Immunology/Inflammation
The immune and inflammation-related pathway including the Toll-like receptors pathway, the B cell receptor signaling pathway, the T cell receptor signaling pathway, etc.
Toll-like receptors (TLRs) play a central role in host cell recognition and responses to microbial pathogens. TLR4 initially recruits TIRAP and MyD88. MyD88 then recruits IRAKs, TRAF6, and the TAK1 complex, leading to early-stage activation of NF-κB and MAP kinases [1]. TLR4 is endocytosed and delivered to intracellular vesicles and forms a complex with TRAM and TRIF, which then recruits TRAF3 and the protein kinases TBK1 and IKKi. TBK1 and IKKi catalyze the phosphorylation of IRF3, leading to the expression of type I IFN [2].
BCR signaling is initiated through ligation of mIg under conditions that induce phosphorylation of the ITAMs in CD79, leading to the activation of Syk. Once Syk is activated, the BCR signal is transmitted via a series of proteins associated with the adaptor protein B-cell linker (Blnk, SLP-65). Blnk binds CD79a via non-ITAM tyrosines and is phosphorylated by Syk. Phospho-Blnk acts as a scaffold for the assembly of the other components, including Bruton’s tyrosine kinase (Btk), Vav 1, and phospholipase C-gamma 2 (PLCγ2) [3]. Following the assembly of the BCR-signalosome, GRB2 binds and activates the Ras-guanine exchange factor SOS, which in turn activates the small GTPase RAS. The original RAS signal is transmitted and amplified through the mitogen-activated protein kinase (MAPK) pathway, which including the serine/threonine-specific protein kinase RAF followed by MEK and extracellular signal related kinases ERK 1 and 2 [4]. After stimulation of BCR, CD19 is phosphorylated by Lyn. Phosphorylated CD19 activates PI3K by binding to the p85 subunit of PI3K and produce phosphatidylinositol-3,4,5-trisphosphate (PIP3) from PIP2, and PIP3 transmits signals downstream [5].
Central process of T cells responding to specific antigens is the binding of the T-cell receptor (TCR) to specific peptides bound to the major histocompatibility complex which expressed on antigen-presenting cells (APCs). Once TCR connected with its ligand, the ζ-chain–associated protein kinase 70 molecules (Zap-70) are recruited to the TCR-CD3 site and activated, resulting in an initiation of several signaling cascades. Once stimulation, Zap-70 forms complexes with several molecules including SLP-76; and a sequential protein kinase cascade is initiated, consisting of MAP kinase kinase kinase (MAP3K), MAP kinase kinase (MAPKK), and MAP kinase (MAPK) [6]. Two MAPK kinases, MKK4 and MKK7, have been reported to be the primary activators of JNK. MKK3, MKK4, and MKK6 are activators of P38 MAP kinase [7]. MAP kinase pathways are major pathways induced by TCR stimulation, and they play a key role in T-cell responses.
Phosphoinositide 3-kinase (PI3K) binds to the cytosolic domain of CD28, leading to conversion of PIP2 to PIP3, activation of PKB (Akt) and phosphoinositide-dependent kinase 1 (PDK1), and subsequent signaling transduction [8].
References
[1] Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors[J]. Nature immunology, 2010, 11(5): 373-384.
[2] Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-650.
[3] Packard T A, Cambier J C. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation[J]. F1000Prime Rep, 2013, 5(40.10): 12703.
[4] Zhong Y, Byrd J C, Dubovsky J A. The B-cell receptor pathway: a critical component of healthy and malignant immune biology[C]//Seminars in hematology. WB Saunders, 2014, 51(3): 206-218.
[5] Baba Y, Matsumoto M, Kurosaki T. Calcium signaling in B cells: regulation of cytosolic Ca 2+ increase and its sensor molecules, STIM1 and STIM2[J]. Molecular immunology, 2014, 62(2): 339-343.
[6] Adachi K, Davis M M. T-cell receptor ligation induces distinct signaling pathways in naive vs. antigen-experienced T cells[J]. Proceedings of the National Academy of Sciences, 2011, 108(4): 1549-1554.
[7] Rincón M, Flavell R A, Davis R A. The Jnk and P38 MAP kinase signaling pathways in T cell–mediated immune responses[J]. Free Radical Biology and Medicine, 2000, 28(9): 1328-1337.
[8] Bashour K T, Gondarenko A, Chen H, et al. CD28 and CD3 have complementary roles in T-cell traction forces[J]. Proceedings of the National Academy of Sciences, 2014, 111(6): 2241-2246.
Targets for Immunology/Inflammation
- Cyclic GMP-AMP Synthase(1)
- Apoptosis(137)
- 5-Lipoxygenase(18)
- TLR(106)
- Papain(2)
- PGDS(1)
- PGE synthase(26)
- SIKs(10)
- IκB/IKK(83)
- AP-1(2)
- KEAP1-Nrf2(47)
- NOD1(1)
- NF-κB(265)
- Interleukin Related(129)
- 15-lipoxygenase(2)
- Others(10)
- Aryl Hydrocarbon Receptor(35)
- CD73(16)
- Complement System(46)
- Galectin(30)
- IFNAR(19)
- NO Synthase(78)
- NOD-like Receptor (NLR)(37)
- STING(84)
- Reactive Oxygen Species(434)
- FKBP(14)
- eNOS(4)
- iNOS(24)
- nNOS(21)
- Glutathione(37)
- Adaptive Immunity(144)
- Allergy(129)
- Arthritis(25)
- Autoimmunity(134)
- Gastric Disease(64)
- Immunosuppressants(27)
- Immunotherapeutics(3)
- Innate Immunity(411)
- Pulmonary Diseases(76)
- Reactive Nitrogen Species(43)
- Specialized Pro-Resolving Mediators(42)
- Reactive Sulfur Species(24)
Products for Immunology/Inflammation
- Cat.No. Product Name Information
-
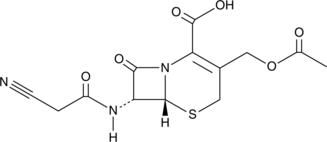
GC10692
Caffeic Acid Phenethyl Ester
NF-κB activation inhibitor
-
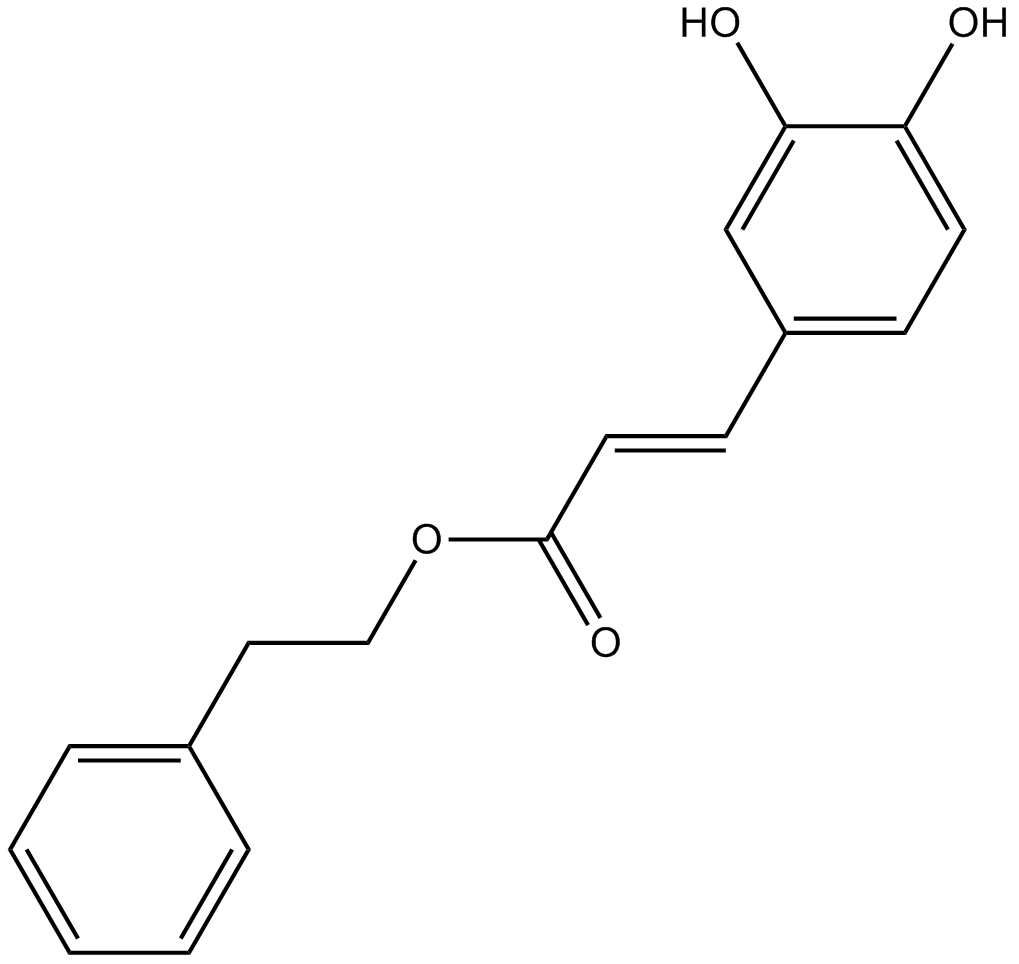
GC47020
Calcium D-Glucarate (hydrate)
The calcium salt form of D-glucaric acid
-
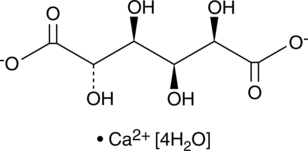
GC43129
CALP1 (trifluoroacetate salt)
CALP1 is an 8-residue calcium-like peptide that interacts with an EF hand motif based on the troponin C superfamily calcium binding site.
