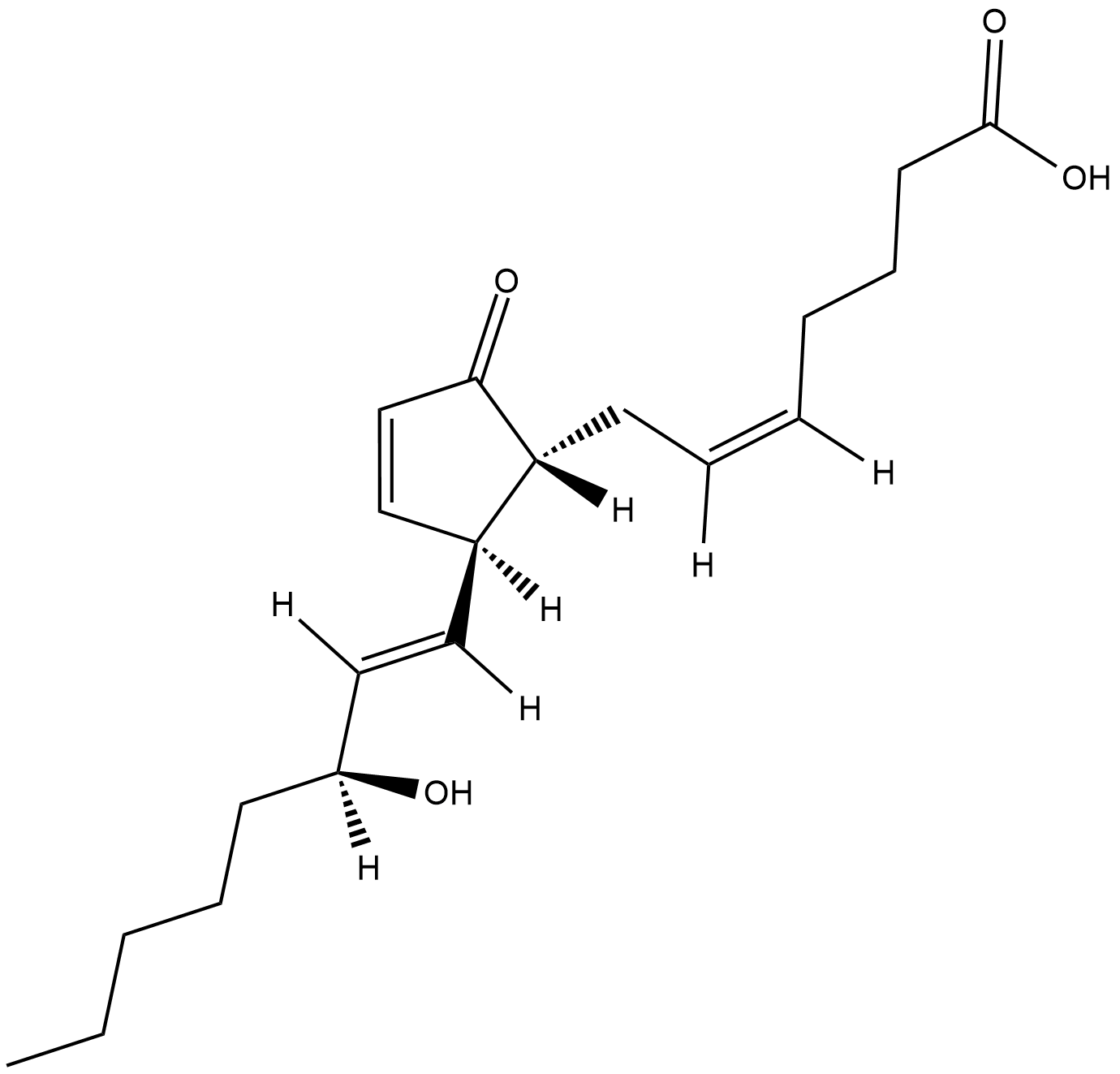
Cell Cycle/Checkpoint
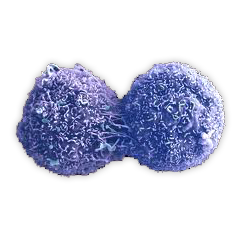
Cell Cycle
Cells undergo a complex cycle of growth and division that is referred to as the cell cycle. The cell cycle consists of four phases, G1 (GAP 1), S (synthesis), G2 (GAP 2) and M (mitosis). DNA replication occurs during S phase. When cells stop dividing temporarily or indefinitely, they enter a quiescent state called G0.
Targets for Cell Cycle/Checkpoint
- ATM/ATR(23)
- Aurora Kinase(17)
- Cdc42(4)
- Cdc7(3)
- Chk(14)
- c-Myc(20)
- CRM1(8)
- Cyclin-Dependent Kinases(77)
- E1 enzyme(1)
- G-quadruplex(11)
- Haspin(6)
- HMTase(1)
- Kinesin(23)
- Ksp(4)
- Microtubule/Tubulin(219)
- Mps1(15)
- Mitotic(7)
- RAD51(16)
- ROCK(60)
- Rho(13)
- PERK(10)
- PLK(33)
- PTEN(6)
- Wee1(7)
- PAK(20)
- Arp2/3 Complex(8)
- Dynamin(10)
- ECM & Adhesion Molecules(40)
- Cholesterol Metabolism(3)
- Endomembrane System & Vesicular Trafficking(26)
- G1(38)
- G2/M(26)
- G2/S(10)
- Genotoxic Stress(18)
- Inositol Phosphates(18)
- Proteolysis(99)
- Cytoskeleton & Motor Proteins(53)
- Cellular Chaperones(8)
Products for Cell Cycle/Checkpoint
- Cat.No. Product Name Information
-
GC44464
NSC 109555
An inhibitor of Chk2
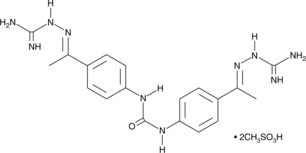
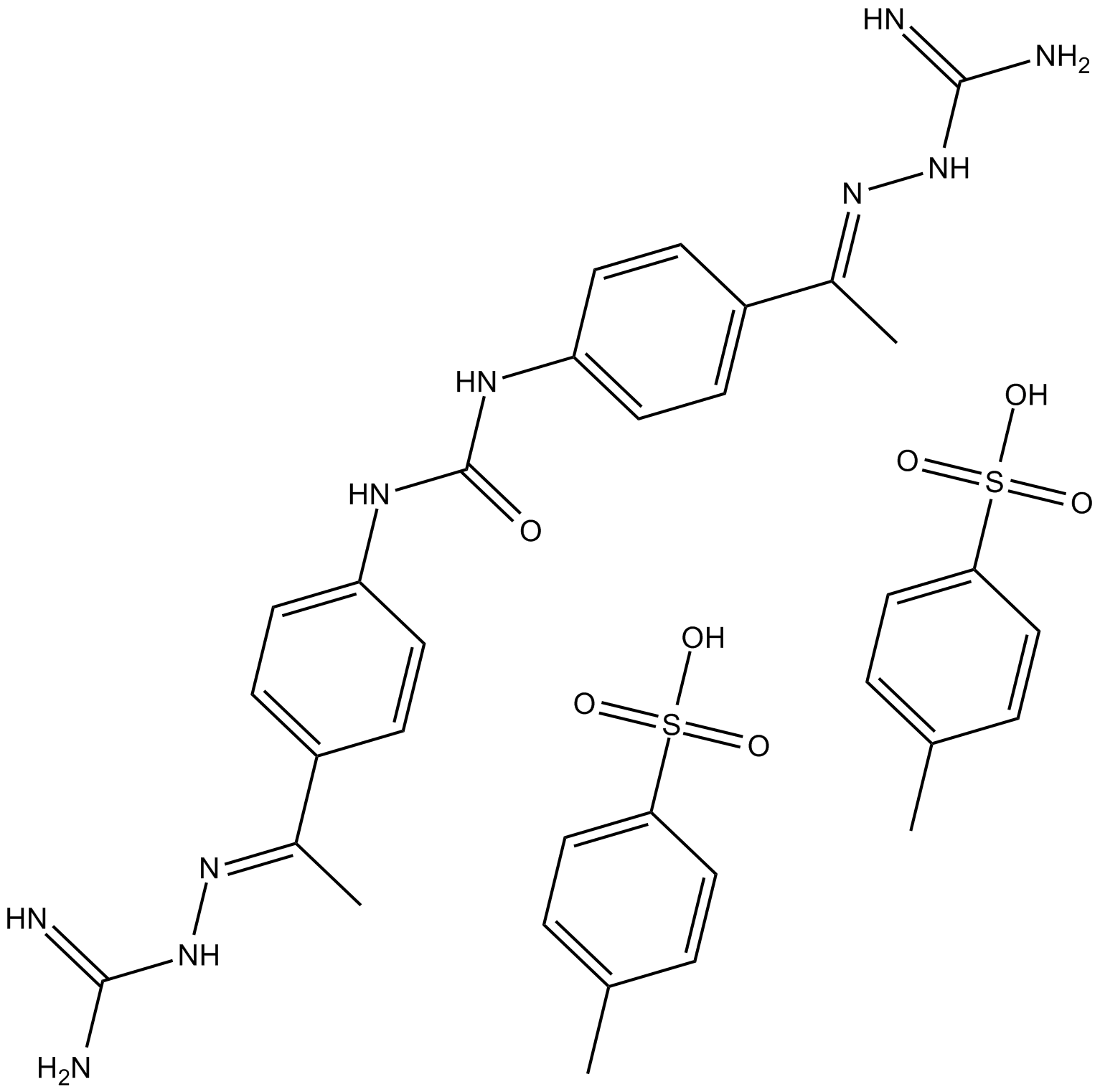
-
GC13448
NSC 109555 ditosylate
Chk2 inhibitor,ATP-competitive
-
GC46190
NSC 15520
An inhibitor of RPA70 dsDNA binding
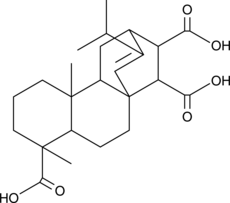
-
GC10069
NSC 23766 trihydrochloride
Selective inhibitor of Rac1-GEF interaction.
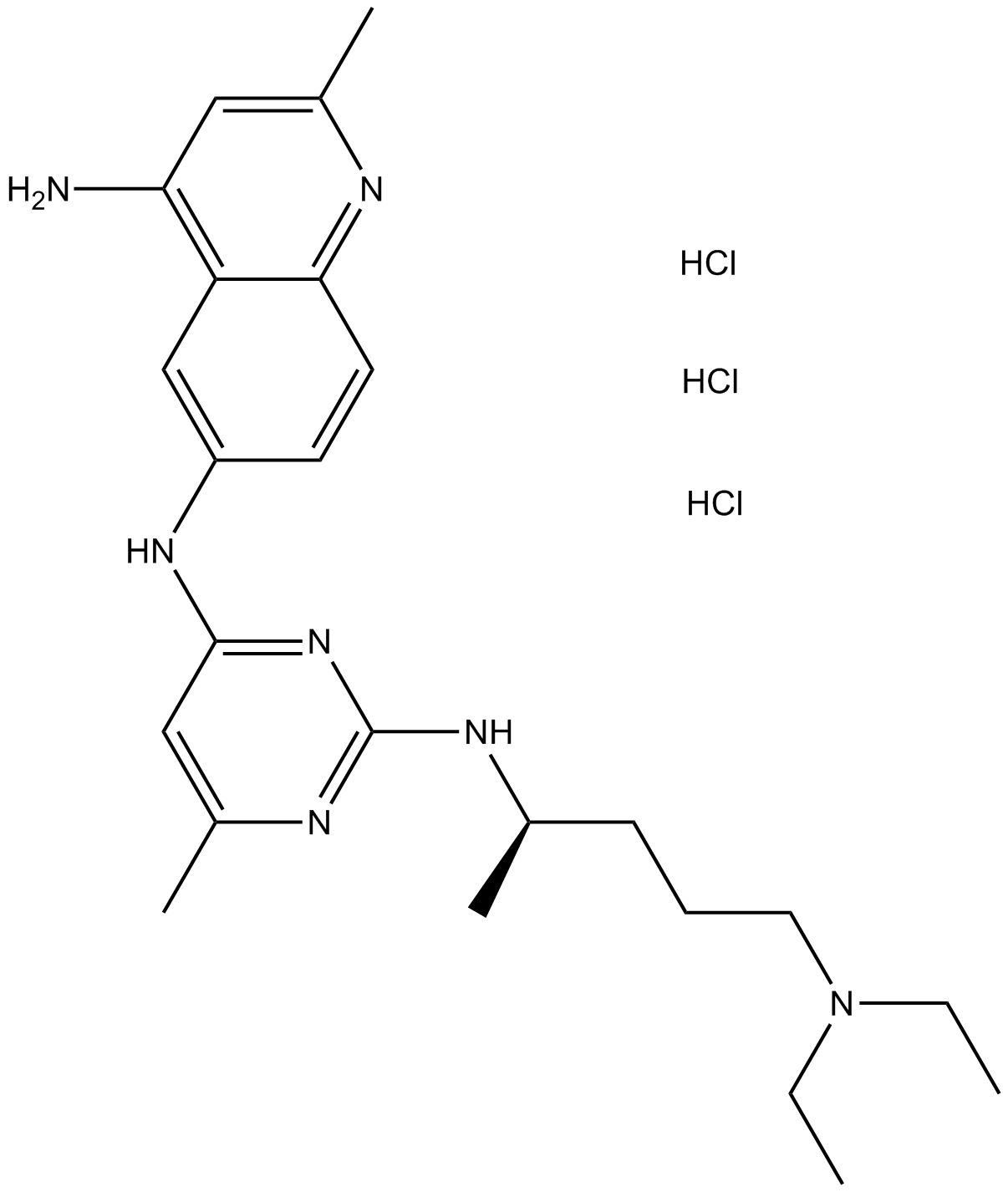
