Cell Cycle/Checkpoint
Cell Cycle
Cells undergo a complex cycle of growth and division that is referred to as the cell cycle. The cell cycle consists of four phases, G1 (GAP 1), S (synthesis), G2 (GAP 2) and M (mitosis). DNA replication occurs during S phase. When cells stop dividing temporarily or indefinitely, they enter a quiescent state called G0.
Targets for Cell Cycle/Checkpoint
- ATM/ATR(26)
- Aurora Kinase(47)
- Cdc42(4)
- Cdc7(4)
- Chk(16)
- c-Myc(20)
- CRM1(8)
- Cyclin-Dependent Kinases(91)
- E1 enzyme(1)
- G-quadruplex(14)
- Haspin(7)
- HMTase(1)
- Kinesin(26)
- Ksp(6)
- Microtubule/Tubulin(243)
- Mps1(15)
- Mitotic(11)
- RAD51(18)
- ROCK(71)
- Rho(13)
- PERK(11)
- PLK(37)
- PTEN(8)
- Wee1(7)
- PAK(21)
- Arp2/3 Complex(8)
- Dynamin(12)
- ECM & Adhesion Molecules(40)
- Cholesterol Metabolism(3)
- Endomembrane System & Vesicular Trafficking(26)
- G1(38)
- G2/M(26)
- G2/S(10)
- Genotoxic Stress(18)
- Inositol Phosphates(18)
- Proteolysis(99)
- Cytoskeleton & Motor Proteins(53)
- Cellular Chaperones(8)
Products for Cell Cycle/Checkpoint
- Cat.No. Product Name Information
-
GC15663
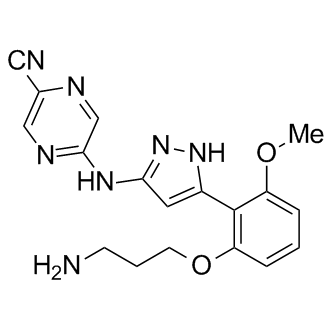
LY2606368
LY2606368 (LY2606368) is a selective, ATP-competitive second-generation checkpoint kinase 1 (CHK1) inhibitor with a Ki of 0.9 nM and an IC50 of <1 nM. LY2606368 inhibits CHK2 (IC50=8 nM) and RSK1 (IC50=9 nM). LY2606368 causes double-stranded DNA breakage and replication catastrophe resulting in apoptosis. LY2606368 shows potent anti-tumor activity.
-
GC16822
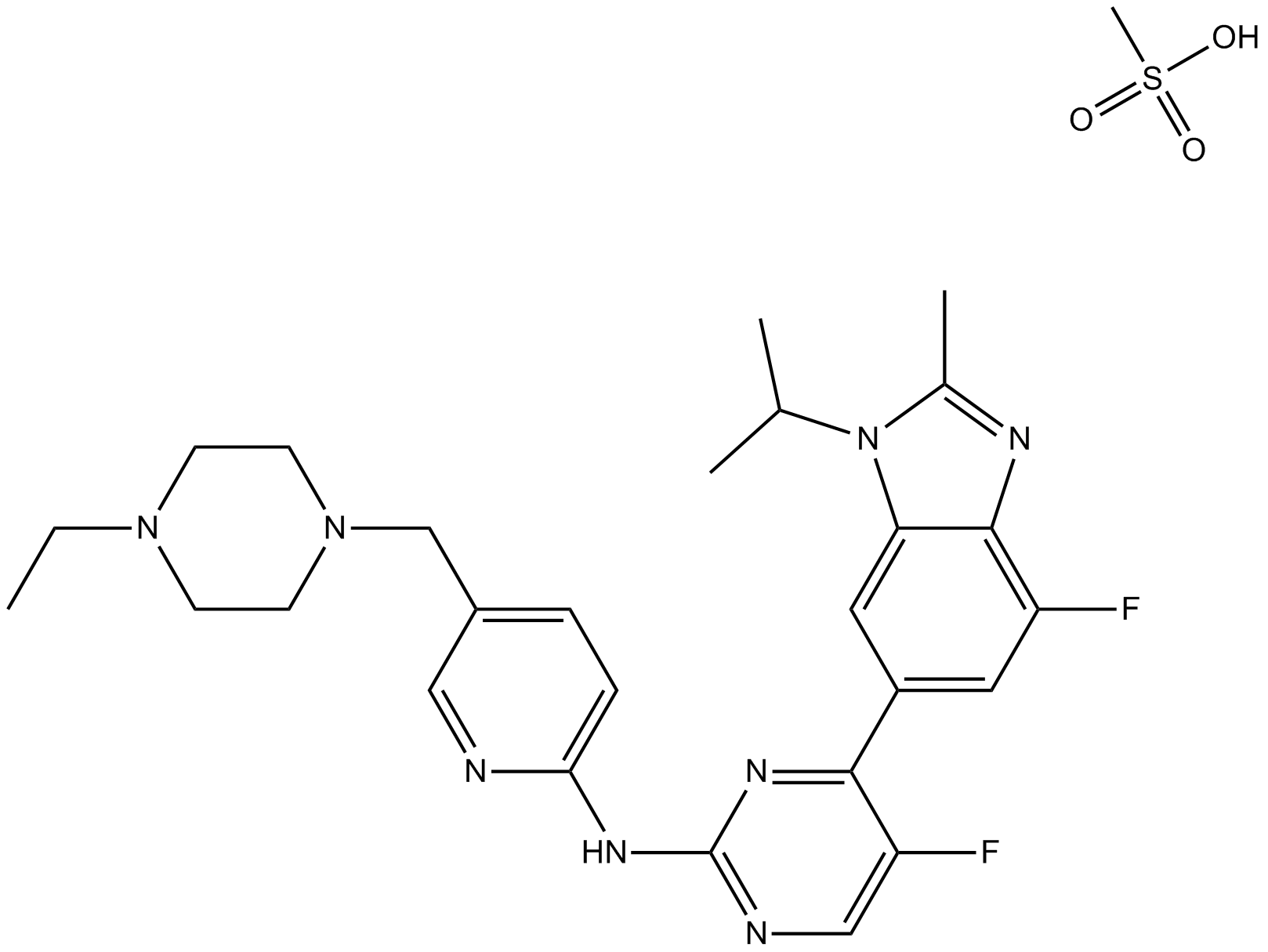
LY2835219
LY2835219 (LY2835219 methanesulfonate) is a selective CDK4/6 inhibitor with IC50s of 2 nM and 10 nM for CDK4 and CDK6, respectively.
-
GC11823
LY2835219 free base
A dual inhibitor of Cdk4 and Cdk6

-
GC11971
LY2857785
CDK9 inhibitor
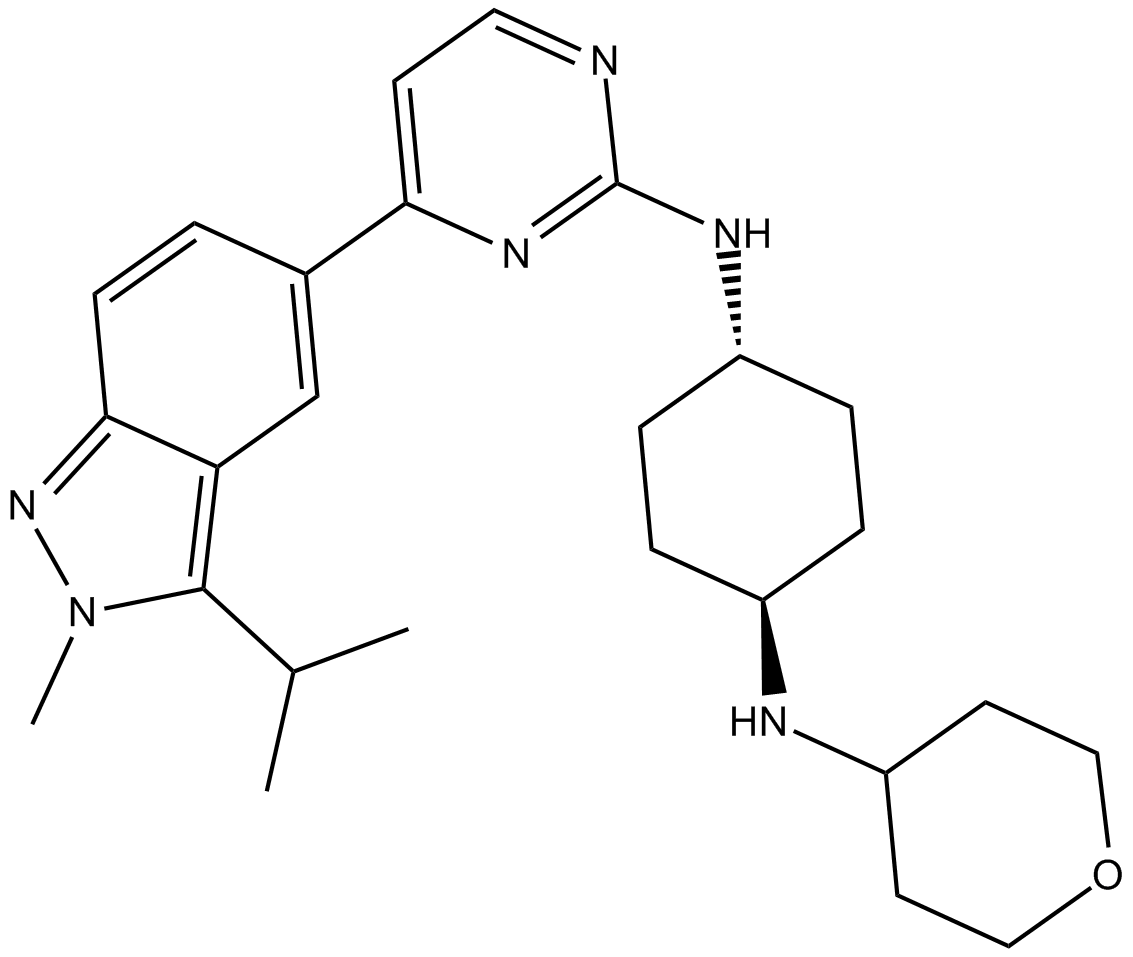
-
GC33057
LY3295668 (AK-01)
LY3295668 (AK-01) (AK-01) is a potent, orally active and highly specific Aurora-A kinase inhibitor, with Ki values of 0.8 nM and 1038 nM for AurA and AurB, respectively.
-
GC44100
LysoFP-NH2
LysoFP-NH2 is the fluorescent form of the lysosomal turn-on fluorescent probe for carbon monoxide (CO) lysoFP-NO2.
-
GC40021
LysoFP-NO2
LysoFP-NO2 is a turn-on fluorescent probe for carbon monoxide (CO) that localizes to the lysosome.
-
GC52358
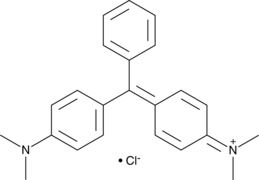
Malachite Green (chloride)
A triphenylmethane dye
-
GC41186
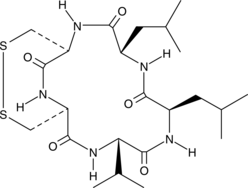
Malformin C
Malformin C is a natural fungus-derived bicyclic pentapeptide that has antibacterial properties, particularly against species of Bacillus.
-
GC61400
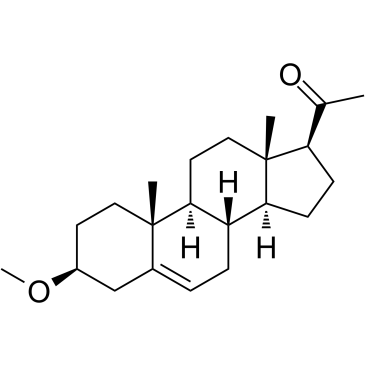
MAP4343
MAP4343 is the 3-methylether derivative of Pregnenolone.
