Proteases
Proteases is a general term for a class of enzymes that hydrolyze protein peptide chains. According to the way they degrade polypeptides, they are divided into two categories: endopeptidases and telopeptidases. The former can cut the large molecular weight polypeptide chain from the middle to form prions and peptones with smaller molecular weights; the latter can be divided into carboxypeptidase and aminopeptidase, which respectively remove the peptide from the free carboxyl terminus or free amino terminus of the polypeptide one by one. Chain hydrolysis produces amino acids.
A general term for a class of enzymes that hydrolyze peptide bonds in proteins. According to the way they hydrolyze polypeptides, they can be divided into endopeptidases and exopeptidases. Endopeptidase cleaves the interior of the protein molecule to form smaller molecular weight peptones and peptones. Exopeptidase hydrolyzes peptide bonds one by one from the end of the free amino group or carboxyl group of protein molecules, and frees amino acids, the former is aminopeptidase and the latter is carboxypeptidase. Proteases can be classified into serine proteases, sulfhydryl proteases, metalloproteases and aspartic proteases according to their active centers and optimum pH. According to the optimum pH value of its reaction, it is divided into acidic protease, neutral protease and alkaline protease. The proteases used in industrial production are mainly endopeptidases.
Proteases are widely found in animal offal, plant stems and leaves, fruits and microorganisms. Microbial proteases are mainly produced by molds and bacteria, followed by yeast and actinomycetes.
Enzymes that catalyze the hydrolysis of proteins. There are many kinds, the important ones are pepsin, trypsin, cathepsin, papain and subtilisin. Proteases have strict selectivity for the reaction substrates they act on. A protease can only act on certain peptide bonds in protein molecules, such as the peptide bonds formed by the hydrolysis of basic amino acids catalyzed by trypsin. Proteases are widely distributed, mainly in the digestive tract of humans and animals, and are abundant in plants and microorganisms. Due to limited animal and plant resources, the industrial production of protease preparations is mainly prepared by fermentation of microorganisms such as Bacillus subtilis and Aspergillus terrestris.
Targets for Proteases
- Caspase(114)
- Aminopeptidase(24)
- ACE(74)
- Calpains(20)
- Carboxypeptidase(10)
- Cathepsin(81)
- DPP-4(31)
- Elastase(26)
- Gamma Secretase(67)
- HCV Protease(59)
- HSP(113)
- HIV Integrase(37)
- HIV Protease(47)
- MMP(228)
- NS3/4a protease(8)
- Serine Protease(18)
- Thrombin(58)
- Urokinase(4)
- Cysteine Protease(0)
- Other Proteases(18)
- Tyrosinases(47)
- 15-PGDH(1)
- Acetyl-CoA Carboxylase(13)
- Acyltransferase(59)
- Aldehyde Dehydrogenase (ALDH)(28)
- Aminoacyl-tRNA Synthetase(9)
- ATGL(1)
- Dipeptidyl Peptidase(56)
- Drug Metabolite(457)
- E1/E2/E3 Enzyme(90)
- Endogenous Metabolite(1636)
- FABP(30)
- Farnesyl Transferase(23)
- Glutaminase(14)
- Glutathione Peroxidase(14)
- Isocitrate Dehydrogenase (IDH)(28)
- Lactate Dehydrogenase(17)
- Lipoxygenase(234)
- Mitochondrial Metabolism(207)
- NEDD8-activating Enzyme(7)
- Neprilysin(12)
- PAI-1(13)
- Ser/Thr Protease(41)
- Tryptophan Hydroxylase(13)
- Xanthine Oxidase(18)
- MALT1(10)
- PCSK9(1)
Products for Proteases
- Cat.No. Product Name Information
-
GC31602
Adrenic Acid (cis-7,10,13,16-Docosatetraenoic acid)
Adrenic Acid (cis-7,10,13,16-Docosatetraenoic acid) (cis-7,10,13,16-Docosatetraenoic acid) is a naturally polyunsaturated fatty acid in the adrenal gland, brain, kidney, and vasculature.
-
GC42742
ADT-OH
ADT-OH is a derivative of anethole dithiolethione (ADT) and synthetic hydrogen sulfide (H2S) donor.
-
GC32016
AE-3763
AE-3763 is a peptide-based human neutrophil elastase inhibitor with an IC50 of 29 nM.
-
GC14502
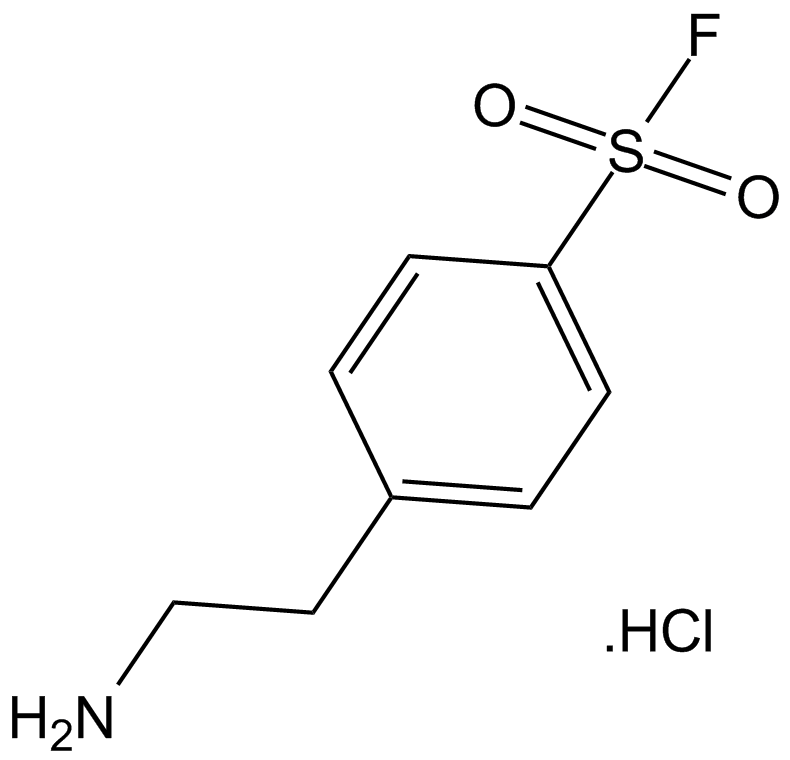
AEBSF.HCl
AEBSF.HCl is a broad-spectrum irreversible inhibitor of serine proteases, which can inhibit chymotrypsin, kallikrein, plasmin, thrombin, trypsin and related thrombolytic enzymes.
-
GC35261
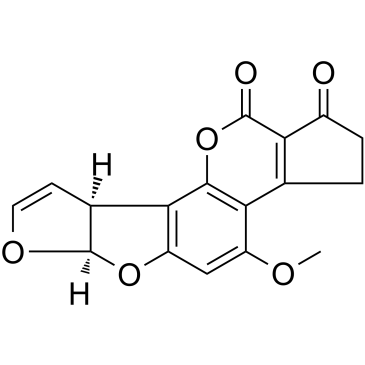
Aflatoxin B1
Aflatoxin B1, as a class of carcinogenic mycotoxins produced by Aspergillus fungi, always lead to the development of hepatocellular carcinoma (HCC) in humans and animals.
-
GC46812
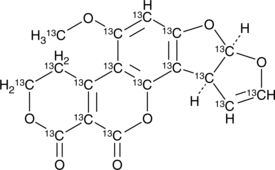
Aflatoxin G1-13C17
An internal standard for the quantification of aflatoxin G1
-
GC35262
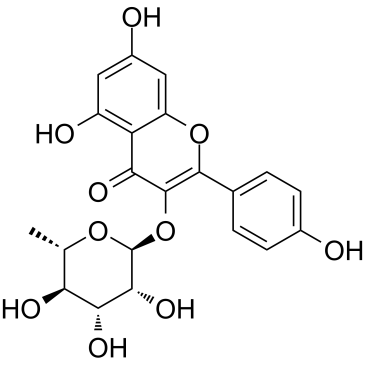
Afzelin
A polyphenolic glycoside flavone with diverse biological activities
-
GC15601
AG-120
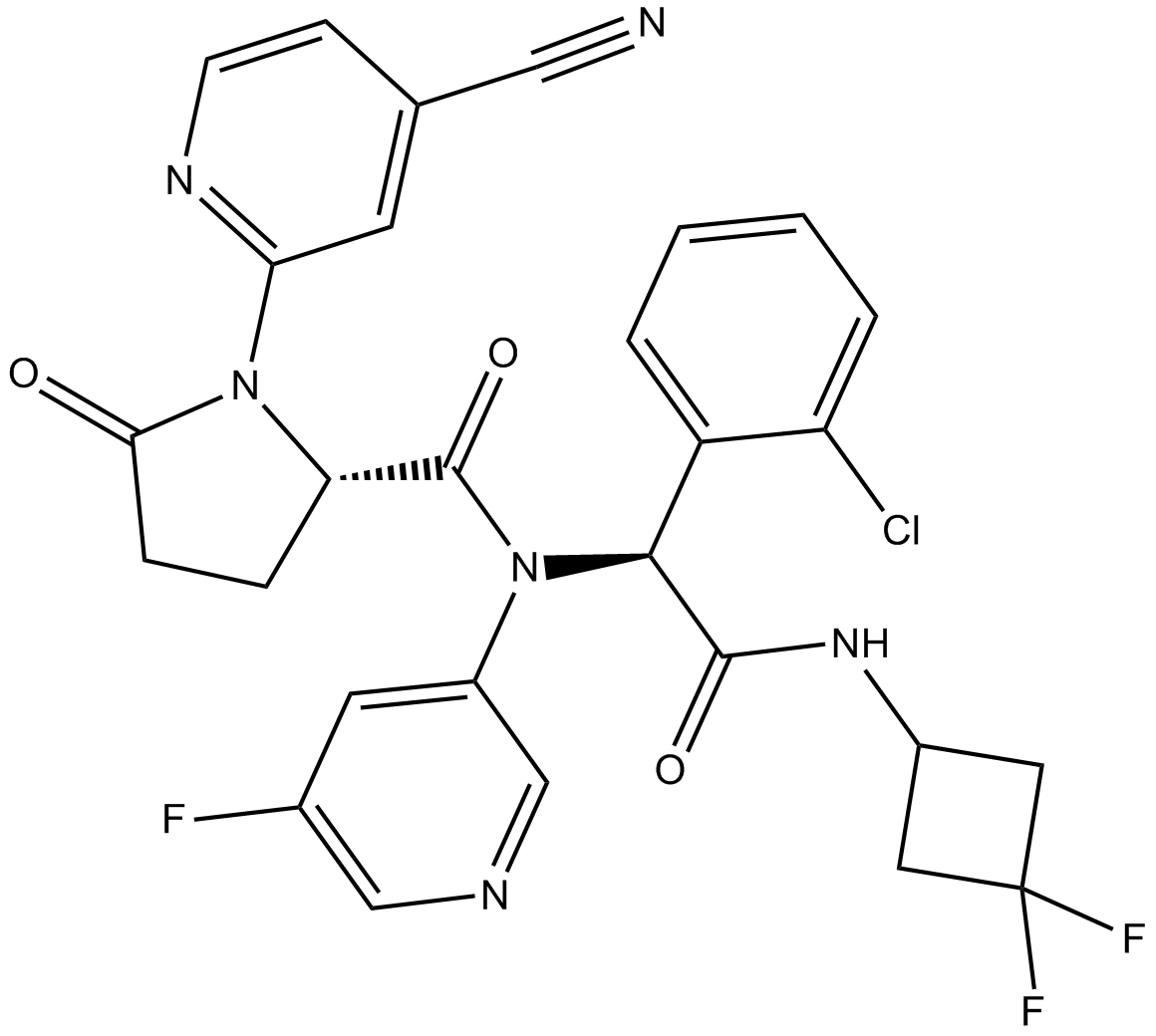
AG-120 (AG-120) is an orally active inhibitor of isocitrate dehydrogenase 1 mutant (mIDH1) enzyme, it exhibits profound d-2-hydroxyglutatrate (2-HG) lowering in vivo.
