Bcl-2 Family
Bcl-2 family proteins are a group of proteins homologous to the Bcl-2 protein and characterized by containing at least one of four conserved Bcl-2 homology (BH) domains (BH1, BH2, BH3 and BH4). Bcl-2 family proteins, consisting of pro-apoptotic and anti-apoptotic molecules, can be classified into the following three subfamilies according to sequence homology within four BH domains: (1) a subfamily shares sequence homology within all four BH domains, such as Bcl-2, Bcl-XL and Bcl-w which are anti-apoptotic; (2) a subfamily shares sequence homology within BH1, BH2 and BH4, such as Bax and Bak which are pro-apoptotic; (3) a subfamily shares sequence homology only within BH3, such as Bik and Bid which are pro-apoptotic.
Products for Bcl-2 Family
- Cat.No. Product Name Information
-
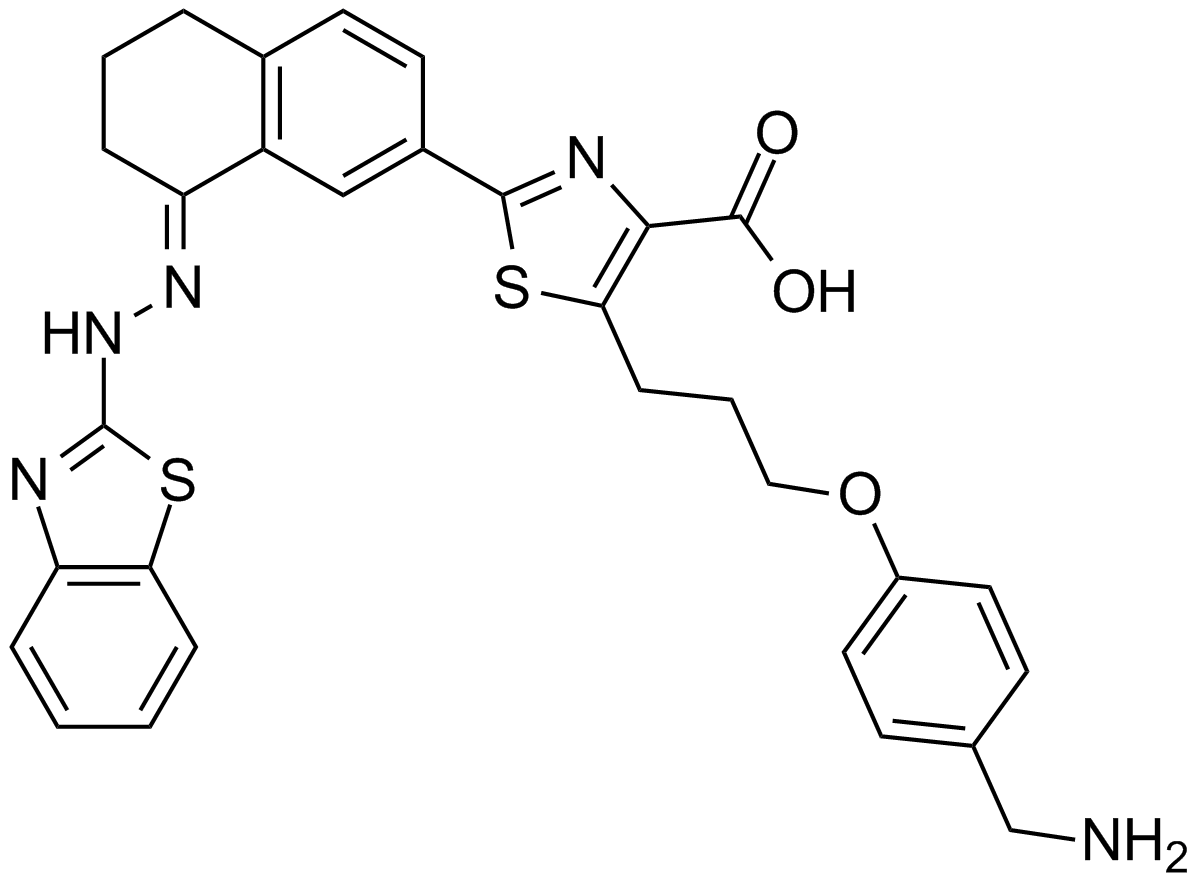
GC17008
(+)-Apogossypol
inhibitor of Bcl-2 family proteins
-
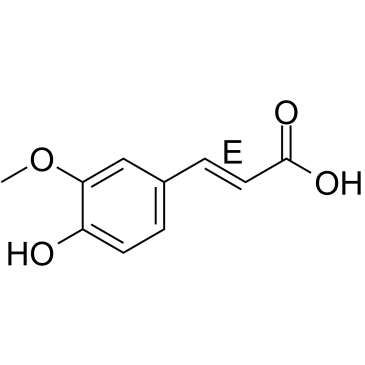
GC34980
(E)-Ferulic acid
(E)-Ferulic acid is a isomer of Ferulic acid which is an aromatic compound, abundant in plant cell walls. (E)-Ferulic acid causes the phosphorylation of β-catenin, resulting in proteasomal degradation of β-catenin and increases the expression of pro-apoptotic factor Bax and decreases the expression of pro-survival factor survivin. (E)-Ferulic acid shows a potent ability to remove reactive oxygen species (ROS) and inhibits lipid peroxidation. (E)-Ferulic acid exerts both anti-proliferation and anti-migration effects in the human lung cancer cell line H1299.
-
