PI3K/Akt/mTOR Signaling
- Akt(129)
- AMPK(75)
- CK2(11)
- DNA-PK(25)
- GSK-3(76)
- MELK(7)
- mTOR(118)
- PI3K(227)
- PI4K(15)
- PDK-1(18)
- PIKfyve(4)
- PKB(1)
- S6 Kinase(12)
Products for PI3K/Akt/mTOR Signaling
- Cat.No. Product Name Information
-
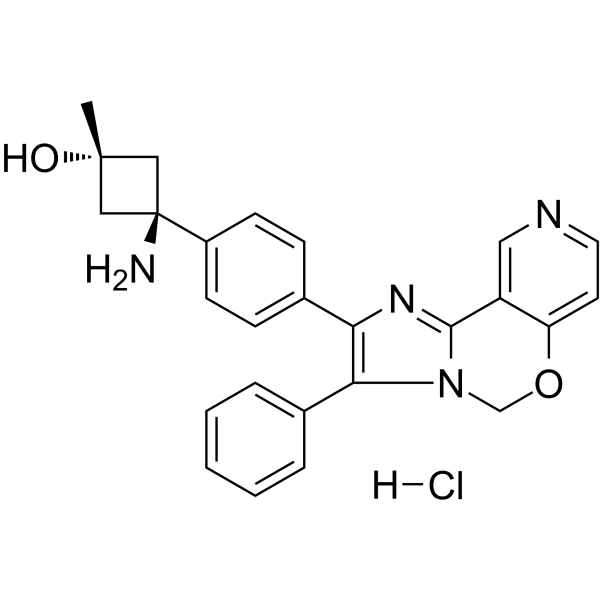
GC62630
TAS-117 hydrochloride
TAS-117 hydrochloride is a potent, selective, orally active allosteric Akt inhibitor (with IC50s of 4.8, 1.6, and 44 nM for Akt1, 2, and 3, respectively). TAS-117 hydrochloride triggers anti-myeloma activities and enhances fatal endoplasmic reticulum (ER) stress induced by proteasome inhibition. TAS-117 hydrochloride induces apoptosis and autophagy.
-
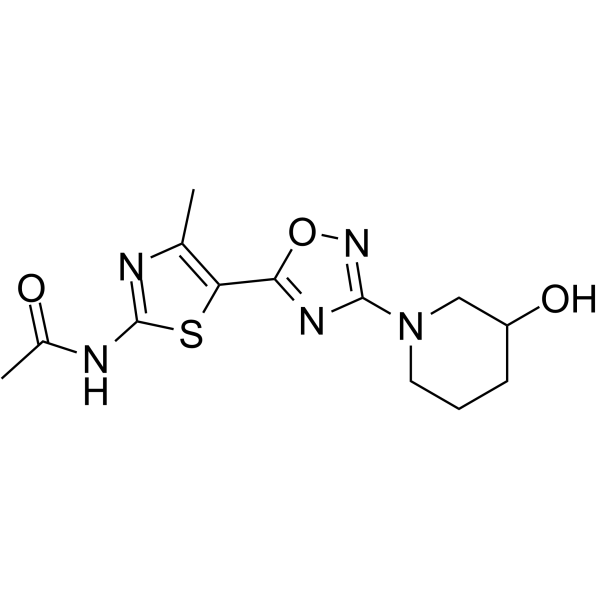
GC62342
TASP0415914
TASP0415914 is a potent and orally active PI3Kγ inhibitor with an IC50 of 29 nM.
-
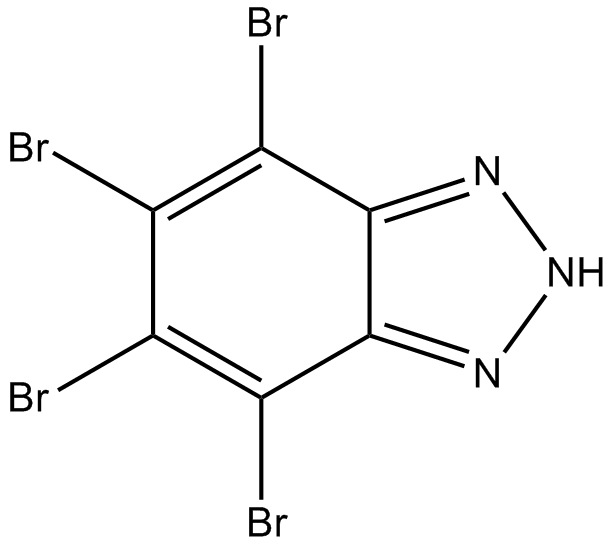
GC12181
TBB
An inhibitor of CK2
-
GC14181
TBCA
CK2 inhibitor
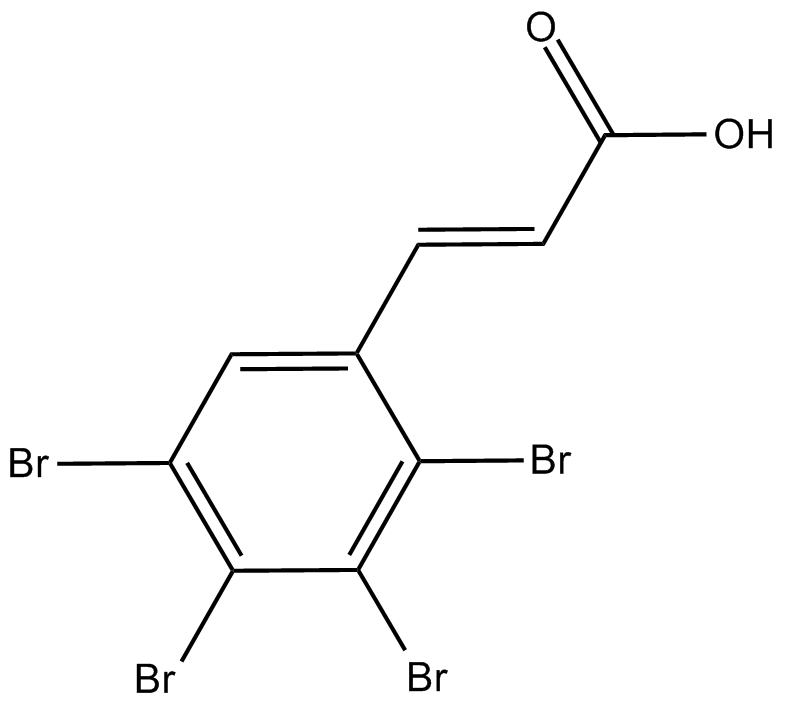
-
GC50332
TC KHNS 11
Potent and selective PI 3-kinase δ inhibitor; orally bioavailable
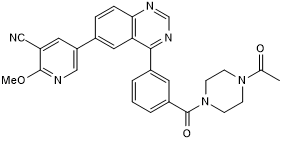
-
GC13576
TC-G 24
GSK-3β inhibitor
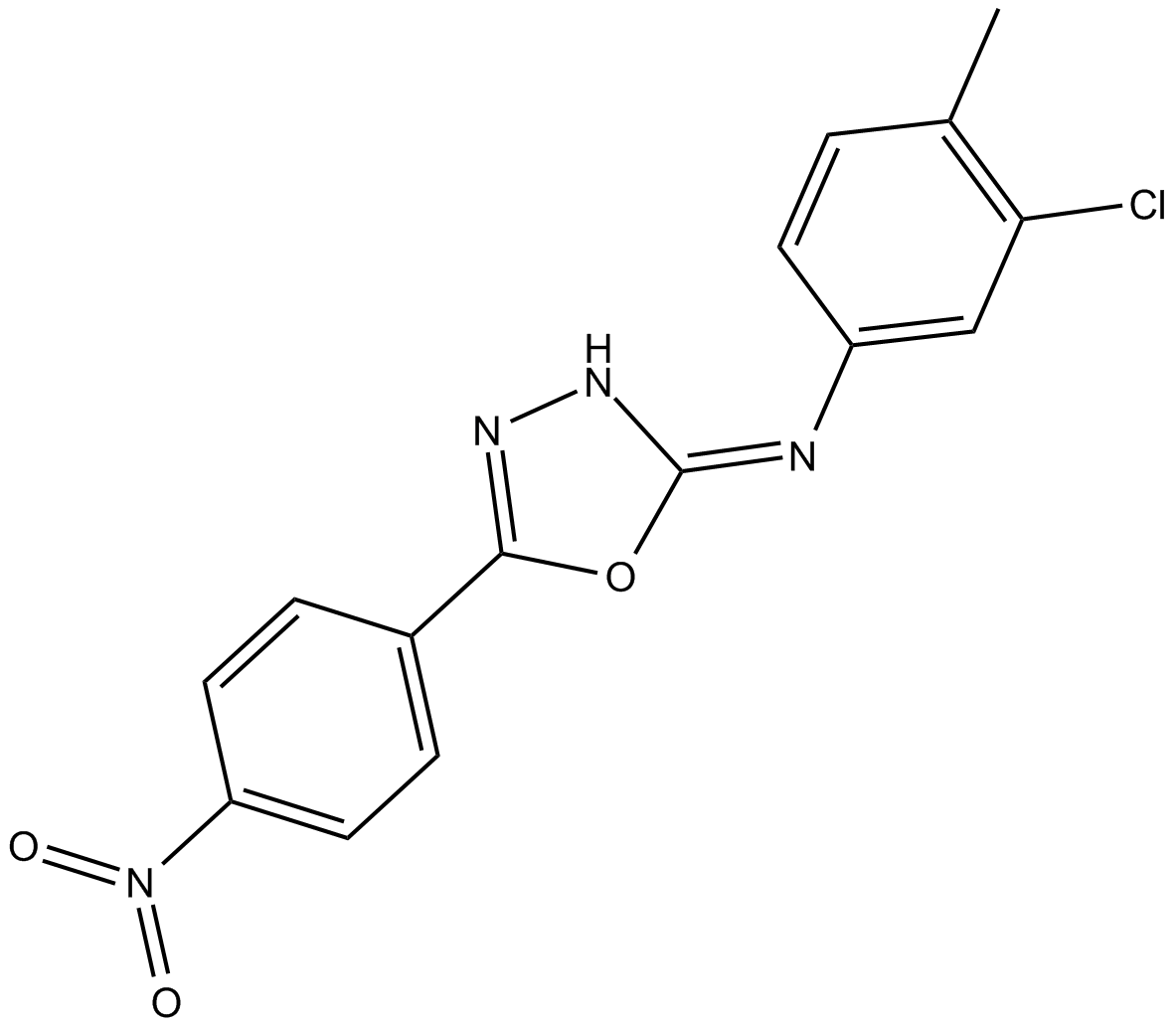
-
GC10440
TCS 183
competitive inhibitor of GSK-3β (Ser9) phosphorylation
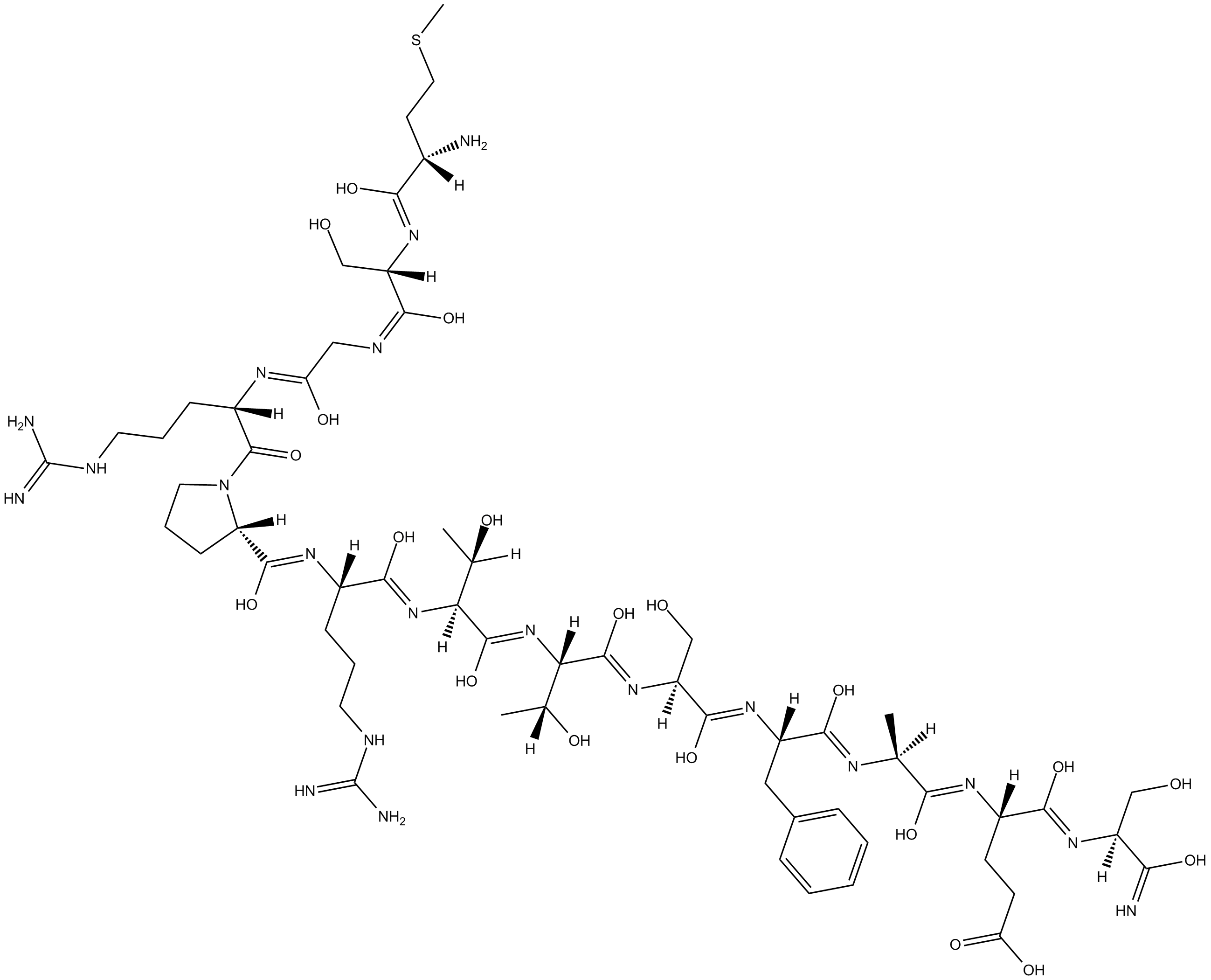
-
GC11515
TCS 184
Scrambled control peptide for use with TCS 183
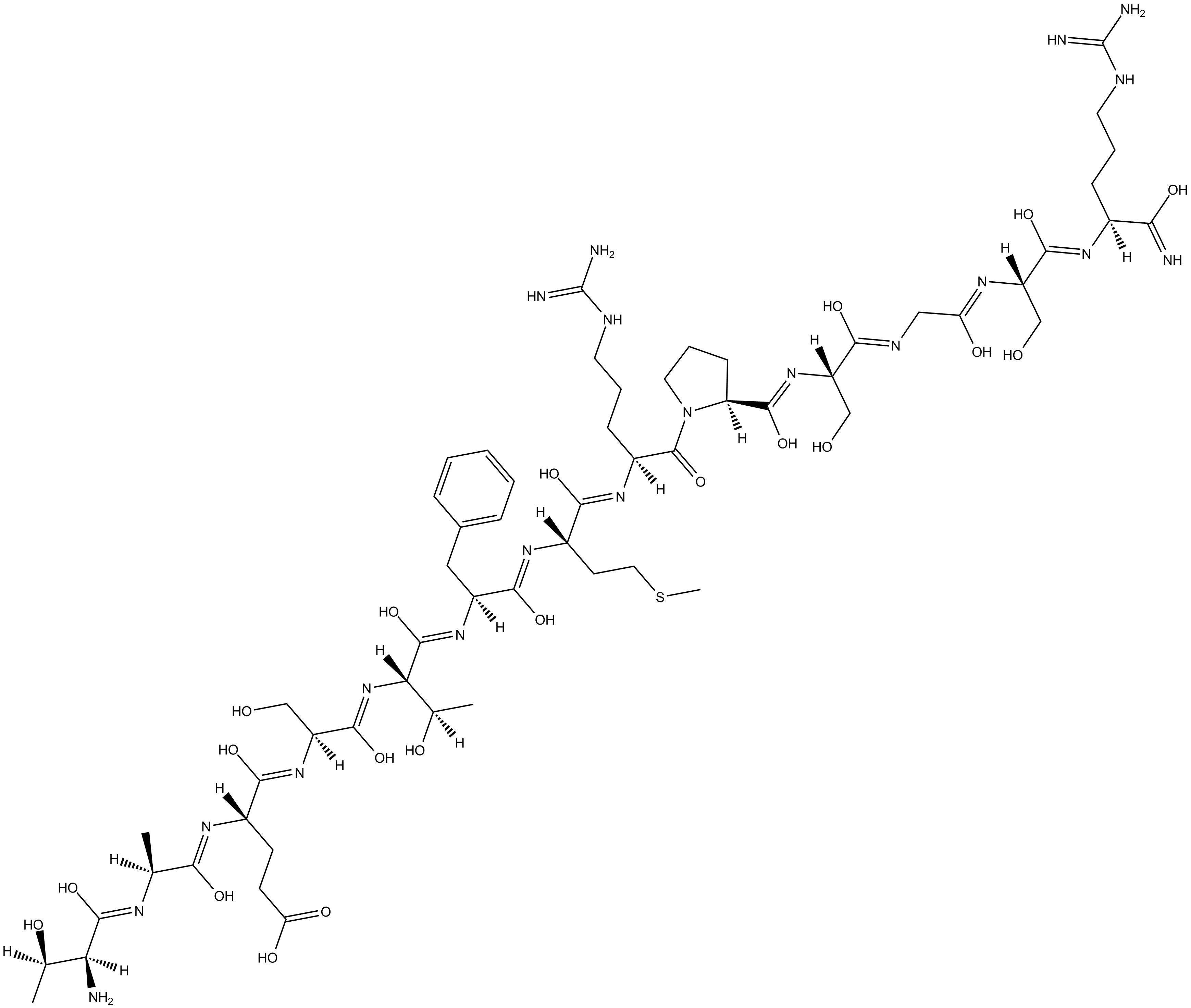
-
GC11627
TCS 2002
TCS 2002 (Compound 9b) is a highly selective, orally bioavailable and potent GSK-3β inhibitor with the IC50 of 35 nM.
