DNA Damage/DNA Repair
- MTH1(4)
- PARP(84)
- ATM/ATR(30)
- DNA Alkylating(20)
- DNA Ligases(3)
- DNA Methyltransferase(25)
- DNA-PK(30)
- HDAC(136)
- Nucleoside Antimetabolite/Analogue(140)
- Telomerase(17)
- Topoisomerase(151)
- tankyrase(5)
- Antifolate(38)
- CDK(250)
- Checkpoint Kinase (Chk)(31)
- CRISPR/Cas9(9)
- Deubiquitinase(72)
- DNA Alkylator/Crosslinker(71)
- DNA/RNA Synthesis(400)
- Eukaryotic Initiation Factor (eIF)(23)
- IRE1(23)
- LIM Kinase (LIMK)(9)
- TOPK(5)
- Casein Kinase(61)
- DNA Intercalating Agents(7)
- DNA/RNA Oxidative Damage(12)
Products for DNA Damage/DNA Repair
- Cat.No. Product Name Information
-
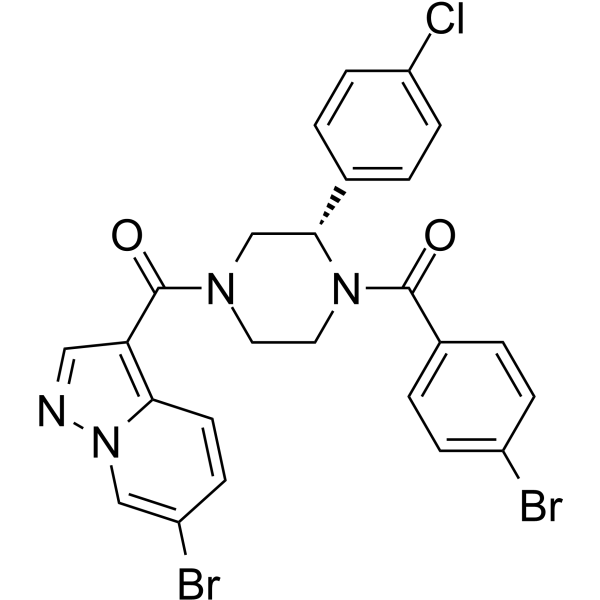
GC35761
CX-5461 dihydrochloride
CX-5461 dihydrochloride is a potent and orally bioavailable inhibitor of Pol I-mediated rRNA synthesis, with IC50s of 142 nM in HCT-116, 113 nM in A375, and 54 nM in MIA PaCa-2 cells, and shows little or no effect on Pol II (IC50 ≥25 μM).
-
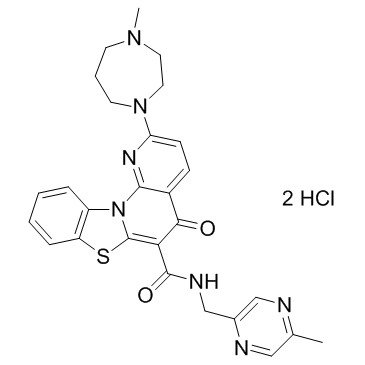
GC32700
CYC065
CYC065 (CYC065) is a second-generation, orally available ATP-competitive inhibitor of CDK2/CDK9 kinases with IC50s of 5 and 26 nM, respectively.
-
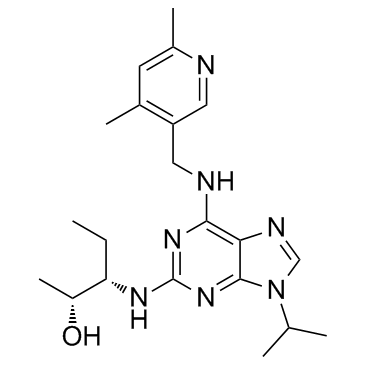
GC34309
Cycloguanil D6 (Chlorguanide triazine D6)
-
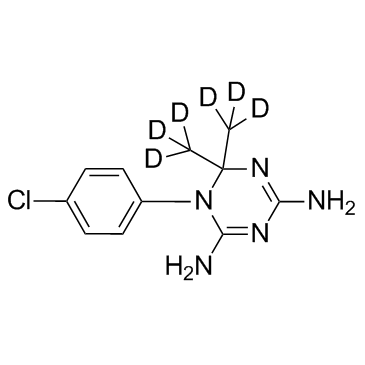
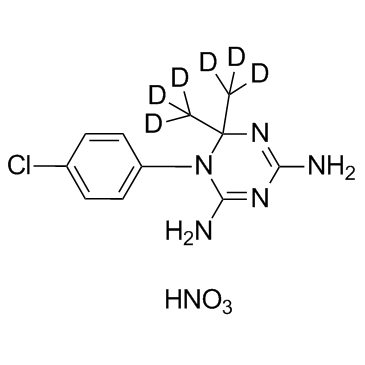
GC35781
Cycloguanil D6 Nitrate
Cycloguanil D6 Nitrate is the deuterium labeled Cycloguanil, which is a dihydrofolate reductase inhibitor.
-
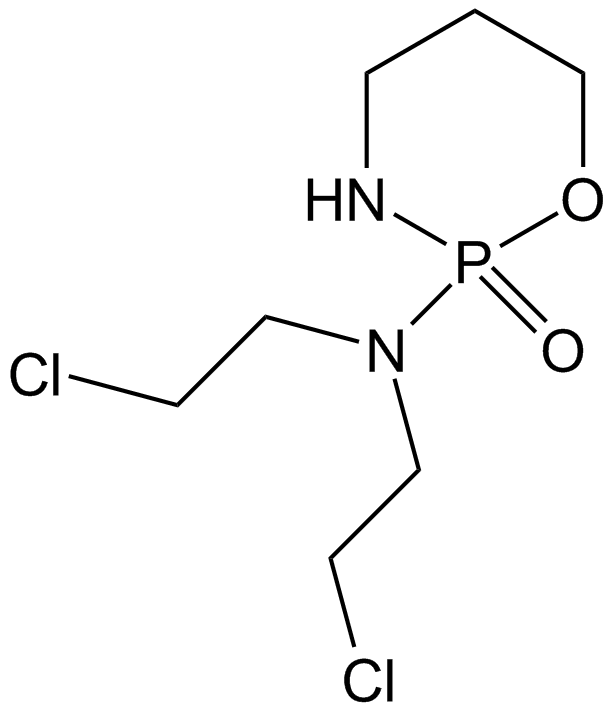
GC11145
Cyclophosphamide
Cyclophosphamide is a frequently used chemotherapy, often in combination with other chemotherapy types, for the treatment of breast cancer, malignant lymphomas, multiple myeloma, and neuroblastoma
-
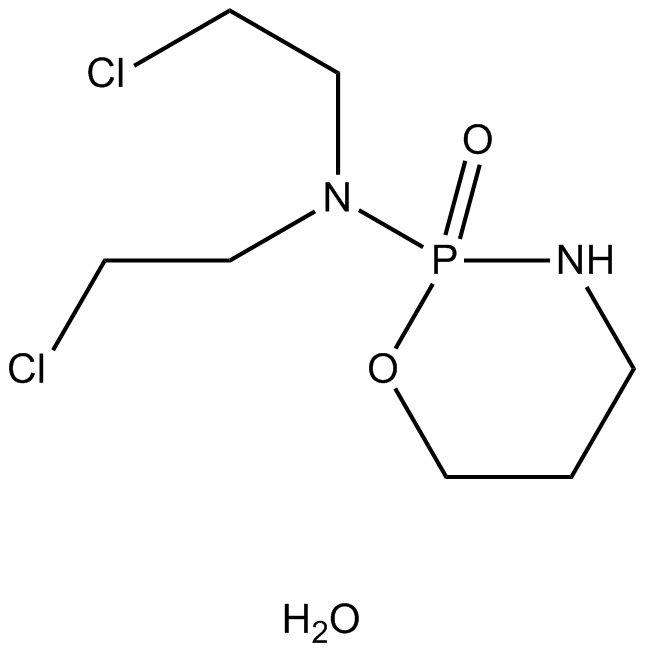
GC14077
Cyclophosphamide monohydrate
An alkylating agent
-
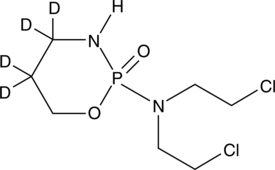
GC47148
Cyclophosphamide-d4
An internal standard for the quantification of cyclophosphamide
-
GC43351
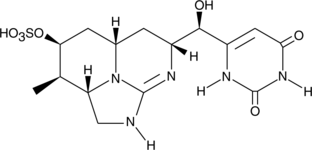
Cylindrospermopsin
Cylindrospermopsin, a tricyclic uracil derivative, is a cyanobacterial toxin that was first discovered in an algal bloom contaminating a local drinking supply on Palm Island in Queensland, Australia after an outbreak of a mysterious disease.
-
GC13070
Cytarabine
Cytotoxic agent, blocks DNA synthesis
-
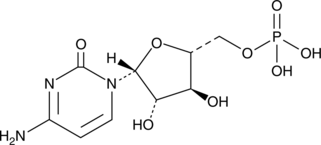
GC49863
Cytarabine 5′-monophosphate
An active metabolite of cytarabine
-
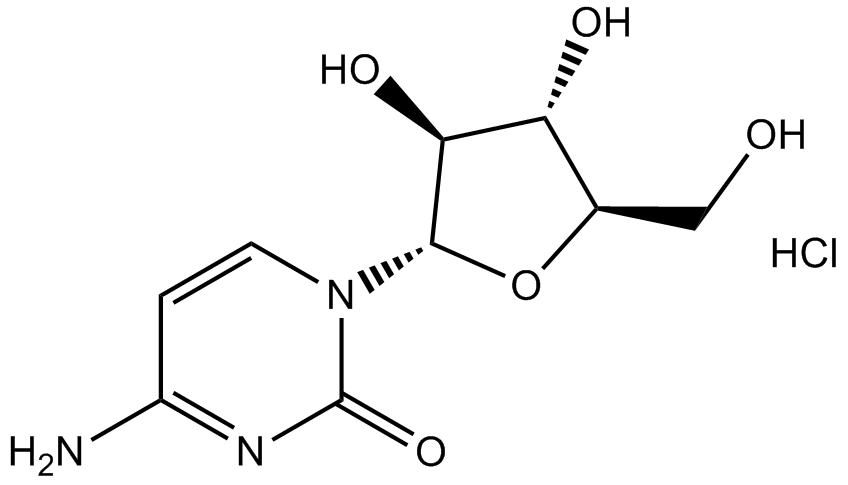
GC14961
Cytarabine hydrochloride
DNA synthsis inhibitor
-
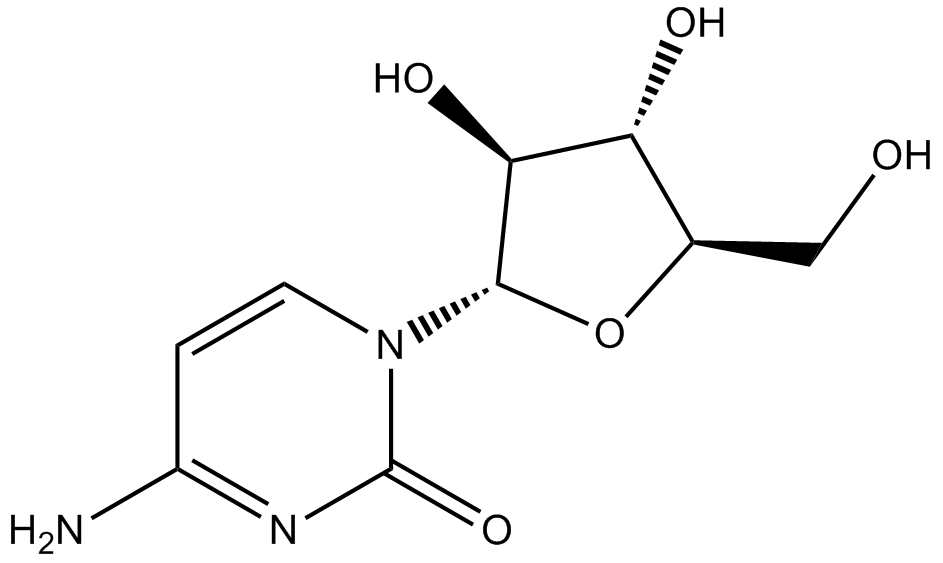
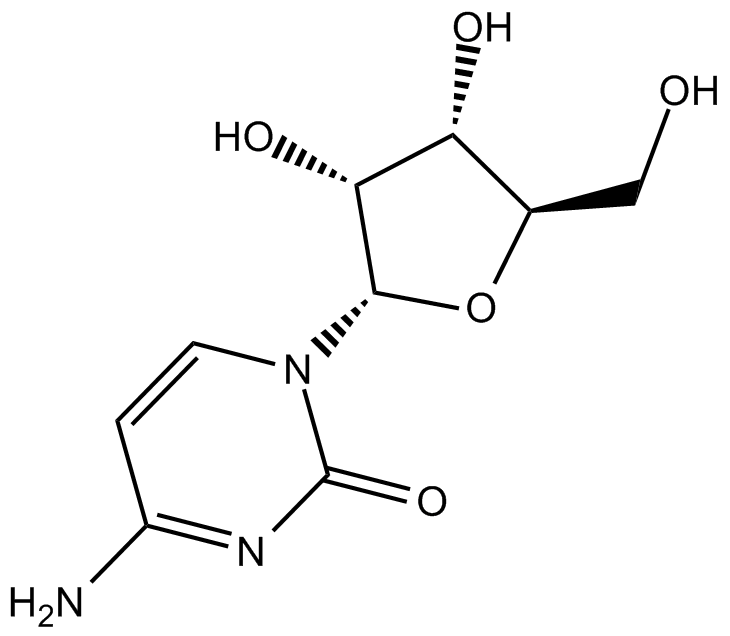
GC13729
Cytidine
pyrimidine nucleoside
-
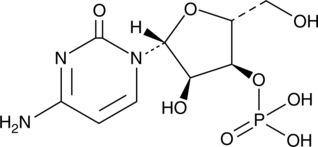
GC49104
Cytidine 3’-monophosphate
A ribonucleotide
-
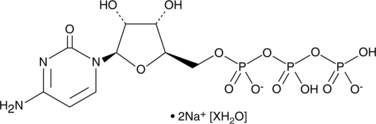
GC47162
Cytidine 5'-triphosphate (sodium salt hydrate)
A pyrimidine nucleoside triphosphate
-
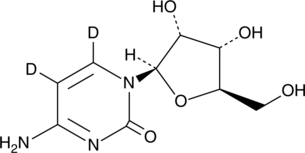
GC49526
Cytidine-d2
An internal standard for the quantification of cytidine
-
GC49464
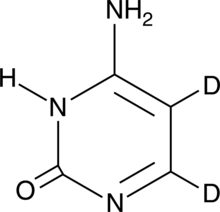
Cytosine-d2
An internal standard for the quantification of cytosine
-
GC39149
D-I03
D-I03 is a selective RAD52 inhibitor with a Kd of 25.8 ?M. D-I03 specifically inhibits RAD52-dependent single-strand annealing (SSA) and D-loop formation with IC50s of 5 ?M and 8 ?M, respectively. D-I03 suppresses growth of BRCA1- and BRCA2-deficient cells and inhibits formation of damage-induced RAD52 foci, but does not effect on RAD51 foci induced by Cisplatin.
