Angiogenesis
Products for Angiogenesis
- Cat.No. Product Name Information
-
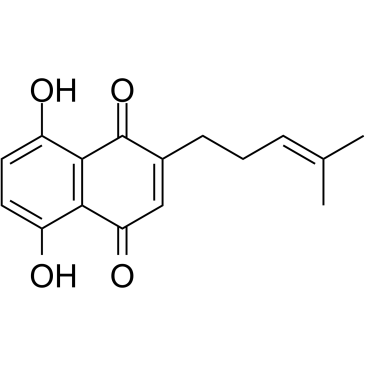
GC38767
Deoxyshikonin
A natural products with anti-tumor activity
-
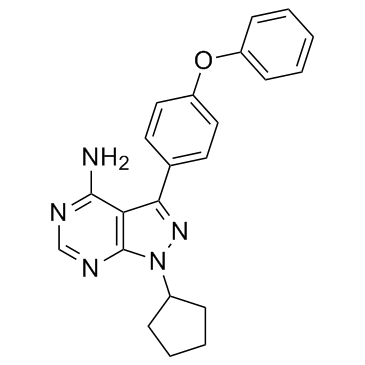
GC32449
Desidustat
An inhibitor of HIF-PH
-
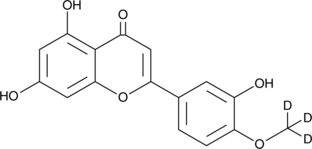
GC47234
Diosmetin-d3
An internal standard for the quantification of diosmetin
-
GC45767
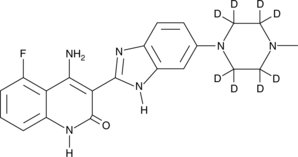
Dovitinib-d8
An internal standard for the quantification of dovitinib
-
GC19128
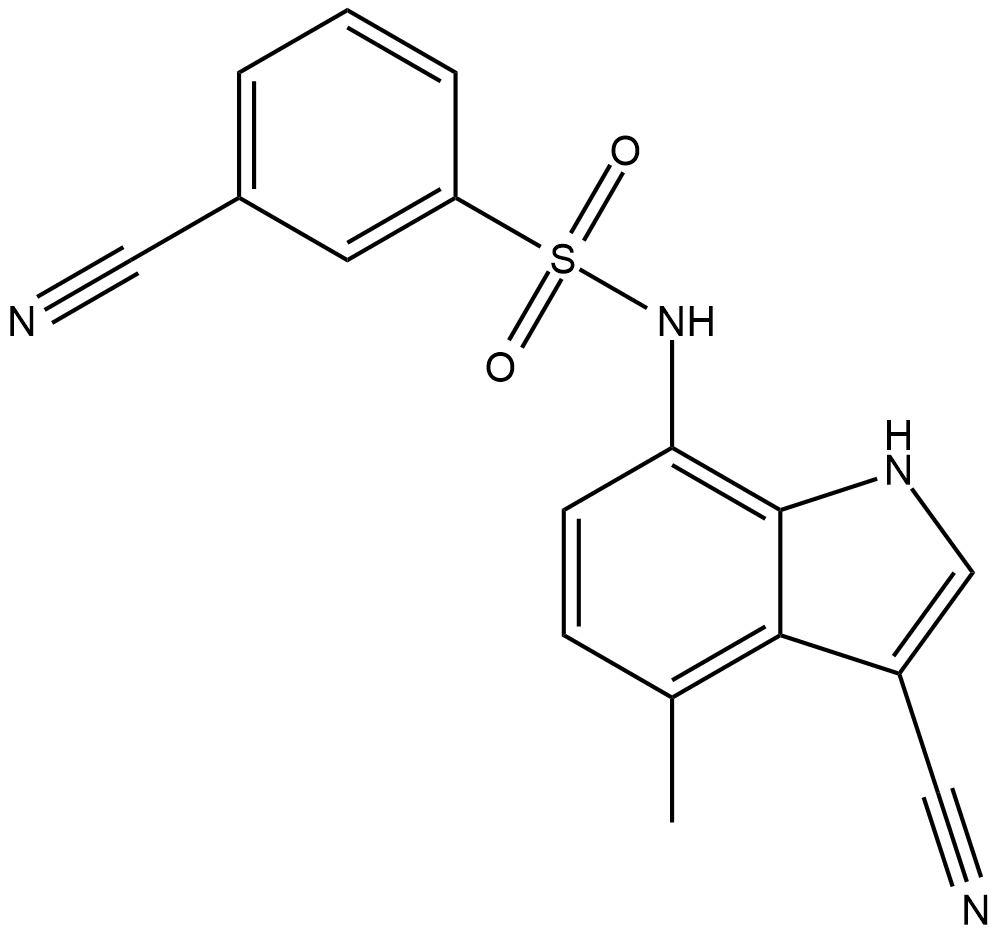
E7820
E7820 is an angiogenesis inhibitor by suppressing integrin a2, a cell adhesion molecule expressed on endothelial cells.
-
GC18236
Echinomycin
An inhibitor of HIF1mediated gene transcription

-
GC62182
Echistatin TFA
Echistatin TFA, the smallest active RGD protein belonging to the family of disintegrins that are derived from snake venoms, is a potent inhibitor of platelet aggregation.

-
GC17236
Echistatin, α1 isoform
Echistatin, α1 isoform, the smallest active RGD protein belonging to the family of disintegrins that are derived from snake venoms, is a potent inhibitor of platelet aggregation.

-
GC33043
EL-102
EL-102 is a hypoxia-induced factor 1 (Hif1α) inhibitor. EL-102 induces apoptosis, inhibits tubulin polymerisation and shows activities against prostate cancer. EL-102 can be used for the research of cancer.
-
GC64819
Elsubrutinib
Elsubrutinib (ABBV-105) is an orally active, potent, selective and irreversible Bruton's tyrosine kinase (BTK) inhibitor。The IC50 of Elsubrutinib for BTK catalytic domain is 0.18 μM.
-
GC33634
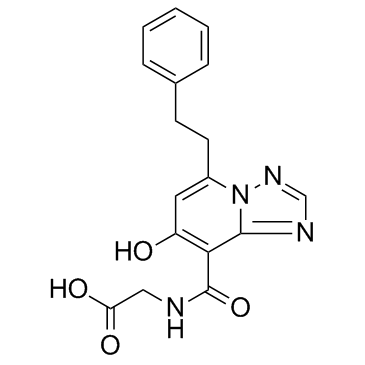
Enarodustat (JTZ-951)
Enarodustat (JTZ-951) is a potent and orally active hypoxia-inducible factor prolyl hydroxylase inhibitor, with an EC50 of 0.22 μM.
-
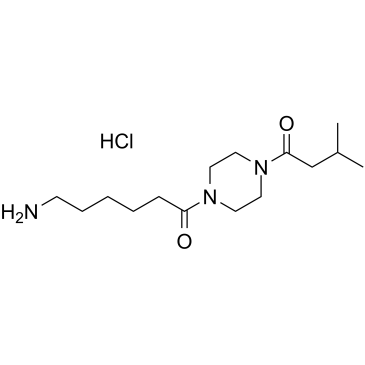
GC60807
ENMD-1068 hydrochloride
Selective protease-activated receptor 2 (PAR2) anatagonist
-
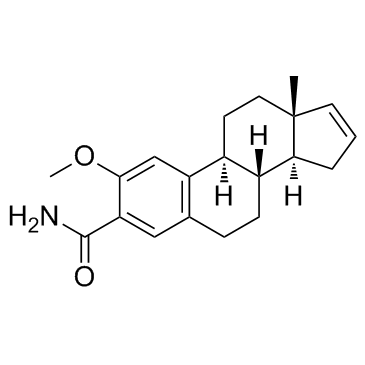
GC33170
ENMD-119 (ENMD 1198)
ENMD-119 (ENMD 1198) (IRC-110160), an orally active microtubule destabilizing agent, is a 2-methoxyestradiol analogue with antiproliferative and antiangiogenic activity. ENMD-119 (ENMD 1198) is suitable for inhibiting HIF-1alpha and STAT3 in human HCC cells and leads to reduced tumor growth and vascularization.
-
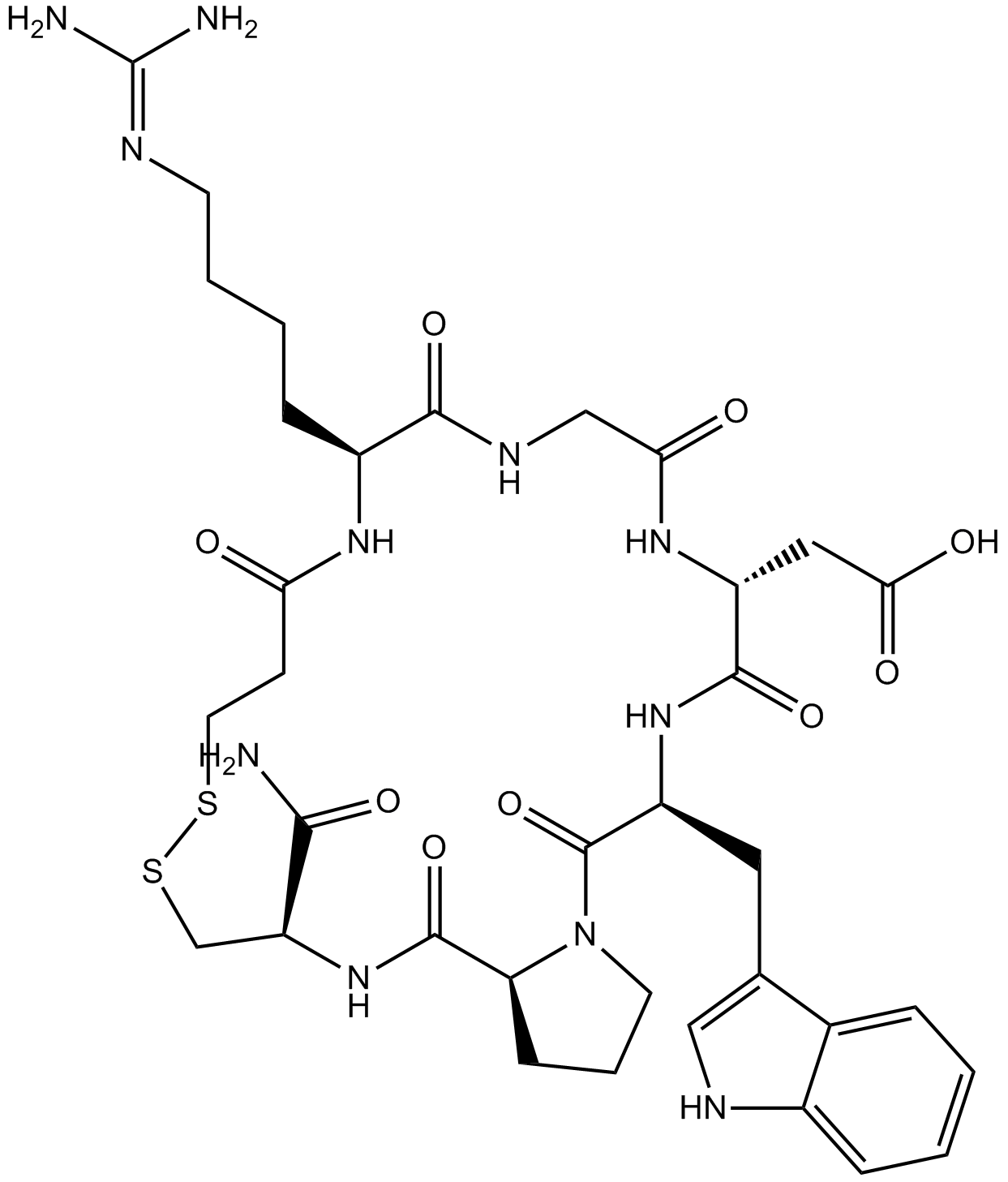
GC12447
Eptifibatide Acetate
Glycoprotein (GP) IIb/IIIa inhibitor
-
GC68283
Etrolizumab

-
GC47328
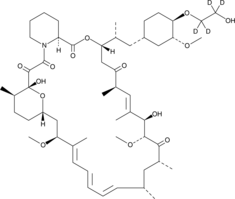
Everolimus-d4
An internal standard for the quantification of everolimus
-
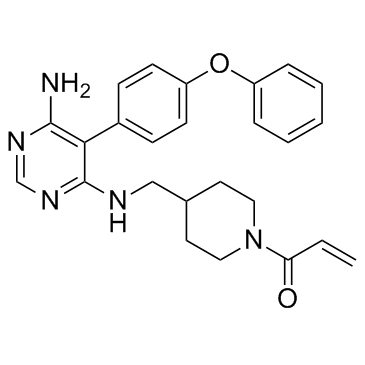
GC34062
Evobrutinib (M2951)
Evobrutinib, as an orally, highly selective, covalent Bruton's tyrosine kinase inhibitor, was well‐tolerated and effective.
