Chromatin/Epigenetics
Epigenetics
Epigenetics means above genetics. It determines how much and whether a gene is expressed without changing DNA sequences. Epigenetic regulations include, 1. DNA methylation: the addition of methyl group to DNA, converting cytosine to 5-methylcytosine, mostly at CpG sites; 2. Histone modifications: posttranslational modificationEpigeneticss of histone proteins including acetylation, methylation, ubiquitylation, phosphorylation and sumoylation; 3. miRNAs: non-coding microRNA downregulating gene expression; 4. Prions: infectious proteins viewed as epigenetic agents capable of inducing a phenotype without changing the genome.
Targets for Chromatin/Epigenetics
- Bromodomain(52)
- Aurora Kinase(89)
- DNA Methyltransferase(40)
- HDAC(218)
- Histone Acetyltransferases(67)
- Histone Demethylases(98)
- Histone Methyltransferase(212)
- HIF(101)
- JAK(178)
- MBT Domains(1)
- PARP(128)
- Pim(35)
- Protein Ser/Thr Phosphatases(41)
- RNA Polymerase(8)
- Sirtuin(84)
- Sphingosine Kinase-2(1)
- Polycomb repressive complex(2)
- SUMOylation(3)
- PAD(18)
- Epigenetic Reader Domain(207)
- MicroRNA(13)
- Protein Arginine Deiminase(12)
- Chromodomain(1)
- Citrullination(15)
- DNA/RNA Demethylation(1)
- DNA/RNA Methylation(6)
- Histone Deacetylation(38)
- Histones/Histone Peptides(7)
- PHD Domains(0)
- Tandem Tudor & Tudor-like Domains(1)
- PRMT(2)
Products for Chromatin/Epigenetics
- Cat.No. Product Name Information
-
GC25580
LLY-284
LLY-284 is the diastereomer of LLY-283, which is a potent and selective SAM-competitive chemical probe for PRMT5. LLY-284 is much less active than LLY-283 and can be used as a negative control for LLY-283.
-
GC16261
LLY507
SMYD2 inhibitor
-
GC13237
LMK 235
A selective inhibitor of HDAC4 and HDAC5
-
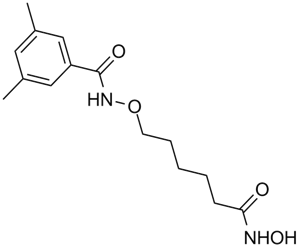
GC11778
Lomeguatrib
Inactivator of O6-methylguanine-DNA methyltransferase
-
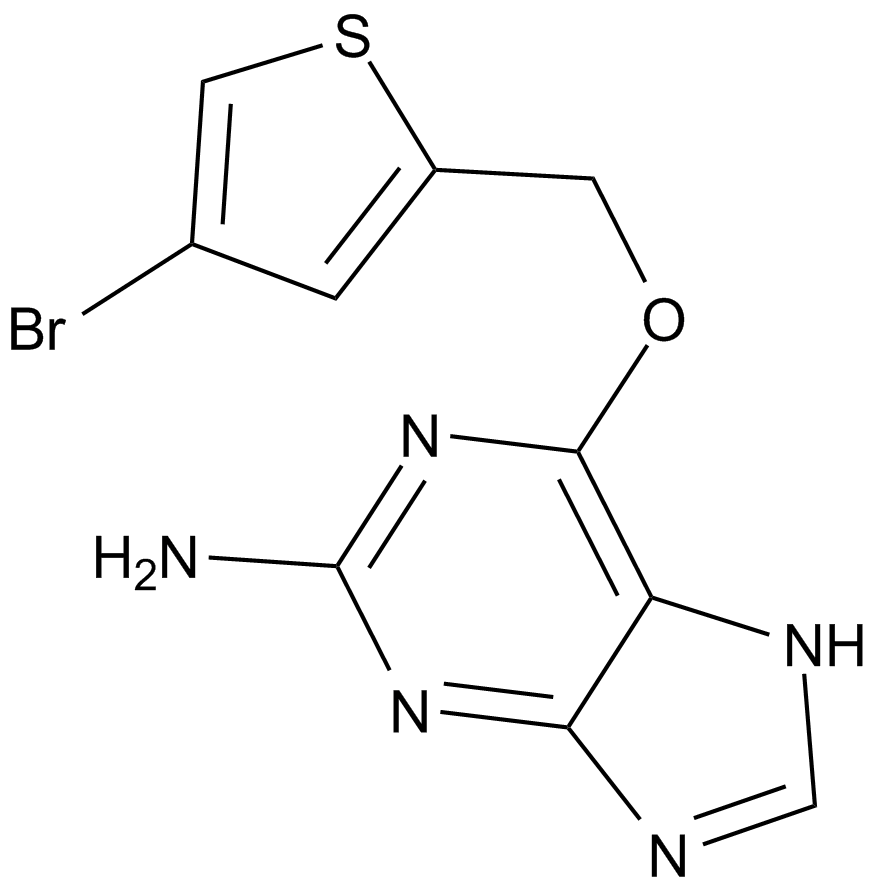
GC64372
Lorpucitinib
Lorpucitinib is a Gut-Restricted JAK Inhibitor for the research of Inflammatory Bowel Disease.
-
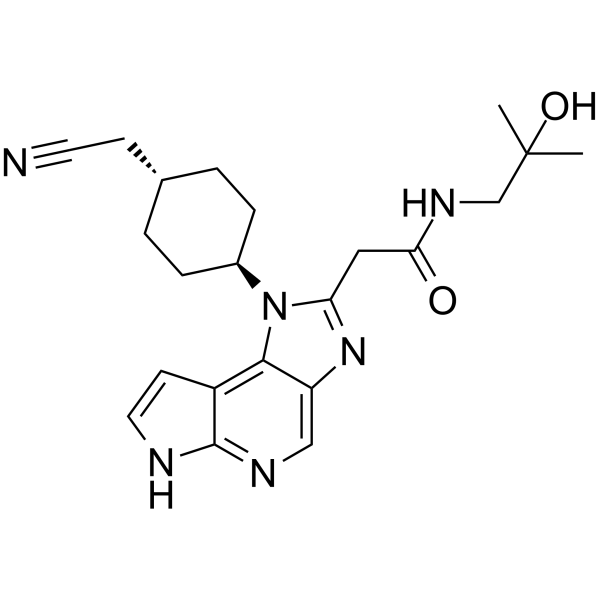
GC18731
LP99
LP99 is a potent inhibitor of the bromodomain containing proteins BRD7 and BRD9 that binds with Kd values of 99 and 909 nM, respectively, as determined by isothermal titration calorimetry.
-
GC36484
LSD1-IN-5
LSD1-IN-5 (Compound 4e) is a potent and reversible inhibitor of lysine-specific demethylase 1 (LSD1), with an IC50 of 121 nM. LSD1-IN-5 increases dimethylated Lys4 of histone H3, shows no effect on expression of LSD1.
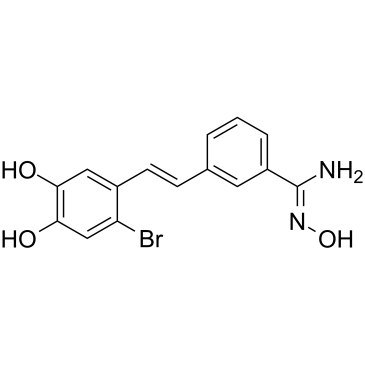
-
GC36485
LSD1-IN-6
LSD1-IN-6 (Compound 4m) is a potent and reversible inhibitor of lysine-specific demethylase 1 (LSD1), with an IC50 of 123 nM. LSD1-IN-6 increases dimethylated Lys4 of histone H3, shows no effect on expression of LSD1.
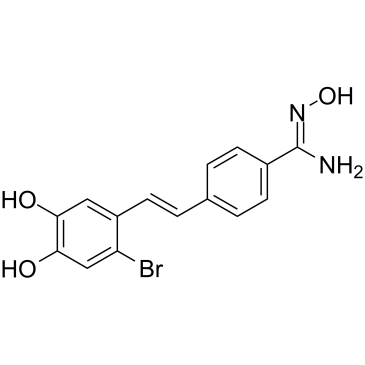
-
GC62434
LSD1/HDAC6-IN-1
LSD1/HDAC6-IN-1 is an orally active dual inhibitor of lysine specific demethylase 1(LSD1)/Histone deacetylase 6 (HDAC6), with anti-tumor activity. LSD1/HDAC6-IN-1 can be used for the research of multiple myeloma (MM).
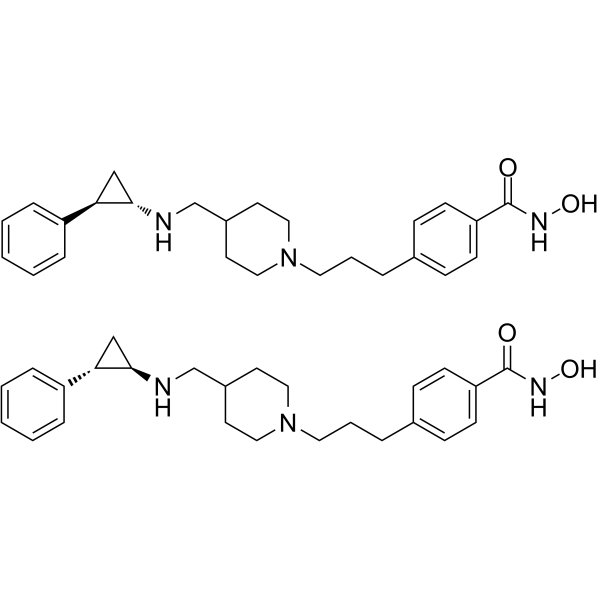
-
GC62591
LT052
LT052 is a highly selective BET BD1 inhibitor with an IC50 of 87.7 nM.
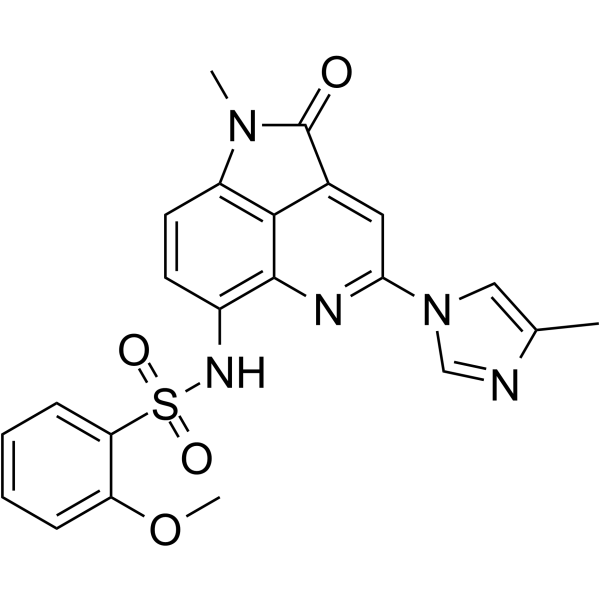
-
GC32724
LW6 (HIF-1α inhibitor)
LW6 (HIF-1α inhibitor) (HIF-1α inhibitor) is a novel HIF-1 inhibitor with an IC50 of 4.4 μM. LW6 (HIF-1α inhibitor) decreases HIF-1α protein expression without affecting HIF-1β expression.
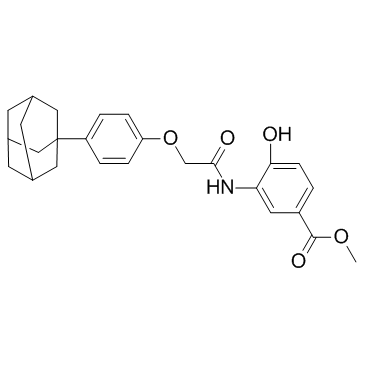
-
GC13848
LY2784544
Potent inhibitor of JAK2
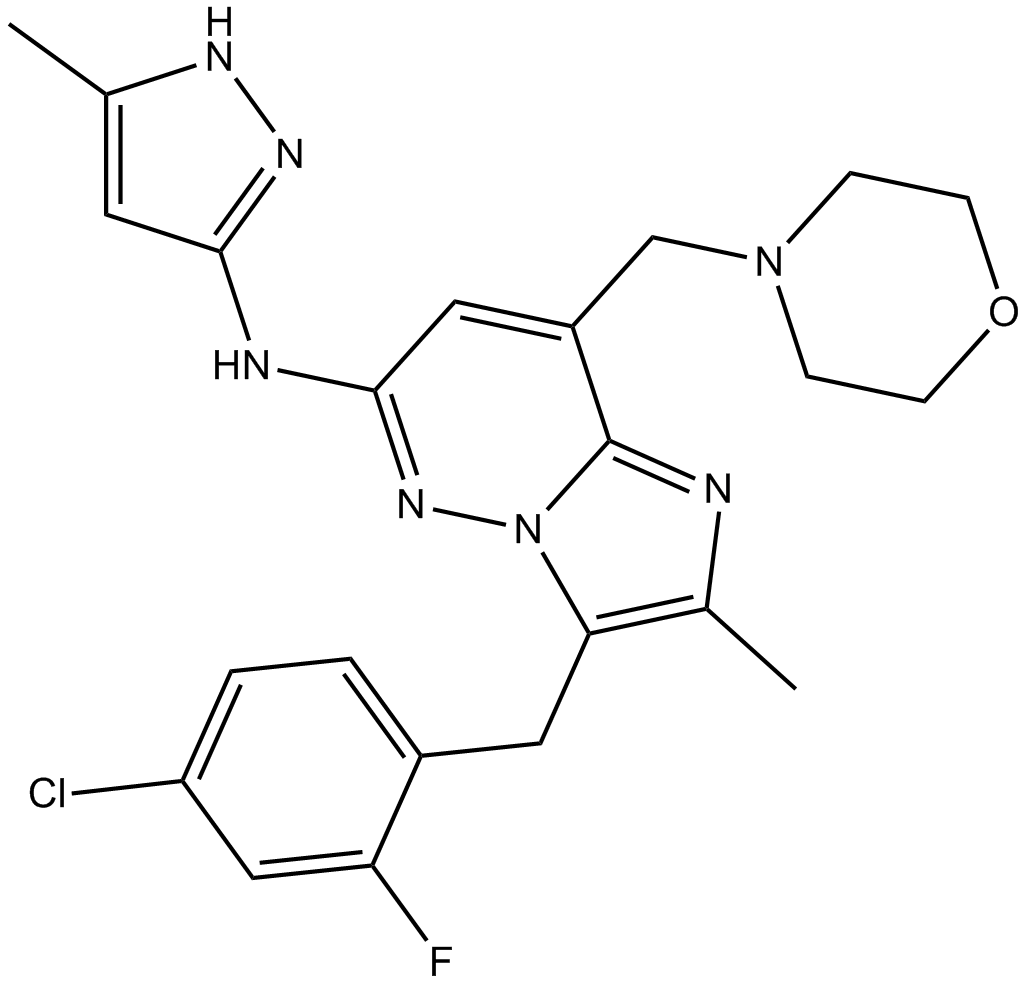
-
GC33057
LY3295668 (AK-01)
LY3295668 (AK-01) (AK-01) is a potent, orally active and highly specific Aurora-A kinase inhibitor, with Ki values of 0.8 nM and 1038 nM for AurA and AurB, respectively.
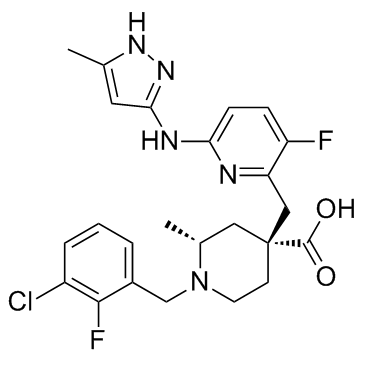
-
GC68405
Lys-CoA TFA
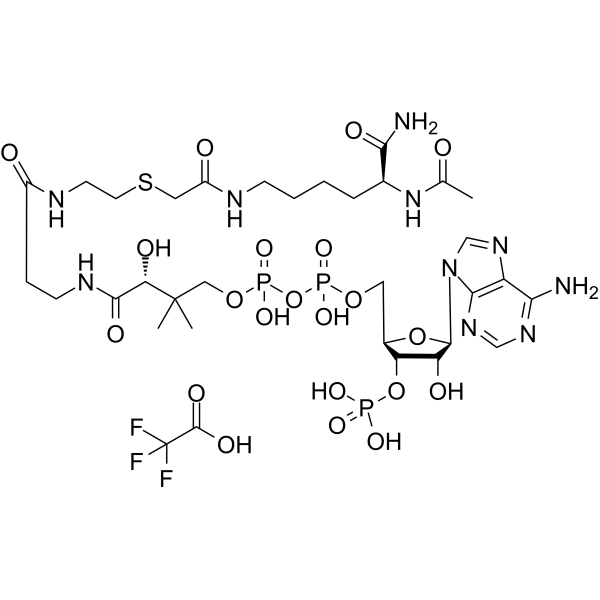
-
GC13534
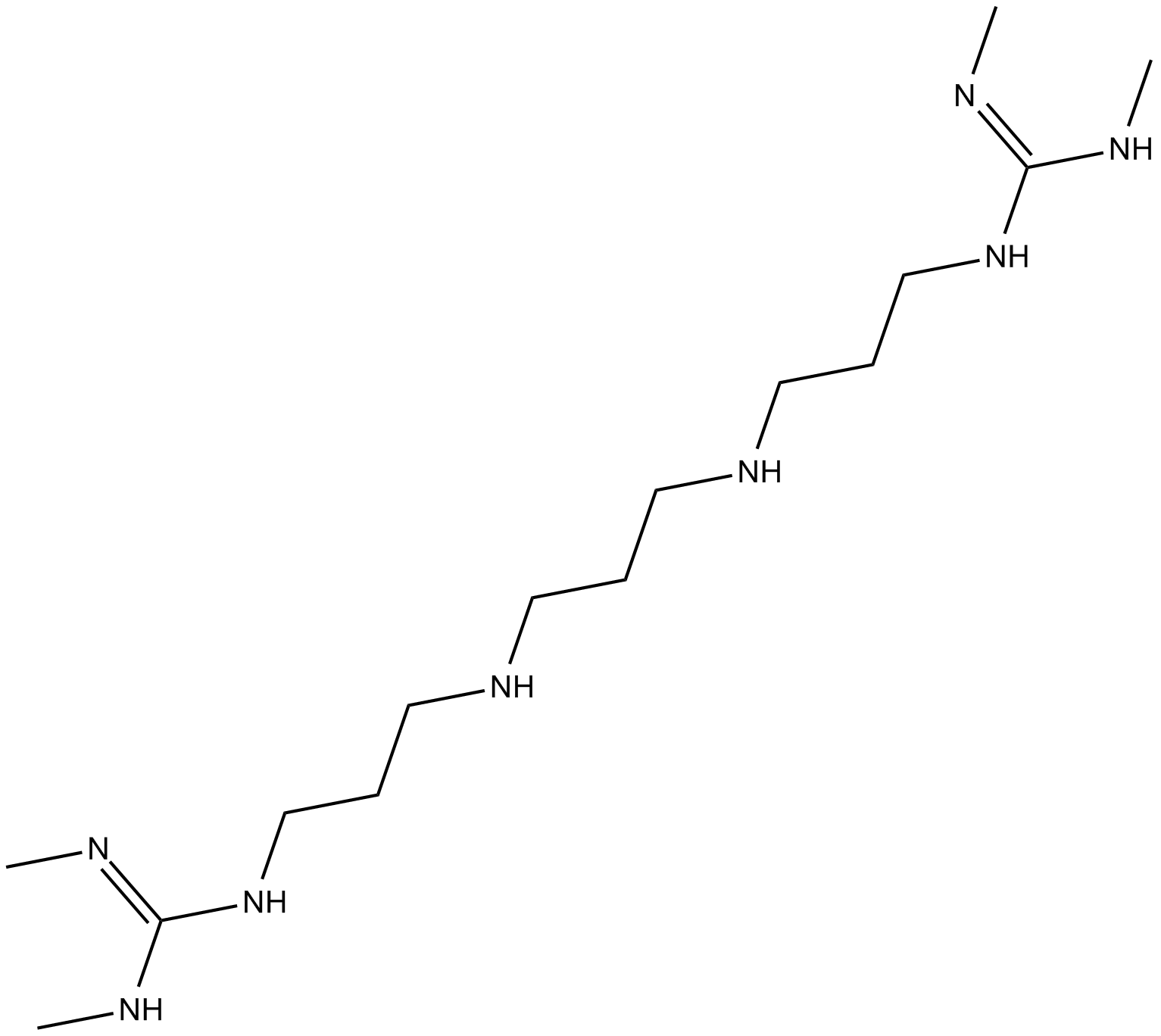
Lysine-specific Demethylase Inhibitor (1C)
lysine-specific demethylase Inhibitor
-
GC47586
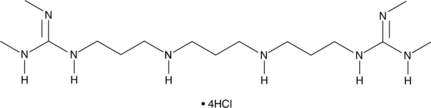
Lysine-specific Demethylase Inhibitor (1C) (hydrochloride)
An LSD1 inhibitor
-
GC34138
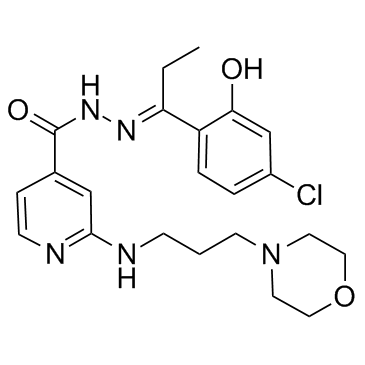
M-110
M-110 is a highly selective, ATP-competitive inhibitor of PIM kinases with a preference for PIM-3 (IC50=47 nM). M-110 inhibits PIM-1 and PIM-2 with similar IC50s of 2.5 μM. M-110 inhibits the proliferation of prostate cancer cell lines with IC50s of 0.6 to 0.9 μM.
