iGluR
iGluR (ionotropic glutamate receptor) is a ligand-gated ion channel that is activated by the neurotransmitter glutamate. iGluR are integral membrane proteins compose of four large subunits that form a central ion channel pore. Sequence similarity among all known glutamate receptor subunits, including the AMPA, kainate, NMDA, and δ receptors.
AMPA receptors are the main charge carriers during basal transmission, permitting influx of sodium ions to depolarise the postsynaptic membrane. NMDA receptors are blocked by magnesium ions and therefore only permit ion flux following prior depolarisation. This enables them to act as coincidence detectors for synaptic plasticity. Calcium influx through NMDA receptors leads to persistent modifications in the strength of synaptic transmission.
Targets for iGluR
Products for iGluR
- Cat.No. Product Name Information
-
GC18033
γDGG
γDGG is a competitive AMPA receptor blocker.
-
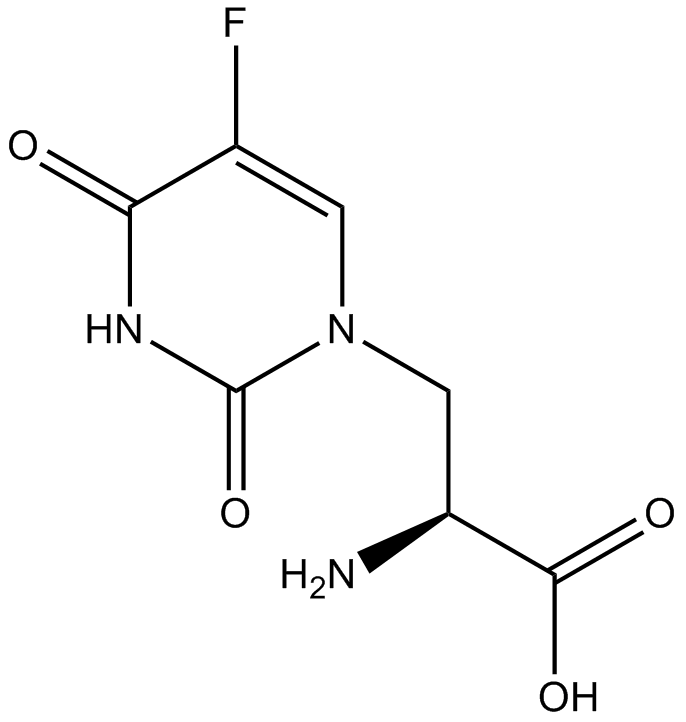
GC11889
(S)-(-)-5-Fluorowillardiine
AMPA receptor agonist
-
GC34998
(S)-(-)-5-Fluorowillardiine hydrochloride
(S)-(-)-5-Fluorowillardiine hydrochloride is a potent and specific AMPAR agonist.

-
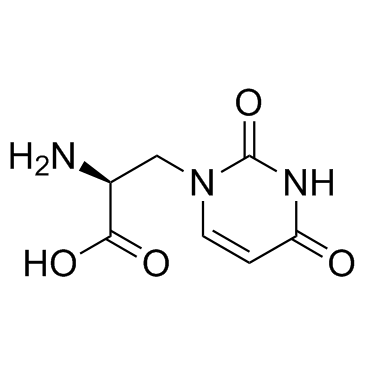
GC30212
(S)-Willardiine ((-)-Willardiine)
(S)-Willardiine ((-)-Willardiine) is a potent agonist of AMPA/kainate receptors with EC50 of 44.8 uM.
-
GC12863
1-BCP
potentiator of AMPA-mediated responses
-
GC33673
24-Hydroxycholesterol
24-Hydroxycholesterol is a natural sterol, which serves as a positive allosteric modulator of N-Methyl-d-Aspartate (NMDA) receptorsR, and a potent activator of the transcription factors LXR.
-
GC31309
6-Methoxy-2-naphthoic acid (Naproxen impurity O)
6-Methoxy-2-naphthoic acid (Naproxen impurity O) is an NMDA receptor modulator extracted from patent WO 2012019106 A2.
-
GC10781
7-Chlorokynurenic acid
NMDA receptor glycine site antagonist
-
GC11395
7-Chlorokynurenic acid sodium salt
NMDA receptor antagonist acting at the glycine site

-
GC68438
AMPA receptor modulator-3

-
GC17695
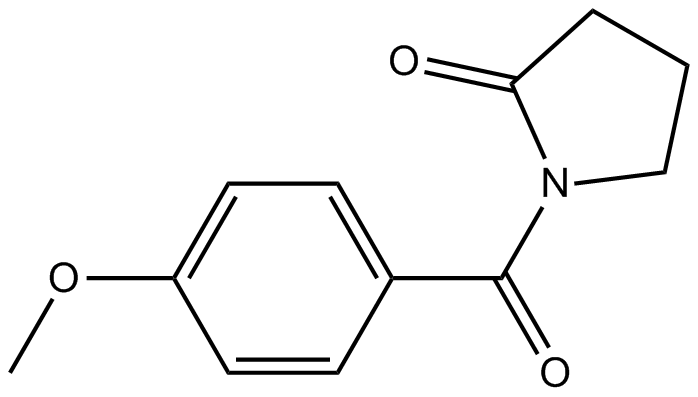
Aniracetam
Nootropic drug for senile dementia
-
GC30911
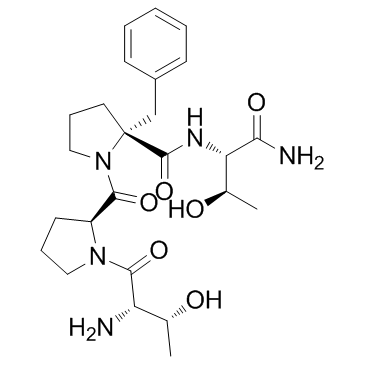
Apimostinel (NRX-1074)
Apimostinel (NRX-1074) (NRX-1074; AGN-241660) is an orally active NMDA receptor partial agonist.
-
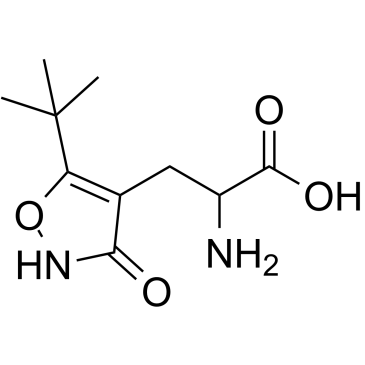
GC38497
ATPA
-
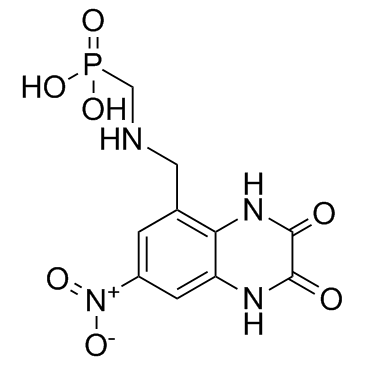
GC33723
Becampanel (AMP 397)
Becampanel (AMP 397) (AMP397) is the first competitive AMPA antagonist and an antiepileptic agent.
-
GC64574
BZAD-01
BZAD-01 is a potent, selective and orally active inhibitor of NMDA NR2B subunit, with a Ki of 72 nM.
-
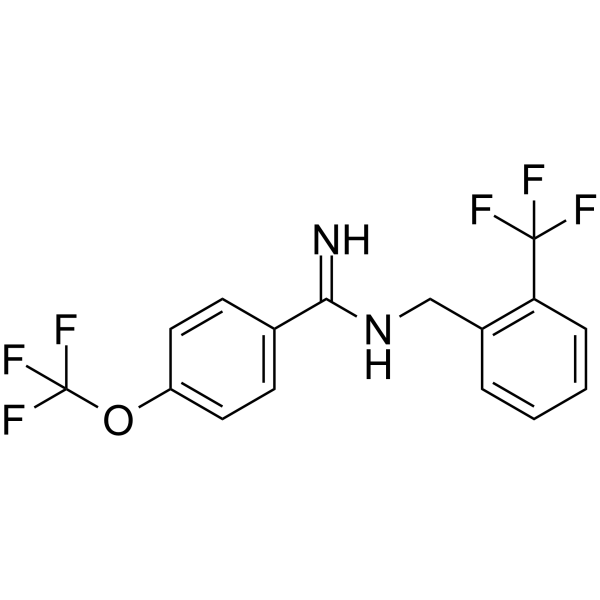
GC18079
CFM-2
AMPA antagonist
-
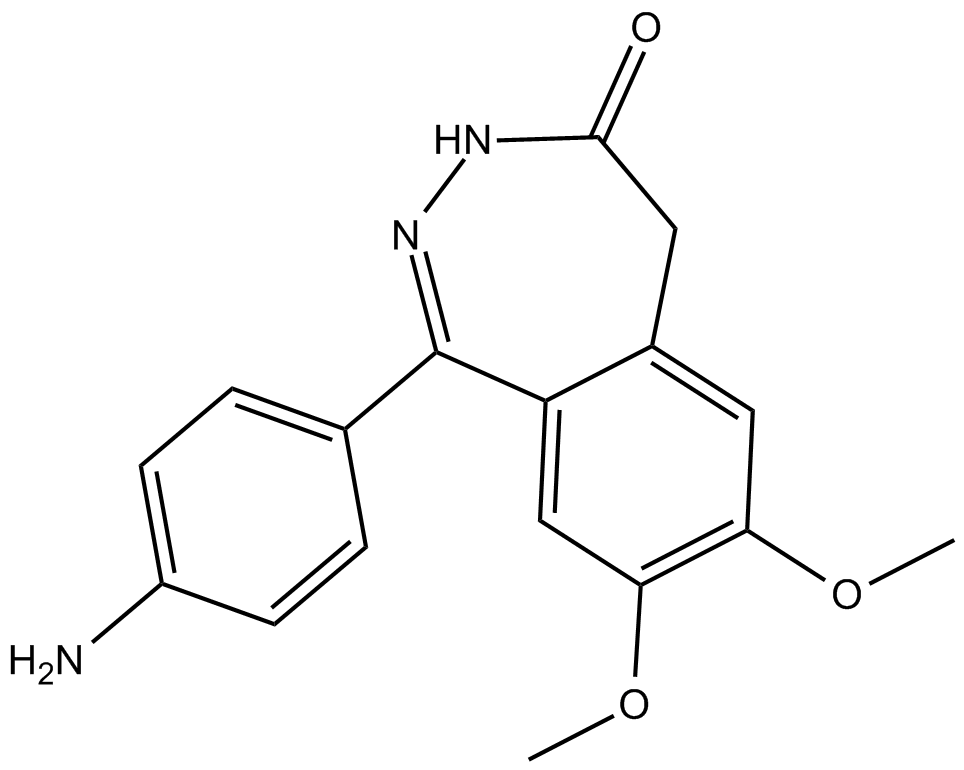
GC38493
CGP 37849

-
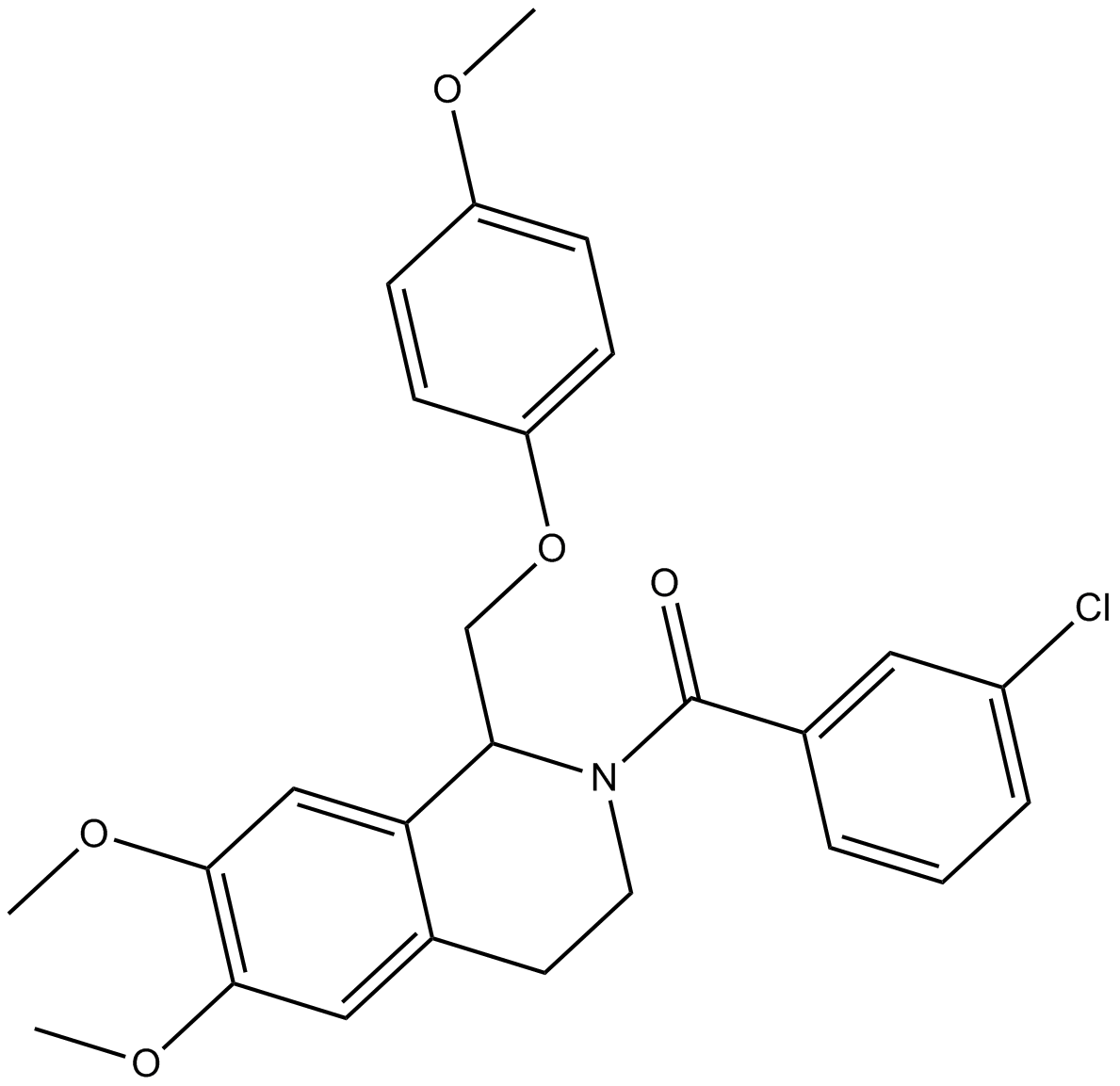
GC14414
CIQ
NMDA receptor potentiator
-
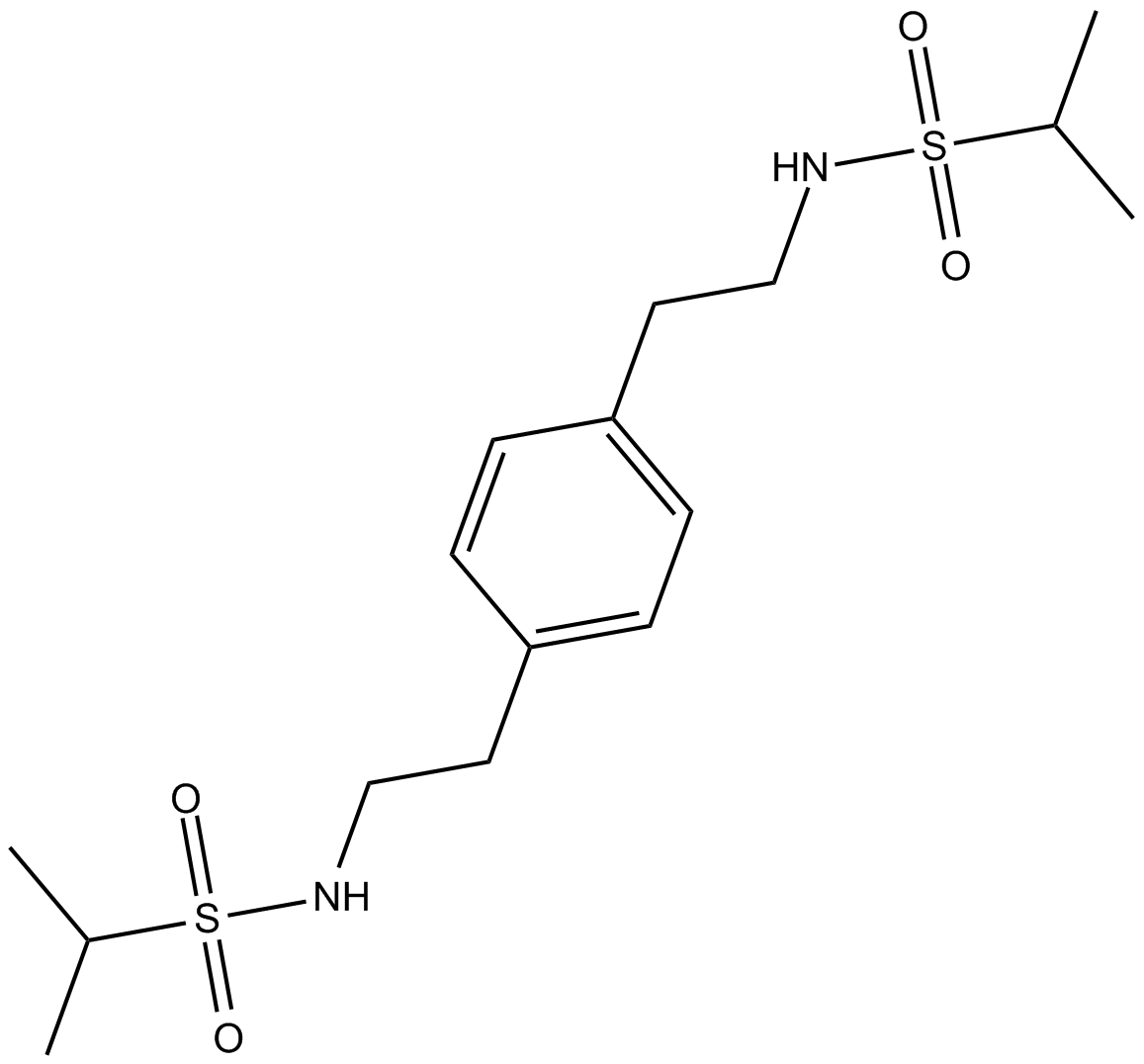
GC13290
CMPDA
positive allosteric modulator of AMPA receptors
-
GC11799
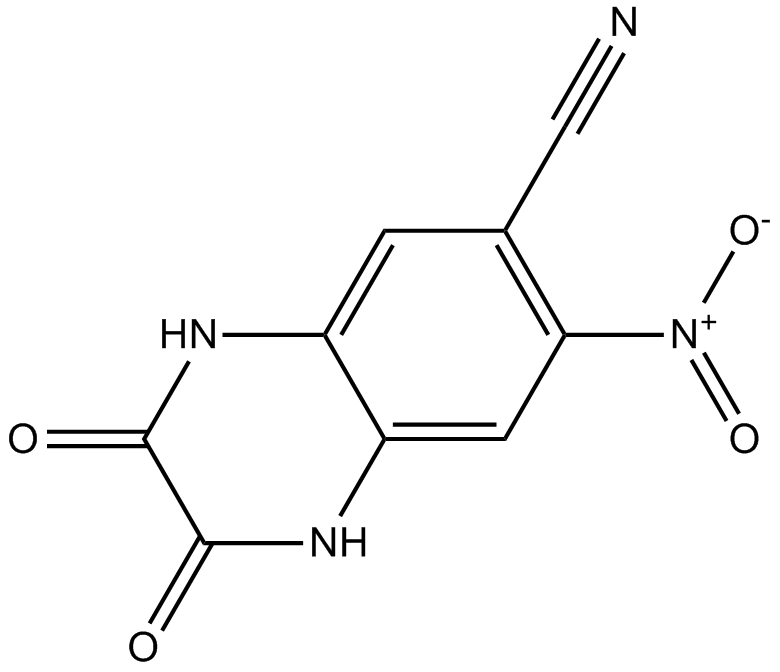
CNQX
CNQX is an effective competitive antagonist of AMPA/kainate receptors, with IC50 values of 0.3µM and 1.5µM, respectively..
