DNA/RNA Synthesis
RNA synthesis, which is also called DNA transcription, is a highly selective process. Transcription by RNA polymerase II extends beyond RNA synthesis, towards a more active role in mRNA maturation, surveillance and export to the cytoplasm.
Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA.DNA ligases discriminate against substrates containing RNA strands or mismatched base pairs at positions near the ends of the nickedDNA. Bleomycin (BLM) exerts its genotoxicity by generating free radicals, whichattack C-4′ in the deoxyribose backbone of DNA, leading to opening of the ribose ring and strand breakage; it is an S-independentradiomimetic agent that causes double-strand breaks in DNA.
First strand cDNA is synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H). Second strand cDNA synthesis is subsequently performed using DNA Polymerase I and RNase H. The remaining overhangs are converted into blunt ends using exonuclease/polymerase activity. After adenylation of the 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure is ligated to prepare the samples for hybridization. Cell cycle and DNA replication are the top two pathways regulated by BET bromodomain inhibition. Cycloheximide blocks the translation of mRNA to protein.
Targets for DNA/RNA Synthesis
Products for DNA/RNA Synthesis
- Cat.No. Product Name Information
-
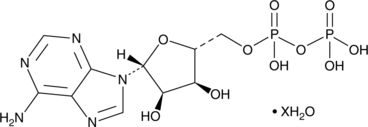
GC49285
Adenosine 5’-methylenediphosphate (hydrate)
An inhibitor of ecto-5’-nucleotidase
-
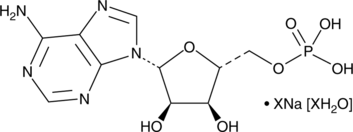
GC42733
Adenosine 5'-monophosphate (sodium salt hydrate)
Adenosine 5'-monophosphate (AMP) is a central nucleotide with functions in metabolism and cell signaling.
-
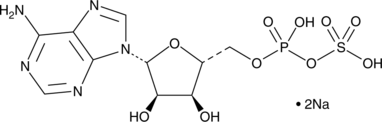
GC42734
Adenosine 5'-phosphosulfate (sodium salt)
Adenosine 5'-phosphosulfate is an ATP and sulfate competitive inhibitor of ATP sulfurylase in humans, S.
-
GC49231
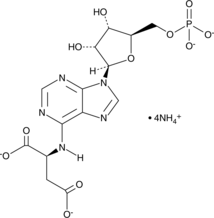
Adenylosuccinic Acid (ammonium salt)
A purine nucleotide and an intermediate in the purine nucleotide cycle
-
GC62470
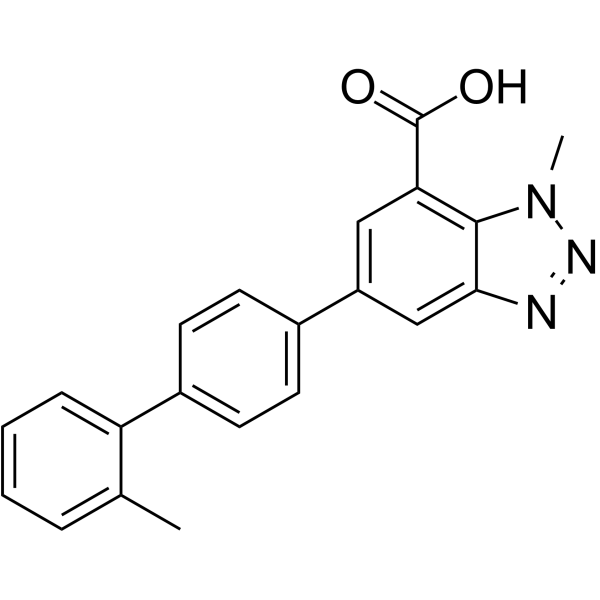
AG-636
AG-636 is a potent, reversible, selective and orally active dihydroorotate dehydrogenase (DHODH) inhibitor with an IC50 of 17 nM. AG-636 has strong anticancer effects.
-
GC49039
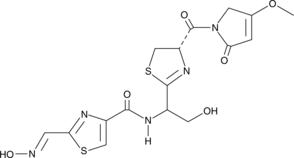
Althiomycin
A thiazole antibiotic
-
GC11559
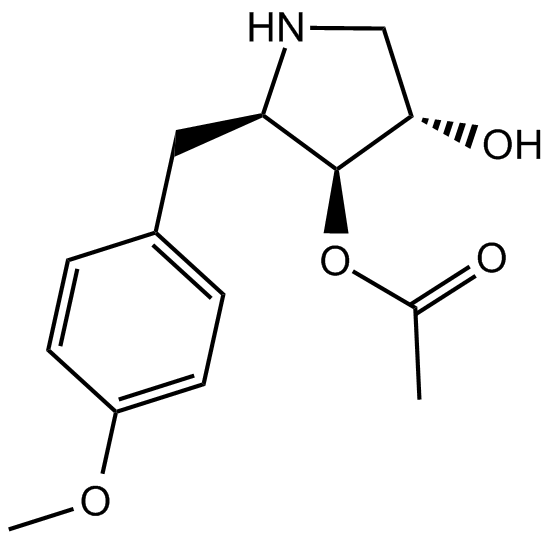
Anisomycin
JNK agonist, potent and specific
-
GC68667
Anticancer agent 73
Anticancer agent 73 (compound CIB-3b) is an anticancer drug that targets TAR RNA binding protein 2 (TRBP) and disrupts its interaction with Dicer. Anticancer agent 73 can rebalance the expression profile of oncogenic or tumor-suppressive miRNAs. Anticancer agent 73 has inhibitory effects on the proliferation and metastasis of hepatocellular carcinoma (HCC) cells both in vitro and in vivo.
-
GC40032
Antipain (hydrochloride)
Antipain (hydrochloride) is a protease inhibitor isolated from Actinomycetes.
-
GC68448
AOH1160
-
GC67876
APE1-IN-1

-
GC38463
ASLAN003
ASLAN003 (ASLAN003) is an orally active and potent Dihydroorotate Dehydrogenase (DHODH) inhibitor with an IC50 of 35 nM for human DHODH enzyme. ASLAN003 inhibits protein synthesis via activation of AP-1 transcription factors. ASLAN003 induces apoptosis and substantially prolongs survival in acute myeloid leukemia (AML) xenograft mice.
-
GC61821
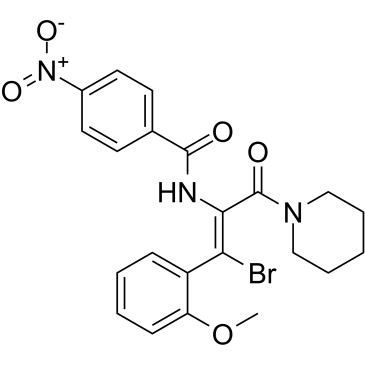
AT-130
AT-130, a phenylpropenamide derivative, is a potent hepatitis B virus (HBV) replication non-nucleoside inhibitor.
-
GC46895
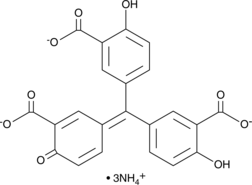
Aurintricarboxylic Acid (ammonium salt)
A protein synthesis inhibitor with diverse biological activities
-
GC40005
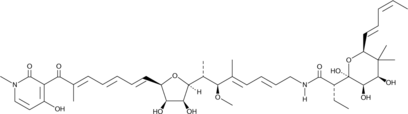
Aurodox
Aurodox is a polyketide antibiotic originally isolated from S.
-
GC60062
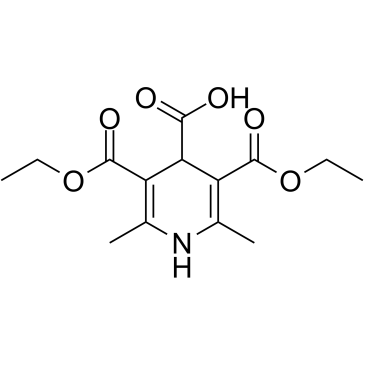
AV-153 free base
AV-153 free base, a 1,4-dihydropyridine (1,4-DHP) derivative, is an antimutagenic. AV-153 free base intercalates to DNA in a single strand break and reduces DNA damage, stimulates DNA repair in human cells in vitro. AV-153 free base interacts with thymine and cytosine and has an influence on poly(ADP)ribosylation. AV-153 free base has anti-cancer activity.
-
GC67960
AVG-233
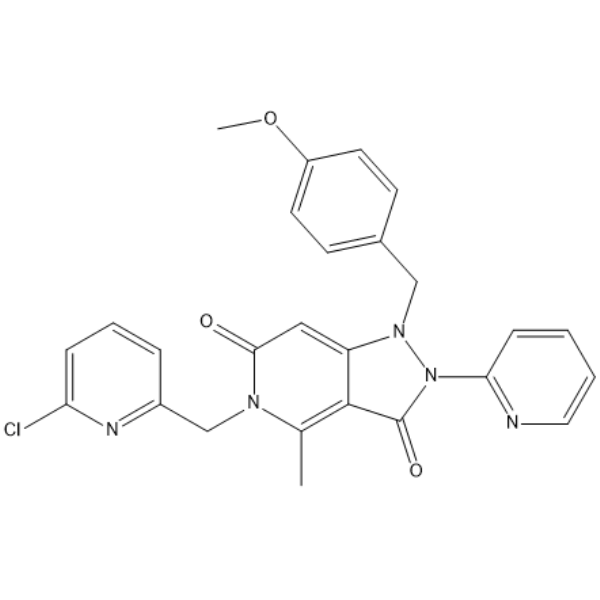
-
GC15033
Azathioprine
purine synthesis and GTP-binding protein Rac1 activation inhibitor
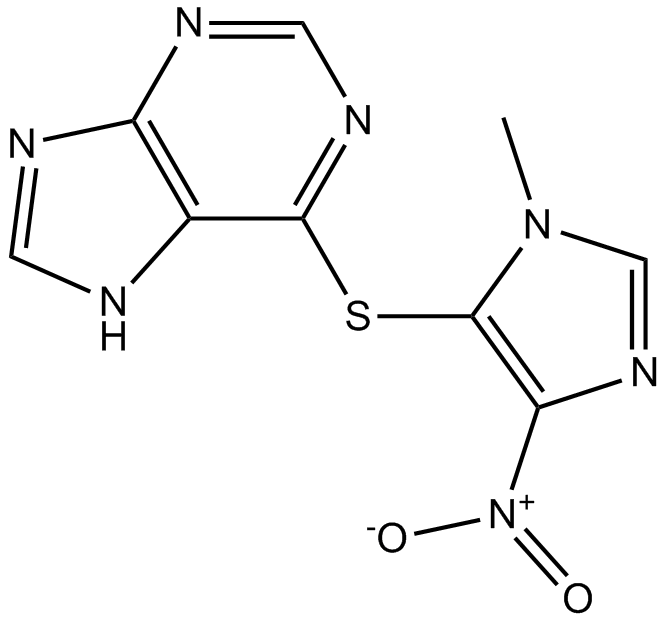
-
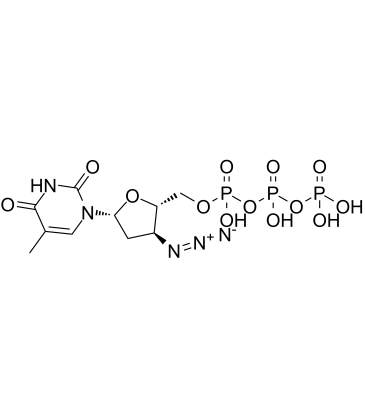
GC60616
AZT triphosphate
AZT triphosphate (3'-Azido-3'-deoxythymidine-5'-triphosphate) is a active triphosphate metabolite of Zidovudine (AZT).
-
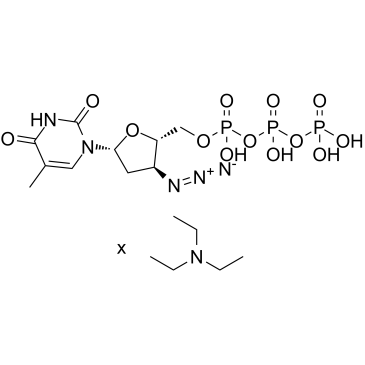
GC60617
AZT triphosphate TEA
AZT triphosphate TEA (3'-Azido-3'-deoxythymidine-5'-triphosphate TEA) is a active triphosphate metabolite of Zidovudine (AZT).
-
GC50656
BAY 707
BAY 707 is a substrate-competitive, highly potent and selective inhibitor of MTH1(NUDT1) with an IC50 of 2.3 nM. BAY 707 has a good pharmacokinetic (PK) profile to other MTH1 compounds and is well-tolerated in mice, but shows a clear lack of in vitro or in vivo anticancer efficacy.
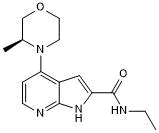
-
GC32838
BAY-2402234
A DHODH inhibitor
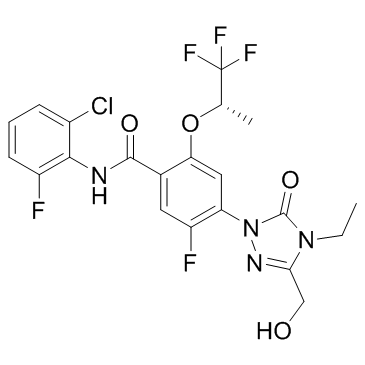
-
GC50572
BC-LI-0186
Leucyl-tRNA synthase (TRS)/Ras-related GTP-binding protein D (RagD) interaction inhibitor
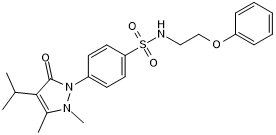
-
GC62863
BCH001
BCH001, a quinoline derivative, is a specific PAPD5 inhibitor.
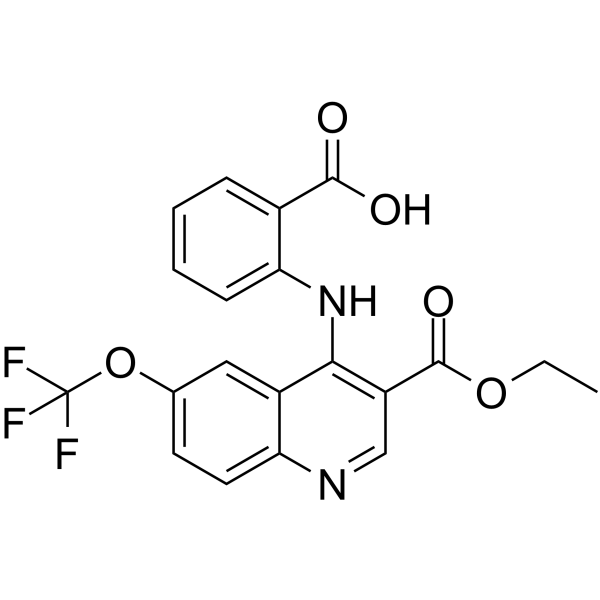
-
GC32333
Beaucage reagent
Beaucage reagent is found to be potent in causing DNA cleavage.
