Apoptosis
As one of the cellular death mechanisms, apoptosis, also known as programmed cell death, can be defined as the process of a proper death of any cell under certain or necessary conditions. Apoptosis is controlled by the interactions between several molecules and responsible for the elimination of unwanted cells from the body.
Many biochemical events and a series of morphological changes occur at the early stage and increasingly continue till the end of apoptosis process. Morphological event cascade including cytoplasmic filament aggregation, nuclear condensation, cellular fragmentation, and plasma membrane blebbing finally results in the formation of apoptotic bodies. Several biochemical changes such as protein modifications/degradations, DNA and chromatin deteriorations, and synthesis of cell surface markers form morphological process during apoptosis.
Apoptosis can be stimulated by two different pathways: (1) intrinsic pathway (or mitochondria pathway) that mainly occurs via release of cytochrome c from the mitochondria and (2) extrinsic pathway when Fas death receptor is activated by a signal coming from the outside of the cell.
Different gene families such as caspases, inhibitor of apoptosis proteins, B cell lymphoma (Bcl)-2 family, tumor necrosis factor (TNF) receptor gene superfamily, or p53 gene are involved and/or collaborate in the process of apoptosis.
Caspase family comprises conserved cysteine aspartic-specific proteases, and members of caspase family are considerably crucial in the regulation of apoptosis. There are 14 different caspases in mammals, and they are basically classified as the initiators including caspase-2, -8, -9, and -10; and the effectors including caspase-3, -6, -7, and -14; and also the cytokine activators including caspase-1, -4, -5, -11, -12, and -13. In vertebrates, caspase-dependent apoptosis occurs through two main interconnected pathways which are intrinsic and extrinsic pathways. The intrinsic or mitochondrial apoptosis pathway can be activated through various cellular stresses that lead to cytochrome c release from the mitochondria and the formation of the apoptosome, comprised of APAF1, cytochrome c, ATP, and caspase-9, resulting in the activation of caspase-9. Active caspase-9 then initiates apoptosis by cleaving and thereby activating executioner caspases. The extrinsic apoptosis pathway is activated through the binding of a ligand to a death receptor, which in turn leads, with the help of the adapter proteins (FADD/TRADD), to recruitment, dimerization, and activation of caspase-8 (or 10). Active caspase-8 (or 10) then either initiates apoptosis directly by cleaving and thereby activating executioner caspase (-3, -6, -7), or activates the intrinsic apoptotic pathway through cleavage of BID to induce efficient cell death. In a heat shock-induced death, caspase-2 induces apoptosis via cleavage of Bid.
Bcl-2 family members are divided into three subfamilies including (i) pro-survival subfamily members (Bcl-2, Bcl-xl, Bcl-W, MCL1, and BFL1/A1), (ii) BH3-only subfamily members (Bad, Bim, Noxa, and Puma9), and (iii) pro-apoptotic mediator subfamily members (Bax and Bak). Following activation of the intrinsic pathway by cellular stress, pro‑apoptotic BCL‑2 homology 3 (BH3)‑only proteins inhibit the anti‑apoptotic proteins Bcl‑2, Bcl-xl, Bcl‑W and MCL1. The subsequent activation and oligomerization of the Bak and Bax result in mitochondrial outer membrane permeabilization (MOMP). This results in the release of cytochrome c and SMAC from the mitochondria. Cytochrome c forms a complex with caspase-9 and APAF1, which leads to the activation of caspase-9. Caspase-9 then activates caspase-3 and caspase-7, resulting in cell death. Inhibition of this process by anti‑apoptotic Bcl‑2 proteins occurs via sequestration of pro‑apoptotic proteins through binding to their BH3 motifs.
One of the most important ways of triggering apoptosis is mediated through death receptors (DRs), which are classified in TNF superfamily. There exist six DRs: DR1 (also called TNFR1); DR2 (also called Fas); DR3, to which VEGI binds; DR4 and DR5, to which TRAIL binds; and DR6, no ligand has yet been identified that binds to DR6. The induction of apoptosis by TNF ligands is initiated by binding to their specific DRs, such as TNFα/TNFR1, FasL /Fas (CD95, DR2), TRAIL (Apo2L)/DR4 (TRAIL-R1) or DR5 (TRAIL-R2). When TNF-α binds to TNFR1, it recruits a protein called TNFR-associated death domain (TRADD) through its death domain (DD). TRADD then recruits a protein called Fas-associated protein with death domain (FADD), which then sequentially activates caspase-8 and caspase-3, and thus apoptosis. Alternatively, TNF-α can activate mitochondria to sequentially release ROS, cytochrome c, and Bax, leading to activation of caspase-9 and caspase-3 and thus apoptosis. Some of the miRNAs can inhibit apoptosis by targeting the death-receptor pathway including miR-21, miR-24, and miR-200c.
p53 has the ability to activate intrinsic and extrinsic pathways of apoptosis by inducing transcription of several proteins like Puma, Bid, Bax, TRAIL-R2, and CD95.
Some inhibitors of apoptosis proteins (IAPs) can inhibit apoptosis indirectly (such as cIAP1/BIRC2, cIAP2/BIRC3) or inhibit caspase directly, such as XIAP/BIRC4 (inhibits caspase-3, -7, -9), and Bruce/BIRC6 (inhibits caspase-3, -6, -7, -8, -9).
Any alterations or abnormalities occurring in apoptotic processes contribute to development of human diseases and malignancies especially cancer.
References:
1.Yağmur Kiraz, Aysun Adan, Melis Kartal Yandim, et al. Major apoptotic mechanisms and genes involved in apoptosis[J]. Tumor Biology, 2016, 37(7):8471.
2.Aggarwal B B, Gupta S C, Kim J H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey.[J]. Blood, 2012, 119(3):651.
3.Ashkenazi A, Fairbrother W J, Leverson J D, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors[J]. Nature Reviews Drug Discovery, 2017.
4.McIlwain D R, Berger T, Mak T W. Caspase functions in cell death and disease[J]. Cold Spring Harbor perspectives in biology, 2013, 5(4): a008656.
5.Ola M S, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis[J]. Molecular and cellular biochemistry, 2011, 351(1-2): 41-58.
What is Apoptosis? The Apoptotic Pathways and the Caspase Cascade
Targets for Apoptosis
- Pyroptosis(15)
- Caspase(51)
- 14.3.3 Proteins(1)
- Apoptosis Inducers(41)
- Bax(8)
- Bcl-2 Family(107)
- Bcl-xL(8)
- c-RET(8)
- IAP(26)
- KEAP1-Nrf2(61)
- MDM2(13)
- p53(108)
- PC-PLC(4)
- PKD(6)
- RasGAP (Ras- P21)(1)
- Survivin(6)
- Thymidylate Synthase(10)
- TNF-α(117)
- Other Apoptosis(884)
- Apoptosis Detection(0)
- Caspase Substrate(0)
- APC(5)
- PD-1/PD-L1 interaction(60)
- ASK1(3)
- PAR4(2)
- RIP kinase(45)
- FKBP(19)
Products for Apoptosis
- Cat.No. Product Name Information
-
GC18701
Hellebrin
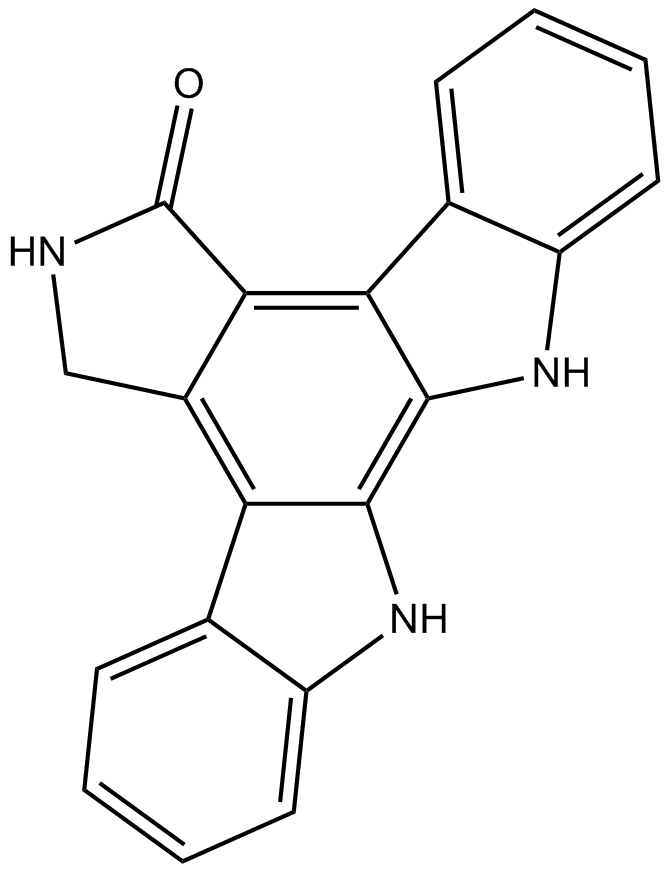
NSC 93134
Hellebrin is a cardiac glycoside that potently inhibits the Na+/K+-ATPase by binding to it and blocking its non-canonical function as a receptor for cardiac glycosides.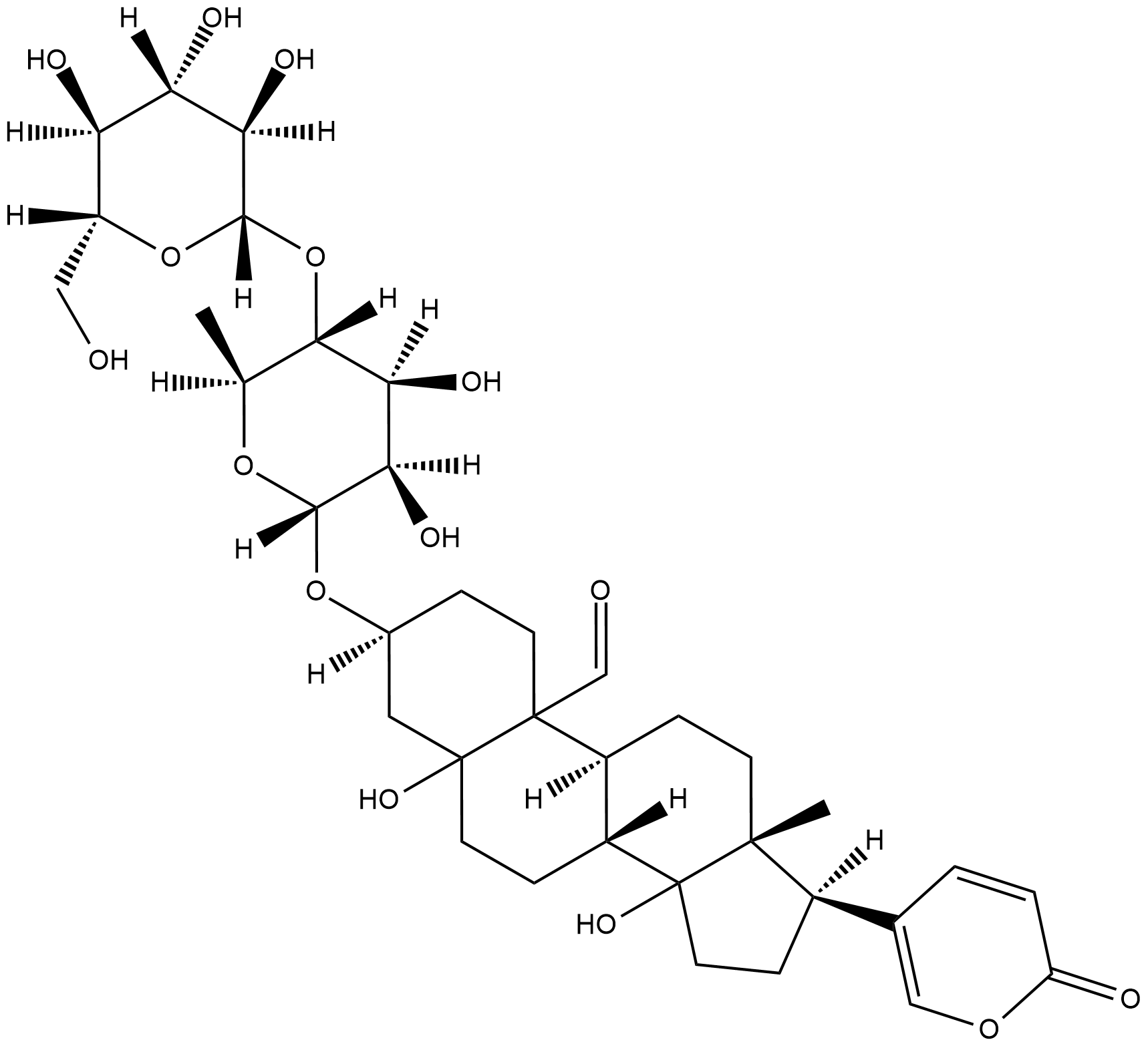
-
GC39266
Hematein
Hematein is a oxidation product of hematoxylin acted as a dye. Hematein is an allosteric casein kinase II inhibitor with an IC50 of 0.74 μM. Hematein inhibits Akt/PKB Ser129 phosphorylation, the Wnt/TCF pathway and increases apoptosis in lung cancer cells.
-
GC63902
Hematoporphyrin monomethyl ether
Hematoporphyrin monomethyl ether, second generation of porphyrin-related photosensitizer, is characterized by its single form, high yield of singlet oxygen, high selectivity, and low toxicity, which has been widely used in the diagnosis and treatment of various tumors, including lung cancer, bladder cancer, and nevus flammeus and brain glioma.
-
GC43816
Heptelidic Acid
Avocettin, BRN 5091359, FO-4443, Koningic Acid
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a key enzyme in carbohydrate metabolism, reversibly catalyzes the conversion of GAP to 1,3-bisphosphoglycerate and NAD+.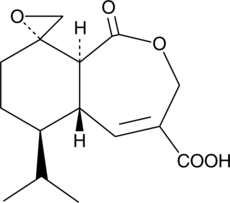
-
GC18796
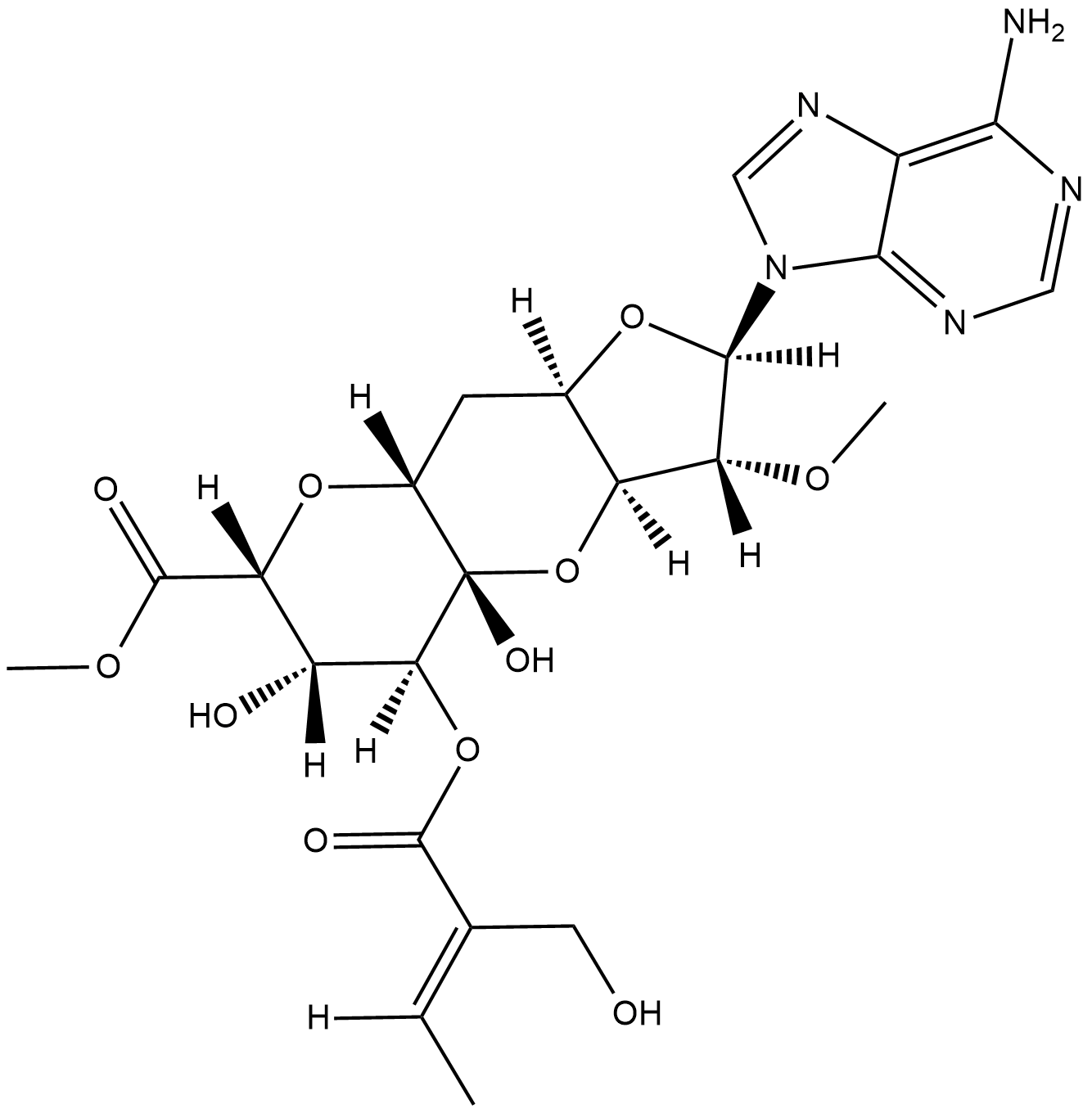
Herbicidin A
Herbicidin A is an adenine nucleoside antibiotic originally isolated from S.