-
GC32553
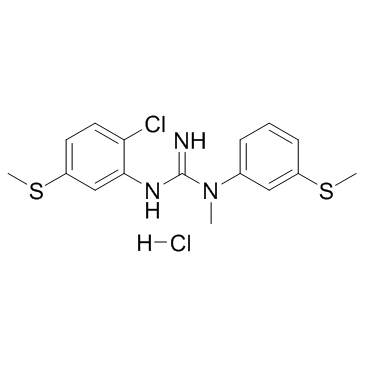
CNS-5161 hydrochloride (CNS 5161A)
CNS-5161 hydrochloride (CNS 5161A) is a novel NMDA ion-channel antagonist that interacts with the NMDA receptor/ion channel site to produce a noncompetitive blockade of the actions of glutamate.
-
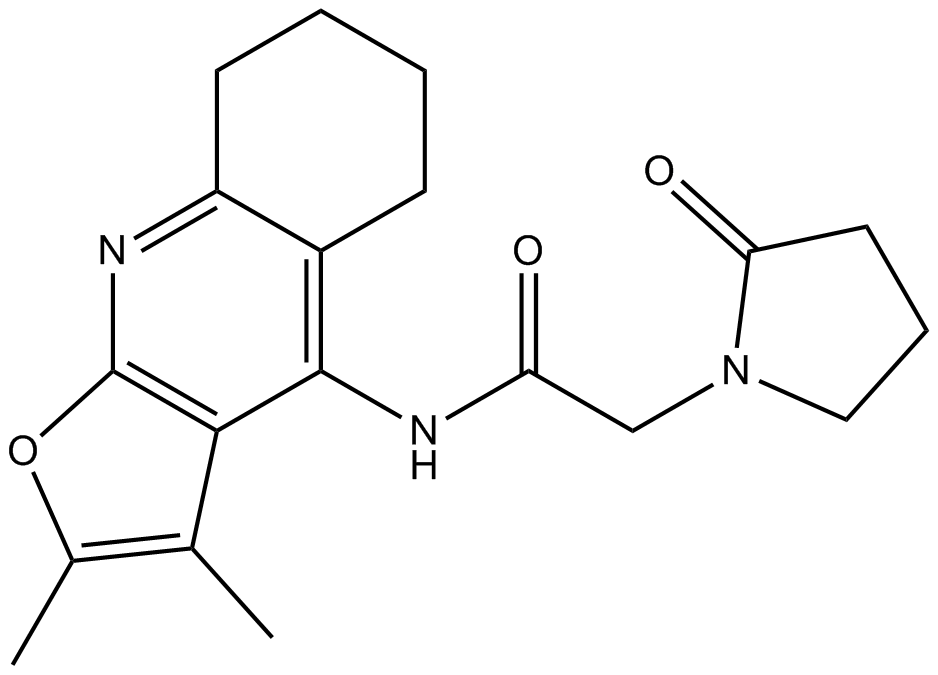
GC16899
Coluracetam
Nootropic agent of the racetam family
-
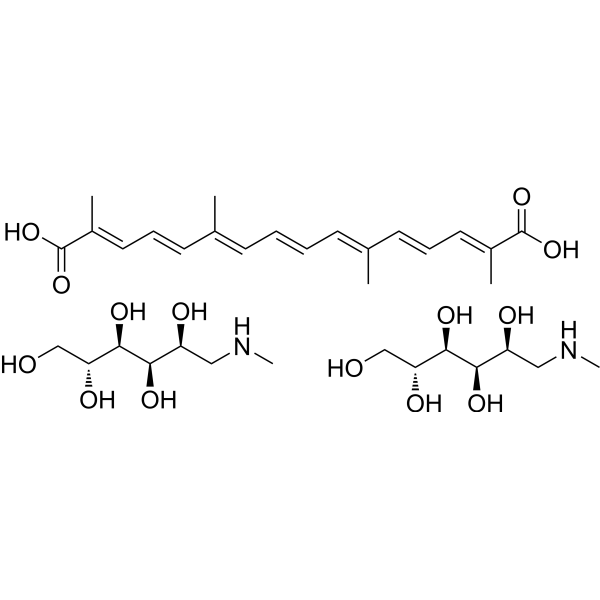
GC70055
Crocetin meglumine
Crocetin (Transcrocetin) meglumine is extracted from saffron (Crocus sativus L.) and is a highly affinity NMDA receptor antagonist.
-
GC14495
CX 546
AMPA receptor potentiator
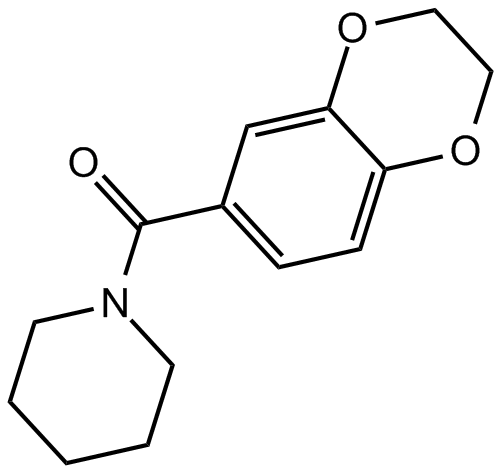
-
GC64244
CX 717
CX 717 is a positive allosteric modulator of AMPA receptor.
-
GC16315
D-AP5
A NMDA antagonist
-
GC63881
DL-Phenylalanine-d5 hydrochloride
-
GC32755
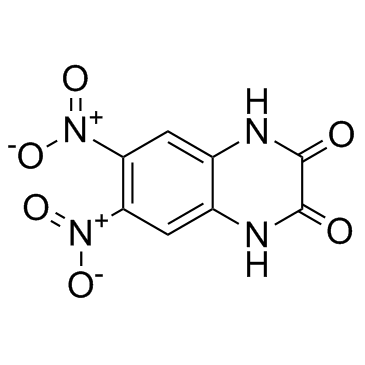
DNQX (FG 9041)
DNQX (FG 9041) (FG 9041), a quinoxaline derivative, is a selective, potent competitive non-NMDA glutamate receptor antagonist (IC50s = 0.5, 2 and 40 μM for AMPA, kainate and NMDA receptors, respectively).
-
GC35917
Dynorphin A (1-10) TFA
-
GC35918
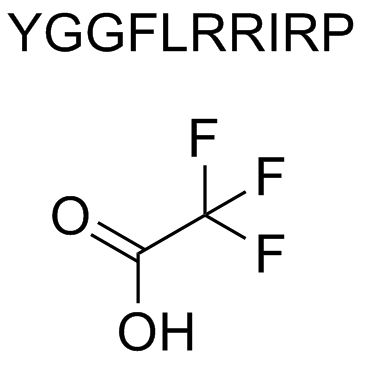
Dynorphin A 1-10
Dynorphin A 1-10 an endogenous opioid neuropeptide, binds to extracellular loop 2 of the κ-opioid receptor.

-
GC15343
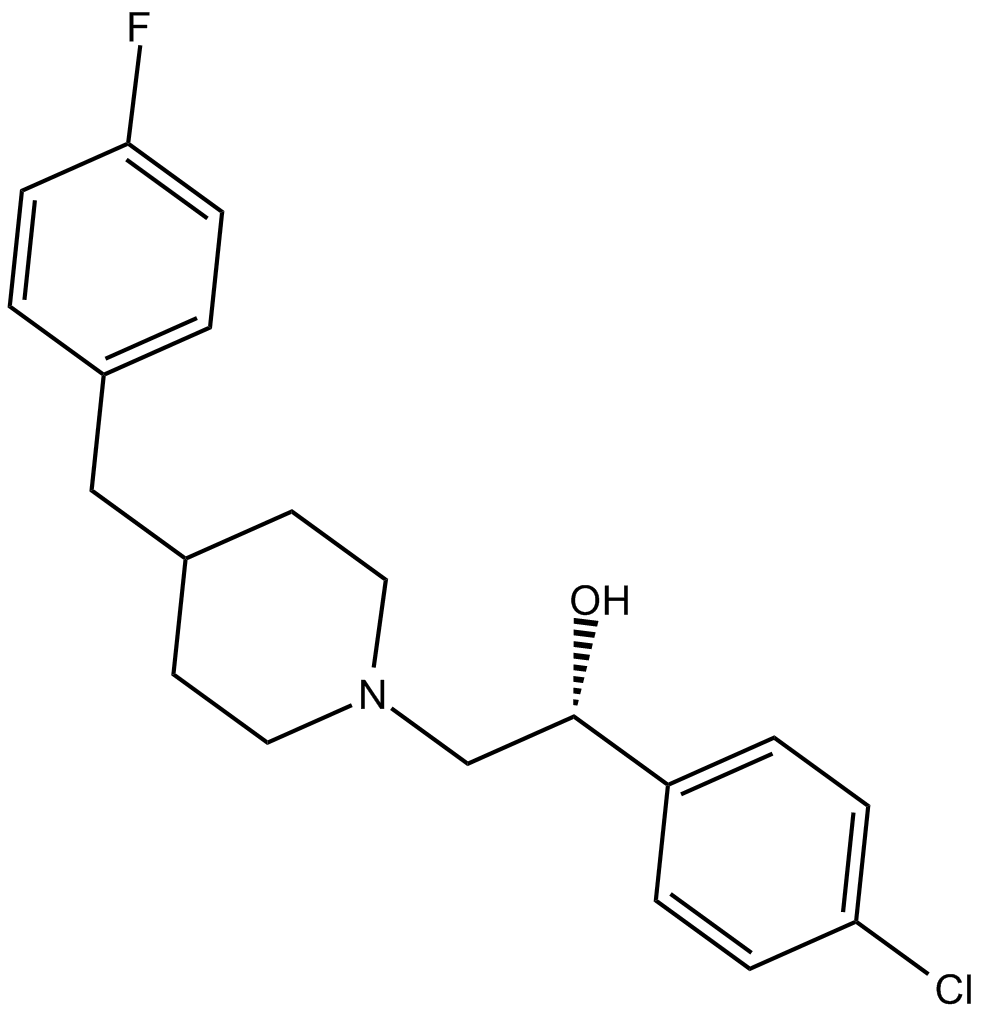
Eliprodil
NMDA receptor antagonist
-
GC36027
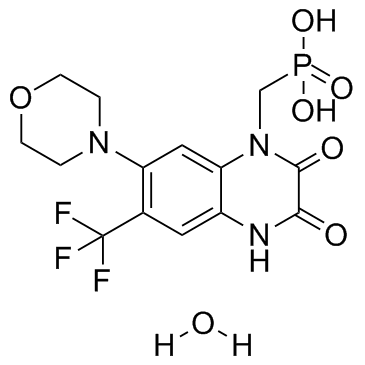
Fanapanel hydrate
Fanapanel hydrate (ZK200775 hydrate) is a highly selective AMPA/kainate antagonist with little activity against NMDA; have Ki values of 3.2 nM, 100 nM, and 8.5 μM against quisqualate, kainate, and NMDA, respectively.
-
GC18223
Farampator
Farampator is a positive allosteric modulator of AMPA receptors that has an EC50 value greater than 32 uM for evoking glutamate currents in isolated pyramidal neurons.
