Chromatin/Epigenetics
Epigenetics
Epigenetics means above genetics. It determines how much and whether a gene is expressed without changing DNA sequences. Epigenetic regulations include, 1. DNA methylation: the addition of methyl group to DNA, converting cytosine to 5-methylcytosine, mostly at CpG sites; 2. Histone modifications: posttranslational modificationEpigeneticss of histone proteins including acetylation, methylation, ubiquitylation, phosphorylation and sumoylation; 3. miRNAs: non-coding microRNA downregulating gene expression; 4. Prions: infectious proteins viewed as epigenetic agents capable of inducing a phenotype without changing the genome.
Targets for Chromatin/Epigenetics
- Bromodomain(52)
- Aurora Kinase(89)
- DNA Methyltransferase(40)
- HDAC(218)
- Histone Acetyltransferases(67)
- Histone Demethylases(98)
- Histone Methyltransferase(212)
- HIF(101)
- JAK(178)
- MBT Domains(1)
- PARP(128)
- Pim(35)
- Protein Ser/Thr Phosphatases(41)
- RNA Polymerase(8)
- Sirtuin(84)
- Sphingosine Kinase-2(1)
- Polycomb repressive complex(2)
- SUMOylation(3)
- PAD(18)
- Epigenetic Reader Domain(207)
- MicroRNA(13)
- Protein Arginine Deiminase(12)
- Chromodomain(1)
- Citrullination(15)
- DNA/RNA Demethylation(1)
- DNA/RNA Methylation(6)
- Histone Deacetylation(38)
- Histones/Histone Peptides(7)
- PHD Domains(0)
- Tandem Tudor & Tudor-like Domains(1)
- PRMT(2)
Products for Chromatin/Epigenetics
- Cat.No. Product Name Information
-
GC43889
I-CBP112 (hydrochloride)
cAMP-responsive element-binding protein binding protein (CREBBP) and E1A-associated protein p300 (EP300) are transcriptional co-activators that modulate DNA replication, DNA repair, cell growth, transformation, and development.
-
GC33042
IACS-9571 (ASIS-P040)
IACS-9571 (ASIS-P040) is a potent and selective inhibitor of TRIM24 and BRPF1, with IC50 of 8 nM for TRIM24, and Kds of 31 nM and 14 nM for TRIM24 and BRPF1, respectively.
-
GC60925
IACS-9571 hydrochloride
IACS-9571 (ASIS-P040) hydrochloride is a potent and selective inhibitor of TRIM24 and BRPF1, with an IC50 of 8 nM for TRIM24, and Kds of 31 nM and 14 nM for TRIM24 and BRPF1, respectively.
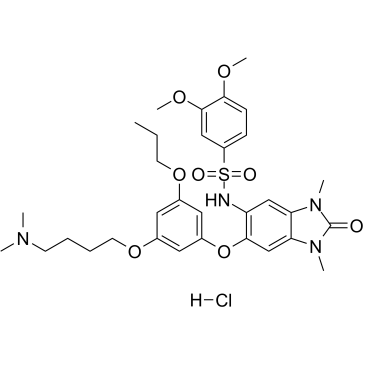
-
GC34437
IACS-9571 Hydrochloride (ASIS-P040 Hydrochloride)
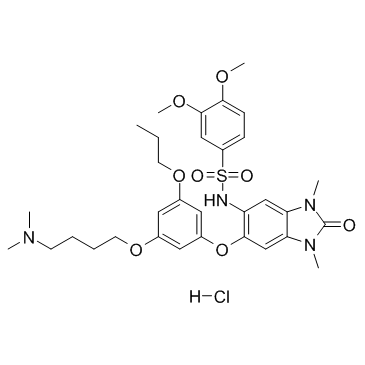
-
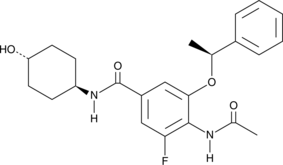
GC48548
iBET-BD2
A BD2 bromodomain inhibitor
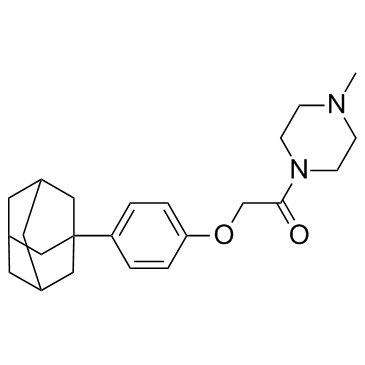
-
GC32948
IDF-11774
A novel HIF-1 inhibitor
-
GC64108
Ifidancitinib
Ifidancitinib (ATI-50002) is a potent and selective inhibitor of JAK kinases 1/3.
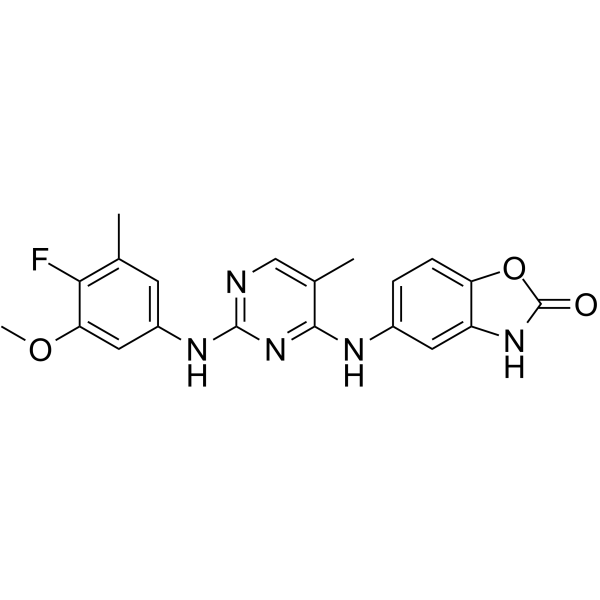
-
GC47450
IL-4 Inhibitor
IL-4 Inhibitor (compound 52) is an IL-4 inhibitor, with an EC50 of 1.81 μM.
-
GC32809
Ilginatinib (NS-018)
Ilginatinib (NS-018) (NS-018) is a highly active and orally bioavailable JAK2 inhibitor, with an IC50 of 0.72 nM, 46-, 54-, and 31-fold selectivity for JAK2 over JAK1 (IC50, 33 nM), JAK3 (IC50, 39 nM), and Tyk2 (IC50, 22 nM).
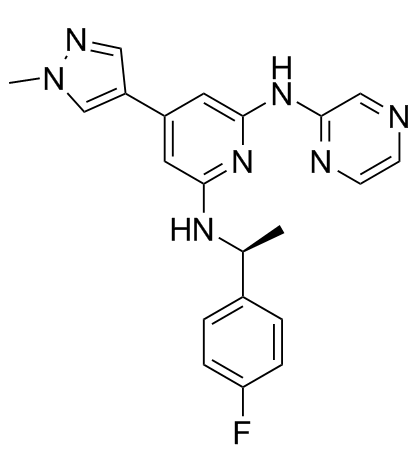
-
GC33037
Ilginatinib hydrochloride (NS-018 hydrochloride)
Ilginatinib hydrochloride (NS-018 hydrochloride) (NS-018 hydrochloride) is a highly active and orally bioavailable JAK2 inhibitor, with an IC50 of 0.72 nM, 46-, 54-, and 31-fold selectivity for JAK2 over JAK1 (IC50, 33 nM), JAK3 (IC50, 39 nM), and Tyk2 (IC50, 22 nM).
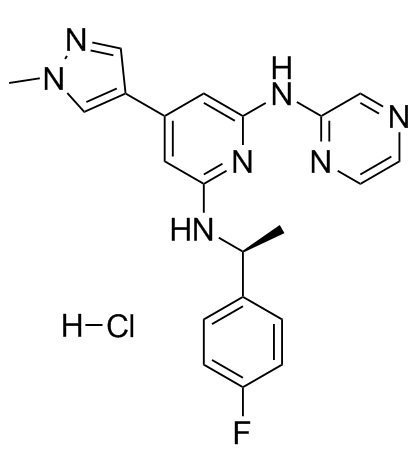
-
GC33018
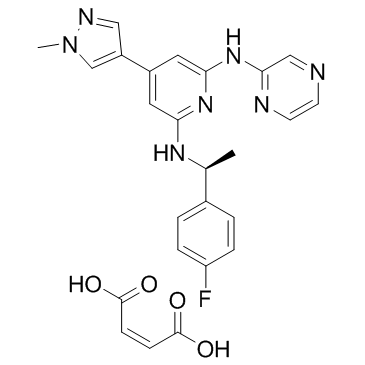
Ilginatinib maleate (NS-018 (maleate))
Ilginatinib maleate (NS-018 (maleate)) (NS-018 maleate) is a highly active and orally bioavailable JAK2 inhibitor, with an IC50 of 0.72 nM, 46-, 54-, and 31-fold selectivity for JAK2 over JAK1 (IC50, 33 nM), JAK3 (IC50, 39 nM), and Tyk2 (IC50, 22 nM).
-
GC34159
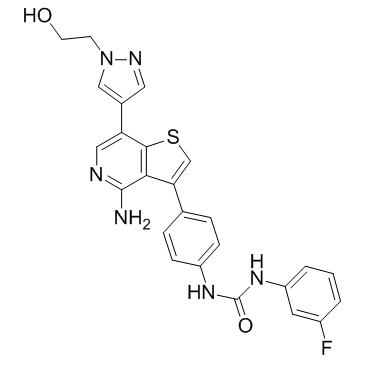
Ilorasertib (ABT-348)
Ilorasertib (ABT-348) (ABT-348) is a potent, orally active and ATP-competitive aurora inhibitor with IC50s of116, 5, 1 nM for aurora A, aurora B, aurora C, respectively. Ilorasertib (ABT-348) also is a potent VEGF, PDGF inhibitor. Ilorasertib (ABT-348) has the potential for the research of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS).
-
GC38519
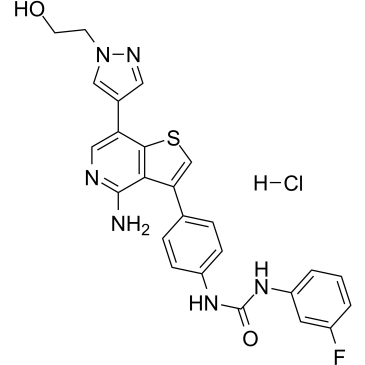
Ilorasertib hydrochloride
Ilorasertib (ABT-348) hydrochloride is a potent, orally active and ATP-competitive aurora inhibitor with IC50s of116, 5, 1 nM for aurora A, aurora B, aurora C, respectively. Ilorasertib hydrochloride also is a potent VEGF, PDGF inhibitor. Ilorasertib hydrochloride has the potential for the research of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS).
-
GC63309
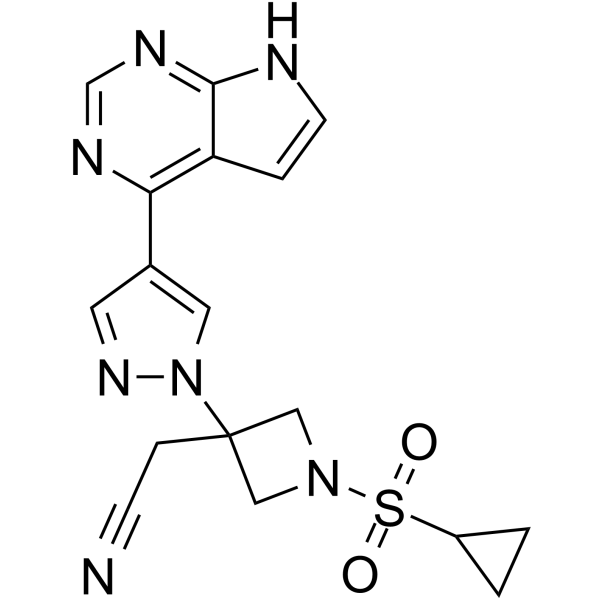
Ilunocitinib
Ilunocitinib (compound 27) is a JAK inhibitor (extracted from patent WO2009114512A1).
-
GC14755
Inauhzin
SIRT1 inhibitor
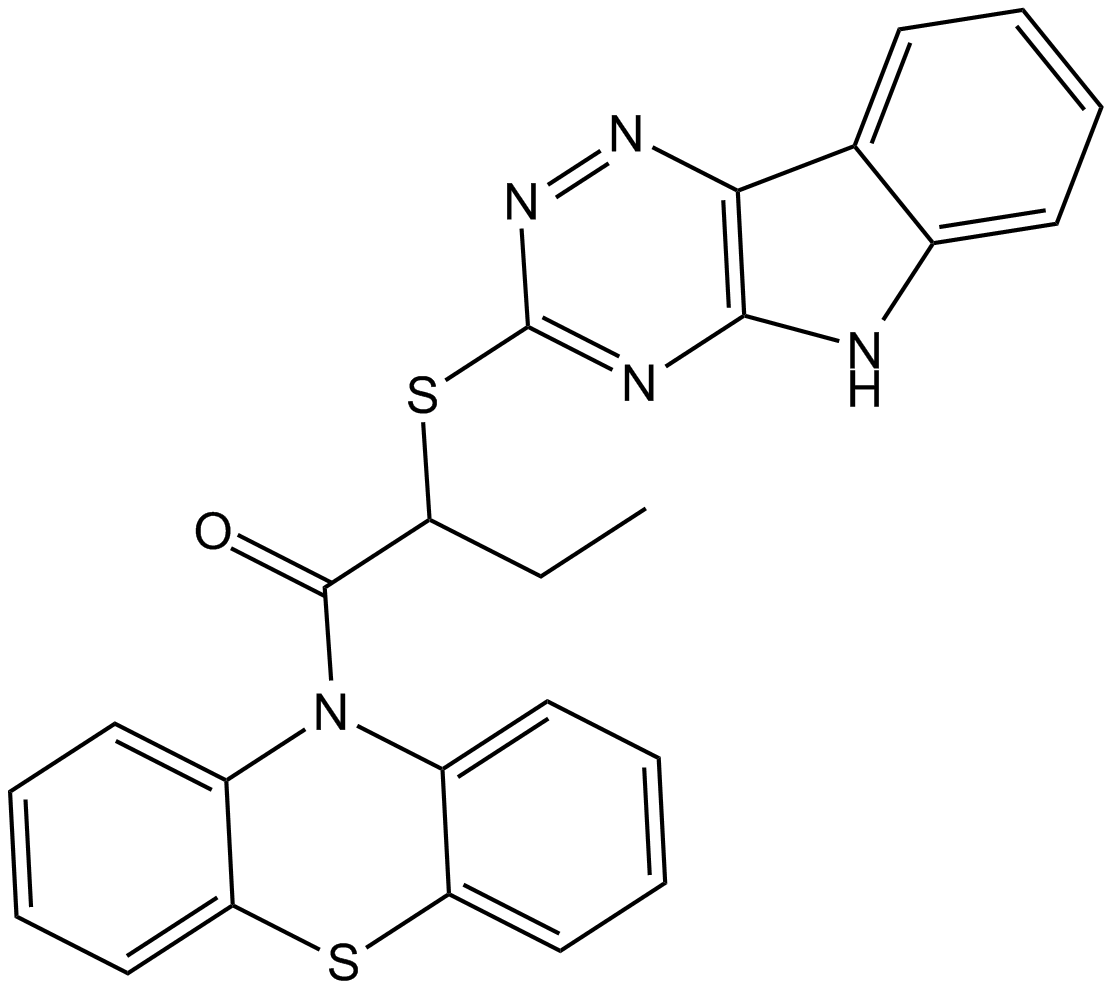
-
GC16117
INCA-6
A cell-permeant inhibitor of NFAT signaling
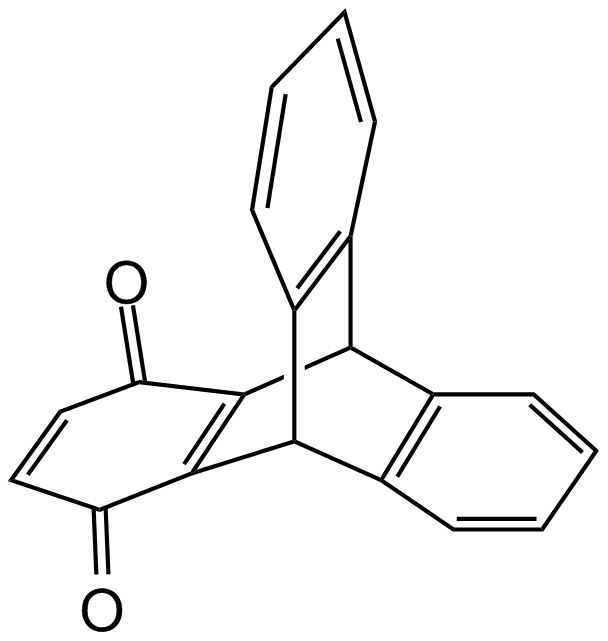
-
GC19200
INCB-057643
INCB-057643 is a novel, orally bioavailable BET inhibitor.