-
GC40067
Herbimycin C
Antibiotic TAN 420D
Herbimycin C is a bacterial metabolite originally isolated from S.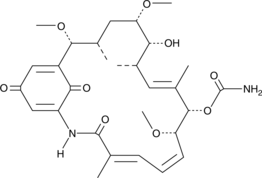
-
GC33092
Hesperin
6-MSITC
An isothiocyanate with diverse biological activities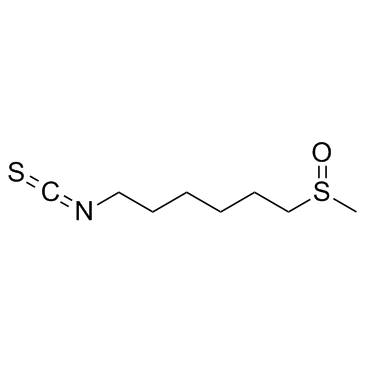
-
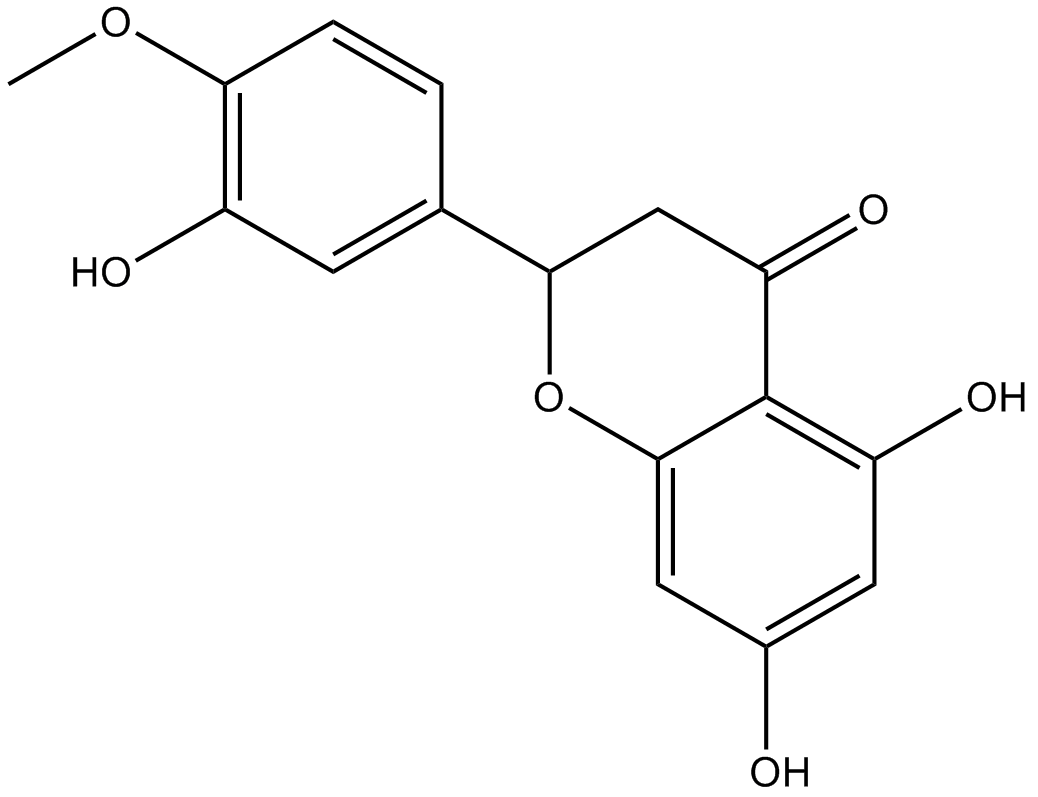
GN10248
Hesperitin
(–)-Hesperetin, (S)-Hesperetin, NSC 57654, (–)-3',5,7-Trihydroxy-4'-methoxyflavanone
-
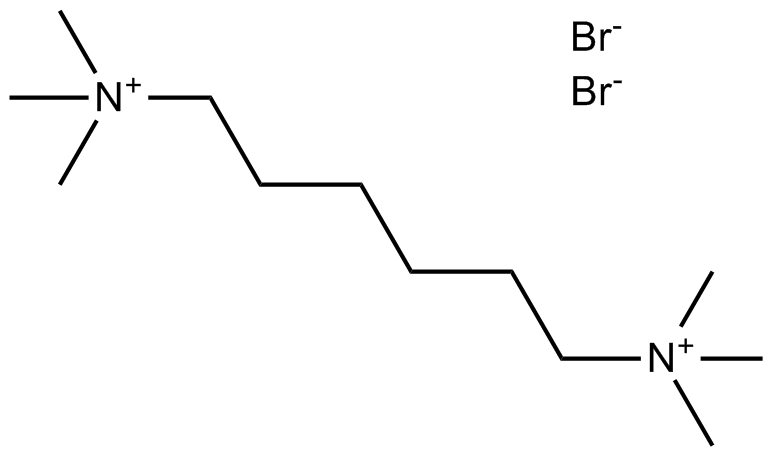
GC15178
Hexamethonium Bromide
Selective antagonist of neuronal-type nicotinic AChR
-
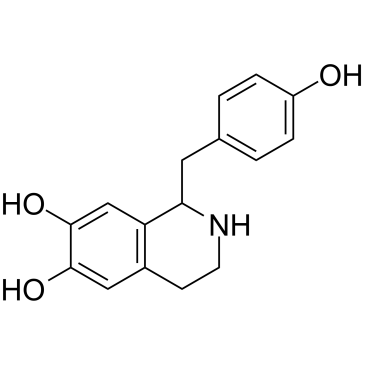
GC38099
Higenamine
Higenamine (Norcoclaurine), a β2-AR agonist, is a key component of the Chinese herb aconite root that prescribes for treating symptoms of heart failure in the oriental Asian countries.
-
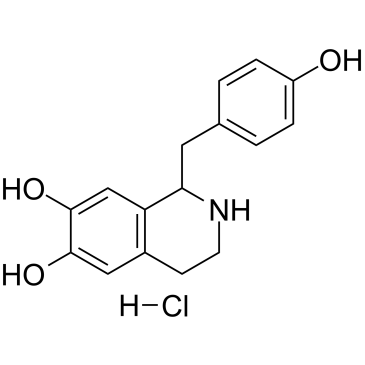
GC38237
Higenamine hydrochloride
Higenamine hydrochloride (Norcoclaurine hydrochloride), a β2-AR agonist, is a key component of the Chinese herb aconite root that prescribes for treating symptoms of heart failure in the oriental Asian countries.
-
GC11302
Hinokitiol
NSC 18804, β-Thujaplicin, 4-isopropyl Tropolone
A tropolone with diverse biological activities
-
GC36229
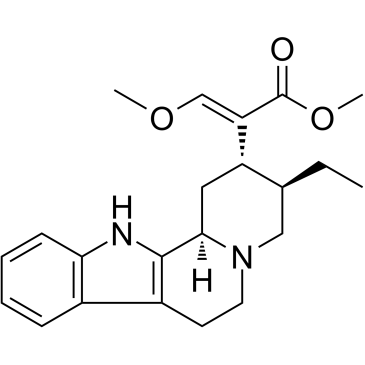
Hirsutine
Hirsutine, an indole alkaloid of Uncaria rhynchophylla, exhibits anti-cancer activity.
-
GC31739
Hispidol ((Z)-Hispidol)
(Z)-Hispidol
Hispidol ((Z)-Hispidol) ((Z)-Hispidol ((Z)-Hispidol)) is a potential therapeutic for inflammatory bowel disease; inhibits TNF-α induced adhesion of monocytes to colon epithelial cells with an IC50 of 0.50 μM.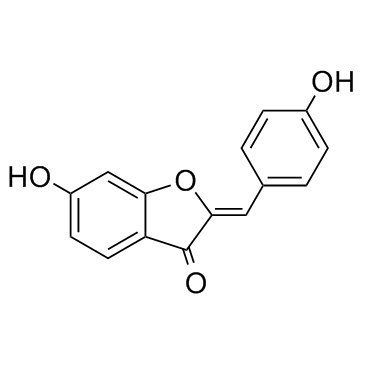
-
GC45473
Histone H2BK12ac (1-22)-GGK-biotin amide (trifluoroacetate salt)

-
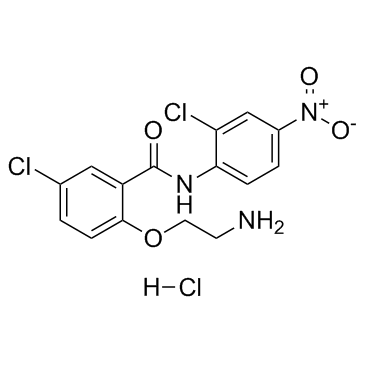
GC32807
HJC0152 hydrochloride
An orally bioavailable inhibitor of STAT3
-
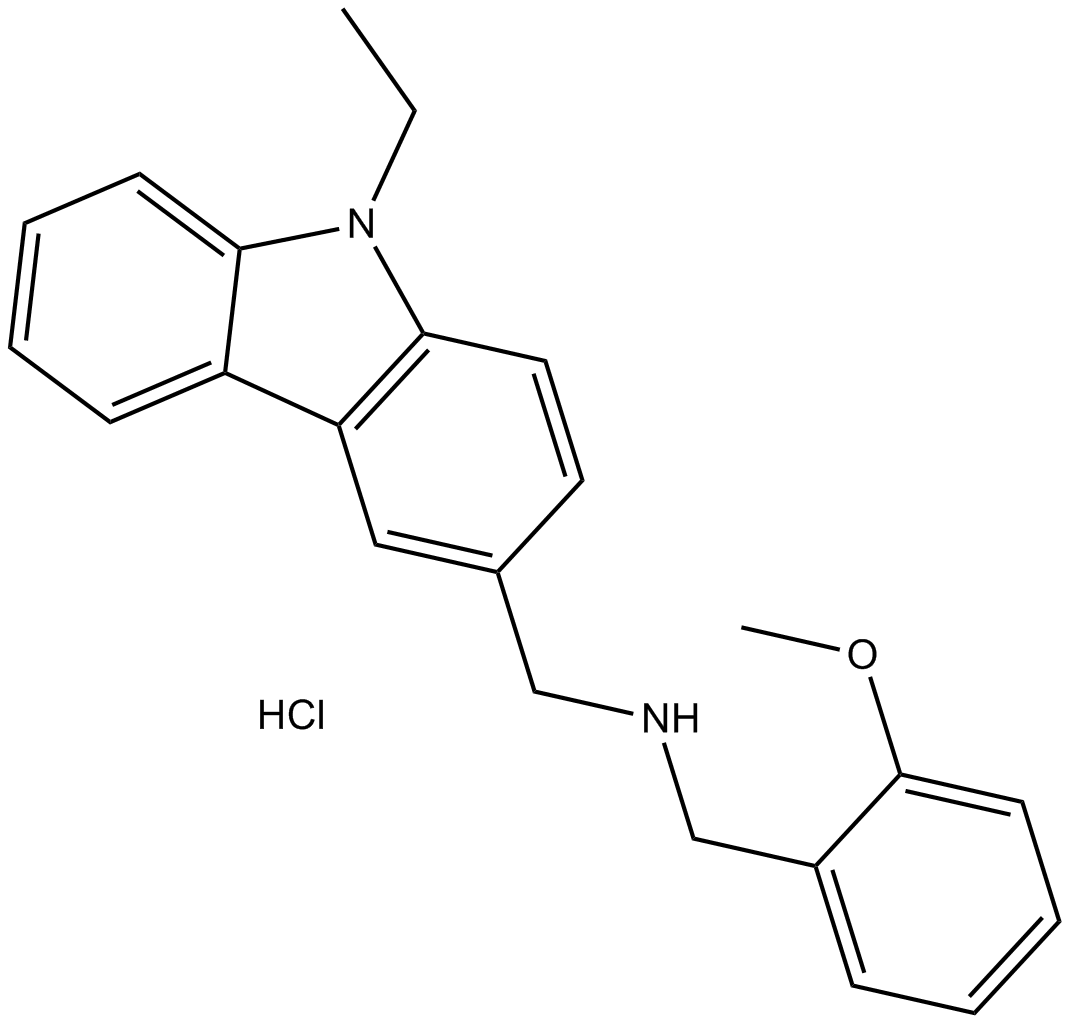
GC17023
HLCL-61
HLCL-61 is a first-in-class inhibitor of protein arginine methyltransferase 5 (PRMT5).
-
GC61608
HLI373 dihydrochloride
HLI373 dihydrochloride is an efficacious Hdm2 inhibitor.
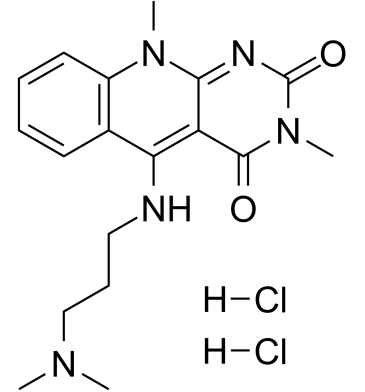
-
GC62594
hnRNPK-IN-1
hnRNPK-IN-1 is a heterogeneous nuclear ribonucleoprotein K (hnRNPK) binding ligand with Kd values of 4.6 μM and 2.6 μM measured with SPR and MST, respectively. hnRNPK-IN-1 inhibits c-myc transcription by disrupting the binding of hnRNPK and c-myc promoter. hnRNPK-IN-1 induces Hela cells apoptosis and has strongly anti-tumor activities.
