Chromatin/Epigenetics
Epigenetics
Epigenetics means above genetics. It determines how much and whether a gene is expressed without changing DNA sequences. Epigenetic regulations include, 1. DNA methylation: the addition of methyl group to DNA, converting cytosine to 5-methylcytosine, mostly at CpG sites; 2. Histone modifications: posttranslational modificationEpigeneticss of histone proteins including acetylation, methylation, ubiquitylation, phosphorylation and sumoylation; 3. miRNAs: non-coding microRNA downregulating gene expression; 4. Prions: infectious proteins viewed as epigenetic agents capable of inducing a phenotype without changing the genome.
Targets for Chromatin/Epigenetics
- Bromodomain(52)
- Aurora Kinase(89)
- DNA Methyltransferase(40)
- HDAC(218)
- Histone Acetyltransferases(67)
- Histone Demethylases(98)
- Histone Methyltransferase(212)
- HIF(101)
- JAK(178)
- MBT Domains(1)
- PARP(128)
- Pim(35)
- Protein Ser/Thr Phosphatases(41)
- RNA Polymerase(8)
- Sirtuin(84)
- Sphingosine Kinase-2(1)
- Polycomb repressive complex(2)
- SUMOylation(3)
- PAD(18)
- Epigenetic Reader Domain(207)
- MicroRNA(13)
- Protein Arginine Deiminase(12)
- Chromodomain(1)
- Citrullination(15)
- DNA/RNA Demethylation(1)
- DNA/RNA Methylation(6)
- Histone Deacetylation(38)
- Histones/Histone Peptides(7)
- PHD Domains(0)
- Tandem Tudor & Tudor-like Domains(1)
- PRMT(2)
Products for Chromatin/Epigenetics
- Cat.No. Product Name Information
-
GC12458
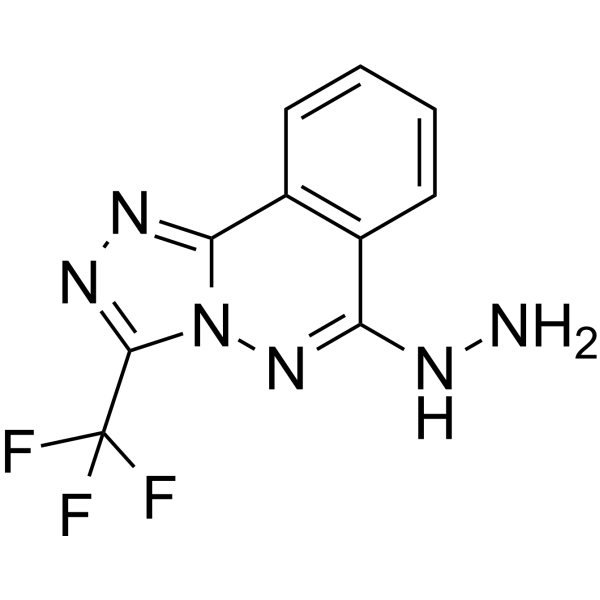
N-Oxalylglycine
cell permeable inhibitor of α-ketoglutarate-dependent enzymes, including JMJD2A, JMJD2C, and JMJD2E.
-
GC40496
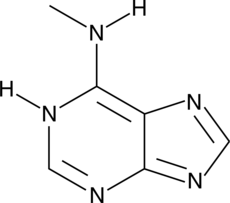
N6-Methyladenine
N6-Methyladenine is a modified purine that is commonly found in genomes of prokaryotes, protists, and plants.
-
GC17676
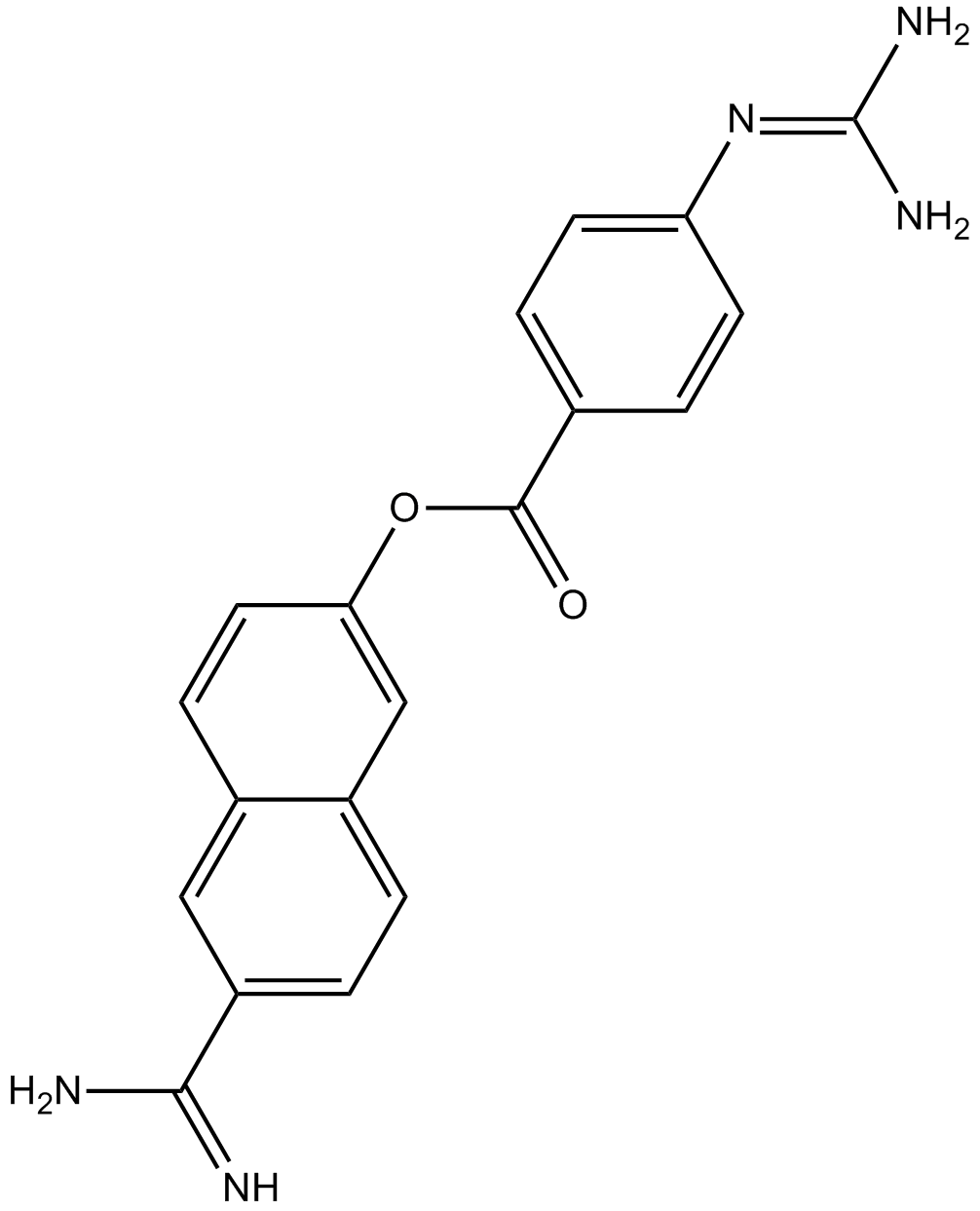
Nafamostat
Broad spectrum serine protease inhibitor, kallikrein inhibitor
-
GC16290
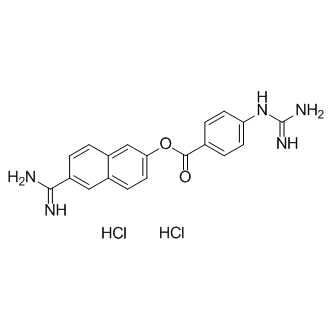
Nafamostat hydrochloride
Synthetic serine protease inhibitor
-
GC36690
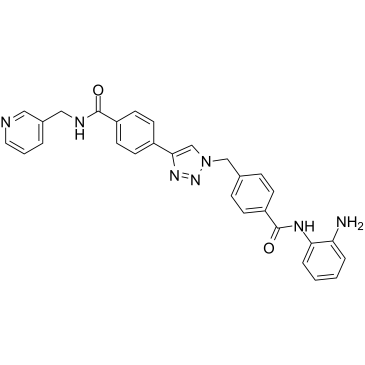
Nampt-IN-3
Nampt-IN-3 (Compound 35) simultaneously inhibit nicotinamide phosphoribosyltransferase (NAMPT) and HDAC with IC50s of 31 nM and 55 nM, respectively. Nampt-IN-3 effectively induces cell apoptosis and autophagy and ultimately leads to cell death.
-
GC14562
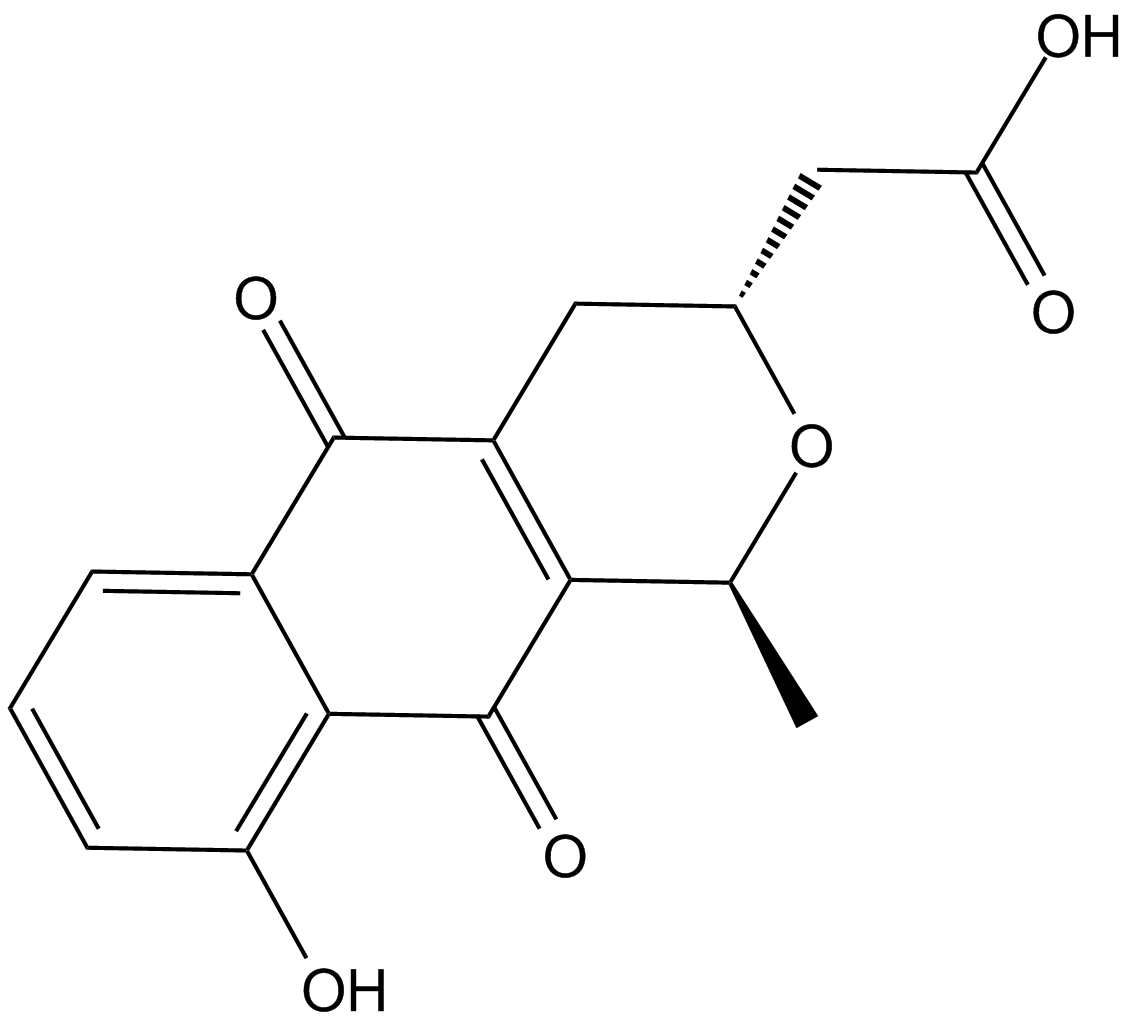
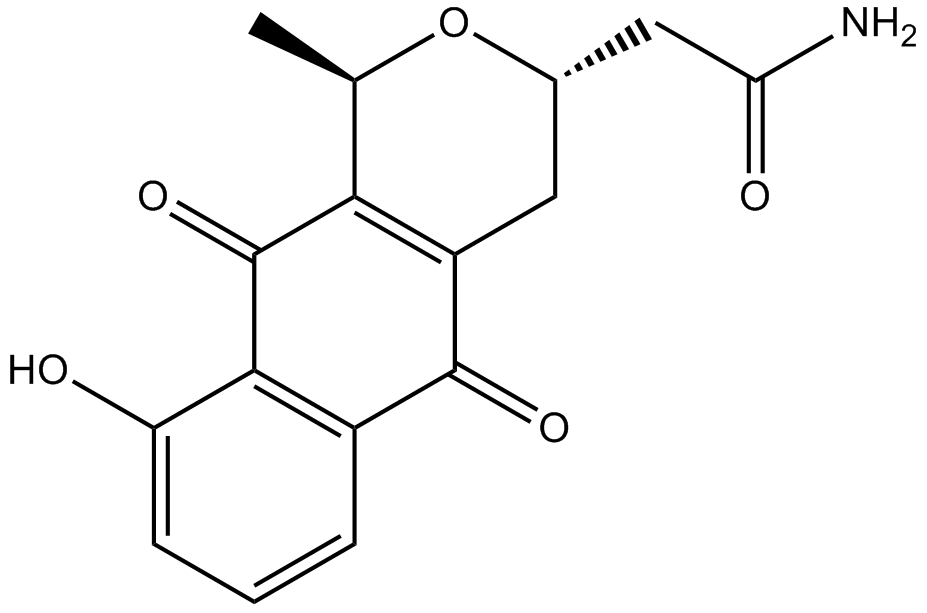
Nanaomycin A
A bacterial metabolite
-
GC10243
Nanaomycin C
Amide of nanaomycin A
-
GC36691
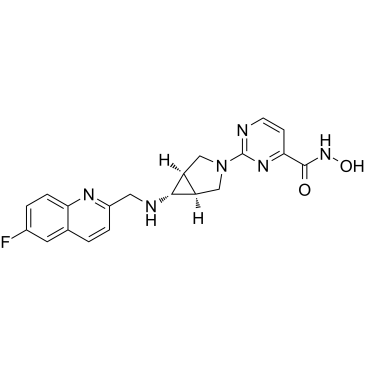
Nanatinostat
Nanatinostat (CHR-3996) is a potent, class I selective and orally active histone deacetylase (HDAC) inhibitor with an IC50 of 8 nM.
-
GC61841
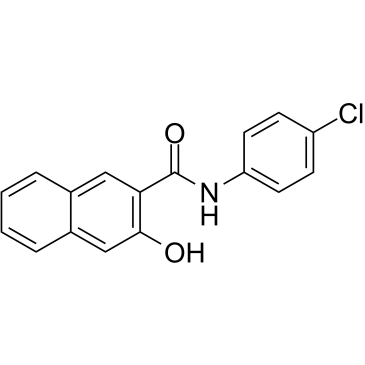
Naphthol AS-E
Naphthol AS-E is a potent and cell-permeable inhibitor of KIX-KID interaction. Naphthol AS-E directly binds to the KIX domain of CBP (Kd:8.6 ?M), blocks the interaction between the KIX domain and the KID domain of CREB with IC50 of 2.26 ?M. Naphthol AS-E can be used for cancer research.
-
GC65138
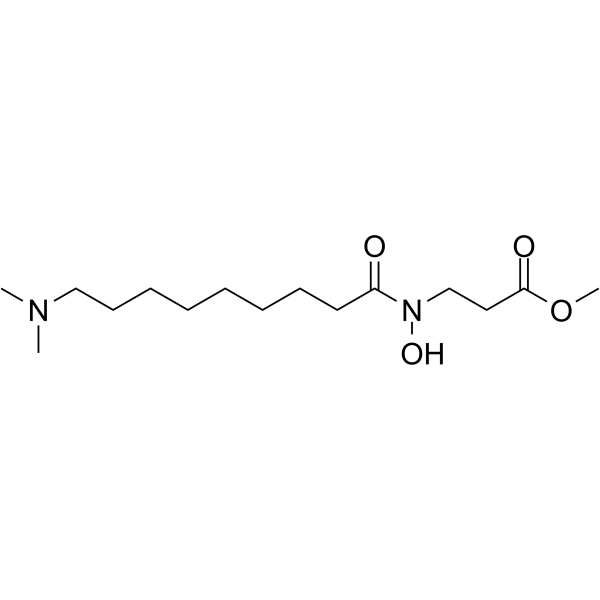
NCDM-32B
NCDM-32B is a potent and selective KDM4 inhibitor that impaires viability and transforming phenotypes of breast cancer.