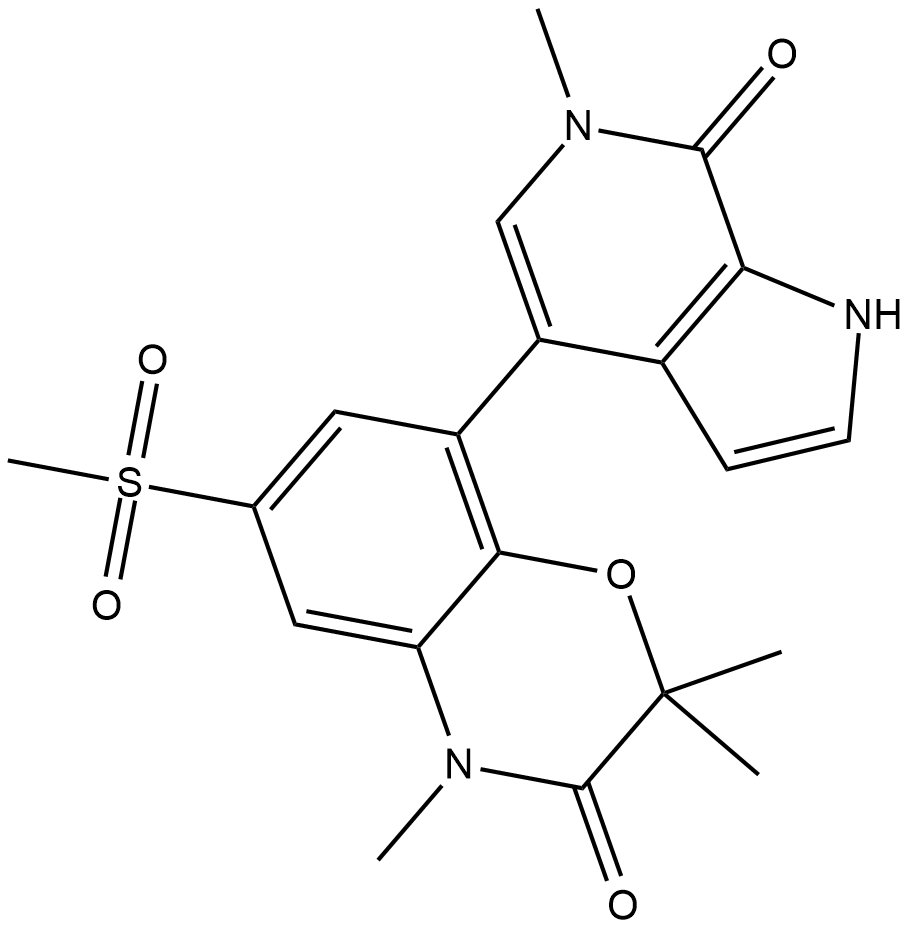
-
GC33026
INCB054329
INCB054329 is a potent BET inhibitor.
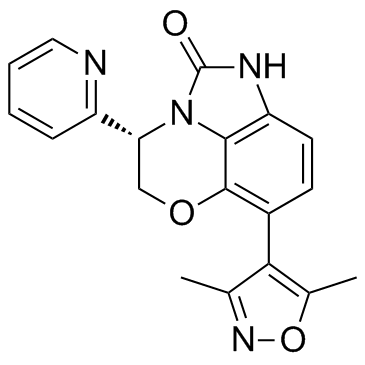
-
GC30644
INCB054329 Racemate
INCB054329 Racemate is a BET protein inhibitor.
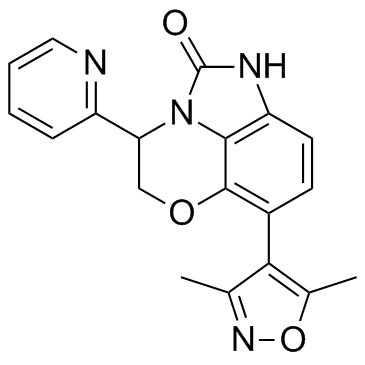
-
GC15353
Iniparib (BSI-201)
A PARP1 inhibitor
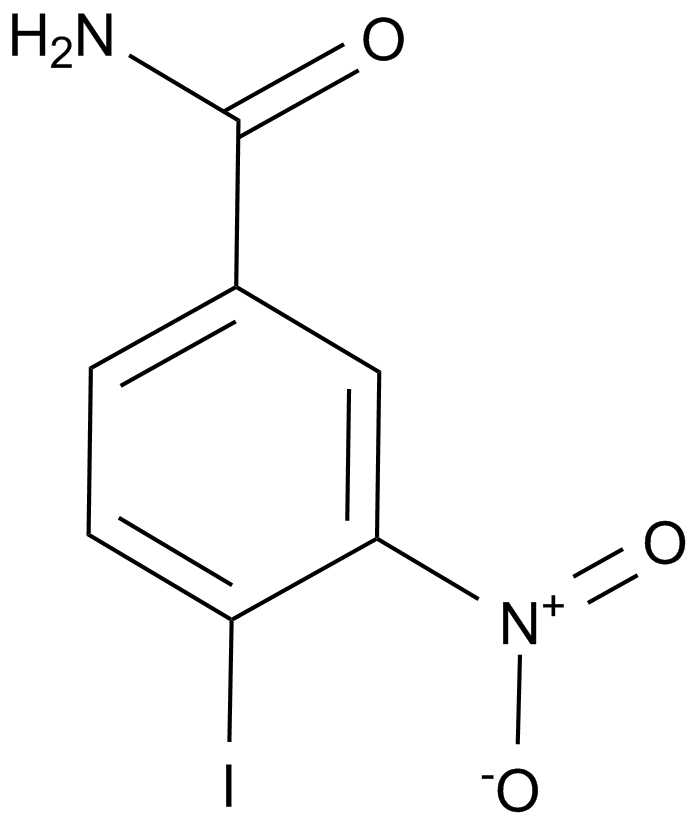
-
GC12496
INO-1001
A PARP inhibitor
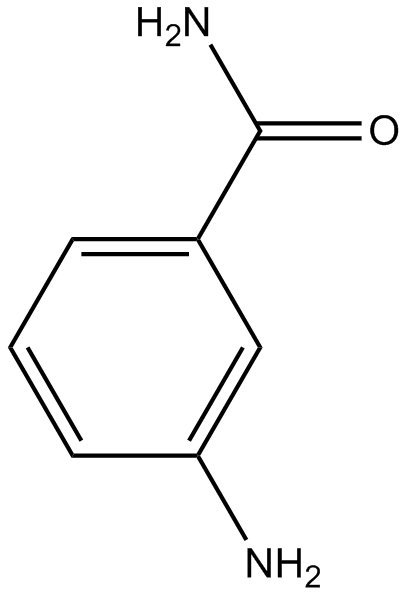
-
GC38385
INO-1001
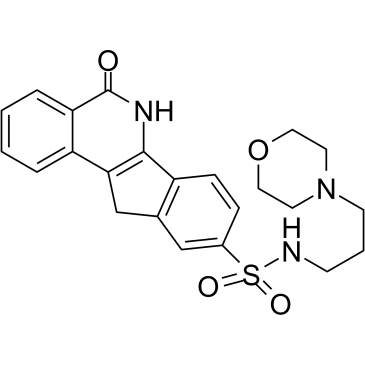
-
GC17754
IOX 1
histone demethylase JMJD inhibitor
-
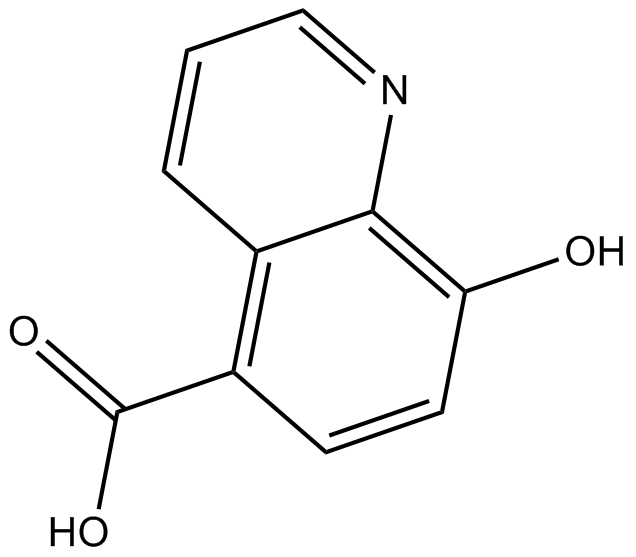
GC13018
IOX2(Glycine)
A selective PHD2 inhibitor
-
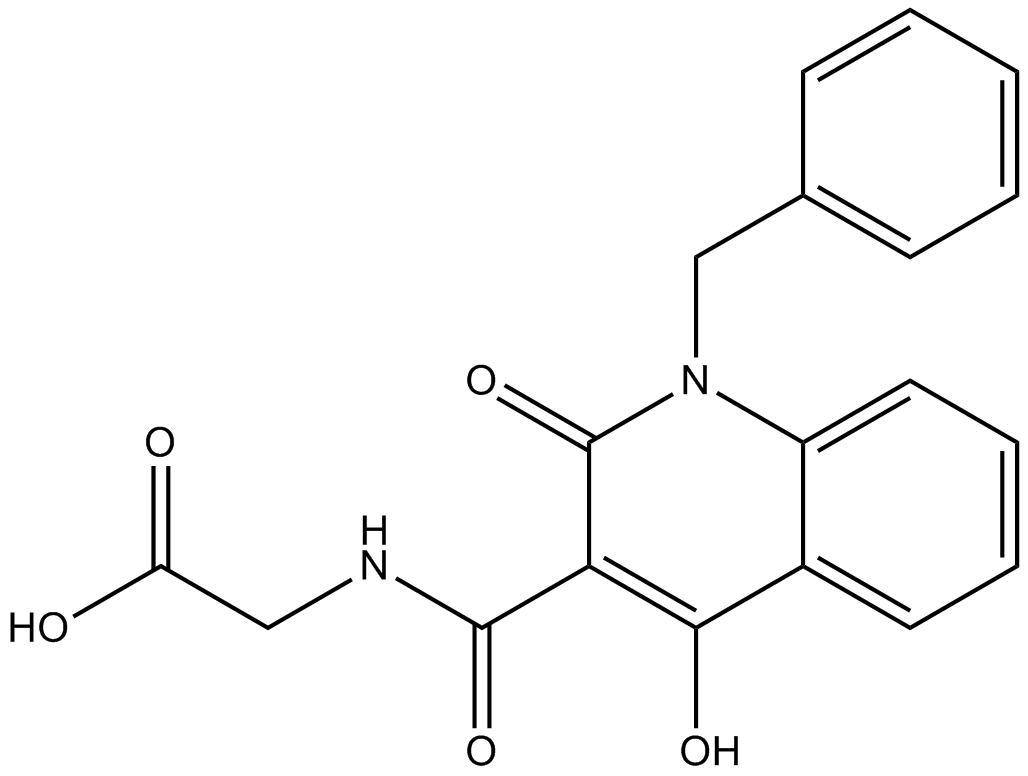
GC12255
IOX4
PHD2 inhibitor
-
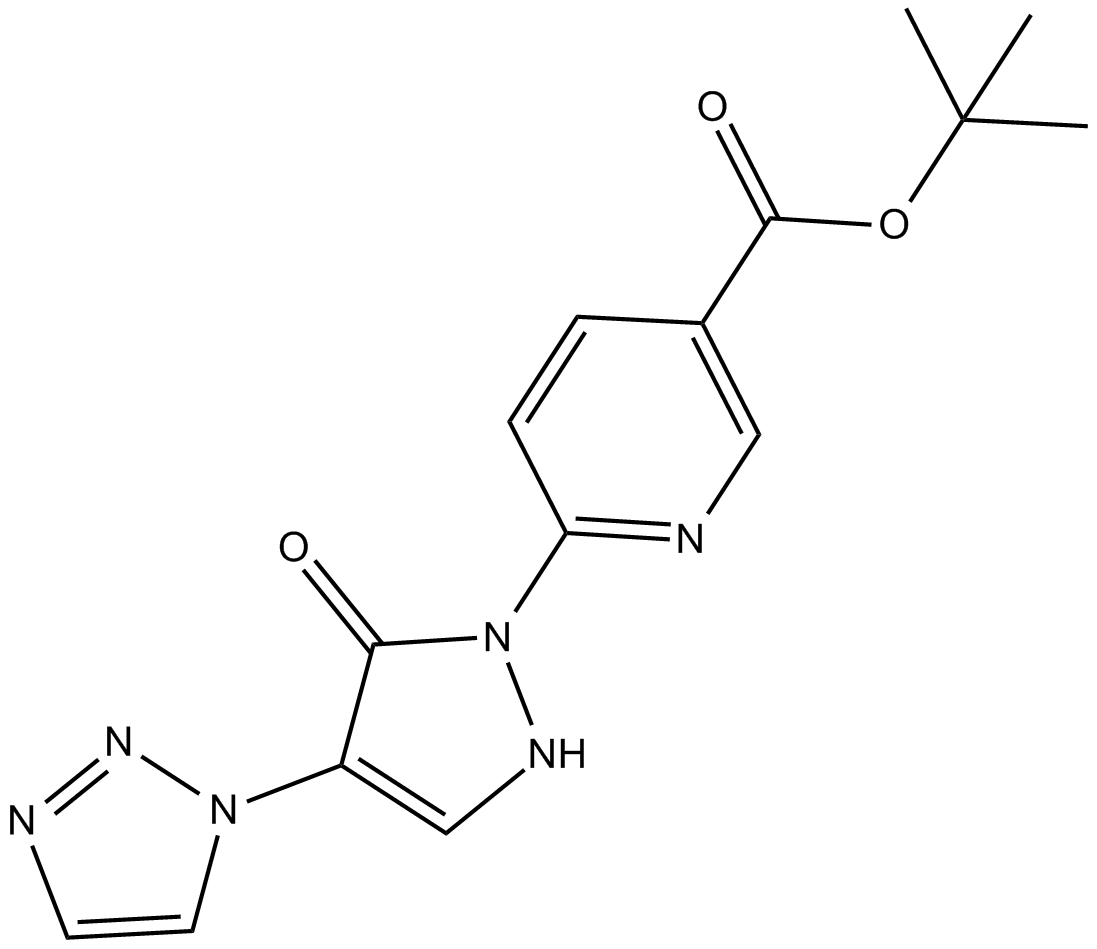
GC50721
iP300w
-
GC10727
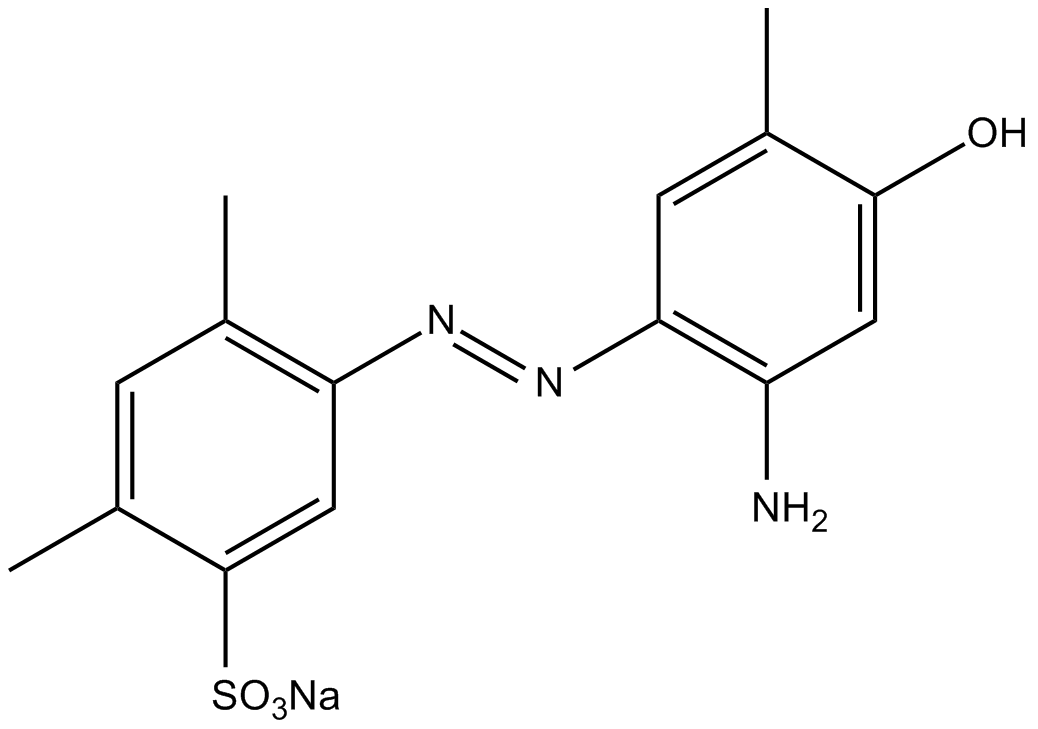
Ischemin sodium salt
CBP bromodomain inhibitor
-
GC19204
Itacitinib
Itacitinib is a potent and selective inhibitor of JAK1, with >20-fold selectivity for JAK1 over JAK2 and >100-fold over JAK3 and TYK2; Itacitinib is used in the research of myelofibrosis.
