Innate Immunity
Products for Innate Immunity
- Cat.No. Product Name Information
-
GC18602
p38 MAPK Inhibitor IV
p38 MAPK Inhibitor IV is a highly specific ATP-competitive p38α MAPK inhibitor with IC50s of 0.13 and 0.55 μM for p38α and p38β MAPK, respectively.
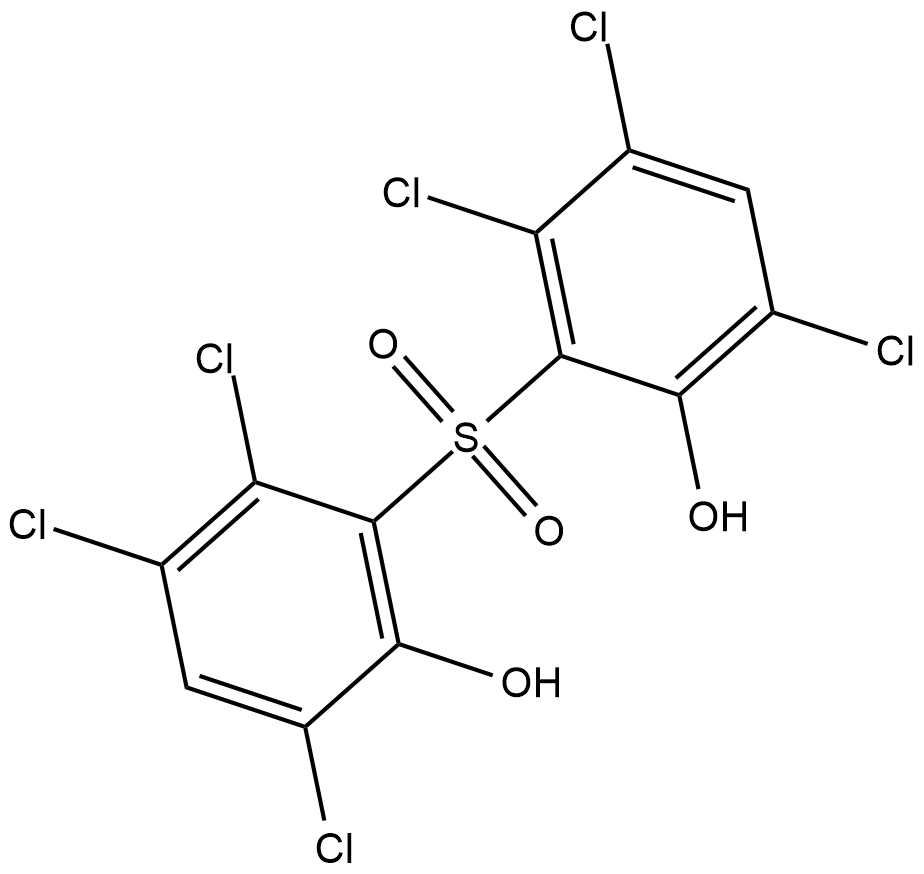
-
GC52342
P4pal10 (trifluoroacetate salt)
A PAR4 and FPR2 antagonist and an FFAR2 agonist
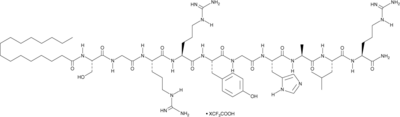
-
GC44540
PAF C-16 Carboxylic Acid
PAF C-16 is a naturally occurring phospholipid produced upon stimulation through two distinct pathways known as the "remodeling" and 'de novo' pathways.
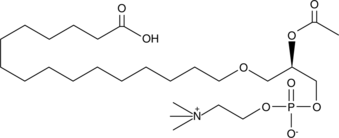
-
GC47855
PAF C-16-d4
An internal standard for the quantification of PAF C16
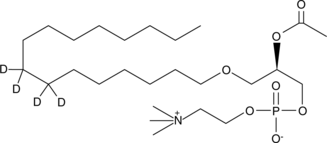
-
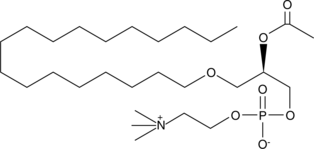
GC44541
PAF C-18
PAF C-18 is a naturally occurring phospholipid produced upon stimulation through two distinct pathways known as the "remodeling" and 'de novo' pathways.
-
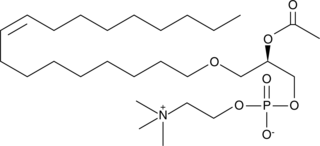
GC41450
PAF C-18:1
PAF C-18:1 is a naturally occurring phospholipid produced by cells upon stimulation and plays a role in the establishment and maintenance of the inflammatory response.