-
GC62205
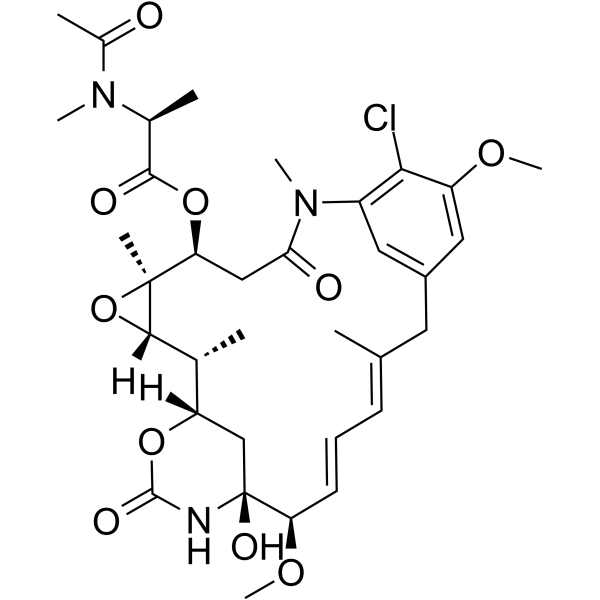
Maytansine
Maytansine is a highly potent microtubule-targeted compound that induces mitotic arrest and kills tumor cells at subnanomolar concentrations.
-
GC32895
Maytansinol (Ansamitocin P-0)
Maytansinol (Ansamitocin P-0) inhibits microtubule assembly and induces microtubule disassembly in vitro.
-
GC50574
MB 0223
Dynamin-related GTPase DRP1 partial inhibitor
-
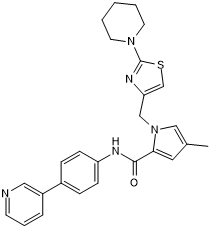
GC36560
Mc-MMAD
Mc-MMAD is a protective group (maleimidocaproyl)-conjugated MMAD. MMAD is a potent tubulin inhibitor. Mc-MMAD is a drug-linker conjugate for ADC.
-
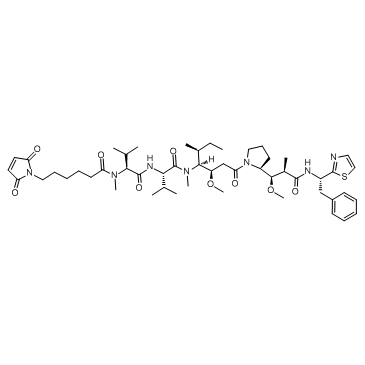
GC36561
Mc-MMAE
Mc-MMAE is a protective group (maleimidocaproyl)-conjugated monomethyl auristatin E (MMAE), which is a potent tubulin inhibitor. Mc-MMAE is a drug-linker conjugate for ADC.
-
GC18363
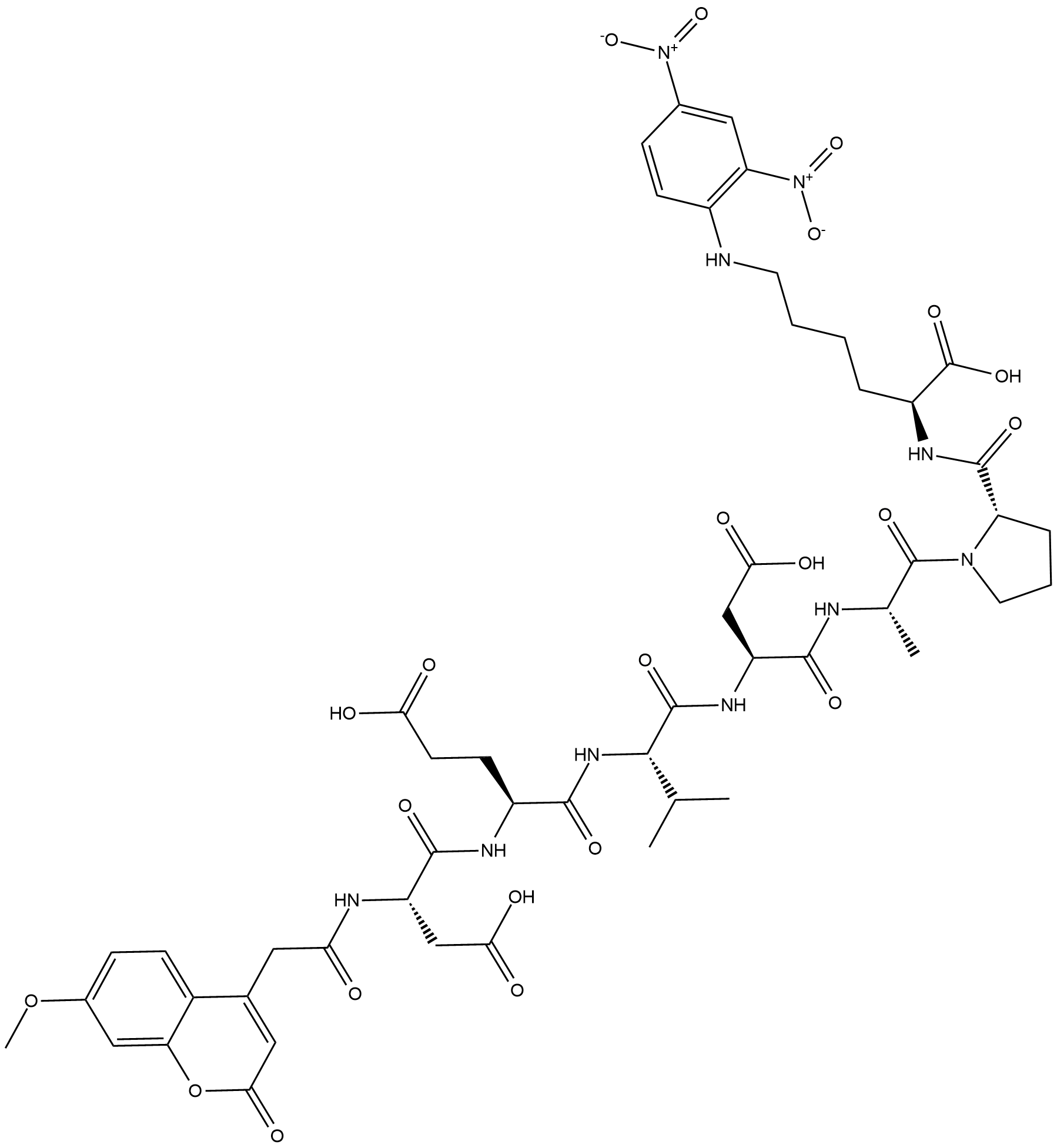
Mca-DEVDAPK(Dnp)-OH
Mca-DEVDAPK(Dnp)-OH is a fluorogenic substrate for caspase-3.
-
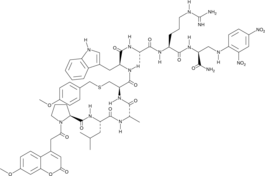
GC44132
Mca-PLAC(p-OMeBz)-WAR(Dpa)-NH2
Mca-PLAC(p-OMeBz)-WAR(Dpa)-NH2 is a fluorogenic substrate for matrix metalloproteinase-14 (MMP-14).
-
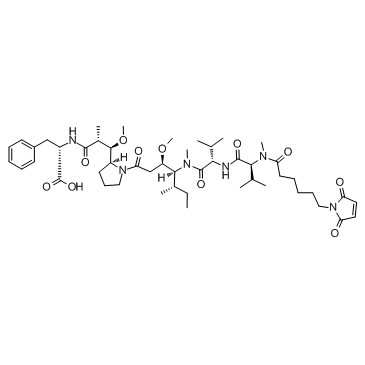
GC34068
McMMAF (Maleimidocaproyl monomethylauristatin F)
McMMAF (Maleimidocaproyl monomethylauristatin F) is a protective group-conjugated MMAF. MMAF is a potent tubulin polymerization inhibitor.
-
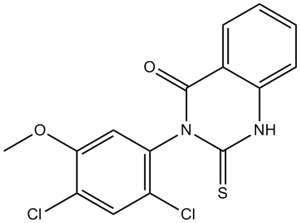
GC10200
Mdivi 1
A mitochondrial division inhibitor
-
GC17649
Mebendazole
broad-spectrum anthelmintic that inhibits intestinal microtubule synthesis
-
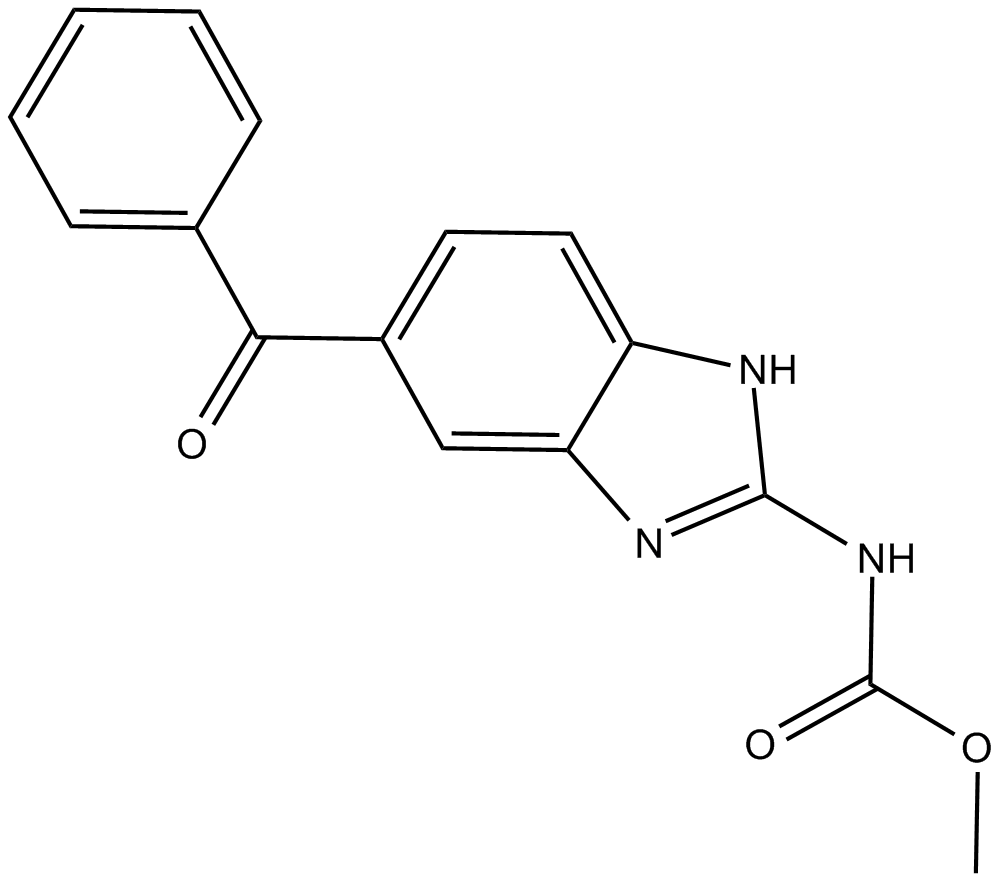
GC49486
Meglutol-d3
An internal standard for the quantification of meglutol
-
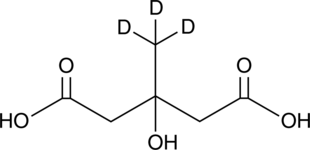
GC47619
Menaquinone 4-d7
An internal standard for the quantification of menaquinone 4
-
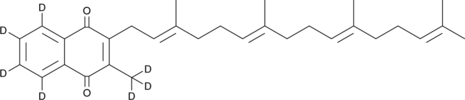
GC19244
Mertansine
Mertansine (DM1) is a microtubulin inhibitor which binds at the tips of microtubules and suppresses the dynamicity of microtubules..
-
GC16607
Mesalamine
5-Aminosalicylic acid (Mesalamine) acts as a specific PPARγ agonist and also inhibits p21-activated kinase 1 (PAK1) and NF-κB.
