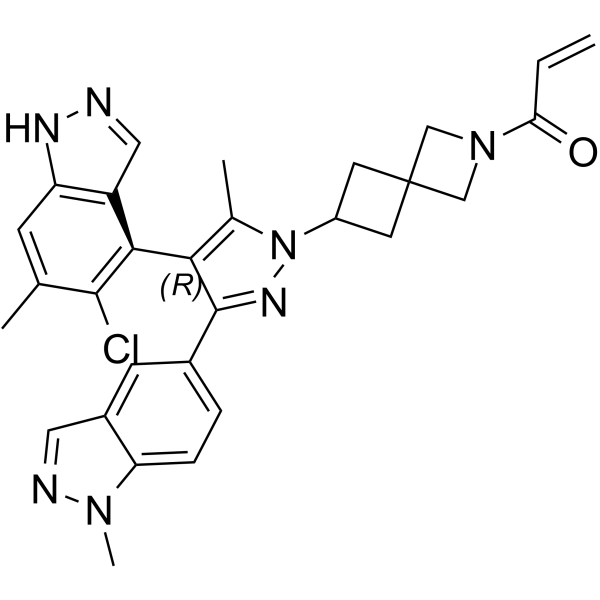
-
GC36398
K-Ras G12C-IN-1
K-Ras G12C-IN-1 is a novel and irreversible inhibitor of mutant K-ras G12C extracted from patent WO 2014152588 A1.
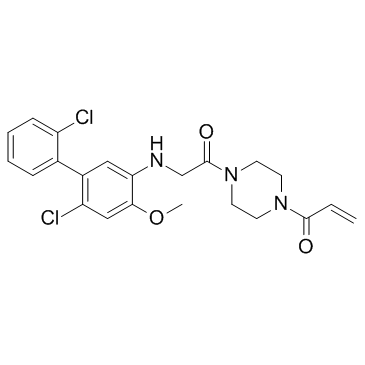
-
GC36399
K-Ras G12C-IN-2
K-Ras G12C-IN-2 is an irreversible covalent K-Ras G12C inhibitor.
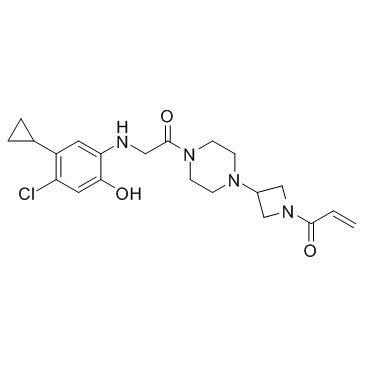
-
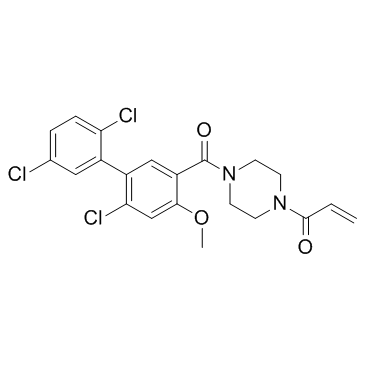
GC36400
K-Ras G12C-IN-3
K-Ras G12C-IN-3 is a novel and irreversible inhibitor of mutant K-ras G12C.
-
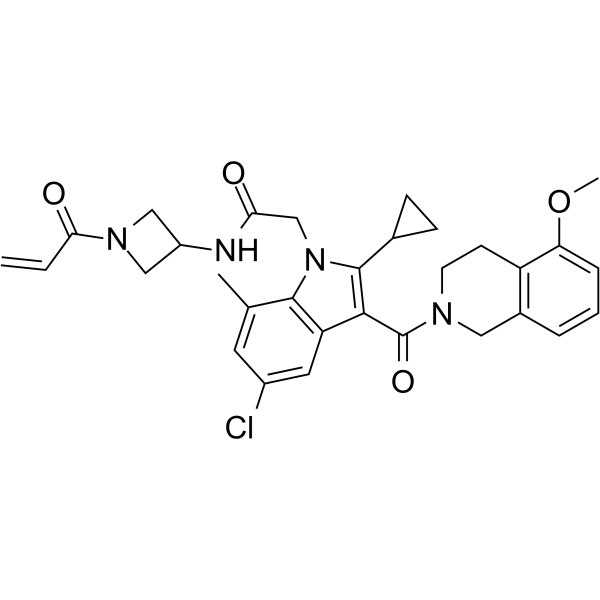
GC62622
K-Ras G12C-IN-4
K-Ras G12C-IN-4, compound 1, is a potent Covalent Inhibitor of KRASG12C.
-
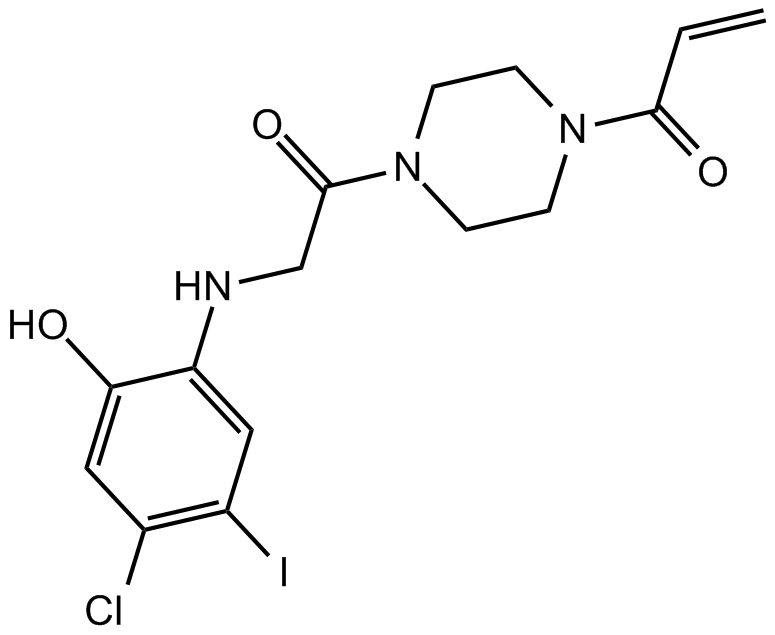
GC12247
K-Ras(G12C) inhibitor 12
allosteric inhibitor of K-Ras(G12C)
-
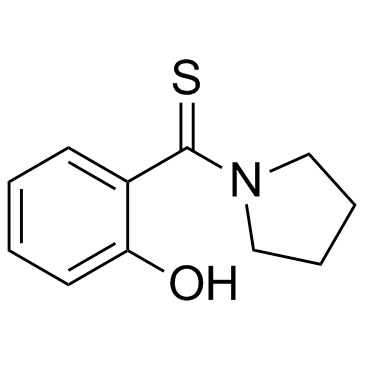
GC33153
K-Ras-IN-1
K-Ras-IN-1 is a K-Ras inhibitor. K-Ras-IN-1 binds to K-Ras (WT), K-Ras (G12D), K-Ras (G12V), and H-Ras. K-Ras-IN-1 has potential for the research of pancreatic, colon and lung carcinomas.
-
GC16922
Kobe0065
Ras inhibitor
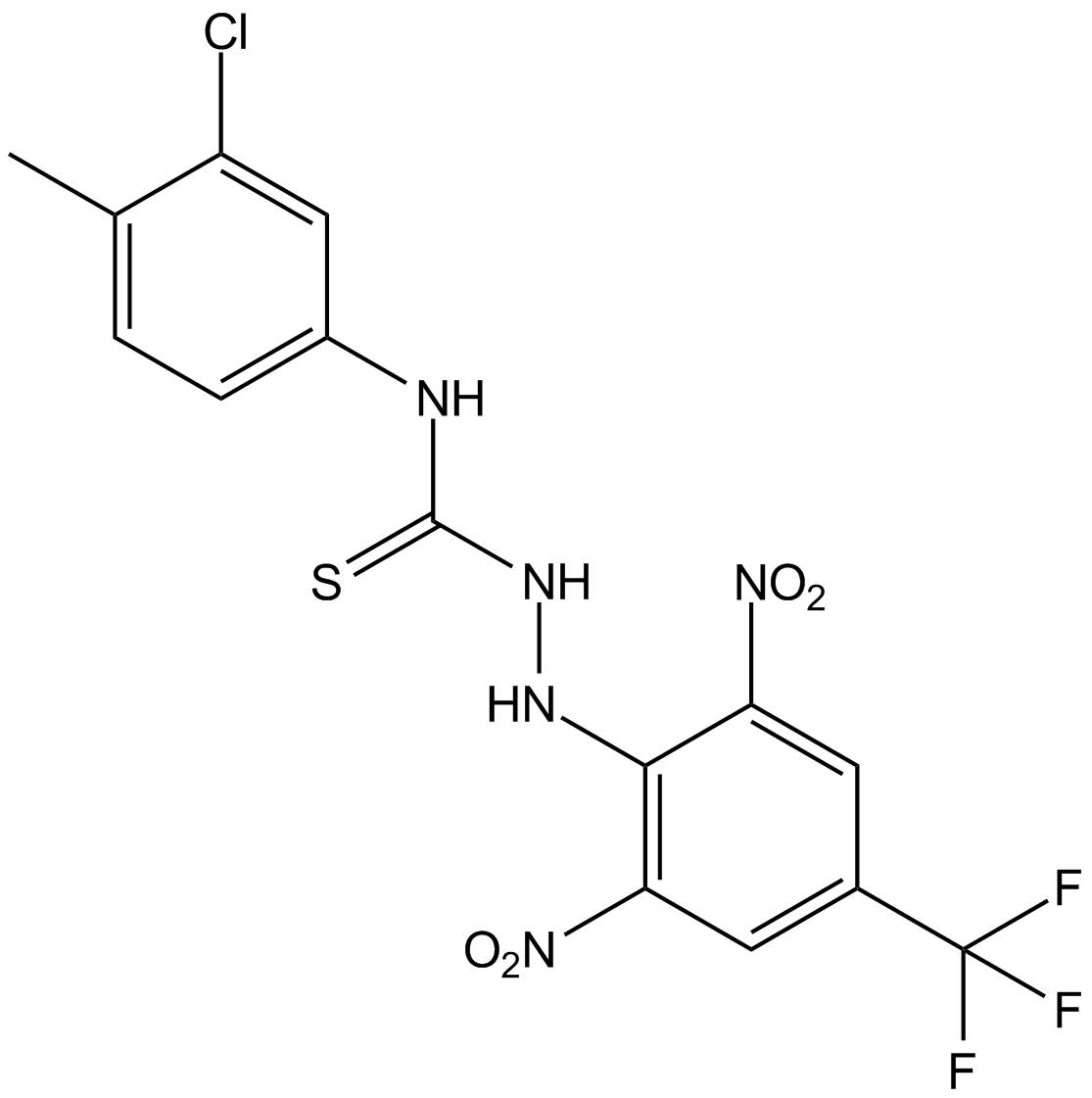
-
GC11204
kobe2602
Ras inhibitor
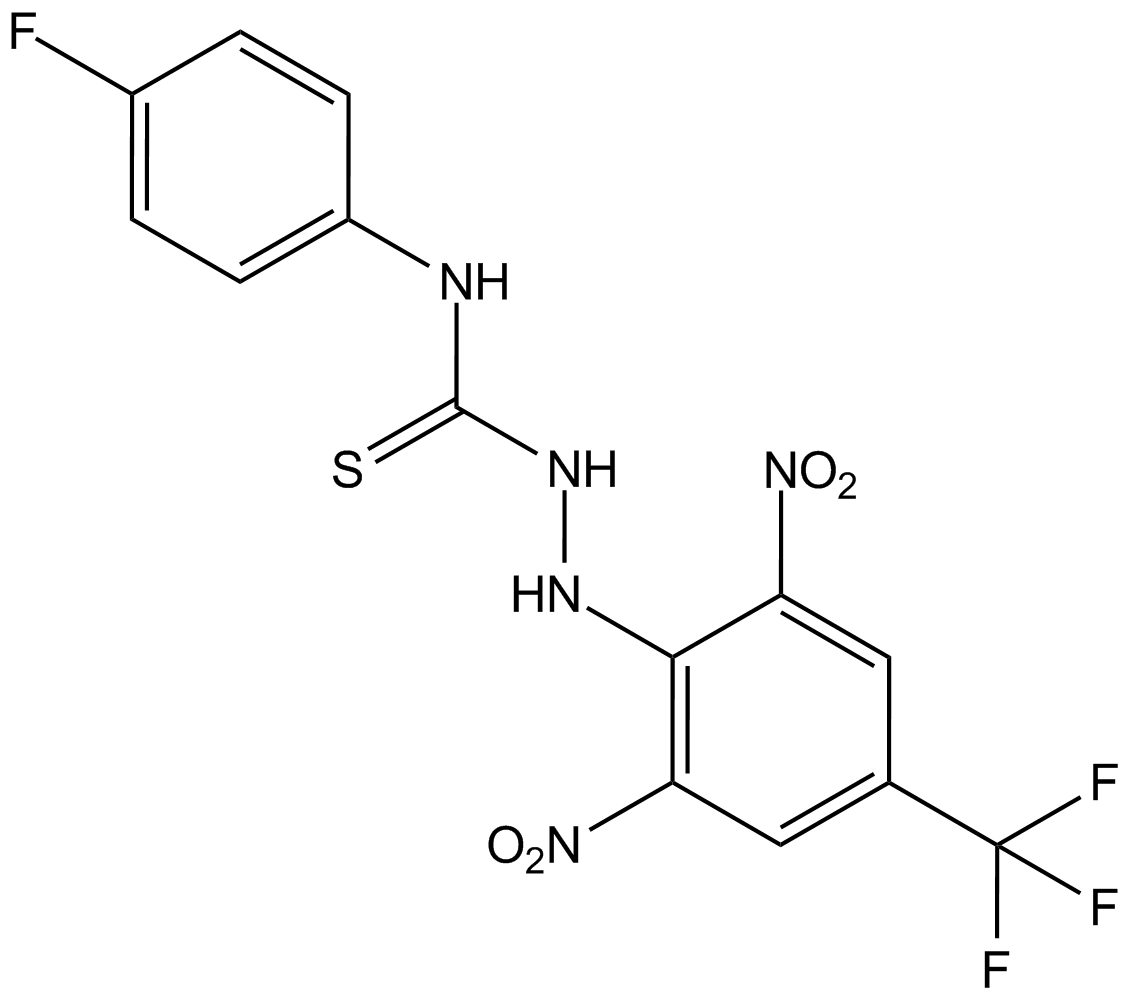
-
GC64558
KRA-533
KRA-533 is a potent KRAS agonist. KRA-533 binds to the GTP/GDP binding pocket in the KRAS protein to prevent GTP cleavage, resulting in the accumulation of constitutively active GTP-bound KRAS that triggers both apoptotic and autophagic cell death pathways in cancer cells.
