Mitochondrial Metabolism
Mitochondria use multiple carbon fuels to produce ATP and metabolites, including pyruvate, which is generated from glycolysis; amino acids such as glutamine; and fatty acids. These carbon fuels feed into the TCA cycle in the mitochondrial matrix to generate the reducing equivalents NADH and FADH2, which deliver their electrons to the electron transport chain. Mitochondria are complex organelles that play an important role in many facets of cellular function, from metabolism to immune regulation and cell death. Mitochondria are actively involved in a wide variety of cellular processes and molecular interactions, such as calcium buffering, lipid flux, and intracellular signaling. It is increasingly recognized that mitochondrial dysfunction is a hallmark of many diseases such as obesity/diabetes, cancer, cardiovascular and neurodegenerative diseases. Mitochondrial metabolism is a key determinant of tumor progression by impacting on functions such as epithelial-to-mesenchymal transition. Mitochondrial metabolism and derived oncometabolites shape the epigenetic landscape to alter aggressiveness features of cancer cells. Changes in mitochondrial metabolism are relevant for the survival of tumors in response to therapy.
Targets for Mitochondrial Metabolism
Products for Mitochondrial Metabolism
- Cat.No. Product Name Information
-
GC62137
IMT1
IMT1 is a first-in-class specific and noncompetitive human mitochondrial RNA polymerase (POLRMT) inhibitor.
-
GC43909
Iromycin A
Iromycin A is a bacterial pyridone metabolite that inhibits nitric oxide synthase (NOS) activity, with selectivity for NOS III (endothelial NOS) over NOS I (neuronal NOS).
-
GC43912
Isoapoptolidin
Isoapoptolidin is a stable derivative of apoptolidin that demonstrates 10-fold reduced F1FO-ATP synthase inhibitory activity compared to apoptolidin (IC50s = 17 and 0.7 μM, respectively).
-
GC43922
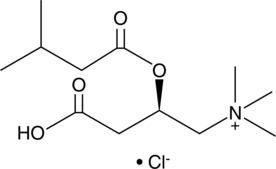
Isovaleryl-L-carnitine (chloride)
Isovaleryl-L-carnitine (chloride), a product of the catabolism of L-leucine, is a potent activator of the Ca2+-dependent proteinase (calpain) of human neutrophils.
-
GC47531
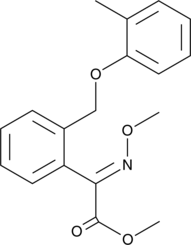
Kresoxim-methyl
A fungicide
-
GC47548
L-Carnitine-d3 (chloride)
L-Carnitine-d3 ((R)-Carnitine-d3) hydrochloride is the deuterium labeled L-Carnitine.

-
GC52094
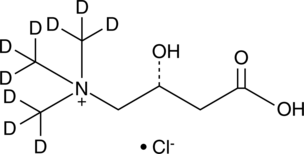
L-Carnitine-d9 (chloride)
An internal standard for the quantification of L-carnitine
-
GC49382
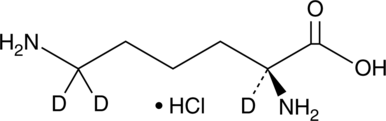
L-Lysine-d3 (hydrochloride)
An internal standard for the quantification of L-lysine
-
GC19221
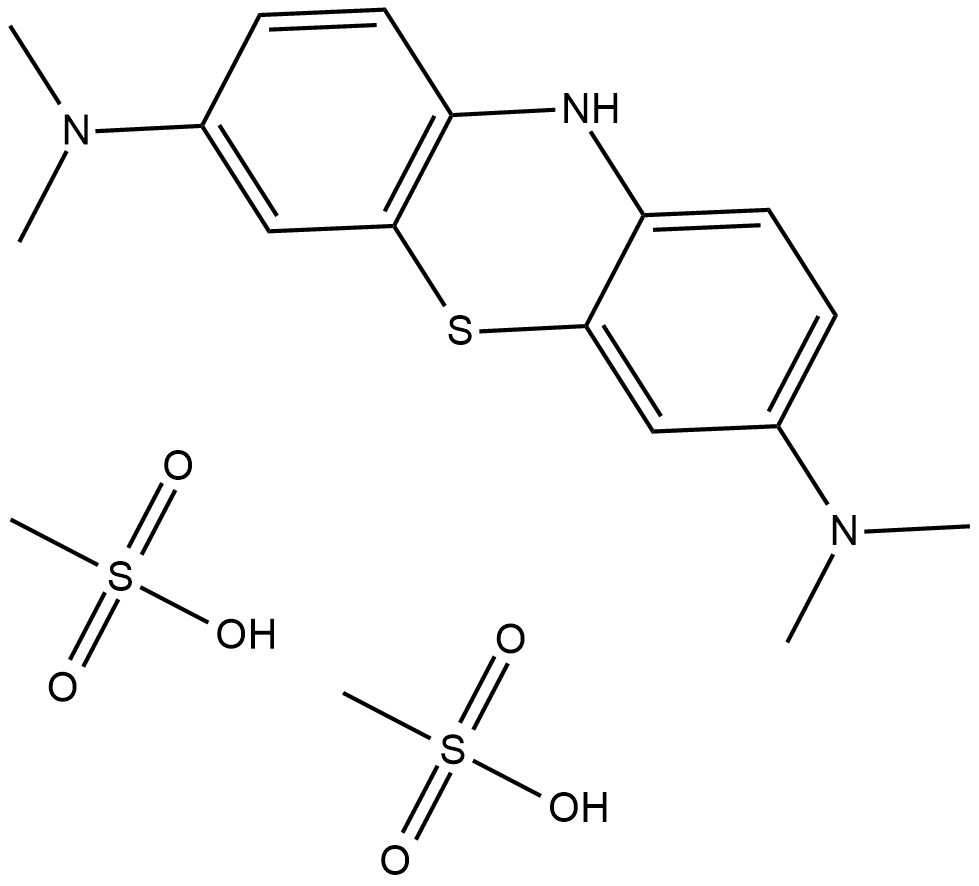
Leucomethylene blue Mesylate
Leucomethylene blue (Mesylate) is a common reduced form of Methylene Blue, Methylene Blue is a member of the thiazine class of dyes.
-
GC63973
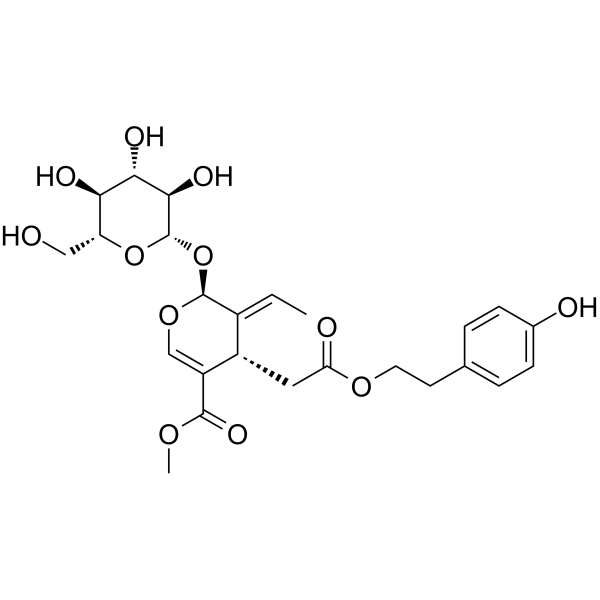
Ligustroside
Ligustroside (Ligstroside), a secoiridoid derivative, has outstanding performance on mitochondrial bioenergetics in models of early Alzheimer's disease (AD) and brain ageing by mechanisms that may not interfere with Aβ production.
