Apoptosis
As one of the cellular death mechanisms, apoptosis, also known as programmed cell death, can be defined as the process of a proper death of any cell under certain or necessary conditions. Apoptosis is controlled by the interactions between several molecules and responsible for the elimination of unwanted cells from the body.
Many biochemical events and a series of morphological changes occur at the early stage and increasingly continue till the end of apoptosis process. Morphological event cascade including cytoplasmic filament aggregation, nuclear condensation, cellular fragmentation, and plasma membrane blebbing finally results in the formation of apoptotic bodies. Several biochemical changes such as protein modifications/degradations, DNA and chromatin deteriorations, and synthesis of cell surface markers form morphological process during apoptosis.
Apoptosis can be stimulated by two different pathways: (1) intrinsic pathway (or mitochondria pathway) that mainly occurs via release of cytochrome c from the mitochondria and (2) extrinsic pathway when Fas death receptor is activated by a signal coming from the outside of the cell.
Different gene families such as caspases, inhibitor of apoptosis proteins, B cell lymphoma (Bcl)-2 family, tumor necrosis factor (TNF) receptor gene superfamily, or p53 gene are involved and/or collaborate in the process of apoptosis.
Caspase family comprises conserved cysteine aspartic-specific proteases, and members of caspase family are considerably crucial in the regulation of apoptosis. There are 14 different caspases in mammals, and they are basically classified as the initiators including caspase-2, -8, -9, and -10; and the effectors including caspase-3, -6, -7, and -14; and also the cytokine activators including caspase-1, -4, -5, -11, -12, and -13. In vertebrates, caspase-dependent apoptosis occurs through two main interconnected pathways which are intrinsic and extrinsic pathways. The intrinsic or mitochondrial apoptosis pathway can be activated through various cellular stresses that lead to cytochrome c release from the mitochondria and the formation of the apoptosome, comprised of APAF1, cytochrome c, ATP, and caspase-9, resulting in the activation of caspase-9. Active caspase-9 then initiates apoptosis by cleaving and thereby activating executioner caspases. The extrinsic apoptosis pathway is activated through the binding of a ligand to a death receptor, which in turn leads, with the help of the adapter proteins (FADD/TRADD), to recruitment, dimerization, and activation of caspase-8 (or 10). Active caspase-8 (or 10) then either initiates apoptosis directly by cleaving and thereby activating executioner caspase (-3, -6, -7), or activates the intrinsic apoptotic pathway through cleavage of BID to induce efficient cell death. In a heat shock-induced death, caspase-2 induces apoptosis via cleavage of Bid.
Bcl-2 family members are divided into three subfamilies including (i) pro-survival subfamily members (Bcl-2, Bcl-xl, Bcl-W, MCL1, and BFL1/A1), (ii) BH3-only subfamily members (Bad, Bim, Noxa, and Puma9), and (iii) pro-apoptotic mediator subfamily members (Bax and Bak). Following activation of the intrinsic pathway by cellular stress, pro‑apoptotic BCL‑2 homology 3 (BH3)‑only proteins inhibit the anti‑apoptotic proteins Bcl‑2, Bcl-xl, Bcl‑W and MCL1. The subsequent activation and oligomerization of the Bak and Bax result in mitochondrial outer membrane permeabilization (MOMP). This results in the release of cytochrome c and SMAC from the mitochondria. Cytochrome c forms a complex with caspase-9 and APAF1, which leads to the activation of caspase-9. Caspase-9 then activates caspase-3 and caspase-7, resulting in cell death. Inhibition of this process by anti‑apoptotic Bcl‑2 proteins occurs via sequestration of pro‑apoptotic proteins through binding to their BH3 motifs.
One of the most important ways of triggering apoptosis is mediated through death receptors (DRs), which are classified in TNF superfamily. There exist six DRs: DR1 (also called TNFR1); DR2 (also called Fas); DR3, to which VEGI binds; DR4 and DR5, to which TRAIL binds; and DR6, no ligand has yet been identified that binds to DR6. The induction of apoptosis by TNF ligands is initiated by binding to their specific DRs, such as TNFα/TNFR1, FasL /Fas (CD95, DR2), TRAIL (Apo2L)/DR4 (TRAIL-R1) or DR5 (TRAIL-R2). When TNF-α binds to TNFR1, it recruits a protein called TNFR-associated death domain (TRADD) through its death domain (DD). TRADD then recruits a protein called Fas-associated protein with death domain (FADD), which then sequentially activates caspase-8 and caspase-3, and thus apoptosis. Alternatively, TNF-α can activate mitochondria to sequentially release ROS, cytochrome c, and Bax, leading to activation of caspase-9 and caspase-3 and thus apoptosis. Some of the miRNAs can inhibit apoptosis by targeting the death-receptor pathway including miR-21, miR-24, and miR-200c.
p53 has the ability to activate intrinsic and extrinsic pathways of apoptosis by inducing transcription of several proteins like Puma, Bid, Bax, TRAIL-R2, and CD95.
Some inhibitors of apoptosis proteins (IAPs) can inhibit apoptosis indirectly (such as cIAP1/BIRC2, cIAP2/BIRC3) or inhibit caspase directly, such as XIAP/BIRC4 (inhibits caspase-3, -7, -9), and Bruce/BIRC6 (inhibits caspase-3, -6, -7, -8, -9).
Any alterations or abnormalities occurring in apoptotic processes contribute to development of human diseases and malignancies especially cancer.
References:
1.Yağmur Kiraz, Aysun Adan, Melis Kartal Yandim, et al. Major apoptotic mechanisms and genes involved in apoptosis[J]. Tumor Biology, 2016, 37(7):8471.
2.Aggarwal B B, Gupta S C, Kim J H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey.[J]. Blood, 2012, 119(3):651.
3.Ashkenazi A, Fairbrother W J, Leverson J D, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors[J]. Nature Reviews Drug Discovery, 2017.
4.McIlwain D R, Berger T, Mak T W. Caspase functions in cell death and disease[J]. Cold Spring Harbor perspectives in biology, 2013, 5(4): a008656.
5.Ola M S, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis[J]. Molecular and cellular biochemistry, 2011, 351(1-2): 41-58.
What is Apoptosis? The Apoptotic Pathways and the Caspase Cascade
Targets for Apoptosis
- Pyroptosis(15)
- Caspase(77)
- 14.3.3 Proteins(3)
- Apoptosis Inducers(71)
- Bax(15)
- Bcl-2 Family(136)
- Bcl-xL(13)
- c-RET(15)
- IAP(32)
- KEAP1-Nrf2(73)
- MDM2(21)
- p53(137)
- PC-PLC(6)
- PKD(8)
- RasGAP (Ras- P21)(2)
- Survivin(8)
- Thymidylate Synthase(12)
- TNF-α(141)
- Other Apoptosis(1144)
- Apoptosis Detection(0)
- Caspase Substrate(0)
- APC(6)
- PD-1/PD-L1 interaction(60)
- ASK1(4)
- PAR4(2)
- RIP kinase(47)
- FKBP(22)
Products for Apoptosis
- Cat.No. Product Name Information
-
GC33043
EL-102
EL-102 is a hypoxia-induced factor 1 (Hif1α) inhibitor. EL-102 induces apoptosis, inhibits tubulin polymerisation and shows activities against prostate cancer. EL-102 can be used for the research of cancer.
-
GC13885
Elesclomol (STA-4783)
An apoptosis inducer
-
GC16827
ELR510444
Novel microtubule disruptor
-
GC48434
Elsinochrome A
A fungal metabolite
-
GC13163
Embelin
A benzoquinone with diverse biological activities
-
GC35980
Emricasan
A pan-caspase inhibitor

-
GC16519
ENMD-2076
A multi-kinase inhibitor
-
GC43610
Enniatin A1
Enniatins are cyclohexadepsipeptides commonly isolated from fungi that are known to have antibiotic properties and to induce apoptosis in several cancer lines.
-
GC16823
Enniatin Complex
ionophore
-
GC11625
Entinostat (MS-275,SNDX-275)
A histone deacetylase inhibitor
-
GC66344
Envafolimab
Envafolimab (ASC 22; KN 035) is a recombinant protein of a humanized single-domain anti- PD-L1 antibody. Envafolimab is created by a fusion of the of anti-PD-L1 domain with Fc fragment of human IgG1 antibody. Envafolimab blocks interaction between PD-L1 and PD-1 with an IC50 value of 5.25nm. Envafolimab shows antitumor activity. Envafolimab has the potential for the research of solid tumors.

-
GC11499
Enzastaurin (LY317615)
Enzastaurin (LY317615) (LY317615) is a potent and selective PKCβ inhibitor with an IC50 of 6 nM, showing 6- to 20-fold selectivity over PKCα, PKCγ and PKCε.
-
GC31390
EP1013 (F1013)
EP1013 (F1013) (F1013) is a broad-spectrum caspase selective inhibitor, used in the research of type 1 diabetes.
-
GC11834
Epibrassinolide
Potential apoptosis inducer;steroidal plant growth stimulant
-
GC35997
Epirubicin
Epirubicin (4'-Epidoxorubicin), a semisynthetic L-arabino derivative of doxorubicin, has an antineoplastic agent by inhibiting Topoisomerase. Epirubicin inhibits DNA and RNA synthesis. Epirubicin is a Forkhead box protein p3 (Foxp3) inhibitor and inhibits regulatory T cell activity.
-
GC11202
Epothilone A
Microtubule stabilizing macrolide
-
GC17240
Epothilone B (EPO906, Patupilone)
Epothilone B (EPO906, Patupilone) is a microtubule stabilizer with a Ki of 0.71μM.
-
GC13383
EPZ004777
A potent inhibitor of DOT1L
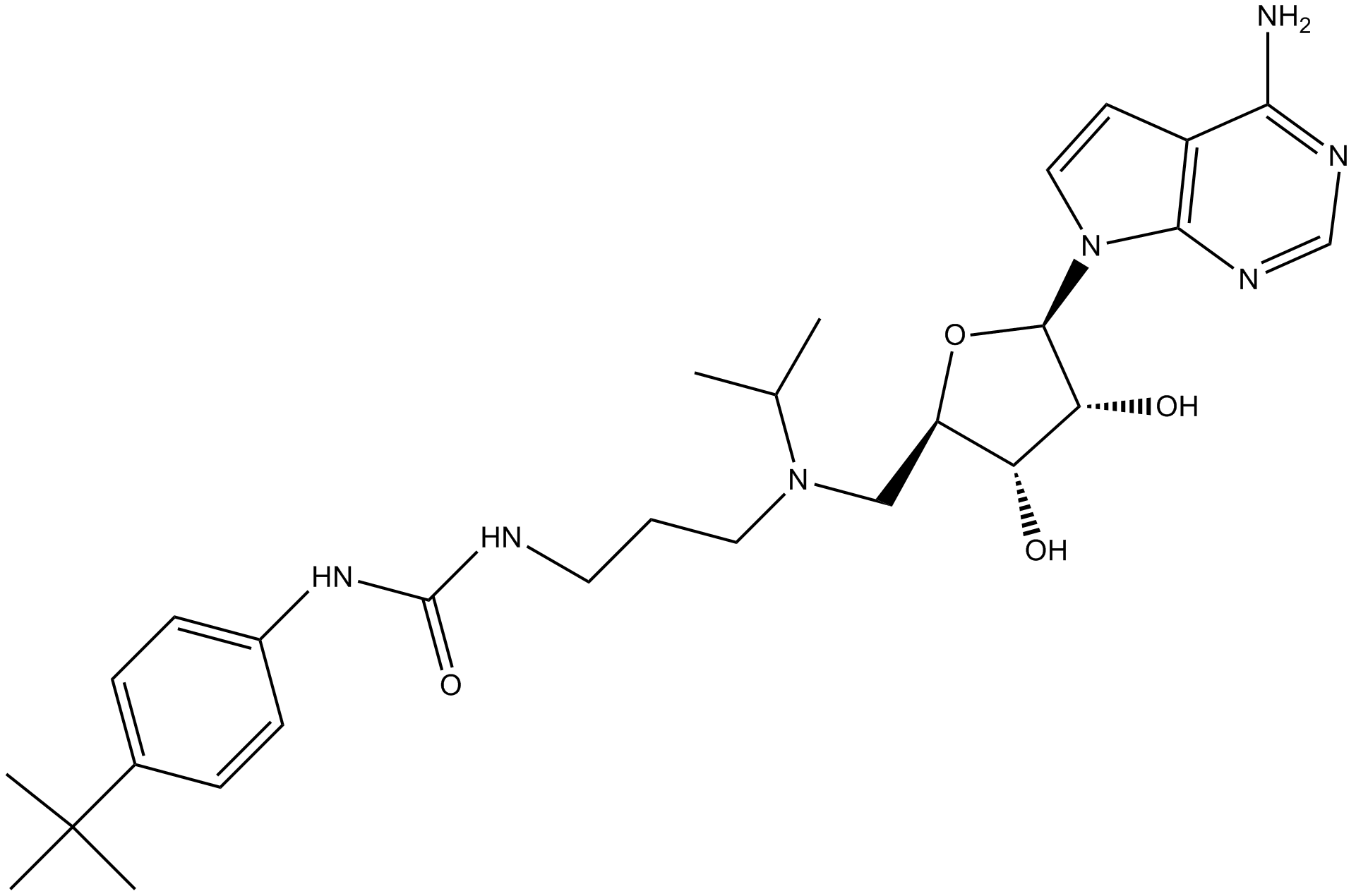
-
GC10389
ERB 041
ERB 041 (ERB-041) is a potent and selective estrogen receptor (ER) β agonist with IC50s of 5.4, 3.1 and 3.7 nM for human, rat and mouse ERβ, respectively. ERB 041 displays >200-fold selectivity for ERβ over ERα. ERB 041 is a potent skin cancer chemopreventive agent that acts by dampening the WNT/β-catenin signaling pathway. ERB 041 induces ovarian cancer apoptosis.
