PPAR
The peroxisome proliferator-activated receptors (PPARs), belonging to the nuclear receptor superfamily, are a group of nuclear receptor proteins that are composed of three isoforms, including PPARγ, PPARα and PPARδ, encoded by separate genes. PPARs have a modular structure characterized by the presence of two highly conserved domains, including the DNA binding domain (DBD) of two zinc fingers and the ligand binding domain (LBD) of 13 α-helices and a small 4-stranded β-sheet. PPARs are ligand-regulated transcription factors controlling gene expression by binding to specific response elements (PPREs) within promoters, where PPARs bind as heterodimers with a retinoid X receptor and interact with cofactors upon binding leading to the increasing of rate of transcription initiation.
Products for PPAR
- Cat.No. Product Name Information
-
GC39271
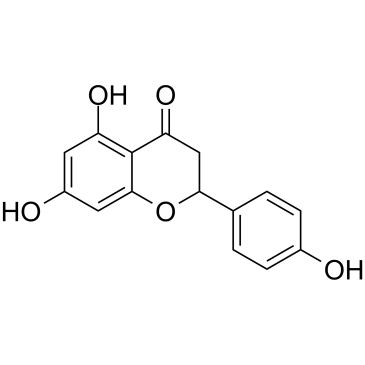
(±)-Naringenin
A citrusderived flavonoid
-
GC46365
1,1'-(Azodicarbonyl)dipiperidine
A Mitsunobu reagent

-
GC48782
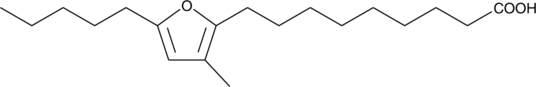
10,13-epoxy-11-methyl-Octadecadienoic Acid
A furan fatty acid
-
GC41867
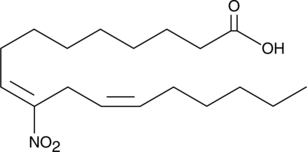
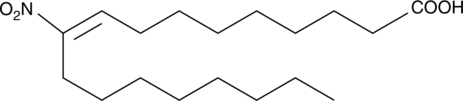
10-Nitrolinoleate
10-Nitrolinoleate is a potent peroxisome proliferator-activated receptor γ (PPARγ) agonist.
-
GC41868
10-Nitrooleate
10-Nitrooleate (CXA-10), a nitro fatty acid, has potential effects in disease states in which oxidative stress, inflammation, fibrosis, and/or direct tissue toxicity play significant roles.
-
GC13960
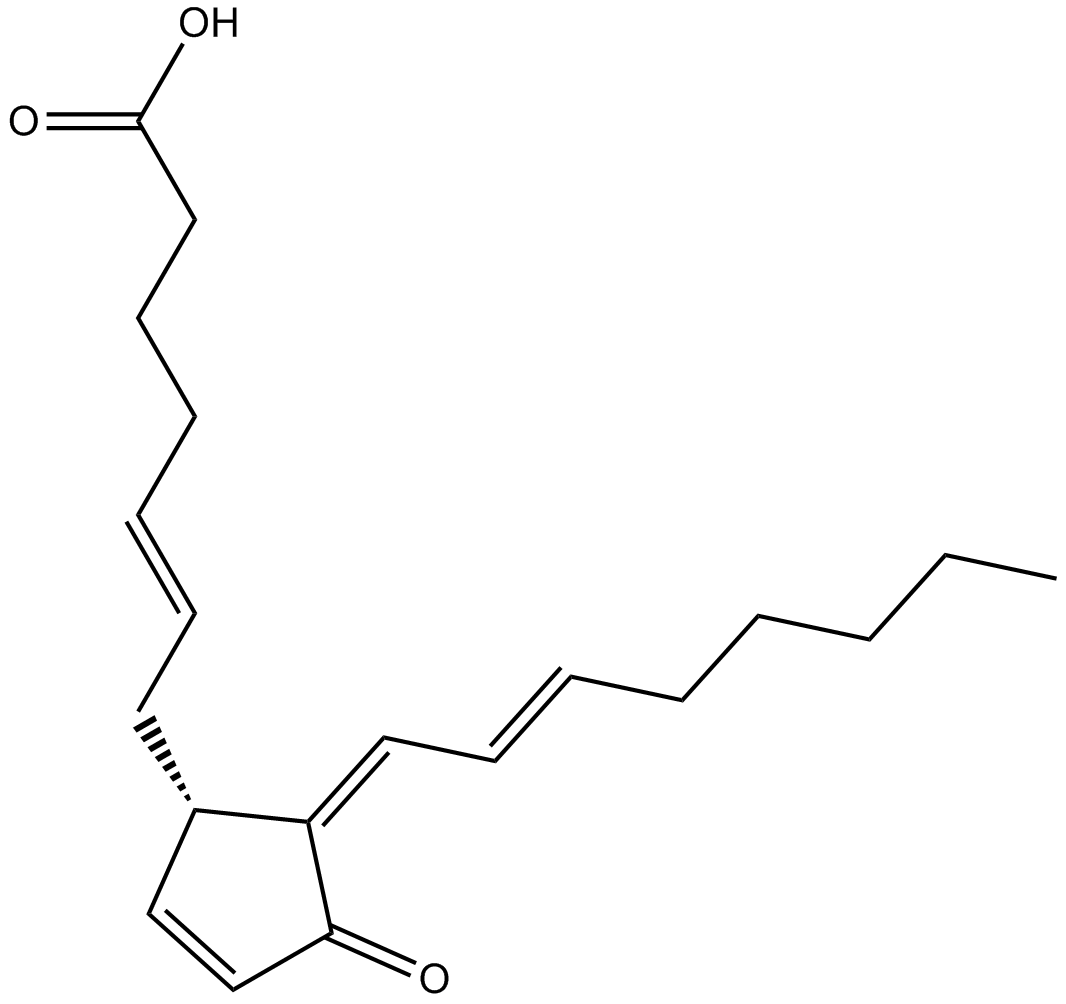
15-deoxy-Δ-12,14-Prostaglandin J2
15-deoxy-Δ-12,14-Prostaglandin J2 (15d-PGJ2) is a cyclopentenone prostaglandin and a metabolite of PGD2.
-
GC46447
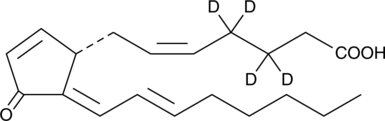
15-deoxy-δ12,14-Prostaglandin J2-d4
An internal standard for the quantification of 15deoxyΔ12,14prostaglandin J2
-
GC65414
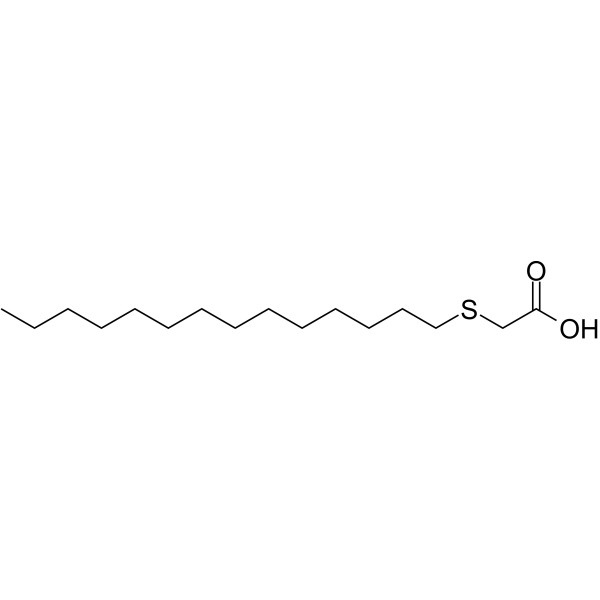
2-(Tetradecylthio)acetic acid
2-Tetradecylthio acetic acid is a pan-peroxisome proliferator activated receptor (pan-PPAR) activator.
-
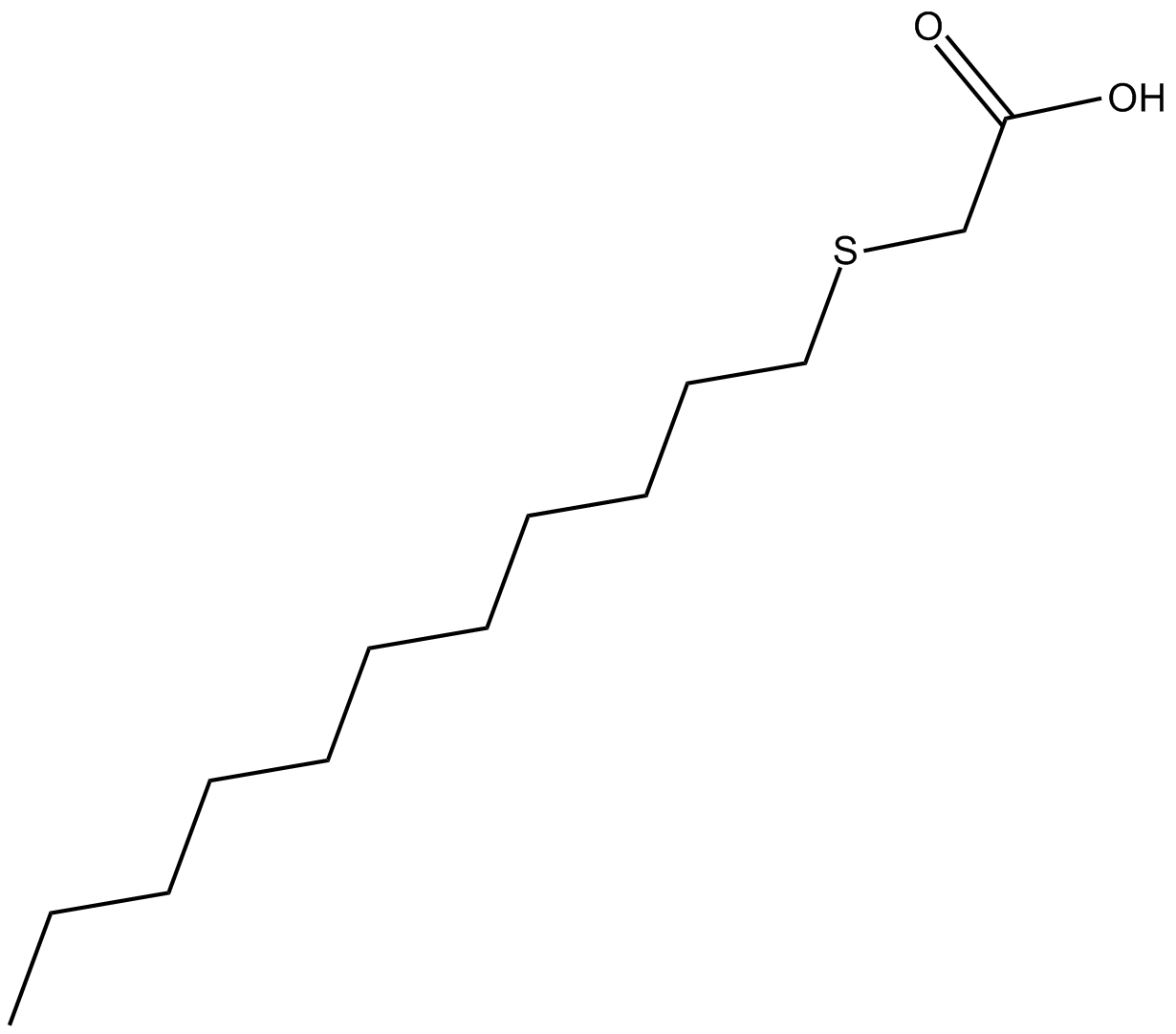
GC12289
3-Thiatetradecanoic Acid
activator of PPAR
-
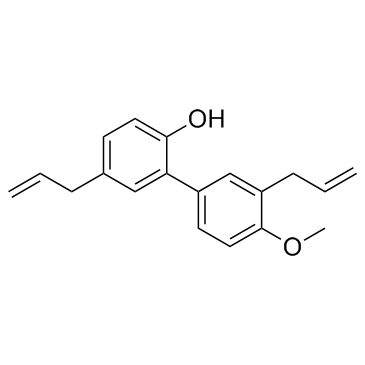
GC35143
4-O-Methyl honokiol
A phenol with diverse biological activities
-
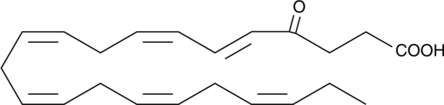
GC41530
4-oxo Docosahexaenoic Acid
4-oxo Docosahexaenoic acid (4-oxo DHA) is a putative metabolite of DHA with antiproliferative and PPARγ agonist activity.
-
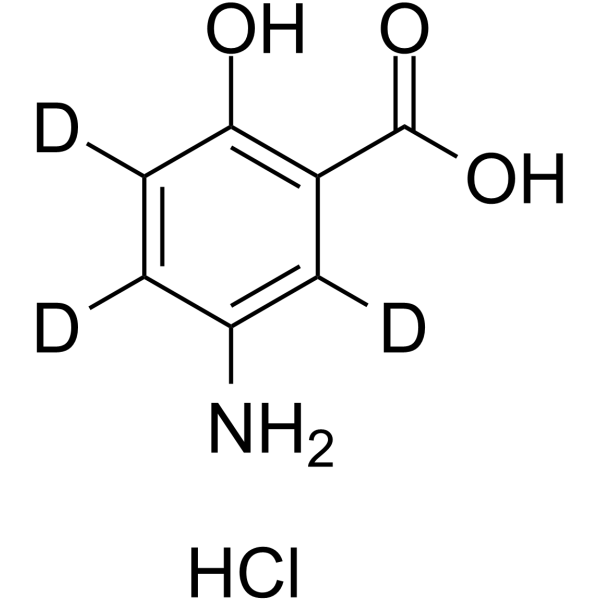
GC63358
5-Aminosalicylic Acid-D3 hydrochloride
-
GC46757
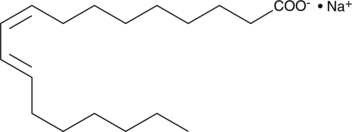
9(Z),11(E)-Conjugated Linoleic Acid (sodium salt)
An isomer of linoleic acid
-
GC42649
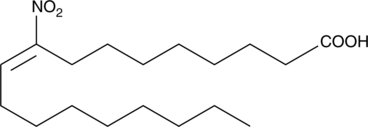
9-Nitrooleate
Nitrated unsaturated fatty acids, such as 10- and 12-nitrolinoleate, cholesteryl nitrolinoleate, and nitrohydroxylinoleate, represent a new class of endogenous lipid-derived signalling molecules.
-
GC42651
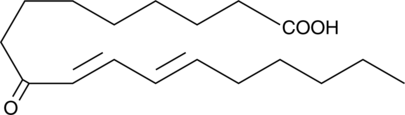
9-oxo-10(E),12(E)-Octadecadienoic Acid
9-oxo-10(E),12(E)-Octadecadienoic acid (9-oxoODA) is a natural agonist, abundant in tomatoes, that activates PPARα at 10-20 μM.
-
GC31693
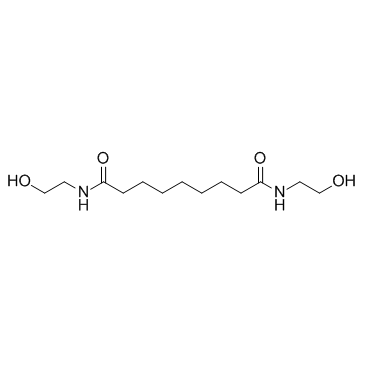
Adelmidrol
Adelmidrol exerts important anti-inflammatory effects that are partly dependent on PPARγ.
