Other Apoptosis
Products for Other Apoptosis
- Cat.No. Product Name Information
-
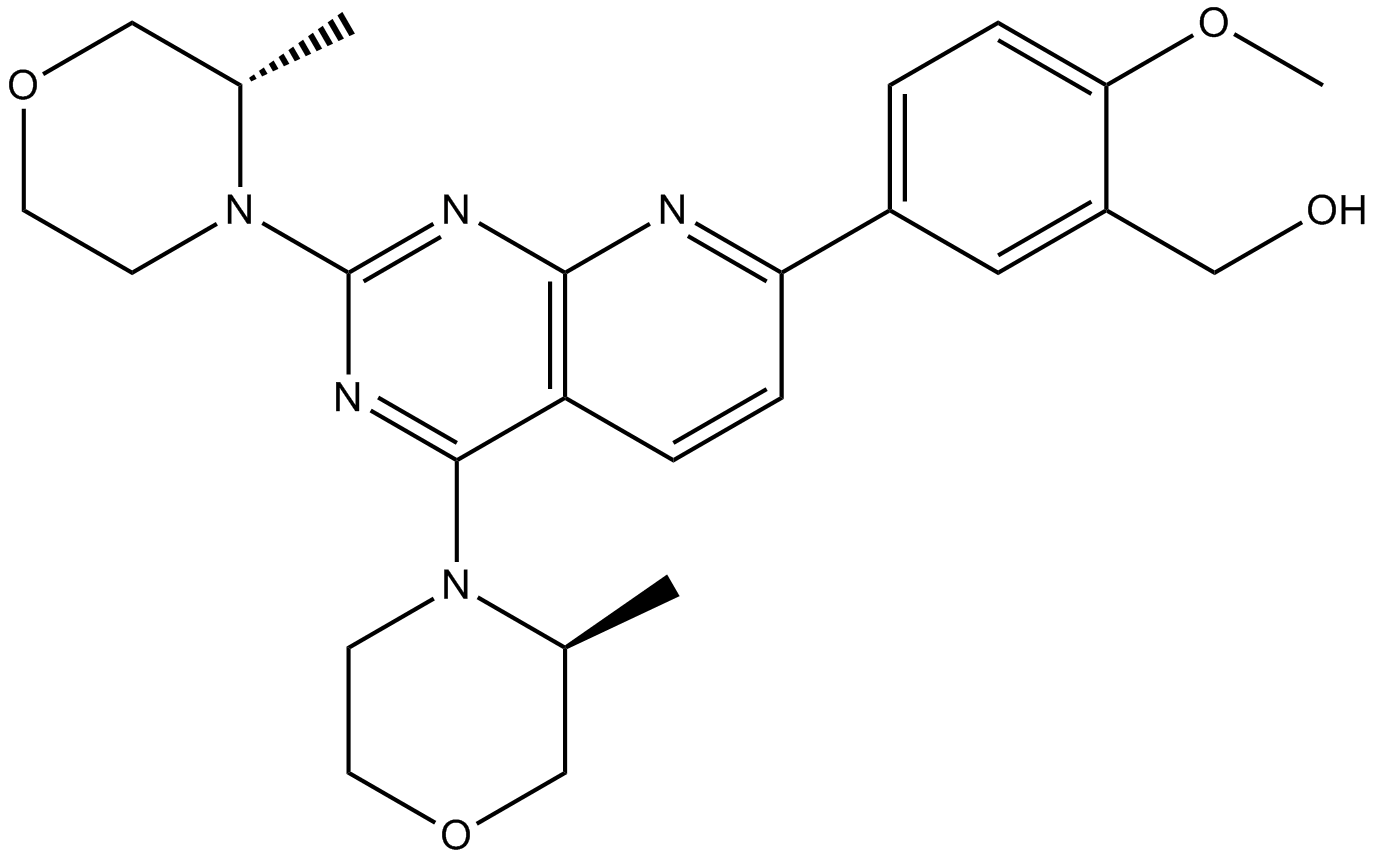
GC10350
TIC10 isomer
Potent Akt/ERK inhibitor
-
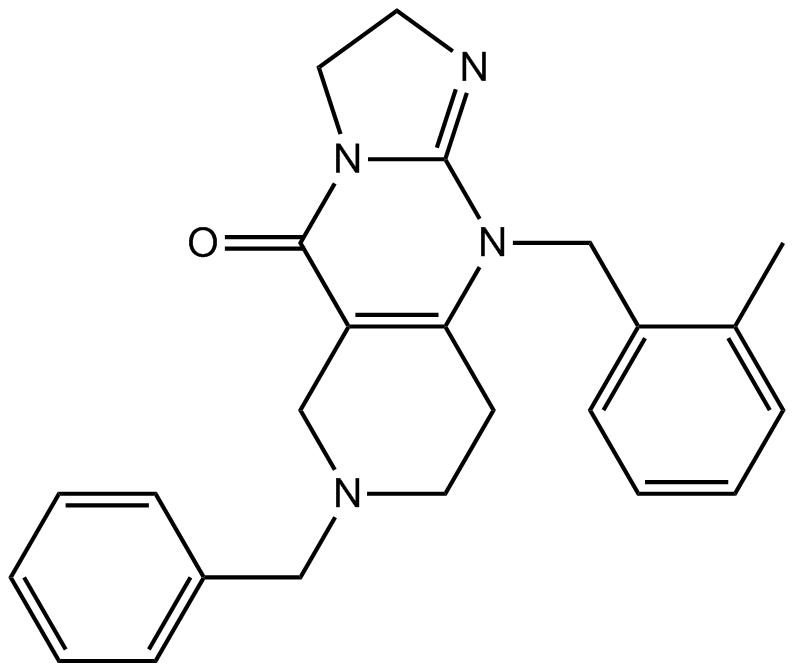
GC45213
α-NETA
Choline acetyltransferase (ChAT) mediates the synthesis of the neurotransmitter acetylcholine from acetyl-CoA and choline.
-
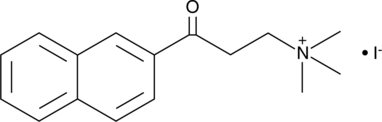
GC46008
(±)-Thalidomide-d4
An internal standard for the quantification of (±)-thalidomide
-
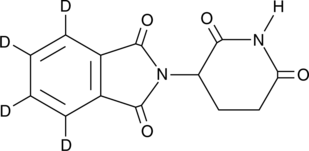
GC45256
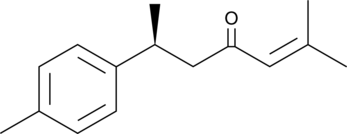
(+)-ar-Turmerone
(+)-ar-Turmerone is an aromatic compound from the rhizomes of C.
-
GC41345
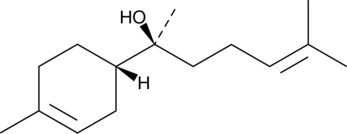
(-)-α-Bisabolol
(-)-α-Bisabolol ((-)-α-Bisabolol), a monocyclic sesquiterpene alcohol, exerts antioxidant, anti-inflammatory, and anti-apoptotic activities.
-
GC40698
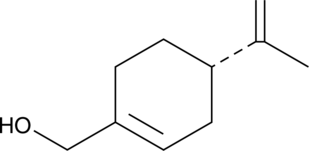
(-)-Perillyl Alcohol
(-)-Perillyl Alcohol is a monoterpene found in lavender, inhibits farnesylation of Ras, upregulates the mannose-6-phosphate receptor and induces apoptosis. Anti-cancer activity.
-
GC34965
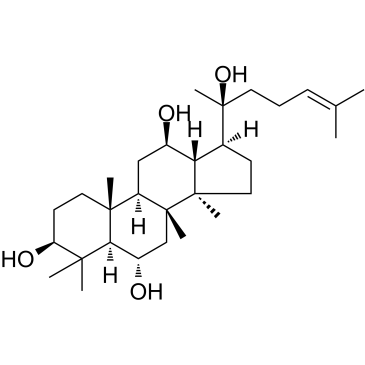
(20S)-Protopanaxatriol
An active ginsenoside metabolite
-
GC34981
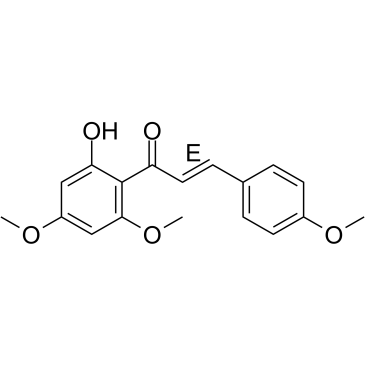
(E)-Flavokawain A
(E)-Flavokawain A, a chalcone extracted from Kava, has anticarcinogenic effect. (E)-Flavokawain A induces apoptosis in bladder cancer cells by involvement of bax protein-dependent and mitochondria-dependent apoptotic pathway and suppresses tumor growth in mice.
-
GC34125
(E)-[6]-Dehydroparadol
(E)-[6]-Dehydroparadol, an oxidative metabolite of [6]-Shogaol, is a potent Nrf2 activator. (E)-[6]-Dehydroparadol can inhibit the growth and induce the apoptosis of human cancer cells.
![(E)-[6]-Dehydroparadol Chemical Structure (E)-[6]-Dehydroparadol Chemical Structure](/media/struct/GC3/GC34125.png)
-
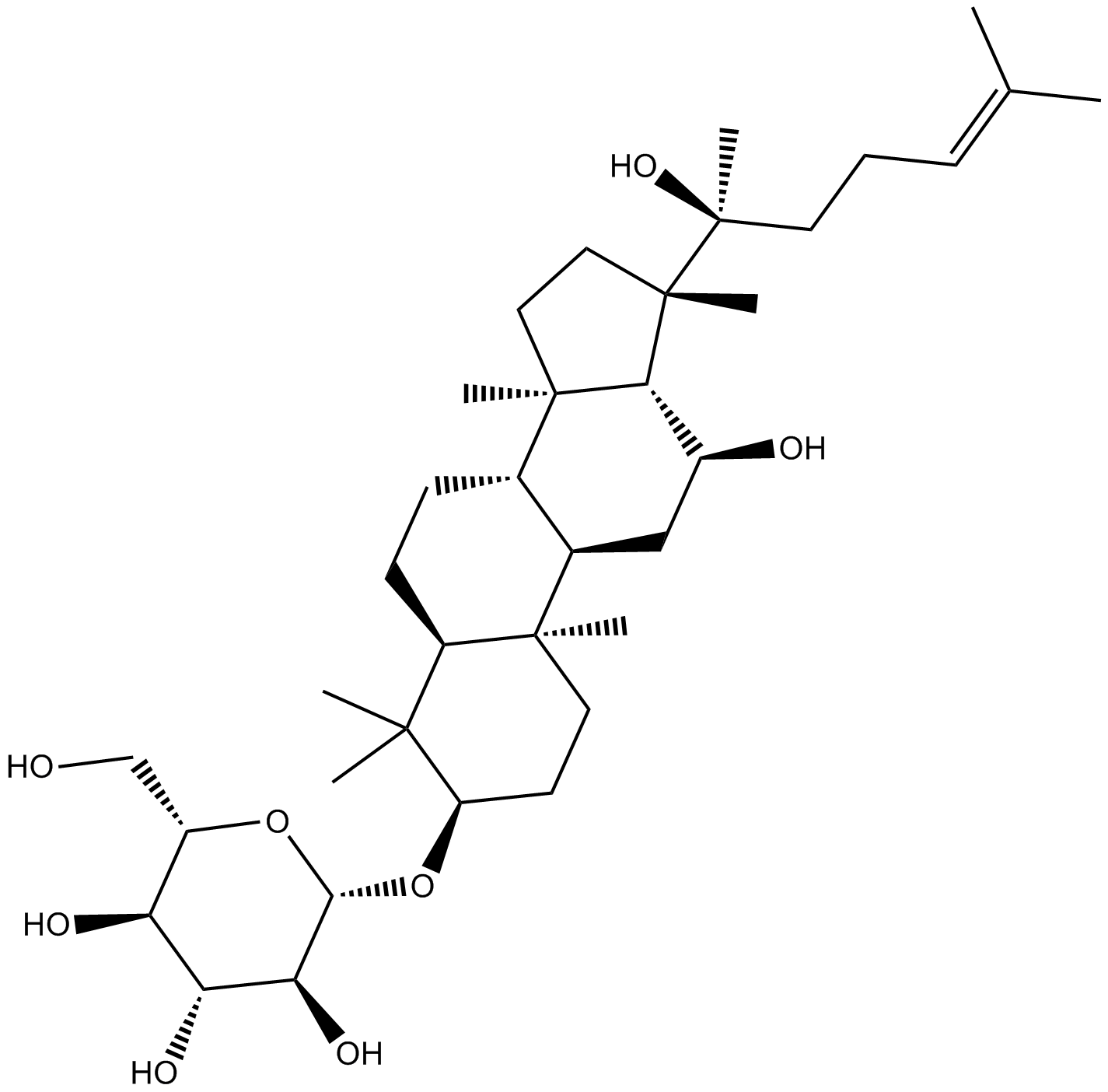
GN10783
(R) Ginsenoside Rh2
-
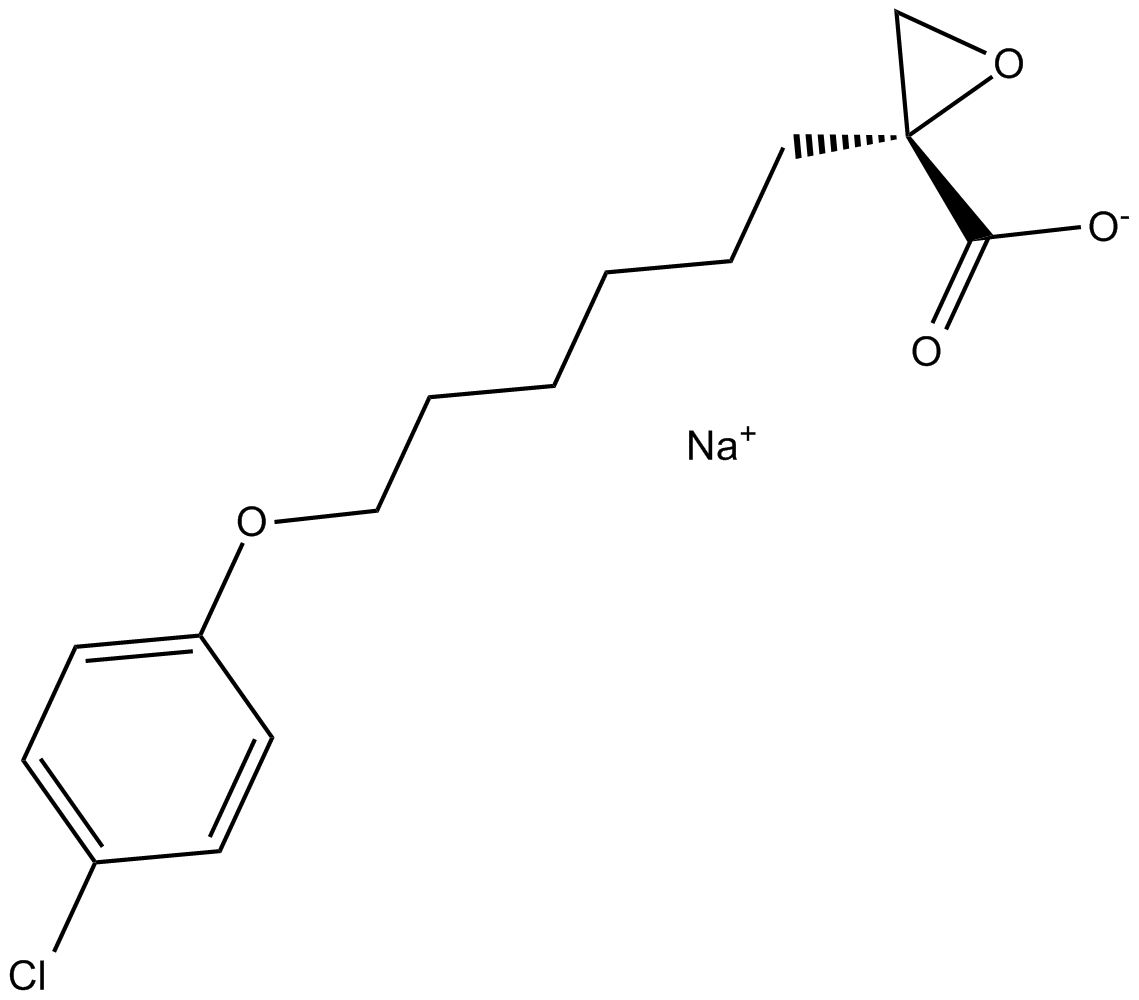
GC15104
(R)-(+)-Etomoxir sodium salt
Etomoxir((R)-(+)-Etomoxir) sodium salt is an irreversible inhibitor of carnitine palmitoyltransferase 1a (CPT-1a), inhibits fatty acid oxidation (FAO) through CPT-1a and inhibits palmitate β-oxidation in human, rat and guinea pig.
-
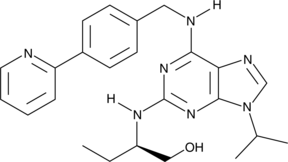
GC41716
(R)-CR8
Cyclin-dependent kinases (CDKs) are key regulators of cell cycle progression and are therefore promising targets for cancer therapy.
