Proteases
Proteases is a general term for a class of enzymes that hydrolyze protein peptide chains. According to the way they degrade polypeptides, they are divided into two categories: endopeptidases and telopeptidases. The former can cut the large molecular weight polypeptide chain from the middle to form prions and peptones with smaller molecular weights; the latter can be divided into carboxypeptidase and aminopeptidase, which respectively remove the peptide from the free carboxyl terminus or free amino terminus of the polypeptide one by one. Chain hydrolysis produces amino acids.
A general term for a class of enzymes that hydrolyze peptide bonds in proteins. According to the way they hydrolyze polypeptides, they can be divided into endopeptidases and exopeptidases. Endopeptidase cleaves the interior of the protein molecule to form smaller molecular weight peptones and peptones. Exopeptidase hydrolyzes peptide bonds one by one from the end of the free amino group or carboxyl group of protein molecules, and frees amino acids, the former is aminopeptidase and the latter is carboxypeptidase. Proteases can be classified into serine proteases, sulfhydryl proteases, metalloproteases and aspartic proteases according to their active centers and optimum pH. According to the optimum pH value of its reaction, it is divided into acidic protease, neutral protease and alkaline protease. The proteases used in industrial production are mainly endopeptidases.
Proteases are widely found in animal offal, plant stems and leaves, fruits and microorganisms. Microbial proteases are mainly produced by molds and bacteria, followed by yeast and actinomycetes.
Enzymes that catalyze the hydrolysis of proteins. There are many kinds, the important ones are pepsin, trypsin, cathepsin, papain and subtilisin. Proteases have strict selectivity for the reaction substrates they act on. A protease can only act on certain peptide bonds in protein molecules, such as the peptide bonds formed by the hydrolysis of basic amino acids catalyzed by trypsin. Proteases are widely distributed, mainly in the digestive tract of humans and animals, and are abundant in plants and microorganisms. Due to limited animal and plant resources, the industrial production of protease preparations is mainly prepared by fermentation of microorganisms such as Bacillus subtilis and Aspergillus terrestris.
Targets for Proteases
- Caspase(88)
- Aminopeptidase(19)
- ACE(69)
- Calpains(11)
- Carboxypeptidase(8)
- Cathepsin(68)
- DPP-4(18)
- Elastase(23)
- Gamma Secretase(46)
- HCV Protease(35)
- HSP(95)
- HIV Integrase(29)
- HIV Protease(32)
- MMP(199)
- NS3/4a protease(4)
- Serine Protease(11)
- Thrombin(46)
- Urokinase(2)
- Cysteine Protease(0)
- Other Proteases(15)
- Tyrosinases(44)
- 15-PGDH(1)
- Acetyl-CoA Carboxylase(13)
- Acyltransferase(53)
- Aldehyde Dehydrogenase (ALDH)(27)
- Aminoacyl-tRNA Synthetase(9)
- ATGL(1)
- Dipeptidyl Peptidase(47)
- Drug Metabolite(453)
- E1/E2/E3 Enzyme(82)
- Endogenous Metabolite(1555)
- FABP(30)
- Farnesyl Transferase(21)
- Glutaminase(14)
- Glutathione Peroxidase(14)
- Isocitrate Dehydrogenase (IDH)(26)
- Lactate Dehydrogenase(17)
- Lipoxygenase(231)
- Mitochondrial Metabolism(201)
- NEDD8-activating Enzyme(6)
- Neprilysin(12)
- PAI-1(12)
- Ser/Thr Protease(40)
- Tryptophan Hydroxylase(11)
- Xanthine Oxidase(16)
- MALT1(10)
- PCSK9(1)
Products for Proteases
- Cat.No. Product Name Information
-
GC39474
7-Ketolithocholic acid
7-KLCA, 7-keto LCA, 7-oxo LCA, 7-keto Lithocholate, 7-oxo Lithocholate, 7-oxo Lithocholic Acid, NSC 226118
7-Ketolithocholic acid (3α-hydroxy-7-oxygen-5β-cholic acid) is a kind of bile acid, 7-Ketolithocholic acid can be absorbed, 7-ketolithocholic acid inhibits the production of endogenous bile acid, and can affect the secretion of biliary cholestero.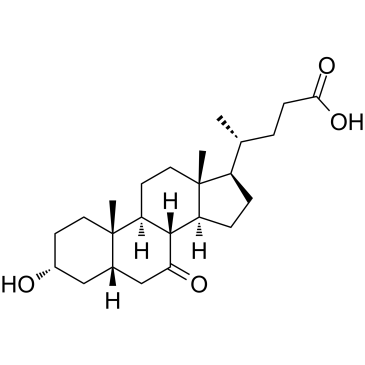
-
GC33616
7-Methylguanine
Epiguanine, 2-Amino-7-methylhypoxanthine, N7-Methylguanine, NSC 193444, NSC 19647
7-Methylguanine is a metabolite of DNA methylation.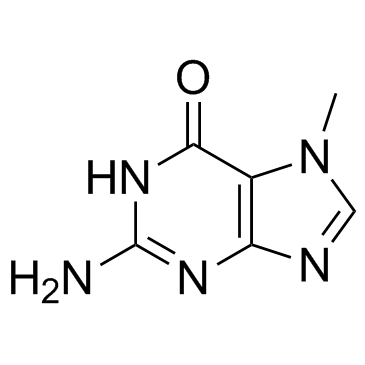
-
GC62817
7-Methylguanosine 5’-diphosphate sodium
7-Methyl-GDP sodium; m7GDP sodium
7-Methylguanosine 5'-diphosphate (7-Methyl-GDP) sodium, a cap analog, can be used in the synthesis of mRNA cap analogues.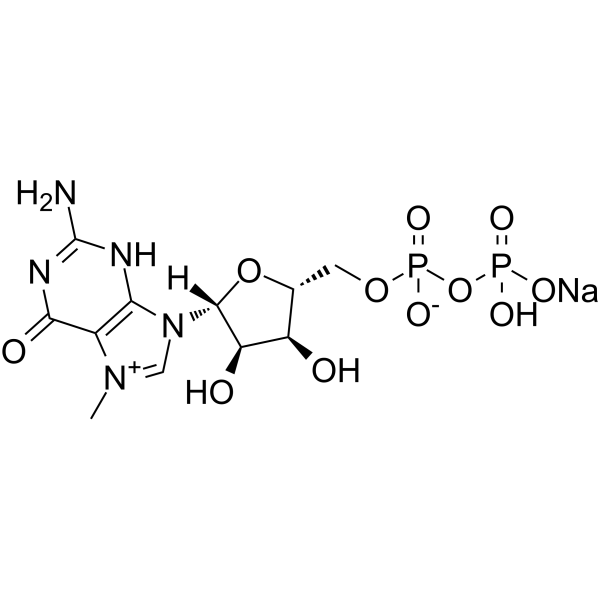
-
GC31617
7-Methylxanthine
7-MX, NSC 7861
7-Methylxanthine, a methyl derivative of xanthine, is one of the purine components in urinary calculi.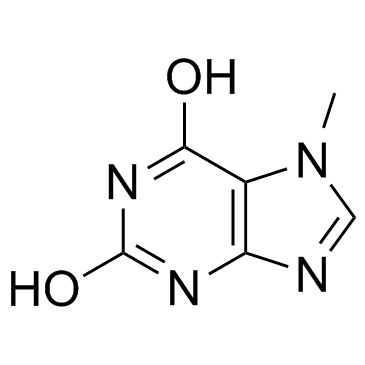
-
GC14433
7α,25-dihydroxy Cholesterol
7α,25-DHC
A GPR183 agonist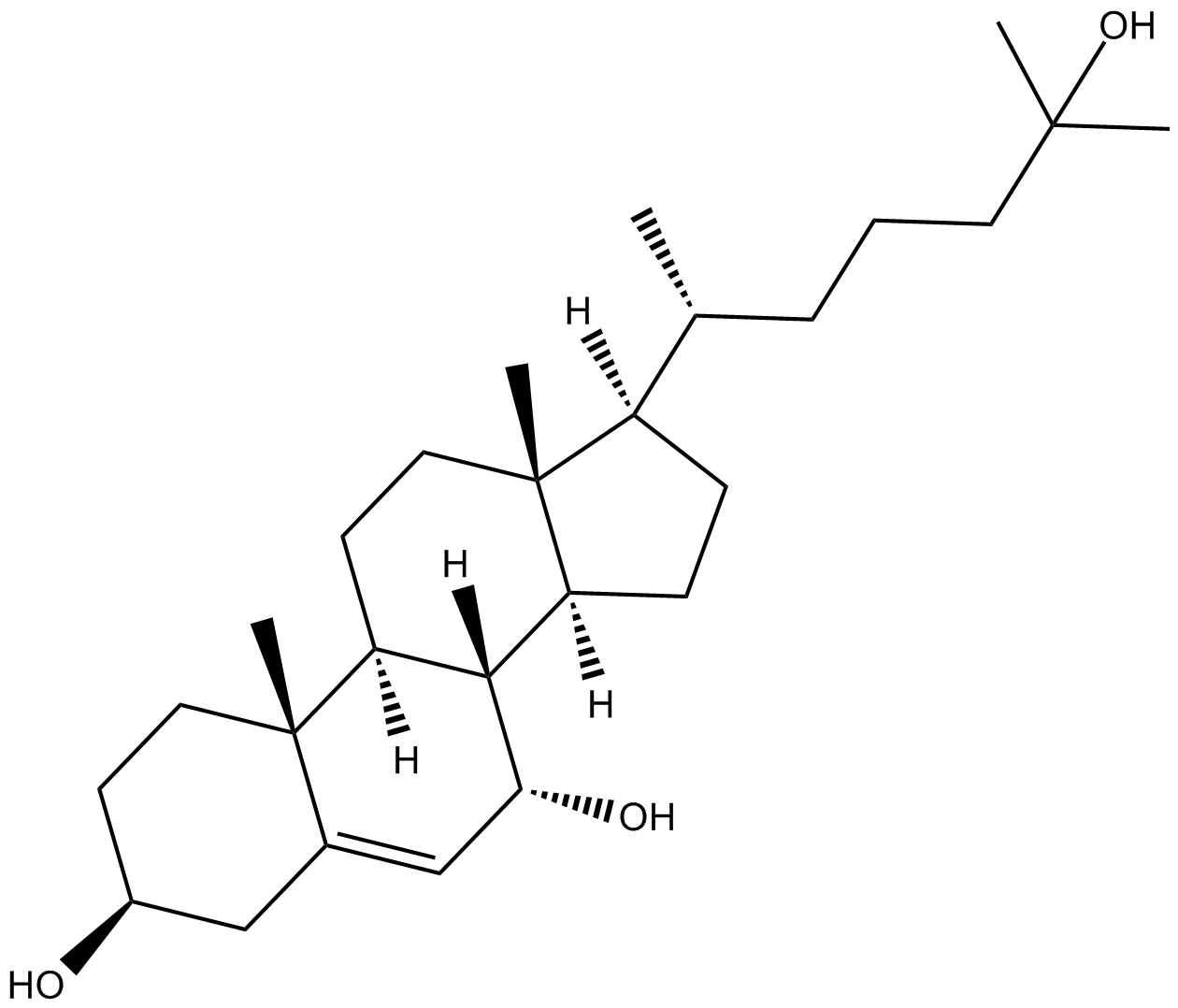
-
GC40462
8(R)-HETE
8(R)-Hydroxyeicosatetraenoic Acid
8(R)-HETE is biosynthesized by lipoxygenation of arachidonic acid in marine invertebrates such as gorgonian corals and starfish.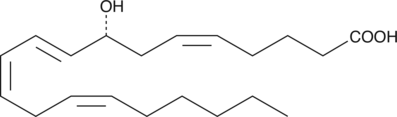
-
GC41136
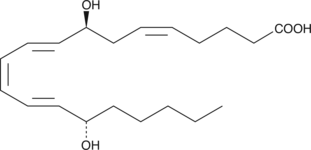
8(S),15(S)-DiHETE
8(S),15(S)-DiHETE is formed when 15(S)-HETE is subjected to further oxidation by 15-LO.
-
GC40382
8(S)-HEPE
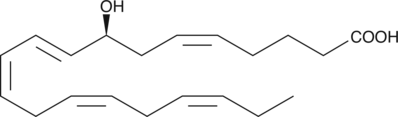
8(S)-HEPE is a monohydroxy fatty acid produced by lipoxygenase oxidation of EPA.
-
GC40463
8(S)-HETE
8(S)-Hydroxyeicosatetraenoic Acid
8(S)-HETE is a major lipoxygenase product in PMA-treated murine epidermis.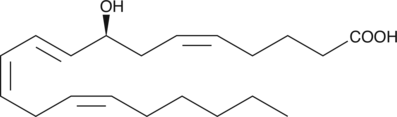
-
GC42619
8(S)-HETrE
8(S)-HETrE is a monohydroxy polyunsaturated fatty acid produced by rabbit neutrophil lipoxygenase when dihomo-γ-linolenic acid is used as a substrate.

-
GC18544
8-dehydro Cholesterol
8-dehydro Cholesterol (8-DHC) is an isomer of the cholesterol precursor 7-DHC .
