DNA/RNA Synthesis
RNA synthesis, which is also called DNA transcription, is a highly selective process. Transcription by RNA polymerase II extends beyond RNA synthesis, towards a more active role in mRNA maturation, surveillance and export to the cytoplasm.
Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA.DNA ligases discriminate against substrates containing RNA strands or mismatched base pairs at positions near the ends of the nickedDNA. Bleomycin (BLM) exerts its genotoxicity by generating free radicals, whichattack C-4′ in the deoxyribose backbone of DNA, leading to opening of the ribose ring and strand breakage; it is an S-independentradiomimetic agent that causes double-strand breaks in DNA.
First strand cDNA is synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H). Second strand cDNA synthesis is subsequently performed using DNA Polymerase I and RNase H. The remaining overhangs are converted into blunt ends using exonuclease/polymerase activity. After adenylation of the 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure is ligated to prepare the samples for hybridization. Cell cycle and DNA replication are the top two pathways regulated by BET bromodomain inhibition. Cycloheximide blocks the translation of mRNA to protein.
Targets for DNA/RNA Synthesis
Products for DNA/RNA Synthesis
- Cat.No. Product Name Information
-
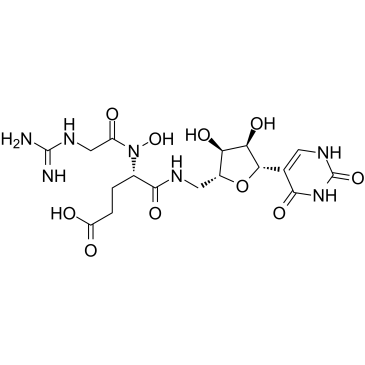
GC52303
Ethyl Mycophenolate
A potential impurity found in commercial preparations of mycophenolate mofetil
-
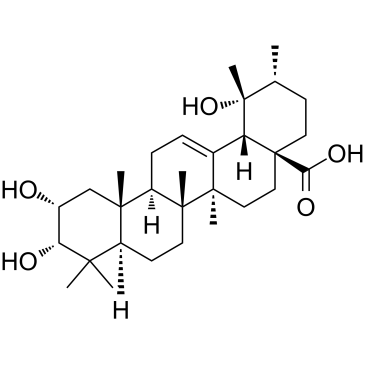
GC38392
Euscaphic acid
Euscaphic acid, a DNA polymerase inhibitor, is a triterpene from the root of the R. alceaefolius Poir. Euscaphic inhibits calf DNA polymerase α (pol α) and rat DNA polymerase β (pol β) with IC50 values of 61 and 108 μM. Euscaphic acid induces apoptosis.
-
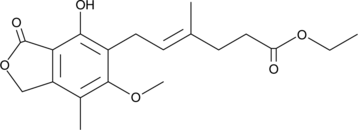
GC46146
FD-211
A δ lactone with anticancer activity
-
GC18014
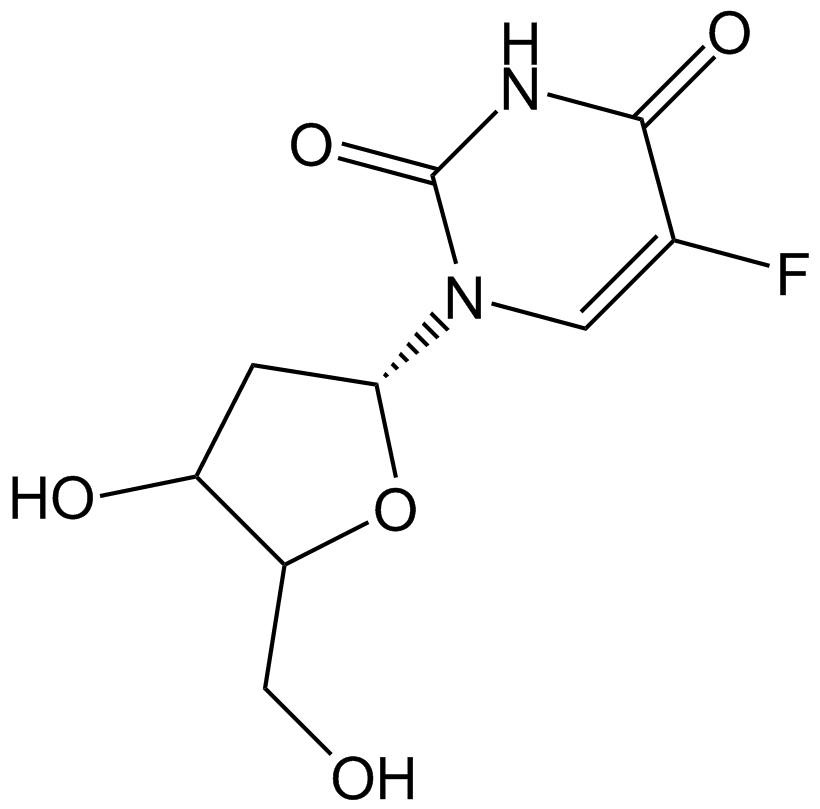
Floxuridine
Antineoplastic antimetabolite
-
GC14144
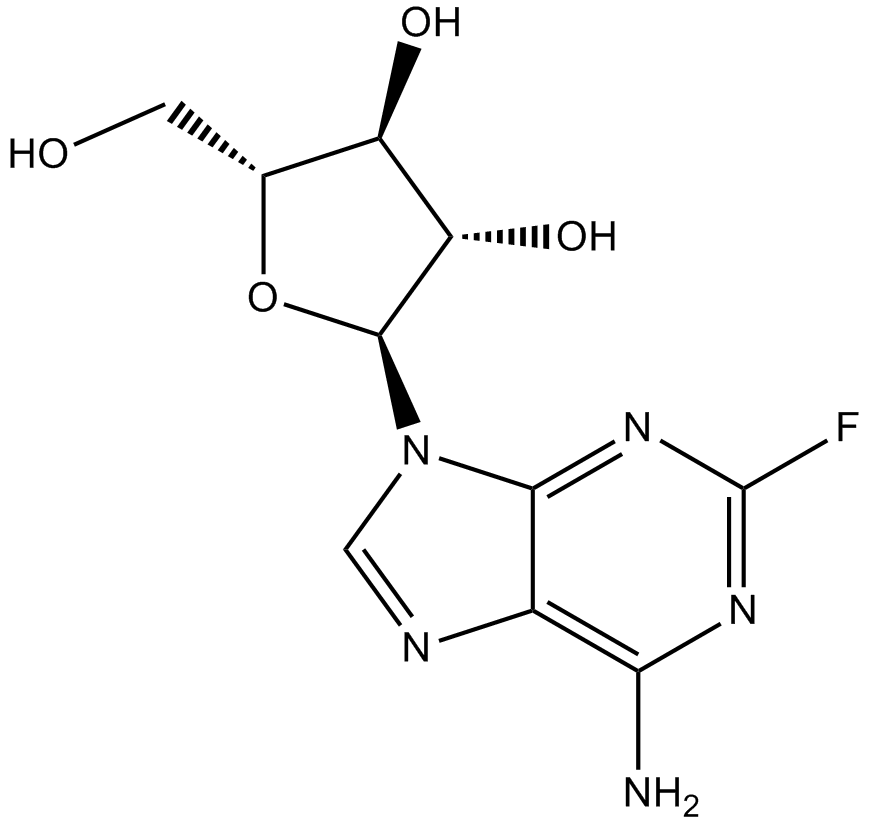
Fludarabine
DNA synthsis inhibitor
-
GC15134
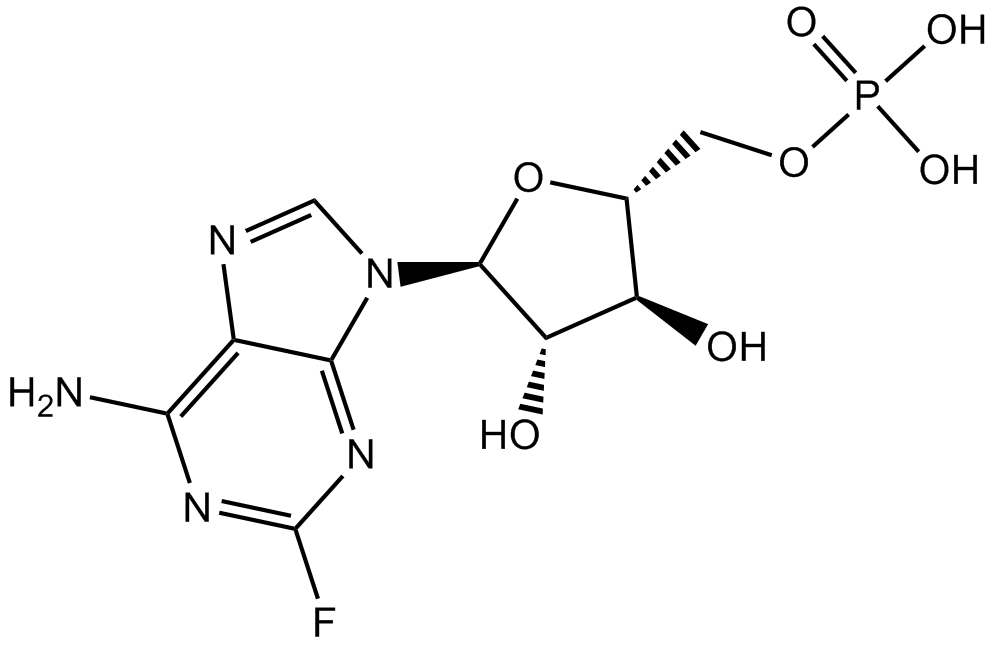
Fludarabine Phosphate (Fludara)
Fludarabine (phosphate) is an analogue of adenosine and deoxyadenosine, which is able to compete with dATP for incorporation into DNA and inhibit DNA synthesis.
-
GC49134
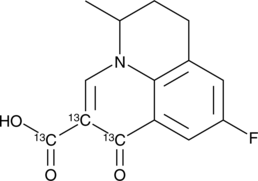
Flumequine-13C3
An internal standard for the quantification of flumequine
-
GC49455
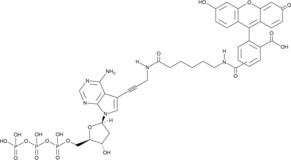
Fluorescein-12-dATP
A fluorescently labeled form of dATP
-
GC49746
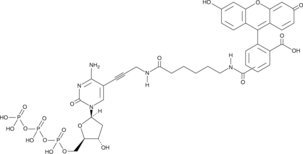
Fluorescein-12-dCTP
A fluorescently labeled form of dCTP
-
GC49456
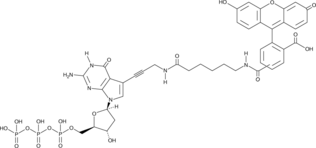
Fluorescein-12-dGTP
A fluorescently labeled form of dGTP
-
GC14466
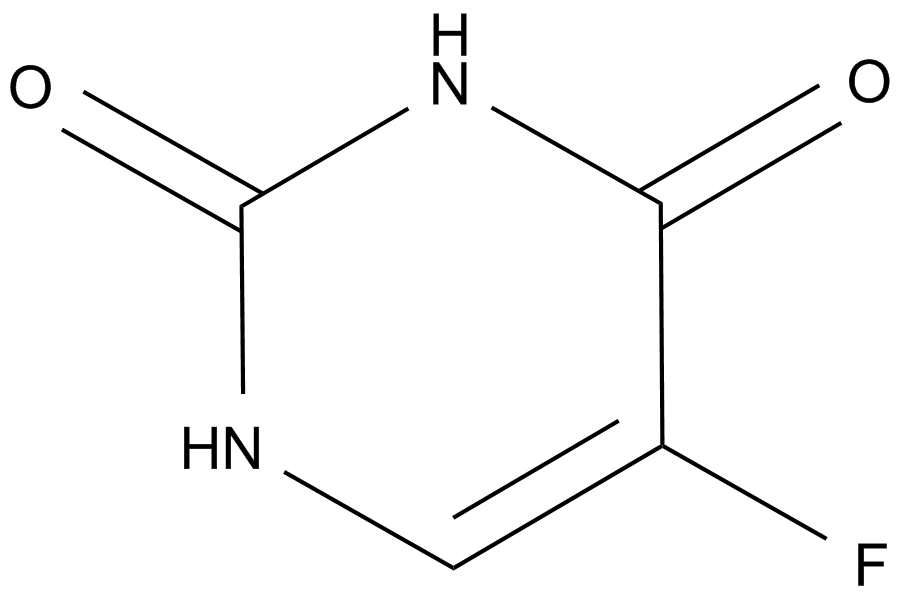
Fluorouracil (Adrucil)
A prodrug form of FdUMP
-
GN10306
Folic acid
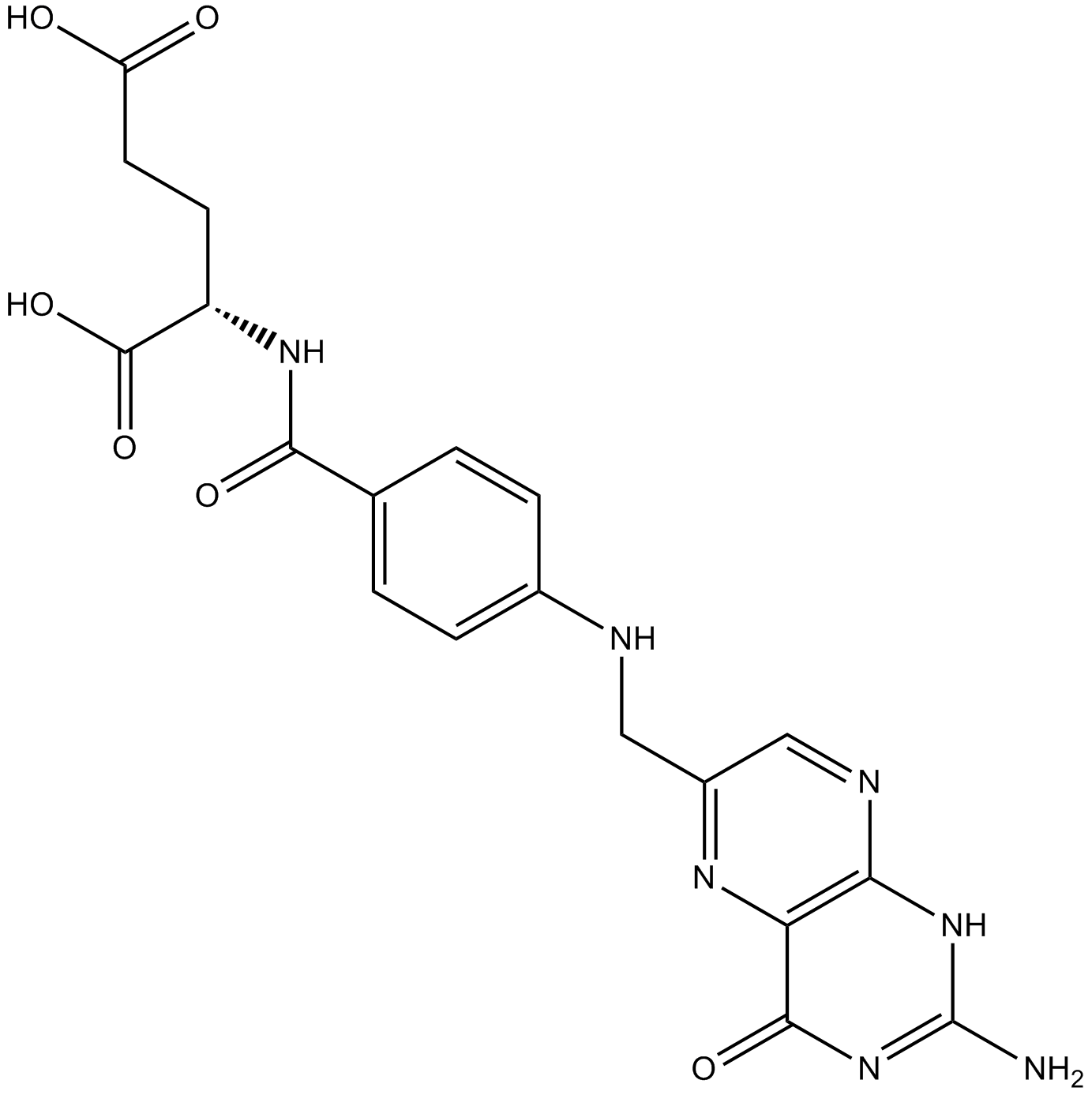
-
GC63952
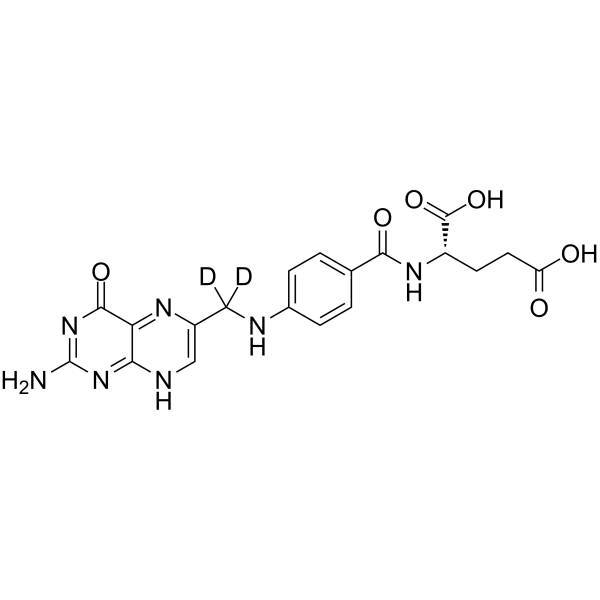
Folic Acid-d2
Folic Acid-d2 is the deuterium labeled Folic acid. Folic acid (Vitamin M; Vitamin B9) is a B vitamin; is necessary for the production and maintenance of new cells, for DNA synthesis and RNA synthesis.
