DNA Damage/DNA Repair
- MTH1(4)
- PARP(84)
- ATM/ATR(30)
- DNA Alkylating(20)
- DNA Ligases(3)
- DNA Methyltransferase(25)
- DNA-PK(30)
- HDAC(136)
- Nucleoside Antimetabolite/Analogue(140)
- Telomerase(17)
- Topoisomerase(151)
- tankyrase(5)
- Antifolate(38)
- CDK(250)
- Checkpoint Kinase (Chk)(31)
- CRISPR/Cas9(9)
- Deubiquitinase(72)
- DNA Alkylator/Crosslinker(71)
- DNA/RNA Synthesis(400)
- Eukaryotic Initiation Factor (eIF)(23)
- IRE1(23)
- LIM Kinase (LIMK)(9)
- TOPK(5)
- Casein Kinase(61)
- DNA Intercalating Agents(7)
- DNA/RNA Oxidative Damage(12)
Products for DNA Damage/DNA Repair
- Cat.No. Product Name Information
-
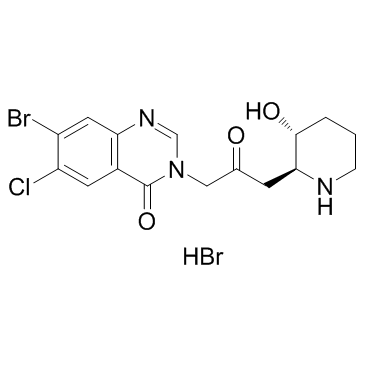
GC31950
Halofuginone hydrobromide (RU-19110 (hydrobromide))
Halofuginone (RU-19110) hydrobromid, a Febrifugine derivative, is a competitive prolyl-tRNA synthetase inhibitor with a Ki of 18.3 nM.
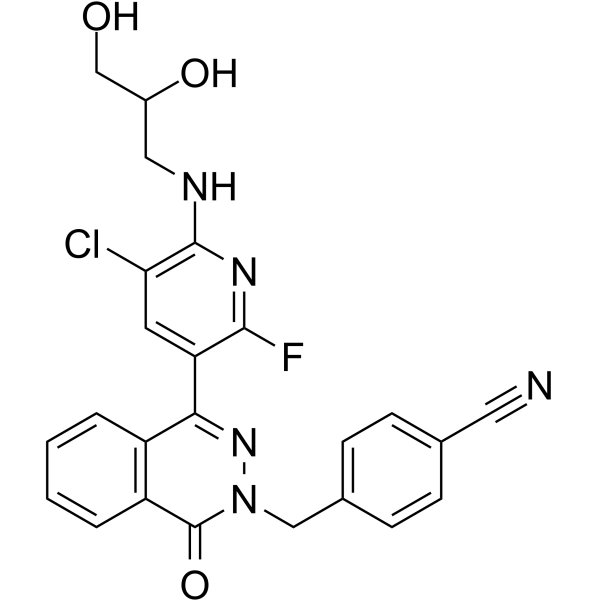
-
GC62328
HBV-IN-4
HBV-IN-4, a phthalazinone derivative, is a potent and orally active HBV DNA replication inhibitor with an IC50 of 14 nM.
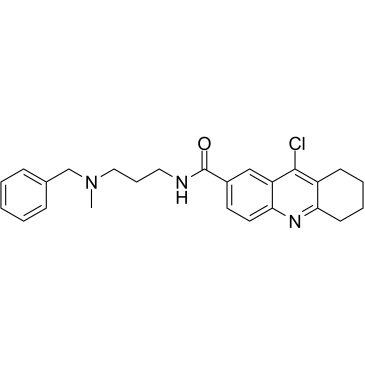
-
GC36211
HBX 19818
HBX 19818 is a specific inhibitor of ubiquitin-specific protease 7 (USP7), with an IC50 of 28.1 μM.
-
GC19190
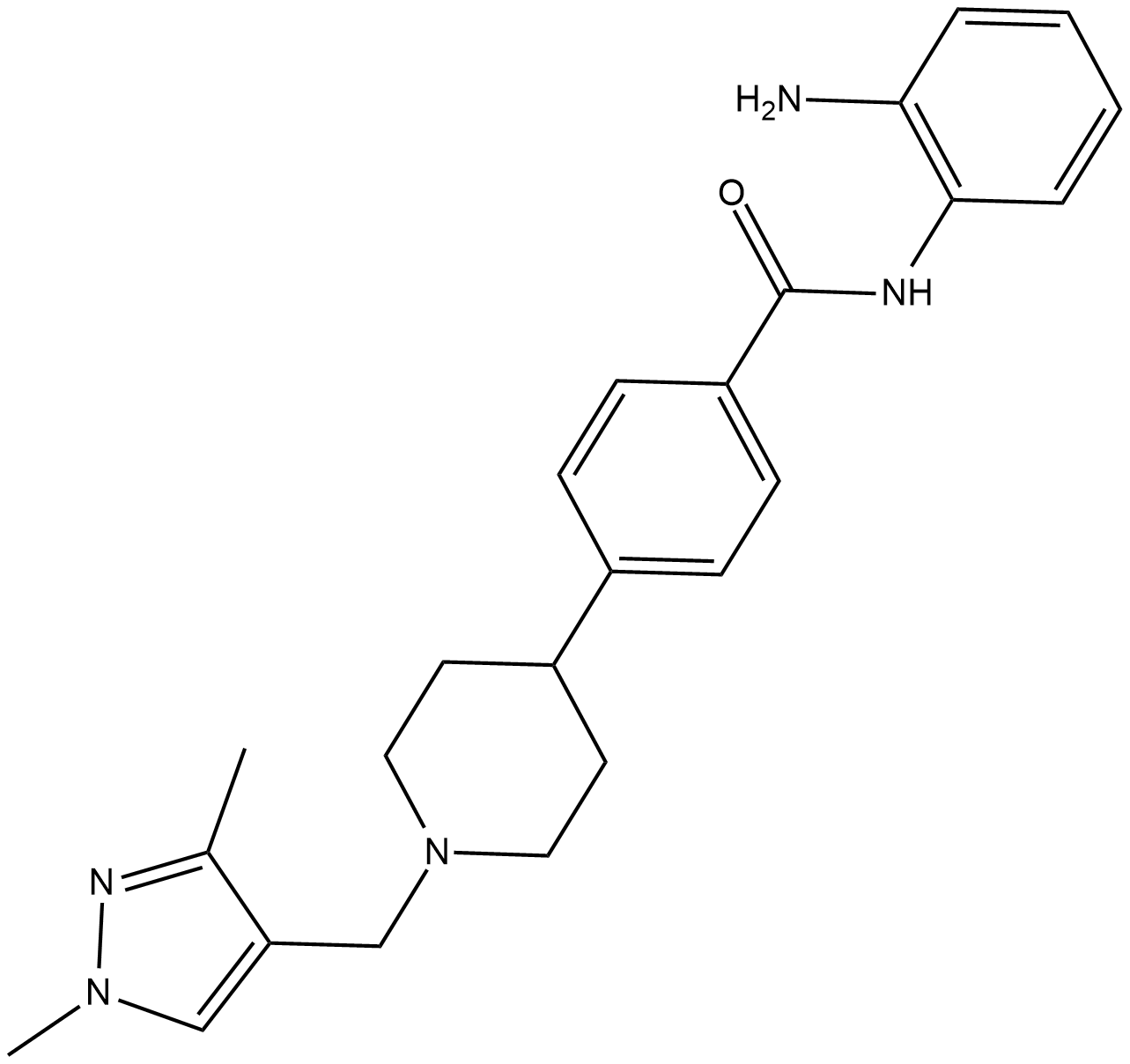
HDAC-IN-4
HDAC-IN-4 is a potent, selective and orally active class I HDAC inhibitor with IC50s of 63 nM, 570 nM and 550 nM for HDAC1, HDAC2 and HDAC3, respectively. HDAC-IN-4 has no activity against HDAC class II. HDAC-IN-4 has antitumor activity.
-
GC66052
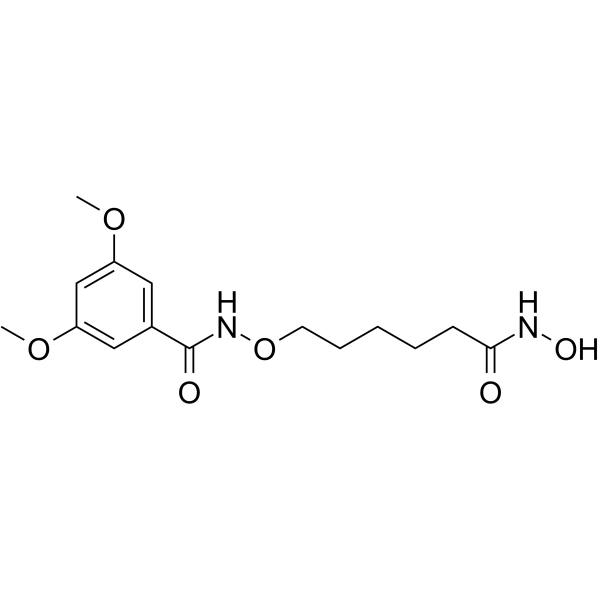
HDAC-IN-40
HDAC-IN-40 is a potent alkoxyamide-based HDAC inhibitor with Ki values of 60 nM and 30 nM for HDAC2 and HDAC6, respectively. HDAC-IN-40 had antitumor effects.
-
GC33395
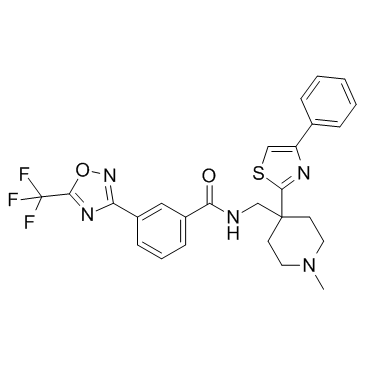
HDAC-IN-5
HDAC-IN-5 is a histone deacetylase (HDAC) inhibitor.
-
GC68010
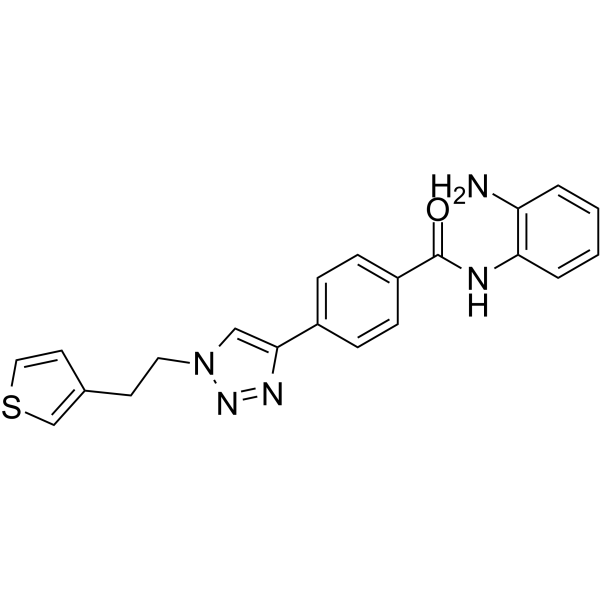
HDAC3-IN-T247
-
GC41085
HDAC6 Inhibitor
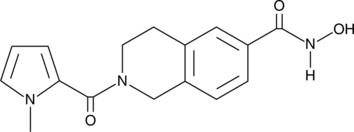
HDAC6 Inhibitor is a potent and selective HDAC6 inhibitor (IC50=36 nM). HDAC6 Inhibitor weakly inhibits other HDAC isoforms. HDAC6 Inhibitor inhibits acyl-tubulin accumulation in cells with an IC50 value of 210 nM.
-
GC33317
HDAC6-IN-1
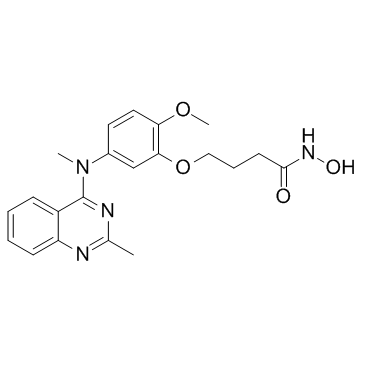
HDAC6-IN-1 is a potent and selective inhibitor for HDAC6 with an IC50 of 17 nM and shows 25-fold and 200-fold selectivity relative to HDAC1 (IC50=422 nM) and HDAC8 (IC50=3398 nM), respectively.
-
GC19189
HDAC8-IN-1
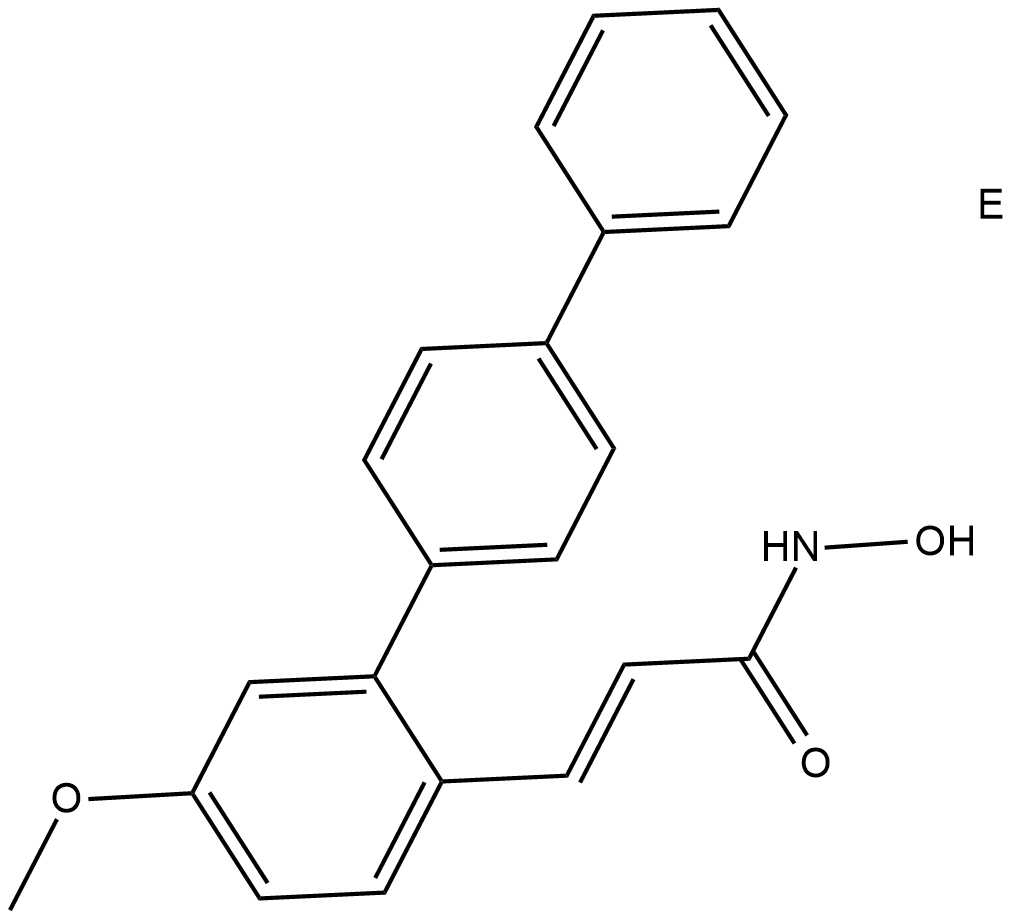
HDAC8-IN-1 is a HDAC8 inhibitor with an IC50 of 27.2 nM.
