Immunology/Inflammation
The immune and inflammation-related pathway including the Toll-like receptors pathway, the B cell receptor signaling pathway, the T cell receptor signaling pathway, etc.
Toll-like receptors (TLRs) play a central role in host cell recognition and responses to microbial pathogens. TLR4 initially recruits TIRAP and MyD88. MyD88 then recruits IRAKs, TRAF6, and the TAK1 complex, leading to early-stage activation of NF-κB and MAP kinases [1]. TLR4 is endocytosed and delivered to intracellular vesicles and forms a complex with TRAM and TRIF, which then recruits TRAF3 and the protein kinases TBK1 and IKKi. TBK1 and IKKi catalyze the phosphorylation of IRF3, leading to the expression of type I IFN [2].
BCR signaling is initiated through ligation of mIg under conditions that induce phosphorylation of the ITAMs in CD79, leading to the activation of Syk. Once Syk is activated, the BCR signal is transmitted via a series of proteins associated with the adaptor protein B-cell linker (Blnk, SLP-65). Blnk binds CD79a via non-ITAM tyrosines and is phosphorylated by Syk. Phospho-Blnk acts as a scaffold for the assembly of the other components, including Bruton’s tyrosine kinase (Btk), Vav 1, and phospholipase C-gamma 2 (PLCγ2) [3]. Following the assembly of the BCR-signalosome, GRB2 binds and activates the Ras-guanine exchange factor SOS, which in turn activates the small GTPase RAS. The original RAS signal is transmitted and amplified through the mitogen-activated protein kinase (MAPK) pathway, which including the serine/threonine-specific protein kinase RAF followed by MEK and extracellular signal related kinases ERK 1 and 2 [4]. After stimulation of BCR, CD19 is phosphorylated by Lyn. Phosphorylated CD19 activates PI3K by binding to the p85 subunit of PI3K and produce phosphatidylinositol-3,4,5-trisphosphate (PIP3) from PIP2, and PIP3 transmits signals downstream [5].
Central process of T cells responding to specific antigens is the binding of the T-cell receptor (TCR) to specific peptides bound to the major histocompatibility complex which expressed on antigen-presenting cells (APCs). Once TCR connected with its ligand, the ζ-chain–associated protein kinase 70 molecules (Zap-70) are recruited to the TCR-CD3 site and activated, resulting in an initiation of several signaling cascades. Once stimulation, Zap-70 forms complexes with several molecules including SLP-76; and a sequential protein kinase cascade is initiated, consisting of MAP kinase kinase kinase (MAP3K), MAP kinase kinase (MAPKK), and MAP kinase (MAPK) [6]. Two MAPK kinases, MKK4 and MKK7, have been reported to be the primary activators of JNK. MKK3, MKK4, and MKK6 are activators of P38 MAP kinase [7]. MAP kinase pathways are major pathways induced by TCR stimulation, and they play a key role in T-cell responses.
Phosphoinositide 3-kinase (PI3K) binds to the cytosolic domain of CD28, leading to conversion of PIP2 to PIP3, activation of PKB (Akt) and phosphoinositide-dependent kinase 1 (PDK1), and subsequent signaling transduction [8].
References
[1] Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors[J]. Nature immunology, 2010, 11(5): 373-384.
[2] Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-650.
[3] Packard T A, Cambier J C. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation[J]. F1000Prime Rep, 2013, 5(40.10): 12703.
[4] Zhong Y, Byrd J C, Dubovsky J A. The B-cell receptor pathway: a critical component of healthy and malignant immune biology[C]//Seminars in hematology. WB Saunders, 2014, 51(3): 206-218.
[5] Baba Y, Matsumoto M, Kurosaki T. Calcium signaling in B cells: regulation of cytosolic Ca 2+ increase and its sensor molecules, STIM1 and STIM2[J]. Molecular immunology, 2014, 62(2): 339-343.
[6] Adachi K, Davis M M. T-cell receptor ligation induces distinct signaling pathways in naive vs. antigen-experienced T cells[J]. Proceedings of the National Academy of Sciences, 2011, 108(4): 1549-1554.
[7] Rincón M, Flavell R A, Davis R A. The Jnk and P38 MAP kinase signaling pathways in T cell–mediated immune responses[J]. Free Radical Biology and Medicine, 2000, 28(9): 1328-1337.
[8] Bashour K T, Gondarenko A, Chen H, et al. CD28 and CD3 have complementary roles in T-cell traction forces[J]. Proceedings of the National Academy of Sciences, 2014, 111(6): 2241-2246.