-
GC65081
CALP1 TFA
CALP1 TFA is a calmodulin (CaM) agonist (Kd of 88 ?M) with binding to the CaM EF-hand/Ca2+-binding site.
-
GC43131
Calpinactam
Calpinactam is a fungal metabolite originally isolated from M.
-
GN10356
Calycosin-7-glucoside
-
GC18922
Camalexin
Camalexin is an alkaloid released by plants of the Brassicaceae family in response to pathogen infection.
-
GC43135
Cambendazol
Cambendazol is one of the most effective agents for the therapy of human strongyloidiasis and .
-
GC43136
Campestanol
Campestanol is a phytosterol found in vegetables, fruits, nuts, and seeds.
-
GC66328
Canakinumab
Canakinumab (ACZ885) is a recombinant human anti-IL-1β monoclonal antibody. Canakinumab shows IC50 values of 43.6 and 40.8 pM for human and marmoset IL-1β, respectively. The mode of action of canakinumab is based on the neutralization of IL-1β signaling, resulting in suppression of inflammation related to disorders of autoimmune origin.

-
GC13816
Cannabidiol dimethyl ether
Pomalidomide - Peg2-azide is a synthetic E3 ligand -linker conjugate containing Pomalidomide based cereblon ligand and 2-unit PEG linker.
-
GC30495
Canthaxanthin (E 161g)
Canthaxanthin (E 161g) is a red-orange carotenoid with various biological activities, such as antioxidant, antitumor properties.
-
GC43138
CAP 3
CAP 3 is a cholic acid-peptide conjugate (CAP) with antibacterial activity.
-
GC65429
Capillarisin
Capillarisin, as a constituent from Artemisiae Capillaris herba, is found to exert anti-inflammatory and antioxidant properties.
-
GC49761
Capsaicin-d3
Capsaicin-d3 ((E)-Capsaicin-d3) is the deuterium labeled Capsaicin. Capsaicin ((E)-Capsaicin), an active component of chili peppers, is a TRPV1 agonist. Capsaicin has pain relief, antioxidant, anti-inflammatory, neuroprotection and anti-cancer effects.
-
GC43139
Capsanthin
Capsanthin is a carotenoid that has been found in red paprika and has diverse biological activities.
-
GC49433
Capsiate
A capsaicin analog with diverse biological activities
-
GC48878
Carbazomycin A
A bacterial metabolite with diverse biological activities
-
GC48893
Carbazomycin B
A bacterial metabolite with diverse biological activities
-
GC48850
Carbazomycin C
A bacterial metabolite with diverse biological activities
-
GC48826
Carbazomycin D
A bacterial metabolite with diverse biological activities
-
GC47037
Carbidopa (hydrate)
Carbidopa ((S)-(-)-Carbidopa) monohydrate, a peripheral decarboxylase inhibitor, can be used for the research of Parkinson's disease.
-
GC49197
Carbinoxamine-d6 (maleate)
An internal standard for the quantification of carbinoxamine
-
GC49147
Carboxyphosphamide
An inactive metabolite of cyclophosphamide
-
GC38059
Cardamonin
A chalconoid with anti-inflammatory and anti-tumor activity
-
GC18069
Cardamonin
Cardamonin ((E)-Cardamomin) is a novel antagonist of hTRPA1 cation channel with an IC50 of 454 nM.
-
GC43142
Cardanol diene
Cardanol diene is a phenol found in cashew nut shell liquid that inhibits tyrosinase with an IC50 value of 52.5 μM in vitro.
-
GC18449
Cardanol monoene
Cardanol monoene is a phenol found in cashew nut shell liquid that reversibly inhibits tyrosinase with an IC50 value of 56 uM in vitro.
-
GC43143
Cardanol triene
Cardanol triene is a phenol found in cashew nut shell liquid that reversibly inhibits tyrosinase with an IC50 value of 40.5 μM in vitro.
-
GC41394
Cardol diene
Cardol diene is a phenol found in cashew nut shell liquid.
-
GC41400
Cardol triene
Cardol triene is a phenol found in cashew nut shell liquid that competitively and irreversibly inhibits mushroom tyrosinase (IC50 = 22.5 μM).
-
GC47040
Carebastine-d5
An internal standard for the quantification of carebastine
-
GC46112
Carnaubadiol
An antiprotozoal triterpene
-
GC47044
Carotenoid Mixture
A mixture of carotenoids
-
GC64110
Carubicin hydrochloride
Carubicin hydrochloride is a microbially-derived compound.
-
GC47046
Carviolin
A fungal metabolite with immunosuppressive and antitrypanosomal activities
-
GC60674
Catalase from Aspergillus niger
Catalase is a key enzyme in the metabolism of H2O2 and reactive oxygen species (ROS), and its expression and localization is markedly altered in tumors.

-
GC64933
Catalase from bovine liver
Catalase is a key enzyme in the metabolism of H2O2 and reactive oxygen species (ROS), and its expression and localization is markedly altered in tumors[1].

-
GC18498
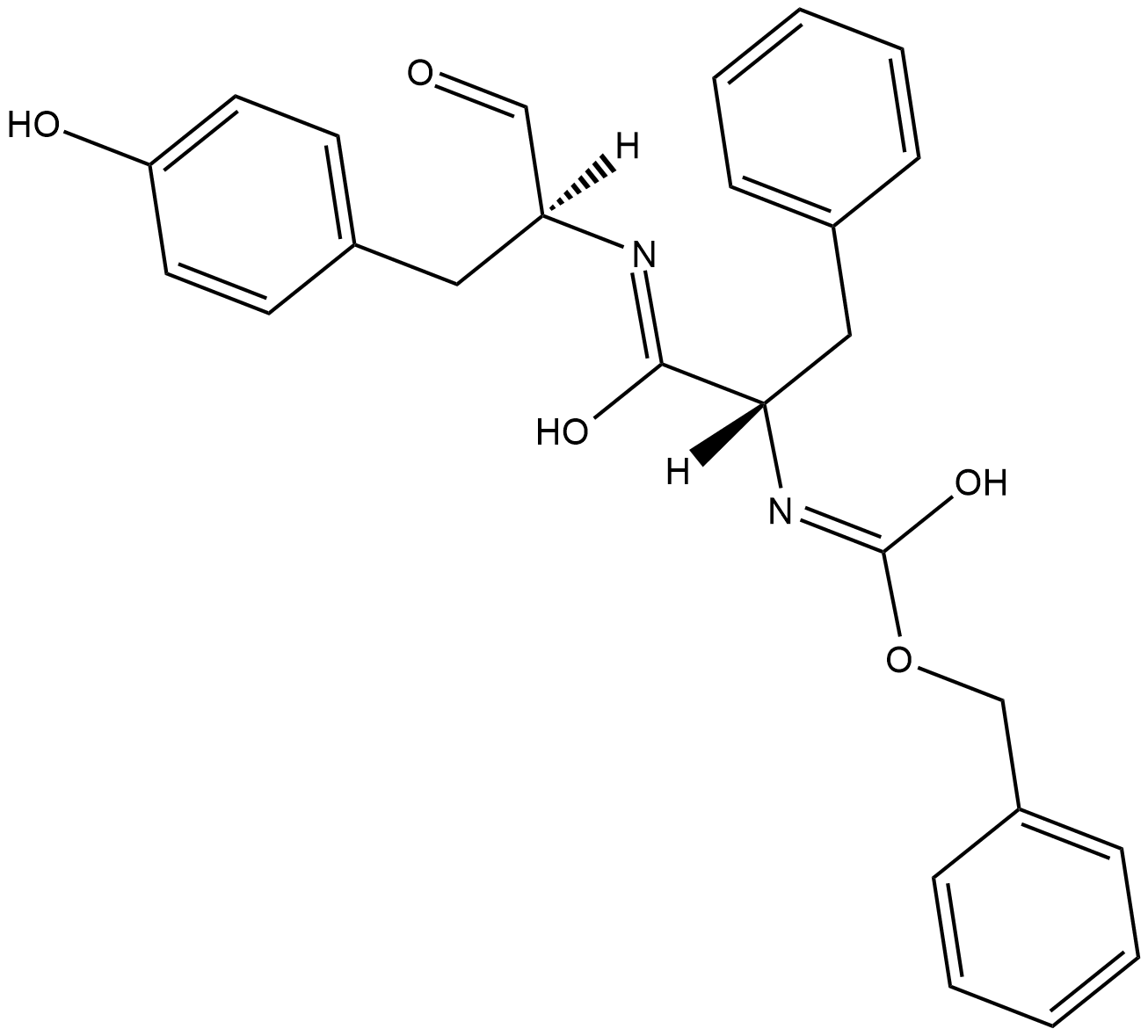
Cathepsin L Inhibitor
Cathepsin L Inhibitor (Z-Phe-Tyr-CHO) is a potent and specific cathepsin L (CTSL) inhibitor.
