Apoptosis
As one of the cellular death mechanisms, apoptosis, also known as programmed cell death, can be defined as the process of a proper death of any cell under certain or necessary conditions. Apoptosis is controlled by the interactions between several molecules and responsible for the elimination of unwanted cells from the body.
Many biochemical events and a series of morphological changes occur at the early stage and increasingly continue till the end of apoptosis process. Morphological event cascade including cytoplasmic filament aggregation, nuclear condensation, cellular fragmentation, and plasma membrane blebbing finally results in the formation of apoptotic bodies. Several biochemical changes such as protein modifications/degradations, DNA and chromatin deteriorations, and synthesis of cell surface markers form morphological process during apoptosis.
Apoptosis can be stimulated by two different pathways: (1) intrinsic pathway (or mitochondria pathway) that mainly occurs via release of cytochrome c from the mitochondria and (2) extrinsic pathway when Fas death receptor is activated by a signal coming from the outside of the cell.
Different gene families such as caspases, inhibitor of apoptosis proteins, B cell lymphoma (Bcl)-2 family, tumor necrosis factor (TNF) receptor gene superfamily, or p53 gene are involved and/or collaborate in the process of apoptosis.
Caspase family comprises conserved cysteine aspartic-specific proteases, and members of caspase family are considerably crucial in the regulation of apoptosis. There are 14 different caspases in mammals, and they are basically classified as the initiators including caspase-2, -8, -9, and -10; and the effectors including caspase-3, -6, -7, and -14; and also the cytokine activators including caspase-1, -4, -5, -11, -12, and -13. In vertebrates, caspase-dependent apoptosis occurs through two main interconnected pathways which are intrinsic and extrinsic pathways. The intrinsic or mitochondrial apoptosis pathway can be activated through various cellular stresses that lead to cytochrome c release from the mitochondria and the formation of the apoptosome, comprised of APAF1, cytochrome c, ATP, and caspase-9, resulting in the activation of caspase-9. Active caspase-9 then initiates apoptosis by cleaving and thereby activating executioner caspases. The extrinsic apoptosis pathway is activated through the binding of a ligand to a death receptor, which in turn leads, with the help of the adapter proteins (FADD/TRADD), to recruitment, dimerization, and activation of caspase-8 (or 10). Active caspase-8 (or 10) then either initiates apoptosis directly by cleaving and thereby activating executioner caspase (-3, -6, -7), or activates the intrinsic apoptotic pathway through cleavage of BID to induce efficient cell death. In a heat shock-induced death, caspase-2 induces apoptosis via cleavage of Bid.
Bcl-2 family members are divided into three subfamilies including (i) pro-survival subfamily members (Bcl-2, Bcl-xl, Bcl-W, MCL1, and BFL1/A1), (ii) BH3-only subfamily members (Bad, Bim, Noxa, and Puma9), and (iii) pro-apoptotic mediator subfamily members (Bax and Bak). Following activation of the intrinsic pathway by cellular stress, pro‑apoptotic BCL‑2 homology 3 (BH3)‑only proteins inhibit the anti‑apoptotic proteins Bcl‑2, Bcl-xl, Bcl‑W and MCL1. The subsequent activation and oligomerization of the Bak and Bax result in mitochondrial outer membrane permeabilization (MOMP). This results in the release of cytochrome c and SMAC from the mitochondria. Cytochrome c forms a complex with caspase-9 and APAF1, which leads to the activation of caspase-9. Caspase-9 then activates caspase-3 and caspase-7, resulting in cell death. Inhibition of this process by anti‑apoptotic Bcl‑2 proteins occurs via sequestration of pro‑apoptotic proteins through binding to their BH3 motifs.
One of the most important ways of triggering apoptosis is mediated through death receptors (DRs), which are classified in TNF superfamily. There exist six DRs: DR1 (also called TNFR1); DR2 (also called Fas); DR3, to which VEGI binds; DR4 and DR5, to which TRAIL binds; and DR6, no ligand has yet been identified that binds to DR6. The induction of apoptosis by TNF ligands is initiated by binding to their specific DRs, such as TNFα/TNFR1, FasL /Fas (CD95, DR2), TRAIL (Apo2L)/DR4 (TRAIL-R1) or DR5 (TRAIL-R2). When TNF-α binds to TNFR1, it recruits a protein called TNFR-associated death domain (TRADD) through its death domain (DD). TRADD then recruits a protein called Fas-associated protein with death domain (FADD), which then sequentially activates caspase-8 and caspase-3, and thus apoptosis. Alternatively, TNF-α can activate mitochondria to sequentially release ROS, cytochrome c, and Bax, leading to activation of caspase-9 and caspase-3 and thus apoptosis. Some of the miRNAs can inhibit apoptosis by targeting the death-receptor pathway including miR-21, miR-24, and miR-200c.
p53 has the ability to activate intrinsic and extrinsic pathways of apoptosis by inducing transcription of several proteins like Puma, Bid, Bax, TRAIL-R2, and CD95.
Some inhibitors of apoptosis proteins (IAPs) can inhibit apoptosis indirectly (such as cIAP1/BIRC2, cIAP2/BIRC3) or inhibit caspase directly, such as XIAP/BIRC4 (inhibits caspase-3, -7, -9), and Bruce/BIRC6 (inhibits caspase-3, -6, -7, -8, -9).
Any alterations or abnormalities occurring in apoptotic processes contribute to development of human diseases and malignancies especially cancer.
References:
1.Yağmur Kiraz, Aysun Adan, Melis Kartal Yandim, et al. Major apoptotic mechanisms and genes involved in apoptosis[J]. Tumor Biology, 2016, 37(7):8471.
2.Aggarwal B B, Gupta S C, Kim J H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey.[J]. Blood, 2012, 119(3):651.
3.Ashkenazi A, Fairbrother W J, Leverson J D, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors[J]. Nature Reviews Drug Discovery, 2017.
4.McIlwain D R, Berger T, Mak T W. Caspase functions in cell death and disease[J]. Cold Spring Harbor perspectives in biology, 2013, 5(4): a008656.
5.Ola M S, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis[J]. Molecular and cellular biochemistry, 2011, 351(1-2): 41-58.
What is Apoptosis? The Apoptotic Pathways and the Caspase Cascade
Targets for Apoptosis
- Pyroptosis(15)
- Caspase(77)
- 14.3.3 Proteins(3)
- Apoptosis Inducers(71)
- Bax(15)
- Bcl-2 Family(136)
- Bcl-xL(13)
- c-RET(15)
- IAP(32)
- KEAP1-Nrf2(73)
- MDM2(21)
- p53(137)
- PC-PLC(6)
- PKD(8)
- RasGAP (Ras- P21)(2)
- Survivin(8)
- Thymidylate Synthase(12)
- TNF-α(141)
- Other Apoptosis(1144)
- Apoptosis Detection(0)
- Caspase Substrate(0)
- APC(6)
- PD-1/PD-L1 interaction(60)
- ASK1(4)
- PAR4(2)
- RIP kinase(47)
- FKBP(22)
Products for Apoptosis
- Cat.No. Product Name Information
-
GC38182
Dauricine
-
GC49913
Davunetide (acetate)
A neuroprotective ADNP-derived peptide
-
GC63364
DB2115 tertahydrochloride
2-Deoxy-2-fluoro-L-fucose, an L-fucose analog, is a fucosylation inhibitor. 2-Deoxy-2-fluoro-L-fucose inhibits de novo synthesis of GDP-fucose in mammalian cells. Fucosylation is a relatively well-defined biomarker for progression in many human cancers; for example, pancreatic and hepatocellular carcinoma.
-
GC62222
DB2313
DB2313 is a potent transcription factor PU.1 inhibitor with an apoptosis of 14 nM. DB2313 disrupts the interaction of PU.1 with target gene promoters. DB2313 induces apoptosis of acute myeloid leukemia (AML) cells, and has anticancer effects.
-
GC10824
DBeQ
P97 ATPase inhibitor
-
GC32719
dBET6
dBET6 is a highly potent, selective and cell-permeable PROTAC connected by ligands for Cereblon and BET, with an IC50 of 14 nM, and has antitumor activity.
-
GC19485
DC661
A potent PPT1 inhibitor

-
GC16795
DCA
DCA is a metabolic regulator in cancer cells' mitochondria with anticancer activity. DCA inhibits PDHK, resulting in decreased lactic acid in the tumor microenvironment. DCA increases reactive oxygen species (ROS) generation and promotes cancer cell apoptosis. DCA also works as NKCC inhibitor.

-
GC14007
DCC-2036 (Rebastinib)
DCC-2036 (Rebastinib) (DCC-2036) is an orally active, non-ATP-competitiveBcr-Abl inhibitor for Abl1WT and Abl1T315I with IC50s of 0.8 nM and 4 nM, respectively. DCC-2036 (Rebastinib) also inhibits SRC, KDR, FLT3, and Tie-2, and has low activity to seen towards c-Kit.
-
GC33934
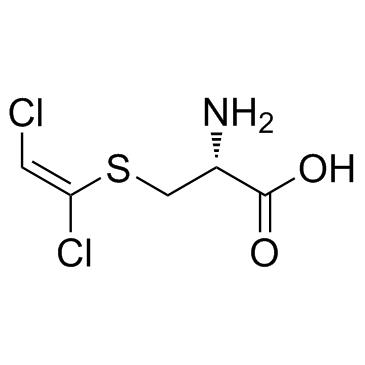
DCVC
DCVC (S-[(1E)-1,2-dichloroethenyl]--L-cysteine) is a bioactive metabolite of trichloroethylene (TCE).
