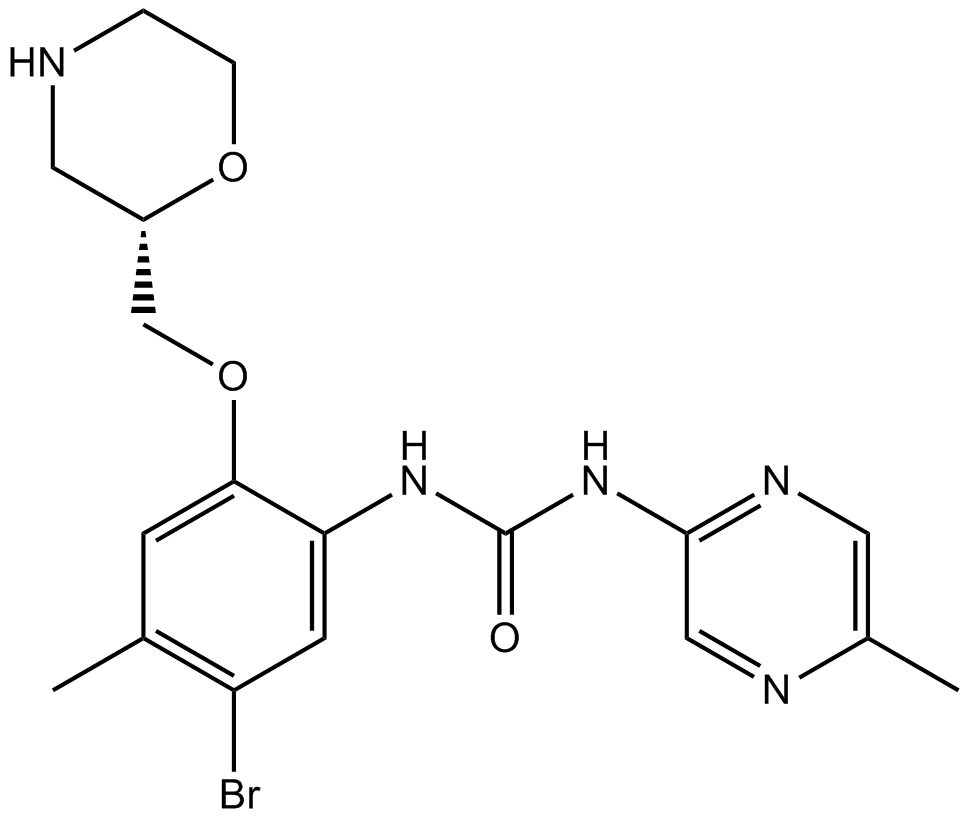
Cell Cycle/Checkpoint
Cell Cycle
Cells undergo a complex cycle of growth and division that is referred to as the cell cycle. The cell cycle consists of four phases, G1 (GAP 1), S (synthesis), G2 (GAP 2) and M (mitosis). DNA replication occurs during S phase. When cells stop dividing temporarily or indefinitely, they enter a quiescent state called G0.
Targets for Cell Cycle/Checkpoint
- ATM/ATR(26)
- Aurora Kinase(47)
- Cdc42(4)
- Cdc7(4)
- Chk(16)
- c-Myc(20)
- CRM1(8)
- Cyclin-Dependent Kinases(91)
- E1 enzyme(1)
- G-quadruplex(14)
- Haspin(7)
- HMTase(1)
- Kinesin(26)
- Ksp(6)
- Microtubule/Tubulin(243)
- Mps1(15)
- Mitotic(11)
- RAD51(18)
- ROCK(71)
- Rho(13)
- PERK(11)
- PLK(37)
- PTEN(8)
- Wee1(7)
- PAK(21)
- Arp2/3 Complex(8)
- Dynamin(12)
- ECM & Adhesion Molecules(40)
- Cholesterol Metabolism(3)
- Endomembrane System & Vesicular Trafficking(26)
- G1(38)
- G2/M(26)
- G2/S(10)
- Genotoxic Stress(18)
- Inositol Phosphates(18)
- Proteolysis(99)
- Cytoskeleton & Motor Proteins(53)
- Cellular Chaperones(8)
Products for Cell Cycle/Checkpoint
- Cat.No. Product Name Information
-
GA21951
H-4-Nitro-Phe-OEt . HCl
A building block

-
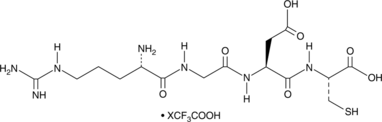
GC49305
H-Arg-Gly-Asp-Cys-OH (trifluoroacetate salt)
An RGD-containing tetrapeptide
-
GA22303
H-D-Pro-Phe-Arg-pNA . 2 HCl
pFR-pNA, chromogenic substrate for the determination of plasma kallikrein-like activity.
CAS Number (net): 64816-19-9.

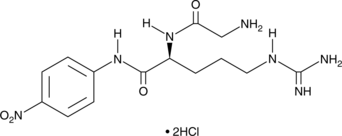
-
GC45471
H-Gly-Arg-pNA (hydrochloride)
-
GC49605
H-Gly-Pro-Hyp-OH (acetate)
A peptide DPP-4 inhibitor
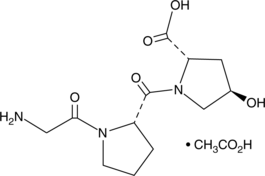
-
GC43803
HA-1077 (hydrochloride)
Fasudil (HA-1077; AT877) dihydrochloride is a nonspecific RhoA/ROCK inhibitor and also has inhibitory effect on protein kinases, with an Ki of 0.33 μM for ROCK1, IC50s of 0.158 μM and 4.58 μM, 12.30 μM, 1.650 μM for ROCK2 and PKA, PKC, PKG, respectively. HA-1077 (hydrochloride) is also a potent Ca2+ channel antagonist and vasodilator.
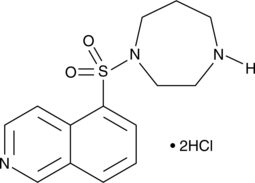
-
GC43805
HAPC-Chol
HAPC-Chol is a cationic cholesterol.
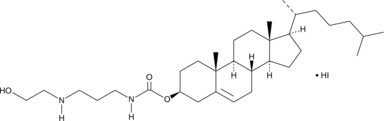
-
GC49855
Harmalol (hydrochloride hydrate)
A β-carboline alkaloid and an active metabolite of harmaline
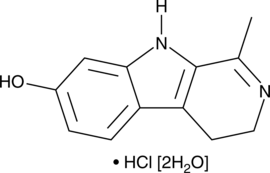
-
GC49915
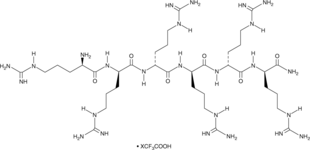
Hexa-D-Arginine (trifluoroacetate salt)
A furin inhibitor
-
GC45639
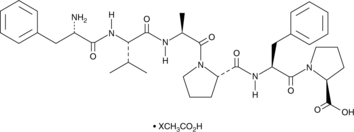
Hexapeptide-11 (acetate)
A synthetic hexapeptide
-
GC43832
Histone H3 (21-44)-GK-biotin amide (trifluoroacetate salt)
Histone H3 (21-44)-GK-biotin is a peptide fragment of histone H3 that corresponds to amino acid residues 22-45 of the human histone H3.3 sequence and is biotinylated via a C-terminal GK linker.
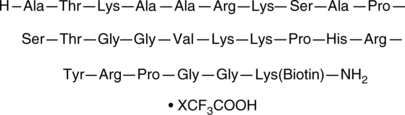
-
GC13928
HMN-214
Plk inhibitor,broad-spectrum anti-tumor agent
-
GC62594
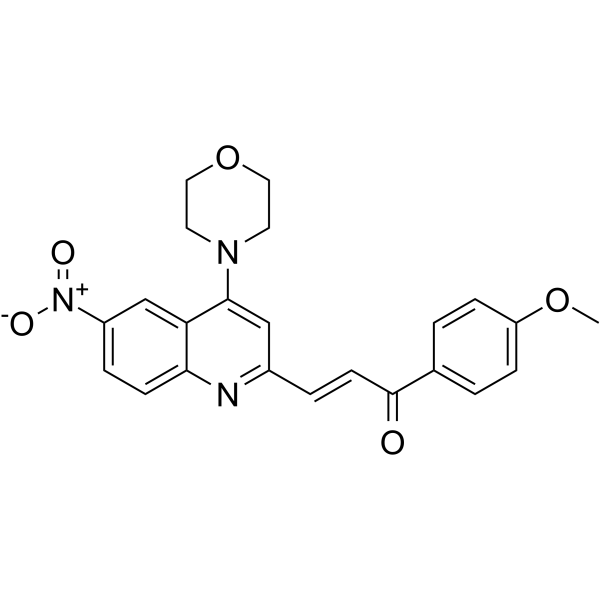
hnRNPK-IN-1
hnRNPK-IN-1 is a heterogeneous nuclear ribonucleoprotein K (hnRNPK) binding ligand with Kd values of 4.6 μM and 2.6 μM measured with SPR and MST, respectively. hnRNPK-IN-1 inhibits c-myc transcription by disrupting the binding of hnRNPK and c-myc promoter. hnRNPK-IN-1 induces Hela cells apoptosis and has strongly anti-tumor activities.
-
GC49742
Hsf1 Monoclonal Antibody (Clone 10H8)
For immunochemical analysis of Hsf1

-
GC47436
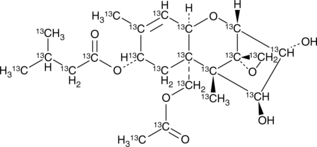
HT-2 Toxin-13C22
An internal standard for the quantification of HT-2 toxin
-
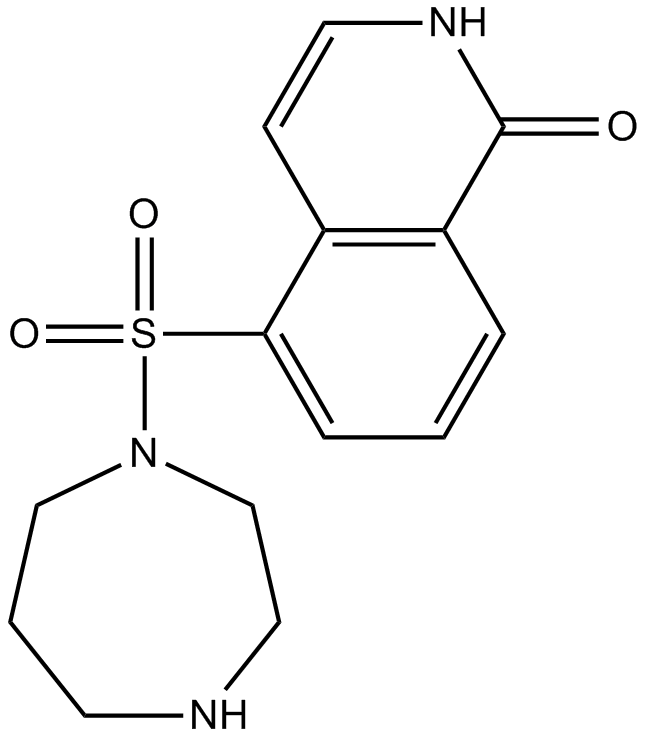
GC10901
Hydroxyfasudil
-
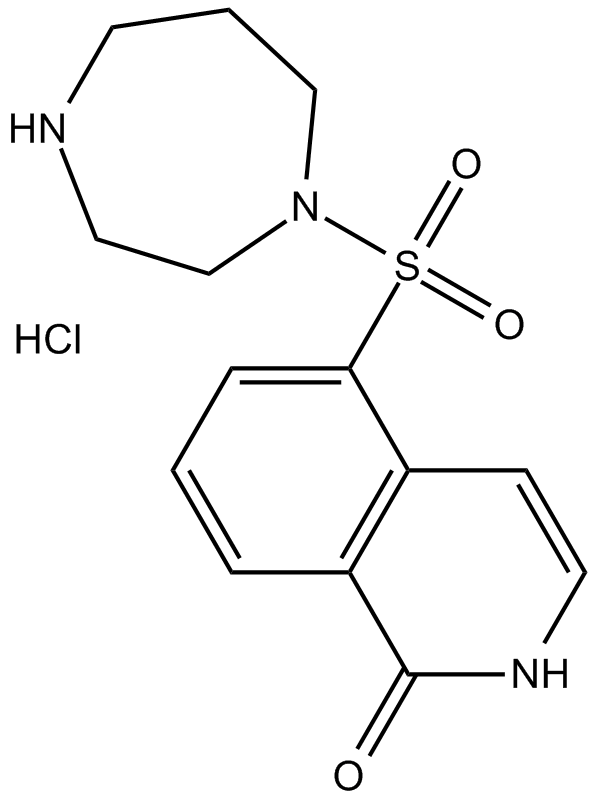
GC10338
Hydroxyfasudil hydrochloride
A ROCK inhibitor
-
GC32912
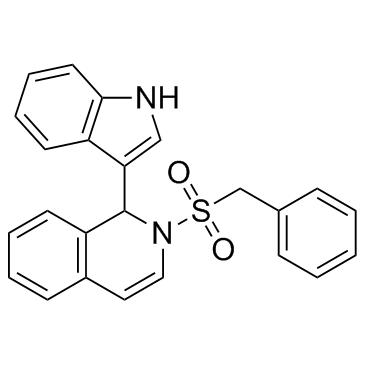
IBR2
IBR2 is a potent and specific RAD51 inhibitor and inhibits RAD51-mediated DNA double-strand break repair. IBR2 disrupts RAD51 multimerization, accelerates proteasome-mediated RAD51 protein degradation, inhibits cancer cell growth and induces apoptosis.
-
GC45743
ICAAc
A solvatochromic fluorophore and pH probe
-
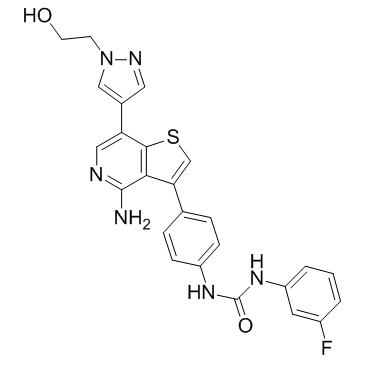
GC34159
Ilorasertib (ABT-348)
Ilorasertib (ABT-348) (ABT-348) is a potent, orally active and ATP-competitive aurora inhibitor with IC50s of116, 5, 1 nM for aurora A, aurora B, aurora C, respectively. Ilorasertib (ABT-348) also is a potent VEGF, PDGF inhibitor. Ilorasertib (ABT-348) has the potential for the research of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS).
-
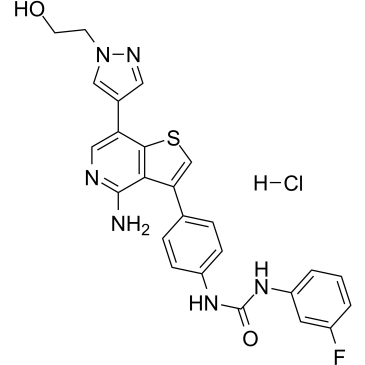
GC38519
Ilorasertib hydrochloride
Ilorasertib (ABT-348) hydrochloride is a potent, orally active and ATP-competitive aurora inhibitor with IC50s of116, 5, 1 nM for aurora A, aurora B, aurora C, respectively. Ilorasertib hydrochloride also is a potent VEGF, PDGF inhibitor. Ilorasertib hydrochloride has the potential for the research of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS).