-
GC13147
AG-221 (Enasidenib)
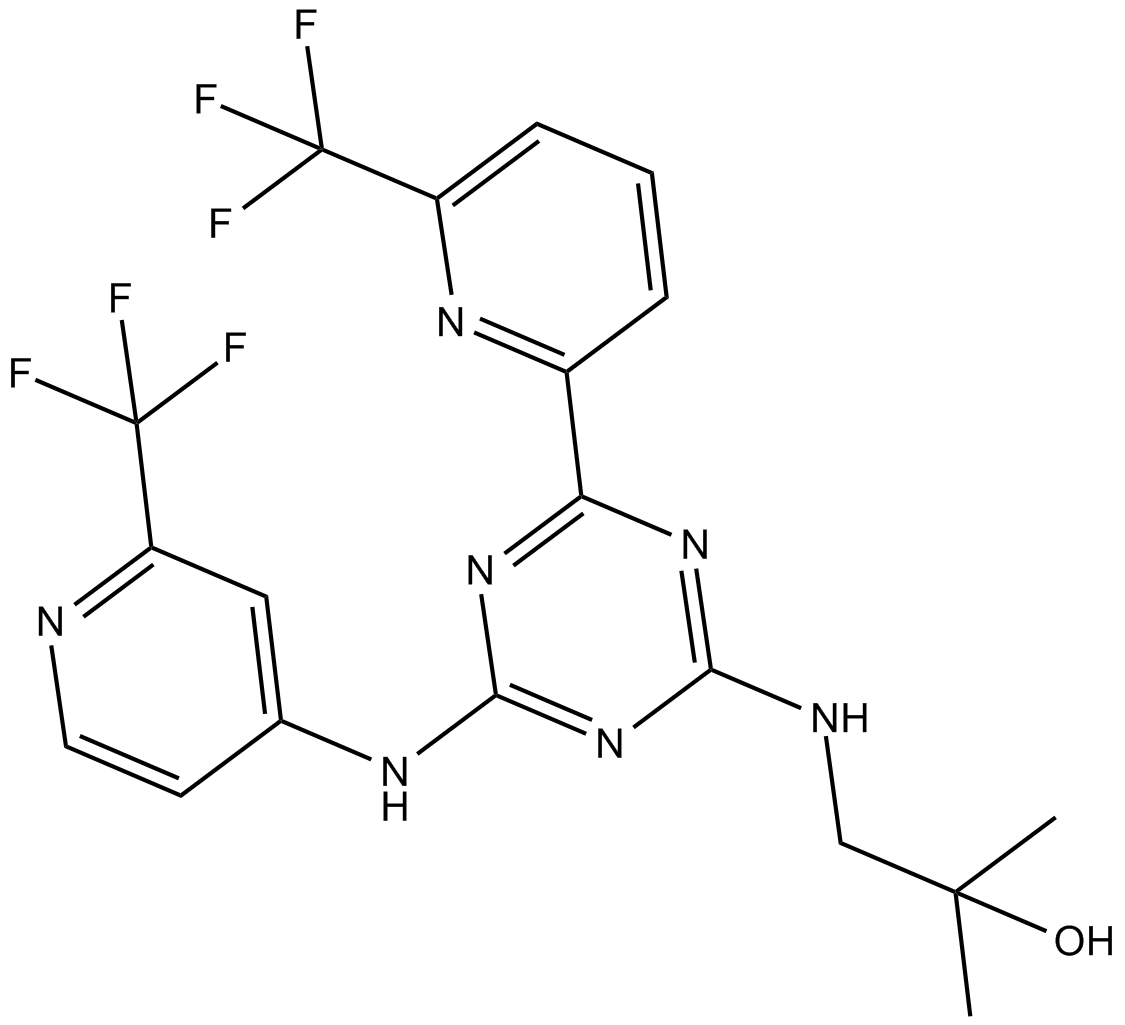
AG-221 (Enasidenib) is an oral, potent, reversible, selective inhibitor of the IDH2 mutant enzymes, with IC50s of 100 and 400 nM against IDH2R140Q and IDH2R172K, respectively.
-
GC63642
Agaric acid
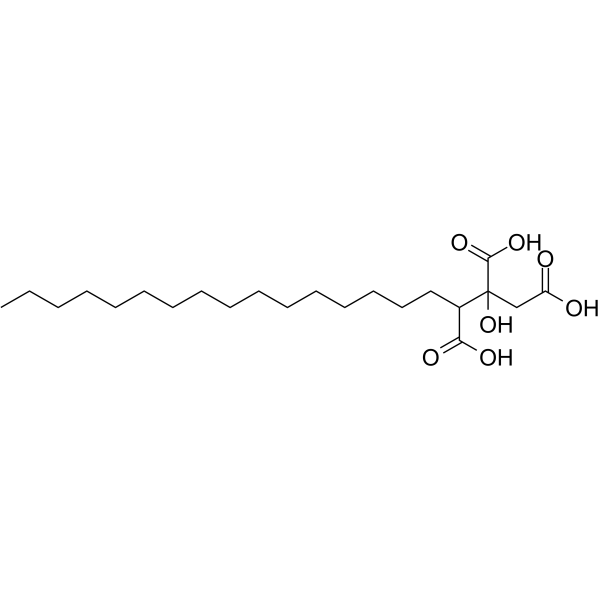
Agaric acid (Agaricinic Acid) is obtained from various plants of the fungous tribe, i.
-
GC10531
AGI-5198
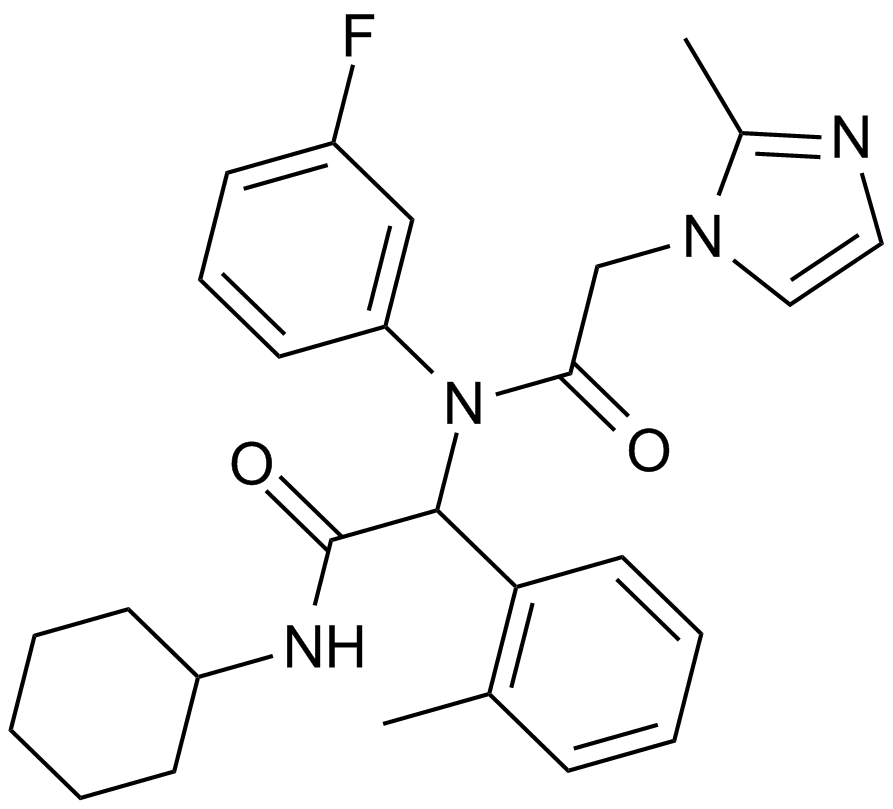
A potent, selective inhibitor of IDH1 mutations
-
GC14757
AGI-6780
A potent, selective inhibitor of mutant IDH2
-
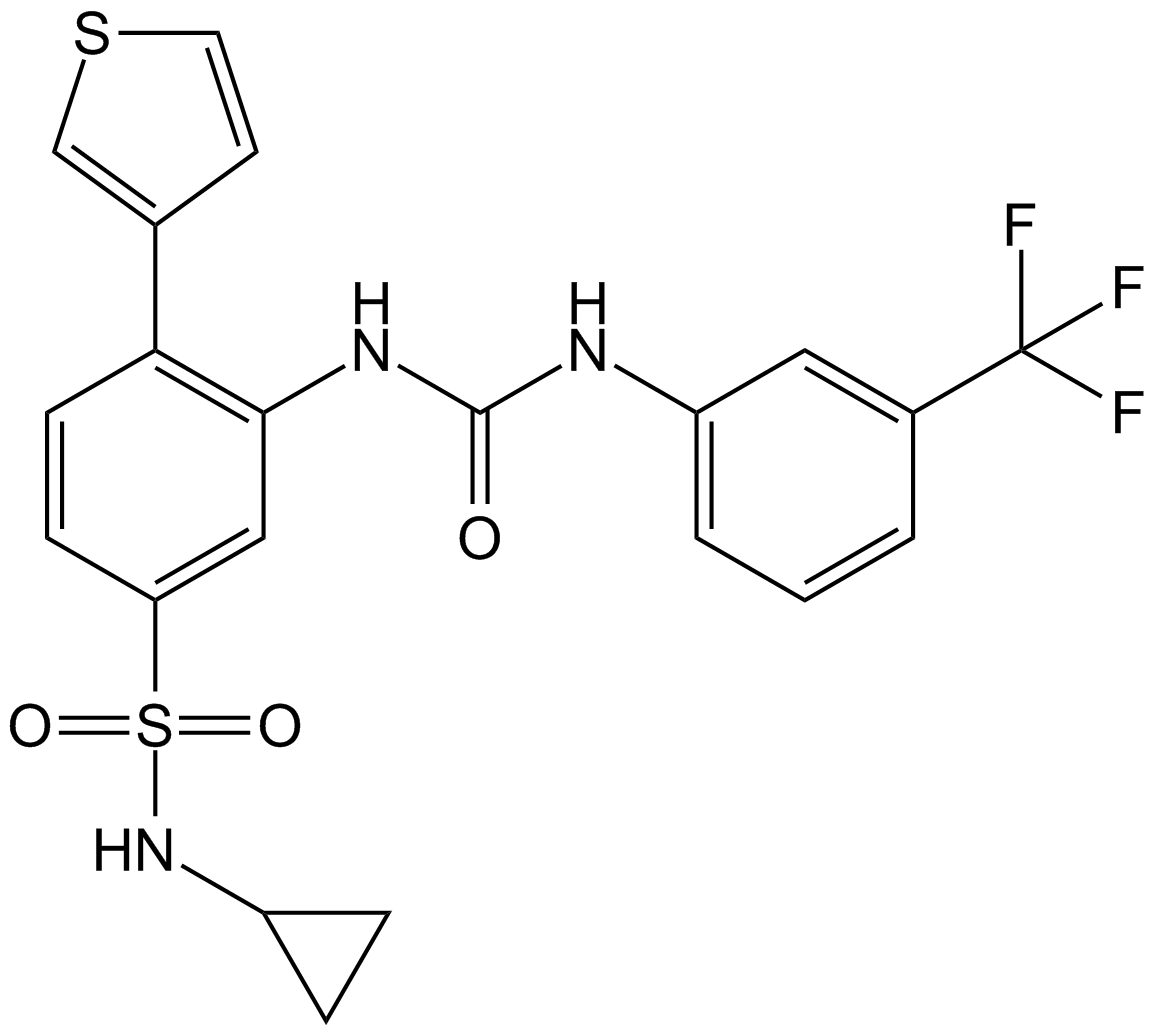
GC16831
Agmatine sulfate
α2-adrenergic receptor ligand
-
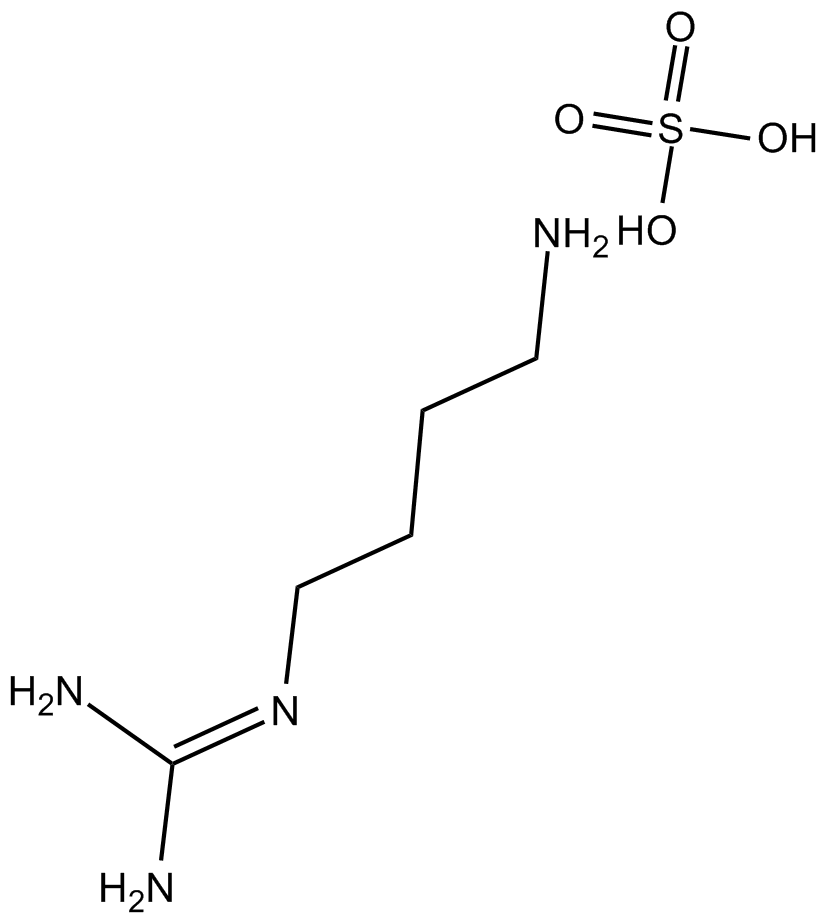
GC16138
AHU-377 hemicalcium salt
AHU-377 hemicalcium salt (AHU-377 hemicalcium salt) is a potent NEP inhibitor with an IC50 of 5 nM.
