CDK
<div class="colum_1 clearer"><p>CDKs (Cyclin-dependent kinases) are serine-threonine kinases first discovered for their role in regulating the cell cycle. They are also involved in regulating transcription, mRNA processing, and the differentiation of nerve cells. CDKs are relatively small proteins, with molecular weights ranging from 34 to 40 kDa, and contain little more than the kinase domain. In fact, yeast cells can proliferate normally when their CDK gene has been replaced with the homologous human gene. By definition, a CDK binds a regulatory protein called a cyclin. Without cyclin, CDK has little kinase activity; only the cyclin-CDK complex is an active kinase.</p><p>There are around 20 Cyclin-dependent kinases (CDK1-20) known till date. CDK1, 4 and 5 are involved in cell cycle, and CDK 7, 8, 9 and 11 are associated with transcription.</p><p>CDK levels remain relatively constant throughout the cell cycle and most regulation is post-translational. Most knowledge of CDK structure and function is based on CDKs of <i>S. pombe</i> (Cdc2), <i>S. cerevisia</i> (CDC28), and vertebrates (CDC2 and CDK2). The four major mechanisms of CDK regulation are cyclin binding, CAK phosphorylation, regulatory inhibitory phosphorylation, and binding of CDK inhibitory subunits (CKIs).</p></div>
Targets for CDK
Products for CDK
- Cat.No. Product Name Information
-
GC33107
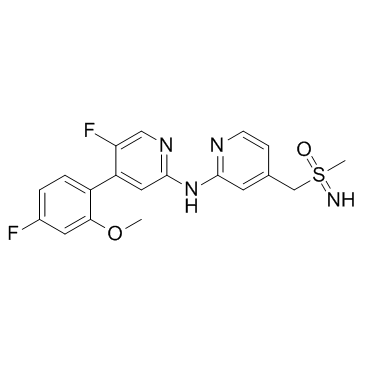
(±)-BAY-1251152
(±)-BAY-1251152 ((±)-BAY-1251152) is a racemic mixture of BAY-1251152. BAY-1251152 is a potent and highly selective PTEF/CDK9 inhibitor.
-
GC32429
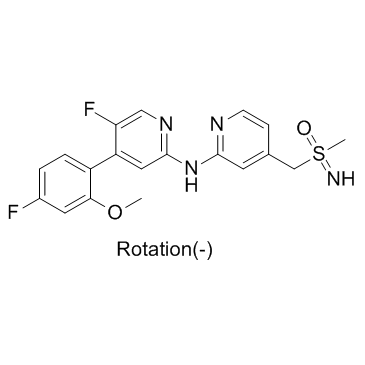
(-)-BAY-1251152
(-)-BAY-1251152 ((-)-BAY-1251152) is an enanthiomer of BAY-1251152 with rotation (-). BAY-1251152 is a potent and highly selective PTEF/CDK9 inhibitor.
-
GC38377
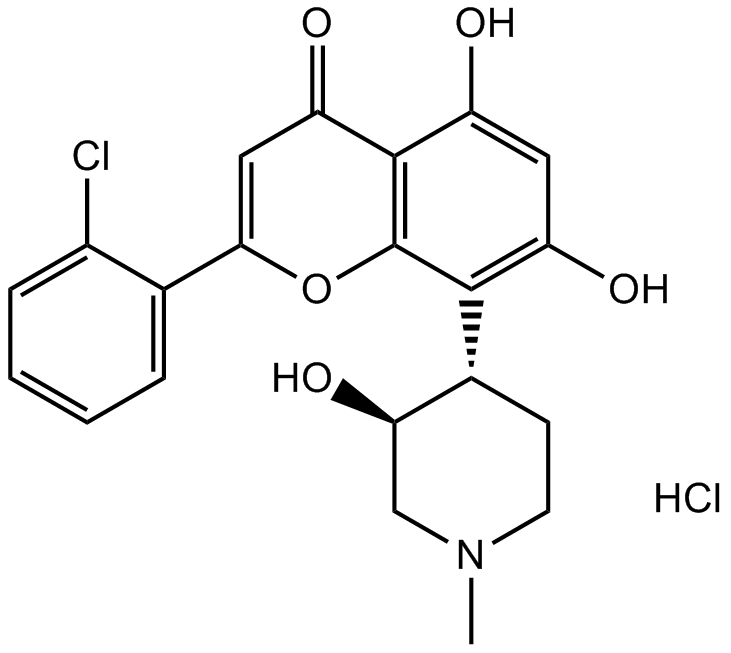
(2S,3R)-Voruciclib hydrochloride
(2S,3R)-Voruciclib hydrochloride is the enantiomer of Voruciclib hydrochloride. (2S,3R)-Voruciclib is an orally active CDK inhibitor.
-
GC64429
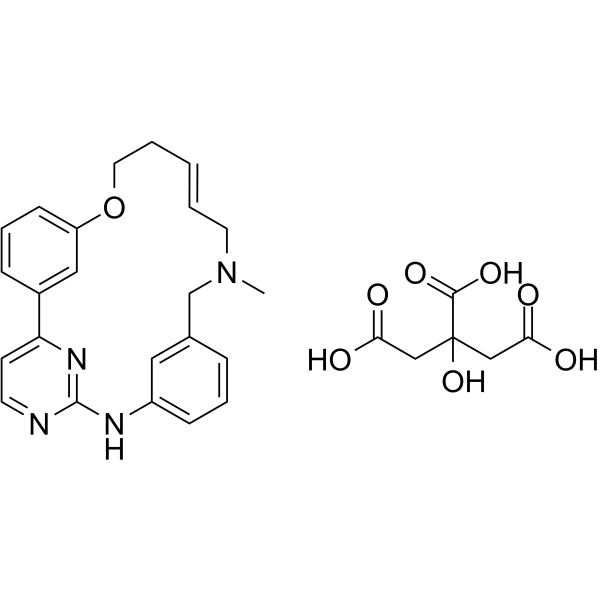
(E/Z)-Zotiraciclib citrate
(E/Z)-Zotiraciclib citrate is a potent CDK2, JAK2, and FLT3 inhibitor.
-
GC63864
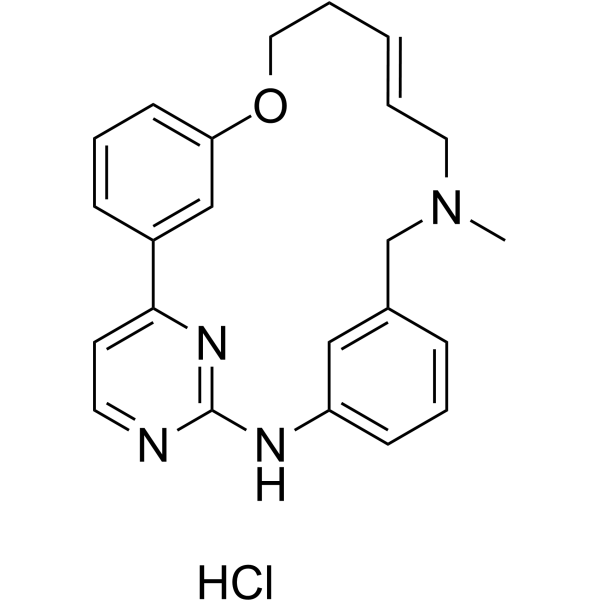
(E/Z)-Zotiraciclib hydrochloride
(E/Z)-Zotiraciclib ((E/Z)-TG02) hydrochloride is a potent CDK2, JAK2, and FLT3 inhibitor.
-
GC41716
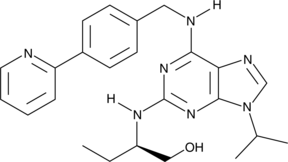
(R)-CR8
Cyclin-dependent kinases (CDKs) are key regulators of cell cycle progression and are therefore promising targets for cancer therapy.
-
GC39281
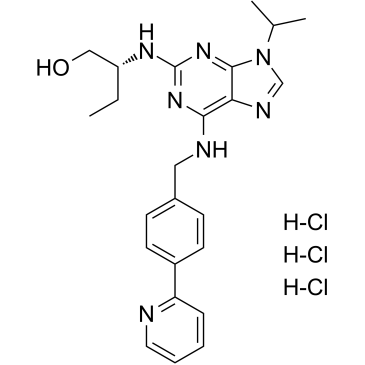
(R)-CR8 trihydrochloride
(R)-CR8 (CR8) trihydrochloride, a second-generation analog of Roscovitine, is a potent CDK1/2/5/7/9 inhibitor.
-
GC34124
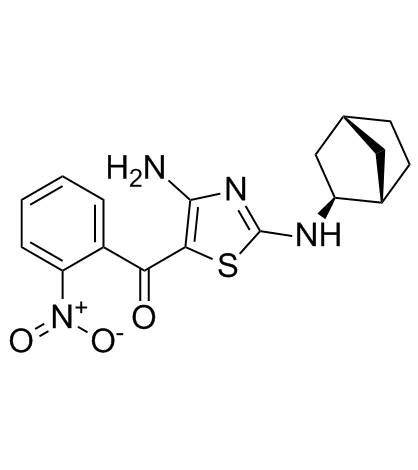
(rel)-MC180295
(rel)-MC180295 ((rel)-(rel)-MC180295) is a potent and selective CDK9-Cyclin T1 inhibitor, with an IC50 of 5 nM, at least 22-fold more selective for CDK9 over other CDKs. (rel)-MC180295 also inhibits GSK-3α and GSK-3β. (rel)-MC180295 ((rel)-(rel)-MC180295) has potent anti-tumor effect.
-
GC65877
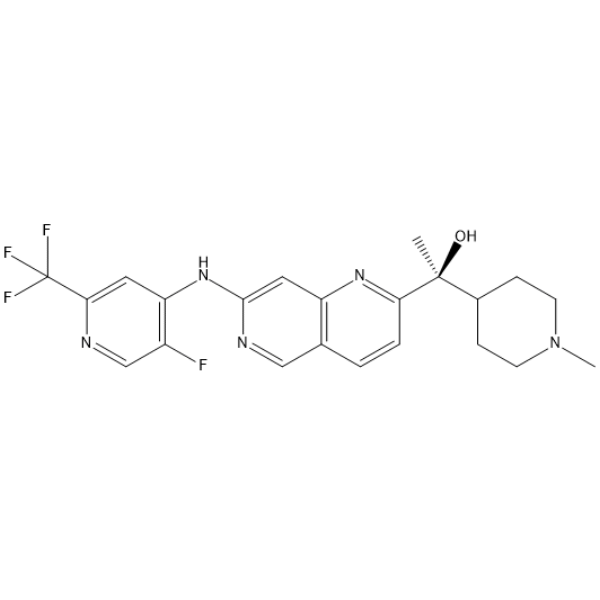
(S)-GFB-12811
(S)-GFB-12811 (compound 596) is a potent and selective CDK5 inhibitor, with an IC50 value less than 10 nM. (S)-GFB-12811 can be used in the research of cell cycle progression, neuronal development, tumorigenesis.
-
GC65997
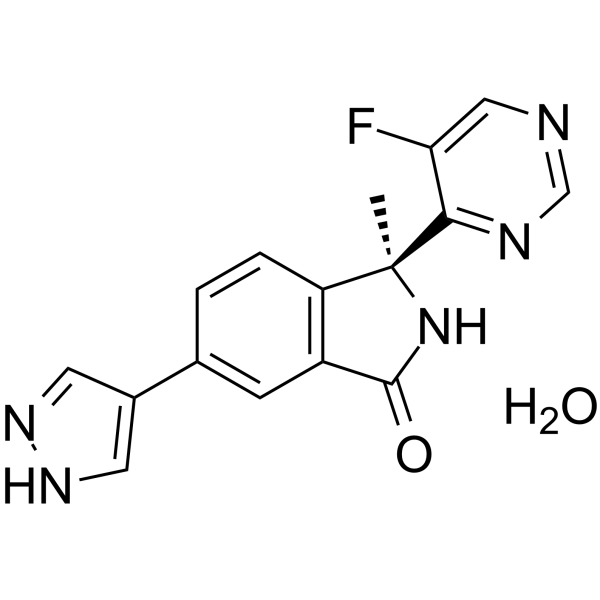
(S)-LY3177833 hydrate
(S)-LY3177833 ((S)-Example 2) hydrate is an orally active CDC7 kinase inhibitor. (S)-LY3177833 hydrate shows broad in vitro anticancer activity.
-
GC64399
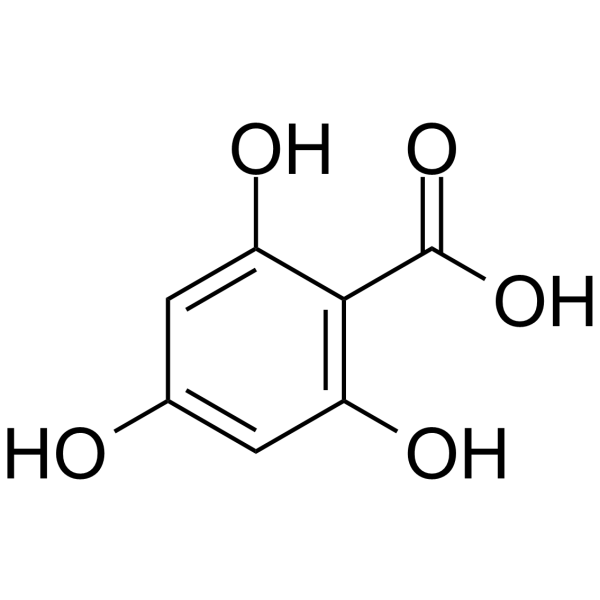
2,4,6-Trihydroxybenzoic acid
2,4,6-Trihydroxybenzoic acid, the flavonoid metabolite, is a CDK inhibitor. 2,4,6-Trihydroxybenzoic acid can be used for the research of cancer.
-
GC35162
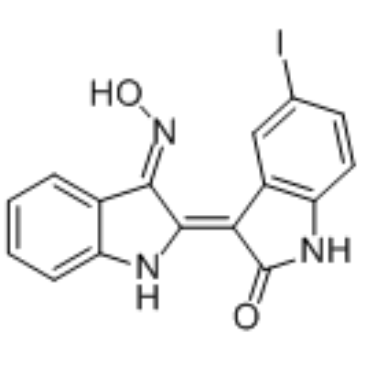
5-Iodo-indirubin-3'-monoxime
5-Iodo-indirubin-3'-monoxime is a potent GSK-3β, CDK5/P25 and CDK1/cyclin B inhibitor, competing with ATP for binding to the catalytic site of the kinase, with IC50s of 9, 20 and 25 nM, respectively.
-
GC60532
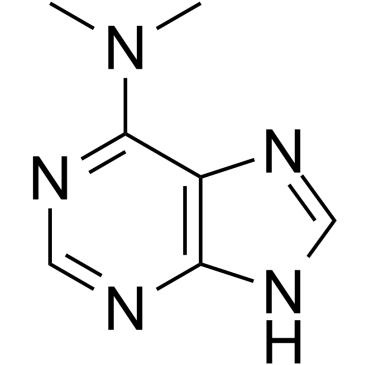
6-(Dimethylamino)purine
6-(Dimethylamino)purine is a dual inhibitor of protein kinase and CDK.
-
GC63728
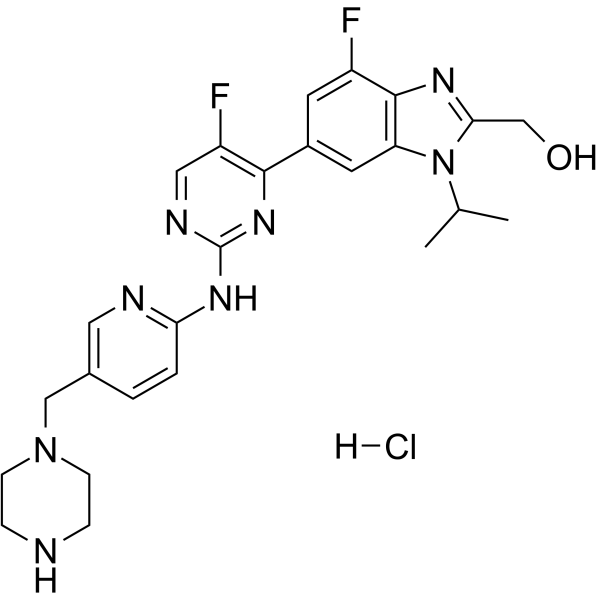
Abemaciclib metabolite M18 hydrochloride
Abemaciclib metabolite M18 (LSN3106729) hydrochloride, the metabolite of Abemaciclib, is a CDK inhibitor with antitumor activity. Abemaciclib metabolite M18 hydrochloride and a CRBN ligand have been used to design PROTAC CDK4/6 degrader.
-
GC38749
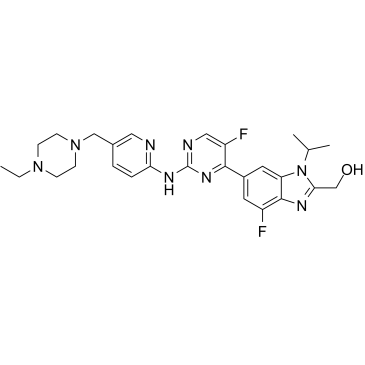
Abemaciclib metabolite M20
Abemaciclib metabolite M20 (LSN3106726), the active metabolite of Abemaciclib, is a selective CDK4/6 inhibitor for the treatment of cancer.
-
GC35218
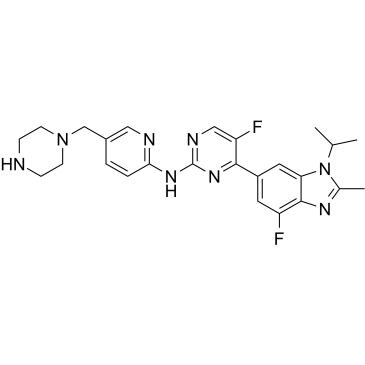
Abemaciclib Metabolites M2
Abemaciclib Metabolites M2 (LSN2839567) is a metabolite of Abemaciclib, acts as a potent CDK4 and CDK6 inhibitor, with IC50s of 1.2 and 1.3 nM, respectively. Anti-cancer activity.