-
GC64971
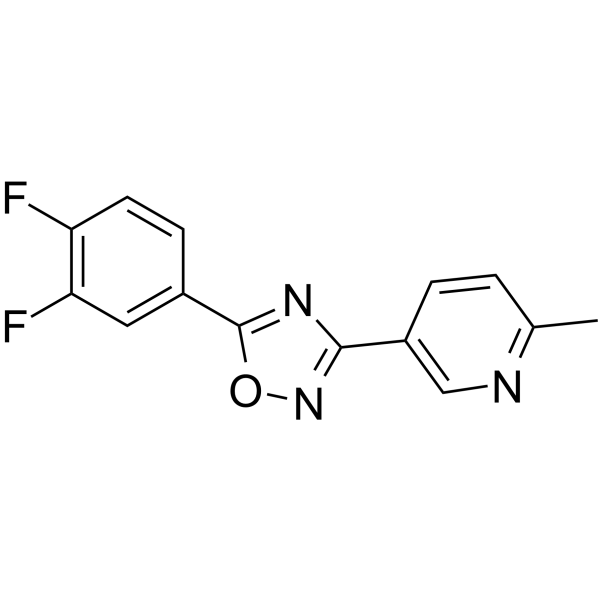
DDO-7263
DDO-7263, a 1,2,4-Oxadiazole derivative, is a potent Nrf2-ARE activator.
-
GC52191
Deacetylanisomycin
Deacetylanisomycin is a potent growth regulator in plants and an inactive derivative of Anisomycin.
-
GC15255
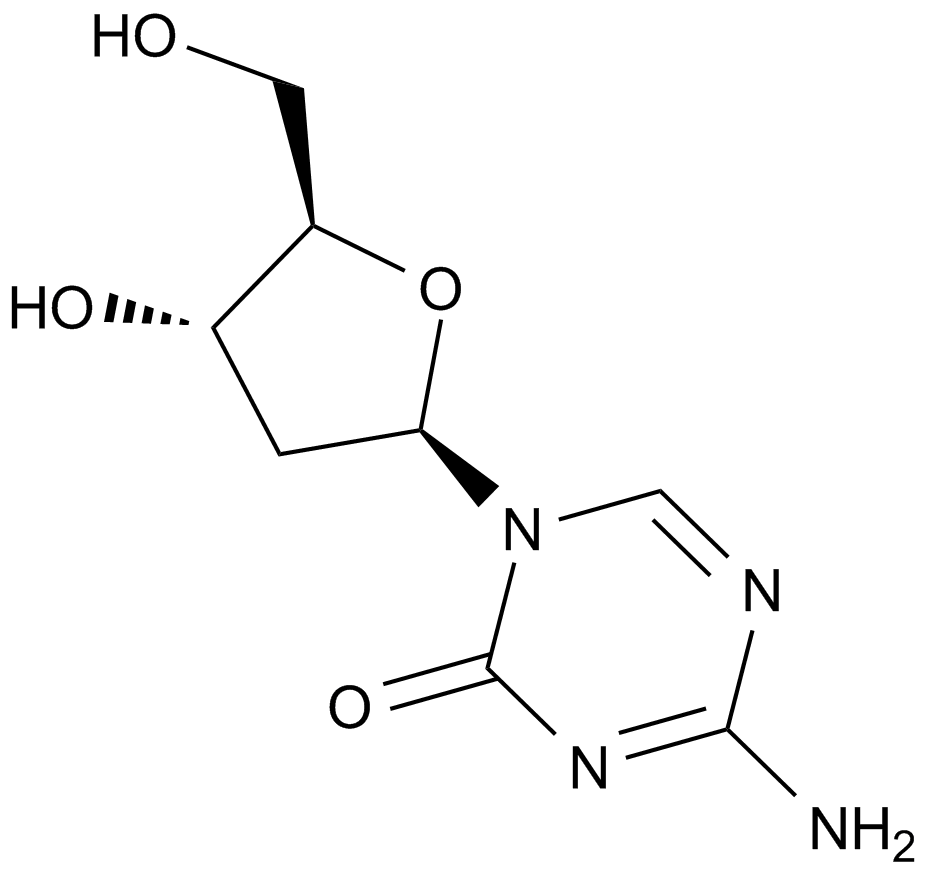
Decitabine(NSC127716, 5AZA-CdR)
Decitabine(DAC) is a deoxycytidine analogue antimetabolite with oral bioactivity and functions as an inhibitor of DNA methyltransferase.
-
GC31892
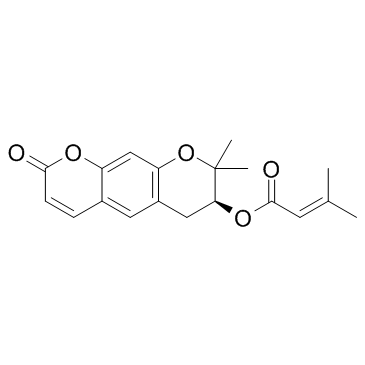
Decursin ((+)-Decursin)
Decursin ((+)-Decursin) ((+)-Decursin ((+)-Decursin)) is a potent anti-tumor agent.
-
GC18556
Degarelix (acetate)
Degarelix is a synthetic gonadotropin-releasing hormone receptor (GNRHR) antagonist (IC50 = 3 nM in HEK293 cells expressing the human receptor).
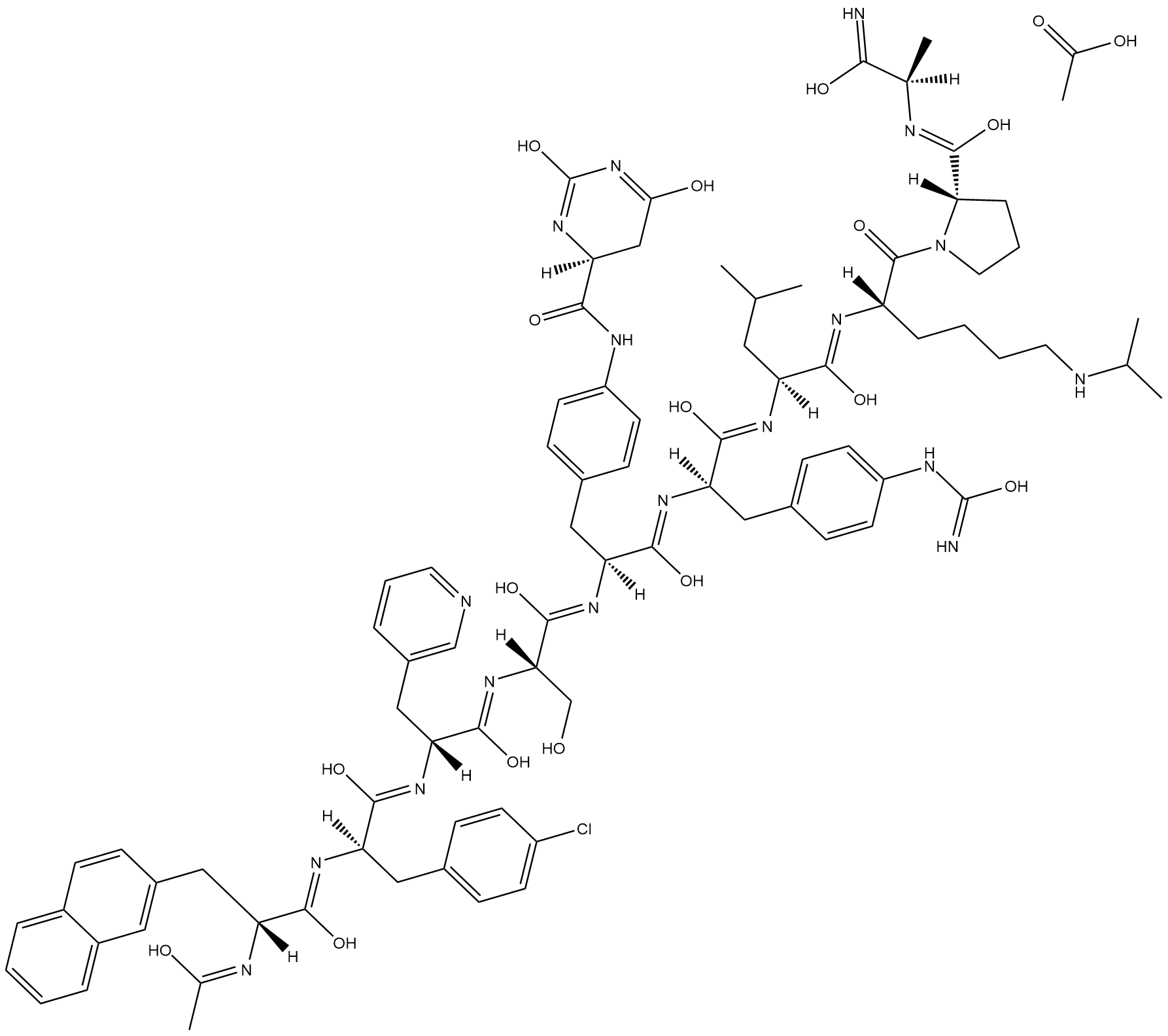
-
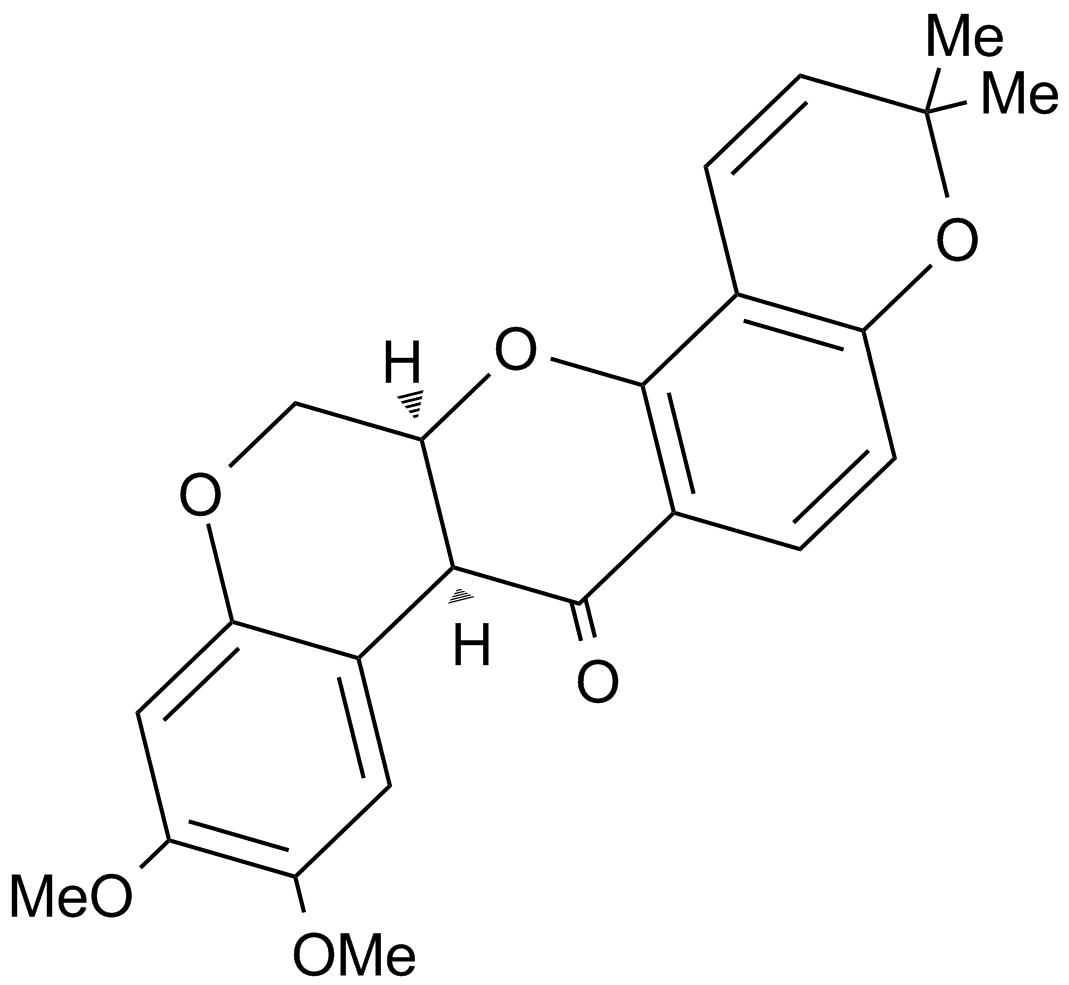
GC15484
Deguelin
A potent antiproliferative rotenoid compound
-
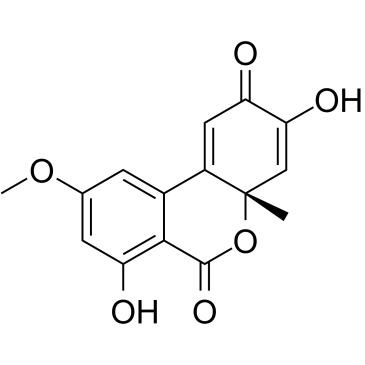
GC35829
Dehydroaltenusin
Dehydroaltenusin is a small molecule selective inhibitor of eukaryotic DNA polymerase α, a type of antibiotic produced by a fungus with an IC50 value of 0.68 μM. The inhibitory mode of action of dehydroaltenusin against