-
GC19410
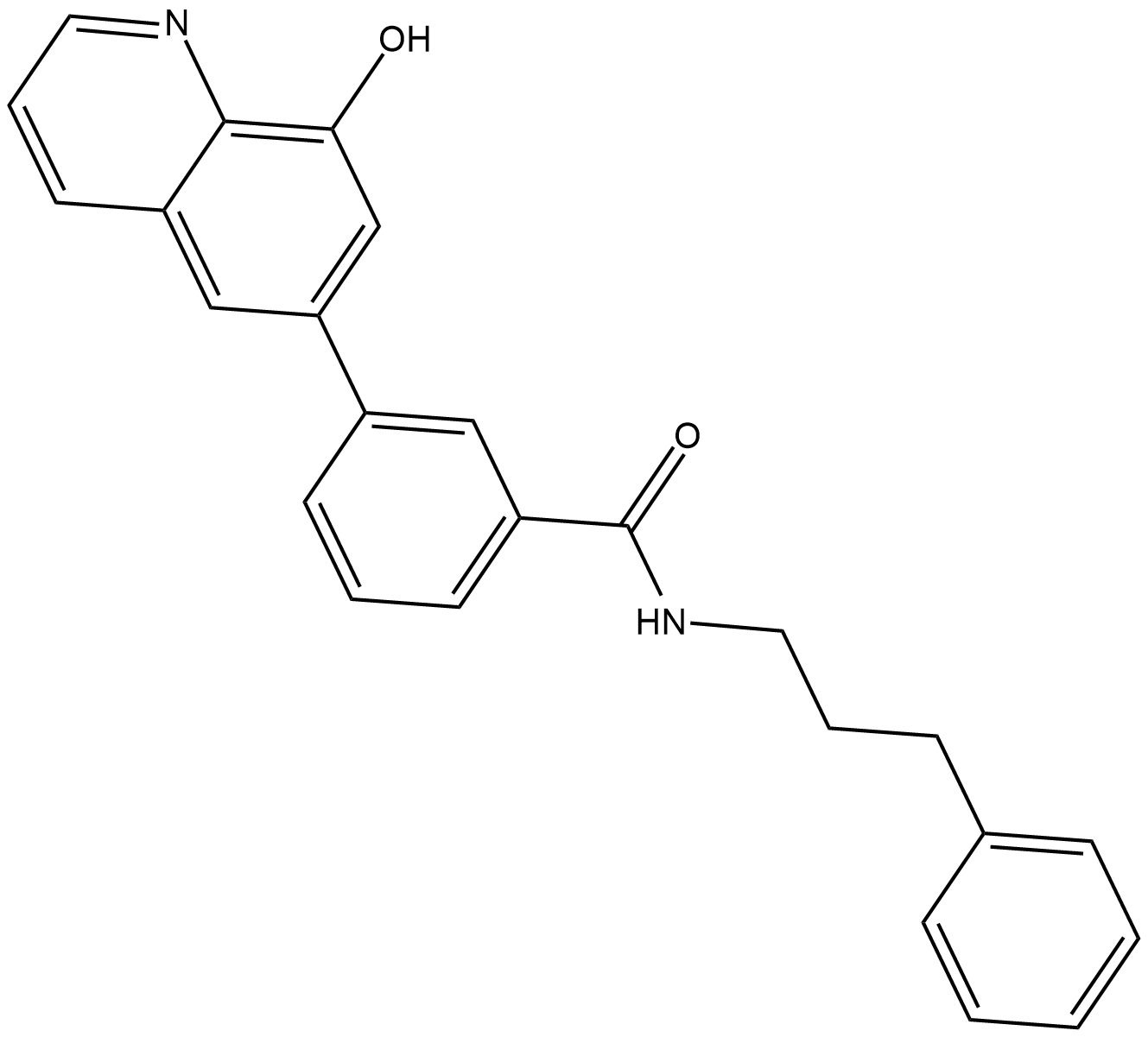
NCGC00244536
NCGC00244536 is a potent KDM4B inhibitor with an IC50 of 10 nM
-
GC32896
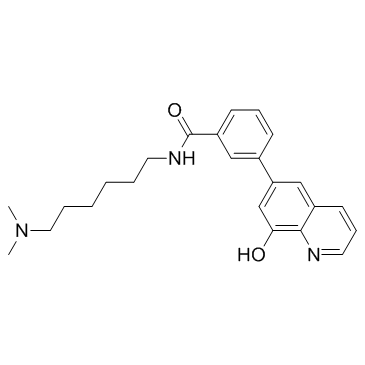
NCGC00247743
NCGC00247743 is a histone lysine demethylase KDM4 inhibitor.
-
GC15536
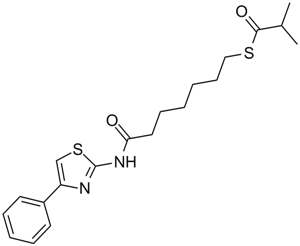
NCH 51
An HDAC inhibitor
-
GC66329
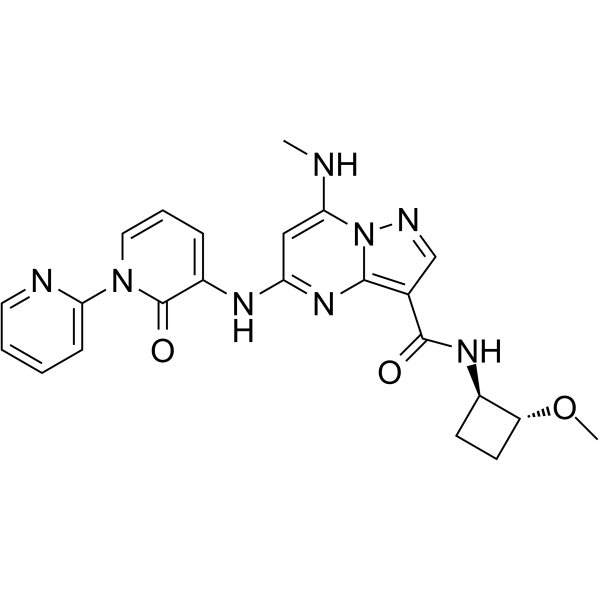
NDI-034858
NDI-034858 is a TYK2 inhibitor, target TYK2 JH2 domain with binding constant Kd of <200 pM.
-
GC65202
Nesuparib
Nesuparib is a potent inhibitor of PARP.
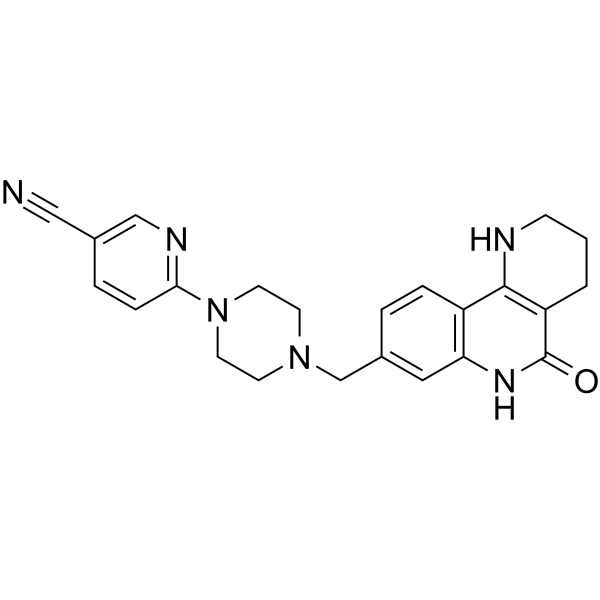
-
GC13764
Nexturastat A
HDAC6 inhibitor,highly potent and selective
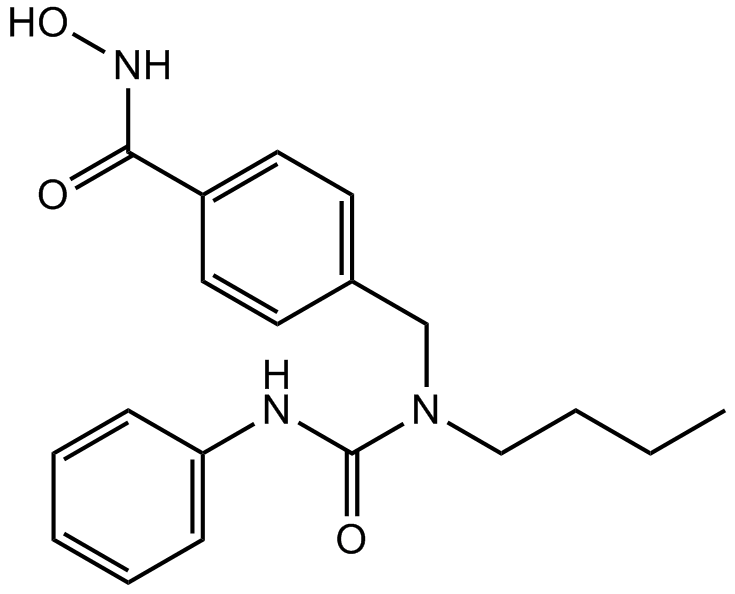
-
GC62620
NHWD-870
NHWD-870 is a potent, orally active and selective BET family bromodomain inhibitor and only binds bromodomains of BRD2, BRD3, BRD4 (IC50=2.7 nM), and BRDT. NHWD-870 has potent tumor suppressive efficacies and suppresses cancer cell-macrophage interaction. NHWD-870 increases tumor apoptosis and inhibits tumor proliferation.
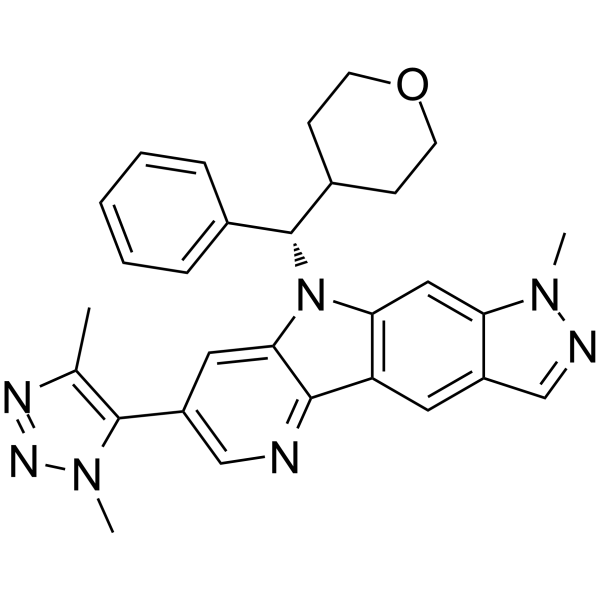
-
GC36734
NI-42
An inhibitor of the BRPF1 bromodomain
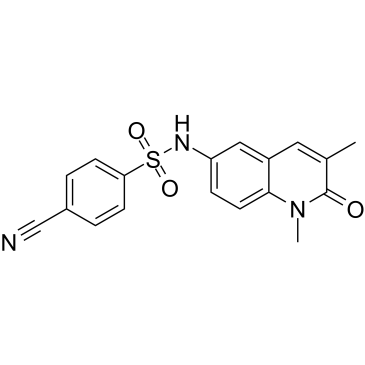
-
GC16763
NI-57
bromodomains of BRPF proteins inhibitor
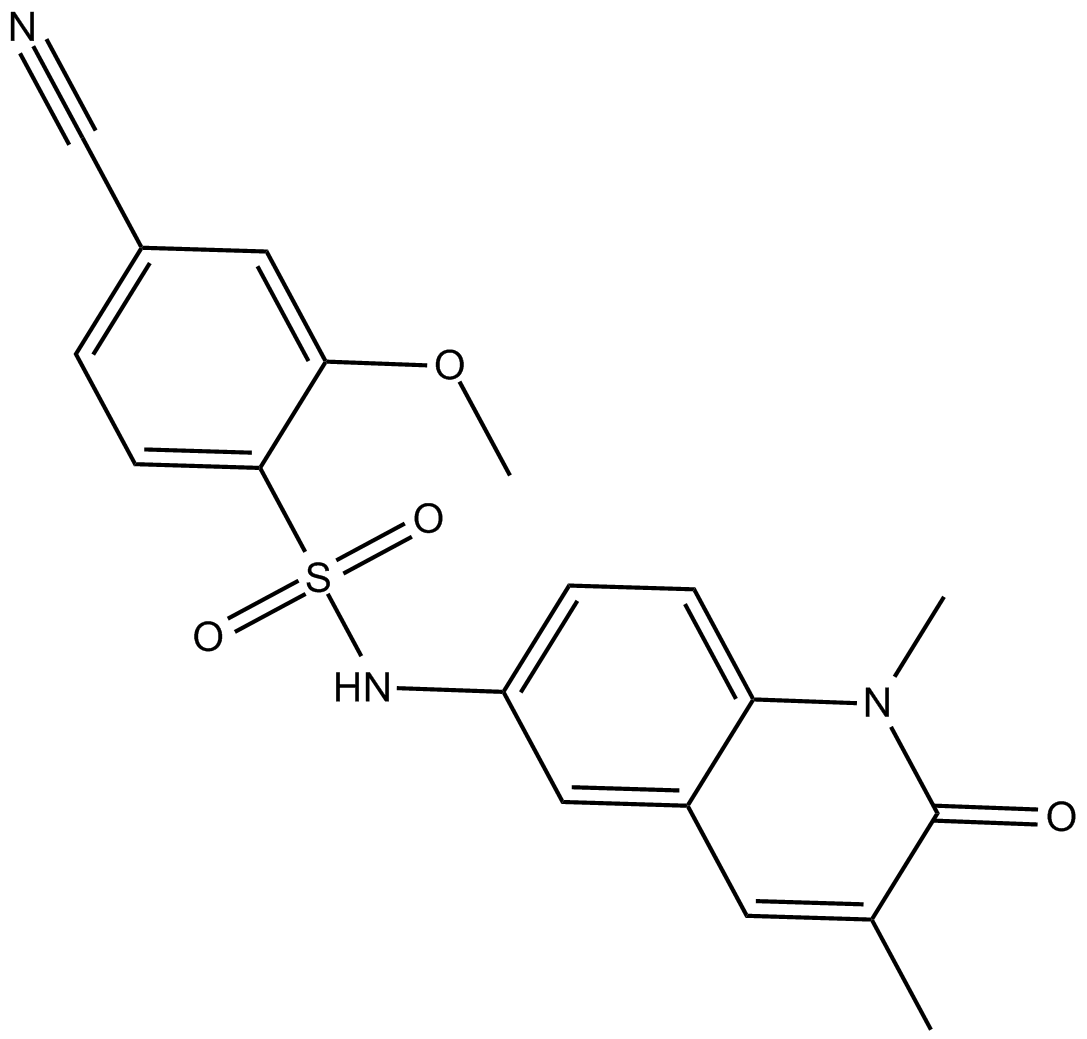
-
GN10347
Nicotinamide (Vitamin B3)
Nicotinamide is an inhibitor of poly(ADP-ribose) polymerase (PARP-1) enzymes.
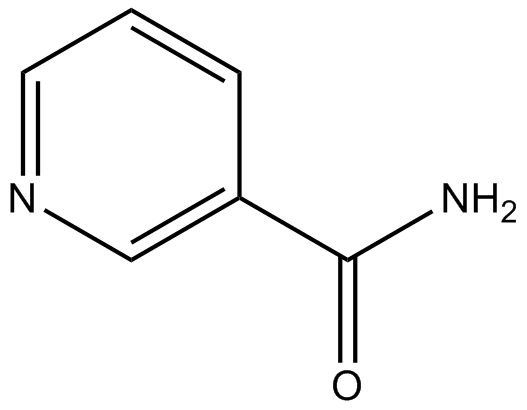
-
GC44401
Nicotinamide riboside
Nicotinamide riboside, a form of vitamin B3 and NAD+ precursor, is converted to bioavailable NAD+, via nicotinamide riboside kinase (NRK) and NMNAT, or by the action of nucleoside phosphorylase and NAM salvage.

-
GC36738
Nicotinamide riboside chloride
A nicotinamide adenine dinucleotide (NAD[+]) precursor vitamin
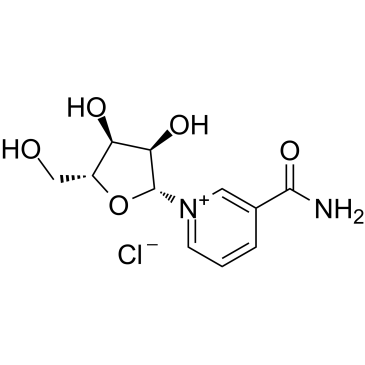
-
GC49270
Nicotinamide-d4
An internal standard for the quantification of nicotinamide
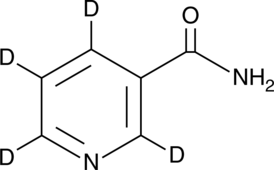
-
GC62248
NiCur
NiCur is a potent and selective CBP histone acetyltransferase (HAT) inhibitor with an IC50 value of 0.35 μΜ. NiCur, which blocks CBP HAT activity and downregulates p53 activation upon genotoxic stress. NiCur can be used for performing mechanistic studies without affecting the expression of target proteins.