-
GC64280
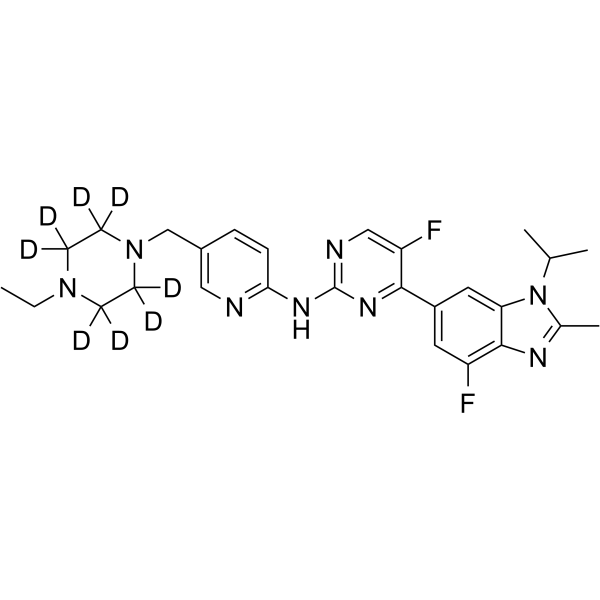
Abemaciclib-d8
Abemaciclib-d8 (LY2835219-d8) is the deuterium labeled Abemaciclib. Abemaciclib (LY2835219) is a selective CDK4/6 inhibitor with IC50 values of 2 nM and 10 nM for CDK4 and CDK6, respectively.
-
GC15841
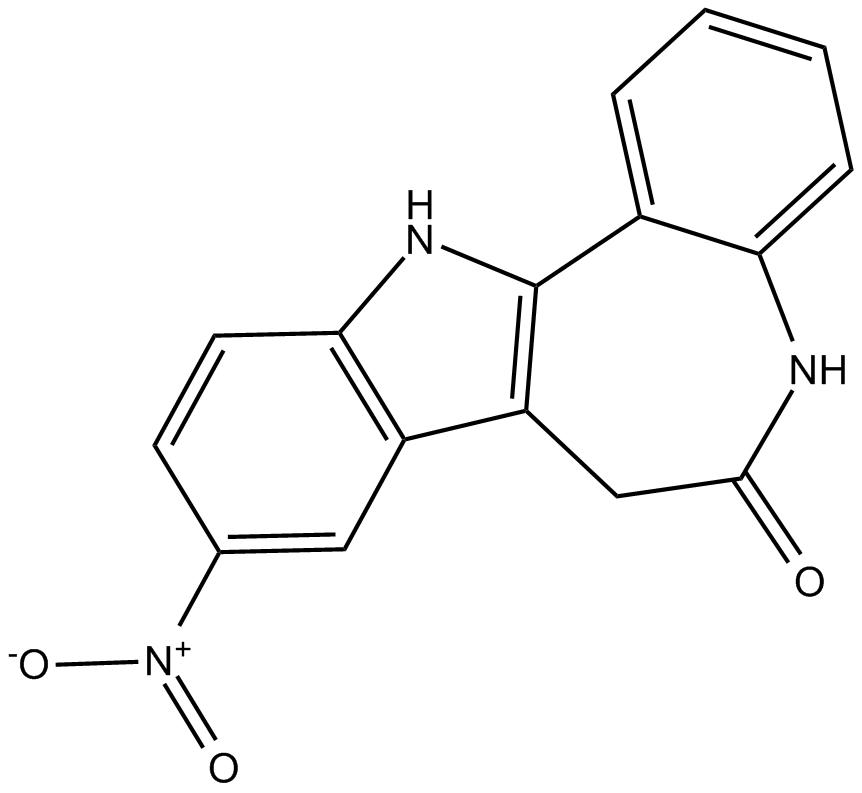
Alsterpaullone
CDKs and GSK3β inhibitor
-
GC14974
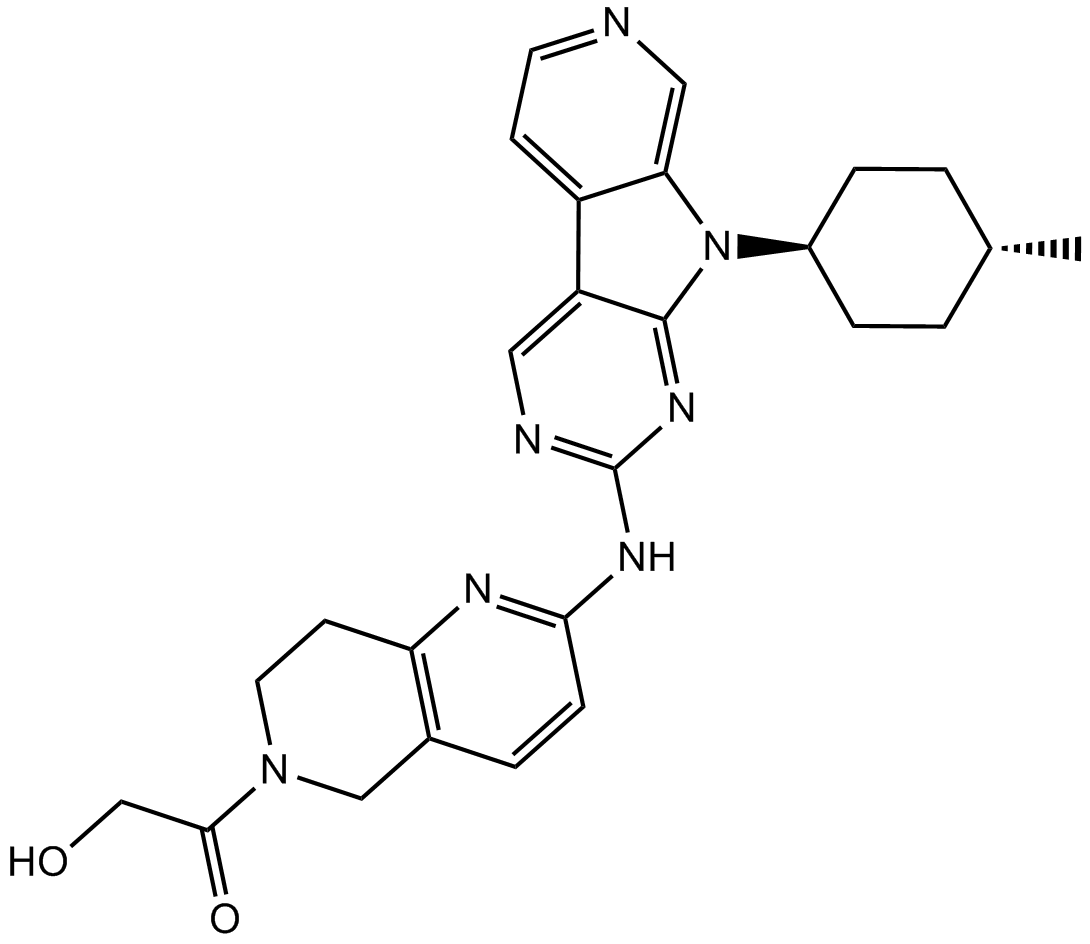
AMG 925
FLT3/CDK4 inhibitor,potent and selective
-
GC35315
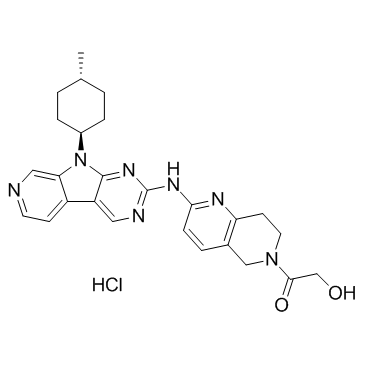
AMG 925 HCl
AMG 925 HCl is a potent, selective, and orally available FLT3/CDK4 dual inhibitor with IC50s of 2±1 nM and 3±1 nM, respectively.
-
GC38736
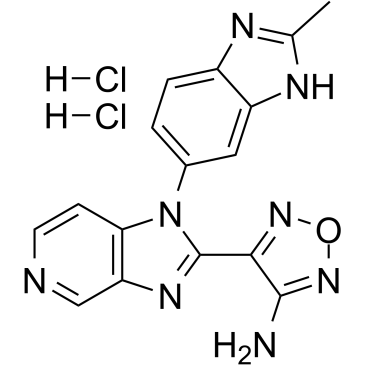
AS2863619
A dual inhibitor of Cdk8 and Cdk19
-
GC38737
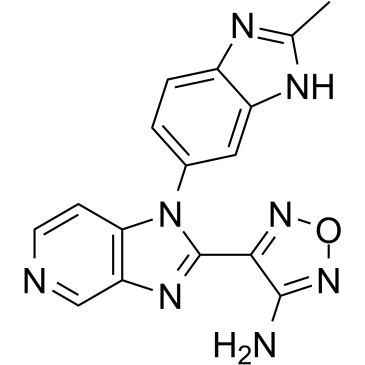
AS2863619 free base
AS2863619 free base enables conversion of antigen-specific effector/memory T cells into Foxp3+ regulatory T (Treg) cells for the treatment of various immunological diseases.
-
GC15870
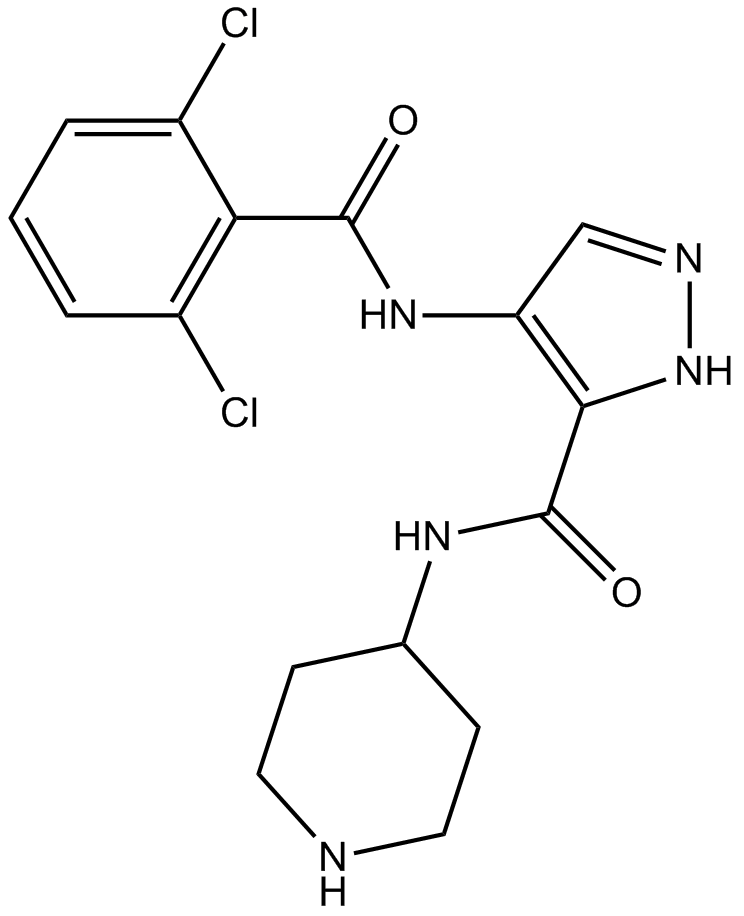
AT7519
Multi-CDK inhibitor
-
GC13998
AT7519 Hydrochloride
A Cdk inhibitor
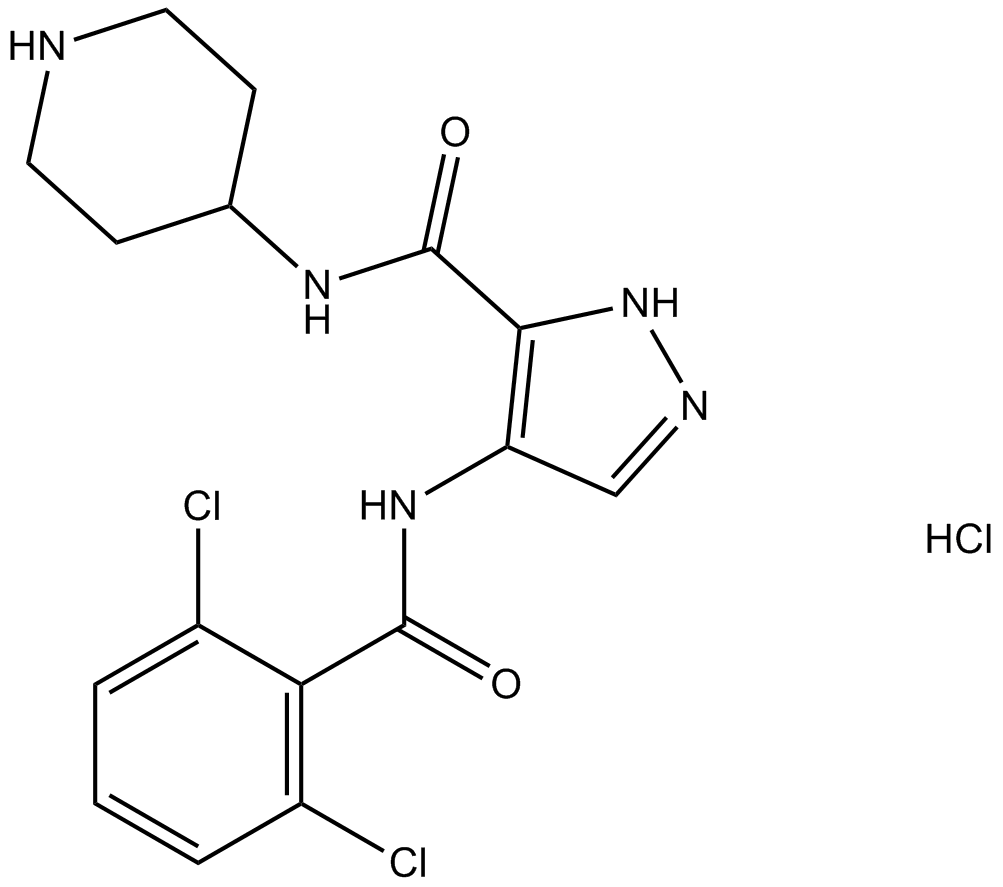
-
GC15597
AT7519 trifluoroacetate
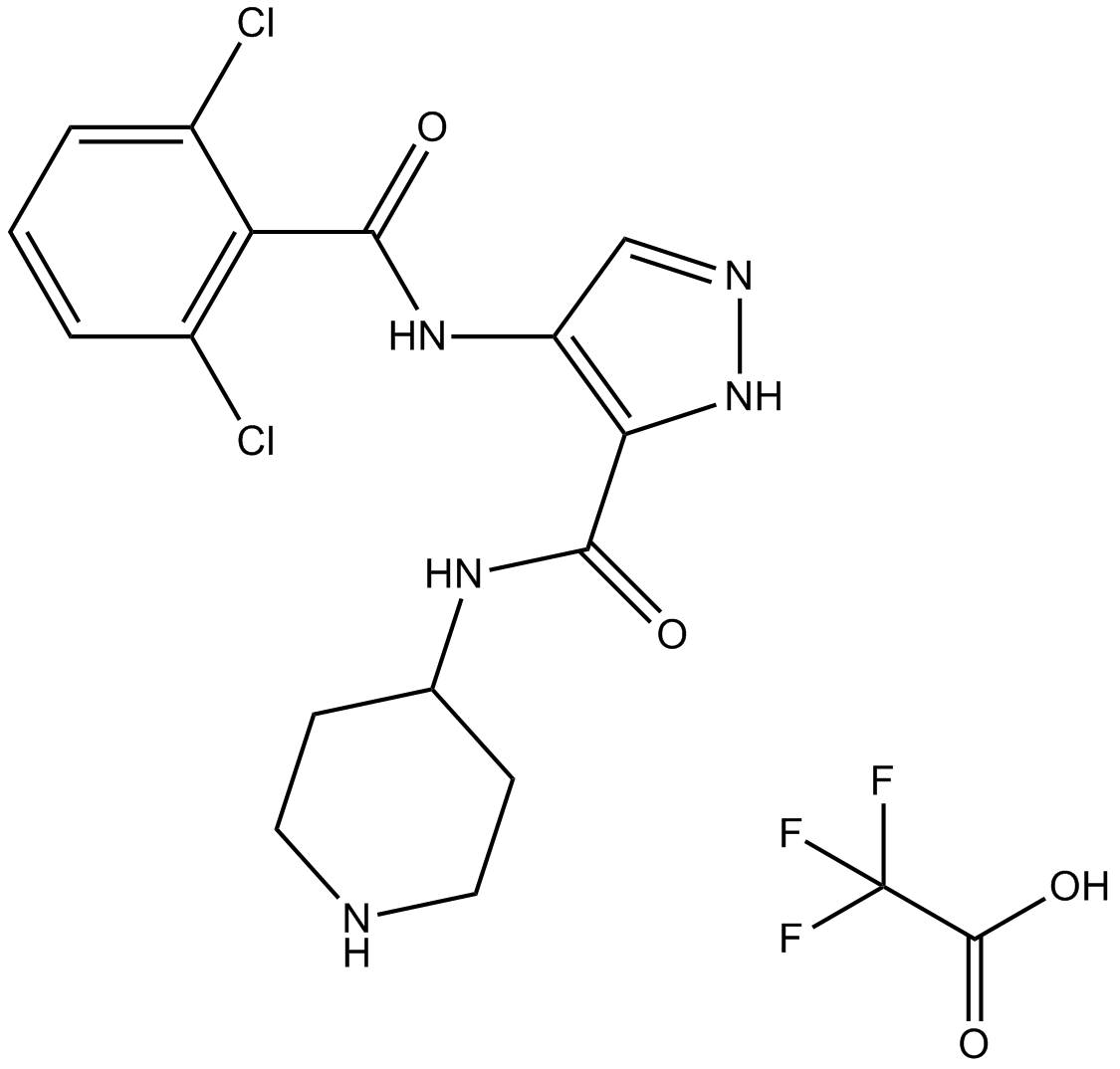
-
GC34422
Atuveciclib (BAY-1143572)
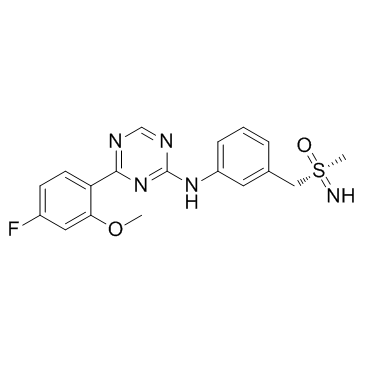
-
GC34059
Atuveciclib Racemate (BAY-1143572 Racemate)
Atuveciclib Racemate (BAY-1143572 Racemate) (BAY-1143572 Racemate) is the racemate mixture of Atuveciclib. Atuveciclib is a potent and highly selective, oral P-TEFb/CDK9 inhibitor which supresses CDK9/CycT1 with an IC50 of 13 nM.