-
GC47693
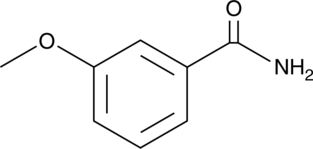
m-Methoxybenzamide
m-Methoxybenzamide (3-MBA), an inhibitor of ADP-ribosyltransferase (ADPRTs) and PARP, inhibits cell division in Bacillus subtilis, leading to filamentation and eventually lysis of cells. m-Methoxybenzamide (3-MBA) enhances in vitro plant growth, microtuberization, and transformation efficiency of blue potato (Solanum tuberosum L. subsp. andigenum).
-
GC64945
M1001
M1001 is a weak hypoxia-inducible factor-2α (HIF-2α) agonist. M1001 can bind to the HIF-2α PAS-B domain, with a Kd of 667 nM. M1001 can be used in chronic kidney disease research.
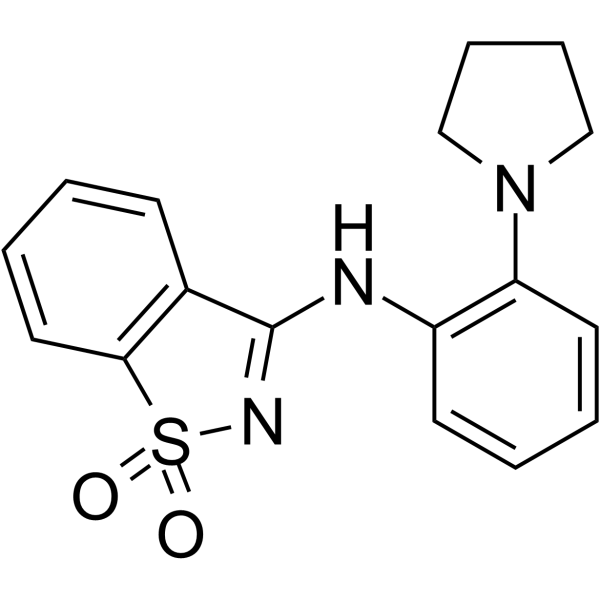
-
GC65046
M1002
M1002 is a hypoxia-inducible factor-2 (HIF-2) agonist, and can enhance the expression of HIF-2 target genes. M1002 shows synergy with prolyl-hydroxylase domain (PHD) inhibitors.
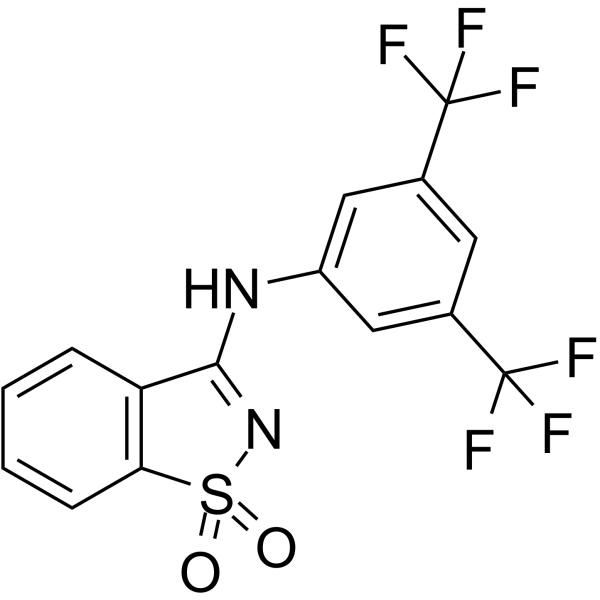
-
GC16456
M344
HDAC inhibitor
-
GC36521
M89
M-89 is a highly potent and specific menin inhibitor, with a Kd of 1.4 nM for binding to menin.
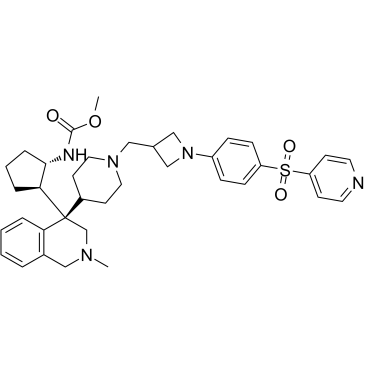
-
GC63056
MAK683-CH2CH2COOH
MAK683-CH2CH2COOH binds to EED (embryonic ectoderm development protein). MAK683-CH2CH2COOH and a VHL ligand for the E3 ubiquitin ligase have been used to design PROTAC EED degrader-1 and PROTAC EED degrader-2.
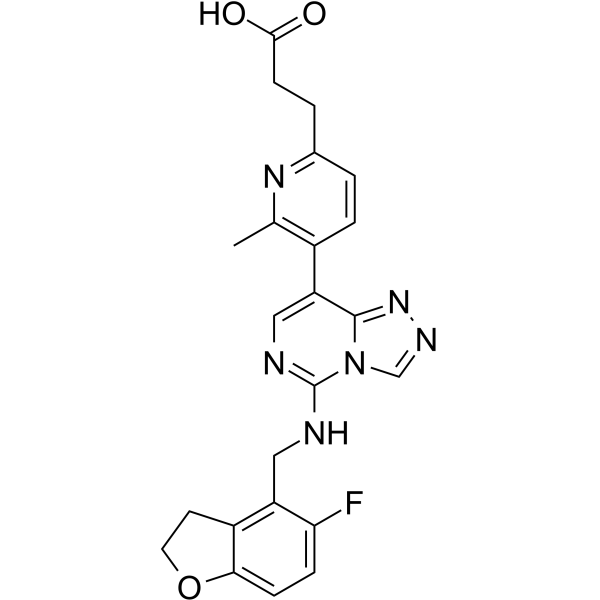
-
GC61029
Marein
A glucoside chalcone with diverse biological activities
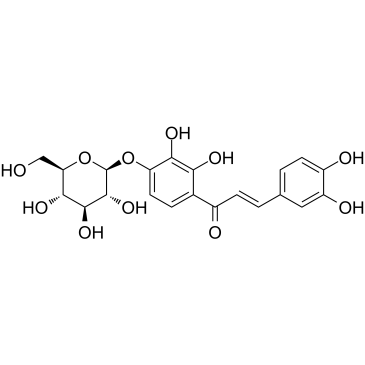
-
GC15965
MC1568
A selective class IIa HDAC inhibitor
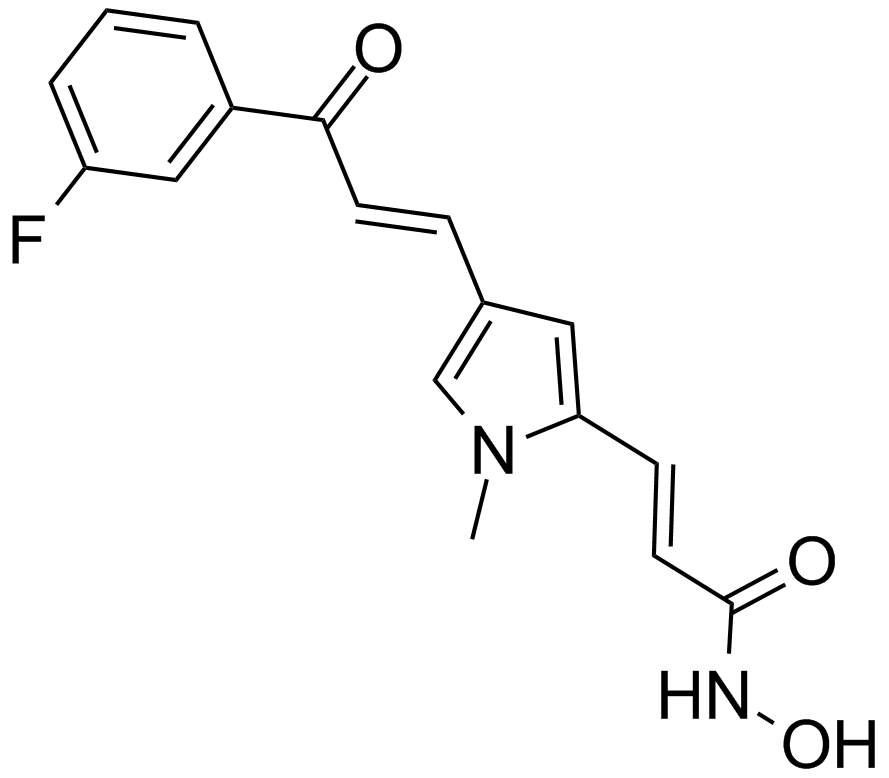
-
GC45906
MC1742
An inhibitor of class I and class IIb HDACs
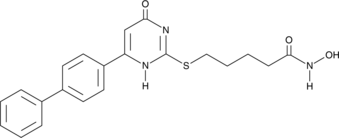
-
GC34431
MC3482
MC3482 is a specific sirtuin5 (SIRT5) inhibitor.
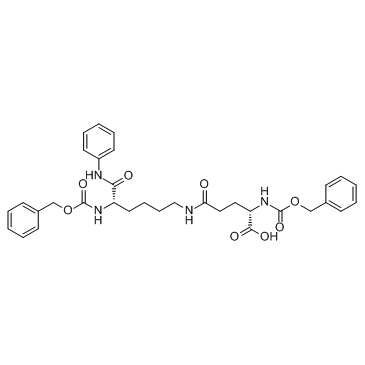
-
GC65254
MC4355
MC4355 is a dual inhibitor of EZH2 and histone deacetylase (HDAC).
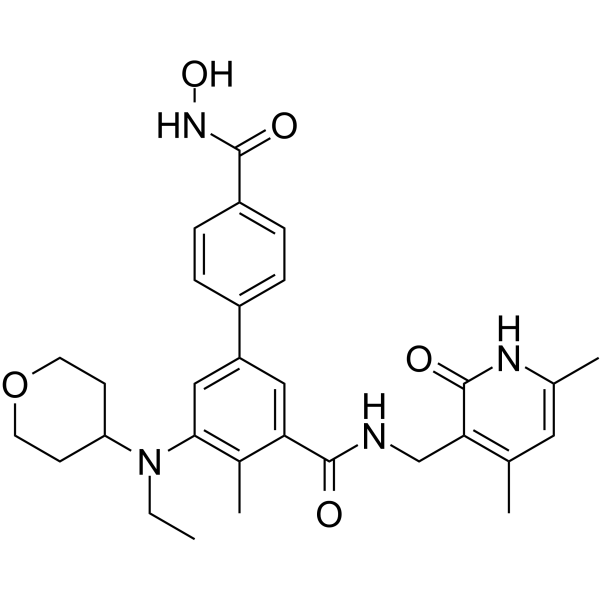
-
GC15049
MCB-613
stimulator of steroid receptor coactivator (SRC)
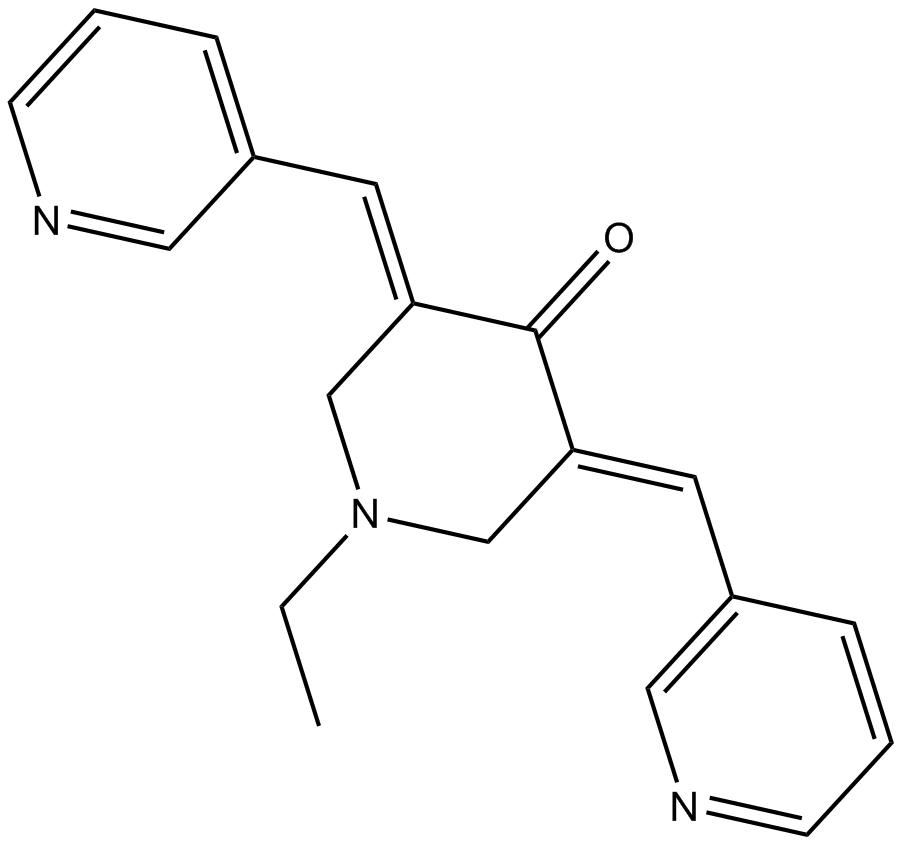
-
GC52188
MDL 800

-
GC13419
ME0328
PARP inhibitor,potent and selective
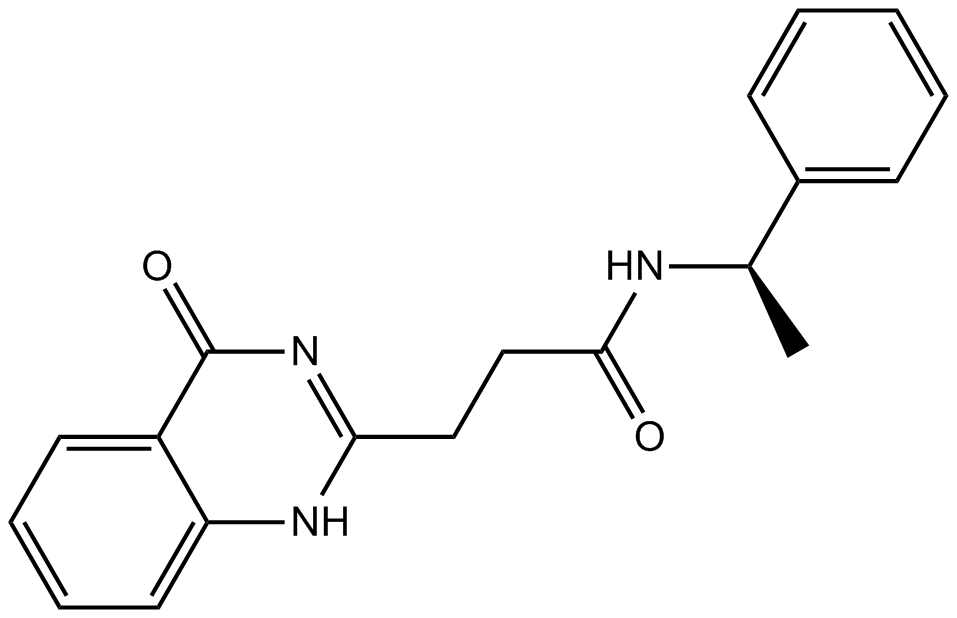
-
GC62252
Mefuparib hydrochloride
Mefuparib hydrochloride (MPH) is an orally active, substrate-competitive and selective PARP1/2 inhibitor with IC50s of 3.2 nM and 1.9 nM, respectively. Mefuparib hydrochloride induces apoptosis and possesses prominent anticancer activity in vitro and in vivo.
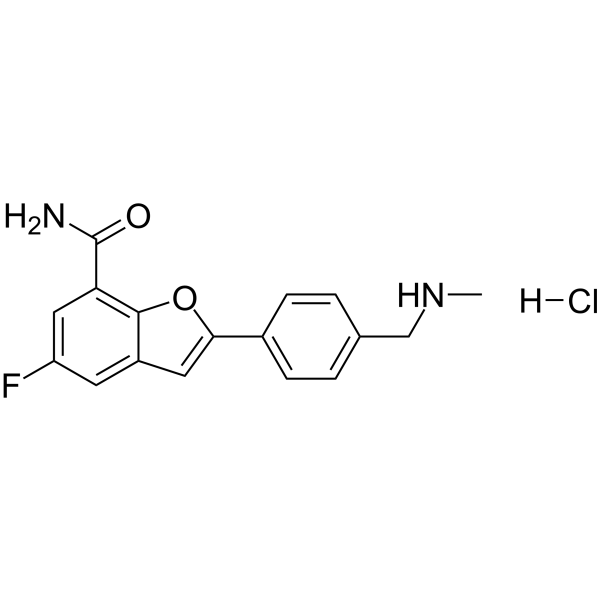
-
GC63064
Menin-MLL inhibitor 19
Menin-MLL inhibitor 19, a potent exo-aza spiro inhibitor of menin-mll interaction, example A17, extracted from patent WO2019120209A1.