-
GC19541
(rac)-Antineoplaston A10
(rac)-Antineoplaston A10 is the racemate of Antineoplaston A10

-
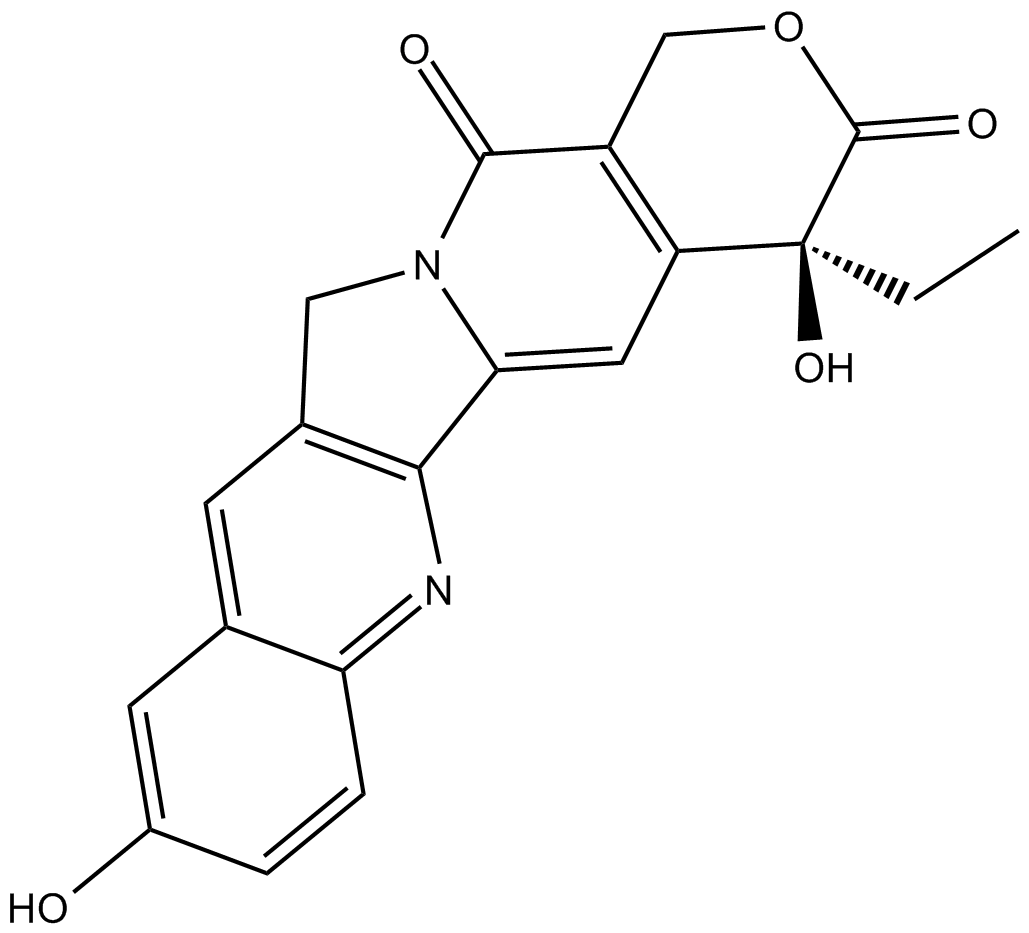
GC10098
(S)-10-Hydroxycamptothecin
inhibitor of topoisomerase I
-
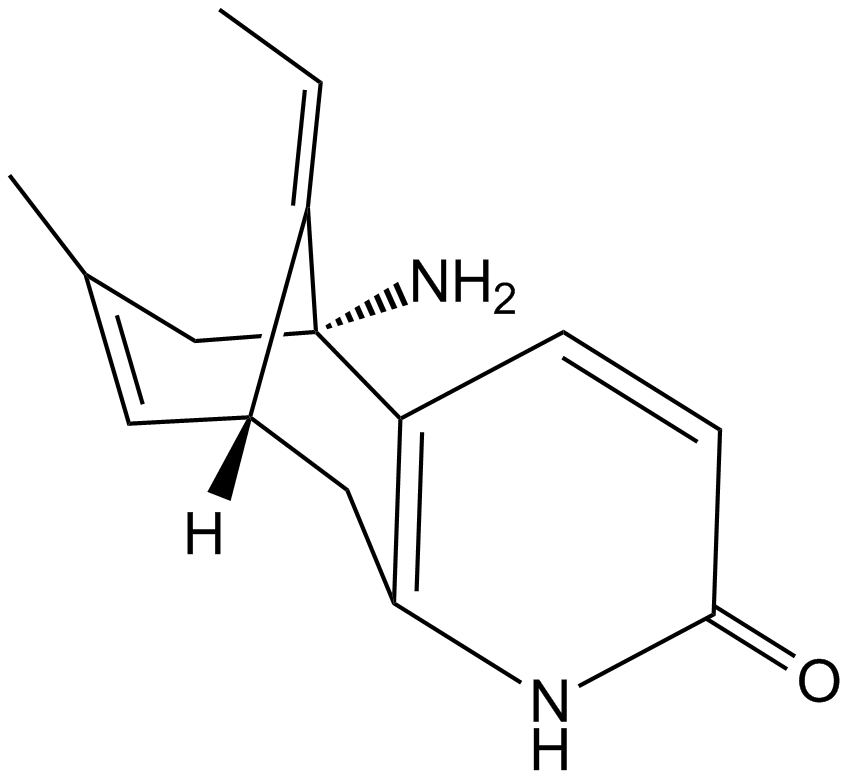
GC11965
(±)-Huperzine A
A neuroprotective AChE inhibitor
-
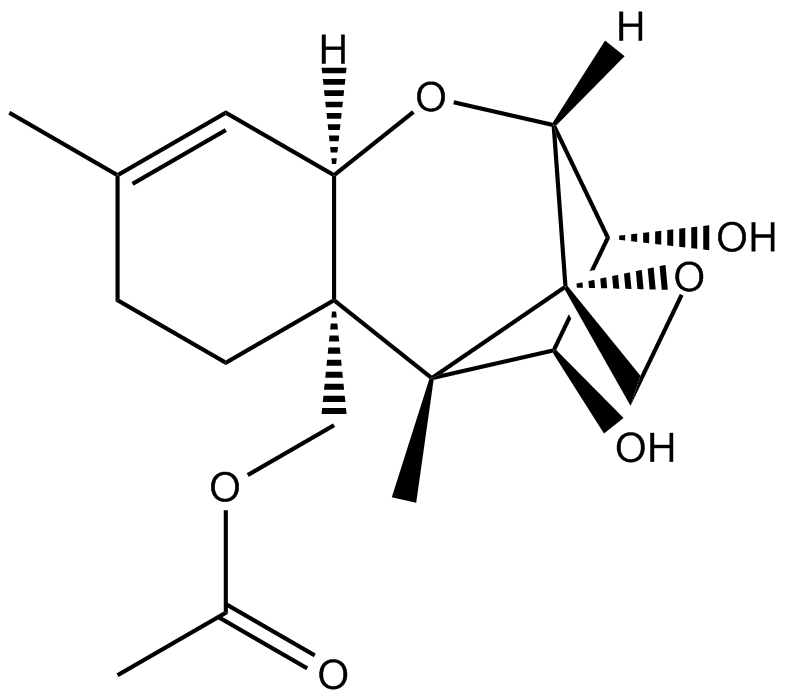
GC11988
15-acetoxy Scirpenol
mycotoxin that induce apoptotic cell death
-
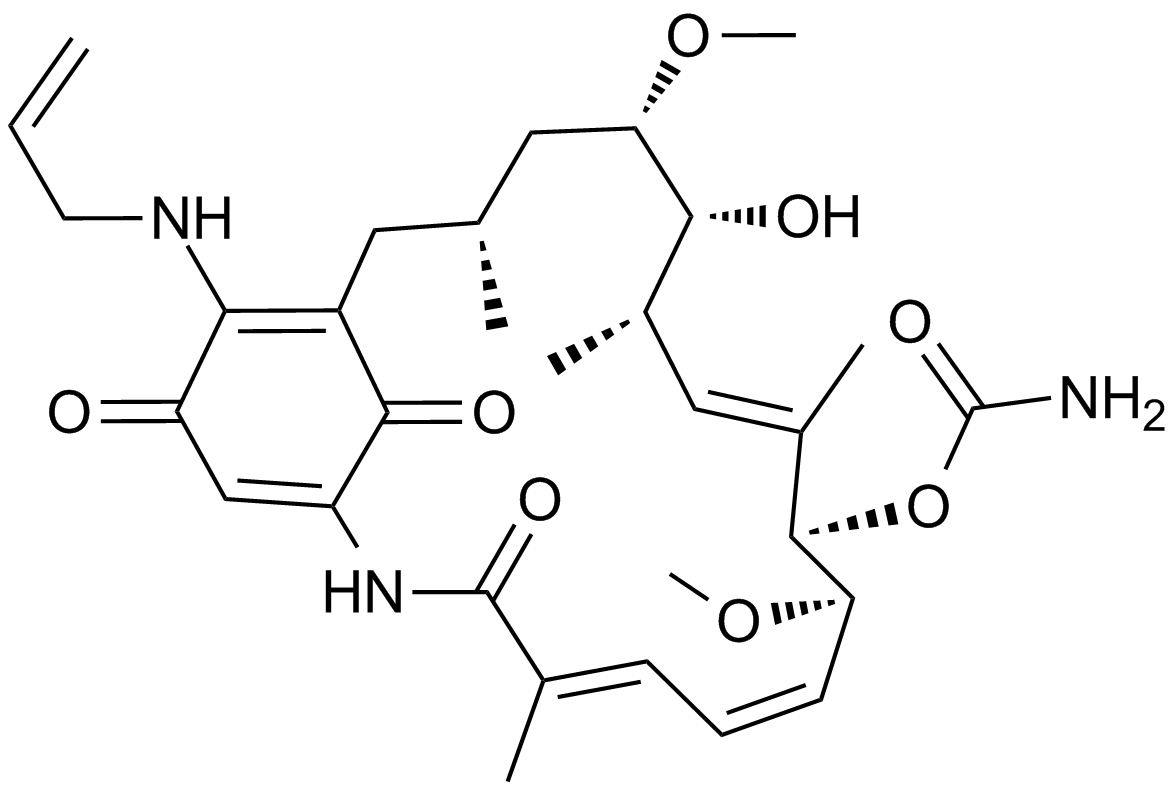
GC11720
17-AAG (KOS953)
An inhibitor of Hsp90
-
GC13044
17-DMAG (Alvespimycin) HCl
17-DMAG (Alvespimycin) HCl (17-DMAG hydrochloride; KOS-1022; BMS 826476) is a potent inhibitor of Hsp90, binding to Hsp90 with EC50 of 62±29 nM.

-
GN10065
2-Atractylenolide
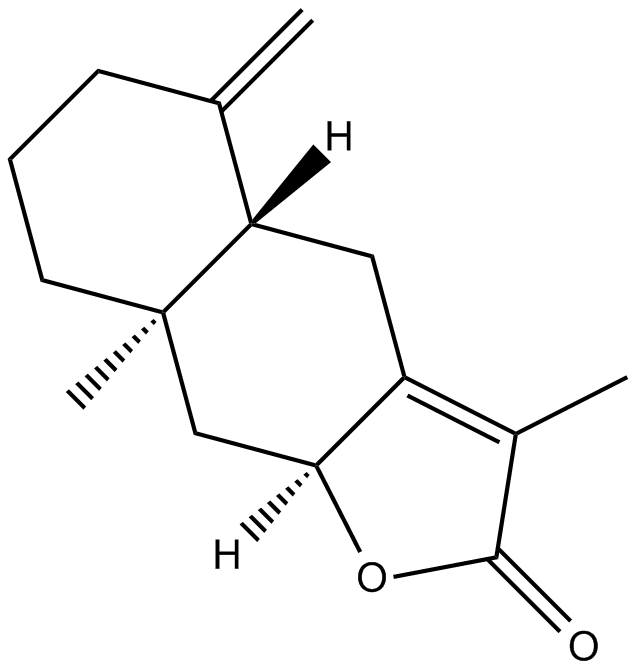
-
GC17430
2-Deoxy-D-glucose
2-Deoxy-D-glucose (2DG), is a glucose analogue, act as competitive glycolytic inhibitor.
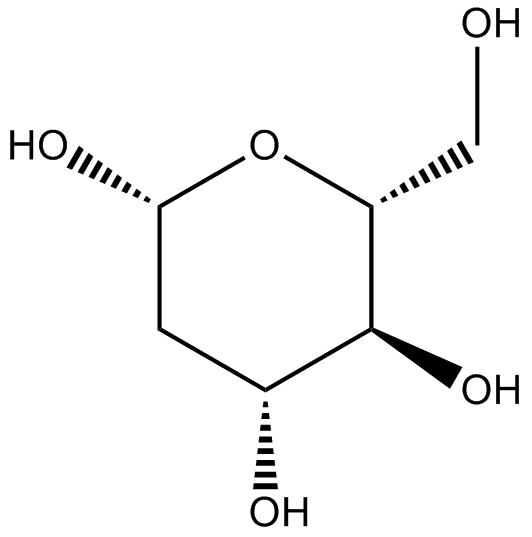
-
GC38318
2-Methoxycinnamaldehyde
2-Methoxycinnamaldehyde (o-Methoxycinnamaldehyde) is a natural compound of Cinnamomum cassia, with antitumor activity. 2-Methoxycinnamaldehyde inhibits proliferation and induces apoptosis by mitochondrial membrane potential (ΔΨm) loss, activation of both caspase-3 and caspase-9. 2-Methoxycinnamaldehyde effectively inhibits platelet-derived growth factor (PDGF)-induced HASMC migration.
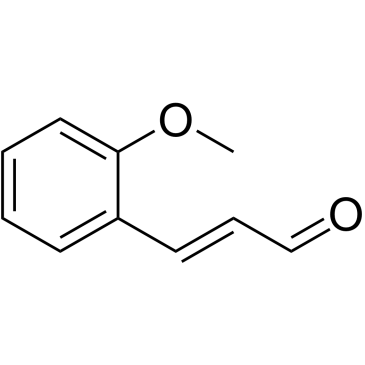
-
GC15084
2-Methoxyestradiol (2-MeOE2)
2-Methoxyestradiol (2-MeOE2/2-Me) is a HIF-1α inhibitor that inhibits HIF-1α accumulation and HIF transcriptional activity. 2-Methoxyestradiol can trigger p53-induced apoptosis and has potential antitumor activity..
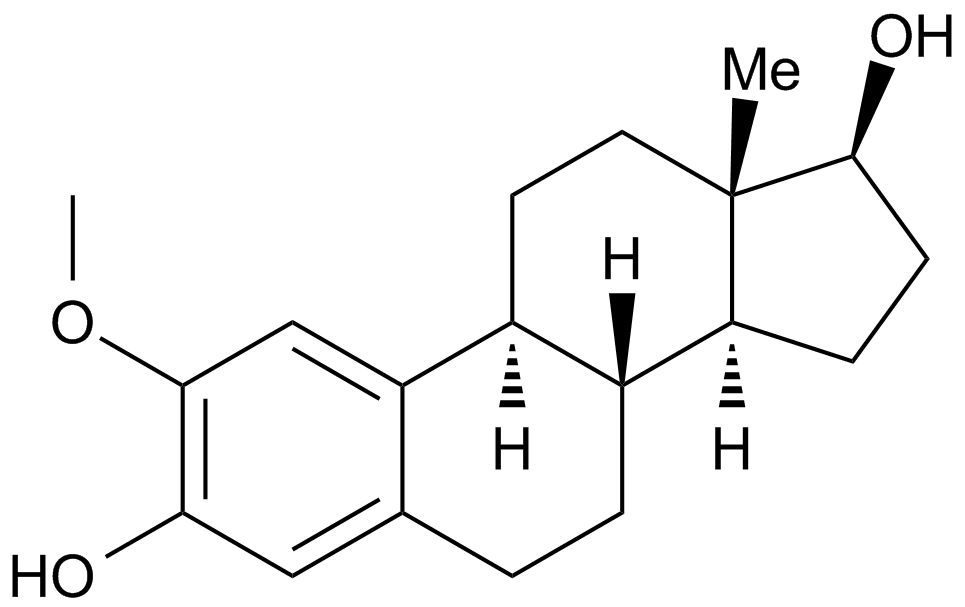
-
GN10800
20(S)-NotoginsenosideR2
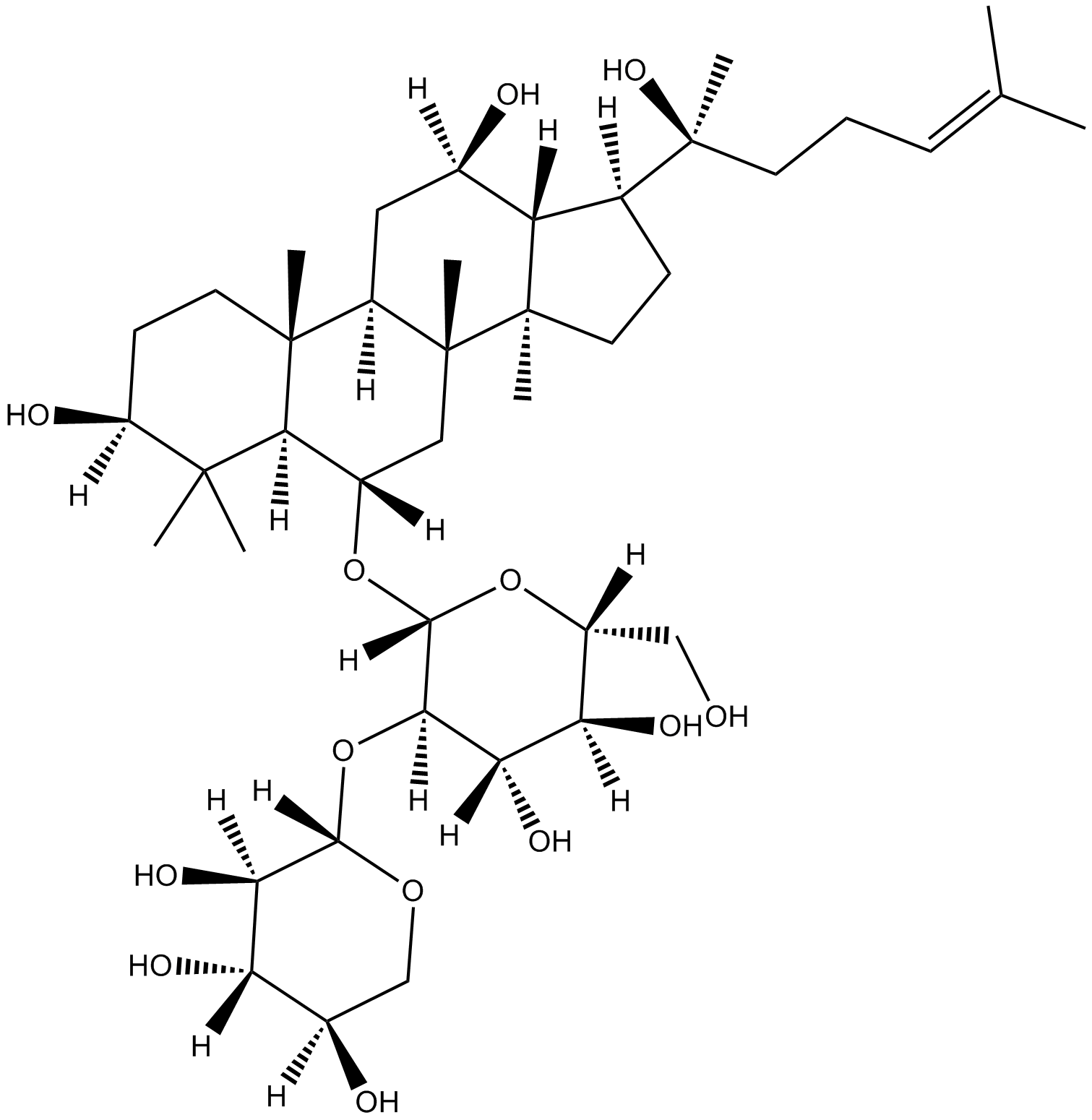
-
GC35112
3'-Hydroxypterostilbene
3'-Hydroxypterostilbene is a Pterostilbene analogue. 3'-Hydroxypterostilbene inhibits the growth of COLO 205, HCT-116 and HT-29 cells with IC50s of 9.0, 40.2 and 70.9 ?M, respectively. 3'-Hydroxypterostilbene significantly down-regulates PI3K/Akt and MAPKs signaling pathways and effectively inhibits the growth of human colon cancer cells by inducing apoptosis and autophagy. 3'-Hydroxypterostilbene can be used for the research of cancer.
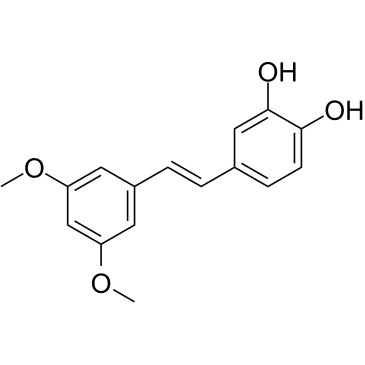
-
GC35106
3-Dehydrotrametenolic acid
3-?Dehydrotrametenolic acid, isolated from the sclerotium of Poria cocos, is a lactate dehydrogenase (LDH) inhibitor. 3-?Dehydrotrametenolic acid promotes adipocyte differentiation in vitro and acts as an insulin sensitizer in vivo. 3-?Dehydrotrametenolic acid induces apoptosis and has anticancer activity.