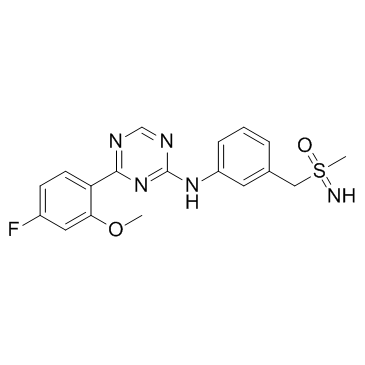
-
GC34421
Atuveciclib S-Enantiomer (BAY-1143572 S-Enantiomer)
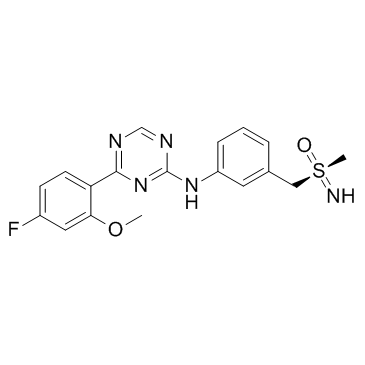
-
GC63449
Avotaciclib
Avotaciclib (BEY1107) is a potent and orally active inhibitor of cyclin dependent kinase 1 (CDK1). Avotaciclib can be used for the research of locally advanced or metastatic pancreatic cancer.
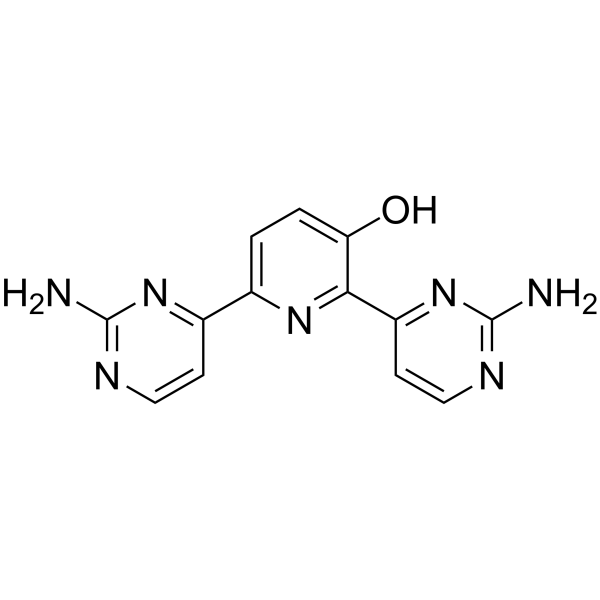
-
GC12438
AZD-5438
Potent CDK1/2/9 inhibitor
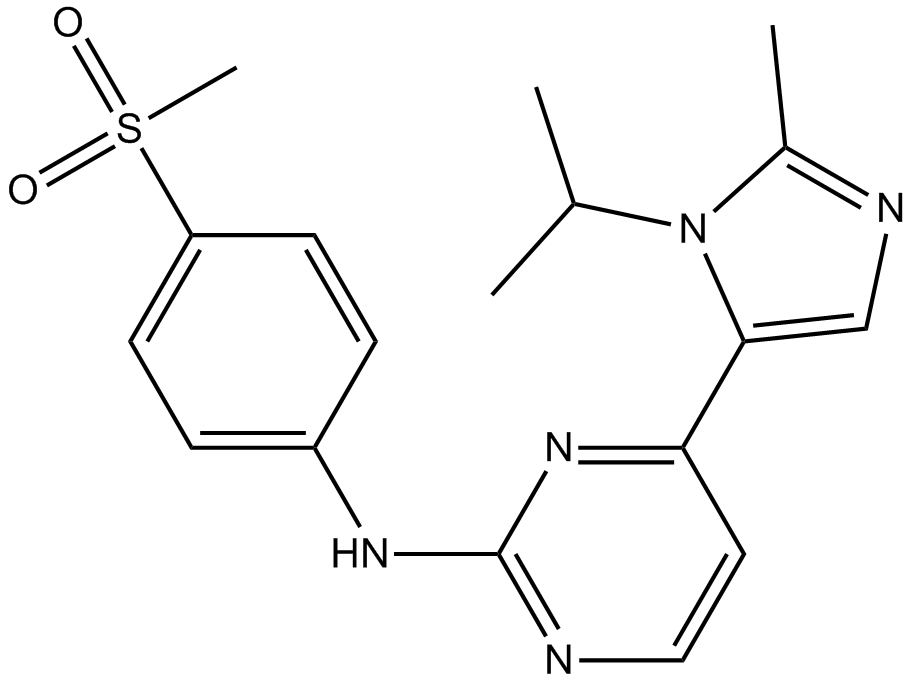
-
GC32717
AZD4573
AZD4573 is a potent and highly selective CDK9 inhibitor (IC50 of <4 nM) that enables transient target engagement for the treatment of hematologic malignancies.
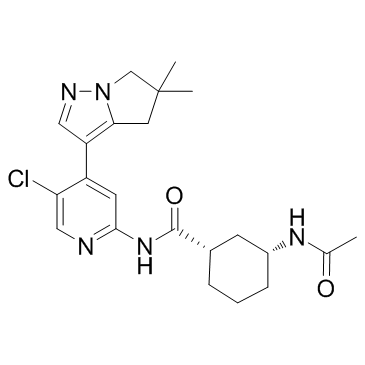
-
GC19067
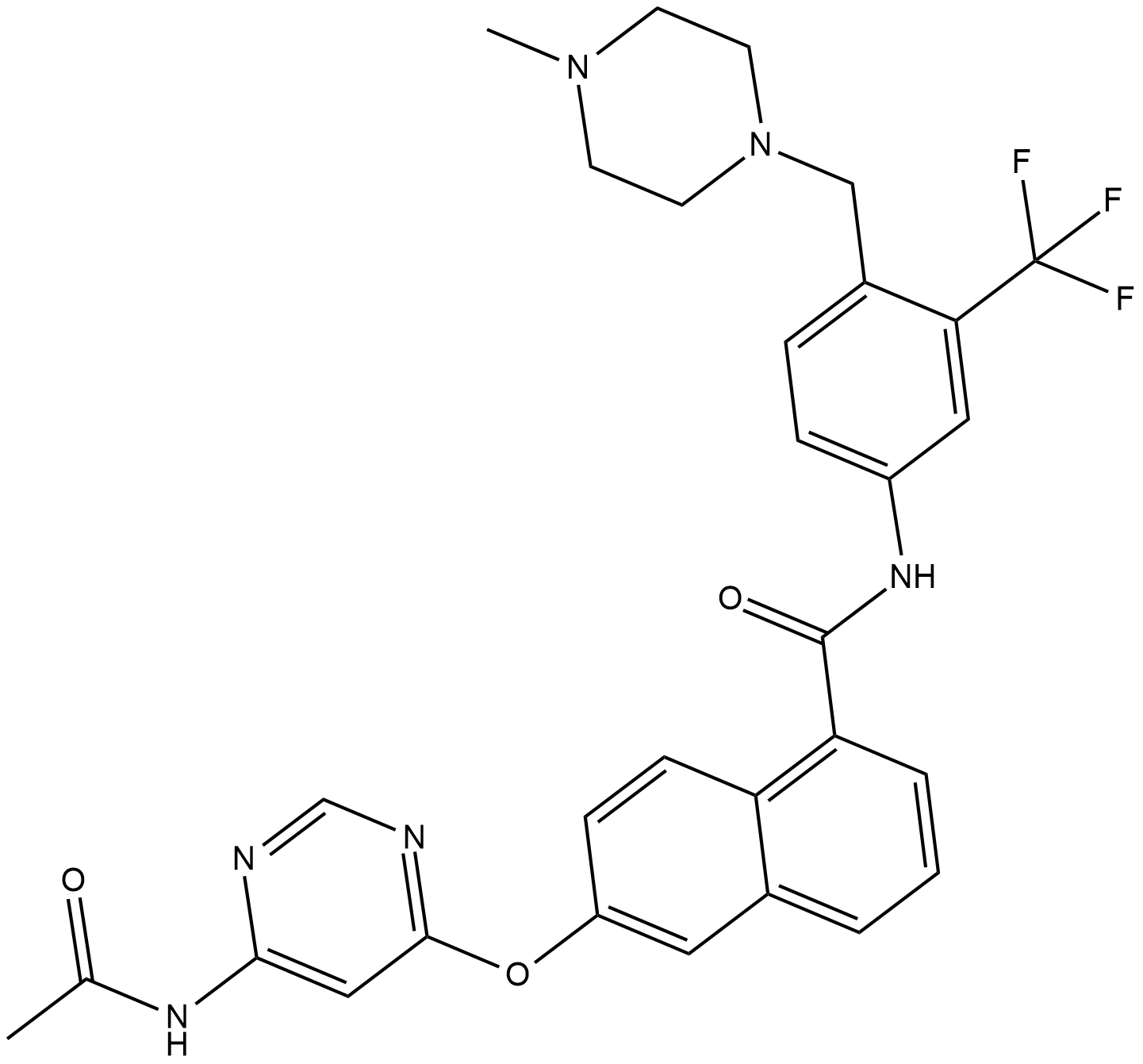
BGG463
BGG463 is a CDK2 inhibitor.
-
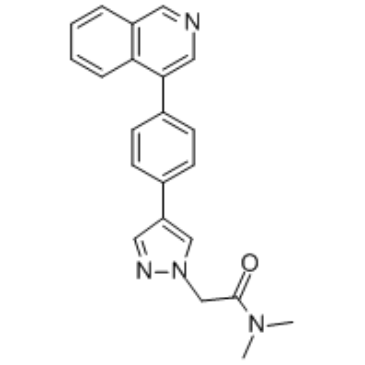
GC35512
BI-1347
BI-1347 is an orally active, selective and potent CDK8 inhibitor (IC50=1.1 nM). BI-1347 shows anti-tumoral activity.
-
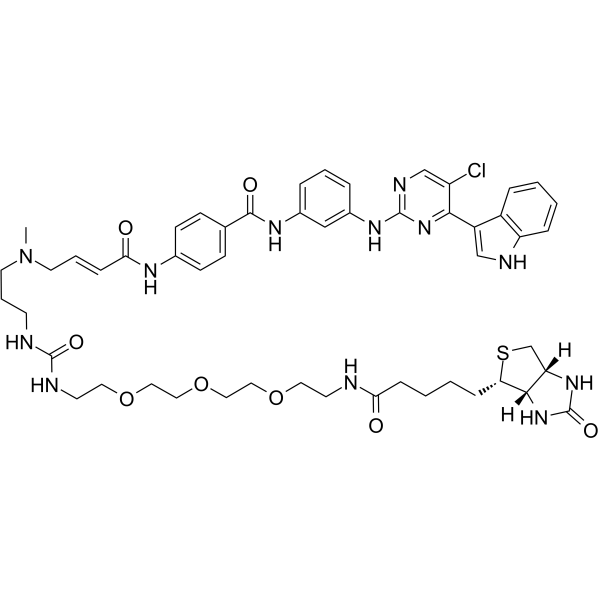
GC62653
bio-THZ1
bio-THZ1 is a biotinylated version of THZ1 and binds irreversibly to CDK7. THZ1 is a selective and potent covalent CDK7 inhibitor with an IC50 of 3.2 nM.
-
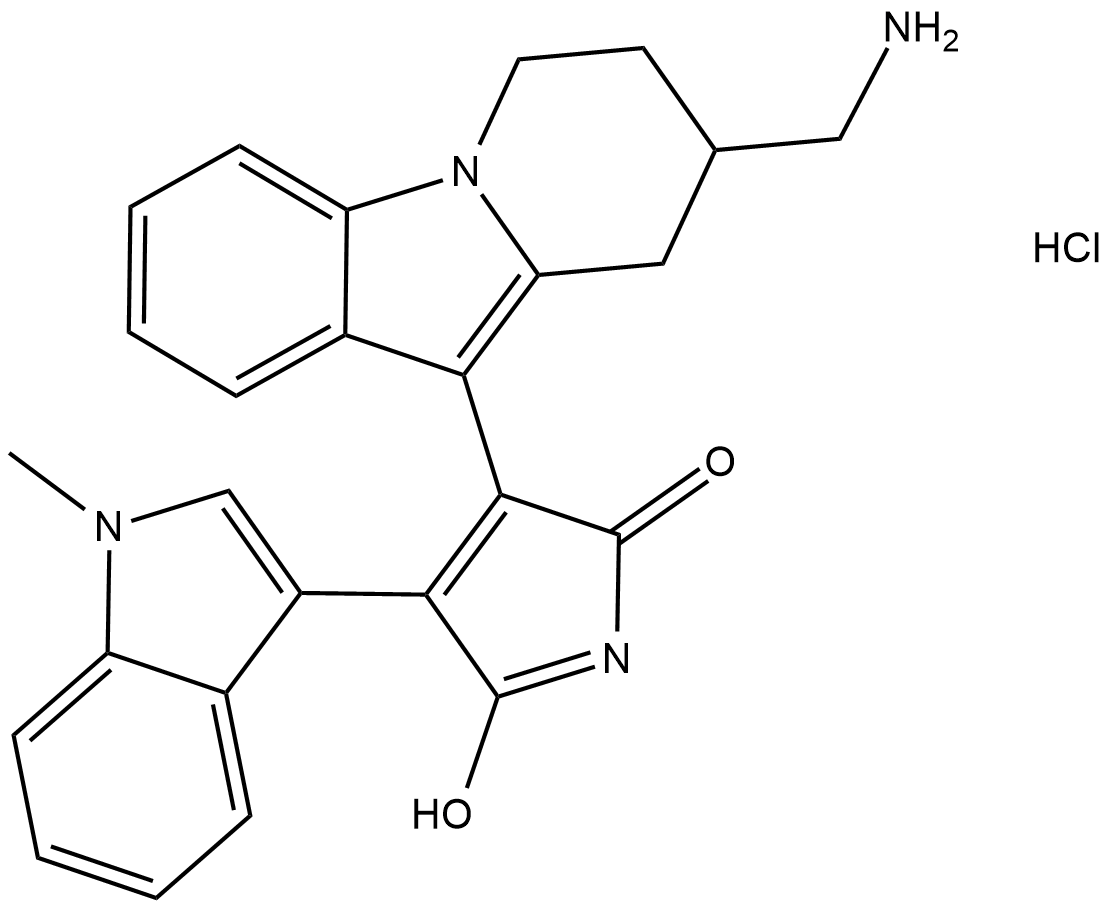
GC18354
Bisindolylmaleimide X (hydrochloride)
Bisindolylmaleimide X is a cell-permeable, reversible, ATP-competitive protein kinase C (PKC) inhibitor (IC50 = 15 nM, rat brain PKC).