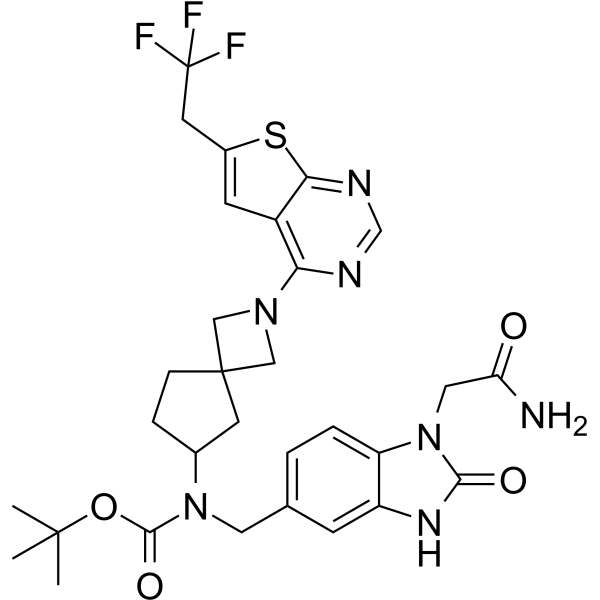
-
GC63538
Menin-MLL inhibitor 20
Menin-MLL inhibitor 20 is an irreversible menin-MLL interaction inhibitor with antitumor activities (WO2020142557A1, Intermediate 6).

-
GC48805
Methapyrilene (hydrochloride)
An H1 receptor antagonist and non-genotoxic carcinogen
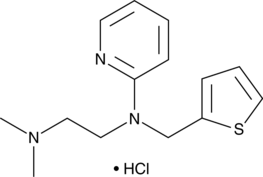
-
GC44184
Methylstat (hydrate)
Methylstat is a methyl ester prodrug of a Jumonji C domain-containing histone demethylase (JMJD) inhibitor that has favorable cell permeability.
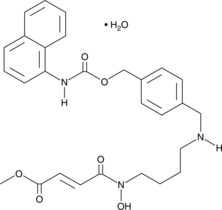
-
GC61058
Metoprine
Metoprine (BW 197U) is a potent histamine N-methyltransferase (HMT) inhibitor.
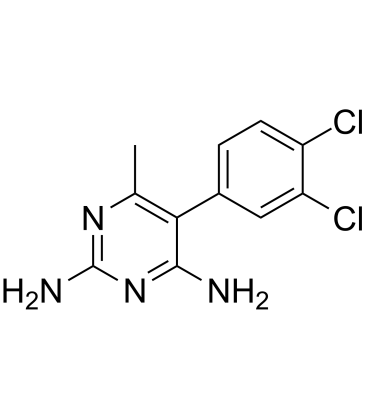
-
GC11439
MG 149
HAT inhibitor
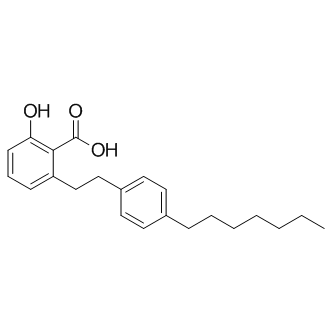
-
GC11949
MI-192
inhibitor of histone deacetylases (HDACs)
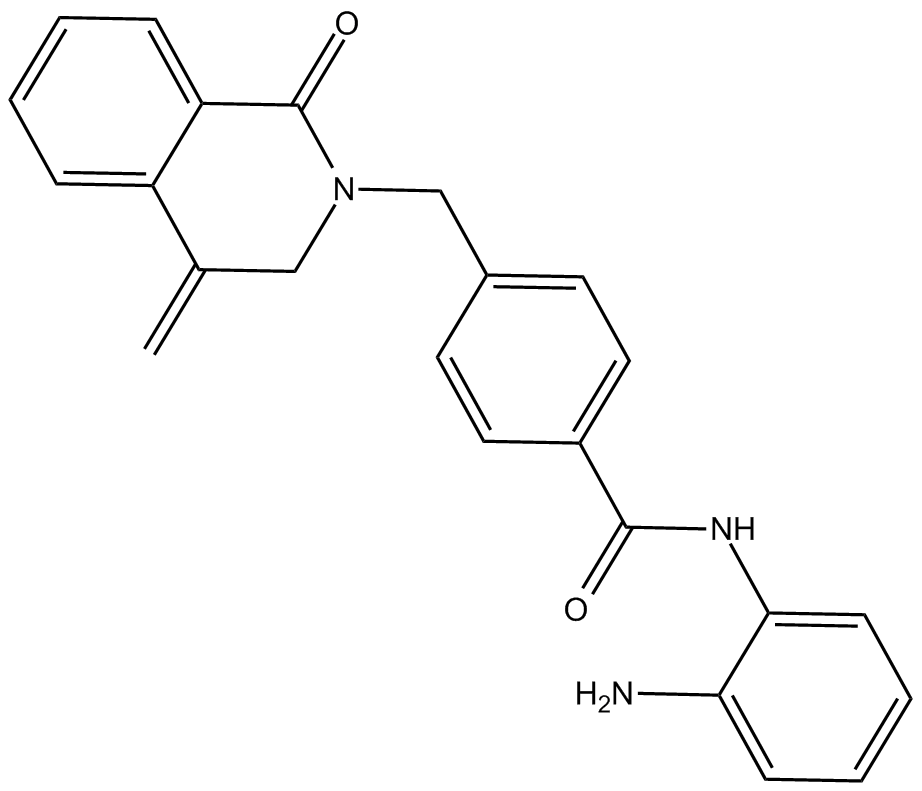
-
GC11418
MI-2
An inhibitor of menin-MLL fusion protein interactions
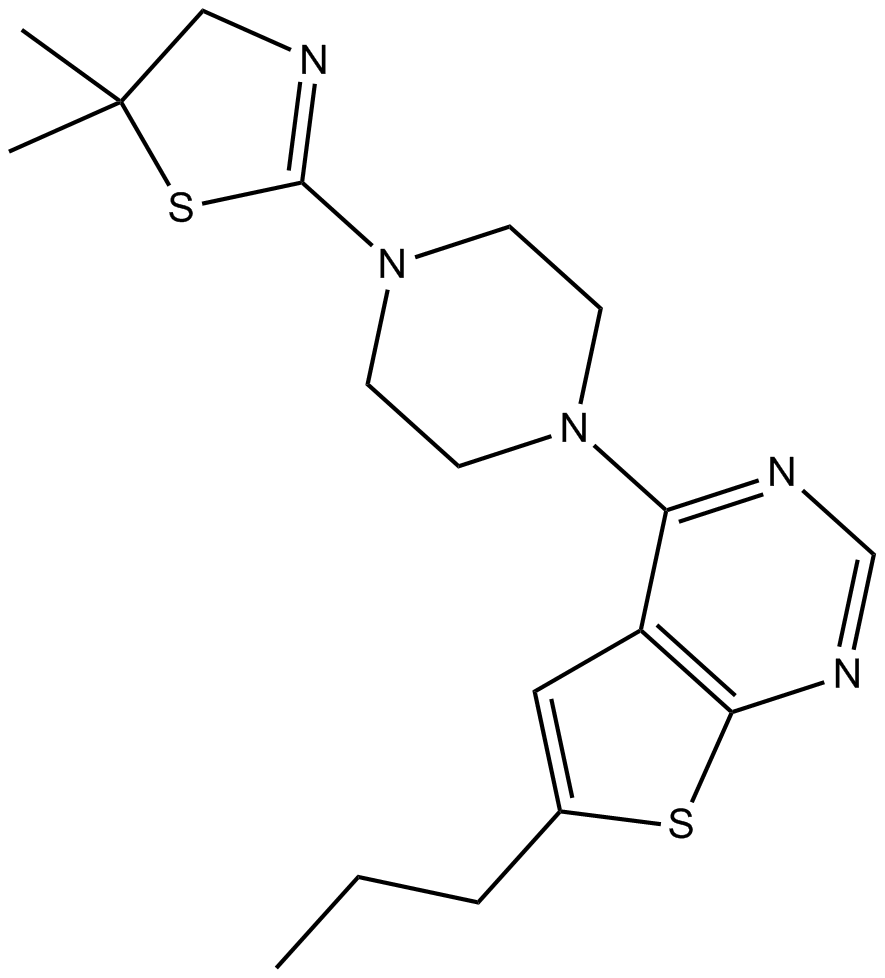
-
GC12199
MI-3
An inhibitor of menin-MLL fusion protein interactions
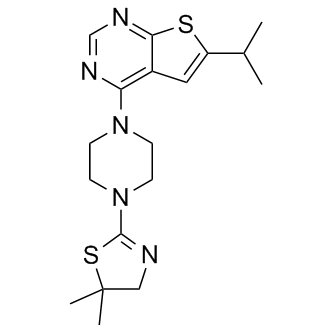
-
GC19246
MI-463
MI-463 is a highly potent and orally bioavailable small molecule inhibitor of the menin-mLL interaction.
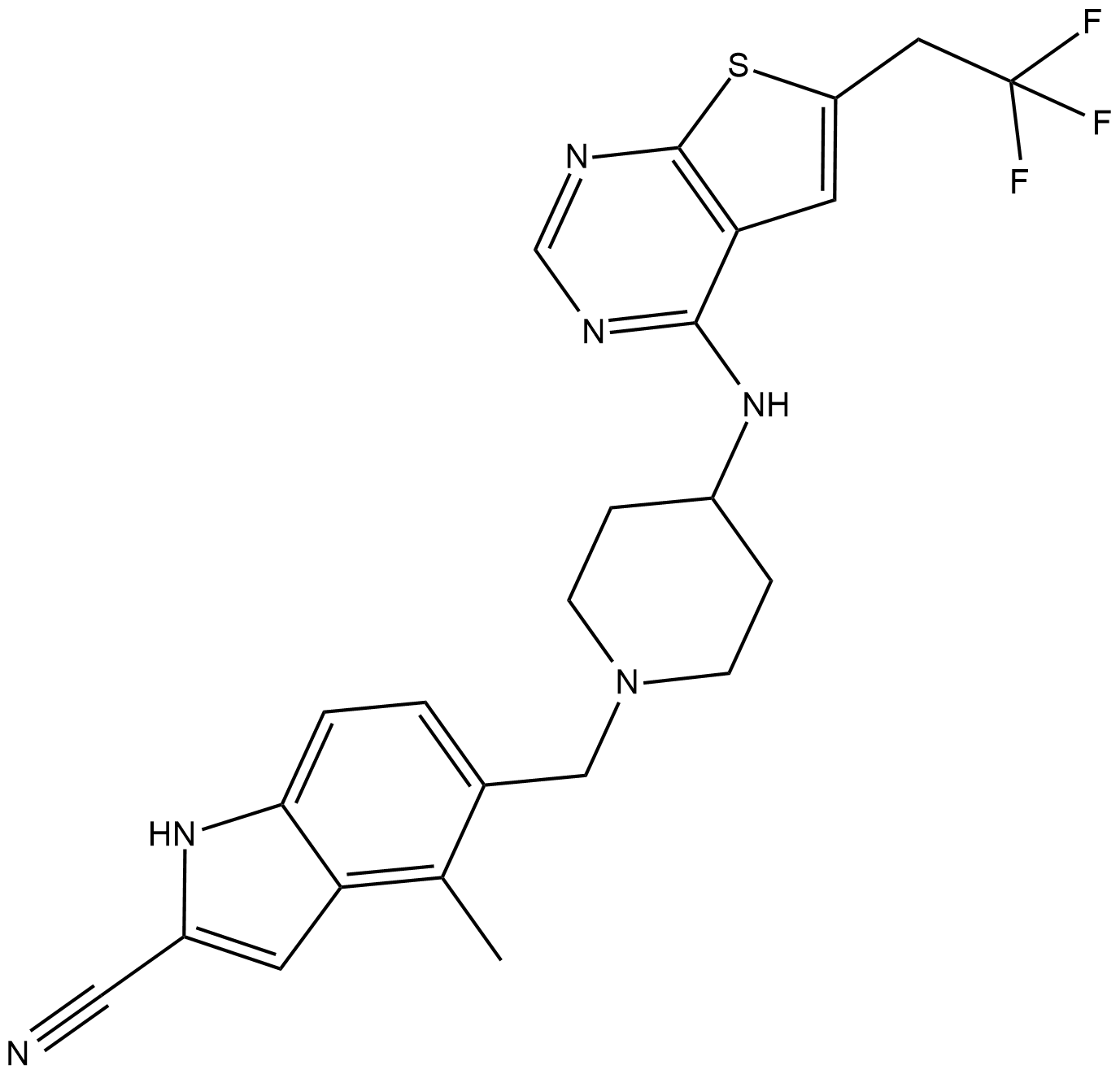
-
GC19247
MI-503
MI-503 is a highly potent and orally bioavailable small molecule inhibitor of the menin-mLL interaction.
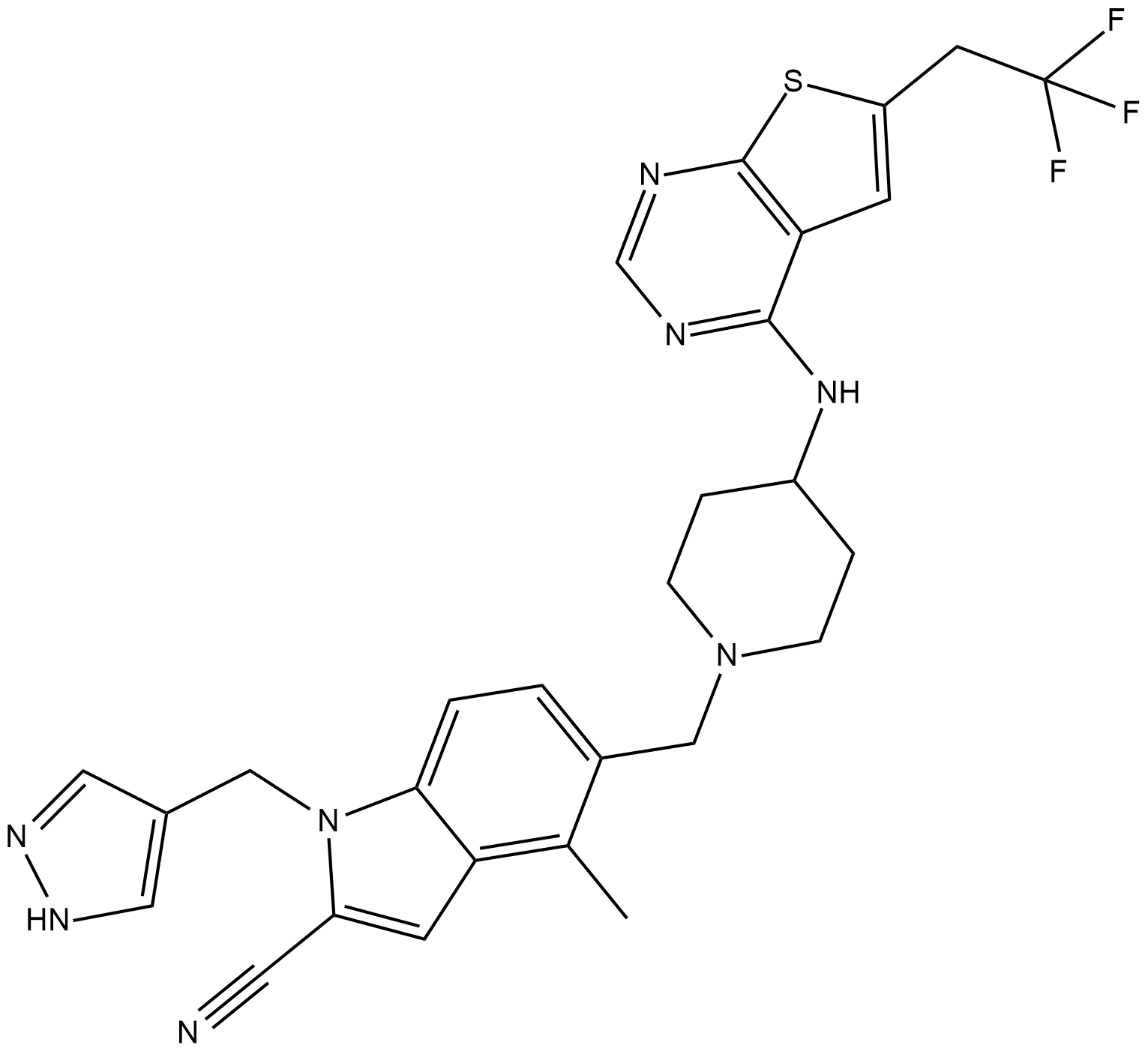
-
GC32798
MI-538
MI-538 is an inhibitor of the interaction between menin and MLL fusion proteins with an IC50 of 21 nM.
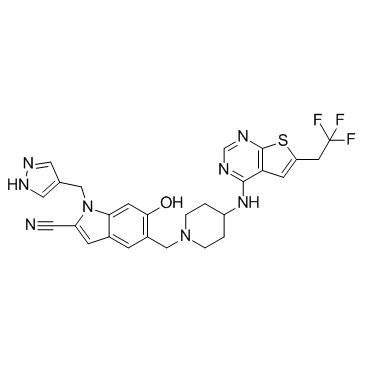
-
GC65581
MIND4-19
MIND4-19 is a potent SIRT2 inhibitor with an IC50 value of 7.0 μM.
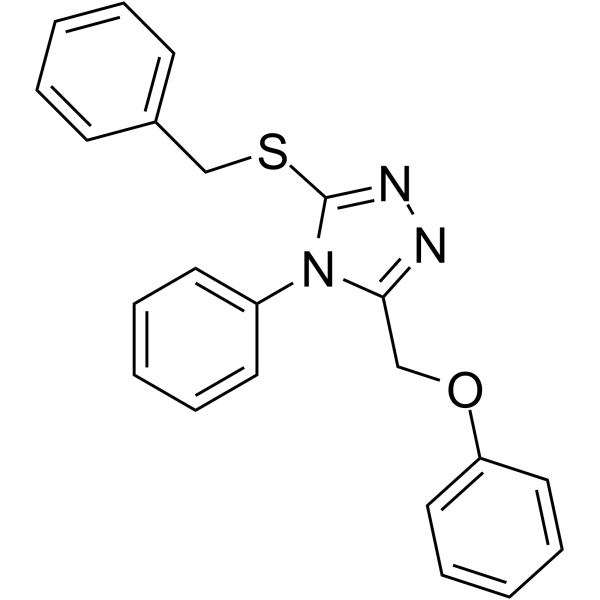
-
GC32929
MIR96-IN-1
MIR96-IN-1 targets the Drosha site in the miR-96 (miRNA-96, microRNA-96) hairpin precursor, inhibiting its biogenesis, derepressing downstream targets, and triggering apoptosis in breast cancer cells. MIR96-IN-1 binds to RNAs with Kds of 1.3, 9.4, 3.4, 1.3 and 7.4 μM for RNA1, RNA2, RNA3, RNA4 and RNA5, respectively.
