PI3K/Akt/mTOR Signaling
- Akt(129)
- AMPK(75)
- CK2(11)
- DNA-PK(25)
- GSK-3(76)
- MELK(7)
- mTOR(118)
- PI3K(227)
- PI4K(15)
- PDK-1(18)
- PIKfyve(4)
- PKB(1)
- S6 Kinase(12)
Products for PI3K/Akt/mTOR Signaling
- Cat.No. Product Name Information
-
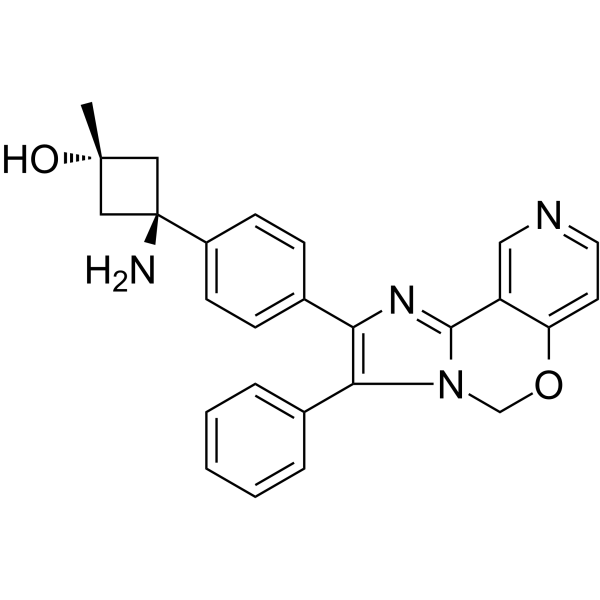
GC44641
PI3-Kinase α Inhibitor 2
Phosphatidylinositol 3-kinase (PI3K) catalyzes the phosphorylation of the 3' hydroxyl position of PIs to produce the second messengers PtdIns-(3,4)-P2 and PtdIns-(3,4,5)-P3.
-
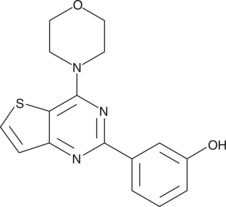
GC44642
PI3-Kinase α Inhibitor 2 (hydrochloride)
A PI3K p110α inhibitor
-
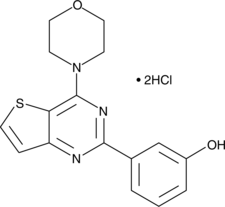
GC62521
PI3Kα-IN-4
PI3Kα-IN-4 is a potent, selective and orally active inhibitor of PI3Kα, with an IC50 of 1.8 nM. PI3Kα-IN-4 has antitumor activity.
-
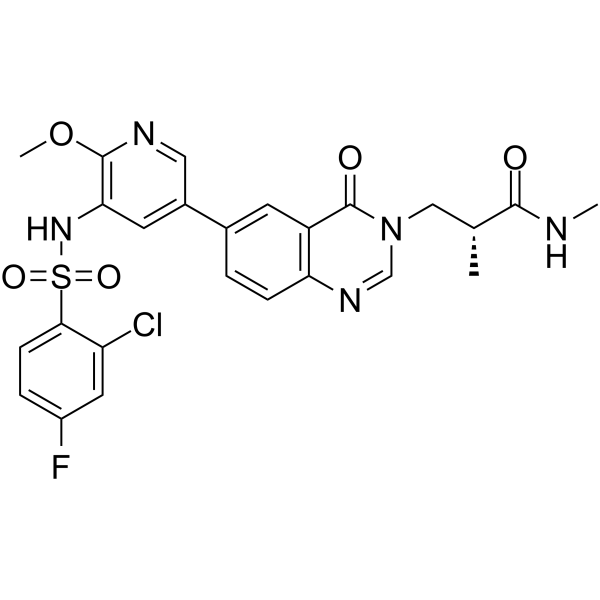
GC36909
PI3Kα/mTOR-IN-1
PI3Kα/mTOR-IN-1 is a potent PI3Kα/mTOR dual inhibitor, with an IC50 of 7 nM for PI3Kα in a cell assay, and Kis of 10.6 nM and 12.5 nM for mTOR and PI3Kα in a cell free assay , respectively.
-
GC36910
PI3Kγ inhibitor 1
PI3Kγ inhibitor 1 is a PI3Kδ and PI3Kγ inhibitor extracted from patent WO2014004470A1, Compound 168 in Table 4, has IC50s of <100 nM.
-
GC36911
PI3kδ inhibitor 1
PI3kδ inhibitor 1 is a potent and selective PI3Kδ inhibitor with an IC50 of 3.8 nM.
-
GC65199
PI3Kδ-IN-1
PI3Kδ-IN-1 is a potent, selective, and efficacious PI3Kδ inhibitor with an IC50 of 1.7 nM.
-
GC31751
PI3Kδ-IN-2
PI3Kδ-IN-2 (YY-20394) is a potent, orally bioavailable and selective inhibitor of PI3Kδ extracted from patent WO 2015055071 A1, compound 10; has an IC50 of 6.4 nM.
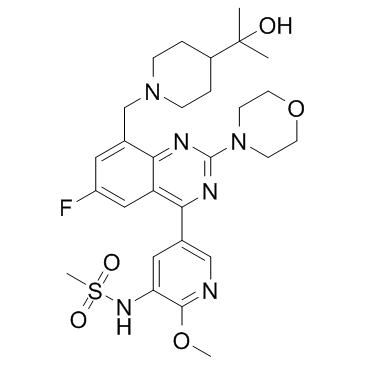
-
GC36907
PI3K-IN-2
PI3K-IN-2 (compound 10) is a potent and orally active PI3Kβ/δ (IC50=7.1/8.6 nM) inhibitor with excellent selectivity versus PI3Kσ and PI3Kγ (IC50=13/190 nM, respectively).
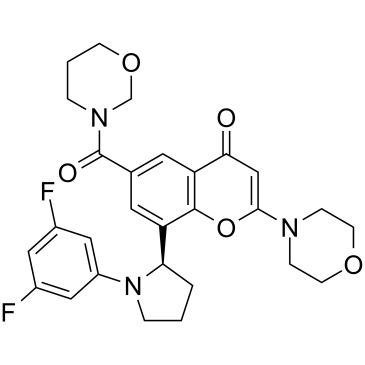
-
GC69707
PI3K-IN-30
PI3K-IN-30 (compound 6d) is an effective PI3K inhibitor with IC50 values of 5.1 nM, 136 nM, 30.7 nM and 8.9 nM for PI3Kα, PI3Kβ, PI3Kγ and PI3Kδ respectively.
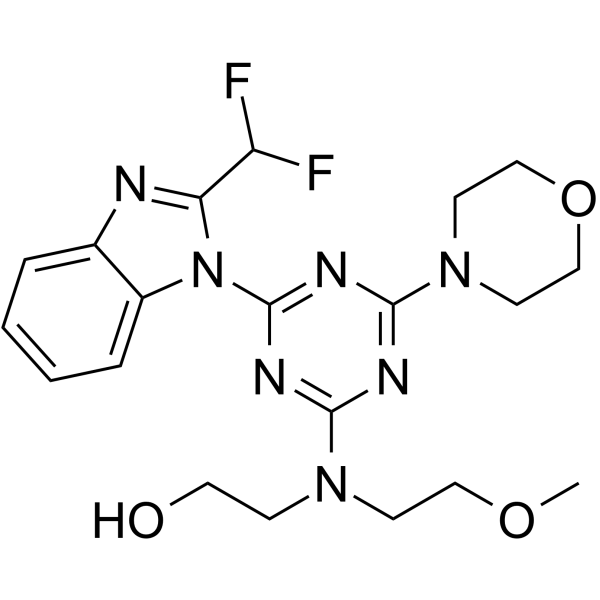
-
GC69708
PI3K-IN-31
PI3K-IN-31 (Compound 6b) is an effective PI3K inhibitor with IC50 values of 3.7 nM, 74 nM, 14.6 nM and 9.9 nM for PI3Kα, PI3Kβ, PI3Kγ and PI3Kδ respectively. It has anti-cancer properties.
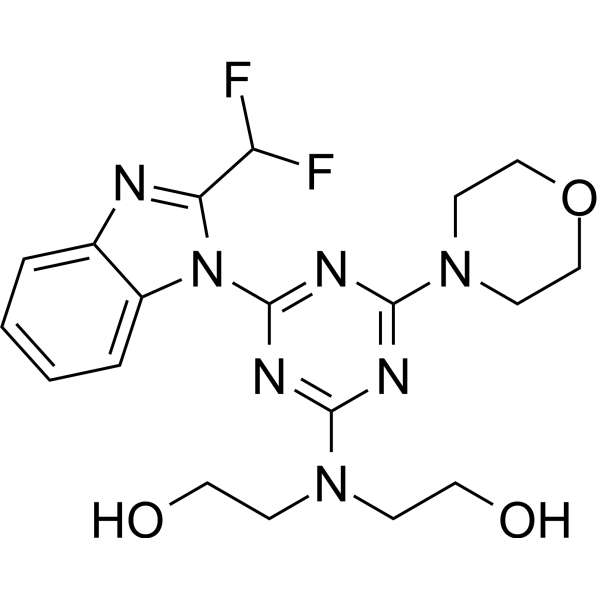
-
GC67954
PI3K-IN-36
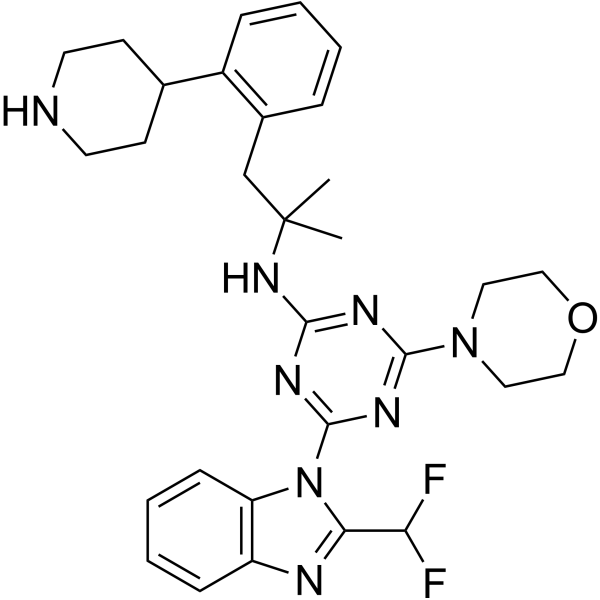
-
GC36908
PI3K-IN-6
PI3K-IN-6 (compound 20a) is an oral active and highly selective phosphoinositide 3-kinase (PI3K) β/δ inhibitor, with IC50 values of 7.8 nM/5.3 nM for PI3K β/δ, respectively. PI3K-IN-6 (compound 20a) has potential top treat phosphatase and tensin homolog (PTEN) feficient tumors.
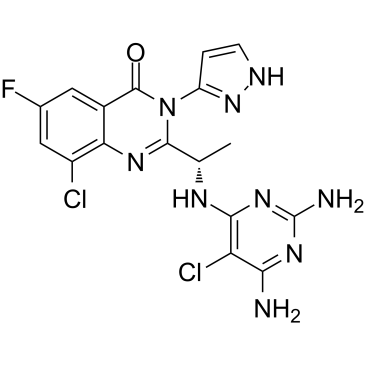
-
GC69706
PI3K/AKT-IN-1
PI3K/AKT-IN-1 is an effective dual inhibitor of PI3K/AKT (with IC50 values of 6.99 μM, 4.01 μM, and 3.36 μM for PI3Kα, PI3Kδ, and AKT, respectively). It has anti-cancer activity by inhibiting the PI3K/AKT pathway and inducing caspase 3-dependent apoptosis.
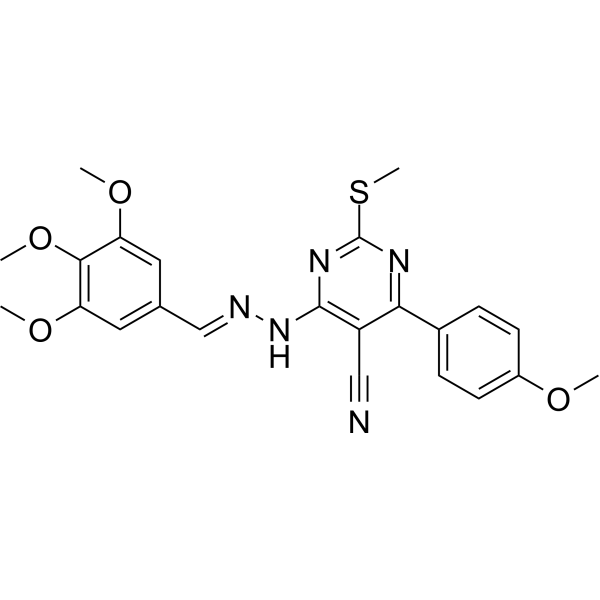
-
GC66463
PI3K/Akt/mTOR-IN-2
PI3K/Akt/mTOR-IN-2 is a PI3K/AKT/mTOR pathway inhibitor. PI3K/Akt/mTOR-IN-2 possess anti-cancer effects and selectivity against MDA-MB-231 cells with IC50 value of 2.29 μM. PI3K/Akt/mTOR-IN-2 can induce cancer cell cycle arrest and apoptosis.
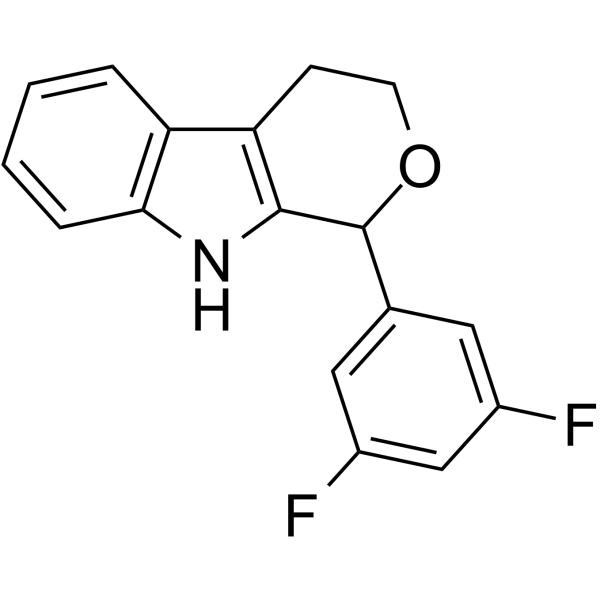
-
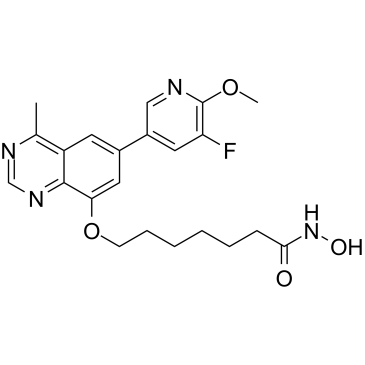
GC36905
PI3K/HDAC-IN-1
PI3K/HDAC-IN-1 is a potent dual inhibitor of PI3K/HDAC, potently inhibits PI3Kδ and HDAC1 with IC50s of 8.1 nM and 1.4 nM, respectively.
-
GC69709
PI3K/mTOR Inhibitor-11
PI3K/mTOR Inhibitor-11 is an orally active inhibitor of PI3K/mTOR (with IC50 values of 3.5, 4.6, and 21.3 nM for PI3Kα, PI3Kδ, and mTOR, respectively). It regulates the PI3K/AKT/mTOR signaling pathway by inhibiting the phosphorylation of AKT and S6 protein. PI3K/mTOR Inhibitor-11 can be used in cancer research.

-
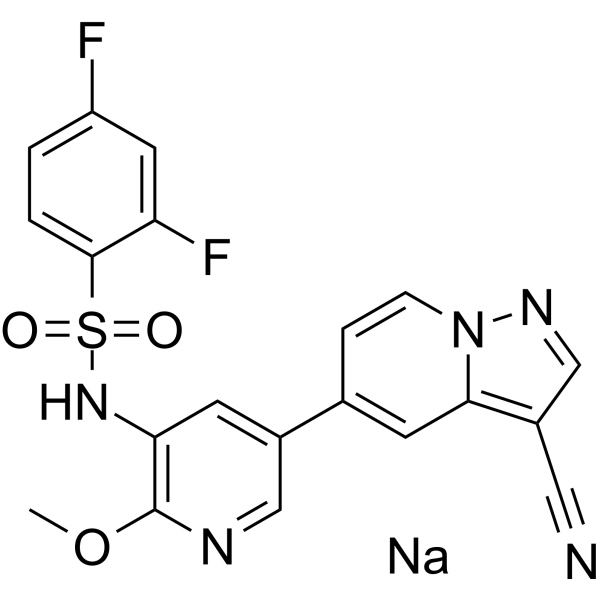
GC69710
PI3K/mTOR Inhibitor-13 sodium
PI3K/mTOR Inhibitor-13 sodium is an orally active dual inhibitor of phosphoinositide 3-kinase (PI3K) and mTOR kinase. PI3K/mTOR Inhibitor-13 sodium has potential applications in sexual disorders, solid tumors, and idiopathic pulmonary fibrosis (IPF).