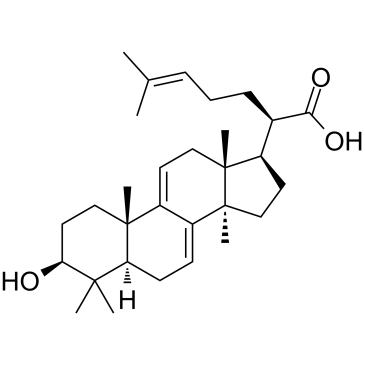
-
GC17394
3-Nitropropionic acid
3-Nitropropionic acid (β-Nitropropionic acid) is an irreversible inhibitor of succinate dehydrogenase.
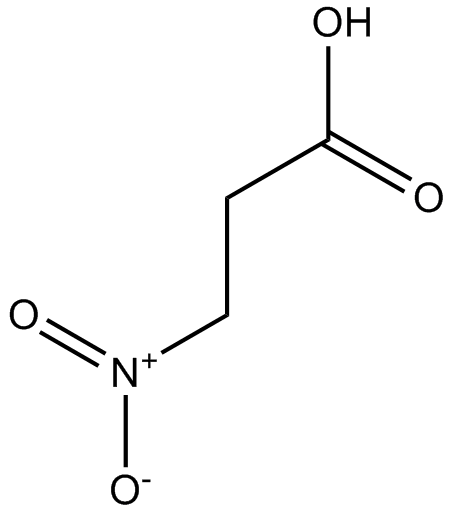
-
GC35099
3-O-Acetyloleanolic acid
3-O-Acetyloleanolic acid (3AOA), an oleanolic acid derivative isolated from the seeds of Vigna sinensis K., induces in cancer and also exhibits anti-angiogenesis activity.
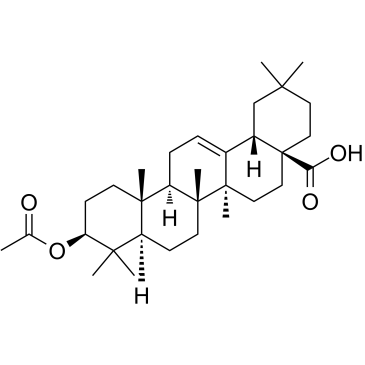
-
GC32767
3BDO
A butyrolactone derivative and autophagy inhibitor
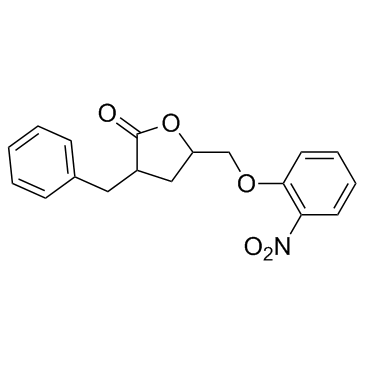
-
GC42346
4-bromo A23187
4-bromo A23187 is a halogenated analog of the highly selective calcium ionophore A23187.
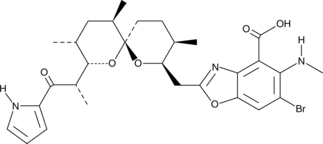
-
GC30896
4-Hydroxybenzyl alcohol
A phenol with diverse biological activities
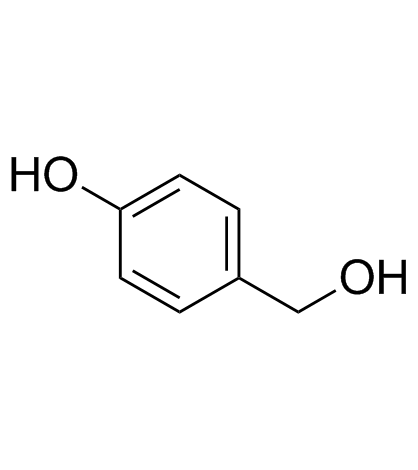
-
GC35138
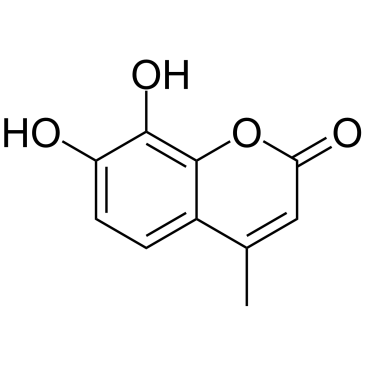
4-Methyldaphnetin
4-Methyldaphnetin is a precursor in the synthesis of derivatives of 4-methyl coumarin. 4-Methyldaphnetin has potent, selective anti-proliferative and apoptosis-inducing effects on several cancer cell lines. 4-Methyldaphnetin possesses radical scavenging property and strongly inhibits membrane lipid peroxidation.
-
GC10468
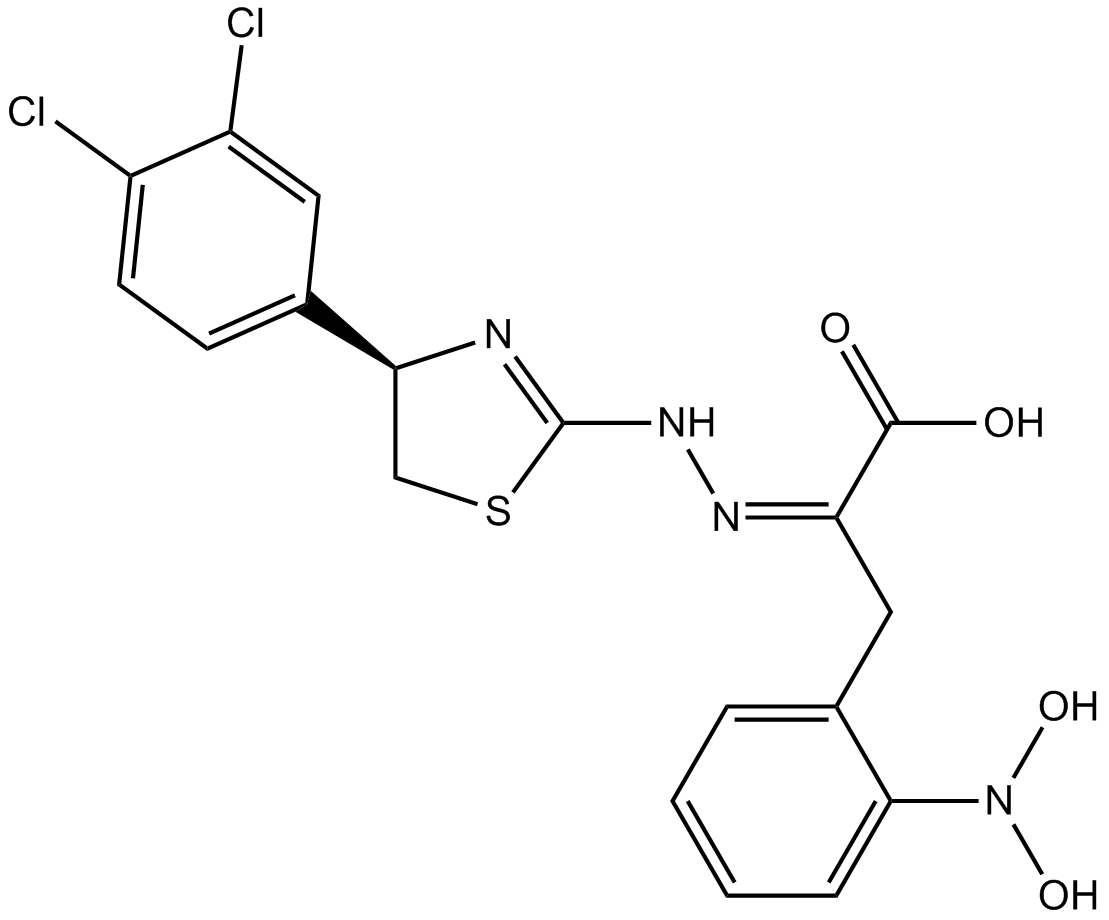
4EGI-1
Competitive eIF4E/eIF4G interaction inhibitor
-
GC35150
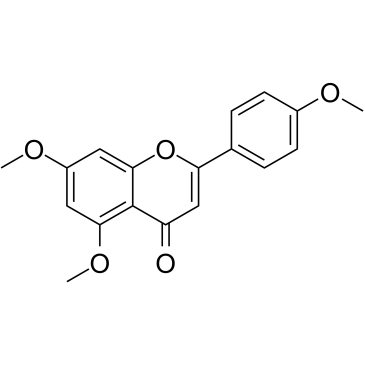
5,7,4'-Trimethoxyflavone
5,7,4'-Trimethoxyflavone is isolated from Kaempferia parviflora (KP) that is a famous medicinal plant from Thailand. 5,7,4'-Trimethoxyflavone induces apoptosis, as evidenced by increments of sub-G1 phase, DNA fragmentation, annexin-V/PI staining, the Bax/Bcl-xL ratio, proteolytic activation of caspase-3, and degradation of poly (ADP-ribose) polymerase (PARP) protein.5,7,4'-Trimethoxyflavone is significantly effective at inhibiting proliferation of SNU-16 human gastric cancer cells in a concentration dependent manner.
-
GC35147
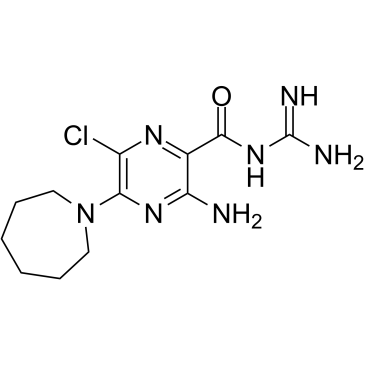
5-(N,N-Hexamethylene)-amiloride
An amiloride derivative with diverse biological activities
-
GC45356
5-Aminolevulinic Acid (hydrochloride)
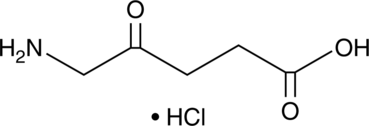
-
GN10093
6-gingerol
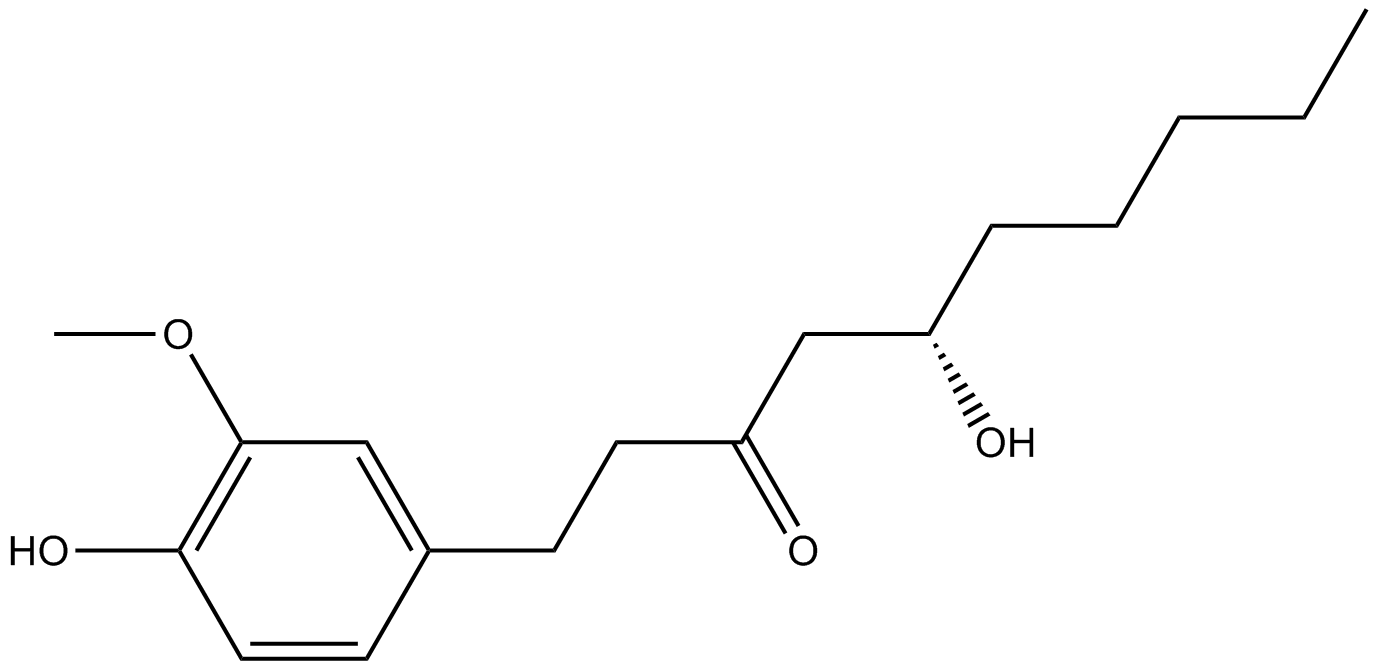
-
GC16853
7,8-Dihydroxyflavone
Tyrosine kinase receptor B (TrkB) agonist
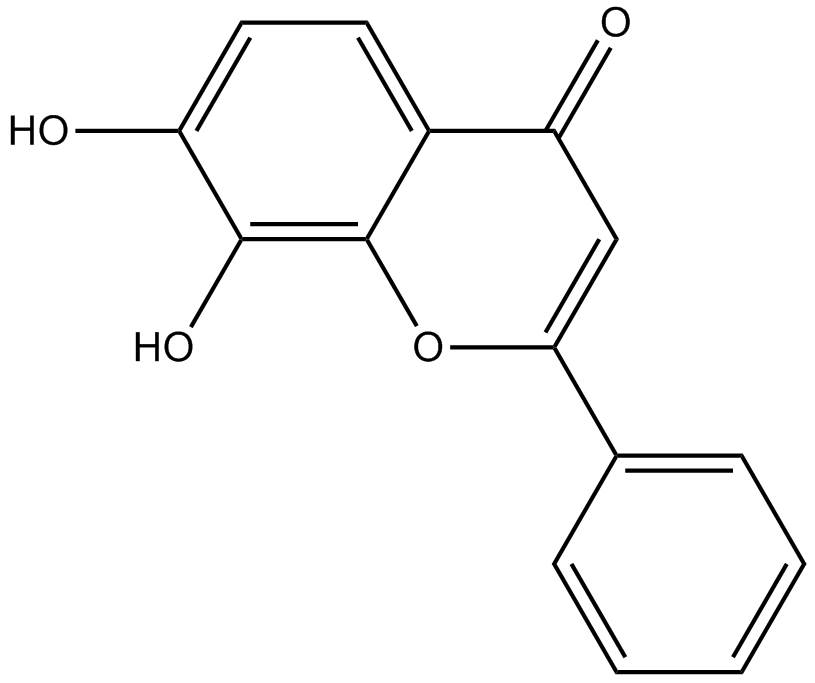
-
GC17119
8-Prenylnaringenin
estrogen receptor inhibitor
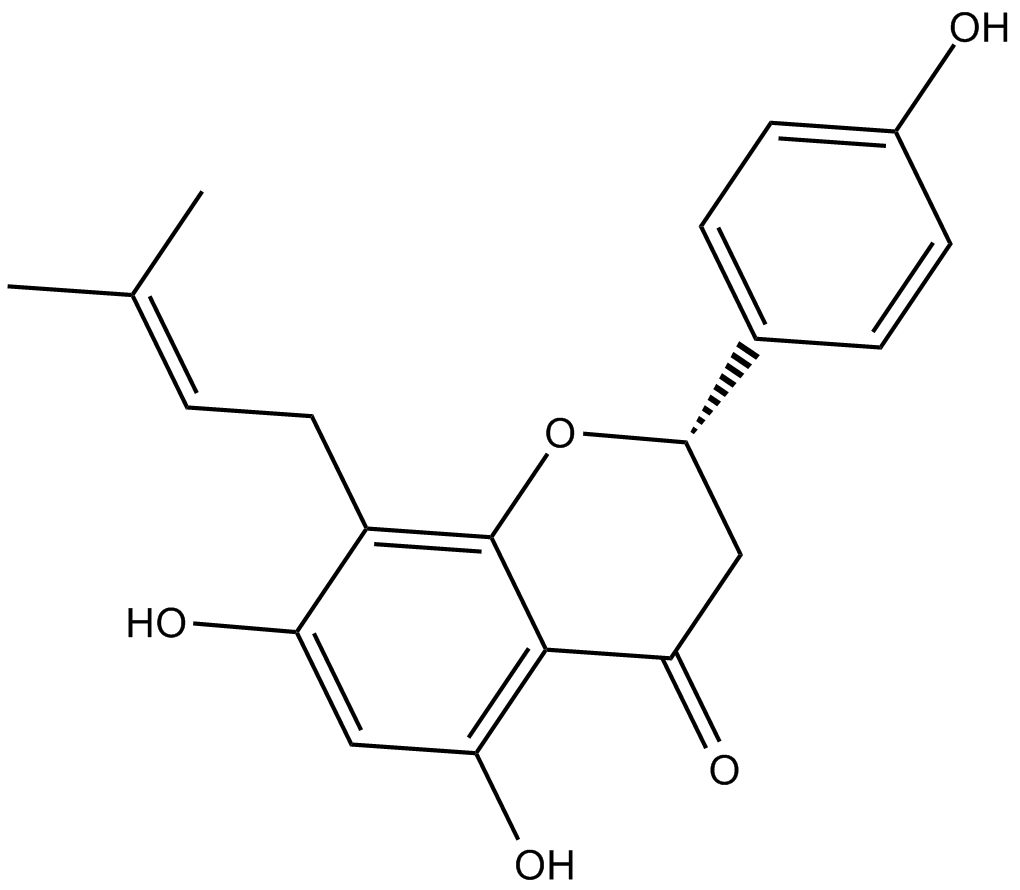
-
GC39152
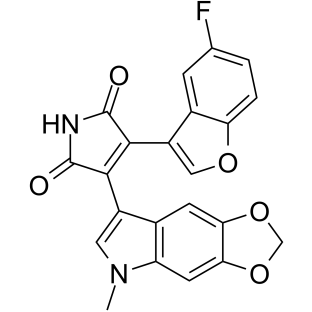
9-ING-41
9-ING-41 is a maleimide-based ATP-competitive and selective glycogen synthase kinase-3β (GSK-3β) inhibitor with an IC50 of 0.71 μM. 9-ING-41 significantly leads to cell cycle arrest, autophagy and apoptosis in cancer cells. 9-ING-41 has anticancer activity and has the potential for enhancing the antitumor effects of chemotherapeutic drugs.