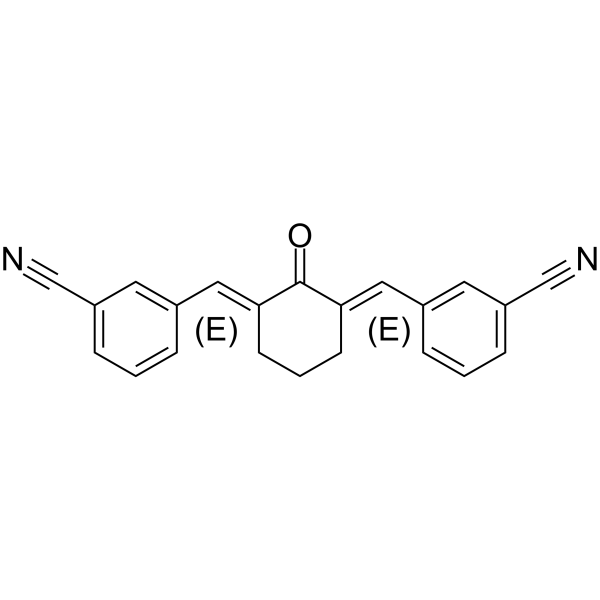
-
GC34120
Niraparib R-enantiomer (MK 4827 (R-enantiomer))
Niraparib R-enantiomer (MK-4827 R-enantiomer) is an excellent PARP1 inhibitor with IC50 of 2.4 nM.
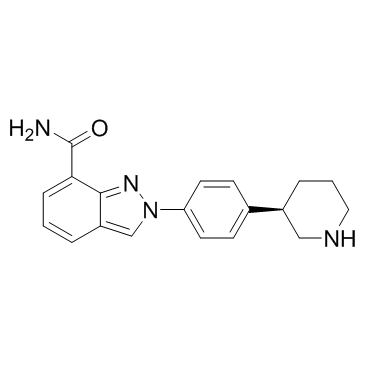
-
GC32964
NKL 22
A selective HDAC1/3 inhibitor
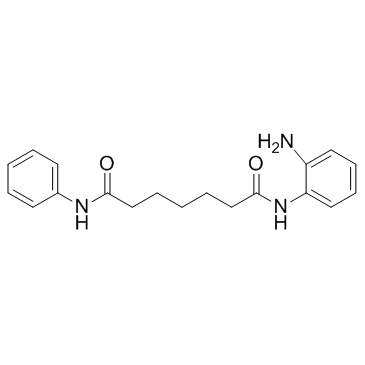
-
GC19264
NMS-P118
NMS-P118 is a potent, orally available, and highly selective PARP-1 Inhibitor for cancer therapy.
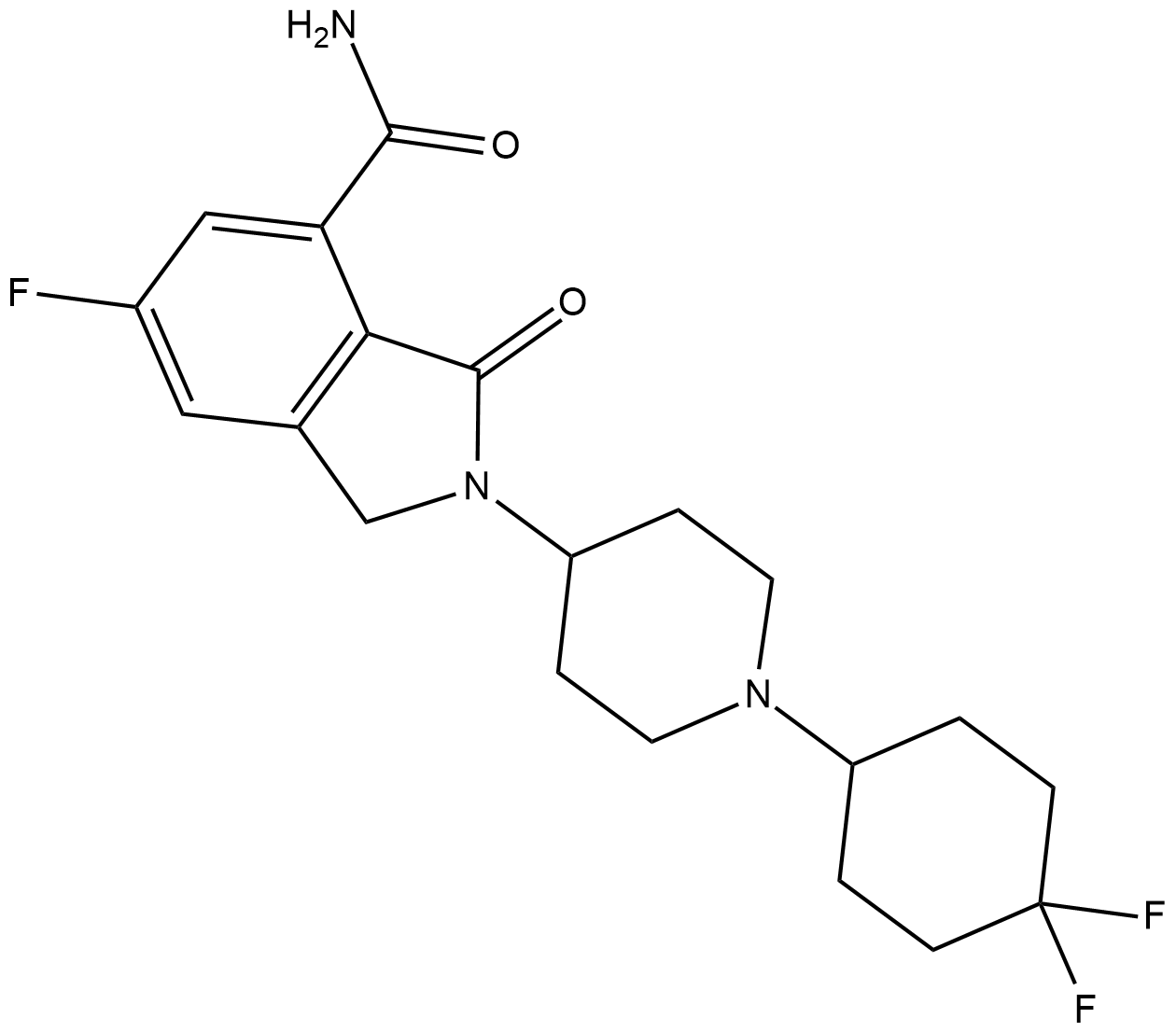
-
GC36751
NMS-P515
NMS-P515 is a potent, orally active and stereospecific PARP-1 inhibitor, with a Kd of 16 nM and an IC50 of 27 nM (in Hela cells). Anti-tumor activity.
-
GC52166
NN-390
NN-390 is a potent and selective HDAC6 inhibitor, with an IC50 of 9.8 nM. NN-390 penetrates the blood-brain barrier (BBB). NN-390 shows study potential in metastatic Group 3 MB (medulloblastoma).

-
GC48460
NR-160
An inhibitor of HDAC6
-
GC17762
NSC 33994
Selective inhibitor of JAK2 (IC50 = 60 nM). Displays no effect on Src and TYK2 tyrosine kinase activity at a concentration of 25 μM.
-
GC15040
NSC 3852
A tumor cell differentiating agent
-
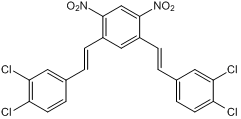
GC50136
NSC 636819
KDM4A/KDM4B inhibitor
-
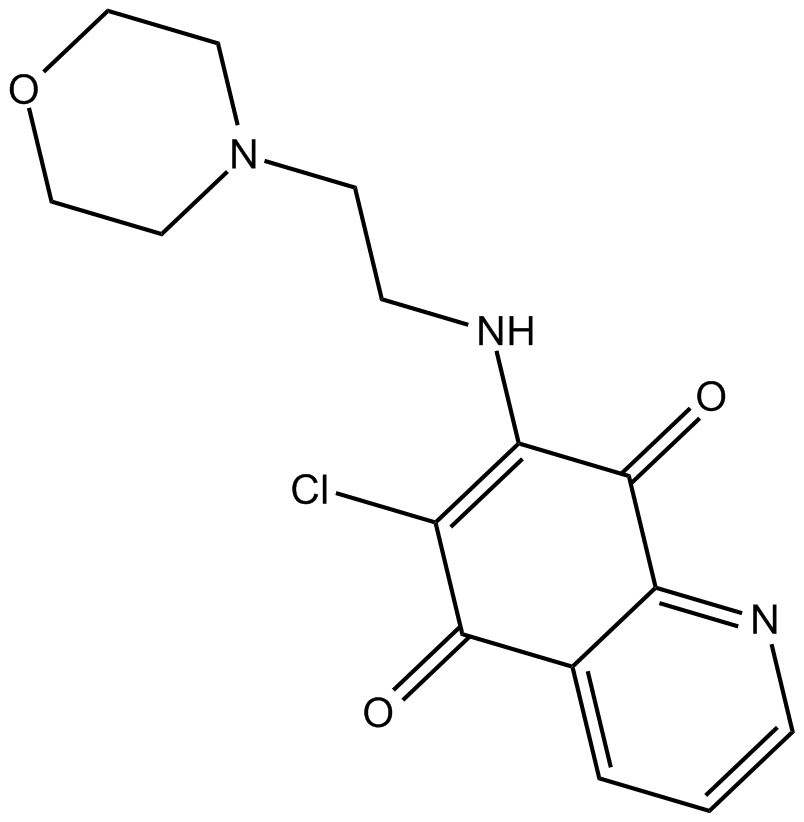
GC14875
NSC 663284
Cdc25 dual specificity phosphatases inhibitor
-
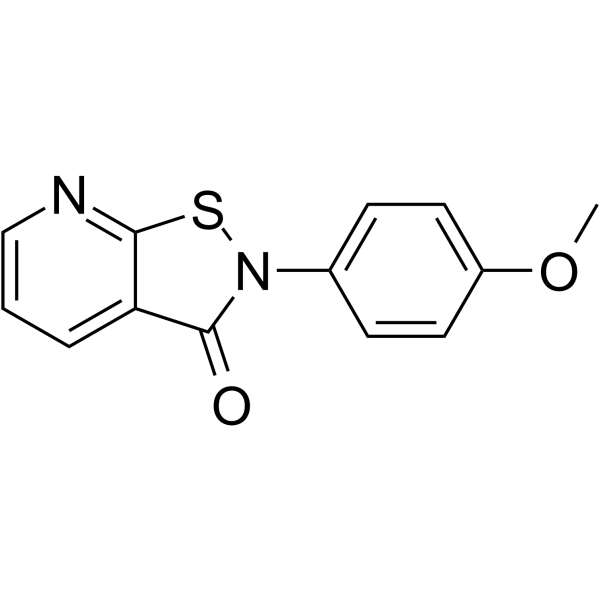
GC66049
NSC 694621
NSC 694621 is a potent PCAF inhibitor, with an IC50 of 5.71 μM (PCAF/H31-21). NSC 694621 exhibits good activity of inhibiting the proliferation of cancer cells.
-
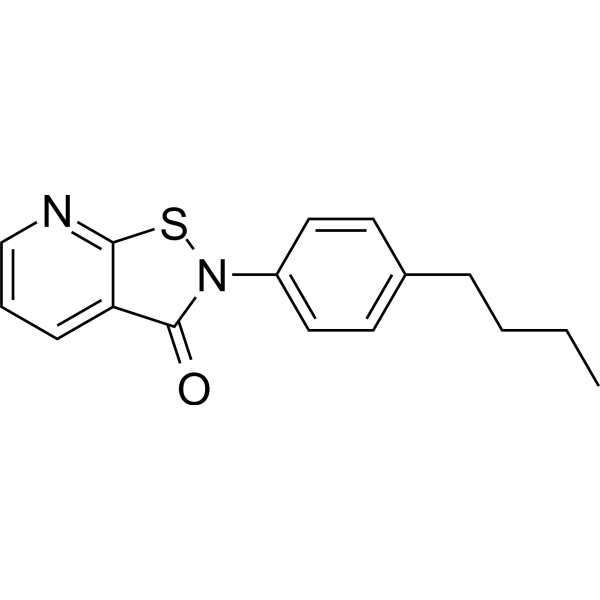
GC69597
NSC 694623
NSC 694623 is an effective inhibitor of histone acetyltransferase (HAT), with an IC50 of 15.9 μM against recombinant HAT p300/CBP-associated factor (PCAF). NSC 694623 has anti-proliferative activity in certain cancer cells and can be used for anticancer research.
-
GC49412
NSC 756093
An inhibitor of the GBP1-Pim-1 protein-protein interaction
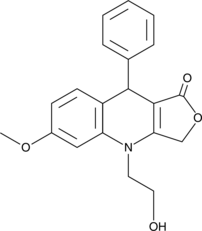
-
GC11561
NSC 95397
Cdc25 dual specificity phosphatases inhibitor
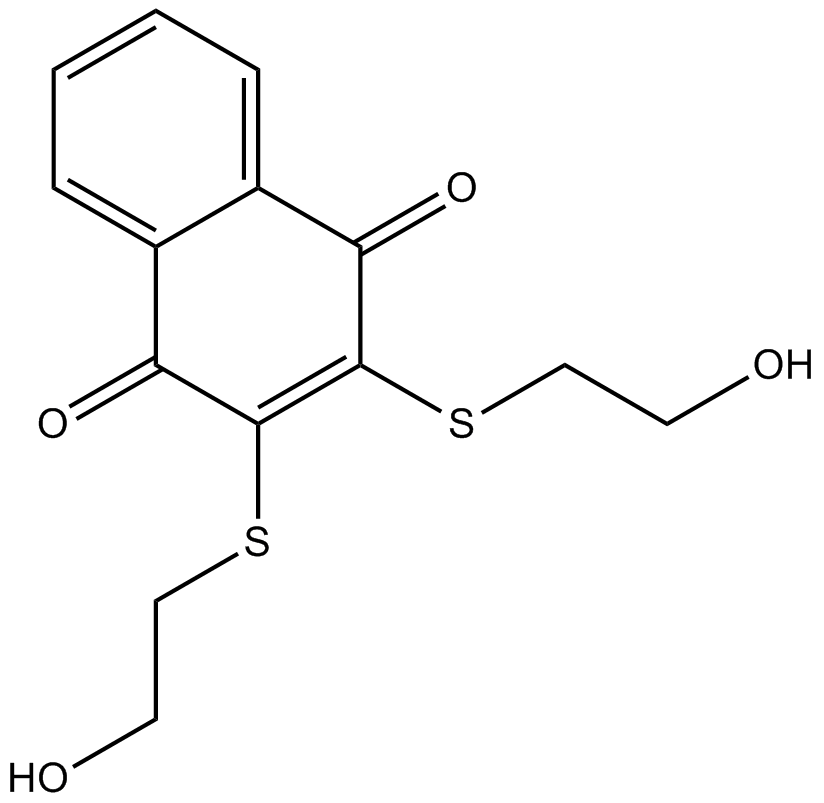
-
GC14103
NSC228155
EGFR activator
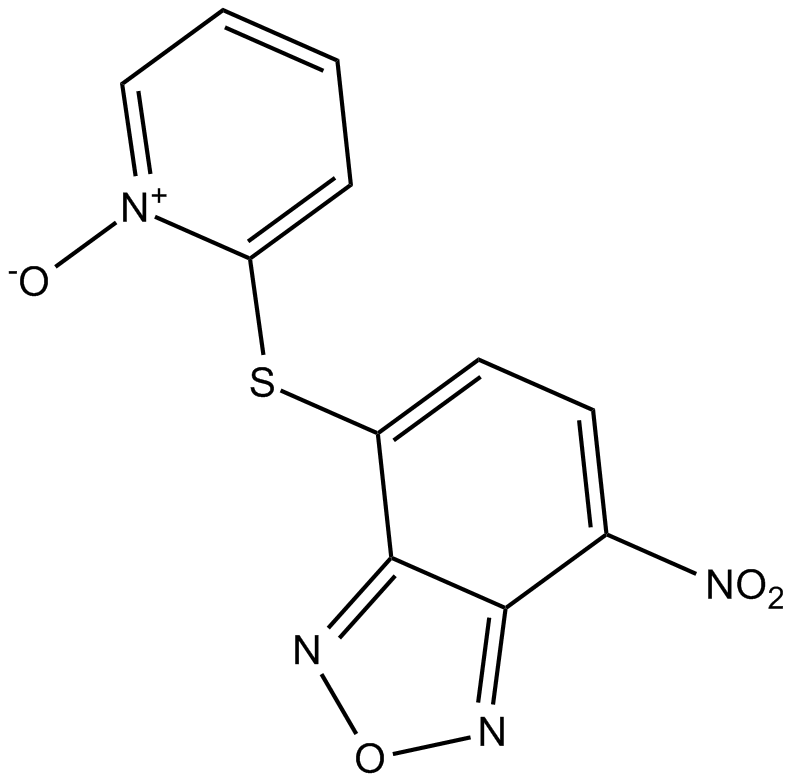
-
GC61142
NSC745885
NSC745885 an effective anti-tumor agent, shows selective toxicity against multiple?cancer?cell lines but not normal cells.?NSC745885 is an effective?down-regulator of EZH2 via proteasome-mediated degradation. NSC745885 provides possibilities for the study of advanced?bladder?and oral squamous cell carcinoma (OSCC) cancers.