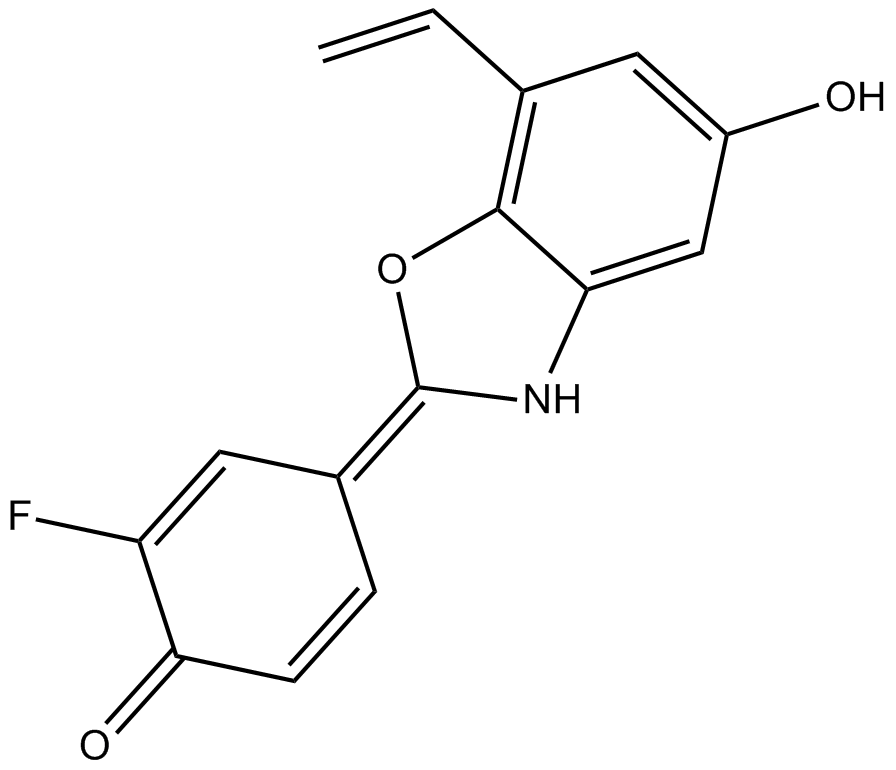
-
GC52516
Erbstatin
A tyrosine kinase inhibitor
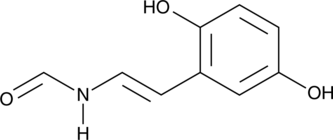
-
GC19142
Erdafitinib
A pan-FGFR tyrosine kinase inhibitor
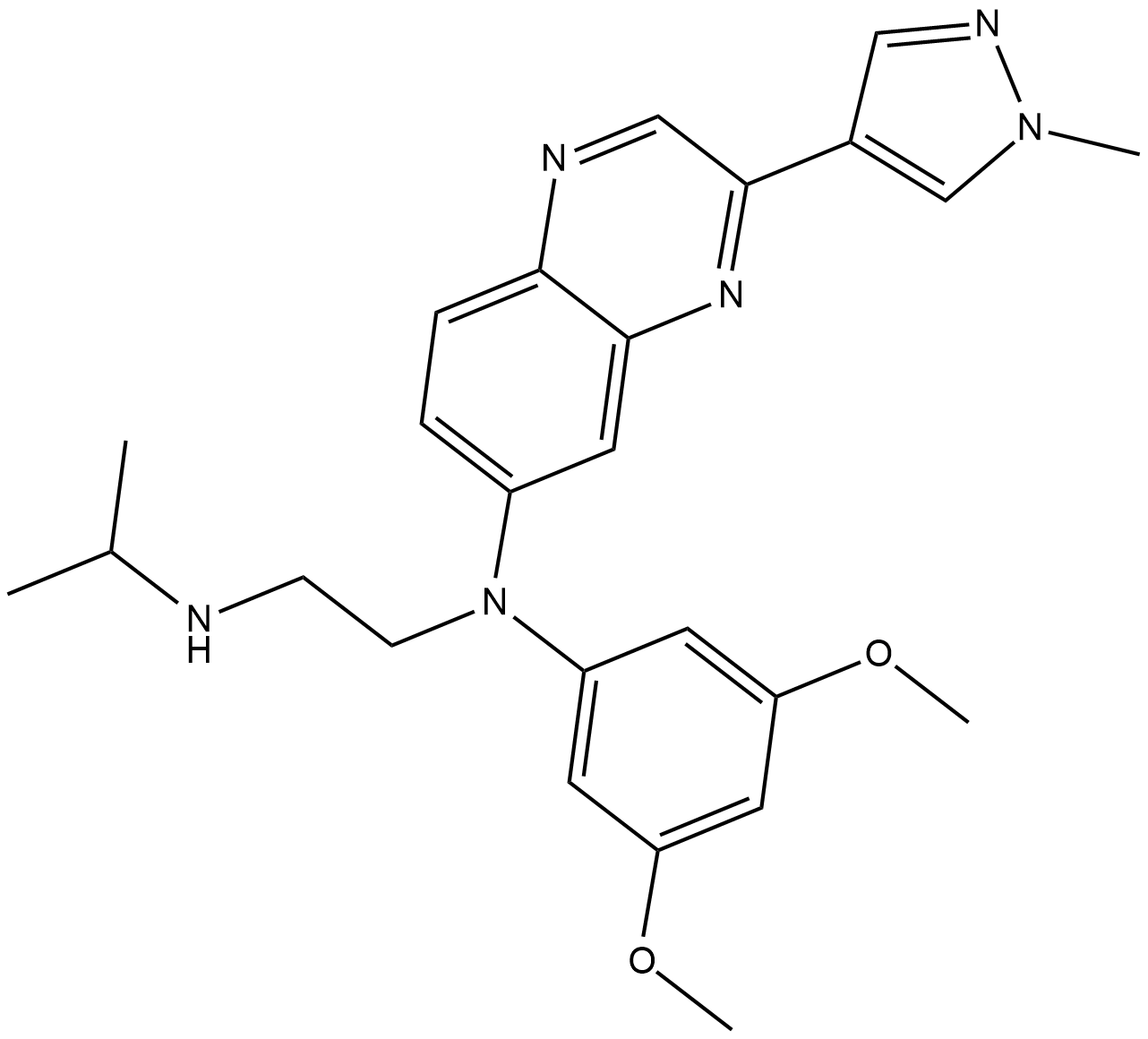
-
GC63845
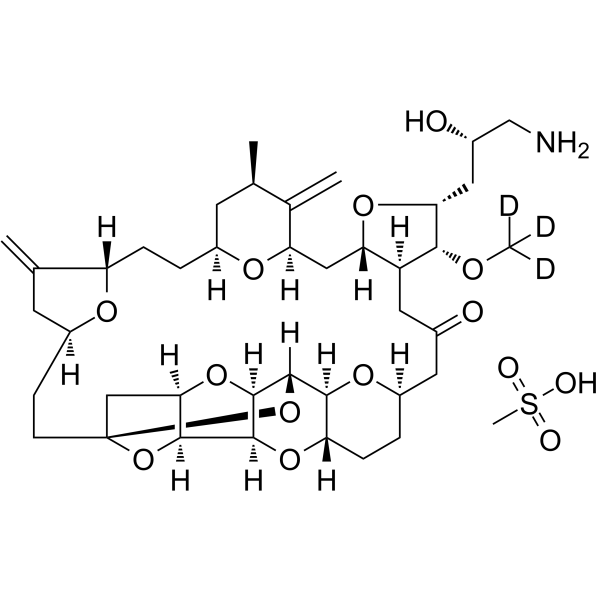
Eribulin-d3 mesylate
Eribulin-d3 mesylate is a deuterium labeled Eribulin mesylate. Eribulin mesylate is a microtubule targeting agent that is used for the research of cancer.
-
GC60153
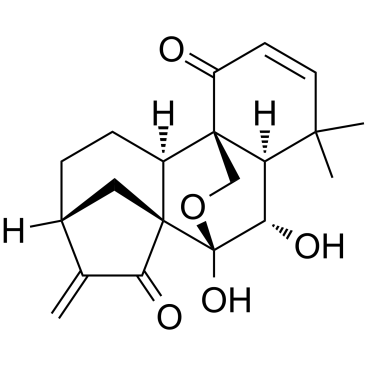
Eriocalyxin B
Eriocalyxin B is an ent-Kaurene diterpenoid isolated from Chinese herb Isodon eriocalyx.
-
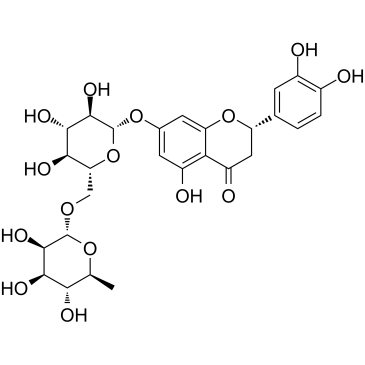
GC38181
Eriocitrin
A flavonoid with antioxidant activity
-
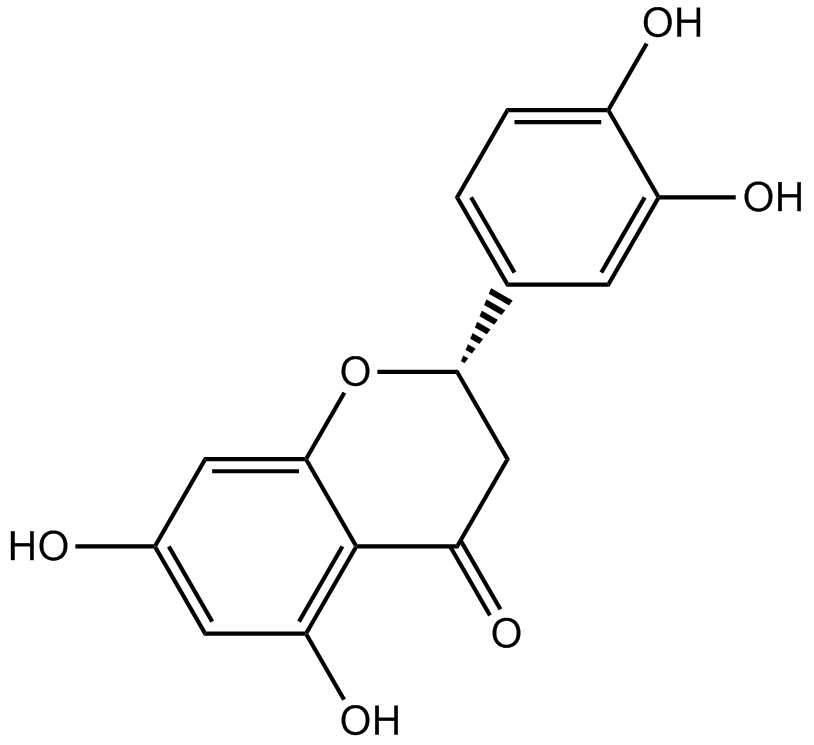
GN10470
Eriodictyol
-
GC62957
Eriodictyol-7-O-glucoside
Eriodictyol-7-O-glucoside (Eriodictyol 7-O-β-D-glucoside), a flavonoid, is a potent free radical scavenger.
