Cell Cycle/Checkpoint
Cell Cycle
Cells undergo a complex cycle of growth and division that is referred to as the cell cycle. The cell cycle consists of four phases, G1 (GAP 1), S (synthesis), G2 (GAP 2) and M (mitosis). DNA replication occurs during S phase. When cells stop dividing temporarily or indefinitely, they enter a quiescent state called G0.
Ziele für Cell Cycle/Checkpoint
- ATM/ATR(26)
- Aurora Kinase(47)
- Cdc42(4)
- Cdc7(4)
- Chk(16)
- c-Myc(20)
- CRM1(8)
- Cyclin-Dependent Kinases(91)
- E1 enzyme(1)
- G-quadruplex(14)
- Haspin(7)
- HMTase(1)
- Kinesin(26)
- Ksp(6)
- Microtubule/Tubulin(243)
- Mps1(15)
- Mitotic(11)
- RAD51(18)
- ROCK(71)
- Rho(13)
- PERK(11)
- PLK(37)
- PTEN(8)
- Wee1(7)
- PAK(21)
- Arp2/3 Complex(8)
- Dynamin(12)
- ECM & Adhesion Molecules(40)
- Cholesterol Metabolism(3)
- Endomembrane System & Vesicular Trafficking(26)
- G1(38)
- G2/M(26)
- G2/S(10)
- Genotoxic Stress(18)
- Inositol Phosphates(18)
- Proteolysis(99)
- Cytoskeleton & Motor Proteins(53)
- Cellular Chaperones(8)
Produkte für Cell Cycle/Checkpoint
- Bestell-Nr. Artikelname Informationen
-
GC43546
Dnp-PLALWAR (trifluoroacetate salt)
Dnp-PLALWAR is a fluorogenic substrate for matrix metalloproteinase-1 (MMP-1) and MMP-8.
-
GC43547
Dnp-PLAYWAR (trifluoroacetate salt)
Dnp-PLAYWAR is a fluorogenic substrate for matrix metalloproteinase-8 (MMP-8) and MMP-26.
-
GC43550
Dnp-PLGMWSR (trifluoroacetate salt)
Dnp-PLGMWSR is a fluorogenic substrate for matrix metalloproteinase-2 (MMP-2) and MMP-9.
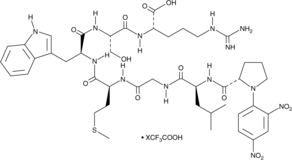
-
GC43552
Dnp-RPLALWRS (trifluoroacetate salt)
Dnp-RPLALWRS is a fluorogenic substrate for matrix metalloproteinase-7 (MMP-7).
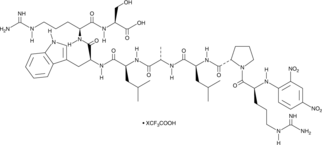
-
GC16684
Docetaxel
Docetaxel ist ein antineoplastisches Medikament, das die Depolymerisation von Mikrotubuli hemmt und die Auswirkungen der Genexpression von bcl-2 und bcl-xL abschwächt.
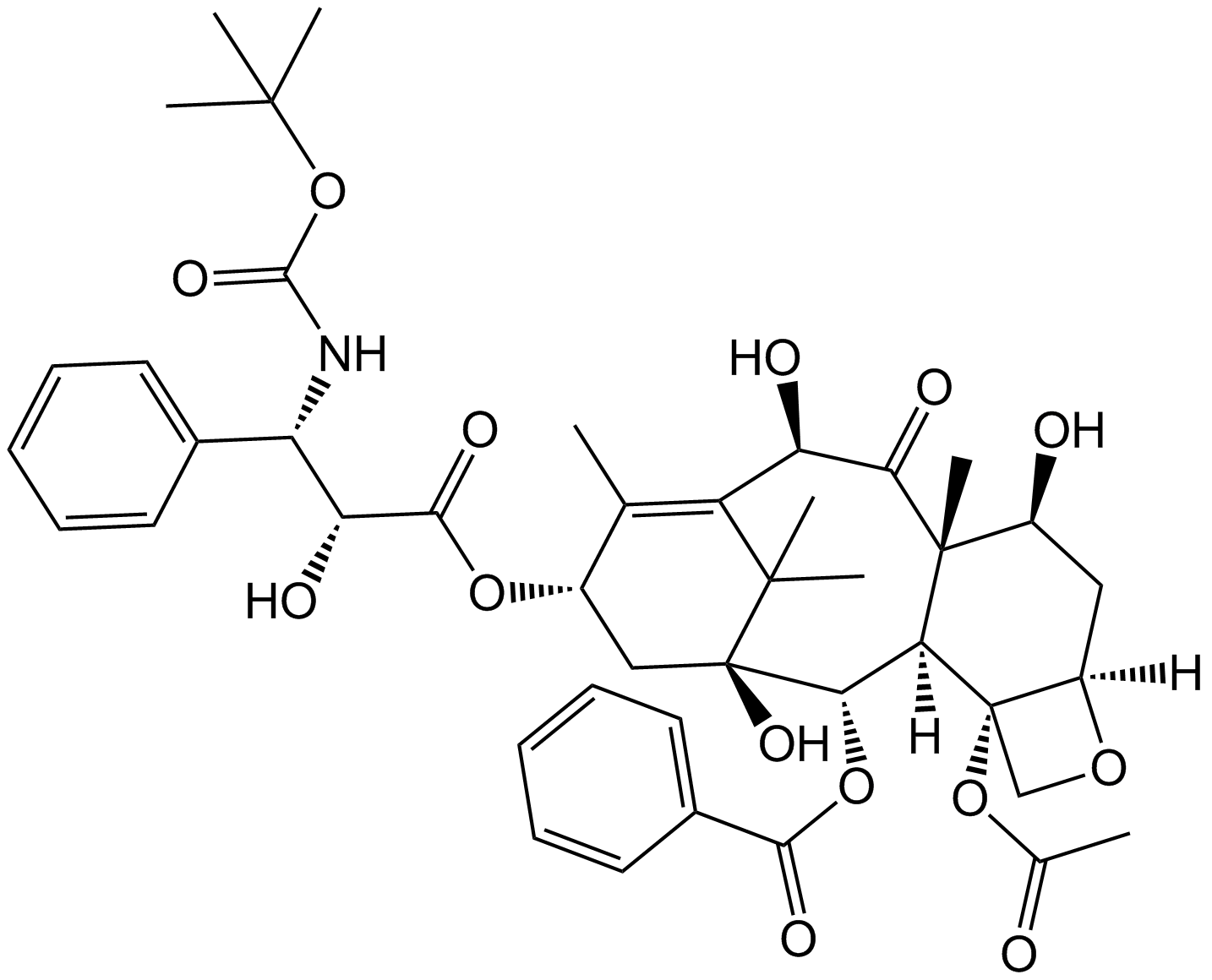
-
GC48968
Docetaxel (hydrate)
An analog of taxol with antitumor properties
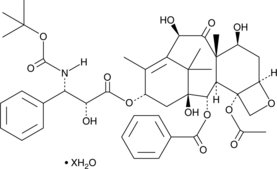
-
GC12550
Docetaxel Trihydrate
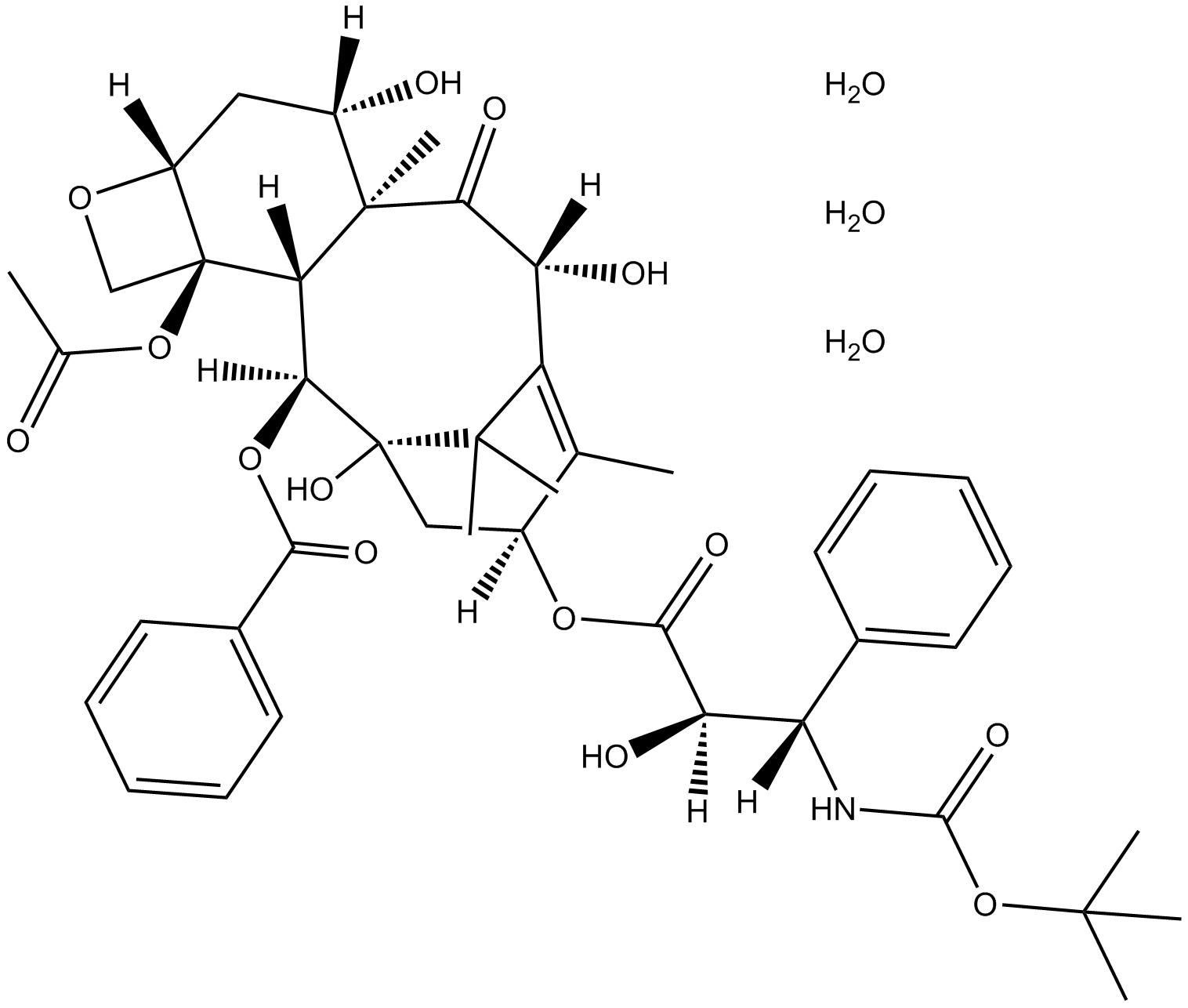
-
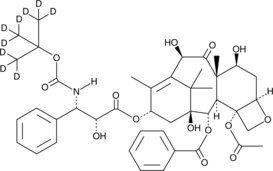
GC47250
Docetaxel-d9
Docetaxel-d9 (RP-56976-d9) ist das mit Deuterium bezeichnete Docetaxel. Docetaxel (RP-56976) ist ein Inhibitor der Mikrotubuli-Depolymerisation mit einem IC50-Wert von 0,2μM. Docetaxel dÄmpft die Wirkungen der bcl-2- und bcl-xL-Genexpression. Docetaxel stoppt den Zellzyklus bei G2/M und fÜhrt zur Zellapoptose. Docetaxel hat Anti-Krebs-AktivitÄt.
-
GC52095
Dodecyltrimethylammonium (bromide)
Dodecyltrimethylammonium(bromid) (DTAB) ist ein Tensid.

-
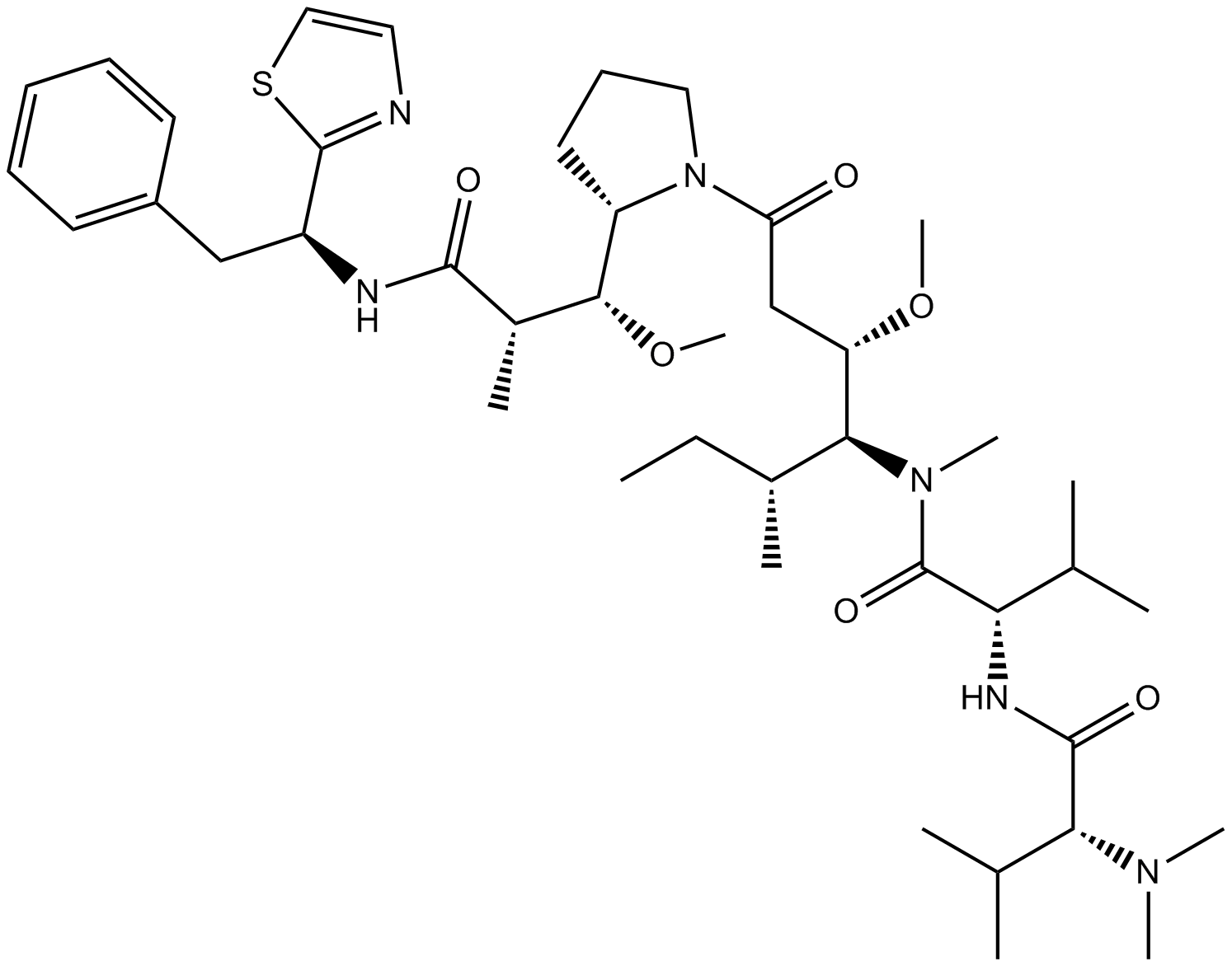
GC16273
Dolastatin 10
Dolastatin 10 (DLS 10) ist ein starkes antimitotisches Peptid, das die Tubulinpolymerisation hemmt.
-
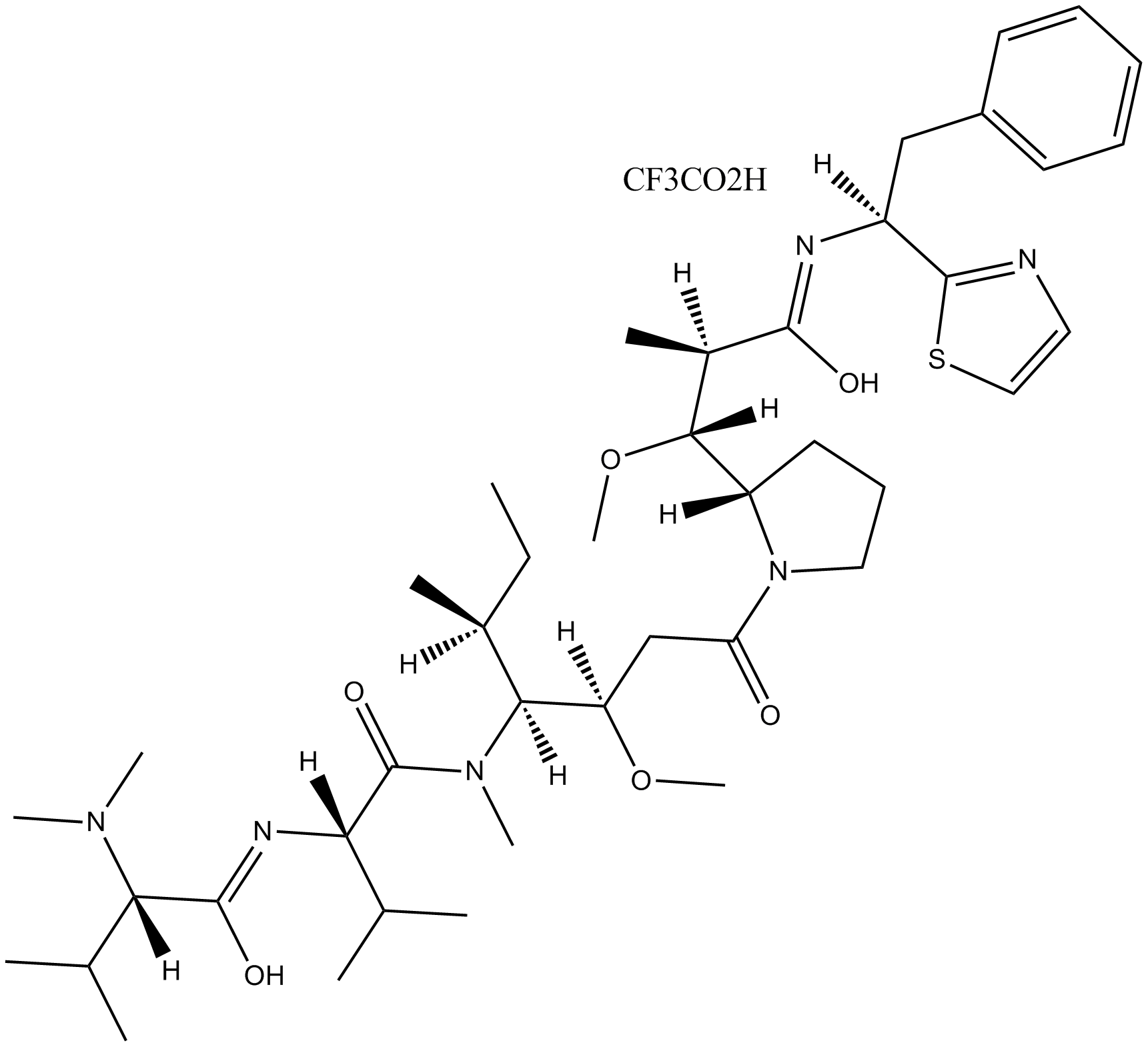
GC10557
Dolastatin 10 trifluoroacetate
Antitumor agent
-
GA21397
Dolastatin 15
Dolastatin 15 (DLS 15), ein von Dolabella auricularia abgeleitetes Depsipeptid, ist ein starkes Antimitosemittel, das strukturell mit dem Antitubulinmittel Dolastatin 10 verwandt ist. Dolastatin 15 induziert Zellzyklusstillstand und Apoptose in multiplen Myelomzellen. Dolastatin 15 kann als ADC-Zytotoxin verwendet werden.