Targets for Immunology/Inflammation
- Cyclic GMP-AMP Synthase(1)
- Apoptosis(137)
- 5-Lipoxygenase(18)
- TLR(106)
- Papain(2)
- PGDS(1)
- PGE synthase(26)
- SIKs(10)
- IκB/IKK(83)
- AP-1(2)
- KEAP1-Nrf2(47)
- NOD1(1)
- NF-κB(265)
- Interleukin Related(129)
- 15-lipoxygenase(2)
- Others(10)
- Aryl Hydrocarbon Receptor(35)
- CD73(16)
- Complement System(46)
- Galectin(30)
- IFNAR(19)
- NO Synthase(78)
- NOD-like Receptor (NLR)(37)
- STING(84)
- Reactive Oxygen Species(434)
- FKBP(14)
- eNOS(4)
- iNOS(24)
- nNOS(21)
- Glutathione(37)
- Adaptive Immunity(144)
- Allergy(129)
- Arthritis(25)
- Autoimmunity(134)
- Gastric Disease(64)
- Immunosuppressants(27)
- Immunotherapeutics(3)
- Innate Immunity(411)
- Pulmonary Diseases(76)
- Reactive Nitrogen Species(43)
- Specialized Pro-Resolving Mediators(42)
- Reactive Sulfur Species(24)
Products for Immunology/Inflammation
- Cat.No. Product Name Information
-
GC49393
all-trans-13,14-Dihydroretinol
A metabolite of all-trans retinoic acid

-
GC45379
Alloxan (hydrate)
-
GC11443
Allylthiourea
Selective inhibitor of ammonia oxidation
-
GP10015
alpha-1 antitrypsin fragment
Protease inhibitor
-
GP10008
alpha-1 antitrypsin fragment 235-243 [Homo sapiens]/[Papio hamadryas]/[Cercopithecus aethiops]
Protease inhibitor
![alpha-1 antitrypsin fragment 235-243 [Homo sapiens]/[Papio hamadryas]/[Cercopithecus aethiops] Chemical Structure alpha-1 antitrypsin fragment 235-243 [Homo sapiens]/[Papio hamadryas]/[Cercopithecus aethiops] Chemical Structure](/media/struct/GP1/GP10008.png)
-
GC35306
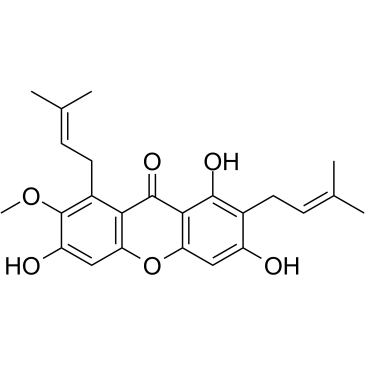
alpha-Mangostin
alpha-Mangostin (α-Mangostin) is a dietary xanthone with broad biological activities, such as antioxidant, anti-allergic, antiviral, antibacterial, anti-inflammatory and anticancer effects. It is an inhibitor of mutant IDH1 (IDH1-R132H) with a Ki of 2.85 μM.
-
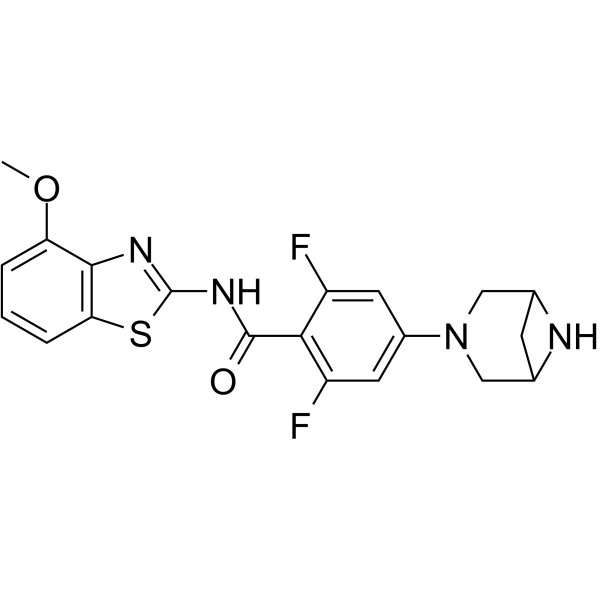
GC67968
ALPK1-IN-2
-
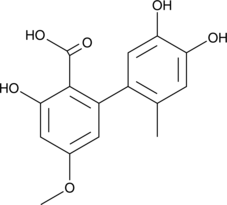
GC40024
Altenusin
Altenusin is a polyphenol fungal metabolite originally isolated from the fungus Alternaria that has diverse biological activities.