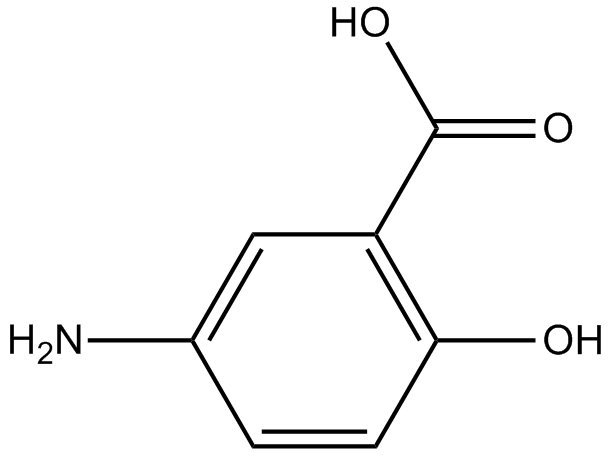
-
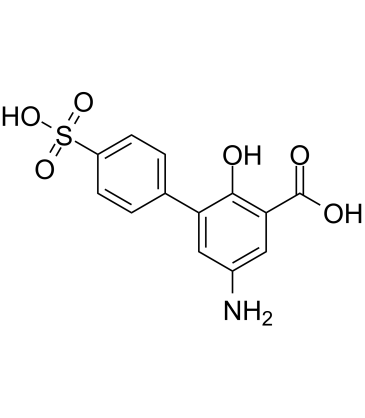
GC61043
Mesalamine impurity P
Mesalamine impurity P is an impurity of Mesalamine.
-
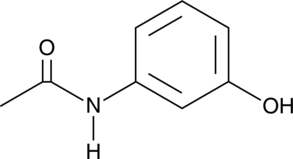
GC49065
Metacetamol
A derivative of acetaminophen
-
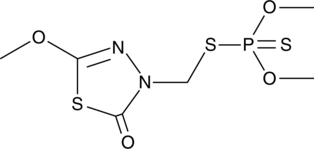
GC47645
Methidathion
An organophosphate insecticide
-
GC47646
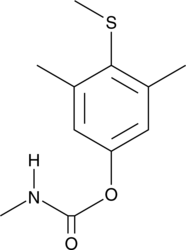
Methiocarb
A carbamate pesticide
-
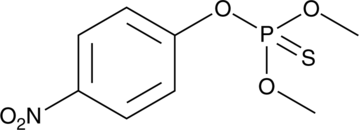
GC47660
Methyl Parathion
An organophosphate insecticide
-
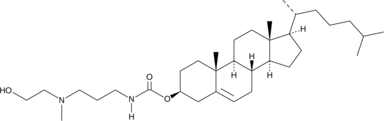
GC40036
MHAPC-Chol
MHAPC-Chol is a cationic cholesterol.
-
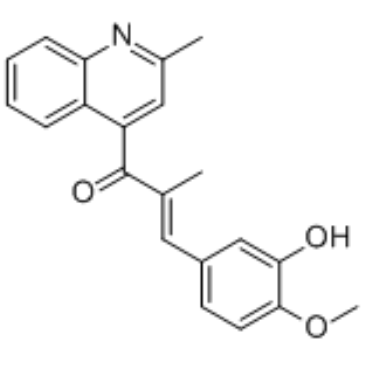
GC36608
Microtubule inhibitor 1
Microtubule inhibitor 1 is an antitumor agent with microtubule polymerization inhibitory activity, with an IC50 value of 9-16 nM in cancer cells.
-
GC13491
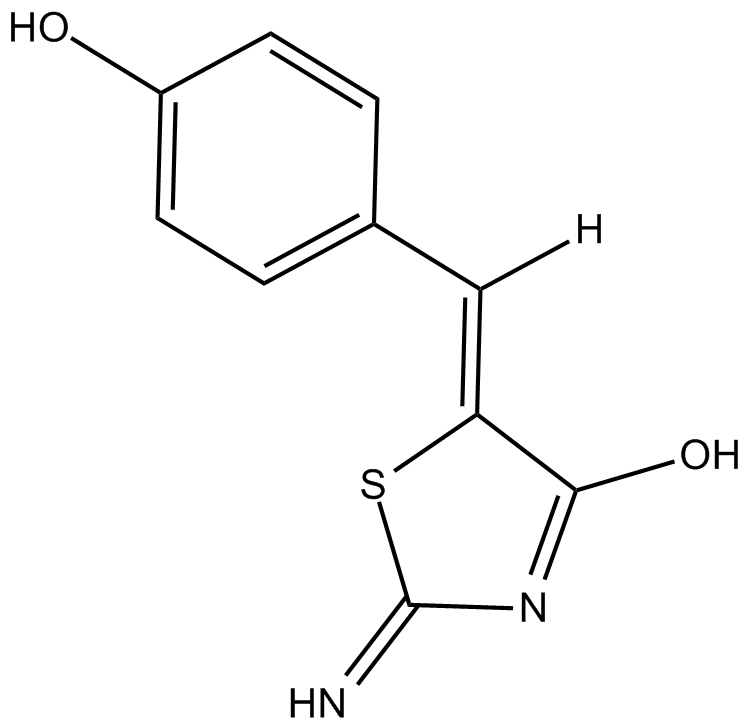
Mirin
Mirin is a small-molecule inhibitor of MRN (Mre11, Rad50, and Nbs1) complex.
-
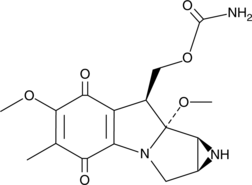
GC44200
Mitomycin A
Mitomycin A is a bacterial metabolite originally isolated from S.
-
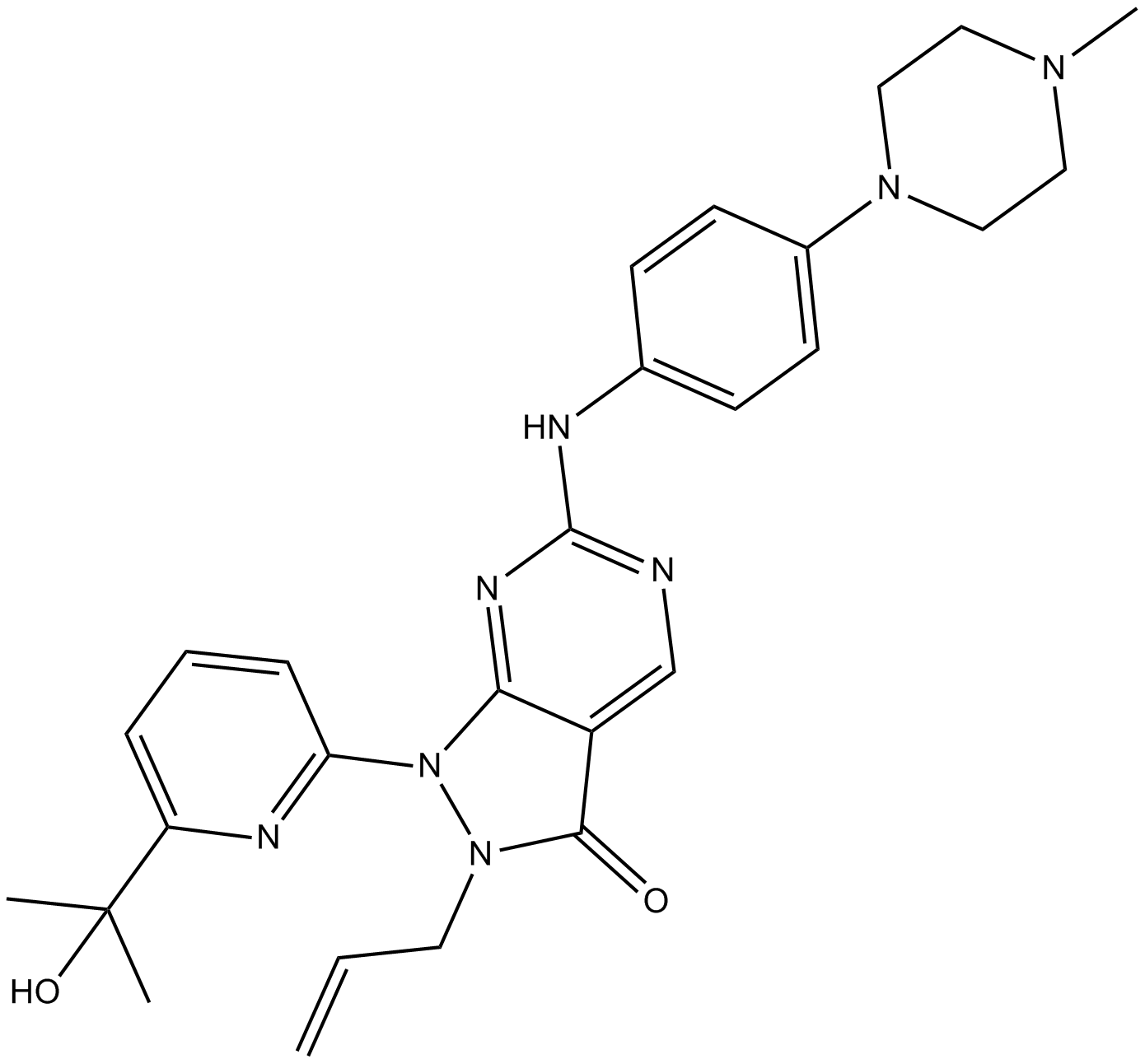
GC16030
MK-1775
MK-1775 (AZD-1775; MK-1775) is a potent Wee1 inhibitor with an IC50 of 5.2 nM.
-
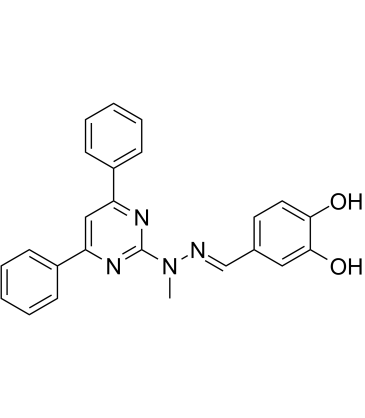
GC61072
MK-28
MK-28 is a potent and selective PERK activator.
-
GC10442
MK-8745
Aurora A inhibitor,potent and selective
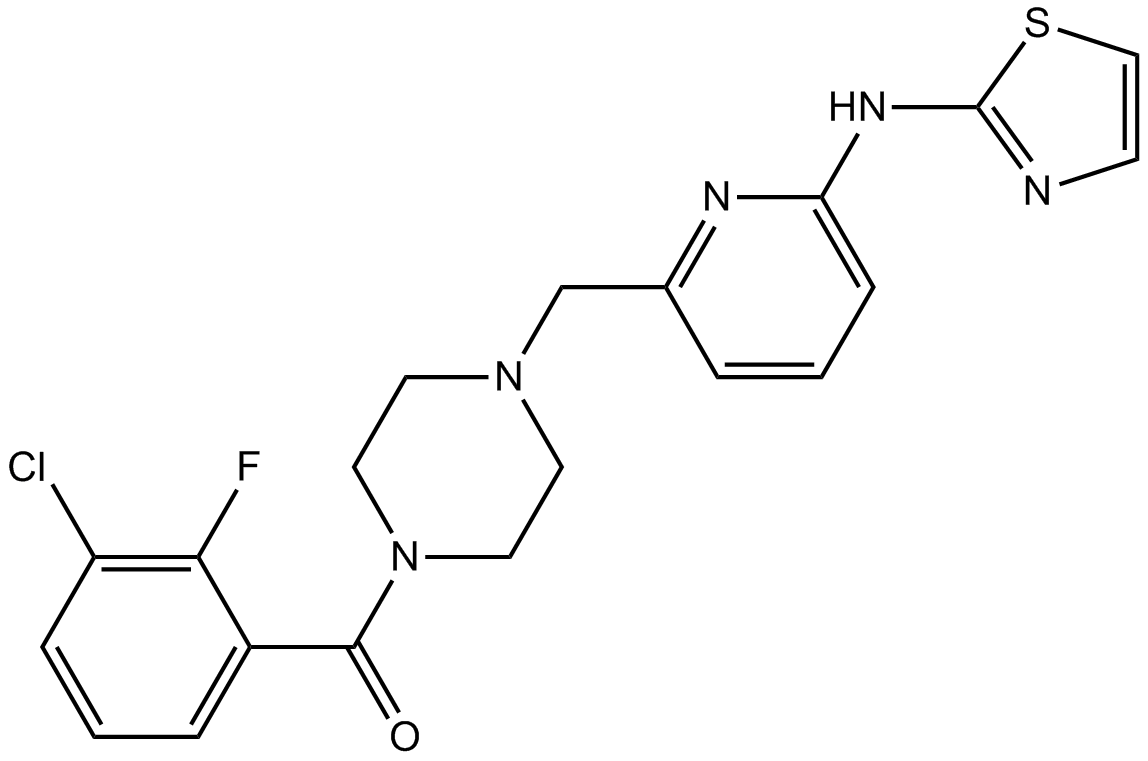
-
GC16374
MK-8776(SCH-900776)
MK-8776(SCH-900776) (MK-8776) is a potent, selective and orally bioavailable inhibitor of checkpoint kinase1 (Chk1) with an IC50 of 3 nM. MK-8776(SCH-900776) shows 50- and 500-fold selectivity over CDK2 and Chk2, respectively.