-
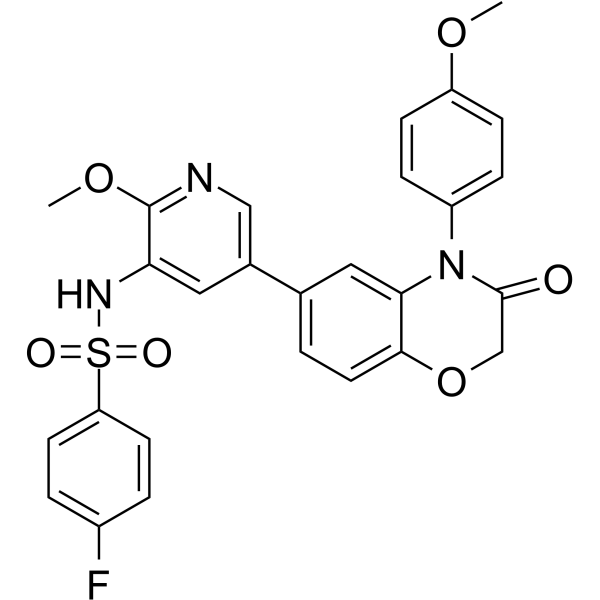
GC66447
PI3K/mTOR Inhibitor-4
PI3K/mTOR Inhibitor-4 is an orally active pan-class I PI3K/mTOR inhibitor. PI3K/mTOR Inhibitor-4 has enzymatic inhibition activity for PI3Kα, PI3Kγ, PI3Kδ and mTOR with IC50 values of 0.63 nM, 22 nM, 9.2 nM and 13.85 nM, respectively. PI3K/mTOR Inhibitor-4 can be used for the research of cancer.
-
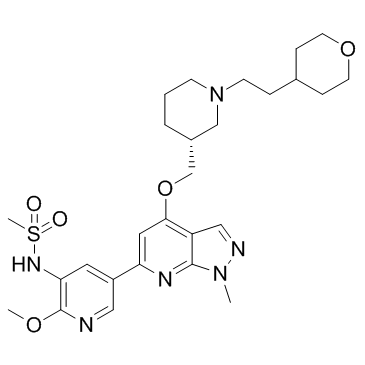
GC36906
PI3Kdelta inhibitor 1
PI3Kdelta inhibitor 1 (Compound 5d) is a potent, selective and orally available PI3Kδ inhibitor with an IC50 of 1.3 nM.
-
GC15508
PI4KIII beta inhibitor 3

-
GC30777
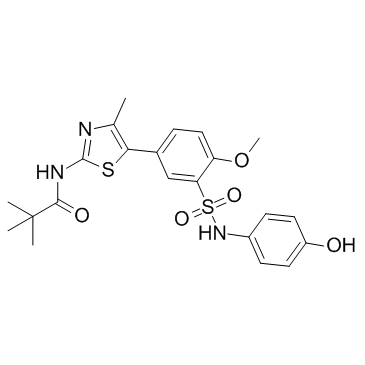
PI4KIIIbeta-IN-10
PI4KIIIbeta-IN-10 is a potent PI4KIIIβ inhibitor with an IC50 of 3.6 nM.
-
GC32847
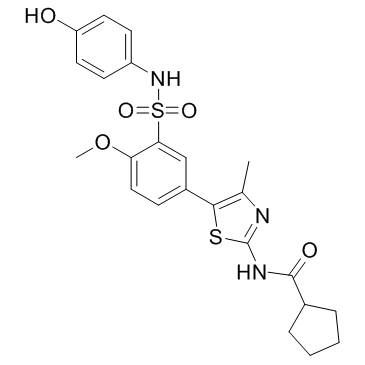
PI4KIIIbeta-IN-9
PI4KIIIbeta-IN-9 is a potent PI4KIIIβ inhibitor with an IC50 of 7 nM. PI4KIIIbeta-IN-9 also inhibits PI3Kδ and PI3Kγ with IC50s of 152 nM and 1046 nM, respectively.
-
GC69991
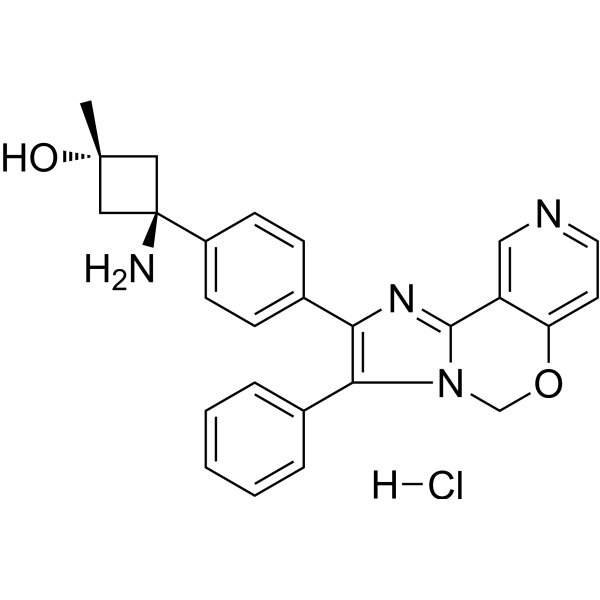
Pifusertib hydrochloride
Pifusertib (TAS-117) hydrochloride is an effective, selective, and orally active isoform Akt inhibitor (with IC50 values of 4.8, 1.6, and 44 nM for Akt1, 2, and 3 respectively). Pifusertib hydrochloride stimulates anti-myeloma activity and enhances lethal endoplasmic reticulum stress induced by proteasome inhibition. Pifusertib hydrochloride induces apoptosis and autophagy in cells.
-
GC62340
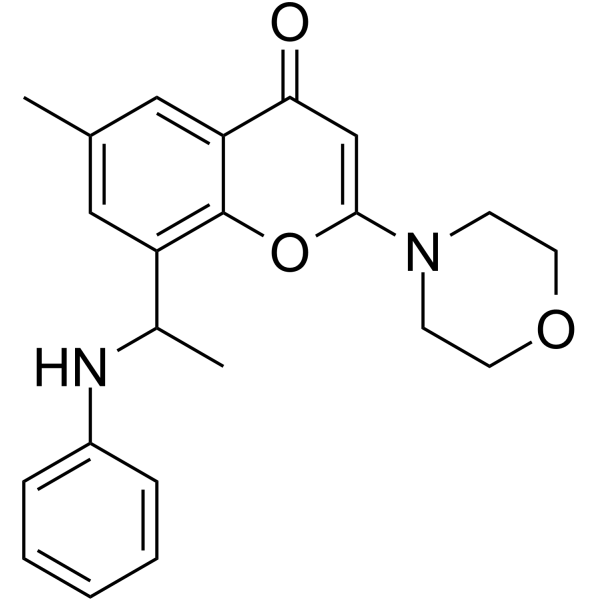
PIK-108
PIK-108 is a non-ATP competitive, allosteric p110β/p110δ selective inhibitor.
-
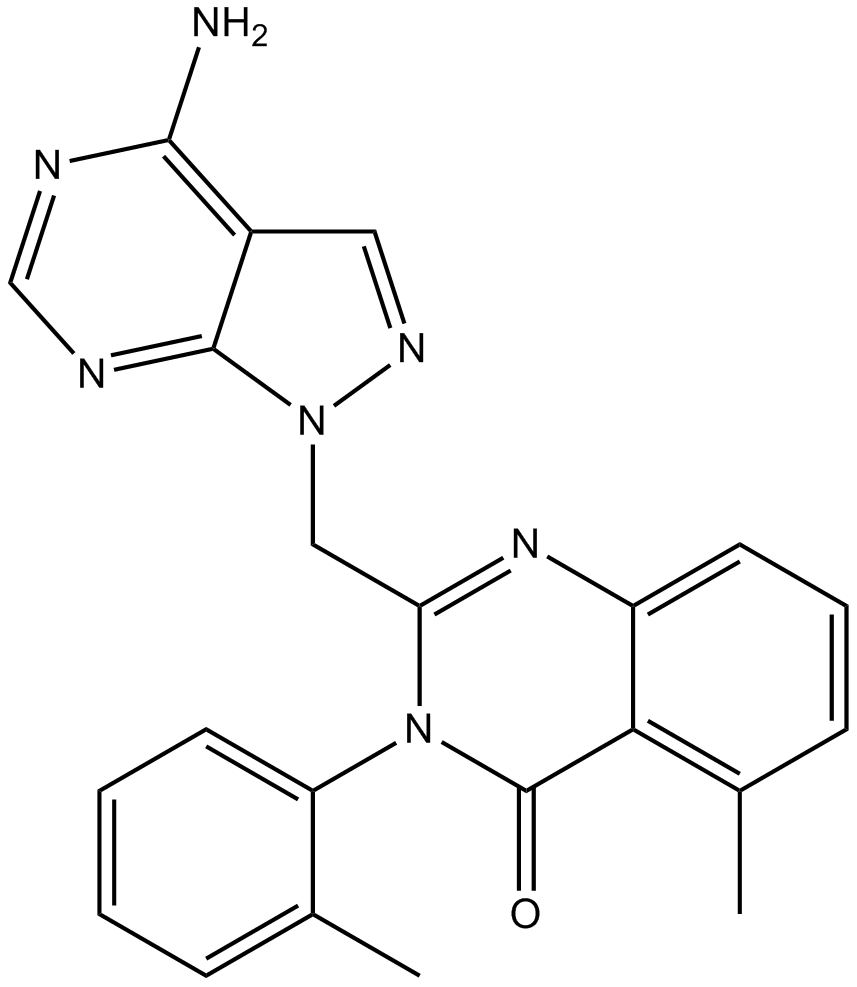
GC15982
PIK-293
PI3K inhibitor
-
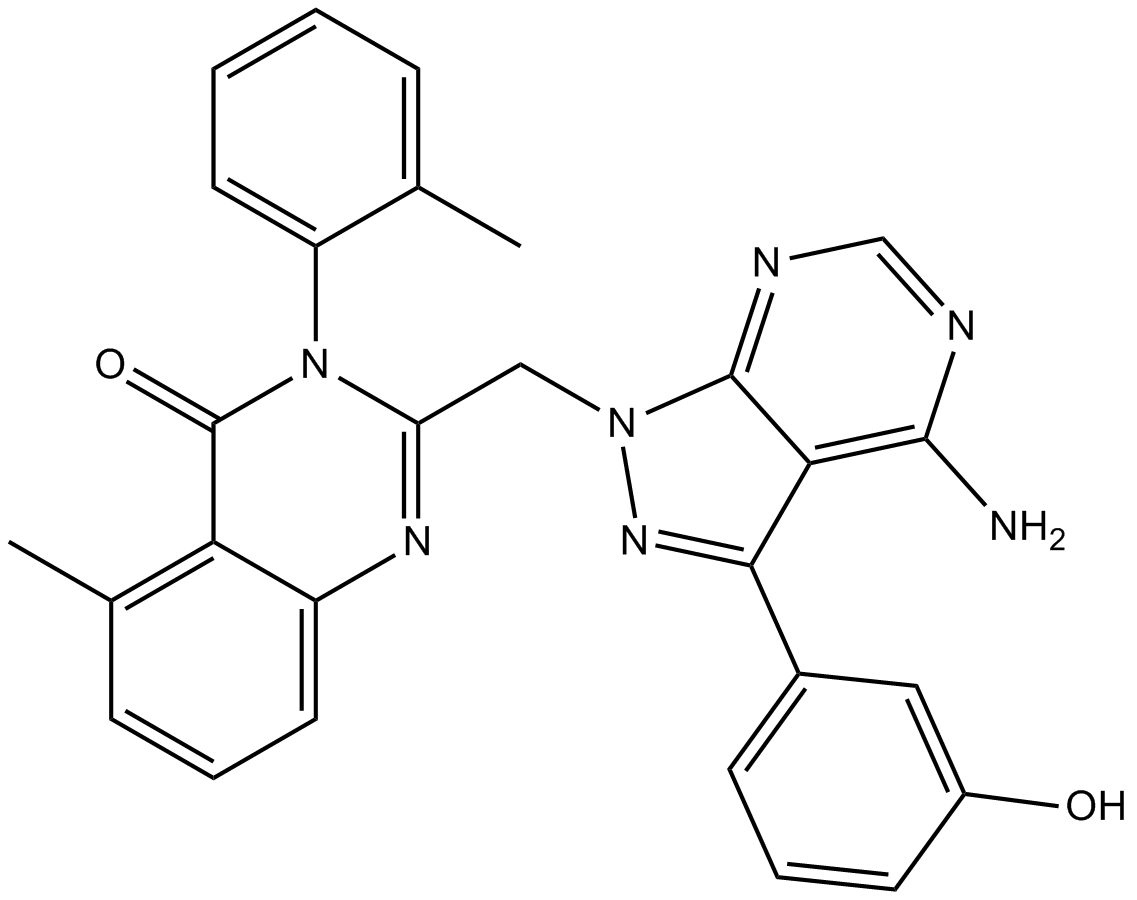
GC13612
PIK-294
highly selective p110δ inhibitor
-
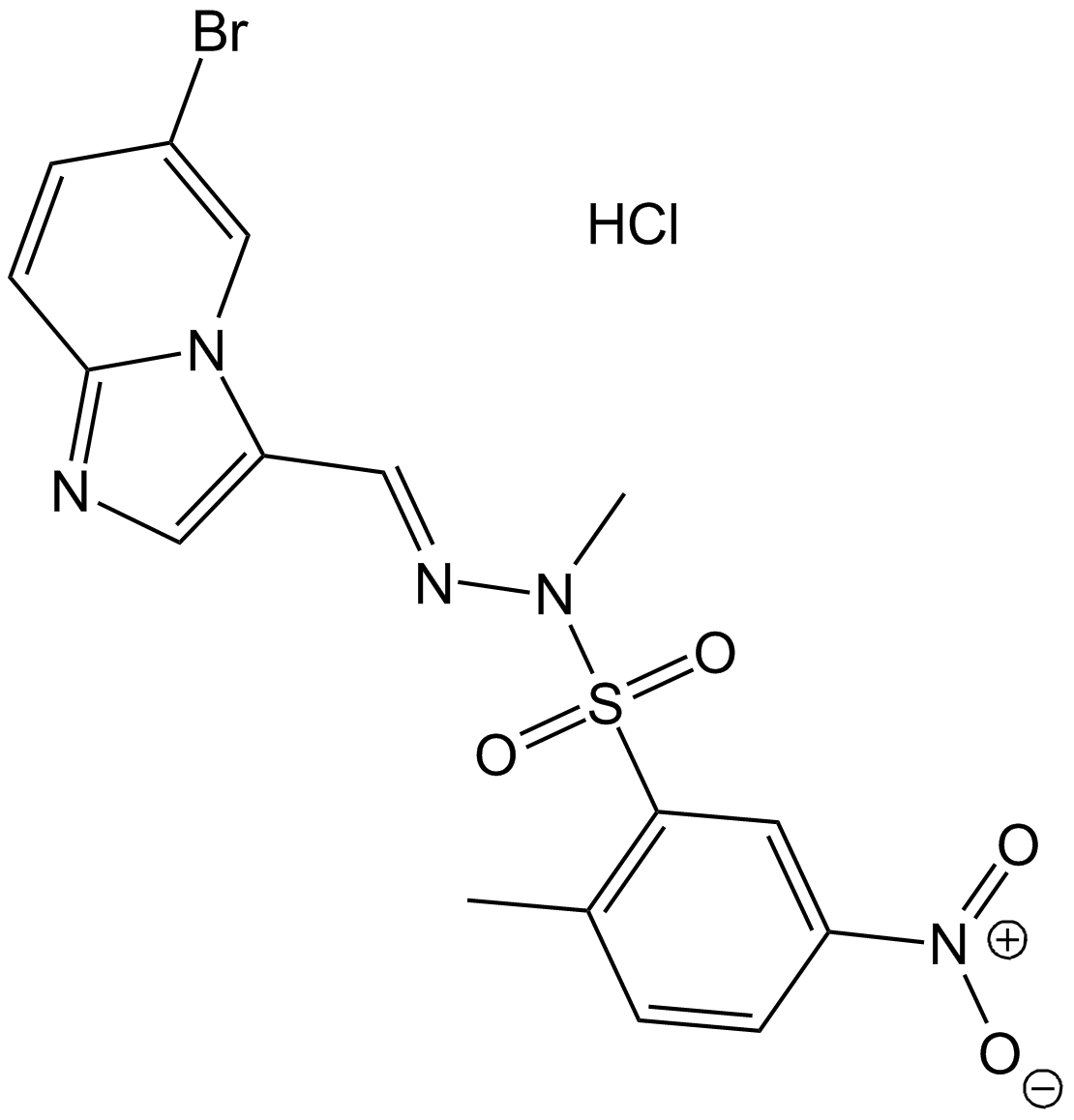
GC16904
PIK-75
PIK-75 is a reversible DNA-PK and p110α-selective inhibitor, which inhibits DNA-PK, p110α and p110γ with IC50s of 2, 5.8 and 76 nM, respectively. PIK-75 inhibits p110α >200-fold more potently than p110β (IC50=1.3 μM). PIK-75 induces apoptosis.