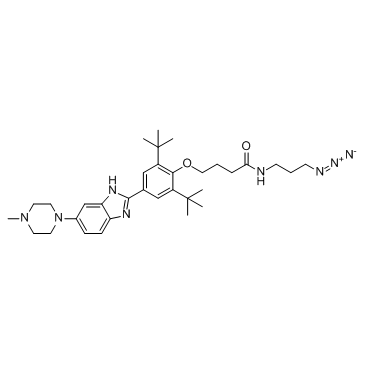
-
GC64487
Miravirsen
Miravirsen (SPC-3649), a β-d-oxy-locked nucleic acid-modified phosphorothioate antisense oligonucleotide, inhibit the biogenesis of miR-122.

-
GC19248
Mivebresib
Mivebresib is a potent and orally available bromodomain and extraterminal domain (BET) bromodomain inhibitor.
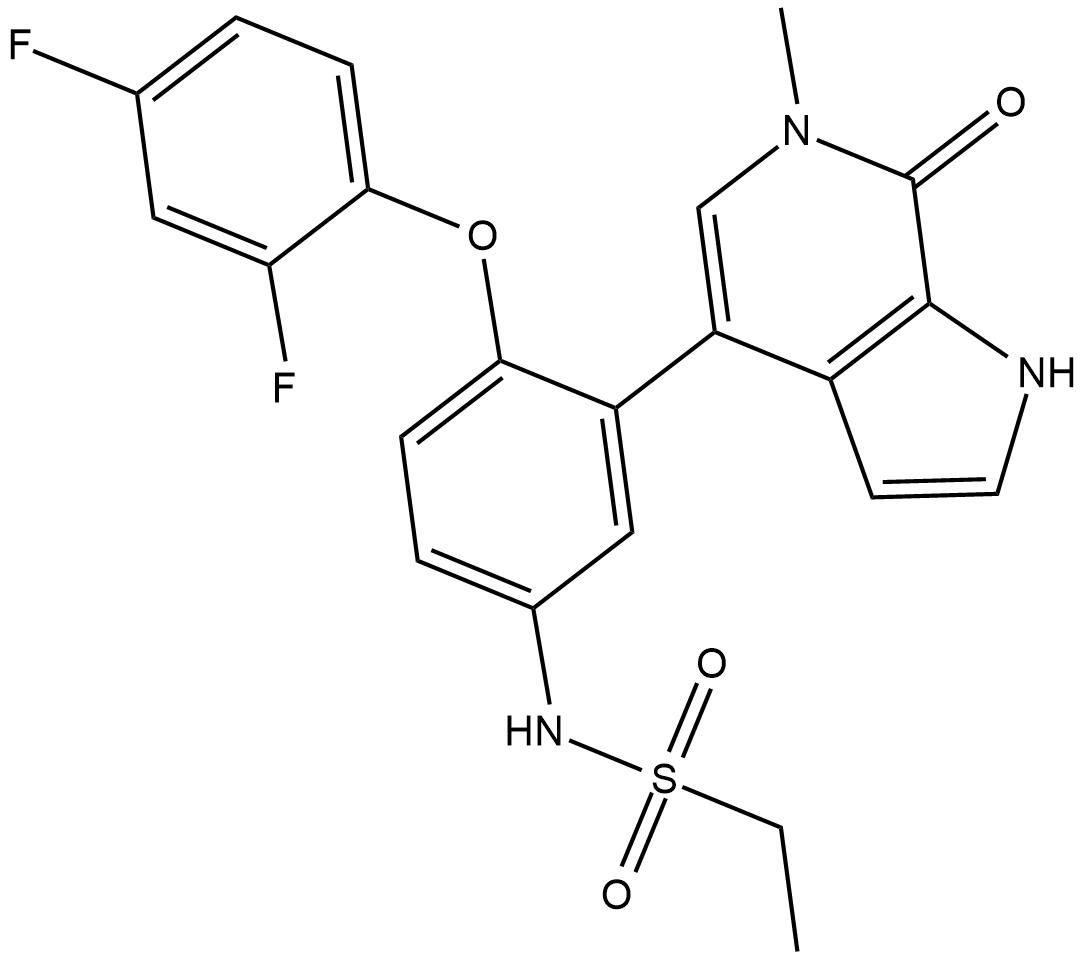
-
GC17802
MK-4827
An orally bioavailable PARP1/2 inhibitor
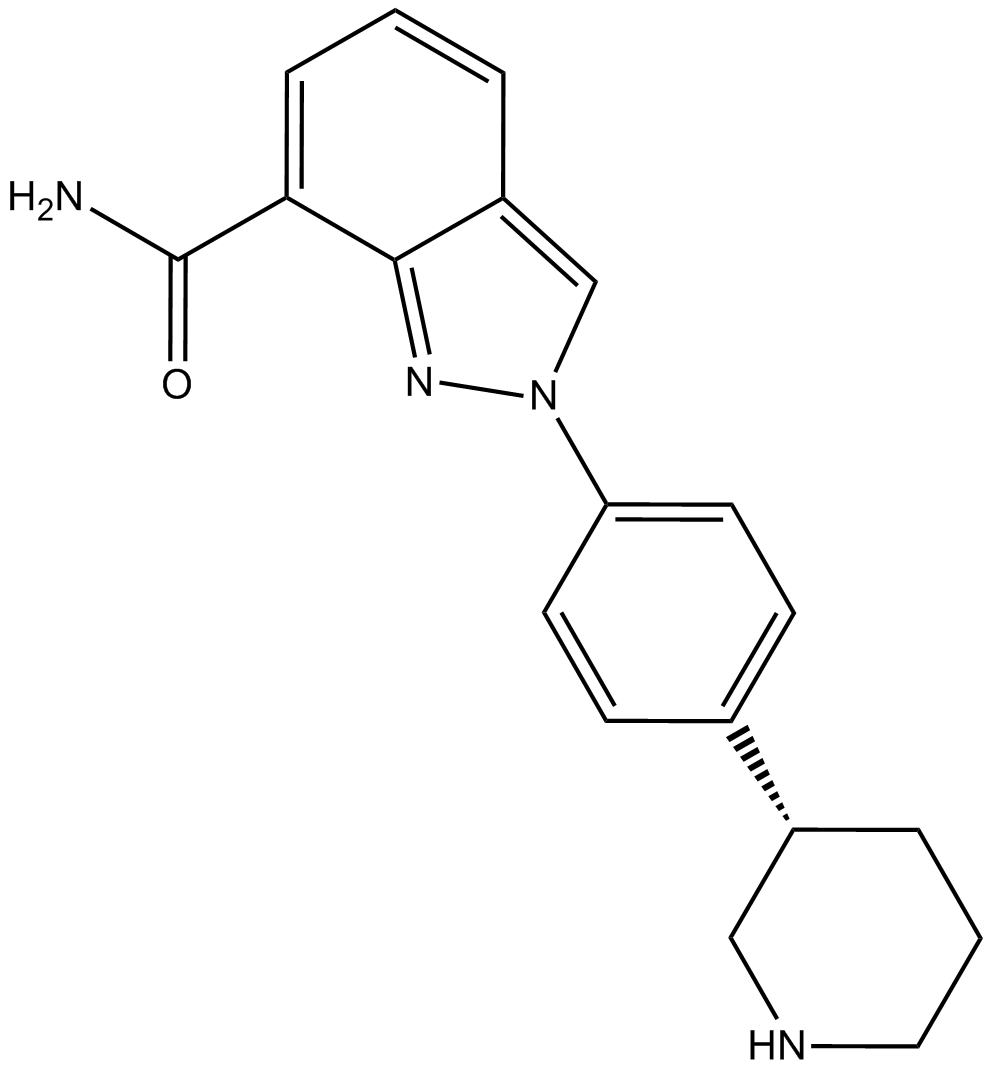
-
GC12756
MK-4827 hydrochloride
MK-4827 hydrochloride (MK-4827 hydrochloride) is a highly potent and orally bioavailable PARP1 and PARP2 inhibitor with IC50s of 3.8 and 2.1 nM, respectively. MK-4827 hydrochloride leads to inhibition of repair of DNA damage, activates apoptosis and shows anti-tumor activity.
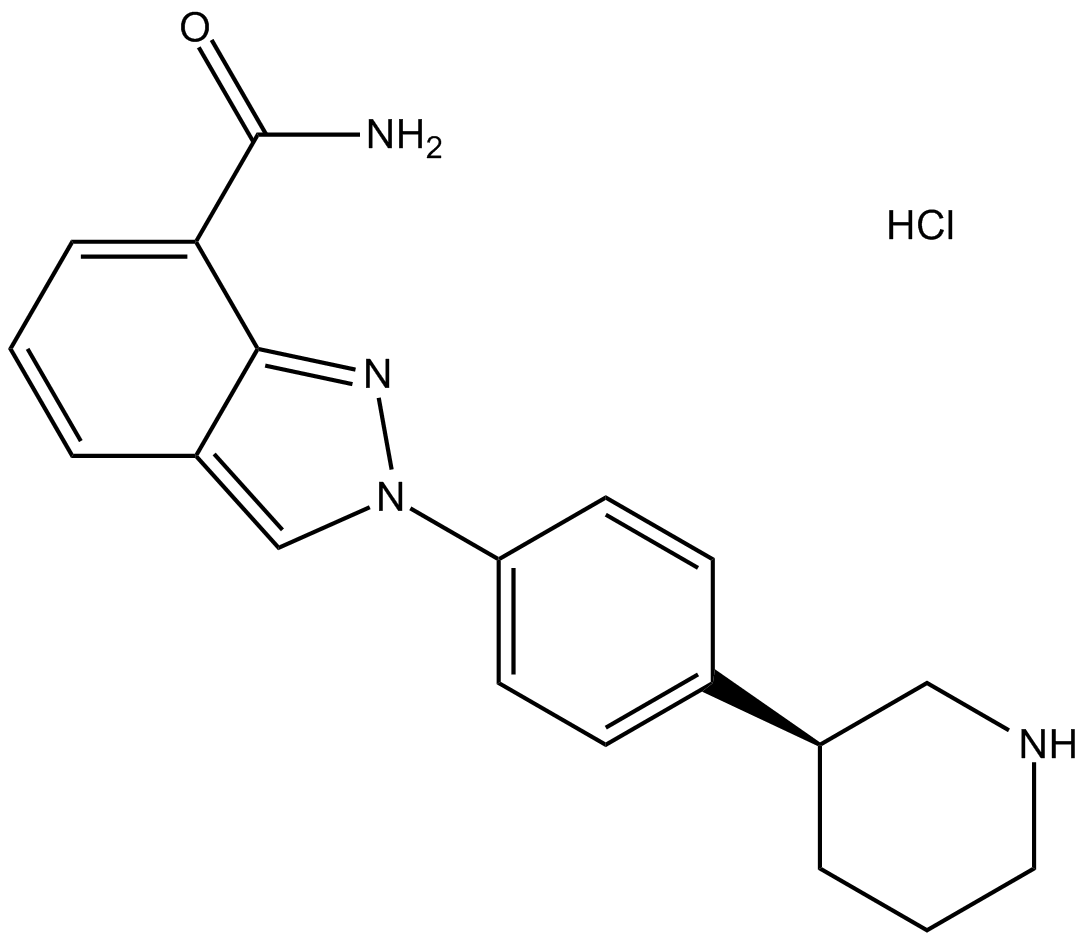
-
GC17052
MK-4827 Racemate
selective inhibitor of PARP1/PARP2
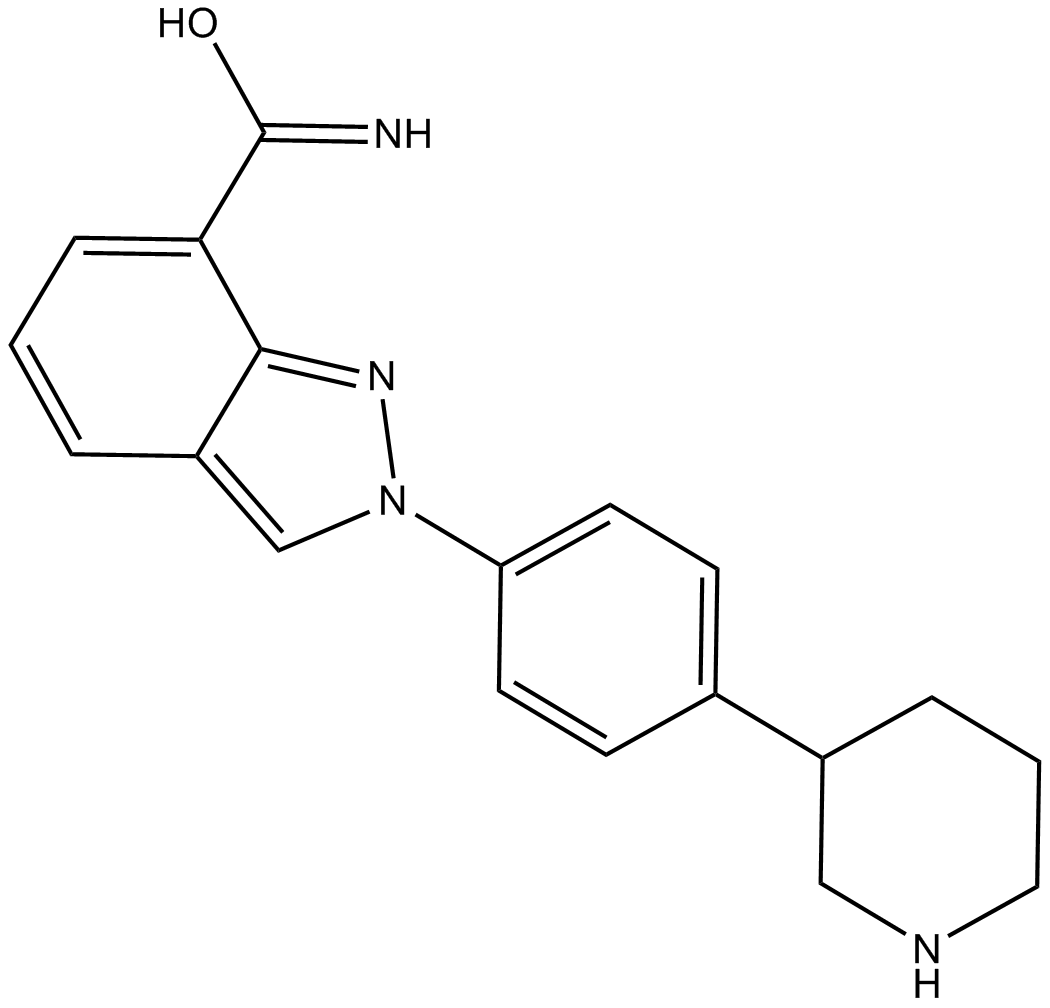
-
GC11537
MK-4827 tosylate
MK-4827 tosylate (MK-4827 tosylate) is a highly potent and orally bioavailable PARP1 and PARP2 inhibitor with an IC50 of 3.8 and 2.1 nM, respectively. MK-4827 tosylate leads to inhibition of repair of DNA damage, activates apoptosis and shows anti-tumor activity.
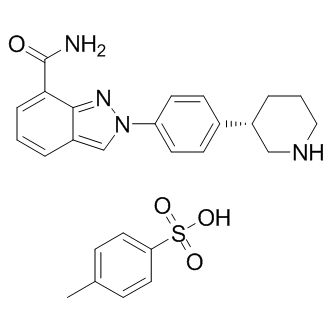
-
GC17162
MK-5108 (VX-689)
A potent inhibitor of Aurora kinases
-
GC19251
MK-8617
MK-8617 is an orally active pan-inhibitor of hypoxia-inducible factor prolyl hydroxylase 1-3 (HIF PHD1-3) with an IC50 of 1 nM for PHD2.