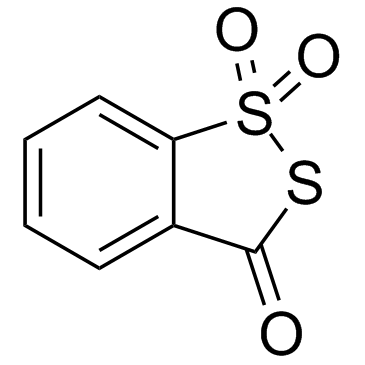
-
GC18094
Blasticidin S HCl
Blasticidin S HCl is a nucleoside antibiotic isolated from Streptomyces griseochromogenes[1].
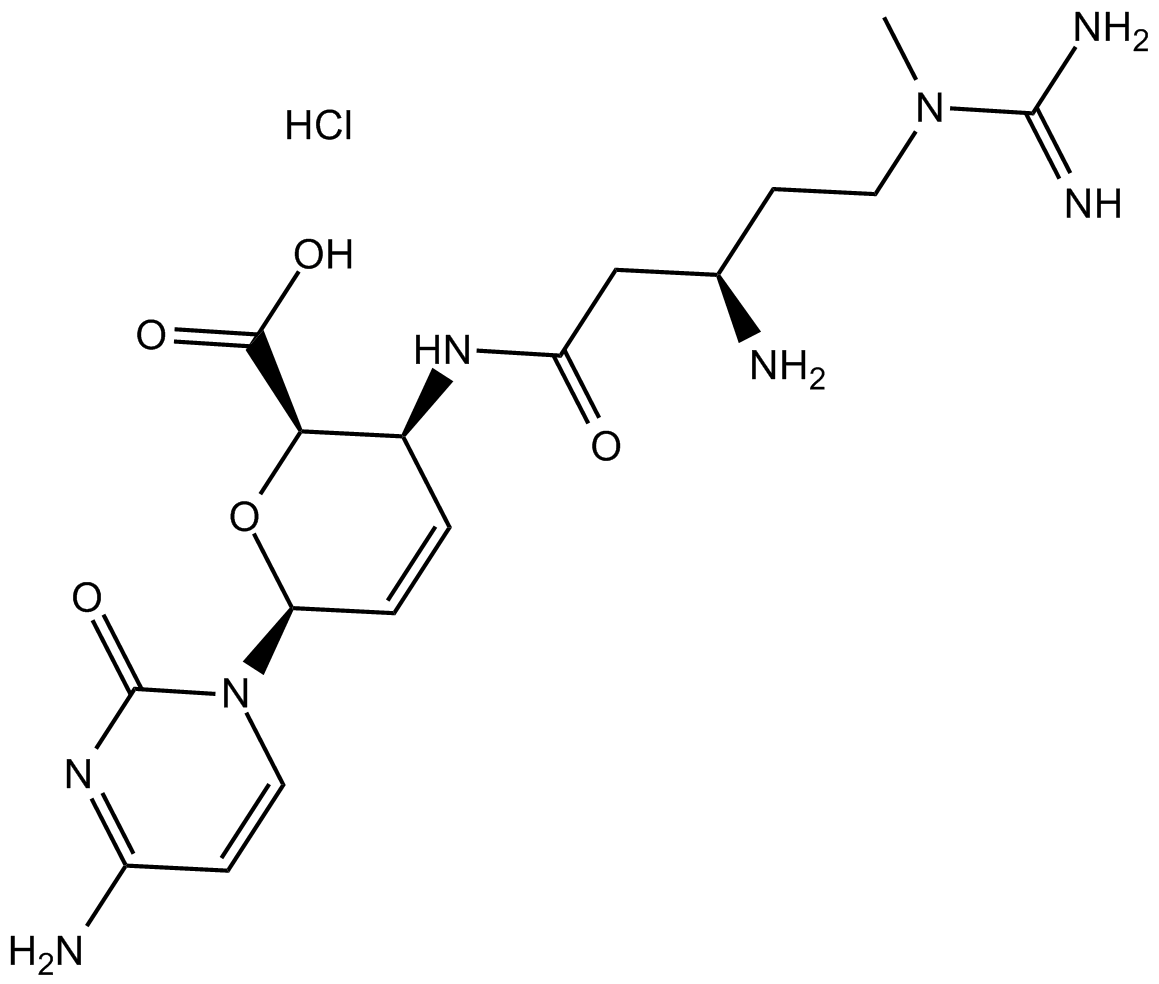
-
GC60089
Bleomycin hydrochloride
Bleomycin hydrochloride is a DNA synthesis inhibitor. Bleomycin hydrochloride is a DNA damaging agent. Bleomycin hydrochloride is an antitumor antibiotic.
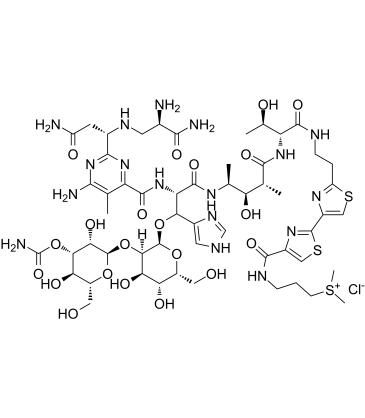
-
GC15819
Bleomycin Sulfate
Bleomycin is produced by Streptomyces verticillis.
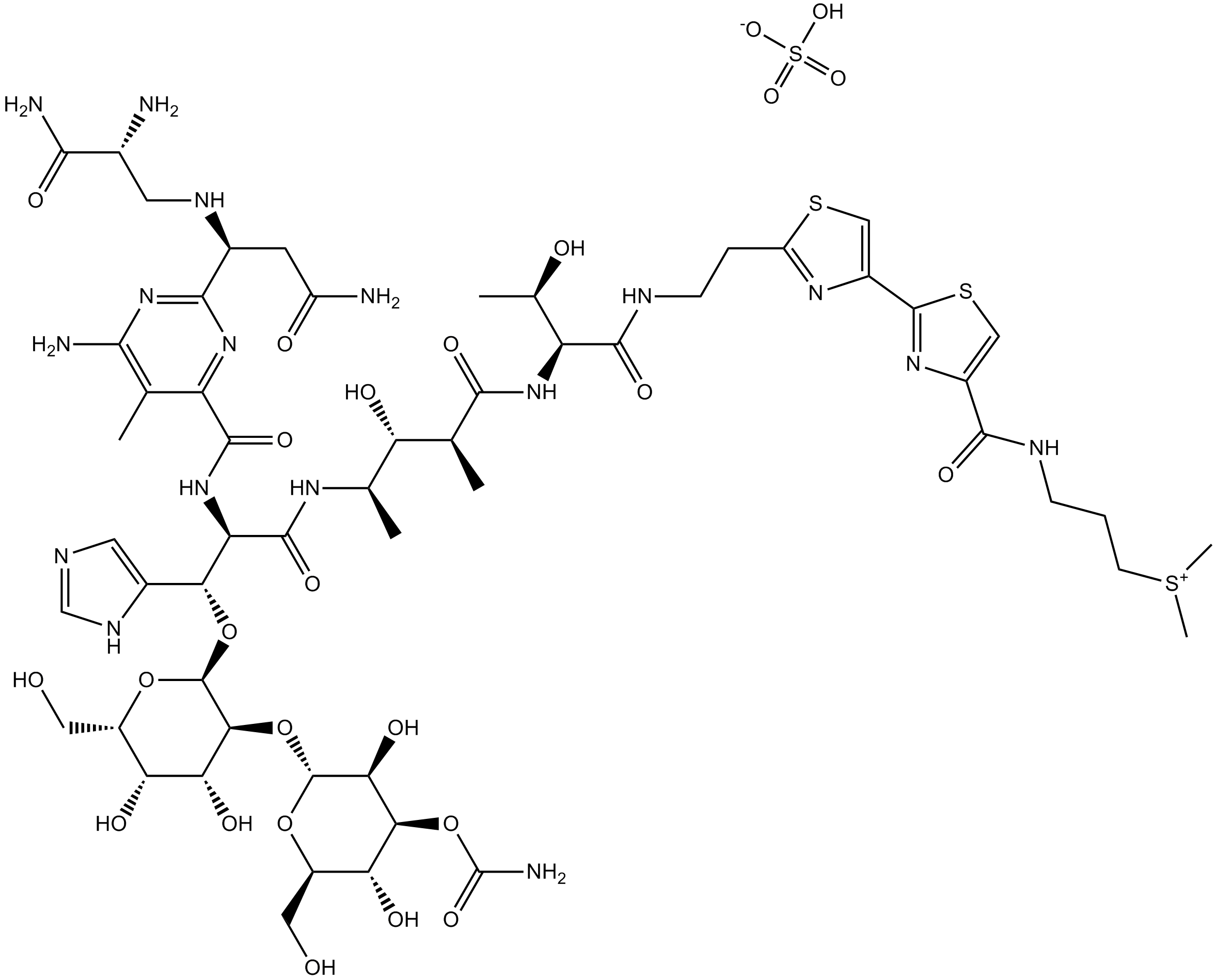
-
GC15190
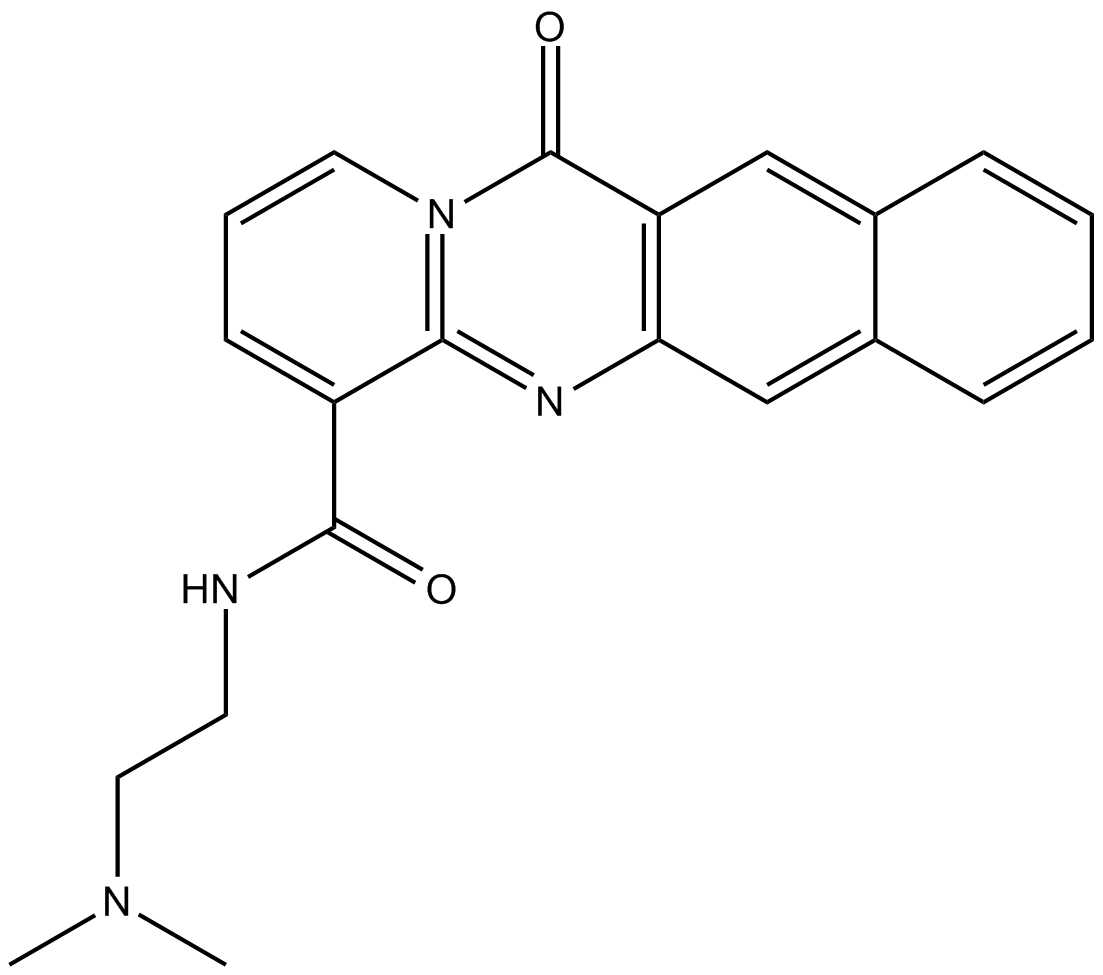
BMH-21
RNA polymerase I Inhibitior
-
GC48365
Bottromycin A2
An antibiotic
-
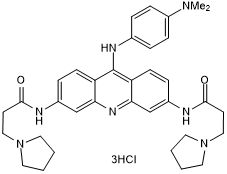
GC50140
BRACO 19 trihydrochloride
Telomerase inhibitor
-
GC62128
Bractoppin
Bractoppin is a inhibitor of phosphopeptide recognition by the human BRCA1 tBRCT domain with IC50 of 74 nM.
-
GC32702
Branaplam (LMI070)
Branaplam (LMI070) (LMI070; NVS-SM1) is a highly potent, selective and orally active survival motor neuron-2 (SMN2) splicing modulator with an EC50 of 20 nM for SMN. Branaplam (LMI070) inhibits human-ether-a-go-go-related gene (hERG) with an IC50 of 6.3 μM. Branaplam (LMI070) elevates full-length SMN protein and extends survival in a severe spinal muscular atrophy (SMA) mouse model.
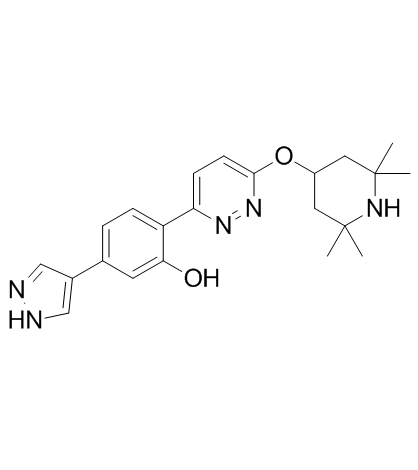
-
GC64383
Branaplam hydrochloride
Branaplam (LMI070; NVS-SM1) hydrochloride is a highly potent, selective and orally active survival motor neuron-2 (SMN2) splicing modulator with an EC50 of 20 nM for SMN. Branaplam hydrochloride inhibits human-ether-a-go-go-related gene (hERG) with an IC50 of 6.3 μM. Branaplam hydrochloride elevates full-length SMN protein and extends survival in a severe spinal muscular atrophy (SMA) mouse model.