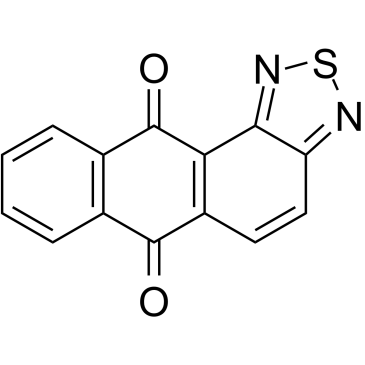
-
GC17775
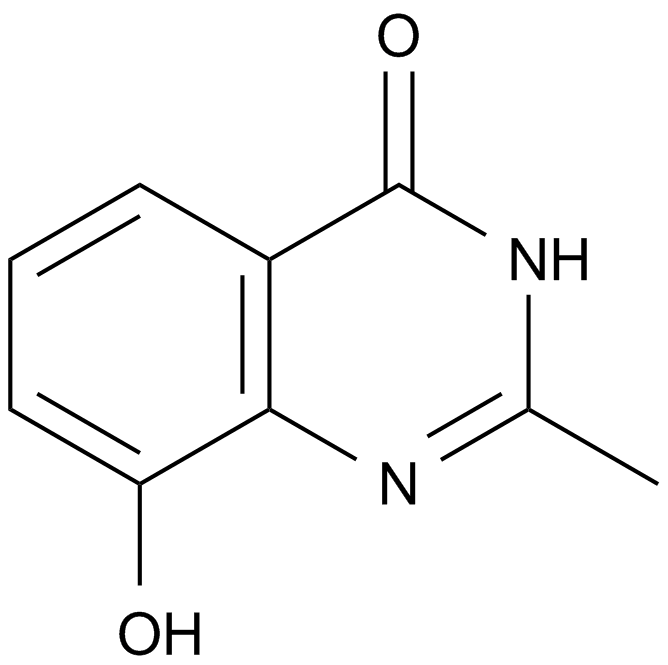
NU 1025
An inhibitor of PARP
-
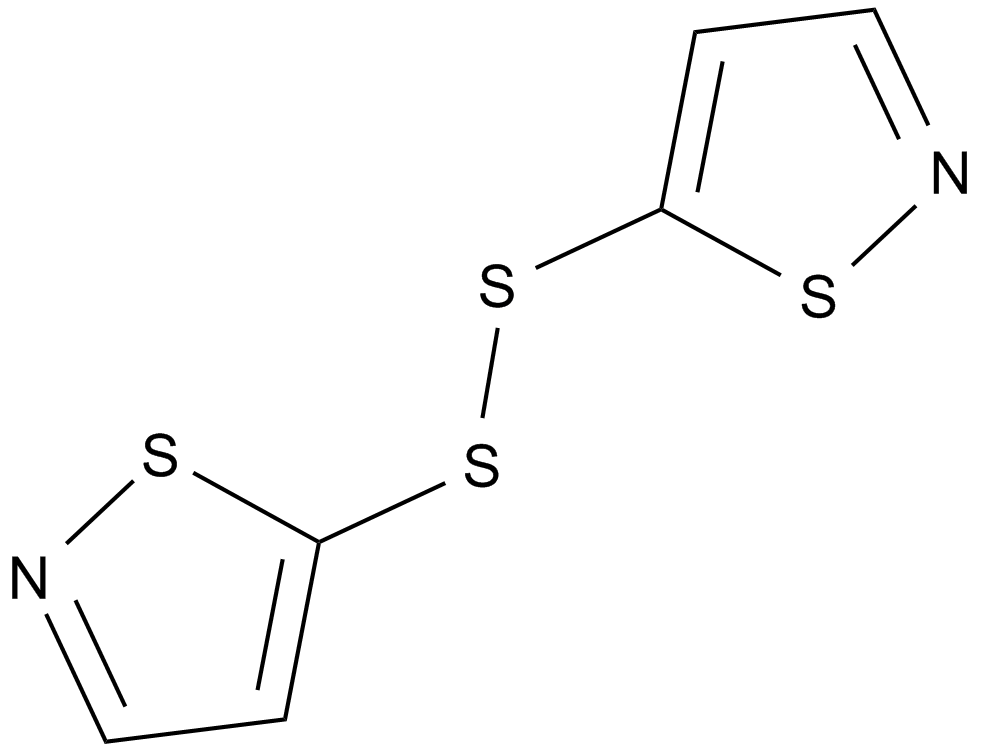
GC11972
NU 9056
Selective KAT5 (Tip60) histone acetyltransferase inhibitor (IC50 values are < 2, 60, 36, and >100 μM for KAT5, p300, pCAF and GCN5, respectively).
-
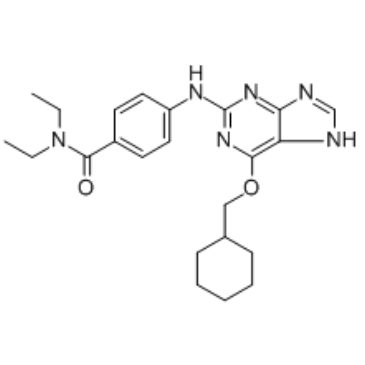
GC34692
NU6140
A Cdk2 inhibitor
-
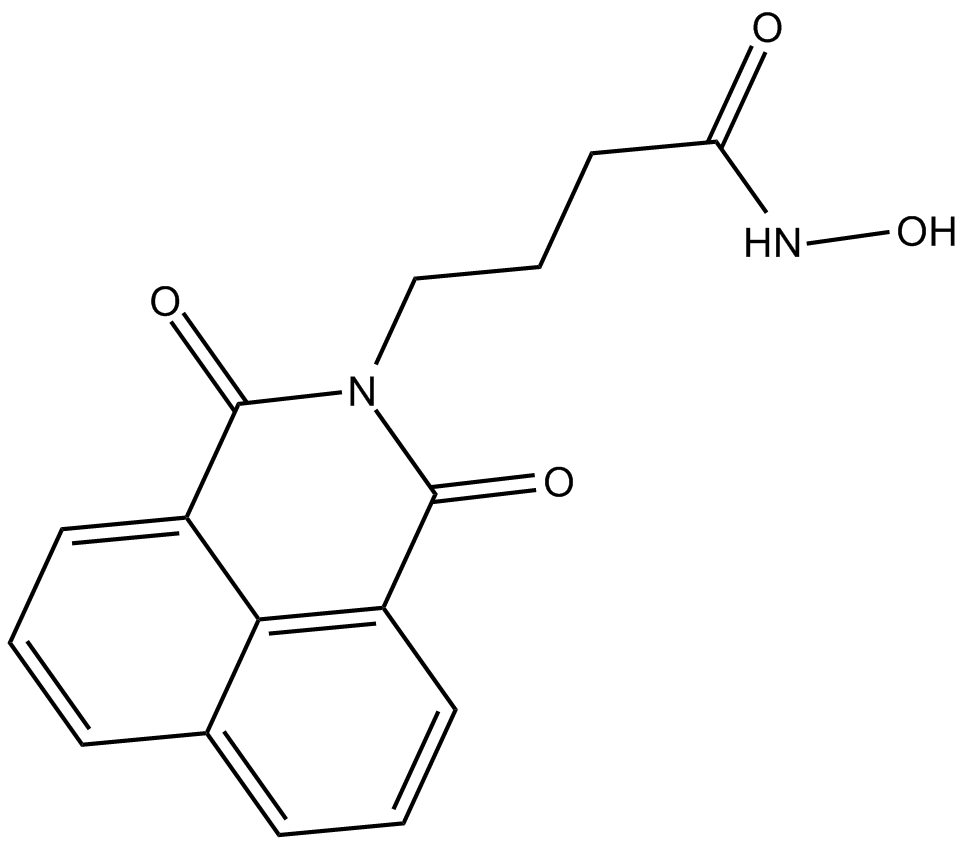
GC13925
Nullscript
negative control of scriptaid, HDAC inhibitor
-
GC64772
NV03
NV03 is a potent and selective antagonist of UHRF1 (Ubiquitin-like with PHD and RING finger domains 1)- H3K9me3 interaction by binding to UHRF1 tandem tudor domain, with a Kd of 2.4 μM. NV03 has anticancer activity.
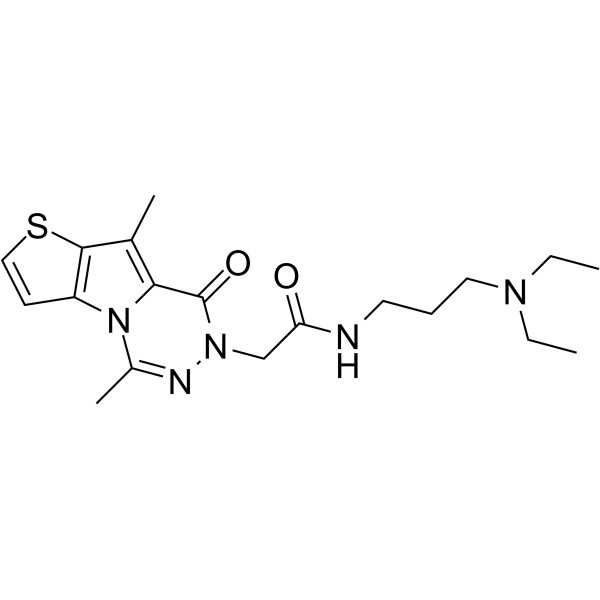
-
GC13229
NVP-BSK805
A potent, selective JAK2 inhibitor
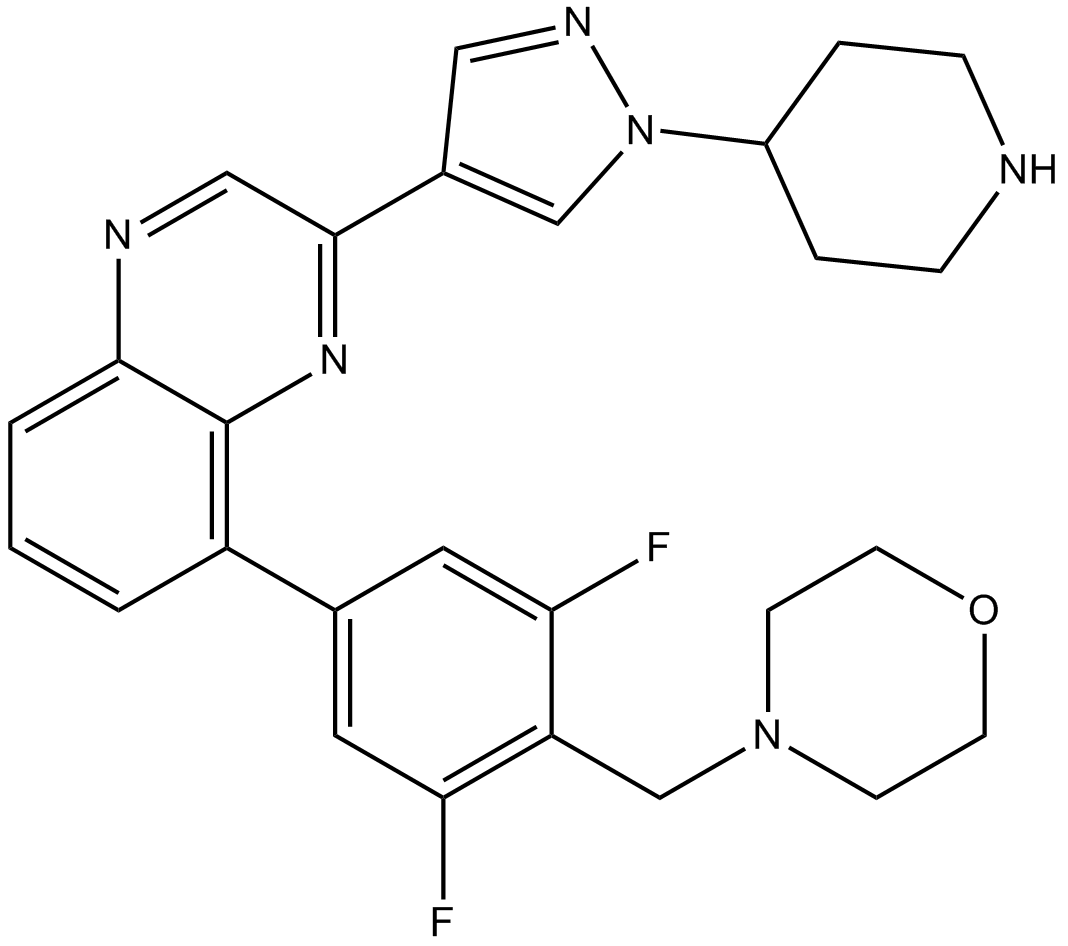
-
GC36783
NVP-BSK805 dihydrochloride
A potent, selective JAK2 inhibitor
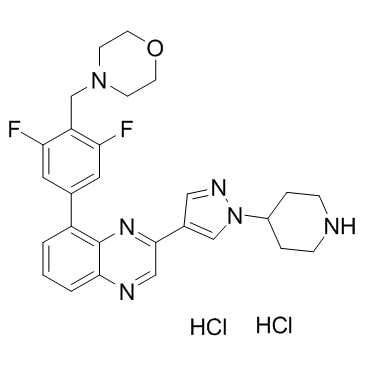
-
GC17555
NVP-TNKS656
TNKS2 inhibitor
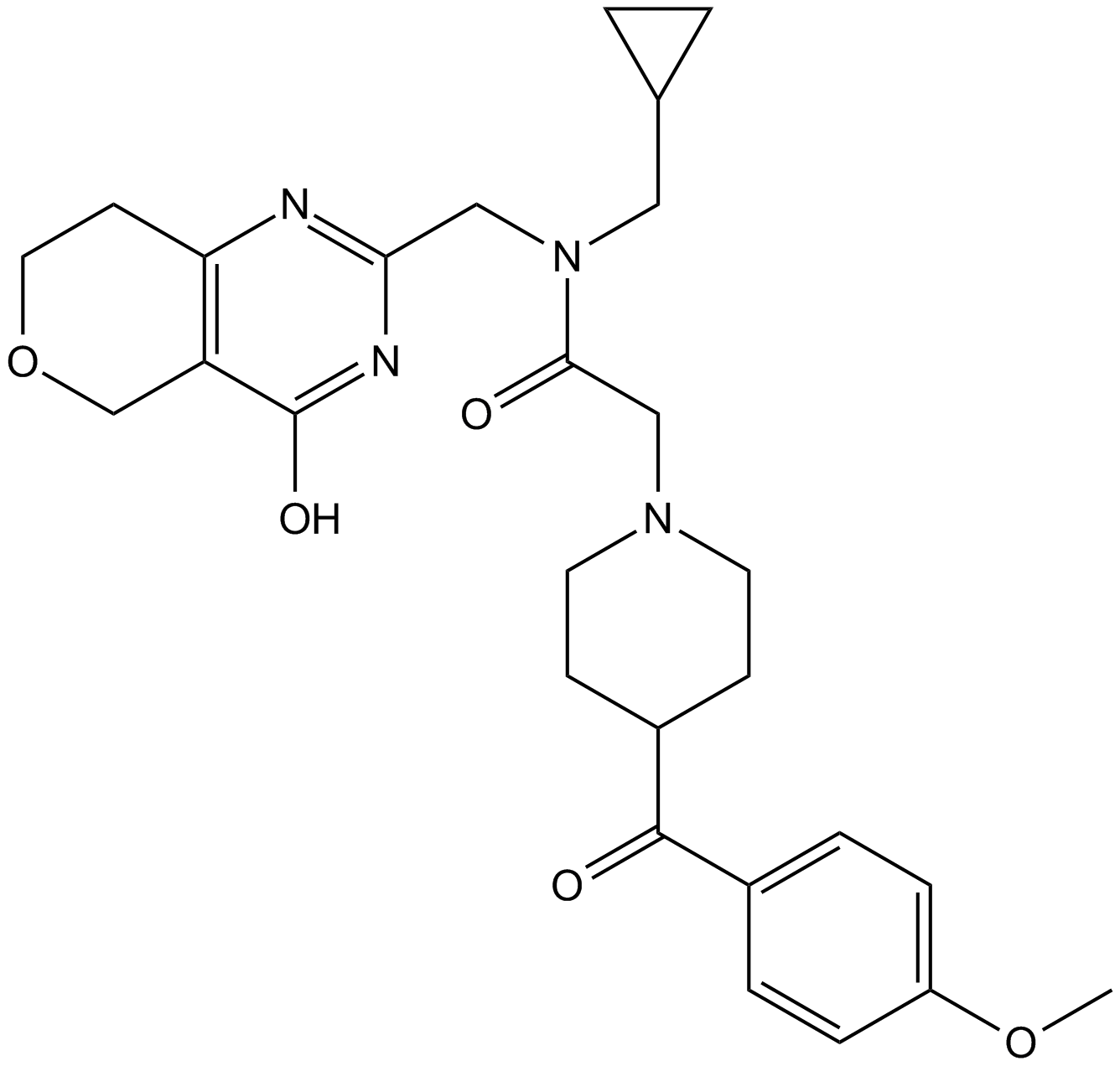
-
GC44477
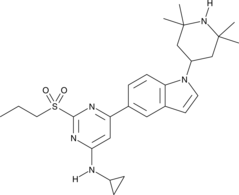
NVS-CECR2-1
NVS-CECR2-1 is a potent inhibitor of CECR2 (cat eye syndrome chromosome region, candidate 2), a component of chromatin complexes that regulate gene expression controlling development.