-
GC65460
HDACs/mTOR Inhibitor 1
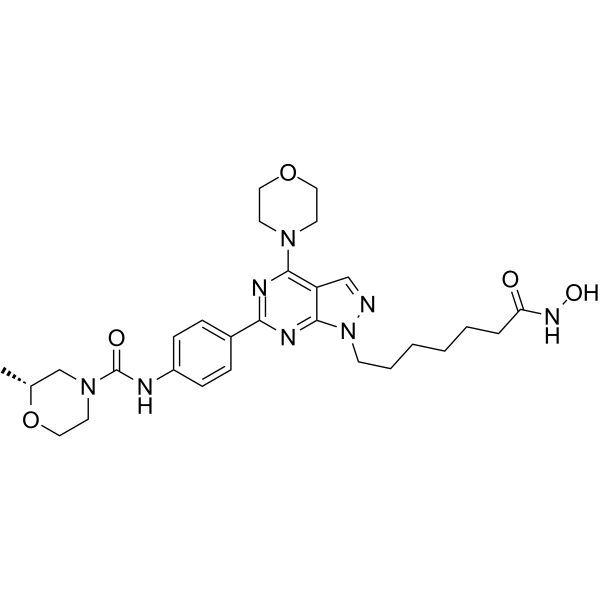
HDACs/mTOR Inhibitor 1 is a dual Histone Deacetylases (HDACs) and mammalian target of Rapamycin (mTOR) target inhibitor for treating hematologic malignancies, with IC50s of 0.19 nM, 1.8 nM, 1.2 nM and >500 nM for HDAC1, HDAC6, mTOR and PI3Kα, respectively. HDACs/mTOR Inhibitor 1 stimulates cell cycle arrest in G0/G1 phase and induce tumor cell apoptosis with low toxicity in vivo.
-
GC39279
hDHODH-IN-1
hDHODH-IN-1 is a human dihydroorotate dehydrogenase (hDHODH) inhibitor.
-
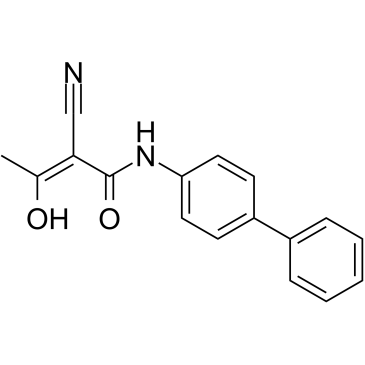
GC48388
Heliquinomycin
A bacterial metabolite with diverse biological activities
-
GC39266
Hematein
Hematein is a oxidation product of hematoxylin acted as a dye. Hematein is an allosteric casein kinase II inhibitor with an IC50 of 0.74 μM. Hematein inhibits Akt/PKB Ser129 phosphorylation, the Wnt/TCF pathway and increases apoptosis in lung cancer cells.
-
GC40103
Herboxidiene
Herboxidiene, as a potent antitumor agent, can target the SF3B subunit of the spliceosome. Herboxidiene also induces both G1 and G2/M cell cycle arrest in a human normal fibroblast cell line WI-38.
-
GC12201
HI TOPK 032
T-LAK-cell-originated protein kinase (TOPK) inhibitor
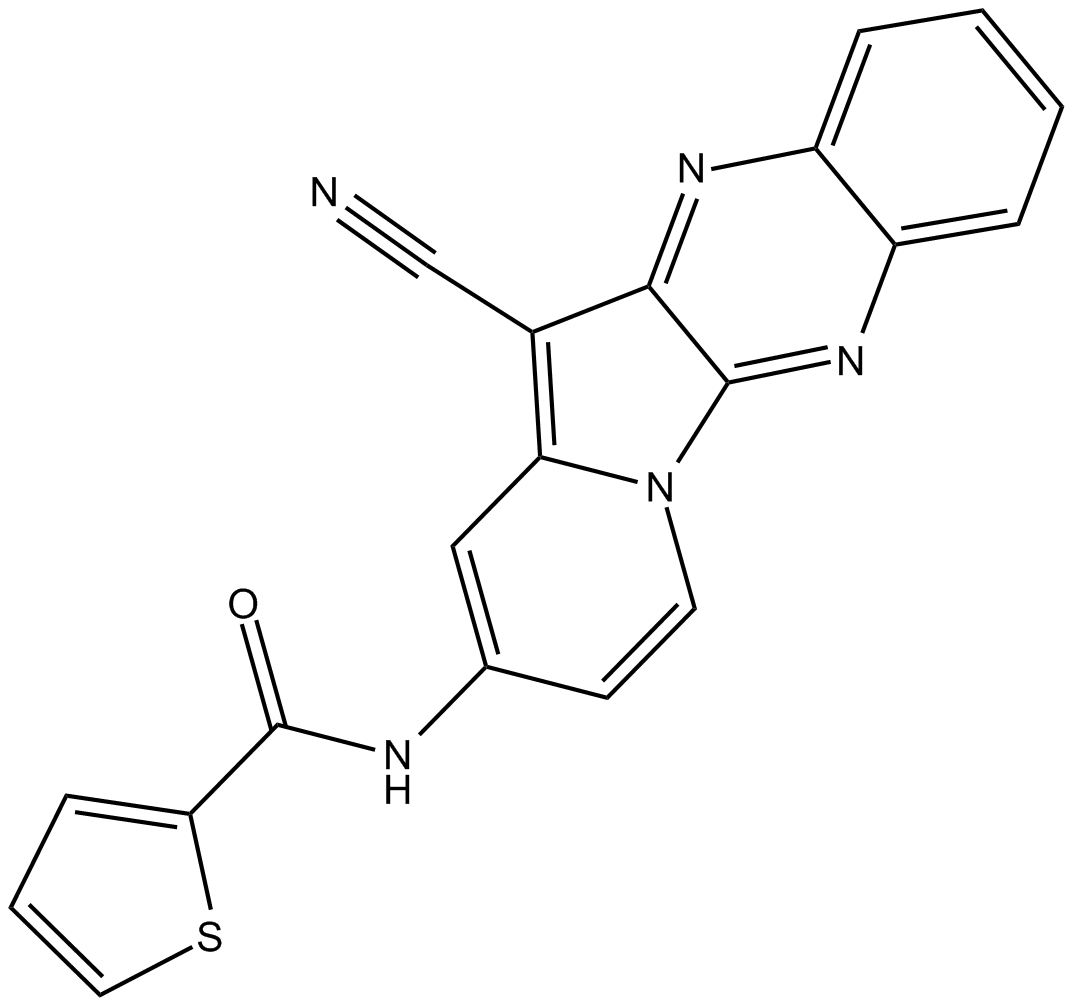
-
GC11302
Hinokitiol
A tropolone with diverse biological activities

-
GC12334
HNHA
HDAC inhibitor
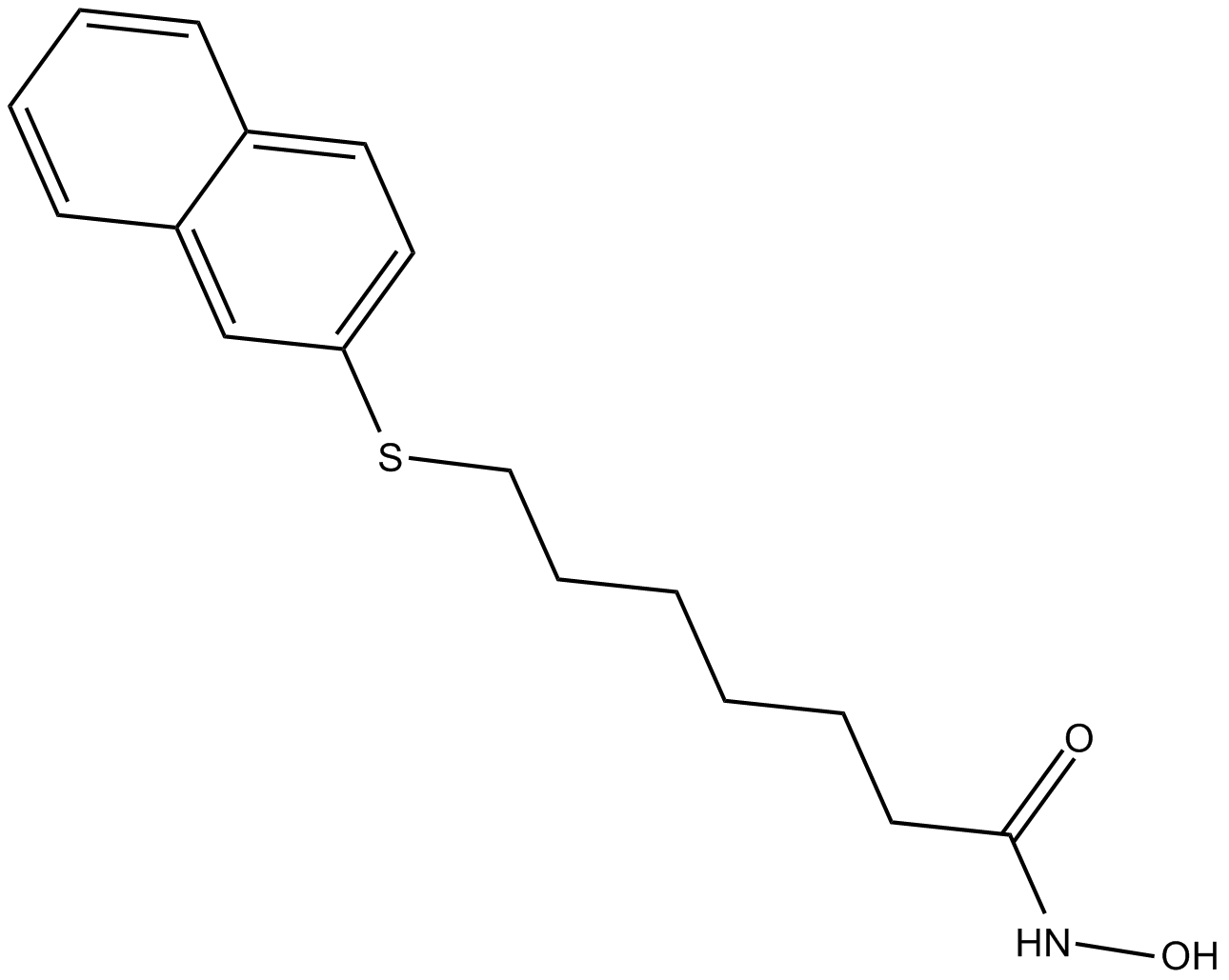
-
GC11574
HPOB
HDAC6 inhibitor, potent and selective
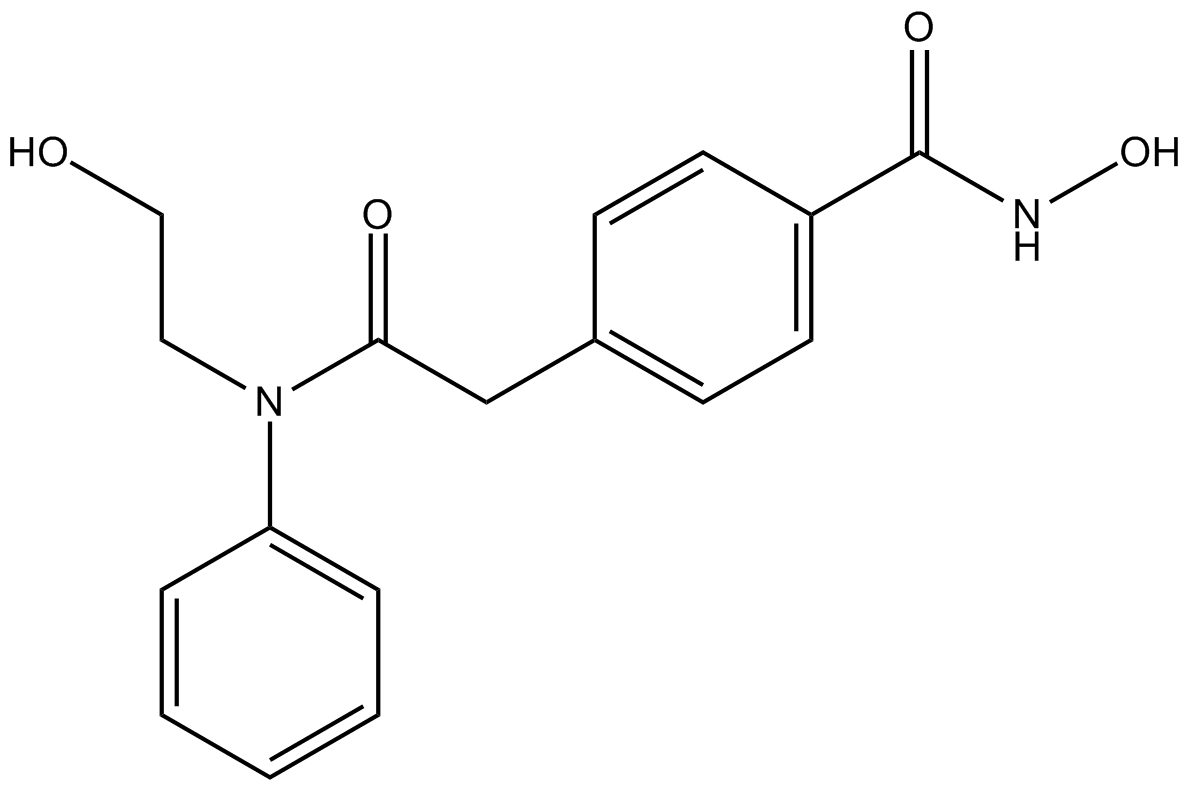
-
GC63854
HQ461
HQ461 is a molecular glue that promotes CDK12-DDB1 interaction to trigger cyclin K degradation. HQ461-mediated degradation of cyclin K impairs CDK12 function, resulting in decreased CDK12 substrate phosphorylation, downregulation of DNA damage response genes, and cell death.
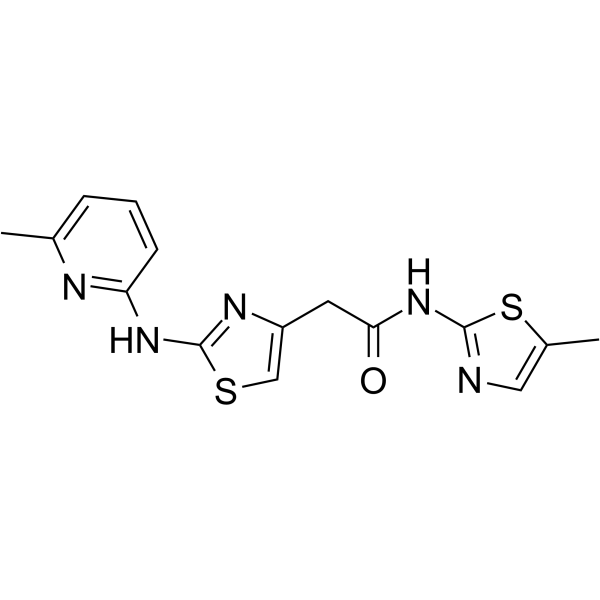
-
GC62572
hSMG-1 inhibitor 11e
hSMG-1 inhibitor 11e is a potent and selective hSMG-1 kinase inhibitor with an IC50 of 900-fold selectivity over mTOR (IC50 of 45 nM), PI3Kα/γ (IC50s of 61 nM and 92 nM) and CDK1/CDK2 (IC50s of 32 μM and 7.1 μM).
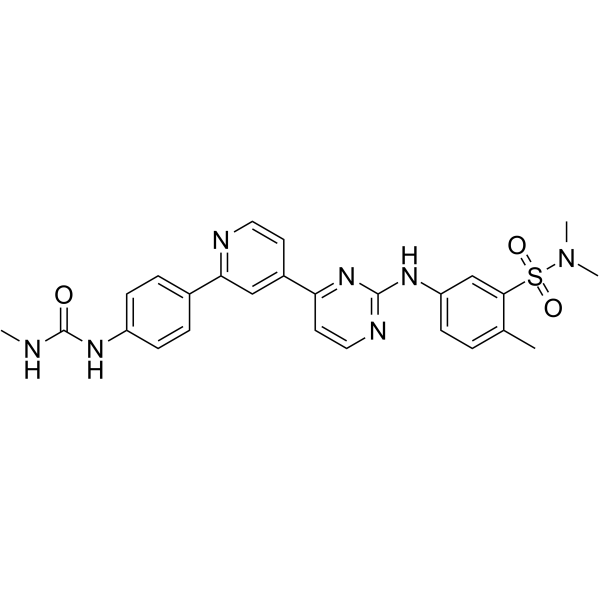
-
GC61925
hSMG-1 inhibitor 11j
hSMG-1 inhibitor 11j, a pyrimidine derivative, is a potent and selective inhibitor of hSMG-1, with an IC50 of 0.11 nM. hSMG-1 inhibitor 11j exhibits >455-fold selectivity for hSMG-1 over mTOR (IC50=50 nM), PI3Kα/γ (IC50=92/60 nM) and CDK1/CDK2 (IC50=32/7.1 μM). hSMG-1 inhibitor 11j can be used for the research of cancer.