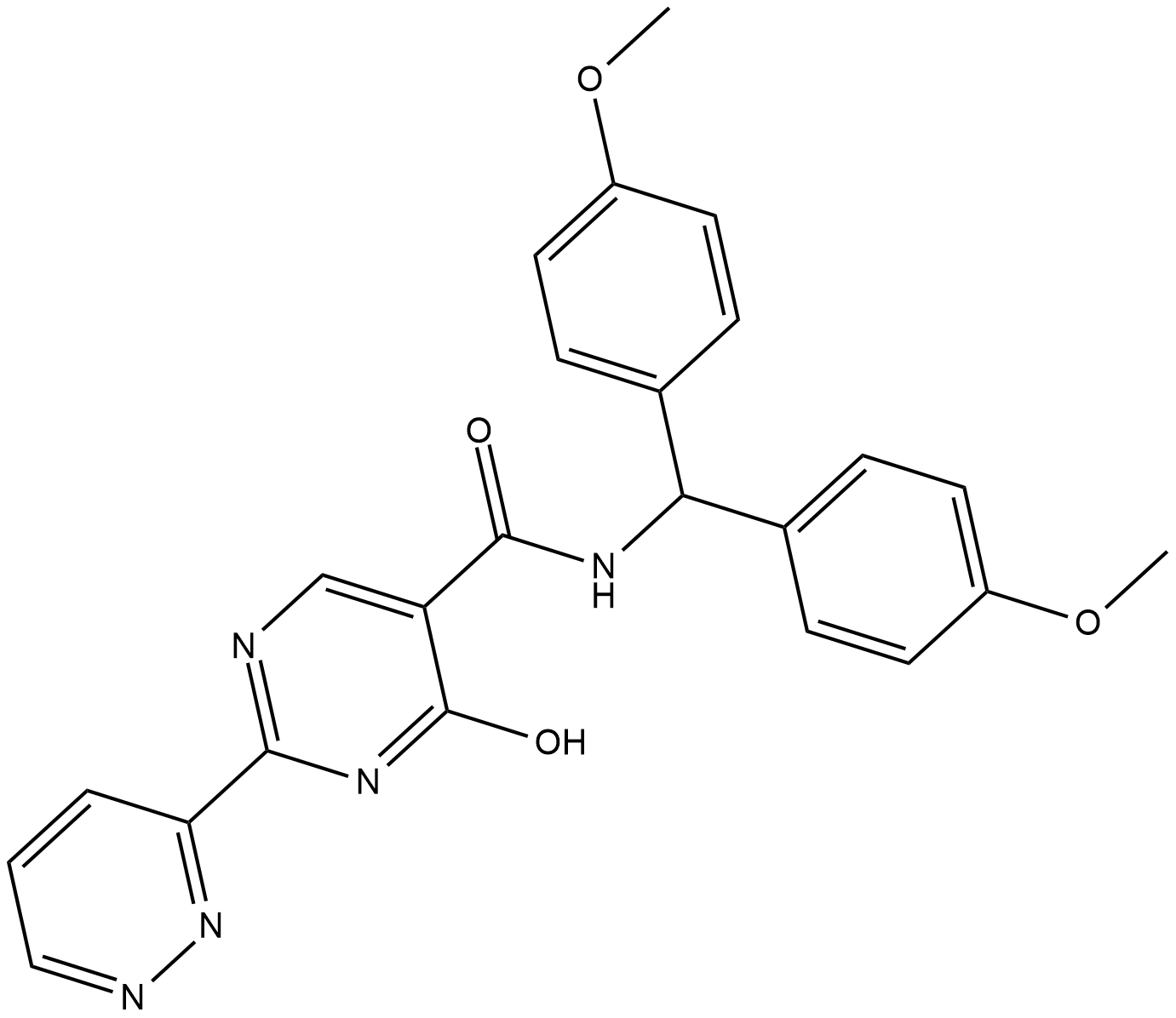
-
GC10442
MK-8745
Aurora A inhibitor,potent and selective
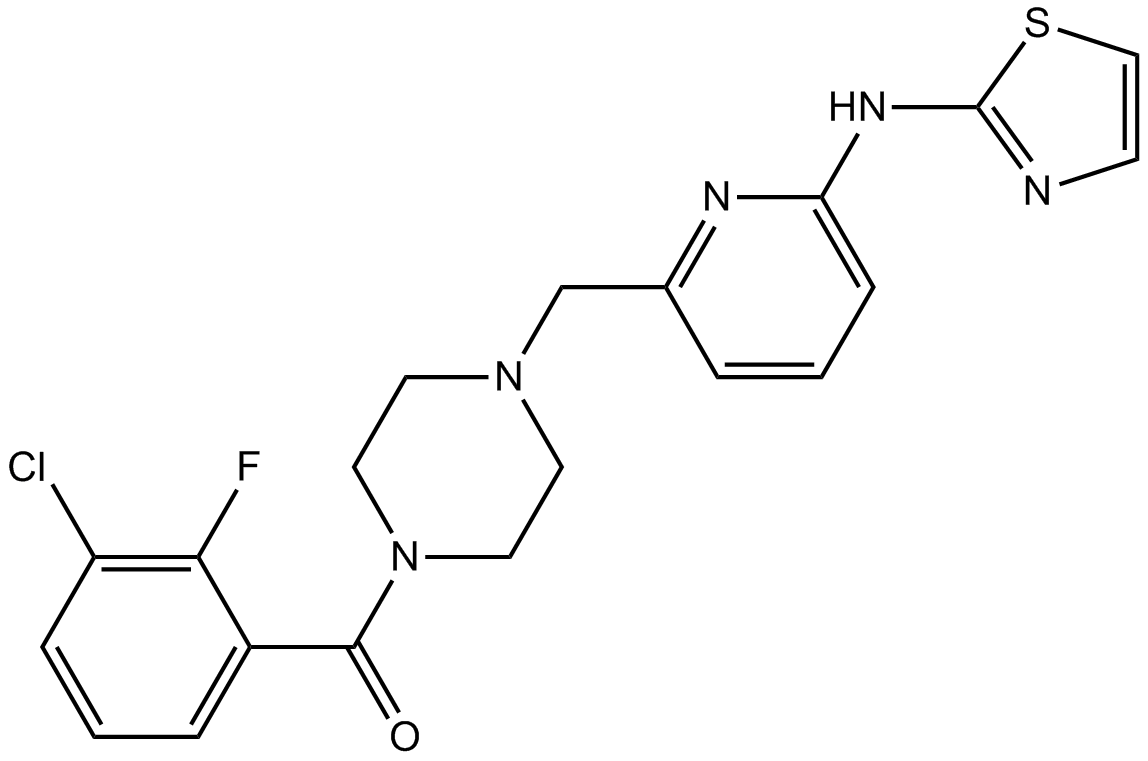
-
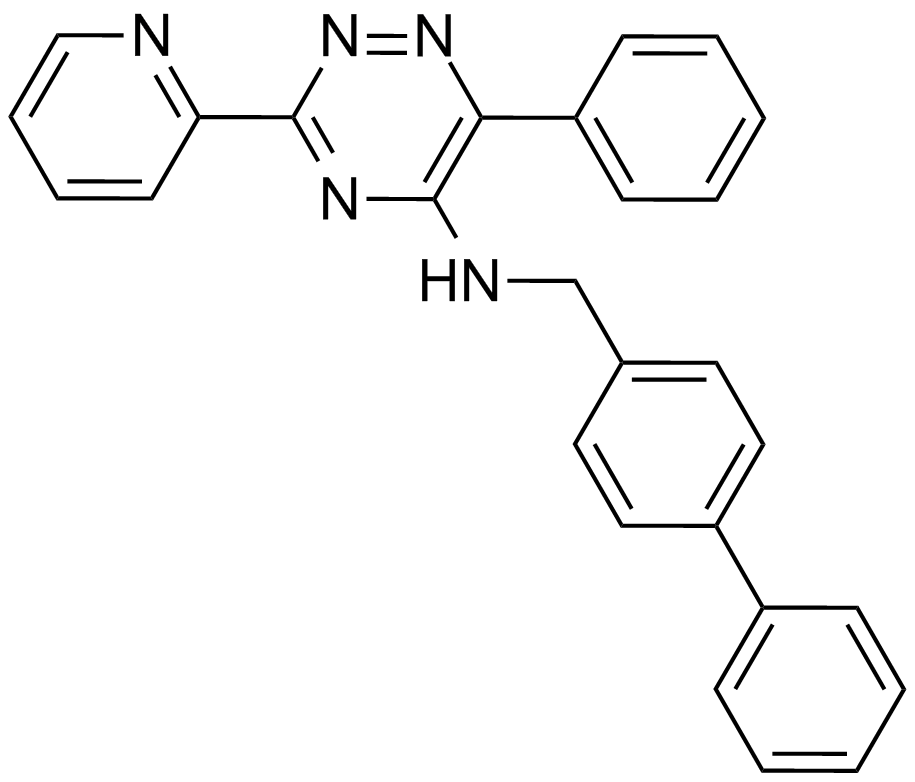
GC14319
ML 228
A HIF pathway activator
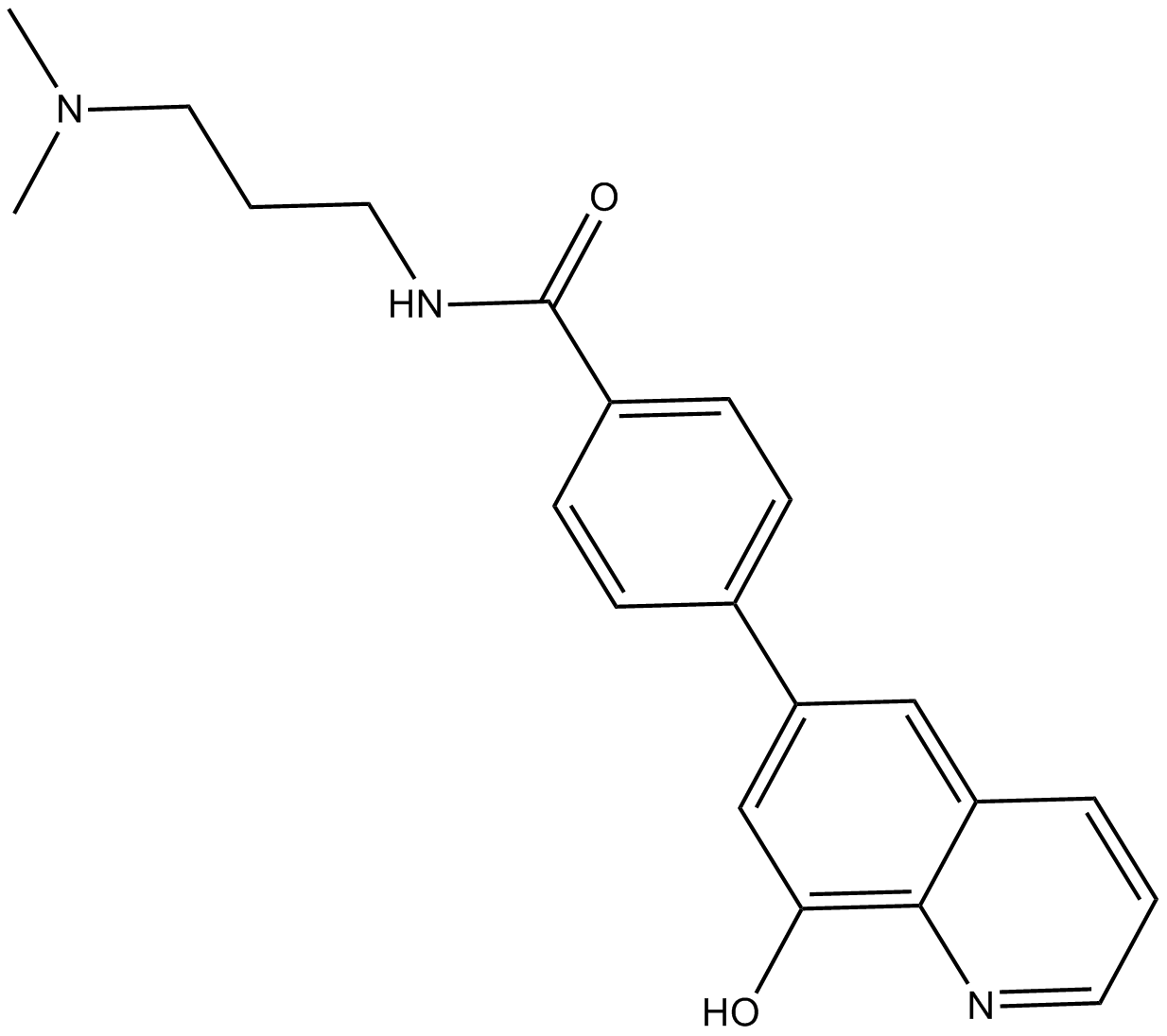
-
GC12557
ML324
JMJD2 demethylase inhibitor, potent and cell-permeable
-
GC12208
MLN8054
An inhibitor of Aurora A kinase
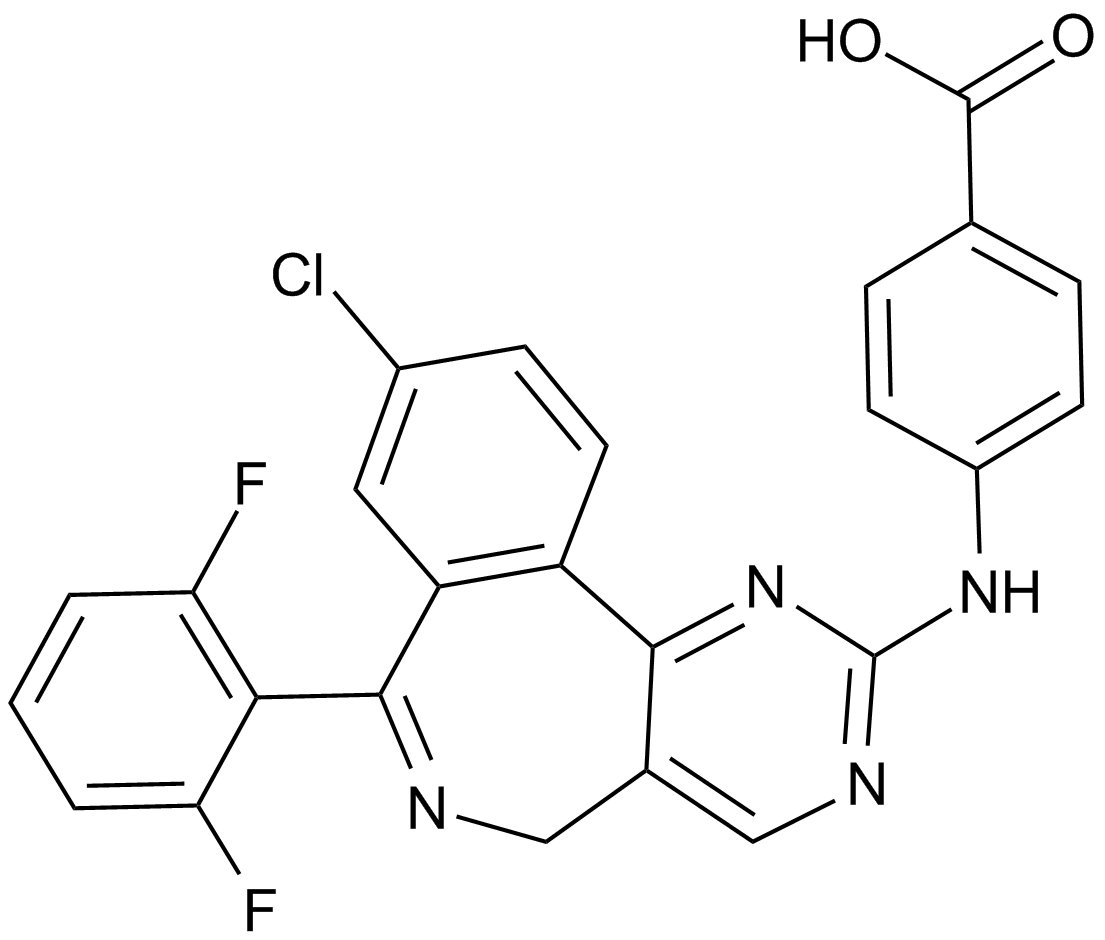
-
GC12690
MLN8237 (Alisertib)
An Aurora A kinase inhibitor
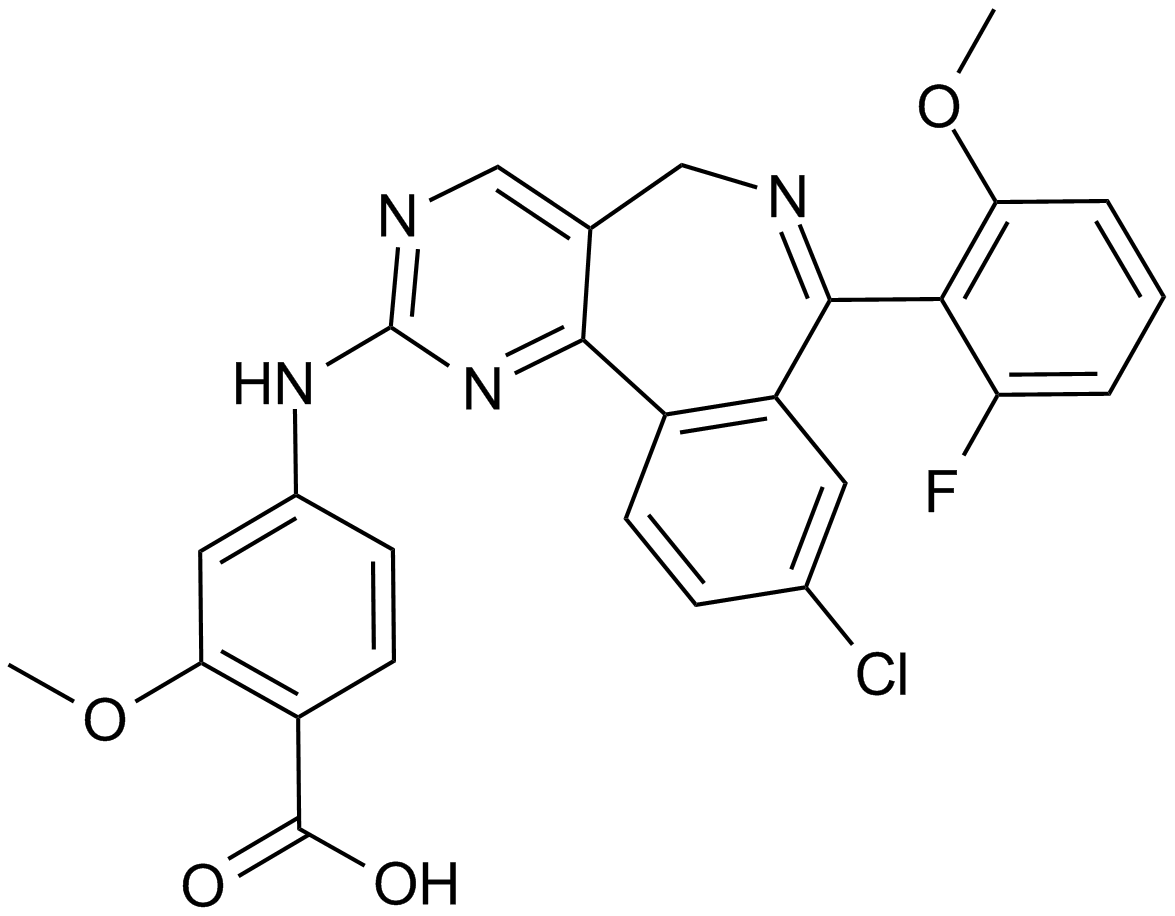
-
GC14652
MM-102
MLL1 inhibitor,high-affinity peptidomimetic
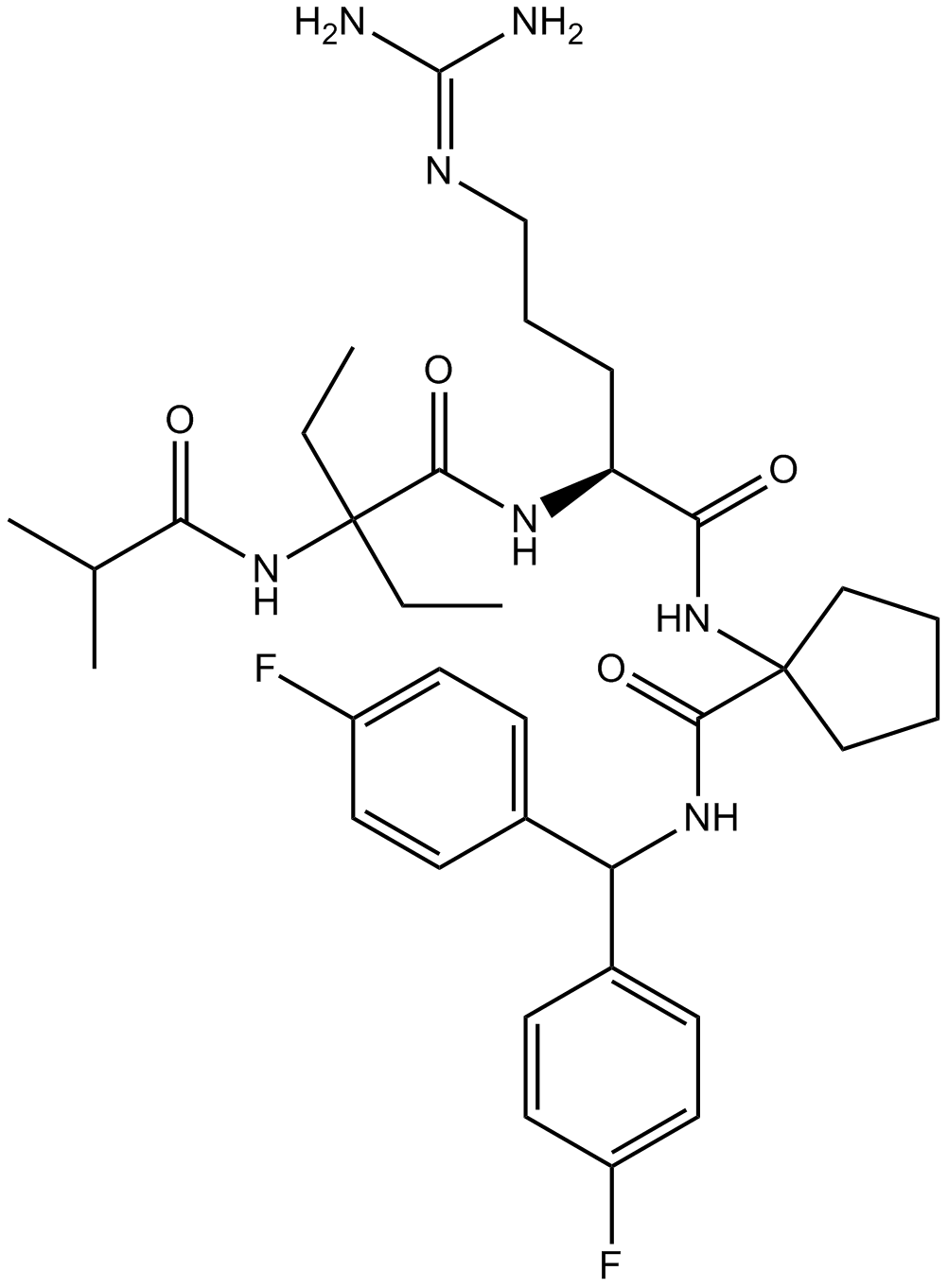
-
GC36632
MM-102 TFA
MM-102 TFA (HMTase Inhibitor IX TFA) is a potent WDR5/MLL interaction inhibitor, achieves IC50 = 2.4 nM with an estimated Ki 200 times more potent than the ARA peptide.