-
GC49126
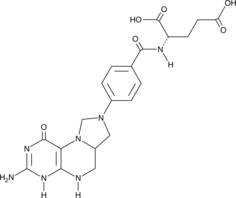
Folitixorin
A reduced form of folate and cofactor for thymidylate synthetase
-
GC14885
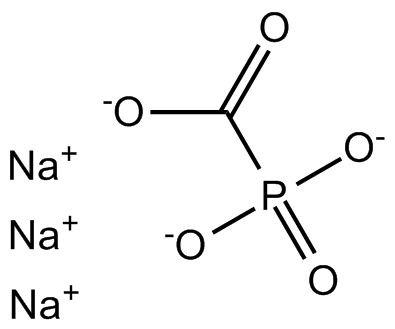
Foscarnet Sodium
Reverse transcriptase inhibitor
-
GC33094
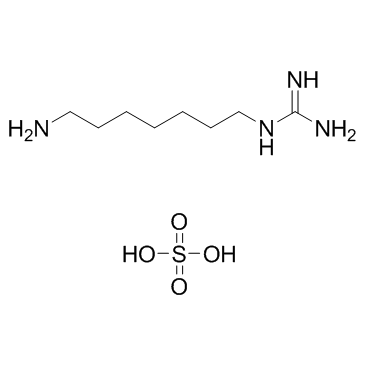
GC7 Sulfate
GC7 Sulfate is a deoxyhypusine synthase (DHPS) inhibitor.
-
GC16805
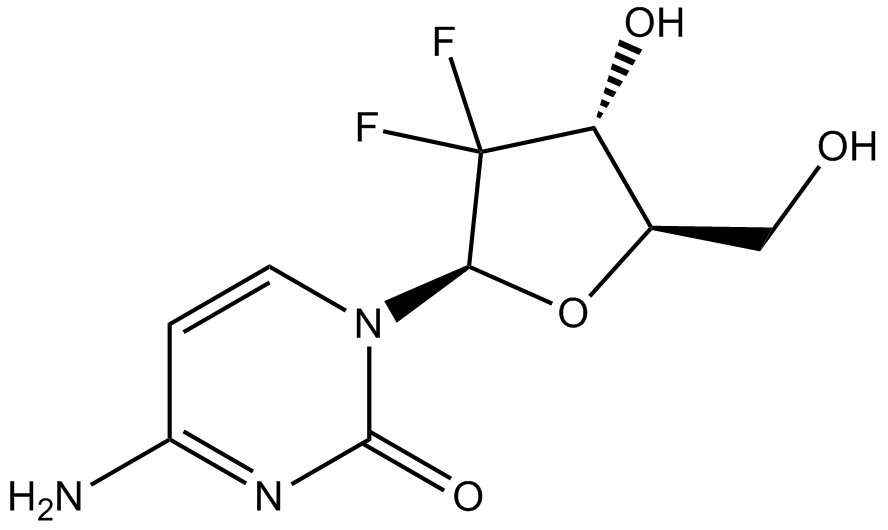
Gemcitabine
An inhibitor of DNA synthesis
-
GC14447
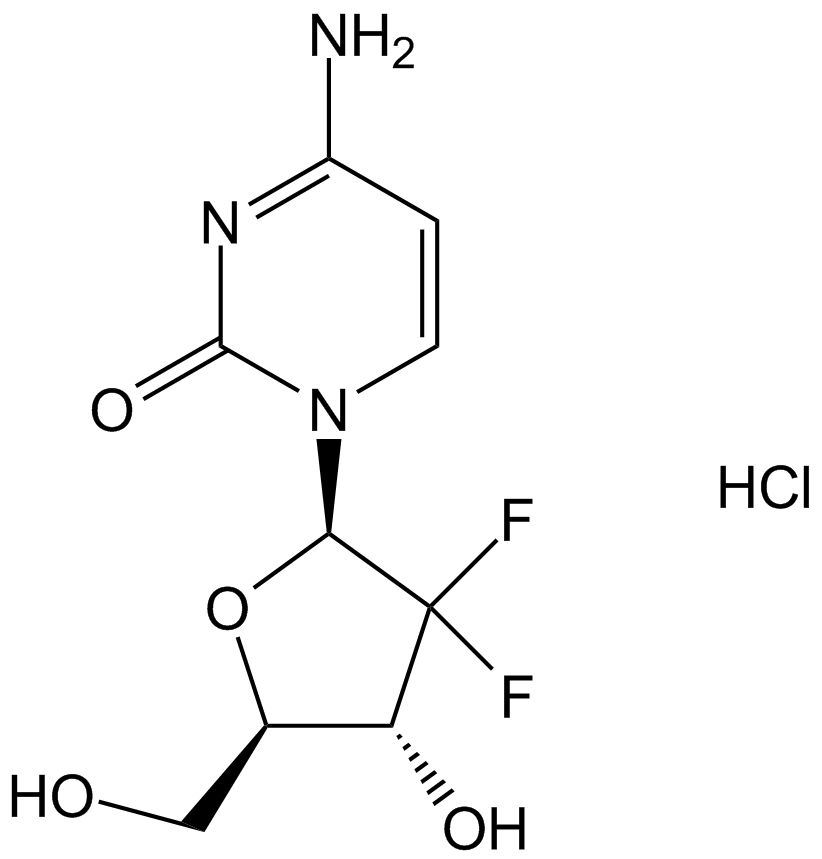
Gemcitabine HCl
Gemcitabine Hydrochloride (LY 188011 Hydrochloride) is a pyrimidine nucleoside analog antimetabolite and an antineoplastic agent. Gemcitabine Hydrochloride inhibits DNA synthesis and repair, resulting in autophagyand apoptosis.
-
GC18840
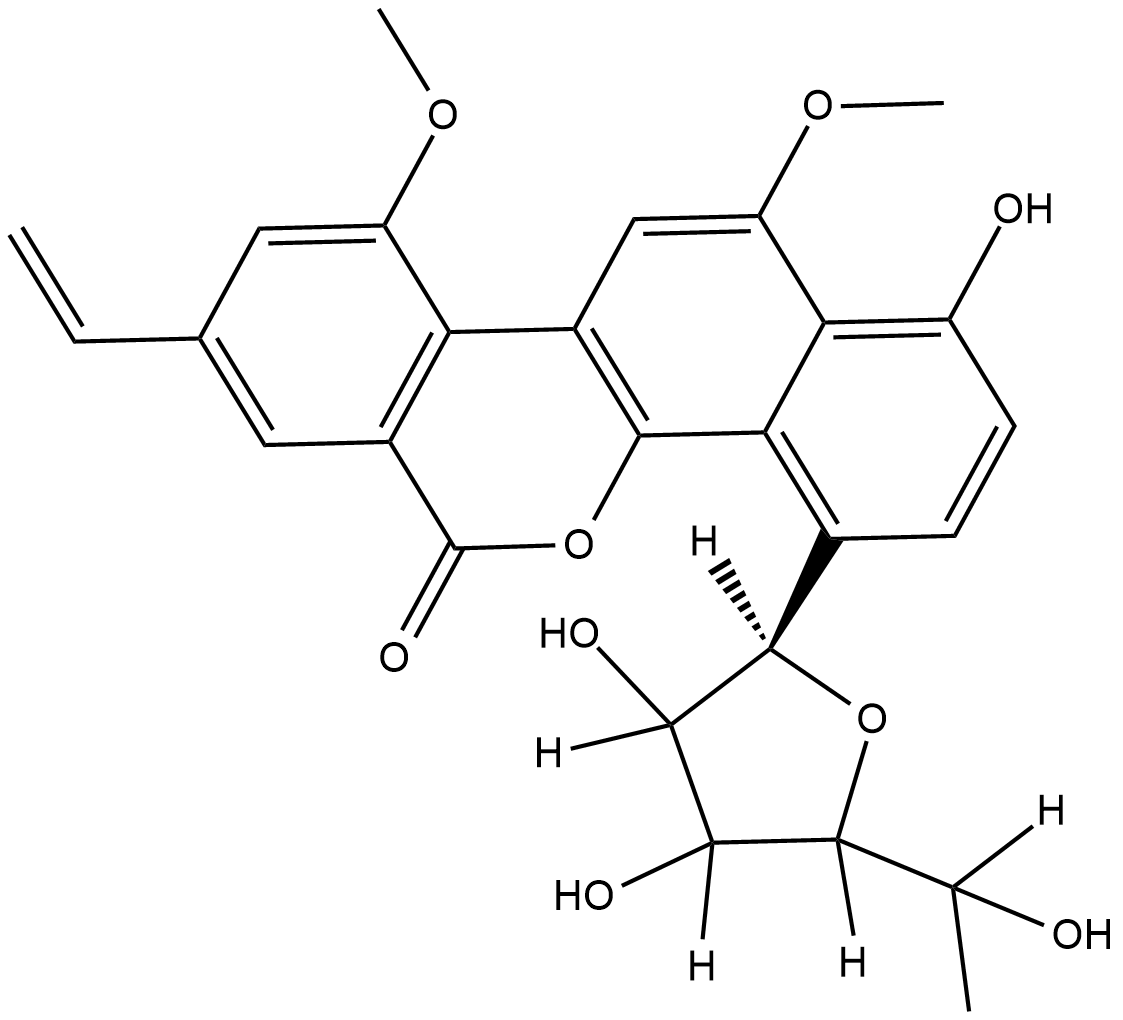
Gilvocarcin V
Gilvocarcin V is an antitumor antibiotic with a coumarin-based aromatic structure that was orignally isolated from the culture broth of S.
-
GC30529
GNE-371
GNE-371 is a potent and selective chemical probe for the second bromodomains of human transcription-initiation-factor TFIID subunit 1 and transcription-initiation-factor TFIID subunit 1-like, with an IC50 of 10 nM for TAF1(2).