-
GN10423
Lipoic acid

-
GC11532
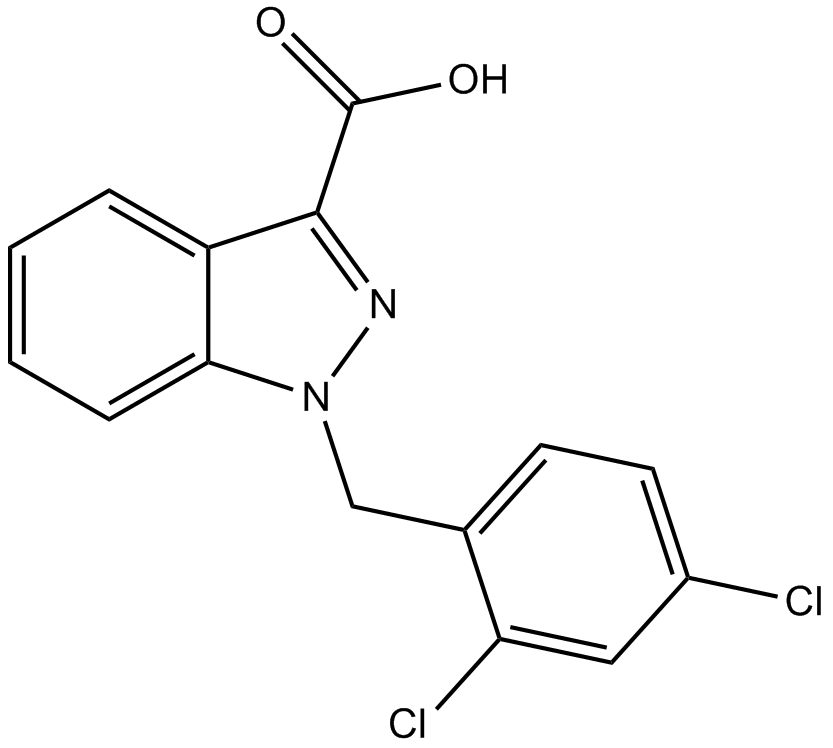
Lonidamine
hexokinase inactivator
-
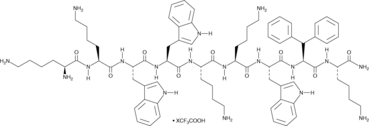
GC48989
LTX-315 (trifluoroacetate salt)
A synthetic cationic amphiphilic peptide
-
GC44093
Lutein
Lutein is a natural yellow carotenoid, which can be found in plants, egg yolks, and in the human retina.
-
GC40007
Malformin A
Malformin A is a cyclopentapeptide fungal metabolite that has been found in A.
-
GC61790
MCU-i4
MCU-i4 blocks the IP3-dependent mitochondrial Ca2+-uptake, maintaining the gatekeeping role of their target.
-
GC66184
Menadiol
Menadiol (Dihydrovitamin K3), a menaquinol analogue, is an electron donor for reversed oxidative phosphorylation in submitochondrial particles.
-
GC47620
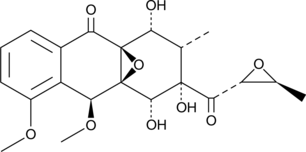
Mensacarcin
A bacterial metabolite with anticancer activity
-
GC47663
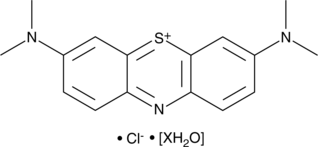
Methylene Blue (hydrate)
A cationic dye with diverse biological activities
-
GC49472
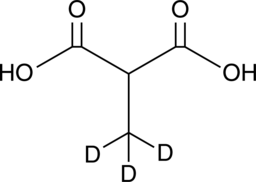
Methylmalonic Acid-d3
Methylmalonic acid-d3 (Methylpropanedioic acid-d3) is the deuterium labeled Methylmalonic acid. Methylmalonic acid (Methylmalonate) is an indicator of Vitamin B-12 deficiency in cancer.
-
GC65989
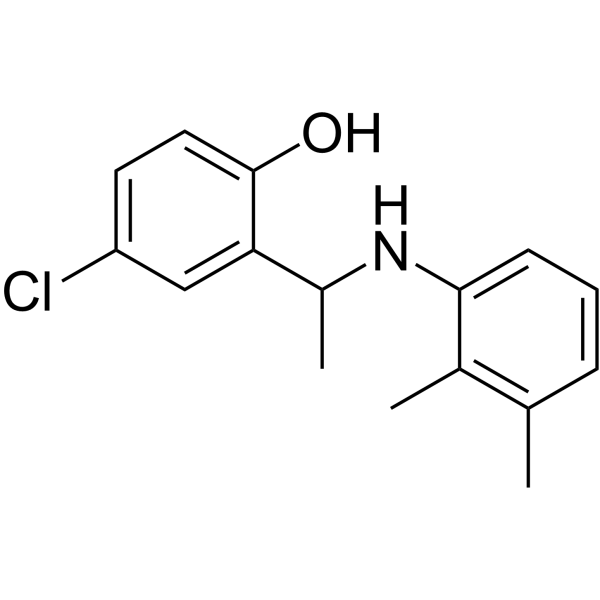
MFI8
MFI8 is a small molecule inhibitor of mitochondrial fusion.
-
GC47685
MIT-PZR
A mitochondria-targeted fluorescent probe
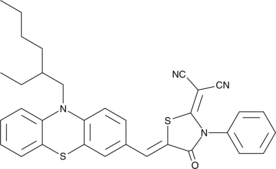
-
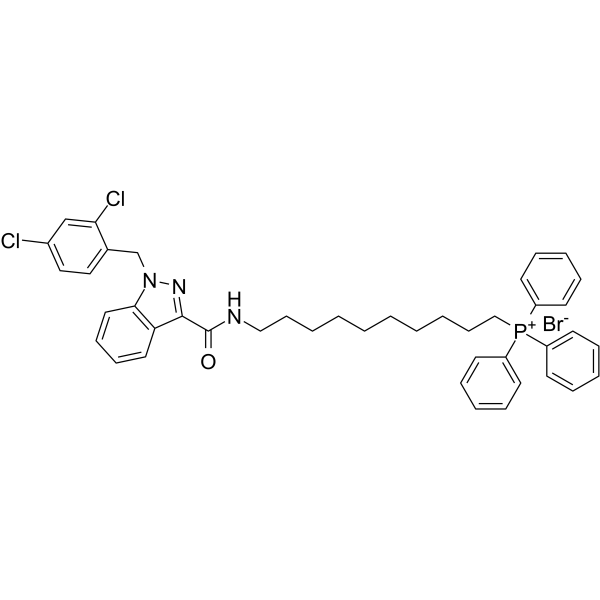
GC62564
Mito-LND
Mito-LND (Mito-Lonidamine) is an orally active and mitochondria-targeted inhibitor of oxidative phosphorylation (OXPHOS). Mito-LND inhibits mitochondrial bioenergetics, stimulates the formation of reactive oxygen species, and induces autophagic cell death in lung cancer cells.
-
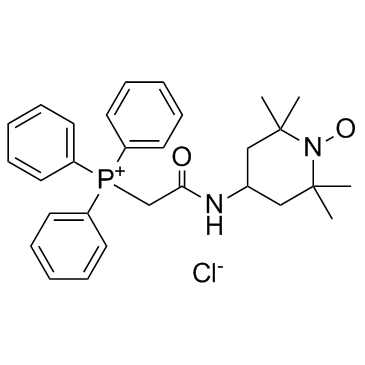
GC31682
Mito-TEMPO
Mito-Tempo is a mitochondria-targeted antioxidant with effective superoxide scavenging properties
-
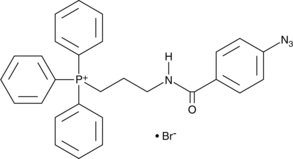
GC44198
MitoA
MitoA is a ratiometric mass spectrometry probe that can be used for assessing changes in H2S within mitochondria in vivo.
-
GC44199
MitoB
MitoB is a ratiometric mass spectrometry probe that can be used for assessing changes in H2O2 within mitochondria in vivo.
-
GC61634
Mitochondrial respiration-IN-1 hydrobromide
Mitochondrial respiration-IN-1 hydrobromide (compound 49) is a potent mitochondrial inhibitor (IC50=8.8 mg/mL) extracted from patent US20110301180A1, compound 49.
-
GC31123
Mitochonic acid 5 (MA-5)
Mitochonic acid 5 (MA-5) binds mitochondria and ameliorates renal tubular and cardiac myocyte damage.
-
GC44201
MitoP
MitoP is a phenol product produced by the reaction of H2O2 with the ratiometric mass spectrometry probe MitoB.
-
GC44202
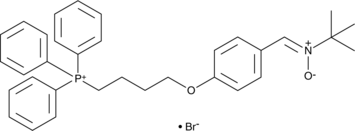
MitoPBN
MitoPBN is a mitochondria-targeted antioxidant.