-
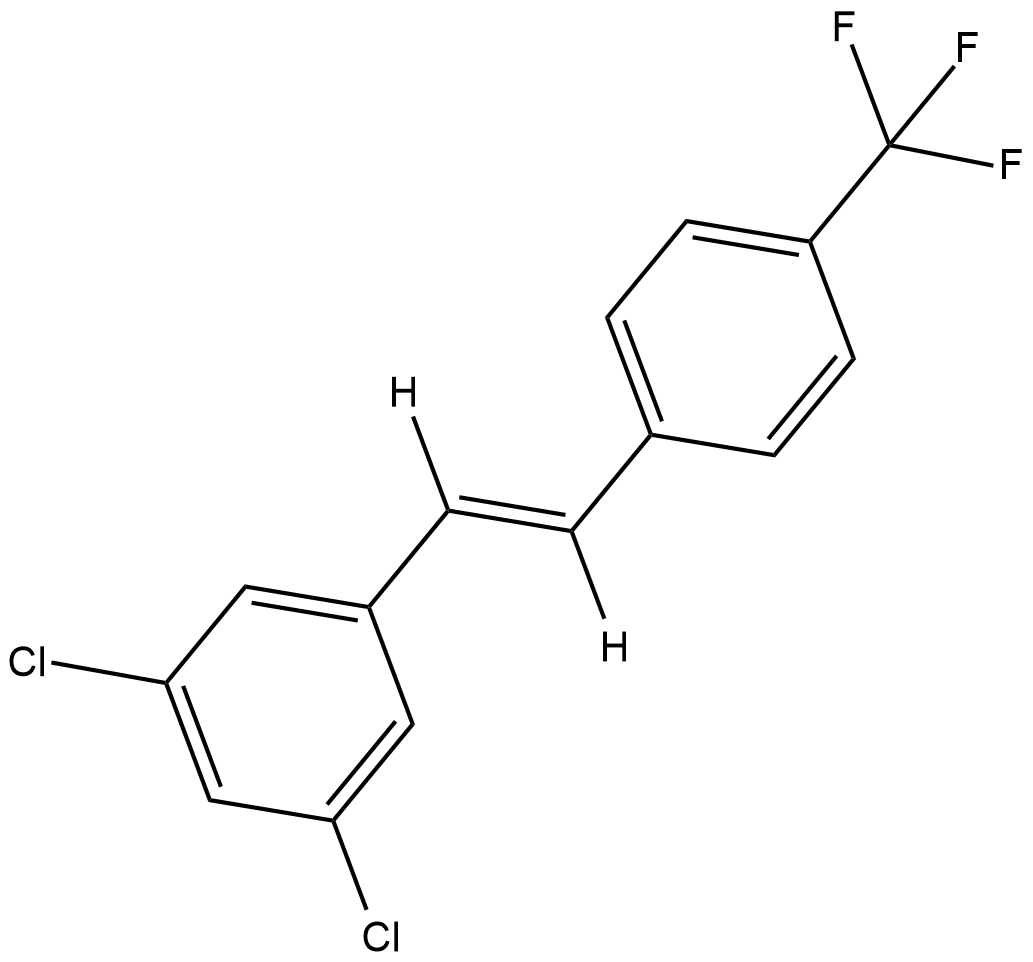
GC18824
CAY10465
The aryl hydrocarbon receptor (AhR) is a ligand-dependent intracellular transcription factor whose ligands include some of the most infamous xenobiotics, including dioxin, benzo[a]pyrene, and numerous polyaromatics from soot and coal tar.
-
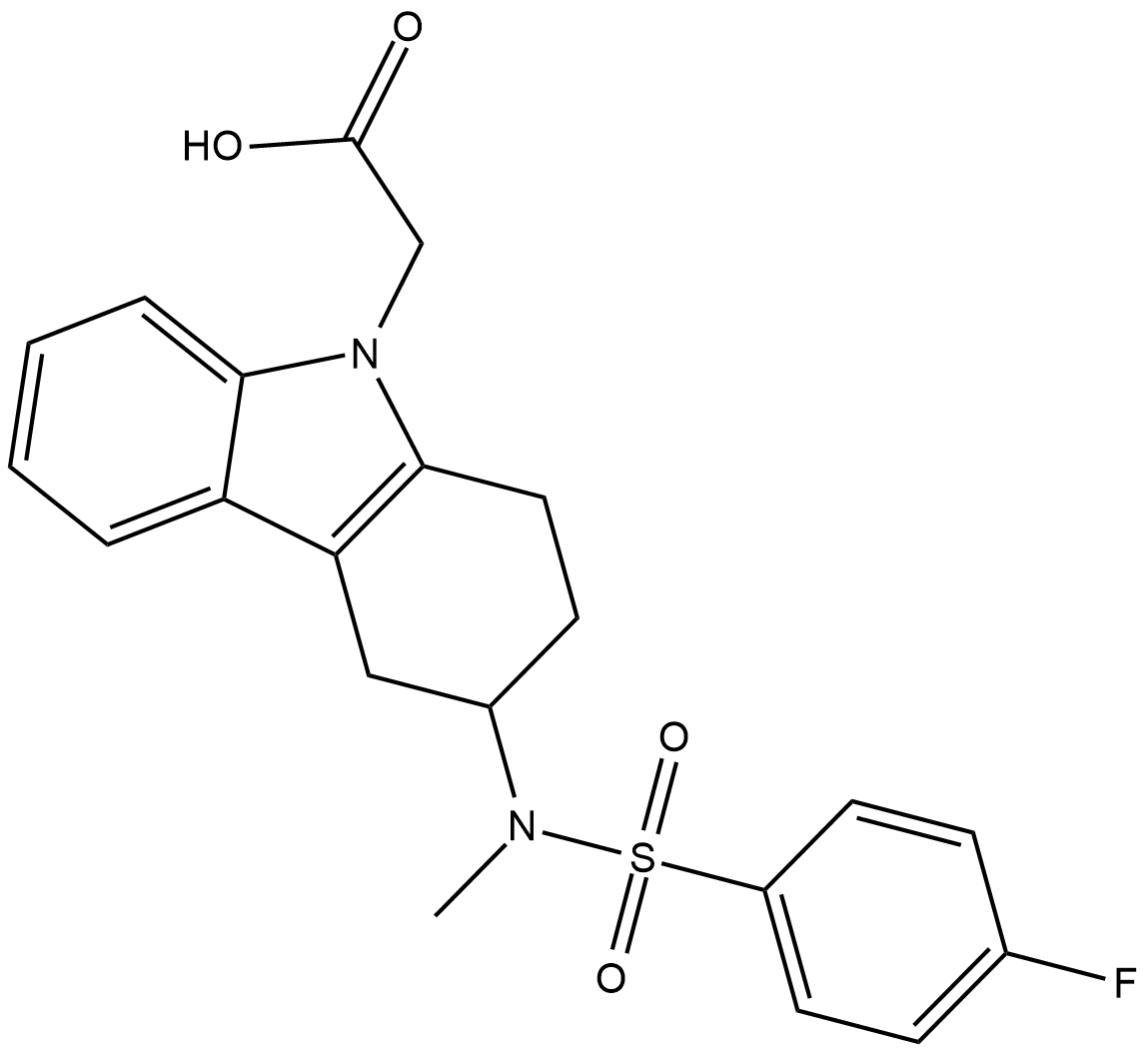
GC18811
CAY10471
The biological effects of prostaglandin D2 are transduced by at least two 7-transmembrane G-protein coupled receptors, designated DP1 and CRTH2/DP2.
-
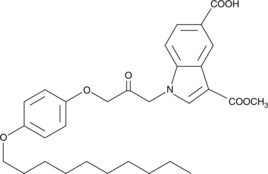
GC43161
CAY10502
Phospholipase A2 (PLA2) catalyzes the hydrolysis of phospholipids at the sn-2 position leading to the production of lysophospholipids and free fatty acids.