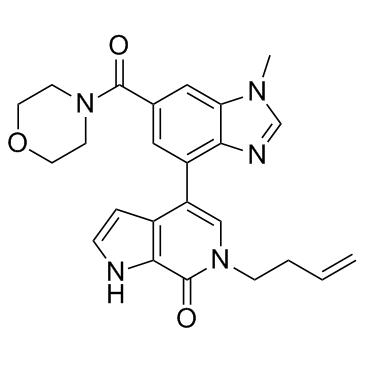
-
GC16949
Guanine
A purine base
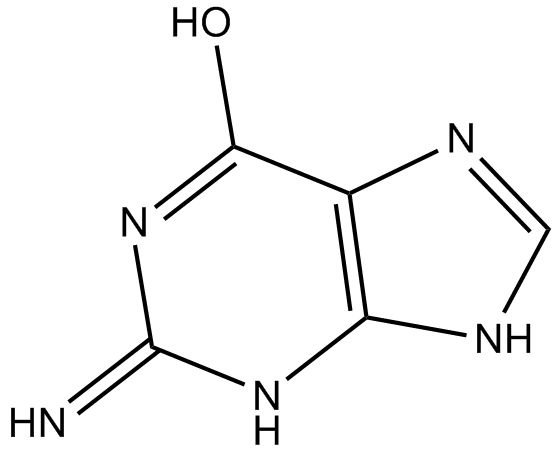
-
GC49033
Guanosine 5’-diphosphate (sodium salt hydrate)
A purine nucleotide
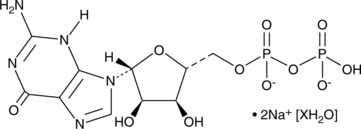
-
GC43798
Guanylyl Imidodiphosphate (lithium salt)
Guanylyl imidodiphosphate (Gpp(NH)p) lithium, a non-hydrolyzable GTP analogue, increases adenylate cyclase activity.
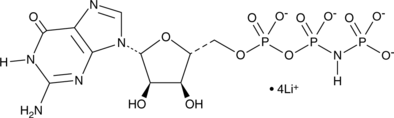
-
GC31650
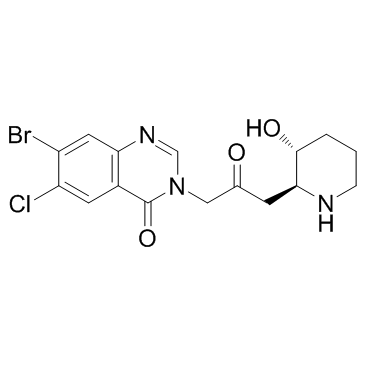
Halofuginone (RU-19110)
Halofuginone (RU-19110) (RU-19110), a Febrifugine derivative, is a competitive prolyl-tRNA synthetase inhibitor with a Ki of 18.3 nM.
-
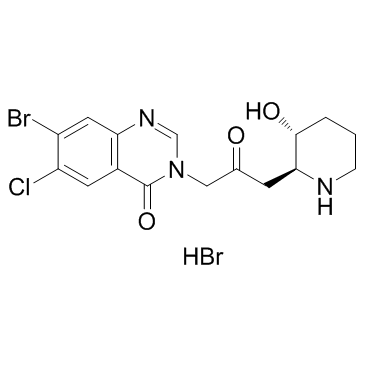
GC31950
Halofuginone hydrobromide (RU-19110 (hydrobromide))
Halofuginone (RU-19110) hydrobromid, a Febrifugine derivative, is a competitive prolyl-tRNA synthetase inhibitor with a Ki of 18.3 nM.
-
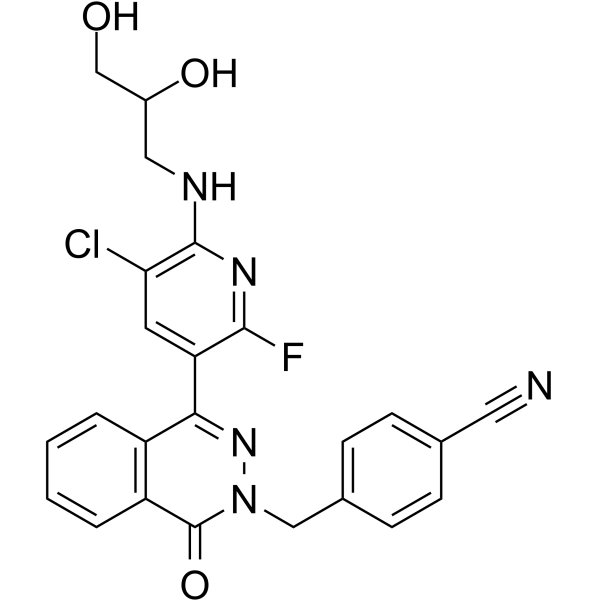
GC62328
HBV-IN-4
HBV-IN-4, a phthalazinone derivative, is a potent and orally active HBV DNA replication inhibitor with an IC50 of 14 nM.
-
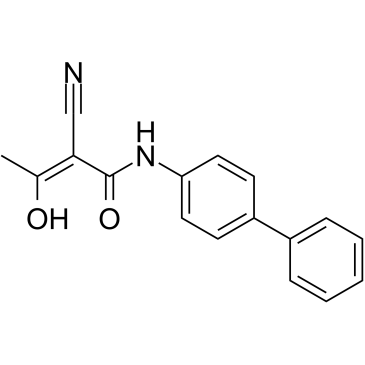
GC39279
hDHODH-IN-1
hDHODH-IN-1 is a human dihydroorotate dehydrogenase (hDHODH) inhibitor.
-
GC48388
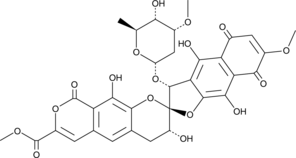
Heliquinomycin
A bacterial metabolite with diverse biological activities
-
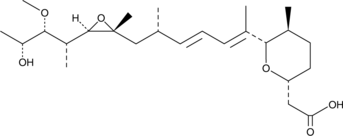
GC40103
Herboxidiene
Herboxidiene, as a potent antitumor agent, can target the SF3B subunit of the spliceosome. Herboxidiene also induces both G1 and G2/M cell cycle arrest in a human normal fibroblast cell line WI-38.
-
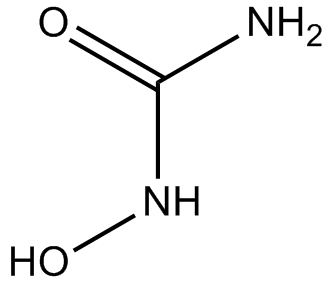
GC16843
Hydroxyurea
DNA synthesis inhibitor
-
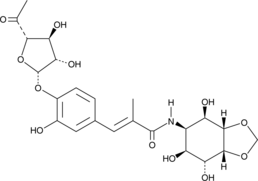
GC49694
Hygromycin A
An antibiotic
-
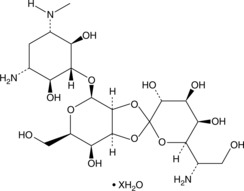
GC48445
Hygromycin B (hydrate)
An aminoglycoside antibiotic
-
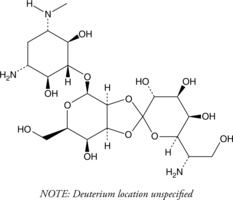
GC49267
Hygromycin B-d4
An internal standard for the quantification of hygromycin
-
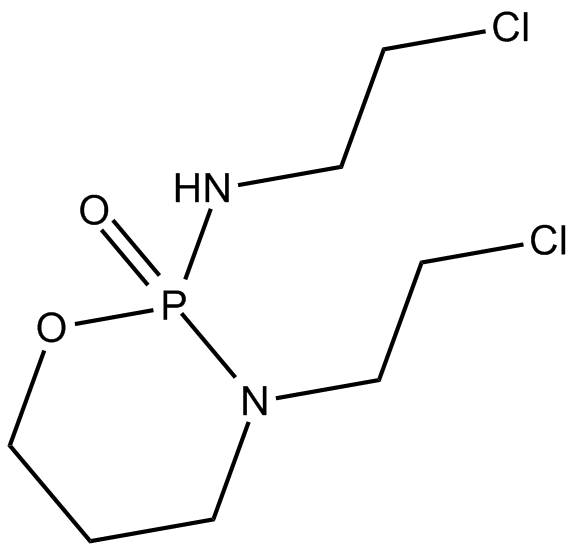
GC16003
Ifosfamide
Cytostatic agent
-
GC62137
IMT1
IMT1 is a first-in-class specific and noncompetitive human mitochondrial RNA polymerase (POLRMT) inhibitor.
-
GC39140
Isopimpinellin
Isopimpinellin, an orally active compound isolated from the roots of Pimpinella saxifrage.
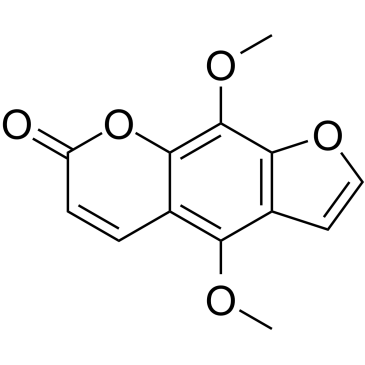
-
GC10834
Isoxanthopterin
interferes with RNA and DNA synthesis
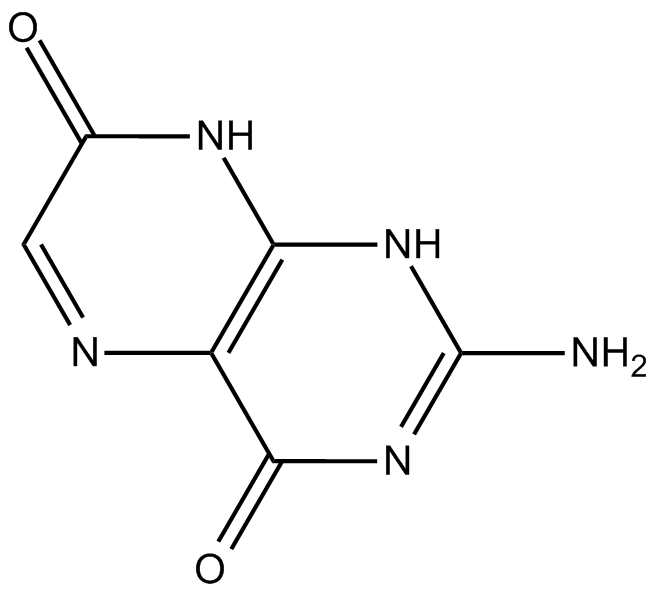
-
GC36367
JH-RE-06
JH-RE-06, a potent REV1-REV7 interface inhibitor (IC50=0.78 μM; Kd=0.42 μM), targets REV1 that interacts with the REV7 subunit of POLζ. JH-RE-06 disrupts mutagenic translesion synthesis (TLS) by preventing recruitment of mutagenic POLζ. JH-RE-06 improves chemotherapy.
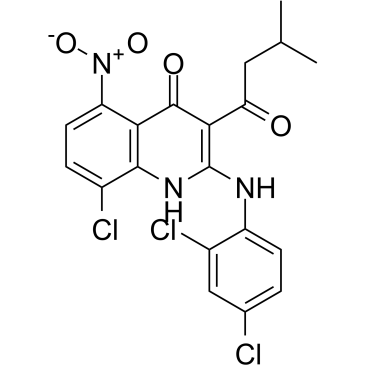
-
GC50117
JTE 607 dihydrochloride
JTE 607 dihydrochloride, a highly selective inflammatory cytokine synthesis inhibitor, protects from endotoxin shock in mice.