-
GC36308
Indibulin
An inhibitor of microtubule assembly
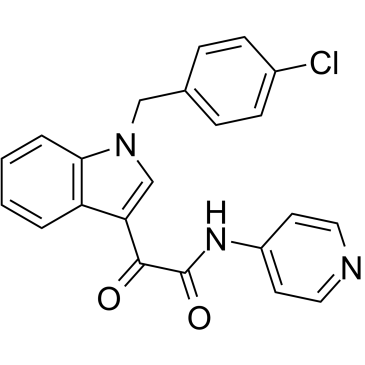
-
GC52513
Indolimine-214
A genotoxic gut microbiota metabolite
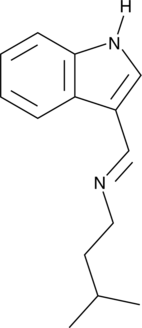
-
GC49460
ING-2 AM
A cell-permeable fluorescent sodium indicator

-
GC10334
INH6
Hec1/Nek2 inhibitor, potent
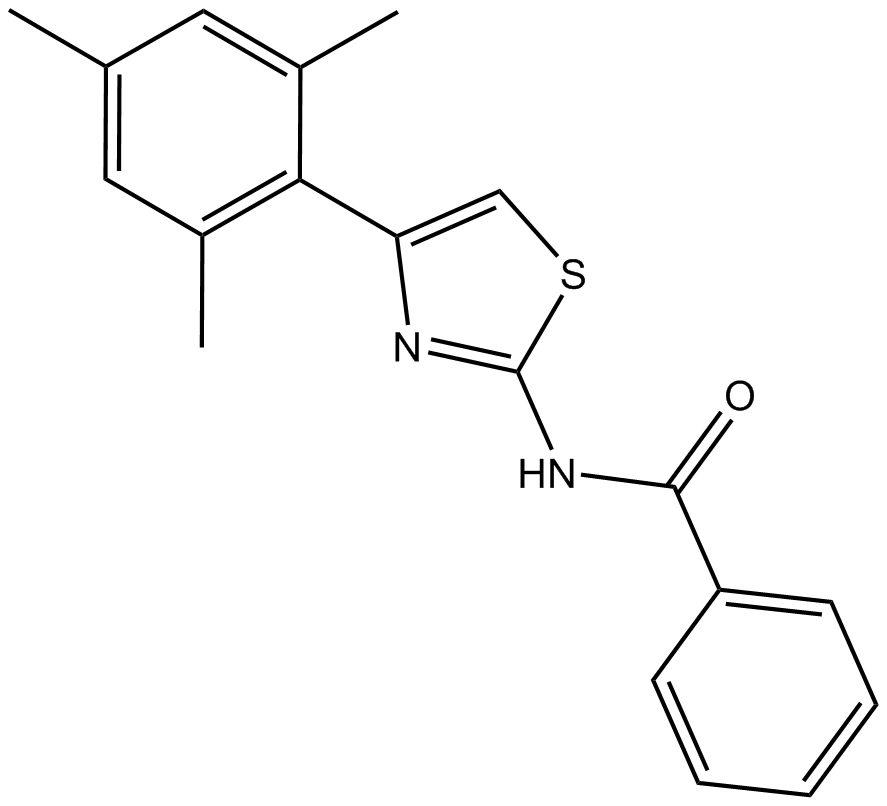
-
GC48387
Inostamycin A
A bacterial metabolite with anticancer activity
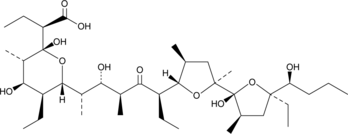
-
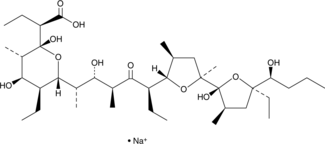
GC52472
Inostamycin A (sodium salt)
A bacterial metabolite with anticancer activity
-
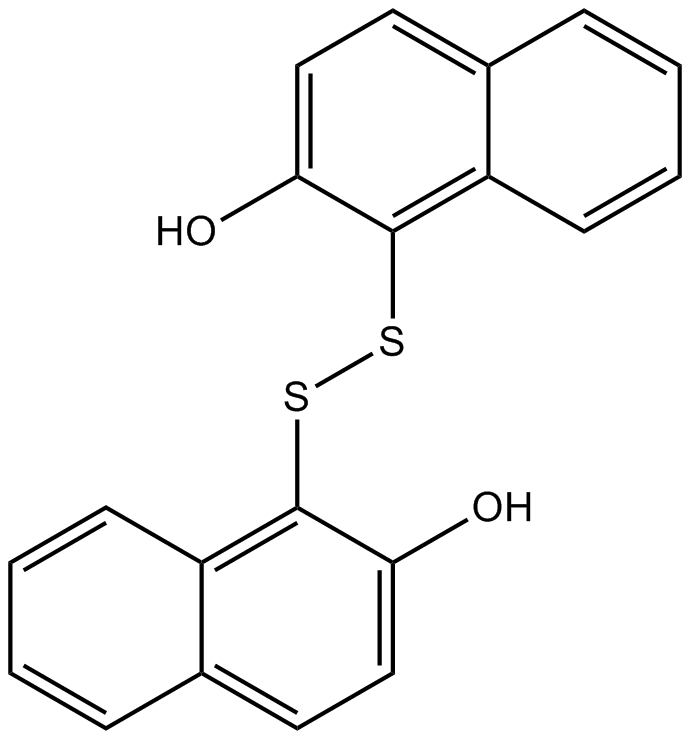
GC16225
IPA-3
Non-ATP competitive Pak1 inhibitor
-
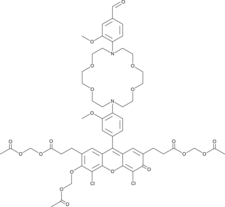
GC49468
IPG-1 AM
A cell-permeable fluorescent potassium indicator
-
GC49469
IPG-1 TMA
A cell-impermeable fluorescent potassium indicator

-
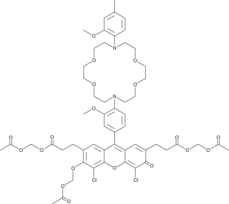
GC49463
IPG-2 AM
A cell-permeable fluorescent potassium indicator
-
GC49466
IPG-2 TMA
A cell-impermeable fluorescent potassium indicator
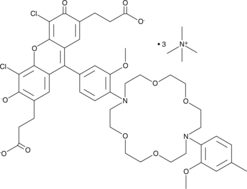
-
GC65188
IRES-C11
IRES-C11 is a spectfic c-MYC internal ribosome entry site (IRES) translation inhibitor. IRES-C11 blocks the interaction of a requisite c-MYC IRES trans-acting factor, heterogeneous nuclear ribonucleoprotein A1, with its IRES. IRES-C11 does not inhibits BAG-1, XIAP and p53 IRESes.
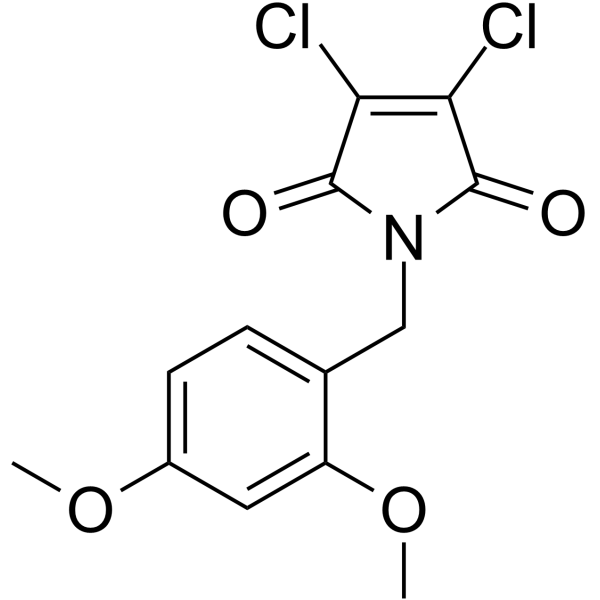
-
GC15215
Iso-Olomoucine
inactive stereoisomer of the Cdk5 inhibitor olomoucine
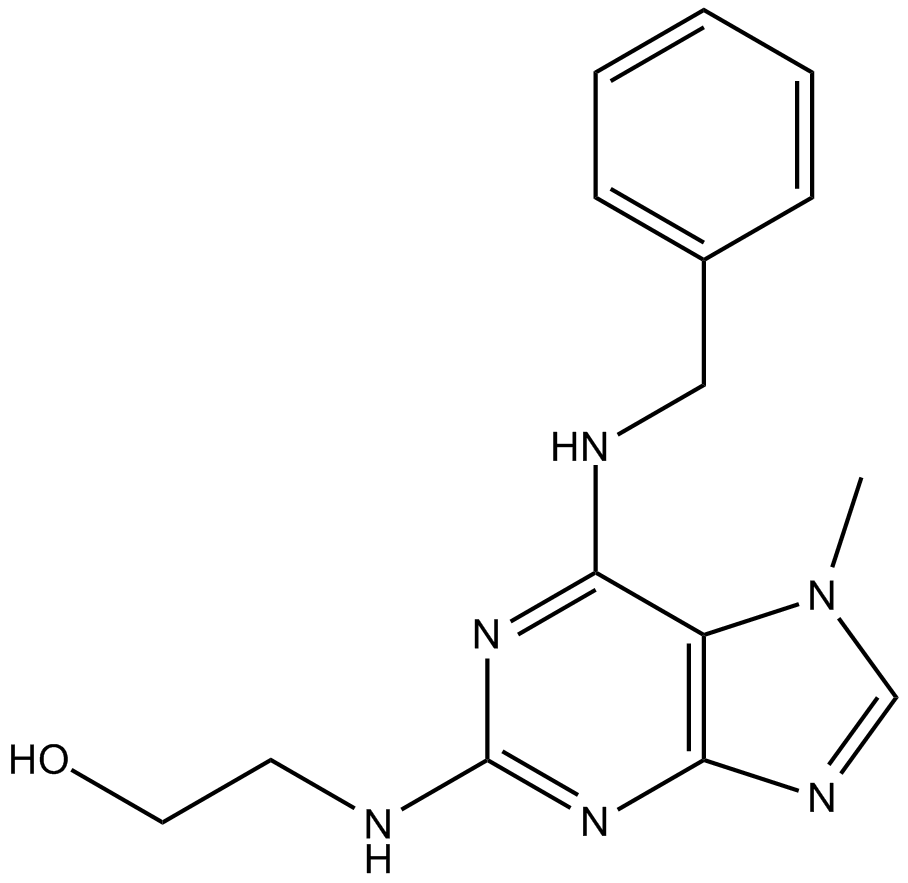
-
GC40901
Isogarcinol
Isogarcinol is a natural polyisoprenylated benzophenone first isolated from plant species in the genus Garcinia.
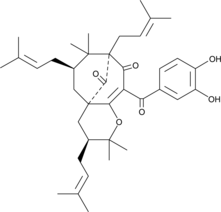
-
GC43917
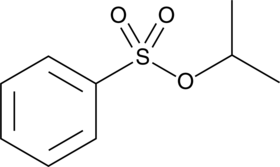
Isopropyl Benzenesulfonate
Isopropyl benzenesulfonate is a sulfonate ester genotoxic impurity found in active pharmaceutical ingredients.
-
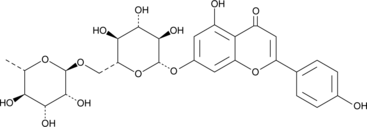
GC49142
Isorhoifolin
A flavonoid glycoside with diverse biological activities
-
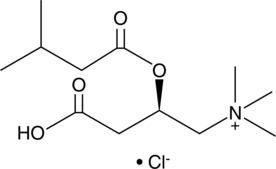
GC43922
Isovaleryl-L-carnitine (chloride)
Isovaleryl-L-carnitine (chloride), a product of the catabolism of L-leucine, is a potent activator of the Ca2+-dependent proteinase (calpain) of human neutrophils.
-
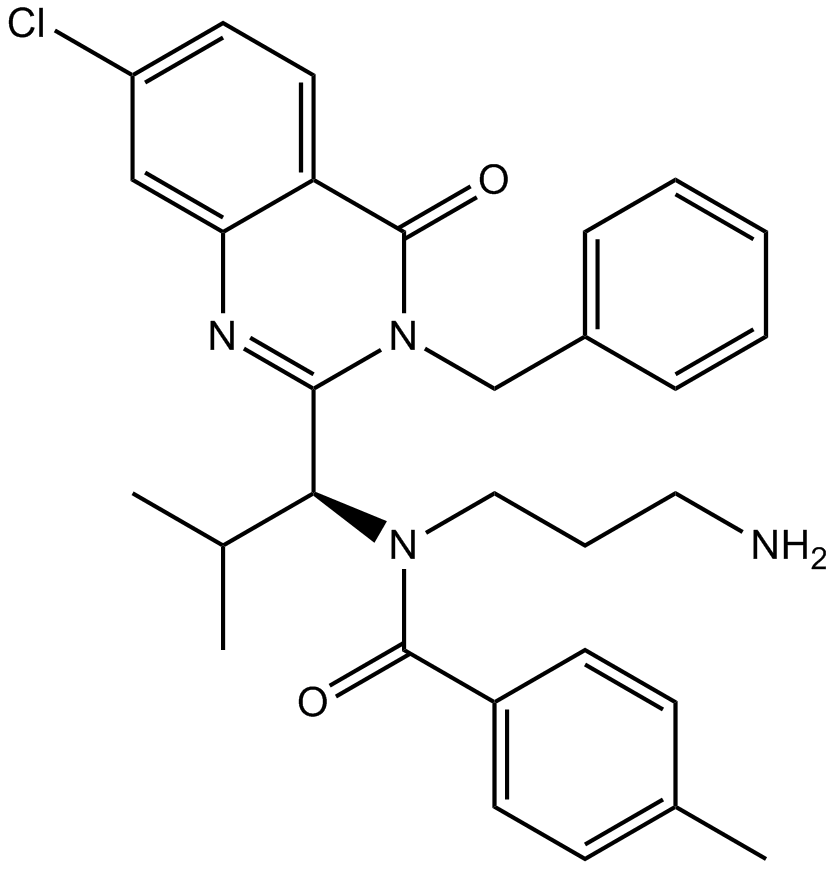
GC13394
Ispinesib (SB-715992)
Ispinesib (SB-715992) is a specific inhibitor of kinesin spindle protein (KSP), with a Ki app of 1.7 nM.