-
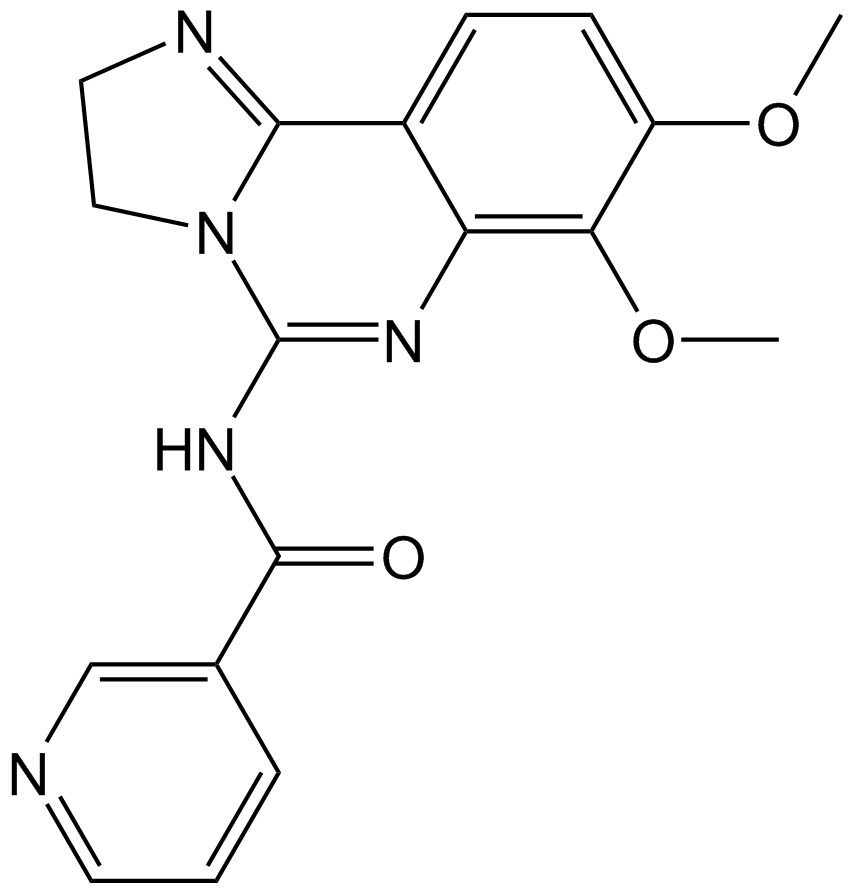
GC17199
PIK-90
PI3K inhibitor,potent selective
-
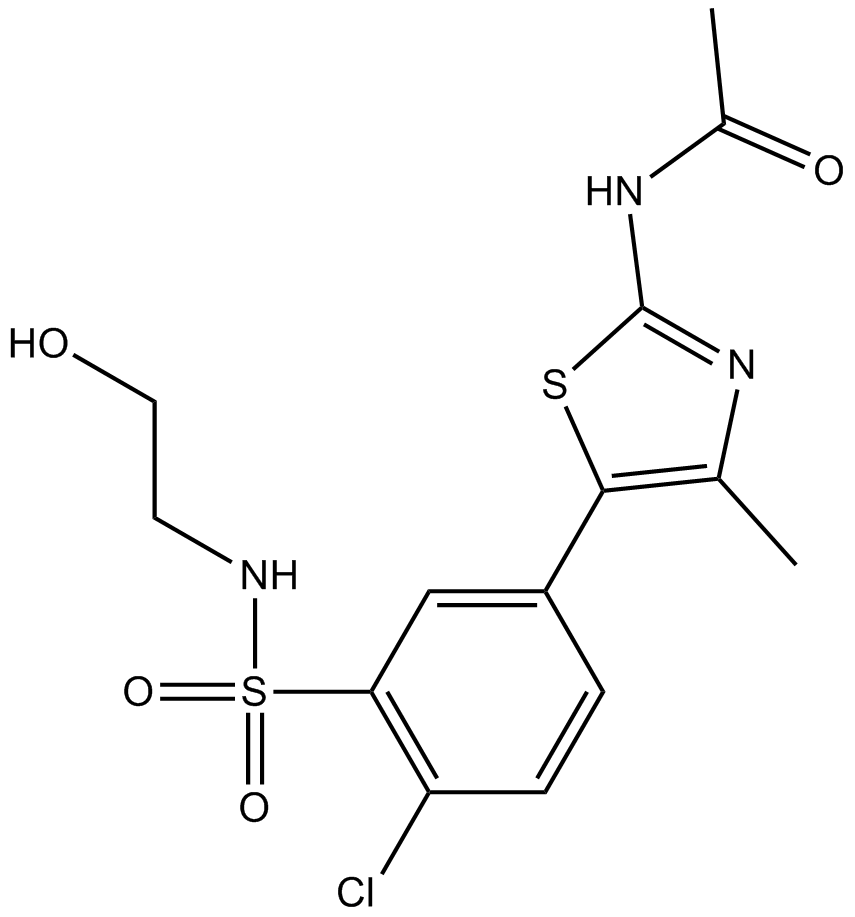
GC13089
PIK-93
PI3Kγ/PI4KIIIβ/PI3Kα inhibitor
-
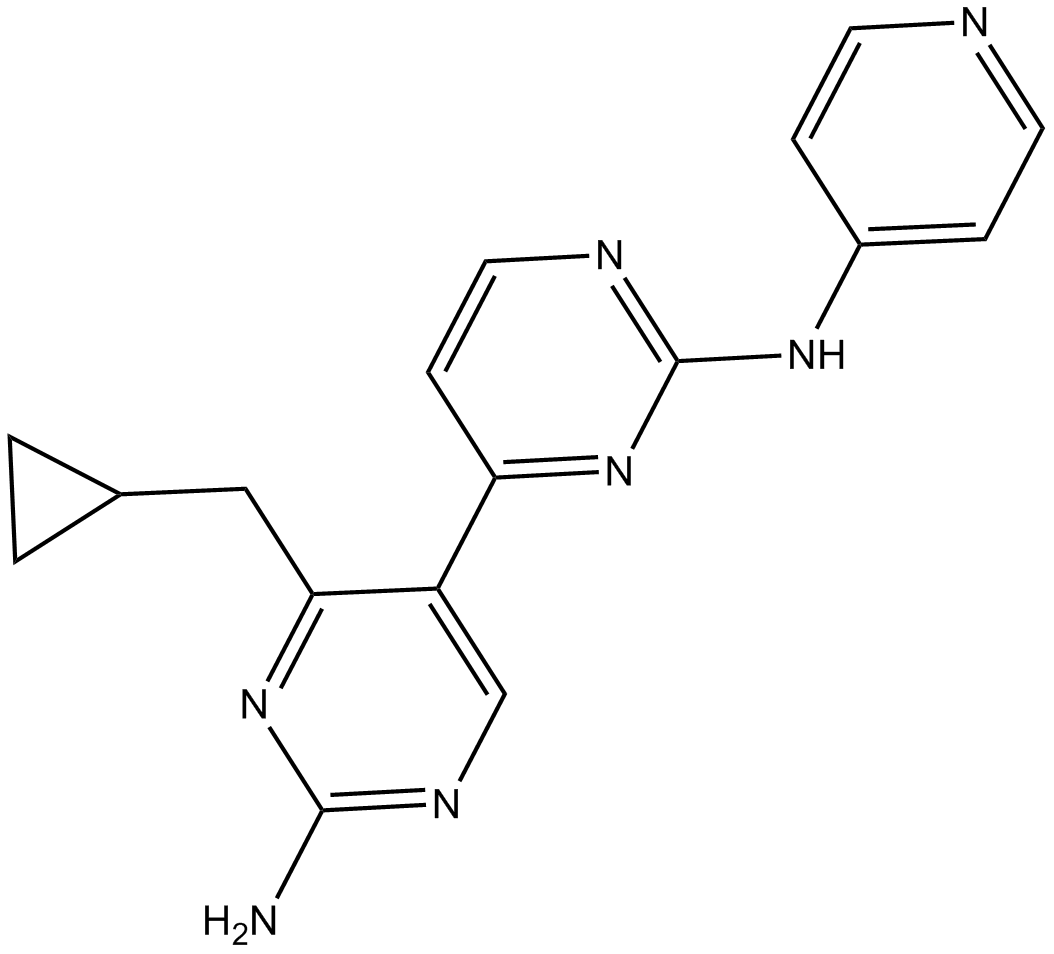
GC14679
PIK-III
PIK-III is a potent and selective inhibitor of VPS34 with an IC50 of 18 nM.
-
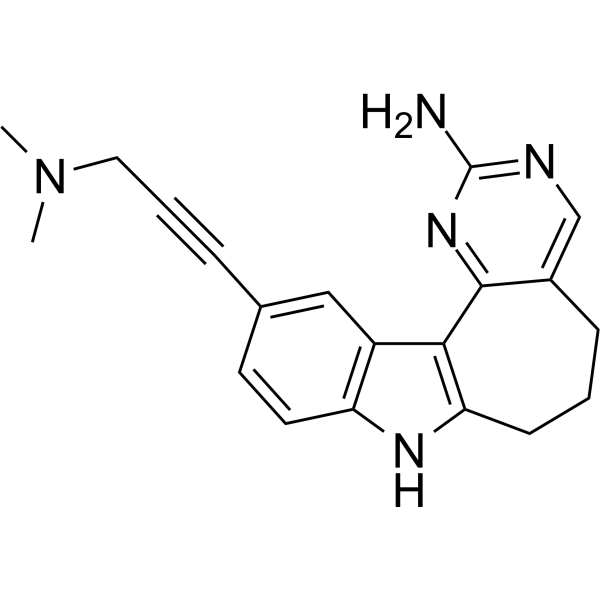
GC69711
PIKfyve-IN-1
PIKfyve-IN-1 is an efficient and cell-active chemical probe that can inhibit phosphoinositide 3-phosphate 5-kinase (PIKfyve) with an IC50 value of 6.9 nM. PIKfyve-IN-1 can be used for the study of PIKfyve in virology.
-
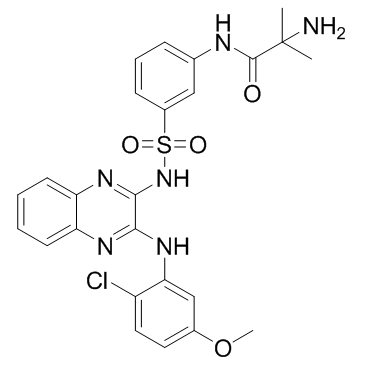
GC36917
Pilaralisib
A class I PI3K inhibitor
-
GC11690
PIT 1
Akt signaling inhibitor
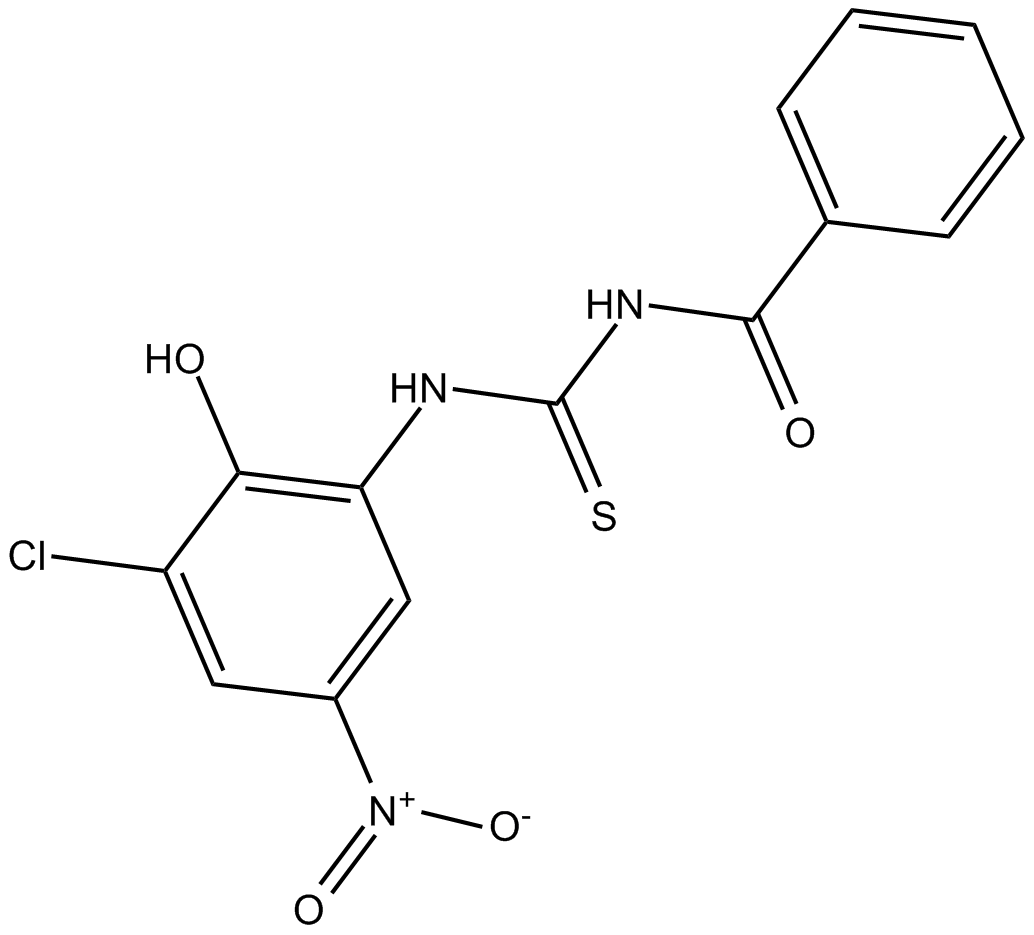
-
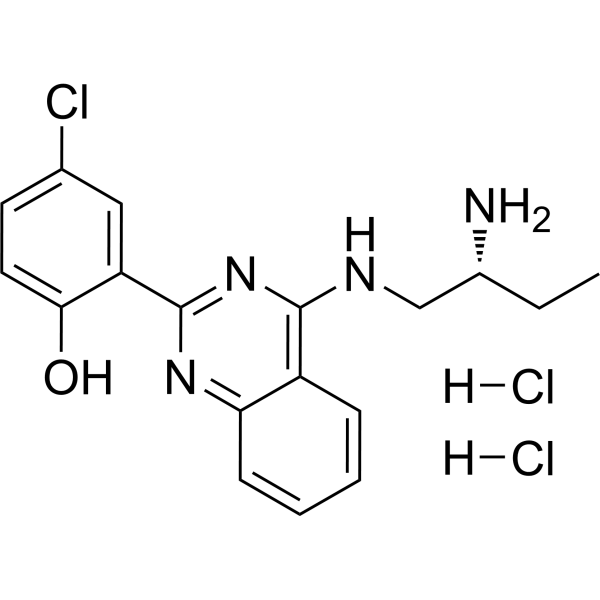
GC69714
PKD-IN-1 dihydrochloride
PKD-IN-1 dihydrochloride (compound 32) is an aminoethylaminobenzyl (AEAA) compound that can be used as a PKD-1 inhibitor. PKD-IN-1 can be used in research on diseases mediated by protein kinase D (PKD).
-
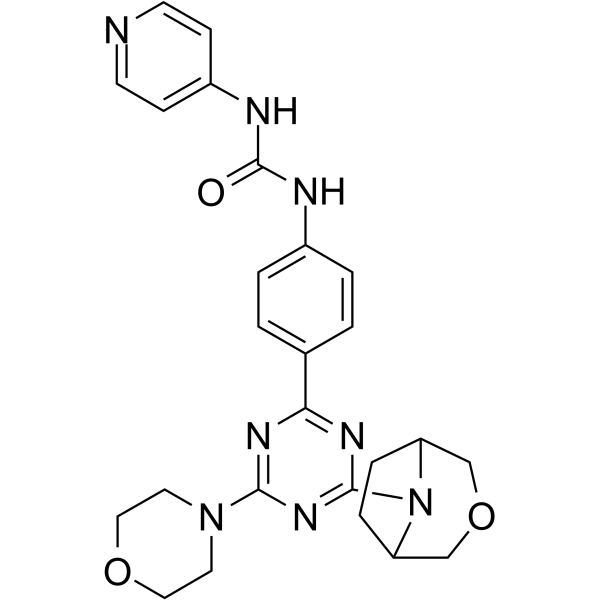
GC62140
PKI-179
PKI-179 is a potent and orally active dual PI3K/mTOR inhibitor, with IC50s of 8 nM, 24 nM, 74 nM, 77 nM, and 0.42 nM for PI3K-α, PI3K-β, PI3K-γ, PI3K-δ and mTOR, respectively. PKI-179 also exhibits activity over E545K and H1047R, with IC50s of 14 nM and 11 nM, respectively. PKI-179 shows anti-tumor activity in vivo.