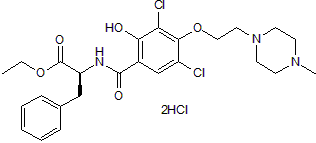
-
GC47521
K22
An antiviral agent
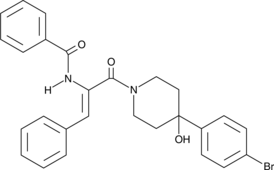
-
GC49381
L-Leucine-d10
An internal standard for the quantification of L-leucine
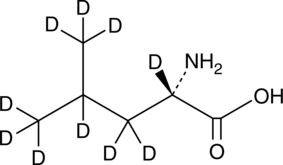
-
GC49368
L-Methionine-d3
An internal standard for the quantification of L-methionine
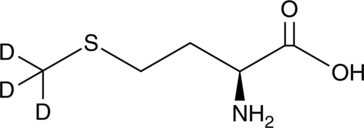
-
GC49384
L-Proline-d3
An internal standard for the quantification of L-proline
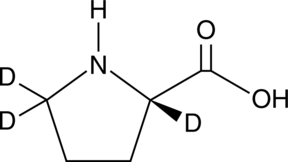
-
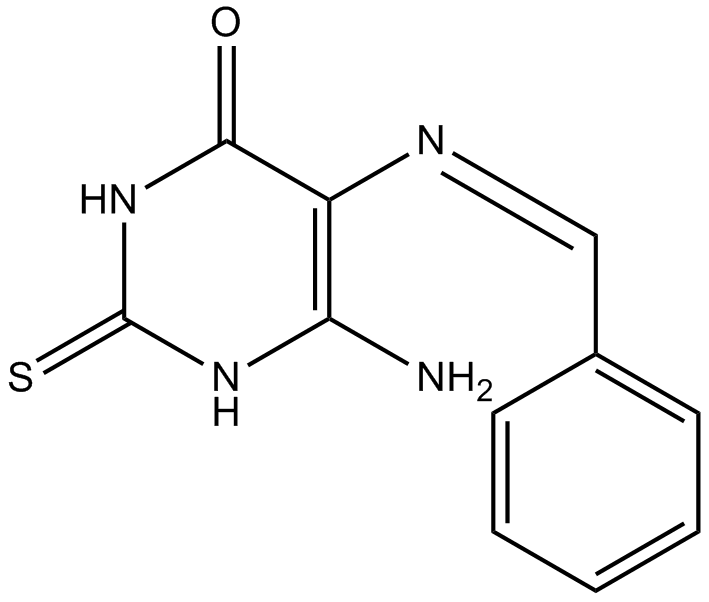
GC12131
L189
inhibitor of human DNA ligases I, III and IV
-
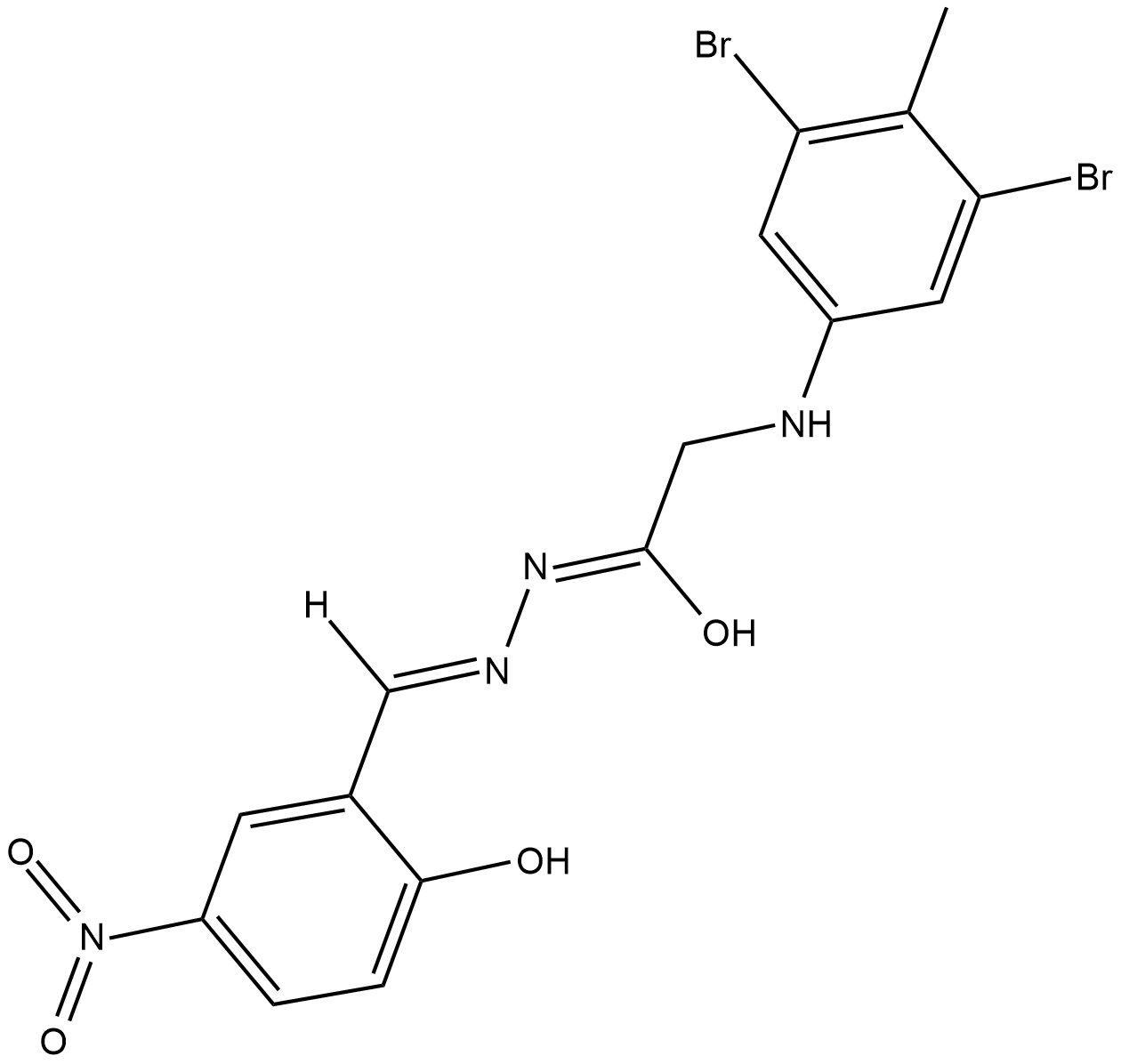
GC18186
L67
L67 is a competitive inhibitor of DNA ligases I and III (IC50s = 10 and 10 uM for human DNA ligase I and human ligase IIIβ).
-
GC67785
L82

-
GC68436
L82-G17

-
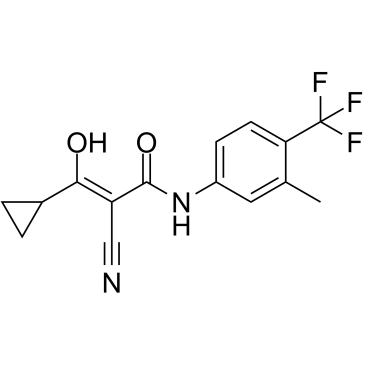
GC39632
Laflunimus
Laflunimus (HR325) is an immunosuppressive agent and an analogue of the Leflunomide-active metabolite A77 1726.
-
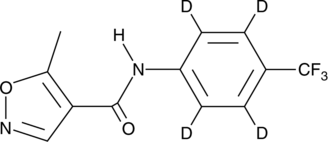
GC47552
Leflunomide-d4
An internal standard for the quantification of leflunomide
-
GC46166
Leoidin
A depsidone
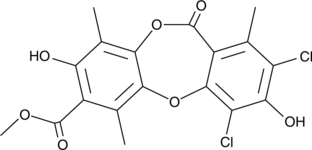
-
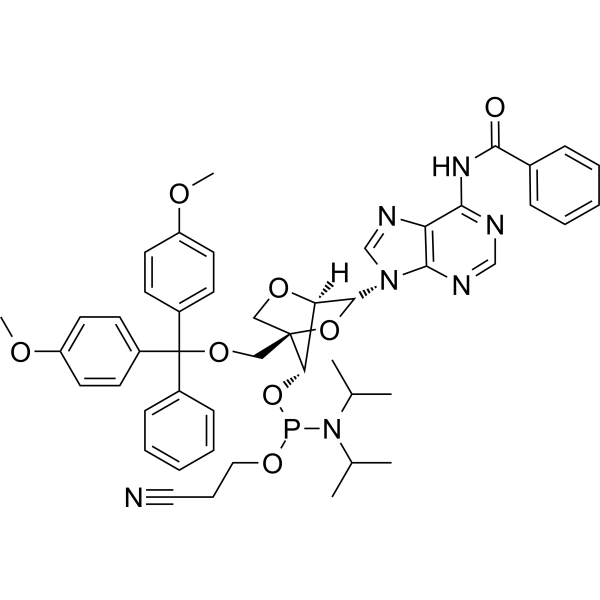
GC66659
LNA-A(Bz) amidite
LNA-A(Bz) amidite can be used for synthesis of ASOs (antisense oligonucleotides).
-
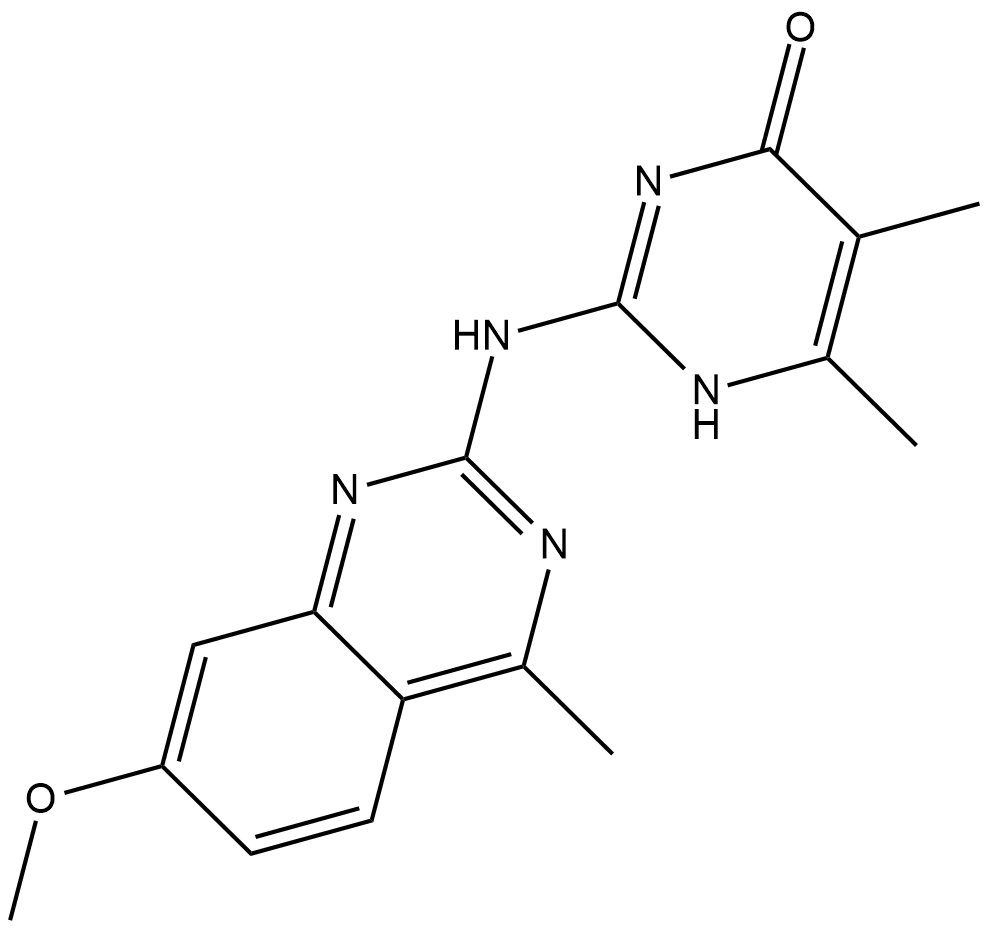
GC19238
Madrasin
Madrasin is a potent and cell penetrant splicing inhibitor that interferes with the early stages of spliceosome assembly.
-
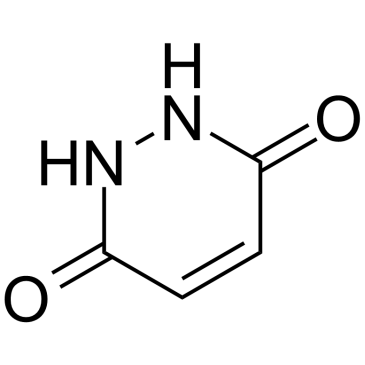
GC60234
Maleic hydrazide
Maleic hydrazide is extensively used as a systemic plant growth regulator and as a herbicide. Maleic hydrazide acts as an inhibitor of the synthesis of nucleic acids and proteins.
-
GC33411
MB-7133
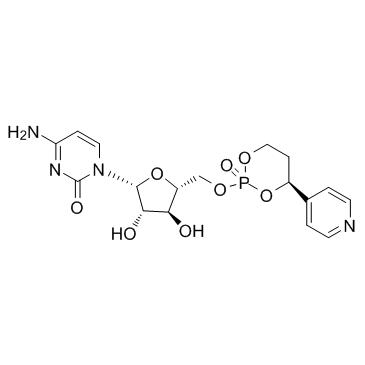
-
GC13704
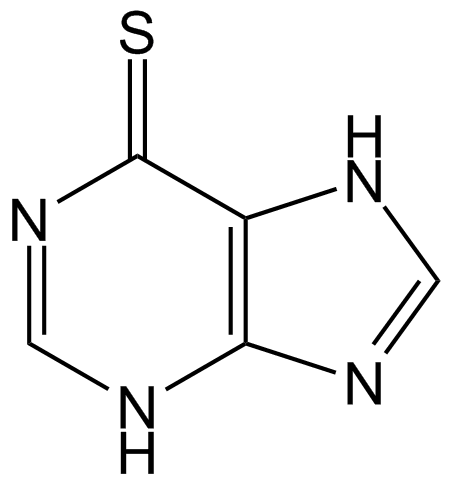
Mercaptopurine (6-MP)
Mercaptopurine (6-MP) is a purine analogue which acts as an antagonist of the endogenous purines and has been widely used as antileukemic agent and immunosuppressive drug.
-
GC38201
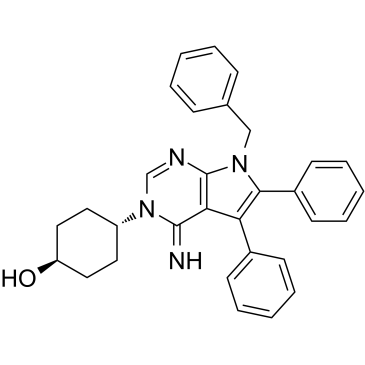
Metarrestin
Metarrestin (ML246) is an orally active, first-in-class and specific perinucleolar compartment inhibitor. Metarrestin disrupts the nucleolar structure and inhibits RNA polymerase (Pol) I transcription, at least in part by interacting with the translation elongation factor eEF1A2. Metarrestin blocks metastatic development and extends survival in mouse cancer models.
-
GC61047
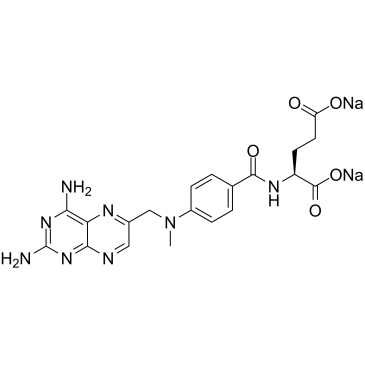
Methotrexate disodium
Methotrexate (Amethopterin) disodium, an antimetabolite and antifolate agent, inhibits the enzyme dihydrofolate reductase, thereby preventing the conversion of folic acid into tetrahydrofolate, and inhibiting DNA synthesis.