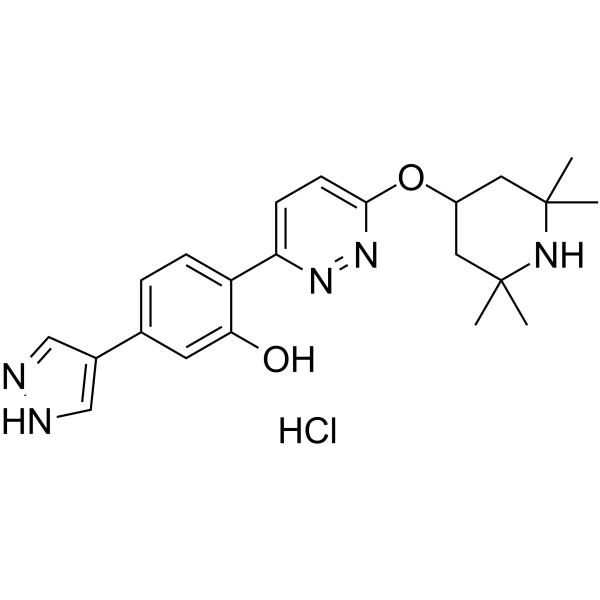
-
GC42978
Bromamphenicol
Bromamphenicol is a dibrominated derivative of the antibiotic chloramphenicol.
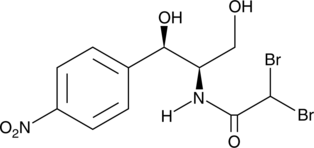
-
GC62136
BVDV-IN-1
BVDV-IN-1 is a non-nucleoside inhibitor (NNI) of bovine viral diarrhea virus (BVDV), with an EC50 of 1.8 μM.
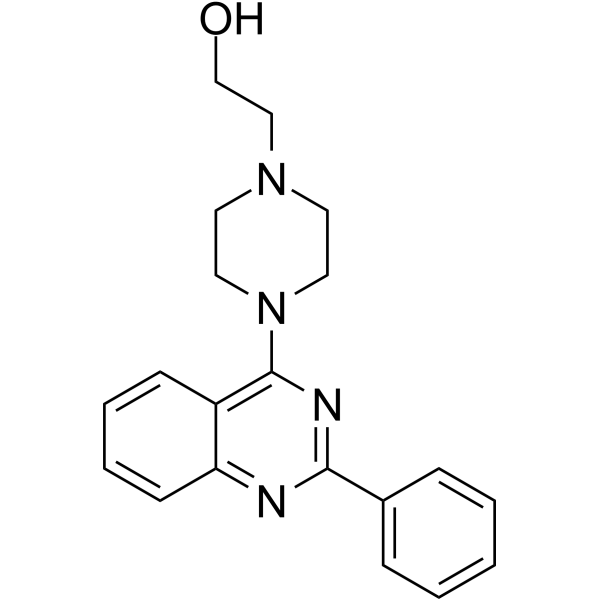
-
GC61735
Bz-rA Phosphoramidite
Bz-rA Phosphoramidite is used for ribonucleotides modification.
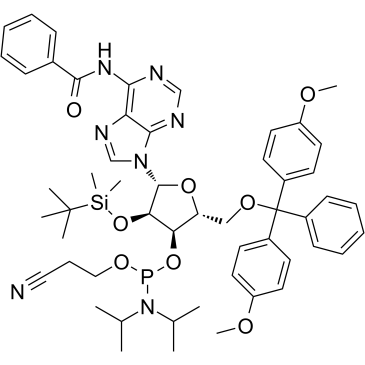
-
GC66885
Bz-rC Phosphoramidite
Bz-rC Phosphoramidite is a phosphinamide monomer that can be used in the preparation of oligonucleotides.
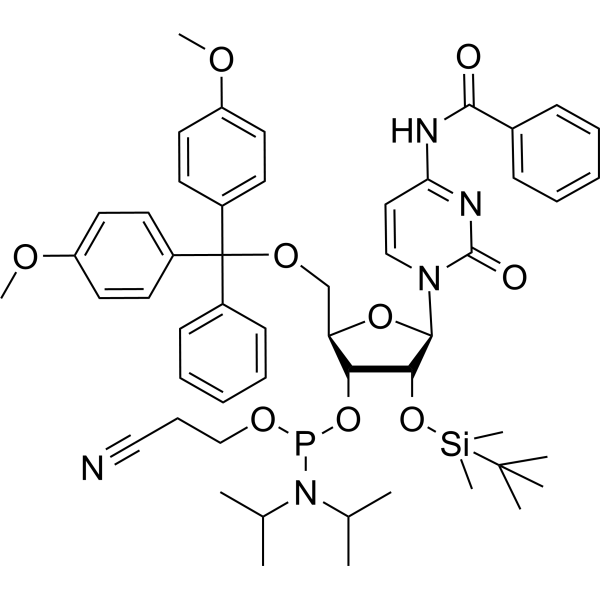
-
GC15866
Capecitabine
DNA, RNA and protein synthesis inhibitor
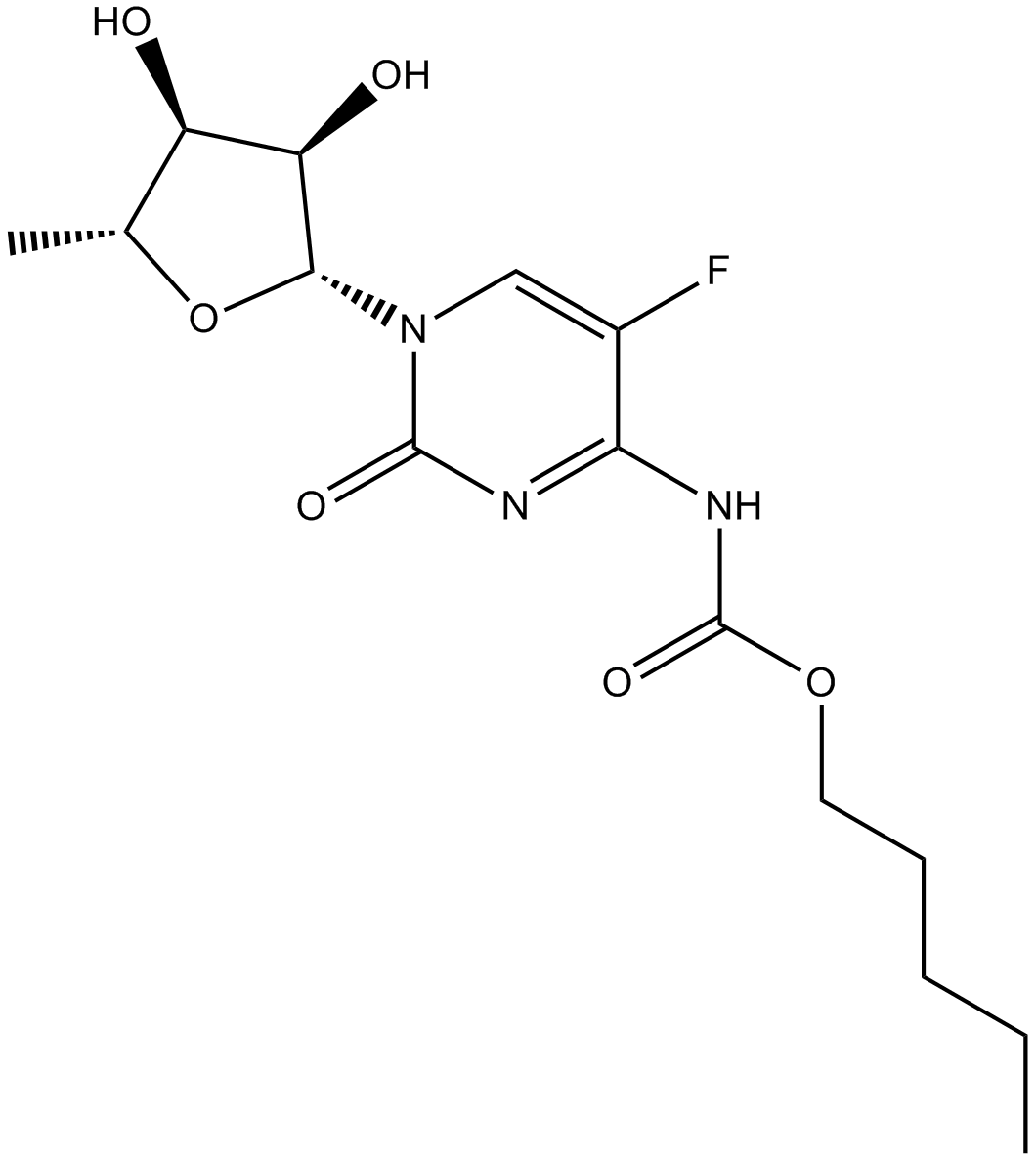
-
GC38896
Caracemide
Caracemide (NSC-253272) inhibits the enzyme ribonucleotide reductase of Escherichia coli.
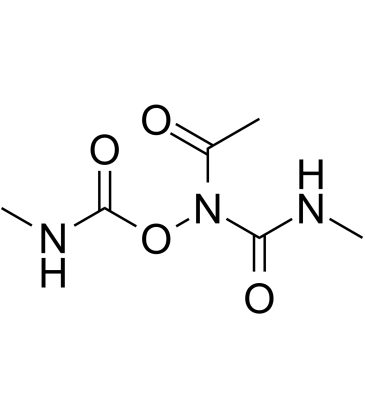
-
GC11207
Carboplatin
Antitumor agent that forms platinum-DNA adducts.
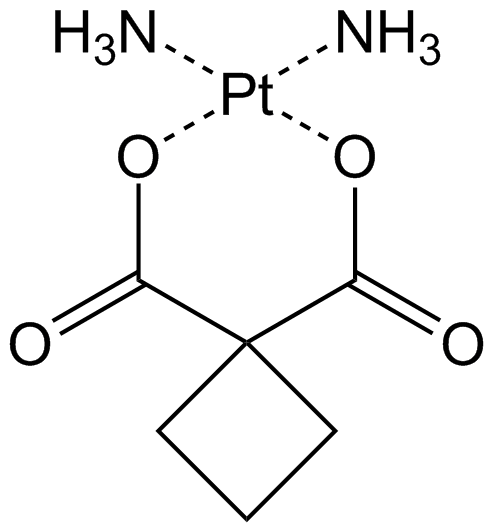
-
GC12748
Carmofur
Cytostatic derivative of fluorouracilm,antineoplatic agent
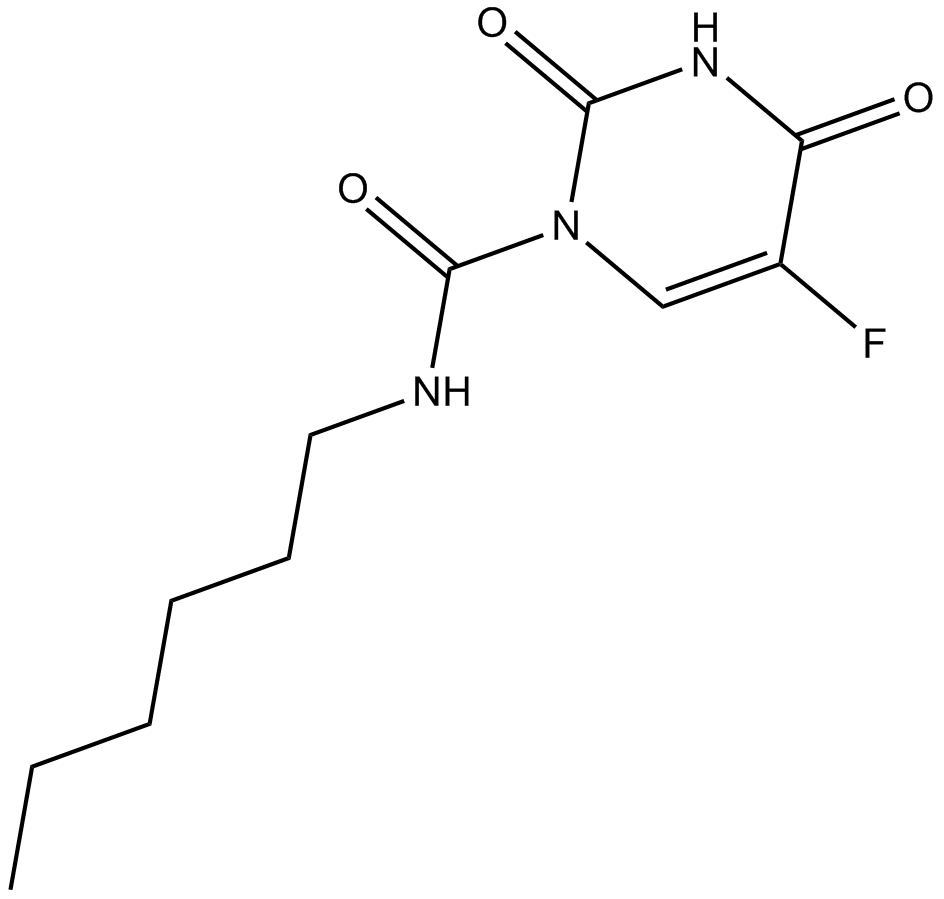
-
GC52006
CB-096
An inhibitor of RAN translation
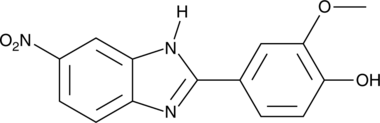
-
GC49307
cDPCP
A DNA-crosslinking agent
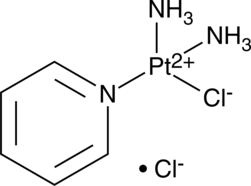
-
GC19098
CeMMEC1
CeMMEC1 is an inhibitor of BRD4, and also has high affinity for TAF1, with an IC50 of 0.9 uM for TAF1, and a Kd of 1.8 uM for TAF1 (2).