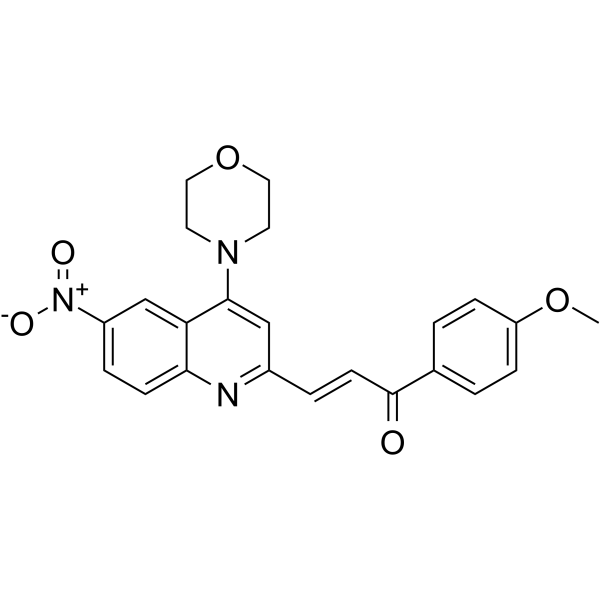
-
GC16208
HO-3867
STAT3 inhibitor, selective
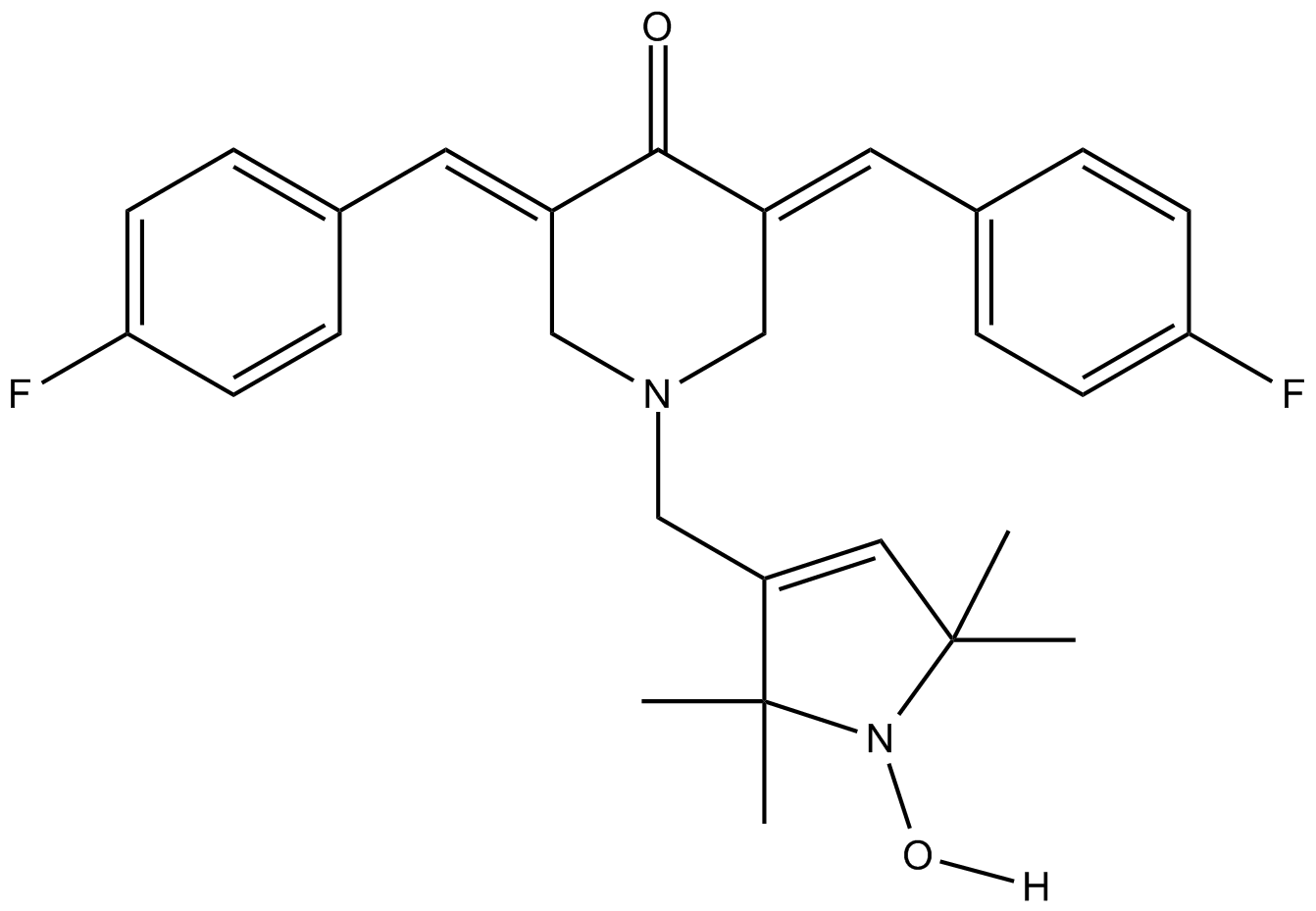
-
GC31692
Homoplantaginin
Hispidulin-7-O-D-glucoside
A flavonoid glycoside with antioxidant and anti-inflammatory activities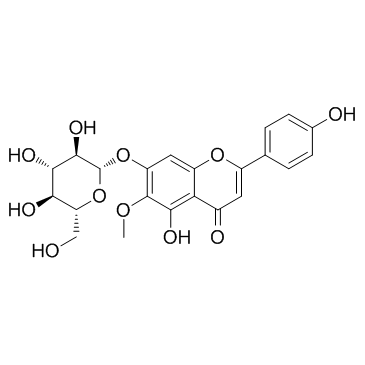
-
GC11574
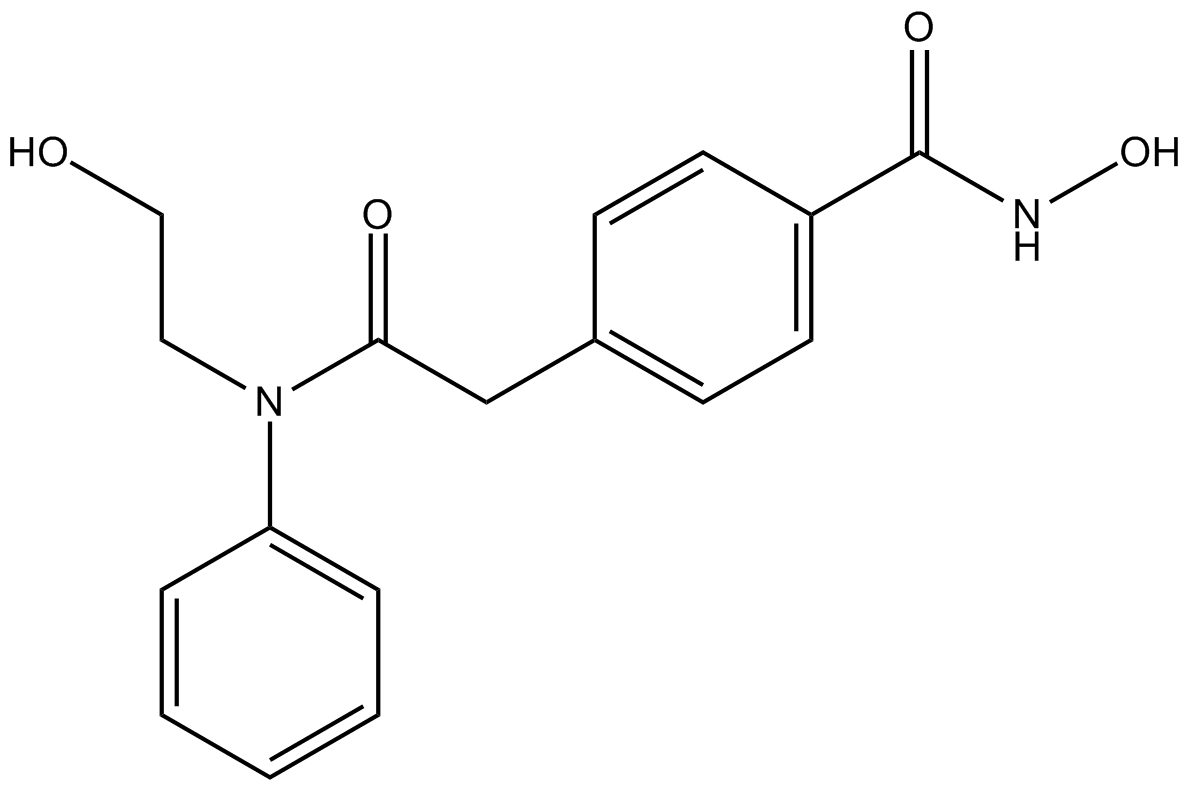
HPOB
HDAC6 inhibitor, potent and selective
-
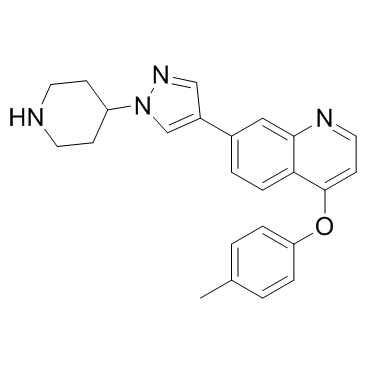
GC32280
HS-1371
A RIPK3 inhibitor
-
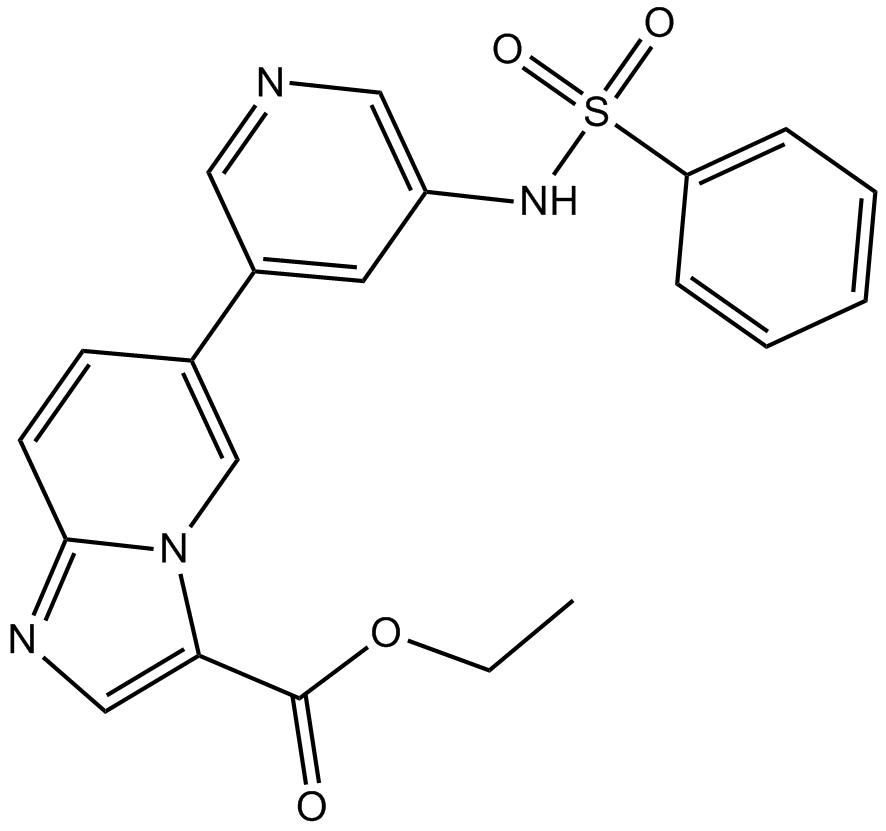
GC16713
HS-173
novel PI3K inhibitor
-
GC11973
HSP990 (NVP-HSP990)
NVP-HSP990
HSP990 (NVP-HSP990) is a potent and selective Hsp90 inhibitor, with IC50 values of 0.6, 0.8, and 8.5 nM for Hsp90α, Hsp90β, and Grp94, respectively.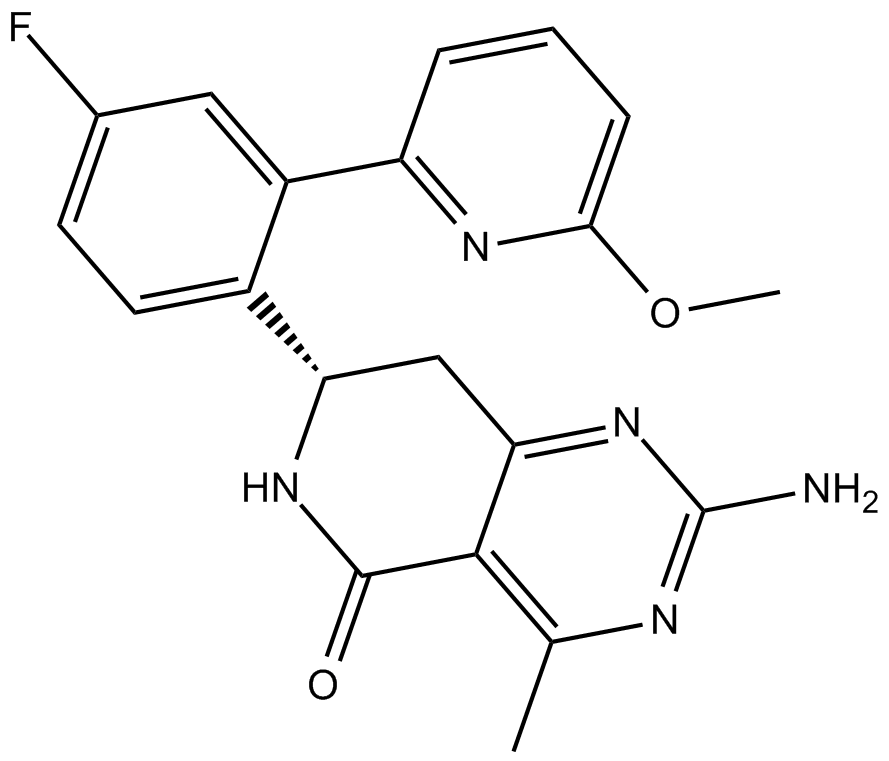
-
GC13074
HU 211
NMDA antagonist, novel and non-competitive
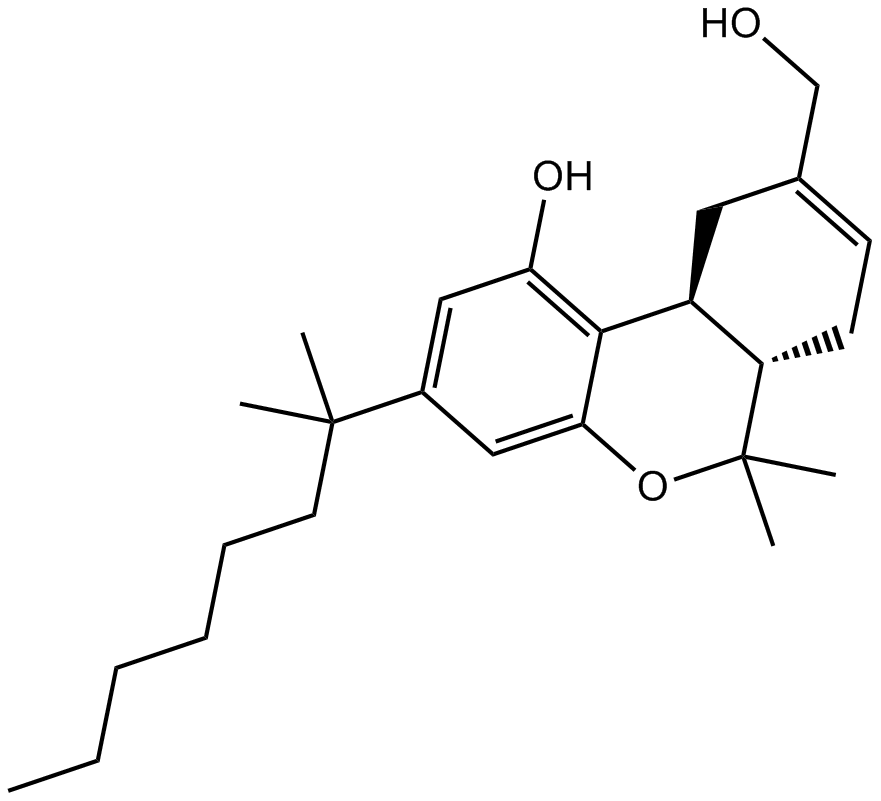
-
GC68404
Human PD-L1 inhibitor IV
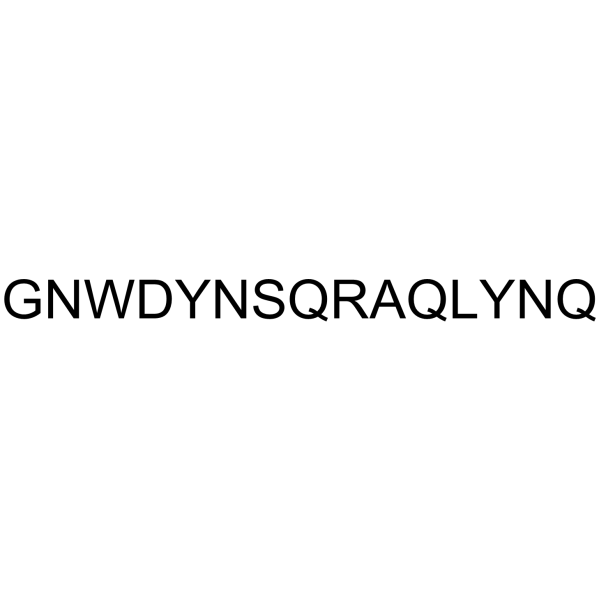
-
GC64485
HXR9 hydrochloride
HXR9 hydrochloride is a cell-permeable peptide and a competitive antagonist of HOX/PBX interaction. HXR9 hydrochloride antagonizes the interaction between HOX and a second transcrip-tion factor (PBX), which binds to HOX proteins in paralogue groups1 to 8. HXR9 hydrochloride selectively decreases cell proliferation and promotes apoptosis in cells with a high level of expression of the HOXA/PBX3 genes, such as MLL-rearranged leukemic cells.