-
GC38447
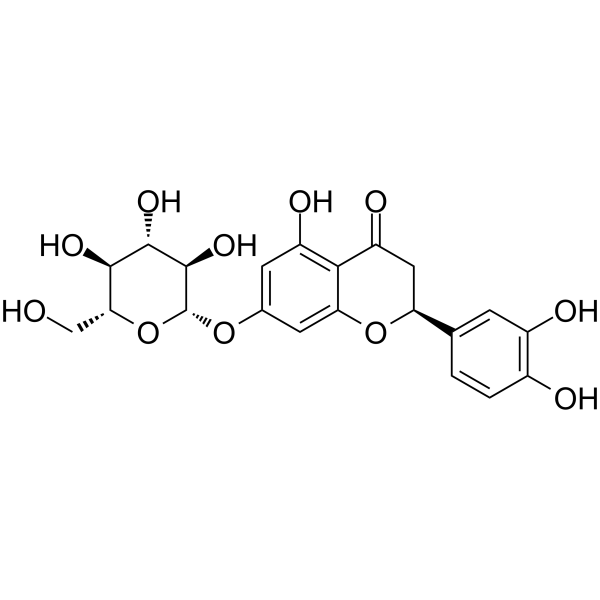
Eriosematin
Eriosematin is a compound from the roots of Flemingia philippinensis with antiproliferative activity and apoptosis-inducing property.
-
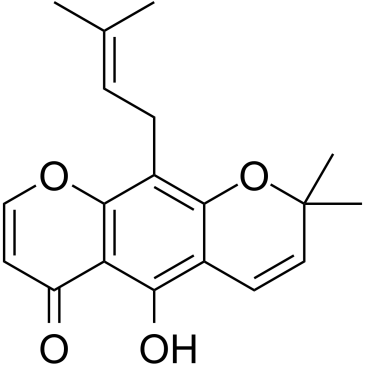
GC43625
Erucin
Erucin is an isothiocyanate derived from glucoerucin, a glucosinolate predominant in arugula (Eruca sativa Mill.) and other cruciferous vegetables.
-
GC11935
Escin
mixture of saponins with anti-inflammatory, vasoconstrictor and vasoprotective effects
-
GC34579
Etanercept
Etanercept, a dimeric fusion protein that binds TNF, acts as a TNF inhibitor.

-
GC38083
Ethoxysanguinarine
Ethoxysanguinarine is a benzophenanthridine alkaloid natural product that is mainly found in Macleaya cordata. Ethoxysanguinarine is an inhibitor of protein phosphatase 2A (CIP2A). Ethoxysanguinarine induces cell apoptosis and inhibits colorectal cancer cells growth.
-
GC61669
Ethyl 3,4-dihydroxybenzoate
Ethyl 3,4-dihydroxybenzoate (Ethyl protocatechuate), an antioxidant, is a prolyl-hydroxylase inhibitor found in the testa of peanut seeds.
-
GC60824
Ethylene dimethanesulfonate
Ethylene dimethane sulfonate is a mild alkylating, non-volatile methanesulfonic diester of ethylene glycol.
-
GC12105
Etidronate
Etidronate (Etidronate) is an orally and intravenously active bisphosphonate.
-
GC15617
Etoposide
Topo II inhibitor
-
GC60826
Etoposide phosphate
Etoposide phosphate (BMY-40481) is a potent anti-cancer chemotherapy agent and a selective topoisomerase II inhibitor?to prevent re-ligation of DNA strands.
-
GC10855
Etretinate
retinoid receptor agonist
-
GC30910
Eucalyptol (1,8-Cineole)
A bicyclic monoterpene with diverse biological activities
-
GC16517
Eugenol
naturally occuring scent chemical
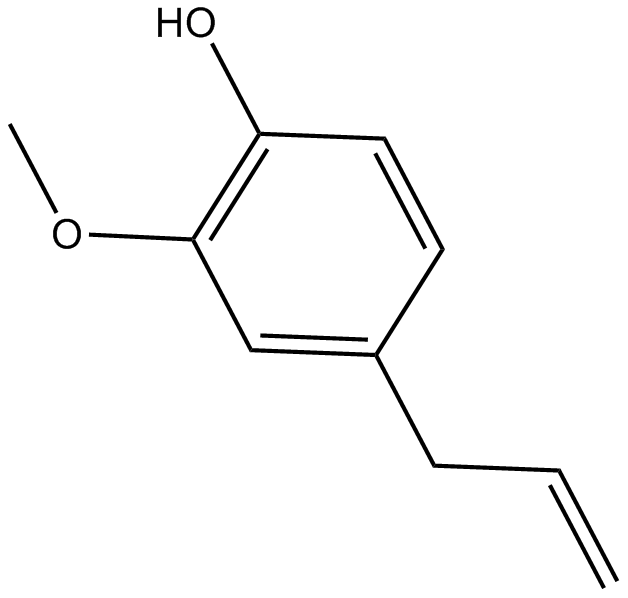
-
GC43643
Eupenifeldin
Eupenifeldin is a pentacyclic bistropolone fungal metabolite originally isolated from E.
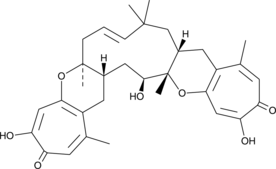
-
GC38165
Euphorbia Factor L1
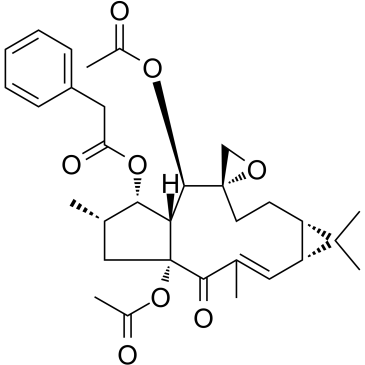
-
GC60157
Euphorbia Factor L2
Euphorbia factor L2, a lathyrane diterpenoid isolated from caper euphorbia seed (the seeds of Euphorbia lathyris L.), has been traditionally applied to treat cancer. Euphorbia factor L2 shows potent cytotoxicity and induces apoptosis via a mitochondrial pathway.
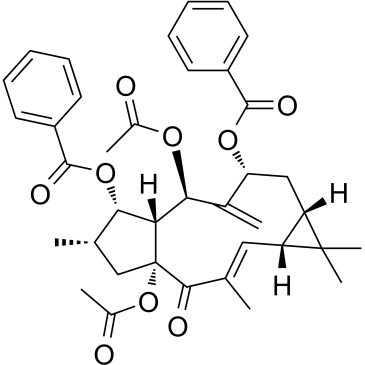
-
GC38392
Euscaphic acid
Euscaphic acid, a DNA polymerase inhibitor, is a triterpene from the root of the R. alceaefolius Poir. Euscaphic inhibits calf DNA polymerase α (pol α) and rat DNA polymerase β (pol β) with IC50 values of 61 and 108 μM. Euscaphic acid induces apoptosis.
-
GC13601
Everolimus (RAD001)
A rapamycin derivative
-
GC15605
Ezetimibe
Cholesterol transport inhibitor
-
GC60159
Ezetimibe ketone
Ezetimibe ketone (EZM-K) is a phase-I metabolite of Ezetimibe.
-
GC66354
Ezetimibe-d4-1
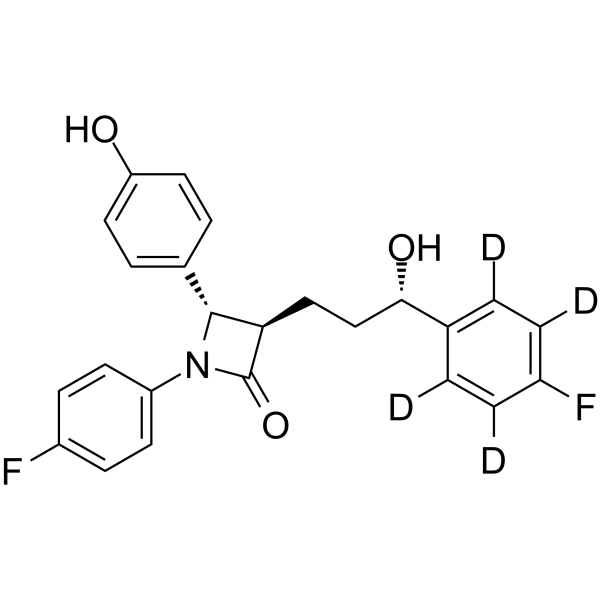
Ezetimibe-d4 is deuterium labeled Ezetimibe. Ezetimibe (SCH 58235) is a potent cholesterol absorption inhibitor. Ezetimibe is a Niemann-Pick C1-like1 (NPC1L1) inhibitor, and is a potent Nrf2 activator.
-
GC14791
F16
potential antitumor agent
-
GC62203
Falcarindiol
Falcarindiol, an orally active polyacetylenic oxylipin, activates PPARγ and increases the expression of the cholesterol transporter ABCA1 in cells.
-
GC38437
Fangchinoline
An alkaloid with diverse biological activities
-
GC60836
Fenobucarb
Fenobucarb is a carbamate insecticide.
-
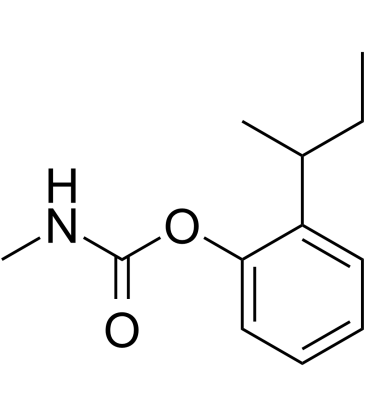
GC14499
Fenoprofen calcium hydrate
nonsteroidal, anti-inflammatory antiarthritic agent
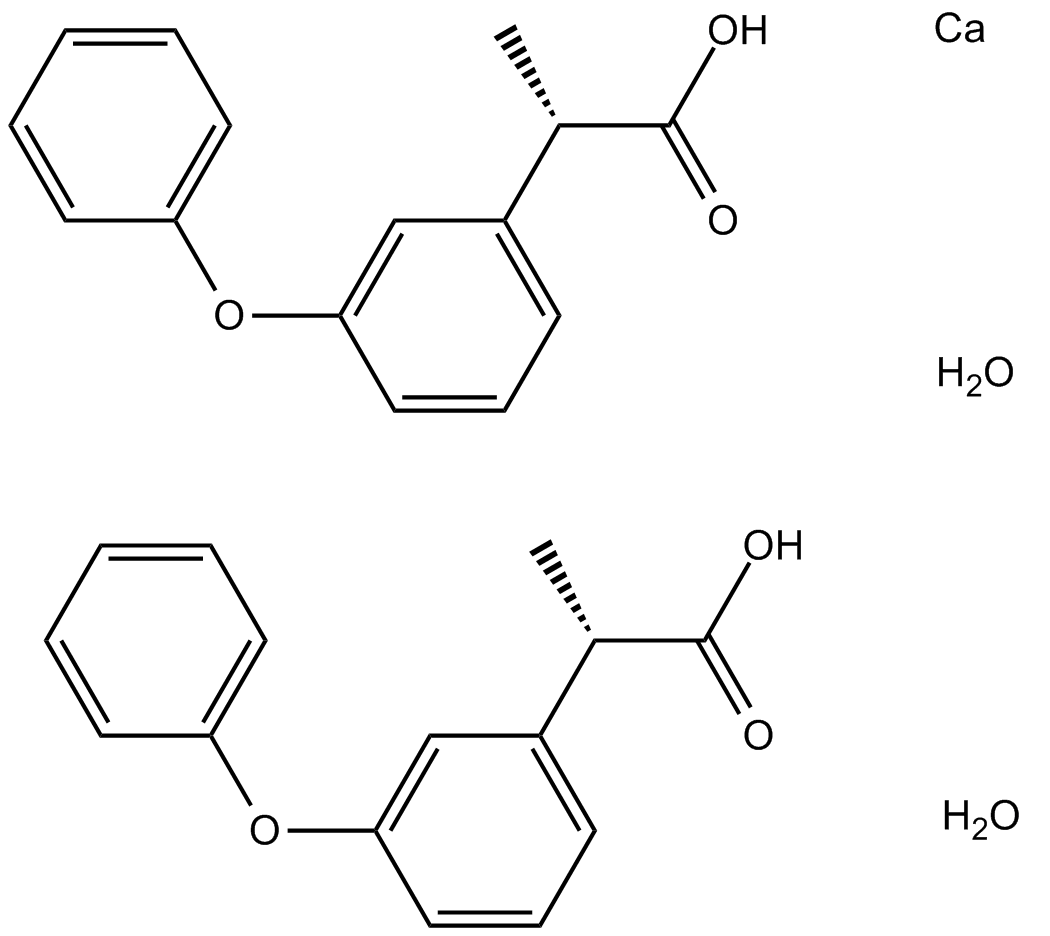
-
GC18641
Ferutinin
Ferutinin is a plant-derived ester of a sesquiterpenic alcohol that acts as an agonist for estrogen receptor (ER) α (IC50 = 33.1 nM) and an agonist/antagonist for ERβ (IC50 = 180.5 nM).