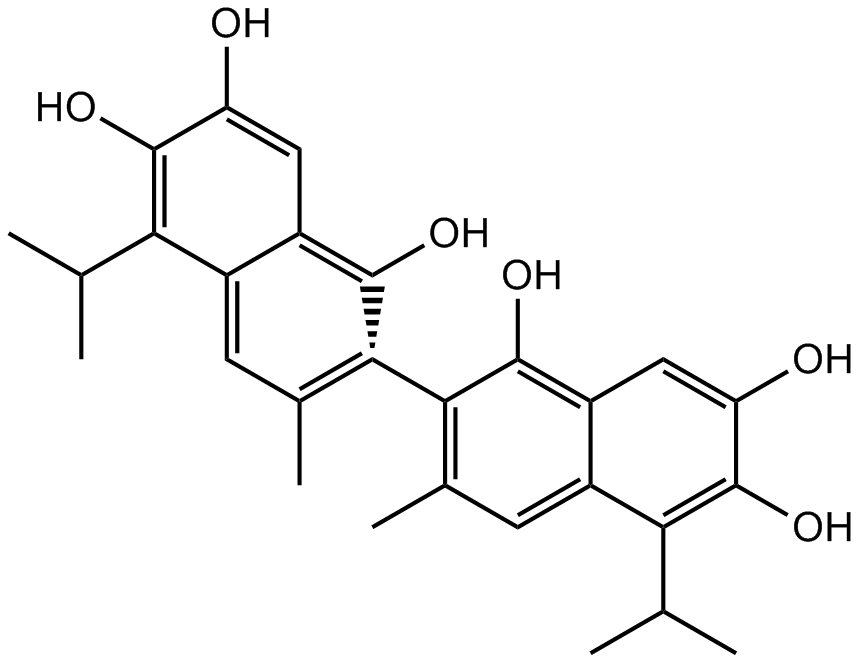
GC34096
(R)-(-)-Gossypol acetic acid (AT-101 (acetic acid))
(R)-(-)-Gossypol acetic acid (AT-101 (acetic acid)) (AT-101 (acetic acid)) is the levorotatory isomer of a natural product Gossypol. AT-101 is determined to bind to Bcl-2, Mcl-1 and Bcl-xL proteins with Kis of 260±30 nM, 170±10 nM, and 480±40 nM, respectively.
-
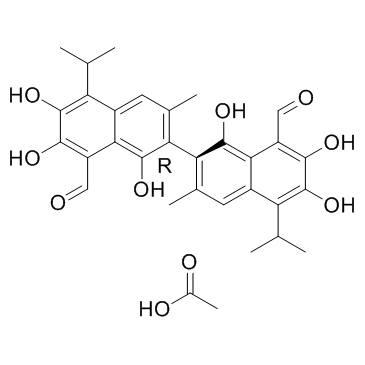
GC35001
(S)-Gossypol acetic acid
(S)-Gossypol is the isomer of a natural product Gossypol. (S)-Gossypol binds to the BH3-binding groove of Bcl-xL and Bcl-2 proteins with high affinity.
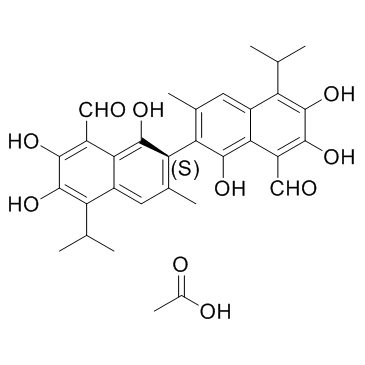
-
GC12258
2,3-DCPE hydrochloride
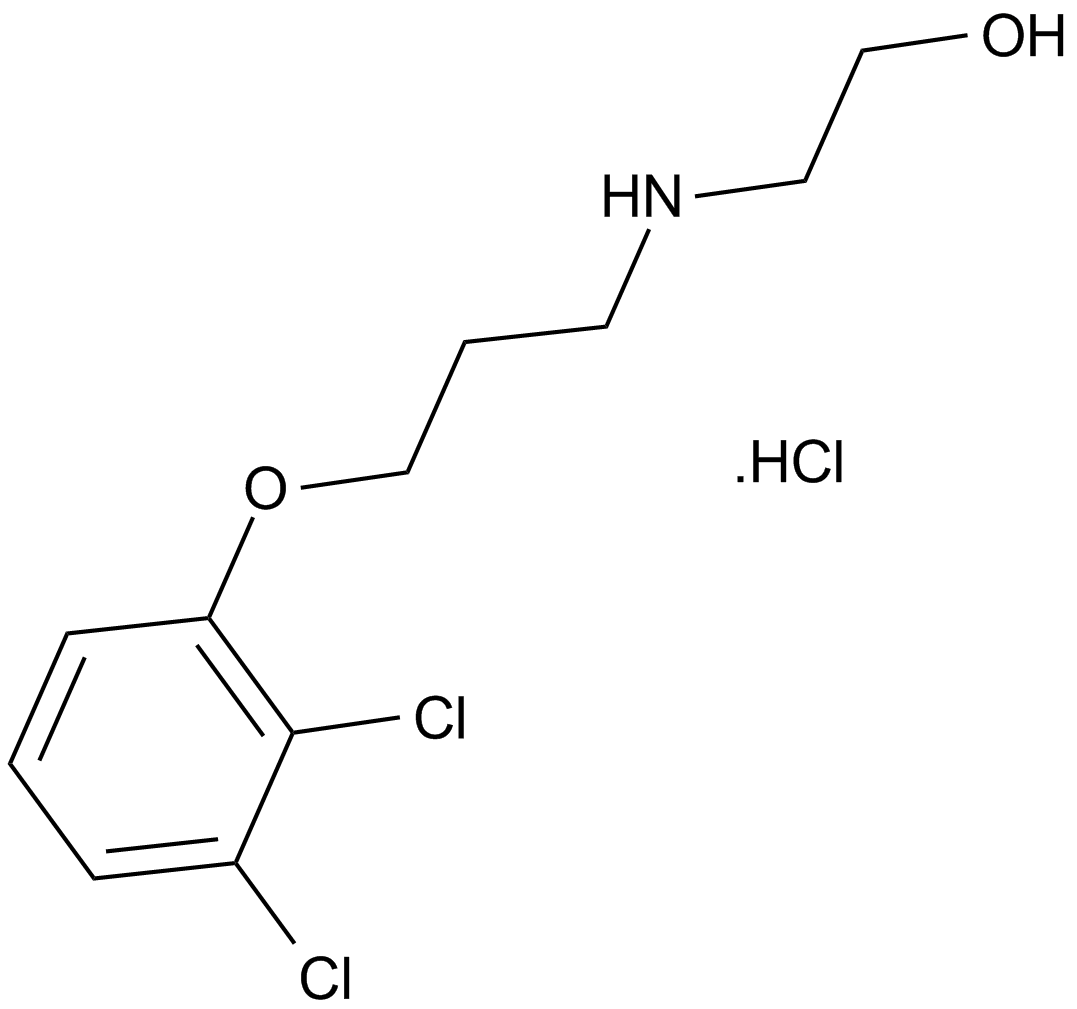
-
GC17512
A-1155463
BCL-XL inhibitor, potent and selective
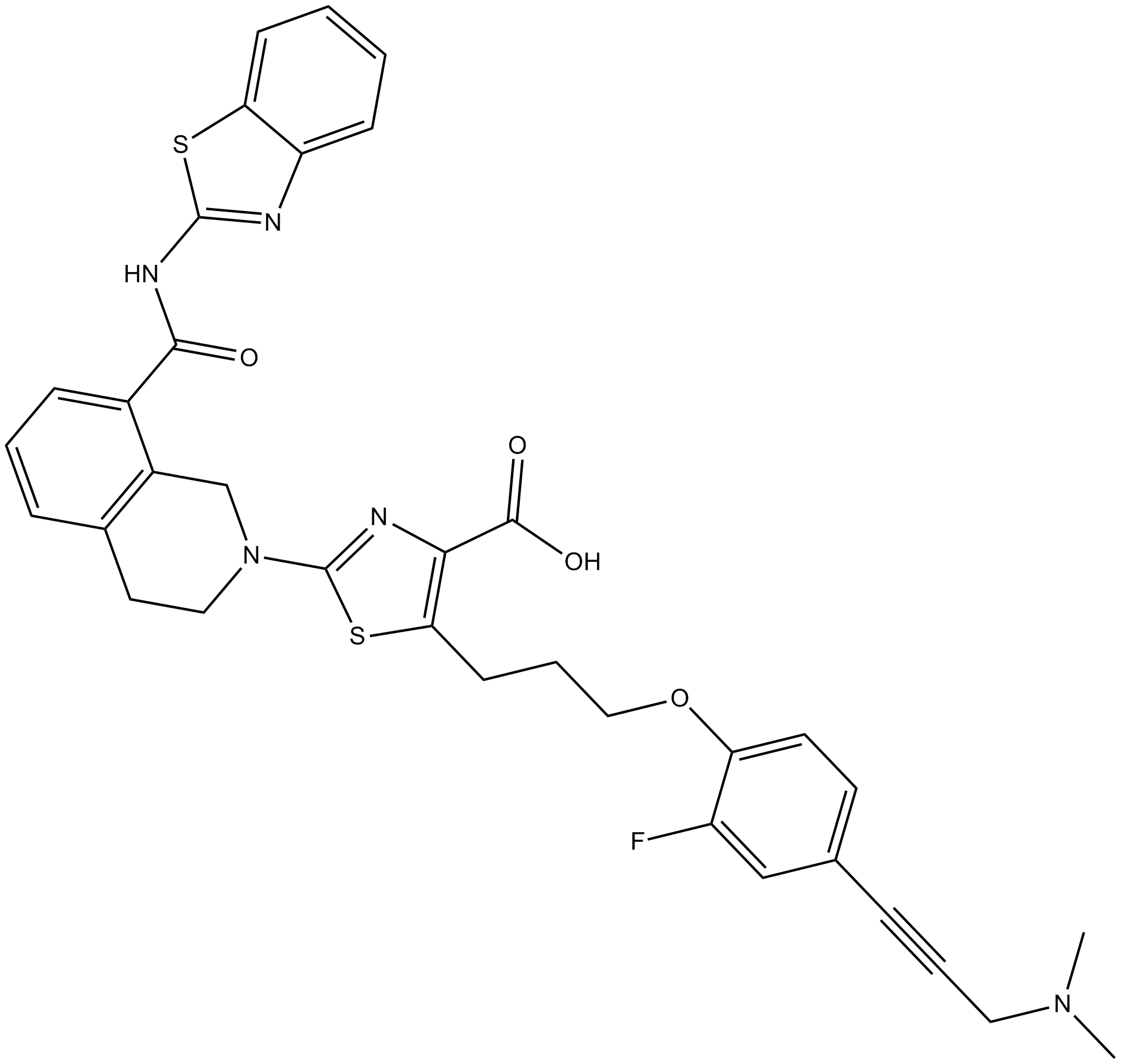
-
GC16278
A-1210477
MCL-1 inhibitor
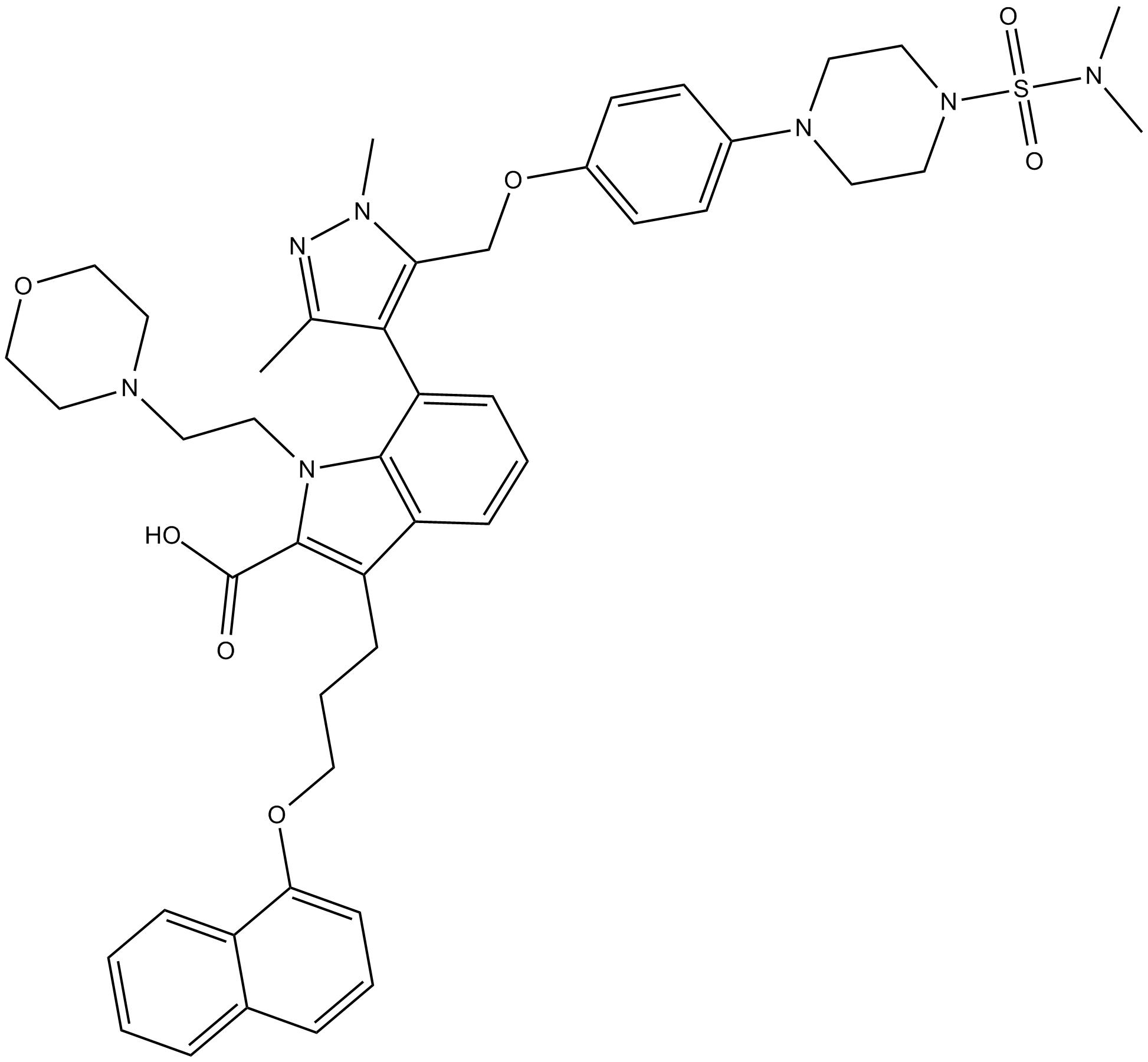
-
GC17513
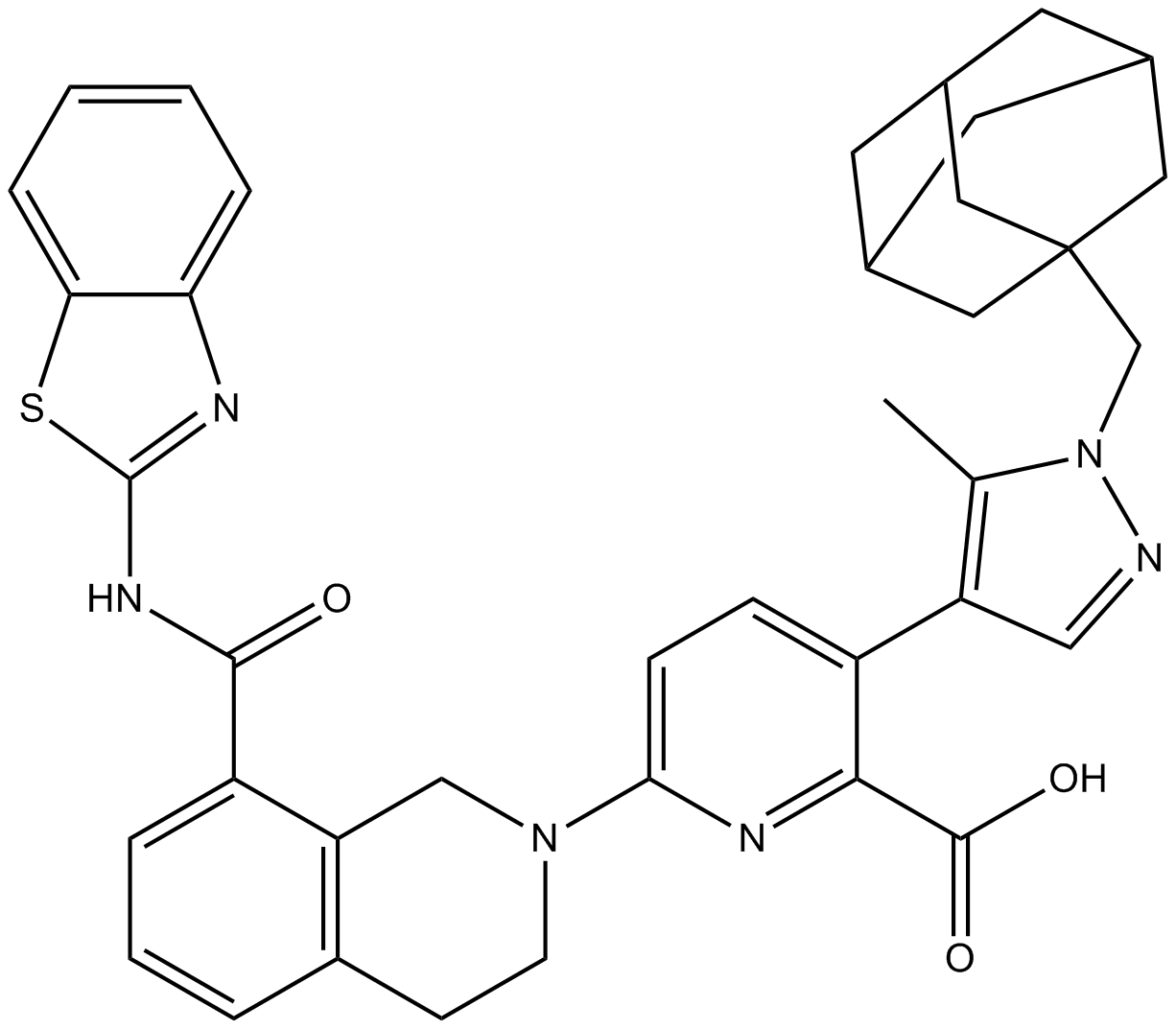
A-1331852
BCL-XL inhibitor, potent and selective
-
GC32981
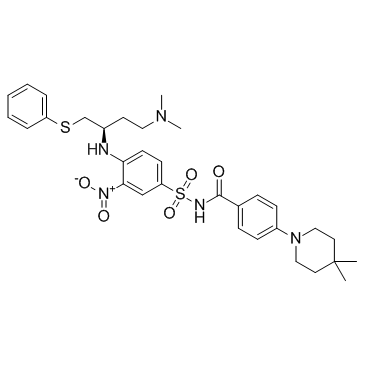
A-385358
A-385358 is a selective inhibitor of Bcl-XL with Kis of 0.80 and 67 nM for Bcl-XL and Bcl-2, respectively.
-
GC64674
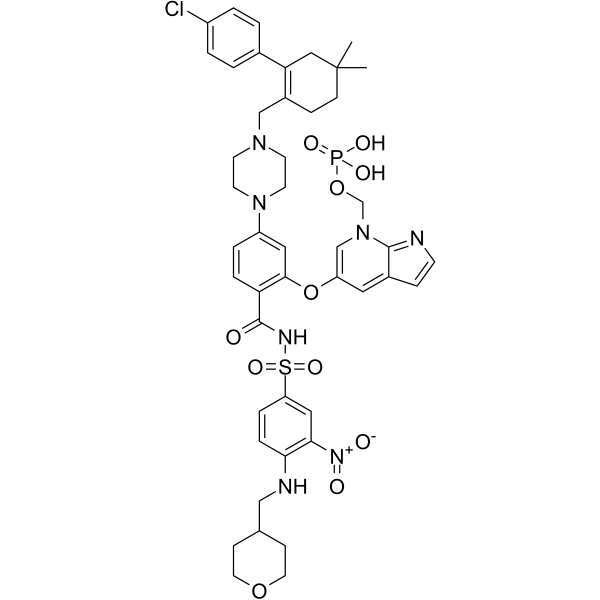
ABBV-167
ABBV-167 is a phosphate prodrug of the BCL-2 inhibitor venetoclax.
-
GC14069
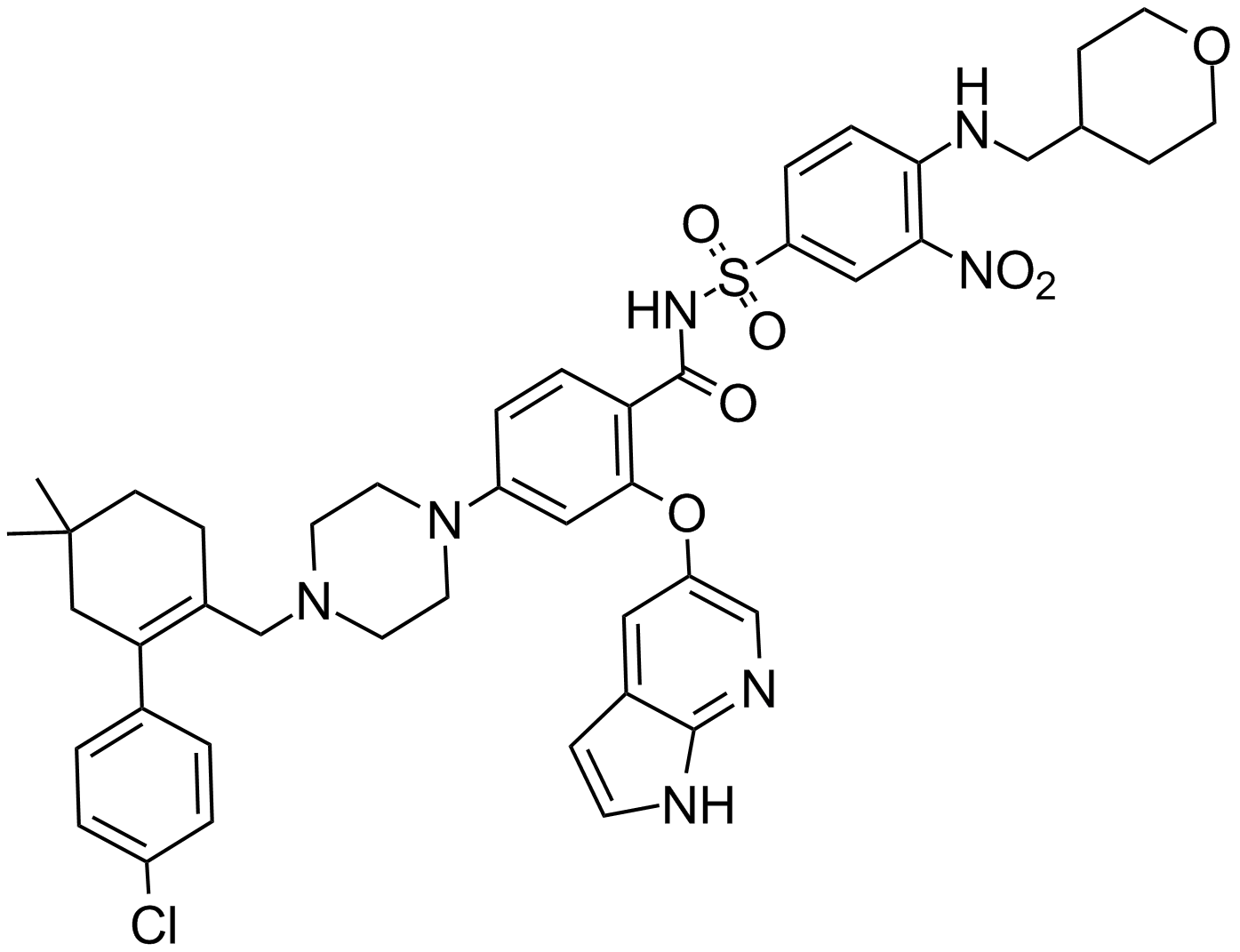
ABT-199
A Bcl-2 inhibitor
-
GC12405
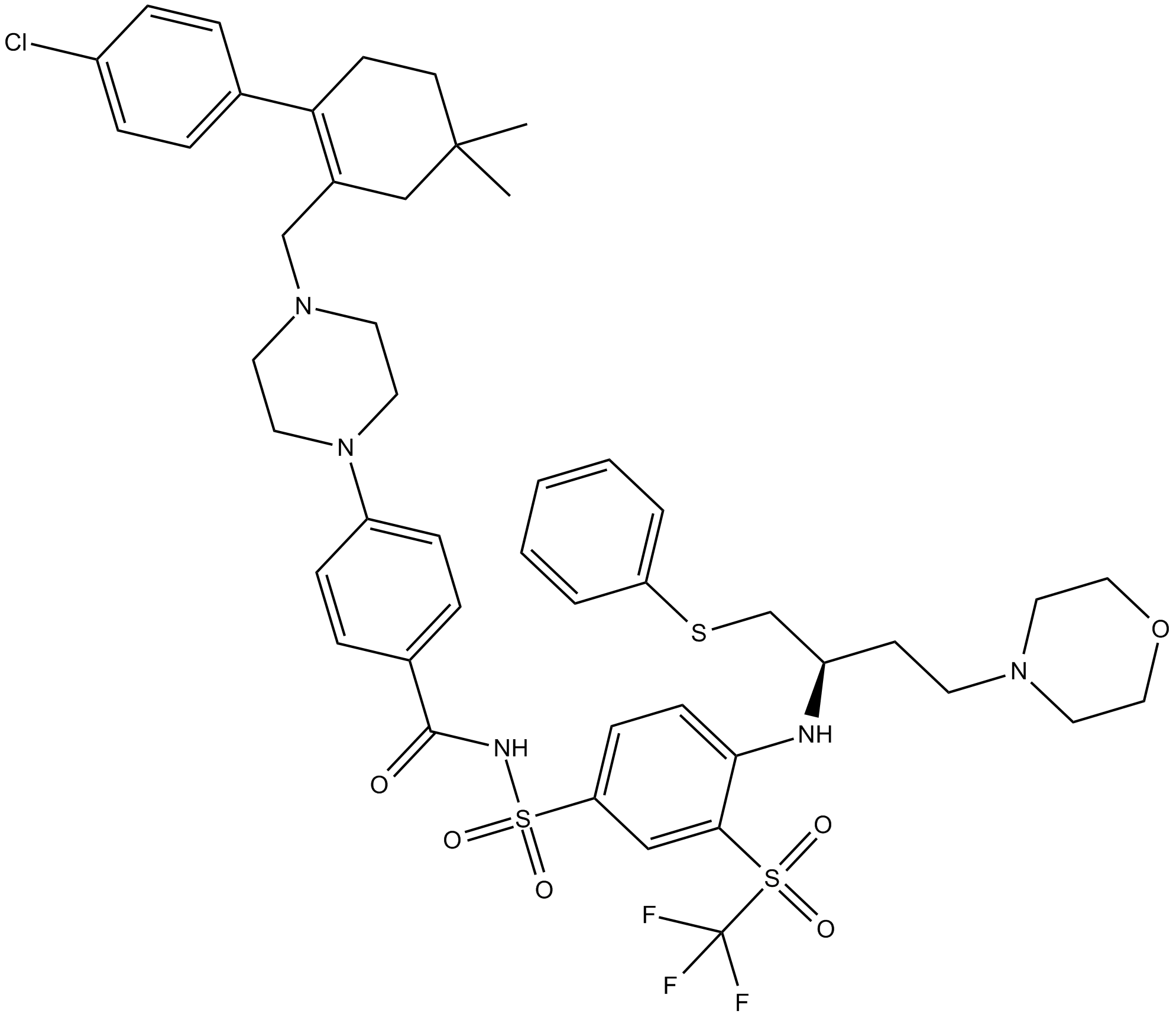
ABT-263 (Navitoclax)
ABT-263 (Navitoclax) is a inhibitor of Bcl-xL, Bcl-2 and Bcl-w, with Ki ≤0.5 nM, ≤1 nM and ≤1 nM respectively.