-
GC69601
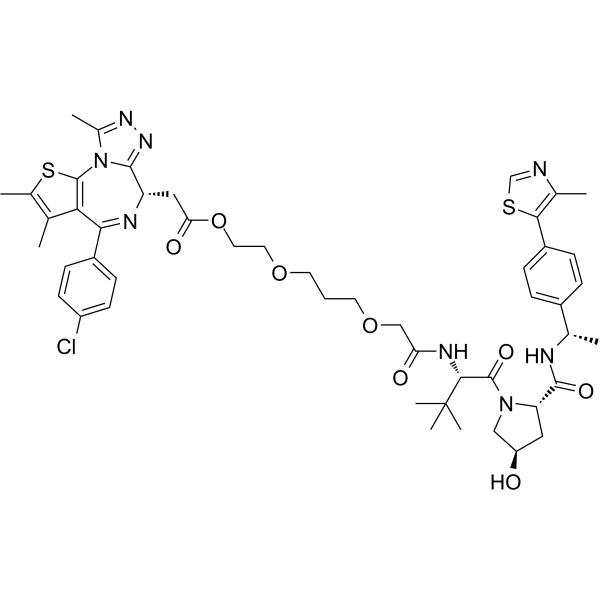
OARV-771
OARV-771 is a VHL-based BET degrader (PROTAC) with improved cell permeability. OARV-771 shows DC50 values of 6, 1, and 4 nM for Brd4, Brd2, and Brd3 respectively.
-
GC31677
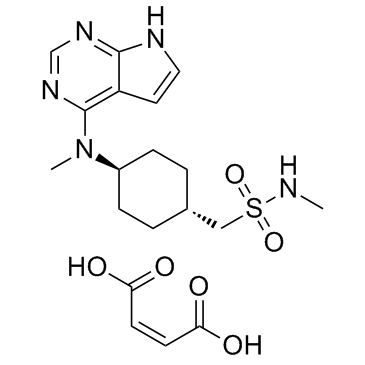
Oclacitinib maleate (PF-03394197 maleate)
Oclacitinib maleate (PF-03394197 maleate) (PF-03394197 maleate) is a novel JAK inhibitor.
-
GC17358
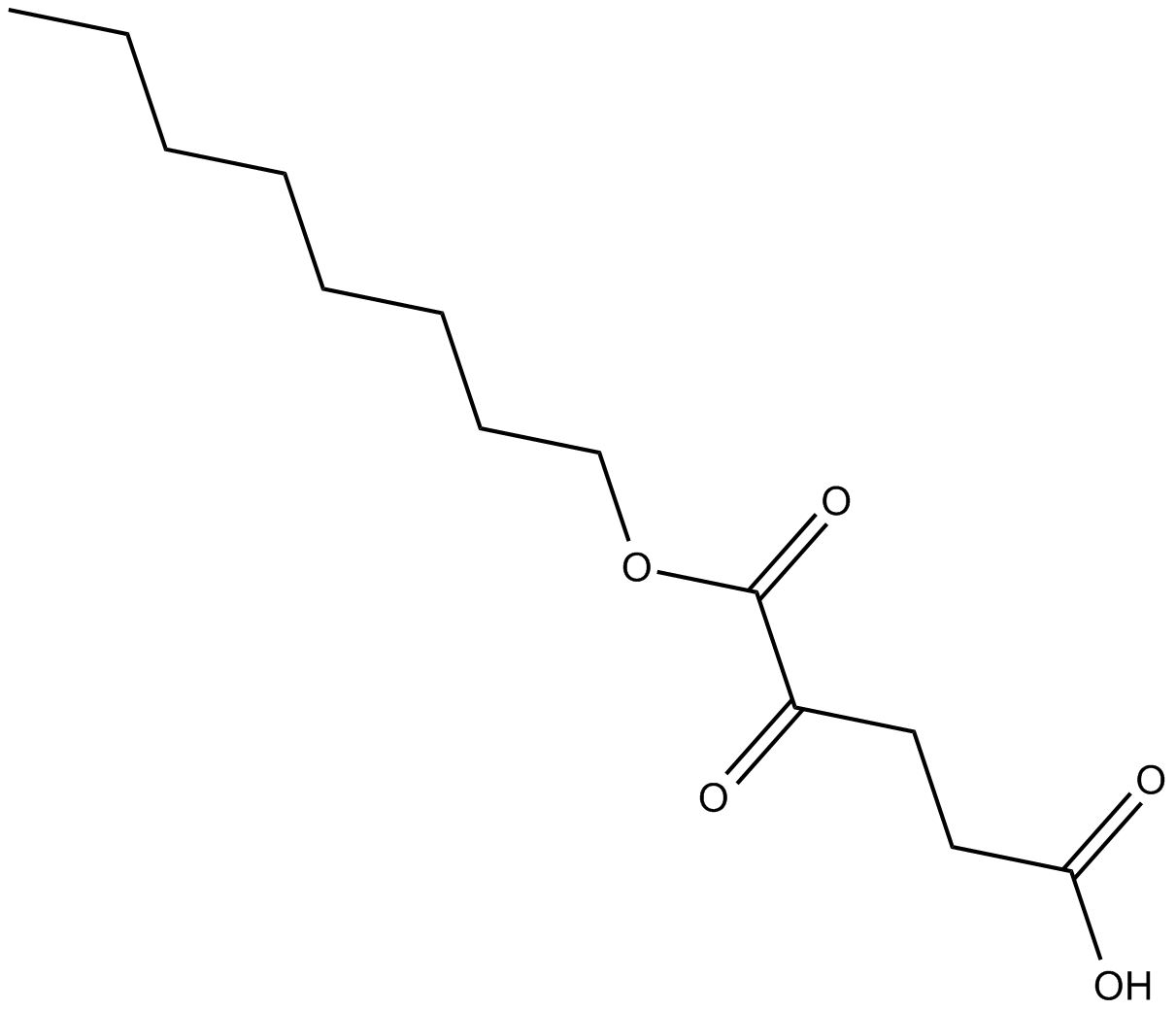
Octyl-α-ketoglutarate
prolyl hydroxylases (PHD) activator
-
GC66067
ODM-207
ODM-207 (BET-IN-4) is a potent BET bromodomain protein (BRD4) inhibitor, with an IC50 of ≤ 1 μM.
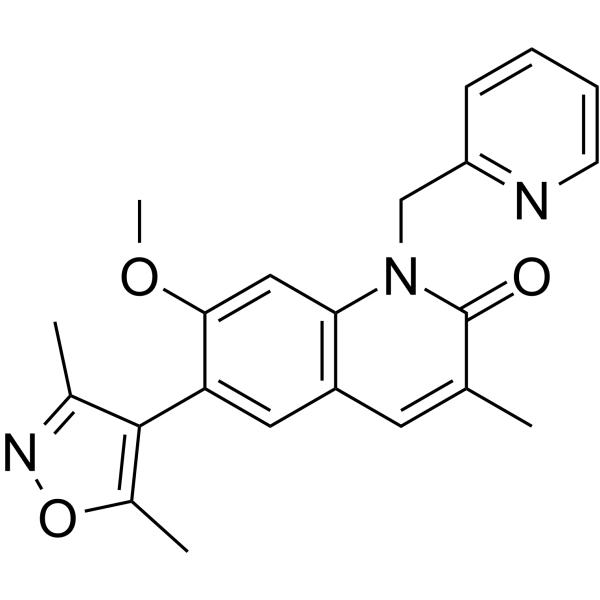
-
GC17482
OF-1
BRPF1B and BRPF2 bromodomain inhibitor
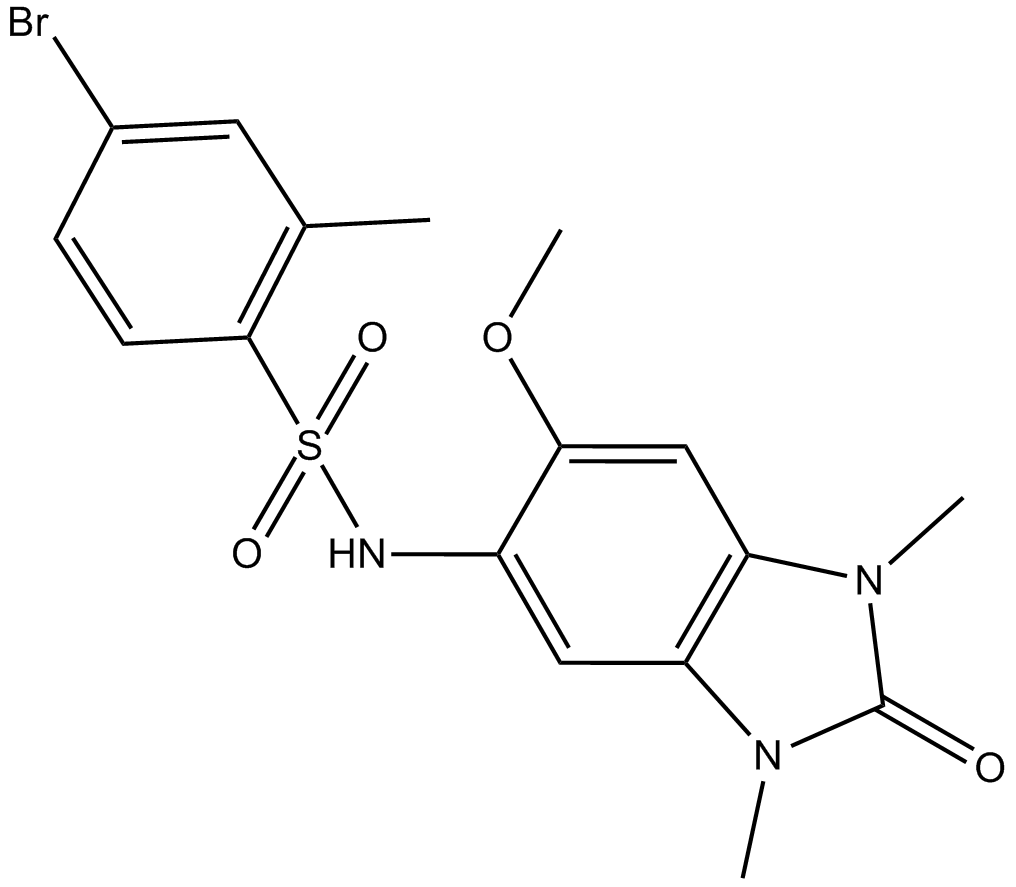
-
GC17314
OG-L002
LSD1 inhibitor,potent and specific
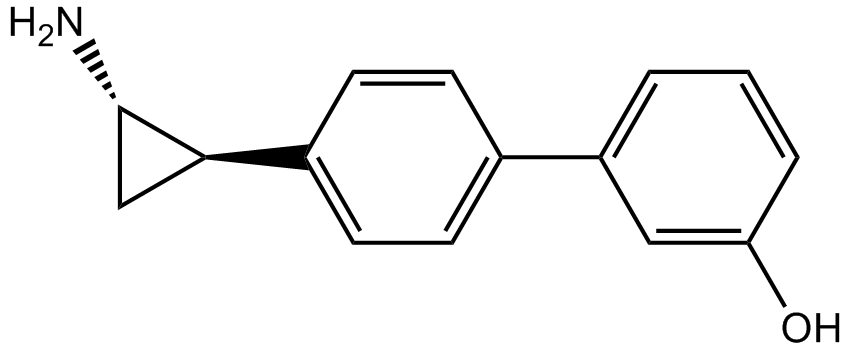
-
GC11792
OG-L002 HCl
LSD1 inhibitor,potent and specific
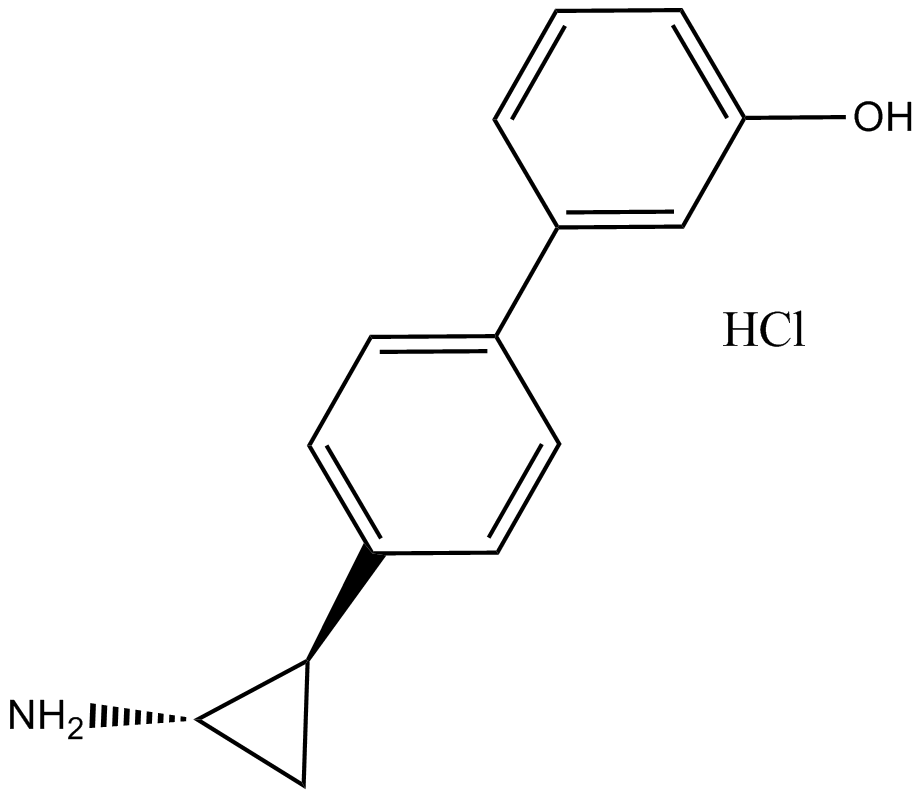
-
GC16397
OICR-9429
Wdr5-MLL interaction antagonist
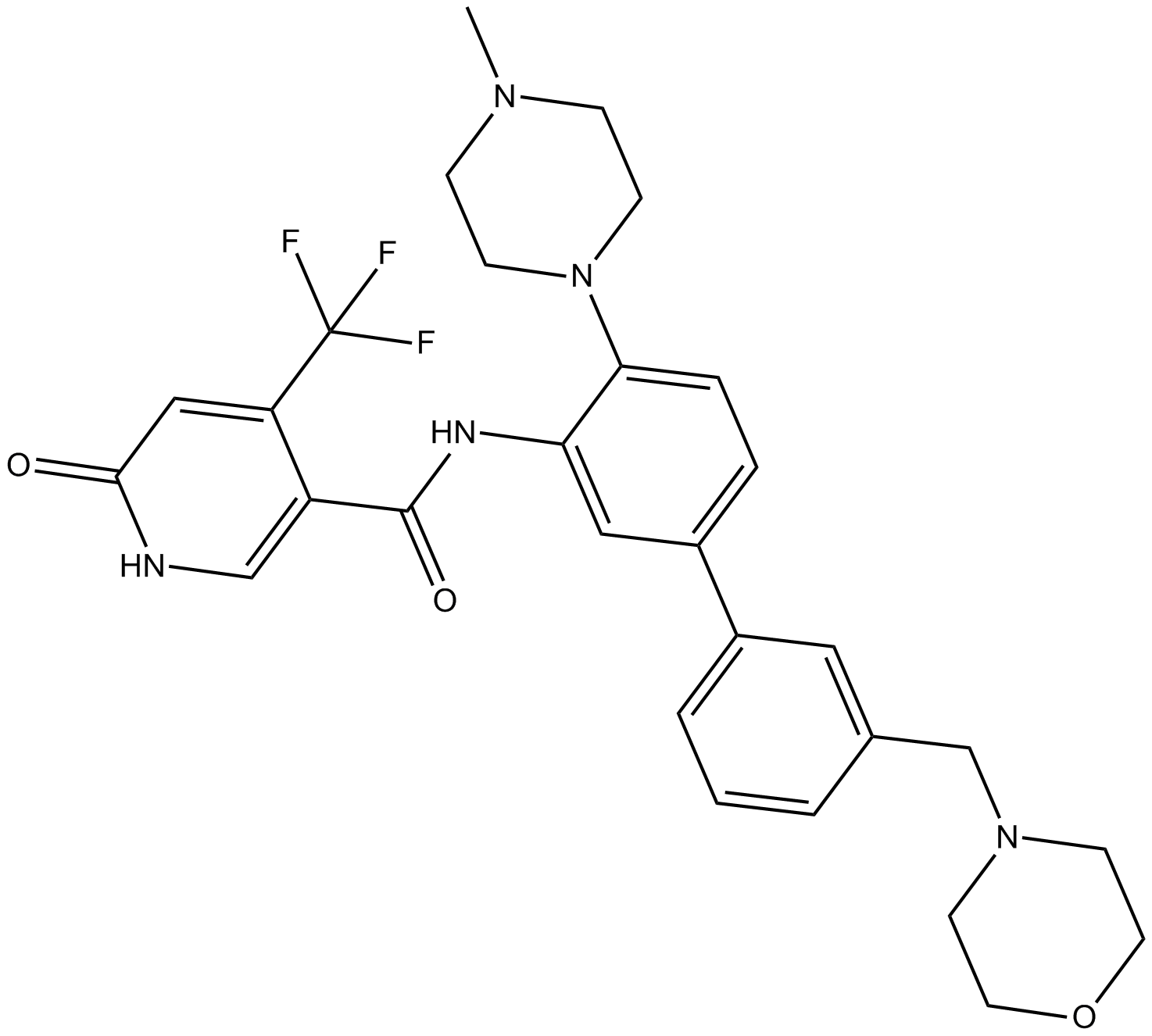
-
GC16958
Okadaic acid
A potent inhibitor of protein phosphatases
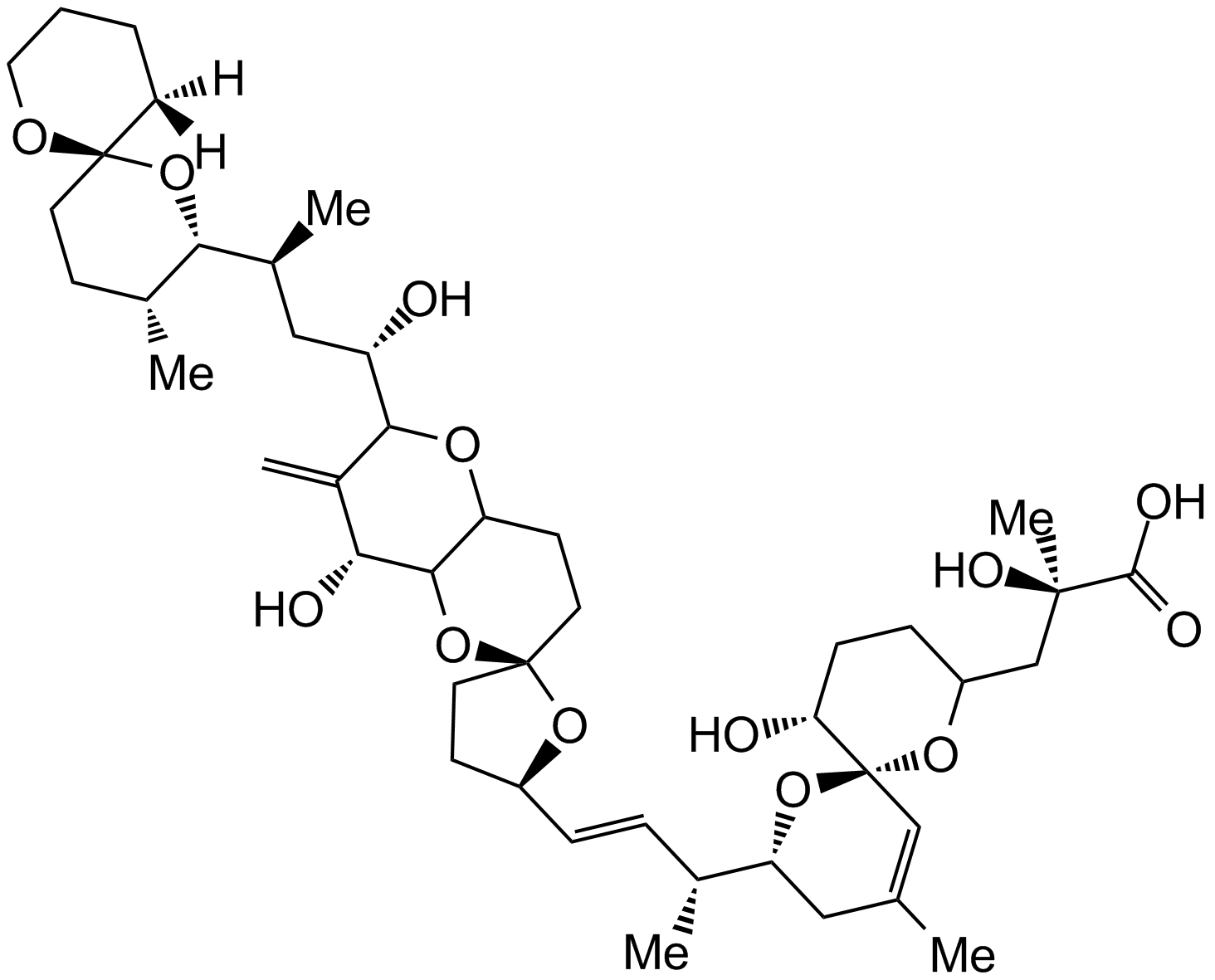
-
GC17580
Olaparib (AZD2281, Ku-0059436)
A PARP inhibitor
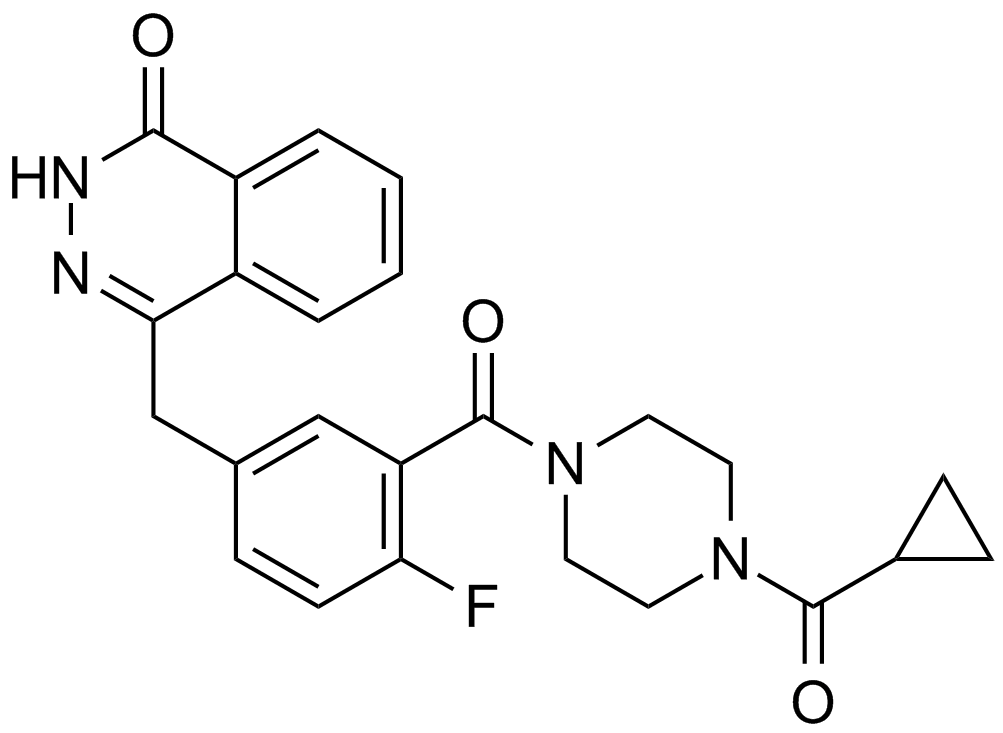
-
GC69618
Olaparib-d8