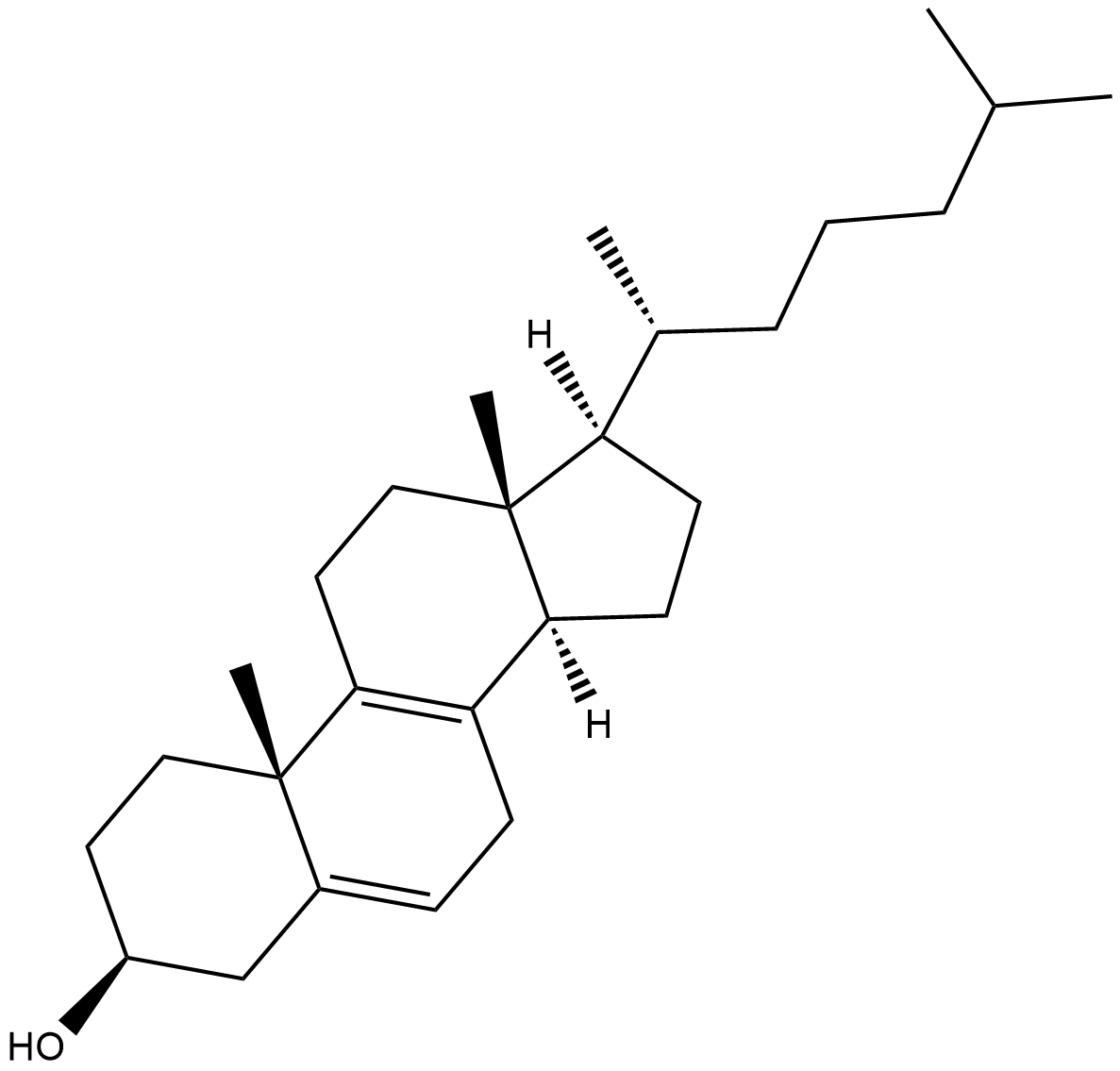
-
GC60541
8-Demethyl Ivabradine
8-Demethyl Ivabradine is a metabolite of Ivabradine.
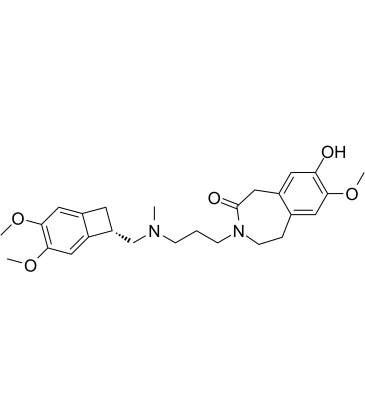
-
GC49017
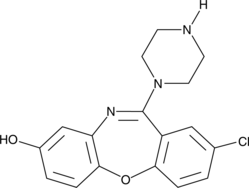
8-hydroxy Amoxapine
A metabolite of amoxapine
-
GC42626
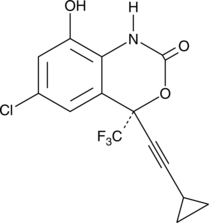
8-hydroxy Efavirenz
8-hydroxy Efavirenz is a major oxidative metabolite of the non-nucleoside reverse transcriptase inhibitor efavirenz.
-
GC18654
8-hydroxy Loxapine
8-OH Loxapine
8-hydroxy Loxapine (8-OH loxapine) is a metabolite formed when loxapine , an atypical antipsychotic, is metabolized by the cytochrome P450 isoform CYP1A2.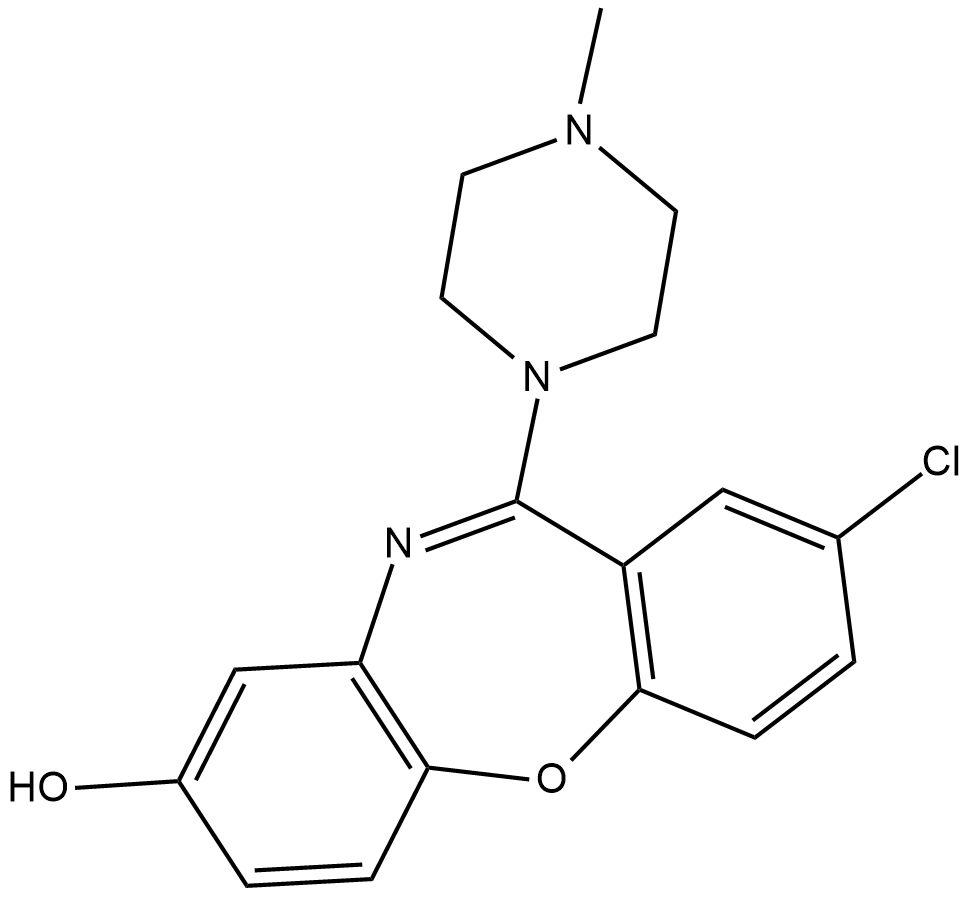
-
GC49544
8-hydroxy Mirtazapine
8-hydroxy-6-Azamianserin
A metabolite of mirtazapine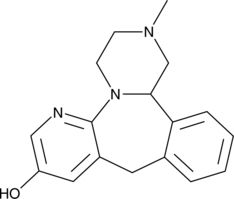
-
GC40926
8-Hydroxy-2'-deoxyguanosine
8-OHdG
8-Hydroxy-2'-deoxyguanosine is produced by oxidative damage of DNA by reactive oxygen and nitrogen species, including hydroxyl radical and peroxynitrite.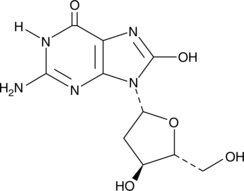
-
GC35204
8-Hydroxyguanine
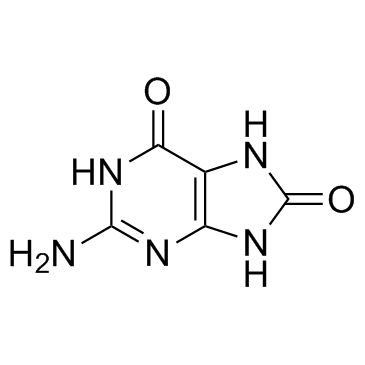
8-Hydroxyguanine is a major pre-mutagenic lesion generated from reactive oxygen species.
-
GC31219
8-Hydroxyguanosine
7,8-Dihydro-8-oxoguanosine, 8-oxo-G, NSC 90393, 8-OHG
Product of oxidative damage to RNA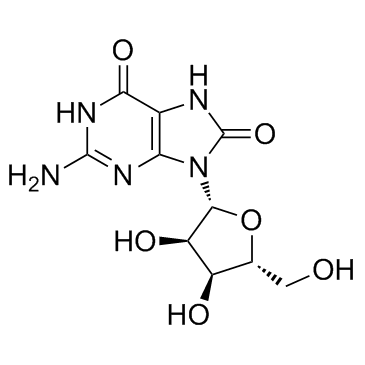
-
GC18515
8-iso Prostaglandin F2α
iPF2α-III, 8-iso-15(S)-Prostaglandin F2α, 8-Isoprostane, 8-epi PGF2α, 15-F2t-Isoprostane
8-iso PGF2α is an isoprostane produced by the non-enzymatic peroxidation of arachidonic acid in membrane phospholipids.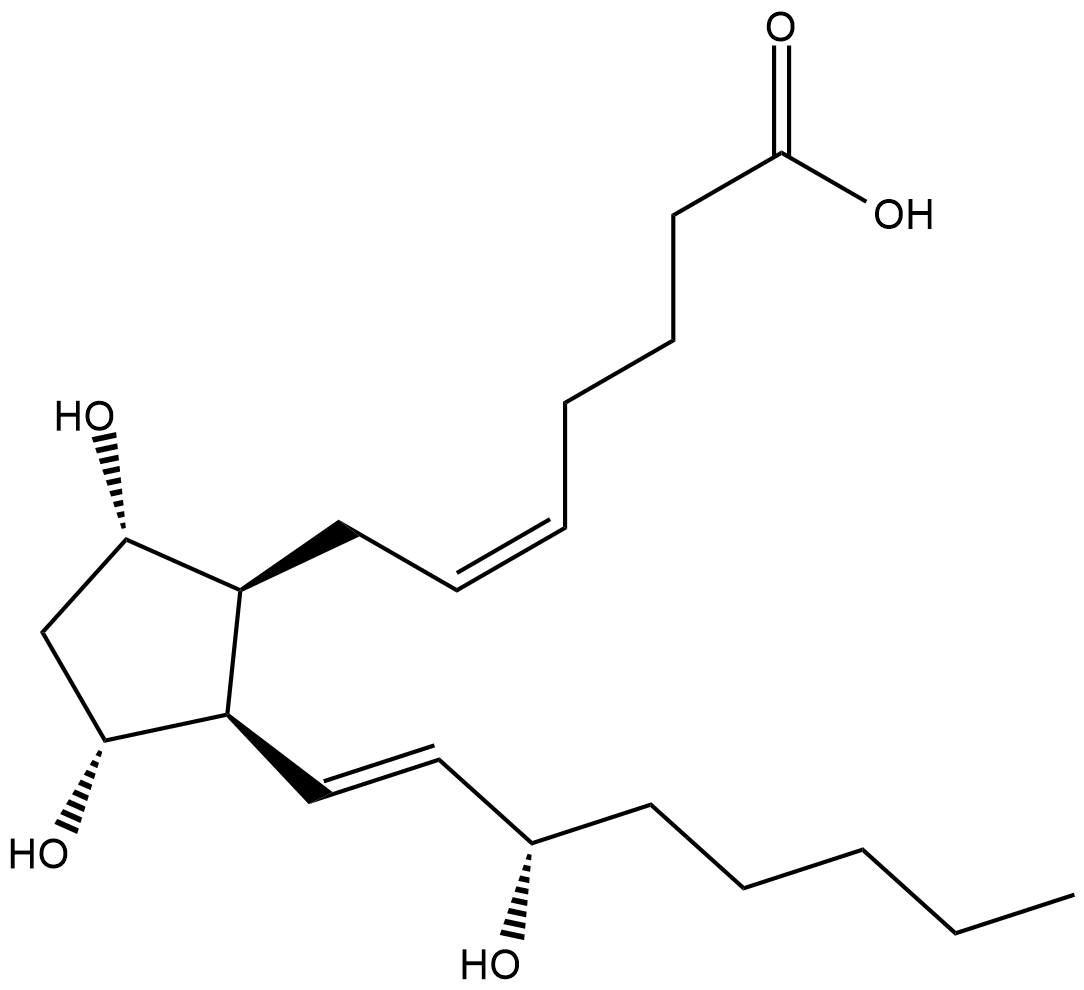
-
GC18994
8-iso Prostaglandin F2β
iPF2α-III, 8-iso-15(S)-Prostaglandin F2α, 8-Isoprostane, 8-epi PGF2α, 15-F2t-Isoprostane
8-iso Prostaglandin F2β (8-iso PGF2β) is an isomer of PGF2α with a non-enzymatic, non-cyclooxygenase origin.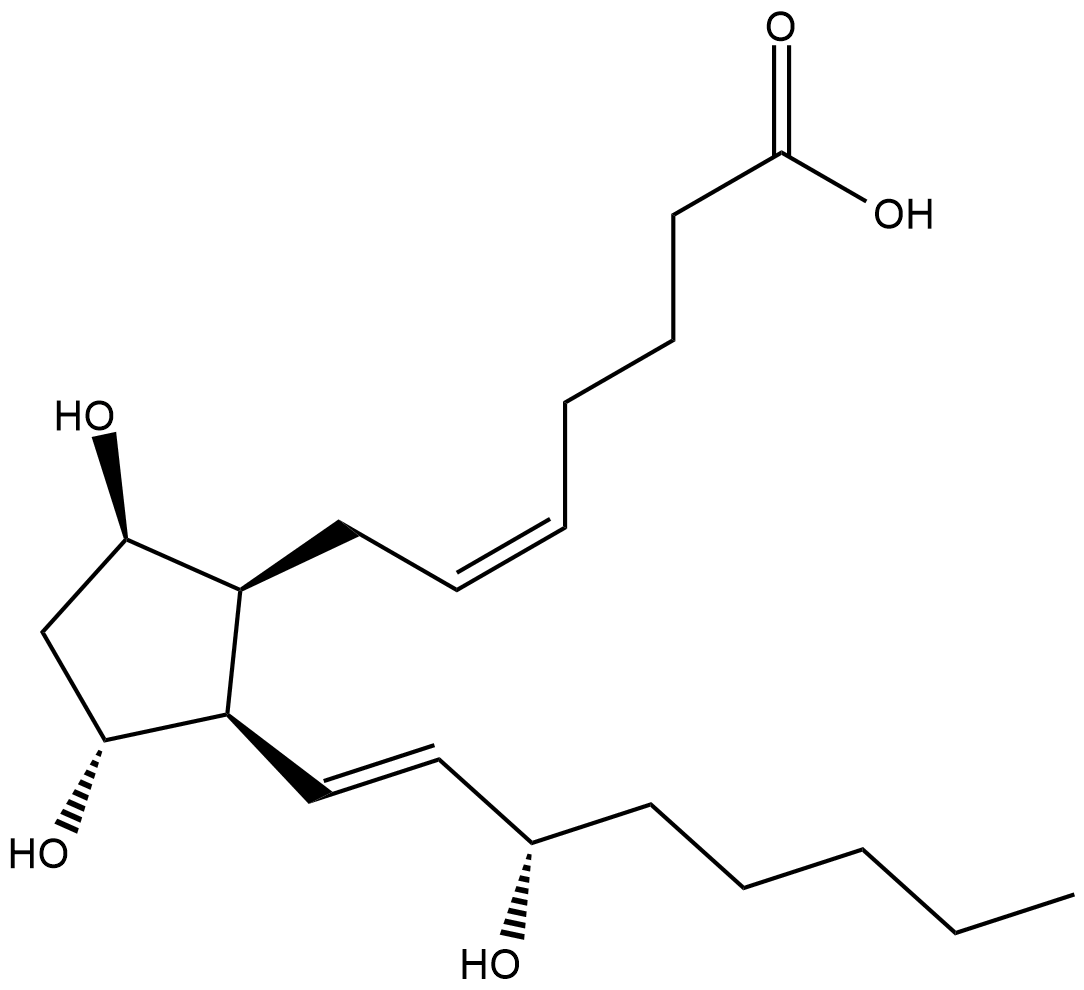
-
GC41642
9(E),11(E),13(E)-Octadecatrienoic Acid
β-Eleostearic Acid, β-ESA
9(E),11(E),13(E)-Octadecatrienoic acid (β-ESA) is a conjugated polyunsaturated fatty acid that is found in plant seed oils and in mixtures of conjugated linolenic acids synthesized by the alkaline isomerization of linolenic acid.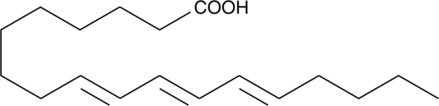
-
GC42633
9(E)-Erythromycin A oxime
(9E)-Erythromycin A oxime is a metabolite of the semisynthetic antibiotic roxithromycin .