-
GC10581
ISRIB
PERK signaling inhibitor
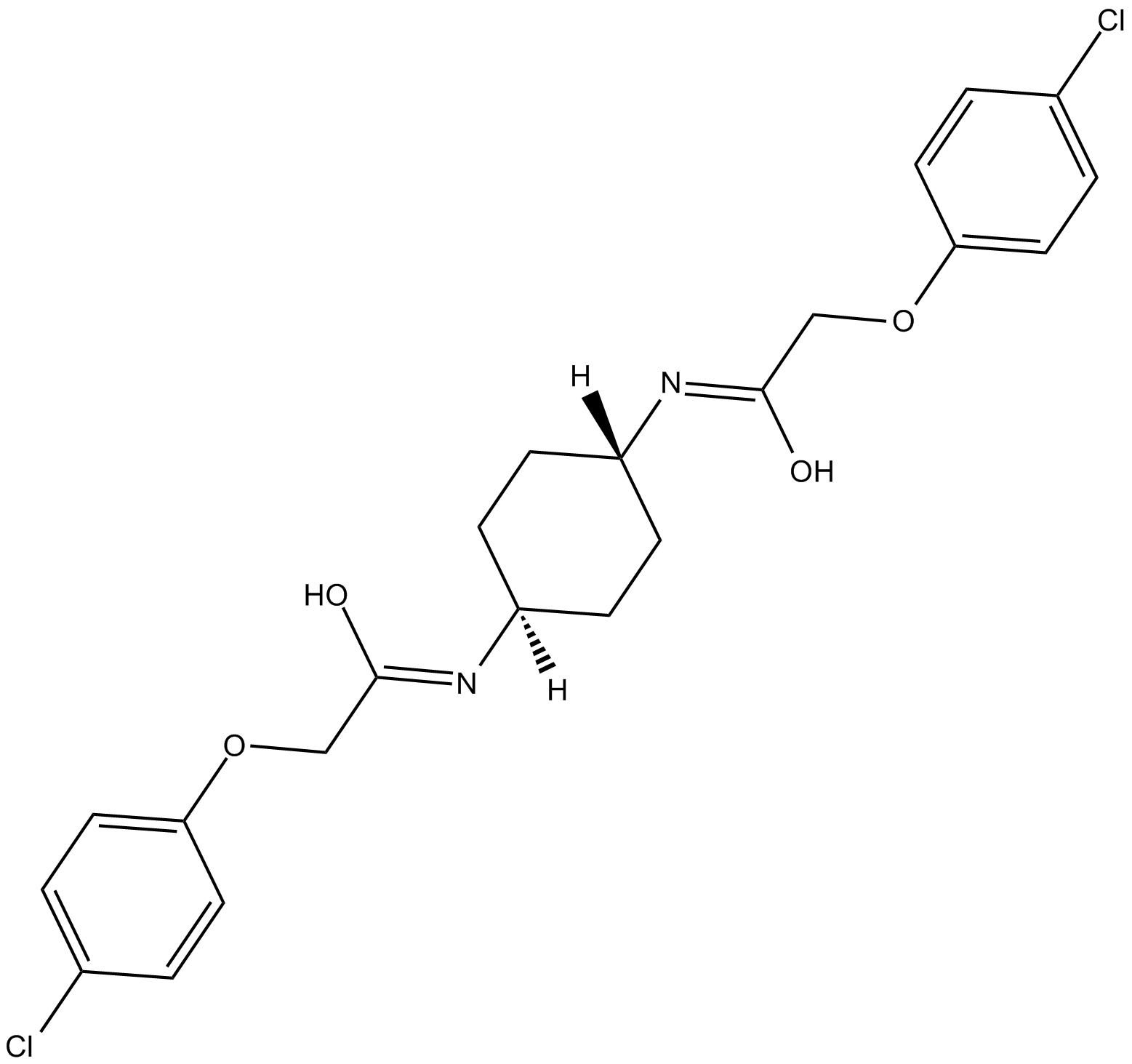
-
GC16869
Ixabepilone
A broad-spectrum anticancer agent
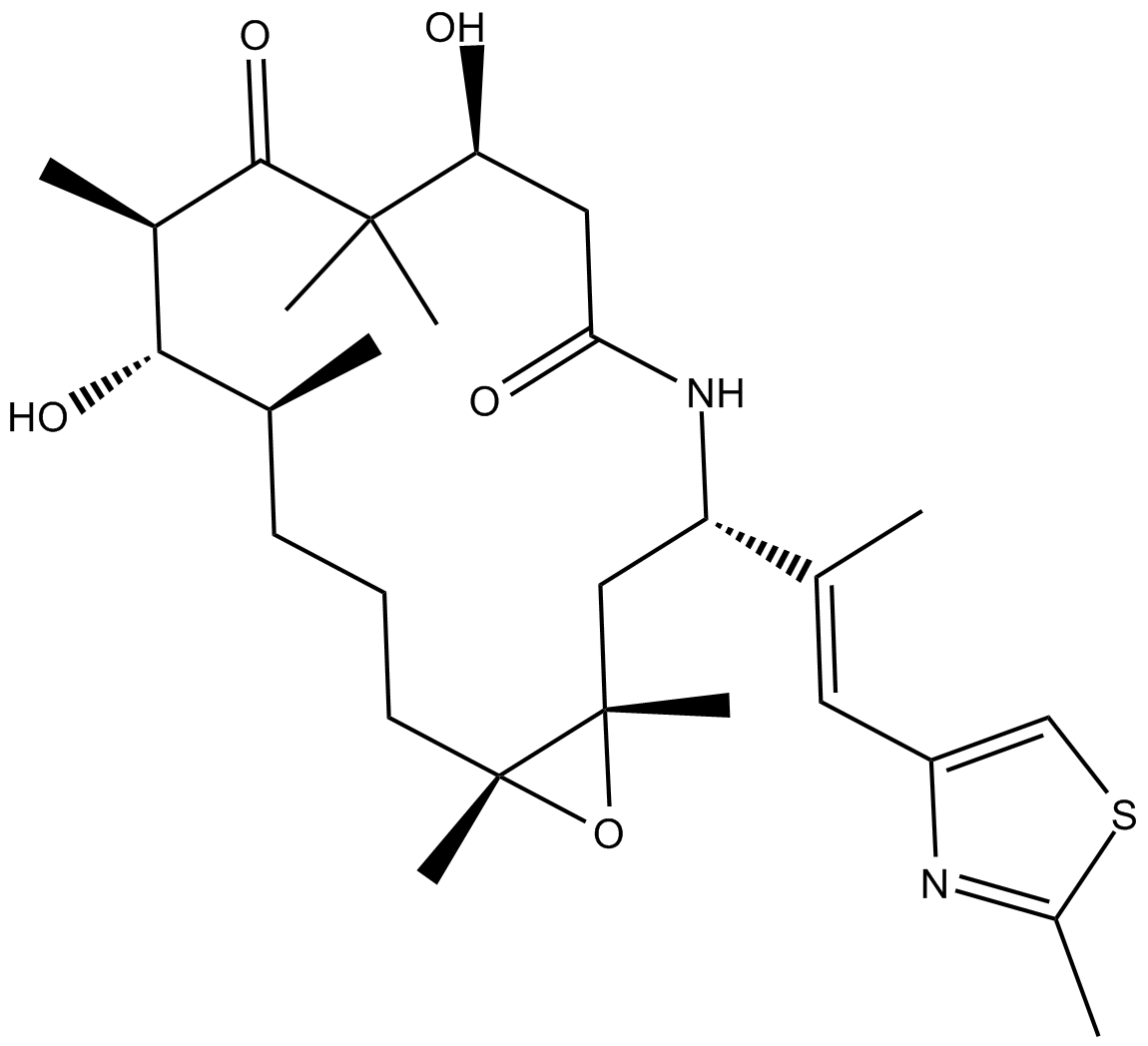
-
GC65331
IZCZ-3
IZCZ-3 is a potent c-MYC transcription inhibitor with antitumor activity.
-
GC41645
Jacaric Acid
Jacaric acid is a conjugated polyunsaturated fatty acid first isolated from seeds of Jacaranda plants.
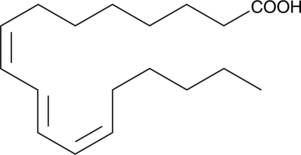
-
GC18699
JCP174
An inhibitor of palmitoyl protein thioesterase-1
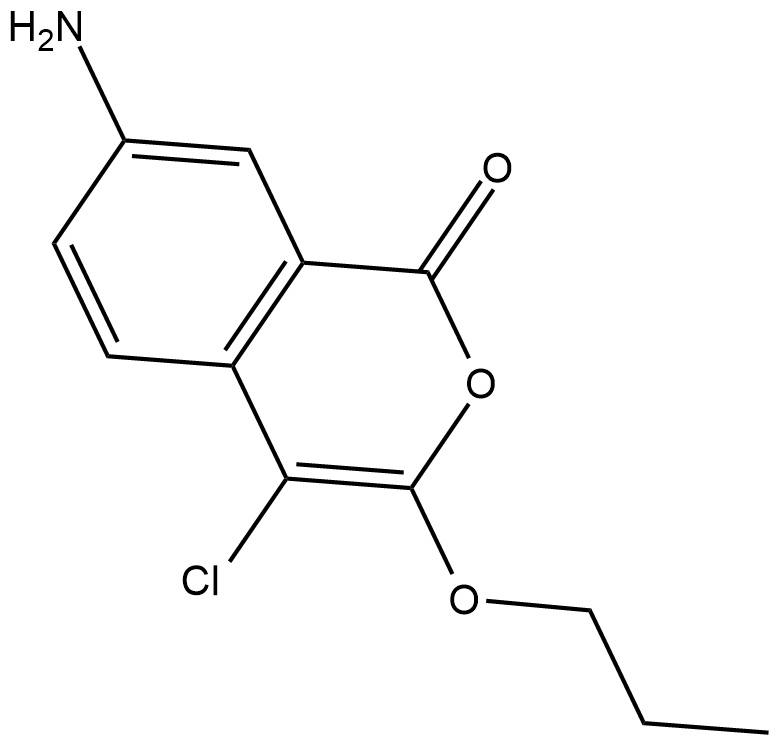
-
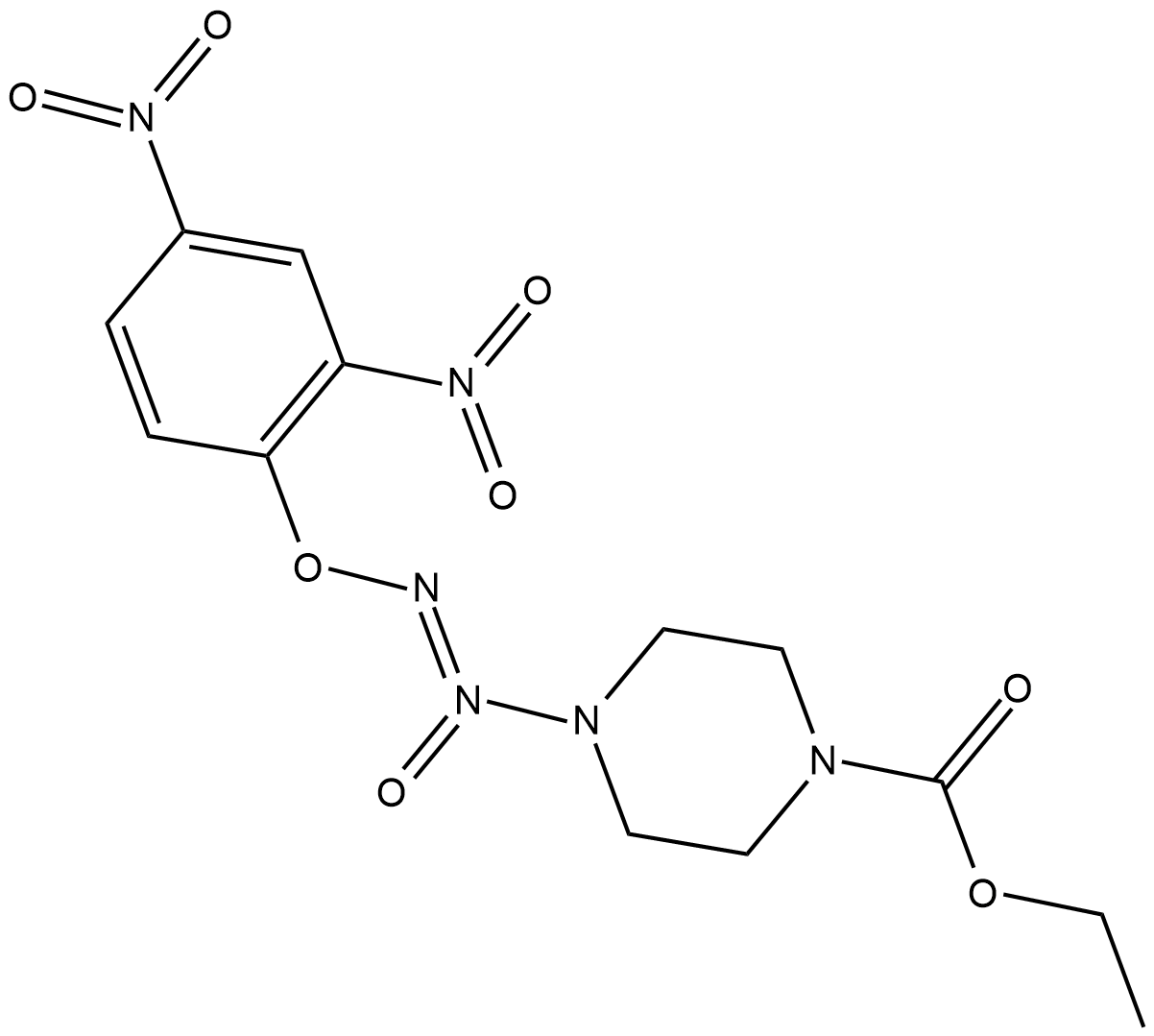
GC18734
JS-K
JS-K is a nitric oxide (NO) donor that reacts with glutathione to generate NO at physiological pH.
-
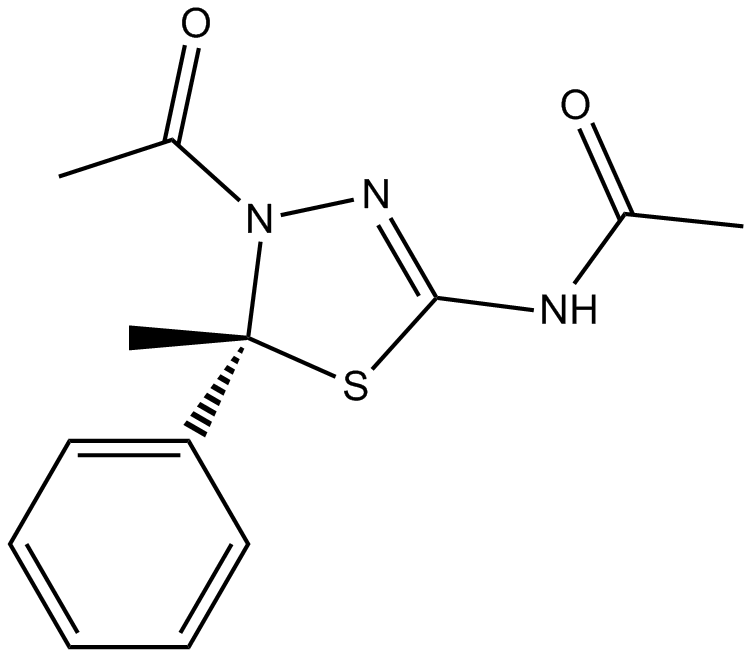
GC16167
K 858
K858 Racemic is an ATP-uncompetitive inhibitor of kinesin Eg5 with an IC50 of 1.3 μM.