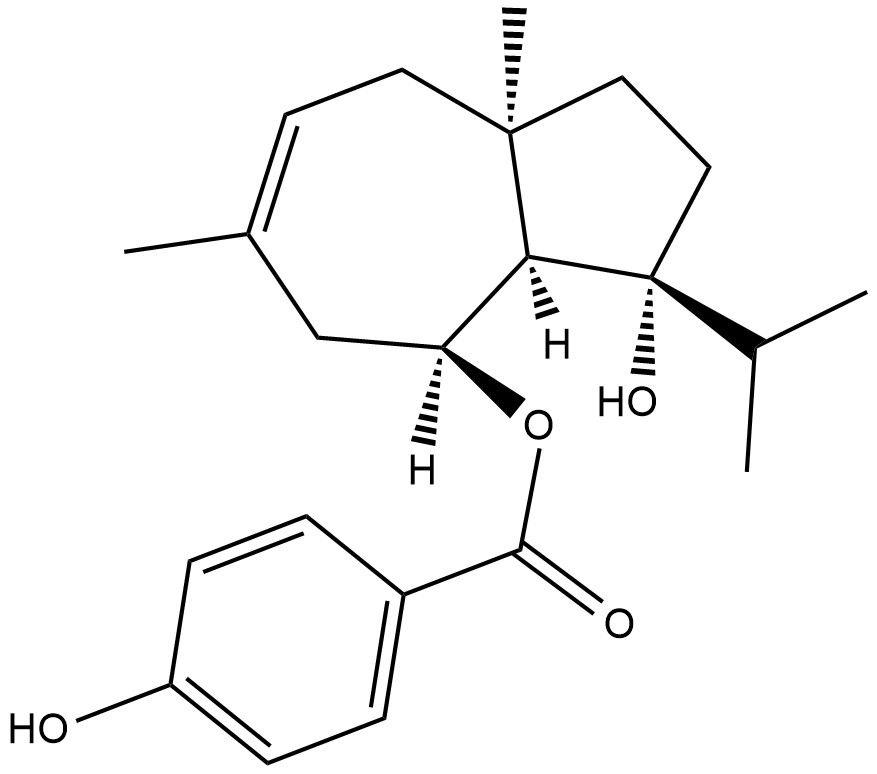
-
GC12940
Fidaxomicin
macrocyclic antibiotic
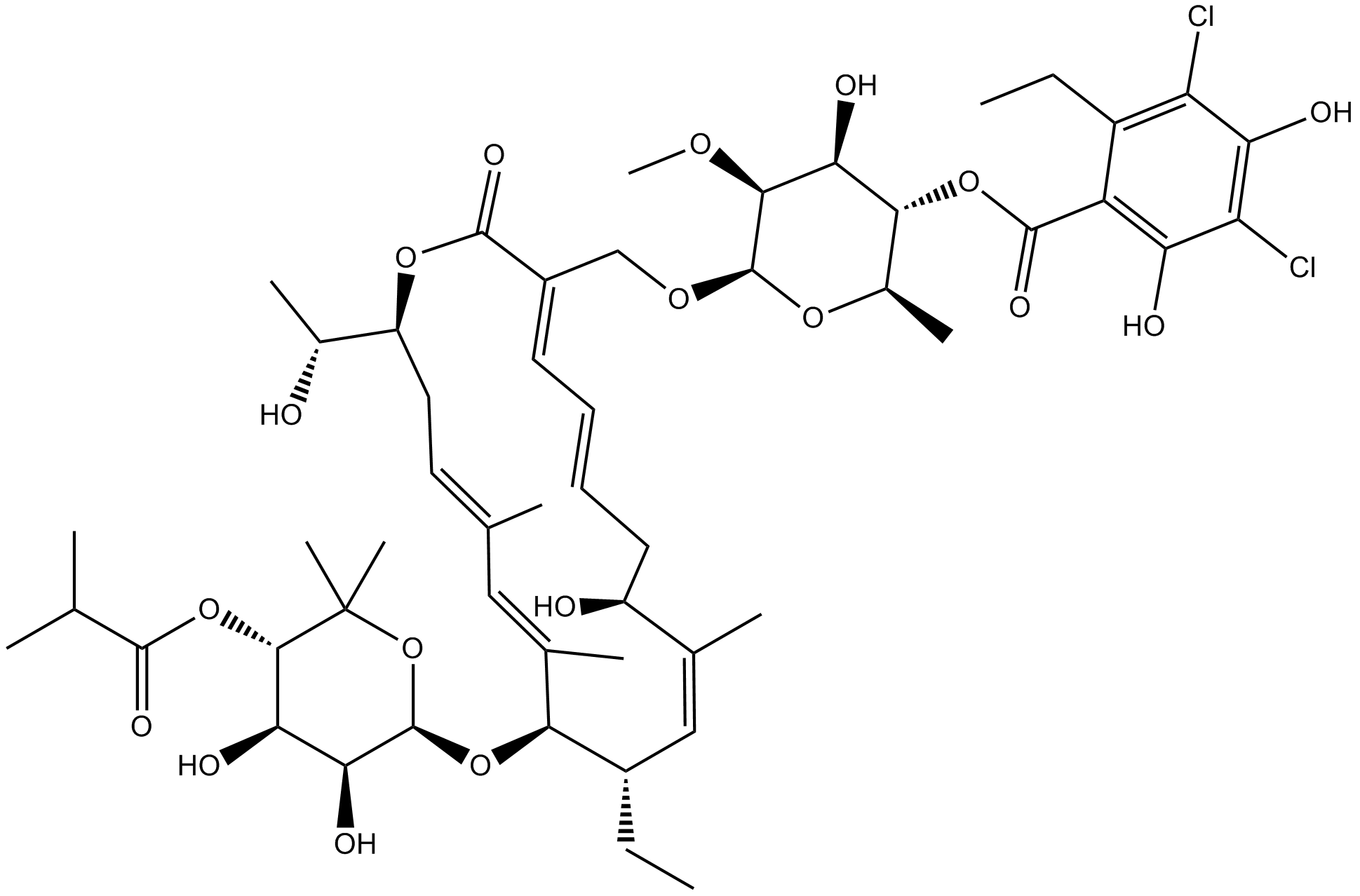
-
GC36046
Fimasartan
Fimasartan(BR-A-657) is a non-peptide angiotensin II receptor antagonist used for the treatment of hypertension and heart failure.
-
GN10030
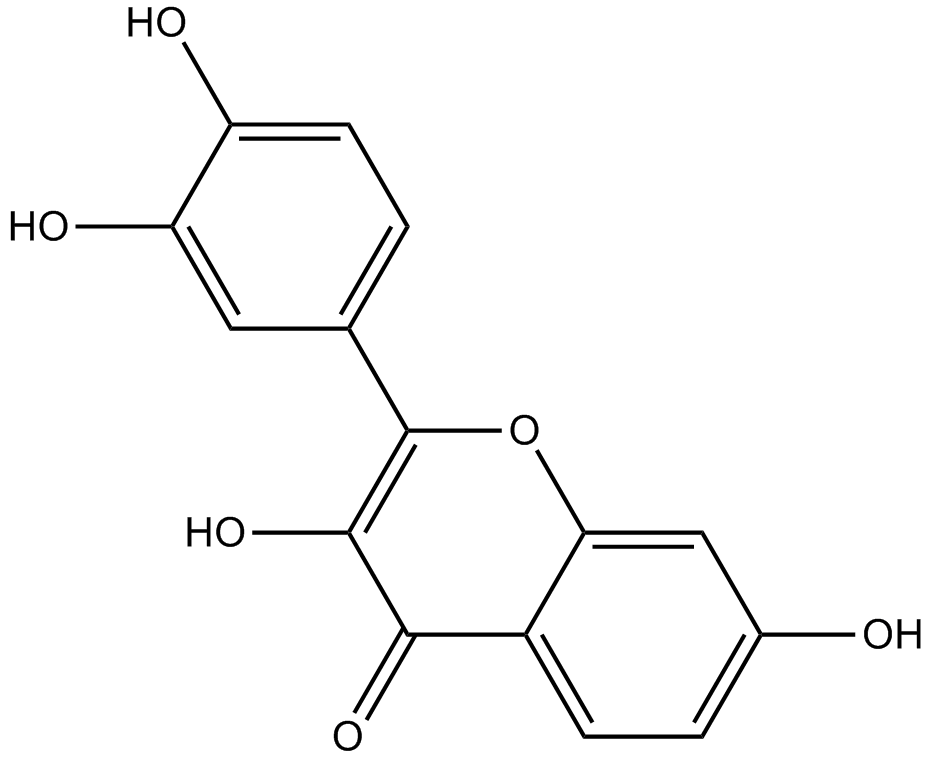
Fisetin
-
GC49344
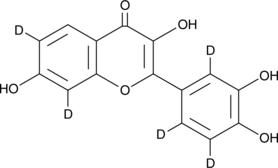
Fisetin-d5
An internal standard for the quantification of fisetin
-
GC60845
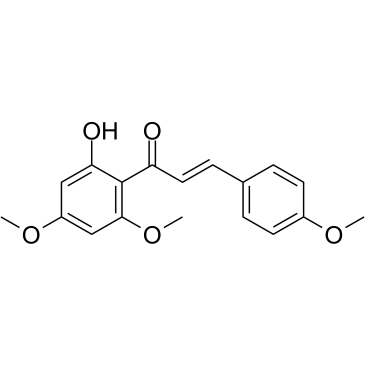
Flavokawain A
Flavokawain A, a proming anticarcinogenic agent, is a chalcone from kava extract with anti-tumor activity. Flavokawain A induces cell apoptosis by involvement of Bax protein-dependent and mitochondria-dependent apoptotic pathway. Flavokawain A has the potential for the study of bladder cancer.
-
GC41261
Flavokawain B
Flavokawain B is a natural chalcone first isolated from extracts of kava roots.
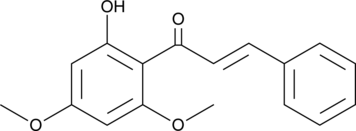
-
GC36050
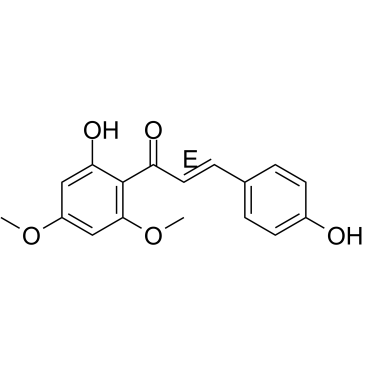
Flavokawain C
Flavokawain C is a natural chalcone found in Kava root. Flavokawain C exerts cytotoxicity against human cancer cell lines, with an IC50 of 12.75 μM for HCT 116 cells.