-
GC36351
Itacitinib adipate
Itacitinib adipate is an orally bioavailable and selective JAK1 inhibitor which has been tested for efficacy and safety in a phase II trial in myelofibrosis.
-
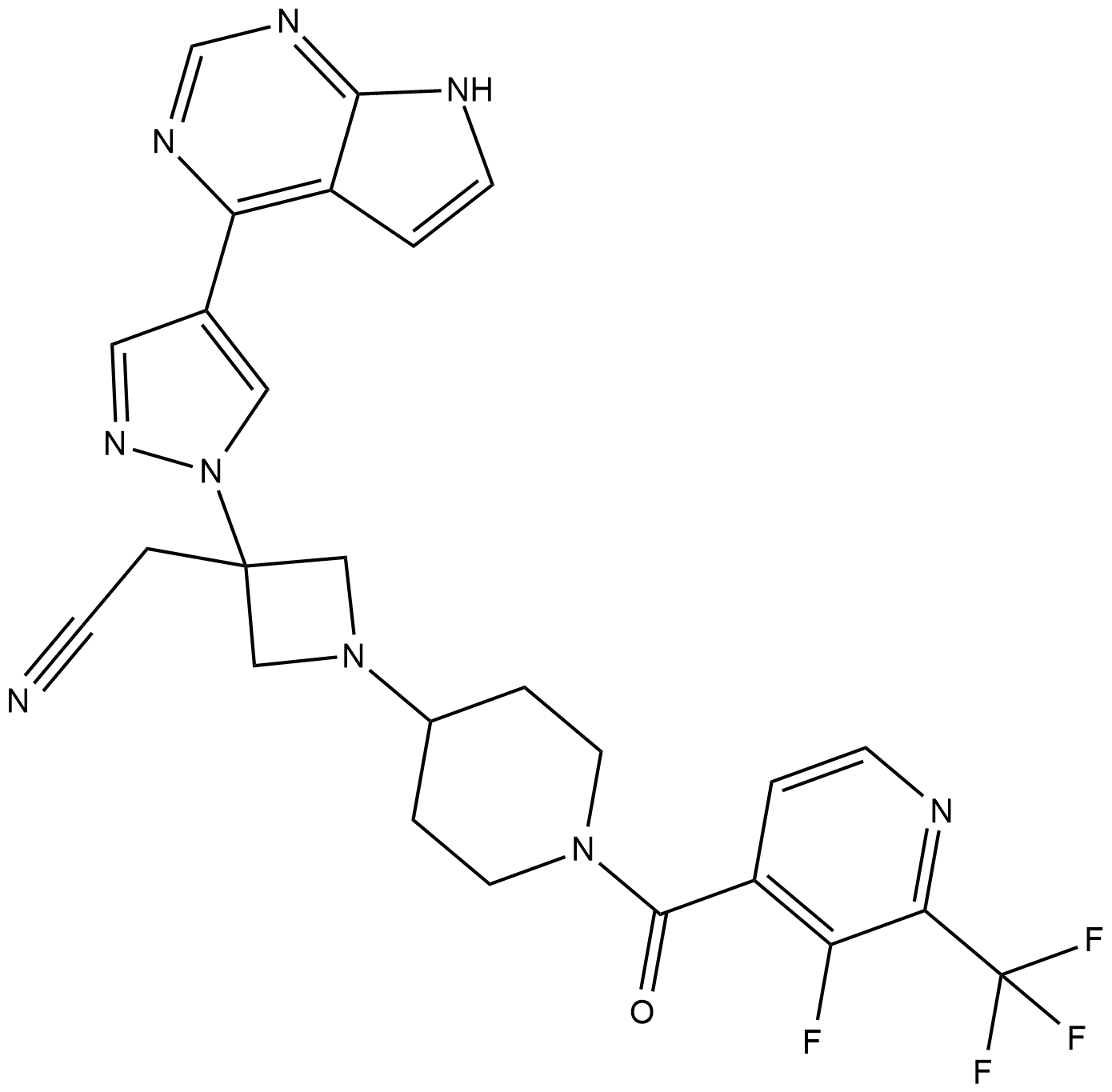
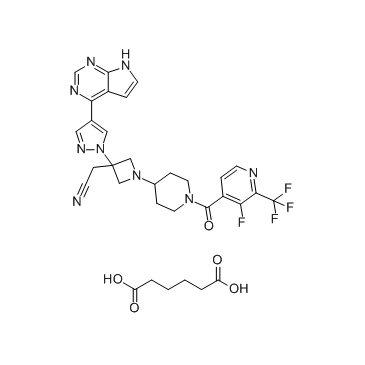
GC63416
Itacnosertib
Itacnosertib (TP-0184) is both inhibitor to JAK2, ACVR1 (ALK2) and ALK5 as described in WO2014151871.
-
GC17836
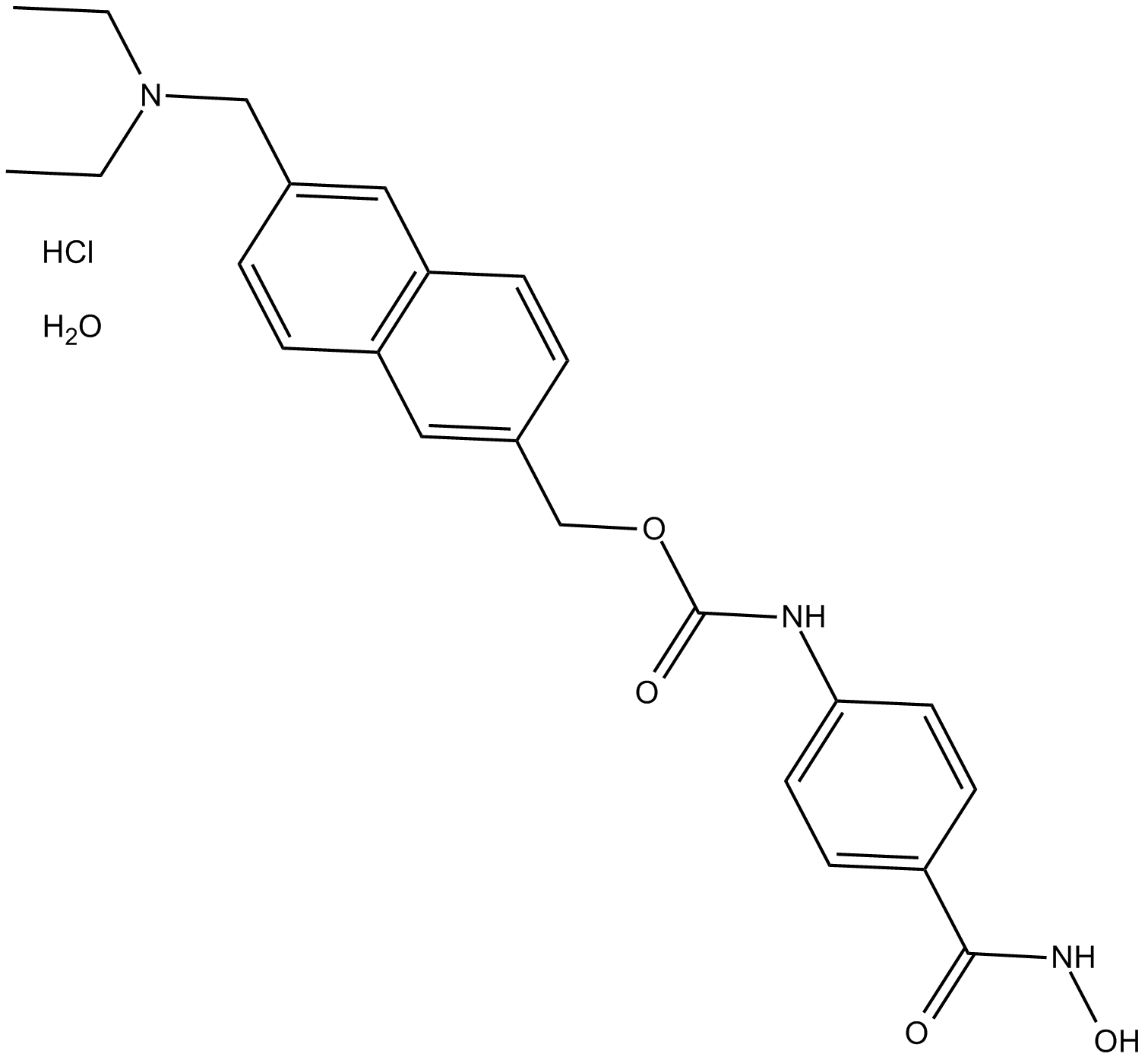
ITF2357 (Givinostat)
HDAC inhibitor with anti-inflammatory and antineoplastic activities
-
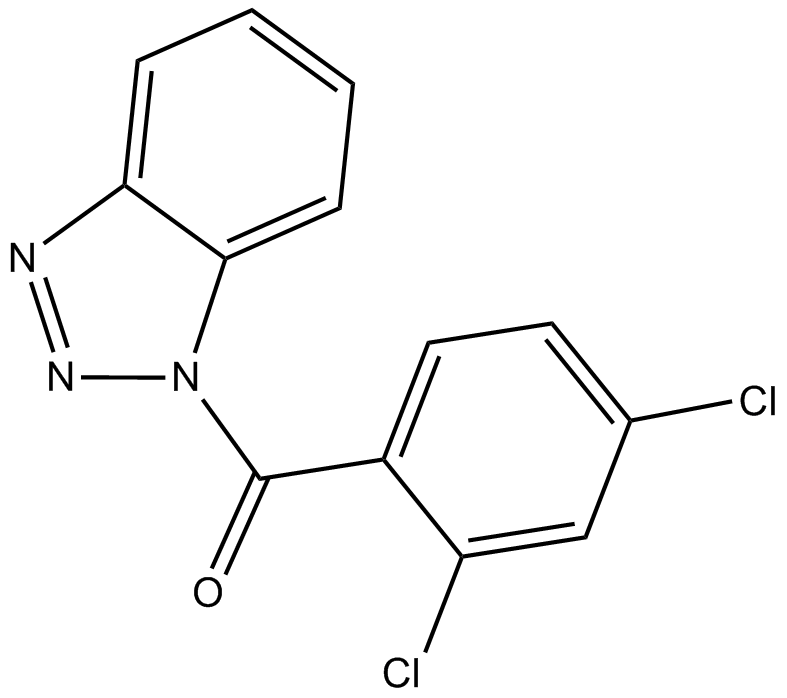
GC14597
ITSA-1 (ITSA1)
ITSA-1 (ITSA1) is an activator of histone deacetylase (HDAC), and counteract trichostatin A (TSA)-induced cell cycle arrest, histone acetylation, and transcriptional activation.
-
GC63703
Ivaltinostat formic
Ivaltinostat (CG-200745) formic is an orally active, potent pan-HDAC inhibitor which has the hydroxamic acid moiety to bind zinc at the bottom of catalytic pocket.

-
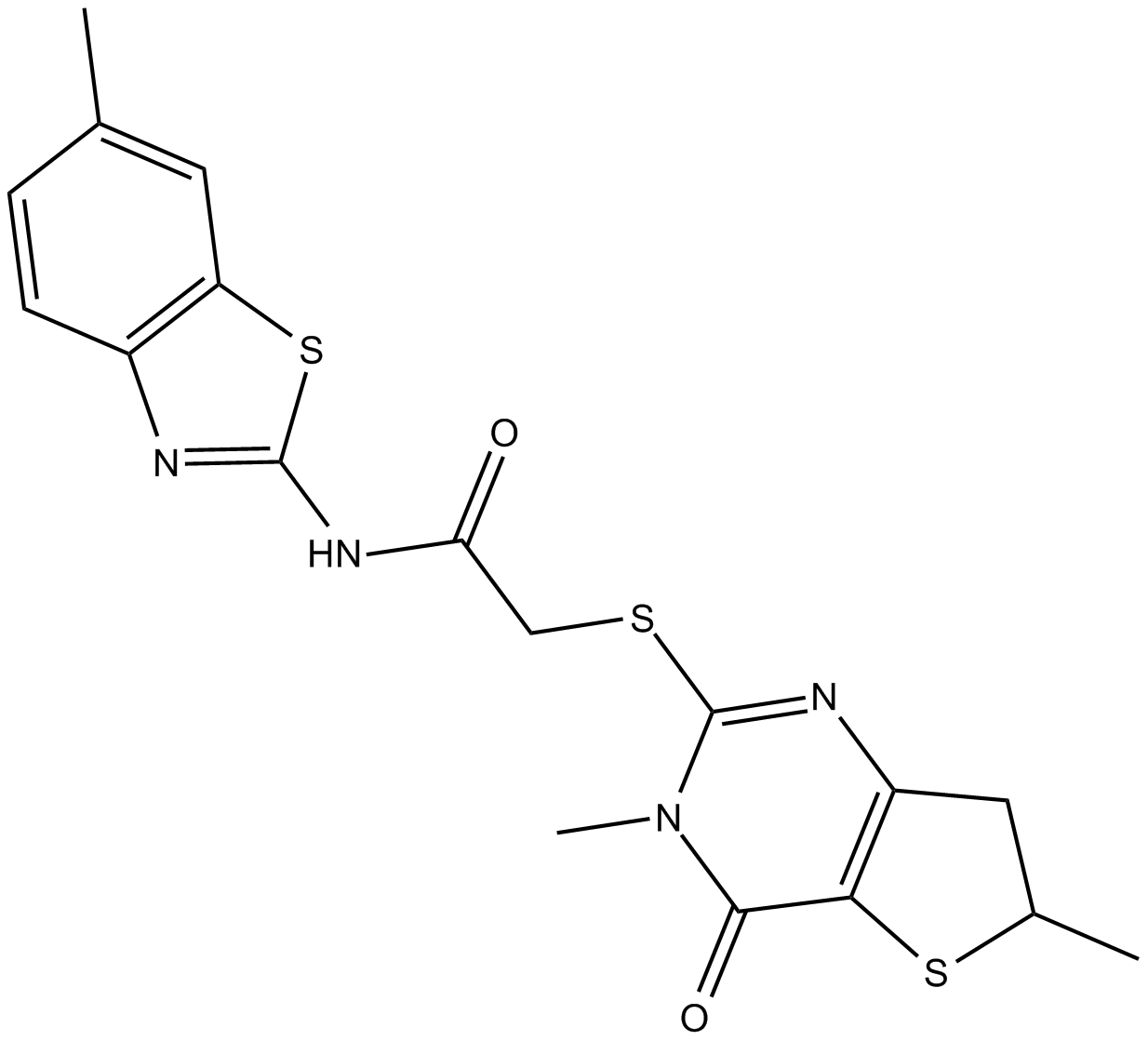
GC15836
IWP 12
Potent inhibitor of Porcupine (PORCN)
-
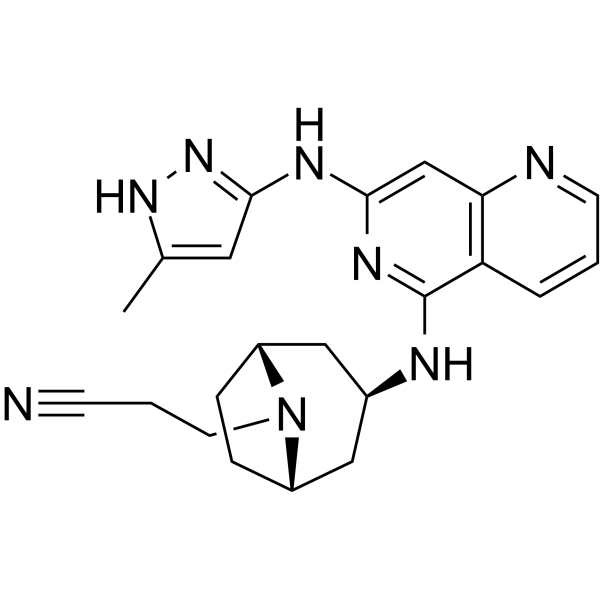
GC62313
Izencitinib
Izencitinib (TD-1473) is an orally active, non-selective and gut-restricted JAK inhibitor.