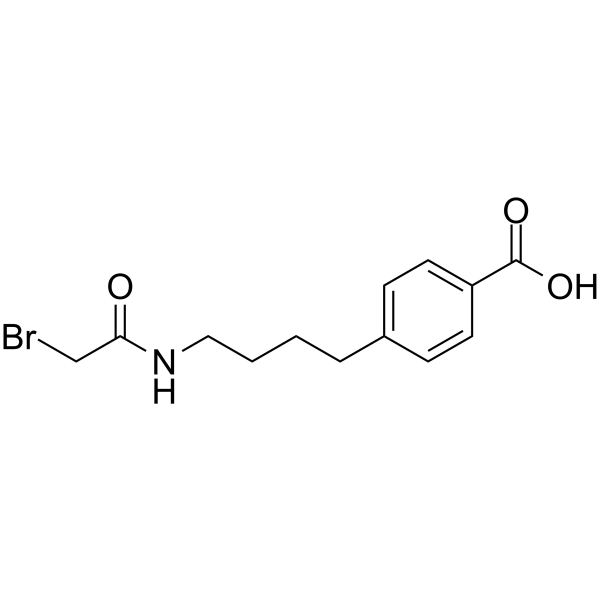
-
GC34198
KRas G12C inhibitor 1
KRas G12C inhibitor 1 is a compound that inhibits KRas G12C, extracted from patent US 20180072723 A1.
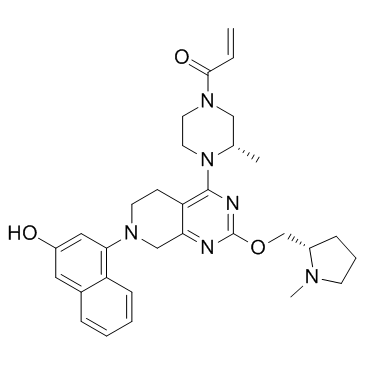
-
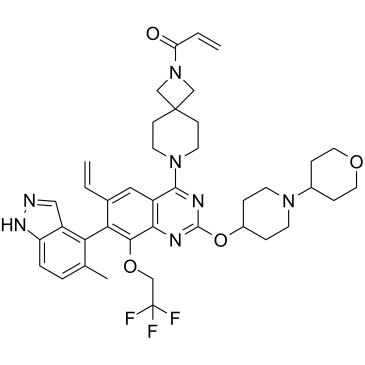
GC36397
KRAS G12C inhibitor 13
KRAS G12C inhibitor 13 is a KRAS G12C inhibitor extracted from patent WO2018143315A1, compound 30.
-
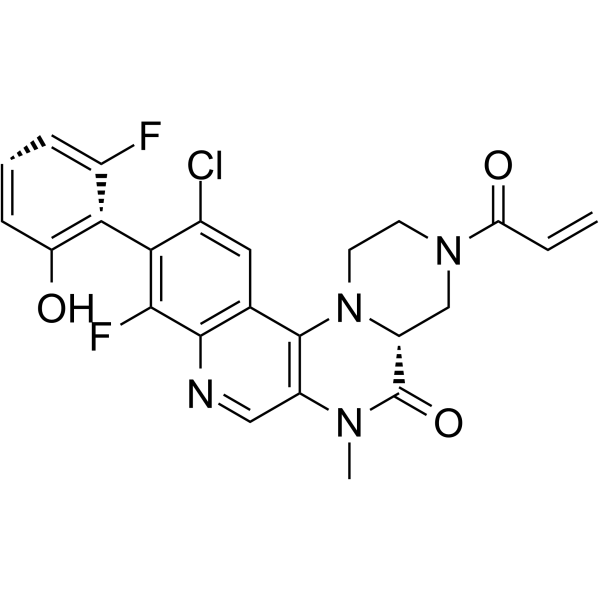
GC64442
KRAS G12C inhibitor 14
KRAS G12C inhibitor 14 is a potent KRAS G12C inhibitor extracted from patent WO2019110751A1, compound 17, has an IC50 of 18 nM.
-
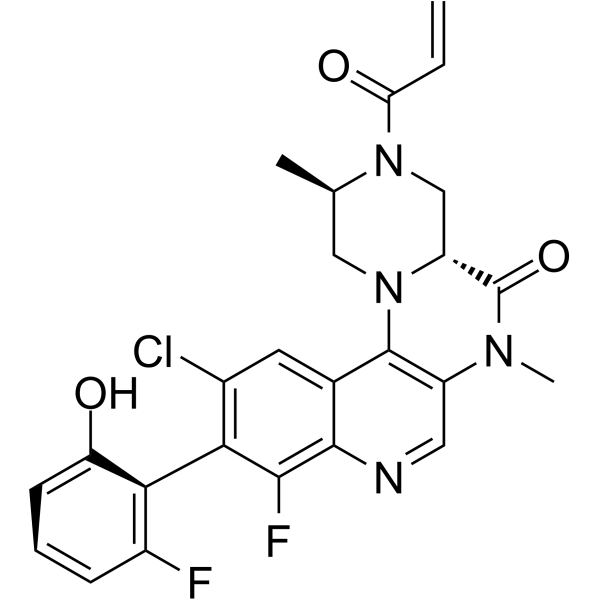
GC63875
KRAS G12C inhibitor 15
KRAS G12C inhibitor 15 is a potent KRAS G12C inhibitor extracted from patent WO2019110751A1, compound 22, has an IC50 of 5 nM.
-
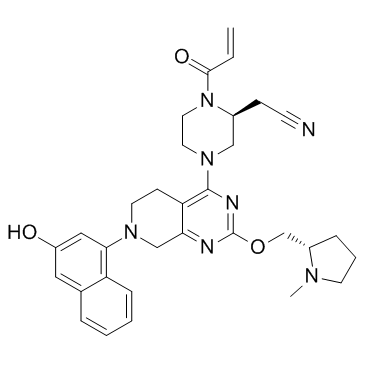
GC34188
KRas G12C inhibitor 2
KRas G12C inhibitor 2 is a compound that inhibits KRas G12C, extracted from patent US 20180072723 A1.
-
GC65904
KRAS G12C inhibitor 28
KRAS G12C inhibitor 28 is a KRAS G12C inhibitor with an IC50 of 57 nM. KRAS G12C inhibitor 28 has antitumor effects (WO2021113595A1; Example 1).
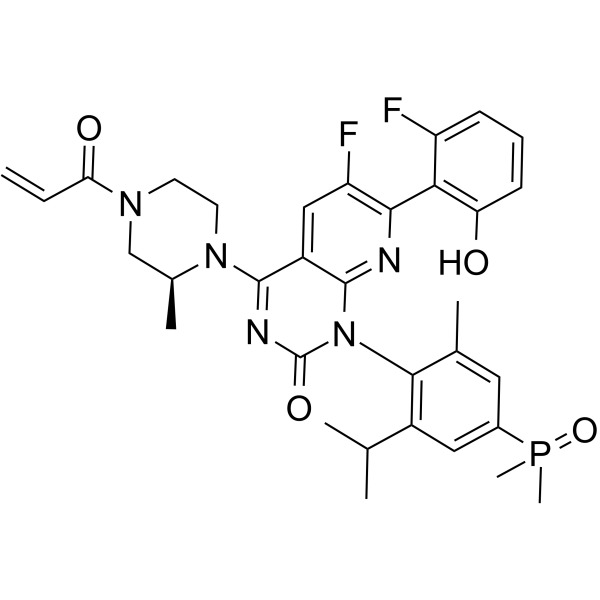
-
GC34196
KRas G12C inhibitor 3
KRas G12C inhibitor 3 is a compound that inhibits KRas G12C, extracted from patent US 20180072723 A1.