-
GC36270
Hydrolyzed Fumonisin B1
Aminopentol
Hydrolyzed Fumonisin B1 (Aminopentol) is the backbone and main hydrolysis product of the mycotoxin Fumonisin B1.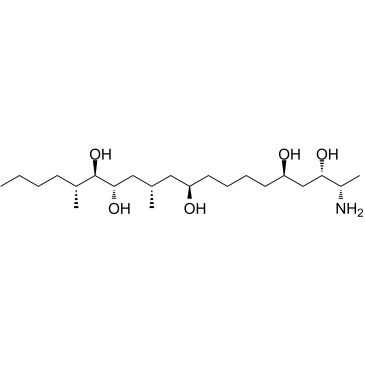
-
GC49130
Hydroxy Celecoxib
An inactive metabolite of celecoxib
-
GC16843
Hydroxyurea
NCI C04831, NSC 32065
DNA synthesis inhibitor
-
GN10217
Hypaconitine
-
GN10302
Hypericin
NSC 407313
-
GC36283
Hypocrellin B
A fungal metabolite
-
GC49479
Hypoxanthine-d4
6-Hydroxypurine-d4
An internal standard for the quantification of hypoxanthine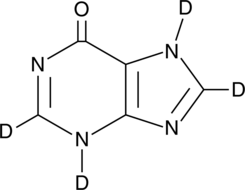
-
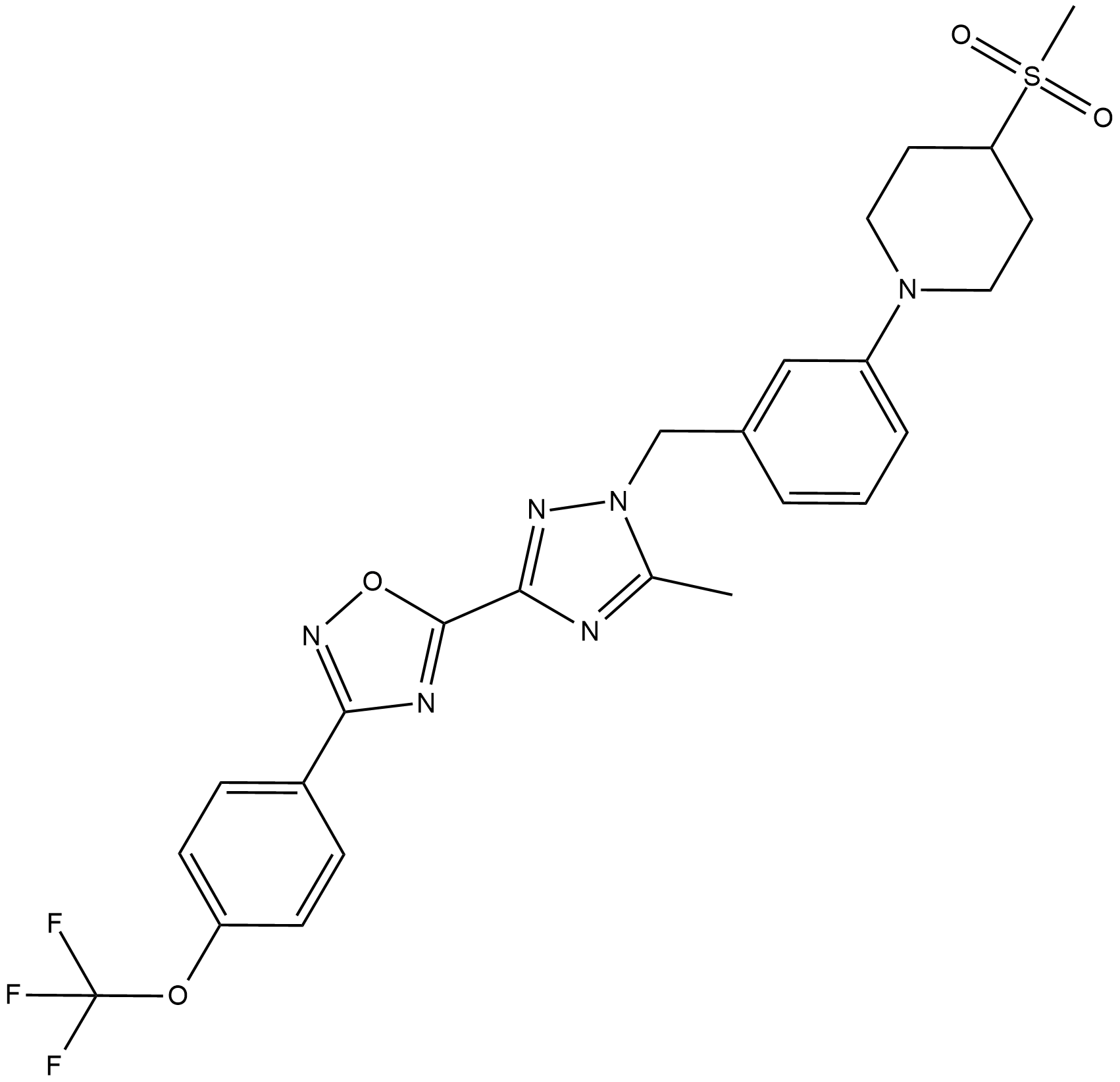
GC19194
IACS-10759
IACS-10759 (IACS-010759) is an oxidative phosphorylation inhibitor, IACS-10759 is a potent inhibitor of complex I of oxidative phosphorylation ( OXPHOS )
-
GC36286
IACS-10759 Hydrochloride
IACS-10759 hydrochloride
IACS-10759 Hydrochlorideis an orally active, potent mitochondrial complex I of oxidative phosphorylation (OXPHOS) inhibitor. IACS-10759 Hydrochlorideinhibits proliferation and induces apoptosis in models of brain cancer and acute myeloid leukemia (AML) reliant on OXPHOS. IACS-10759 Hydrochloride has the potential for relapsed/refractory AML and solid tumors research.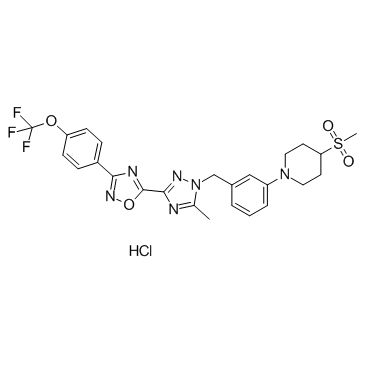
-
GC19197
IB-MECA
CF-101, IB-MECA
IB-MECA (IB-MECA) is a first-in-class, orally active and selective A3 adenosine receptor (A3AR) agonist.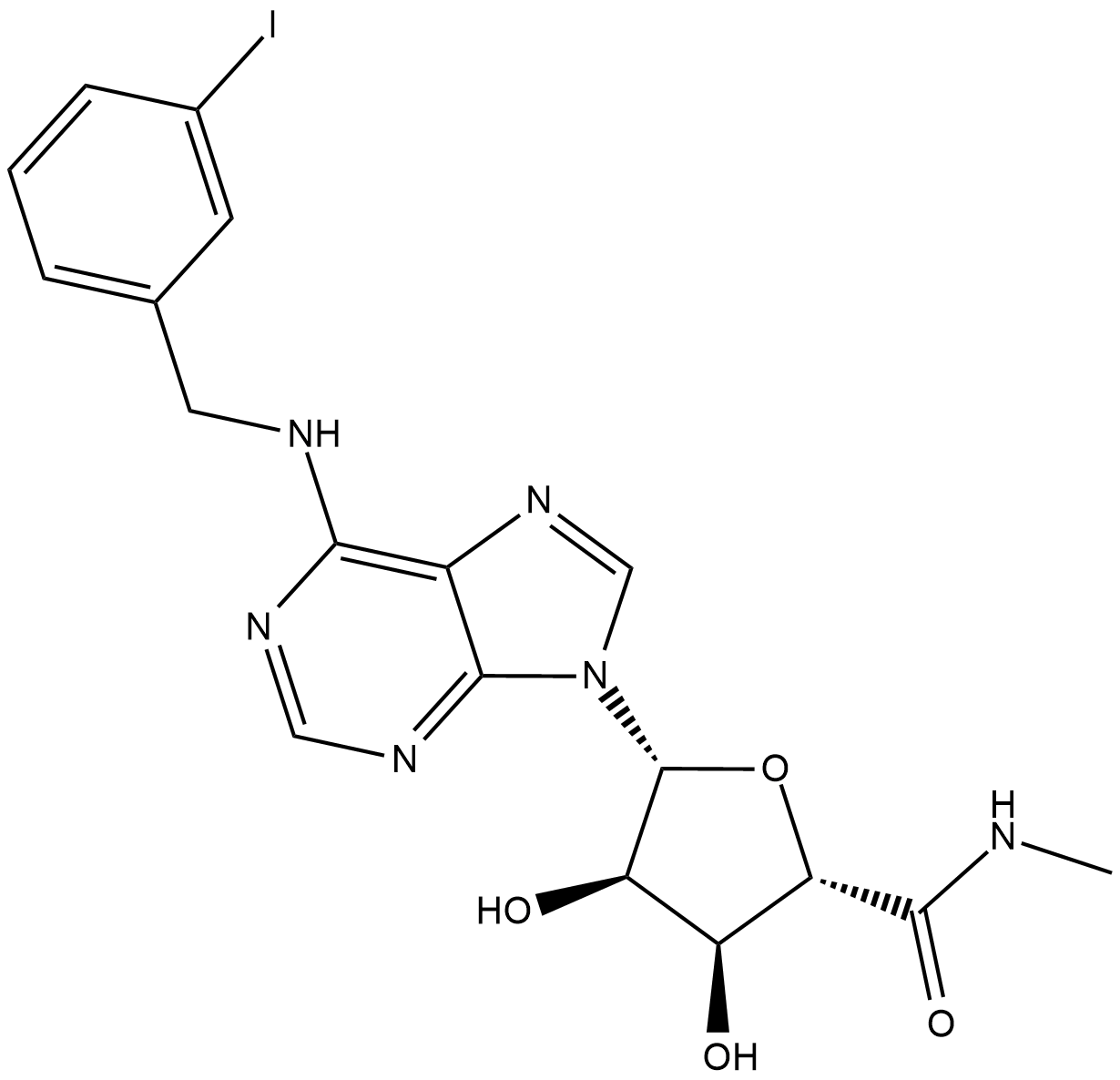
-
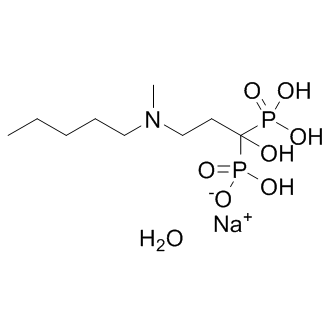
GC13246
Ibandronate sodium
Ibandronate sodium is a highly potent nitrogen-containing bisphosphonate used for the treatment of osteoporosis.
-
GC19195
Iberdomide
CC-220
Iberdomide (CC-220) is a cereblon modulator with an IC50 of 60 nM.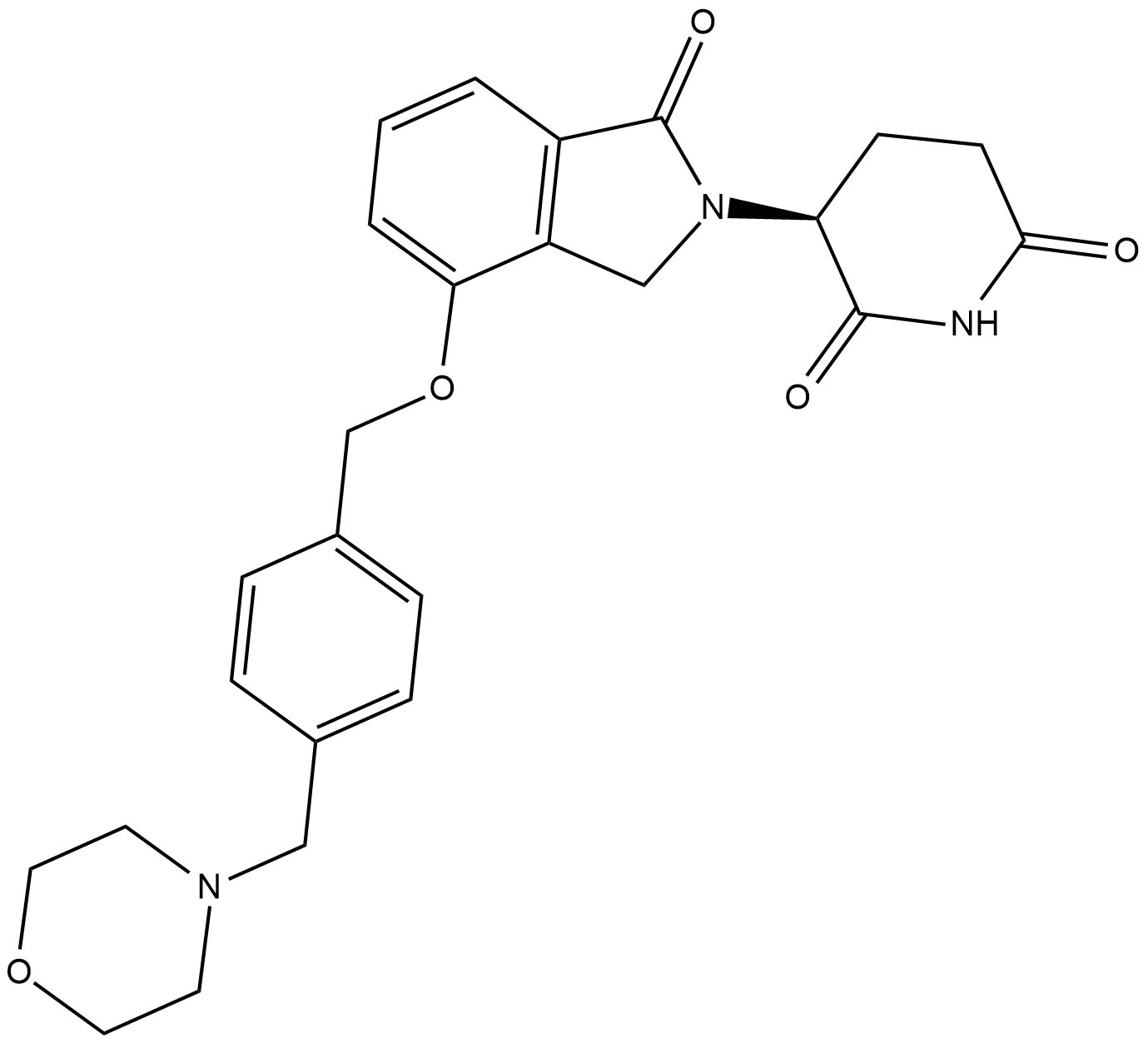
-
GC17287
Iberin
NSC 321801
induces the expression of phase II detoxification enzymes, including quinone reductase and glutathione S-transferase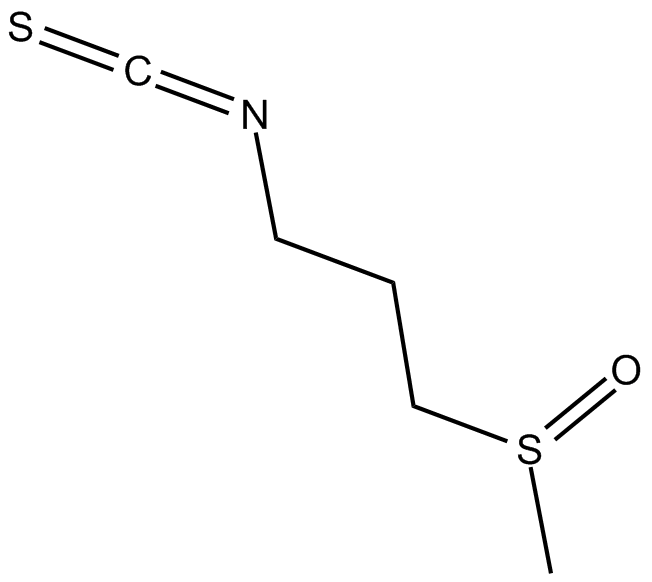
-
GC14503
IC261
SU5607
CK1 inhibitor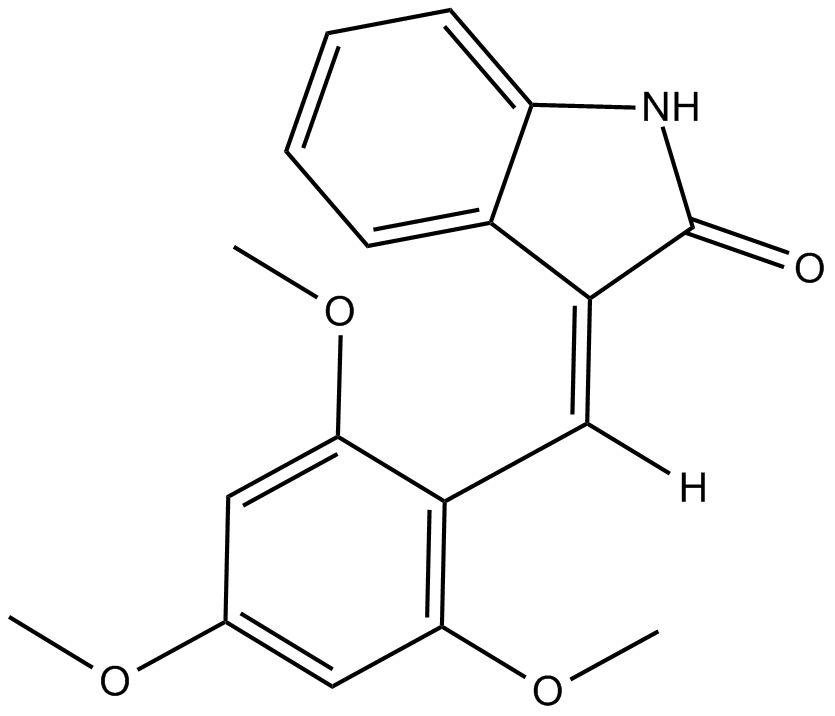
-
GC38096
Icaritin
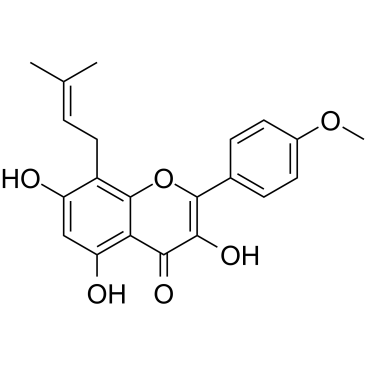
Icaritin is a prenylflavonoid derivative obtained from the Epimedium genus.
-
GC32762
Icaritin (Anhydroicaritin)
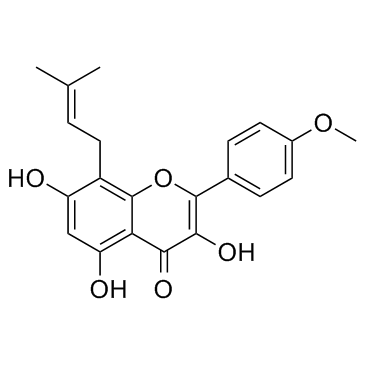
Icaritin (Anhydroicaritin) (Anhydroicaritin) is a prenylflavonoid derivative from Epimedium brevicornuMaxim. and potently inhibits proliferation of K562 cells (IC50 of 8 μM) and primary CML cells (IC50 of 13.4 μM for CML-CP and 18 μM for CML-BC). Icaritin (Anhydroicaritin) can regulate MAPK/ERK/JNK and JAK2/STAT3 /AKT signalings, also enhances osteogenesis[3.