-
GC12886
NSC 625987
Cyclin-dependent kinase (cdk) 4 inhibitor
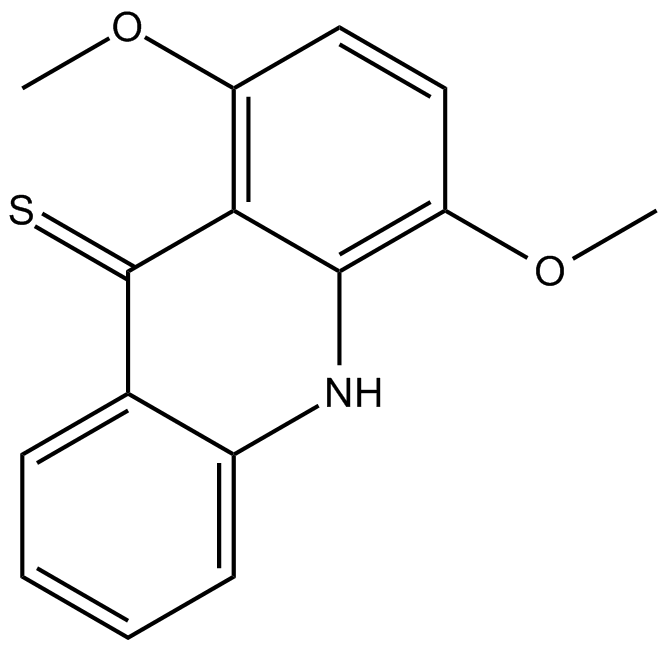
-
GC11634
NSC 693868
CDKs and GSK-3 inhibitor
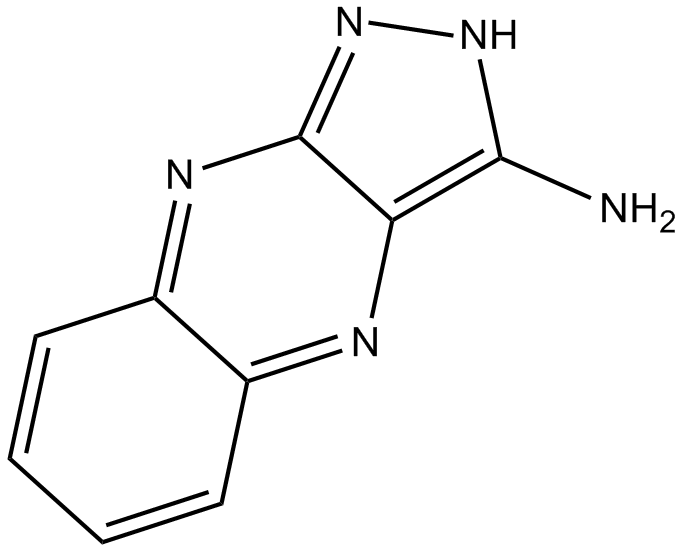
-
GC49412
NSC 756093
An inhibitor of the GBP1-Pim-1 protein-protein interaction
-
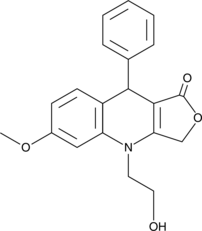
GC15963
Nu 6027
ATR/CDK inhibitor, potent and selective
-
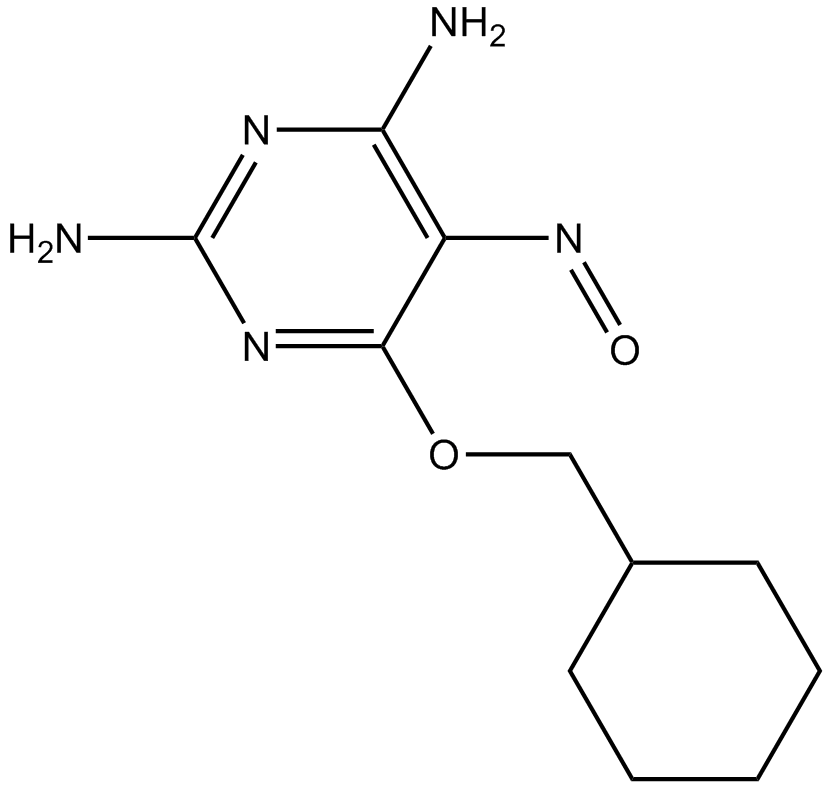
GC18278
NU 6102
Cyclin-dependent kinases (CDKs) play a key role in regulating cell division by phosphorylating distinct substrates in different phases of the cell cycle.
-
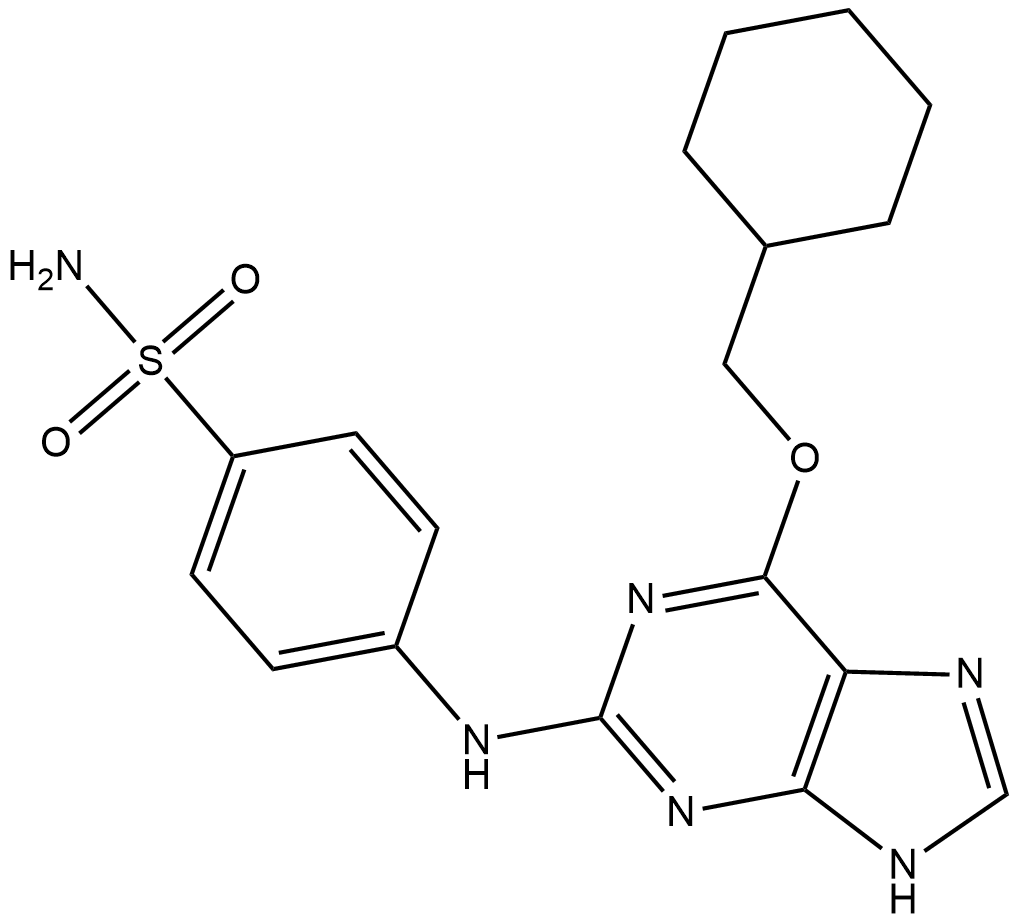
GC34692
NU6140
A Cdk2 inhibitor
-
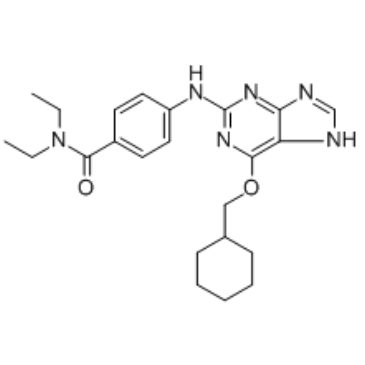
GC16959
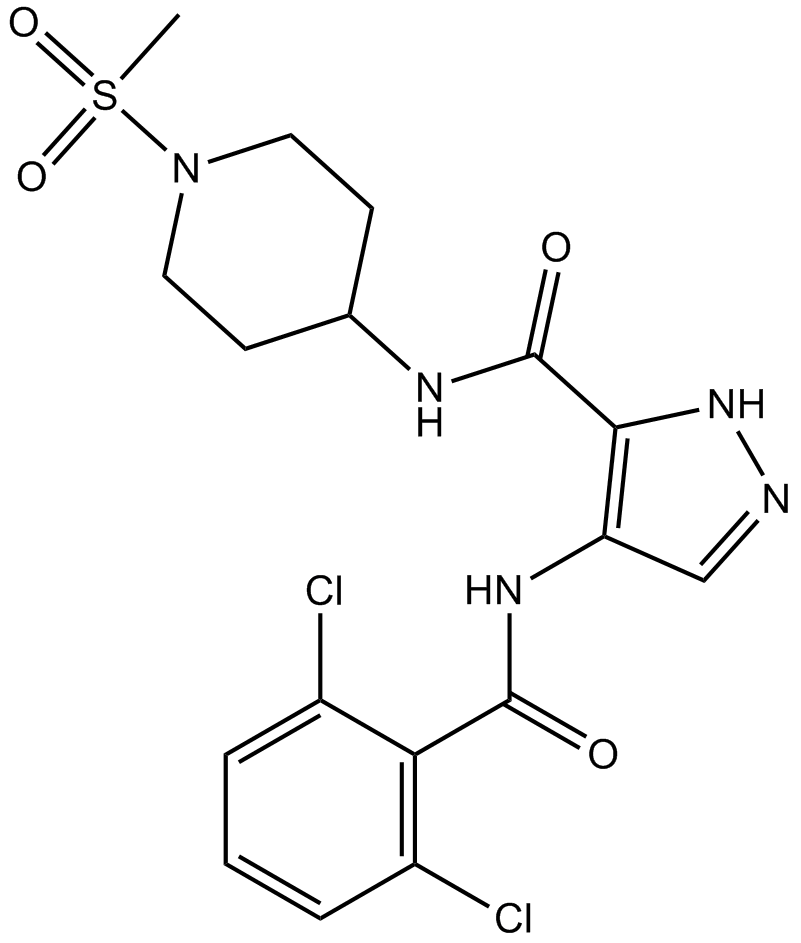
NVP-LCQ195
-
GC32830
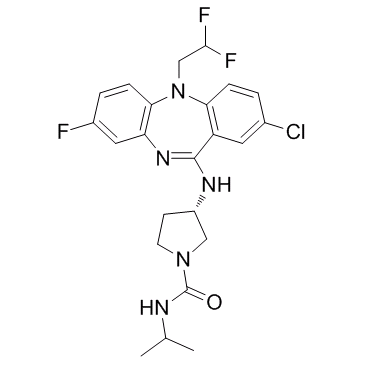
NVS-PAK1-1
An allosteric inhibitor of PAK1
-
GC44478
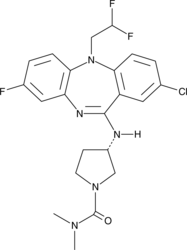
NVS-PAK1-C
NVS-PAK1-C is a negative control for the allosteric PAK1 inhibitor NVS-PAK1-1.
-
GC61870
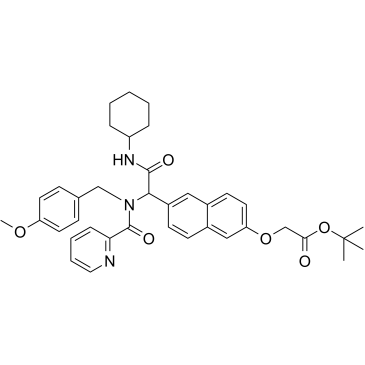
NY2267
NY2267 is a disruptor of Myc-Max interaction, with an IC50 of 36.5 μM. NY2267 inhibits Myc- and Jun-induced transcriptional activation.
-
GC49186
O-Demethyl Apremilast
An active metabolite of apremilast
-
GC18727
O-methyl Sterigmatocystin
O-methyl Sterigmatocystin (OMST) is a xanthone found in several fungal species including A.
-
GC47814
Ochratoxin A-13C20
An internal standard for the quantification of ochratoxin A
-
GC44495
OH-C-Chol
OH-C-Chol is a cationic cholesterol derivative.