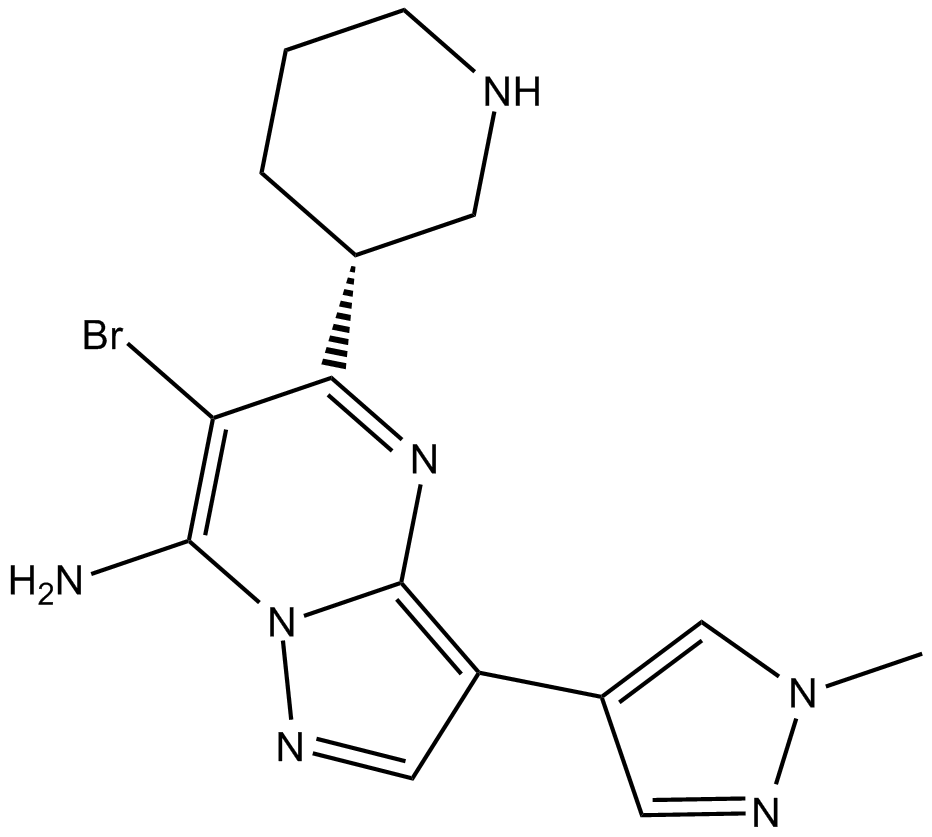
-
GC65143
MKC-1
MKC-1 (Ro-31-7453) is an orally active and potent cell cycle inhibitor with broad antitumor activity. MKC-1 inhibits the Akt/mTOR pathway. MKC-1 arrests cellular mitosis and induces cell apoptosis by binding to a number of different cellular proteins including tubulin and members of the importin β family.
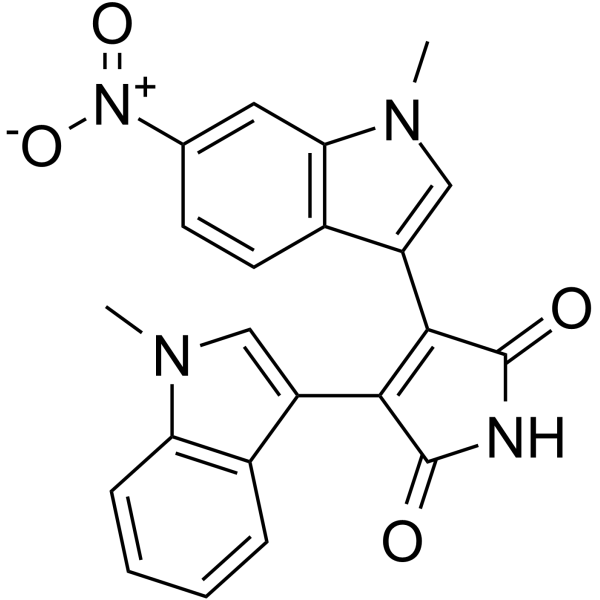
-
GC14800
ML 141
Cdc42 GTPase inhibitor
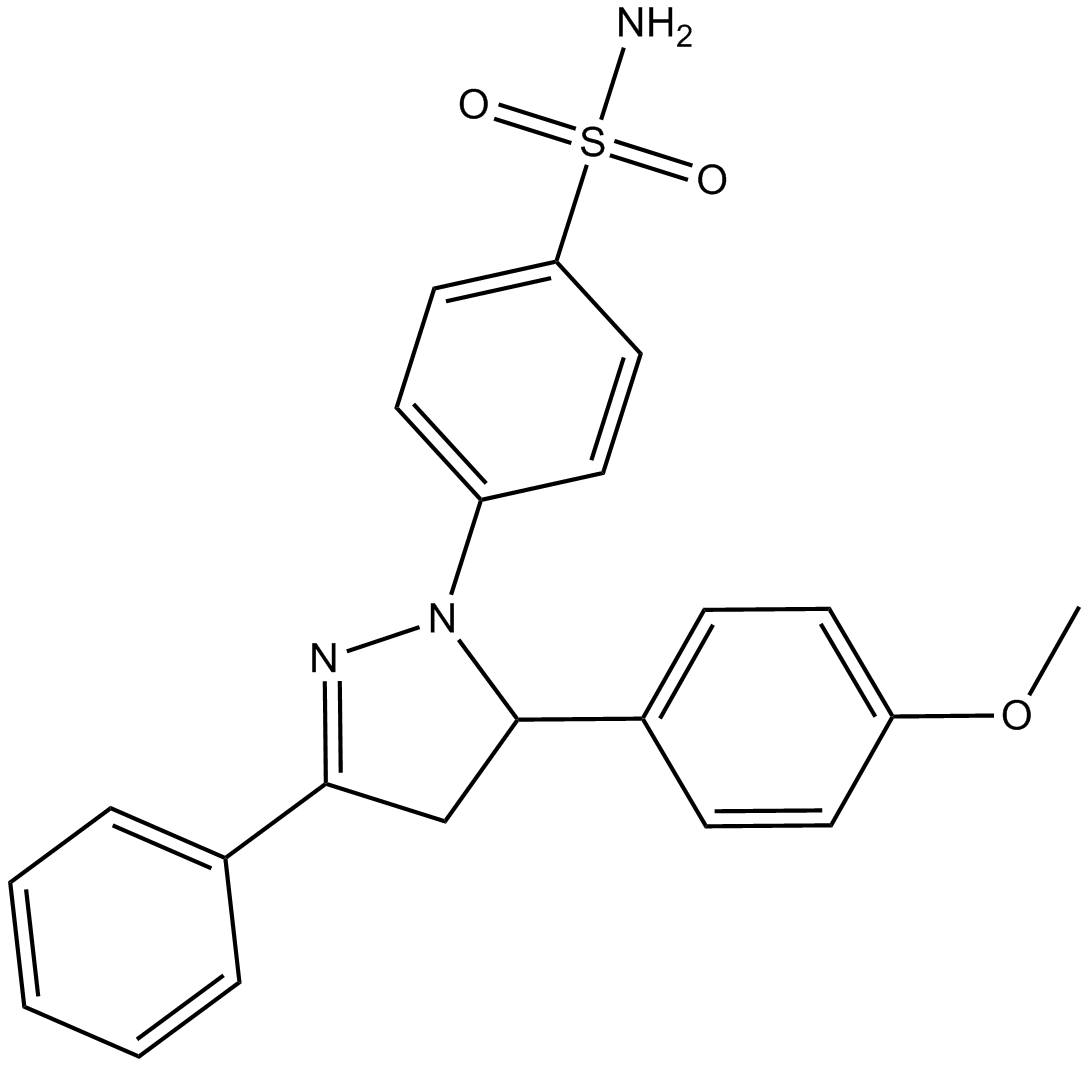
-
GC14162
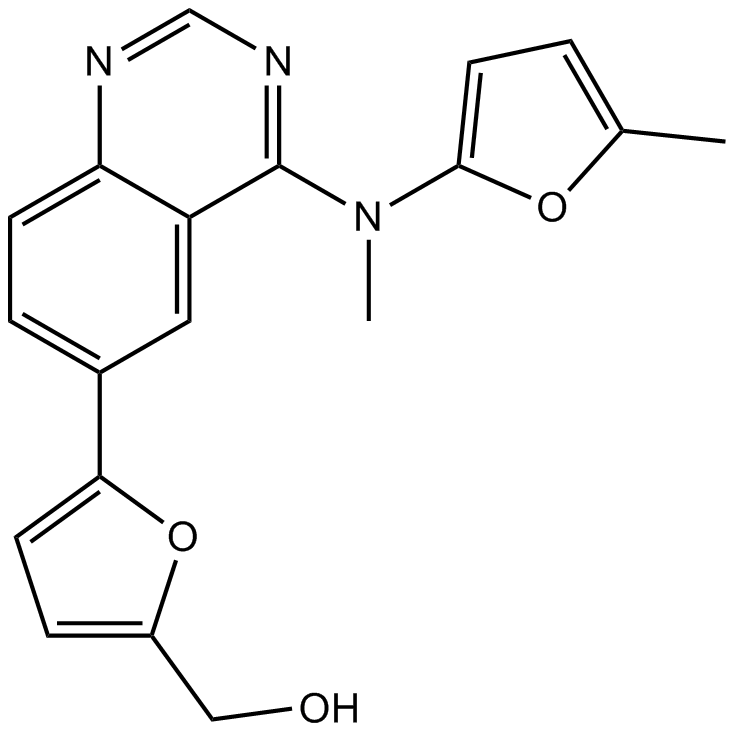
ML167
Clk4 inhibitor,highly selective
-
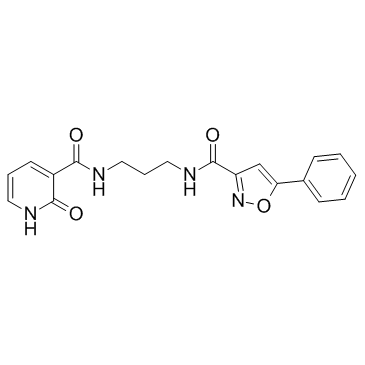
GC32785
ML327
ML327 is a blocker of MYC which can also de-repress E-cadherin transcription and reverse Epithelial-to-Mesenchymal Transition (EMT).