-
GC43165
CAY10512
The nuclear factor-κB (NF-κB) regulates the expression of numerous genes involved in immunity and inflammation, cellular stress responses, growth, and apoptosis.
-
GC18856
CAY10526
Prostaglandin E2 (PGE2) is the major PG synthesized at sites of inflammation and plays an important role in different inflammatory diseases.
-
GC43176
CAY10575
CAY10575 (Compound 8) is an IKK2 inhibitor with an IC50 of 0.075 μM.
-
GC41044
CAY10583
Leukotriene B4 (LTB4) promotes a number of leukocyte functions including aggregation, stimulation of ion fluxes, superoxide anion production, chemotaxis, and chemokinesis.
-
GC41601
CAY10591
CAY10591 is a potent activator of Sirt1 and suppresses TNF-α in a dose-dependent manner.
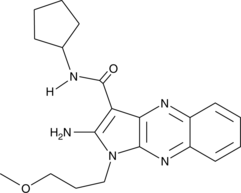
-
GC43181
CAY10597
The biological effects of prostaglandin D2 (PGD2) are transduced by at least two 7-transmembrane G protein-coupled receptors, designated DP1 and CRTH2/DP2.
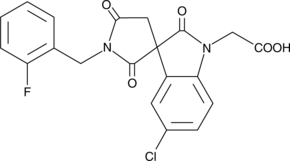
-
GC18877
CAY10606
5-Lipoxygenase (5-LO) initiates the synthesis of leukotrienes (LTs) from arachidonic acid, primarily in certain leukocyte populations.
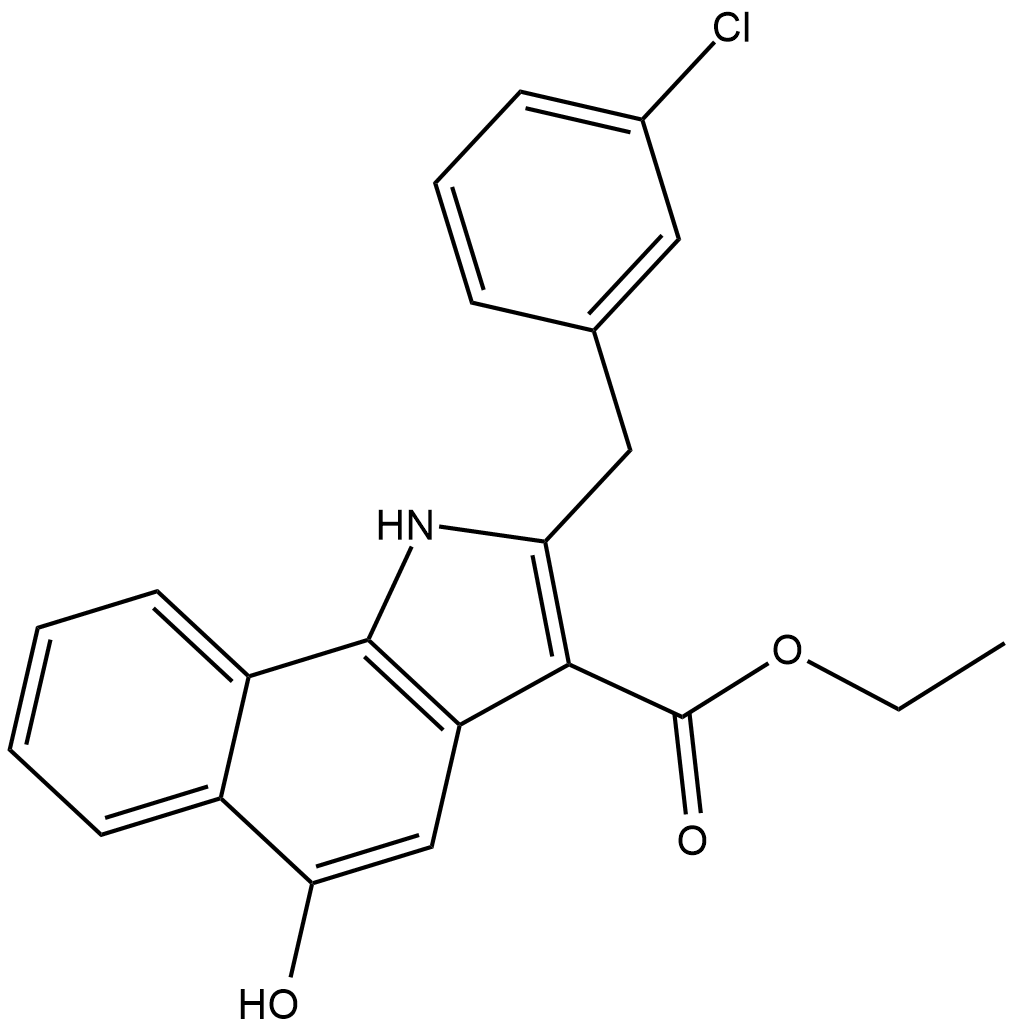
-
GC18883
CAY10614
Lipid A is a lipid-rich component of endotoxin, also known as lipopolysaccharide (LPS), which in turn is part of the outer membrane of Gram-negative bacteria.
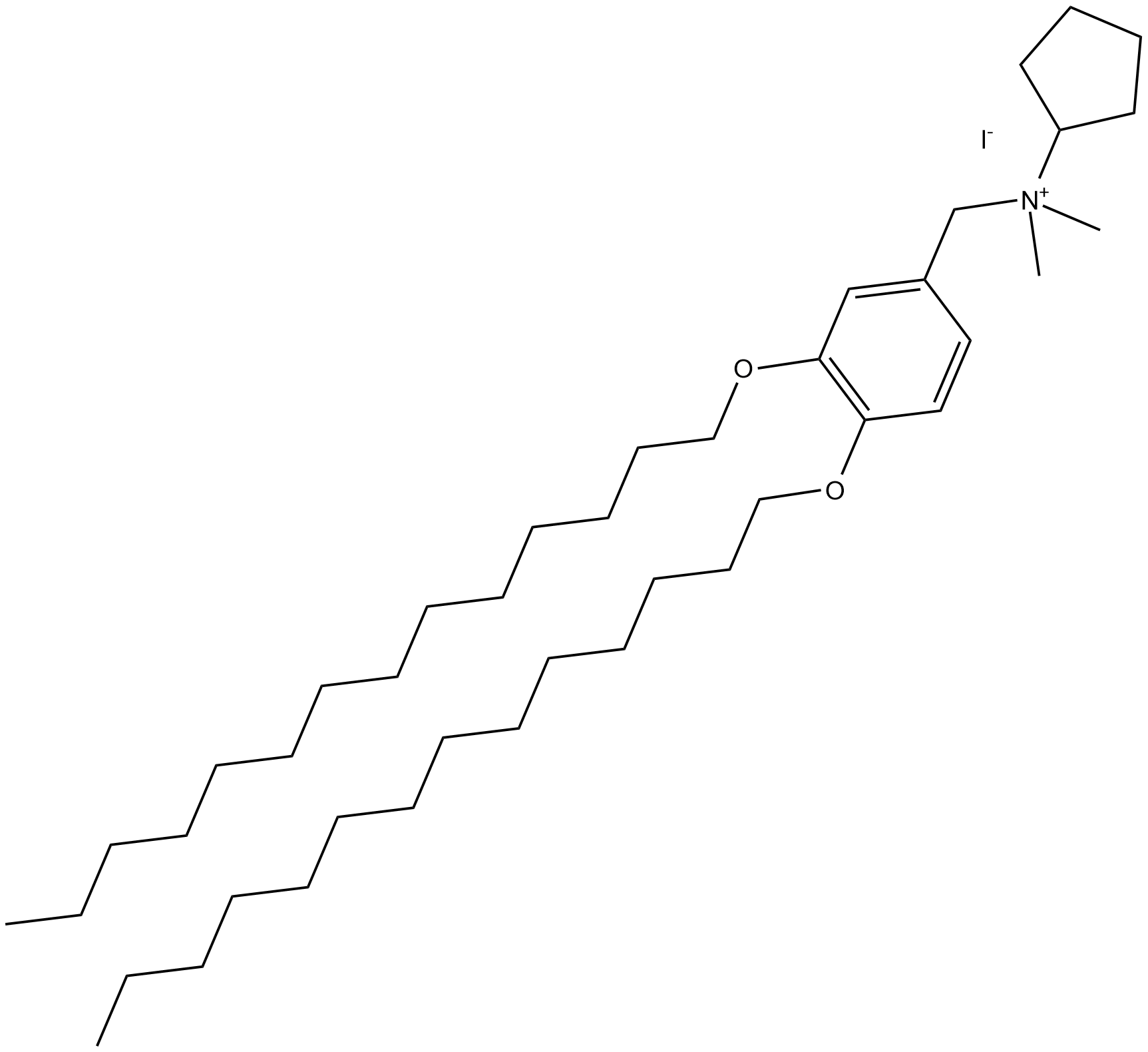
-
GC18887
CAY10640
CAY10640 (compound TCPU ) is a potent and oral active inhibitor of sEH (soluble epoxide hydrolase) with IC50s of 0.4 and 5.3 nM in human and murine, respectively.