-
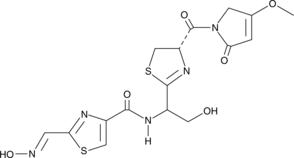
GC49039
Althiomycin
A thiazole antibiotic
-
GC46835
Alyssin
An isothiocyanate with diverse biological activities
-
GC49638
Ambrisentan-d3
An internal standard for the quantification of ambrisentan
-
GC48611
Ambroxol-d5
An internal standard for the quantification of ambroxol
-
GC42780
Ambuic Acid
Ambuic acid is a cyclohexanone originally isolated from Pestalotiopsis and Monochaetia species that has phytopathogenic antifungal, quorum sensing inhibitory, and antibacterial activities.

-
GN10484
Amentoflavone
-
GC46840
Amicoumacin B
An amicoumacin with quorum-sensing inhibitory activity
-
GC15727
Aminoguanidine hydrochloride
iNOS inhibitor
-
GC65226
Aminopicoline
Aminopicoline (Ascensil) is a potent and nonselective inhibitor of NO synthase (NOS) isoenzymes (iNOS, nNOS, eNOS).
-
GC18274
Amiprofos-methyl
Amiprofos-methyl (APM) is a phosphoric amide herbicide.
-
GC49336
AMK (hydrochloride)
An active metabolite of melatonin
-
GC31319
Amlexanox (AA673)
Amlexanox (AA673) (AA673; Amoxanox; CHX3673) is a specific inhibitor of IKKε and TBK1, and inhibits the IKKε and TBK1 activity determined by MBP phosphorylation with an IC50 of approximately 1-2 μM.
-
GC45725
Amodiaquine-d10
An internal standard for the quantification of amodiaquine
-
GC40636
Amorfrutin A
Amorfrutin A is an isoprenoid-substituted benzoic acid natural product found in the fruit of A.
-
GC52059
AMOZ
-
GC41406
AMP-Deoxynojirimycin
The lipid messenger ceramide is converted to glucosylceramide by glucosylceramide synthase (GCS).
-
GC52406
AMPR-22 (trifluoroacetate salt)
An antimicrobial peptide
-
GC63932
Amsilarotene
Amsilarotene (TAC-101; Am 555S), an orally active synthetic retinoid, has selective affinity for retinoic acid receptor α (RAR-α) binding with Ki of 2.4, 400 nM for RAR-α and RAR-β. Amsilarotene induces the apoptotic of human gastric cancer, hepatocellular carcinoma and ovarian carcinoma cells. Amsilarotene can be used for the research of cancer.
-
GC15010
AMT hydrochloride
iNOS inhibitor
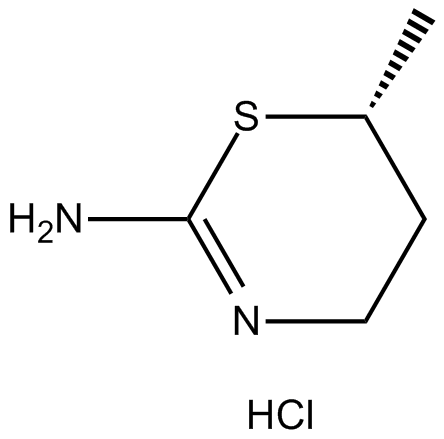
-
GC48339
Amycolatopsin A
A macrolide polyketide with antimycobacterial and anticancer activities
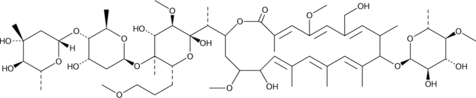
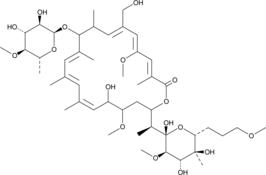
-
GC48350
Amycolatopsin C
A polyketide macrolide with antimycobacterial and anticancer activities
-
GP10099
amyloid A protein fragment [Homo sapiens]
Apolipoproteins related to HDL in plasma
![amyloid A protein fragment [Homo sapiens] Chemical Structure amyloid A protein fragment [Homo sapiens] Chemical Structure](/media/struct/GP1/GP10099.png)
-
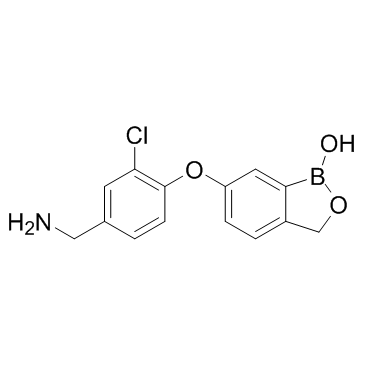
GC32057
AN-3485
AN-3485 is a benzoxaborole analog, Toll-Like Receptor(TLR) inhibitor with IC50 values ranging from 18 to 580 nM.
