Immunology/Inflammation
The immune and inflammation-related pathway including the Toll-like receptors pathway, the B cell receptor signaling pathway, the T cell receptor signaling pathway, etc.
Toll-like receptors (TLRs) play a central role in host cell recognition and responses to microbial pathogens. TLR4 initially recruits TIRAP and MyD88. MyD88 then recruits IRAKs, TRAF6, and the TAK1 complex, leading to early-stage activation of NF-κB and MAP kinases [1]. TLR4 is endocytosed and delivered to intracellular vesicles and forms a complex with TRAM and TRIF, which then recruits TRAF3 and the protein kinases TBK1 and IKKi. TBK1 and IKKi catalyze the phosphorylation of IRF3, leading to the expression of type I IFN [2].
BCR signaling is initiated through ligation of mIg under conditions that induce phosphorylation of the ITAMs in CD79, leading to the activation of Syk. Once Syk is activated, the BCR signal is transmitted via a series of proteins associated with the adaptor protein B-cell linker (Blnk, SLP-65). Blnk binds CD79a via non-ITAM tyrosines and is phosphorylated by Syk. Phospho-Blnk acts as a scaffold for the assembly of the other components, including Bruton’s tyrosine kinase (Btk), Vav 1, and phospholipase C-gamma 2 (PLCγ2) [3]. Following the assembly of the BCR-signalosome, GRB2 binds and activates the Ras-guanine exchange factor SOS, which in turn activates the small GTPase RAS. The original RAS signal is transmitted and amplified through the mitogen-activated protein kinase (MAPK) pathway, which including the serine/threonine-specific protein kinase RAF followed by MEK and extracellular signal related kinases ERK 1 and 2 [4]. After stimulation of BCR, CD19 is phosphorylated by Lyn. Phosphorylated CD19 activates PI3K by binding to the p85 subunit of PI3K and produce phosphatidylinositol-3,4,5-trisphosphate (PIP3) from PIP2, and PIP3 transmits signals downstream [5].
Central process of T cells responding to specific antigens is the binding of the T-cell receptor (TCR) to specific peptides bound to the major histocompatibility complex which expressed on antigen-presenting cells (APCs). Once TCR connected with its ligand, the ζ-chain–associated protein kinase 70 molecules (Zap-70) are recruited to the TCR-CD3 site and activated, resulting in an initiation of several signaling cascades. Once stimulation, Zap-70 forms complexes with several molecules including SLP-76; and a sequential protein kinase cascade is initiated, consisting of MAP kinase kinase kinase (MAP3K), MAP kinase kinase (MAPKK), and MAP kinase (MAPK) [6]. Two MAPK kinases, MKK4 and MKK7, have been reported to be the primary activators of JNK. MKK3, MKK4, and MKK6 are activators of P38 MAP kinase [7]. MAP kinase pathways are major pathways induced by TCR stimulation, and they play a key role in T-cell responses.
Phosphoinositide 3-kinase (PI3K) binds to the cytosolic domain of CD28, leading to conversion of PIP2 to PIP3, activation of PKB (Akt) and phosphoinositide-dependent kinase 1 (PDK1), and subsequent signaling transduction [8].
References
[1] Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors[J]. Nature immunology, 2010, 11(5): 373-384.
[2] Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity[J]. Immunity, 2011, 34(5): 637-650.
[3] Packard T A, Cambier J C. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation[J]. F1000Prime Rep, 2013, 5(40.10): 12703.
[4] Zhong Y, Byrd J C, Dubovsky J A. The B-cell receptor pathway: a critical component of healthy and malignant immune biology[C]//Seminars in hematology. WB Saunders, 2014, 51(3): 206-218.
[5] Baba Y, Matsumoto M, Kurosaki T. Calcium signaling in B cells: regulation of cytosolic Ca 2+ increase and its sensor molecules, STIM1 and STIM2[J]. Molecular immunology, 2014, 62(2): 339-343.
[6] Adachi K, Davis M M. T-cell receptor ligation induces distinct signaling pathways in naive vs. antigen-experienced T cells[J]. Proceedings of the National Academy of Sciences, 2011, 108(4): 1549-1554.
[7] Rincón M, Flavell R A, Davis R A. The Jnk and P38 MAP kinase signaling pathways in T cell–mediated immune responses[J]. Free Radical Biology and Medicine, 2000, 28(9): 1328-1337.
[8] Bashour K T, Gondarenko A, Chen H, et al. CD28 and CD3 have complementary roles in T-cell traction forces[J]. Proceedings of the National Academy of Sciences, 2014, 111(6): 2241-2246.
Targets for Immunology/Inflammation
- Cyclic GMP-AMP Synthase(1)
- Apoptosis(137)
- 5-Lipoxygenase(18)
- TLR(106)
- Papain(2)
- PGDS(1)
- PGE synthase(26)
- SIKs(10)
- IκB/IKK(83)
- AP-1(2)
- KEAP1-Nrf2(47)
- NOD1(1)
- NF-κB(265)
- Interleukin Related(129)
- 15-lipoxygenase(2)
- Others(10)
- Aryl Hydrocarbon Receptor(35)
- CD73(16)
- Complement System(46)
- Galectin(30)
- IFNAR(19)
- NO Synthase(78)
- NOD-like Receptor (NLR)(37)
- STING(84)
- Reactive Oxygen Species(434)
- FKBP(14)
- eNOS(4)
- iNOS(24)
- nNOS(21)
- Glutathione(37)
- Adaptive Immunity(144)
- Allergy(129)
- Arthritis(25)
- Autoimmunity(134)
- Gastric Disease(64)
- Immunosuppressants(27)
- Immunotherapeutics(3)
- Innate Immunity(411)
- Pulmonary Diseases(76)
- Reactive Nitrogen Species(43)
- Specialized Pro-Resolving Mediators(42)
- Reactive Sulfur Species(24)
Products for Immunology/Inflammation
- Cat.No. Product Name Information
-
GC48458
Betulinic glycine amide
A derivative of betulinic acid
-
GC48520
Betulonaldehyde
A pentacyclic triterpenoid
-
GC46100
BHBM
An acylhydrazone with antifungal activity
-
GC18200
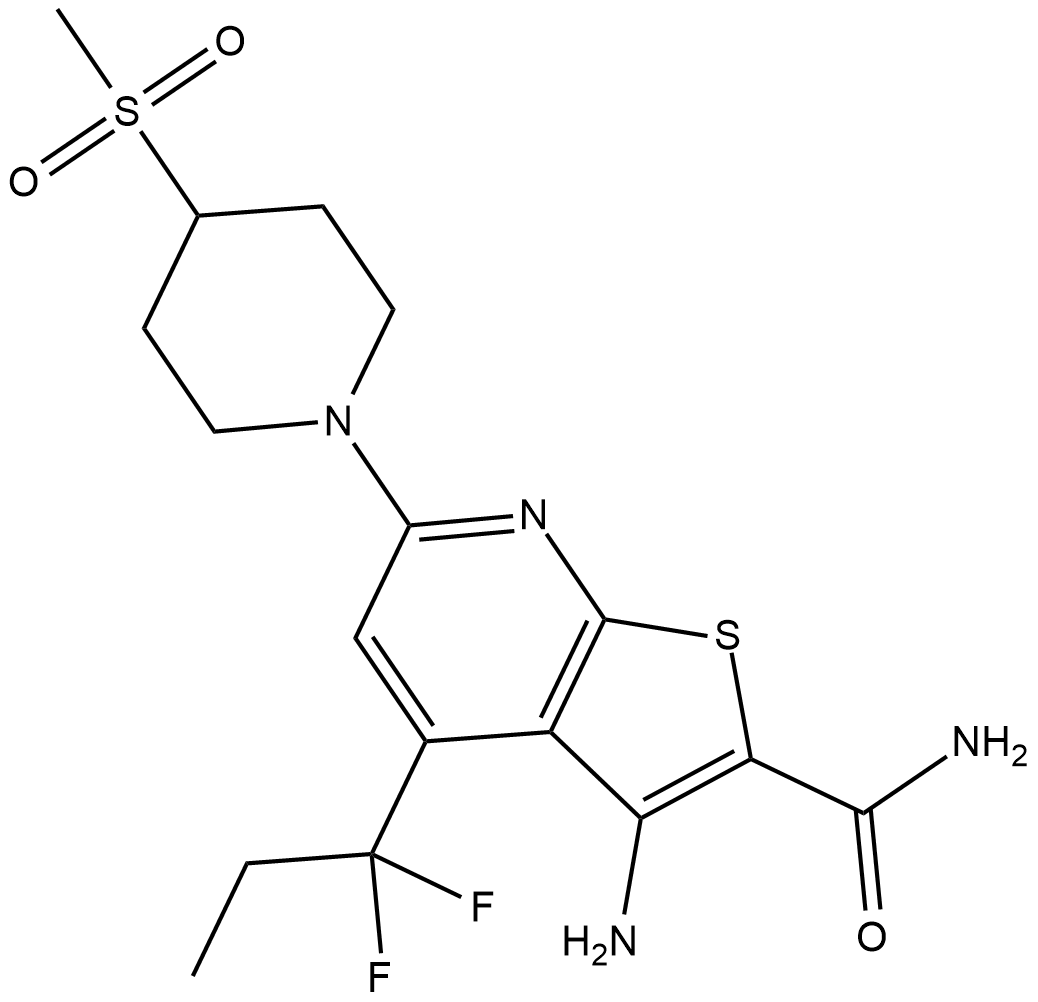
BI-605906
BI-605906 is an inhibitor of inhibitor of kappa B kinase subunit β (IKKβ; IC50 = 380 nM).