-
GN10413
Agrimol B
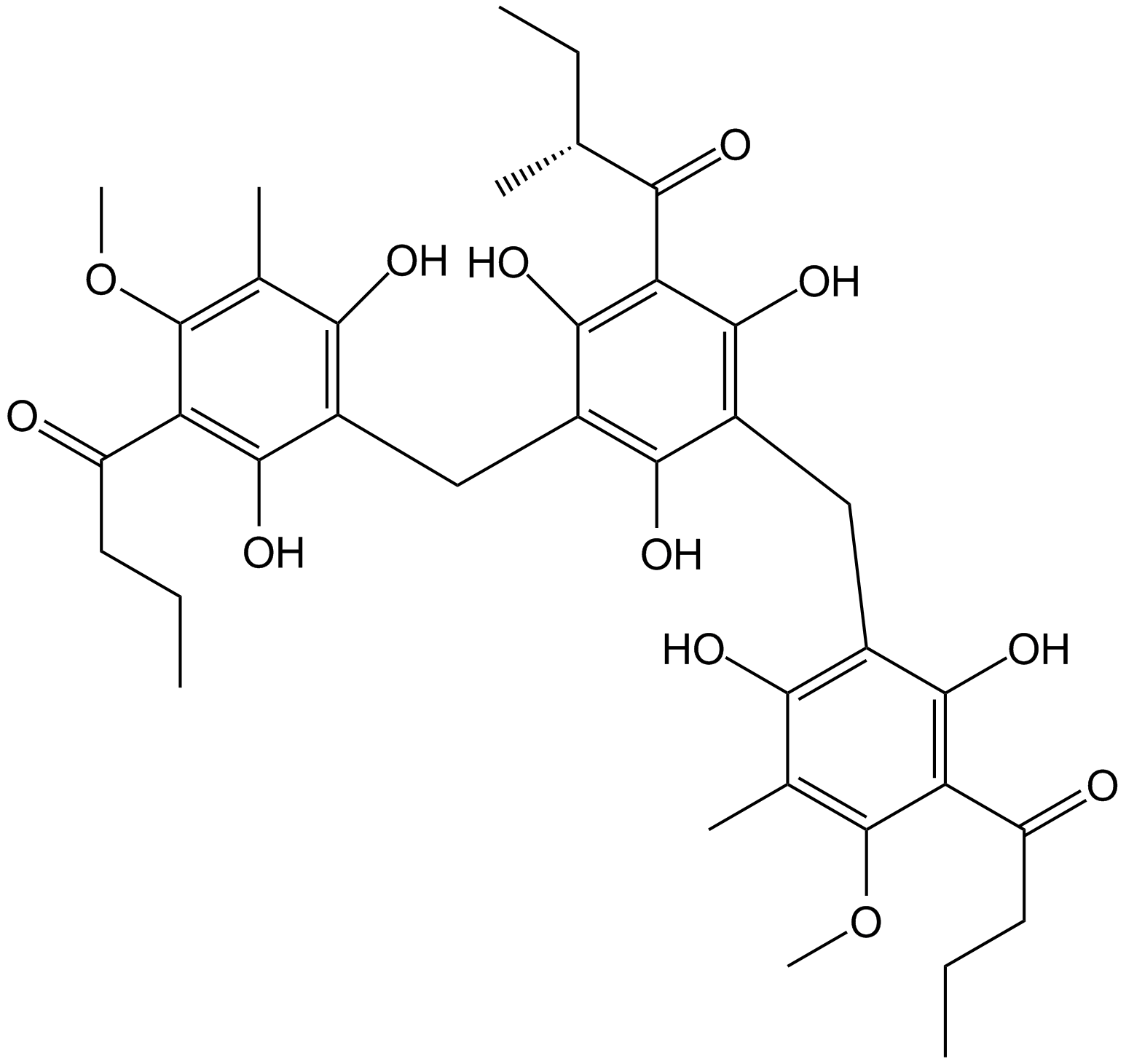
-
GC35281
Aleglitazar
Aleglitazar (R1439) is a potent dual PPARα/γ agonist, with IC50s of 38 nM and 19 nM for human PPARa and PPARγ, respectively.
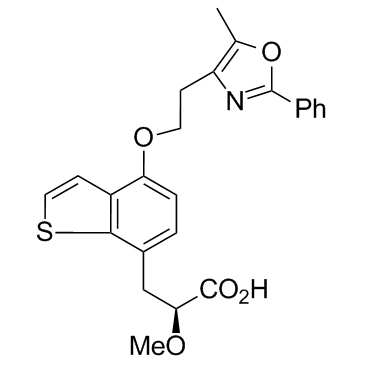
-
GC35308
Alpinetin
Alpinetin is a flavonoid isolated from Alpinia katsumadai Hayata, activates activates PPAR-γ, with potent anti-inflammatory activity.
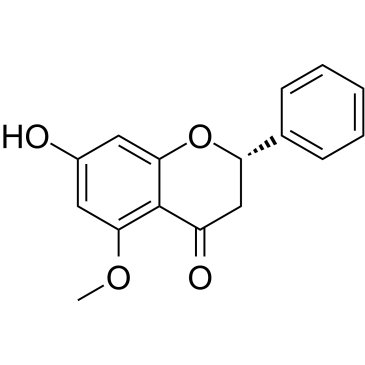
-
GC62836
AMG131
AMG131 (INT131), a potent and highly selective PPARγ partial agonist, binds to PPARγ and displaces Rosiglitazone with a Ki of ~10 nM.
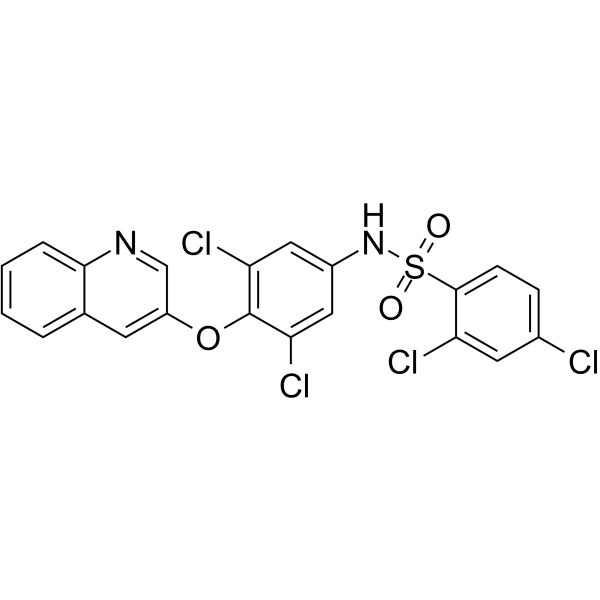
-
GC40636
Amorfrutin A
Amorfrutin A is an isoprenoid-substituted benzoic acid natural product found in the fruit of A.
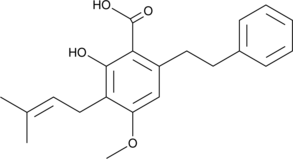
-
GC42791
Amorfrutin B
Amorfrutin B is a partial agonist of the peroxisome proliferator-activated receptor γ (PPARγ; Ki = 19 nM and EC50 = 73 nM) that was first isolated from A.
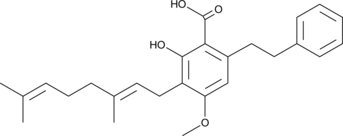
-
GC35349
Angeloylgomisin H
Angeloylgomisin H, as a major lignin extract of Schisandra rubriflora, has the potential to improve insulin-stimulated glucose uptake by activating PPAR-γ.
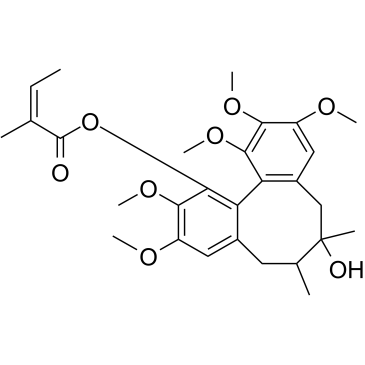
-
GC35358
Ankaflavin
Ankaflavin, isolated from Monascus-Fermented red rice, is an orally active PPARγ agonist.
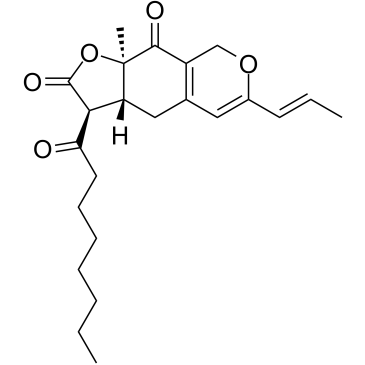
-
GC31462
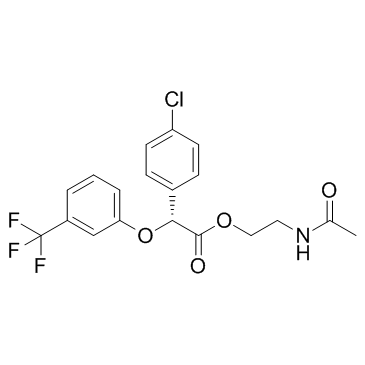
Arhalofenate (MBX 102)
Arhalofenate (MBX 102) (MBX 102) is a selective partial agonist of peroxisome proliferator-activated receptor (PPAR)-γ, used for the treatment of type 2 diabetes.
-
GC31350
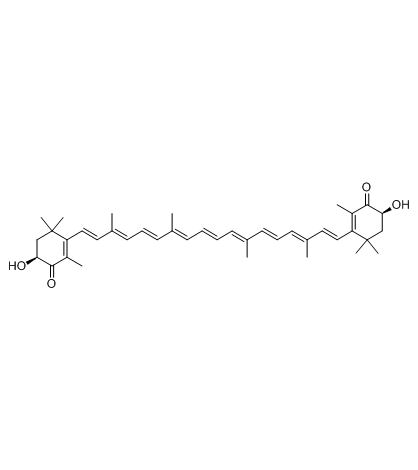
Astaxanthin
Astaxanthin, the red dietary carotenoid, is an orally effective and potent antioxidant.
-
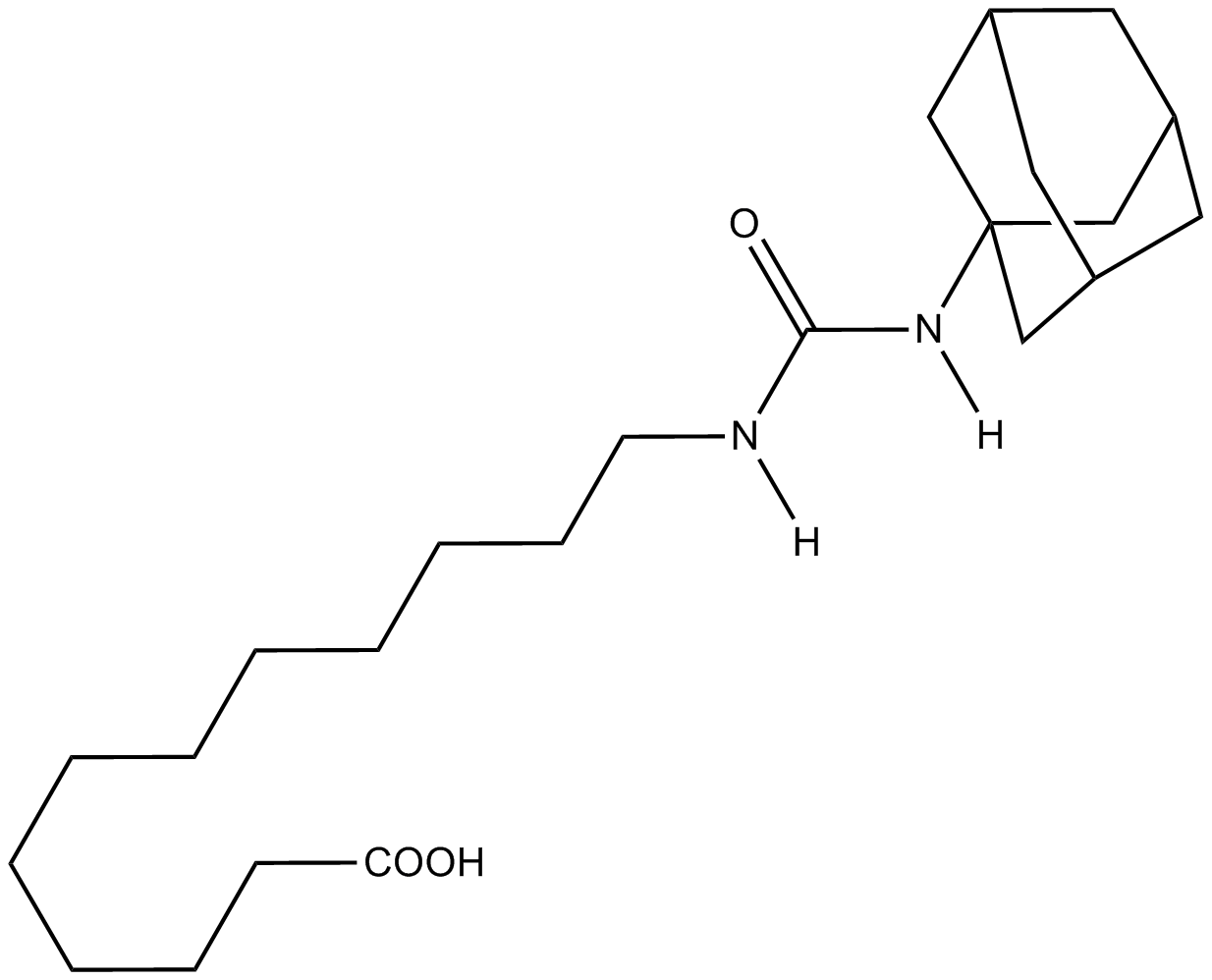
GC10381
AUDA
Potent epoxide hydrolase inhibitor/PPARα activator
-
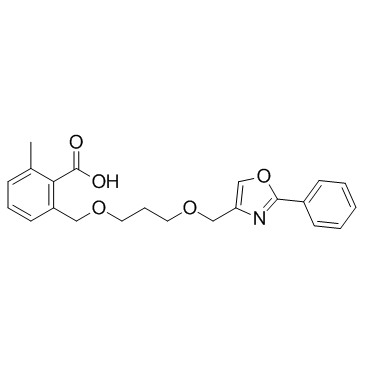
GC31452
AVE-8134
AVE-8134 is a potent PPARα agonist, with EC50 values of 100 and 3000 nM for human and rodent PPARα receptor, respectively.
-
GC64099
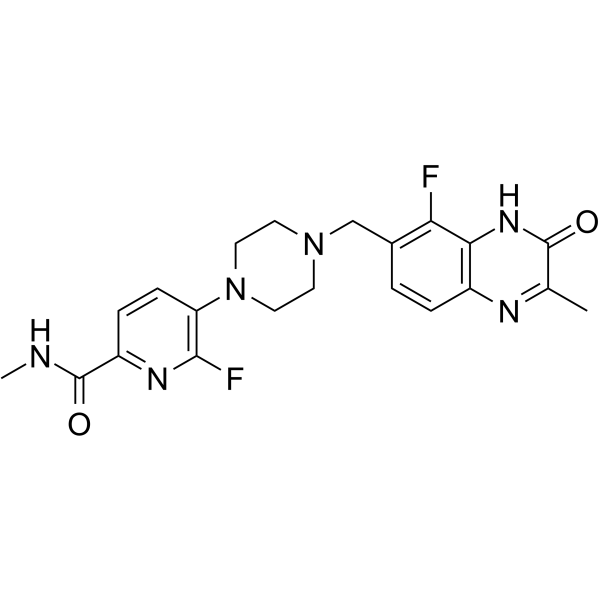
AZD-9574
AZD-9574 is a potent and brain penetrant PARP1 inhibitor and shows >8000-fold selectivity for PARP1 compared to PARP2/3/5a/6.
-
GC42890
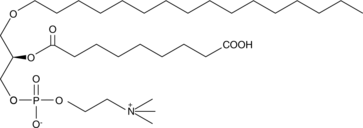
Azelaoyl PAF
Oxidized low-density lipoprotein (oxLDL) particles contain low molecular weight species which promote the differentiation of monocytes via the nuclear receptor PPARγ.