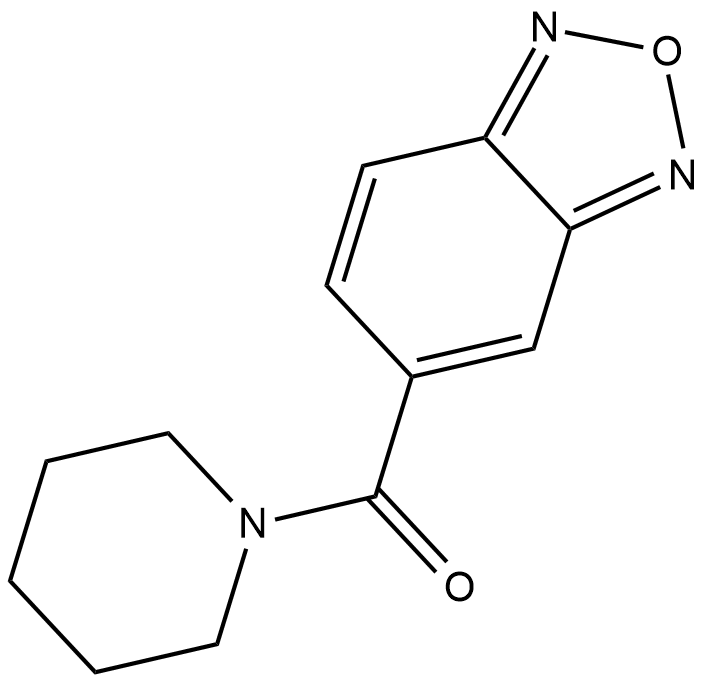
-
GC10522
Felbamate
antagonist at the NMDA-associated glycine binding site
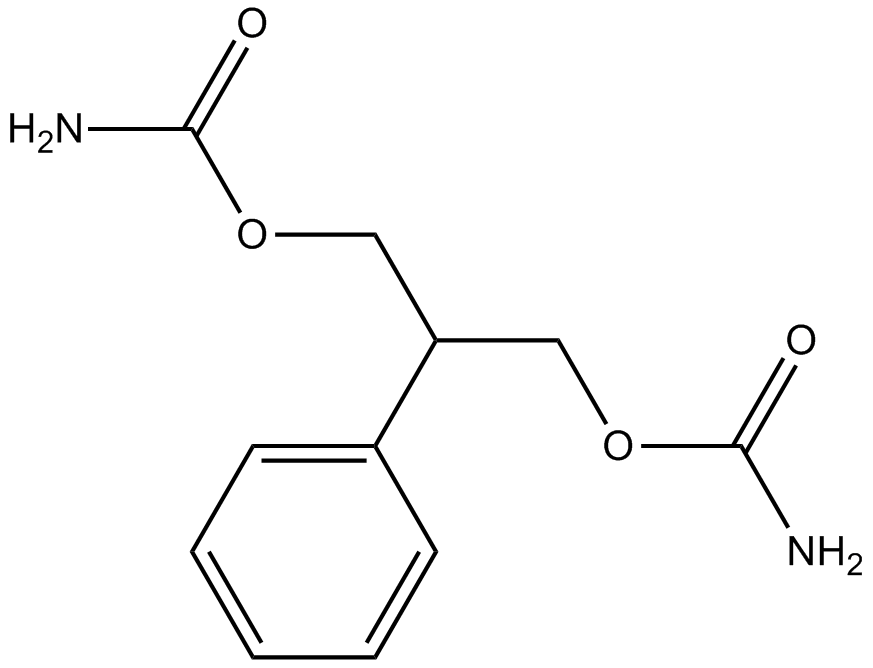
-
GC15363
Felbamate hydrate
N-methyl-D-aspartate (NMDA) inhibitor
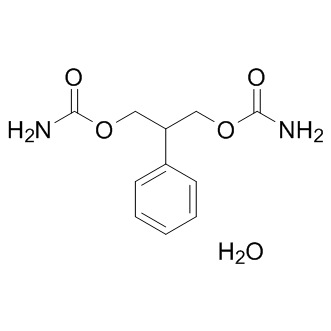
-
GC36060
Flupirtine
Flupirtine(D 9998) is a selective neuronal potassium channel opener that also has NMDA receptor antagonist properties.
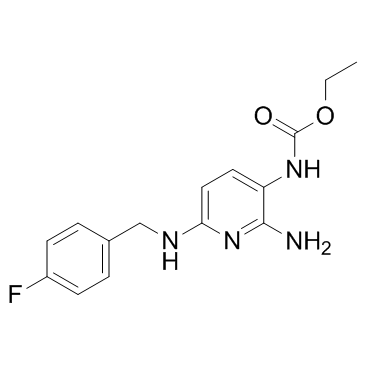
-
GC11470
Flupirtine maleate
NMDA receptor antagonist
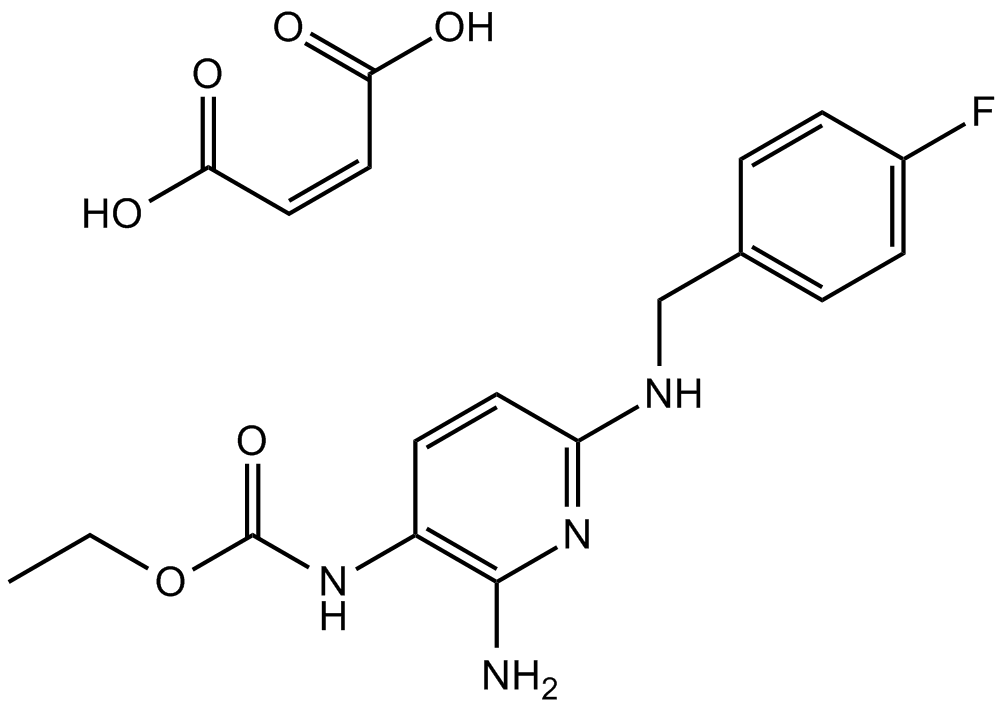
-
GC64245
Glycine-13C2
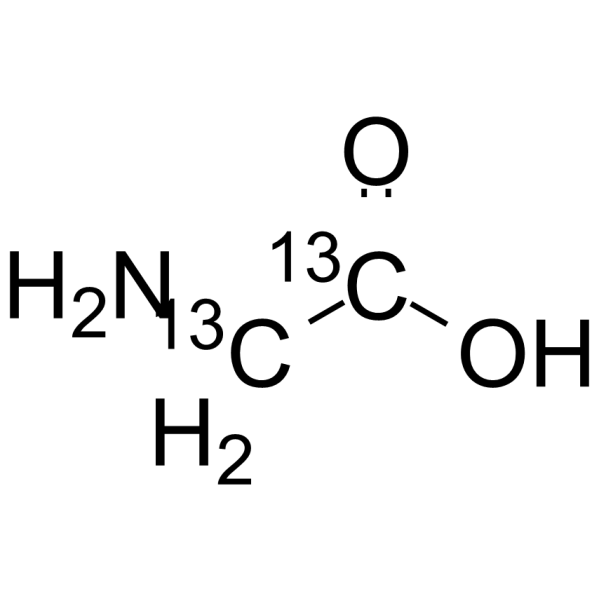
-
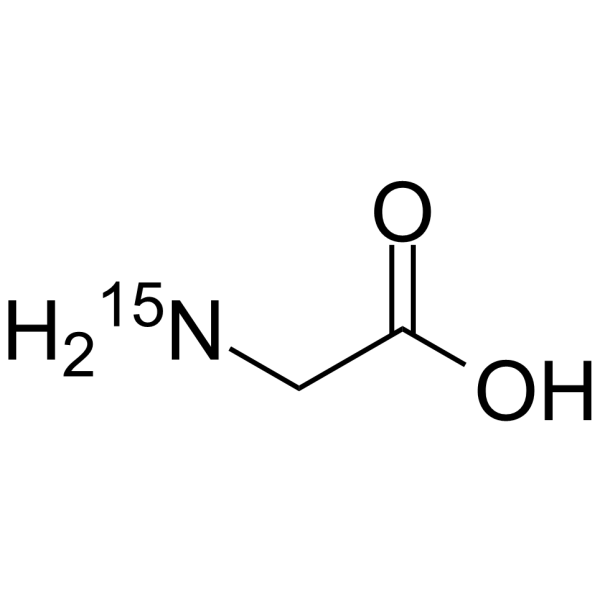
GC64249
Glycine-15N
-
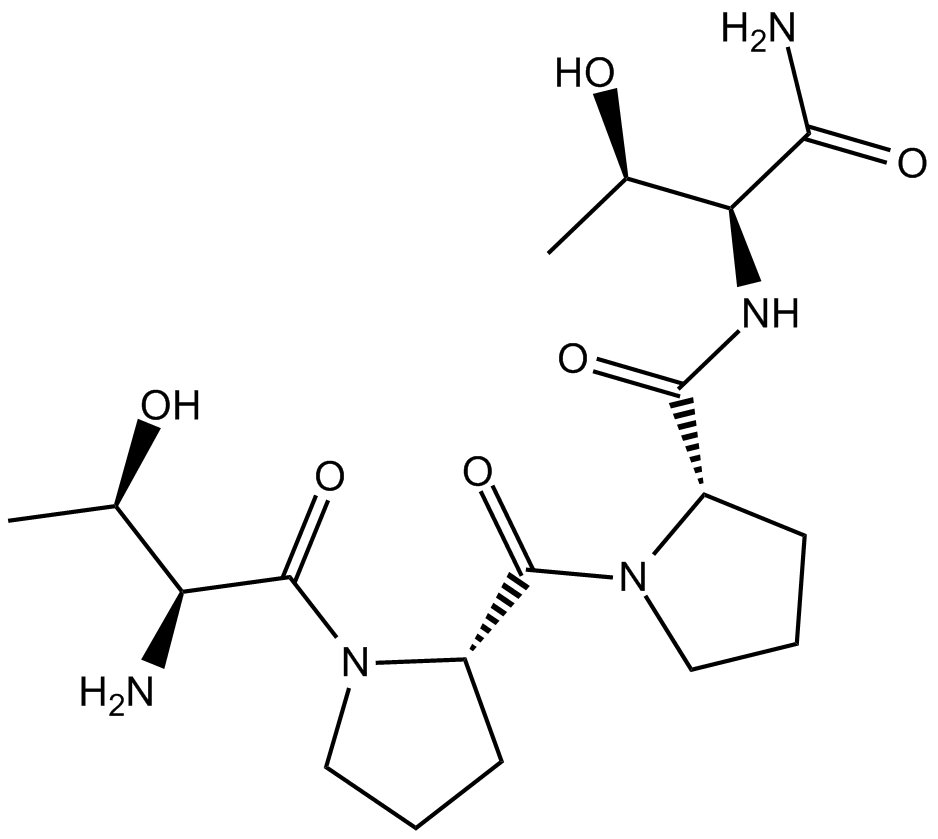
GC14121
GLYX 13
GLYX 13 (GLYX-13) is an N-methyl-D-aspartate receptor (NMDAR) modulator that has characteristics of a glycine site partial agonist.
-
GC31270
GNE 0723
GNE 0723 is a brain permeable positive allosteric modulator of NMDAR, with an EC50 of 21 nM for GluN2A, 7.4 and 6.2 μM for GluN2C and GluN2D, respectively.
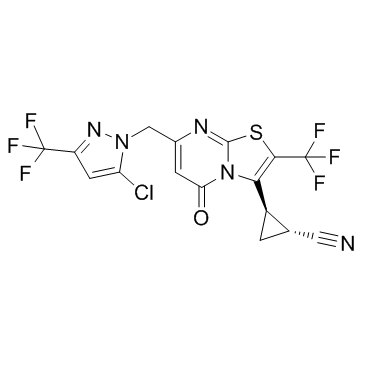
-
GC31076
GNE 5729
GNE 5729 is a brain permeable positive allosteric modulator of NMDAR, with an EC50 of 37 nM for GluN2A, 4.7 and 9.5 μM for GluN2C and GluN2D, respectively.
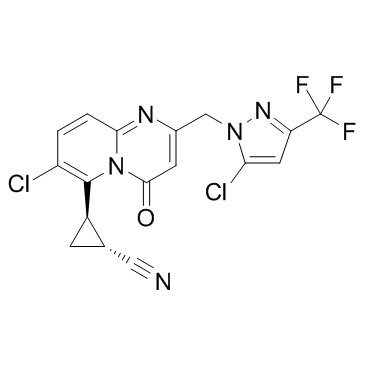
-
GC31019
GV-196771A
GV-196771A is the sodium salt form of GV196771, is an NMDA receptor antagonist.