-
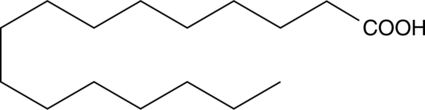
GC49024
Palmitic Acid MaxSpec® Standard
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
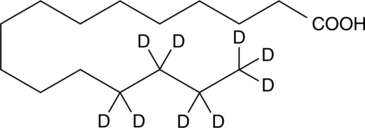
GC49023
Palmitic Acid-d9 MaxSpec® Standard
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
GC49061
Pam2CSK4 (trifluoroacetate salt)
A bacterial lipopeptide and TLR2 agonist
-
GC49422
PAR2 (1-6) amide (human) (trifluoroacetate salt)
A peptide agonist of PAR2
-
GC44579
PCTR1
Protectin conjugates in tissue regeneration 1 (PCTR1) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid.
-
GC44580
PCTR2
Protein conjugates in tissue regeneration 2 (PCTR2) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid.
-
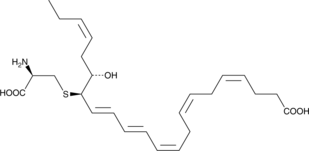
GC44581
PCTR3
Protein conjugates in tissue regeneration 3 (PCTR3) is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid.
-
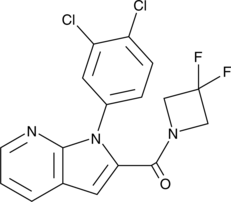
GC47931
PDE4B Inhibitor
A PDE4B inhibitor
-
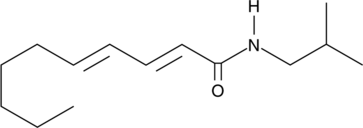
GC44590
Pellitorine
An amide alkaloid with diverse biological activities
-
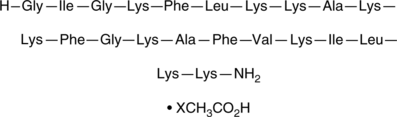
GC52127
Pexiganan (acetate)
-
GC44633
Phosphatidylserines (bovine)
Phosphatidylserine is a naturally occurring phospholipid that comprises 2-10% of total phospholipids in mammals and is enriched in the central nervous system, particularly the retina.
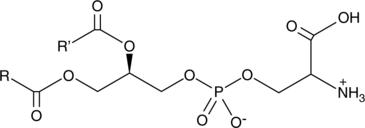
-
GC18522
Phosphatidylserines (sodium salt)
Phosphatidylserine is a naturally occurring phospholipid that comprises 2-10% of total phospholipids in mammals and is enriched in the central nervous system, particularly the retina.
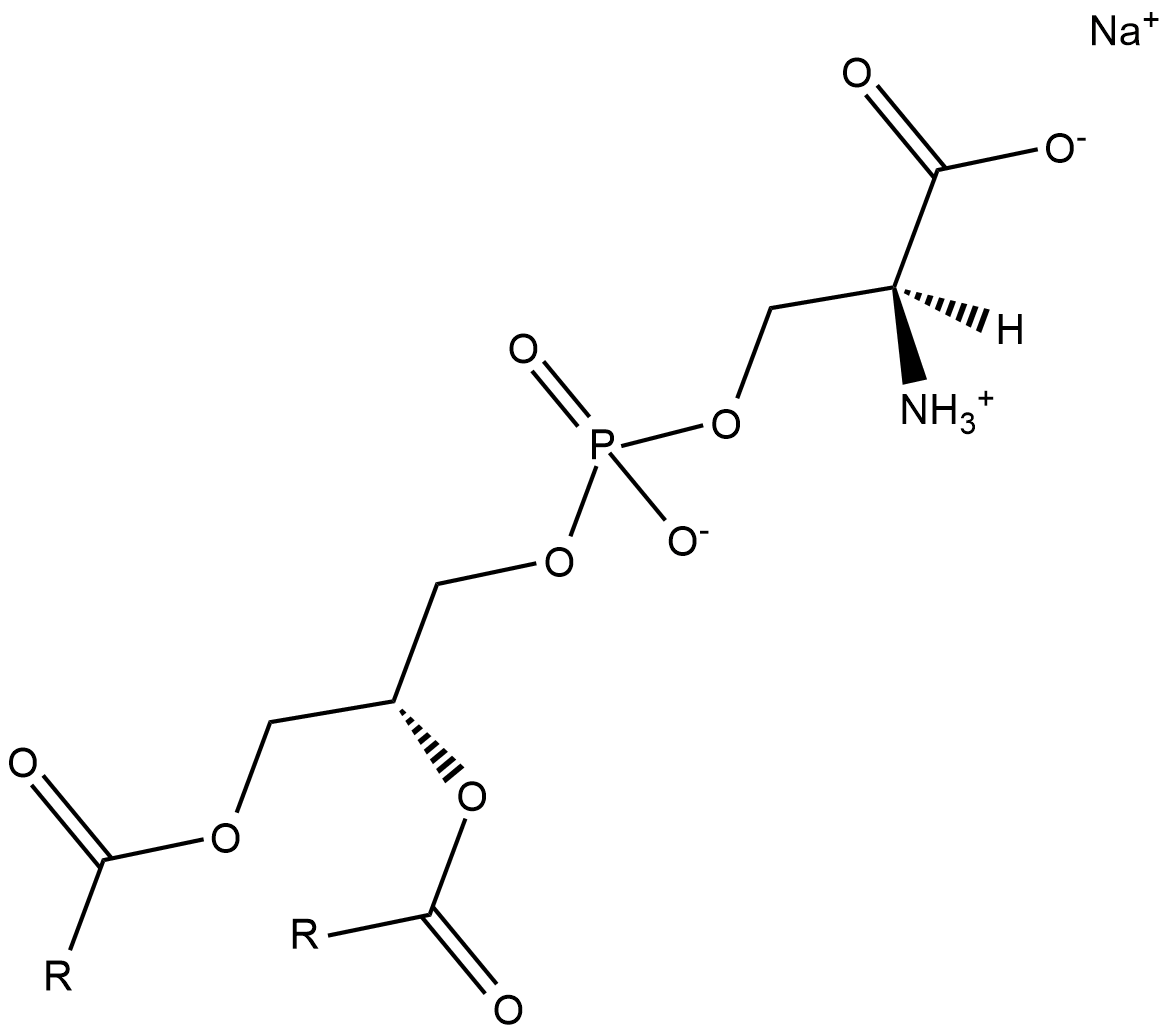
-
GC49265
PKI (14-22) amide (myristoylated) (trifluoroacetate salt)
A PKA inhibitor
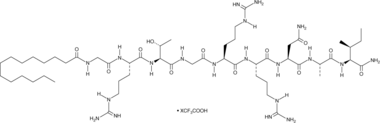
-
GC47963
Plerixafor-d4
An internal standard for the quantification of plerixafor
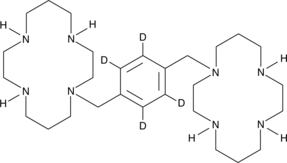
-
GC47964
pNPS-DHA
An anti-allergic DHEA derivative
-
GC52270
Pranoprofen-13C-d3
An internal standard for the quantification of pranoprofen