-
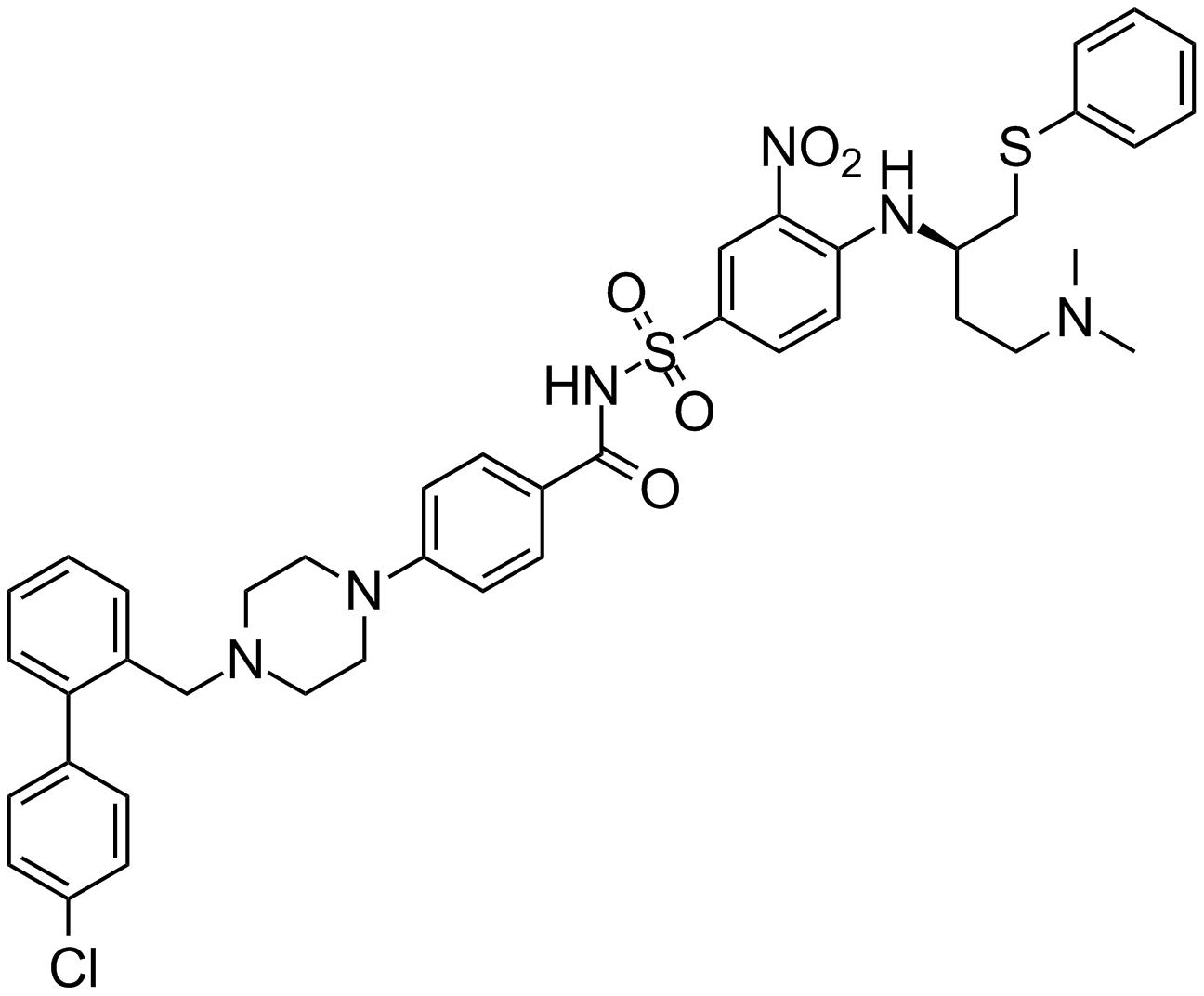
GC17234
ABT-737
An inhibitor of anti-apoptotic Bcl-2 proteins
-
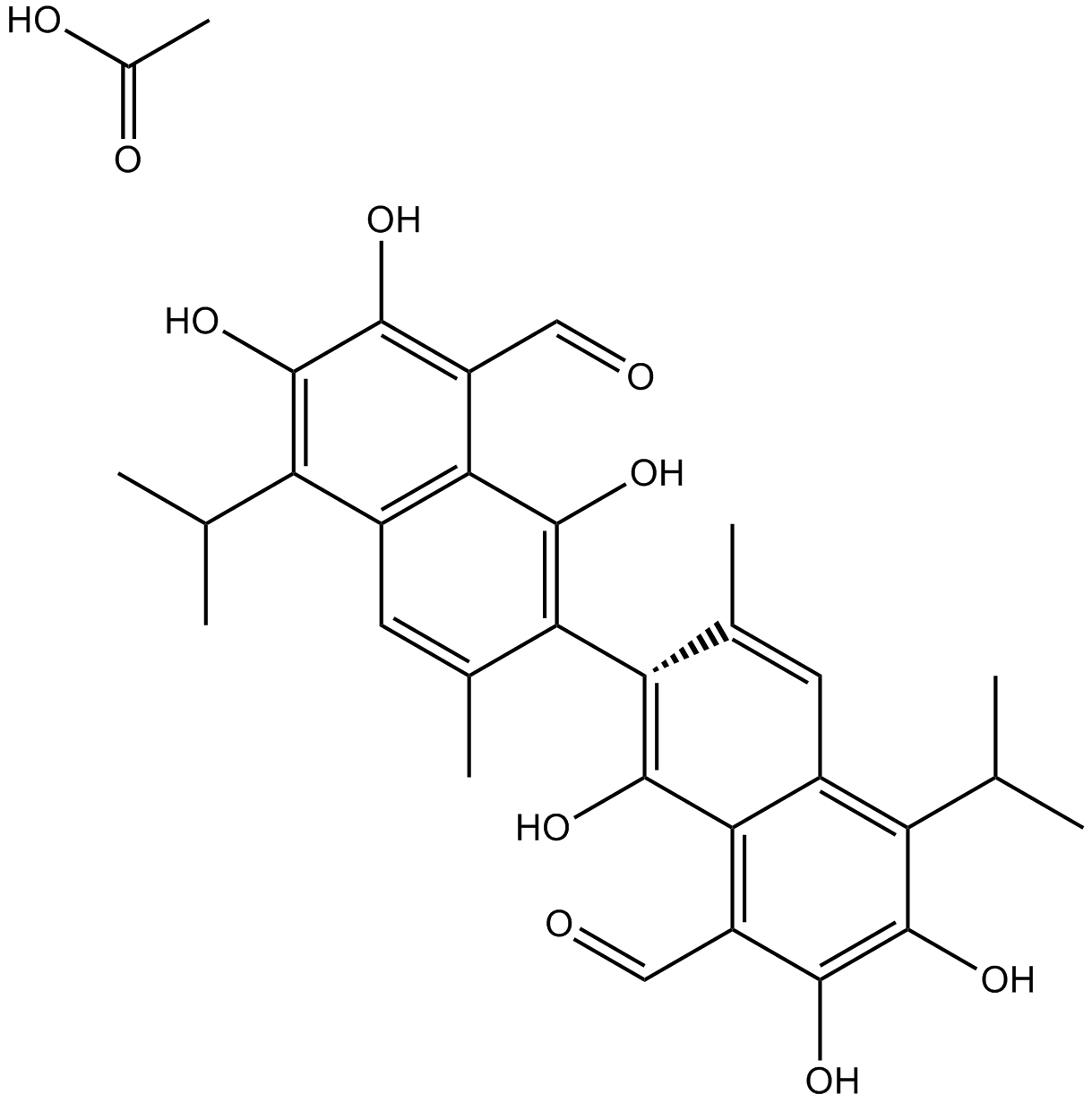
GN10341
Acetate gossypol
-
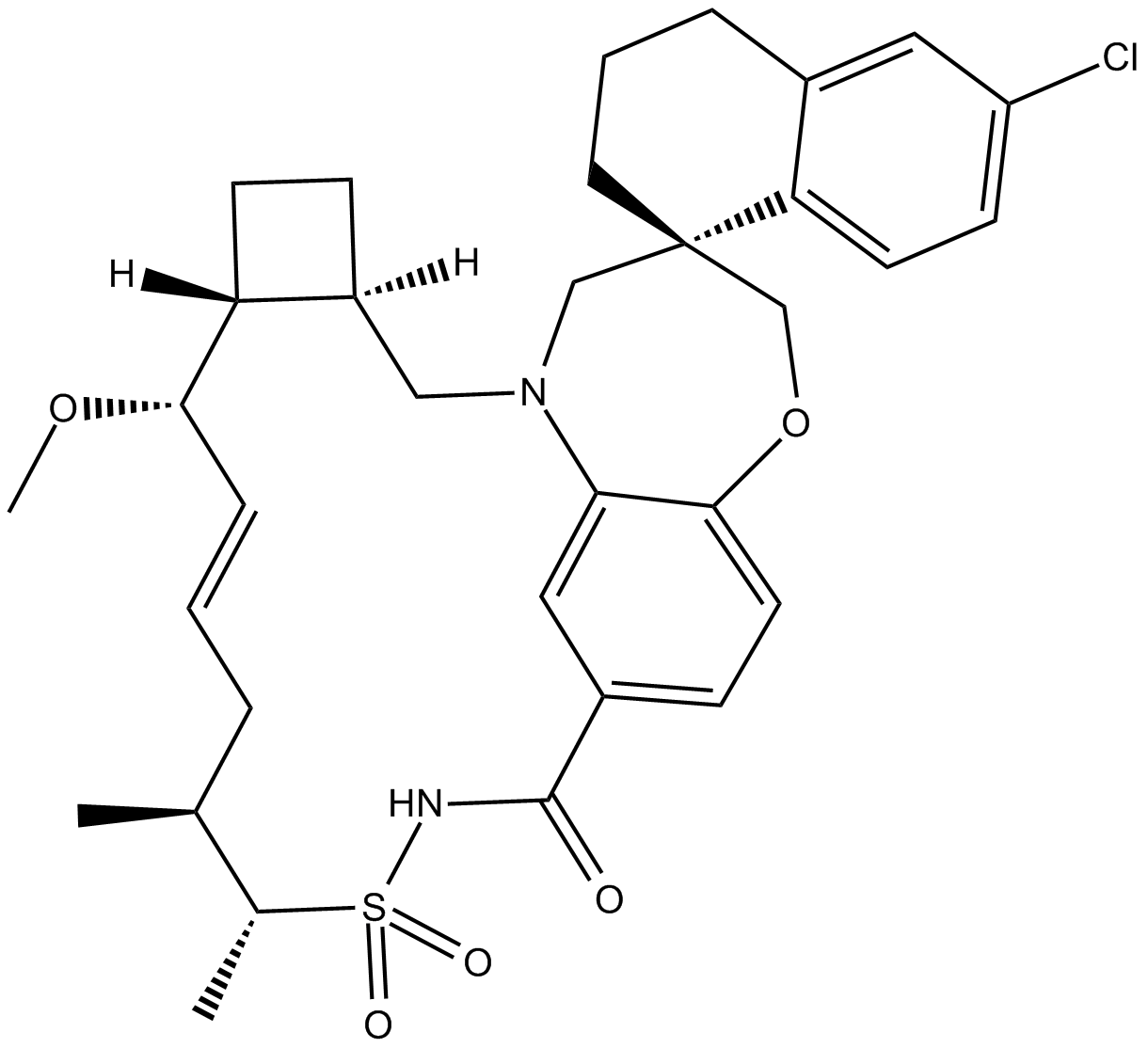
GC19452
AMG-176
AMG-176 (AMG-176) is a potent, selective and orally active MCL-1 inhibitor, with a Ki of 0.13 nM.
-
GC14080
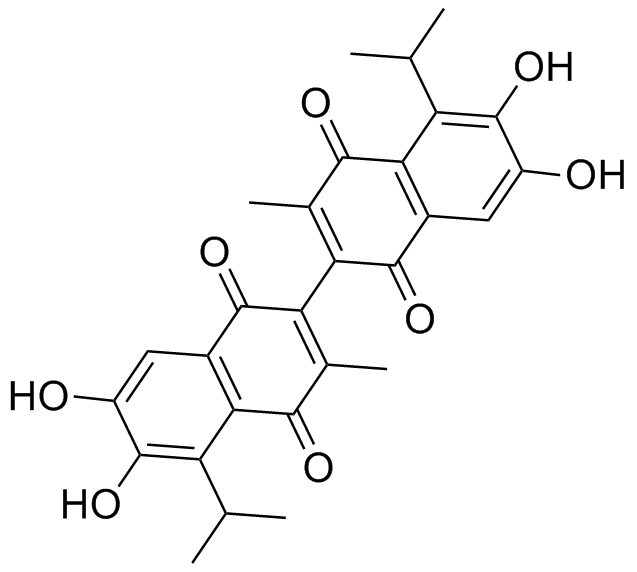
Apogossypolone (ApoG2)
-
GC11106
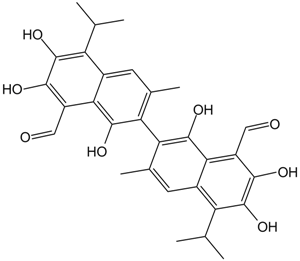
AT-101
-
GC33247
AZD-5991
An Mcl-1 inhibitor

-
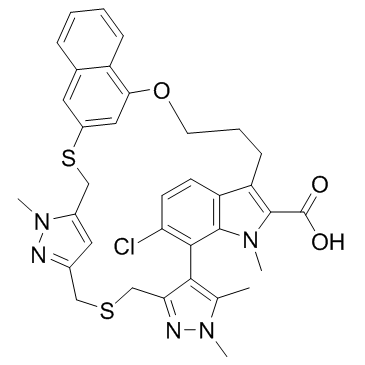
GC33283
AZD-5991 Racemate
AZD-5991 Racemate is the racemate of AZD-5991. AZD-5991 Racemate is a Mcl-1 inhibitor with an IC50 of <3 nM in FRET assay.
-
GC33239
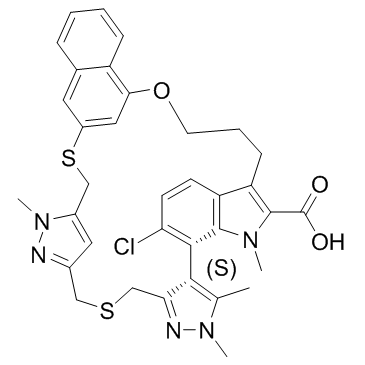
AZD-5991 S-enantiomer
AZD-5991 S-enantiomer is the less active enantiomer of AZD-5991. AZD-5991 S-enantiomer is a Mcl-1 inhibitor with an IC50 of 6.3 μM in FRET assay and a Kd of 0.98 μM in surface plasmon resonance (SPR) assay.
-
GC33255
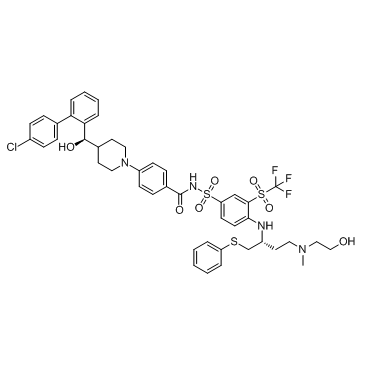
AZD4320
AZD4320 is a novel BH3-mimicking dual BCL2/BCLxL inhibitor with IC50s of 26 nM, 17 nM, and 170 nM for KPUM-MS3, KPUM-UH1, and STR-428 cells, respectively.
-
GC34263
Bak BH3
Bak BH3 is derived from the BH3 domain of Bak, can antagonize the function of Bcl-xL in cells.

-
GC12053
BAM7
A direct activator of Bax
-
GC12763
Bax channel blocker
-
GC16023
Bax inhibitor peptide P5
Bax inhibitor
-
GC17195
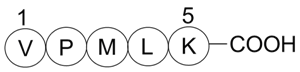
Bax inhibitor peptide V5
A Bax inhibitor
-
GC16695
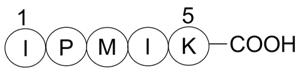
Bax inhibitor peptide, negative control
Peptide inhibit Bax translocation to mitochondria
-
GC63325
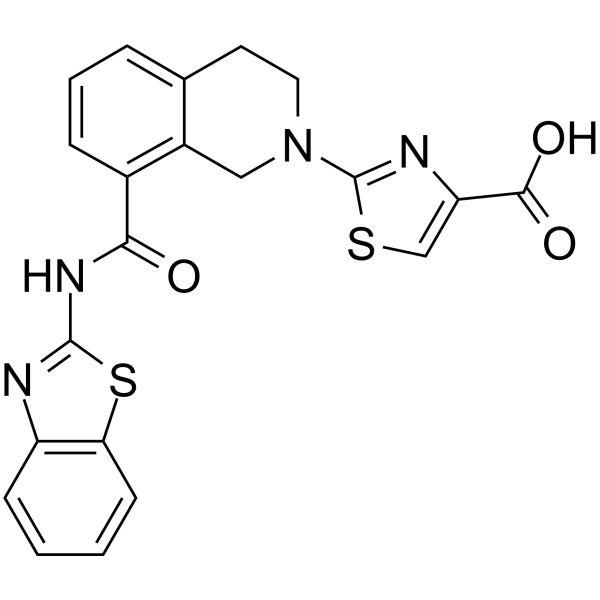
Bcl-xL antagonist 2
Bcl-xL antagonist 2 is a potent, selective, and orally active antagonist of BCL-XL with an IC50 and Ki of 0.091 μM and 65 nM, respectively. Bcl-xL antagonist 2 promotes the apoptosis of cancer cells. Bcl-xL antagonist 2 has the potential for the research of the chronic lymphocytic leukemia (CLL) and non-Hodgkin’s lymphoma (NHL).
-
GC62599
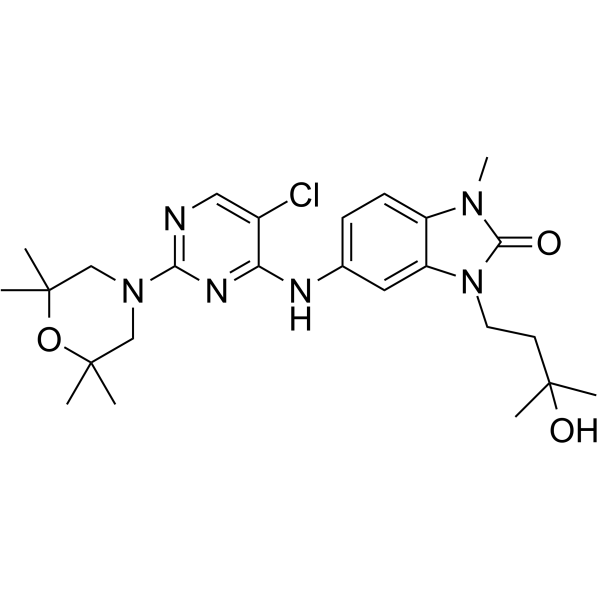
BCL6-IN-4
BCL6-IN-4 is a potent B-cell lymphoma 6 (BCL6) inhibitor with an IC50 of 97 nM. BCL6-IN-4 has anti-tumor activities.
-
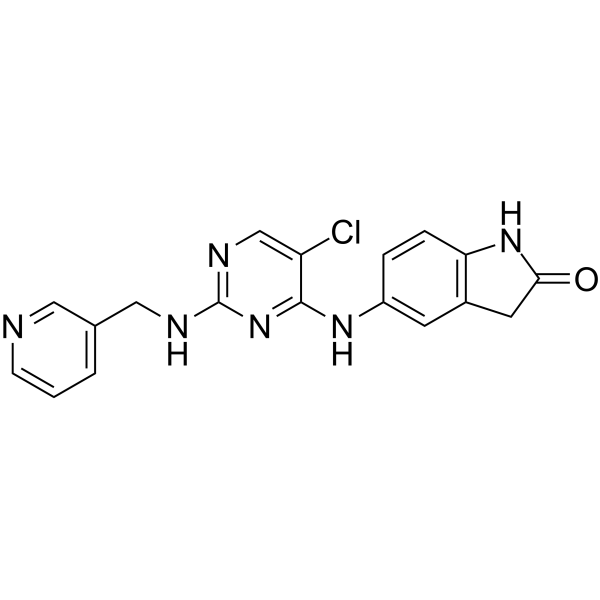
GC68012
BCL6-IN-7
-
GC10721
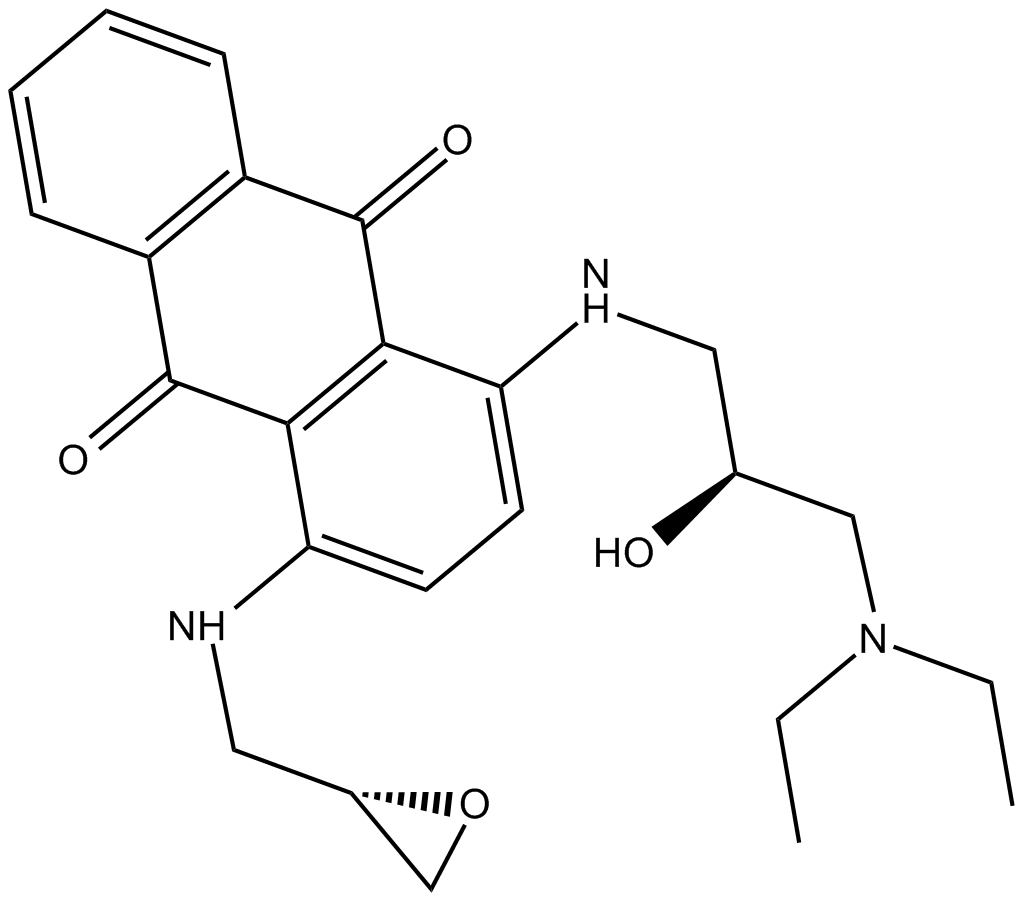
BDA-366
BDA-366 is a potent Bcl2 antagonist (Ki = 3.3 nM), binding Bcl2-BH4 domain with high affinity and selectivity. BDA-366 induces conformational change in Bcl2 that abrogates its antiapoptotic function, converting it from a survival molecule to a cell death inducer. BDA-366 suppresses growth of lung cancer cells.