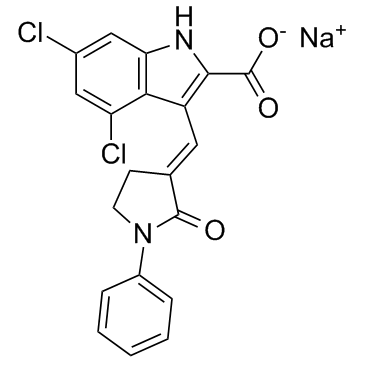
-
GC14842
GYKI 53655 hydrochloride
AMPA and kainate receptor antagonist
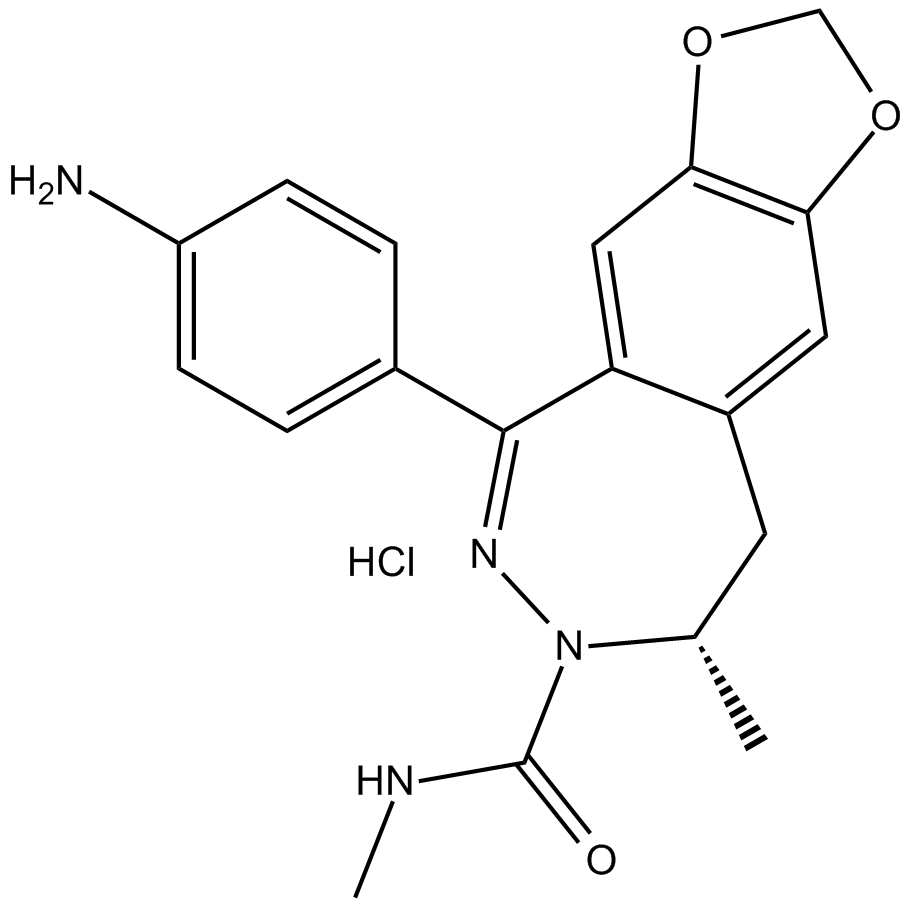
-
GC11715
Ibotenic acid
NMDA and metabotropic glutamate receptor agonist

-
GC30847
IC87201
An inhibitor of the nNOS-PSD-95 protein-protein interaction
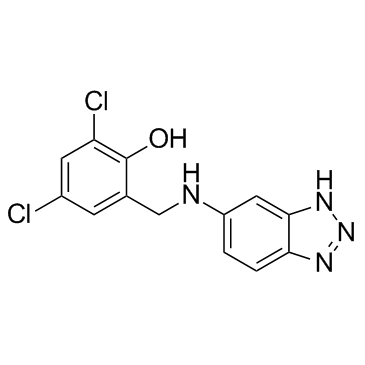
-
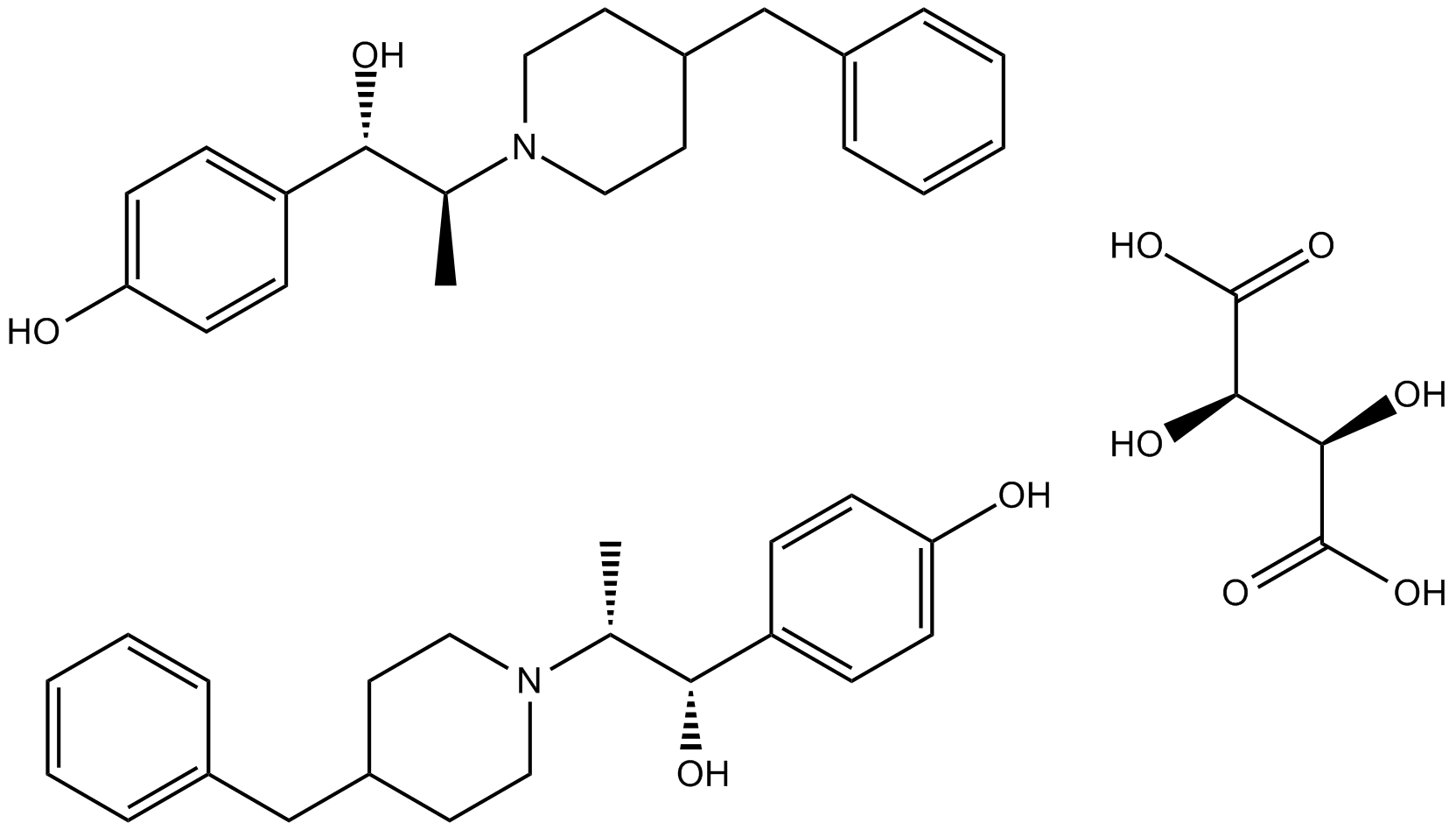
GC12521
Ifenprodil Tartrate
NMDA receptor antagonist
-
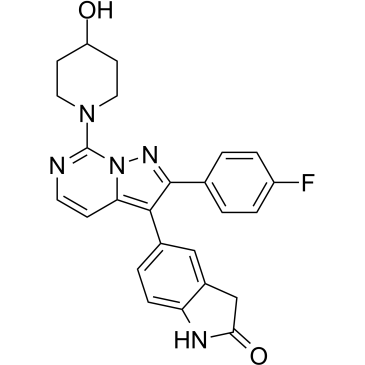
GC34638
JNJ-61432059
JNJ-61432059 is an oral active and selective negative modulator of AMPAR associated with trans-membrane AMPAR regulatory protein (TARP) γ-8, with a pIC50 of 9.7 for GluA1/γ-8.
-
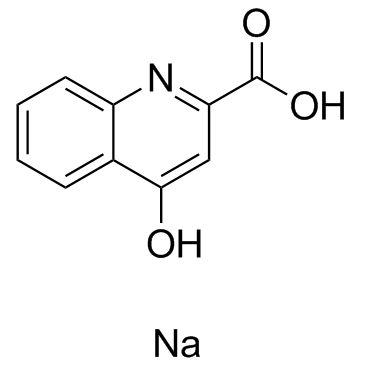
GC36411
Kynurenic acid sodium
-
GC14810
L-701,324
NMDA receptor antagonist
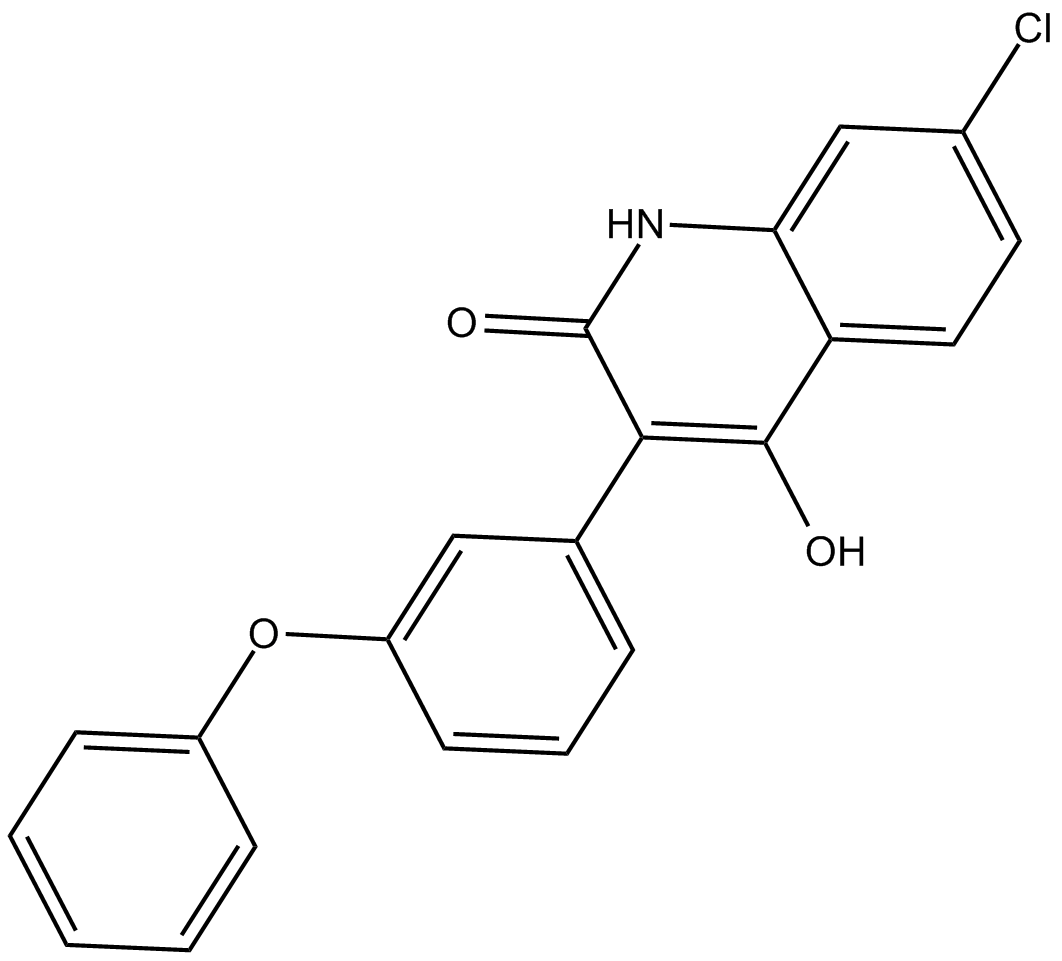
-
GC30788
L-Glutamic acid monosodium salt (Monosodium glutamate)
L-Glutamic acid monosodium salt (Monosodium glutamate) acts as an excitatory transmitter and an agonist at all subtypes of glutamate receptors (metabotropic, kainate, NMDA, and AMPA).
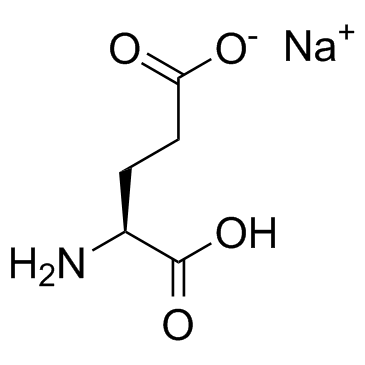
-
GC64352
L-Glutamic acid-15N
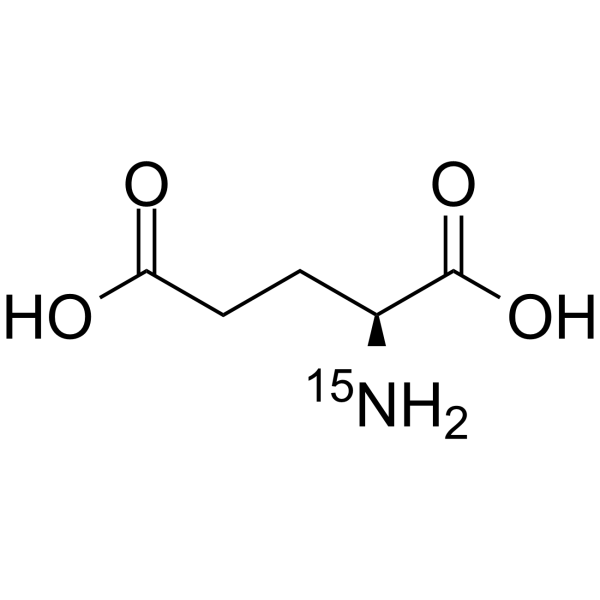
-
GC65095
L-Glutamic acid-d5
L-Glutamic acid-d5 is the deuterium labeled L-Glutamic acid.
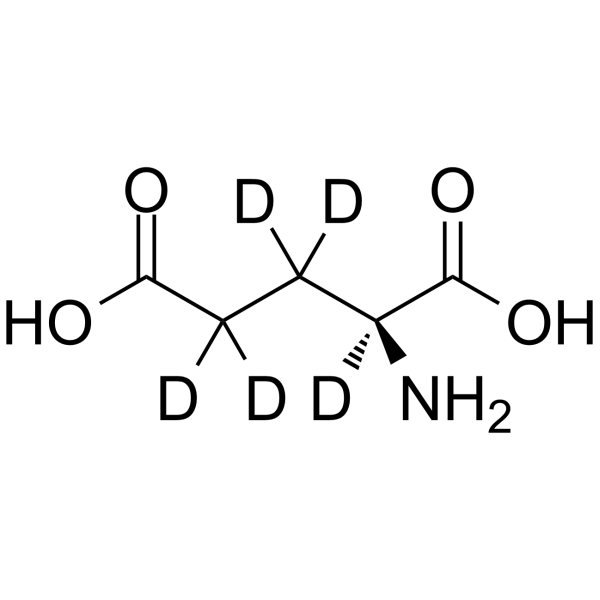
-
GC66843
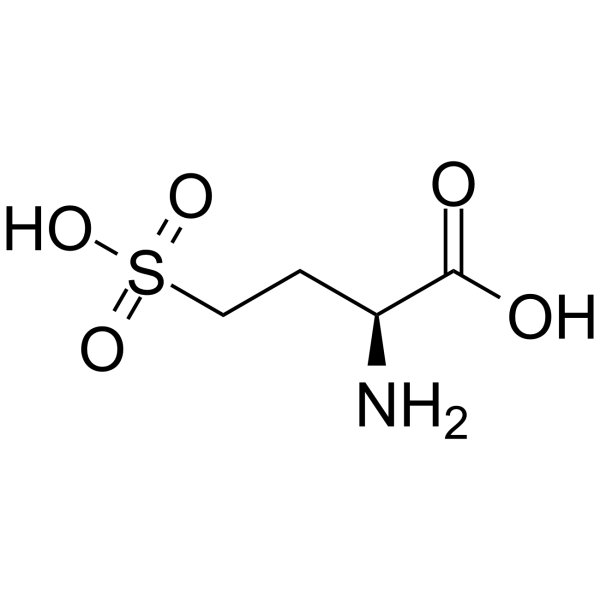
L-Homocysteic acid
L-Homocysteic acid (L-HCA) is an endogenous excitatory amino acid that acts as a NMDA receptor agonist (EC50: 14 μM). L-Homocysteic acid is neurotoxic, and can be used in the research of neurological disorders.