-
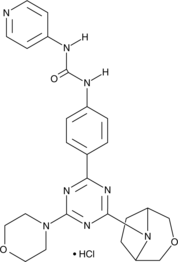
GC44658
PKI-179 (hydrochloride)
PKI-179 is an orally bioavailable dual inhibitor of PI3K and mammalian target of rapamycin (mTOR).
-
GC17104
PKI-402
PI3K inhibitor,selective, reversible and ATP-competitive
-
GN10592
Platycodin D
-
GN10312
Polygalasaponin F
-
GN10709
Polyphyllin A
-
GC33115
Pomiferin (NSC 5113)
Pomiferin (NSC 5113) (NSC 5113) acts as an potential inhibitor of HDAC, with an IC50 of 1.05 μM, and also potently inhibits mTOR (IC50, 6.2 μM).
-
GC11003
PP121
Dual inhibitor of tyrosine and phosphoinositide kinases
-
GC15904
PP242
PP242 (PP 242) is a selective and ATP-competitive mTOR inhibitor with an IC50 of 8 nM. PP242 inhibits both mTORC1 and mTORC2 with IC50s of 30 nM and 58 nM, respectively.
-
GC33385
PQR-530
A dual inhibitor of PI3Kα and mTOR
-
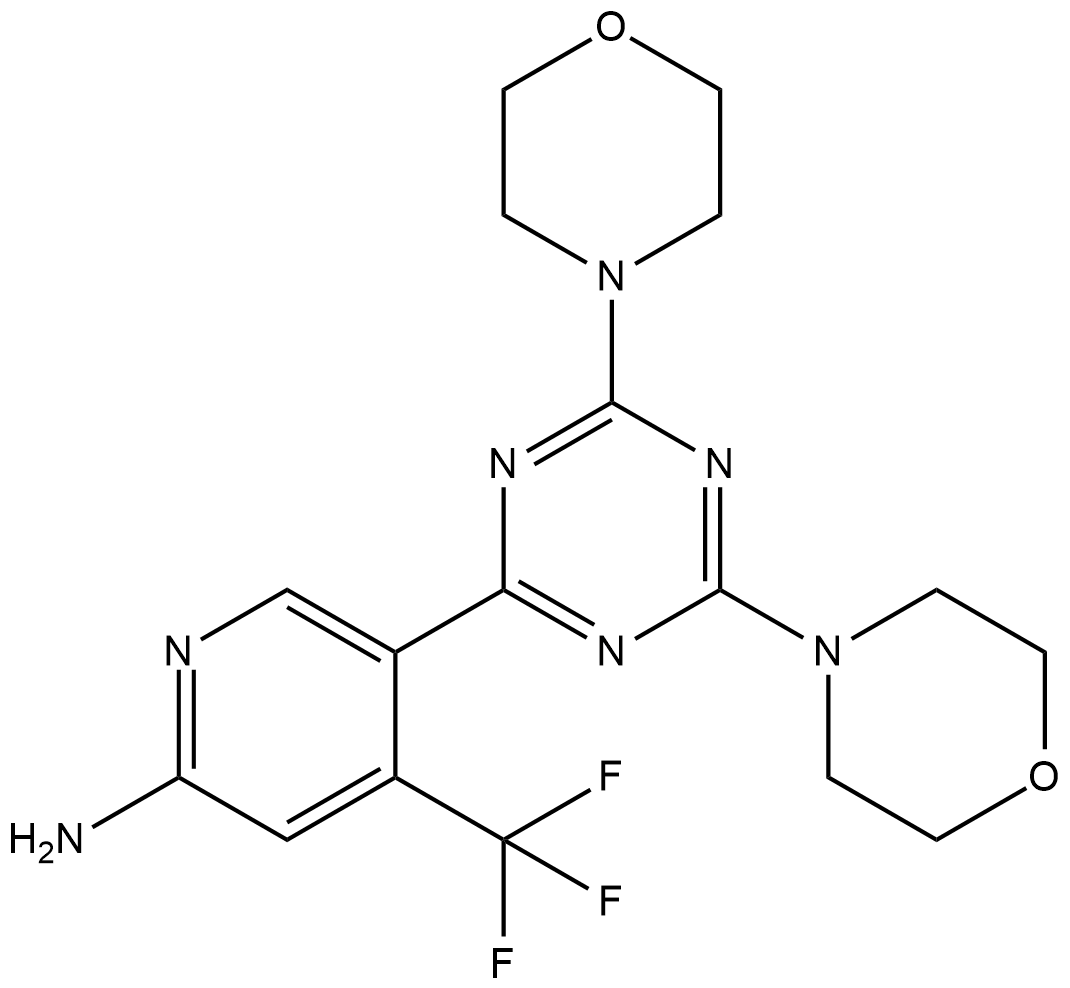
GC19300
PQR309
PQR309 (PQR309) is a potent, brain-penetrant, orally bioavailable, pan-class I PI3K/mTOR inhibitor with IC50s of 33 nM, 451 nM, 661 nM, 708 nM and 89 nM for PI3Kα, PI3Kδ, PI3Kβ, PI3Kγ and mTOR, respectively. PQR309 is an mTORC1 and mTORC2 inhibitor.
-
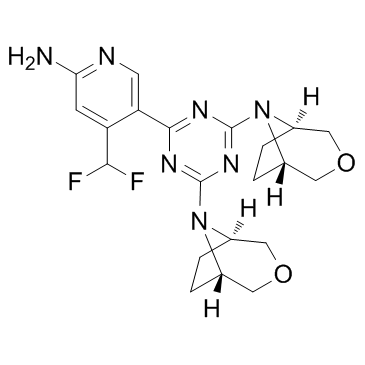
GC32891
PQR620
PQR620 is an orally bioavailable and selective brain penetrant inhibitor of mTORC1/2.
-
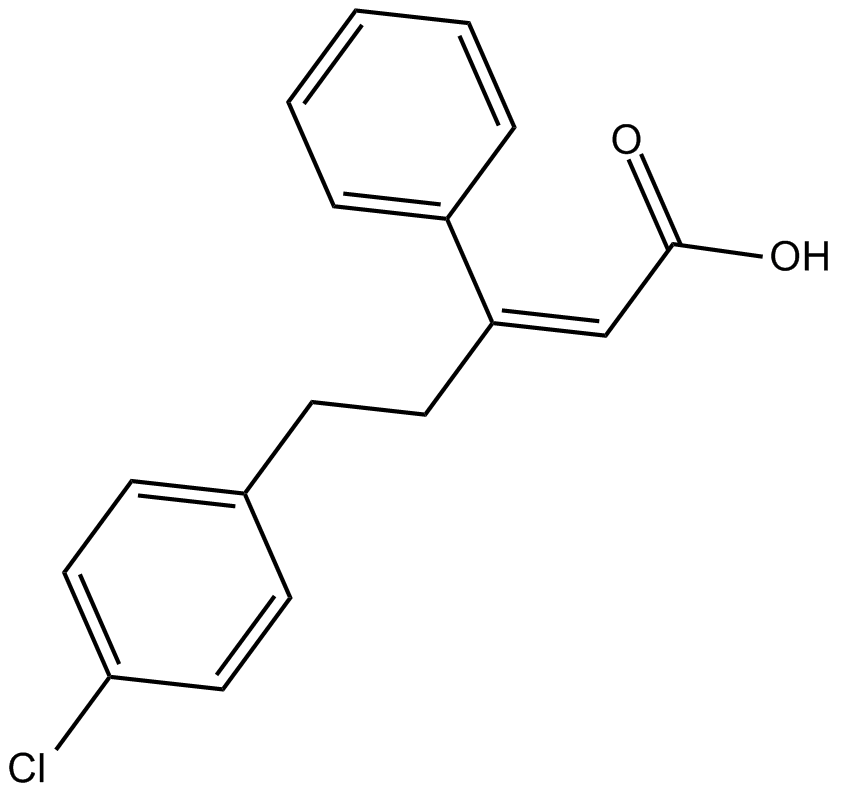
GC13920
PS 48
PDK1 activator
-
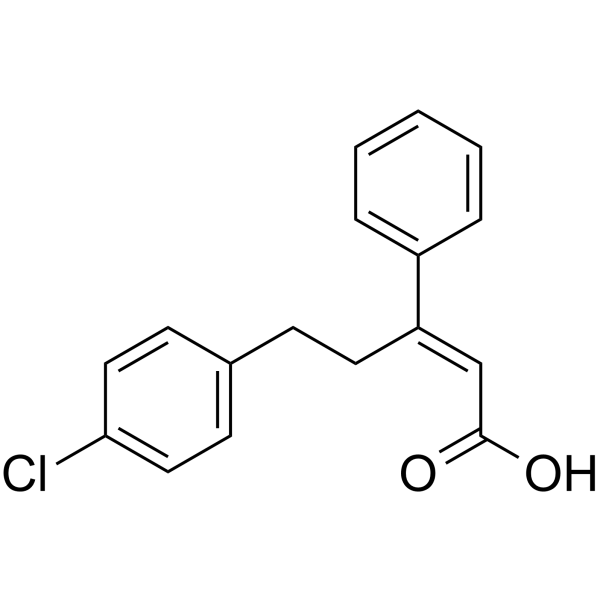
GC63155
PS47
PS47 is an inactive E-isomer of PS48.
-
GC17204
PT 1
AMPK activator, selective
-
GC12889
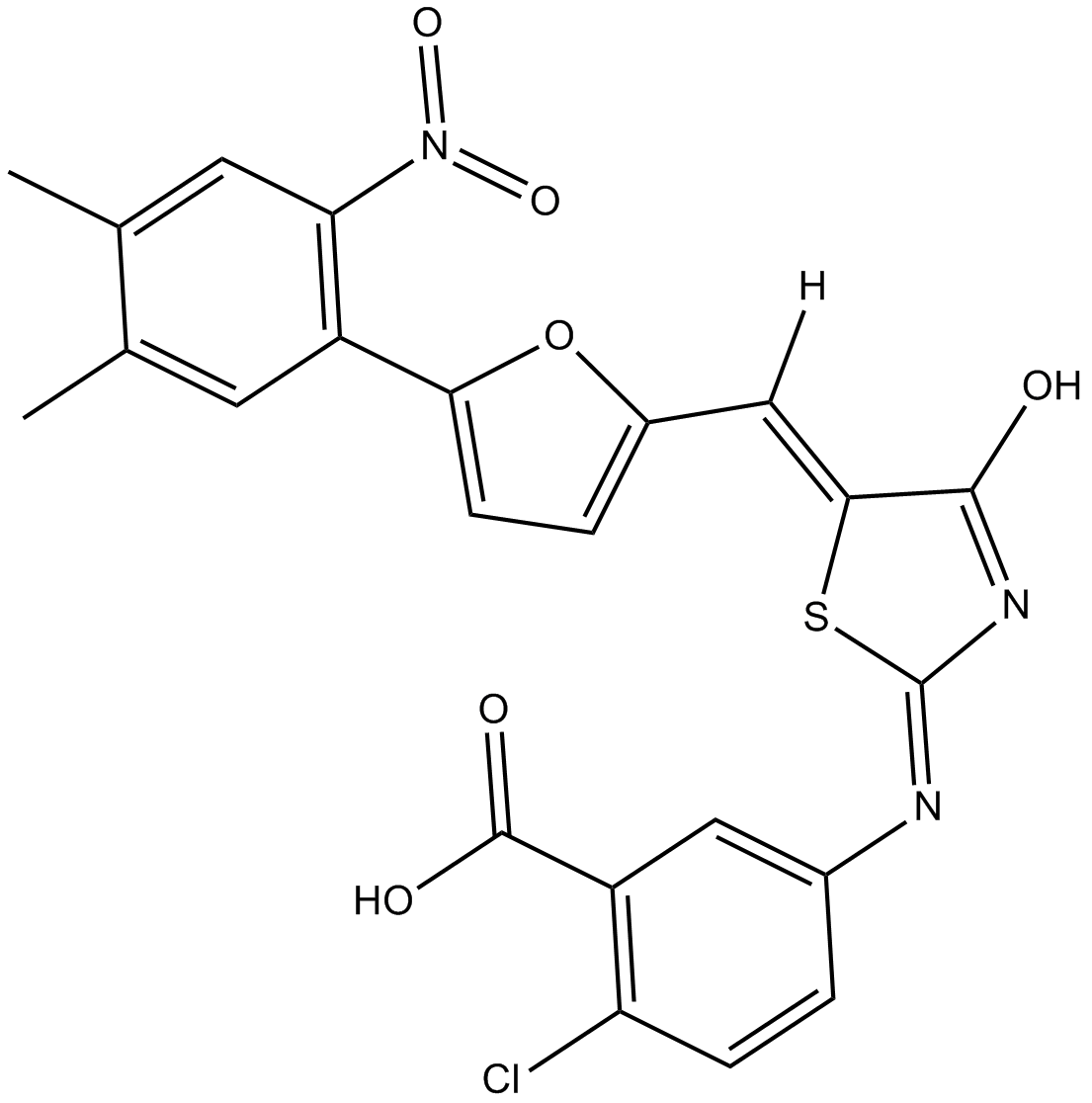
PX 866
PX 866 (PX-866), an improved Wortmannin analogue, is an oral, irreversible, and pan-isoform inhibitor of PI3K (IC50=0.1 nM (p110α), 1.0 nM (p120γ), 2.9 nM (p110δ)). Antitumor activity.