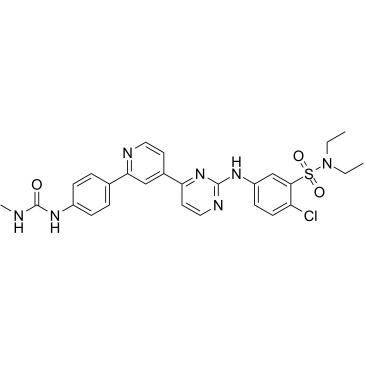
-
GC16843
Hydroxyurea
DNA synthesis inhibitor
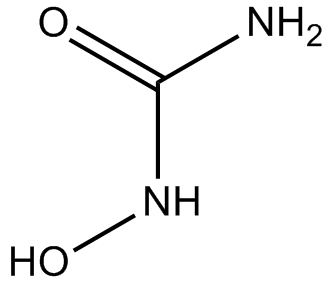
-
GC49694
Hygromycin A
An antibiotic
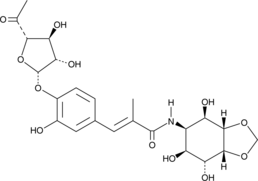
-
GC48445
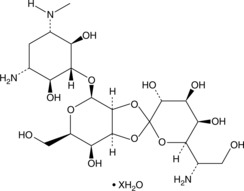
Hygromycin B (hydrate)
An aminoglycoside antibiotic
-
GC49267
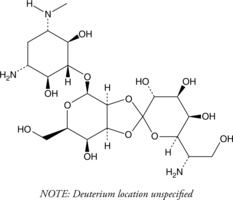
Hygromycin B-d4
An internal standard for the quantification of hygromycin
-
GC14503
IC261
CK1 inhibitor
-
GC14969
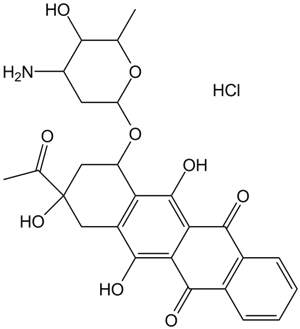
Idarubicin HCl
Idarubicin HCl is an anthracycline antileukemic drug.
-
GC16003
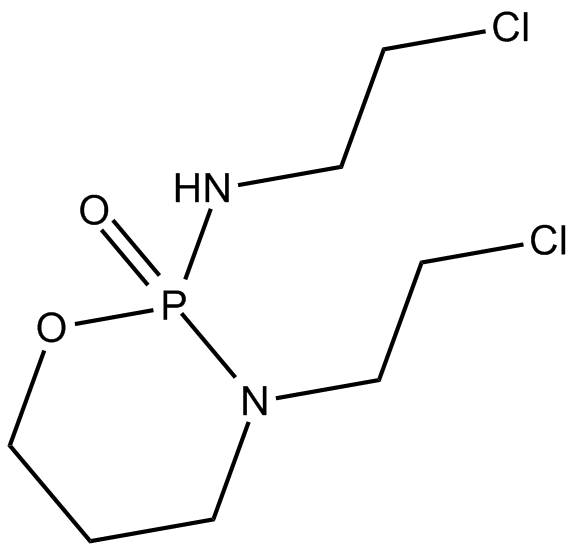
Ifosfamide
Cytostatic agent
-
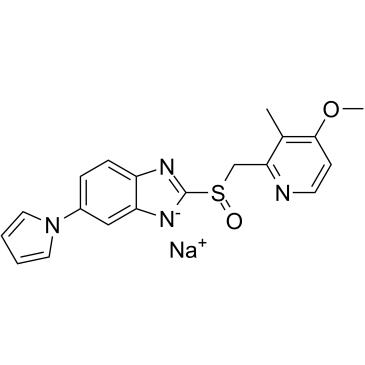
GC60929
Ilaprazole sodium
Ilaprazole (IY-81149) sodium is an orally active proton pump inhibitor.
-
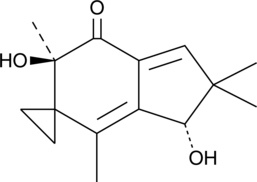
GC41340
Illudin M
Illudin M is a cytotoxic sesquiterpene from the fungus O.
-
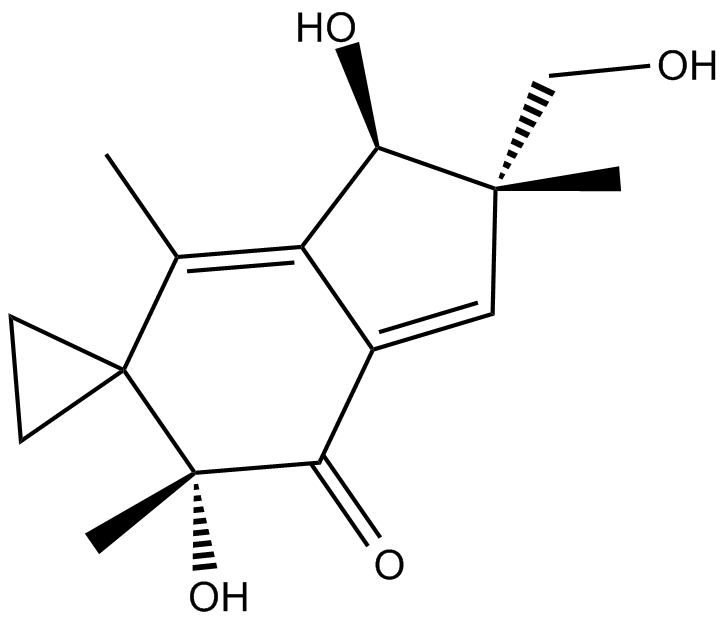
GC18100
Illudin S
potent antitumor sesquiterpene
-
GC62137
IMT1
IMT1 is a first-in-class specific and noncompetitive human mitochondrial RNA polymerase (POLRMT) inhibitor.
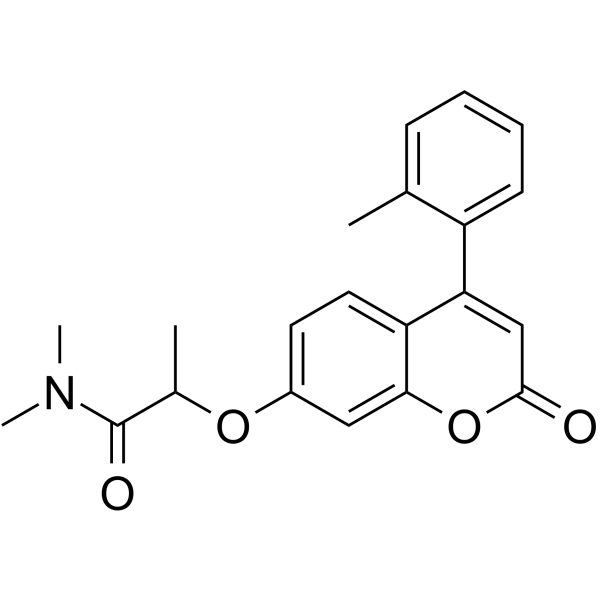
-
GC65939
Indimitecan
Indimitecan (LMP776) is a topoisomerase I (Top1) inhibitor with anticancer activities.

-
GC36312
Indirubin-3'-monoxime-5-sulphonic acid
Indirubin-3'-monoxime-5-sulphonic acid is a potent and selective inhibitor of CDK1, CDK5, and GSK-3β with IC50s of 5 nM, 7 nM, and 80 nM, respectively.
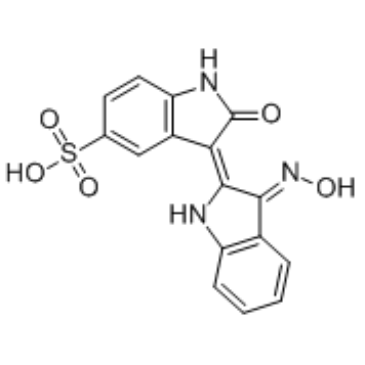
-
GC36313
Indirubin-5-sulfonate
Indirubin-5-sulfonate is a cyclin-dependent kinase (CDK) inhibitor, with IC50 values of 55 nM, 35 nM, 150 nM, 300 nM and 65 nM for CDK1/cyclin B, CDK2/cyclin A, CDK2/cyclin E, CDK4/cyclin D1, and CDK5/p35, respectively. Indirubin-5-sulfonate also shows inhibitory activity against GSK-3β.
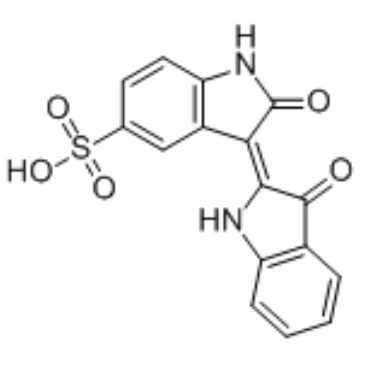
-
GC52513
Indolimine-214
A genotoxic gut microbiota metabolite
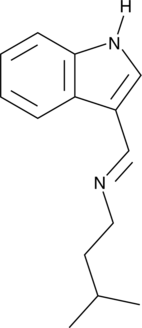
-
GC33205
Indotecan (LMP-400)
Indotecan (LMP-400) (LMP-400) is a potent topoisomerase 1(Top1) inhibitor with IC50 values of 300, 1200, 560 nM for P388, HCT116, MCF-7 cell lines, respectively.
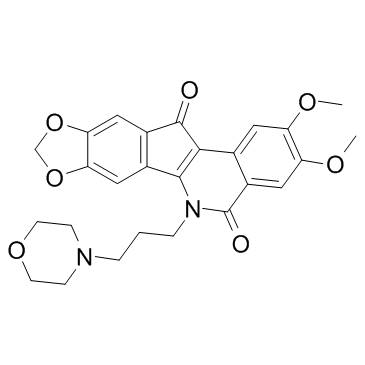
-
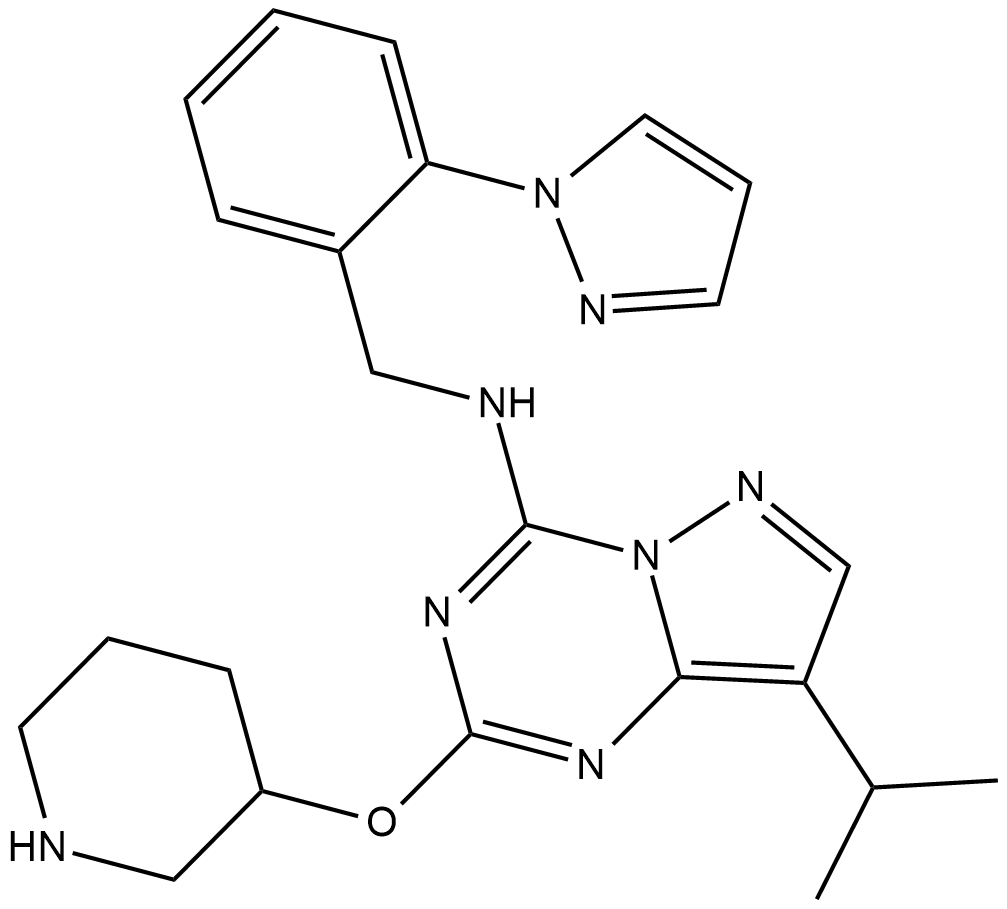
GC38385
INO-1001
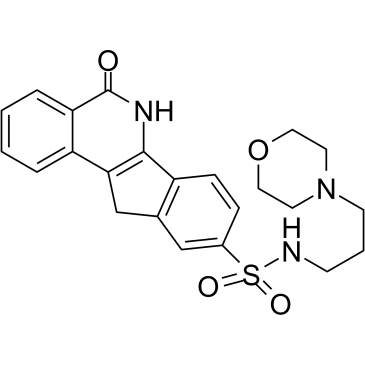
-
GC33165
Intoplicine
Intoplicine (RP 60475), an antitumor derivative in the 7H-benzo[e]pyrido[4,3-b]indole series, is a DNA topoisomerase I and II inhibitor. Intoplicine strongly binds DNA (KA = 2 x 105 /M) and thereby increases the length of linear DNA.
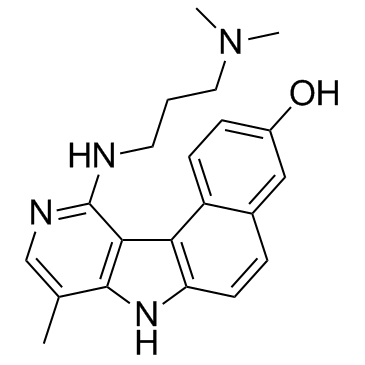
-
GC63354
Intoplicine dimesylate
Intoplicine (RP 60475) dimesylate, an antitumor derivative in the 7H-benzo[e]pyrido[4,3-b]indole series, is a DNA topoisomerase I and II inhibitor. Intoplicine dimesylate strongly binds DNA (KA = 2 x 105 /M) and thereby increases the length of linear DNA.
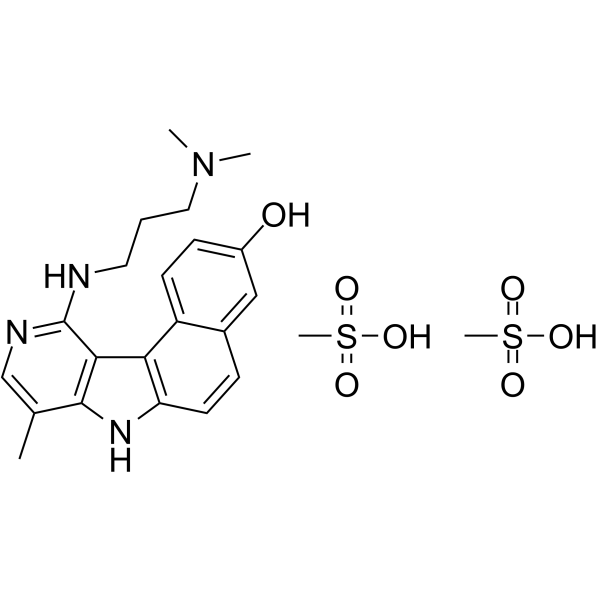
-
GC63023
Ipivivint
Ipivivint (compound 38) is a potent CDC-like kinase (CLK) inhibitor with EC50s of 1 nM, 7 nM for CLK2 and CLK3, respectively. Ipivivint inhibits Wnt pathway (EC50=13 nM).