-
GC41211
Anacardic Acid Diene
Anacardic acid diene is a polyunsaturated form of anacardic acid that has been found in cashew nut shell liquid.
-
GC41531
Anacardic Acid Triene
Anacardic acid triene is a polyunsaturated form of anacardic acid that has been found in cashew nut shell liquid.
-
GC39254
Anatabine dicitrate
Anatabine dicitrate is a tobacco alkaloid that can cross the blood-brain barrier.
-
GN10718
Andrographolide
-
GC35340
Andropanolide
Andrographolide (Andro) is a small antagonist for NF-κB activation by covalent modifying reduced cysteine 62 of p50.
-
GN10685
Anemarsaponin B
-
GC19428
Angeli’s Salt
A classical nitroxyl (NO-) donor
-
GC39284
ANI-7
ANI-7 is an activator of aryl hydrocarbon receptor (AhR) pathway. ANI-7 inhibits the growth of multiple cancer cells, and potently and selectively inhibits the growth of MCF-7 breast cancer cells with a GI50 of 0.56 μM. ANI-7 induces CYP1-metabolizing mono-oxygenases by activating AhR pathway, and also induces DNA damage, checkpoint Kinase 2 (Chk2) activation, S-phase cell cycle arrest, and cell death in sensitive breast cancer cell lines.
-
GC49419
Aniline-d5
An internal standard for the quantification of aniline
-
GC42815
Ansatrienin A
Ansatrienin A is an ansamycin antibiotic and antifungal agent first isolated from S.
-
GC49259
Antagonist G (trifluoroacetate salt)