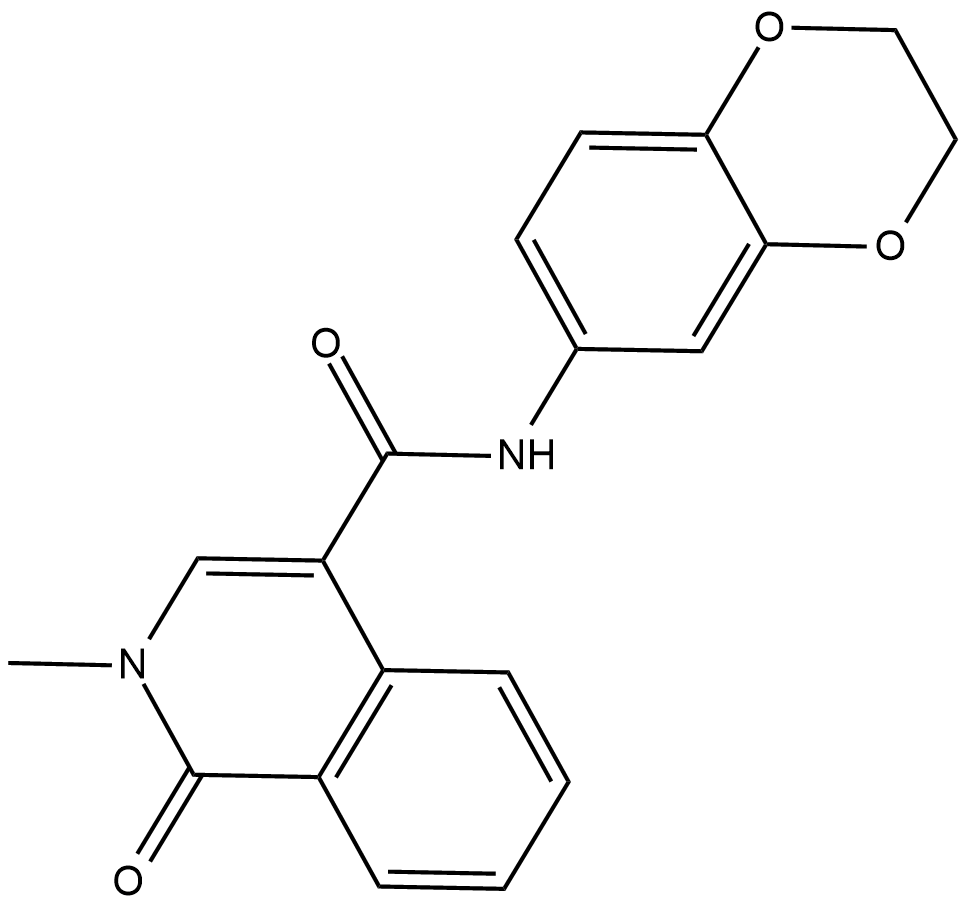
-
GC19099
CeMMEC13
CeMMEC13 is a potent inhibitor of TAF1 (2) bromodomain, with an IC50 of 2.1 uM.
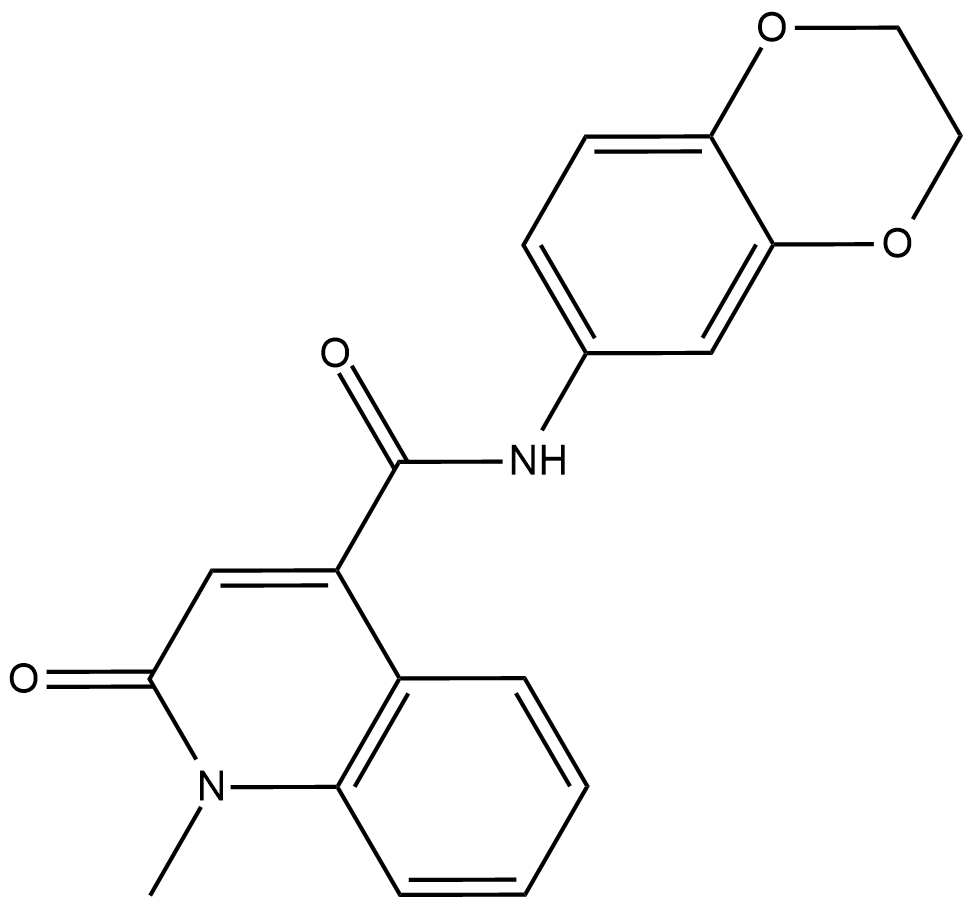
-
GC32250
Chebulinic acid
An ellagitannin with diverse biological activities
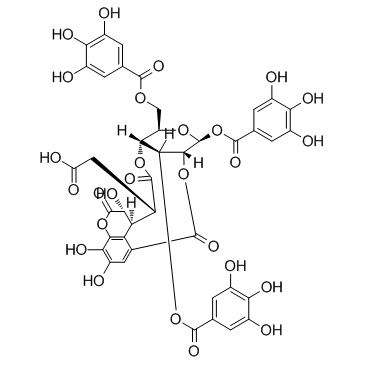
-
GC18640
cis-9,10-Methyleneoctadecanoic Acid methyl ester
cis-9,10-Methyleneoctadecanoic acid methyl ester is a cyclopropane fatty acid methyl ester.
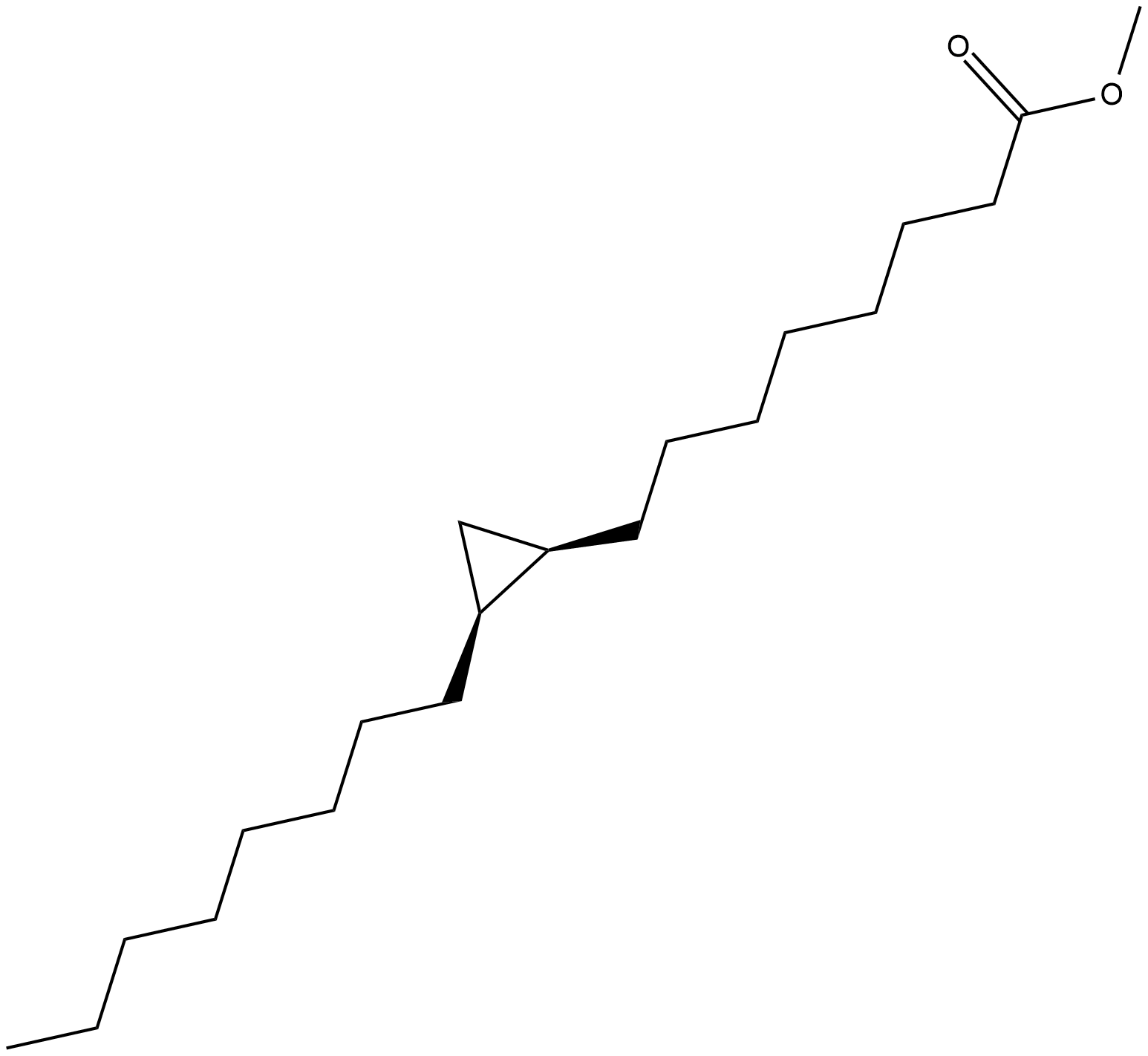
-
GC52351
Citrullinated α-Enolase (R8 + R14) (1-19)-biotin Peptide
A biotinylated and citrullinated α-enolase peptide

-
GC10509
Cladribine
Apoptosis inducer in CLL cells
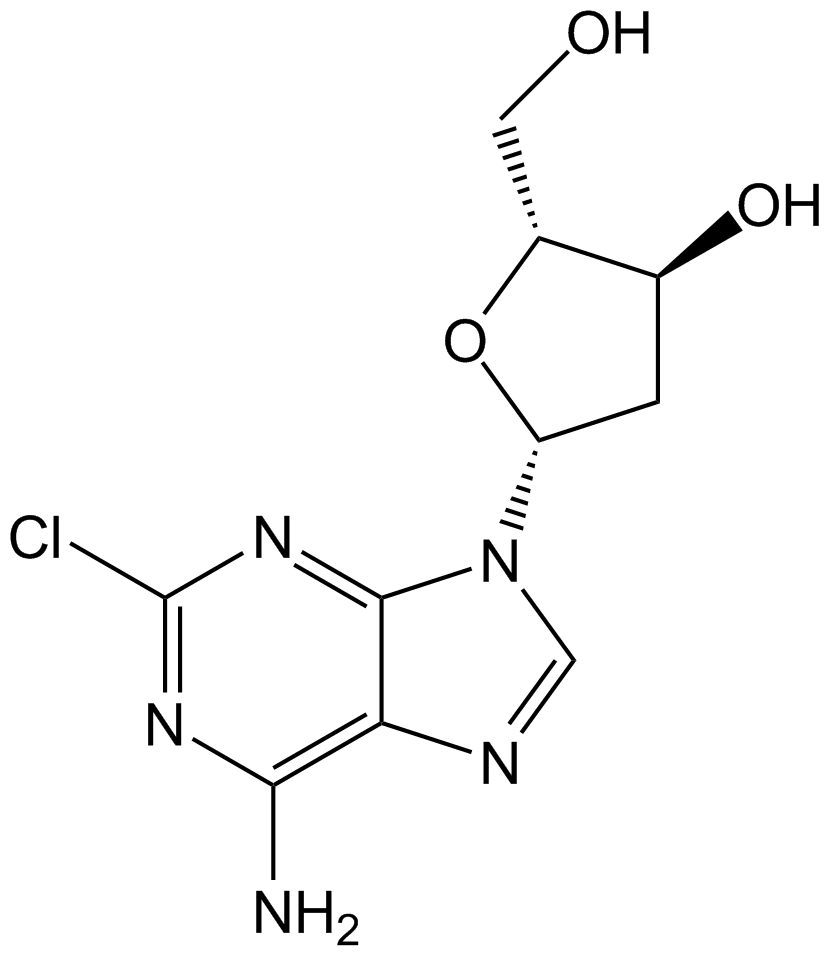
-
GC49852
Clindamycin (hydrochloride hydrate)
Clindamycin (hydrochloride hydrate) is an oral protein synthesis inhibitory agent that has the ability to suppress the expression of virulence factors in Staphylococcus aureus at sub-inhibitory concentrations (sub-MICs).
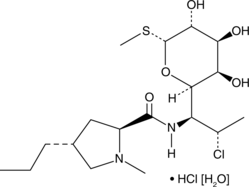
-
GC52171
Clindamycin-d3 (hydrochloride)
Clindamycin-d3 (hydrochloride) is the deuterium labeled Clindamycin.
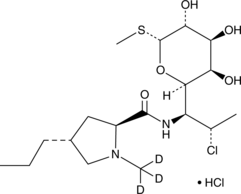
-
GC15219
Clofarabine
Antimetabolite,inhibit DNA polymerase and ribonucleotide reductase
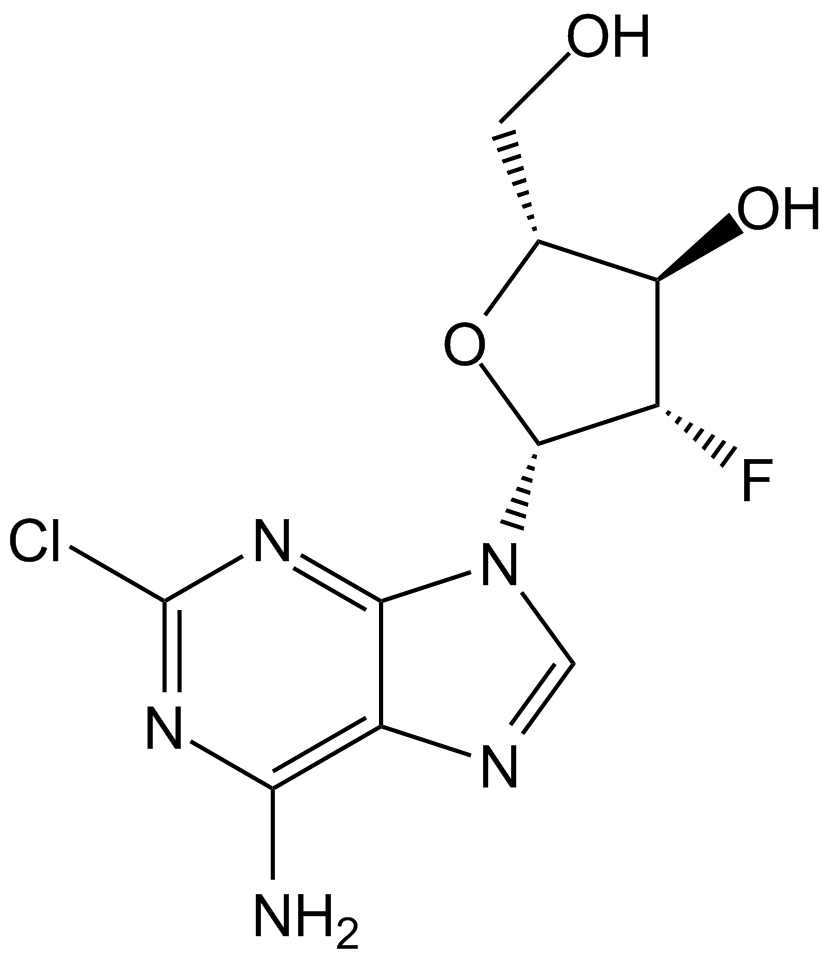
-
GC19110
COH29
COH29 is a potent ribonucleotide reductase (RNR) inhibitor with anticancer activity.