Olaparib-d8 is the deuterated form of Olaparib (AZD2281). Olaparib is an orally effective PARP inhibitor that inhibits PARP-1 and PARP-2 with IC50 values of 5 and 1 nM, respectively. Olaparib is also an activator of autophagy and mitophagy.
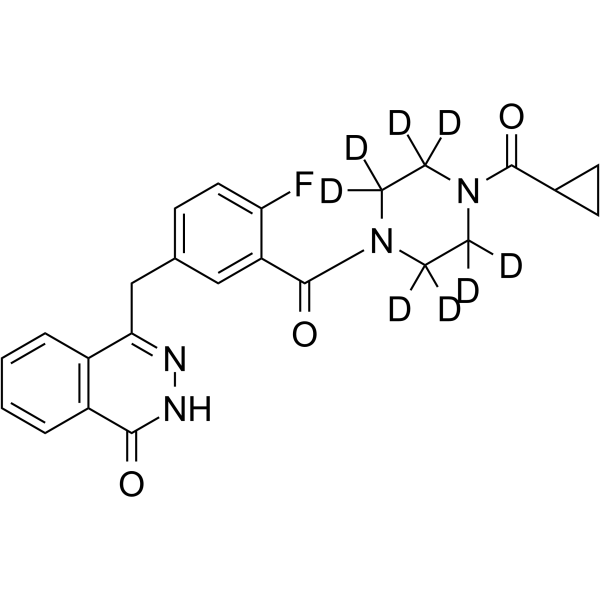
-
GC49815
Oleuropein aglycone
A polyphenol with diverse biological activities
-
GC12821
Oltipraz
Nrf2 activator;An antischistosomal agent
-
GC67906
OM-153

-
GN10420
Ophiopogonin D'
-
GN10114
Oroxin A

-
GC41625
Oroxylin A
Oroxylin A is a flavonoid that has been found in S.
-
GC11360
ORY-1001
Selective inhibitor of KDM1A.
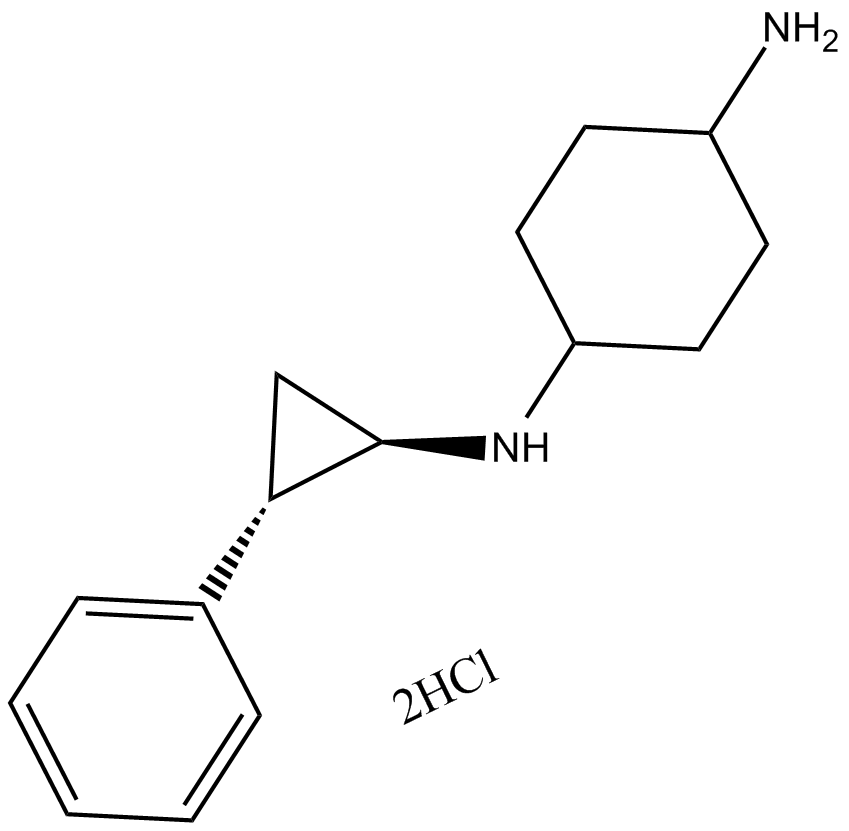
-
GC36818
ORY-1001(trans)
Iadademstat (ORY-1001) dihydrochloride is a selective irreversible lysine (K)-specific demethylase 1A (KDM1A/LSD1) inhibitor.
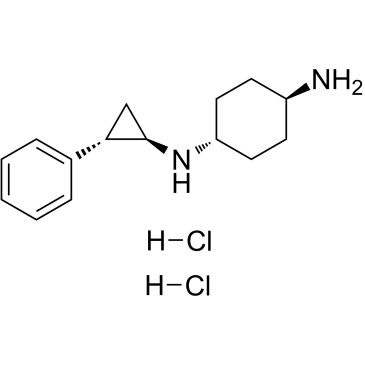
-
GC32745
OSS_128167
OSS_128167 is a potent selective silencing regulatory protein 6 (SIRT6) inhibitor with an IC50 of 89 μM.
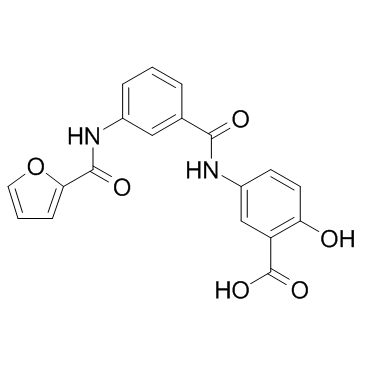
-
GC69636
OTS193320
OTS193320 is an imidazopyridine compound, a SUV39H2 methyltransferase activity inhibitor. OTS193320 reduces the global histone H3 lysine 9 trimethylation level in breast cancer cells and induces apoptosis cell death. Compared to single-agent OTS193320 or DOX, the combination of OTS193320 with Doxorubicin (DOX; A) can lead to a decrease in γ-H2AX levels and cancer cell viability.