-
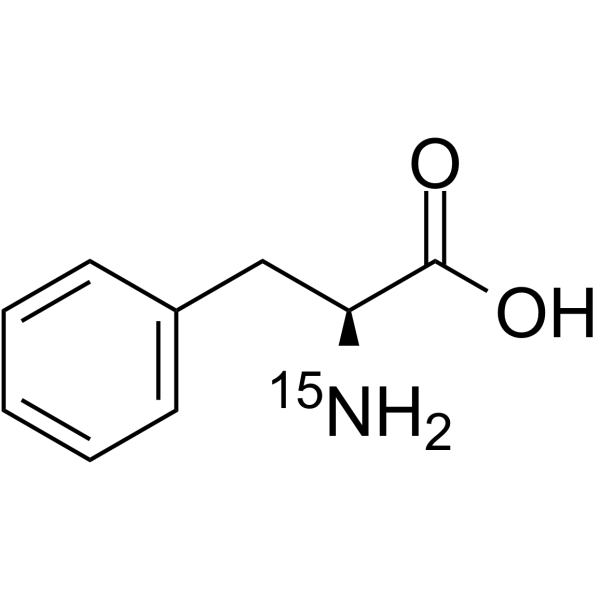
GC64148
L-Phenylalanine-15N
-
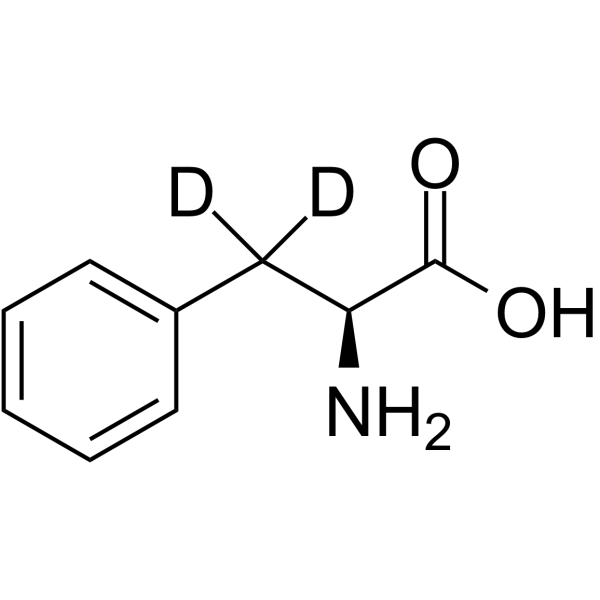
GC65050
L-Phenylalanine-d2
-
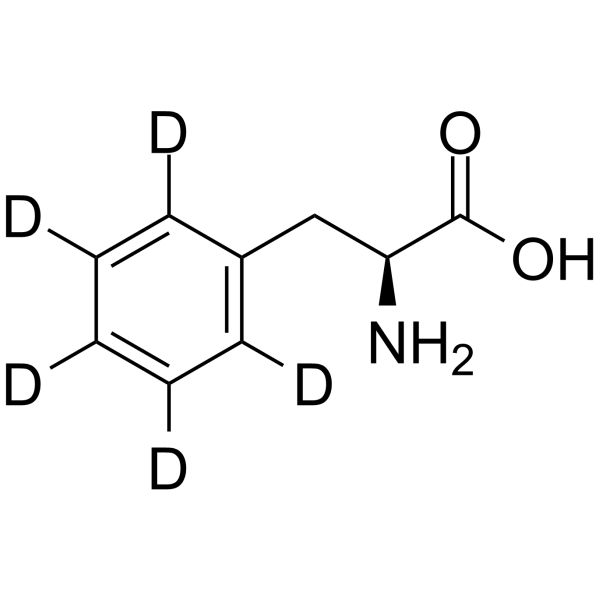
GC63982
L-Phenylalanine-d5
-
GC64290
L-Phenylalanine-d8

-
GC11124
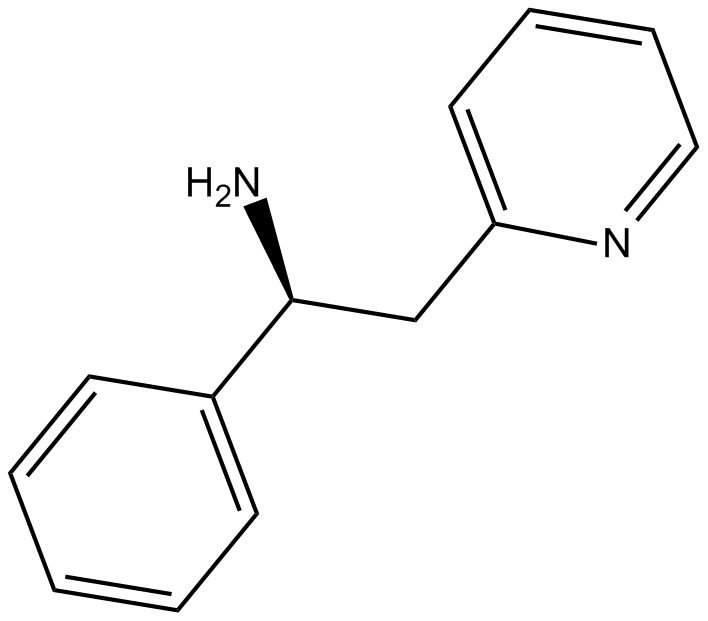
Lanicemine
voltage-dependent NMDA channel blocker
-
GC36510
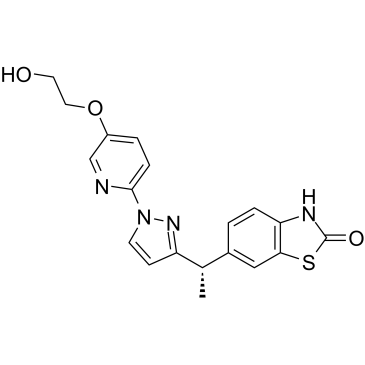
LY3130481
LY3130481 is an AMPA receptor antagonist that is dependent upon transmembrane AMPA receptor regulatory protein (TARP) γ-8, selective inhibits AMPA/TARP γ-8 with an IC50 of 65 nM.
-
GC36515
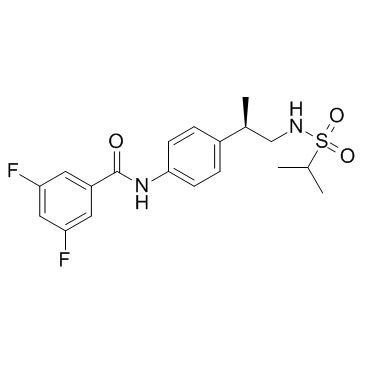
LY450108
LY450108 is a potent AMPA receptor potentiator.
-
GC13010
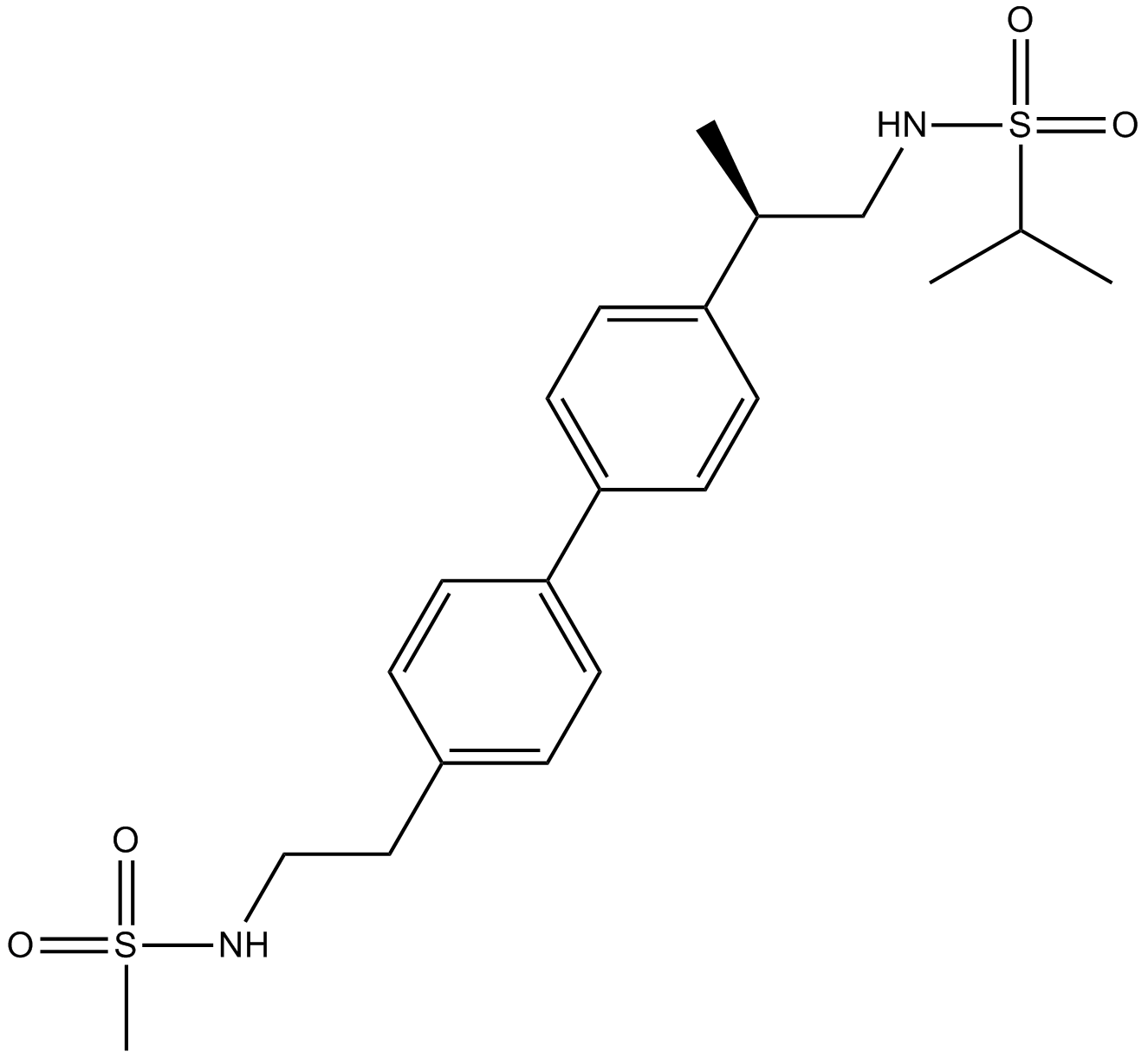
LY451395
LY451395 (LY451395) is a potent and highly selective potentiator of the AMPA receptors.
-
GC30951
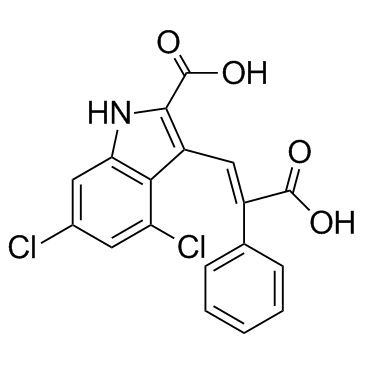
MDL 105519
MDL 105519 is a potent and selective antagonist of glycine binding to the NMDA receptor.
-
GC14039
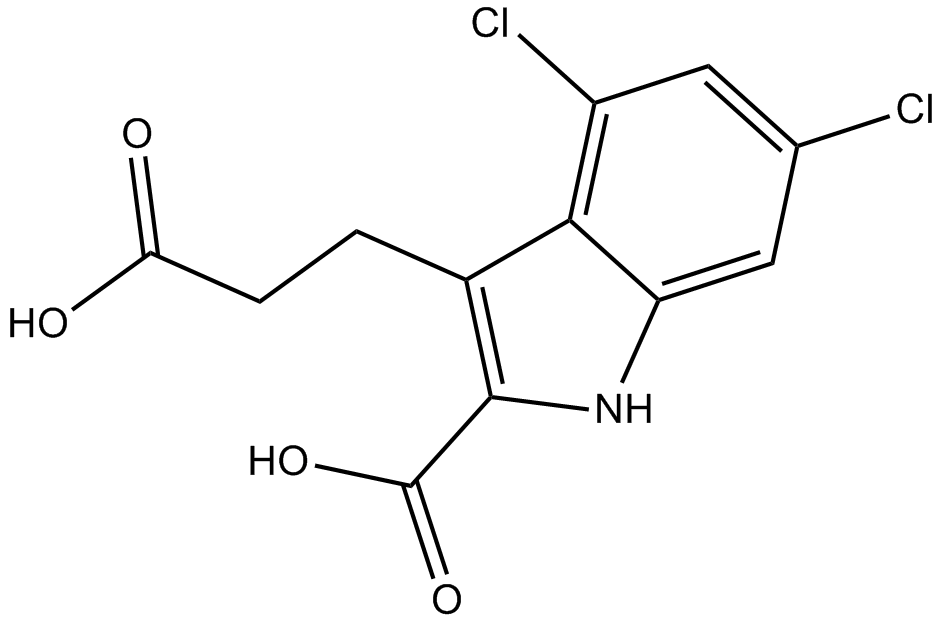
MDL-29951
Glycine antagonist of NMDA receptor activation
-
GC10443
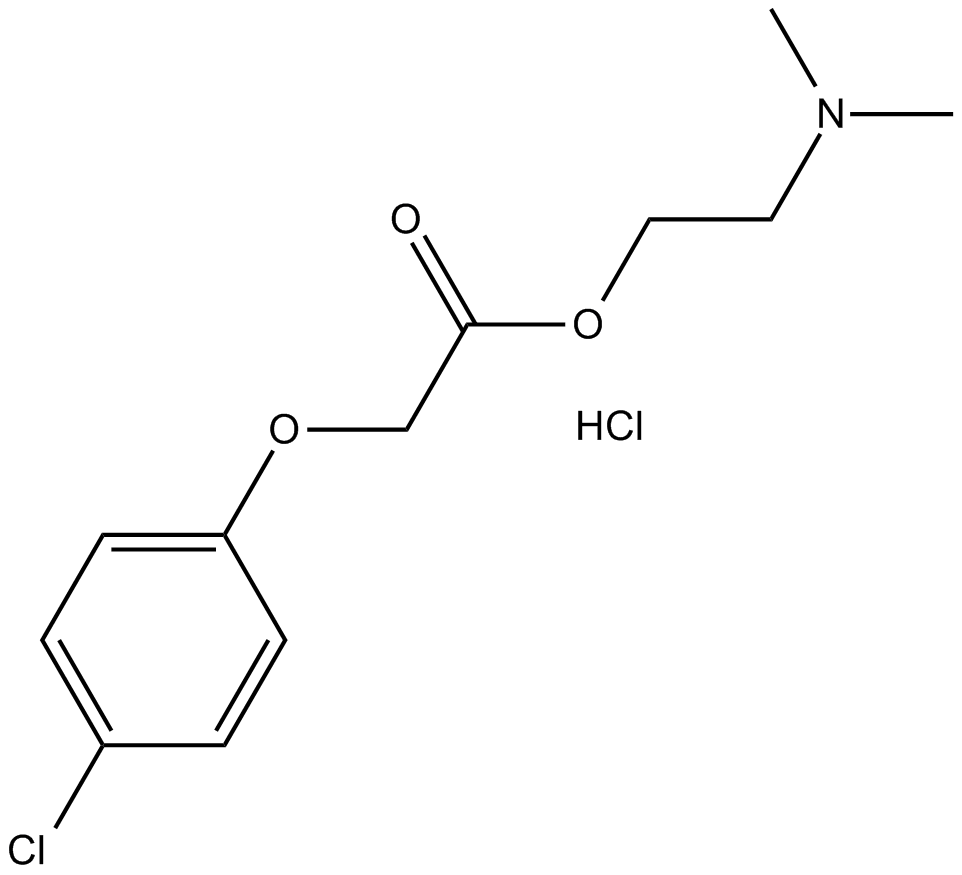
Meclofenoxate hydrochloride
Drug for senile dementia and AD treatment
-
GC10198
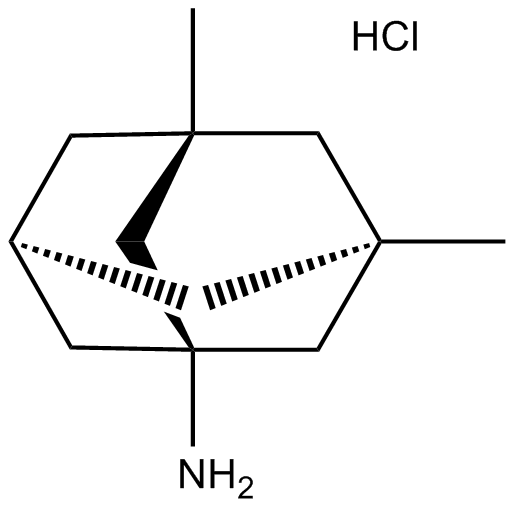
Memantine hydrochloride
NMDA receptor antagonist
-
GC30973
Mephenesin
Mephenesin is an NMDA receptor antagonist, is a centrally acting muscle relaxant.