-
GC62626
ICCB-19 hydrochloride
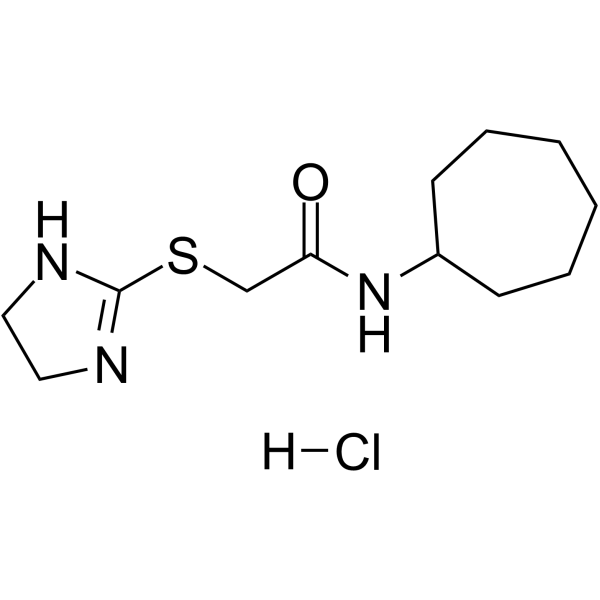
ICCB-19 hydrochloride is a TRADD (TNFRSF1A associated via death domain) inhibitor.
-
GC61414
ICCB280
ICCB280 is a potent inducer of C/EBPα. ICCB280 exhibits anti-leukemic properties including terminal differentiation, proliferation arrest, and apoptosis through activation of C/EBPα and affecting its downstream targets (such as C/EBPε, G-CSFR and c-Myc).
-
GC19198
iCRT3
iCRT3 is an inhibitor of both Wnt and β-catenin-responsive transcription.
-
GC19411
IITZ-01
IITZ-01 is a potent lysosomotropic autophagy inhibitor with single-agent antitumor activity, with an IC50 of 2.62 μM for PI3Kγ.
-
GC43894
IKK2 Inhibitor VI
5-Phenyl-2-ureidothiophene-3-carboxylic Acid Amide
Inhibitor of NF-κB kinase 2 (IKK2, also known as IKKβ) acts as part of an IKK complex in the canonical NF-κB pathway, phosphorylating inhibitors of NF-κB (IκBs) to initiate signaling.
-
GC38566
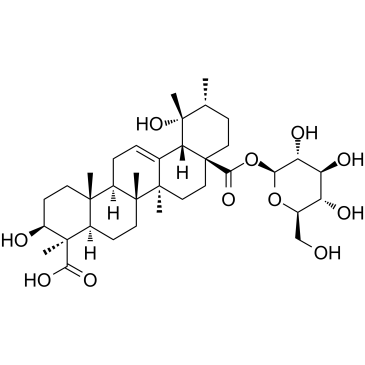
Ilexsaponin A
Ilexsaponin A, isolated from the root of Ilex pubescens, attenuates ischemia-reperfusion-induced myocardial injury through anti-apoptotic pathway.
-
GC41340
Illudin M
DR-15977, (-)-Illudin M, NSC 400978, NSC 626370
Illudin M is a cytotoxic sesquiterpene from the fungus O.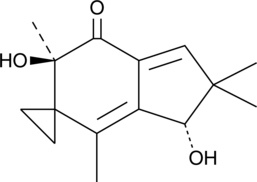
-
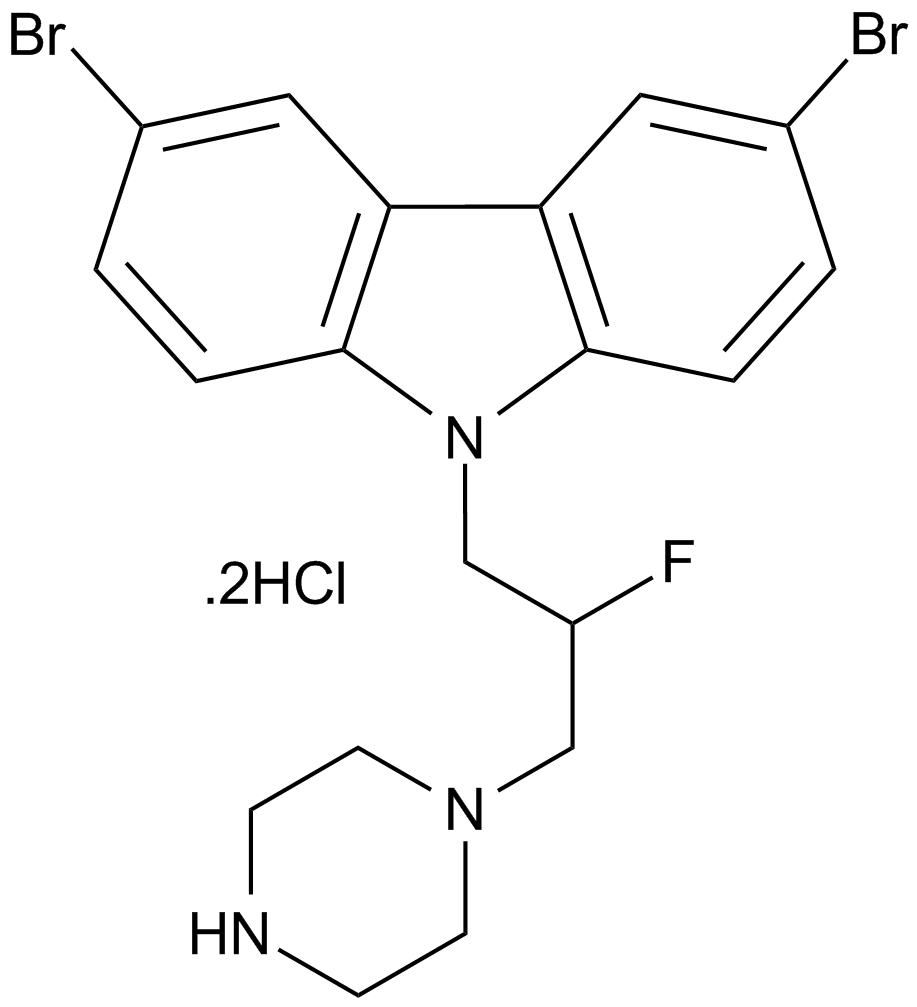
GC12046
iMAC2
-
GC65168
Imifoplatin
PT-112
Imifoplatin (PT-112) is a platinum-based agent belonging to the phosphaplatin family. Imifoplatin exhibits antineoplastic activity.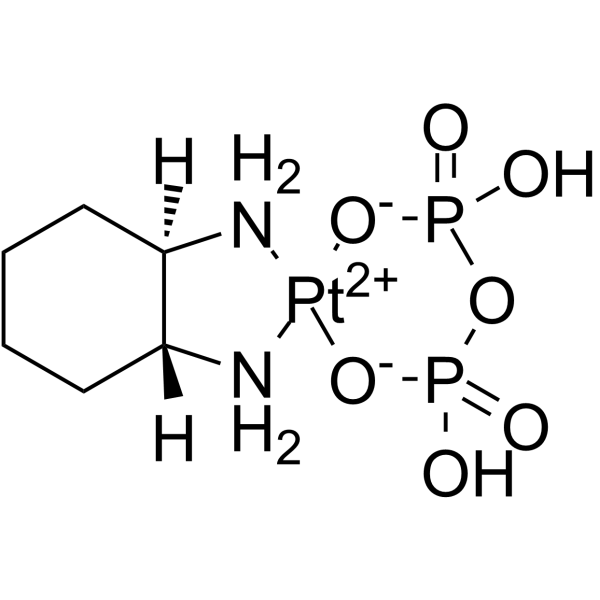
-
GC19199
Importazole
IPZ
Importazole is a small molecule inhibitor of the nuclear transport receptor importin-β.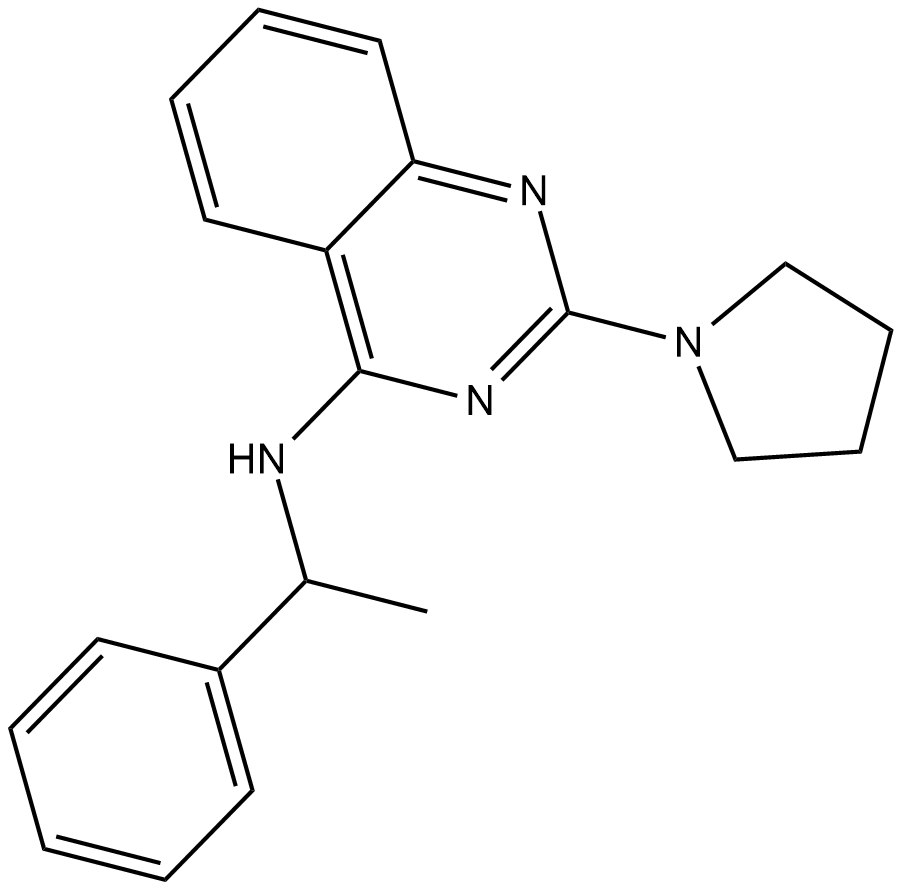
-
GC14755
Inauhzin
INZ
SIRT1 inhibitor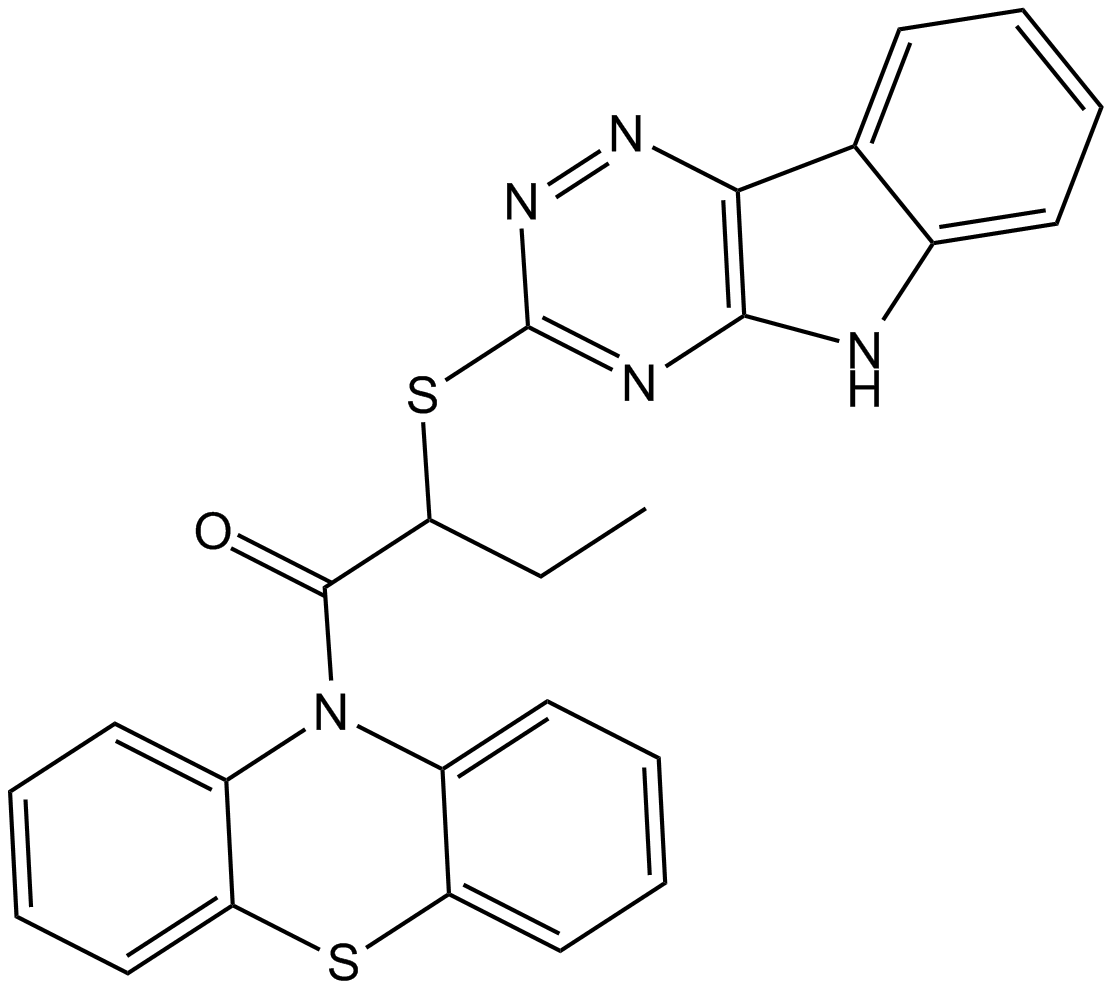
-
GC19200
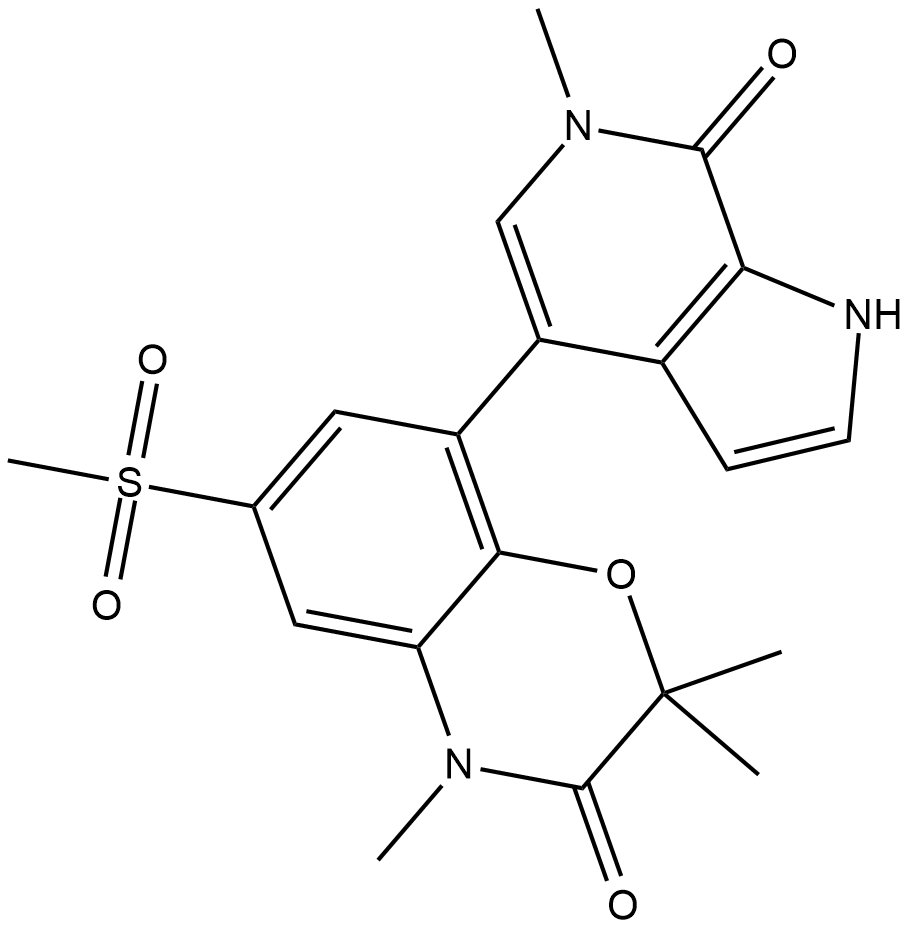
INCB-057643
INCB-057643 is a novel, orally bioavailable BET inhibitor.