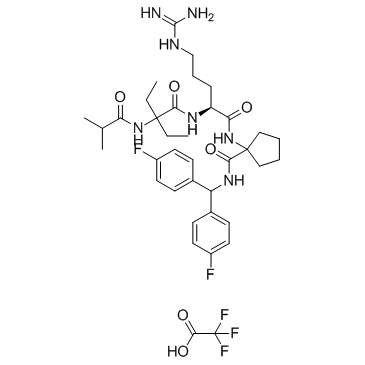
-
GC67758
MM-401 TFA
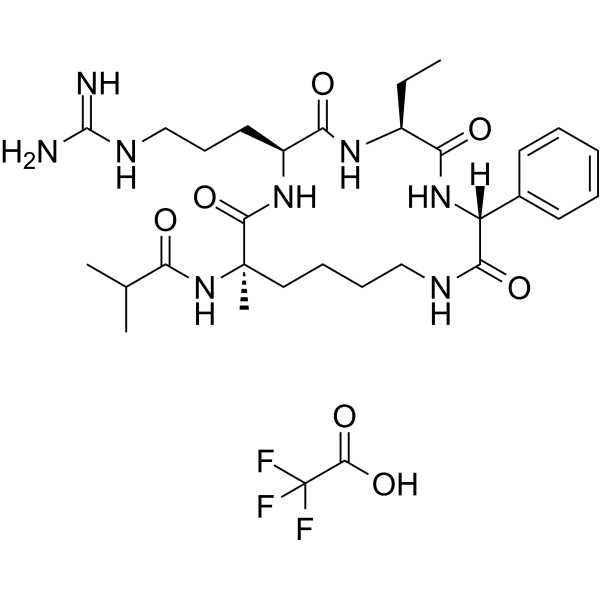
-
GC33278
MM-589
MM-589 is a potent inhibitor of WD repeat domain 5 (WDR5) and mixed lineage leukemia (MLL) protein-protein interaction. MM-589 binds to WDR5 with an IC50 of 0.90 nM and inhibits the MLL H3K4 methyltransferase activity with an IC50 of 12.7 nM.
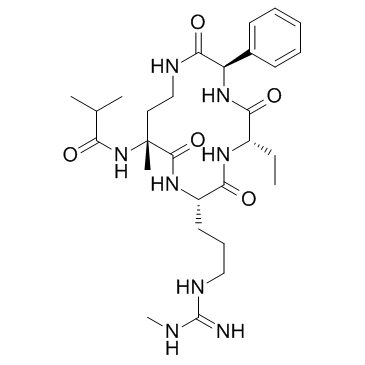
-
GC65037
MM-589 TFA
MM-589 TFA is a potent inhibitor of WD repeat domain 5 (WDR5) and mixed lineage leukemia (MLL) protein-protein interaction. MM-589 binds to WDR5 with an IC50 of 0.90 nM and inhibits the MLL H3K4 methyltransferase activity with an IC50 of 12.7 nM.
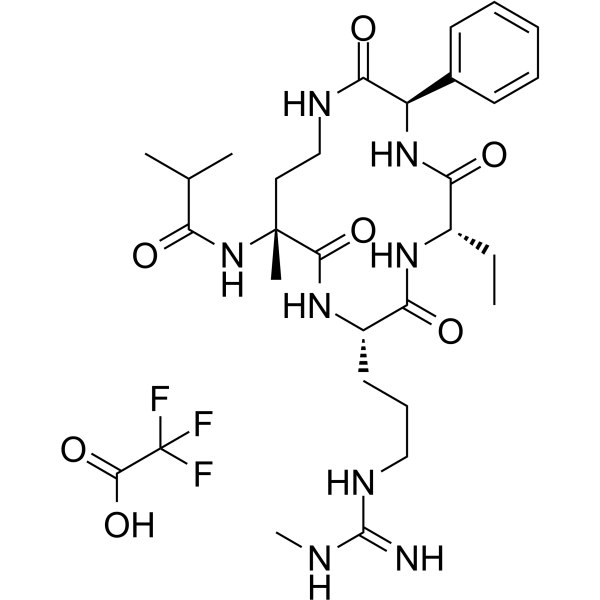
-
GC16914
MN 64
tankyrase inhibitor
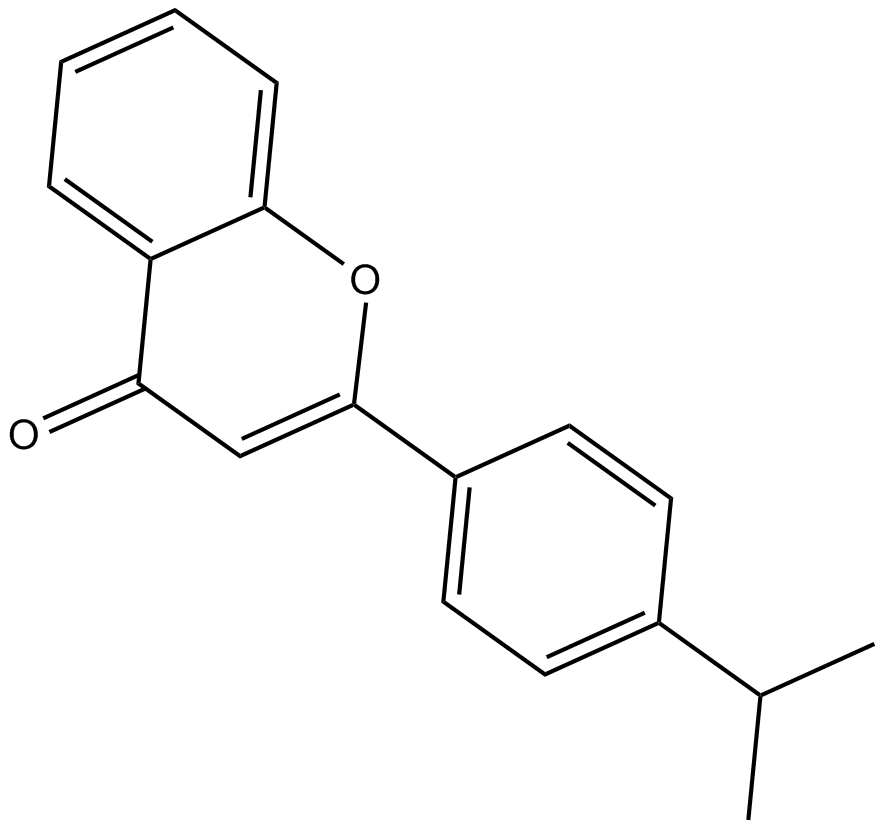
-
GC13055
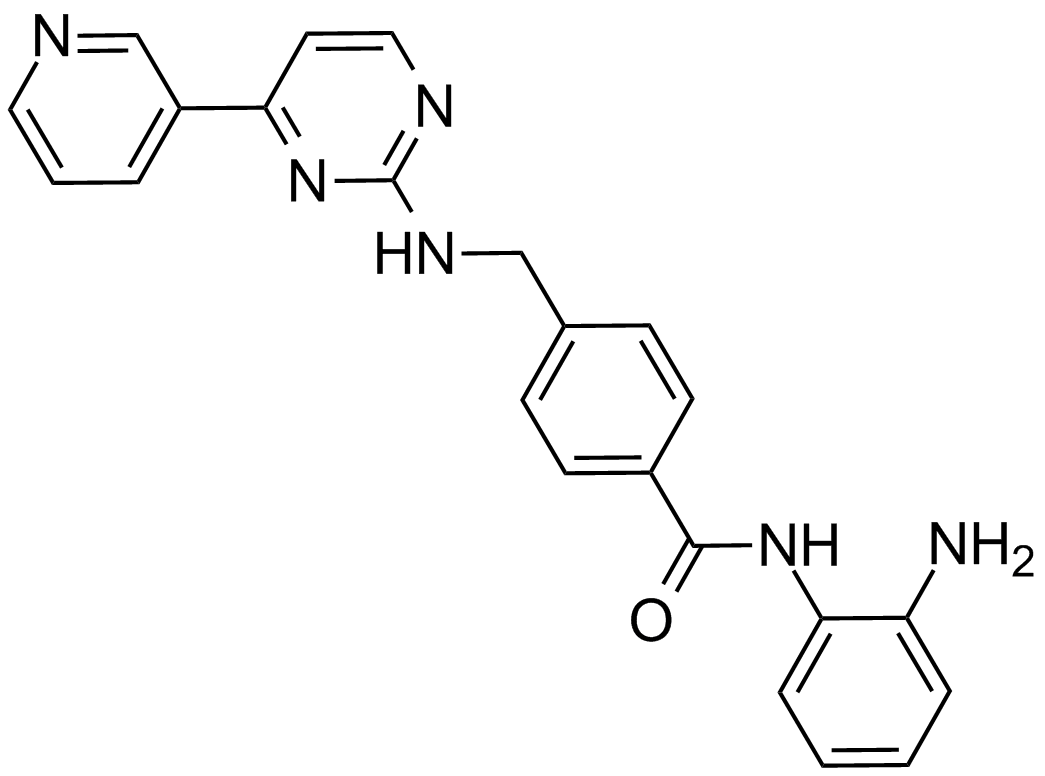
Mocetinostat (MGCD0103, MG0103)
An orally available HDAC inhibitor
-
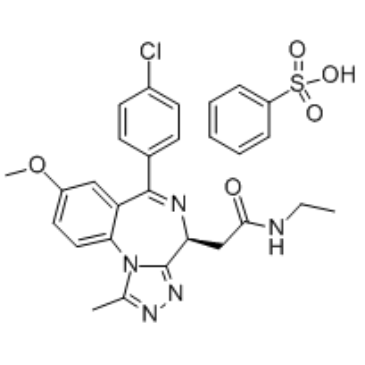
GC34675
Molibresib besylate
Molibresib besylate (GSK 525762C; I-BET 762 besylate) is a BET bromodomain inhibitor with IC50 of 32.5-42.5 nM.
-
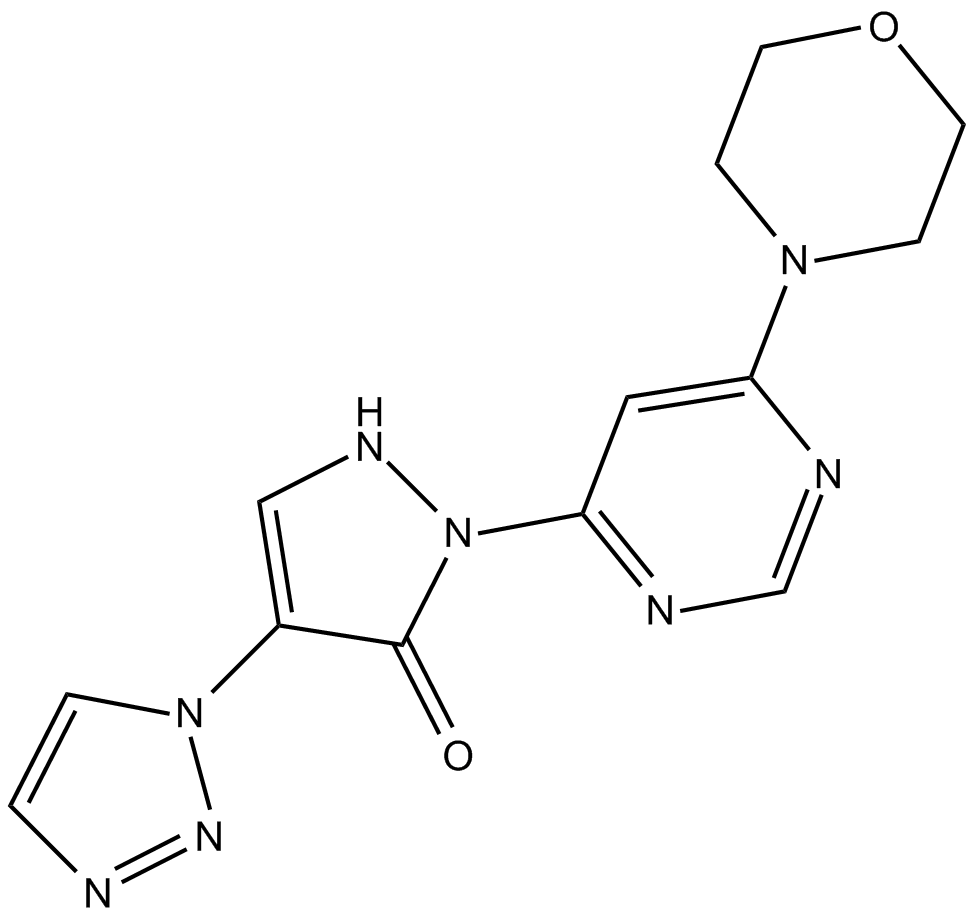
GC10046
Molidustat (BAY85-3934)
Molidustat (BAY85-3934) (BAY 85-3934) is a novel inhibitor of hypoxia-inducible factor prolyl hydroxylase (HIF-PH) with mean IC50 values of 480 nM for PHD1, 280 nM for PHD2, and 450 nM for PHD3.
-
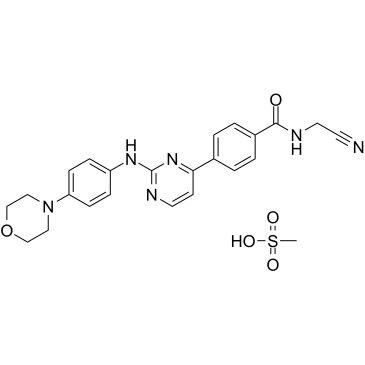
GC36646
Momelotinib Mesylate
Momelotinib Mesylate (CYT387 Mesylate) is an ATP-competitive inhibitor of JAK1/JAK2 with IC50 of 11 nM/18 nM, appr 10-fold selectivity versus JAK3.
-
GC62406
Moracin O
Moracin O is a 2-arylbenzofuran isolated from the Mori Cortex Radicis.
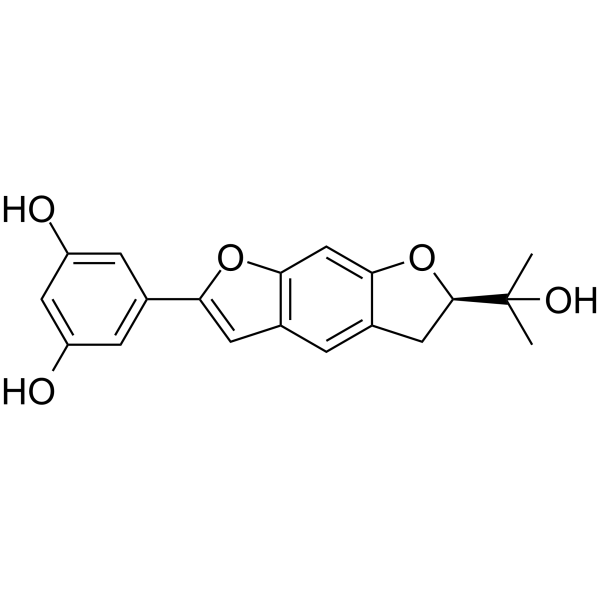
-
GC33371
MOZ-IN-2
An inhibitor of KAT6A/MOZ
-
GC64729
MPT0B390
MPT0B390 is an arylsulfonamide-based derivative with potent HDAC inhibitory ability. MPT0B390, TIMP3 inducer, inhibits tumor growth, metastasis and angiogenesis. MPT0B390 shows antiproliferative activity against human colon cancer cell line HCT116 with the GI50 of 0.03 μM.
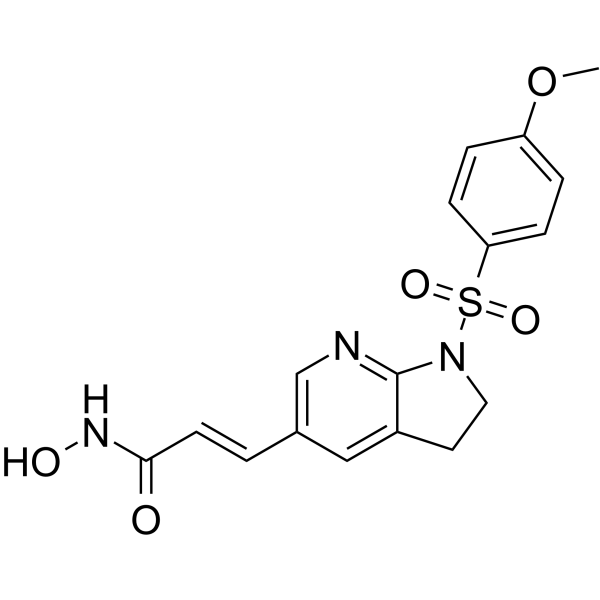
-
GC65965
MPT0E028
MPT0E028 is an orally active and selective HDAC inhibitor with IC50s of 53.0 nM, 106.2 nM, 29.5 nM for HDAC1, HDAC2 and HDAC6, respectively. MPT0E028 reduces the viability of B-cell lymphomas by inducing apoptosis and possesses potent direct Akt targeting ability and reduces Akt phosphorylation in B-cell lymphoma. MPT0E028 has good anticancer activity.
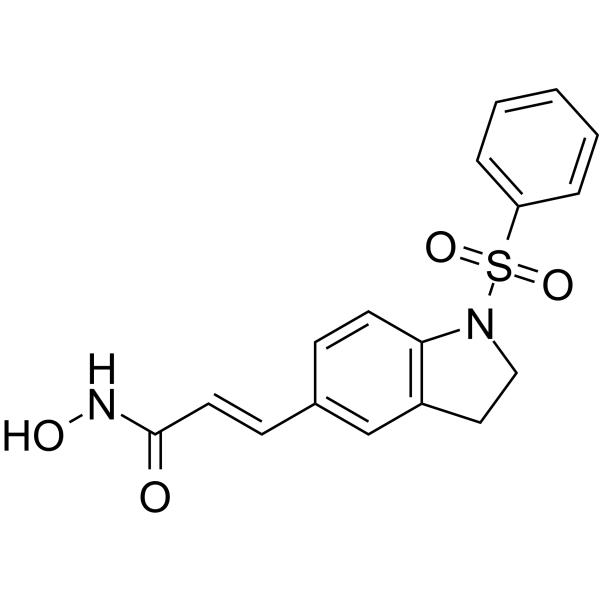
-
GC62308
MPT0G211
MPT0G211 is a potent, orally active and selective HDAC6 inhibitor (IC50=0.291?nM).