-
GC47276
D-threo-PPMP (hydrochloride)
A GlyCer synthetase inhibitor
-
GC13202
D4476
Inhibitor of CK1 and ALK5
-
GC14485
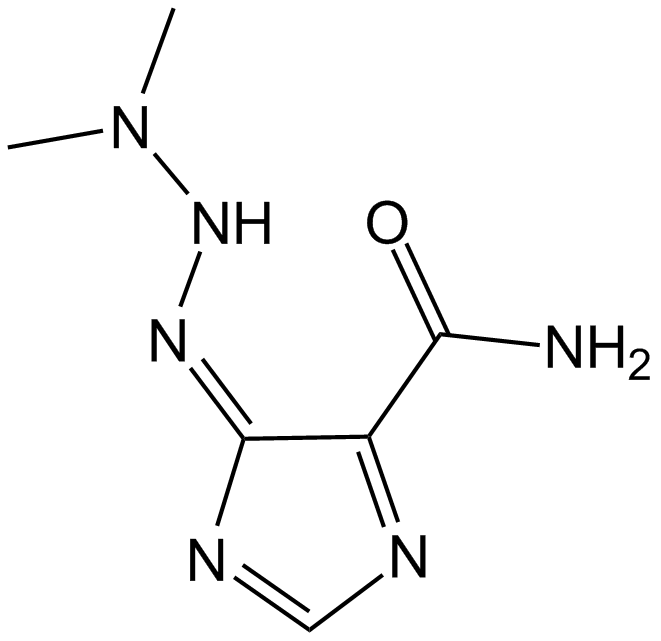
Dacarbazine
Antineoplastic( malignant melanoma and sarcomas)
-
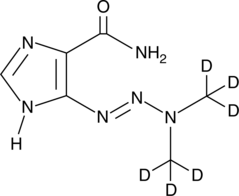
GC47167
Dacarbazine-d6
An internal standard for the quantification of dacarbazine
-
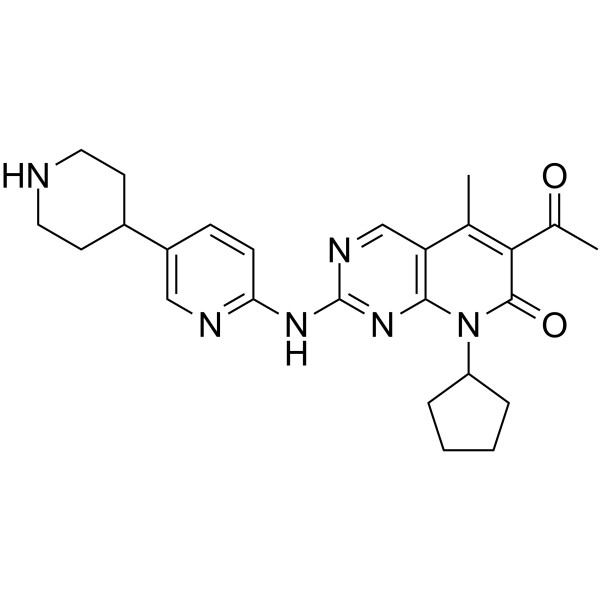
GC63419
Dalpiciclib
Dalpiciclib (SHR-6390) is an orally active and highly selective inhibitor of CDK4 and 6 with IC50 values of 12.4?nM and 9.9?nM, respectively. Dalpiciclib shows antitumor activity against breast cancer and esophageal squamous cell carcinoma.
-
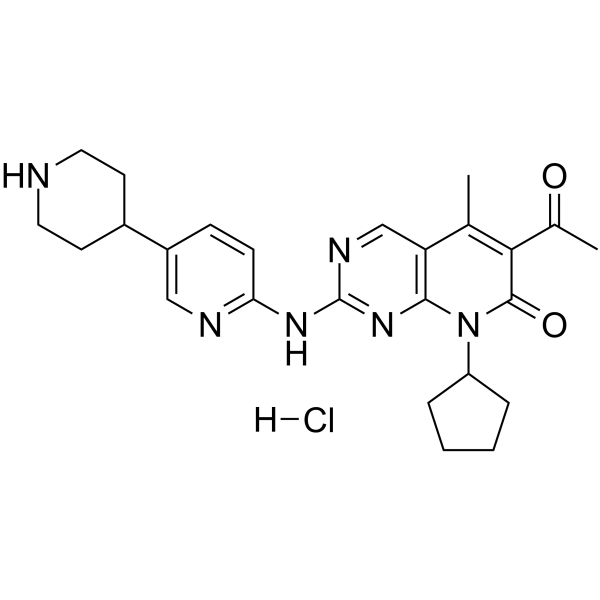
GC65369
Dalpiciclib hydrochloride
Dalpiciclib (SHR-6390) hydrochloride is an orally active and highly selective inhibitor of CDK4 and 6 with IC50 values of 12.4?nM and 9.9?nM, respectively. Dalpiciclib hydrochloride shows antitumor activity against breast cancer and esophageal squamous cell carcinoma.
-
GC10214
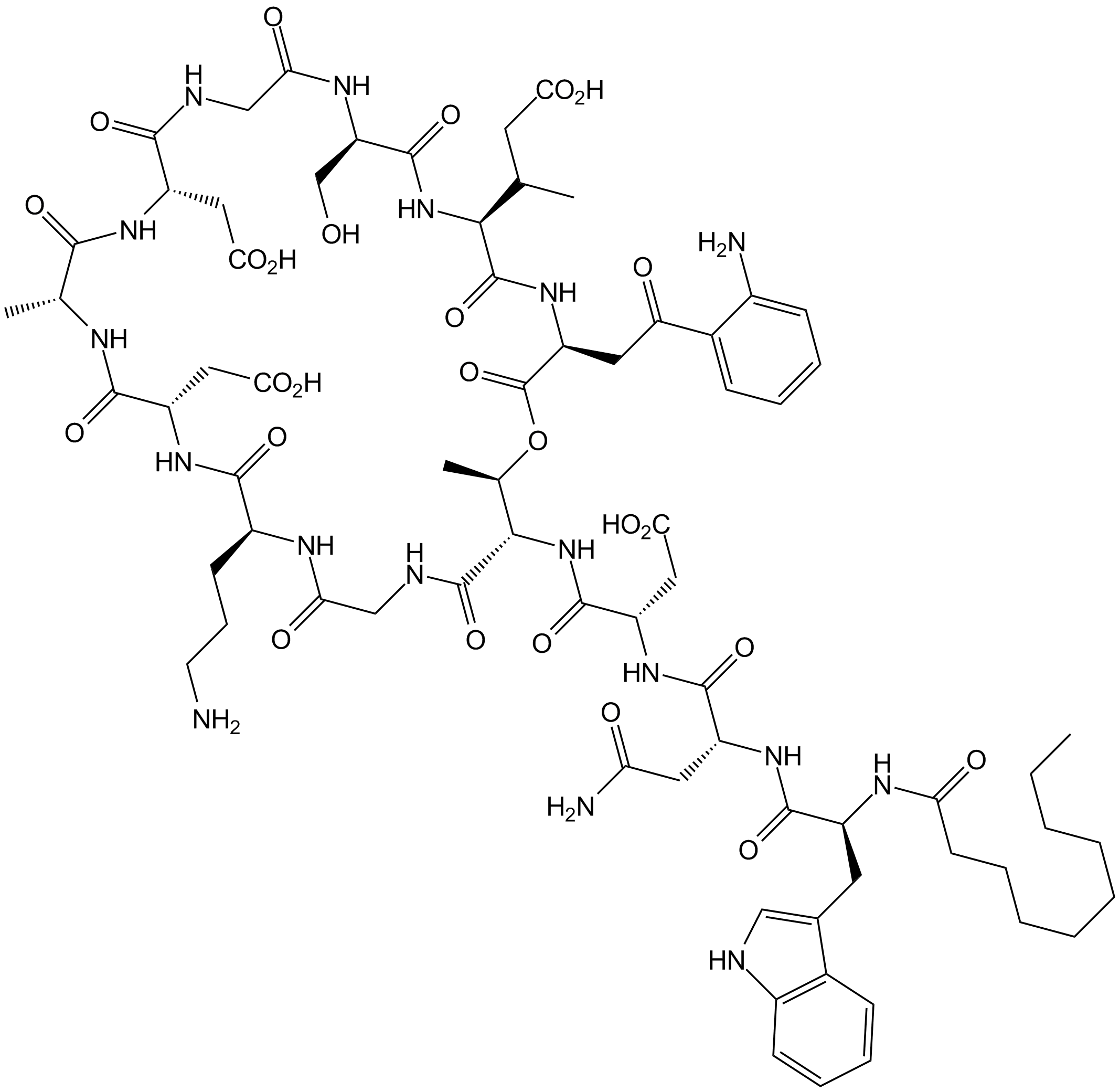
Daptomycin
Calcium-dependent antibiotic
-
GC33124
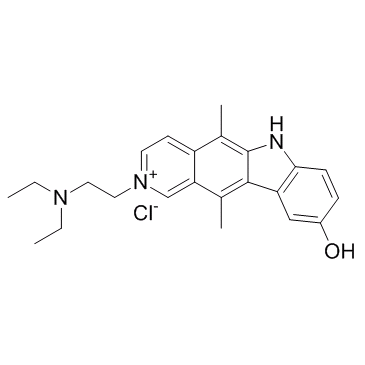
Datelliptium chloride
Datelliptium chloride is a DNA-intercalating agent derived from ellipticine, with anti-tumor activities.