-
GC48501
Preterramide C
Preterramide C is a fungal metabolite.
-
GC41170
Prostaglandin D2 methyl ester
Prostaglandin D2 (PGD2) is the major eicosanoid product of mast cells and is produced in large quantities by hematopoietic PGD synthase during allergic and asthmatic anaphylaxis.
-
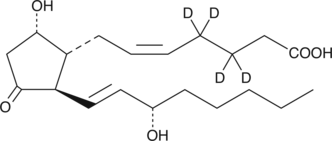
GC47988
Prostaglandin D2-d4
An internal standard for the quantification of prostaglandin D2
-
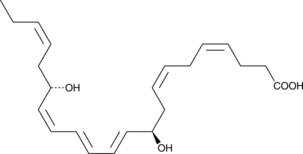
GC40981
Protectin D1
Protectin D1 is a specialized pro-resolving mediator (SPM) synthesized from docosahexaenoic acid.
-
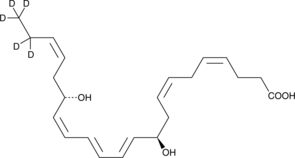
GC48993
Protectin D1-d5
An internal standard for the quantification of protectin D1
-
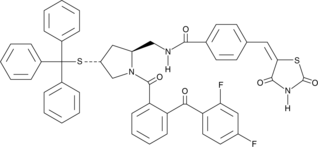
GC44794
Pyrrophenone
The group IVA phospholipase A2 (PLA2), known as calcium-dependent cytosolic PLA2 (cPLA2), selectively releases arachidonic acid (AA) from membrane phospholipids, playing a central role in initiating the synthesis of prostaglandins (PGs) and leukotrienes (LTs).
-
GC40004
Quinolactacin A
Quinolactacin A is a TNF production inhibitor.
-
GC44802
R-8507
R-8507 is a small molecule antagonist of the TNF-α type 1 receptor (TNF-αRI).
-
GC48461
rac-trans-4-hydroxy Glyburide
An active metabolite of glyburide
-
GC48035
RCTR1
A specialized pro-resolving mediator
-
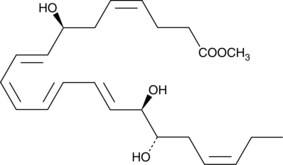
GC41231
Resolvin D2 methyl ester
Resolvin D2 (RvD2) is a lipid mediator biosynthesized by the sequential oxygenation of docosahexaenoic acid by 15- and 5-lipoxygenase.
-
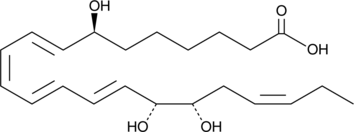
GC49279
Resolvin D2 n-3 DPA
A specialized pro-resolving mediator
-
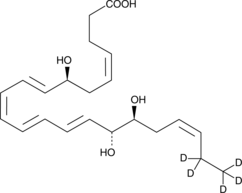
GC48042
Resolvin D2-d5
An internal standard for the quantification of RvD2
-
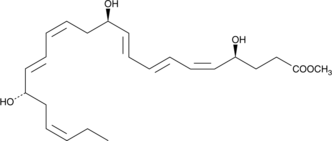
GC41232
Resolvin D3 methyl ester
Resolvin D3 methyl ester is a methyl ester version of the free acid that may act as a lipophilic prodrug form that will alter its distribution and pharmacokinetic properties.