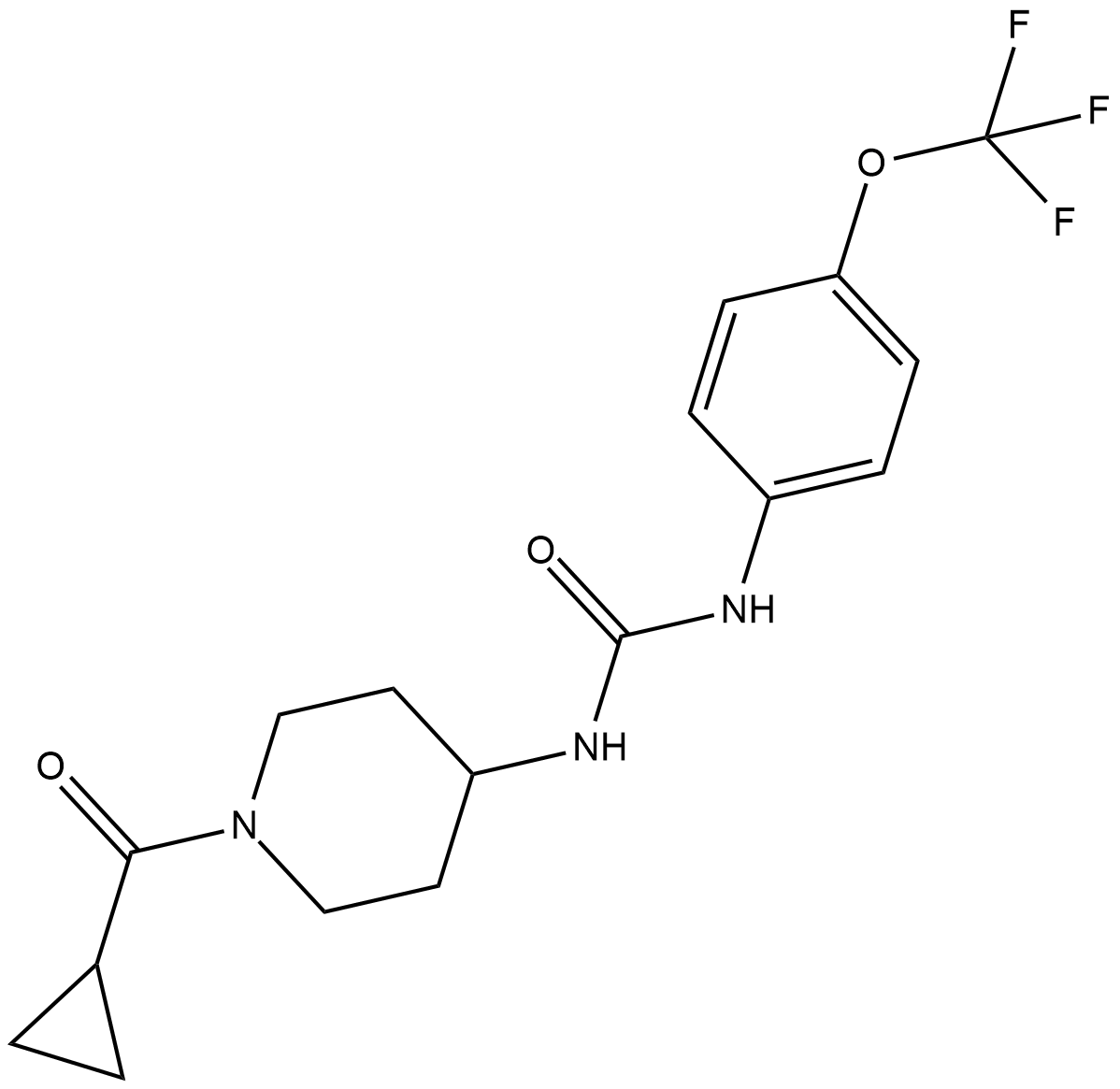
-
GC47049
CAY10641
An inactive alcohol derivative of a highly potent cPLA2 inhibitor
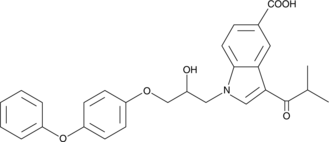
-
GC43186
CAY10648
Prostaglandin D2 (PGD2) evokes its effects through two receptors, DP1 and CRTH2/DP2.
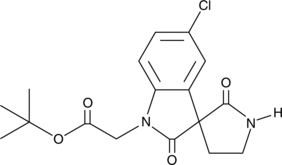
-
GC18910
CAY10654
The gram-negative pathogen Pseudomonas aeruginosa uses N-acylated-L-homoserine lactones (AHLs) in quorum sensing to modulate transcriptional activators, including LasR, and regulate the expression of virulence factors.
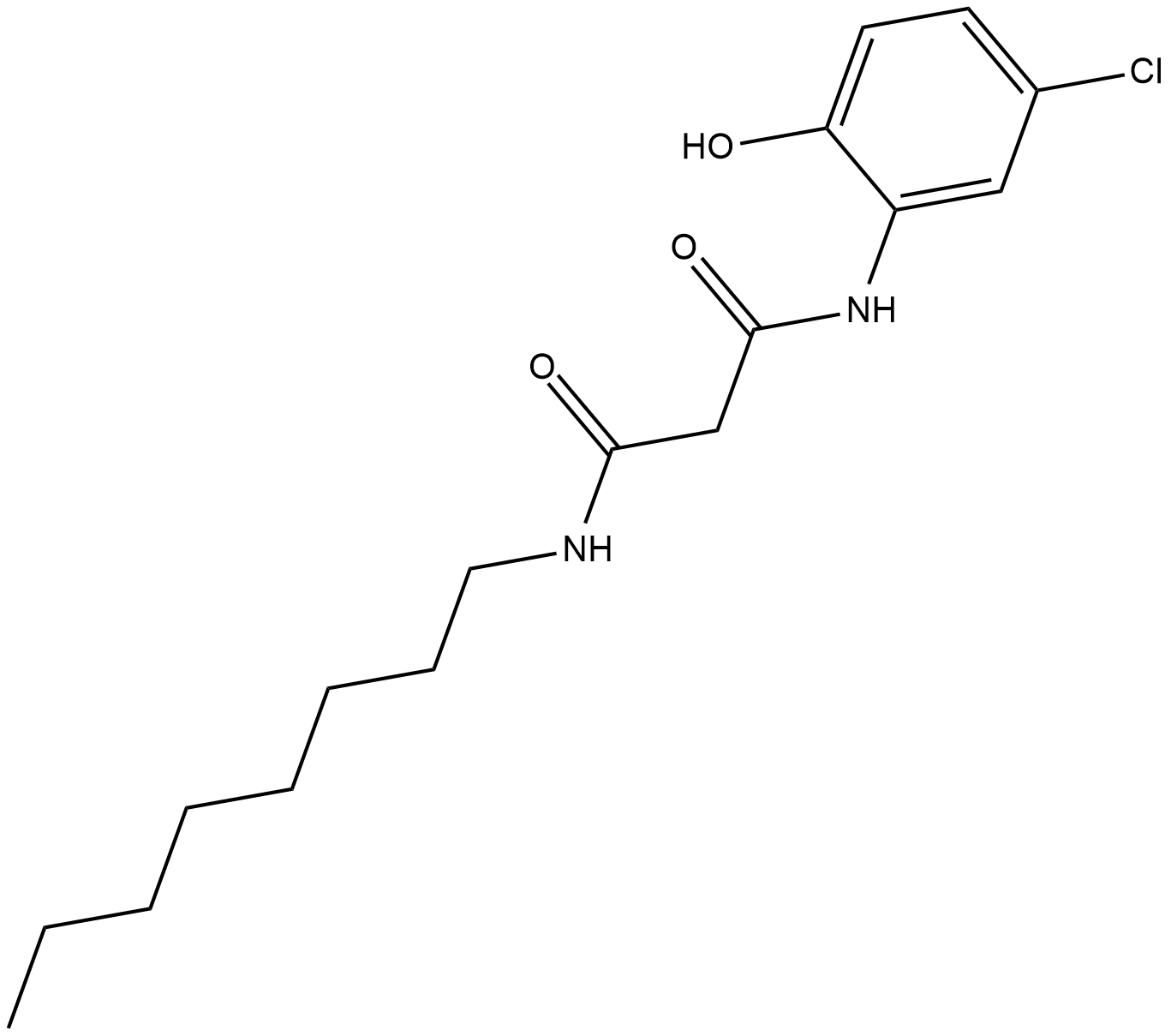
-
GC18782
CAY10657
The NF-κB/Rel family of transcription factors induce and coordinate the expression of pro-inflammatory genes including cytokines, chemokines, interferons, MHC proteins, growth factors, and cell adhesion molecules.