A neuropeptide antagonist
-
GP10124
Anti-Inflammatory Peptide 1
-
GC45383
Antibiotic PF 1052
-
GC49360
Antimycin A Complex
A bacterial metabolite complex
-
GC52128
AOD-9604
-
GC52380
AOD-9604 (acetate)
A synthetic lipolytic peptide
-
GC34172
AP1867
AP1867 is a synthetic FKBP12F36V-directed ligand.
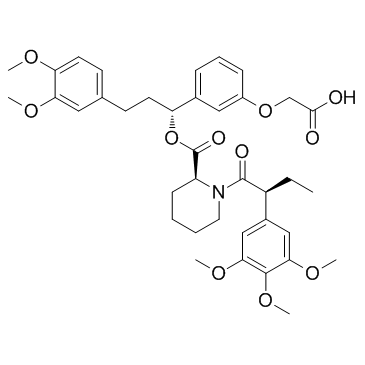
-
GC15586
AP1903
AP1903 (AP1903) is a dimerizer agent that acts by cross-linking the FKBP domains. AP1903 (AP1903) dimerizes the Caspase 9 suicide switch and rapidly induces apoptosis.
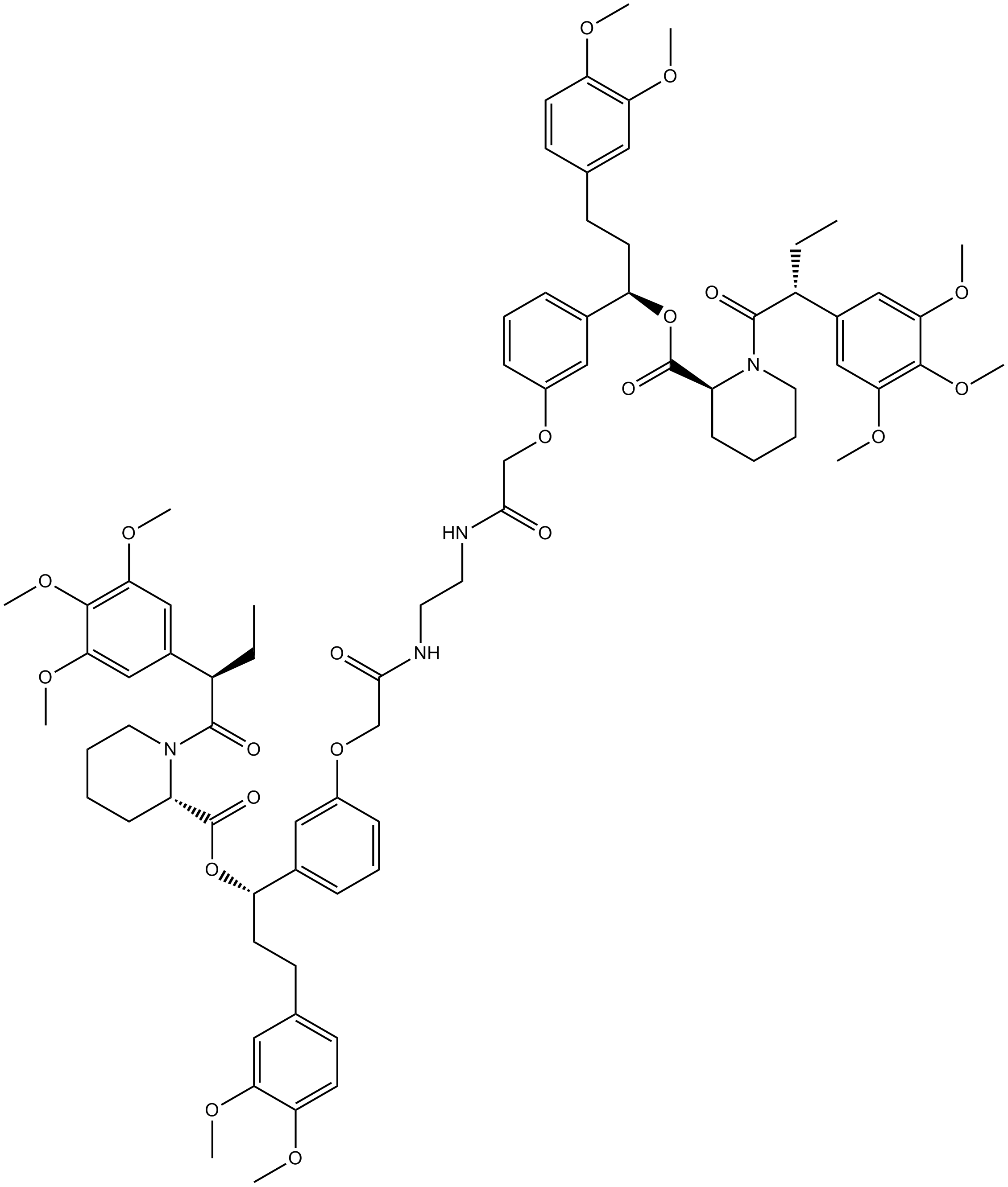
-
GC14498
AP20187
Dimerizer,synthetic and cell-permeable
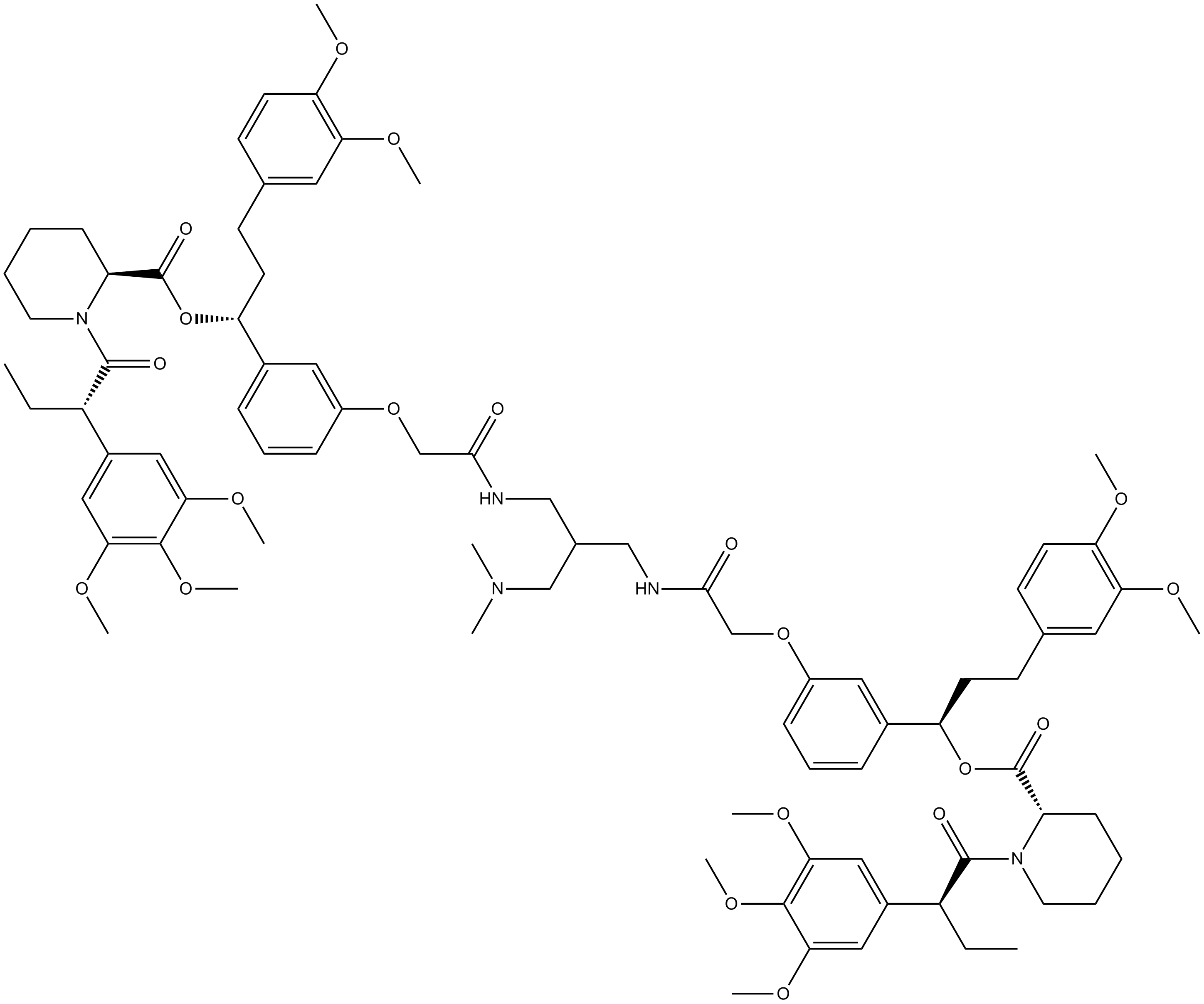
-
GC42821
AP219
AP39 is a compound used to increase the levels of hydrogen sulfide (H2S) within mitochondria.
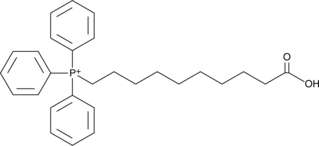
-
GC42823
AP39
AP39 is a compound used to increase the levels of hydrogen sulfide (H2S) within mitochondria.
-
GC52427
Apelin-12 (human, mouse, rat, bovine) (acetate)
An endogenous agonist of the APJ receptor

-
GC42825
APF
APF is an aromatic amino-fluorescein derivative and fluorescent probe for highly reactive radicals.
-
GN10509
Apigenin-7-O-β-D-glucopyranoside
-
GC46862
Apigenin-d5
An internal standard for the quantification of apigenin
-
GC42826
Apigeninidin (chloride)
Apigeninidin is a natural 3-deoxyanthocyanidin that can be isolated from leaves of sorghum.