mammalian pol α activity is competitive with respect to the DNA template primer (Ki=0.23 ?M) and non-competitive with respect to the 2'-deoxyribonucleoside 5'-triphosphate substrate (Ki=0.18 ?M). Dehydroaltenusin arrests the cancer cell cycle at the S-phase and triggers apoptosis. Dehydroaltenusin possesses anti-tumor activity against human adenocarcinoma tumor in vivo.
-
GN10040
Dehydrocorydaline
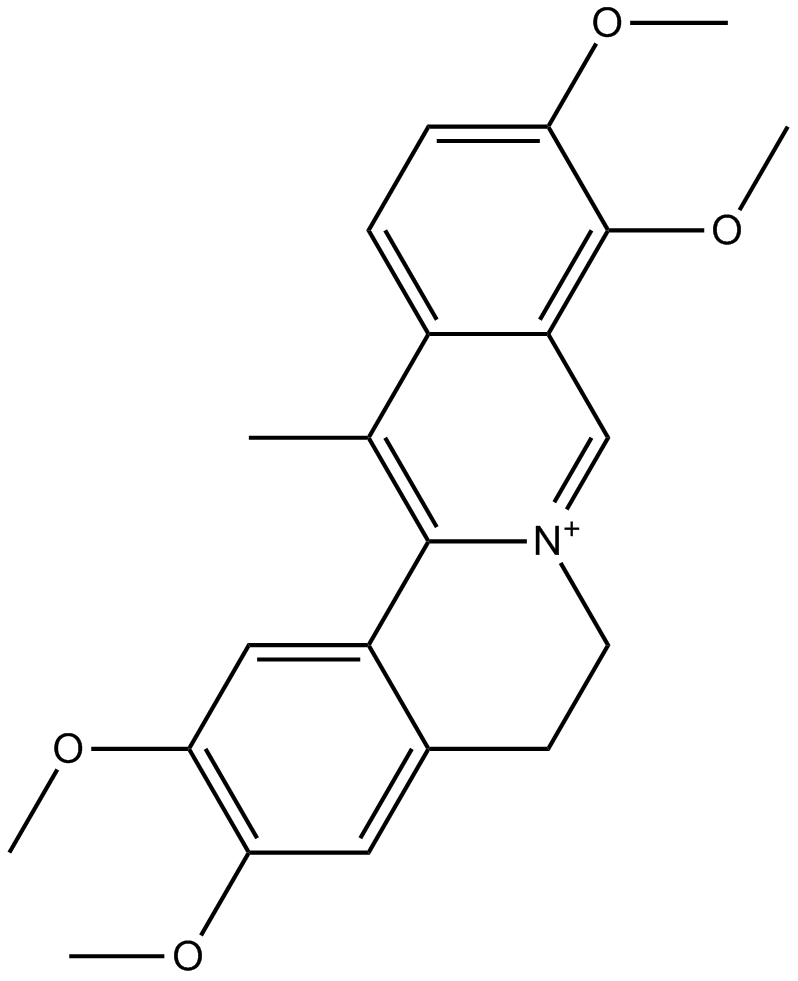
-
GC35834
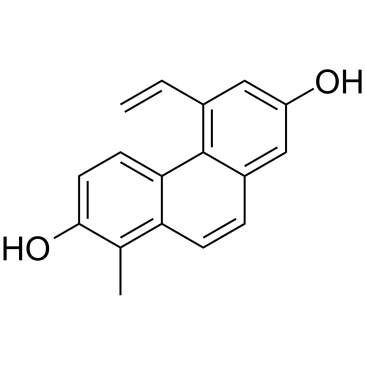
Dehydroeffusol
Dehydroeffusol is a phenanthrene from medicinal herb Juncus effuses. Dehydroeffusol inhibits gastric cancer cell growth and tumorigenicity by selectively inducing tumor-suppressive endoplasmic reticulum stress and a moderate apoptosis. It shows very low toxicity.
-
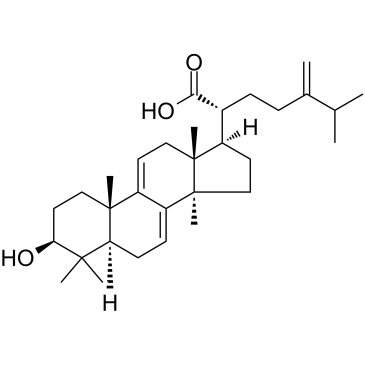
GC38452
Dehydrotrametenolic acid
Dehydrotrametenolic acid is a sterol isolated from the sclerotium of Poria cocos.
-
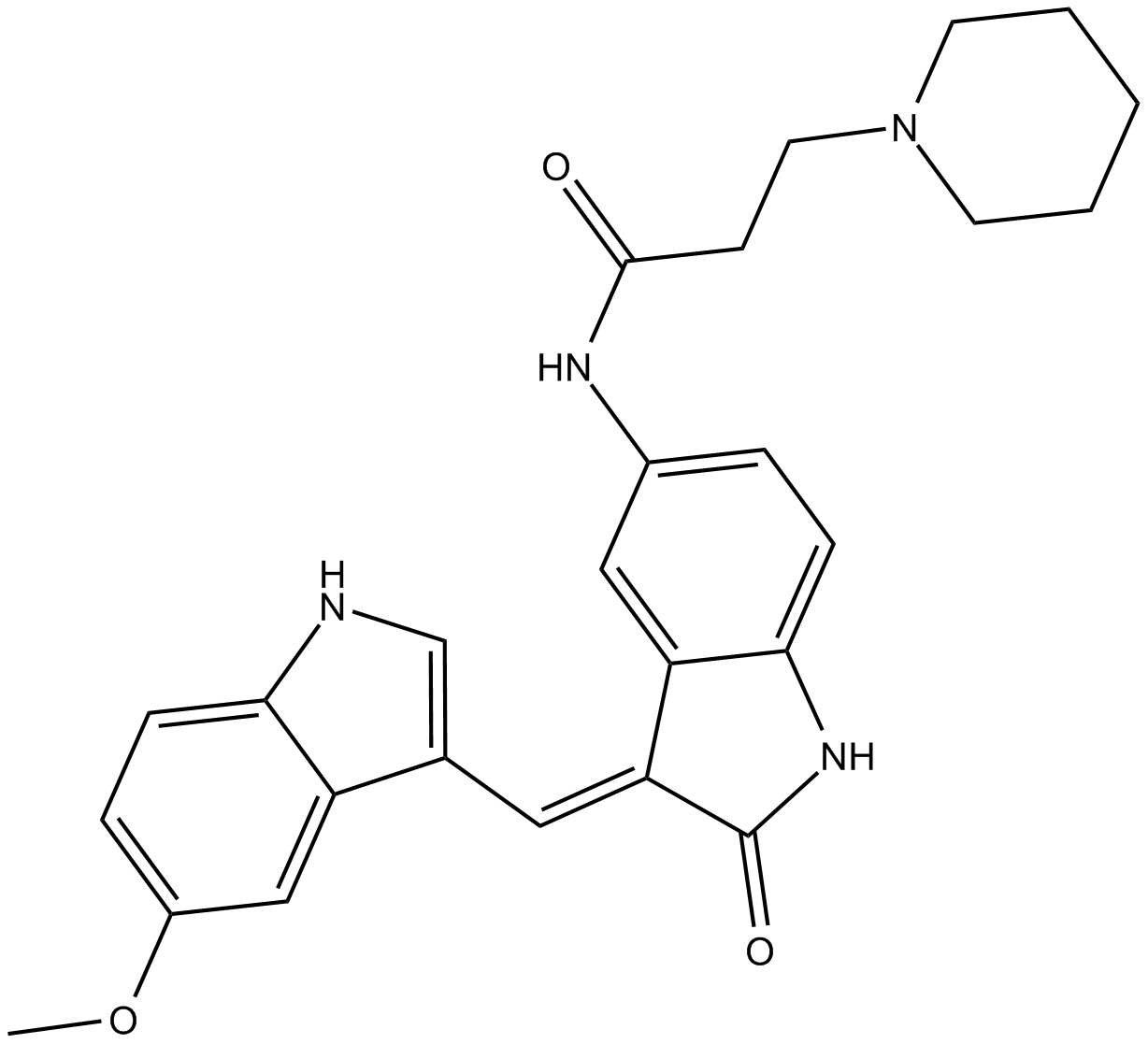
GC16214
DEL-22379
inhibitor of the dimerization of ERK
-
GC43406
Delphinidin (chloride)
Delphinidin (chloride) is an anthocyanidin, a natural plant pigment which serves as the precursor of certain anthocyanins that provide the blue-red colors of flowers, fruits, and red wine.