-
GC63828
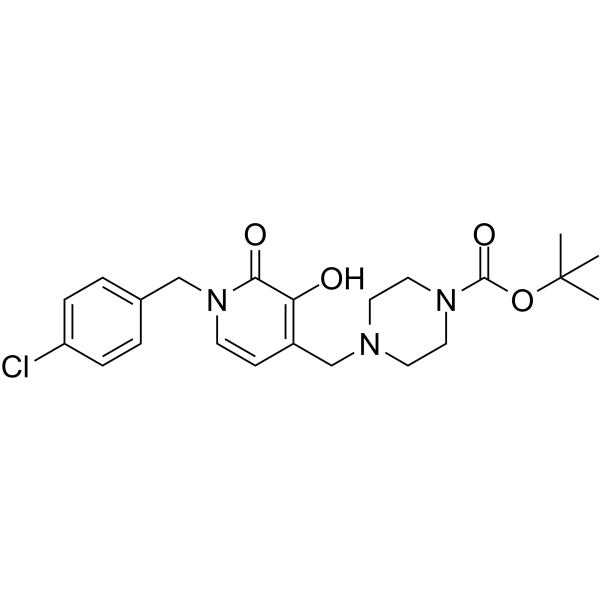
Izilendustat
Izilendustat is a potent inhibitor of prolyl hydroxylase which can stabilize hypoxia inducible factor- 1 alpha (HIF- lα) and hypoxia inducible factor-2 (HIF-2).
-
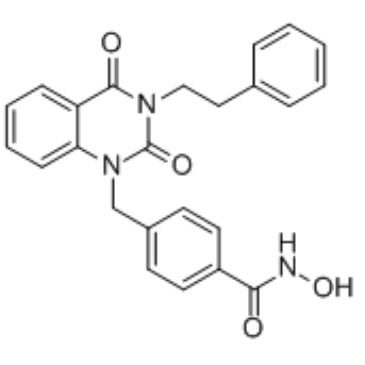
GC34629
J22352
J22352 is a PROTAC (proteolysis-targeting chimeras)-like and highly selective HDAC6 inhibitor with an IC50 value of 4.7 nM. J22352 promotes HDAC6 degradation and induces anticancer effects by inhibiting autophagy and eliciting the antitumor immune response in glioblastoma cancers, and leading to the restoration of host antitumor activity by reducing the immunosuppressive activity of PD-L1.
-
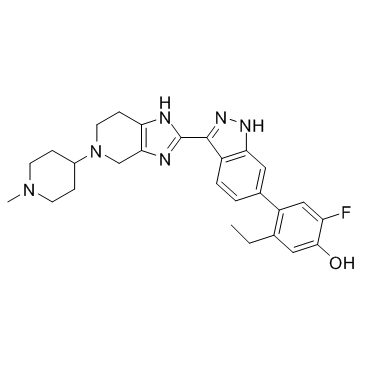
GC31866
JAK inhibitor 1
JAK inhibitor 1 is an inhibitor of JAK extracted from patent US20170121327A1, compound example 283.
-
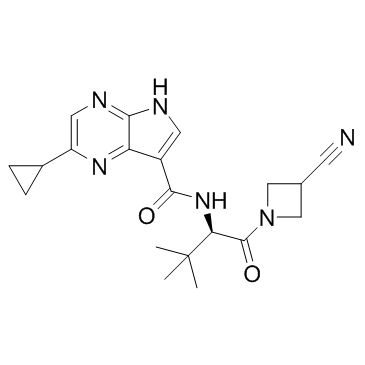
GC31999
JAK-IN-1
JAK-IN-1 is a JAK1/2/3 inhibitor with IC50s of 0.26, 0.8 and 3.2 nM, respectively.
-
GC36366
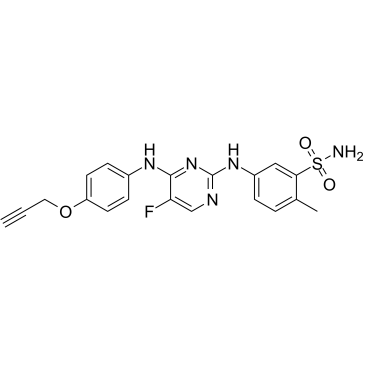
JAK-IN-10
JAK-IN-10 is a JAK inhibitor.
-
GC63448
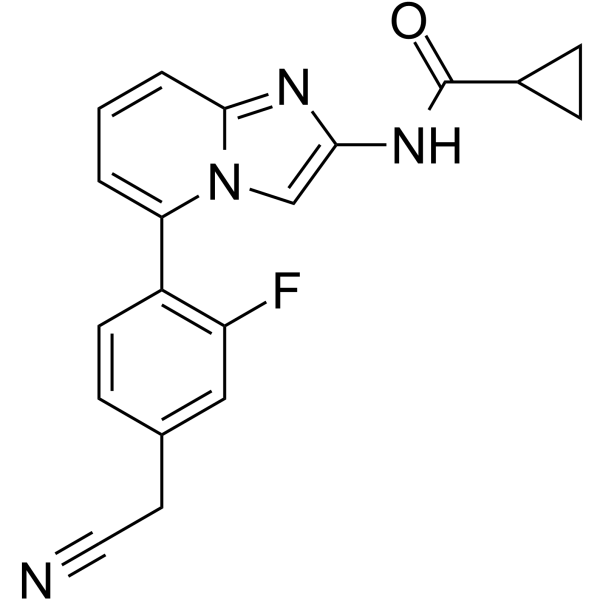
JAK-IN-14
JAK-IN-14 is a potent and selective JAK1 inhibitor, with an IC50 of <5 μM.
-
GC68000
JAK-IN-23
-
GC65453
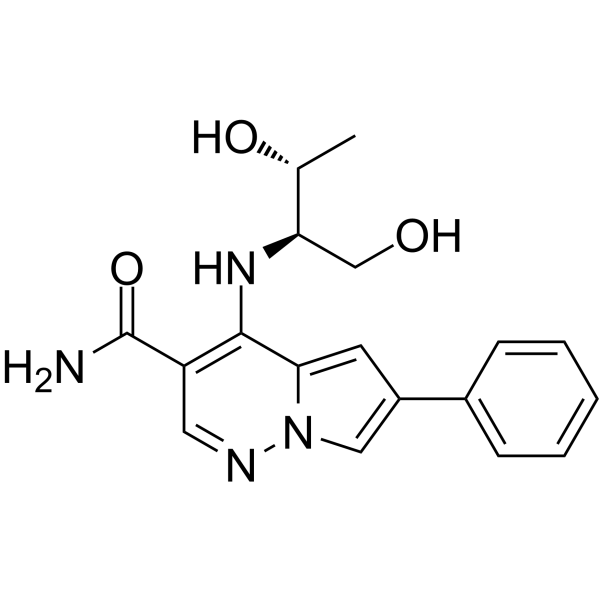
JAK-IN-3
JAK-IN-3 (compound 22) is a potent JAK inhibitor, with IC50 values of 3 nM, 5 nM, 34 nM and 70 nM for JAK3, JAK1, TYK2 and JAK2, respectively.
-
GC64959
JAK/HDAC-IN-1
JAK/HDAC-IN-1 is a potent JAK2/HDAC dual inhibitor, exhibits antiproliferative and proapoptotic activities in several hematological cell lines. JAK/HDAC-IN-1 shows IC50s of 4 and 2 nM for JAK2 and HDAC, respectively.
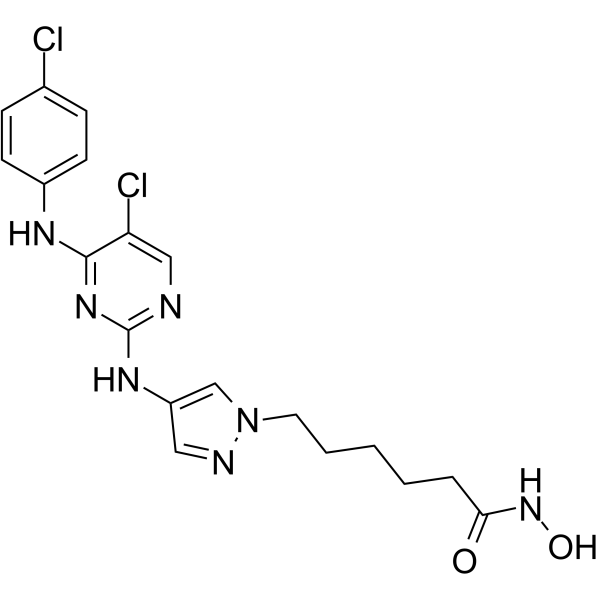
-
GC33036
JAK1-IN-3
JAK1-IN-3 (AZD4205) is a selective JAK1 inhibitor, with an IC50 of 73 nM, weakly inhibits JAK2 (IC50>14.7 μM), and shows little inhibition on JAK3 (IC50>30 μM).
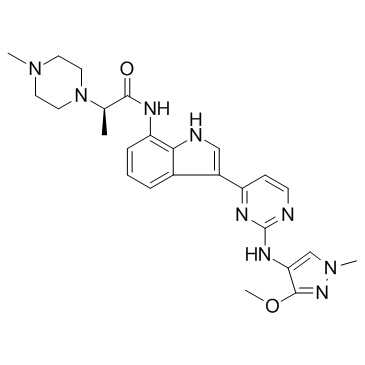
-
GC33258
JAK1-IN-4
JAK1-IN-4 is a potent and selective JAK1 inhibitor, with IC50s of 85 nM, 12.8 μM and >30 μM for JAK1, JAK2, and JAK3, respectively. JAK1-IN-4 inhibits STAT3 phosphorylation in NCI-H 1975 cells (IC50, 227 nM).
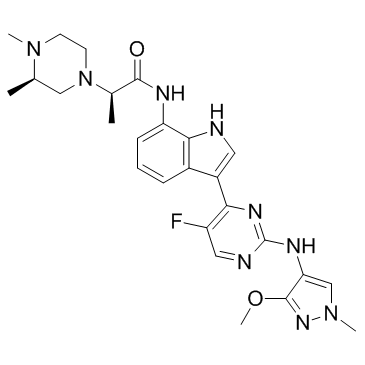
-
GC36363
JAK1-IN-7
JAK1-IN-7 (JAK1-IN-7) is a Janus-associated kinase 1 (JAK1) inhibitor extracted from patent WO2018134213A1, Example 63, has an anti-inflammatory effect.
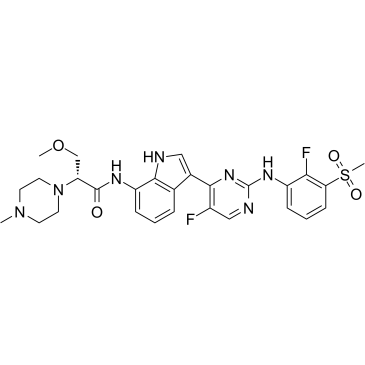
-
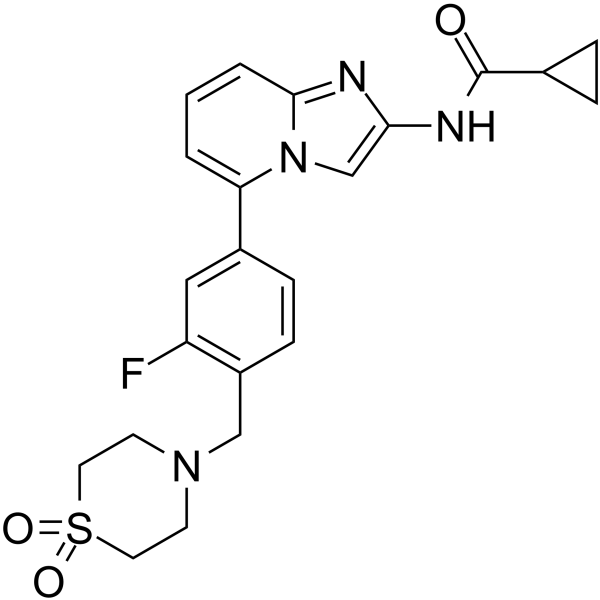
GC63029
JAK1-IN-8
JAK1-IN-8, a potent JAK1 inhibitor (IC50<500 nM), compound 28, extracted from patent WO2016119700A1.
-
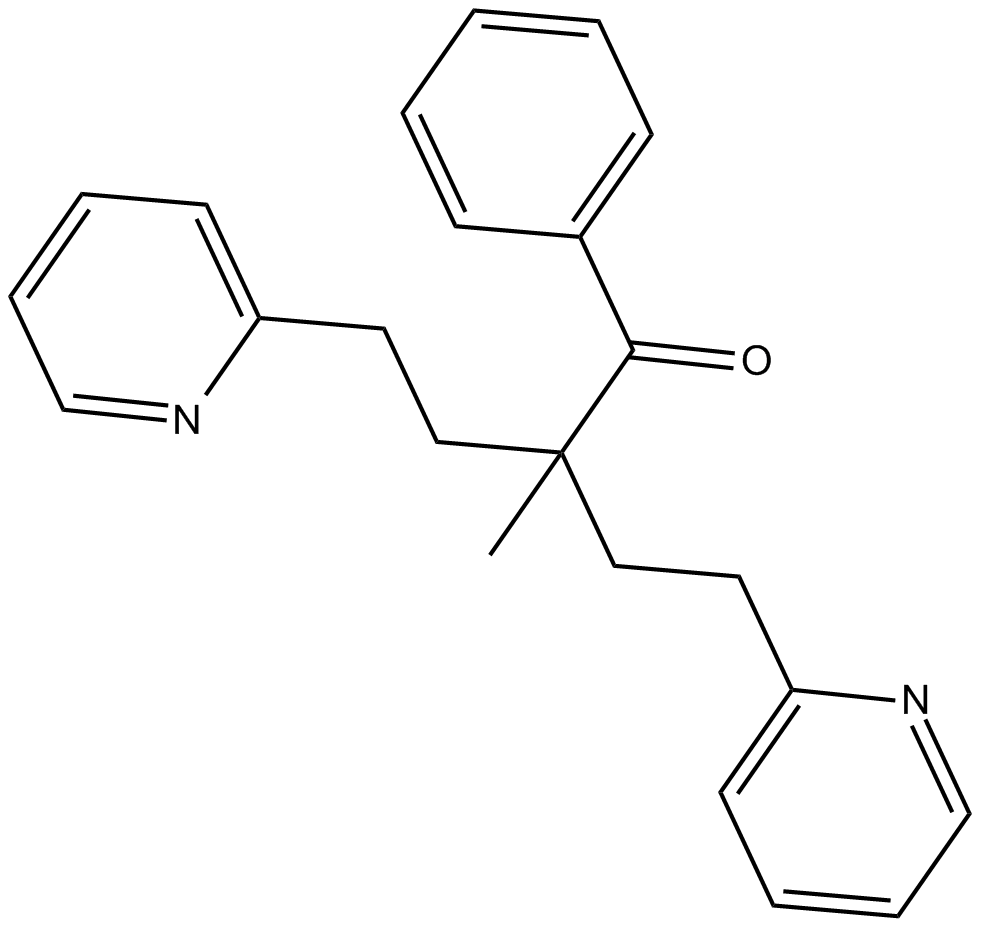
GC13826
JAK2 Inhibitor V, Z3
A selective inhibitor of the autophosphorylation of wild type and V617F mutant forms of JAK
-
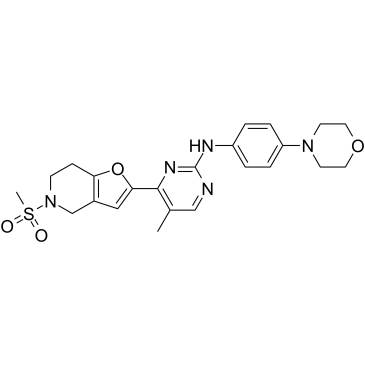
GC36364
JAK2-IN-4
JAK2-IN-4 (compound 16h) is a selective JAK2/JAK3 inhibitor, with IC50 values of 0.7 nM and 23.2 nM for JAK2 and JAK3, respectively.