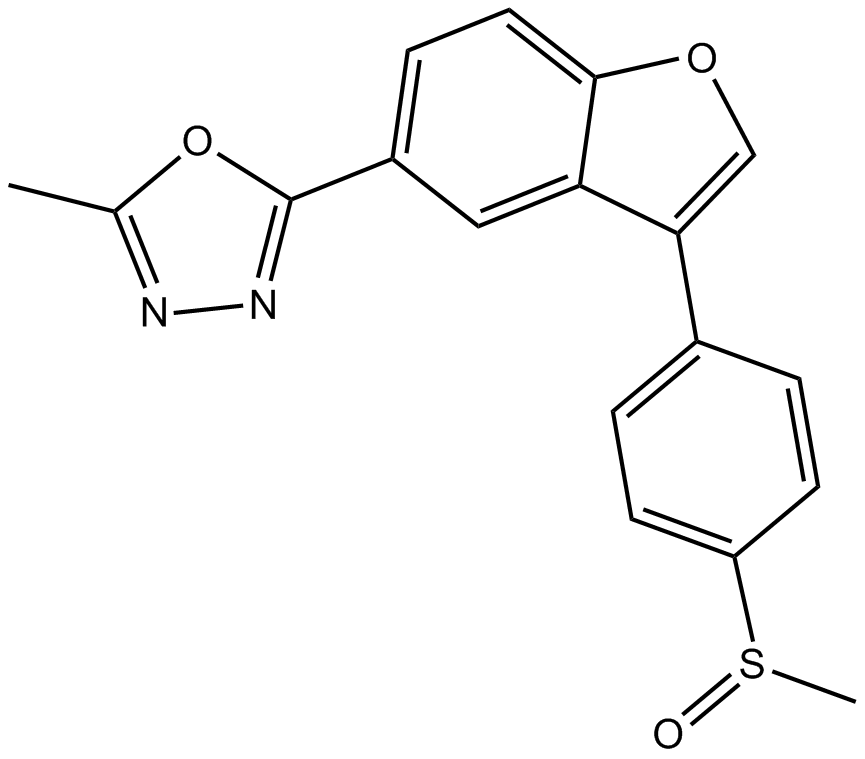
-
GC16584
TCS 21311
A JAK3 inhibitor
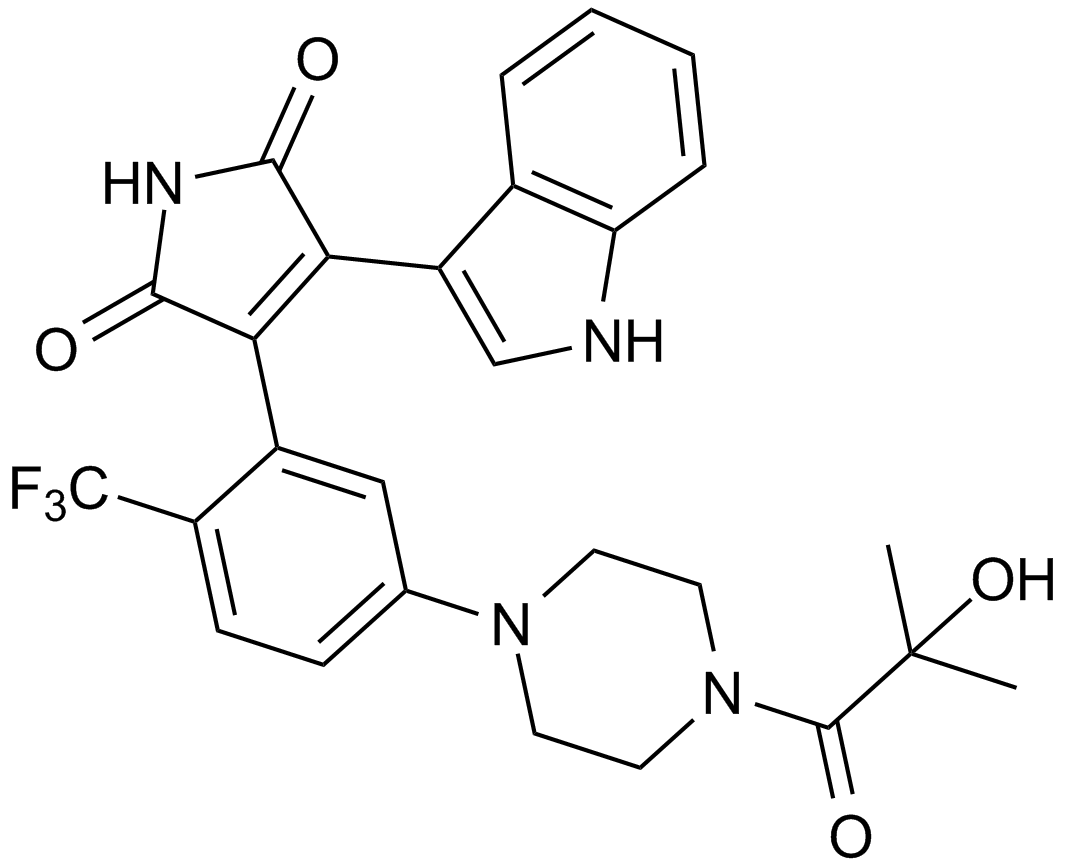
-
GC62191
TD52
TD52, an Erlotinib derivative, is an orally active, potent cancerous inhibitor of protein phosphatase 2A (CIP2A) inhibitor. TD52 mediates the apoptotic effect in triple-negative breast cancer (TNBC) cells via regulating the CIP2A/PP2A/p-Akt signalling pathway. TD52 indirectly reduced CIP2A by disturbing Elk1 binding to the CIP2A promoter. TD52 has less p-EGFR inhibition and has potent anti-cancer activity.
-
GC64936
TD52 dihydrochloride
TD52 dihydrochloride, an Erlotinib derivative, is an orally active, potent cancerous inhibitor of protein phosphatase 2A (CIP2A) inhibitor. TD52 dihydrochloride mediates the apoptotic effect in triple-negative breast cancer (TNBC) cells via regulating the CIP2A/PP2A/p-Akt signalling pathway. TD52 dihydrochloride indirectly reduced CIP2A by disturbing Elk1 binding to the CIP2A promoter. TD52 dihydrochloride has less p-EGFR inhibition and has potent anti-cancer activity.
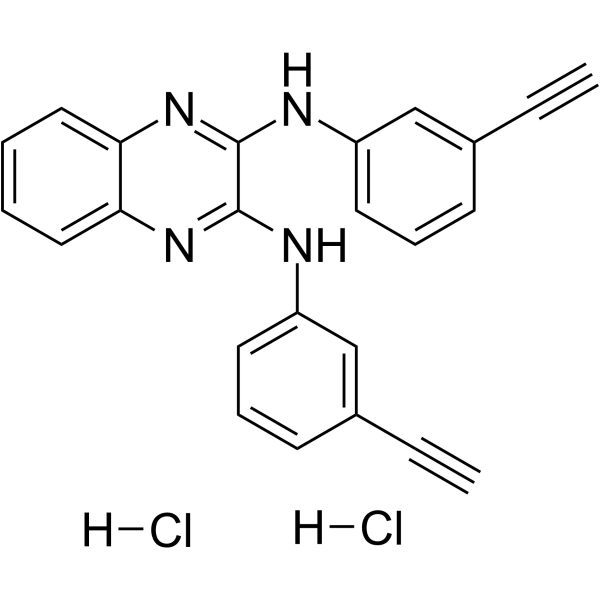
-
GC14840
TDZD-8
GSK-3β inhibitor
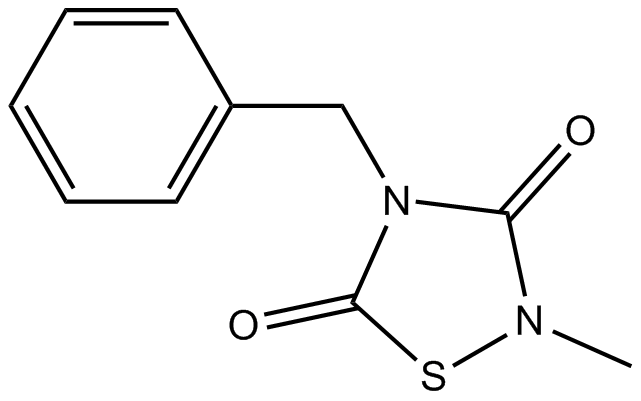
-
GC12573
Temsirolimus
A MTOR inhibitor
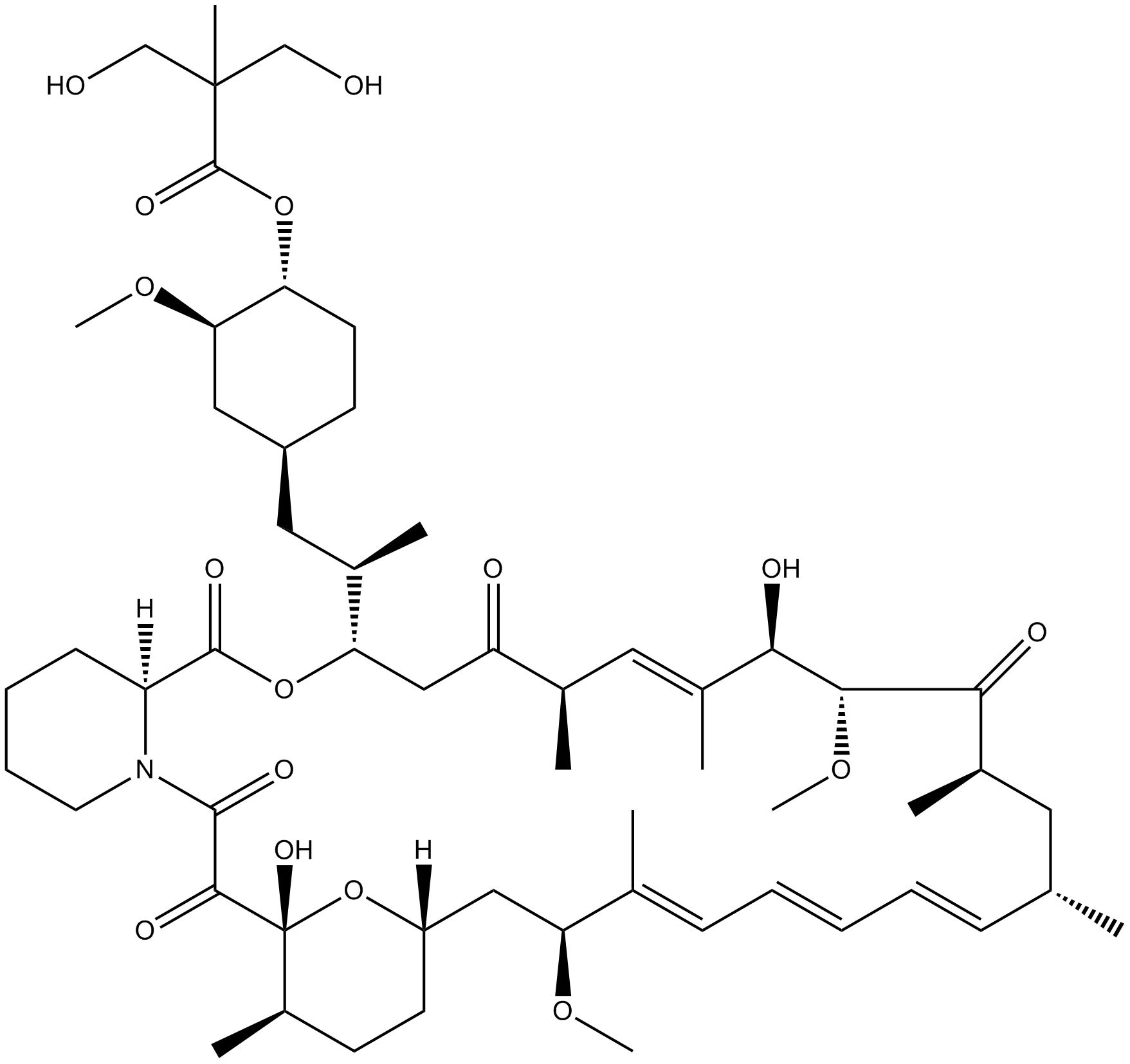
-
GC32828
Tenalisib (RP6530)
Tenalisib (RP6530) (RP6530) is a novel, potent, and selective PI3Kδ and PI3Kγ inhibitor with IC50 values of 25 and 33 nM, respectively.
