DNA Damage/DNA Repair
- MTH1(4)
- PARP(58)
- ATM/ATR(27)
- DNA Alkylating(19)
- DNA Ligases(3)
- DNA Methyltransferase(18)
- DNA-PK(27)
- HDAC(94)
- Nucleoside Antimetabolite/Analogue(136)
- Telomerase(14)
- Topoisomerase(133)
- tankyrase(5)
- Antifolate(32)
- CDK(233)
- Checkpoint Kinase (Chk)(29)
- CRISPR/Cas9(9)
- Deubiquitinase(62)
- DNA Alkylator/Crosslinker(64)
- DNA/RNA Synthesis(388)
- Eukaryotic Initiation Factor (eIF)(22)
- IRE1(22)
- LIM Kinase (LIMK)(9)
- TOPK(5)
- Casein Kinase(53)
- DNA Intercalating Agents(7)
- DNA/RNA Oxidative Damage(12)
Products for DNA Damage/DNA Repair
- Cat.No. Product Name Information
-
GC44348
N-desmethyl Bendamustine
nor-Bendamustine
N-desmethyl Bendamustine is an active metabolite of the DNA alkylating agent bendamustine.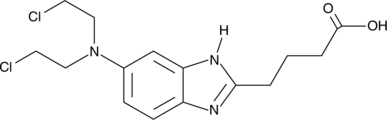
-
GC39498
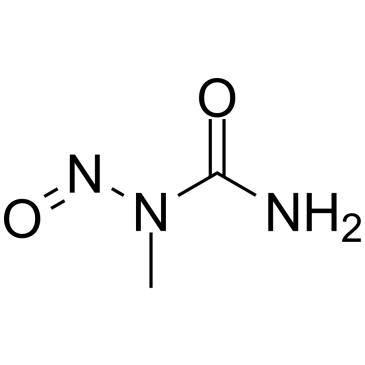
N-Nitroso-N-methylurea
N-Nitroso-N-methylurea (NMU;MNU;NMH) is a potent carcinogen, mutagen and teratogenand. N-Nitroso-N-methylurea is a direct-acting alkylating agent that interacts with DNA. N-Nitroso-N-methylurea targets multiple animal organs to cause various cancer and/or degenerative disease. N-Nitroso-N-methylurea is also a precursor in the synthesis of diazomethane.
-
GC19566
N-Nitrosodiethylamine (950mg/mL)
N-Nitrosodiethylamine (Diethylnitrosamine) is a potent hepatocarcinogenic dialkylnitrosoamine. N-Nitrosodiethylamine is mainly present in tobacco smoke, water, cheddar cheese, cured, fried meals and many alcoholic beverages. N-Nitrosodiethylamine is responsible for the changes in the nuclear enzymes associated with DNA repair/replication. N-Nitrosodiethylamine results in various tumors in all animal species. The main target organs are the nasal cavity, trachea, lung, esophagus and liver.
-
GC69577
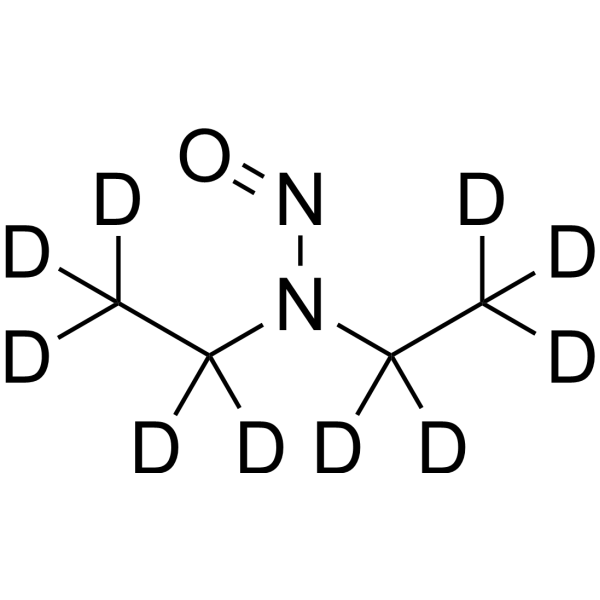
N-Nitrosodiethylamine-d10
Diethylnitrosamine-d10
N-Nitrosodiethylamine-d10 is the deuterated form of N-Nitrosodiethylamine. N-Nitrosodiethylamine (Diethylnitrosamine) is a potent carcinogen belonging to the group of nitrosamines. It is mainly found in tobacco smoke, water, cheddar cheese, pickled foods, fried foods and many alcoholic beverages. N-Nitrosodiethylamine is responsible for changes in nucleases involved in DNA repair/replication. It can cause various tumors in all animals with major target organs being the nasal cavity, trachea, lungs, esophagus and liver.
-
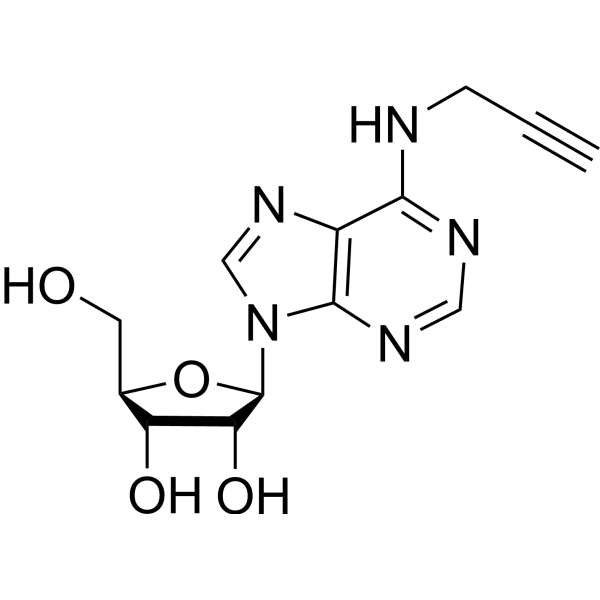
GC69589
N-Propargyladenosine
N-Propargyladenosine is an adenosine analogue. Adenosine analogues are mostly used as smooth muscle vasodilators and have also been shown to inhibit cancer progression. Some popular products in this series include Adenosine phosphate, Acadesine, Clofarabine, Fludarabine phosphate, and Vidarabine.
-
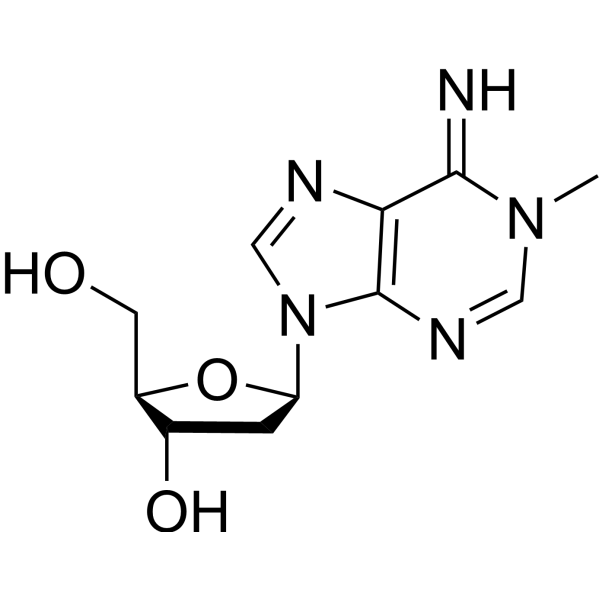
GC69525
N1-Methyl-2'-deoxyadenosine
N1-Methyl-2'-deoxyadenosine, DNA adduct is a purine nucleoside analogue. Purine nucleoside analogues have broad anti-tumor activity and target inert lymphoid malignancies. The anticancer mechanism in this process depends on inhibiting DNA synthesis, inducing cell apoptosis, etc.
-
GC66075
N1-Methylpseudouridine-5′-triphosphate trisodium
1-Methylpseudouridine-5′-triphosphate trisodium
N1-Methylpseudouridine-5′-triphosphate (1-Methylpseudouridine-5′-triphosphate) trisodium is a nucleobase-modified nucleotide, used for synthesizing mRNA with reduced immunogenicity and improved stability.
-
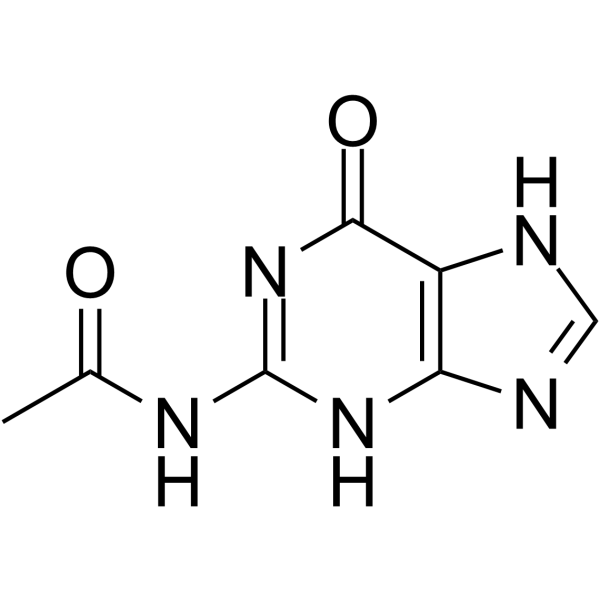
GC66325
N2-Acetylguanine
N2-Acetylguanine is a C2-modified guanine. N2-Acetylguanine binds GR (guanine-guanine riboswitch) with an Kd value of 300 nM. N2-Acetylguanine modulate transcriptional termination. N2-Acetylguanine has the potential for the research of antimicrobial agent.
-
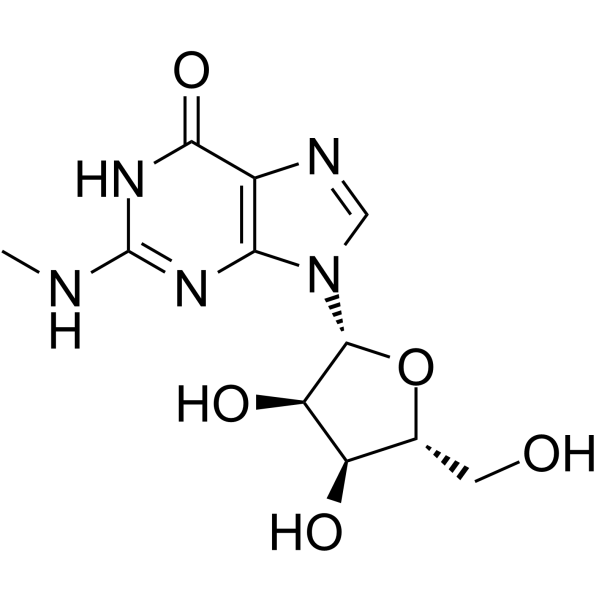
GC65127
N2-Methylguanosine
N2-methylguanosine is a modified nucleoside that occurs at several specific locations in many tRNA's.
-
GC36681
N6,N6-Dimethyladenosine
6-N,N-Dimethyladenosine, 6-DMA, m62A
N6,N6-Dimethyladenosine is a modified ribonucleoside previously found in rRNA, and also exhibits in mycobacterium bovis Bacille Calmette-Guérin tRNA.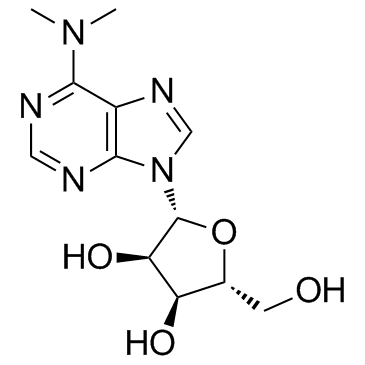
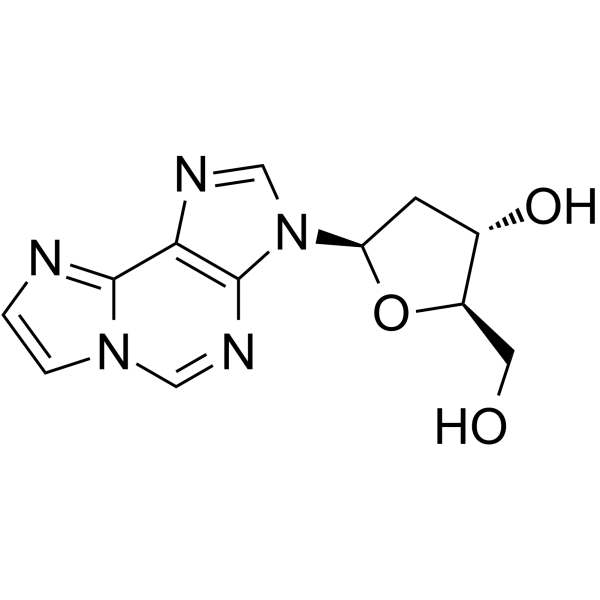
-
GC65241
N6-Etheno 2'-deoxyadenosine
N6-Etheno 2'-deoxyadenosine is a reactive oxygen species (ROS)/reactive nitrogen species (RNS)-induced DNA oxidation product, used as a biomarker to evaluate chronic inflammation and lipid peroxidation in animal or human tissues.
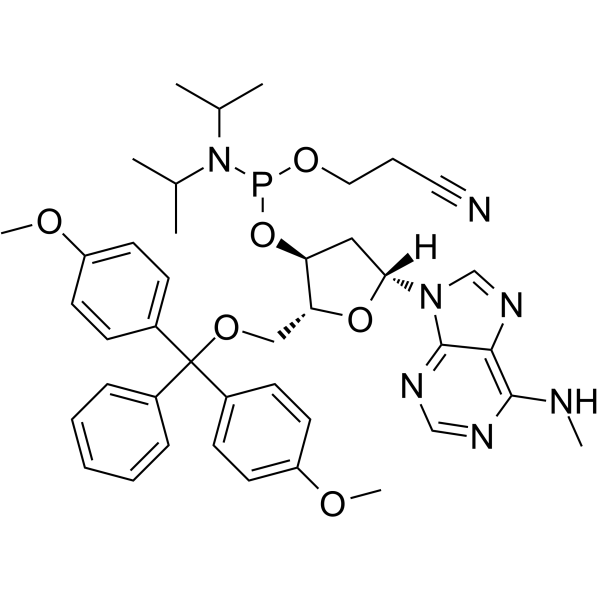
-
GC65561
N6-Methyl-dA phosphoramidite
N6-Methyl-dA phosphoramidite can be used in the synthesis of oligodeoxyribonucleotides.
-
GC34156
Namitecan (ST-1968)
ST-1968
Namitecan (ST-1968) is a potent topoisomerase I inhibitor, with antitumor property.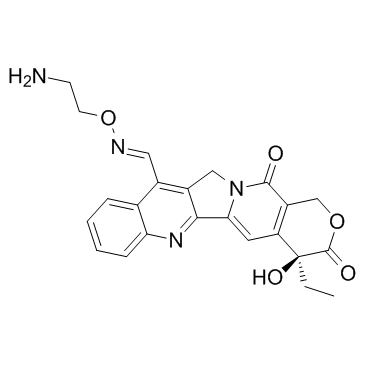
-
GC36690
Nampt-IN-3
Nampt-IN-3 (Compound 35) simultaneously inhibit nicotinamide phosphoribosyltransferase (NAMPT) and HDAC with IC50s of 31 nM and 55 nM, respectively. Nampt-IN-3 effectively induces cell apoptosis and autophagy and ultimately leads to cell death.
-
GC36691
Nanatinostat
CHR-3996
Nanatinostat (CHR-3996) is a potent, class I selective and orally active histone deacetylase (HDAC) inhibitor with an IC50 of 8 nM.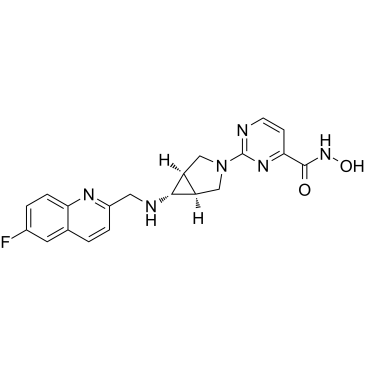
-
GC36708
NCC007
NCC007 is a dual casein kinase Iα (CKIα) and δ (CKIδ) inhibitor with IC50s of 1.8 and 3.6 μM, respectively.
-
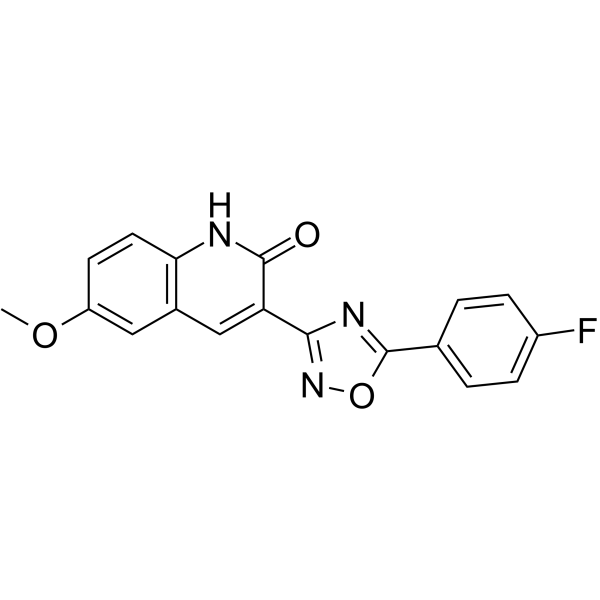
GC63653
NCGC00029283
NCGC00029283 is a werner syndrome helicase-nuclease (WRN) helicase inhibitor with IC50s of 2.3 μM, 12.5 μM, and 3.4 μM for WRN, BLM and FANCJ helicase, respectively.