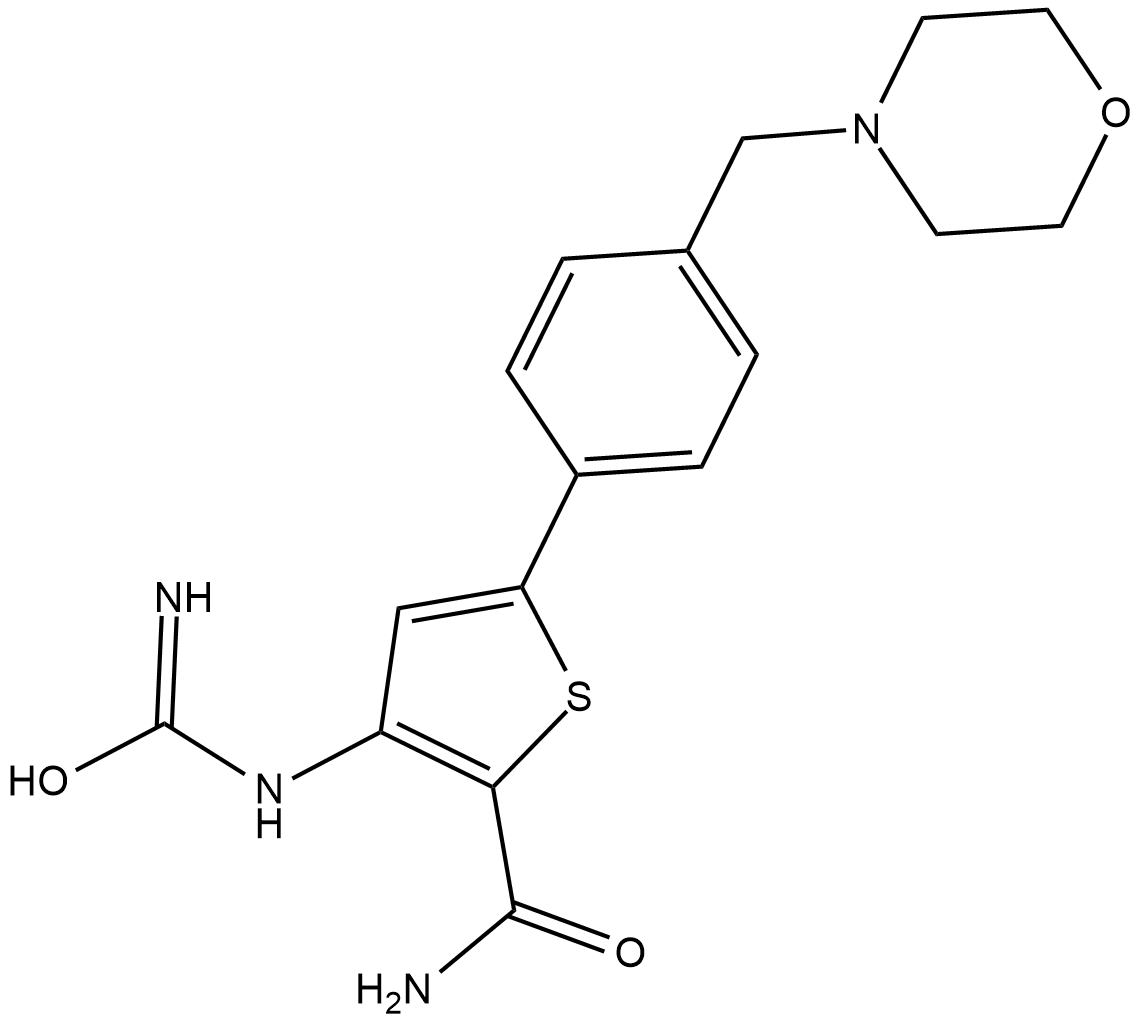
-
GC18904
CAY10678
Microsomal prostaglandin E synthase-1 (mPGES-1) converts the COX product PGH2 into the biologically active PGE2.
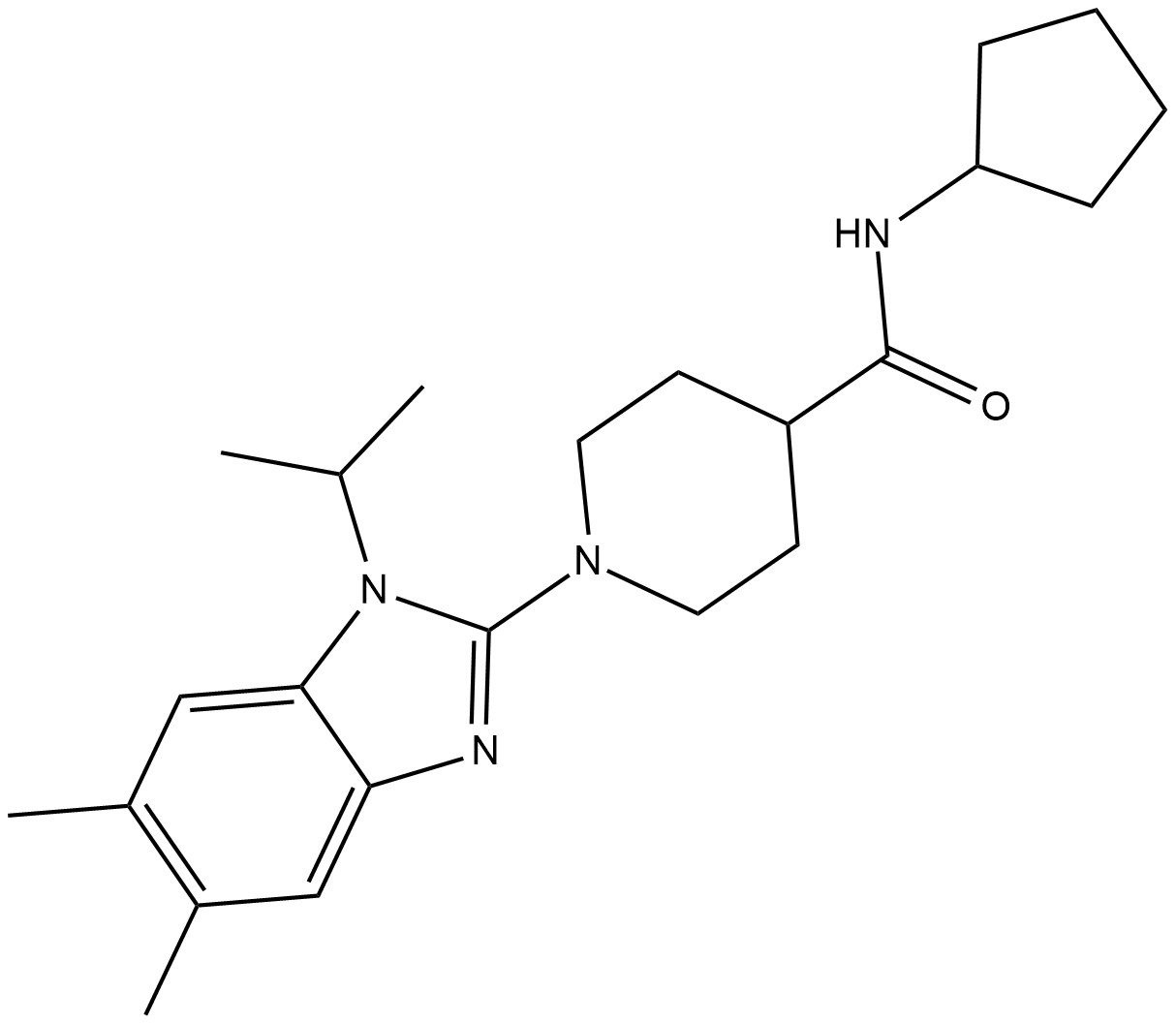
-
GC43189
CAY10681
Inactivation of the tumor suppressor p53 commonly coincides with increased signaling through NF-κB in cancer.
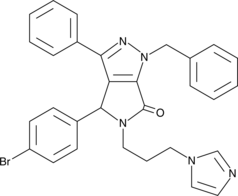
-
GC43190
CAY10682
(±)-Nutlin-3 blocks the interaction of p53 with its negative regulator Mdm2 (IC50 = 90 nM), inducing the expression of p53-regulated genes and blocking the growth of tumor xenografts in vivo.
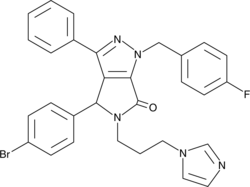
-
GC43196
CAY10704
CAY10704 is a potent inhibitor of hepatitis C virus (HCV) infection (EC50 = 17 nM) that displays low cytotoxicity of virally-infected human hepatoma Huh7.5.1 cells (CC50 = 21.3 μM).