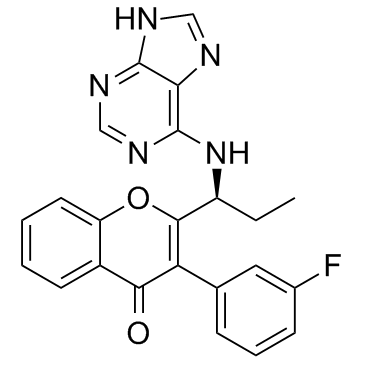
-
GC15151
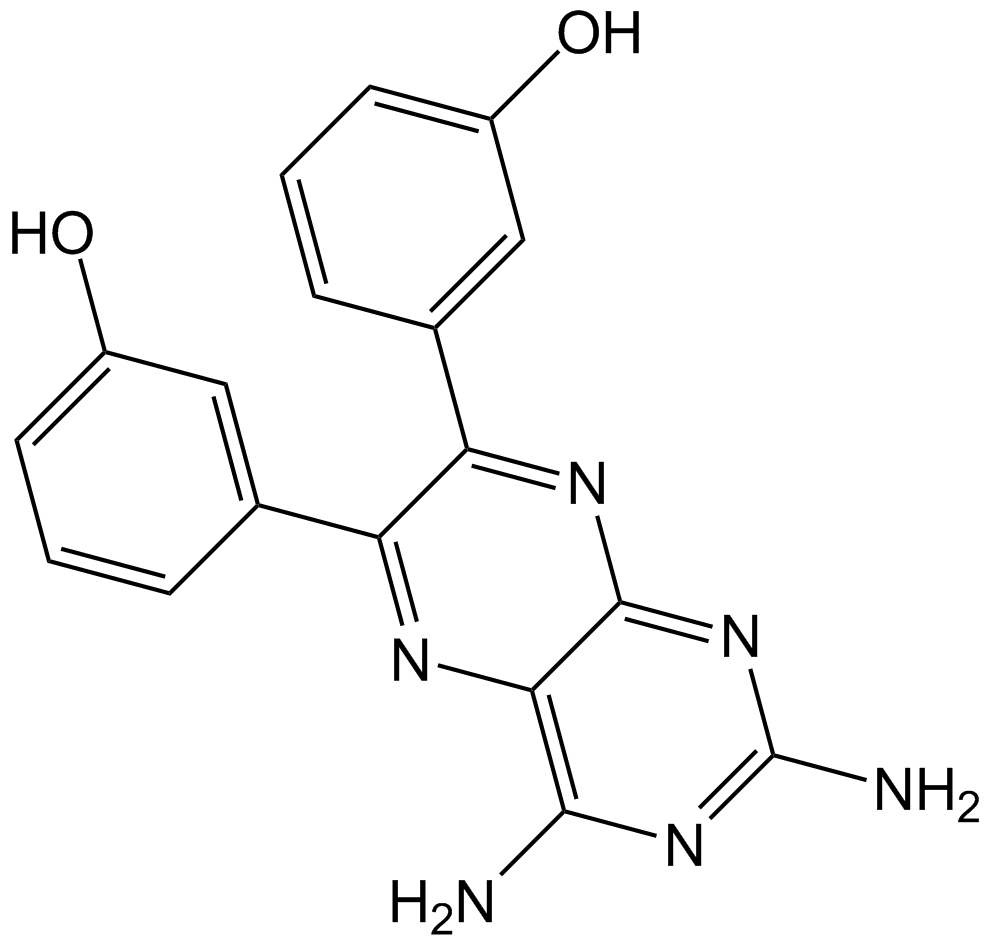
TG100-115
PI3Kγ/PI3Kδ inhibitor
-
GC16231
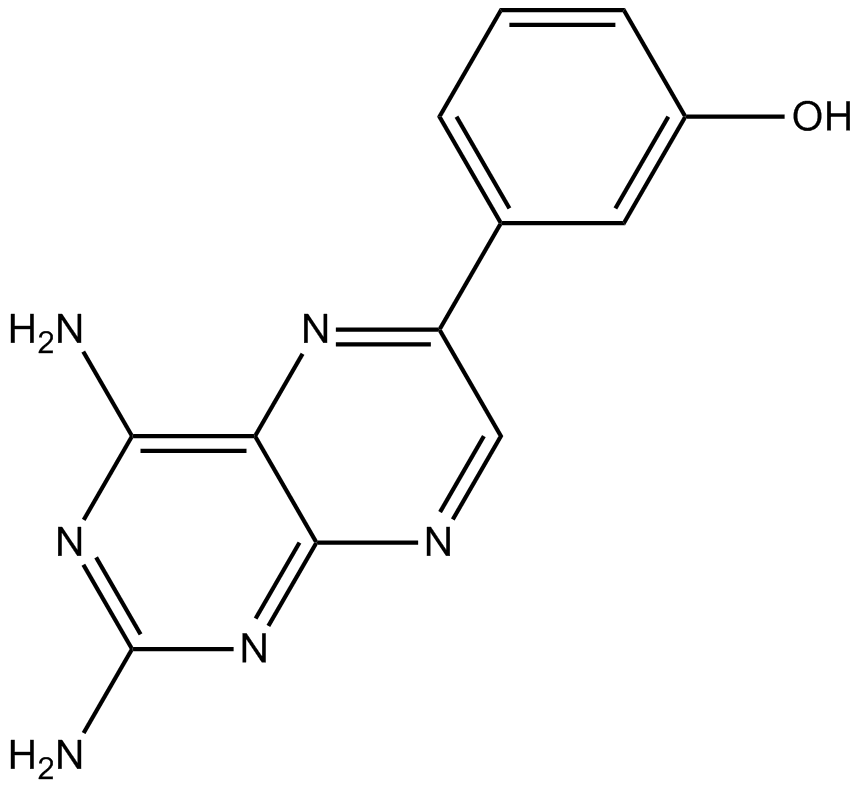
TG100713
pan-PI3K inhibitor
-
GC41573
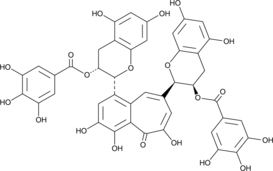
Theaflavin 3,3'-digallate
Theaflavin-3,3'-digallate (TFDG) is a major polyphenol found in black tea with diverse biological activities.
-
GC14465
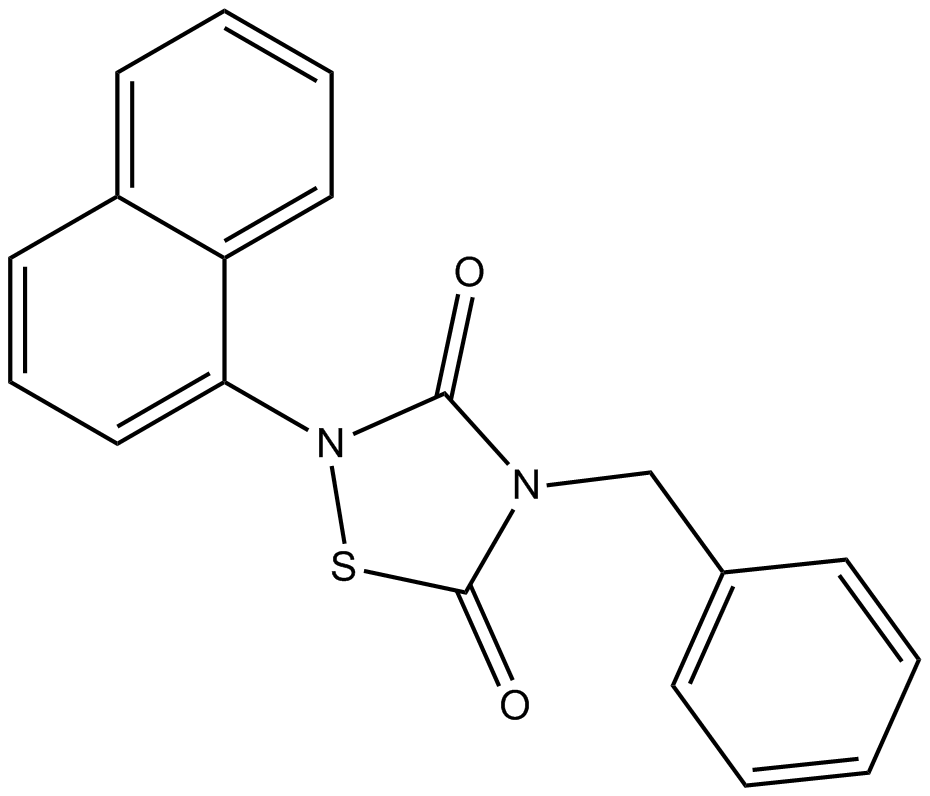
Tideglusib
non-ATP-competitive GSK-3β inhibitor
-
GC61336
TMBIM6 antagonist-1
TMBIM6 antagonist-1, a potential TMBIM6 antagonist, prevents TMBIM6 binding to mTORC2, decreases mTORC2 activity, and also regulates TMBIM6-leaky Ca2+.
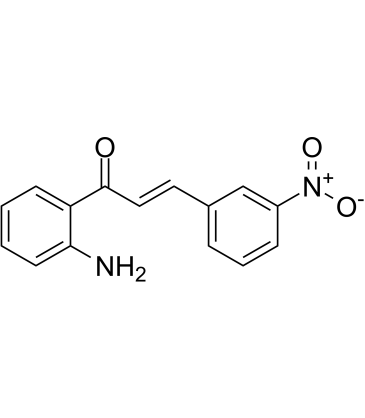
-
GC10131
Torin 1
MTOR inhibitor,potent and selective
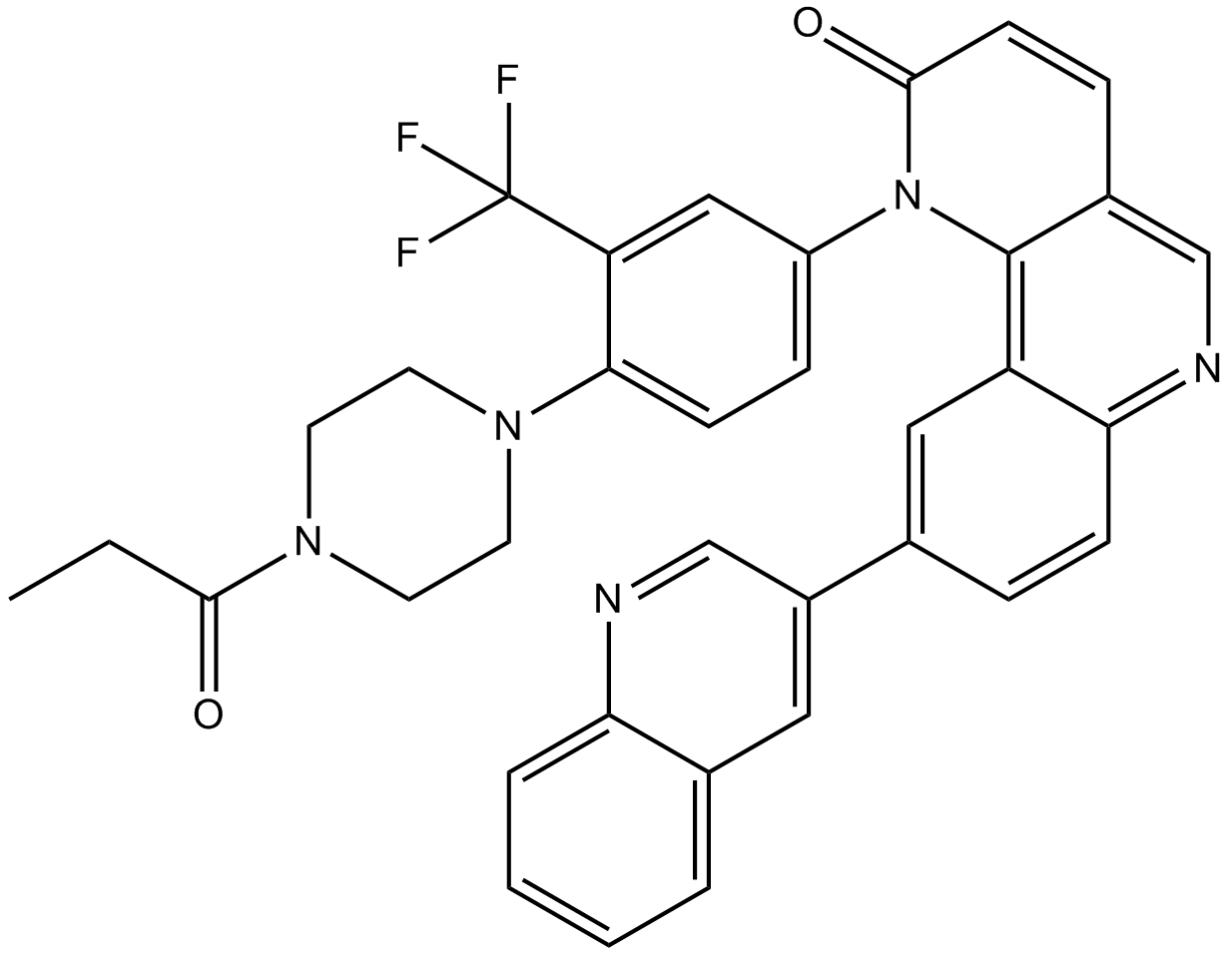
-
GC13858
Torin 2
MTOR inhibitor,highly potent and selective
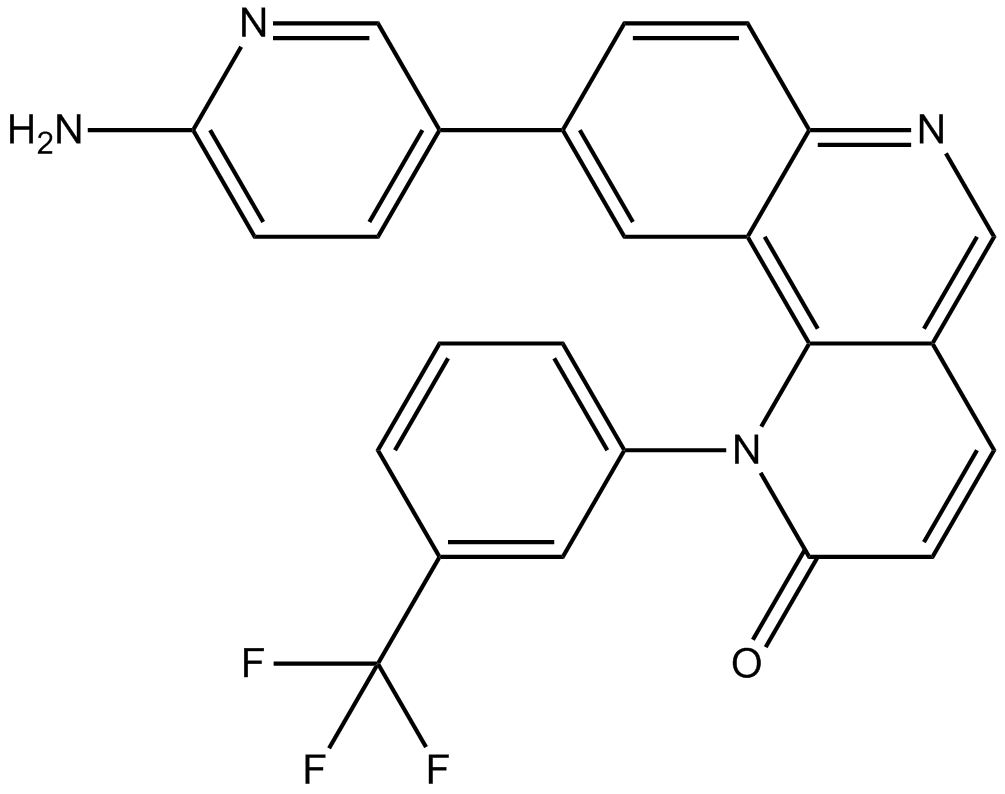
-
GC15392
Triciribine
Akt inhibitor,highly selective
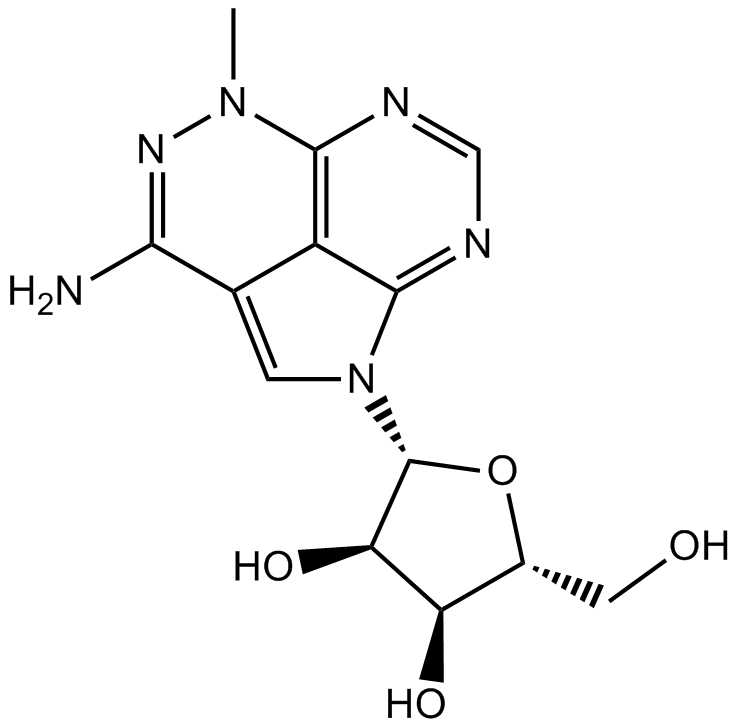
-
GC10649
TTP 22
A CK2 inhibitor
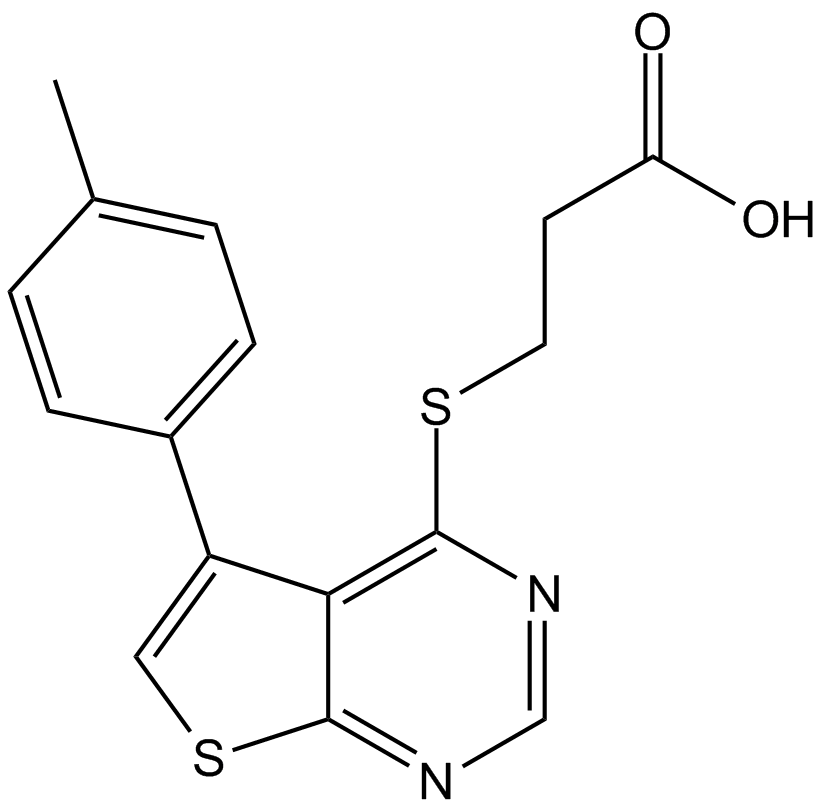
-
GC17868
TWS119
GSK-3β inhibitor