-
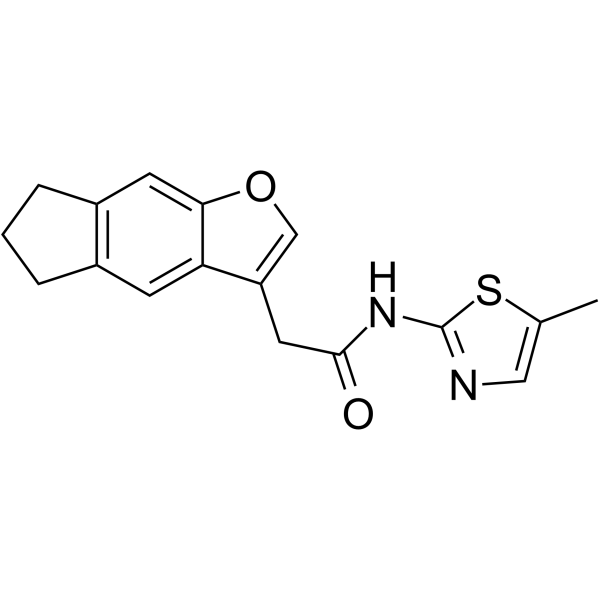
GC64393
NCT02
NCT02 is a cyclin K degrader. NCT02 induces ubiquitination of cyclin K (CCNK) and proteasomal degradation of CCNK and its complex partner CDK12. NCT02 has the potential for the research of metastatic colorectal cancer (CRC).
-
GC15786
Nedaplatin
NSC 375101
DNA synthesis inhibitor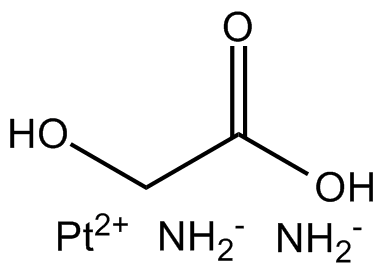
-
GC10591
Nelarabine
GW 506U78, Nelzarabine
Prodrug of ara-G for T-LBL/T-ALL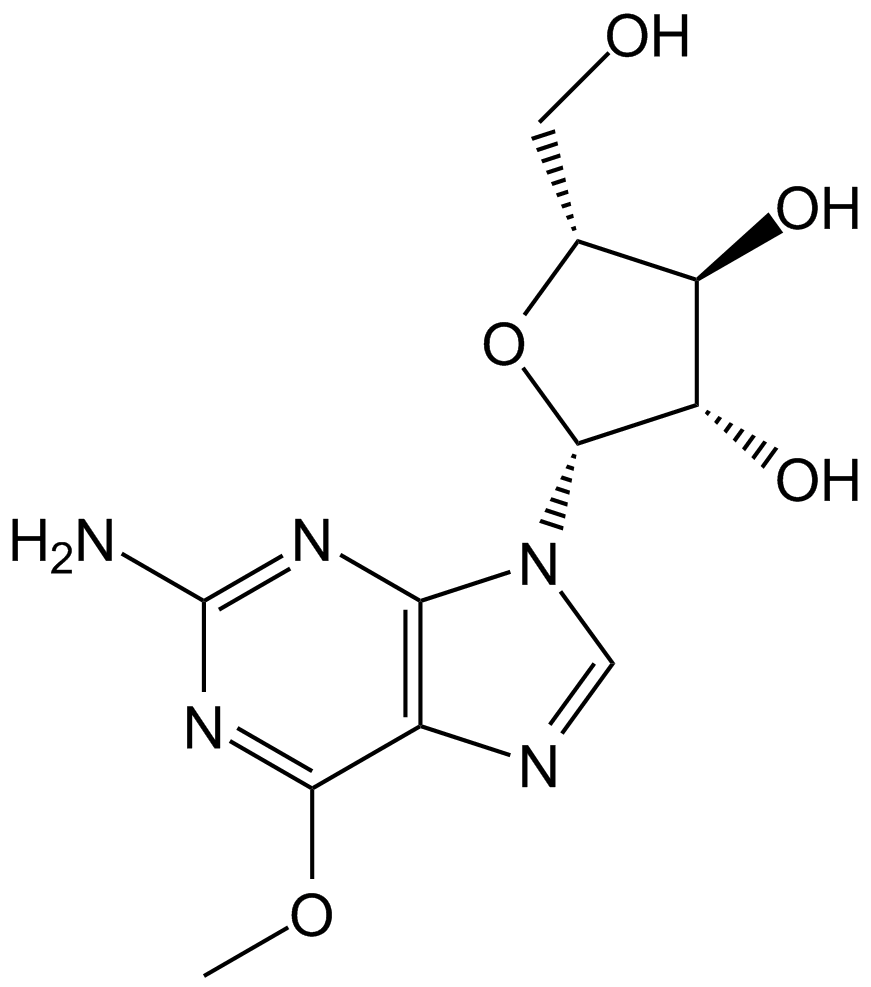
-
GC45523
Nemorosone
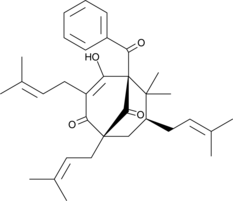
-
GC64493
Neocarzinostatin
Zinostatin; Vinostatin
Neocarzinostatin, a potent DNA-damaging, anti-tumor antibiotic, recognizes double-stranded DNA bulge and induces DNA double strand breaks (DSBs).

-
GC60268
Neoxanthin
Neoxanthin is a major xanthophyll carotenoid and a precursor of the plant hormone abscisic acid in dark green leafy vegetables. Neoxanthin is a potent antioxidant and light-harvesting pigment. Neoxanthin induces apoptosis and has anticancer actions.
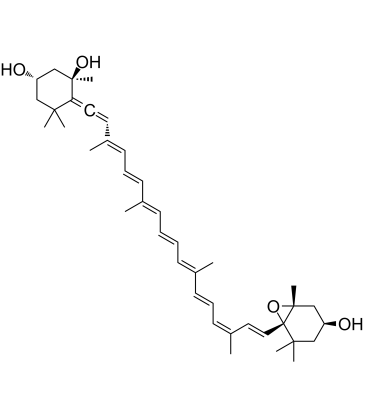
-
GC65202
Nesuparib
JPI-547/OCN-201
Nesuparib is a potent inhibitor of PARP.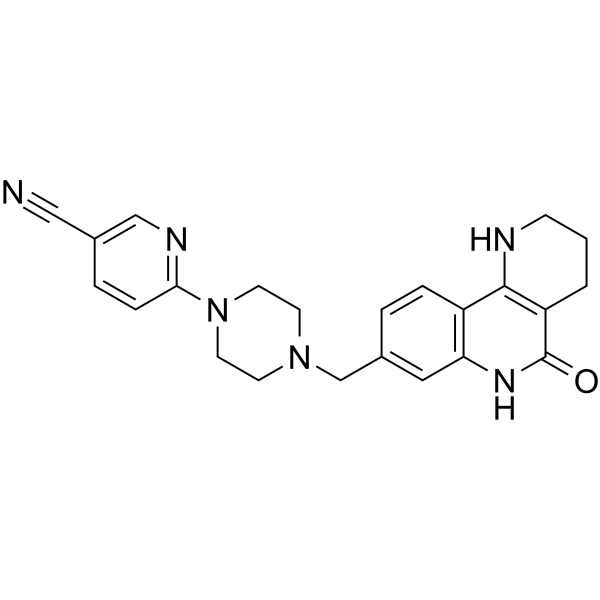
-
GC13764
Nexturastat A
HDAC6 inhibitor,highly potent and selective
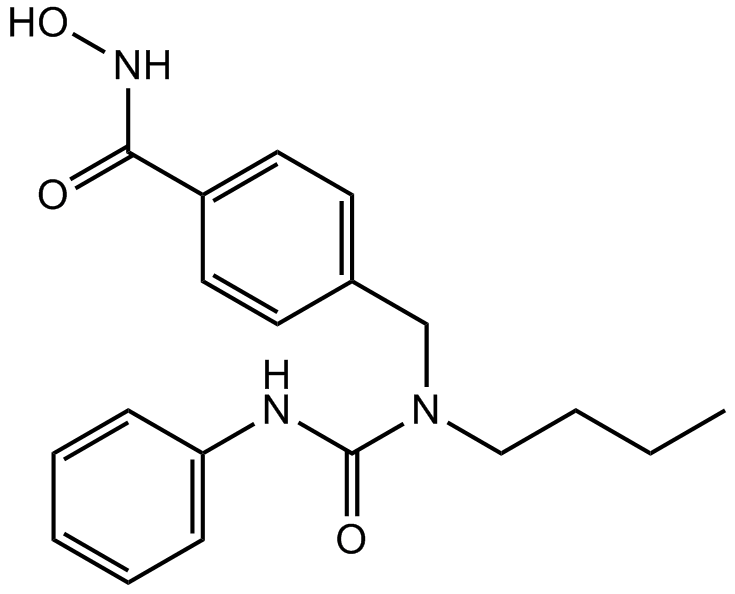
-
GC36732
NG 52
Compound 52
NG 52 is a potent, cell-permeable, selective, ATP-compatible and orally active Cdc28p and Pho85p kinase inhibitor with IC50s of 7 μM and 2 μM, respectively. NG 52 also inhibits the activity of phosphoglycerate kinase 1 (PGK1) with an IC50 of 2.5 μM. NG 52 is inactive against yeast kinases Kin28p, Srb10, and Cak1p.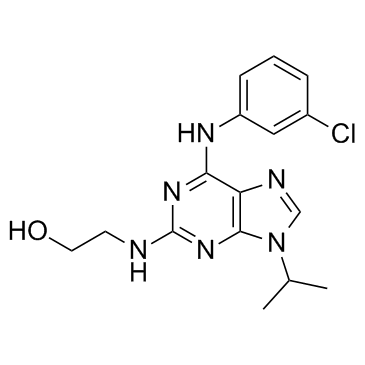
-
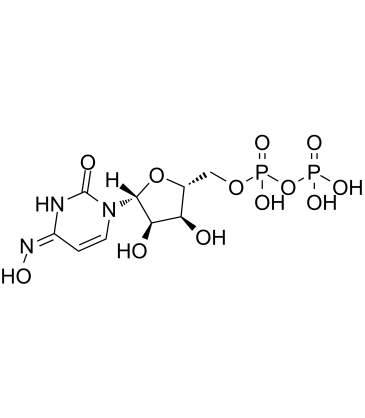
GC61956
NHC-diphosphate
NHC-diphosphate is an active phosphorylated?intracellular metabolite?of β-d-N4-Hydroxycytidine (NHC) as a diphosphate form.
-
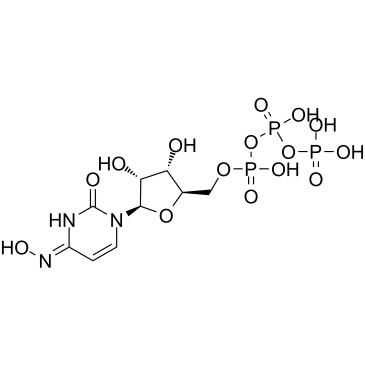
GC61130
NHC-triphosphate
NHC-triphosphate is an active phosphorylated intracellular metabolite of β-d-N4-Hydroxycytidine (NHC) as a triphosphate form.
-
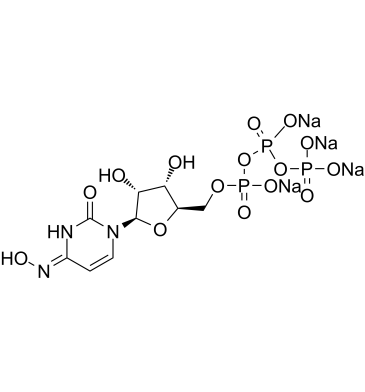
GC61961
NHC-triphosphate tetrasodium
NHC-triphosphate tetrasodium is an active phosphorylated intracellular metabolite of β-d-N4-Hydroxycytidine (NHC) as a triphosphate form.
-
GC36743
Nimustine hydrochloride
ACNU, NSC-D 245382
Nimustine hydrochloride (ACNU) is a DNA cross-linking and DNA alkylating agent, which induces DNA replication blocking lesions and DNA double-strand breaks and inhibits DNA synthesis, commonly used in chemotherapy for glioblastomas.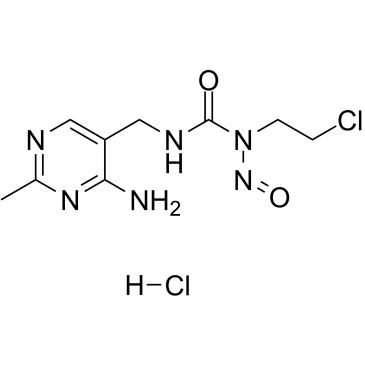
-
GC34120
Niraparib R-enantiomer (MK 4827 (R-enantiomer))
MK 4827 (R-enantiomer)
Niraparib R-enantiomer (MK-4827 R-enantiomer) is an excellent PARP1 inhibitor with IC50 of 2.4 nM.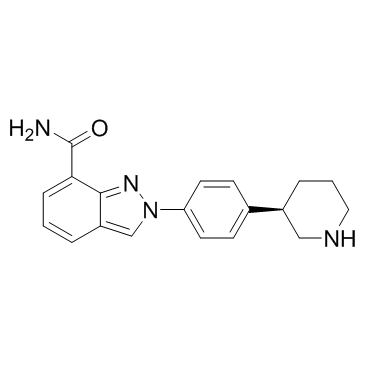
-
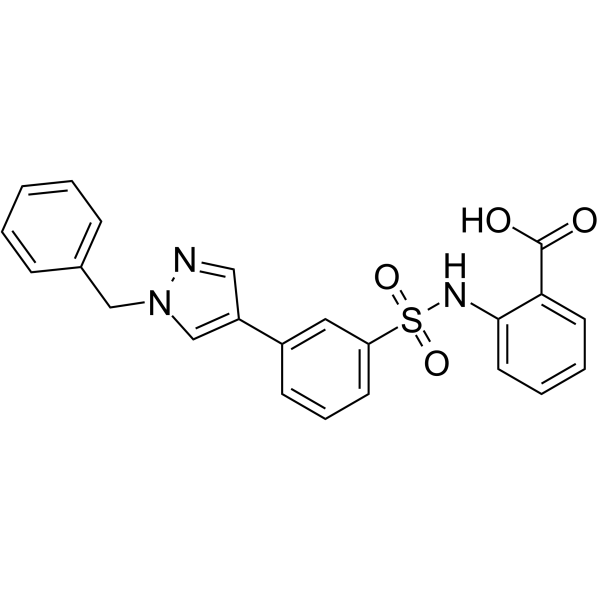
GC64559
NITD-2
NITD-2, a dengue virus (DENV) polymerase inhibitor, inhibits the DENV RdRp-mediated RNA elongation.
-
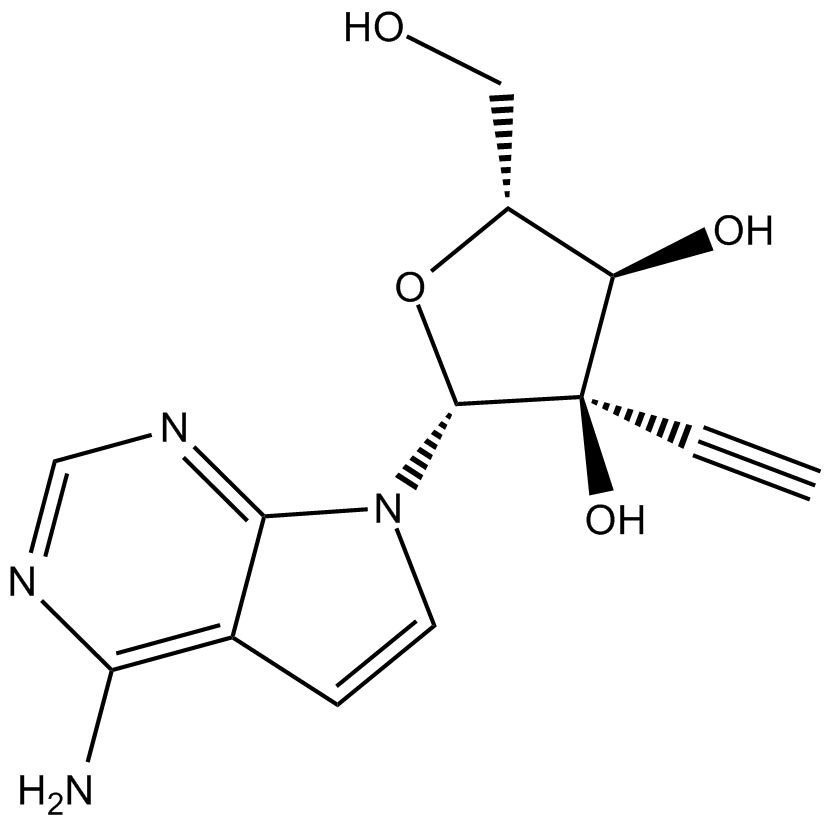
GC19506
NITD008
7-Deaza-2'-C-acetylene-adenosine
NITD008 is a potent and selective flaviviruse inhibitor which can inhibit Dengue Virus Type 2 (DENV-2) with an EC50 of 0.64 μM.