-
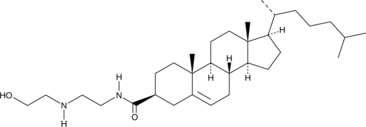
GC45534
OH-Chol
-
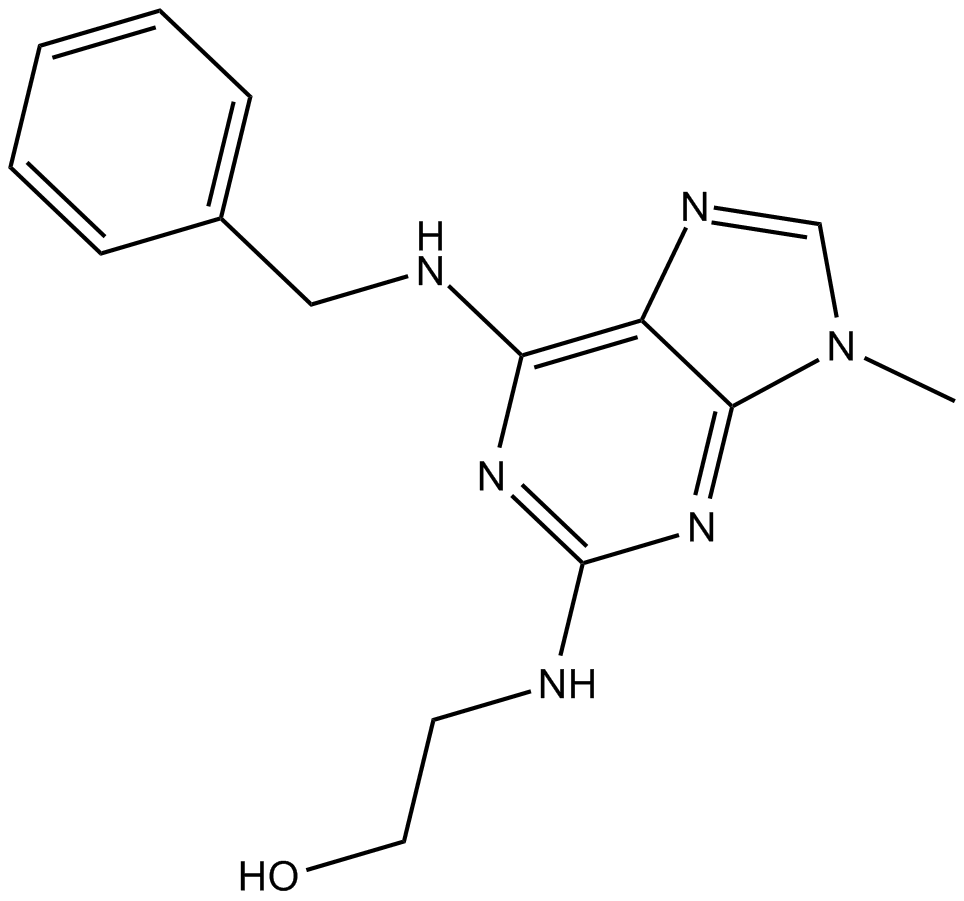
GC16520
Olomoucine
cdk inhibitor
-
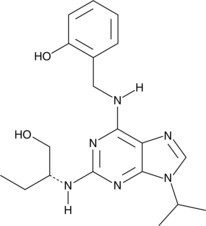
GC48652
Olomoucine II
A CDK inhibitor
-
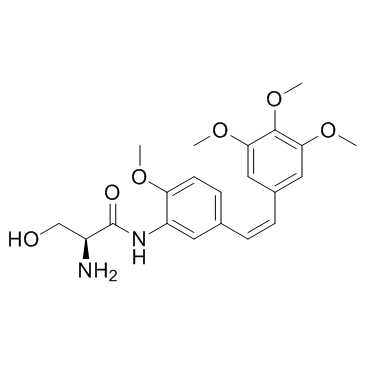
GC32551
Ombrabulin (AC-7700)
Ombrabulin (AC-7700) (AVE8062) is a derivative of CA-4 phosphate, which is known to exhibit antivascular effects through selective disruption of the tubulin cytoskeleton of endothelial cells.
-
GC34022
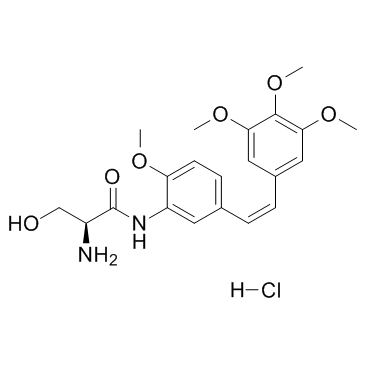
Ombrabulin hydrochloride (AVE8062)
Ombrabulin hydrochloride (AVE8062) is a derivative of CA-4 phosphate, which is known to exhibit antivascular effects through selective disruption of the tubulin cytoskeleton of endothelial cells.
-
GC62174
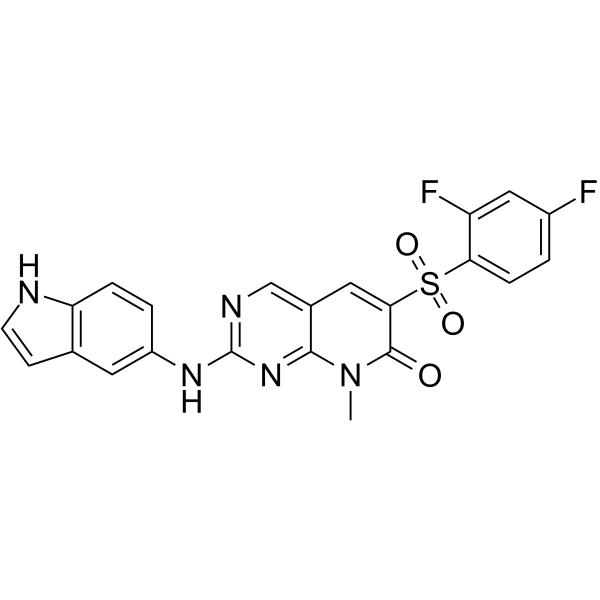
ON1231320
ON1231320 is a highly specific polo like kinase 2 (PLK2) inhibitor with an IC50 of 0.31 ?M. ON1231320 blocks tumor cell cycle progression in the G2/M phase in mitosis, causing apoptotic cell death. ON1231320, an arylsulfonyl pyrido-pyrimidinone, has antitumor activity.
-
GC15607
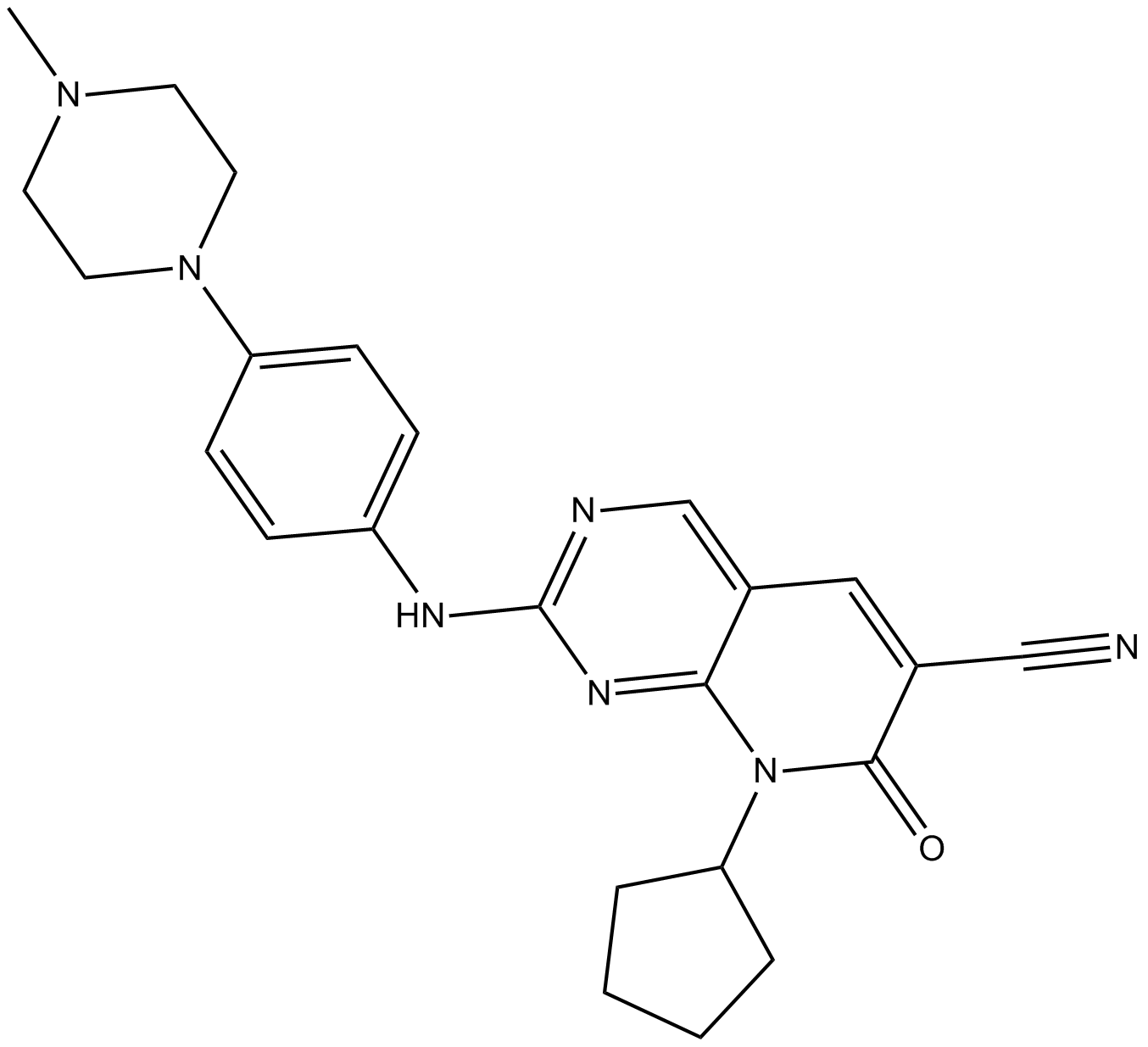
ON123300
multi-targeted kinase inhibitor,inhibits CDK4, Ark5, PDGFRβ, FGFR1, RET, and Fyn
-
GN10732
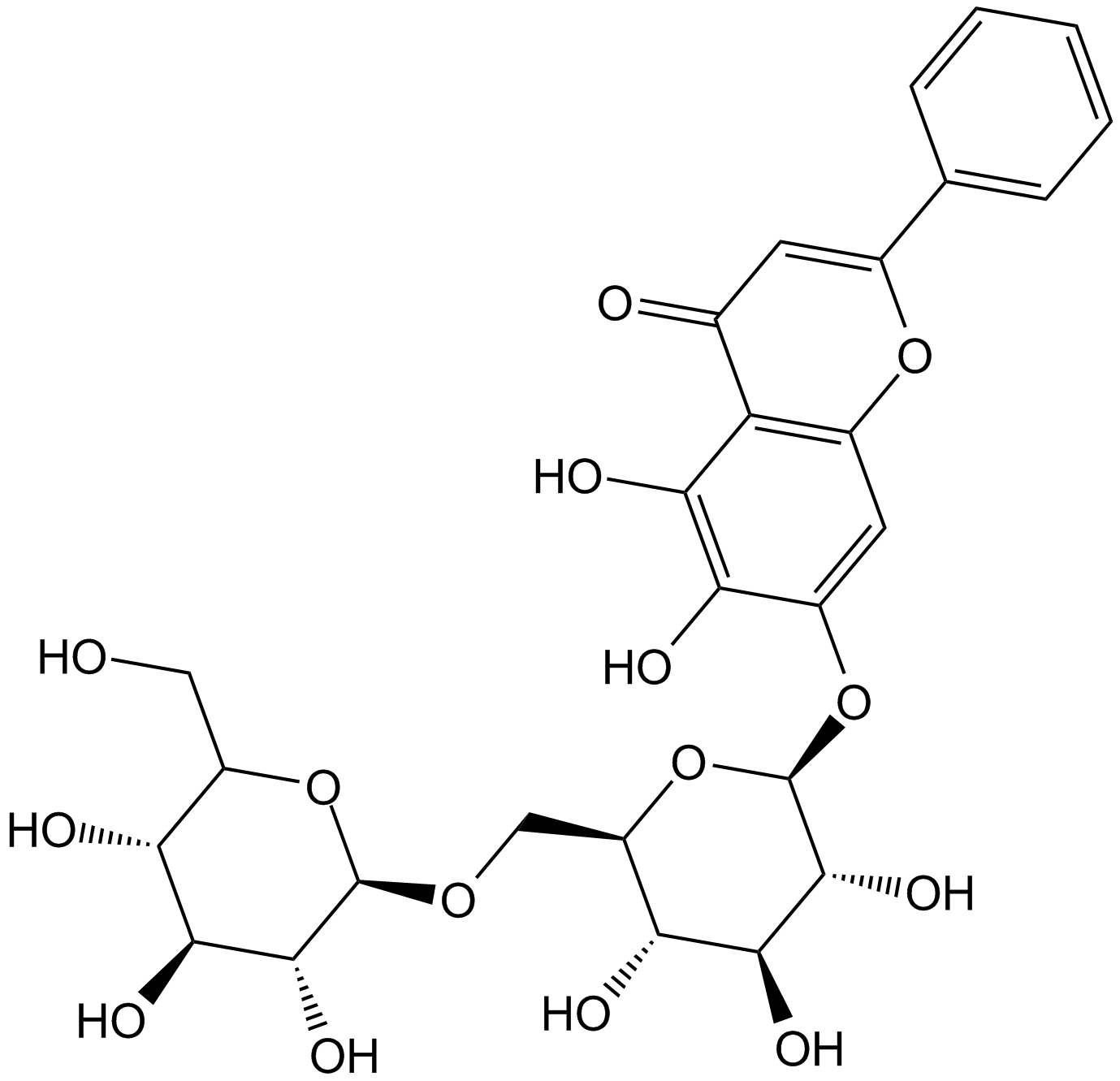
Oroxin B
-
GC64626
Oryzalin
Oryzalin is a dinitroaniline herbicide, binding to plant tubulin and inhibits microtubule (MT) polymerization in vitro.
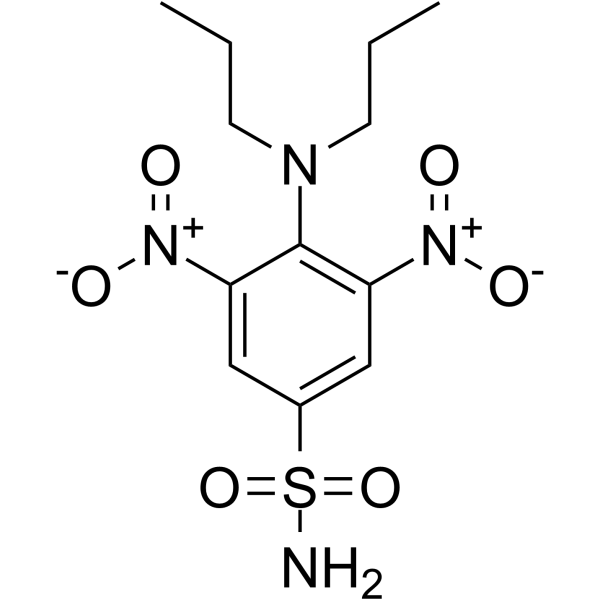
-
GC33163
OSIP-486823 (OSIP 486823)
OSIP-486823 (OSIP 486823) is a novel microtubule-interfering agent with distinct biological effects on both protein kinase G (PKG) and microtubules.

-
GC50116
OXA 06 dihydrochloride
Potent ROCK inhibitor
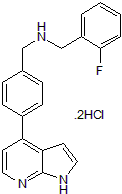
-
GC65897
OXi8007
OXi8007 is a water-soluble phosphate prodrug of OXi8006, a tubulin-binding compound.