-
GN10529
Demethoxycurcumin
-
GC38160
Demethylzeylasteral
A nortriterpenoid with diverse biological activities
-
GC43408
Deoxycholic Acid (sodium salt hydrate)
Deoxycholic acid (cholanoic acid) sodium hydrate,a bile acid, is a by-product of intestinal metabolism, that activates the G protein-coupled bile acid receptorTGR5.
-
GC47187
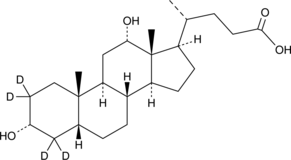
Deoxycholic Acid-d4
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
GC68306
Deoxynyboquinone
-
GC38564
Deoxypodophyllotoxin
Deoxypodophyllotoxin (DPT), a derivative of podophyllotoxin, is a lignan with potent antimitotic, anti-inflammatory and antiviral properties isolated from rhizomes of Sinopodophullumhexandrum (Berberidaceae).
-
GC16077
Deracoxib
Selective COX-2 inhibitor
-
GC34157
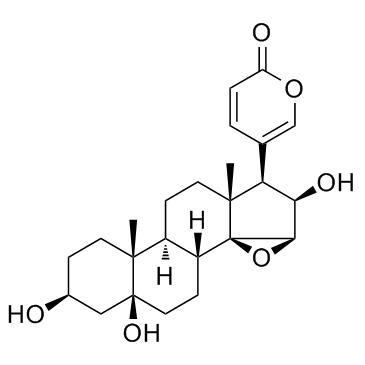
Desacetylcinobufotalin (Deacetylcinobufotalin)
Desacetylcinobufotalin (Deacetylcinobufotalin) is a natural compound; apoptosis inducer and shows the marked inhibition effect to HepG2 cells and the IC50 value is 0.0279μmol/ml.
-
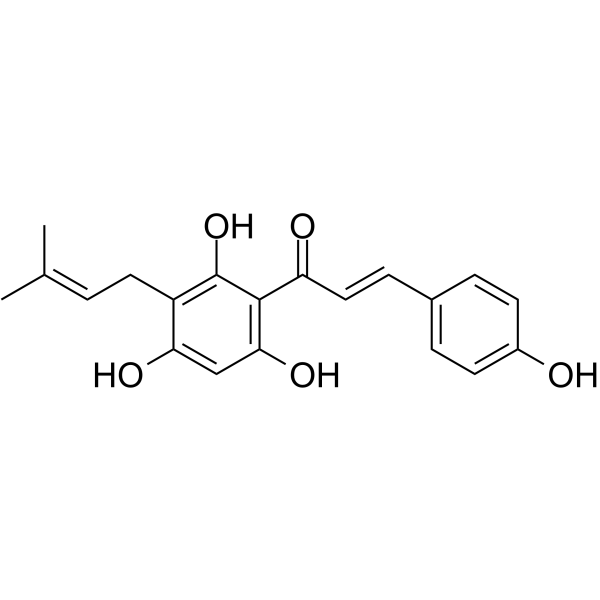
GC62927
Desmethylxanthohumol
Desmethylxanthohumol is a prenylated hydroxychalcone isolated from hop cones (Humulus lupulus L.). Desmethylxanthohumol is a powerful apoptosis inducing agent. Desmethylxanthohumol has antiplasmodial, antiproliferative, and antioxidant bioactivities.
-
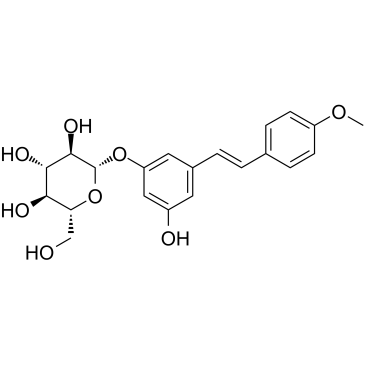
GC38221
Desoxyrhaponticin
Desoxyrhaponticin is a stilbene glycoside from the Tibetan nutritional food Rheum tanguticum Maxim. Desoxyrhaponticin is a Fatty acid synthase (FASN) inhibitor, and has apoptotic effect on human cancer cells.
-
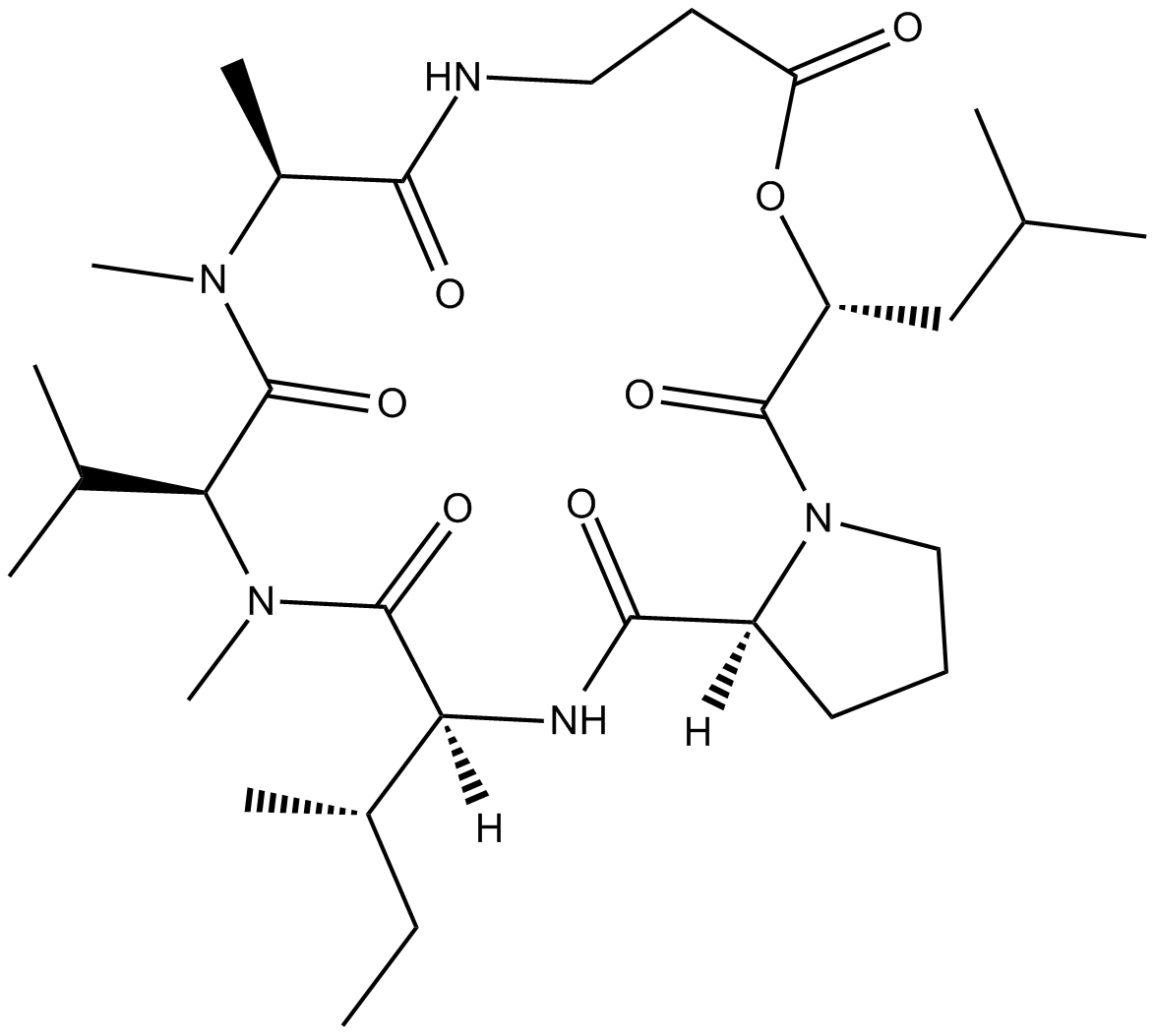
GC10661
Destruxin B
insecticidal and phytotoxic activity;induces apoptosis
-
GC10520
Dextran sulfate sodium salt (M.W 200000)
Dextran sulfate sodium salt is a is a polymer of anhydroglucose with the molecular weight range of 35000-45000.