-
GC52165
Minosaminomycin
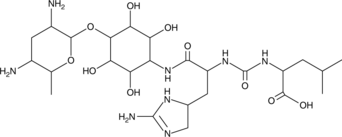
-
GC15060
Mithramycin A
A DNA-binding antitumor antibiotic
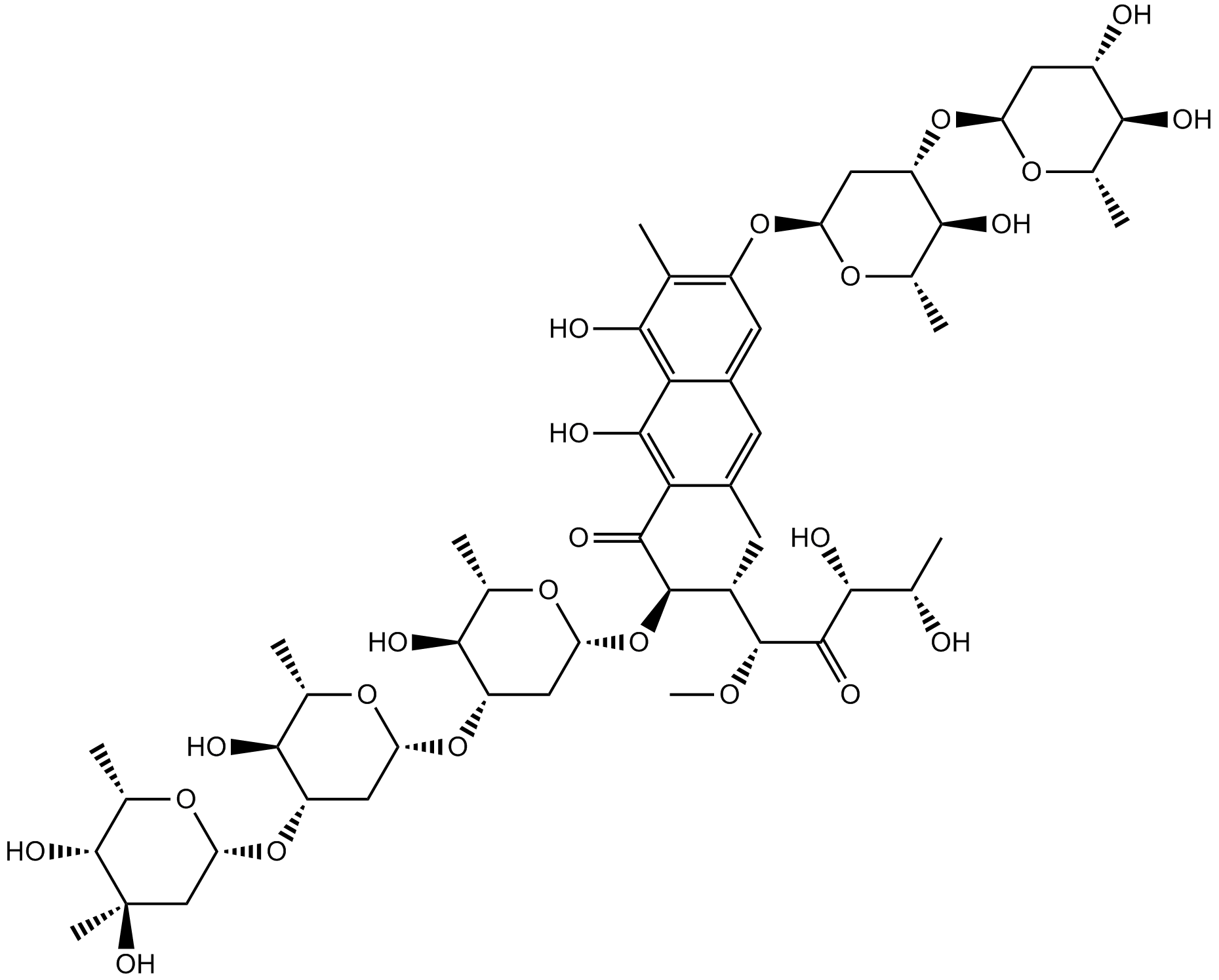
-
GC12353
Mitomycin C
Mitomycin C is an antibiotic isolated from Streptomyces Caespitosus or Streptomyces Lavendulae. Mitomycin C inhibits DNA synthesis by forming covalent mitomycin C-DNA adducts with DNA, with an EC50 value of 0.14 μM in PC3 cells.
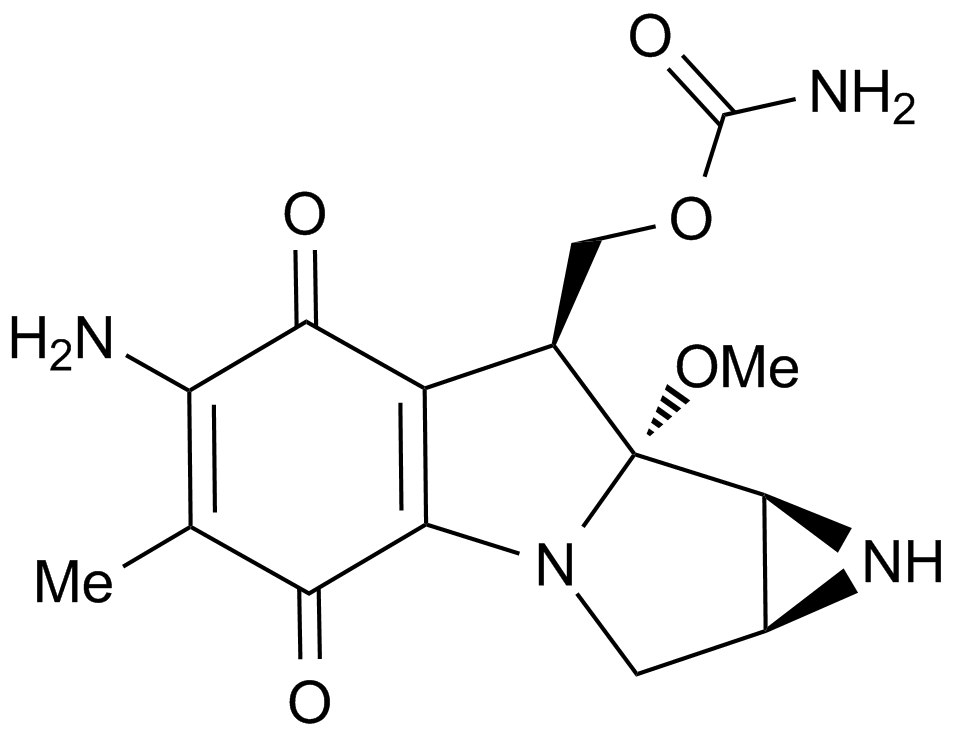
-
GC67716
Mitonafide
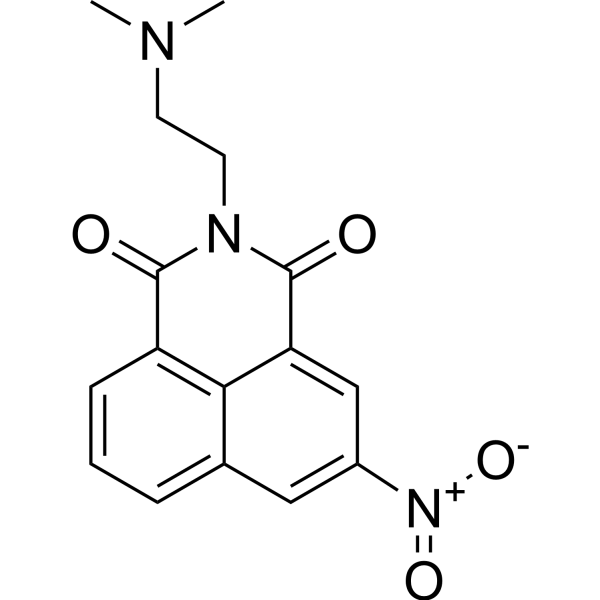
-
GC12786
ML216
BLM helicase inhibitor
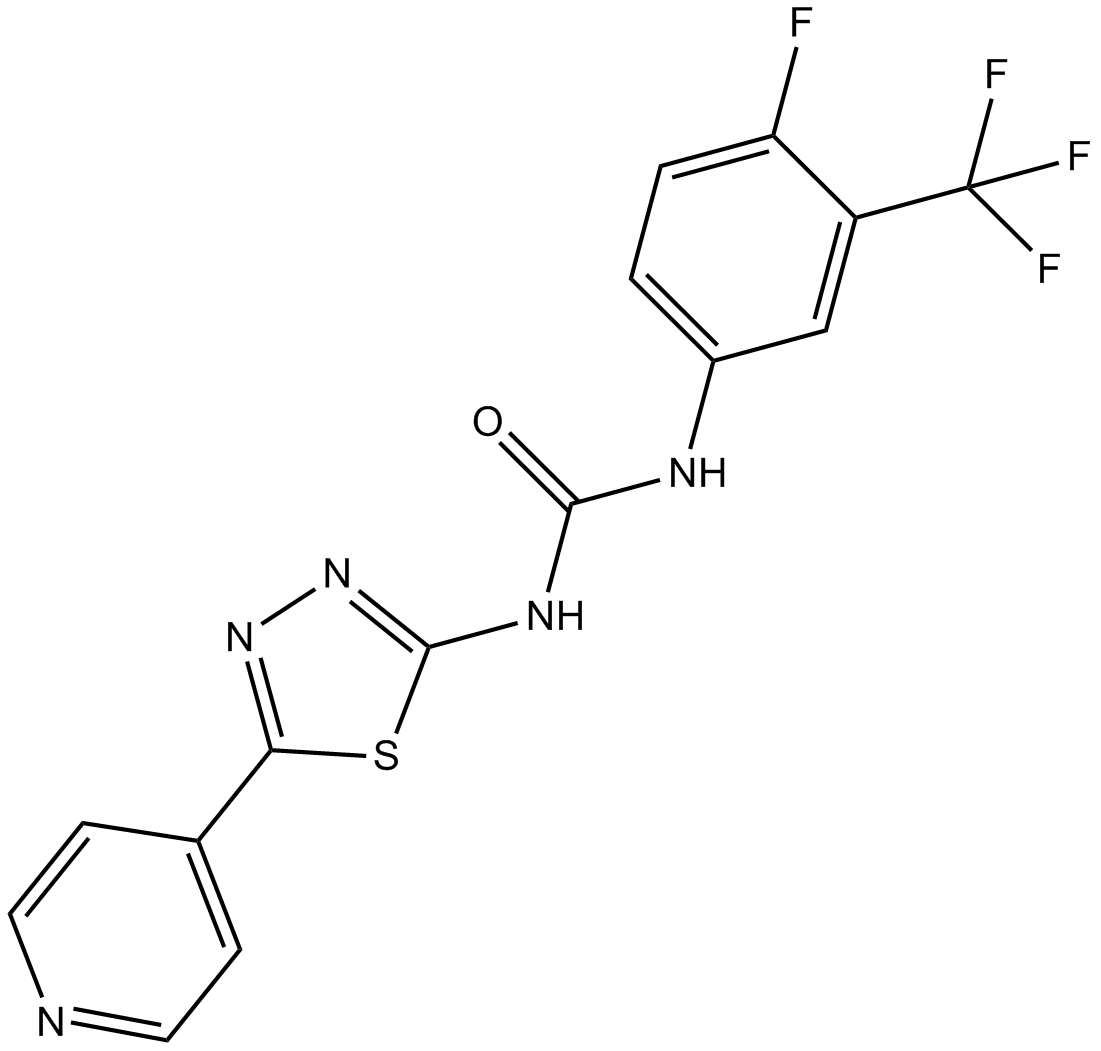
-
GC64666
ML372
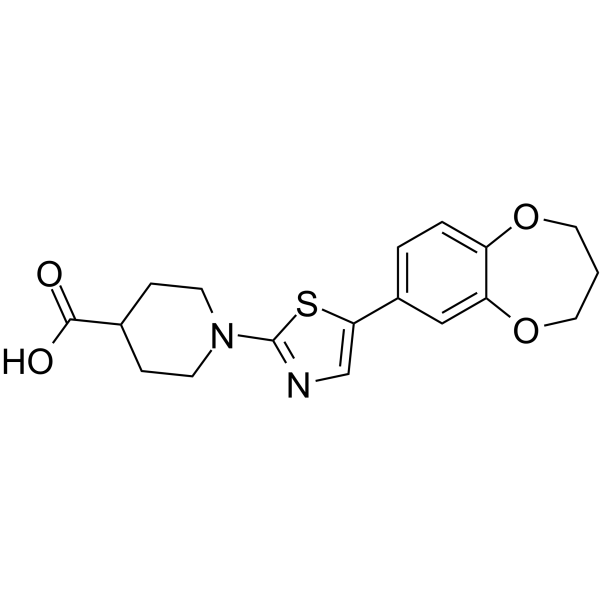
-
GC15238
Mupirocin
isoleucyl t-RNA synthetase inhibitor
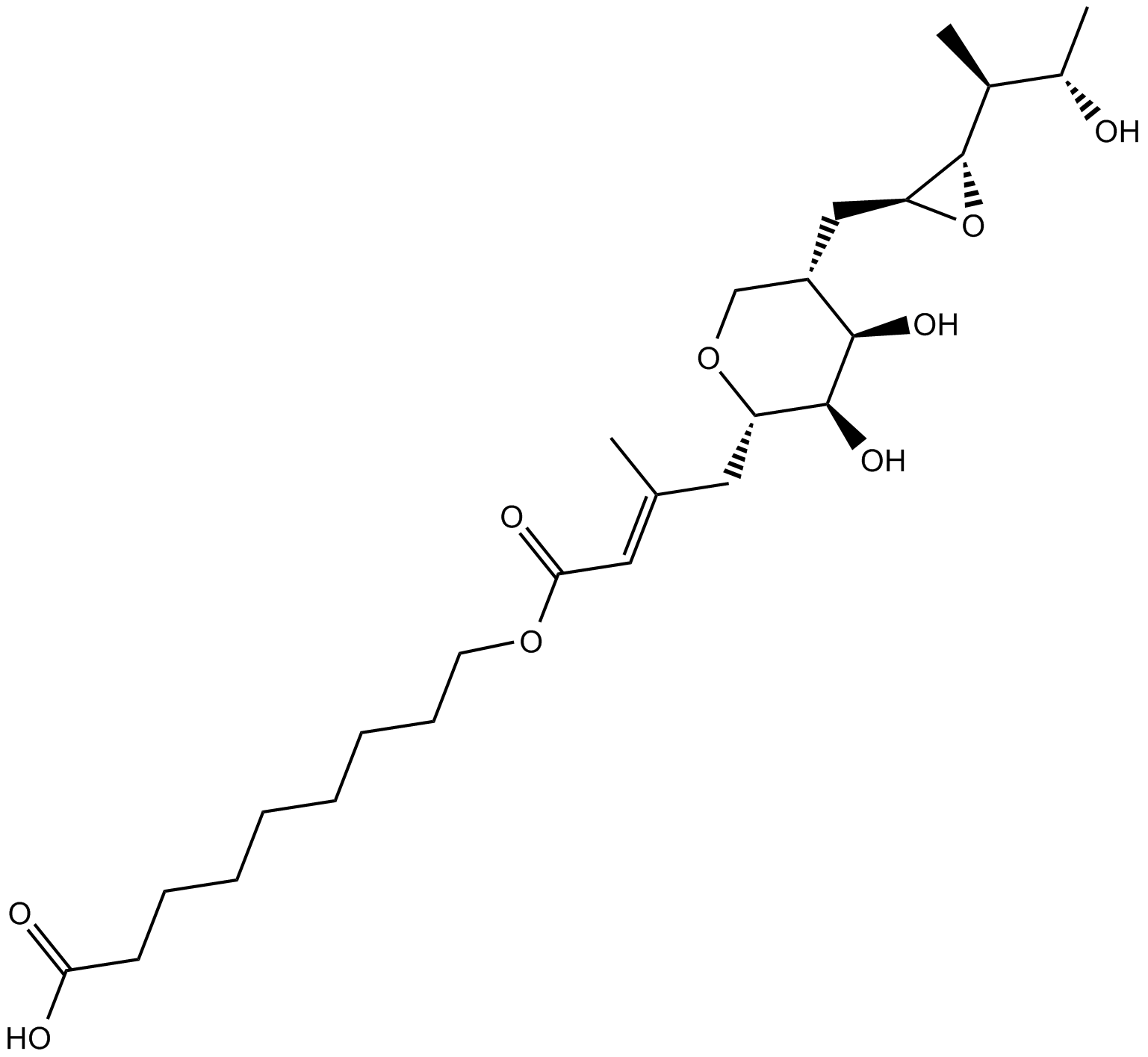
-
GC12423
Mycophenolate mofetil hydrochloride
Immunosuppresant drug
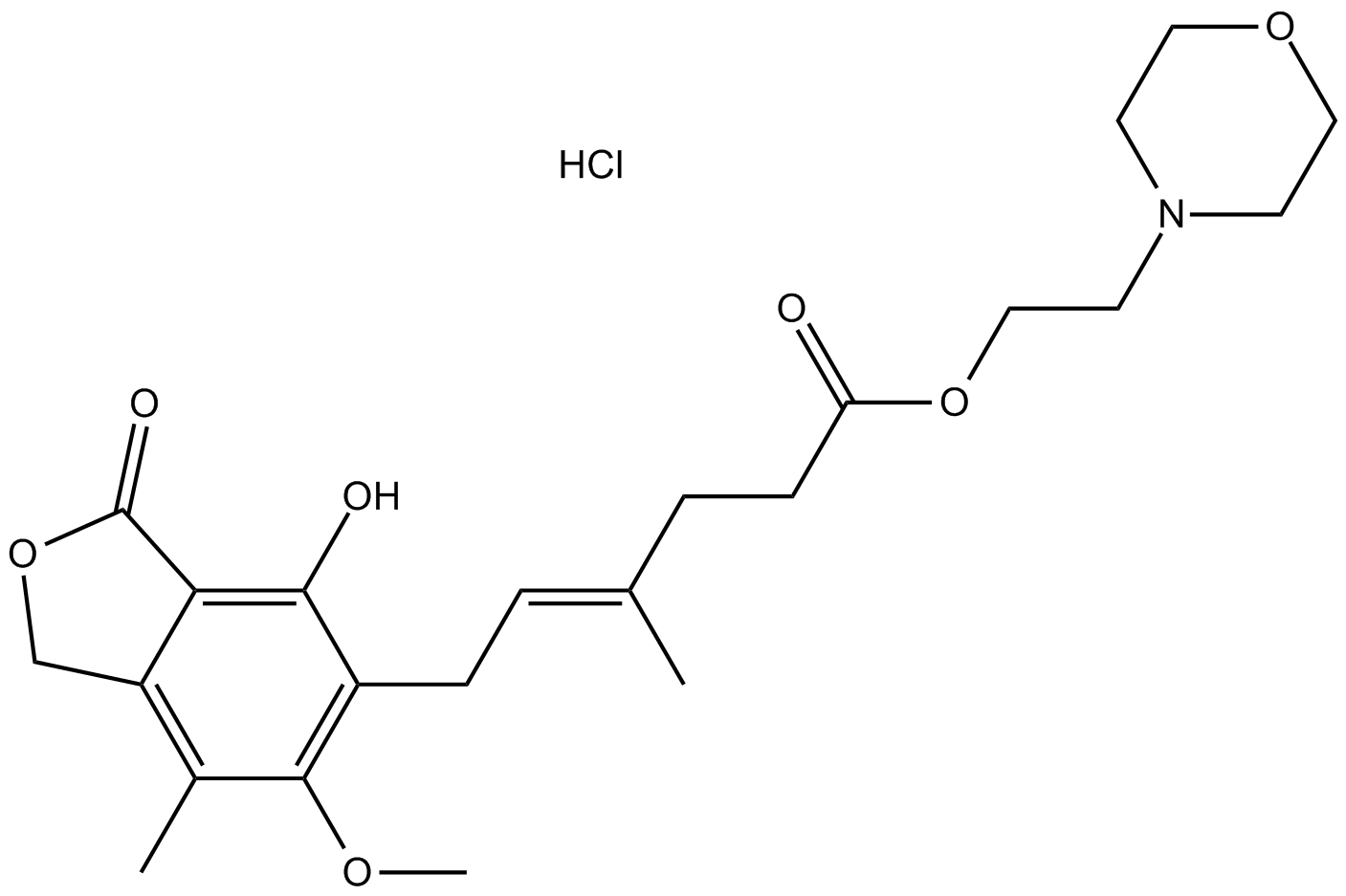
-
GC40683
N-(2-hydroxyethyl)-Naphthalimide
N-(2-hydroxyethyl)-Naphthalimide is an N-substituted 1,8-naphthalimide used as a fluorescent probe and as a precursor for protection of amine groups.
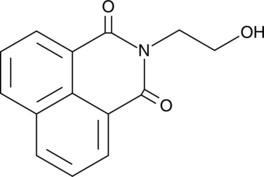
-
GC19566
N-Nitrosodiethylamine (950mg/mL)
N-Nitrosodiethylamine (Diethylnitrosamine) is a potent hepatocarcinogenic dialkylnitrosoamine. N-Nitrosodiethylamine is mainly present in tobacco smoke, water, cheddar cheese, cured, fried meals and many alcoholic beverages. N-Nitrosodiethylamine is responsible for the changes in the nuclear enzymes associated with DNA repair/replication. N-Nitrosodiethylamine results in various tumors in all animal species. The main target organs are the nasal cavity, trachea, lung, esophagus and liver.
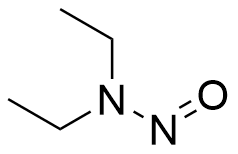
-
GC69577
N-Nitrosodiethylamine-d10
N-Nitrosodiethylamine-d10 is the deuterated form of N-Nitrosodiethylamine. N-Nitrosodiethylamine (Diethylnitrosamine) is a potent carcinogen belonging to the group of nitrosamines. It is mainly found in tobacco smoke, water, cheddar cheese, pickled foods, fried foods and many alcoholic beverages. N-Nitrosodiethylamine is responsible for changes in nucleases involved in DNA repair/replication. It can cause various tumors in all animals with major target organs being the nasal cavity, trachea, lungs, esophagus and liver.