-
GN10035
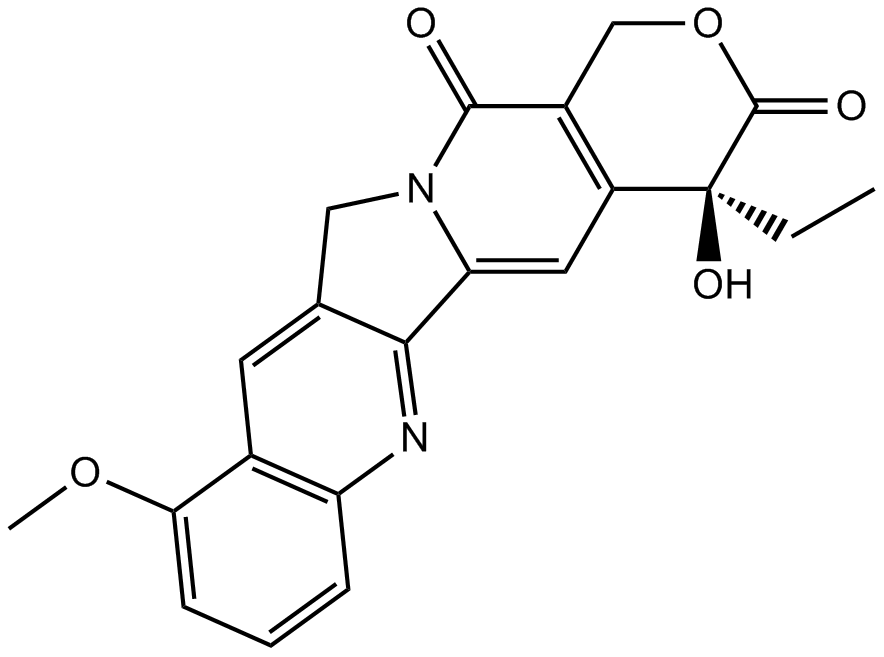
9-Methoxycamptothecin
-
GC11200
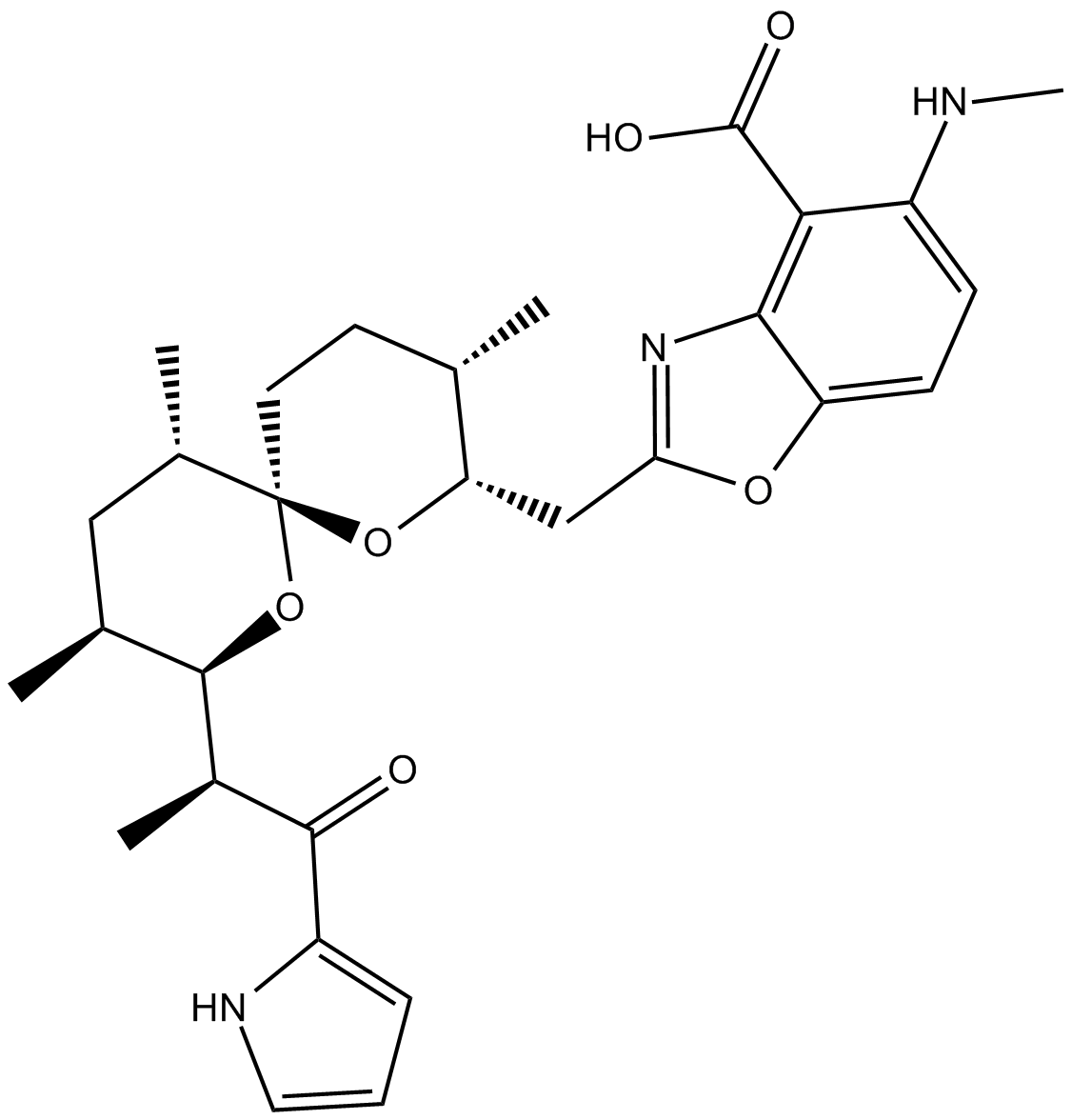
A23187
A23187, free acid is a Ca2+ ionophore
-
GC35216
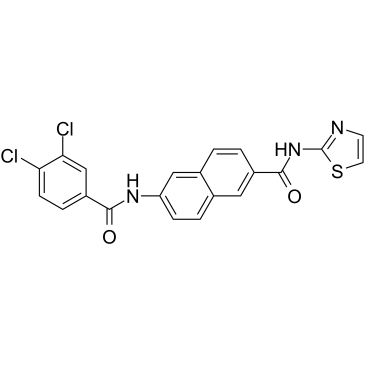
AAPK-25
AAPK-25 is a potent and selective Aurora/PLK dual inhibitor with anti-tumor activity, which can cause mitotic delay and arrest cells in a prometaphase, reflecting by the biomarker histone H3Ser10 phosphorylation and followed by a surge in apoptosis. AAPK-25 targets Aurora-A, -B, and -C with Kd values ranging from 23-289 nM, as well as PLK-1, -2, and -3 with Kd values ranging from 55-456 nM.
-
GC13805
Abacavir
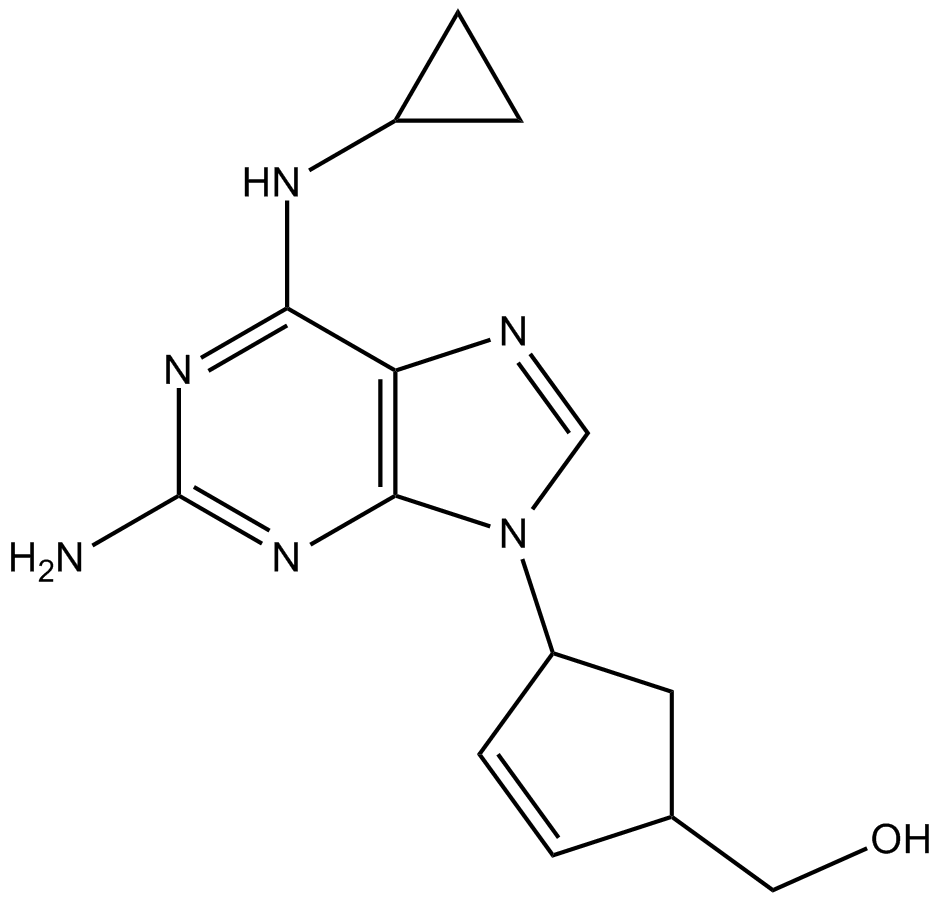
-
GC35227
ACBI1
ACBI1 is a potent and cooperative SMARCA2, SMARCA4 and PBRM1 degrader with DC50s of 6, 11 and 32 nM, respectively. ACBI1 is a PROTAC degrader. ACBI1 shows anti-proliferative activity. ACBI1 induces apoptosis.
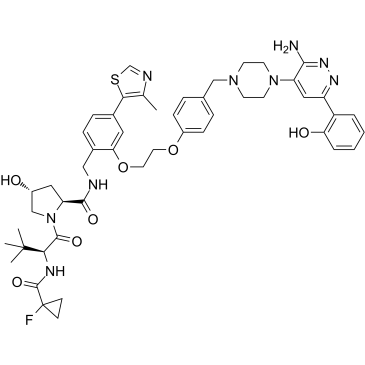
-
GC11786
Acetylcysteine
Acetylcysteine is the N-acetyl derivative of CYSTEINE.
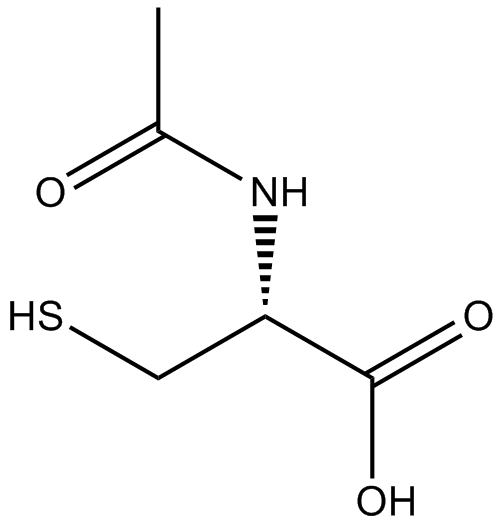
-
GC17094
Acitretin
Metabolite of etretinate
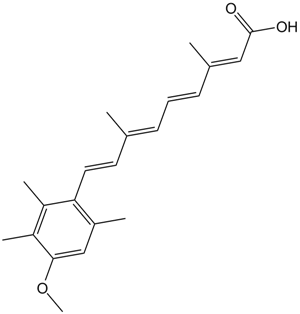
-
GC35242
Actein
Actein is a triterpene glycoside isolated from the rhizomes of Cimicifuga foetida. Actein suppresses cell proliferation, induces autophagy and apoptosis through promoting ROS/JNK activation, and blunting AKT pathway in human bladder cancer. Actein has little toxicity in vivo.
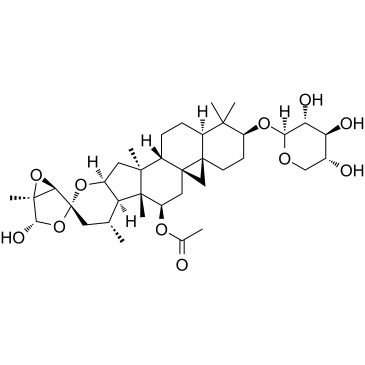
-
GC16350
Actinonin
Peptidomimetic antibiotic that inhibits aminopeptidases
-
GC10610
Adapalene
RARβ and RARγ agonist

-
GC13959
Adarotene
An atypical retinoid
-
GC11892
AEE788 (NVP-AEE788)
AEE788 (NVP-AEE788) is an inhibitor of the EGFR and ErbB2 with IC50 values of 2 and 6 nM, respectively.
-
GC13168
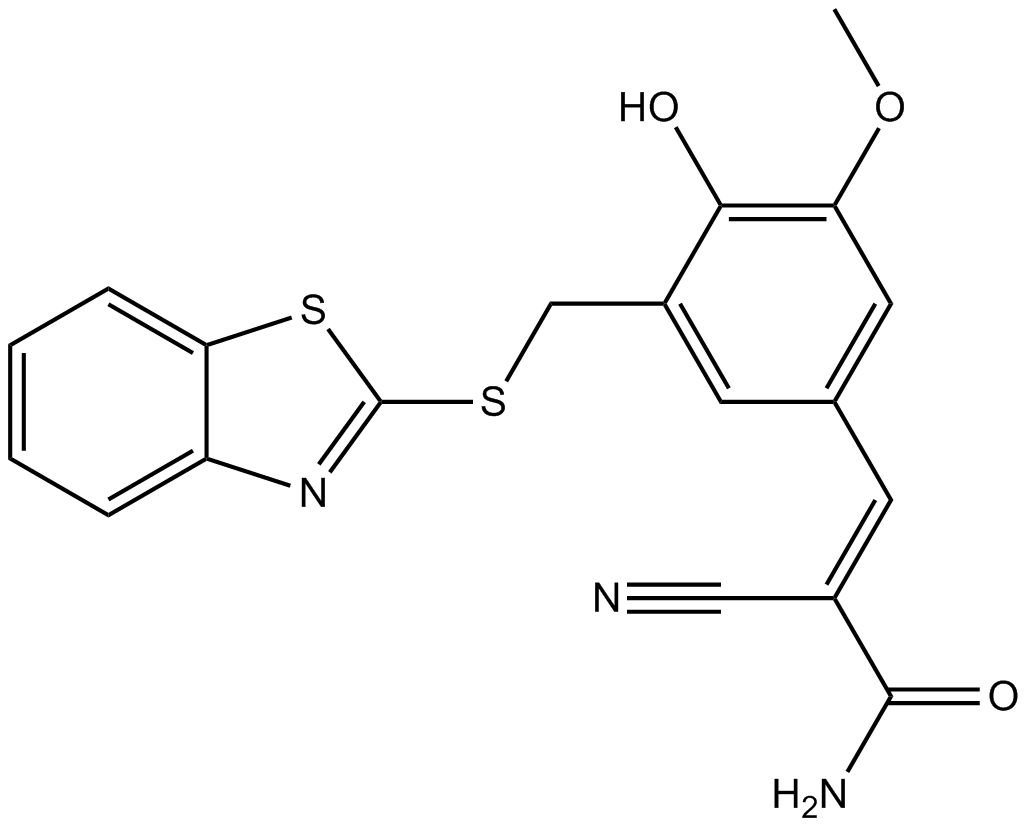
AG 825
Selective ErbB2 inhibitor
-
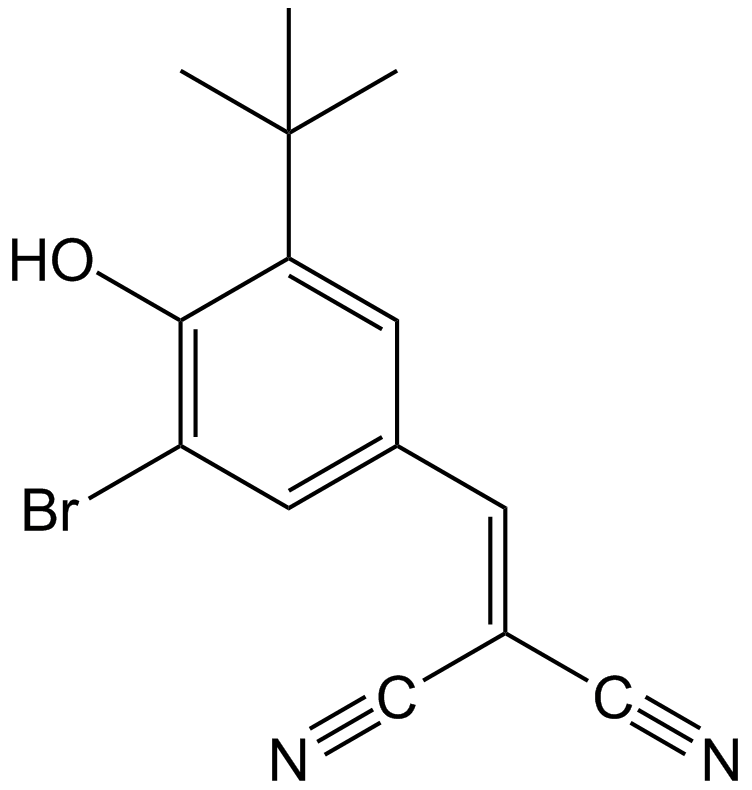
GC13697
AG-1024
Selective IGF-1R inhibitor
-
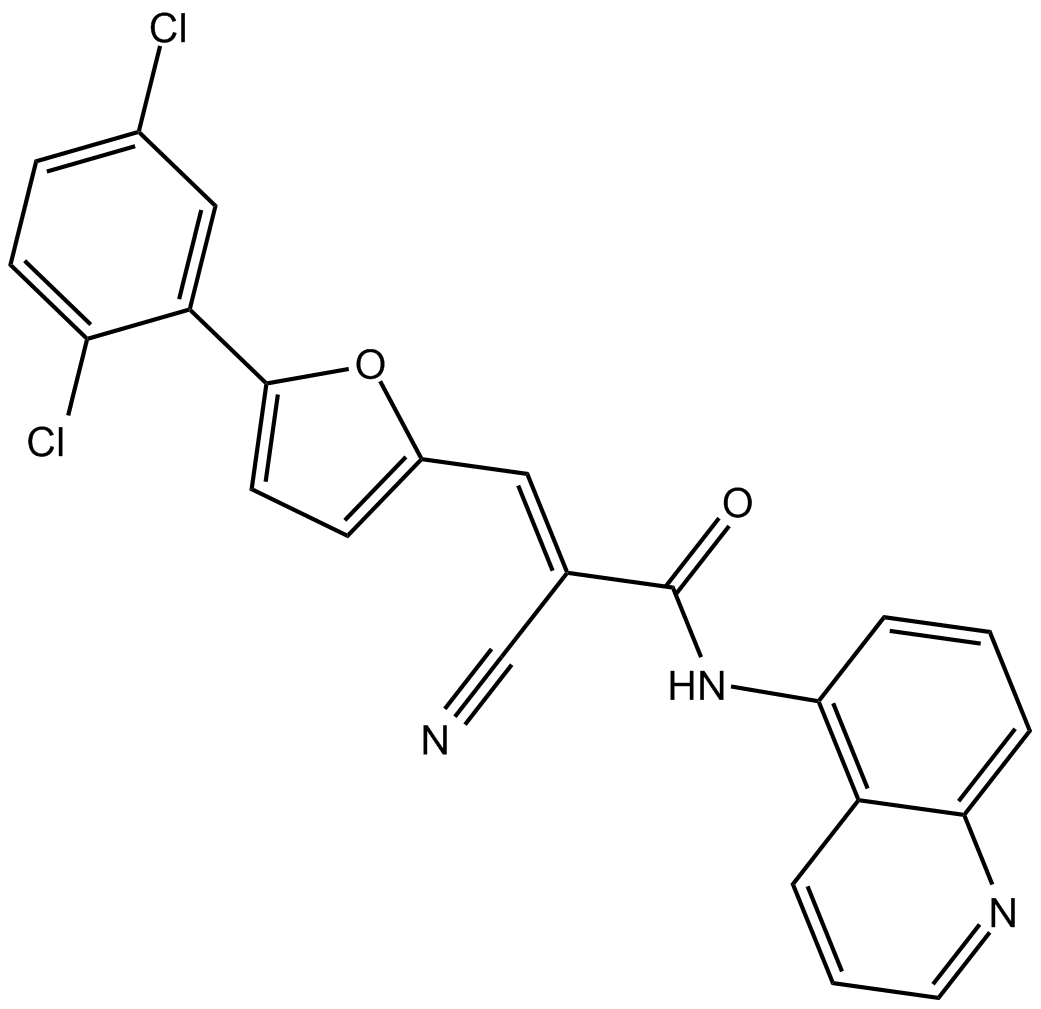
GC17881
AGK 2
AGK2 is a selective SIRT2 inhibitor, with an IC50 of 3.