-
GC38250
Dextran sulfate sodium salt (MW 16000-24000)
A sulfated polysaccharide

-
GC38252
Dextran sulfate sodium salt (MW 35000-45000)
A sulfated polysaccharide

-
GC38253
Dextran sulfate sodium salt (MW 4500-5500)
A sulfated polysaccharide

-
GC38251
Dextran sulfate sodium salt (MW 450000-550000)
A sulfated polysaccharide

-
GC43436
Diacetylcercosporin
Diacetylcercosporin is a perylenequinone produced by Cercospora and Septoria that has diverse biological activities.
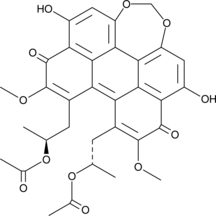
-
GC45992
Diallyl Tetrasulfide
An organosulfur compound with diverse biological activities
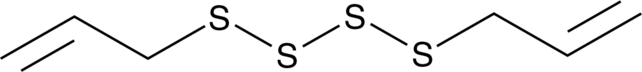
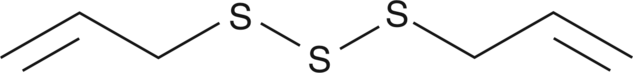
-
GC43439
Diallyl Trisulfide
Hydrogen sulfide (H2S) is an endogenously-produced gaseous second messenger that can regulate many physiological processes.
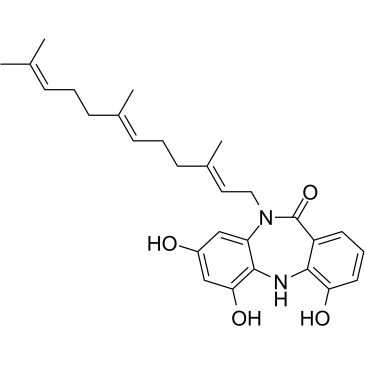
-
GC35859
Diazepinomicin
Diazepinomicin (TLN-4601) is a secondary metabolite produced by Micromonospora sp. Diazepinomicin (TLN-4601) inhibits the EGF-induced Ras-ERK MAPK signaling pathway and induces apoptosis. An anti-tumor agent for K-Ras mutant models.
-
GC61419
Dibenzoylmethane
Dibenzoylmethane, a minor ingredient in licorice, activates Nrf2 and prevents various cancers and oxidative damage.
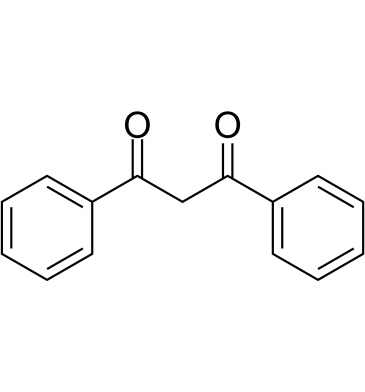
-
GC64139
Dibromoacetic acid
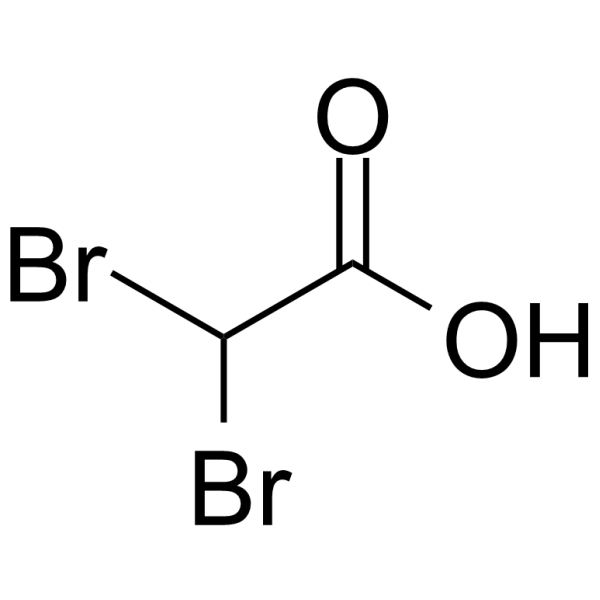
-
GC14622
Diclofenac Potassium
nonsteroidal anti-inflammatory drug
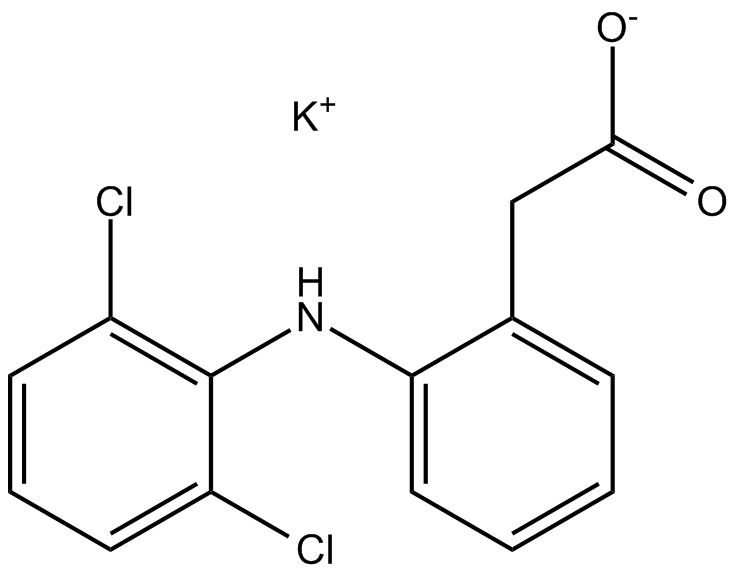
-
GN10751
Dictamnine
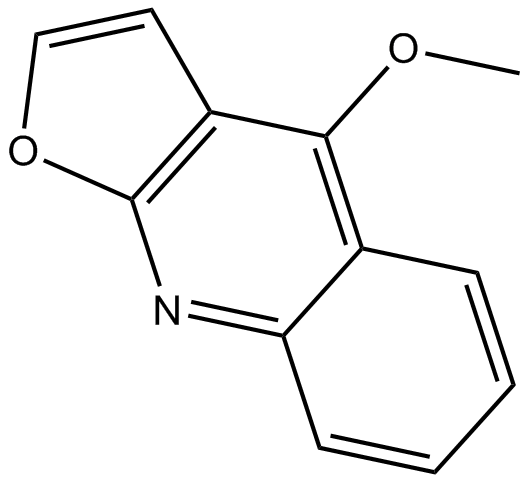
-
GC49153
Didemnin B
Didemnin B is a cyclic depsipeptide produced by marine tunicates that specifically binds the GTP-bound conformation of EEF1A.
-
GC39808
Didesmethylrocaglamide
Didesmethylrocaglamide, a derivative of Rocaglamide, is a potent eukaryotic initiation factor 4A (eIF4A) inhibitor. Didesmethylrocaglamide has potent growth-inhibitory activity with an IC50 of 5 nM. Didesmethylrocaglamide suppresses multiple growth-promoting signaling pathways and induces apoptosis in tumor cells. Antitumor activity.
-
GC38179
Didymin
A flavonoid with diverse biological activities
-
GC15578
Dienogest
Orally active synthetic progesterone
-
GC43452
Diffractaic Acid
Diffractaic acid is a lichen metabolite that has been found in P.
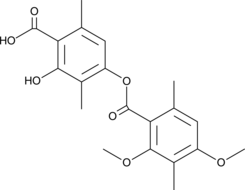
-
GC64375
Difopein TFA
Difopein (TFA), a specific and competitive inhibitor of 14-3-3 protein (a highly conserved eukaryotic regulatory molecule), blocking the ability of 14-3-3 to bind to target proteins and inhibits 14-3-3/Ligand interactions. Difopein (TFA) leads to induction of apoptosis and enhances the ability of cisplatin to kill cells.

-
GC49842
Digoxigenin Bisdigitoxoside
A Na+/K+-ATPase inhibitor
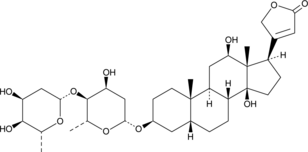
-
GN10056
Dihydroartemisinin
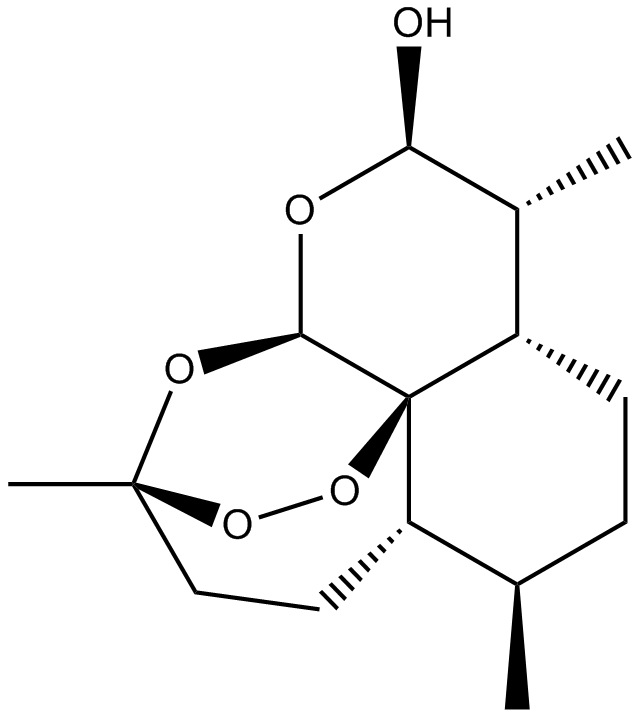
-
GC38617
Dihydrokaempferol
A flavone with diverse biological activities
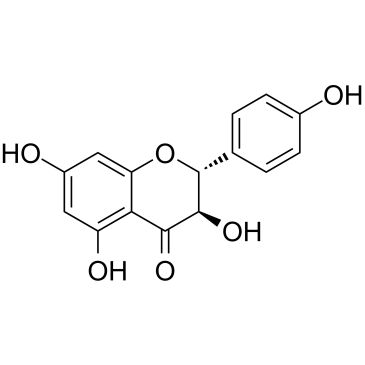
-
GC43462
Dihydrolipoic Acid
Dihydrolipoic acid (DHLA) is a dithiol-containing carboxylic acid that is the reduced form of α-lipoic acid.