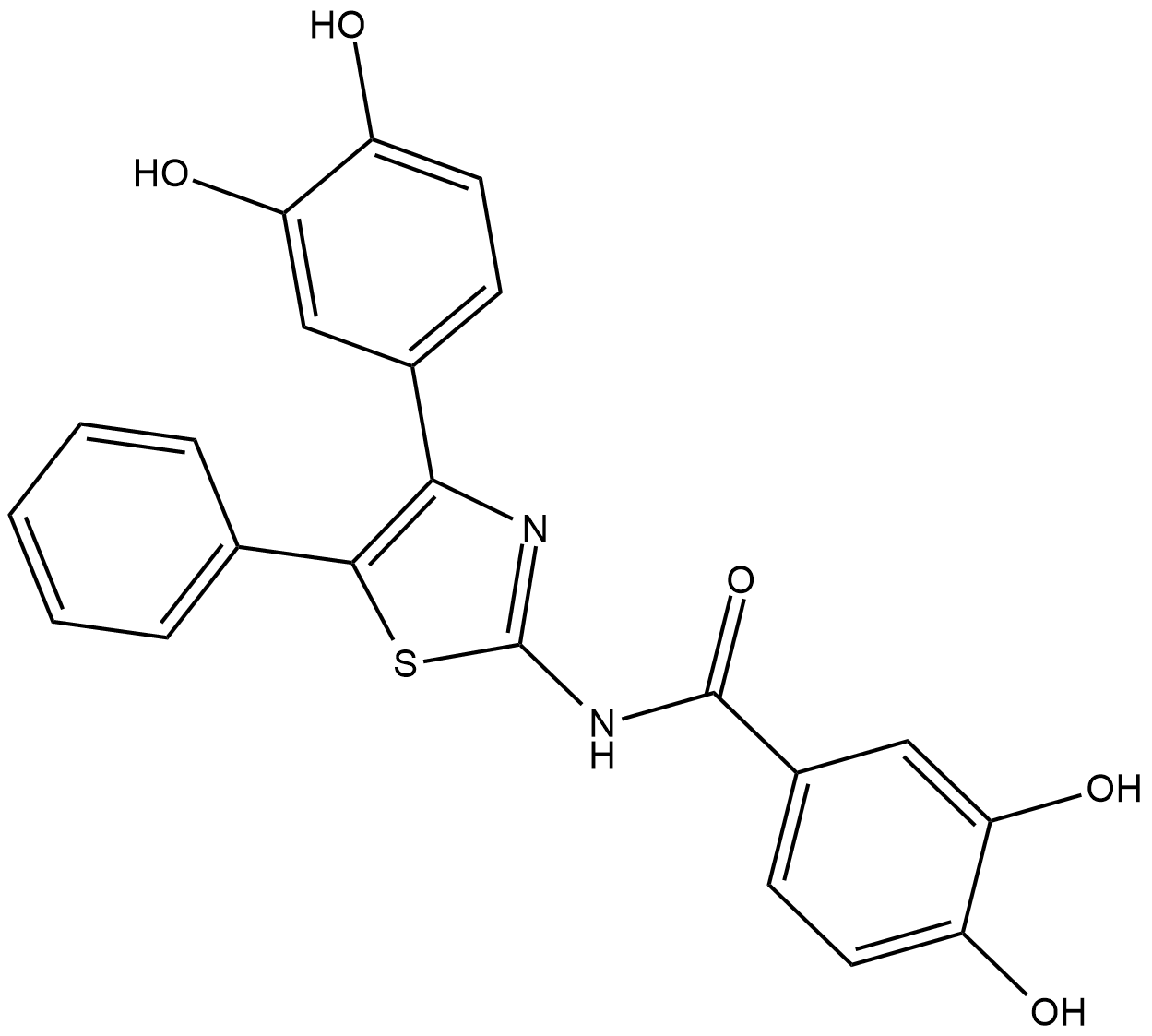
-
GC17231
CRT0044876
APE1 inhibitor, potent and selective
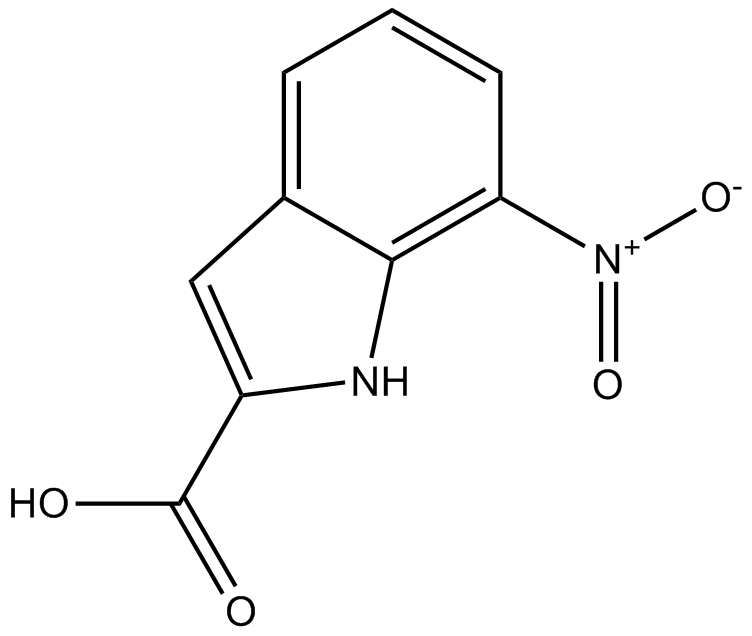
-
GC14404
CX-5461
Pol I-mediated rRNA synthesis inhibitor
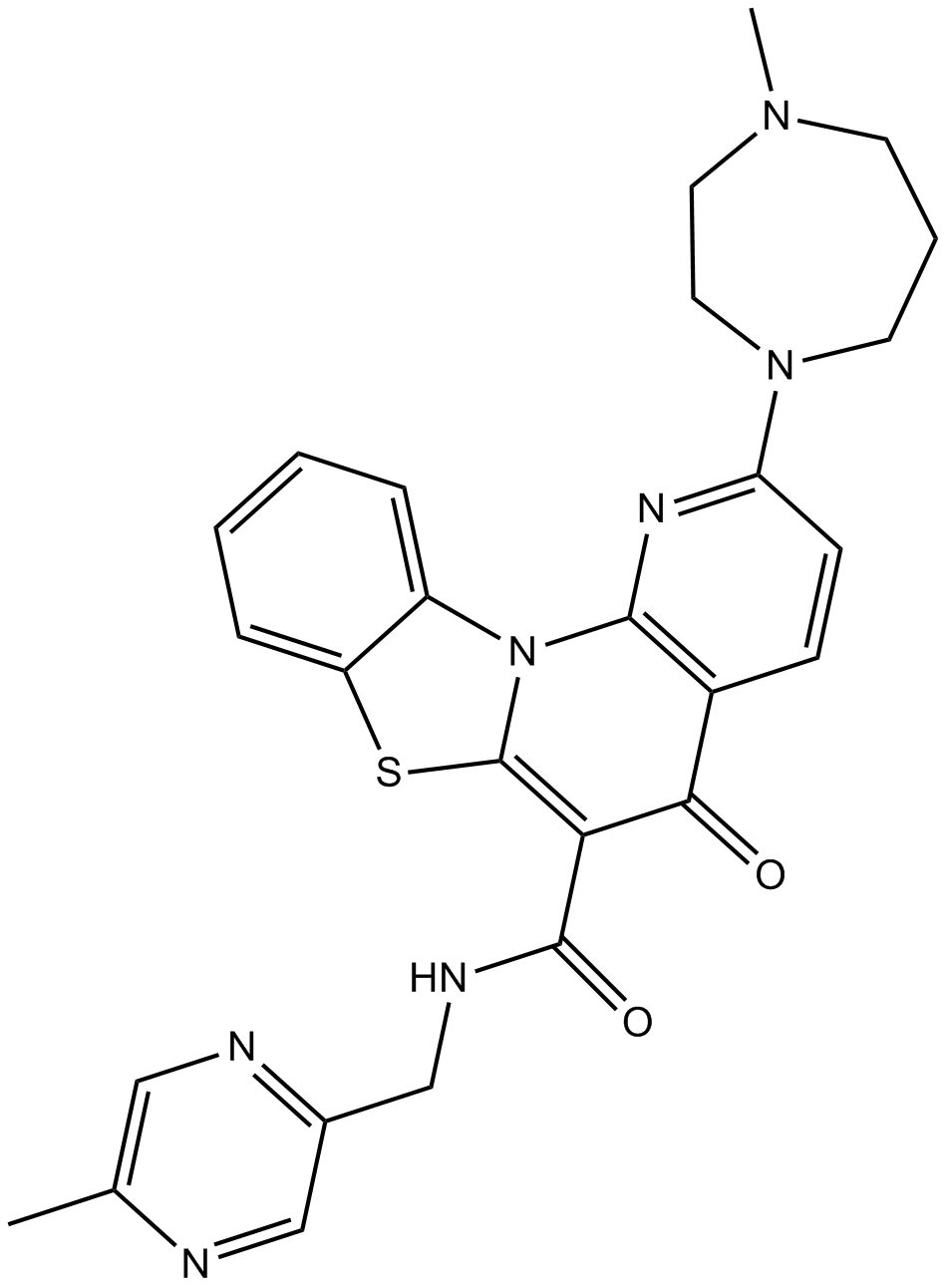
-
GC35761
CX-5461 dihydrochloride
CX-5461 dihydrochloride is a potent and orally bioavailable inhibitor of Pol I-mediated rRNA synthesis, with IC50s of 142 nM in HCT-116, 113 nM in A375, and 54 nM in MIA PaCa-2 cells, and shows little or no effect on Pol II (IC50 ≥25 μM).
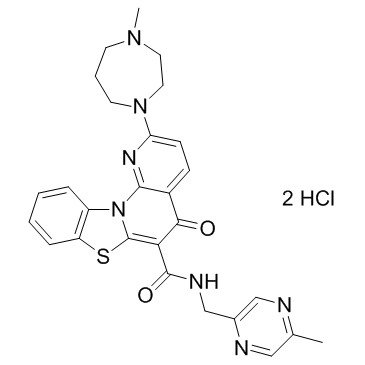
-
GC11145
Cyclophosphamide
Cyclophosphamide is a frequently used chemotherapy, often in combination with other chemotherapy types, for the treatment of breast cancer, malignant lymphomas, multiple myeloma, and neuroblastoma
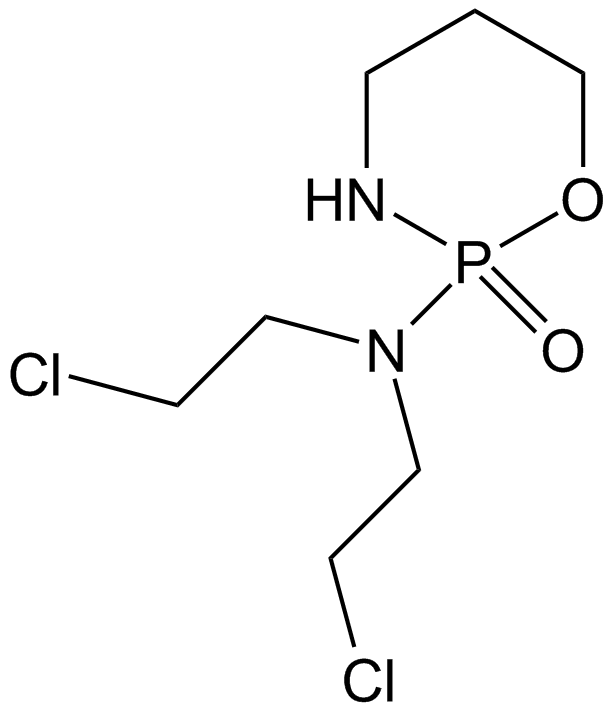
-
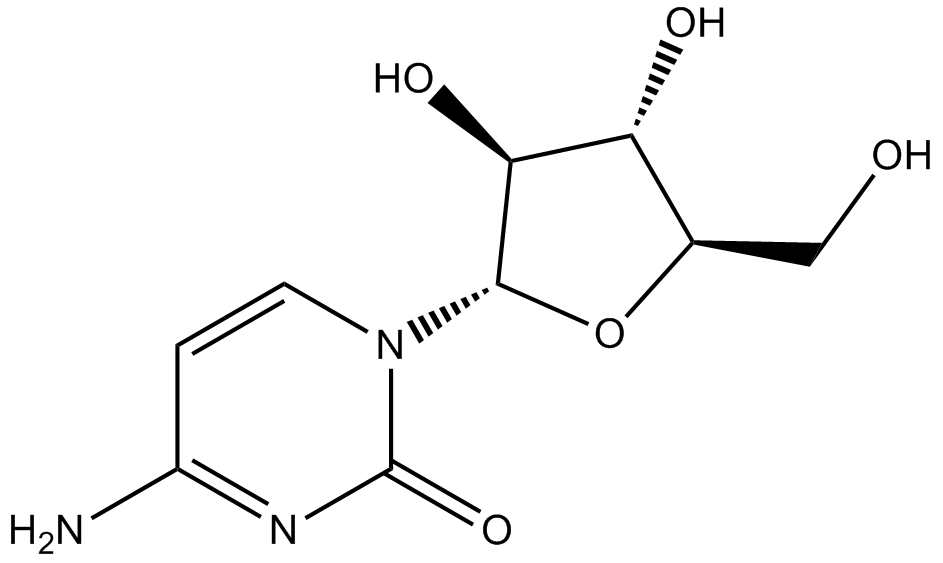
GC13070
Cytarabine
Cytotoxic agent, blocks DNA synthesis
-
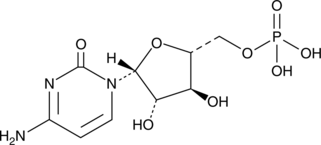
GC49863
Cytarabine 5′-monophosphate
An active metabolite of cytarabine
-
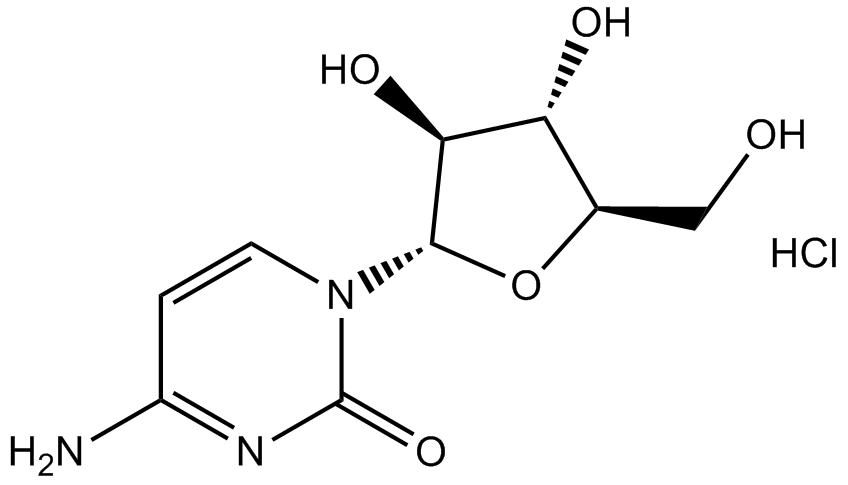
GC14961
Cytarabine hydrochloride
DNA synthsis inhibitor
-
GC49104
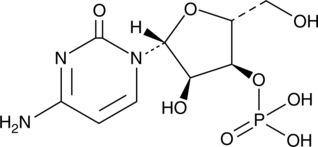
Cytidine 3’-monophosphate
A ribonucleotide
-
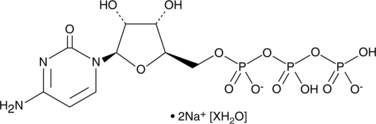
GC47162
Cytidine 5'-triphosphate (sodium salt hydrate)
A pyrimidine nucleoside triphosphate
-
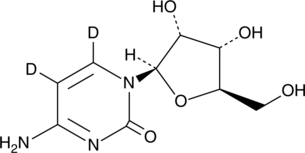
GC49526
Cytidine-d2
An internal standard for the quantification of cytidine
-
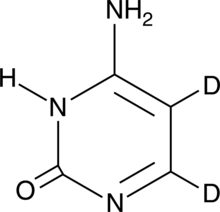
GC49464
Cytosine-d2
An internal standard for the quantification of cytosine
-
GC39149
D-I03
D-I03 is a selective RAD52 inhibitor with a Kd of 25.8 ?M. D-I03 specifically inhibits RAD52-dependent single-strand annealing (SSA) and D-loop formation with IC50s of 5 ?M and 8 ?M, respectively. D-I03 suppresses growth of BRCA1- and BRCA2-deficient cells and inhibits formation of damage-induced RAD52 foci, but does not effect on RAD51 foci induced by Cisplatin.