-
GC35373
Apilimod
Apilimod (STA 5326) is a potent IL-12/IL-23 inhibitor, and strongly inhibits IL-12 with IC50s of 1 nM and 2 nM, in IFN-γ/SAC-stimulated human PBMCs and SAC-treated monkey PBMCs, respectively.
-
GC35374
Apilimod mesylate
Apilimod (STA 5326) mesylate is a potent IL-12/IL-23 inhibitor, and strongly inhibits IL-12 with IC50s of 1 nM and 2 nM, in IFN-γ/SAC-stimulated human PBMCs and SAC-treated monkey PBMCs, respectively.
-
GC65004
Apostatin-1
Apostatin-1 (Apt-1) is a potent TRADD inhibitor.
-
GC48988
Apramycin (sulfate hydrate)
An aminoglyco
side antibiotic 
-
GC46867
Apremilast-d5
An internal standard for the quantification of apremilast
-
GC49776
Apricitabine
-
GC31661
APY0201
APY0201 is a potent PIKfyve inhibitor, which inhibits the conversion of PtdIns3P to PtdIns(3,5)P2 in the presence of in the presence of [33P]ATP with an IC50 of 5.2 nM.
-
GC48393
Aquastatin A
A fungal metabolite with diverse biological activities
-
GC14231
AR-C 102222
iNOS inhibitor
-
GC38735
AR-C102222 hydrochloride
AR-C102222 hydrochloride is a potent, competitive, orally active and highly selective inducible nitric oxide synthase (iNOS) inhibitor, with an IC50 of 37 nM.
-
GC42832
Arachidic Acid (sodium salt)
Arachidic acid is a long-chain saturated fatty acid that has been found in peanut butter and anaerobic fungi.