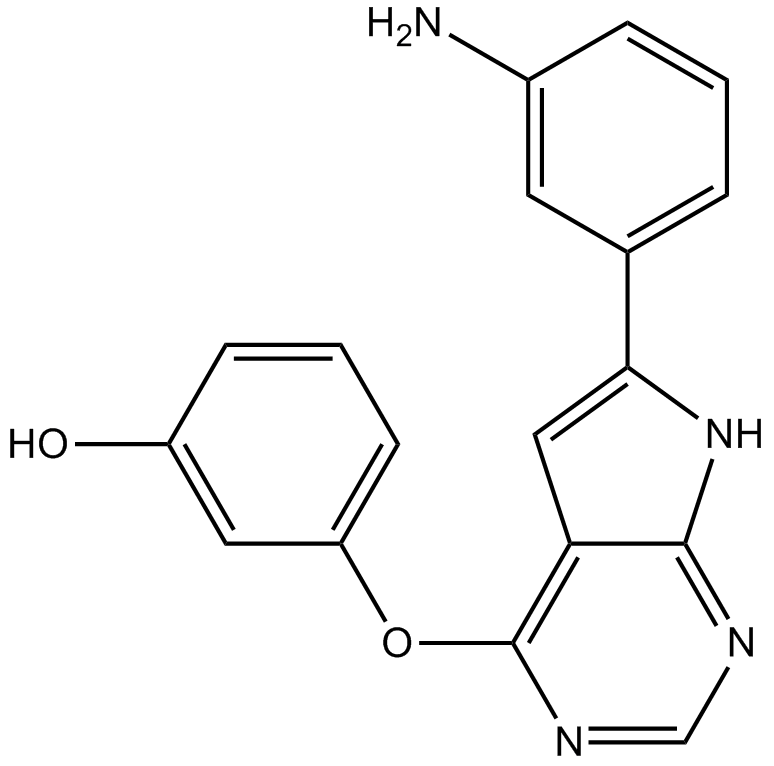
-
GC18164
UCB9608
PI4KIIIβ inhibitor
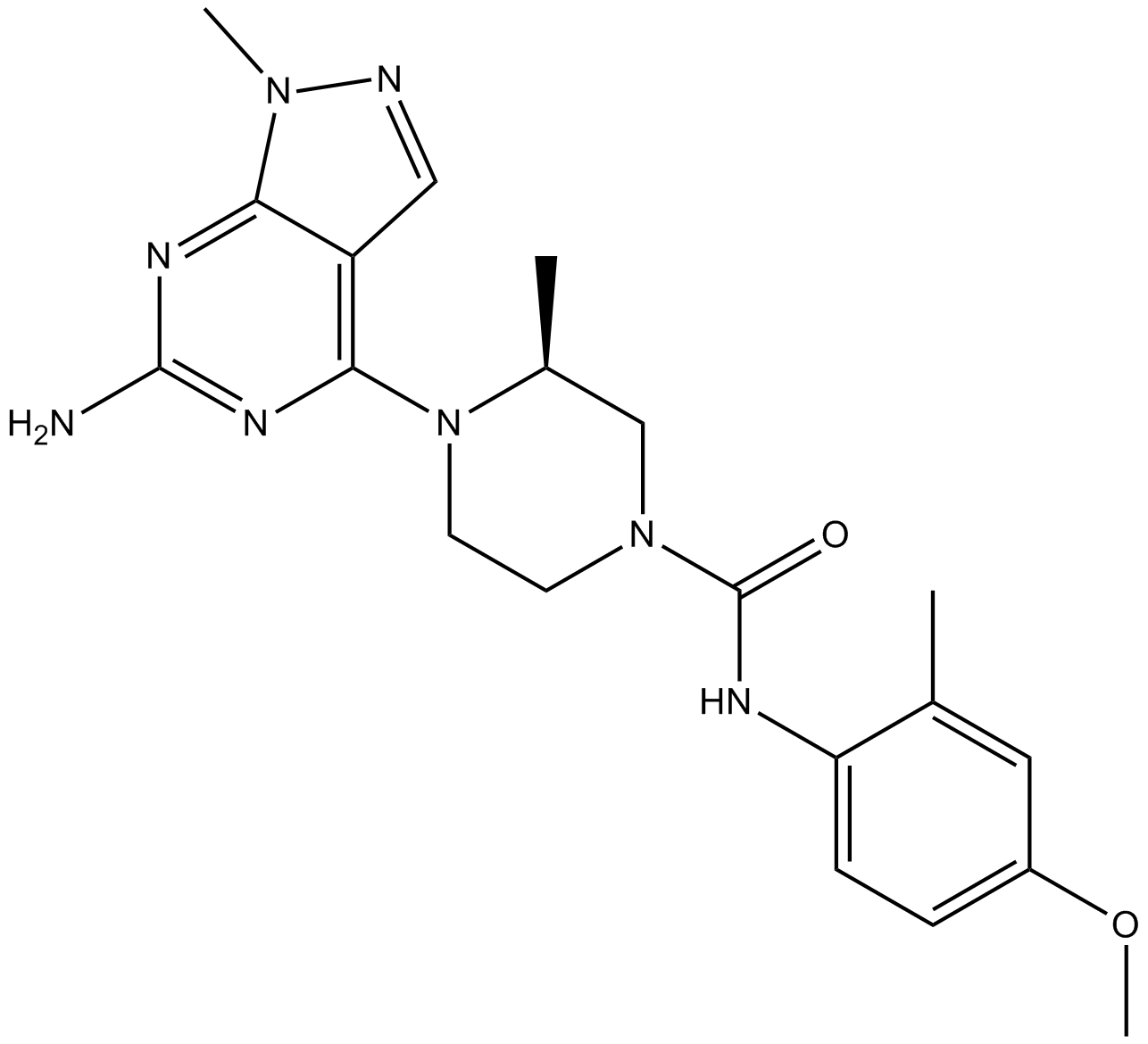
-
GC10857
UCN-01
inhibitor of Akt, protein kinase C, PDK1 and cyclin-dependent kinases
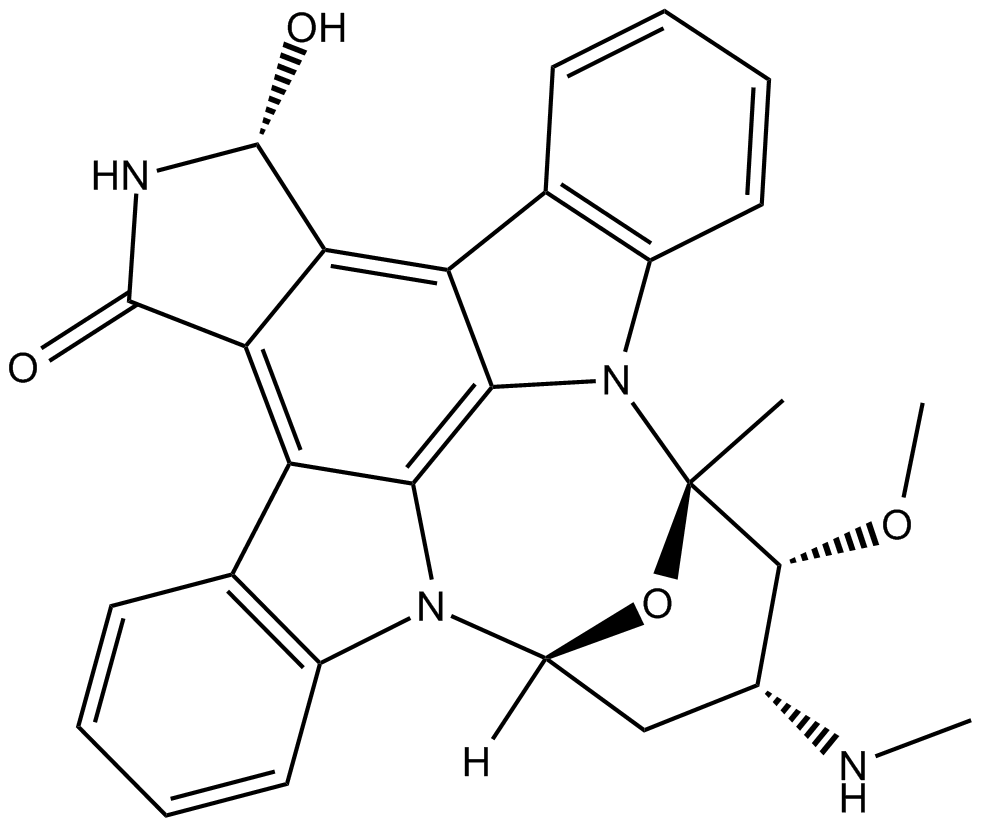
-
GC65454
UCT943
UCT943 is a next-generation Plasmodium falciparum PI4K inhibitor.
-
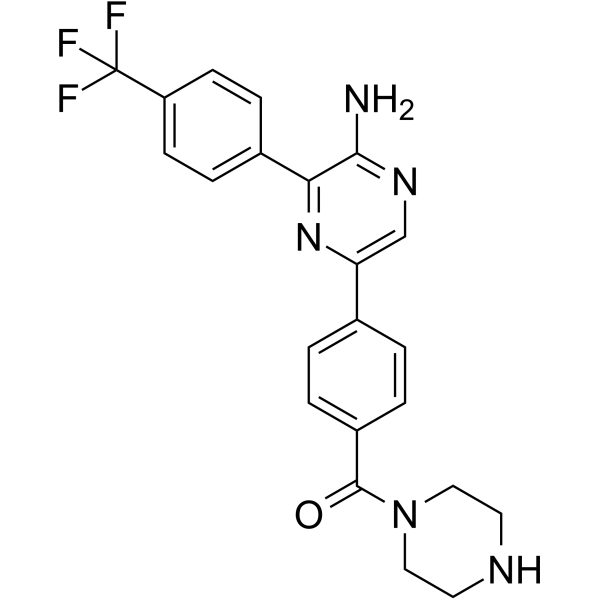
GC19355
Umbralisib
TGR-1202 is a PI3Kδ inhibitor, with IC50 and EC50 of 22.2 nM and 24.3 nM, respectively.
-
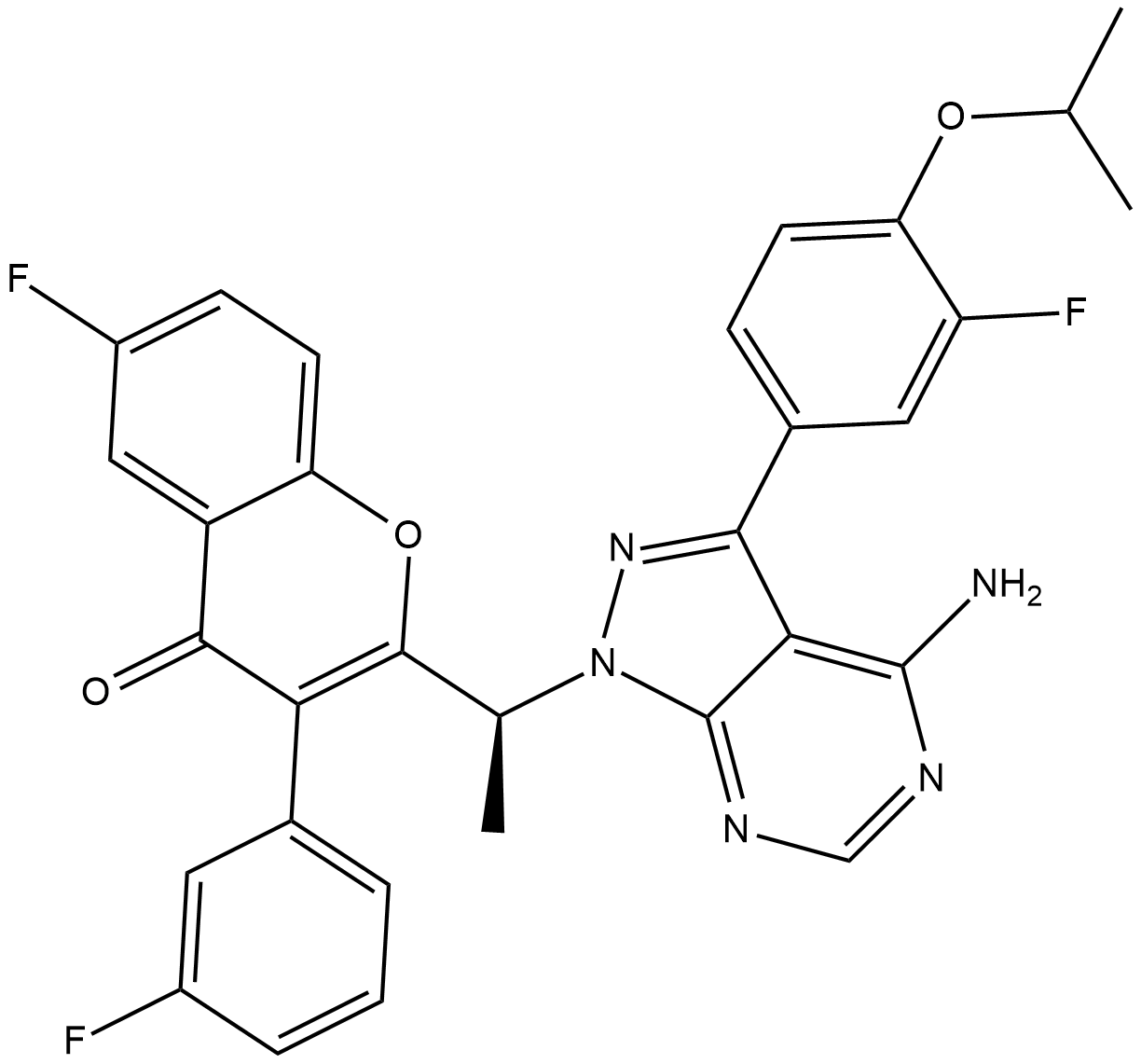
GC37854
Umbralisib hydrochloride
Umbralisib (TGR-1202) hydrochloride is an orally active, potent and selective dual PI3Kδ and casein kinase-1-ε (CK1ε) inhibitor, with EC50 of 22.2 nM and 6.0 μM, respectively. Umbralisib hydrochloride exhibits unique immunomodulatory effects on chronic lymphocytic leukemia (CLL) T cells. Umbralisib hydrochloride can be used for haematological malignancies reseach.
-
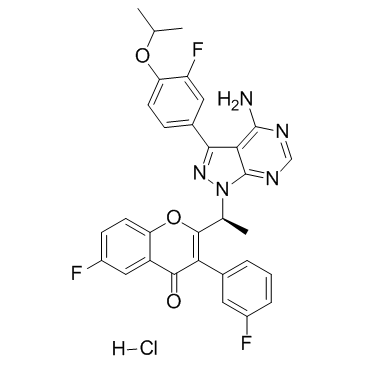
GC37855
Umbralisib R-enantiomer
Umbralisib R-enantiomer (TGR-1202 R-enantiomer) is a PI3Kδ inhibitor, which is the less active enantiomer of TGR-1202.