-
GC43197
CAY10711
CAY10711 is a substituted diamine that produces rapid bactericidal activity against both Gram-positive and Gram-negative bacteria, including methicillin-resistant S.
-
GC43205
CAY10731
CAY10731 is a fluorescent probe for hydrogen sulfide (H2S).
-
GC40757
CAY10734
An S1P1 receptor agonist
-
GC47052
CAY10742
An oxadiazole antibiotic
-
GC47054
CAY10748
A STING agonist
-
GC47062
CAY10766
An antiviral compound
-
GC48427
CAY10774
A PD-1/PD-L1 interaction inhibitor
-
GC49139
CAY10784
A STAT3 inhibitor
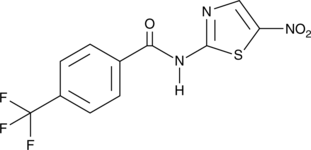
-
GC49758
CAY10788
A CysLT1 receptor antagonist
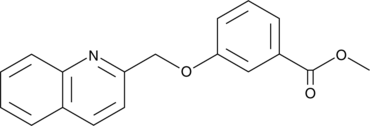
-
GC52307
CAY10790
A CysLT1 receptor antagonist and GPBAR1 agonist
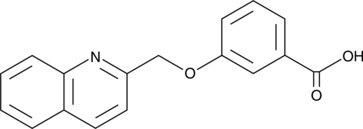
-
GC52245
CAY10792
An anticancer agent
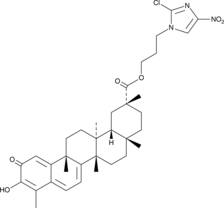
-
GC49664
CB-1158
An arginase inhibitor
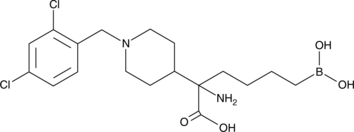
-
GC15394
CBL0137 (hydrochloride)
curaxin that activates p53 and inhibits NF-κB
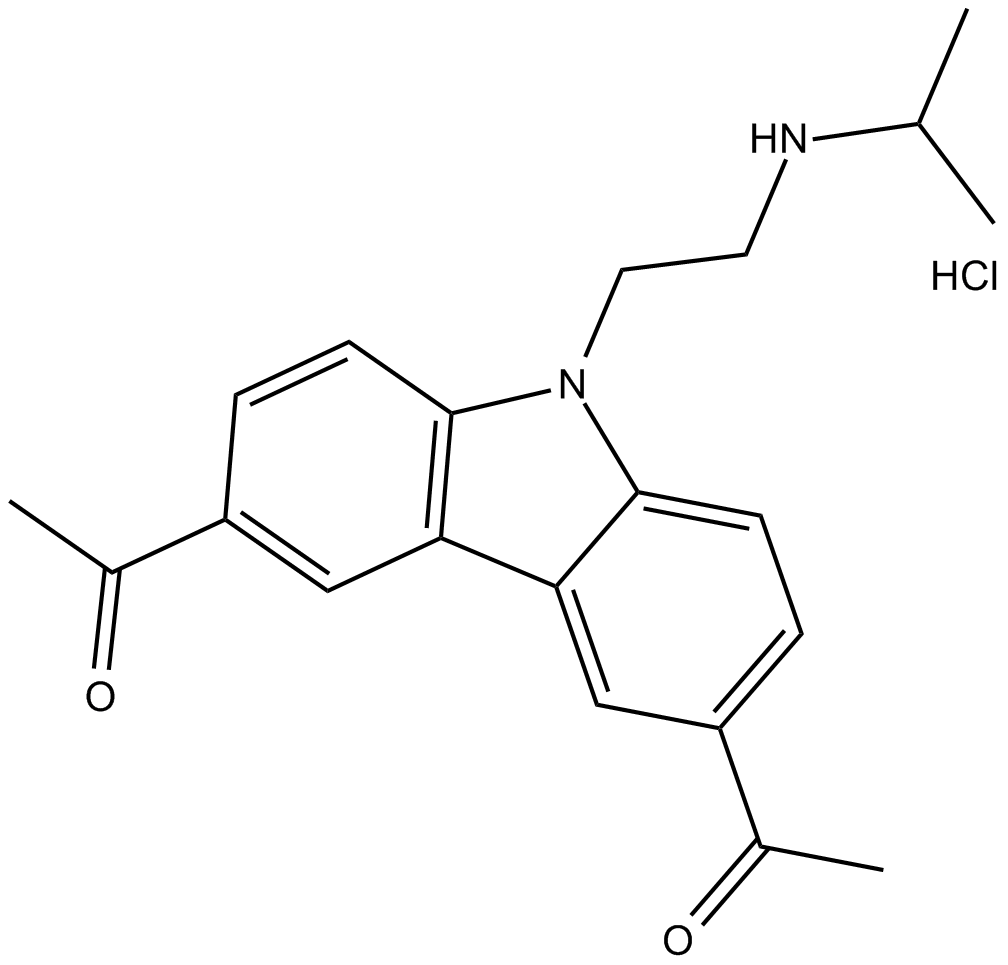
-
GC14727
CCCP
Carbonylcyanide-3-chlorophenylhydrazone (CCCP) is a protonophore, which causes uncoupling of proton gradient in the inner mitochondrial membrane, thus inhibiting the rate of ATP synthesis.
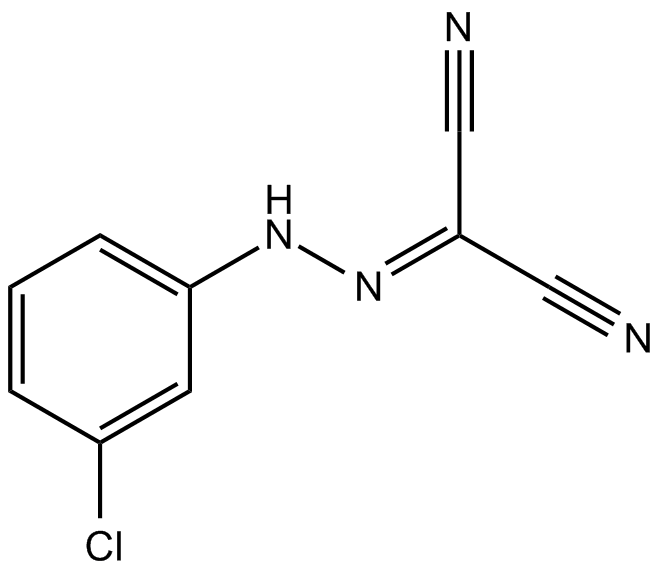
-
GC43212
CCG-232601
CCG-232601 is an inhibitor of the Rho/MRTF/SRF transcriptional pathway and a derivative of CCG-203971.
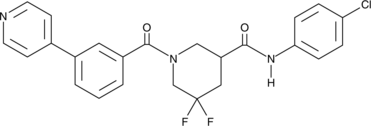
-
GC31914
CD73-IN-1
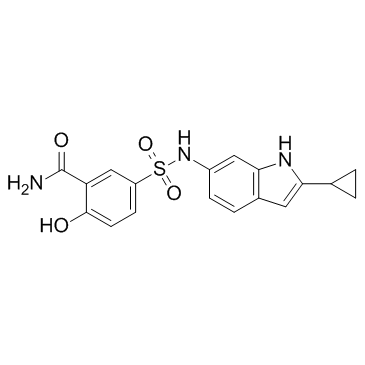
CD73-IN-1 is an inhibitor of CD73 which can be used in the treatment of cancer extracted from patent WO 2017153952 A1, example 80.
-
GC62331
CD73-IN-3
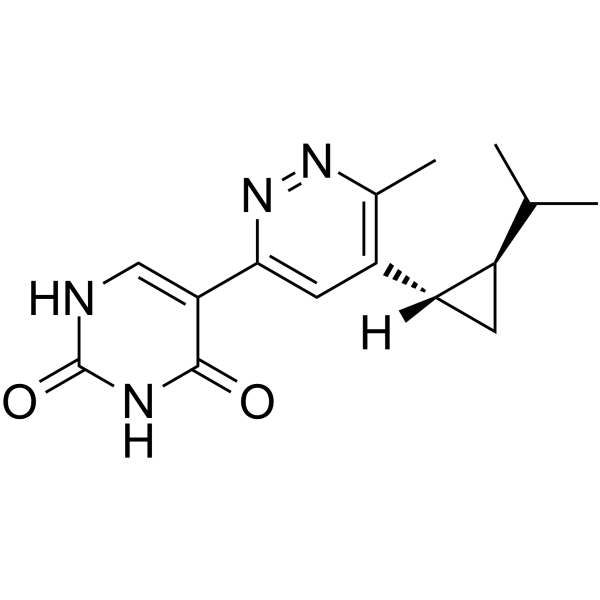
CD73-IN-3 is a potent CD73 inhibitor (IC50=7.3 nM in Calu6 human cell assay). CD73-IN-3, example 2 extracted from patent WO2019168744 A1, has the potential for cancer research.