-
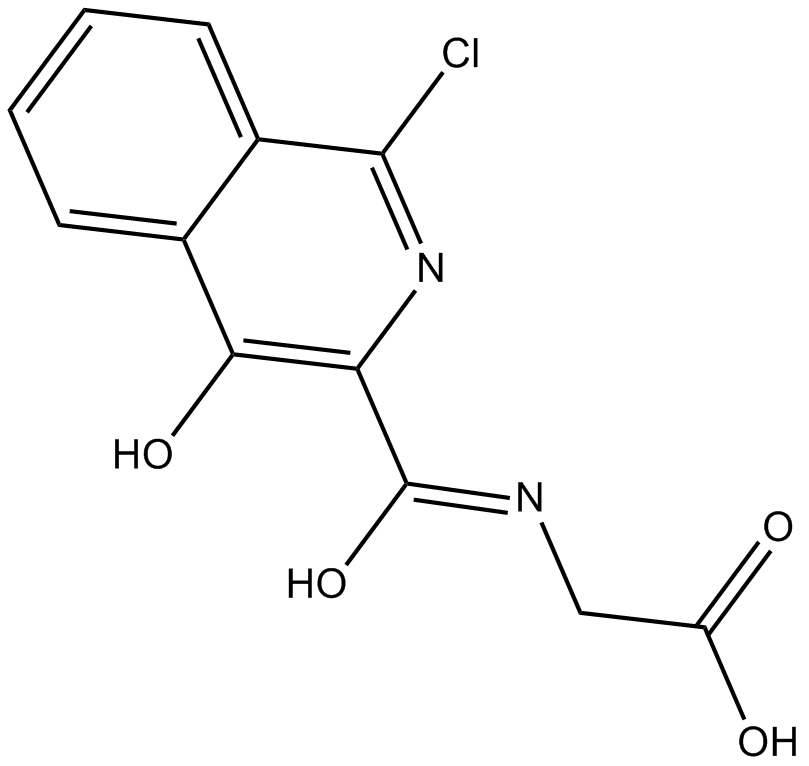
GC16638
FG2216
HIF-prolyl hydroxylase inhibitor
-
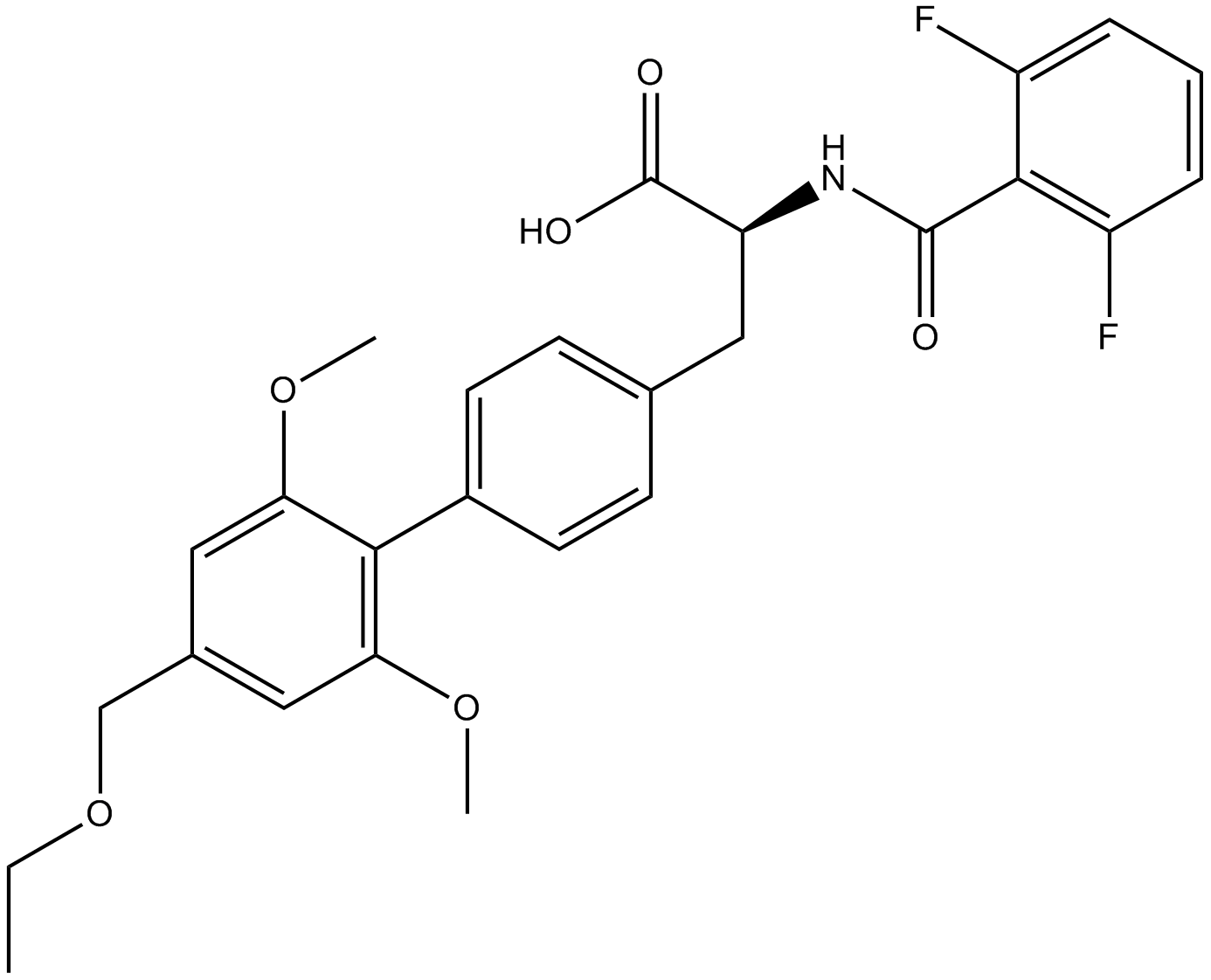
GC14804
Firategrast
-
GC32562
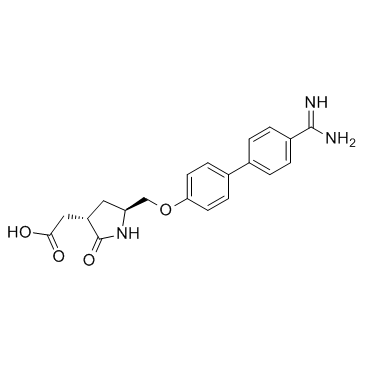
Fradafiban (BIBU-52)
Fradafiban (BIBU-52) is a nonpeptide platelet glycoprotein IIb/IIIa antagonist, which binds to the human platelet GP IIb/IIIa complex with a Kd value of 148 nM.
-
GC38044
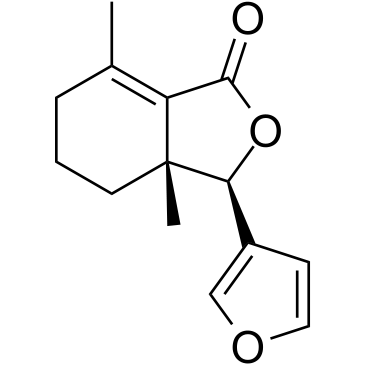
Fraxinellone
-
GC61501
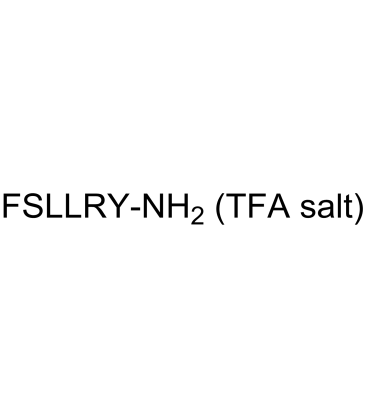
FSLLRY-NH2 TFA
FSLLRY-NH2 TFA is a protease-activated receptor 2 (PAR2) inhibitor.
-
GC43727
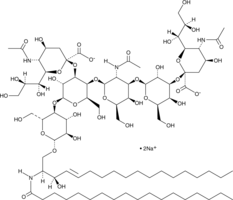
Ganglioside GD1a mixture (sodium salt)
Ganglioside GD1a is a sialic acid-containing glycosphingolipid found in brain, erythrocytes, bone marrow, testis, spleen, and liver.
-
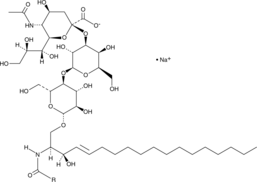
GC43732
Ganglioside GM3 Mixture (sodium salt)
Ganglioside GM3 is a monosialoganglioside that demonstrates antiproliferative and proapoptotic effects in tumor cells by modulating cell adhesion, proliferation, and differentiation.
-
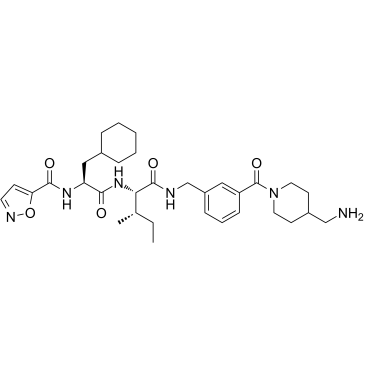
GC36125
GB-110
GB-110 is a potent, orally active, and nonpeptidic protease activated receptor 2 (PAR2) agonist.
-
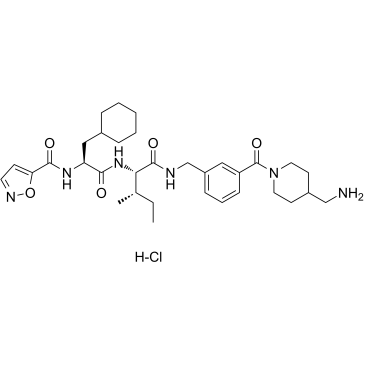
GC38329
GB-110 hydrochloride
GB-110 hydrochloride is a potent, orally active, and nonpeptidic protease activated receptor 2 (PAR2) agonist.
-
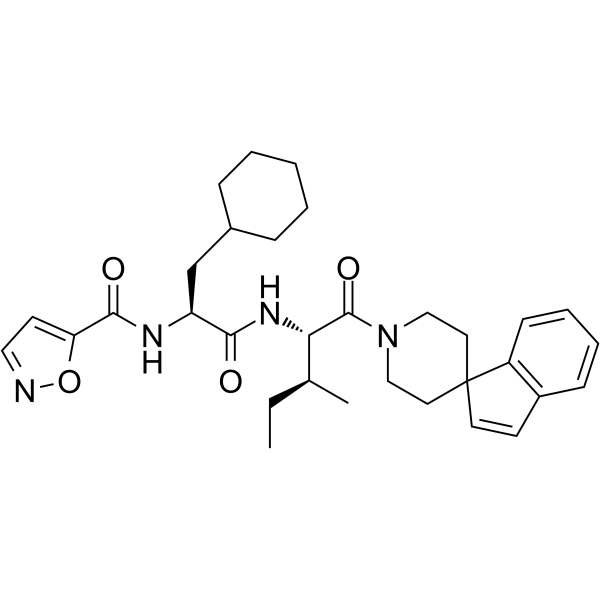
GC65439
GB-88
GB-88 is an oral, selective non-peptide antagonist of PAR2, inhibits PAR2 activated Ca2+ release with an IC50 of 2 ?M.
-
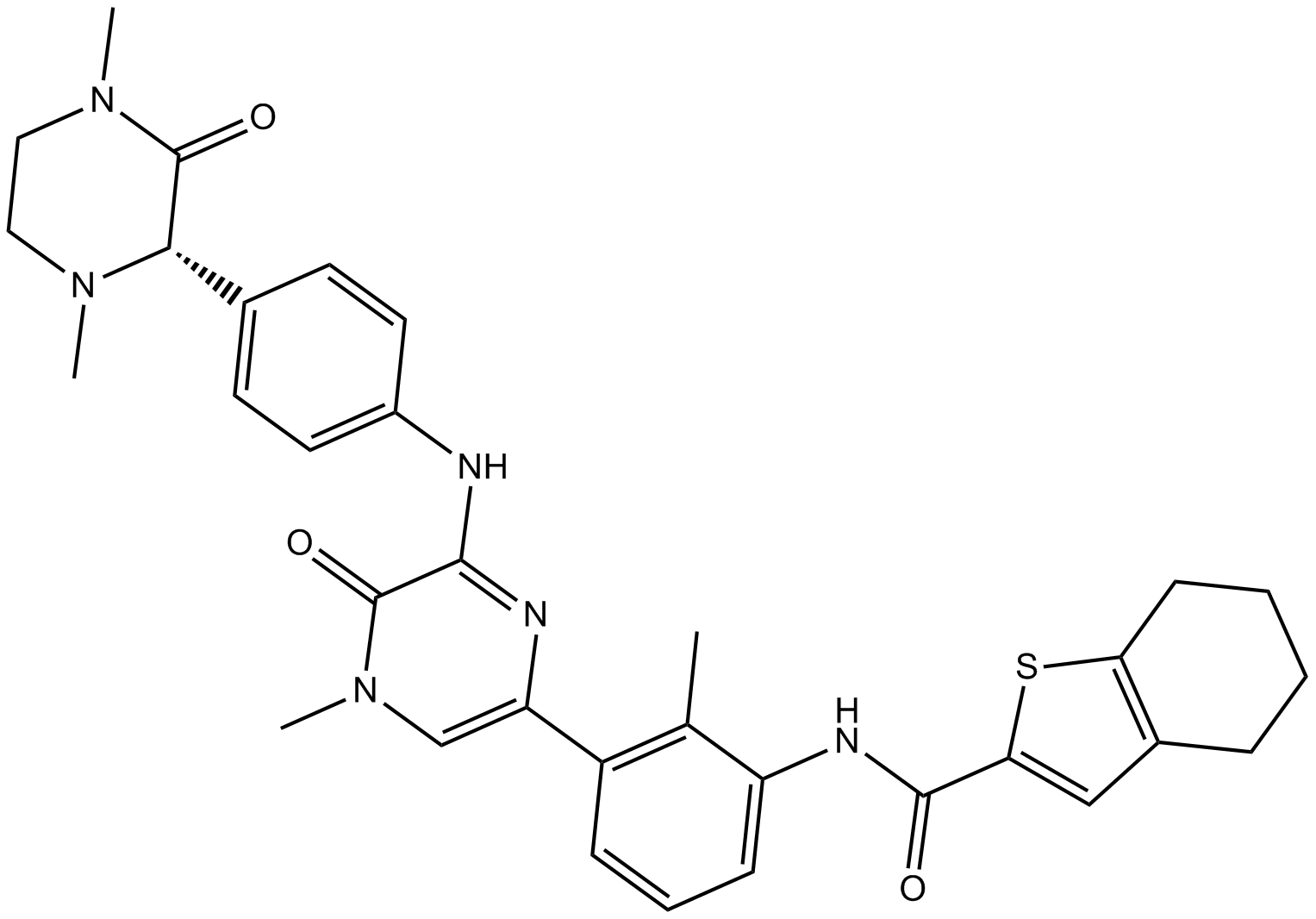
GC11995
GDC-0834
GDC-0834 is the S-enantiomer of GDC-0834.
-
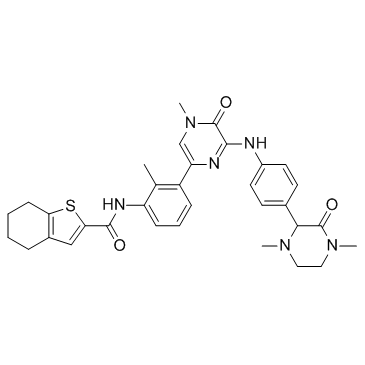
GC36127
GDC-0834 Racemate
Racemic form of GDC-0834
-
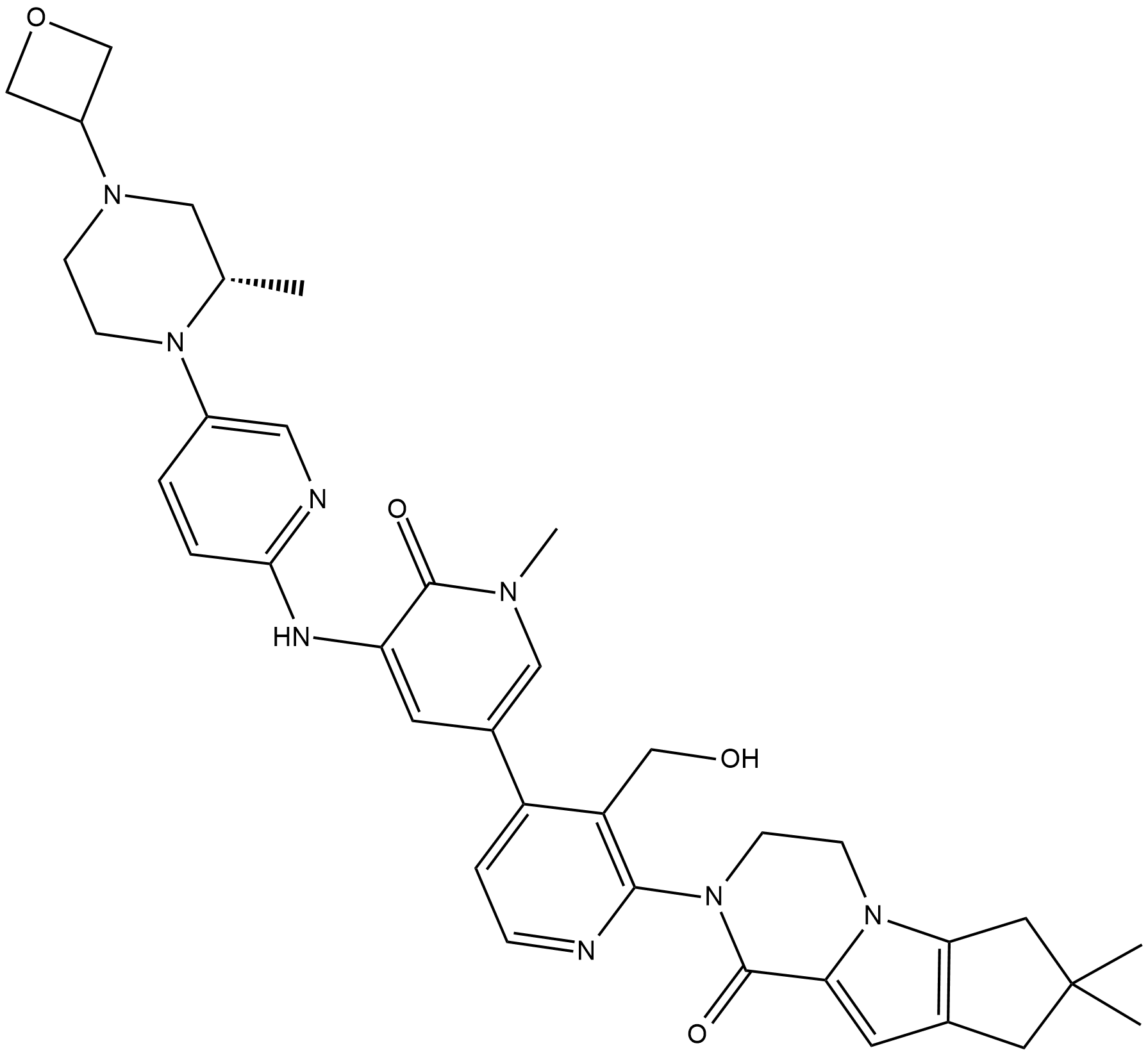
GC19162
GDC-0853
GDC-0853 (GDC-0853) is a potent, selective, orally available, and noncovalent bruton's tyrosine kinase (Btk) inhibitor with Kis of 0.91 nM, 1.6, 1.3, 12.6, and 3.4 nM for WT Btk, and the C481S, C481R, T474I, T474M mutants. GDC-0853 has the potential for rheumatoid arthritis and systemic lupus erythematosus research.