-
GC40542
9(R)-HODE
9(R)-Hydroxyoctadecadienoic Acid
9(R)-HODE is one of several monohydroxylated products of linoleic acid.
-
GC40029
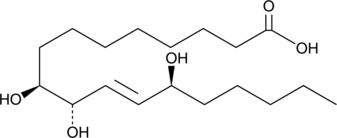
9(S),10(S),13(S)-TriHOME
9(S),10(S),13(S)-TriHOME is an oxylipin derived from linoleic acid.
-
GC46753
9(S),12(S),13(S)-TriHOME
(–)-Pinellic Acid, 9S,12S,13S-Pinellic Acid
An oxylipin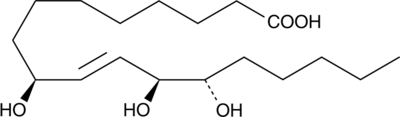
-
GC40383
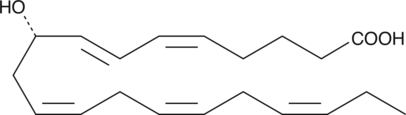
9(S)-HEPE
9(S)-HEPE is a monohydroxy fatty acid derived from EPA.
-
GC19460
9(S)-HODE
9(S)-HODE is produced by the lipoxygenation of linoleic acid in both plants and animals.

-
GC40250
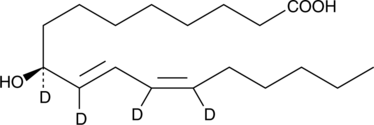
9(S)-HODE-d4 MaxSpec® Standard
9(S)-HODE-d4 is intended for use as an internal standard for the quantification of 9(S)-HODE by GC- or LC-mass spectrometry.
-
GC42636
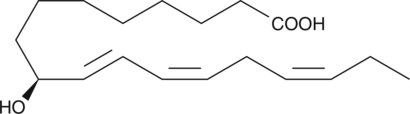
9(S)-HOTrE
9(S)-HOTrE is a monohydroxy polyunsaturated fatty acid produced by the action of 5-lipoxygenase on α-linolenic acid.
-
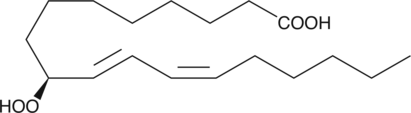
GC40357
9(S)-HpODE
9(S)-HpODE is produced by the action of arachidonate 5-LO on linoleic acid.
-
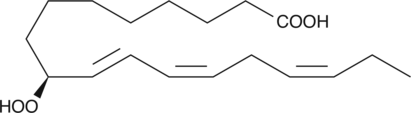
GC42637
9(S)-HpOTrE
9(S)-HpOTrE is a monohydroperoxy polyunsaturated fatty acid produced by the action of 5-lipoxygenase (5-LO) on α-linolenic acid.
-
GC18389
9-cis Retinal
9-cis Retinaldehyde, 9-cis Vitamin A aldehyde
9-cis Retinal is a natural retinoid that is produced by oxidation of 9-cis retinol by cis-retinol dehydrogenase (cRDH).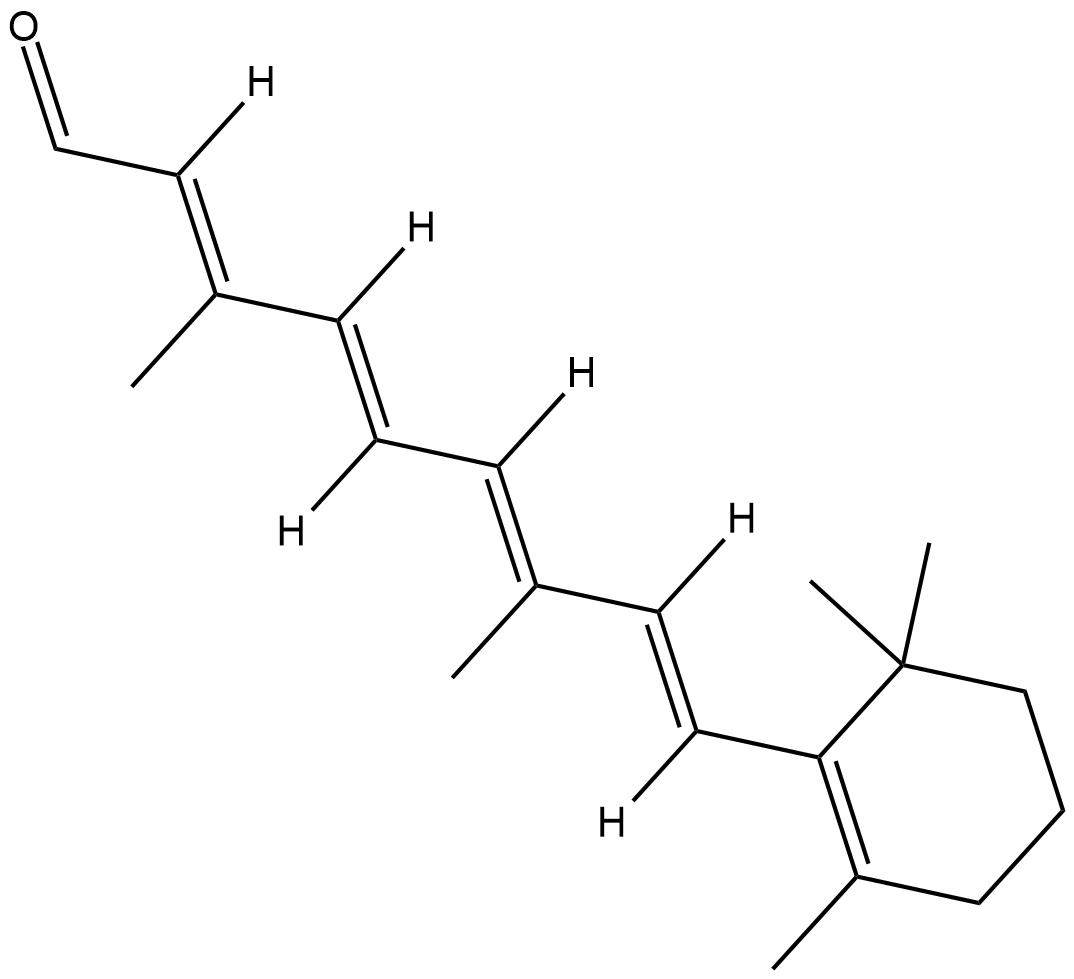
-
GC38883
9-Ethyladenine
9-Ethyladenine is a partially effective inhibitor of APRT (adenine phosphoribosyltransferase).
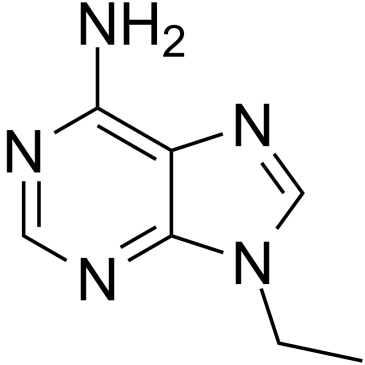
-
GC42651
9-oxo-10(E),12(E)-Octadecadienoic Acid
9-oxo ODA
9-oxo-10(E),12(E)-Octadecadienoic acid (9-oxoODA) is a natural agonist, abundant in tomatoes, that activates PPARα at 10-20 μM.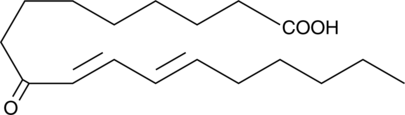
-
GC42653
9-OxoOTrE
9KOTE, 9KOTrE
9-OxoOTrE is produced by the oxidation of 9-HpOTrE.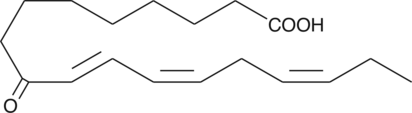
-
GC45960
9c(i472)
15-LOX-1 Inhibitor i472
9c(i472) is a potent inhibitor of 15-LOX-1 (15-lipoxygenase-1) with an IC50 value of 0.19 μM.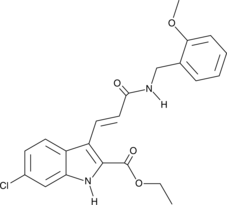
-
GC62859
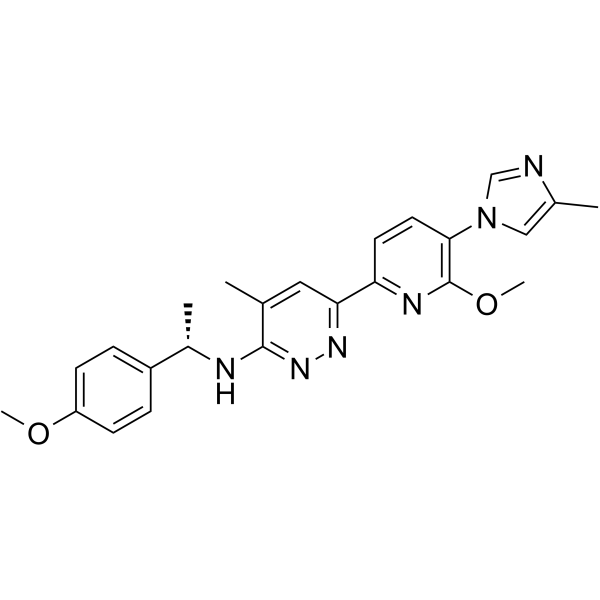
Aβ42-IN-2
Aβ42-IN-2 is a γ-secretase modulator extracted from patent WO2016070107, compound example 36.
-
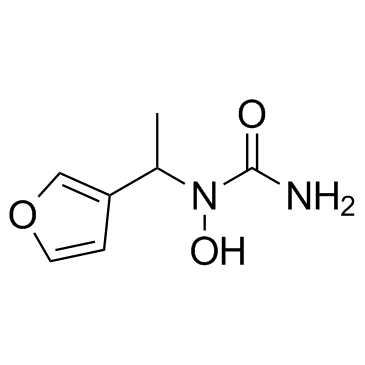
GC31938
A-69412
A-69412 is a reversible, specific inhibitor of the 5-lipoxygenase (5-LO).