-
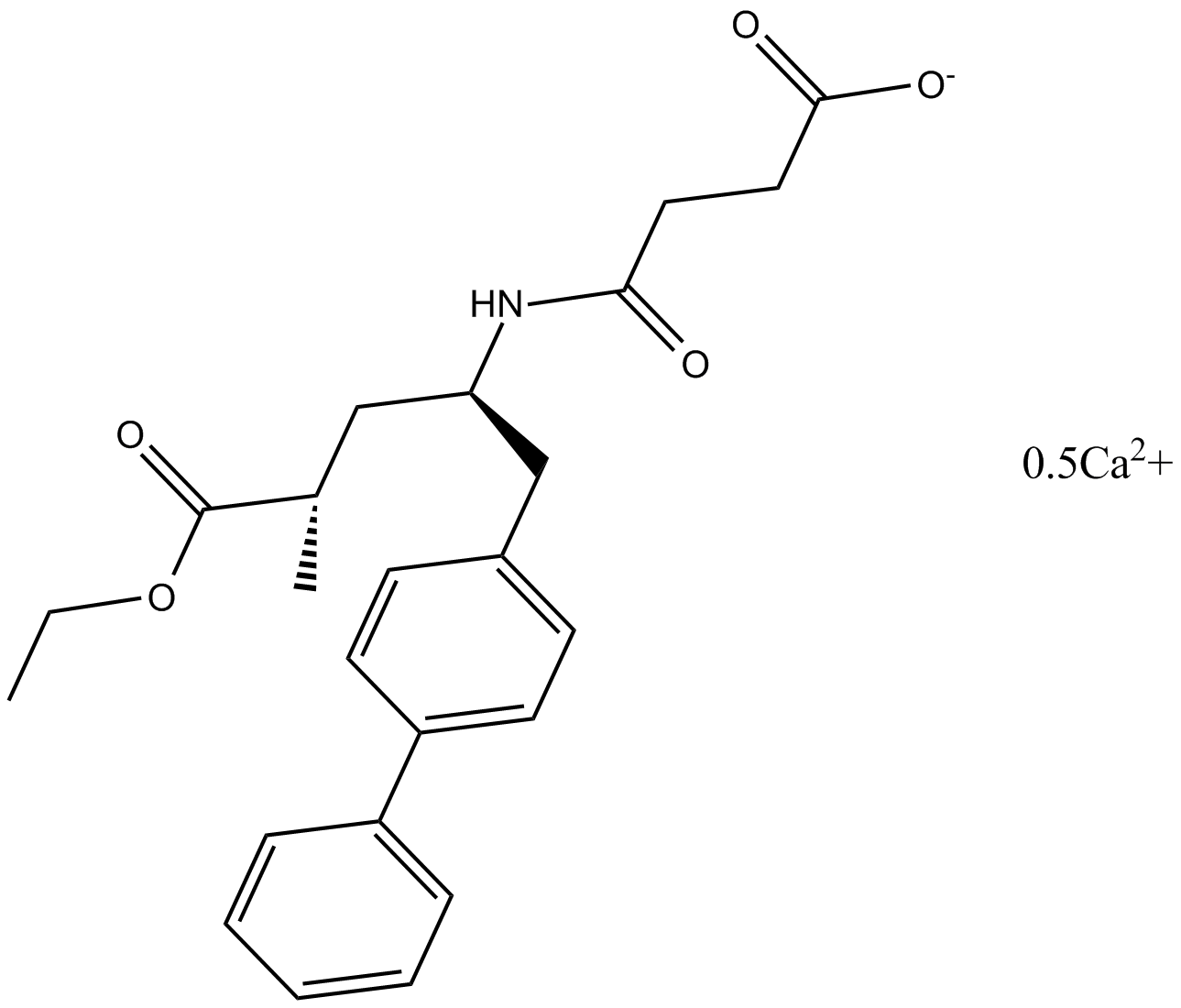
GC10146
AHU-377(Sacubitril)
AHU-377(Sacubitril) (AHU-377) is a potent and orally active NEP (neprilysin) inhibitor with an IC50 of 5 nM.
-
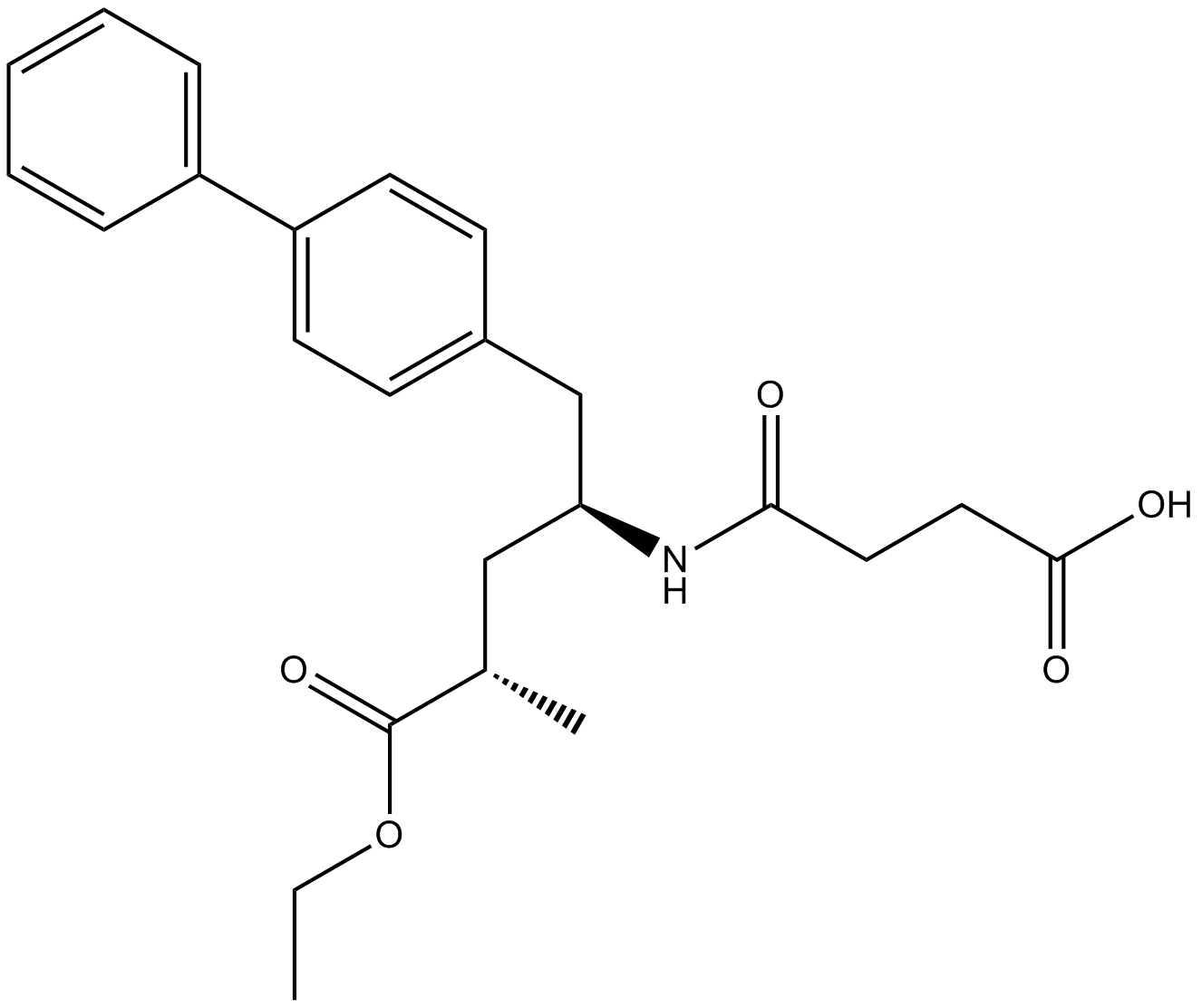
GC46820
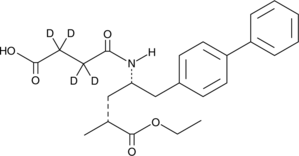
AHU377-d4
AHU377-d4 (AHU-377-d4) is the deuterium labeled Sacubitril.
-
GC49773
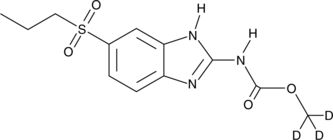
Albendazole sulfone-d3
An internal standard for the quantification of albendazole sulfone
-
GC16597
Alda 1
ALDH2 activator
-
GC64063
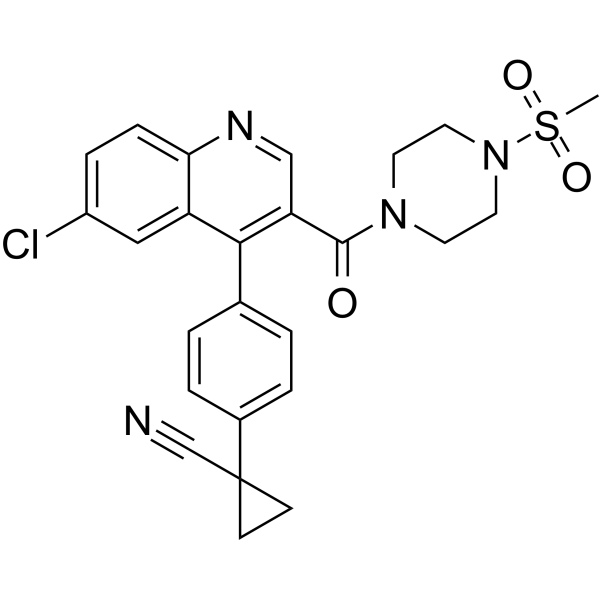
ALDH1A1-IN-2
ALDH1A1-IN-2 is a potent inhibitor of aldehyde dehydrogenase 1a1 (aldh1a1). Aldehyde dehydrogenases (ALDH) constitute a family of enzymes that play a critical role in oxidizing various cytotoxic xenogenic and biogenic aldehydes. ALDH1A1-IN-2 has the potential for the research of cancer, inflammation, or obesity (extracted from patent WO2019089626A1, compound 295).