-
GC17866
INCB28060
Capmatinib, INC 280
INCB28060 (INC280; INCB28060) is a potent, orally active, selective, and ATP competitive c-Met kinase inhibitor (IC50=0.13 nM). INCB28060 can inhibit phosphorylation of c-MET as well as c-MET pathway downstream effectors such as ERK1/2, AKT, FAK, GAB1, and STAT3/5. INCB28060 potently inhibits c-MET-dependent tumor cell proliferation and migration and effectively induces apoptosis. Antitumor activity. INCB28060 is largely metabolized by CYP3A4 and aldehyde oxidase.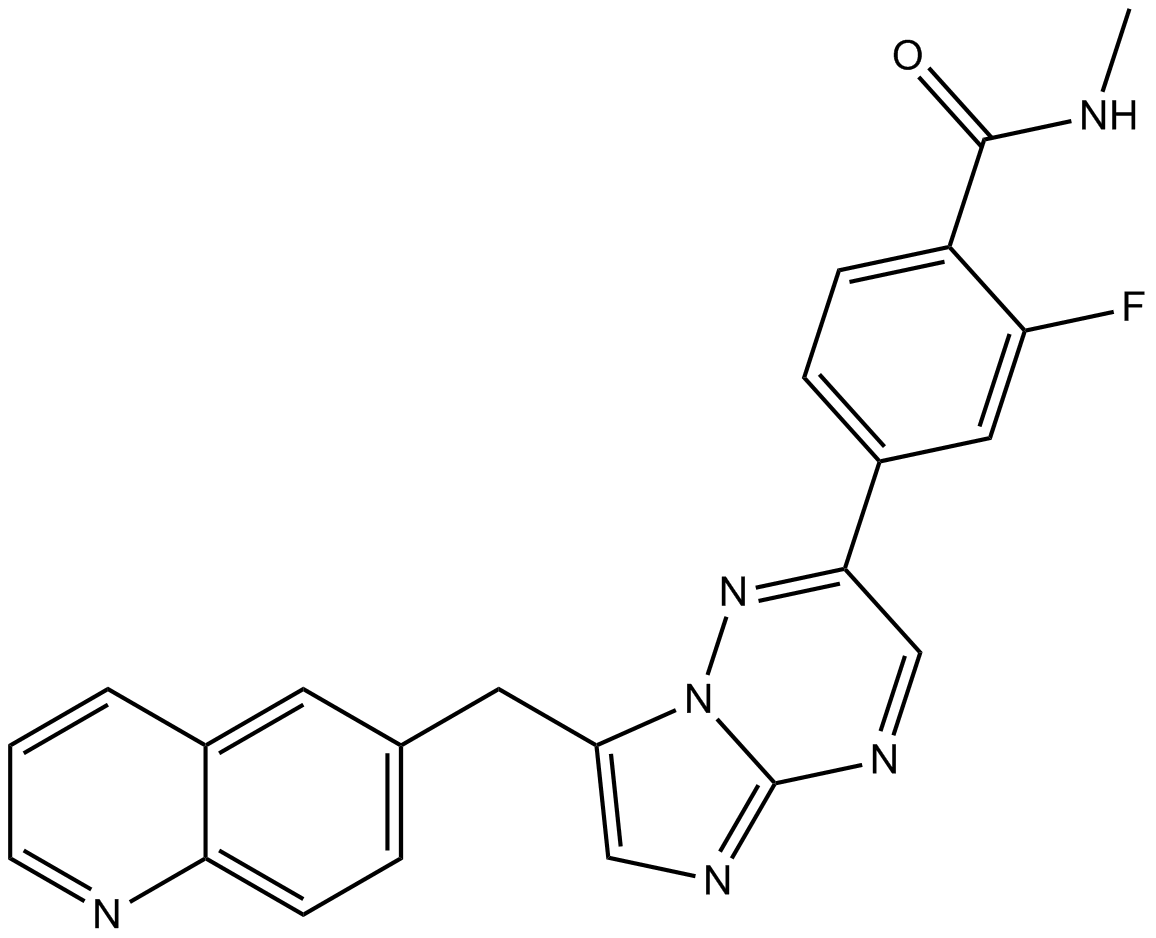
-
GC38213
Incensole Acetate
Incensol Acetate
Incensole acetate is a main constituent of Boswellia carterii resin, has neuroprotective effects against neuronal damage in traumatic and ischemic head injury.
-
GC36308
Indibulin
D-24851, ZIO 301
An inhibitor of microtubule assembly
-
GC12975
Indirubin
C.I. 73200, Couroupitine B, Indigopurpurin, NSC 105327
A cyclin-dependent kinases and GSK-3β inhibitor
-
GC49670
Indium (III) thiosemicarbazone 5b
An anticancer agent
-
GC61948
Inecalcitol
Inecalcitol (TX 522), a unique vitamin D3 analog, is an orally active vitamin D receptor (VDR) agonist with a Kd of 0.53 nM. Inecalcitol can induce cell apoptosis and has potent anticancer activities.
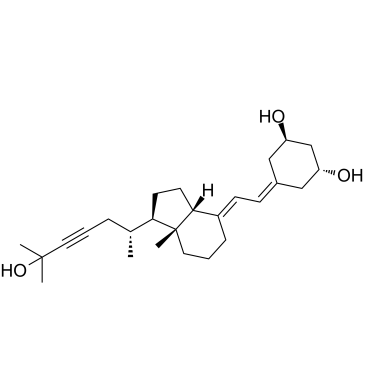
-
GC19533
Infliximab
Infliximab is a chimeric monoclonal IgG1 antibody that specifically binds to TNF-α.

-
GC61776
Ingenol 3,20-dibenzoate
Ingenol 3,20-dibenzoate is a potent protein kinase C (PKC) isoform-selective agonist.

-
GC10334
INH6
Hec1/Nek2 inhibitor, potent
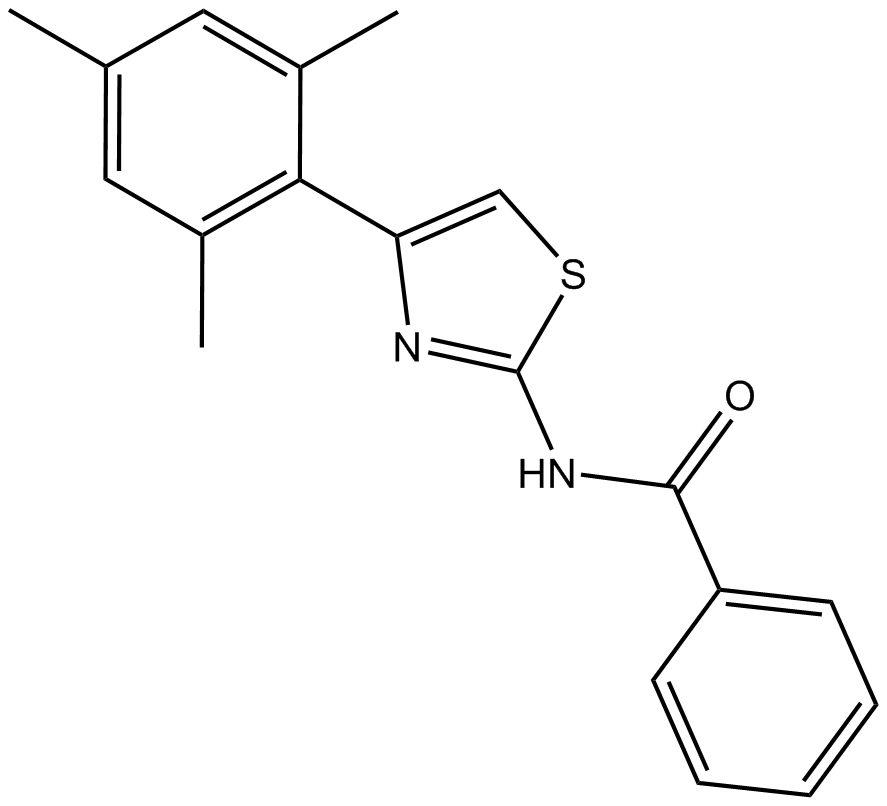
-
GC48387
Inostamycin A
A bacterial metabolite with anticancer activity
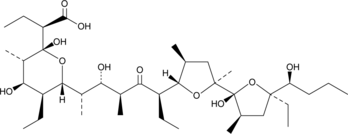
-
GC52472
Inostamycin A (sodium salt)
Inostamycin
A bacterial metabolite with anticancer activity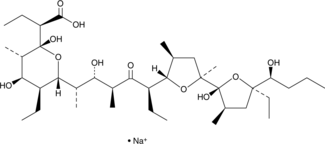
-
GC15148
Ionomycin calcium salt
Lonomycin is a selective calcium ionophore derived from S. conglobatus that mobilizes intracellular calcium stores.
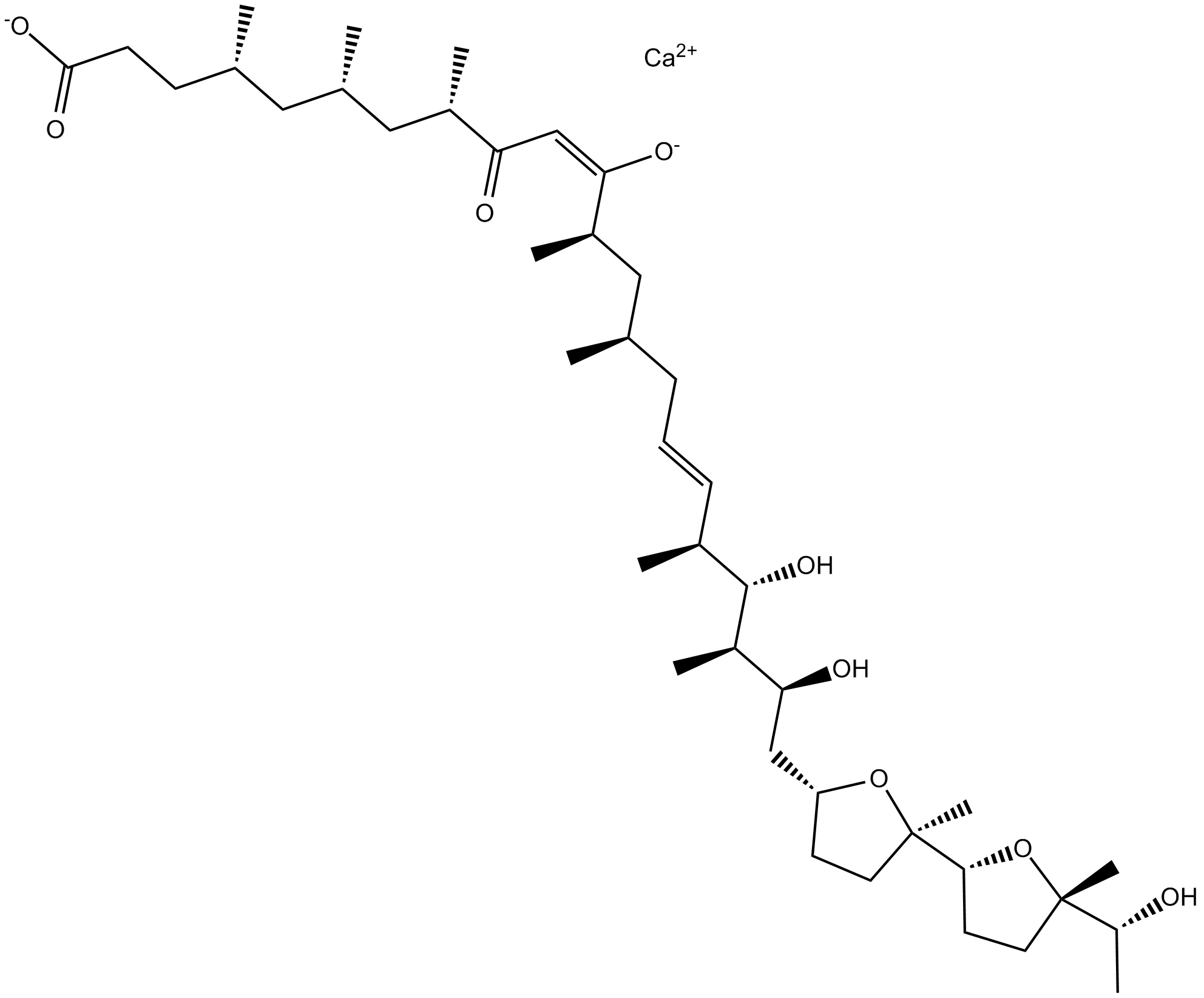
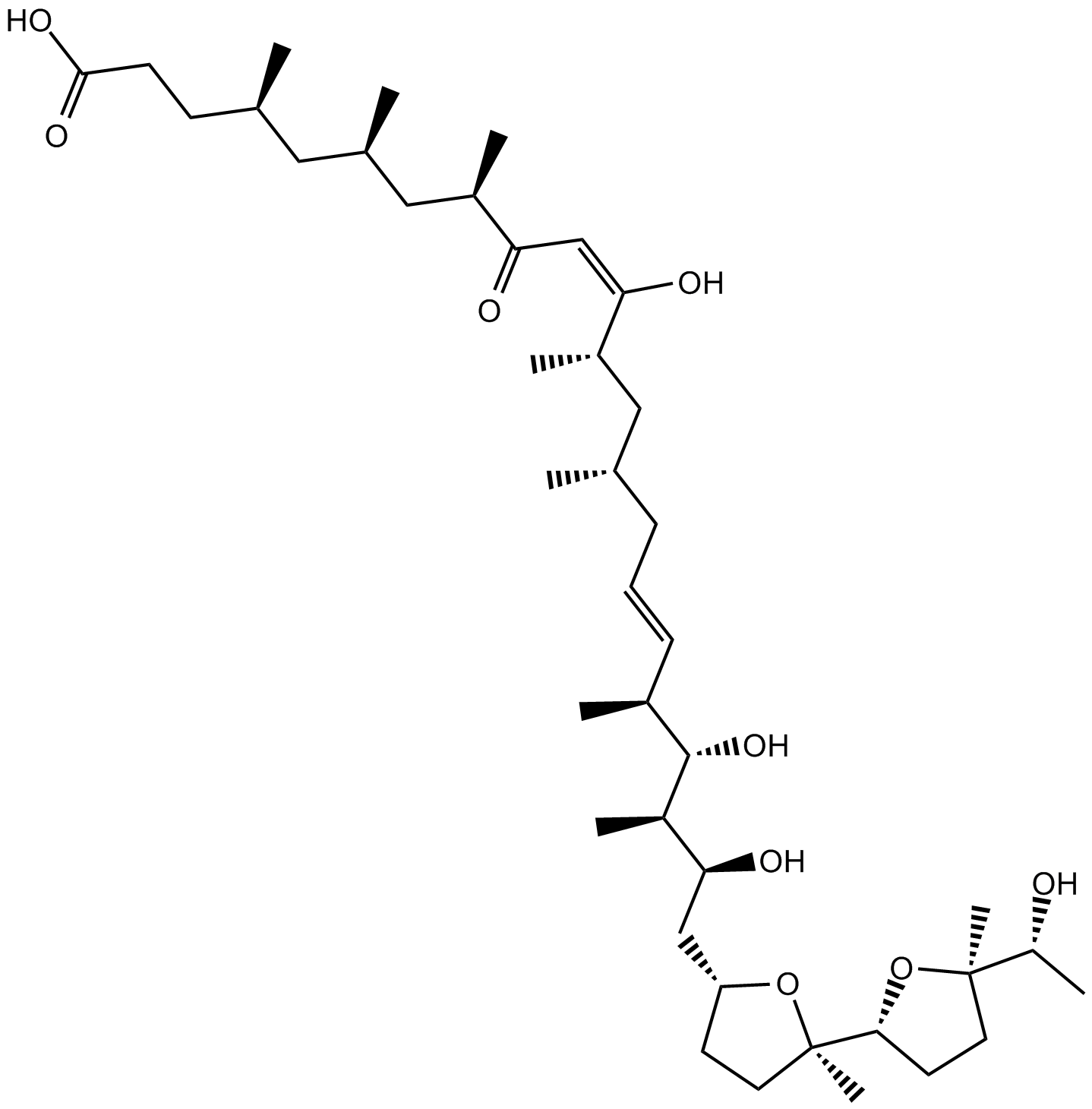
-
GC15446
Ionomycin free acid
Ionomycin free acid (SQ23377) is a potent, selective calcium ionophore and an antibiotic produced by Streptomyces conglobatus.
-
GC46027
IP7e
Isoxazolo-pyridinone 7e
An activator of Nurr1 signaling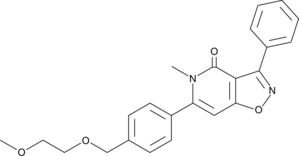
-
GC52175
IQA
CGP 029482
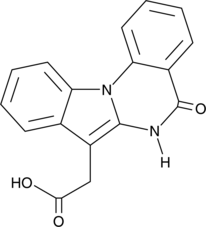
-
GC10467
Irbesartan
BMS 186295, SR 47436
Angiotensin II inhibitor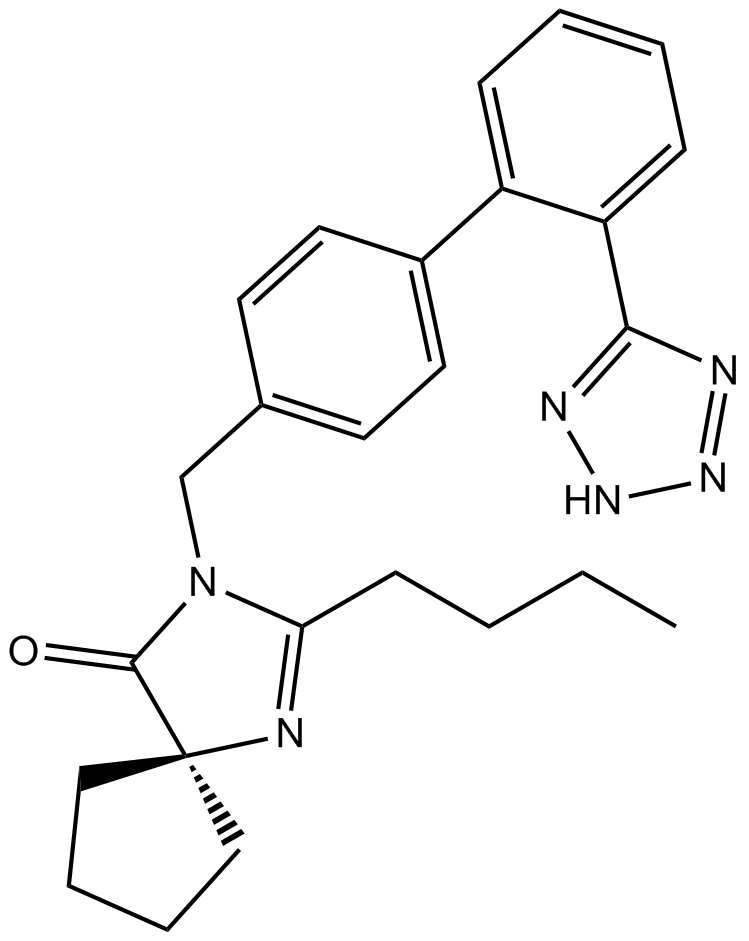
-
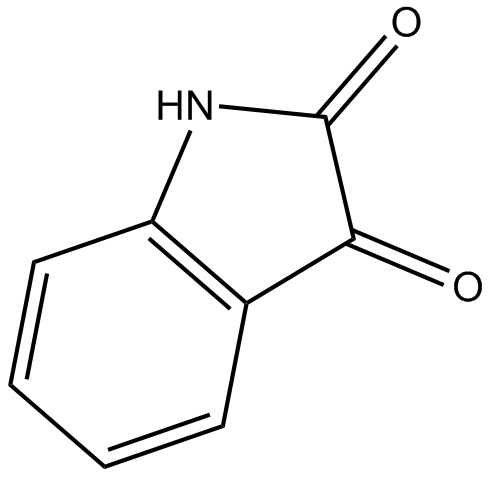
GC12962
Isatin
endogenous monoamine oxidase inhibitor
-
GC63386
Isatuximab
Isatuximab is a monoclonal antibody targeting the transmembrane receptor and ectoenzyme CD38, a protein highly expressed on hematological malignant cells, including those in multiple myeloma (MM). Isatuximab has antitumor activity via multiple biological mechanisms, including antibody-dependent cellular-mediated cytotoxicity, complement-dependent cytotoxicity, antibody-dependent cellular phagocytosis, and direct induction of apoptosis without crosslinking. Isatuximab also directly inhibits CD38 ectoenzyme activity, which is implicated in many cellular functions.