-
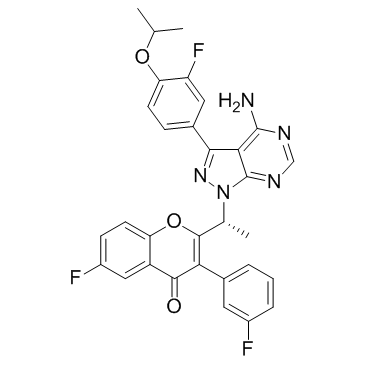
GC37859
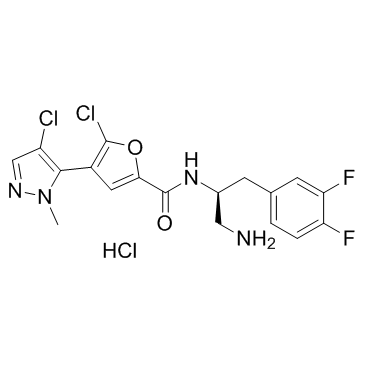
Uprosertib hydrochloride
Uprosertib hydrochloride (GSK2141795 hydrochloride) is a potent and selective pan-Akt inhibitor with IC50 values of 180/328/38 nM for Akt1/Akt2/Akt3, respectively.
-
GC38679
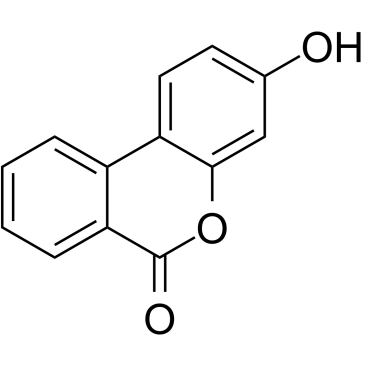
Urolithin B
A metabolite of ellagic acid
-
GC64231
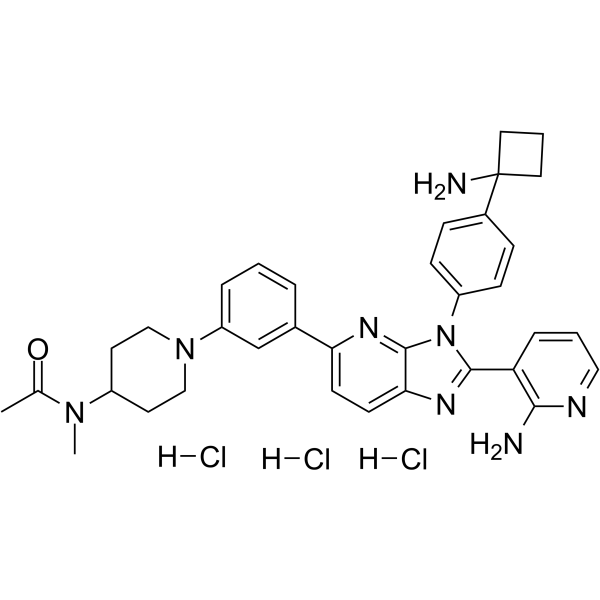
Vevorisertib trihydrochloride
Vevorisertib (ARQ 751) trihydrochloride is a selective, allosteric, pan-AKT and AKT1-E17K mutant inhibitors. Vevorisertib trihydrochloride potently inhibit phosphorylation of AKT. Vevorisertib trihydrochloride has Kd values of 1.2 nM and 8.6 nM for AKT1 and AKT1-E17K, respectively. Vevorisertib trihydrochloride has IC50 values of 0.55, 0.81, and 1.3 nM for AKT1, AKT2, and AKT3, respectively. Vevorisertib trihydrochloride can be used for the research of cancer.
-
GC41527
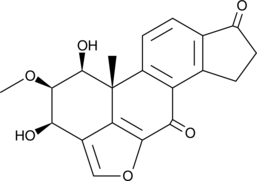
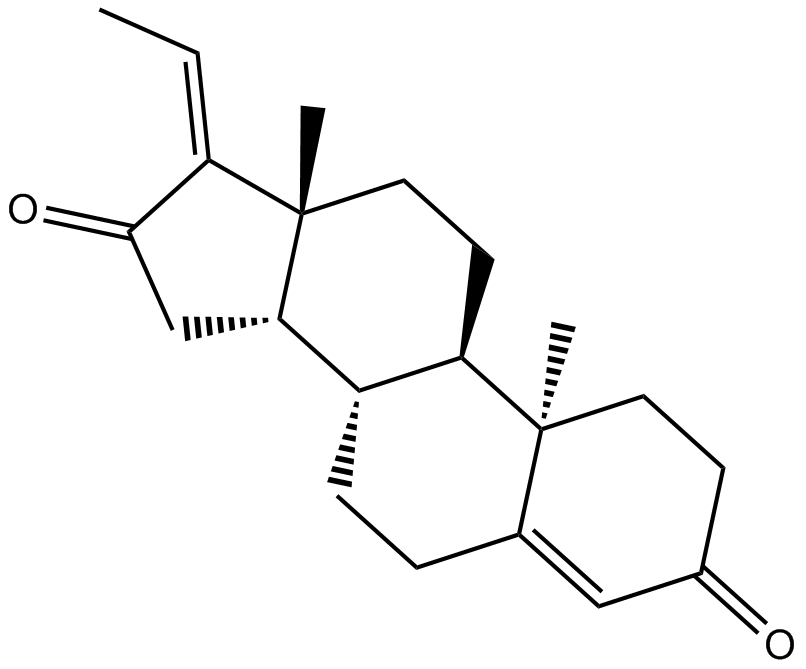
Viridiol
Viridiol is a fungal steroid that demonstrates phytotoxic activity.
-
GC37922
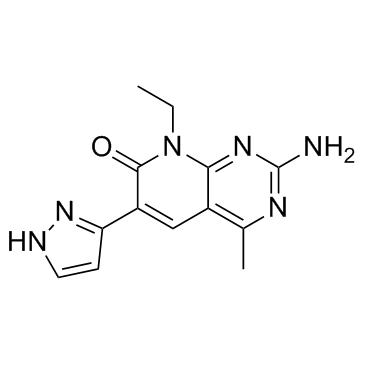
Voxtalisib
A dual inhibitor of PI3K and mTORC
-
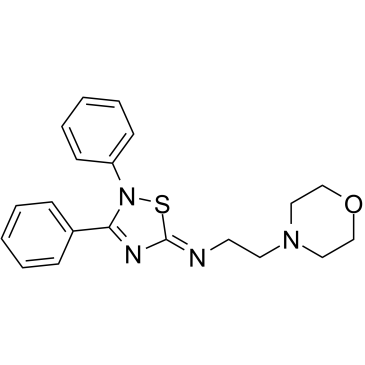
GC37923
VP3.15
VP3.15 is a potent, orally bioavailable and CNS-penetrant dual phosphodiesterase (PDE)7- glycogen synthase kinase (GSK)3 inhibitor, with IC50s of 1.59 μM and 0.88 μM for PDE7 and GSK-3, respectively.
-
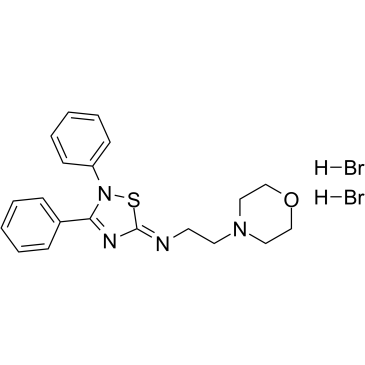
GC37924
VP3.15 dihydrobromide
VP3.15 dihydrobromide is a potent, orally bioavailable and CNS-penetrant dual phosphodiesterase (PDE)7- glycogen synthase kinase (GSK)3 inhibitor, with IC50s of 1.59 μM and 0.88 μM for PDE7 and GSK-3, respectively.
-
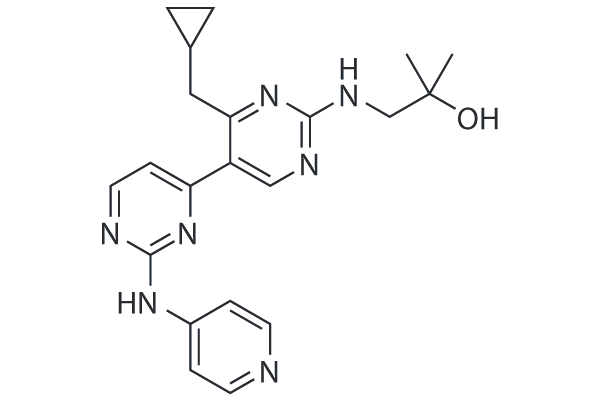
GC26044
VPS34 inhibitor 1 (Compound 19)
VPS34 inhibitor 1 (Compound 19, PIK-III analogue) is a potent and selective inhibitor of VPS34 with an IC50 of 15 nM.
-
GC32932
Vps34-IN-2
Vps34-IN-2 is a novel, potent and selective inhibitor of Vps34 with IC50s of 2 and 82 nM on the Vps34 enzymatic assay and the GFP-FYVE cellular assay, respectively. Vps34-IN-2 shows antiviral activity against SARS-CoV-2 (IC50 of 3.1 μM), HCoV-229E (IC50 of 0.7 μM) and HCoV-OC43.
-
GC10436
VPS34-IN1
Vps34 inhibitor
-
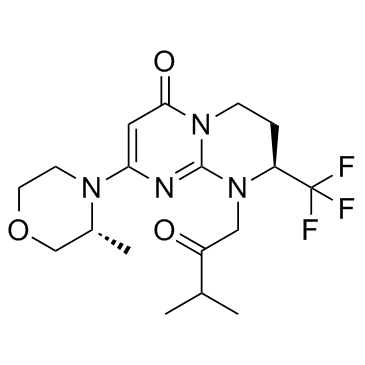
GC17771
VS-5584 (SB2343)
A selective PI3K/mTOR kinase inhibitor
-
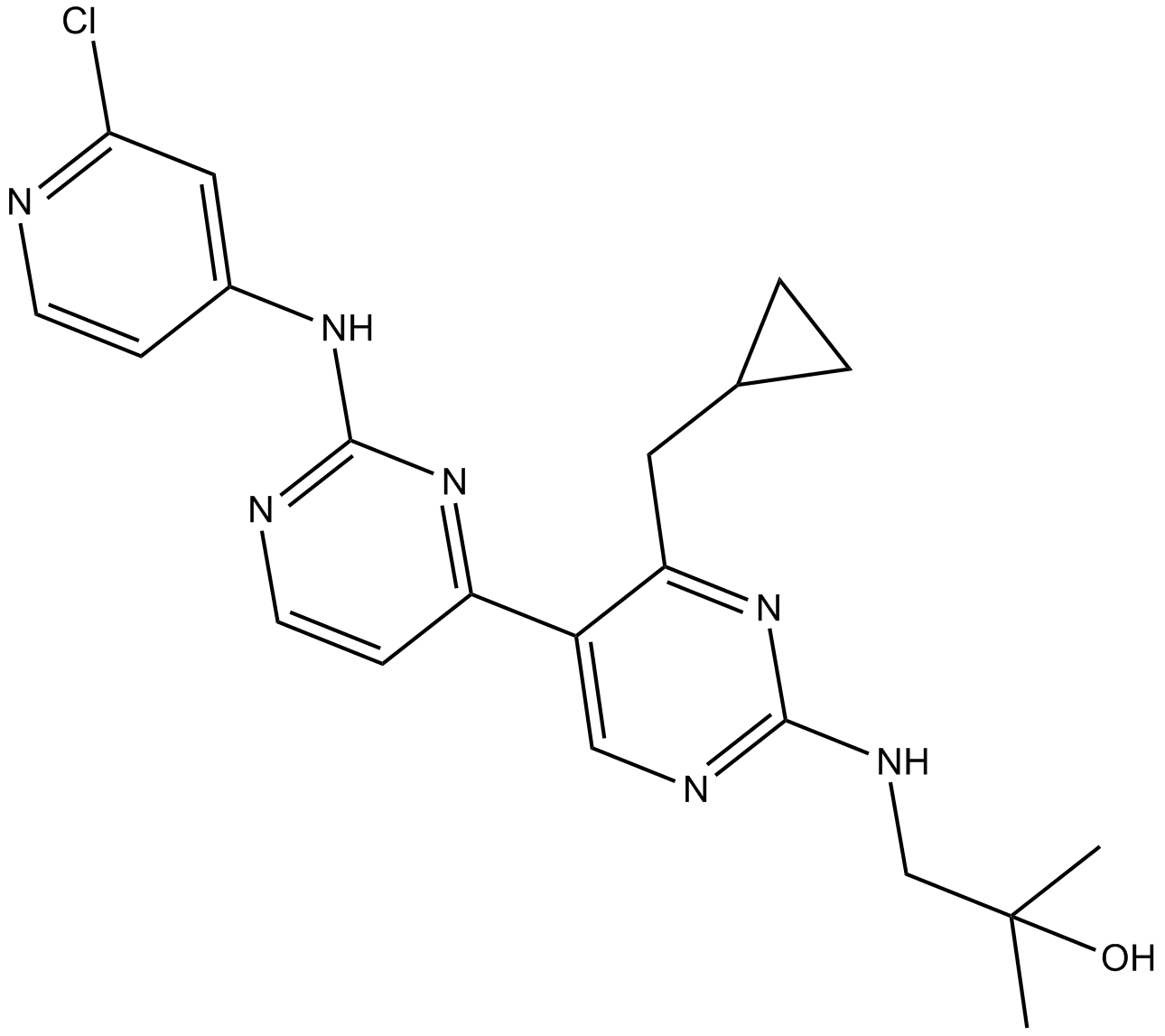
GC48256
VS-5584 analog
VS-5584 analog is a dimethyl analog of VS-5584 which is an potent and selective mTOR/PI3K dual inhibitor with pyrido [2,3-d] pyrimidine structure.
-
GC70138
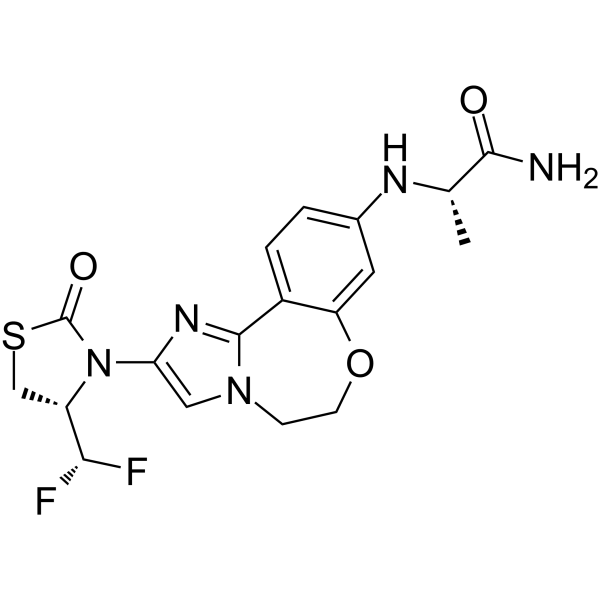
Vulolisib
Vulolisib is an effective and orally active phosphoinositide 3-kinase (PI3K) inhibitor with IC50 values of 0.2 nM, 168 nM, 90 nM, and 49 nM for PI3Kα, PI3Kβ, PI3Kγ, and PI3Kδ respectively. It has anti-proliferative activity against cancer cells and also exhibits anti-tumor activity.
-
GC33325
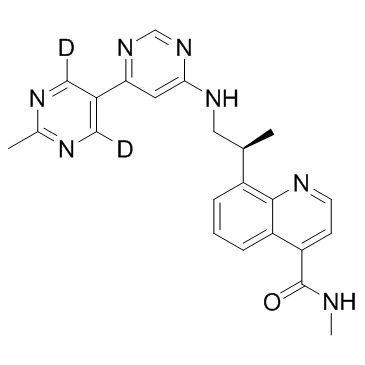
VX-984 (M9831)
VX-984 (M9831) (M9831) is an orally active, potent, selective and BBB-penetrated DNA-PK inhibitor. VX-984 (M9831) efficiently inhibits NHEJ (non-homologous end joining) and increases DSBs (DNA double-strand breaks). VX-984 (M9831) can be used for glioblastomas (GBM) and non-small cell lung cancer (NSCLC) research.
-
GC26049
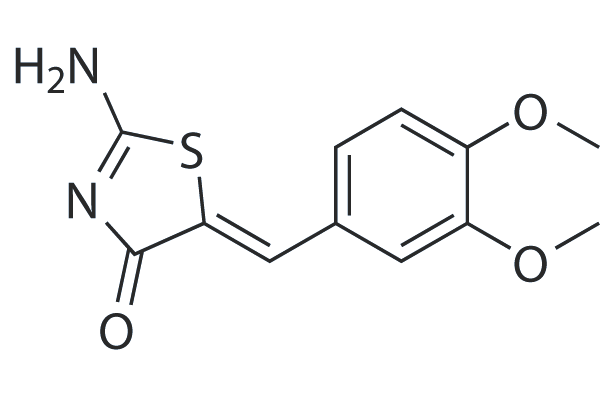
WAY-119064
WAY-119064 is a potent glycogen synthase kinase-3β (GSK-3β) inhibitor with an IC50 value of 80.5 nM.