-
GC43567
DPC-AJ1951 (trifluoroacetate salt)
DPC-AJ1951 is a peptide agonist of the parathyroid hormone (PTH)/PTH-related peptide receptor (PPR; EC50 = 0.15 nM in HEK293 cells expressing human PPR).
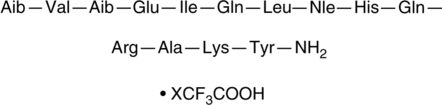
-
GC64927
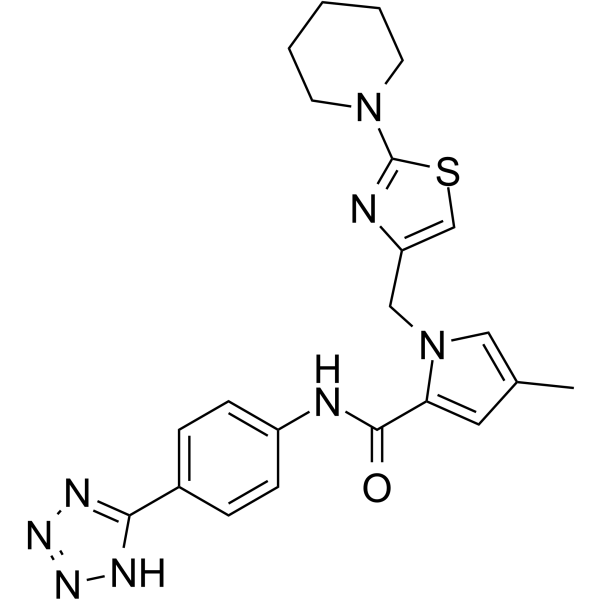
Drp1-IN-1
Drp1-IN-1 (comp A-7) ist ein Inhibitor des Dynamin-1-Ähnlichen Proteins (Drp1) mit einem IC50-Wert von 0,91 μM.
-
GC50020
Dynamin inhibitory peptide, myristoylated
DynaMin inhibitory peptide, myristoylated ist ein DynaMin-Inhibitor, der die Bindung von Amphiphysin an Dynamin stÖrt.
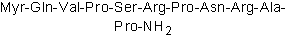
-
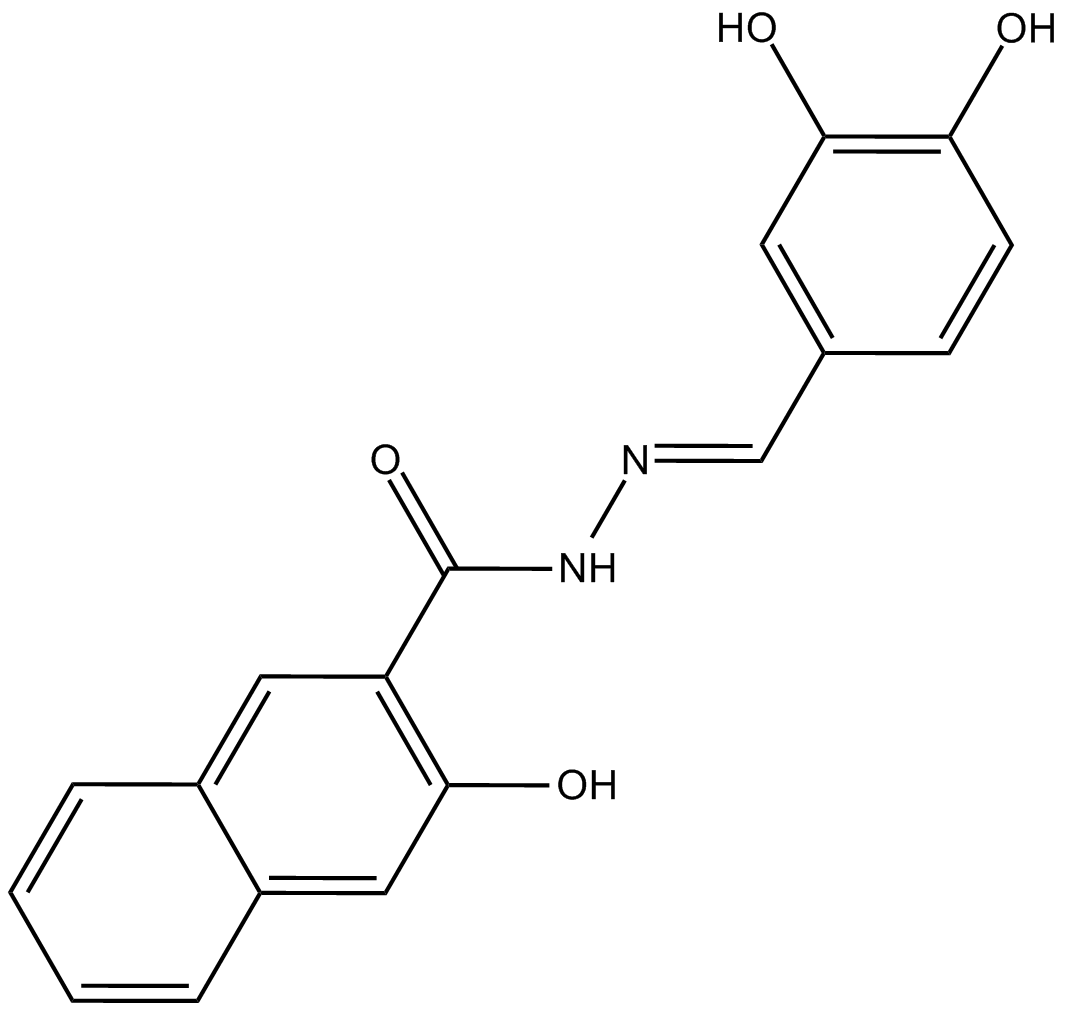
GC10395
Dynasore
Dynasore, als GTPase-Inhibitor, kann die Aktivität von Dynamin schnell und reversibel hemmen, was Endozytose verhindert.
-
GC17208
Dyngo-4a
Hydroxy Dynasore (Dyngo-4a), ein strukturelles Analogon von Dynasore, ist ein in seiner Potenz verbesserter Dynamin-Inhibitor mit geringer ZytotoxizitÄt und unspezifischer Bindung mit IC50-Werten von 0,38 &7#956;M und 2,3&7#8956;M fÜr Gehirn-Dynamin I bzw. rekombinantes Ratten-Dynamin II .
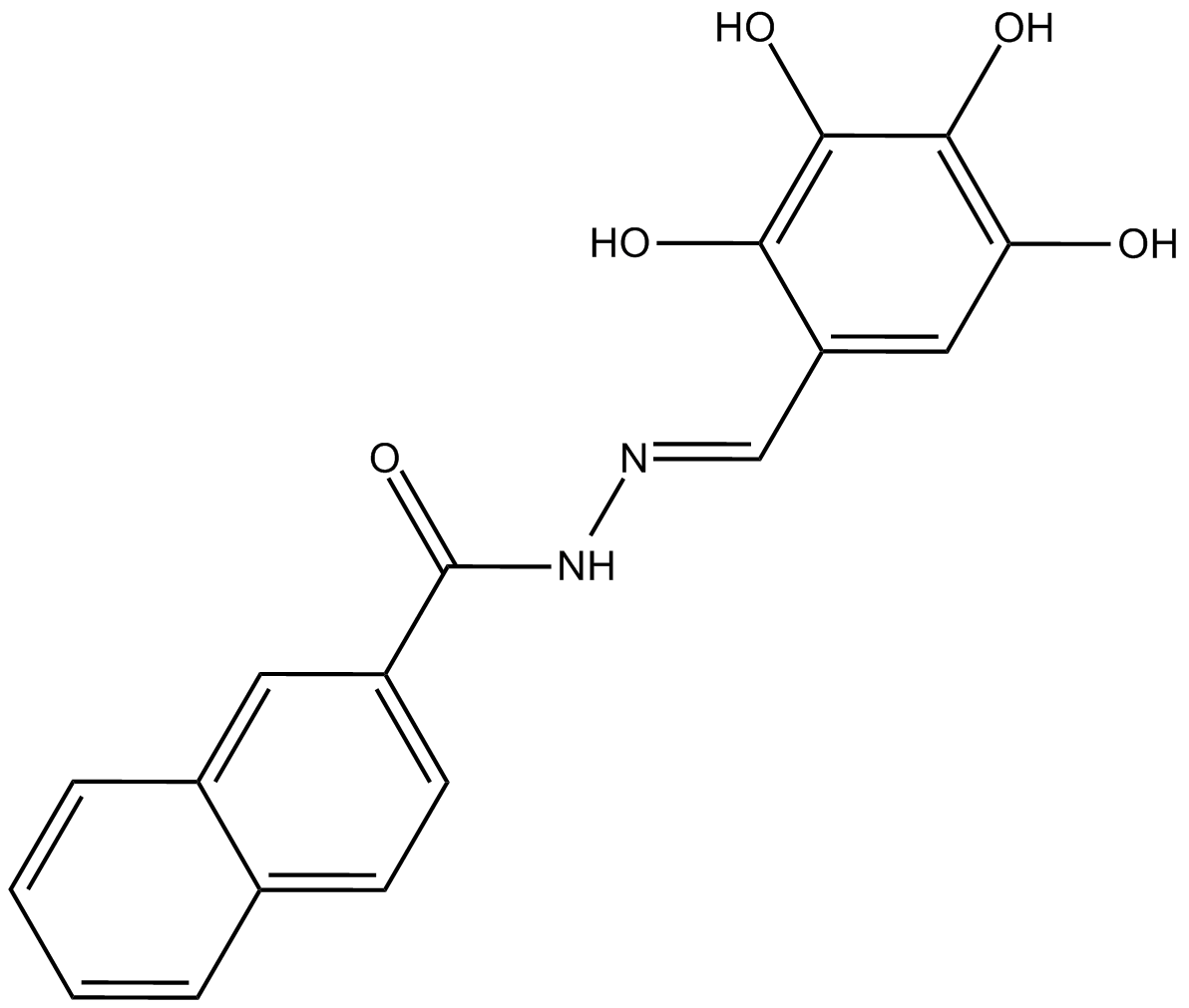
-
GC66056
Dynole 2-24
Dynole 2-24 ist ein Indol-basierter Dynamin-GTPase-Inhibitor (IC50=0,56 μ M fÜr Dynamin I). Dynole 2-24 ist nicht toxisch und zeigt eine erhÖhte Potenz gegen Dynamin I und II in vitro und in Zellen (IC\u0026sub5;\u0026sub0;(CME)=1,9 μ M). Dynole 2-24 zeigt auch eine 4,4-fache SelektivitÄt fÜr Dynamin I. Dynole 2-24 ist ein aktiver zellinterner Inhibitor der Clathrin-vermittelten Endozytose. CME: Clathrin-vermittelte Endozytose
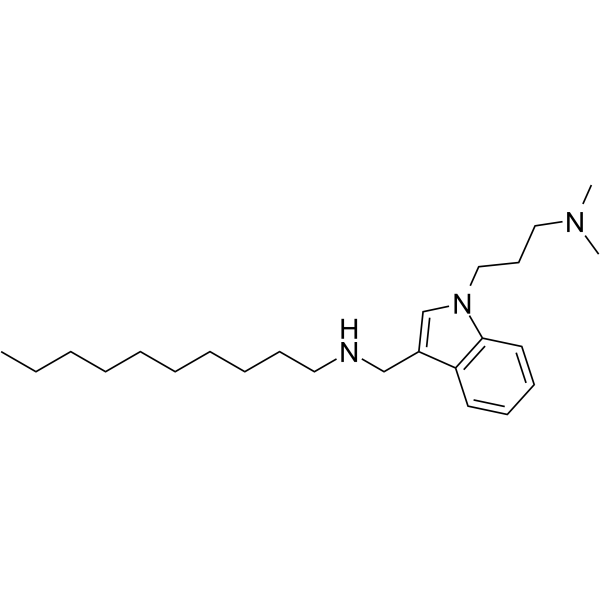
-
GC47279
Echinosporin
A bacterial metabolite with antibacterial and anticancer activities
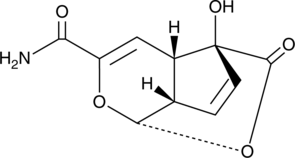
-
GC35964
Eg5 Inhibitor V, trans-24
Eg5 Inhibitor V, trans-24 ist ein potenter und spezifischer Kinesin-Eg5-Inhibitor mit einem IC50 von 0,65 μM und kann in der Krebsforschung eingesetzt werden.
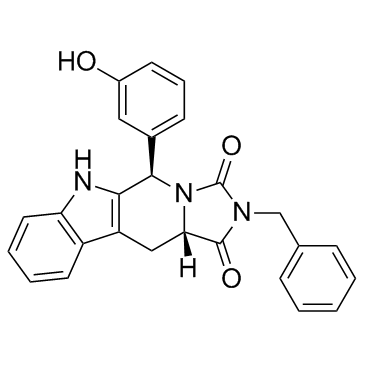
-
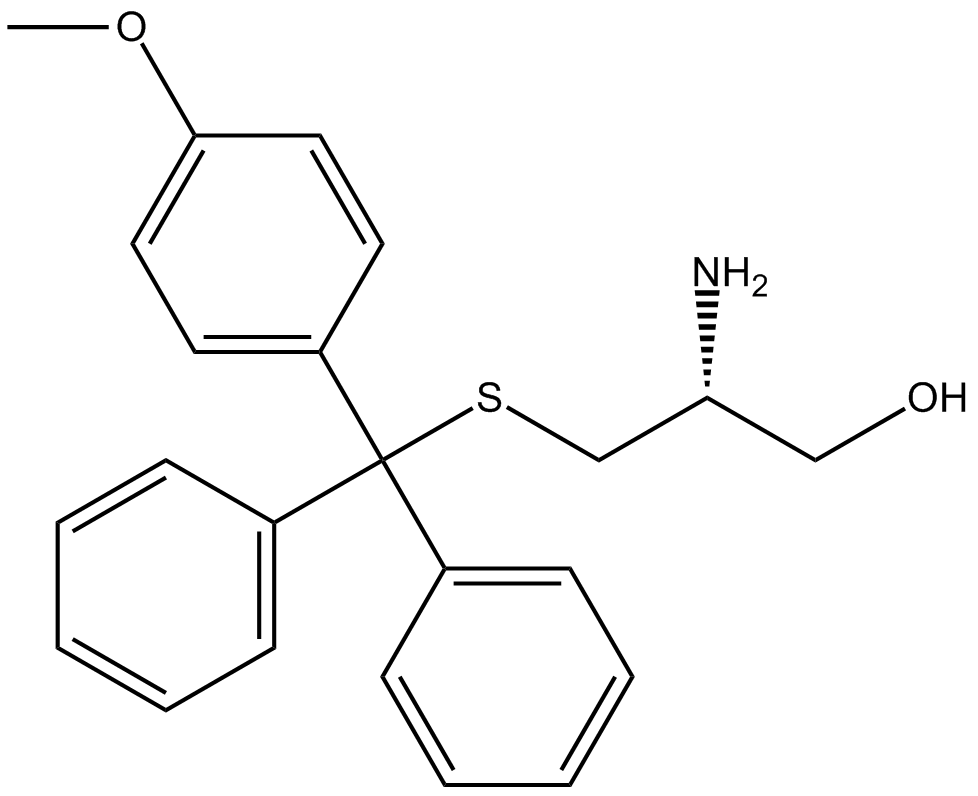
GC10681
Eg5-I
potent inhibitor of Eg5
-
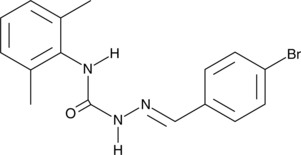
GC43588
EGA
EGA ist ein selektiver Inhibitor von Endosomol-Transportwegen, die von mehreren Toxinen und Viren ausgenutzt werden.
-
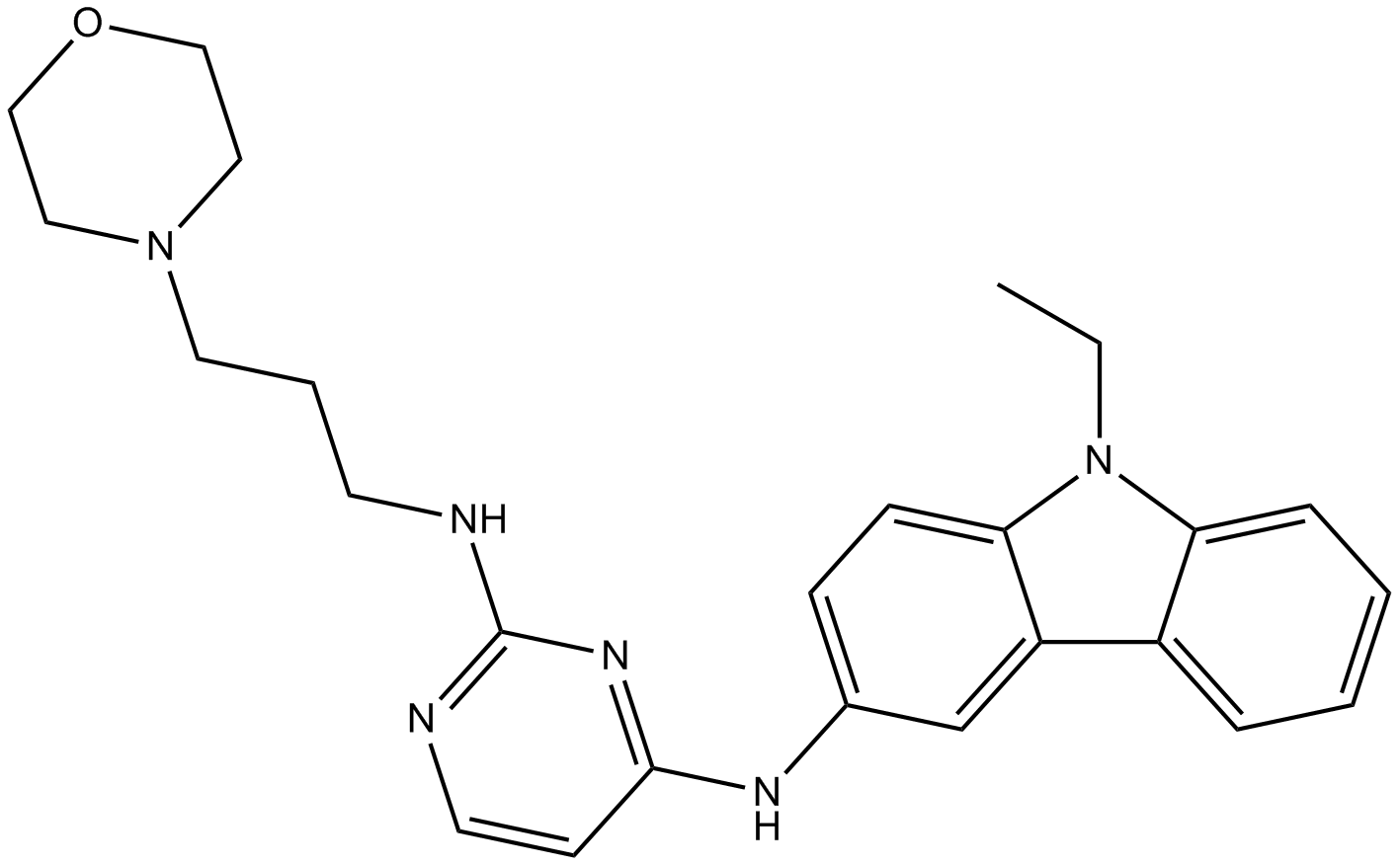
GC10030
EHop-016
Rac1/Rac3 GTPase-Inhibitor, stark und spezifisch.