-
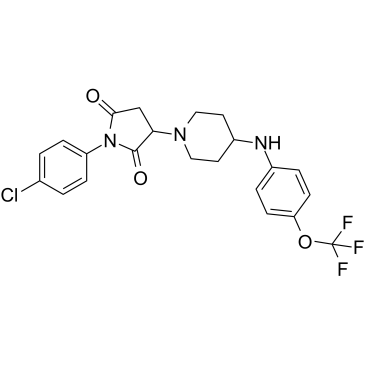
GC39620
AKOS-22
-
GC11589
AKT inhibitor VIII
A potent inhibitor of Akt1 and Akt2

-
GC35275
AKT-IN-3
AKT-IN-3 (compound E22) is a potent, orally active low hERG blocking Akt inhibitor, with 1.4 nM, 1.2 nM and 1.7 nM for Akt1, Akt2 and Akt3, respectively. AKT-IN-3 (compound E22) also exhibits good inhibitory activity against other AGC family kinases, such as PKA, PKC, ROCK1, RSK1, P70S6K, and SGK. AKT-IN-3 (compound E22) induces apoptosis and inhibits metastasis of cancer cells.
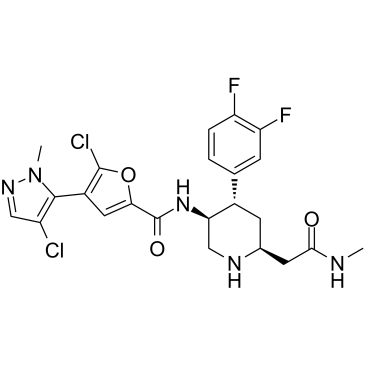
-
GC16597
Alda 1
ALDH2 activator
-
GC35288
Alkannin
A naphthoquinone with diverse biological activities
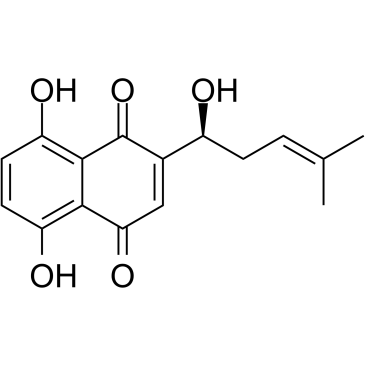
-
GC32127
Alofanib (RPT835)
Alofanib (RPT835) (RPT835) is a potent and selective allosteric inhibitor of fibroblast growth factor receptor 2 (FGFR2).

-
GC14314
Aloperine
An alkaloid
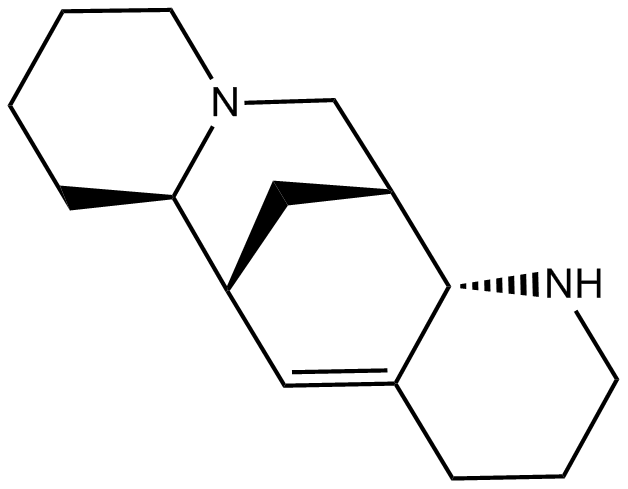
-
GC35306
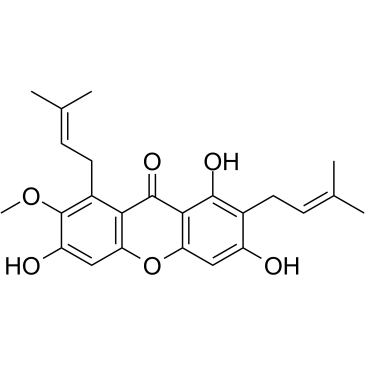
alpha-Mangostin
alpha-Mangostin (α-Mangostin) is a dietary xanthone with broad biological activities, such as antioxidant, anti-allergic, antiviral, antibacterial, anti-inflammatory and anticancer effects. It is an inhibitor of mutant IDH1 (IDH1-R132H) with a Ki of 2.85 μM.
-
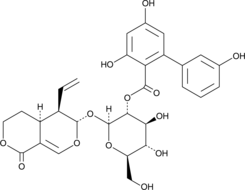
GC42776
Amarogentin
A secoiridoid glycoside with diverse biological activities
-
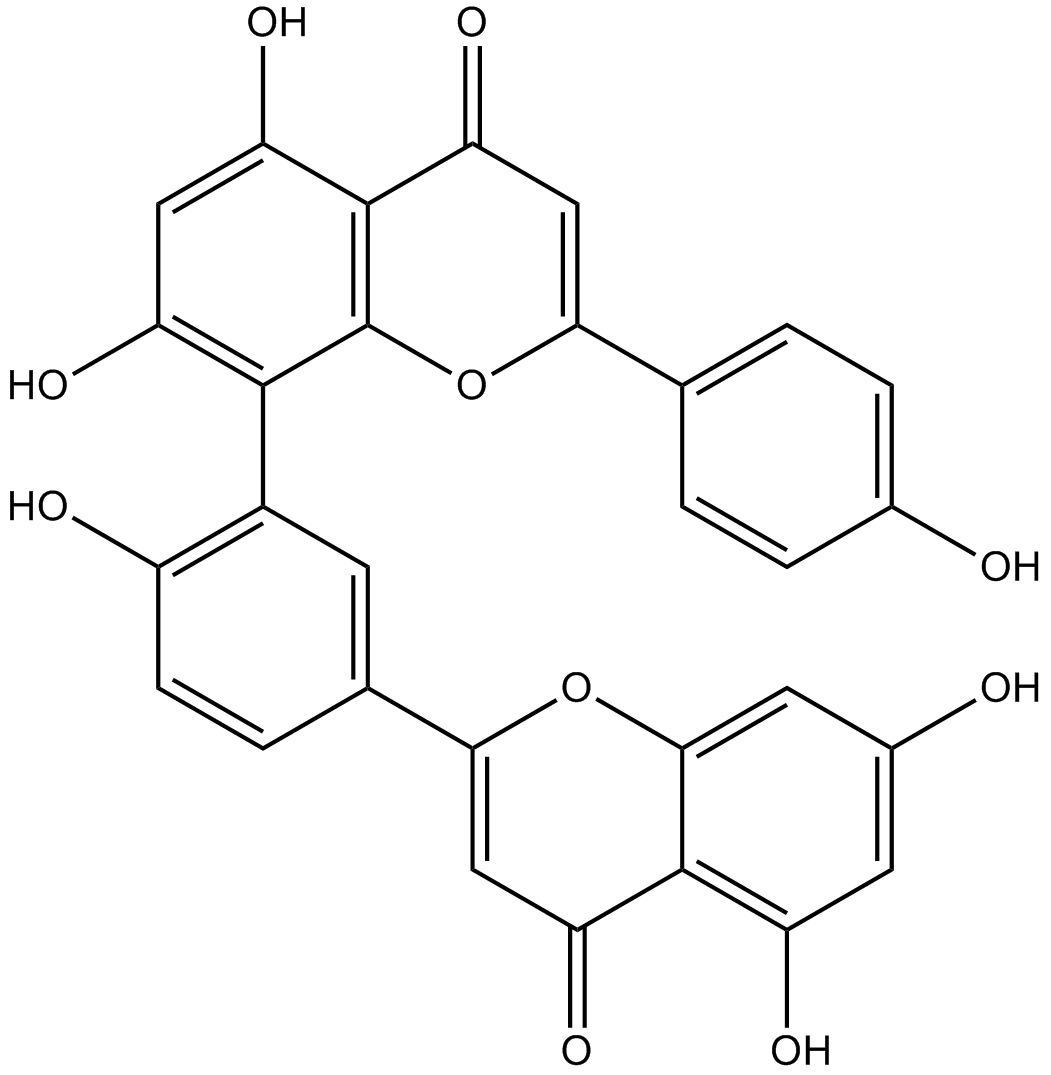
GN10484
Amentoflavone
-
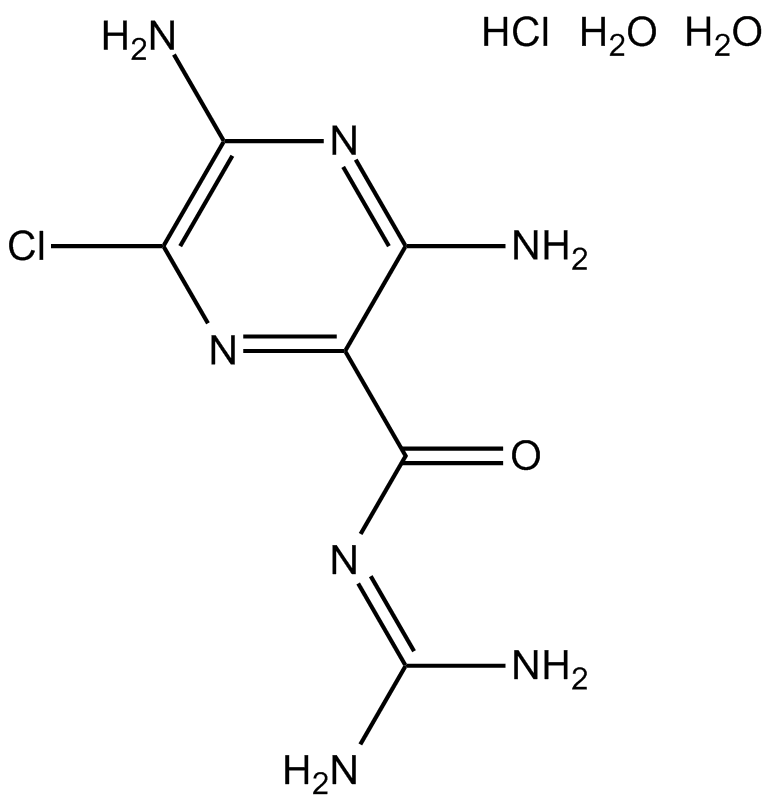
GC12051
Amiloride HCl dihydrate
Amiloride HCl dihydrate (MK-870 hydrochloride dihydrate) is an inhibitor of both epithelial sodium channel (ENaC[1]) and urokinase-type plasminogen activator receptor (uTPA[2]).
-
GC16391
Amuvatinib (MP-470, HPK 56)
A multi-targeted RTK inhibitor
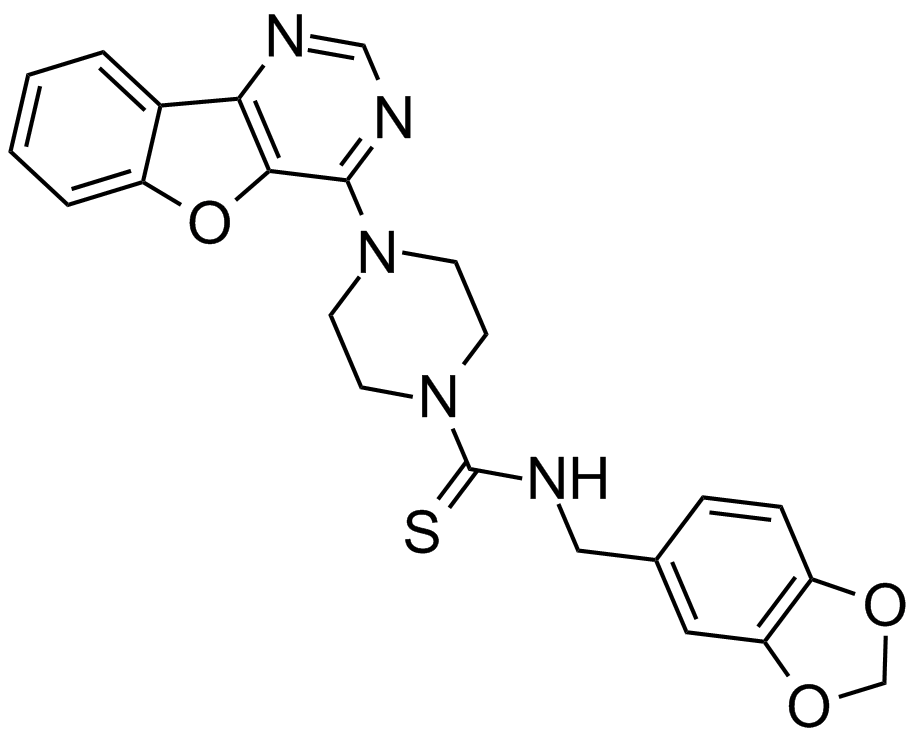
-
GN10045
Angelicin
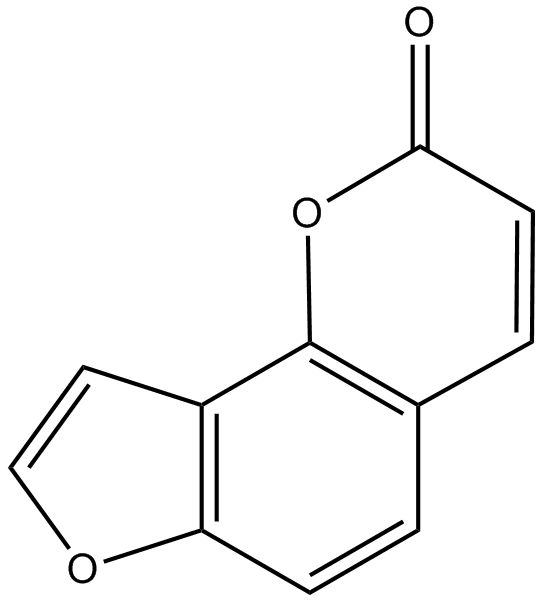
-
GC11559
Anisomycin
JNK agonist, potent and specific
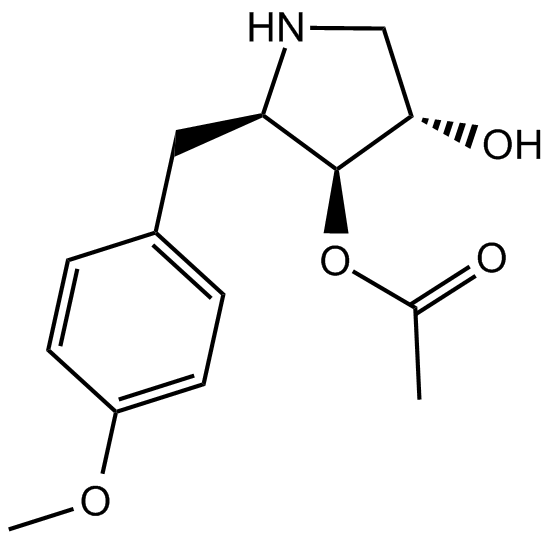
-
GC35361
Antineoplaston A10
Antineoplaston A10, a naturally occurring substance in human body, is a Ras inhibitor potentially for the treatment of glioma, lymphoma, astrocytoma and breast cancer.
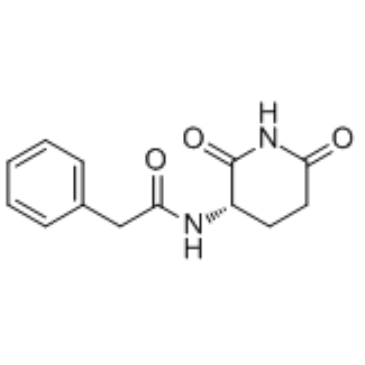
-
GC15586
AP1903
AP1903 (AP1903) is a dimerizer agent that acts by cross-linking the FKBP domains. AP1903 (AP1903) dimerizes the Caspase 9 suicide switch and rapidly induces apoptosis.
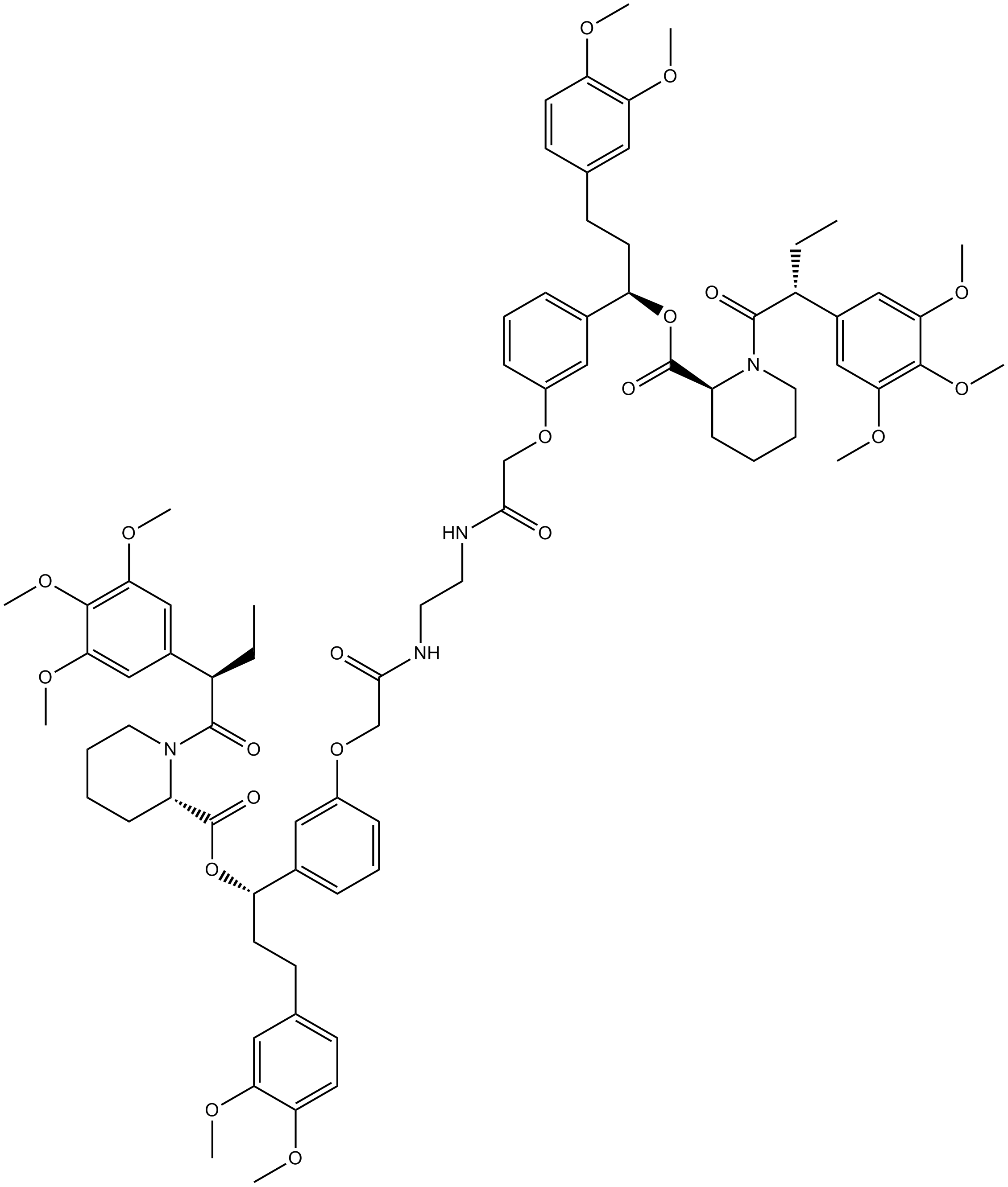
-
GC35367
APG-115
APG-115 (APG-115) is an orally active MDM2 protein inhibitor binding to MDM2 protein with IC50 and Ki values of 3.8 nM and 1 nM, respectively. APG-115 blocks the interaction of MDM2 and p53 and induces cell-cycle arrest and apoptosis in a p53-dependent manner.