-
GC60076
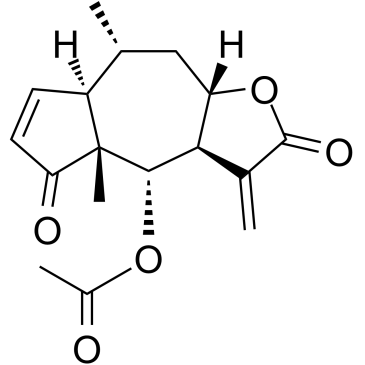
Bigelovin
Bigelovin, a sesquiterpene lactone isolated from Inula helianthus-aquatica, is a selective retinoid X receptor α agonist. Bigelovin suppresses tumor growth through inducing apoptosis and autophagy via the inhibition of mTOR pathway regulated by ROS generation.
-
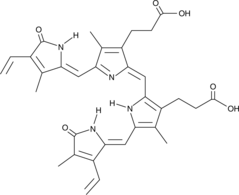
GC49708
Biliverdin (technical grade)
A bile pigment
-
GC49513
Bim/BOD (IN) Polyclonal Antibody
For immunodetection of Bim-
related proteins 
-
GC52355
BimS BH3 (51-76) (human) (trifluoroacetate salt)
A Bim-derived peptide

-
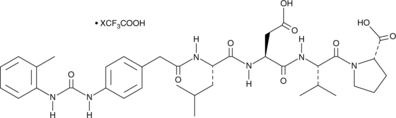
GC49724
BIO-1211 (trifluoroacetate salt)
A peptide inhibitor of α4β1 integrin
-
GC49200
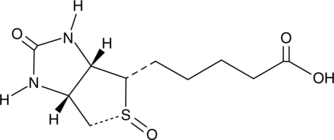
Biotin (R)-Sulfoxide
A metabolite of biotin
-
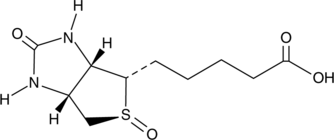
GC49170
Biotin (S)-sulfoxide
An inactive metabolite of biotin
-
GC52326
Biotin-PEG4-LL-37 (human) (trifluoroacetate salt)
A biotinylated and pegylated form of LL-37

-
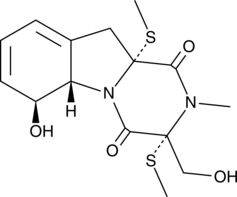
GC42941
Bis(methylthio)gliotoxin
Bis(methylthio)gliotoxin is a fungal metabolite originally isolated from G.
-
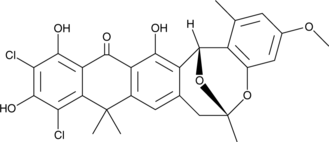
GC42943
Bischloroanthrabenzoxocinone
Bacterial type II fatty acid synthesis (FAS-II) is mediated by a series of enzymes, each of which may be targeted by potential antibiotics.
-
GC68308
Bisdemethoxycurcumin-d8
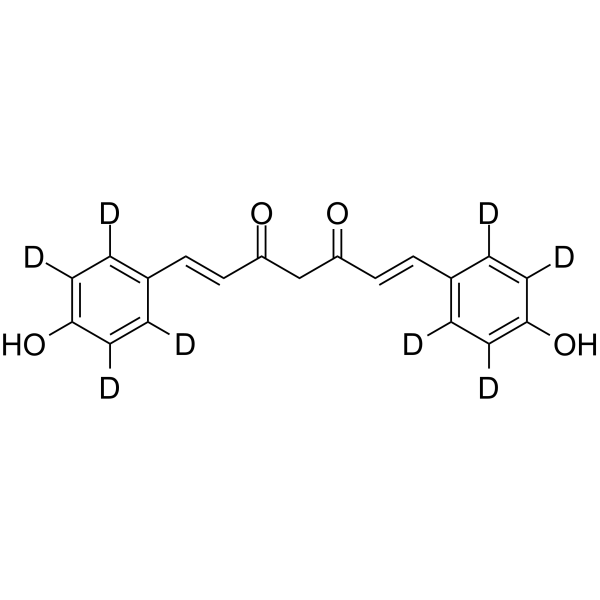
-
GC18716
Bisindolylmaleimide XI (hydrochloride)
Bisindolylmaleimide XI (BIM XI) is a selective, cell-permeable protein kinase C (PKC) inhibitor that displays 10-fold greater selectivity for PKCα (IC50 = 9 nM) and 4-fold greater selectivity for PKCβI (IC50 = 28 nM) over Ca2+-independent PKCε (IC50 = 108 nM).
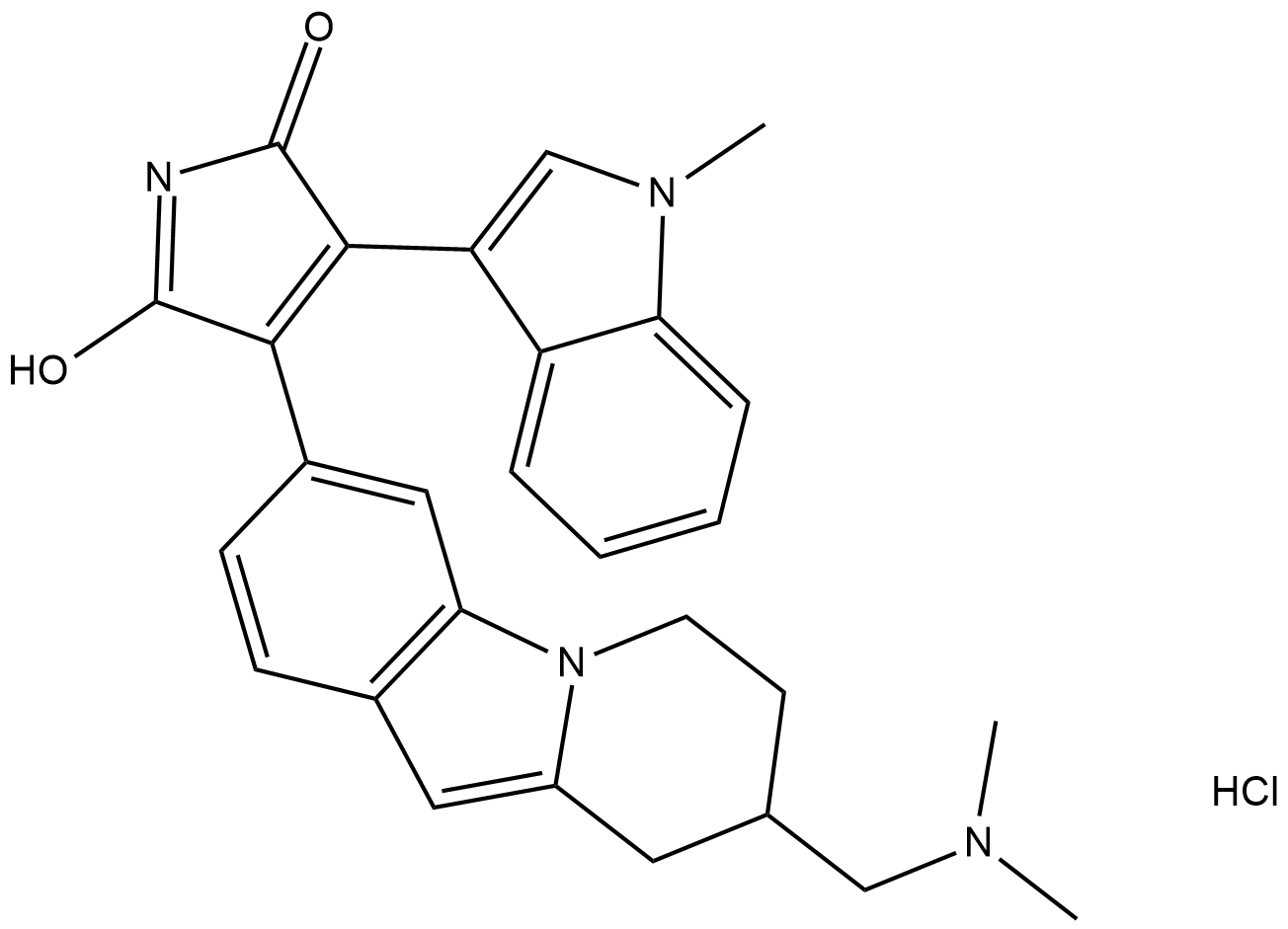
-
GC17707
Bismuth Subsalicylate
Prostaglandin G/H Synthase 1/2 inhibitor
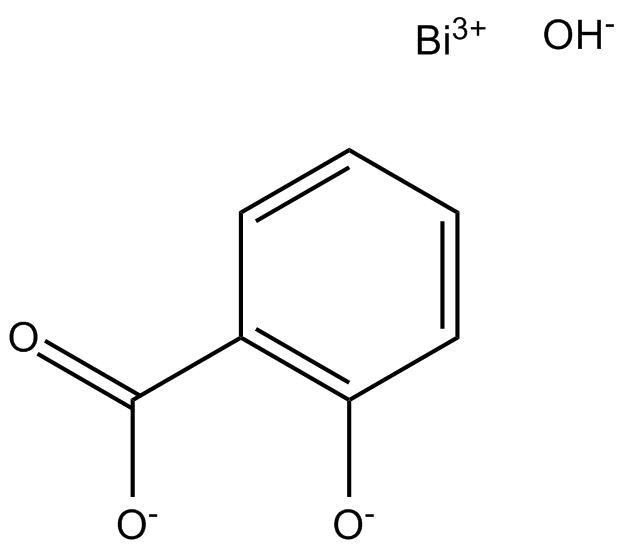
-
GC49041
Bisucaberin
A siderophore with anticancer activity
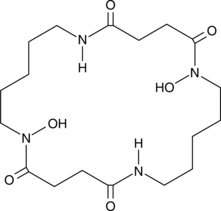
-
GC66485
Bis[3,4,6-trichloro-2-(pentyloxycarbonyl)phenyl] oxalate