-
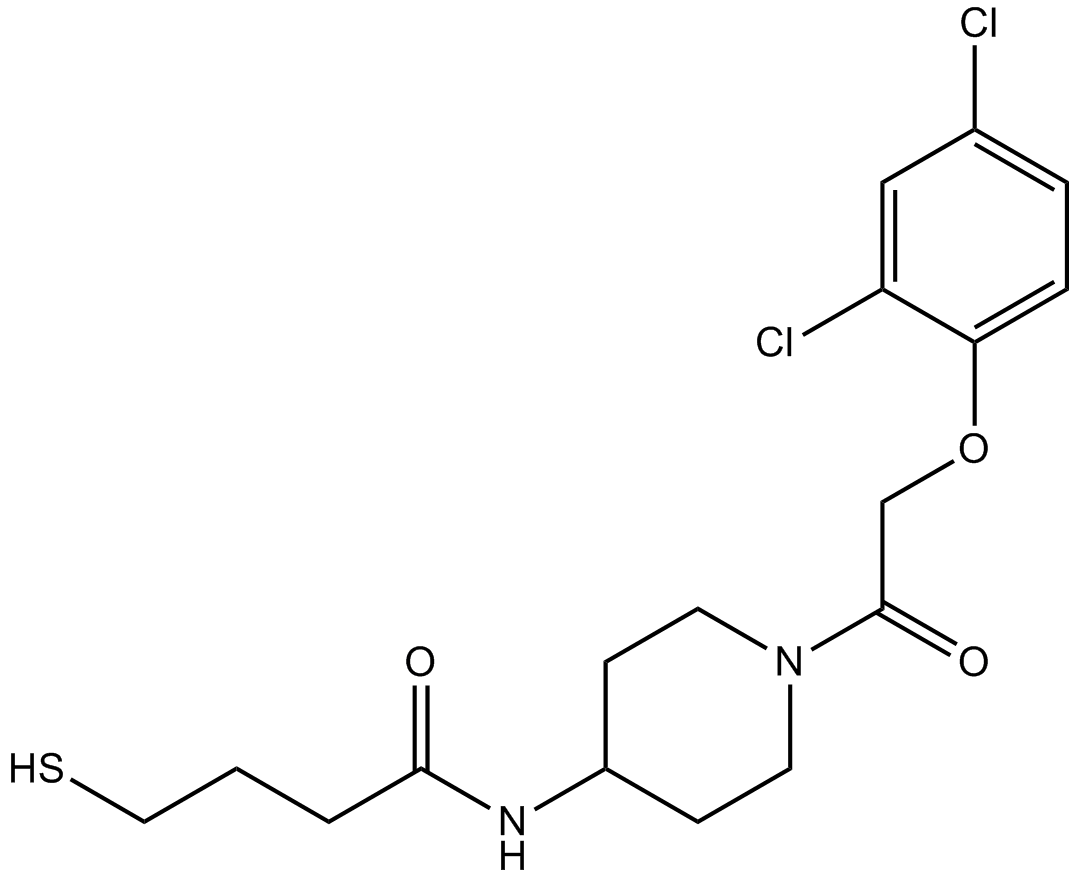
GC17388
K-Ras(G12C) inhibitor 6
K-Ras (G12C) inhibitor
-
GC14230
K03861
K03861 (K03861) is a type II CDK2 inhibitor with Kd of 8.2 nM. K03861 (K03861) inhibits CDK2 activity by competing with binding of activating cyclins.
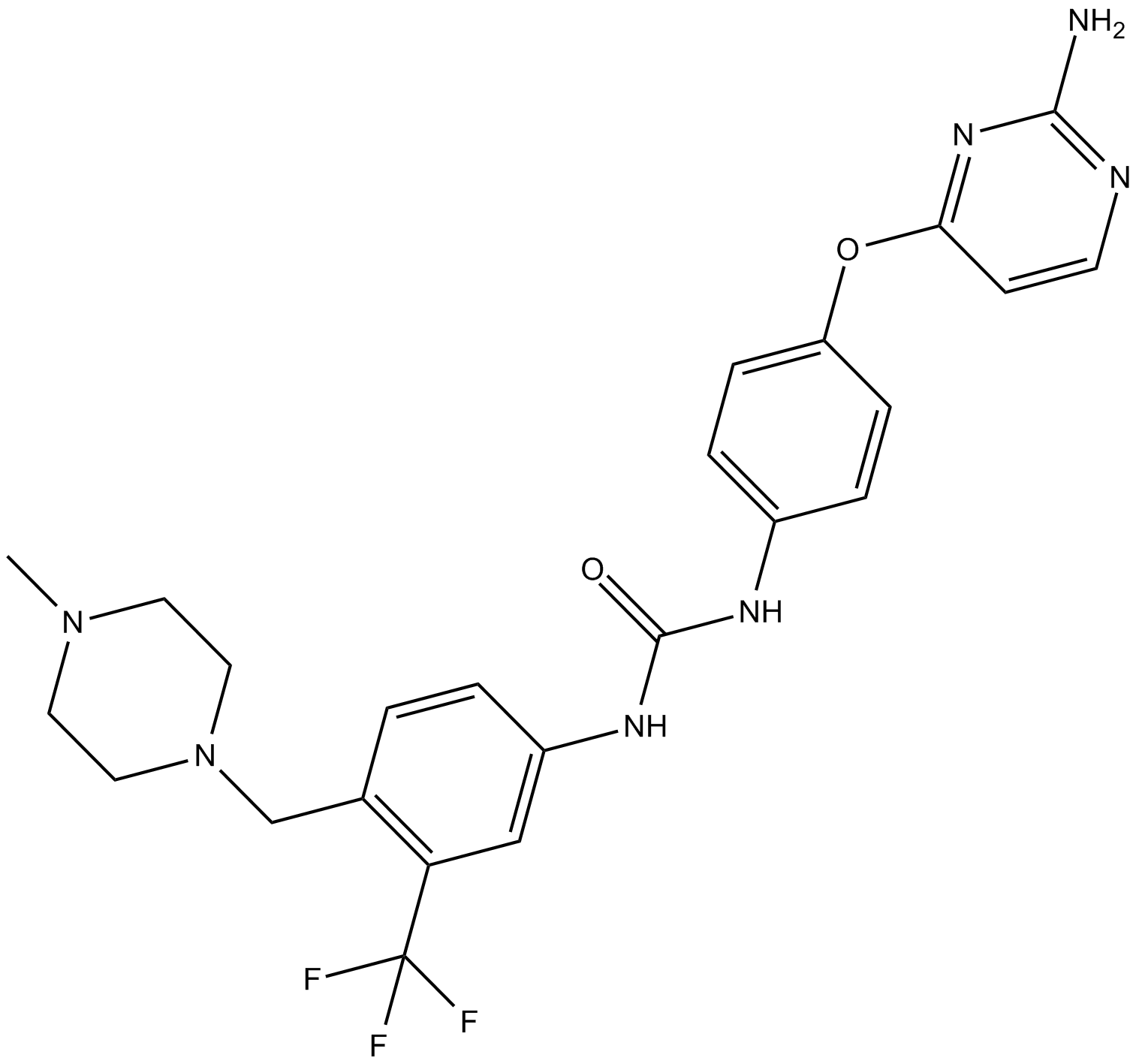
-
GC43994
KAPA (hydrochloride)
Biotin is a growth factor that plays an important role in numerous reactions that are critical to microorganisms, plants, and animals.
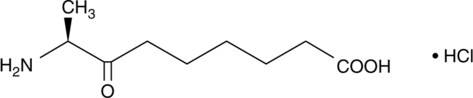
-
GC43995
Kazusamycin B
Kazusamycin B is a bacterial metabolite originally isolated from Streptomyces.
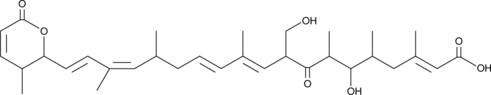
-
GC14182
Kenpaullone
CDK1/cyclin B and GSK-3β inhibitor
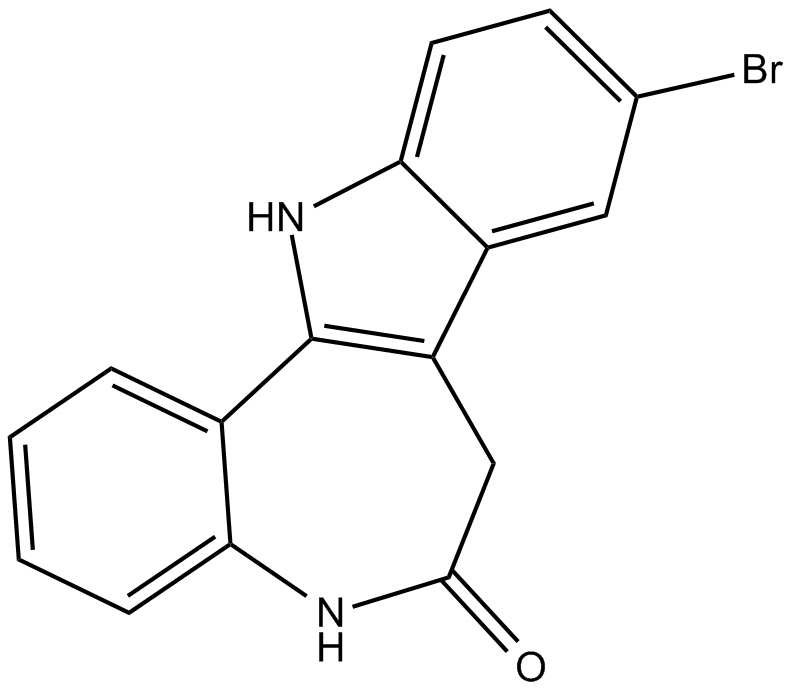
-
GC14589
KF 38789
Selective inhibitor of P-selectin-mediated cell adhesion
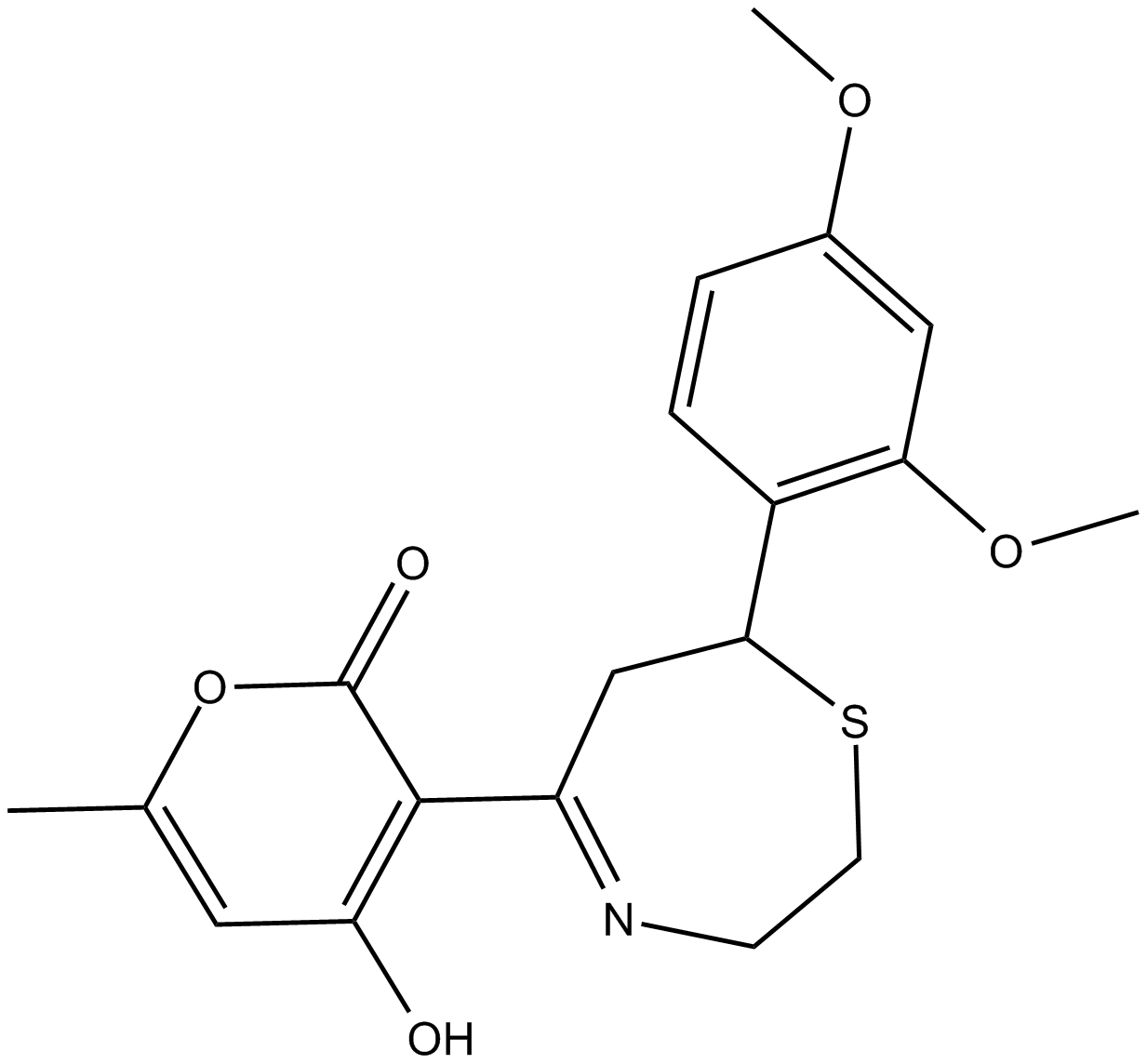
-
GC16603
Kif15-IN-1
potent Kif15 kinesin inhibitor
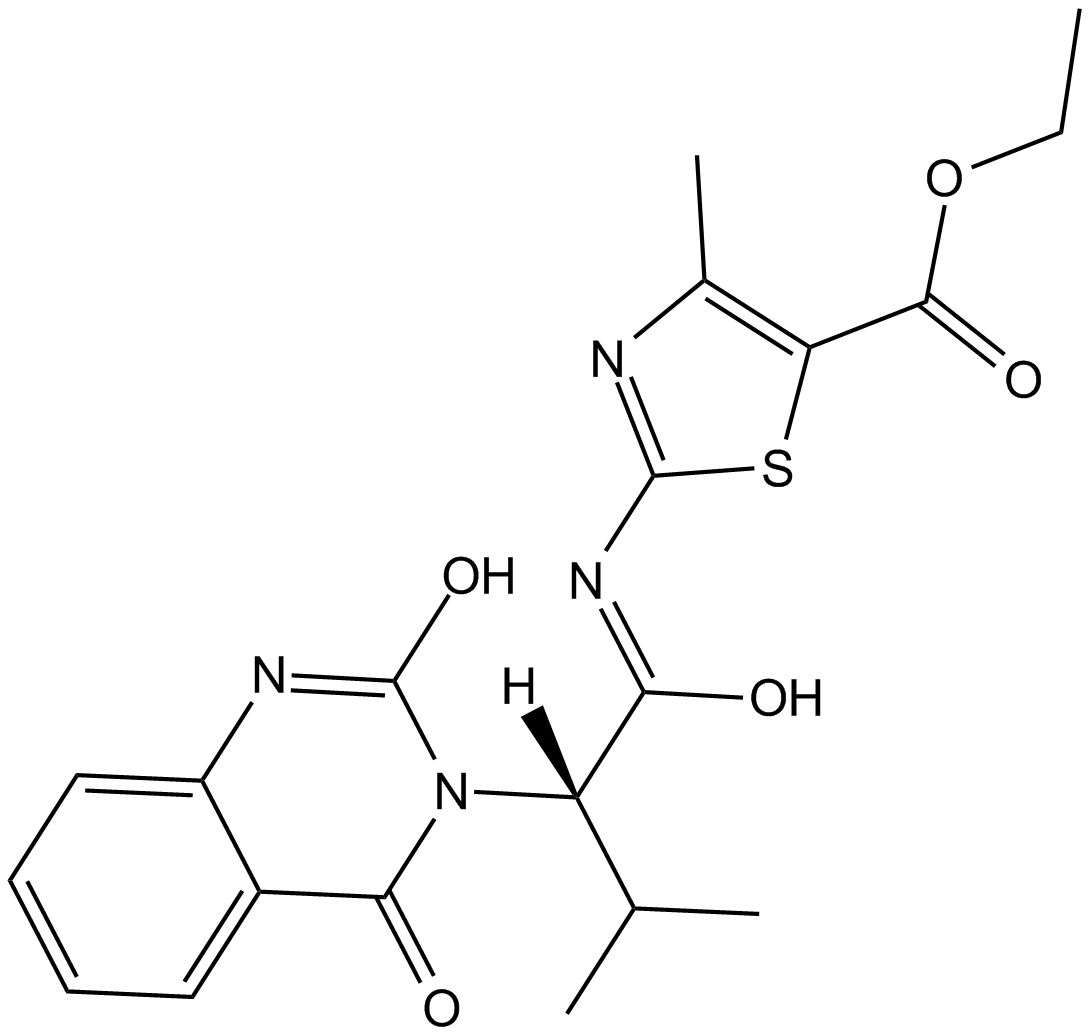
-
GC17994
Kif15-IN-2
potent Kif15 kinesin inhibitor
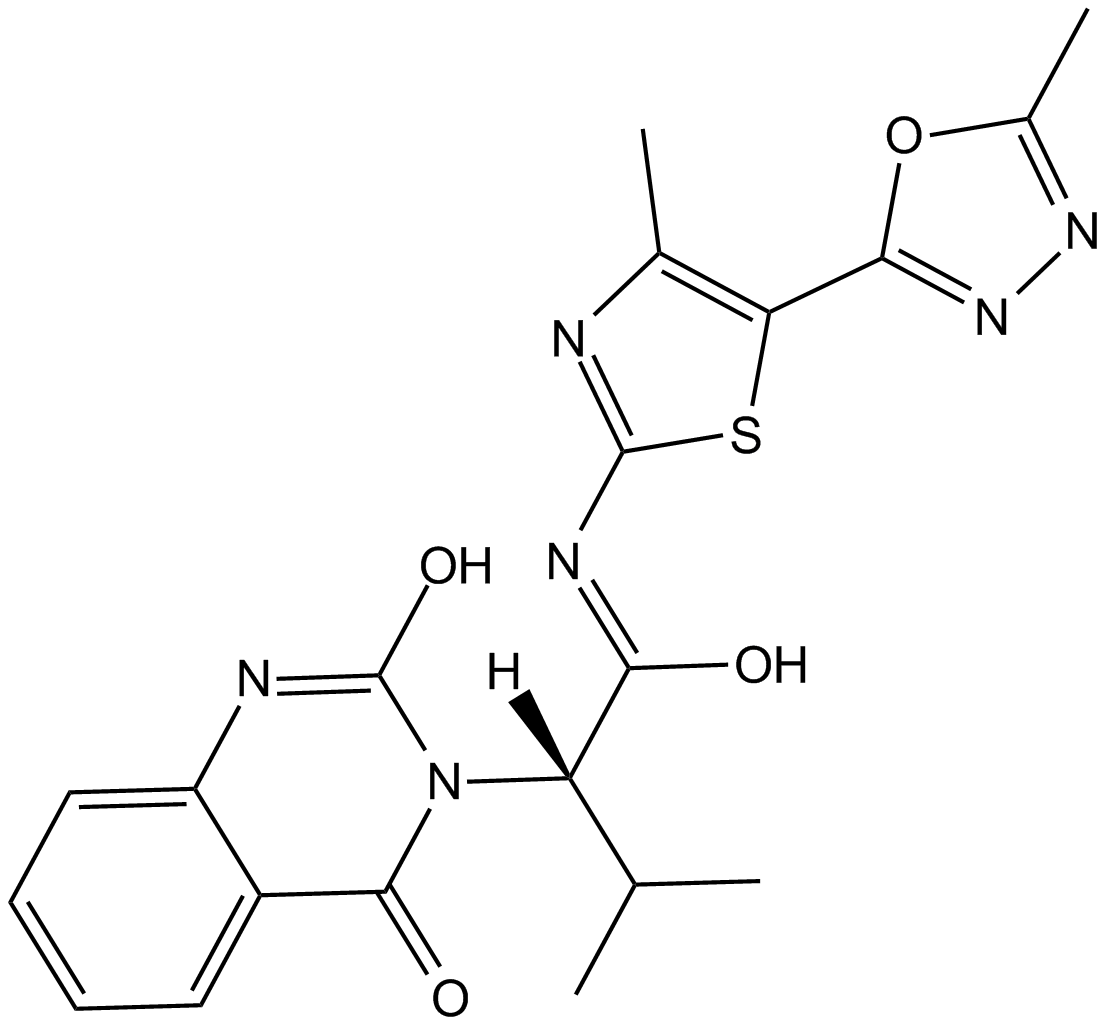
-
GC64085
KIF18A-IN-1
KIF18A-IN-1 is a mitotic kinesin KIF18A inhibitor extracted from patent WO2021026098A1 example 100-13.