-
GC13829
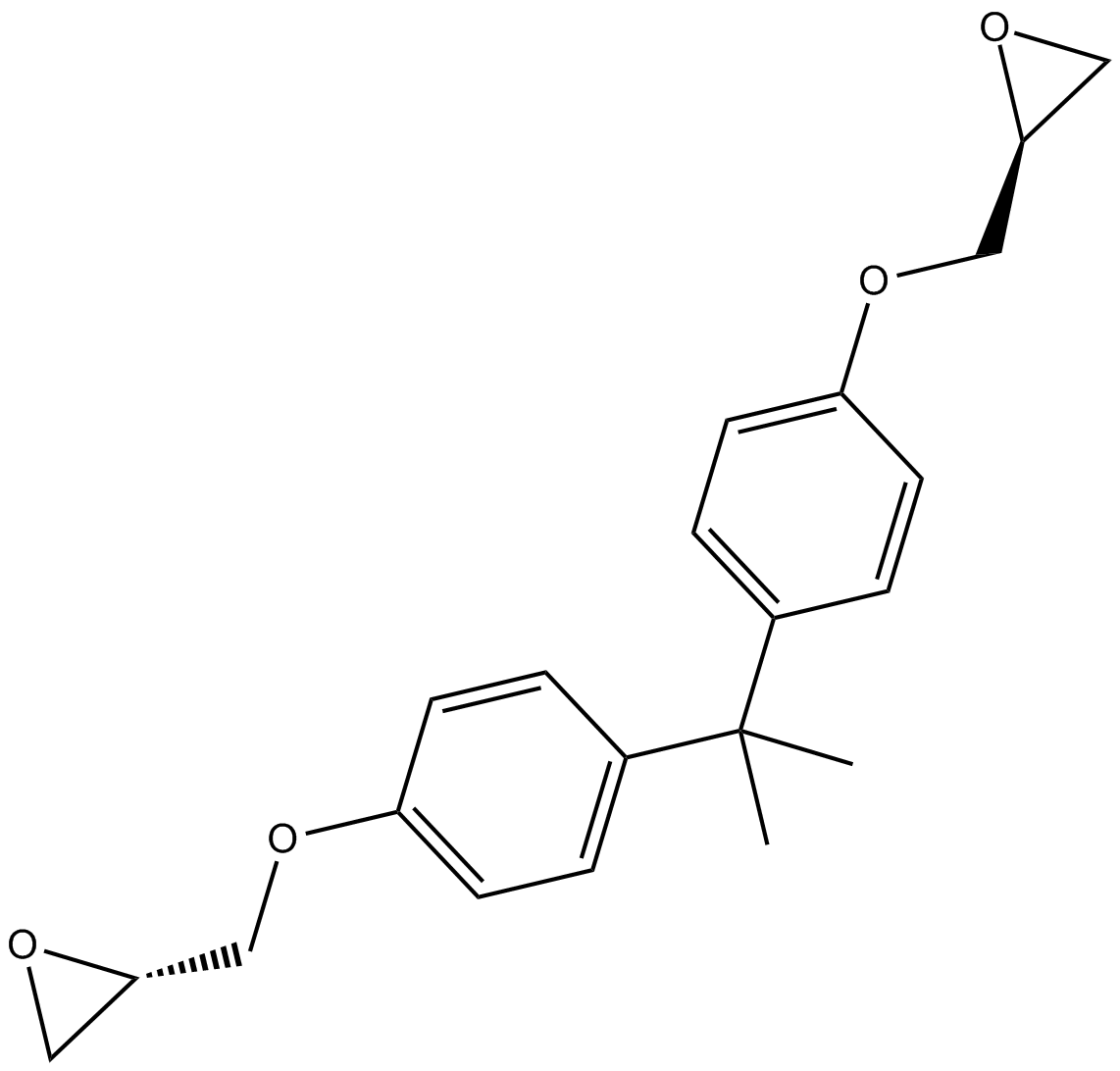
BADGE
PPARγ antagonist
-
GC12894
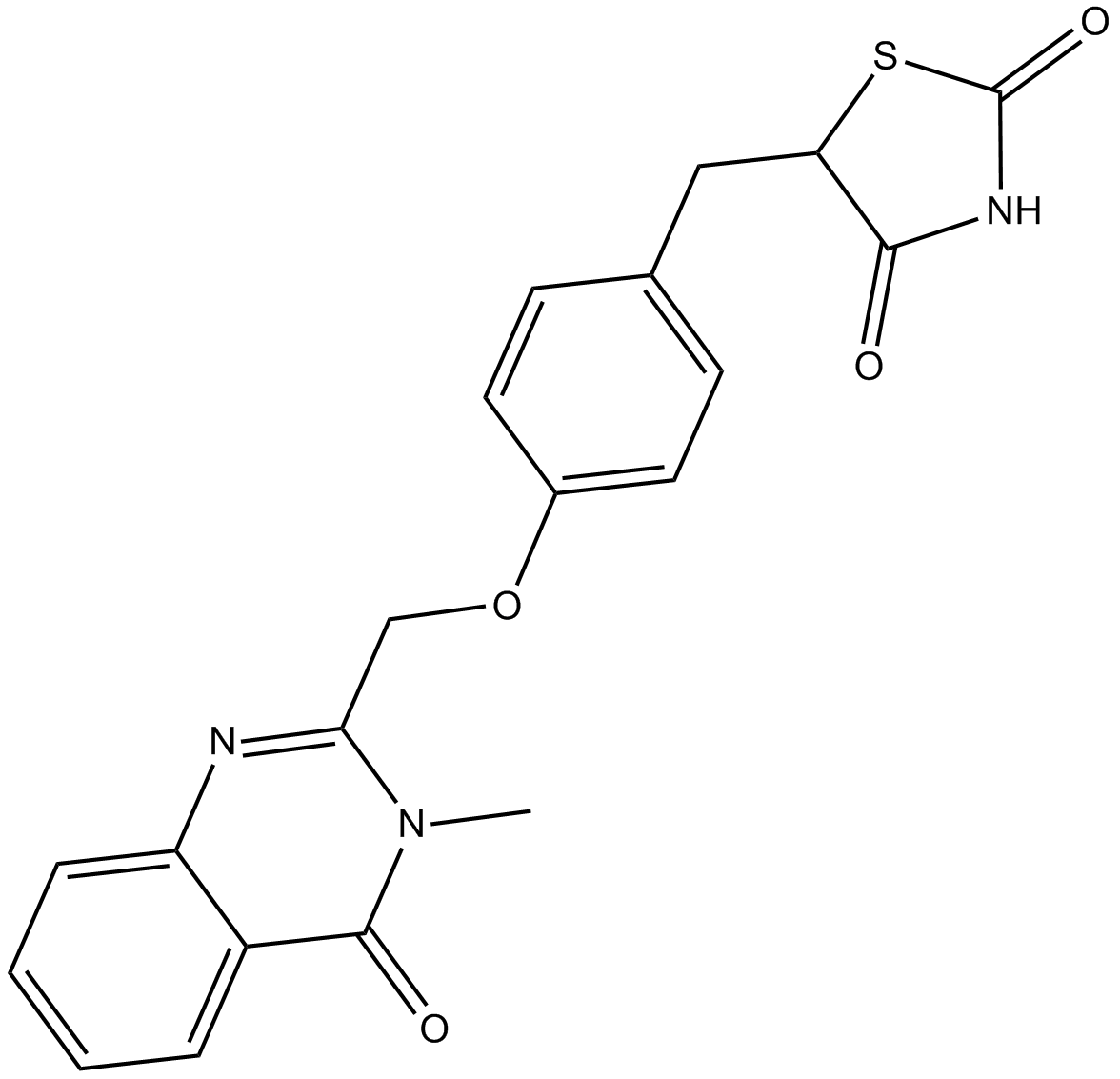
Balaglitazone
A partial agonist of PPARγ
-
GC14543
Bezafibrate
Lipid-lowering agent
-
GC45726
Bezafibrate-d4
An internal standard for the quantification of bezafibrate
-
GC35518
Bilobetin
A biflavonoid with diverse biological activities
-
GC15995
BMS 687453
PPARα agonist
-
GC64699
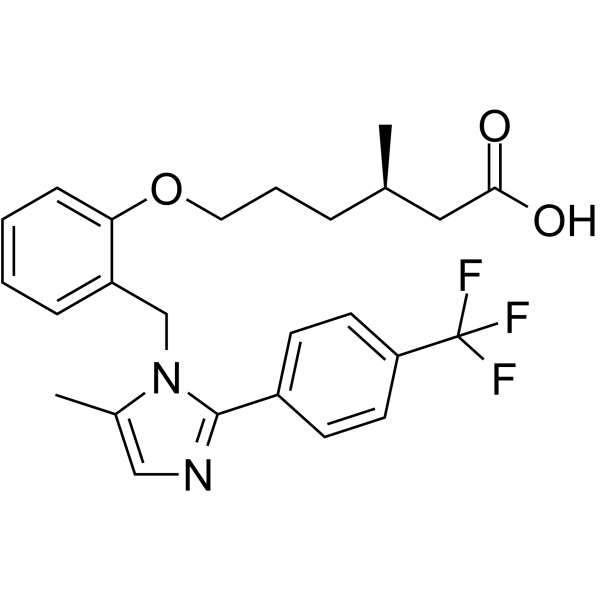
Bocidelpar
Bocidelpar is a modulator of peroxisome proliferator-activated receptor delta (PPAR-δ).
-
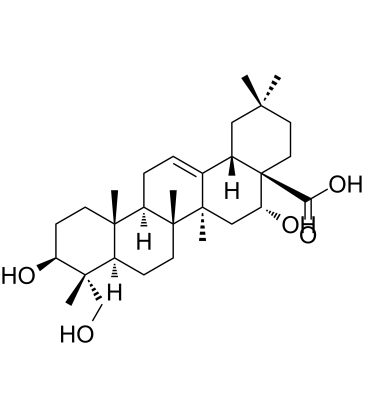
GC60675
Caulophyllogenin
Caulophyllogenin is a triterpene saponin extracted from M.
-
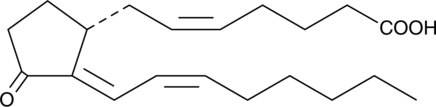
GC40384
CAY10410
CAY10410 is an analog of prostaglandin D2/prostaglandin J2 (PGD2/PGJ2) with structural modifications intended to give it PPARγ ligand activity and resistance to metabolism.
-
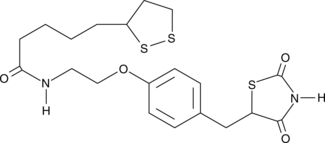
GC43162
CAY10506
Anti-diabetic drugs of the thiazolidinedione (TZD) structural class as well as α-lipoic acid activate peroxisome proliferator-activated receptor γ (PPARγ), a nuclear transcription factor that controls glucose metabolism and lipid homeostasis.
-
GC40764
CAY10514
CAY10514 is an aromatic analog of 8(S)-HETE.
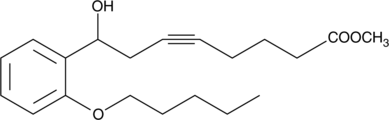
-
GC43174
CAY10573
The peroxisome proliferator-activated receptors (PPARs) are lipid-activated transcription factors often used as drug targets for the treatment of metabolic disorders.
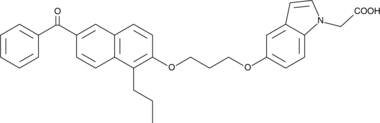
-
GC43180
CAY10592
Peroxisome proliferator-activated receptors (PPARs) α, δ, γ are ligand-activated nuclear transcription factors involved in the regulation of energy homeostasis as well as insulin sensitivity and glucose metabolism.
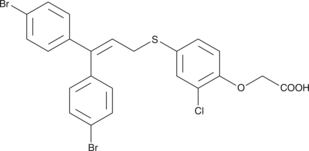
-
GC18876
CAY10599
Thiazolidinediones (TZDs) are a group of structurally related peroxisome proliferator-activated receptor γ (PPARγ) agonists with antidiabetic actions in vivo.
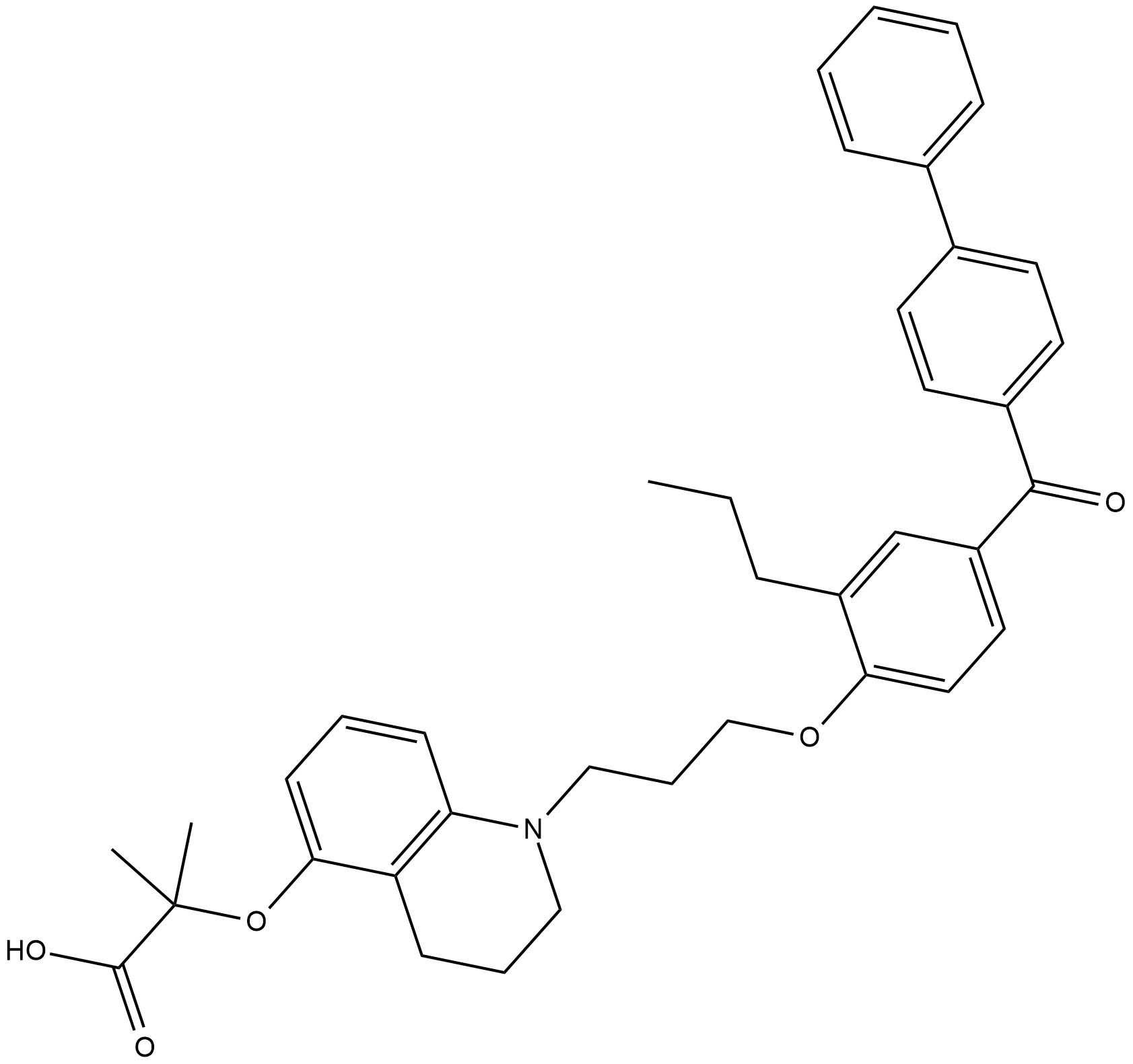
-
GC47063
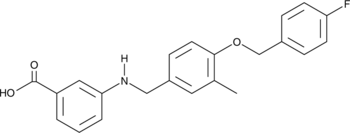
CAY10767
A PPARα agonist
-
GC48426
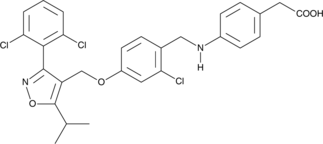
CAY10771
A dual agonist of FXR and PPARδ
-
GC32723
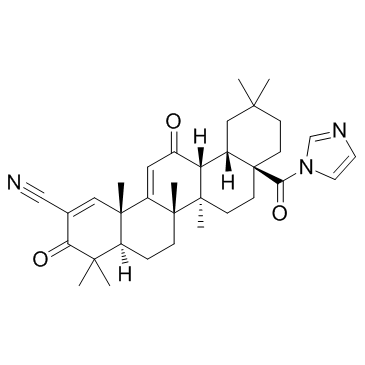
CDDO-Im (RTA-403)
CDDO-Im (RTA-403) (RTA-403) is an activator of Nrf2 and PPAR, with Kis of 232 and 344 nM for PPARα and PPARγ.
-
GC61805
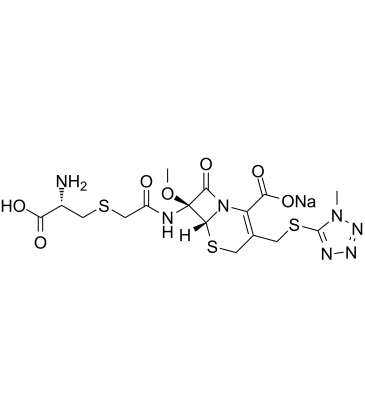
Cefminox sodium
Cefminox sodium (MT-141) is a semisynthetic cephamycin, which exhibits a broad spectrum of antibacterial activity.