-
GC64767
ISIS 104838
ISIS?104838?is an antisense oligonucleotide drug that reduces the production of tumor necrosis factor (TNF-alpha),?a substance that contributes to joint pain and swelling in rheumatoid arthritis.

-
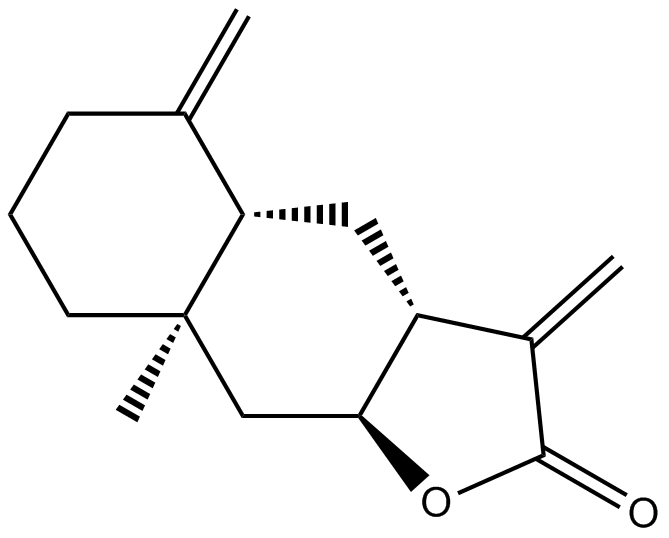
GN10666
Isoalantolactone
(+)-Isoalantolactone, NSC 241036
-
GC13795
Isobavachalcone
Corylifolinin, IBC
A chalcone and flavonoid with diverse biological activities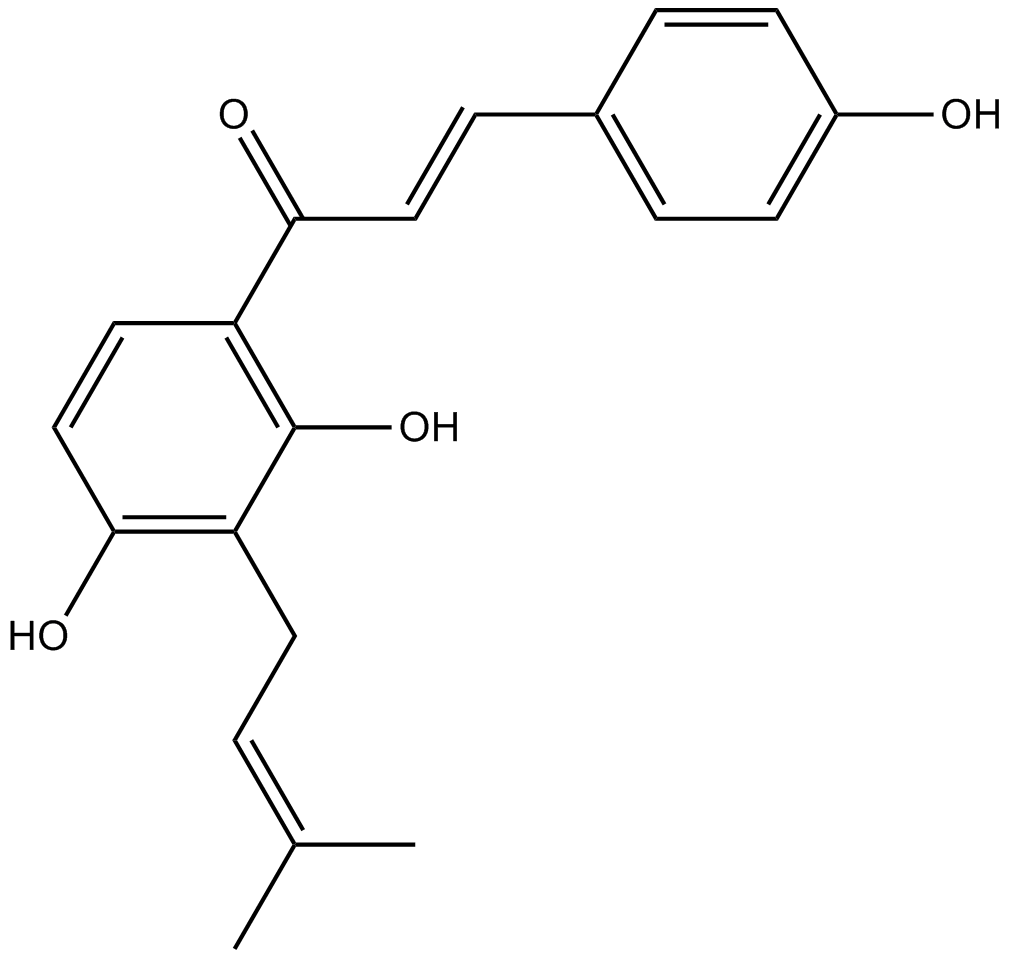
-
GC48900
Isocurcumenol
(+)-Isocurcumenol
A sesquiterpene with diverse biological activities
-
GC40901
Isogarcinol
Cambogin
Isogarcinol is a natural polyisoprenylated benzophenone first isolated from plant species in the genus Garcinia.
-
GC38220
Isoliensinine
-
GN10088
Isoliquiritigenin
GU 17, ILQ, ISL
-
GC60949
Isolongifolene
Isolongifolene ((-)-Isolongifolene) is a tricyclic sesquiterpene isolated from Murraya koenigii.
-
GC48618
Isonanangenine B
SF002-96-1
A drimane sesquiterpene lactone
-
GC36343
Isosilybin A
Isosilybin A, a flavonolignan isolated from silymarin, has anti-prostate cancer (PCA) activity. Isosilybin A inhibits proliferation and induces G1 phase arrest and apoptosis in cancer cells, which activates apoptotic machinery in PCA cells via targeting Akt-NF-κB-androgen receptor (AR) axis.
-
GC36344
Isosilybin B
Isosilybin B, a flavonolignan isolated from silymarin, has anti-prostate cancer (PCA) activity via inhibiting proliferation and inducing G1 phase arrest and apoptosis. Isosilybin B causes androgen receptor (AR) degradation.
-
GC43922
Isovaleryl-L-carnitine (chloride)
CAR 5:0, C5:0 Carnitine, L-Carnitine isovaleryl ester, L-Isovalerylcarnitine
Isovaleryl-L-carnitine (chloride), a product of the catabolism of L-leucine, is a potent activator of the Ca2+-dependent proteinase (calpain) of human neutrophils.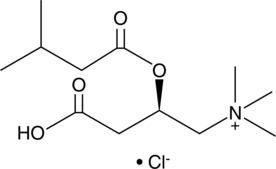
-
GC13394
Ispinesib (SB-715992)
SB-715992
Ispinesib (SB-715992) is a specific inhibitor of kinesin spindle protein (KSP), with a Ki app of 1.7 nM.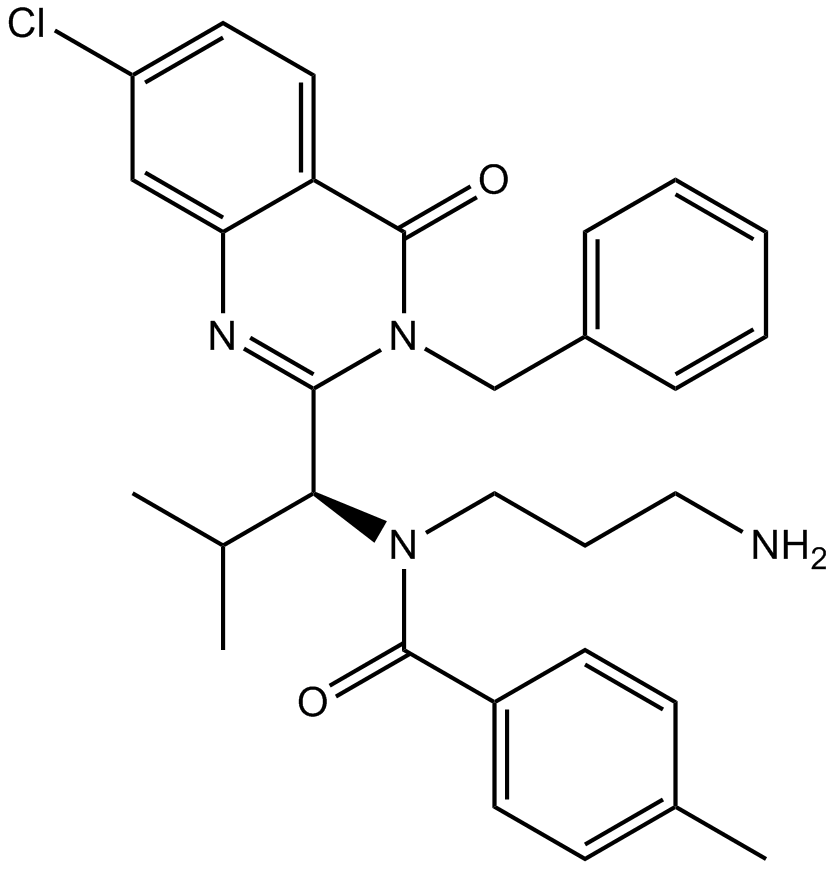
-
GC16869
Ixabepilone
Azaepothilone B, BMS-247550
A broad-spectrum anticancer agent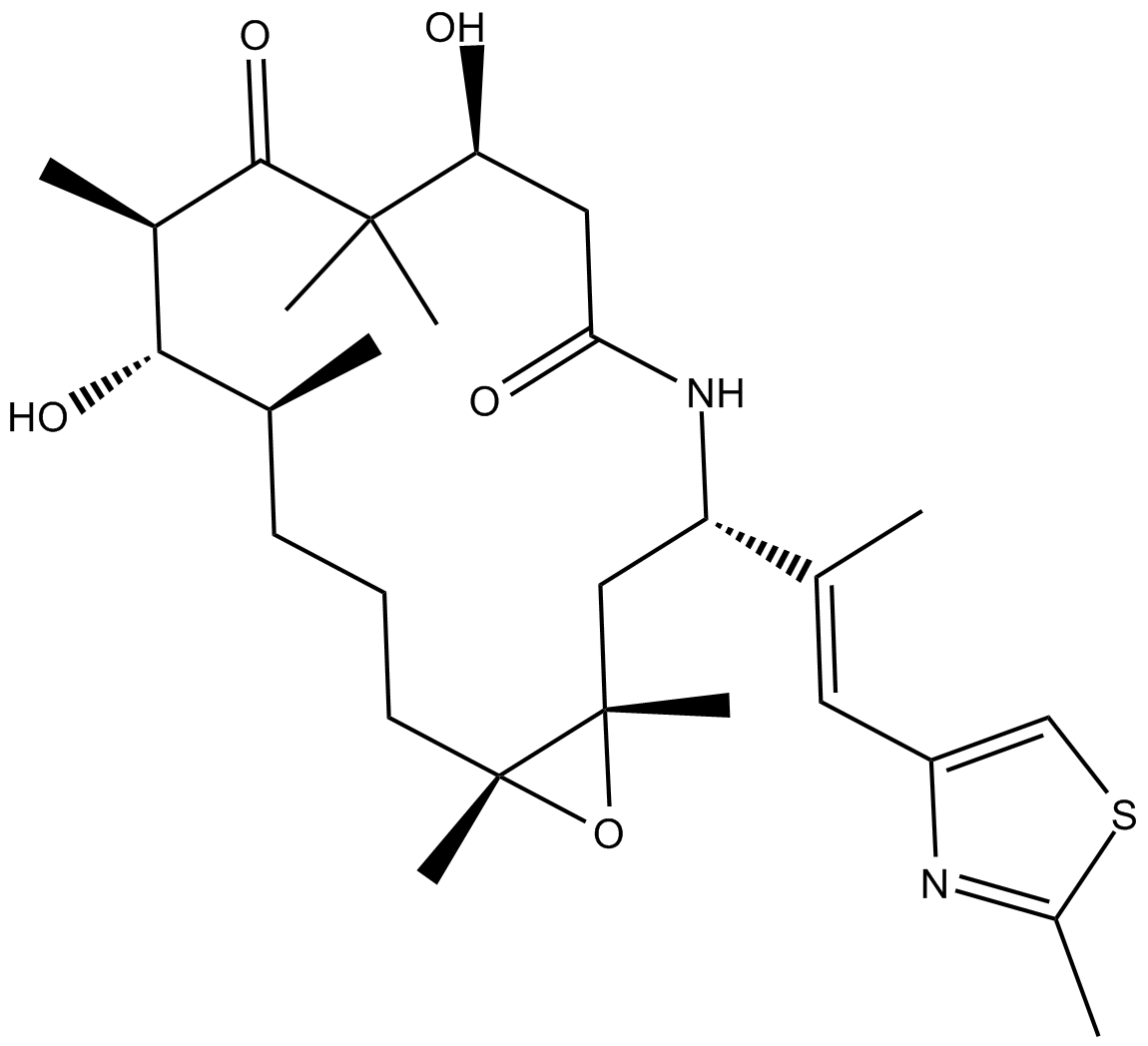
-
GC41645
Jacaric Acid
8(Z),10(E),12(Z)-Octadecatrienoic Acid
Jacaric acid is a conjugated polyunsaturated fatty acid first isolated from seeds of Jacaranda plants.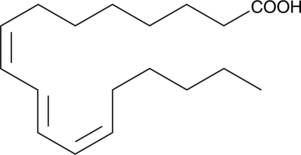
-
GN10645
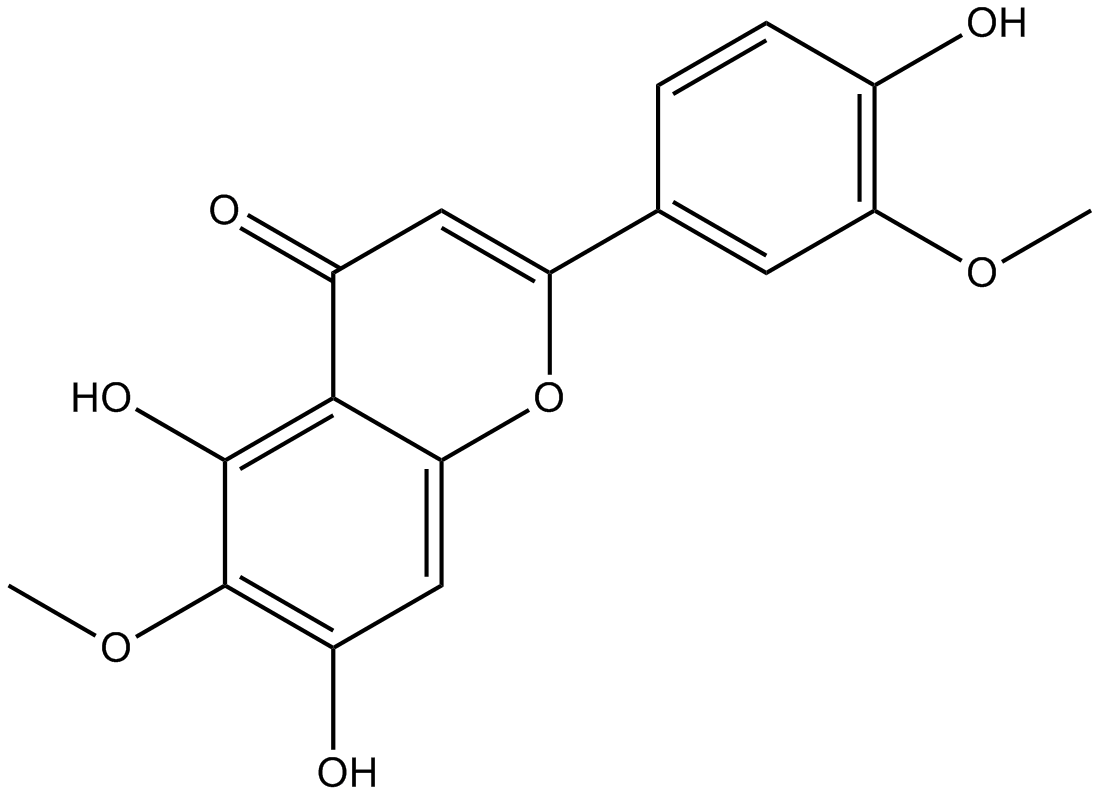
Jaceosidin
-
GC62665
JAK2/FLT3-IN-1 TFA
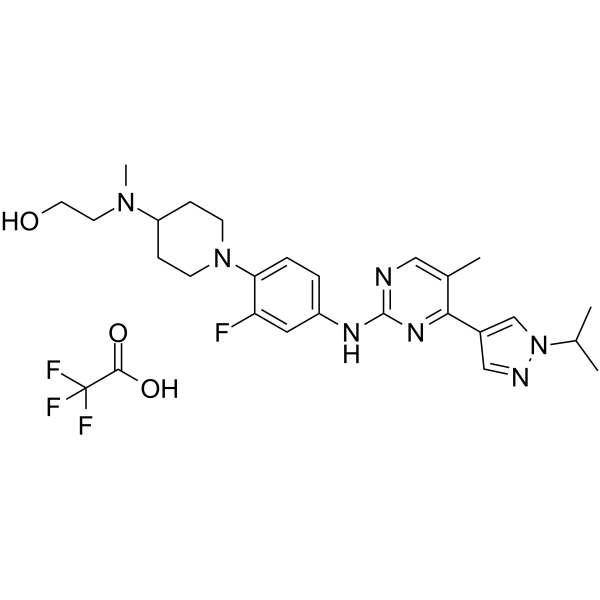
JAK2/FLT3-IN-1 (TFA) is a potent and orally active dual JAK2/FLT3 inhibitor with IC50 values of 0.7 nM, 4 nM, 26 nM and 39 nM for JAK2, FLT3, JAK1 and JAK3, respectively. JAK2/FLT3-IN-1 (TFA) has anti-cancer activity.