-
GC11175
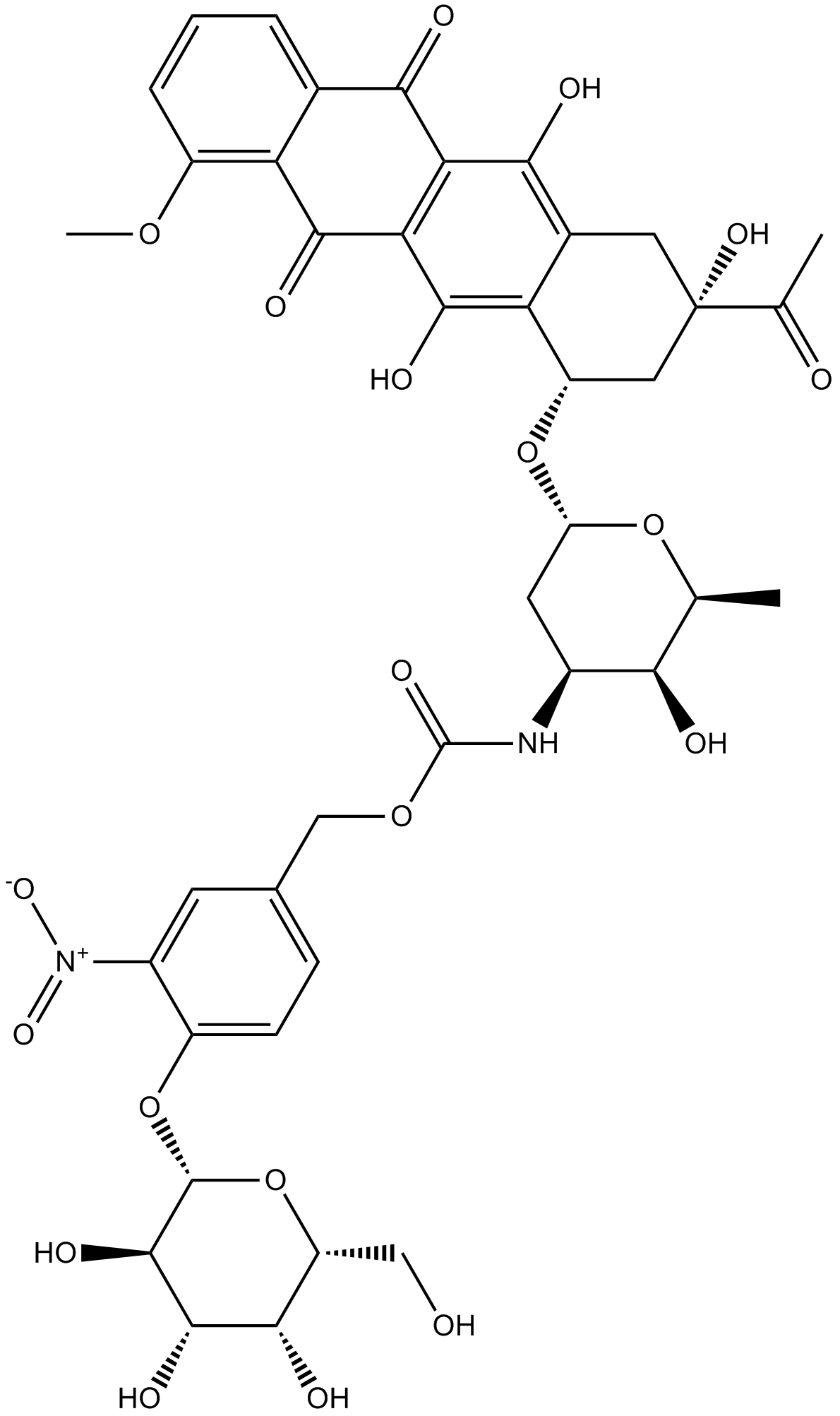
Daun02
-
GC12828
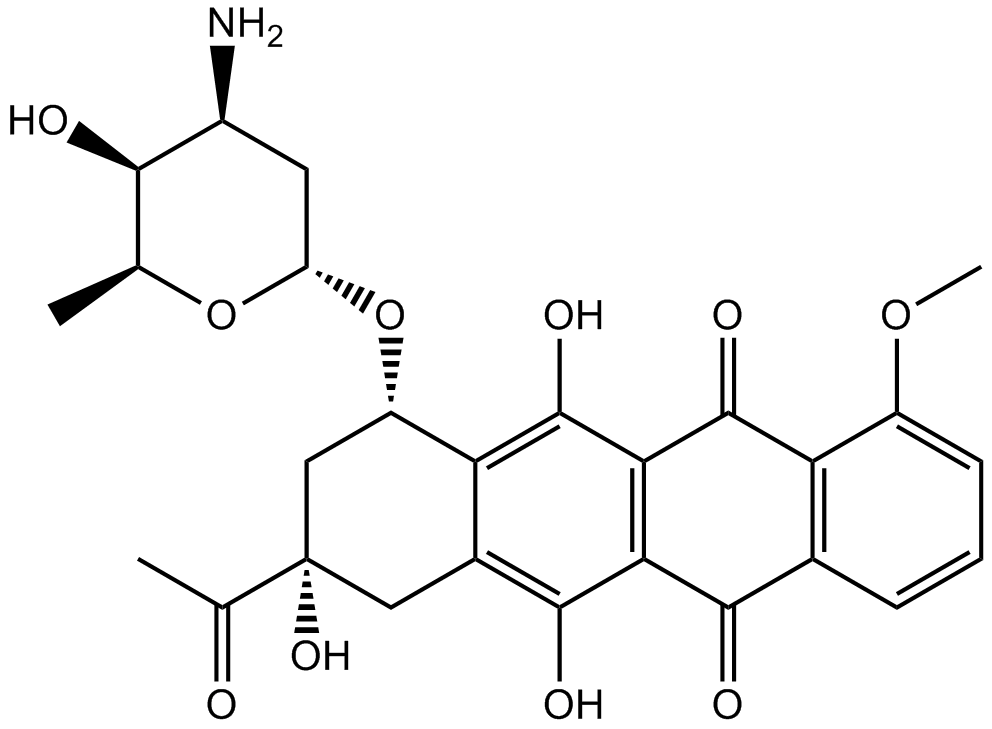
Daunorubicin
DNA topoisomerase II inhibitor
-
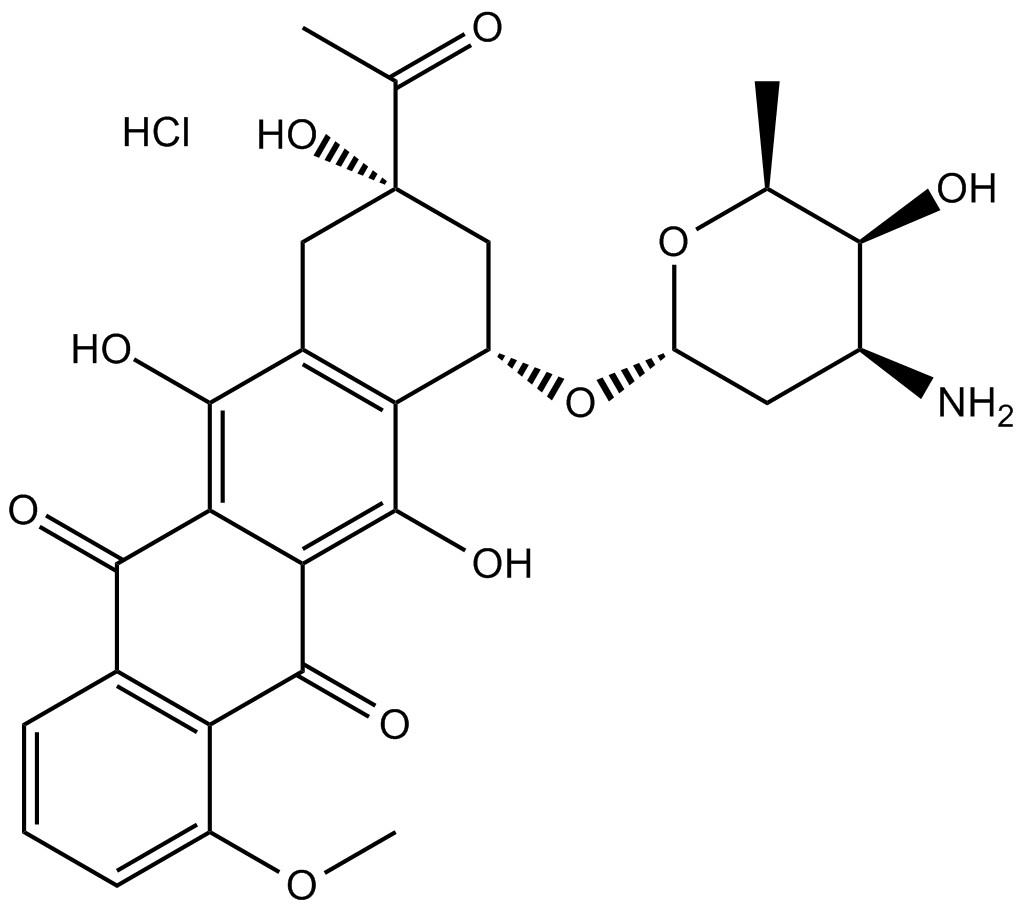
GC10354
Daunorubicin HCl
Daunorubicin (Daunomycin) hydrochloride is a topoisomerase II inhibitor with potent anti-tumor activity.
-
GC33148
DC-05
DC-05 is a DNA methyltransferase 1 (DNMT1) inhibitor, with an IC50 and a Kd of 10.3 μM and 1.09 μM, respectively.
-
GC67886
DC-U4106

-
GC32872
DC_517
DC_517 is a DNA methyltransferase 1 (DNMT1) inhibitor, with an IC50 and a Kd of 1.7 μM and 0.91 μM, respectively.
-
GC67970
DD-03-156
-
GC65440
ddCTP trisodium
ddCTP trisodium is one of 2',3'-dideoxyribonucleoside 5'-triphosphates (ddNTPs) that acts as chain-elongating inhibitor of DNA polymerase for DNA sequencing.
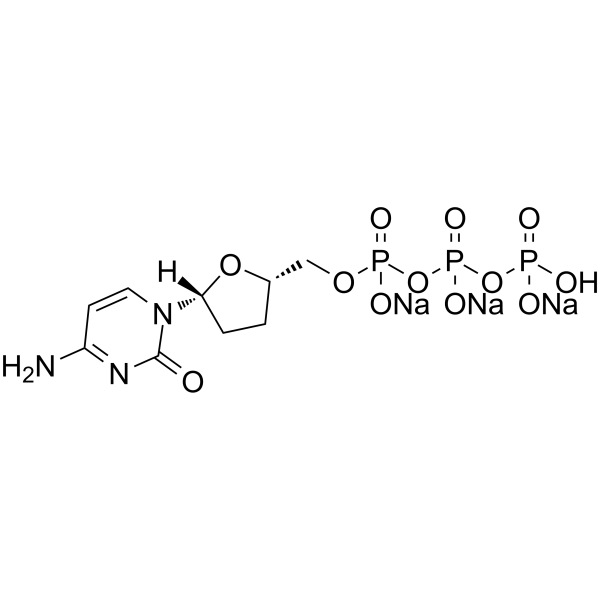
-
GC65295
ddGTP trisodium
ddGTP (2′,3′-Dideoxyguanosine 5′-triphosphate) trisodium is one of 2',3'-dideoxyribonucleoside 5'-triphosphates (ddNTPs) that acts as chain-elongating inhibitor of DNA polymerase for DNA sequencing.
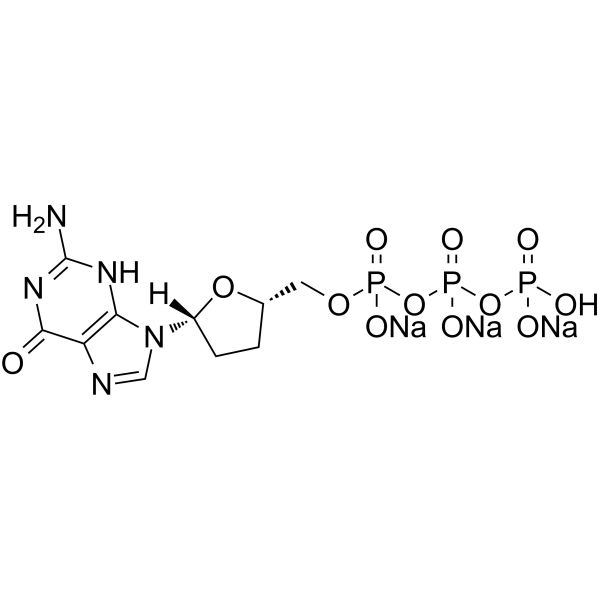
-
GC64020
ddTTP
ddTTP is one of 2',3'-dideoxyribonucleoside 5'-triphosphates (ddNTPs) that acts as chain-elongating inhibitor of DNA polymerase for DNA sequencing.
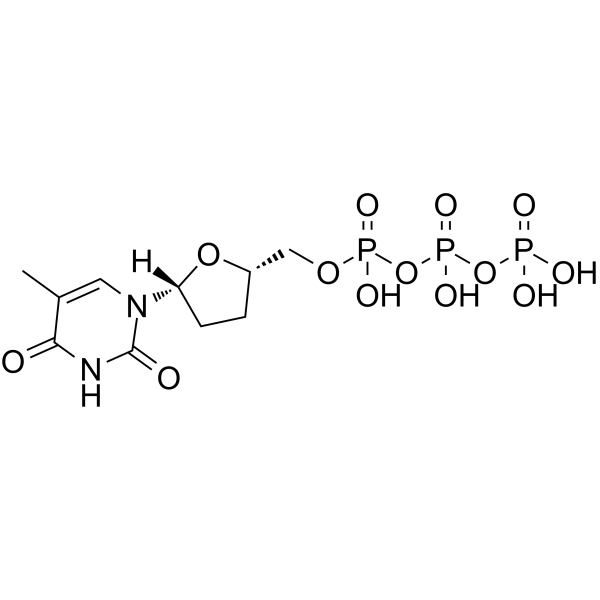
-
GC18666
Debromohymenialdisine
Damaged DNA in humans is detected by sensor proteins that transmit a signal through checkpoint kinases (Chks) Chk1 and Chk2.
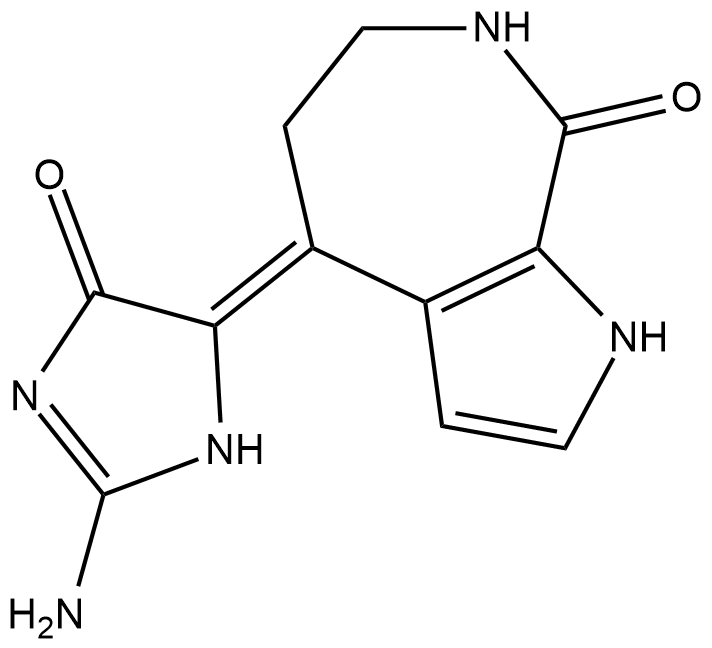
-
GC17412
Deferasirox Fe3+ chelate
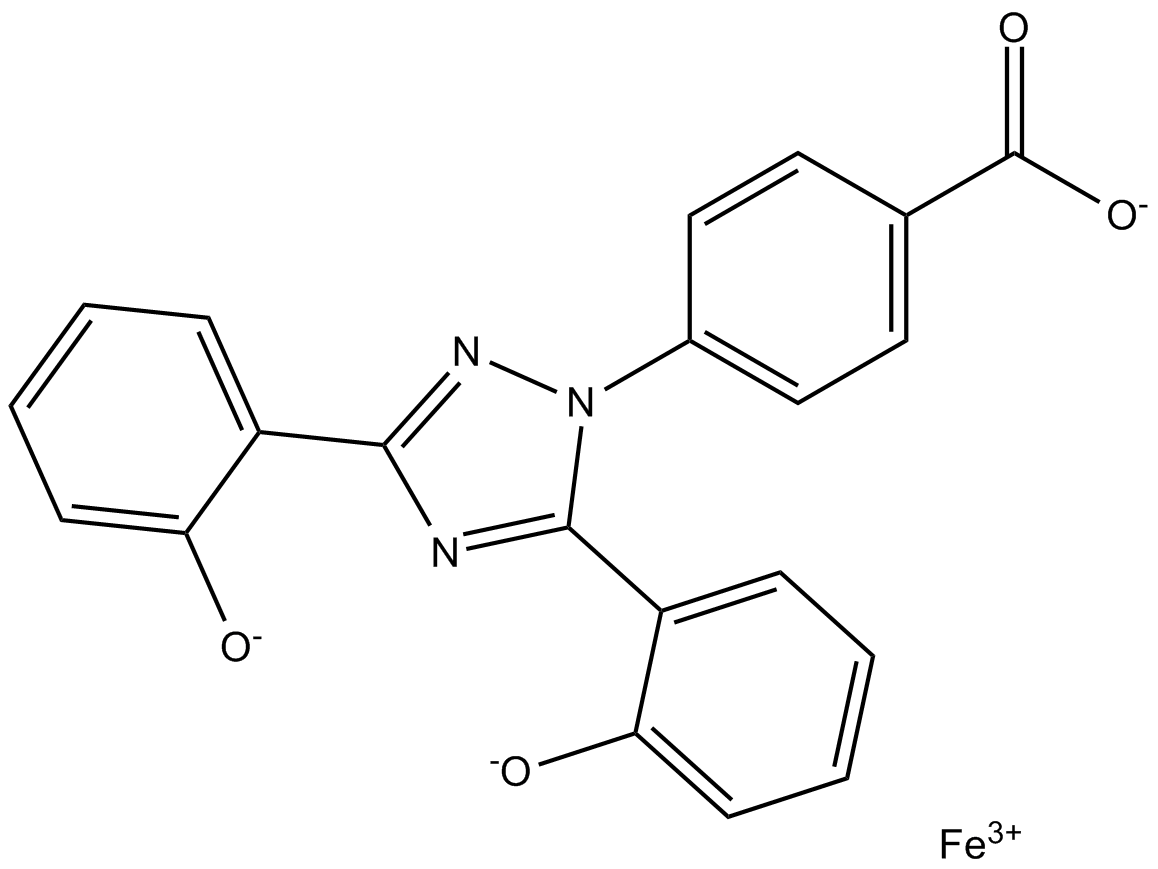
-
GC35829
Dehydroaltenusin
Dehydroaltenusin is a small molecule selective inhibitor of eukaryotic DNA polymerase α, a type of antibiotic produced by a fungus with an IC50 value of 0.68 μM. The inhibitory mode of action of dehydroaltenusin against mammalian pol α activity is competitive with respect to the DNA template primer (Ki=0.23 ?M) and non-competitive with respect to the 2'-deoxyribonucleoside 5'-triphosphate substrate (Ki=0.18 ?M). Dehydroaltenusin arrests the cancer cell cycle at the S-phase and triggers apoptosis. Dehydroaltenusin possesses anti-tumor activity against human adenocarcinoma tumor in vivo.