-
GC64808
KIF18A-IN-2
KIF18A-IN-2 is a potent KIF18A inhibitor (IC50=28 nM). KIF18A-IN-2 causes significant mitotic arrest and increases the number of mitotic cells in tumor tissues. KIF18A-IN-2 can be used for researching cancer.
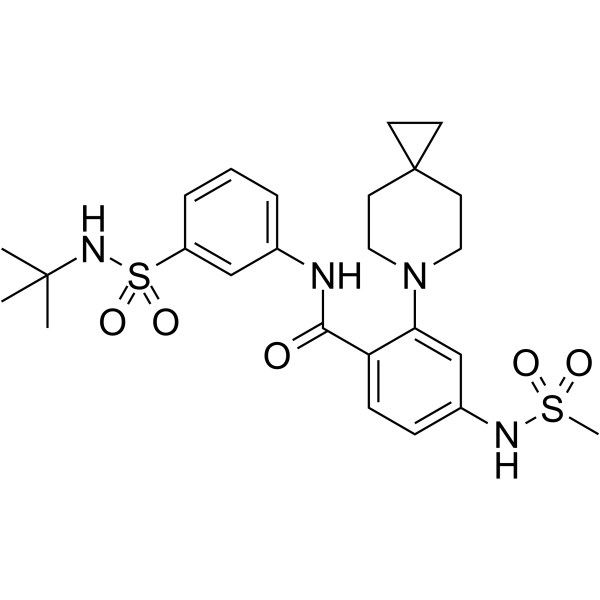
-
GC64857
KIF18A-IN-3
KIF18A-IN-3 is a potent KIF18A inhibitor (IC50=61 nM). KIF18A-IN-3 causes significant mitotic arrest and increases the number of mitotic cells in tumor tissues. KIF18A-IN-3 can be used for researching cancer.
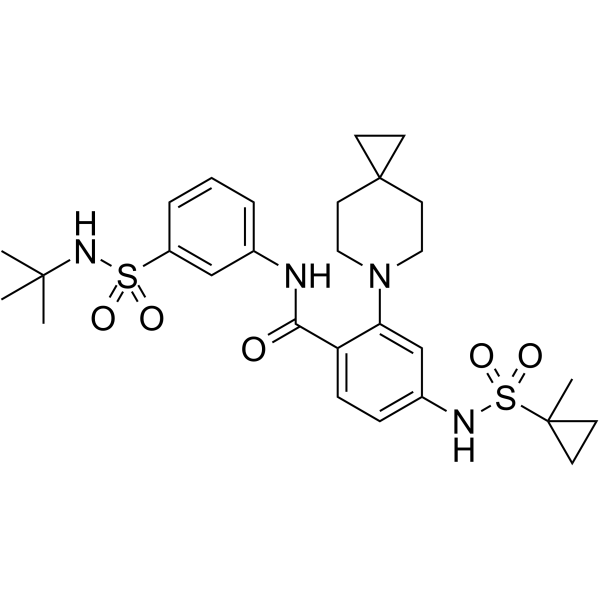
-
GC69333
KIF18A-IN-7
KIF18A-IN-7 (Compound 22) is an orally active inhibitor of KIF18A that inhibits the microtubule-dependent ATPase activity of KIF18A with an IC50 value of 9.4 nM.
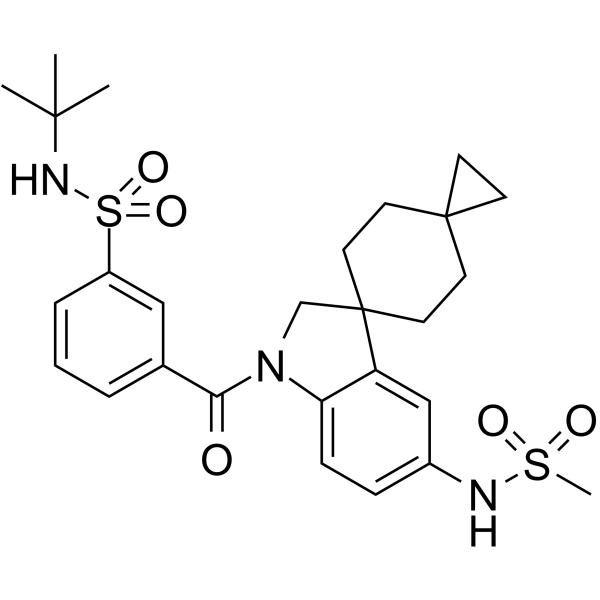
-
GC14413
KJ Pyr 9
c-Myc inhibitor, cell-permeable
-
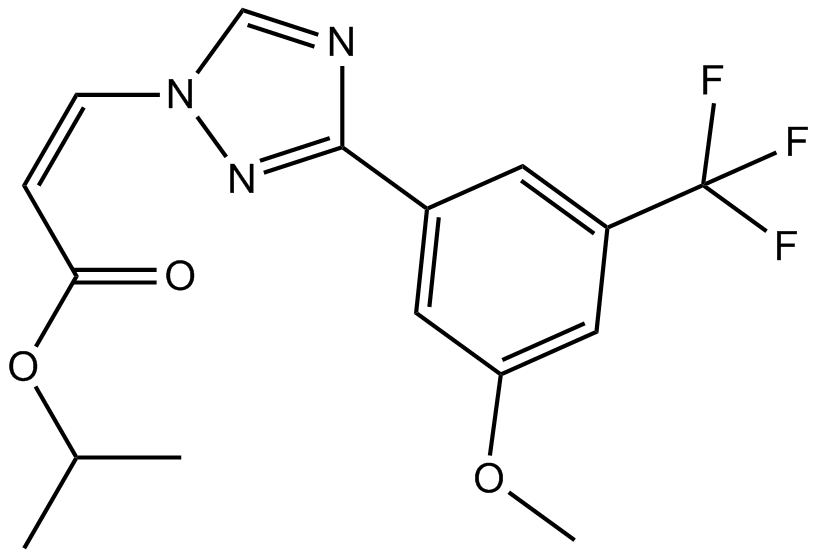
GC14517
KPT-185
CRM1 inhibitor,selective and irrversible
-
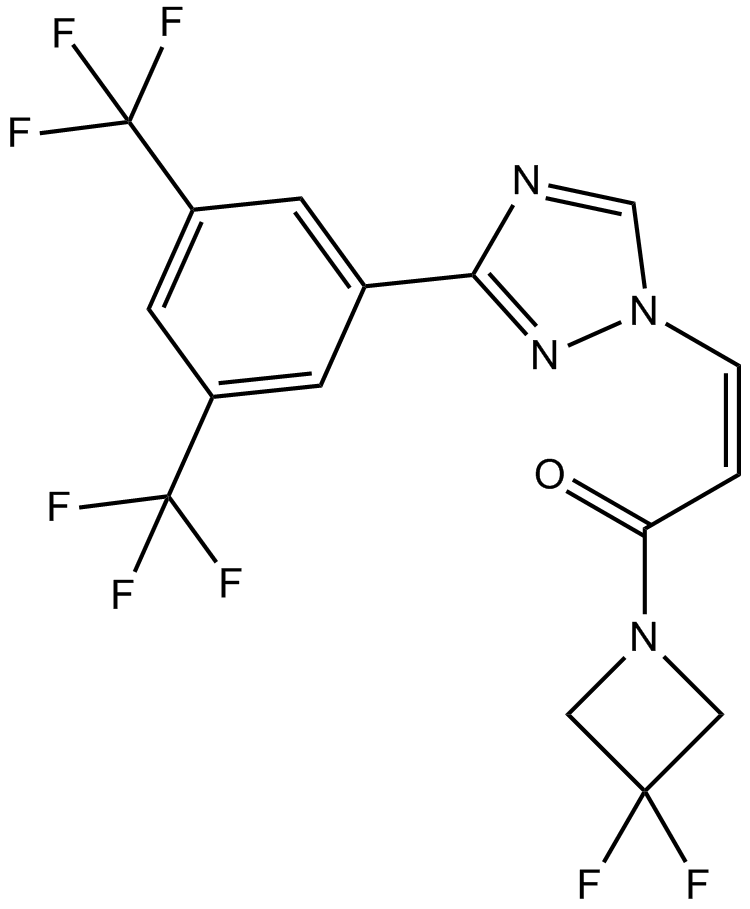
GC12142
KPT-276
inhibitor of nuclear export (SINE) and CRM1, orally bioavailable
-
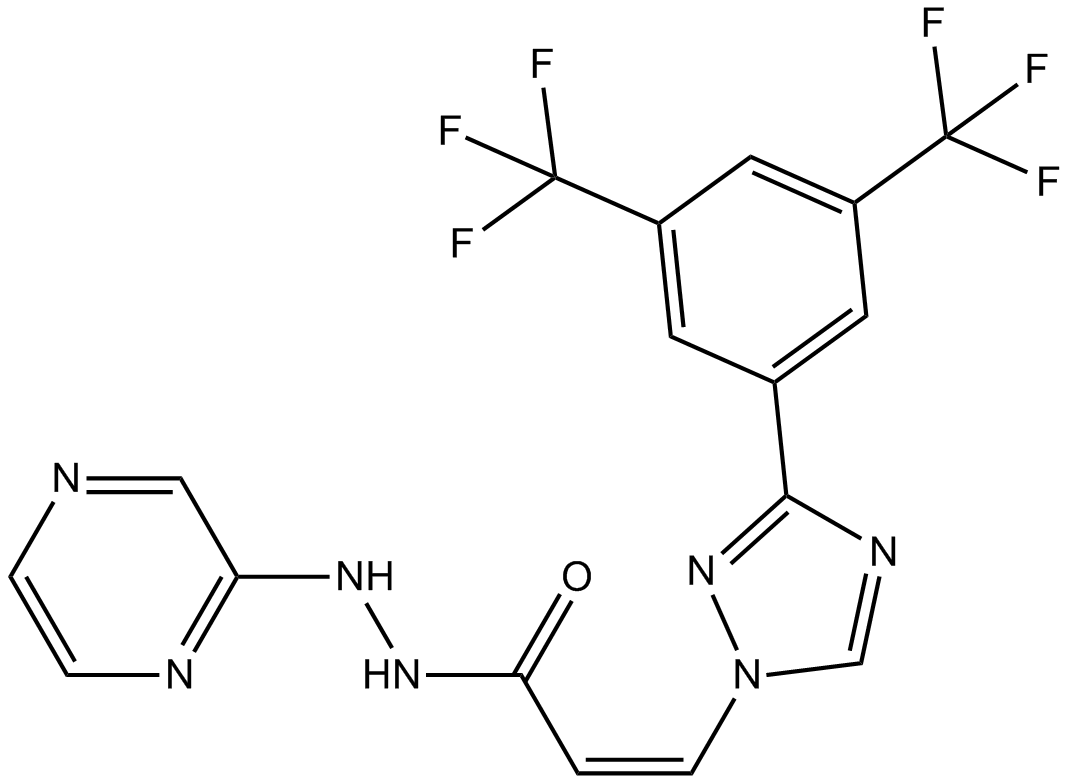
GC12467
KPT-330
CRM1 inhibitor, orally bioavailable and selective
-
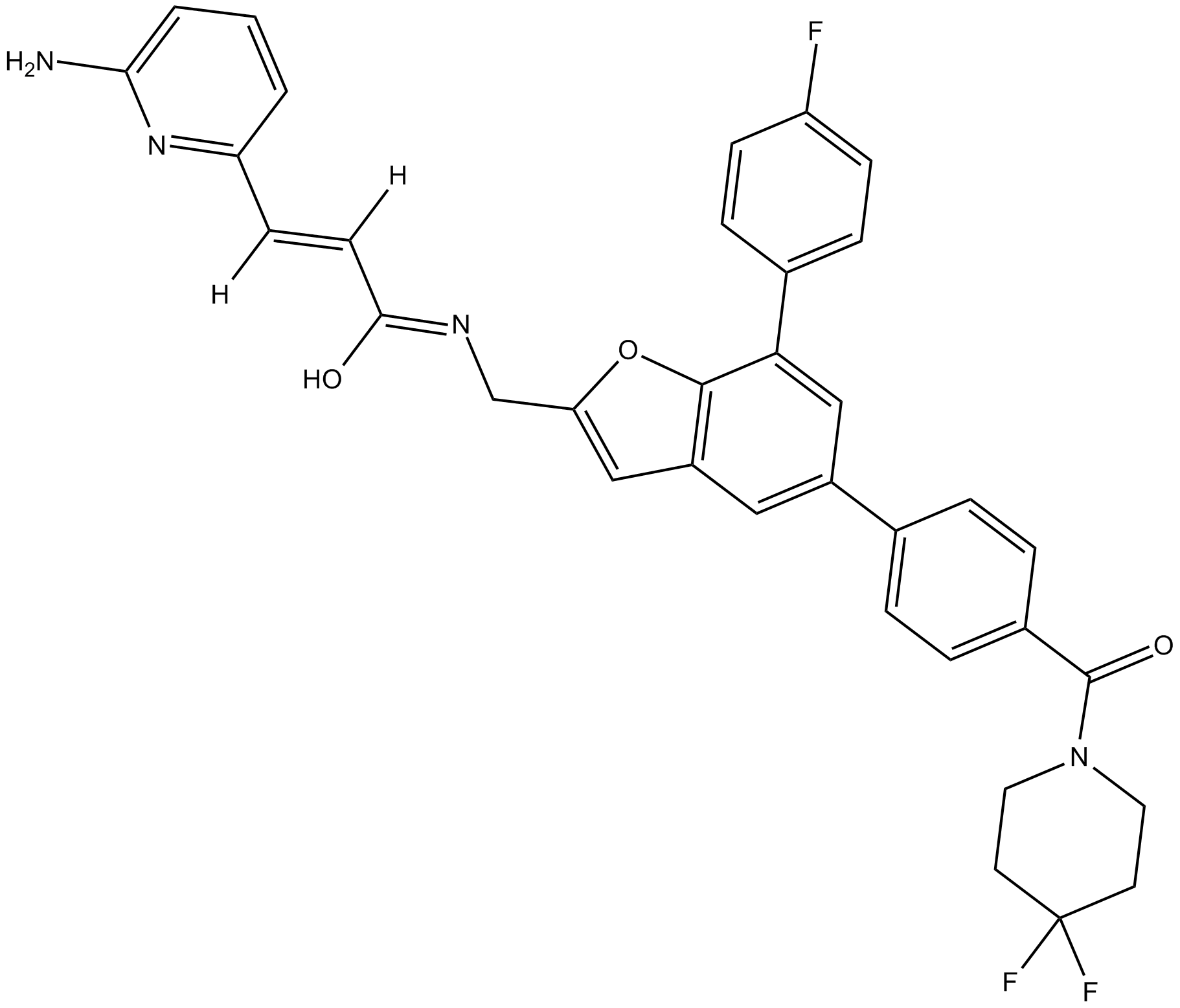
GC14615
KPT-9274
Orally acitve allosteric inhibitor of PAK4
-
GC18909
KRIBB3
KRIBB3 is an Hsp27 and microtubule inhibitor that inhibits migration and invasion of MDA-MB-231 cells in vitro in an Hsp27-dependent manner.