-
GC12865
BMS265246
CDK1/2 inhibitor,potent and selective
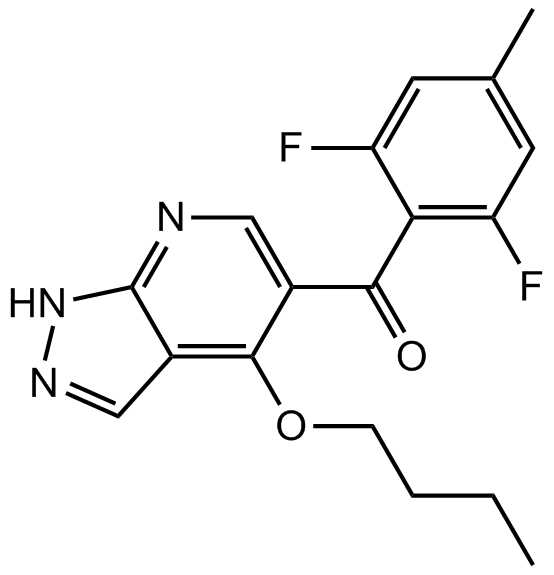
-
GC11040
Borrelidin
threonyl-tRNA synthetase (ThrRS) inhibitor
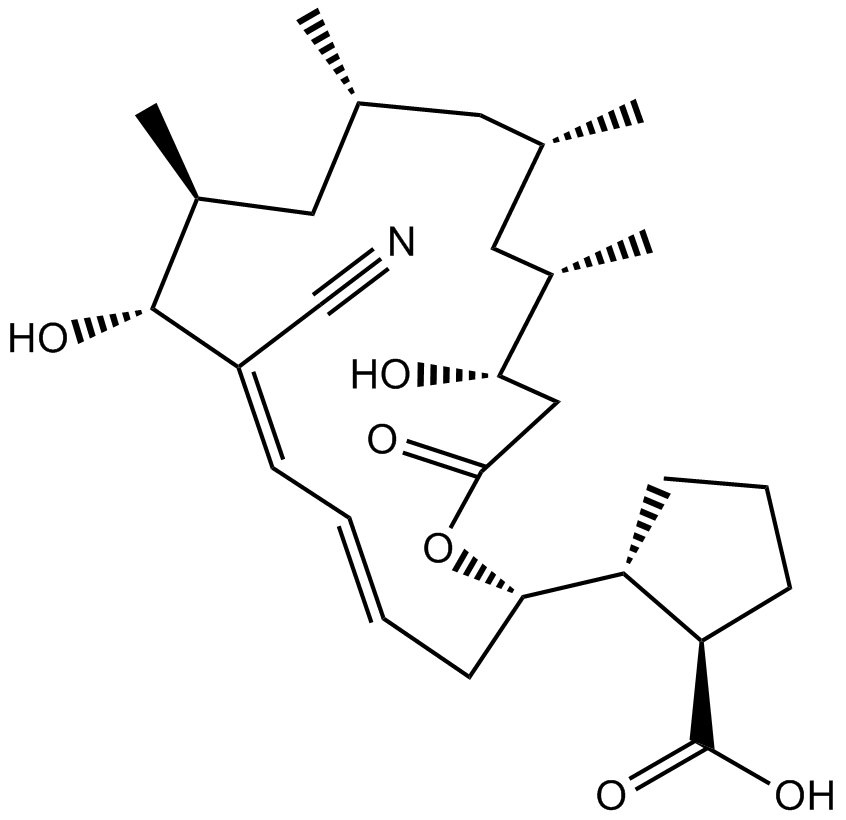
-
GC50452
BRD 6989
Cdk8 inhibitor; enhances IL-10 production
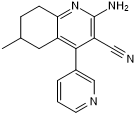
-
GC50217
BS 181 dihydrochloride
Selective cdk7 inhibitor
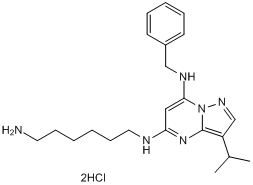
-
GC12001
BS-181
A selective Cdk7 inhibitor
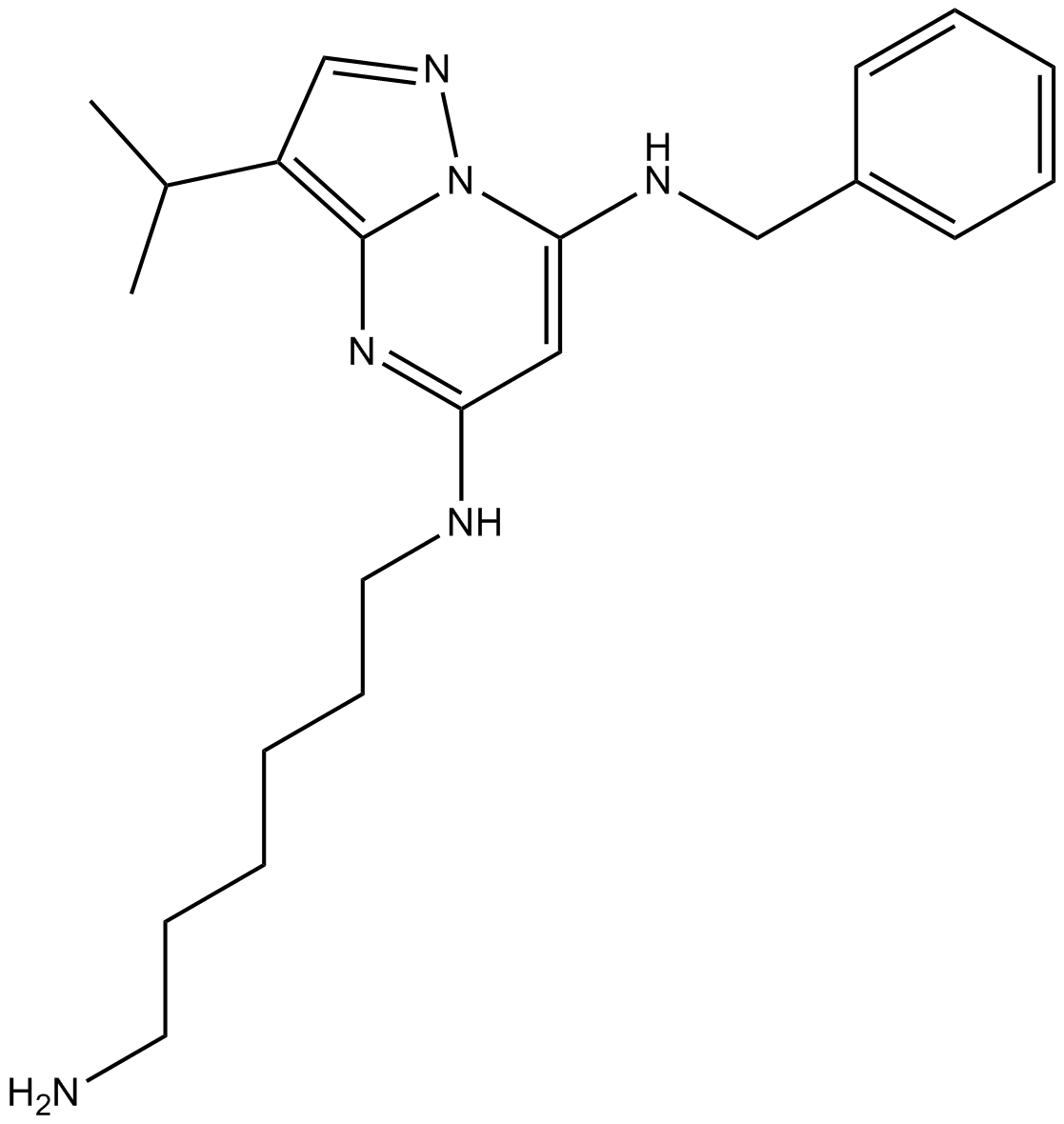
-
GC13690
BS-181 HCl
BS-181 HCl is a highly selective CDK7 inhibitor with IC50 of 21 nM, and > 40-fold selective for CDK7 than CDK1, 2, 4, 5, 6, or 9.
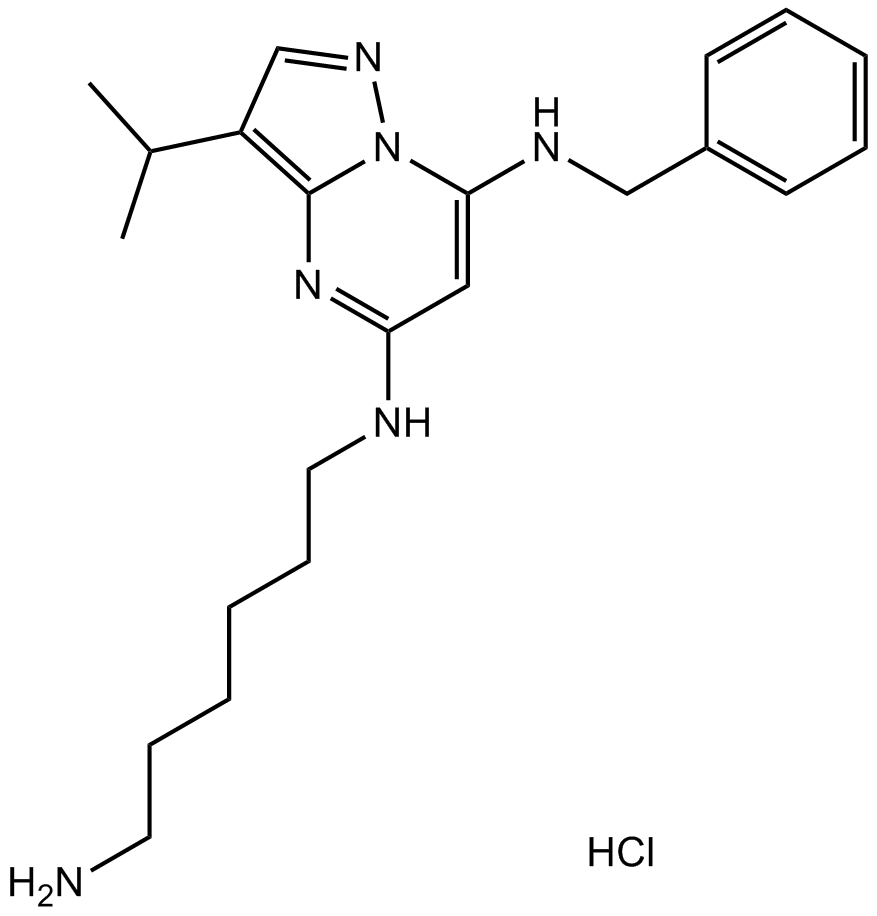
-
GC65128
BSJ-03-123
BSJ-03-123 is a PROTAC connected by ligands for Cereblon and CDK as a potent and novel CDK6-selective small-molecule degrader.
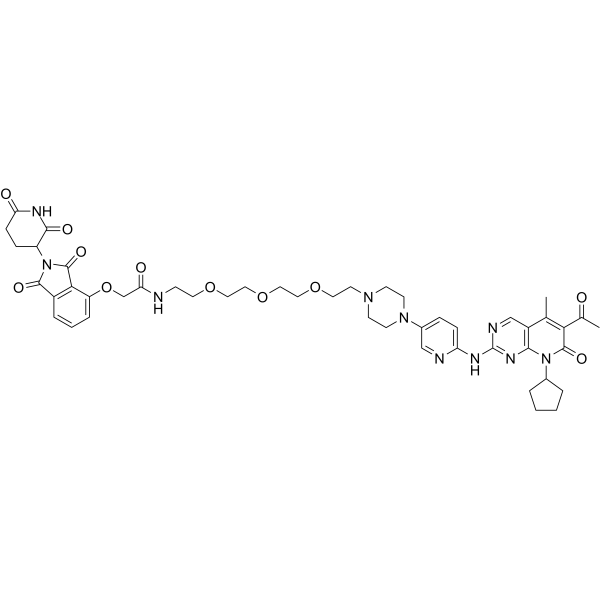
-
GC50615
BSJ-03-204
Selective Cdk4/6 degrader
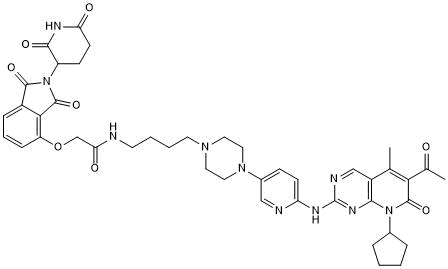
-
GC62263
BSJ-4-116
BSJ-4-116 is a PROTAC connected by ligands for Cereblon and CDK. BSJ-4-116 is a highly potent and selective CDK12 degrader (PROTAC) with an IC50 of 6 nM. BSJ-4-116 downregulates DDR genes through a premature termination of transcription, primarily through increasing poly(adenylation). BSJ-4-116 exhibits potent antiproliferative effects, alone and in combination with the poly(ADP-ribose) polymerase inhibitor Olaparib.
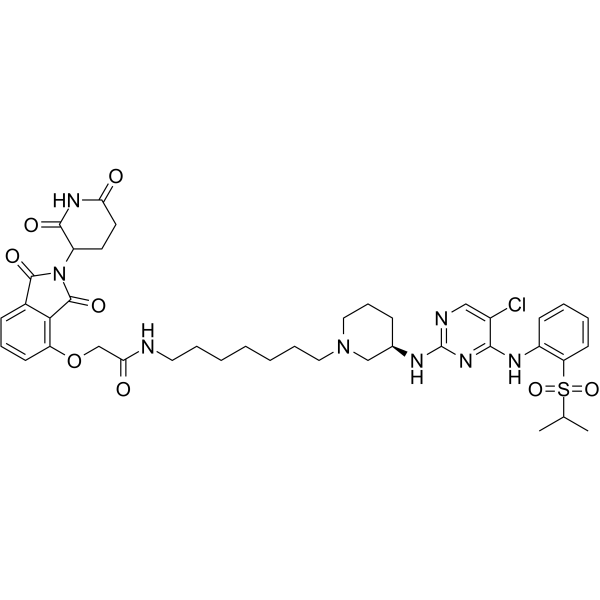
-
GC30977
Ca2+ channel agonist 1
Ca2+ channel agonist 1 is an agonist of N-type Ca2+ channel and an inhibitor of Cdk2, with EC50s of 14.23 μM and 3.34 μM, respectively, and is used as a potential treatment for motor nerve terminal dysfunction.
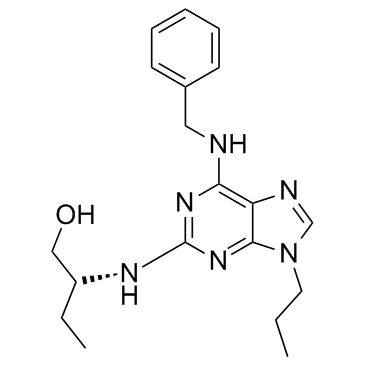
-
GC62442
Casein Kinase inhibitor A51
Casein Kinase inhibitor A51 is a potent and orally active casein kinase 1α (CK1α) inhibitor. Casein Kinase inhibitor A51 induces leukemia cell apoptosis, and has potent anti-leukemic activities.
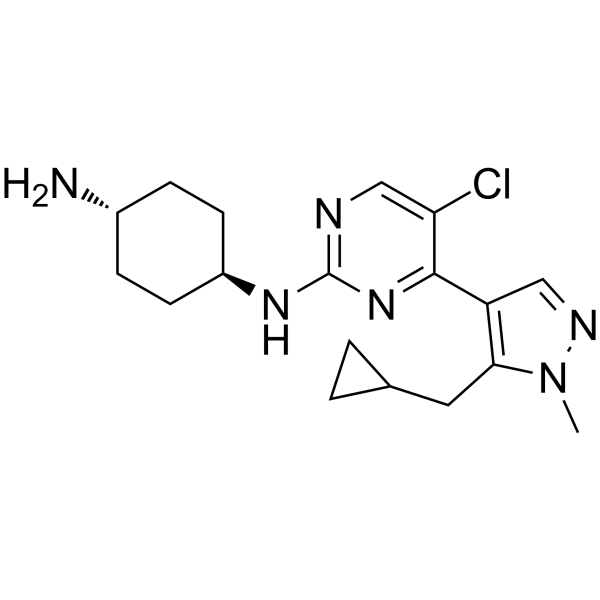
-
GC43175
CAY10574
CAY10574 is a potent, ATP-competitive CDK9/cyclin T1 inhibitor with an IC50 of 0.35 μM. CAY10574 exhibits a 38-fold selectivity for CDK9/cyclin T over other CDK/cyclin complexes. Antitumor activity.
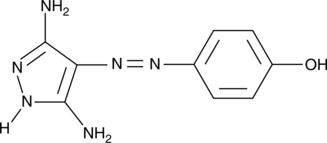
-
GC19089
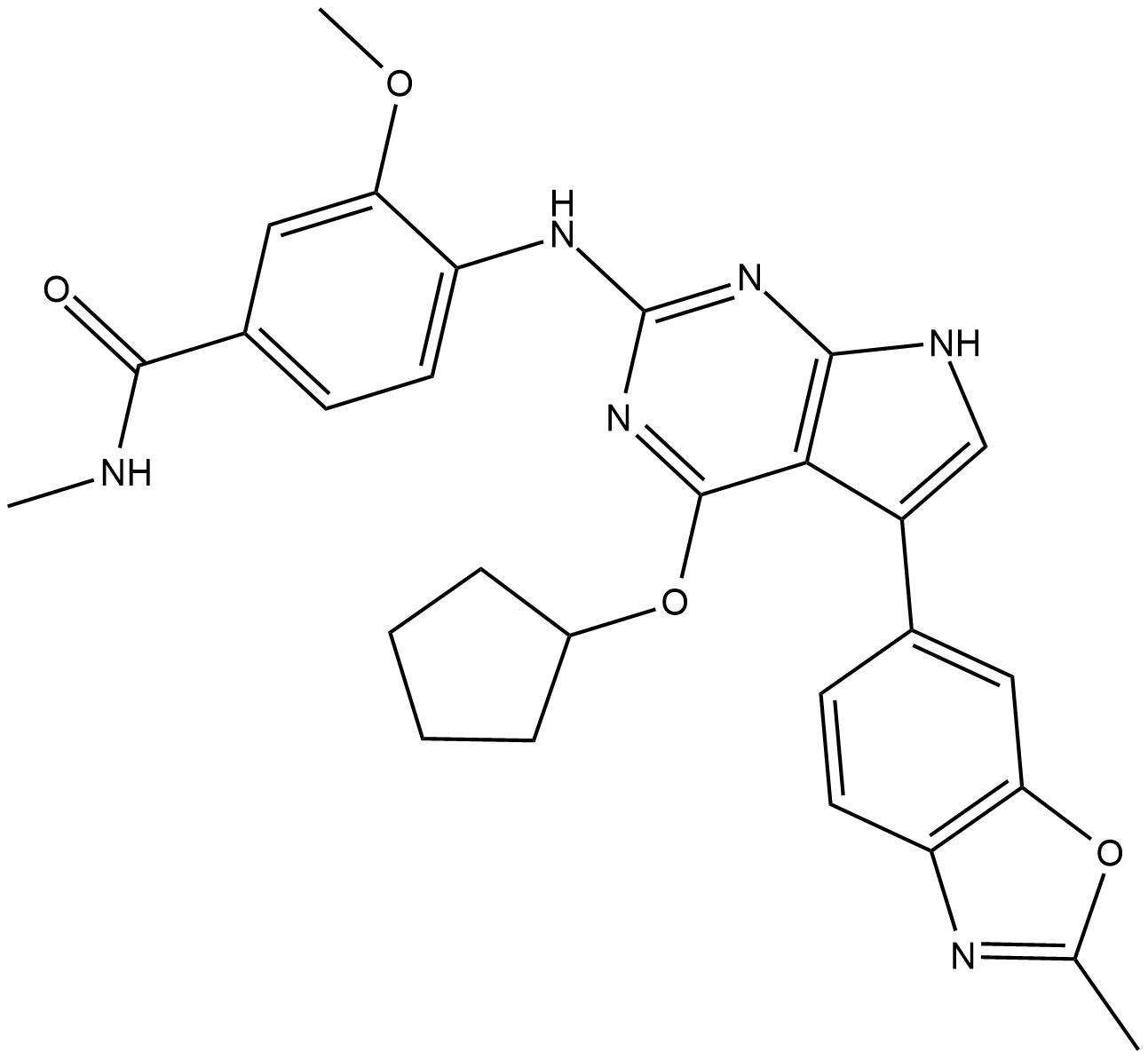
CC-671
CC-671 is a dual TTK protein kinase/CDC2-like kinase (CLK2) inhibitor with IC50s of 0.005 and 0.006 uM for TTK and CLK2, respectively.
-
GC32741
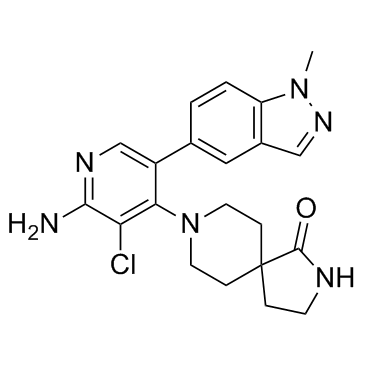
CCT-251921
CCT-251921 is a potent, selective, and orally bioavailable CDK8 inhibitor with an IC50 of 2.3 nM.