-
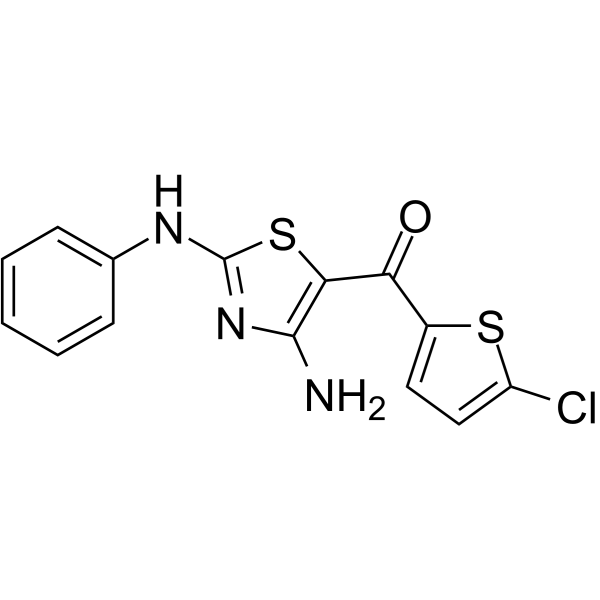
GC65405
JAK2-IN-6
JAK2-IN-6, a multiple-substituted aminothiazole derivative, is a potent and selective JAK2 inhibitor with an IC50 of 22.86 μg/mL. JAK2-IN-6 shows no activity against JAK1 and JAK3. JAK2-IN-6 has anti-proliferative effect against cancer cells.
-
GC62500
JAK2-IN-7
JAK2-IN-7 is a selective JAK2 inhibitor with IC50s of 3, 11.7, and 41 nM for JAK2, SET-2, and Ba/F3V617F cells, respectively. JAK2-IN-7 possesses >14-fold selectivity over JAK1, JAK3, FLT3. JAK2-IN-7 stimulates cell cycle arrest in the G0/G1 phase and induces tumor cellapoptosis. Antitumor activities.
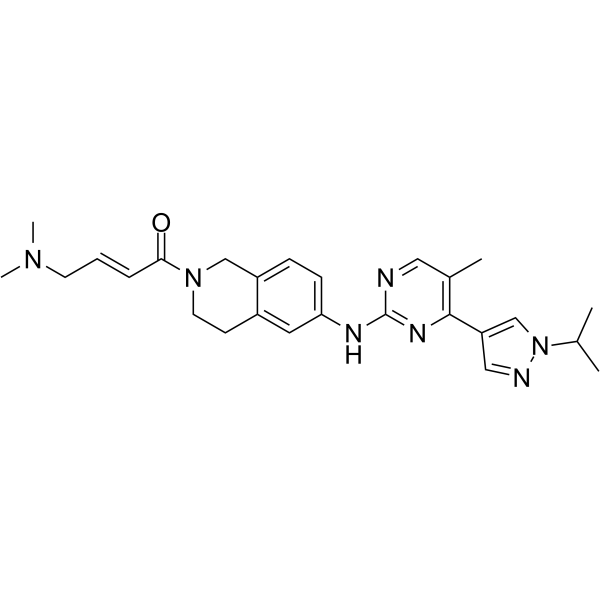
-
GC62665
JAK2/FLT3-IN-1 TFA
JAK2/FLT3-IN-1 (TFA) is a potent and orally active dual JAK2/FLT3 inhibitor with IC50 values of 0.7 nM, 4 nM, 26 nM and 39 nM for JAK2, FLT3, JAK1 and JAK3, respectively. JAK2/FLT3-IN-1 (TFA) has anti-cancer activity.
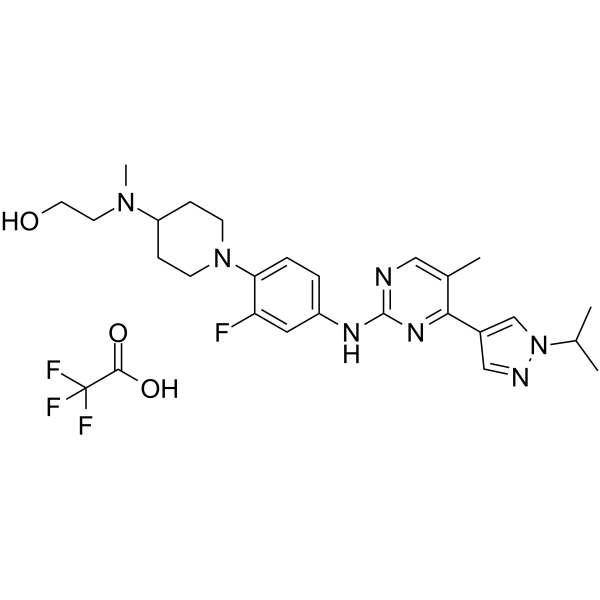
-
GC36365
JAK3 covalent inhibitor-1
JAK3 covalent inhibitor-1 is a potent and selective janus kinase 3 (JAK3) covalent inhibitor with an IC50 of 11 nM and shows 246-fold selectivity vs other JAKs.
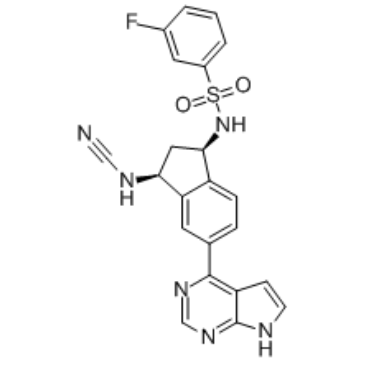
-
GC33046
JAK3-IN-1
JAK3-IN-1 is a potent, selective and orally active JAK3 inhibitor with an IC50 of 4.8 nM. JAK3-IN-1 shows over 180-fold more selective for JAK3 than JAK1 (IC50 of 896 nM) and JAK2 (IC50 of 1050 nM).