Bis[3,4,6-trichloro-2-(pentyloxycarbonyl)phenyl] oxalate is a fluorescent dye that can be used for generation of chemiluminescence.
![Bis[3,4,6-trichloro-2-(pentyloxycarbonyl)phenyl] oxalate Chemical Structure Bis[3,4,6-trichloro-2-(pentyloxycarbonyl)phenyl] oxalate Chemical Structure](/media/struct/GC6/GC66485.png)
-
GC35529
Bixin
Bixin (BX), isolated from the seeds of Bixa orellana, is a carotenoid, possessing anti-inflammatory, anti-tumor and anti-oxidant activities.
-
GC65428
BLM-IN-1
BLM-IN-1 (compound 29) is an effective Bloom syndrome protein (BLM) inhibitor, with a strong BLM binding KD of 1.81 μM and an IC50 of 0.95 μM for BLM. Induces DNA damage response, as well as apoptosis and proliferation arrest in cancer cells.
-
GC45951
BLX3887
A 15-LO-1 inhibitor
-
GC41397
BMPO
BMPO is a cyclic nitrone spin trap agent, it is a water-soluble white solid which makes BMPO purification easier than other spin trap agents.
-
GC42953
BMS 345541 (trifluoroacetate salt)
BMS 345541 is a cell permeable inhibitor of the IκB kinases IKKα and IKKβ (IC50s = 4 and 0.3 μM).
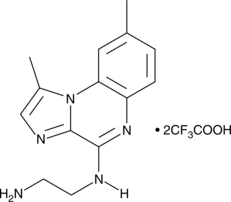
-
GC32028
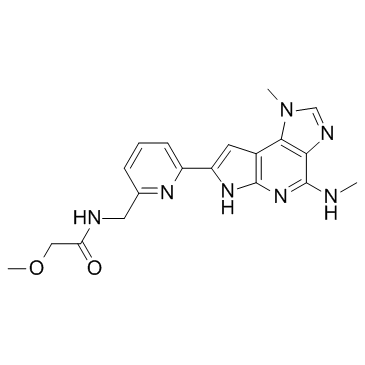
BMS-066
BMS-066 is an IKKβ/Tyk2 pseudokinase inhibitor, with IC50s of 9 nM and 72 nM, respectively.
-
GC13926
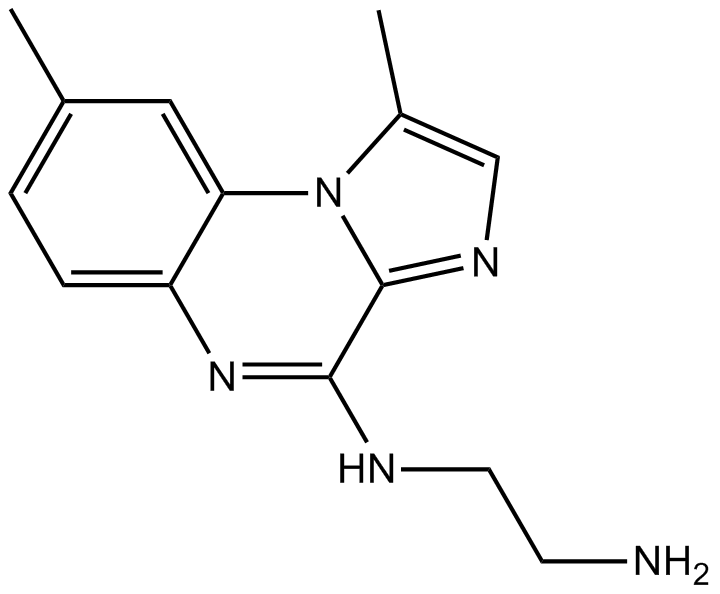
BMS-345541(free base)
BMS-345541(free base) is a selective inhibitor of the catalytic subunits of IKK (IKK-2 IC50=0.3 μM, IKK-1 IC50=4 μM). BMS-345541(free base) binds at an allosteric site of IKK.
-
GC42957
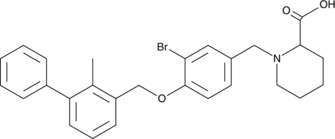
BMS-8
BMS-8 is a small molecule inhibitor of the interaction between programmed death protein 1 (PD-1) and its ligand programmed cell death ligand 1 (PD-L1) with an IC50 value of 146 nM in a homogeneous time-resolved fluorescence (HTRF) binding assay.
-
GC62507
BMS-986299
BMS-986299 (compound 112) is a first-in-class NLRP3 inflammasome agonist with an EC50 of 1.28 μM.
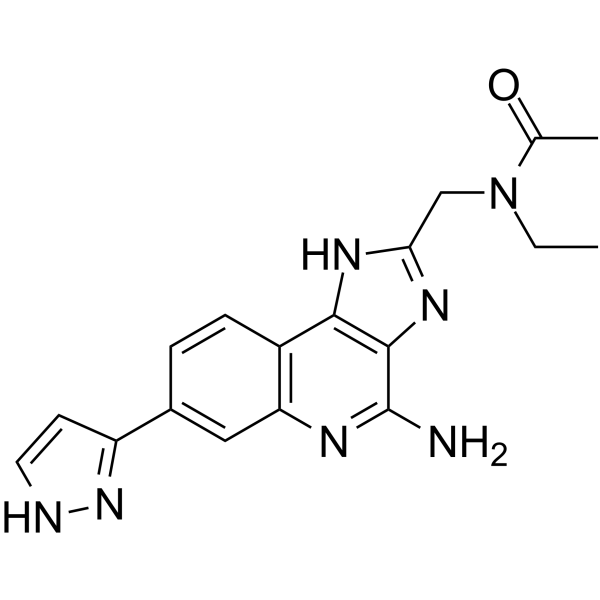
-
GC48495
BMS-P5
BMS-P5 is a specific and orally active peptidylarginine deiminase 4 (PAD4) inhibitor. BMS-P5 blocks MM-induced NET formation and delays progression of MM in a syngeneic mouse model.