-
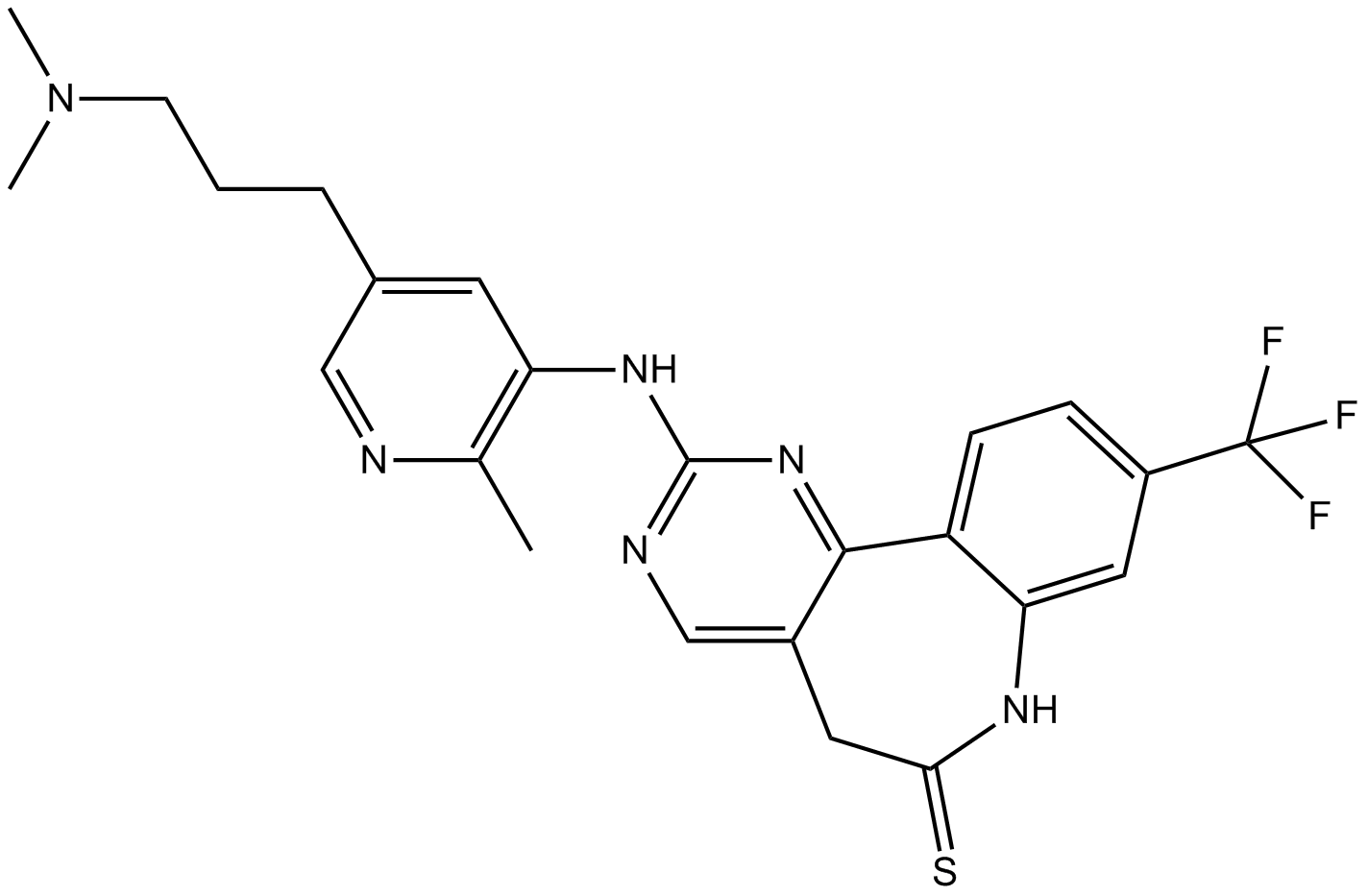
GC10163
MLN0905
Potent PLK1 inhibitor
-
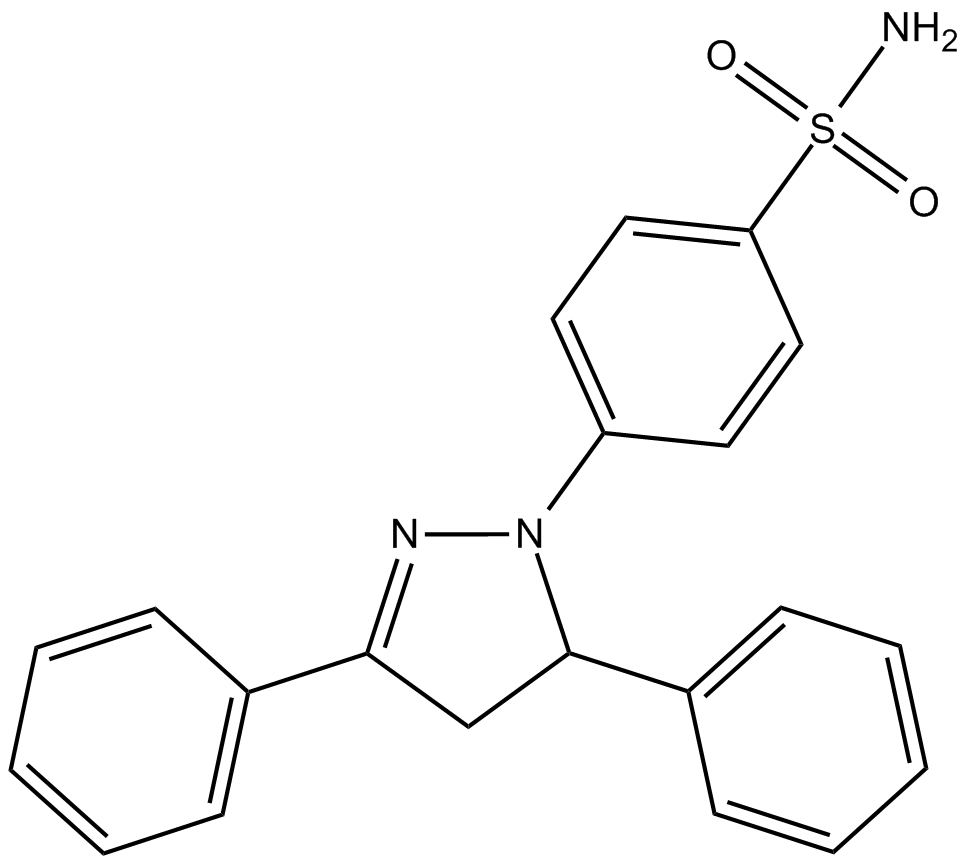
GC13096
MLS-573151
Cdc42 inhibitor
-
GC64591
MM41
MM41 is a potent stabilizer of human telomeric and gene promoter DNA quadruplexes. MM41 is potently against the MIA PaCa-2 pancreatic cancer cell line with IC50 value of <10 nM.

-
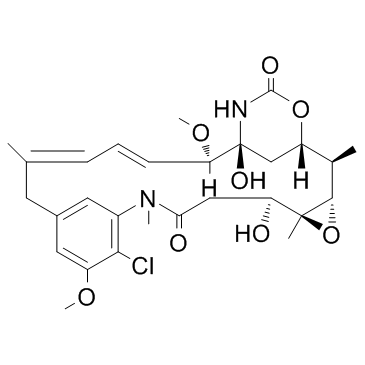
GC15798
MMAD
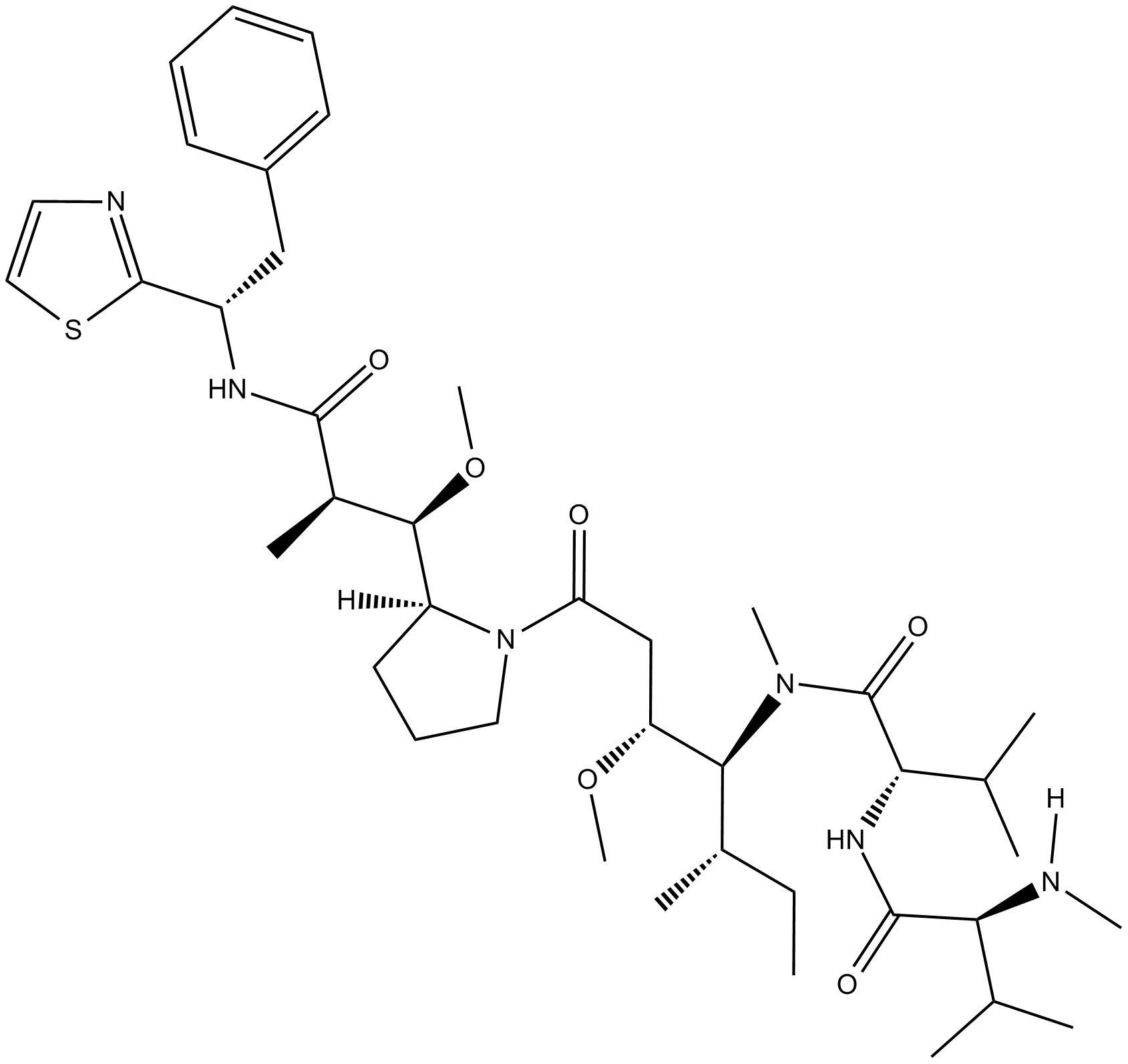
-
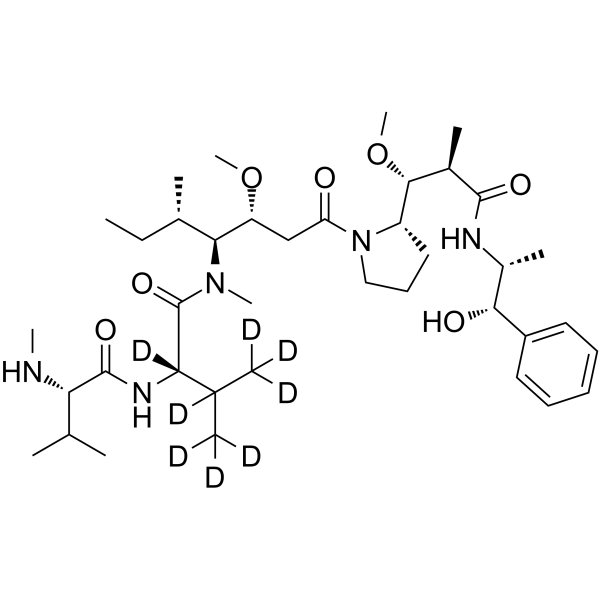
GC63461
MMAE-d8
MMAE-d8 (Monomethyl auristatin E-d8) is a deuterated labeled MMAE, a potent mitotic inhibitor and a tubulin inhibitor.
