Immunology/Inflammation
The immune and inflammation-related pathway including the Toll-like receptors pathway, the B cell receptor signaling pathway, the T cell receptor signaling pathway, etc.
Toll-like receptors (TLRs) play a central role in host cell recognition and responses to microbial pathogens. TLR4 initially recruits TIRAP and MyD88. MyD88 then recruits IRAKs, TRAF6, and the TAK1 complex, leading to early-stage activation of NF-κB and MAP kinases [1]. TLR4 is endocytosed and delivered to intracellular vesicles and forms a complex with TRAM and TRIF, which then recruits TRAF3 and the protein kinases TBK1 and IKKi. TBK1 and IKKi catalyze the phosphorylation of IRF3, leading to the expression of type I IFN [2].
BCR signaling is initiated through ligation of mIg under conditions that induce phosphorylation of the ITAMs in CD79, leading to the activation of Syk. Once Syk is activated, the BCR signal is transmitted via a series of proteins associated with the adaptor protein B-cell linker (Blnk, SLP-65). Blnk binds CD79a via non-ITAM tyrosines and is phosphorylated by Syk. Phospho-Blnk acts as a scaffold for the assembly of the other components, including Bruton’s tyrosine kinase (Btk), Vav 1, and phospholipase C-gamma 2 (PLCγ2) [3]. Following the assembly of the BCR-signalosome, GRB2 binds and activates the Ras-guanine exchange factor SOS, which in turn activates the small GTPase RAS. The original RAS signal is transmitted and amplified through the mitogen-activated protein kinase (MAPK) pathway, which including the serine/threonine-specific protein kinase RAF followed by MEK and extracellular signal related kinases ERK 1 and 2 [4]. After stimulation of BCR, CD19 is phosphorylated by Lyn. Phosphorylated CD19 activates PI3K by binding to the p85 subunit of PI3K and produce phosphatidylinositol-3,4,5-trisphosphate (PIP3) from PIP2, and PIP3 transmits signals downstream [5].
Central process of T cells responding to specific antigens is the binding of the T-cell receptor (TCR) to specific peptides bound to the major histocompatibility complex which expressed on antigen-presenting cells (APCs). Once TCR connected with its ligand, the ζ-chain–associated protein kinase 70 molecules (Zap-70) are recruited to the TCR-CD3 site and activated, resulting in an initiation of several signaling cascades. Once stimulation, Zap-70 forms complexes with several molecules including SLP-76; and a sequential protein kinase cascade is initiated, consisting of MAP kinase kinase kinase (MAP3K), MAP kinase kinase (MAPKK), and MAP kinase (MAPK) [6]. Two MAPK kinases, MKK4 and MKK7, have been reported to be the primary activators of JNK. MKK3, MKK4, and MKK6 are activators of P38 MAP kinase [7]. MAP kinase pathways are major pathways induced by TCR stimulation, and they play a key role in T-cell responses.
Phosphoinositide 3-kinase (PI3K) binds to the cytosolic domain of CD28, leading to conversion of PIP2 to PIP3, activation of PKB (Akt) and phosphoinositide-dependent kinase 1 (PDK1), and subsequent signaling transduction [8].
References
[1] Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors[J]. Nature immunology, 2010, 11(5): 373-384.
[2] Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-650.
[3] Packard T A, Cambier J C. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation[J]. F1000Prime Rep, 2013, 5(40.10): 12703.
[4] Zhong Y, Byrd J C, Dubovsky J A. The B-cell receptor pathway: a critical component of healthy and malignant immune biology[C]//Seminars in hematology. WB Saunders, 2014, 51(3): 206-218.
[5] Baba Y, Matsumoto M, Kurosaki T. Calcium signaling in B cells: regulation of cytosolic Ca 2+ increase and its sensor molecules, STIM1 and STIM2[J]. Molecular immunology, 2014, 62(2): 339-343.
[6] Adachi K, Davis M M. T-cell receptor ligation induces distinct signaling pathways in naive vs. antigen-experienced T cells[J]. Proceedings of the National Academy of Sciences, 2011, 108(4): 1549-1554.
[7] Rincón M, Flavell R A, Davis R A. The Jnk and P38 MAP kinase signaling pathways in T cell–mediated immune responses[J]. Free Radical Biology and Medicine, 2000, 28(9): 1328-1337.
[8] Bashour K T, Gondarenko A, Chen H, et al. CD28 and CD3 have complementary roles in T-cell traction forces[J]. Proceedings of the National Academy of Sciences, 2014, 111(6): 2241-2246.
Targets for Immunology/Inflammation
- Apoptosis(303)
- 5-Lipoxygenase(18)
- Cyclic GMP-AMP Synthase(2)
- TLR(98)
- Papain(1)
- PGDS(1)
- PGE synthase(24)
- SIKs(10)
- IκB/IKK(62)
- AP-1(3)
- KEAP1-Nrf2(42)
- NOD1(1)
- NF-κB(240)
- Interleukin Related(147)
- 15-lipoxygenase(2)
- Others(10)
- Aryl Hydrocarbon Receptor(33)
- CD73(16)
- Complement System(52)
- Galectin(31)
- IFNAR(21)
- NO Synthase(75)
- NOD-like Receptor (NLR)(46)
- STING(101)
- Reactive Oxygen Species(446)
- FKBP(11)
- eNOS(5)
- iNOS(28)
- nNOS(20)
- Glutathione(54)
- Adaptive Immunity(216)
- Allergy(161)
- Arthritis(32)
- Autoimmunity(185)
- Gastric Disease(96)
- Immunosuppressants(37)
- Immunotherapeutics(5)
- Innate Immunity(579)
- Pulmonary Diseases(119)
- Reactive Nitrogen Species(54)
- Specialized Pro-Resolving Mediators(50)
- Reactive Sulfur Species(26)
- BCL6(0)
- CD20(0)
- CD22(0)
- CD28(0)
- PSMA(0)
- FAP(0)
Products for Immunology/Inflammation
- Cat.No. Product Name Information
-
GC40202
7α-hydroxy Cholesterol-d7
7α-hydroxycholesterol-d7
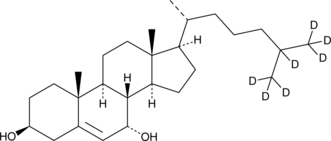
7α-hydroxy Cholesterol-d7 is intended for use as an internal standard for the quantification of 7α-hydroxy cholesterol by GC- or LC-MS.