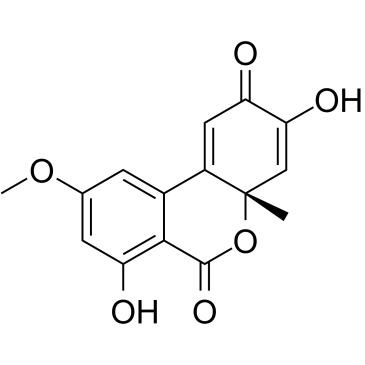
-
GC34123
Deoxycytidine triphosphate (dCTP)
Deoxycytidine triphosphate (dCTP) (dCTP) is a nucleoside triphosphate that can be used for DNA synthesis.
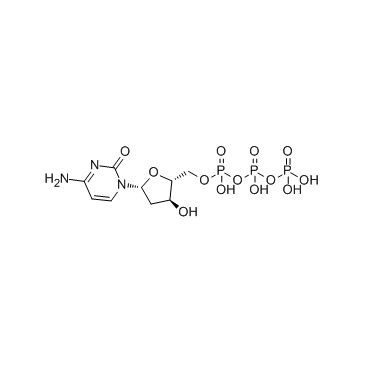
-
GC12634
Deoxyguanosine 5-triphosphate
Deoxyguanosine triphosphate (dGTP) trisodium salt is a nucleotide precursor in cells for DNA synthesis.
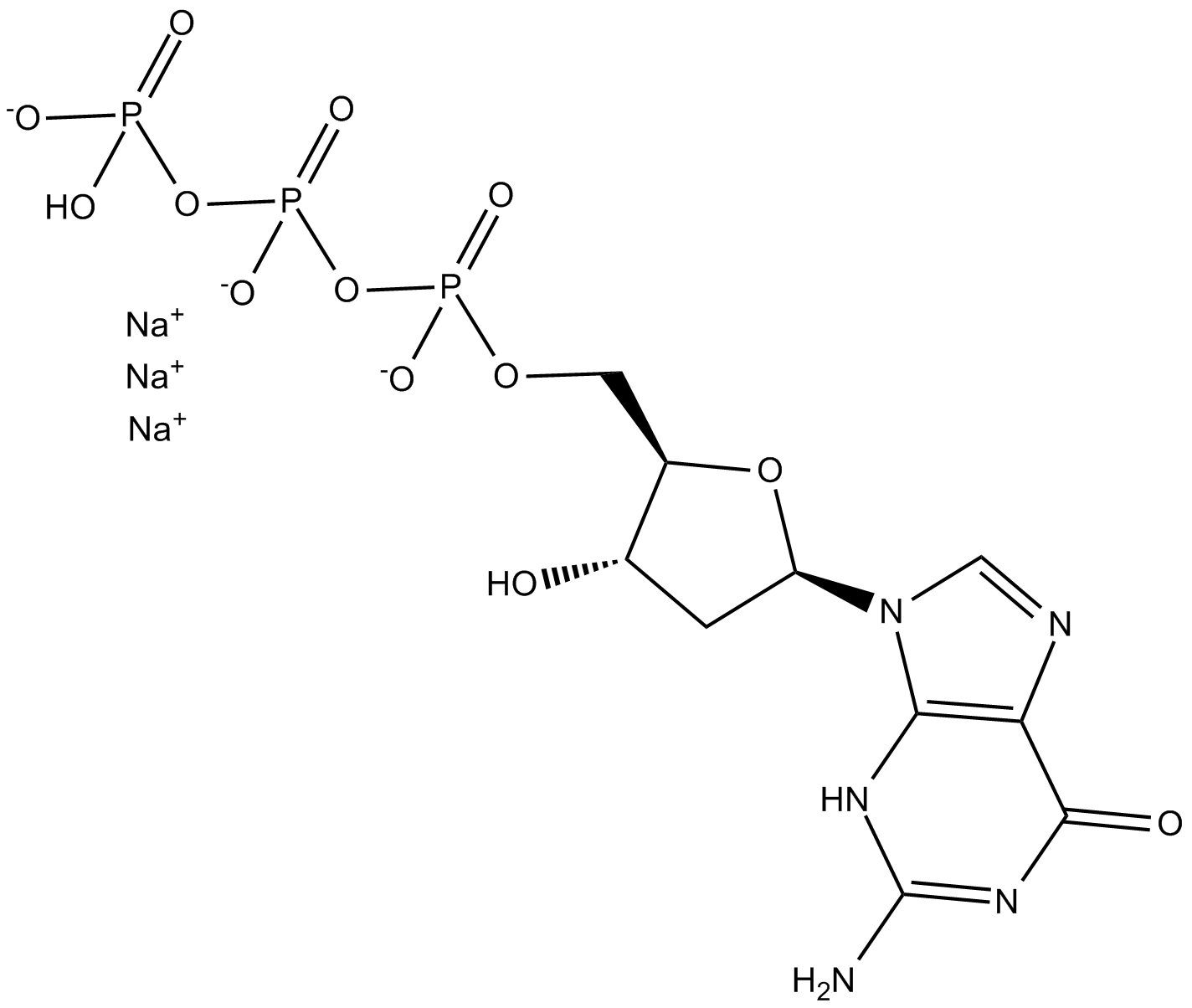
-
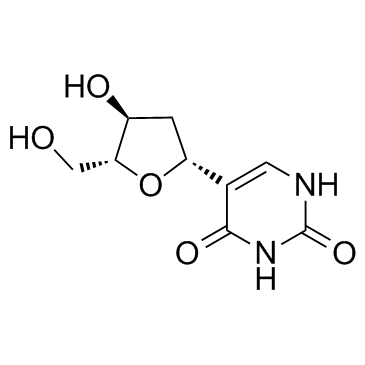
GC33494
Deoxypseudouridine
Deoxypseudouridine is a nucleoside analog.
-
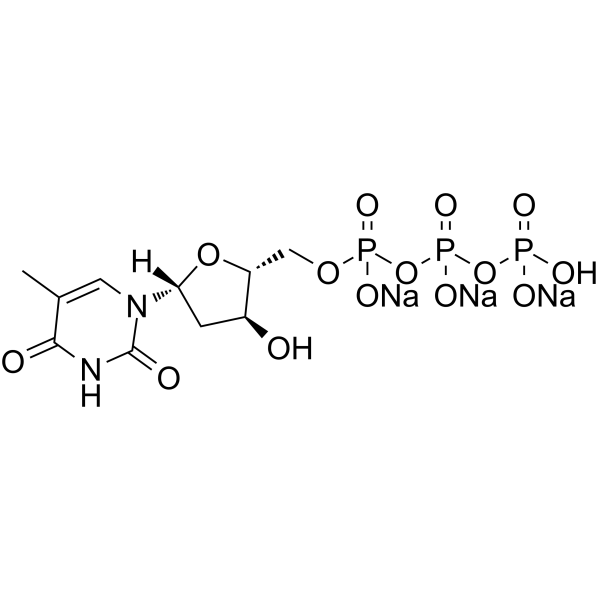
GC62925
Deoxythymidine-5’-triphosphate trisodium salt
Deoxythymidine-5'-triphosphate (dTTP) trisodium is one of the four nucleoside triphosphates used in the synthesis of DNA.
-
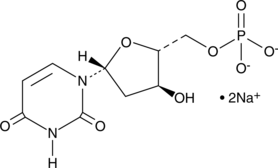
GC49225
Deoxyuridine 5′-monophosphate (sodium salt)
Deoxyuridine 5′-monophosphate (sodium salt) is reductively methylated to dTMP (2'-deoxythymidine 5'-monophosphate) by bisubstrate enzyme thymidylate synthase (TS).
-
GC38482
Desmethylglycitein
Desmethylglycitein (4',6,7-Trihydroxyisoflavone), a metabolite of daidzein, sourced from Glycine max with antioxidant, and anti-cancer activities.Desmethylglycitein binds directly to CDK1 and CDK2 in vivo, resulting in the suppresses CDK1 and CDK2 activity. Desmethylglycitein is a direct inhibitor of protein kinase C (PKC)α, against solar UV (sUV)-induced matrix matrix metalloproteinase 1 (MMP1). Desmethylglycitein binds to PI3K in an ATP competitive manner in the cytosol, where it inhibits the activity of PI3K and downstream signaling cascades, leading to the suppression of adipogenesis in 3T3-L1 preadipocytes.

-
GC65617
DHFR-IN-3
DHFR-IN-3 is a dihydrofolate reductase (DHFR) inhibitor with the IC50 values of 19 μM and 12 μM in rat liver and P. carinii DHFR, respectively.

-
GC60763
DHODH-IN-11
DHODH-IN-11 (Compound 14b) is a Leflunomide derivative and a weak dihydroorotate dehydrogenase (DHODH) inhibitor with a pKa of 5.03.
-
GC39234
DHODH-IN-2
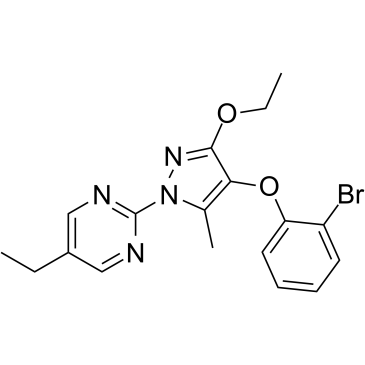
-
GC39374
DHODH-IN-5
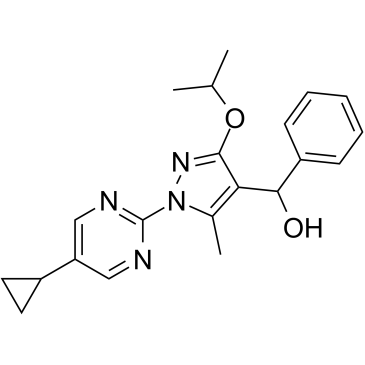
-
GC32343
Diaveridine (EGIS-5645)
Diaveridine (EGIS-5645) (EGIS-5645) is a dihydrofolate reductase (DHFR) inhibitor with a Ki of 11.5 nM for the wild type DHFR and also an antibacterial agent.
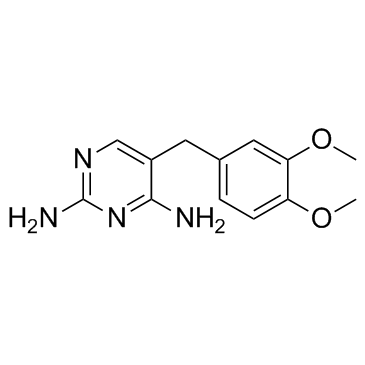
-
GC49022
Didanosine-d2
An internal standard for the quantification of didanosine
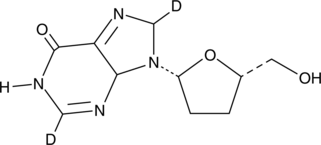
-
GC39808
Didesmethylrocaglamide
Didesmethylrocaglamide, a derivative of Rocaglamide, is a potent eukaryotic initiation factor 4A (eIF4A) inhibitor. Didesmethylrocaglamide has potent growth-inhibitory activity with an IC50 of 5 nM. Didesmethylrocaglamide suppresses multiple growth-promoting signaling pathways and induces apoptosis in tumor cells. Antitumor activity.