-
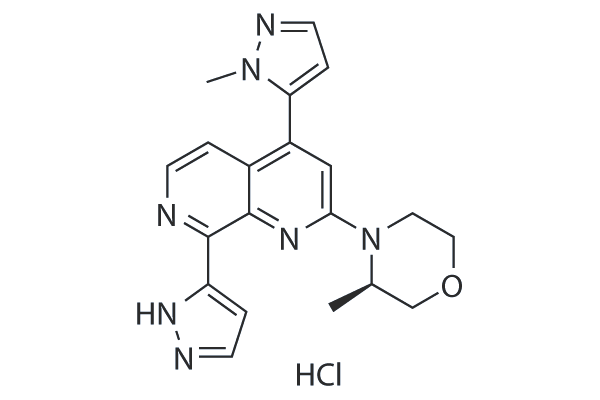
GC25372
Elimusertib (BAY-1895344) hydrochloride
Elimusertib (BAY-1895344) hydrochloride is a potent, highly selective and orally available ATR inhibitor with an IC50 of 7 nM.
-
GC16827
ELR510444
ELR510444 ist ein neuartiger Disruptor fÜr Mikrotubuli; hemmt die MDA-MB-231-Zellproliferation mit IC50 von 30,9 nM; kein Substrat fÜr den P-Glykoprotein-Medikamententransporter und behÄlt seine AktivitÄt in βIII-Tubulin-Überexprimierenden Zelllinien bei.
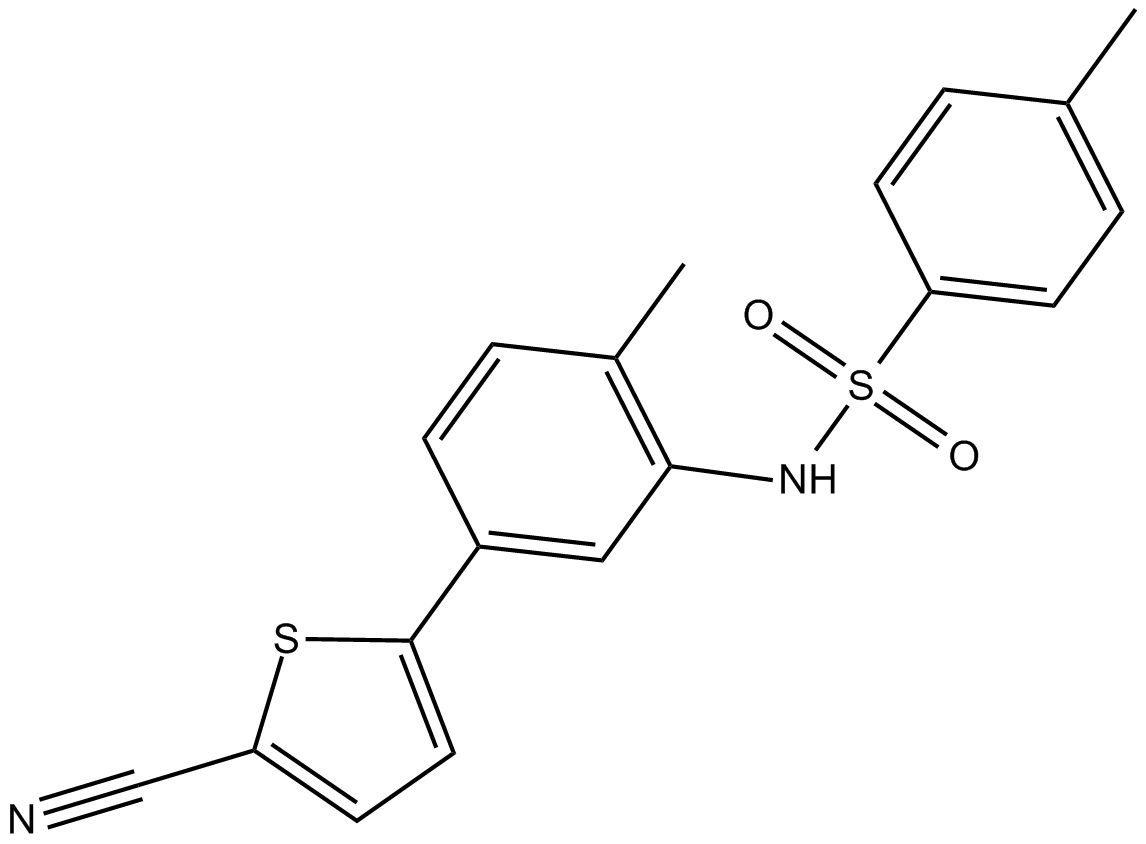
-
GC19466
Eltanexor (KPT-8602)
A second-generation exportin-1 inhibitor
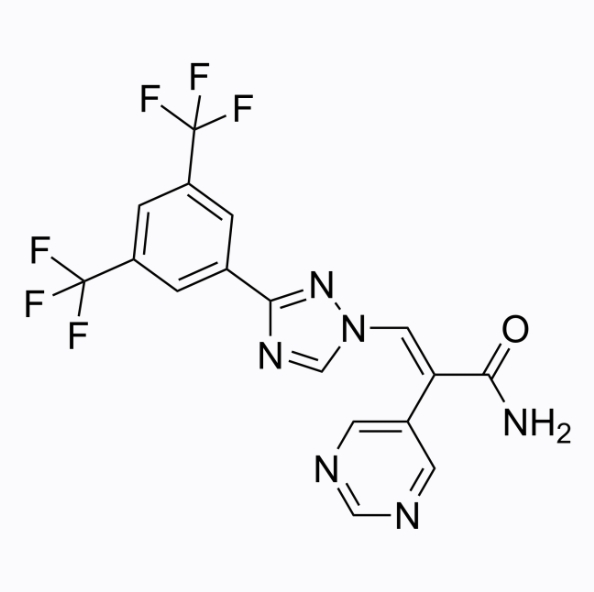
-
GC19213
Eltanexor Z-isomer
Eltanexor Z-Isomer (KPT-8602 Z-Isomer) ist das weniger aktive Isomer von KPT-8602.
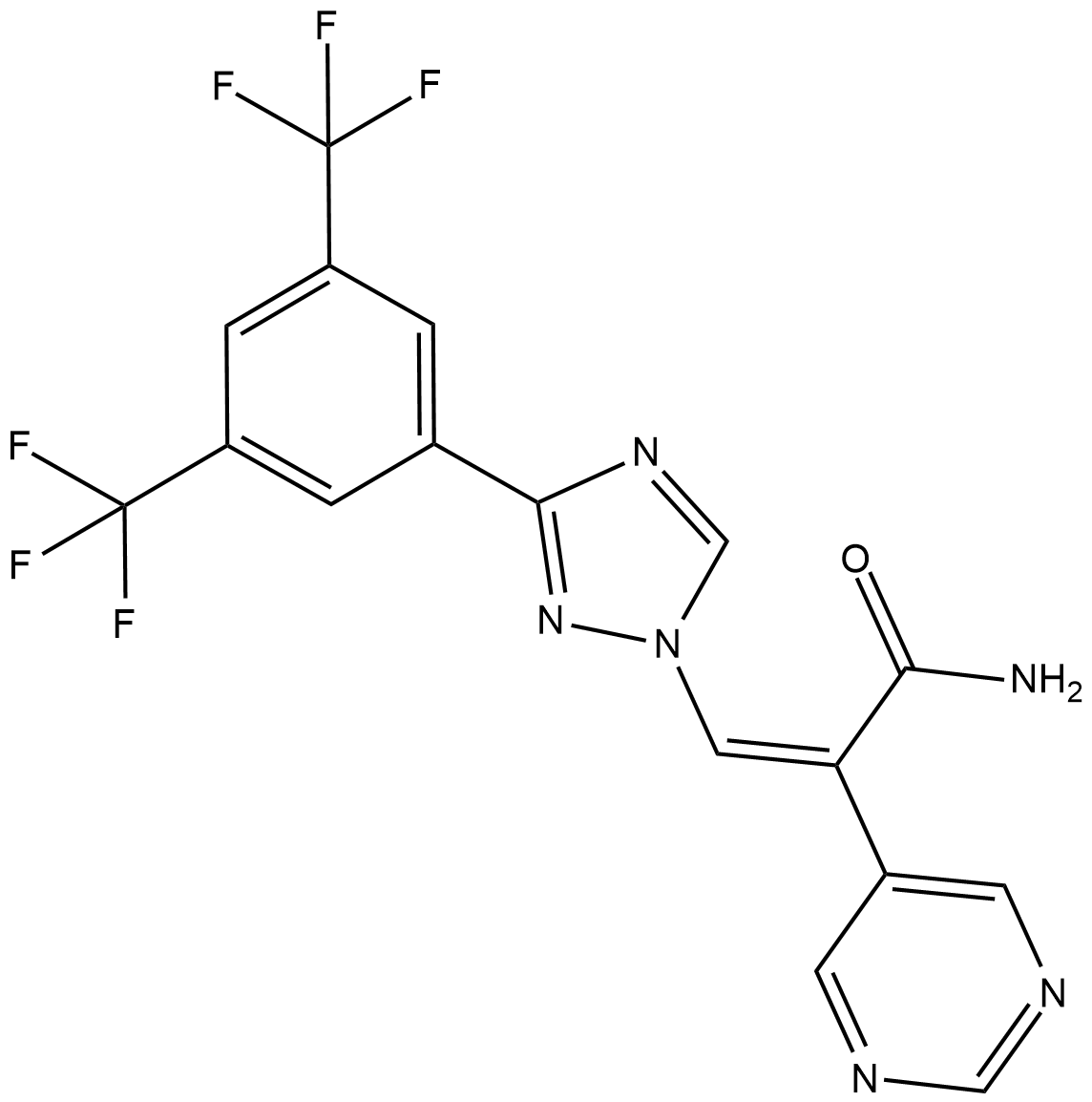
-
GC18165
EMD534085
EMD534085 ist ein potenter und selektiver Inhibitor des mitotischen Kinesin-5 mit einem IC50 von 8 nM.
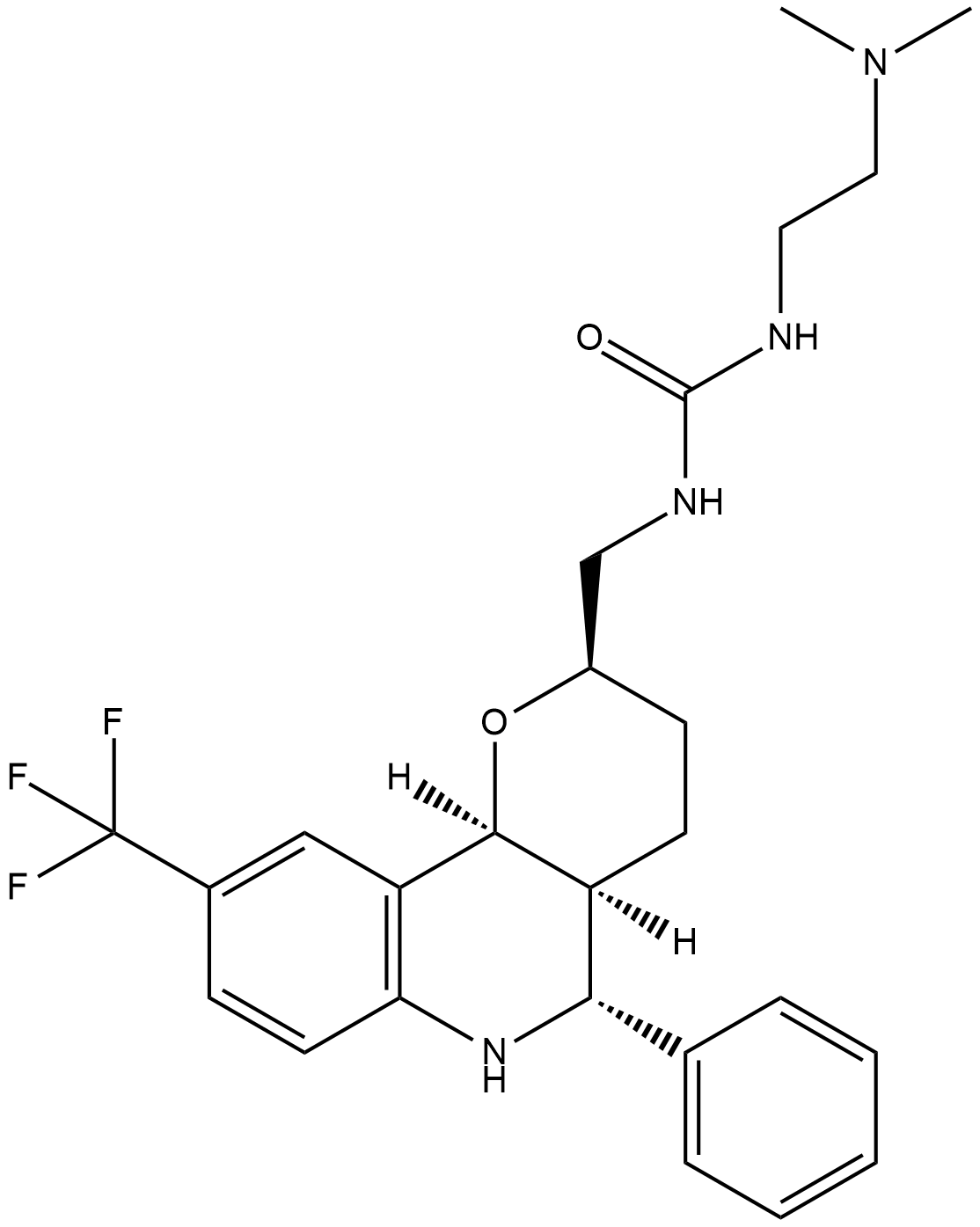
-
GC34073
Empesertib (BAY 1161909)
Empesertib (BAY 1161909) (BAY 1161909) ist ein potenter Mps1-Inhibitor mit einem IC50 von < 1 nM.
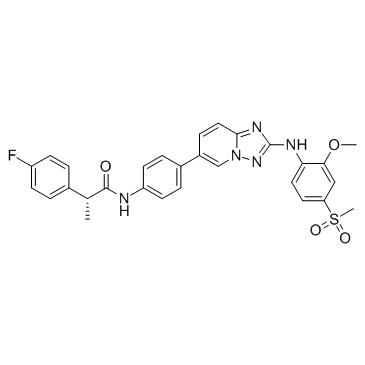
-
GC49124
EN219
EN219 ist ein mÄßig selektiver synthetischer kovalenter Ligand gegen ein N-terminales Cystein (C8) von RNF114 mit einem IC50 von 470 nM. EN219 hemmt die RNF114-vermittelte Autoubiquitinierung und p21-Ubiquitinierung.
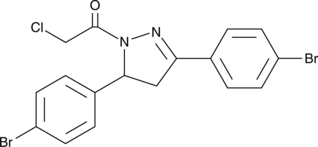
-
GC62287
EN4
EN4 ist ein kovalenter Ligand, der auf Cystein 171 (C171) von MYC abzielt. EN4 ist selektiv fÜr c-MYC gegenÜber N-MYC und L-MYC. EN4 hemmt die MYC-TranskriptionsaktivitÄt, reguliert MYC-Targets herunter und beeintrÄchtigt die Tumorentstehung.
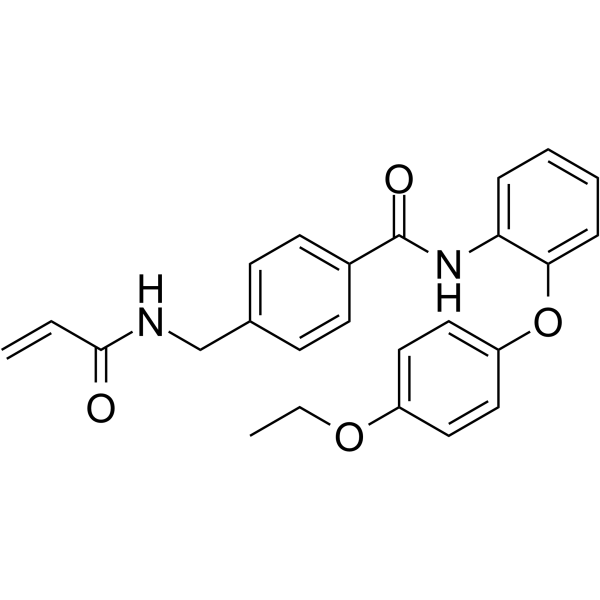
-
GC19010
Endosidin 2
Ein zellpermeabler Inhibitor der Exozytose und des endosomalen Recyclings.
-
GC43605
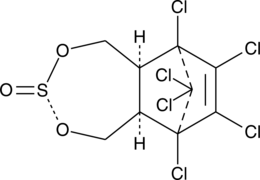
Endosulfan II
Endosulfan II is an organochlorine insecticide and a stereoisomer of endosulfan I.