-
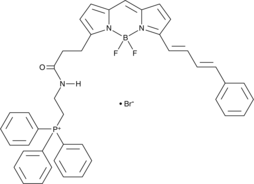
GC44203
MitoPerOx
MitoPerOx is a ratiometric fluorescent probe that can be used to assess changes in lipid peroxidation within mitochondria.
-
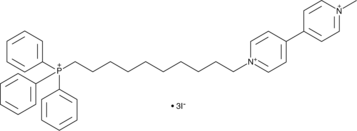
GC44204
MitoPQ
MitoPQ is comprised of a triphenylphosphonium lipophilic cation conjugated to the redox cycler paraquat.
-
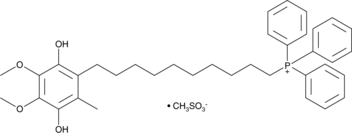
GC44205
Mitoquinol
Mitoquinol is a ubiquinone derivative that specifically accumulates in mitochondria due to the covalent attachment of the cation triphenylphosphonium.
-
GC49222
Mitoquinol-d15
An internal standard for the quantification of mitoquinol
-
GC36619
MitoTam bromide, hydrobromide
MitoTam bromide, hydrobromide, a Tamoxifen derivative, is an electron transport chain (ETC) inhibitor. MitoTam bromide, hydrobromide reduces mitochondrial membrane potential in senescent cells and affects mitochondrial morphology. MitoTam bromide, hydrobromide is an effective anticancer agent, suppresses respiratory complexes (CI-respiration) and disrupts respiratory supercomplexes (SCs) formation in breast cancer cells.
-
GC36620
MitoTam iodide, hydriodide
MitoTam iodide, hydriodide is a Tamoxifen derivative, an electron transport chain (ETC) inhibitor, spreduces mitochondrial membrane potential in senescent cells and affects mitochondrial morphology.MitoTam iodide, hydriodide is an effective anticancer agent, suppresses respiratory complexes (CI-respiration) and disrupts respiratory supercomplexes (SCs) formation in breast cancer cells. MitoTam iodide, hydriodide causes apoptosis.
-
GC44206
MitoTEMPO (hydrate)
MitoTEMPO is a mitochondria-targeted superoxide dismutase mimetic that possesses superoxide and alkyl radical scavenging properties.
-
GC44207
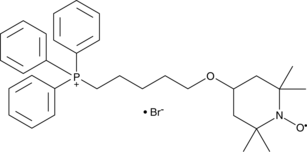
MitoTEMPOL
MitoTEMPOL is a mitochondria-targeting superoxide dismutase mimetic that reduces mitochondrial O2- to H2O2.
-
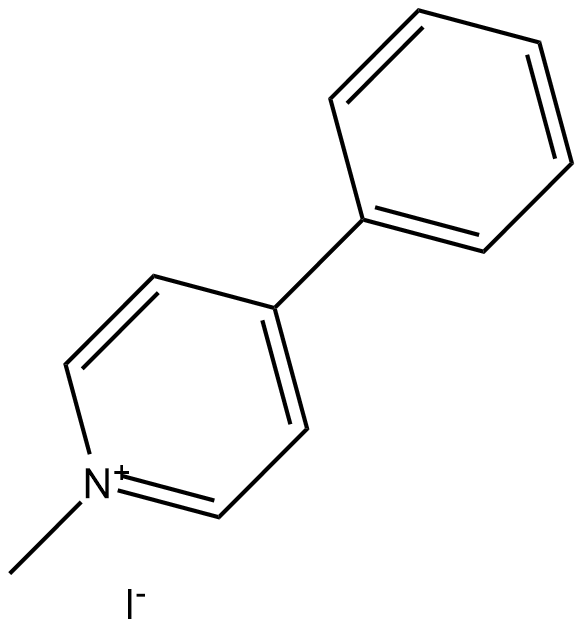
GC18188
MPP+ Iodide
MPP+ Iodide (1-methyl-4-phenylpyridinium iodide) is a toxic metabolite of the neurotoxin MPTP, and has successfully induced Parkinson-like syndromes in an in vitro model by selectively destroying dopaminergic neurons in substantia nigra.
-
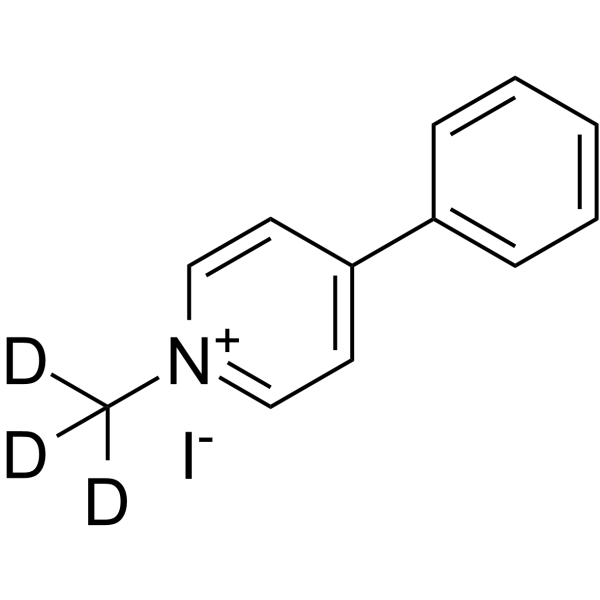
GC66401
MPP+-d3(iodide)
MPP+-d3(iodide) is deuterium labeled MPP+ (iodide). MPP+ iodide, a toxic metabolite of the neurotoxin MPTP, causes symptom of Parkinson's disease in animal models by selectively destroying dopaminergic neurons in substantia nigra. MPP+ iodide is taken up by the dopamine transporter into dopaminergic neurons where it exerts its neurotoxic action on mitochondria by affecting complex I of the respiratory chain. MPP+ iodide is also a high affinity substrate for the serotonin transporter (SERT).
-
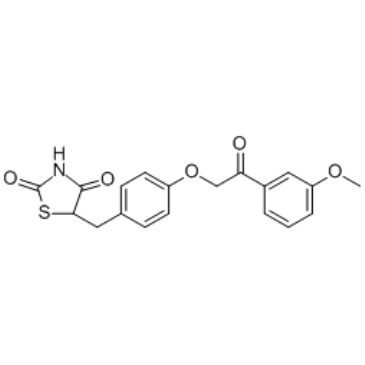
GC36658
MSDC-0602
MSDC-0602 (MSDC-0602), a PPARγ-sparing thiazolidinedione (TZD), interacts with the mitochondrial pyruvate carrier (MPC) and inhibits its activity and has the potential for type 2 diabetes study with reducing risk of PPARγ-mediated side effects.
-
GC48399
MTP 131 (acetate)
A mitochondria-targeted peptide antioxidant
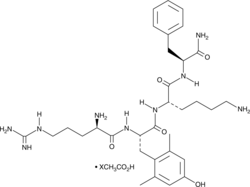
-
GC48526
MXS
A fluorescent probe for hypochlorous acid
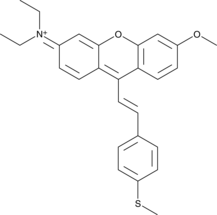
-
GC49399
N-Desbutyl Dronedarone (hydrochloride)
Debutyldronedarone (SR35021) hydrochloride, the main metabolite of Dronedarone, is a selective thyroid hormone receptor α1 (TRα1) inhibitor.
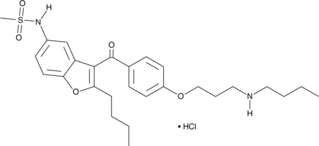
-
GC44416
N-methyl Mesoporphyrin IX
A ferrochelatase inhibitor and turn-on biosensor for DNA
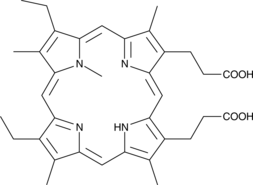
-
GC47789
N-Oleoyl Glutamine
An endogenous N-acyl amine
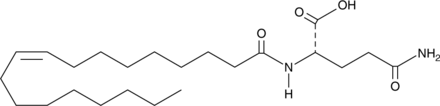
-
GC44441
N-Oleoyl Leucine
N-Oleoyl leucine is an N-acyl amide generated by PM20D1 that uncouples mitochondrial respiration independent of uncoupling protein 1 (UCP1) in vitro.