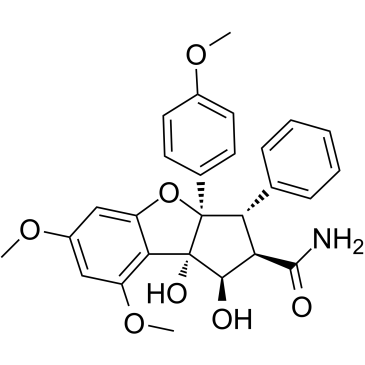
-
GC12266
Didox
synthetic antioxidant
-
GC41112
Dihydroxy Melphalan
Dihydroxy melphalan is an inactive degradation product of melphalan.
-
GC17648
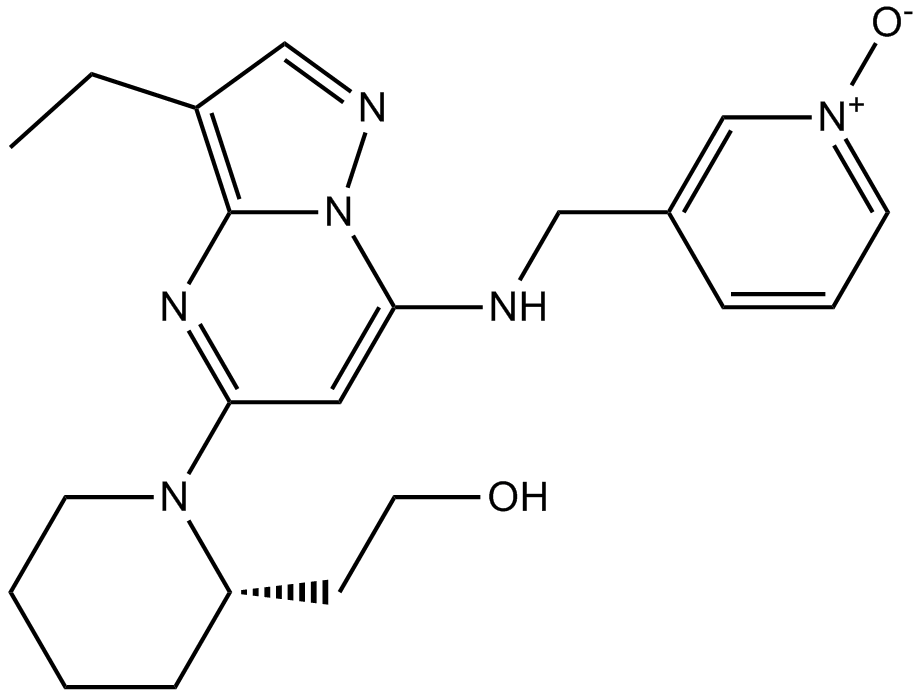
Dinaciclib(SCH727965)
Dinaciclib(SCH727965) (SCH 727965) is a potent inhibitor of CDK, with IC50s of 1 nM, 1 nM, 3 nM, and 4 nM for CDK2, CDK5, CDK1, and CDK9, respectively.
-
GC31684
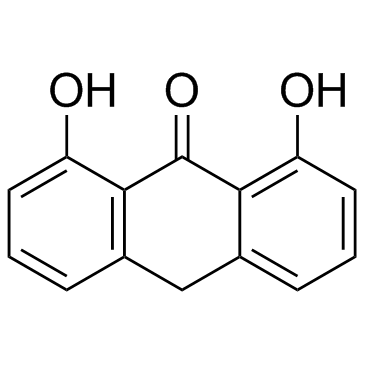
Dithranol (Anthralin)
Dithranol (Anthralin) (Anthralin) is an anthraquinone derivative, with potent anti-psoriatic effects.
-
GC19495
DL-Folinic Acid (calcium salt)
DL-Folinic Acid (calcium salt) (Leucovorin calcium) is a biological folic acid and is generally administered along with methotrexate (MTX) as a rescue agent to decrease MTX-induced toxicity.

-
GC17837
DMAT
A cell-permeable inhibitor of CK2
-
GC16582
DMNB
DNA-dependent protein kinase (DNA-PK) inhibitor
-
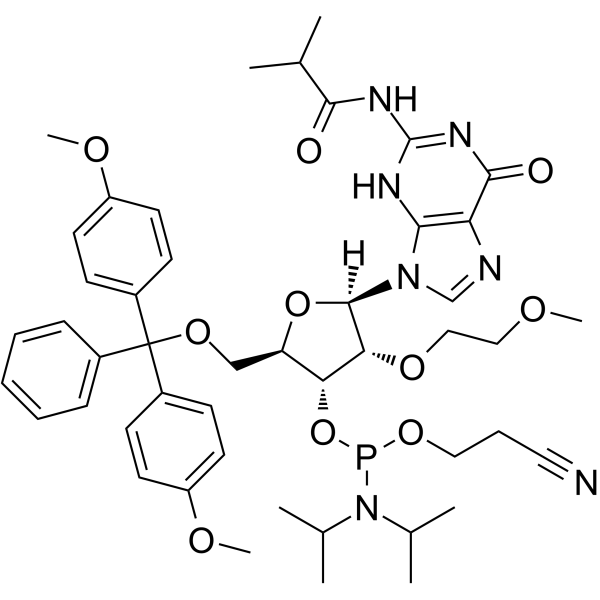
GC66210
DMT-2'O-MOE-rG(ib) Phosphoramidite
DMT-2'O-MOE-rG(ib) Phosphoramidite (1g), belonging to the amide family of trivalent phosphate H3PO3, is a derivative of nucleotides and guanosine and can be used in the stereochemical synthesis of phosphorothioate oligonucleotides.
-
GC61708
Dmt-2'fluoro-da(bz) amidite
Dmt-2'fluoro-da(bz) amidite, an uniformly modified 2'-deoxy-2'-fluoro phosphorothioate oligonucleotide, is a nuclease-resistant antisense compound with high affinity and specificity for RNA targets.
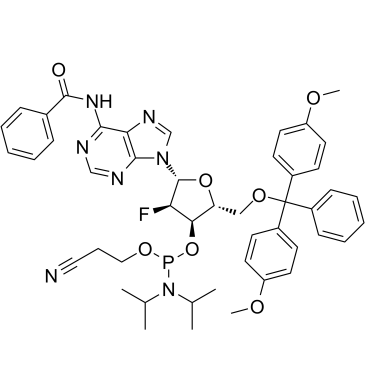
-
GC62150
DMT-dC(ac) Phosphoramidite
DMT-dC(ac) Phosphoramidite is a modified phosphoramidite monomer, which can be used for the oligonucleotide synthesis.
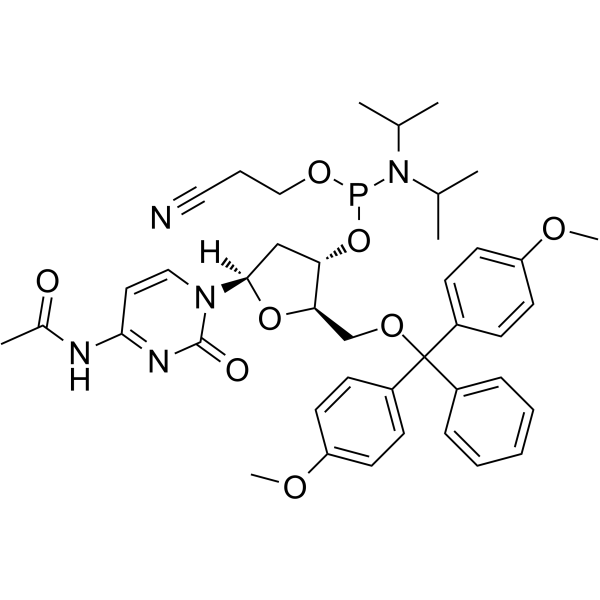
-
GC61916
DMT-dC(bz) Phosphoramidite
DMT-dC(bz) Phosphoramidite is typically used in the synthesis of DNA.
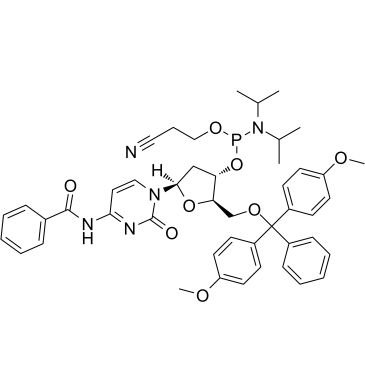
-
GC62538
DMT-dG(dmf) Phosphoramidite
DMT-dG(dmf) Phosphoramidite is a phosphinamide monomer that can be used in the preparation of oligonucleotides
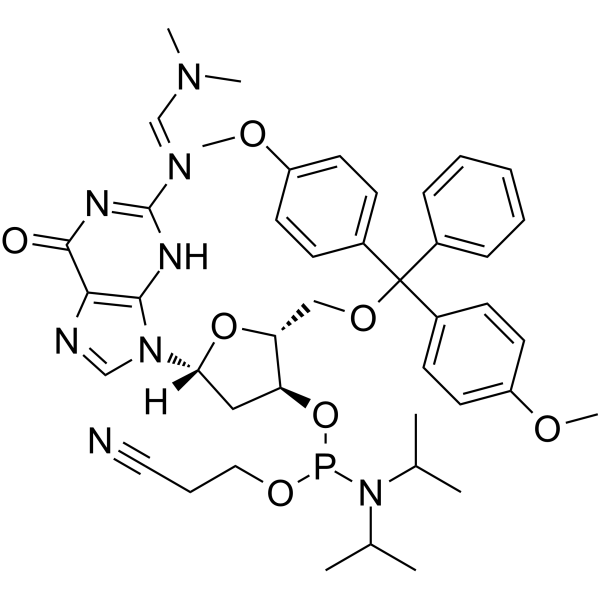
-
GC61912
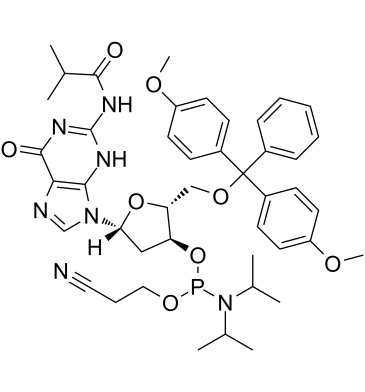
DMT-dG(ib) Phosphoramidite
DMT-dG(ib) Phosphoramidite is typically used in the synthesis of DNA.
-
GC61913
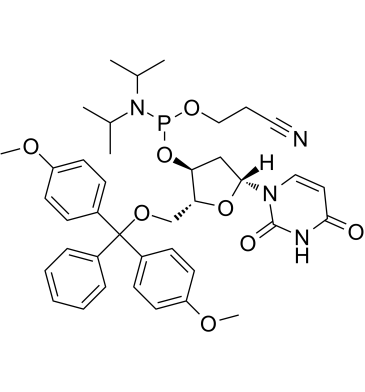
DMT-dU-CE Phosphoramidite
DMT-dU-CE Phosphoramidite is a nucleoside molecule that can be used in DNA synthesis and DNA sequencing.
-
GC66711
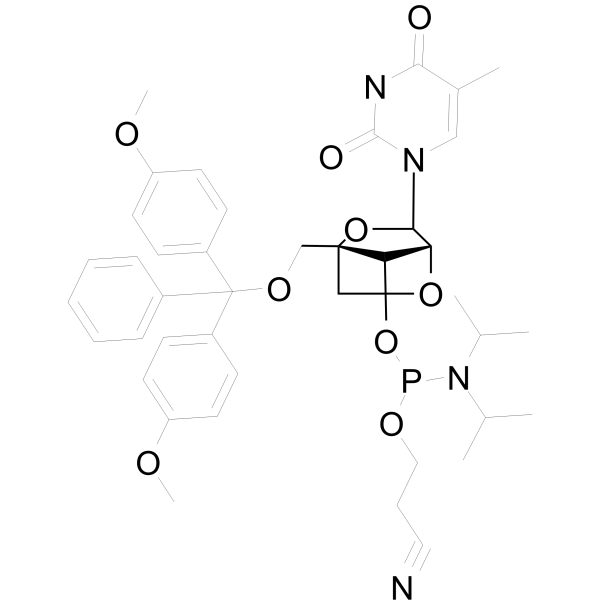
DMTr-LNA-5MeU-3-CED-phosphoramidite
DMTr-LNA-5MeU-3-CED-phosphoramidite is a nucleoside derivative.
-
GC35888
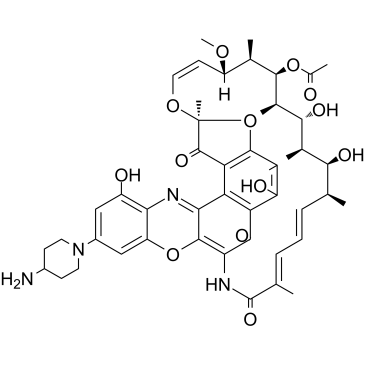
DNA31
DNA31 is a potent RNA polymerase inhibitor.
-
GC65546
DNMT3A-IN-1
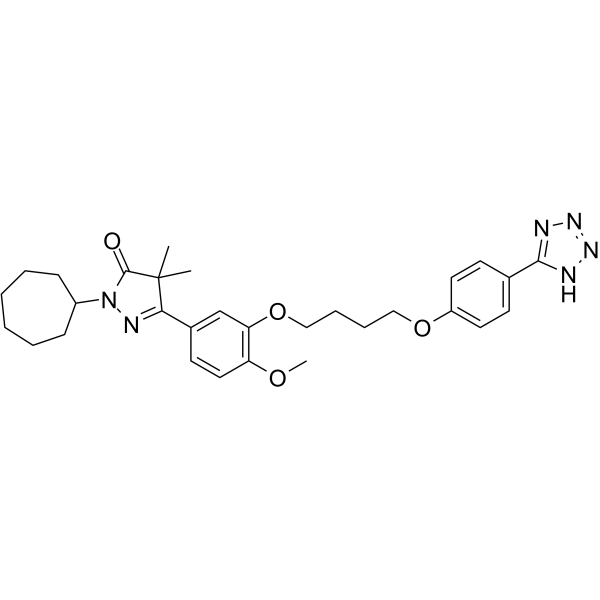
DNMT3A-IN-1 is a potent and selective DNMT3A inhibitor.