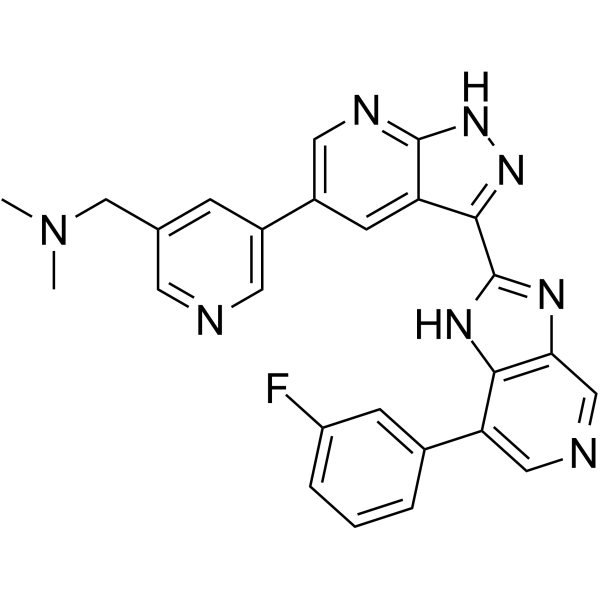
-
GC52175
IQA
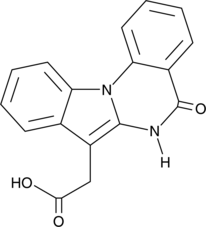
-
GC63513
IRE1α kinase-IN-1
IRE1α kinase-IN-1 is a highly selective IRE1α (ERN1) inhibitor, with an IC50 of 77 nM.
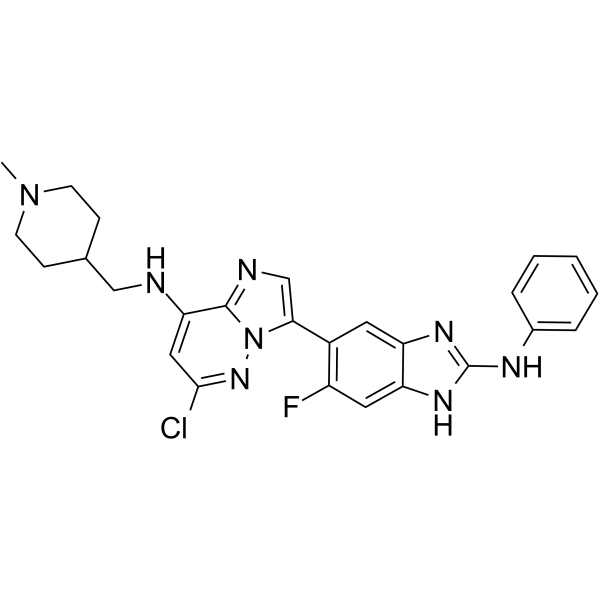
-
GC64232
IRE1α kinase-IN-2
IRE1α kinase-IN-2 is a potent IRE1α kinase inhibitor, with an EC50 of 0.82 μM. IRE1α kinase-IN-2 inhibits IRE1α kinase autophosphorylation (IC50=3.12 μM). IRE1α kinase-IN-2 inhibits XBP1 mRNA splicing in the WT cell lines.
-
GC11473
Irinotecan
Topoisomerase I inhibitor
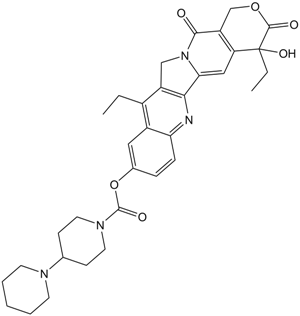
-
GC11048
Irinotecan HCl Trihydrate
Irinotecan HCl Trihydrate ((+)-Irinotecan HCl Trihydrate) is a topoisomerase I inhibitor with antitumor activity.
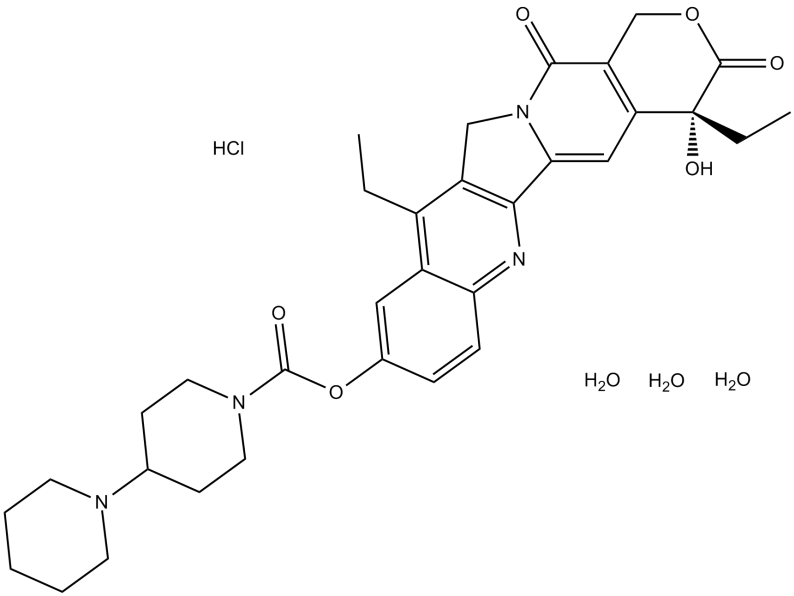
-
GC36329
Irinotecan hydrochloride
Irinotecan hydrochloride ((+)-Irinotecan hydrochloride) is a topoisomerase I inhibitor mainly used to treat colon cancer and rectal cancer.
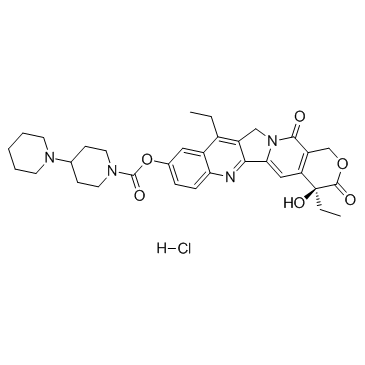
-
GC30554
Isocytosine
Isocytosine is a non-natural nucleobase and an isomer of cytosine.
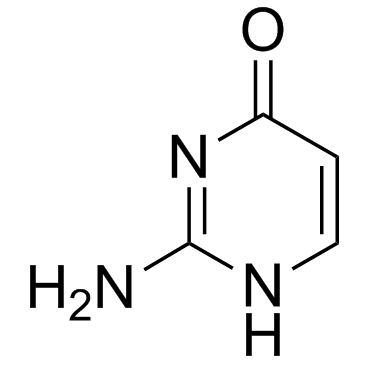
-
GC33662
Isoguanine
An isomer of guanine
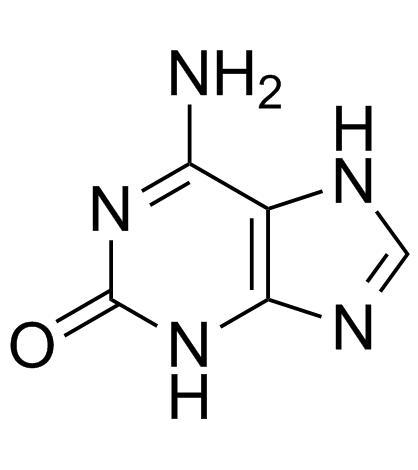
-
GC39140
Isopimpinellin
Isopimpinellin, an orally active compound isolated from the roots of Pimpinella saxifrage.
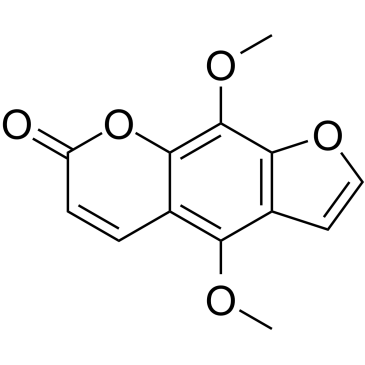
-
GC10834
Isoxanthopterin
interferes with RNA and DNA synthesis
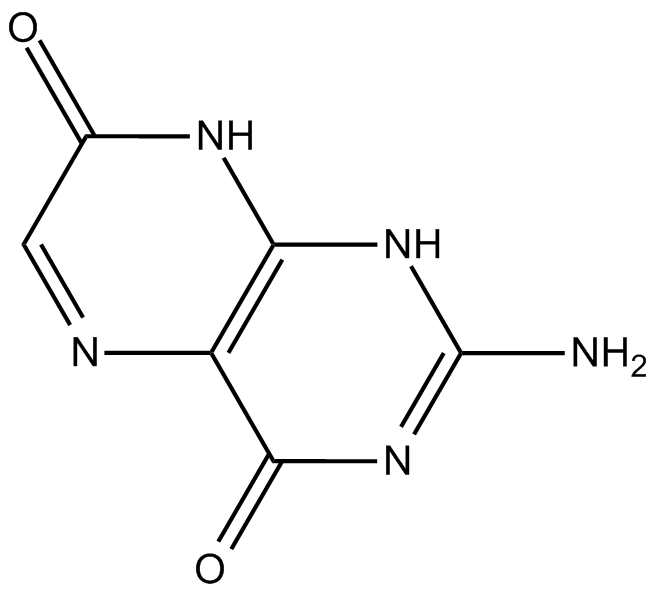
-
GC14597
ITSA-1 (ITSA1)
ITSA-1 (ITSA1) is an activator of histone deacetylase (HDAC), and counteract trichostatin A (TSA)-induced cell cycle arrest, histone acetylation, and transcriptional activation.
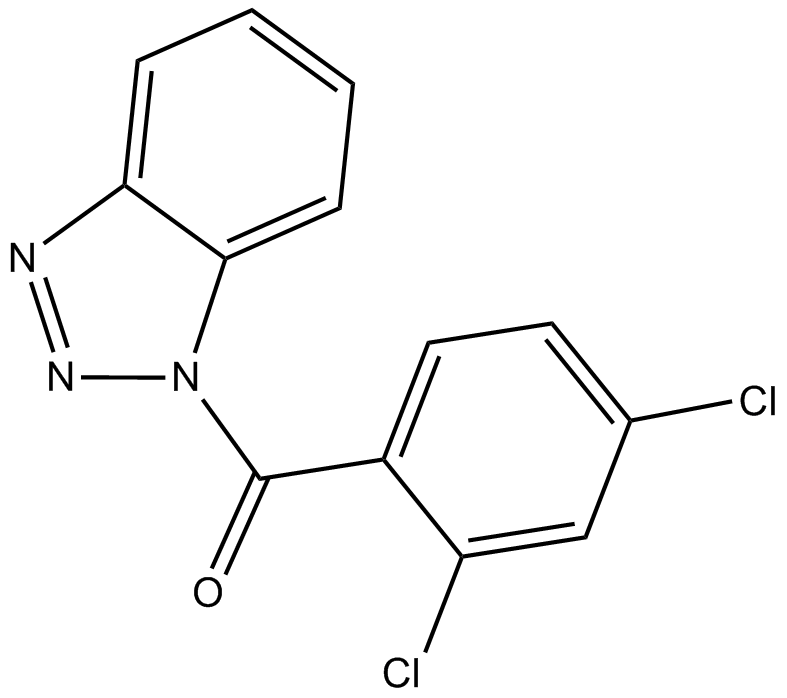
-
GC14797
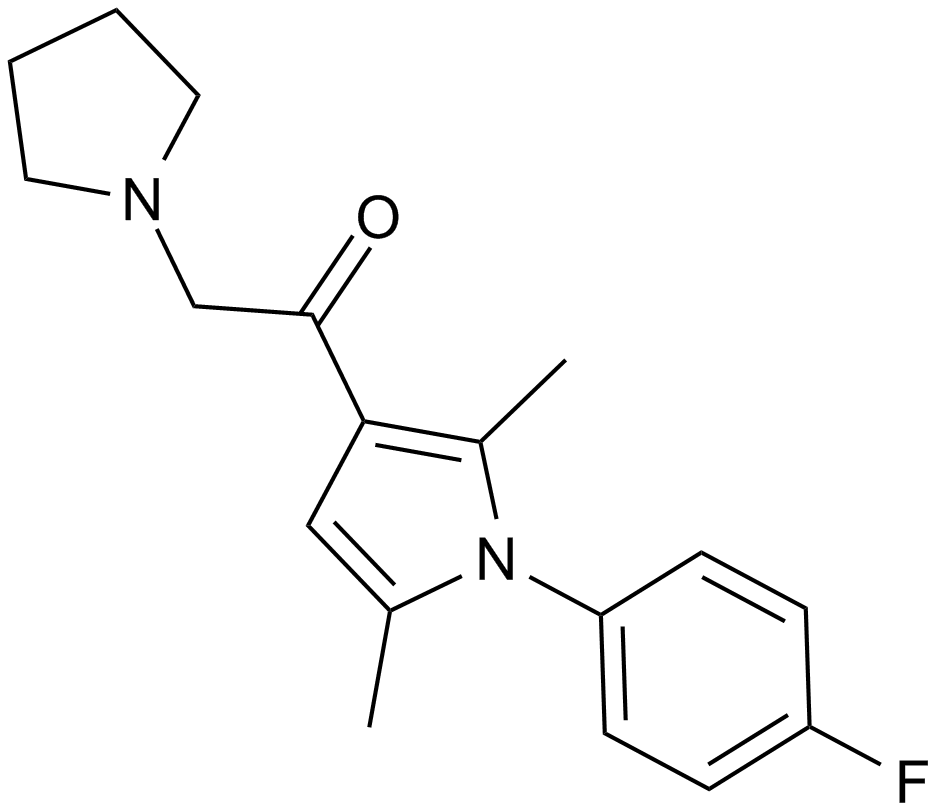
IU1
A deubiquitinating enzyme inhibitor
-
GC63027
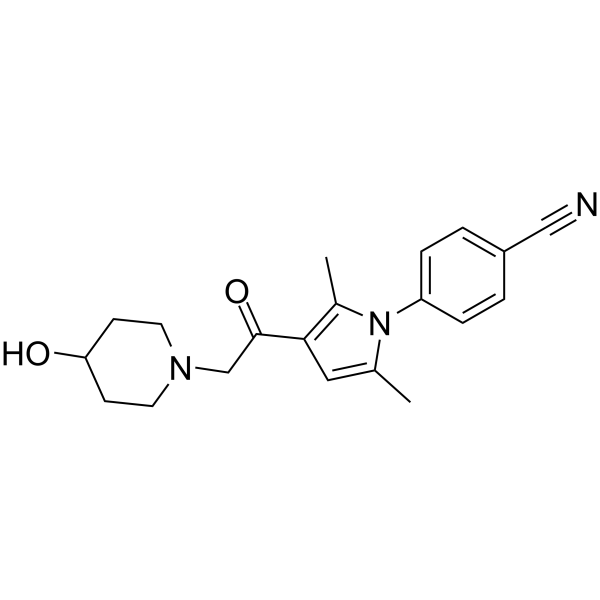
IU1-248
IU1-248, a derivative of IU1, is a potent and selective USP14 inhibitor with an IC50 of 0.83?μM.
-
GC64956
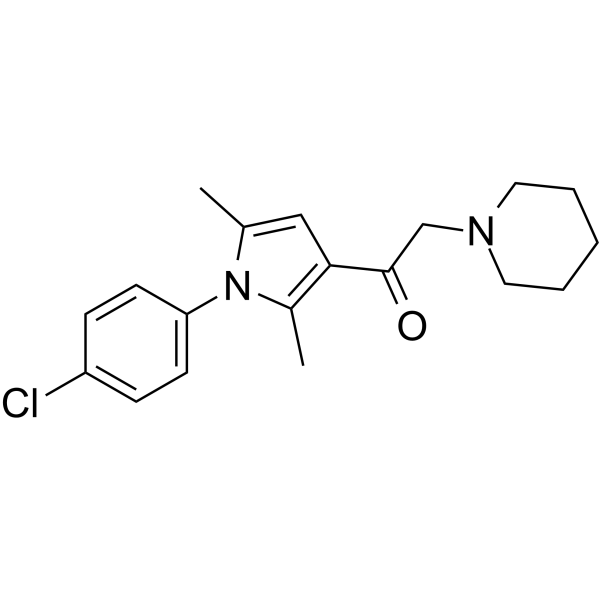
IU1-47
IU1-47 is a potent and specific USP14 inhibitor with an IC50 of 0.6 μM.