-
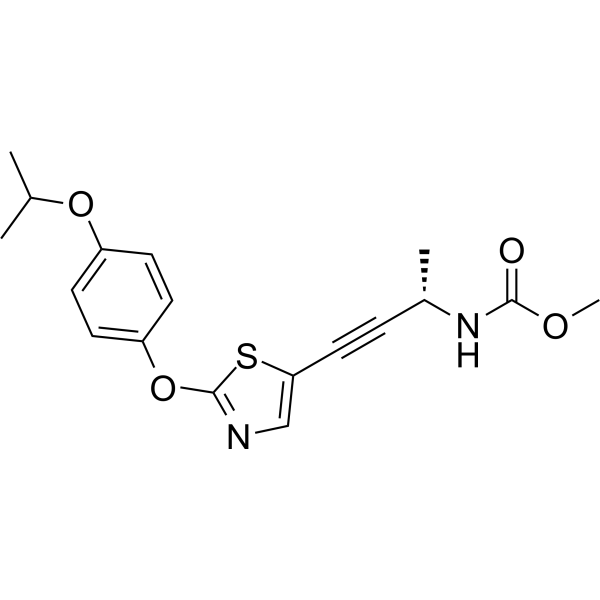
GC68587
A-908292
A-908292 is an effective and selective inhibitor of acetyl-CoA carboxylase 2 (ACC2), with an IC50 value of 23 nM for hACC2. A-908292 can be used in the study of fatty acid metabolism.
-
GC15614
A922500
(1R,2R)2(4'(3phenylureido)biphenylcarbonyl)cyclopentanecarboxylic acid
A diacylglycerol acyltransferase 1 inhibitor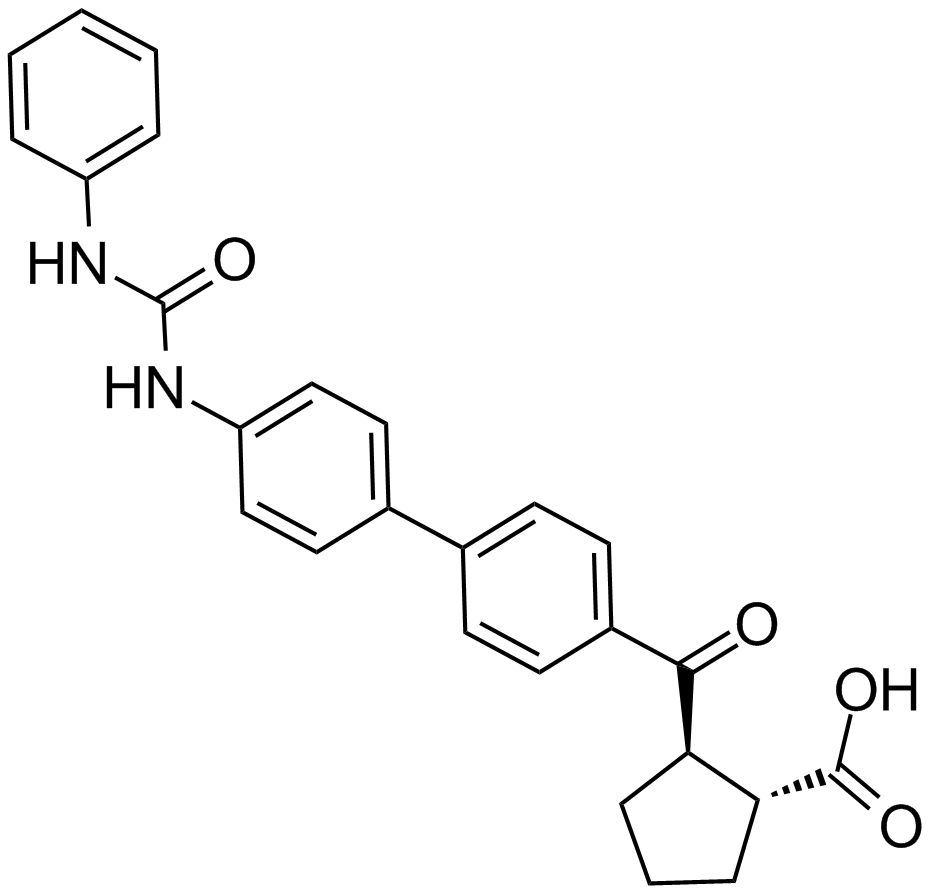
-
GC42665
AAF-CMK (trifluoroacetate salt)
NAlaAlaPheCMK, Tripeptidyl Peptidase Inhibitor II
Tripeptidyl peptidase II (TPPII) is a serine peptidase of the subtilisin-type which removes tripeptides from the free NH2 terminus of oligopeptides.
-
GC42667
Abacavir Carboxylate
Abacavir carboxylate is an inactive metabolite of the HIV-1 reverse transcriptase inhibitor abacavir.
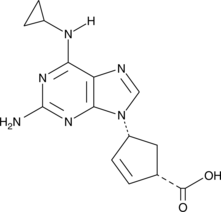
-
GC46769
Abametapir
5,5′-Dimethyl-2,2′-dipyridyl
A building block and an insecticide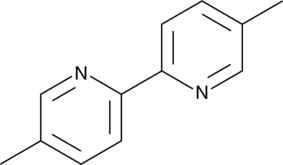
-
GC35219
Abiraterone metabolite 1
3β-OH-5α-Abi
Abiraterone metabolite 1 is a 5β-reduced metabolite of abiraterone. Abiraterone, a steroidal drug, inhibits CYP17A1, blocks androgen synthesis and prolongs survival in prostate cancer.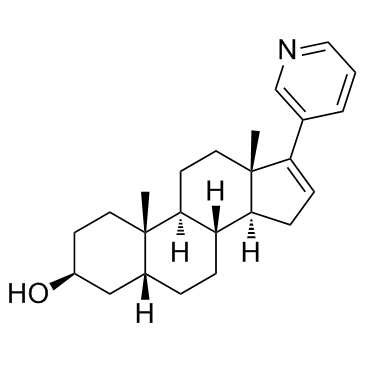
-
GC67766
Abiraterone sulfate

-
GC35221
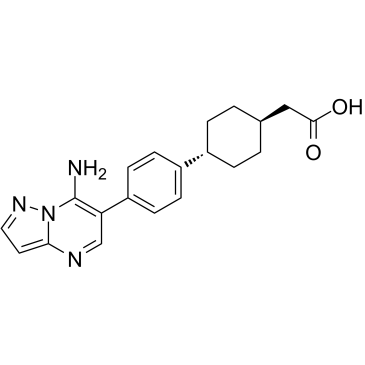
ABT-046
ABT-046 is a potent, selective, and orally active acyl CoA:diacylglycerol acyltransferase 1 (DGAT-1) inhibitor with IC50s of both 8 nM against human and mouse DGAT-1.
-
GC60548
ABT-100
ABT-100 is a potent, highly selective and orally active farnesyltransferase inhibitor. ABT-100 inhibits cell proliferation (IC50s of 2.2 nM, 3.8 nM, 5.9 nM, 6.9 nM, 9.2 nM, 70 nM and 818 nM for EJ-1, DLD-1, MDA-MB-231, HCT-116, MiaPaCa-2, PC-3, and DU-145 cells, respectively), increases apoptosis and decreases angiogenesis. ABT-100 possesses broad-spectrum antitumor activity.
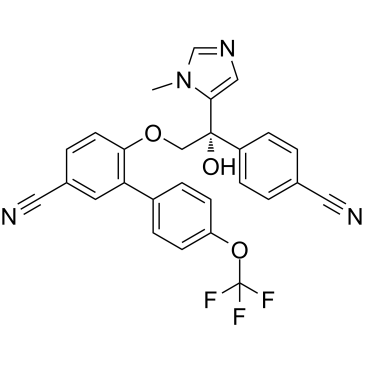
-
GC38264
Ac-Ala-OH
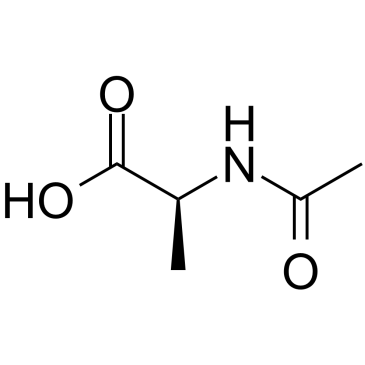
-
GA11174
Ac-Arg-OH.2H2O
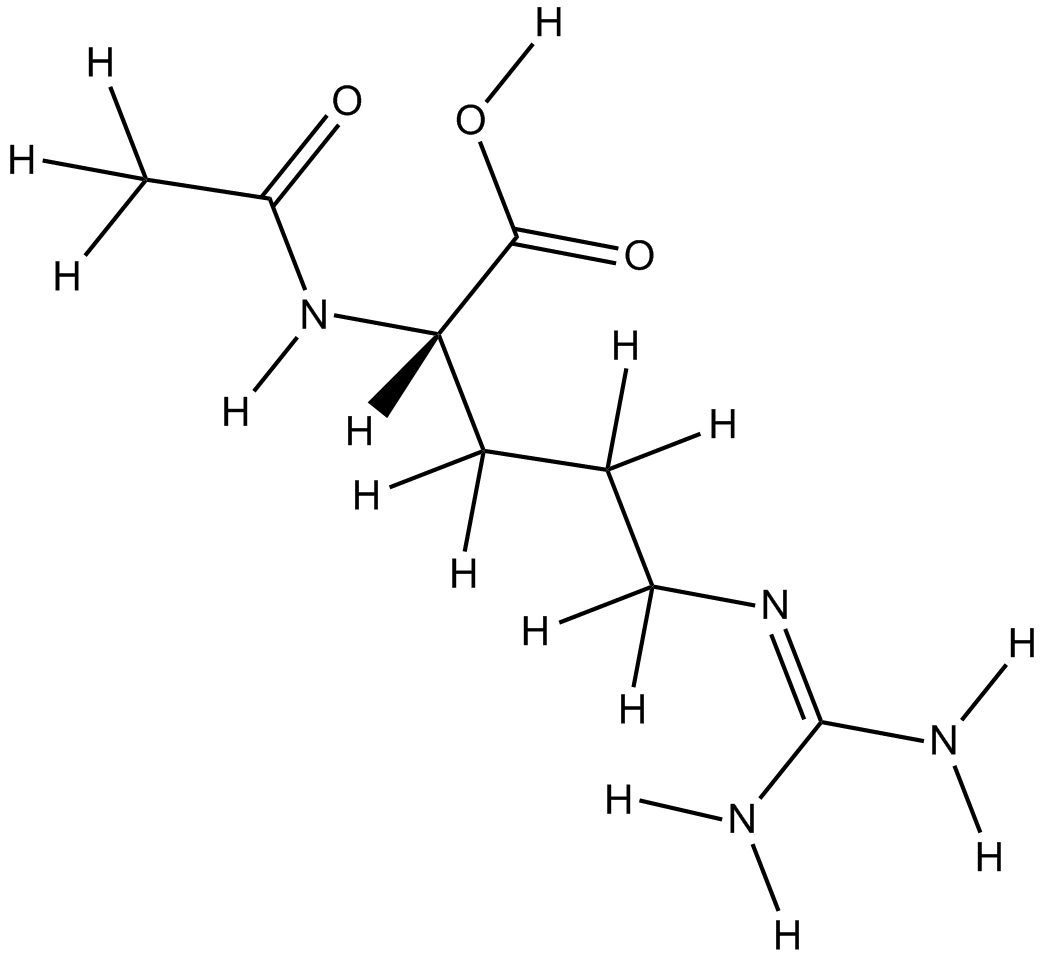
-
GA20489
Ac-Asn-OH
Ac-Asn-OH is an endogenous metabolite.

-
GC17602
Ac-DEVD-AFC
N-Acetyl-Asp-Glu-Val-Asp-7-amido-4-Trifluoromethylcoumarin,Caspase-3 Substrate (Fluorogenic)
fluorogenic substrate for activated caspase-3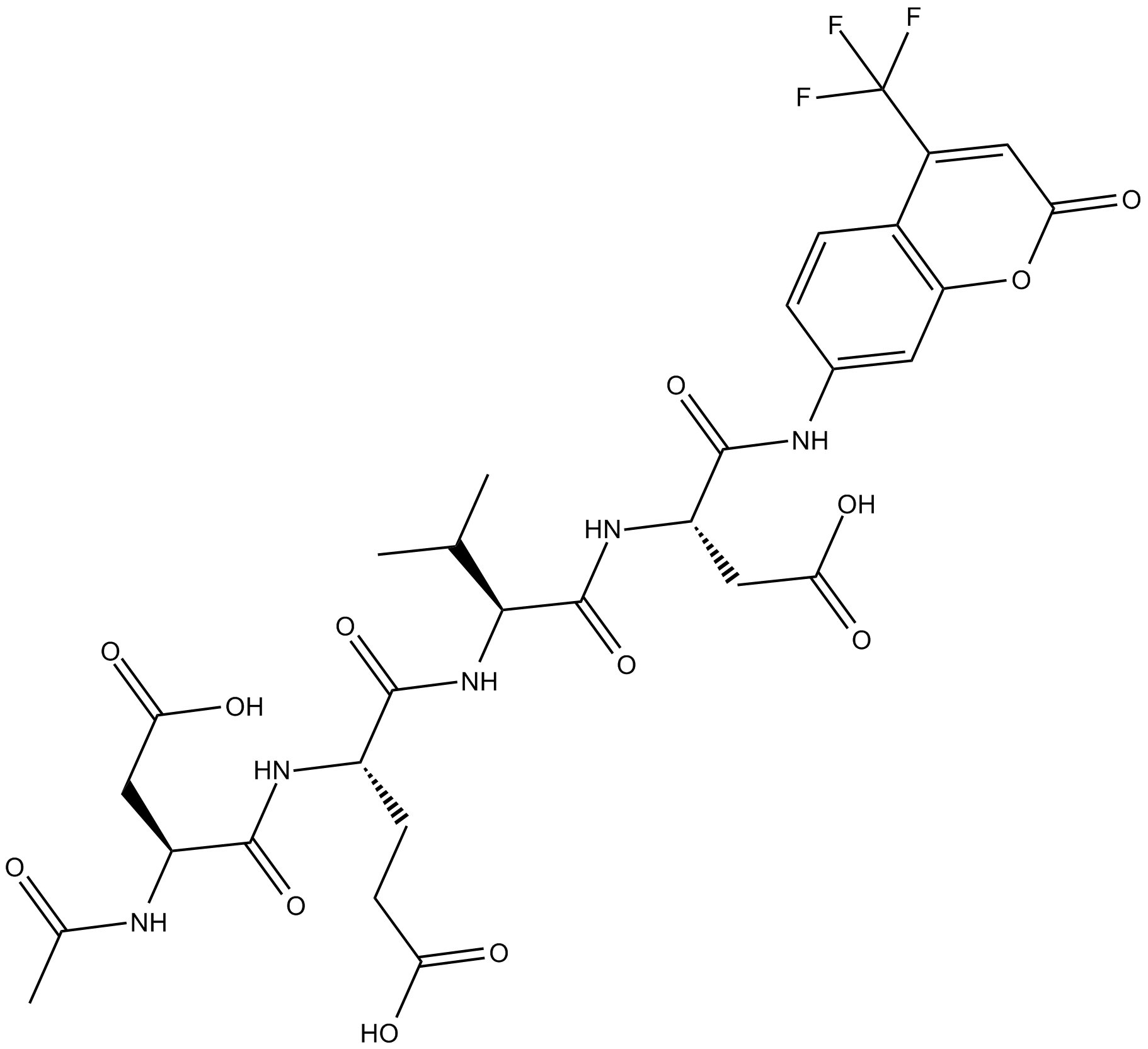
-
GC32695
Ac-DEVD-CHO
Ac-DEVD-CHO is a Caspase-3 inhibitor with an IC50 value of 0.016μM.