-
GC47291
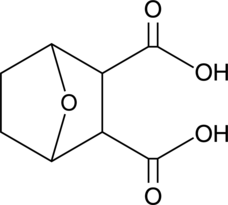
Endothall
An herbicide
-
GC33125
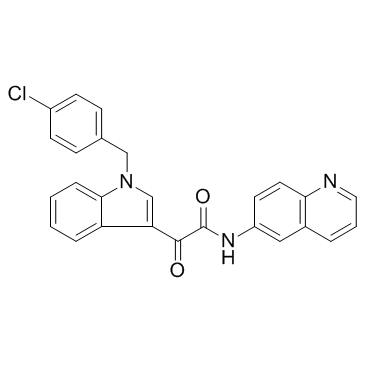
Entasobulin
Entasobulin ist ein β-Tubulin-Polymerisationsinhibitor mit potenzieller AntikrebsaktivitÄt.
-
GC43614
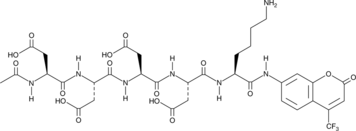
Enteropeptidase Fluorogenic Substrate
Enteropeptidase fluorogenic substrate is a substrate for enteropeptidase that contains a 7-amino-4-trifluoromethylcoumarin (AFC) moiety.
-
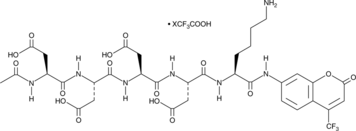
GC48995
Enteropeptidase Fluorogenic Substrate (trifluoroacetate salt)
A fluorogenic substrate of enteropeptidase
-
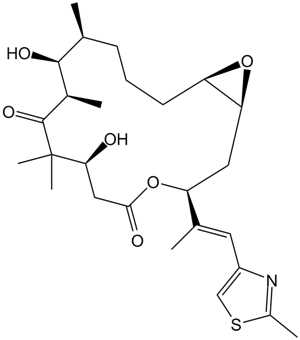
GC11202
Epothilone A
Epothilon A ist ein kompetitiver Inhibitor der Bindung von [3H]-Paclitaxel an Tubulinpolymere mit einem Ki von 0,6-1,4 μM.
-
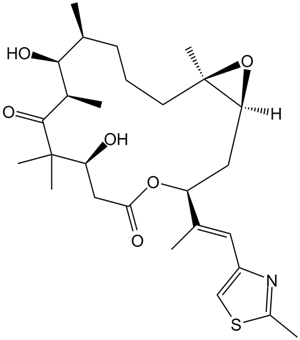
GC17240
Epothilone B (EPO906, Patupilone)
Epothilone B (EPO906, Patupilone) ist ein Mikrotubuli-Stabilisator mit einem Ki von 0,71&7#956; M.
-
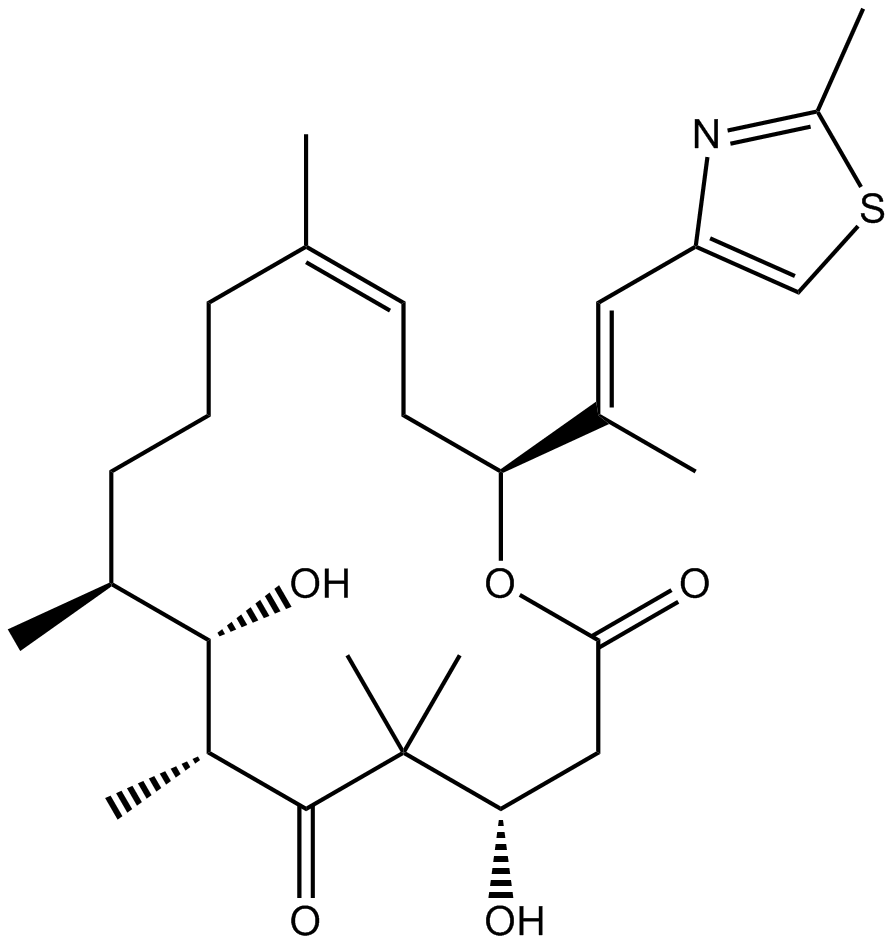
GC17440
Epothilone D
A microtubule-stabilizing agent
-
GC14354
Eribulin
Eribulin (E7389) ist ein Mikrotubuli-Targeting-Wirkstoff, der fÜr die Erforschung von metastasierendem Brustkrebs verwendet wird. Eribulin hemmt die Proliferation von Krebszellen, indem es Mikrotubuli-Proteine und Mikrotubuli bindet.
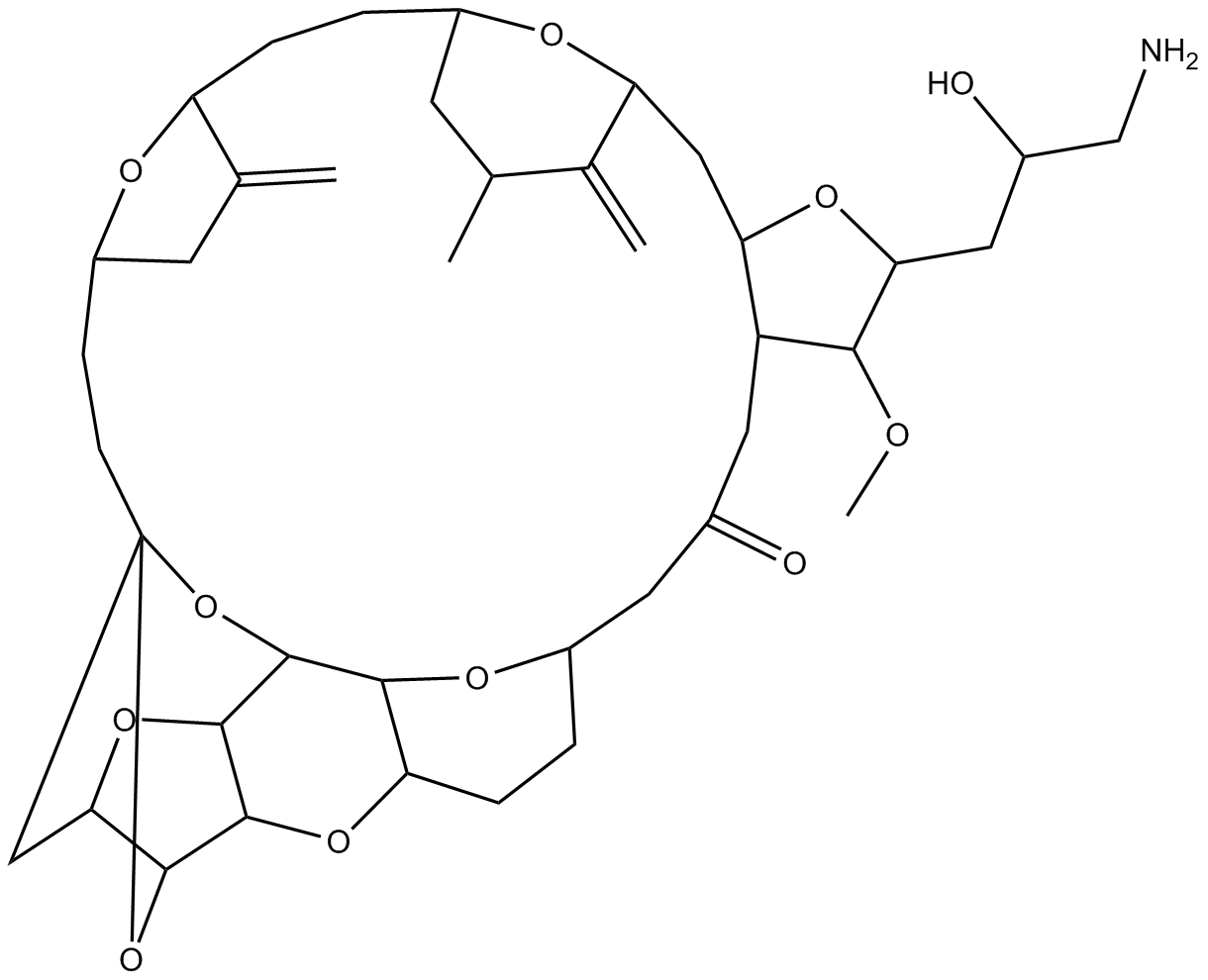
-
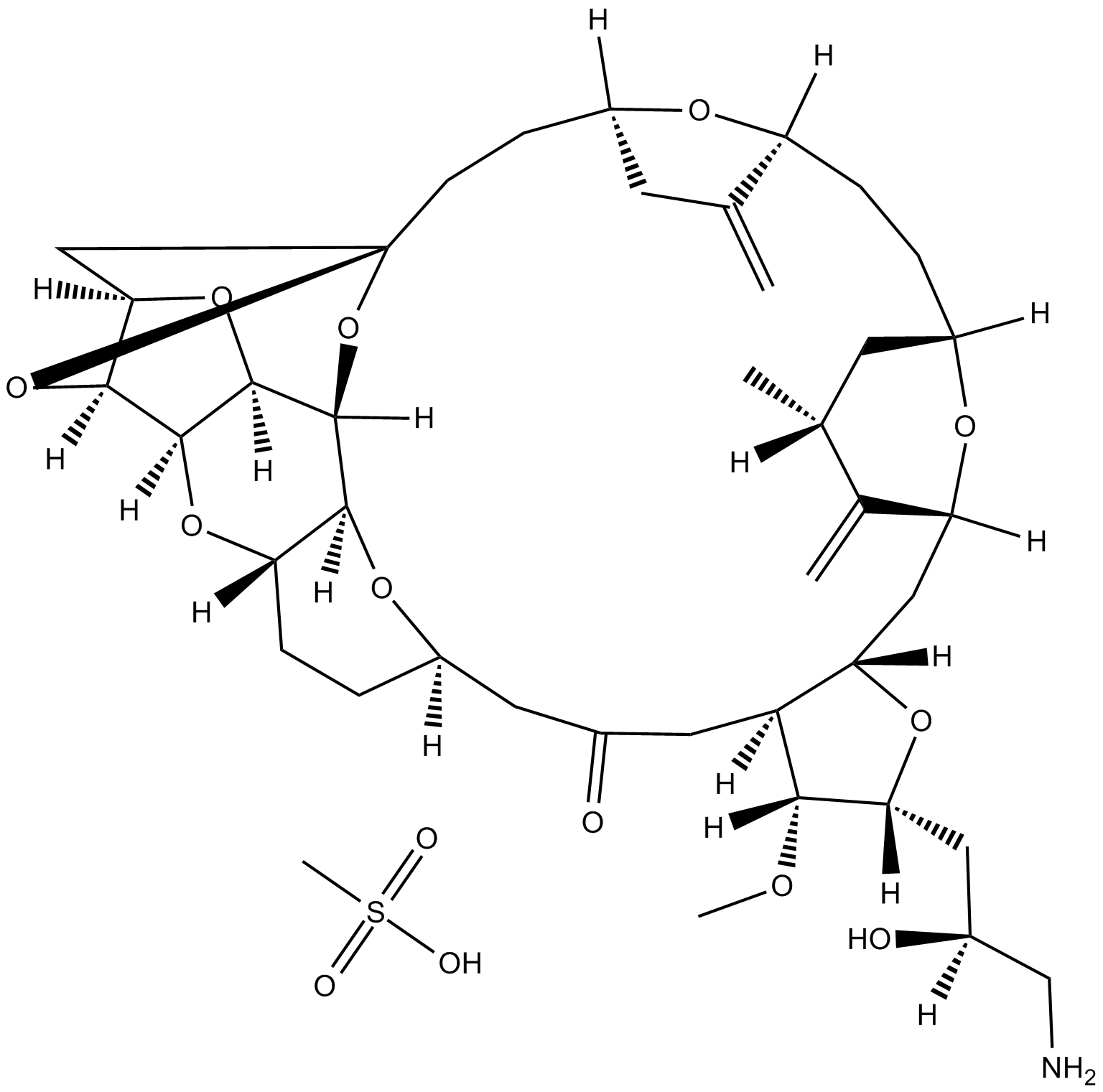
GC12483
Eribulin mesylate
Synthetisches Analogon von Halichondrin B
-
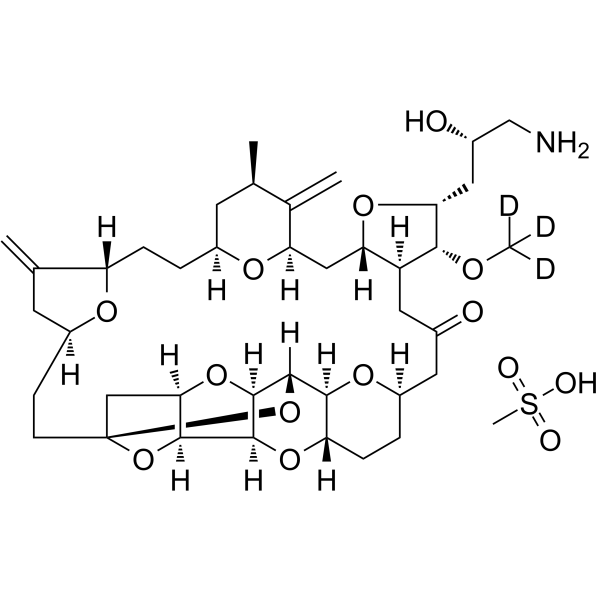
GC63845
Eribulin-d3 mesylate
Eribulin-d3-Mesylat ist ein Deuterium-markiertes Eribulin-Mesylat. Eribulinmesylat ist ein Wirkstoff, der auf Mikrotubuli abzielt und in der Krebsforschung eingesetzt wird.
-
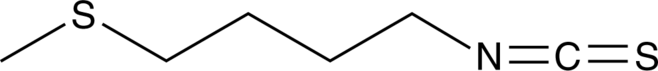
GC43625
Erucin
Erucin (ERU) ist ein Isothiocyanat, das besonders hÄufig in Rucola vorkommt.
-
GC67333
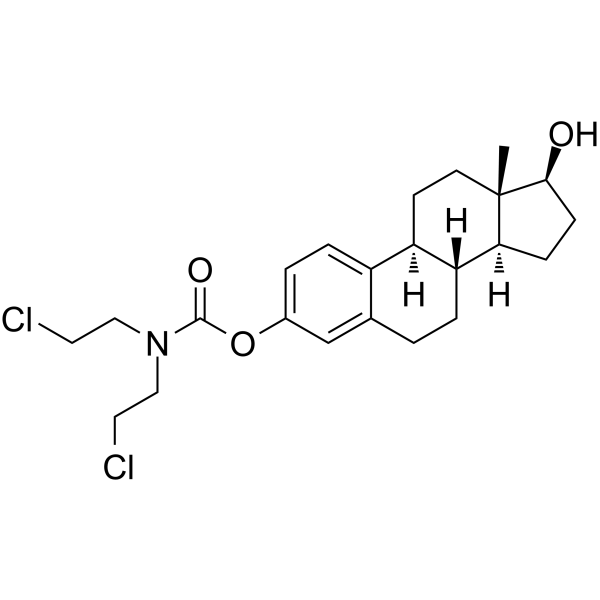
Estramustine
Estramustin ist ein antineoplastisches Mittel. Estramustin depolymerisiert Mikrotubuli durch Bindung an Tubulin 1, zeigt antimitotische Aktivität mit einem IC50-Wert von ~16 μ M für die Mitose von DU 145-Zellen. Estramustin blockiert Zellen bei der Mitose in Prostatatumor-Xenotransplantaten.
-
GC32816
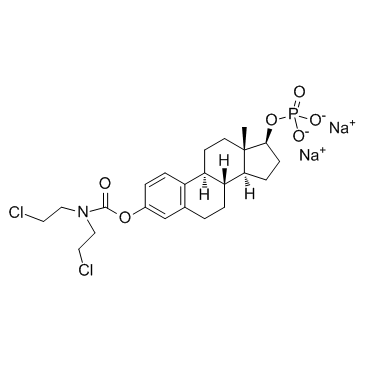
Estramustine phosphate sodium
Estramustinphosphat-Natrium, ein Estradiol-Analogon, ist ein oral wirksames Chemotherapeutikum gegen Mikrotubuli.
-
GC52061
Ethylene Glycol monododecyl ether
A nonionic surfactant

-
GC10196
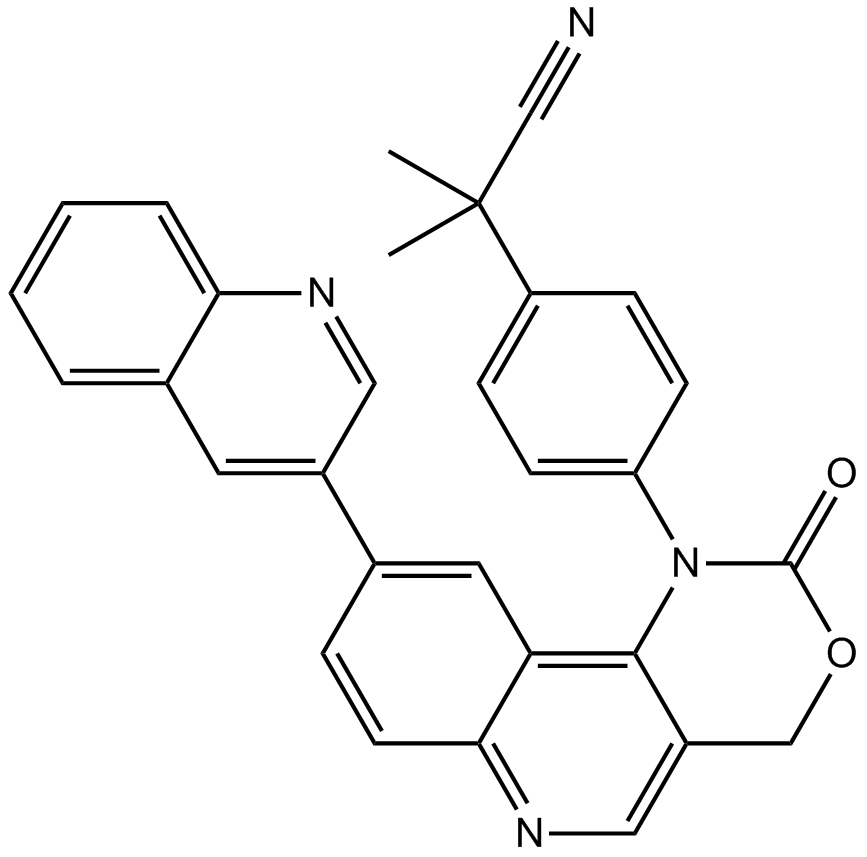
ETP-46464
ETP-46464 ist ein wirksamer mTOR- und ATR-Inhibitor mit IC50-Werten von 0,6 bzw. 14 nM.