-
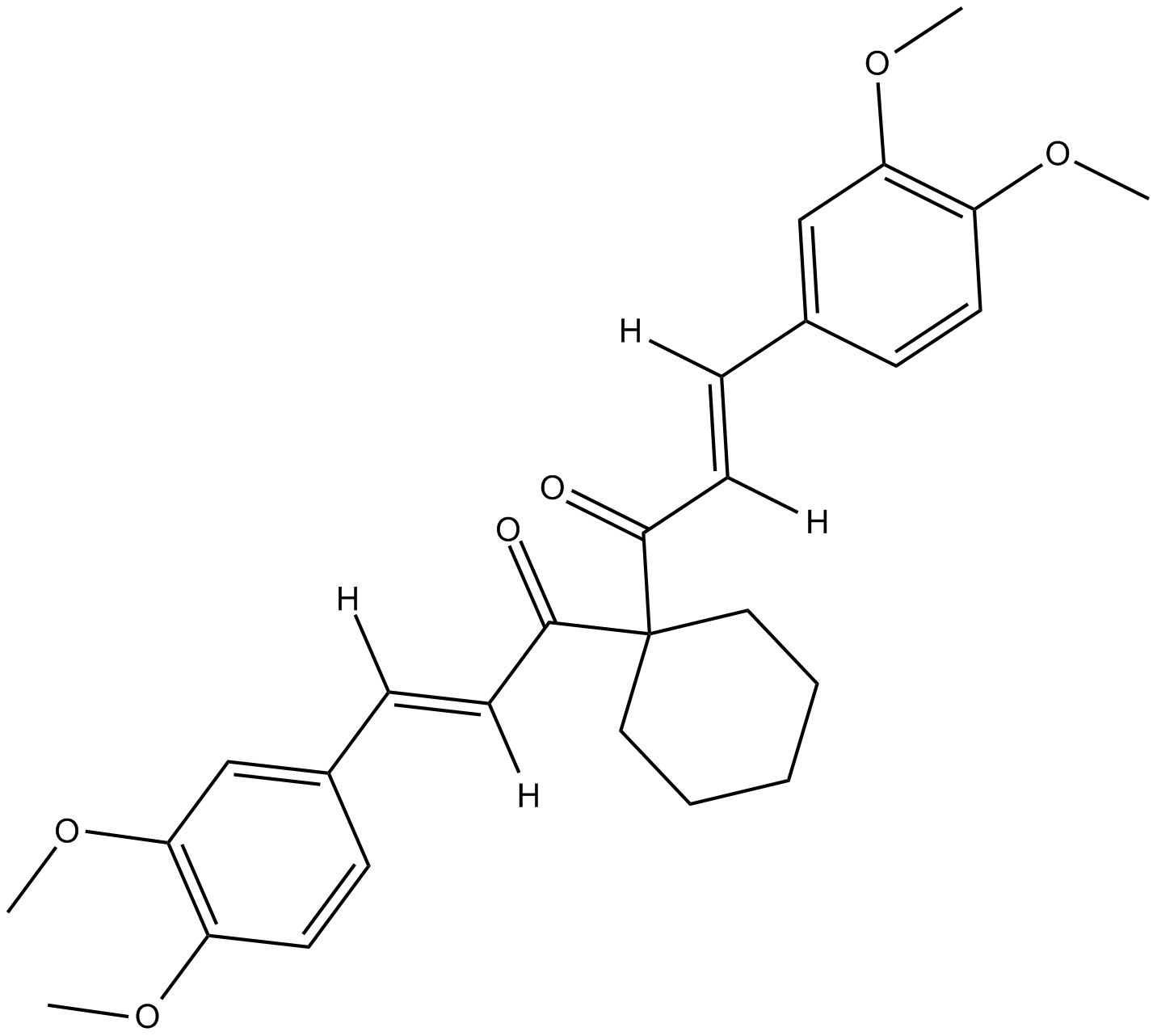
GC16875
FLLL32
STAT3 inhibitor
-
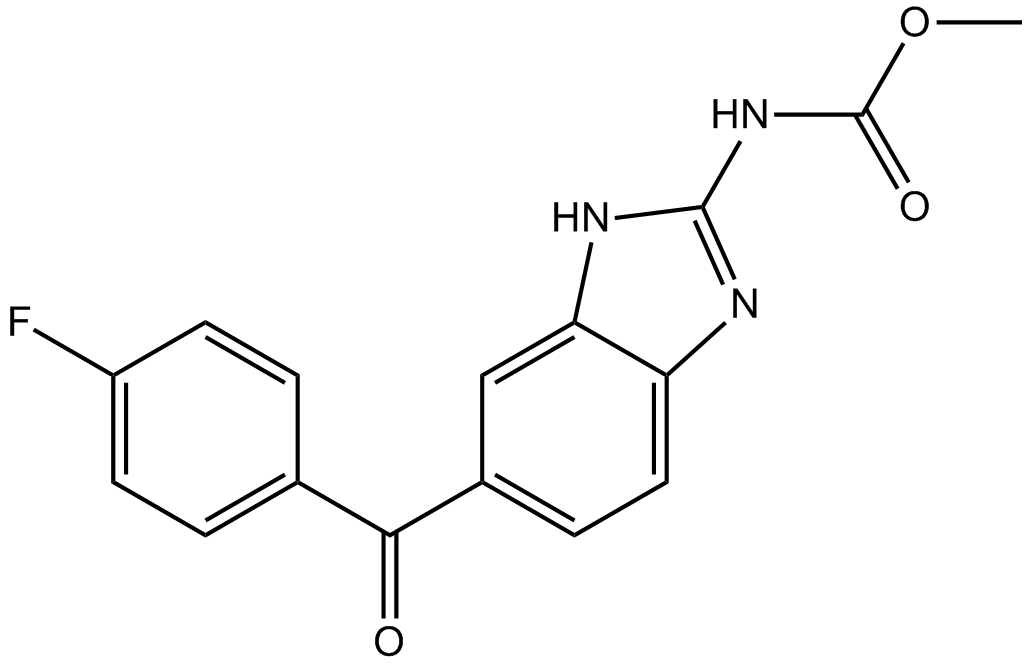
GC13210
Flubendazole
Autophagy activator
-
GC14144
Fludarabine
DNA synthsis inhibitor
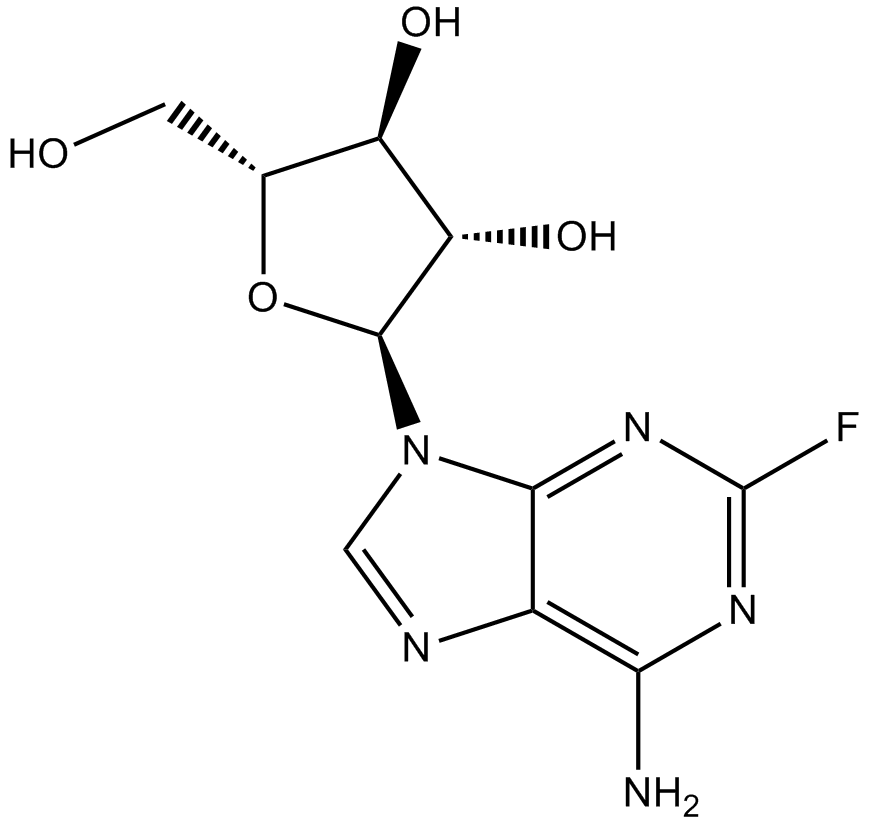
-
GC15134
Fludarabine Phosphate (Fludara)
Fludarabine (phosphate) is an analogue of adenosine and deoxyadenosine, which is able to compete with dATP for incorporation into DNA and inhibit DNA synthesis.
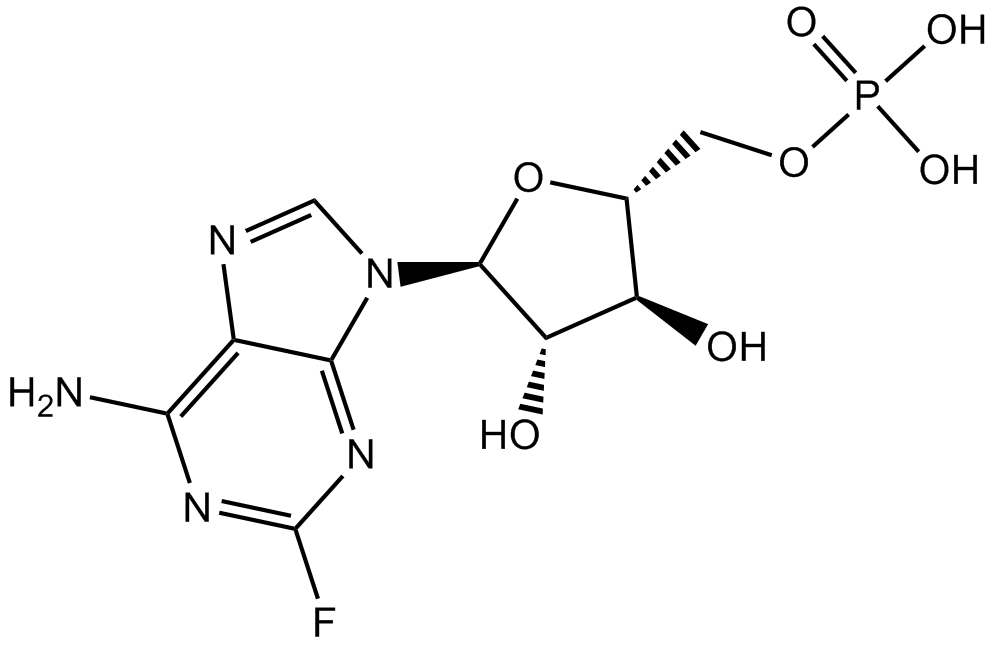
-
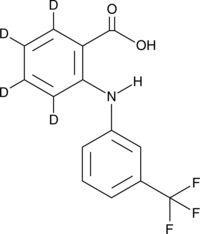
GC48827
Flufenamic Acid-d4
An internal standard for the quantification of flufenamic acid
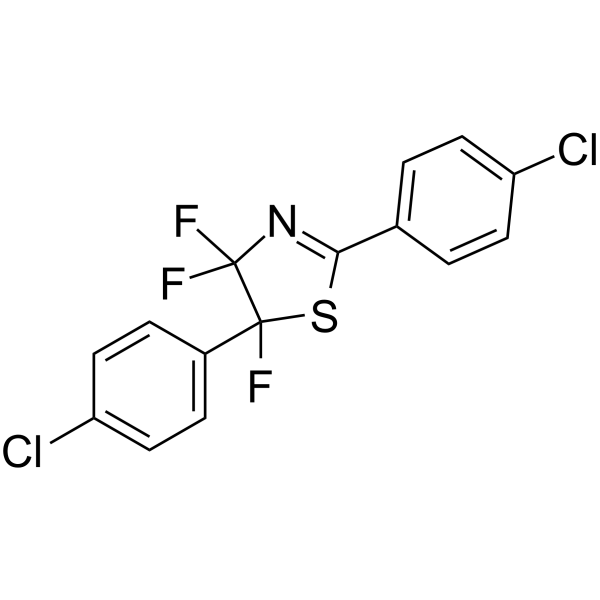
-
GC62167
Fluorizoline
Fluorizoline selectively and directly binds to prohibitin 1 (PHB1) and 2 (PHB2), and induces apoptosis. Fluorizoline reduces chronic lymphocytic leukemia (CLL) cell viability through the upregulation of NOXA and BIM. Fluorizoline exerts antitumor action in a p53-independent manner.