-
GC44408
Nitrosobenzene
NSC 66479
Nitrosobenzene is a spin trap that has been used in the study of oxidative DNA damage and nitroso-compound-induced respiratory burst in neutrophils.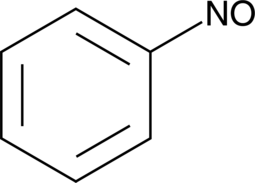
-
GC32964
NKL 22
Histone Deacetylase Inhibitor IV
A selective HDAC1/3 inhibitor
-
GC19263
NKP-1339
KP1339
NKP-1339(IT-139) is a ruthenium(iii) coordination anticancer compound based on target to transferrin.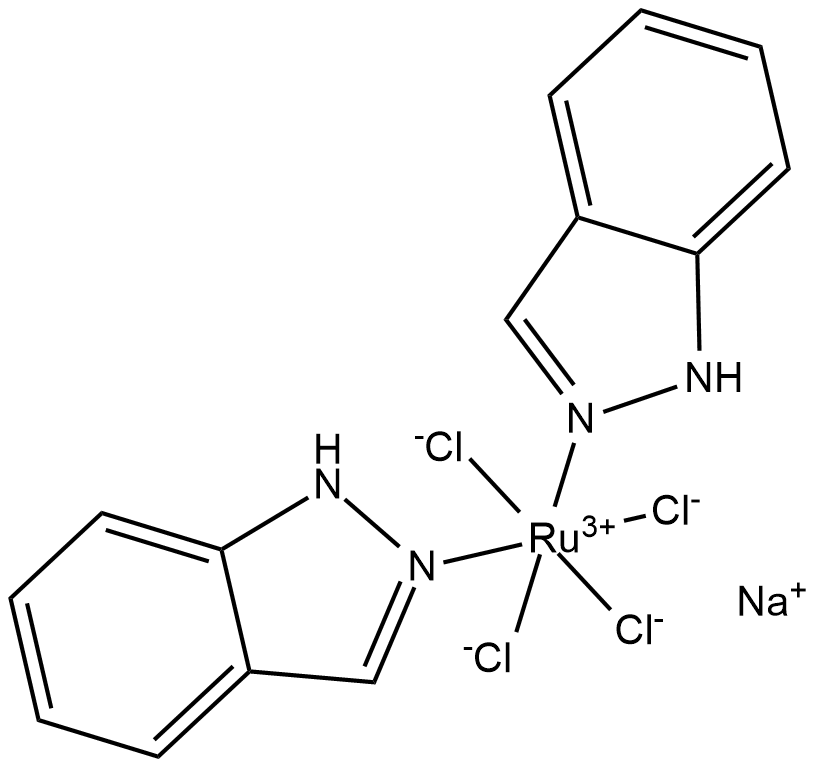
-
GC19264
NMS-P118
NMS-P118 is a potent, orally available, and highly selective PARP-1 Inhibitor for cancer therapy.
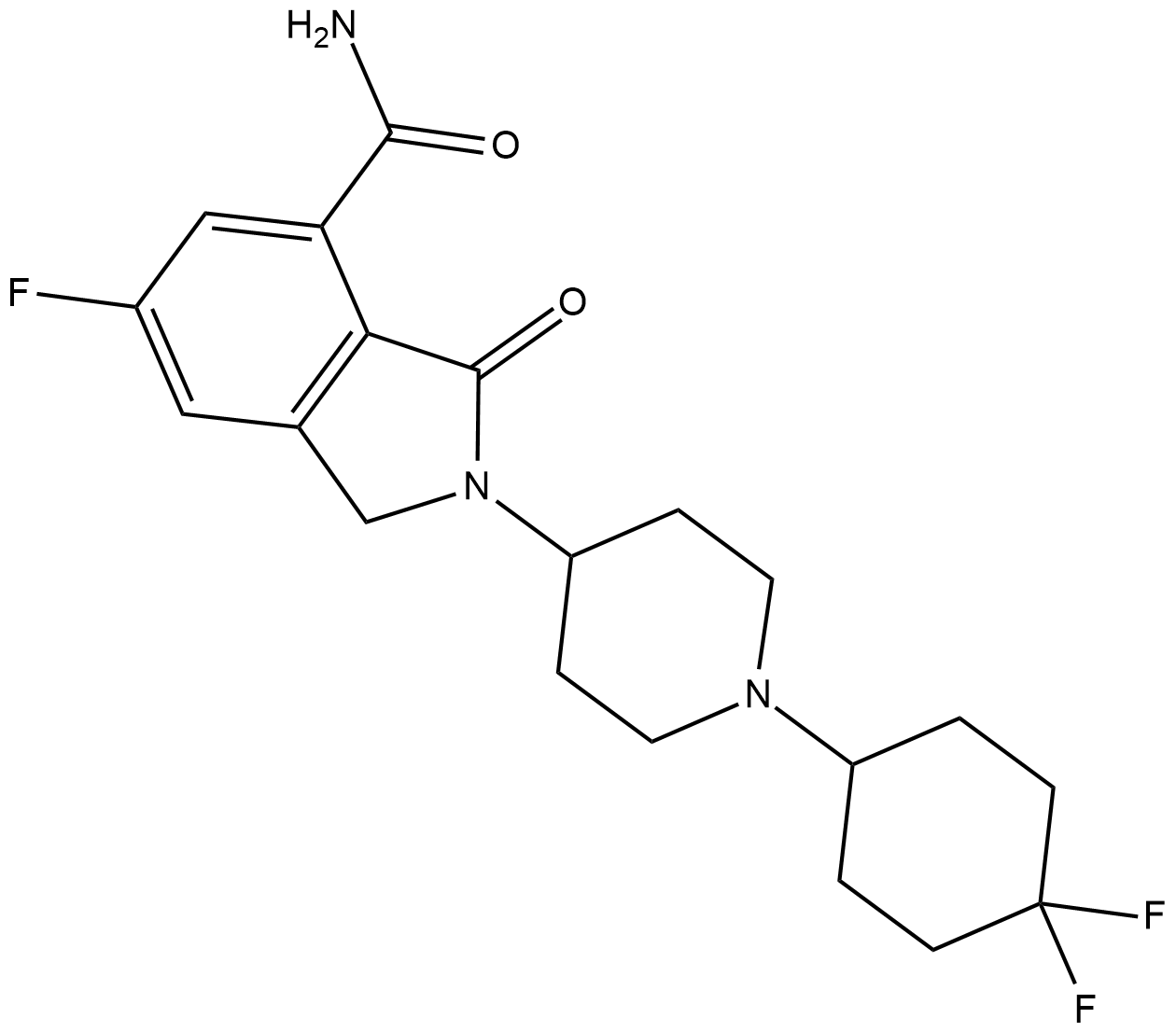
-
GC36751
NMS-P515
NMS-P515 is a potent, orally active and stereospecific PARP-1 inhibitor, with a Kd of 16 nM and an IC50 of 27 nM (in Hela cells). Anti-tumor activity.
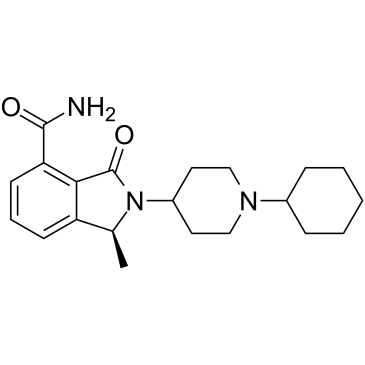
-
GC14075
Nocodazole
NSC 238159, Oncodazole, R 17934
A tubulin production inhibitor,anti-neoplastic agent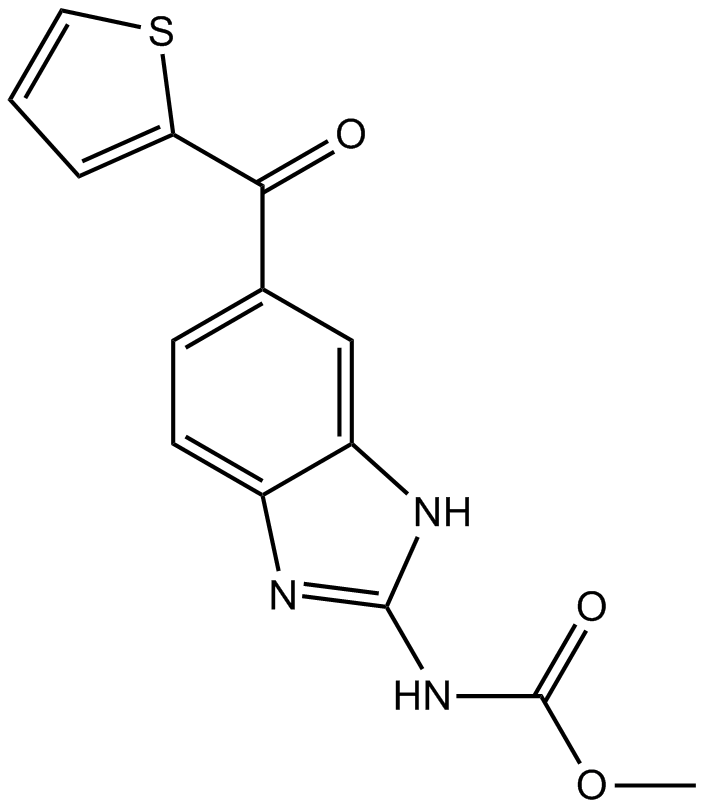
-
GC44438
Nogalamycin
NSC 70845, U 15167
Nogalamycin is an anthracycline originally isolated from S.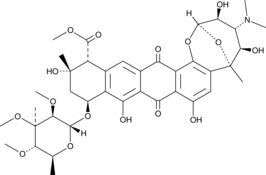
-
GC62679
NSAH
NSAH is a reversible and competitive nonnucleoside ribonucleotide reductase (RR) inhibitor, with cell-free IC50 of 32 μM and cell-based IC50 of ~250 nM, respectively.
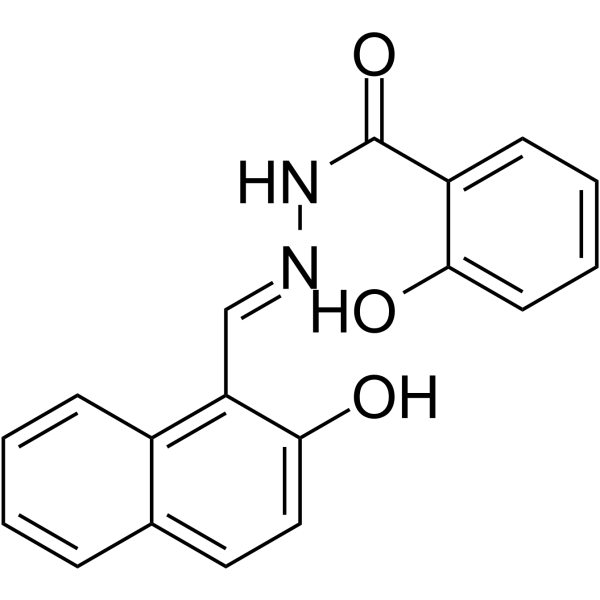
-
GC63874
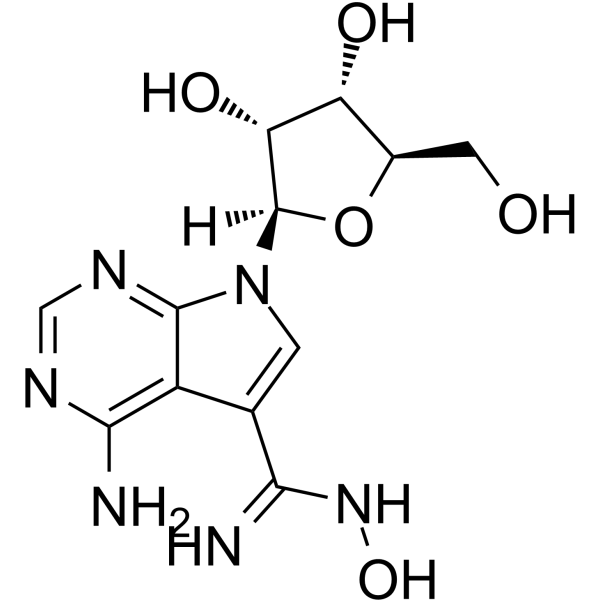
NSC 107512
NSC 107512 is a potent inhibitor of cyclin-dependent kinase 9 (CDK9). NSC 107512 is a class of sangivamycin-like molecules (SLM). NSC 107512 inhibits growth and induces apoptosis of multiple myeloma tumors.
-
GC12475
NSC 632839 hydrochloride
F6, Ubiquitin Isopeptidase Inhibitor II
A deubiquitylase and deSUMOylase inhibitor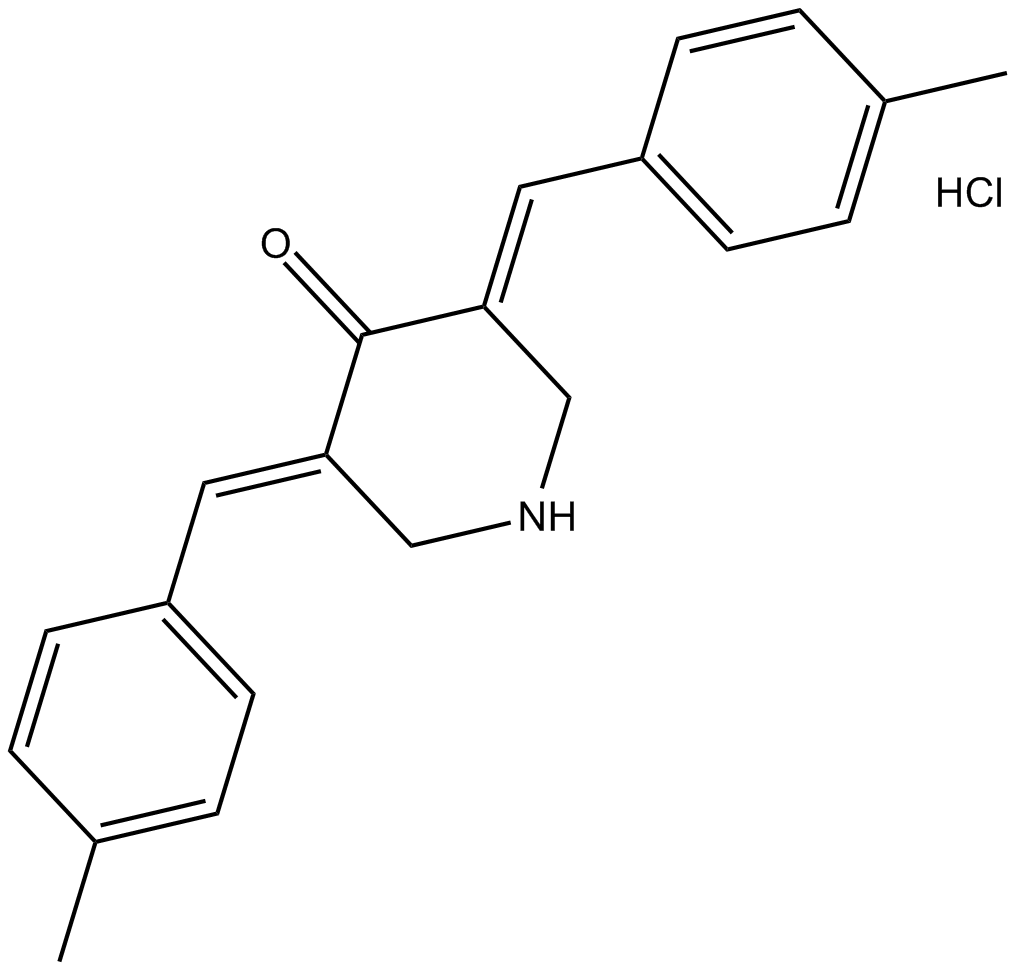
-
GC15503
NSC 687852 (b-AP15)
b-AP15
An inhibitor of the deubiquitinases USP14 and UCHL5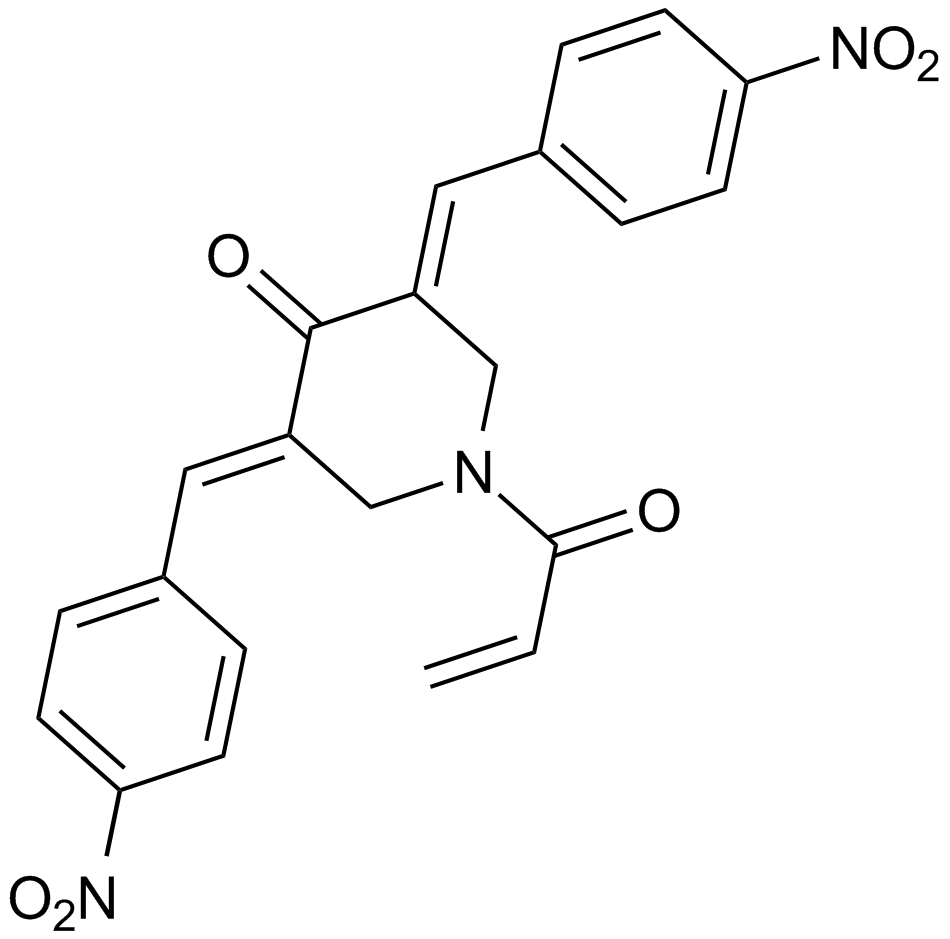
-
GC69598
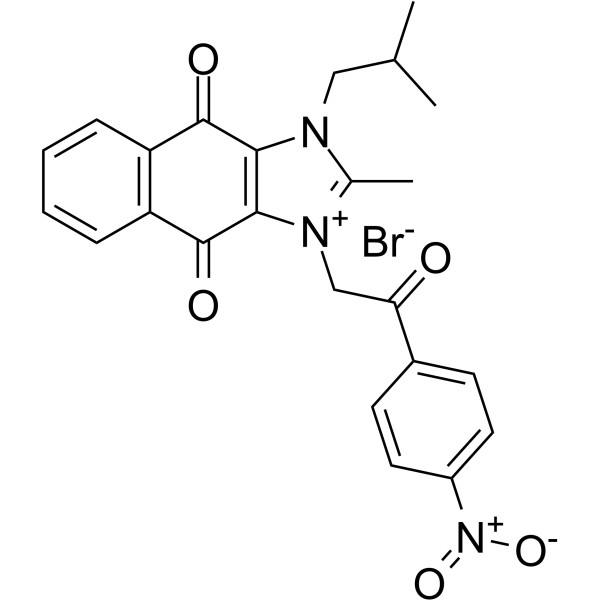
NSC 80467
NSC 80467 is a DNA damaging agent that selectively inhibits survivin. NSC 80467 preferentially inhibits DNA synthesis and induces two markers of DNA damage, γH2AX and pKAP1.