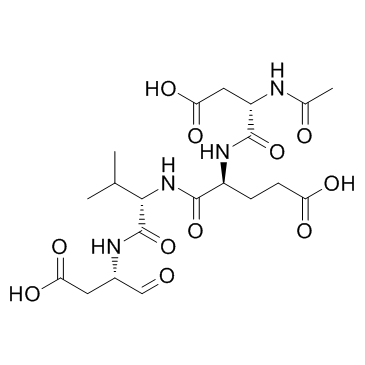
-
GC10951
Ac-DEVD-CMK
Ac-Asp-Glu-Val-Asp-CMK,Caspase-3 Inhibitor III
cell-permeable, and irreversible inhibitor of caspase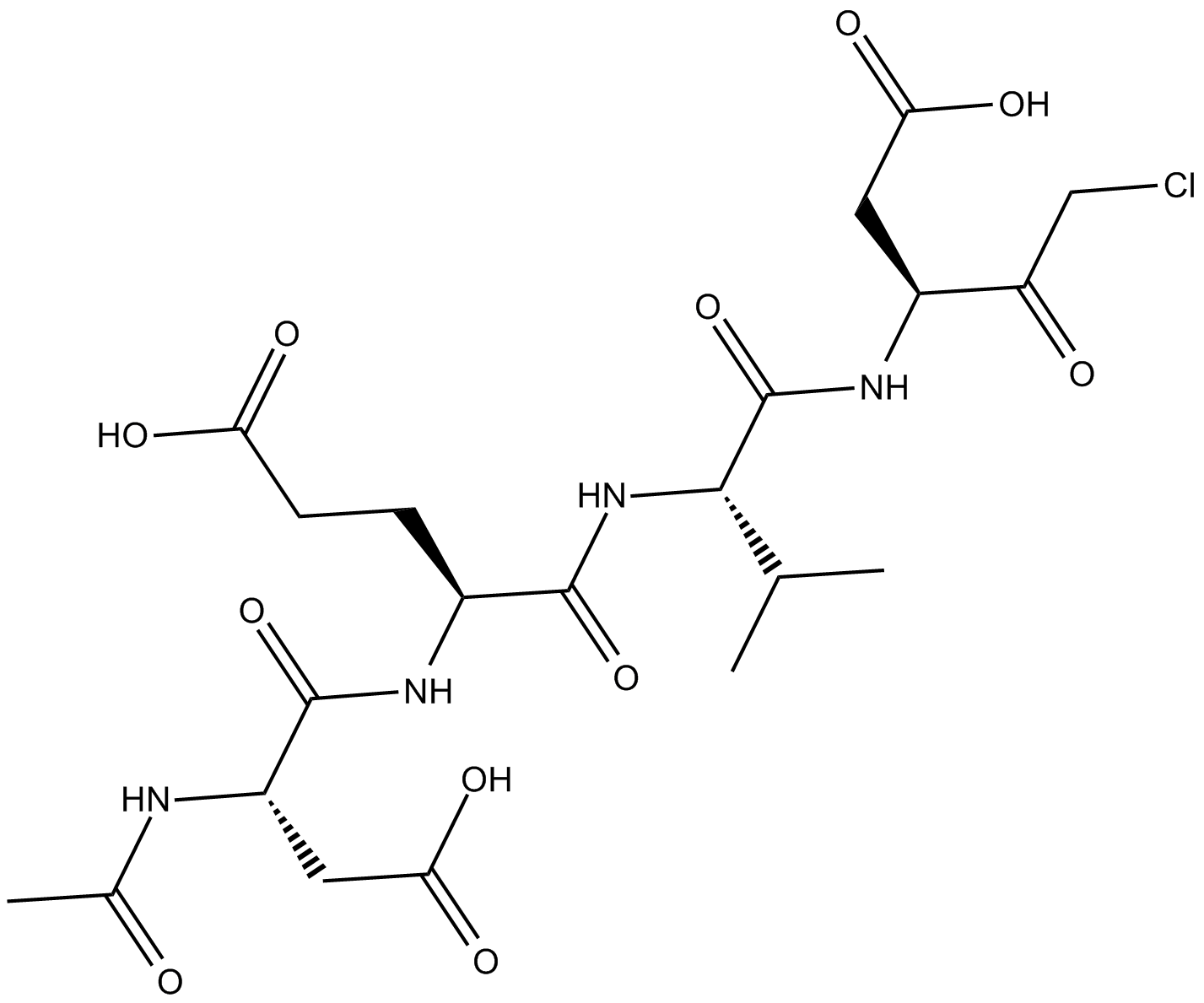
-
GA10446
Ac-DL-Trp-OH
-
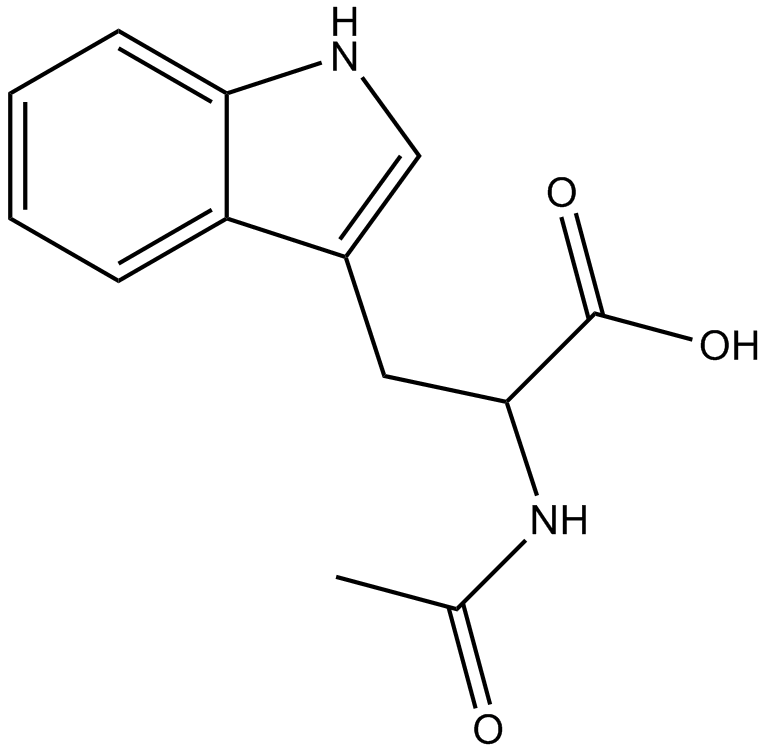
GC60558
Ac-FLTD-CMK
Ac-FLTD-CMK, a gasdermin D (GSDMD)-derived inhibitor, is a specific inflammatory caspases inhibitor.
-
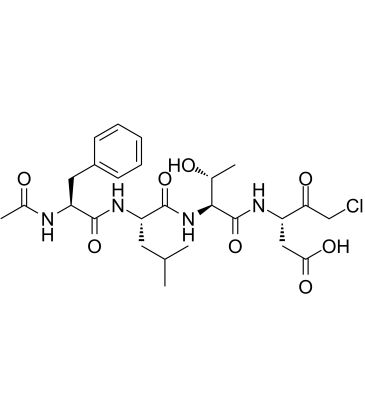
GC49704
Ac-FLTD-CMK (trifluoroacetate salt)
Ac-Phe-Leu-Thr-Asp-CMK
An inhibitor of caspase-1, -4, -5, and -11
-
GA10356
Ac-Gly-OH
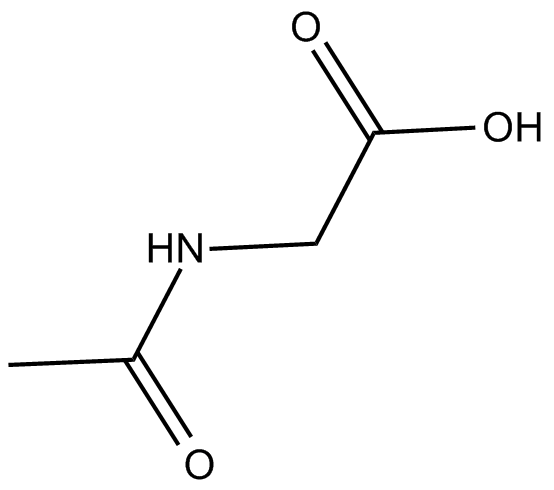
-
GA10825
Ac-His-OH
Ac-His-OH

-
GC10258
Ac-IEPD-AFC
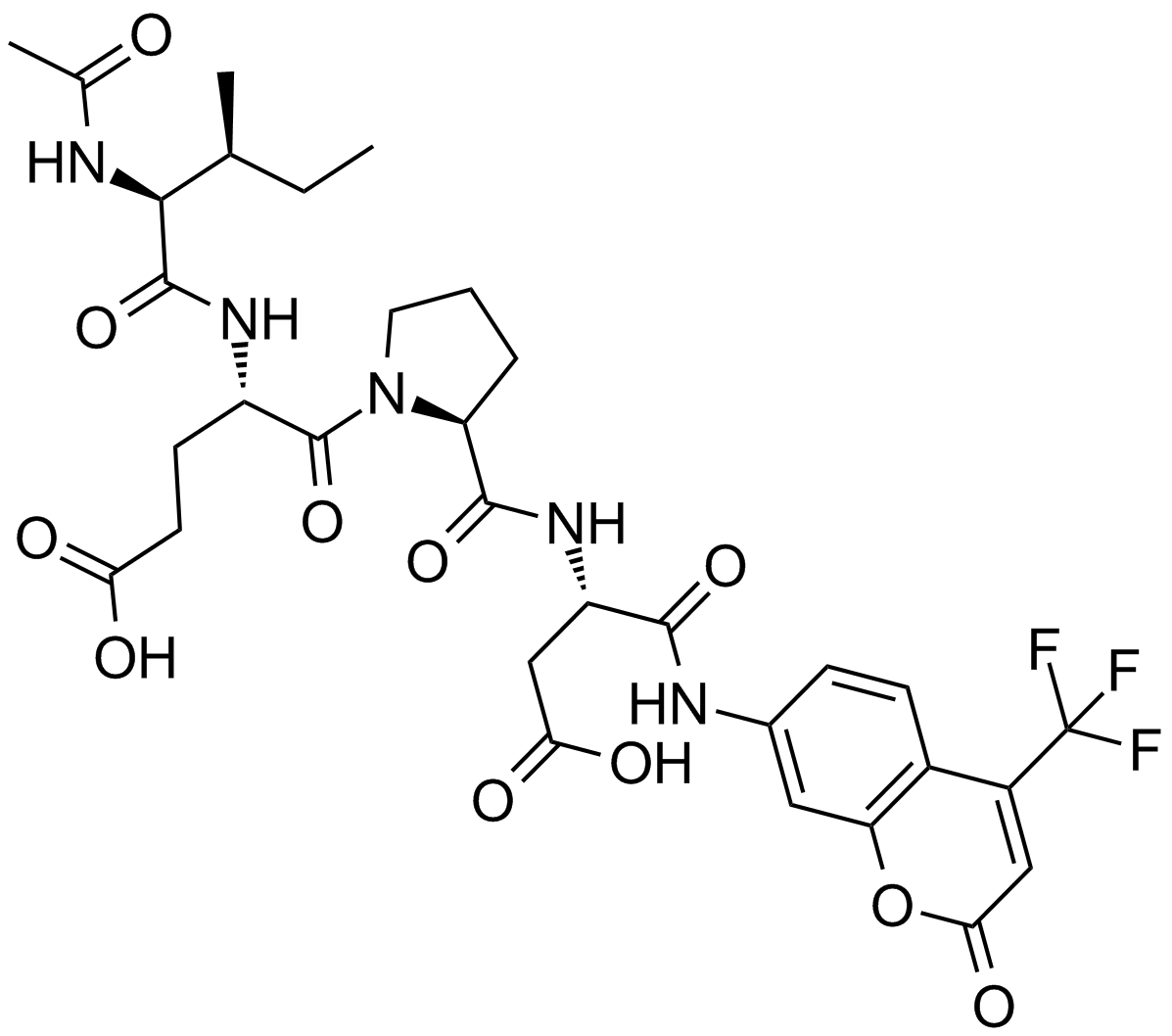
-
GC10968
Ac-LEHD-AFC
A caspase-4, -5, and -9 fluorogenic substrate
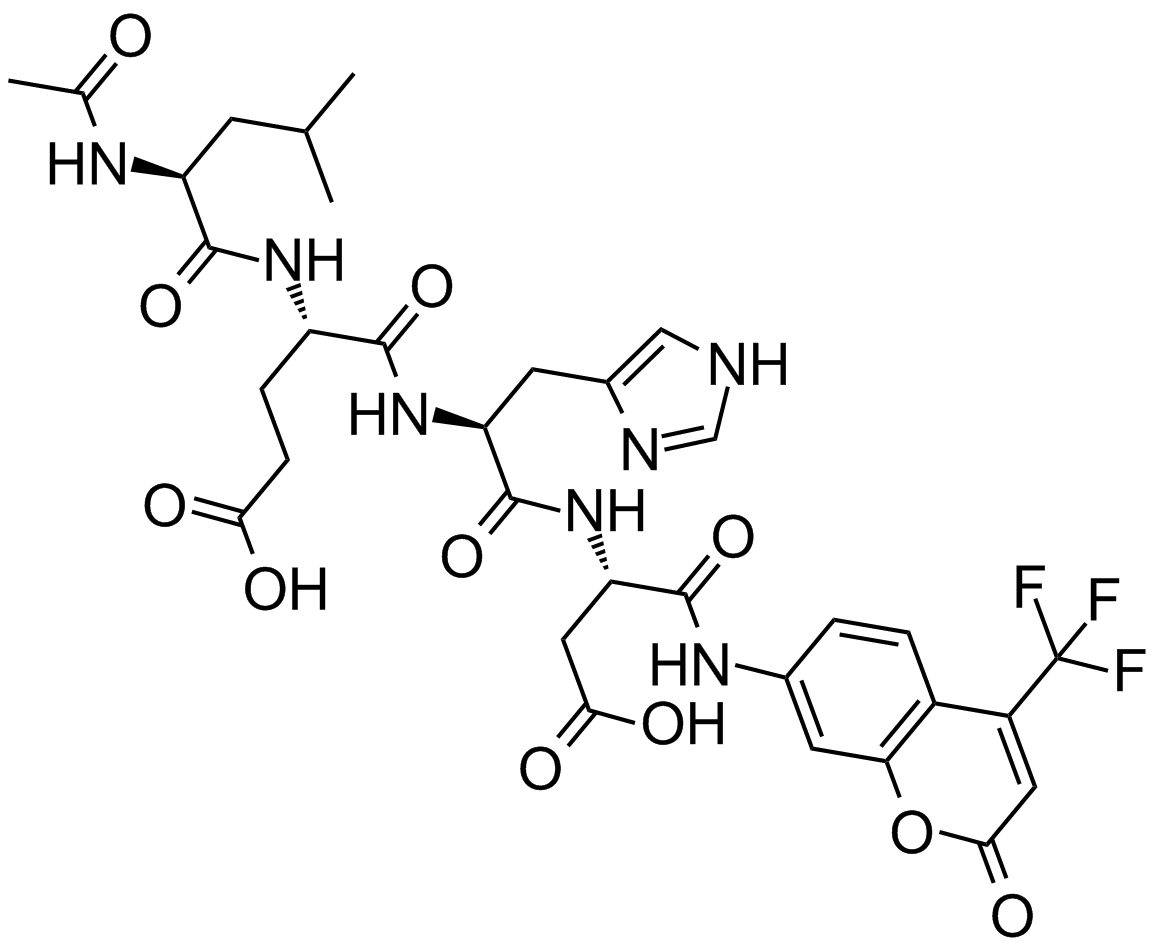
-
GA20612
Ac-Lys-OH
Ac-Lys-OH is an endogenous metabolite.