-
GC63460
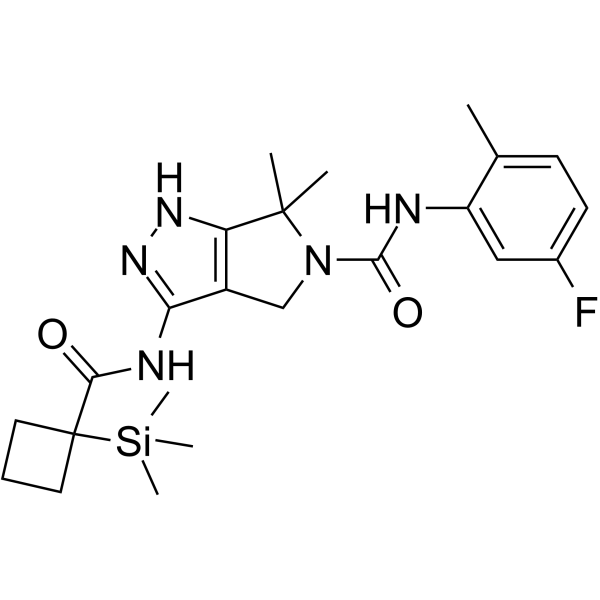
IV-361
IV-361 is an orally active and selective CDK7 inhibitor (Ki≤50 nM). IV-361 has anti-cancer activity (US20190256531A1).
-
GC16454
IWP-2
IWP-2 is a inhibitor of the Wnt signaling pathway, with an IC50 value of 27 nM. IWP-2 is also an ATP-competitive inhibitor of CK1δ, with an IC50 value of 40 nM.
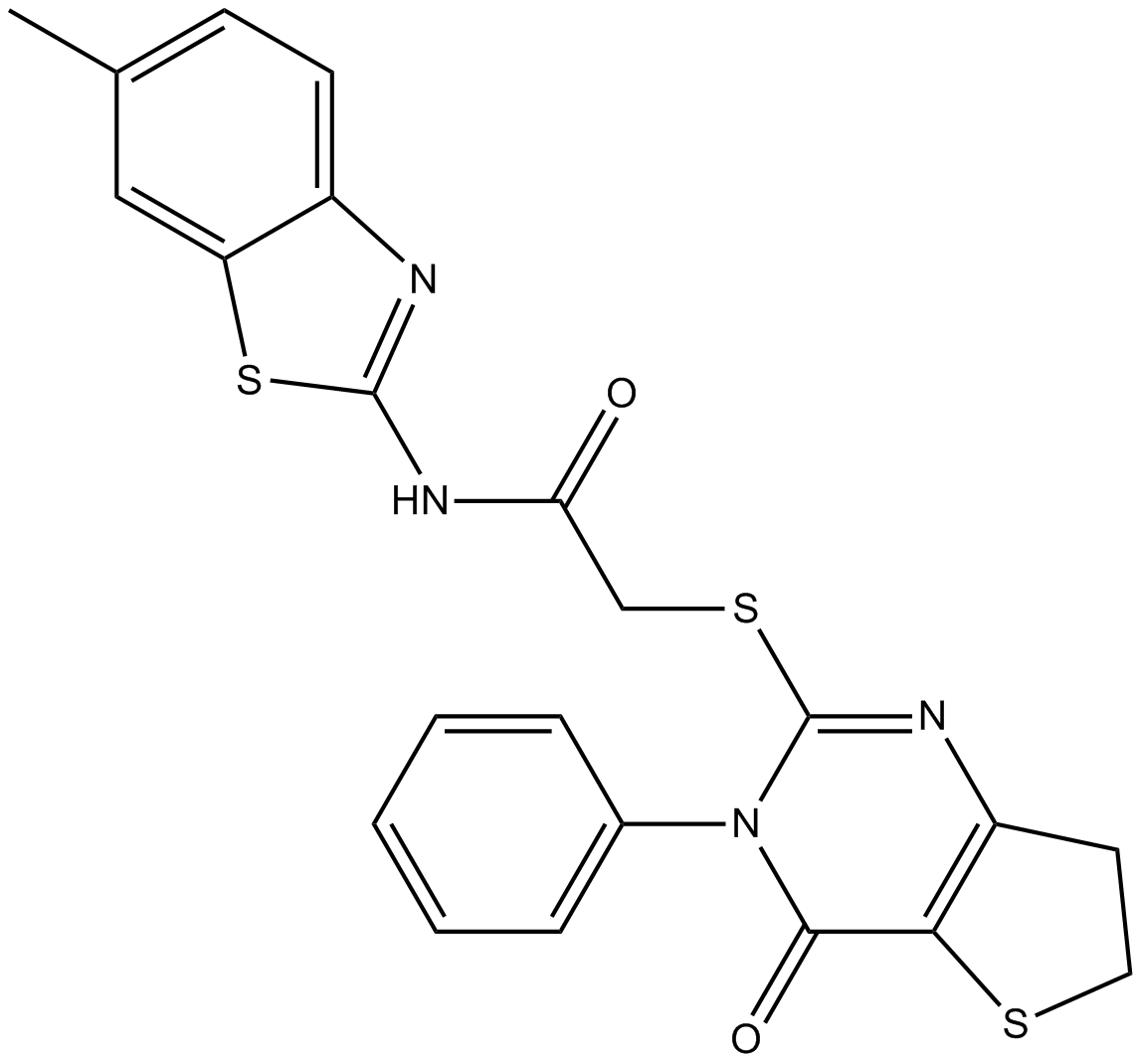
-
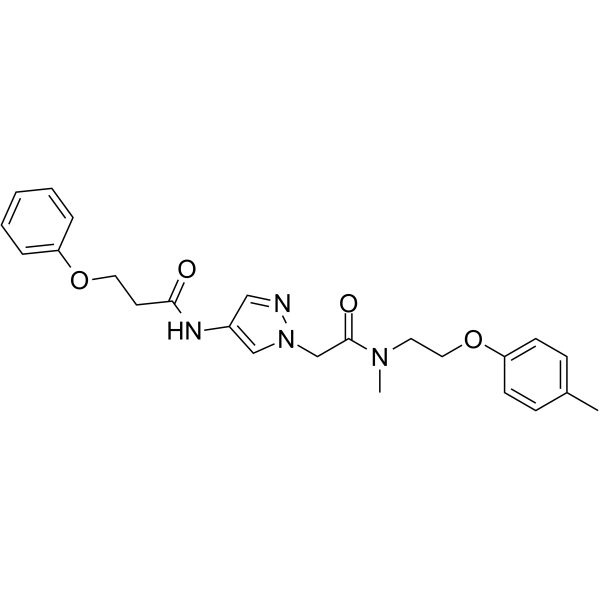
GC63785
IXA4
IXA4 is a highly selective, non-toxic IRE1/XBP1s activator.
-
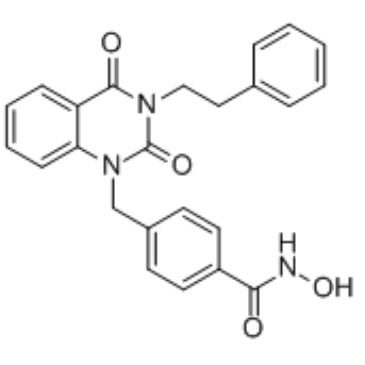
GC34629
J22352
J22352 is a PROTAC (proteolysis-targeting chimeras)-like and highly selective HDAC6 inhibitor with an IC50 value of 4.7 nM. J22352 promotes HDAC6 degradation and induces anticancer effects by inhibiting autophagy and eliciting the antitumor immune response in glioblastoma cancers, and leading to the restoration of host antitumor activity by reducing the immunosuppressive activity of PD-L1.
-
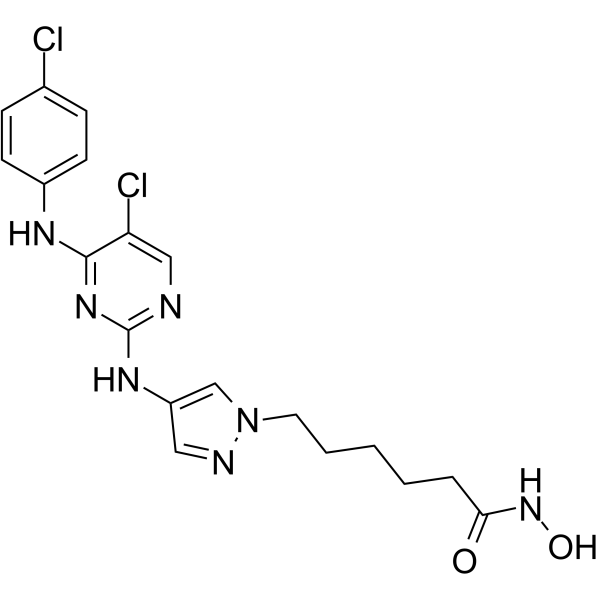
GC64959
JAK/HDAC-IN-1
JAK/HDAC-IN-1 is a potent JAK2/HDAC dual inhibitor, exhibits antiproliferative and proapoptotic activities in several hematological cell lines. JAK/HDAC-IN-1 shows IC50s of 4 and 2 nM for JAK2 and HDAC, respectively.
-
GC36367
JH-RE-06
JH-RE-06, a potent REV1-REV7 interface inhibitor (IC50=0.78 μM; Kd=0.42 μM), targets REV1 that interacts with the REV7 subunit of POLζ. JH-RE-06 disrupts mutagenic translesion synthesis (TLS) by preventing recruitment of mutagenic POLζ. JH-RE-06 improves chemotherapy.
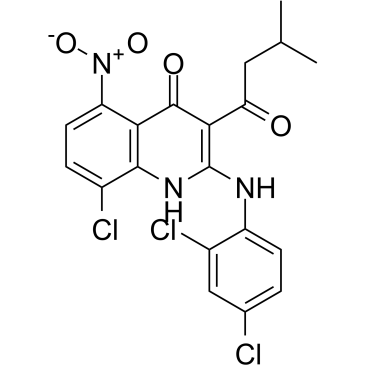
-
GC65471
JH-XI-10-02
JH-XI-10-02 is a PROTAC connected by ligands for Cereblon and CDK. JH-XI-10-02 is a highly potent and selective PROTAC CDK8 degrader, with an IC50 of 159 nM. JH-XI-10-02 causes proteasomal degradation, does not affect CDK8 mRNA levels. JH-XI-10-02 shows no effect on CDK19.
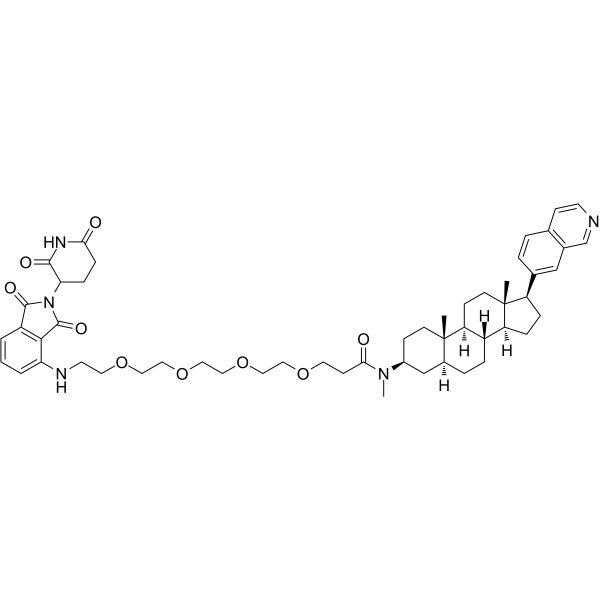
-
GC65577
JH-XVI-178
JH-XVI-178 is a highly potent and selective inhibitor of CDK8/19 that displays low clearance and moderate oral pharmacokinetic properties.

-
GC12612
JNJ-7706621
A dual inhibitor of CDKs and Aurora kinases
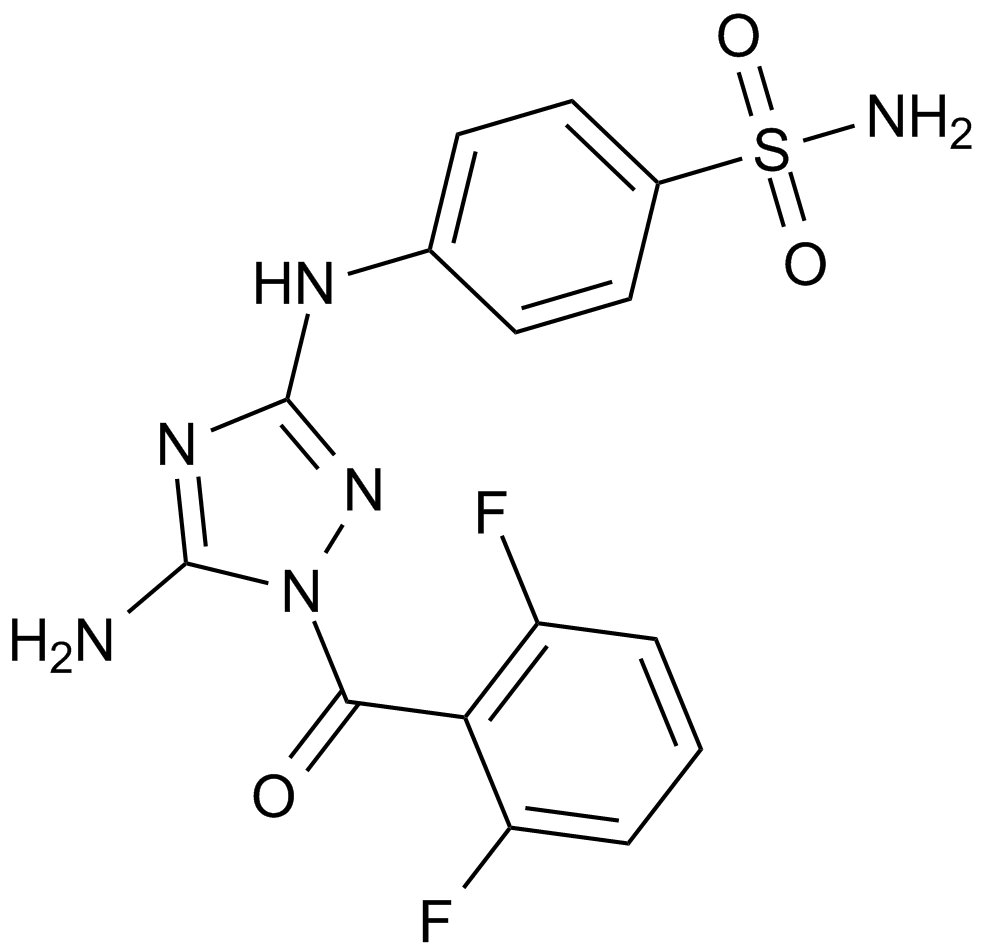
-
GC18734
JS-K
JS-K is a nitric oxide (NO) donor that reacts with glutathione to generate NO at physiological pH.