-
GC18136
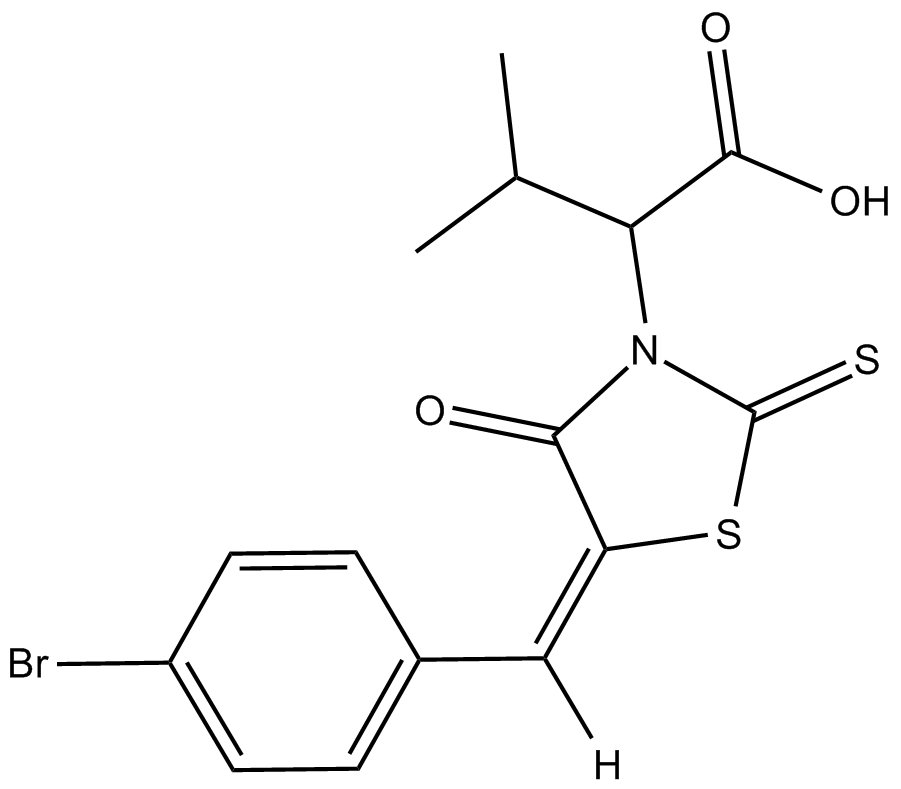
BH3I-1
Bcl-2 or Bcl-XL inhibitor
-
GC15987
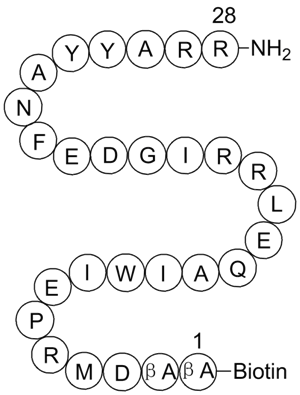
BIM, Biotinylated
Bim peptide fragment with a biotin moiety attached
-
GC33407
BM 957
BM 957 is a potent Bcl-2 and Bcl-xL inhibitor, with Kis of 1.2, <1 nM and IC50s of 5.4, 6.0 nM respectively.
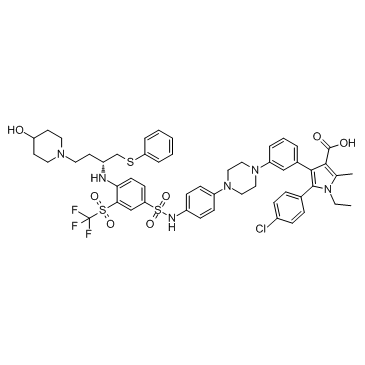
-
GC13498
BM-1074
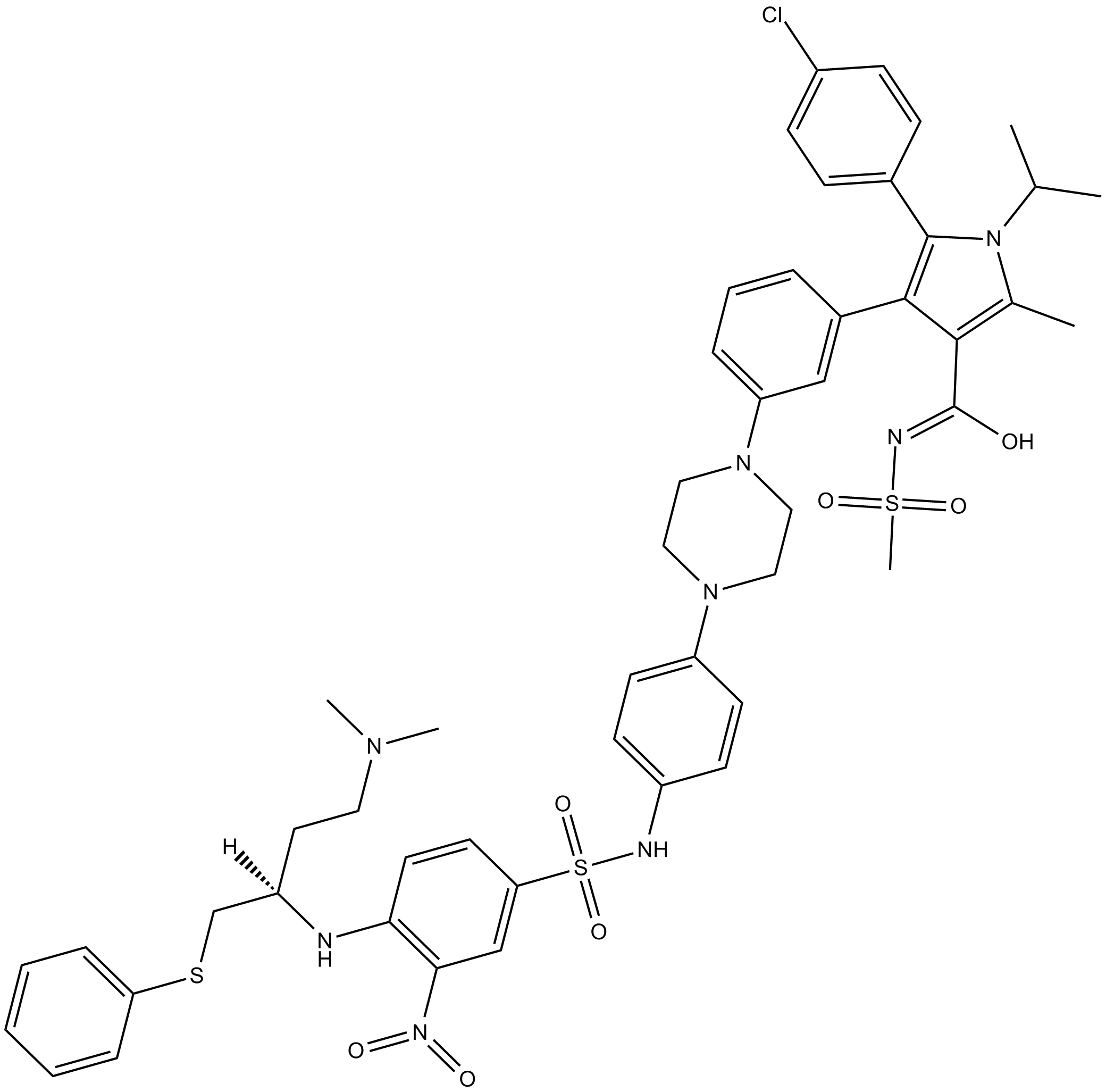
-
GC62871
BM-1244
BM-1244 (APG-1252-M1) is a potent Bcl-xL/Bcl-2 inhibitor with Kis of 134 and 450 nM for Bcl- xL and Bcl-2, respectively. BM-1244 inhibits senescent fibroblasts (SnCs) with an EC50 of 5 nM. (From patent WO2019033119A1).
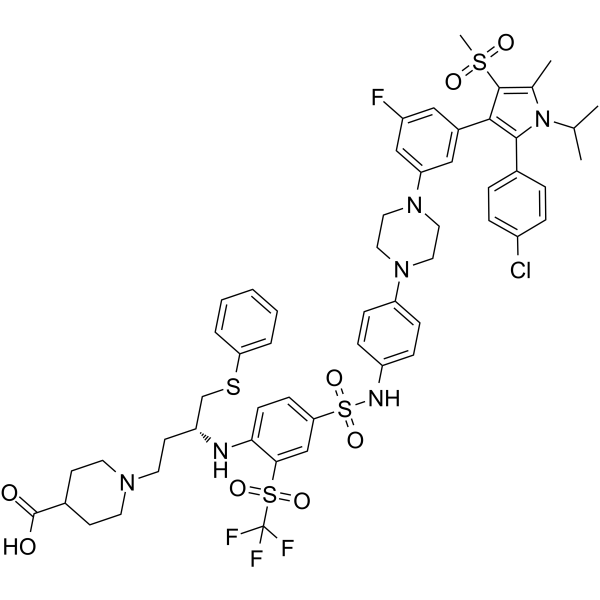
-
GC38014
BT2
An Mcl-1 inhibitor
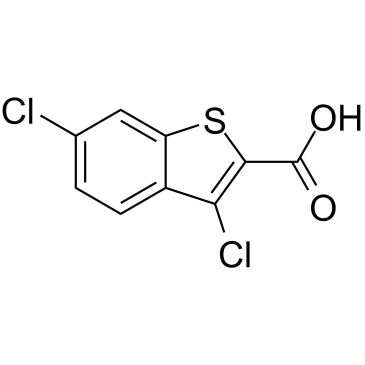
-
GC31806
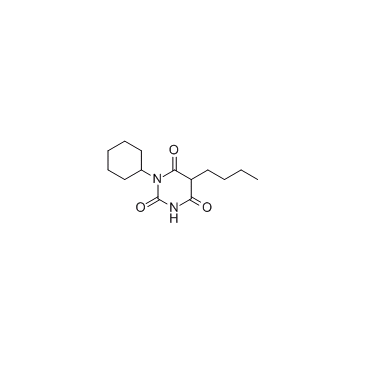
Bz 423 (BZ48)
Bz 423 (BZ48) is a pro-apoptotic 1,4-benzodiazepine with therapeutic properties in murine models of lupus demonstrating selectivity for autoreactive lymphocytes, and activates Bax and Bak.
-
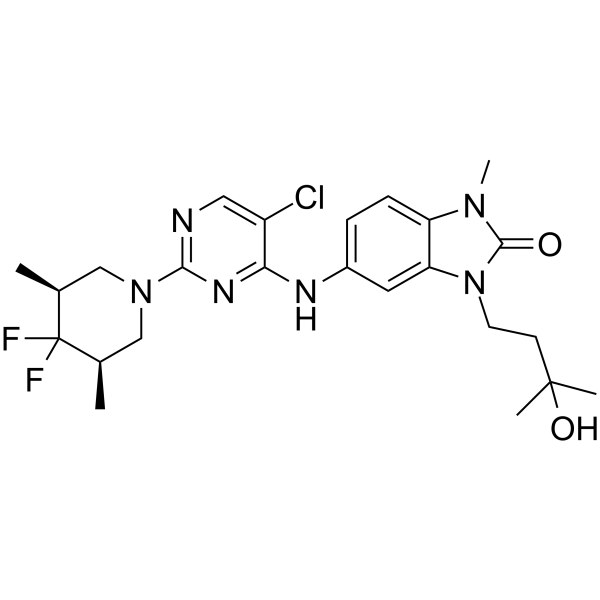
GC62561
CCT369260
CCT369260 (compound 1) is an orally avtive B-cell lymphoma 6 (BCL6) inhibitor with anti-tumor activity. CCT369260 (compound 1) exhibits an IC50 of 520 nM.
-
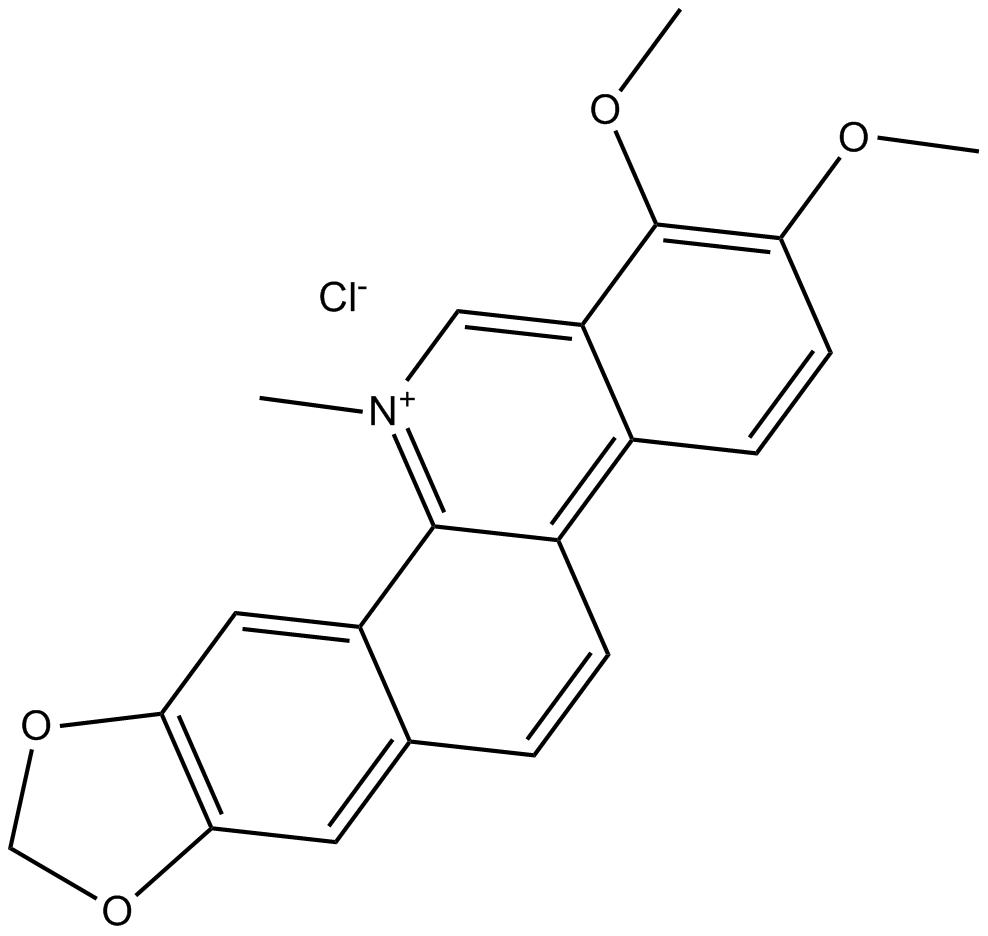
GN10463
Chelerythrine
-
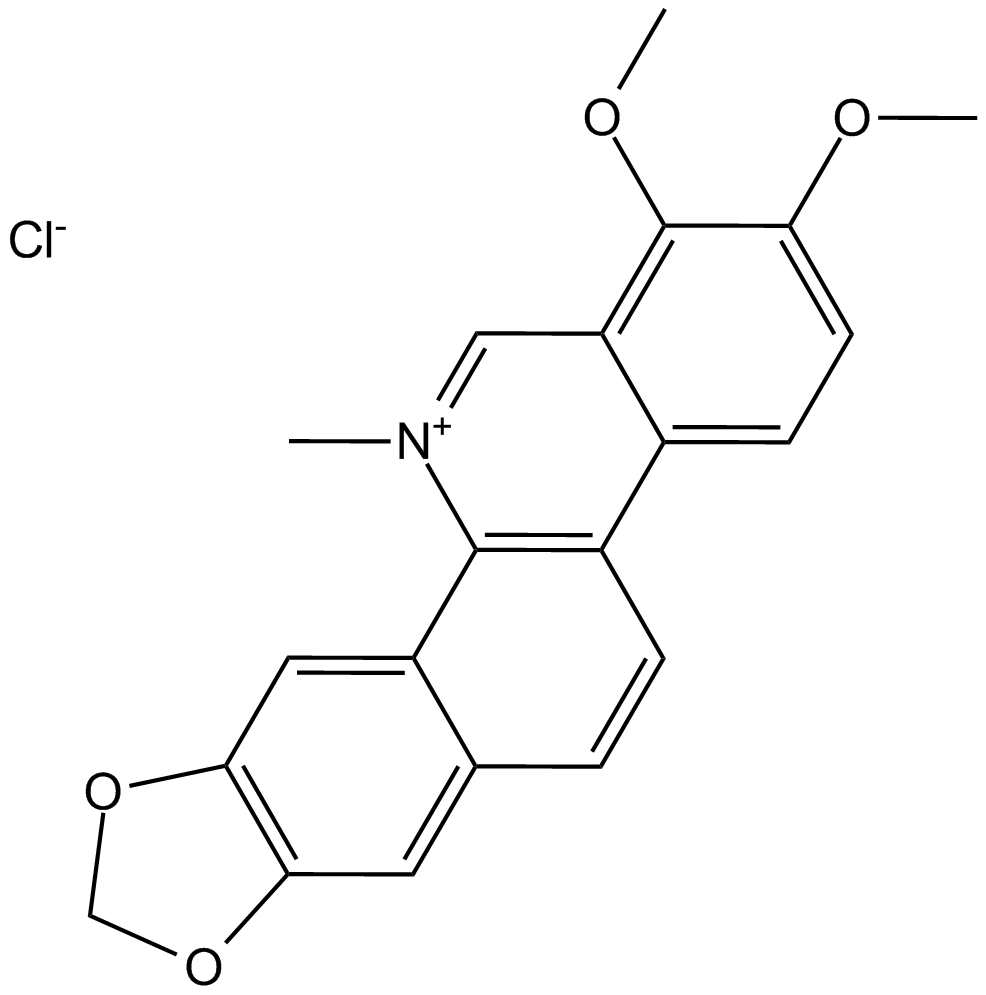
GC13065
Chelerythrine Chloride
Potent inhibitor of PKC and Bcl-xL
-
GN10219
Ciwujianoside-B
-
GC60111
Clitocine
Clitocine, an adenosine nucleoside analog isolated from mushroom, is a potent and efficacious readthrough agent. Clitocine acts as a suppressor of nonsense mutations and can induce the production of p53 protein in cells harboring p53 nonsense-mutated alleles. Clitocine can induce apoptosis in multidrug-resistant human cancer cells by targeting Mcl-1. Anticancer activity.