-
GC33377
KSI-3716
KSI-3716 is a potent c-Myc inhibitor that blocks c-MYC/MAX binding to target gene promoters. KSI-3716 is an effective intravesical chemotherapy agent for bladder cancer.
-
GC33404
KU 59403
KU 59403 is a potent ATM inhibitor, with IC50 values of 3 nM, 9.1 μM and 10 μM for ATM, DNA-PK and PI3K, respectively.
-
GC12054
KU-60019
ATM kinase inhibitor,potent and selective
-
GC47532
KUS121
A VCP modulator
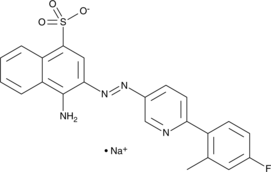
-
GC14288
KX2-391
KX2-391 (KX2-391) is an inhibitor of Src that targets the peptide substrate site of Src, with GI50 of 9-60 nM in cancer cell lines.
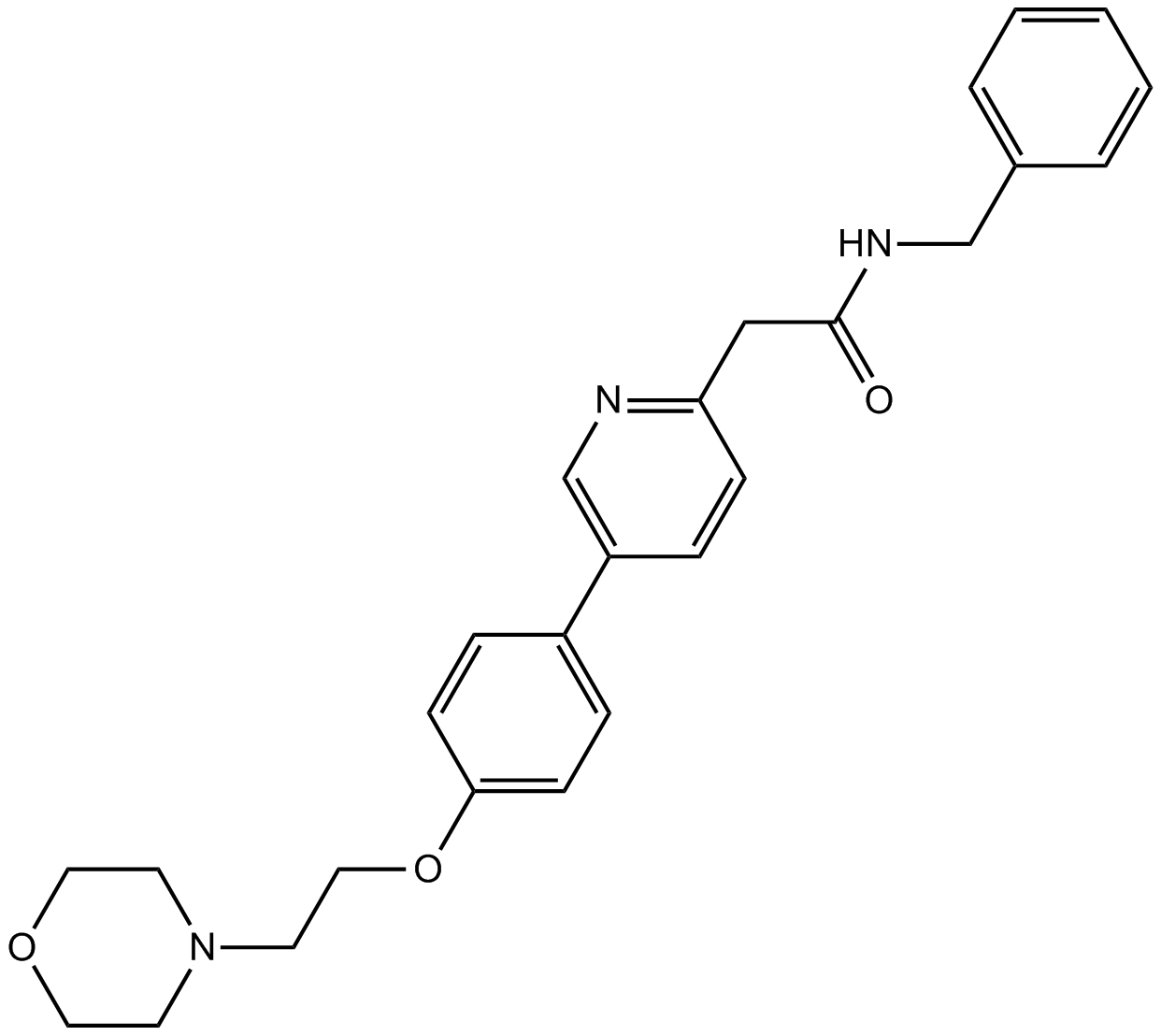
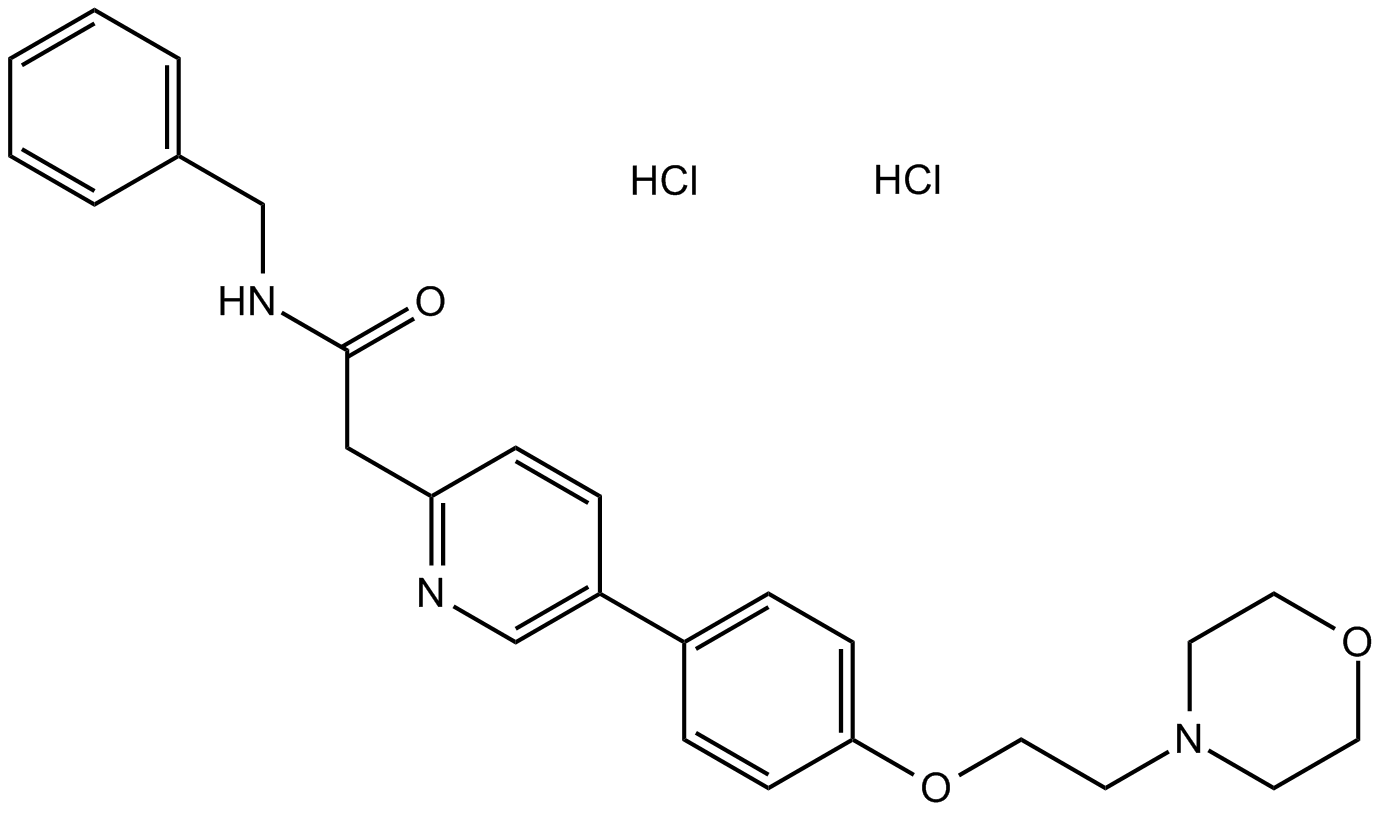
-
GC10222
KX2-391 dihydrochloride
A Src kinase inhibitor
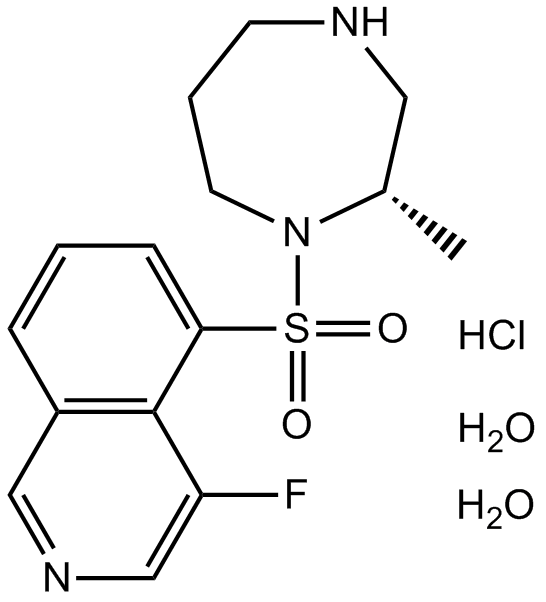
-
GC12817
K–115 hydrochloride dihydrate
K-115 hydrochloride dihydrate (K-115) is a specific inhibitor of ROCK, with IC50s of 19 and 51 nM for ROCK2 and ROCK1, respectively.
-
GC44020
L-681,217
L-681,217 is an antibiotic originally isolated from S.
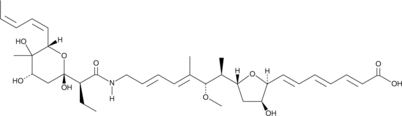
-
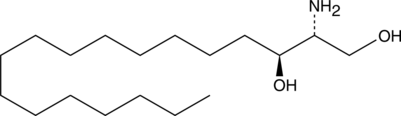
GC40793
L-erythro Sphinganine (d18:0)
L-erythro Sphinganine (d18:0) is a synthetic bioactive sphingolipid and stereoisomer of sphinganine (d18:0) and D-threo sphinganine.
-
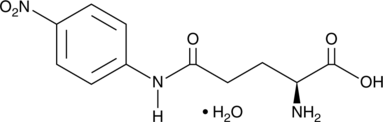
GC49547
L-Glutamic Acid γ-p-Nitroanilide (hydrate)
A colorimetric substrate for γ-glutamyl transpeptidase
-
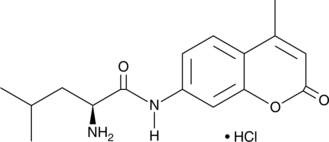
GC49205
L-Leu-AMC (hydrochloride)
L-Leucine-7-amido-4-methylcoumarin (Leu-AMC) hydrochloride is a bright blue fluorogenic peptidyl substrate for LAP3 (leucine aminopeptidase).
-
GC44081
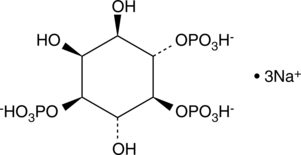
L-myo-Inositol-1,4,5-triphosphate (sodium salt)
Ins(1,4,5)P3 is an isomer of the biologically important D-myo-inositol-1,4,5-triphosphate.
-
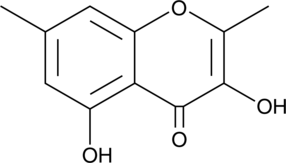
GC41520
Lachnone A
Lachnone A is a chromone fungal metabolite originally isolated from Lachnum.
-
GC49471
LAMP1 Monoclonal Antibody (Clone Ly1C6)
For immunochemical analysis of LAMP1

-
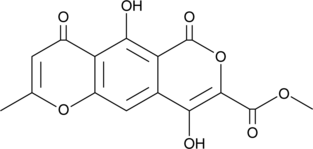
GC41272
Lateropyrone
Lateropyrone is a fungal metabolite produced by Fusarium species.