-
GC43689
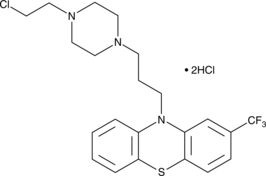
Fluphenazine-N-2-chloroethane (hydrochloride)
Fluphenazine is a traditional antipsychotic compound that tightly binds the dopamine D2 receptor (Ki = 0.55 nM) and also reversibly inhibits calmodulin at micromolar concentrations.
-
GC16880
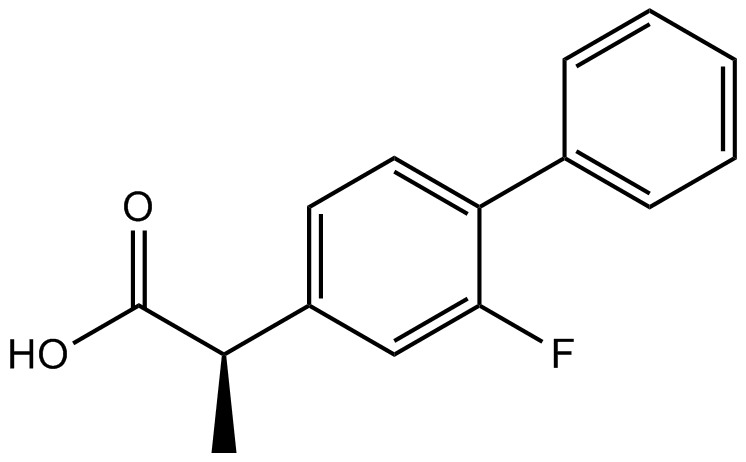
Flurbiprofen
Cyclooxygenase inhibitors
-
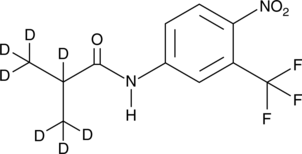
GC48831
Flutamide-d7
An internal standard for the quantification of flutamide
-
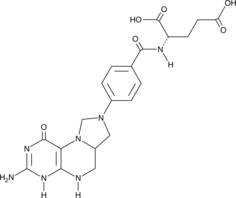
GC49126
Folitixorin
A reduced form of folate and cofactor for thymidylate synthetase
-
GN10527
Formononetin
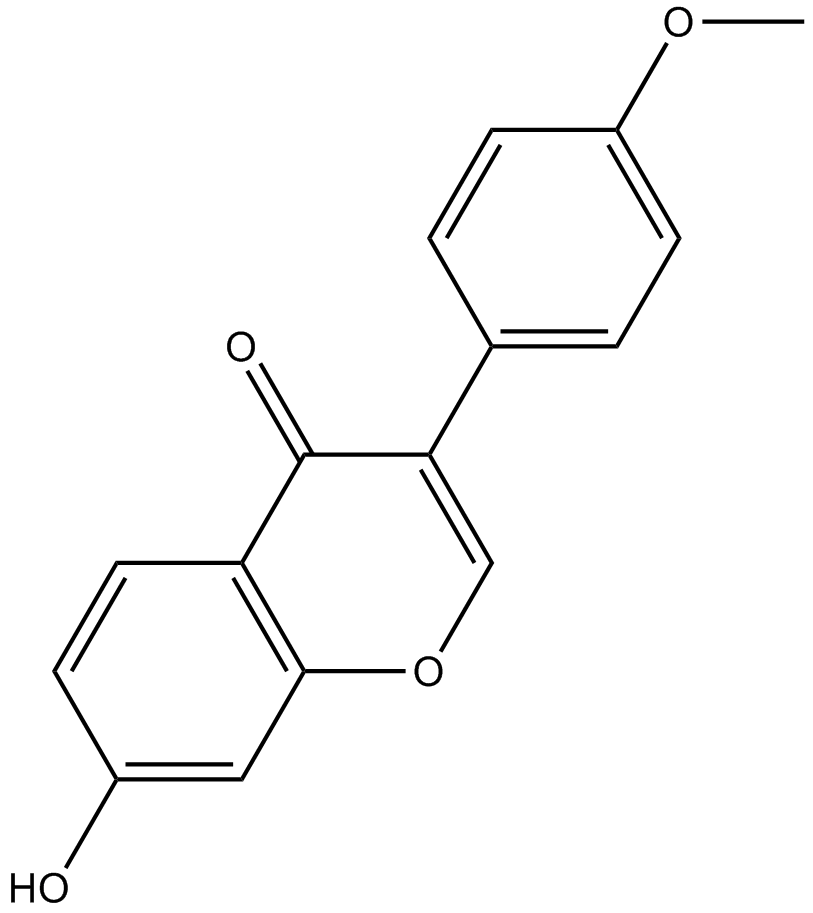
-
GC36065
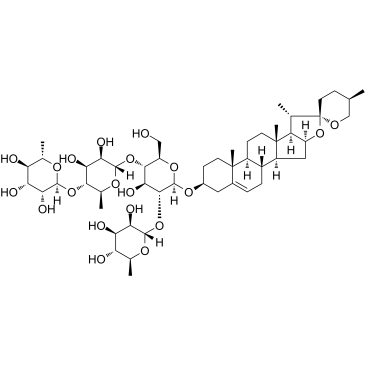
Formosanin C
Formosanin C is a diosgenin saponin isolated from Paris formosana Hayata and an immunomodulator with antitumor activity. Formosanin C induces apoptosis.
-
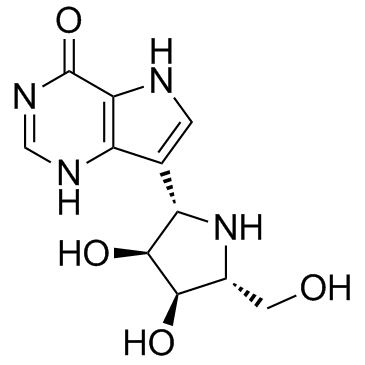
GC33029
Forodesine (BCX-1777 freebase)
Forodesine (BCX-1777 freebase) (BCX-1777) is a highly potent and orally active purine nucleoside phosphorylase (PNP) inhibitor with IC50 values ranging from 0.48 to 1.57 nM for human, mouse, rat, monkey and dog PNP. Forodesine (BCX-1777 freebase) is a potent human lymphocyte proliferation inhibitor. Forodesine (BCX-1777 freebase) could induce apoptosis in leukemic cells by increasing the dGTP levels.
-
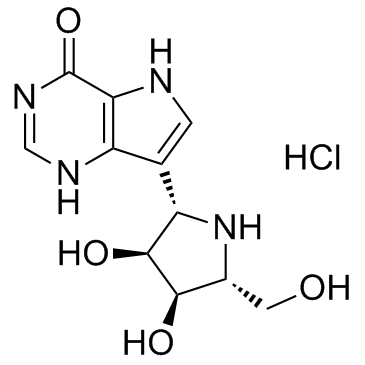
GC32708
Forodesine hydrochloride (BCX-1777)
Forodesine hydrochloride (BCX-1777) (BCX-1777 hydrochloride) is a highly potent and orally active purine nucleoside phosphorylase (PNP) inhibitor with IC50 values ranging from 0.48 to 1.57 nM for human, mouse, rat, monkey and dog PNP. Forodesine hydrochloride (BCX-1777) is a potent human lymphocyte proliferation inhibitor. Forodesine hydrochloride (BCX-1777) could induce apoptosis in leukemic cells by increasing the dGTP levels.
-
GN10727
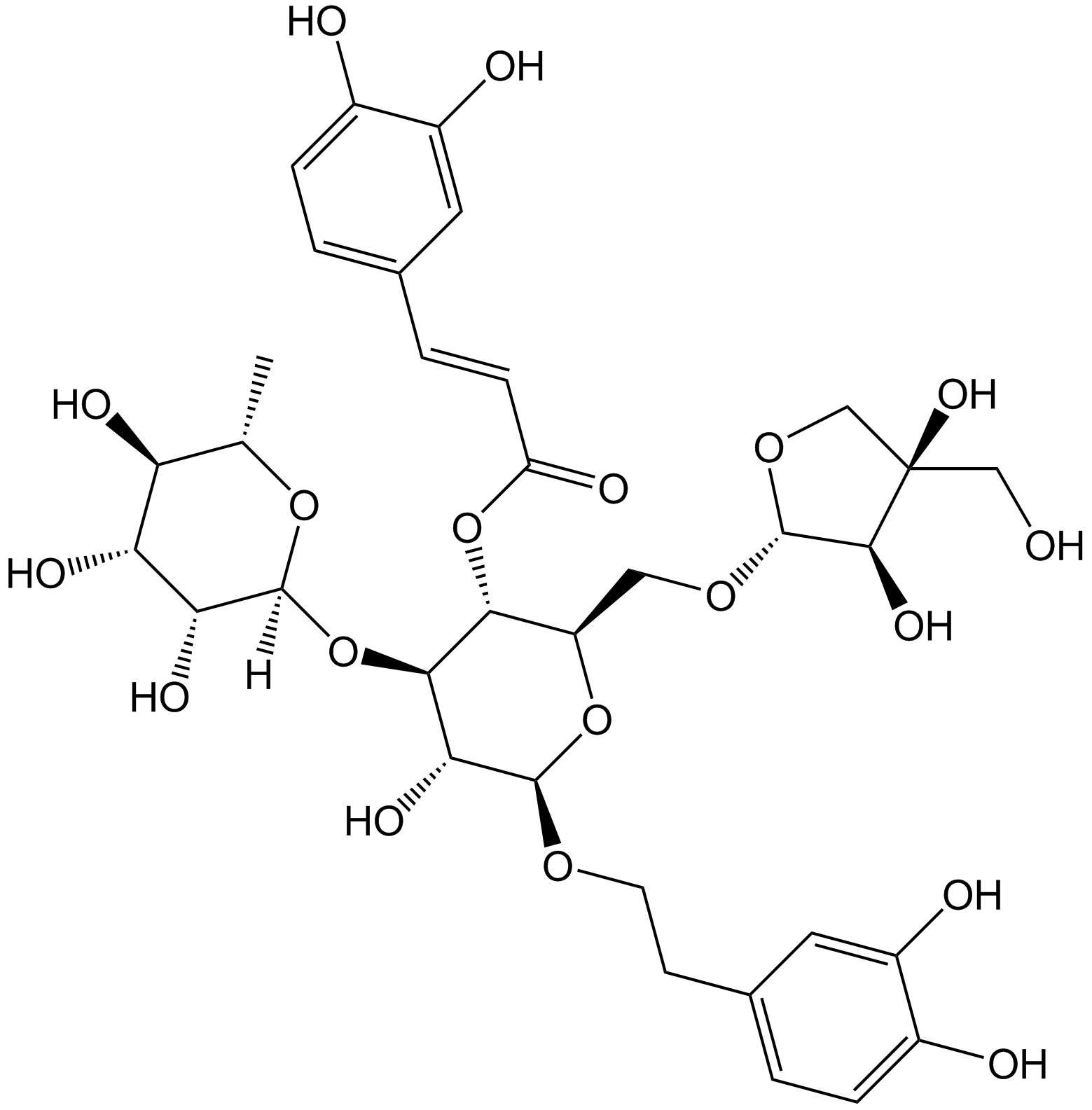
Forsythoside B
-
GC12308
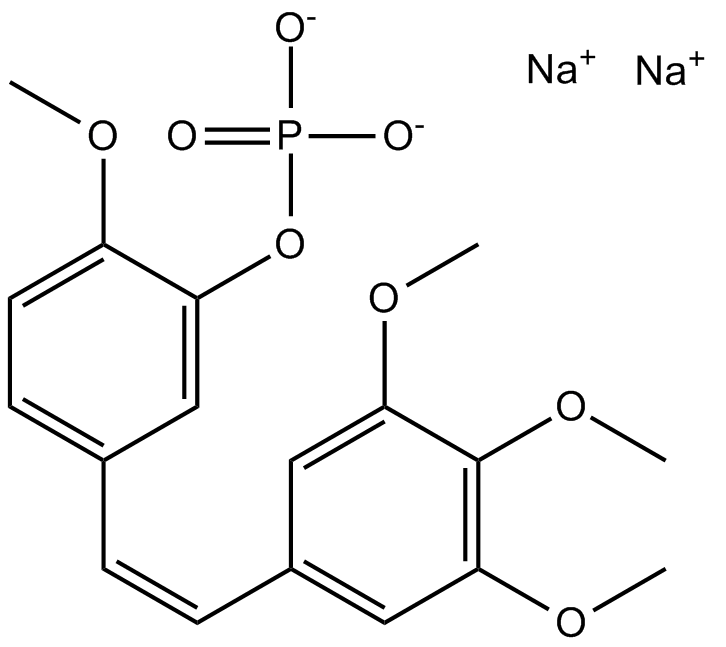
Fosbretabulin (Combretastatin A4 Phosphate (CA4P)) Disodium
Fosbretabulin (Combretastatin A4 Phosphate (CA4P)) Disodium (CA 4DP) is a tubulin destabilizing agent. Fosbretabulin (Combretastatin A4 Phosphate (CA4P)) Disodium is the Combretastatin A4 prodrug that selectively targets endothelial cells, induces regression of nascent tumour neovessels, reduces tumour blood flow and causes central tumour necrosis.