-
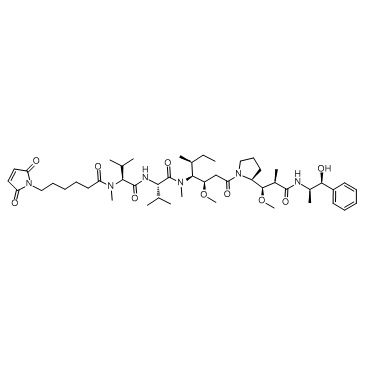
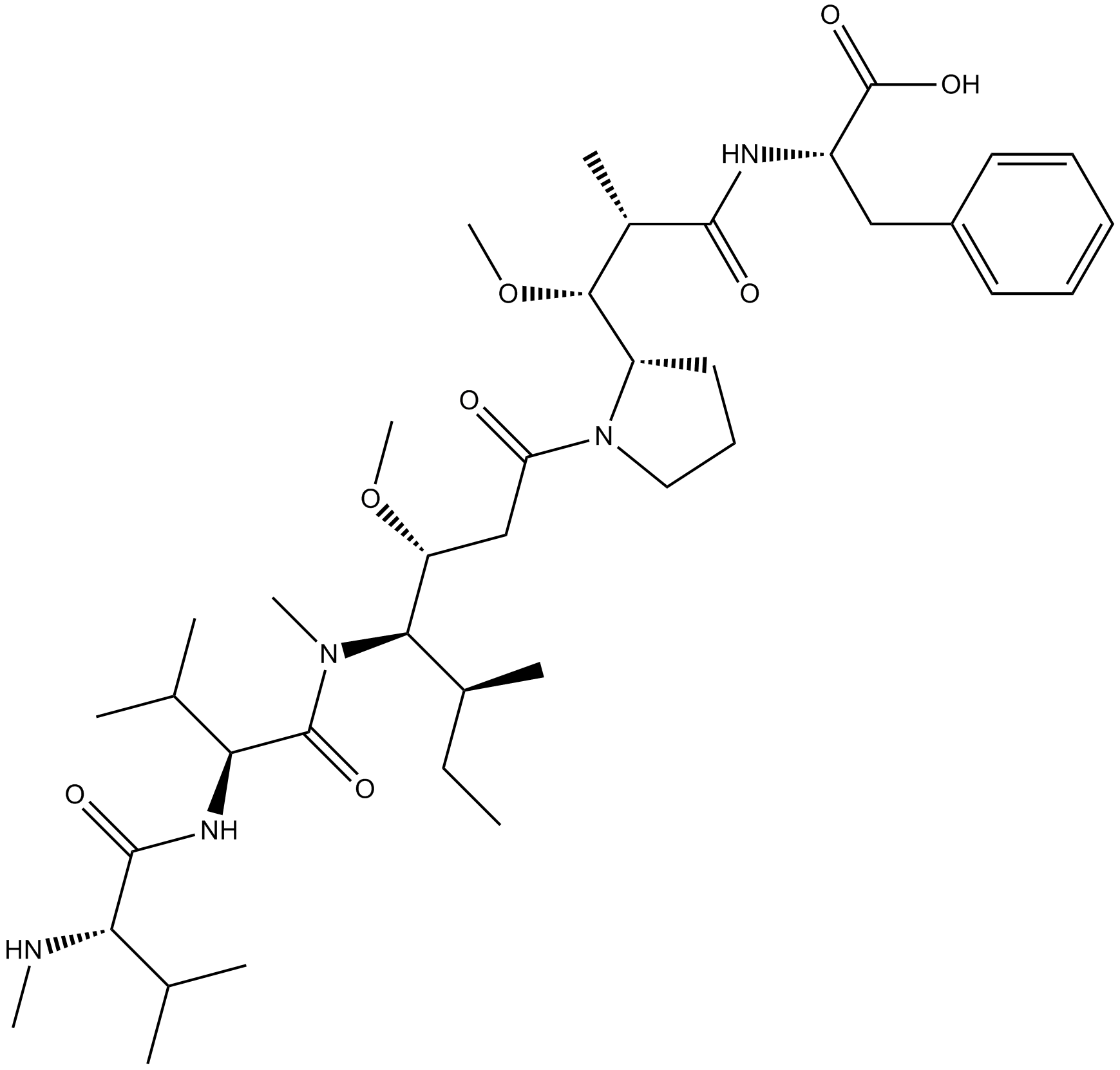
GC10034
MMAF
Anti-mitotic/anti-tubulin/antineoplastic agent
-
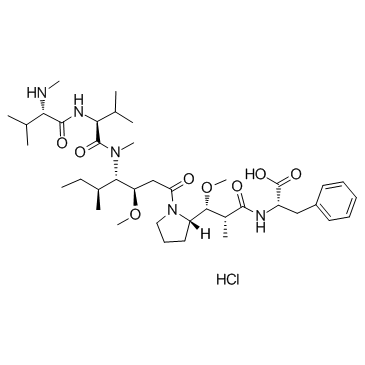
GC36633
MMAF Hydrochloride
MMAF (Monomethylauristatin F) hydrochloride is a potent tubulin polymerization inhibitor and is used as a antitumor agent. MMAF hydrochloride is widely used as a cytotoxic component of antibody-drug conjugates (ADCs) such as Vorsetuzumab mafodotin and SGN-CD19A.
-
GC38397
MMAF sodium
MMAF sodium (Monomethylauristatin F sodium) is a potent tubulin polymerization inhibitor and is used as a antitumor agent. MMAF sodium (Monomethylauristatin F sodium) is widely used as a cytotoxic component of antibody-drug conjugates (ADCs) such as Vorsetuzumab mafodotin and SGN-CD19A.
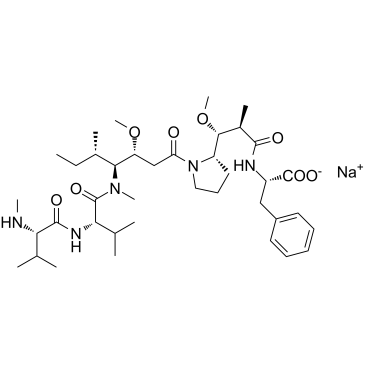
-
GC63774
MMAF-d8 hydrochloride
D8-MMAF hydrochloride is a deuterated form of MMAF hydrochloride, which is a microtubule disrupting agent.
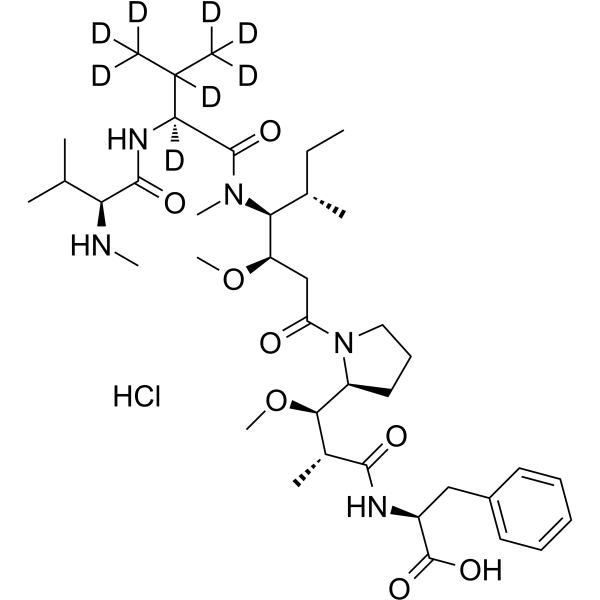
-
GC44235
MMP Inhibitor I (trifluoroacetate salt)
MMP inhibitor I is a peptide inhibitor of matrix metalloproteinase-1 (MMP-1), MMP-2, and MMP-3 (IC50s = 1.3, 30, and 150 μM, respectively).
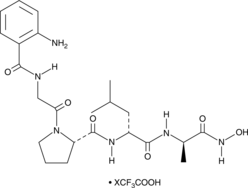
-
GC18218
MMP-3 Inhibitor
MMP-3 inhibitor is a peptide inhibitor of matrix metalloproteinase-3 (MMP-3) with a Ki value of 95 nM.
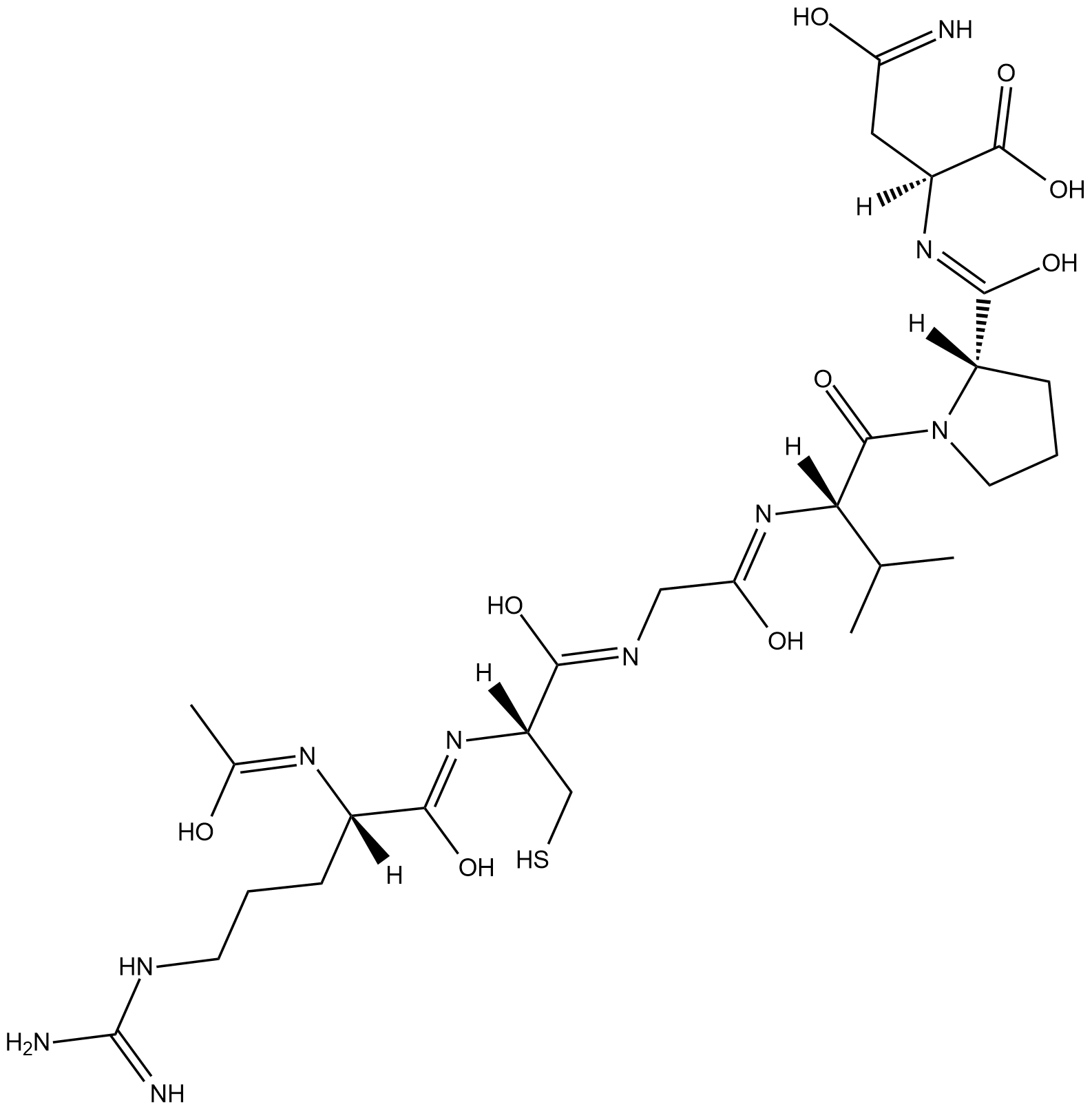
-
GC40624
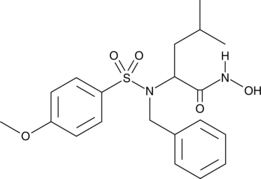
MMP-3 Inhibitor VIII
Matrix metalloproteinases (MMPs) belong to a family of proteases that play a crucial role in tissue remodeling and repair by degrading extracellular matrix proteins to enable cell migration.