-
GA20680
Ac-Val-OH

-
GC31548
ACAT-IN-1 cis isomer
ACAT-IN-1 cis isomer is a potent ACAT inhibitor with an IC50 of 100 nM.
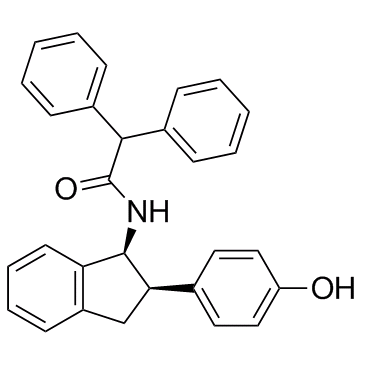
-
GC42690
Acecainide (hydrochloride)
N-Acetylprocainamide
Acecainide is an active metabolite of the class III antiarrhythmic procainamide.
-
GC35228
Aceglutamide
Aceglutamide, Acetylglutamine, N-acetyl Gln, NSC 186896
Aceglutamide (α-N-Acetyl-L-glutamine) is a psychostimulant and nootropic, used to improve memory and concentration.
-
GC46779
Acequinocyl
A naphthoquinone acaricide
-
GC38317
Acetamide
Acetamide is used as an intermediate in the synthesis of methylamine, thioacetamide, and insecticides, and as a plasticizer in leather, cloth and coatings. Acetamide has carcinogenicity.
-
GC12917
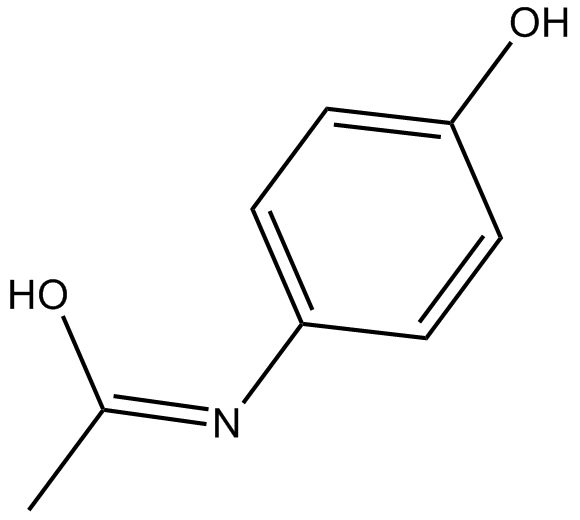
Acetaminophen
4-Acetamidophenol, APAP, 4'-Hydroxyacetanilide, NSC 3991, NSC 109028, Paracetamol
A COX inhibitor
-
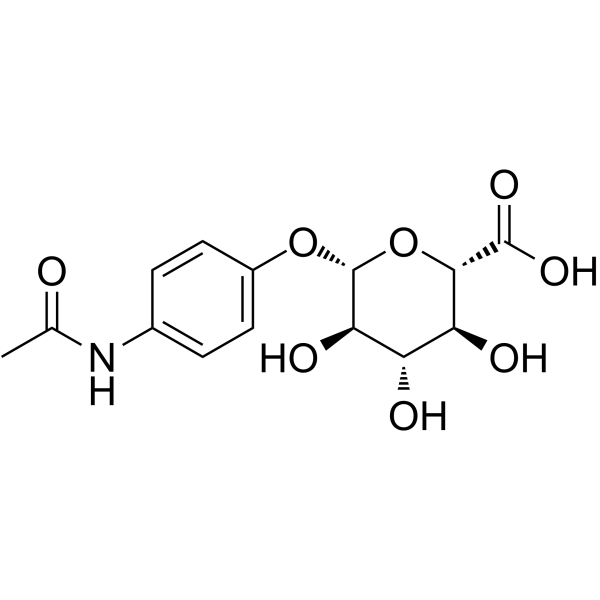
GC62824
Acetaminophen glucuronide
Acetaminophen glucuronide (APAP-glu) is an inactive glucuronide metabolite of Acetaminophen.
-
GC42693
Acetaminophen Glucuronide (sodium salt)
4-Acetamidophenol Glucuronide, APAP Glucuronide, 4'-Hydroxyacetanilide Glucuronide, Paracetamol Glucuronide
Acetaminophen glucuronide is an inactive metabolite of the analgesic and antipyretic agent acetaminophen.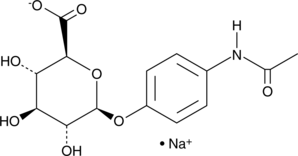
-
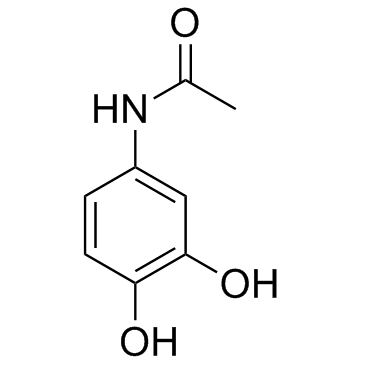
GC33558
Acetaminophen metabolite 3-hydroxy-acetaminophen (3-Hydroxyacetaminophen)
3-hydroxy-acetaminophen is a metabolite of Acetaminophen, which is a pain medicine.
-
GC42694
Acetaminophen sulfate (potassium salt)
4-Acetamidophenol sulfate, APAP sulfate, 4'-Hydroxyacetanilide sulfate, Paracetamol sulfate
Acetaminophen sulfate is a metabolite of acetaminophen.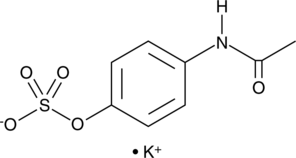
-
GC64137
Acetaminophen-d3
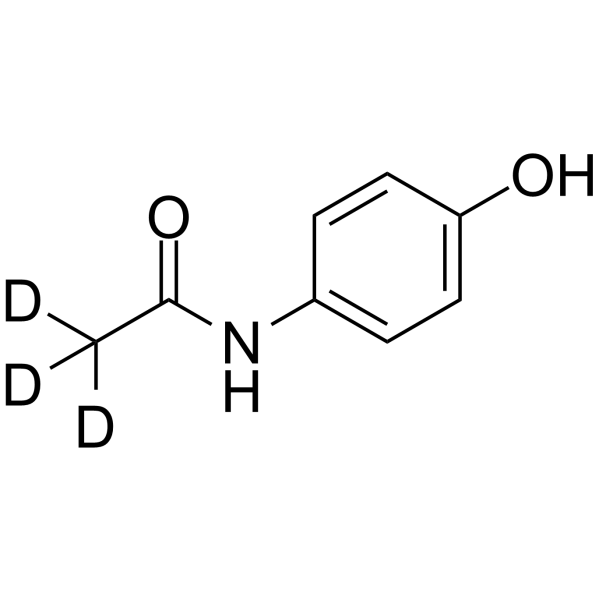
-
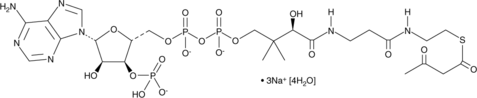
GC42695
Acetoacetyl Coenzyme A (sodium salt hydrate)
Acetoacetyl-CoA
Acetoacetyl coenzyme A (acetoacetyl-CoA) is a precursor to HMG-CoA in the isoprenoid pathway.
-
GC42697
Acetyl Coenzyme A (sodium salt)
AcetylCoA
Acetyl-coenzyme A (Acetyl-CoA) trisodium is a membrane-impermeant central metabolic intermediate, participates in the TCA cycle and oxidative phosphorylation metabolism.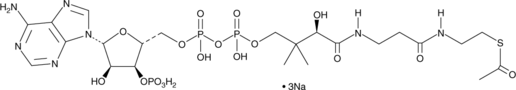
-
GC60555
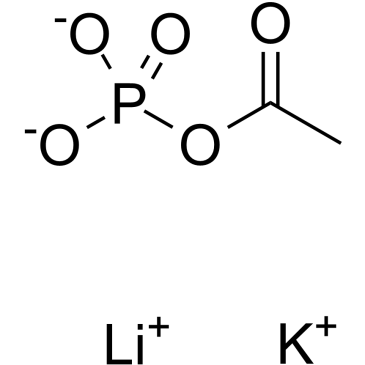
Acetyl phosphate(lithium potassium)
Acetyl phosphate(lithium potassium) is an endogenous metabolite.