-
GC16301
Cetaben
unique,PPARα-independent peroxisome proliferator
-
GC31552
Chiglitazar
Chiglitazar (Carfloglitazar) is a PPARα/γ dual agonist, with EC50s of 1.2, 0.08, 1.7 μM for PPARα, PPARγ and PPARδ, respectively.
-
GC11170
Choline Fenofibrate
-
GC17135
Ciglitazone
PPARγ agonist
-
GN10678
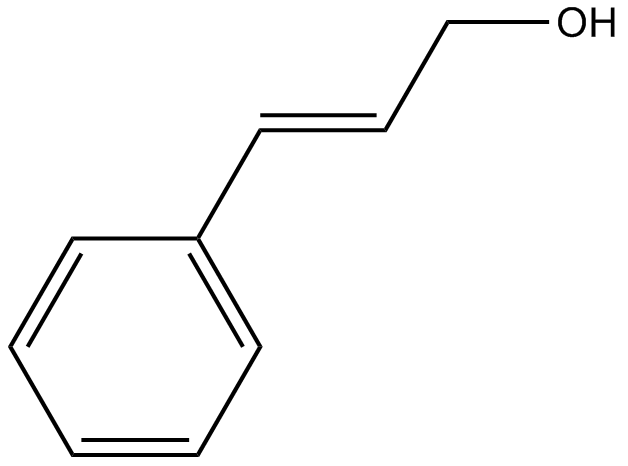
Cinnamyl alcohol
-
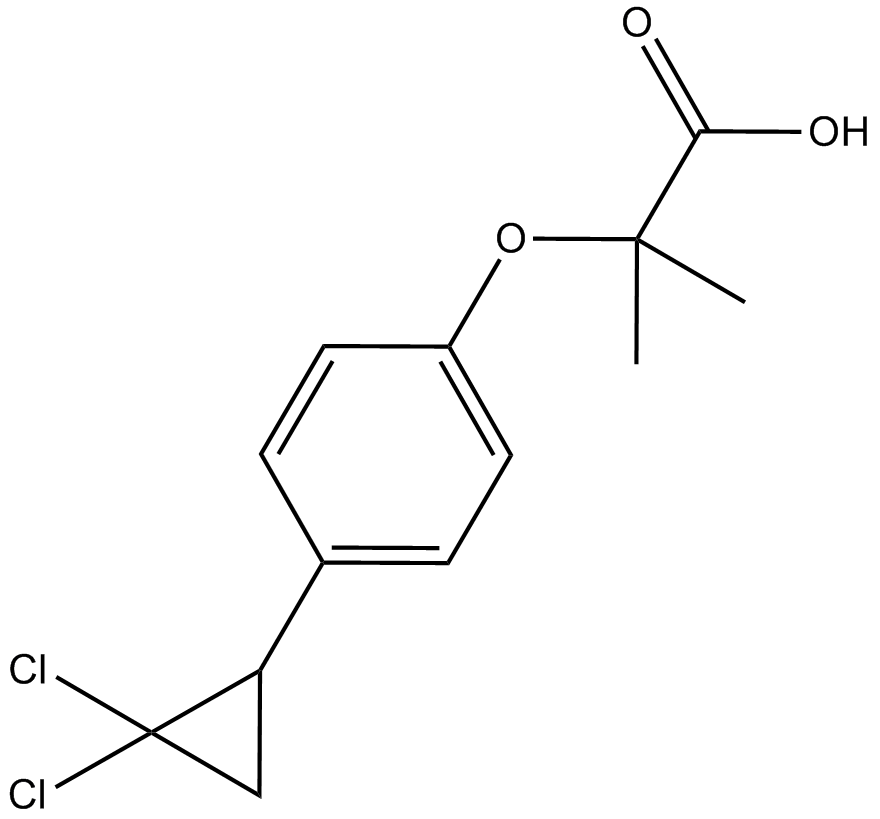
GC13633
Ciprofibrate
A selective PPARα agonist
-
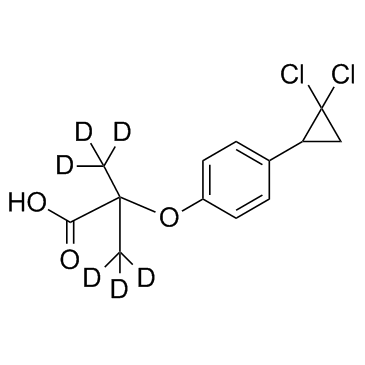
GC35698
Ciprofibrate D6
Ciprofibrate D6 is deuterium labeled Ciprofibrate.
-
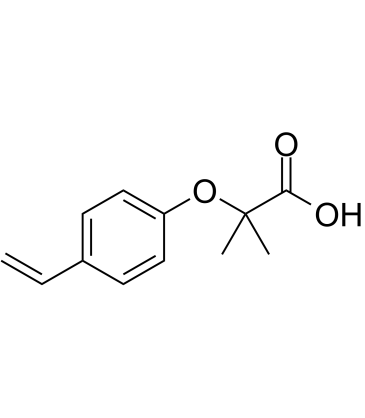
GC60710
Ciprofibrate impurity A
Ciprofibrate impurity A is an impurity of Ciprofibrate. Ciprofibrate (Win35833) is a potent peroxisome proliferator, increases the phosphorylation level of the PPARalpha.
-
GC17377
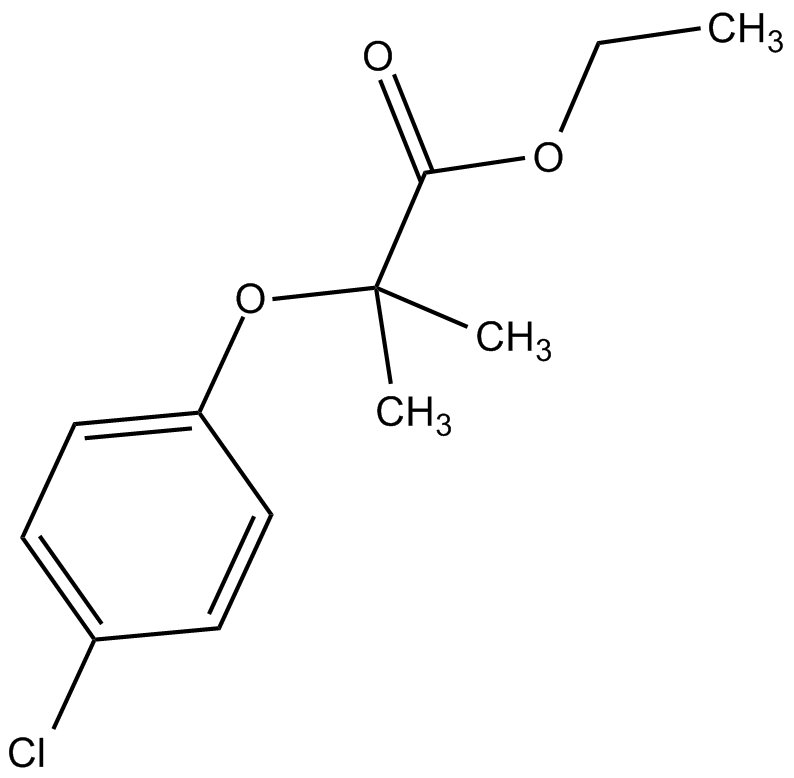
Clofibrate
PPAR agonist
-
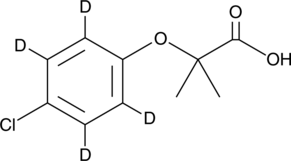
GC49087
Clofibric Acid-d4
An internal standard for the quantification of clofibric acid
-
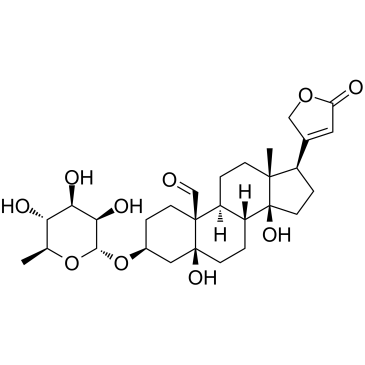
GC39701
Convallatoxin
Convallatoxin is a cardiac glycoside isolated from Adonis amurensis Regel et Radde.
-
GC15673
CP 775146
PPARα agonist
-
GN10293
Daidzein
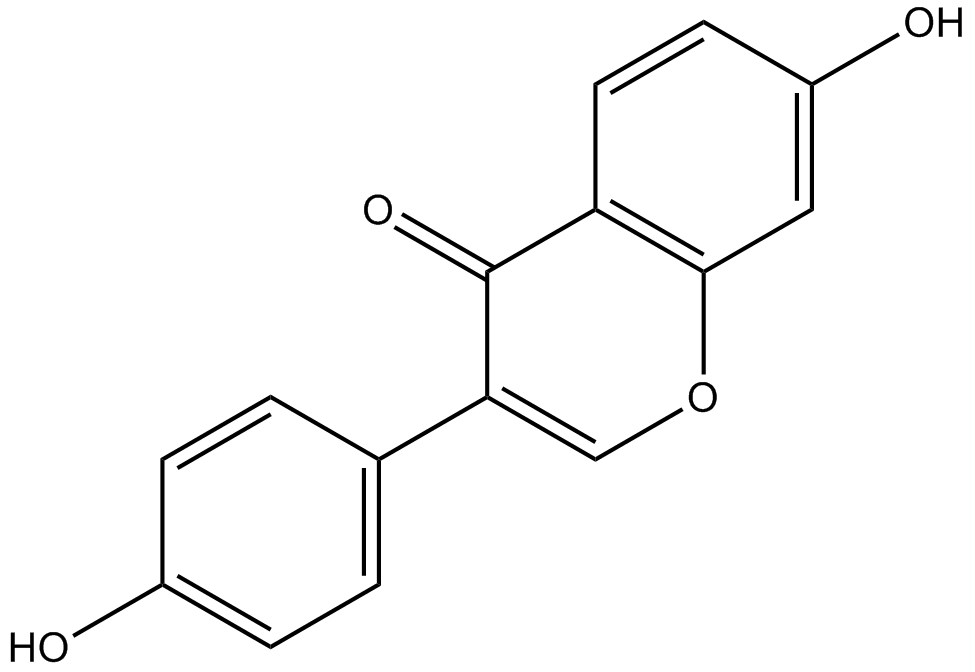
-
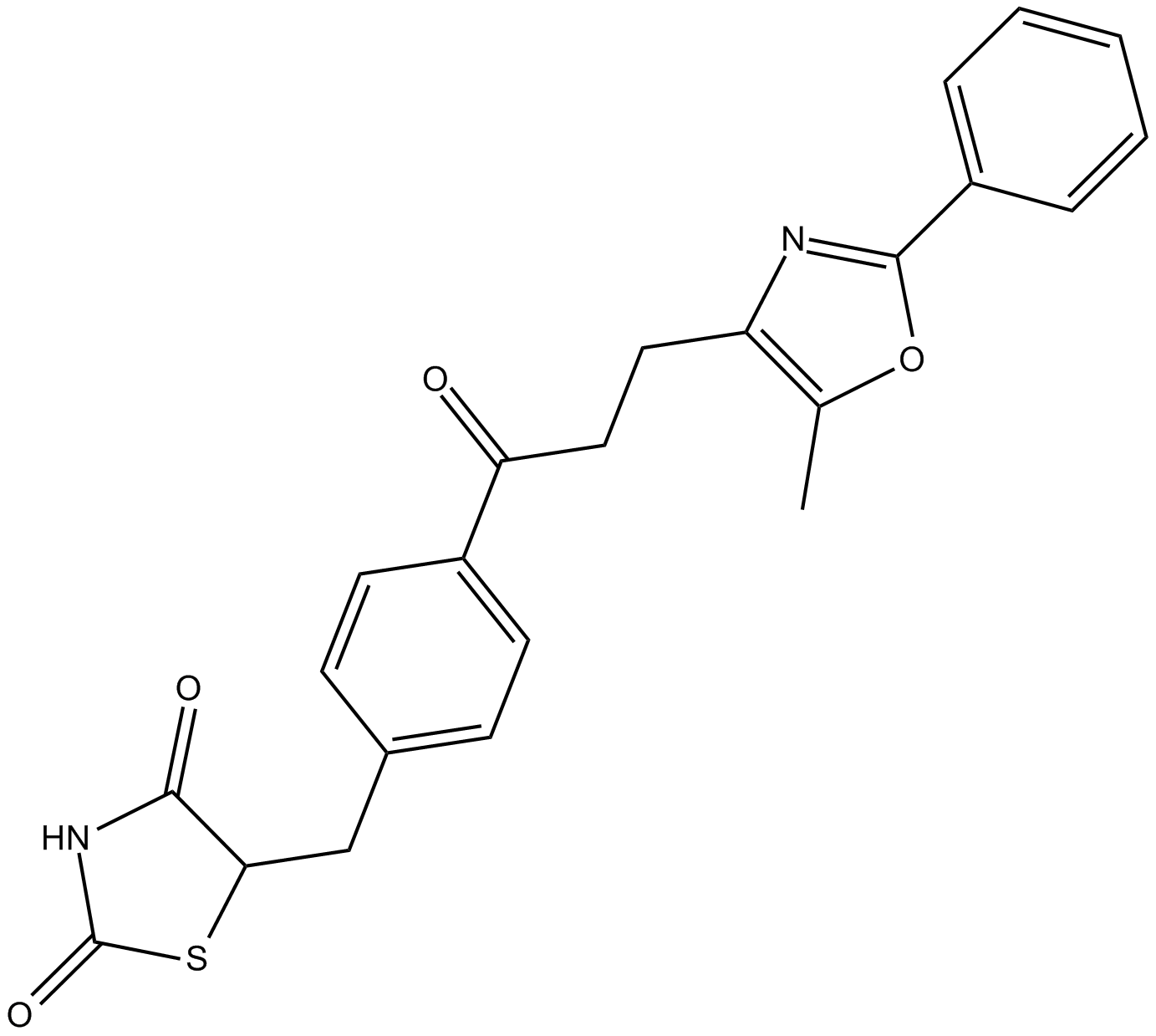
GC11477
Darglitazone
PPARγ agonist with antidiabetic actions
-
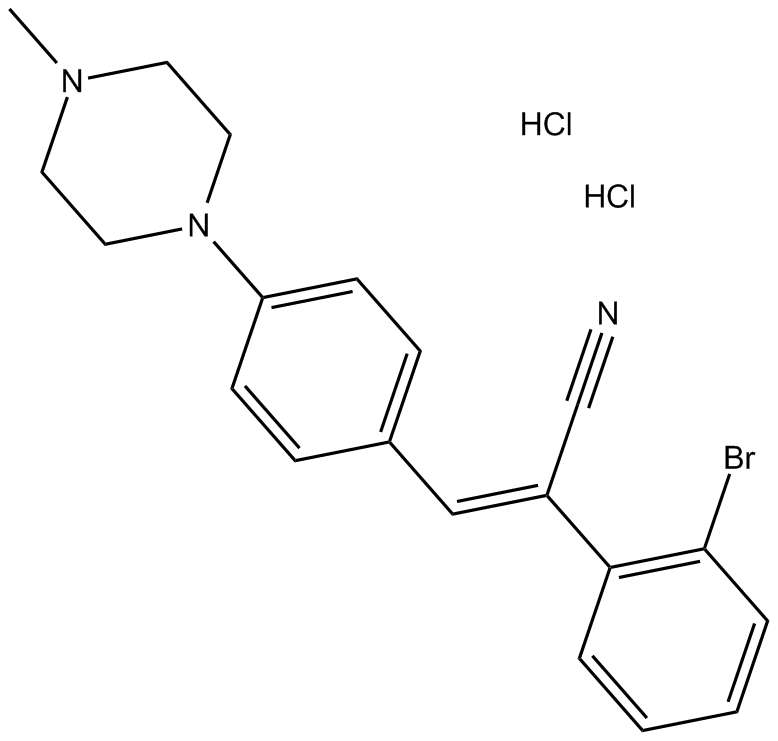
GC10190
DG-172 (hydrochloride)
DG-172 (hydrochloride) is a selective PPARβ/δ antagonist, with an IC50 of 27 nM.