-
GC67782
ALDH1A2-IN-1
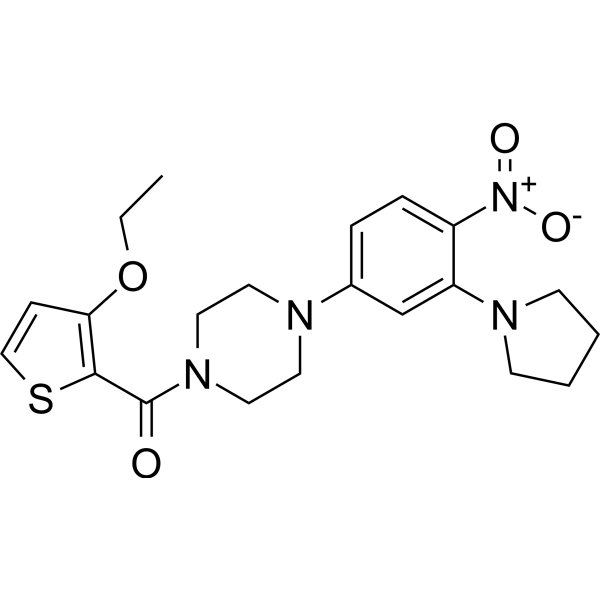
-
GC68464
ALDH1A3-IN-1

-
GC68462
ALDH1A3-IN-2
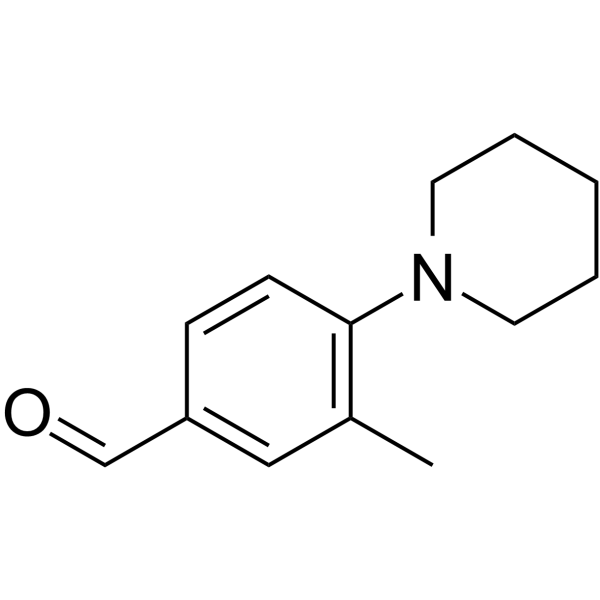
-
GC64618
ALDH1A3-IN-3
ALDH1A3-IN-3 (compound 16) is a potent inhibitor of ALDH1A3, with an IC50 of 0.26 μM. ALDH1A3-IN-3 is also a good ALDH3A1 substrate. ALDH1A3-IN-3 can be used for the research of prostate cancer.
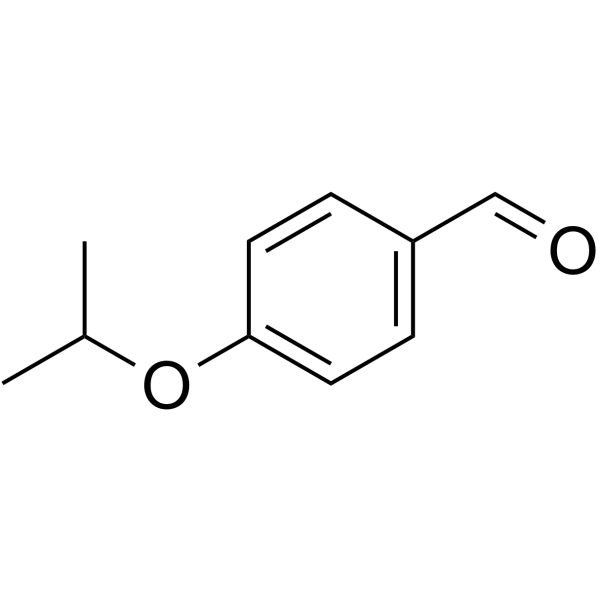
-
GC41390
Aldosterone
Aldosterone is a steroid hormone secreted by the adrenal cortex and is the principle mineralocorticoid controlling sodium and potassium balance
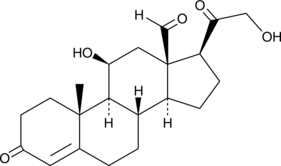
-
GC65281
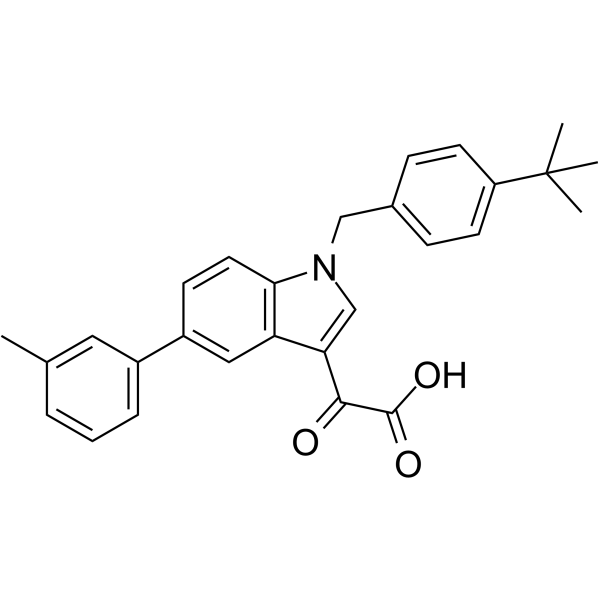
Aleplasinin
Aleplasinin is an orally active, potent, BBB-penetrated and selectiveSERPINE1 (PAI-1, Plasminogen activator inhibitor-1) inhibitor.
-
GC65044
Alirocumab (anti-PCSK9)
Alirocumab (anti-PCSK9) is a human monoclonal antibody inhibiting proprotein convertase subtilisin/kexin type 9 (PCSK9).
-
GC19584
Alkaline Phosphatase
10-50units/mg protein(37℃,pH 9.8)

-
GC14511
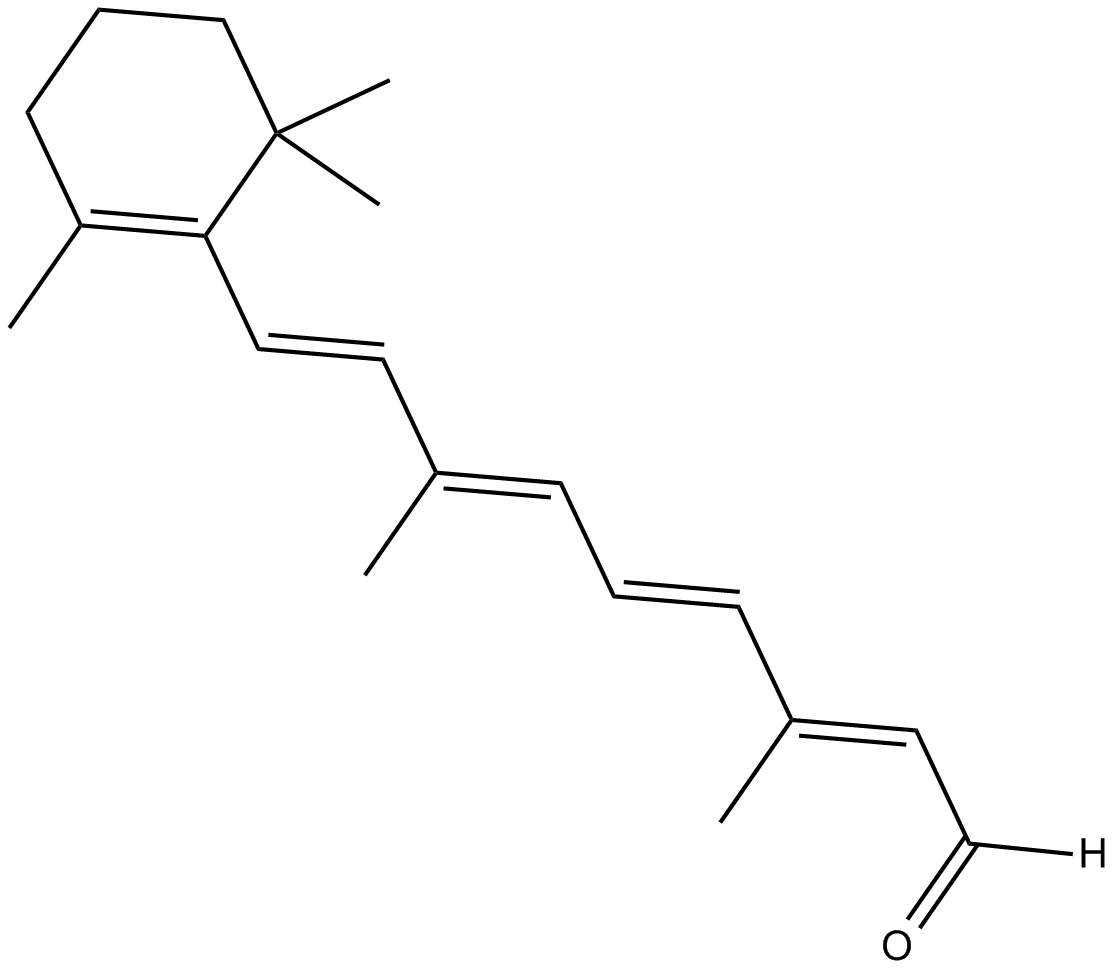
all-trans Retinal
converted to retinoic acid in vivo
-
GC42769
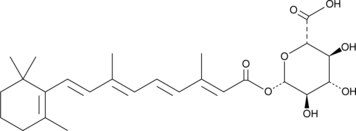
all-trans Retinoyl β-D-Glucuronide
all-trans Retinoyl β-D-glucuronide is a metabolite of all-trans retinoic acid formed by the UDP-glucuronosyltransferase (UGT) system.