-
GC44818
Resolvin D4
Resolvin D4 (RvD4) is a specialized pro-resolving mediator derived from docosahexaenoic acid.
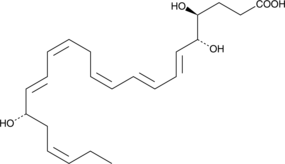
-
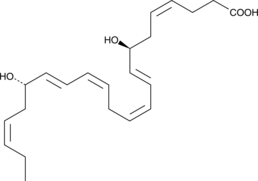
GC40982
Resolvin D5
Resolvins are a family of potent lipid mediators derived from both eicosapentaenoic acid and docosahexaenoic acid.
-
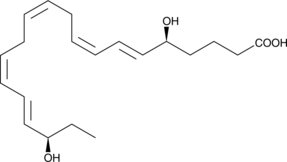
GC48972
Resolvin E2
A specialized pro-resolving mediator
-
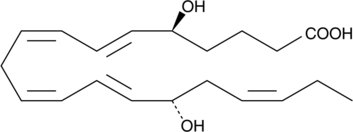
GC48045
Resolvin E4
A specialized pro-resolving mediator
-
GC44820
Resveratrol-3-O-Sulfate (sodium salt)
Resveratrol-3-O-sulfate (sodium) is the metabolite of Resveratrol.
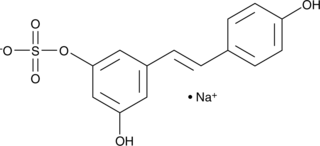
-
GC41189
Ricinelaidic Acid
Ricinelaidic acid is a 12-hydroxy fatty acid and an antagonist of leukotriene B4 (LTB4) receptors (Ki = 2 μM in porcine neutrophil membranes).
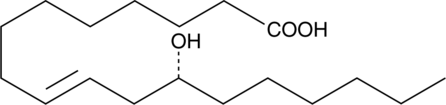
-
GC44839
Ridaifen-B
An analog of tamoxifen
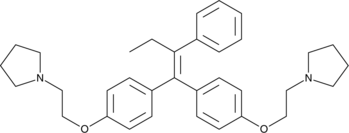
-
GC48048
Rifampicin-d3
An internal standard for the quantification of rifampicin
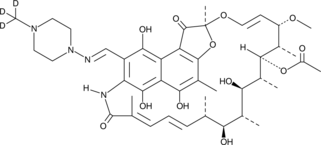
-
GC44851
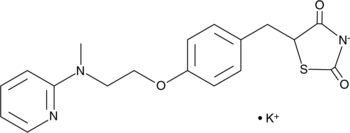
Rosiglitazone (potassium salt)
Rosiglitazone (BRL 49653) potassium is an orally active selective PPARγ agonist (EC50: 60 nM, Kd: 40 nM).
-
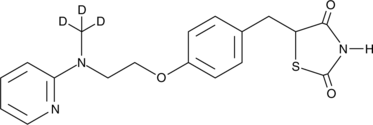
GC48059
Rosiglitazone-d3
An internal standard for the quantification of rosiglitazone
-
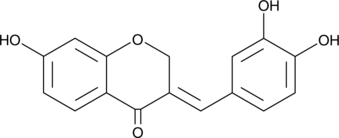
GC41626
Sappanone A
Sappanone A is a homoisoflavanone with strong antioxidant and anti-inflammatory activities.
-
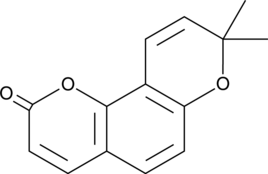
GC46221
Seselin
An angular pyranocoumarin with diverse biological activities
-
GC18821
SGA360
SGA360 is a selective modulator of the aryl hydrocarbon receptor (AhR), which is a ligand-dependent transcription factor that mediates the toxicity of certain xenobiotics and polyaromatic hydrocarbons.