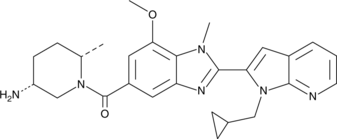
-
GC12399
BMS345541 hydrochloride
A selective inhibitor of IKKα and IKKβ
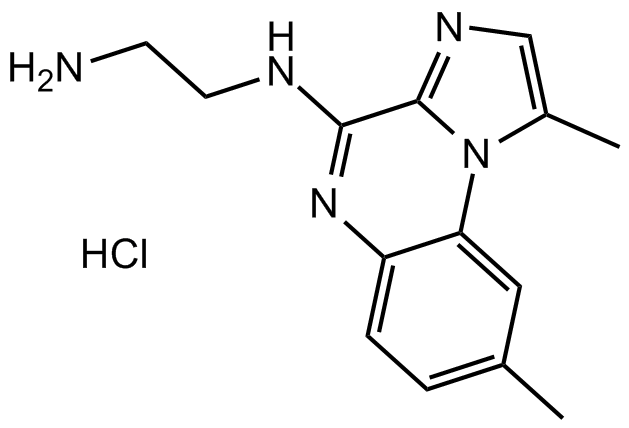
-
GA20967
Boc-D-His-OH
An amino acid building block

-
GA20972
Boc-D-Leu-OSu
An amino acid-containing building block

-
GC46937
Boc-Glu-OBzl
An amino acid-containing building block
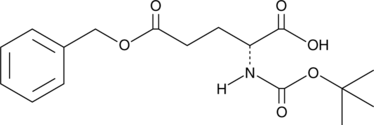
-
GC46938
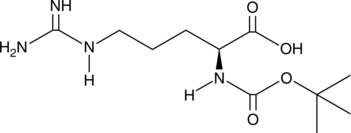
Boc-L-Arg-OH
An amino acid building block
-
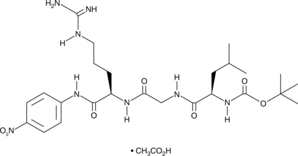
GC49717
Boc-LGR-pNA (acetate)
A chromogenic substrate for endotoxins
-
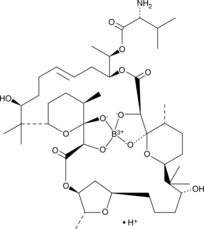
GC42967
Boromycin
Boromycin is a boron-containing macrolide antibiotic that has been found in Streptomyces.
-
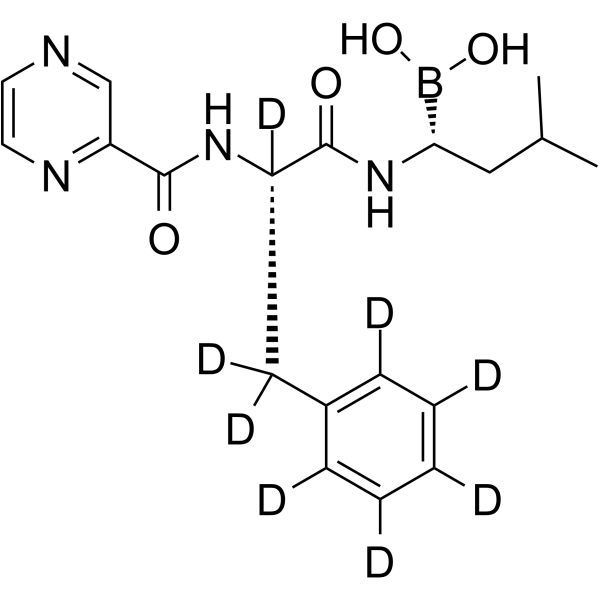
GC65010
Bortezomib-d8
Bortezomib-d8 (PS-341-d8) is the deuterium labeled Bortezomib. Bortezomib (PS-341) is a reversible and selective proteasome inhibitor, and potently inhibits 20S proteasome (Ki=0.6 nM) by targeting a threonine residue. Bortezomib disrupts the cell cycle, induces apoptosis, and inhibits NF-κB. Bortezomib is the first proteasome inhibitor anticancer agent. Anti-cancer activity.
-
GC40009
Bostrycin
Bostrycin is an anthraquinone originally isolated from B.
-
GC48365
Bottromycin A2
An antibiotic
-
GC46946
BPC 157 (acetate)
A pentadecapeptide with diverse biological activities
-
GC52240
BPH-1358
An inhibitor of UPPS and FPPS
-
GC13506
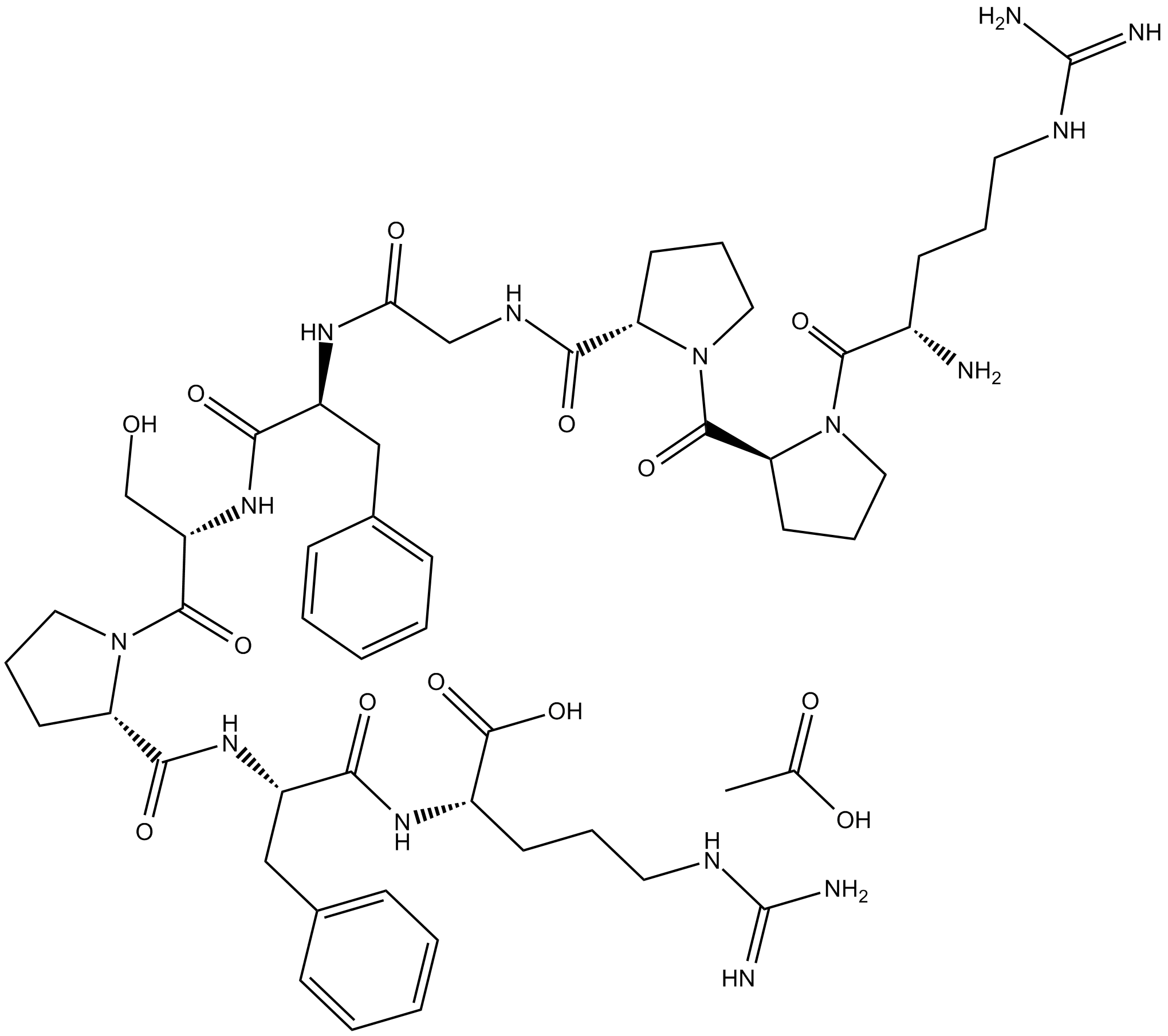
Bradykinin (acetate)
inflammatory mediator
-
GC52145
Bradykinin (human, mouse, rat, bovine) (acetate)

-
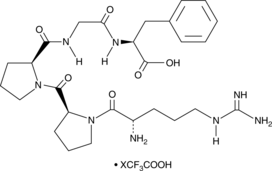
GC52424
Bradykinin Fragment (1-5) (trifluoroacetate salt)
A metabolite of bradykinin
-
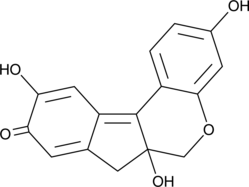
GC52101
Brazilein
Brazilein is an important immunosuppressive component isolated from Caesalpinia sappan L.