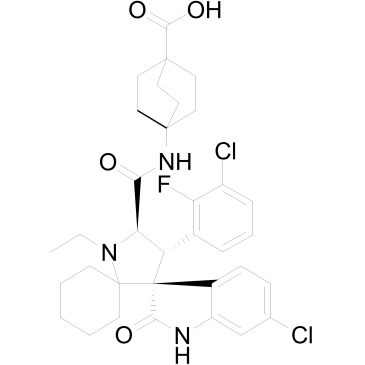
-
GC16237
Apocynin
Selective NADPH-oxidase inhibitor
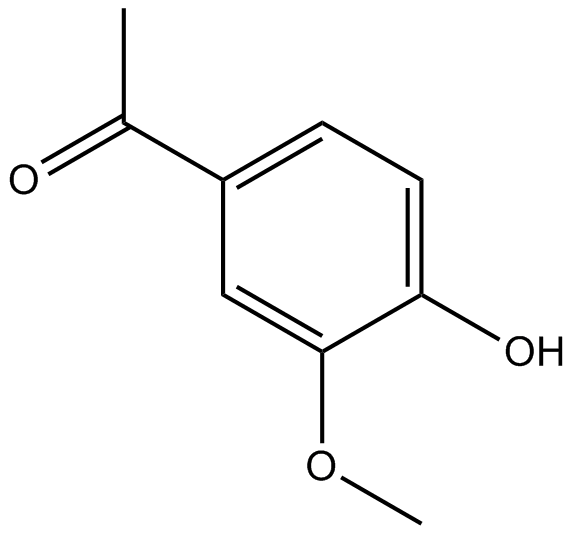
-
GC14209
Apoptosis Activator 2
An activator of caspases
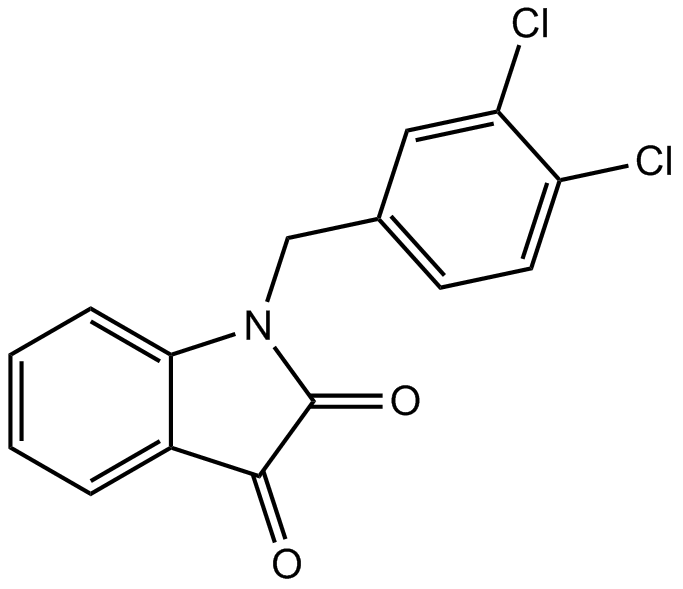
-
GC14411
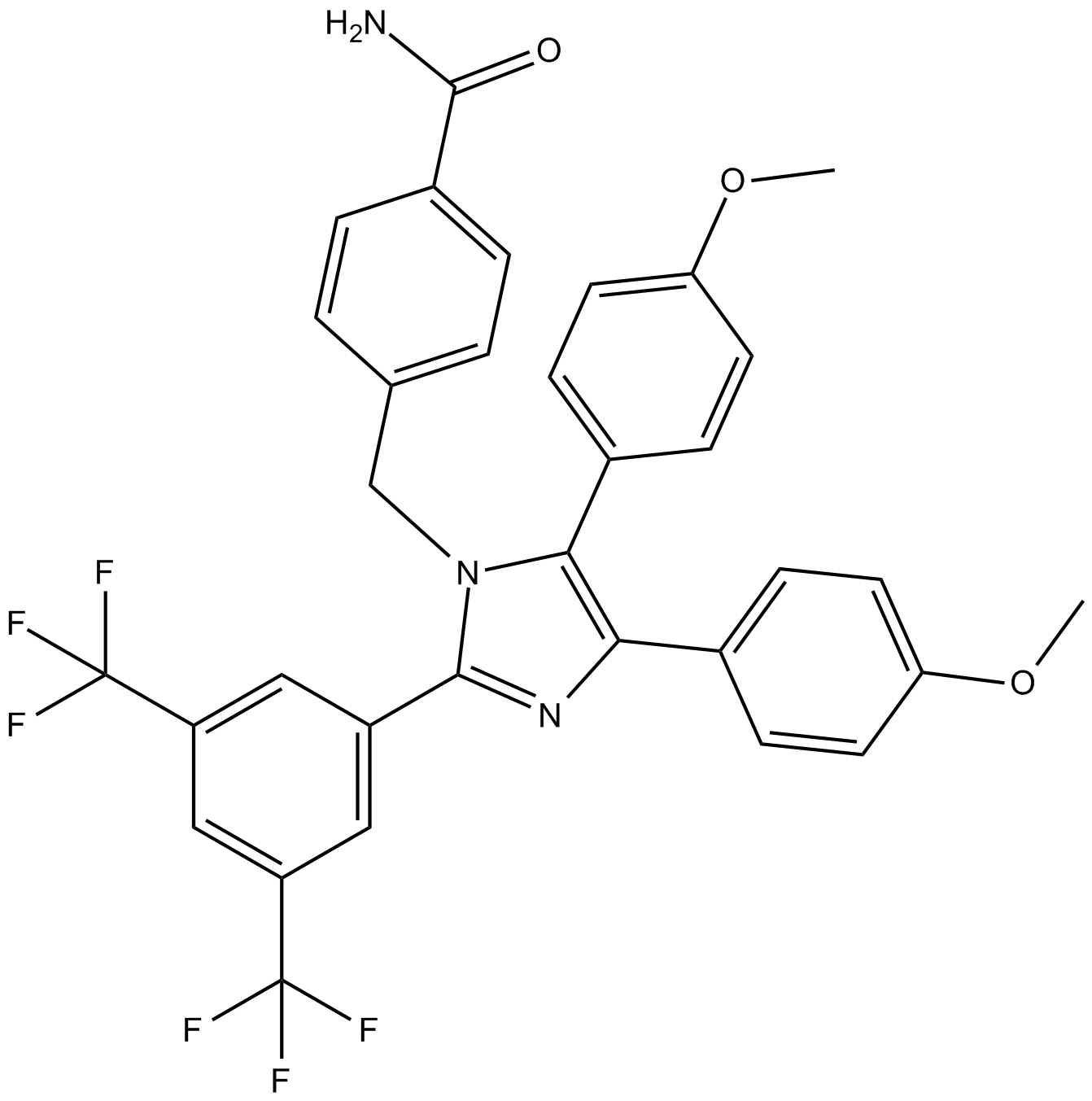
Apoptozole
inhibitor of heat shock protein 70 (Hsp70)
-
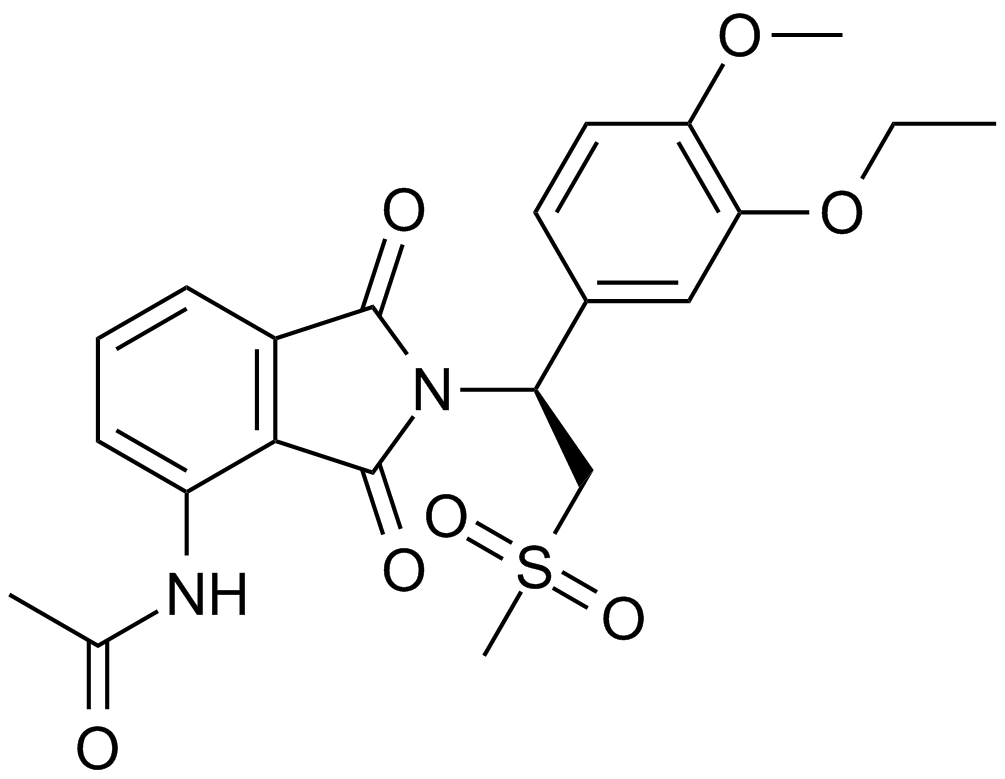
GC10420
Apremilast (CC-10004)
An orally available PDE4 inhibitor
-
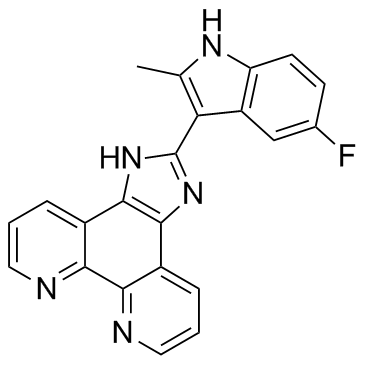
GC32692
APTO-253 (LOR-253)
APTO-253 (LOR-253) (LOR-253) is a small molecule that inhibits c-Myc expression, stabilizes G-quadruplex DNA, and induces cell cycle arrest and apoptosis in acute myeloid leukemia cells.
-
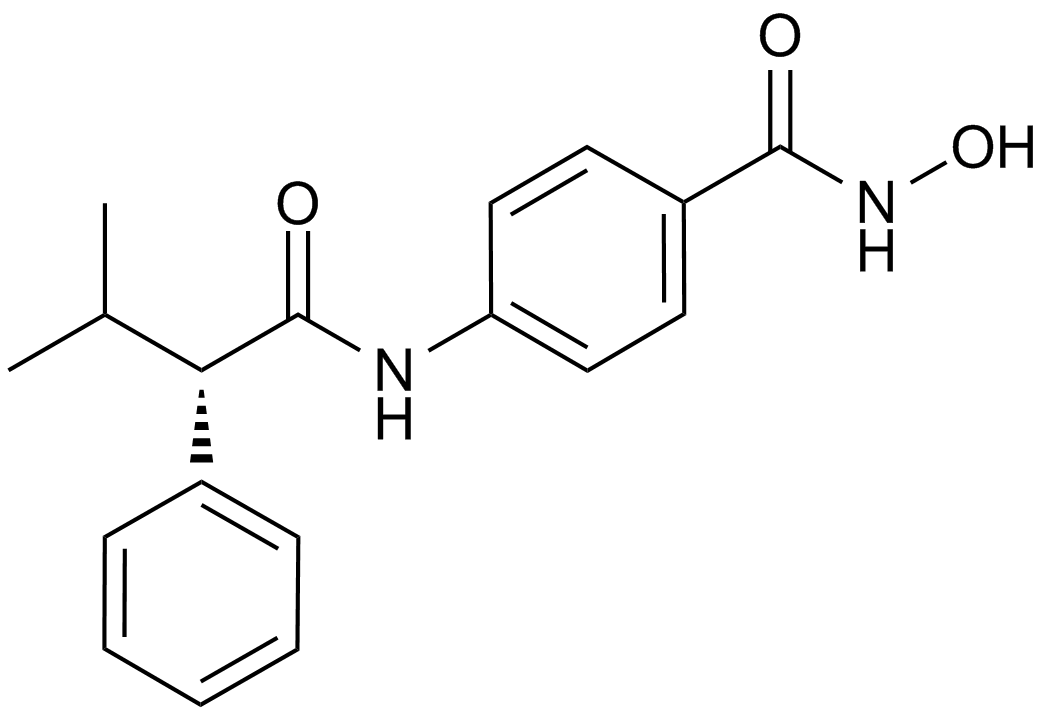
GC14590
AR-42 (OSU-HDAC42)
HDAC inhibitor,novel and potent
-
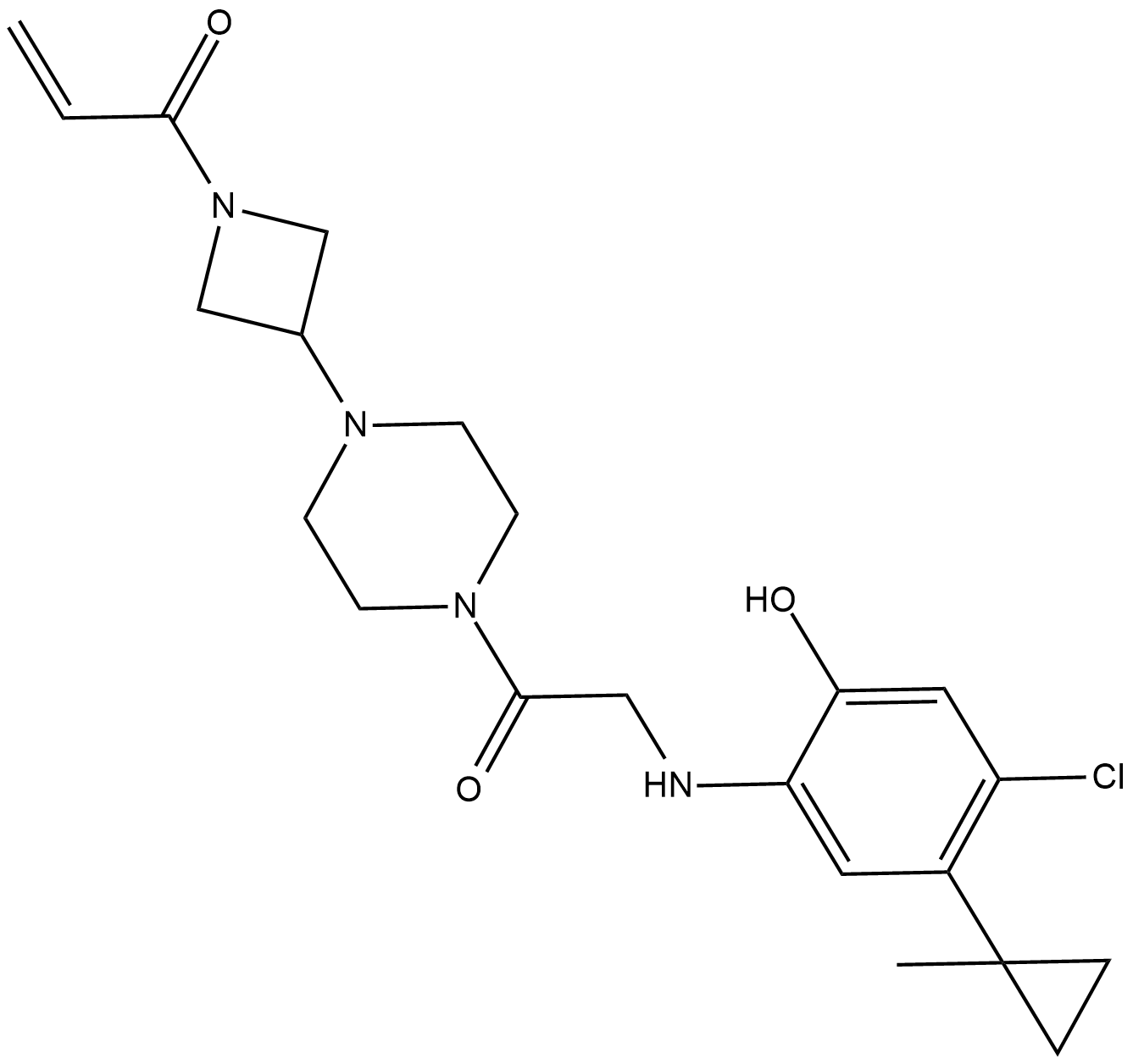
GC19037
ARS-853
ARS-853 is a selective, covalent KRASG12C inhibitor with an IC50 of 2.5 uM.