-
GC10583
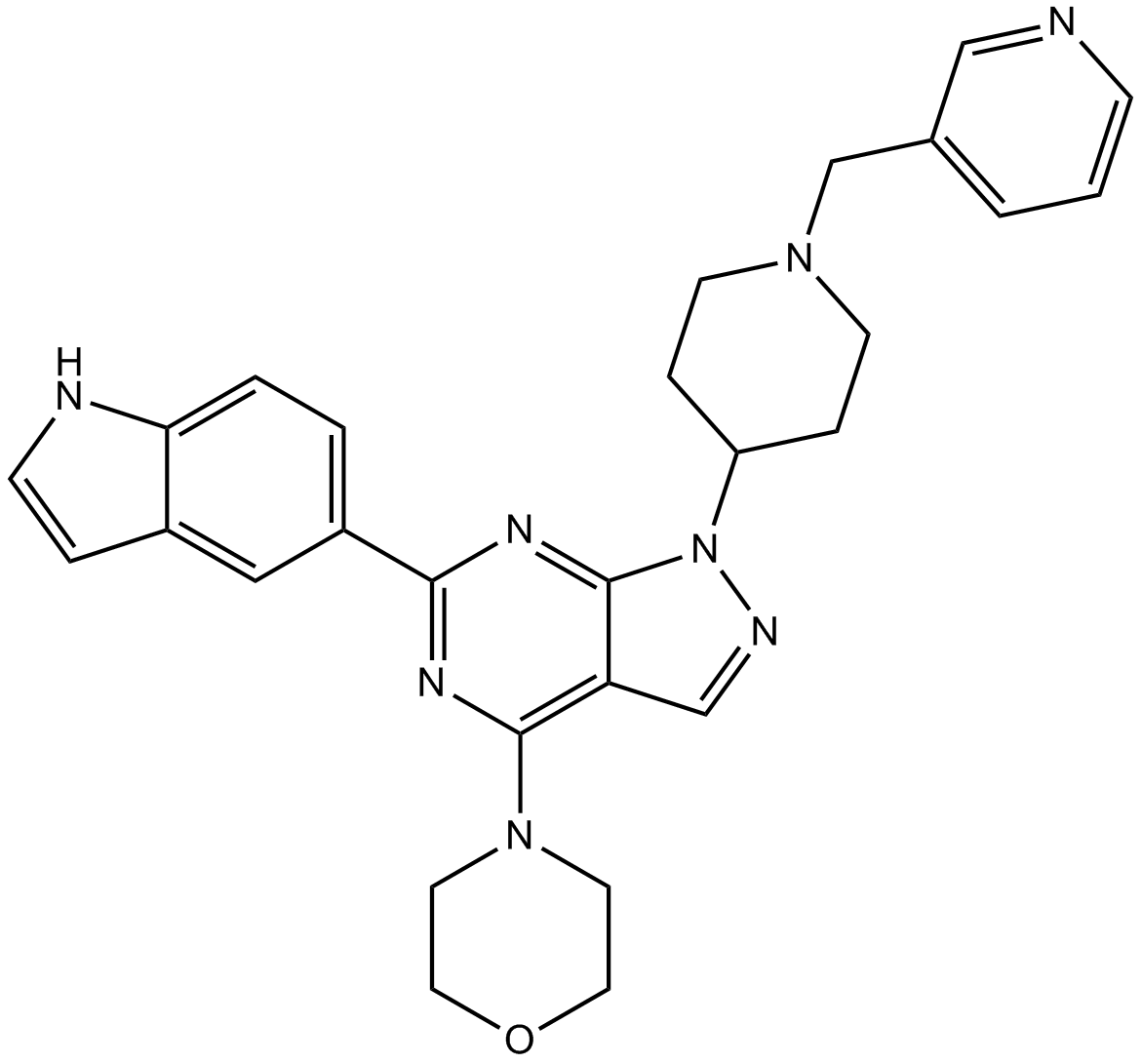
WAY-600
MTOR inhibitor
-
GC12338
Wortmannin
Wortmannin is a highly potent direct inhibitor of PI3-kinase specificity originally derived from fungi (1,2).
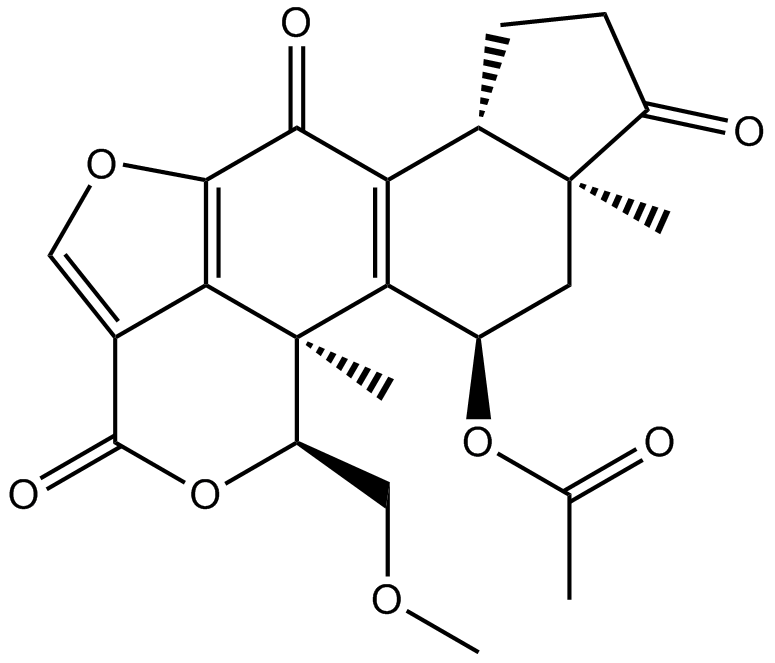
-
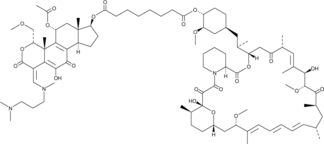
GC45160
Wortmannin-Rapamycin Conjugate
Phosphoinositide 3-kinase (PI3K) and mammalian target of rapamycin (mTOR) act synergistically in promoting cancer.
-
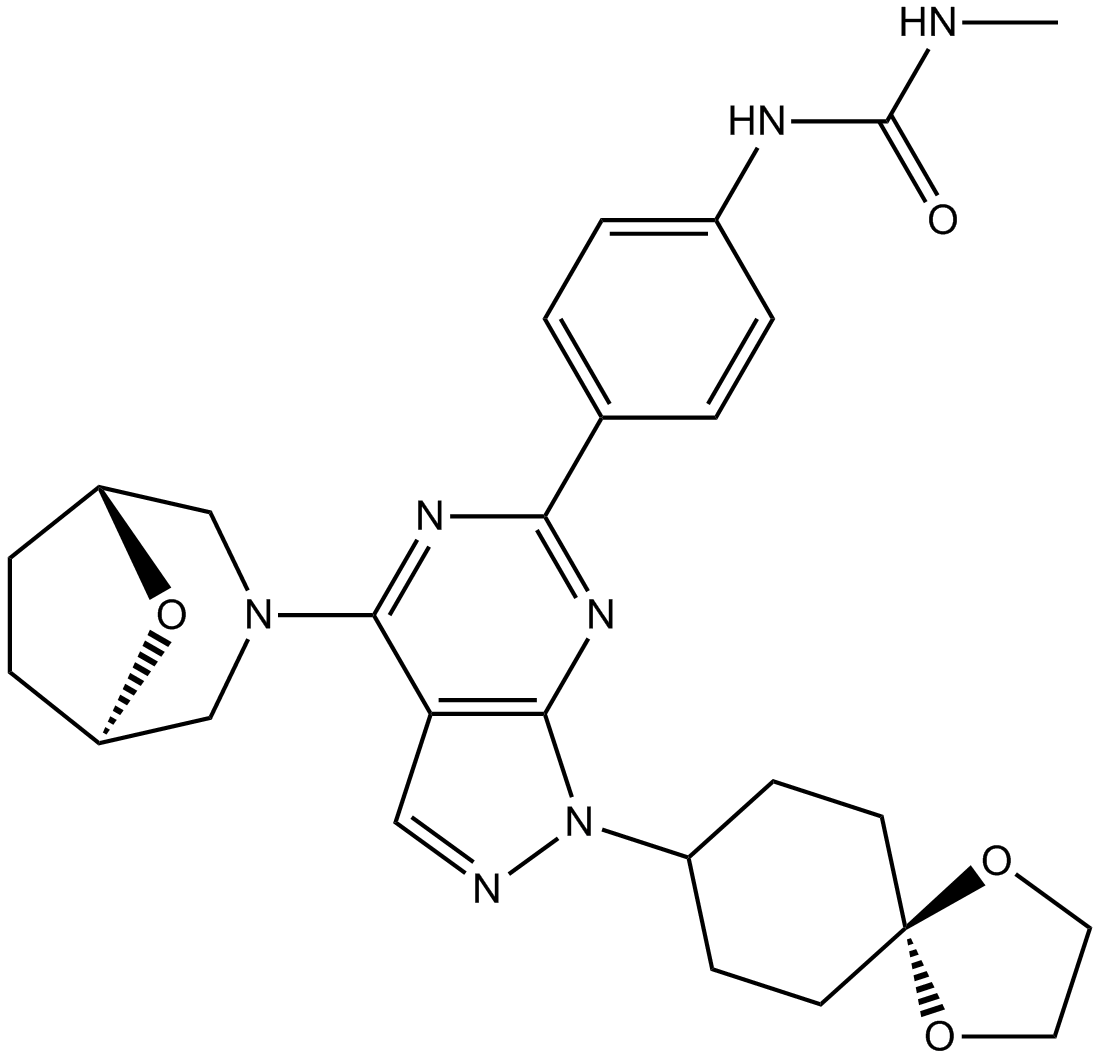
GC14675
WYE-125132 (WYE-132)
WYE-125132 (WYE-132) (WYE-125132) is a highly potent, ATP-competitive, and specific mTOR kinase inhibitor (IC50: 0.19±0.07 nM; >5,000-fold selective versus PI3Ks). WYE-125132 (WYE-132) (WYE-125132) inhibits mTORC1 and mTORC2.
-
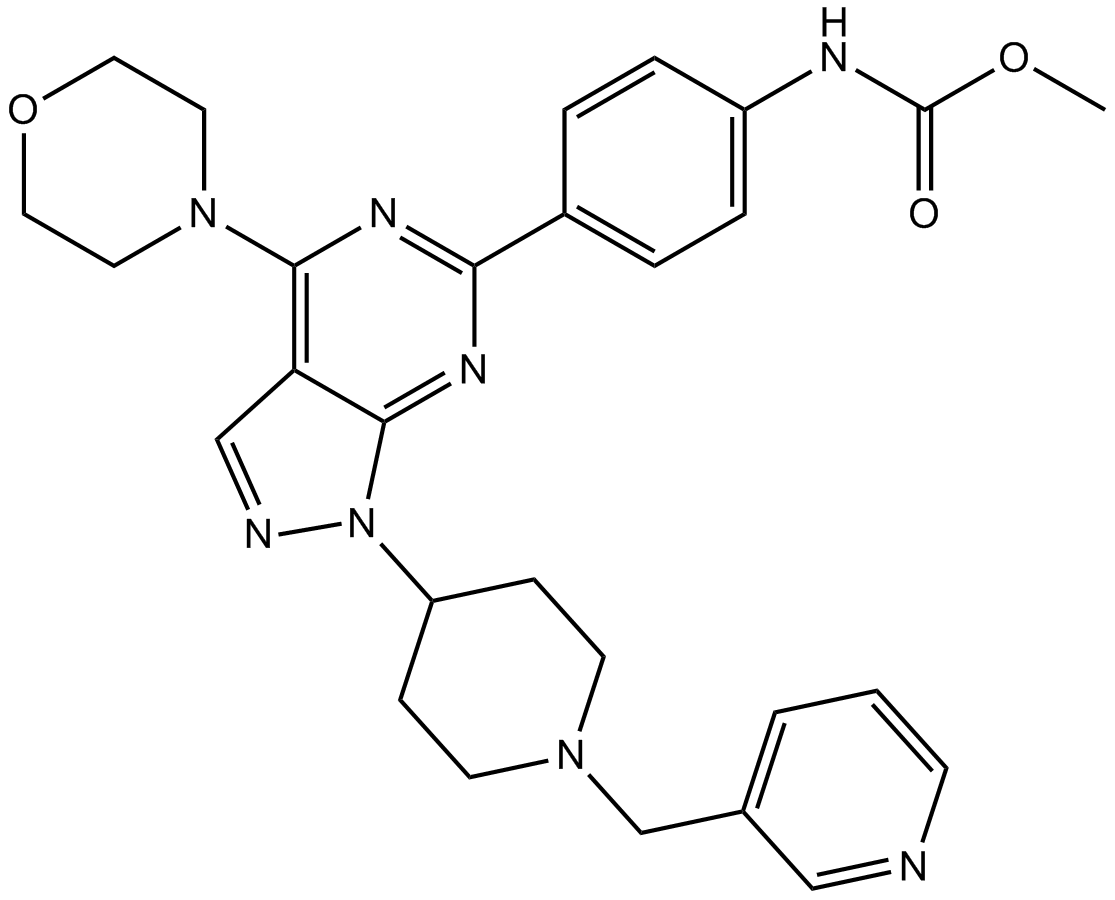
GC13321
WYE-687
MTOR inhibitor,ATP-competitive and selective
-
GC38473
WYE-687 dihydrochloride
WYE-687 dihydrochloride is an ATP-competitive mTOR inhibitor with an IC50 of 7 nM. WYE-687 dihydrochloride concurrently inhibits activation of mTORC1 and mTORC2. WYE-687 also inhibits PI3Kα and PI3Kγ with IC50s of 81 nM and 3.11 μM, respectively.
-
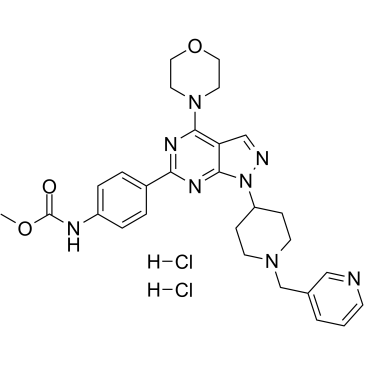
GC16549
WZ4003
NUAK1/2 inhibitor, potent and selective
-
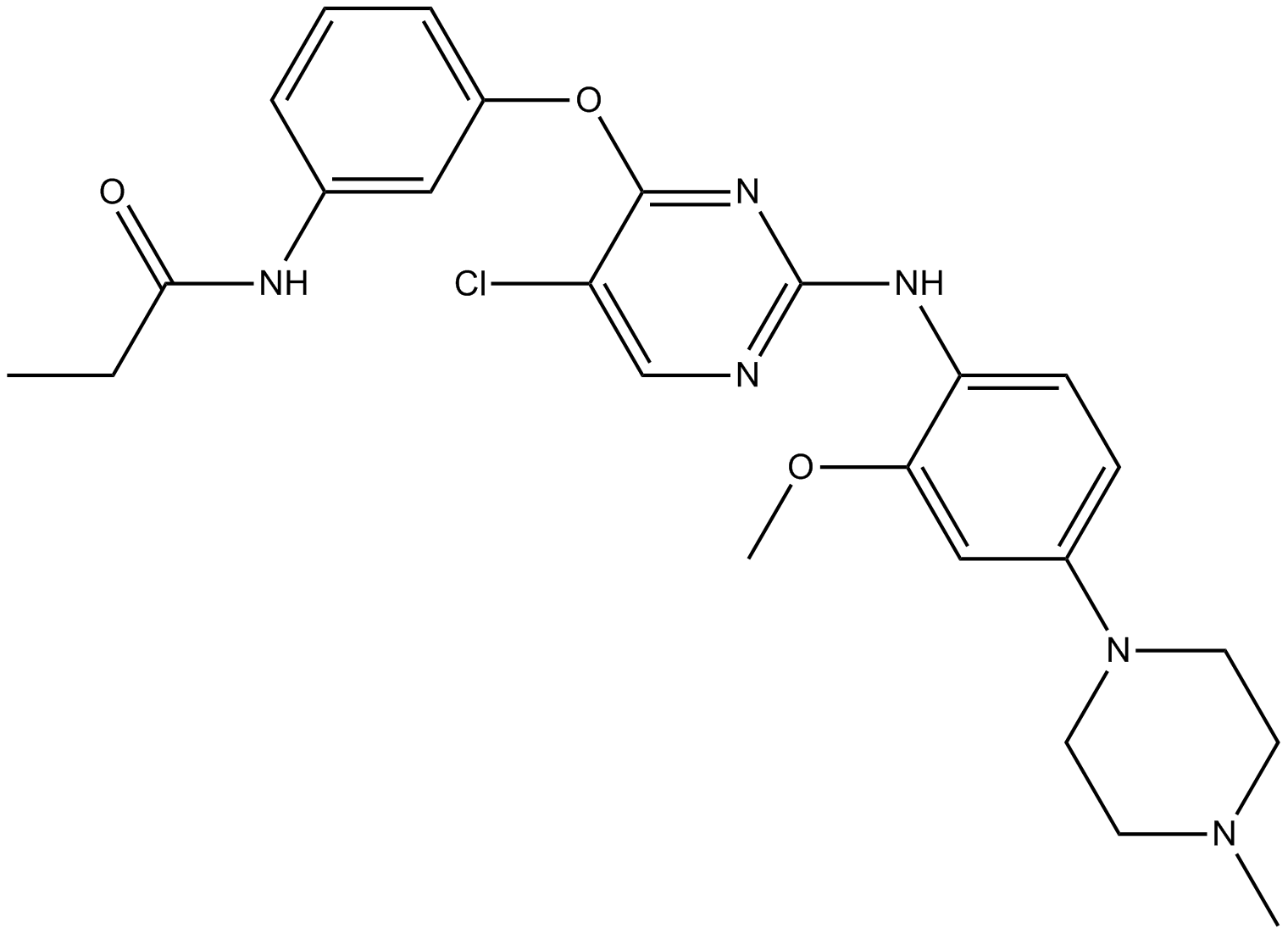
GC61382
Xanthoangelol
Xanthoangelol, extracted from Angelica keiskei, suppresses obesity-induced inflammatory responses.