-
GC49206
7α-hydroxy Dehydroepiandrosterone
7α-hydroxy DHEA
An active metabolite of dehydroepiandrosterone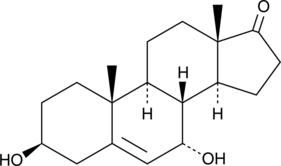
-
GC46740
7β,27-dihydroxy Cholesterol
7β,27-DHC
An oxysterol and agonist of RORγ and RORγt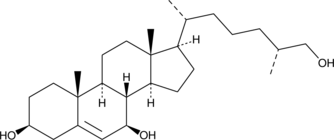
-
GC52471
7(S),10(S)-DiHOME
DHOE, DOD
An antibacterial hydroxy fatty acid
-
GC46732
7(S),17(S)-dihydroxy-8(E),10(Z),13(Z),15(E),19(Z)-Docosapentaenoic Acid
7(S),17(S)hydroxy DPA
A metabolite of DPA with antiinflammatory properties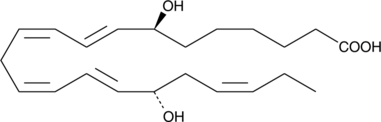
-
GC48769
7,10-dihydroxy-8(E)-Octadecenoic Acid
An antibacterial hydroxy fatty acid
-
GC46733
7,12-Dimethylbenz[a]anthracene
DMBA
7,12-Dimethylbenz[a]anthracene has carcinogenic activity as a polycyclic aromatic hydrocarbon (PAH). 7,12-Dimethylbenz[a]anthracene is used to induce tumor formation in various rodent models.![7,12-Dimethylbenz[a]anthracene Chemical Structure 7,12-Dimethylbenz[a]anthracene Chemical Structure](/media/struct/GC4/GC46733.png)
-
GC35184
7,3',4'-Tri-O-methylluteolin
7,3',4'-Tri-O-methylluteolin (5-Hydroxy-3',4',7-trimethoxyflavone), a flavonoid compound, possesses potent anti-inflammatory effects in LPS-induced macrophage cell line mediated by inhibition of release of inflammatory mediators, NO, PGE2, and pro-inflammatory cytokines.
-
GC46080
7,3',4'-Trihydroxyflavone
3’,4’,7-Trihydroxyflavone, 5-Deoxyluteolin
7,3',4'-Trihydroxyflavone is a flavonoid aglycon compound isolated from broad bean pods.
-
GC35185
7,4'-Dihydroxyflavone
7,4'-DHF
7,4'-Dihydroxyflavone (7,4'-DHF) is a flavonoid isolated from Glycyrrhiza uralensis, the eotaxin/CCL11 inhibitor, has the ability to consistently suppress eotaxin production and prevent dexamethasone (Dex)‐paradoxical adverse effects on eotaxin production.
-
GC45673
7,8-Dihydroneopterin
D-erythro-7,8-Dihydroneopterin
An antioxidant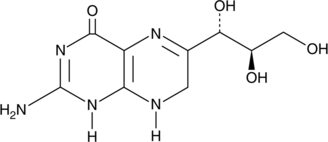
-
GC49887
7-(β-Hydroxyethyl)theophylline-d6
Etophylline-d6, Hydroxyethyltheophylline-d6
An internal standard for the quantification of 7-(β-hydroxyethyl)theophylline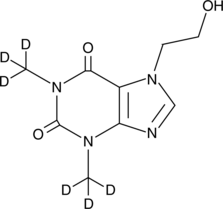
-
GC45707
7-Azido-4-methylcoumarin
AzMC
A fluorescent H2S probe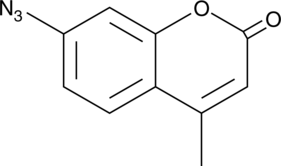
-
GC40978
7-epi Maresin 1
7(S)MaR1, 7(S)Maresin 1
7-epi Maresin 1 is the inactive 7(S) epimer of Maresin 1, which contains a 7(R) hydroxyl group.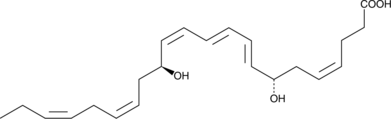
-
GC49312
7-Fluoro-2,1,3-benzoxadiazole-4-sulfonate (ammonium salt)
Ammonium 7-fluorobenzo-2-oxa-1,3-diazole-4-sulfonate, 7-Fluorobenzofurazan-4-Sulfonic Acid, SBD-F
7-Fluoro-2,1,3-benzoxadiazole-4-sulfonate (ammonium salt) is a fluorescent label.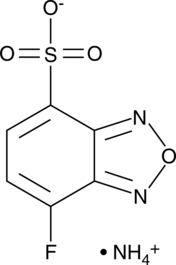
-
GC49051
7-hydroxy Methotrexate
NSC 380962, 7-hydroxy MTX
A metabolite of methotrexate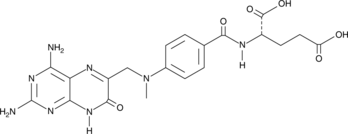
-
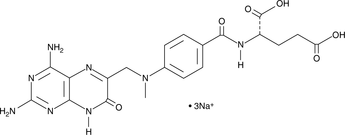
GC42608
7-hydroxy Methotrexate (sodium salt)
7-hydroxy MTX
7-hydroxy Methotrexate (7-hydroxy MTX) is a phase I metabolite of MTX, which is converted by hepatic aldehyde oxidases.
-
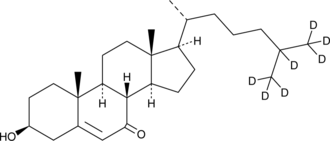
GC46241
7-keto Cholesterol-d7
7-oxo Cholesterol-d7
7-keto Cholesterol is a bioactive sterol and a major oxysterol component of oxidized LDL
-
GC48880
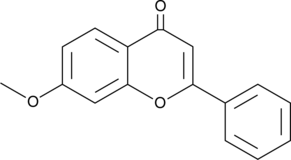
7-Methoxyflavone
A flavone with diverse biological activities
-
GC13982
7-NINA
non-selective NOS inhibitor
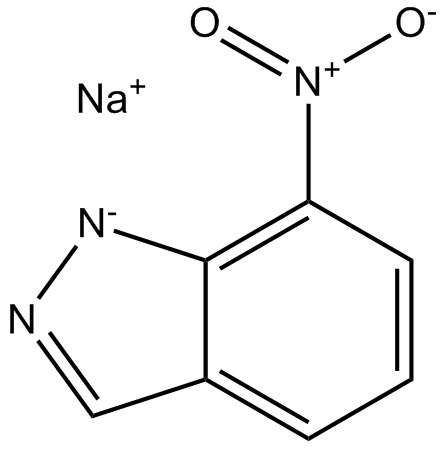
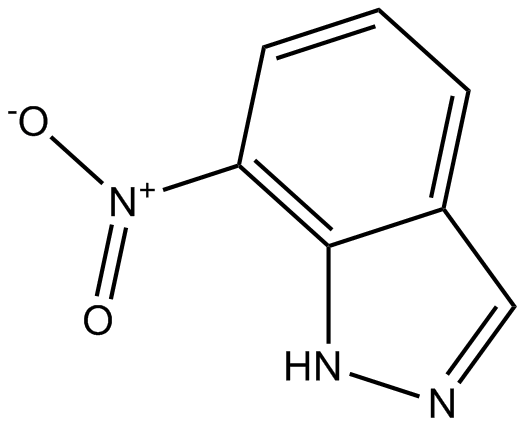
-
GC12309
7-Nitroindazole
nNOS inhibitor
-
GC71920
7-O-Methylaloesinol
7-O-Methylaloesinol is a chromone derivative.