-
GC64103
NSC639828
NSC639828 is a potent inhibitor of DNA polymerase α with an IC50 of 70 μM. NSC639828 has high antitumor activity. NSC639828 has the potential for researching cancer disease.
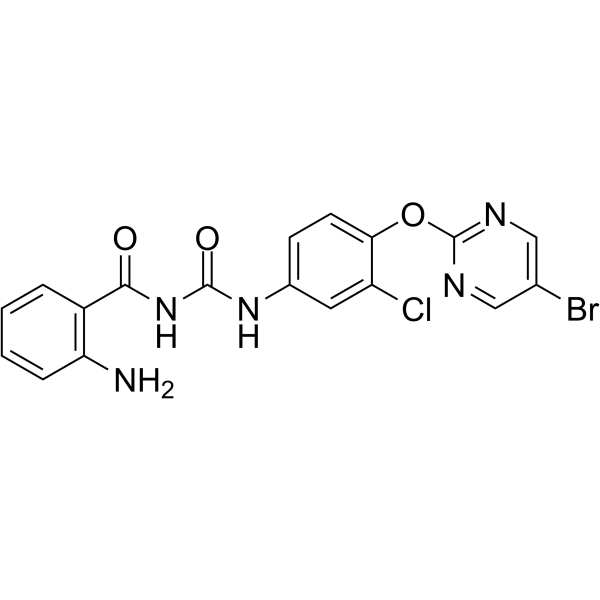
-
GC34111
NSC95682 (6-Bromo-2-hydroxy-3-methoxybenzaldehyde)
NSC95682 (6-Bromo-2-hydroxy-3-methoxybenzaldehyde) (NSC95682) is an IRE-1α inhibitor with an IC50 of 0.08 μM, extracted from patent WO 2008154484 A1, IRE-lα inhibitor compound 3-5.
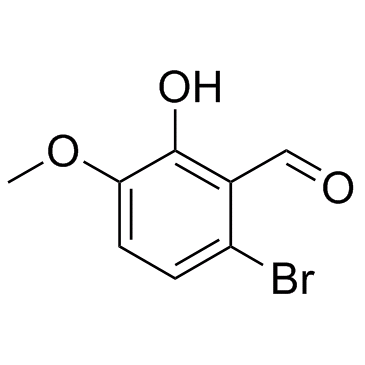
-
GC12332
NU 7026
DNAPK Inhibitor II, LY293646
DNPK inhibitor,ATP-competitive and potent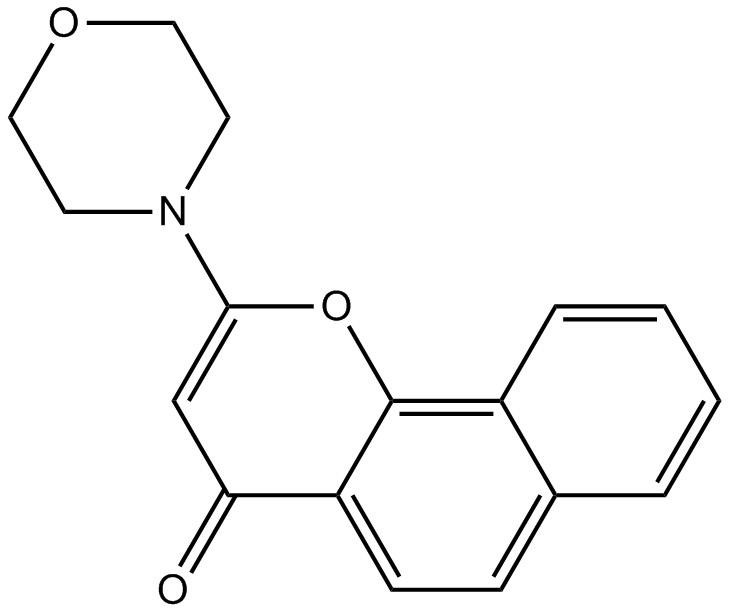
-
GC19267
NU2058
O6-(Cyclohexylmethyl)guanine
NU2058 is a guanine-based CDK inhibitor with IC50 of 17 uM and 26 uM for CDK2 and CDK1.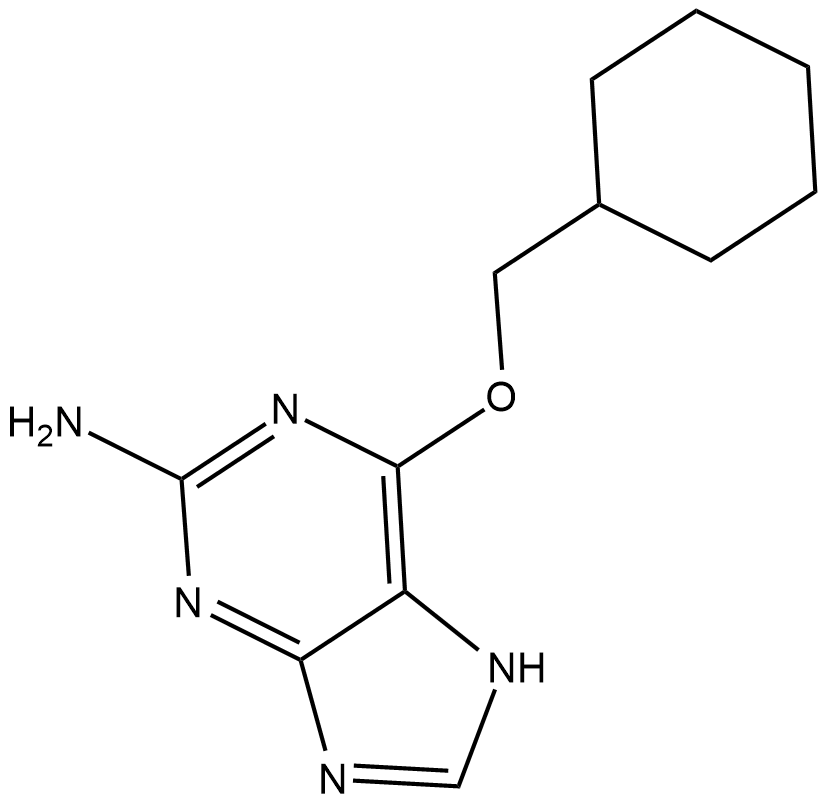
-
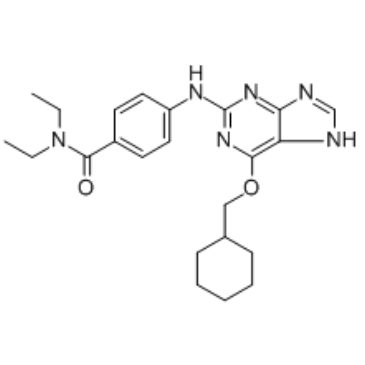
GC34692
NU6140
A Cdk2 inhibitor
-
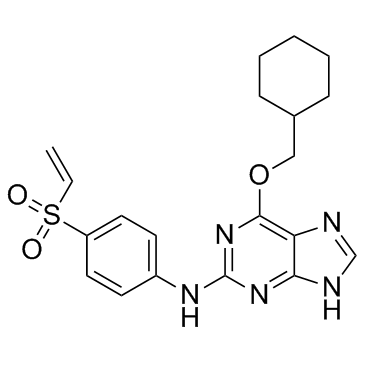
GC32829
NU6300
NU6300 is a covalent, irreversible and ATP-competitive CDK2 inhibitor with an IC50 value of 0.16 μM. NU6300 can be used for the research of eukaryotic cell cycle- and transcription-related.
-
GC11251
NU7441 (KU-57788)
KU 57788; NU-7441;KU57788;NU7441;NU 7441
NU7441 (KU-57788) (NU7441) is a highly potent and selective DNA-PK inhibitor with an IC50 of 14 nM. NU7441 (KU-57788) is an NHEJ pathway inhibitor. NU7441 (KU-57788) also inhibits PI3K and mTOR with IC50s of 5.0 and 1.7 μM, respectively.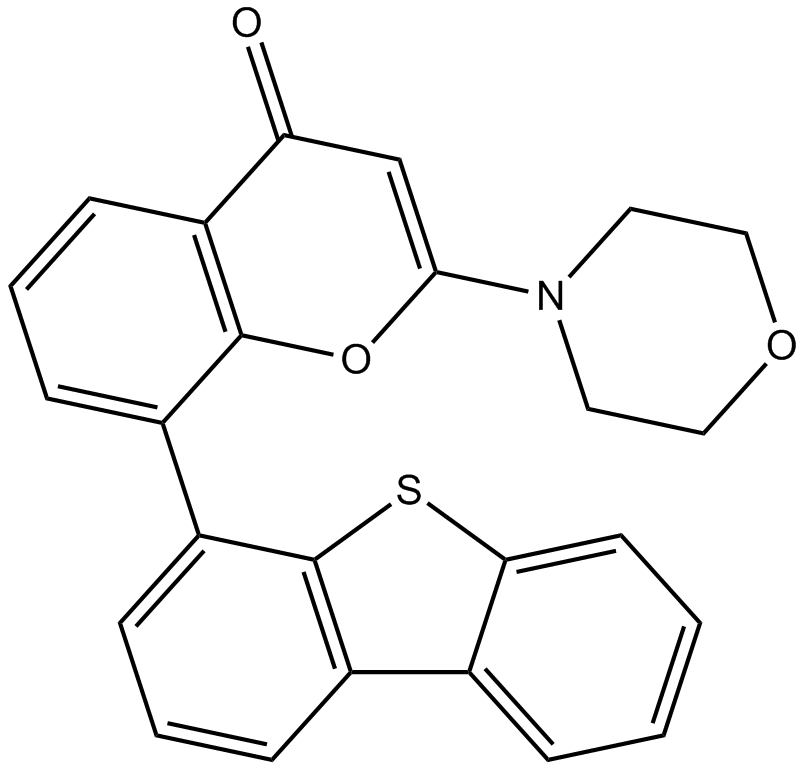
-
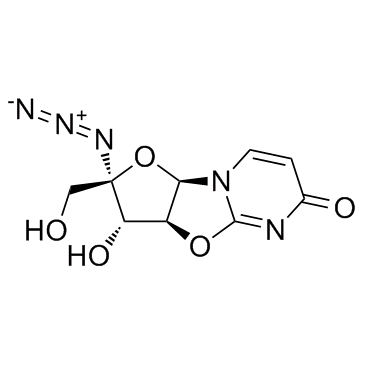
GC33956
Nucleoside-Analog-1
Nucleoside-Analog-1 is a 4′-Azidocytidine analogue against Hepatitis C virus replication.
-
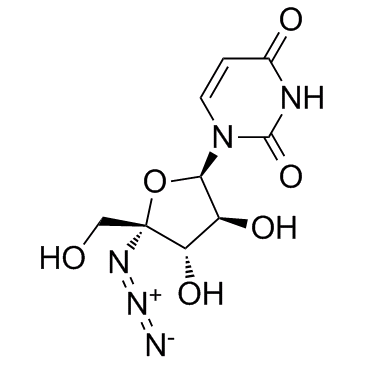
GC33959
Nucleoside-Analog-2
Nucleoside-Analog-2 is a 4'-Azidocytidine analogue against Hepatitis C virus (HCV) replication.
-
GC13925
Nullscript
negative control of scriptaid, HDAC inhibitor
-
GC64293
Nusinersen
Nusinersen is an antisense oligonucleotide drug that modifies pre–messenger RNA splicing of the SMN2 gene and thus promotes increased production of full-length SMN protein.

-
GC34056
NVP-2
NVP-2 is a potent and selective ATP-competitive cyclin dependent kinase 9 (CDK9) probe, inhibits CDK9/CycT activity with an IC50 of 0.514 nM. NVP-2 displays inhibitory effcts on CDK1/CycB, CDK2/CycA and CDK16/CycY kinases with IC50 values of 0.584 ?M, 0.706 ?M, and 0.605 ?M, respectively. NVP-2 induces cell apoptosis.
-
GC16959
NVP-LCQ195
-
GC17555
NVP-TNKS656
TNKS656
TNKS2 inhibitor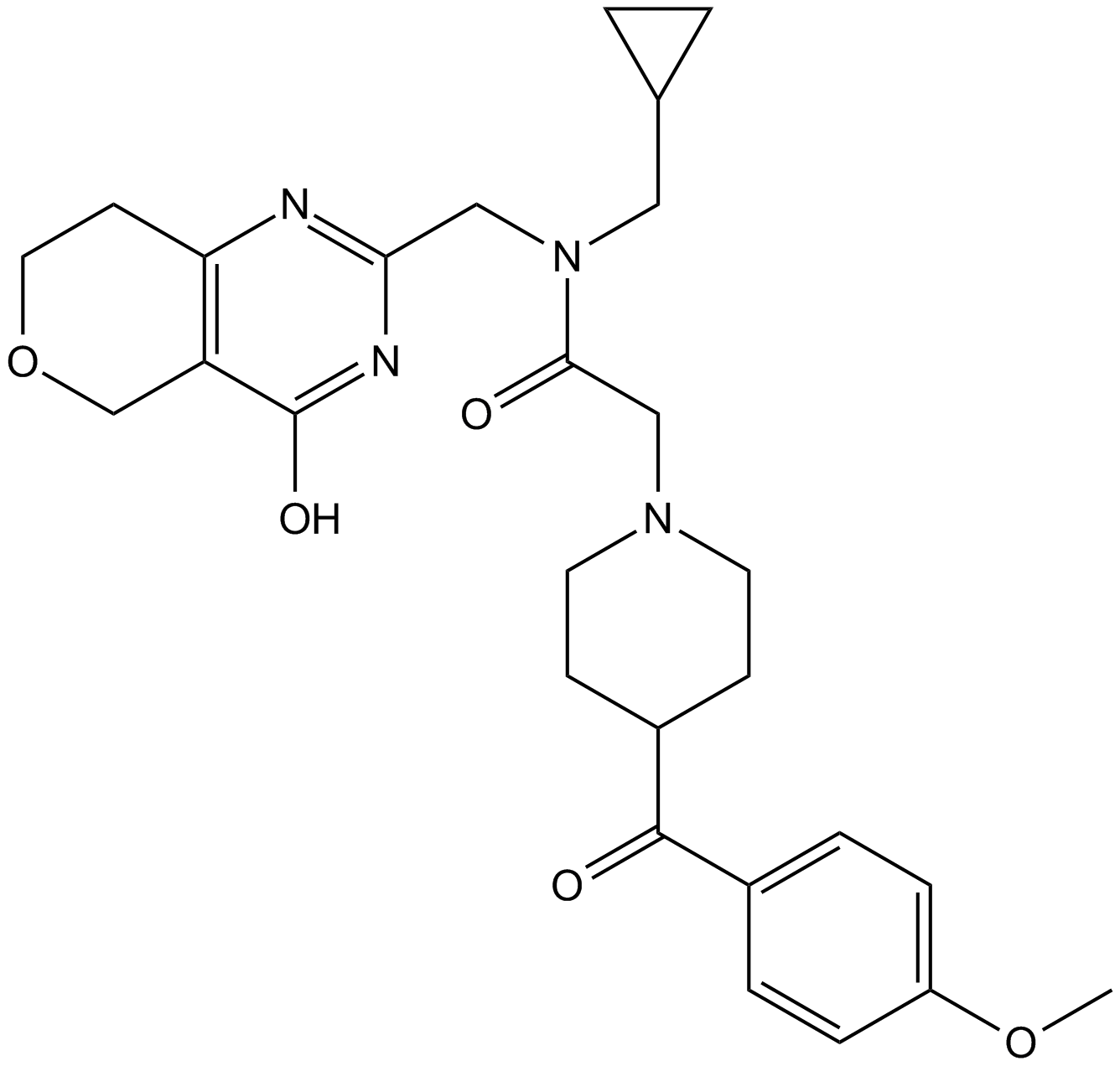
-
GC63123
NVS-SM2
NVS-SM2 is a potent, orally active and brain-penetrant SMN2 splicing enhancer with an EC50 of 2 nM for SMN.