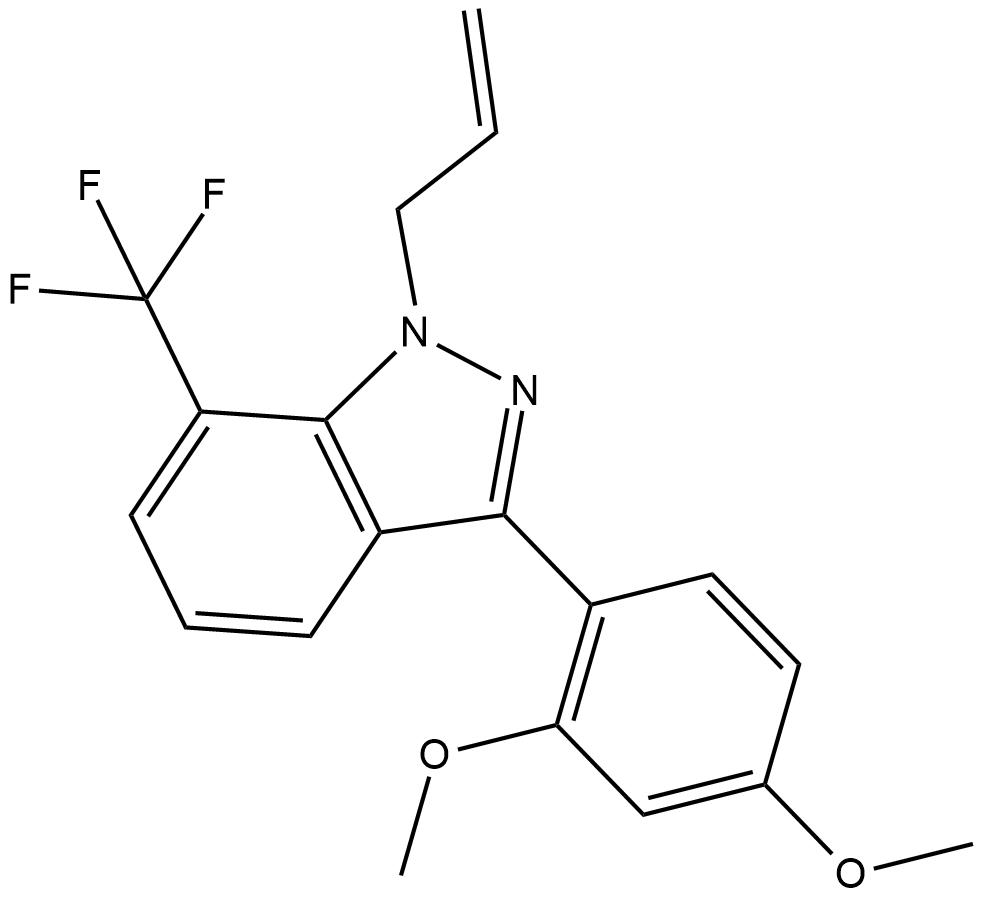
-
GC49333
SH-42
An inhibitor of DHCR24

-
GC49002
Sinigrin (hydrate)
A glucosinolate with diverse biological activities
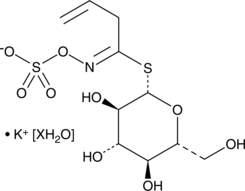
-
GC49049
SMU127
An agonist of TLR1/2
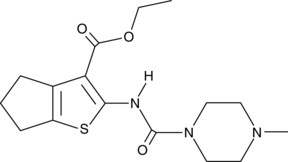
-
GC49400
SN-011
A STING antagonist
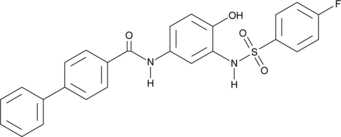
-
GC44910
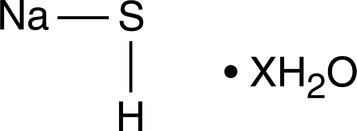
Sodium Hydrogen Sulfide (hydrate)
Hydrogen sulfide (H2S) is, like nitric oxide, an important gaseous mediator that has significant effects on the immunological, neurological, cardiovascular and pulmonary systems of mammals.
-
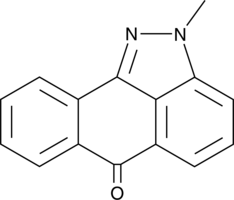
GC44919
SP 600125, negative control
SP 600125 is a selective, cell-permeable, reversible inhibitor of JNK isoforms 1-3 with Ki values of 0.19 μM.
-
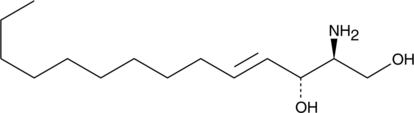
GC44930
Sphingosine (d14:1)
Sphingosine (d14:1) is a bioactive sphingolipid that has been found in B.
-
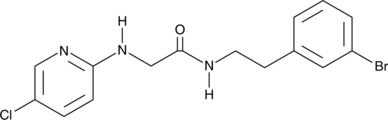
GC48098
SR 12343
An IKKβ NBD mimetic
-
GC48099
SR 12460
An IKKβ NBD mimetic
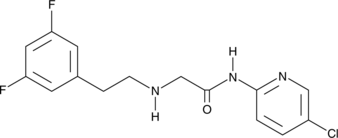
-
GC45567
SR 1903
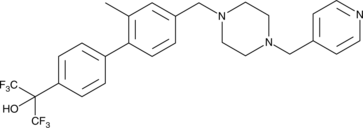
-
GC61293
SR-717
SR-717 is a stable cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) mimetic, Antitumor activity.
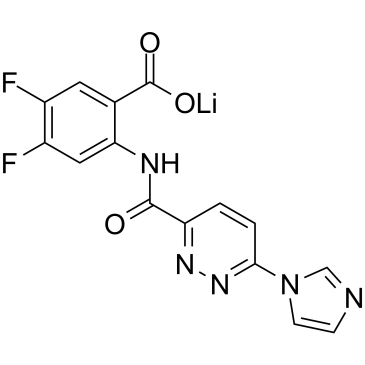
-
GC45831
SRI-011381
A TGF-β signaling activator
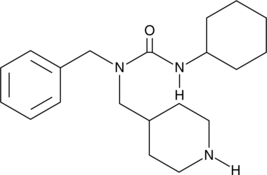
-
GC40922
SRS11-92
SRS11-92 is a ferroptosis inhibitor and a derivative of ferrostatin-1.