-
GC18539
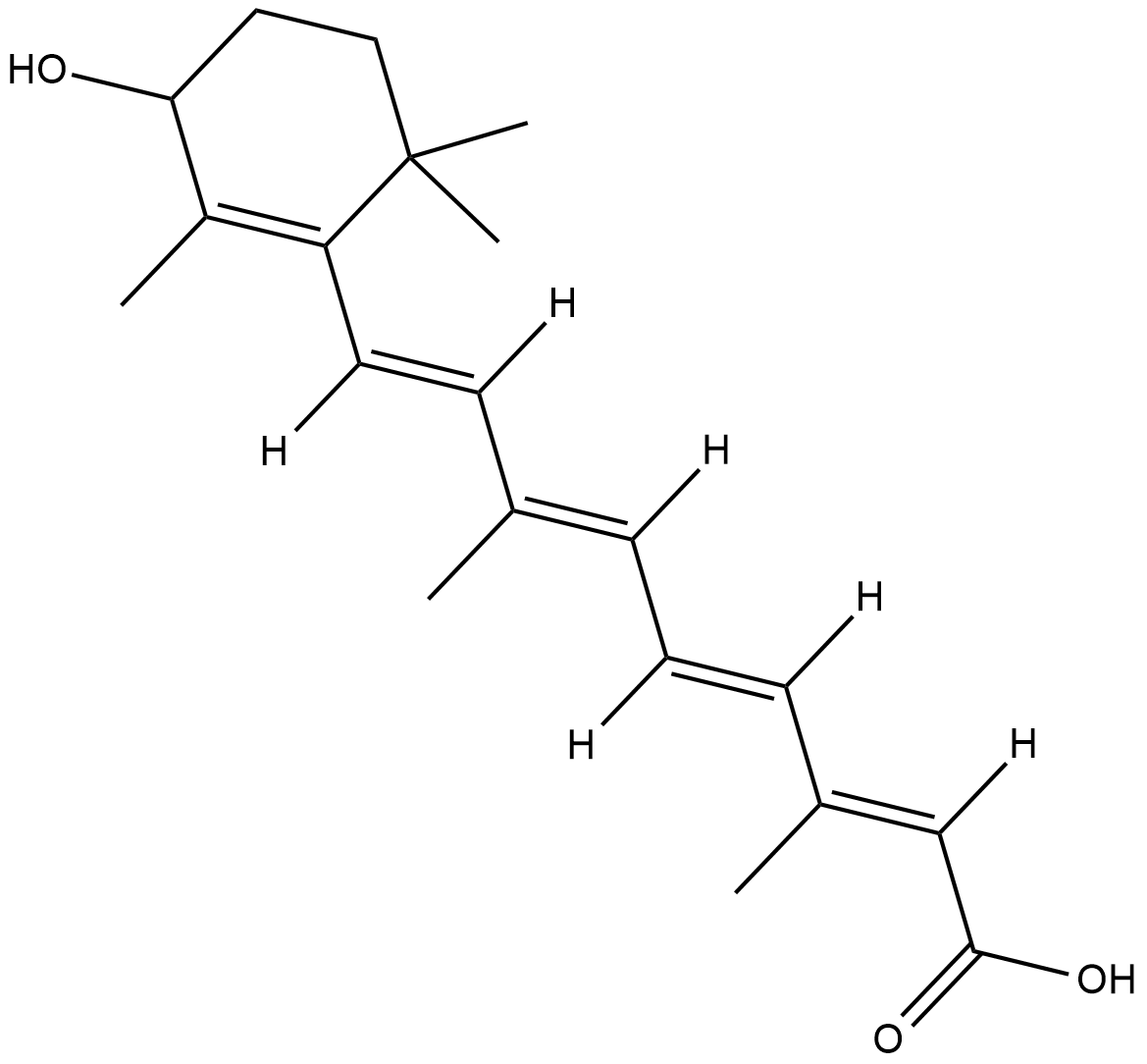
all-trans-4-hydroxy Retinoic Acid
all-trans-4-hydroxy Retinoic acid is a metabolite of all-trans retinoic acid formed by the cytochrome P450 (CYP) isoforms CYP26A1, B1, and C1.
-
GC31420
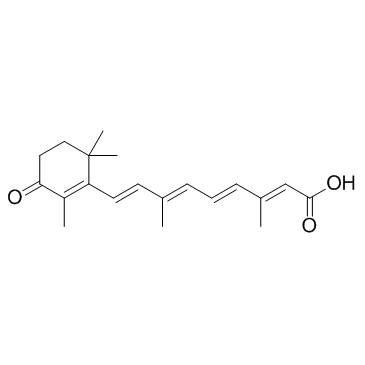
all-trans-4-Oxoretinoic acid (all-trans 4-Keto Retinoic Acid)
all-trans-4-Oxoretinoic acid (all-trans 4-Keto Retinoic Acid), an active metabolite of vitamin A, induces gene transcription via binding to nuclear retinoic acid receptors (RARs).
-
GC61487
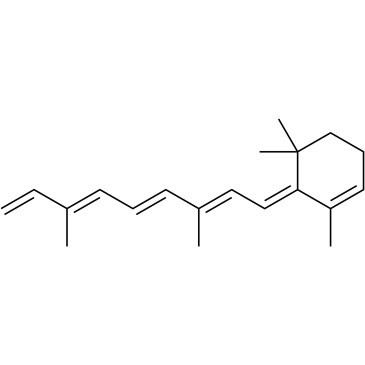
all-trans-Anhydro Retinol
all-trans-Anhydro Retinol (Anhydrovitamin A) is a metabolite of Vitamin A.
-
GC60574
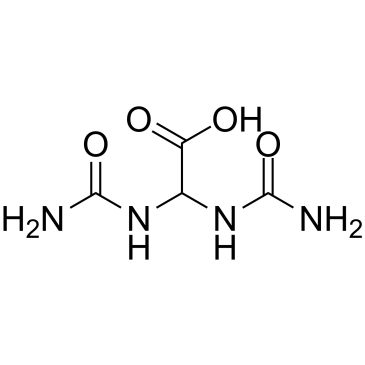
Allantoic acid
Allantoic acid is a degradative product of uric acid and associated with purine metabolism.
-
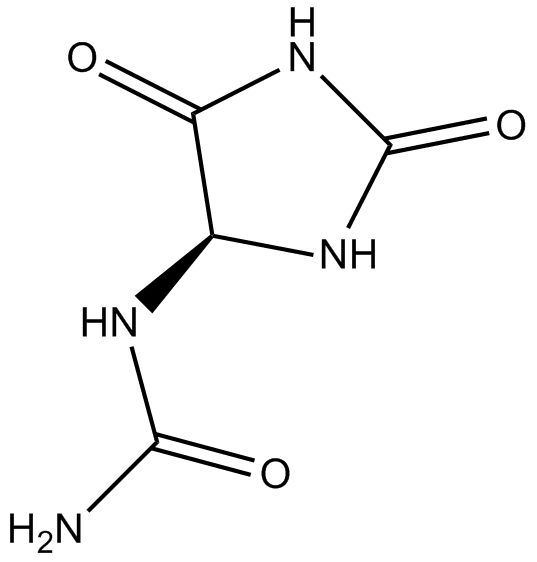
GN10492
Allantoin
-
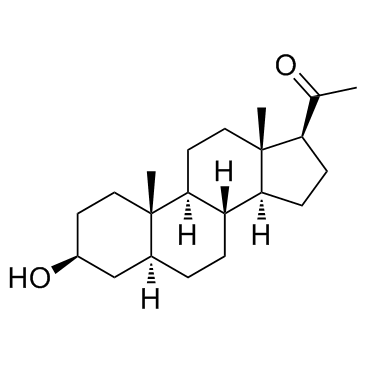
GC33807
Alloepipregnanolone
An Analytical Reference Standard
-
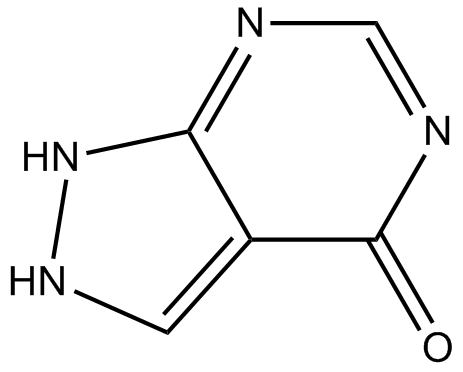
GC15322
Allopurinol
Xanthine oxidase inhibitor
-
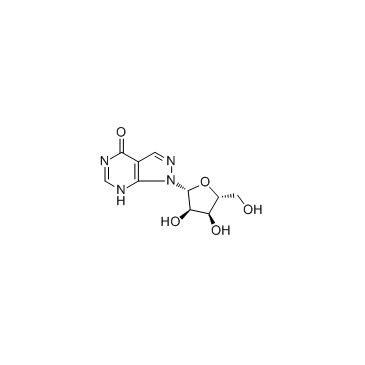
GC35295
Allopurinol riboside
A ribonucleoside
-
GC38734
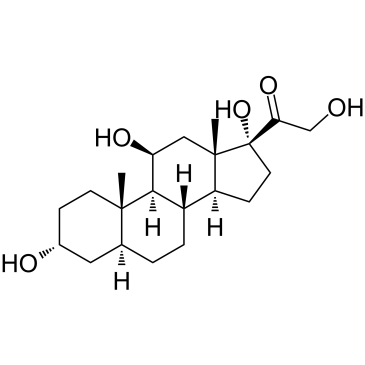
Allotetrahydrocortisol
Allotetrahydrocortisol (5a-Tetrahydrocortisol) is a metabolite of Cortisol.
-
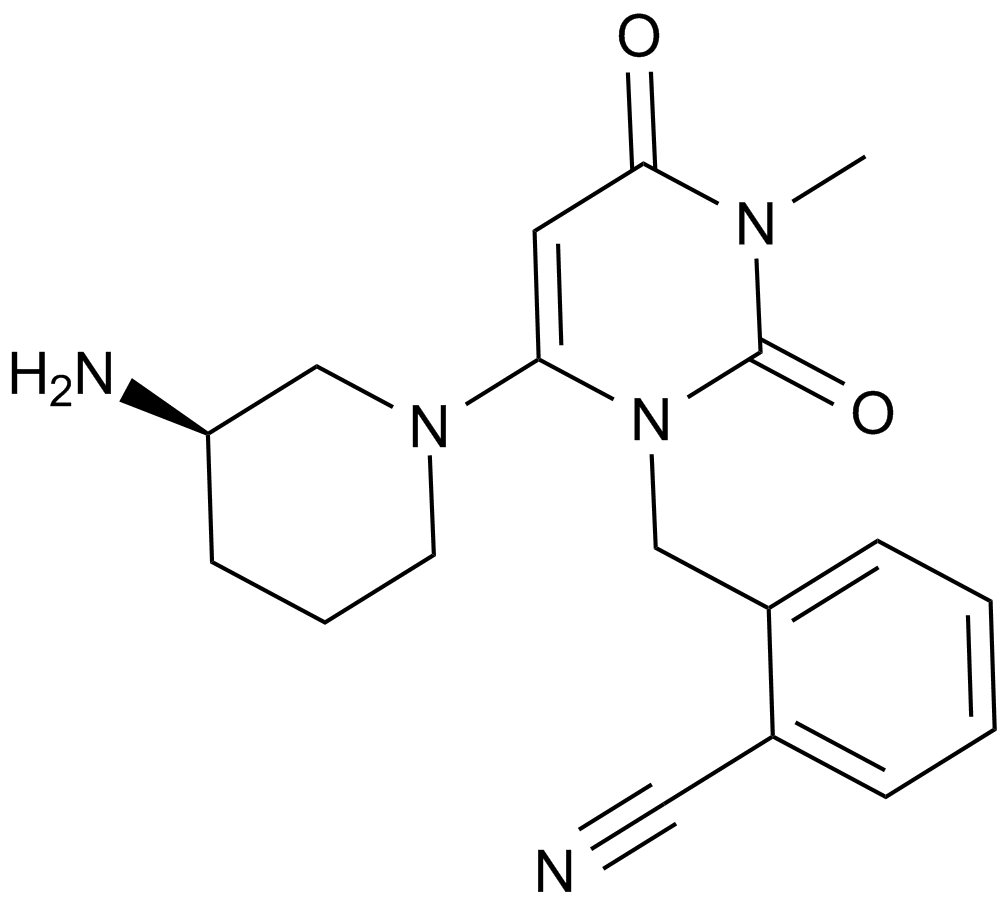
GC15130
Alogliptin (SYR-322)
-
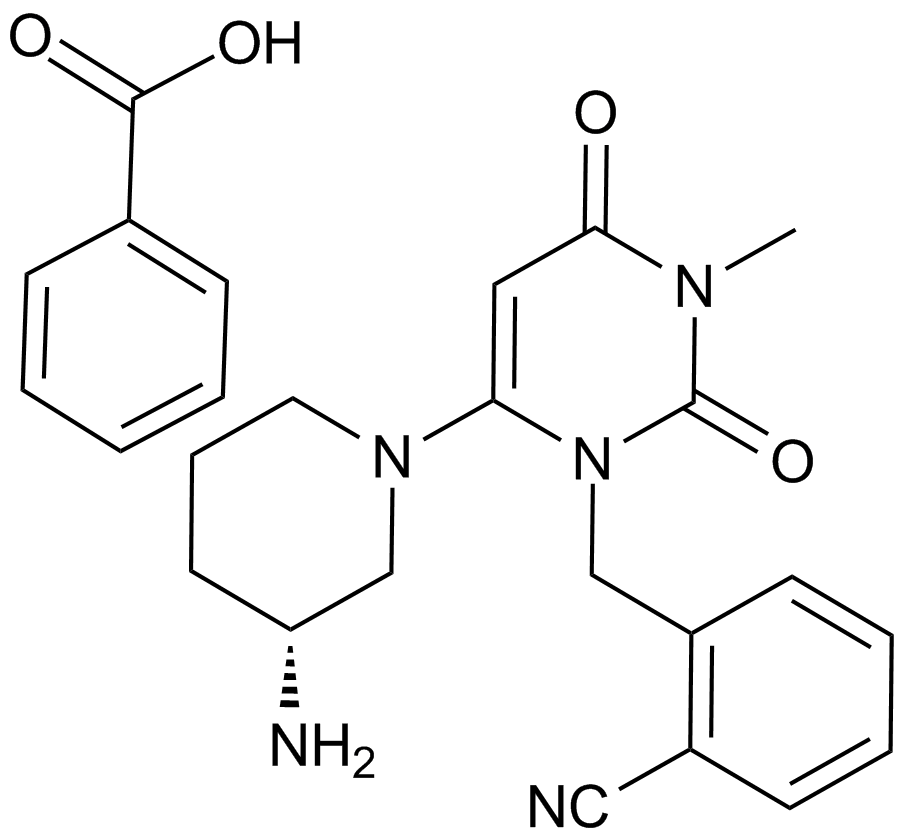
GC13949
Alogliptin Benzoate
A DPP-4 inhibitor
-
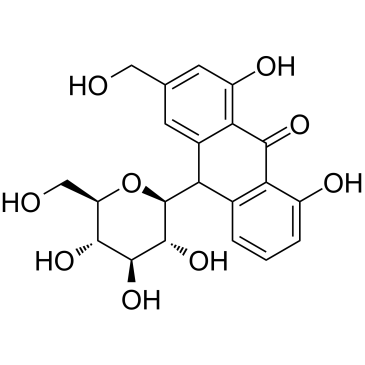
GC35300
Aloin(mixture of A&B)
Aloin (mixture of A&B) is anthraquinone derivative isolated from Aloe vera.