-
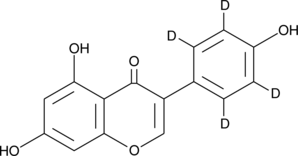
GC47398
Genistein-d4
An internal standard for the quantification of genistein
-
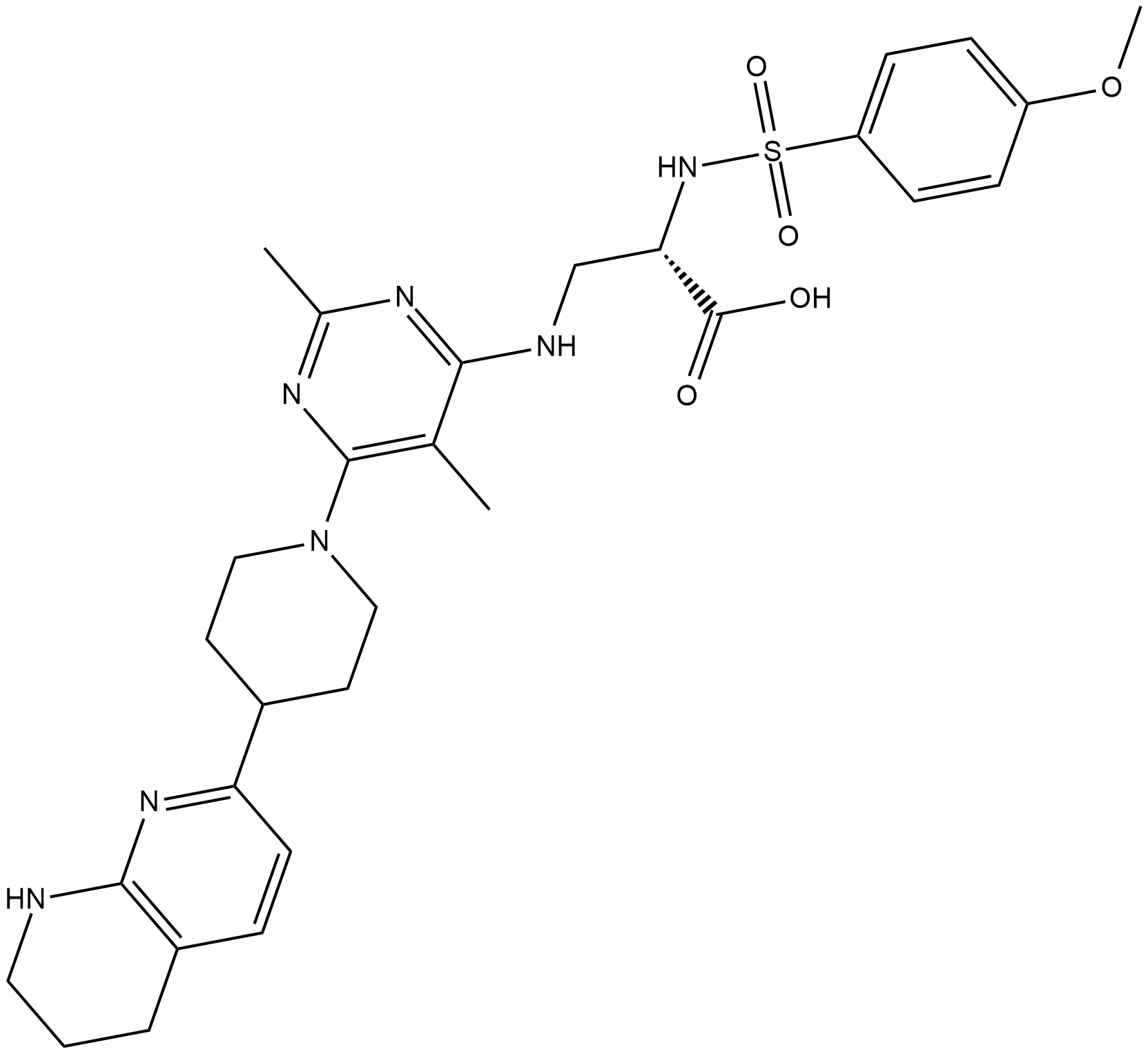
GC19167
GLPG0187
An αν?integrin receptor antagonist
-
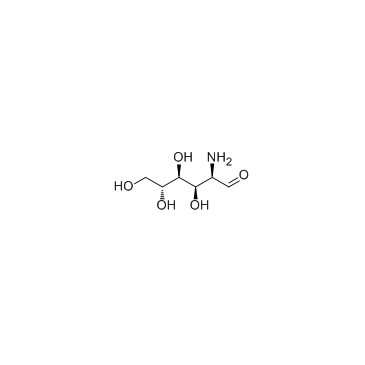
GC30263
Glucosamine (D-Glucosamine)
Glucosamine (D-Glucosamine) (D-Glucosamine (D-Glucosamine)) is an amino sugar and a prominent precursor in the biochemical synthesis of glycosylated proteins and lipids, is used as a dietary supplement.
-
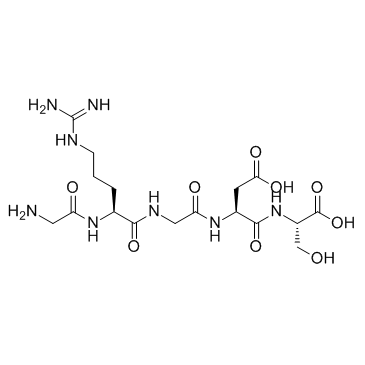
GC30223
Gly-Arg-Gly-Asp-Ser
Gly-Arg-Gly-Asp-Ser is a pentapeptide that forms the cell-binding domain of a glycoprotein, osteopontin.
-
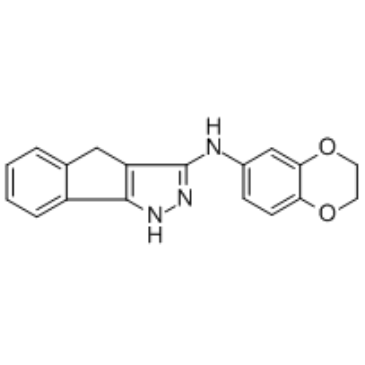
GC34595
GN44028
A HIF-1 inhibitor
-
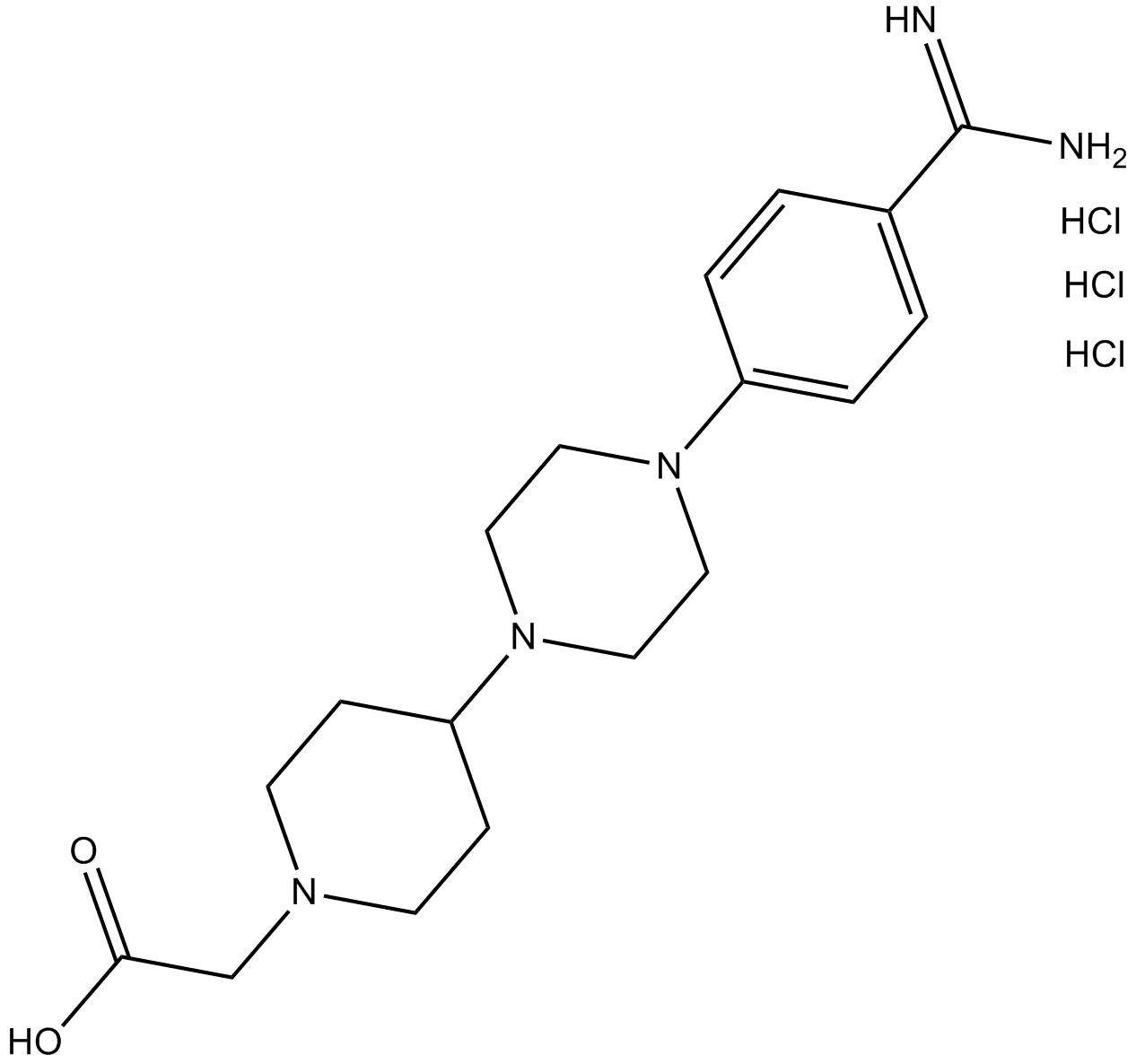
GC13951
GR 144053 trihydrochloride
platelet fibrinogen receptor glycoprotein IIb/IIIa (GpIIb/IIIa) antagonist
-
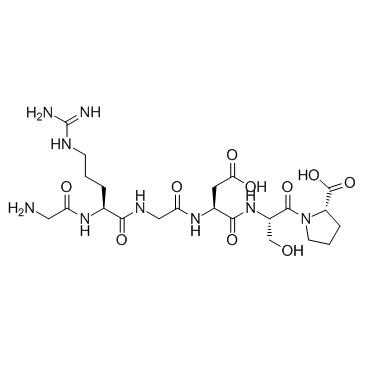
GC33323
GRGDSP
GRGDSP, a synthetic linear RGD peptide, is an integrin inhibitor.
-
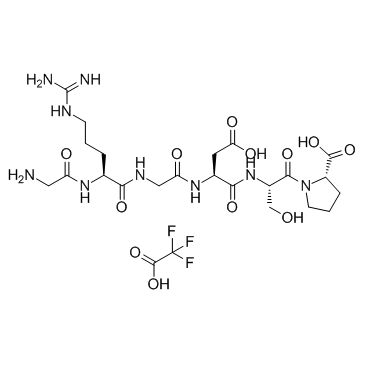
GC34265
GRGDSP TFA
GRGDSP (TFA) is an integrin inhibitor.
-
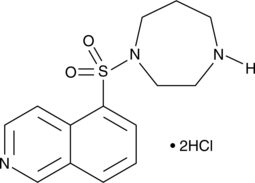
GC43803
HA-1077 (hydrochloride)
Fasudil (HA-1077; AT877) dihydrochloride is a nonspecific RhoA/ROCK inhibitor and also has inhibitory effect on protein kinases, with an Ki of 0.33 μM for ROCK1, IC50s of 0.158 μM and 4.58 μM, 12.30 μM, 1.650 μM for ROCK2 and PKA, PKC, PKG, respectively. HA-1077 (hydrochloride) is also a potent Ca2+ channel antagonist and vasodilator.
-
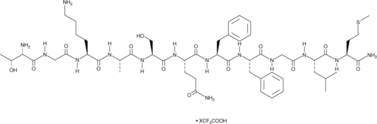
GC49882
Hemokinin 1 (human) (trifluoroacetate salt)
A peptide agonist of NK1 receptors
-
GC39146
HIF-1 inhibitor-1
An inhibitor of HIF-1 signaling

-
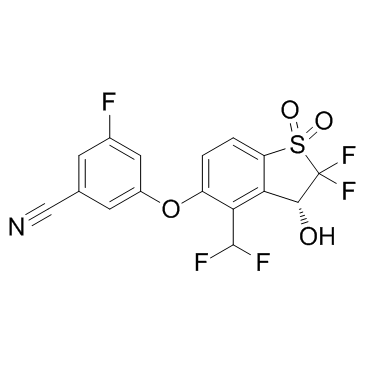
GC31358
HIF-2α-IN-1
HIF-2α-IN-1 is a HIF-2α inhibitor has an IC50 of less than 500 nM in HIF-2α scintillation proximity assay.
-
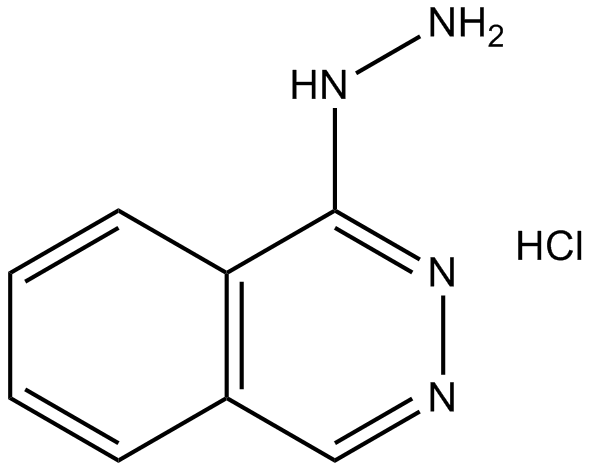
GC11767
Hydralazine HCl
Hydralazine HCl is a orally active antihypertensive agent, reduces peripheral resistance directly by relaxing the smooth muscle cell layer in arterial vessel.
-
GC64979
I-191
I-191 is a potent, selective protease-activated receptor 2 (PAR2) antagonist.

-
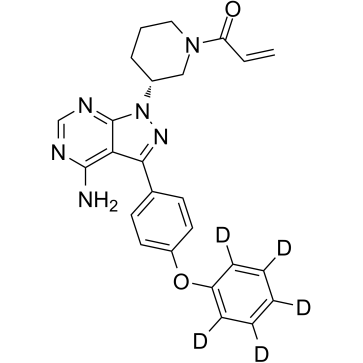
GC39194
Ibrutinib D5
Ibrutinib D5 (PCI-32765 D5) is a deuterium labeled Ibrutinib. Ibrutinib is a selective, irreversible Btk inhibitor.
-
GC60197
Ibrutinib deacryloylpiperidine
Ibrutinib deacryloylpiperidine (IBT4A) is an impurity of Ibrutinib. Ibrutinib is a selective, irreversible Btk inhibitor with an IC50 of 0.5 nM.
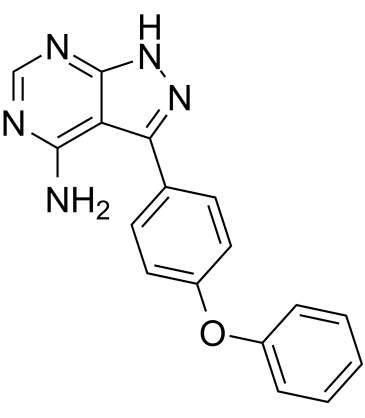
-
GC36289
Ibrutinib-biotin
Ibrutinib-biotin is a probe that consists of Ibrutinib linked to biotin via a long chain linker, extracted from patent WO2014059368A1 Compound 1-5, has an IC50 of 0.755-1.02 nM for BTK.