-
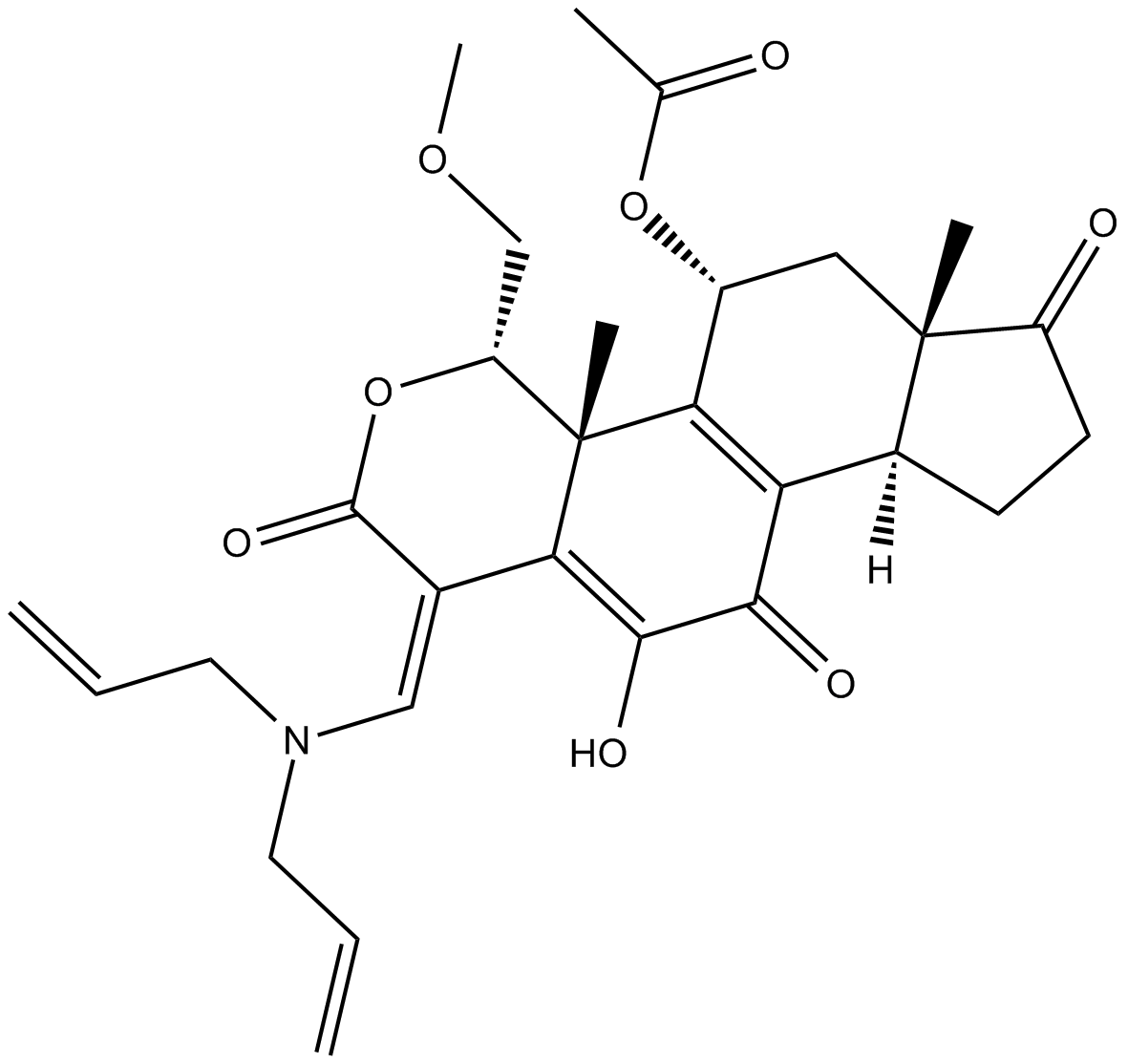
GC44781
PX-13-17OH
A PI3K inhibitor
-
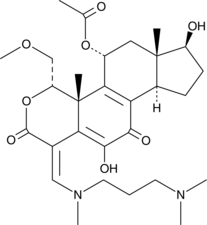
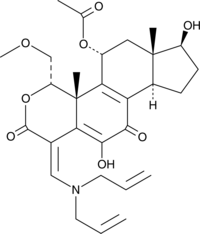
GC44782
PX-866-17OH
A PI3K inhibitor
-
GC18032
QL-IX-55
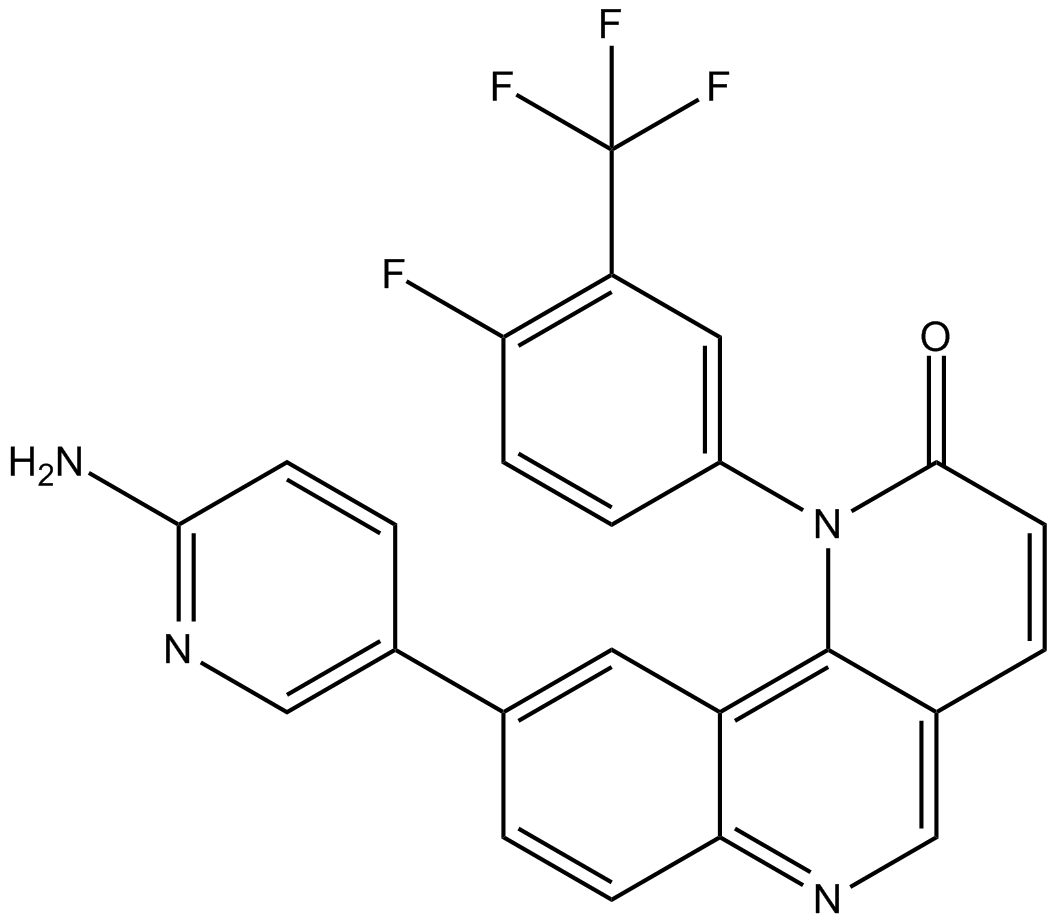
-
GN10266
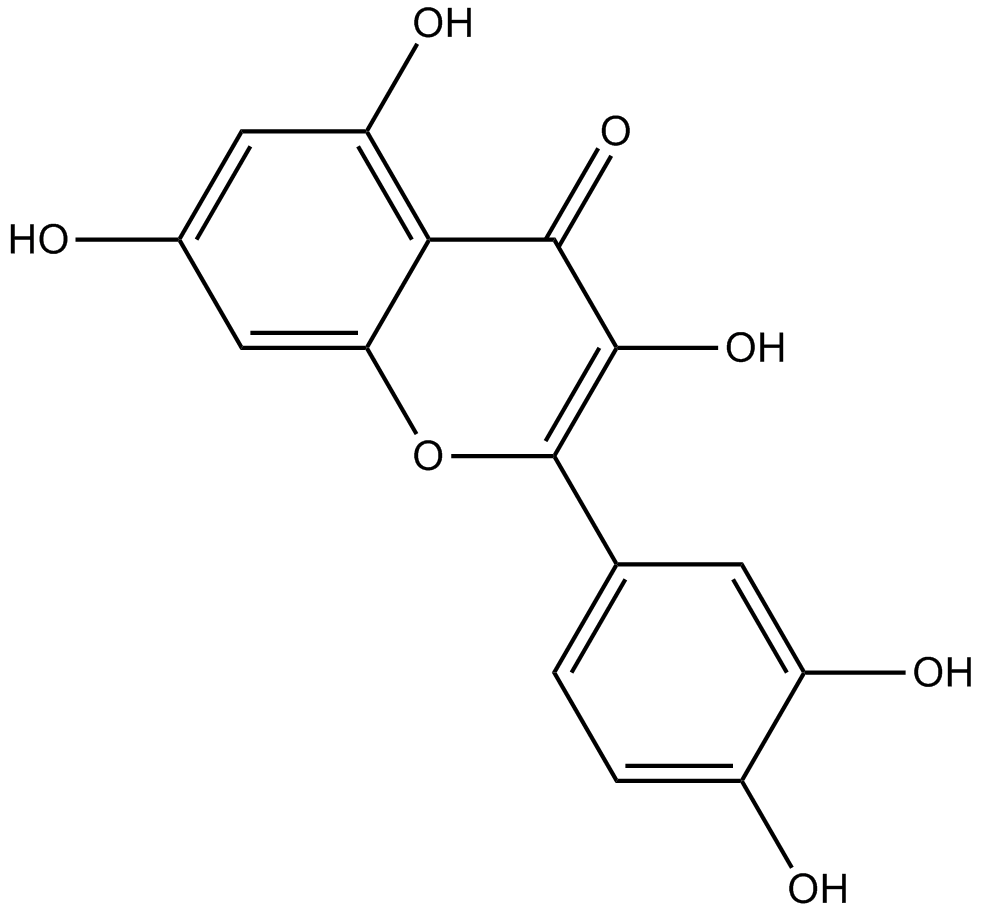
Quercetin
Quercetin is an important dietary flavonoid, present in vegetables, fruits, seeds, nuts, tea and red wine.
-
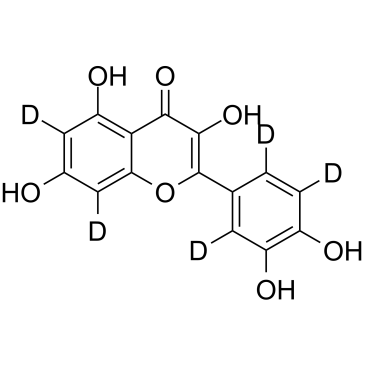
GC61227
Quercetin D5
Quercetin D5 is a deuterium labeled Quercetin. Quercetin, a natural flavonoid, is a stimulator of recombinant SIRT1 and also a PI3K inhibitor with IC50 of 2.4 μM, 3.0 μM and 5.4 μM for PI3K γ, PI3K δ and PI3K β, respectively.
-
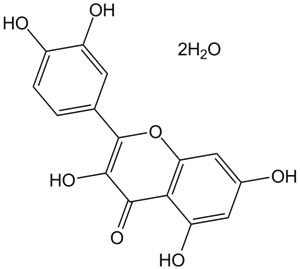
GC15665
Quercetin dihydrate
PLA2 and PI 3-kinase inhibitor
-
GC38401
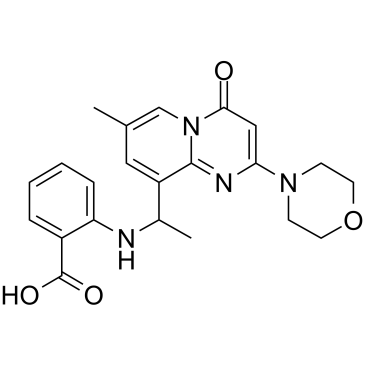
rac-AZD 6482
rac-AZD 6482 ((Rac)-KIN-193) is the racemate of AZD 6482. AZD 6482 is a potent and selective p110β inhibitor with an IC50 of 0.69 nM.
-
GC11607
Rapalink-1
third-generation mTOR inhibitor

-
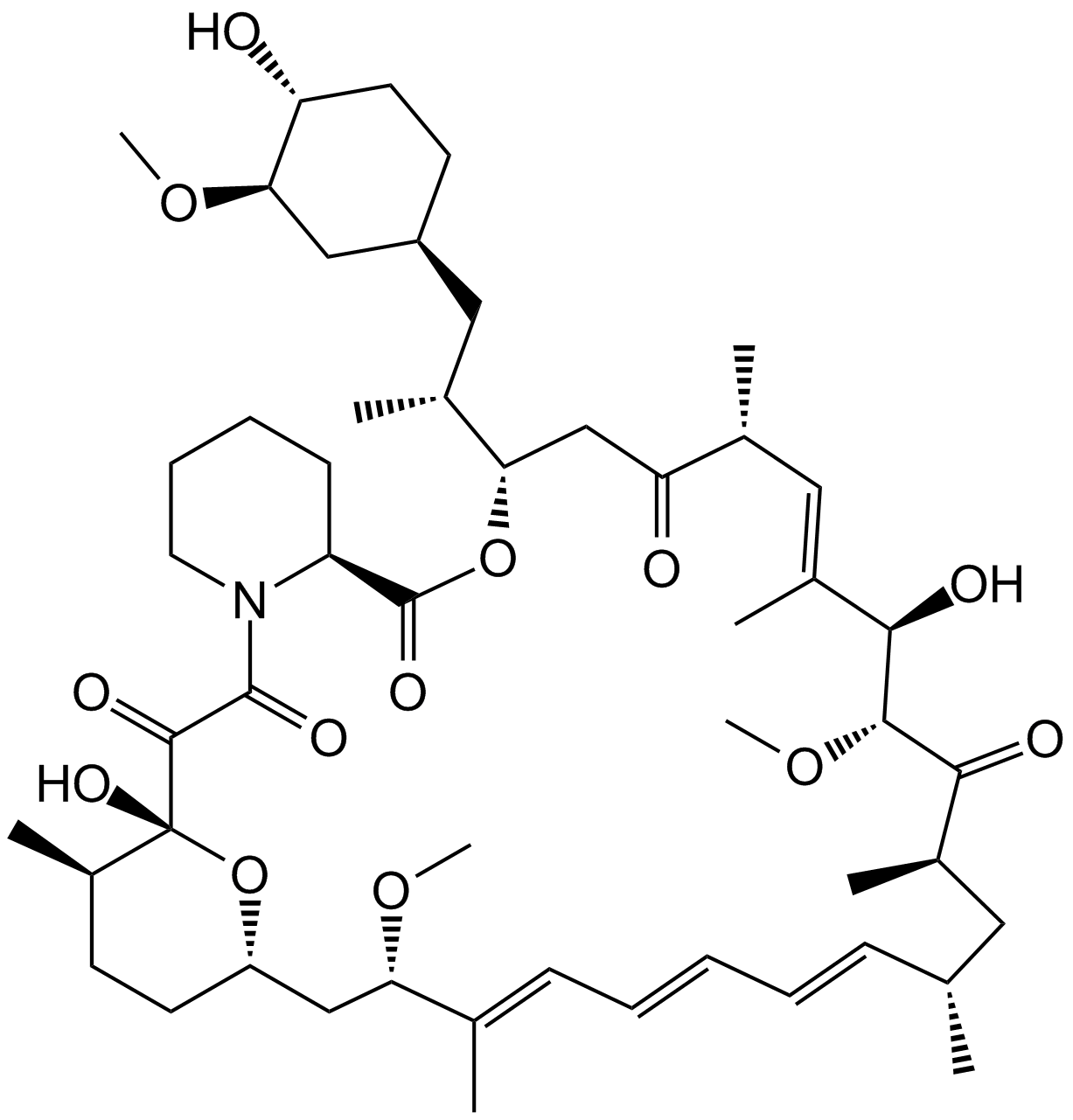
GC15031
Rapamycin (Sirolimus)
An allosteric inhibitor of mTORC1
-
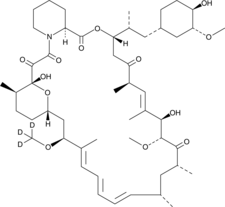
GC48027
Rapamycin-d3
An internal standard for the quantification of rapamycin
-
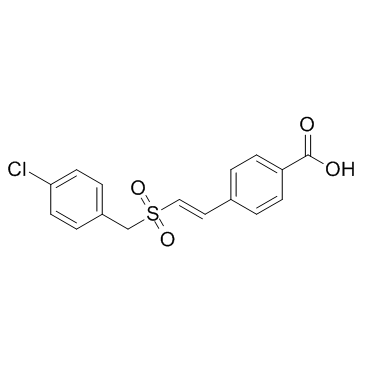
GC33130
Recilisib (Ex-RAD)
Recilisib (Ex-RAD) (ON 01210) is a radioprotectant, which can activate AKT, PI3K activities in cells.
-
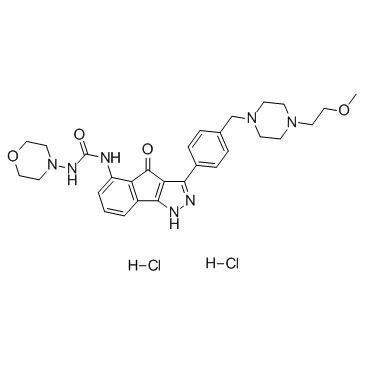
GC37522
RGB-286638
A multi-kinase inhibitor
-
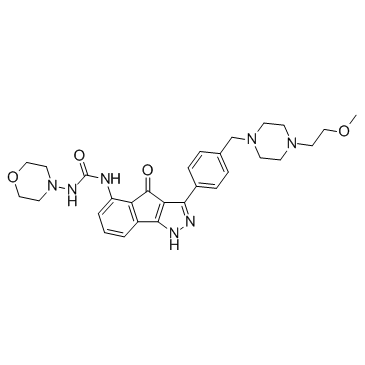
GC37523
RGB-286638 free base
A multi-kinase inhibitor
-
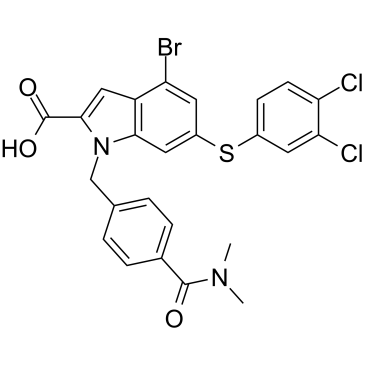
GC39292
Rheb inhibitor NR1
Rheb inhibitor NR1 is a Rheb inhibitor with an IC50 of 2.1??M in the Rheb-IVK assay.