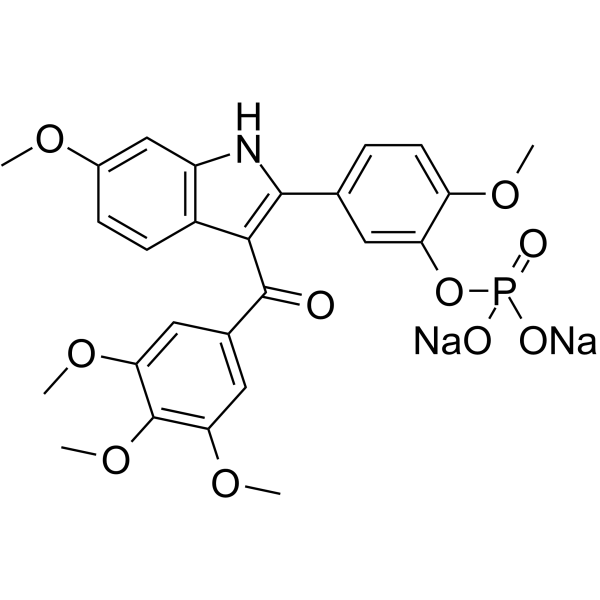
-
GC45538
Oxychlororaphine
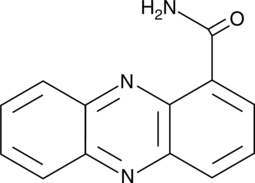
-
GA23337
Oxythiamine . HCl
Thiamine antagonist and inhibitor of saccharomyces cerevisiae transketolase. RaÏs et al. indicated that oxythiamine inhibits the growth of Ehrlich's tumor cells.

-
GC40256
p-APMSF (hydrochloride)
p-APMSF is an irreversible inhibitor of serine proteases with Ki values of 1.02, 1.18, 1.5, and 1.54 μM for bovine trypsin, human thrombin, bovine plasmin, and bovine Factor Xa, respectively.
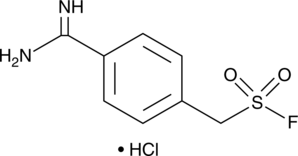
-
GC50599
P110
Dynamin-related protein 1 (Drp1) inhibitor; cell-permeable
-
GC12511
Paclitaxel (Taxol)
A potent mitotic inhibitor
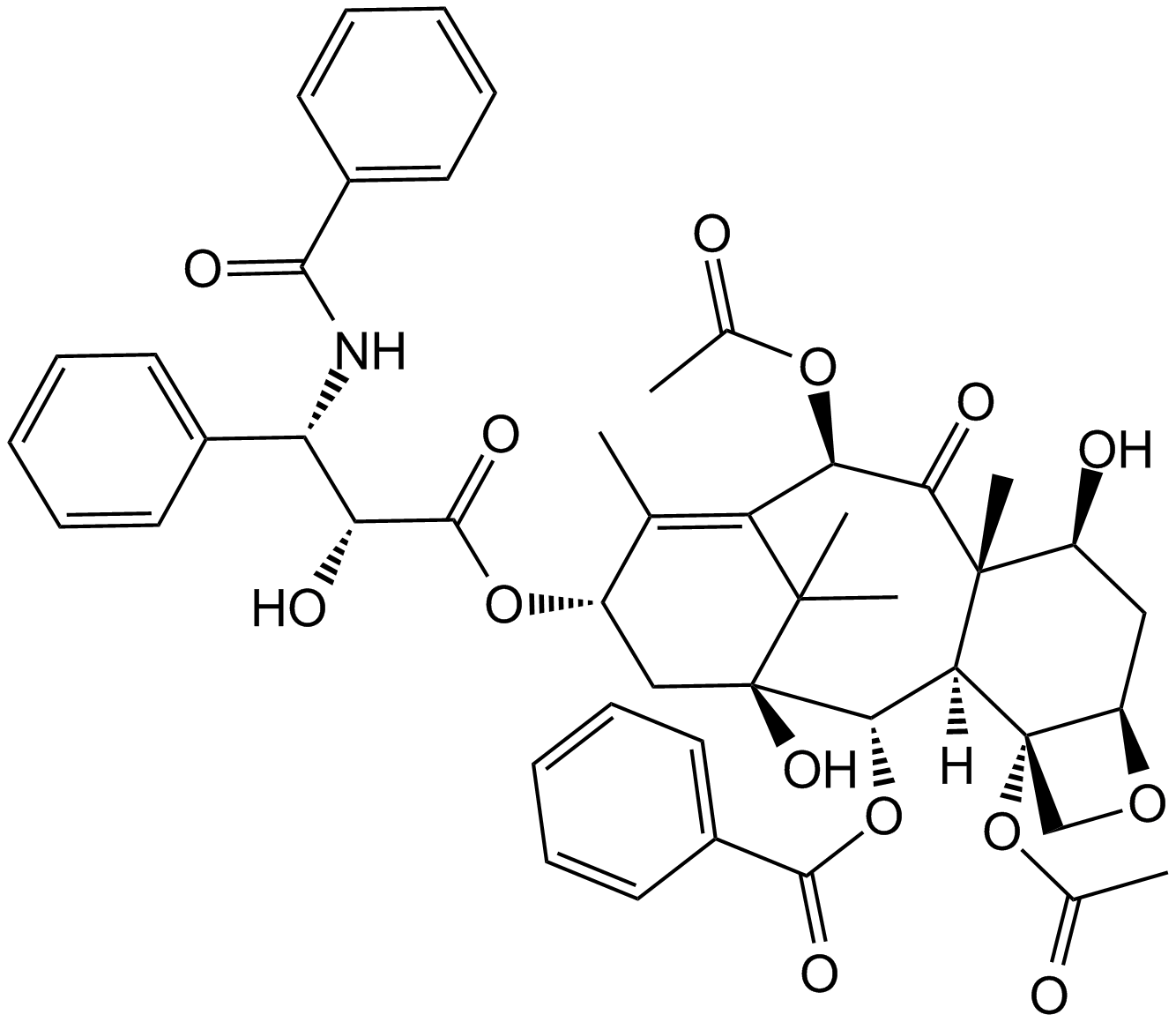
-
GC49404
Paclitaxel C
A taxoid and an inhibitor of microtubule depolymerization
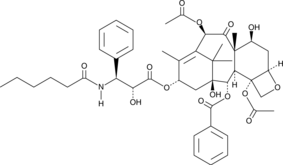
-
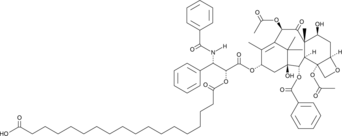
GC45758
Paclitaxel octadecanedioate
A prodrug form of paclitaxel
-
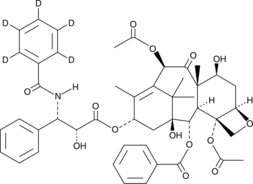
GC47853
Paclitaxel-d5
An internal standard for the quantification of paclitaxel
-
GC33421
PAK-IN-1
PAK-IN-1 is a PAK inhibitor that displays group II selectivity. PAK-IN-1 inhibits PAK4, PAK5 and PAK6 with IC50s of 7.5, 36, 126 nM, respectively.
-
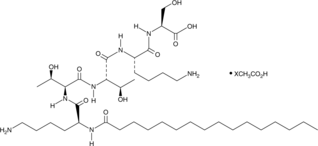
GC46196
Pal-KTTKS (acetate)
A lipidated pentapeptide
-
GC15421
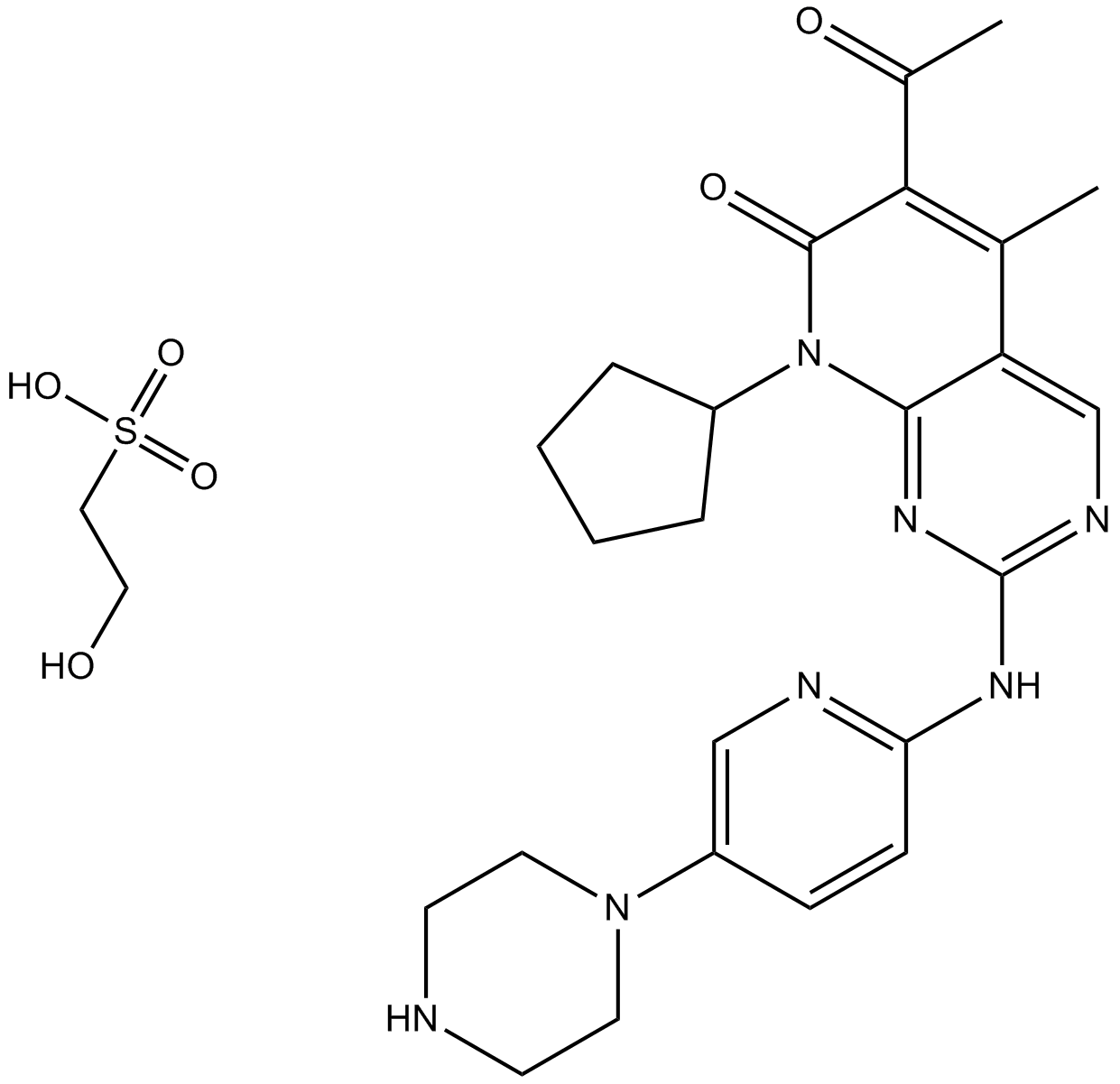
Palbociclib (PD0332991) Isethionate
Palbociclib (PD 0332991) isethionate is an orally active selective CDK4 and CDK6 inhibitor with IC50 values of 11 and 16 nM, respectively. Palbociclib (PD0332991) Isethionate has potent anti-proliferative activity and induces cell cycle arrest in cancer cells, which can be used in the research of HR-positive and HER2-negative breast cancer and hepatocellular carcinoma.