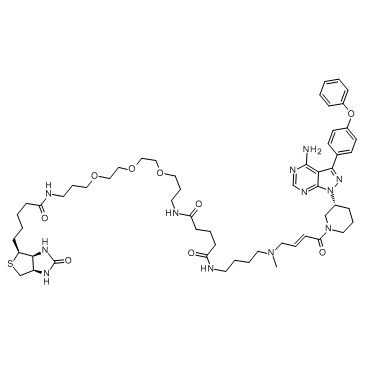
-
GC32948
IDF-11774
A novel HIF-1 inhibitor
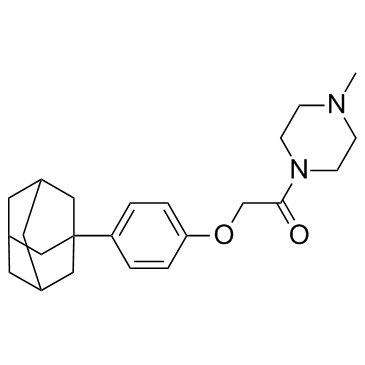
-
GC34301
ILK-IN-2
ILK-IN-2 (OSU-T315 analog) is a ILK inhibitor.
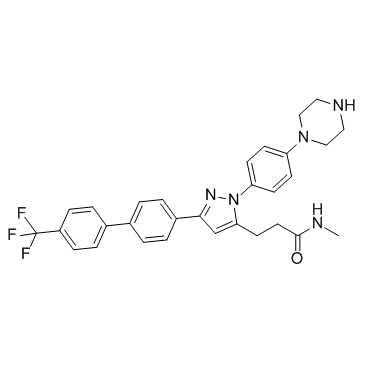
-
GC60200
ILK-IN-3
ILK-IN-3 is an integrin linked kinase inhibitor with antitumor activity.
-
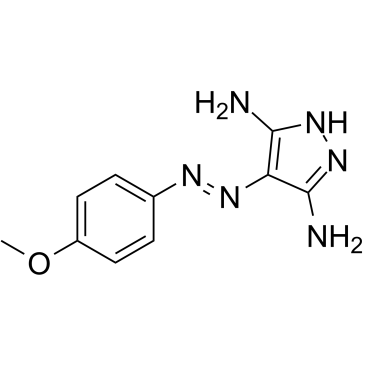
GC47454
IMS 2186
An anti-choroidal neovascularization agent
-
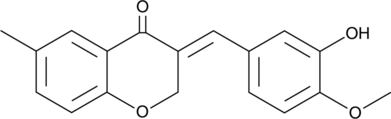
GC32967
Integrin Antagonists 27
Integrin Antagonists 27 is a small molecule integrin αvβ3 antagonist with binding affinity of 18 nM, as s novel anticancer agent.
-
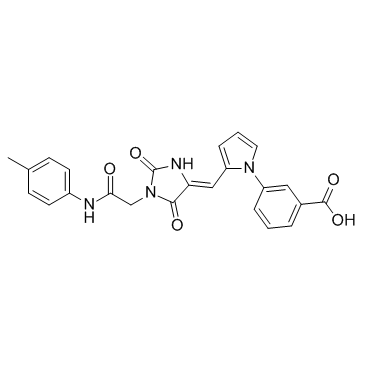
GC61613
Integrin modulator 1
Integrin modulator 1 is a potent and selective α4β1 integrin agonist, with an IC50 of 9.8 nM for RGD-binding α4β1.
-
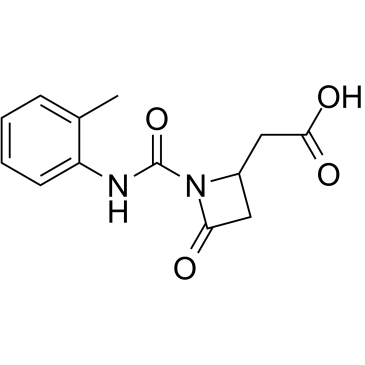
GC12255
IOX4
PHD2 inhibitor
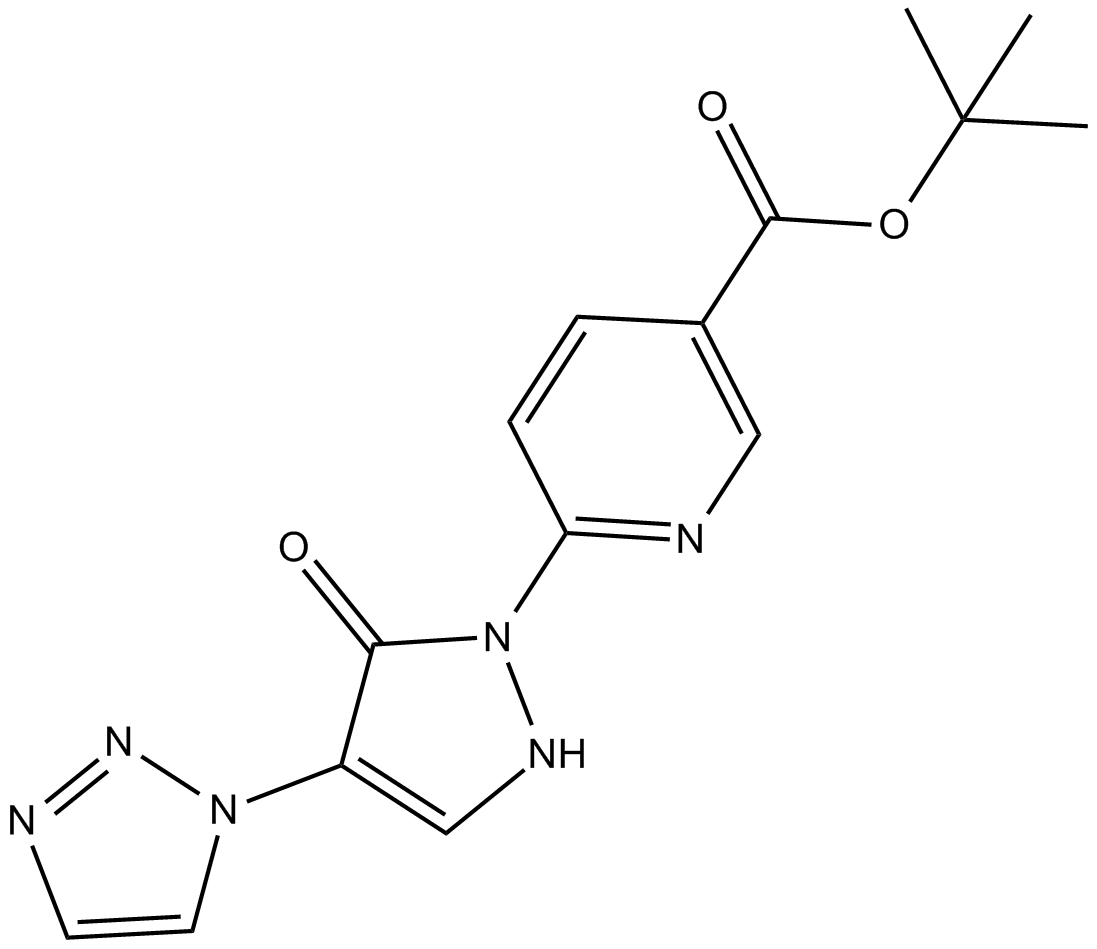
-
GC32787
iRGD peptide (c(CRGDKGPDC))
iRGD peptide (c(CRGDKGPDC)) is a 9-amino acid cyclic peptide, triggers tissue penetration of drugs by first binding to av integrins, then proteolytically cleaved in the tumor to produce CRGDK/R to interact with neuropilin-1, and has tumor-targeting and tumor-penetrating properties.
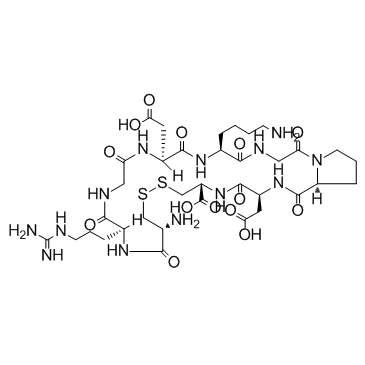
-
GC38801
Irigenin