-
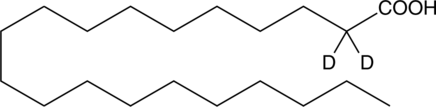
GC45384
Arachidic Acid-d2
-
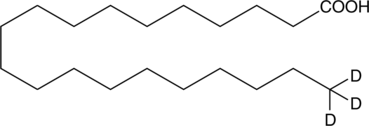
GC46869
Arachidic Acid-d3
An internal standard for the quantification of arachidic acid
-
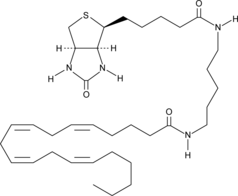
GC42837
Arachidonic Acid-biotin
Virtually all cellular arachidonic acid is esterified in membrane phospholipids where its presence is tightly regulated through multiple interconnected pathways.
-
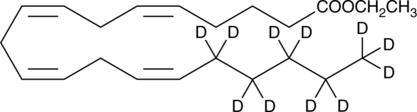
GC52514
Arachidonic Acid-d11 ethyl ester
An internal standard for the quantification of arachidonic acid ethyl ester
-
GC46872
Arachidonic Acid-d5
An internal standard for the quantification of arachidonic acid
-
GC46878
Aranciamycin
A fungal metabolite with diverse biological activities
-
GC48472
Aranciamycin A
An antibiotic
-
GC40116
Aranorosin
Aranorosin is a fungal metabolite originally isolated from P.
-
GC69582
ARC186
ARC 186 is a nucleic acid adapter that serves as an efficient complement inhibitor by blocking the activation of C5 catalyzed by convertase.

-
GC65163
Ardisiacrispin B
Ardisiacrispin B displays cytotoxic effects in multi-factorial drug resistant cancer cells via ferroptotic and apoptotic cell death.
-
GC35385
Arglabin
A sesquiterpene lactone
-
GC52332
Arimoclomol
A co-inducer of heat shock proteins
-
GN10579
Aristolochic Acid A
-
GC46005
Arjunolic Acid
A triterpene with diverse biological activities
-
GC14802
ARL 17477 dihydrochloride
Selective nNOS inhibitor
-
GC35394
Armepavine
Armepavine, an active compound from Nelumbo nucifera, exerts not only anti-inflammatory effects on human peripheral blood mononuclear cells, but also immunosuppressive effects on T lymphocytes and on lupus nephritic mice.
-
GC61796
Armillarisin A
Armillarisin A has the potential for the ulcerative colitis (UC) study.
-
GC33070
ARN-3236
An inhibitor of SIK2
-
GC49103
Aromadendrene
A sesquiterpene with diverse biological activities

-
GC46881
Artemether-d3
An internal standard for the quantification of artemether
-
GC46882
Artemisinin-d3
An internal standard for the quantification of artemisinin
-
GC45790
Artesunate-d4
An internal standard for the quantification of artesunate
-
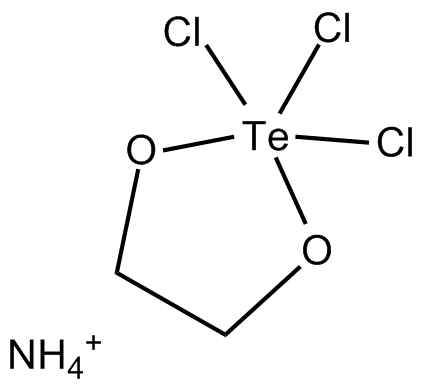
GC17659
AS 101
AS 101 (AS101), an immunomodulatory tellurium compound, is a potent IL-1β inhibitor.
-
GC46883
AS-2077715
An inhibitor of fungal complex III
-
GC35401
Asatone
Asatone is an active component isolated from Radix et Rhizoma Asari, with anti-inflammatory effect via activation of NF-κB and donwn regulation of p-MAPK (ERK, JNK and p38) pathways.
-
GC12070
Ascorbic acid
An electron donor
-
GN10534
Asiaticoside
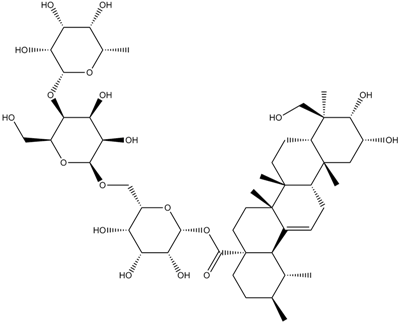
-
GC18978
Aspartocin D
Aspartocin D is a lipopeptide antibiotic originally isolated from S.