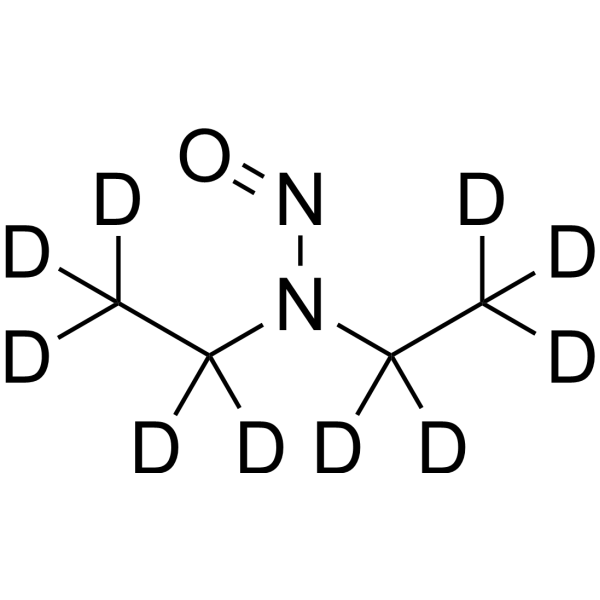
-
GC66325
N2-Acetylguanine
N2-Acetylguanine is a C2-modified guanine. N2-Acetylguanine binds GR (guanine-guanine riboswitch) with an Kd value of 300 nM. N2-Acetylguanine modulate transcriptional termination. N2-Acetylguanine has the potential for the research of antimicrobial agent.
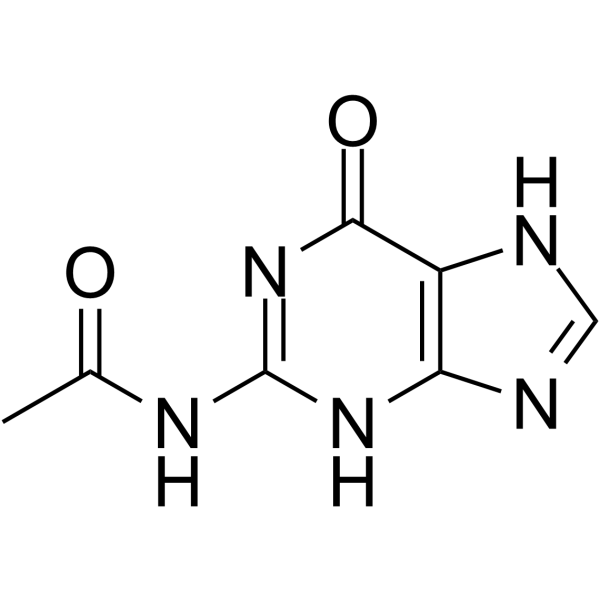
-
GC65561
N6-Methyl-dA phosphoramidite
N6-Methyl-dA phosphoramidite can be used in the synthesis of oligodeoxyribonucleotides.
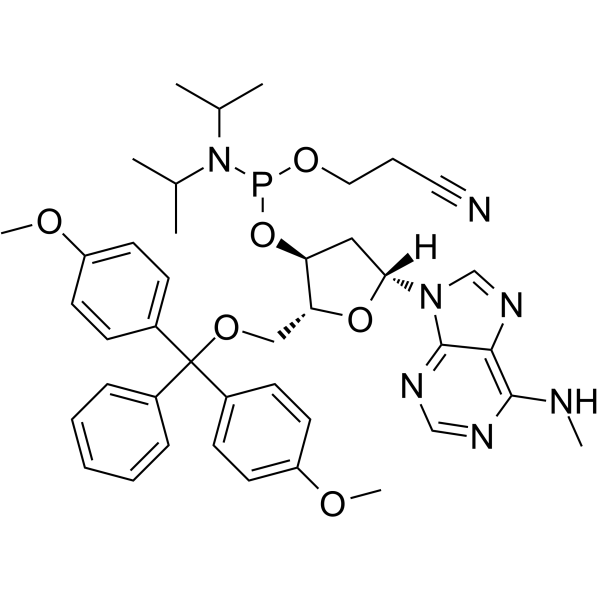
-
GC63653
NCGC00029283
NCGC00029283 is a werner syndrome helicase-nuclease (WRN) helicase inhibitor with IC50s of 2.3 μM, 12.5 μM, and 3.4 μM for WRN, BLM and FANCJ helicase, respectively.
-
GC15786
Nedaplatin
DNA synthesis inhibitor
-
GC10591
Nelarabine
Prodrug of ara-G for T-LBL/T-ALL
-
GC64493
Neocarzinostatin
Neocarzinostatin, a potent DNA-damaging, anti-tumor antibiotic, recognizes double-stranded DNA bulge and induces DNA double strand breaks (DSBs).

-
GC60268
Neoxanthin
Neoxanthin is a major xanthophyll carotenoid and a precursor of the plant hormone abscisic acid in dark green leafy vegetables. Neoxanthin is a potent antioxidant and light-harvesting pigment. Neoxanthin induces apoptosis and has anticancer actions.
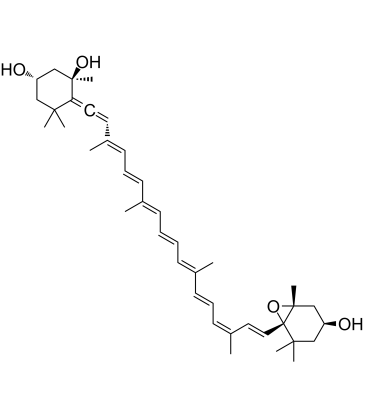
-
GC36743
Nimustine hydrochloride
Nimustine hydrochloride (ACNU) is a DNA cross-linking and DNA alkylating agent, which induces DNA replication blocking lesions and DNA double-strand breaks and inhibits DNA synthesis, commonly used in chemotherapy for glioblastomas.
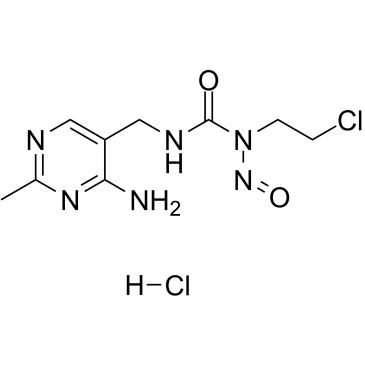
-
GC64559
NITD-2
NITD-2, a dengue virus (DENV) polymerase inhibitor, inhibits the DENV RdRp-mediated RNA elongation.