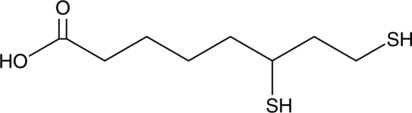
-
GC38620
Dihydrorotenone
Dihydrorotenone, a natural pesticide, is a potent mitochondrial inhibitor.
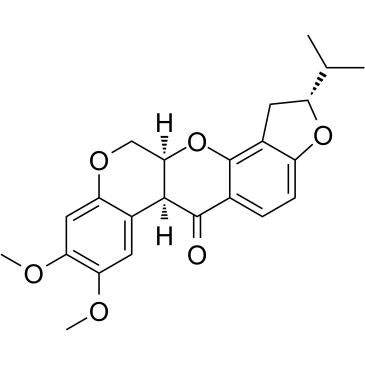
-
GC41112
Dihydroxy Melphalan
Dihydroxy melphalan is an inactive degradation product of melphalan.
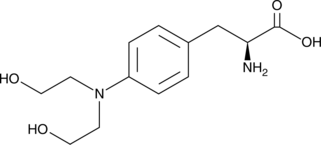
-
GC43463
DiIC1(5)
DiIC1(5) is a signal-off fluorescent probe for the detection of mitochondrial membrane potential disruption.
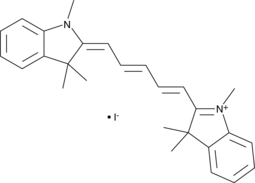
-
GC38772
DIM-C-pPhOH
A Nur77 antagonist
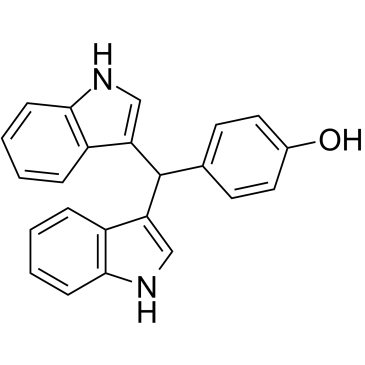
-
GC16590
Dimethyl Fumarate
nuclear factor (erythroid-derived)-like 2 (Nrf2) pathway activator
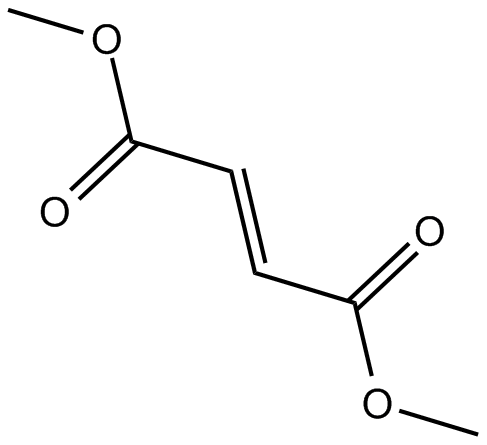
-
GC62936
Dimethyl fumarate D6
Dimethyl fumarate D6 is a deuterium labeled Dimethyl fumarate.
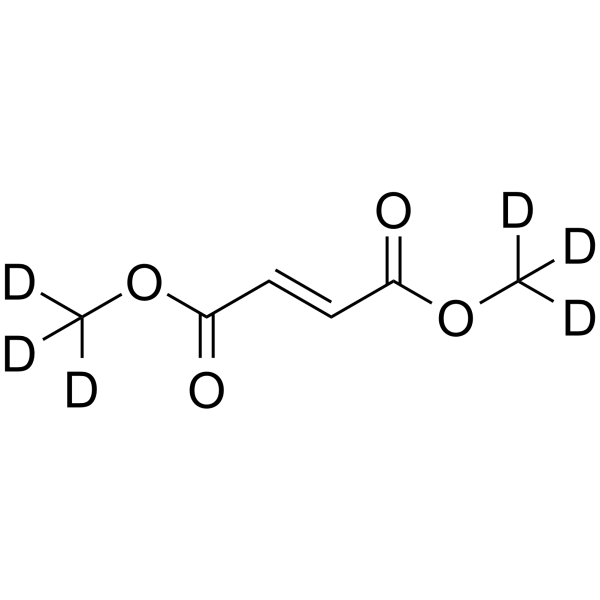
-
GC25351
Dimethyl itaconate
Dimethyl itaconate can reprogram neurotoxic to neuroprotective primary astrocytes through the regulation of LPS-induced Nod-like receptor protein 3 (NLRP3) inflammasome and nuclear factor 2/heme oxygenase-1 (NRF2/HO-1) pathways.
-
GN10531
Dimethylfraxetin
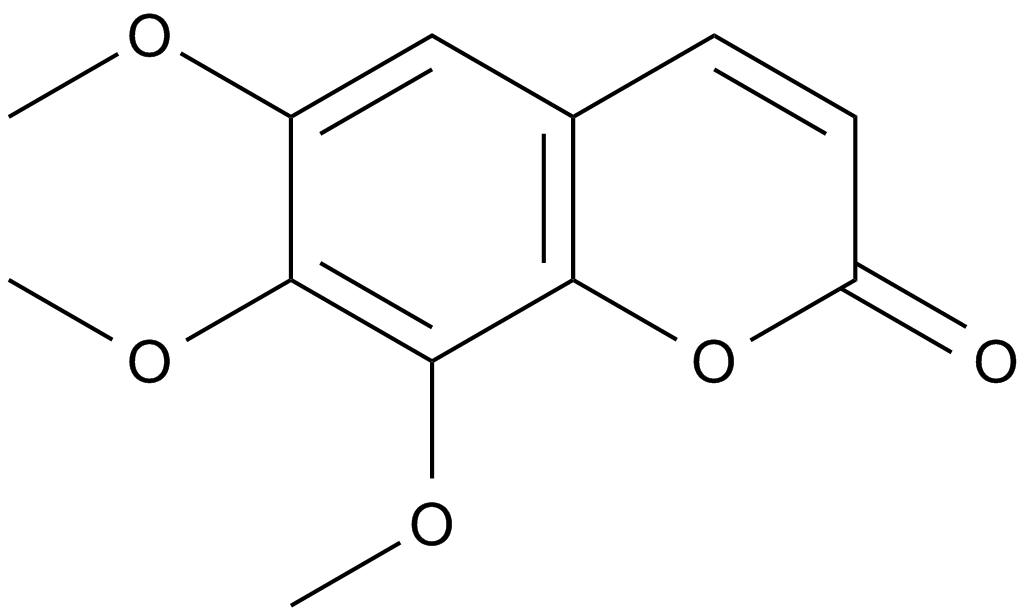
-
GC17648
Dinaciclib(SCH727965)
Dinaciclib(SCH727965) (SCH 727965) is a potent inhibitor of CDK, with IC50s of 1 nM, 1 nM, 3 nM, and 4 nM for CDK2, CDK5, CDK1, and CDK9, respectively.