-
GC25428
Foscenvivint (ICG-001)
Foscenvivint (ICG-001) antagonizes Wnt/β-catenin/TCF-mediated transcription and specifically binds to CREB-binding protein (CBP) with IC50 of 3 μM, but is not the related transcriptional coactivator p300. ICG-001 induces apoptosis.

-
GC62645
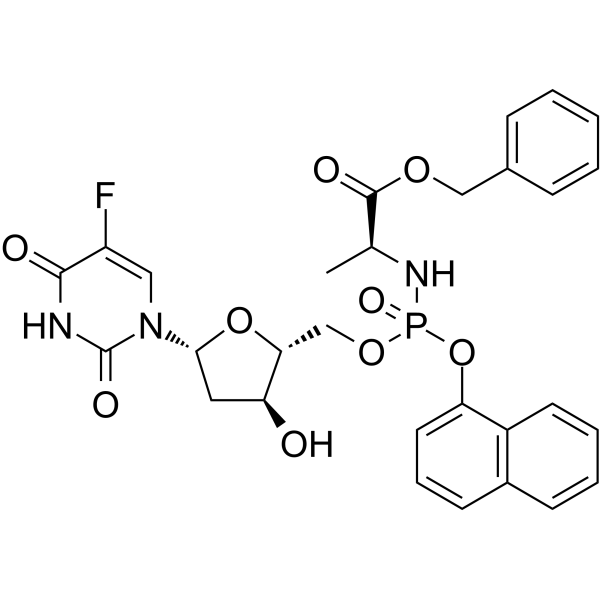
Fosifloxuridine nafalbenamide
Fosifloxuridine nafalbenamide (NUC-3373), a pyrimidine nucleotide analogue, is a Thymidylate synthase inhibitor. Fosifloxuridine nafalbenamide has anticancer activity. Fosifloxuridine nafalbenamide has the potential to evoke a host immune response and enhance immunotherapy.
-
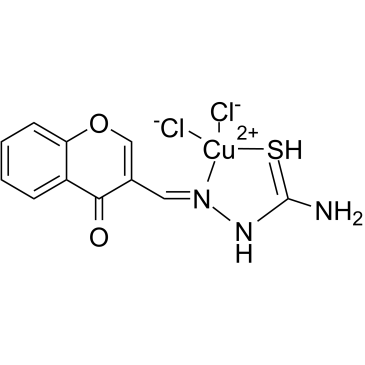
GC38551
FPA-124
-
GC18652
FQI 1
An inhibitor of Late SV40 Factor
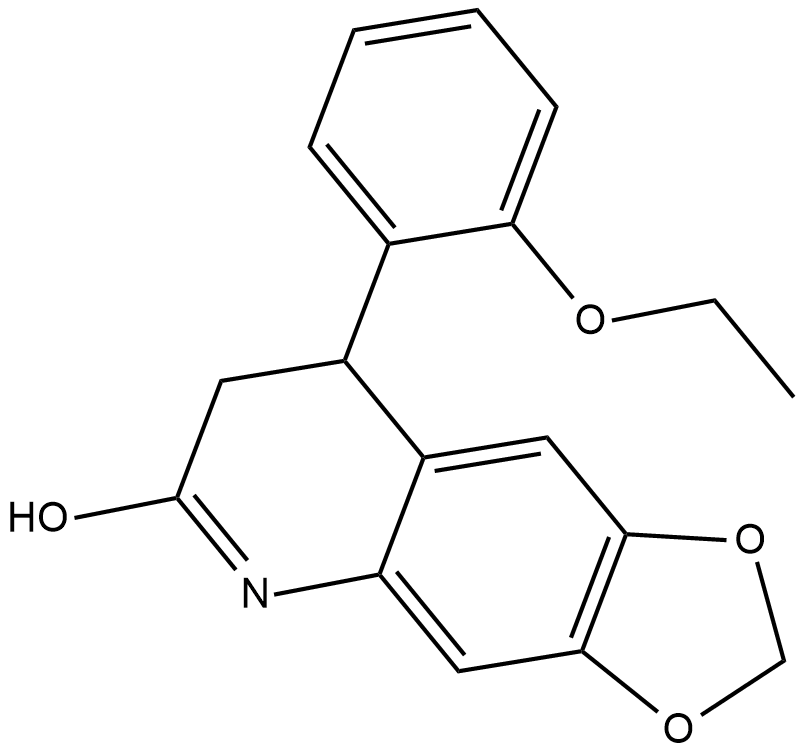
-
GC10647
FR 180204
ERK inhibitor
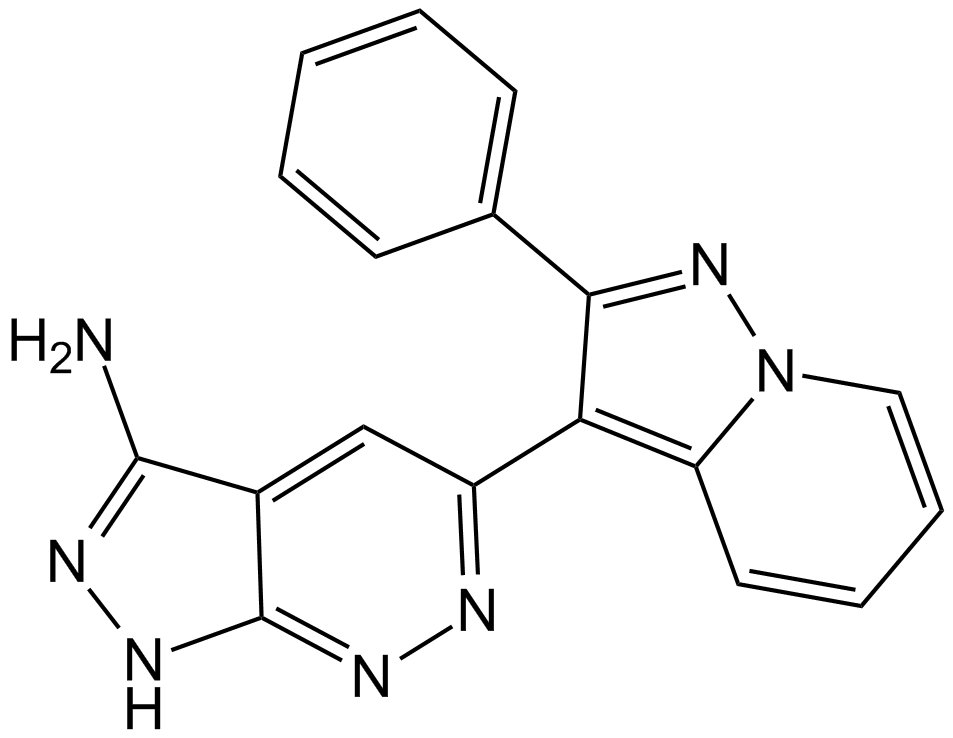
-
GC38044
Fraxinellone
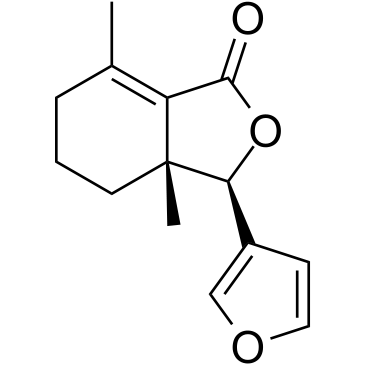
-
GC16310
FTI 277 HCl
FTI 277 HCl is an inhibitor of farnesyl transferase (FTase); a highly potent Ras CAAX peptidomimetic which antagonizes both H- and K-Ras oncogenic signaling.
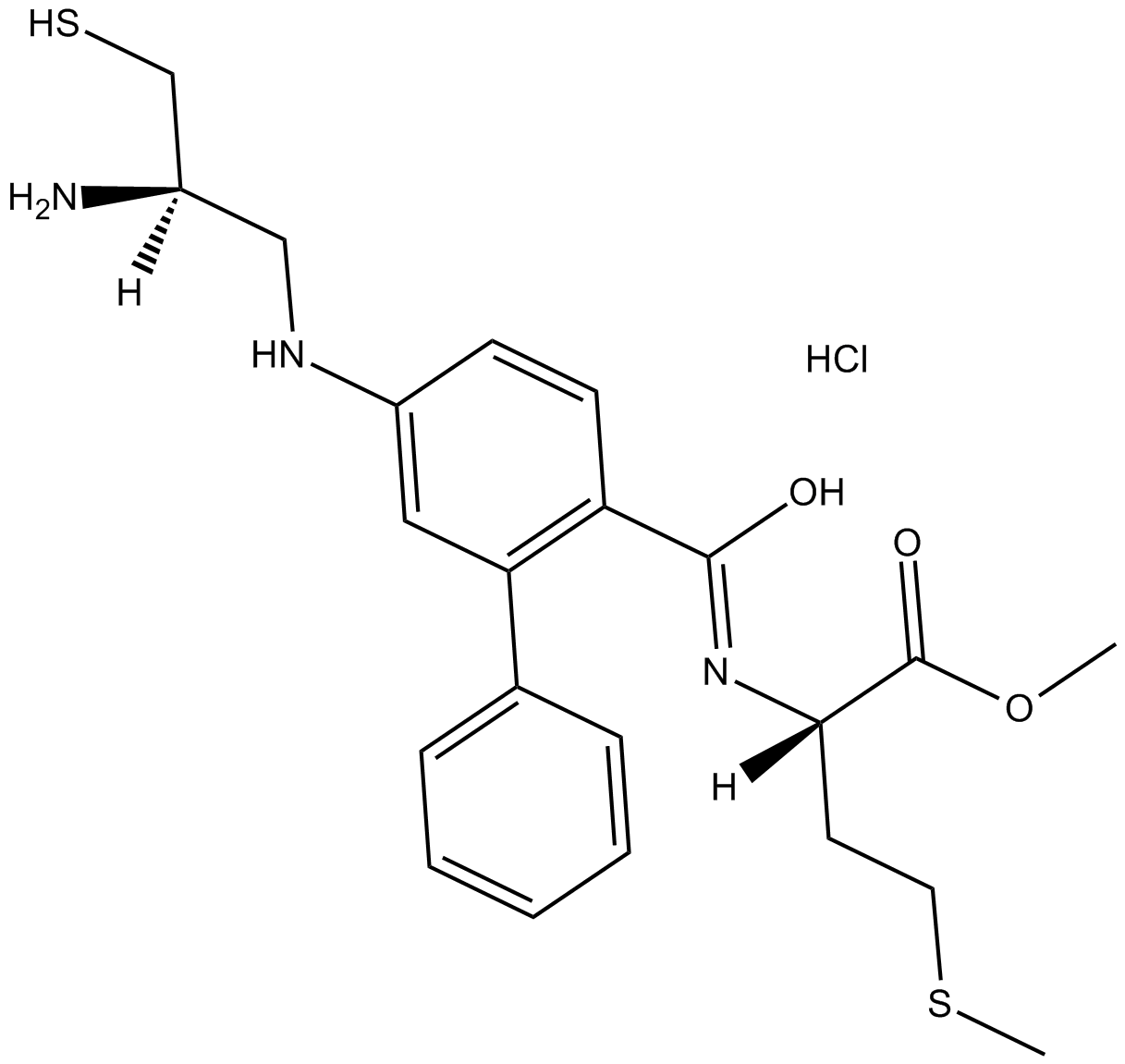
-
GC52288
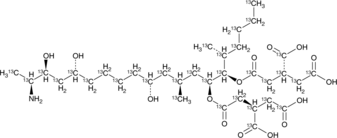
Fumonisin B1-13C34
An internal standard for the quantification of fumonisin B1
-
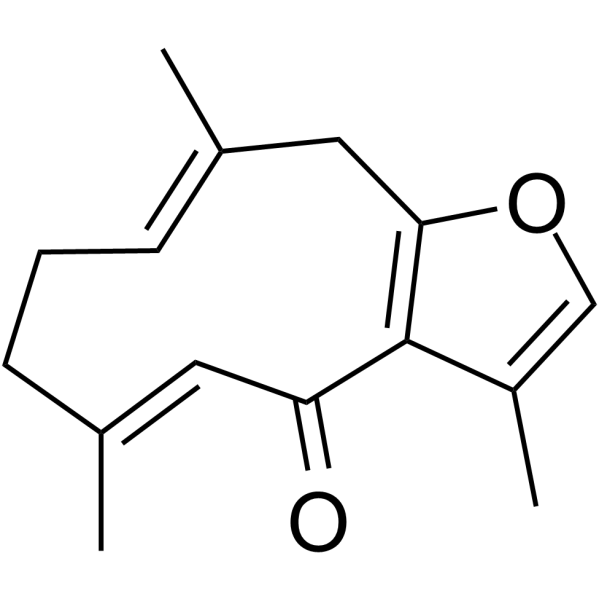
GC62981
Furanodienone
Furanodienone is one of the major bioactive constituents derived from Rhizoma Curcumae. Furanodienone induced apoptosis.