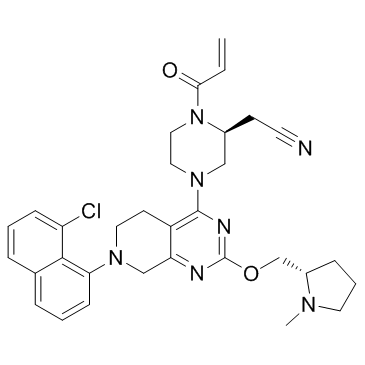
-
GC34192
KRas G12C inhibitor 4
KRas G12C inhibitor 1 is a compound that inhibits KRas G12C, extracted from patent US 20180072723 A1.
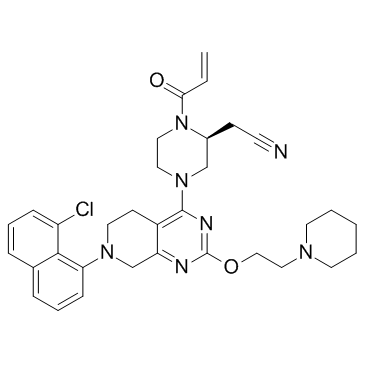
-
GC34162
KRAS G12C inhibitor 5
KRAS G12C inhibitor 5 is a KRas G12C inhibitor extracted from patent WO2017201161A1, Compound example 147.
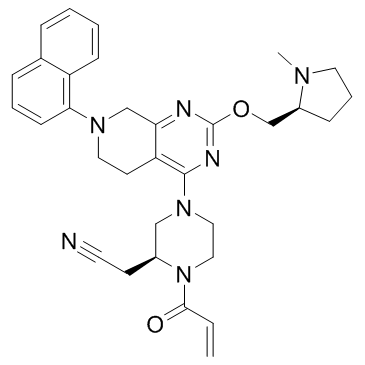
-
GC62227
KRAS G12D inhibitor 1
KRAS G12D inhibitor 1 (example 243) is a KRAS G12D inhibitor, with an IC50 of 0.8 nM for KRAS G12D-mediated ERK phosphorylation.
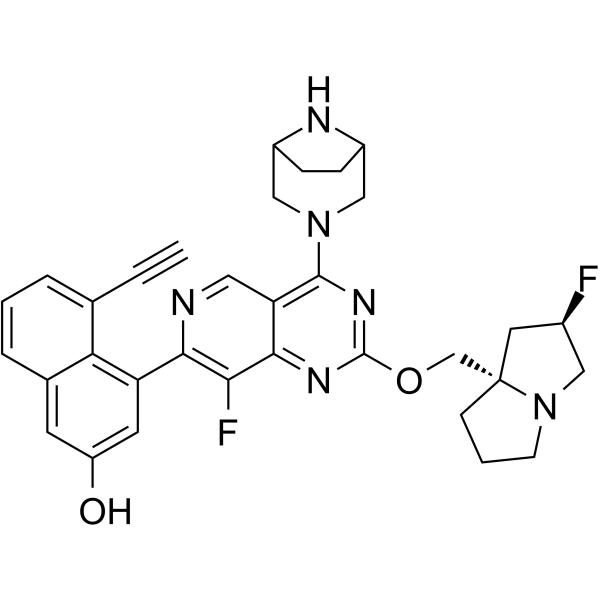
-
GC69339
KRAS G12D inhibitor 14
KRAS G12D inhibitor 14 is an effective inhibitor of KRAS G12D, with a binding affinity (KD) to the KRAS G12D protein of 33 nM. It reduces the activity of KRAS G12D (KRAS G12D-GTP), but does not reduce that of KRAS G13D.
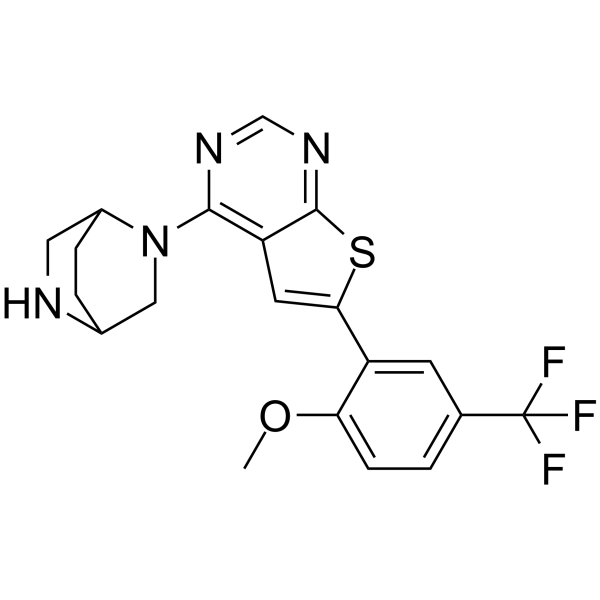
-
GC62486
KRAS inhibitor-10
KRAS inhibitor-10 selectively and effectively inhibit RAS proteins, and particularly KRAS proteins. KRAS inhibitor-10 is an orally active anti-cancer agent and can be used for cancer research, such as pancreatic cancer, breast cancer, multiple myeloma, leukemia and lung cancer. KRAS inhibitor-10 is a?tetrahydroisoquinoline compound (compound 11) extracted from patent WO2021005165 A1.
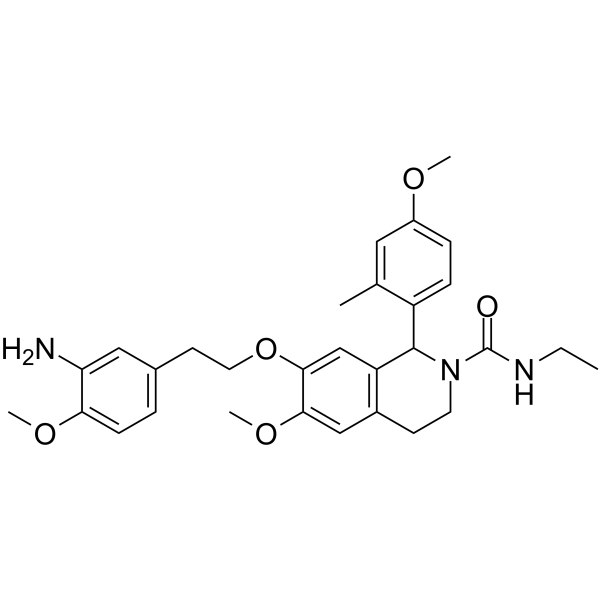
-
GC60969
KRAS inhibitor-9
KRAS inhibitor-9, a potent KRAS inhibitor (Kd=92 μM), blocks the formation of GTP-KRAS and downstream activation of KRAS. KRAS inhibitor-9 binds to KRAS G12D, KRAS G12C and KRAS Q61H protein with a moderate binding affinity. KRAS inhibitor-9 causes G2/M cell cycle arrest and induces apoptosis. KRAS inhibitor-9 selectively inhibits the proliferation of NSCLC cells with KRAS mutation but not normal lung cells.
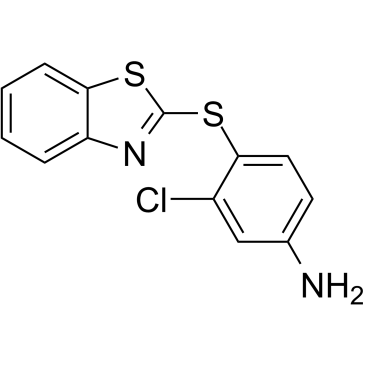
-
GC64447
KRpep-2d
KRpep-2d is a potent K-Ras inhibitor and inhibits proliferation of K-Ras-driven cancer cells. KRpep-2d can be used for cancer research.

-
GC65032
KRpep-2d TFA
KRpep-2d (TFA) is a potent K-Ras inhibitor and inhibits proliferation of K-Ras-driven cancer cells. KRpep-2d can be used for cancer research.
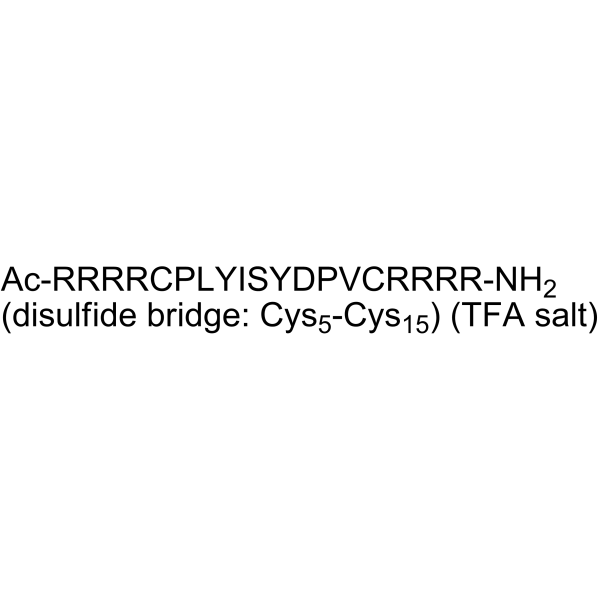
-
GC61766
LC-2
LC-2 is a potent and first-in-class von Hippel-Lindau-based PROTAC capable of degrading endogenous KRAS G12C, with DC50s between 0.25 and 0.76 μM. LC-2 covalently binds KRAS G12C with a MRTX849 warhead and recruits the E3 ligase VHL, inducing rapid and sustained KRAS G12C degradation leading to suppression of MAPK signaling in both homozygous and heterozygous KRAS G12C cell lines.