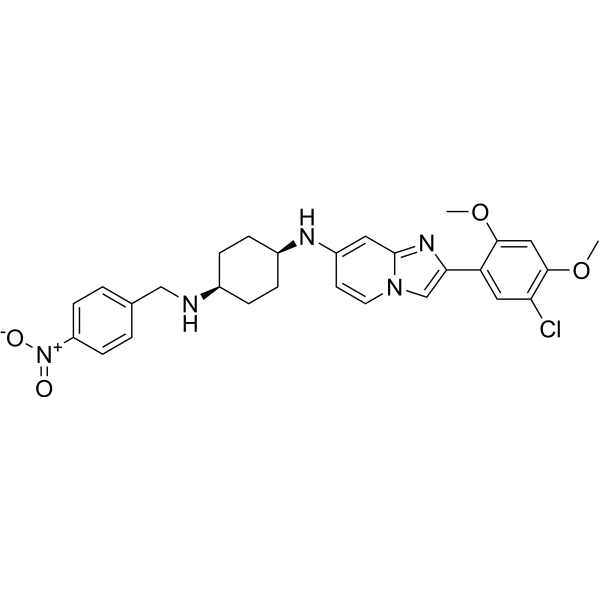
-
GC17973
OTX-015
A BRD2, BRD3, and BRD4 inhibitor
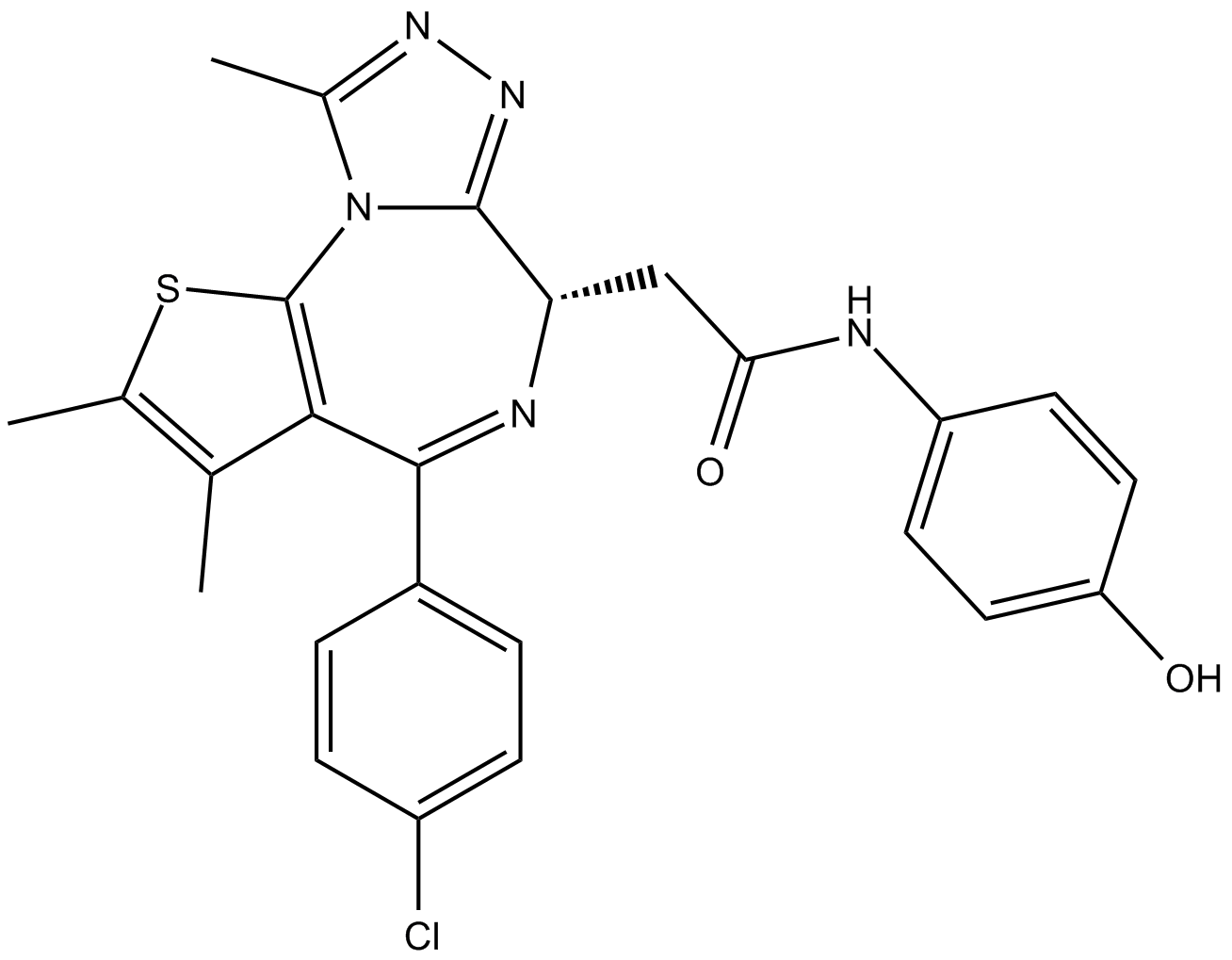
-
GC45808
OUL35
An inhibitor of PARP10
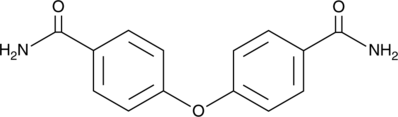
-
GC17472
Oxamflatin
HADC inhibitor
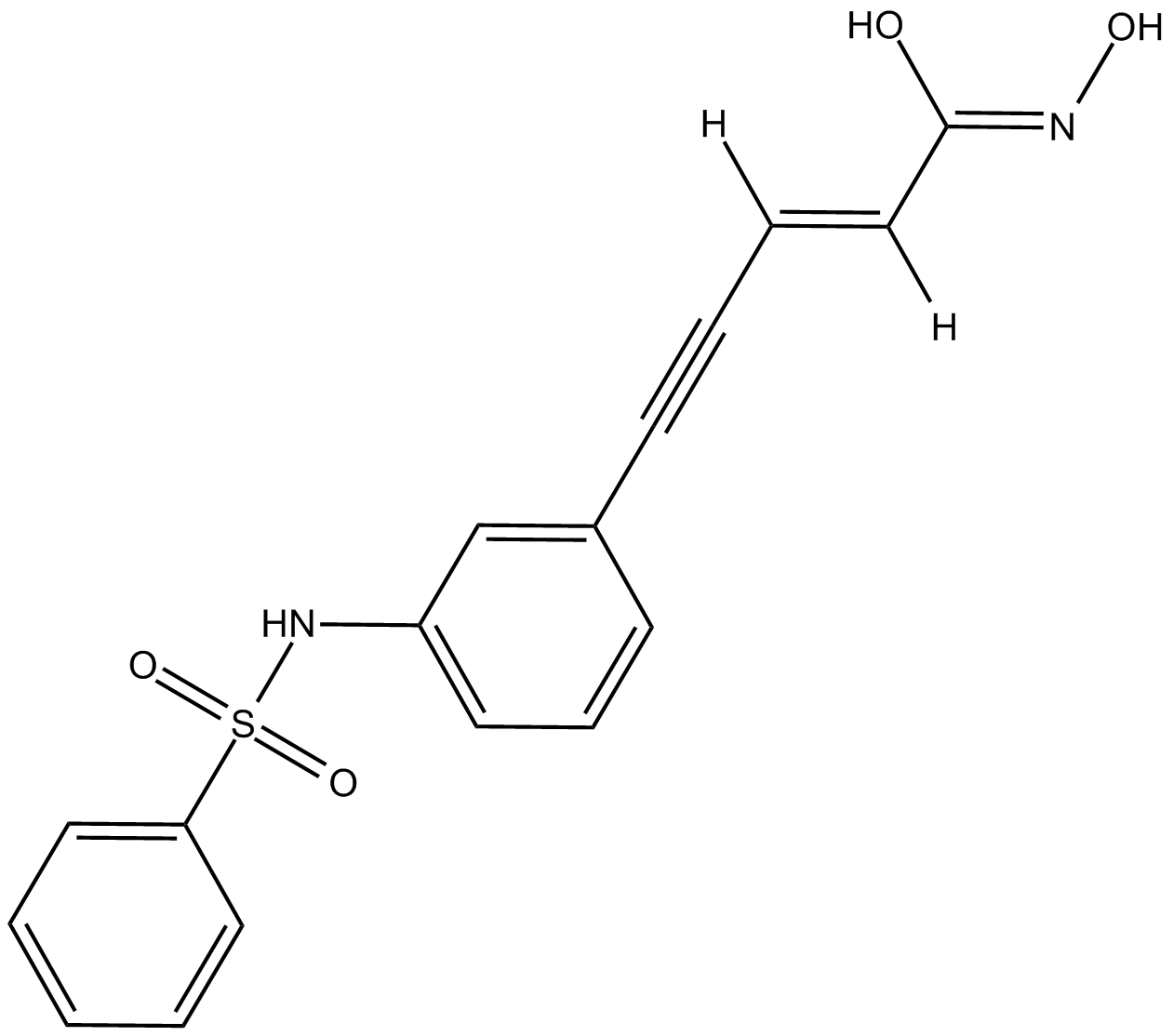
-
GC14655
OXF BD 02
Selective inhibitor of the first bromodomain of BRD4
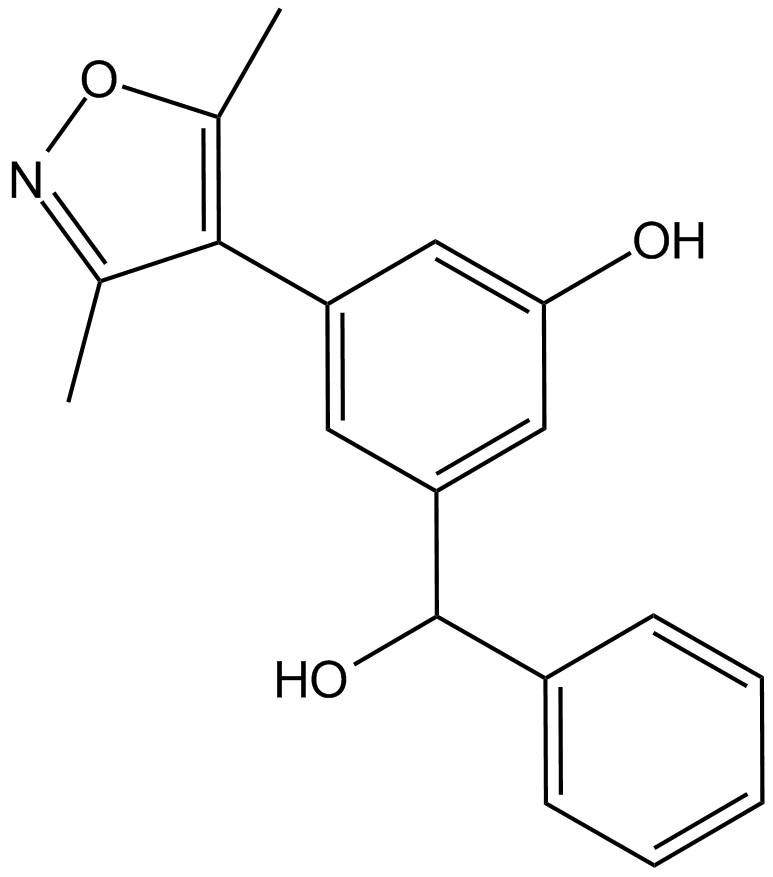
-
GC39417
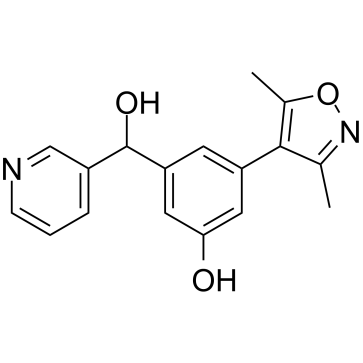
OXFBD04
OXFBD04 is a potent and selective BRD4 inhibitor with an IC50 of 166?nM. OXFBD04 is a potent BET bromodomain ligand with additional modest affinity for the CREBBP bromodomain. OXFBD04 has anti-cancer activity.
-
GC41329
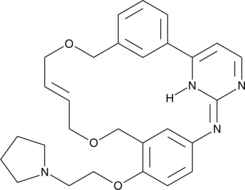
Pacritinib
FMS-like tyrosine kinase 3 (FLT3) and Janus kinase 2 (JAK2) are tyrosine kinases that mediate cytokine signaling and are frequently mutated in cancers, particularly acute myeloid leukemia.
-
GC67873
PAD-IN-2
-
GC64367
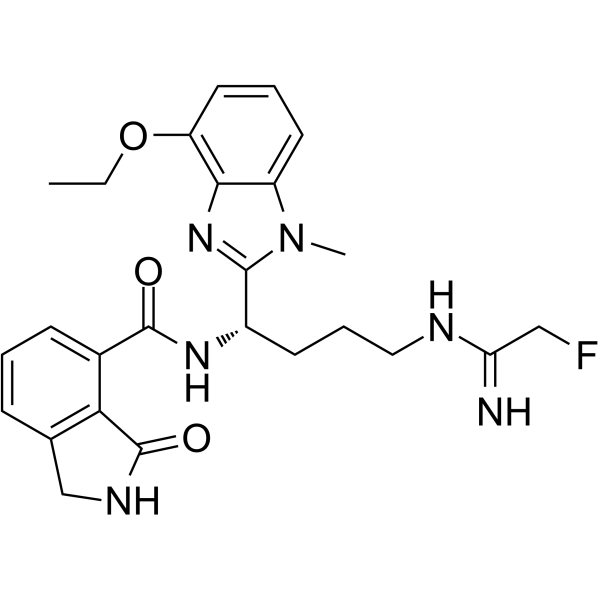
PAD2-IN-1
PAD2-IN-1, a benzimidazole-based derivative, is a potent and selective protein arginine deiminase 2 (PAD2) inhibitor.
-
GC66335
PAD2-IN-1 hydrochloride
PAD2-IN-1 hydrochloride, a benzimidazole-based derivative, is a potent and selective protein arginine deiminase 2 (PAD2) inhibitor. PAD2-IN-1 hydrochloride shows superior selectivity for PAD2 over PAD4 (95-fold) and PAD3 (79-fold).
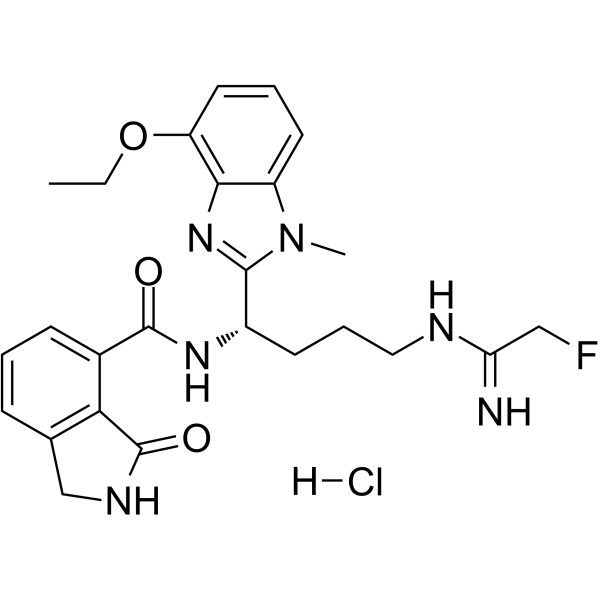
-
GN10452
Paeoniflorin
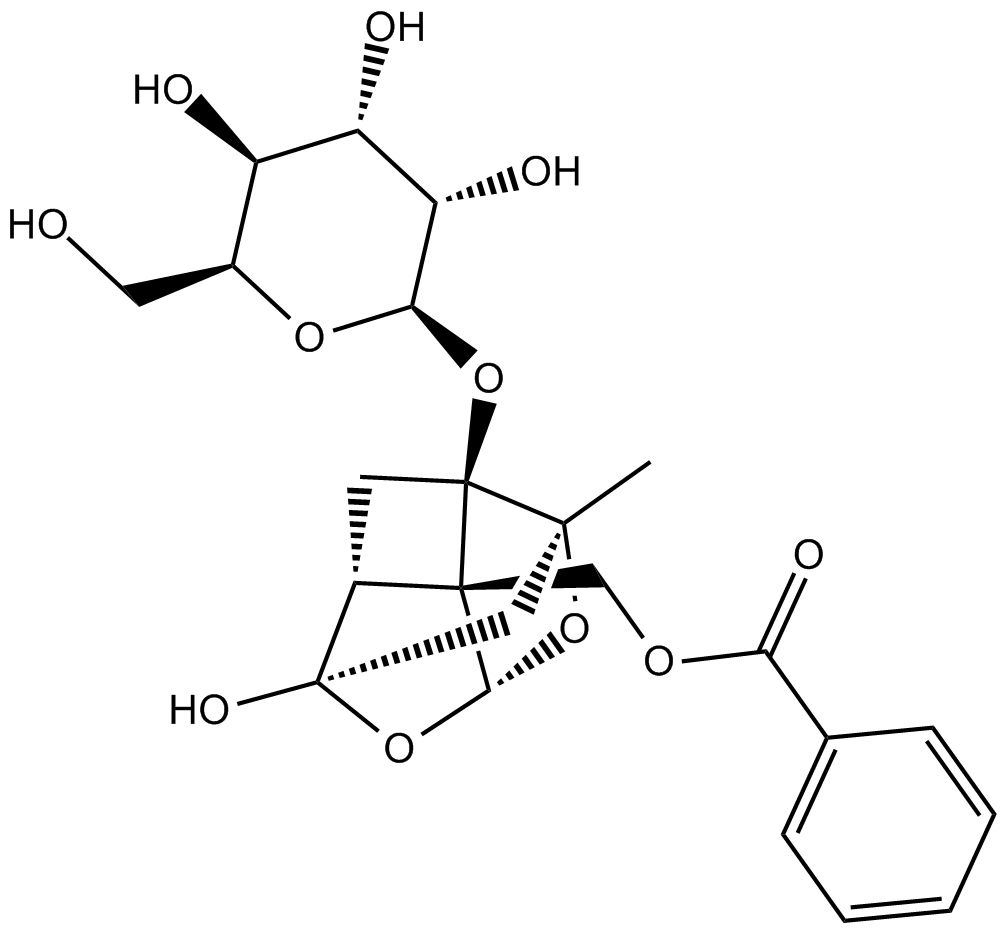
-
GC34071
Pamiparib (BGB-290)
Pamiparib (BGB-290) (BGB-290) is an orally active, potent, highly selective PARP inhibitor, with IC50 values of 0.9 nM and 0.5 nM for PARP1 and PARP2, respectively. Pamiparib (BGB-290) has potent PARP trapping, and capability to penetrate the brain, and can be used for the research of various cancers including the solid tumor.
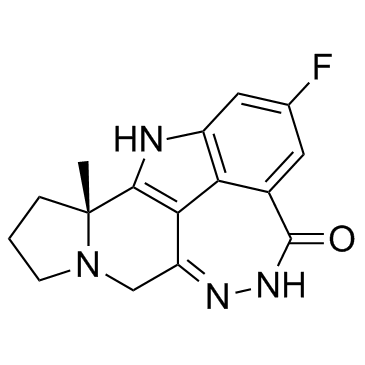
-
GC12257
Panobinostat (LBH589)
A pan-HDAC inhibitor
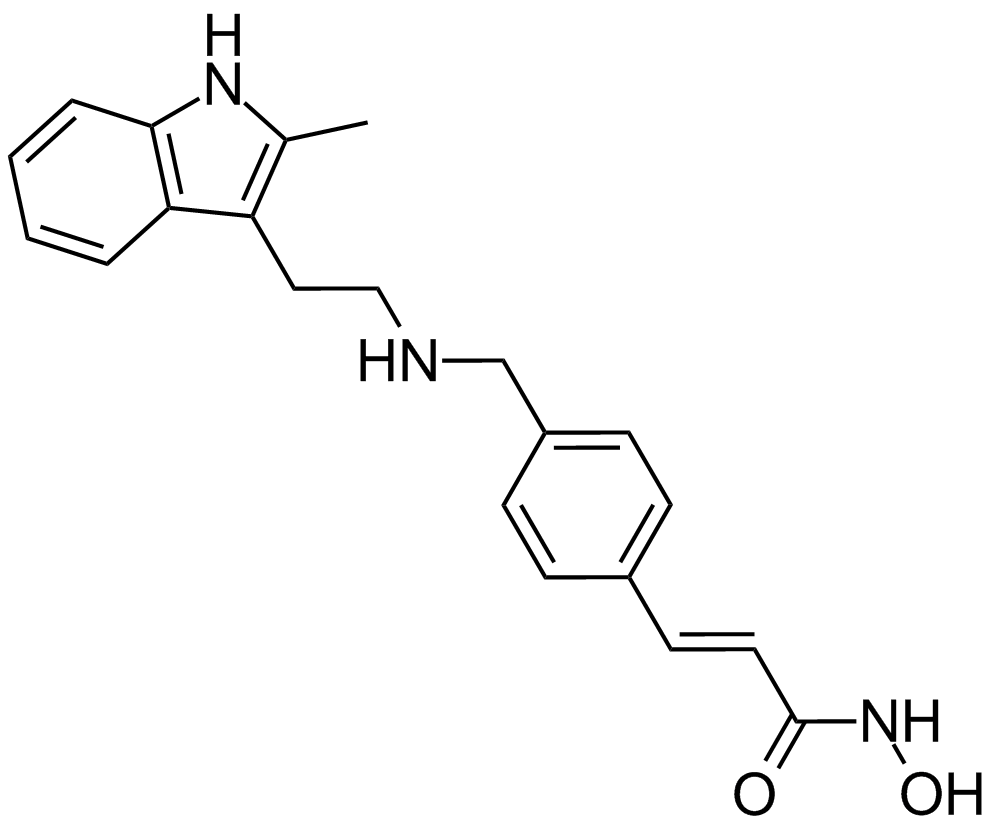
-
GC36855
Paris saponin VII
Paris saponin VII (Chonglou Saponin VII) is a steroidal saponin isolated from the roots and rhizomes of Trillium tschonoskii Maxim. Paris saponin VII-induced apoptosis in K562/ADR cells is associated with Akt/MAPK and the inhibition of P-gp. Paris saponin VII attenuates mitochondrial membrane potential, increases the expression of apoptosis-related proteins, such as Bax and cytochrome c, and decreases the protein expression levels of Bcl-2, caspase-9, caspase-3, PARP-1, and p-Akt. Paris saponin VII induces a robust autophagy in K562/ADR cells and provides a biochemical basis in the treatment of leukemia.