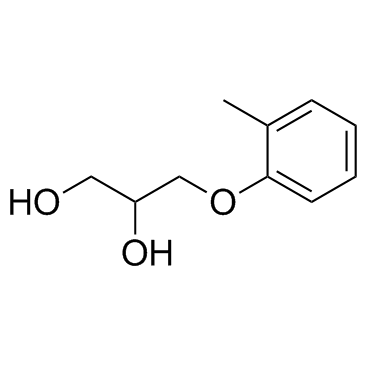
-
GC31036
MRZ 2-514
MRZ 2-514 is an antagonist of the strychnine-insensitive modulatory site of the NMDA receptor (glycineB), with Ki of 33 μM.
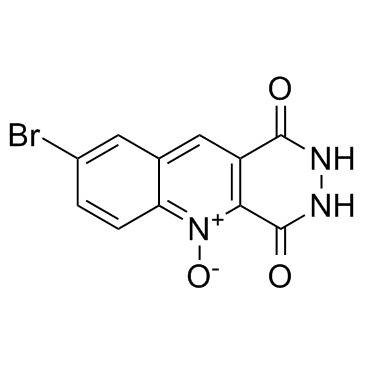
-
GC33668
Naspm (1-Naphthylacetyl spermine)
Naspm (1-Naphthylacetyl spermine) (1-Naphthyl acetyl spermine), a synthetic analogue of Joro spider toxin, is a calcium permeable AMPA (CP-AMPA) receptors antagonist.
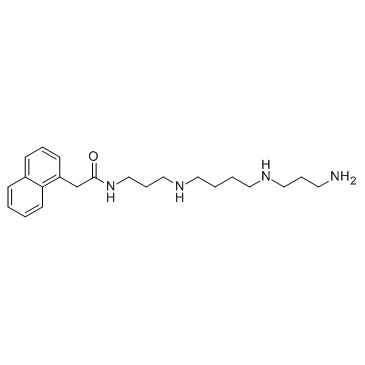
-
GC13349
Naspm trihydrochloride
Ca2+-permeable AMPA receptor antagonist
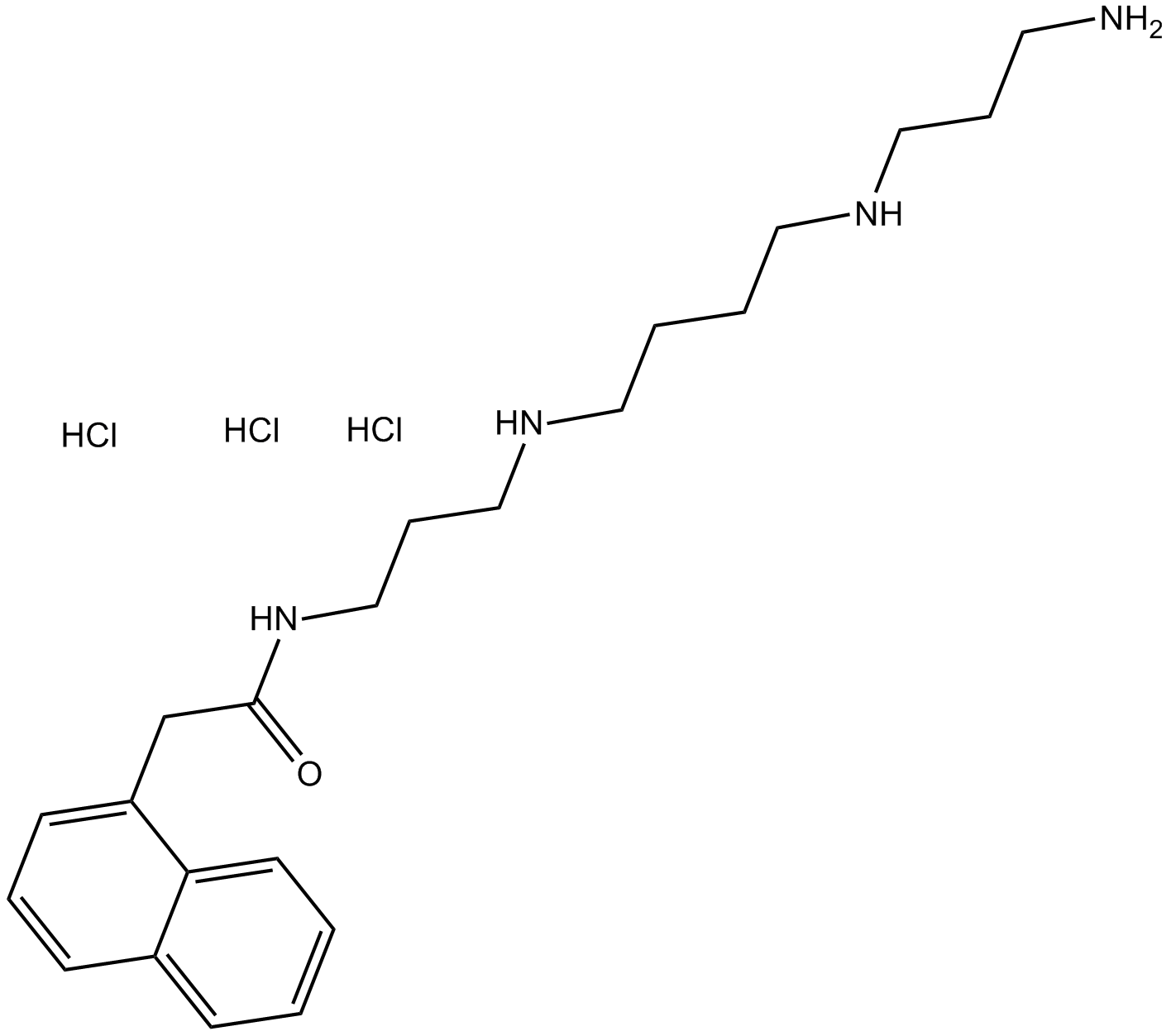
-
GC14156
NBQX
NBQX is a highly selective and competitive AMPA receptor antagonist.
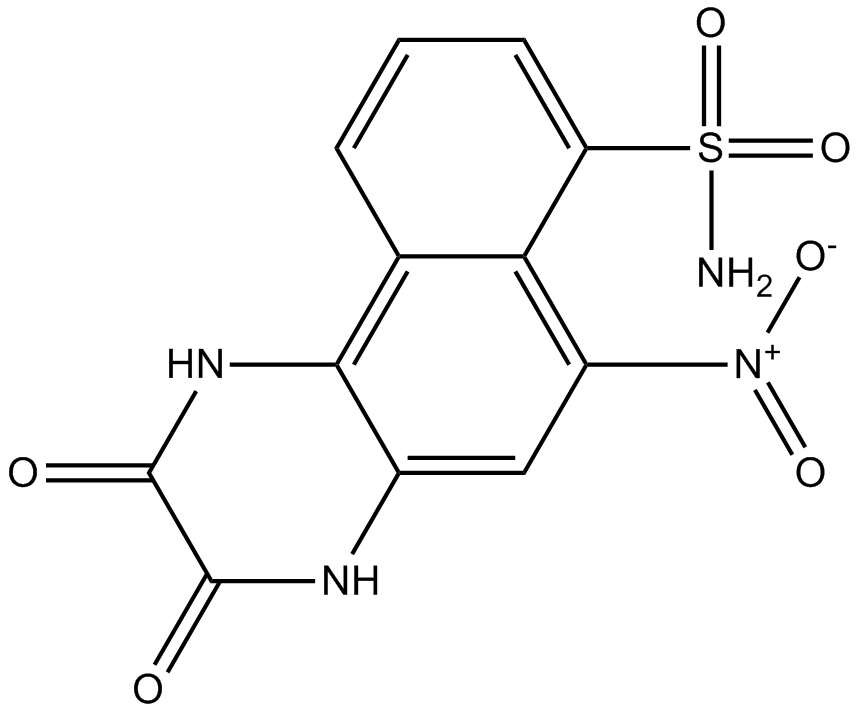
-
GC30860
Neu2000
Neu2000 (Salfaprodil free base) is an NR2B-selective and uncompetitive antagonist of N-methyl-D-aspartate (NMDA).
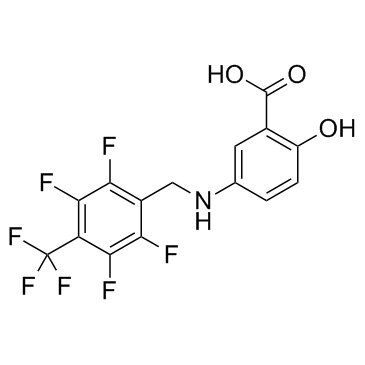
-
GC17456
NMDA (N-Methyl-D-aspartic acid)
NMDA (N-Methyl-D-aspartic acid) is a specific agonist forNMDA (N-Methyl-D-aspartic acid)receptor mimicking the action of glutamate, the neurotransmitter which normally acts at that receptor.
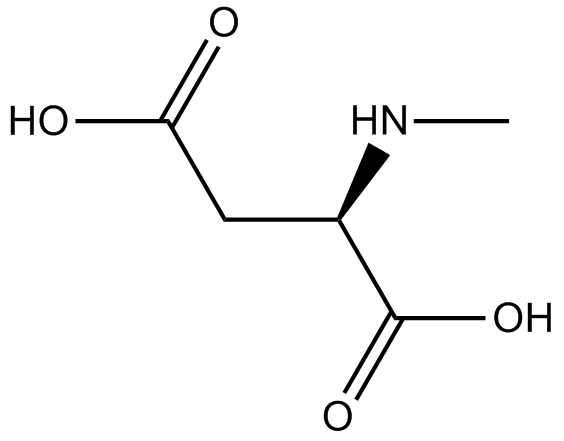
-
GC34352
NMDAR antagonist 1
NMDAR antagonist 1 is a potent and orally bioavailable NR2B-selective NMDAR antagonist.
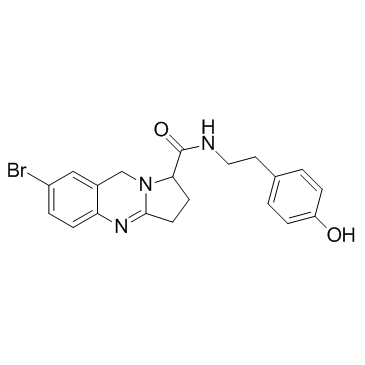
-
GC64521
NMDAR/TRPM4-IN-2 free base
NMDAR/TRPM4-IN-2 free base (compound 8) is a potent NMDAR/TRPM4 interaction interface inhibitor.
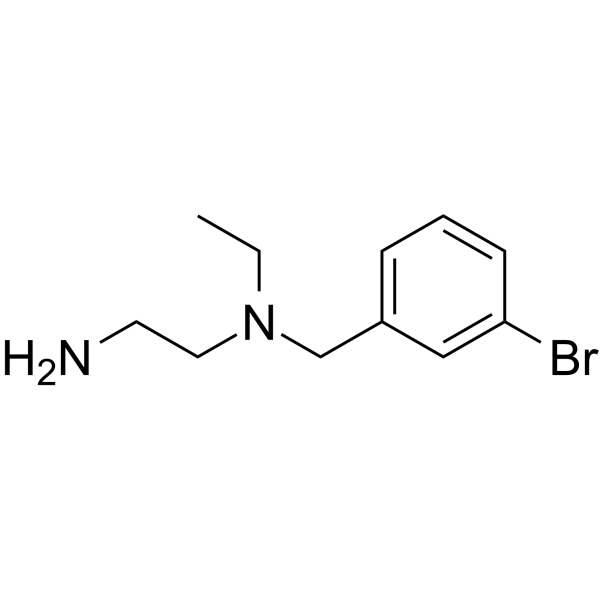
-
GC64133
NS-102
NS-102 is a selective kainate (GluK2) receptor antagonist.
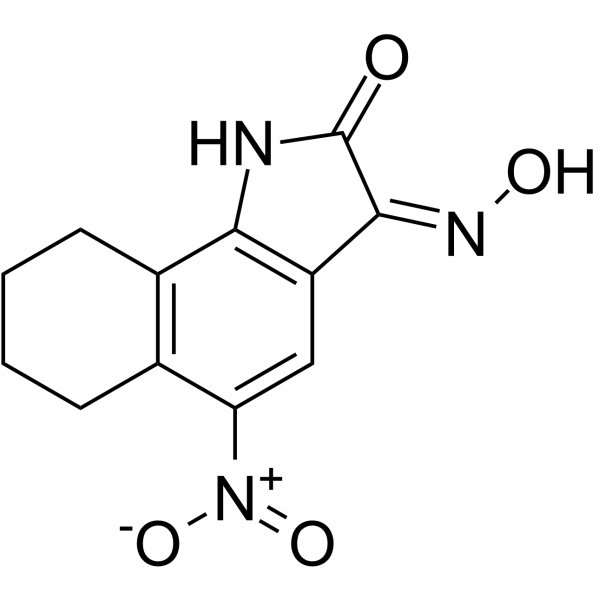
-
GC33740
NT 13 (TPPT)
NT 13 (TPPT) (TPPT) is a tetrapeptide having the amino acid sequence L-threonyl-L-prolyl-L-prolyl-L-threonine amide.
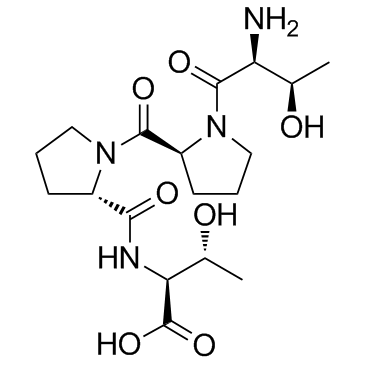
-
GC30852
Org-26576
Org-26576 is a AMPA receptor positive allosteric modulator.