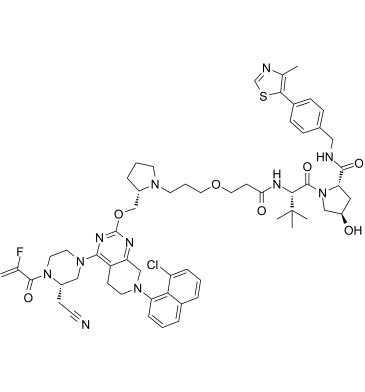
-
GC10265
Manumycin A
farnesyltransferase inhibitor
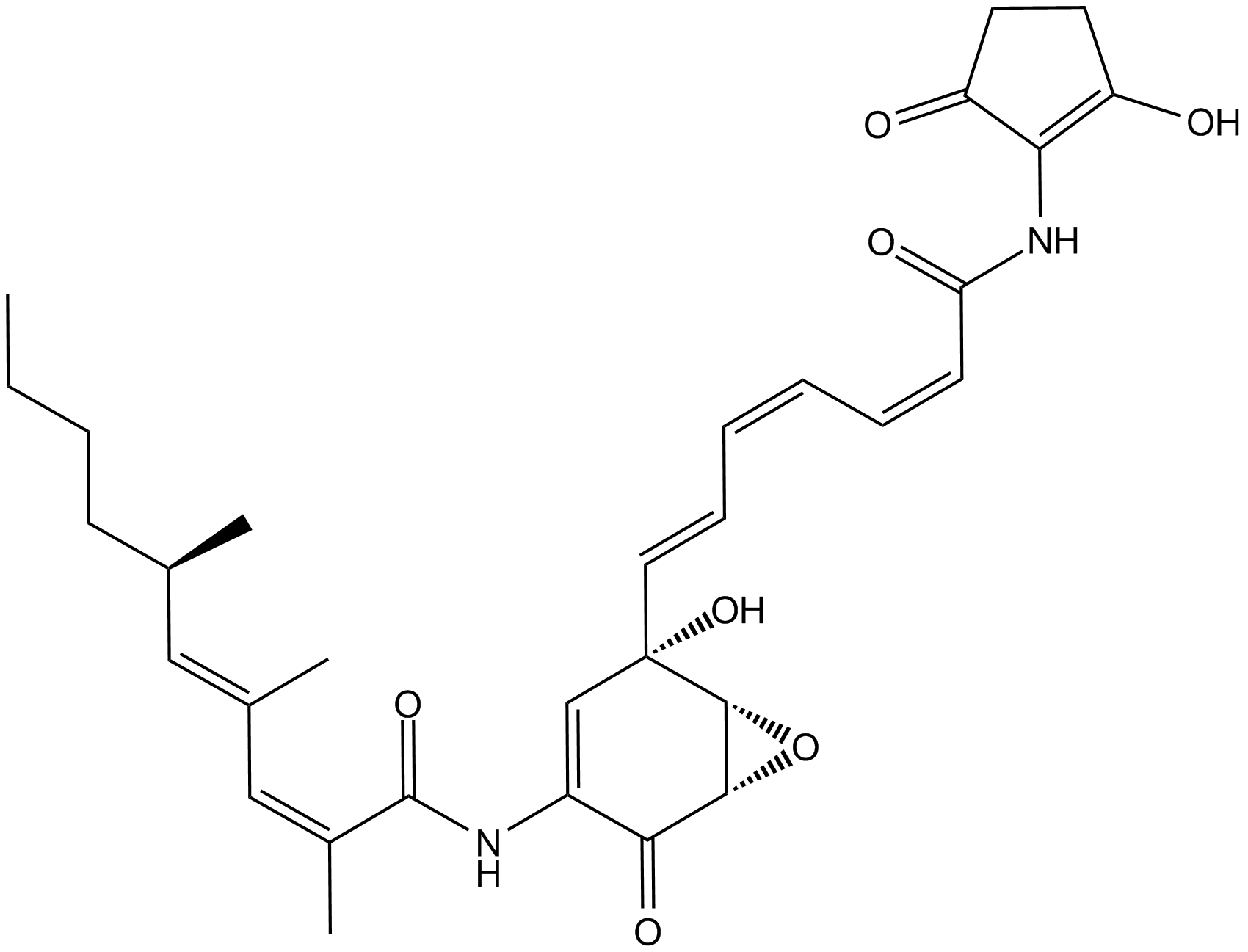
-
GC32970
MBQ-167
MBQ-167 is a dual Rac/Cdc42 inhibitor, with IC50s of 103 nM for Rac 1/2/3 and 78 nM for Cdc42 in MDA-MB-231 cells, respectively.
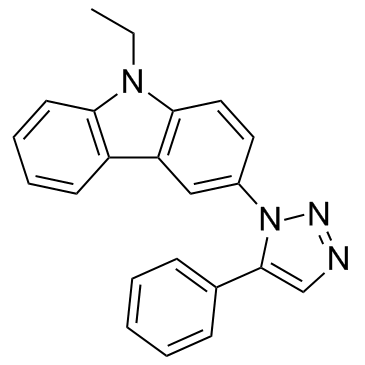
-
GC36599
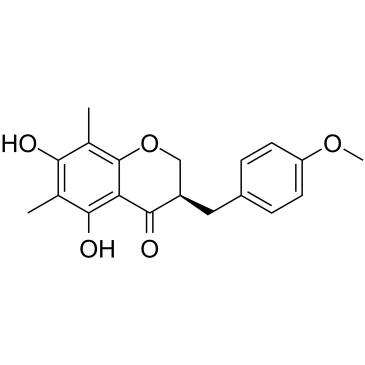
Methylophiopogonanone B
An isoflavone with diverse biological activities
-
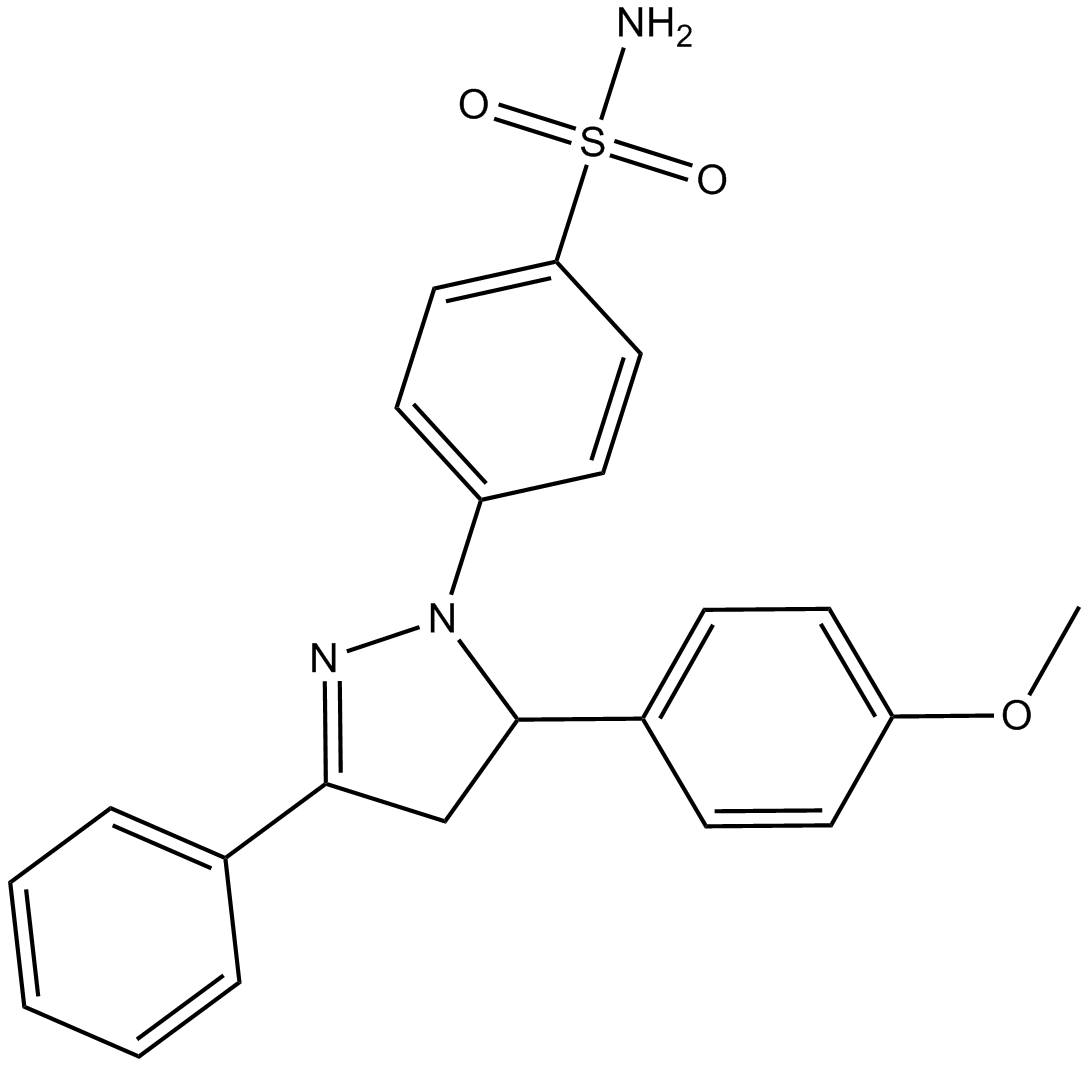
GC14800
ML 141
Cdc42 GTPase inhibitor
-
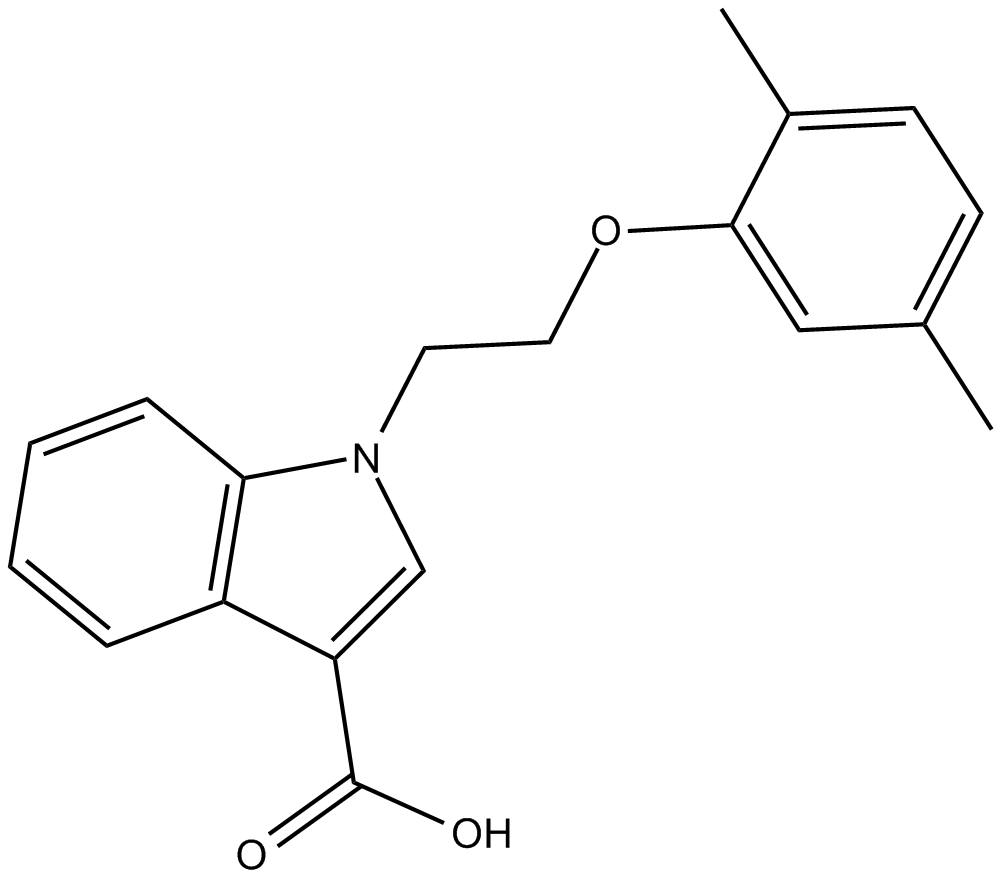
GC13315
ML-098
activator of the GTP-binding protein Rab7
-
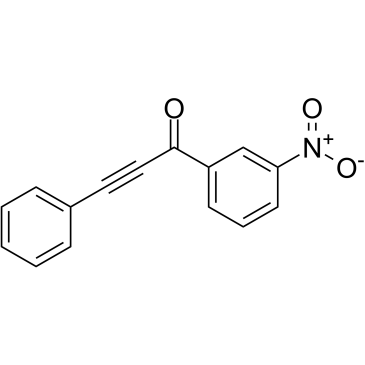
GC61076
MLS000532223
MLS000532223 is a high affinity, selective inhibitor of Rho family GTPases, with EC50 values ranging from 16 μM to 120 μM. MLS000532223 prevents GTP binding to several GTPases.