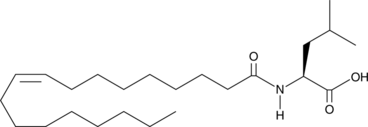
-
GC44444
N-Oleoyl Valine
N-Oleoyl valine is an endogenous N-acyl amine that acts as an antagonist at the transient receptor potential vanilloid type 3 (TRPV3) receptor, which is involved in thermoregulation.
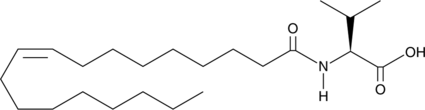
-
GC47739
NADH (sodium salt hydrate)
A reduced form of NAD+
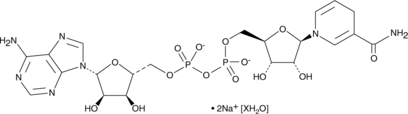
-
GC44308
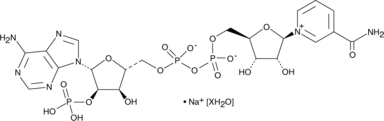
NADP+ (sodium salt hydrate)
NADP+ (sodium salt hydrate), a β-Nicotinamide adenine dinucleotide phosphate sodium salt, is a redox cofactor.
-
GC47747
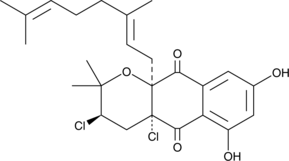
Napyradiomycin A1
A fungal metabolite with diverse biological activities
-
GC40669
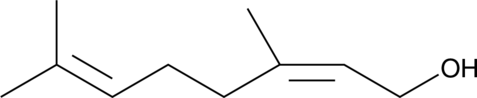
Nerol
Nerol is a monoterpene and isomer of geraniol that has been found in a variety of plants, including Cannabis.
-
GC49511
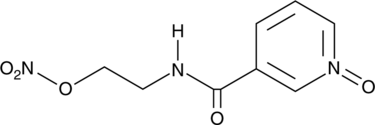
Nicorandil N-oxide
A metabolite of nicorandil
-
GC52214
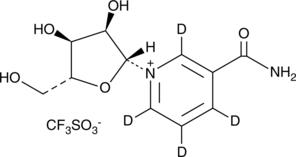
Nicotinamide riboside-d4 (triflate)
An internal standard for the quantification of nicotinamide riboside
-
GC31646
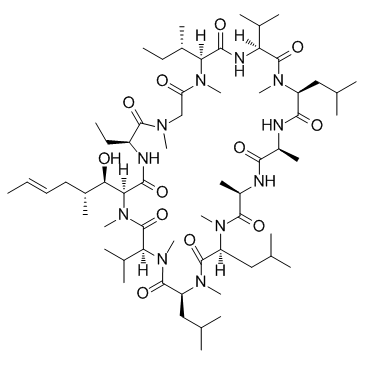
NIM811 ((Melle-4)cyclosporin)
NIM811 ((Melle-4)cyclosporin) ((Melle-4)cyclosporin; SDZ NIM811 ((Melle-4)cyclosporin)) is an orally bioavailable mitochondrial permeability transition and cyclophilin dual inhibitor, which exhibits potent in vitro activity against hepatitis C virus (HCV) .
-
GC39411
NL-1
NL-1 is a mitoNEET inhibitor with antileukemic effect. NL-1 inhibits REH and REH/Ara-C cells growth with IC50s of 47.35 ?M and 56.26 ?M, respectively. NL-1-mediated death in leukemic cells requires the activation of the autophagic pathway.
-
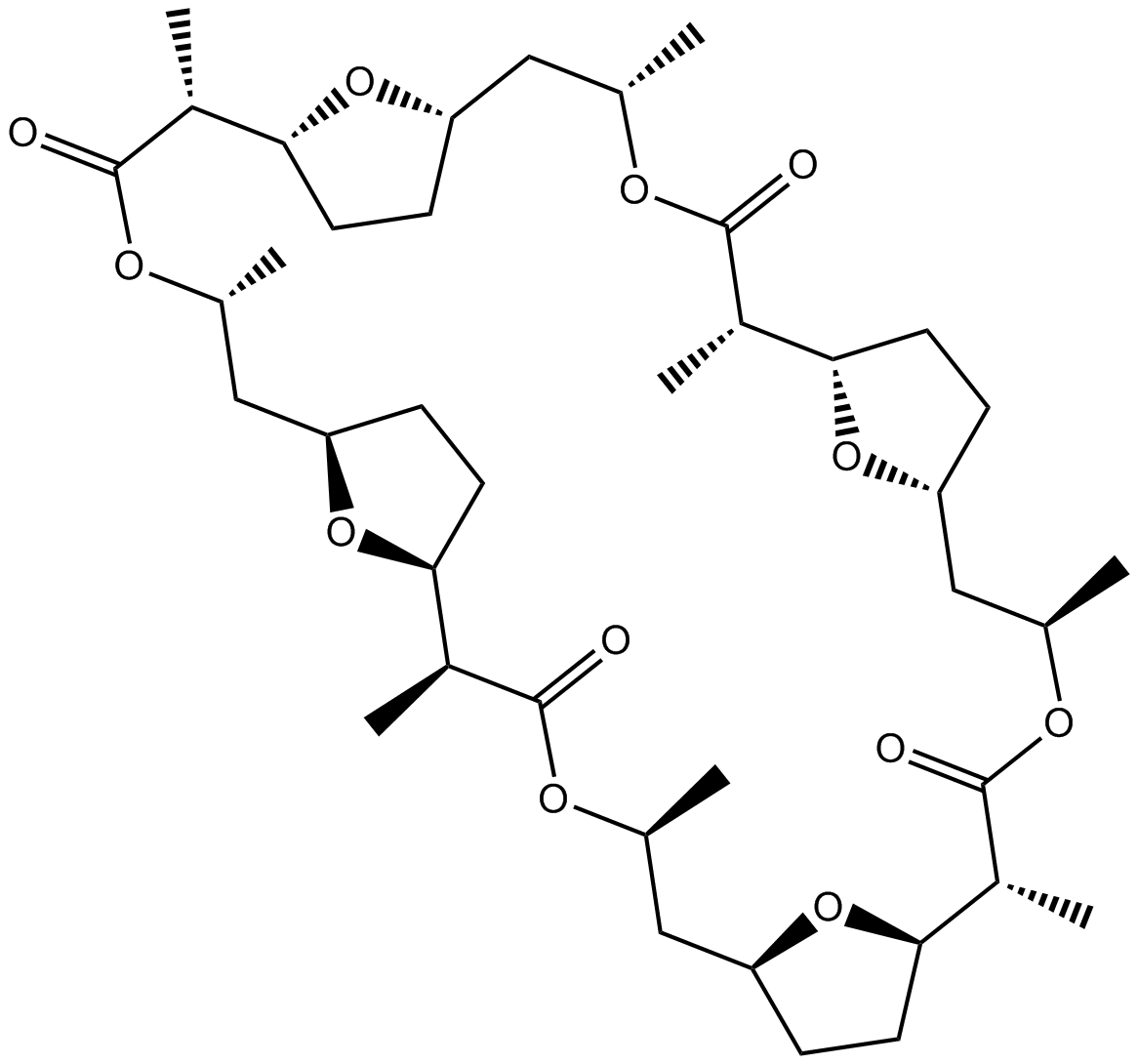
GC12090
Nonactin
Monovalent cation ionophore that displays selectivity for K+ and NH4+
-
GC52223
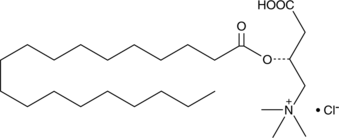
Nonadecanoyl-L-carnitine (chloride)
A long-chain acylcarnitine
-
GC52236
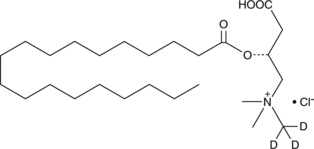
Nonadecanoyl-L-carnitine-d3 (chloride)
An internal standard for the quantification of nonadecanoyl-L-carnitine
-
GC44464
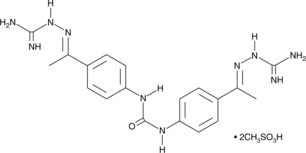
NSC 109555
An inhibitor of Chk2
-
GC44488
Octanoyl Coenzyme A (sodium salt)
Octanoyl coenzyme A (octanoyl-CoA) is a medium-chain acyl CoA and a metabolic intermediate in mitochondrial fatty acid β-oxidation.