-
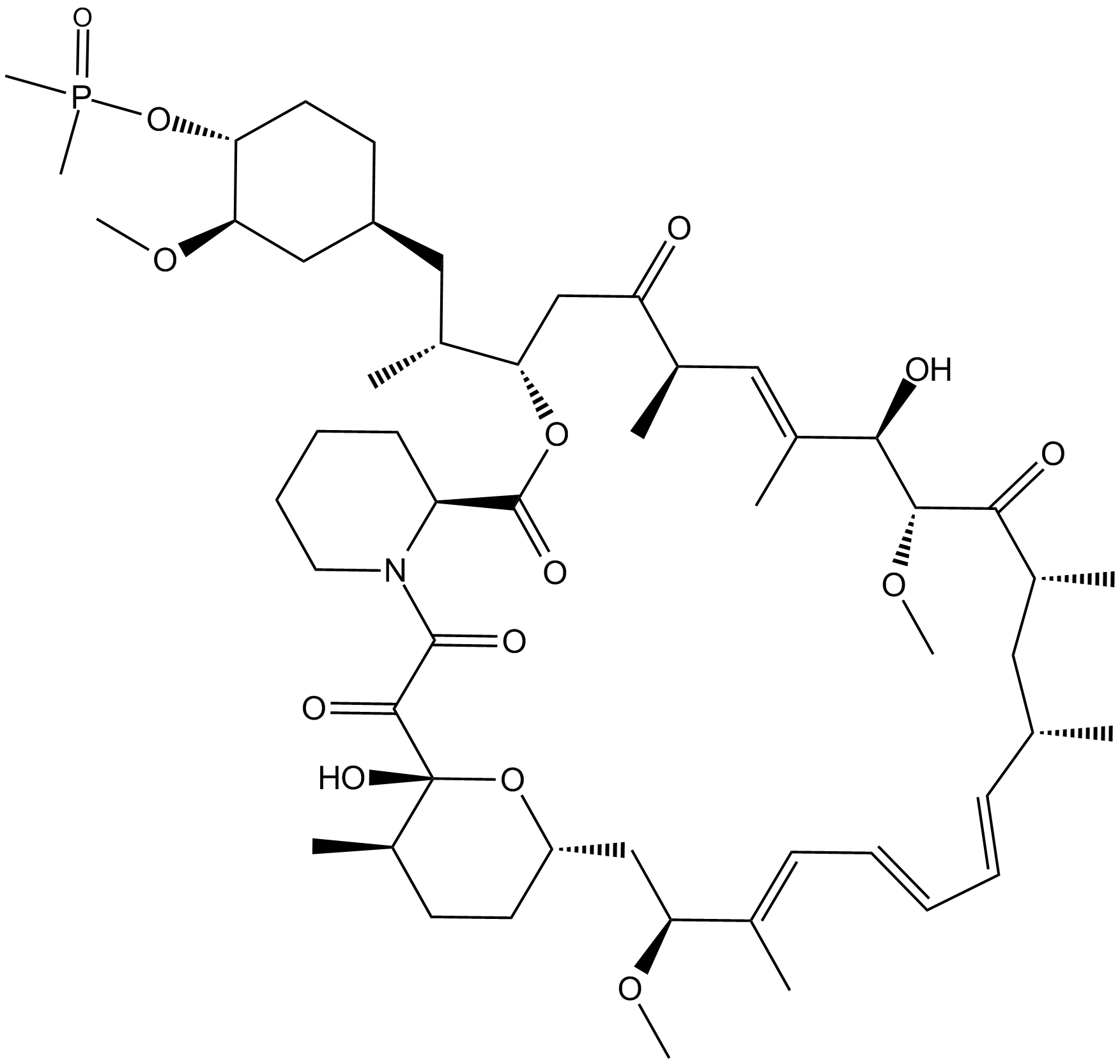
GC13071
Ridaforolimus (Deforolimus, MK-8669)
Ridaforolimus (Deforolimus, MK-8669) (MK-8669) is a potent and selective mTOR inhibitor; inhibits ribosomal protein S6 phosphorylation with an IC50 of 0.2 nM in HT-1080 cells.
-
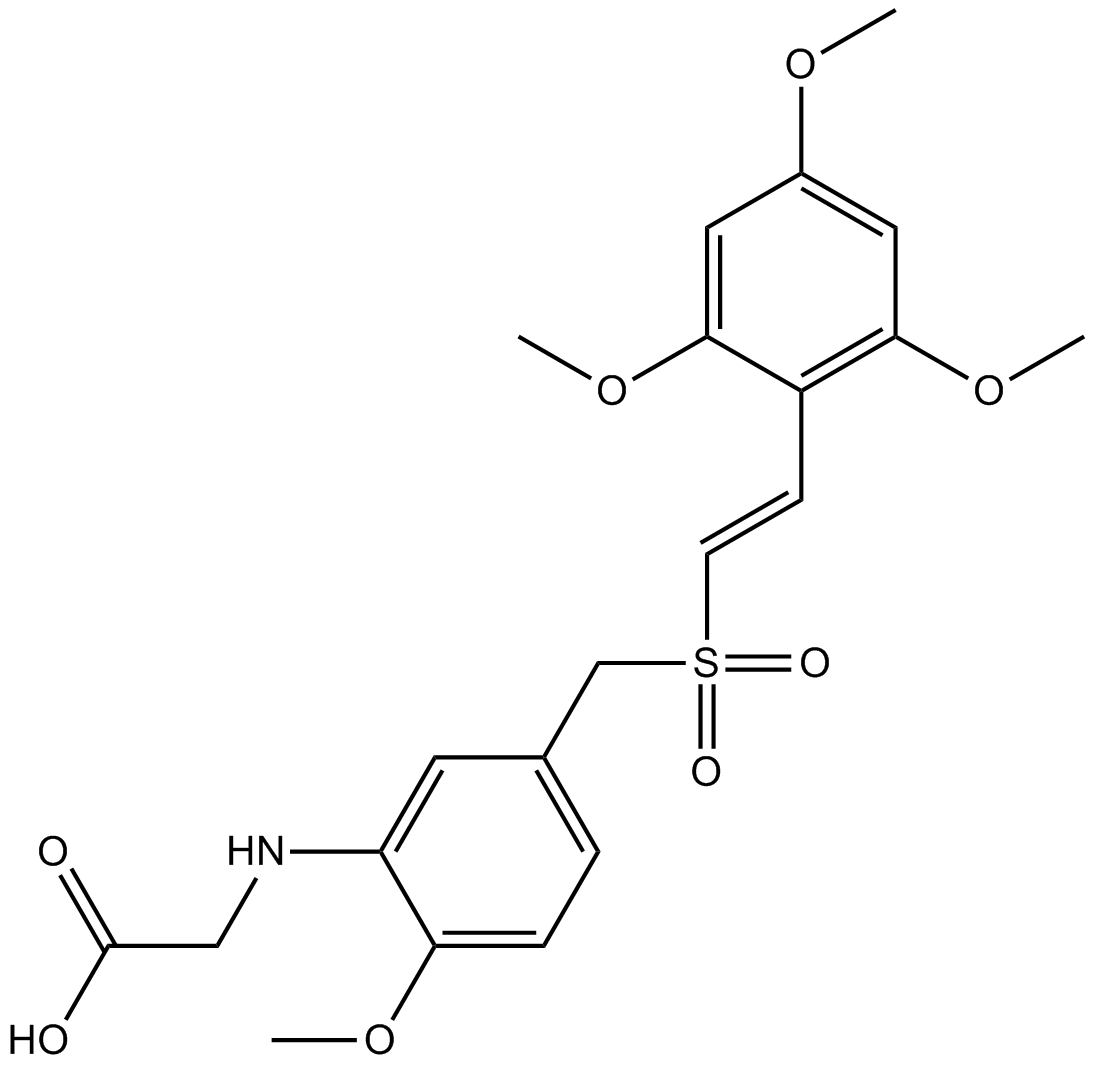
GC12415
Rigosertib
PI3K/PLK1 inhibitor
-
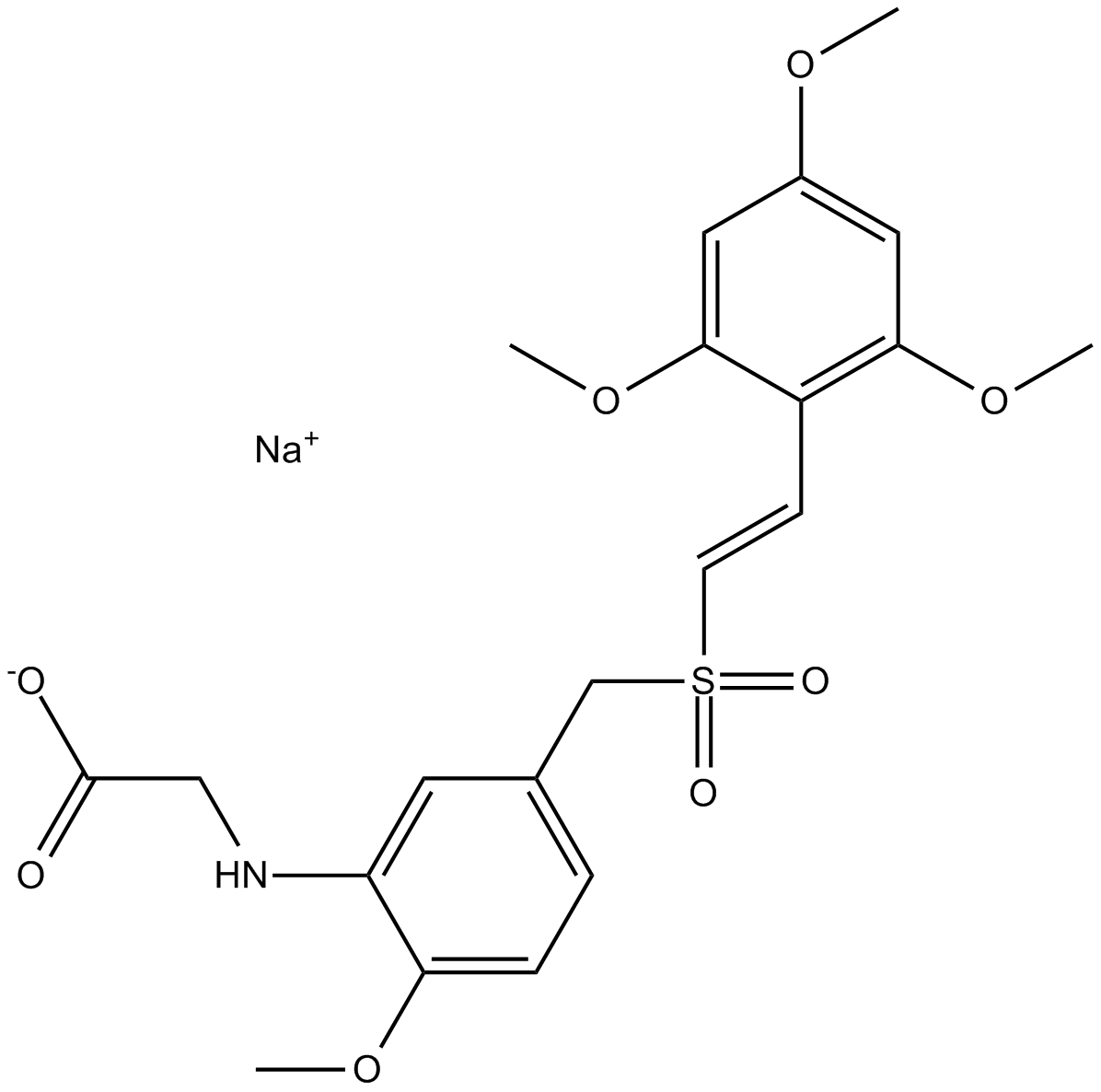
GC13580
Rigosertib sodium
Non-ATP-competitive inhibitor of PLK1
-
GC62649
RMC-5552
RMC-5552 is a potent and selective inhibitor of mTORC1. RMC-5552 inhibits phosphorylation of mTORC1 pS6K and p4EBP1 with IC50s of 0.14 nM and 0.48 nM, respectively. RMC-5552 has anti-cancer activity.

-
GC63894
RMC-6272
RMC-6272 (RM-006) is a bi-steric mTORC1-selective inhibitor. RMC-6272 exhibits potent and selective (> 10-fold) inhibition of mTORC1 over mTORC2. RMC-6272 shows improved inhibition of mTORC1 in comparison to Rapamycin, and induces more cell death in TSC2 null tumors.

-
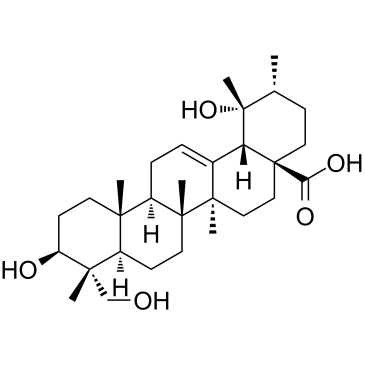
GC38609
Rotundic acid
Rotundic acid, a triterpenoid obtained from I.
-
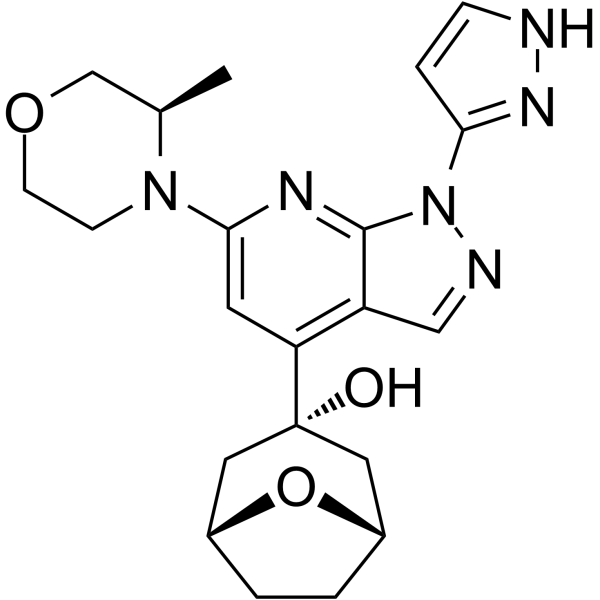
GC64278
RP-3500
RP-3500 (ATR inhibitor 4) is an orally active, selective ATR kinase inhibitor (ATRi) with an IC50 of 1.00 nM in biochemical assays. RP-3500 shows 30-fold selectivity for ATR over mTOR (IC50=120 nM) and >2,000-fold selectivity over ATM, DNA-PK, and PI3Kα kinases. RP-3500 has potent antitumor activity.
-
GC12324
RSVA 405
AMPK activator
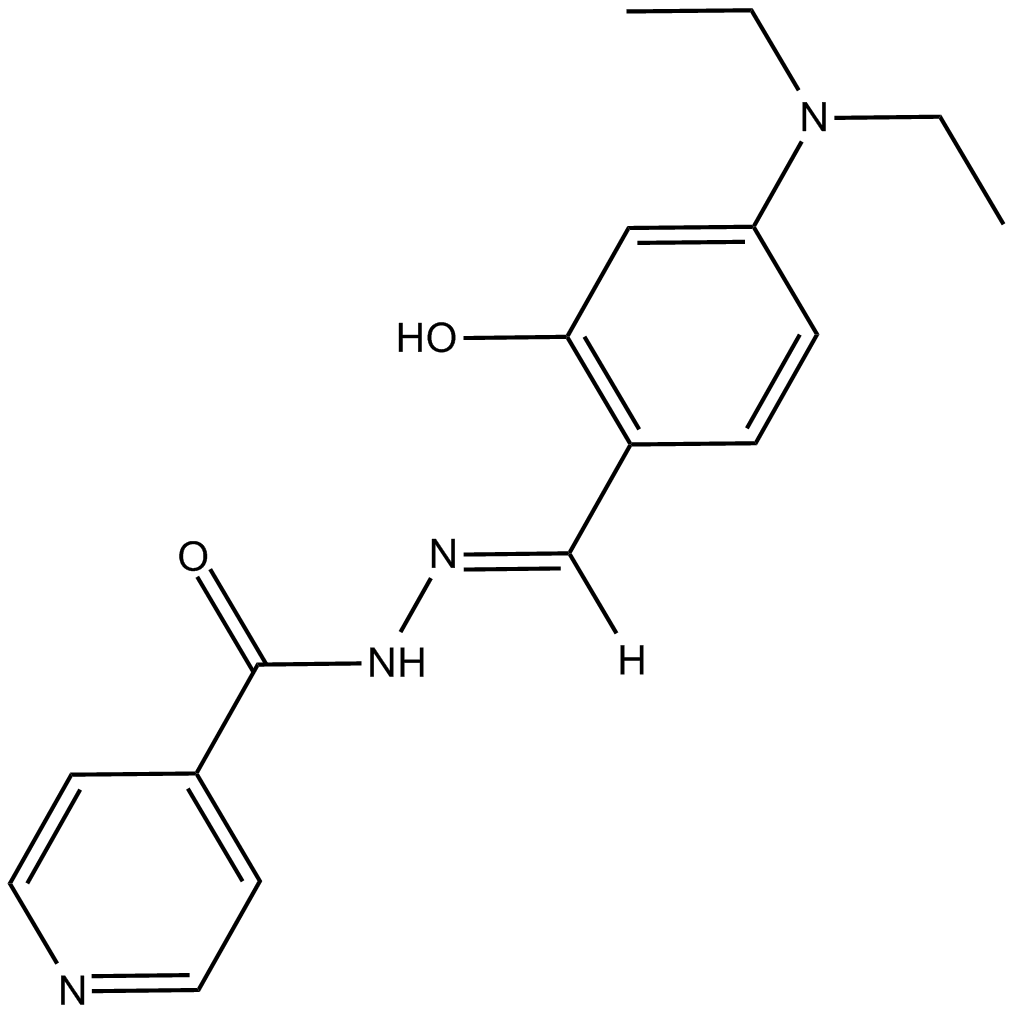
-
GC18852
S14161
D-Cyclins regulate the cell cycle by acting in a complex with cyclin dependent kinases (CDKs) to promote phosphorylation of the retinoblastoma protein and initiate cellular progression from the G1 to the S phase.