-
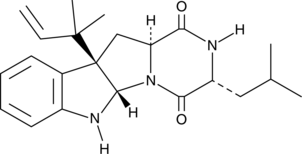
GC40824
Brevicompanine B
Brevicompanine B is a fungal metabolite originally isolated from P.
-
GC35553
Brevifolincarboxylic acid
Brevifolincarboxylic acid is extracted from Polygonum capitatum, has inhibitory effect on the aryl hydrocarbon receptor (AhR).
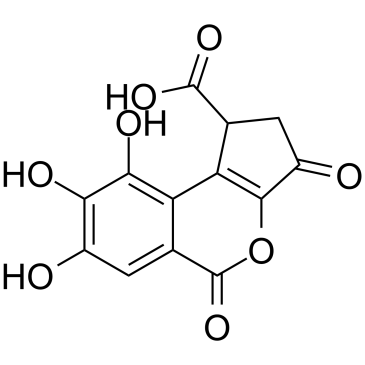
-
GC68807
Briakinumab
Briakinumab (ABT-874) is a fully human monoclonal antibody that targets and neutralizes IL-12/23p40. Briakinumab specifically targets and neutralizes both IL-12 and IL-23. It is being studied for its potential use in rheumatoid arthritis, inflammatory bowel disease, and multiple sclerosis.

-
GC67748
Brodalumab

-
GC42978
Bromamphenicol
Bromamphenicol is a dibrominated derivative of the antibiotic chloramphenicol.
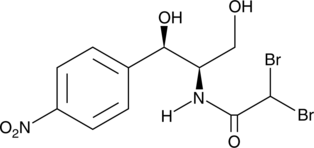
-
GC16921
Bromhexine HCl
Bromhexine HCl is a potent and specific TMPRSS2 protease inhibitor with an IC50 of 0.75 μM.
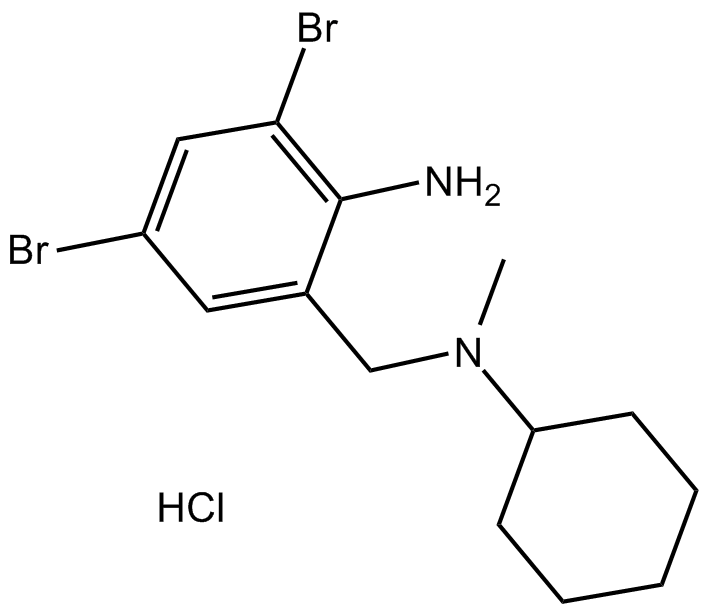
-
GC46024
Bromodiphenhydramine (hydrochloride)
A histamine H1 receptor antagonist
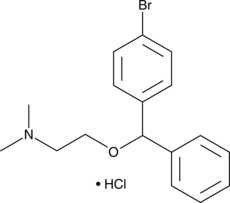
-
GC16345
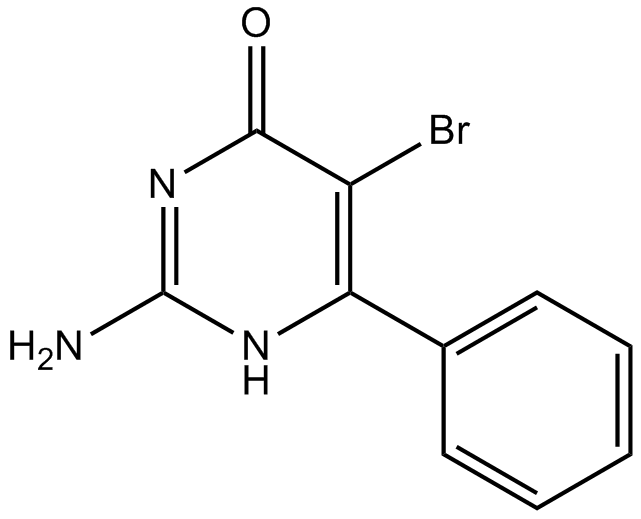
Bropirimine
Bropirimine is a synthetic agonist for toll-like receptor 7 (TLR7). Bropirimine inhibits differentiation of osteoclast precursor cells into osteoclasts via TLR7-mediated production of IFN-β. Bropirimine is an orally active immunomodulator that has demonstrated anticancer activity in transitional cell carcinoma in situ (CIS) in both the bladder and upper urinary tract.
-
GC34070
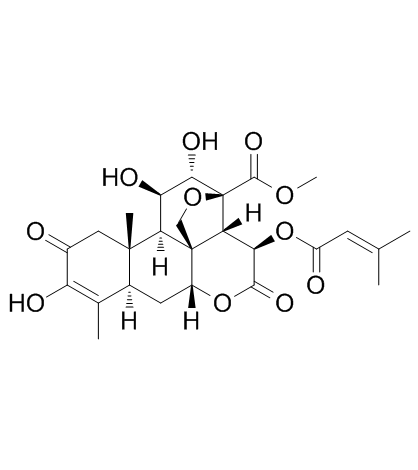
Brusatol (NSC 172924)
Brusatol (NSC 172924) (NSC172924) is a unique inhibitor of the Nrf2 pathway that sensitizes a broad spectrum of cancer cells to Cisplatin and other chemotherapeutic agents. Brusatol (NSC 172924) enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Brusatol (NSC 172924) can be developed into an adjuvant chemotherapeutic agent. Brusatol (NSC 172924) increases cellular apoptosis.
-
GC62878
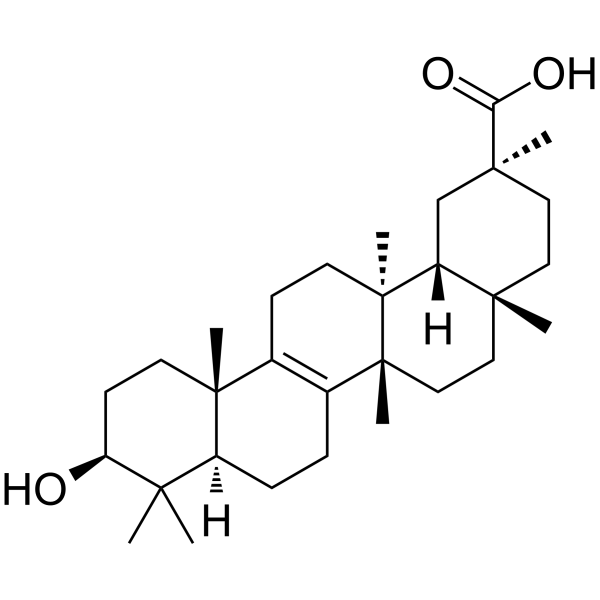
Bryonolic acid
Bryonolic acid is an active triterpenoid compound with immunomodulatory, anti-inflammatory, antioxidant and anticancer activities.
-
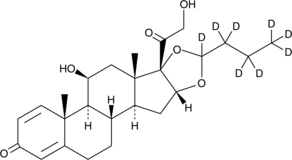
GC46956
Budesonide-d8
An internal standard for the quantification of budesonide
-
GC33134
Bufotalin
Bufotalin is a steroid lactone isolated from Venenum Bufonis with potently antitumor activities. Bufotalin induces cancer cell apoptosis and also induces endoplasmic reticulum (ER) stress activation.
-
GC46960
Buprofezin
An insecticide
-
GC48376
Burnettramic Acid A
A fungal metabolite with diverse biological activities
-
GC49136
Butenafine-13C-d3 (hydrochloride)
An internal standard for the quantification of butenafine
-
GC30067
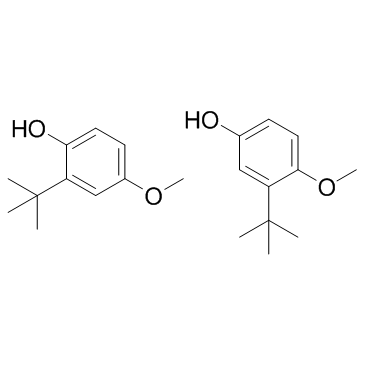
Butylhydroxyanisole (Butylated hydroxyanisole)
Butylhydroxyanisole (Butylated hydroxyanisole) (Butylated hydroxyanisole) is an antioxidant used as a food additive preservative.