-
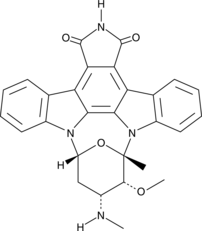
GC42616
7-oxo Staurosporine
BMY 41950, RK-1409
7-oxo Staurosporine is an antibiotic originally isolated from S.
-
GC52211
7-[1-(1H)-Tetrazolylacetamido]desacetoxycephalosporanic Acid
Cefazolin Impurity H
A potential impurity in commercial preparations of cefazolin![7-[1-(1H)-Tetrazolylacetamido]desacetoxycephalosporanic Acid Chemical Structure 7-[1-(1H)-Tetrazolylacetamido]desacetoxycephalosporanic Acid Chemical Structure](/media/struct/GC5/GC52211.png)
-
GC41136
8(S),15(S)-DiHETE
8(S),15(S)-DiHETE is formed when 15(S)-HETE is subjected to further oxidation by 15-LO.
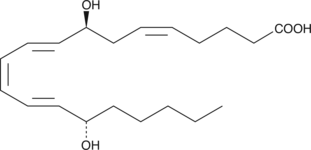
-
GC42623
8-Bromoguanosine
2-Amino-8-bromo-6-hydroxypurine riboside, NSC 79211, NSC 174257
8-Bromoguanosine is a brominated derivative of guanosine.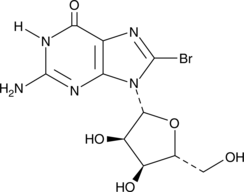
-
GC35199
8-Deoxygartanin
8-Deoxygartanin, a prenylated xanthones from G.
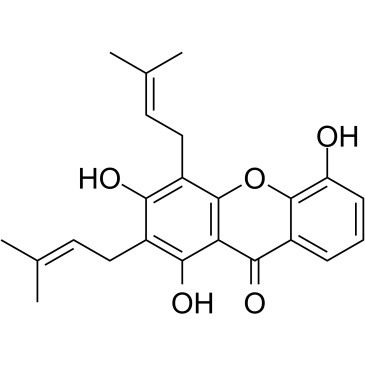
-
GC42627
8-Hydroxyguanine (hydrochloride)
NSC 22720, 8Oxoguanine
8-Hydroxyguanine is produced by oxidative degradation of DNA by hydroxyl radical.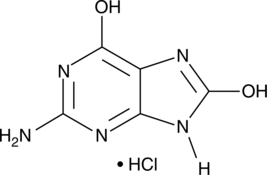
-
GC18426
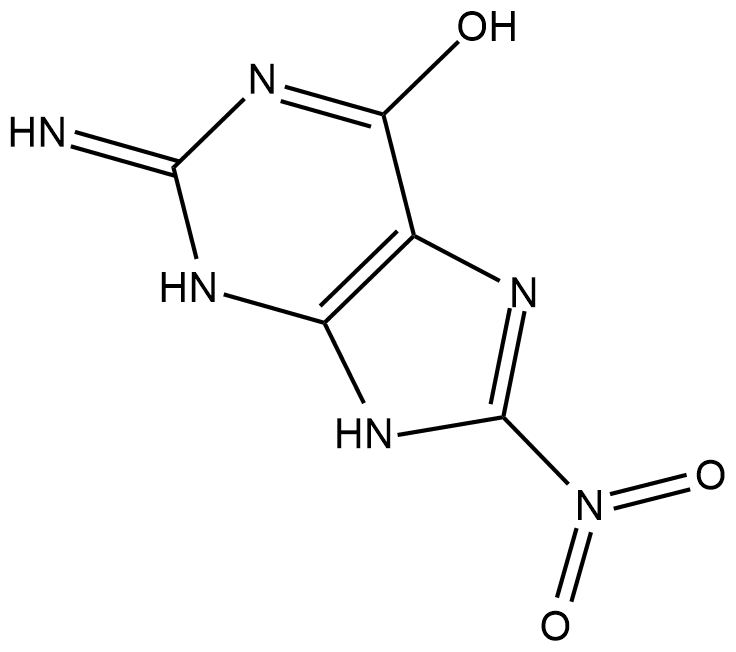
8-Nitroguanine
8-Nitroguanine is a nitrative guanine derivative formed by oxidative damage to the guanine base in DNA by reactive nitrogen species (RNS) during inflammation and in vitro by reaction of DNA with peroxynitrite and other RNS reagents.