-
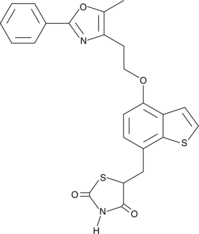
GC41492
Edaglitazone
Edaglitazone is an agonist of peroxisome proliferator-activated receptor γ (PPARγ).
-
GC38015
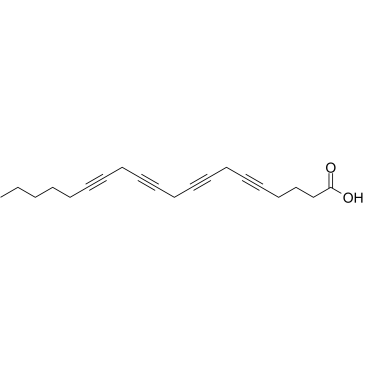
Eicosatetraynoic acid
A nonspecific inhibitor of cyclooxygenases and lipoxygenases
-
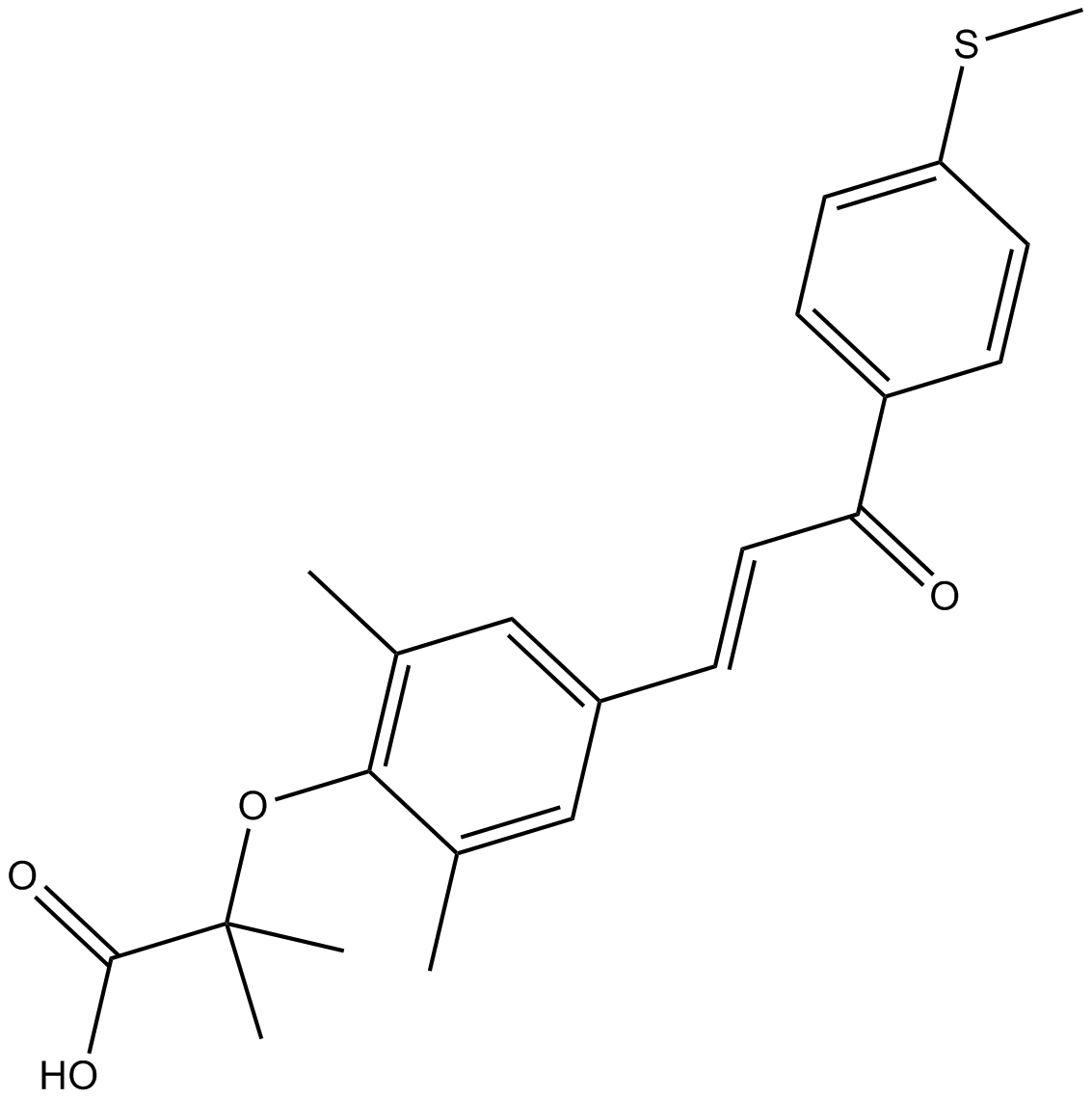
GC18151
Elafibranor (GFT505)
Elafibranor (GFT505) (GFT505) is a PPARα/δ agonist with EC50s of 45 and 175 nM, respectively.
-
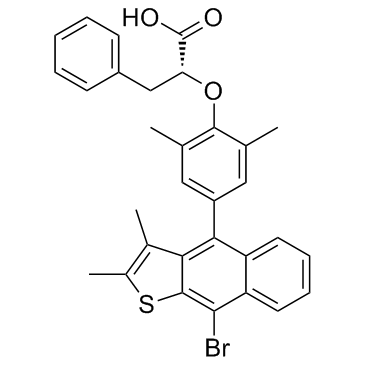
GC33816
Ertiprotafib (PTP 112)
Ertiprotafib (PTP 112) is an inhibitor of PTP1B, IkB kinase β (IKK-β), and a dual PPARα and PPARβ agonist, with an IC50 of 1.6 μM for PTP1B, 400 nM for IKK-β, an EC50 of ~1 μM for PPARα/PPARβ.
-
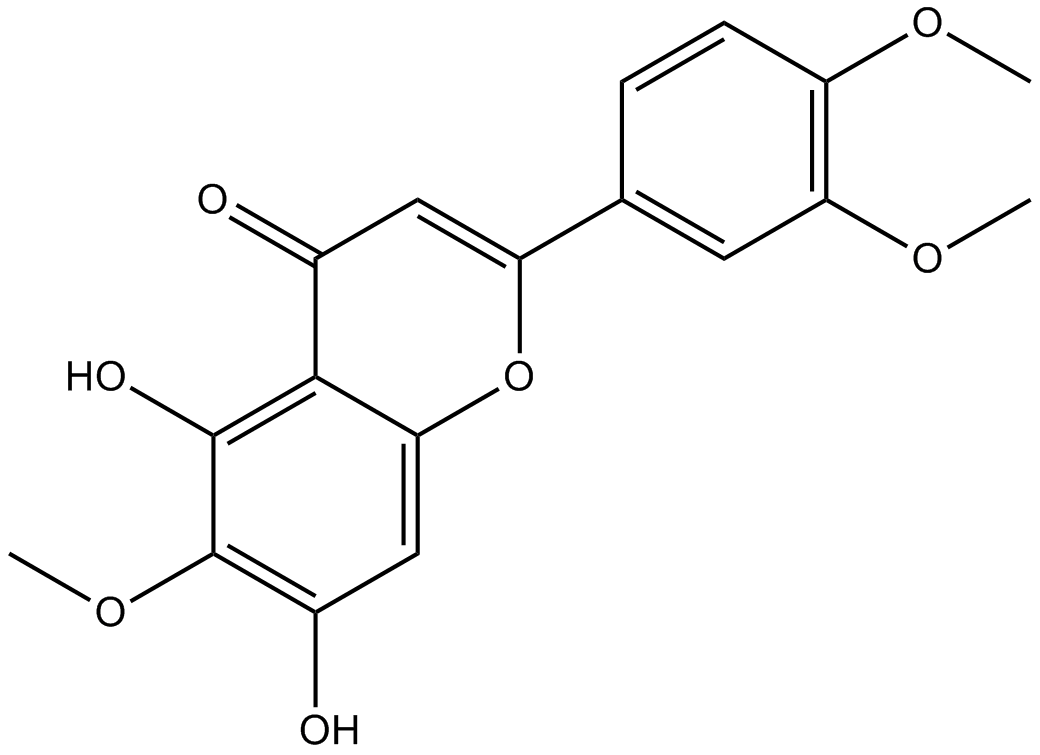
GN10511
Eupatilin
-
GC62203
Falcarindiol
Falcarindiol, an orally active polyacetylenic oxylipin, activates PPARγ and increases the expression of the cholesterol transporter ABCA1 in cells.
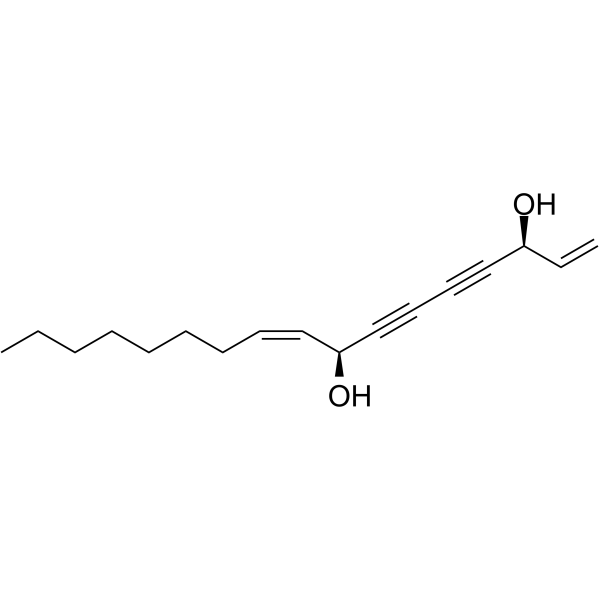
-
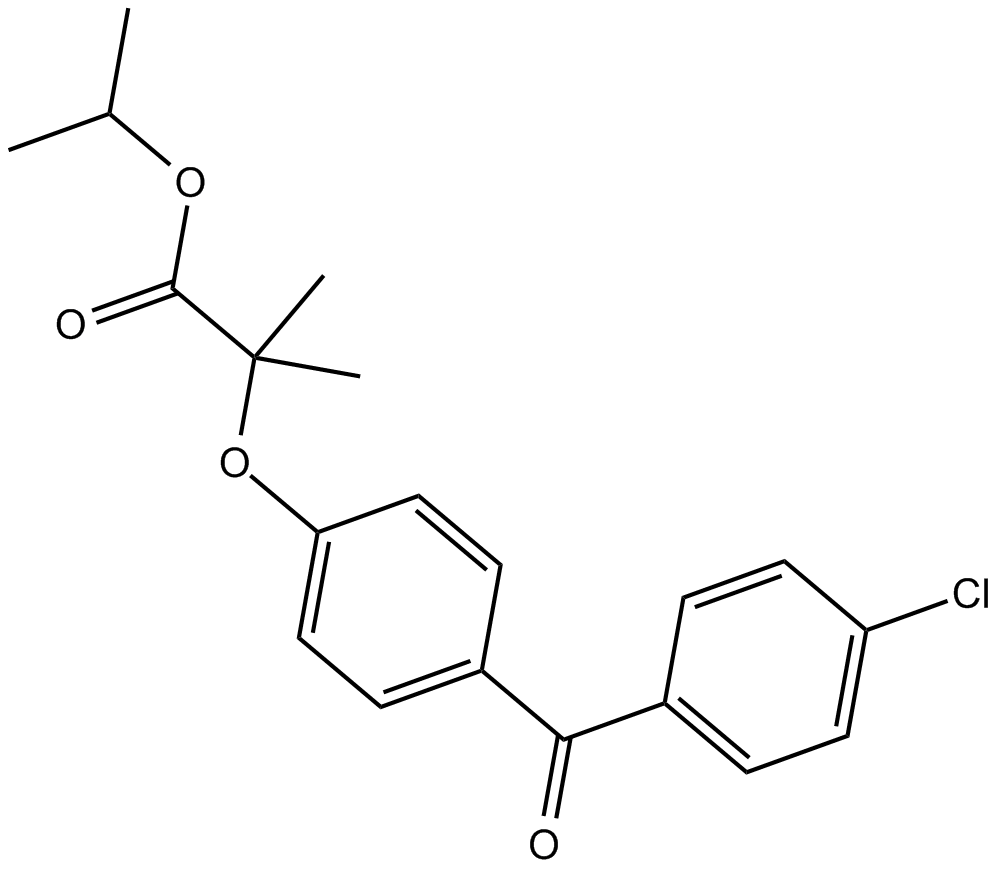
GC12250
Fenofibrate
PPARα agonist
-
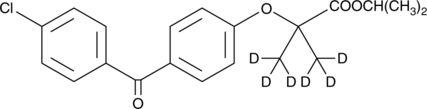
GC47337
Fenofibrate-d6
An internal standard for the quantification of fenofibrate
-
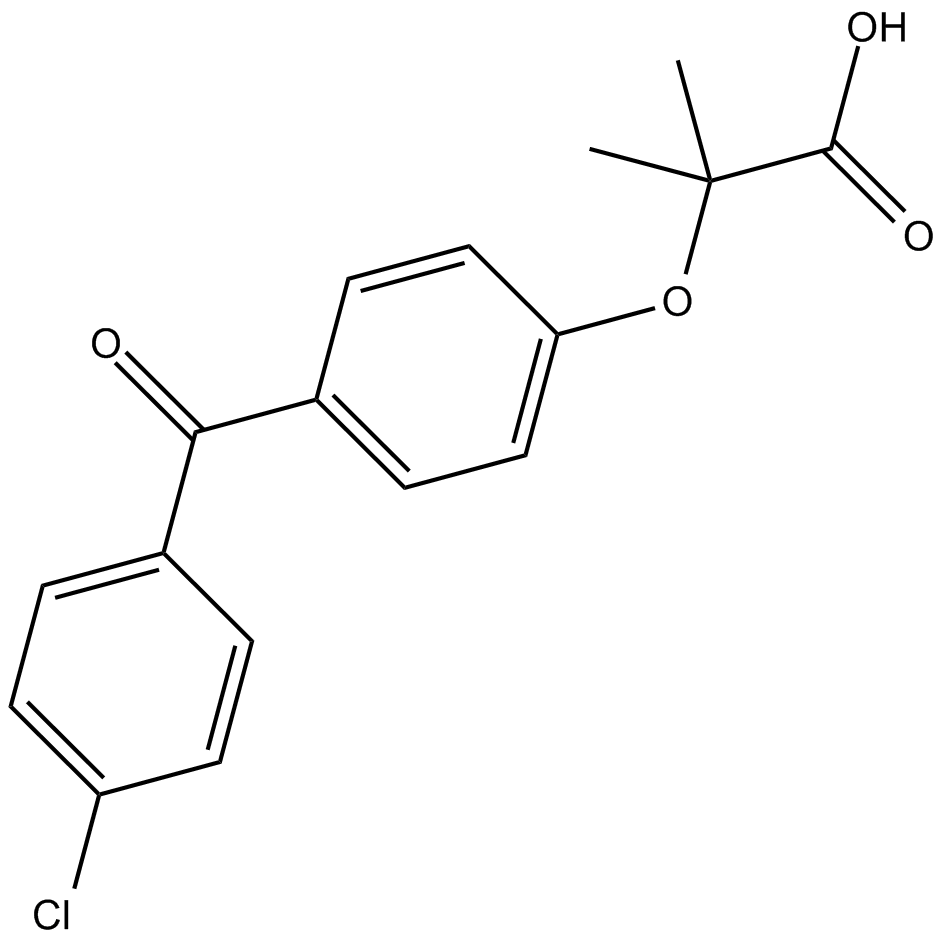
GC14584
Fenofibric acid
ppar inhibitor, lipid-lowering agent
-
GC12134
FH535
An inhibitor of β-catenin signaling
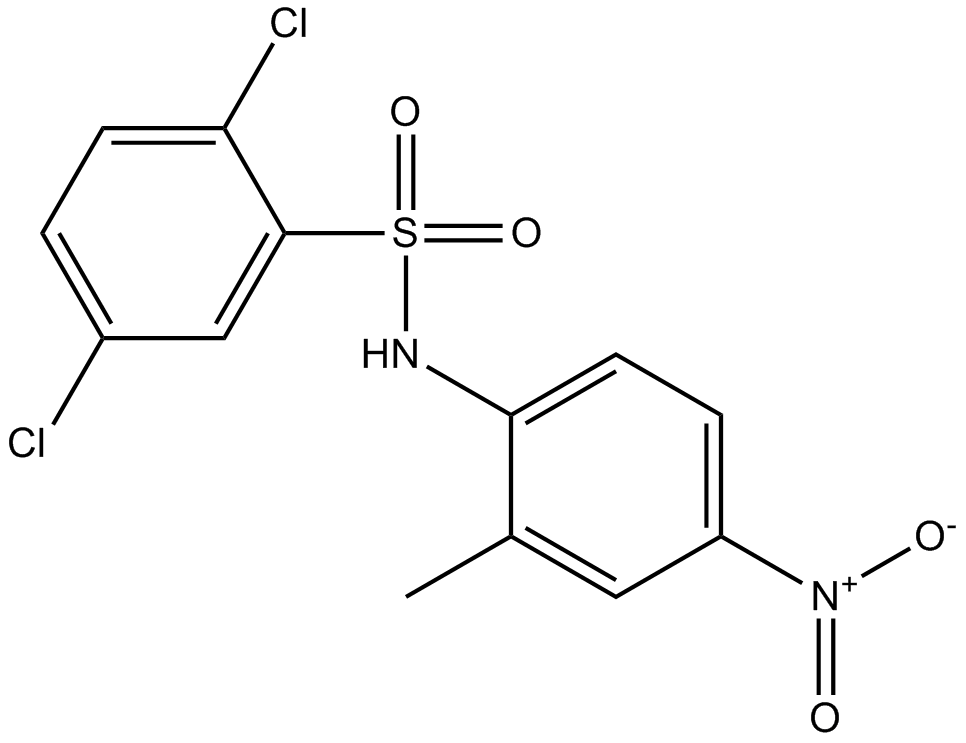
-
GN10030
Fisetin
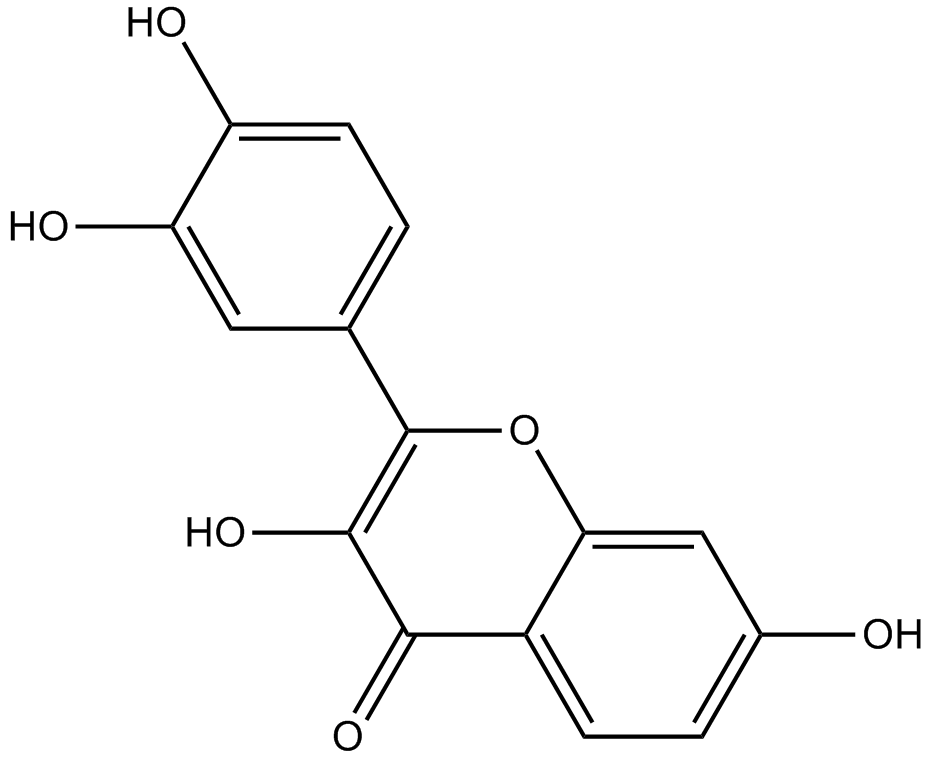
-
GC62974
FK614
FK614 is an orally active, non-thiazolidinedione (TZD) type, and selective PPARγ modulator (SPPARM).