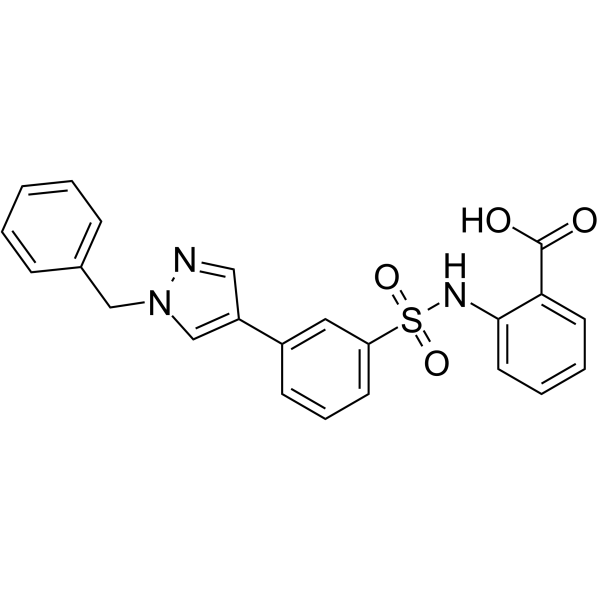
-
GC19506
NITD008
NITD008 is a potent and selective flaviviruse inhibitor which can inhibit Dengue Virus Type 2 (DENV-2) with an EC50 of 0.64 μM.
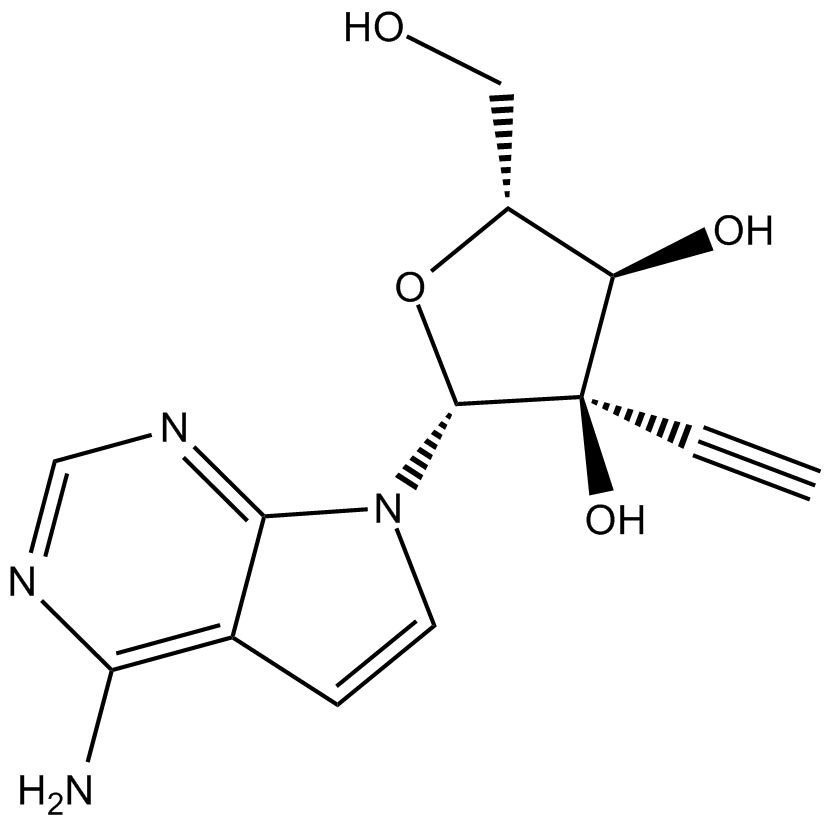
-
GC19263
NKP-1339
NKP-1339(IT-139) is a ruthenium(iii) coordination anticancer compound based on target to transferrin.
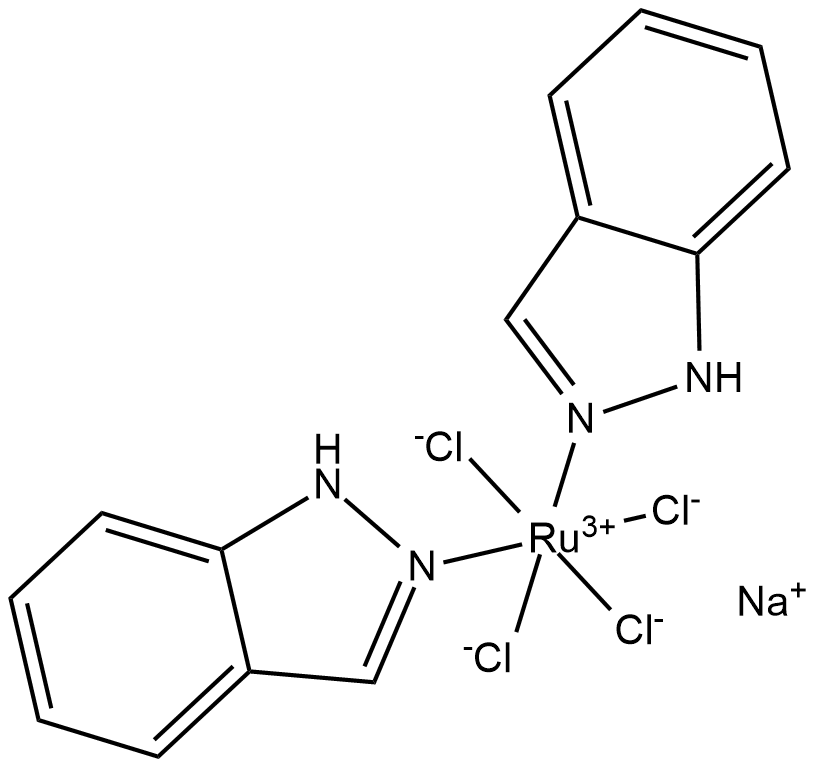
-
GC62679
NSAH
NSAH is a reversible and competitive nonnucleoside ribonucleotide reductase (RR) inhibitor, with cell-free IC50 of 32 μM and cell-based IC50 of ~250 nM, respectively.
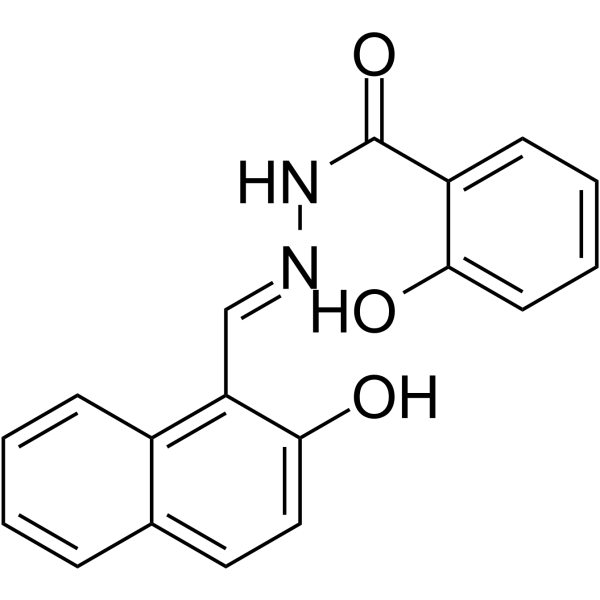
-
GC69598
NSC 80467
NSC 80467 is a DNA damaging agent that selectively inhibits survivin. NSC 80467 preferentially inhibits DNA synthesis and induces two markers of DNA damage, γH2AX and pKAP1.
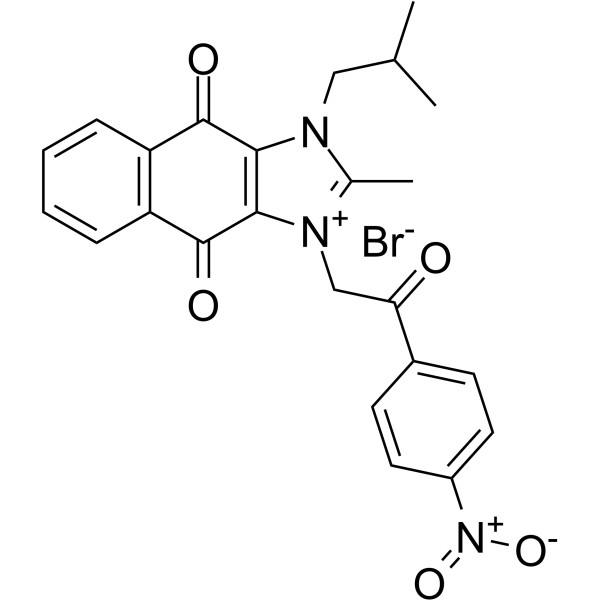
-
GC64103
NSC639828
NSC639828 is a potent inhibitor of DNA polymerase α with an IC50 of 70 μM. NSC639828 has high antitumor activity. NSC639828 has the potential for researching cancer disease.
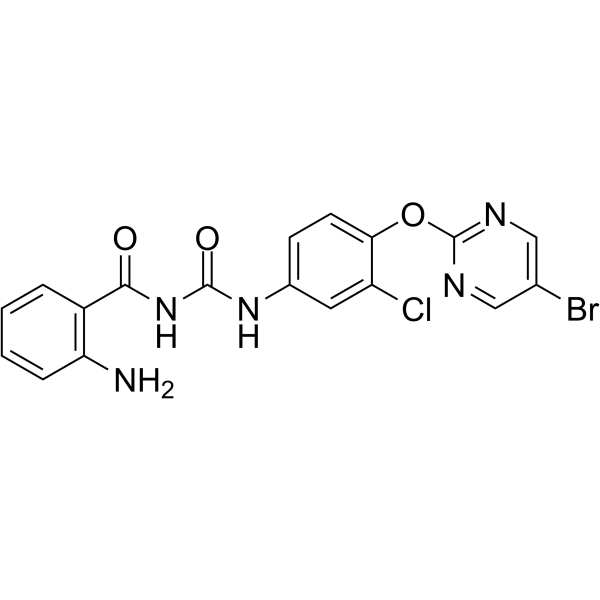
-
GC64293
Nusinersen
Nusinersen is an antisense oligonucleotide drug that modifies pre–messenger RNA splicing of the SMN2 gene and thus promotes increased production of full-length SMN protein.

-
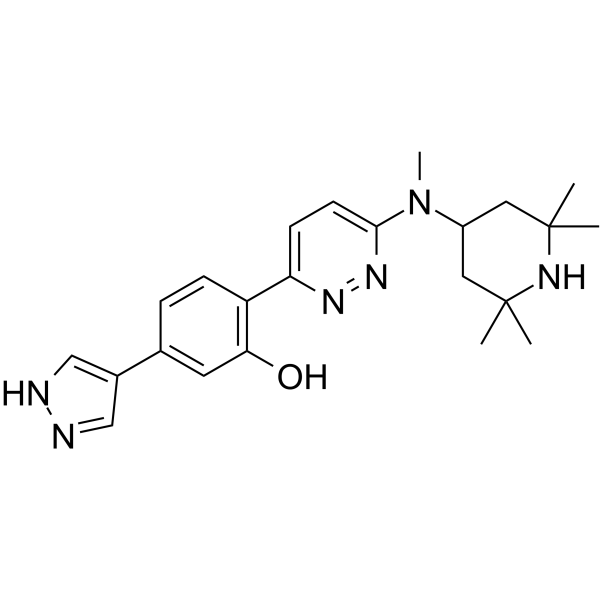
GC63123
NVS-SM2
NVS-SM2 is a potent, orally active and brain-penetrant SMN2 splicing enhancer with an EC50 of 2 nM for SMN.
-
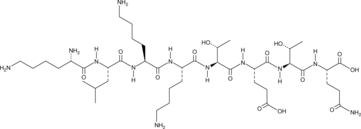
GC49743
Octapeptide-2
A thymosin β4-derived peptide
-
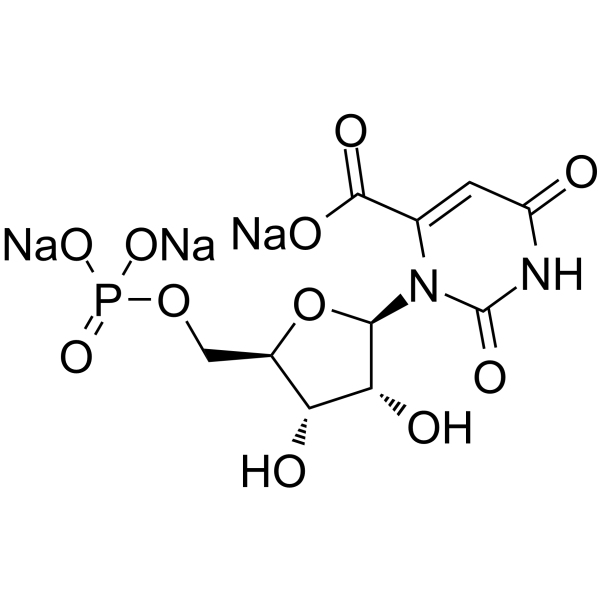
GC64296
Orotidine 5′-monophosphate trisodium
Orotidine 5'-monophosphate trisodium is a pyrimidine nucleotide.
-
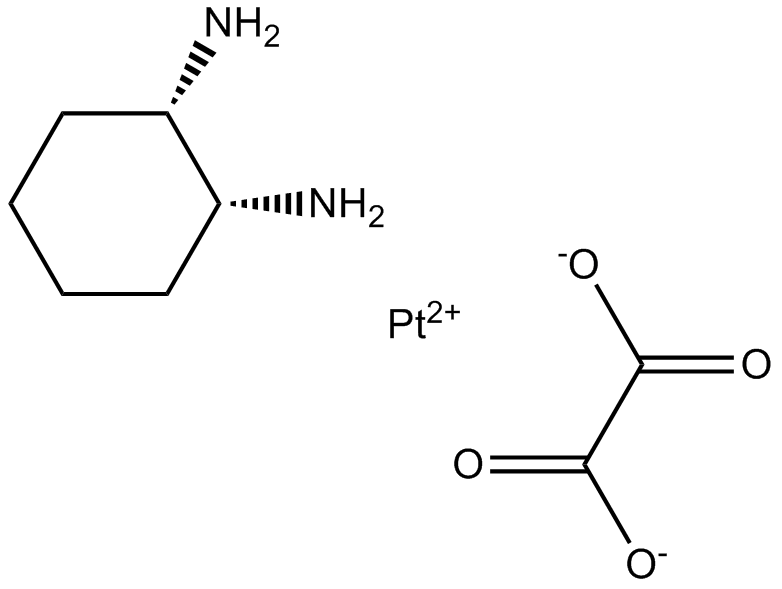
GC17716
Oxaliplatin
Oxaliplatin is a cytotoxic chemotherapy drug used to treat cancer.