-
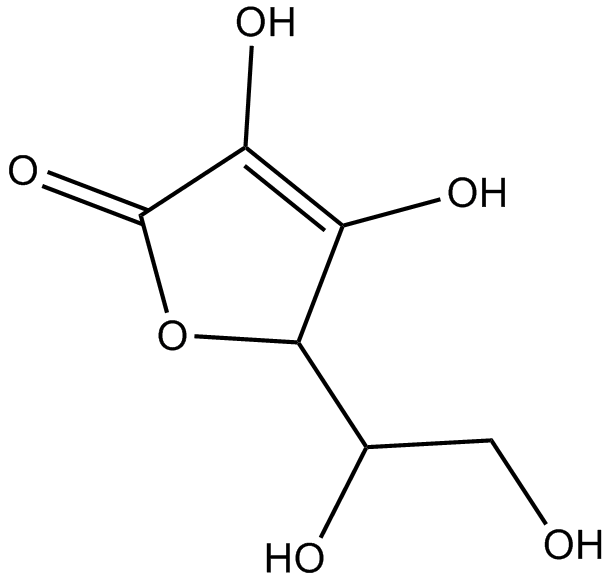
GC12070
Ascorbic acid
An electron donor
-
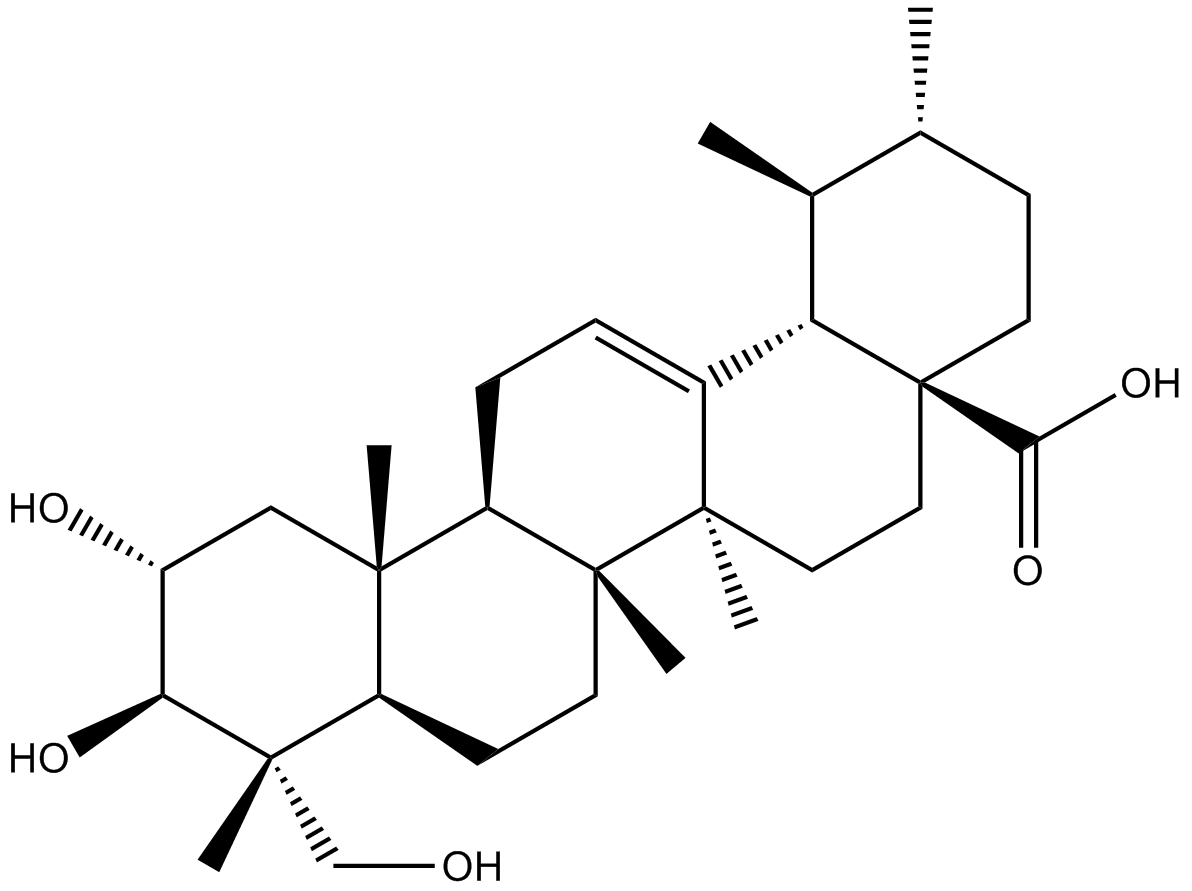
GN10702
Asiatic acid
-
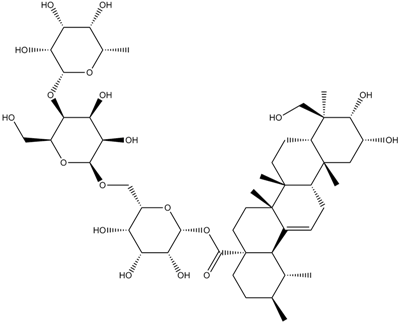
GN10534
Asiaticoside
-
GC19041
ASK1-IN-1
ASK1-IN-1 is a potent, orally available and selective ATP-competitive inhibitor of apoptosis signal-regulating kinase 1 (ASK1) with an IC50 of 2.87 nM.
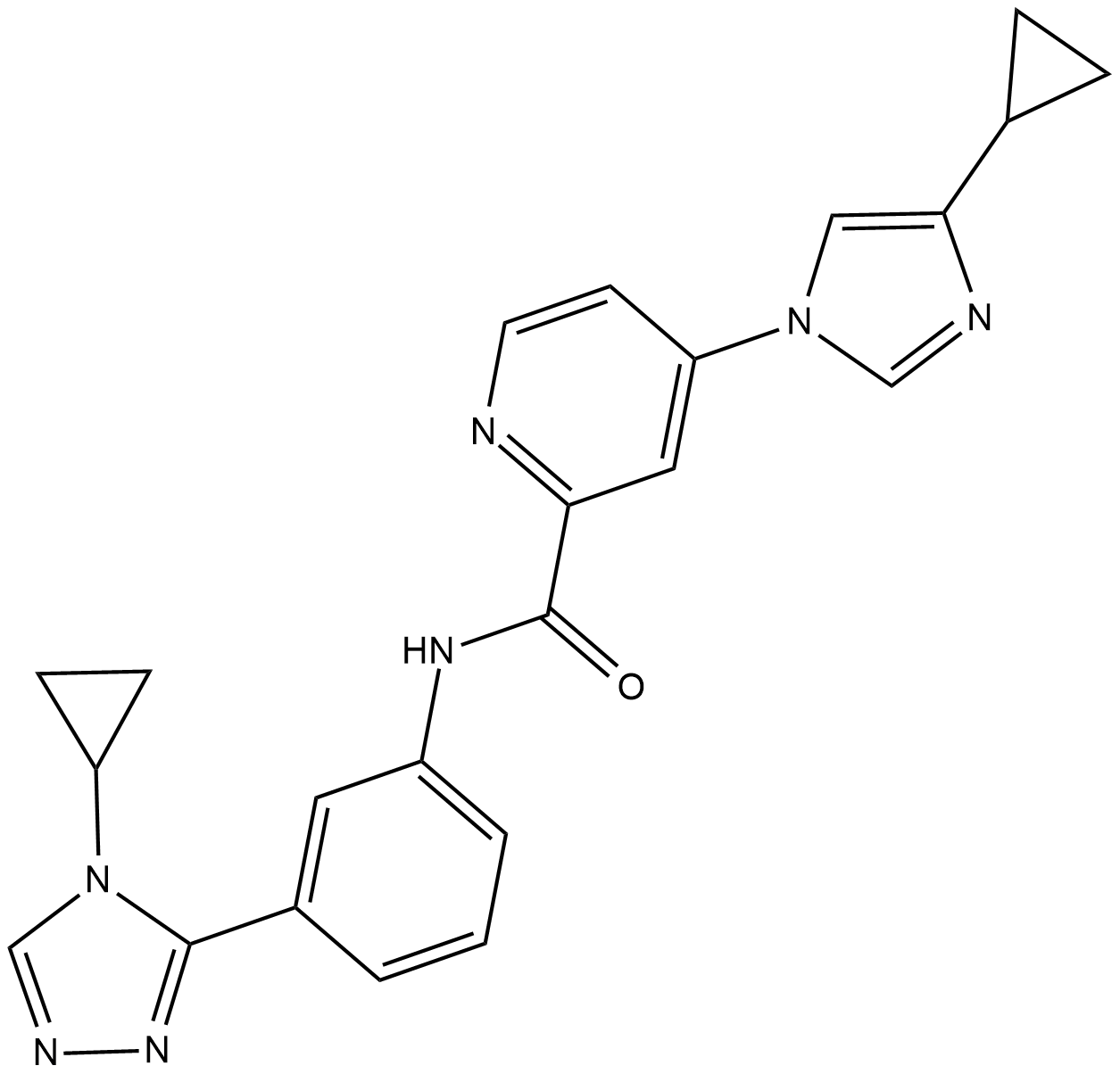
-
GN10064
Asperosaponin VI
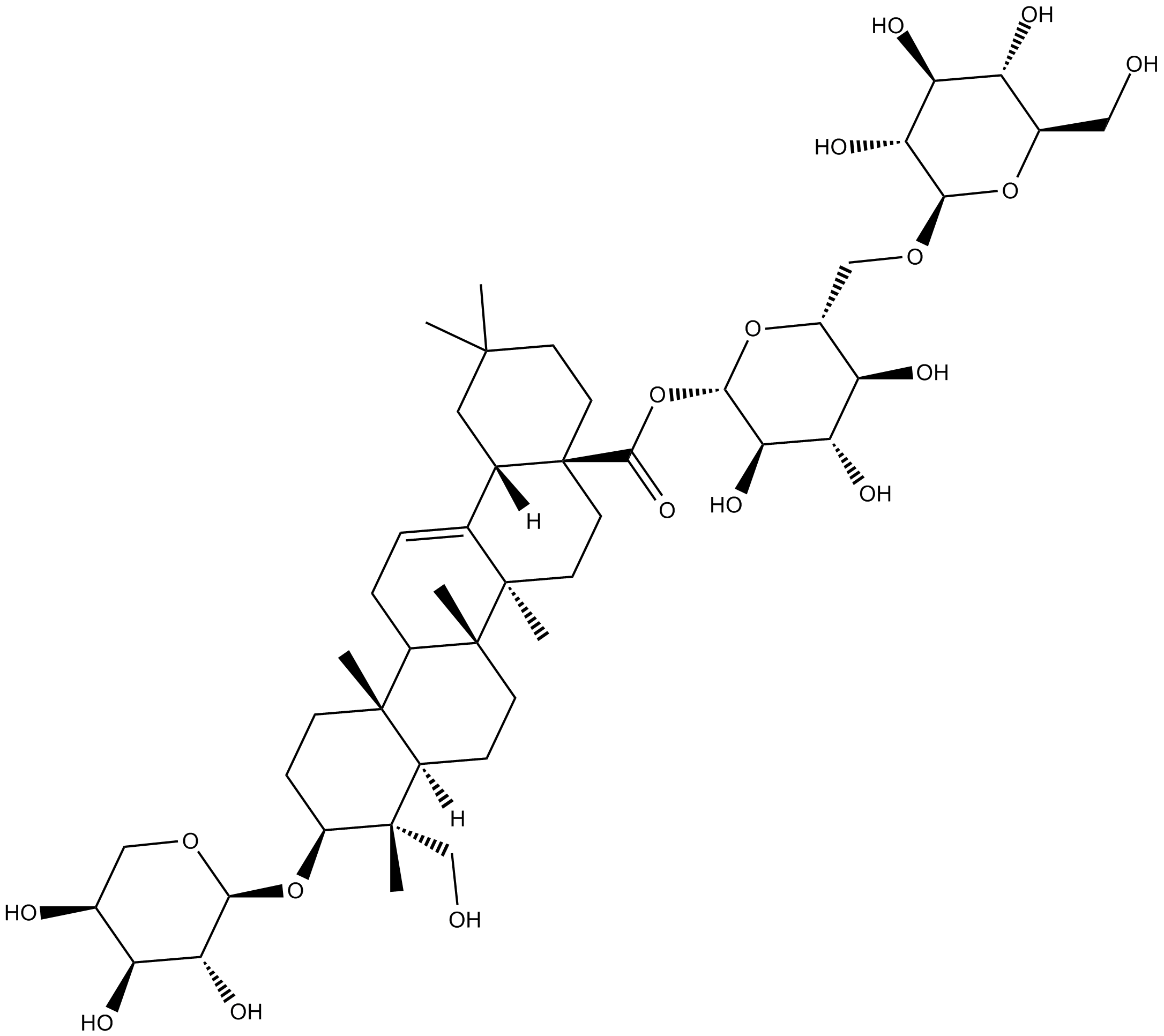
-
GN10561
astragalin
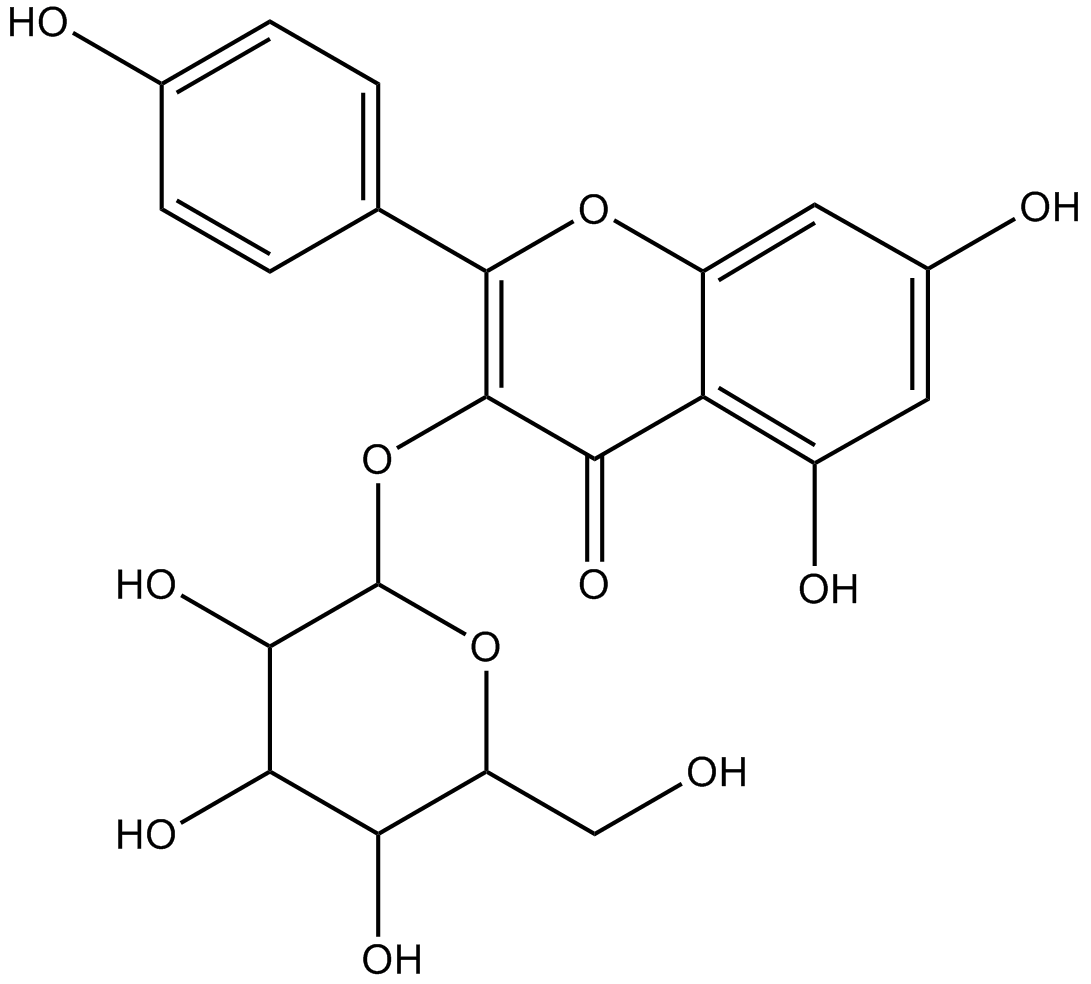
-
GC18109
Astragaloside A
anti-hypertension, positive inotropic action, anti-inflammation, and anti-myocardial injury
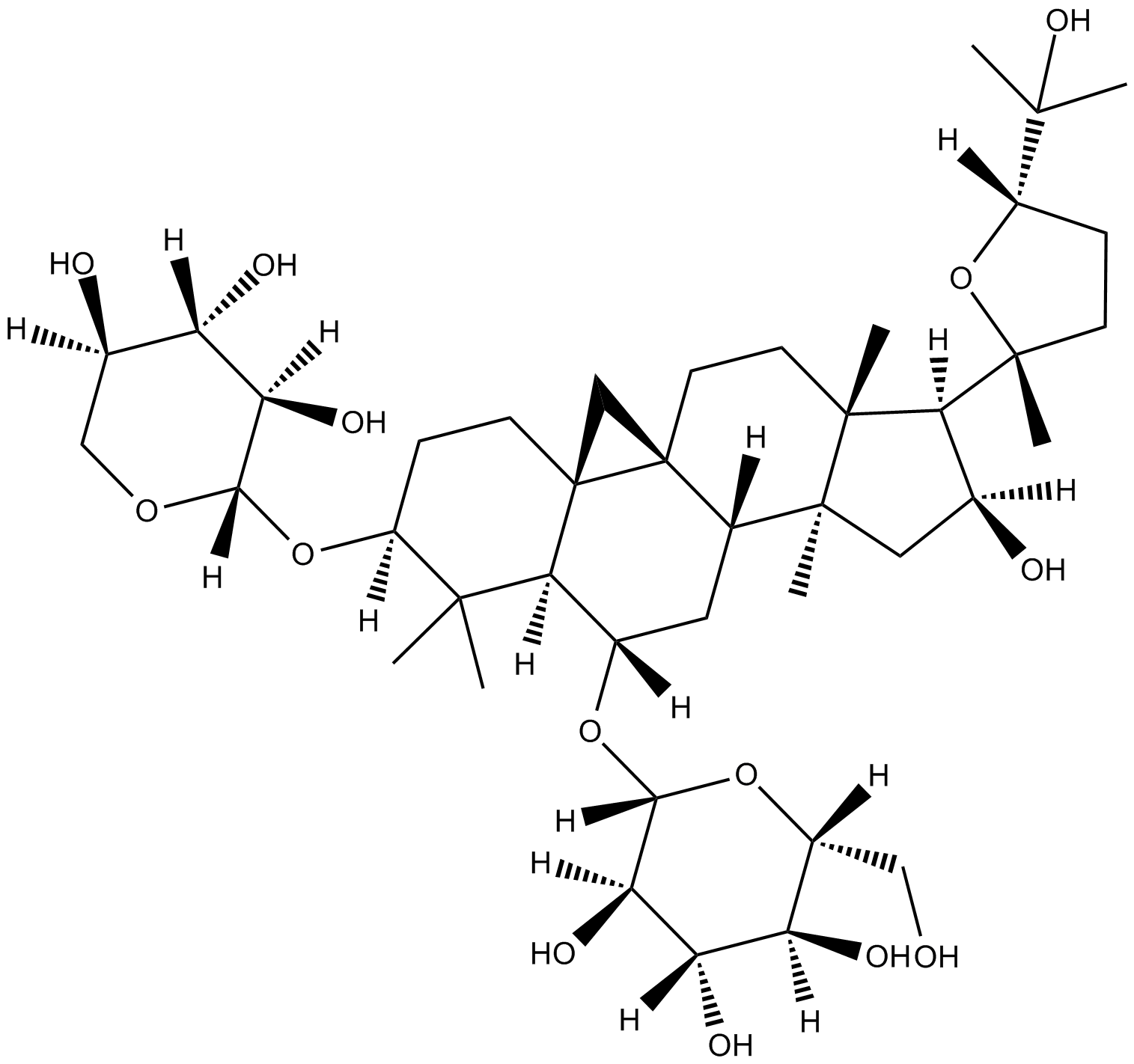
-
GC35415
Astramembrangenin
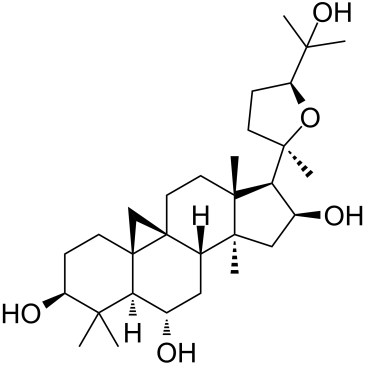
-
GC15870
AT7519
Multi-CDK inhibitor
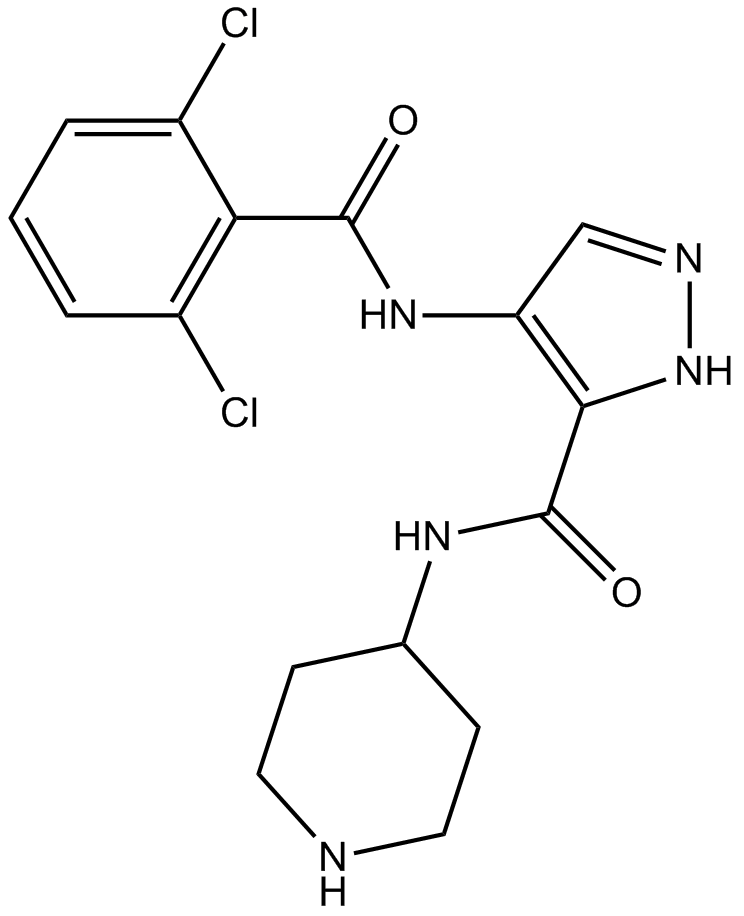
-
GC13998
AT7519 Hydrochloride
A Cdk inhibitor
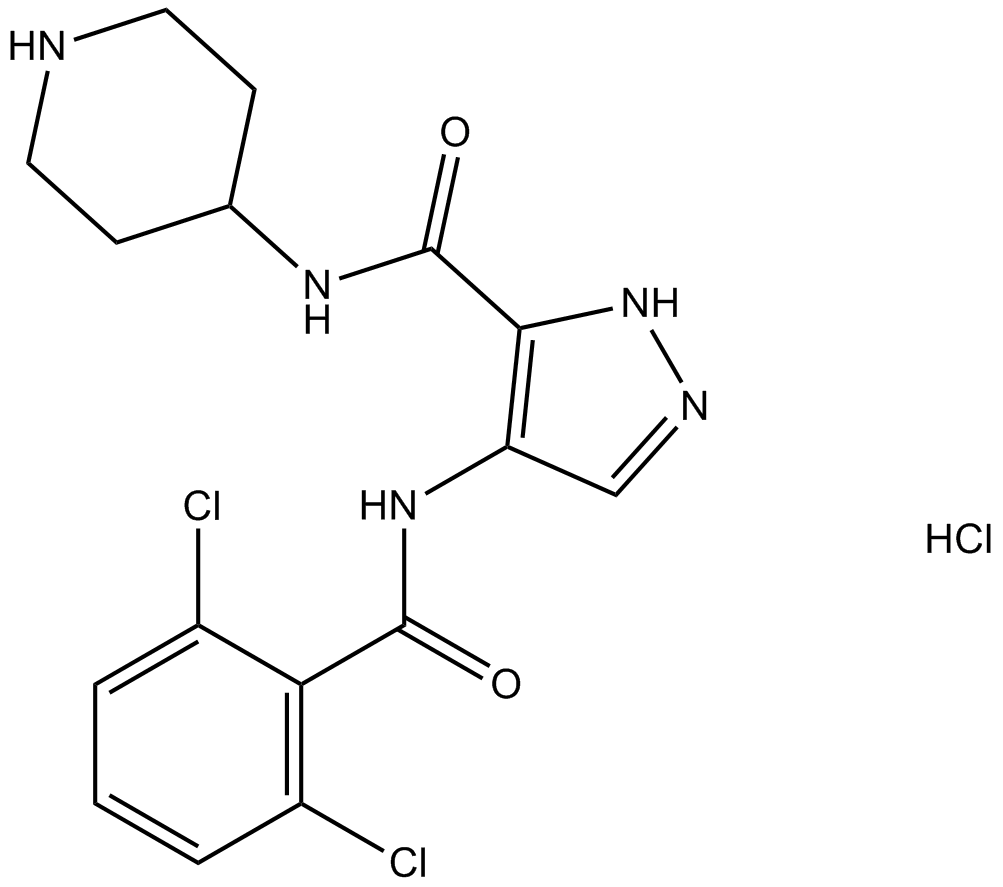
-
GC10638
AT9283
A broad spectrum kinase inhibitor