-
GC35893
Domatinostat
Domatinostat (4SC-202 free base) is a selective class I HDAC inhibitor with IC50 of 1.20 μM, 1.12 μM, and 0.57 μM for HDAC1, HDAC2, and HDAC3, respectively. It also displays inhibitory activity against Lysine specific demethylase 1 (LSD1).
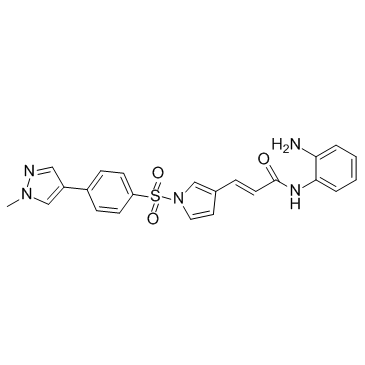
-
GC11099
Doxifluridine
oral prodrug of the antineoplastic agent 5-fluorouracil (5-FU)
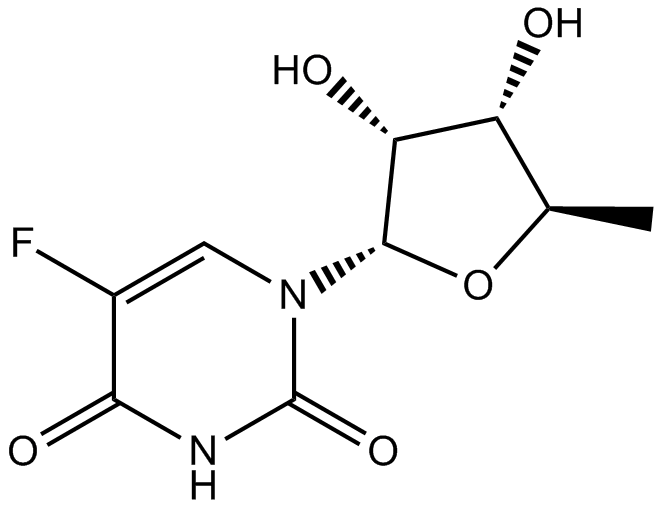
-
GC15250
DOXO-EMCH
Prodrug of doxorubicin
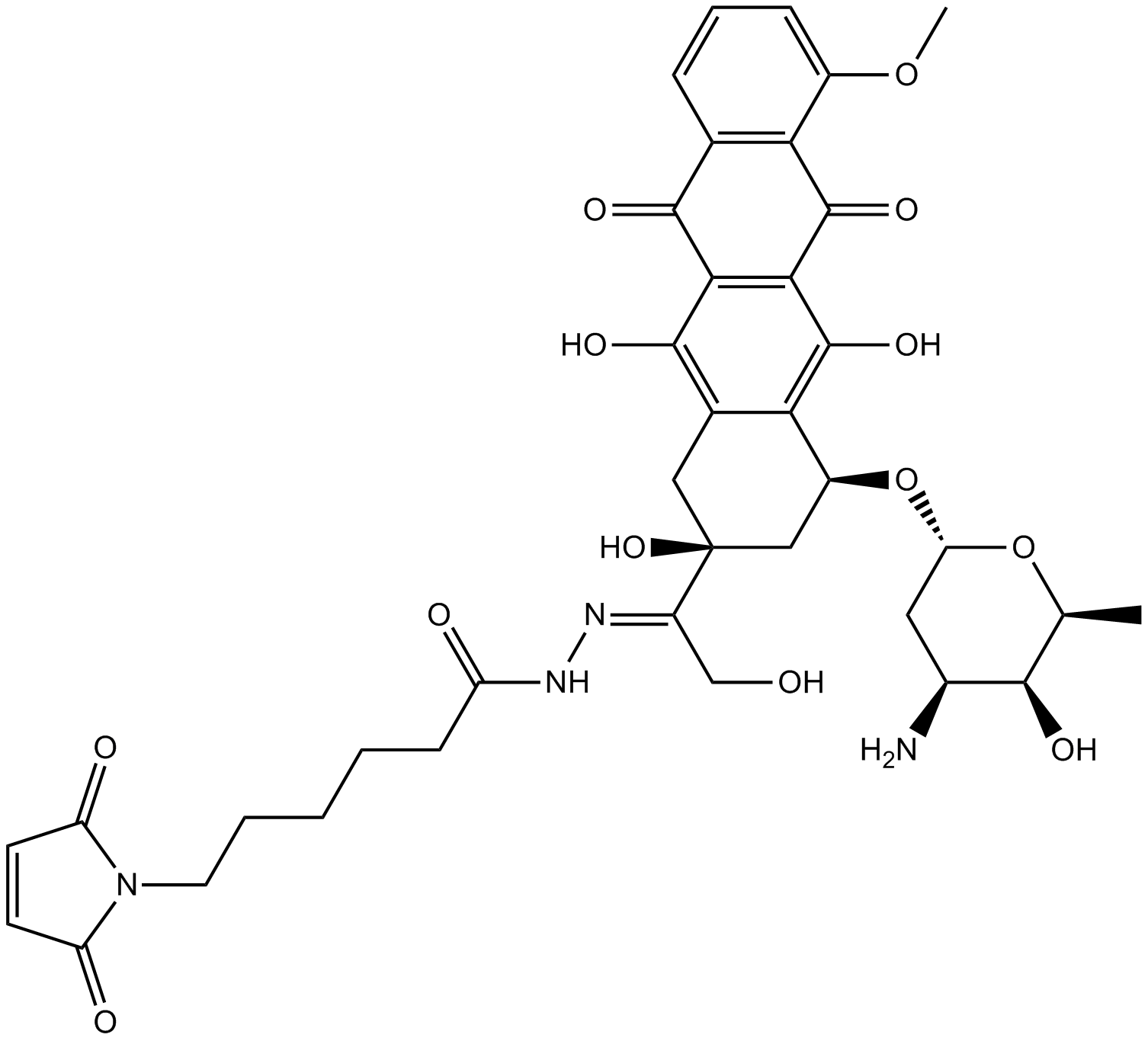
-
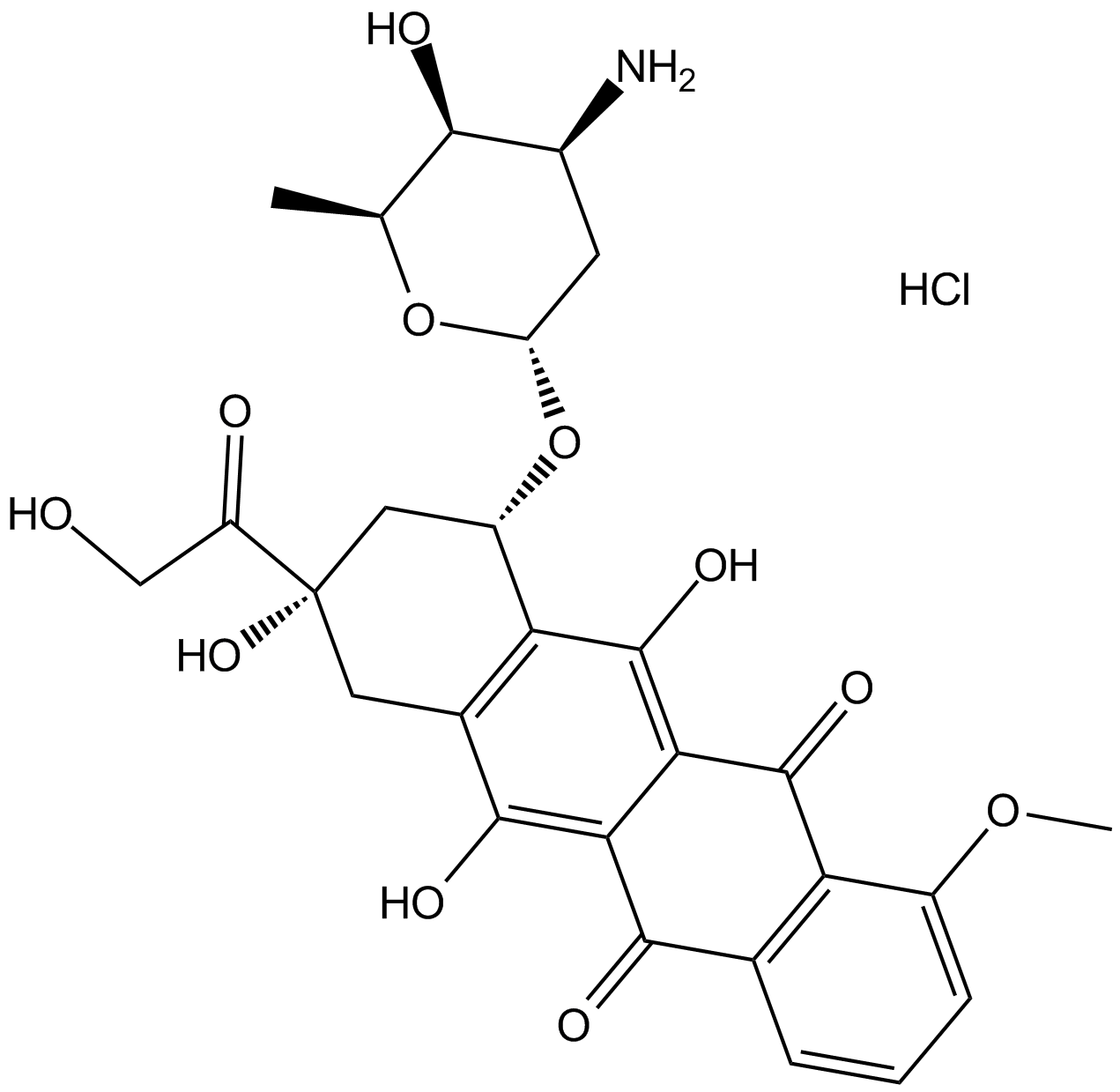
GC17567
Doxorubicin (Adriamycin) HCl
Doxorubicin (Hydroxydaunorubicin) hydrochloride, a cytotoxic anthracycline antibiotic, is an anti-cancer chemotherapy agent.
-
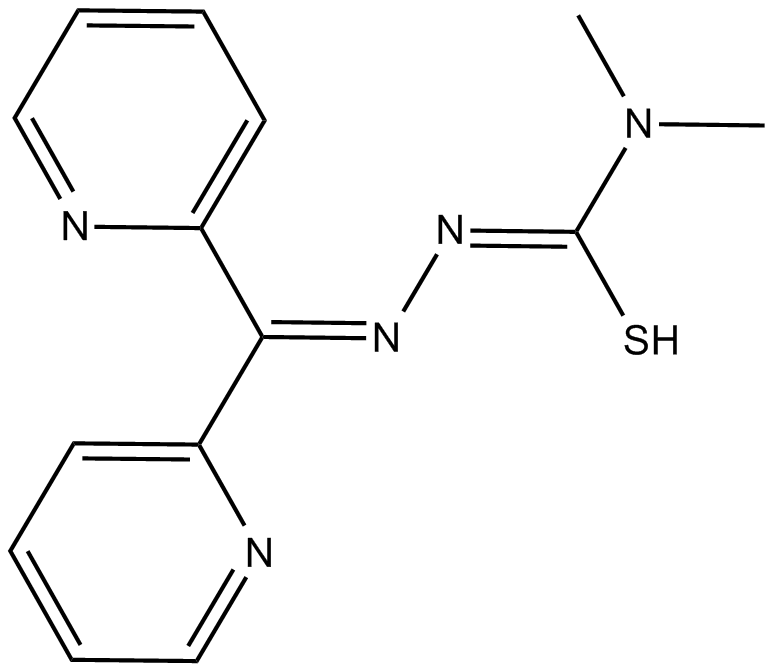
GC15399
Dp44mT
iron chelator and anticancer agent
-
GC15294
DPQ
DPQ is a potent inhibitor of PARPs, inhibiting PARP1 with an IC50 value of 40 nM.
-
GC38774
DRF-1042
DRF-1042 is an orally active derivative of Camptothecin. DRF-1042 acts to inhibit DNA topoisomerase I. DRF-1042 shows good anticancer activity against a panel of human cancer cell lines including multi-drug resistance (MDR) phenotype.

-
GC64715
DS96432529
DS96432529 is a potent and orally active bone anabolic agent through CDK8 inhibition.
-
GC38202
DTP3 TFA
DTP3 TFA is a potent and selective GADD45β/MKK7 (growth arrest and DNA-damage-inducible β/mitogen-activated protein kinase kinase 7) inhibitor. DTP3 TFA targets an essential, cancer-selective cell-survival module downstream of the NF-κB pathway.
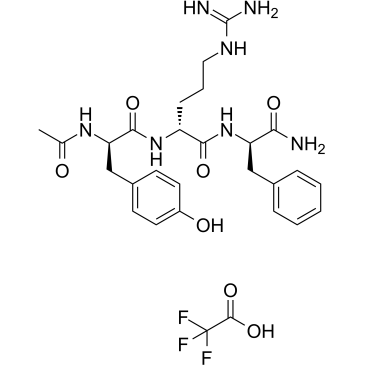
-
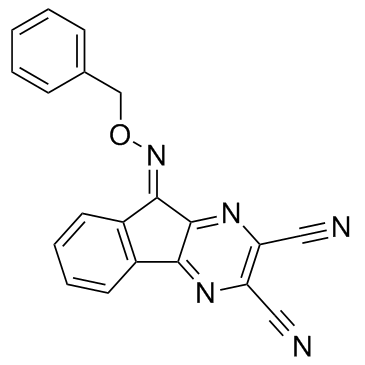
GC35906
DUBs-IN-1
DUBs-IN-1 is an active inhibitor of ubiquitin-specific proteases (USPs), with an IC50 of 0.85 μM for USP8.