-
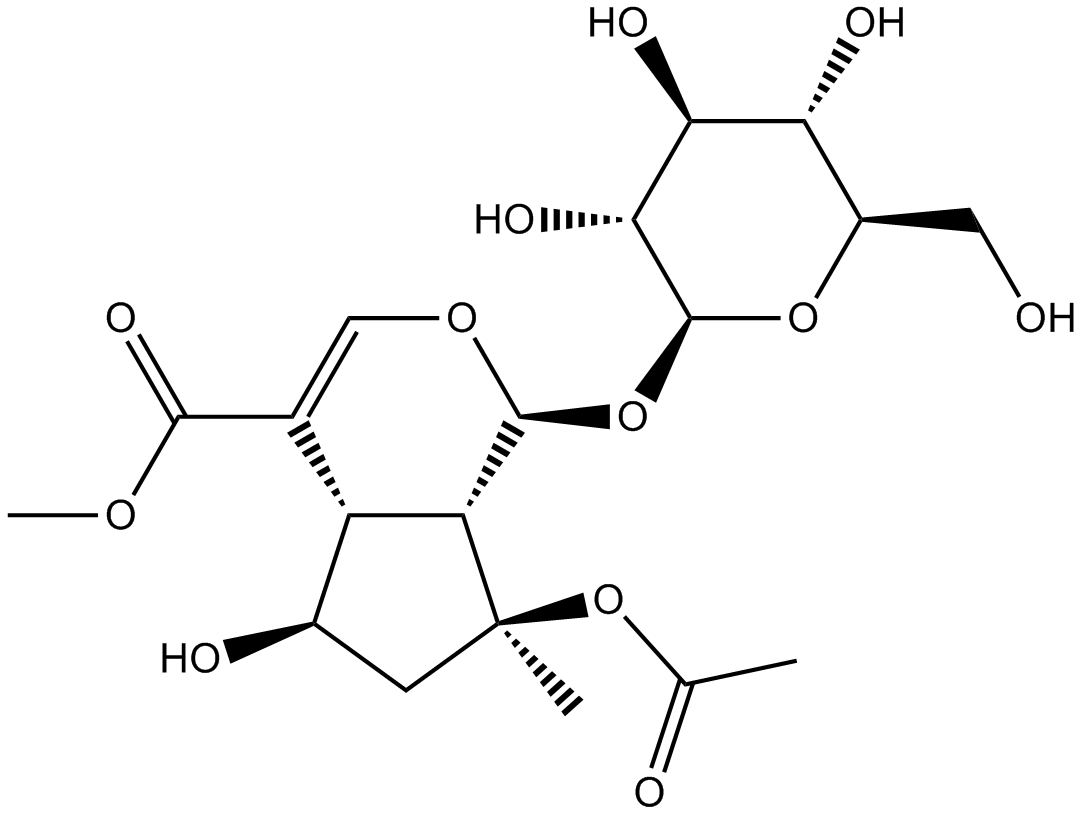
GN10212
8-O-Acetyl shanzhiside methyl ester
-
GC49275
8-Oxycoptisine
8-Oxocoptisine
8-Oxycoptisine is a natural protoberberine alkaloid with anti-cancer activity.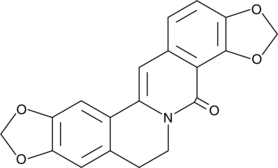
-
GC46748
9(E),11(E)-12-nitro Conjugated Linoleic Acid
9(E),11(E)-12-nitro CLA, 12-NO2-CLA
A nitrated fatty acid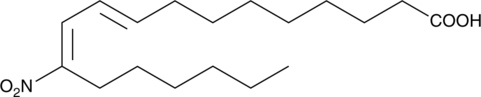
-
GC46749
9(E),11(E)-9-nitro Conjugated Linoleic Acid
9E,11E-9-nitro CLA
A nitrated fatty acid
-
GC42633
9(E)-Erythromycin A oxime
(9E)-Erythromycin A oxime is a metabolite of the semisynthetic antibiotic roxithromycin .
-
GC46753
9(S),12(S),13(S)-TriHOME
(–)-Pinellic Acid, 9S,12S,13S-Pinellic Acid
An oxylipin
-
GC40324
9(S)-PAHSA
9-PAHSA is a newly identified endogenous lipid that belongs to a collection of branched fatty acid esters of hydroxy fatty acids (FAHFAs).
-
GC46757
9(Z),11(E)-Conjugated Linoleic Acid (sodium salt)
cis-9,trans-11-Conjugated Linoleic Acid, 9(Z),11(E)-CLA
An isomer of linoleic acid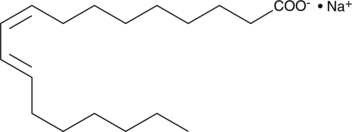
-
GC42642
9,10-Anthracenediyl-bis(methylene)dimalonic Acid
ABMDMA
9,10-Anthracenediyl-bis(methylene)dimalonic acid (ABMDMA) is a reagent used to detect singlet oxygen generation.
-
GC70659
9-cis-Retinoic acid-d5
9-cis-Retinoic acid-d5 is the deuterium labeled 9-cis-Retinoic acid.

-
GC42644
9-Deazaguanine
NSC 344522
9-Deazaguanine is an analog of guanine that acts as an inhibitor of purine nucleoside phosphorylase (PNP; Kd = 160 nM).
-
GC42648
9-Methylstreptimidone
Antibiotic TS 885, NSC 248958
9-Methylstreptimidone is a microbial metabolite originally isolated from Streptomyces sp.
-
GC42649
9-Nitrooleate
9Nitrooleic Acid, 9nitro9transOctadecenoic Acid
Nitrated unsaturated fatty acids, such as 10- and 12-nitrolinoleate, cholesteryl nitrolinoleate, and nitrohydroxylinoleate, represent a new class of endogenous lipid-derived signalling molecules.
-
GC42653
9-OxoOTrE
9KOTE, 9KOTrE
9-OxoOTrE is produced by the oxidation of 9-HpOTrE.
-
GC40325
9-PAHSA
Branched fatty acid esters of hydroxy fatty acids (FAHFAs) are newly identified endogenous lipids regulated by fasting and high-fat feeding and associated with insulin sensitivity.
-
GC49237
93-O17O
A cationic lipidoid
-
GC49238
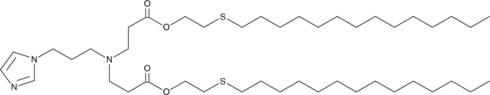
93-O17S
A cationic lipidoid
-
GC45960
9c(i472)
15-LOX-1 Inhibitor i472
9c(i472) is a potent inhibitor of 15-LOX-1 (15-lipoxygenase-1) with an IC50 value of 0.19 μM.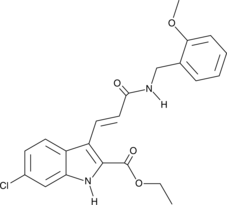
-
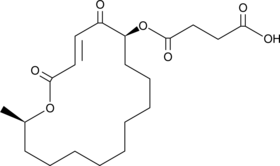
GC45930
A 26771B
A macrolide antibiotic
-
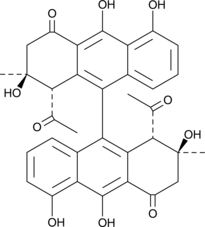
GC42660
A-39183A
A-39183A is an active component of the A-39183 antibiotic complex produced by aerobic fermentation of Streptomyces NRRL 12049.