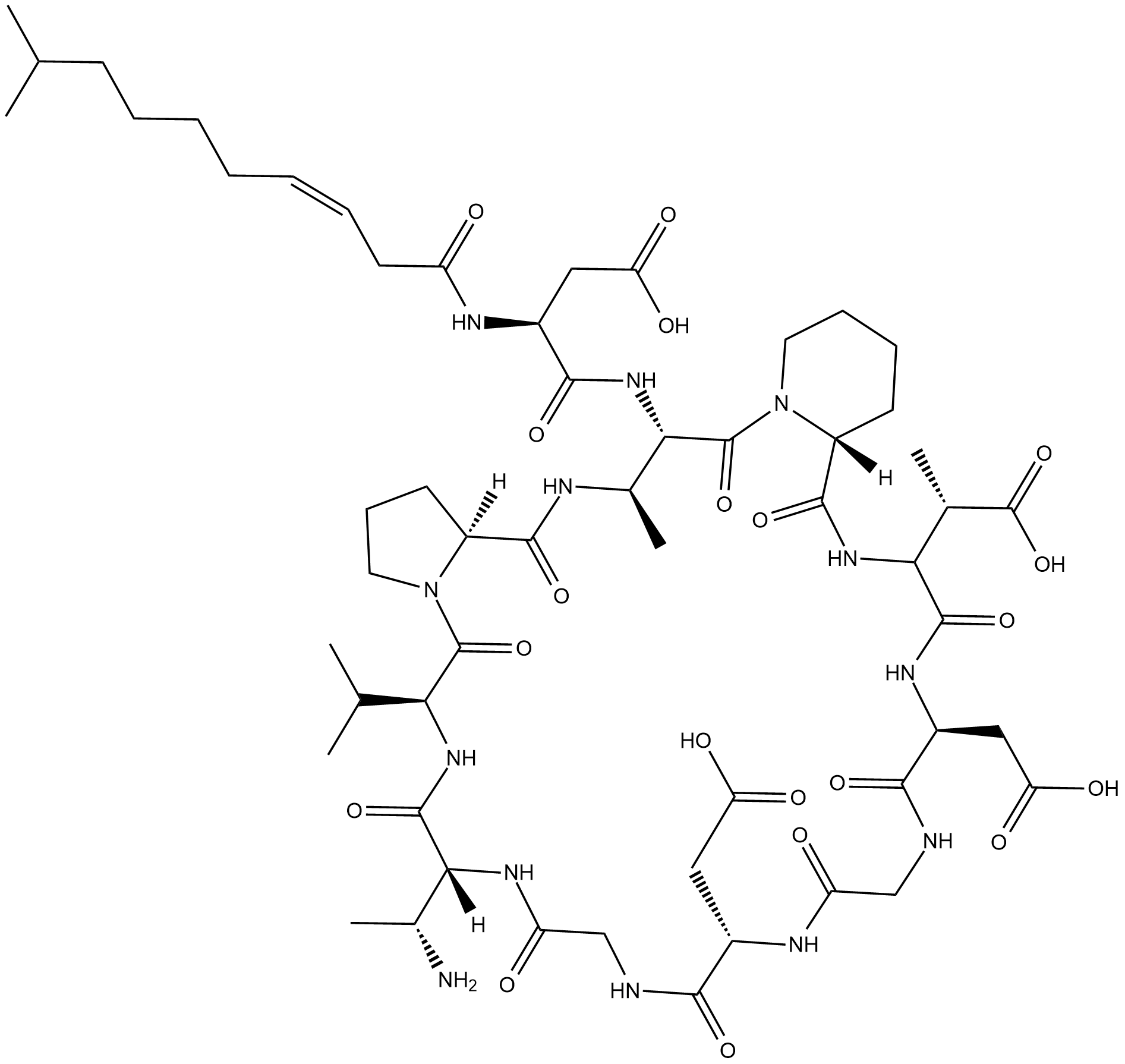
-
GC46089
Asperfuran
A fungal metabolite
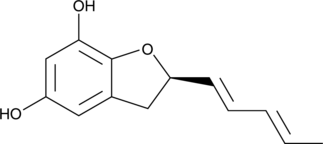
-
GC42858
Aspergillin PZ
Aspergillin PZ is a fungal metabolite originally isolated from A.
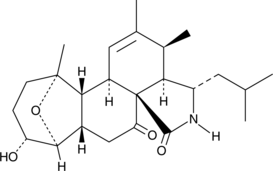
-
GC40682
Asperlactone
Asperlactone is a nematicidal, insecticidal, antibacterial, and antifungal polyketide metabolite produced from A.
-
GC35411
Asperuloside
An iridoid glycoside with diverse biological activities