-
GC49457
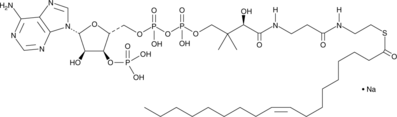
Oleoyl-Coenzyme A (sodium salt)
An acyl-CoA thioester
-
GC44506
Oligomycin D
Oligomycin D is a macrolide antibiotic produced by several species of Streptomyces that inhibits the mitochondrial F1FO-ATPase and is used to uncouple oxidative phosphorylation from electron transport.
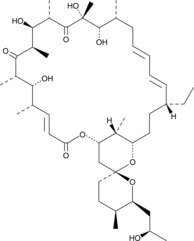
-
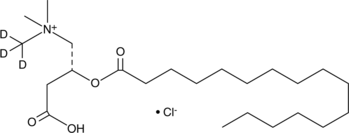
GC47868
Palmitoyl-L-carnitine-d3 (chloride)
An internal standard for the quantification of palmitoyl-L-carnitine
-
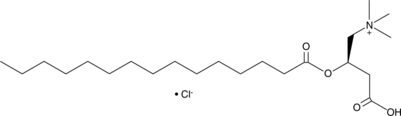
GC52306
Pentadecanoyl-L-carnitine (chloride)
A CB receptor agonist
-
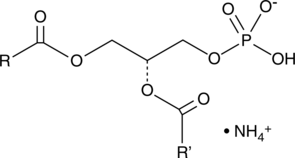
GC40252
Phosphatidic Acids (egg) (ammonium salt)
Phosphatidic acid is a phospholipid and an intermediate in glycerolipid biosynthesis.
-
GC18522
Phosphatidylserines (sodium salt)
Phosphatidylserine is a naturally occurring phospholipid that comprises 2-10% of total phospholipids in mammals and is enriched in the central nervous system, particularly the retina.
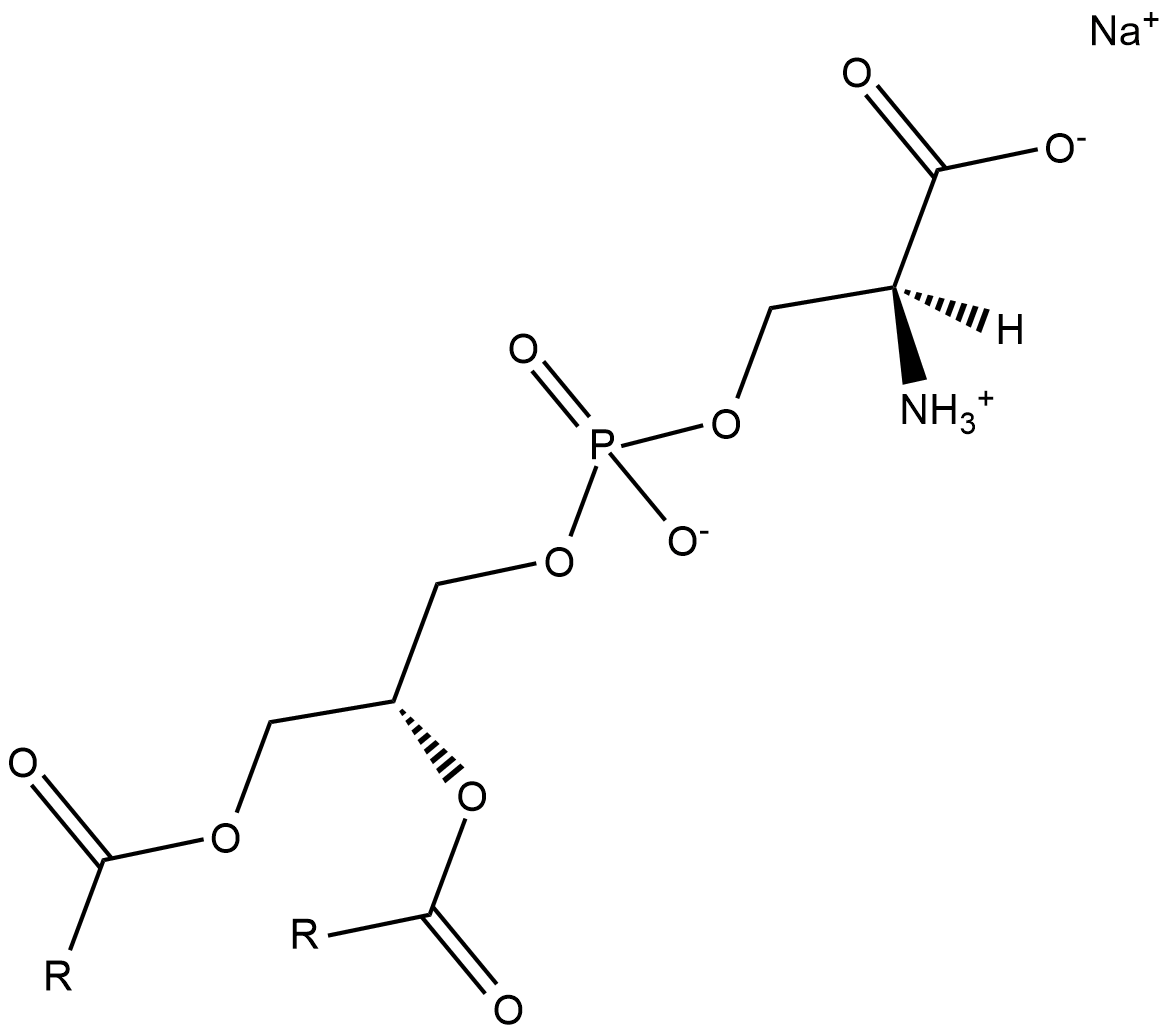
-
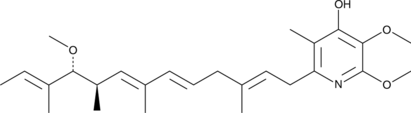
GC48528
Piericidin B
A bacterial metabolite with insecticidal and antimicrobial activities
-
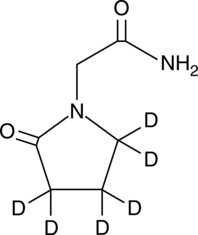
GC49192
Piracetam-d6
An internal standard for the quantification of piracetam
-
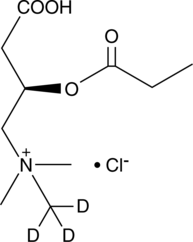
GC47980
Propionyl-L-carnitine-d3 (chloride)