-
GC42661
A-54556B
A-54556 Factor B
A-54556B is a natural acyldepsipeptide (ADEP) antibiotic isolated from the fermentation broth of S.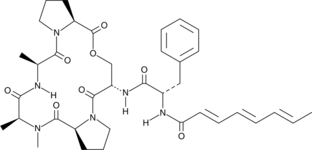
-
GC18936
A-83016F
Antibiotic A-83016F
A-83016F is an aurodox antibiotic isolated from the actinomycete species A83016.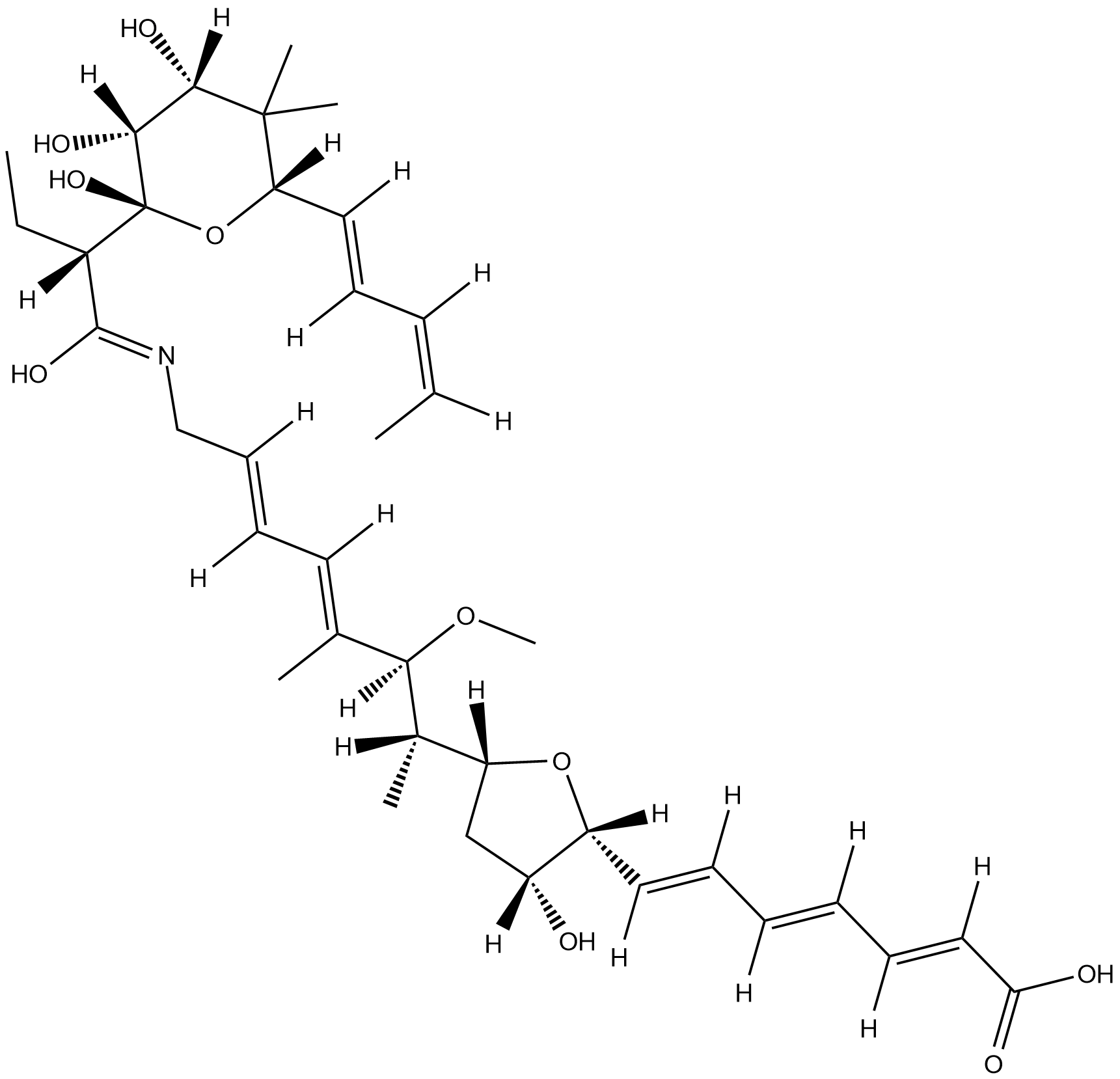
-
GC49309
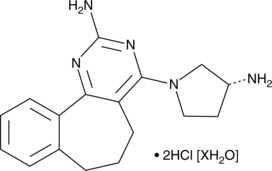
A-943931 (hydrochloride hydrate)
A histamine H4 receptor antagonist
-
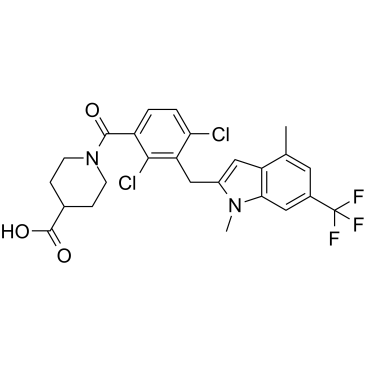
GC35214
A-9758
A-9758 is a RORγ ligand and a potent, selective RORγt inverse agonist (IC50=5 nM), and exhibits robust potency against IL-17A release.
-
GP10060
a-MSH, amide
Ac-Ser-Tyr-Ser-Met-Glu-His-Phe-Arg-Trp-Gly-Lys-Pro-Val-amide, α-MSH, amide
a-MSH (α-Melanocyte-Stimulating Hormone), amide engages the melanocortin-1 receptor and activates cyclic AMP (cAMP) signaling via a G-protein transporter, can synthesize melanin.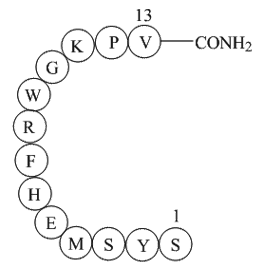
-
GC19496
AAPH
AAPH is a water-soluble azo compound

-
GC42666
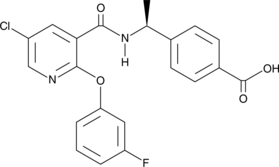
AAT-008
AAT-008 is an orally bioavailable and potent antagonist of the prostaglandin E2 (PGE2) receptor subtype 4 (EP4; IC50 = 16.3 nM in a human EP4 functional assay).
-
GC65293
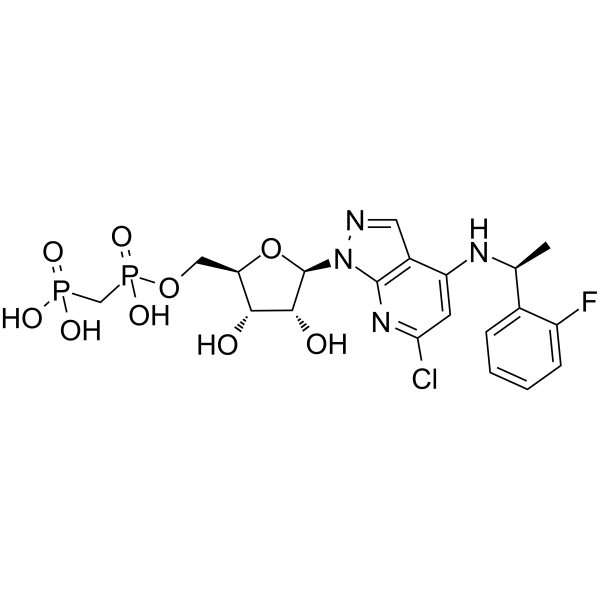
AB-680
AB-680 is a highly potent, reversible and selective inhibitor of CD73 (an ecto-nucleotidase), with a Ki of 4.9 pM for hCD73, displays >10,000-fold selectivity over related ecto-nucleotidases CD39. Anti-tumor activity.