-
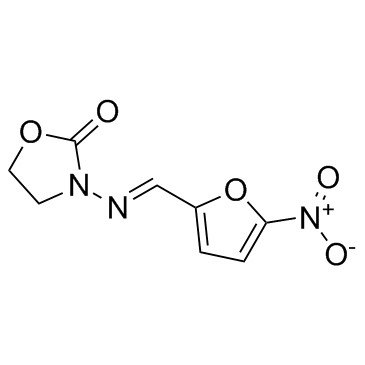
GC32138
Furazolidone
A nitrofuran antiprotozoal and antibacterial agent
-
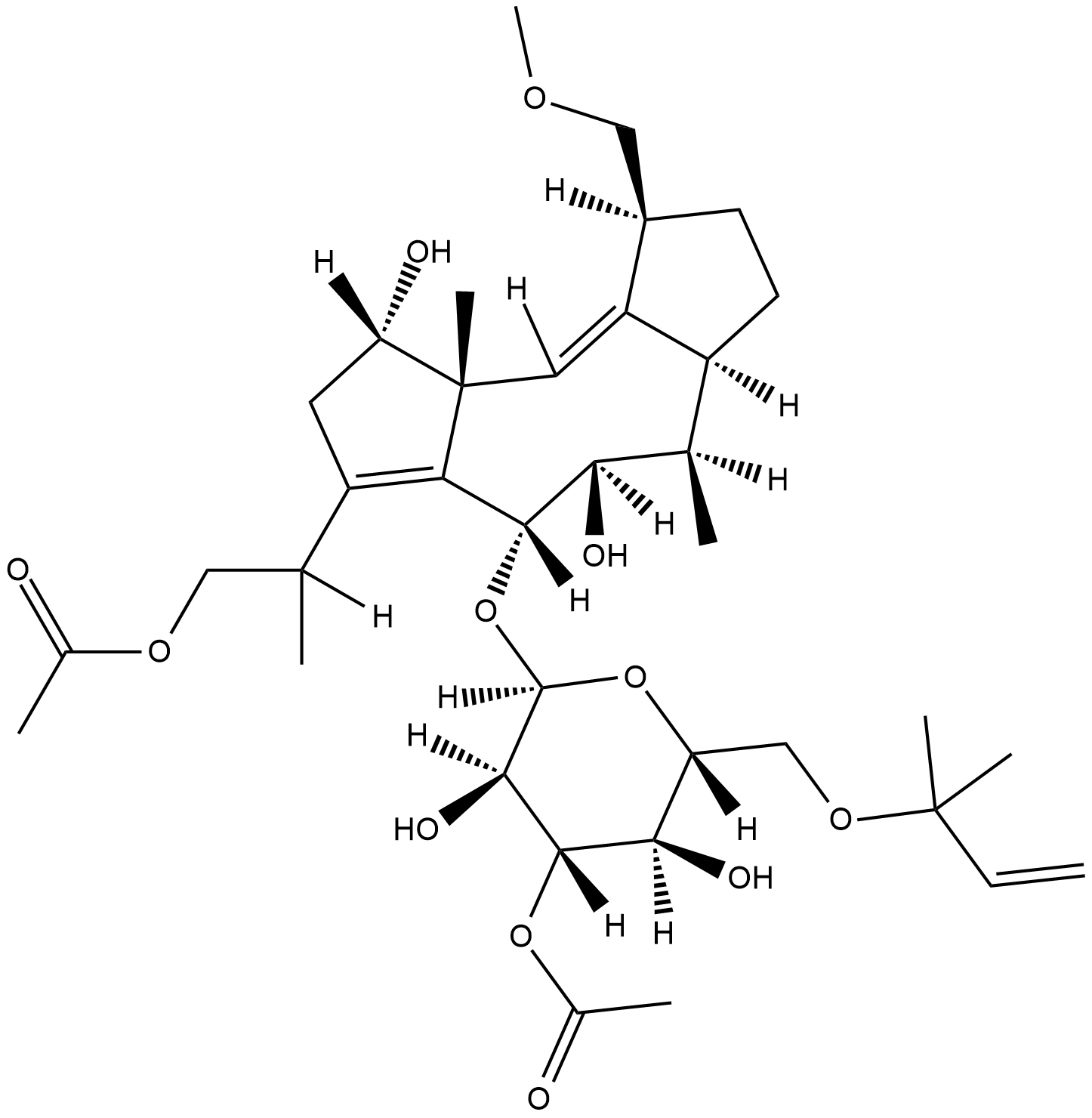
GC18611
Fusicoccin
Fusicoccin is a phytotoxin originally isolated from F.
-
GC39382
FW1256
FW1256 is a phenyl analogue and a slow-releasing hydrogen sulfide (H2S) donor.
-
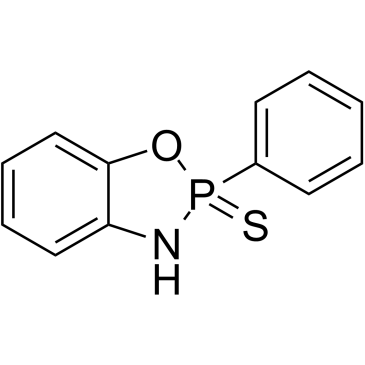
GC12178
G-749
FLT3 inhibitor
-
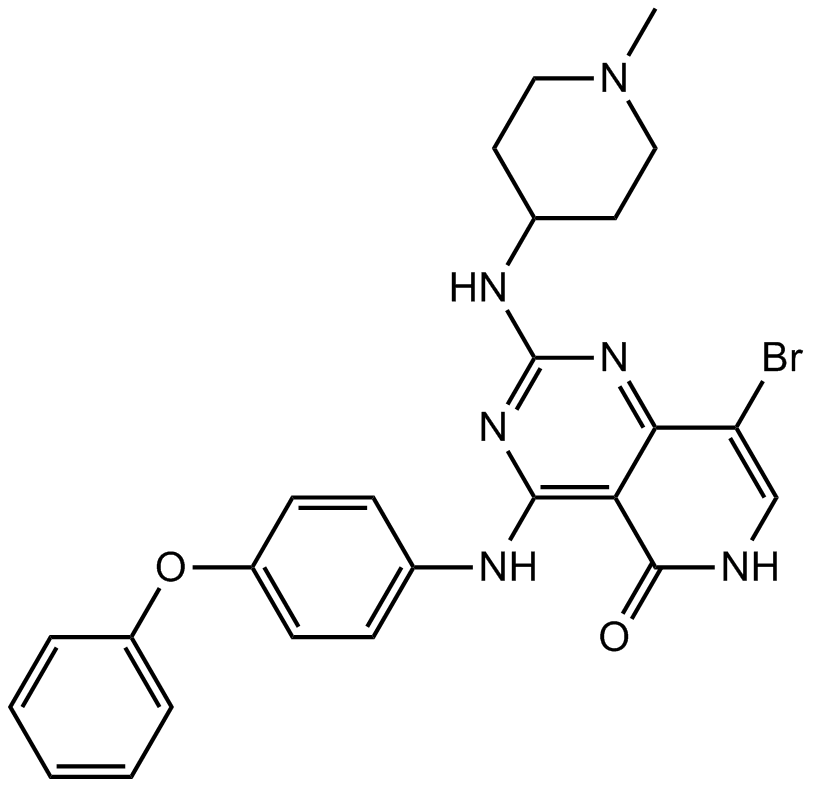
GC62246
G5-7
G5-7, an orally active and allosteric JAK2 inhibitor, selectively inhibits JAK2 mediated phosphorylation and activation of EGFR (Tyr1068) and STAT3 by binding to JAK2. G5-7 induces cell cycle arrest, apoptosis and possesses antiangiogenic effect. G5-7 has the potential for glioma study.
-
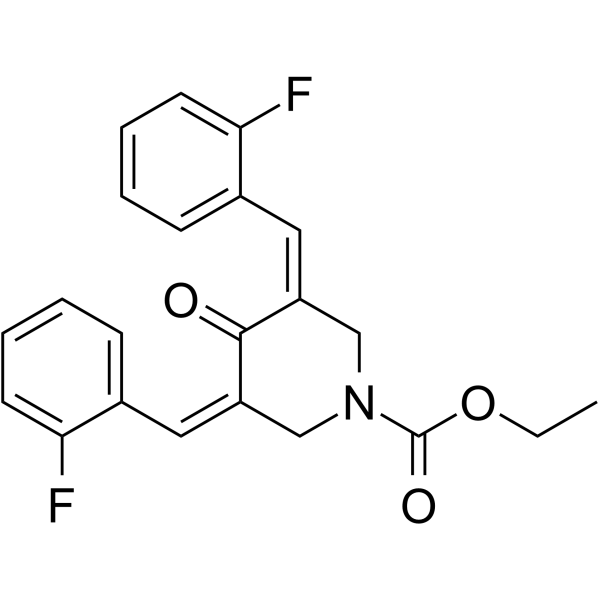
GC43723
Galactosylsphingosine (d18:1)
Galactosylsphingosine (d18:1) (Galactosylsphingosine), a substrate of the galactocerebrosidase (GALC) enzyme, is a potential biomarker for Krabbe disease.
-
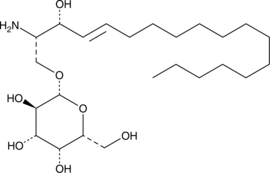
GC36103
Galanthamine hydrobromide
-
GC38610
Galgravin
Galgravin is an active compound in Nectandra megapotamica, with anti-inflammatory activity. Galgravin displays in vitro cytotoxic activity and induce apoptosis in leukemia cells.
-
GN10388
Gallic acid

-
GC61436
Gallic acid hydrate
Gallic acid (3,4,5-Trihydroxybenzoic acid) hydrate is a natural polyhydroxyphenolic compound and an free radical scavenger to inhibit cyclooxygenase-2 (COX-2).
-
GC12139
Gambogic Acid
A xanthonoid with anticancer activity
-
GC36106
Gamma-glutamylcysteine (TFA)
Gamma-glutamylcysteine (γ-Glutamylcysteine) TFA, an intermediate in glutathione (GSH) synthesis, is a dipeptide served as an essential cofactor for the antioxidant enzyme glutathione peroxidase (GPx).
-
GC17655
Ganetespib (STA-9090)
-
GC43729
Ganglioside GD3 Mixture (sodium salt)
Ganglioside GD3 is synthesized by the addition of two sialic acid residues to lactosylceramide and can serve as a precursor to the formation of more complex gangliosides by the action of glycosyl- and sialyltransferases.
-
GC47392
Ganglioside GM1 Mixture (ovine) (ammonium salt)
A mixture of ganglioside GM1