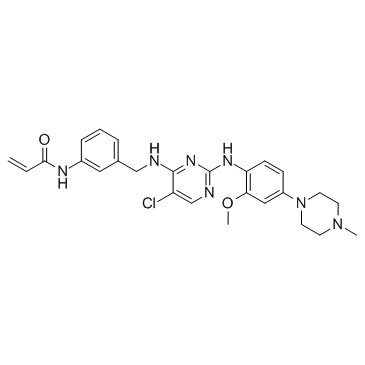
-
GC31902
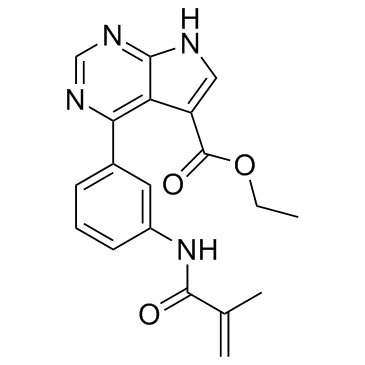
JAK3-IN-6
JAK3-IN-6 is a potent, selective irreversible Janus Associated Kinase 3 (JAK3) inhibitor, with an IC50 of 0.15 nM.
-
GC33382
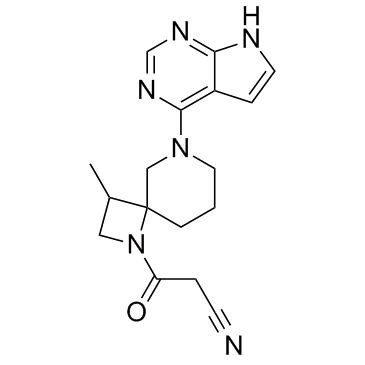
JAK3-IN-7
JAK3-IN-7 is a potent and selective JAK3 inhibitor extracted from patent WO2011013785A1, has an IC50 of <0.01 μM.
-
GC14671
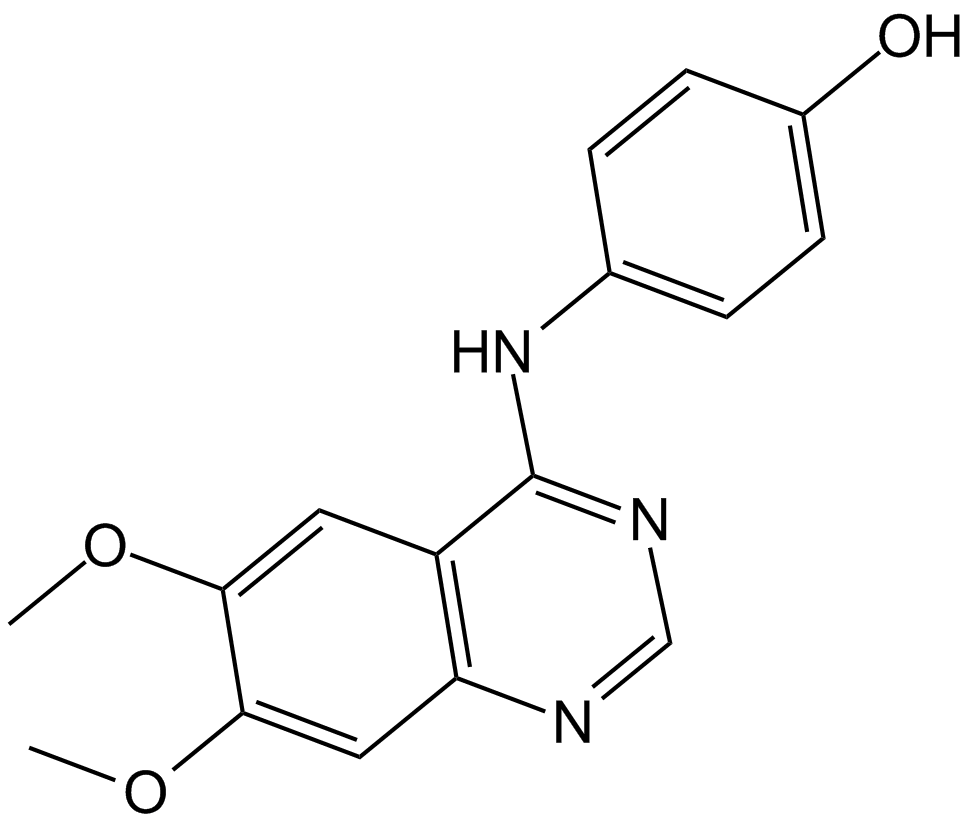
JANEX-1
A selective JAK3 inhibitor
-
GC14686
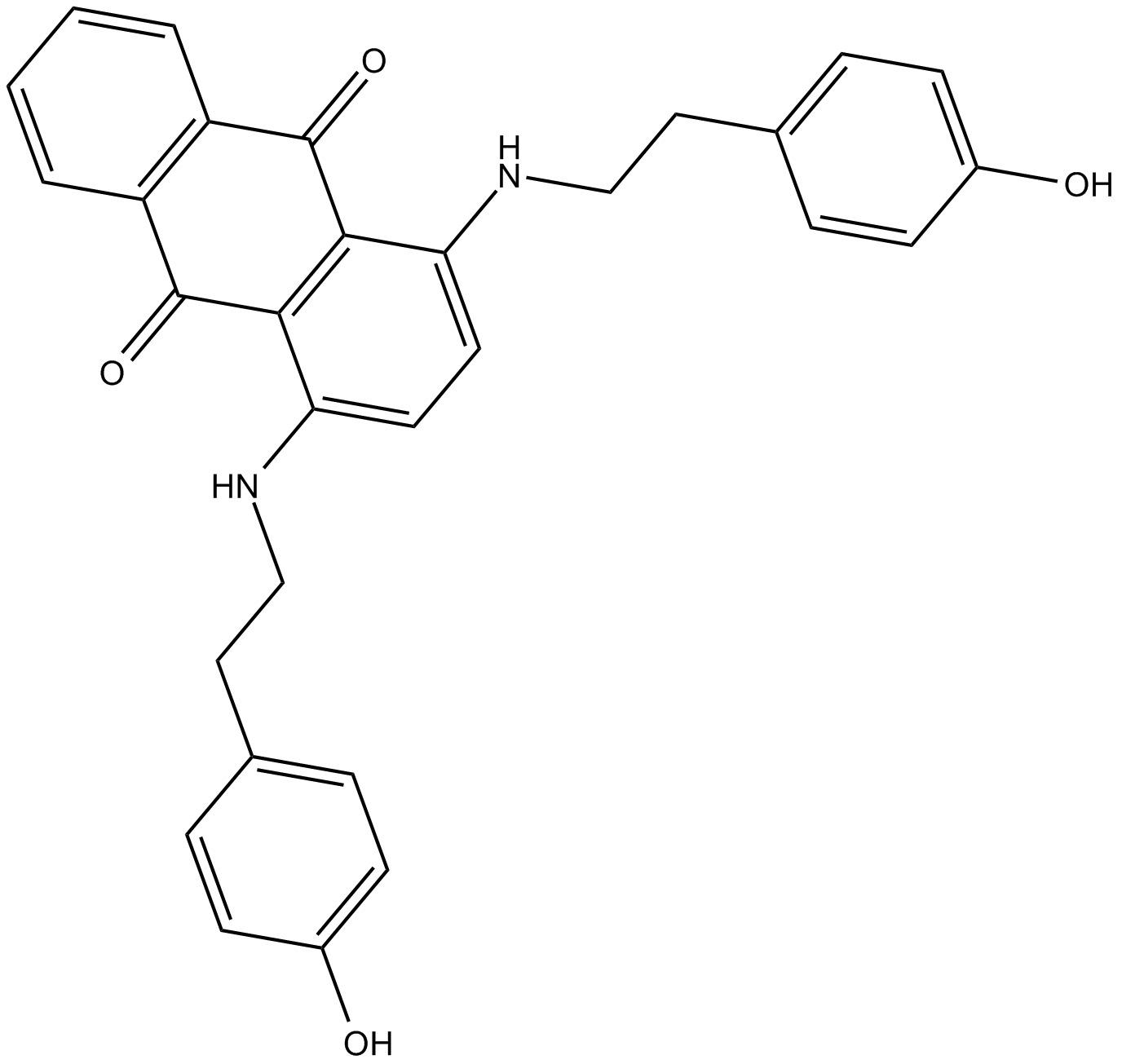
JFD00244
inhibitor of SIRT2
-
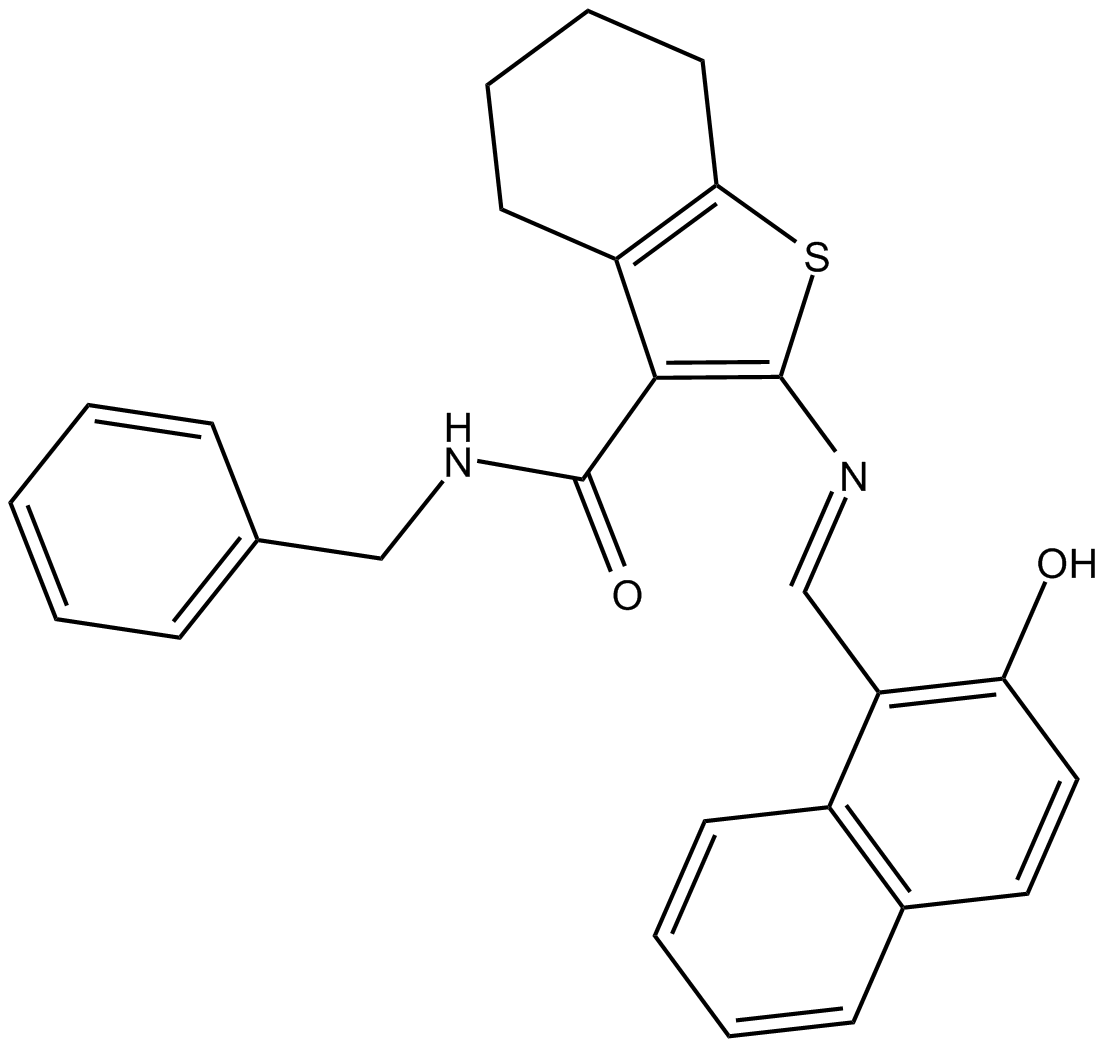
GC13062
JGB1741
SIRT1 inhibitor
-
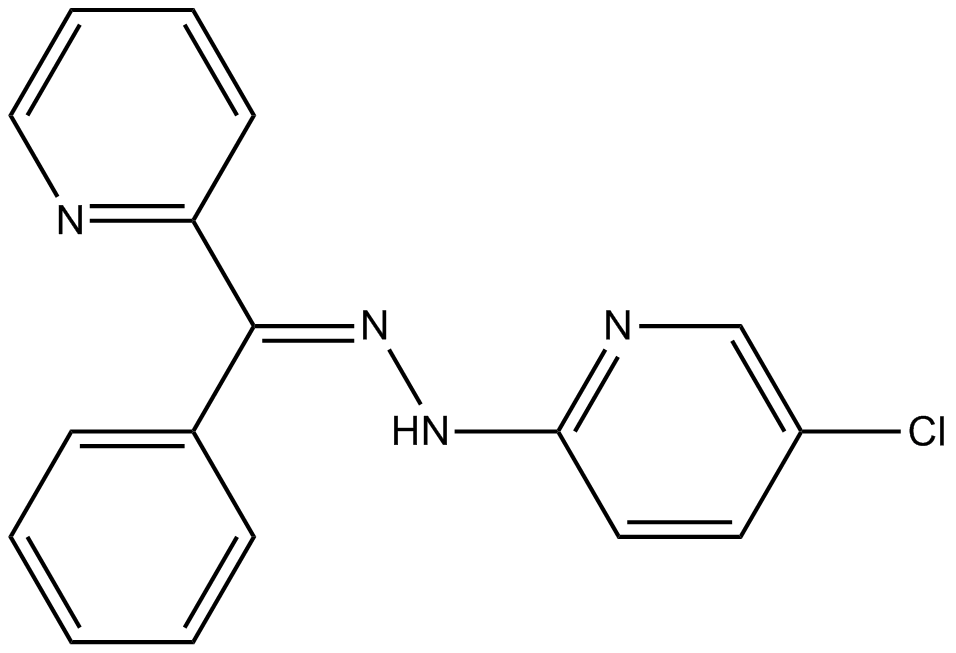
GC15603
JIB-04
Jumonji histone demethylase inihibitor
-
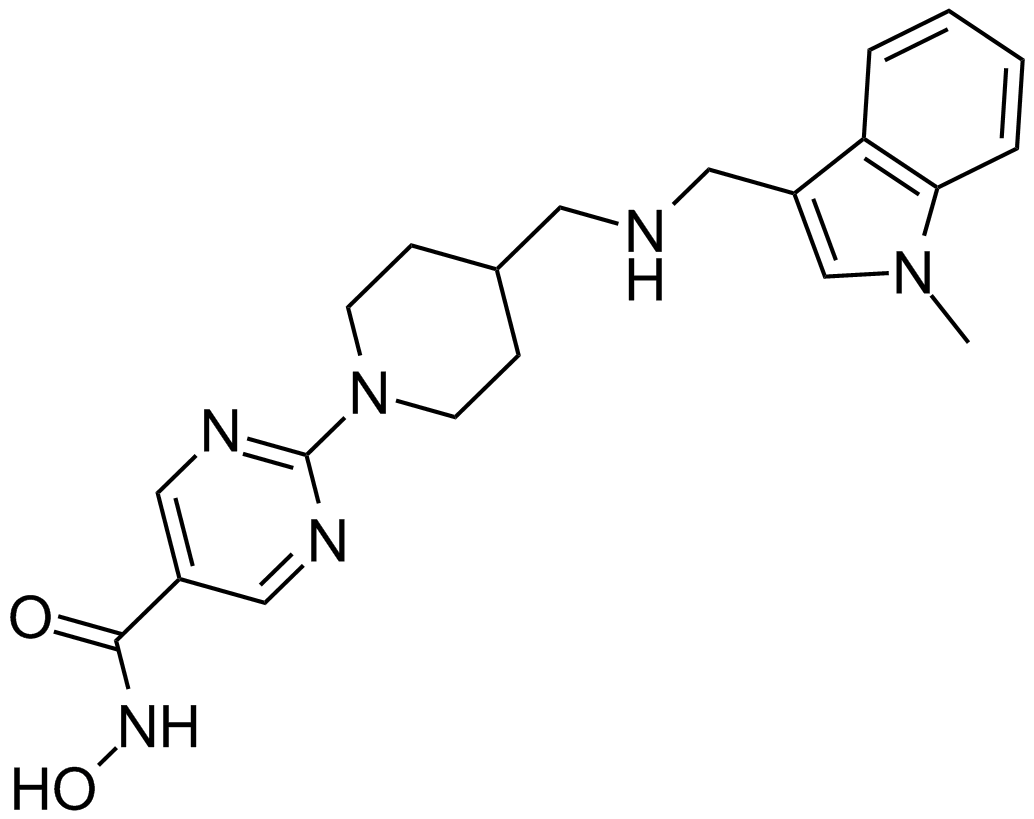
GC15476
JNJ-26481585
A pan-HDAC inhibitor
-
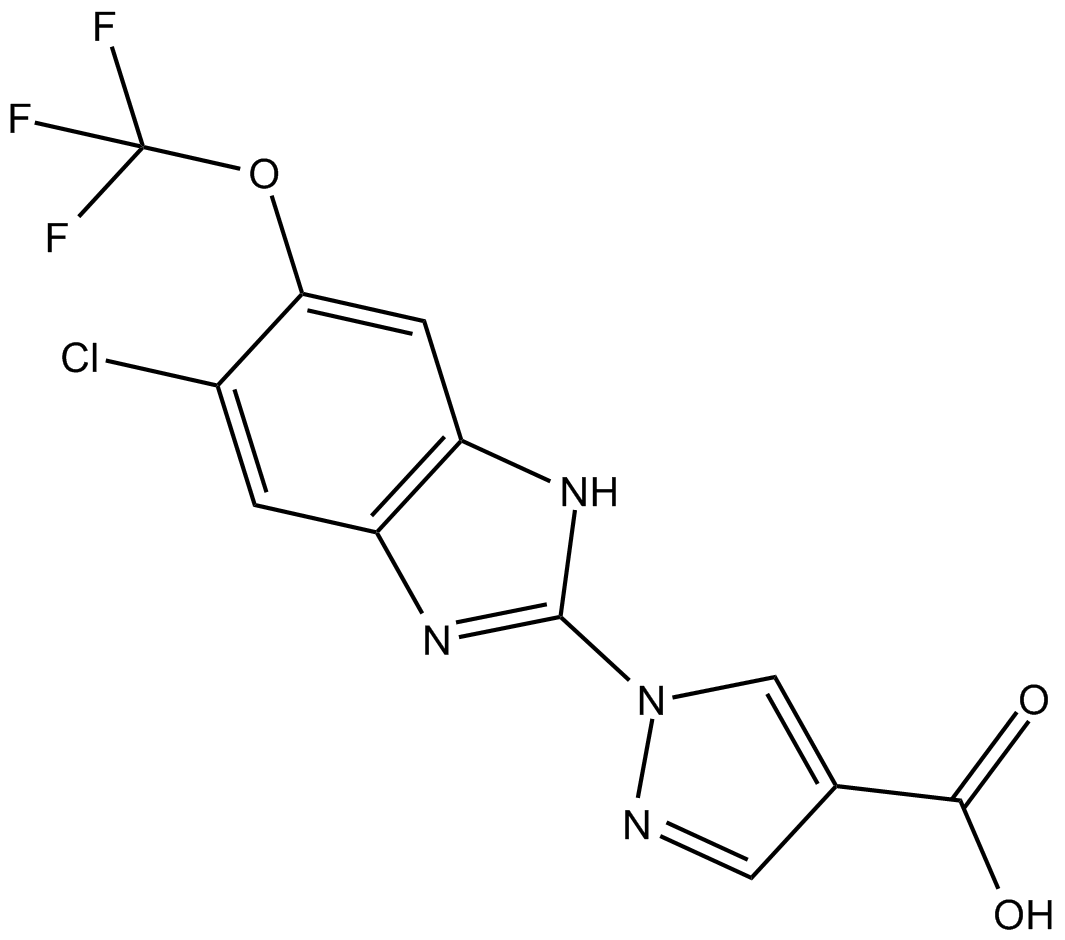
GC15379
JNJ-42041935
Hypoxia-inducible factor (HIF) prolyl hydroxylase (PHD) inhibitor
-
GC19465
JNJ-64619178
JNJ-64619178 is a PRMT5 inhibitor

-
GC12612
JNJ-7706621
A dual inhibitor of CDKs and Aurora kinases
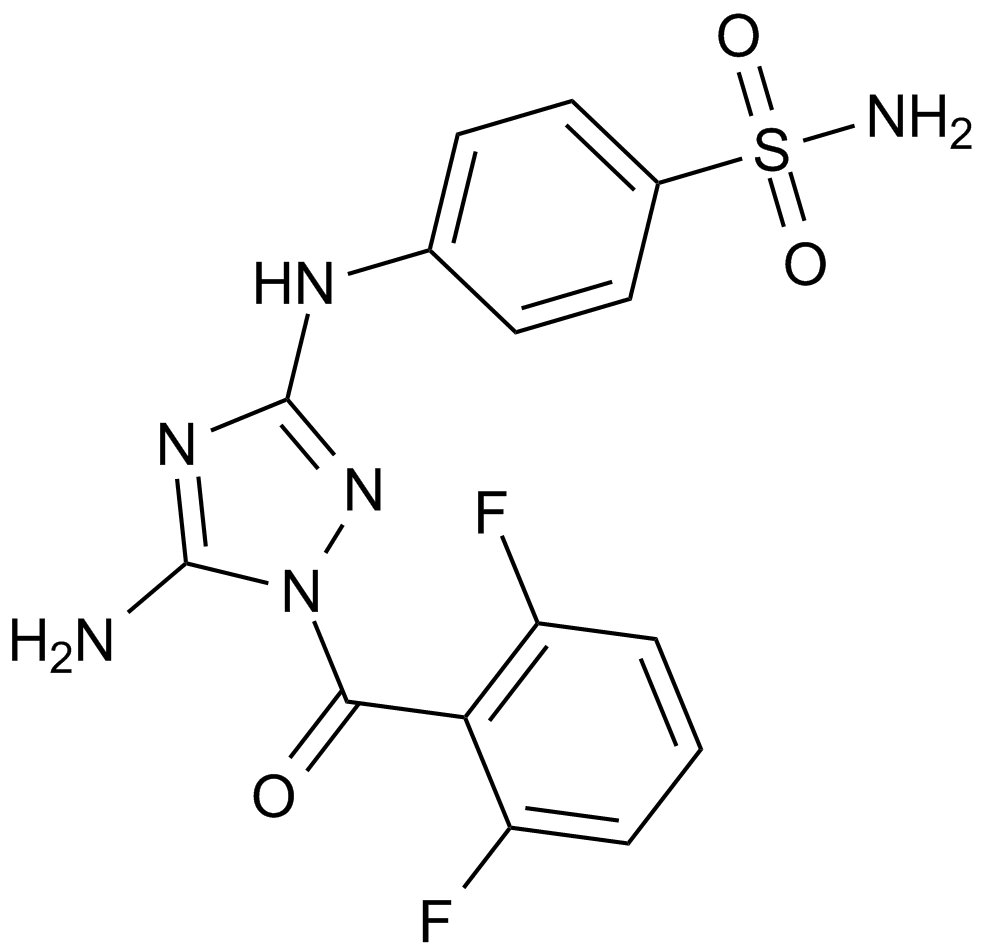
-
GC19210
JQ-1 carboxylic acid
JQ-1 carboxylic acid is a highly potent, selective and cell-permeable BRD4 inhibitor with IC50s of 77 nM and 33 nM for BRD4(1) and BRD4(2), respectively.
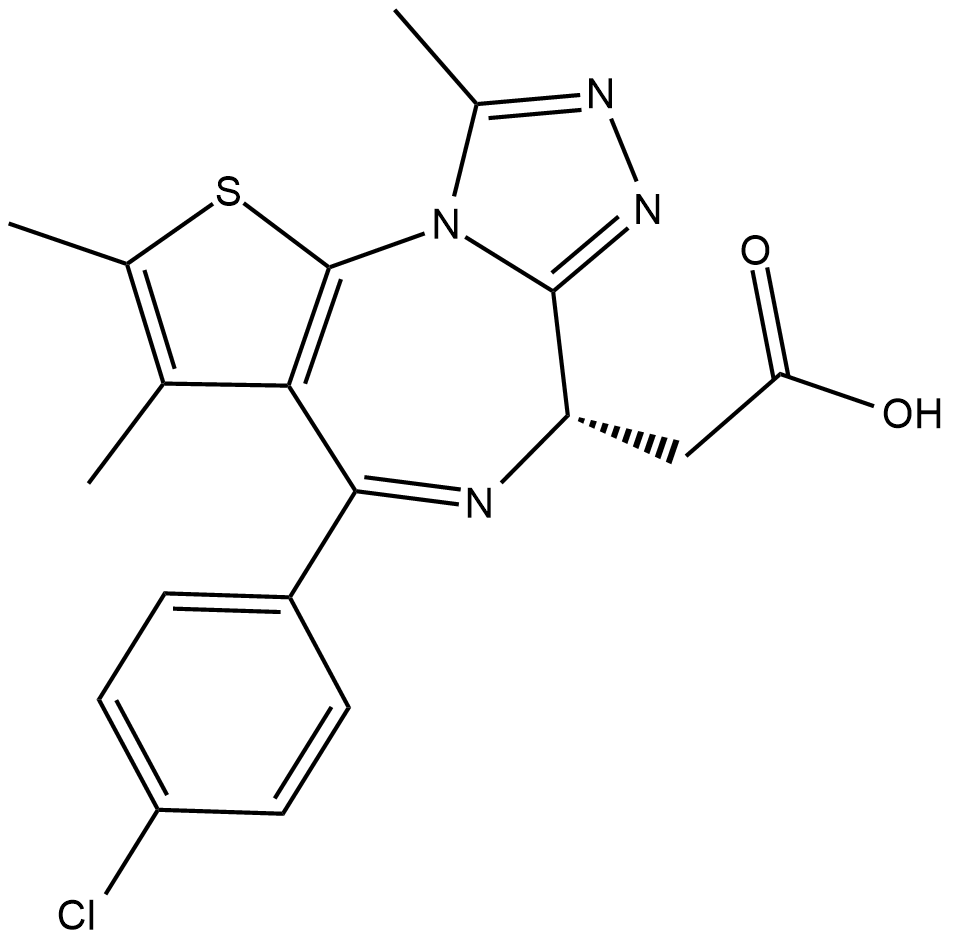
-
GC19211
JQEZ5
JQEZ5 is a novel and potent EZH2 inhibitor.