-
GC35628
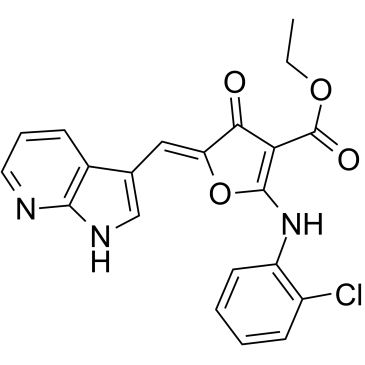
Cdc7-IN-1
Cdc7-IN-1 (Compound 13) is a highly potent, selective and ATP competitive inhibitor of Cdc7 kinase, with an IC50 value of 0.6 nM at 1 mM ATP and with slow off-rate characteristics. Cdc7-IN-1 potently inhibits Cdc7 activity in cancer cells, and effectively induces cell death.
-
GC62427
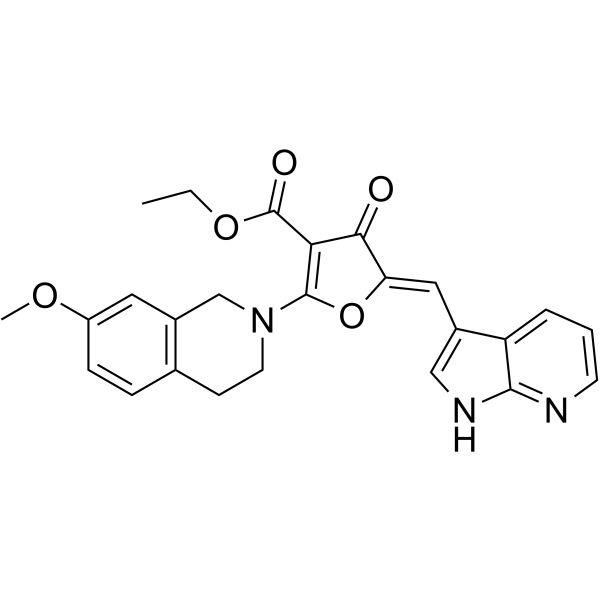
Cdc7-IN-5
Cdc7-IN-5 (compound I-B) is a potent Cdc7 kinase inhibitor extracted from patent WO2019165473A1, compound I-B. Cdc7 is a serine-threonine protein kinase enzyme which is essential for the initiation of DNA replication in the cell cycle.
-
GC12425
CDK inhibitor II
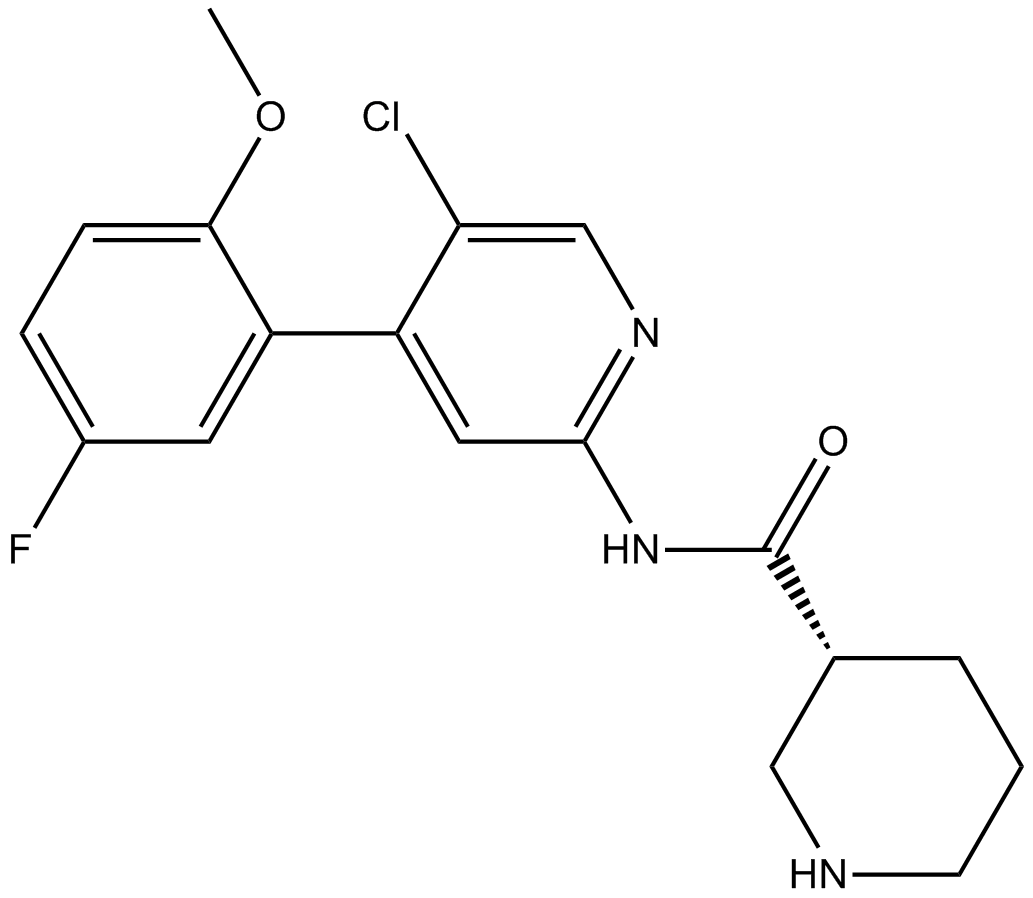
-
GC38899
CDK ligand for PROTAC hydrochloride
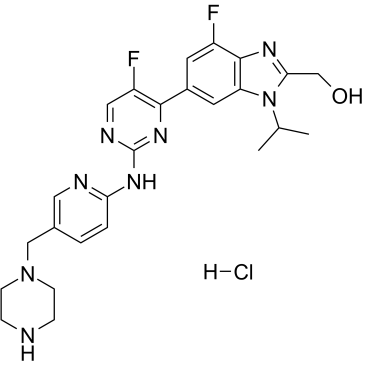
-
GC62168
CDK-IN-6
CDK-IN-6, a class of pyrazolo[1,5-a]pyrimidine compound, is a CDK inhibitor with anticancer activities.
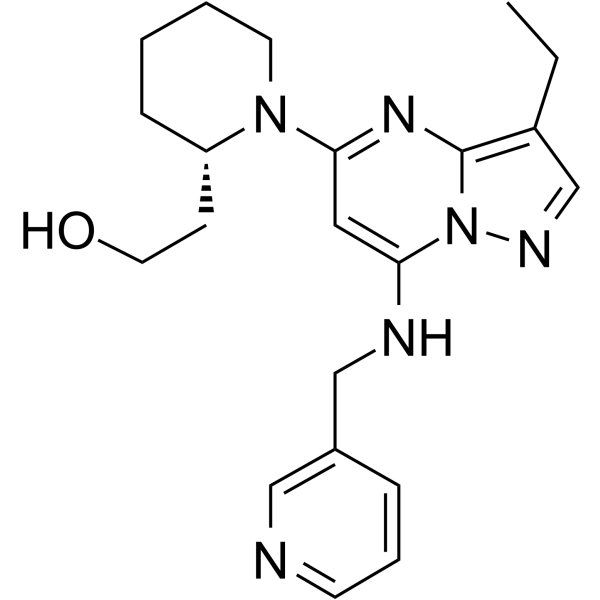
-
GC63499
CDK12-IN-2
CDK12-IN-2 is a potent, selective and nanomolar CDK12 inhibitor (IC50=52 nM) with good physicochemical properties. CDK12-IN-2 is also a strong CDK13 inhibitor due to CDK13 is the closest homologue of CDK12. CDK12-IN-2 shows excellent kinase selectivity for CDK12 over CDK2, 9, 8, and 7. CDK12-IN-2 inhibits the phosphorylation of Ser2 in the C-terminal domain of RNA polymerase II. CDK12-IN-2 can be used an excellent chemical probe for functional studies of CDK12.
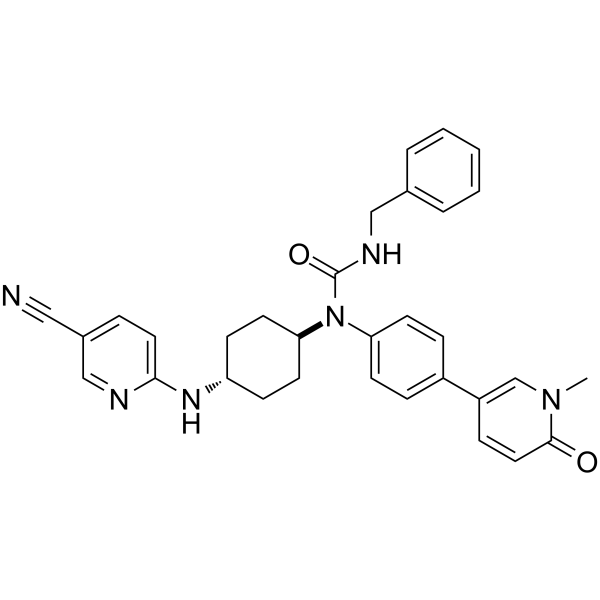
-
GC35632
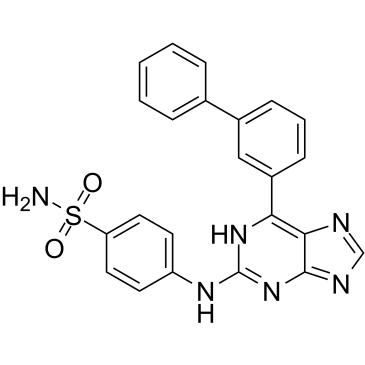
CDK2-IN-4
CDK2-IN-4 is a potent and selective CDK2 inhibitor with an IC50 of 44 nM for CDK2/cyclin A, shows 2,000-fold selectivity over CDK1/cyclin B (IC50=86 uM).
-
GC65190
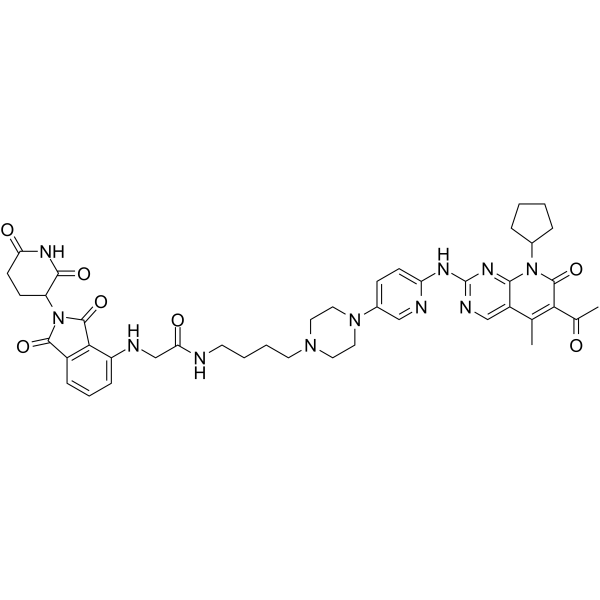
CDK4/6-IN-11
CDK4/6-IN-11 is a potent PROTAC CDK4/6 degrader.
-
GC35633
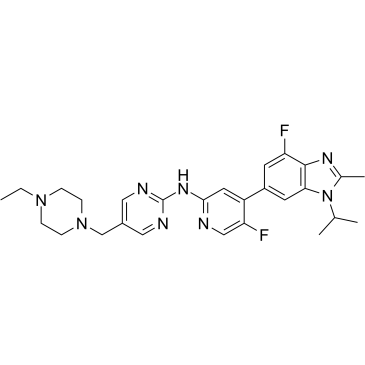
CDK4/6-IN-2
CDK4/6-IN-2 is a potent CDK4 and CDK6 inhibitor extracted from patent US20180000819A1, Compound 1, has IC50s of 2.7 and 16 nM for CDK4 and CDK6, respectively.
-
GC35634
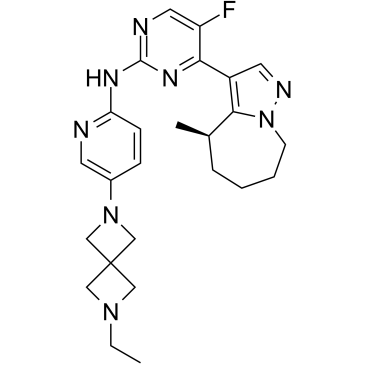
CDK4/6-IN-3
CDK4/6-IN-3 is a brain-penetrant CDK4/CDK6 inhibitor with Kis of <0.3 nM and 2.2 nM, respectively. CDK4/6-IN-3 inhibits CDK1 with a Ki of 110 nM. CDK4/6-IN-3 can be used for the treatment of glioblastoma.
-
GC64802
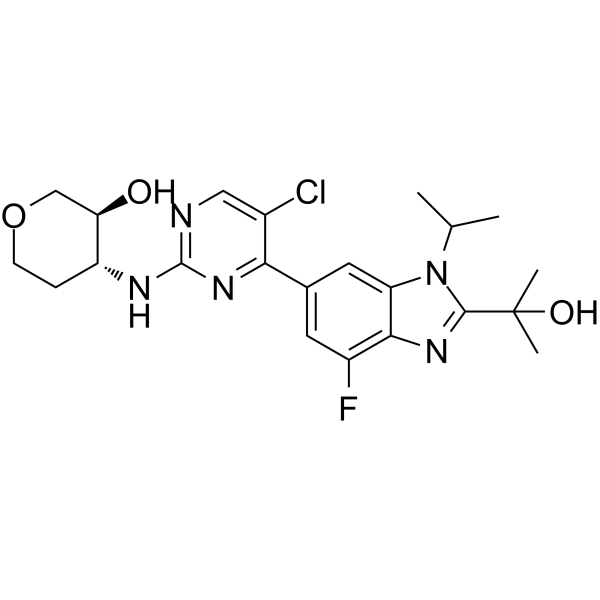
CDK4/6-IN-6
CDK4/6-IN-6 (example A94) is a potent CDK4/CDK6 inhibitor with a Ki of 0.6 nM and 13.9 nM for CDK4/Cyclin D1 and CDK6/Cyclin D3, respectively.
-
GC60680
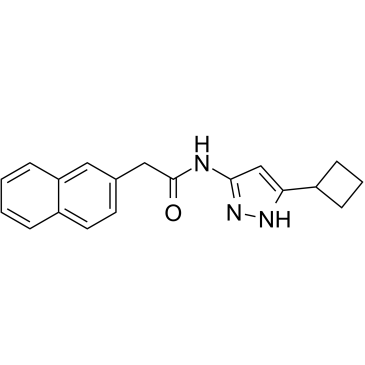
CDK5 inhibitor 20-223
CDK5 inhibitor 20-223 is a potent CDK2 and CDK5 inhibitor with IC50s of 6.0 and 8.8 nM, respectively. CDK5 inhibitor 20-223 is an effective anti-colorectal cancer (CRC) agent.
-
GC66009
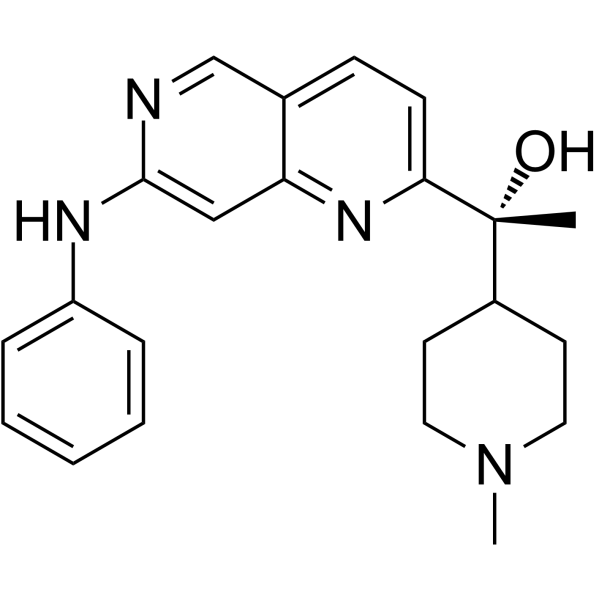
CDK5-IN-3
CDK5-IN-3 (compound 11) is a potent and selective CDK5 inhibitor, with IC50s of 0.6 nM and 18 nM for CDK5/p25 and CDK2/CycA, respectively. CDK5-IN-3 can be used for the research of autosomal dominant polycystic kidney disease (ADPKD).
-
GC63980
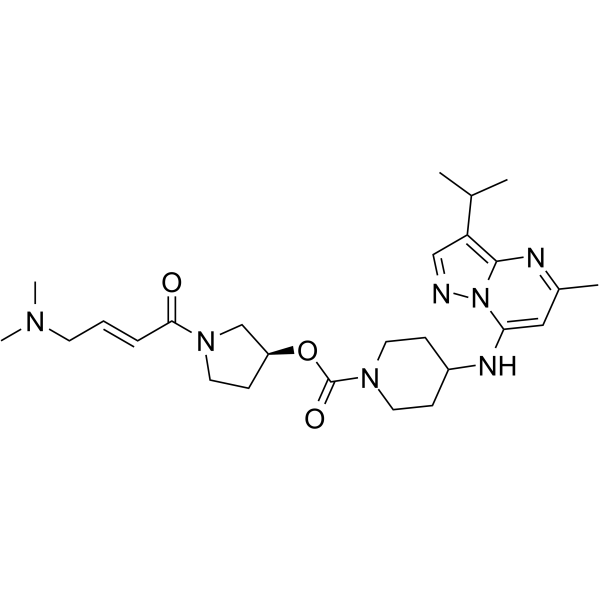
CDK7-IN-2
CDK7-IN-2 is a potent inhibitor of CDK7. CDK7 is implicated in both temporal control of the cell cycle and transcriptional activity. CDK7 is implicated in the transcriptional initiation process by phosphorylation of Rbpl subunit of RNA Polymerase II (RNAPII). CDK7 has the potential for the research of cancer disease, in particular aggressive and hard- to-treat cancers (extracted from patent WO2019099298A1, compound 1).