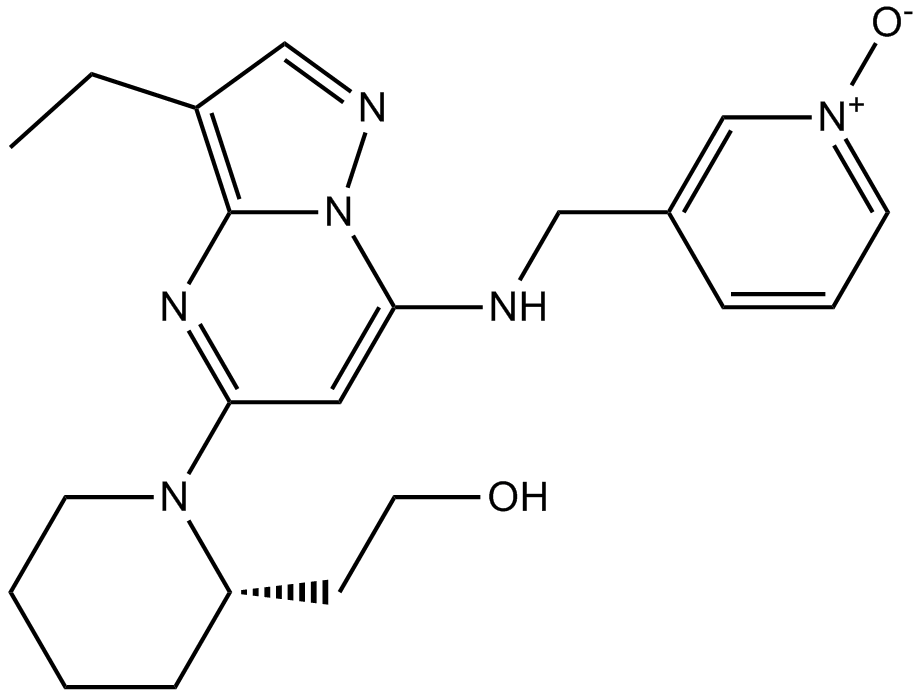
-
GN10284
Dioscin
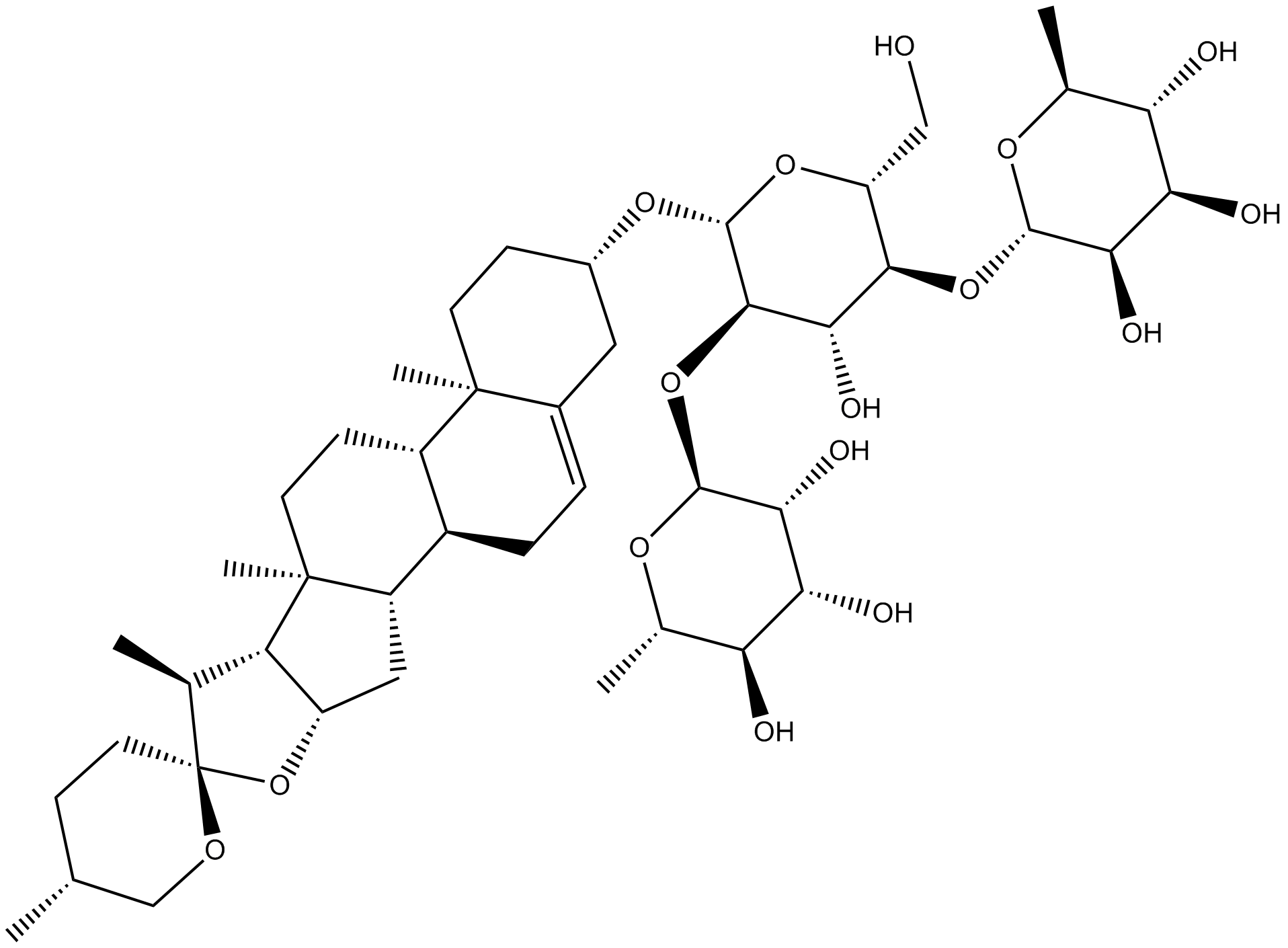
-
GC38408
Diosgenin glucoside
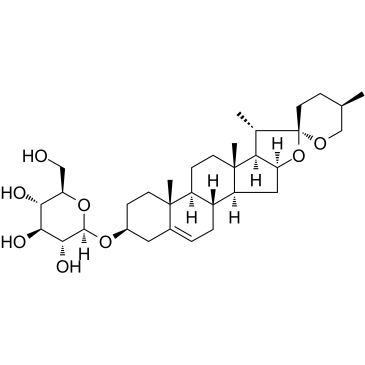
-
GC60782
Disitertide TFA
Disitertide (P144) TFA is a peptidic transforming growth factor-beta 1 (TGF-β1) inhibitor specifically designed to block the interaction with its receptor. Disitertide (P144) TFA is also a PI3K inhibitor and an apoptosis inducer.

-
GC10801
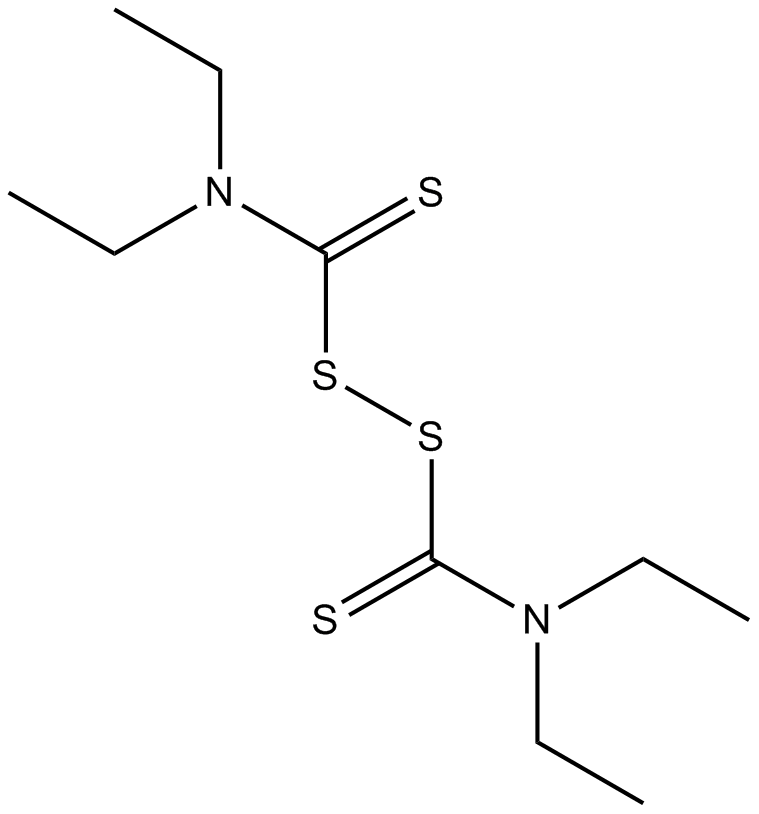
Disulfiram
An irreversible inhibitor of aldehyde dehydrogenase
-
GC39295
DJ001
DJ001 is a highly specific, selective and non-competitive protein tyrosine phosphatase-σ (PTPσ) inhibitor with an IC50 of 1.43 μM.
-
GC49164
DM4 (hydrate)
A derivative of maytansine
-
GC35882
dMCL1-2
dMCL1-2 is a potent and selective PROTAC of myeloid cell leukemia 1 (MCL1) (Bcl-2 family member) based on Cereblon, which binds to MCL1 with a KD of 30 nM. dMCL1-2 activats the cellular apoptosis machinery by degradation of MCL1.
-
GC40192
DMPAC-Chol
DMPAC-Chol is a cationic cholesterol derivative that has been used in liposome formation for gene transfection.
-
GC61466
DMU-212
DMU-212 is a methylated derivative of Resveratrol, with antimitotic, anti-proliferative, antioxidant and apoptosis promoting activities. DMU-212 induces mitotic arrest via induction of apoptosis and activation of ERK1/2 protein. DMU-212 has orally active.
-
GC16684
Docetaxel
Docetaxel is a taxane class of anti-mitotic chemotherapeutic agents with an IC50 of 0.2 µM.
-
GA21397
Dolastatin 15
Dolastatin 15 originally isolated from the sea hare Dolabella auricularia belongs to a family of antimitotic and antineoplastic depsipeptides inducing apoptosis in various malignant cell types. It was shown to inhibit the growth of the P388 lymphocytic leukemia cell line efficiently, with an ED?? value of 2.4 ng/mL.