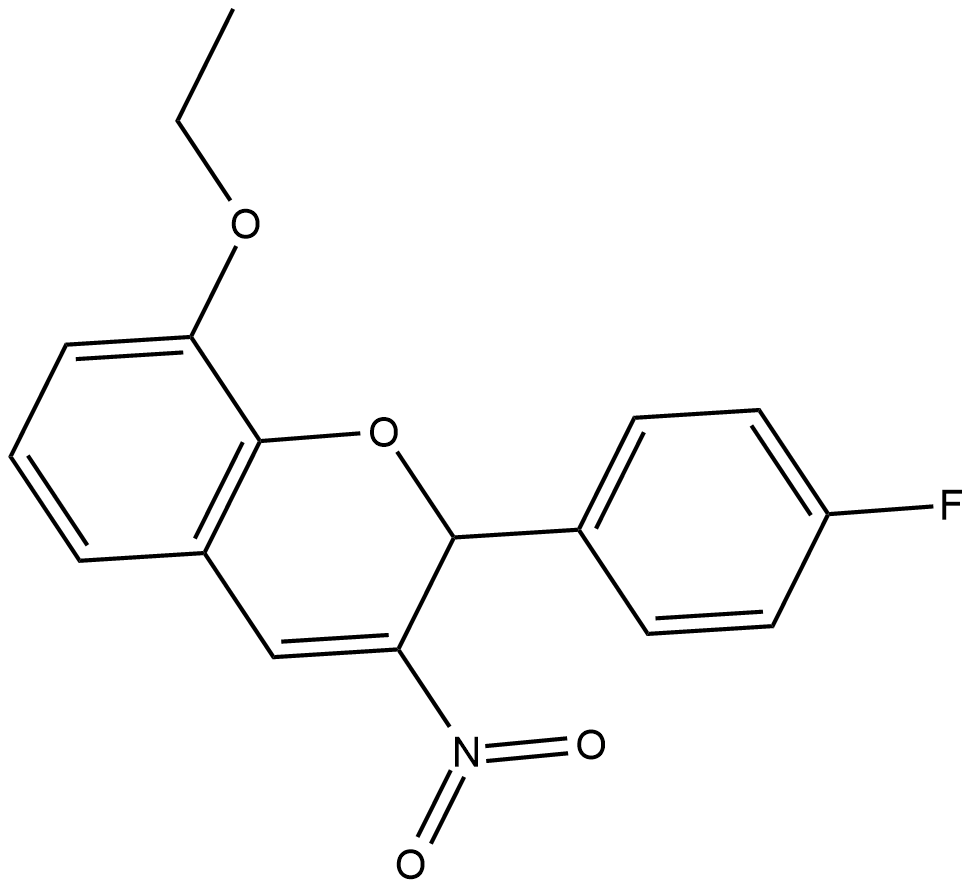
-
GP10104
S6 Kinase Substrate Peptide 32
Measures the activity of kinases that phosphorylate ribosomal protein S6.
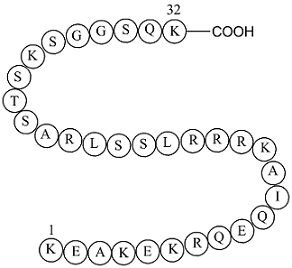
-
GN10127
Salidroside
Salidroside is a glycoside with multiple biological activities and has pharmacological effects such as anti-cancer, antioxidant, anti-aging, anti-diabetic, anti-diabetic, anti-hypertensive, anti-inflammatory, and immune regulation.
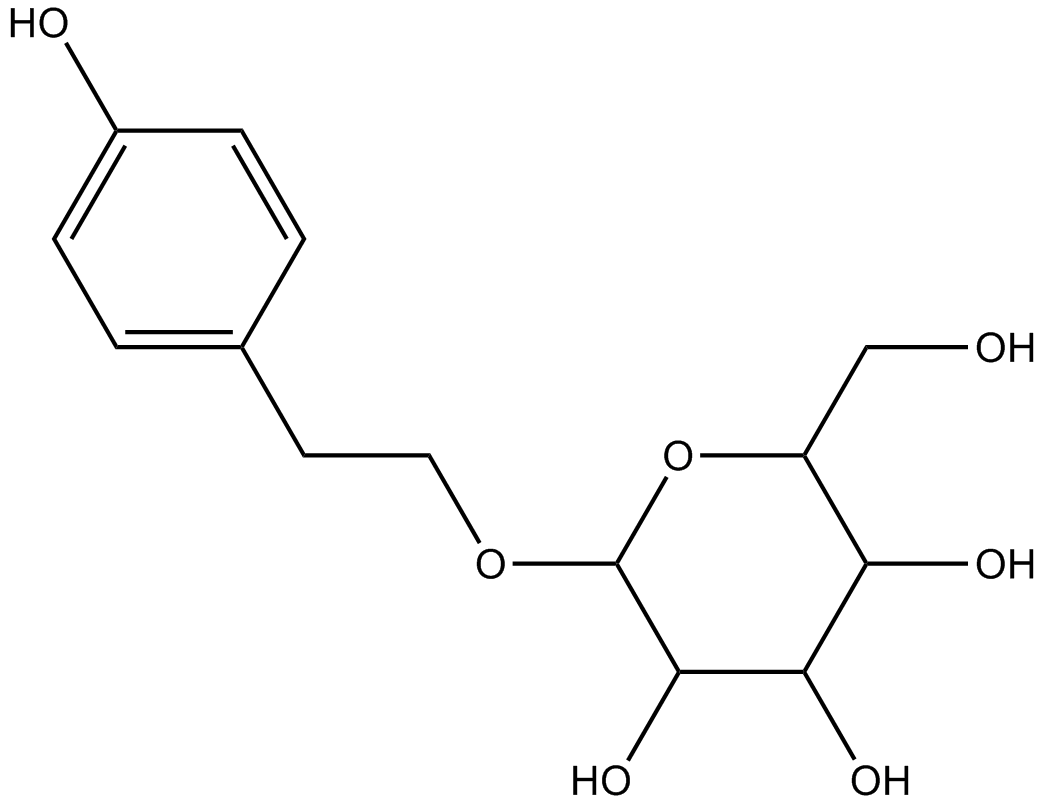
-
GC12364
SAMS Peptide
SAMS Peptide peptide is a specific substrate for the AMP-activated protein kinase (AMPK).
-
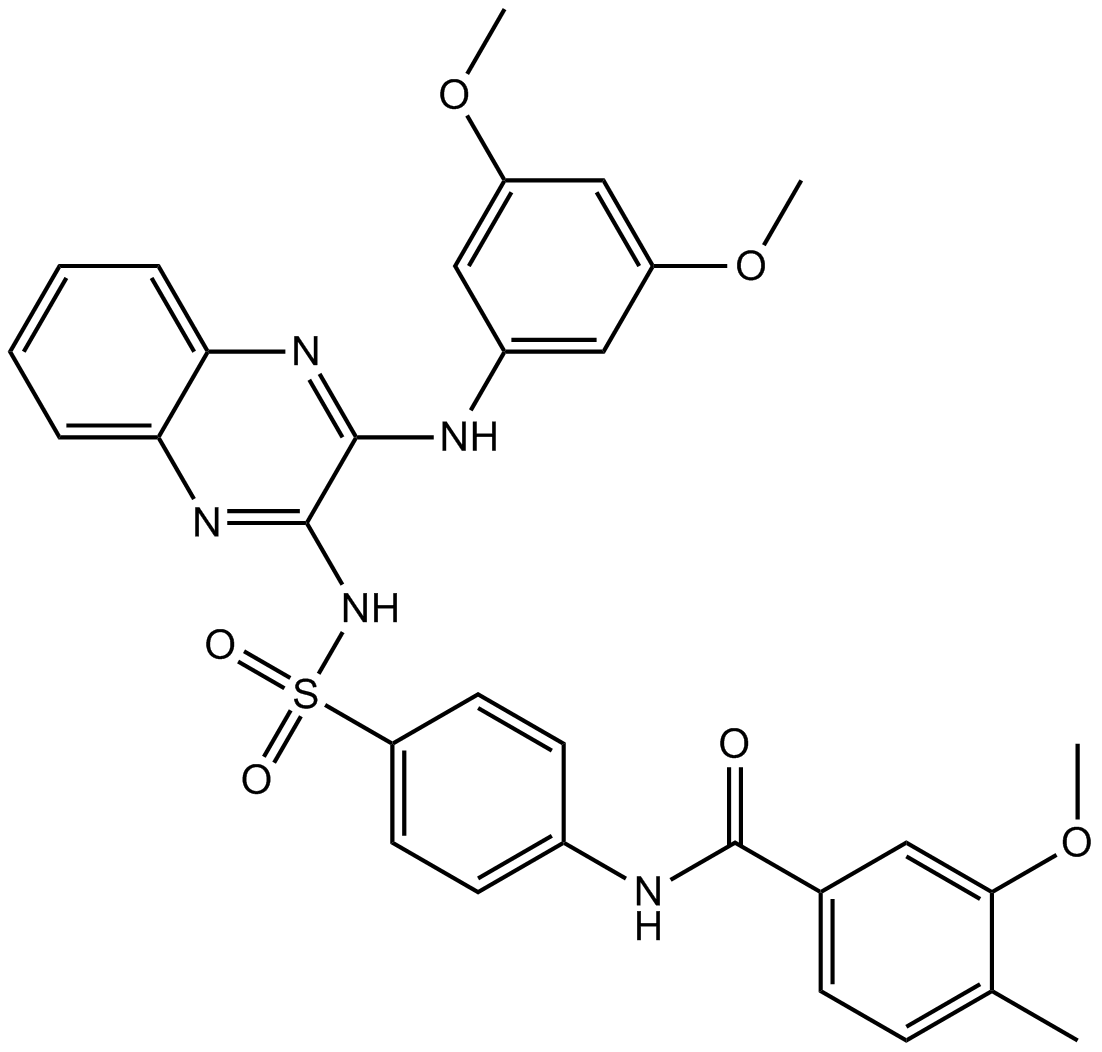
GC14884
SAR245409 (XL765)
SAR245409 (XL765) (XL-147 derivative 1) is a potent inhibitor of PI3K. SAR245409 (XL765) (25 μM) blocks PI3K/Akt signaling pathways.
-
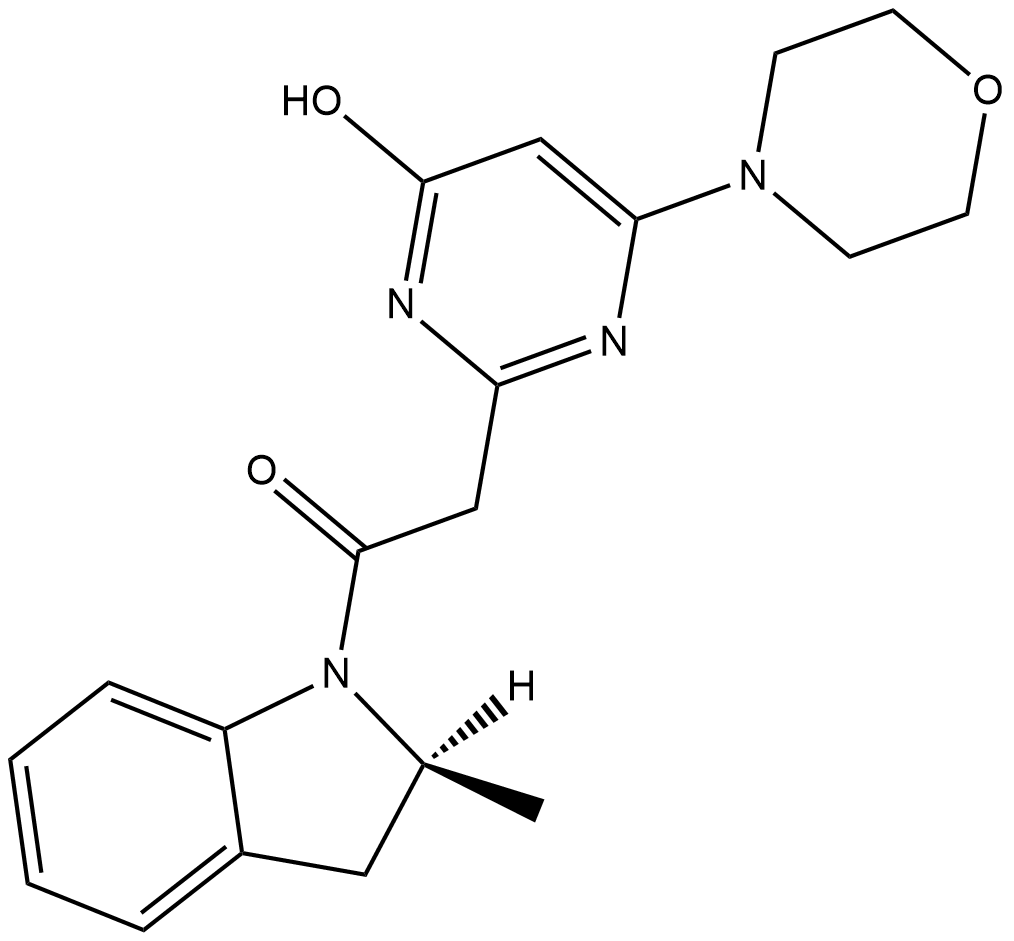
GC18479
SAR260301
SAR260301 is an inhibitor of phosphatidylinositol 3-kinase (PI3K) isoform β (IC50 = 52 nM).
-
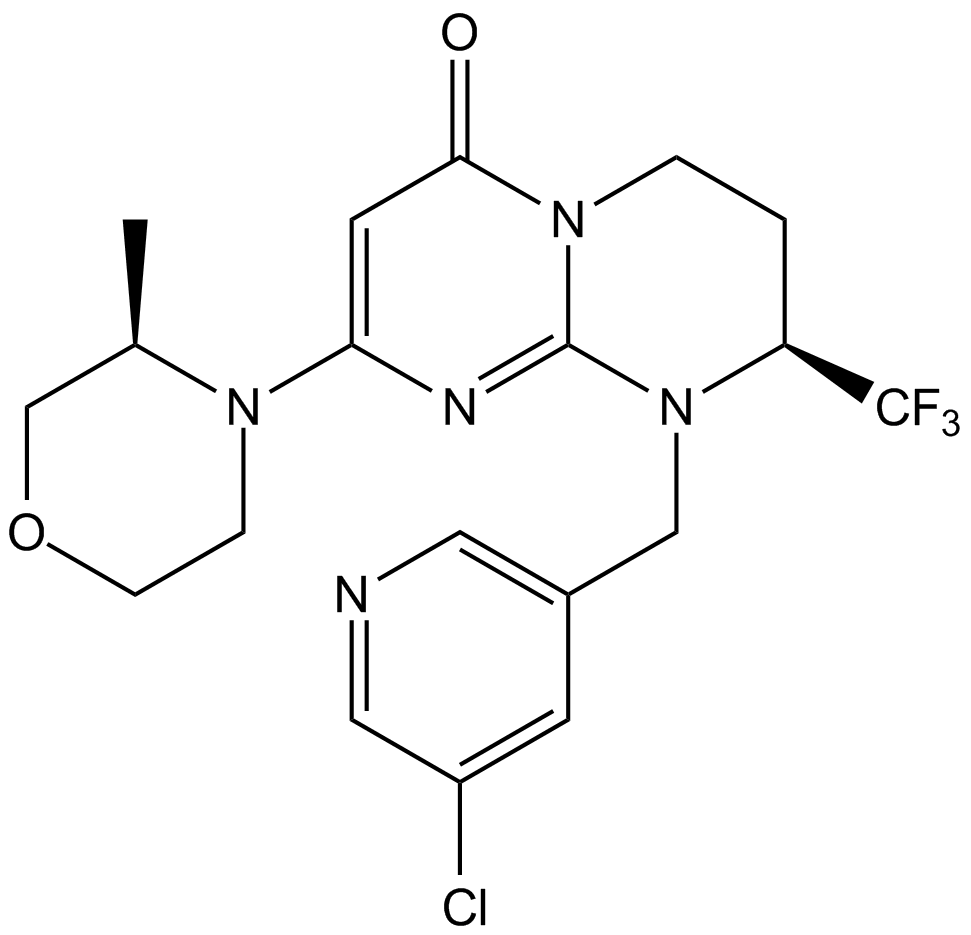
GC11471
SAR405
SAR405 is a Vps34 inhibitor (IC50=1.2 nM),SAR405 inhibits autophagy caused by mTOR inhibition.