-
GC62596
CDK7-IN-3
CDK7-IN-3 (CDK7-IN-3) is an orally active, highly selective, noncovalent CDK7 inhibitor with a KD of 0.065 nM. CDK7-IN-3 shows poor inhibition on CDK2 (Ki=2600 nM), CDK9 (Ki=960 nM), CDK12 (Ki=870 nM). CDK7-IN-3 induces apoptosis in tumor cells and has antitumor activity.
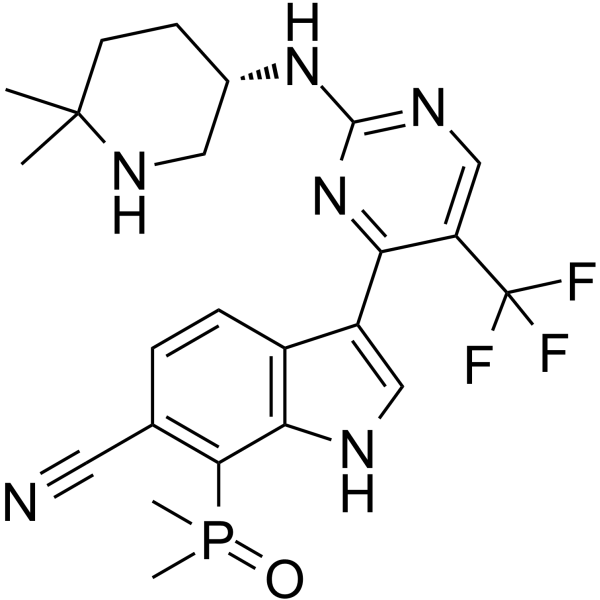
-
GC65434
CDK7/12-IN-1
CDK7/12-IN-1 is a selective CDK7/12 inhibitor with IC50s of 3 and 277 nM for CDK7 and CDK 12, respectively. CDK7 and CDK12 inhibition is an effective strategy to inhibit tumour growth.
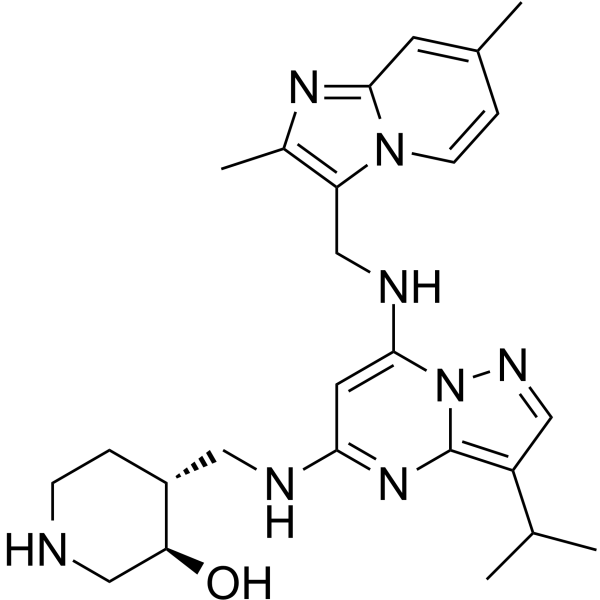
-
GC60681
CDK7/9 tide
CDK7/9 tide is peptide substrate for CDK7 or CDK9.

-
GC33180
CDK8-IN-1
CDK8-IN-1 is a potent and selective CDK8 inhibitor with an IC50 of 3 nM.
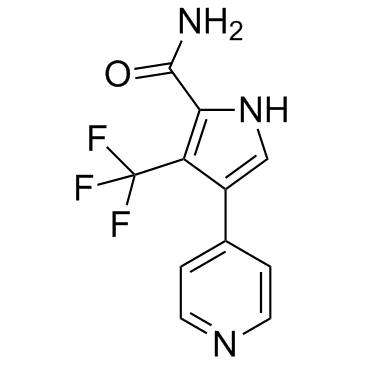
-
GC30466
CDK8-IN-3
CDK8-IN-3 is an inhibitor of CDK8 extracted from patent WO2016041618A1, compound example 1.7.
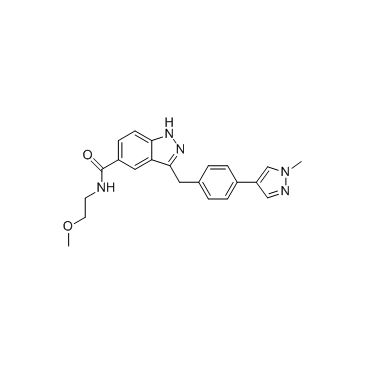
-
GC33366
CDK8-IN-4
CDK8-IN-4 is an inhibitor of CDK8 extracted from patent WO2014090692A1, compound example 16, with an IC50 of 0.2 nM.
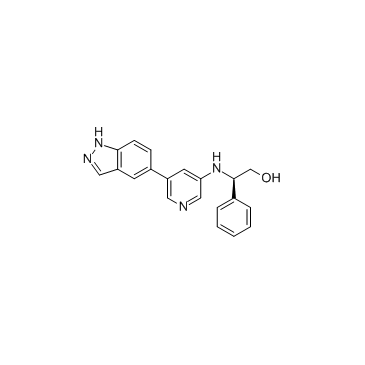
-
GC35635
CDK9 Antagonist-1
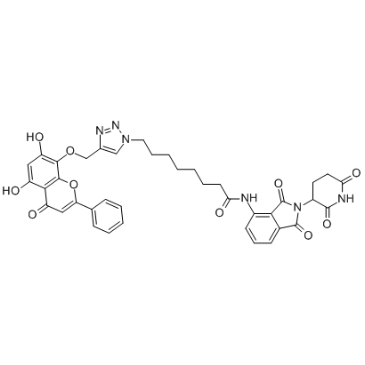
-
GC12871
CDK9 inhibitor
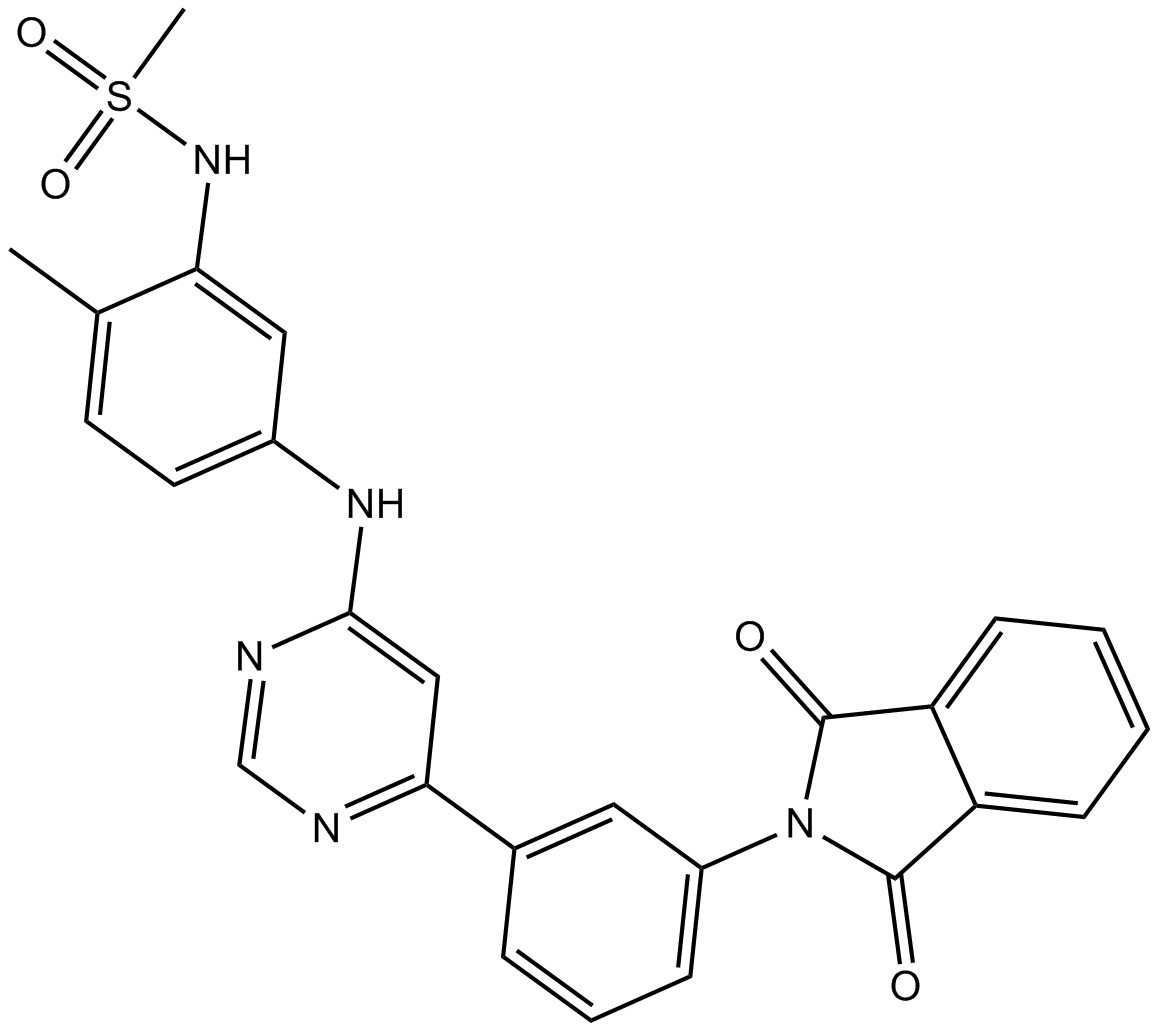
-
GC17359
CDK9 inhibitor 2
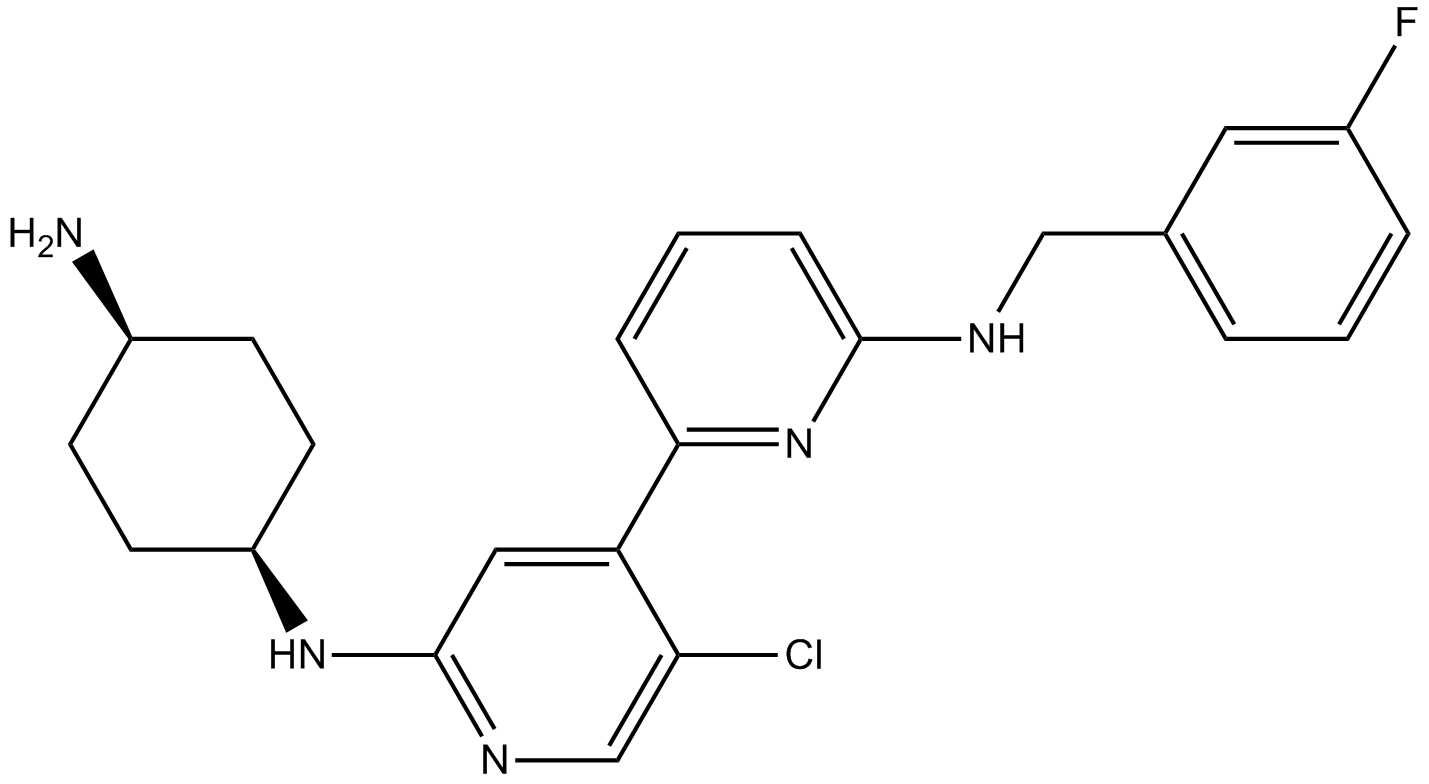
-
GC66475
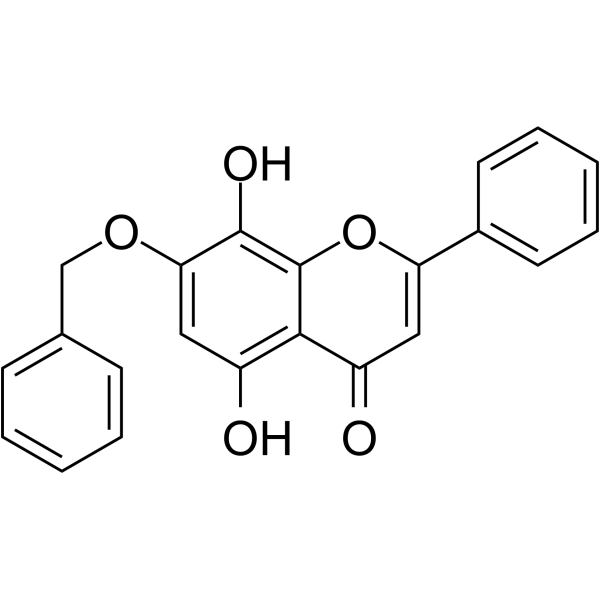
CDK9-IN-10
CDK9-IN-10 is a potent CDK9 inhibitor. CDK9-IN-10 is the ligand for the PROTAC CDK9 degrader-2 (HY-112811).
-
GC65262
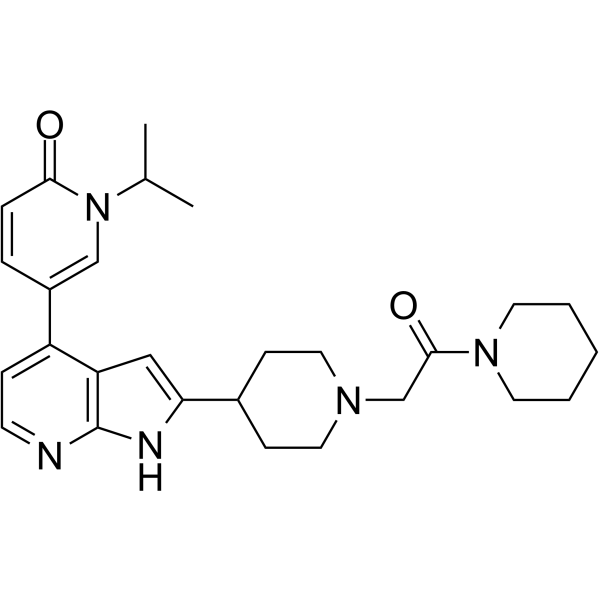
CDK9-IN-13
CDK9-IN-13 (compound 38) is potent and selective CDK9 inhibitor, with an IC50 of <3 nM. CDK9-IN-13 exhibits short half-lives in rodents.
-
GC64446
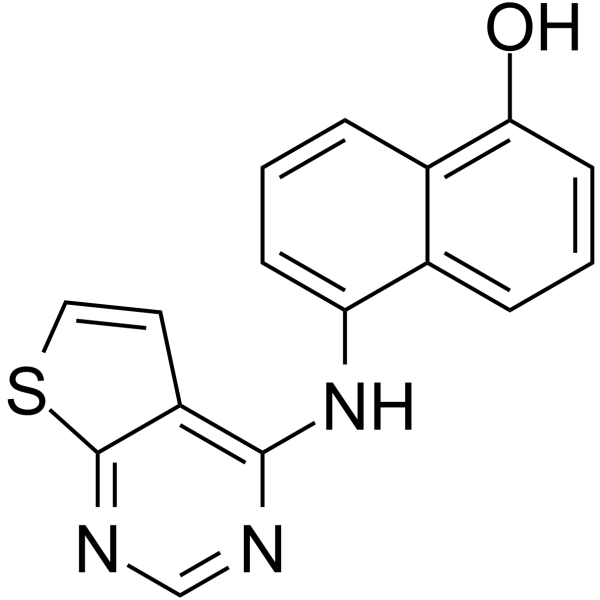
CDK9-IN-15
CDK9-IN-15 (compound 50) is a potent CDK9 inhibitor.
-
GC35636
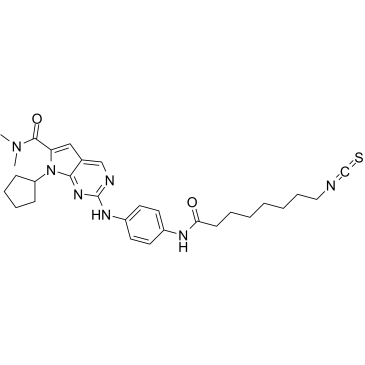
CDK9-IN-7
CDK9-IN-7 (compound 21e) is a selective, highly potent, and orally active CDK9/cyclin T inhibitor (IC50=11 nM), which exhibits more potent over other CDKs (CDK4/cyclinD=148 nM; CDK6/cyclinD=145 nM). CDK9-IN-7 shows antitumor activity without obvious toxicity. CDK9-IN-7 induces NSCLC cell apoptosis, arrests the cell cycle in the G2 phase, and suppresses the stemness properties of NSCLC.