-
GC49251
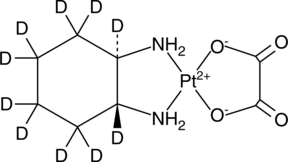
Oxaliplatin-d10
An internal standard for the quantification of oxaliplatin
-
GC14107
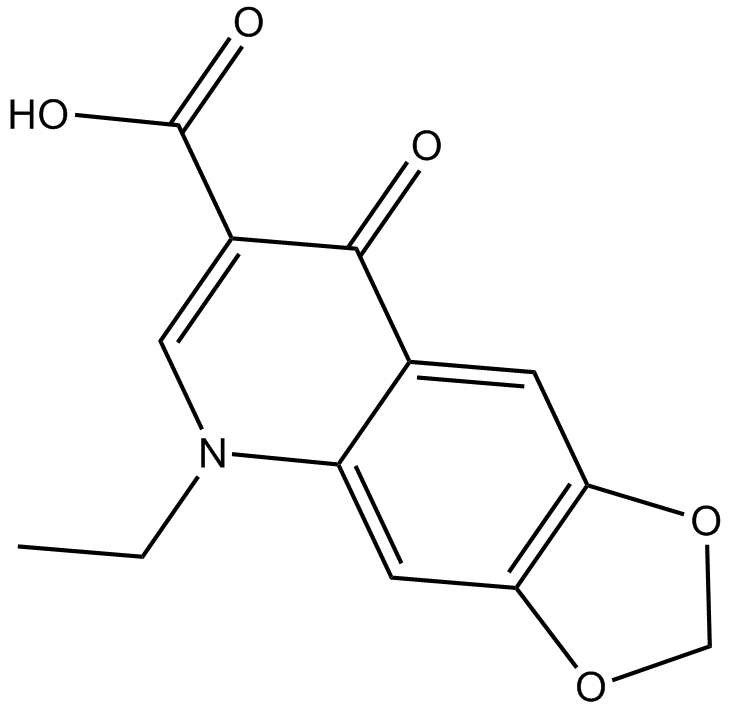
Oxolinic acid
quinolone antibiotic that inhibits bacterial DNA gyrase
-
GC17232
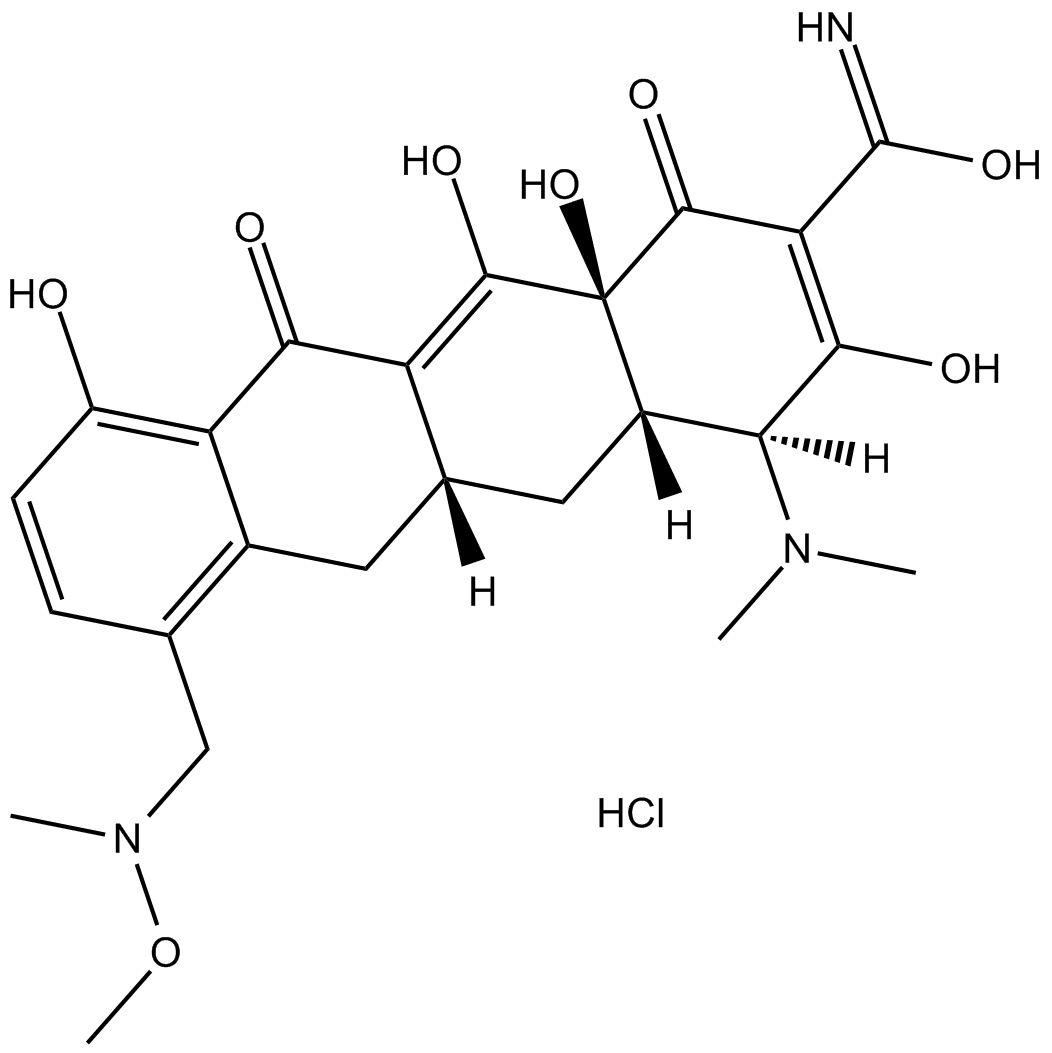
P005672 hydrochloride
P005672 hydrochloride is a narrow-spectrum tetracycline-class antibiotic.
-
GC14849
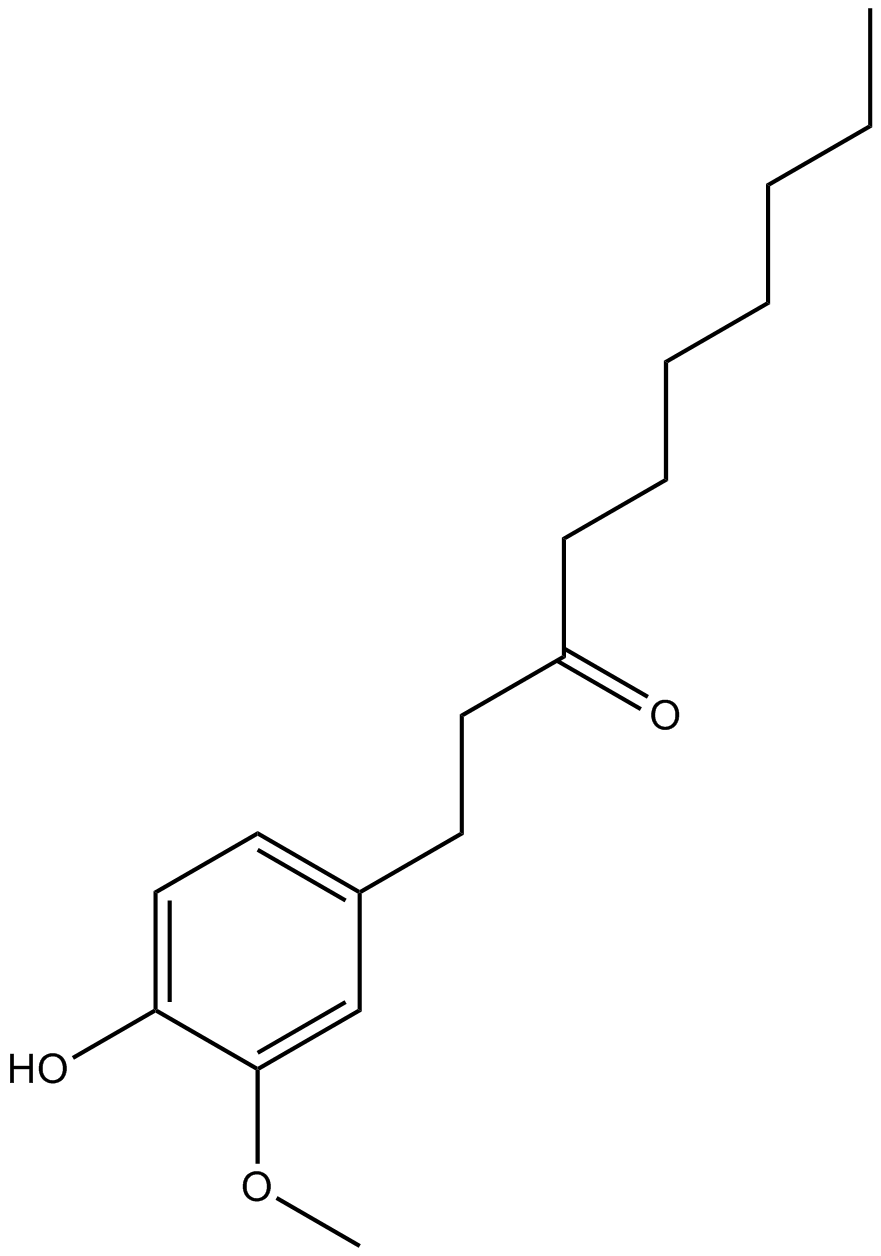
Paradol
A phenolic ketone with diverse biological activities
-
GC65891
PCLX-001
PCLX-001 is an orally acitve, small-molecule, dual N-myristoyltransferase (NMT) inhibitor, with IC50s of 5 nM (NMT1) and 8 nM (NMT2), respectively. PCLX-001 exhibits anti-tumor activity and inhibits early B-cell receptor (BCR) signaling, can be used to B-cell malignancies research.
-
GC44591
Pemetrexed (sodium salt hydrate)
Pemetrexed (sodium salt hydrate) is a novel antifolate, the Ki values of the pentaglutamate of LY231514 are 1.3, 7.2, and 65 nM for inhibits thymidylate synthase (TS), dihydrofolate reductase (DHFR), and glycinamide ribonucleotide formyltransferase (GARFT), respectively.
-
GC30695
Phen-DC3
Phen-DC3 is a G-quadruplex (G4) specific ligand which can inhibit FANCJ and DinG helicases with IC50s of 65±6 and 50±10 nM, respectively.
-
GC14066
Procaine
Sodium channel inhibitor
-
GC44685
Procaine (hydrochloride)
Procaine (hydrochloride) is an analytical reference standard categorized as a local anesthetic that is used as an adulterant.
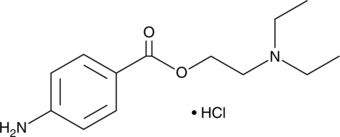
-
GC11793
Procarbazine HCl
Procarbazine HCl is an orally active alkylating agent, with anticancer activity. Procarbazine HCl can be used in Hodgkin's disease research.
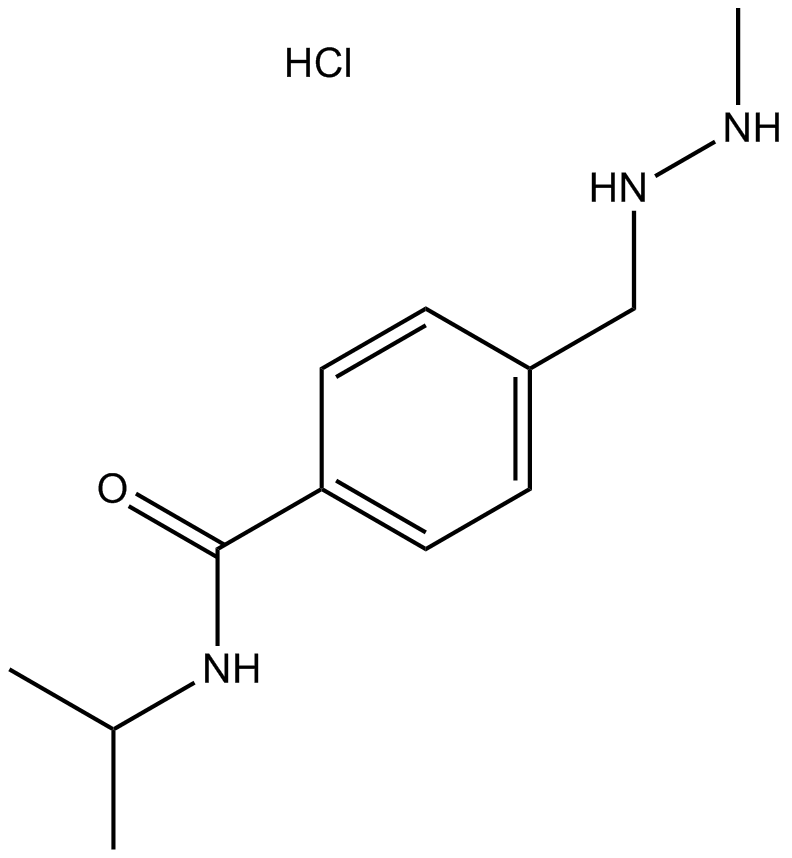
-
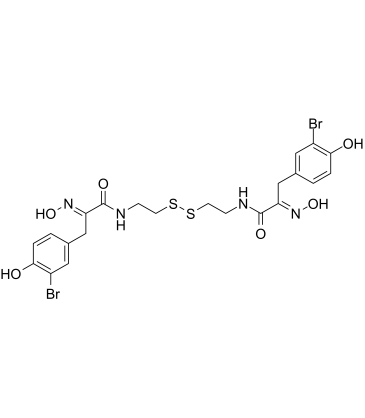
GC60310
Psammaplin A
Psammaplin A, a marine metabolite, is a potent inhibitor of HDAC and DNA methyltransferases.
-
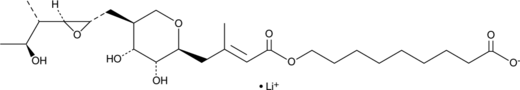
GC49053
Pseudomonic Acid (lithium salt)
An antibiotic
-
GC38466
Pseudouridimycin
Pseudouridimycin (PUM), an antibiotic, is a selective bacterial RNA polymerase (RNAP) inhibitor.