-
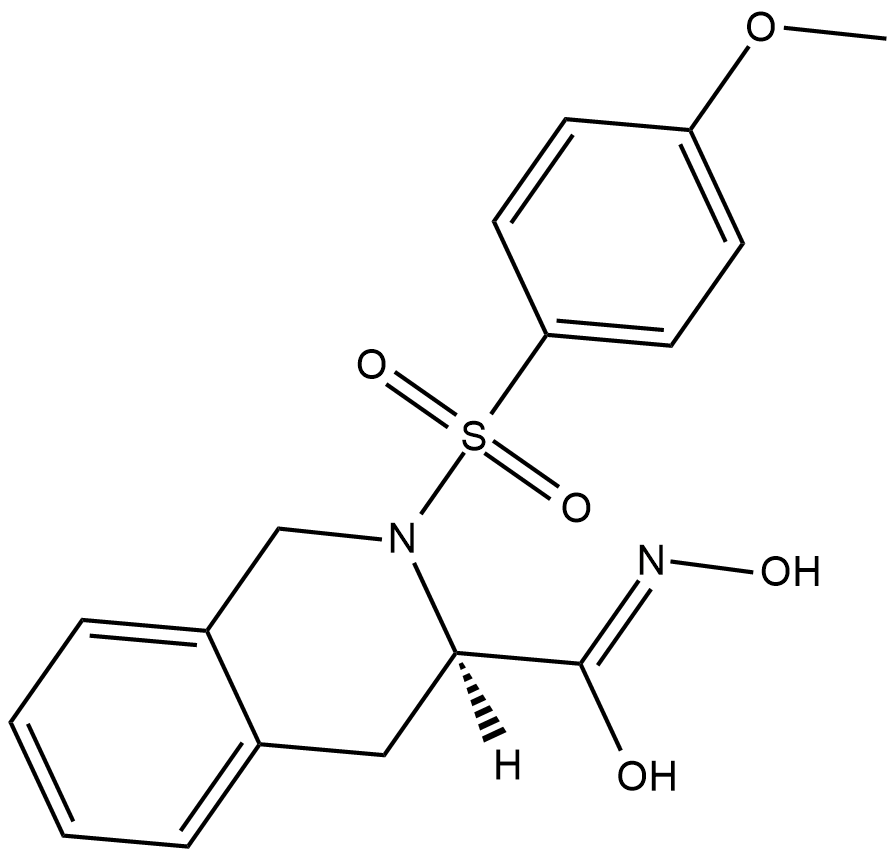
GC18616
MMP-8 Inhibitor I
MMP-8 Inhibitor I is a selective inhibitor of the neutrophil collagenase matrix metalloproteinase-8 (MMP-8) with an IC50 value of 4 nM.
-
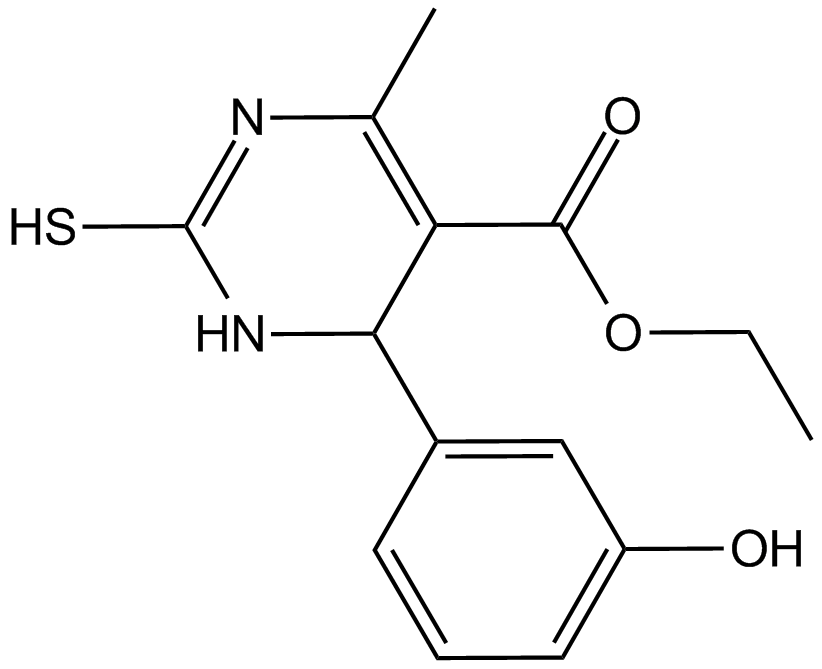
GC14929
Monastrol
Eg5 inhibitor
-
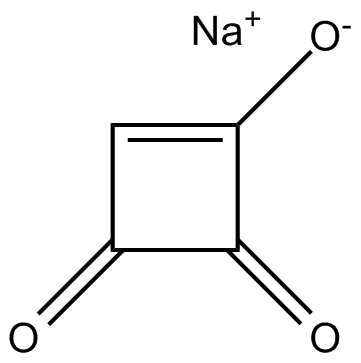
GC15592
Moniliformin (sodium salt)
induces mitotic arrest at the metaphase stage
-
GC17276
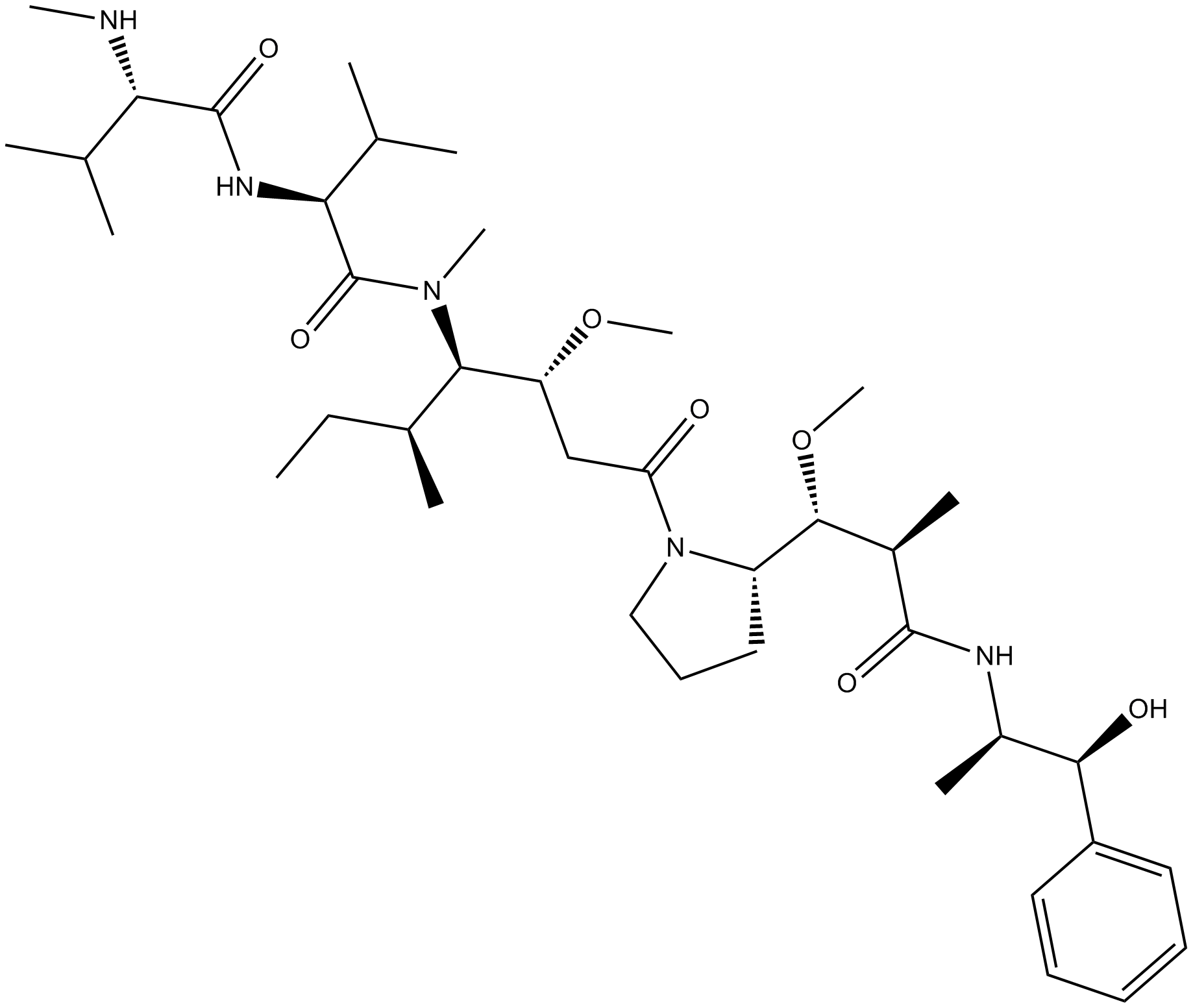
Monomethyl auristatin E
A potent antimitotic compound
-
GC45744
monoMICAAc
A solvatochromic fluorescent pH probe
-
GC15285
MPC 6827 hydrochloride
MPC 6827 hydrochloride (MPC-6827 hydrochloride) is a blood brain barrier permeable microtubule-disrupting agent, with potent and broad-spectrum in vitro and in vivo cytotoxic activities. MPC 6827 hydrochloride (MPC-6827 hydrochloride) exhibits potent anticancer activity in human MX-1 breast and other mouse xenograft cancer models. MPC 6827 hydrochloride (MPC 6827 hydrochloride) is a promising candidate for the treatment of multiple cancer types.