An internal standard for the quantification of propionyl-L-carnitine
-
GC48030
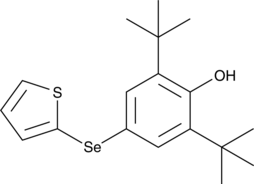
RC574
An inhibitor of ferroptosis
-
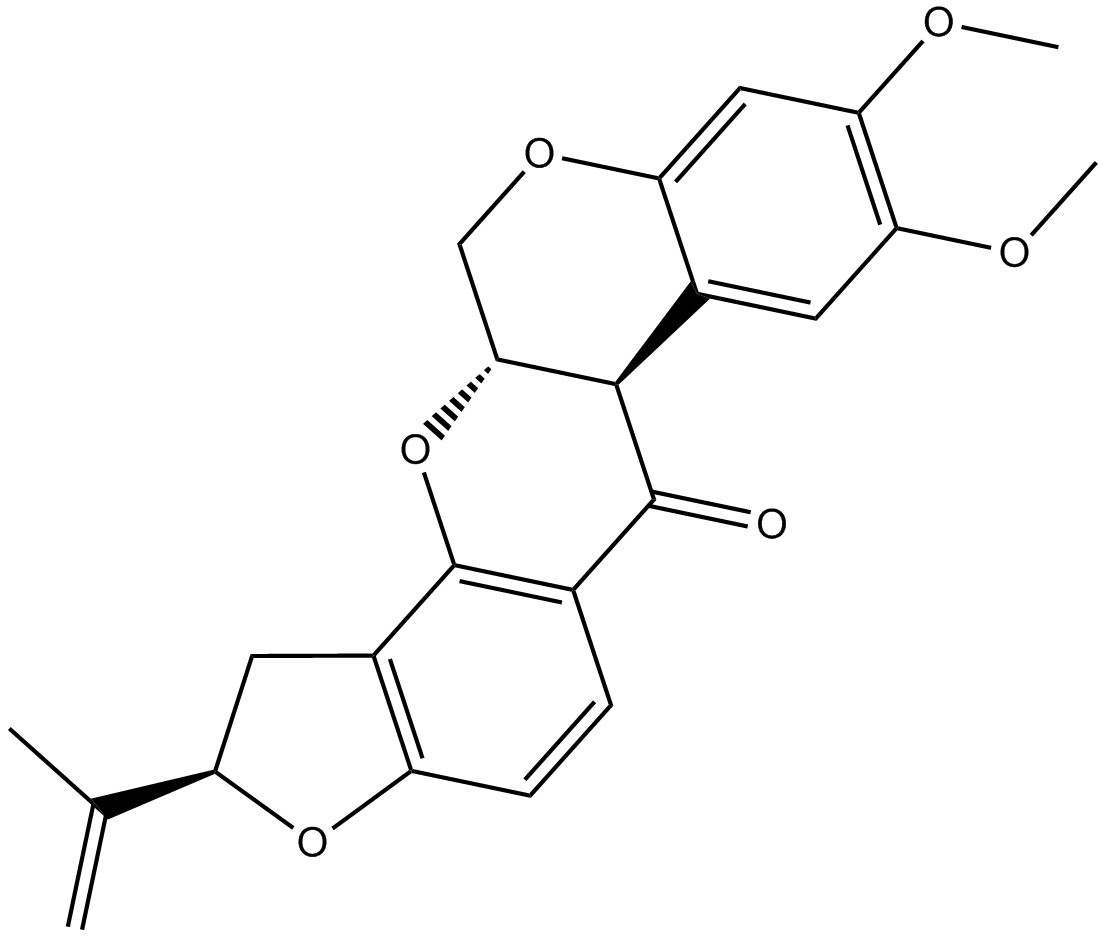
GC16775
Rotenone
inhibitor of the mitochondrial complex I electron transport chain
-
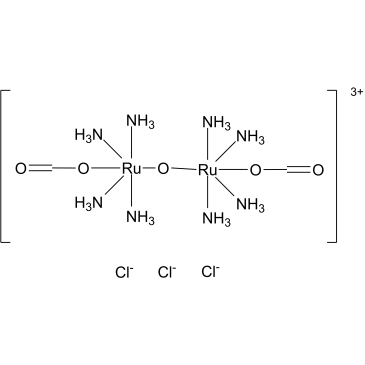
GC60330
Ru360
-
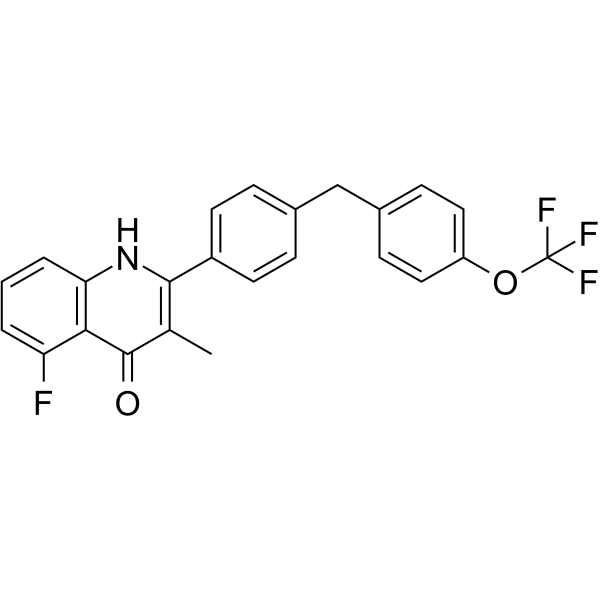
GC64669
RYL-552
RYL-552, a mitochondrial electron transport chain (ETC) inhibitor, is a P.
-
GC44860
S1QEL1.1
S1QEL1.1 is a small molecule that suppresses superoxide production during reverse electron transport through the IQ site of the mitochondrial respiratory complex I (IC50 = 70 nM) without affecting oxidative phosphorylation.
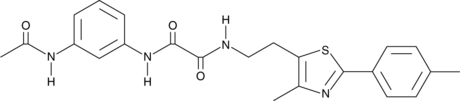
-
GC44862
S3QEL-2
S3QEL-2 is a cell-permeable, selective suppressor of electron leak from mitochondrial respiratory complex III, inhibiting site IIIQo superoxide production (IC50 = 1.7 μM) without altering normal bioenergetics function.
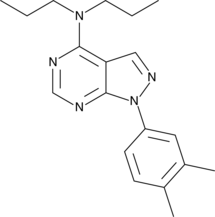
-
GC48547
Sartorypyrone D
A fungal metabolite
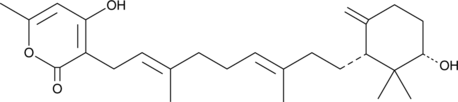
-
GN10091
Schaftoside
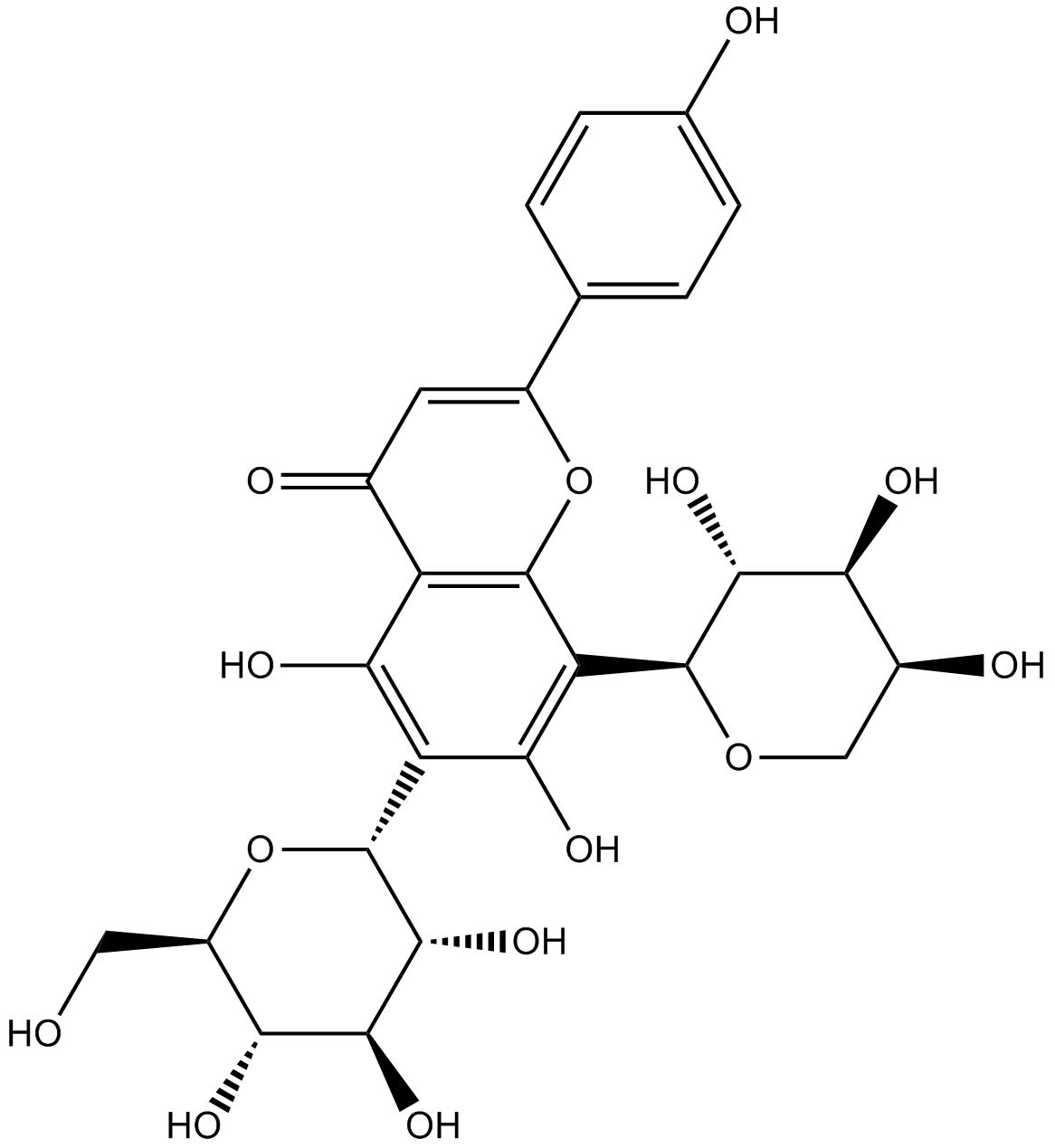
-
GC30459
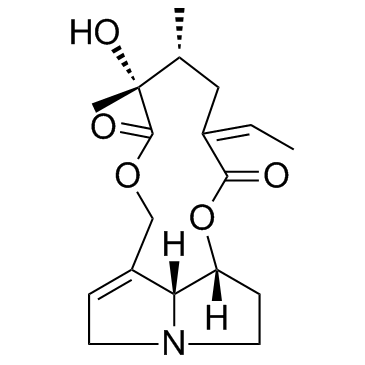
Senecionine (Senecionan-11,16-dione, 12-hydroxy-)
Senecionine (Senecionan-11,16-dione, 12-hydroxy-) (Senecionan-11,16-dione, 12-hydroxy-) is a pyrrolizidine alkaloid isolated from Senecio vulgaris.