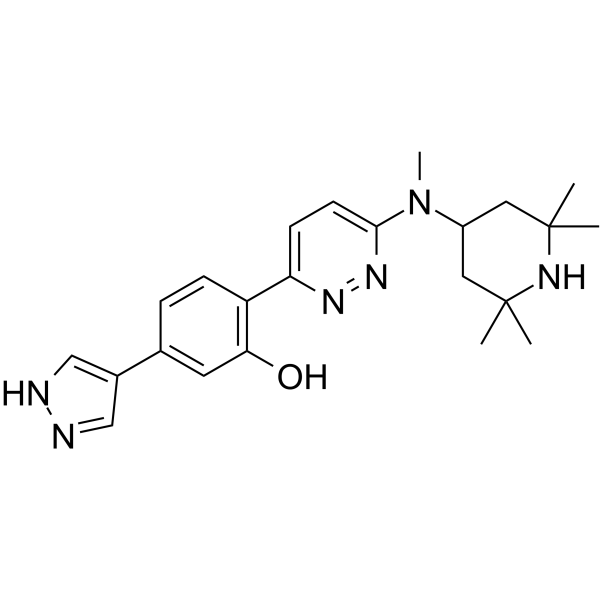
-
GC62501
OBI-3424
TH-3424
OBI-3424 (TH-3424) is a prodrug that is selectively converted by AKR1C3 (aldo-keto reductase 1C3) to a potent DNA-alkylating agent. OBI-3424 can be used for hepatocellular carcinoma, castrate-resistant prostate cancer, and acute lymphoblastic leukemia (ALL) research.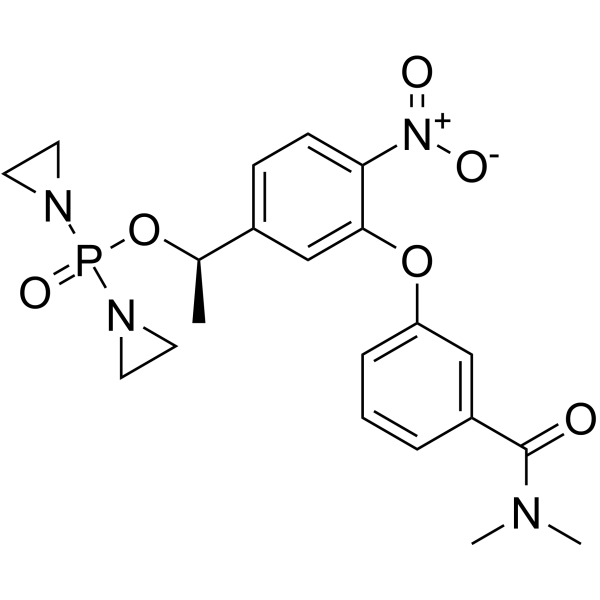
-
GC49743
Octapeptide-2
KLKKTETQ
A thymosin β4-derived peptide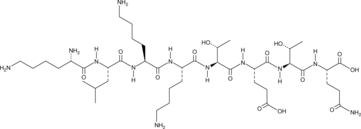
-
GC69618
Olaparib-d8
AZD2281-d8; KU0059436-d8
Olaparib-d8 is the deuterated form of Olaparib (AZD2281). Olaparib is an orally effective PARP inhibitor that inhibits PARP-1 and PARP-2 with IC50 values of 5 and 1 nM, respectively. Olaparib is also an activator of autophagy and mitophagy.
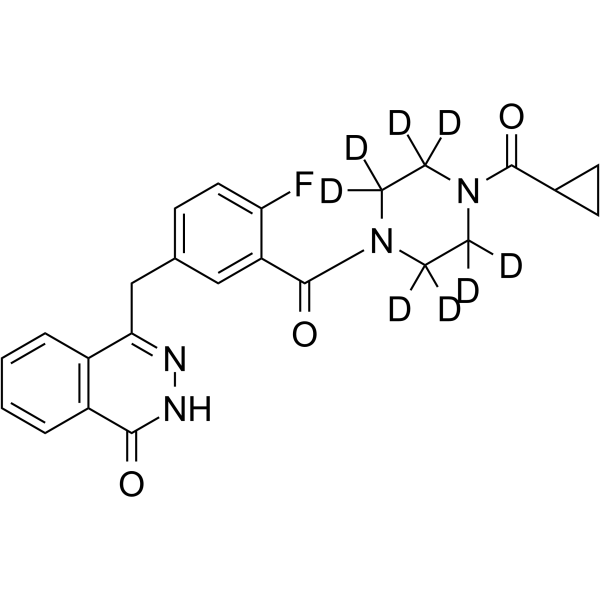
-
GC67906
OM-153

-
GC33353
ON-013100
ON-013100, an antineoplastic drug, acts a mitotic inhibitor that could inhibit Cyclin D1 expression.
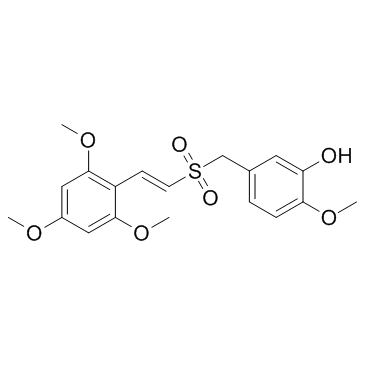
-
GC15607
ON123300
multi-targeted kinase inhibitor,inhibits CDK4, Ark5, PDGFRβ, FGFR1, RET, and Fyn
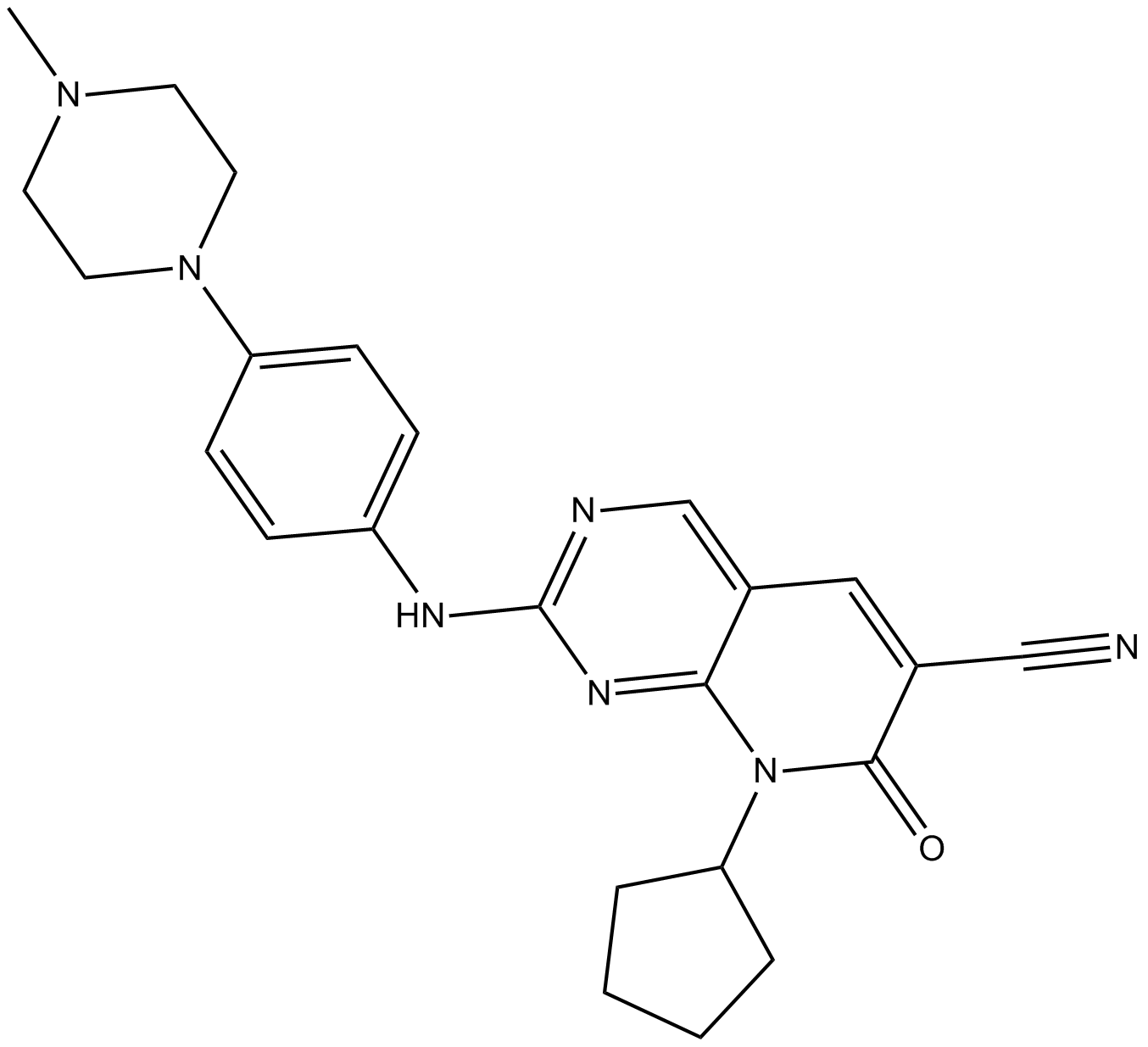
-
GC69633
Orobol
Orobol is a major soy isoflavone with various pharmacological activities, including anti-aging and anti-obesity effects. Orobol inhibits CK1ε, VEGFR2, MAP4K5, MNK1, MUSK, TOPK and TNIK (IC50=1.24-4.45 μM). Orobol also inhibits PI3K subtypes (for PI3K α/β/γ/K/δ, IC50=3.46-5.27 μM).
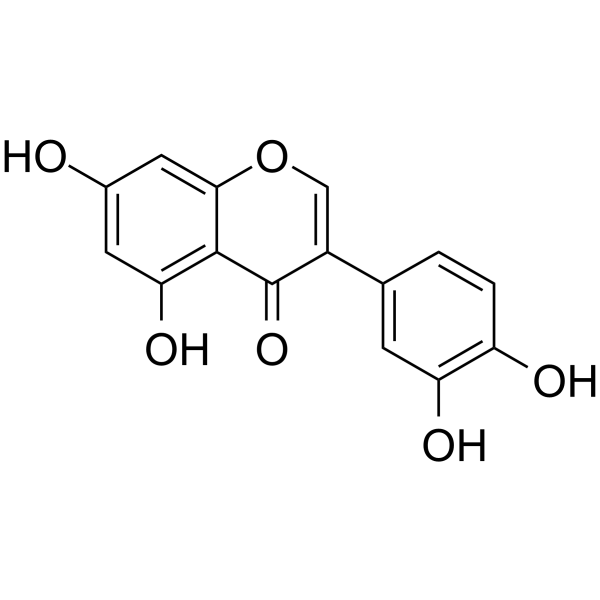
-
GC11528
Orotic acid
Pyrimidinecarboxylic acid
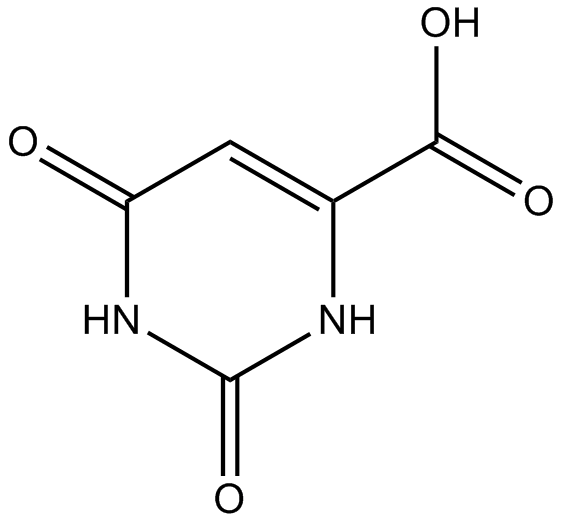
-
GC64296
Orotidine 5′-monophosphate trisodium
Orotidine monophosphate trisodium; Orotidylic acid trisodium
Orotidine 5'-monophosphate trisodium is a pyrimidine nucleotide.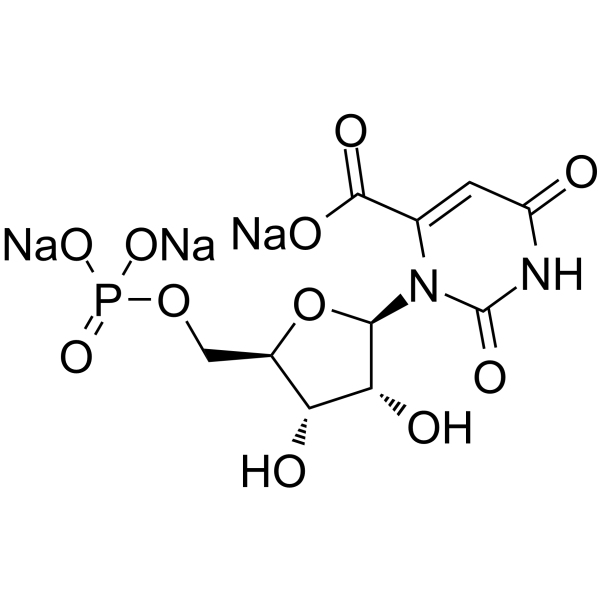
-
GC11360
ORY-1001
Iadademstat, RG-6016
Selective inhibitor of KDM1A.
-
GC12304
OTS514
TOPK inhibitor,highly potent
-
GC16511
OTS964
OTS964 is an orally active, high affinity and selective TOPK (T-lymphokine-activated killer cell-originated protein kinase) inhibitor with an IC50 of 28 nM. OTS964 is also a potent inhibitor of the cyclin-dependent kinase CDK11, which binds to CDK11B with a Kd of 40 nM.
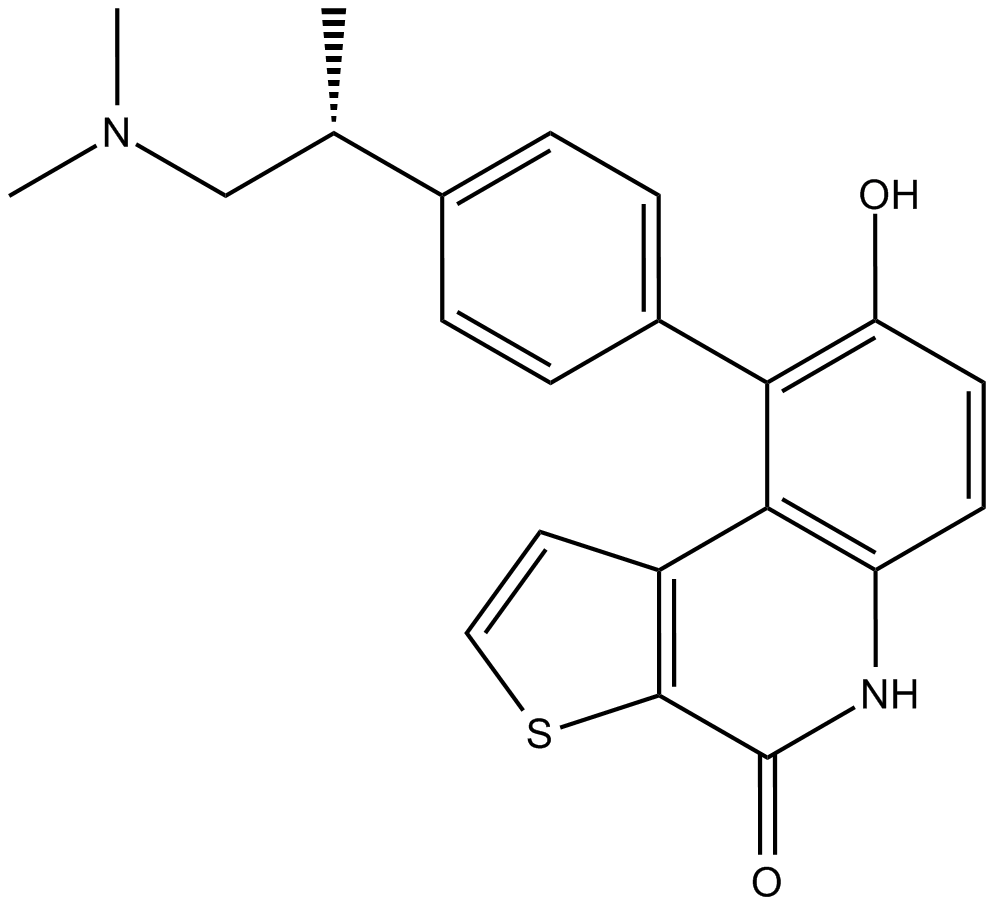
-
GC69637
OTUB1/USP8-IN-1
OTUB1/USP8-IN-1 is an effective dual inhibitor of OTUB1 and USP8, with IC50 values of 0.17 and 0.28 nM, respectively. It can be used for cancer research.