-
GC38885
alpha-D-glucose
alpha-D-glucose is an endogenous metabolite.
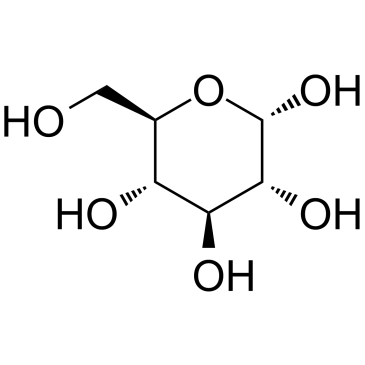
-
GC19750
alpha-L-Rhamnose
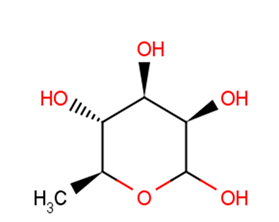
-
GC17347
Alprostadil
Alprostadil (Alprostadil) is a prostanoid receptor ligand, with Kis of 1.1 nM, 2.1 nM, 10 nM, 33 nM and 36 nM for mouse EP3, EP4, EP2, IP and EP1, respectively.
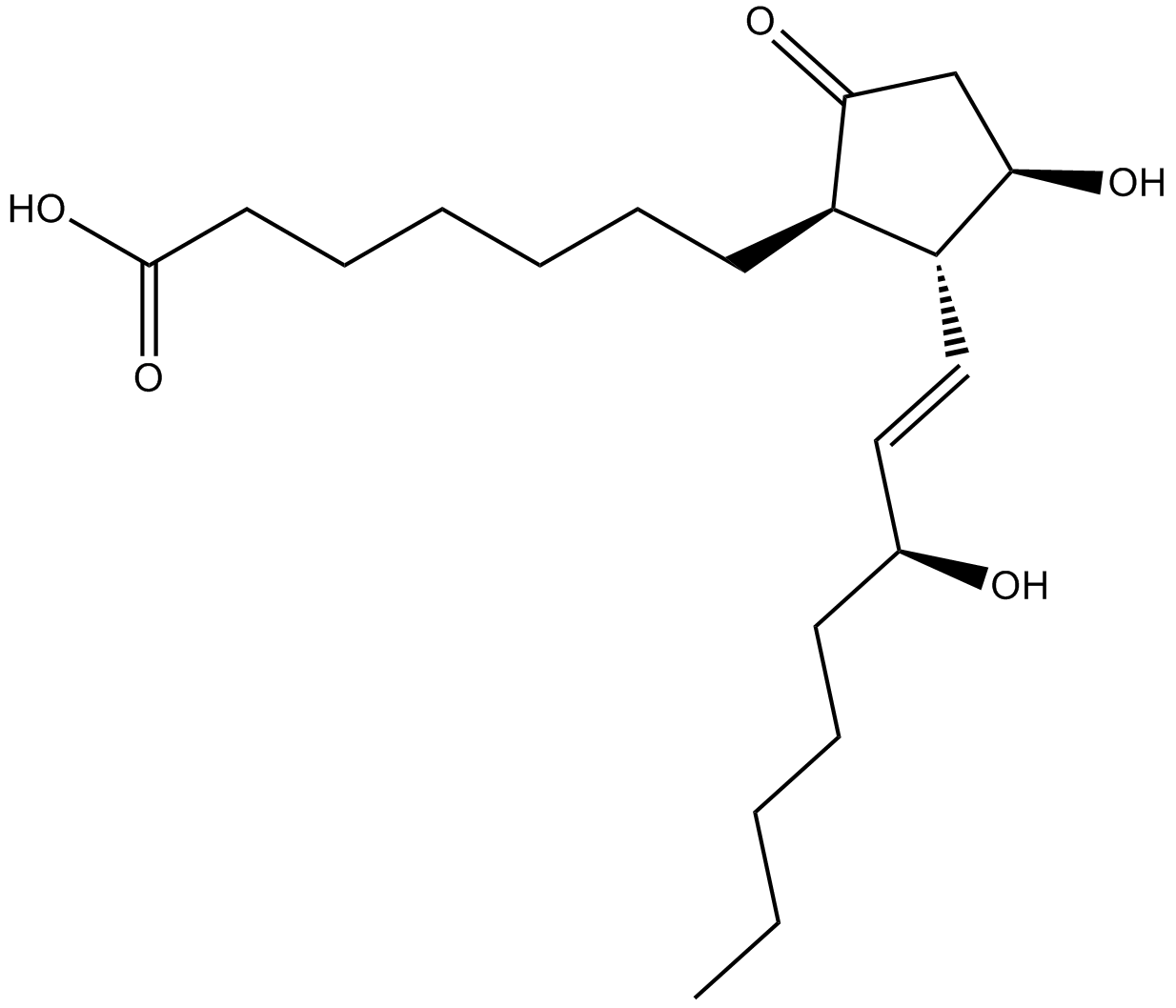
-
GC18437
Alternariol monomethyl ether
Alternariol monomethyl ether, isolated from the roots of Anthocleista djalonensis (Loganiaceae), is an important taxonomic marker of the plant species.
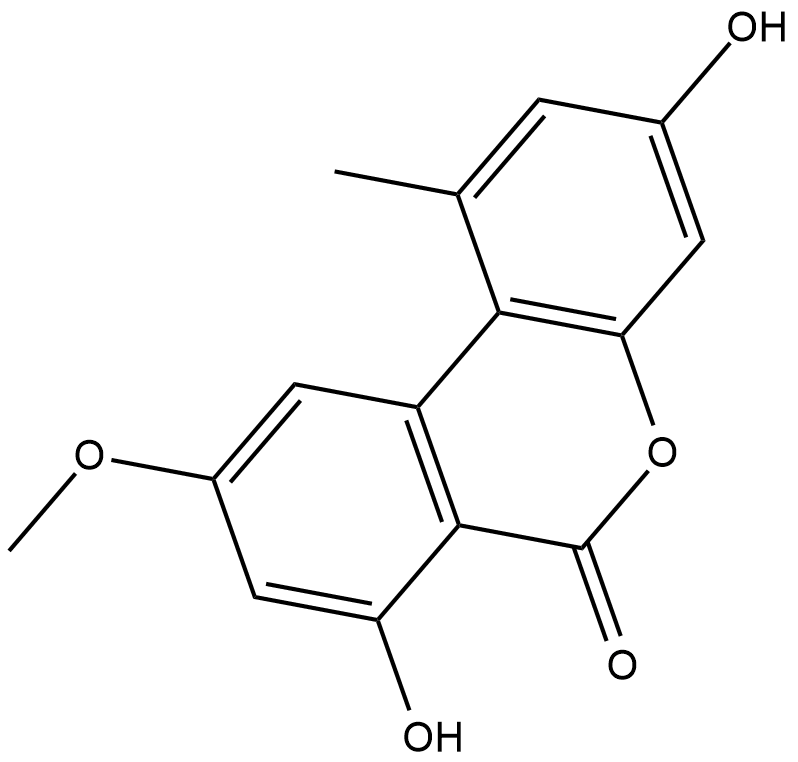
-
GC14785
Alvelestat
NE inhibitor
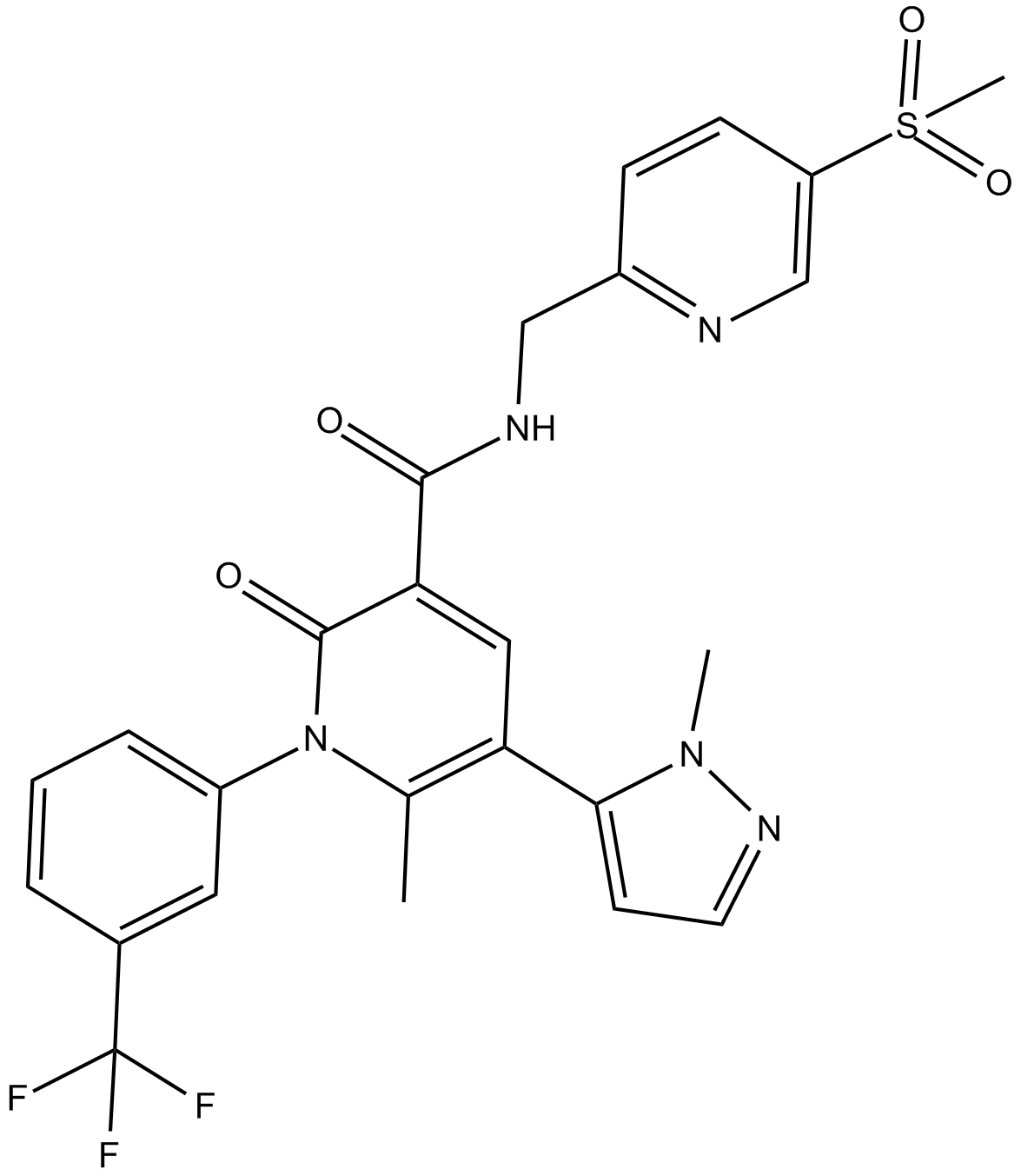
-
GC33356
AM-8735
AM-8735 is a potent and selective MDM2 inhibitor with an IC50 of 25 nM.