-
GC61274
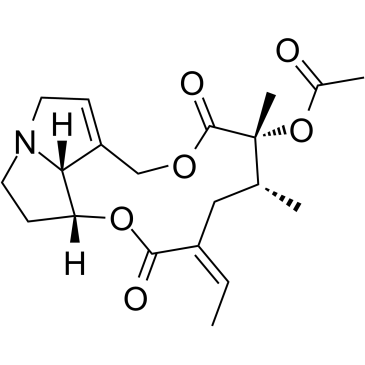
Senecionine acetate
Senecionine acetate (O-Acetylsenecionine) is a pyrrolizidine alkaloid.
-
GC48077
SHS4121705
A mitochondrial uncoupler
-
GC48894
SIH
SIH is an antituberculous compound.
-
GC31491
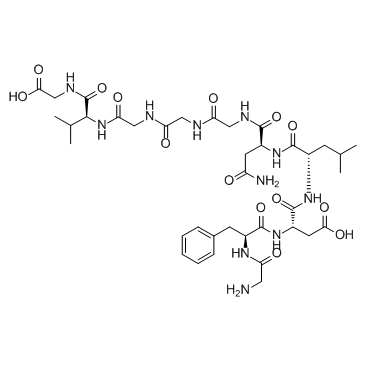
Speract
Speract, a sea urchin egg peptide that regulates sperm motility, also stimulates sperm mitochondrial metabolism.
-
GC63669
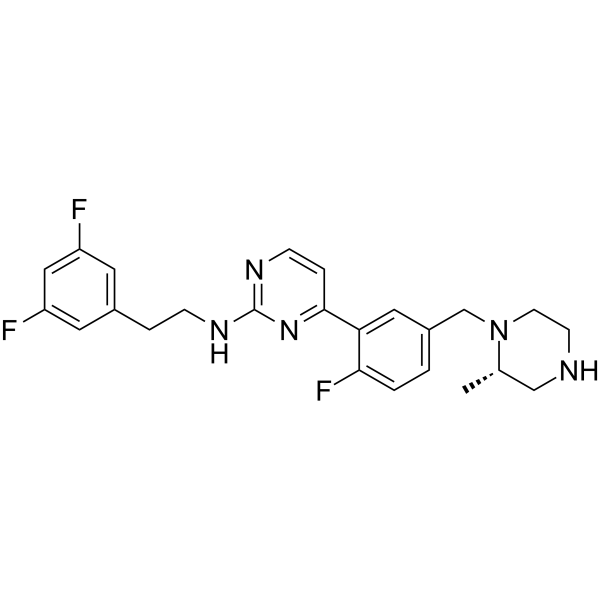
T0467
T0467 activates parkin mitochondrial translocation in a PINK1-dependent manner in vitro.
-
GC39512
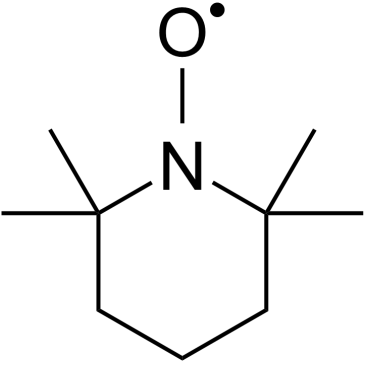
Tempo
Tempo is a classic nitroxide radical and is a selective scavenger of ROS that dismutases superoxide in the catalytic cycle. Tempo induces DNA-strand breakage. Tempo can be used as an organocatalyst for the oxidation of primary alcohols to aldehydes. Tempo has mutagenic and antioxidant effects.
-
GC45020
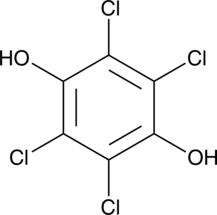
Tetrachlorohydroquinone
Tetrachlorohydroquinone (TCHQ) is a metabolite of the organochlorine biocide pentachlorophenol.
-
GC48924
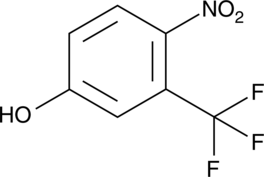
TFM
A piscicide
-
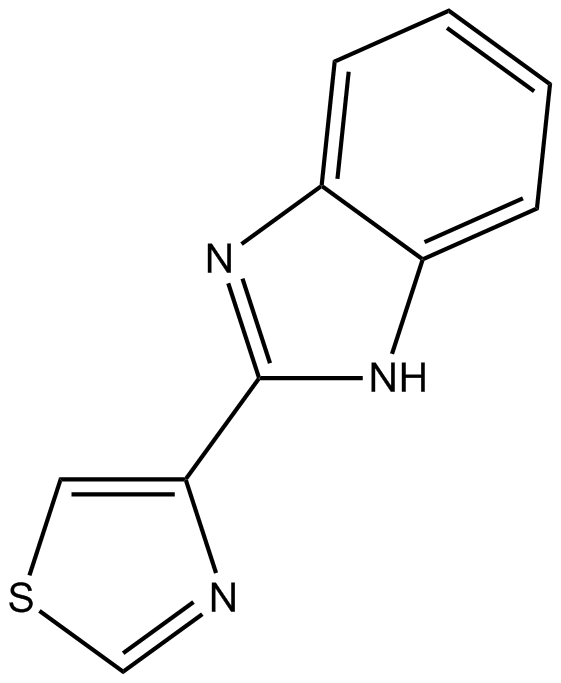
GC17228
Thiabendazole
Fungicide
-
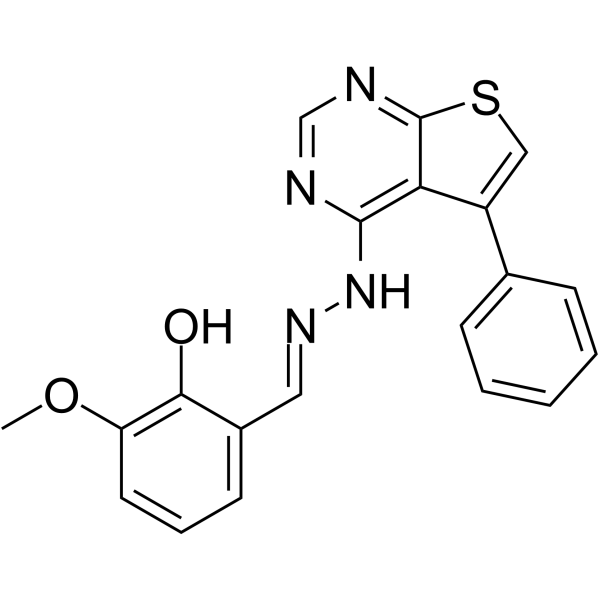
GC64688
THP104c
THP104c is a mitochondrial fission inhibitor.