-
GC11099
Doxifluridine
oral prodrug of the antineoplastic agent 5-fluorouracil (5-FU)
-
GC18439
Doxorubicinone
A metabolite of doxorubicin
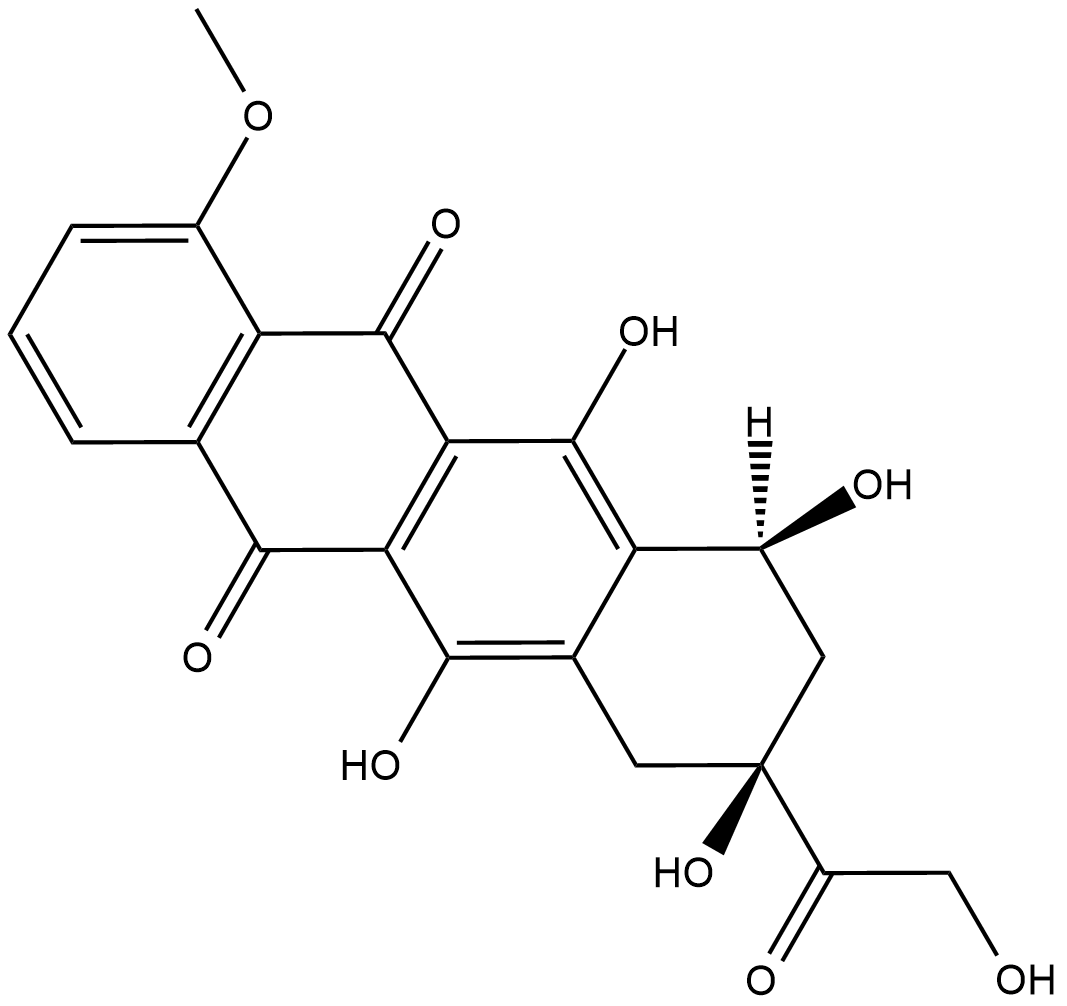
-
GC15399
Dp44mT
iron chelator and anticancer agent
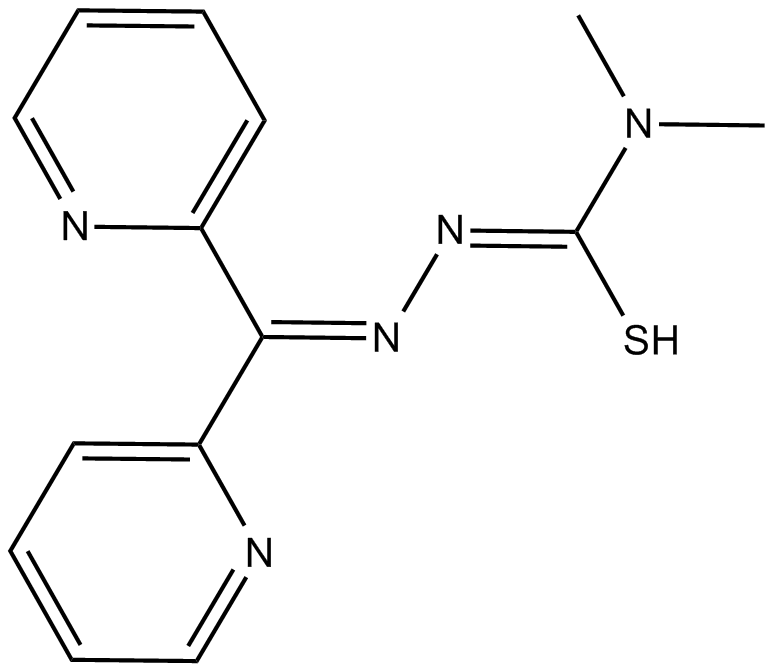
-
GC33384
DPBQ
DPBQ activates p53 and triggers apoptosis in a polyploid-specific manner, but does not inhibit topoisomerase or bind DNA. DPBQ elicits expression and phosphorylation of p53 and this effect is specific to tetraploid cells.
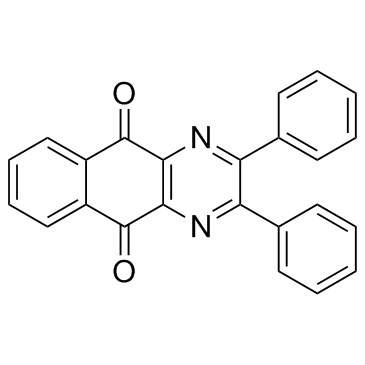
-
GC35898
Dracorhodin perchlorate
A pro-apoptotic compound
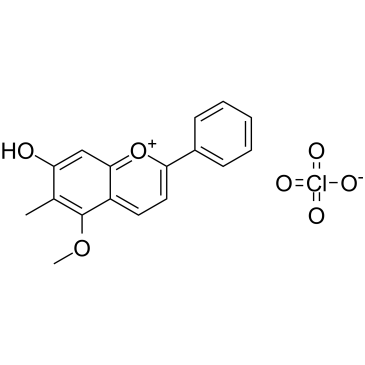
-
GC39707
Droloxifene
A selective estrogen receptor modulator
-
GC13706
Droxinostat
An inhibitor of HDAC3, HDAC6, and HDAC8
-
GC35908
Duocarmycin A
Duocarmycin A, which is one of well-known antitumor antibiotics, is a DNA alkylator and efficiently alkylates adenine N3 at the 3′ end of AT-rich sequences in the DNA. Duocarmycin A, as a chemotherapeutic agent, results HLC-2 cells typically apoptotic changes, including chromatin condensation, sub-G1 accumulation in DNA histogram pattern, and decrease in procaspase-3 and 9 levels.
-
GC35913
Durvalumab
A humanized anti-PD-L1 monoclonal antibody

-
GC10893
Dutasteride
5-alpha-reductase inhibitor
-
GC62590
E64FC26
E64FC26 is a potent pan-inhibitor of the protein disulfide isomerase (PDI) family, with IC50s of 1.9, 20.9, 25.9, 16.3, and 25.4 μM against PDIA1, PDIA3, PDIA4, TXNDC5, and PDIA6, respectively. E64FC26 shows anti-myeloma activity.
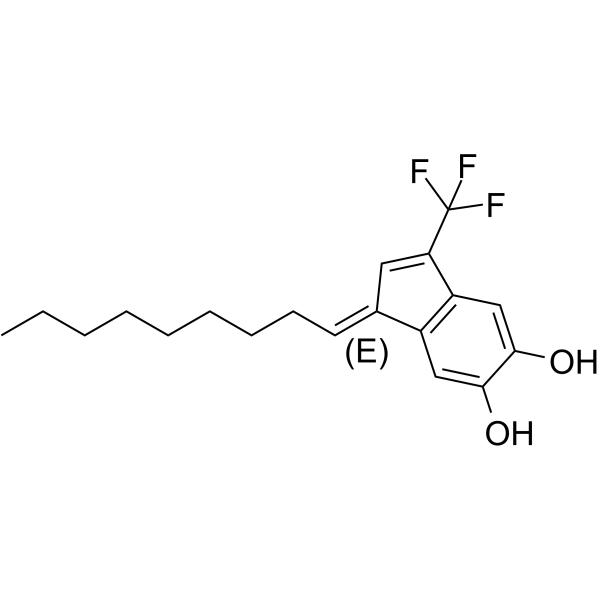
-
GC39377
EB-3D
EB-3D is a potent and selective choline kinase α (ChoKα) inhibitor, with an IC50 of 1 μM for ChoKα1. EB-3D exerts effects on ChoKα expression, AMPK activation, apoptosis, endoplasmic reticulum stress and lipid metabolism. EB-3D exhibits a potent antiproliferative activity in a panel of T-leukemia cell lines. Anti-cancer activity.
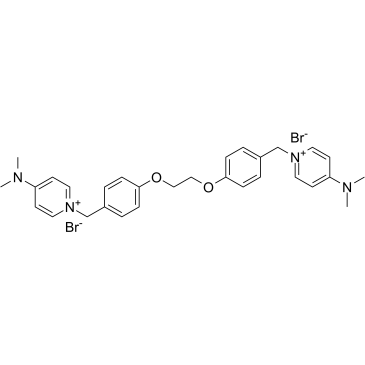
-
GC50282
EC 19
Synthetic retinoid; induces differentiation of stem cells
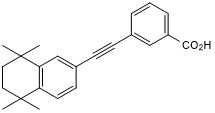
-
GC39174
EC359
EC359 is a potent, selective, high affinity and orally active leukemia inhibitory factor receptor (LIFR) inhibitor with a Kd of 10.2 nM, which directly interacts with LIFR to effectively block LIF/LIFR interactions.
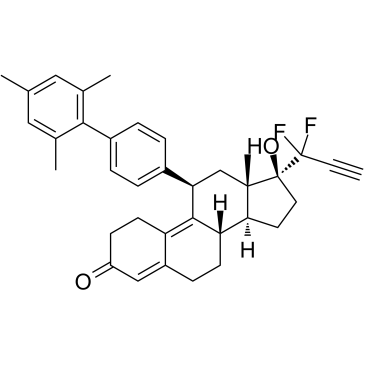
-
GC38149
Ecabet sodium
An antiulcerative and gastroprotective agent
-
GN10504
Echinocystic acid
-
GC47279
Echinosporin
A bacterial metabolite with antibacterial and anticancer activities
-
GC63871
Echitamine chloride
Echitamine chloride is the major monoterpene indole alkaloid present in Alstonia with potent anti-tumour activity. Echitamine chloride induces DNA fragmentation and cells apoptosis. Echitamine chloride inhibits pancreatic lipase with an IC50 of 10.92 μM.
-
GC63470
Eclitasertib
Eclitasertib (DNL-758) is a potent receptor-interacting protein kinase 1 (RIPK1) inhibitor with an IC50 of <1 ?Μ (From patent WO2017136727A2, example 42).
-
GC33156
Ecteinascidin 770 (Ecteinascidine 770)
Ecteinascidin 770 (Ecteinascidine 770) (ET-770) is a 1,2,3,4-tetrahydroisoquinoline alkaloid with potent anti-cancer activities; inhibits U373MG cells with an IC50 of 4.83 nM.
-
GC12838
Edaravone
A radical scavenger and antioxidant
-
GC62562
EGFR-IN-11
EGFR-IN-11 is a fourth-generation EGFR-tyrosine kinase inhibitor (EGFR-TKI) with an IC50 of 18 nM for triple mutant EGFRL858R/T790M/C797S. EGFR-IN-11 significantly suppresses the EGFR phosphorylation, induce the apoptosis, and arrest cell cycle at G0/G1.
-
GC14756
EI1
EZH2 inhibitor
-
GC64864
EJMC-1
EJMC-1 is a moderately potent TNF-α inhibitor with an IC50 value of 42 μM.