-
GC69498
MRTF/SRF-IN-1
MRTF/SRF-IN-1 (example 41) is an inhibitor of myocardin-related transcription factor and serum response factor (MRTF/SRF). MRTF/SRF-IN-1 can be used for research on cancer prevention and fibrosis.
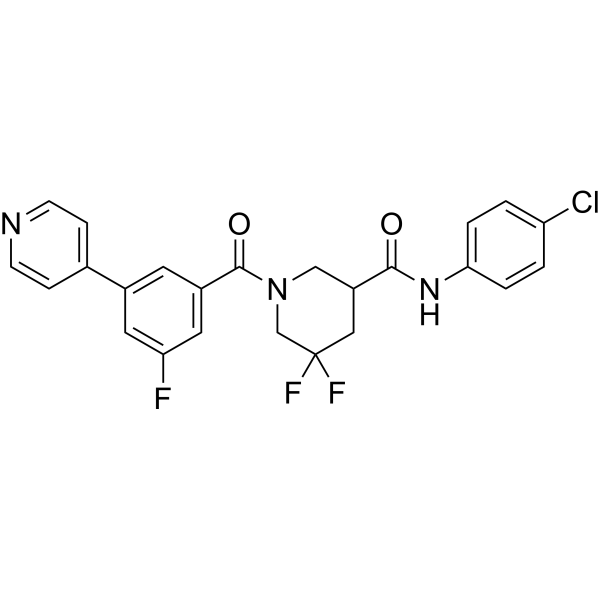
-
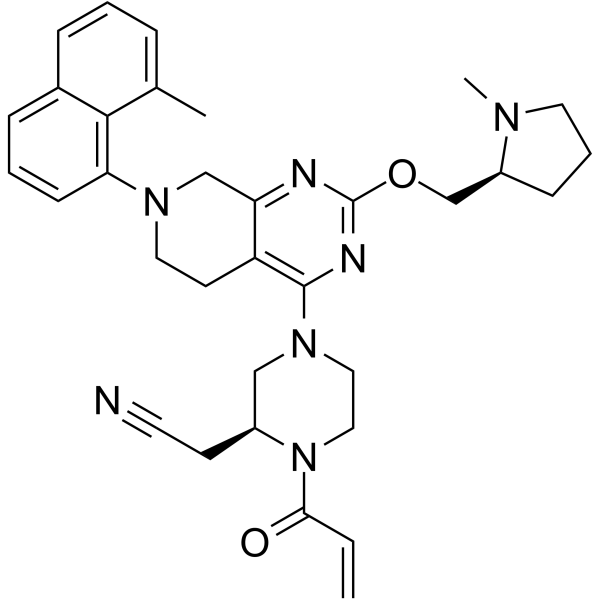
GC64964
MRTX-1257
MRTX-1257 is a selective, irreversible, covalent and orally active KRAS G12C inhibitor, with an IC50 of 900 pM for KRAS dependent ERK phosphorylation in H358 cells.
-
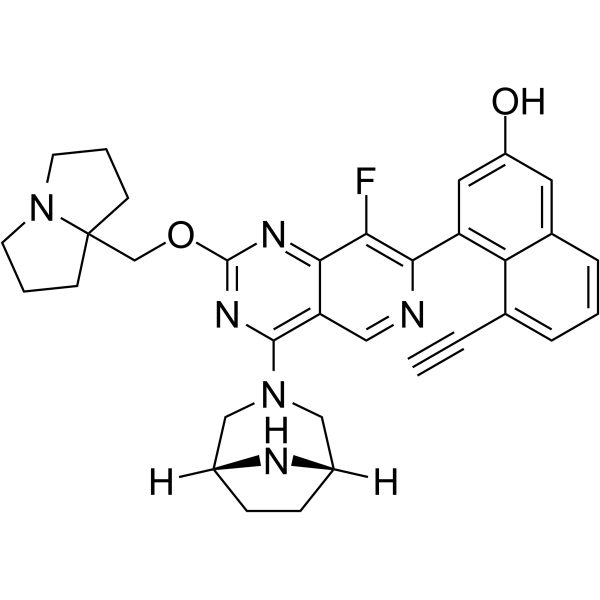
GC64452
MRTX-EX185
MRTX-EX185 is a potent inhibitor of GDP-loaded KRAS and KRAS(G12D), with an IC50 of 90 nM for KRAS(G12D). MRTX-EX185 also binds GDP-loaded HRAS.
-
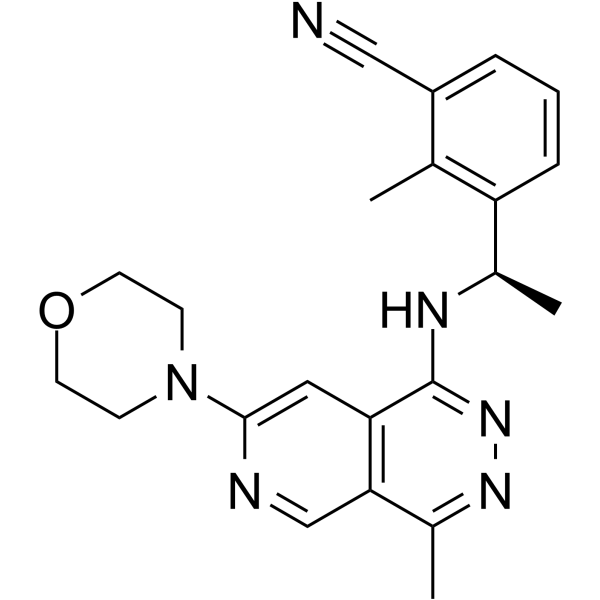
GC64365
MRTX0902
MRTX0902 is an orally active and potent SOS1 inhibitor with an IC50 of 46 nM (WO2021127429A1; Example 12-10).
-
GC62699
MRTX1133
MRTX1133 is an exceptionally potent and selective KRASG12D inhibitor with high affinity (<2nM).
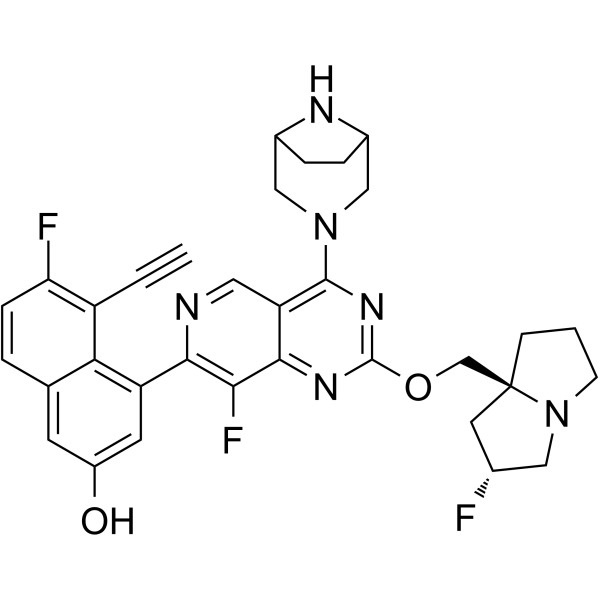
-
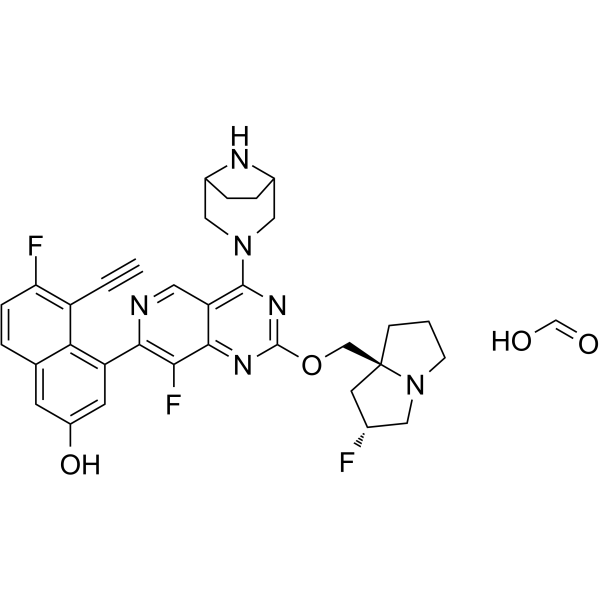
GC63079
MRTX1133 formic
-
GC38400
MRTX849
MRTX849 (MRTX849) is a potent, orally-available, and mutation-selective covalent inhibitor of KRAS G12C with potential antineoplastic activity. MRTX849 covalently binds to KRAS G12C at the cysteine at residue 12, locks the protein in its inactive GDP-bound conformation, and inhibits KRAS-dependent signal transduction.
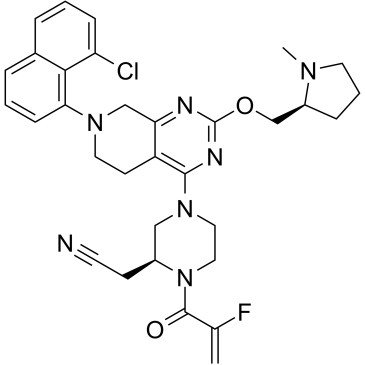
-
GC10069
NSC 23766 trihydrochloride
Selective inhibitor of Rac1-GEF interaction.