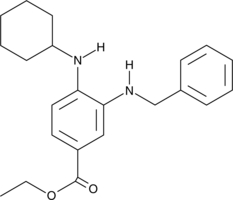
-
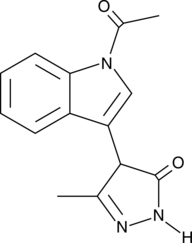
GC44948
StA-IFN-1
StA-IFN-1 is an inhibitor of the interferon (IFN) induction pathway with an IC50 value of 4.1 μM in a GFP reporter assay for IFN induction similar to TPCA-1, which specifically inhibits the IKKβ component of the IFN induction pathway.
-
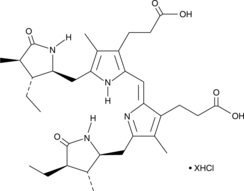
GC49844
Stercobilin (hydrochloride)
A fecal pigment and metabolite of bilirubin
-
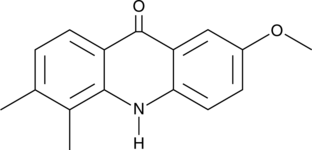
GC48109
STING Agonist 12b
A STING agonist
-
GC48110
STING Agonist 1a
STING Agonist 1a (1a) is a specific stimulator of interferon genes (STING) agonist.
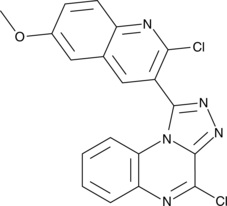
-
GC45570
STING Agonist C11
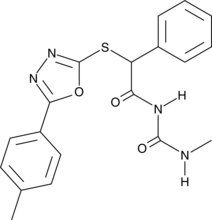
-
GC46227
STING18
A competitive STING ligand
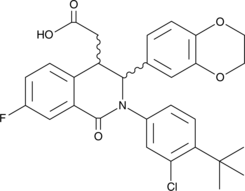
-
GC44956
Streptochlorin
Streptochlorin is a bacterial metabolite originally isolated from Streptomyces sp.
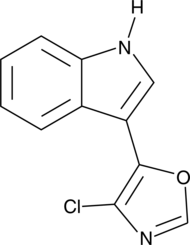
-
GC52496
Sulfatide (bovine) (sodium salt)
A mixture of isolated bovine sulfatides
-
GC44968
Sulfatides (bovine) (sodium salt)
Sulfatides are endogenous sulfoglycolipids with various biological activities in the central and peripheral nervous systems, pancreas, and immune system.
-
GC18453
Sulochrin
Sulochrin is a fungal metabolite produced by A.
-
GC48764
Sydowinin B
A xanthone polyketide with immunosuppressant activity
-
GC49047
T-5342126
A TLR4 antagonist
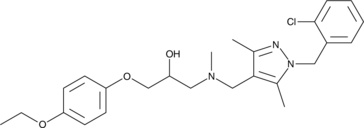
-
GC48447
TAS 205
An inhibitor of H-PGDS
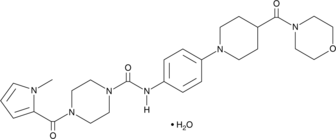
-
GC44999
Tauro-α-muricholic Acid (sodium salt)
Tauro-α-muricholic acid (TαMCA) is an antagonist of the farnesoid X receptor (FXR; IC50 = 28 μM) and a taurine-conjugated form of the murine-specific primary bile acid α-muricholic acid.
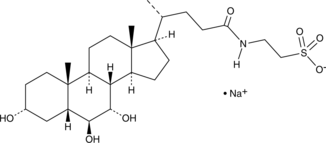
-
GC45004
TC 14012 (trifluoroacetate salt)
C-X-C chemokine receptor type 4 (CXCR4) is a receptor for the stromal cell-derived factor-1 which is designated as chemokine ligand 12 (CXCL12).
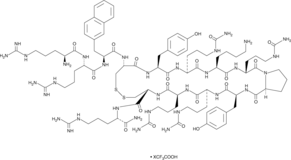
-
GC49105
Tebipenem (hydrate)
A carbenapenem antibiotic
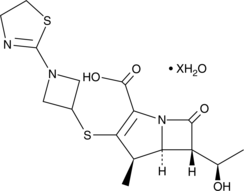
-
GC45012
Tenofovir diphosphate (sodium salt)
Tenofovir diphosphate is an inhibitor of HIV reverse transcriptase (Kis = 0.022 and 1.55 μM for RNA and DNA, respectively) and hepatitis B virus (HBV) polymerase (Ki = 0.18 μM).