-
GC42667
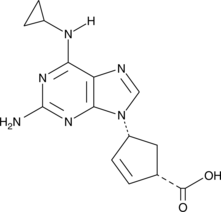
Abacavir Carboxylate
Abacavir carboxylate is an inactive metabolite of the HIV-1 reverse transcriptase inhibitor abacavir.
-
GC46767
Abacavir-d4
An internal standard for the quantification of abacavir
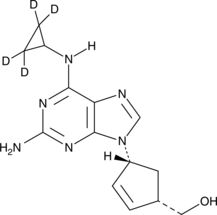
-
GC49766
Abafungin
BAY-W 6341
Abafungin, a antifungal agent, inhibitis the transmethylation at the C-24 position of the sterol side chain, catalyzed by the enzyme sterol-C-24-methyltransferase.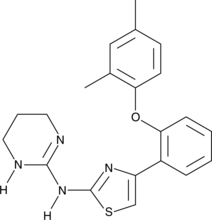
-
GC42668
ABC34
ABC34 is an inactive control probe for JJH260, the inhibitor of androgen-induced gene 1 (AIG1), an enzyme that hydrolyzes fatty acid esters of hydroxy fatty acids (FAHFAs).
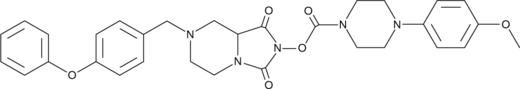
-
GC68595
Abrezekimab
UCB4144; VR 942
Abrezekimab (VR 942) contains CDP7766, which is a humanized, high-affinity, neutralizing anti-human IL-13 antibody fragment that can bind to IL-13. Abrezekimab blocks the binding of IL-13 to the IL-13Rα1 subunit. Abrezekimab can be used for research on asthma.

-
GC49745
ABT-263-d8
Navitoclax-d8
ABT-263-d8 is the deuterium labeled Navitoclax. Navitoclax (ABT-263) is a potent and orally active Bcl-2 family protein inhibitor that binds to multiple anti-apoptotic Bcl-2 family proteins, such as Bcl-xL, Bcl-2 and Bcl-w, with a Ki of less than 1 nM.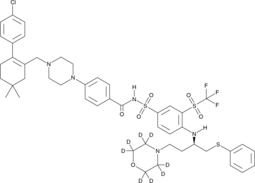
-
GC70733
ABT-510 acetate
ABT-510 acetate is an anti-angiogenic TSP peptide (Thrombospondin-1 analogue) that induces apoptosis and inhibits ovarian tumour growth in an orthotopic, syngeneic model of epithelial ovarian cancer.

-
GC42683
Abz-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-EDDnp
Abz-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-EDDnp is a fluorescence-quenched peptide substrate for human neutrophil elastase (kcat/Km = 531 mM-1s-1).
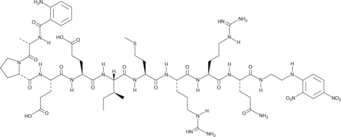
-
GC52499
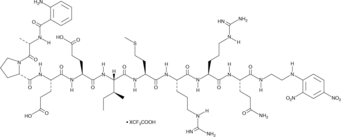
Abz-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-EDDnp (trifluoroacetate salt)
A sensitive substrate for neutrophil elastase
-
GC42684
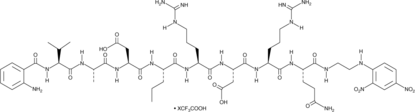
Abz-Val-Ala-Asp-Nva-Arg-Asp-Arg-Gln-EDDnp (trifluoroacetate salt)
Abz-Val-Ala-Asp-Nva-Arg-Asp-Arg-Gln-EDDnp is a fluorescence-quenched peptide substrate for human proteinase 3 (kcat/Km = 1,570 mM-1s-1).
-
GC42685
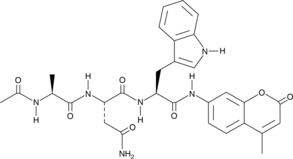
Ac-ANW-AMC
Ac-Ala-Asn-Trp-AMC
Ac-ANW-AMC is a fluorogenic substrate for the β5i/LMP7 subunit of the 20S immunoproteasome.
-
GC49704
Ac-FLTD-CMK (trifluoroacetate salt)
Ac-Phe-Leu-Thr-Asp-CMK
An inhibitor of caspase-1, -4, -5, and -11
-
GA20621
Ac-muramic acid
MurNAc
A component of bacterial cell walls. For a protected derivative of MurNAc see Q-1005.
-
GC52372
Ac-VDVAD-AFC (trifluoroacetate salt)
N-Acetyl-Val-Asp-Val-Ala-Asp-AFC, N-Acetyl-Val-Asp-Val-Ala-Asp-7-amino-4-Trifluoromethylcoumarin, Caspase-2 Substrate (Fluorogenic)
A fluorogenic substrate for caspase-2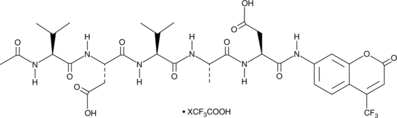
-
GC15598
Ac2-26
inhibitor of leukocyte extravasation
-
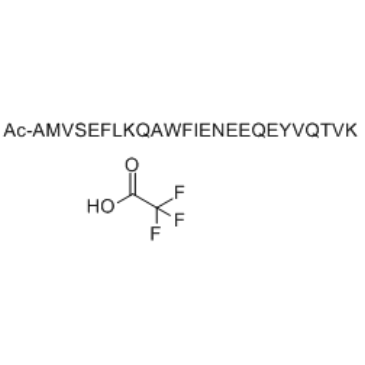
GC49263
Ac2-26 (human) (ammonium salt)
Ac-AMVSEFLKQAWFIENEEQEYVQTVK-OH, Ac-ANX-A12-26, N-Acetyl-AMVSEFLKQAWFIENEEQEYVQTVK
An annexin A1-mimetic peptide
-
GC35224
Ac2-26 TFA
Ac2-26 TFA, an active N-terminal peptide of annexin A1 (AnxA1), attenuates ischemia-reperfusion-induced acute lung injury.