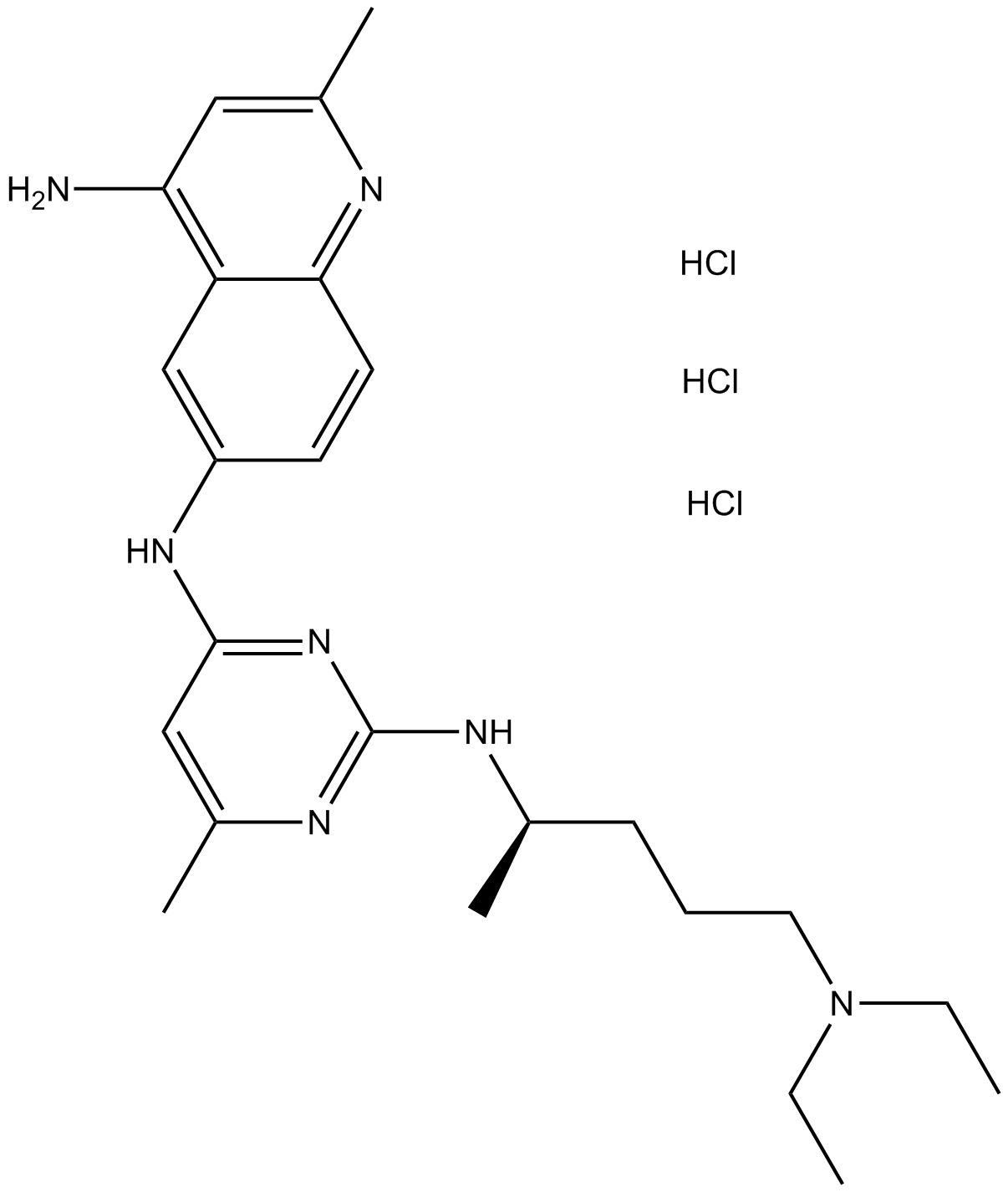
-
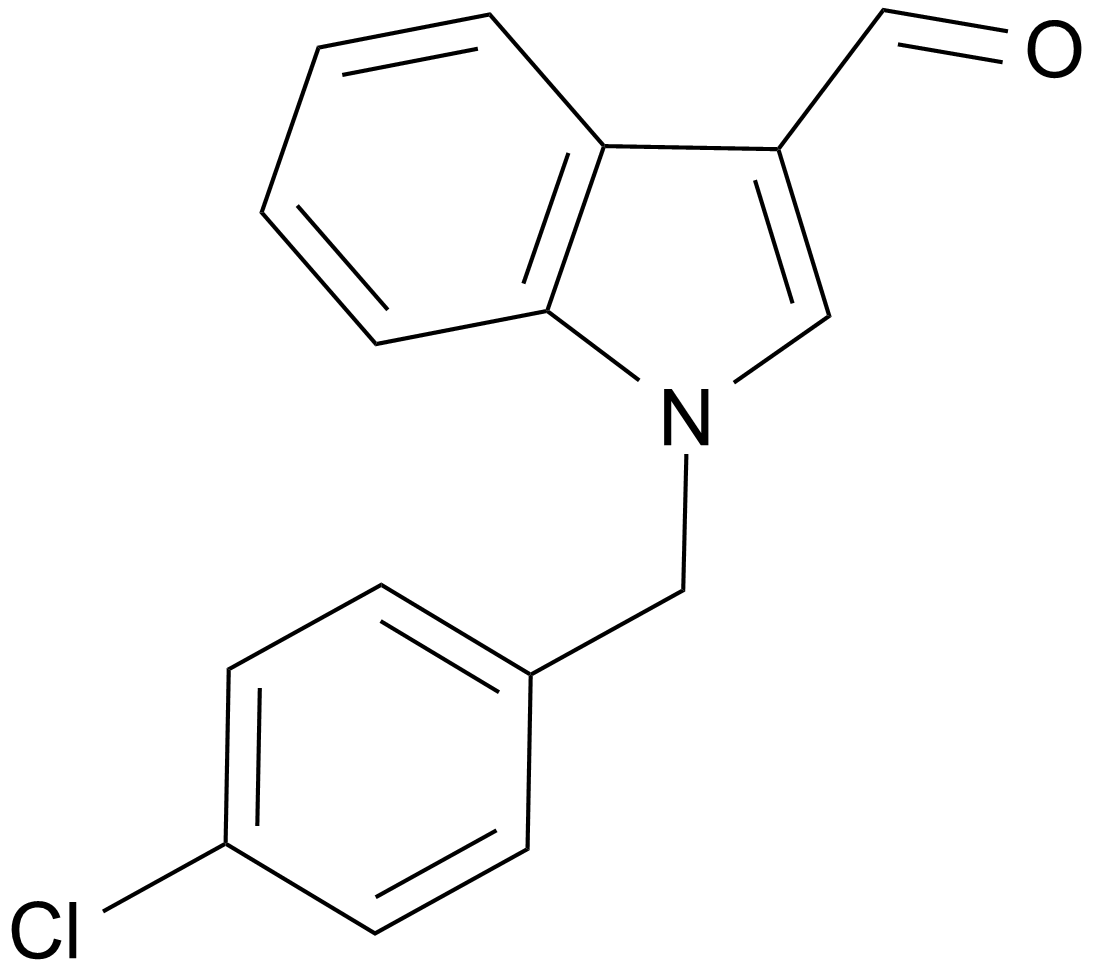
GC14860
Oncrasin 1
An anticancer agent
-
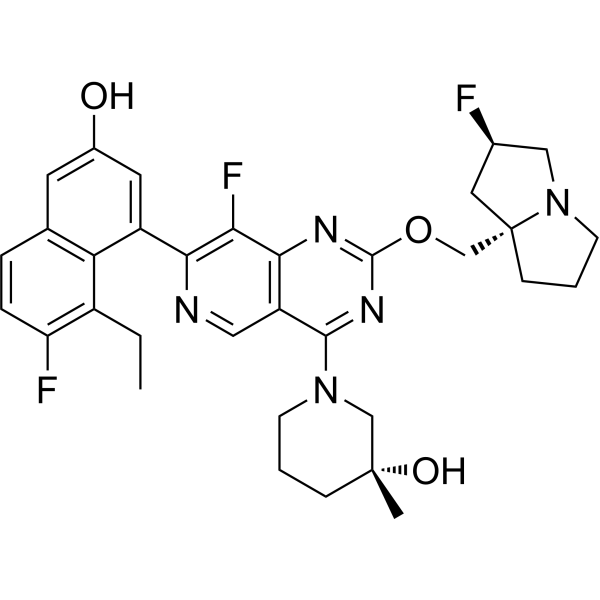
GC69653
Pan KRas-IN-1
Pan KRas-IN-1 is a pan-KRas inhibitor that can be used for studying cancer resistance to KRas G12C inhibitors.
-
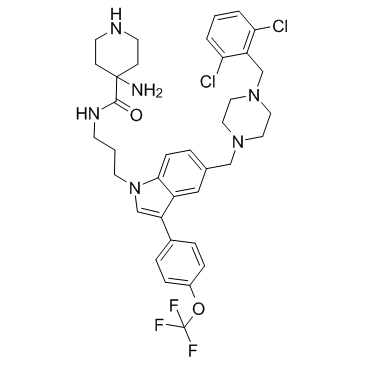
GC32716
Pan-RAS-IN-1
Pan-RAS-IN-1 is a pan-Ras inhibitor that disrupts the interaction of Ras proteins and their effectors.
-
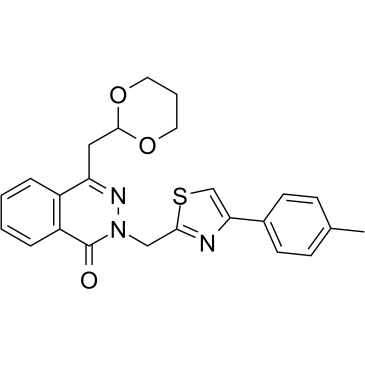
GC36901
PHT-7.3
PHT-7.3 is a selective inhibitor of connector enhancer of kinase suppressor of Ras 1 (Cnk1) pleckstrin homology (PH) domain (Kd=4.7 μM). PHT-7.3 inhibits mut-KRas, but not wild-type KRas cancer cell and tumor growth and signaling. PHT-7.3 has antitumor activity.
-
GC38942
PROTAC K-Ras Degrader-1
PROTAC K-Ras Degrader-1 (Compound 518) is potent K-Ras degrader based on Cereblon E3 ligand, exhibits ≥70% degradation efficacy in SW1573 cells.