Irigenin is a is a lead compound, and mediates its anti-metastatic effect by specifically and selectively blocking α9β1 and α4β1 integrins binding sites on C-C loop of Extra Domain A (EDA). Irigenin shows anti-cancer properties. It sensitizes TRAIL-induced apoptosis via enhancing pro-apoptotic molecules in gastric cancer cells.
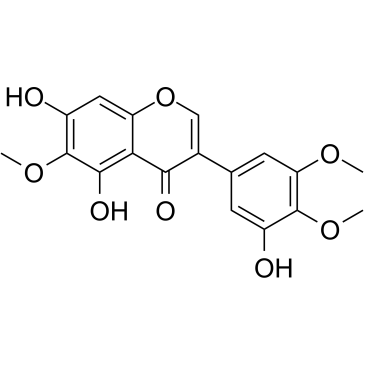
-
GC15379
JNJ-42041935
Hypoxia-inducible factor (HIF) prolyl hydroxylase (PHD) inhibitor
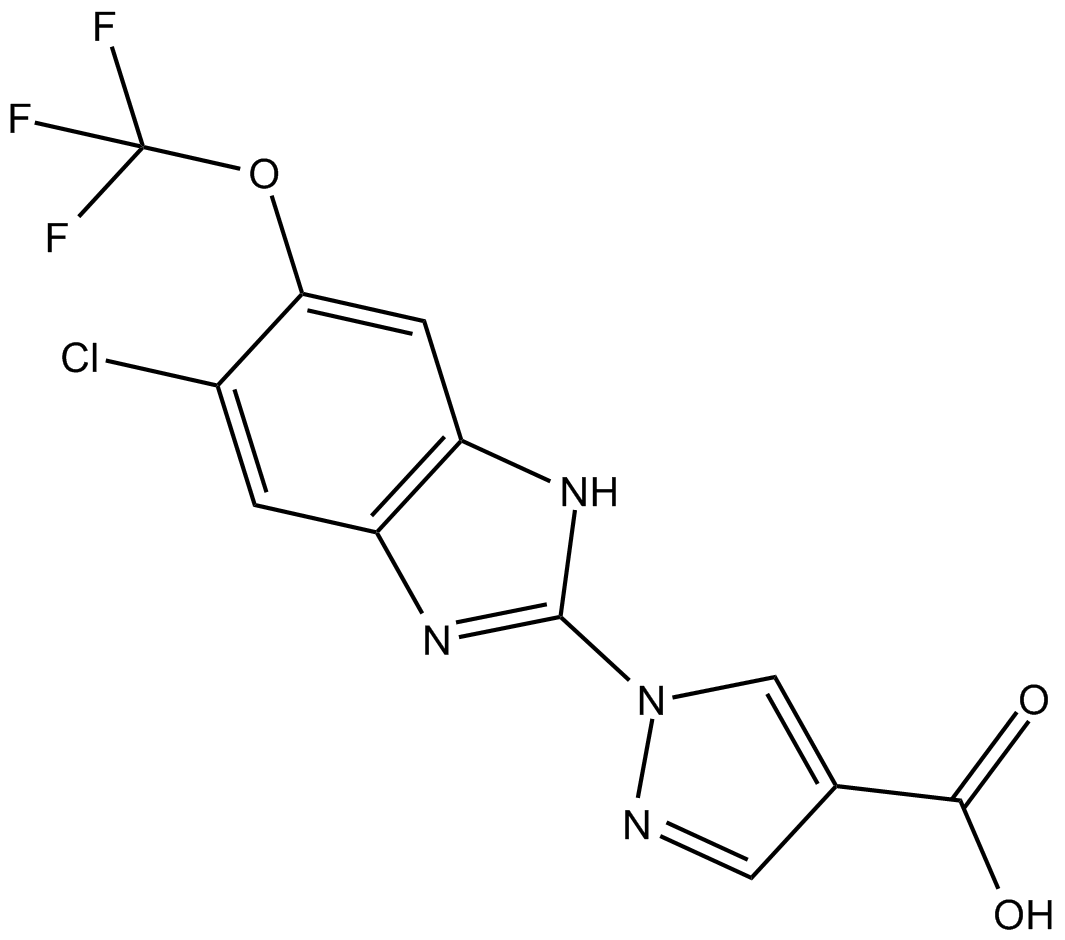
-
GC50642
LDV
α4β1 (VLA-4) ligand

-
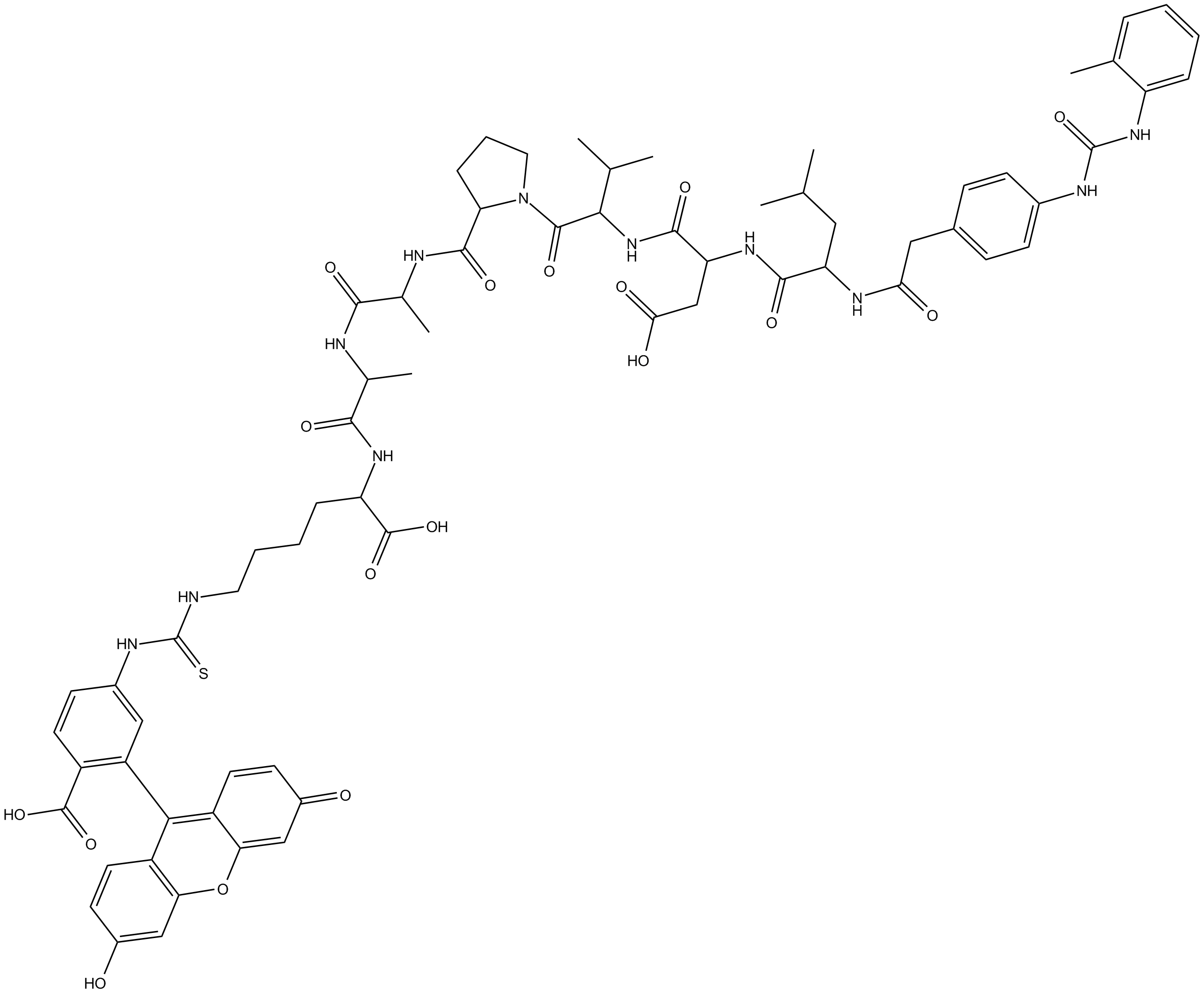
GC16190
LDV FITC
fluorescent ligand that binds to the α4β1 integrin (VLA-4)
-
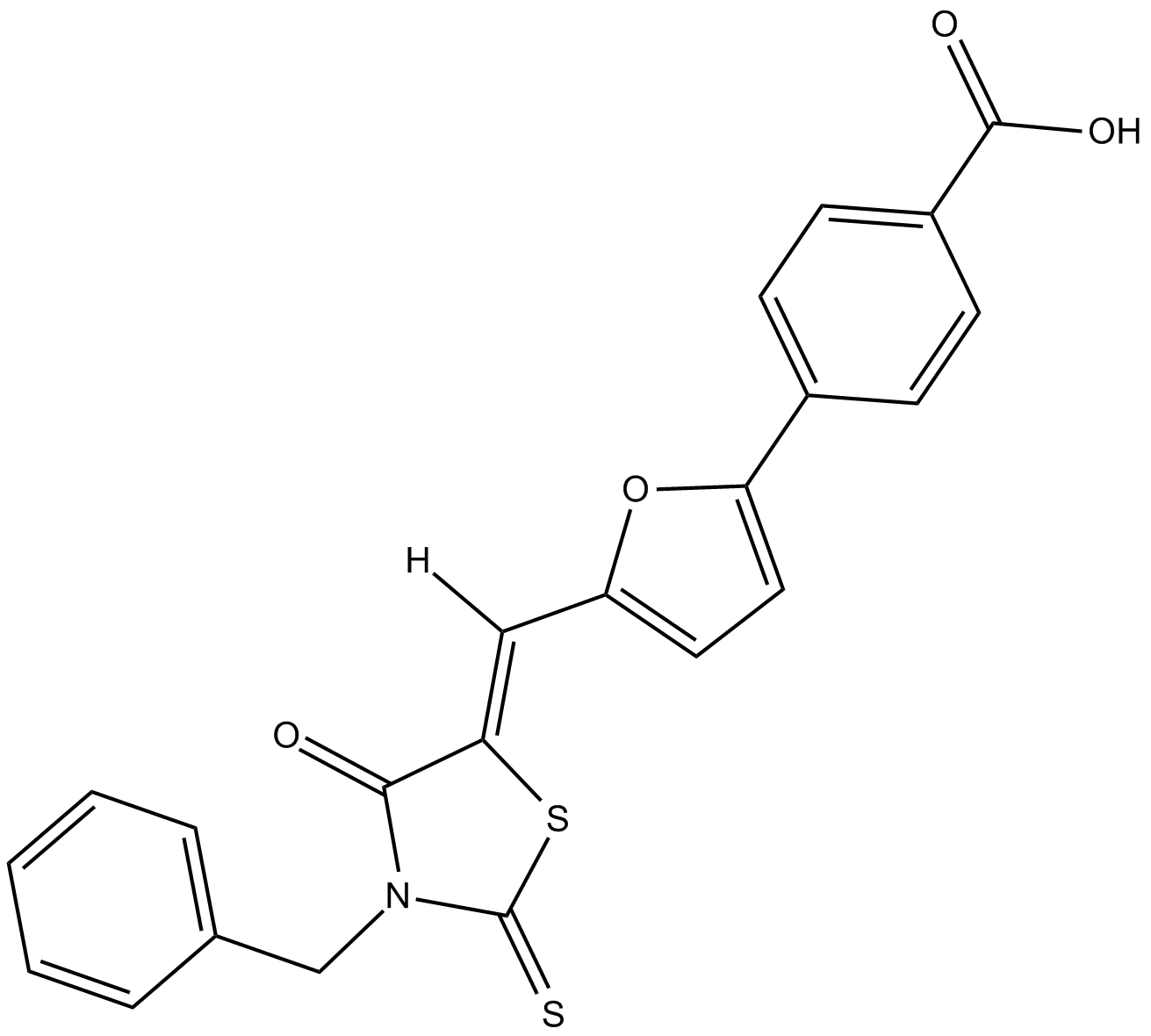
GC17263
Leukadherin 1
CD11b/CD18 activator
-
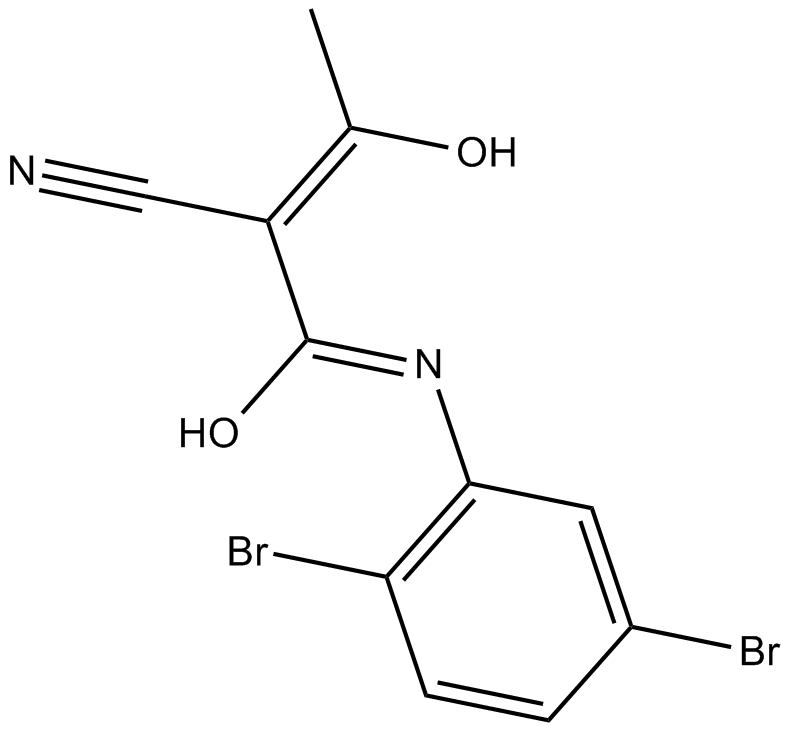
GC14857
LFM-A13
BTK-specific tyrosine kinase inhibitor
-
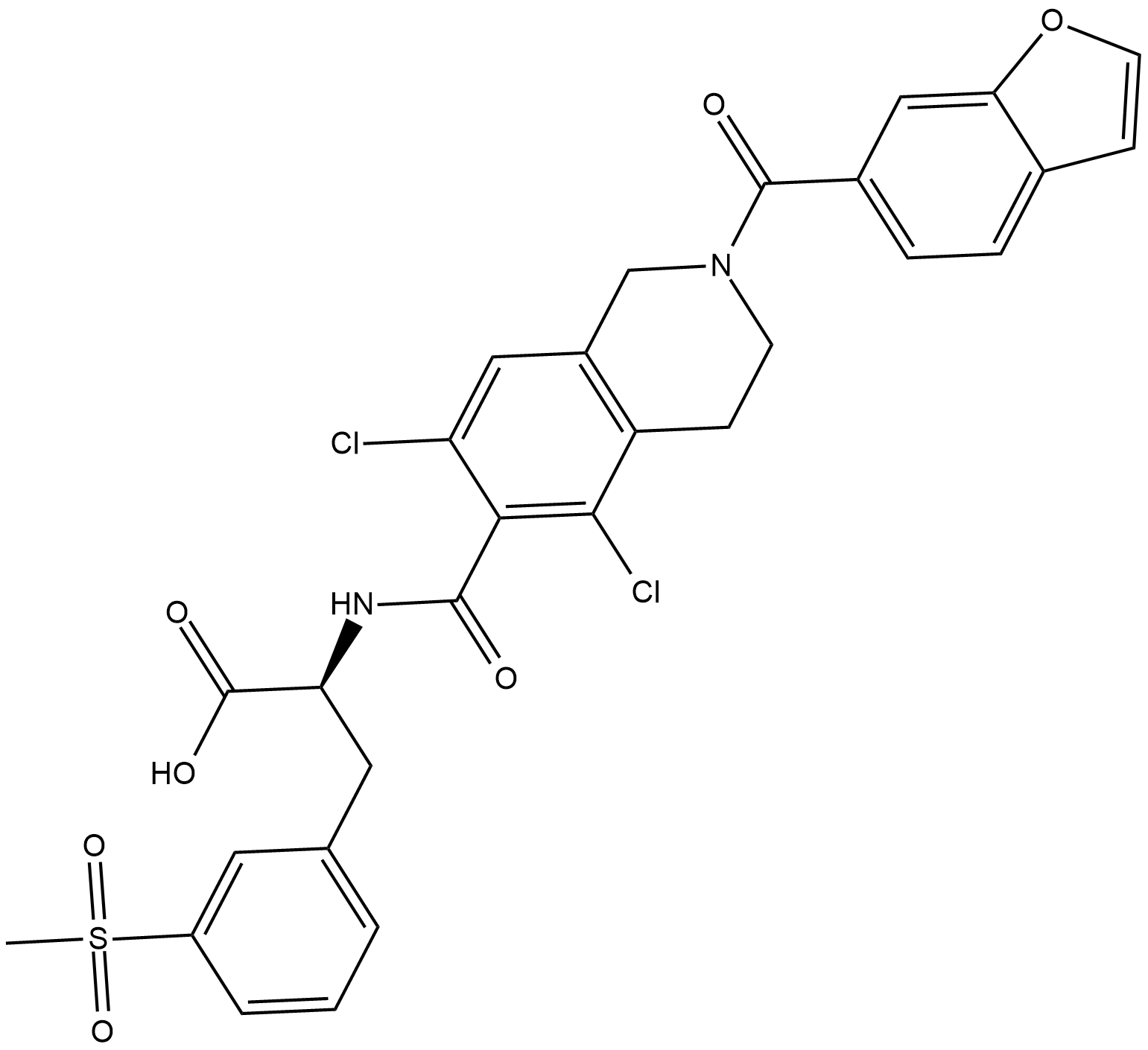
GC19222
Lifitegrast
Lifitegrast is an integrin lymphocyte function-associated antigen-1 (LFA-1) antagonist; inhibits Jurkat T cell attachment to ICAM-1 with an IC50 of 2.98 nM.
-
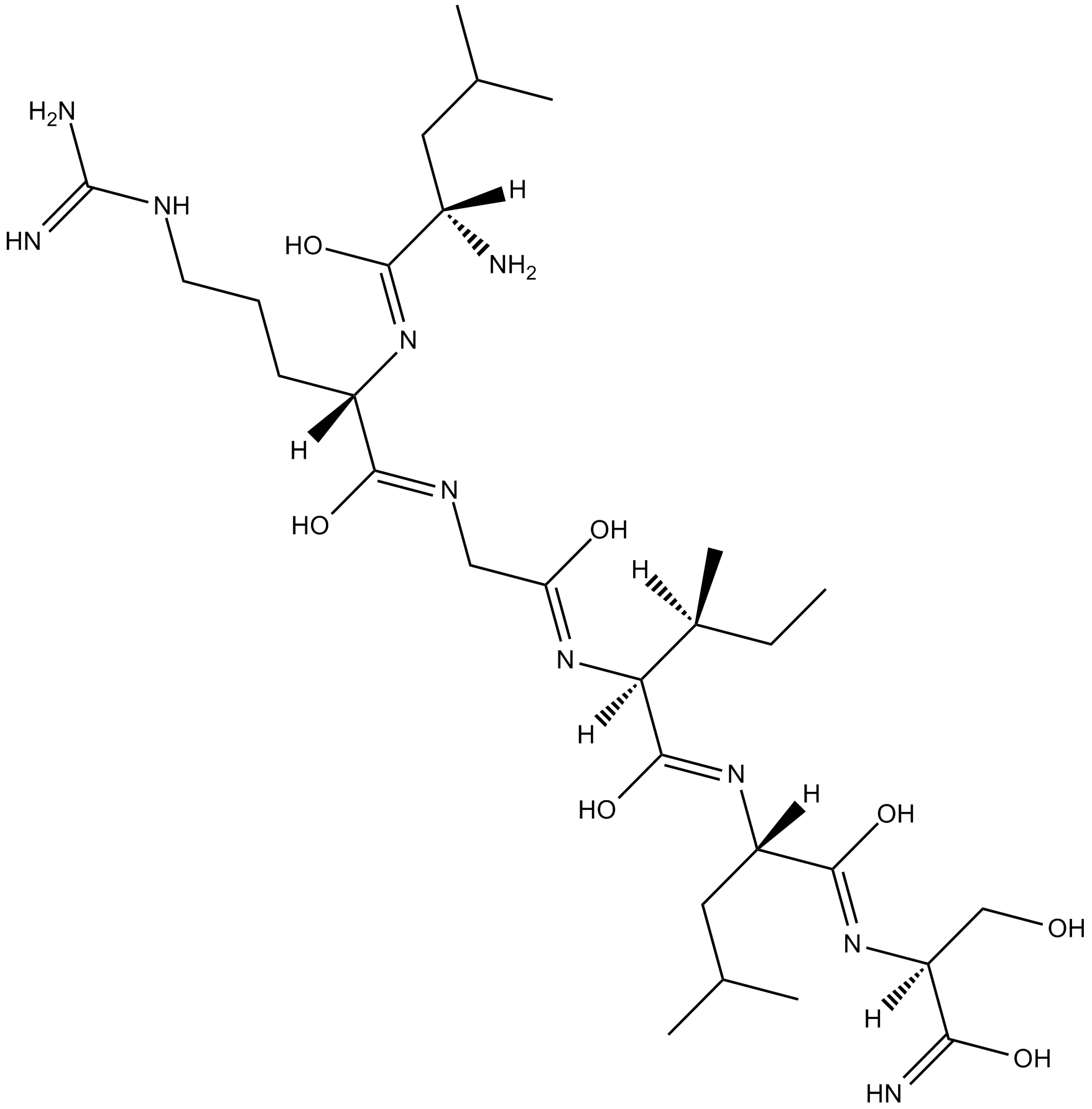
GC13227
LRGILS-NH2
Protease-activated receptor agonist
-
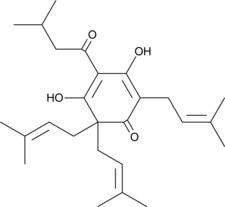
GC47582
Lupulone
A beta-acid
-
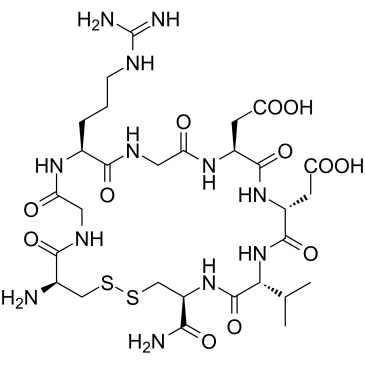
GC32724
LW6 (HIF-1α inhibitor)
LW6 (HIF-1α inhibitor) (HIF-1α inhibitor) is a novel HIF-1 inhibitor with an IC50 of 4.4 μM. LW6 (HIF-1α inhibitor) decreases HIF-1α protein expression without affecting HIF-1β expression.
-
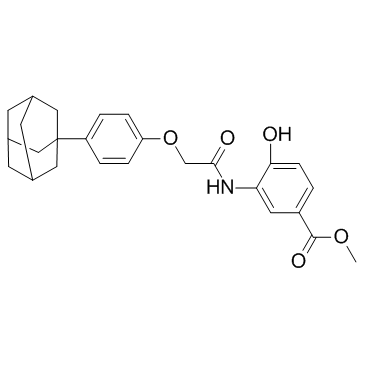
GC36500
LXW7
LXW7, a cyclic peptide containing Arg-Gly-Asp (RGD), is an integrin αvβ3 inhibitor.