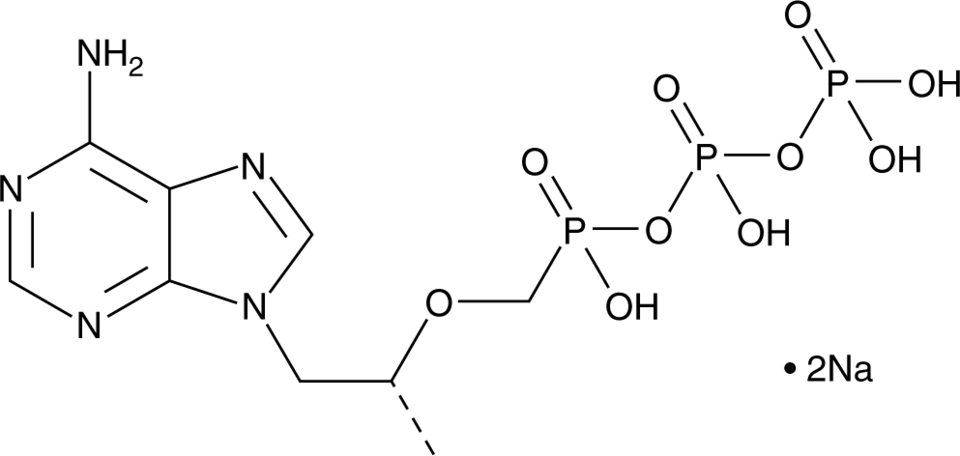
-
GC49386
Tepoxalin
A dual inhibitor of COX and 5-LO
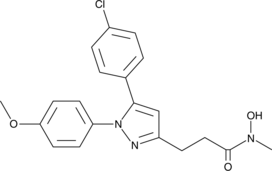
-
GC40627
tetranor-Misoprostol
tetranor-Misoprostol is a metabolite of misoprostol.
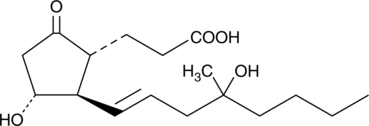
-
GC19001
Tetromycin B
Tetromycin B is a cysteine protease inhibitor with Ki values of 0.62, 1.42, 32.5, and 1.59 μM for rhodesain, falcipain-2, cathepsin L, and cathepsin B, respectively.
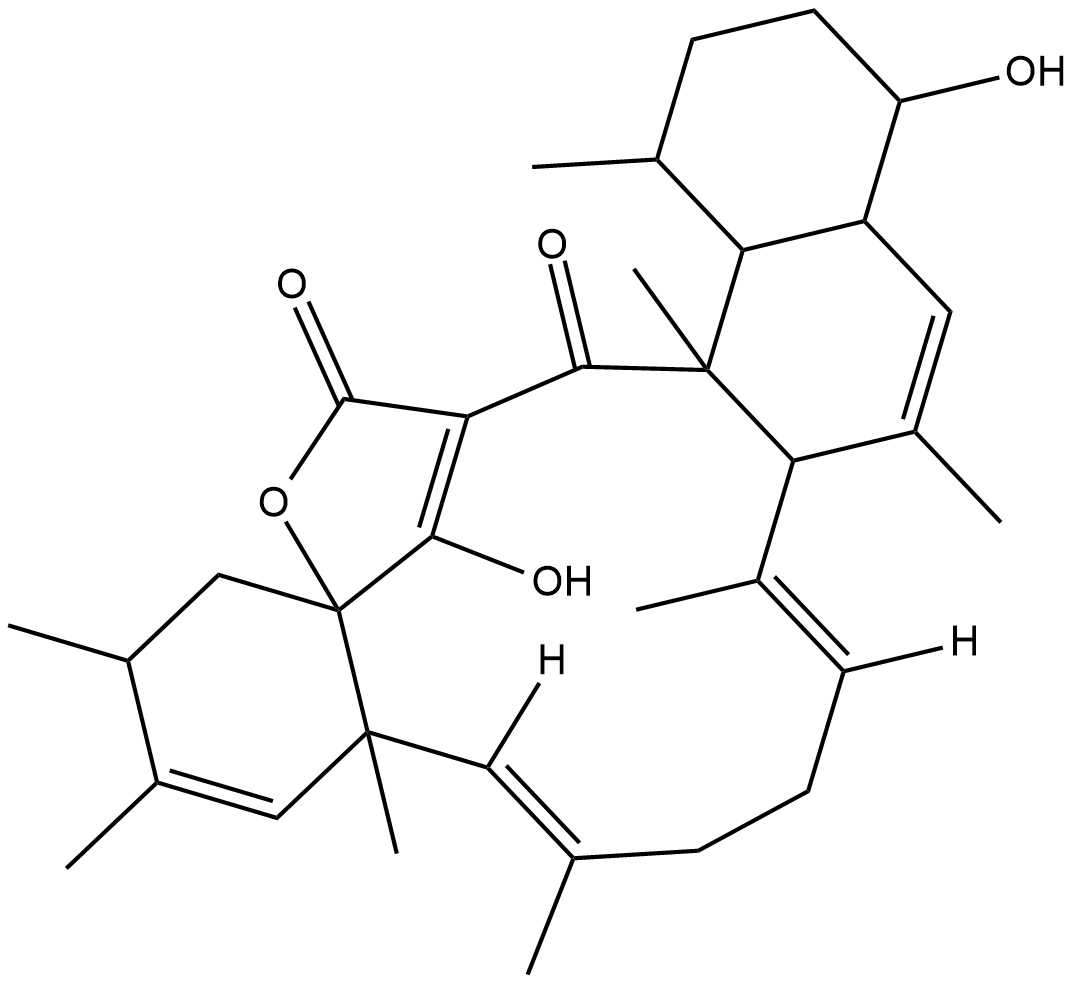
-
GC45030
TG6-129
Prostaglandin E2 evokes distinct responses through four different 'E prostanoid' (EP) receptors.