-
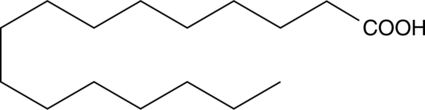
GC49024
Palmitic Acid MaxSpec® Standard
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
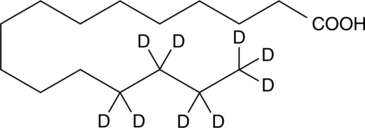
GC49023
Palmitic Acid-d9 MaxSpec® Standard
A quantitative analytical standard guaranteed to meet MaxSpec® identity, purity, stability, and concentration specifications
-
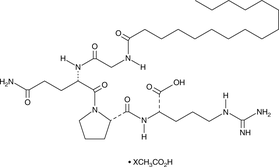
GC49014
Palmitoyl Tetrapeptide-7 (acetate)
A cosmeceutical peptide
-
GC15940
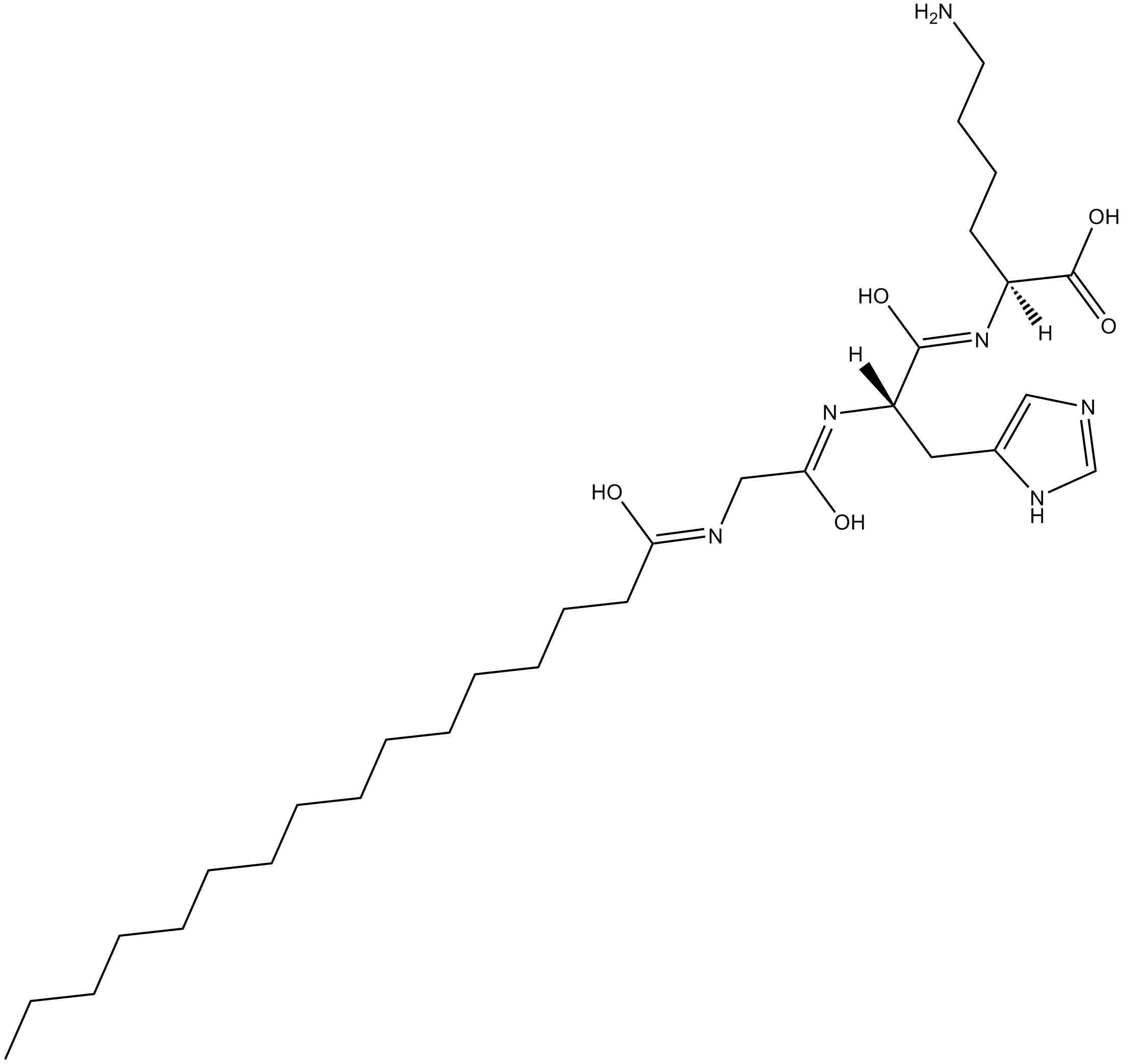
PalMitoyl Tripeptide-1
A form of GHK containing palmitic acid
-
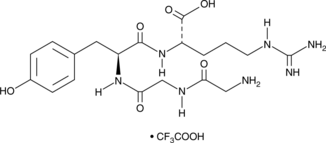
GC49702
Papain Inhibitor (trifluoroacetate salt)
A peptide inhibitor of papain
-
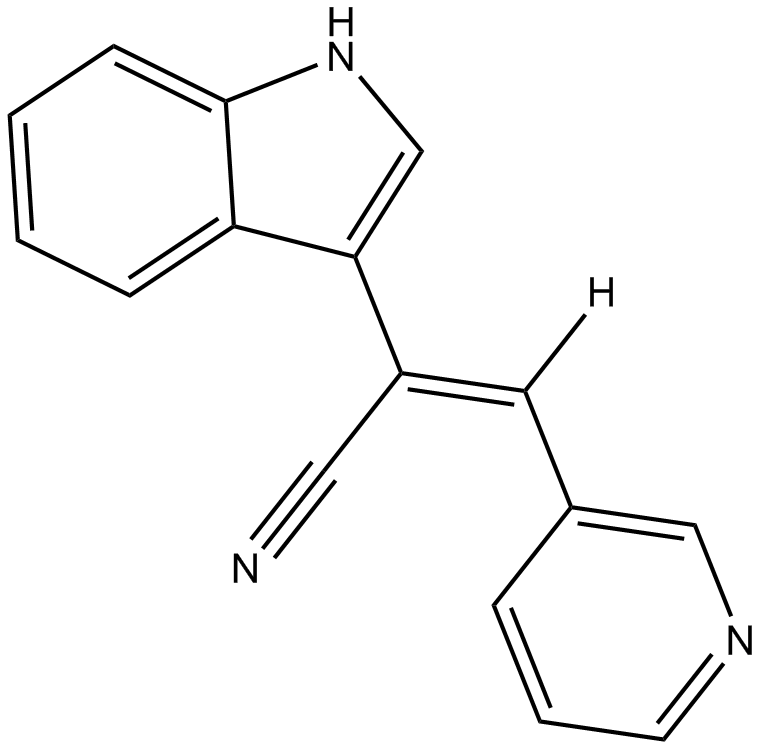
GC12788
Paprotrain
mitotic kinesin-like protein 2 (MKLP-2) inhibitor
-
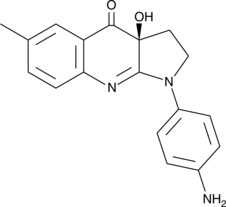
GC45944
para-amino-Blebbistatin
A more stable and less phototoxic form of (–)-blebbistatin with enhanced water solubility
-
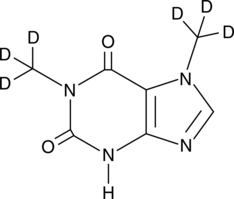
GC47923
Paraxanthine-d6
An internal standard for the quantification of paraxanthine
-
GC32134
Parbendazole (SKF 29044)
Parbendazole (SKF 29044) is a potent inhibitor of microtubule assembly, destabilizes tubulin, with an EC50 of 530nM, and exhibits a broad-spectrum anthelmintic activity.
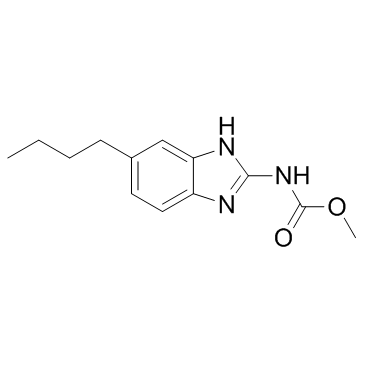
-
GC40085
Pazopanib-d6
Pazopanib-d6 is intended for use as an internal standard for the quantification of pazopanib by GC- or LC-MS.
-
GC33002
PBOX 6
PBOX 6 is a pyrrolo-1,5-benzoxazepine (PBOX) compound, acts as a microtubule-depolymerizing agent and an apoptotic agent.
-
GC15173
PD 0332991 (Palbociclib)
PD 0332991 (Palbociclib) (PD 0332991) is an orally active selective CDK4 and CDK6 inhibitor with IC50 values of 11 and 16 nM, respectively. PD 0332991 (Palbociclib) has potent anti-proliferative activity and induces cell cycle arrest in cancer cells, which can be used in the research of HR-positive and HER2-negative breast cancer and hepatocellular carcinoma.
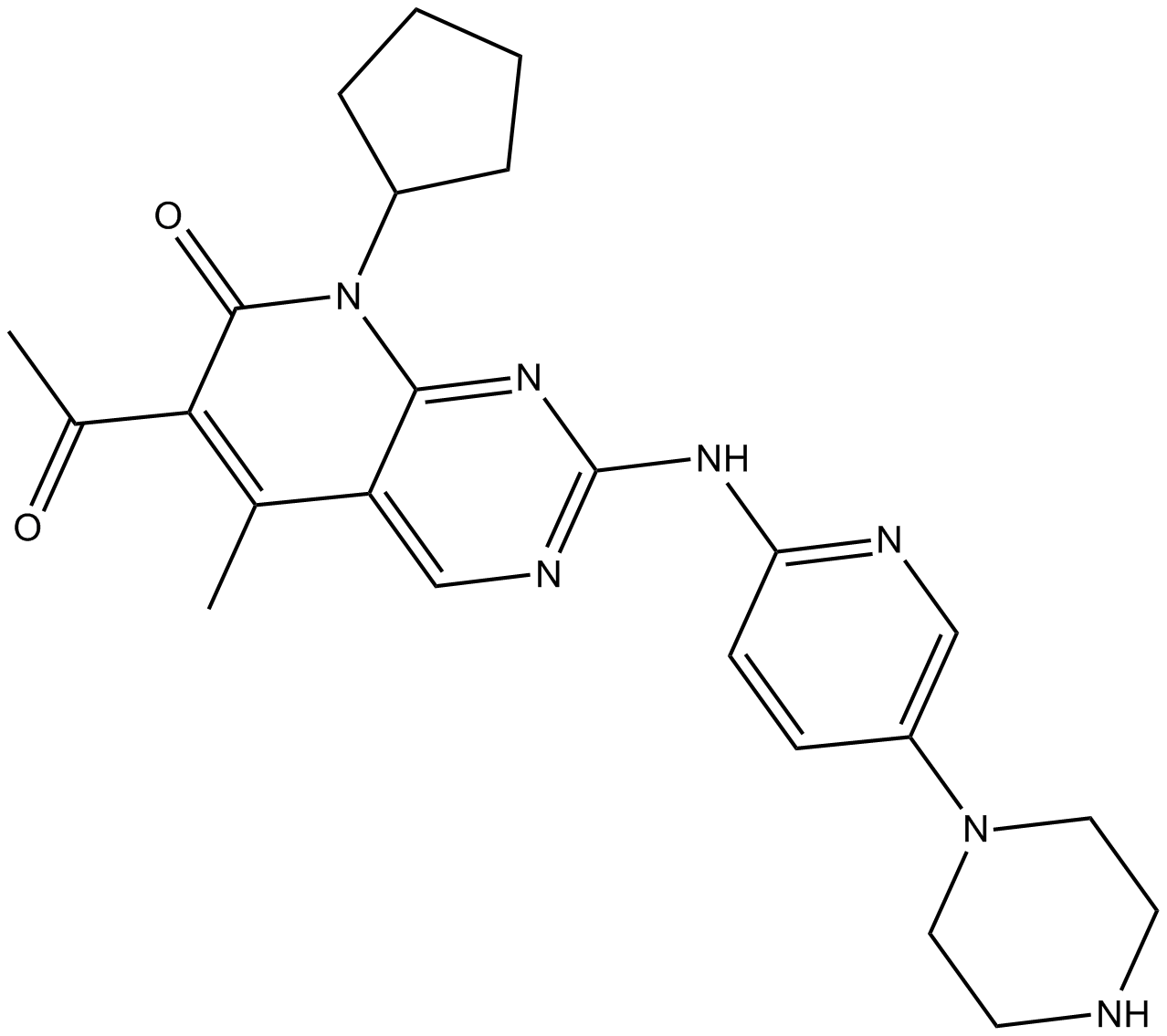
-
GC17935
PD 0332991 (Palbociclib) HCl
Palbociclib (PD 0332991) monohydrochloride is an orally active selective CDK4 and CDK6 inhibitor with IC50 values of 11 and 16 nM, respectively. PD 0332991 (Palbociclib) HCl has potent anti-proliferative activity and induces cell cycle arrest in cancer cells, which can be used in the research of HR-positive and HER2-negative breast cancer and hepatocellular carcinoma.
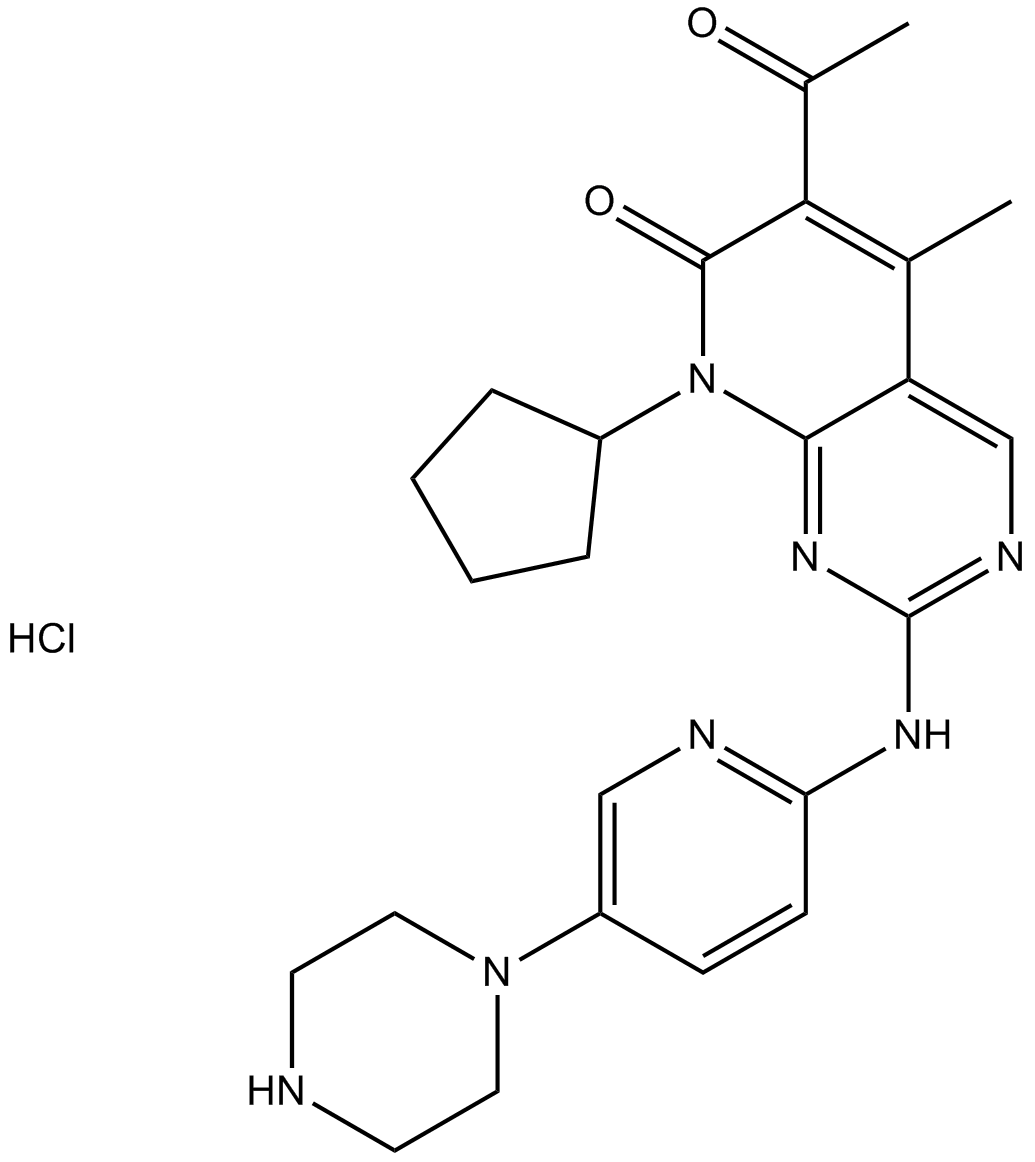
-
GC50045
PD 166285 dihydrochloride
PD 166285 dihydrochloride, a substrate of P-gp, is a WEE1 inhibitor and a weak Myt1 inhibitor with IC50 values of 24 and 72 nM, respectively. PD 166285 dihydrochloride exhibits an IC50 of 3.433 μM for Chk1.
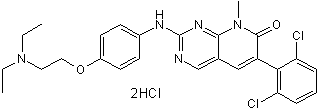
-
GC44584
PD 168368
PD 168368 is a competitive antagonist of neuromedin B (NMB) receptors (Kis = 15-45 nM for rat and human receptors expressed in various cell lines).