-
GN10040
Dehydrocorydaline
-
GC10661
Destruxin B
insecticidal and phytotoxic activity;induces apoptosis
-
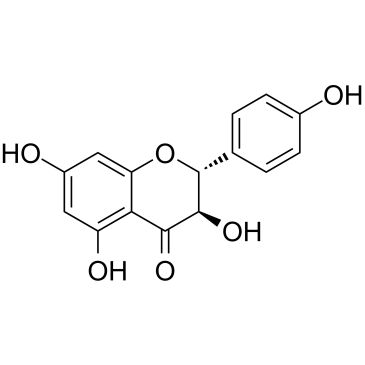
GC38617
Dihydrokaempferol
A flavone with diverse biological activities
-
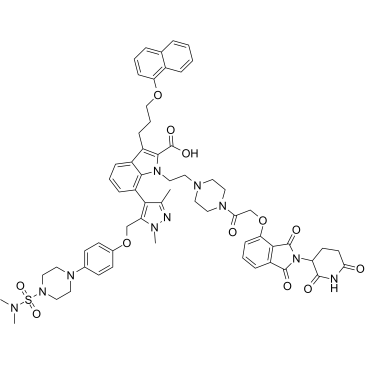
GC35882
dMCL1-2
dMCL1-2 is a potent and selective PROTAC of myeloid cell leukemia 1 (MCL1) (Bcl-2 family member) based on Cereblon, which binds to MCL1 with a KD of 30 nM. dMCL1-2 activats the cellular apoptosis machinery by degradation of MCL1.
-
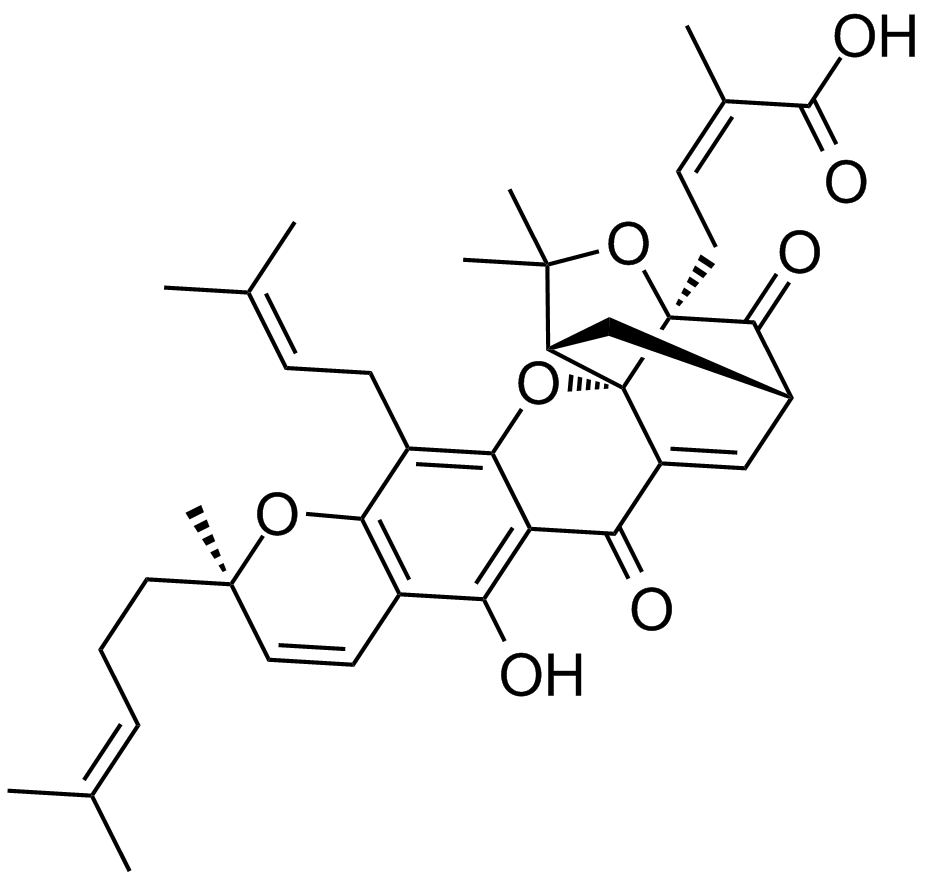
GC12139
Gambogic Acid
A xanthonoid with anticancer activity
-
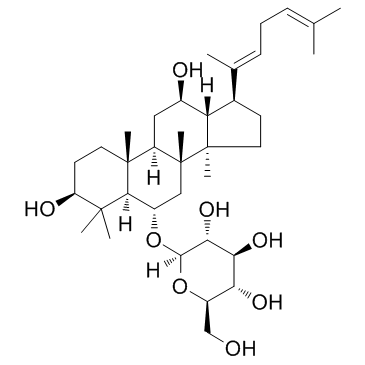
GC32998
Ginsenoside Rh4
-
GC34134
Glycocholic acid
A glycine-conjugated form of cholic acid
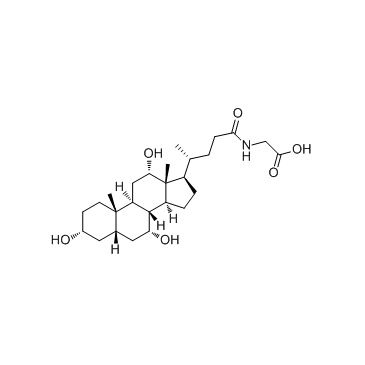
-
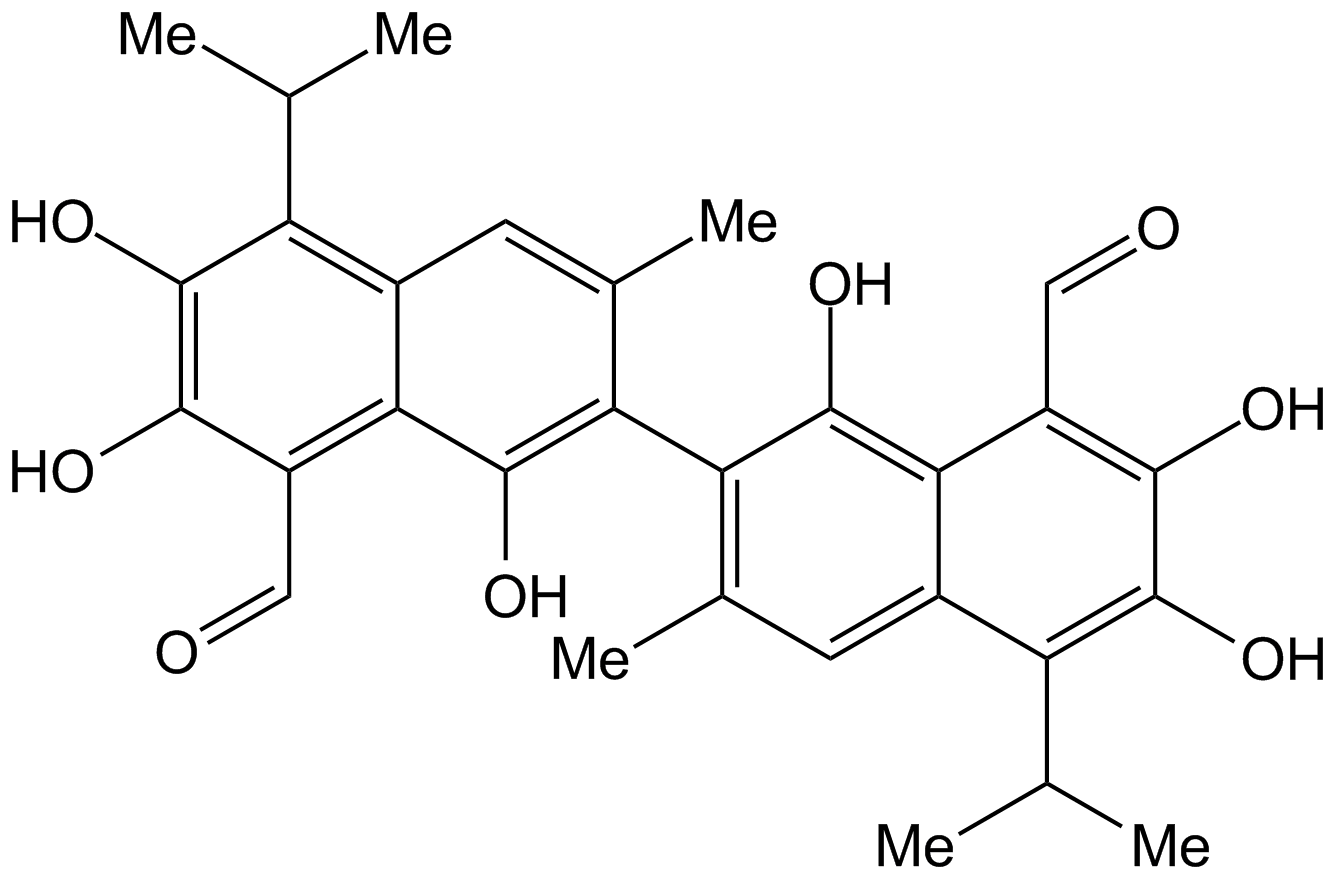
GN10082
Gossypol
-
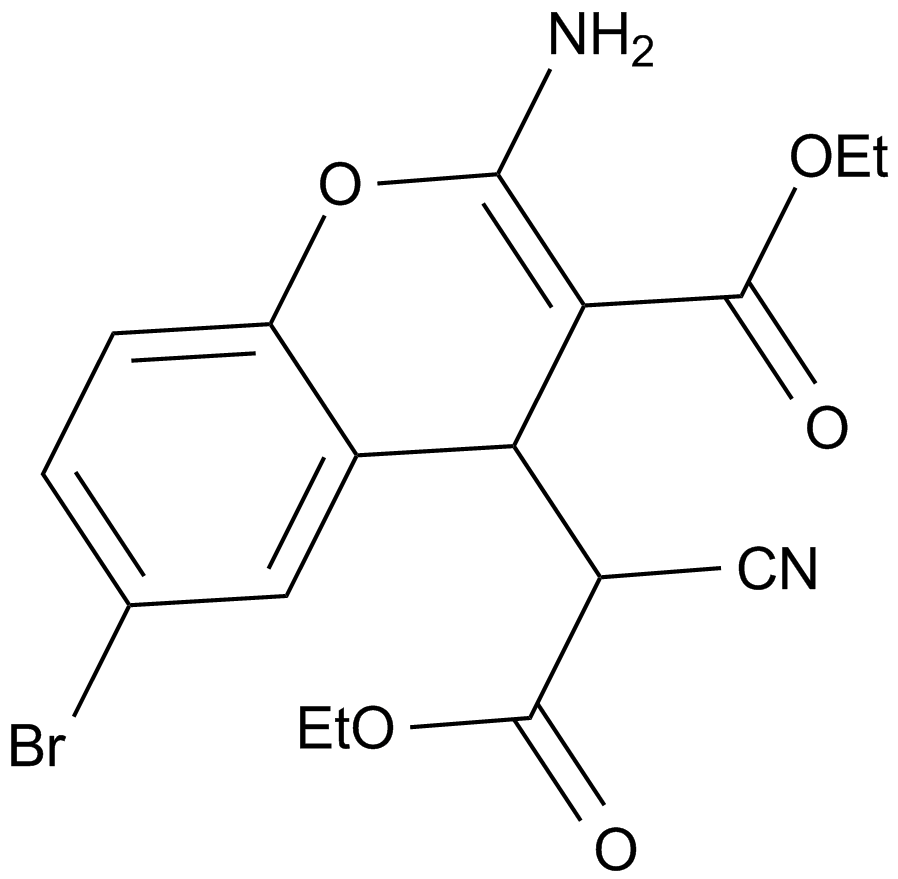
GC11510
HA14-1
A Bcl-2 inhibitor
-
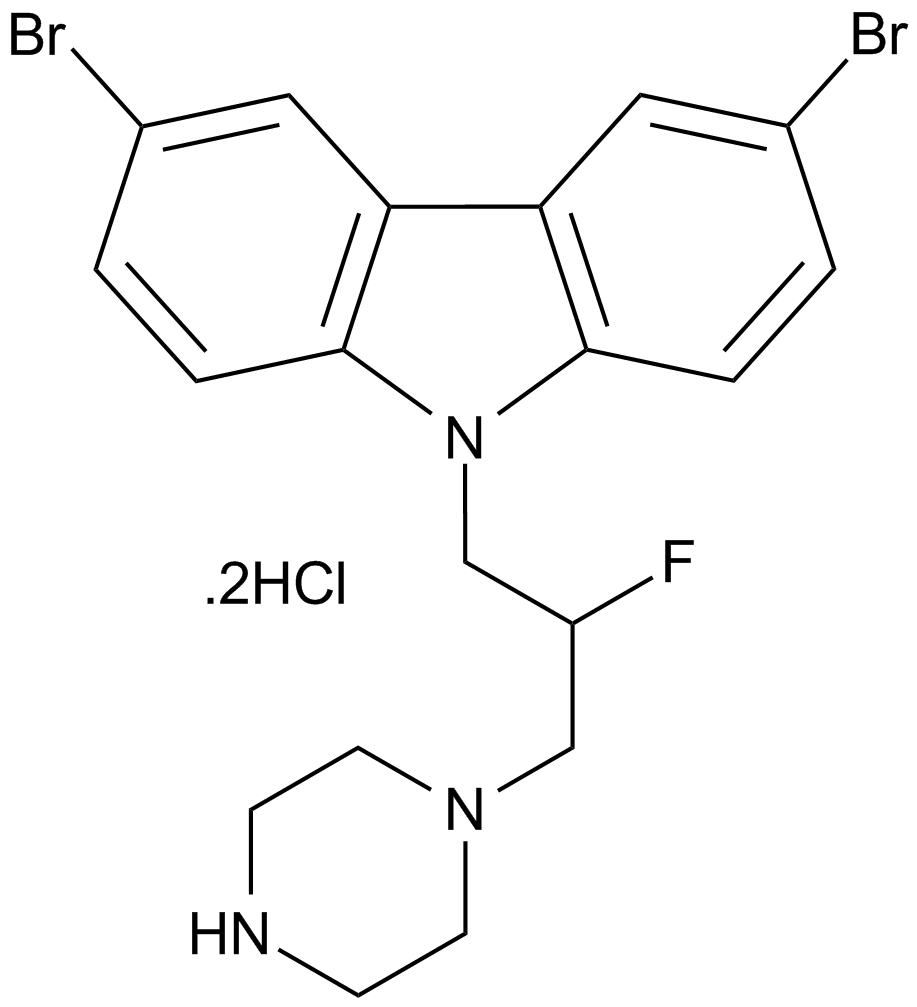
GC12046
iMAC2
-
GN10645
Jaceosidin
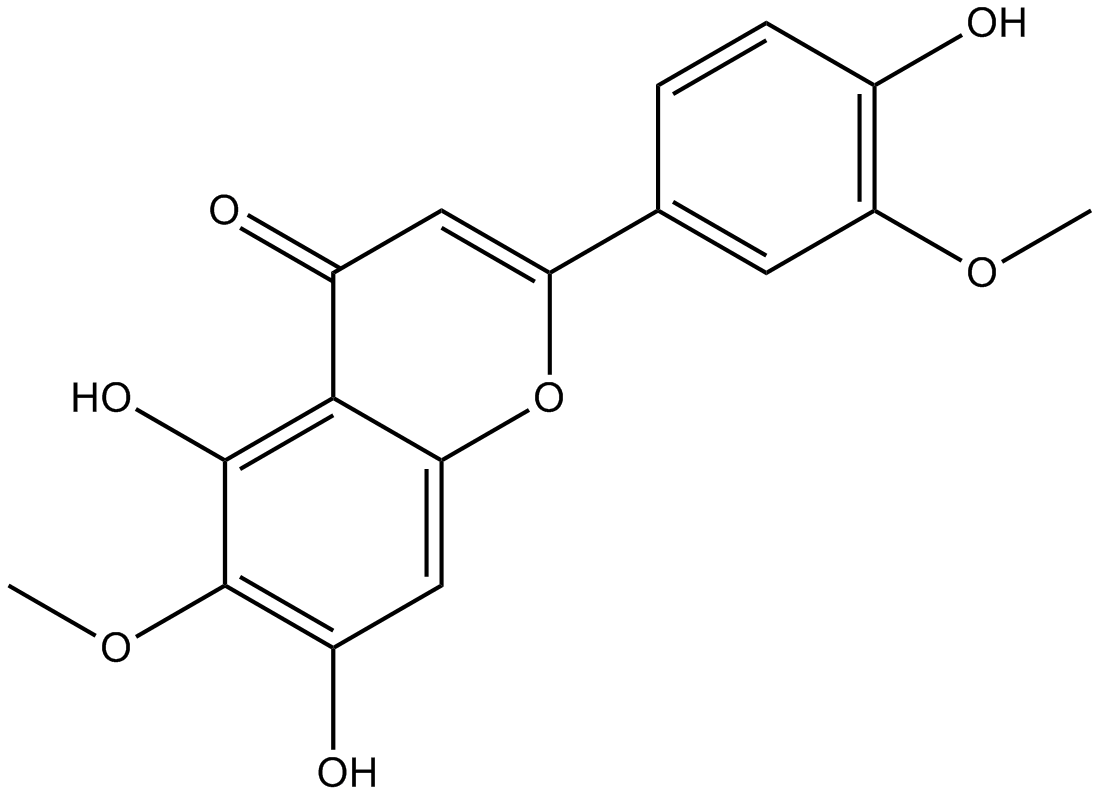
-
GC64467
Lisaftoclax
Lisaftoclax (compound 6) is a dual Bcl-2 and Bcl-xl inhibitor with anti-tumor activity, extracted from patent WO2018027097A1. Lisaftoclax exhibits IC50 values of 2 nM and 5.9 nM for Bcl-2 and Bcl-xl, respectively.