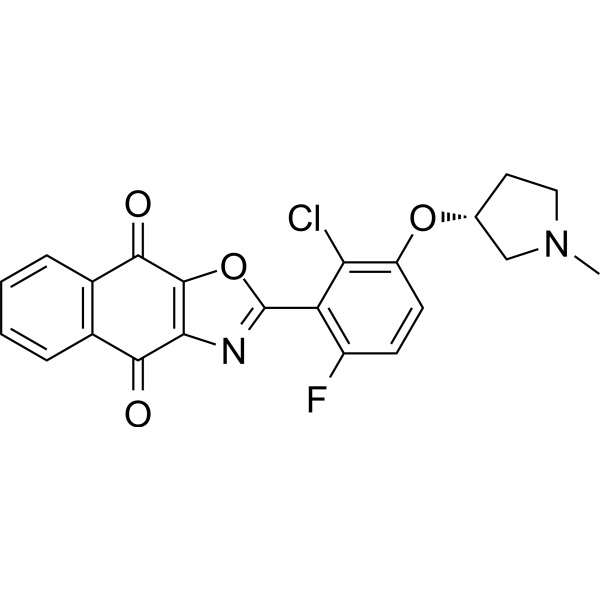
-
GC17716
Oxaliplatin
Lipoxal, NSC 266046, RP 54780
Oxaliplatin is a cytotoxic chemotherapy drug used to treat cancer.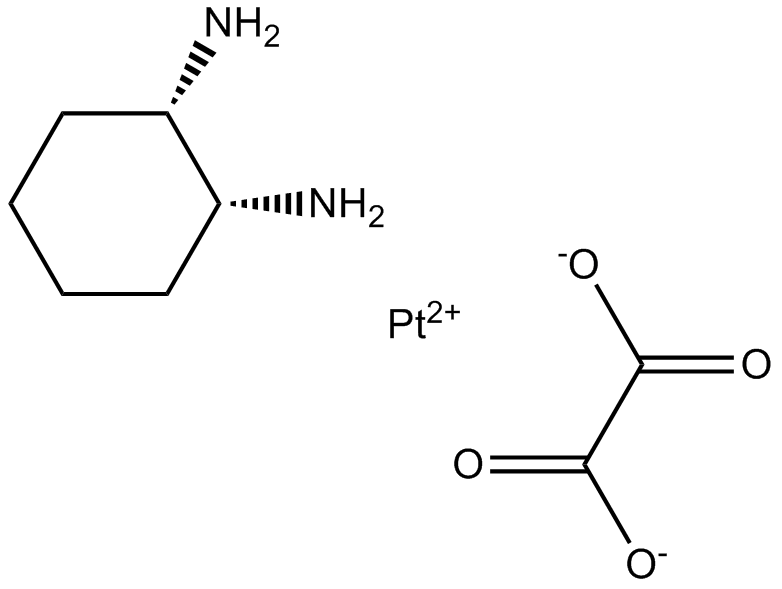
-
GC49251
Oxaliplatin-d10
Lipoxal-d10
An internal standard for the quantification of oxaliplatin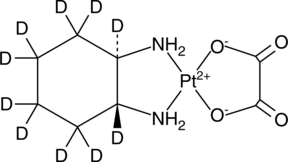
-
GC17472
Oxamflatin
Metacept 3
HADC inhibitor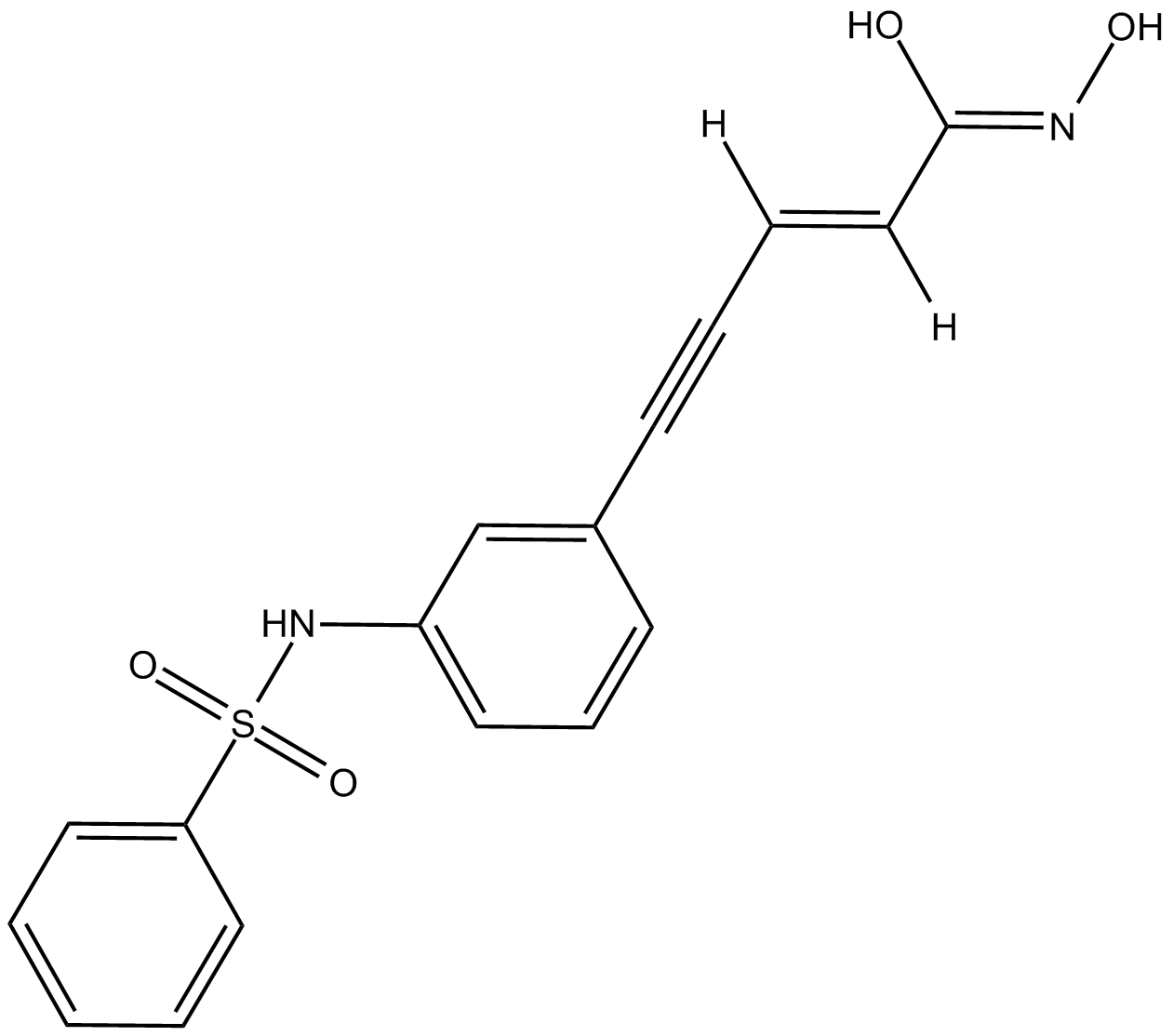
-
GC14107
Oxolinic acid
NSC 110364, Urinox
quinolone antibiotic that inhibits bacterial DNA gyrase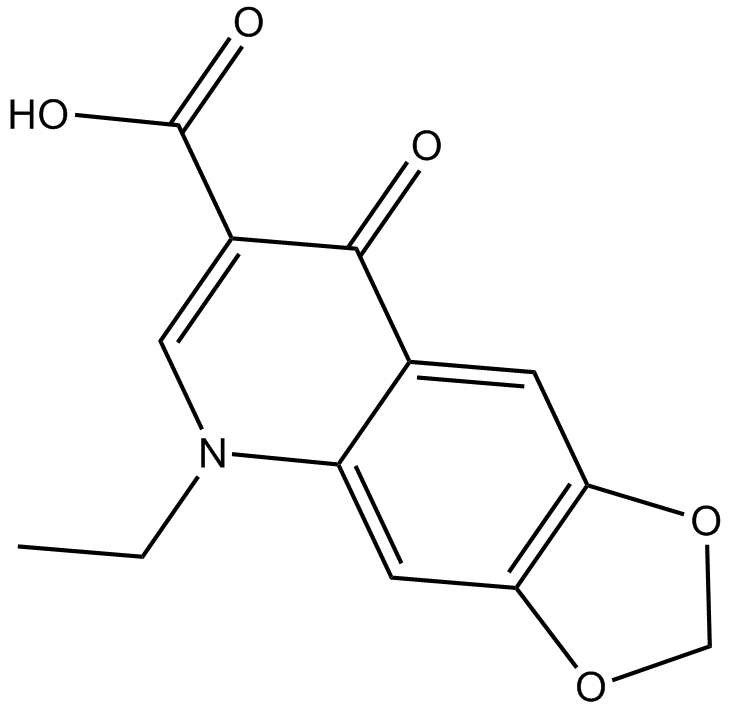
-
GC10379
P 22077
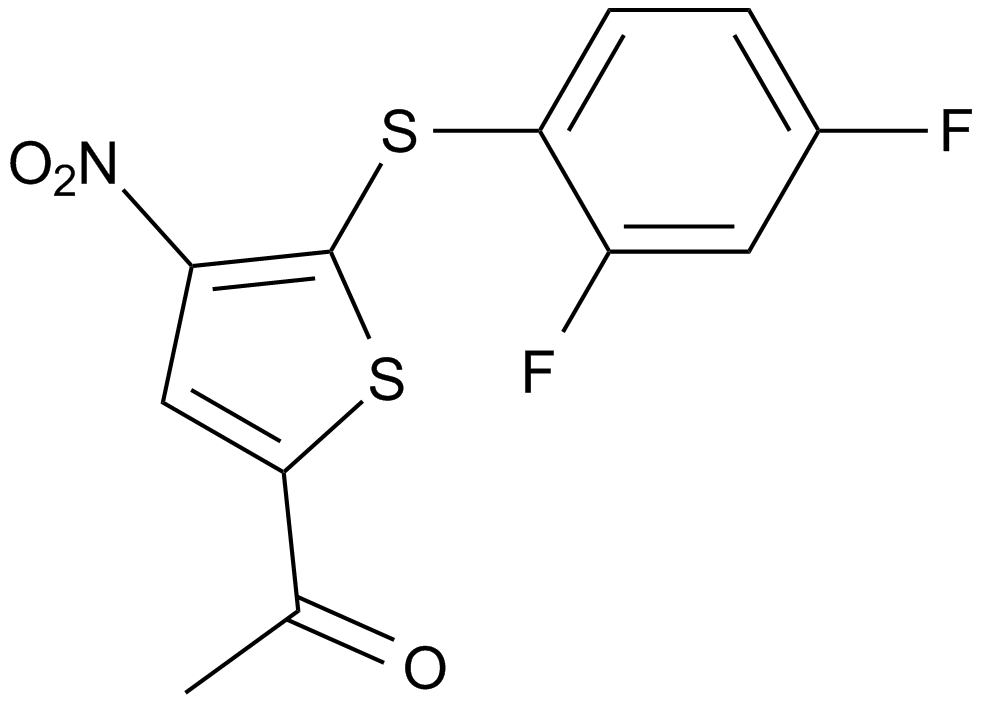
-
GC12067
P005091
P005091,P5091
P005091 is a potent and selective ubiquitin-specific proteinase 7 (USP7) inhibitor with an EC50 value of 4.2 μM.
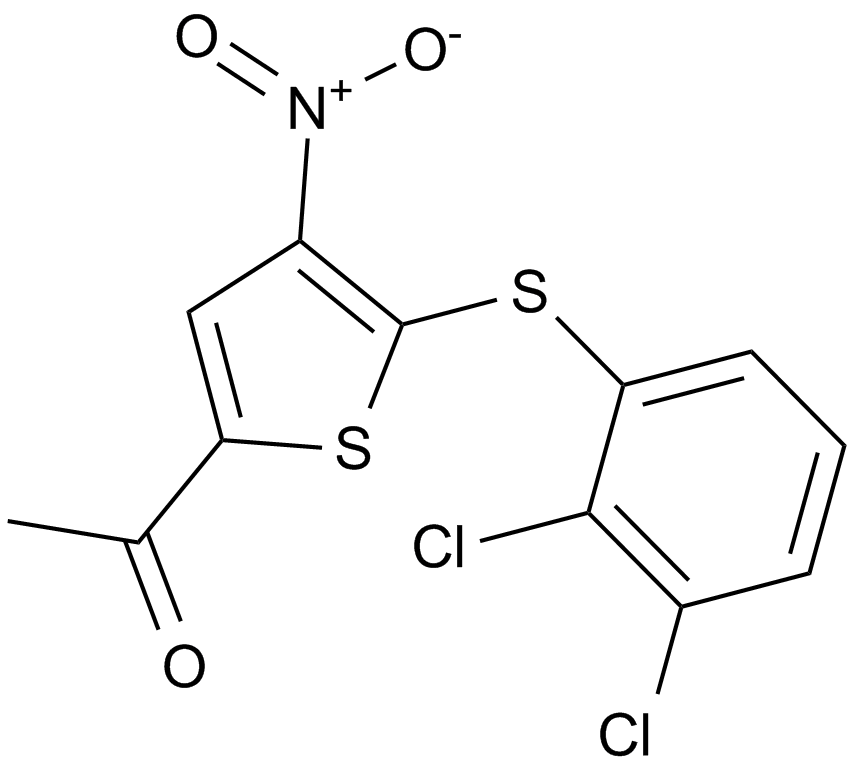
-
GC17232
P005672 hydrochloride
Sarecycline hydrochloride
P005672 hydrochloride is a narrow-spectrum tetracycline-class antibiotic.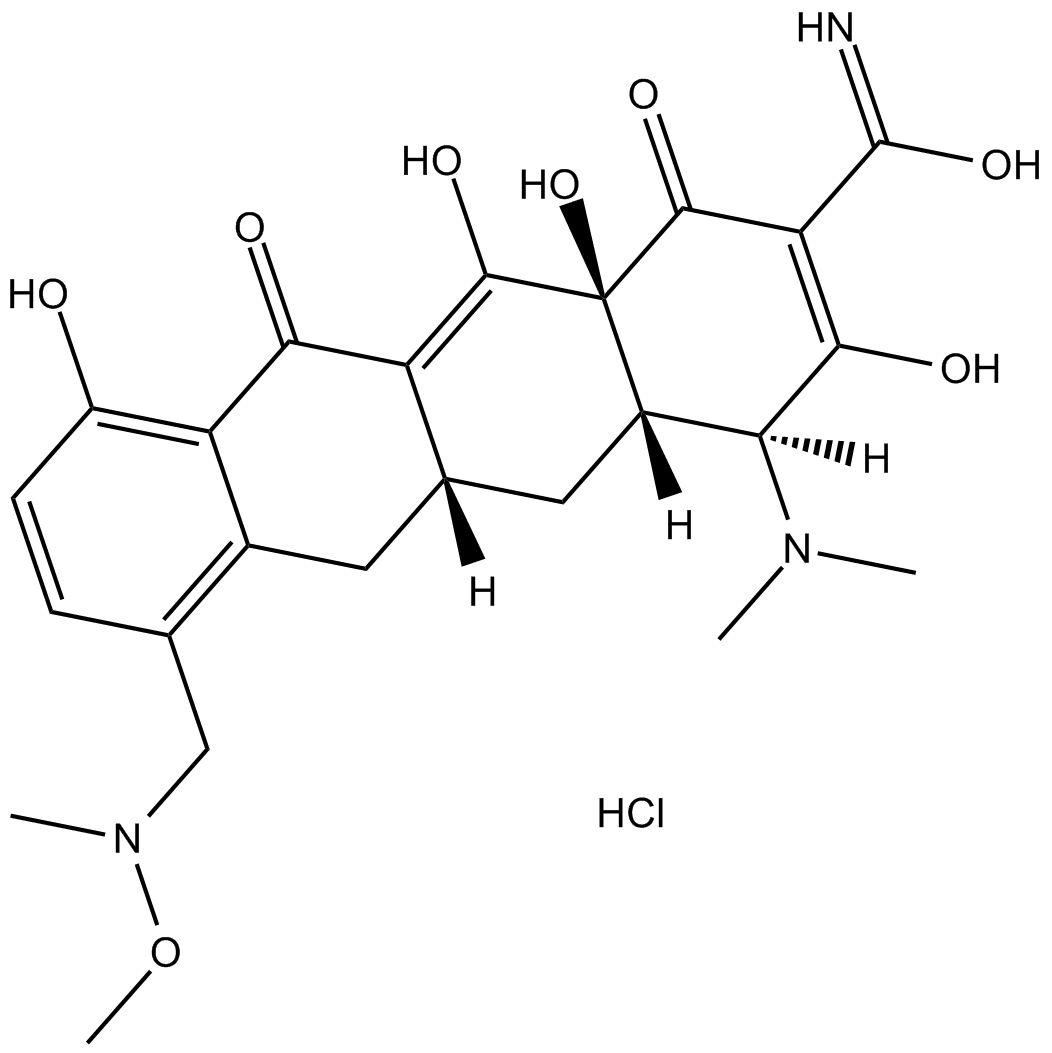
-
GC12011
P276-00
P276-00
P276-00 (P276-00) is a potent cyclin-dependent kinase (CDK) inhibitor, which inhibits CDK9-cyclinT1, CDK4-cyclin D1, and CDK1-cyclinB with IC50s of 20 nM, 63 nM, and 79 nM, respectively.P276-00 (P276-00) shows antitumor activity on cisplatin-resistant cells.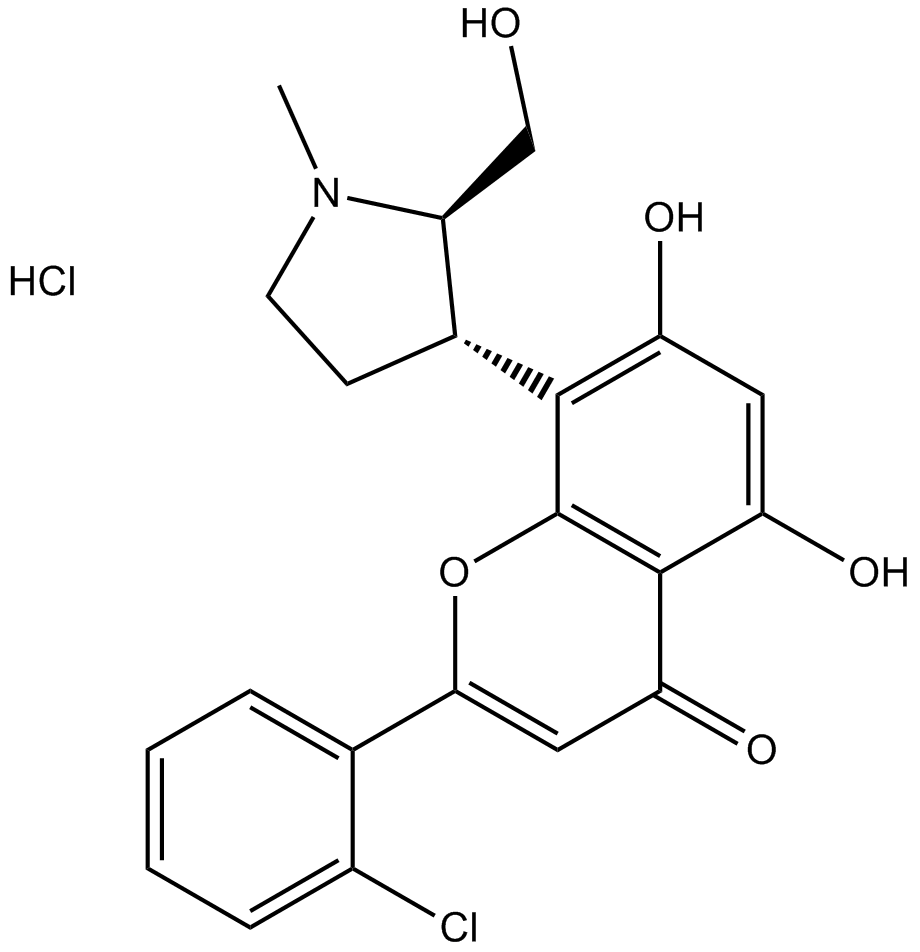
-
GC15421
Palbociclib (PD0332991) Isethionate
Palbociclib Isethionate
Palbociclib (PD 0332991) isethionate is an orally active selective CDK4 and CDK6 inhibitor with IC50 values of 11 and 16 nM, respectively. Palbociclib (PD0332991) Isethionate has potent anti-proliferative activity and induces cell cycle arrest in cancer cells, which can be used in the research of HR-positive and HER2-negative breast cancer and hepatocellular carcinoma.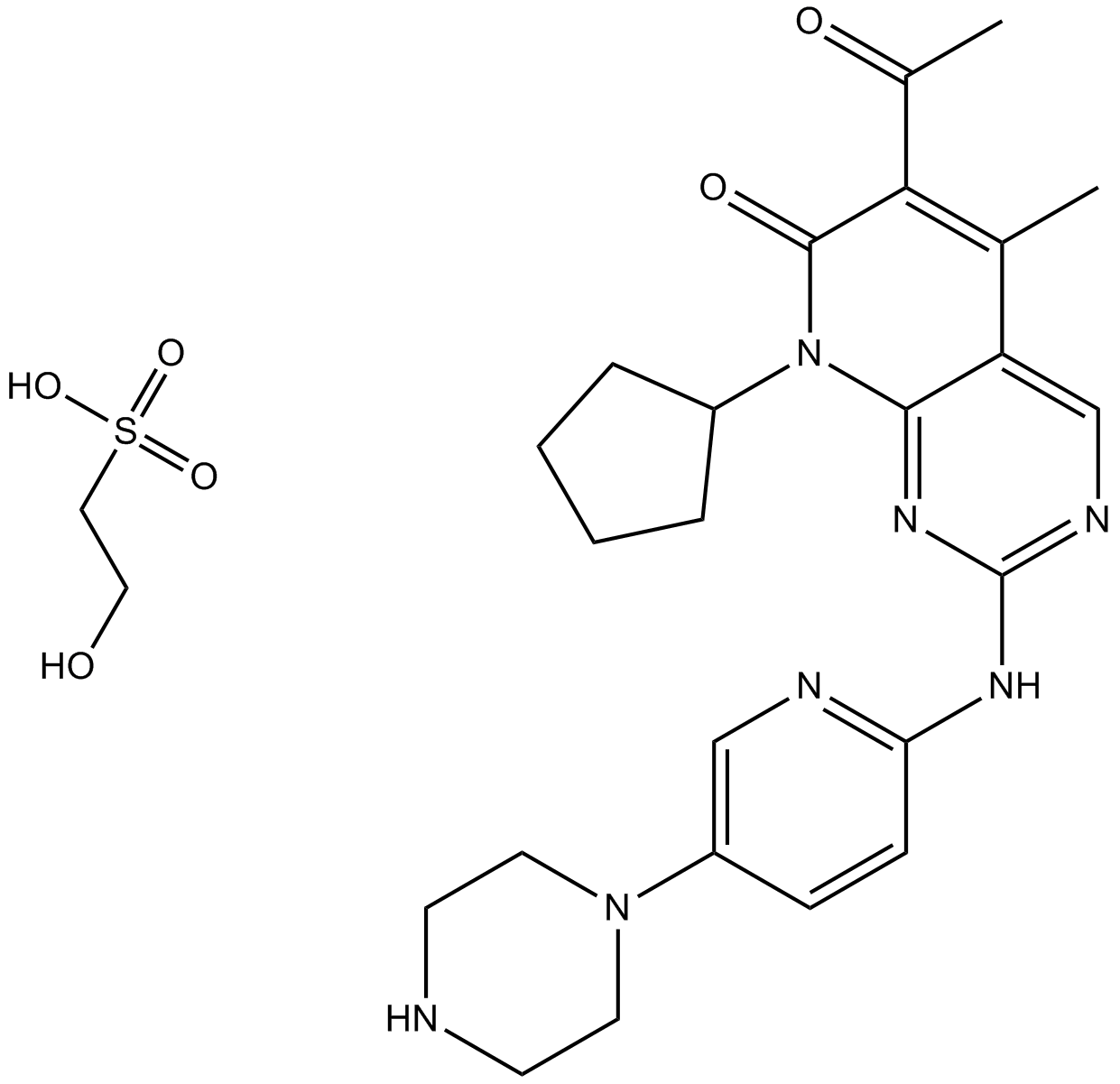
-
GC14762
Palifosfamide
Isophosphoramide Mustard, NSC 297900
An active metabolite of ifosfamide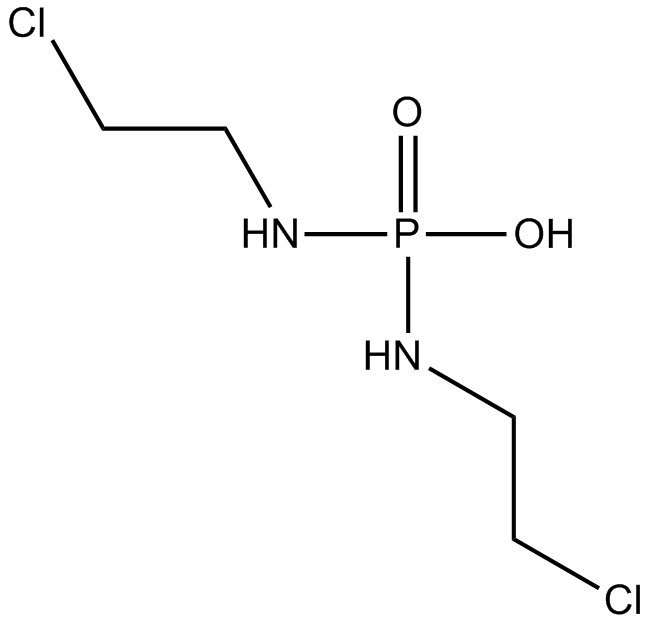