-
GC18693
Fascaplysin (chloride)
Fascaplysin (Chlorid) ist ein antimikrobieller und zytotoxischer roter Farbstoff, der aus dem Meeresschwamm (Fascaplysin (Chlorid)opsis sp.

-
GC13869
Fasudil
Fasudil (HA-1077; AT877) ist ein unspezifischer RhoA/ROCK-Inhibitor und hat auch eine hemmende Wirkung auf Proteinkinasen mit einem Ki von 0,33 μM fÜr ROCK1, IC50s von 0,158 μM und 4,58 μM, 12,30 μM, 1,650 μM fÜr ROCK2 und PKA , PKC bzw. PKG. Fasudil ist auch ein potenter Ca2+-Kanal-Antagonist und Vasodilatator.

-
GC14289
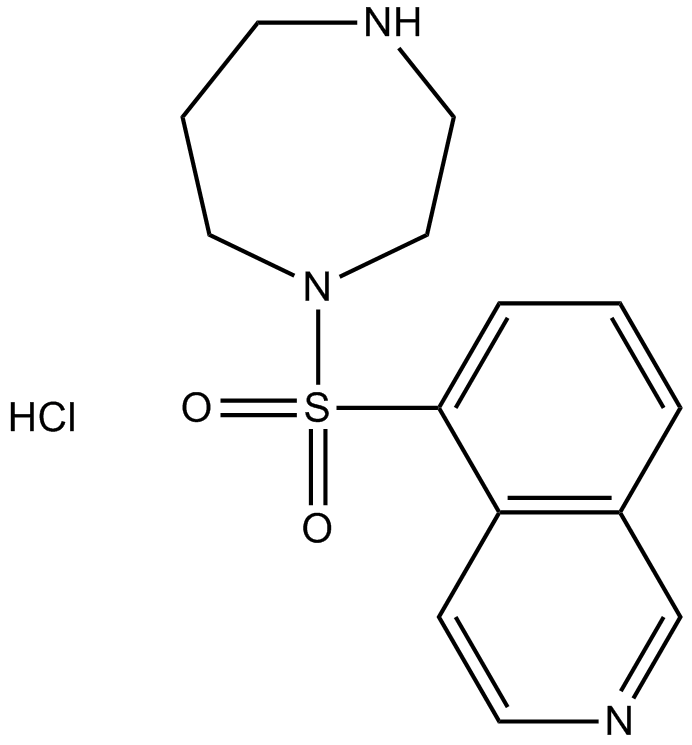
Fasudil (HA-1077) HCl
Fasudil (HA-1077; AT877) Hydrochloride is a nonspecific RhoA/ROCK inhibitor and also has inhibitory effect on protein kinases, with an Ki of 0.33 μM for ROCK1, IC50s of 0.158 μM and 4.58 μM, 12.30 μ ;M, 1.650 μM fÜr ROCK2 bzw. PKA, PKC, PKG. Fasudil (HA-1077) HCl ist auch ein potenter Ca2+-Kanal-Antagonist und Vasodilatator.
-
GC12319
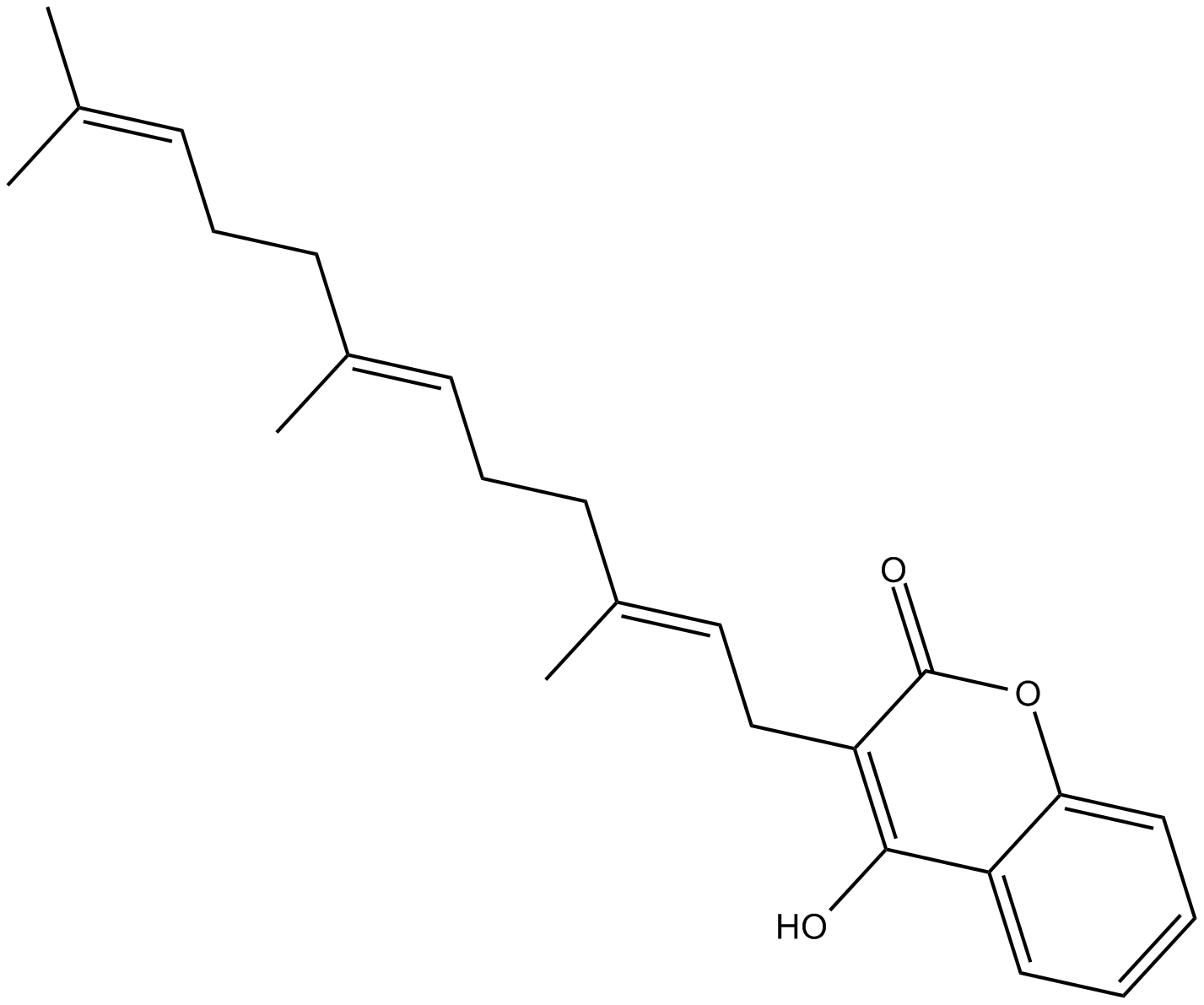
Ferulenol
Ferulenol, ein Sesquiterpen-prenyliertes Cumarinderivat, hemmt spezifisch die Succinat-Ubichinonreduktase auf der Ebene des Ubichinonzyklus.
-
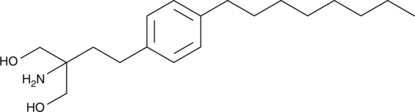
GC43666
Fingolimod
Fingolimod (freie Base FTY720) ist ein Antagonist von Sphingosin-1-phosphat (S1P) mit einer IC50 von 0,033 nM in K562- und NK-Zellen.
-
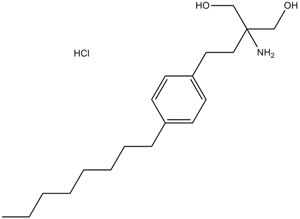
GC14807
Fingolimod(FTY720)
Fingolimod (FTY720) HCl (FTY720), ein Analogon von Sphingosin, ist ein potenter Modulator der Sphingosin-1-Phosphat-(S1P)-Rezeptoren.
-
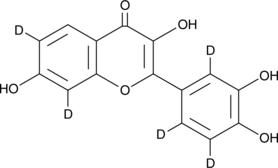
GC49344
Fisetin-d5
An internal standard for the quantification of fisetin
-
GC49695
FKBP52 Monoclonal Antibody (Clone Hi52C)
For immunochemical analysis of FKBP52

-
GC43674
FlAsH-EDT2
FlAsH-EDT2 ist ein Proteinmarkierungsreagenz.
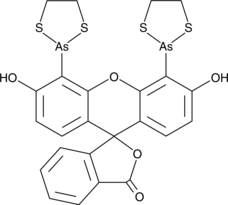
-
GC16063
Flavopiridol
An inhibitor of cyclin-dependent kinases
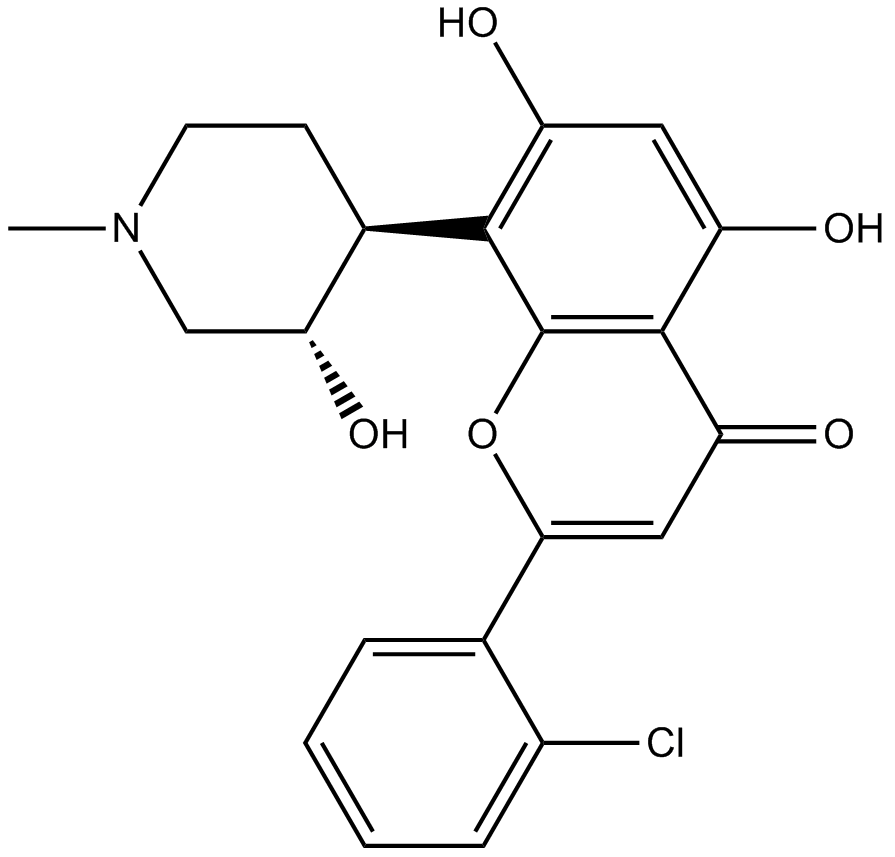
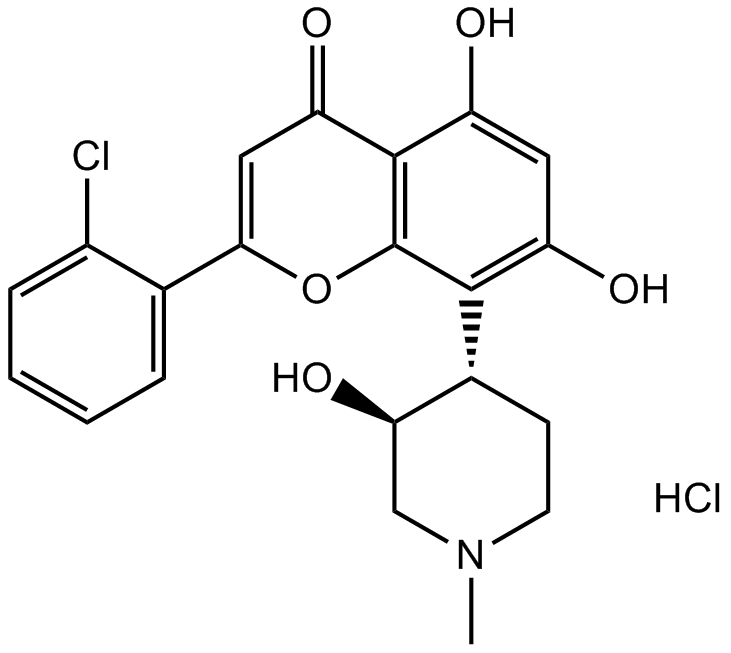
-
GC16425
Flavopiridol hydrochloride
Flavopiridolhydrochlorid (Alvocidibhydrochlorid) ist ein breiter Inhibitor von CDK, der mit ATP konkurriert, um CDKs einschließlich CDK1, CDK2, CDK4 mit IC50-Werten von 30, 170 bzw. 100 nM zu hemmen.
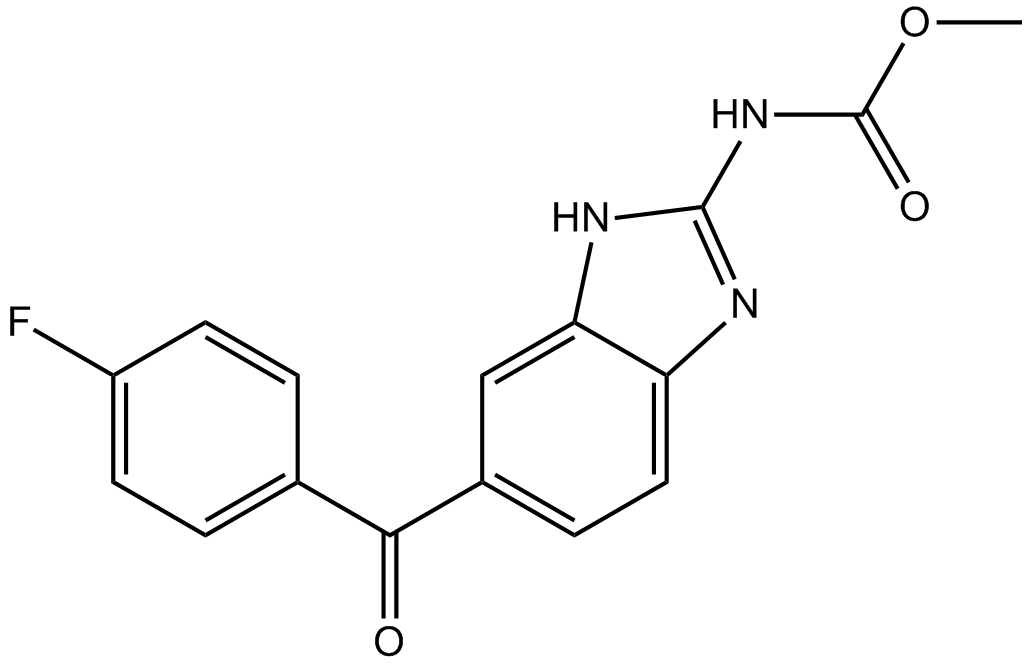
-
GC13210
Flubendazole
Flubendazol ist ein sicheres und wirksames Anthelminthikum, das weithin als Anthelminthikum fÜr Menschen, Nagetiere und WiederkÄuer verwendet wird.
-
GC16727
Flutax 1
A fluorescent taxol derivative used for direct imaging of the microtubule cytoskeleton
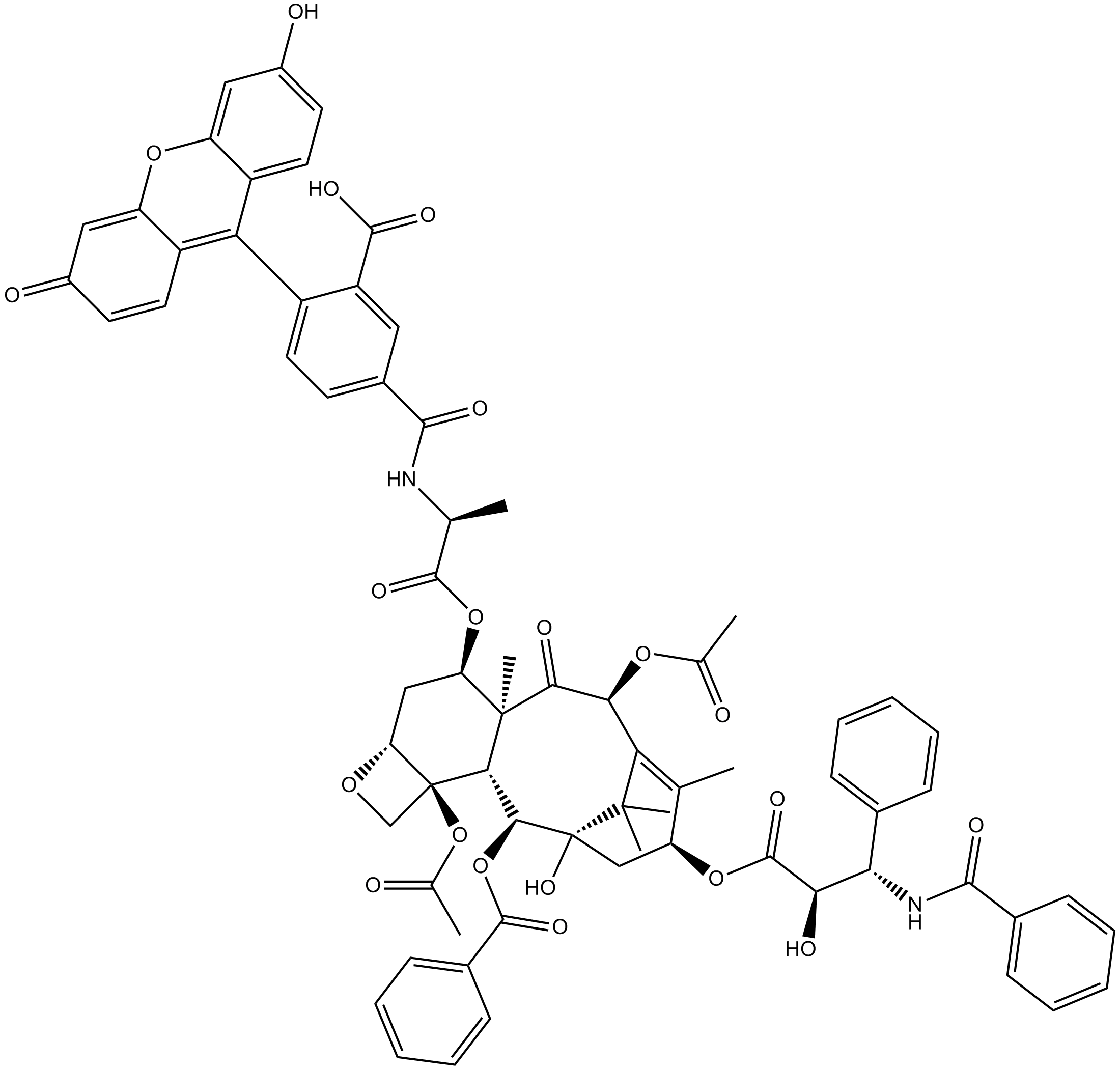
-
GC50397
Flutax 2
Green fluorescent taxol derivative; binds microtubules
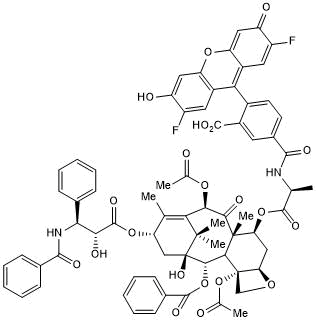
-
GC66242
Flutax-2 (5/6-mixture)
Flutax-2 (5/6-Mischung) ist ein aktives fluoreszierendes Derivat von Taxol. Flutax-2 (5/6-Mischung) bindet an polymerisierte α,&7#946;-Tubulin-Dimere. Flutax-2 (5/6-Mischung) ist in der Lage, Mikrotubuli intakter T. gallinae- und T. fetus-Trophozoiten zu stabilisieren.