-
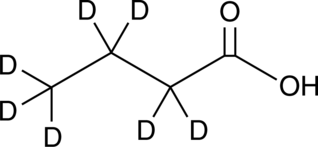
GC46104
Butyric Acid-d7
An internal standard for the quantification of sodium butyrate
-
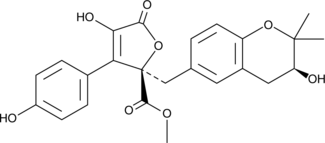
GC46106
Butyrolactone V
A fungal metabolite
-
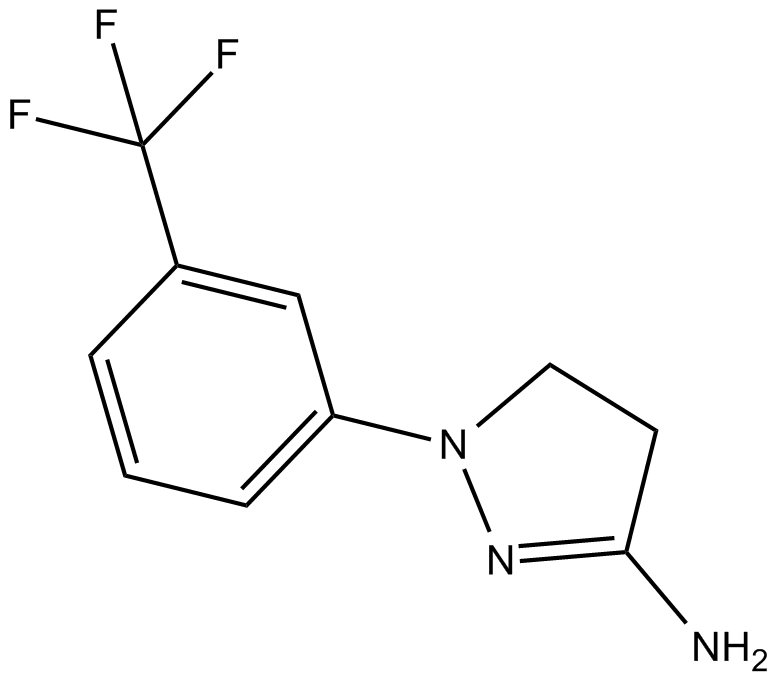
GC14601
BW 755C
dual inhibitor of 5-lipoxygenase (5-LO) and cyclooxygenase (COX) pathways
-
GC10709
BW-B 70C
5-Lipoxygenase inhibitor,potent and selective
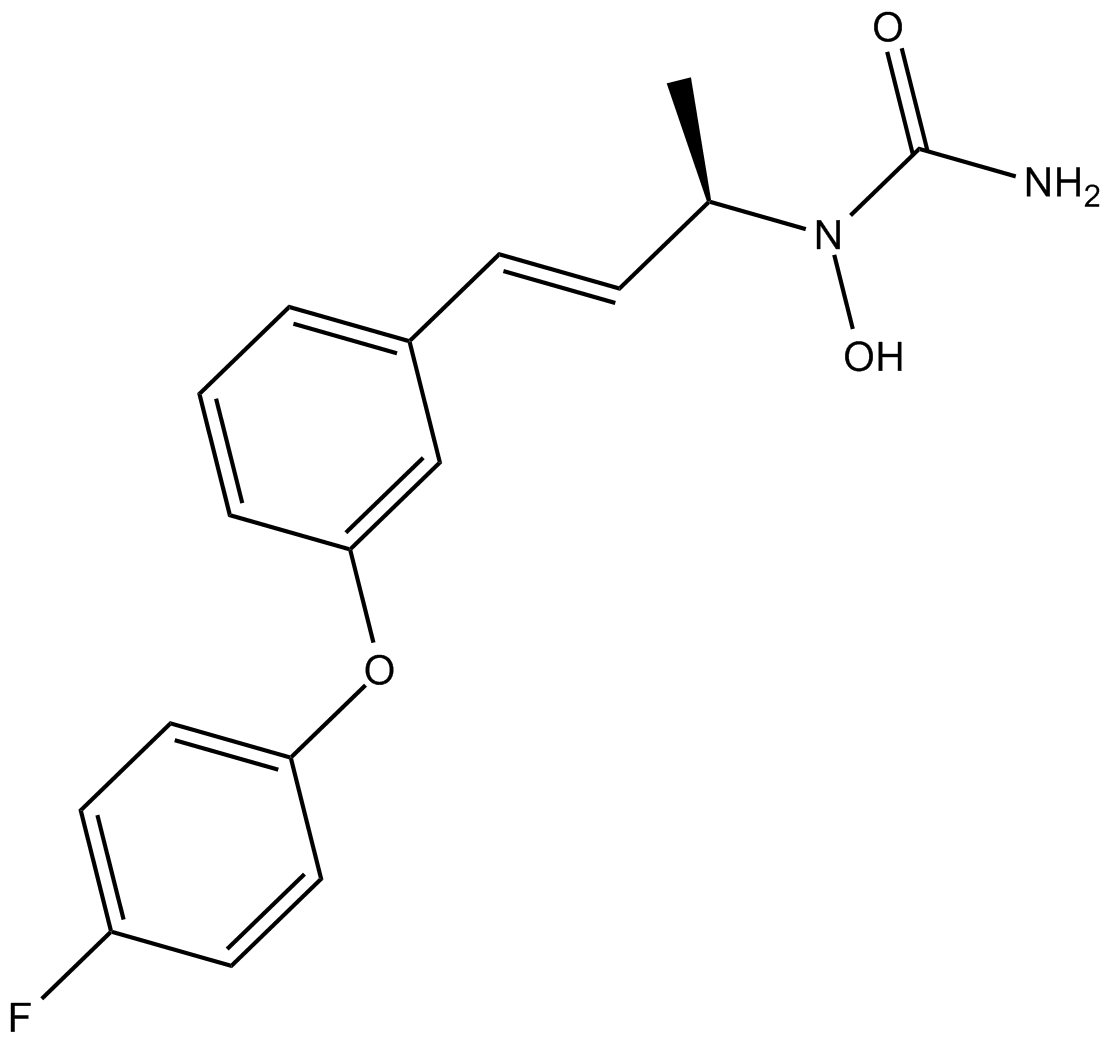
-
GC19432
BXL-628
BXL-628 (BXL-628) is a selective, orally active vitamin D receptor (VDR) agonist.

-
GC12325
BYK 191023 dihydrochloride
INOS inhibitor,potent and selective
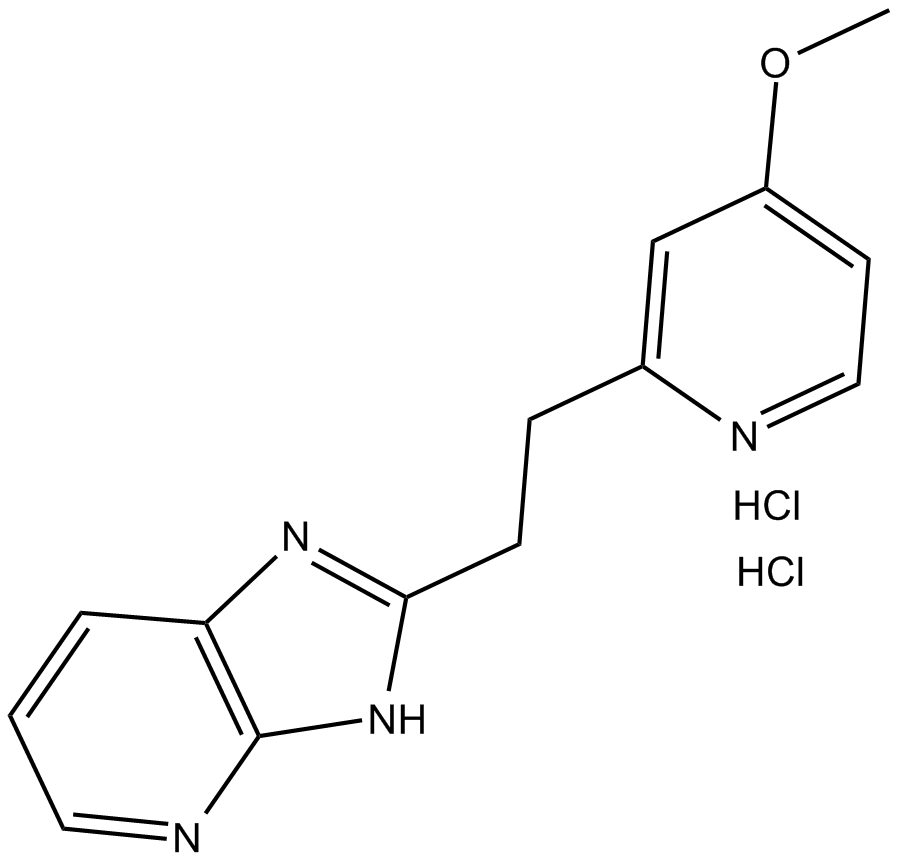
-
GA21221
Bz-Nle-Lys-Arg-Arg-AMC
Bz-Nle-Lys-Arg-Arg-AMC is a fluorogenic tetra-peptide substrate for yellow fever virus (YFV) non-structural 3 (NS3).