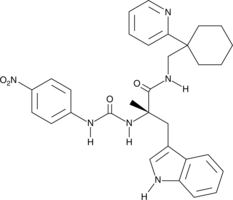
-
GC38216
PD 407824
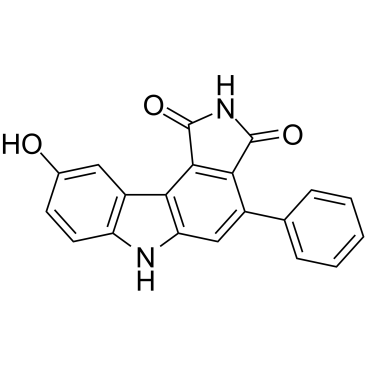
-
GC32853
PD0166285
PD0166285, a substrate of P-gp, is a WEE1 inhibitor and a weak Myt1 inhibitor with IC50 values of 24 and 72 nM, respectively. PD0166285 exhibits an IC50 of 3.433 μM for Chk1.
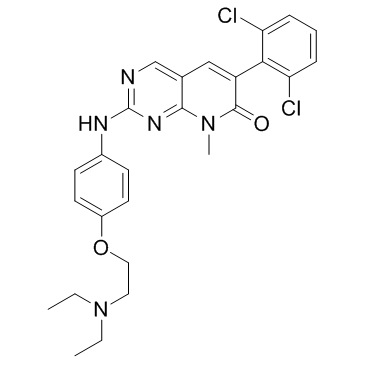
-
GC30801
PE859
PE859 is a potent inhibitor of both tau and Aβ aggregation with IC50 values of 0.66 and 1.2 μM, respectively.
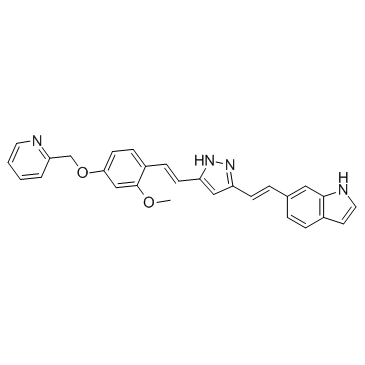
-
GC40622
Penicillide
Penicillide is a fungal metabolite originally isolated from Penicillium that has diverse biological activities.
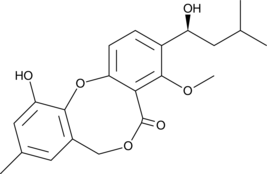
-
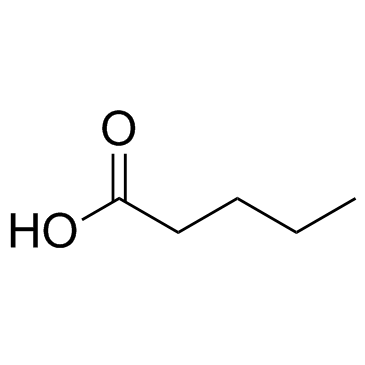
GC33901
Pentanoic acid
-
GC50439
Peptide5
Connexin43 mimetic peptide

-
GC69687
PERK-IN-2
PERK-IN-2 is a highly efficient PERK inhibitor with an IC50 value of 0.2 nM.

-
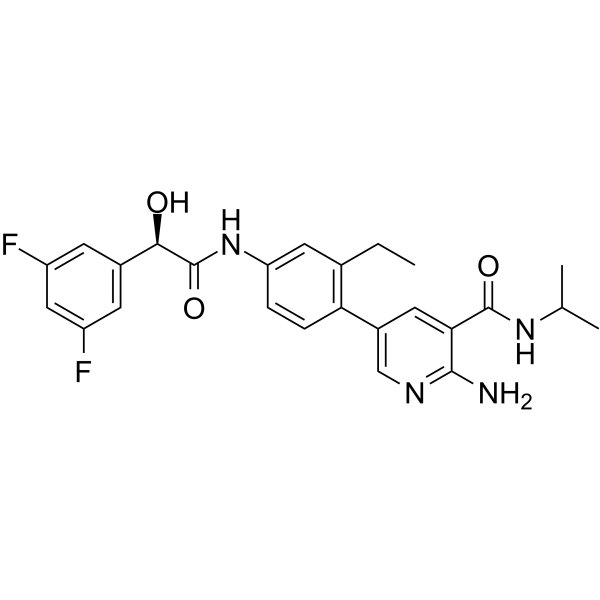
GC64866
PERK-IN-5
PERK-IN-5 is a highly potent, selectively and orally bioavailable PERK inhibitor (IC50s of 2 and 9 nM for PERK and p-eIF2α, respectively). PERK-IN-5 can significantly inhibit tumor growth in the 786-O renal cell carcinoma xenograft tumor model.
-
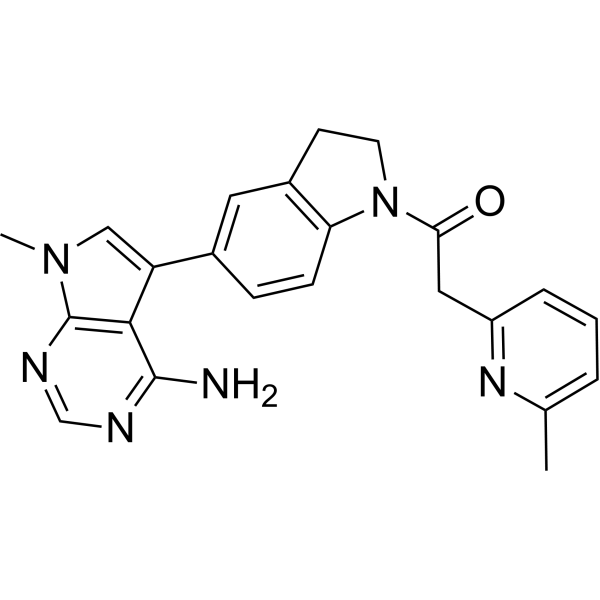
GC69688
PERK-IN-6
PERK-IN-6 (Compound 5) is a PERK inhibitor with an IC50 of 2.5 nM.
-
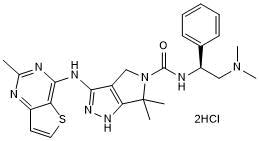
GC50314
PF 3758309 dihydrochloride
Potent PAK4 inhibitor; orally available
-
GC11944
PF 4800567 hydrochloride
casein kinase 1ε inhibitor
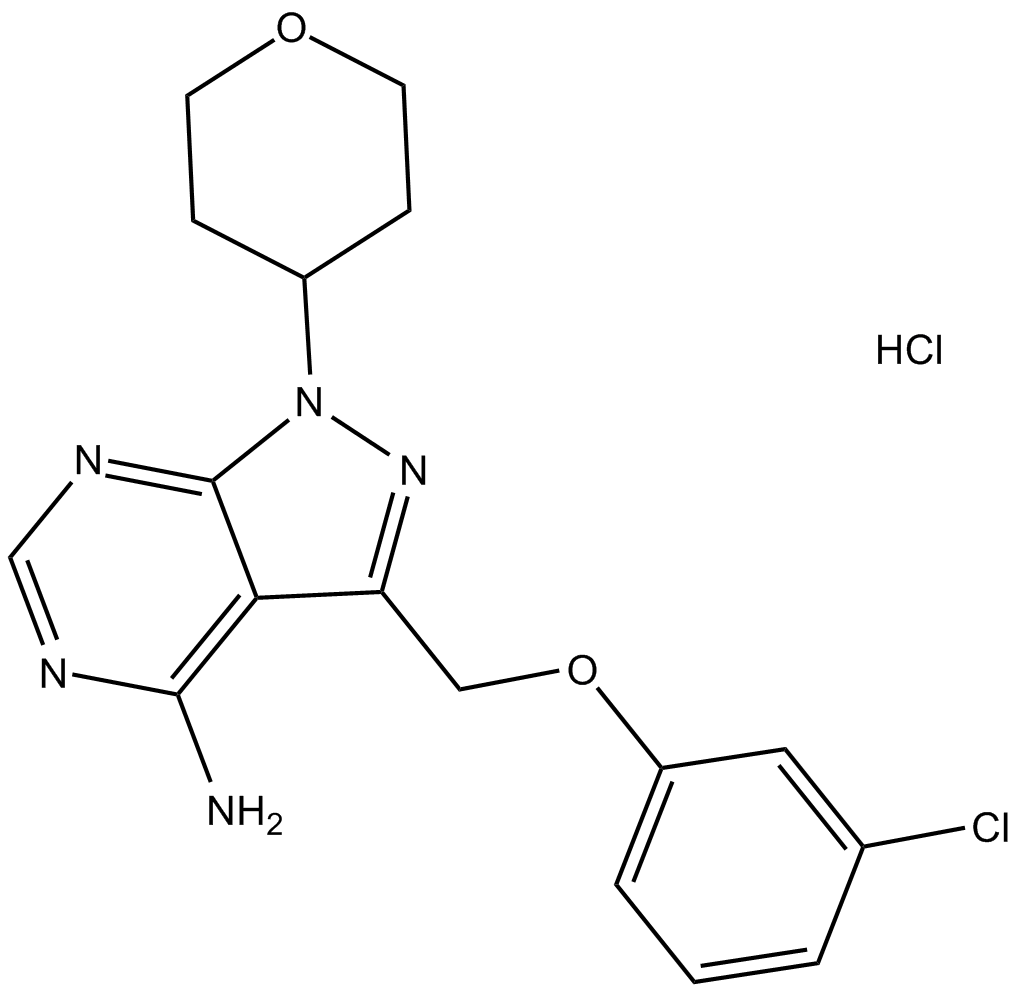
-
GC33136
PF-06380101
PF-06380101 (Aur0101), an auristatin microtubule inhibitor, is a cytotoxic Dolastatin 10 analogue. PF-06380101 (Aur0101) shows excellent potencies in tumor cell proliferation assays and differential ADME properties when compared to other synthetic auristatin analogues that are used in the preparation of ADCs.
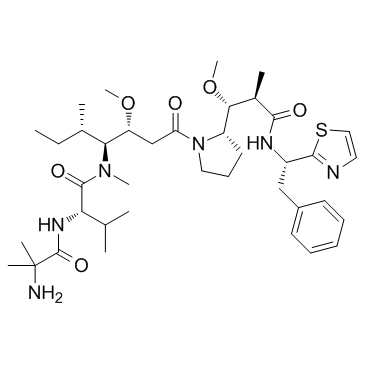
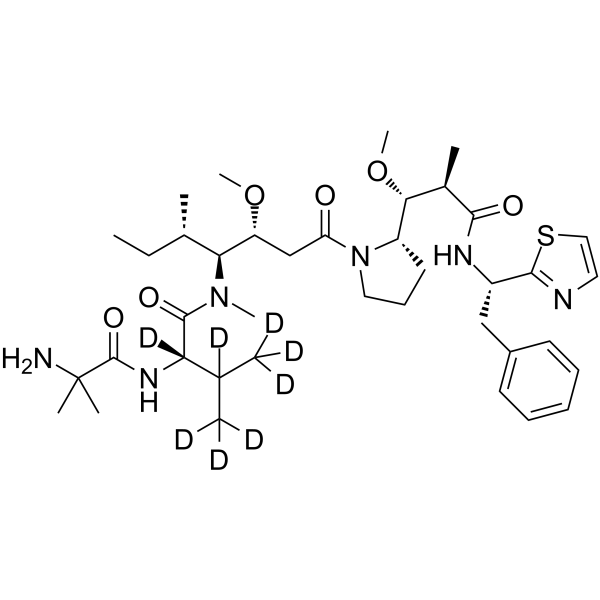
-
GC63740
PF-06380101-d8
PF-06380101 d8 (Aur0101 d8) is a deuterium labeled PF-06380101. PF-06380101, an Auristatin microtubule inhibitor, is a cytotoxic Dolastatin 10 analogue.
-
GC34297
PF-2771
PF-2771 is a potent and selective centromere protein E (CENP-E) inhibitor, inhibiting CENP-E motor activity with an IC50 of 16.1 nM; PF-2771 is used as an anticancer agent.