-
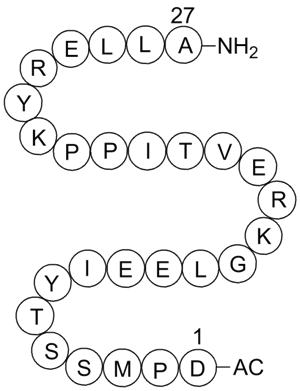
GC12152
Acetyl-Calpastatin (184-210) (human)
-
GC46789
Acetyl-L-carnitine-d3 (chloride)
ALCAR-d3, L-Acetylcarnitine-d3, C2:0 Carnitine-d3, CAR 2:0-d3, L-Carnitine acetyl ester-d3
An internal standard for the quantification of L-acetylcarnitine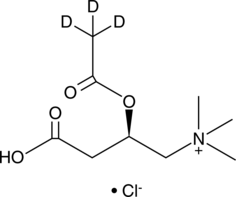
-
GC68097
Acetyl-L-carnitine-d3 hydrochloride
O-Acetyl-L-carnitine-d3 hydrochloride

-
GC14142
Acetylcholine Chloride
ACh
Major transmitter at many nervous sites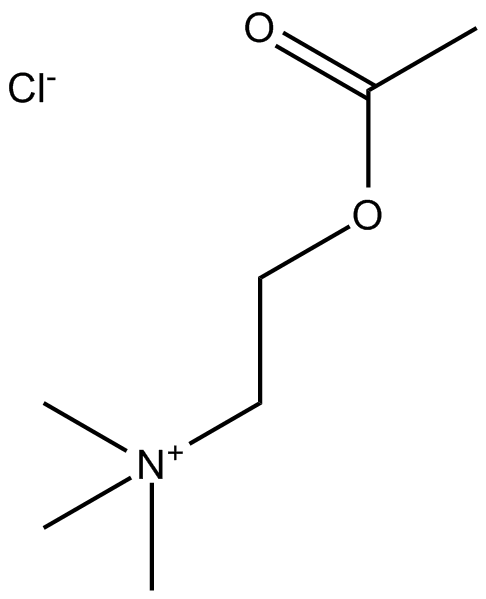
-
GC11786
Acetylcysteine
N-acetylcysteine; N-acetyl-L-cysteine; NAC; Acetadote
Acetylcysteine is the N-acetyl derivative of CYSTEINE.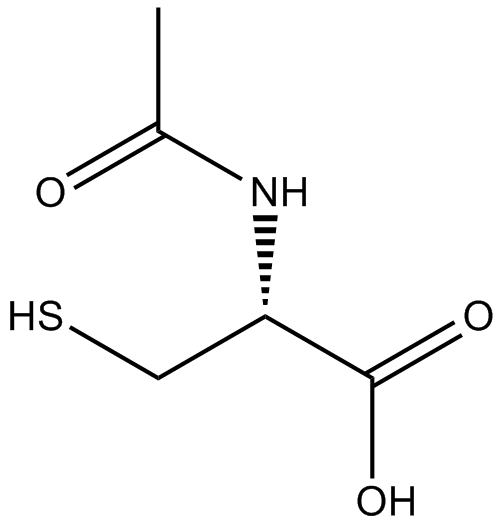
-
GC32305
ACH-806 (GS9132)
GS9132
ACH-806 (GS9132) is an NS4A antagonist which can inhibit Hepatitis C Virus (HCV) replication with an EC50 of 14 nM.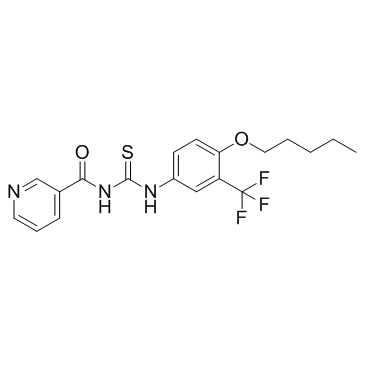
-
GC49406
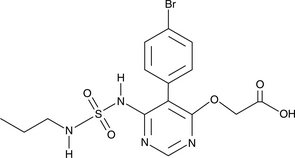
ACT-373898
A metabolite of macitentan
-
GC16350
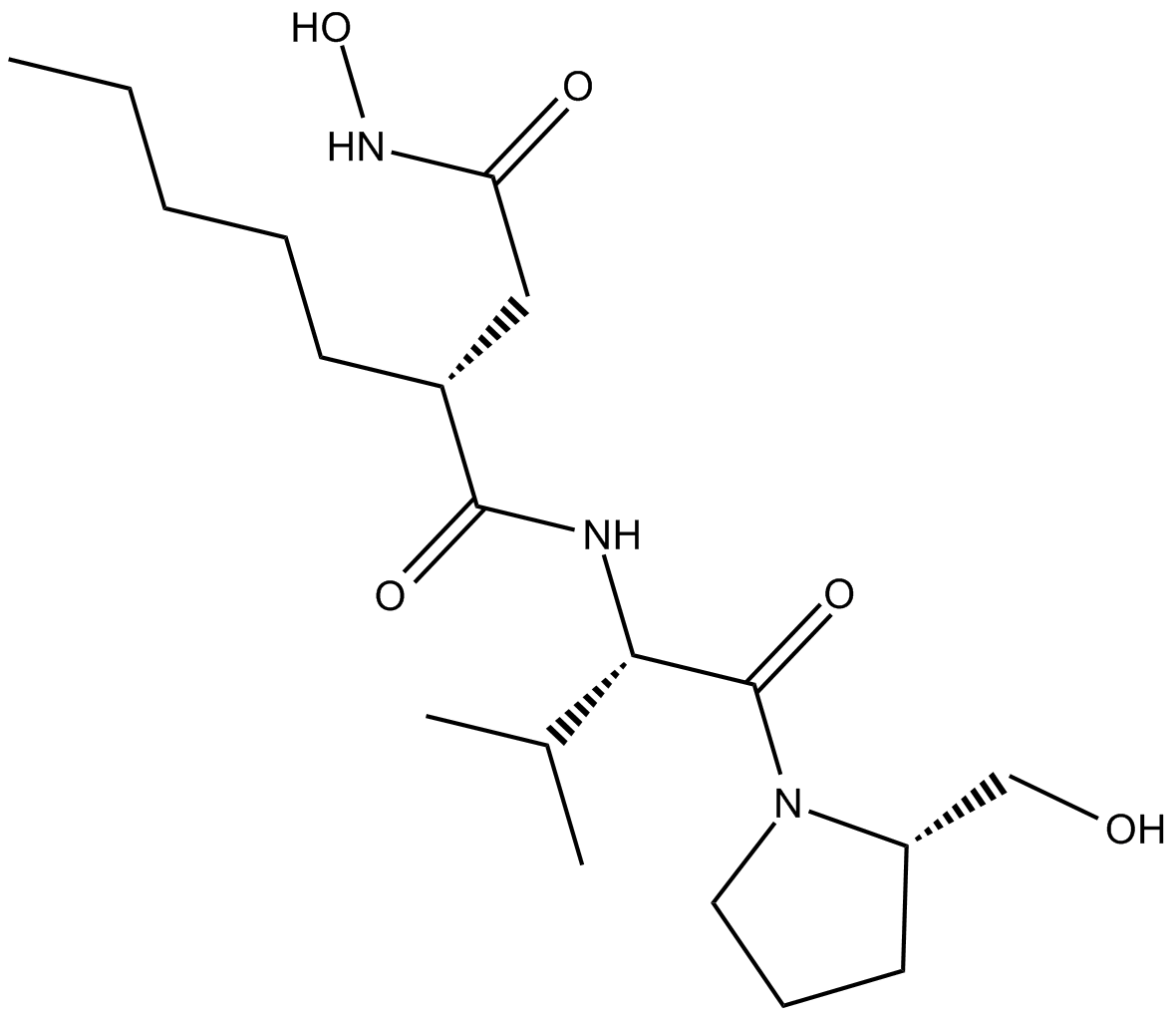
Actinonin
(-)-Actinonin,Ro 06-1467
Peptidomimetic antibiotic that inhibits aminopeptidases
-
GC46798
Adapalene-d3
An internal standard for the quantification of adapalene
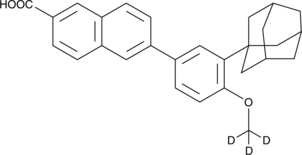
-
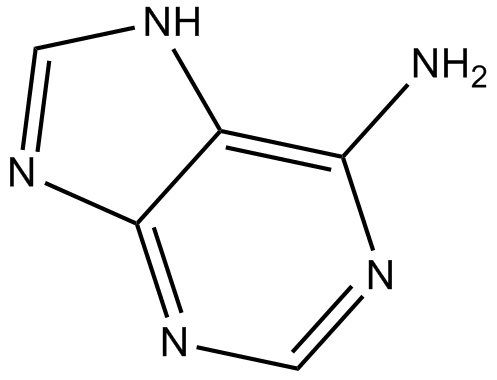
GC13432
Adenine
High affinity adenine receptor agonist
-
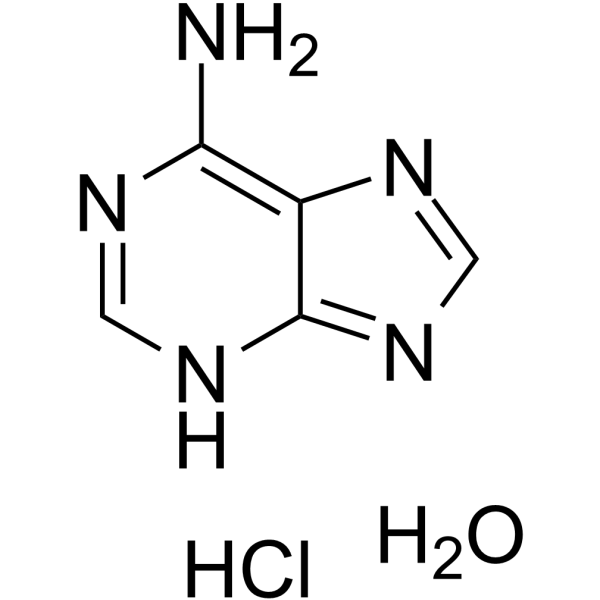
GC62828
Adenine monohydrochloride hemihydrate
Adenine monohydrochloride hemihydrate is an endogenous metabolite.
-
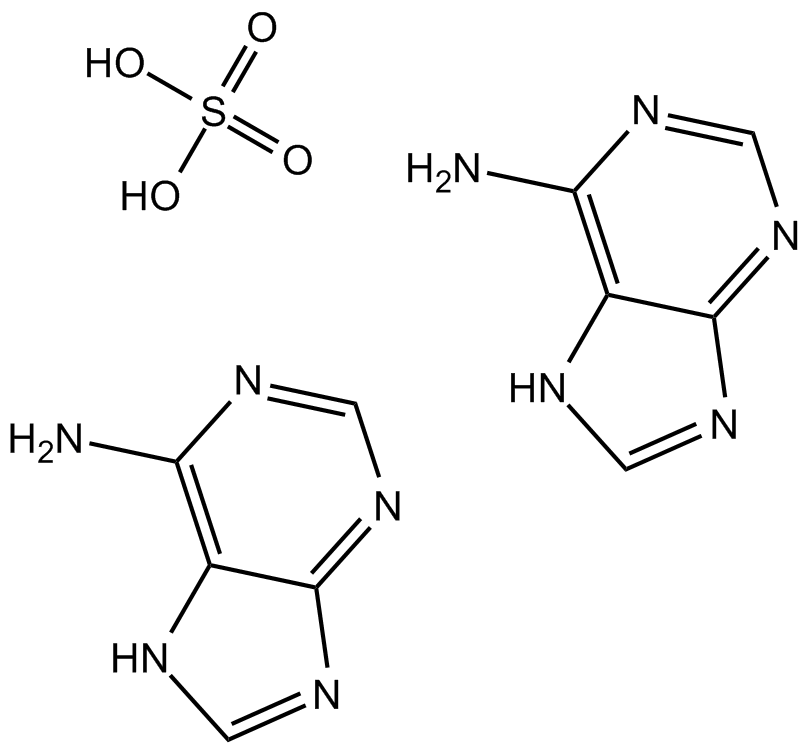
GC17278
Adenine sulfate
Adenine sulfate (6-Aminopurine hemisulfate), a purine, is one of the four nucleobases in the nucleic acid of DNA. Adenine sulfate acts as a chemical component of DNA and RNA. Adenine sulfate also plays an important role in biochemistry involved in cellular respiration, the form of both ATP and the cofactors (NAD and FAD), and protein synthesis.
-
GC14106
Adenosine
NSC 7652
nucleoside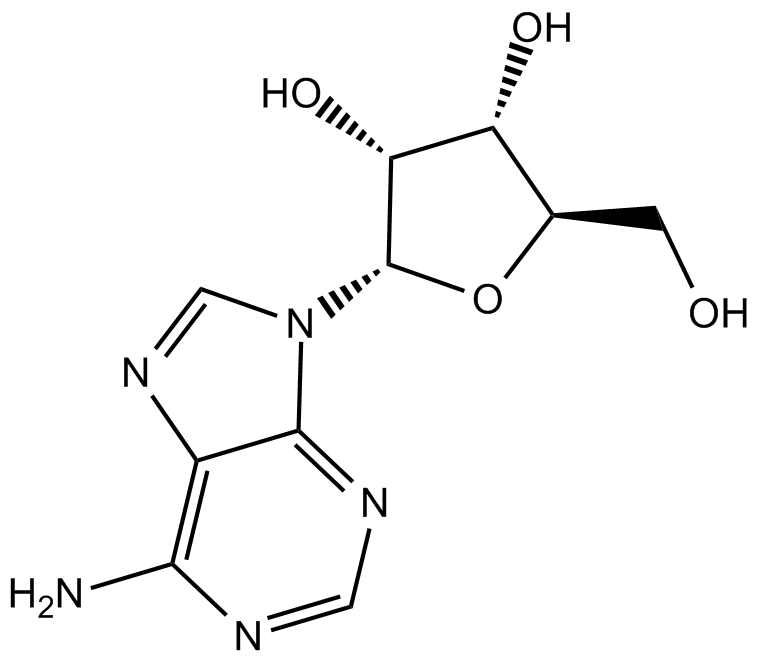
-
GC19630
Adenosine 2′-monophosphate
Adenosine 2′-monophosphate (2'-AMP) is converted by extracellular 2',3'-CAMP.