-
GC34071
Pamiparib (BGB-290)
BGB-290
Pamiparib (BGB-290) (BGB-290) is an orally active, potent, highly selective PARP inhibitor, with IC50 values of 0.9 nM and 0.5 nM for PARP1 and PARP2, respectively. Pamiparib (BGB-290) has potent PARP trapping, and capability to penetrate the brain, and can be used for the research of various cancers including the solid tumor.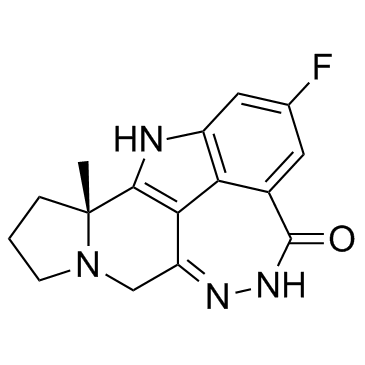
-
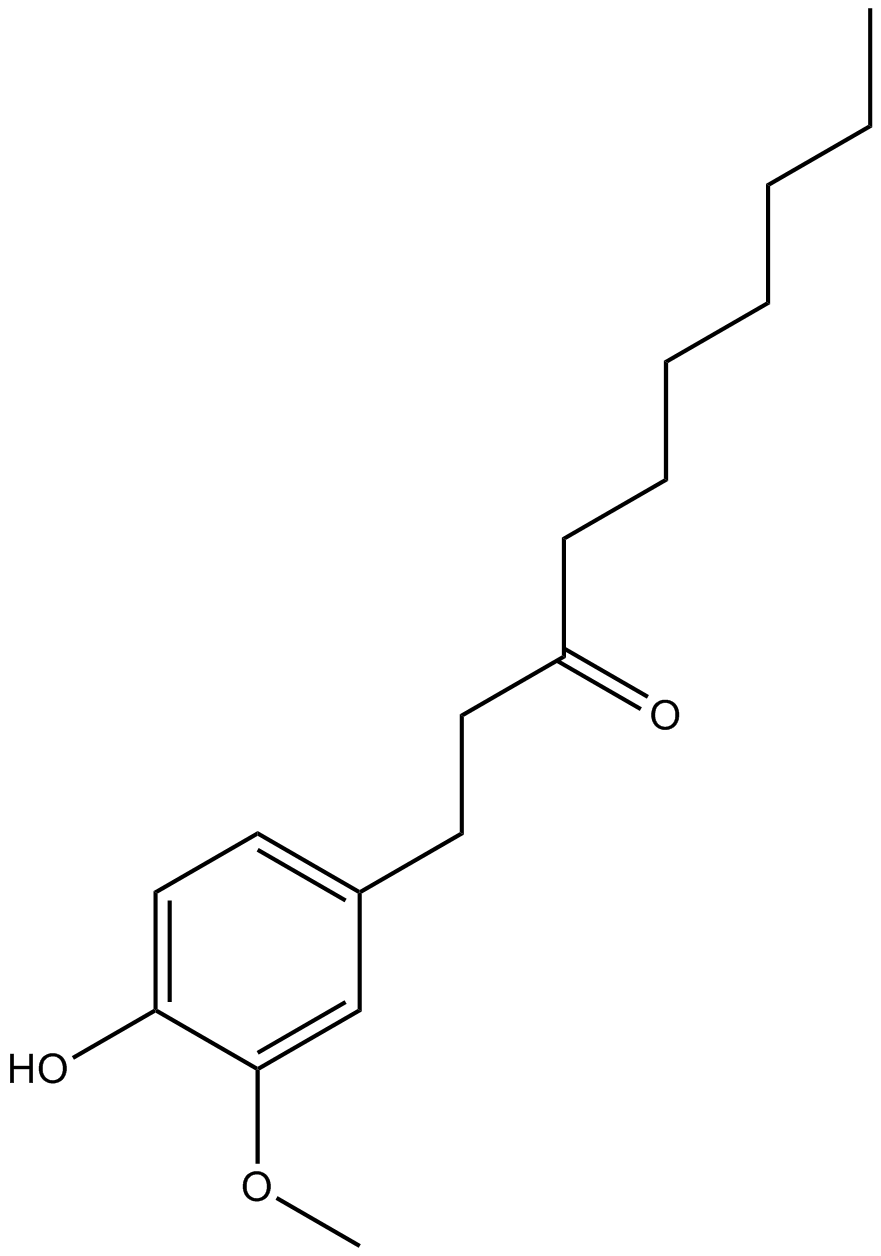
GC14849
Paradol
A phenolic ketone with diverse biological activities
-
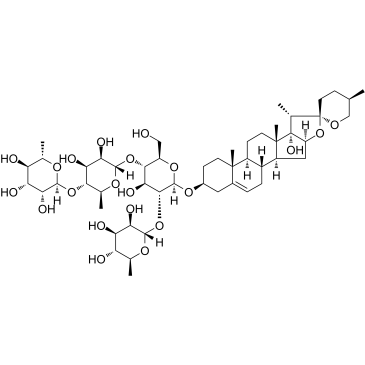
GC36855
Paris saponin VII
Paris saponin VII (Chonglou Saponin VII) is a steroidal saponin isolated from the roots and rhizomes of Trillium tschonoskii Maxim. Paris saponin VII-induced apoptosis in K562/ADR cells is associated with Akt/MAPK and the inhibition of P-gp. Paris saponin VII attenuates mitochondrial membrane potential, increases the expression of apoptosis-related proteins, such as Bax and cytochrome c, and decreases the protein expression levels of Bcl-2, caspase-9, caspase-3, PARP-1, and p-Akt. Paris saponin VII induces a robust autophagy in K562/ADR cells and provides a biochemical basis in the treatment of leukemia.
-
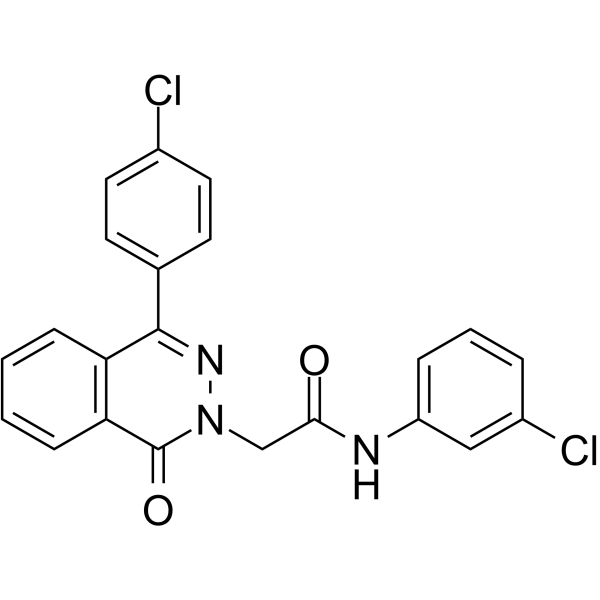
GC64579
PARP-1-IN-2
PARP-1-IN-2 (compound 11g) is a potent and BBB-penetrated PARP1 inhibitor, with an IC50 of 149 nM. PARP1-IN-2 shows significantly potent anti-proliferative activity against Human lung adenocarcinoma epithelial cell line A549. PARP1-IN-2 can induce A549 cells apoptosis.
-
GC65927
PARP-2-IN-1
PARP-2-IN-1 is a potent and selective PARP-2 inhibitor with an IC50 of 11.5 nM.
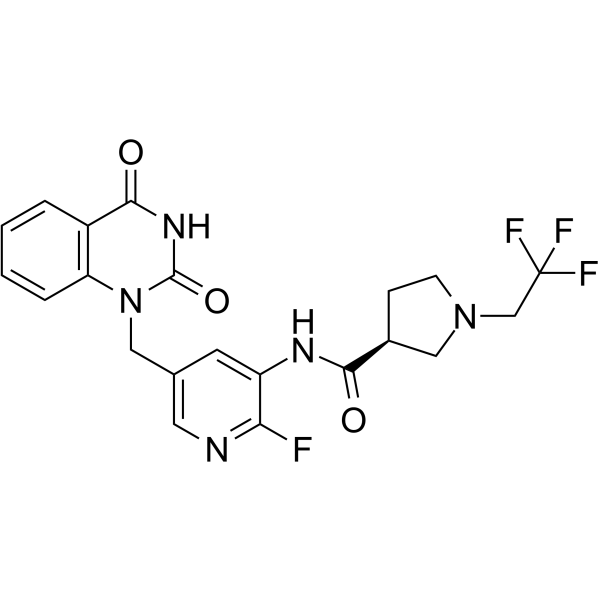
-
GC68006
PARP1-IN-11
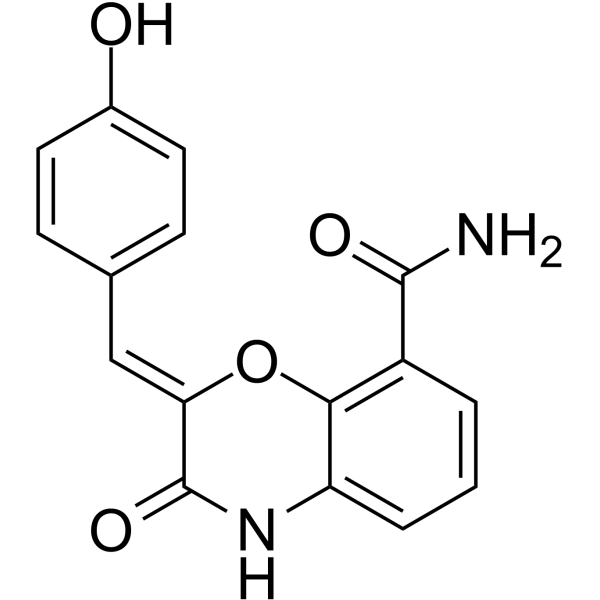
-
GC69660
PARP1-IN-7
PARP1-IN-7 is an inhibitor of poly ADP-ribose polymerase-1 (PARP1), used as an anticancer agent.