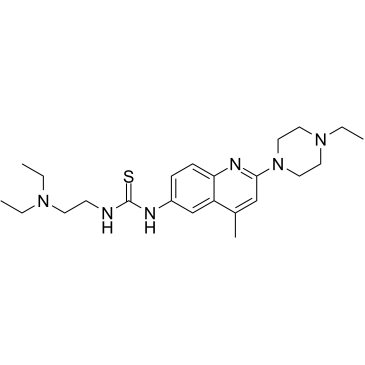
-
GC47276
D-threo-PPMP (hydrochloride)
A GlyCer synthetase inhibitor
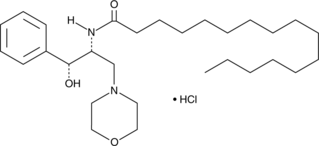
-
GC14485
Dacarbazine
Antineoplastic( malignant melanoma and sarcomas)
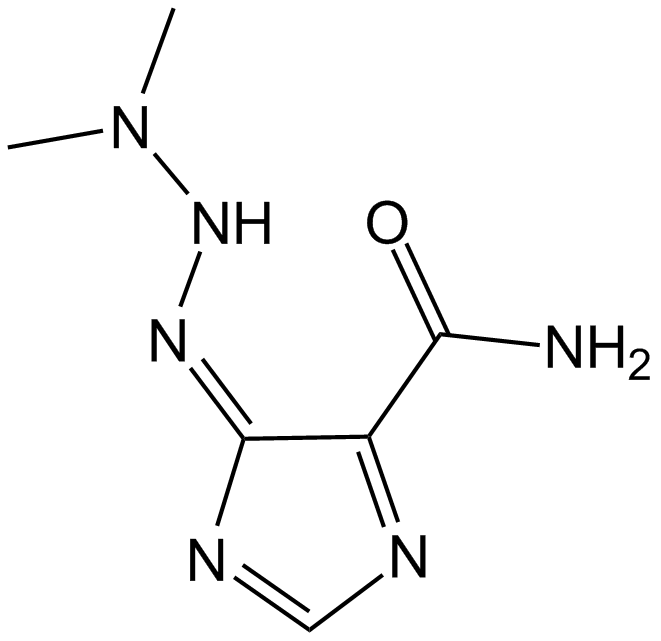
-
GC10214
Daptomycin
Calcium-dependent antibiotic
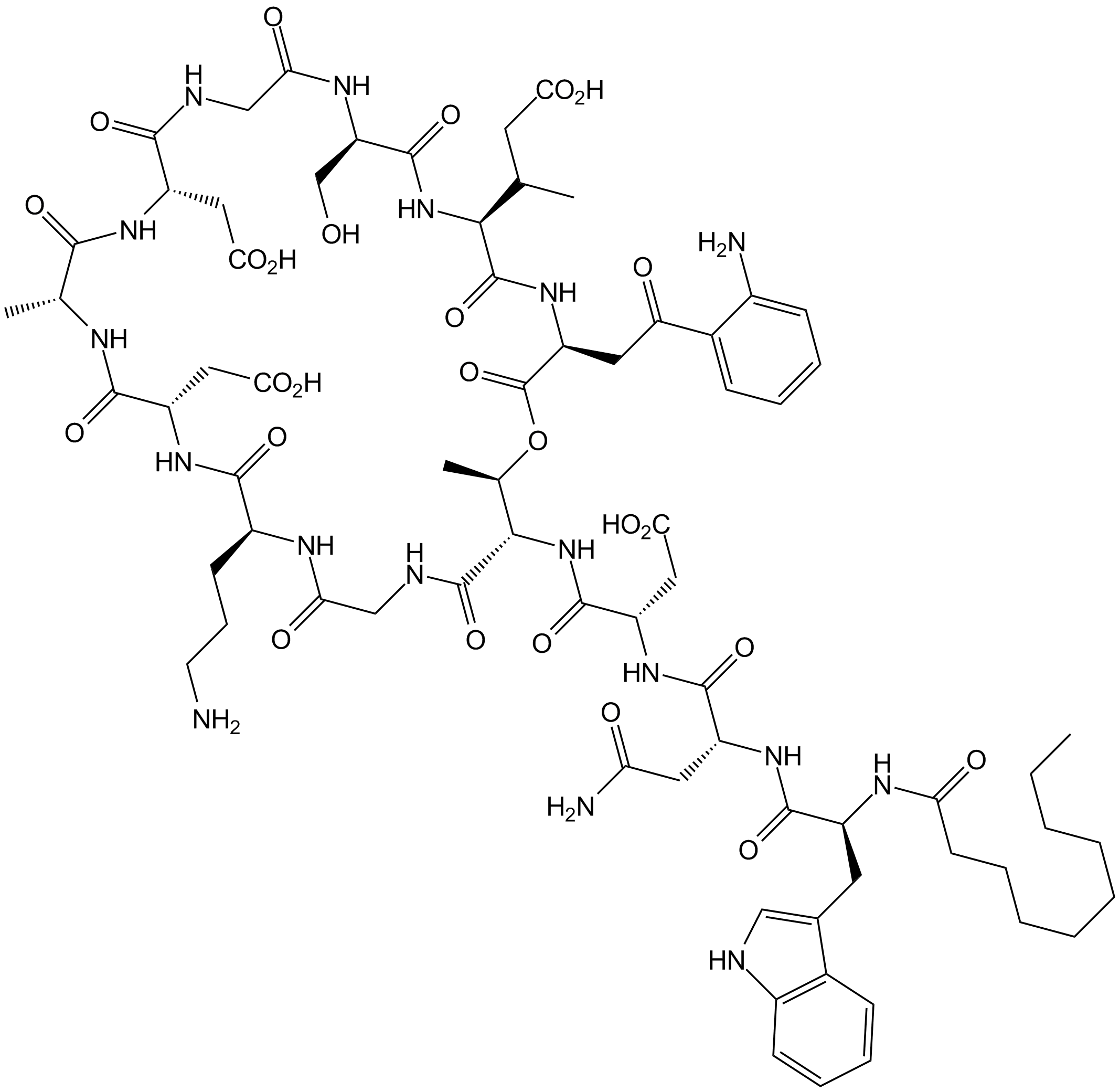
-
GC33124
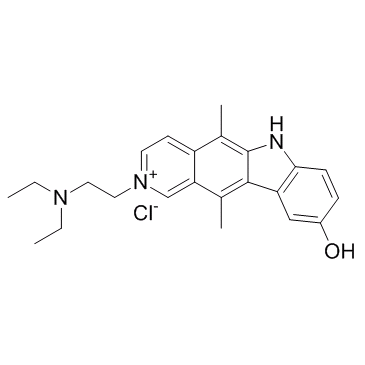
Datelliptium chloride
Datelliptium chloride is a DNA-intercalating agent derived from ellipticine, with anti-tumor activities.
-
GC12828
Daunorubicin
DNA topoisomerase II inhibitor
-
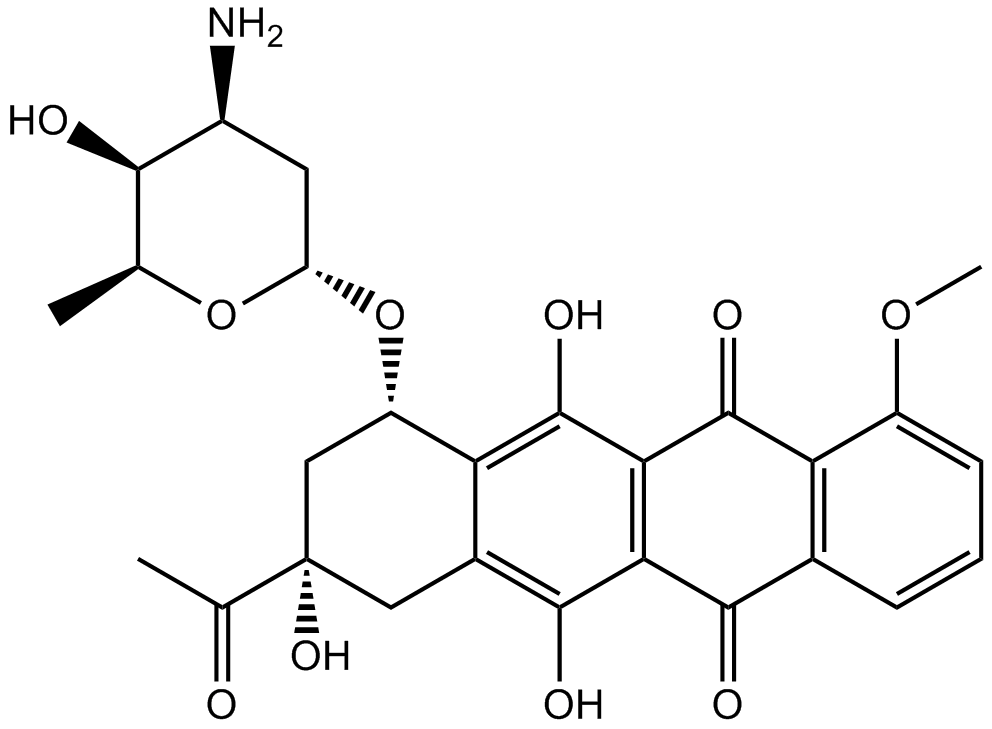
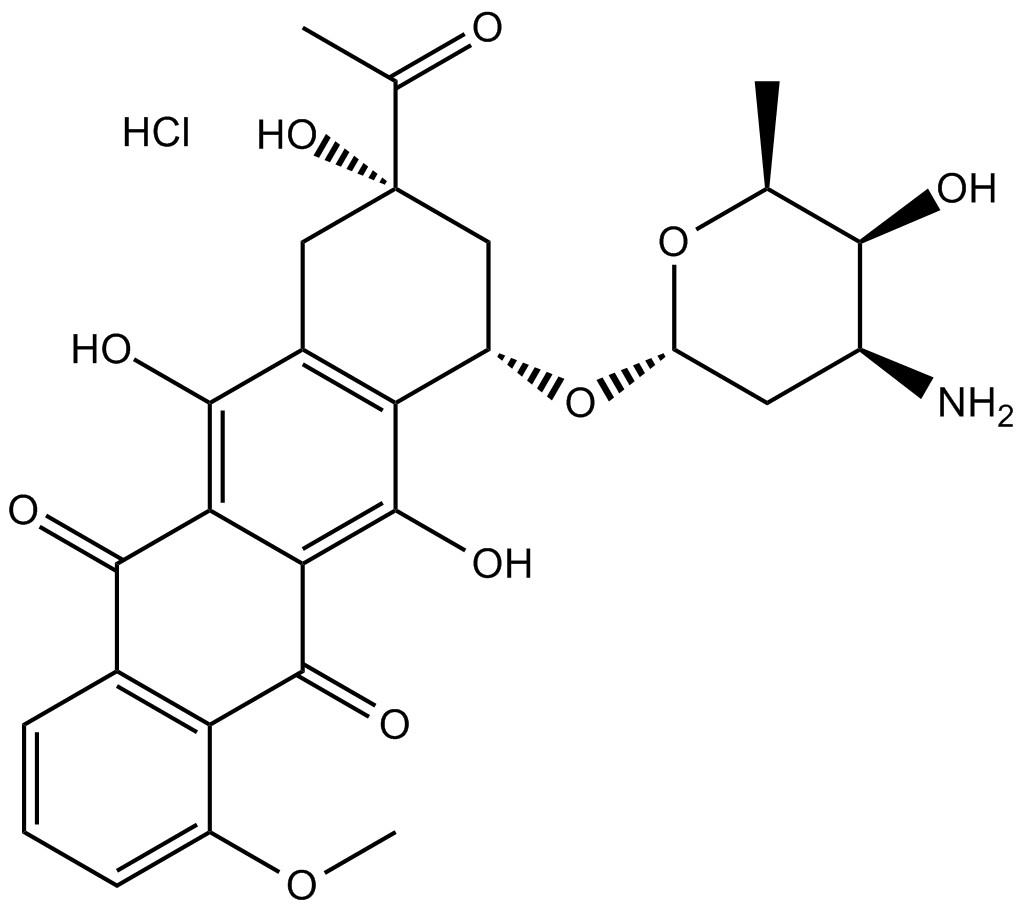
GC10354
Daunorubicin HCl
Daunorubicin (Daunomycin) hydrochloride is a topoisomerase II inhibitor with potent anti-tumor activity.