-
GC46983
C-170
C-170 (C-170) is a potent and covalent STING inhibitor.
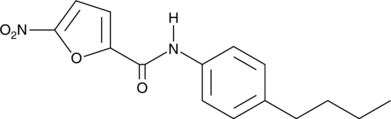
-
GC46984
C-171
C-171 is an inhibitor of stimulator of interferon genes (STING).
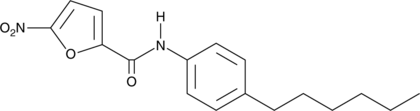
-
GC33823
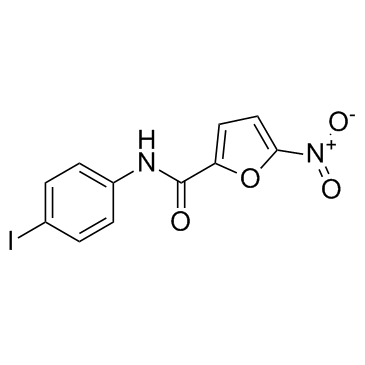
C-176 (STING inhibitor 1)
C-176 (STING inhibitor 1) is a strong and covalent mouse STING inhibitor.
-
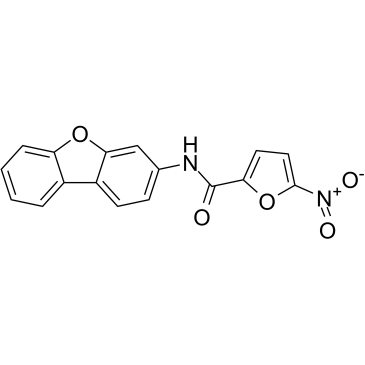
GC38494
C-178
A STING inhibitor
-
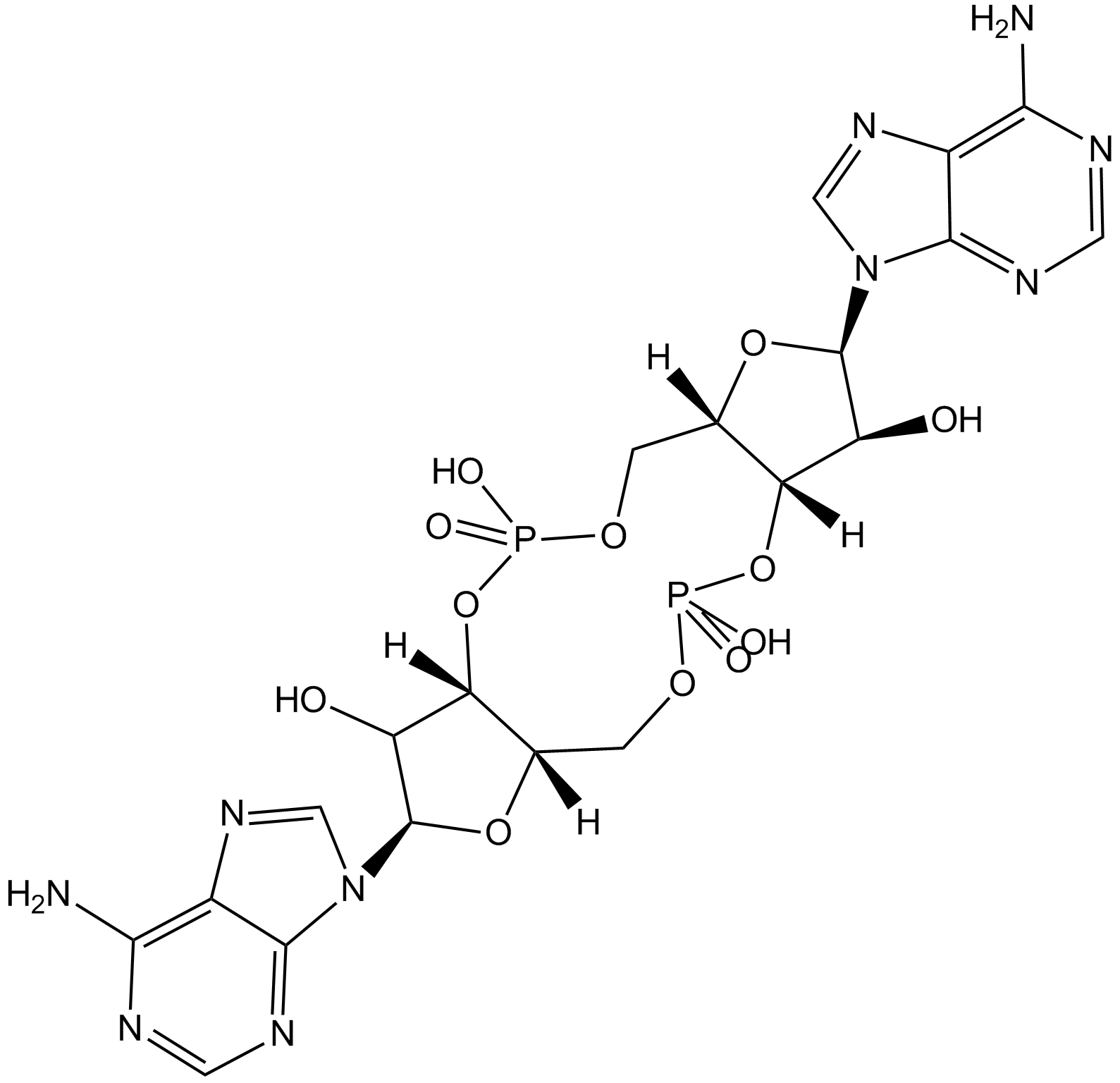
GC13643
c-di-AMP
C-di-AMP is a STING agonist, which binds to the transmembrane protein STING, thereby activating the TBK3-IRF3 signaling pathway, subsequently triggering the production of type I IFN and TNF.
-
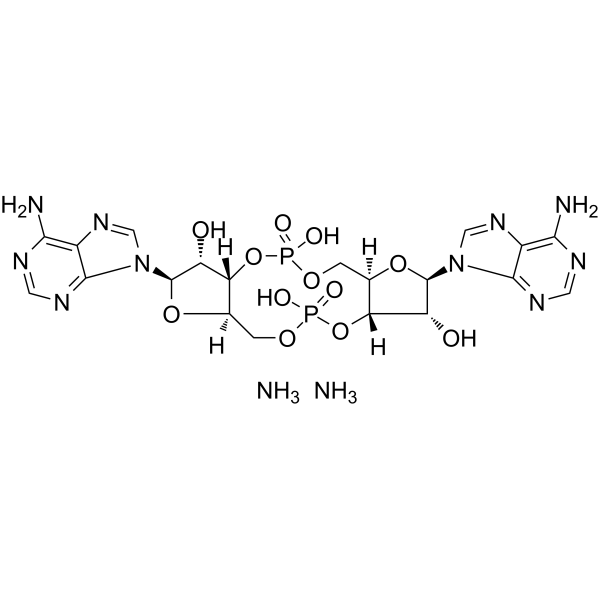
GC62893
c-di-AMP diammonium
c-di-AMP diammonium is a STING agonist, which binds to the transmembrane protein STING thereby activating the TBK3-IRF3 signaling pathway, subsequently triggering the production of type I IFN and TNF.