-
GC62452
CD73-IN-4
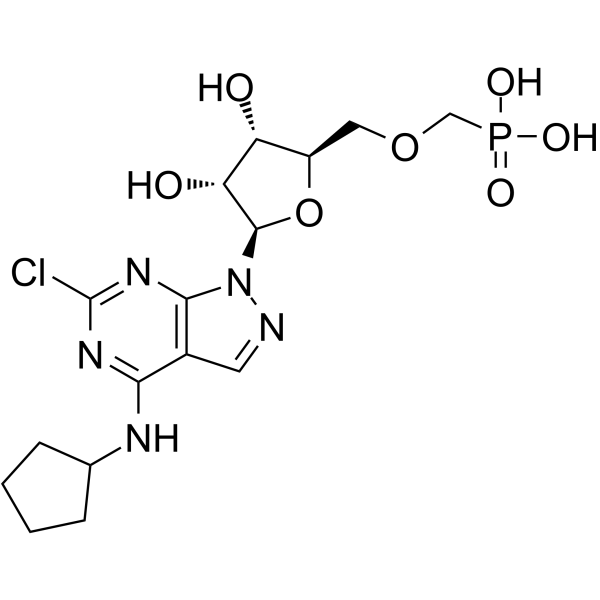
CD73-IN-4 is a potent and selective methylenephosphonic acid CD73 inhibitor, with an IC50 of 2.6 nM for human CD73.
-
GC68386
CD73-IN-5
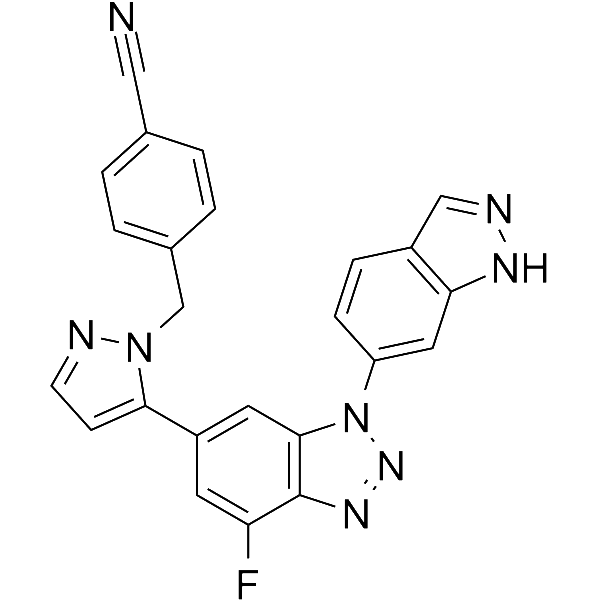
-
GC49584
CD74 Monoclonal Antibody (Clone PIN1)
For immunochemical analysis of CD74

-
GC35629
CDDO-dhTFEA
CDDO-dhTFEA (RTA dh404) is a synthetic oleanane triterpenoid compound which potently activates Nrf2 and inhibits the pro-inflammatory transcription factor NF-κB.
-
GC35630
CDDO-EA
CDDO-EA is an NF-E2 related factor 2/antioxidant response element (Nrf2/ARE) activator.

-
GC32723
CDDO-Im (RTA-403)
CDDO-Im (RTA-403) (RTA-403) is an activator of Nrf2 and PPAR, with Kis of 232 and 344 nM for PPARα and PPARγ.
-
GC61865
Cearoin
Cearoin increases autophagy and apoptosis through the production of ROS and the activation of ERK.
-
GC52378
Cecropin A (trifluoroacetate salt)
An antimicrobial peptide

-
GC52369
Cecropin B (trifluoroacetate salt)
An antimicrobial peptide

-
GC48489
Cefadroxil-d4 (trifluoroacetate salt)
An internal standard for the quantification of cefadroxil
-
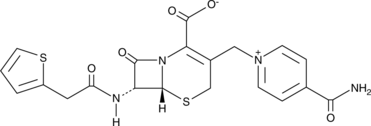
GC49274
Cefalonium
A cephalosporin antibiotic
-
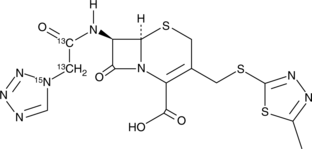
GC43222
Cefazolin-13C2,15N
Cefazolin-13C2,15N is intended for use as an internal standard for the quantification of cefazolin by GC- or LC-MS.
-
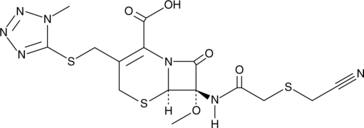
GC47068
Cefmetazole
A cephamycin antibiotic
-
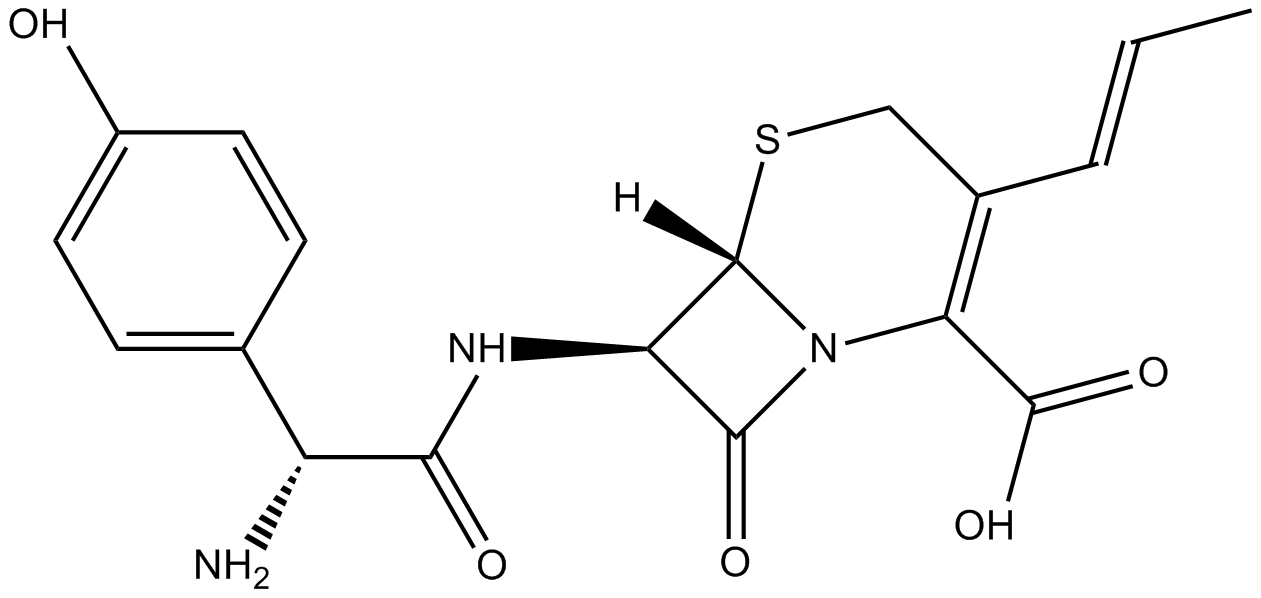
GC16155
Cefprozil
antibiotic
-
GC48443
Cefsulodin (sodium salt hydrate)
A βlactam antibiotic
-
GC47069
Cefuroxime-d3
An internal standard for the quantification of cefuroxime
-
GC15083
Celastrol
A triterpenoid antioxidant
-
GC49152
Celecoxib Carboxylic Acid
An inactive metabolite of celecoxib
-
GC43225
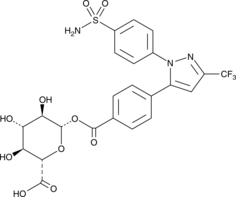
Celecoxib Carboxylic Acid Acyl-β-D-Glucuronide
Celecoxib carboxylic acid acyl-β-D-glucuronide is a phase II metabolite of celecoxib.
-
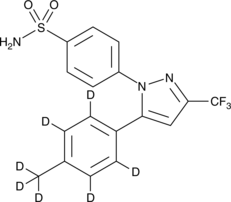
GC47070
Celecoxib-d7
An internal standard for the quantification of celecoxib
-
GC18392
Cellocidin
Cellocidin is an antibiotic originally isolated from S.
-
GC66410
Cemdisiran
Cemdisiran is an N-acetylgalactosamine (GalNAc) conjugated siRNA for the treatment of complement-mediated diseases by suppressing liver production of complement 5 (C5) protein.
-
GC47071
CENTA
A colorimetric β-lactamase substrate
-
GC49593
Cephacetrile
Cephacetrile (Cephacetrile) is a broad-spectrum antibiotic that is effective in gram-positive and gram-negative bacterial infection.