-
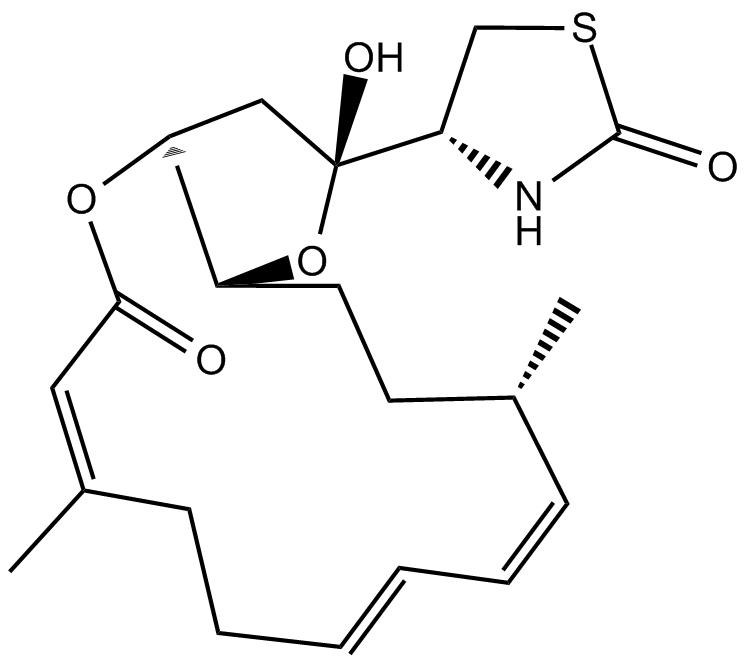
GC15671
Latrunculin A
A reversible inhibitor of actin assembly
-
GC50052
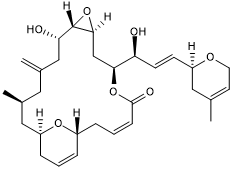
Laulimalide
Microtubule stabilzing agent
-
GC50214
LCB 03-0110 dihydrochloride
Potent c-SRC kinase inhibitor; also inhibits DDR2, BTK and Syk
-
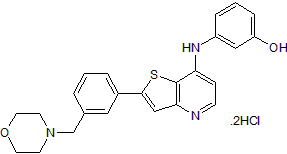
GC39209
LCH-7749944
LCH-7749944 (GNF-PF-2356) is a potent PAK4 inhibitor with an IC50 of 14.93 μM. LCH-7749944 effectively suppresses the proliferation of human gastric cancer cells through downregulation of PAK4/c-Src/EGFR/cyclin D1 pathway and induces apoptosis.
-
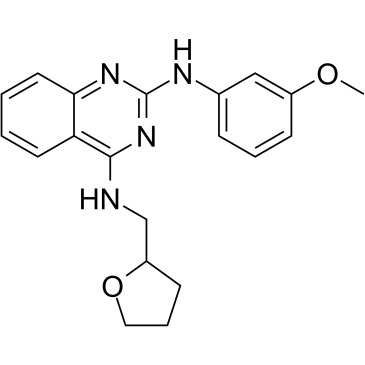
GC17067
LDC000067
CDK9 inhibitor, novel and highly specific
-
GC33086
LDN-192960
LDN-192960 is an inhibitor of Haspin and Dual-specificity Tyrosine-regulated Kinase 2 (DYRK2) with IC50s of 10 nM and 48 nM, respectively.
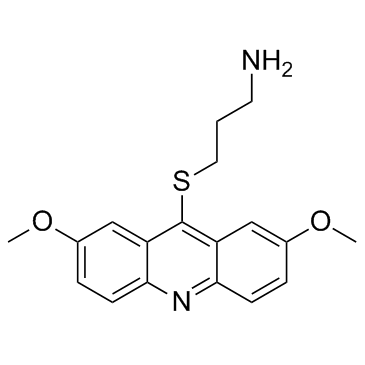
-
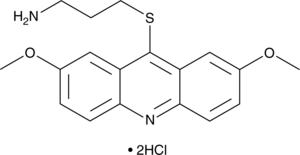
GC44047
LDN-192960 (hydrochloride)
LDN-192960 is an inhibitor of haspin protein kinase and dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 (DYRK2) with IC50 values of 10 and 48 nM, respectively.
-
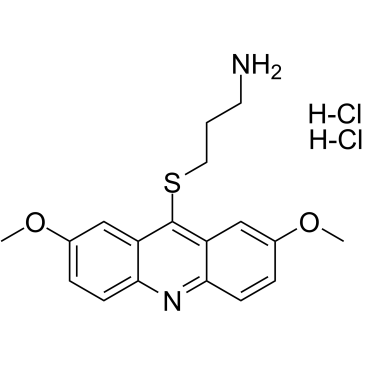
GC38403
LDN-192960 hydrochloride
LDN-192960 hydrochloride is an inhibitor of Haspin and Dual-specificity Tyrosine-regulated Kinase 2 (DYRK2) with IC50s of 10 nM and 48 nM, respectively.
-
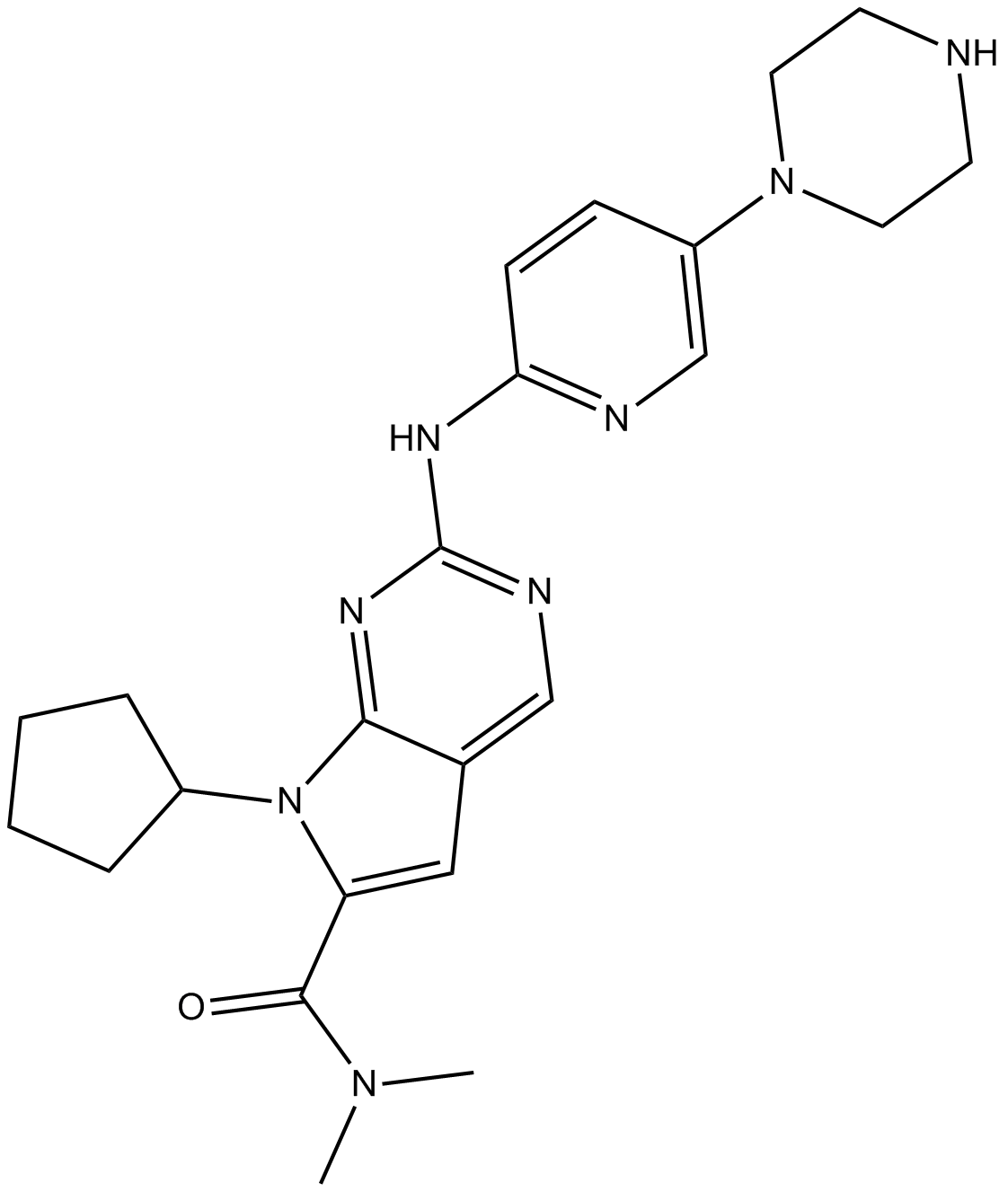
GC10842
LEE011
LEE011 (LEE01) is a highly specific CDK4/6 inhibitor with IC50 values of 10 nM and 39 nM, respectively, and is over 1,000-fold less potent against the cyclin B/CDK1 complex.
-
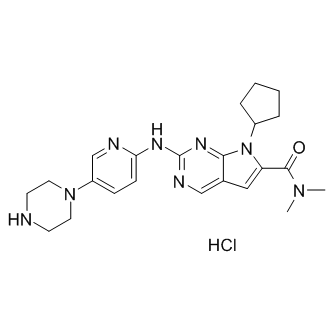
GC15922
LEE011 hydrochloride
LEE011 hydrochloride (LEE011 hydrochloride) is a highly specific CDK4/6 inhibitor with IC50 values of 10 nM and 39 nM, respectively, and is over 1,000-fold less potent against the cyclin B/CDK1 complex.
-
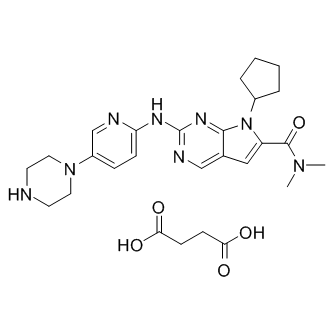
GC15377
LEE011 succinate
LEE011 succinate (LEE011 succinate) is a highly specific CDK4/6 inhibitor with IC50 values of 10 nM and 39 nM, respectively, and is over 1,000-fold less potent against the cyclin B/CDK1 complex.
-
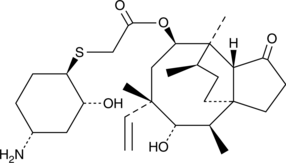
GC49751
Lefamulin
Lefamulin (BC-3781) is an orally active antibiotic.
-
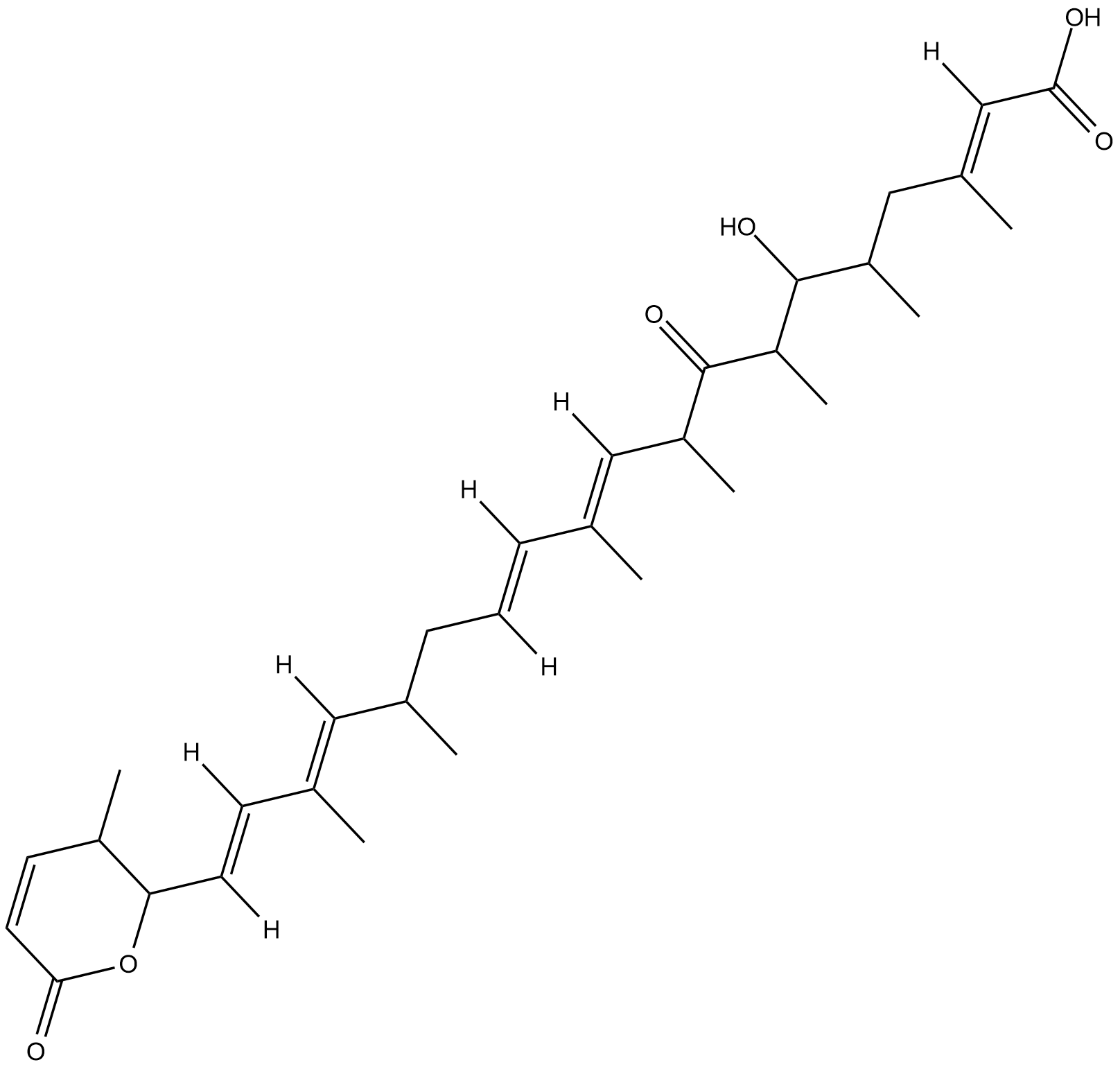
GC18345
Leptomycin A
Leptomycin A is an inhibitor of CRM1 (exportin 1) that blocks CRM1 interaction with nuclear export signals, preventing the nuclear export of a broad range of proteins.