-
GC65247
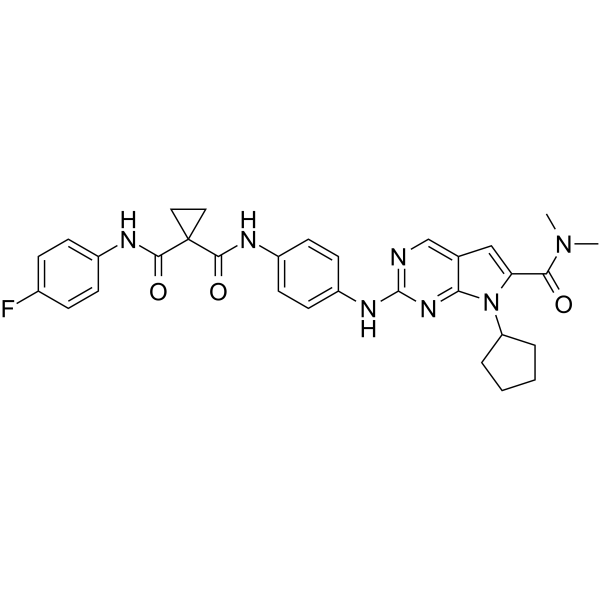
CDK9-IN-8
CDK9-IN-8 is a highly effective and selective CDK9 inhibitor with an IC50 of 12 nM.
-
GC19096
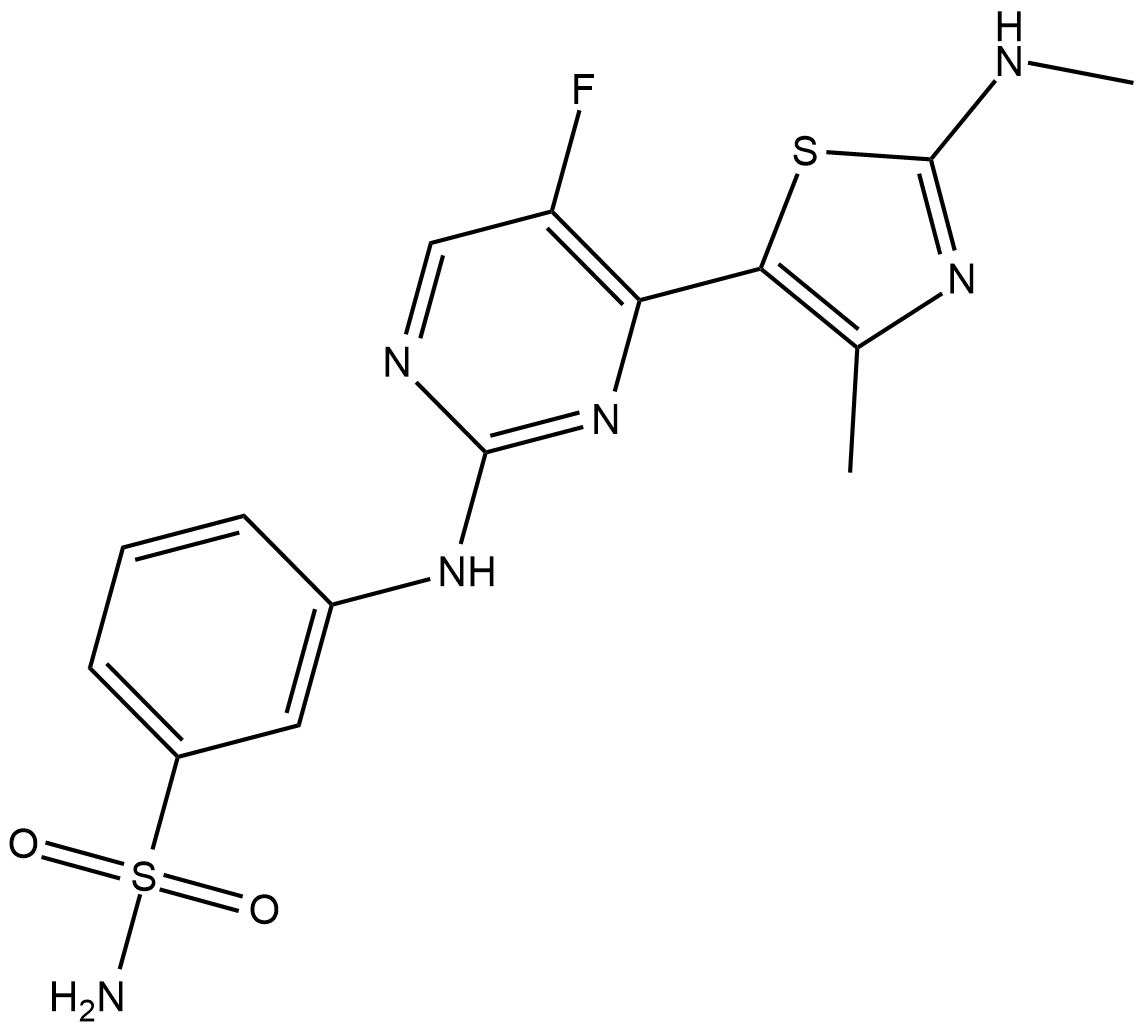
CDKI-73
CDKI-73 is a potent CDK9 inhibitor with Ki of 4 nM; shows selective toxicity to CLL cells(LD50=80 nM) versus normal B cell and normal CD34+ cell(LD50>20 uM).
-
GC65878
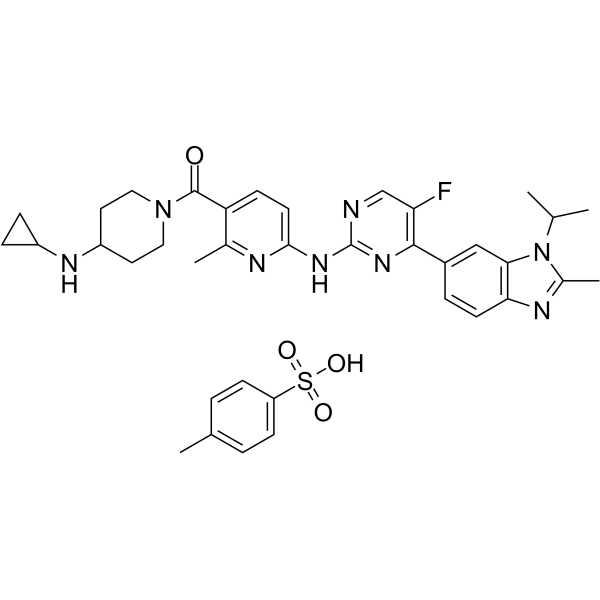
Cimpuciclib tosylate
Cimpuciclib tosylate is a selective CDK4 inhibitor (IC50: 0.49 nM) that has anti-tumor activity.
-
GC63476
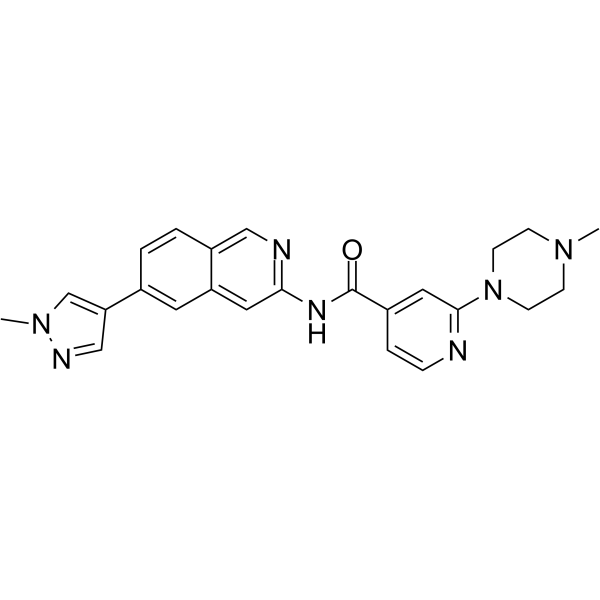
Cirtuvivint
Cirtuvivint (SM08502) is a potent and orally active CDC-like kinase (CLK) inhibitor. Cirtuvivint can be used for solid tumors research.
-
GC63800
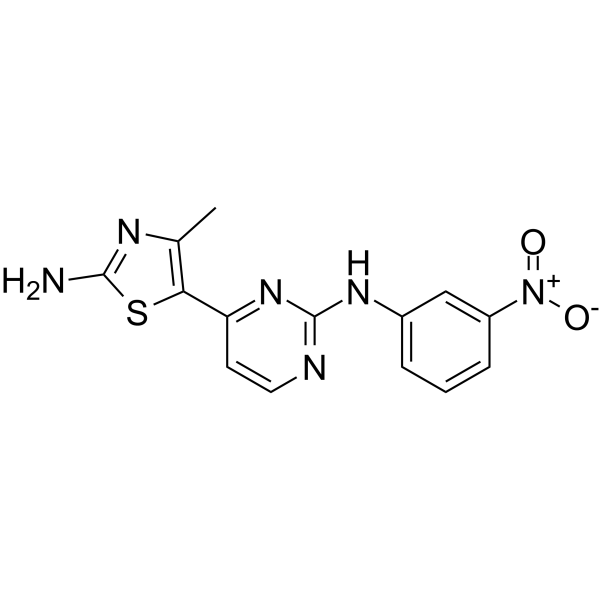
CK7
CK7, a Cdk2/9 inhibitor, can be used for the synthesis of Nek1 inhibitor BSc5231 and BSc5367.
-
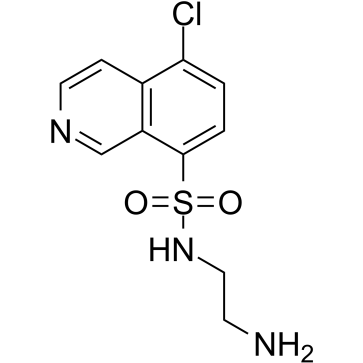
GC38755
CKI-7
CKI-7 is a potent and ATP-competitive casein kinase 1 (CK1) inhibitor with an IC50 of 6 μM and a Ki of 8.5 μM. CKI-7 is a selective Cdc7 kinase inhibitor. CKI-7 also inhibits SGK, ribosomal S6 kinase-1 (S6K1) and mitogen- and stress-activated protein kinase-1 (MSK1). CKI-7 has a much weaker effect on casein kinase II and other protein kinases.
-
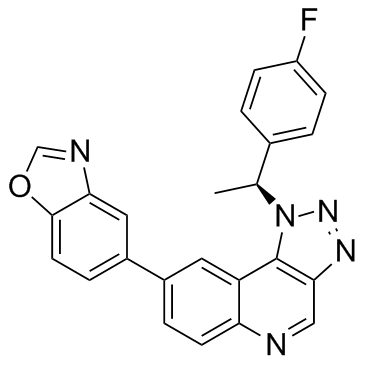
GC30245
CLK1-IN-1
CLK1-IN-1 is a potent and selective of Cdc2-like kinase 1 (CLK1) inhibitor, with an IC50 of 2 nM.
-
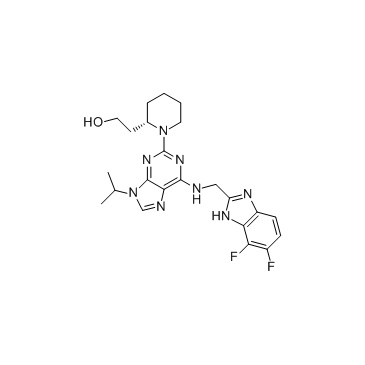
GC33327
CMPD 7
CMPD 7 is a potent and selective CDK12 inhibitor with an IC50 of 491 nM in enzymatic assay.
-
GC35739
CP-10
CP-10 is a PROTAC connected by ligands for Cereblon and CDK, with highly selective, specific, and remarkable CDK6 degradation (DC50=2.1 nM). It inhibits proliferation of several haematopoietic cancer cells with impressive potency including multiple myeloma, and can still degrades mutated and overexpressed CDK6.
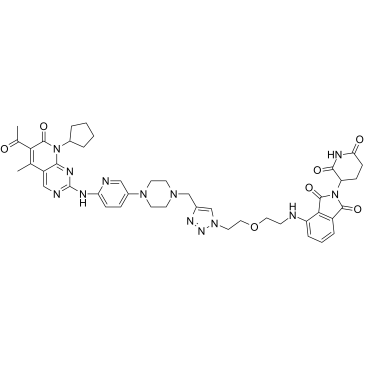
-
GC68455
CP681301
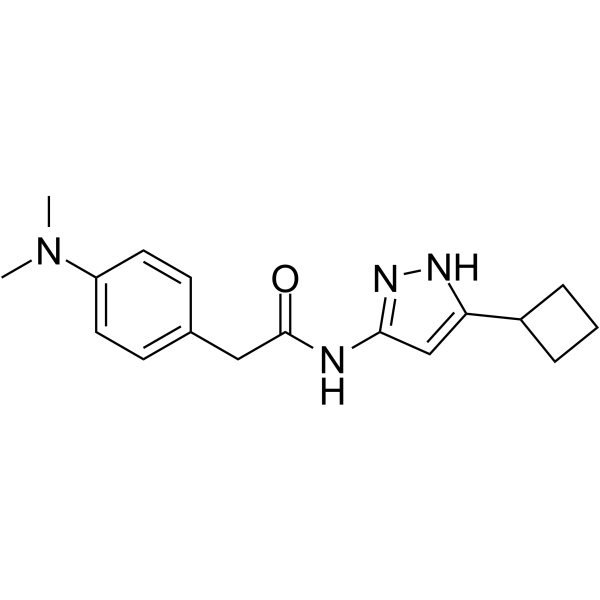
-
GC65023
CTX-712
CTX-712 is a potent inhibitor of cdc2-like kinase (CLK). CTX-712 inhibits CLK kinase activity, and thus inhibits cancer survival and cancer cell growth. CTX-712 has the potential for the research of cancer disease (extracted from patent JPWO2017188374A1, compound 286).