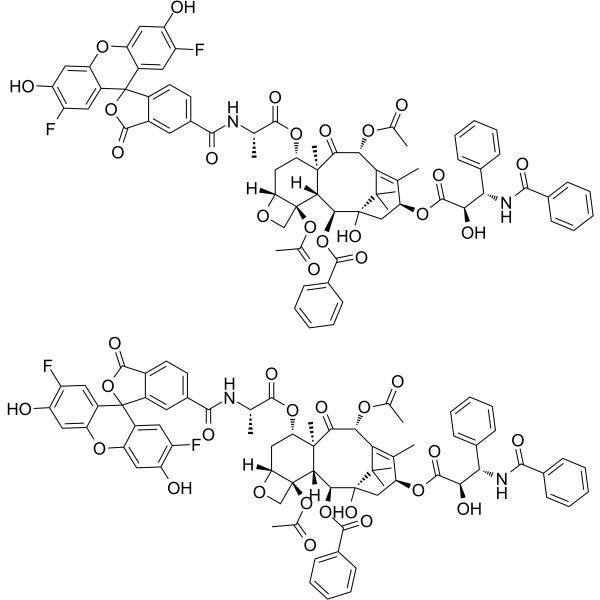
-
GC63596
Fmoc-MMAE
Fmoc-MMAE ist ein Schutzgruppen-konjugiertes Monomethyl-Auristatin E (MMAE), das ein potenter Tubulin-Inhibitor ist. Fmoc-MMAE kann bei der Synthese von ADC verwendet werden.
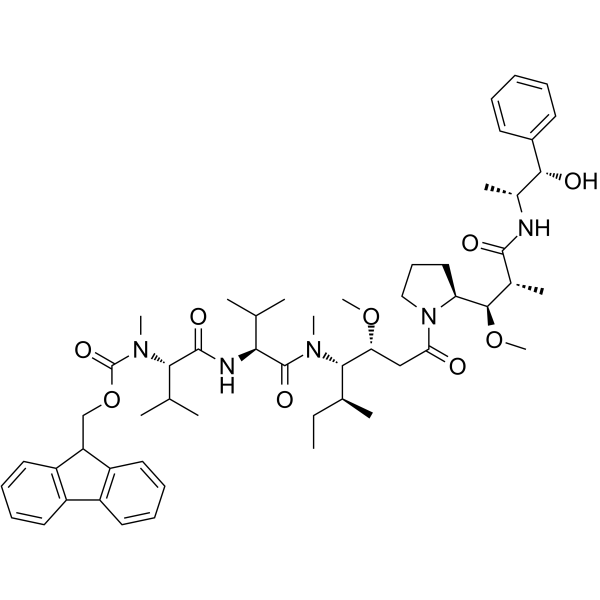
-
GC12308
Fosbretabulin (Combretastatin A4 Phosphate (CA4P)) Disodium
Fosbretabulin (Combretastatin A4 Phosphat (CA4P)) Dinatrium (CA 4DP) ist ein Tubulin destabilisierendes Mittel. Fosbretabulin (Combretastatin A4 Phosphat (CA4P)) Dinatrium ist das Combretastatin A4 Prodrug, das selektiv auf Endothelzellen abzielt, die Regression von neu entstehenden TumorgefÄßen induziert, den Blutfluss des Tumors reduziert und eine zentrale Tumornekrose verursacht.
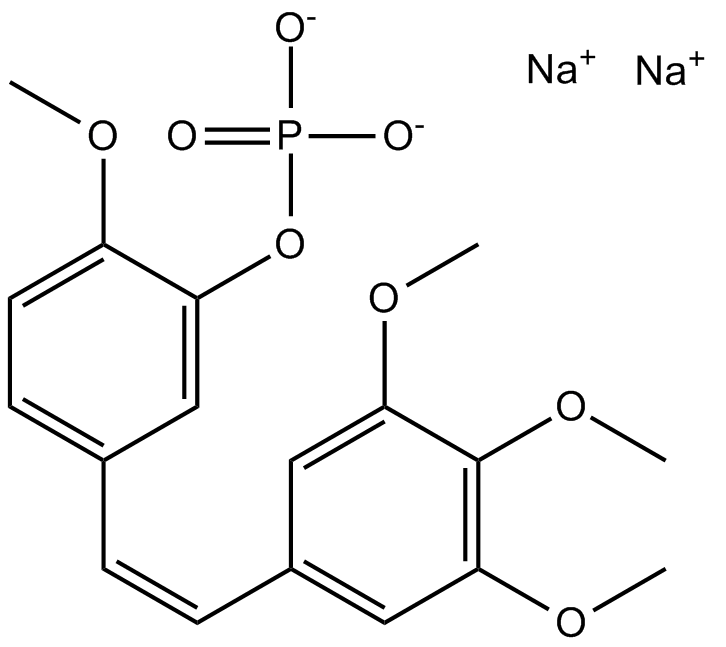
-
GC49089
FR900359
A cyclic depsipeptide and an inhibitor of Gαq, Gα11, and Gα14
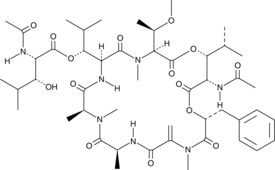
-
GC10251
FRAX1036
FRAX1036 ist ein PAK-Inhibitor mit Kis von 23,3 nM, 72,4 nM und 2,4 μM fÜr PAK1, PAK2 bzw. PAK4.
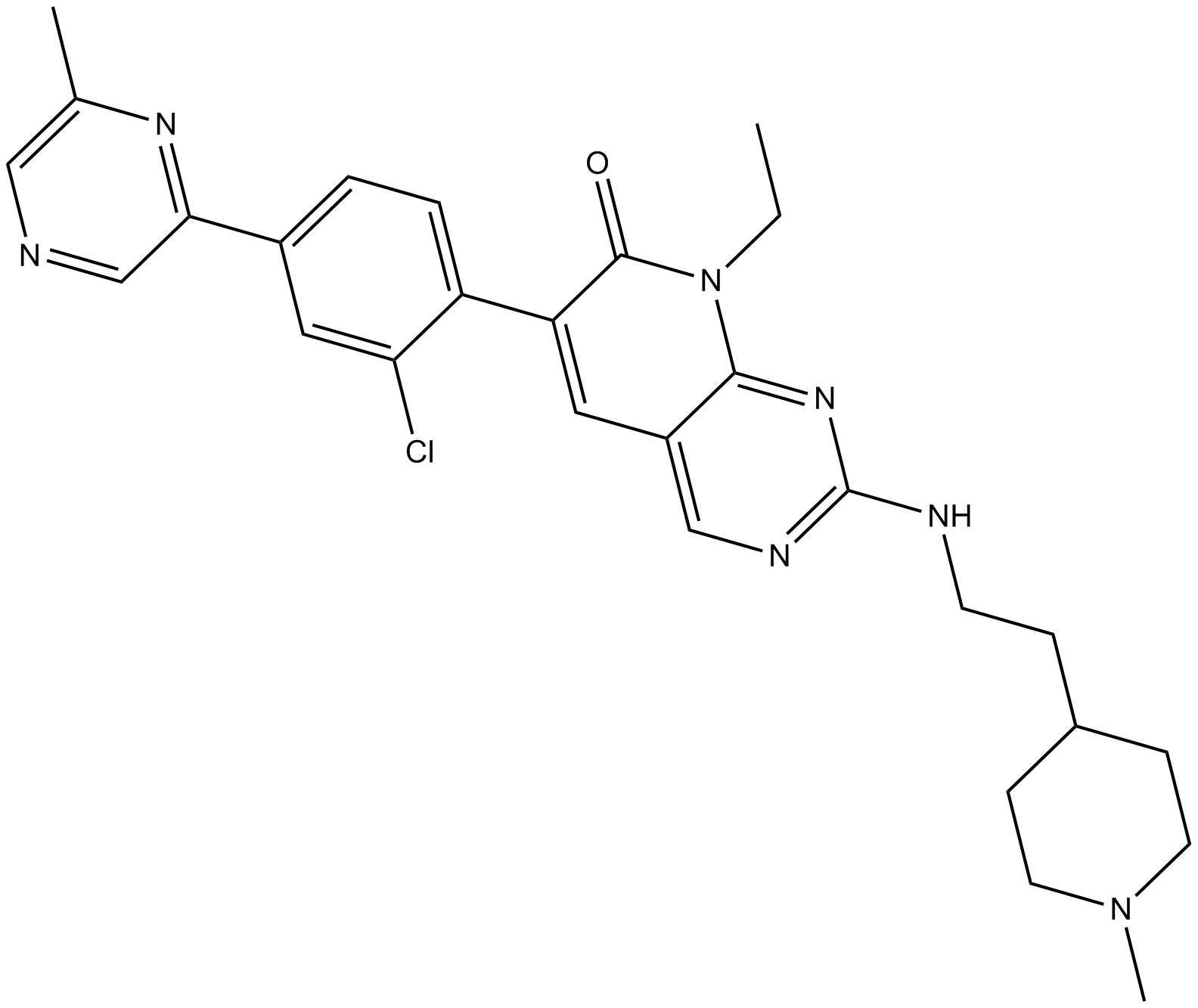
-
GC16075
FRAX486
FRAX486 ist ein Inhibitor der p21-aktivierten Kinase (PAK) mit IC50-Werten von 14, 33 und 39 nM fÜr PAK1, PAK2 bzw. PAK3.
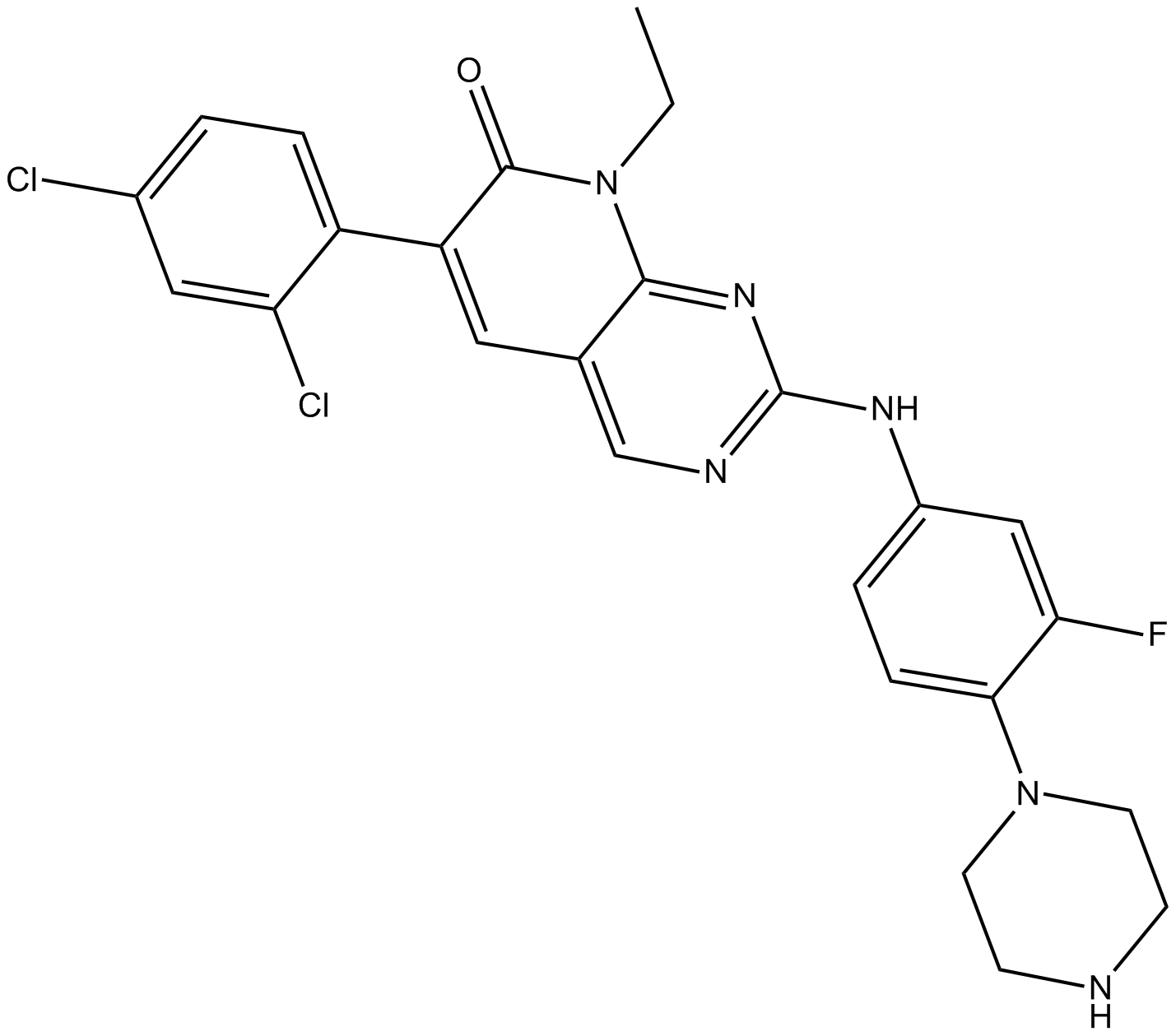
-
GC12261
FRAX597
FRAX597 ist ein potenter Inhibitor der p21-aktivierten Kinasen (PAKs) der Gruppe I mit IC50-Werten von 8, 13 und 19 nM fÜr PAK1, 2 und 3.
-
GC52288
Fumonisin B1-13C34
An internal standard for the quantification of fumonisin B1
-
GC47378
Fumonisin B2-13C34
An internal standard for the quantification of fumonisin B2
-
GC18720
G-5555
G-5555 ist ein potenter Inhibitor der p21-aktivierten Kinase 1 (PAK1) mit Kis von 3,7 nM und 11 nM fÜr PAK1 bzw. PAK2.
-
GC36095
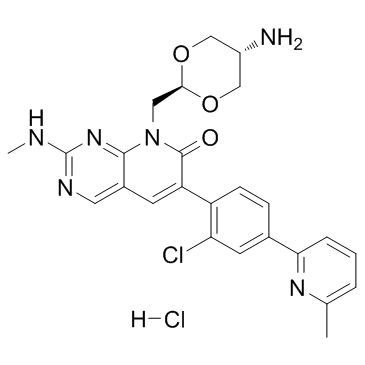
G-5555 hydrochloride
G-5555-Hydrochlorid ist ein potenter und selektiver Inhibitor der p21-aktivierten Kinase 1 (PAK1) mit einem Ki von 3,7 nM.
-
GC40016
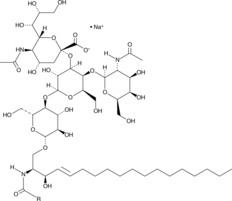
Ganglioside GM2 Mixture (sodium salt)
Ganglioside GM2 is a glycosphingolipid component of cellular membranes, primarily the plasma membrane.
-
GC43732
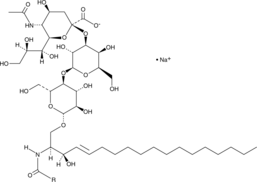
Ganglioside GM3 Mixture (sodium salt)
Ganglioside GM3 is a monosialoganglioside that demonstrates antiproliferative and proapoptotic effects in tumor cells by modulating cell adhesion, proliferation, and differentiation.
-
GC49322
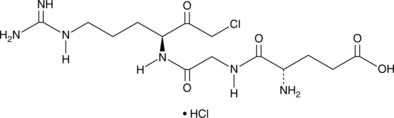
GGACK (hydrochloride)
A uPA and factor Xa inhibitor
-
GC38783
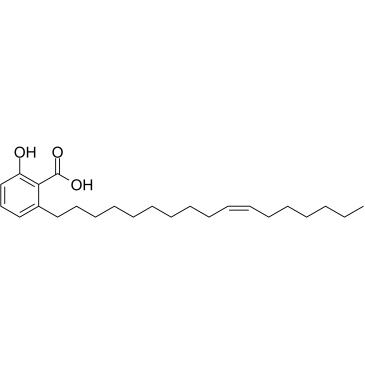
Ginkgolic acid C17:1
GinkgolsÄure C17:1, extrahiert aus Ginkgo-Biloba-BlÄttern, unterdrÜckt die konstitutive und induzierbare STAT3-Aktivierung durch Induktion von PTEN und SHP-1-Tyrosinphosphatase. GinkgolsÄure C17:1 hat krebshemmende Wirkungen.
-
GC49437
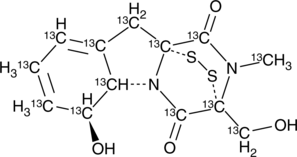
Gliotoxin-13C13
An internal standard for the quantification of gliotoxin
-
GC41563
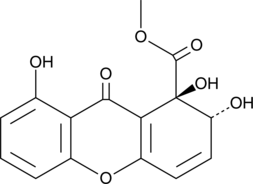
Globosuxanthone A
Globosuxanthon A ist ein Dihydroxanthenon mit offensichtlicher antimykotischer AktivitÄt gegenÜber Fusarium graminearum, Fusarium solani und Botrytis cinerea mit MHK-Werten von 4, 8 bzw. 16 μg/ml.