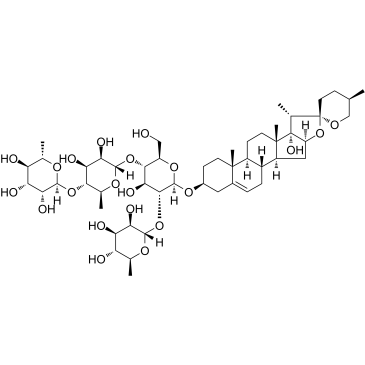
-
GC64579
PARP-1-IN-2
PARP-1-IN-2 (compound 11g) is a potent and BBB-penetrated PARP1 inhibitor, with an IC50 of 149 nM. PARP1-IN-2 shows significantly potent anti-proliferative activity against Human lung adenocarcinoma epithelial cell line A549. PARP1-IN-2 can induce A549 cells apoptosis.
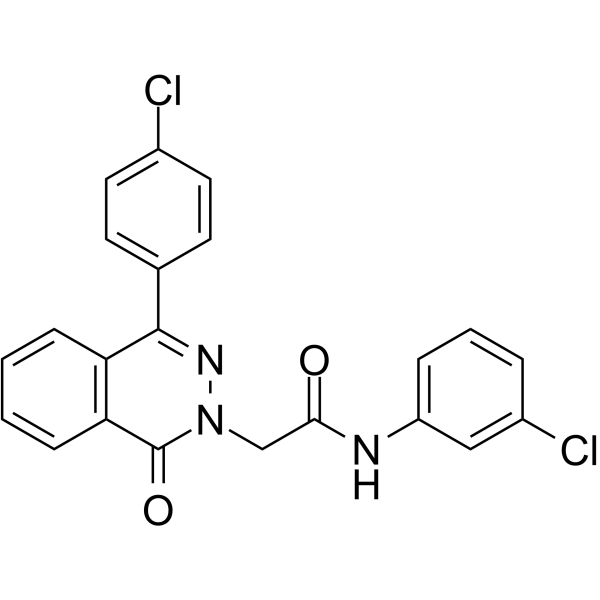
-
GC65927
PARP-2-IN-1
PARP-2-IN-1 is a potent and selective PARP-2 inhibitor with an IC50 of 11.5 nM.
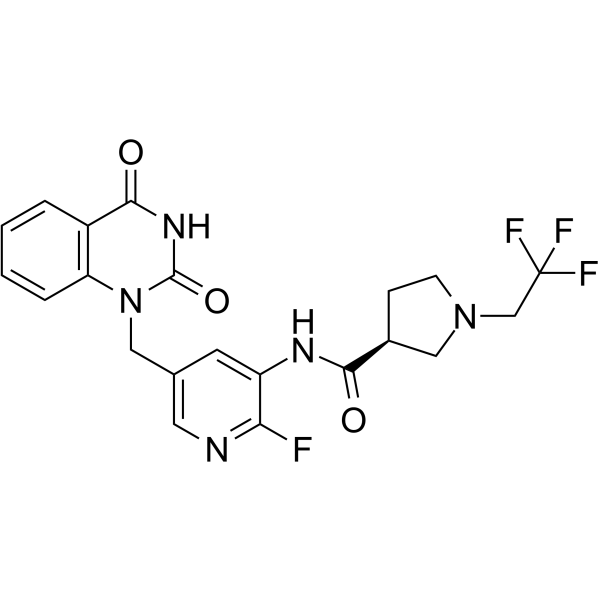
-
GC68006
PARP1-IN-11
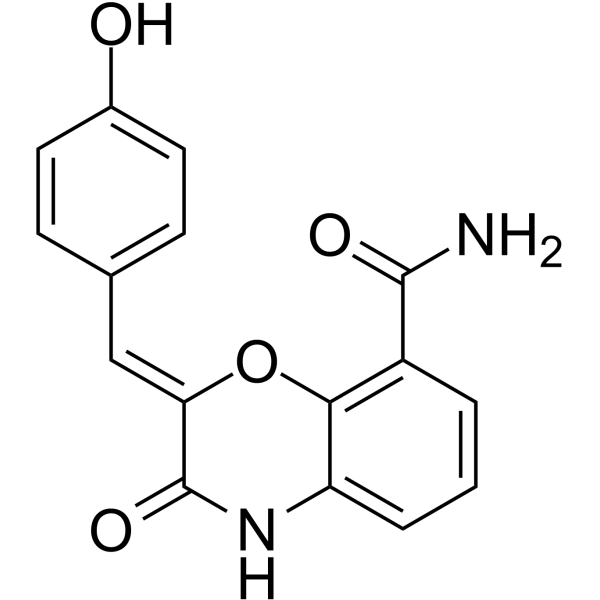
-
GC62275
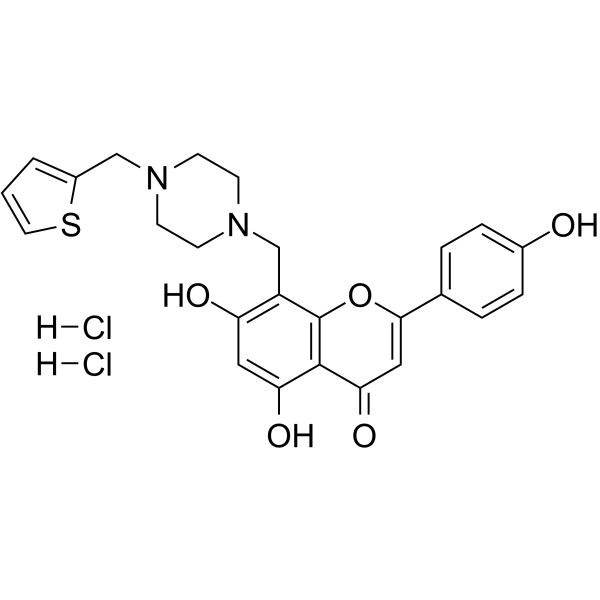
PARP1-IN-5 dihydrochloride
PARP1-IN-5 dihydrochloride is a low toxicity, orally active, potent and selective PARP-1 inhibitor (IC50 =14.7 nM). PARP1-IN-5 dihydrochloride can be used for the research of cancer.
-
GC69660
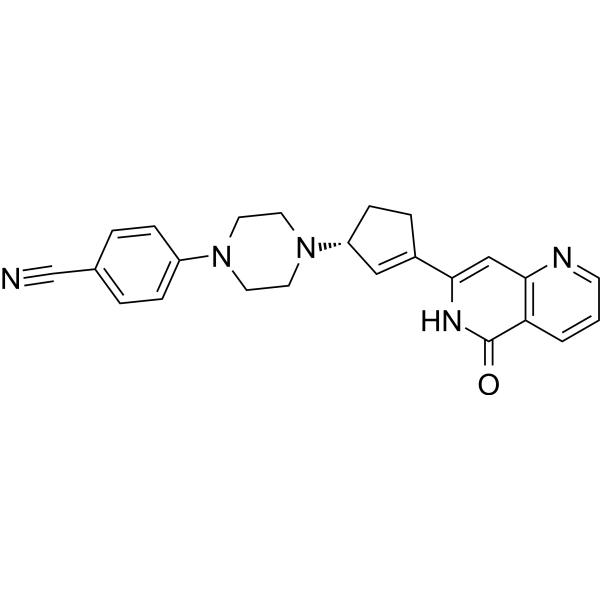
PARP1-IN-7
PARP1-IN-7 is an inhibitor of poly ADP-ribose polymerase-1 (PARP1), used as an anticancer agent.
-
GC64578
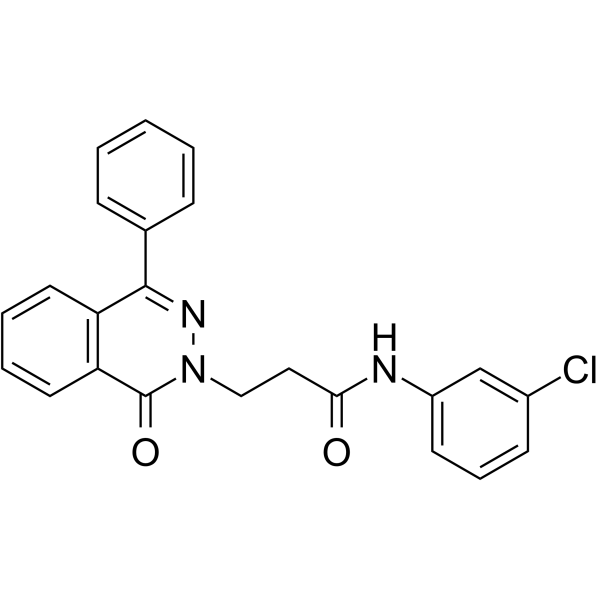
PARP1-IN-8
PARP1-IN-8 (compound 11c) is a potent and BBB-penetrated PARP1 inhibitor, with an IC50 of 97 nM. PARP1-IN-8 shows significantly potent anti-proliferative activity against Human lung adenocarcinoma epithelial cell line A549.
-
GC69657
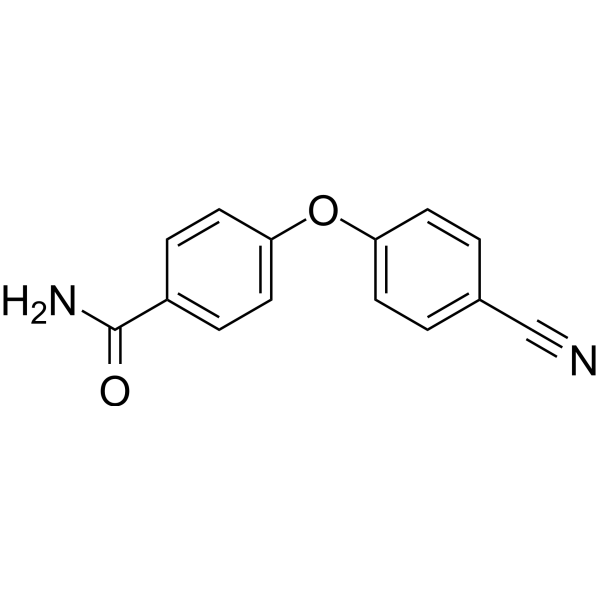
PARP10-IN-2
PARP10-IN-2 is an effective inhibitor of mono-ADP-ribosyltransferase PARP10, with an IC50 of 3.64 μM for human PARP10. It also inhibits PARP2 and PARP15, with IC50 values of 27 μM and 11 μM for human PARP2 and human PARP15, respectively.
-
GC69658
PARP10-IN-3
PARP10-IN-3 is a selective mono-ADP-ribosyltransferase PARP10 inhibitor with an IC50 of 480 nM for human PARP10. It also inhibits PARP2 and PARP15, with IC50 values of 1.7 μM for both human PARP2 and human PARP15.
-
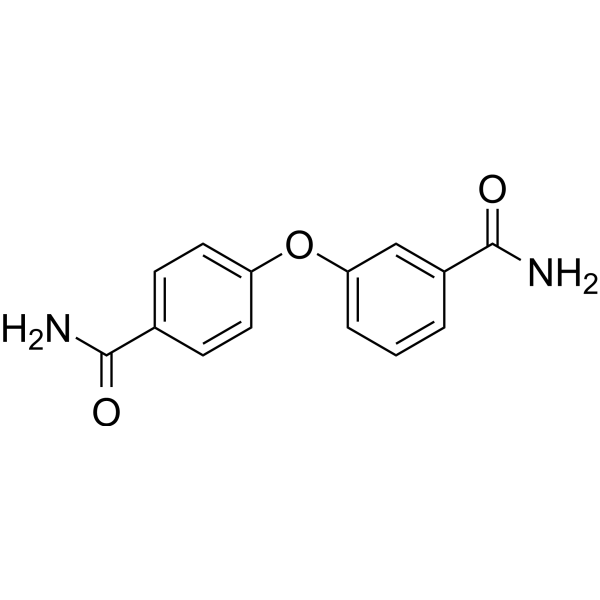
GC68035
PARP10/15-IN-1
-
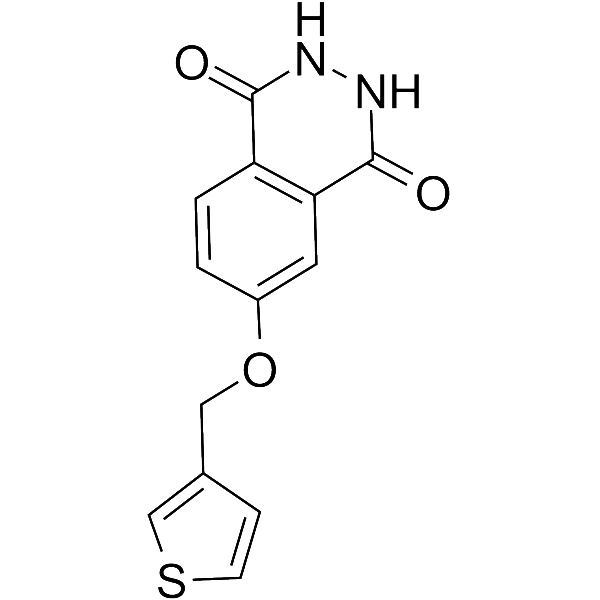
GC69655
PARP10/15-IN-2
PARP10/15-IN-2 (Compound 8h) is an effective dual inhibitor of PARP10 and PARP15, with IC50 values of 0.15 μM and 0.37 μM, respectively. It can enter cells and prevent apoptosis.
-
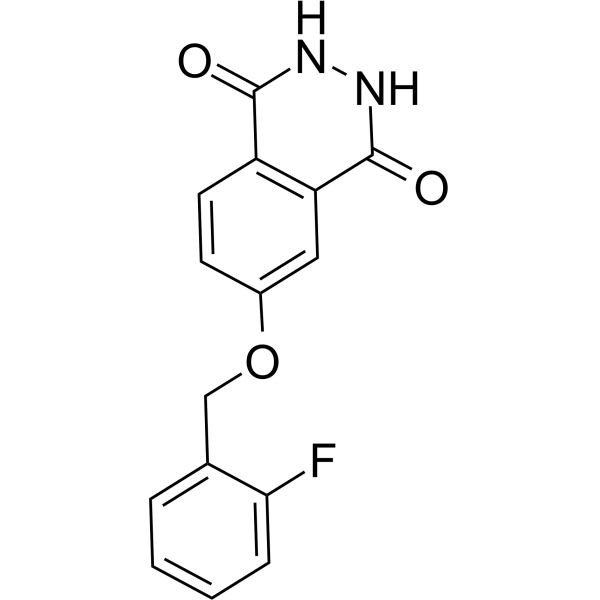
GC69656
PARP10/15-IN-3
PARP10/15-IN-3 (Compound 8a) is an effective dual inhibitor of PARP10 and PARP15, with IC50 values of 0.14 μM and 0.40 μM, respectively. It can enter cells and prevent apoptosis.
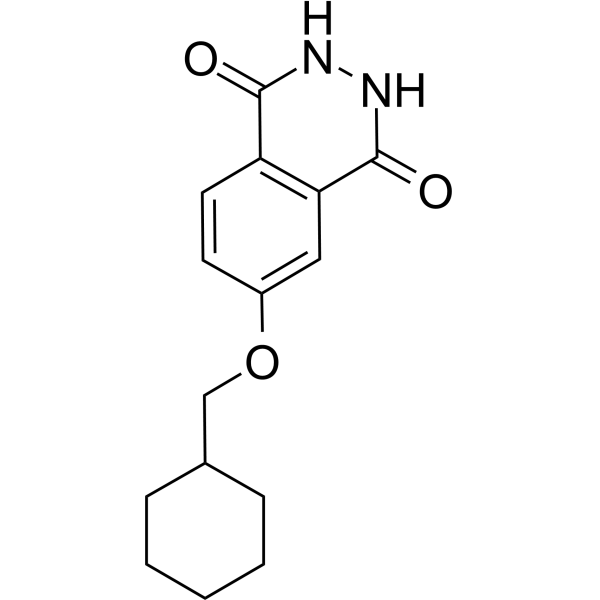
-
GC69659
PARP11 inhibitor ITK7
PARP11 inhibitor ITK7 (ITK7) is an effective and selective inhibitor of PARP11. It can effectively inhibit PARP11 with an IC50 value of 14 nM. PARP11 inhibitor ITK7 can be used for research on cellular localization.
-
GC39302
PARP14 inhibitor H10
PARP14 inhibitor H10, compound H 10, is a selective inhibitor against PARP14 (IC50=490 nM), over other PARPs (≈24 fold over PARP1). PARP14 inhibitor H10 induces caspase-3/7-mediated cell apoptosis.
-
GC69661
PARP7-IN-14
PARP7-IN-14 (I-1) is an effective selective PARP7 inhibitor with an IC50 value of 7.6 nM. PARP7-IN-14 has anti-cancer activity.
-
GN10357
Parthenolide
-
GC15301
PBIT
JARID1 family demethylases inhibitor
-
GC69669
PCAF-IN-2
PCAF-IN-2 (compound 17) is an effective inhibitor of PCAF, with an IC50 value of 5.31 μM. PCAF-IN-2 exhibits anti-tumor activity and induces apoptosis and cell cycle arrest at the G2/M phase.