-
GC64445
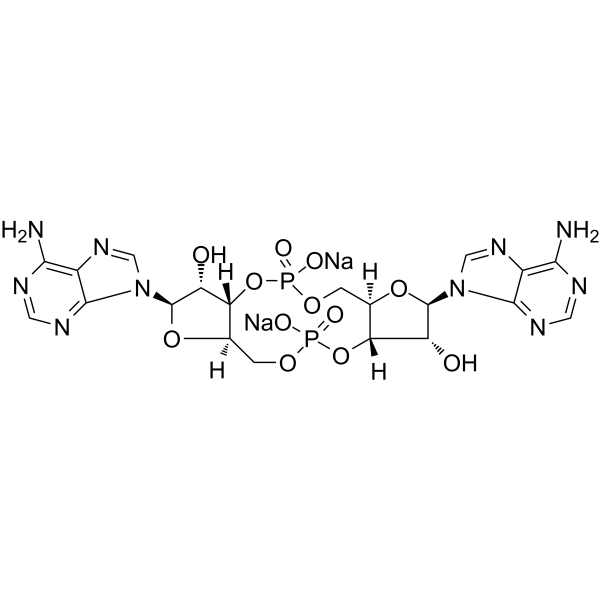
c-di-AMP disodium
c-di-AMP (Cyclic diadenylate) sodium is a STING agonist, which binds to the transmembrane protein STING thereby activating the TBK3-IRF3 signaling pathway, subsequently triggering the production of type I IFN and TNF.
-
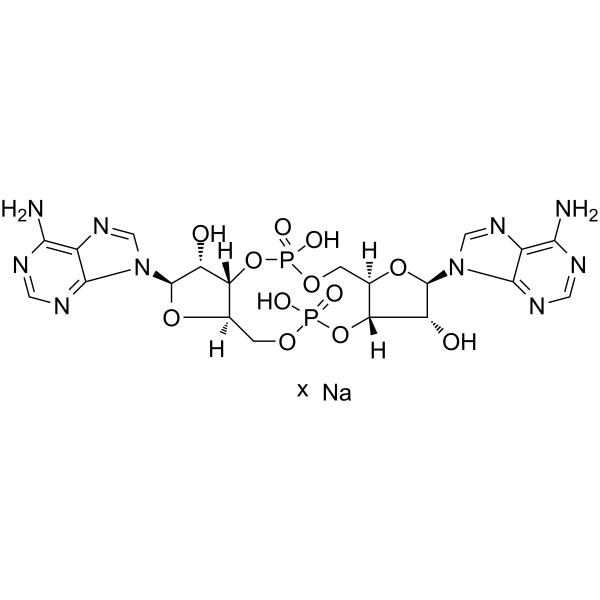
GC62198
c-di-AMP sodium
C-di-AMP is a STING agonist, which binds to the transmembrane protein STING, thereby activating the TBK3-IRF3 signaling pathway, subsequently triggering the production of type I IFN and TNF.
-
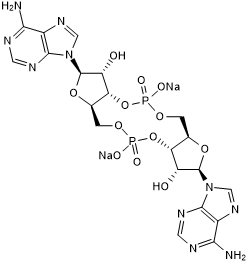
GC50687
c-Di-AMP sodium salt
-
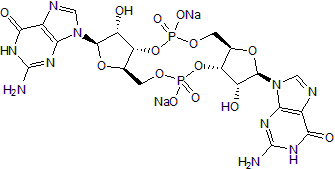
GC50292
c-Di-GMP sodium salt
Endogenous STING and DDX41 agonist; activates STING-dependent signaling
-
GC43007
C12 Galactosylceramide (d18:1/12:0)
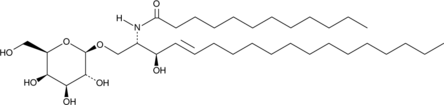
C12 Galactosylceramide is a bioactive sphingolipid.
-
GC43031
C16 Galactosylceramide (d18:1/16:0)
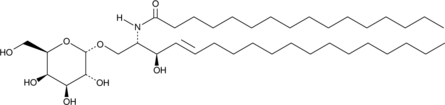
C16 Galactosylceramide is a glycosphingolipid that contains a galactose moiety attached to a ceramide acylated with palmitic acid .
-
GC43032
C16 Globotriaosylceramide (d18:1/16:0)
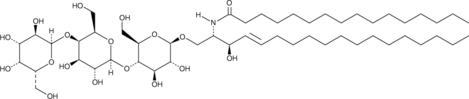
C16 globotriaosylceramide is an endogenous sphingolipid found in mammalian cell membranes that is synthesized from C16 lactosylceramide.
-
GC43049
C18 Globotriaosylceramide (d18:1/18:0)
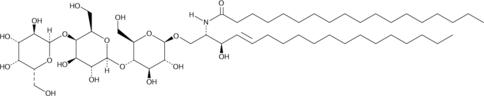
C18 globotriaosylceramide is an endogenous sphingolipid found in mammalian cell membranes that is synthesized from lactosylceramide.
-
GC43052
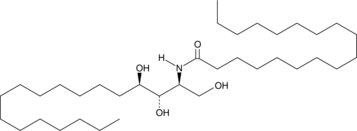
C18 Phytoceramide (t18:0/18:0)
C18 Phytoceramide (t18:0/18:0) (Cer(t18:0/18:0)) is a bioactive sphingolipid found in S.
-
GC40141
C18 Phytoceramide-d3 (t18:0/18:0-d3)
C18 Phytoceramide-d3 (t18:0/18:0-d3) is intended for use as an internal standard for the quantification of C18 phytoceramide (t18:0/18:0) by GC- or LC-MS.

-
GC43060
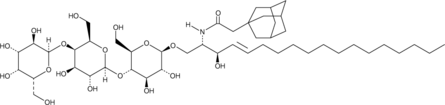
C2 Adamantanyl Globotriaosylceramide (d18:1/2:0)
C2 Adamantanyl globotriaosylceramide (AdaGb3) is a bioactive sphingolipid and water-soluble form of globotriaosylceramide that contains an adamantanyl group in place of the fatty acyl chain.
-
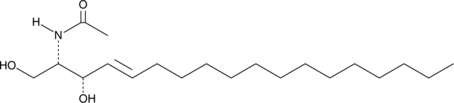
GC40709
C2 L-threo Ceramide (d18:1/2:0)
C2 L-threo Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
-
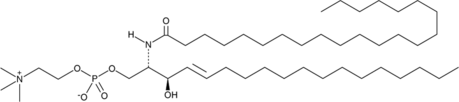
GC43071
C22 Sphingomyelin (d18:1/22:0)
C22 Sphingomyelin is a naturally occurring form of sphingomyelin.
-
GC45834
C24 (2'(R)-hydroxy) Phytoceramide (t18:0/24:0)
A bioactive sphingolipid

-
GC43076
C24 Phytosphingosine (t18:0/24:0)
C24 Phytosphingosine (t18:0/24:0) is a phytoceramide, which is a family of sphingolipids found in the intestine, kidney, and extracellular spaces of the stratum corneum of the mammalian epidermis.
-
GC43077
C24:1 3'-sulfo Galactosylceramide (d18:1/24:1(15Z))
C24:1 3'-sulfo Galactosylceramide is a member of the sulfatide class of glycolipids.
-
GC47007
C26 Sphingomyelin (d18:1/26:0)
A sphingolipid
-
GC33820
C29
C29 is a Toll-like receptor 2 (TLR2) inhibitor.
-
GC35575
C3a (70-77) TFA
-
GC31928
C3a 70-77 (Complement 3a (70-77))
C3a 70-77 (Complement 3a (70-77)) is an octapeptide corresponding to the COOH terminus of C3a, exhibits the specificity and 1 to 2% biologic activities of C3a.
-
GC43084
C4 Ceramide (d18:1/4:0)
C4 Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
-
GC40688
C6 D-threo Ceramide (d18:1/6:0)
C6 D-threo Ceramide is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides., C6 D-threo Ceramide is cytotoxic to U937 cells in vitro (IC50 = 18 μM).
-
GC40690
C6 L-threo Ceramide (d18:1/6:0)
C6 L-threo Ceramide (d18:1/6:0) is a bioactive sphingolipid and cell-permeable analog of naturally occurring ceramides.
-
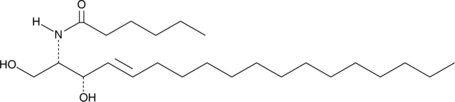
GC43110
C8 Galactosylceramide (d18:1/8:0)
C8 Galactosylceramide is a synthetic C8 short-chain derivative of known membrane microdomain-forming sphingolipids.

-
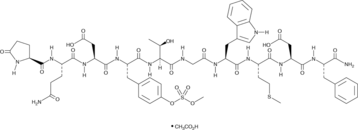
GC43113
Caerulein (acetate)
Caerulein is an oligopeptide originally isolated from skin extracts of H.
-
GC40352
Cafestol
Cafestol is a natural diterpene which is abundant in unfiltered coffee.