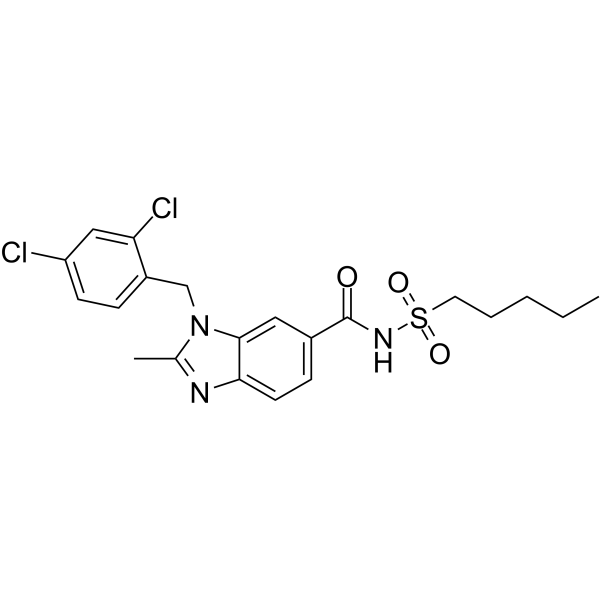
-
GC33734
Fonadelpar (NPS-005)
Fonadelpar (NPS-005) is a PPARδ agonist, used in the research of neuroparalytic keratopathy.
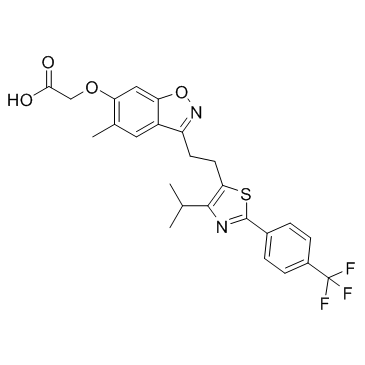
-
GC15085
G3335
G3335 is a selective, reversible and cell-permeable PPARγ with a Kd of ~8 μM.
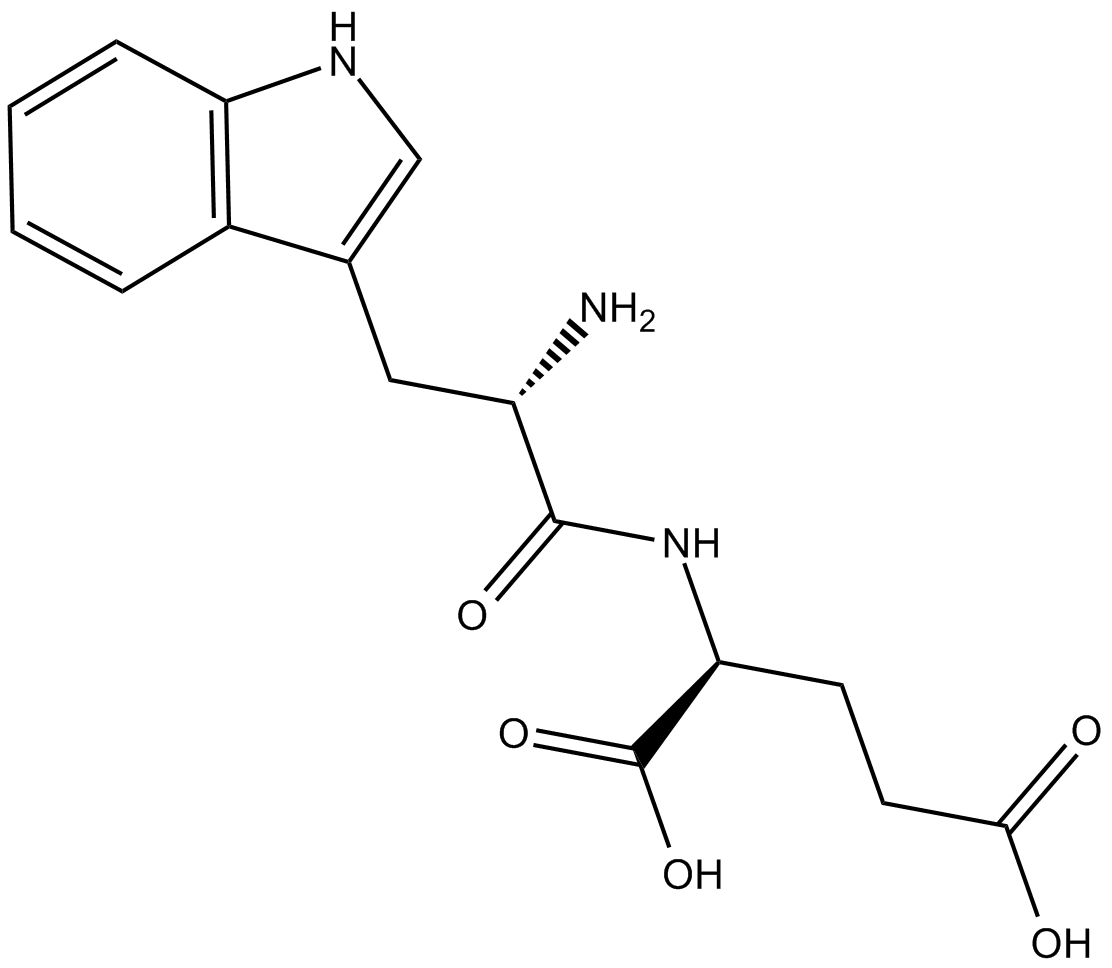
-
GC43723
Galactosylsphingosine (d18:1)
Galactosylsphingosine (d18:1) (Galactosylsphingosine), a substrate of the galactocerebrosidase (GALC) enzyme, is a potential biomarker for Krabbe disease.
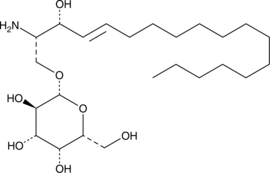
-
GC14915
Gemfibrozil
PPARα activator
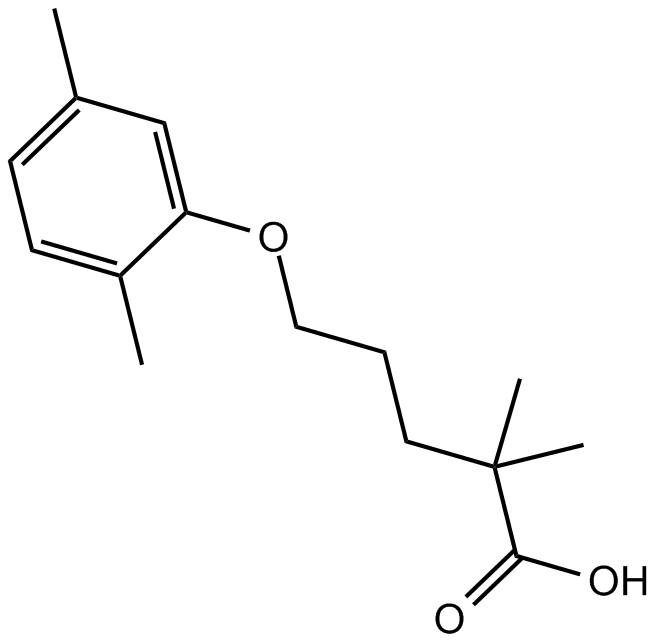
-
GC43743
Gemfibrozil 1-O-β-Glucuronide
Gemfibrozil 1-O-β-glucuronide is a major metabolite of gemfibrozil that is formed when gemfibrozil undergoes glucuronidation by UDP-glucuronosyltransferase (UGT) 2B7.
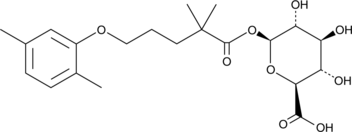
-
GC47397
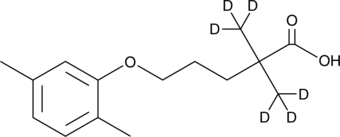
Gemfibrozil-d6
An internal standard for the quantification of gemfibrozil
-
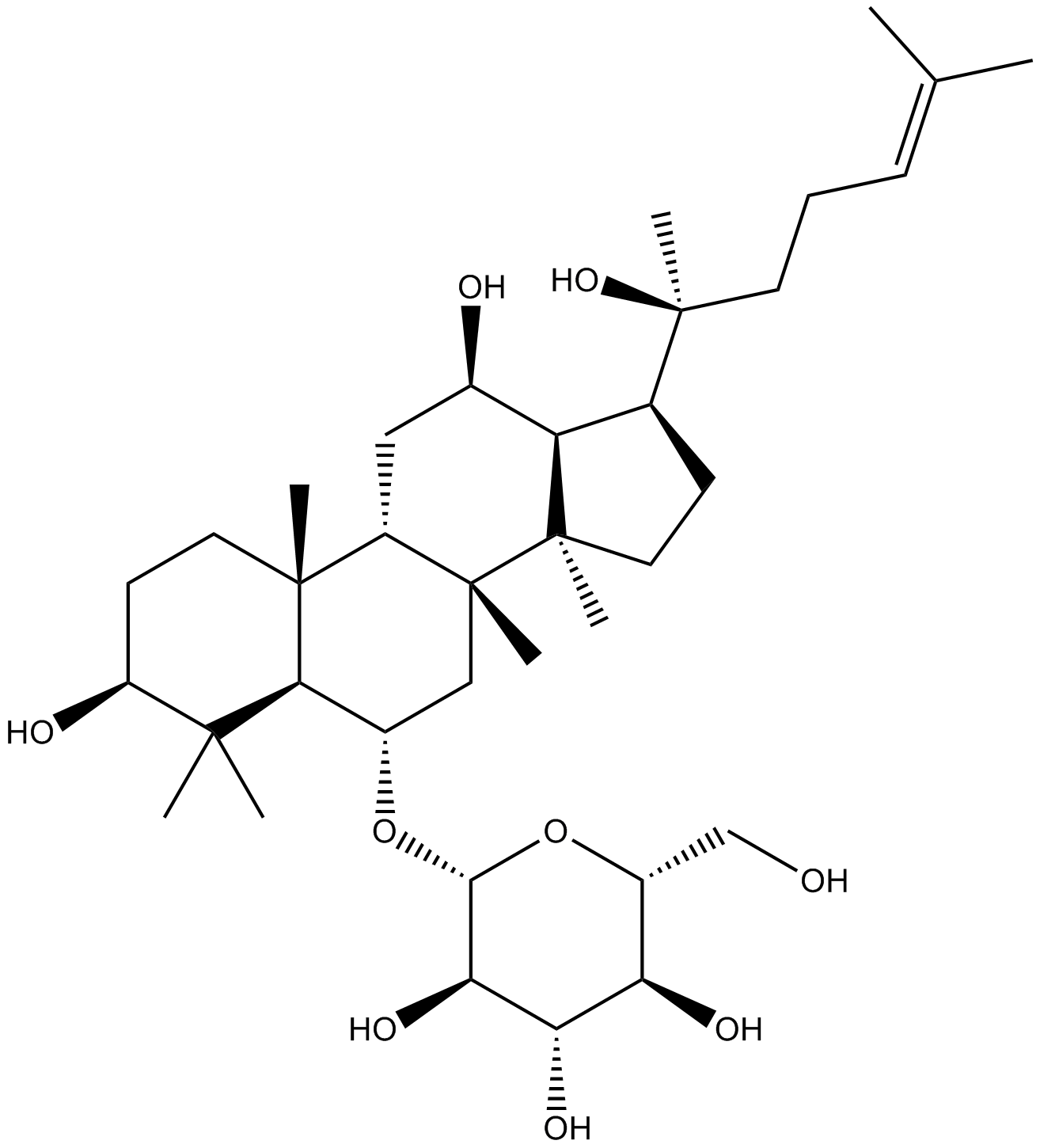
GN10538
Ginsenoside Rh1
-
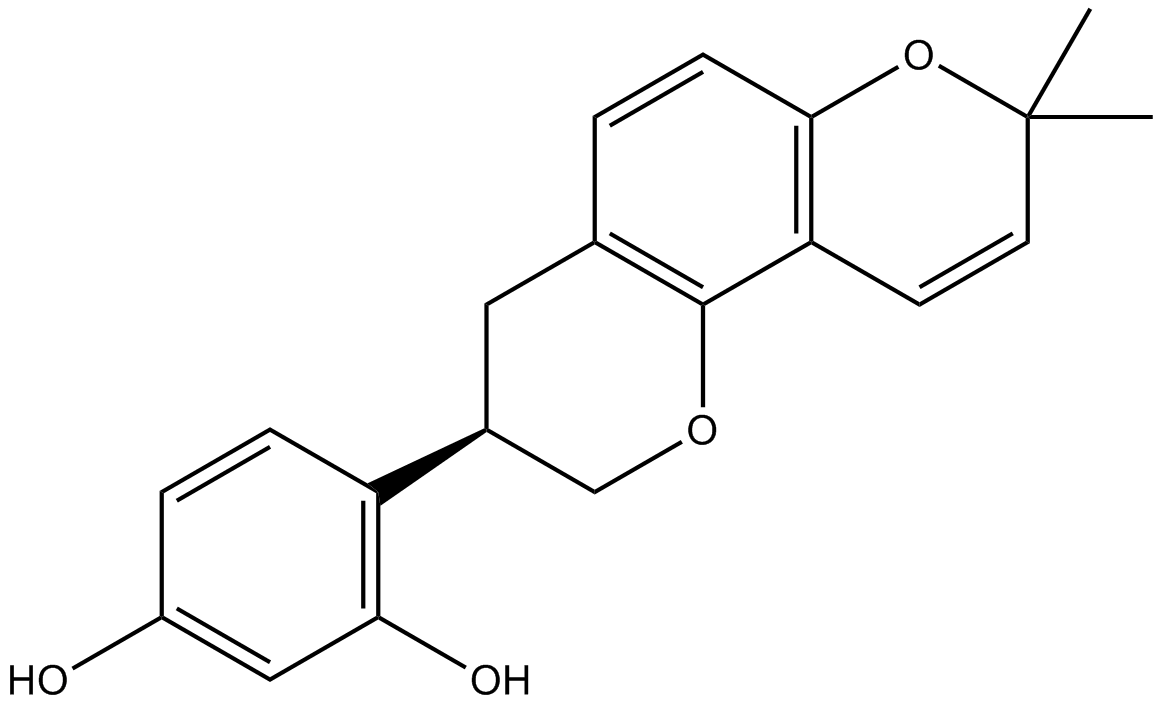
GN10098
Glabridin
-
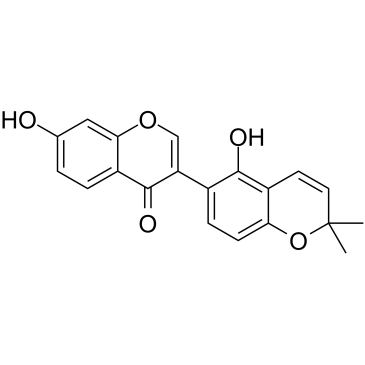
GC36146
Glabrone
Glabrone is an isoflavone isolated from Glycyrrhiza glabra roots.