-
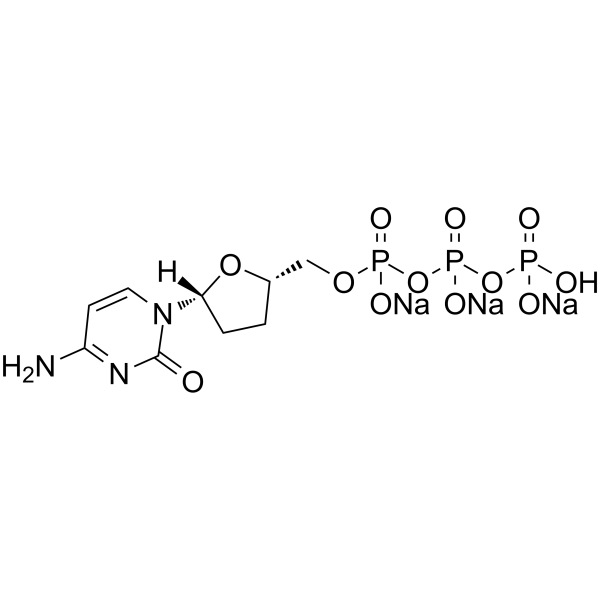
GC65440
ddCTP trisodium
ddCTP trisodium is one of 2',3'-dideoxyribonucleoside 5'-triphosphates (ddNTPs) that acts as chain-elongating inhibitor of DNA polymerase for DNA sequencing.
-
GC65295
ddGTP trisodium
ddGTP (2′,3′-Dideoxyguanosine 5′-triphosphate) trisodium is one of 2',3'-dideoxyribonucleoside 5'-triphosphates (ddNTPs) that acts as chain-elongating inhibitor of DNA polymerase for DNA sequencing.
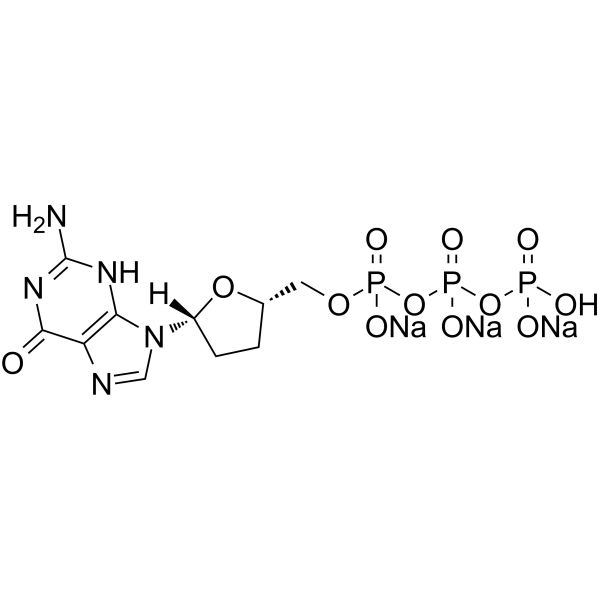
-
GC64020
ddTTP
ddTTP is one of 2',3'-dideoxyribonucleoside 5'-triphosphates (ddNTPs) that acts as chain-elongating inhibitor of DNA polymerase for DNA sequencing.
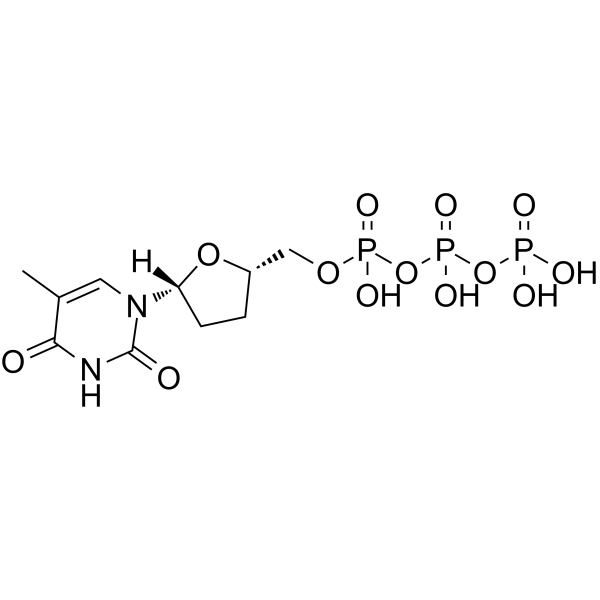
-
GC35829
Dehydroaltenusin
Dehydroaltenusin is a small molecule selective inhibitor of eukaryotic DNA polymerase α, a type of antibiotic produced by a fungus with an IC50 value of 0.68 μM. The inhibitory mode of action of dehydroaltenusin against mammalian pol α activity is competitive with respect to the DNA template primer (Ki=0.23 ?M) and non-competitive with respect to the 2'-deoxyribonucleoside 5'-triphosphate substrate (Ki=0.18 ?M). Dehydroaltenusin arrests the cancer cell cycle at the S-phase and triggers apoptosis. Dehydroaltenusin possesses anti-tumor activity against human adenocarcinoma tumor in vivo.
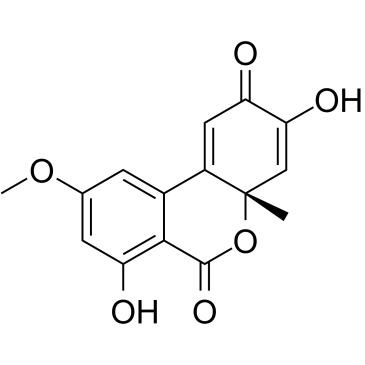
-
GC34123
Deoxycytidine triphosphate (dCTP)
Deoxycytidine triphosphate (dCTP) (dCTP) is a nucleoside triphosphate that can be used for DNA synthesis.
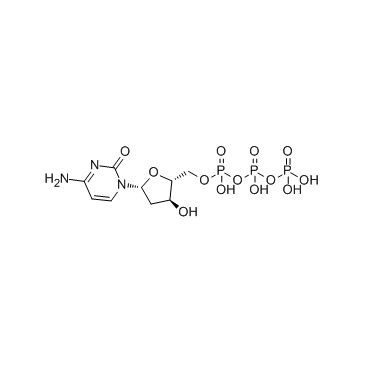
-
GC12634
Deoxyguanosine 5-triphosphate
Deoxyguanosine triphosphate (dGTP) trisodium salt is a nucleotide precursor in cells for DNA synthesis.
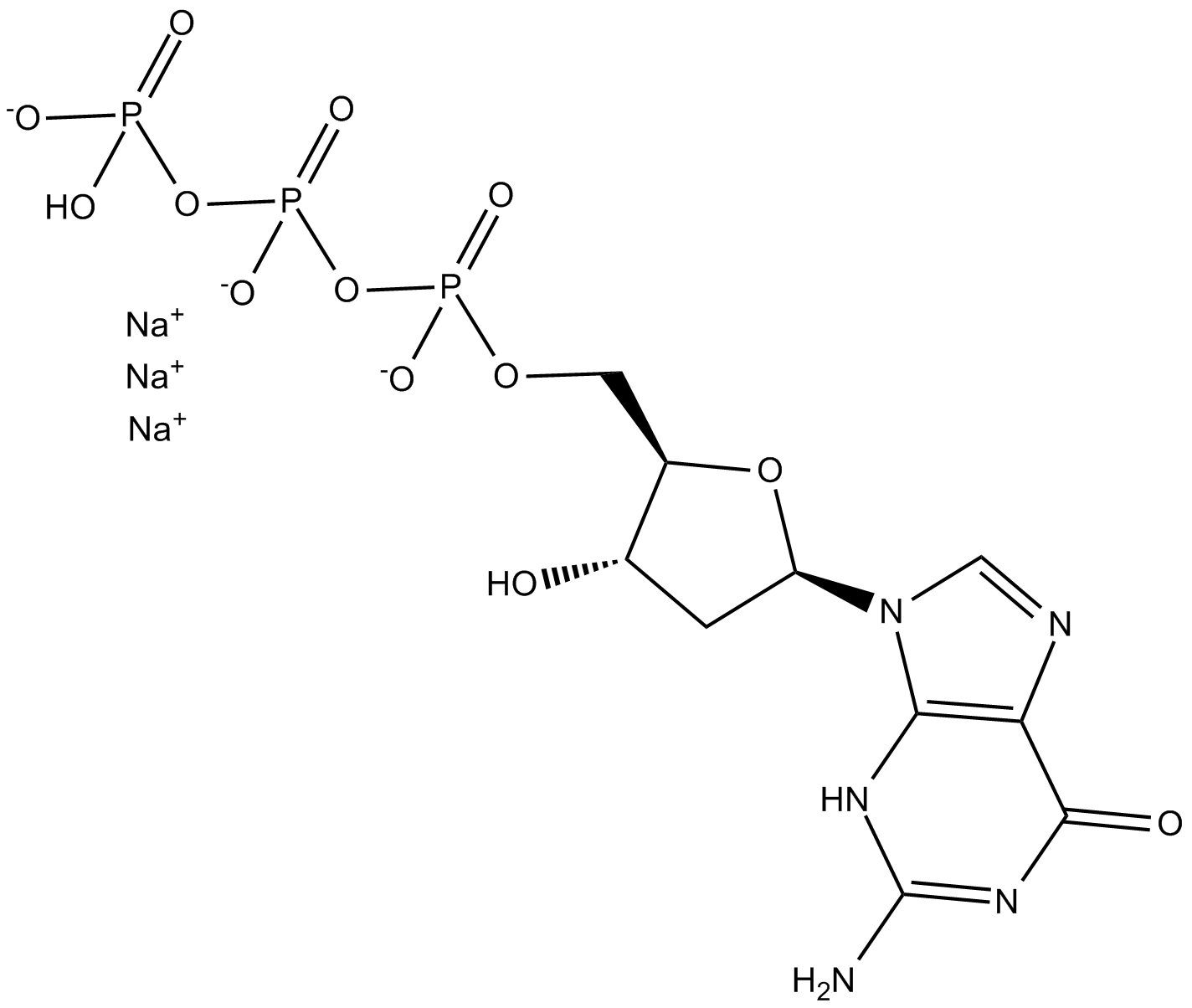
-
GC62925
Deoxythymidine-5’-triphosphate trisodium salt
Deoxythymidine-5'-triphosphate (dTTP) trisodium is one of the four nucleoside triphosphates used in the synthesis of DNA.
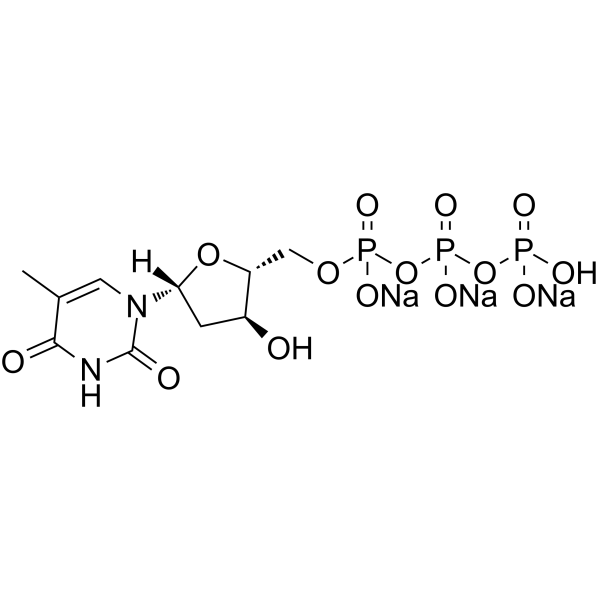
-
GC49225
Deoxyuridine 5′-monophosphate (sodium salt)
Deoxyuridine 5′-monophosphate (sodium salt) is reductively methylated to dTMP (2'-deoxythymidine 5'-monophosphate) by bisubstrate enzyme thymidylate synthase (TS).
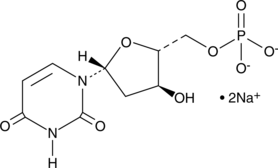
-
GC60763
DHODH-IN-11
DHODH-IN-11 (Compound 14b) is a Leflunomide derivative and a weak dihydroorotate dehydrogenase (DHODH) inhibitor with a pKa of 5.03.
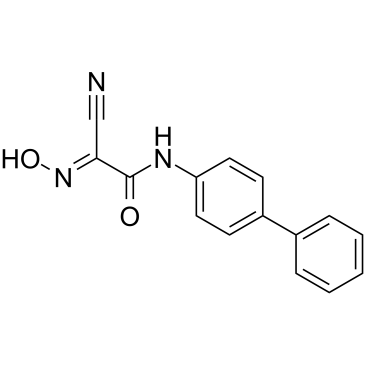
-
GC39234
DHODH-IN-2
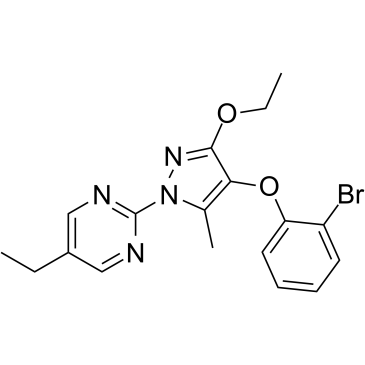
-
GC39374
DHODH-IN-5
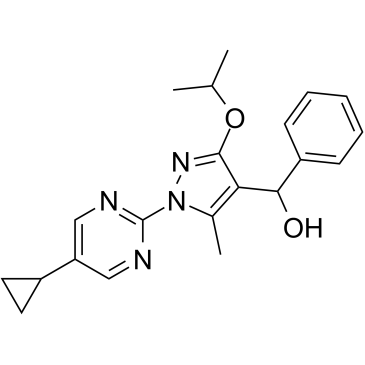
-
GC49022
Didanosine-d2
An internal standard for the quantification of didanosine
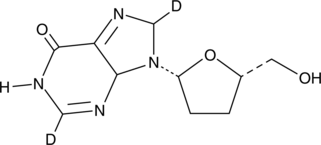
-
GC12266
Didox
synthetic antioxidant
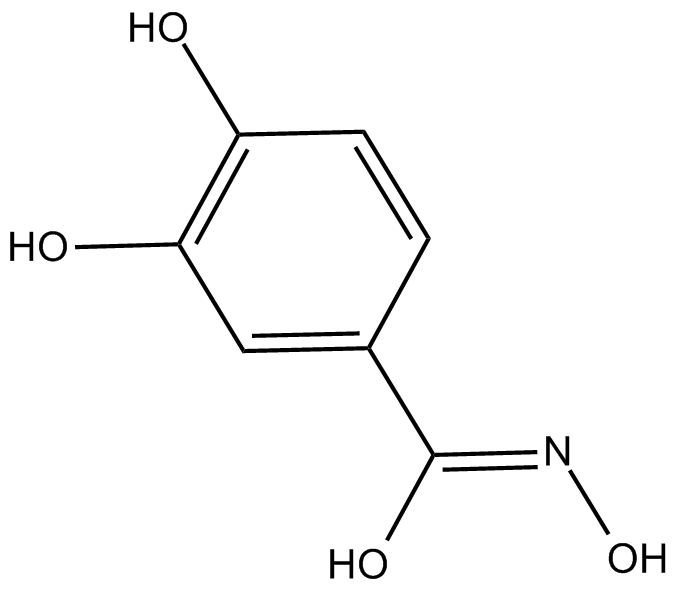
-
GC31684
Dithranol (Anthralin)
Dithranol (Anthralin) (Anthralin) is an anthraquinone derivative, with potent anti-psoriatic effects.