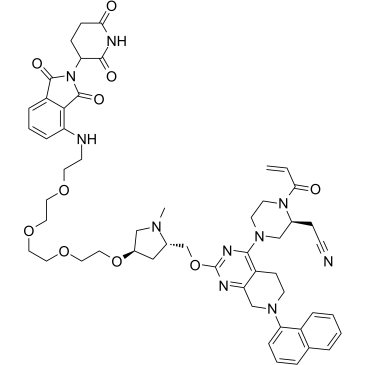
-
GC10015
Rac1 Inhibitor W56
inhibitor of Rac1 interaction with Rac1-specific GEFs TrioN, GEF-H1 and Tiam1
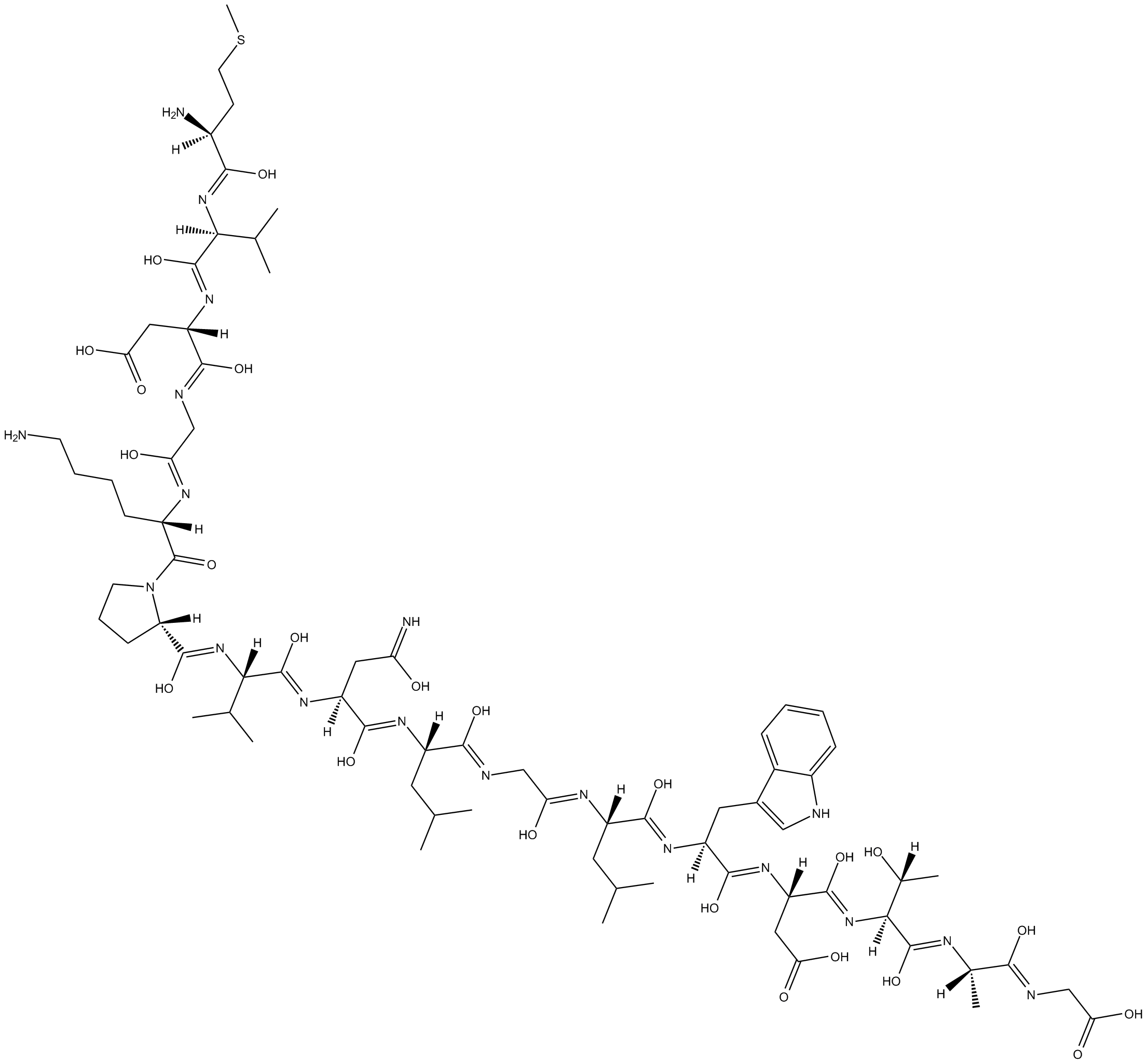
-
GC62421
RAS inhibitor Abd-7
RAS inhibitor Abd-7, a potent RAS-binding compound (Kd=51 nM), is a RAS-effector protein-protein interaction (PPI) inhibitor. RAS inhibitor Abd-7 interacts with RAS inside the cells, prevents RAS-effector interactions and inhibits endogenous RAS-dependent signaling. RAS inhibitor Abd-7 impairs the PPI of various mutant KRAS proteins with PI3K, CRAF and RALGDS as well as NRAS Q61H and HRAS G12V.
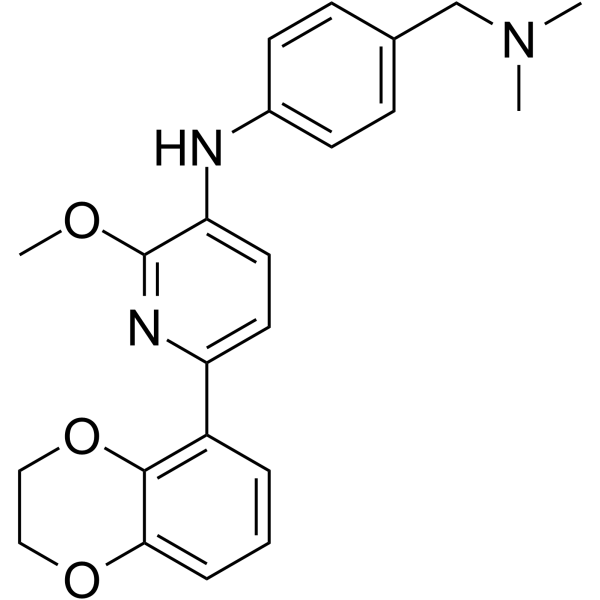
-
GC62504
RAS/RAS-RAF-IN-1
RAS/RAS-RAF-IN-1 is a potent RAS and RAS-RAF inhibitor. RAS/RAS-RAF-IN-1 has a KD of 5.0 μΜ-15 μΜ for cyclophilin A (CYPA) binding affinity. RAS/RAS-RAF-IN-1 has antitumor activity.
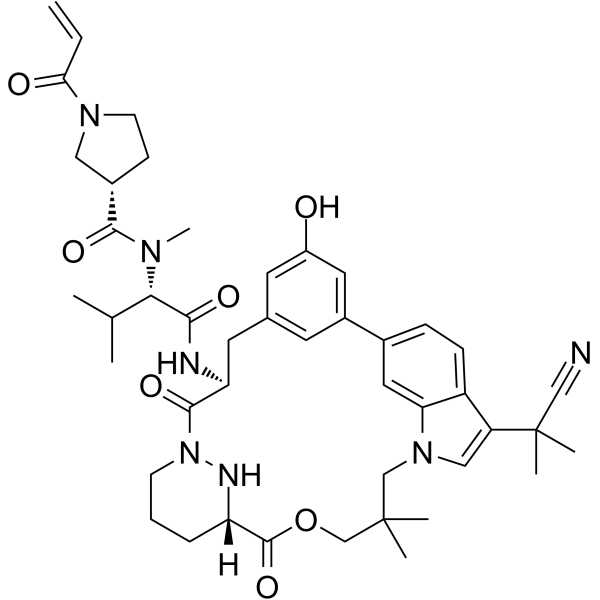
-
GC63647
Rasarfin
Rasarfin is a dual Ras and ARF6 inhibitor.
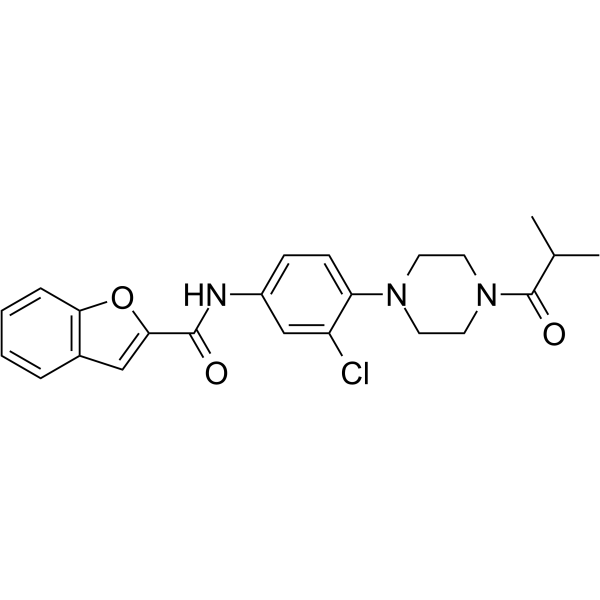
-
GC66001
RBC10
RBC10 is an anti-cancer agent. RBC10 inhibits the binding of Ral to its effector RALBP1. RBC10 also inhibits Ral-mediated cell spreading of murine embryonic fibroblasts and anchorage-independent growth of human cancer cell lines.
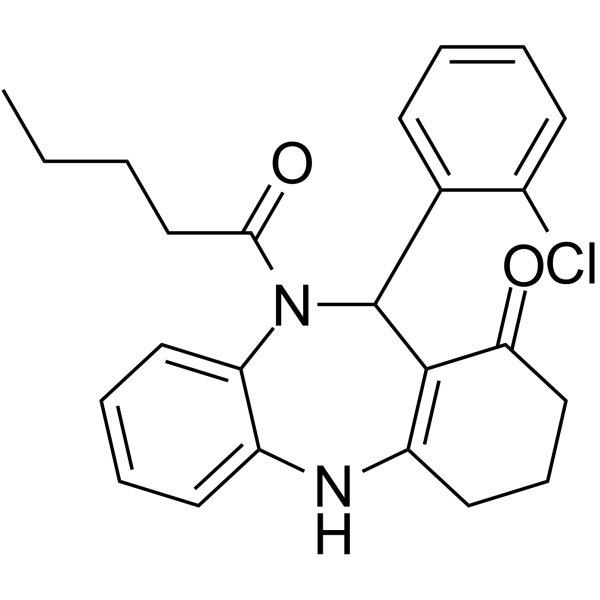
-
GC10906
RBC8
Ral GTPase inhibitor
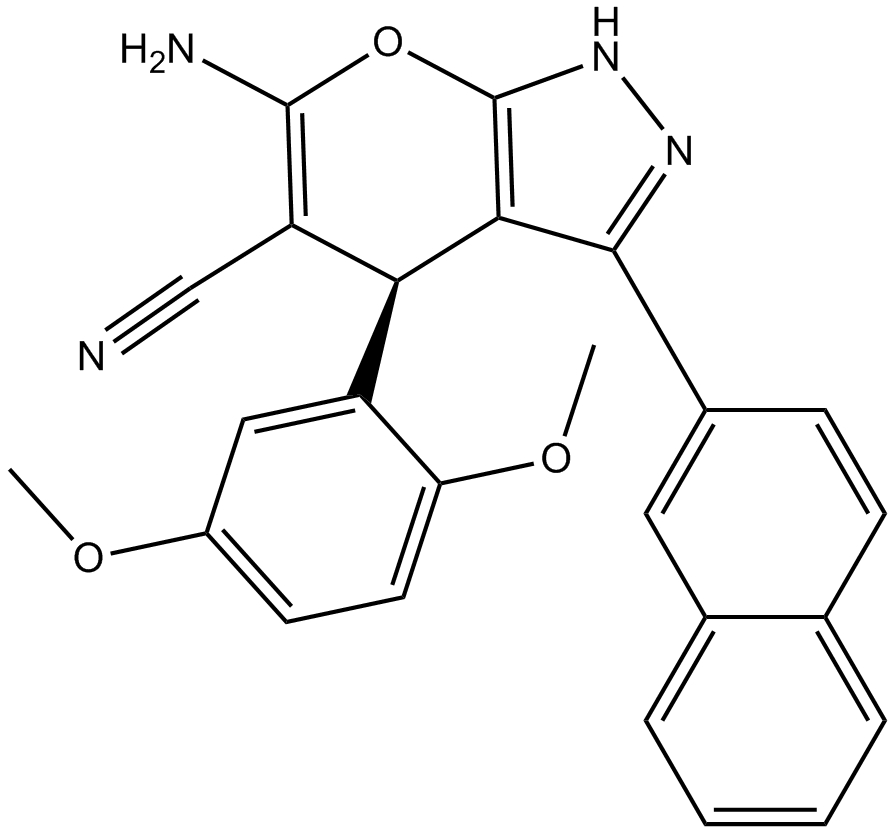
-
GC33167
Rhosin
Rhosin is a potent, specific RhoA subfamily Rho GTPases inhibitor, which specifically binds to RhoA to inhibit RhoA-GEF interaction with a Kd of ~ 0.4 uM, and does not interact with Cdc42 or Rac1, nor the GEF, LARG. Rhosin induces cell apoptosis. Rhosin promotes stress resiliency through enhancing D1-MSN plasticity and reducing hyperexcitability.