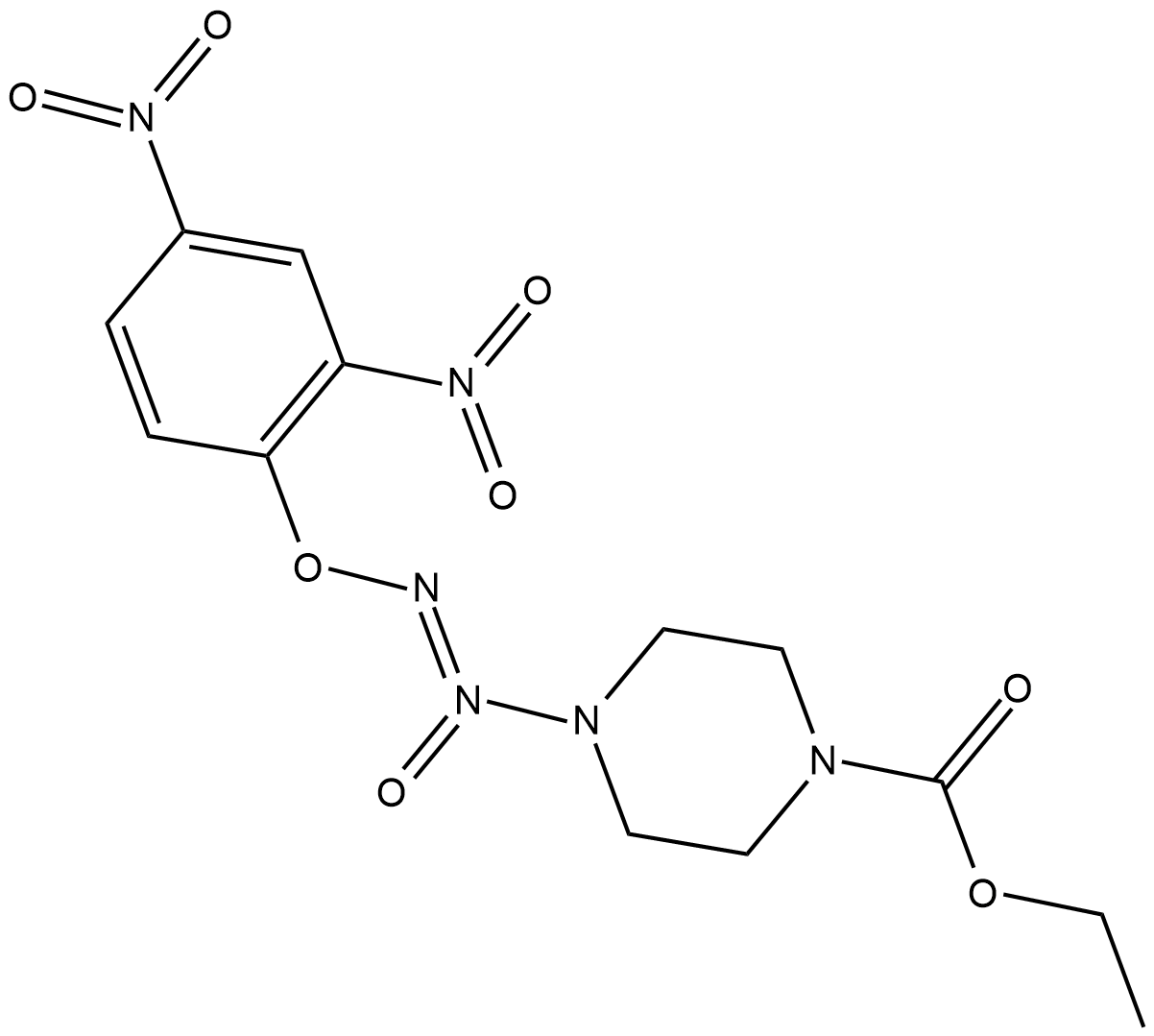
-
GC34204
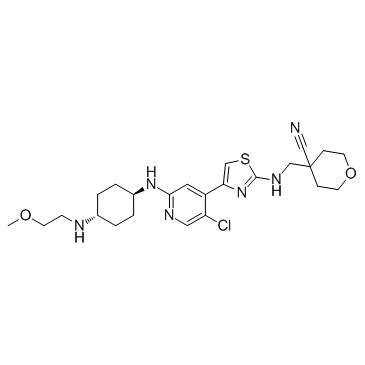
JSH-150
JSH-150 is a highly selective and potent CDK9 inhibitor with an IC50 of 1 nM.
-
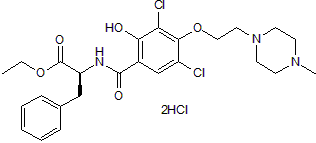
GC50117
JTE 607 dihydrochloride
JTE 607 dihydrochloride, a highly selective inflammatory cytokine synthesis inhibitor, protects from endotoxin shock in mice.
-
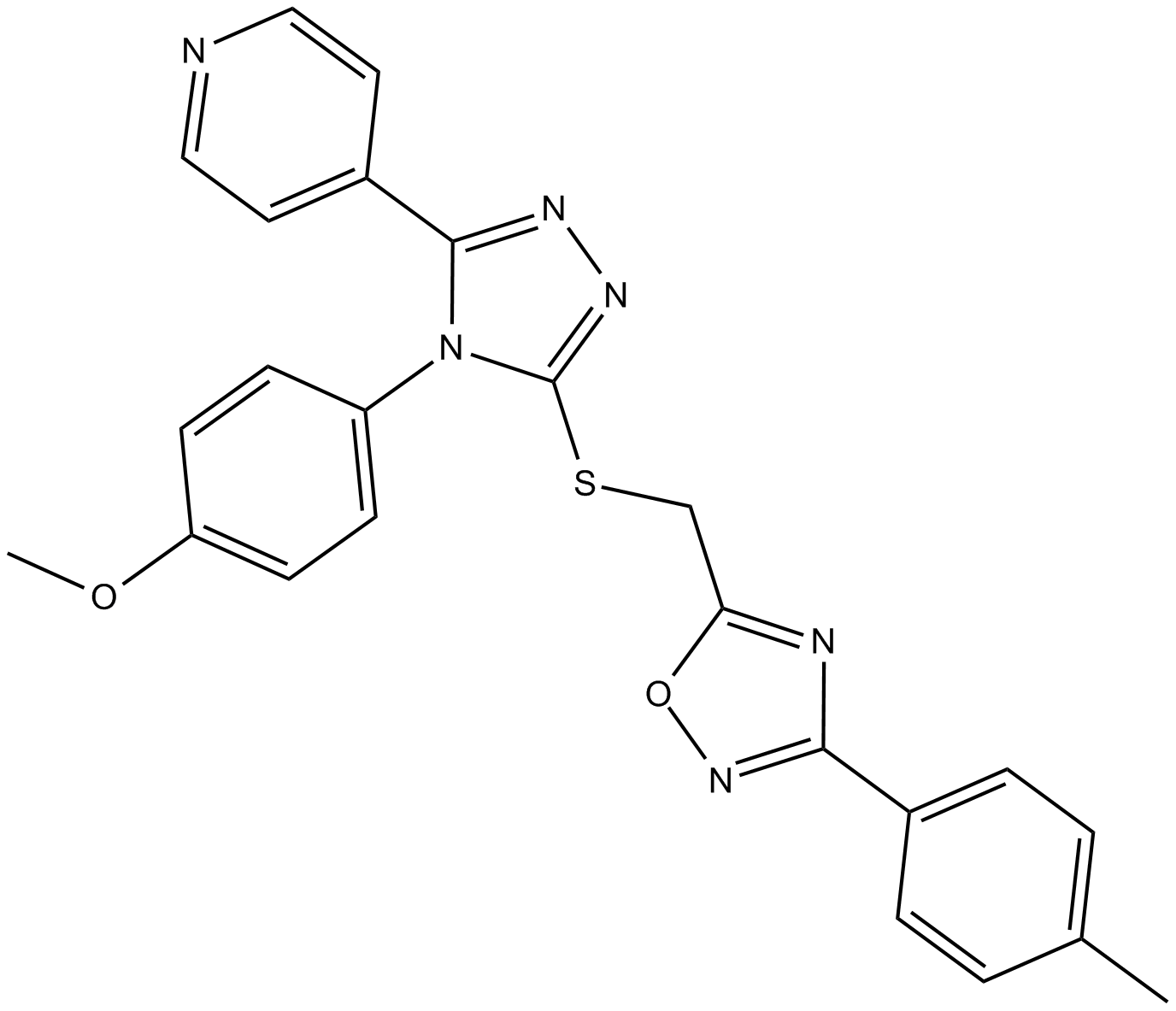
GC13978
JW 74
inhibitor of the catalytic PARP domain of TNKS1/2
-
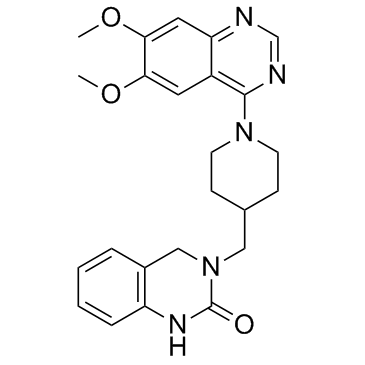
GC34195
K-756
K-756 is a direct and selective tankyrase (TNKS) inhibitor, which inhibits the ADP-ribosylation activity of TNKS1 and TNKS2 with IC50s of 31 and 36 nM, respectively.
-
GC41384
K-TMZ
K-TMZ is a DNA alkylating agent.
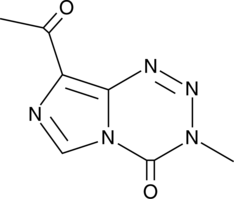
-
GC14230
K03861
K03861 (K03861) is a type II CDK2 inhibitor with Kd of 8.2 nM. K03861 (K03861) inhibits CDK2 activity by competing with binding of activating cyclins.
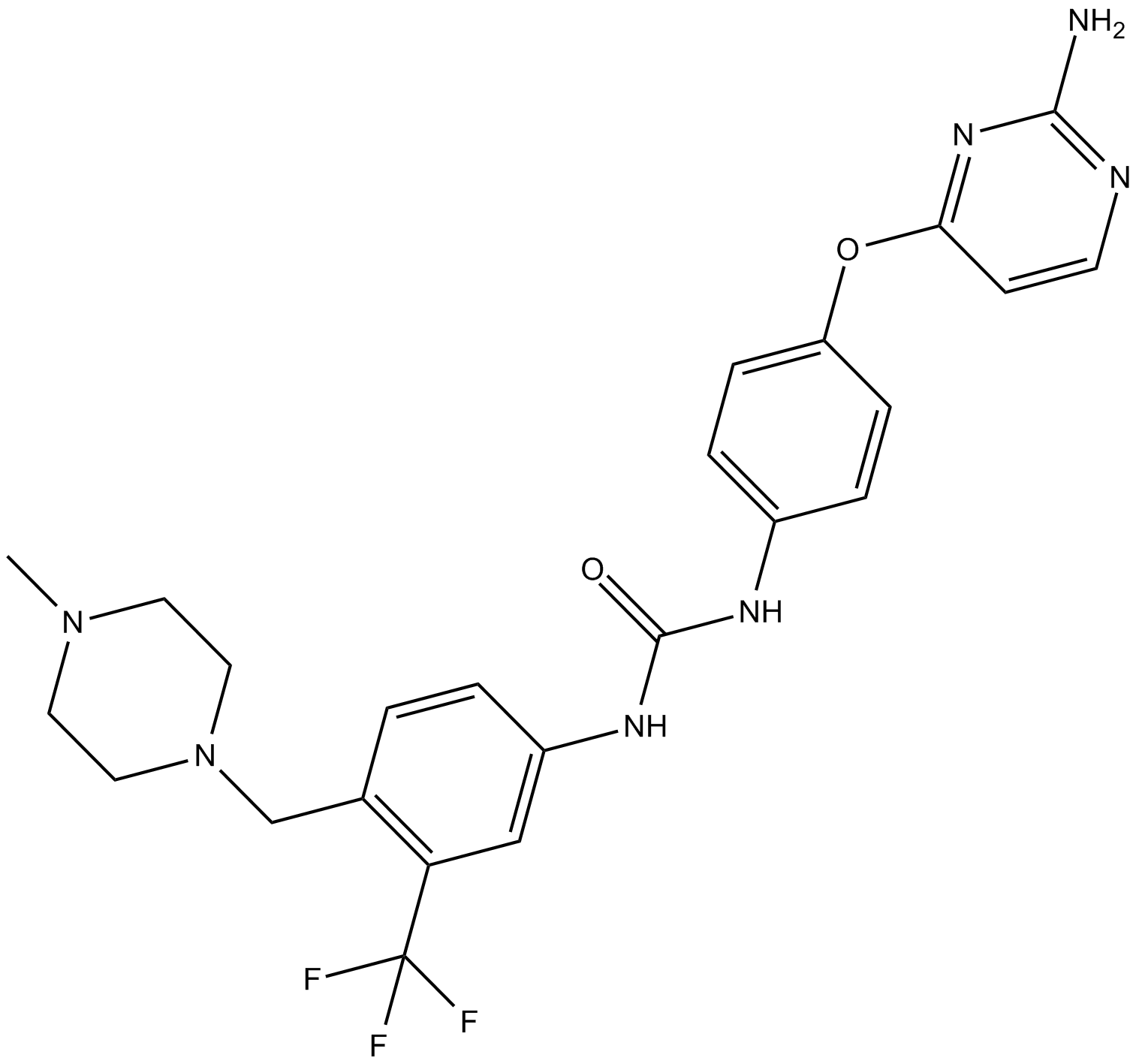
-
GC47521
K22
An antiviral agent
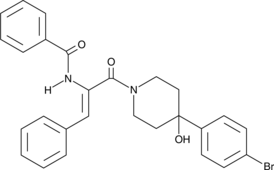
-
GC34144
Karenitecin (Cositecan)
Karenitecin (Cositecan) (Cositecan) is a topoisomerase I inhibitor, with potent anti-cancer activity.
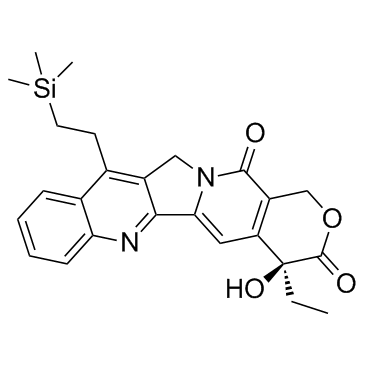
-
GC62392
KB-0742 dihydrochloride
KB-0742 dihydrochloride is a potent, selective and orally active CDK9 inhibitor with an IC50 of 6 nM for CDK9/cyclin T1. KB-0742 dihydrochloride is selective for CDK9/cyclin T1 with >50-fold selectivity over other CDK kinases. KB-0742 dihydrochloride has potent anti-tumor activity.
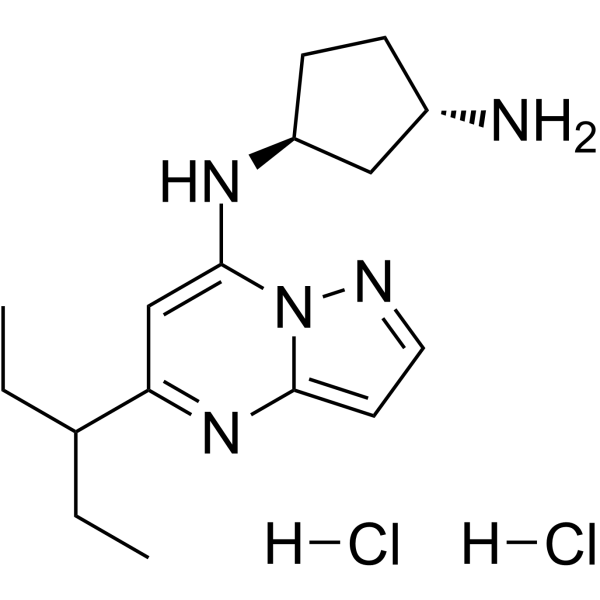
-
GC14182
Kenpaullone
CDK1/cyclin B and GSK-3β inhibitor
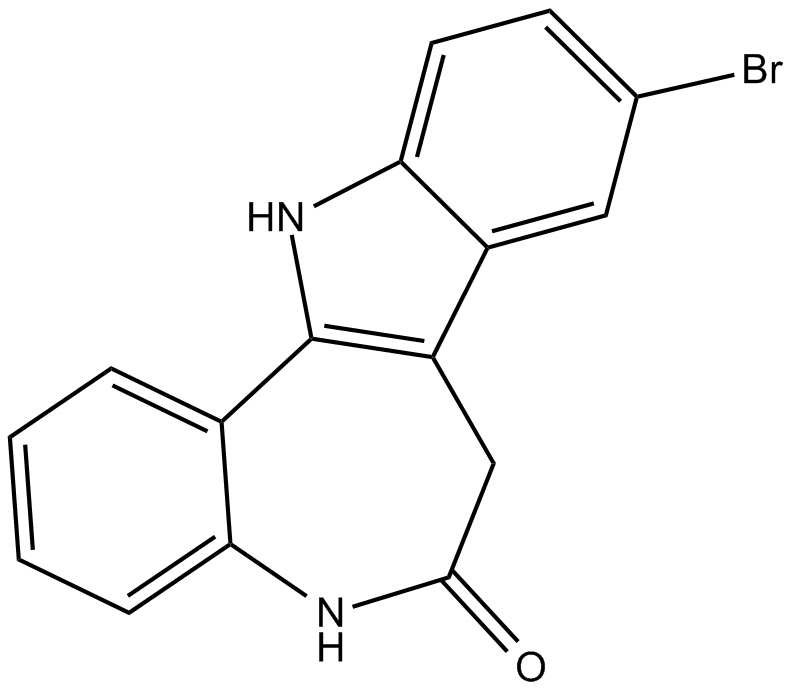
-
GC11774
KH CB19
inhibitor of CLK1 and CLK4, potent and selective
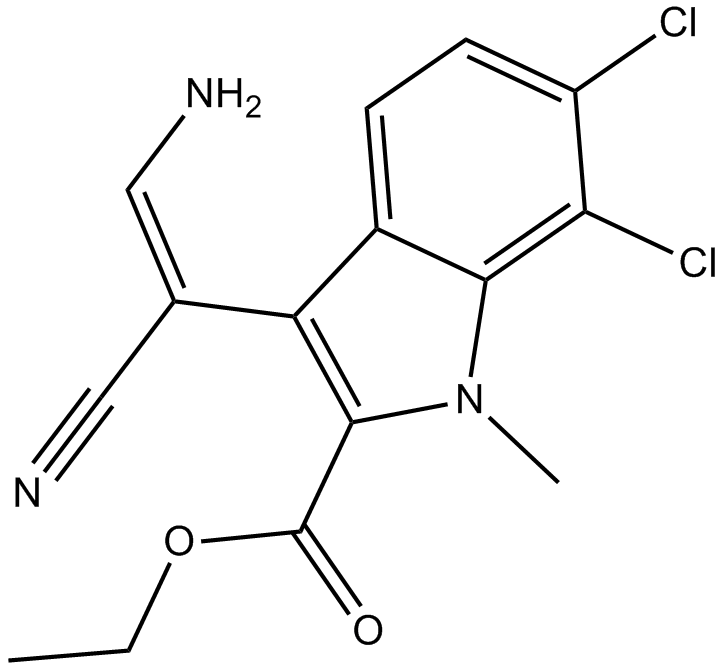
-
GC63943
KH-CB20
KH-CB20, an E/Z mixture, is a potent and selective inhibitor of CLK1 and the closely related isoform CLK4, with an IC50 of 16.5 nM for CLK1.