-
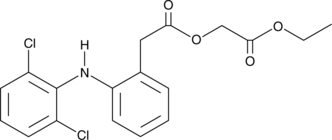
GC42691
Aceclofenac ethyl ester
A potential impurity found in commercial preparations of aceclofenac
-
GC42692
Aceclofenac methyl ester
Aceclofenac Impurity D
A potential impurity in commercial preparations of aceclofenac
-
GC49911
Acetyl Hexapeptide-38 (trifluoroacetate salt)
A hexapeptide
-
GC42700
Acetyl Pentapeptide-1
Acetyl pentapeptide-1 is a pentapeptide that decreases IL-8 secretion in human keratinocytes when used in combination with acetyl hexapeptide-36 and acetyl hexapeptide-38.

-
GC18443
Acetyl-6-formylpterin
Ac-6-FP, 2-Acetamido-6-formylpteridin-4-one, NSC 129965
Acetyl-6-formylpterin is an inhibitor of mucosal-associated invariant T (MAIT) cell activation.
-
GC11786
Acetylcysteine
N-acetylcysteine; N-acetyl-L-cysteine; NAC; Acetadote
Acetylcysteine is the N-acetyl derivative of CYSTEINE.
-
GC17416
ACHP
IκB kinase inhibitor
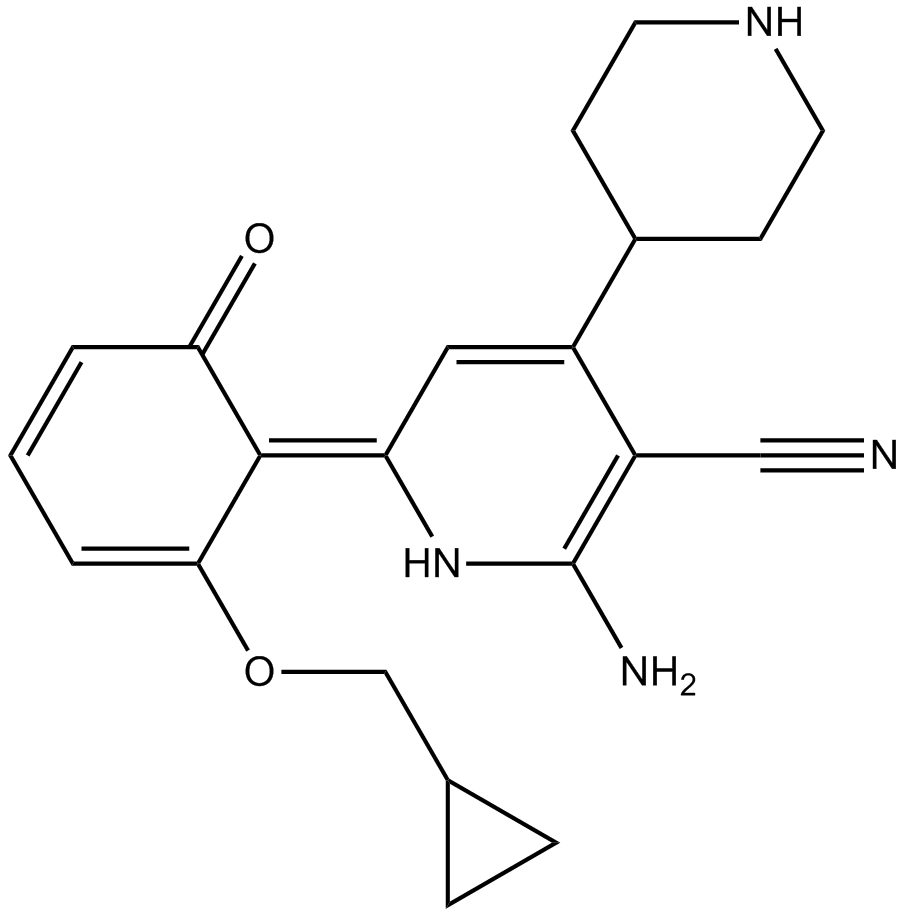
-
GN10536
Aconine
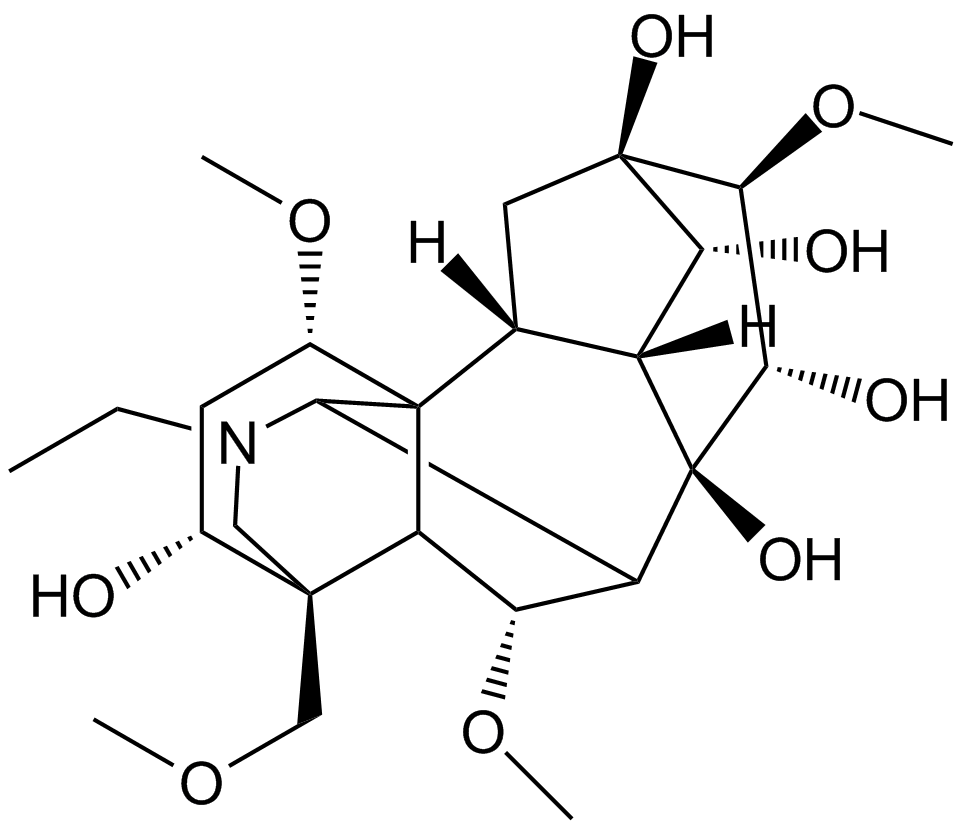
-
GC49804
Acridine
2,3,5,6-Dibenzopyridine, 10-Azaanthracene, Dibenzopyridine, NSC 3408
An azaarene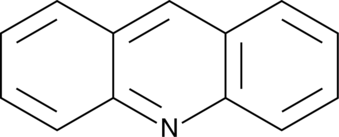
-
GC40645
Actarit
4-Acetylaminophenylacetic Acid, MS-932, NSC 170317
Actarit is an orally active immunomodulator that reduces symptoms in animal models and clinical trials of rheumatoid arthritis.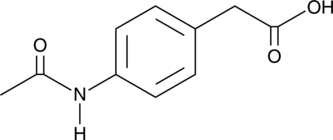
-
GC70584
Actinomycin X2
Actinomycin X2 (Actinomycin V), produced by many Streptomyces sp.