-
GC14531
Glycerophospho-N-Oleoyl Ethanolamine
precursor of oleoyl ethanolamide (OEA)

-
GC11916
GQ-16
partial agonist for PPARγ
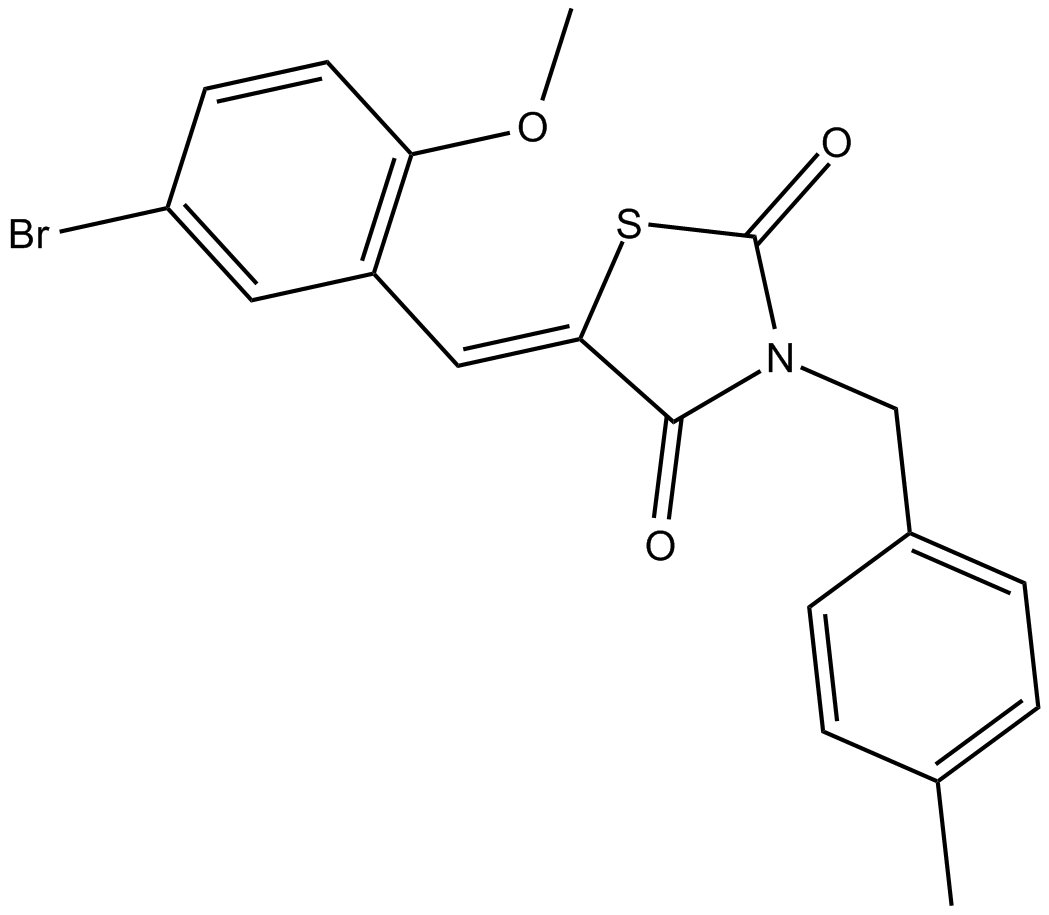
-
GC17824
GSK 0660
PPARδ antagonist
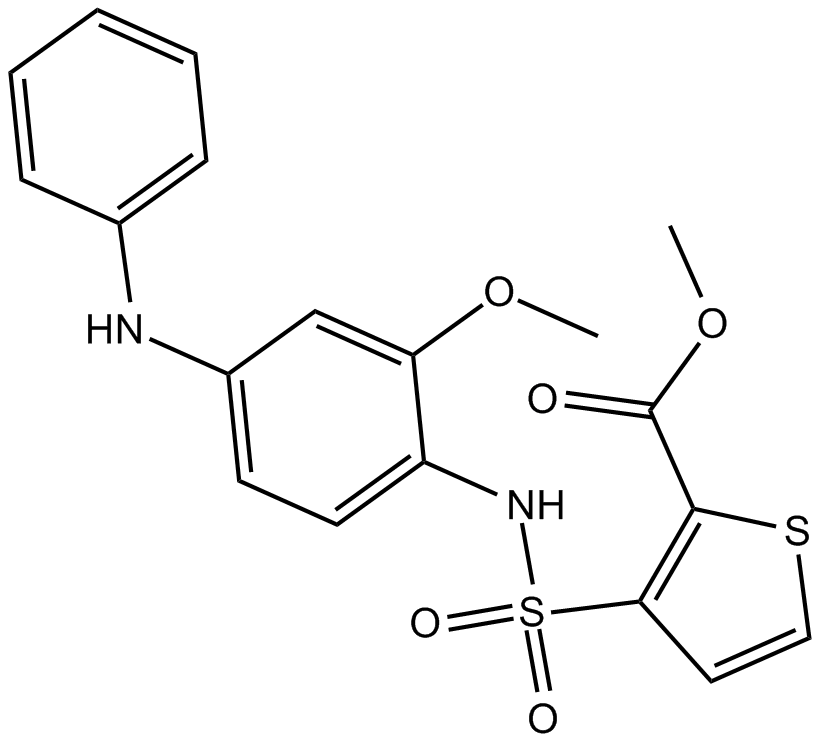
-
GC31471
GSK376501A
GSK376501A is a selective peroxisome proliferator-activated receptor gamma (PPARγ) modulator for the treatment of type 2 diabetes mellitus.
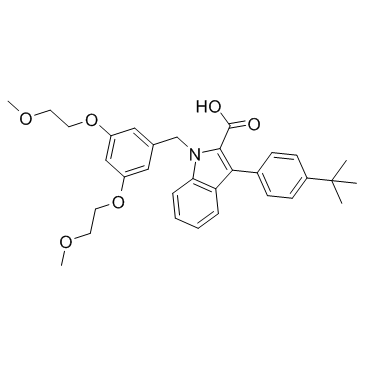
-
GC16009
GSK3787
A selective, irreversible PPARβ/δ antagonist
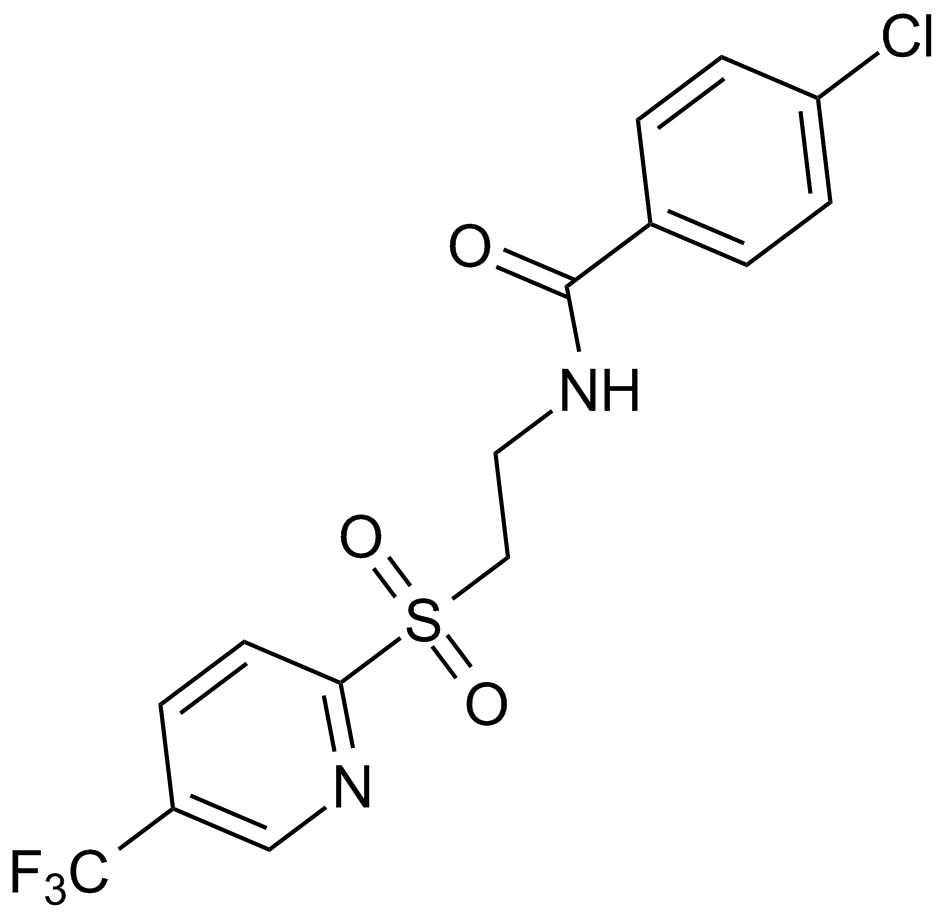
-
GC50019
GW 1929 hydrochloride
Selective PPARγ agonist. Orally active
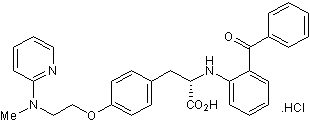
-
GC11732
GW 590735
potent and selective agonist of PPARα
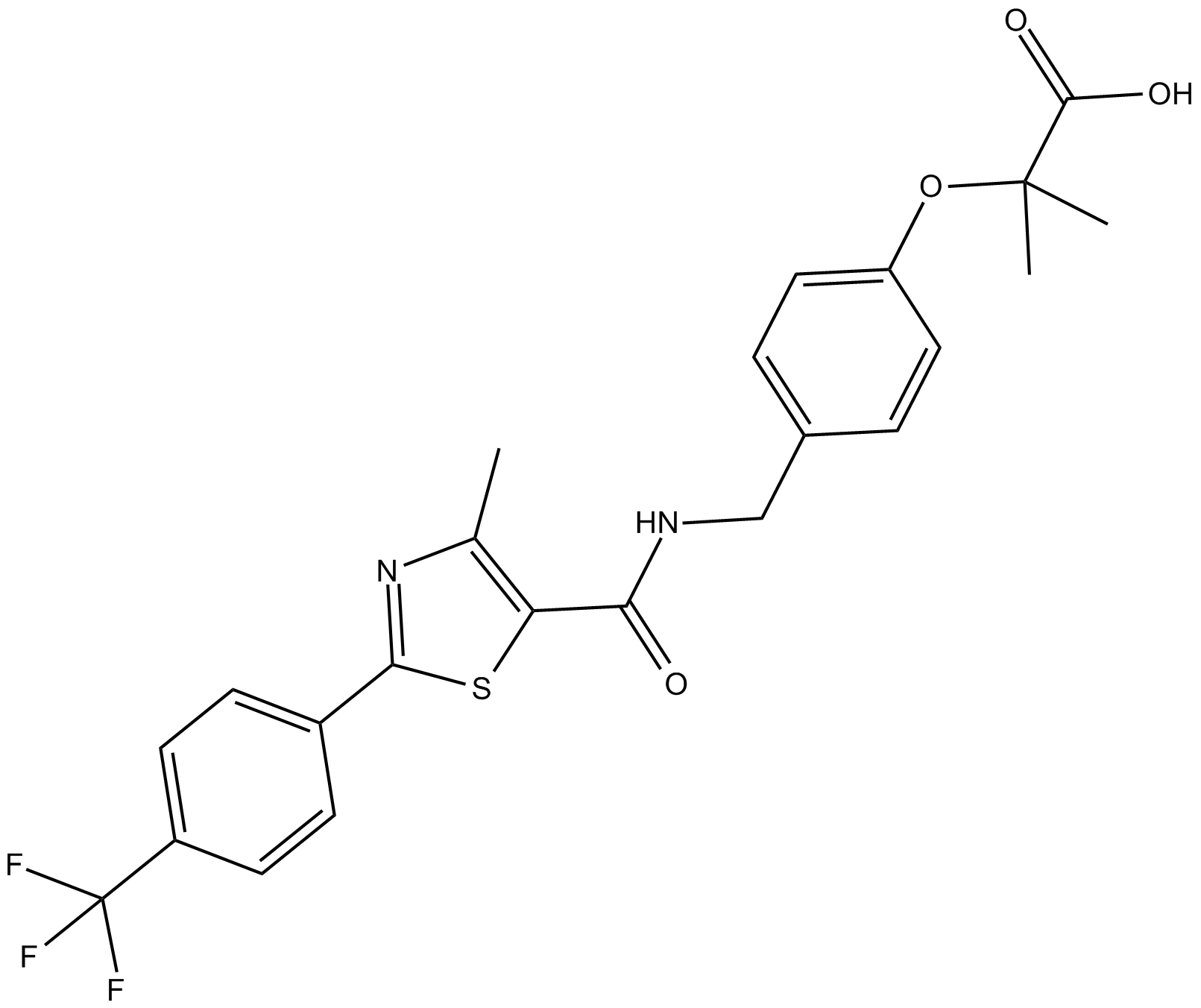
-
GC14187
GW 6471
PPARα antagonist
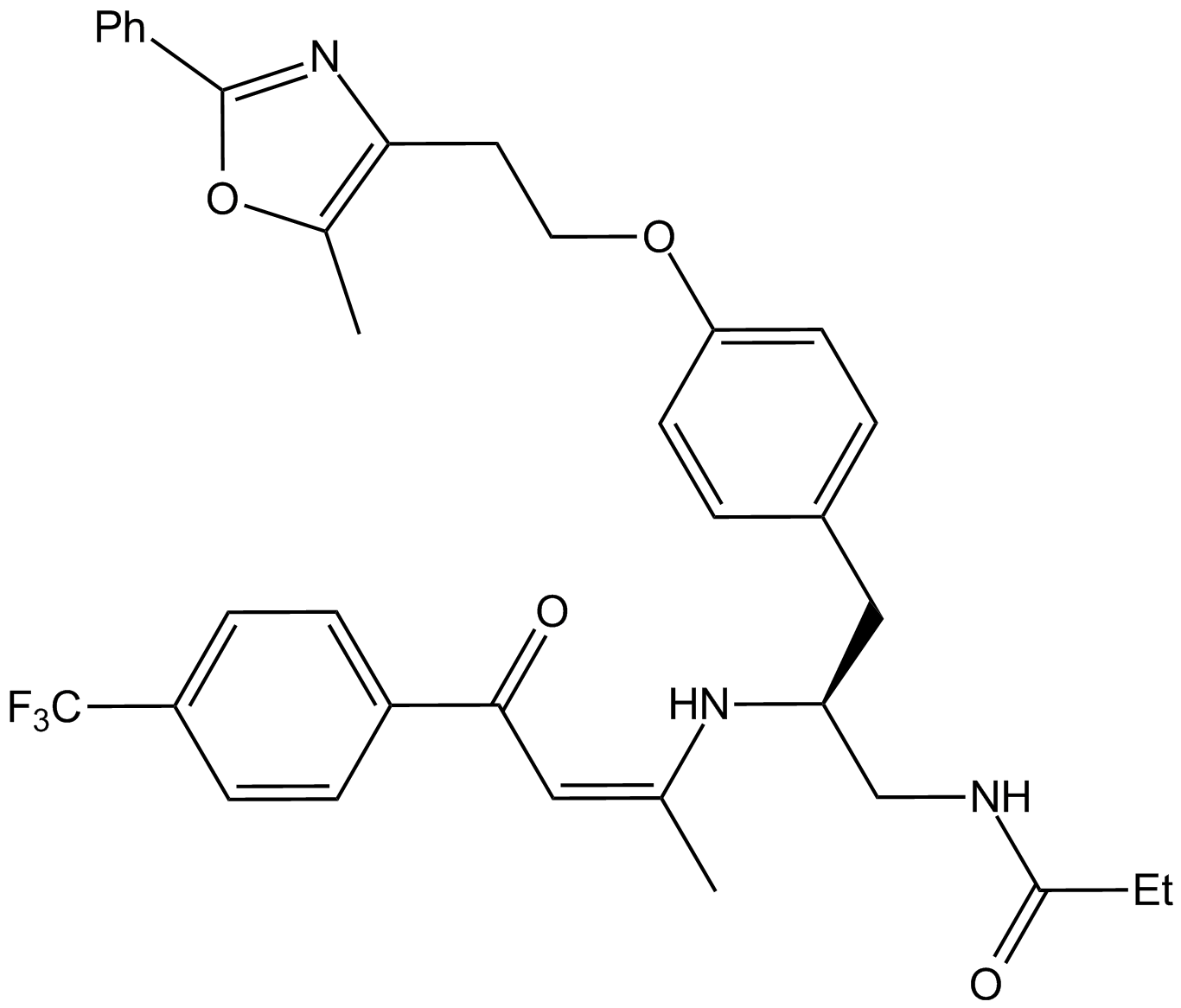
-
GC18024
GW 7647
PPARα agonist
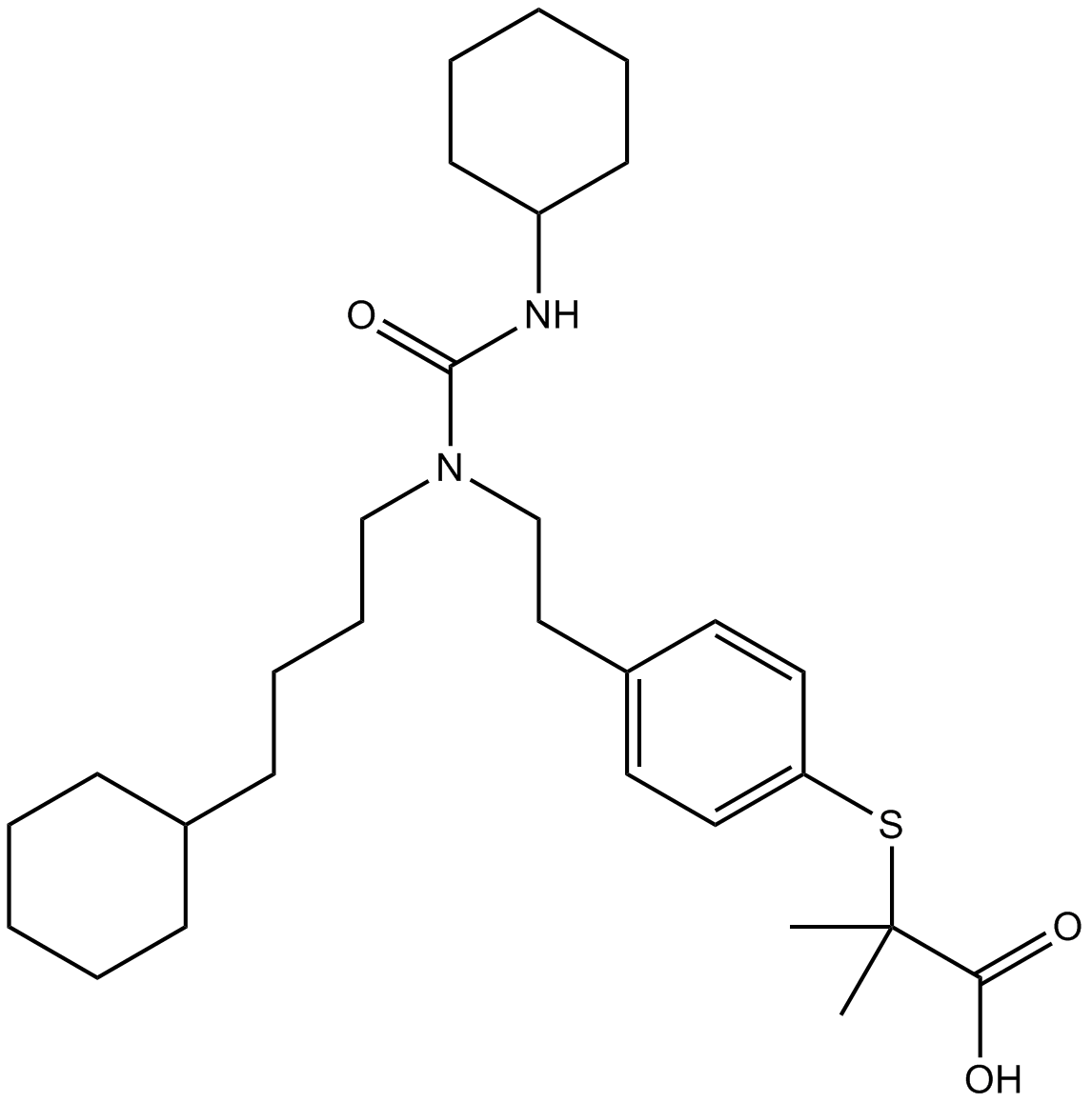