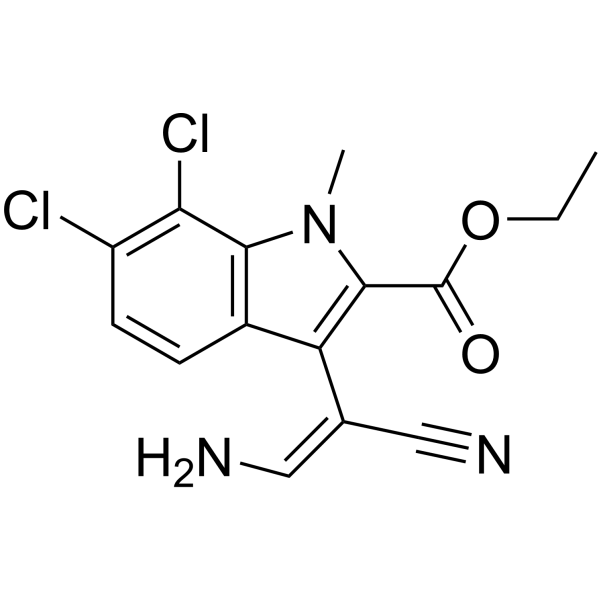
-
GC64238
KIRA-7
KIRA-7, an imidazopyrazine compound, binds the IRE1α kinase (IC50 of 110 nM) to allosterically inhibit its RNase activity.
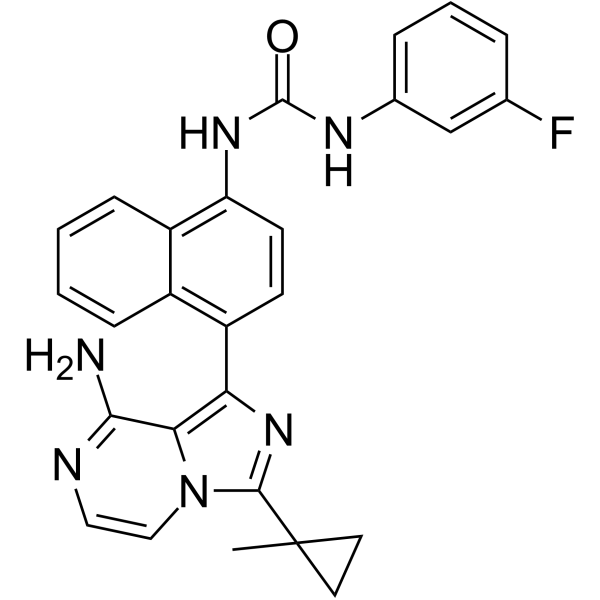
-
GC19212
KIRA6
KIRA6 allosterically inhibits IRE1α RNase kinase activity with an IC50 of 0.6 uM.
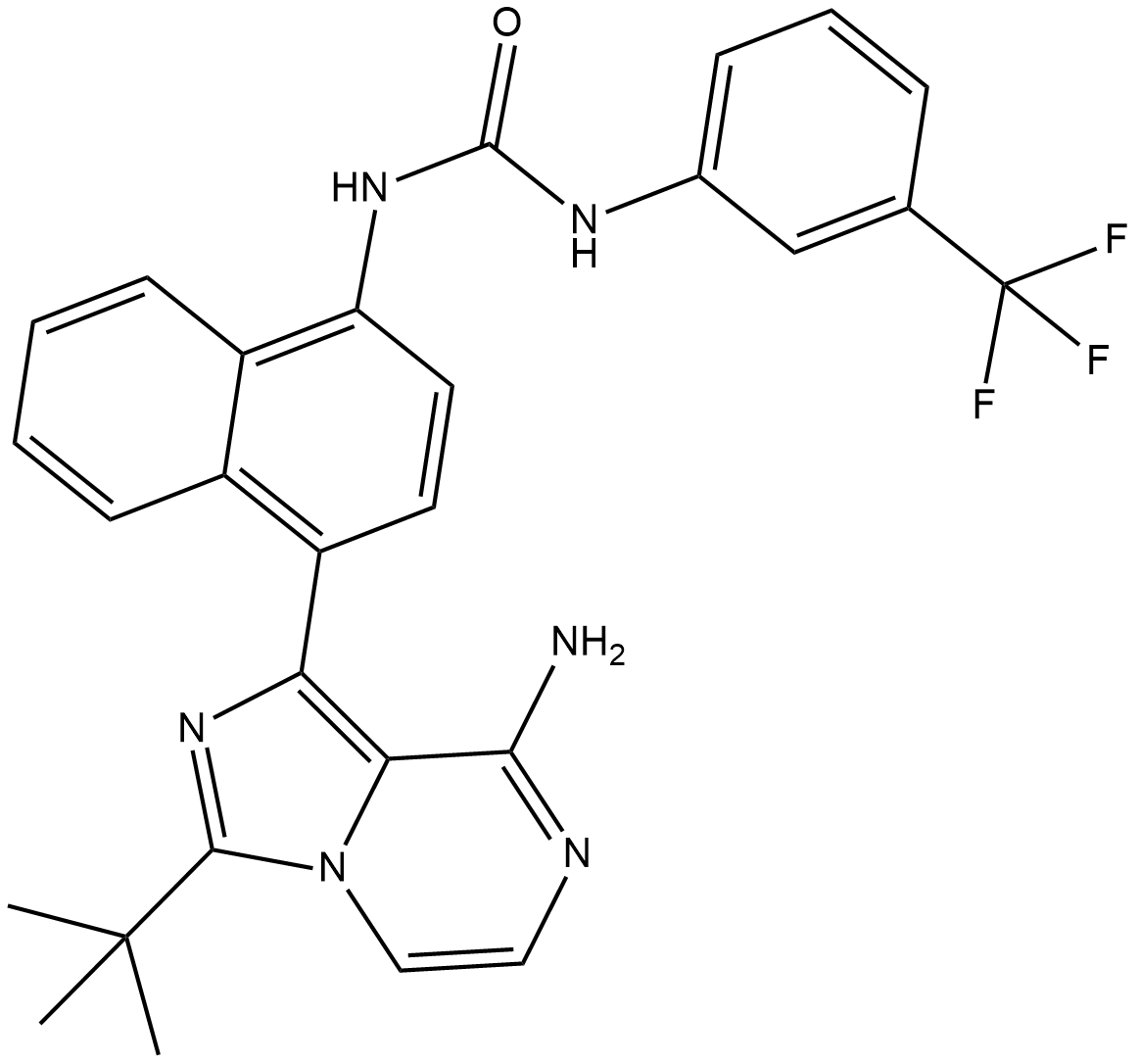
-
GC31879
Kira8
Kira8 (AMG-18) is a mono-selective IRE1α inhibitor that allosterically attenuates IRE1α RNase activity with an IC50 of 5.9 nM.
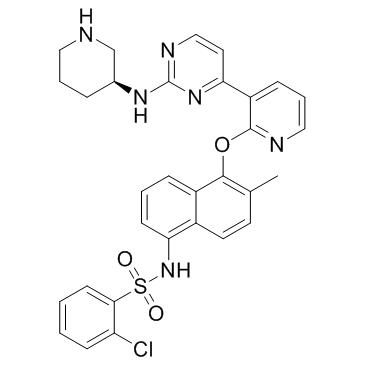
-
GC65907
KSQ-4279
KSQ-4279 (USP1-IN-1, Formula I) is a USP1 and PARP inhibitor (extracted from patent WO2021163530).
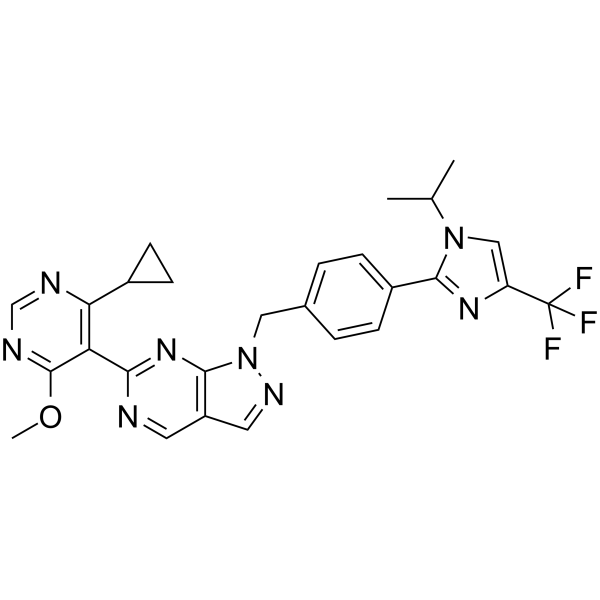
-
GC25552
KT-531
KT-531 (KT531) is a potent, selective HDAC6 inhibitor with IC50 of 8.5 nM, displays 39-fold selectivity over other HDAC isoforms.

-
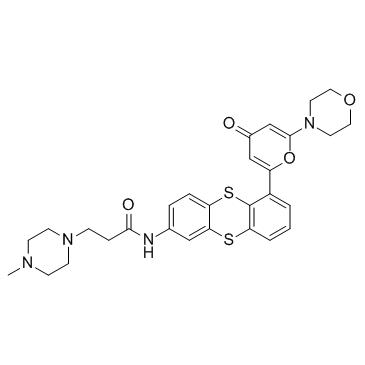
GC33404
KU 59403
KU 59403 is a potent ATM inhibitor, with IC50 values of 3 nM, 9.1 μM and 10 μM for ATM, DNA-PK and PI3K, respectively.