-
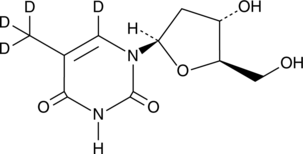
GC49529
Thymidine-d4
An internal standard for the quantification of thymidine
-
GC48192
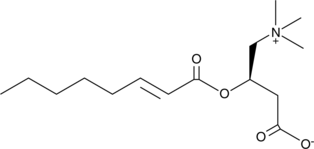
trans-2-Octenoyl-L-carnitine
A synthetic carnitine
-
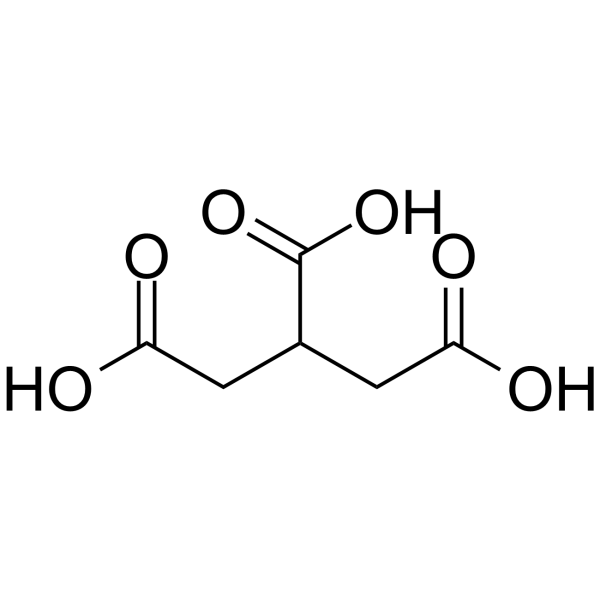
GC63238
Tricarballylic acid
Tricarballylic acid, a conjugate acid of a?tricarballylate, is a competitive inhibitor of the enzyme aconitate hydratase (aconitase; EC 4.2.1.3) with a Ki value of 0.52 mM.
-
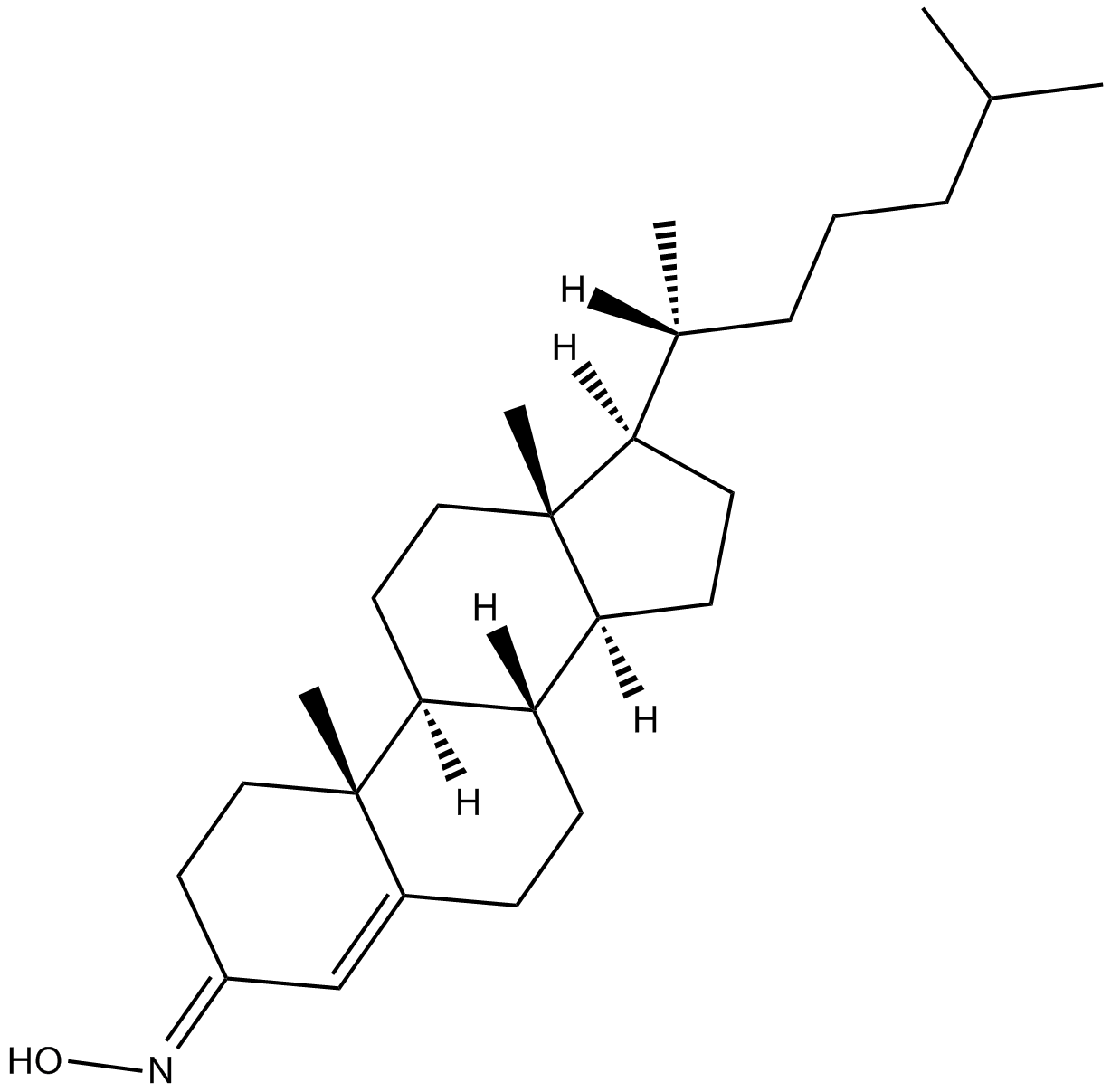
GC13726
TRO 19622
TRO 19622 (TRO 19622) is a mitochondrial-targeted neuroprotective compound with mean EC50 value for increasing cell survival is 3.2±0.2 μM.
-
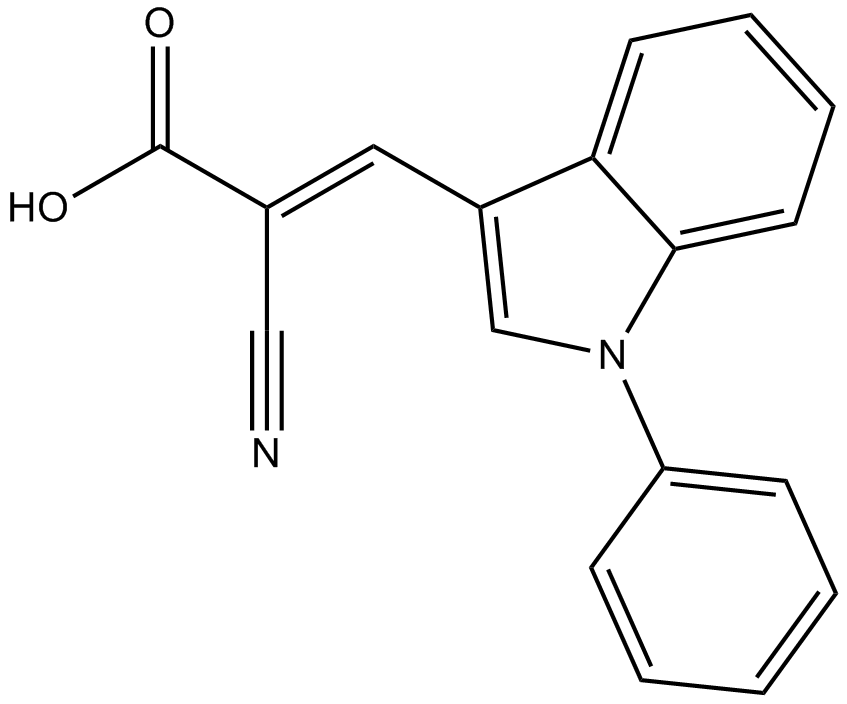
GC11865
UK-5099
An inhibitor of the mitochondrial pyruvate carrier
-
GC30512
Vatiquinone (EPI-743)
Vatiquinone (EPI-743) (Vatiquinone; α-Tocotrienol quinone; PTC-743; NCT04378075) is a potent cellular oxidative stress protectant, inhibits ferroptosis in cells, which could be used for the study for mitochondrial diseases.
-
GC48398
WST-1
A water-soluble and cell-permeable tetrazolium dye