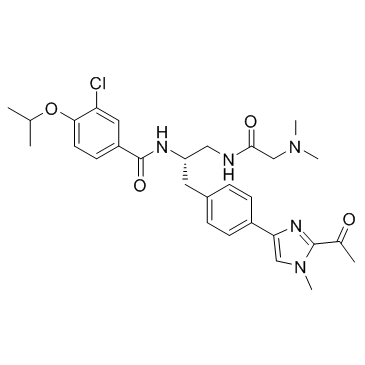
-
GC17214
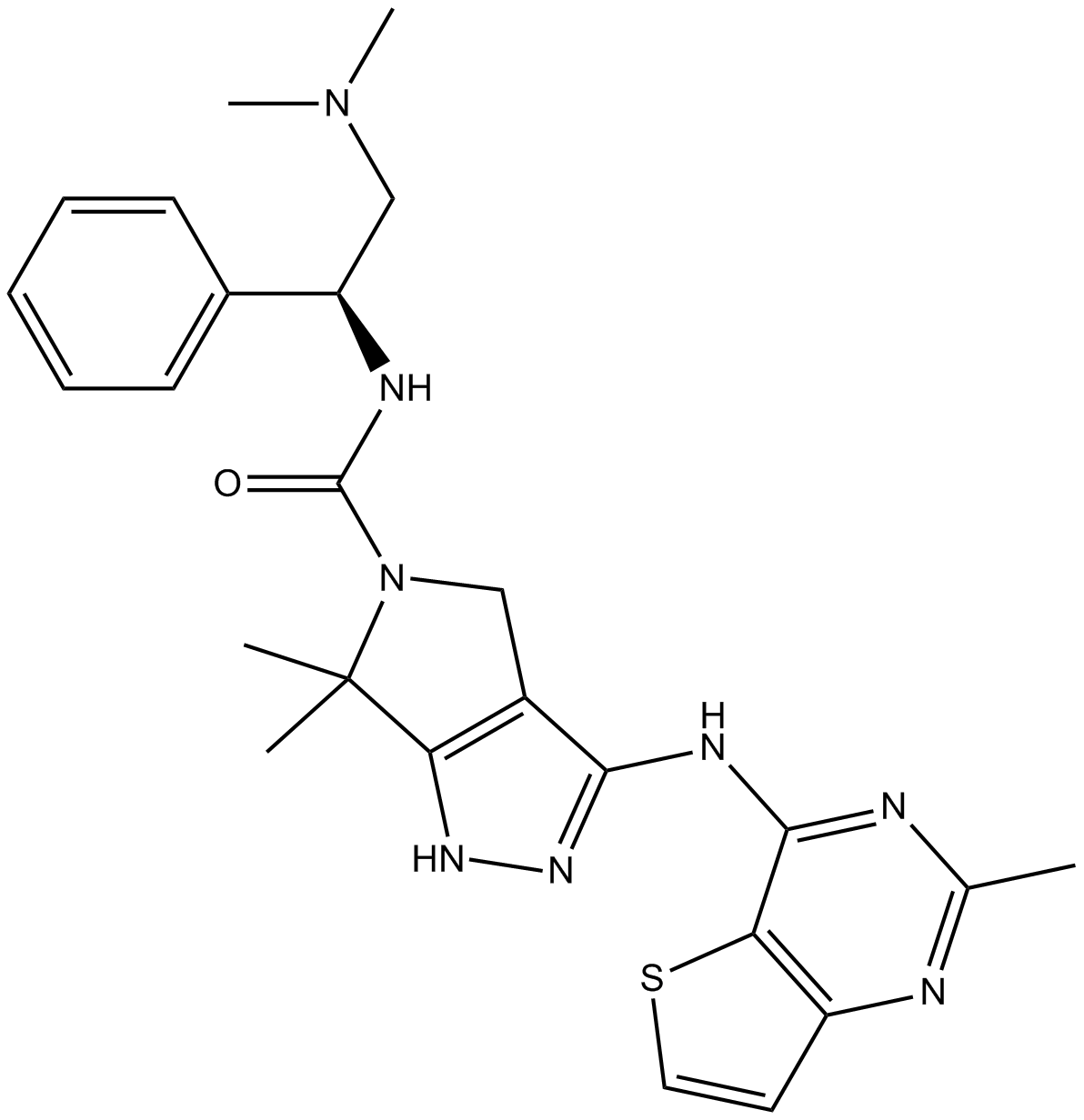
PF-3758309
An inhibitor of PAK4
-
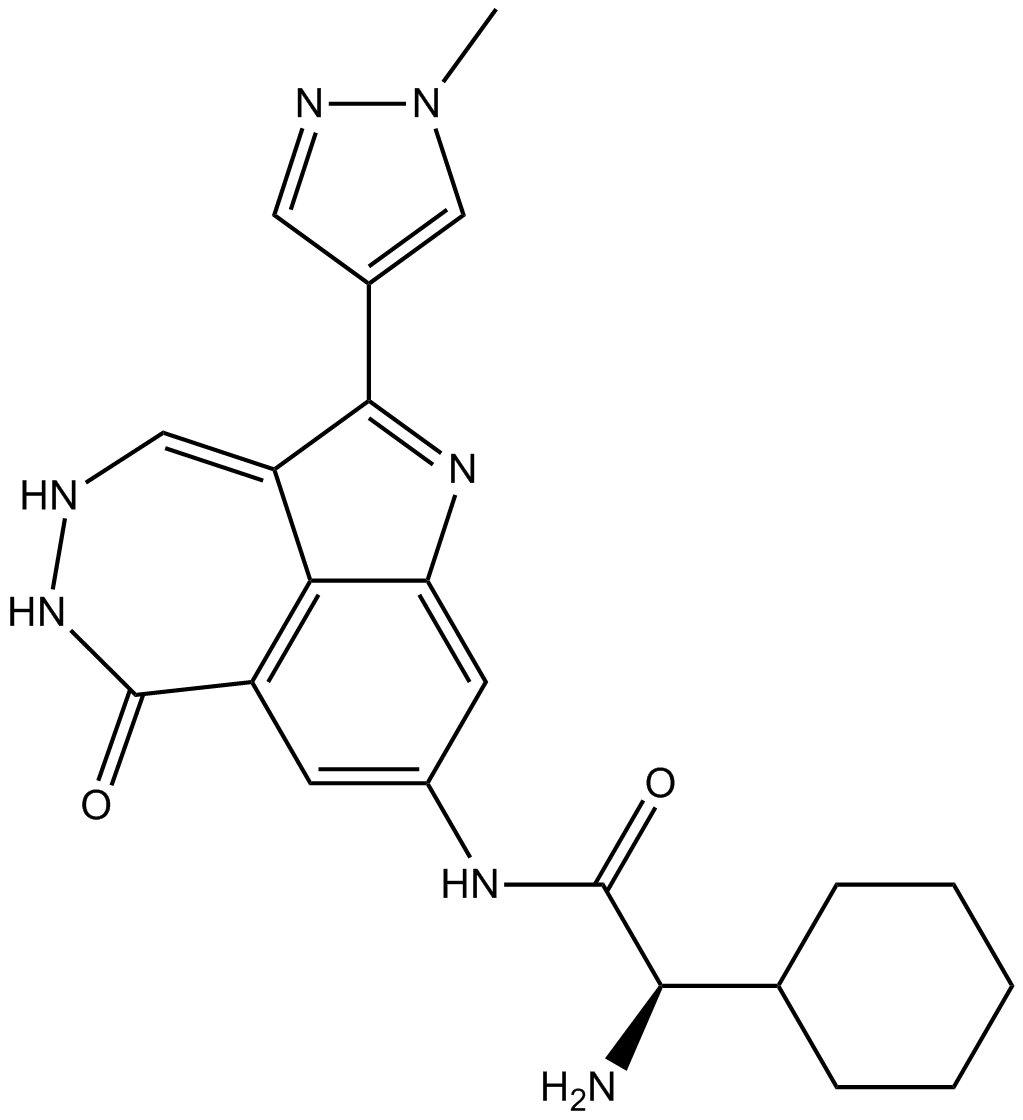
GC14234
PF-477736
Chk1 inhibitor
-
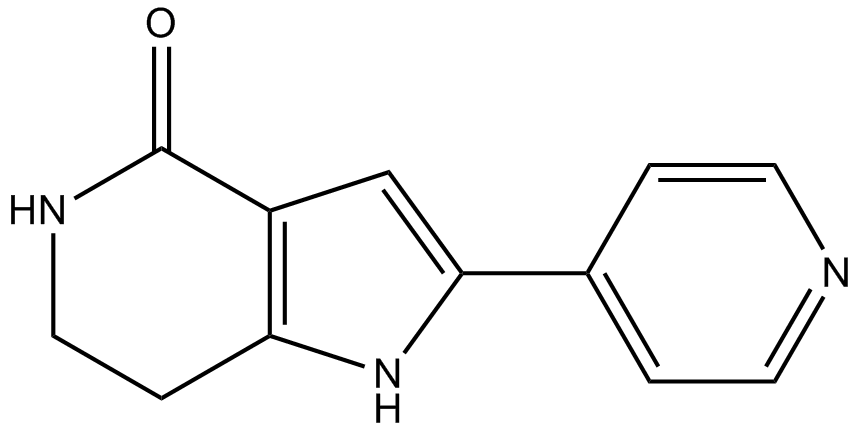
GC11324
PHA-767491
A potent Cdc7 kinase inhibitor
-
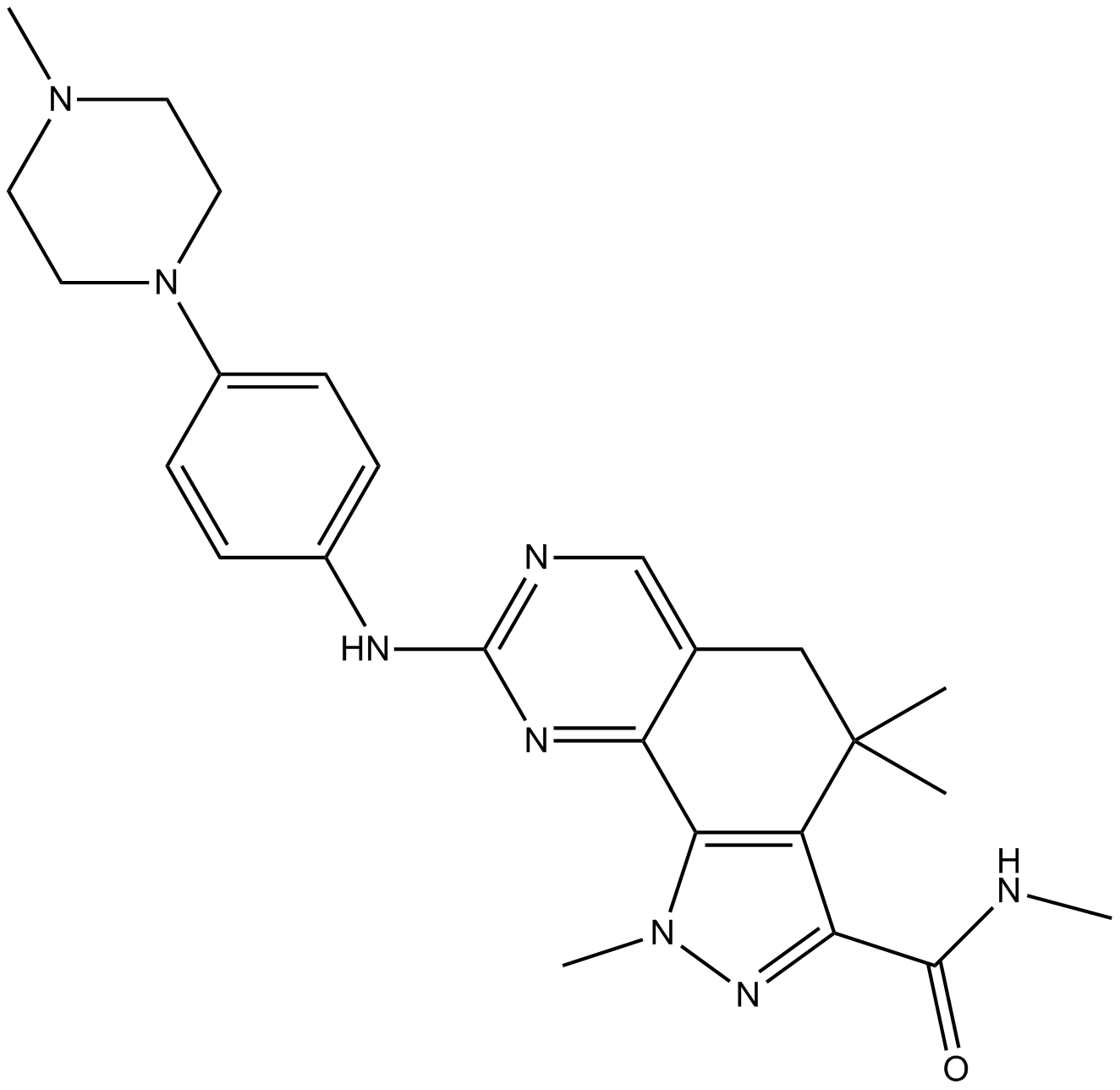
GC15588
PHA-848125
PHA-848125 (PHA-848125) is a potent, ATP-competitive and dual inhibitor of CDK and Tropomyosin receptor kinase (TRK), with IC50s of 45, 150, 160, 363, 398 nM and 53 nM for cyclin A/CDK2, cyclin H/CDK7, cyclin D1/CDK4, cyclin E/CDK2, cyclin B/CDK1 and TRKA, respectively.
-
GC44618
Phalloidin-AMCA Conjugate
Phalloidin-AMCA conjugate is a blue fluorophore that specifically labels filamentous actin (F-actin).
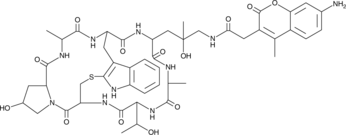
-
GC44620
Phalloidin-Tetramethylrhodamine Conjugate
Phalloidin-retramethylrhodamine conjugate is an orange fluorescent probe (ex/em = 546/575 nm) that selectively binds to F-actin.
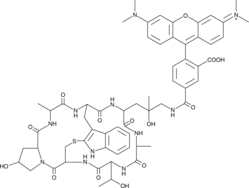
-
GC40265
Pheleuin
Pheleuin is a pyrazinone derivative synthesized from a dipeptide aldehyde.
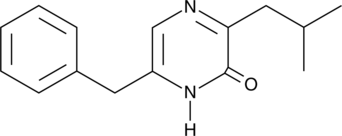
-
GC32743
Phen-DC3 Trifluoromethanesulfonate (Phen-DC3 Triflate)
Phen-DC3 Trifluoromethanesulfonate (Phen-DC3 Triflate) is a G-quadruplex (G4) specific ligand which can inhibit FANCJ and DinG helicases with IC50s of 65±6 and 50±10 nM, respectively.