-
GC40865
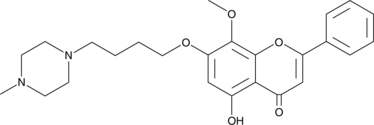
LYG-202
LYG-202 is a synthetic flavonoid with anticancer and anti-angiogenic activities.
-
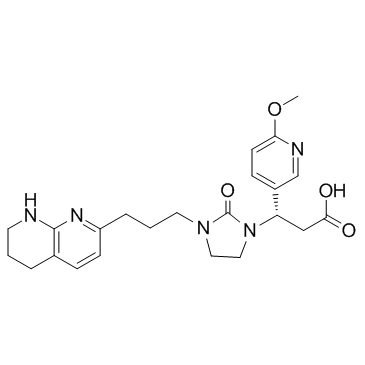
GC32055
MK-0429 (L-000845704)
MK-0429 (L-000845704) (L-000845704) is an orally active, potent, selective and nonpeptide pan-integrin antagonist with IC50 values of 1.6 nM, 2.8 nM, 0.1 nM, 0.7 nM, 0.5 nM and 12.2 nM for αvβ1, αvβ3, αvβ5, αvβ6, αvβ8 and α5β1, respectively.
-
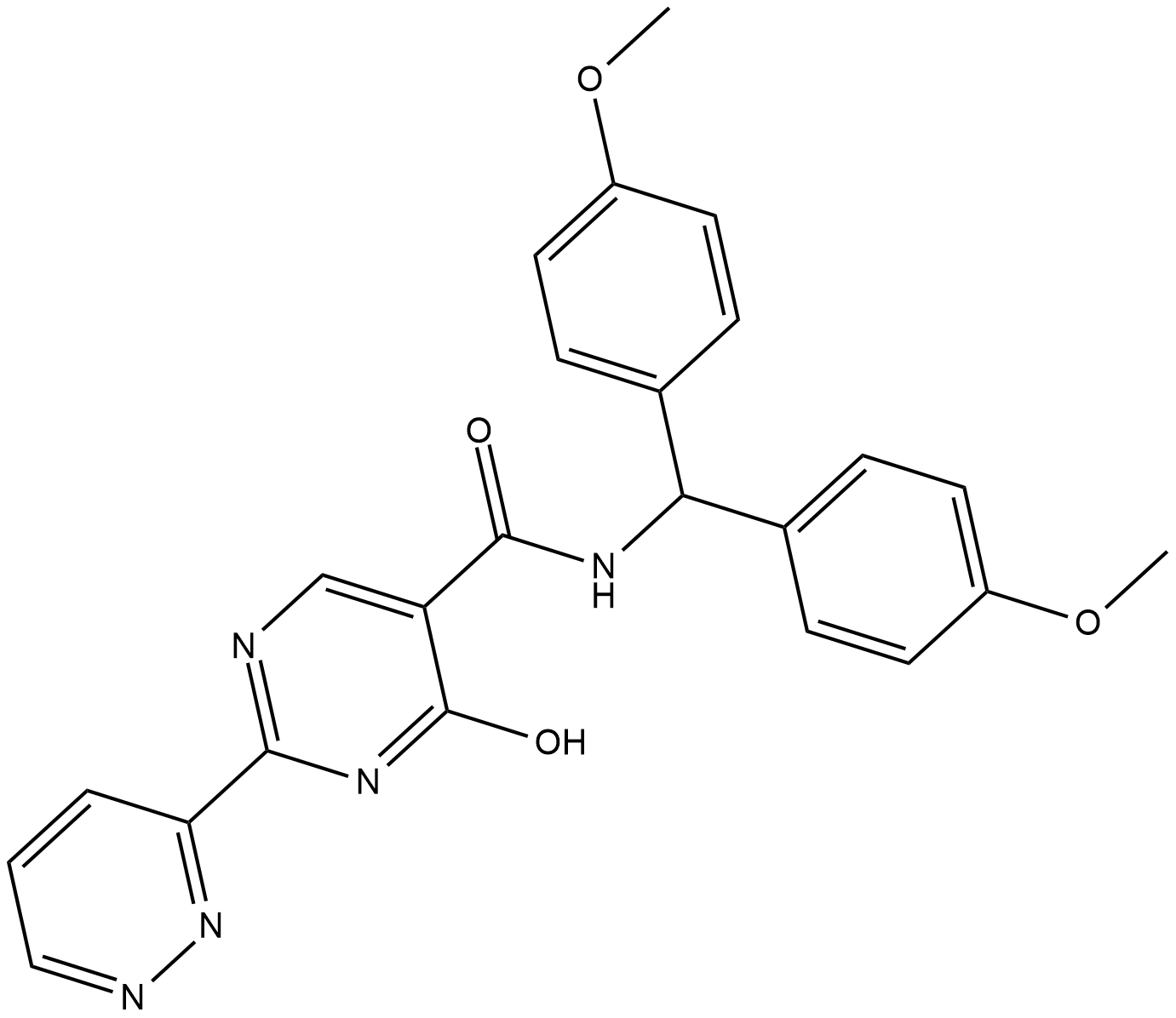
GC19251
MK-8617
MK-8617 is an orally active pan-inhibitor of hypoxia-inducible factor prolyl hydroxylase 1-3 (HIF PHD1-3) with an IC50 of 1 nM for PHD2.
-
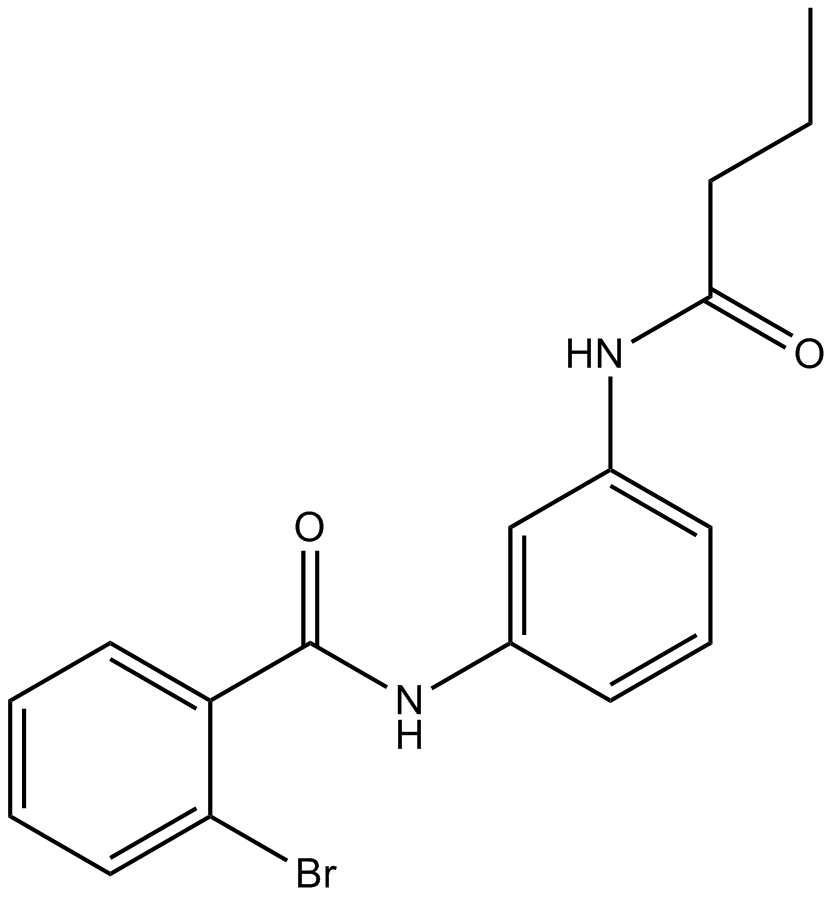
GC13004
ML161
Reversible inhibitor of PAR1-mediated platelet activation
-
GC18608
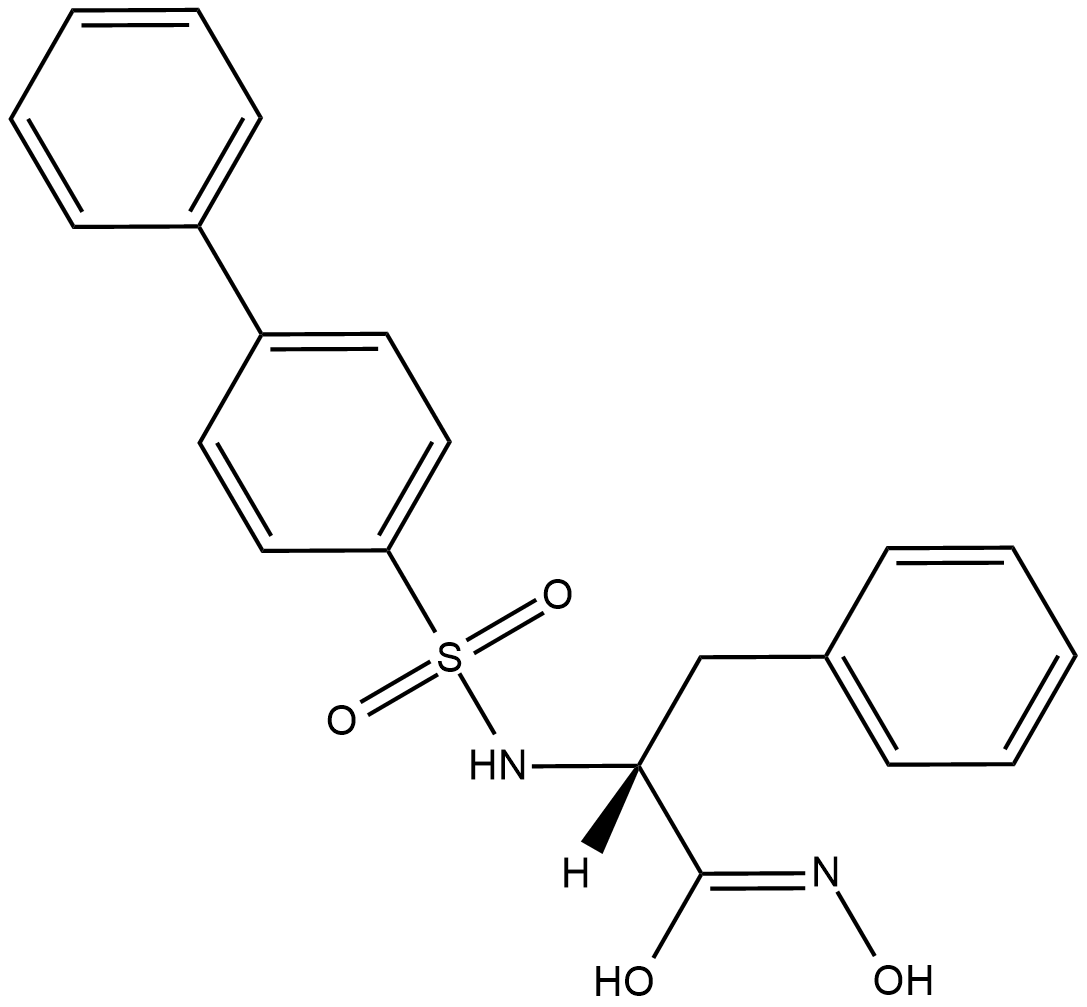
MMP-2/MMP-9 Inhibitor II
MMP-2/MMP-9 inhibitor II is a dual inhibitor of matrix metalloproteinase-2 (MMP-2) and MMP-9 (IC50s = 17 and 30 nM, respectively).
-
GC18616
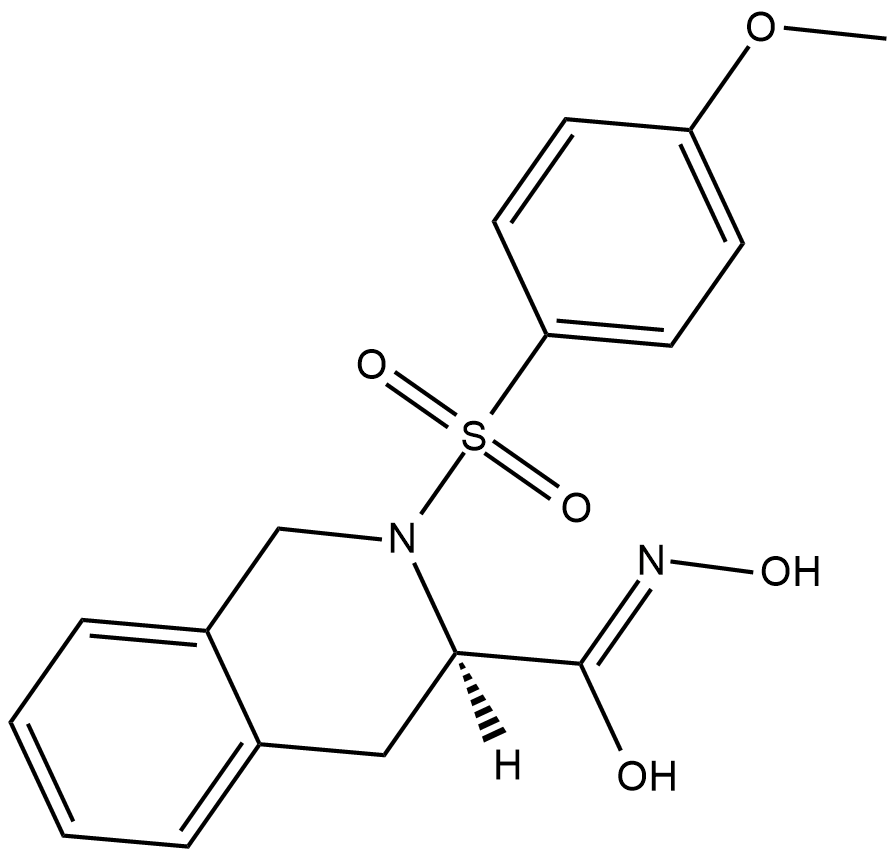
MMP-8 Inhibitor I
MMP-8 Inhibitor I is a selective inhibitor of the neutrophil collagenase matrix metalloproteinase-8 (MMP-8) with an IC50 value of 4 nM.
-
GC10048
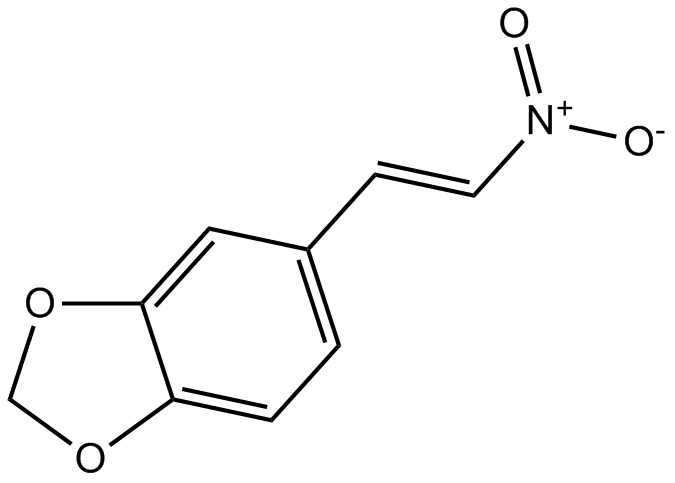
MNS
Inhibitor of Src/Syk tyrosine kinases
-
GC10046
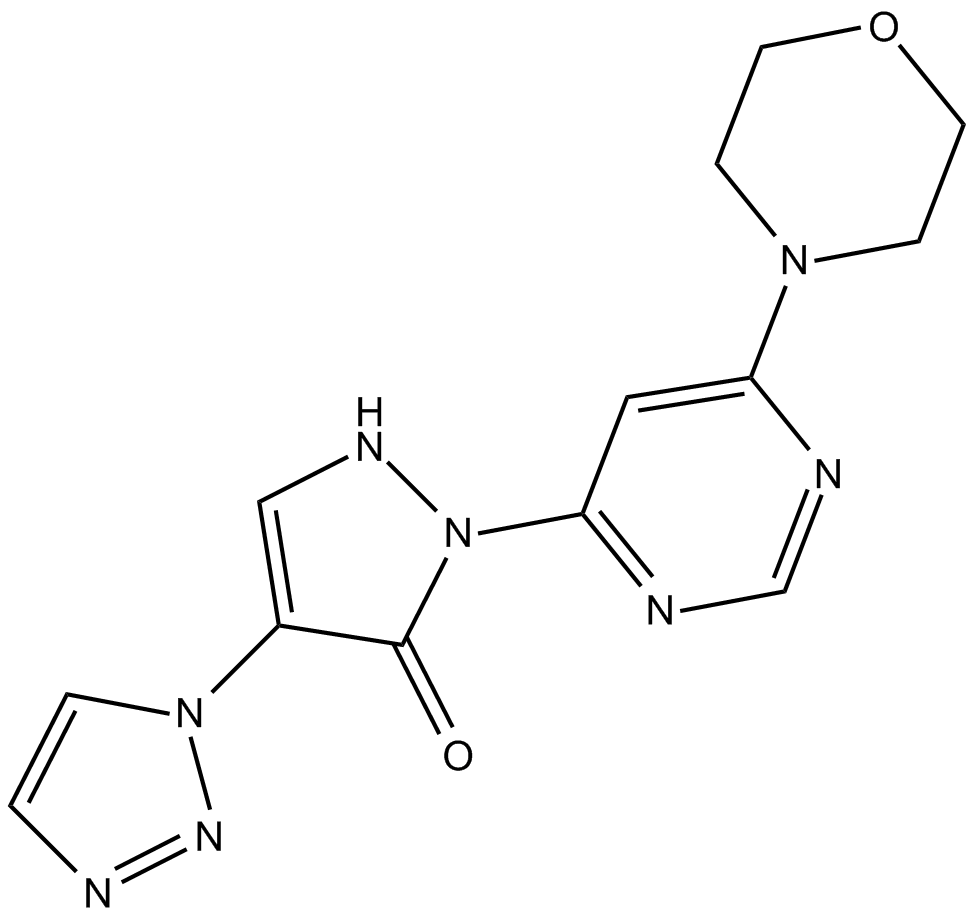
Molidustat (BAY85-3934)
Molidustat (BAY85-3934) (BAY 85-3934) is a novel inhibitor of hypoxia-inducible factor prolyl hydroxylase (HIF-PH) with mean IC50 values of 480 nM for PHD1, 280 nM for PHD2, and 450 nM for PHD3.