-
GC45668
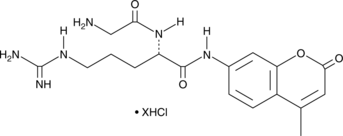
Gly-Arg-AMC (hydrochloride)
A fluorogenic substrate for cathepsin C
-
GC49687
Goralatide (acetate)
A tetrapeptide regulator of hematopoiesis
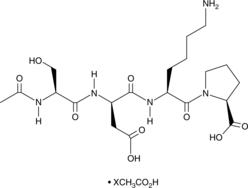
-
GC48720
Griseofulvic Acid
A metabolite of griseofulvin
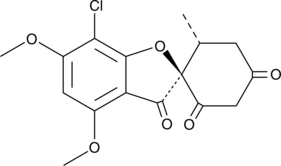
-
GC13484
Griseofulvin
Griseofulvin (Gris-PEG; Grifulvin) ist ein spirozyklisches Pilz-Naturprodukt, das zur Behandlung von Pilz-Dermatophyten verwendet wird; Antimykotisches Medikament.
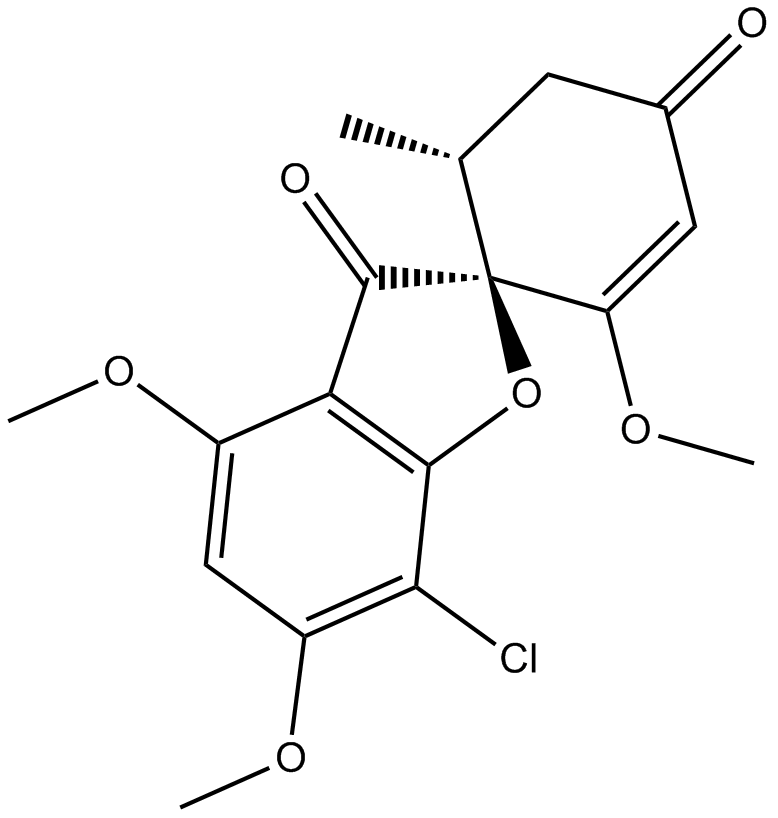
-
GC48688
Griseofulvin-d3
An internal standard for the quantification of griseofulvin
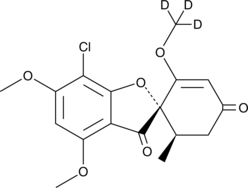
-
GC49744
GRP94 Monoclonal Antibody (Clone 9G10)
For immunochemical detection of GRP94

-
GC38791
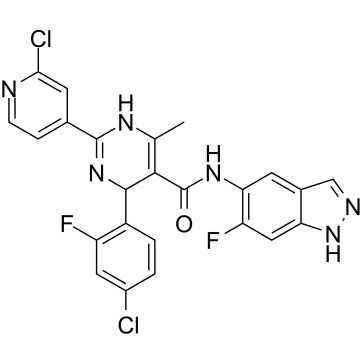
GSK-25
GSK-25 ist ein potenter, selektiver und oral bioverfÜgbarer ROCK1-Inhibitor (IC50=7 nM).
-
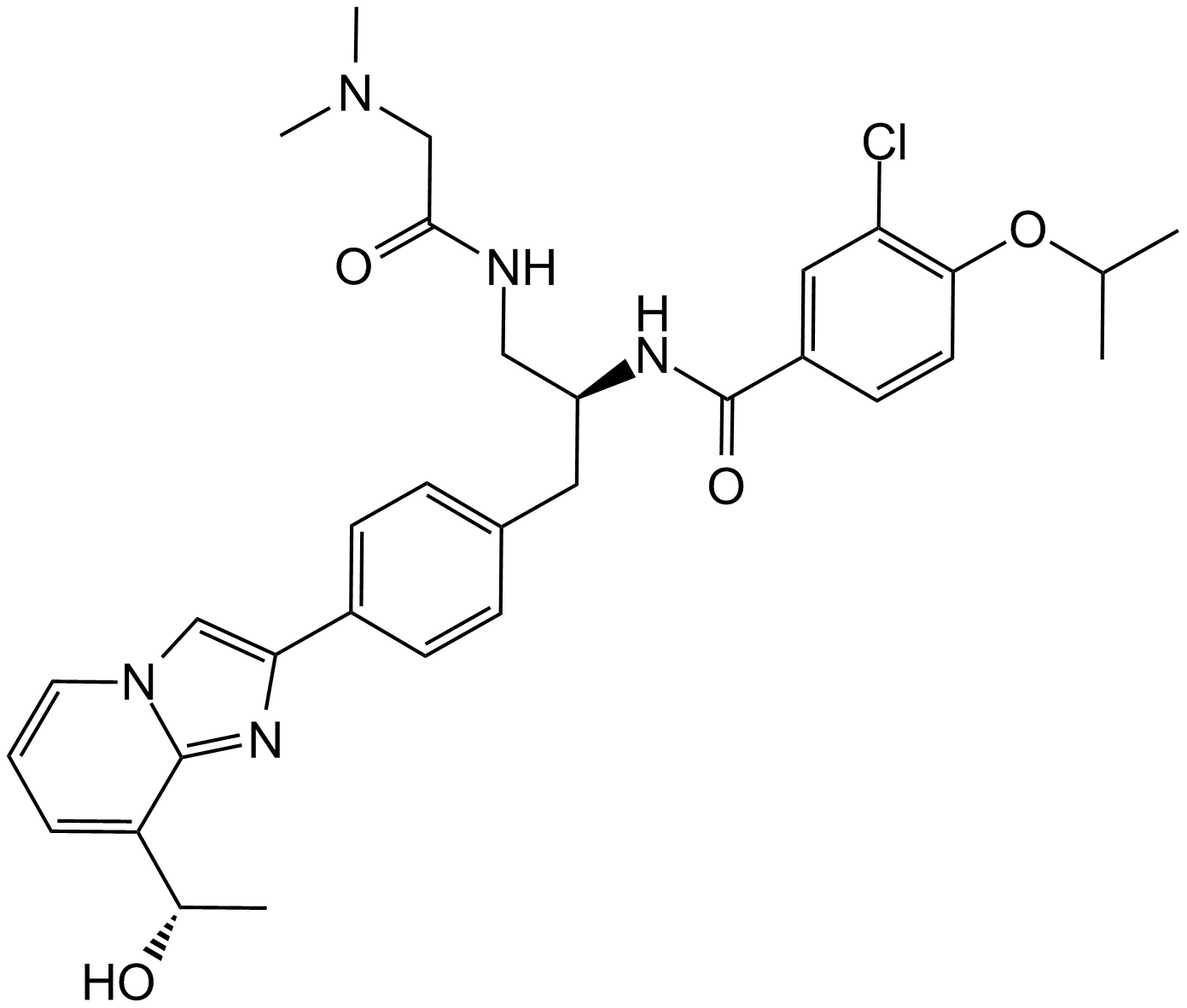
GC15659
GSK-923295
An inhibitor of CENP-E
-
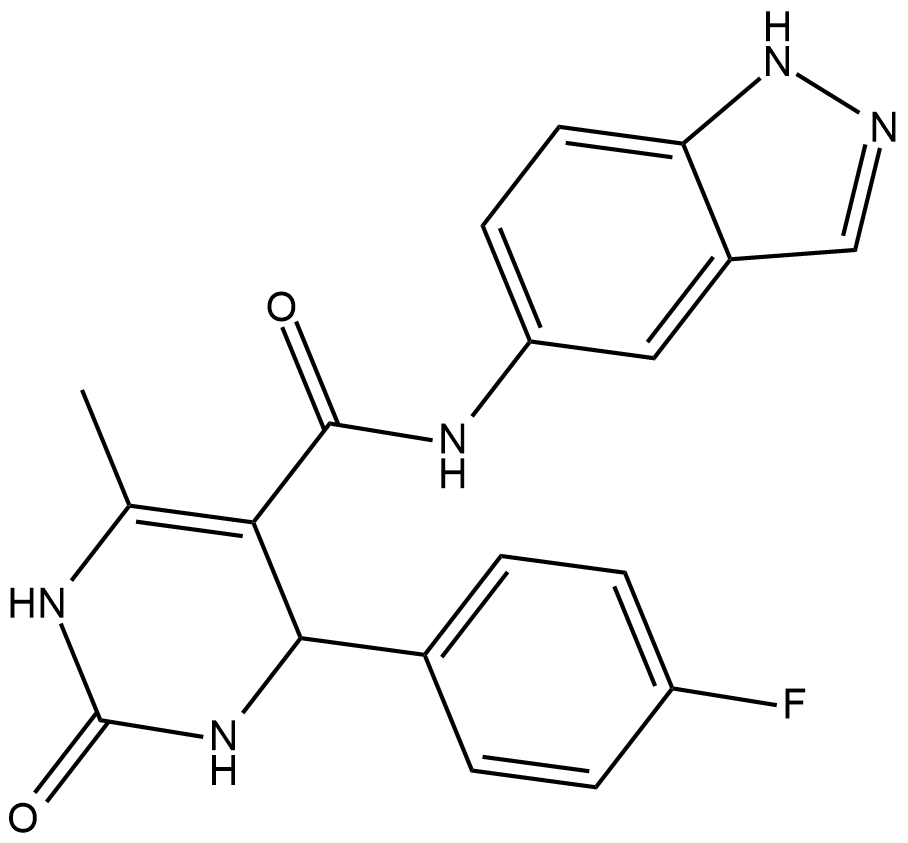
GC19177
GSK180736A
GSK180736A ist ein potenter Hemmer der Rho-assoziierten Coiled-Coil-Kinase 1 (ROCK1) mit einem IC50 von 100 nM.
-
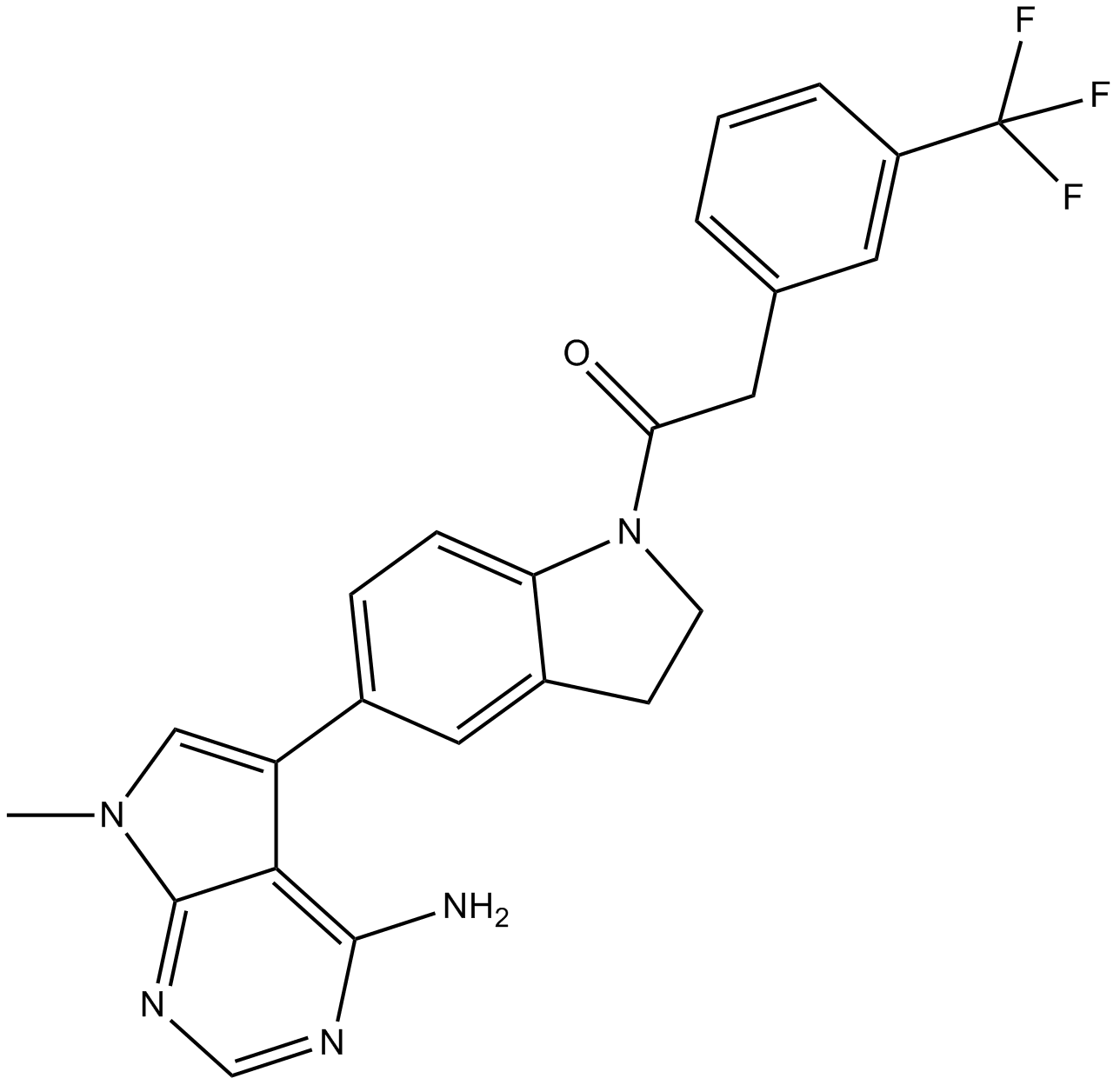
GC17450
GSK2606414
Ein selektiver PERK-Inhibitor
-
GC11068
GSK2656157
GSK2656157 ist ein selektiver und ATP-kompetitiver Inhibitor der Proteinkinase R (PKR)-Ähnlichen endoplasmatischen Retikulumkinase (PERK) mit einem IC50 von 0,9 nM.
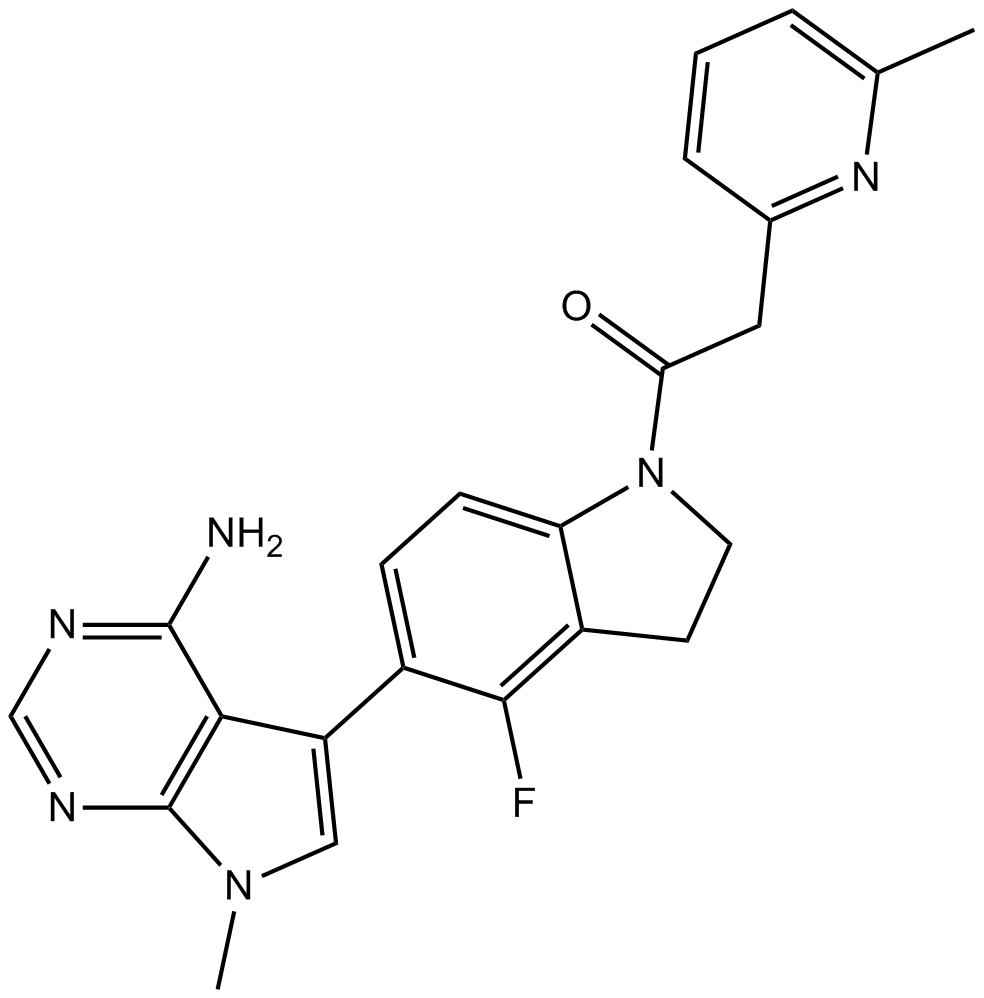
-
GC17936
GSK269962A
GSK269962A (GSK 269962) ist ein potenter ROCK-Inhibitor mit IC50-Werten von 1,6 und 4 nM fÜr rekombinantes humanes ROCK1 bzw. ROCK2.