-
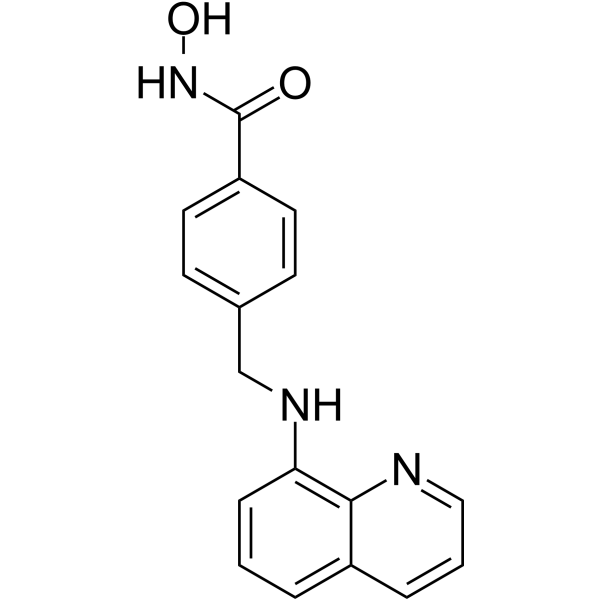
GC61468
MR837
MR837 is an inhibitor of NSD2-PWWP1. MR837 can bind with human nuclear receptor binding SET domain protein 2 (PWWP domain).
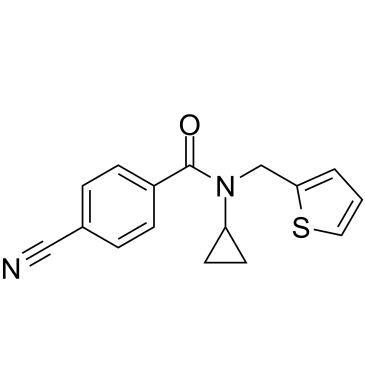
-
GC50576
MRK 740
Potent PRDM9 inhibitor
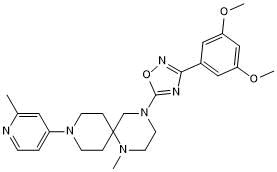
-
GC63558
MRTX-1719
MRTX-1719 is a potent first-in-class selective inhibitor of the PRMT5/MTA complex, with an IC50 of less than 10 nM in PRMT5/MTA MTAPDEL SDMA cells.
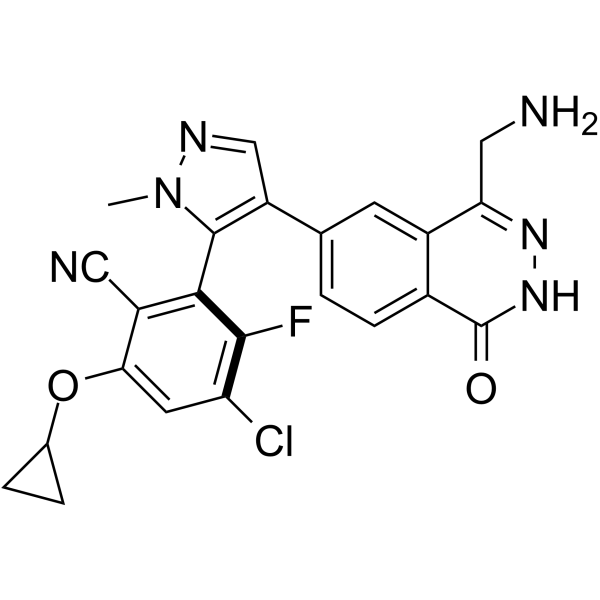
-
GC69499
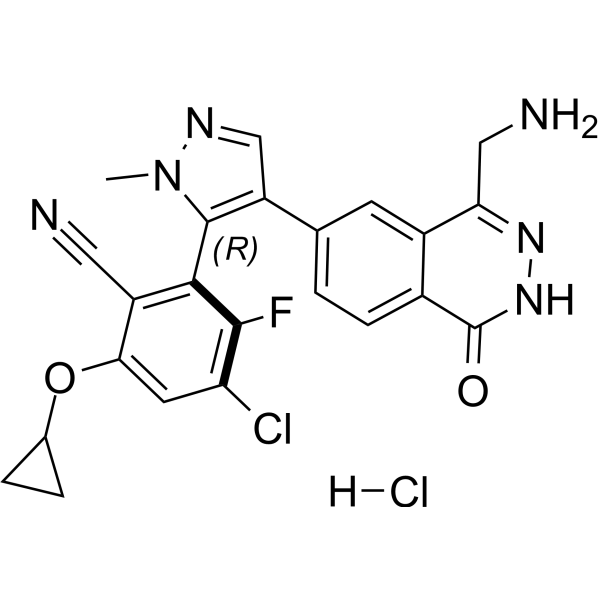
MRTX-1719 hydrochloride
MRTX-1719 hydrochloride is an effective, novel, and selective inhibitor of the PRMT5/MTA complex. Its IC50 value for PRMT5/MTA MTAPDEL SDMA cell lines is <10 nM.
-
GC62715
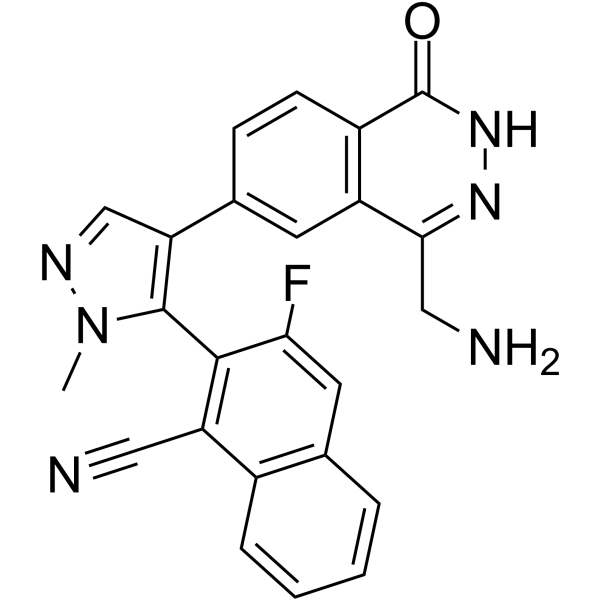
MRTX9768
MRTX9768 is a potent, selective, orally active, first-in-class PRMT5-MTA complex inhibitor.
-
GC63080
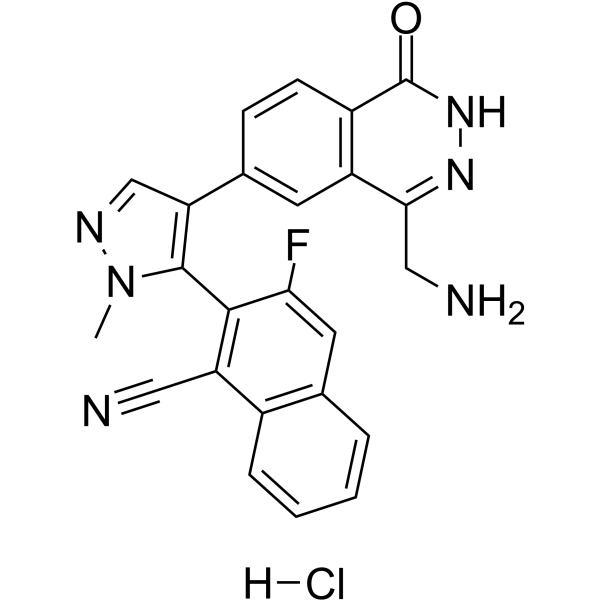
MRTX9768 hydrochloride
MRTX9768 hydrochloride is a potent, selective, orally active, first-in-class PRMT5-MTA complex inhibitor.
-
GC40791
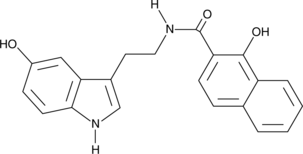
MS-1020
MS-1020 is a cell-permeable inhibitor of janus kinase 3 (JAK3), strongly inhibiting constitutive autophosphorylation of JAK3 in L540 cells when used at 30-50 μM.
-
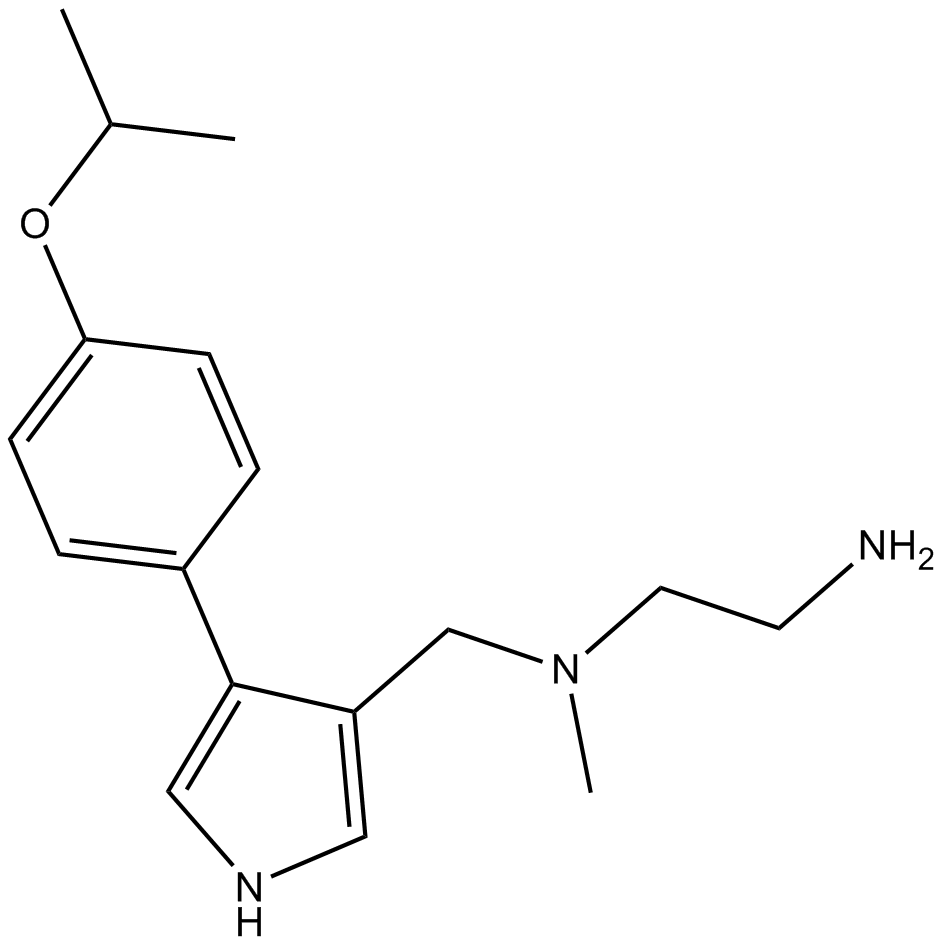
GC10099
MS023
MS023 is a potent, selective, and cell-active inhibitor of type I protein arginine methyltransferases (PRMTs).
-
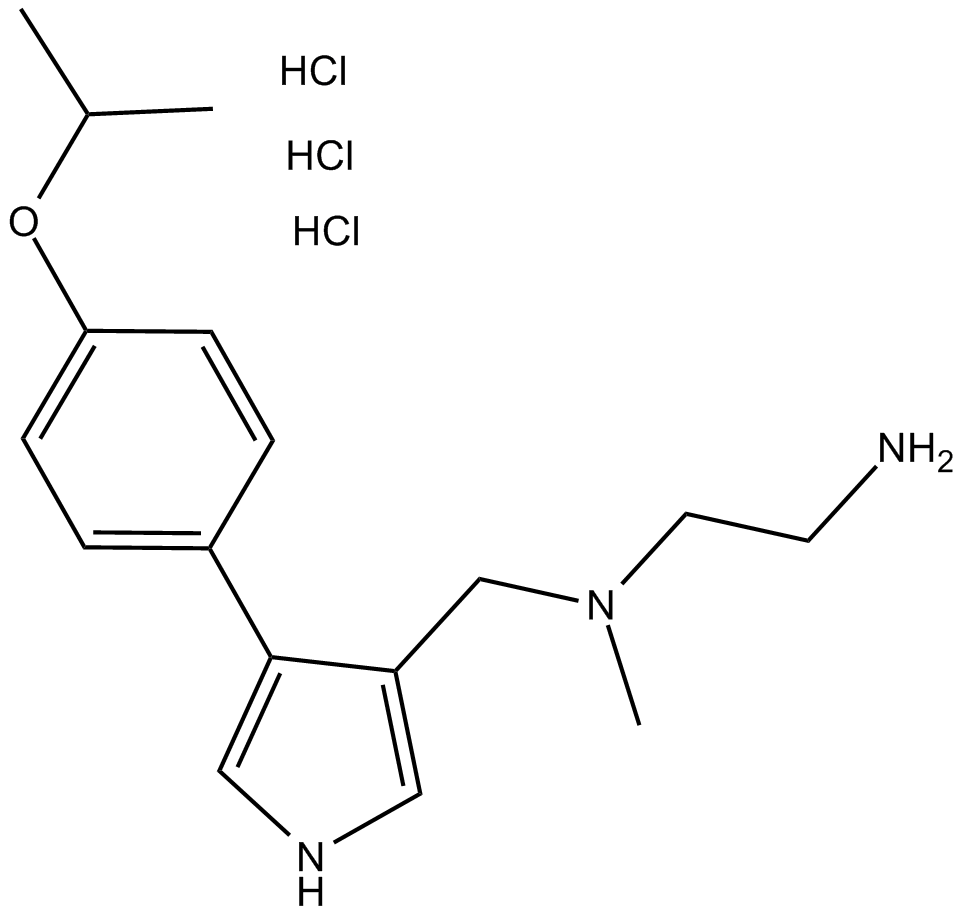
GC16432
MS023 (hydrochloride)
type I PRMTs inhibitor
-
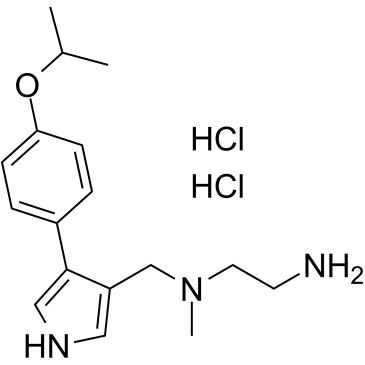
GC36655
MS023 dihydrochloride
MS023 dihydrochloride is a potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases (PRMTs) inhibitor, with IC50s of 30, 119, 83, 4 and 5 nM for PRMT1, PRMT3, PRMT4, PRMT6, and PRMT8, respectively.
-
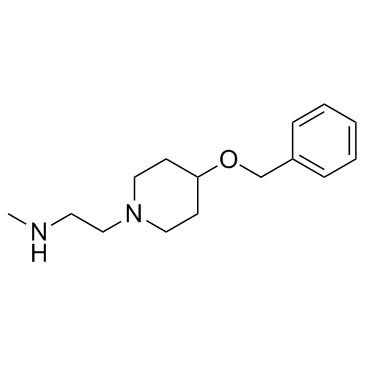
GC36656
MS049
MS049 is a potent, selective, and cell-active dual inhibitor of PRMT4 and PRMT6 with IC50s of 34 nM and 43 nM, respectively. MS049 reduces levels of Med12me2a and H3R2me2a in HEK293 cells. MS049 is not toxic and does not affect the growth of HEK293 cells.
-
GC14240
MS049 (hydrochloride)
MS049 (hydrochloride) is a potent, selective, and cell-active dual inhibitor of PRMT4 and PRMT6 with IC50s of 34 nM and 43 nM, respectively. MS049 (hydrochloride) reduces levels of Med12me2a and H3R2me2a in HEK293 cells. MS049 (hydrochloride) is not toxic and does not affect the growth of HEK293 cells.
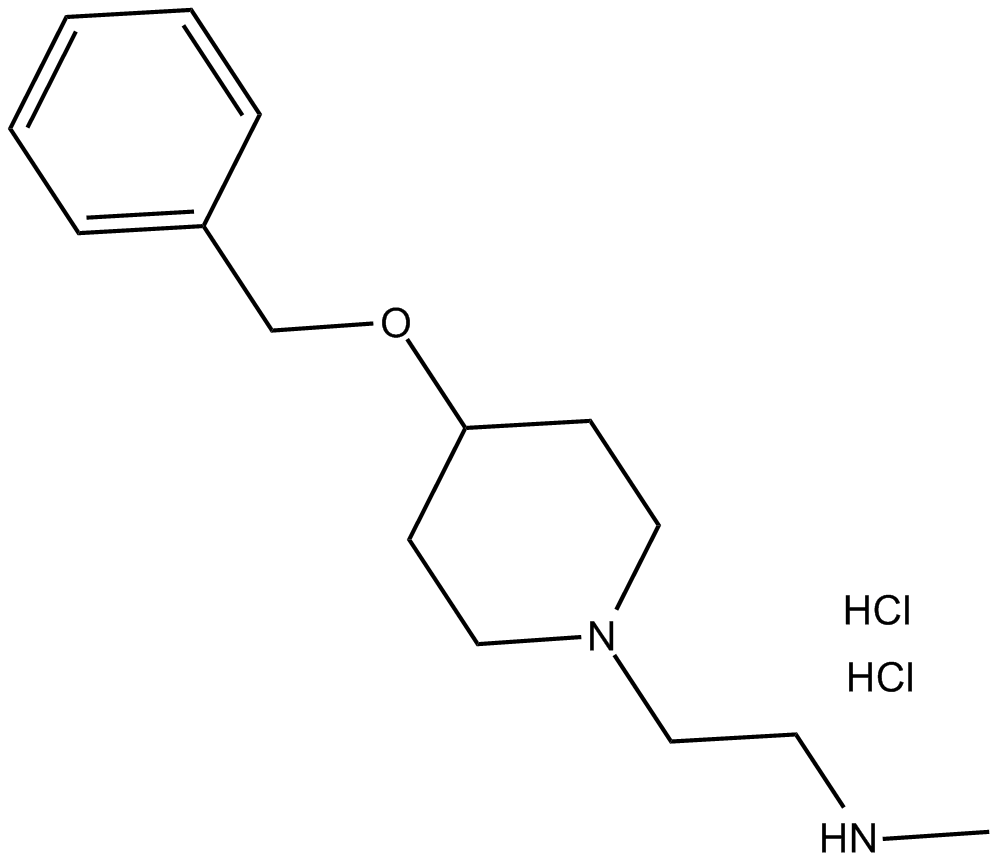
-
GC36657
MS31
MS31 is a potent, highly affinity and selective fragment-like methyllysine reader protein spindlin 1 (SPIN1) inhibitor. MS31 potently inhibits the interactions between SPIN1 and H3K4me3 (IC50=77 nM, AlphaLISA; 243 nM, FP). MS31 selectively binds Tudor domain II of SPIN1 (Kd=91 nM). MS31 potently inhibits binding of trimethyllysine-containing peptides to SPIN1. MS31 is not toxic to nontumorigenic cells.