-
GC36661
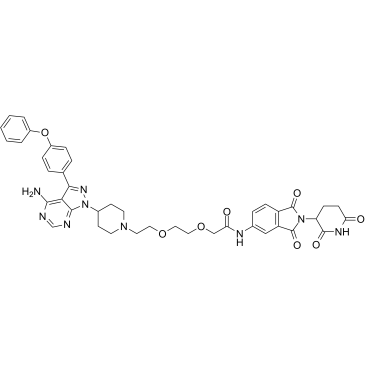
MT-802
MT-802 is a potent BTK degrader based on Cereblon ligand, with a DC50 of 1 nM.
-
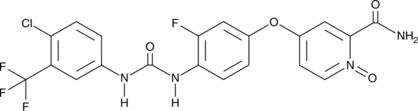
GC49686
N-desmethyl Regorafenib N-oxide
An active metabolite of regorafenib
-
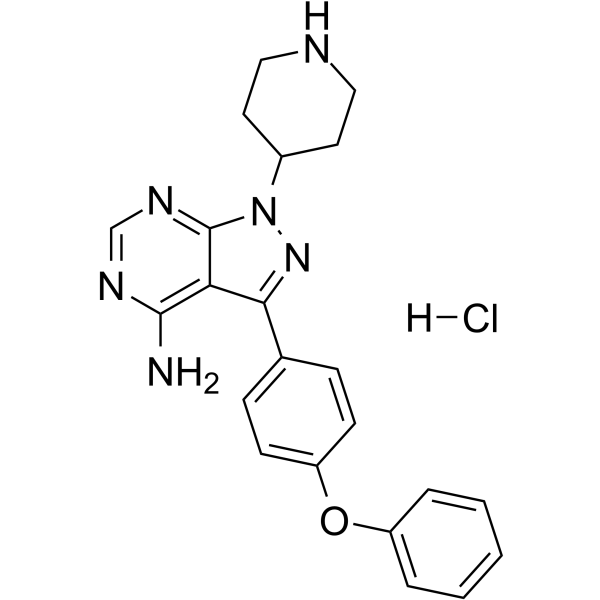
GC62255
N-piperidine Ibrutinib hydrochloride
N-piperidine Ibrutinib hydrochloride (Compound 1) is a reversible Ibrutinib derivative. N-piperidine Ibrutinib hydrochloride is a potent BTK inhibitor with IC50s of 51.0 and 30.7 nM for WT BTK and C481S BTK, respectively. N-piperidine Ibrutinib hydrochloride can be used as a BTK ligand in the synthesis of a series of PROTACs, such as SJF620. SJF620 is a potent PROTAC BTK degrader with a DC50 of 7.9 nM.
-
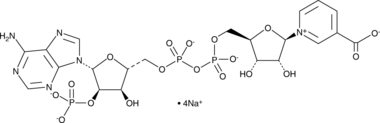
GC44290
NAADP (sodium salt)
Nicotinic acid adenine dinucleotide phosphate (NAADP) is a secondary messenger that induces calcium mobilization.
-
GC61109
Natalizumab
Natalizumab is a recombinant, humanized IgG4 monoclonal antibody, binds to α4β1-integrin and blocks its interaction with vascular cell adhesion molecule-1 (VCAM-1).

-
GC44465
NSC 12
NSC 12 is an extracellular trap for fibroblast growth factor 2 (FGF2) that binds FGF2 (Kd = 51 μM) and interferes with its interaction with FGFR1, without affecting the ability of FGF2 to bind heparin or heparin sulfate proteoglycans (HSPGs).
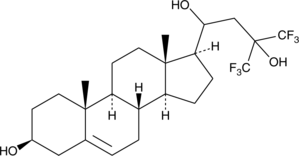
-
GC69600
NX-2127
NX-2127 is an orally effective BTK inhibitor that can induce the degradation of mutated BTKC481S in cells. NX-2127 inhibits the proliferation of TMD8 cells with the BTKC481S mutation more effectively than Ibrutinib. NX-2127 catalyzes the degradation of Ikaros (IKZF1) and Aiolos (IKZF3), corresponding to concentrations of 25 nM and 54 nM, respectively. NX-2127 stimulates T cell activation and increases IL-2 production in primary human T cells.
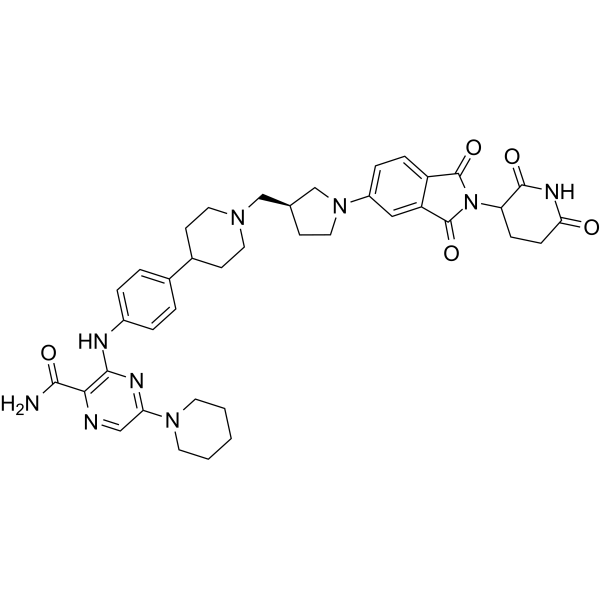
-
GC49349
O-Desethyl Sildenafil
A metabolite of sildenafil
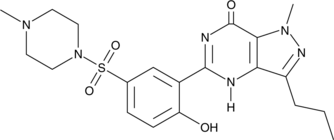
-
GC16107
Obtustatin
An integrin α1β1 inhibitor
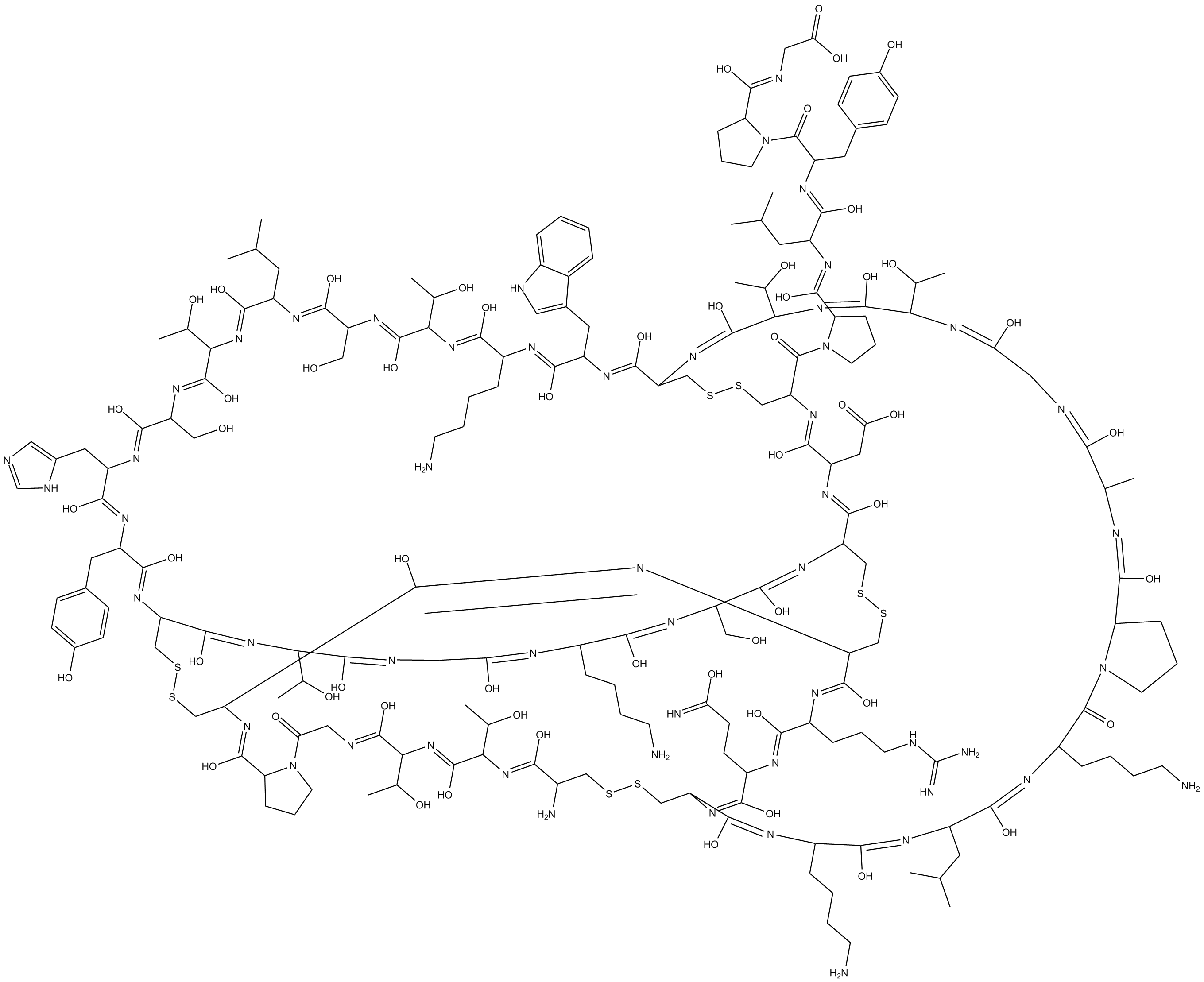
-
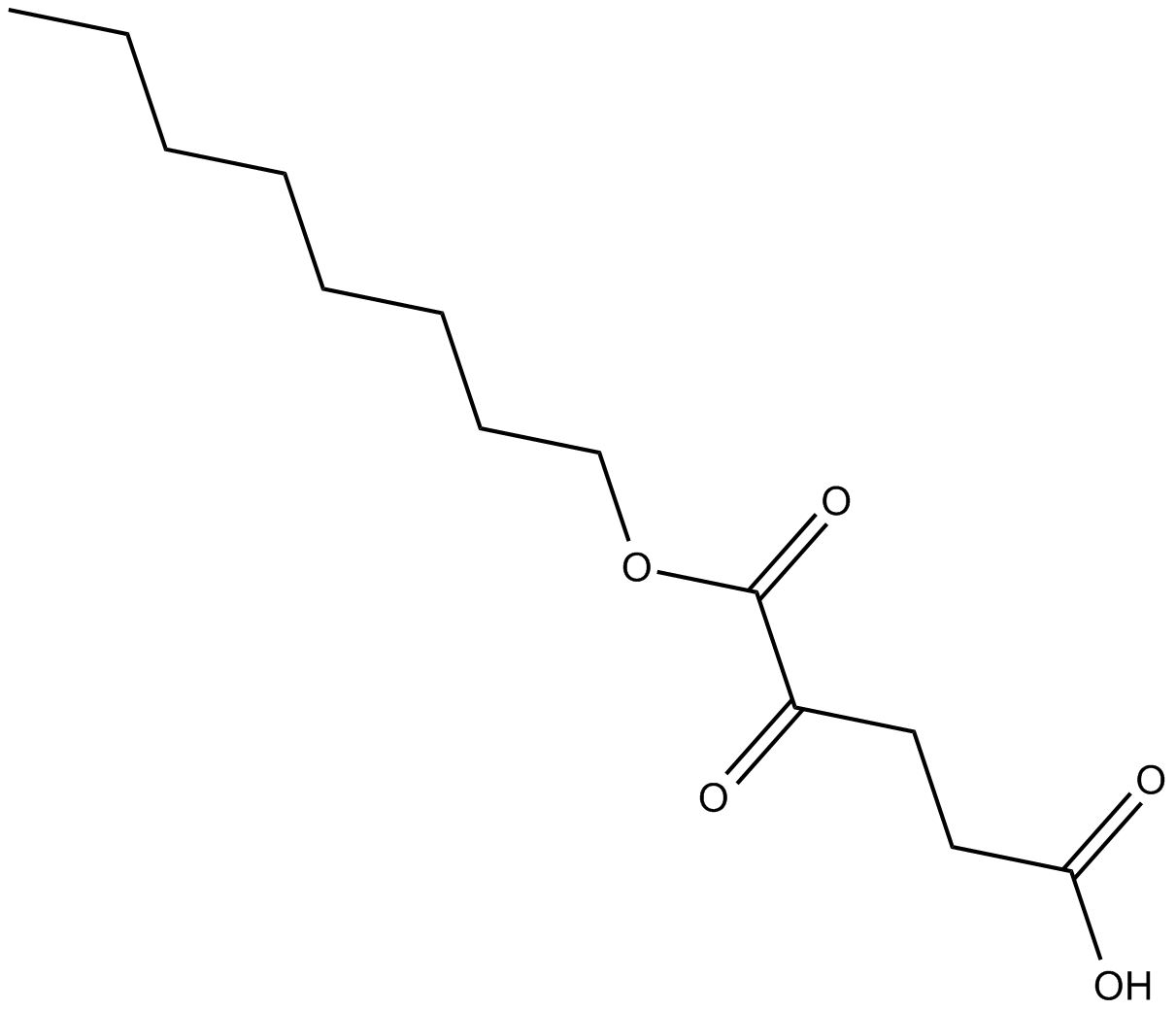
GC17358
Octyl-α-ketoglutarate
prolyl hydroxylases (PHD) activator
-
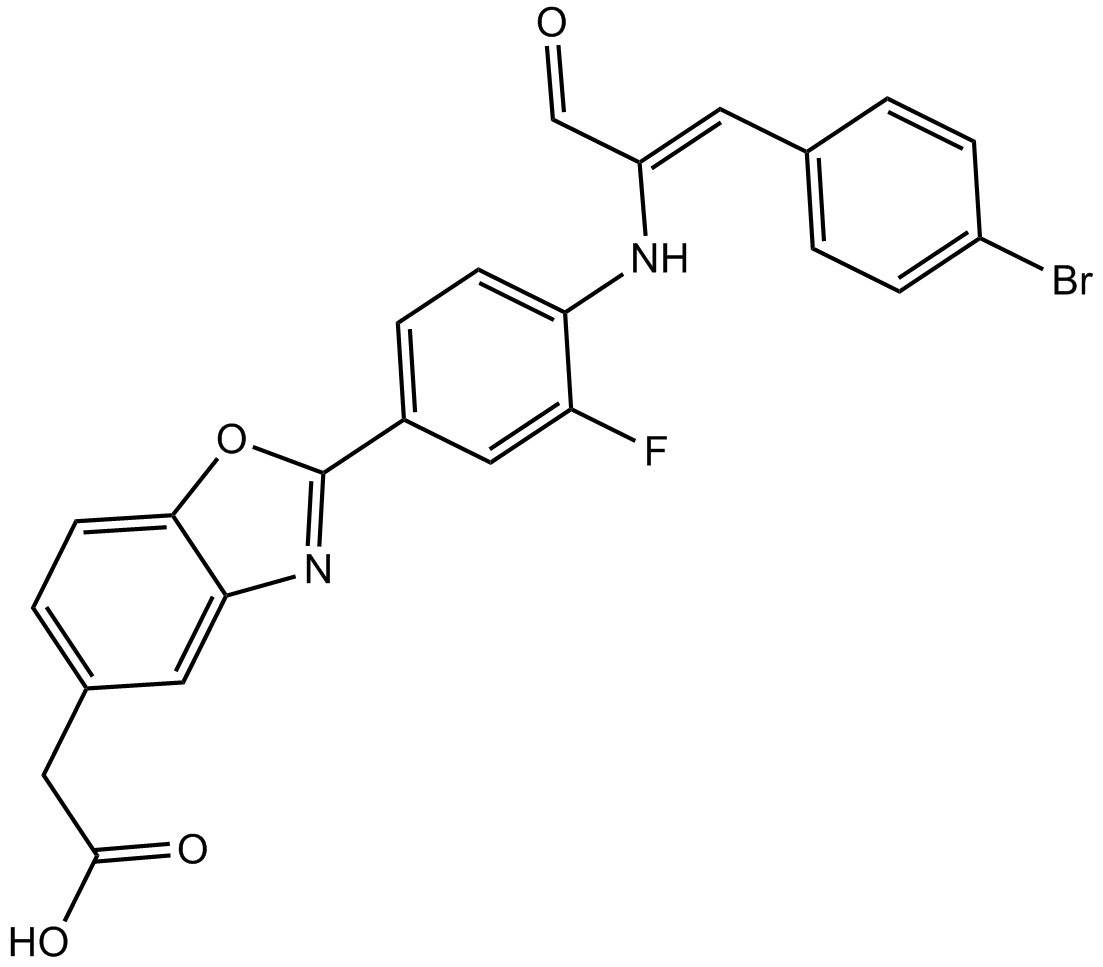
GC15199
OGT 2115
Heparanase inhibitor
-
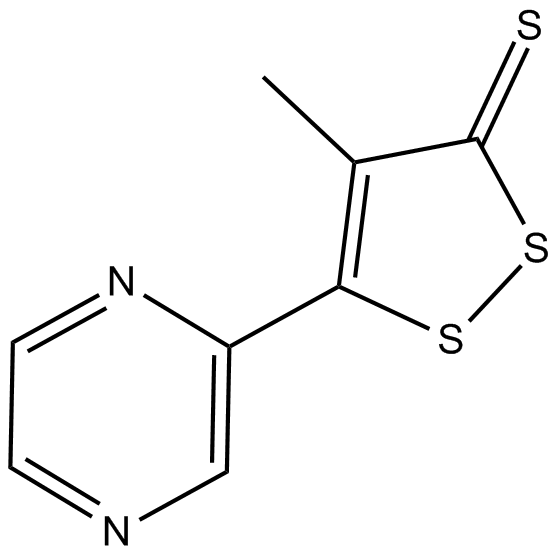
GC12821
Oltipraz
Nrf2 activator;An antischistosomal agent