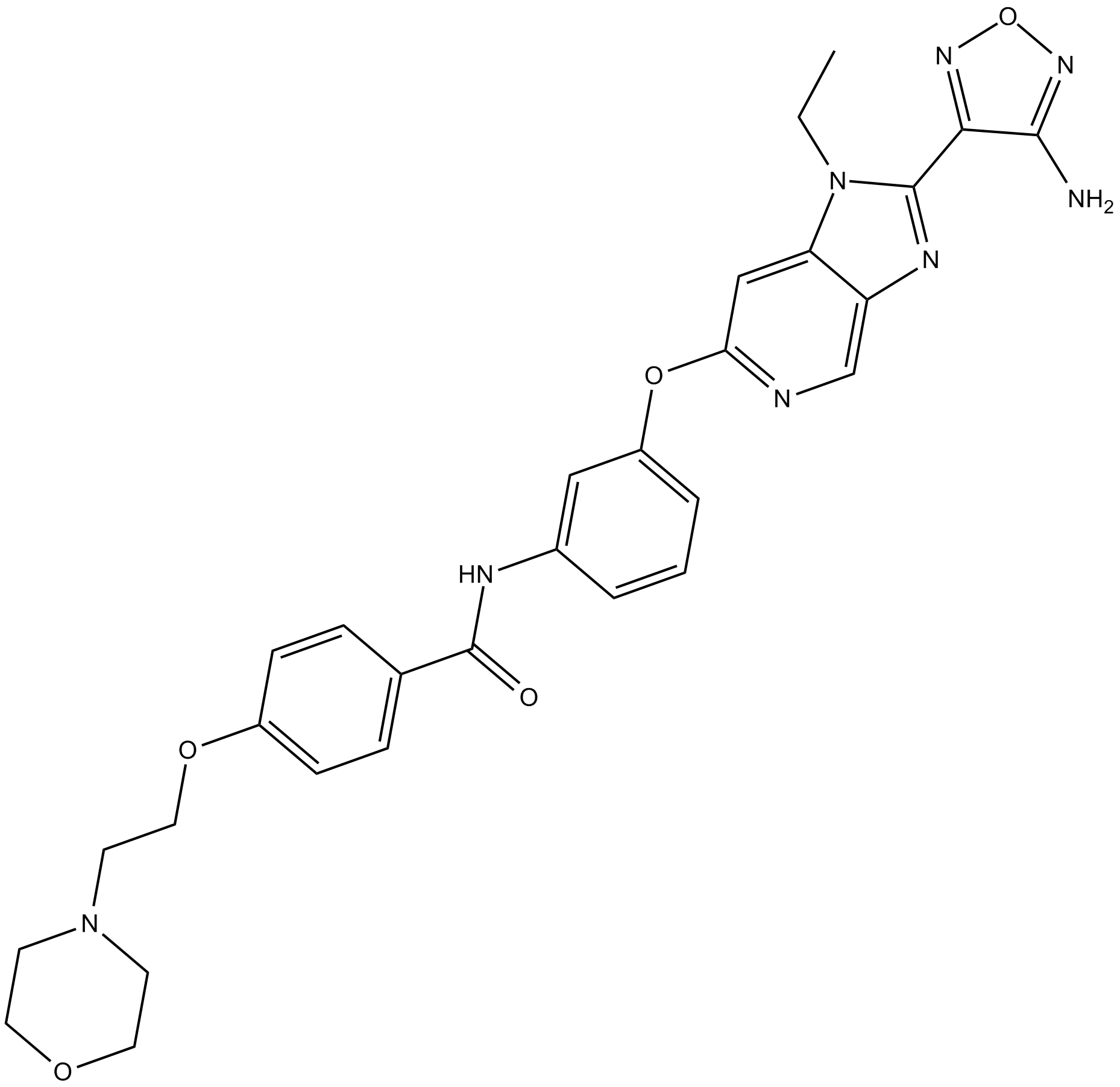
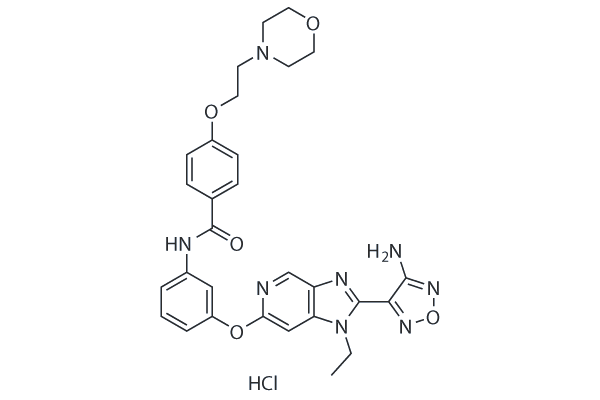
-
GC25482
GSK269962A HCl
GSK269962A HCl (GSK269962B, GSK269962) is a selective ROCK(Rho-associated protein kinase) inhibitor with IC50 values of 1.6 and 4 nM for ROCK1 and ROCK2, respectively.
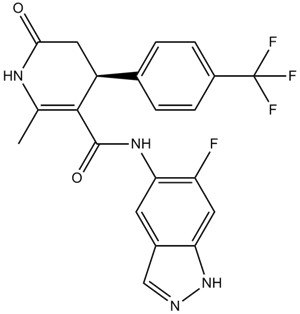
-
GC18119
GSK429286A
GSK429286A ist ein selektiver Inhibitor von ROCK1 mit einem IC50-Wert von 14 nM.
-
GC12335
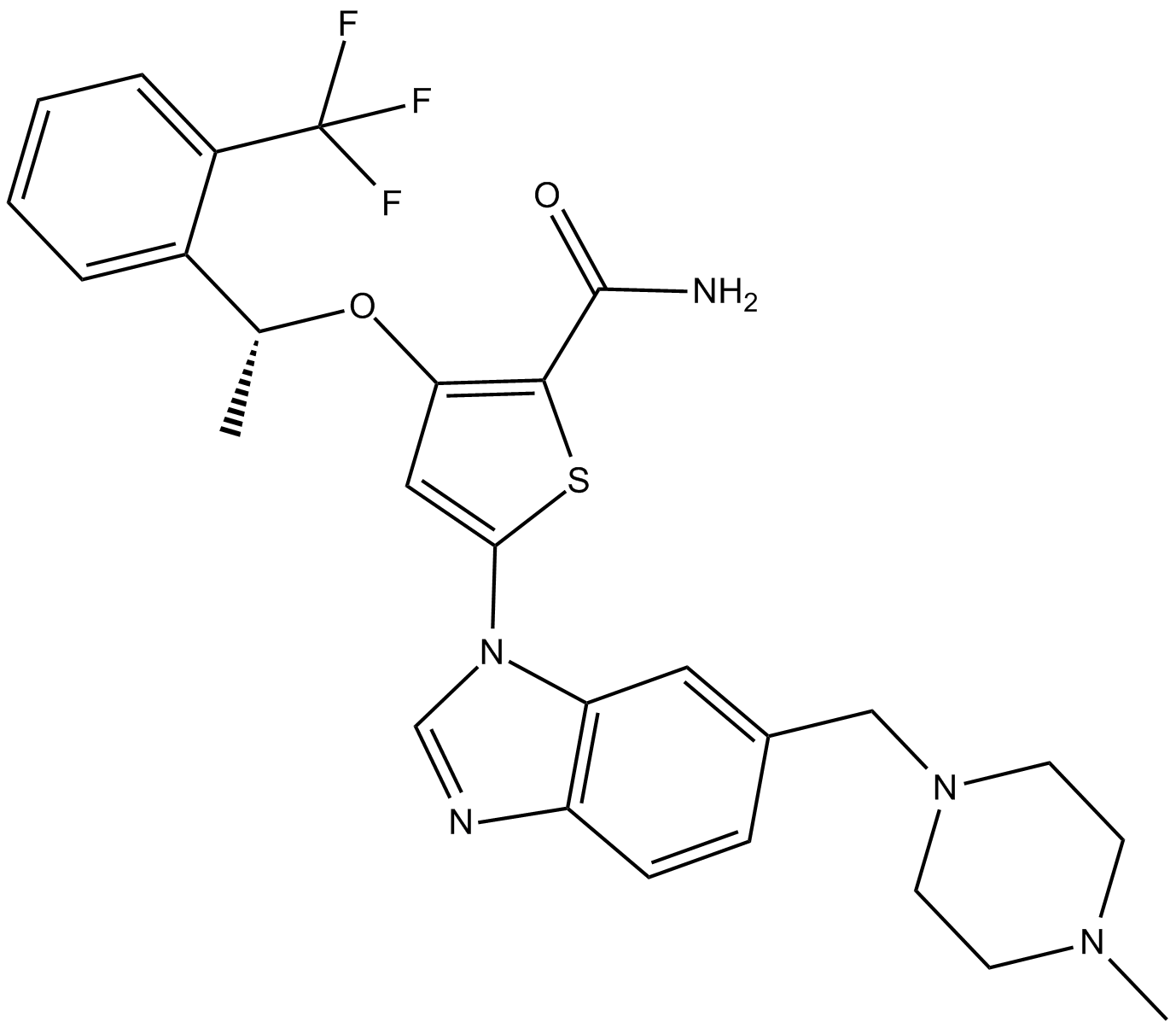
GSK461364
GSK461364 ist ein selektiver, reversibler und ATP-kompetitiver Inhibitor der Polo-Ähnlichen Kinase 1 (PLK1) mit einem Ki-Wert von 2,2 nM.
-
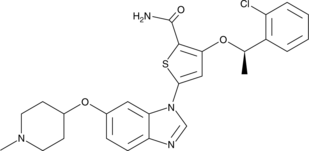
GC49280
GSK579289A
A Plk1 inhibitor
-
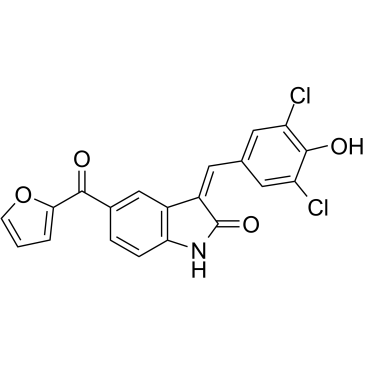
GC60891
GW406108X
GW406108X ist ein spezifischer Kif15 (Kinesin-12)-Inhibitor mit einem IC50-Wert von 0,82 uM in ATPase-Assays. GW406108X, ein potenter Autophagie-Inhibitor, zeigt eine kompetitive Hemmung von ATP gegen ULK1 mit einem pIC50 von 6,37 (427 nM). GW406108X hemmt die AktivitÄt der ULK1-Kinase und blockiert den autophagischen Fluss, ohne die vorgeschalteten Signalkinasen mTORC1 und AMPK zu beeintrÄchtigen.
-
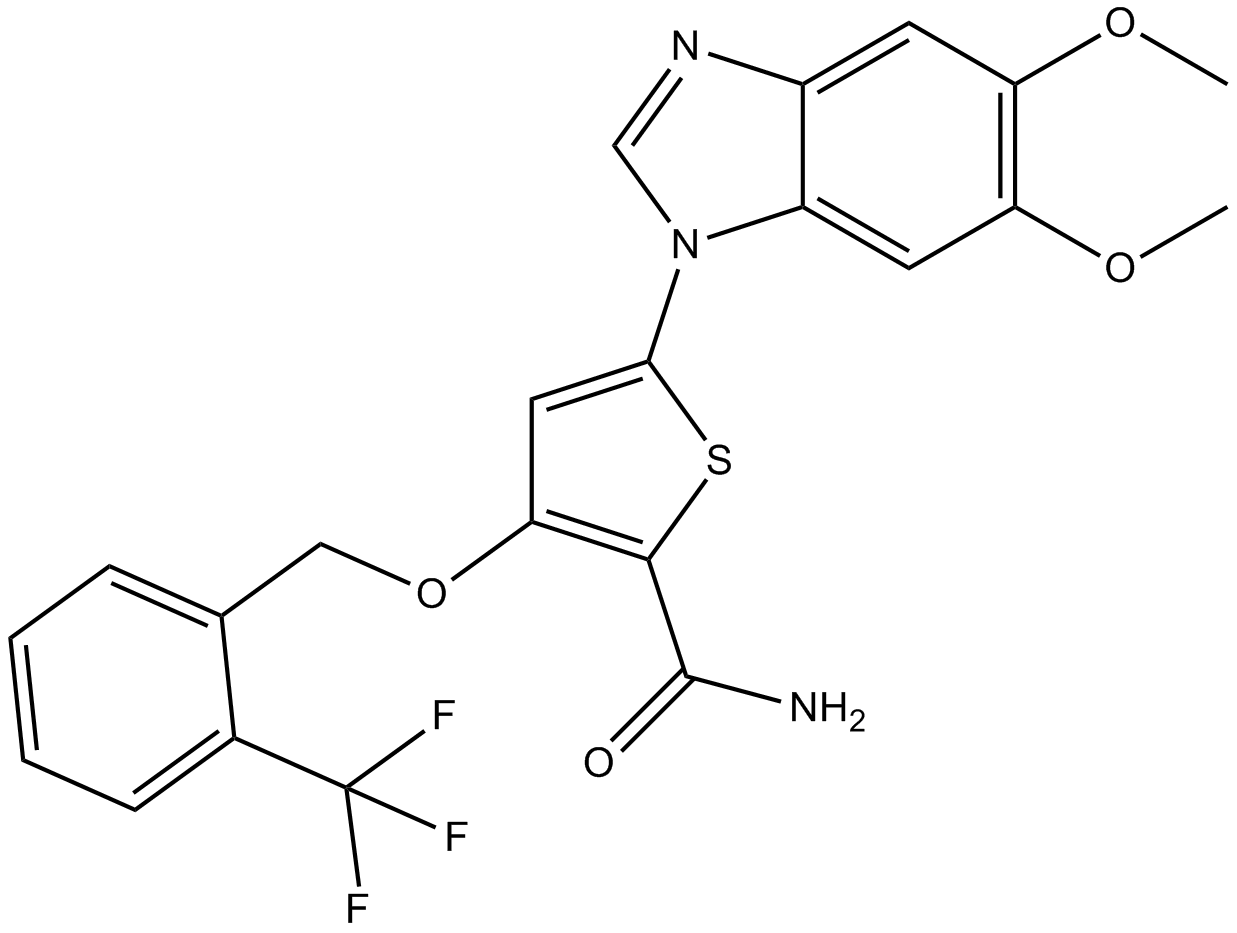
GC11621
GW843682X
A reversible polo-like kinase inhibitor
-
GC16545
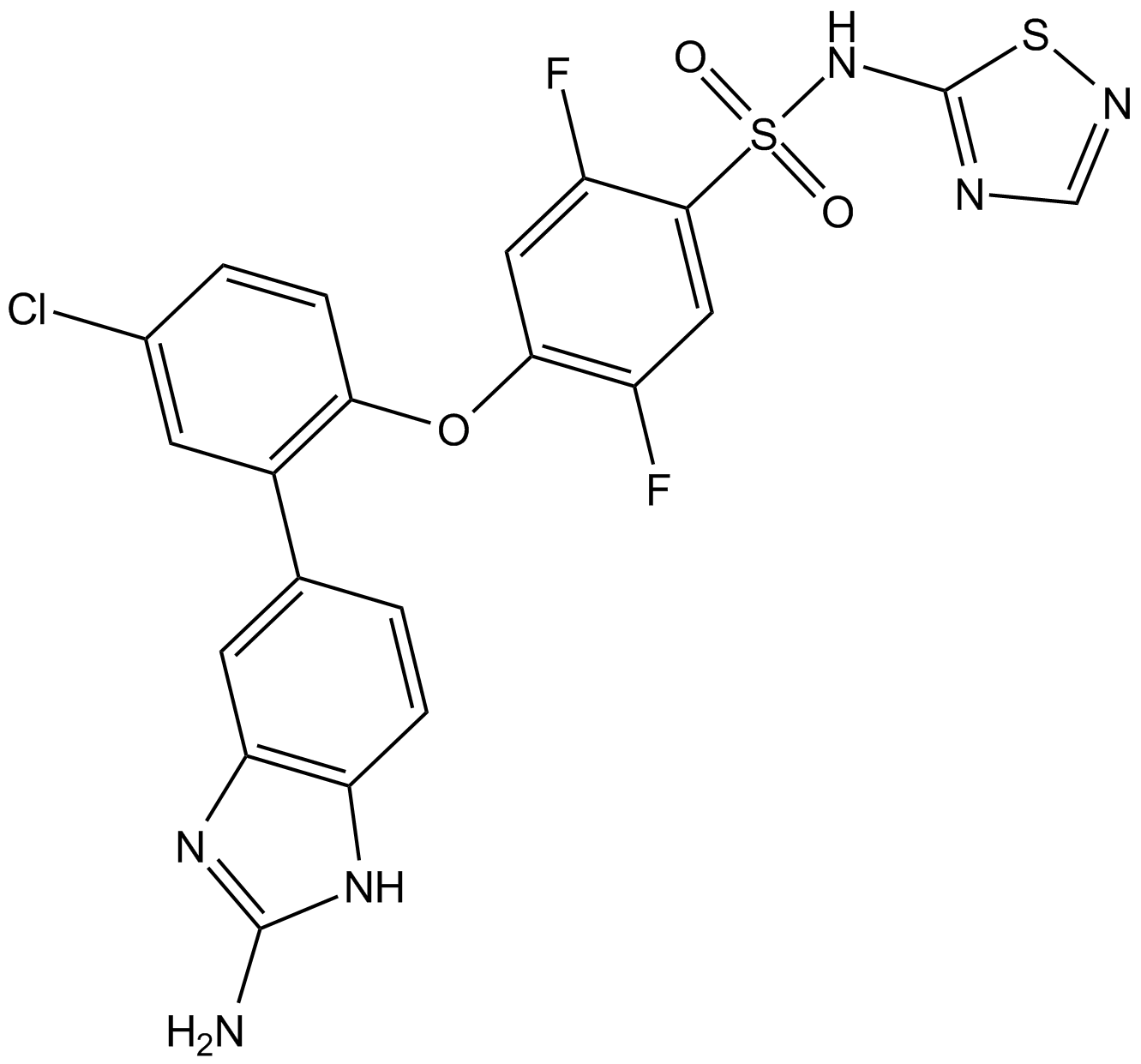
GX-674
GX-674 ist ein potenter, zustandsabhÄngiger, Isoform-selektiver spannungsgesteuerter Natriumkanal 1.7 (Nav1.7)-Antagonist mit einem IC50 von 0,1 nM bei -40 mV.
-
GC36207
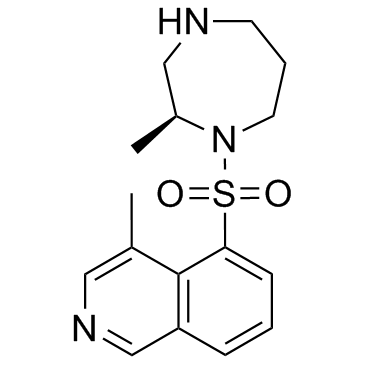
H-1152
H-1152 ist ein membrandurchlÄssiger und selektiver ROCK-Inhibitor mit einem Ki-Wert von 1,6 nM und einem IC50-Wert von 12 nM fÜr ROCK2.
-
GC36208
H-1152 dihydrochloride
H-1152-Dihydrochlorid ist ein membrangÄngiger und selektiver ROCK-Inhibitor mit einem Ki-Wert von 1,6 nM und einem IC50-Wert von 12 nM fÜr ROCK2.