-
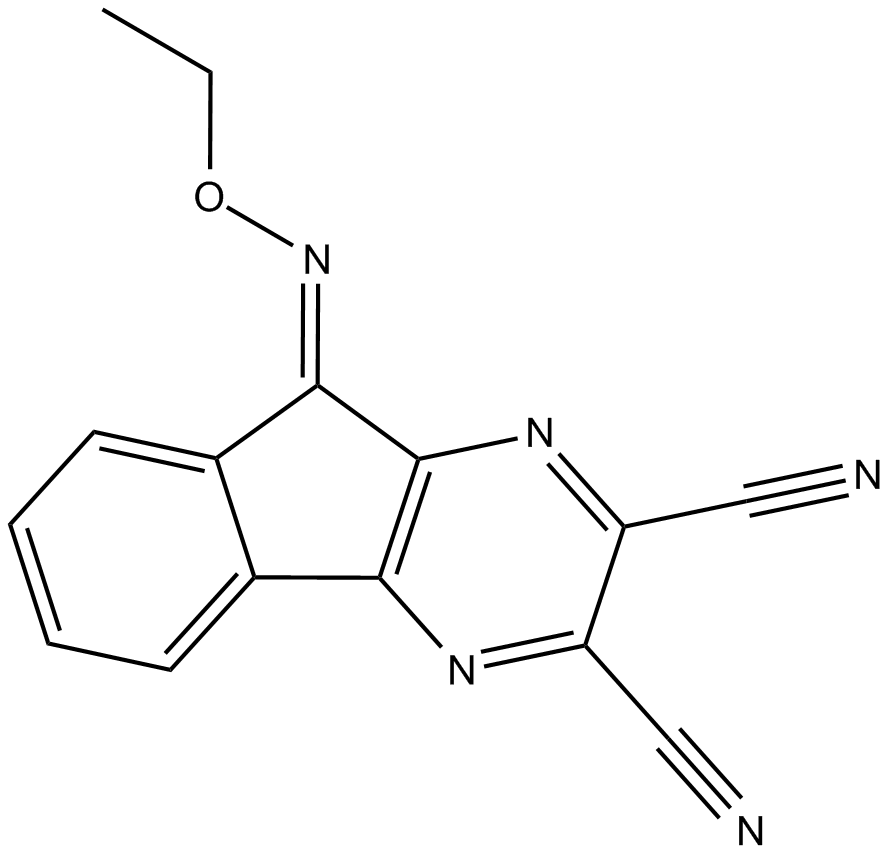
GC17125
DUBs-IN-2
DUBs-IN-2 is a potent deubiquitinase inhibitor with an IC50 of 0.28 μM for USP8.
-
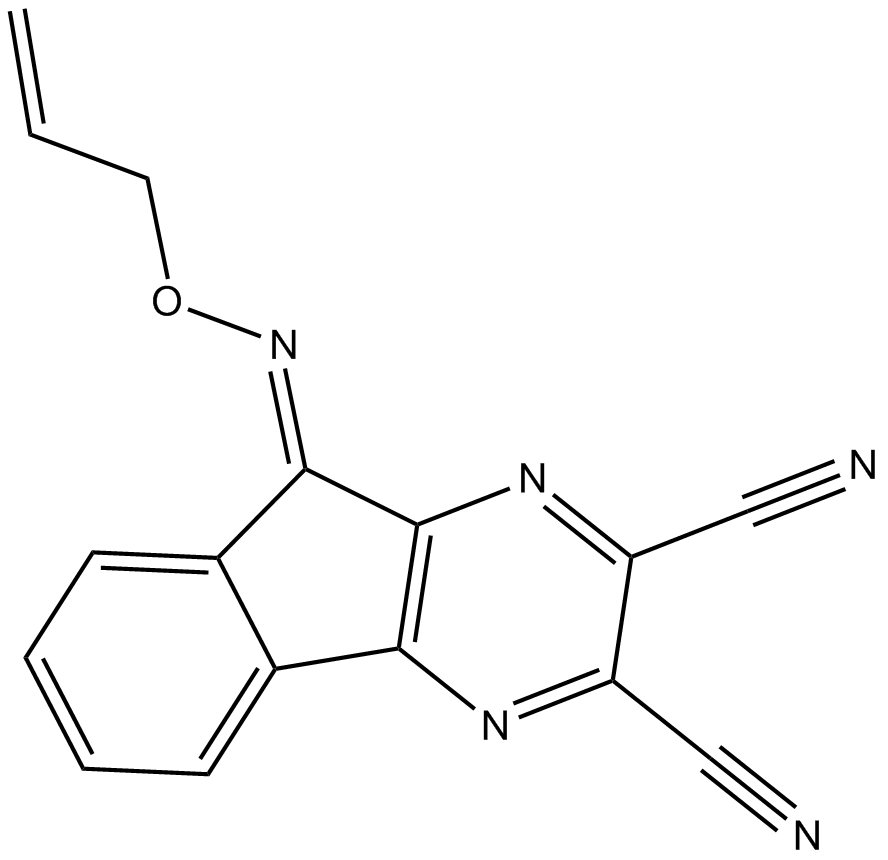
GC10356
DUBs-IN-3
DUBs-IN-3 is a potent deubiquitinase (USP) enzyme inhibitor extracted from reference compound 22c with an IC50 of 0.56 μM for USP8.
-
GC35907
Duocarmycin
Duocarmycin, a DNA minor-groove alkylator, is an antibody drug conjugates (ADCs) toxin. Duocarmycin is based on its characteristic curved indole structure and a spirocyclopropylcyclohexadienone electrophile to act anticancer activity.
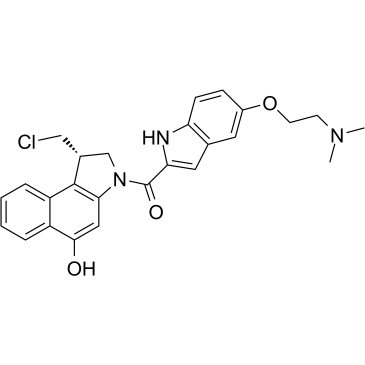
-
GC38080
Duocarmycin Analog
Duocarmycin Analog is an analog of Duocarmycin, and used as an DNA alkylator and ADC cytotoxin.
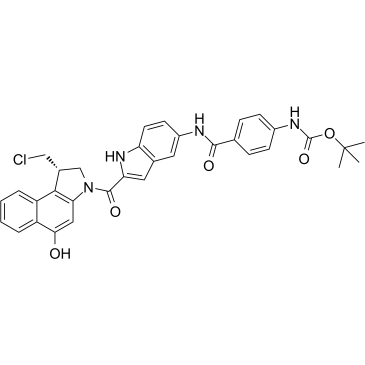
-
GC35909
Duocarmycin GA
Duocarmycin GA is an antibody drug conjugates (ADCs) toxin. Duocarmycin is a DNA alkylating agent that binds in the minor groove. Duocarmycin GA can be used against multi-drug resistant cell lines.
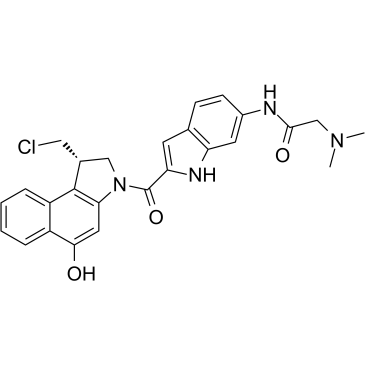
-
GC35910
Duocarmycin MA
Duocarmycin MA is an antibody drug conjugates (ADCs) toxin. Duocarmycin is a DNA alkylating agent that binds in the minor groove. Duocarmycin MA can be used against multi-drug resistant cell lines.
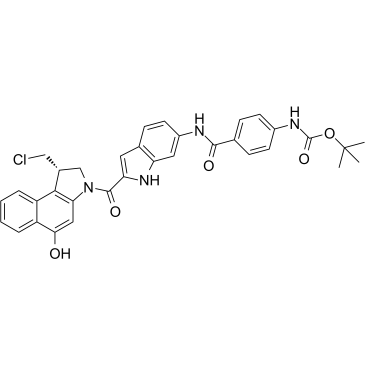
-
GC35911
Duocarmycin MB
Duocarmycin MB is an antibody drug conjugates (ADCs) toxin. Duocarmycin is a DNA alkylating agent that binds in the minor groove. Duocarmycin MB can be used against multi-drug resistant cell lines.
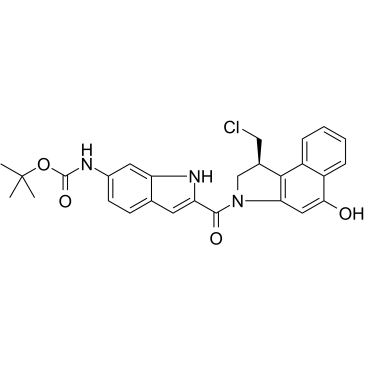
-
GC32187
Duocarmycin TM
Duocarmycin TM (CBI-TMI) is a potent antitumor antibiotic. Duocarmycin TM induces a sequence-selective alkylation of duplex DNA.
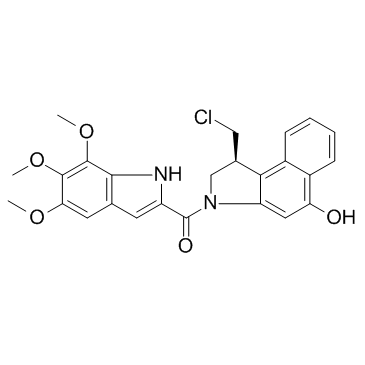
-
GC32926
Dxd (Exatecan derivative)
Dxd (Exatecan derivative) (Exatecan derivative for ADC) is a potent DNA topoisomerase I inhibitor, with an IC50 of 0.31 μM, used as a conjugated drug of HER2-targeting ADC (DS-8201a).
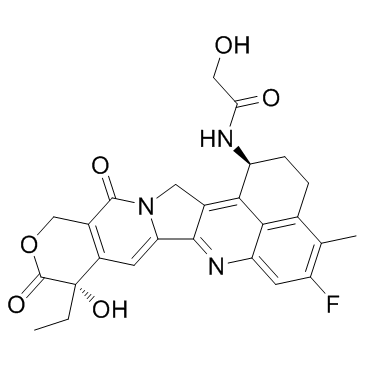
-
GC62212
Dxd-D5
Dxd-d5 (Exatecan-d5 derivative for ADC) is a deuterium labeled Dxd. Dxd is a potent DNA topoisomerase I inhibitor, with an IC50 of 0.31 μM, used as a conjugated drug of HER2-targeting ADC (DS-8201a) .
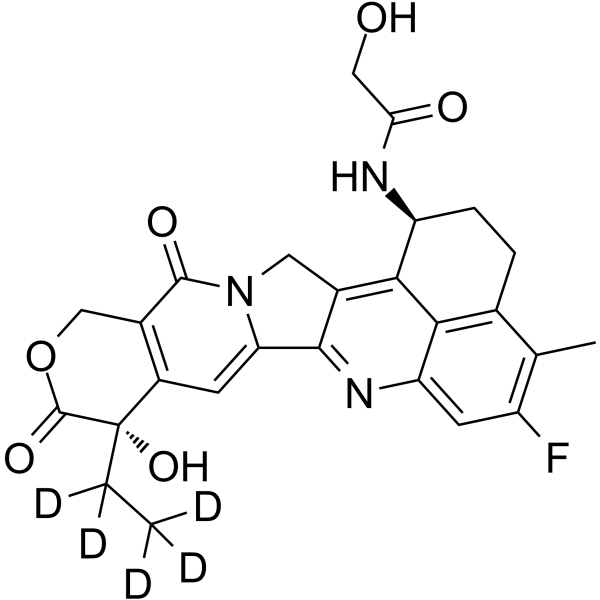
-
GC10620
E3330
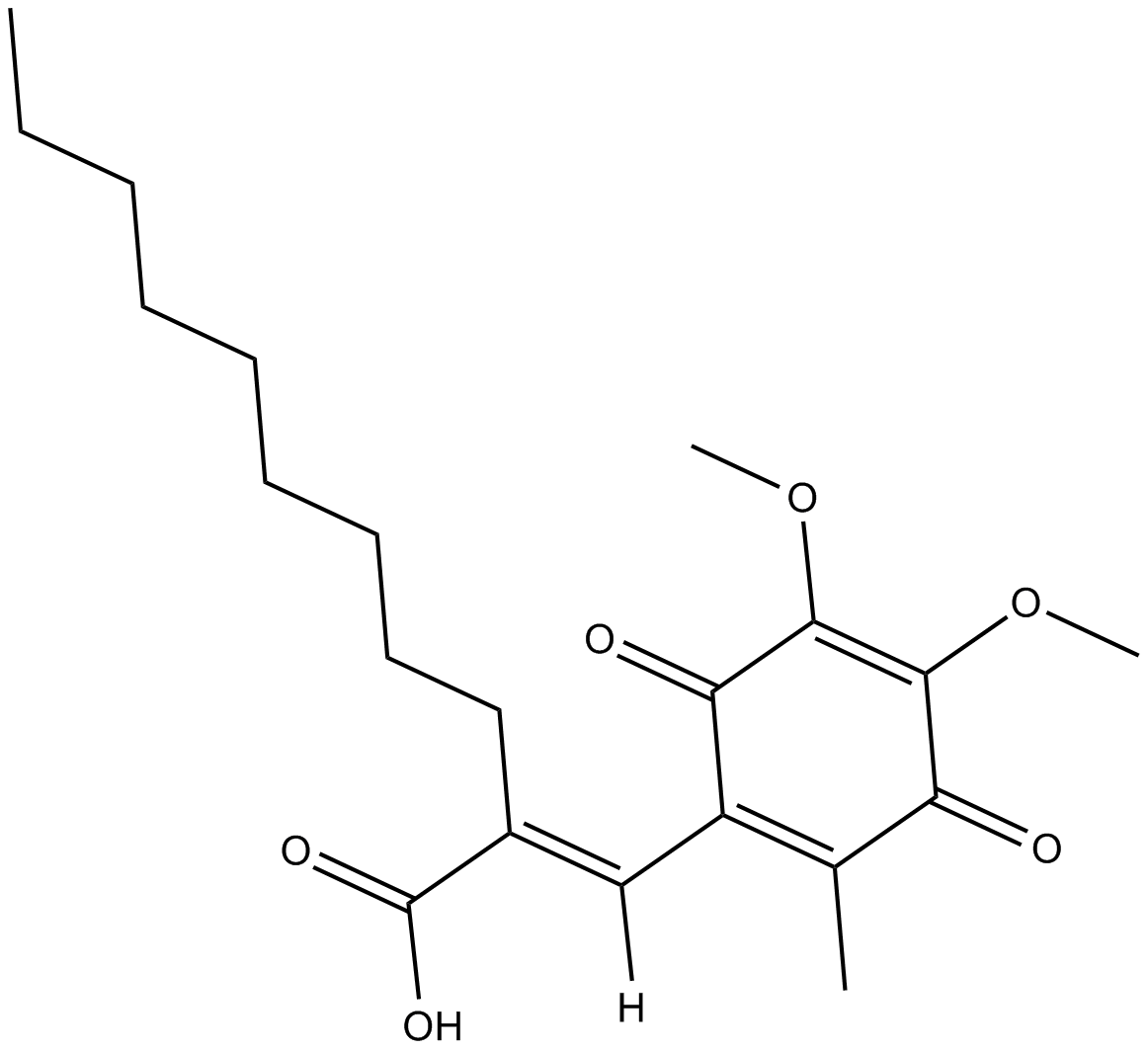
E3330 is a small-molecule inhibitor of apurinic-apyrimidinic endonuclease/redox effector factor (APE1/Ref-1) redox domain function (IC50, 50 µmol/L).