-
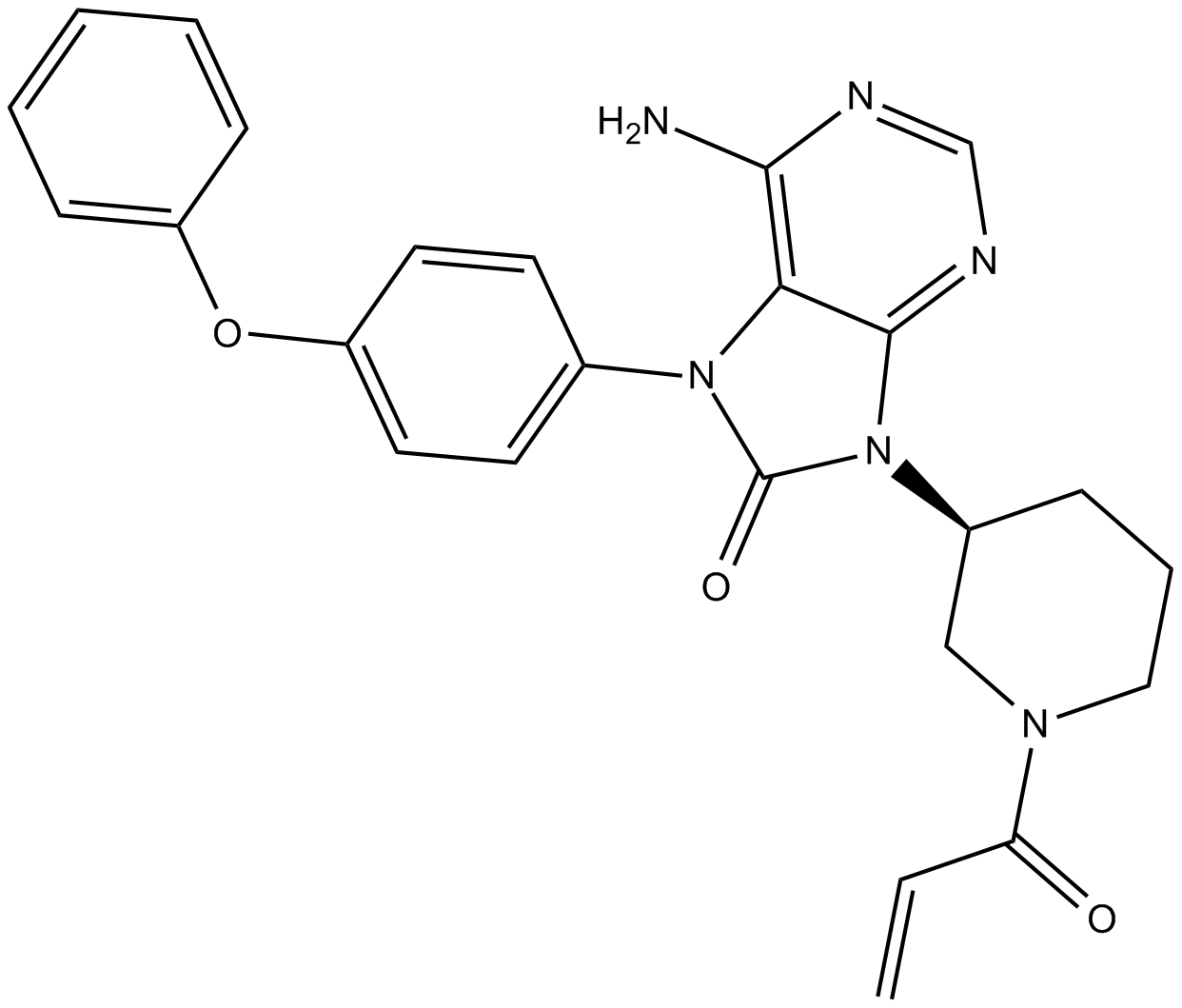
GC13219
ONO-4059
ONO-4059 is the analog of ONO-4059, ONO-4059 is a highly potent and selective Btk inhibitor.
-
GC69630
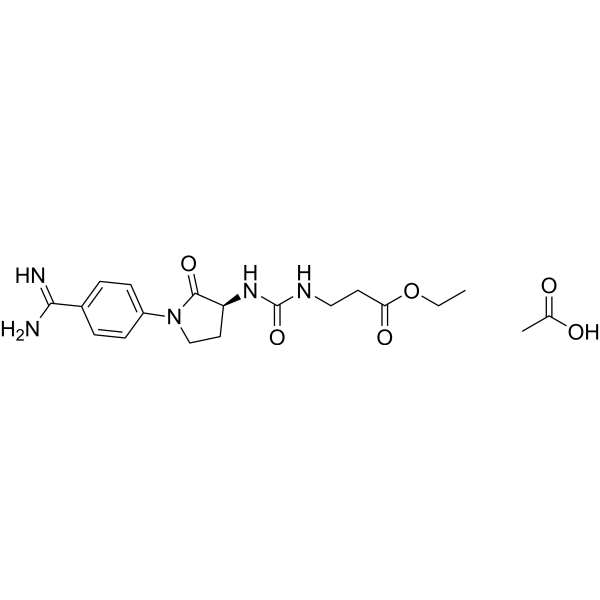
Orbofiban acetate
Orbofiban acetate is an orally active platelet membrane glycoprotein IIb/IIIa antagonist that inhibits platelet aggregation.
-
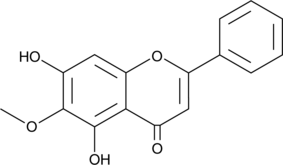
GC41625
Oroxylin A
Oroxylin A is a flavonoid that has been found in S.
-
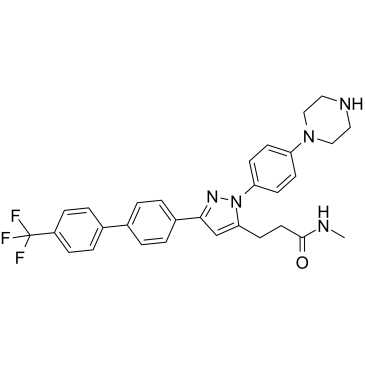
GC36821
OSU-T315
OSU-T315 (ILK-IN-1) is a small Integrin-linked kinase (ILK) inhibitor with an IC50 of 0.6 μM, inhibiting PI3K/AKT signaling by dephosphorylation of AKT-Ser473 and other ILK targets (GSK-3β and myosin light chain). OSU-T315 abrogates AKT activation by impeding AKT localization in lipid rafts and triggers caspase-dependent apoptosis in an ILK-independent manner. OSU-T315 causes cell death through apoptosis and autophagy.
-
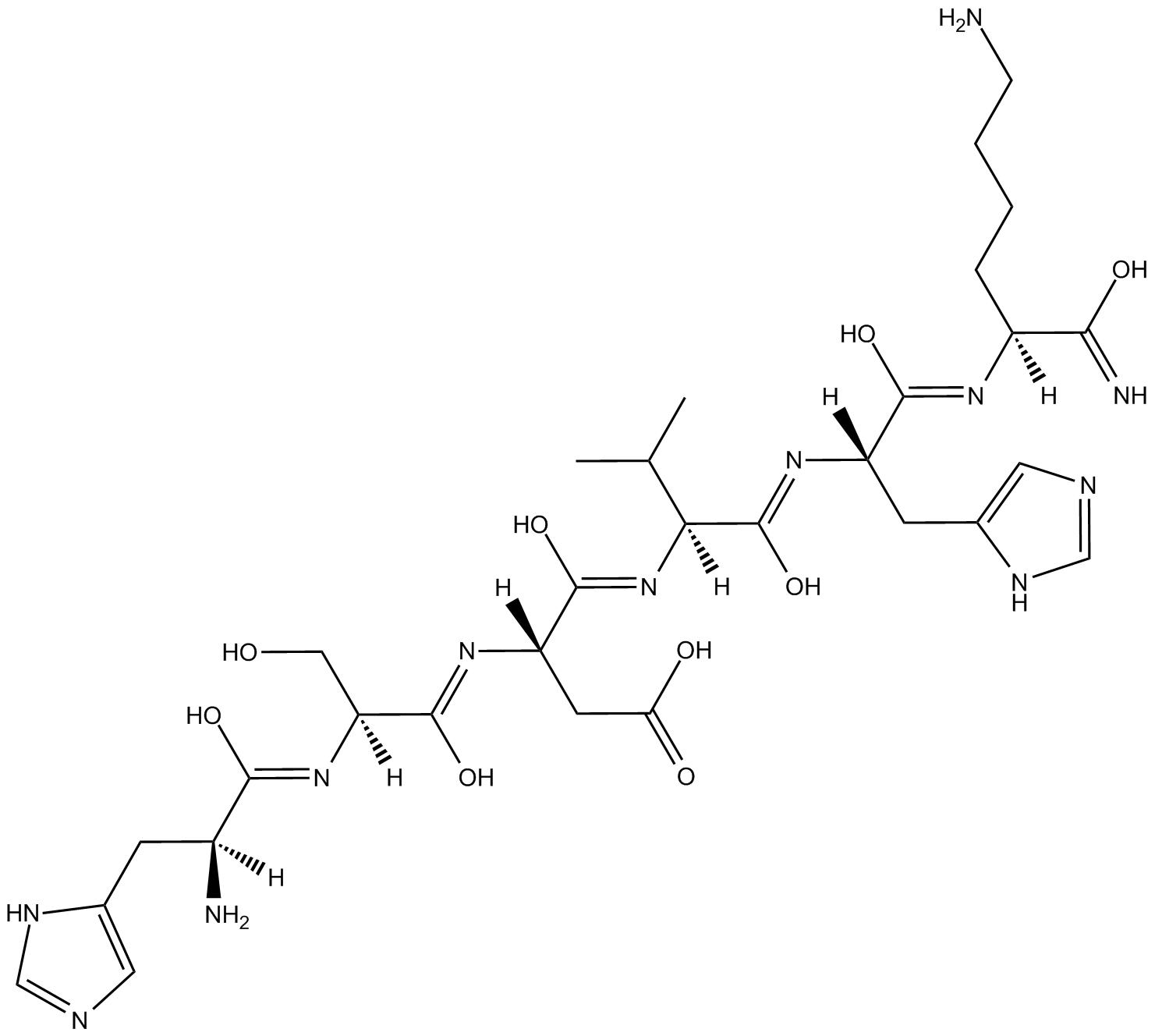
GC11753
P11
P11 is an antagonist of the integrin αvβ3-vitronectin interaction, with an IC50 of 1.74 pg/mL (2.414 pM).
-
GC11951
PAR 4 (1-6)
PAR4 agonist
-
GC62101
PAR-2-IN-1
PAR-2-IN-1 is a protease-activated receptor-2 (PAR2) signaling pathway inhibitor with anti-inflammatory and anticancer effects.
-
GA23350
PAR-3 (1-6) amide (human)
PAR-3 (1-6) amide (human) is a proteinase-activated receptor (PAR-3) agonist peptide.

-
GC33599
PAR-4 Agonist Peptide, amide TFA (PAR-4-AP (TFA))
PAR-4 Agonist Peptide, amide TFA (PAR-4-AP (TFA)) (PAR-4-AP TFA; AY-NH2 TFA) is a proteinase-activated receptor-4 (PAR-4) agonist, which has no effect on either PAR-1 or PAR-2 and whose effects are blocked by a PAR-4 antagonist.
-
GC15817
PAR-4 Agonist Peptide, amide (AY-NH2)
PAR-4 Agonist Peptide, amide(AY-NH2) (PAR-4-AP; AY-NH2) is a proteinase-activated receptor-4 (PAR-4) agonist, which has no effect on either PAR-1 or PAR-2 and whose effects are blocked by a PAR-4 antagonist.
-
GC38134
Parmodulin 2
Reversible inhibitor of PAR1mediated platelet activation
-
GC69663
Parstatin(mouse) TFA
Parstatin (mouse) TFA is a peptide agonist of the PAR-1 thrombin receptor with cell-penetrating properties and an effective inhibitor of angiogenesis.

-
GC40085
Pazopanib-d6
Pazopanib-d6 is intended for use as an internal standard for the quantification of pazopanib by GC- or LC-MS.
-
GC36861
PCI 29732
A multi-kinase inhibitor