-
GC13779
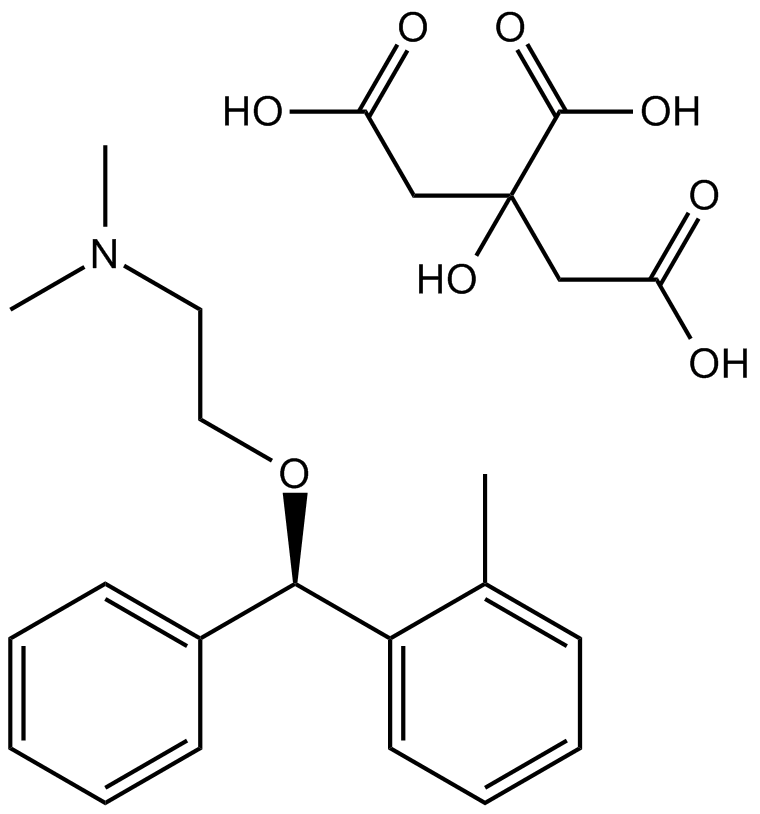
Orphenadrine Citrate
Antiparkinsonian and analgesic drug
-
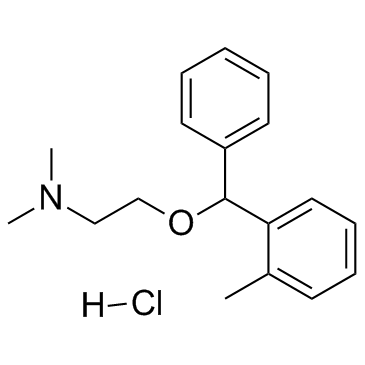
GC33701
Orphenadrine hydrochloride
A muscarinic acetylcholine receptor antagonist
-
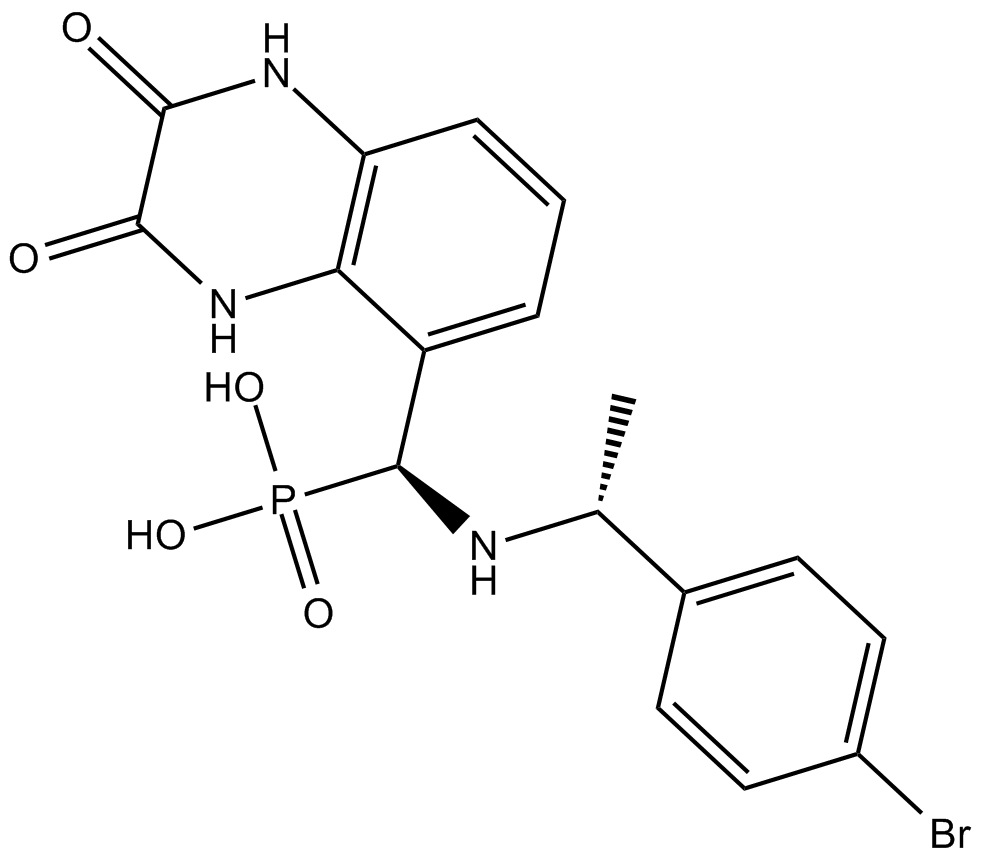
GC16562
PEAQX
NMDA antagonist
-
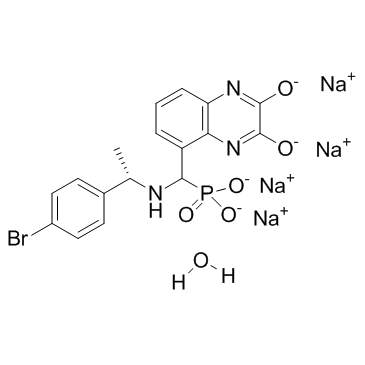
GC36865
PEAQX tetrasodium hydrate
PEAQX (NVP-AAM077) tetrasodium hydrate is a potent, selective and orally active NMDA antagonist, with IC50 values of 270 nM and 29600 nM for hNMDAR 1A/2B and hNMDAR 1A/2B, respectively.
-
GC13007
PEPA
Novel allosteric potentiator of AMPA receptor desensitization
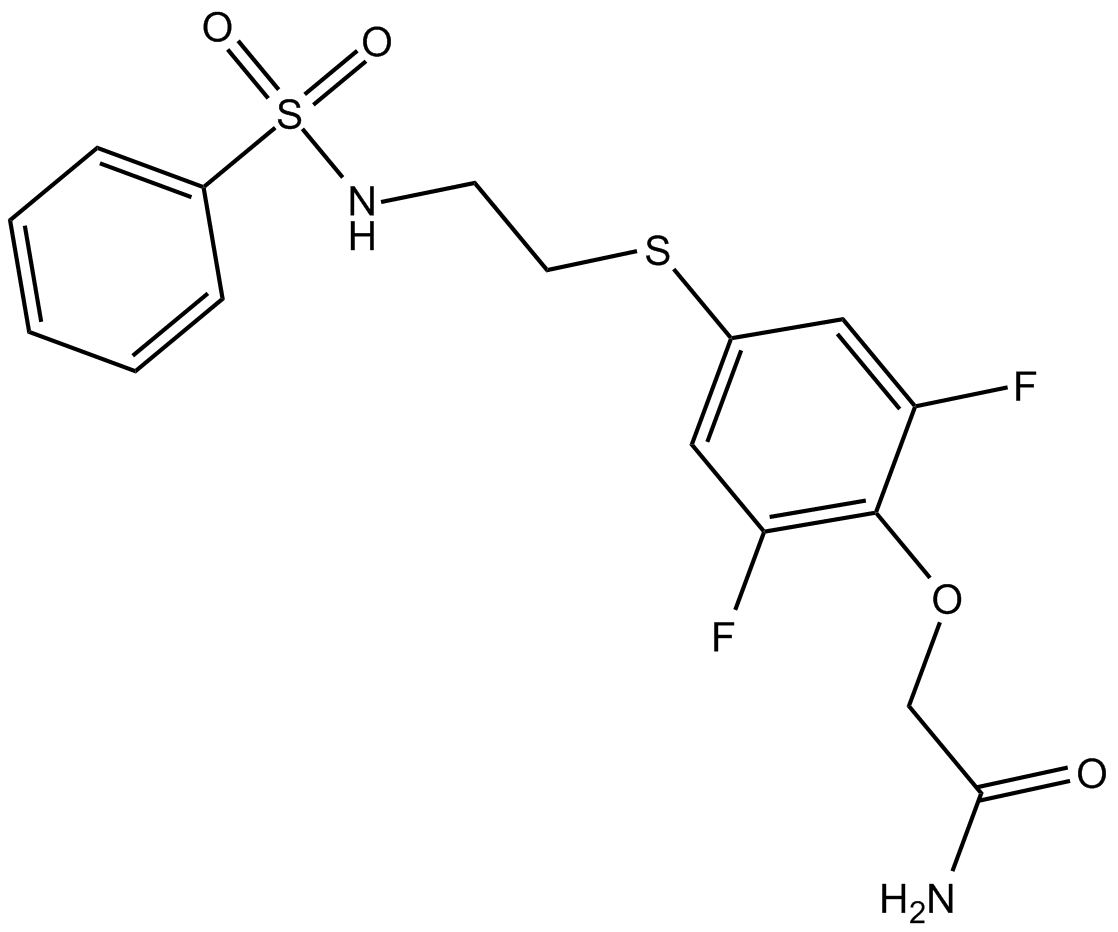
-
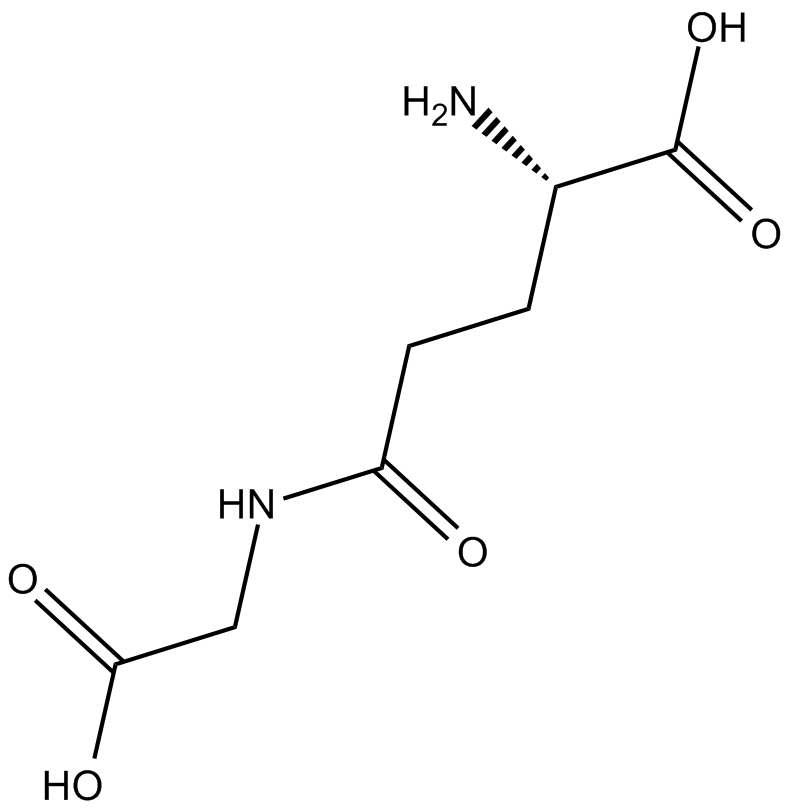
GC36876
Perzinfotel
Perzinfotel (EAA-090) is a potent, selective, and competitive NMDA receptor antagonist with neuroprotective effects.
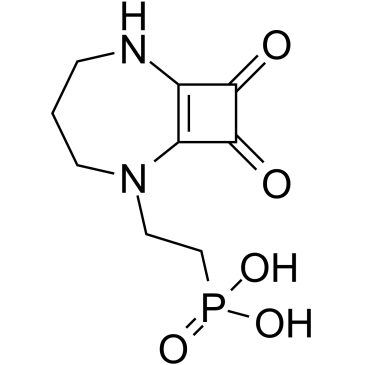
-
GC64929
Pesampator
Pesampator (PF-04958242) is a potent and highly selective positive allosteric modulator of AMPA receptor (an AMPA potentiator) with an EC50 of 310 nM and a Ki of 170 nM.
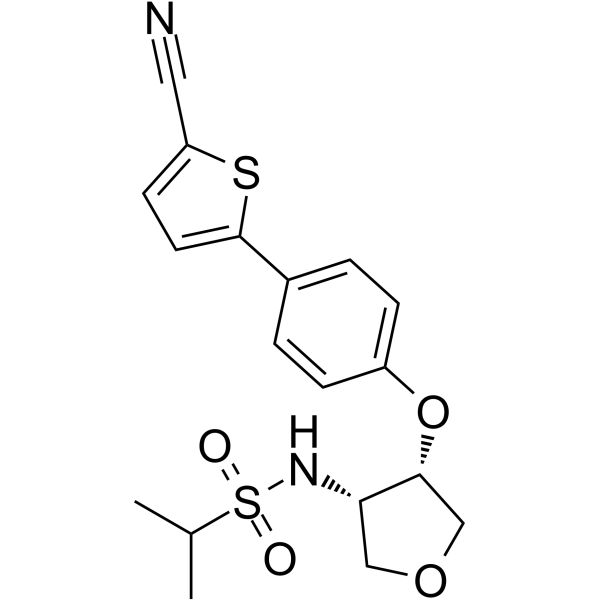
-
GC14492
PF 4778574
positive allosteric modulator of AMPA receptors

-
GC12703
Philanthotoxin 74
Philanthotoxin 74 (PhTx 74) is an AMPAR antagonist; inhibits GluR3 and GluR1 with IC50s of 263 and 296 nM, respectively.
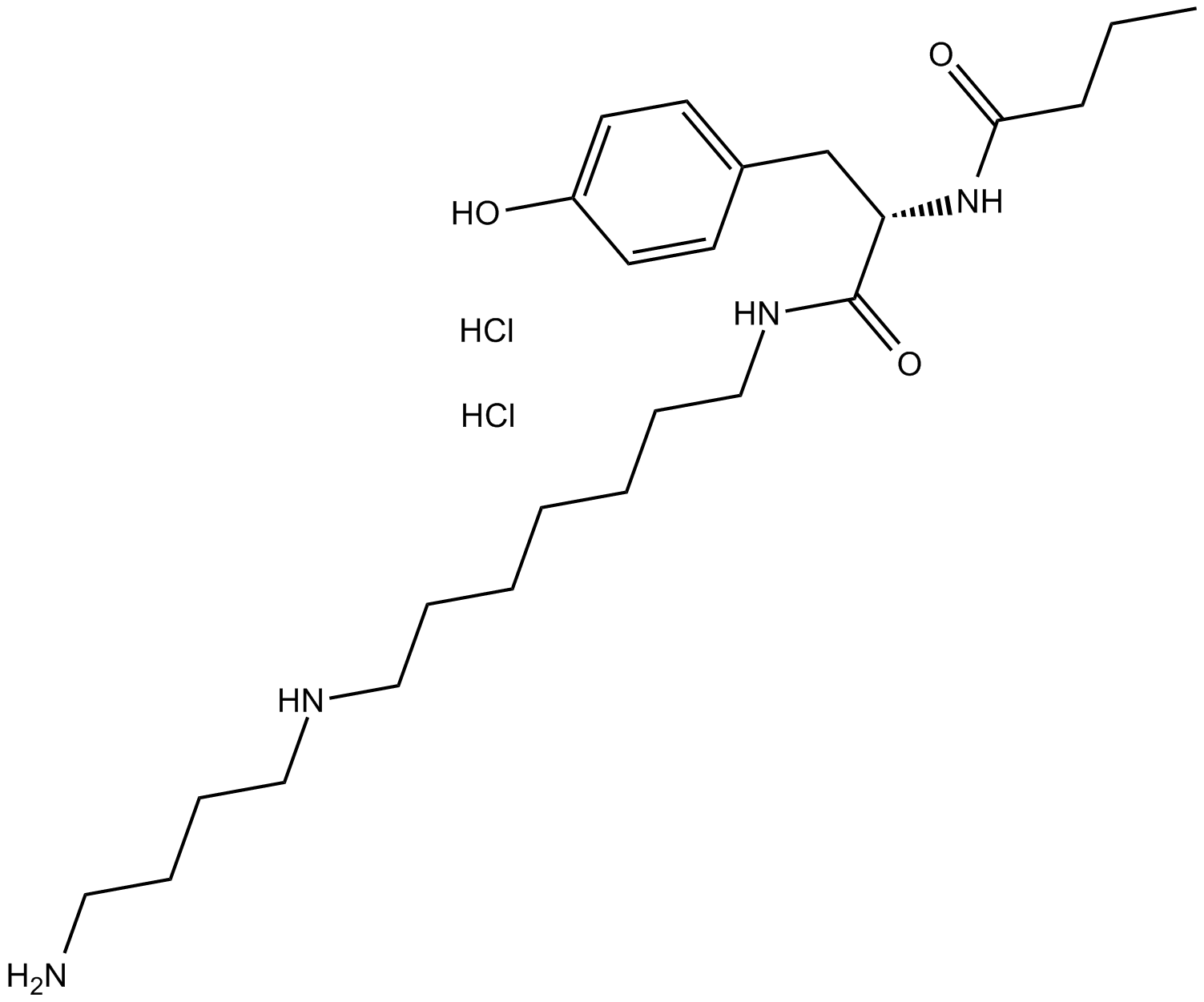
-
GC13469
Piracetam
cyclic derivative of the neurotransmitter gamma-aminobutyric acid (GABA)
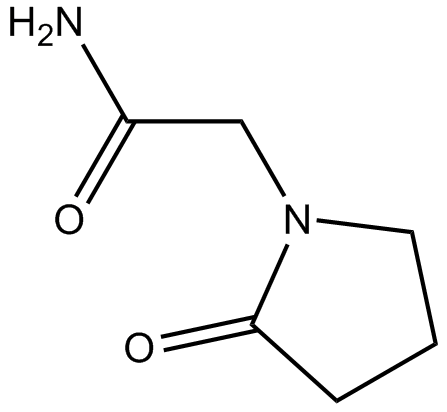
-
GC65954
Plazinemdor
Plazinemdor is a N-methyl-D-aspartate(NMDA) receptor positive allosteric modulator. Plazinemdor can be uses in the research of psychiatric, neurological, and neurodevelopmental disorders, as well as diseases of the nervous system..