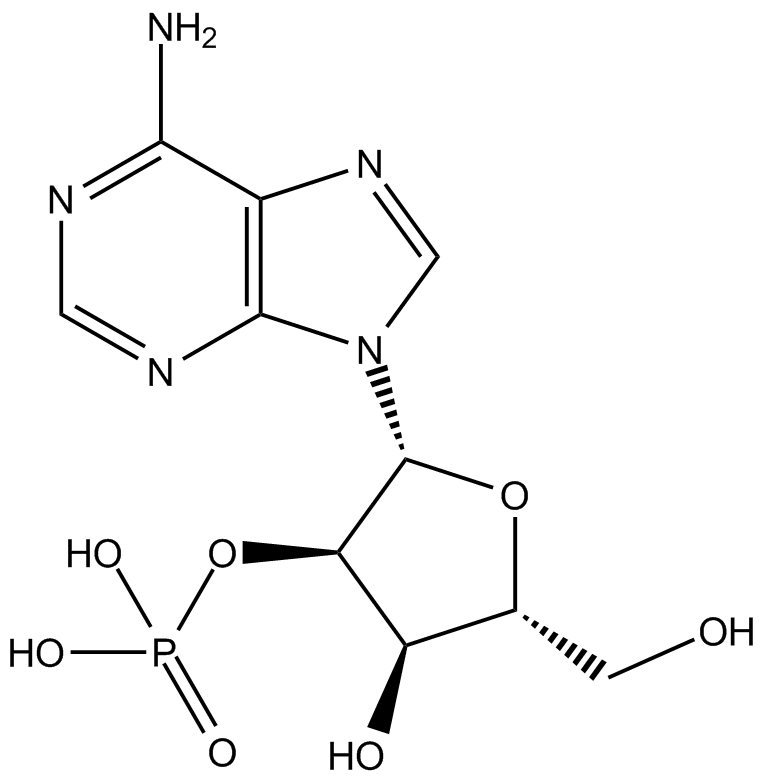
-
GC10880
Adenosine 3-5-cyclic monophosphate
Adenosine 3',5'-cyclic monophosphate, cAMP, Cyclic adenosine monophosphate, NSC 94017, NSC 143670
A second messenger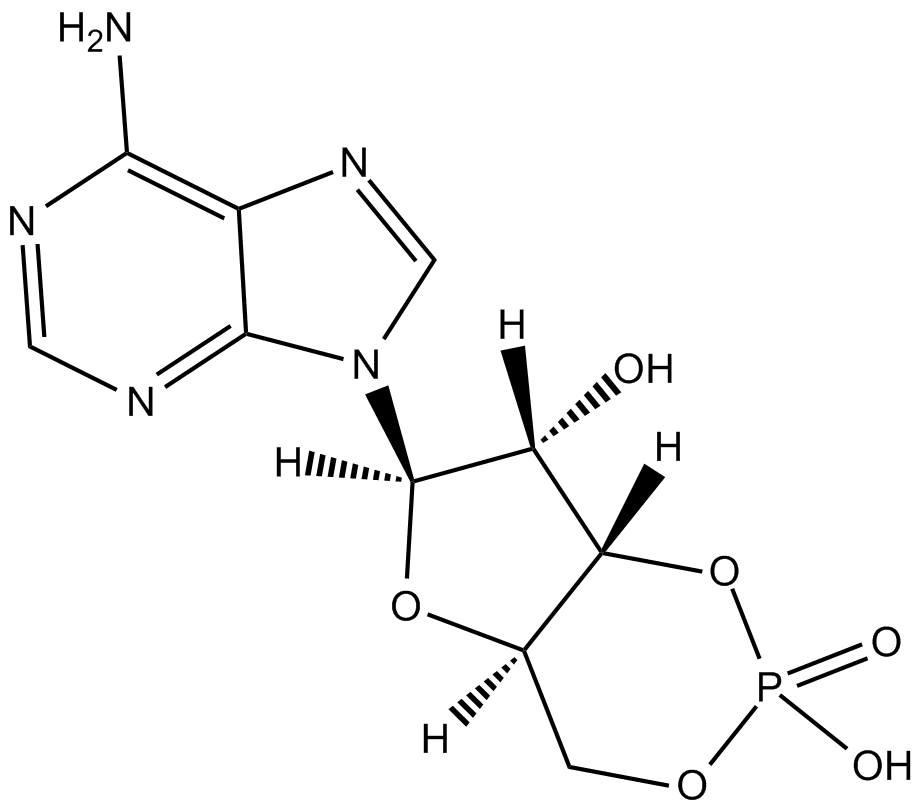
-
GC17558
Adenosine 5'-triphosphate disodium salt hydrate
ATP disodium salt hydrate
Adenosine 5'-triphosphate disodium salt hydrate (Adenosine 5'-triphosphatedisodium salt hydrate) is a central component of energy storage and metabolism in vivo, provides the metabolic energy to drive metabolic pumps and serves as a coenzyme in cells.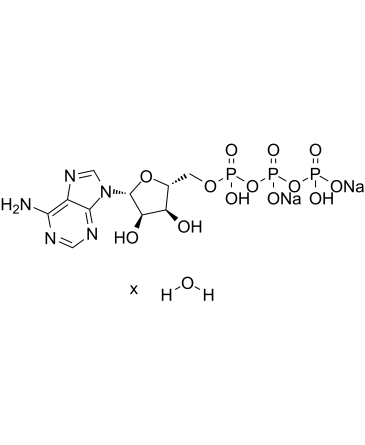
-
GC13172
Adenosine 5-monophosphate
AMP
Adenosine 5-monophosphate is a key cellular metabolite regulating energy homeostasis and signal transduction.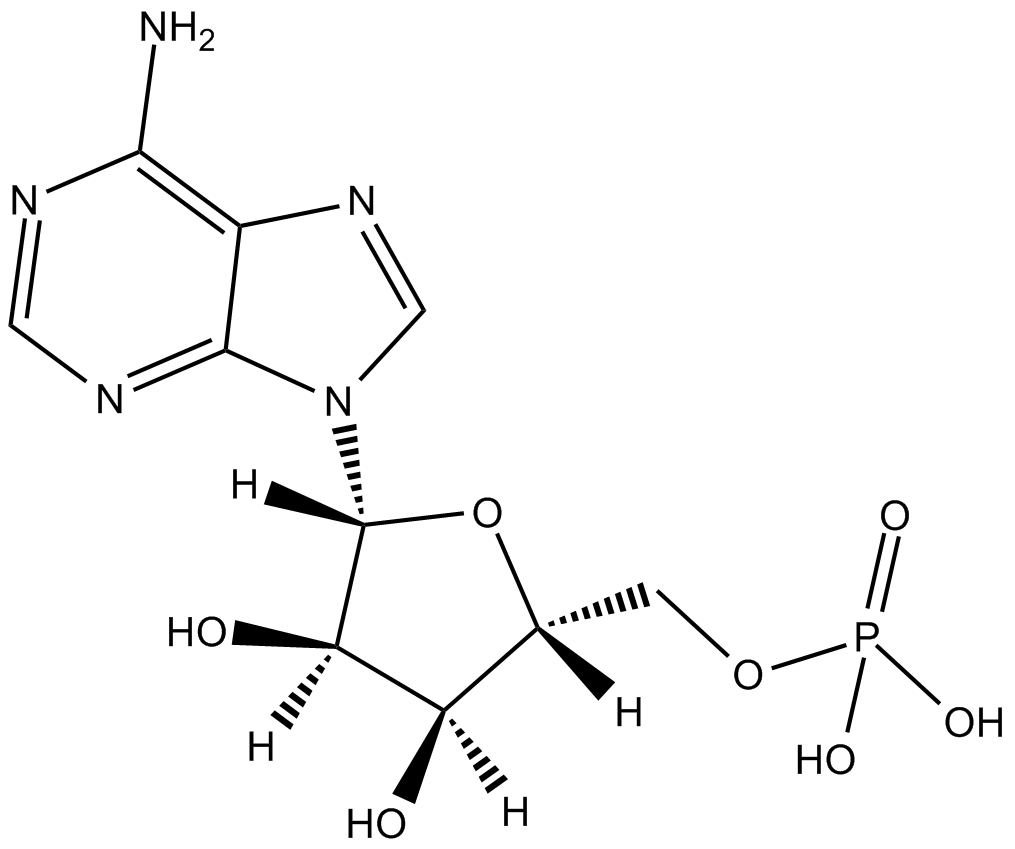
-
GC11969
Adenosine-5'-diphosphate
Adenosine Pyrophosphate,ADP,5′-ADP
Agonist of purinergic receptors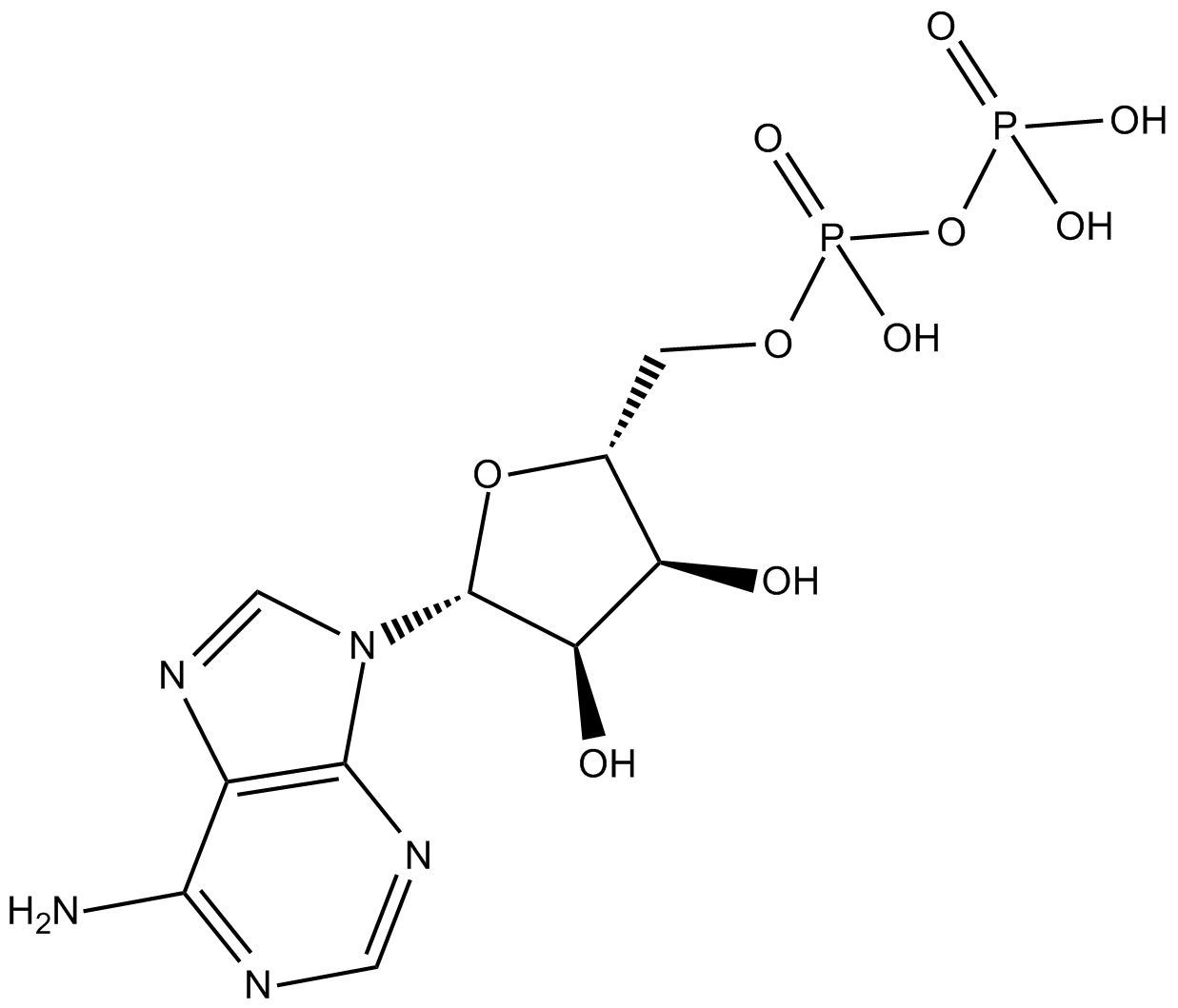
-
GC64916
Adenylosuccinic acid tetraammonium
Adenylosuccinate tetraammonium; Aspartyl adenylate tetraammonium
Adenylosuccinic acid tetraammonium (Adenylosuccinate; Aspartyl adenylate) is a purine ribonucleoside monophosphate and plays a role in nucleotide cycle metabolite.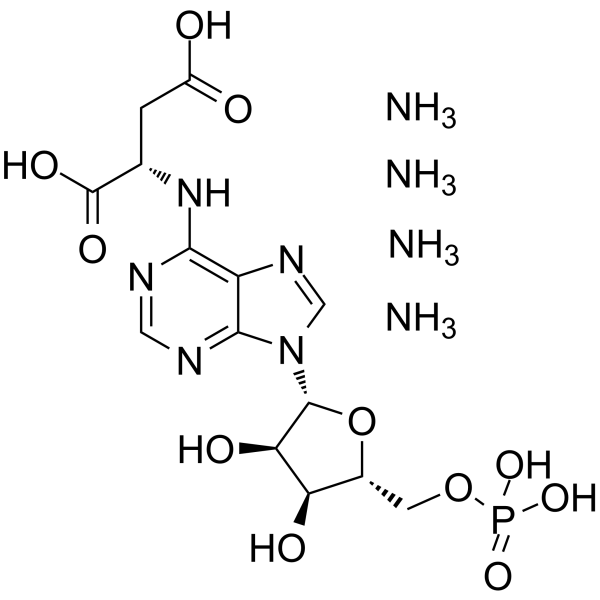
-
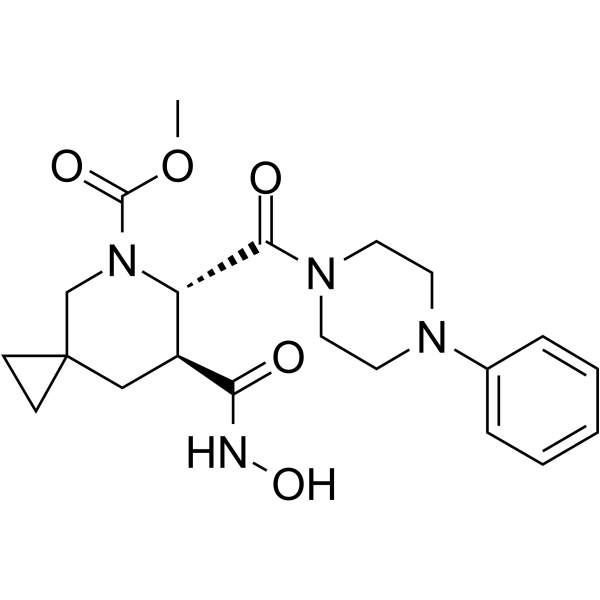
GC63661
Aderbasib
Aderbasib (INCB007839) is a potent, orally active and target specific low nanomolar hydroxamate-based inhibitor of ADAM10 and ADAM17. Aderbasib exhibits robust antineoplastic activity and can be used for cancer research, including diffuse large B-cell non-Hodgkin lymphoma, HER2+?breast cancer, gliomas, et al.
-
GC33805
Adipic acid
Adipic acid is found to be associated with HMG-CoA lyase deficiency, carnitine-acylcarnitine translocase deficiency, malonyl-Coa decarboxylase deficiency, and medium Chain acyl-CoA dehydrogenase deficiency, which are inborn errors of metabolism.
-
GC19021
Adjudin
AF-2364
Adjudin is an extensively studied male contraceptive with a superior mitochondria-inhibitory effect.
-
GC49216
ADL 08-0011 (hydrochloride)
An active metabolite of alvimopan