-
GC64209
E7016
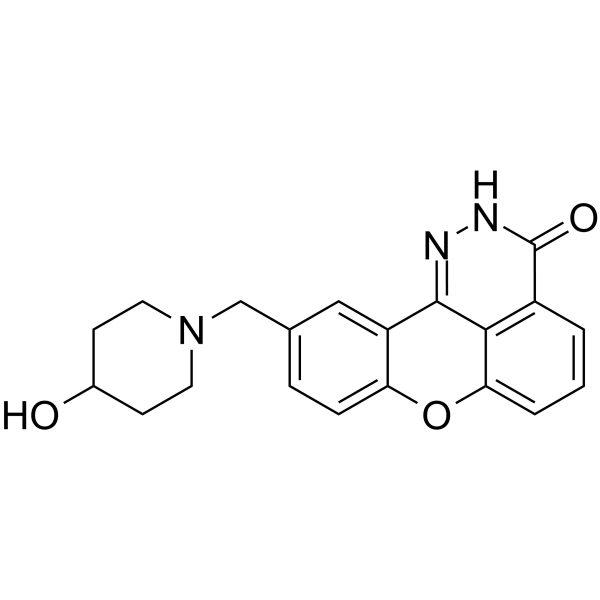
E7016 (GPI 21016) is an orally available PARP inhibitor. E7016 can enhance tumor cell radiosensitivity in vitro and in vivo through the inhibition of DNA repair. E7016 acts as a potential anticancer agent.
-
GC18172
E7449
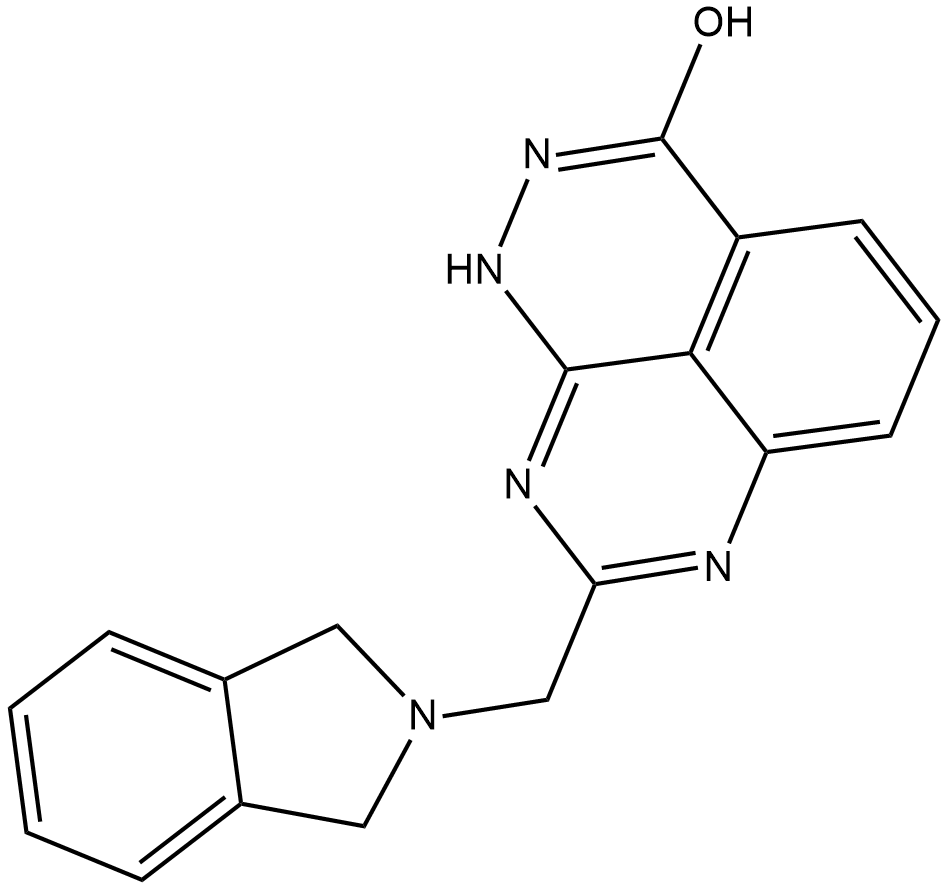
E7449 is an inhibitor of poly(ADP-ribose) polymerase 1 (PARP1) and PARP2 (IC50s = 1 and 1.2 nM, respectively) as well as tankyrase (TNKS) 1/2 (IC50s = 50-100 nM).
-
GC32401
EC0489
EC0489, a conjugate of folic acid and desacetyl vinblastine hydrazide, is a high-affinity ligand for the folate receptor (FR). Refractory or metastatic Tumor. Small molecule-drug conjugate (SMDC).
-
GC64682
Eciruciclib
Eciruciclib is an antineoplastic and potent cyclin dependent kinase (CDK) inhibitor.
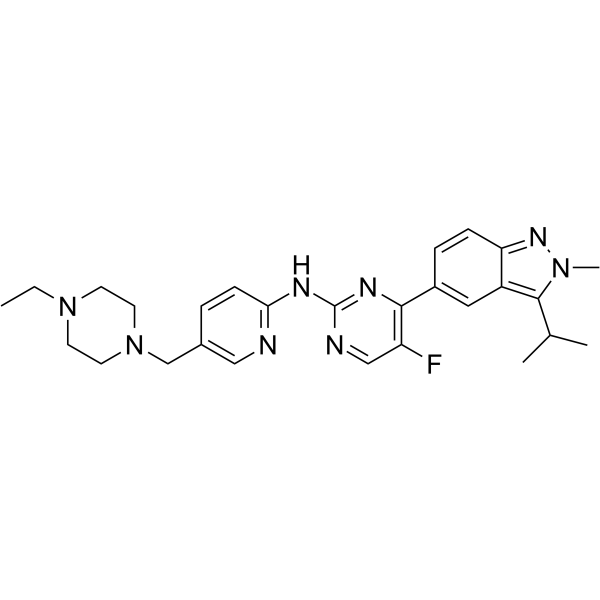
-
GC19129
EDO-S101
EDO-S101 is a pan HDAC inhibitor; inhibits HDAC1, HDAC2 and HDAC3 with IC50 values of 9, 9 and 25 nM, respectively.
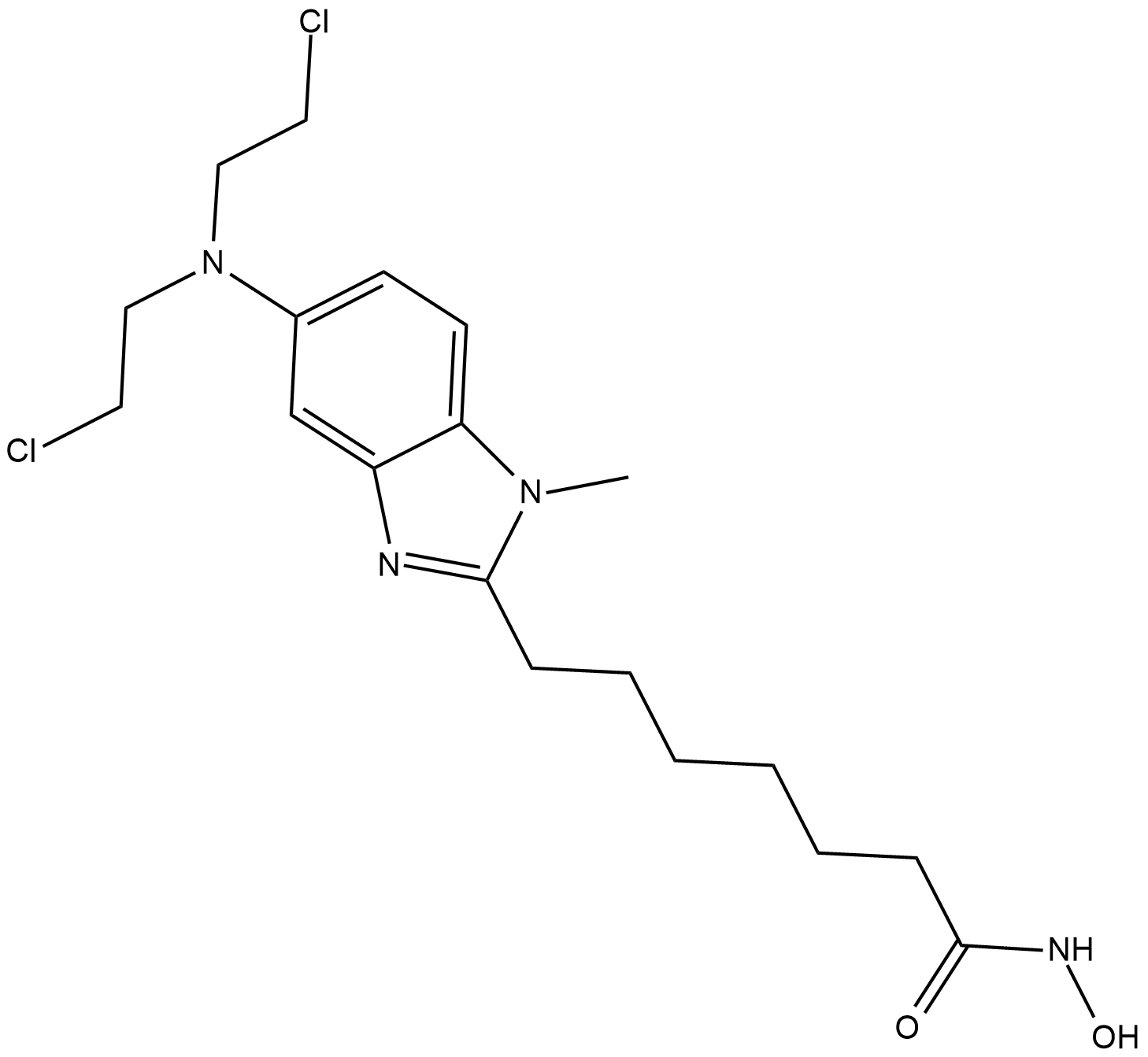
-
GC33128
Edotecarin (J 107088)
Edotecarin (J 107088) is a potent inhibitor of topoisomerase I that can induces single-strand DNA cleavage, with IC50 of 50 nM.
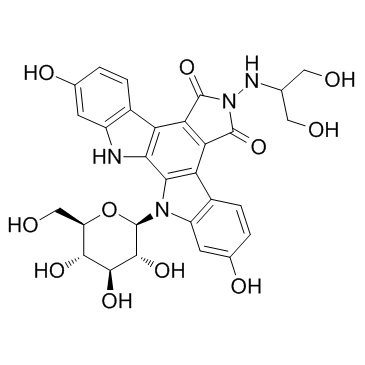
-
GC62607
EFdA-TP tetraammonium
EFdA-TP tetraammonium is a potent nucleoside reverse transcriptase (RT) inhibitor.
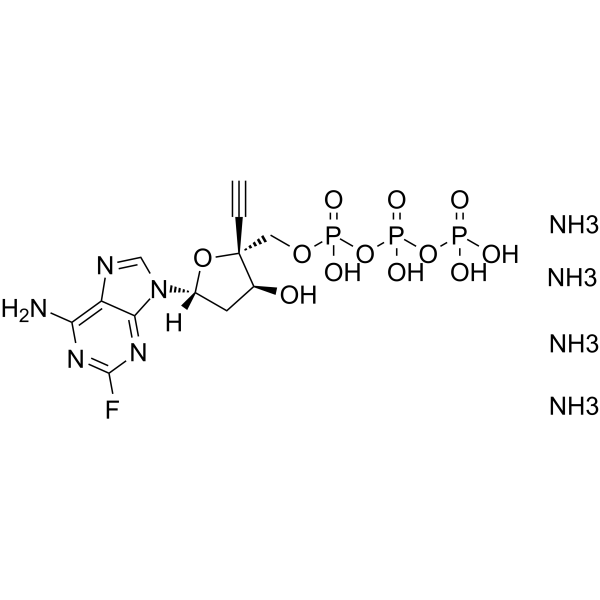
-
GC38330
EHT 5372
-
GC18620
EIDD-1931
A ribonucleoside analog with antiviral activity
-
GC35969
eIF4A3-IN-1
eIF4A3-IN-1 (compound 53a) is a selective eukaryotic initiation factor 4A3 (eIF4A3) inhibitor (IC50=0.26 μM; Kd=0.043 μM), which binds to a non-ATP binding site of eIF4A3 and shows significant cellular nonsense-mediated RNA decay (NMD) inhibition at 10 and 3 μM and can be as a probe for further study of eIF4A3, the exon junction complex (EJC), and NMD.
-
GC65540
eIF4A3-IN-2
eIF4A3-IN-2 is a highly selective and noncompetitive eukaryotic initiation factor 4A-3 (eIF4A3) inhibitor with an IC50 of 110 nM.