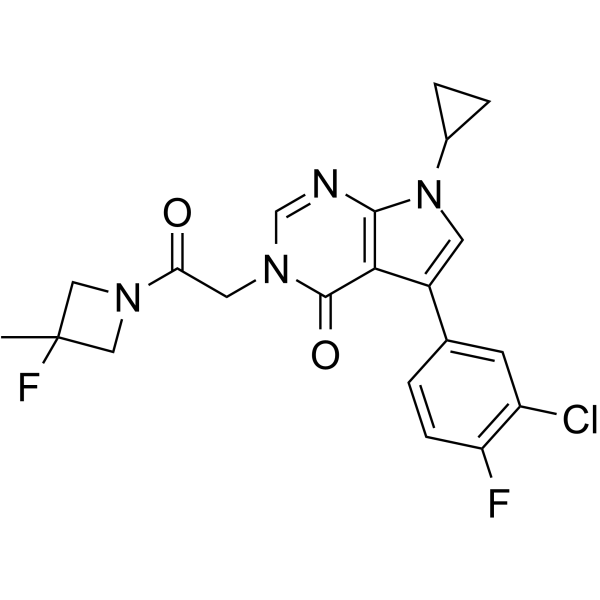
-
GC30946
Procyclidine hydrochloride ((±)-Procyclidine hydrochlorid)
Procyclidine (Tricyclamol, (±)-Procyclidine) hydrochloride , an anticholinergic agent, is a muscarinic receptor antagonist that also has the properties of an N-methyl-D-aspartate (NMDA) antagonist.
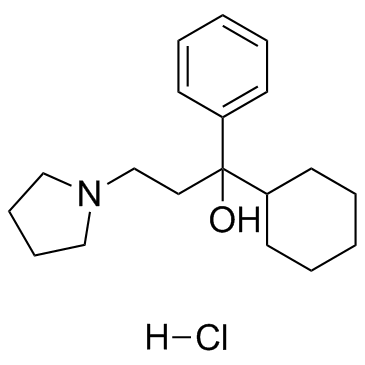
-
GC16776
QNZ 46
NR2C/NR2D-selective NMDA receptor antagonist
-
GC14547
Quinolinic acid
NMDA agonist
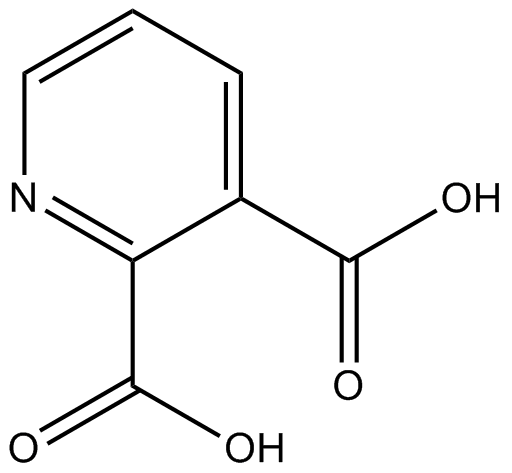
-
GC34753
Radiprodil
An NMDA receptor antagonist
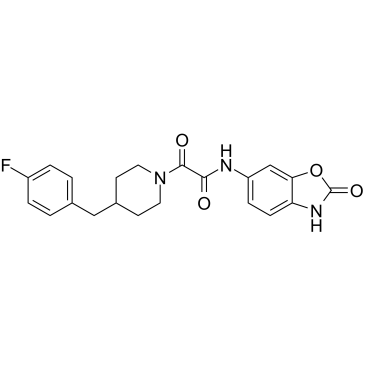
-
GC33492
Rapastinel Trifluoroacetate (GLYX-13 Trifluoroacetate)
Rapastinel Trifluoroacetate (GLYX-13 Trifluoroacetate) (GLYX-13 Trifluoroacetate) is an NMDA receptor modulator with glycine-site partial agonist properties.
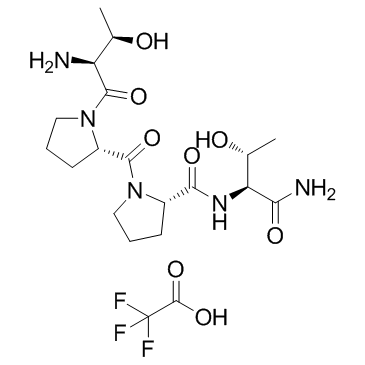
-
GC30835
Rislenemdaz (MK-0657)
Rislenemdaz (MK-0657) (CERC-301) is an orally bioavailable and selective N-methyl-D-aspartate (NMDA) receptor subunit 2B (GluN2B) antagonist with Ki and IC 50 of 8.1 nM and 3.6 nM, respectively.
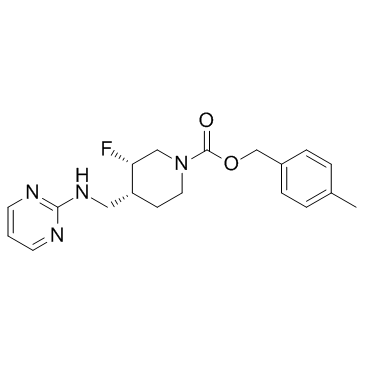
-
GC19310
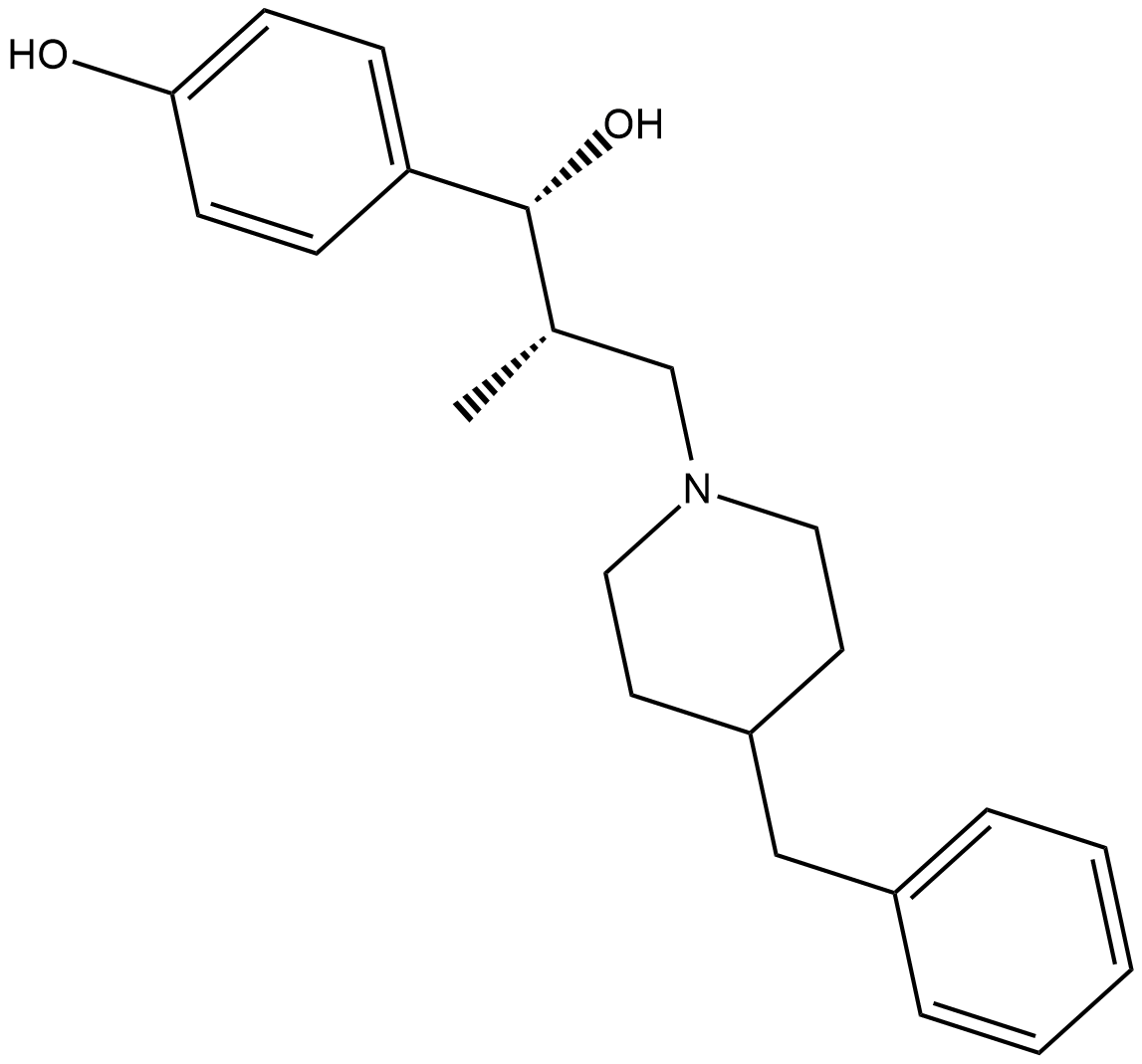
Ro 25-6981
Ro 25-6981 is a potent and selective activity-dependent blocker of NMDA receptors containing the NR2B subunit.
-
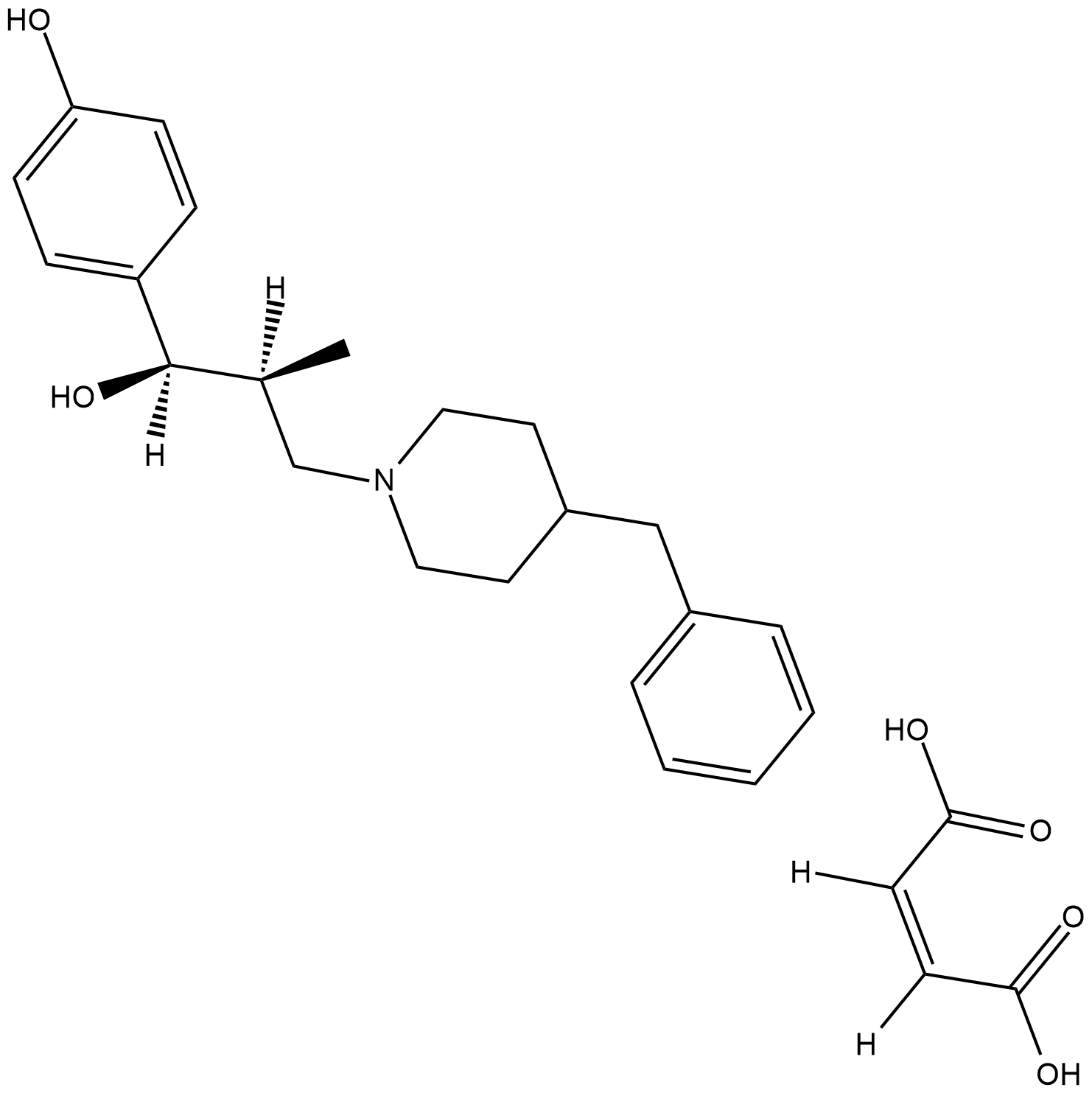
GC18410
Ro 25-6981 (maleate)
Ro 25-6981 (maleate) is a potent, selective activity-dependent blocker of NMDA receptors containing the NR2B subunit (IC50s = 9 nM and 52 μM for cloned receptor subunit combinations NR1C/NR2B and NR1C/NR2A, respectively).
-
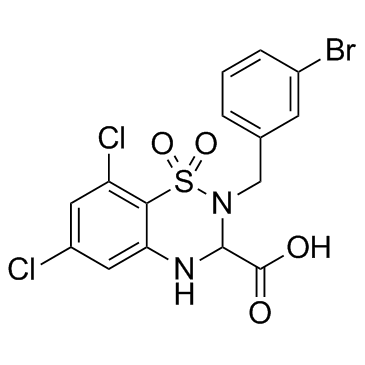
GC31039
RPR104632
RPR104632 is a specific antagonist of NMDA receptor, with potent neuroprotective properties.
-
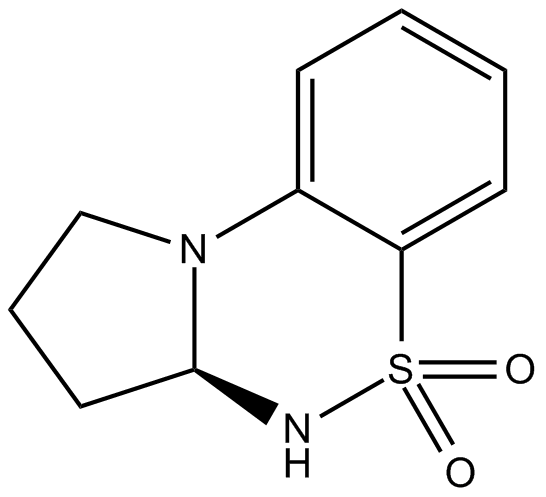
GC13684
S 18986
positive allosteric modulator of AMPA receptors
-
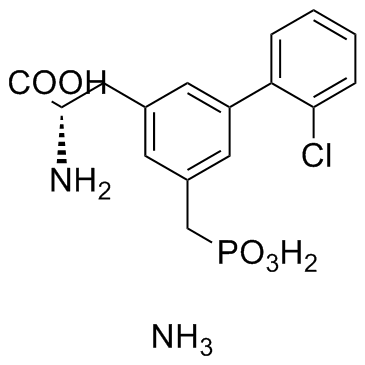
GC37612
SDZ 220-581 Ammonium salt
SDZ 220-581 Ammonium salt is an orally active, potent, competitive NMDA receptor antagonist with pKi value of 7.7.