-
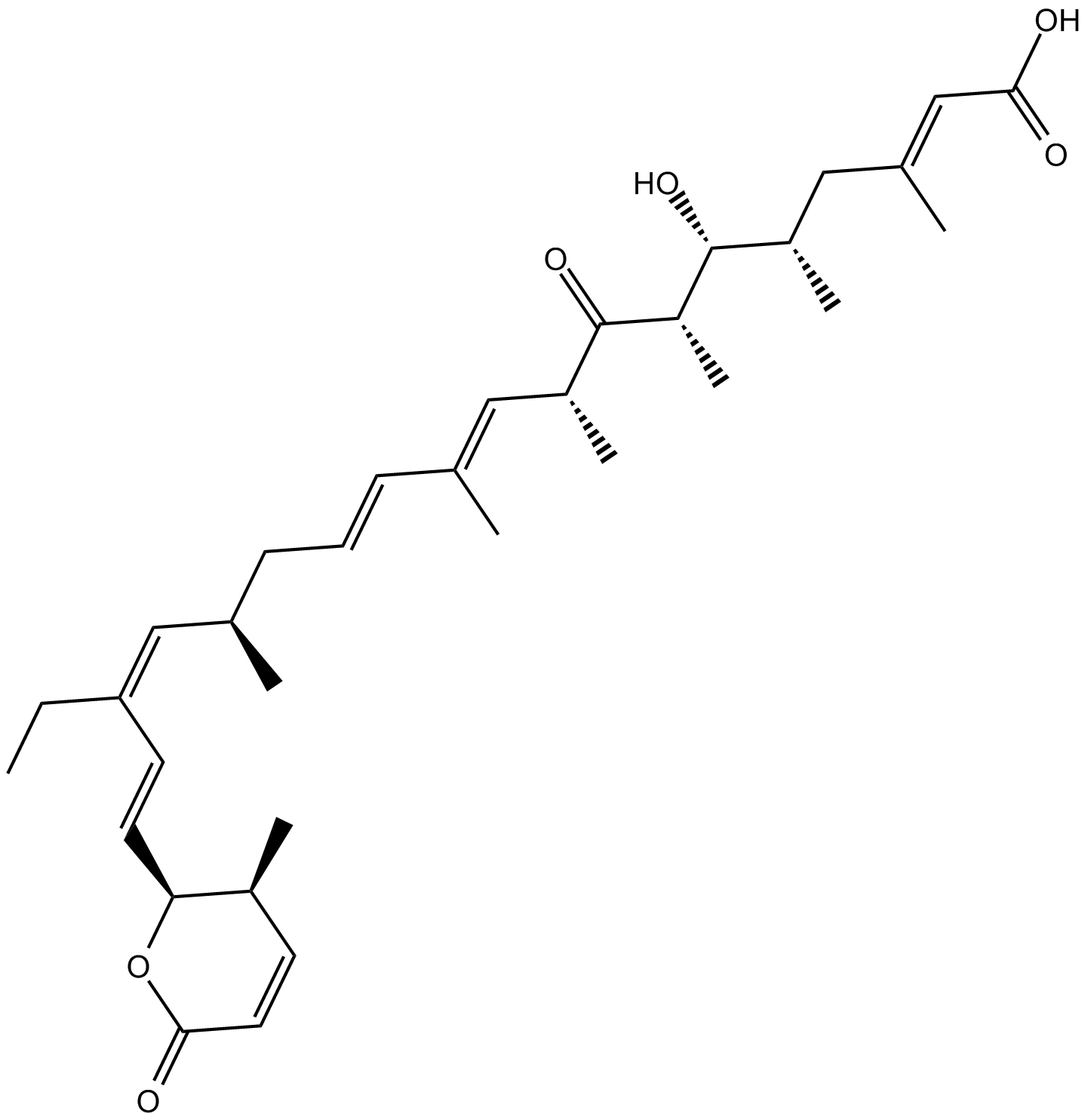
GC10954
Leptomycin B
Leptomycin B (LMB) is an inhibitor of chromosome region maintenance 1 (CRM1) that is one of the nuclear protein exporters.
-
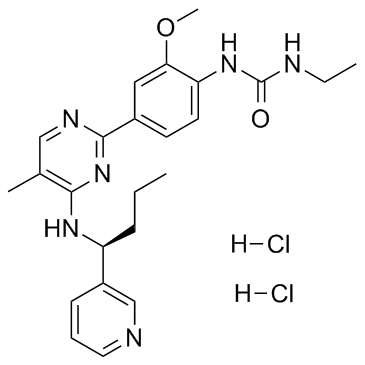
GC33175
Lexibulin dihydrochloride (CYT-997 dihydrochloride)
Lexibulin dihydrochloride (CYT-997 dihydrochloride) (CYT-997 dihydrochloride) is a potent and orally active tubulin polymerisation inhibitor with IC50s of 10-100 nM in cancer cell lines; with potent cytotoxic and vascular disrupting activity in vitro and in vivo. Lexibulin dihydrochloride (CYT-997 dihydrochloride) induces cell apoptosis and induces mitochondrial ROS generation in GC cells.
-
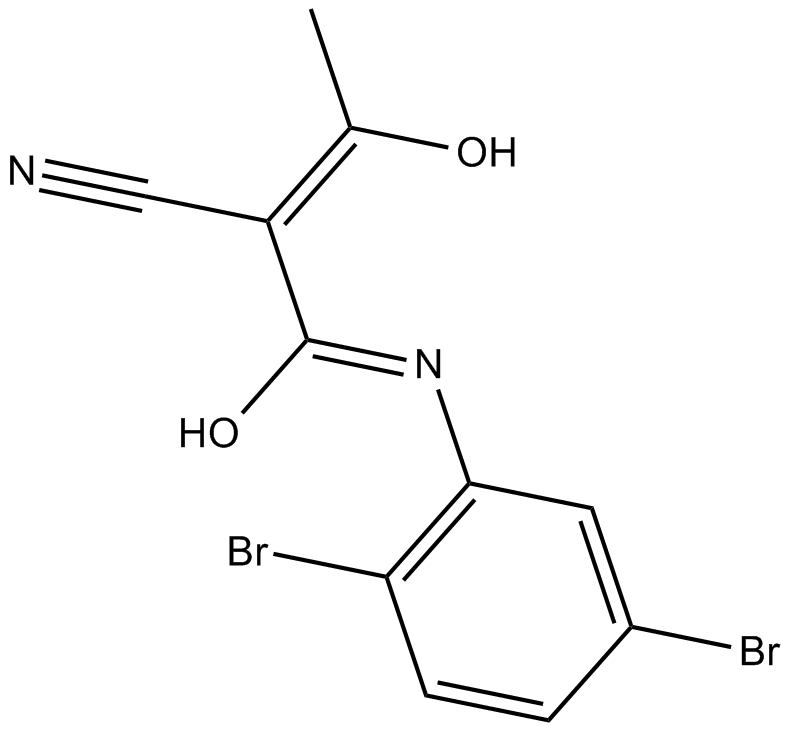
GC14857
LFM-A13
BTK-specific tyrosine kinase inhibitor
-
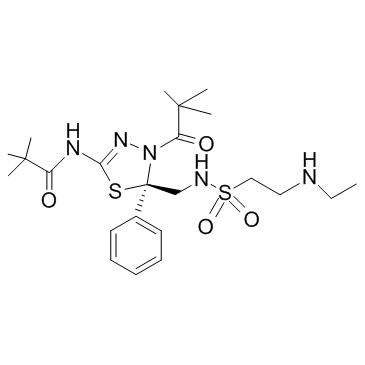
GC31803
Litronesib (LY-2523355)
Litronesib (LY-2523355) (LY2523355) is a selective mitosis-specific kinesin Eg5 inhibitor, with antitumor activity.
-
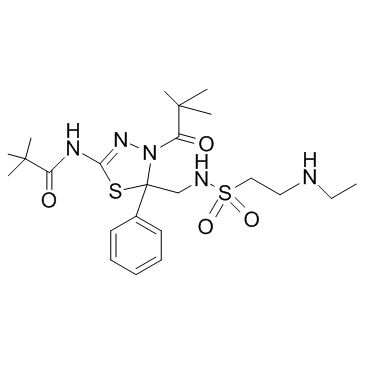
GC30393
Litronesib Racemate (LY-2523355 Racemate)
Litronesib Racemate (LY-2523355 Racemate) (LY2523355 Racemate) is the racemate of litronesib.
-
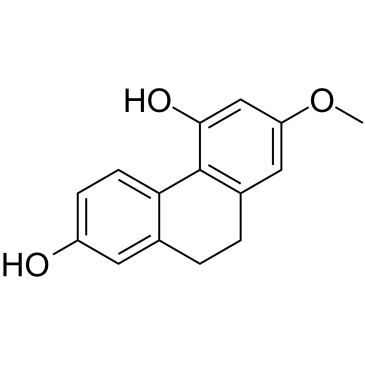
GC61014
Lusianthridin
Lusianthridin, a pure compound from Dendrobium venustum, have an anti-migratory effect. Lusianthridin enhances c-Myc degradation through the inhibition of Src-STAT3 signaling.
-
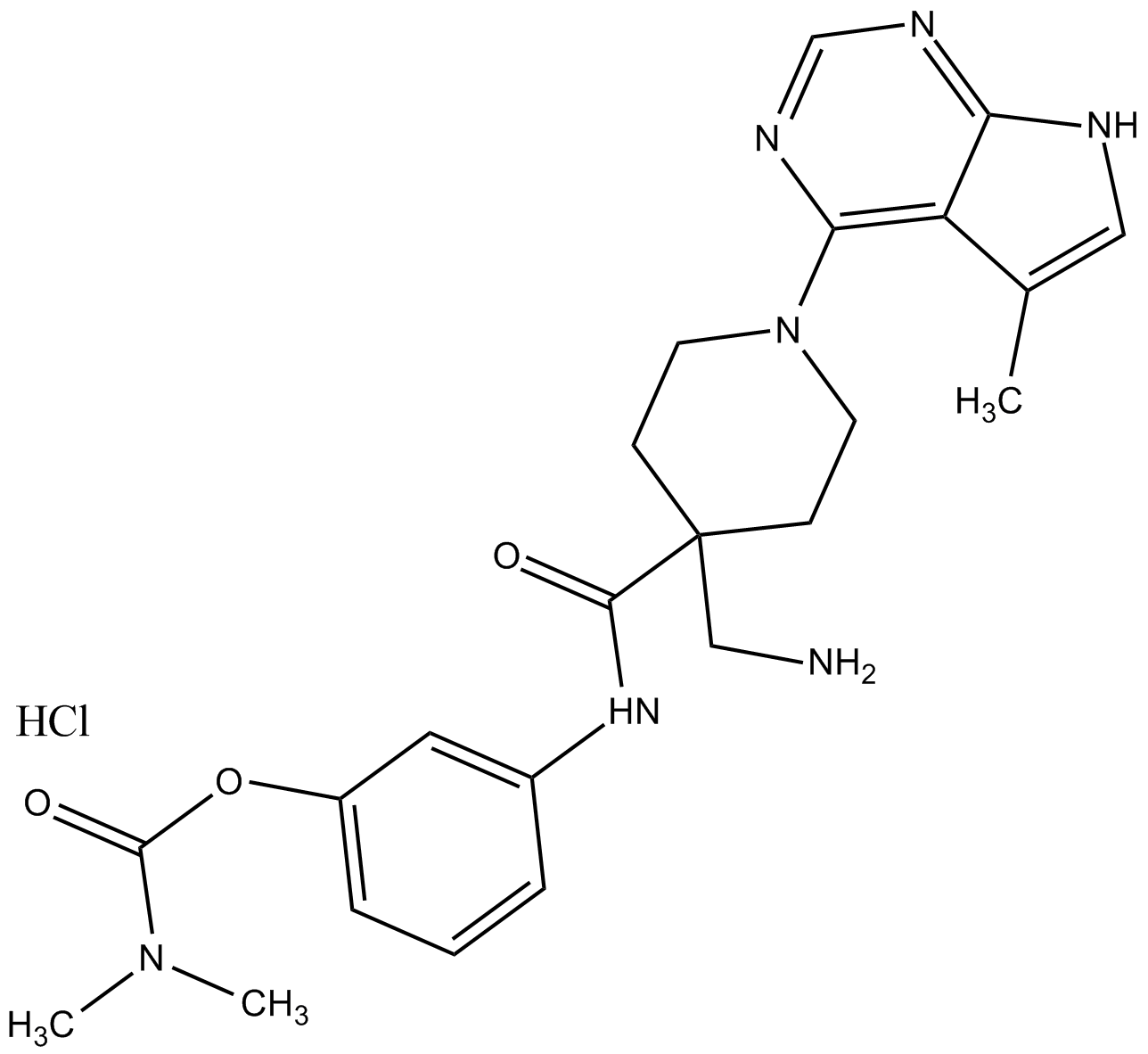
GC12402
LX7101 HCL
LX7101 HCL is a potent inhibitor of LIMK and ROCK2 with IC50 values of 24, 1.6 and 10 nM for LIMK1, LIMK2 and ROCK2, respectively; also inhibits PKA with an IC50 less than 1 nM.
-
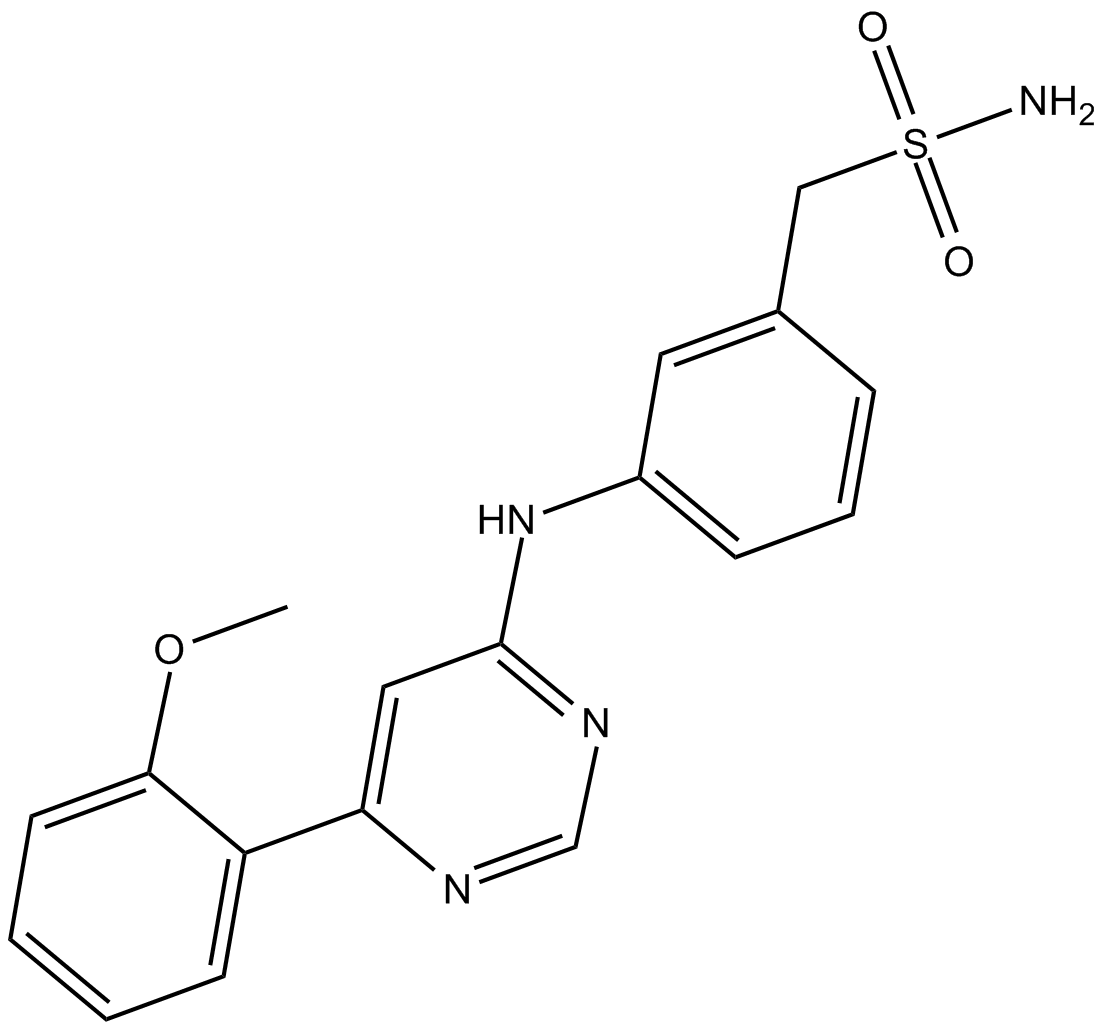
GC15741
LY2603618
LY2603618 (LY2603618) is a potent and selective inhibitor of Chk1 with an IC50 of 7 nM.