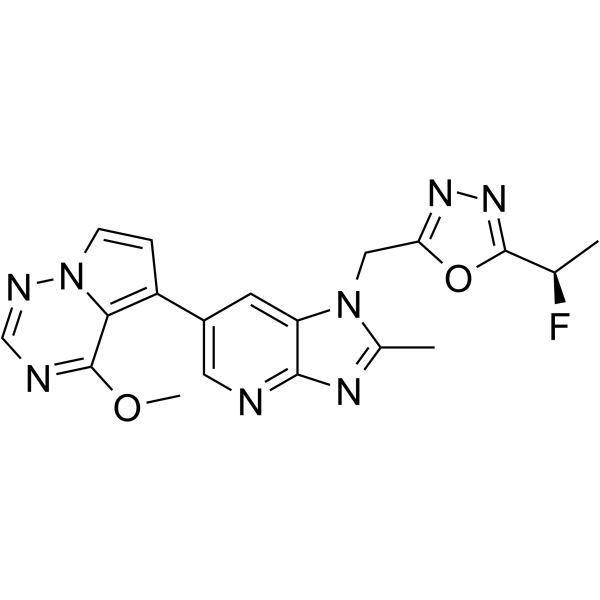
-
GN10526
Cucurbitacin E
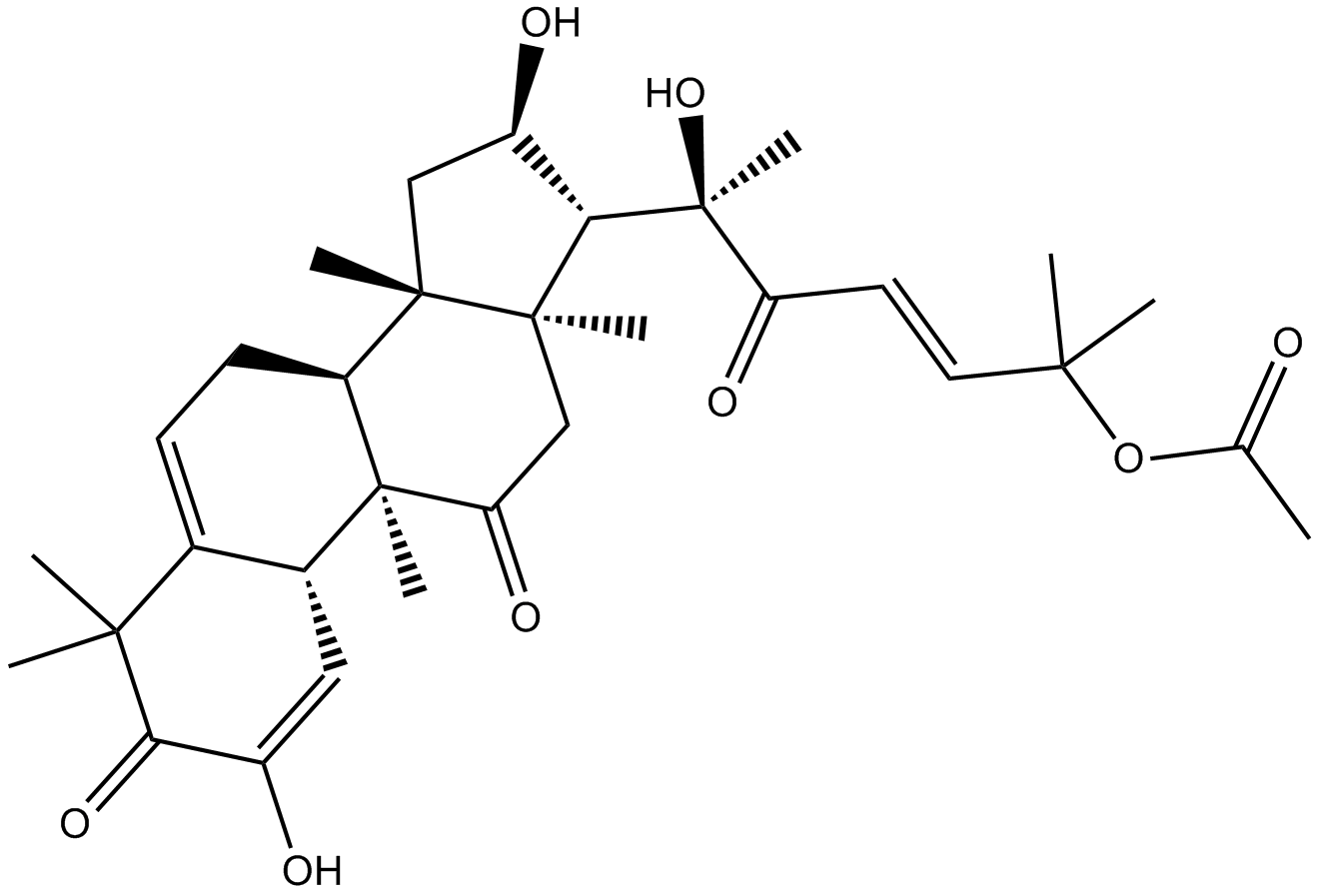
-
GC18028
CVT-313
A Cdk2 inhibitor
-
GC32700
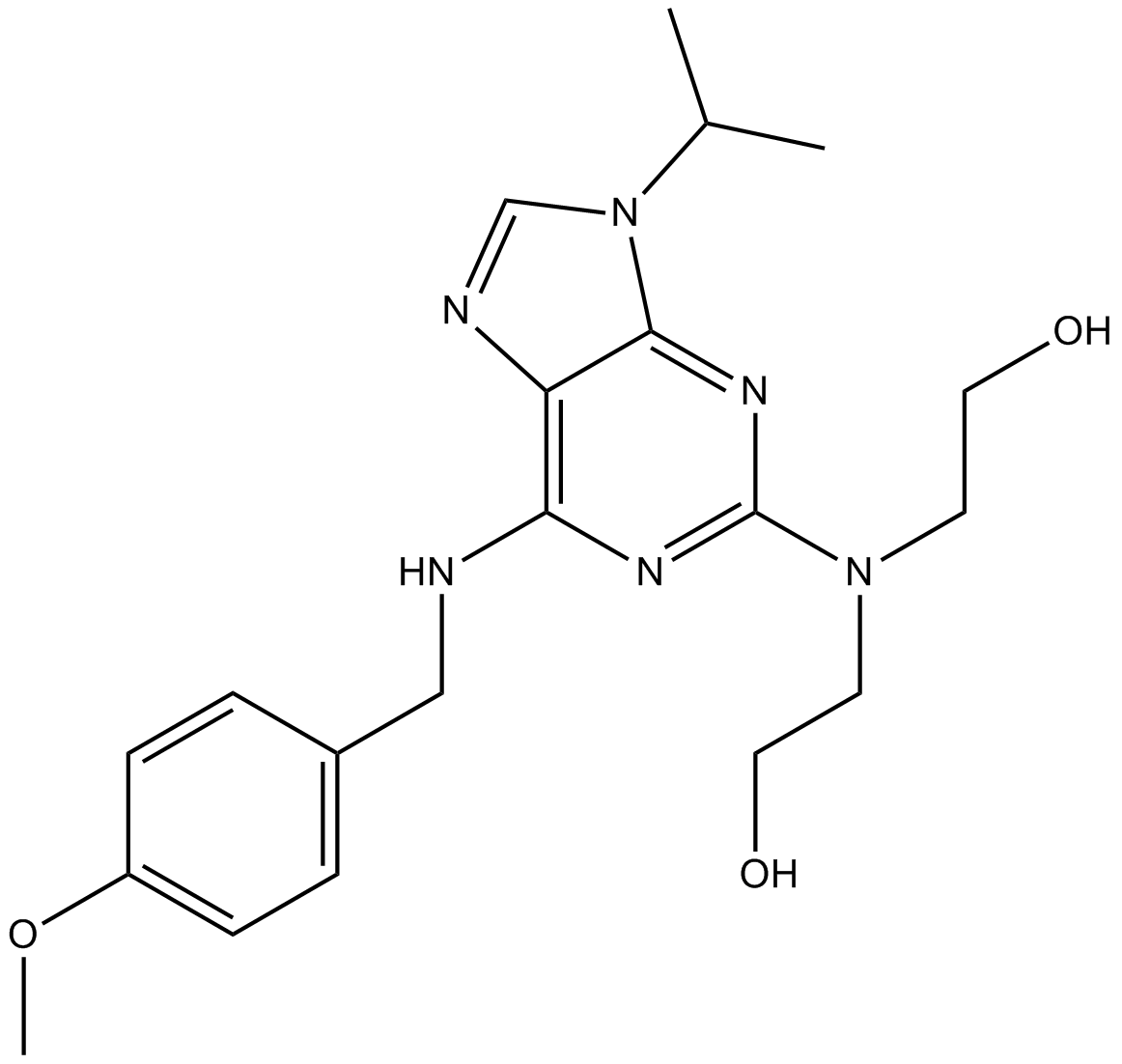
CYC065
CYC065 (CYC065) is a second-generation, orally available ATP-competitive inhibitor of CDK2/CDK9 kinases with IC50s of 5 and 26 nM, respectively.
-
GC63419
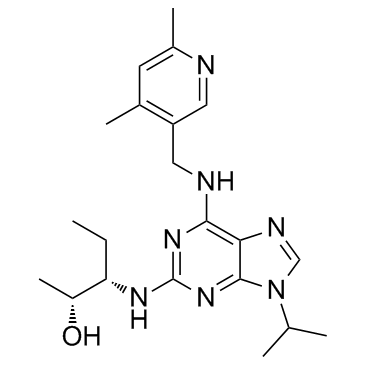
Dalpiciclib
Dalpiciclib (SHR-6390) is an orally active and highly selective inhibitor of CDK4 and 6 with IC50 values of 12.4?nM and 9.9?nM, respectively. Dalpiciclib shows antitumor activity against breast cancer and esophageal squamous cell carcinoma.
-
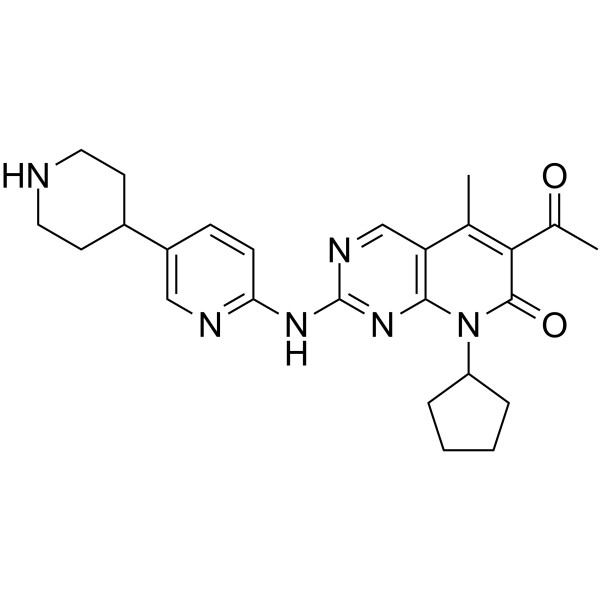
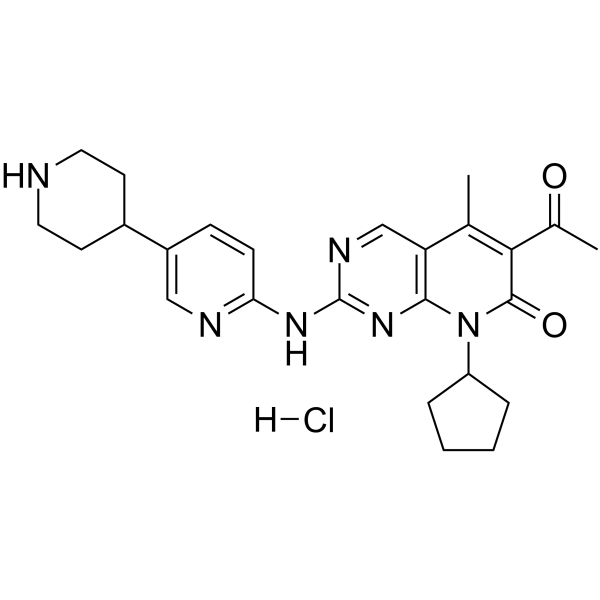
GC65369
Dalpiciclib hydrochloride
Dalpiciclib (SHR-6390) hydrochloride is an orally active and highly selective inhibitor of CDK4 and 6 with IC50 values of 12.4?nM and 9.9?nM, respectively. Dalpiciclib hydrochloride shows antitumor activity against breast cancer and esophageal squamous cell carcinoma.
-
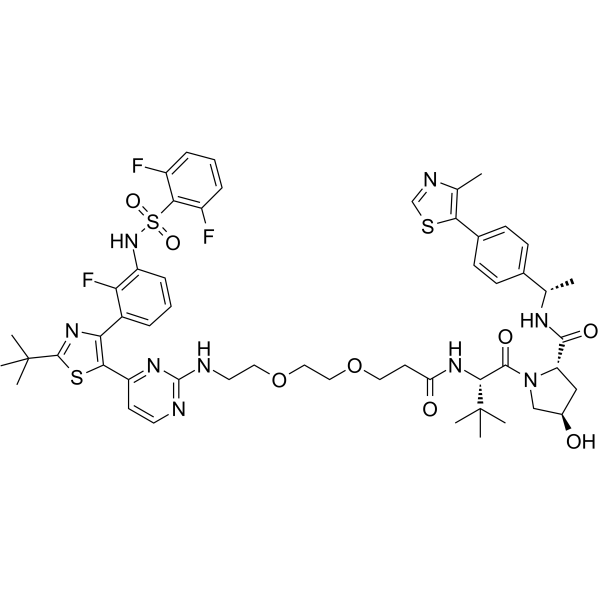
GC67970
DD-03-156
-
GC38482
Desmethylglycitein
Desmethylglycitein (4',6,7-Trihydroxyisoflavone), a metabolite of daidzein, sourced from Glycine max with antioxidant, and anti-cancer activities.Desmethylglycitein binds directly to CDK1 and CDK2 in vivo, resulting in the suppresses CDK1 and CDK2 activity. Desmethylglycitein is a direct inhibitor of protein kinase C (PKC)α, against solar UV (sUV)-induced matrix matrix metalloproteinase 1 (MMP1). Desmethylglycitein binds to PI3K in an ATP competitive manner in the cytosol, where it inhibits the activity of PI3K and downstream signaling cascades, leading to the suppression of adipogenesis in 3T3-L1 preadipocytes.

-
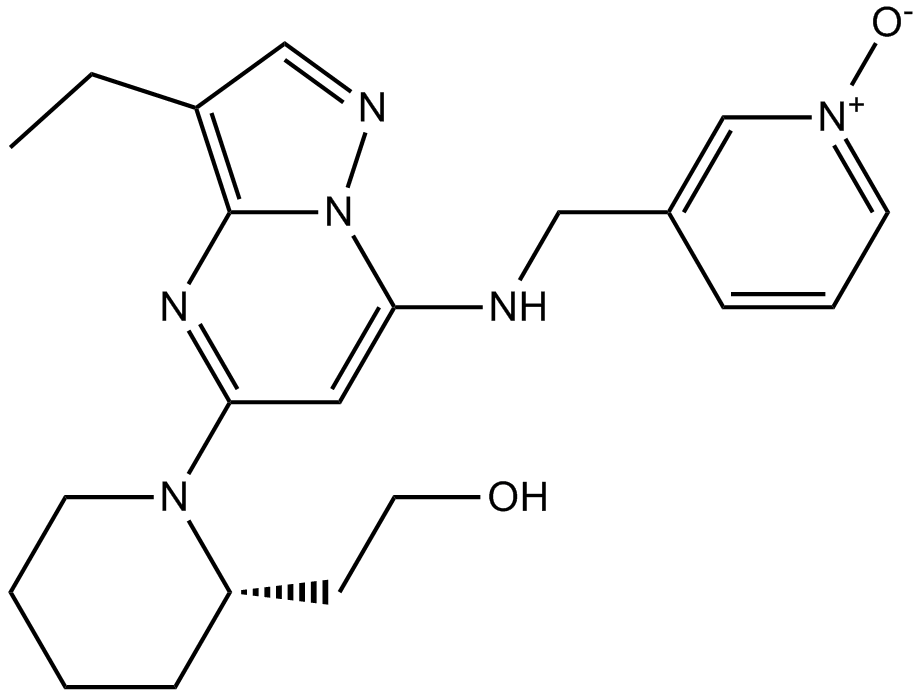
GC17648
Dinaciclib(SCH727965)
Dinaciclib(SCH727965) (SCH 727965) is a potent inhibitor of CDK, with IC50s of 1 nM, 1 nM, 3 nM, and 4 nM for CDK2, CDK5, CDK1, and CDK9, respectively.
-
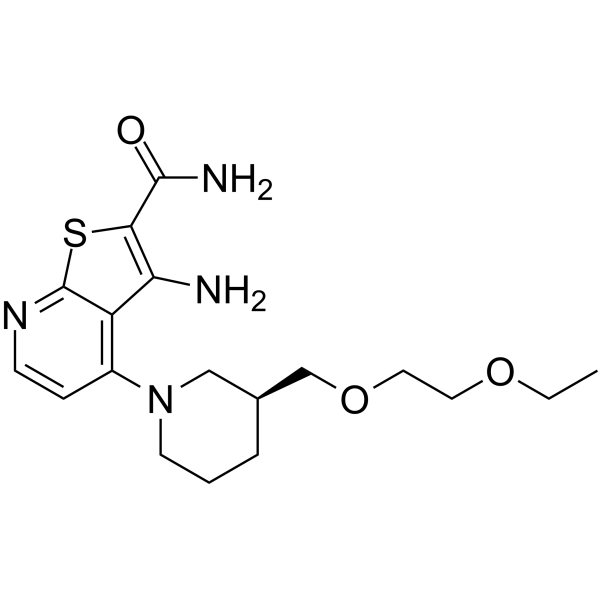
GC64715
DS96432529
DS96432529 is a potent and orally active bone anabolic agent through CDK8 inhibition.
-
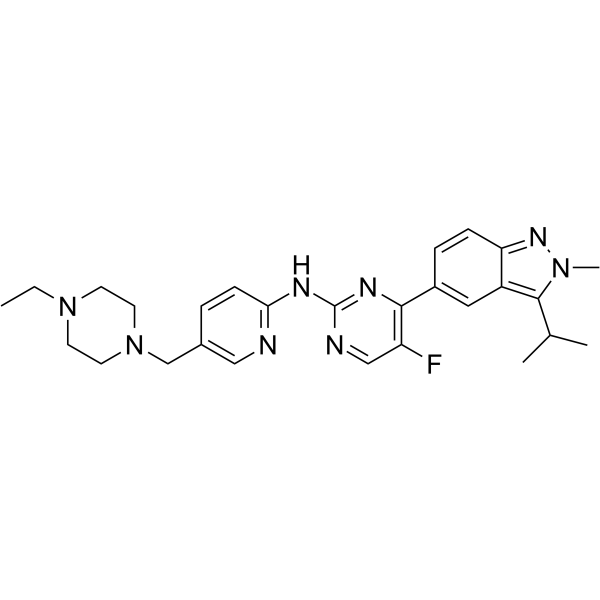
GC64682
Eciruciclib
Eciruciclib is an antineoplastic and potent cyclin dependent kinase (CDK) inhibitor.
-
GC38330
EHT 5372
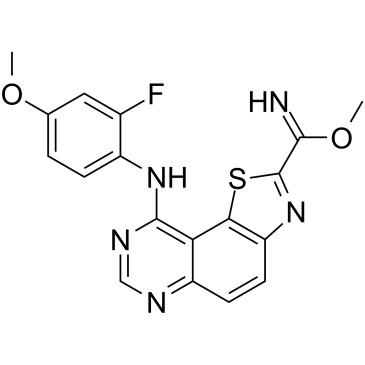
-
GC16063
Flavopiridol
An inhibitor of cyclin-dependent kinases
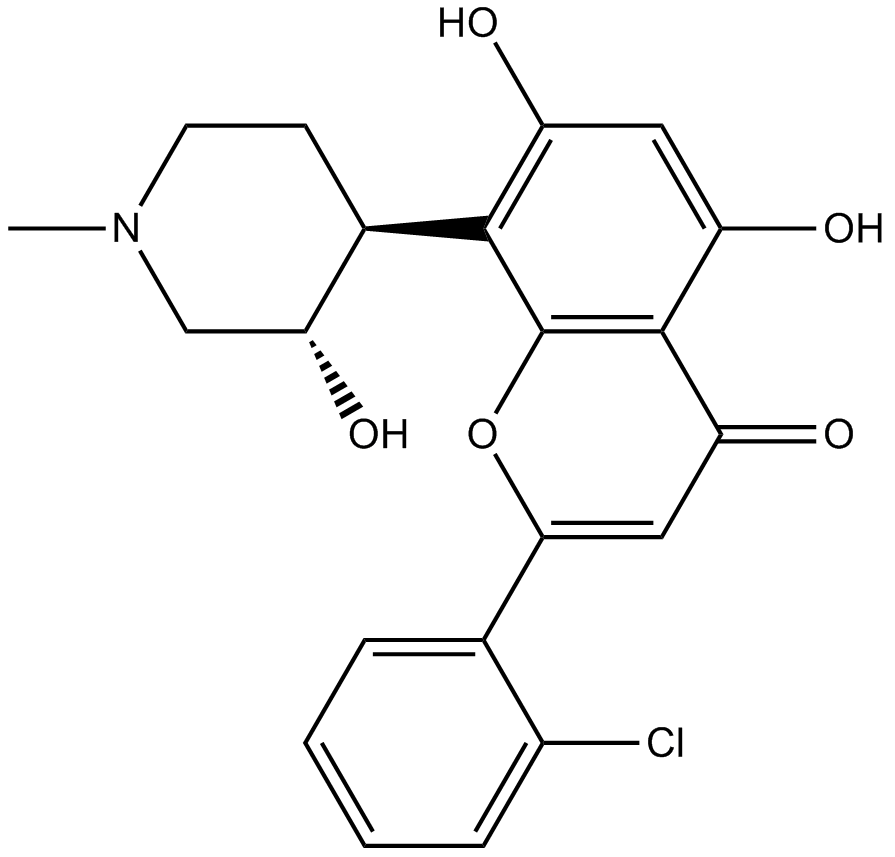
-
GC16425
Flavopiridol hydrochloride
CDK inhibitor, potent and selective