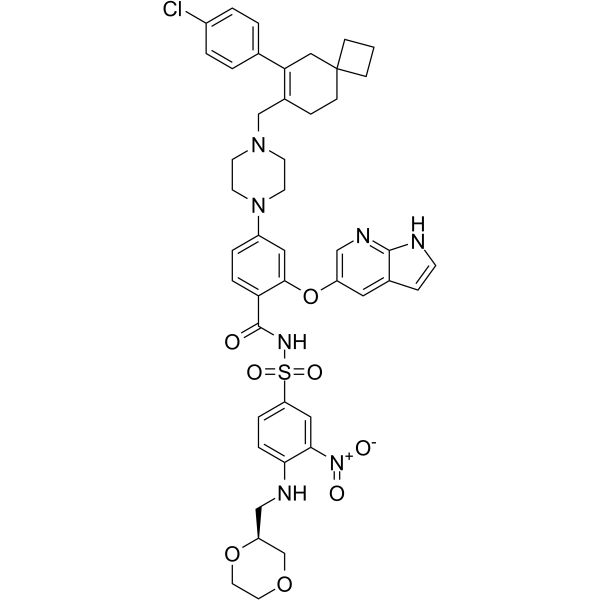
-
GC15480
Marinopyrrole A
Marinopyrrole A (Marinopyrrole A) is a novel and specific Mcl-1 inhibitor with an IC50 value of 10.1 μM, and shows >8 fold selectivity than BCL-xl (IC50 > 80 μM).
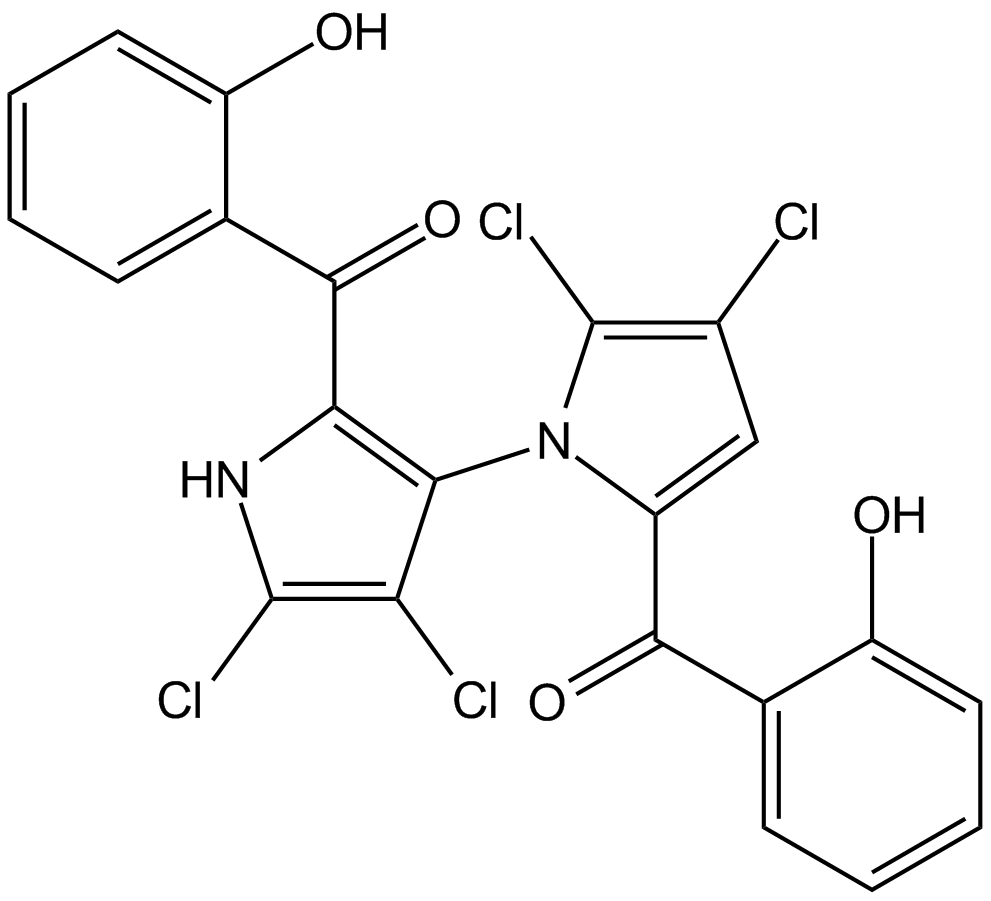
-
GC66017
Mcl-1 inhibitor 6
Mcl-1 inhibitor 6 is an orally active, selective myeloid cell leukemia 1 (Mcl-1) protein inhibitor with a Kd of 0.23 nM and a Ki of 0.02 μM. Mcl-1 inhibitor 6 possesses superior selectivity over other Bcl-2 family members (Bcl-2, Bcl2A1, Bcl-xL, and Bcl-w, Kd>10 μM). Mcl-1 inhibitor 6 is a potent antitumor agent.

-
GC38927
MCL-1/BCL-2-IN-2
MCL-1/BCL-2-IN-2 (Compound 6) is a potent and selective Mcl-1 and Bcl-2 dual inhibitor.
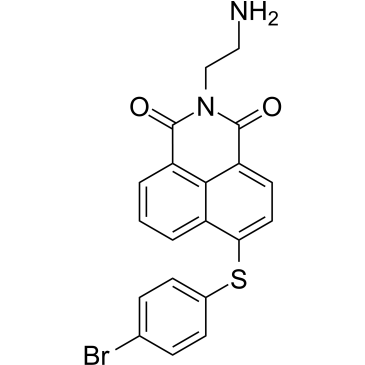
-
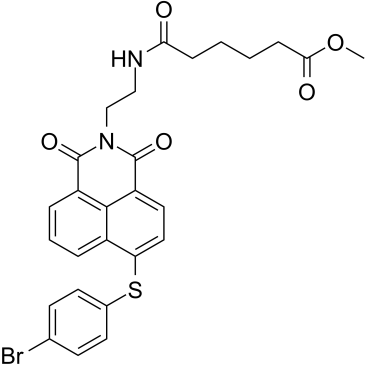
GC38928
MCL-1/BCL-2-IN-3
MCL-1/BCL-2-IN-3 (Compound 2) is a potent and selective Mcl-1 and Bcl-2 dual inhibitor with IC50s of 5.95 and 4.78 μM, respectively.
-
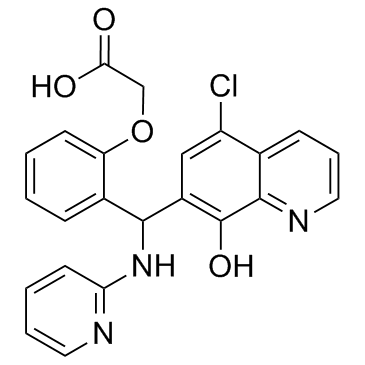
GC36556
Mcl1-IN-1
Mcl1-IN-1 is an inhibitor of myeloid cell factor 1 (Mcl-1) (IC50=2.4 ?M).
-
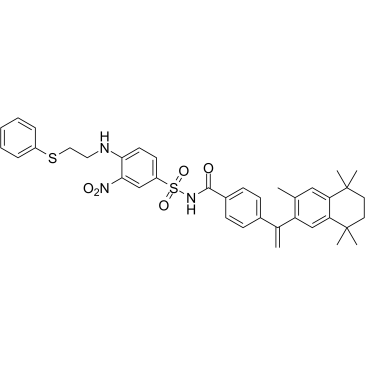
GC36557
Mcl1-IN-11
Mcl1-IN-11 (Compound G) is a selective Mcl-1 inhibitor, less potent at Bcl-2, with Kis of 0.06 and 4.2 μM, respectively.
-
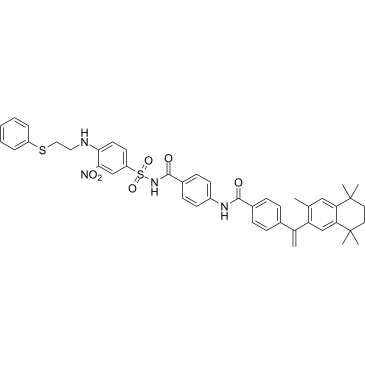
GC36558
Mcl1-IN-12
Mcl1-IN-12 (Compound F) is a selective Mcl-1 inhibitor, less potent at Bcl-2, with Kis of 0.29 and 3.1 μM, respectively. Anti-tumor activity.
-
GC33035
Mcl1-IN-2
Mcl1-IN-2 is an inhibitor of myeloid cell factor 1 (Mcl-1). Mcl1-IN-2 is a non-competitive Delhi metallo-β-lactamase (NDM-1) inhibitor. The IC50s of Mcl1-IN-2 against metallo-β-lactamases NDM-1, IMP-4, ImiS and L1 are 0.4637 μM, 3.980 μM, 0.2287 μM and 1.158 μM, respectively.
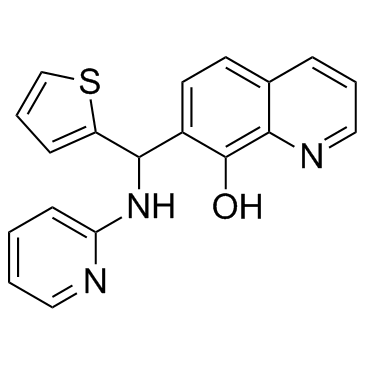
-
GC33347
Mcl1-IN-3
Mcl1-IN-3 is an inhibitor of Mcl1 extracted from patent WO2015153959A2, compound example 57; has an IC50 and Ki of 0.67 and 0.13 μM, respectively.
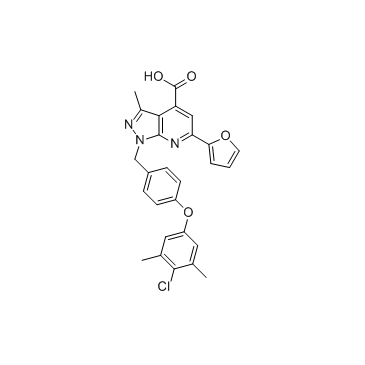
-
GC33364
Mcl1-IN-4
Mcl1-IN-4 is an inhibitor of Mcl1 with an IC50 of 0.2 μM.
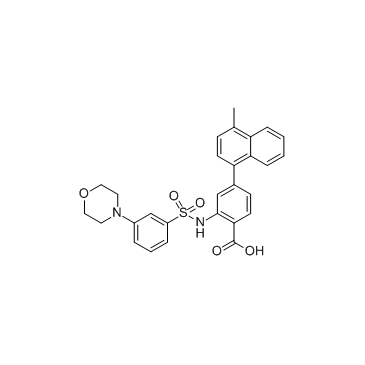
-
GC38811
Mcl1-IN-8
Mcl1-IN-8 (Compound 8) is an orally active Mcl-1-PUMA interface inhibitor, with a Ki of 0.3 μM. Mcl1-IN-8 exhibits dual activity on reduce PUMA-dependent apoptosis while deactivating Mcl-1-mediated anti-apoptosis in cancer cells.
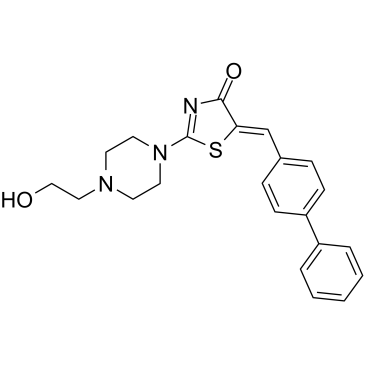
-
GC36559
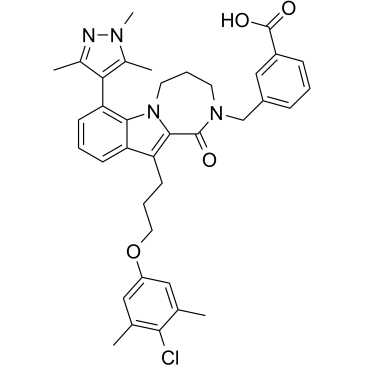
Mcl1-IN-9
Mcl1-IN-9 is a potent myeloid cell leukemia-1 (Mcl-1) Inhibitor with an IC50 of 446 nM in reengineered BCR-ABL+ B-ALL cells and a Ki of 0.03 nM.
-
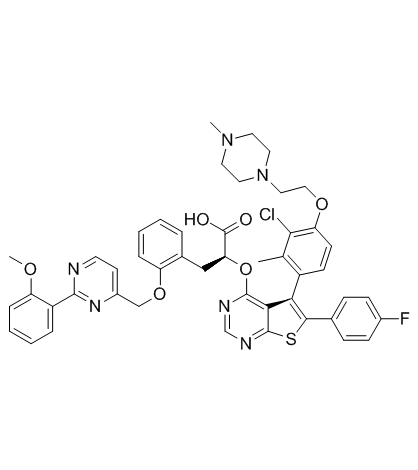
GC33295
MIK665 (S-64315)
MIK665 (S-64315) (S-64315), derived from S63845, is a myeloid cell leukemia sequence 1 (MCL1) inhibitor. MIK665 (S-64315) has an IC50 of 1.81 nM for MCL1.
-
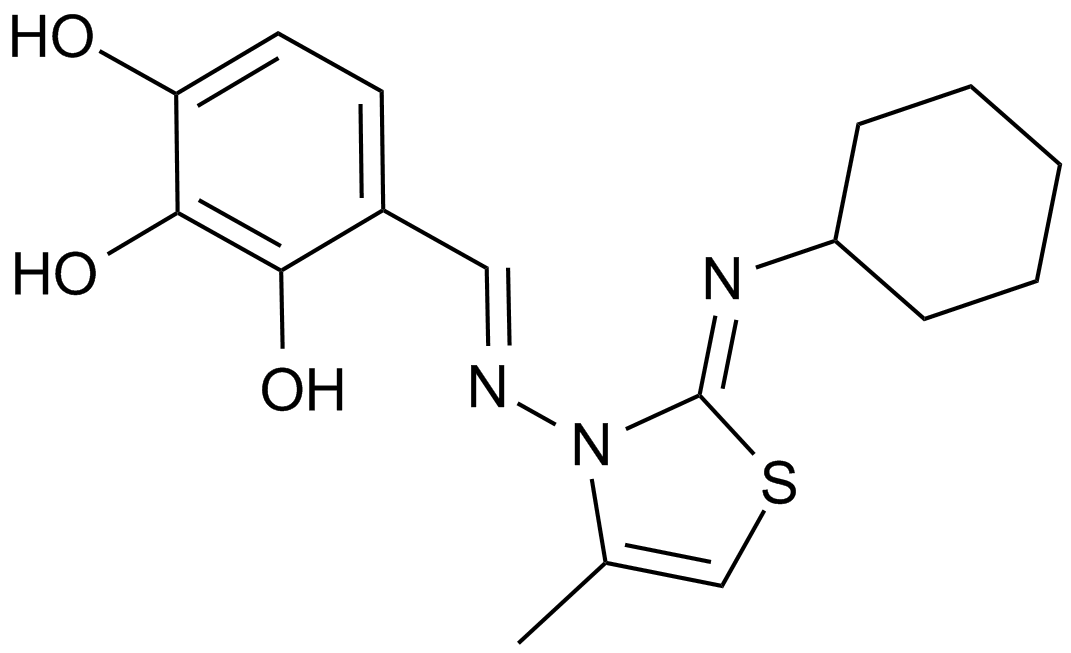
GC15400
MIM1
An Mcl-1 inhibiting molecule
-
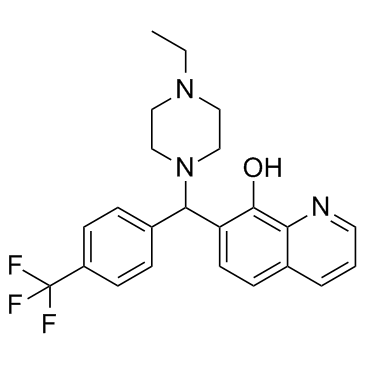
GC33312
ML311
An Mcl-1 inhibitor
-
GC61096
MSN-125
MSN-125 is a potent Bax and Bak oligomerization inhibitor. MSN-125 prevents mitochondrial outer membrane permeabilization (MOMP) with an IC50 of 4 μM. MSN-125 potently inhibits Bax/Bak-mediated apoptosis in HCT-116, BMK Cells, and primary cortical neurons, protects primary neurons against glutamate excitotoxicity.
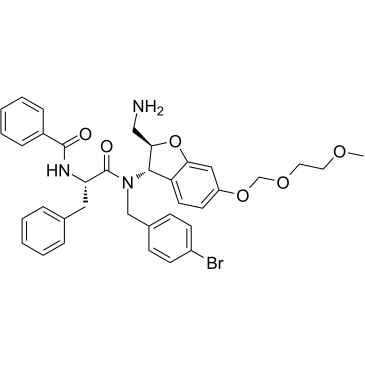
-
GC63083
MSN-50
MSN-50 is a Bax and Bak oligomerization inhibitor.