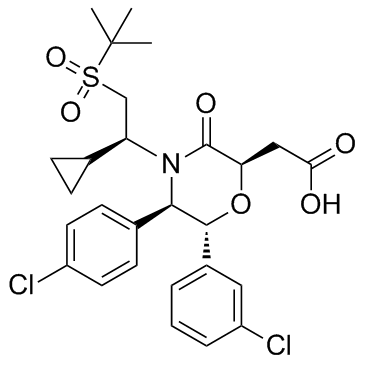
-
GC42777
Amastatin (hydrochloride)
Amastatin is a slow, tight binding, competitive aminopeptidase (AP) inhibitor, first described as an inhibitor of human serum AP-A (glutamyl AP; IC50 = 0.54 μg/ml) but not of AP-B (arginine AP).
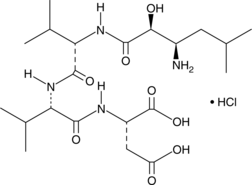
-
GC48611
Ambroxol-d5
An internal standard for the quantification of ambroxol
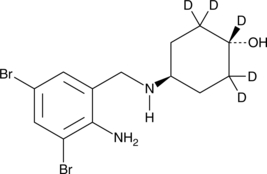
-
GC15828
AMG232
AMG232 (AMG 232) is a potent, selective and orally available inhibitor of p53-MDM2 interaction, with an IC50 of 0.6 nM. AMG232 binds to MDM2 with a Kd of 0.045 nM.
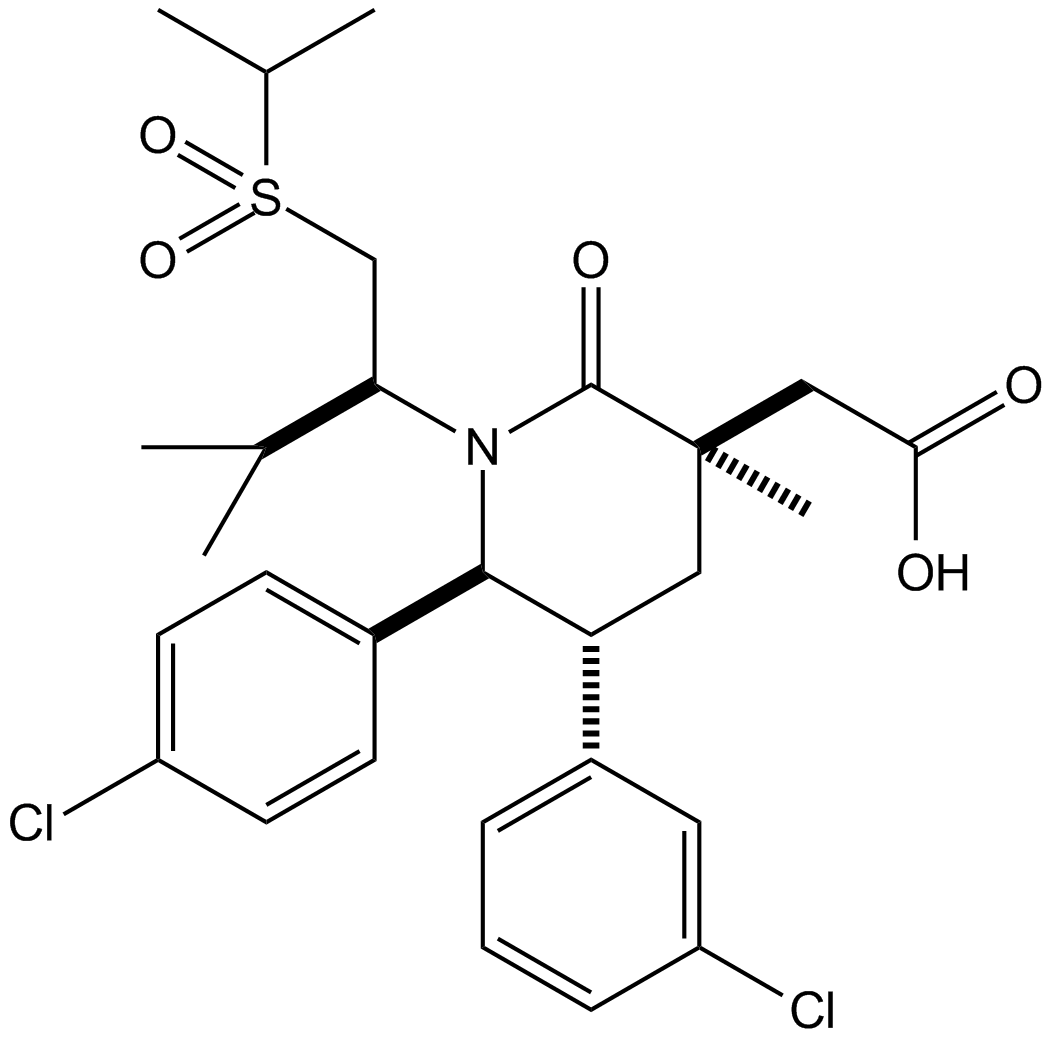
-
GC32149
Aminoacyl tRNA synthetase-IN-1
Aminoacyl tRNA synthetase-IN-1 is a bacterial aminoacyl tRNA synthetase (aaRS) inhibitor.
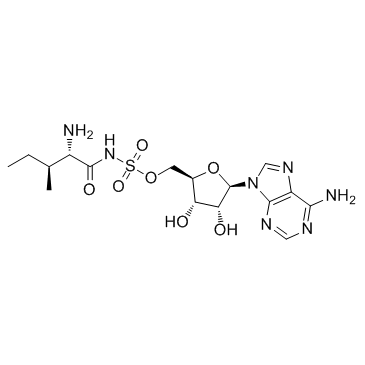
-
GC35320
Aminoadipic acid
Aminoadipic acid is an intermediate in the metabolism of lysine and saccharopine.
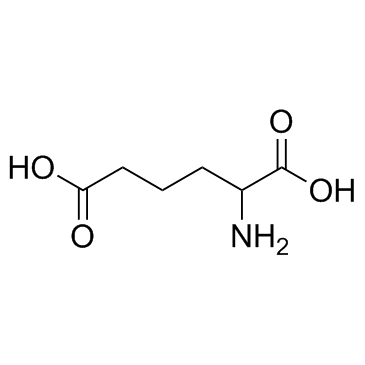
-
GC61563
Aminohexylgeldanamycin
Aminohexylgeldanamycin (AHGDM), a Geldanamycin derivative, is a potent HSP90 inhibitor. Aminohexylgeldanamycin shows antiangiogenic and antitumor activities.
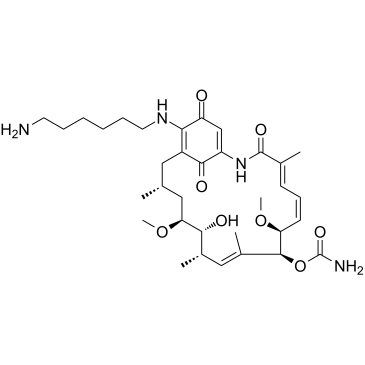
-
GC35321
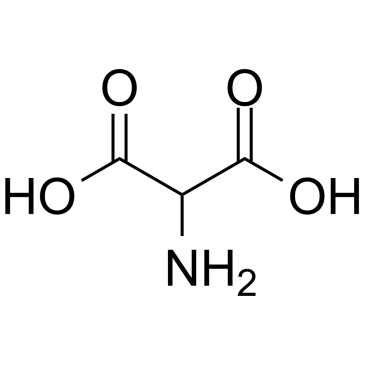
Aminomalonic acid
Aminomalonic acid is an amino endogenous metabolite, acts as a strong inhibitor of L-asparagine synthetase from Leukemia 5178Y/AR (Ki= 0.0023 M) and mouse pancreas (Ki= 0.0015 M) in vitro. Aminomalonic acid is a potential biomarker to discriminate between different stages of melanoma metastasis.
-
GC42788
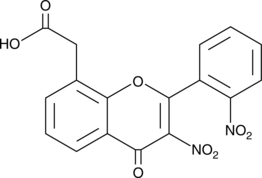
Aminopeptidase N Inhibitor
Aminopeptidase N (AP-N) inhibitor is a reversible inhibitor of AP-N/CD13 (IC50 = 25 μM).
-
GC67997
Aminopeptidase-IN-1

-
GC18274
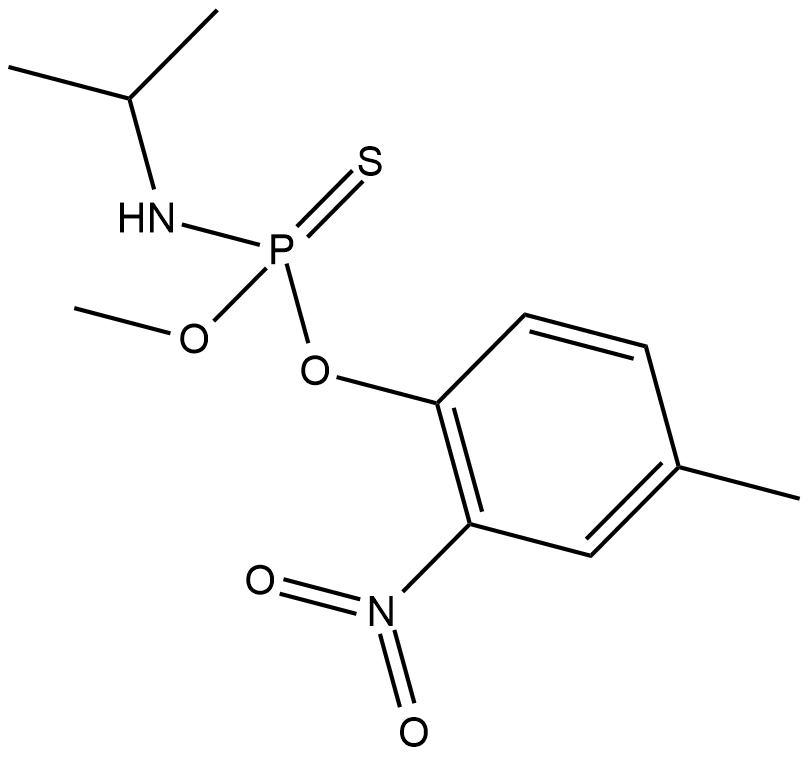
Amiprofos-methyl
Amiprofos-methyl (APM) is a phosphoric amide herbicide.