-
GC41316
Actinopyrone A
(+)-Actinopyrone A, SS 1538A
Actinopyrone A is a pyrone isolated from S.
-
GC48842
Actiphenol
Actinophenol, NSC 58413
A bacterial metabolite with antiviral activity
-
GC46797
Acyclovir-d4
ACV-d4
An internal standard for the quantification of acyclovir
-
GC46798
Adapalene-d3
An internal standard for the quantification of adapalene
-
GC46805
Adefovir-d4
PMEA-d4
An internal standard for the quantification of adefovir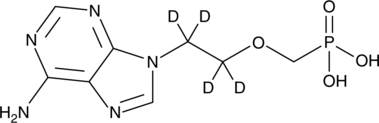
-
GC31693
Adelmidrol
Adelmidrol exerts important anti-inflammatory effects that are partly dependent on PPARγ.
-
GC49285
Adenosine 5’-methylenediphosphate (hydrate)
Adenosine 5'-(α,β-methylene)diphosphate, AMP-CP, APCP, 5'-APCP
An inhibitor of ecto-5’-nucleotidase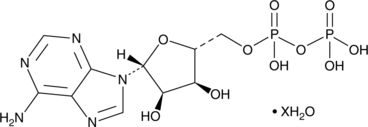
-
GC60040
ADH-503
(Z)-Leukadherin-1 choline
ADH-503 ((Z)-Leukadherin-1 choline) is an orally active and allosteric CD11b agonist. ADH-503 leads to the repolarization of tumor-associated macrophages, reduction in the number of tumor-infiltrating immunosuppressive myeloid cells, and enhances dendritic cell responses.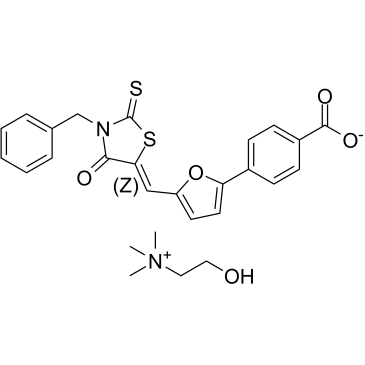
-
GC71405
ADS032
ADS032 is a dual inhibitor of NLRP1 and NLRP3 that can rapidly, reversibly and stably inhibit inflammasome formation.

-
GC42742
ADT-OH
ACS1
ADT-OH is a derivative of anethole dithiolethione (ADT) and synthetic hydrogen sulfide (H2S) donor.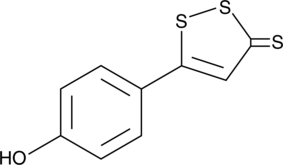
-
GC31649
ADU-S100 (ML RR-S2 CDA)
MIW815; ML RR-S2 CDA
ADU-S100 (ML RR-S2 CDA) (MIW815), an activator of stimulator of interferon genes (STING), leads to potent and systemic tumor regression and immunity.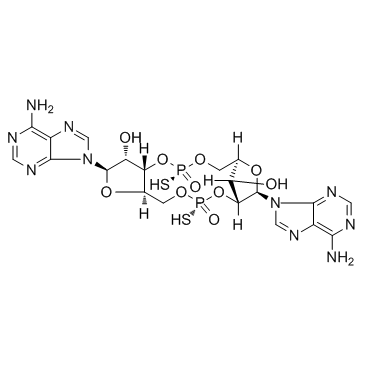
-
GC39161
ADU-S100 disodium salt
MIW815 disodium salt; ML RR-S2 CDA disodium salt
ADU-S100 disodium salt (MIW815 disodium salt) is an activator of stimulatory interferon gene (STING) protein with a Kd of 4.61±0.42μM.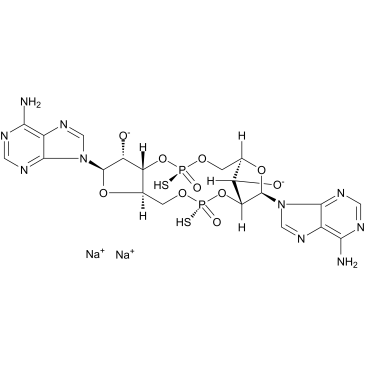
-
GC42743
AEM1
Cancer cell survival appears partly dependent on antioxidative enzymes, whose expression is regulated by the Keap1-Nrf2 pathway, to quench potentially toxic reactive oxygen species generated by their metastatic transformation.
-
GC63485
Afimetoran
BMS-986256
Afimetoran is a toll-like receptor antagonist, which can be used in the research of inflammatory and autoimmune diseases.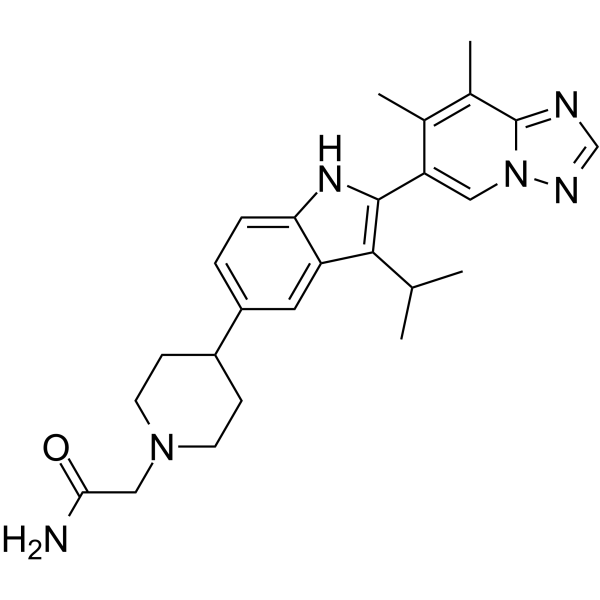
-
GC66396
Agatolimod
ODN 2006; CpG 7909; PF-3512676
Agatolimod (ODN 2006), a class B ODN (oligodeoxynucleotide), is a TLR9 agonist. Agatolimod is also an optimal CpG sequence for humans. Agatolimod stimulates very strong production of NO2 and IL-6 in HD11 cells. Agatolimod can be used for breast cancer research. Sequence: 5'-tcgtcgttttgtcgttttgtcgtt-3'.