-
GC34634
JG-98
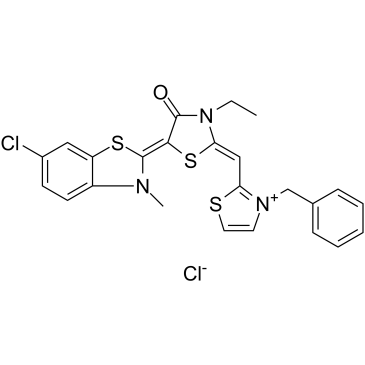
JG-98, an allosteric heat shock protein 70 (Hsp70) inhibitor, which binds tightly to a conserved site on Hsp70 and disrupts the Hsp70-Bag3 interaction. JG-98 shows anti-cancer activities affecting both cancer cells and tumor-associated macrophages.
-
GC15603
JIB-04
JHDM Inhibitor VII, NSC 693627
Jumonji histone demethylase inihibitor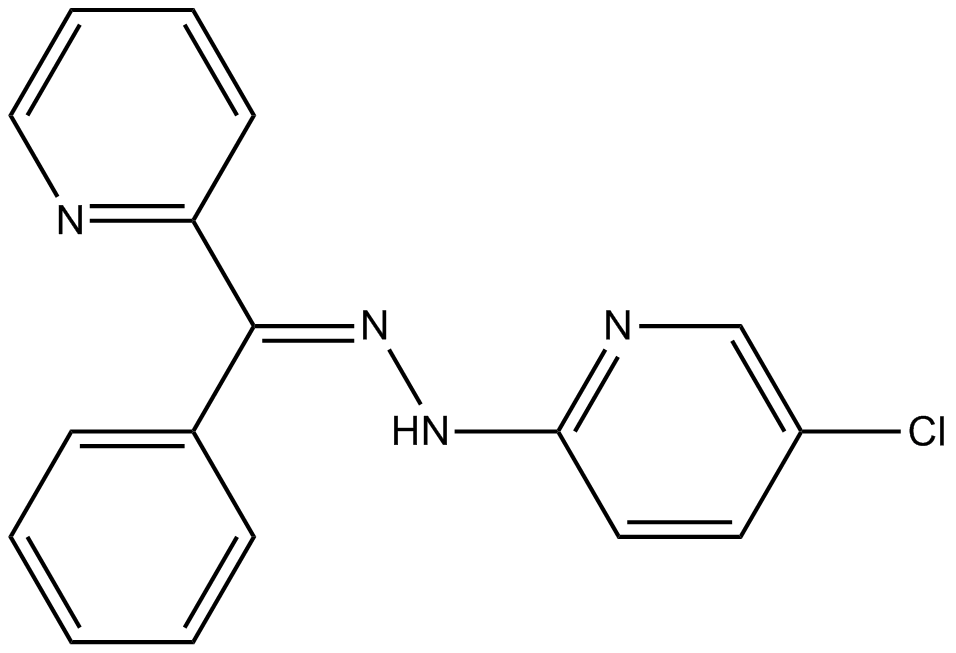
-
GC12117
JNJ-26854165 (Serdemetan)
Serdemetan
An antagonist of MDM2 action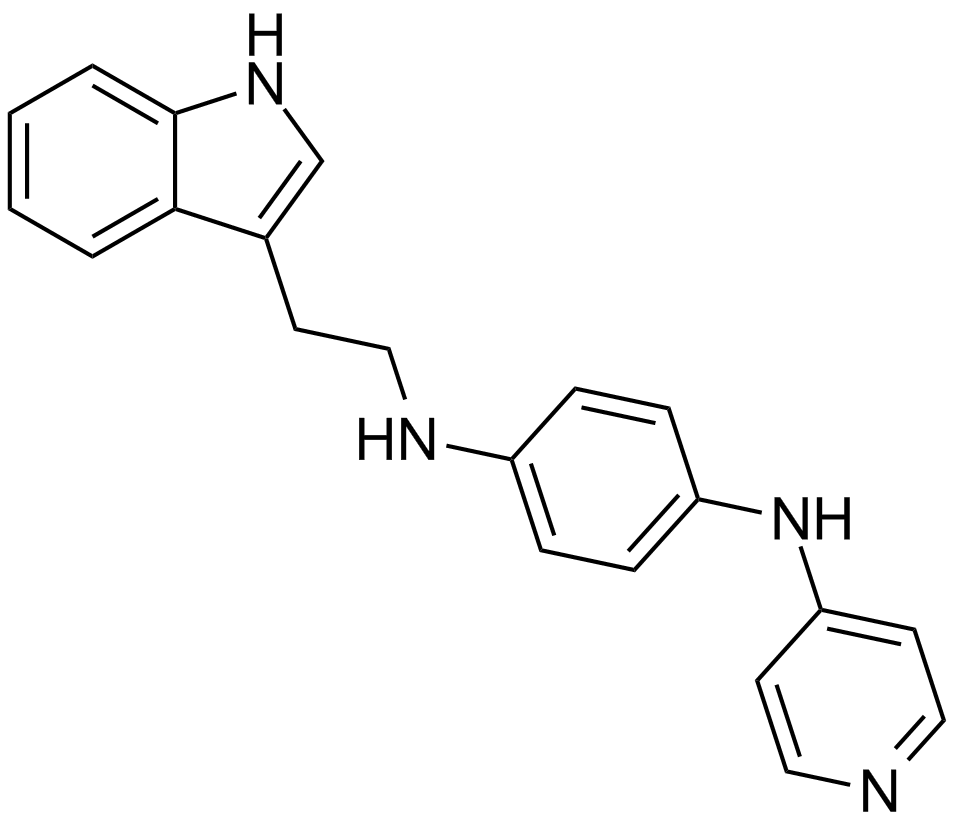
-
GC12612
JNJ-7706621
JNJ7706621, JNJ 7706621
A dual inhibitor of CDKs and Aurora kinases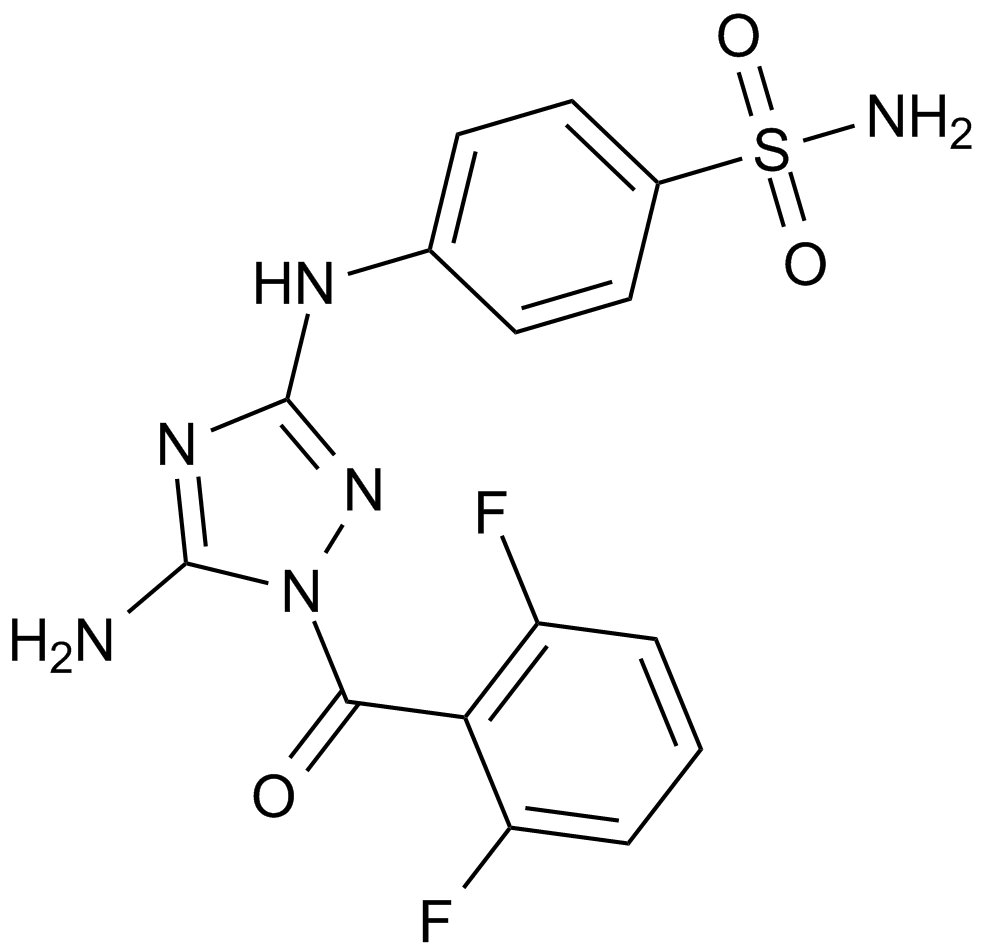
-
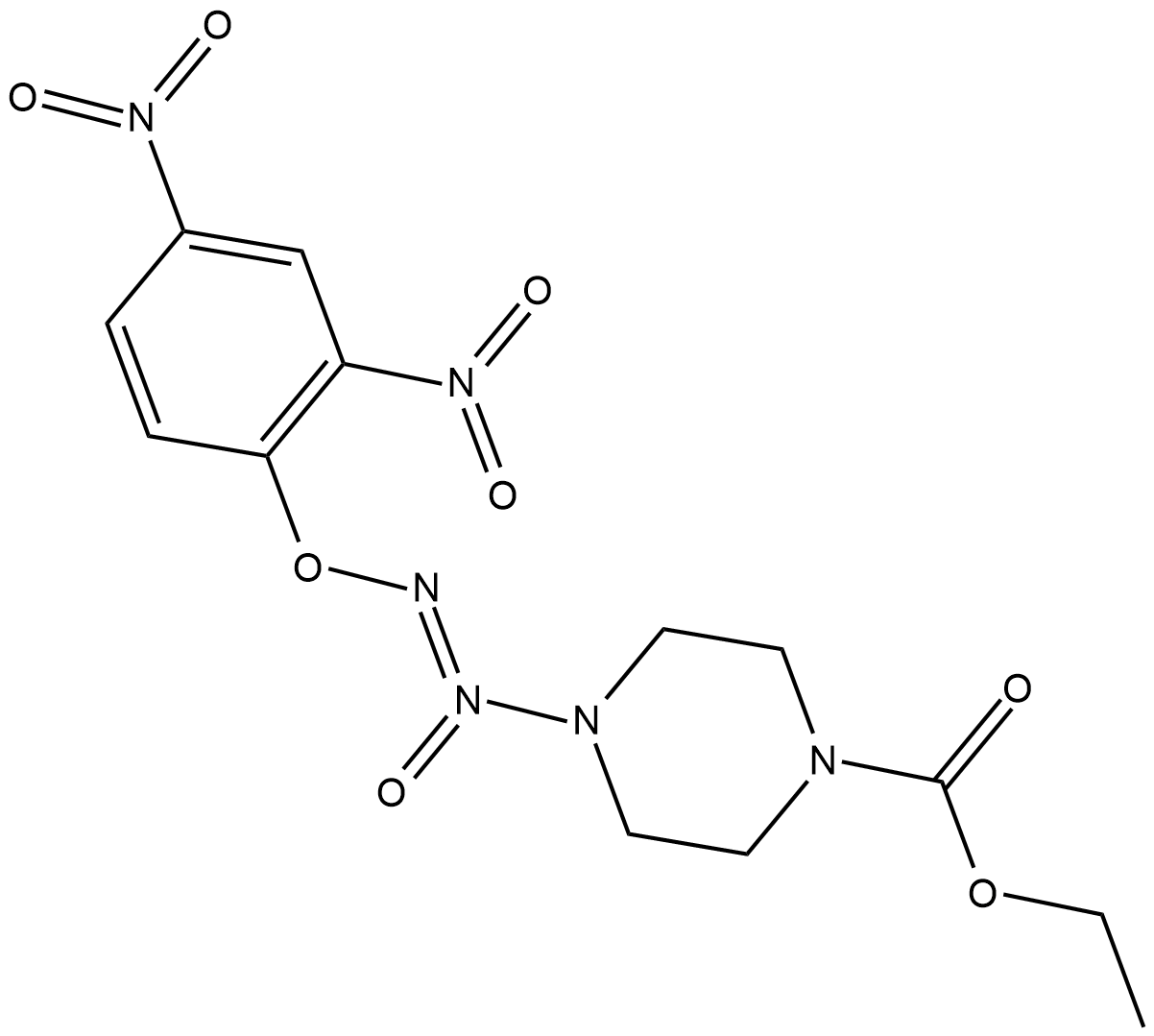
GC18734
JS-K
JS-K is a nitric oxide (NO) donor that reacts with glutathione to generate NO at physiological pH.
-
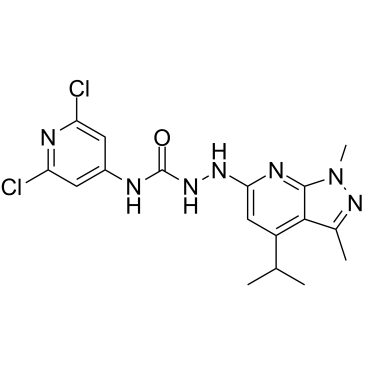
GC38082
JTE-013
A selective S1P2 receptor antagonist
-
GC50280
JX 06
NSC 402538
Potent and selective pyruvate dehydrogenase kinase (PDK) 1/2/3 inhibitor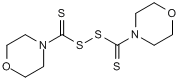
-
GC16167
K 858
K858 Racemic is an ATP-uncompetitive inhibitor of kinesin Eg5 with an IC50 of 1.3 μM.
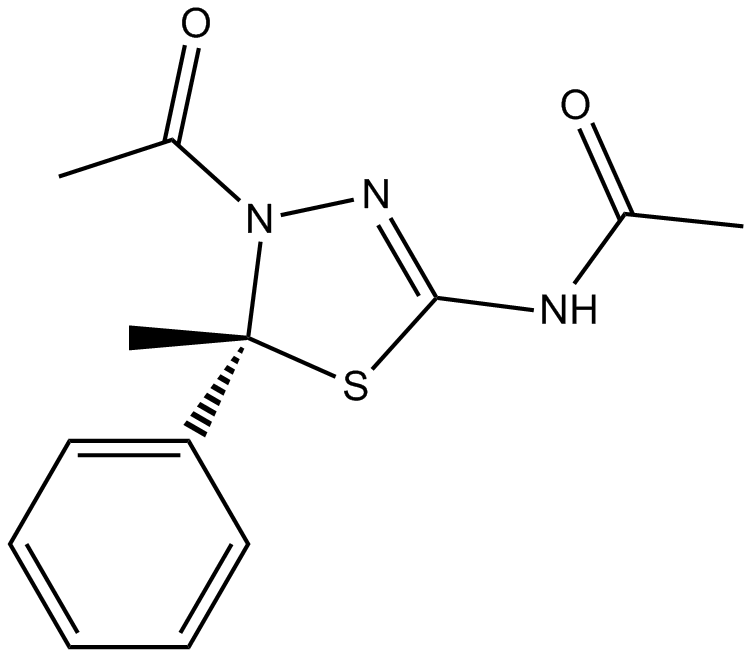
-
GC15281
K-252c
Staurosporinone
Protein kinase inhibitor