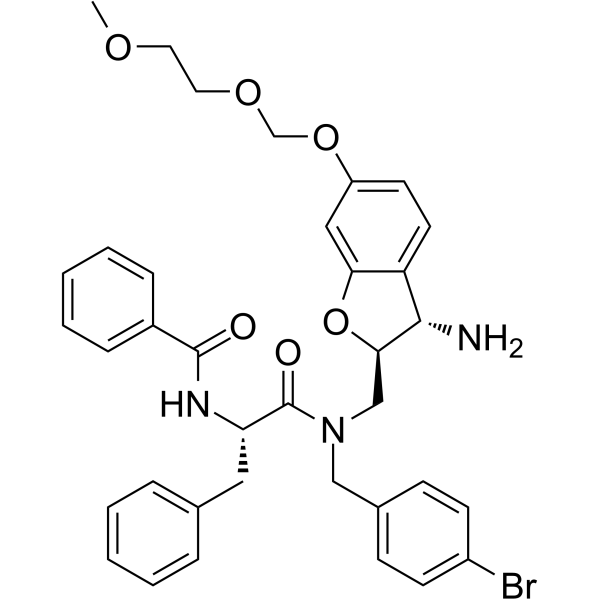
-
GC16938
Muristerone A
An ecdysteroid receptor agonist
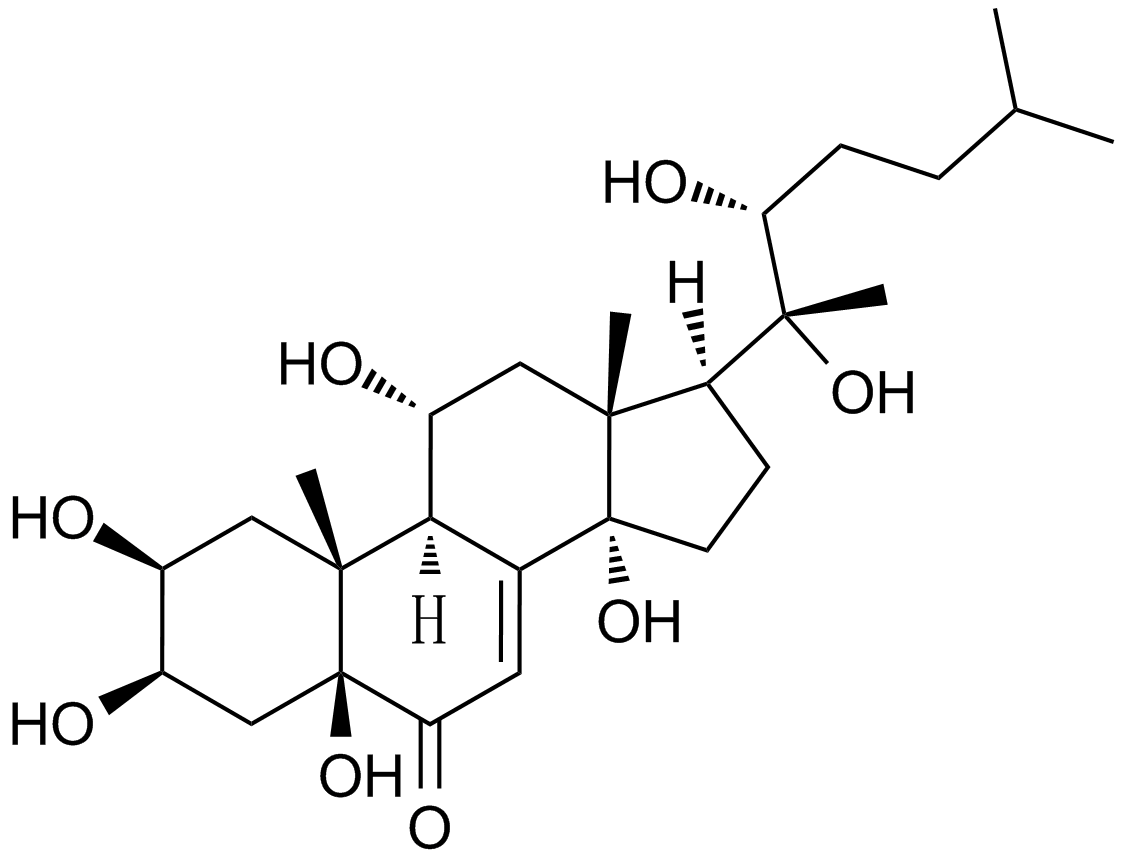
-
GC62707
Murizatoclax
Murizatoclax (AMG 397) is a potent, selective and orally active inhibitor of myeloid leukemia 1 (MCL-1) inhibitor, with a Ki of 15 pM. Murizatoclax competitive binds to the BH3-binding groove of MCL1 with pro-apoptotic BCL-2 family members. Murizatoclax can be used for the research of cancer.
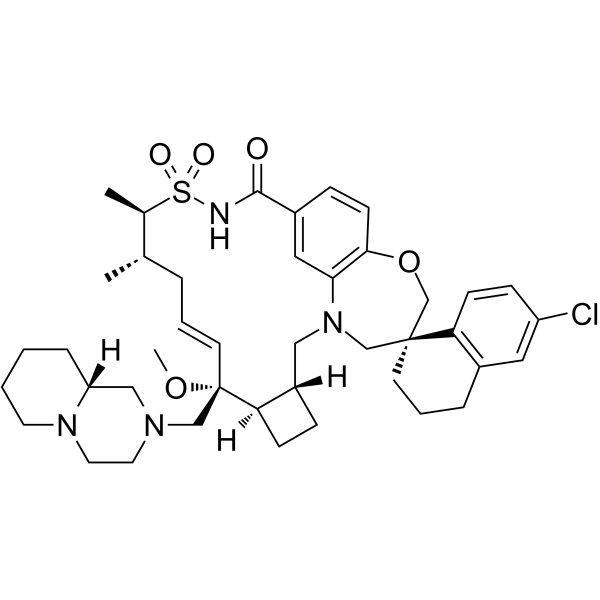
-
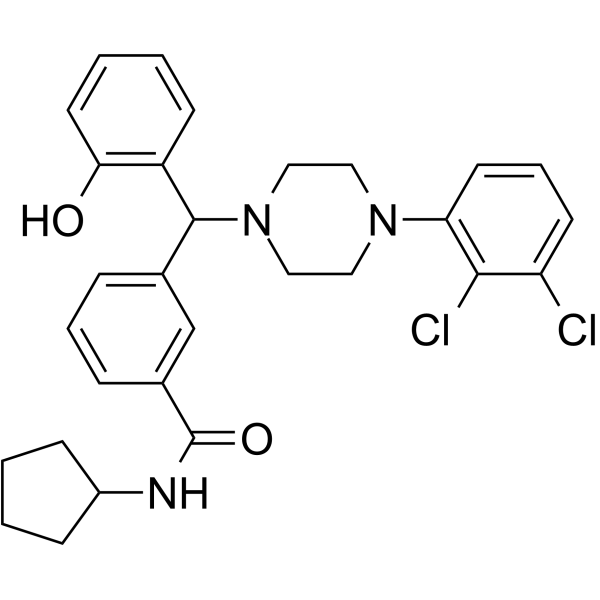
GC68019
NPB
-
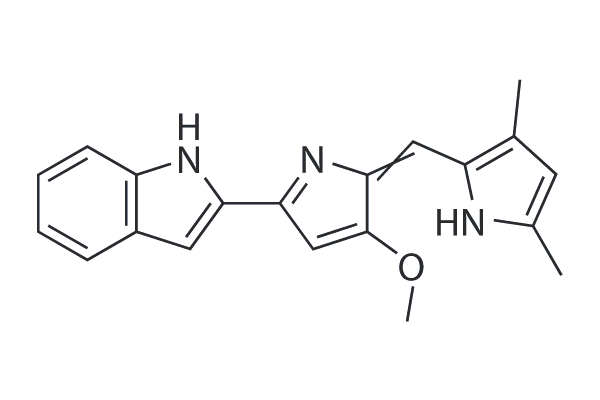
GC25673
Obatoclax (GX15-070)
-
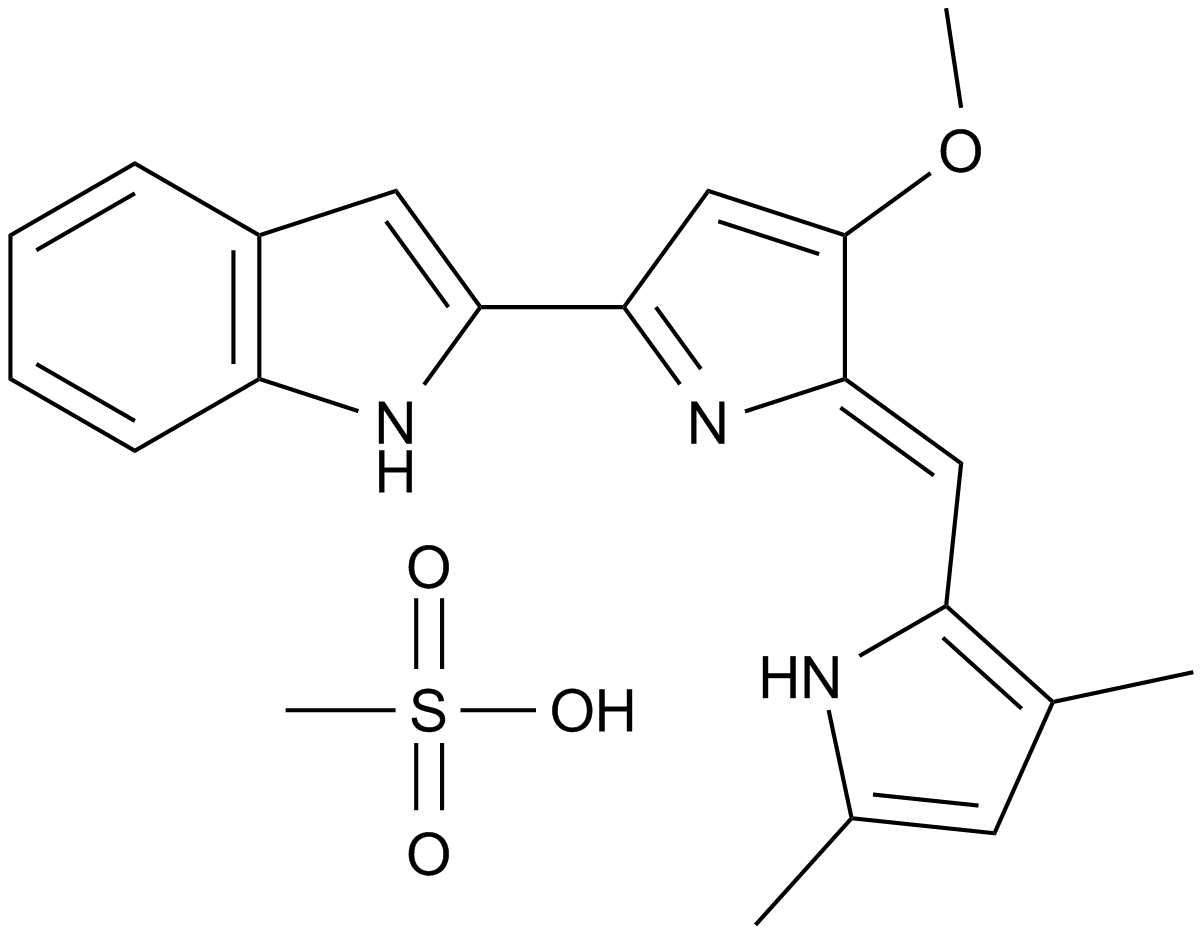
GC11569
Obatoclax mesylate (GX15-070)
An antagonist of pro-survival Bcl-2 proteins
-
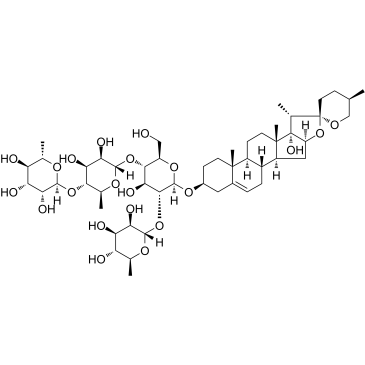
GC36855
Paris saponin VII
Paris saponin VII (Chonglou Saponin VII) is a steroidal saponin isolated from the roots and rhizomes of Trillium tschonoskii Maxim. Paris saponin VII-induced apoptosis in K562/ADR cells is associated with Akt/MAPK and the inhibition of P-gp. Paris saponin VII attenuates mitochondrial membrane potential, increases the expression of apoptosis-related proteins, such as Bax and cytochrome c, and decreases the protein expression levels of Bcl-2, caspase-9, caspase-3, PARP-1, and p-Akt. Paris saponin VII induces a robust autophagy in K562/ADR cells and provides a biochemical basis in the treatment of leukemia.
-
GC62505
Pelcitoclax
Pelcitoclax (APG-1252) is a potent Bcl-2/Bcl-xl inhibitor with antineoplastic and pro-apoptotic effects.
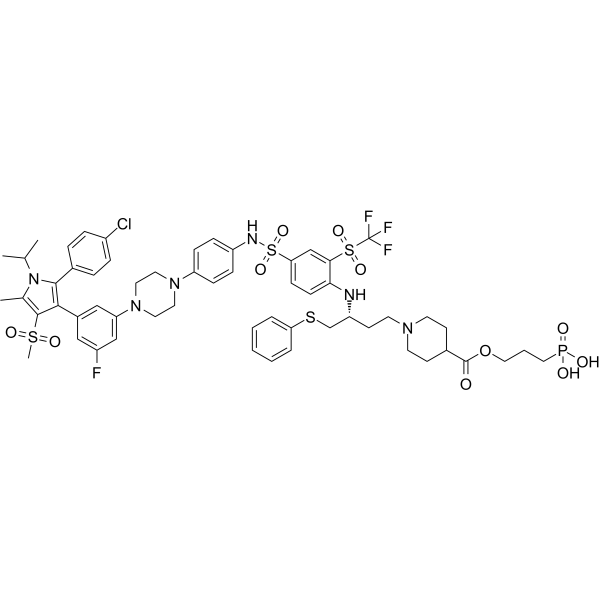
-
GC63769
PROTAC Bcl-xL degrader-2
PROTAC Bcl-xL degrader-2 is a potent Bcl-xL (Bcl-2 family member) degrader based on von Hippel-Lindau ligand, with an IC50 of 0.6 nM.

-
GC38941
PROTAC Bcl2 degrader-1
PROTAC Bcl2 degrader-1 (Compound C5) is a PROTAC based on Cereblon ligand, which potently and selectively induces the degradation of Bcl-2 (IC50, 4.94 μM; DC50, 3.0 μM) and Mcl-1 (IC50, 11.81 μM) by introducing the E3 ligase cereblon (CRBN)-binding ligand pomalidomide to Mcl-1/Bcl-2 dual inhibitor Nap-1.
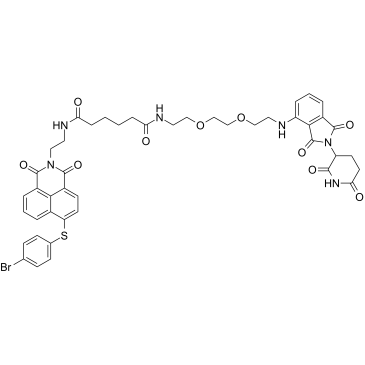
-
GC38943
PROTAC Mcl1 degrader-1
PROTAC Mcl1 degrader-1 (compound C3), a proteolysis targeting chimera (PROTAC) based on Cereblon ligand, is a potently and selectively Mcl-1 (Bcl-2 family member) inhibitor with an IC50 of 0.78 μM. PROTAC Mcl1 degrader-1 induces the ubiquitination and proteasomal degradation of Mcl-1 by introducing the E3 ligase cereblon (CRBN)-binding ligand pomalidomide to Mcl-1 inhibitor S1-6 with μM-range affinity.
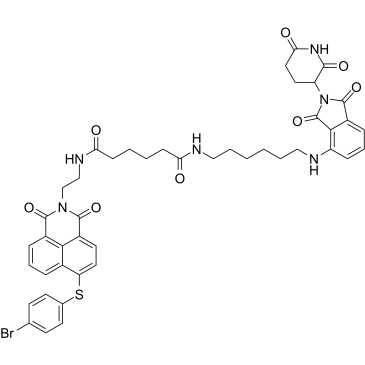
-
GC34389
PUMA BH3
PUMA BH3 is a p53 upregulated modulator of apoptosis (PUMA) BH3 domain peptide, acts as a direct activator of Bak, with a Kd of 26 nM.