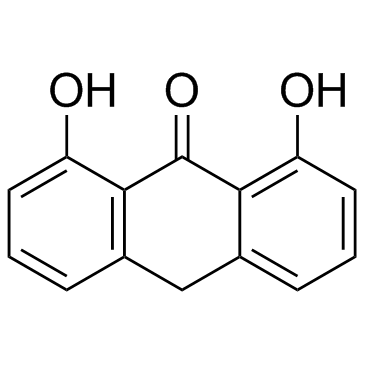
-
GC66210
DMT-2'O-MOE-rG(ib) Phosphoramidite
DMT-2'O-MOE-rG(ib) Phosphoramidite (1g), belonging to the amide family of trivalent phosphate H3PO3, is a derivative of nucleotides and guanosine and can be used in the stereochemical synthesis of phosphorothioate oligonucleotides.
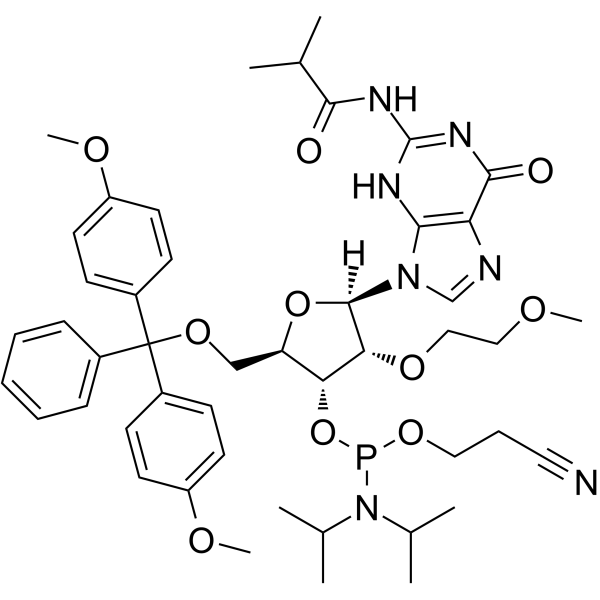
-
GC61708
Dmt-2'fluoro-da(bz) amidite
Dmt-2'fluoro-da(bz) amidite, an uniformly modified 2'-deoxy-2'-fluoro phosphorothioate oligonucleotide, is a nuclease-resistant antisense compound with high affinity and specificity for RNA targets.
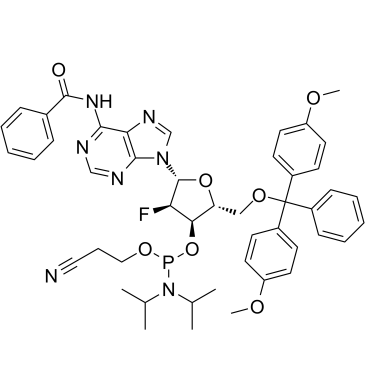
-
GC62150
DMT-dC(ac) Phosphoramidite
DMT-dC(ac) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
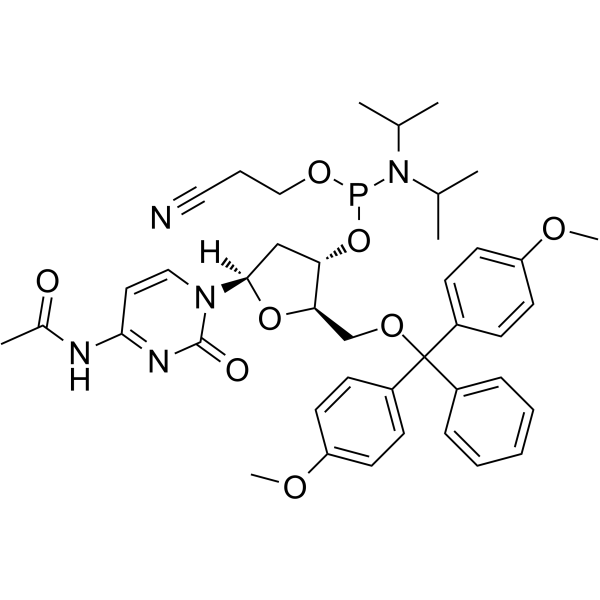
-
GC61916
DMT-dC(bz) Phosphoramidite
DMT-dC(bz) Phosphoramidite is typically used in the synthesis of DNA.
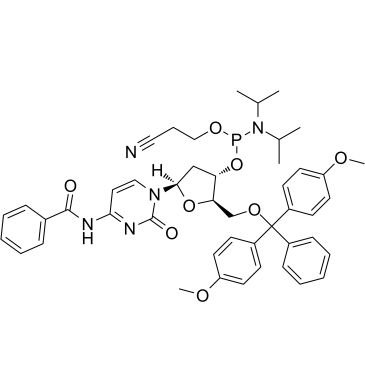
-
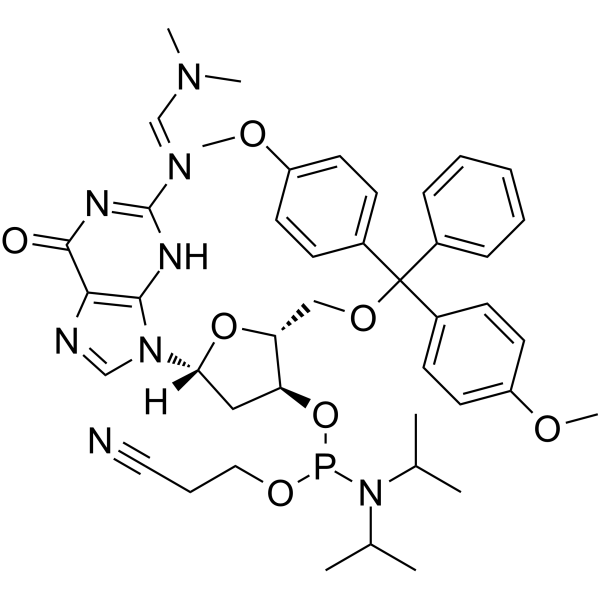
GC62538
DMT-dG(dmf) Phosphoramidite
DMT-dG(dmf) Phosphoramidite is a phosphinamide monomer that can be used in the preparation of oligonucleotides
-
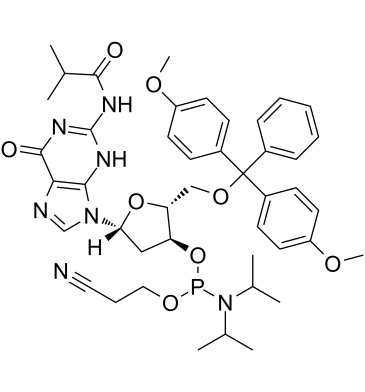
GC61912
DMT-dG(ib) Phosphoramidite
DMT-dG(ib) Phosphoramidite is typically used in the synthesis of DNA.
-
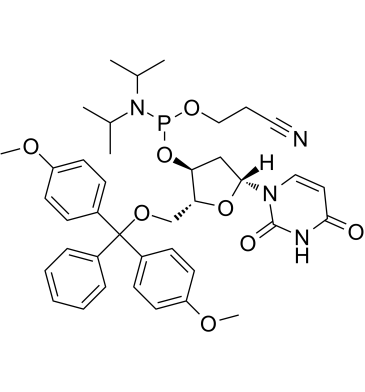
GC61913
DMT-dU-CE Phosphoramidite
DMT-dU-CE Phosphoramidite is a nucleoside molecule that can be used in DNA synthesis and DNA sequencing.
-
GC35888
DNA31
DNA31 is a potent RNA polymerase inhibitor.
-
GC15294
DPQ
DPQ is a potent inhibitor of PARPs, inhibiting PARP1 with an IC50 value of 40 nM.
-
GC38202
DTP3 TFA
DTP3 TFA is a potent and selective GADD45β/MKK7 (growth arrest and DNA-damage-inducible β/mitogen-activated protein kinase kinase 7) inhibitor. DTP3 TFA targets an essential, cancer-selective cell-survival module downstream of the NF-κB pathway.
-
GC10620
E3330
E3330 is a small-molecule inhibitor of apurinic-apyrimidinic endonuclease/redox effector factor (APE1/Ref-1) redox domain function (IC50, 50 µmol/L).
-
GC62607
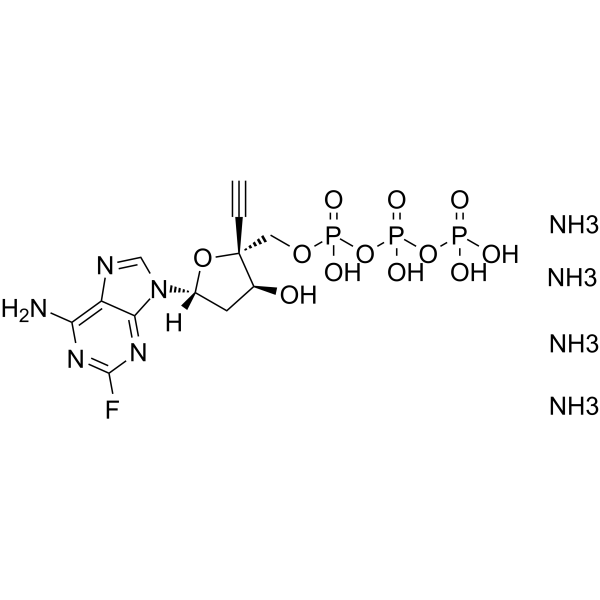
EFdA-TP tetraammonium
EFdA-TP tetraammonium is a potent nucleoside reverse transcriptase (RT) inhibitor.
-
GC11654
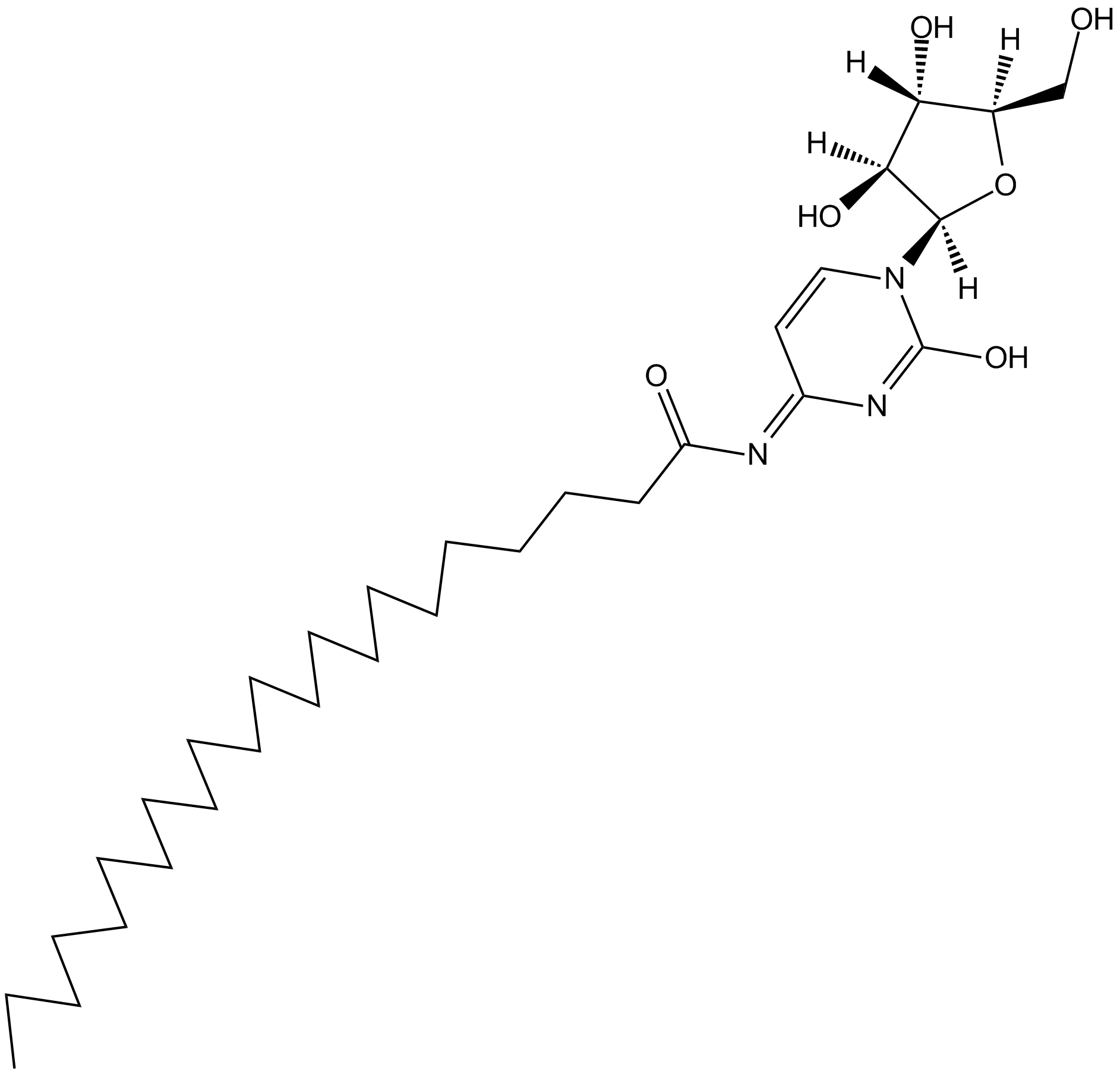
Enocitabine
nucleoside analog used as chemotherapy
-
GC48372
Erythromycin 2'-Propionate
An antibiotic