-
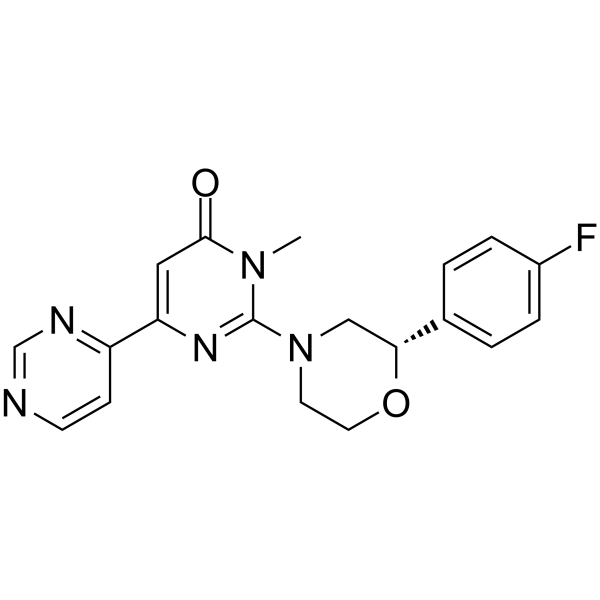
GC63181
SAR502250
SAR502250 is a potent, selective, ATP competitive, orally active and brain-penetrant inhibitor of GSK3, with an IC50 of 12 nM for human GSK-3β.
-
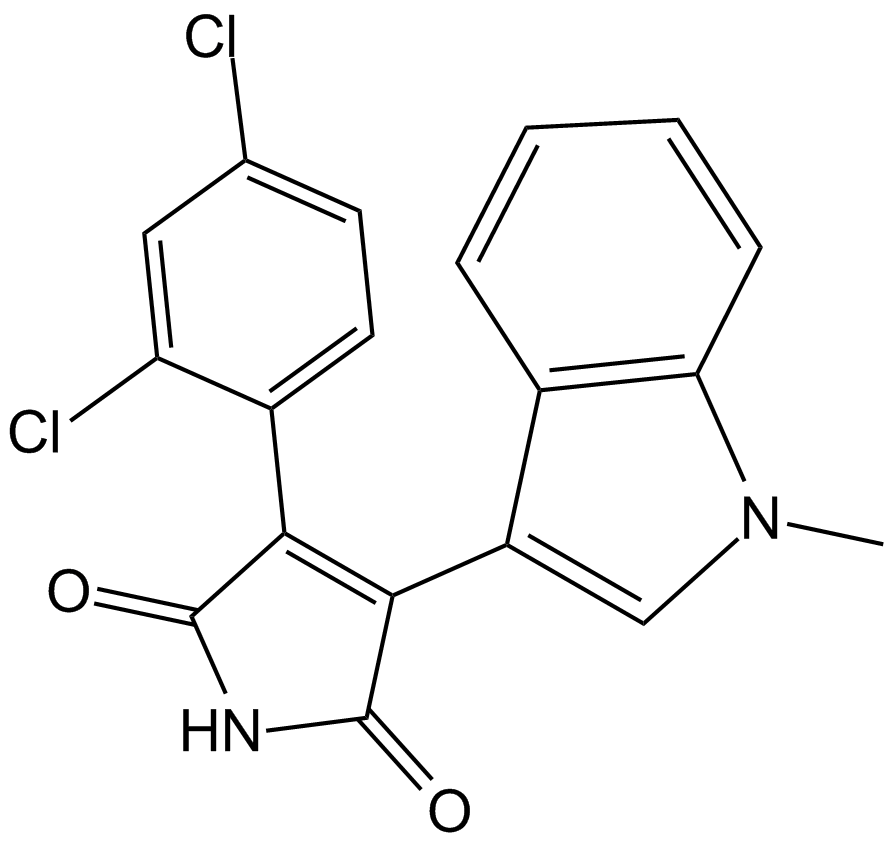
GC10463
SB 216763
SB 216763 stands out as a powerful, selective, and ATP-competitive inhibitor of GSK-3, effectively targeting both GSK-3α and GSK-3β with an impressive IC50 of 34.3 nM[1-3].
-
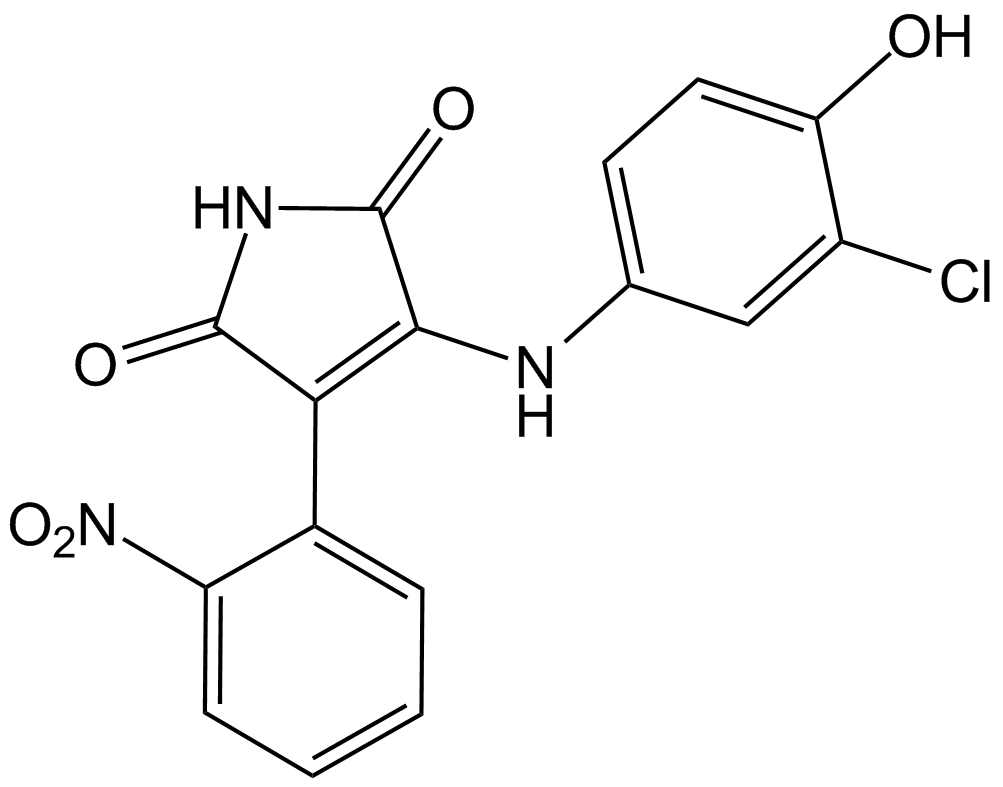
GC13028
SB 415286
A selective inhibitor of GSK-3
-
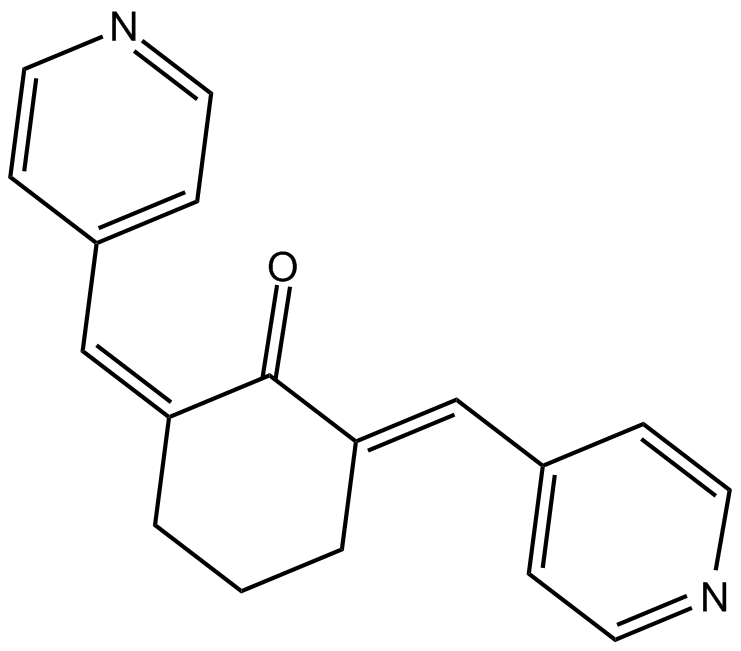
GC11985
SC 66
Akt inhibitor
-
GC11645
SC 79
SC 79 is an activator of osmotic Akt phosphorylation in the brain and an inhibitor of AKT-PH domain translocation.
-
GN10624
Scutellarin
-
GC25913
Selective PI3Kδ Inhibitor 1 (compound 7n)
Selective PI3Kδ Inhibitor 1 (compound 7n) is an inhibitor of PI3Kδ with an IC50 of 0.9 nM and >1000-fold selectivity against other class I PI3K isoforms [PI3K α/γ/β=3670/1460/21300 nM].

-
GC31666
Seletalisib (UCB5857)
Seletalisib (UCB5857) (UCB5857) is potent and selective PI3Kδ inhibitor with an IC50 of 12 nM.
-
GC37633
SF1126
SF1126 is a relevant pan and dual first-in-class PI3K/BRD4 inhibitor, has antitumor and anti-angiogenic activity. SF1126 is an RGDS-conjugated LY294002 prodrug, which is designed to exhibit increased solubility and bind to specific integrins within the tumor compartment. SF1126 induces cell apoptosis.
-
GC19328
SF2523
SF2523 is a highly selective and potent inhibitor of PI3K with IC50s of 34 nM, 158 nM, 9 nM, 241 nM and 280 nM for PI3Kα, PI3Kγ, DNA-PK, BRD4 and mTOR, respectively.
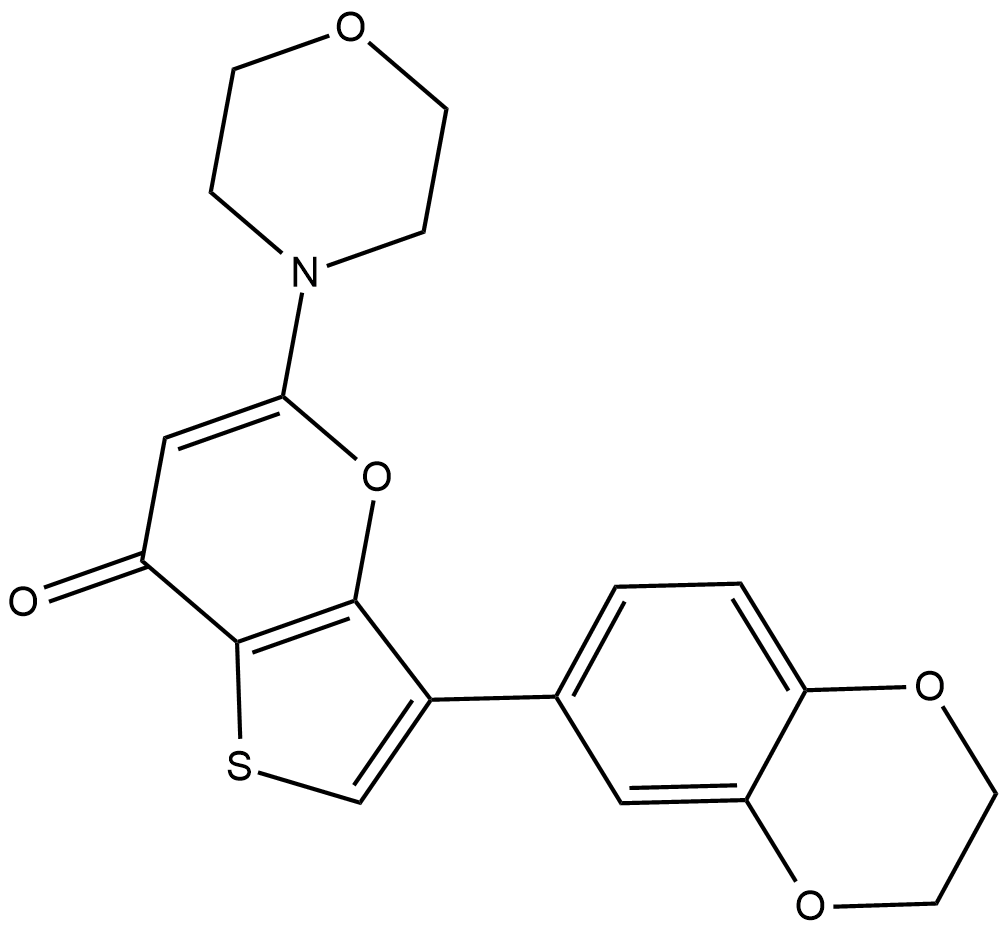
-
GC60339
SKI V
SKI V is a noncompetitive and potent non-lipid sphingosine kinase (SPHK; SK) inhibitor with an IC50 of 2 μM for GST-hSK. SKI V potently inhibits PI3K with an IC50 of 6 μM for hPI3k. SKI V decreases formation of the mitogenic second messenger sphingosine-1-phosphate (S1P). SKI V induces apoptosis and has antitumor activity.
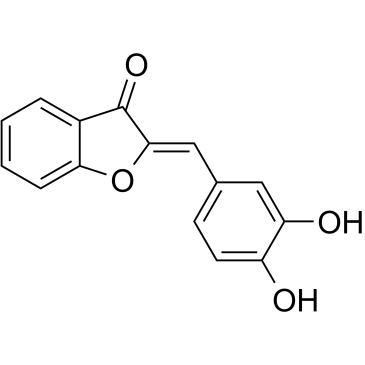
-
GC34053
SOLENOPSIN
Solenopsin is an ATP-competitive AKT inhibitor with IC50 value of 10 μM .
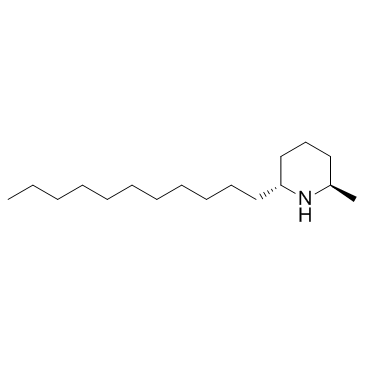
-
GN10681
Sophocarpine
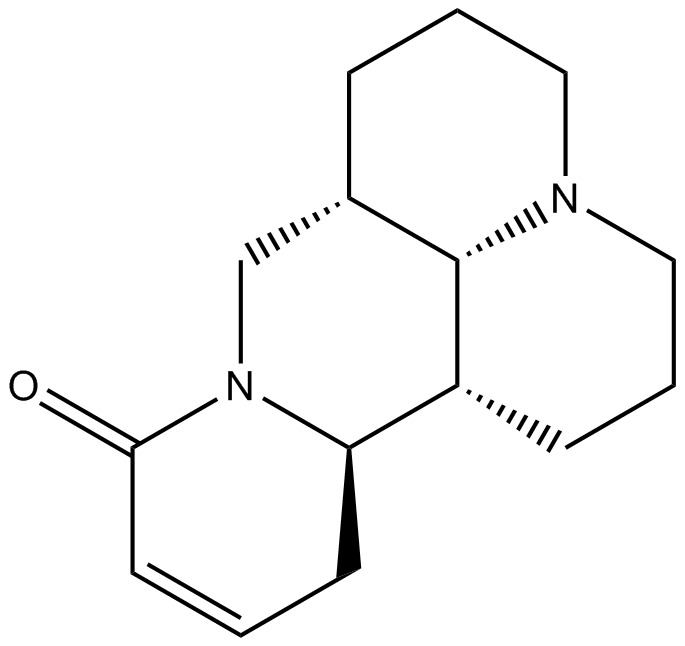
-
GC64223
Sophocarpine monohydrate
Sophocarpine (monohydrate) is one of the significant alkaloid extracted from the traditional herb medicine Sophora flavescens which has many pharmacological properties such as anti-virus, anti-tumor, anti-inflammatory.
-
GC62456
SRX3207
SRX3207 is an orally active and first-in-class dual Syk/PI3K inhibitor, with IC50 values of 10.7 nM and 861 nM for Syk and PI3Kα, respectively. SRX3207 relieves tumor immunosuppression.
-
GC65249
STL127705
STL127705 (Compound L) is a potent Ku 70/80 heterodimer protein inhibitor with an IC50 of 3.5 μM. STL127705 interferes the binding of Ku70/80 to DNA and by inhibits the activation of the DNA-PKCS kinase. STL127705 shows antiproliferative and anticancer activity. STL127705 induces apoptosis.
-
GC37694
STO-609
A CaMKK inhibitor
-
GC17500
SU6656
Src tyrosine kinases inhibitor
-
GC32199
T-00127_HEV1
T-00127_HEV1 is a phosphatidylinositol 4-kinase III beta (PI4KB) inhibitor with an IC50 of 60 nM.
-
GC63210
TAS-117
TAS-117 is a potent, selective, orally active allosteric Akt inhibitor (with IC50s of 4.8, 1.6, and 44 nM for Akt1, 2, and 3, respectively). TAS-117 triggers anti-myeloma activities and enhances fatal endoplasmic reticulum (ER) stress induced by proteasome inhibition. TAS-117 induces apoptosis and autophagy.