-
GC12902
Pyridoclax
Mcl-1 inhibitor
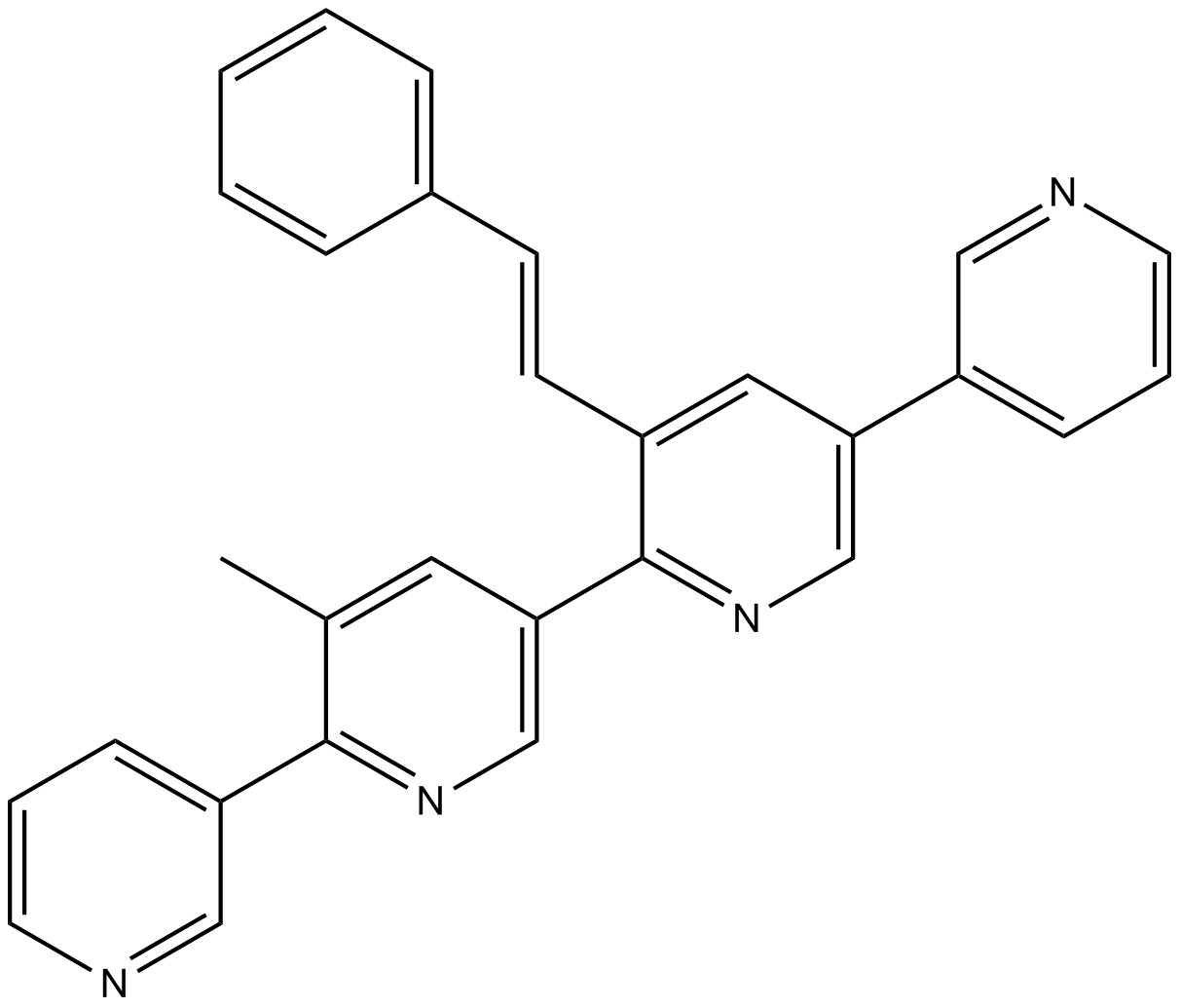
-
GC11140
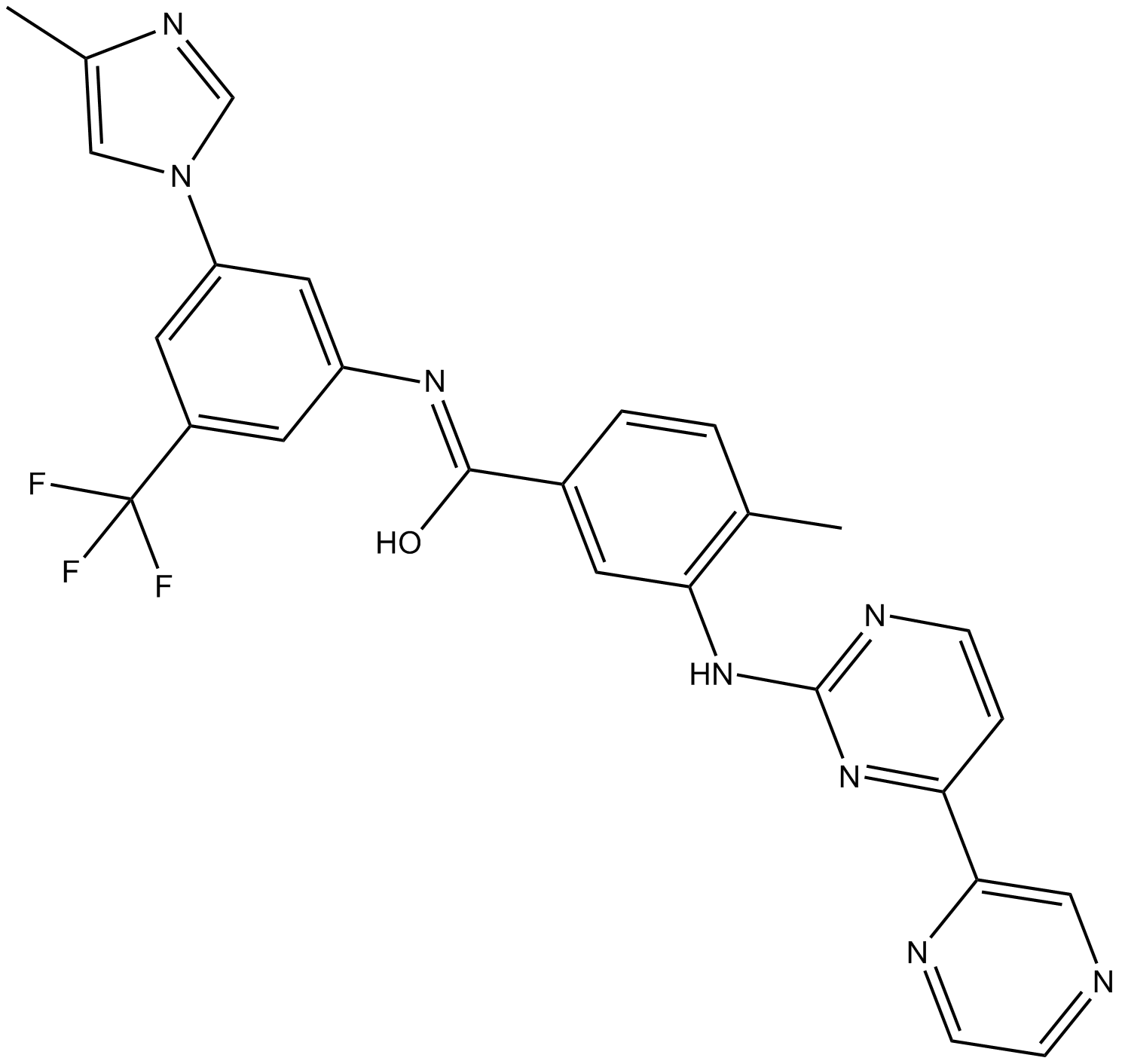
Radotinib(IY-5511)
Bcr-Abl tyrosine kinase inhibitor
-
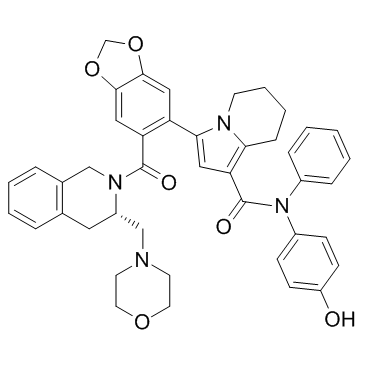
GC32947
S55746 (BLC201)
S55746 (BLC201) (BCL201) is a potent, orally active and selective BCL-2 inhibitor, with a Ki of 1.3 nM and a Kd of 3.9 nM. S55746 (BLC201) (BCL201) has antitumor activity with low toxicity.
-
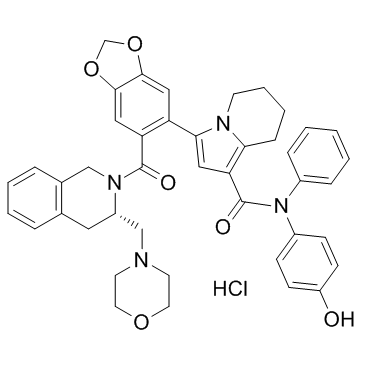
GC33401
S55746 hydrochloride (BLC201 (hydrochloride))
S55746 hydrochloride (BLC201 (hydrochloride)) (BCL201 hydrochloride) is a potent, orally active and selective BCL-2 inhibitor, with a Ki of 1.3 nM and a Kd of 3.9 nM. S55746 hydrochloride (BLC201 (hydrochloride)) (BCL201 hydrochloride) has antitumor activity with low toxicity.
-
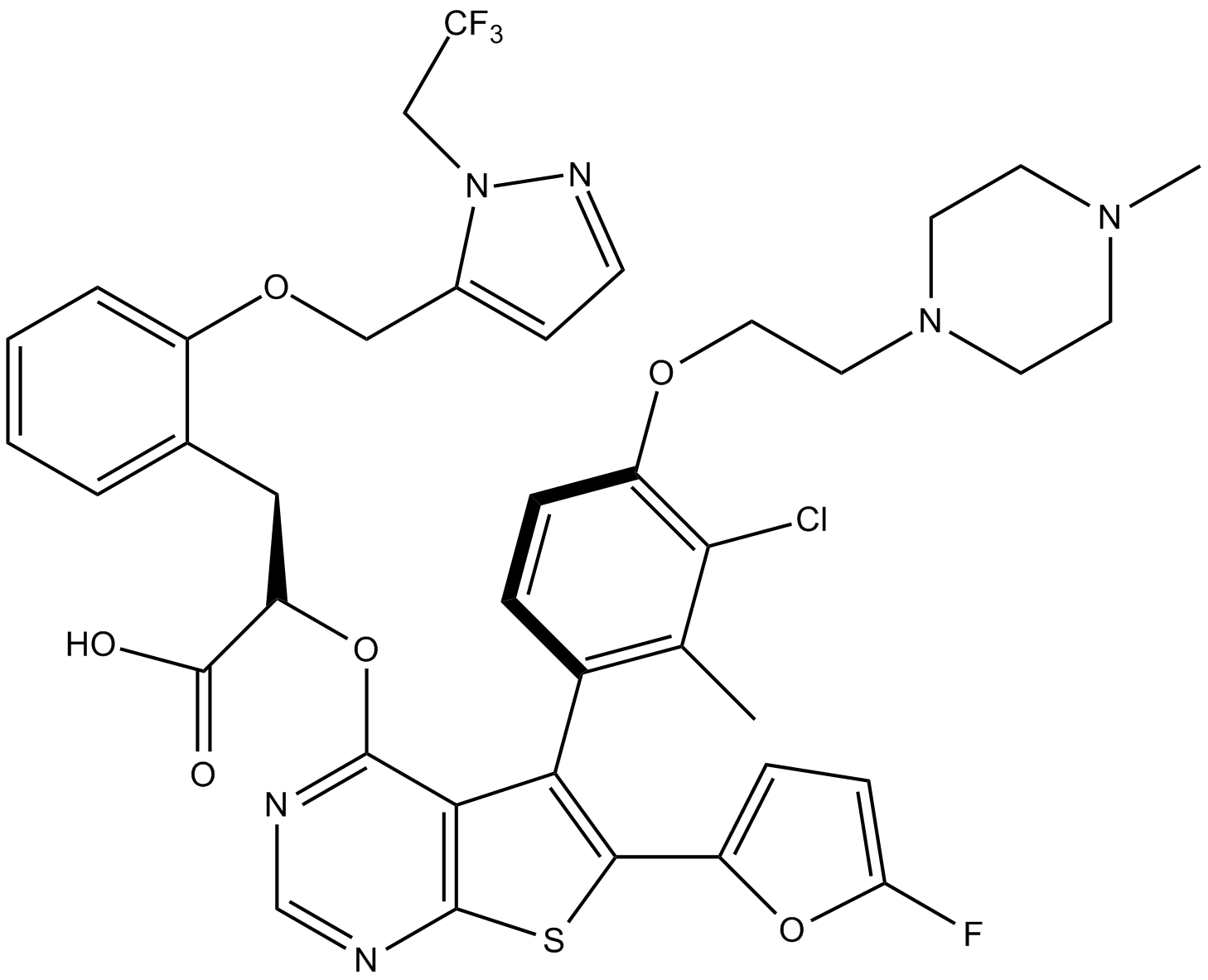
GC12621
S63845
MCL1 inhibitor
-
GC62554
S65487
S65487 (VOB560), a potent and selective BCL-2 inhibitor, is a prodrug of S55746. S65487 is also active on BCL-2 mutations, such as G101V and D103Y. S65487 has poor affinity with MCL-1, BFL-1 and BCL-XL. S65487 induces apoptosis and has anticaner activities.
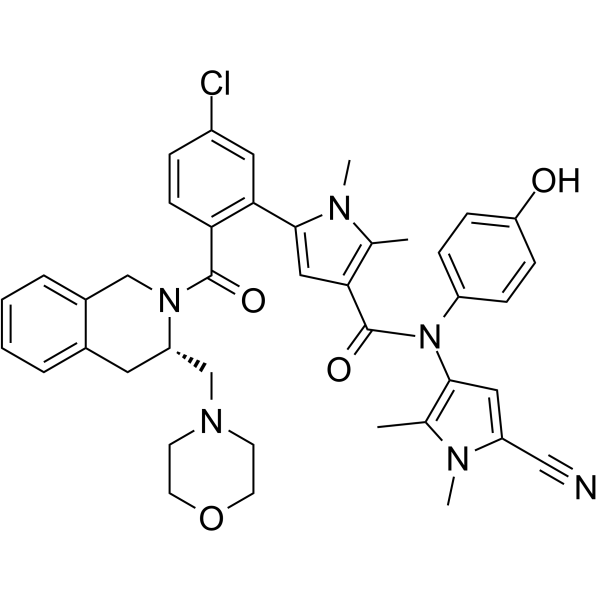
-
GC63426
S65487 hydrochloride
S65487 (VOB560) hydrochloride, a potent and selective Bcl-2 inhibitor, is a prodrug of S55746. S65487 hydrochloride is also active on BCL-2 mutations, such as G101V and D103Y. S65487 hydrochloride has poor affinity with MCL-1, BFL-1 and BCL-XL. S65487 hydrochloride induces apoptosis and has anticaner activities.
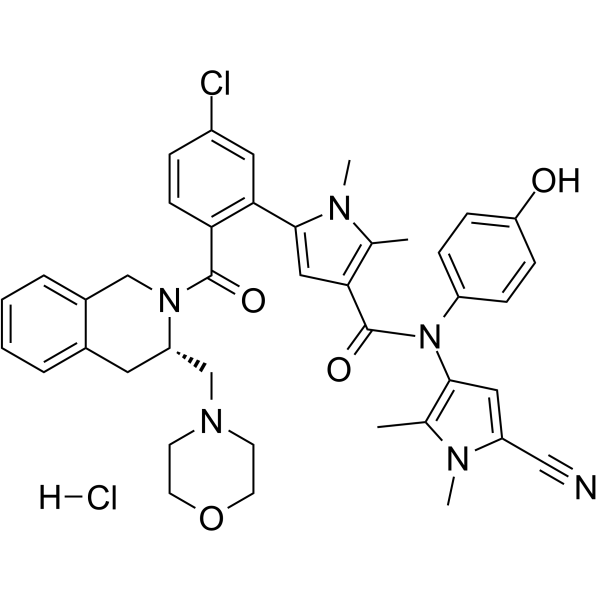
-
GC63531
S65487 sulfate
S65487 (VOB560) sulfate, a potent and selective Bcl-2 inhibitor, is a prodrug of S55746. S65487 sulfate is also active on BCL-2 mutations, such as G101V and D103Y. S65487 sulfate has poor affinity with MCL-1, BFL-1 and BCL-XL. S65487 sulfate induces apoptosis and has anticaner activities.
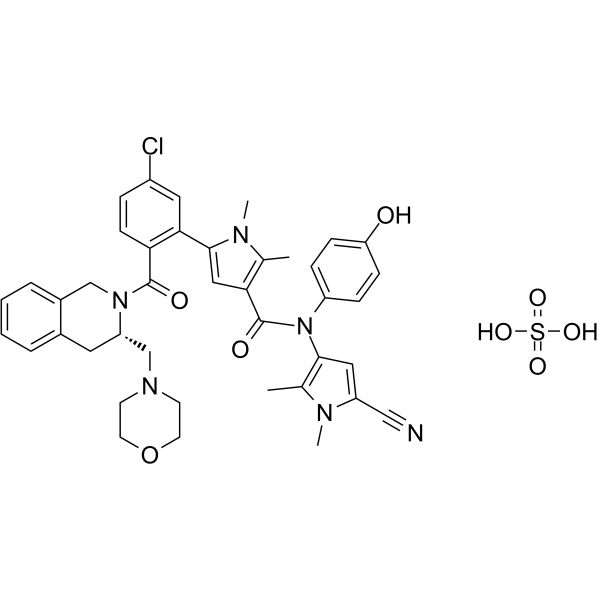
-
GC10153
Sabutoclax
A pan-Bcl-2 inhibitor
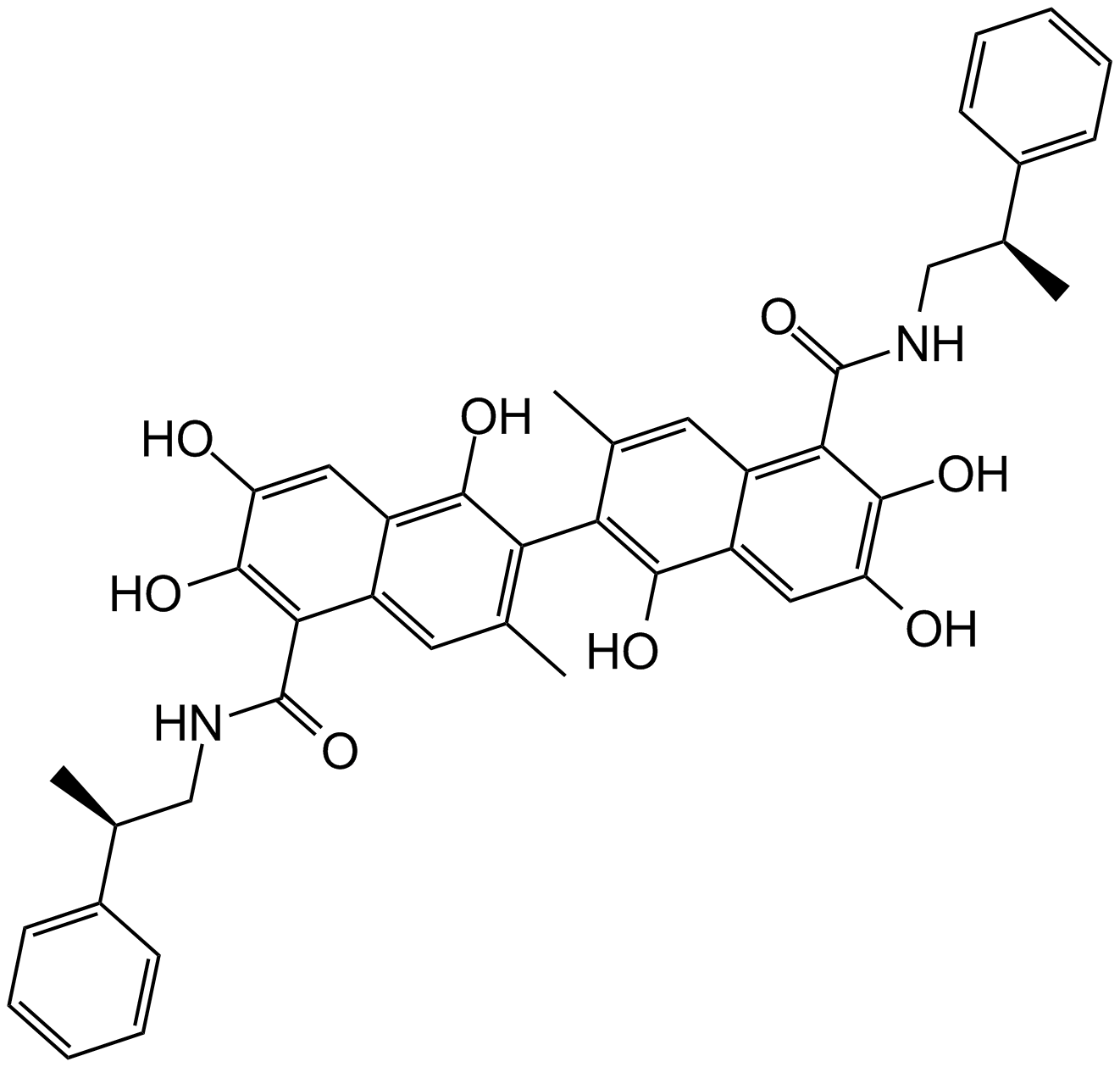
-
GC10567
TCPOBOP
constitutive androstane receptor (CAR) agonist
-
GC37780
Thevetiaflavone
Thevetiaflavone could upregulate the expression of Bcl?2 and downregulate that of Bax and caspase?3.
-
GC66021
TP-021
TP-021 (BCL6-IN-8c) is a potent and orally active B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor with an IC50 of 0.10 μM in cell-free enzyme-linked immunosorbent assay.
-
GC12649
TW-37
An inhibitor of the Bcl-2 family proteins
-
GC15805
UMI-77
Mcl-1 inhibitor, novel
-
GC70114
Venetoclax-d8
Venetoclax-d8 is the deuterated form of Venetoclax. Venetoclax (ABT-199; GDC-0199) is a highly efficient, selective and orally effective Bcl-2 inhibitor with a Ki value less than 0.01 nM. Venetoclax can induce autophagy.
-
GC33052
VU0661013
VU661013 is a potent and selective MCL-1 inhibitor.
-
GC11921
WEHI-539
A selective Bcl-xL inhibitor