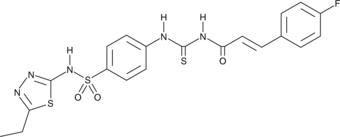
-
GC48163
Theophylline-d6
An internal standard for the quantification of theophylline
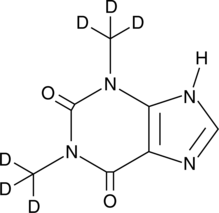
-
GC49503
Thujopsene
A sesquiterpene with diverse biological activities
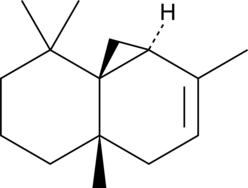
-
GC45750
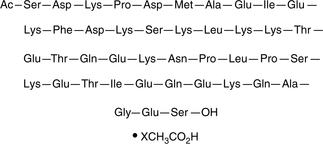
Thymosin β4 (human, mouse, rat, porcine, bovine) (acetate)
An actin-sequestering peptide
-
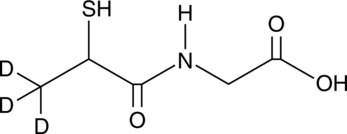
GC49062
Tiopronin-d3
An internal standard for the quantification of tiopronin
-
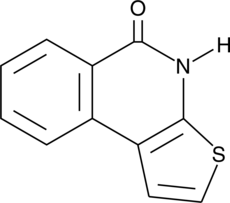
GC45057
TIQ-A
Poly(ADP-ribose) polymerase 1 (PARP1) is a critical DNA repair enzyme involved in DNA single-strand break repair via the base excision repair pathway.
-
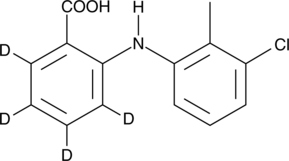
GC45965
Tolfenamic Acid-d4
An internal standard for the quantification of tolfenamic acid
-
GC45063
Toll-Like Receptor 7 Ligand II
Toll-Like Receptor 7 Ligand II (CL-087) is a potent, oral actively TLR7 agonist.
-
GC49150
Tpl2 Kinase Inhibitor (hydrochloride)
A Tpl2 inhibitor
-
GC48202
Treprostinil (diethanolamine salt)
Treprostinil (UT-15C) diethanolamine is a potent EP2, DP1 and IP agonist with Ki values of 3.6, 4.4, 32.1, 212, 826, 2505 and 4680 nM for EP2, DP1, IP, EP1, EP4, EP3 and FP, respectively.
-
GC49685
Trifluoroacetyl Tripeptide-2 (acetate)
A peptide inhibitor of pancreatic and neutrophil elastases
-
GC49884
Trigonelline-d3 (chloride)
An internal standard for the quantification of trigonelline
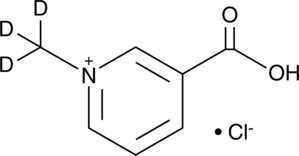
-
GC41638
TSI-01
Platelet-activating factor (PAF) is a pro-inflammatory phospholipid mediator that is rapidly synthesized by lyso-PAF acetyltransferase (lyso-PAFAT) in response to extracellular stimuli.
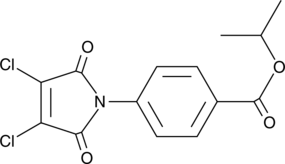
-
GC52412
TT-232 (trifluoroacetate salt)
A synthetic peptide derivative of somatostatin
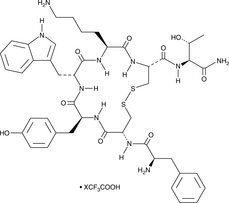
-
GC49099
Tuftsin (human) (trifluoroacetate salt)
A phagocytosis-stimulating peptide
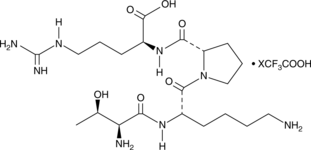
-
GC49835
Tumulosic Acid
Tumulosic Acid, a triterpenoid, inhibits KLK5 protease activity (IC50= 14.84 μM).
-
GC48987
UDP (sodium salt hydrate)
An agonist of P2Y6
-
GC52364
Vimentin (139-159)-biotin Peptide
A biotinylated vimentin peptide