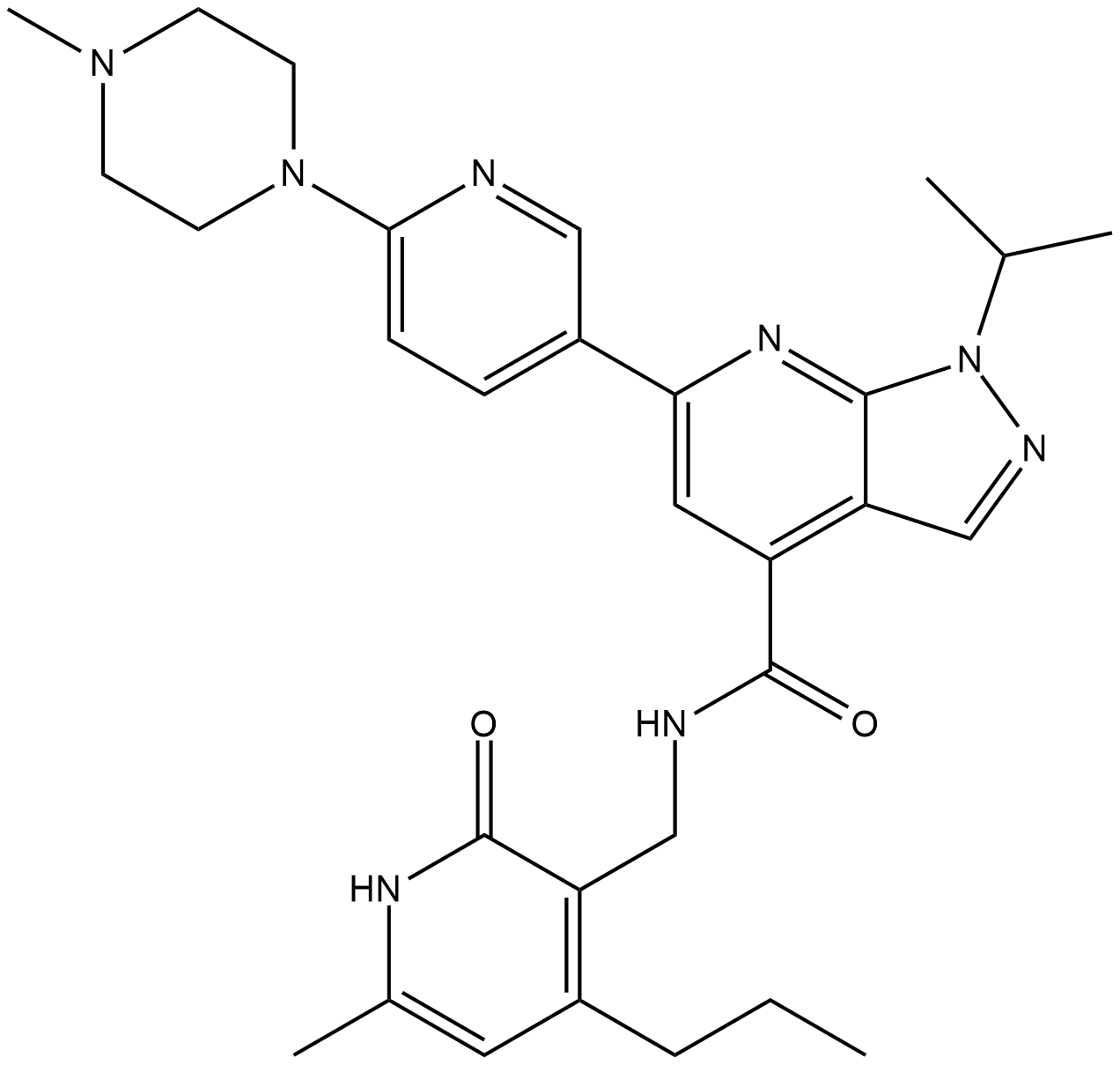
-
GC64841
JQKD82 trihydrochloride
JQKD82 (JADA82) trihydrochloride is a cell-permeable and selective KDM5 inhibitor. JQKD82 trihydrochloride increases H3K4me3 and can be used for the research of multiple myeloma.
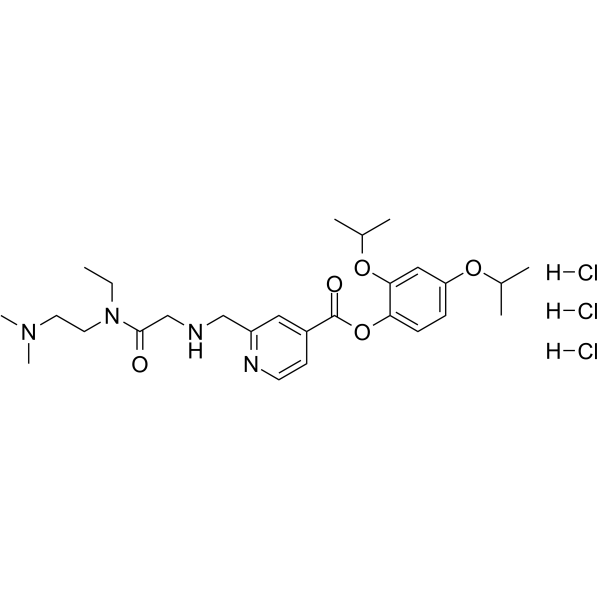
-
GC34195
K-756
K-756 is a direct and selective tankyrase (TNKS) inhibitor, which inhibits the ADP-ribosylation activity of TNKS1 and TNKS2 with IC50s of 31 and 36 nM, respectively.
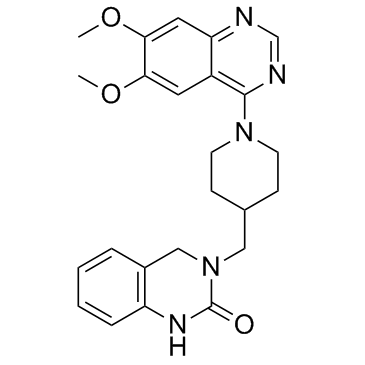
-
GC45489
K-Biotin-W-Histone H2B (108-125) (trifluoroacetate salt)

-
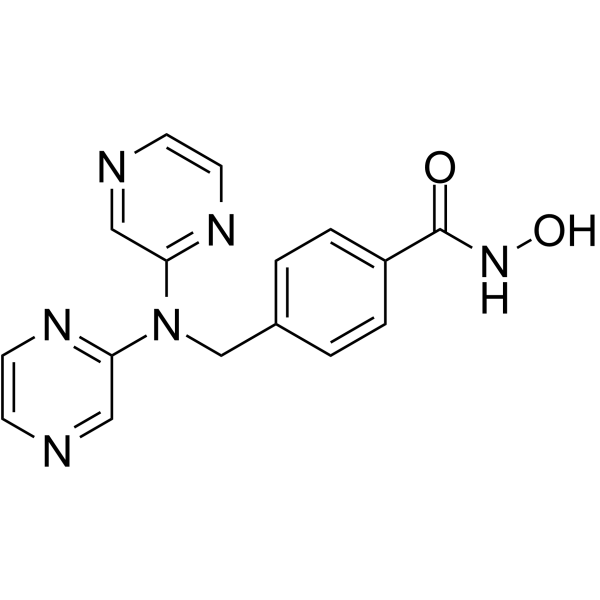
GC62430
KA2507
KA2507 is a potent, orally active and selective HDAC6 inhibitor, with an IC50 of 2.5 nM. KA2507 shows antitumor activities and immune modulatory effects in preclinical models.
-
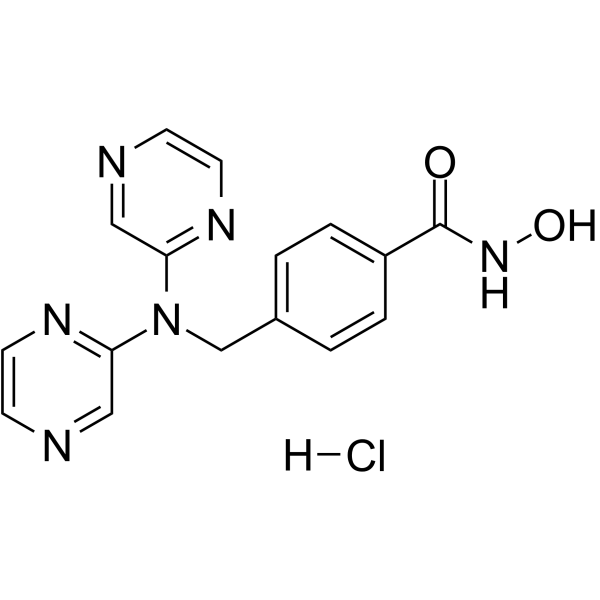
GC62374
KA2507 monohydrochloride
KA2507 hydrochloride is a potent and highly selective inhibitor of HDAC6 (IC50=2.5 nM) with no significant toxicities.
-
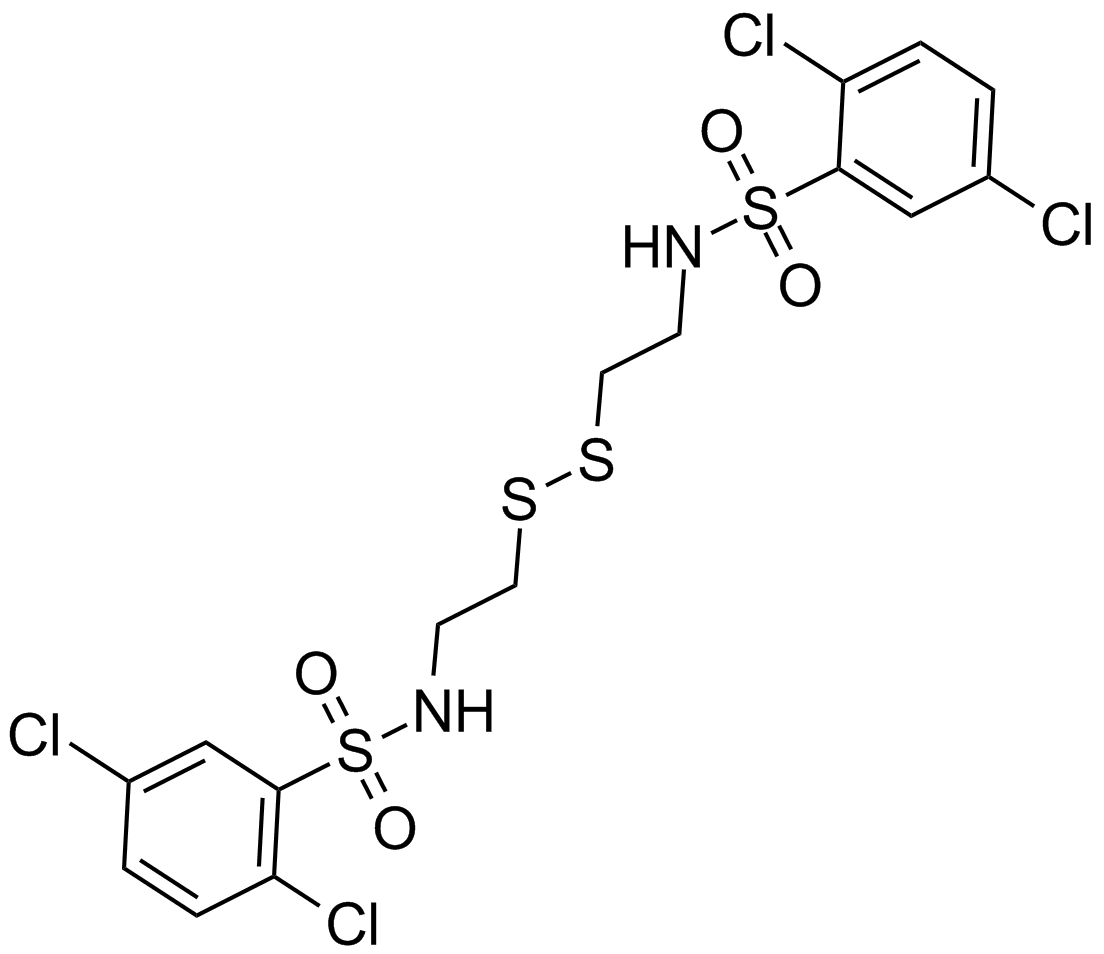
GC13141
KC7F2
An inhibitor of HIF-1a protein synthesis
-
GC13435
KD 5170
An inhibitor of class I and II HDACs
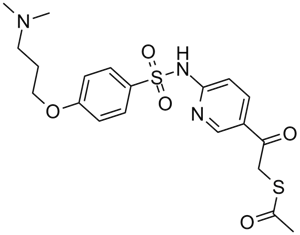
-
GC36388
KDM2A/7A-IN-1
KDM2A/7A-IN-1 is a first-in-class, selective and cell-permeable inhibitor of histone lysine demethylases KDM2A/7A, with an IC50 of 0.16?μM for KDM2A, exhibits 75 fold selevtivity over other JmjC lysine demethylases, and is inactive on methyl transferases, and histone acetyl transferases.
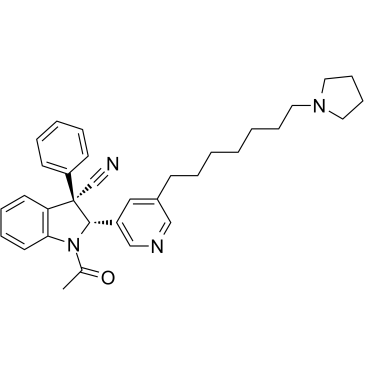
-
GC69327
KDM2B-IN-4
KDM2B-IN-4 is a histone demethylase KDM2B inhibitor. It can be used for cancer research. For more information, please refer to compound 182b in patent document WO2016112284A1.
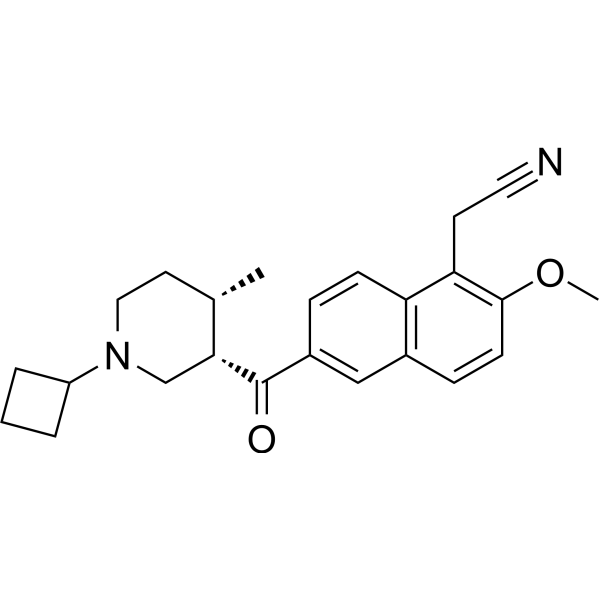
-
GC36389
KDM4-IN-2
KDM4-IN-2 (Compound 19a) is a potent and selective KDM4/KDM5 dual inhibitor with Kis of 4 and 7?nM for KDM4A and KDM5B, respectively.
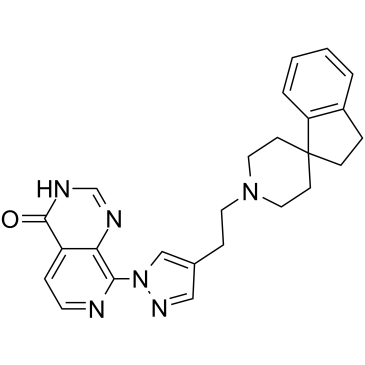
-
GC31887
KDM4D-IN-1
KDM4D-IN-1 is a new histone lysine demethylase 4D (KDM4D) inhibitor with an IC50 value of 0.41±0.03 μM.
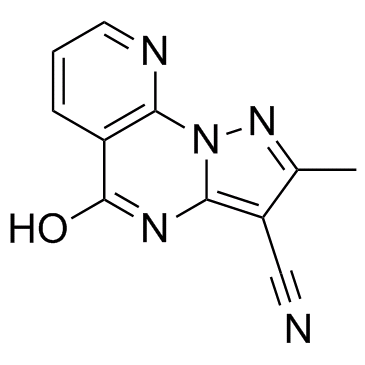
-
GC32790
KDM5-IN-1
KDM5-IN-1 is a potent, selective and orally bioavailable KDM5 inhibitor with an IC50 of 15.1 nM.
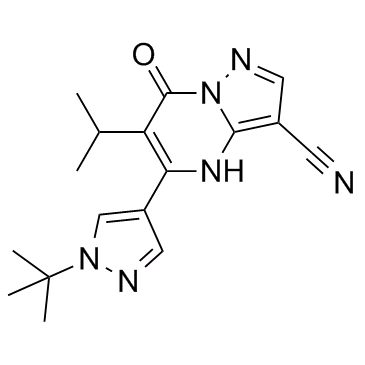
-
GC32842
KDM5A-IN-1
KDM5A-IN-1 is a potent, orally bioavailable pan-histone lysine demethylases 5 (KDM5) inhibitor with IC50s of 45 nM, 56 nM and 55 nM for KDM5A, KDM5B and KDM5C, respectively, and with an EC50 value of 960 nM for PC9 H3K4Me3. KDM5A-IN-1 is significantly less potent against other KDM5B enzymes (1A, 2B, 3B, 4C, 6A, 7B).
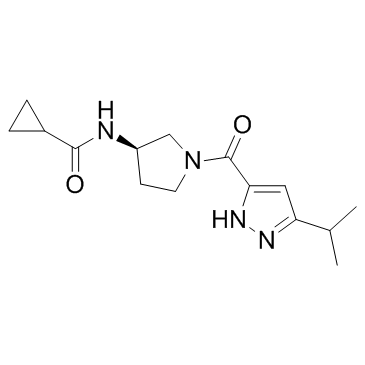
-
GC38709
KDOAM-25 citrate
KDOAM-25 citrate is a potent and highly selective histone lysine demethylases 5 (KDM5) inhibitor with IC50s of 71 nM, 19 nM, 69 nM, 69 nM for KDM5A, KDM5B, KDM5C, KDM5D, respectively. KDOAM-25 citrate increases global H3K4 methylation at transcriptional start sites and impairs proliferation in multiple myeloma MM1S cells.