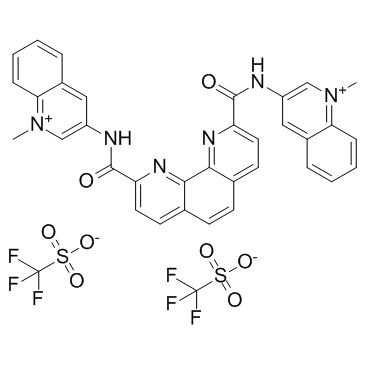
-
GC49015
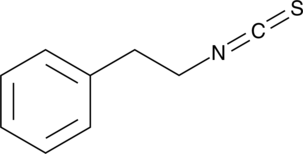
Phenethyl isothiocyanate
An isothiocyanate with anticancer activity
-
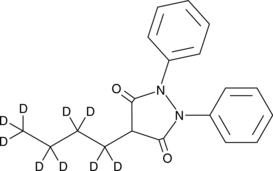
GC49128
Phenylbutazone-d9
An internal standard for the quantification of phenylbutazone
-
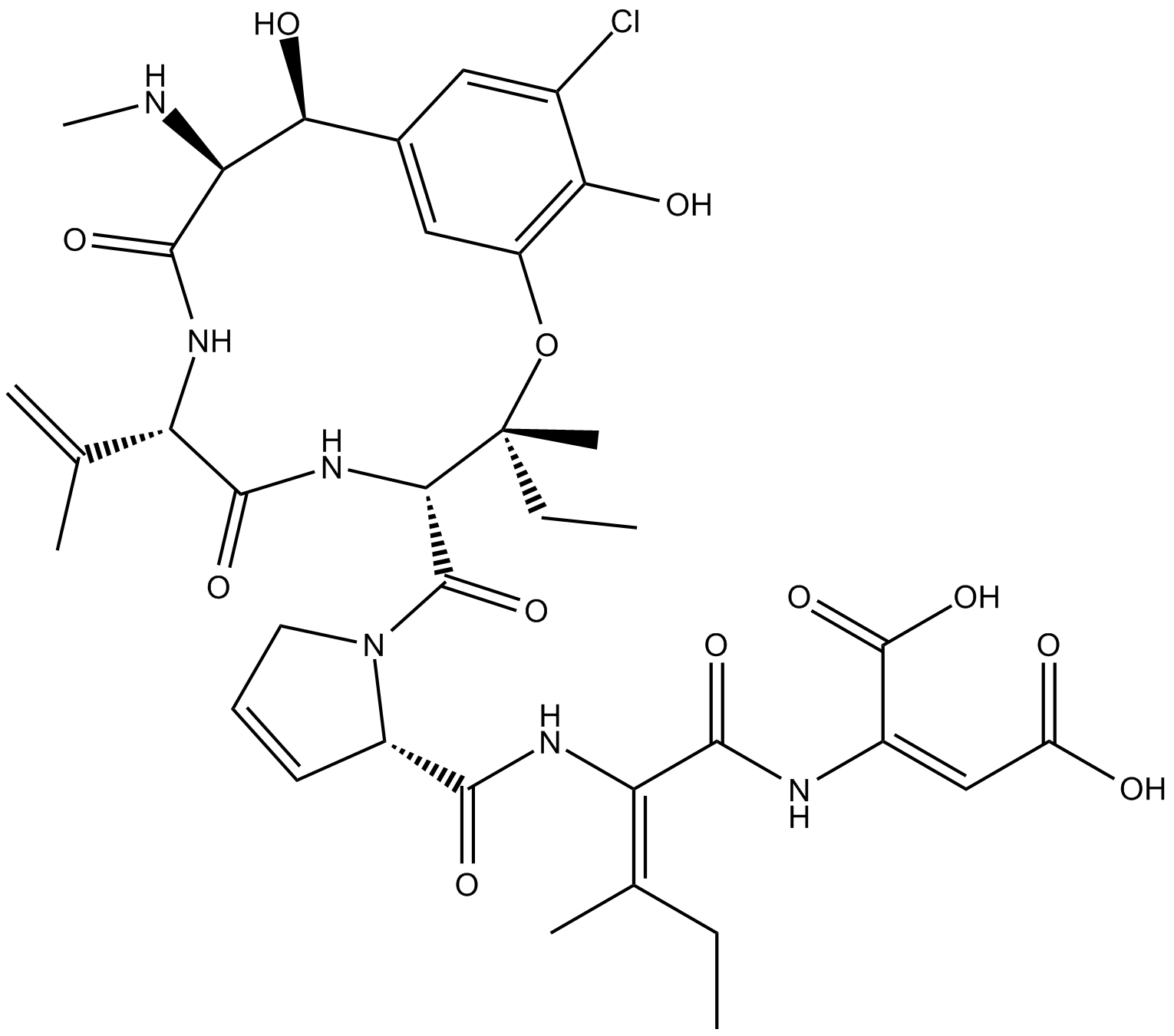
GC15514
Phomopsin A
cyclic hexapeptide mycotoxin that binds β-tubulin
-
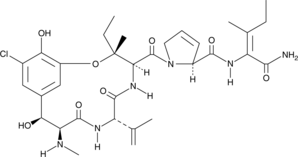
GC48394
Phomopsinamine
An inhibitor of microtubule polymerization
-
GC44633
Phosphatidylserines (bovine)
Phosphatidylserine is a naturally occurring phospholipid that comprises 2-10% of total phospholipids in mammals and is enriched in the central nervous system, particularly the retina.
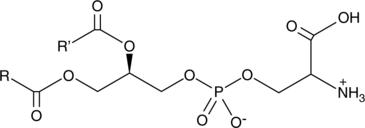
-
GC18522
Phosphatidylserines (sodium salt)
Phosphatidylserine is a naturally occurring phospholipid that comprises 2-10% of total phospholipids in mammals and is enriched in the central nervous system, particularly the retina.
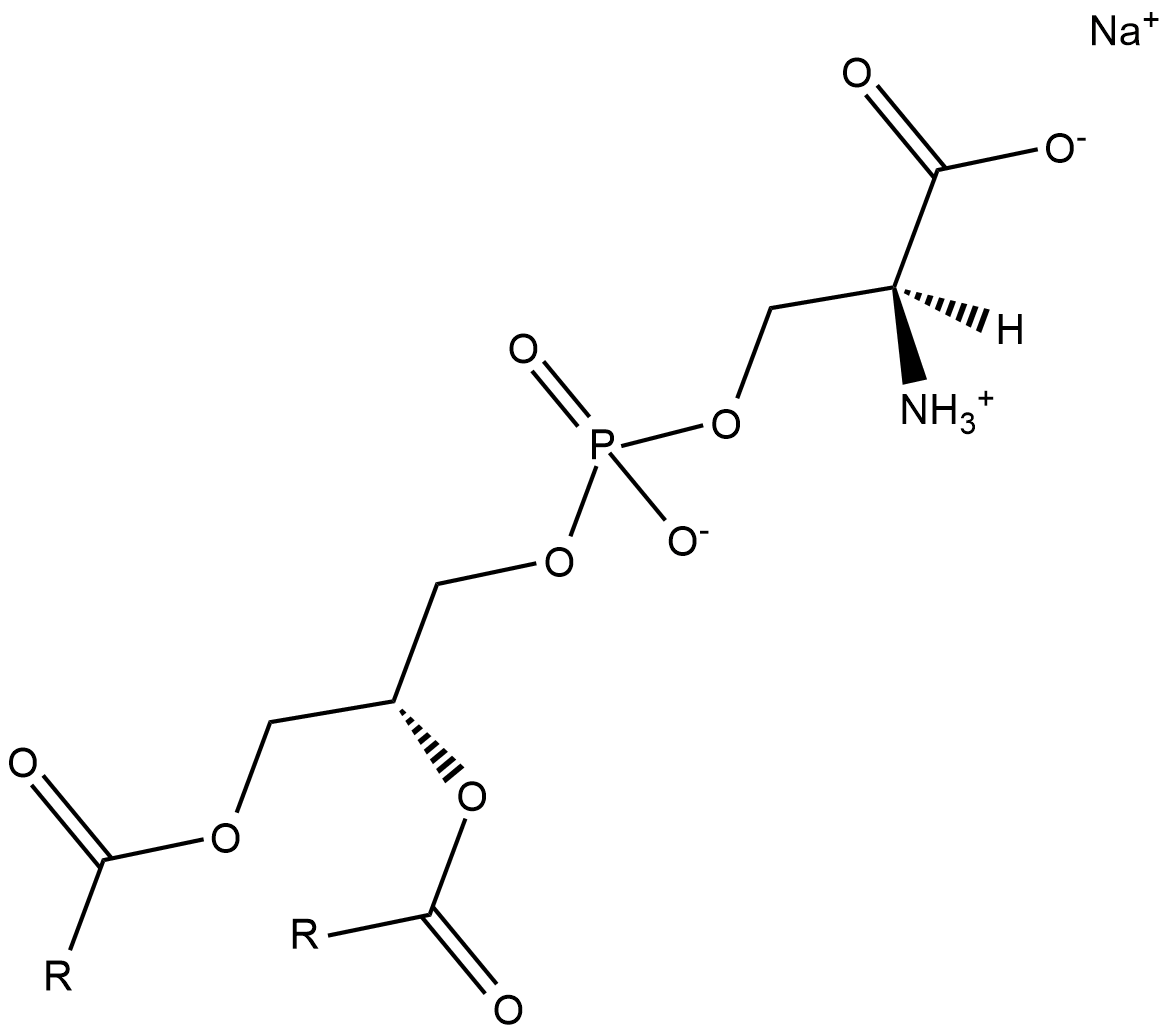
-
GC38456
Pironetin
An inhibitor of microtubule assembly
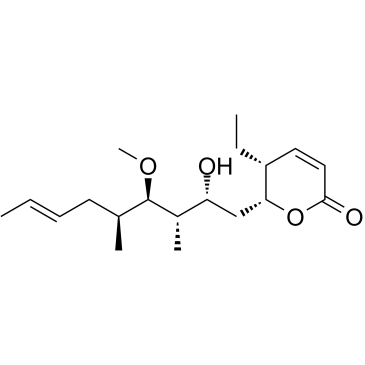
-
GC44652
PK7242 (maleate)
The protein p53, often called the 'guardian of the genome,' is a transcription factor that is activated in response to cellular stress (low oxygen levels, heat shock, DNA damage, etc.) and acts to prevent further proliferation of the stressed cell by promoting cell cycle arrest or apoptosis.
-
GC45846
PKUMDL-LC-101-D04
An allosteric activator of GPX4
-
GC10235
Plinabulin (NPI-2358)
Plinabulin (NPI-2358) (NPI-2358) is a vascular disrupting agen (VDA) against tubulin-depolymerizing with an IC50 of 9.8 nM against HT-29 cells. Plinabulin (NPI-2358) binds the colchicine binding site of β-tubulin preventing polymerization and has potent inhibitory to tumor cells.
-
GC65189
PLK4-IN-1
PLK4-IN-1 (Example A6) is a PLK4 inhibitor, with an IC50 of ≤ 0.1 μM.
-
GC69718
Plogosertib
Plogosertib (CYC140) is a selective, potent, orally active ATP-competitive PLK1 inhibitor (IC50: 3 nM). Plogosertib is an anti-cancer agent with anti-proliferative activity and can be used for research on various tumors including esophageal cancer, gastric cancer, leukemia, non-small cell lung cancer, ovarian cancer and squamous cell carcinoma.
-
GC46019
Pluviatolide
A lignan
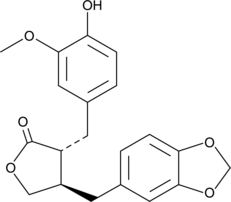
-
GC17977
PM 102
heparin antagonist
-
GC10521
Podophyllotoxin
A lignan with diverse biological activities
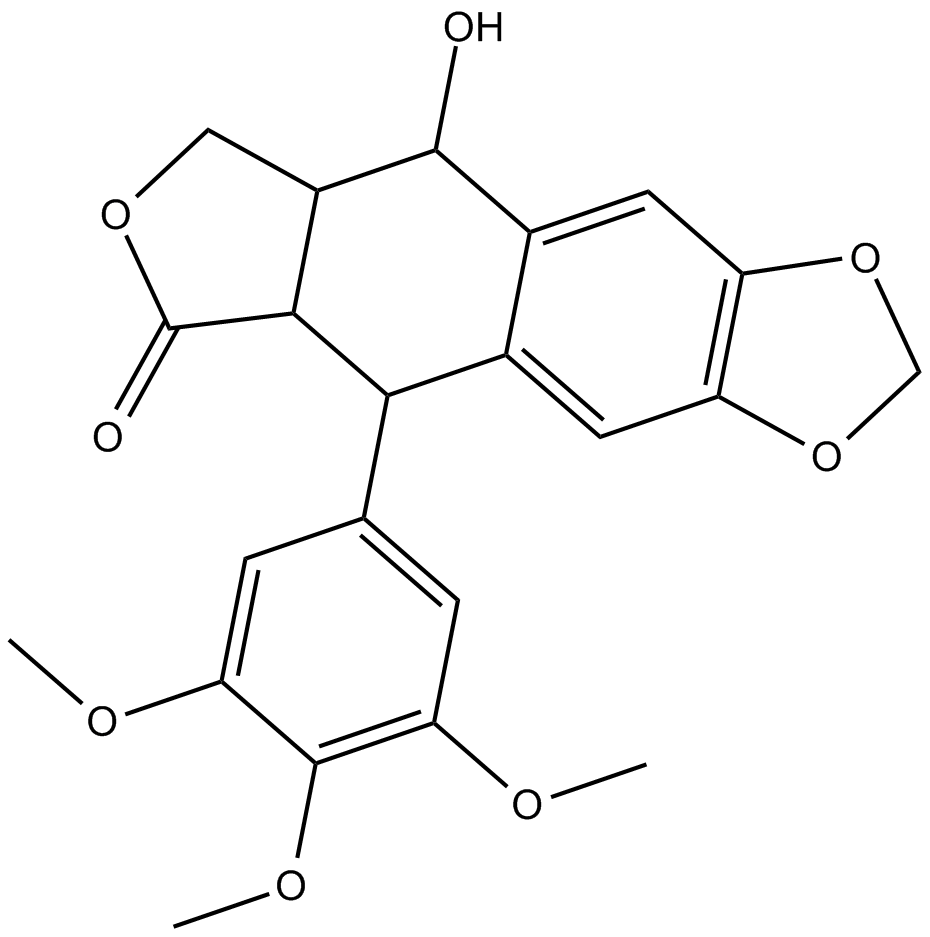
-
GC49077
Podophyllotoxin-d6
An internal standard for the quantification of podophyllotoxin and picropodophyllotoxin
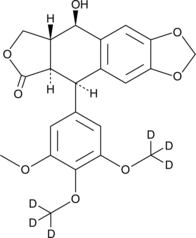
-
GC64705
Podophyllotoxone
Podophyllotoxone is isolated from the roots of Dysosma versipellis and has anti-cancer activities.Podophyllotoxone is able to inhibit the tubulin polymerization.

-
GC14735
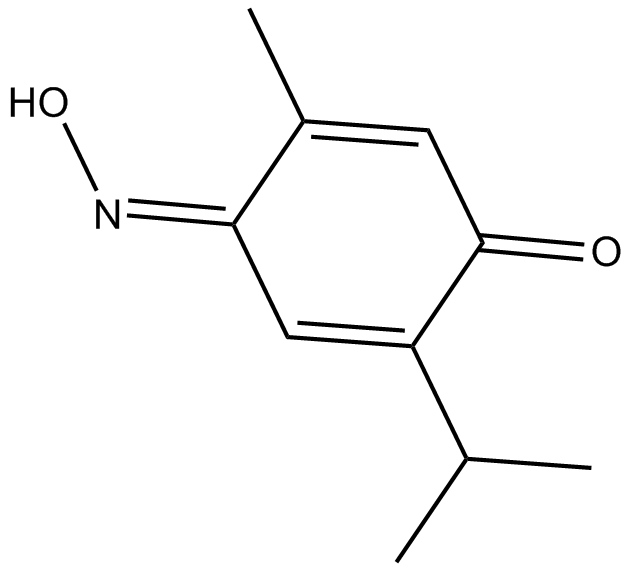
Poloxime
polo-like kinase 1 (PLK) analogue
-
GC13543
Poloxin
An inhibitor of the Plk1 polo-box domain

-
GC45680
Prodan
A solvatochromic fluorescent probe
-
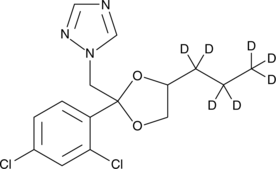
GC47979
Propiconazole-d7
An internal standard for the quantification of propiconazole
-
GC18353
Prostaglandin A2
A naturally occurring prostaglandin with antiviral/antitumor activity