-
GC52059
AMOZ
-
GC39477
AMP-PCP disodium
AMP-PCP disodium is an ATP analogue and can bind to Hsp90 N-terminal domain with a Kd value of 3.8 μM. AMP-PCP disodium binding favors the formation of the active homodimer of Hsp90.
-
GC14152
Amprenavir (agenerase)
A selective HIV protease inhibitor
-
GC33484
Ampyrone (4-Aminoantipyrine)
-
GP10057
Amyloid β-Peptide (10-20) (human)
-
GC41211
Anacardic Acid Diene
Anacardic acid diene is a polyunsaturated form of anacardic acid that has been found in cashew nut shell liquid.
-
GC41531
Anacardic Acid Triene
Anacardic acid triene is a polyunsaturated form of anacardic acid that has been found in cashew nut shell liquid.
-
GC31341
Anagliptin (SK-0403)
Anagliptin (SK-0403) (SK-0403) is a highly selective, potent, orally active inhibitor of dipeptidyl peptidase 4 (DPP-4), with an IC50 of 3.8 nM, and less selective at DPP-8 and DDP-9 with IC50s of 68 nM and 60 nM, respectively.
-
GC35339
Anandamide
An immune modulator in the central nervous system
-
GC46854
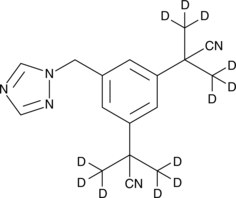
Anastrozole-d12
An internal standard for the quantification of anastrozole
-
GC42806
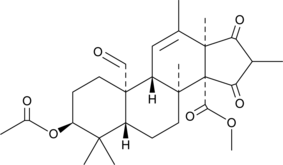
Andrastin A
Andrastin A is a meroterpenoid farnesyltransferase inhibitor.
-
GC30842
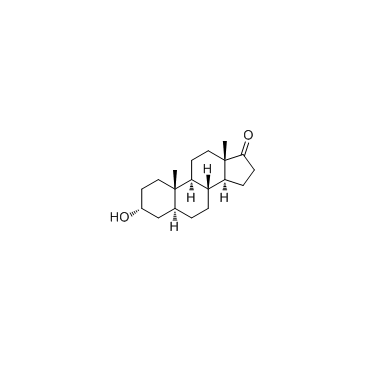
Androsterone (5α-Androstan-3α-ol-17-one)
An androgenic steroid
-
GP10077
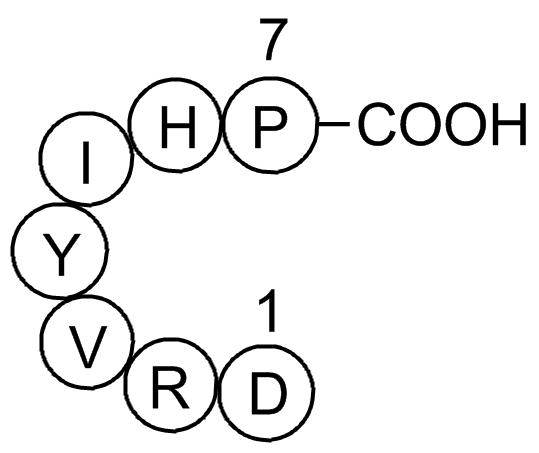
Angiotensin (1-7)
Ang-(1-7) (H - Asp - Arg - Val - Tyr - Ile - His - Pro - OH) is an endogenous peptide fragment that can be produced from Ang I or Ang II via endo- or carboxy-peptidases respectively[1].
-
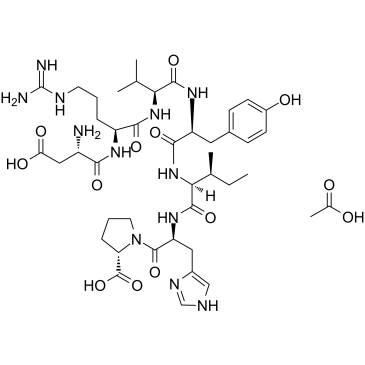
GC61496
Angiotensin (1-7) (acetate)
Angiotensin 1-7 (Ang-(1-7)) acetate is an endogenous heptapeptide from the renin-angiotensin system (RAS) with a cardioprotective role due to its anti-inflammatory and anti-fibrotic activities in cardiac cells.
-
GA20760
Angiotensin A (1-7)
Angiotensin A (1-7), a member of the renin-angiotensin system (RAS), a vasoactive peptide, is an endogenous ligand of the G protein-coupled receptor MrgD.

-
GC35355
Angiotensin III

-
GC35356
Angiotensin III TFA

-
GC32604
Angiotensinogen (1-14), human
Angiotensinogen (1-14), human is a fragment of the renin substrate angiotensinogen.

-
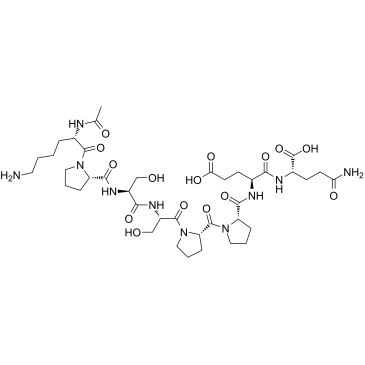
GC61511
Angstrom6
Angstrom6 (A6 Peptide) is an 8 amino-acid peptide derived from single-chain urokinase plasminogen activator (scuPA) and interferes with the uPA/uPAR cascade and abrogates downstream effects. Angstrom6 binds to CD44 resulting in the inhibition of migration, invasion, and metastasis of tumor cells, and the modulation of CD44-mediated cell signaling.
-
GC14937
Anguizole
-
GC30722
Anserine
Anserine, a methylated form of Carnosine, is an orally active, natural Histidine-containing dipeptide found in skeletal muscle of vertebrates.
-
GC42818
Antimycin A1
Antimycin A, an antibiotic produced by Streptomyces species that demonstrates antifungal, insecticidal, nematocidal, and piscicidal properties, is a mixture of Antimycins A1, A2, A3, and A4.
-
GC18621
Antimycin A2
Antimycin A2 is an active component of the antimycin A antibiotic complex.
-
GC42820
Antimycin A4
Antimycin A4 is an active component of the antimycin A antibiotic complex that is more polar than antimycin A1, antimycin A2, and antimycin A3.
-
GC35361
Antineoplaston A10
Antineoplaston A10, a naturally occurring substance in human body, is a Ras inhibitor potentially for the treatment of glioma, lymphoma, astrocytoma and breast cancer.
-
GC40032
Antipain (hydrochloride)
Antipain (hydrochloride) is a protease inhibitor isolated from Actinomycetes.
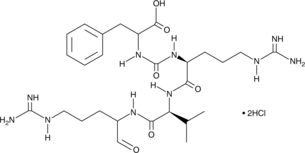
-
GC42821
AP219
AP39 is a compound used to increase the levels of hydrogen sulfide (H2S) within mitochondria.
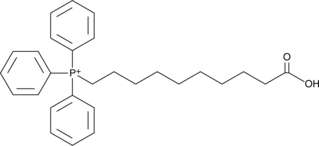
-
GC42823
AP39
AP39 is a compound used to increase the levels of hydrogen sulfide (H2S) within mitochondria.
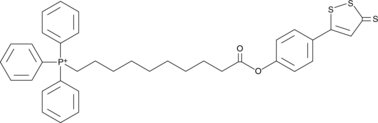
-
GC65035
Apatorsen
Apatorsen?is?an?antisense?oligonucleotide?designed?to?bind?to?Hsp27?mRNA,?resulting?in?the?inhibition of?the?production?of?Hsp27?protein.

-
GC35367
APG-115
APG-115 (APG-115) is an orally active MDM2 protein inhibitor binding to MDM2 protein with IC50 and Ki values of 3.8 nM and 1 nM, respectively. APG-115 blocks the interaction of MDM2 and p53 and induces cell-cycle arrest and apoptosis in a p53-dependent manner.
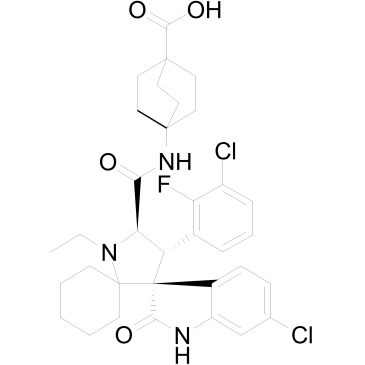
-
GC35369
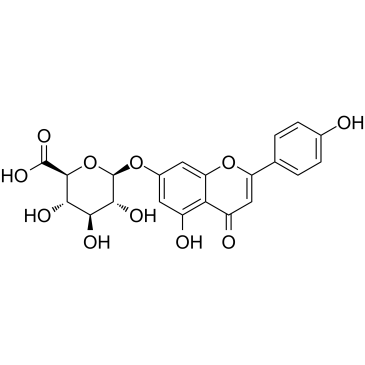
Apigenin-7-glucuronide
Apigenin-7-glucuronide could inhibit Matrix Metalloproteinases (MMP) activities, with IC50s of 12.87, 22.39, 17.52, 0.27 μM for MMP-3, MMP-8, MMP-9, MMP-13, respectively.
-
GC13130
Apixaban
An inhibitor of Factor Xa
-
GC62845
Apoatropine hydrochloride
-
GC14209
Apoptosis Activator 2
An activator of caspases
-
GC10160
Apoptosis Inhibitor
A cell-permeable inhibitor of caspase-3 activation
-
GC14411
Apoptozole
inhibitor of heat shock protein 70 (Hsp70)
-
GC49843
Aposcopolamine
A tropane alkaloid and an active metabolite of scopolamine
-
GC60591
Apovincaminic acid hydrochloride salt
Apovincaminic acid hydrochloride salt is an orally active and brain-penetrant main active metabolite of Vinpocetine (VP).
-
GC35377
Apratastat
An inhibitor of ADAM17 and MMPs
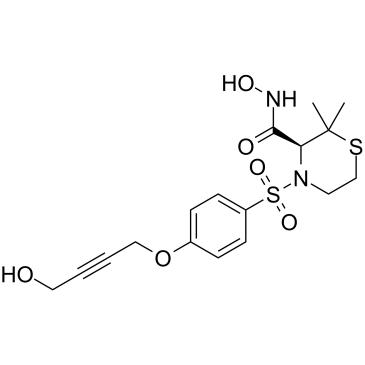
-
GC12900
Aprotinin
A serine protease inhibitor
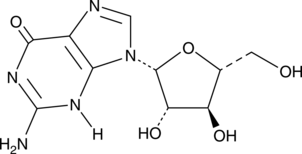
-
GC45385
Ara-G
-
GC61691
Arabinose
Arabinose is an endogenous metabolite.
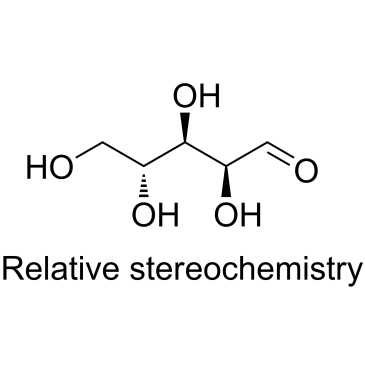
-
GC35379
Arachidic acid
A longchain saturated fatty acid