-
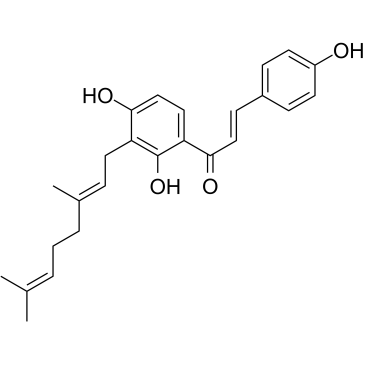
GC12709
XL147
XL147 (XL147 analogue) is a representative and selective PI3Kα inhibitor extracted from patent WO2012006552A1, Compound 147 in Table 1.
-
GC16481
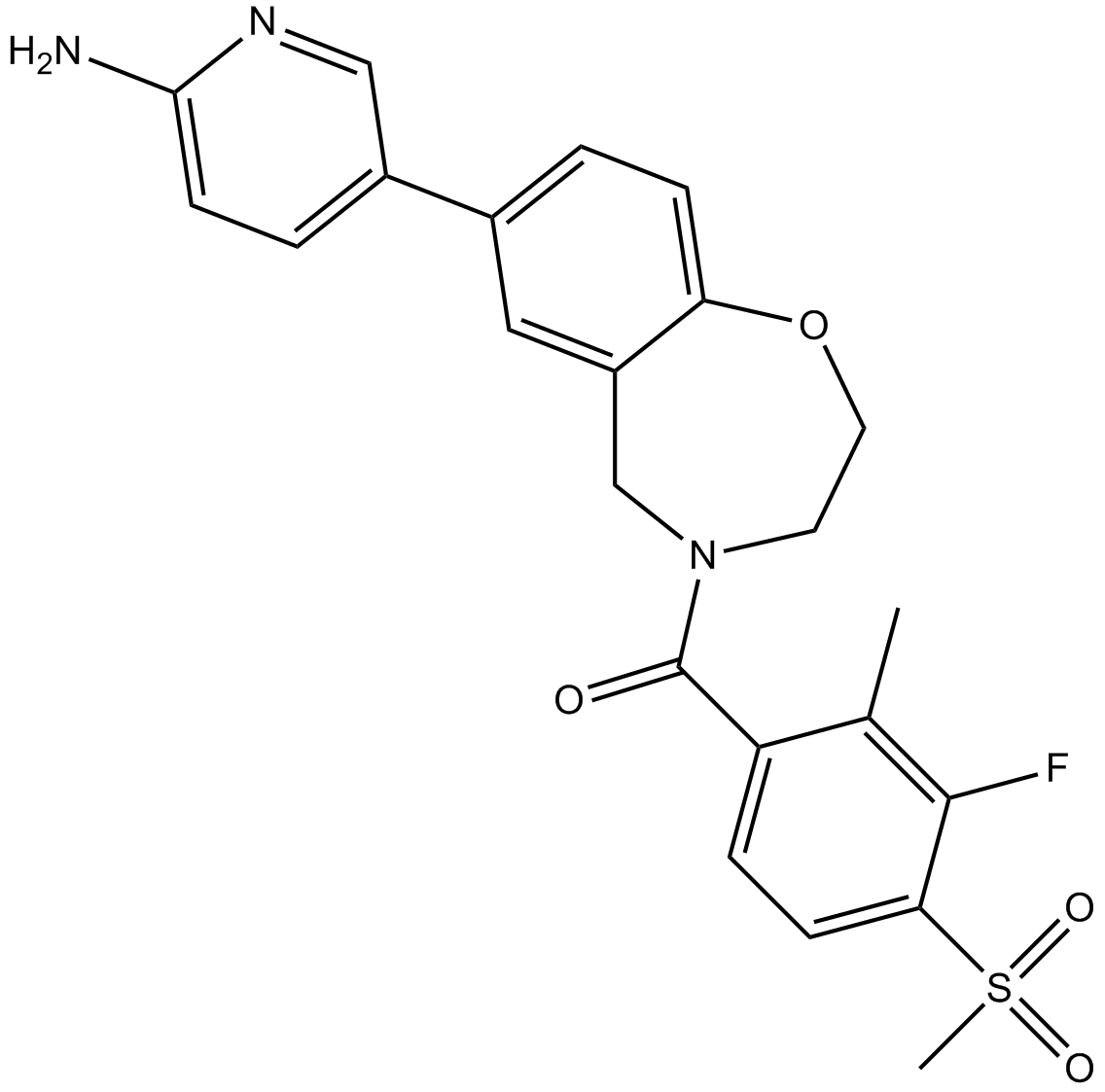
XL388
An mTOR inhibitor
-
GC12336
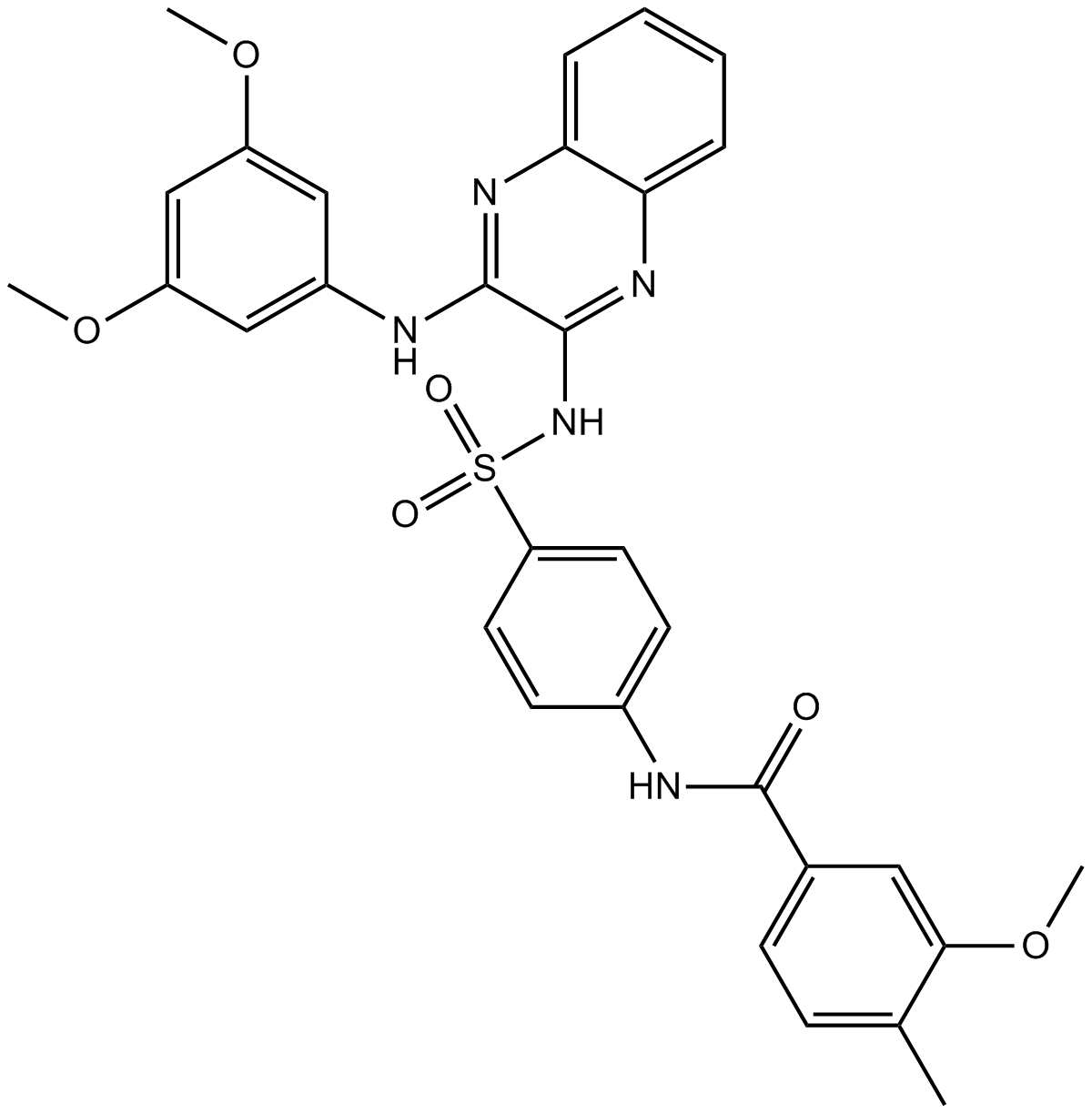
XL765
PI3K/mTOR inhibitor
-
GC64992
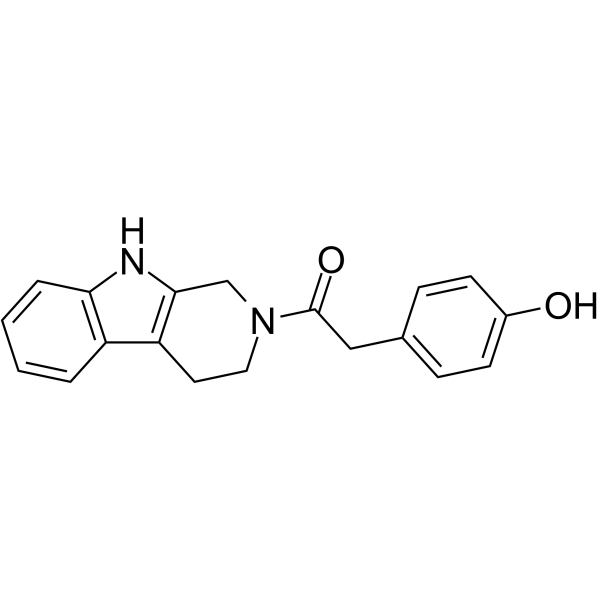
YH-306
YH-306 is an antitumor agent. YH-306 suppresses colorectal tumour growth and metastasis via FAK pathway. YH-306 significantly inhibits the migration and invasion of colorectal cancer cells. YH-306 potently suppresses uninhibited proliferation and induces cell apoptosis. YH-306 suppresses the activation of FAK, c-Src, paxillin, and PI3K, Rac1 and the expression of MMP2 and MMP9. YH-306 also inhibita actin-related protein (Arp2/3) complex-mediated actin polymerization.
-
GC18208
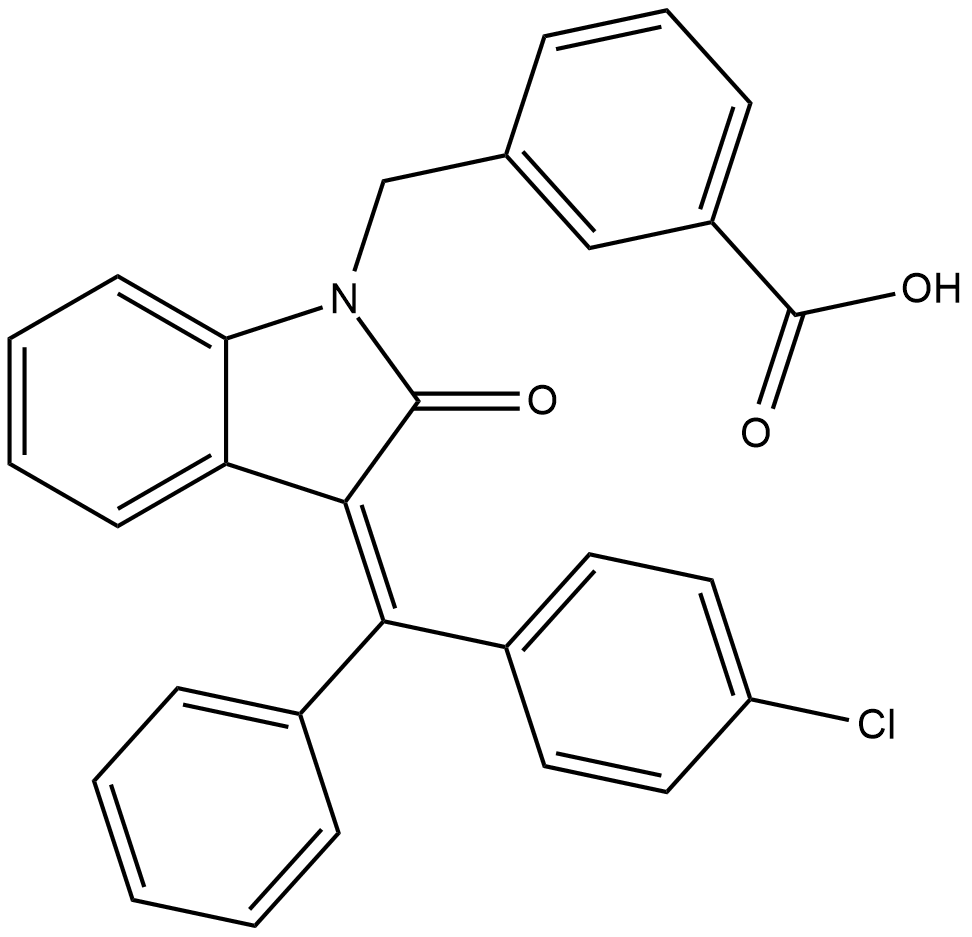
YLF-466D
YLF-466D is an orally bioavailable activator of AMP-activated protein kinase (AMPK), an enzyme involved in regulation of cellular energy homeostasis.