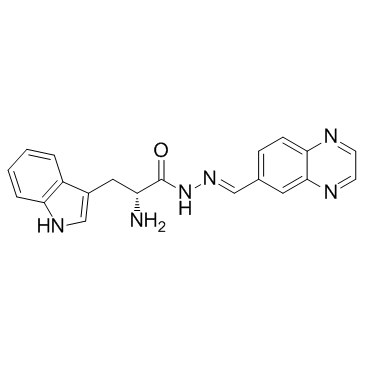
-
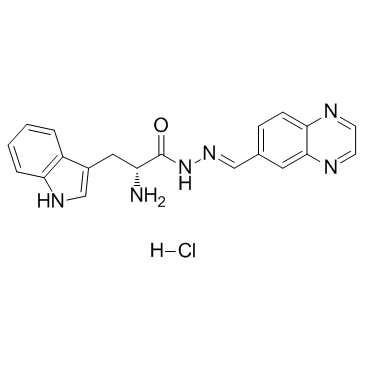
GC32729
Rhosin hydrochloride
Rhosin hydrochloride is a potent, specific RhoA subfamily Rho GTPases inhibitor. Rhosin hydrochloride specifically binds to RhoA to inhibit RhoA-GEF interaction with a Kd of ~ 0.4 uM, and does not interact with Cdc42 or Rac1, nor the GEF, LARG. Rhosin hydrochloride induces cell apoptosis. Rhosin hydrochloride promotes stress resiliency through enhancing D1-MSN plasticity and reducing hyperexcitability.
-
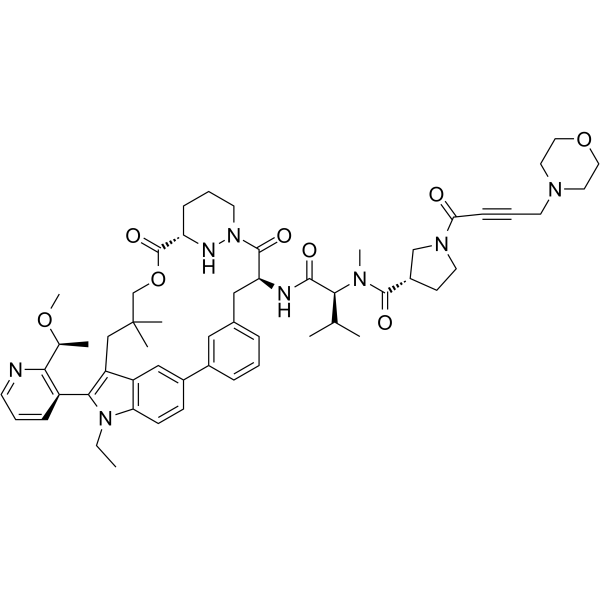
GC63562
RM-018
RM-018 is a potent, functionally distinct tricomplex KRASG12C active-state inhibitor. RM-018 retains the ability to bind and inhibit KRASG12C/Y96D and could overcome resistance. RM-018 binds specifically to the GTP-bound, active [“RAS(ON)”] state of KRASG12C.
-
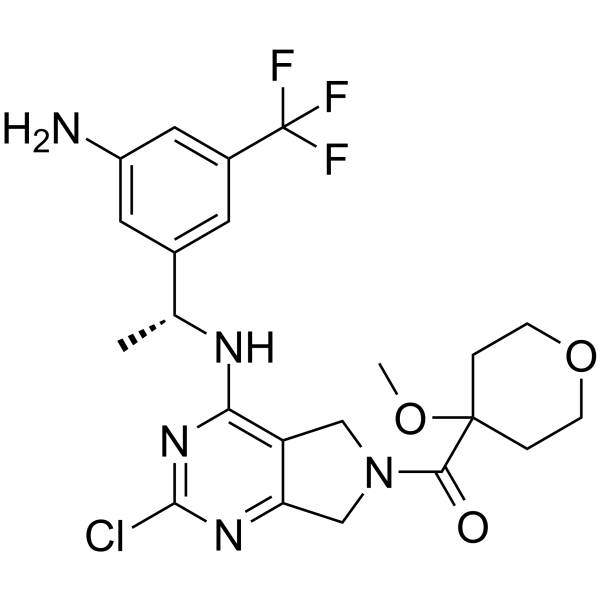
GC62494
RMC-0331
RMC-0331 (RM-023) is a potent, selective and orally bioavailable SOS1 inhibitor. RMC-0331 is an in vivo tool compound that blocks RAS activation via disruption of the RAS-SOS1 interaction.
-
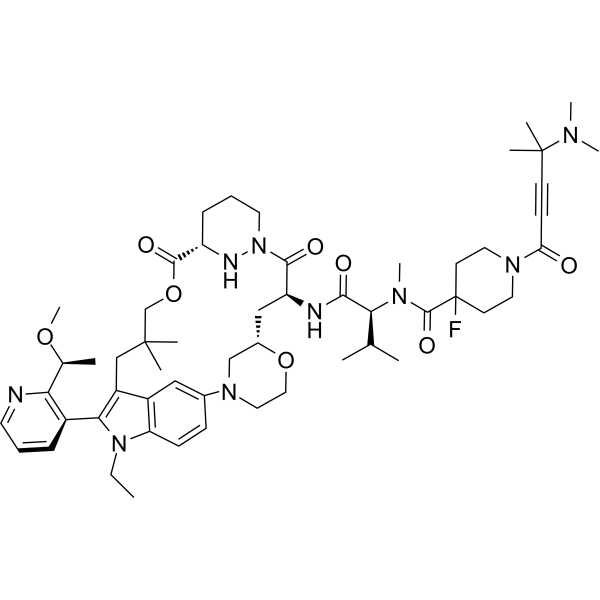
GC69822
RMC-6291
RMC-6291 is an orally effective covalent inhibitor of KRASG12C(ON). RMC-6291 forms a tri-complex within tumor cells between KRASG12C(ON) and cyclophilin A (CypA). Therefore, RMC-6291 blocks the signaling of KRASG12C(ON) by spatially obstructing the binding of RAS effectors. RMC-6291 produces profound and sustained inhibition of the RAS pathway activity in KRASG12C tumor models.
-
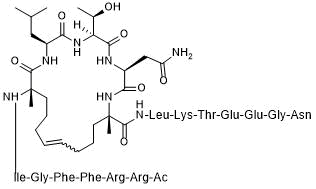
GC50609
SAH-SOS1A
KRas/SOS1 interaction inhibitor
-
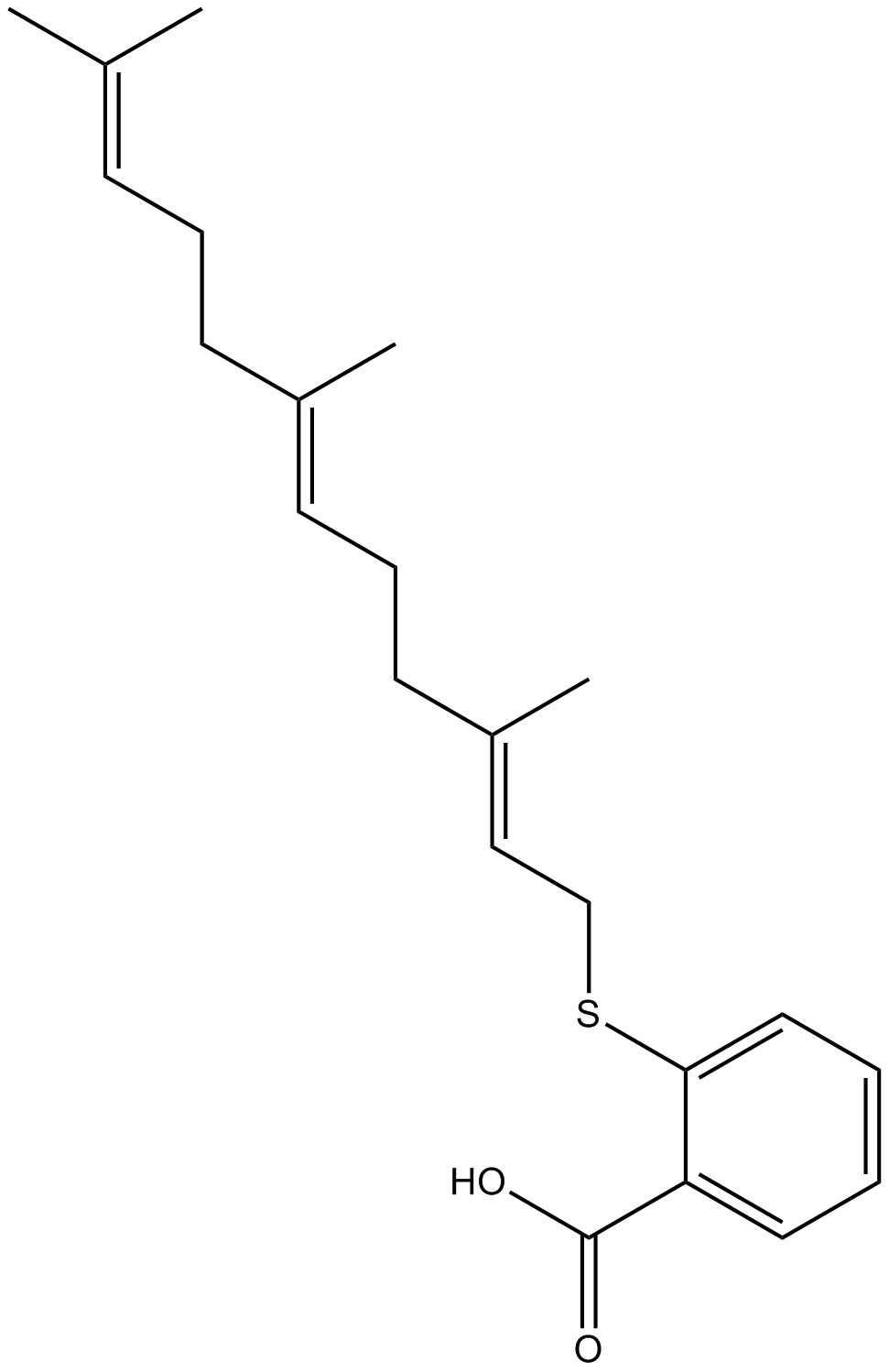
GC10528
Salirasib
A Ras inhibitor with anti-cancer and anti-atherogenic activity
-
GC65263
SOS1-IN-11
SOS1-IN-11 is a potent SOS1 inhibitor with an IC50 value of 30 nM.