-
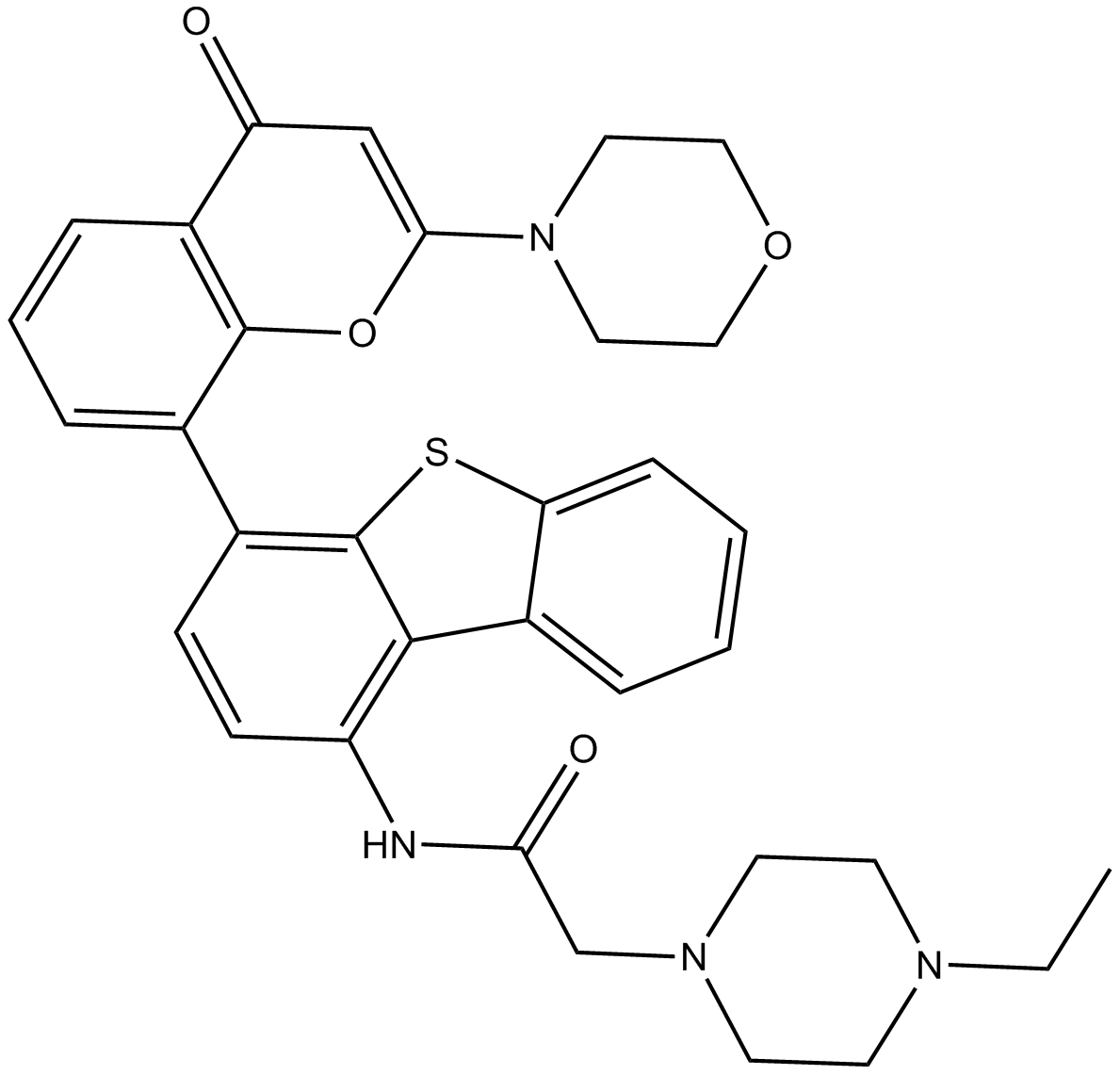
GC14346
KU-0060648
Dual DNA-PK/PI3-K inhibitor, ATP-competitive
-
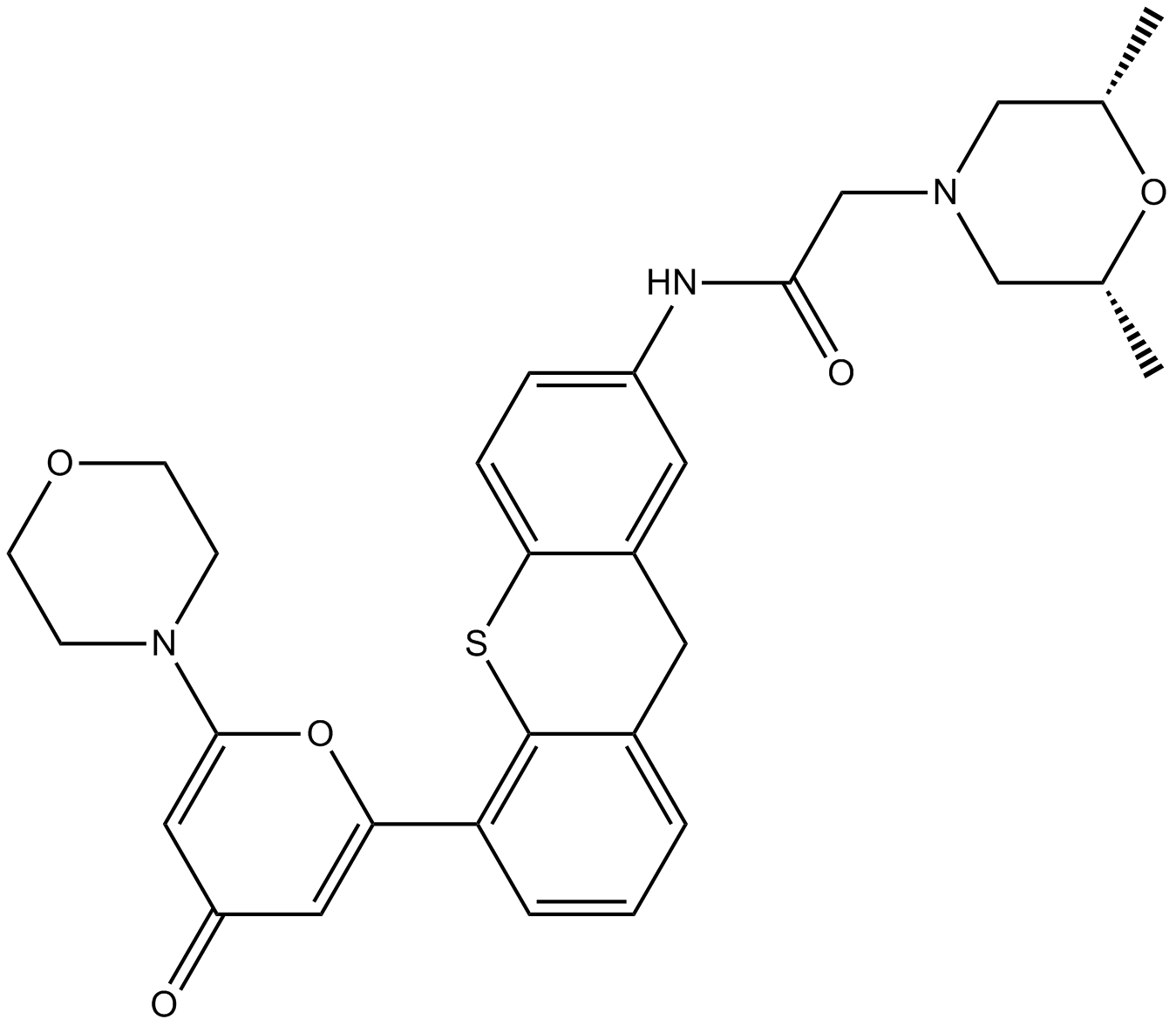
GC12054
KU-60019
ATM kinase inhibitor,potent and selective
-
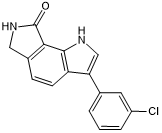
GC50532
KuWal151
Potent and selective CLK inhibitor
-
GC15743
L-755,507
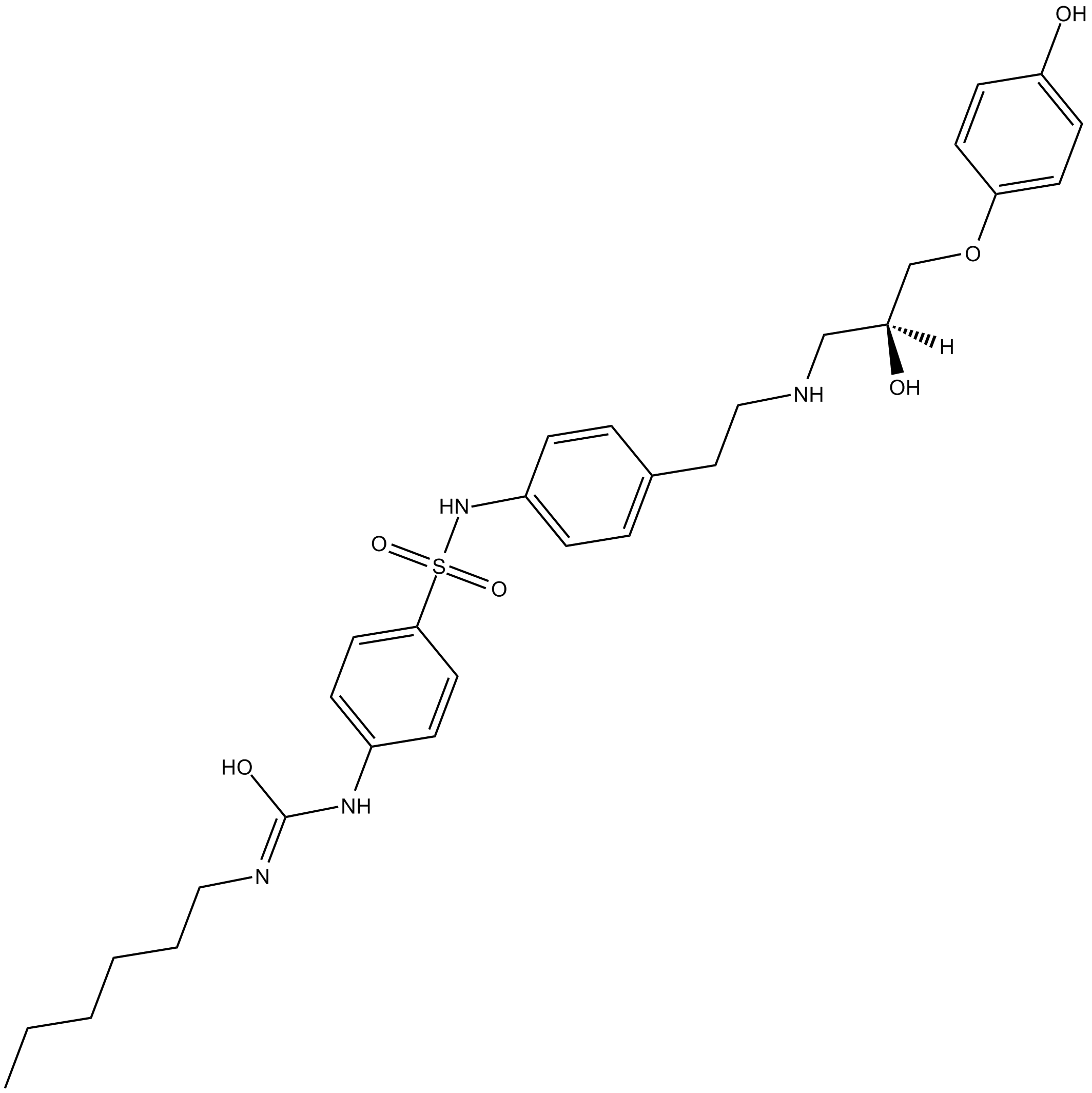
β3 adrenergic receptor agonist
-
GC66622
L-Guanosine
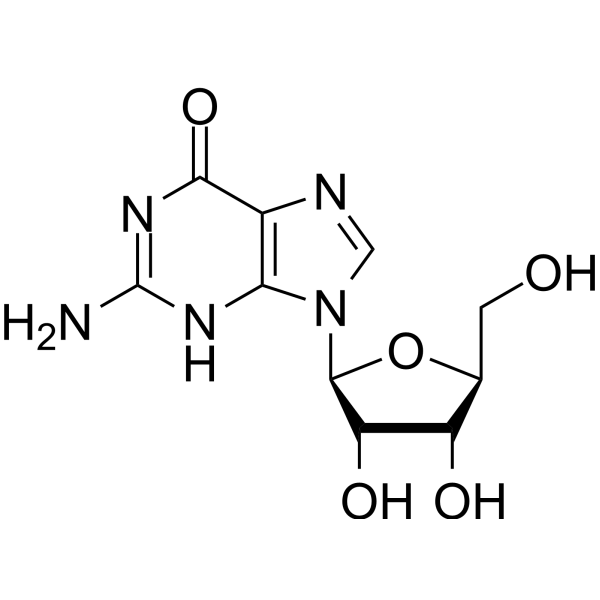
L-Guanosine is the L-configuration of Guanosine . Guanosine is a purine nucleoside with anti-herpesvirus activity.
-
GC49381
L-Leucine-d10
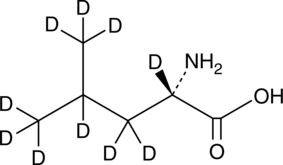
An internal standard for the quantification of L-leucine
-
GC49368
L-Methionine-d3
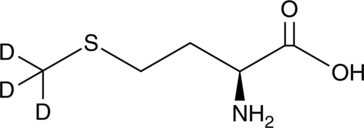
An internal standard for the quantification of L-methionine
-
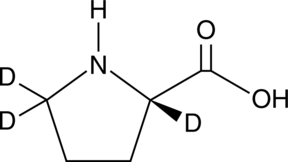
GC49384
L-Proline-d3
An internal standard for the quantification of L-proline
-
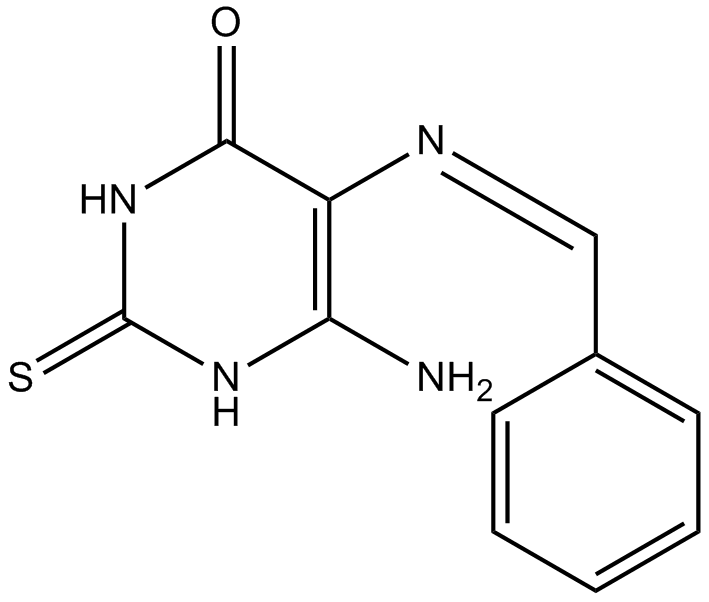
GC12131
L189
inhibitor of human DNA ligases I, III and IV
-
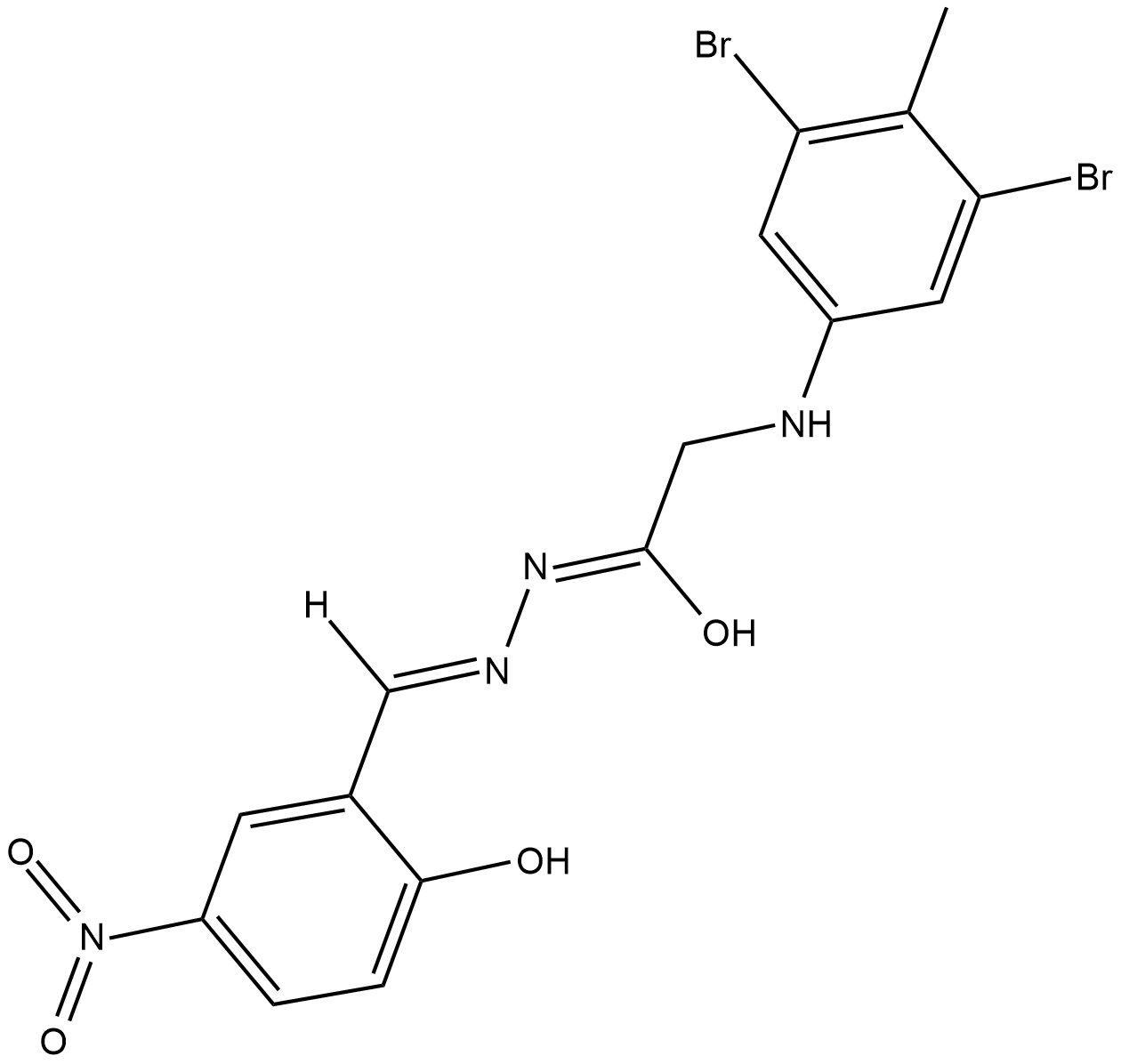
GC18186
L67
L67 is a competitive inhibitor of DNA ligases I and III (IC50s = 10 and 10 uM for human DNA ligase I and human ligase IIIβ).
-
GC67785
L82

-
GC68436
L82-G17

-
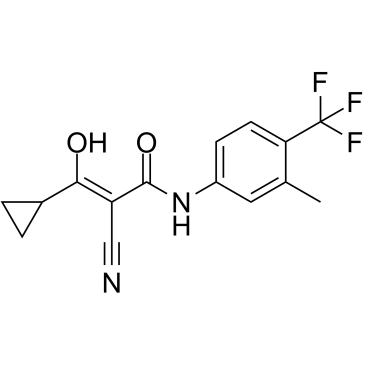
GC39632
Laflunimus
Laflunimus (HR325) is an immunosuppressive agent and an analogue of the Leflunomide-active metabolite A77 1726.
-
GC62460
LCAHA
LCAHA (LCA hydroxyamide) is a deubiquitinase USP2a inhibitor with IC50s of 9.7 μM and 3.7μM in Ub-AMC Assay and Di-Ub Assay, respectively. LCAHA destabilizes Cyclin D1 and induces G0/G1 arrest by inhibiting deubiquitinase USP2a.
-
GC17067
LDC000067
CDK9 inhibitor, novel and highly specific
-
GC19219
LDC4297
LDC4297 is a potent and selective CDK7 inhibitor with an IC50 of 0.13 nM.