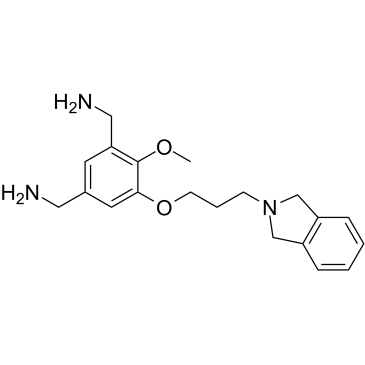
-
GC39252
MS31 trihydrochloride
MS31 trihydrochloride is a potent, highly affinity and selective fragment-like methyllysine reader protein spindlin 1 (SPIN1) inhibitor. MS31 trihydrochloride potently inhibits the interactions between SPIN1 and H3K4me3 (IC50=77 nM, AlphaLISA; 243 nM, FP). MS31 trihydrochloride selectively binds Tudor domain II of SPIN1 (Kd=91 nM). MS31 trihydrochloride potently inhibits binding of trimethyllysine-containing peptides to SPIN1, and is not toxic to nontumorigenic cells.
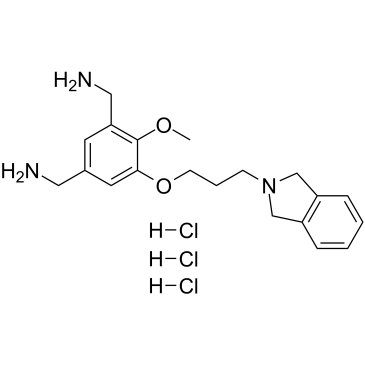
-
GC44250
MS351
MS351 is an antagonist of chromobox 7 (CBX7) that acts by binding the CBX7 chromodomain.
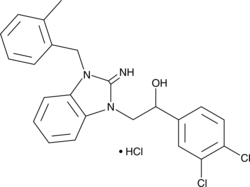
-
GC12630
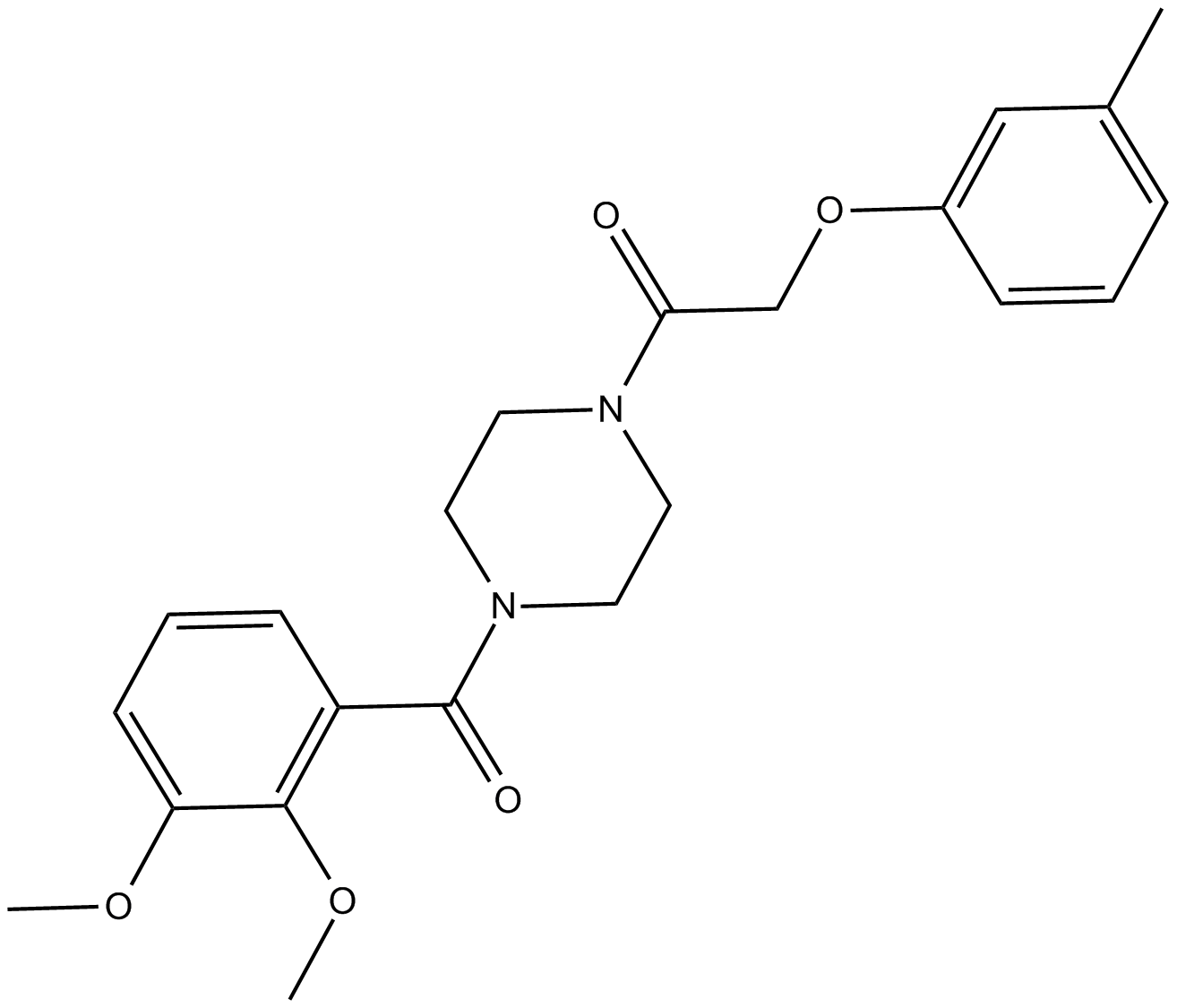
MS37452
competitive inhibitor of CBX7 chromodomain binding to H3K27me3
-
GC64999
MS402
MS402 is a BD1-selective BET BrD inhibitor with Kis of 77 nM, 718 nM, 110 nM, 200 nM, 83 nM, and 240 nM for BRD4(BD1), BRD4(BD2), BRD3(BD1), BRD3(BD2), BRD2(BD1) and BRD2(BD2), respectively.

-
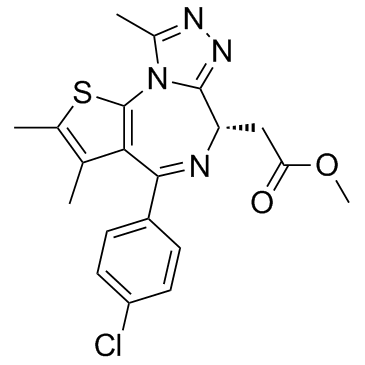
GC31881
MS417 (GTPL7512)
MS417 (GTPL7512) is a selective BET-specific BRD4 inhibitor, binds to BRD4-BD1 and BRD4-BD2 with IC50s of 30, 46 nM and Kds of 36.1, 25.4 nM, respectively, with weak selectivity at CBP BRD (IC50, 32.7 μM).
-
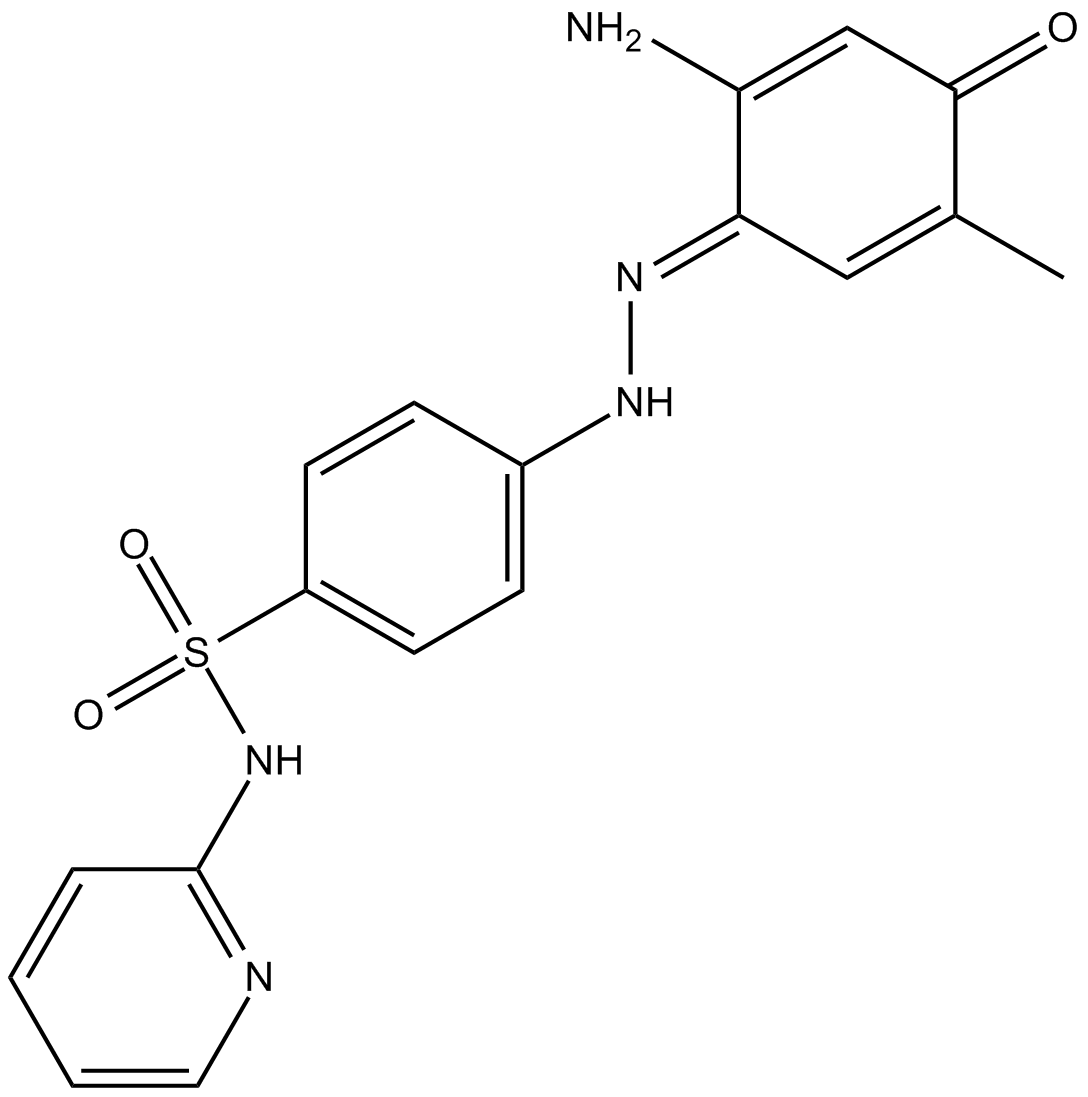
GC13148
MS436
BRD4 inhibitor
-
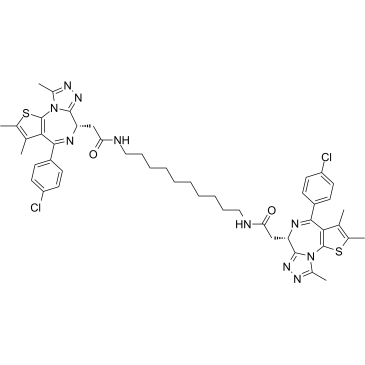
GC38474
MS645
MS645 is a bivalent BET bromodomains (BrD) inhibitor with a Ki of 18.4 nM for BRD4-BD1/BD2. MS645 spatially constrains bivalent inhibition of BRD4 BrDs resulting in a sustained repression of BRD4 transcriptional activity in solid-tumor cells.
-
GC64295
MS67
MS67 is a potent and selective WD40 repeat domain protein 5 (WDR5) degrader with a Kd of 63 nM. MS67 is inactive against other protein methyltransferases, kinases, GPCRs, ion channels, and transporters. MS67 shows potent acticancer effects.

-
GC69504
MS8815
MS8815 is a selective PROTAC degrader of zeste homolog 2 (EZH2). It has inhibitory activity against EZH2 with an IC50 value of 8.6 nM. MS8815 can be used for research on triple-negative breast cancer (TNBC).
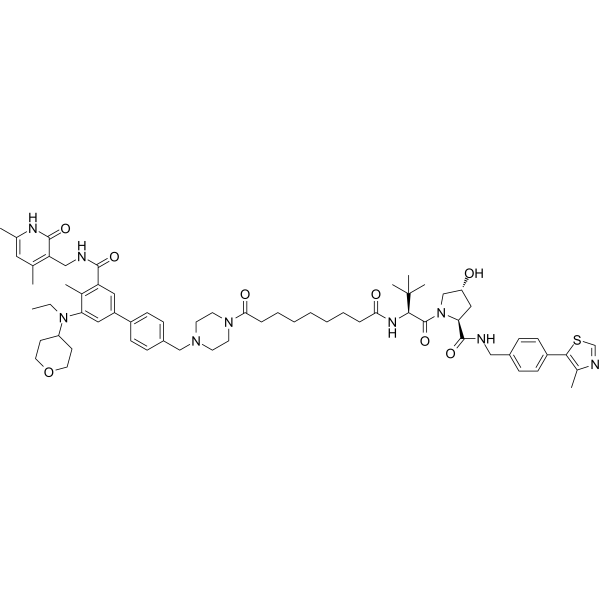
-
GC18729
MZ1
MZ1 is a hybrid compound that drives the selective proteasomal degradation of bromodomain-containing protein 4 (BRD4).
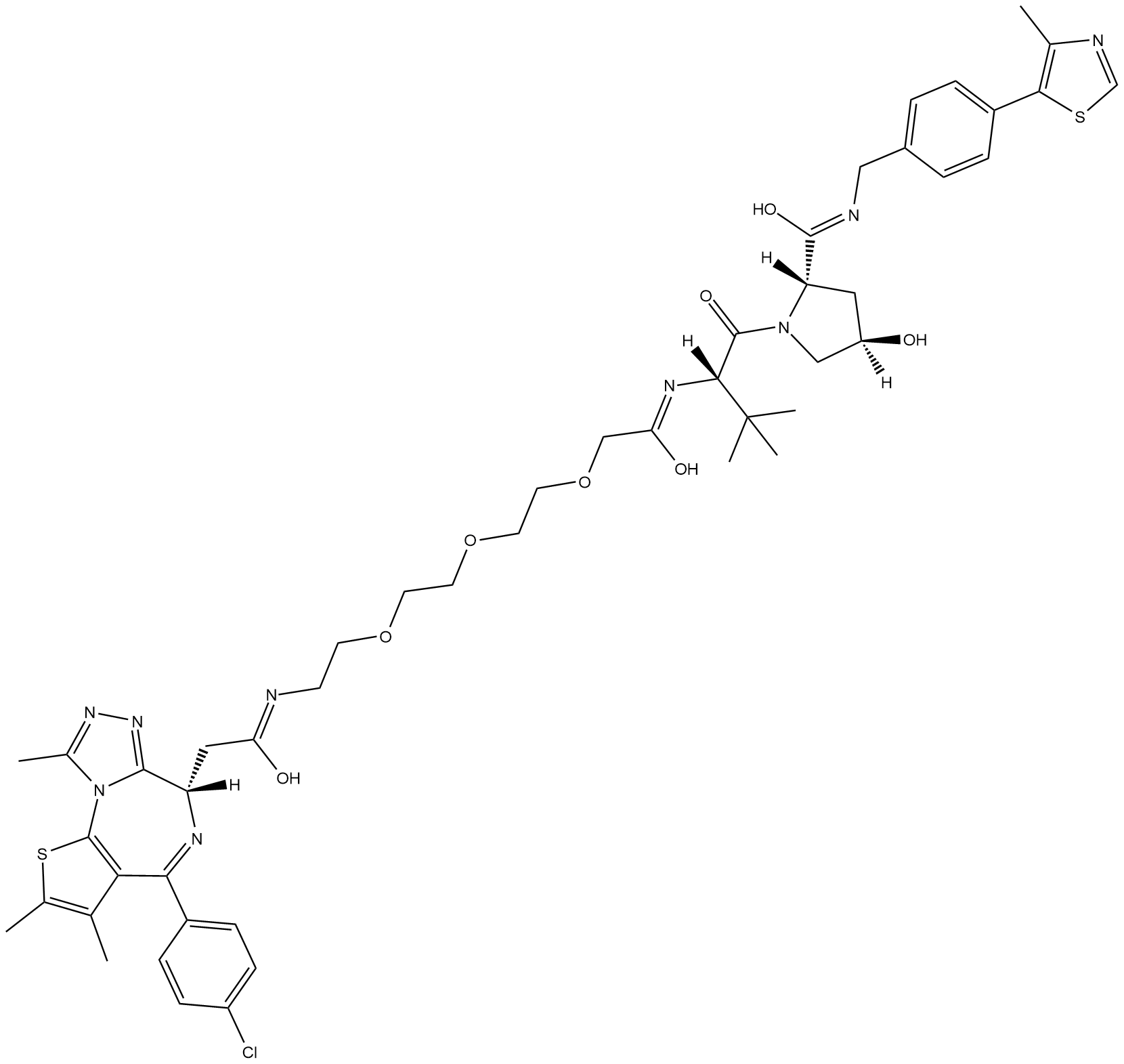
-
GC33102
MZP-54
MZP-54 is a PROTAC connected by ligands for von Hippel-Lindau and BRD3/4, with a Kd of 4 nM for Brd4BD2.
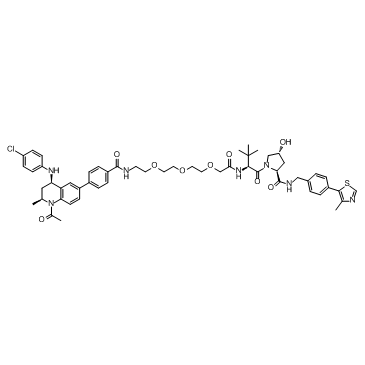
-
GC33363
MZP-55
MZP-55 is a PROTAC connected by ligands for von Hippel-Lindau and BRD3/4, with a Kd of 8 nM for Brd4BD2.
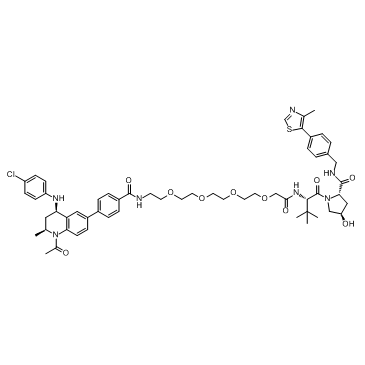
-
GC62154
N-Descyclopropanecarbaldehyde Olaparib
N-Descyclopropanecarbaldehyde Olaparib is an analogue of Olaparib containing DOTA moiety. N-Descyclopropanecarbaldehyde Olaparib is a CRBN-based ligand for synthesizing novel dual EGFR and PARP PROTAC, DP-C-4. N-Descyclopropanecarbaldehyde Olaparib can be radiolabeled F-18 or fluorophore for positron emission tomography (PET) or optical imaging in several types of tumor.