-
GC43800
GW 9578
Peroxisome proliferator-activated receptor α (PPARα) is a ligand-activated transcription factor found predominantly in the liver that is involved in the regulation of lipid homeostasis.
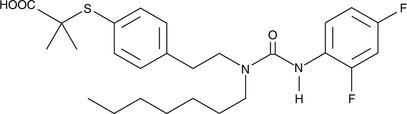
-
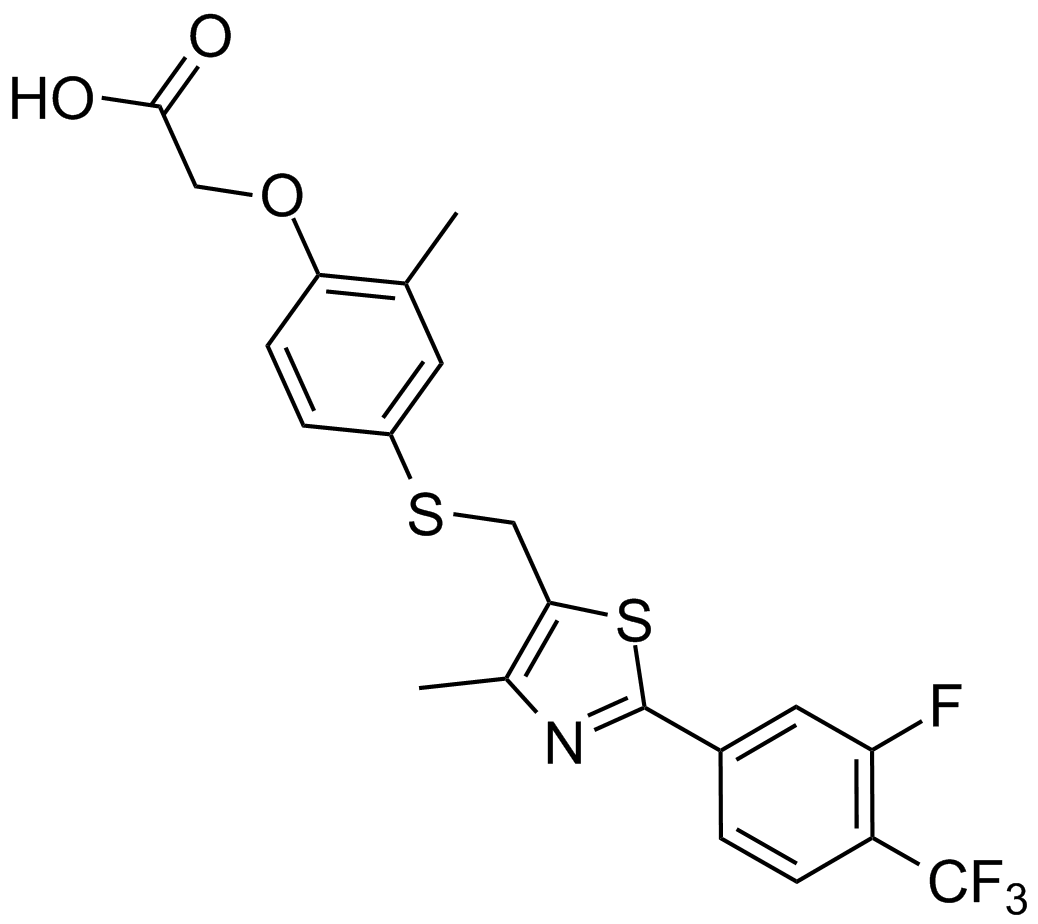
GC14983
GW0742
A selective agonist of PPARδ
-
GC11423
GW1929
A non-thiazolidinedione activator of PPARγ

-
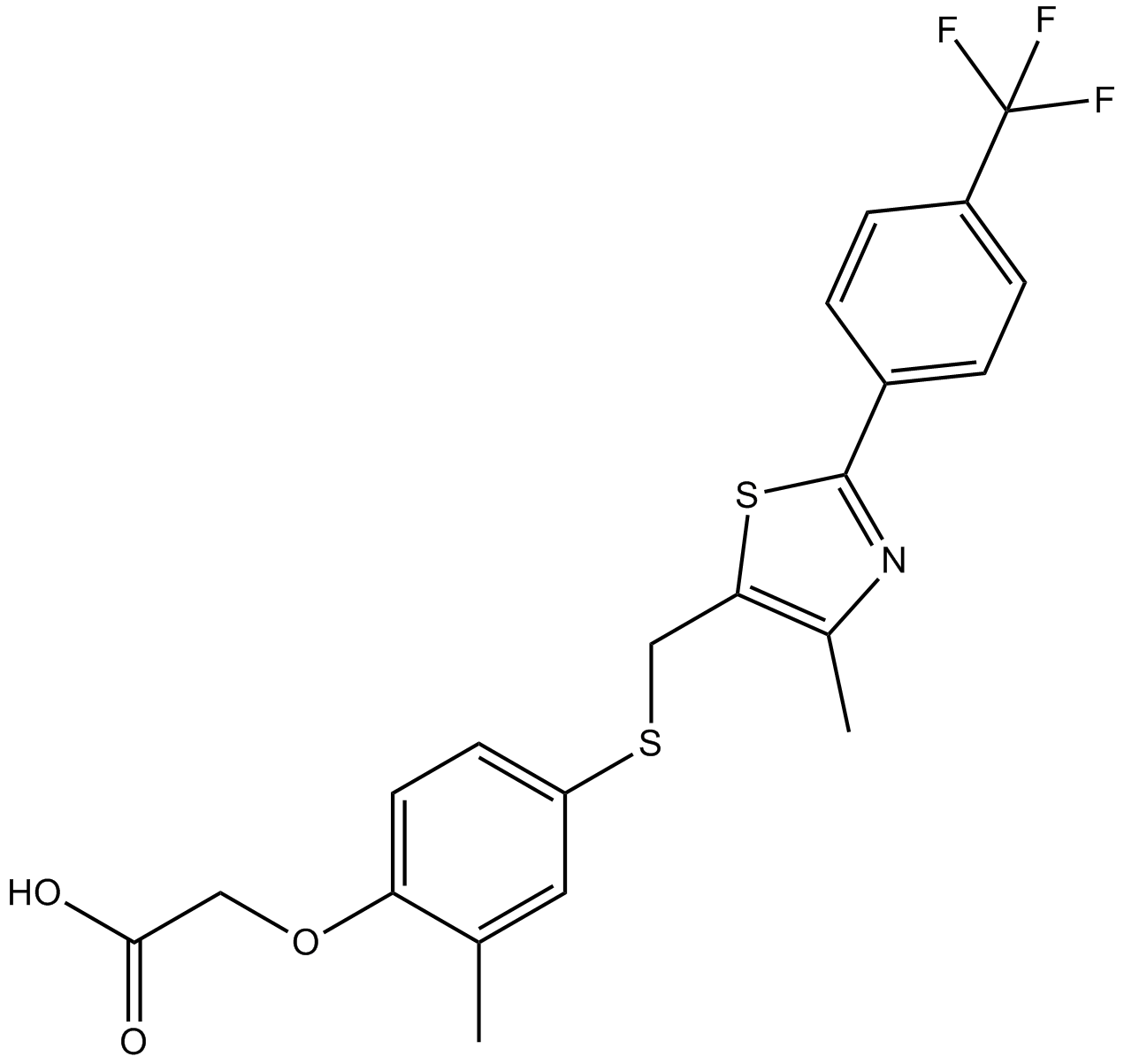
GC15318
GW501516
A selective PPARδ-agonist with anti-obesity activity
-
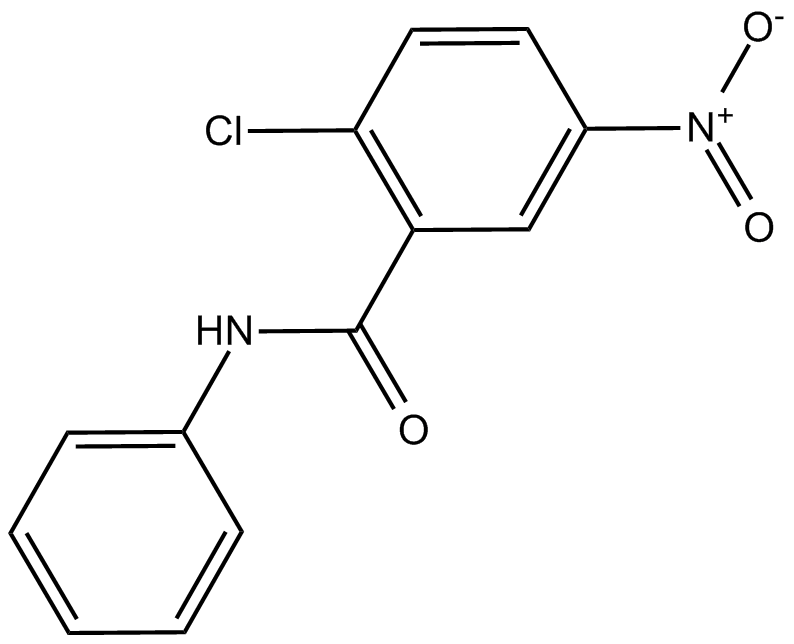
GC13969
GW9662
GW9662 is a specific inhibitor of peroxisome proliferator activated receptor-gamma (PPAR-gamma) with an IC50 of 3.
-
GN10762
Gypenoside XLIX

-
GC40558
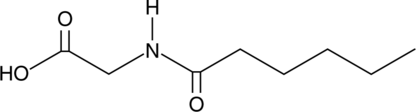
Hexanoyl Glycine
Hexanoyl glycine is an acylated amino acid that is used as a urinary biomarker for several indications.