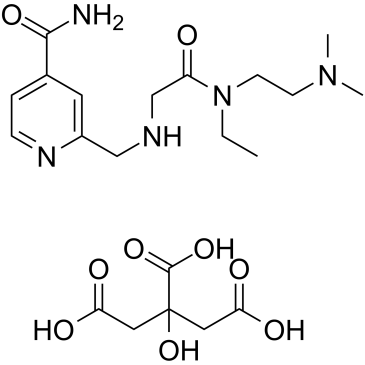
-
GC31654
KG-501 (Naphthol AS-E phosphate)
KG-501 (Naphthol AS-E phosphate) is a CREB inhibitor, with an IC50 of 6.89 μM.
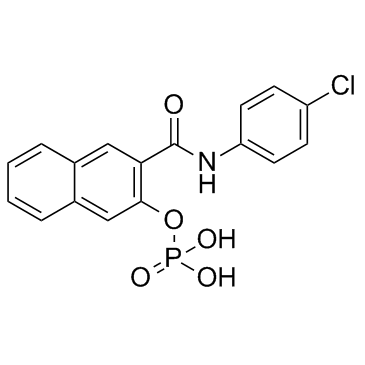
-
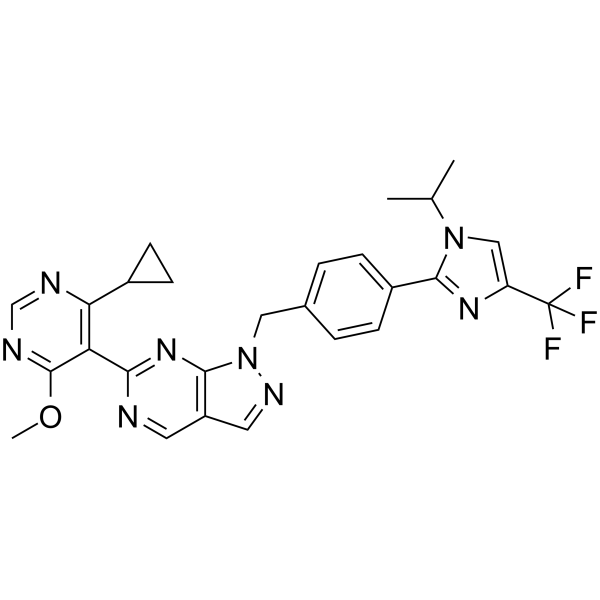
GC65907
KSQ-4279
KSQ-4279 (USP1-IN-1, Formula I) is a USP1 and PARP inhibitor (extracted from patent WO2021163530).
-
GC25552
KT-531
KT-531 (KT531) is a potent, selective HDAC6 inhibitor with IC50 of 8.5 nM, displays 39-fold selectivity over other HDAC isoforms.

-
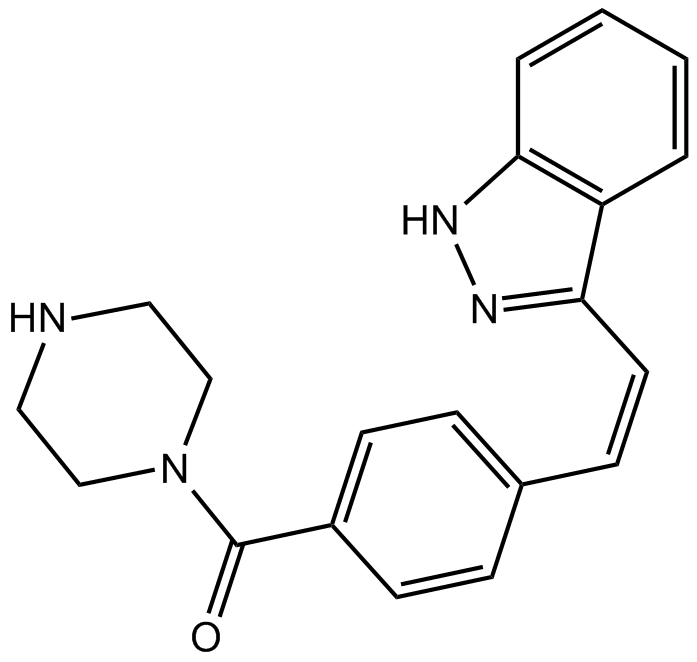
GC14592
KW 2449
A multi-kinase inhibitor
-
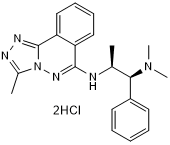
GC50395
L Moses dihydrochloride
High affinity and selective PCAF bromodomain inhibitor
-
GC33183
L-45 (L-Moses)
L-45 (L-Moses) (L-45) is the first potent, selective, and cell-active p300/CBP-associated factor (PCAF) bromodomain (Brd) inhibitor with a Kd of 126 nM.
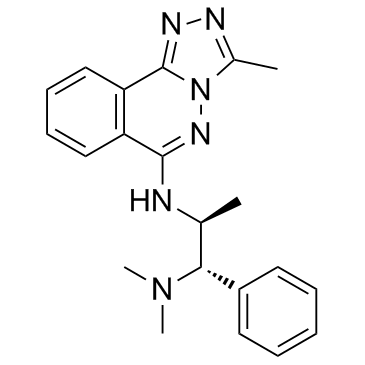
-
GC34377
L-45 dihydrochloride (L-Moses dihydrochloride)
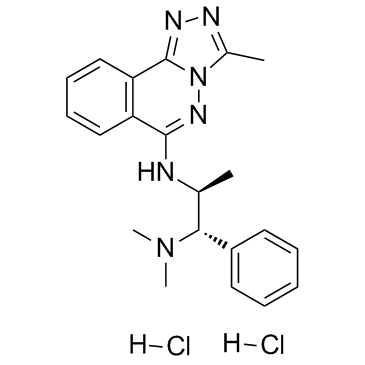
-
GC47579
L-Pyrohomoglutamic Acid
An amino acid building block
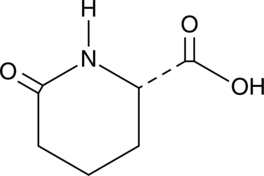
-
GC15731
L002
p300 inhibitor
-
GC65289
Lademirsen
Lademirsen (SAR339375; RG-012) is a single stranded, chemically modified oligonucleotide that binds to and inhibits the function of miR-21.

-
GC15114
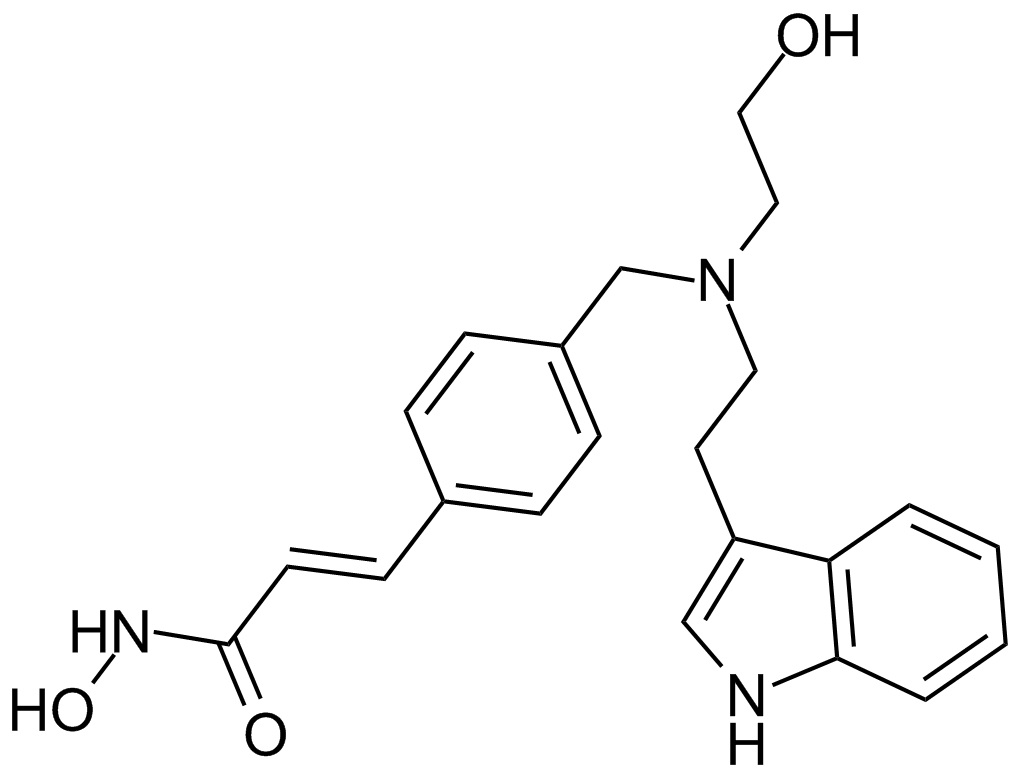
LAQ824 (NVP-LAQ824,Dacinostat)
A hydroxamate-based HDAC inhibitor
-
GC12189
LB-100
protein phosphatase 2A(PP2A)inhibitor
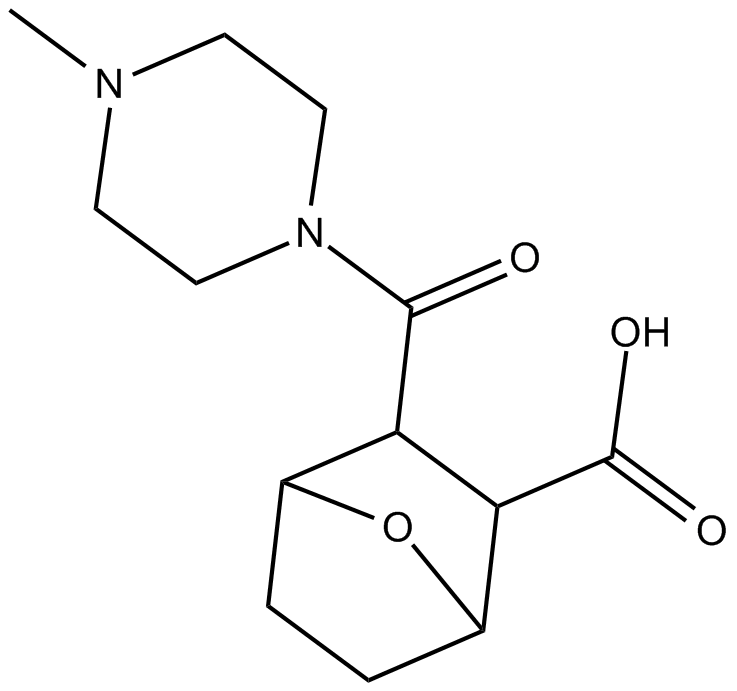
-
GC14857
LFM-A13
BTK-specific tyrosine kinase inhibitor
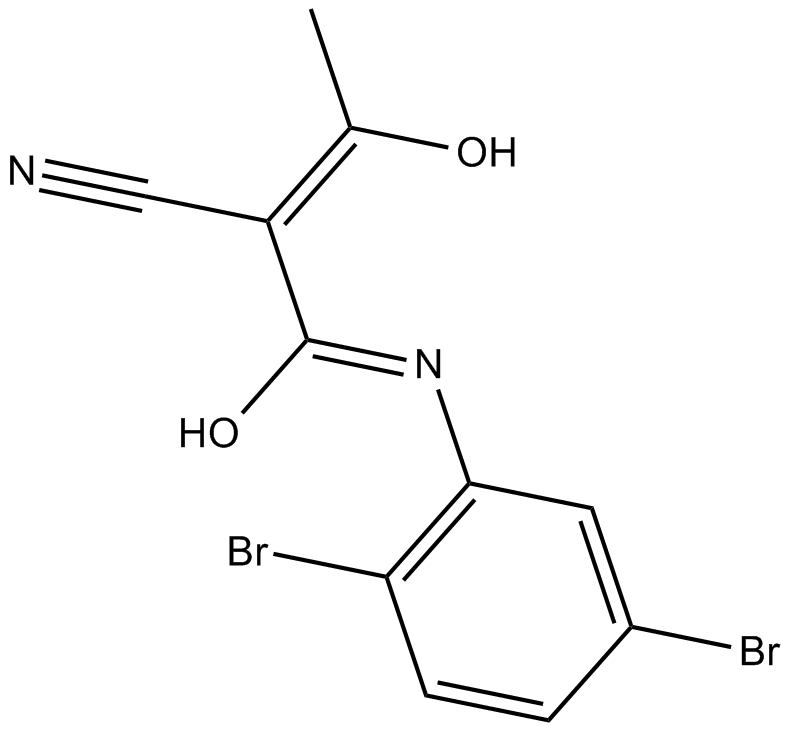
-
GC14362
Lin28 1632
Lin28 1632 (compound 1632) is a potent antagonist of Lin28/pre-let-7 interaction.
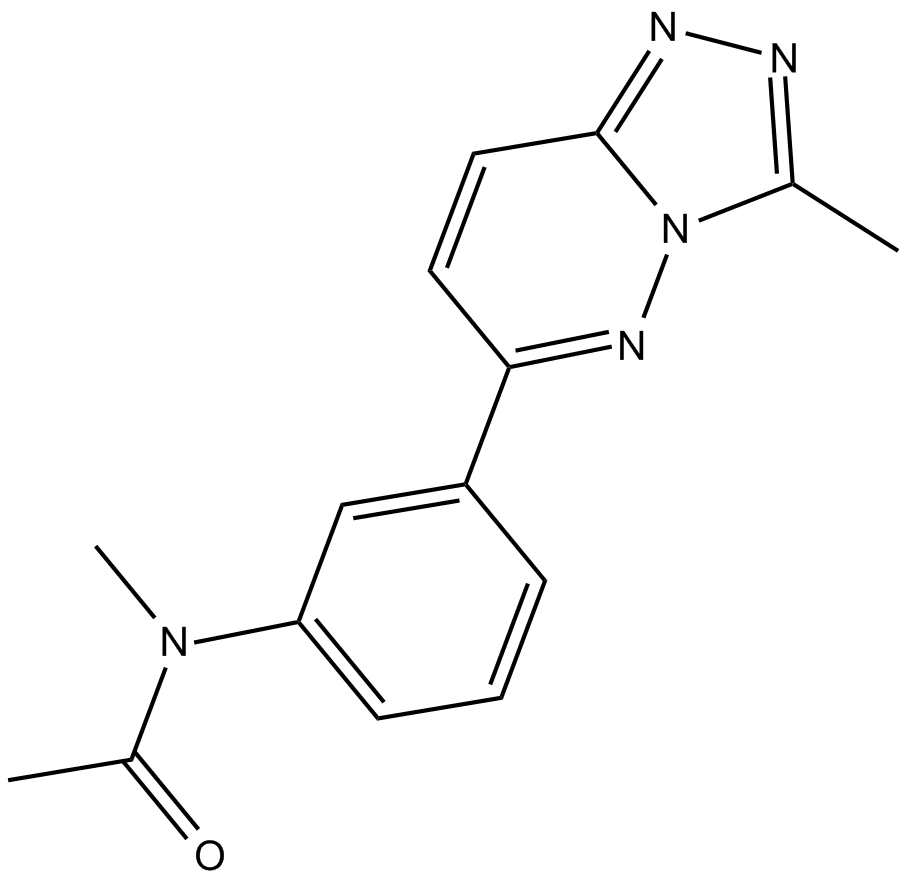
-
GC36464
LIN28 inhibitor LI71
LIN28 inhibitor LI71 is a potent and cell-permeable LIN28 inhibitor, which abolishes LIN28-mediated oligouridylation with an IC50 of 7 uM. LIN28 inhibitor LI71 directly binds the cold shock domain (CSD) to suppress LIN28’s activity against let-7 in leukemia cells and embryonic stem cells.
-
GC30501
Lin28-let-7a antagonist 1
Lin28-let-7a antagonist 1 shows a clear antagonistic effect against the Lin28-let-7a interaction with an IC50 of 4.03 μM for Lin28A-let-7a-1 interaction.
-
GC13154
LKB1(AAK1 dual inhibitor)
-
GC33184
LLY-283
A PRMT5 inhibitor
-
GC25580
LLY-284
LLY-284 is the diastereomer of LLY-283, which is a potent and selective SAM-competitive chemical probe for PRMT5. LLY-284 is much less active than LLY-283 and can be used as a negative control for LLY-283.