-
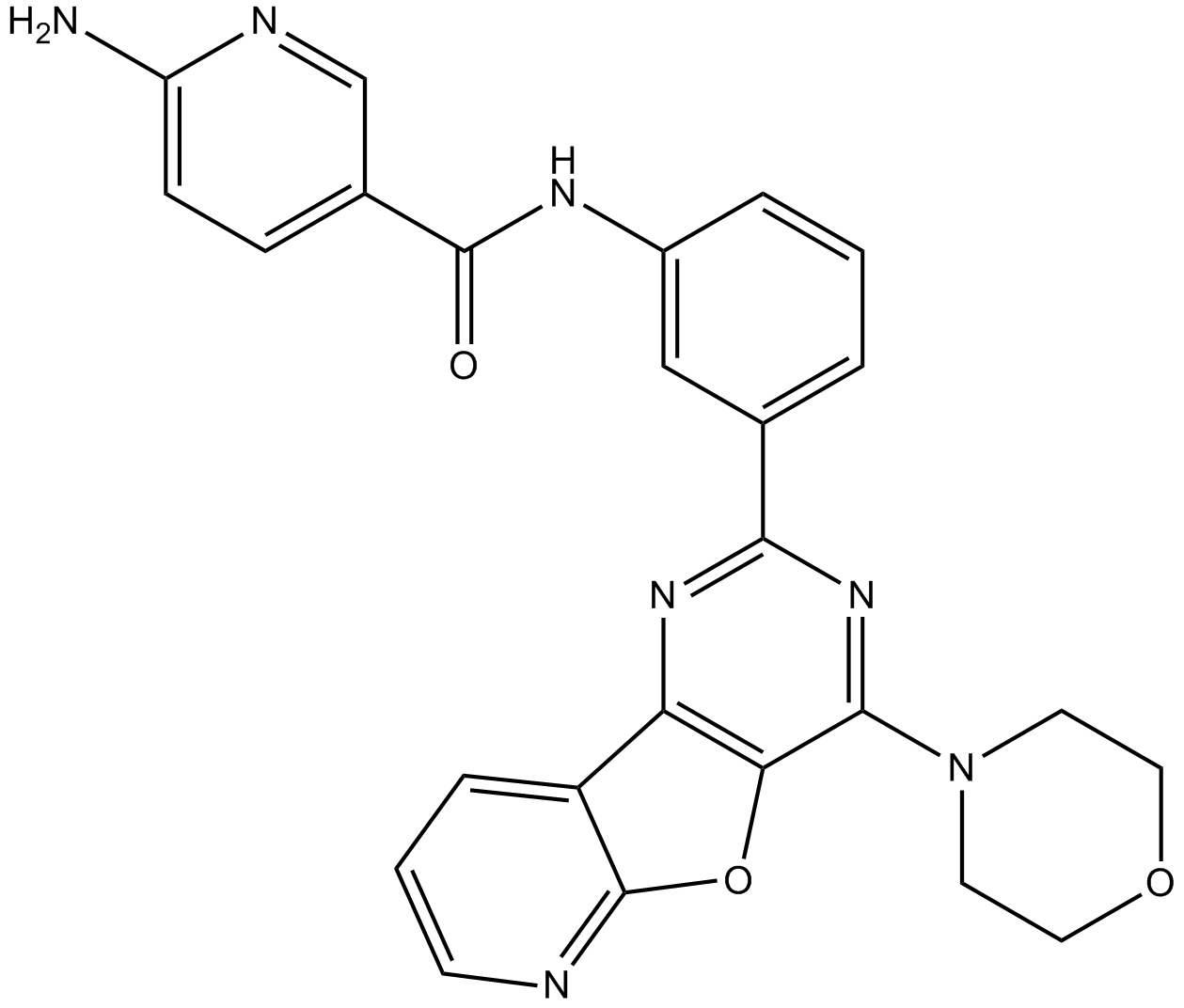
GC10982
YM201636
PIKfyve inhibitor,potent and selective
-
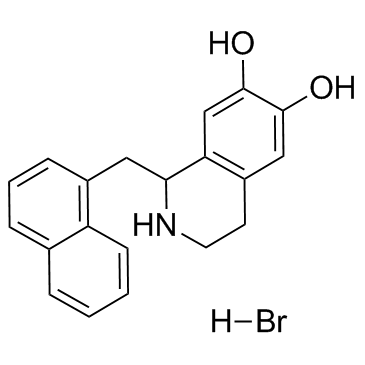
GC37954
YS-49
YS-49 is a PI3K/Akt (a downstream target of RhoA) activator, to reduce RhoA/PTEN activation in the 3-methylcholanthrene-treated cells.
-
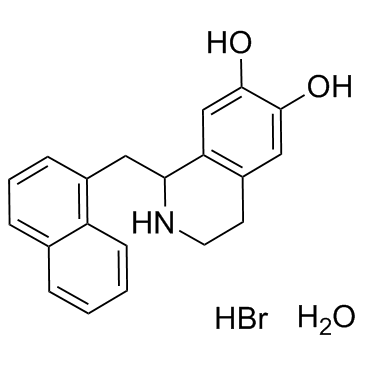
GC37955
YS-49 monohydrate
YS-49 (monohydrate) is a PI3K/Akt (a downstream target of RhoA) activator, to reduce RhoA/PTEN activation in the 3-methylcholanthrene-treated cells.
-
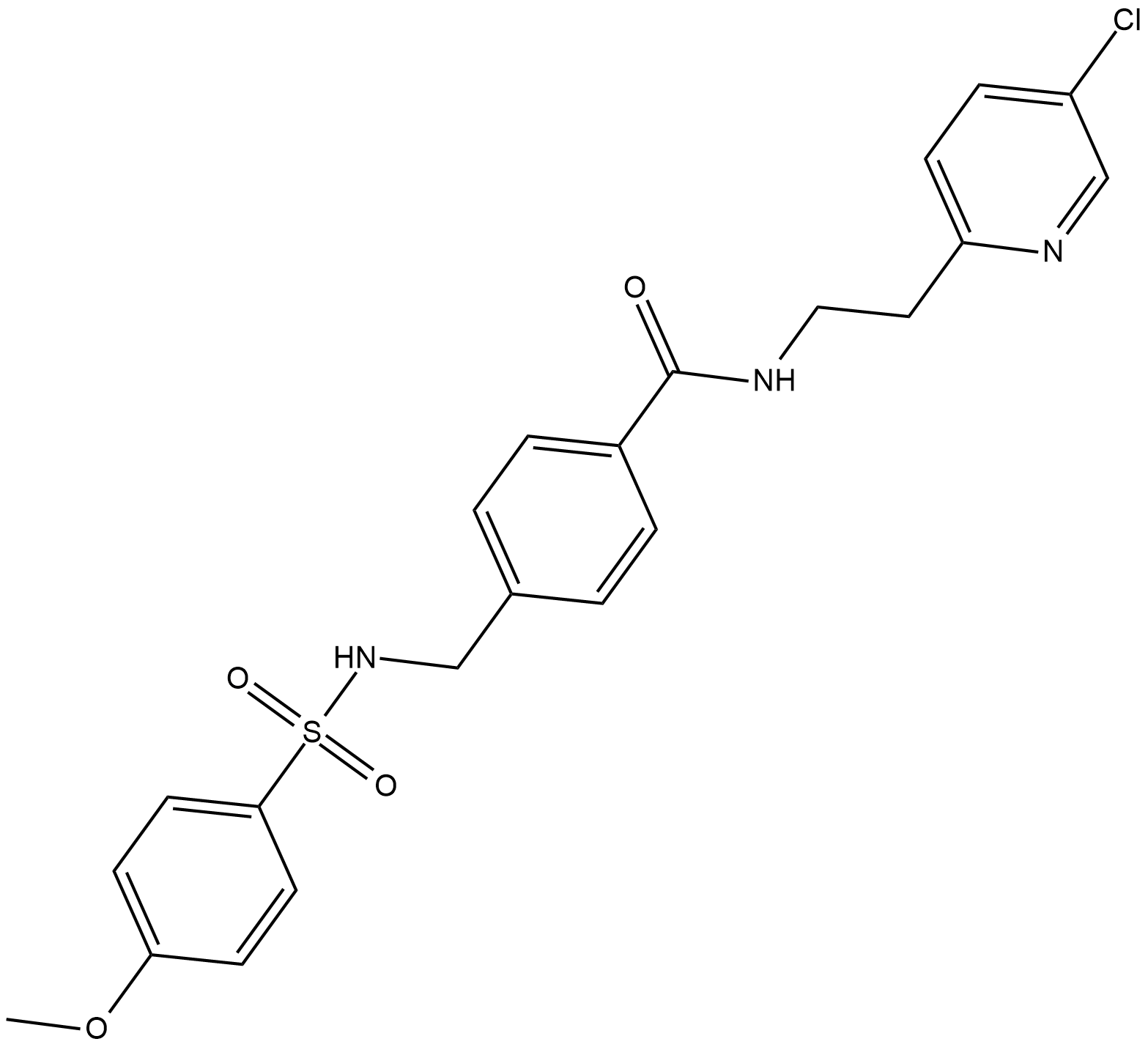
GC19390
YU238259
YU238259 is an inhibitor of homology-dependent DNA repair (HDR), used for cancer research.
-
GC10195
Z-Guggulsterone
Z-Guggulsterone suppresses angiogenesis in vitro and in vivo with IC50 values of 1740, 1000, 220 and > 50000 nM for glucocorticoid, mineralocorticoid, androgen and farnesoid X receptors .
-
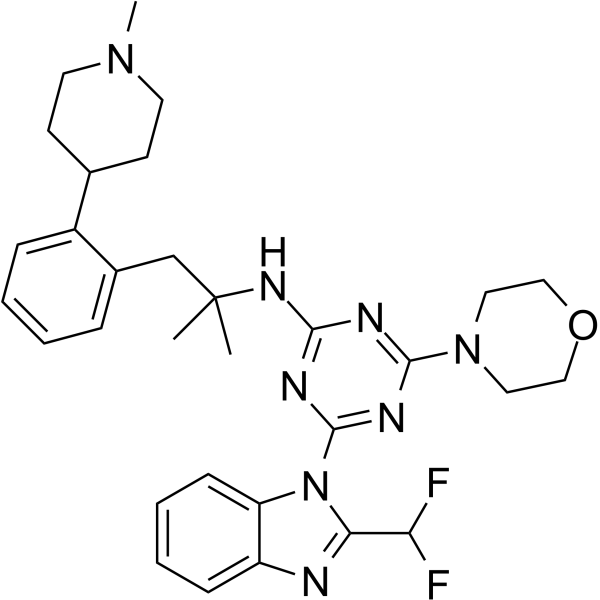
GC62480
Zandelisib
Zandelisib (ME-401) is a phosphatidylinositol 3-kinase (PI3K) inhibitor extracted from patent WO2019183226 A1, Compound Example 1. Zandelisib selectively inhibits p110δ with an IC50 of 3.5 nM. Zandelisib functions as an antineoplastic.
-
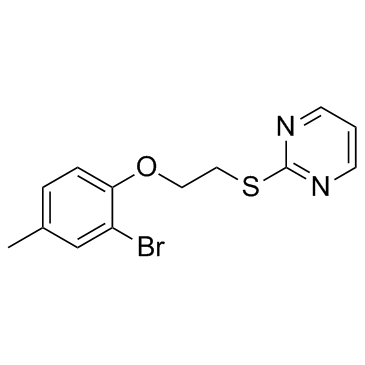
GC37970
ZLN024
An allosteric activator of AMPK
-
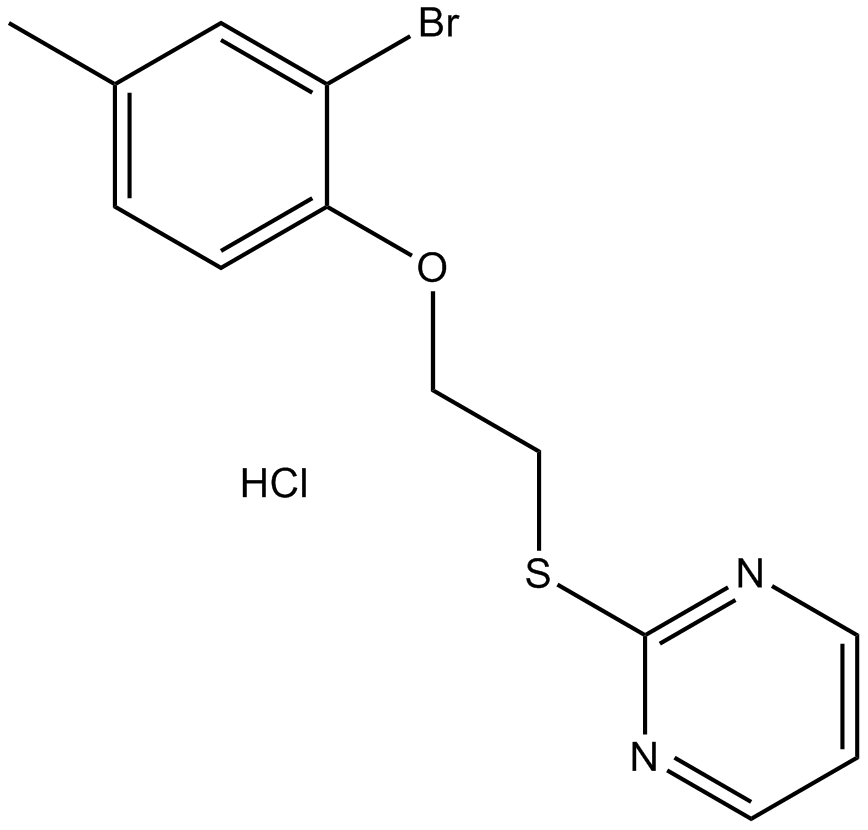
GC12162
ZLN024 hydrochloride
novel AMPK allosteric activator
-
GC19540
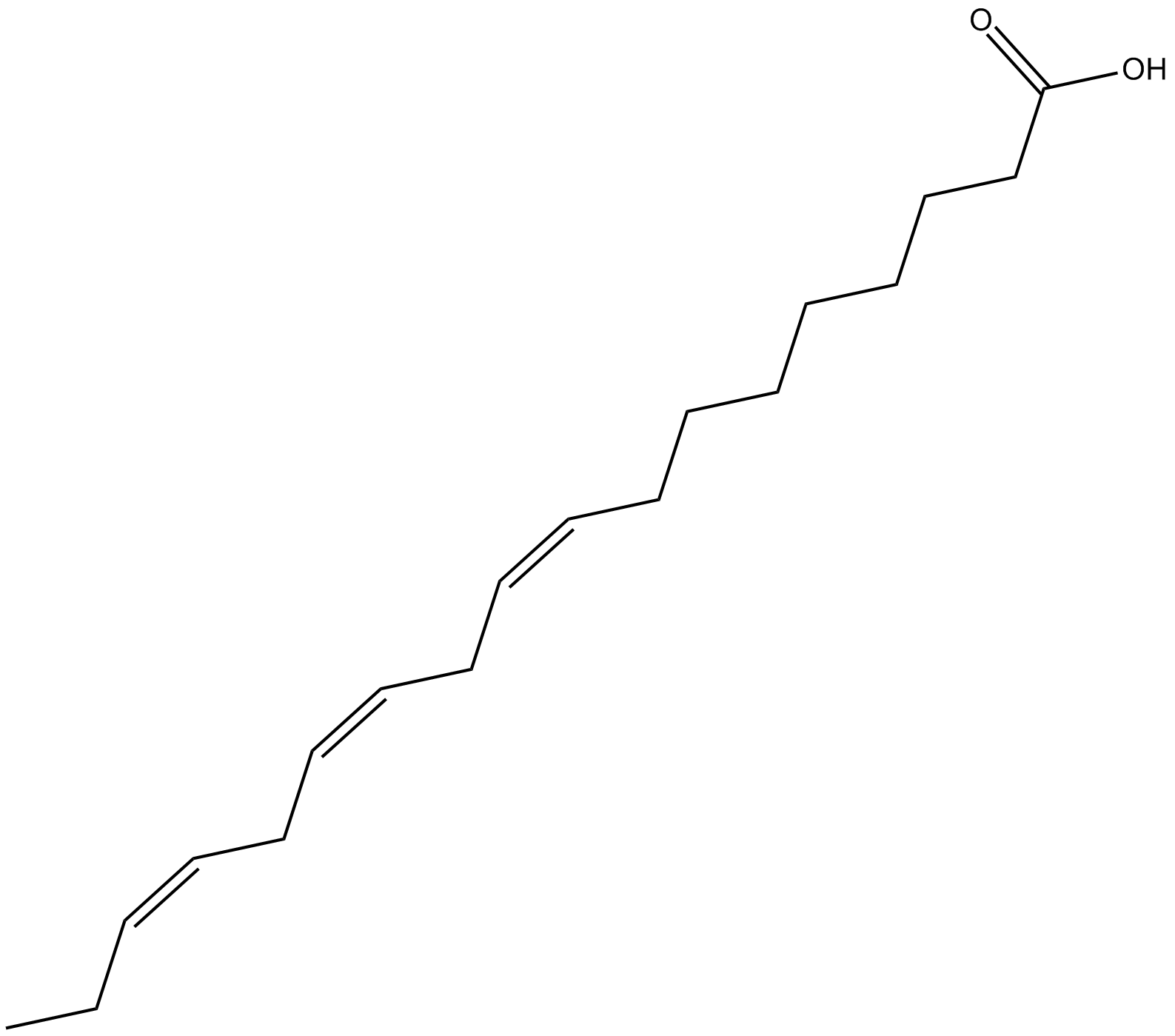
α-Linolenic Acid
An essential fatty acid found in leafy green vegetables