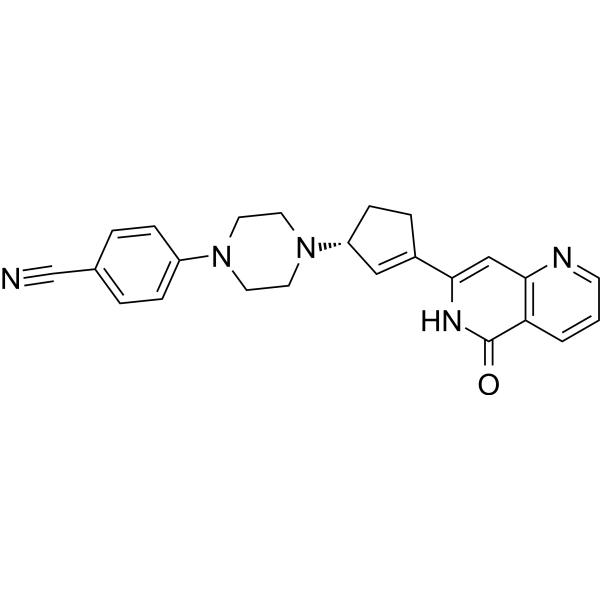
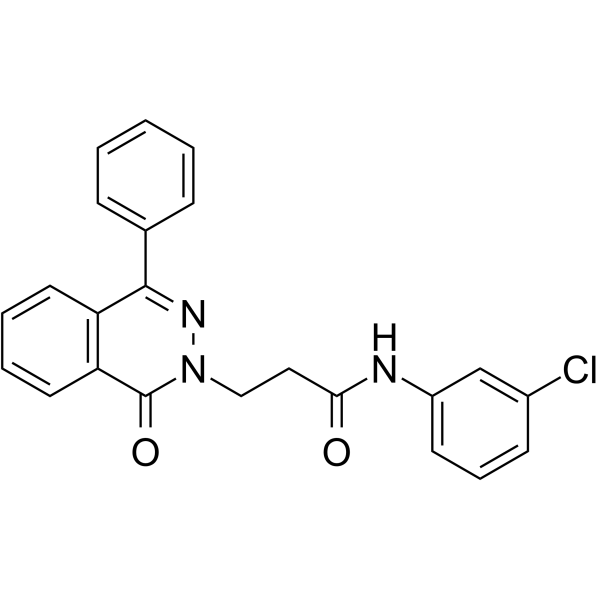
-
GC64578
PARP1-IN-8
PARP1-IN-8 (compound 11c) is a potent and BBB-penetrated PARP1 inhibitor, with an IC50 of 97 nM. PARP1-IN-8 shows significantly potent anti-proliferative activity against Human lung adenocarcinoma epithelial cell line A549.
-
GC69657
PARP10-IN-2
PARP10-IN-2 is an effective inhibitor of mono-ADP-ribosyltransferase PARP10, with an IC50 of 3.64 μM for human PARP10. It also inhibits PARP2 and PARP15, with IC50 values of 27 μM and 11 μM for human PARP2 and human PARP15, respectively.
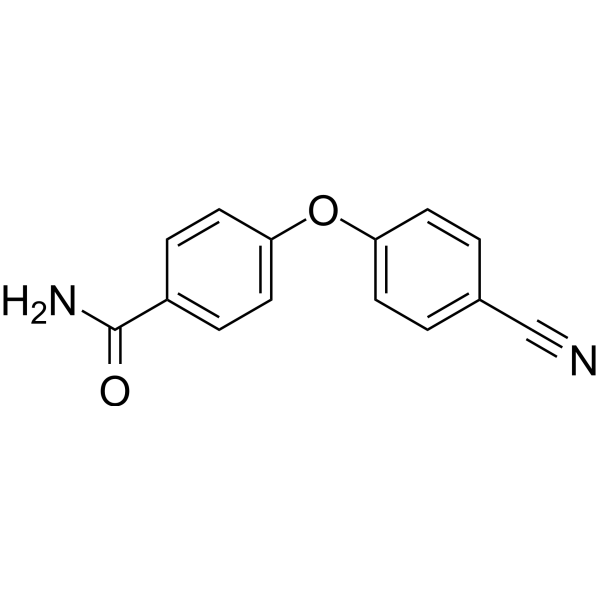
-
GC69658
PARP10-IN-3
PARP10-IN-3 is a selective mono-ADP-ribosyltransferase PARP10 inhibitor with an IC50 of 480 nM for human PARP10. It also inhibits PARP2 and PARP15, with IC50 values of 1.7 μM for both human PARP2 and human PARP15.
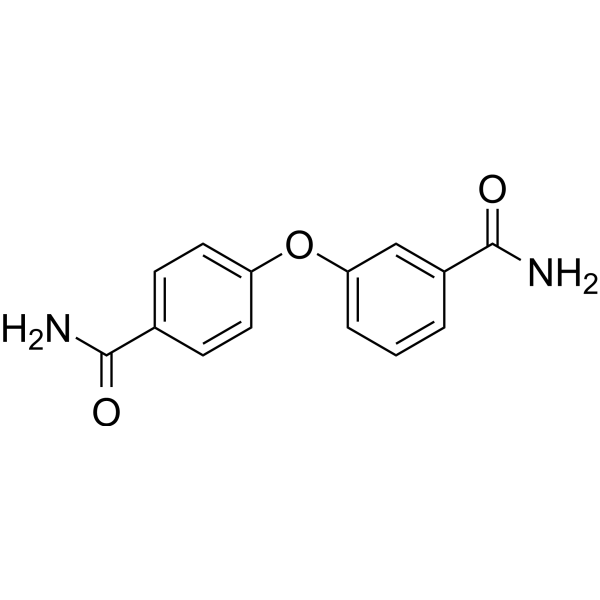
-
GC68035
PARP10/15-IN-1
-
GC69655
PARP10/15-IN-2
PARP10/15-IN-2 (Compound 8h) is an effective dual inhibitor of PARP10 and PARP15, with IC50 values of 0.15 μM and 0.37 μM, respectively. It can enter cells and prevent apoptosis.
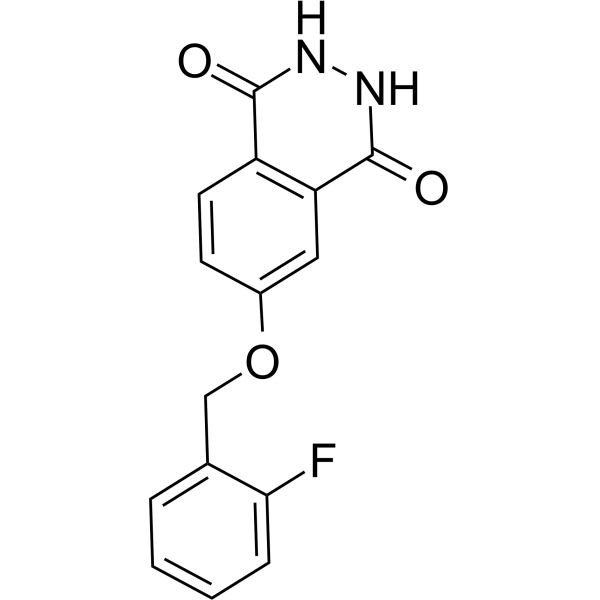
-
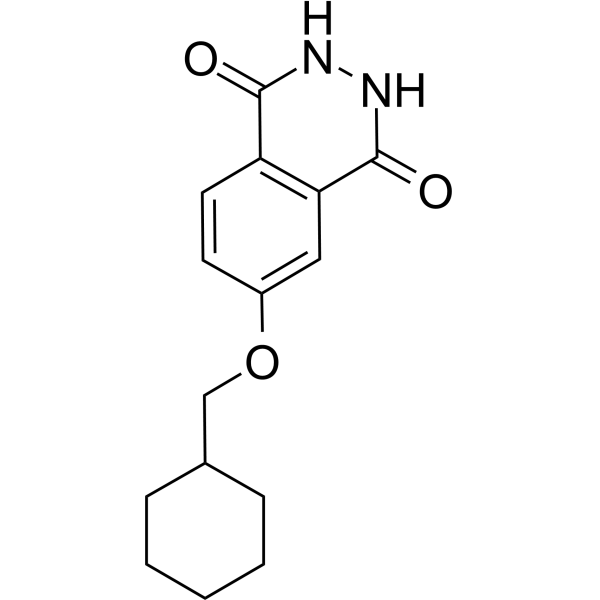
GC69656
PARP10/15-IN-3
PARP10/15-IN-3 (Compound 8a) is an effective dual inhibitor of PARP10 and PARP15, with IC50 values of 0.14 μM and 0.40 μM, respectively. It can enter cells and prevent apoptosis.
-
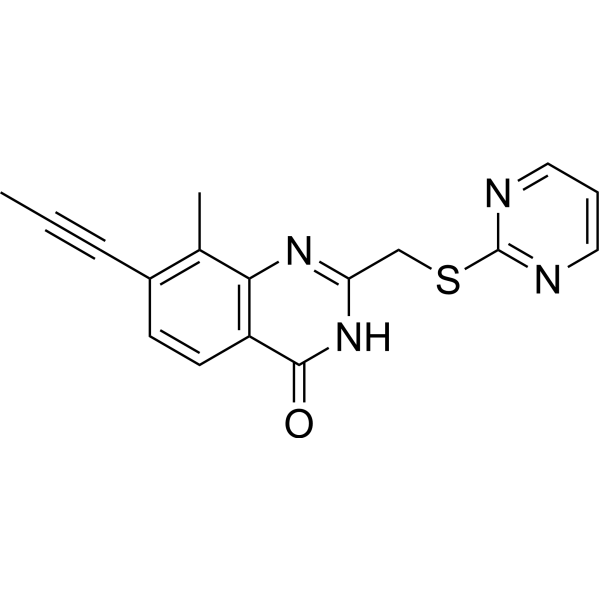
GC69659
PARP11 inhibitor ITK7
ITK7
PARP11 inhibitor ITK7 (ITK7) is an effective and selective inhibitor of PARP11. It can effectively inhibit PARP11 with an IC50 value of 14 nM. PARP11 inhibitor ITK7 can be used for research on cellular localization.
-
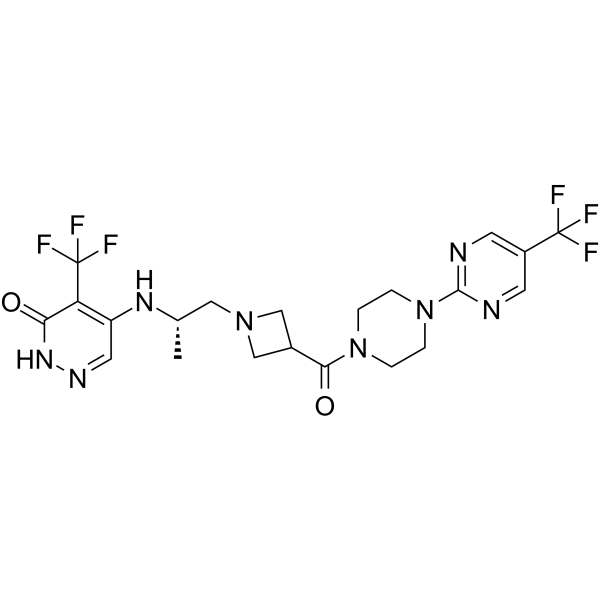
GC69661
PARP7-IN-14
PARP7-IN-14 (I-1) is an effective selective PARP7 inhibitor with an IC50 value of 7.6 nM. PARP7-IN-14 has anti-cancer activity.
-
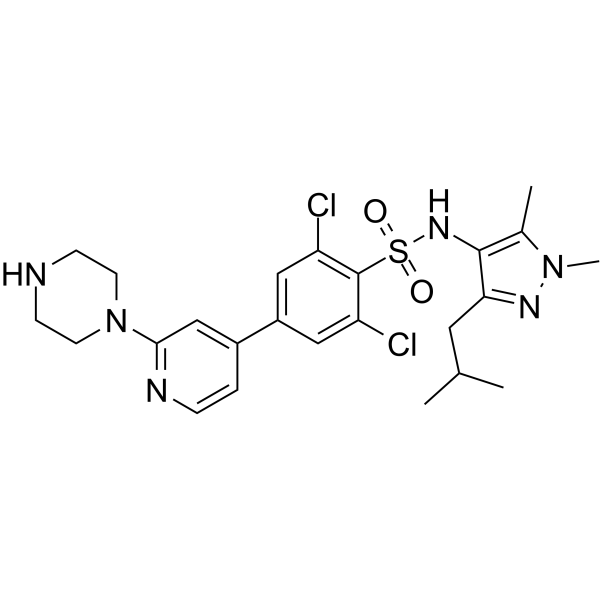
GC65891
PCLX-001
PCLX-001 is an orally acitve, small-molecule, dual N-myristoyltransferase (NMT) inhibitor, with IC50s of 5 nM (NMT1) and 8 nM (NMT2), respectively. PCLX-001 exhibits anti-tumor activity and inhibits early B-cell receptor (BCR) signaling, can be used to B-cell malignancies research.
-
GC17935
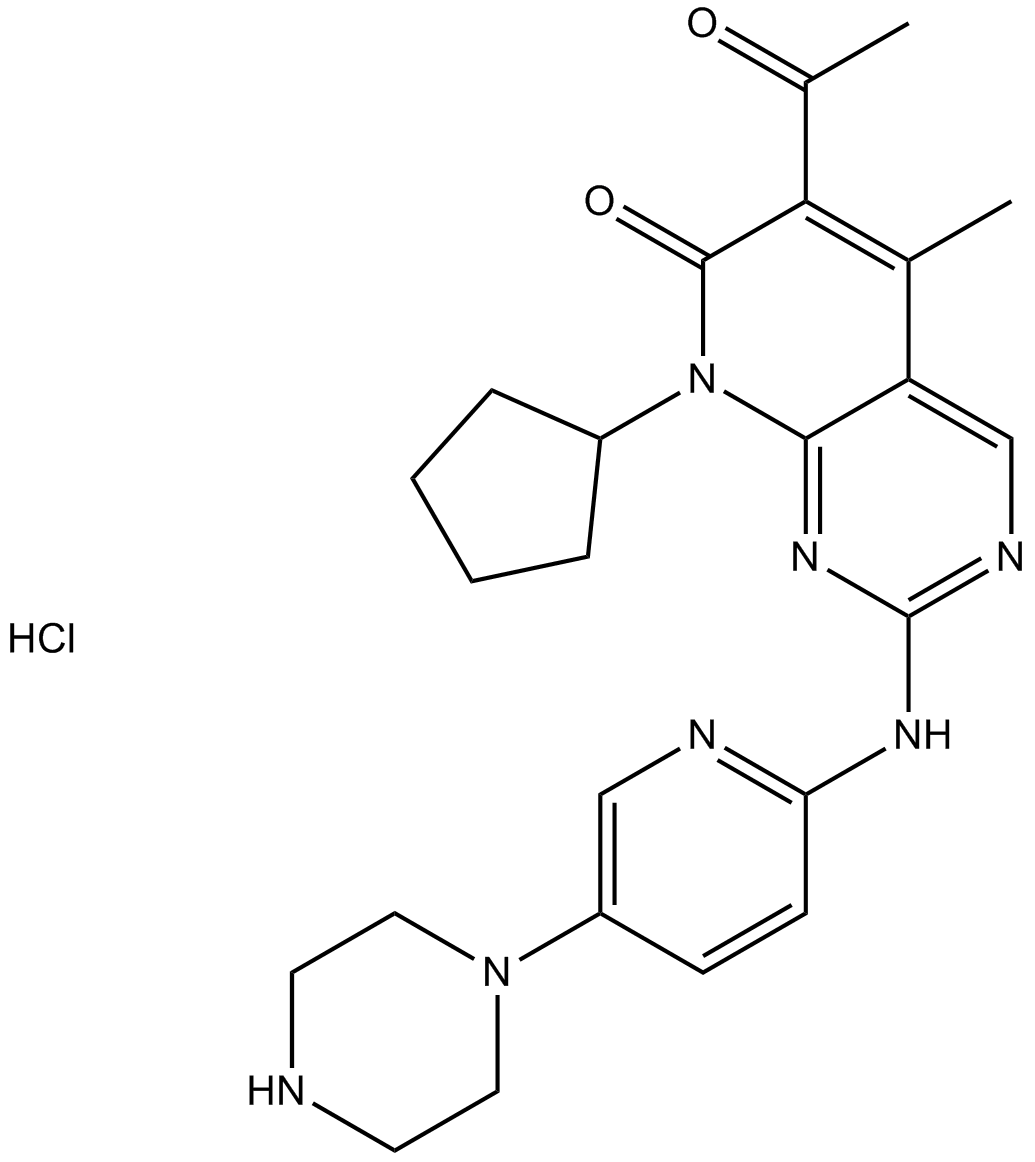
PD 0332991 (Palbociclib) HCl
PD0332991;PD-0332991;PD 0332991
Palbociclib (PD 0332991) monohydrochloride is an orally active selective CDK4 and CDK6 inhibitor with IC50 values of 11 and 16 nM, respectively. PD 0332991 (Palbociclib) HCl has potent anti-proliferative activity and induces cell cycle arrest in cancer cells, which can be used in the research of HR-positive and HER2-negative breast cancer and hepatocellular carcinoma.