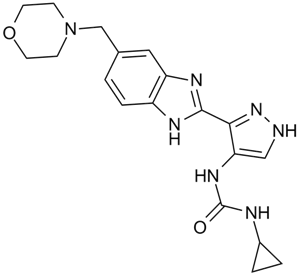
-
GC18133
ATB-346
ATB-346 (ATB-346), an orally active non-steroidal anti-inflammatory drug (NSAID), inhibits cyclooxygenase-1 and 2 (COX-1 and 2).
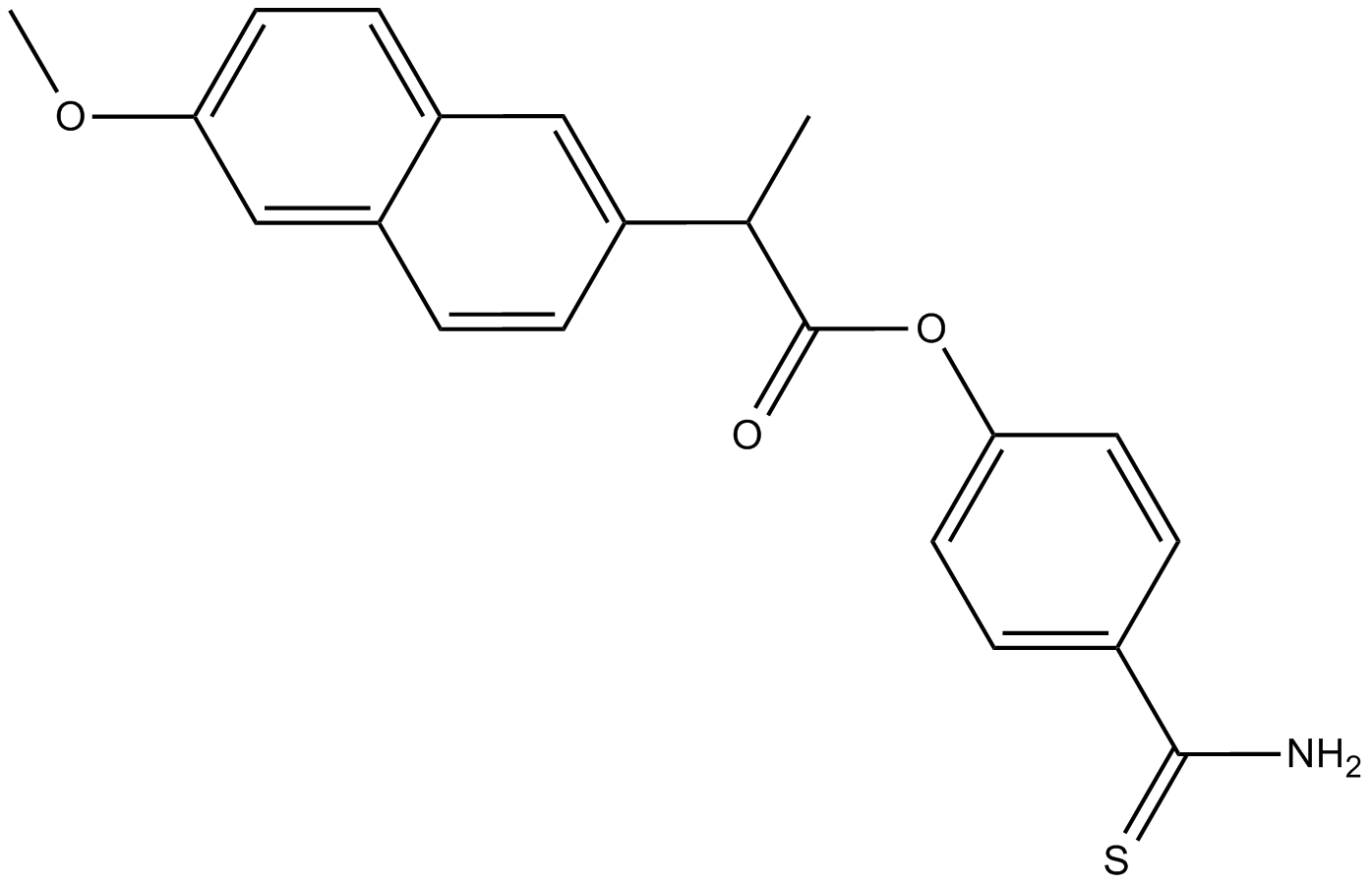
-
GN10394
Atractylenolide III

-
GC13332
Aurora A Inhibitor I
A potent and selective inhibitor of Aurora A kinase

-
GC15295
AUY922 (NVP-AUY922)
An Hsp90 inhibitor
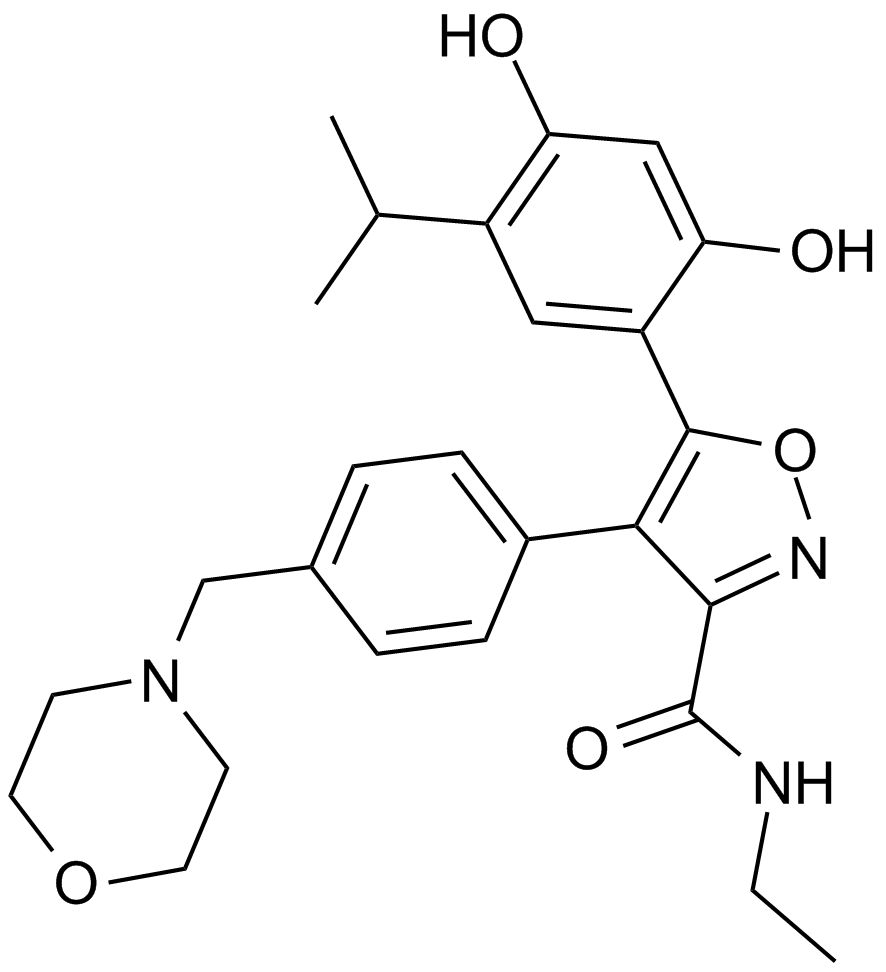
-
GC17045
AXL1717
A potent and selective inhibitor of IGF-1R
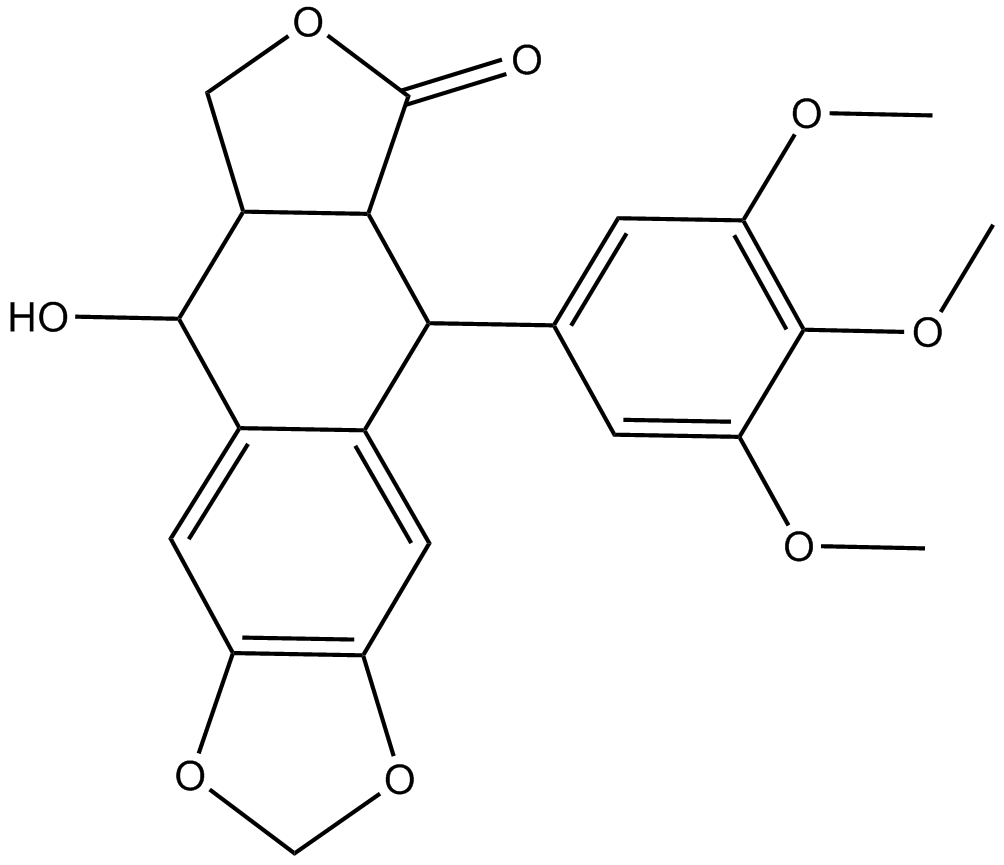
-
GC15055
AZ 628
Raf kinases,potent and ATP-competitive

-
GC13433
AZ 960
A JAK2 inhibitor
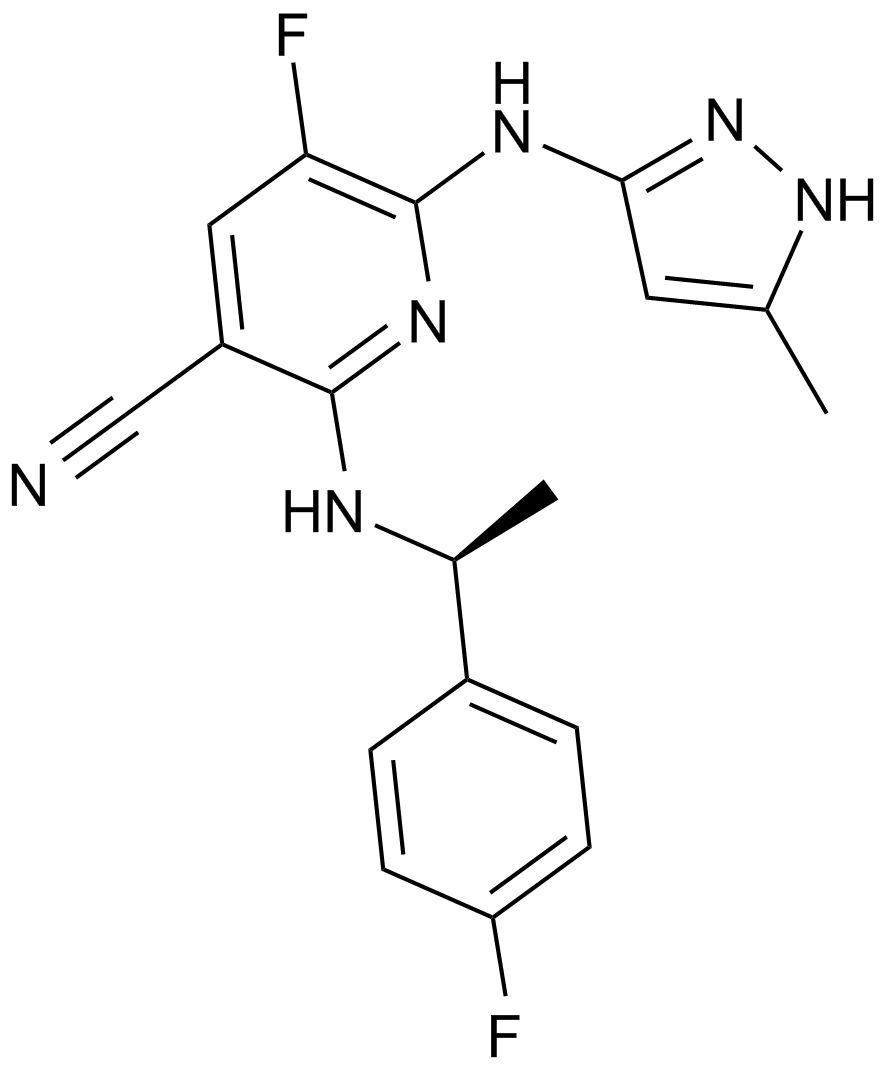
-
GC15033
Azathioprine
purine synthesis and GTP-binding protein Rac1 activation inhibitor
-
GC12660
AZD1208
A pan-Pim kinase inhibitor
-
GC13029
AZD2014
AZD2014 (AZD2014) is an ATP competitive mTOR inhibitor with an IC50 of 2.81 nM. AZD2014 inhibits both mTORC1 and mTORC2 complexes.
-
GC16380
AZD8055
MTOR inhibitor