-
GC10083
MPI-0479605
Mps1 inhibitor,selective and ATP competitive
-
GC11292
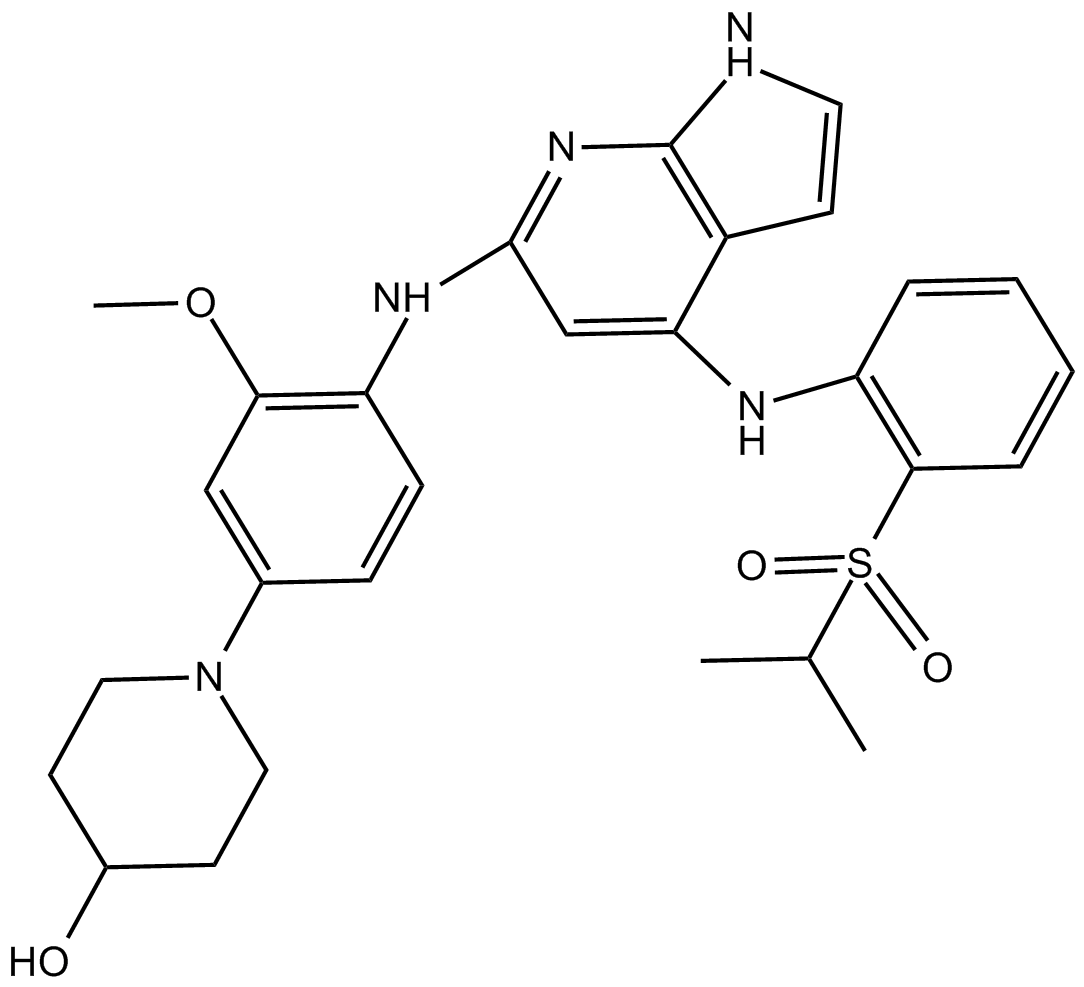
Mps1-IN-1
Mps1 kinase inhibitor
-
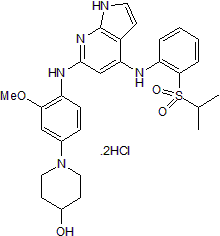
GC50110
Mps1-IN-1 dihydrochloride
Selective Mps1 kinase inhibitor
-
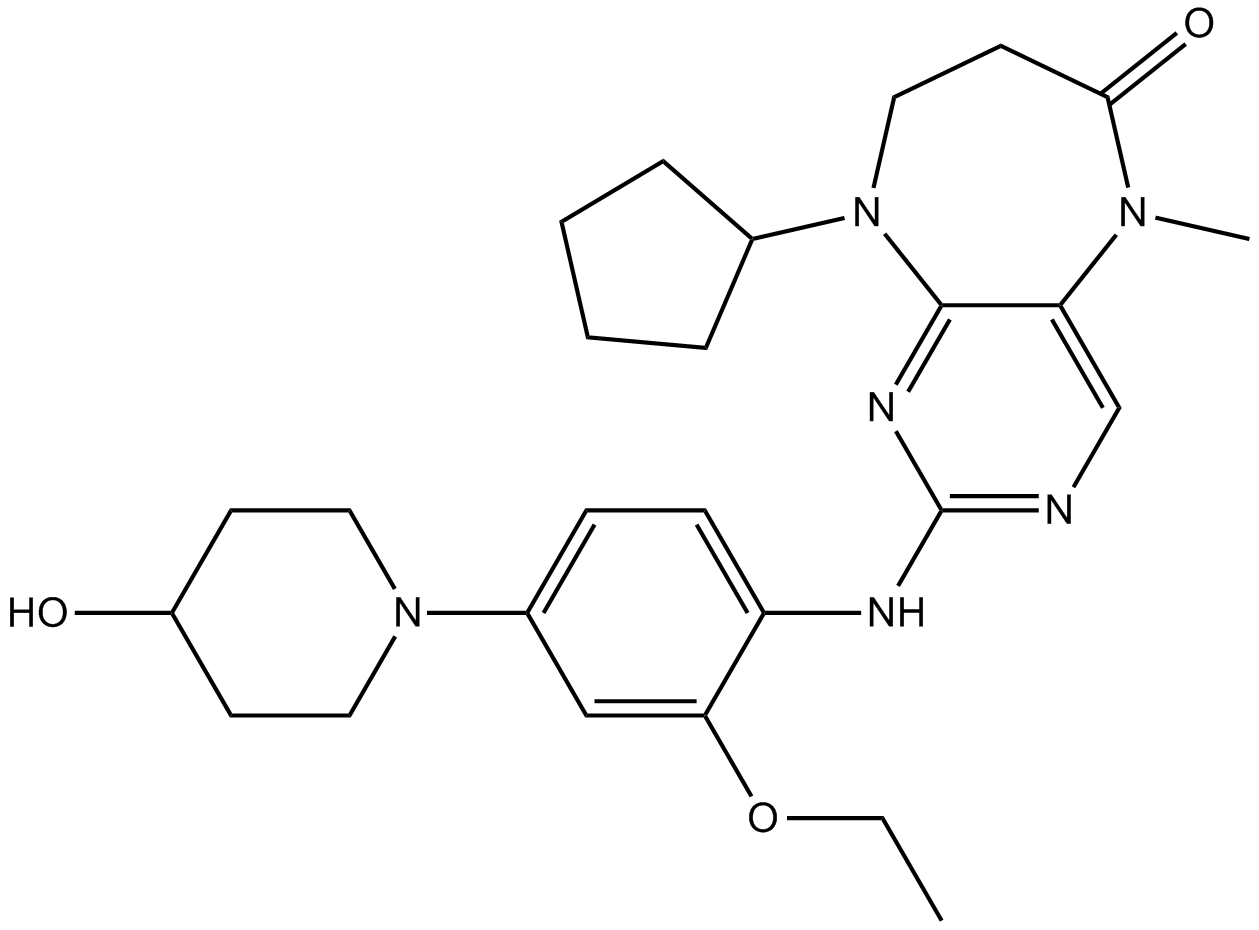
GC15745
Mps1-IN-2
Mps1 kinase inhibitor
-
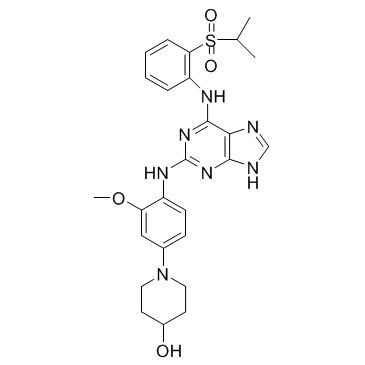
GC36651
Mps1-IN-3
Mps1-IN-3 is a potent and selective MPS1 kinase inhibitor, with an IC50 of 50 nM.
-
GC65339
Mps1-IN-3 hydrochloride
Mps1-IN-3 hydrochloride is a potent and selective Mps1 inhibitor with an IC50 value of 50 nM. Mps1-IN-3 hydrochloride can inhibit the proliferation of glioblastoma cells, and effectively sensitizes glioblastomas to Vincristine in orthotopic glioblastoma xenograft model.
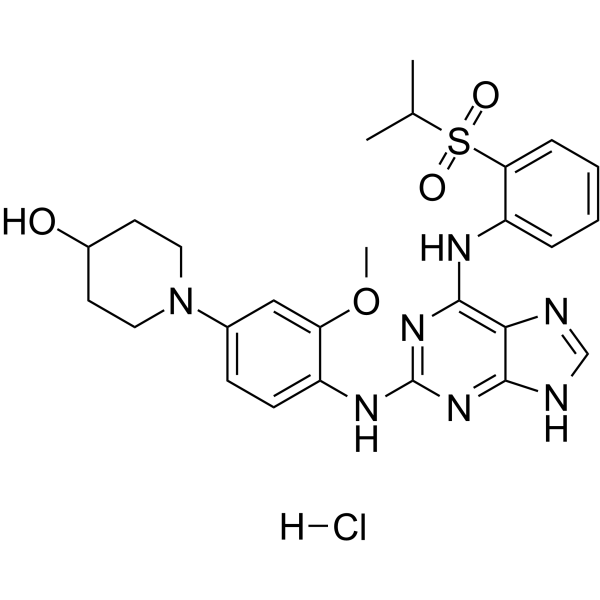
-
GC40048
MPS1/TTK Inhibitor
MPS1/TTK inhibitor is an inhibitor of monopolar spindle 1 (MPS1/TTK; IC50 = 5.8 nM), a kinase involved in mitotic spindle checkpoint signaling that is overexpressed in certain cancerous tumors.
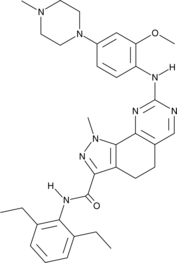
-
GC41564
MPT0B014
An inhibitor of tubulin polymerization
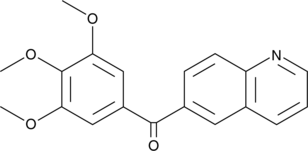
-
GC36652
MPT0B392
MPT0B392, an orally active quinoline derivative, induces c-Jun N-terminal kinase (JNK) activation, leading to apoptosis. MPT0B392 inhibits tubulin polymerization and triggers induction of the mitotic arrest, followed by mitochondrial membrane potential loss and caspases cleavage by activation of JNK and ultimately leads to apoptosis. MPT0B392 is demonstrated to be a novel microtubule-depolymerizing agent and enhances the cytotoxicity of sirolimus in sirolimus-resistant acute leukemic cells and the multidrug resistant cell line.
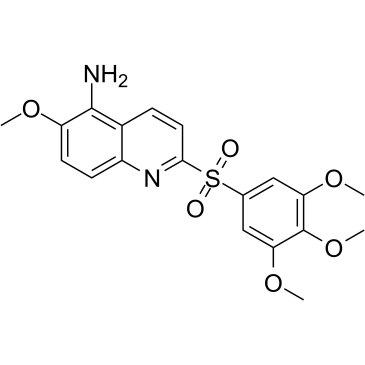
-
GC62657
MRIA9
MRIA9 is an ATP-competitive, pan Salt-Inducible kinase (SIK) and PAK2/3 inhibitor, with IC50 values of 516 nM, 180 nM and 127 nM for SIK1, SIK2 and SIK3, respectively.
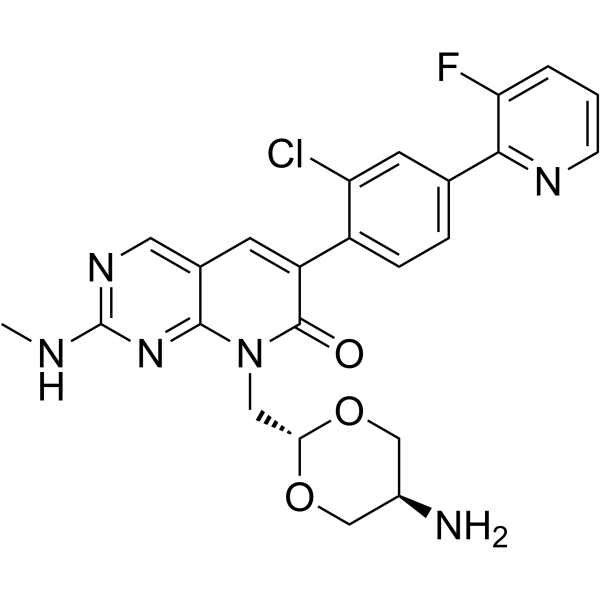
-
GC45515
MX1013
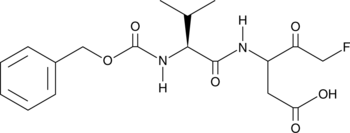
-
GC66457
MY-875
MY-875 is a competitive microtubulin polymerization inhibitor with an IC50 value of 0.92 μM. MY-875 inhibits microtubulin polymerization by targeting colchicine binding sites and activates the Hippo pathway. MY-875 induces apoptosis and has anticancer activity.
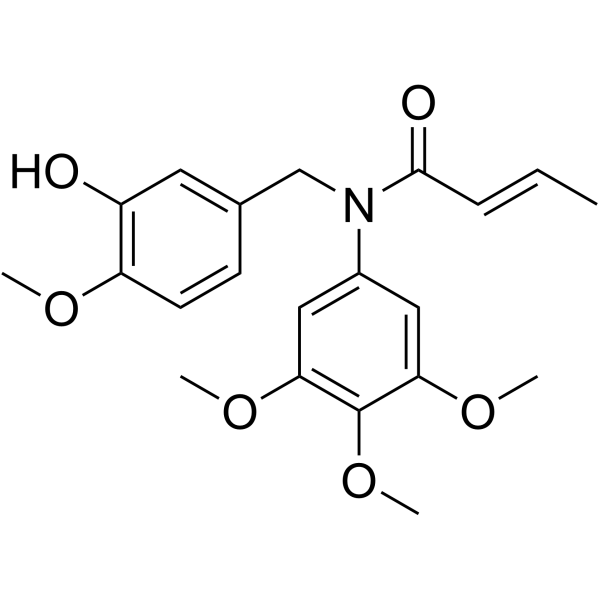
-
GC69516
MY-943
MY-943 is an effective tubulin polymerization and LSD1 inhibitor with anti-cancer activity. MY-943 induces G2/M phase arrest and cell apoptosis, and inhibits cell migration, making it suitable for gastric cancer research.
-
GC38589
MYCi361
MYCi361 (NUCC-0196361) is a MYC inhibitor with the Kd of 3.2 μM for binding to MYC. MYCi361 (NUCC-0196361) suppresses tumor growth and enhances anti-PD1 immunotherapy.
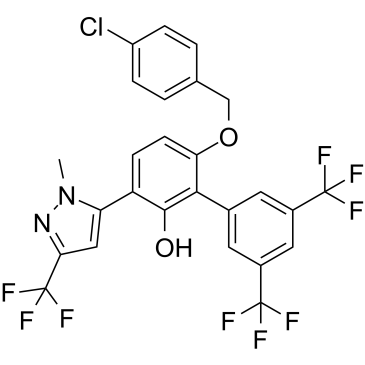
-
GC38588
MYCi975
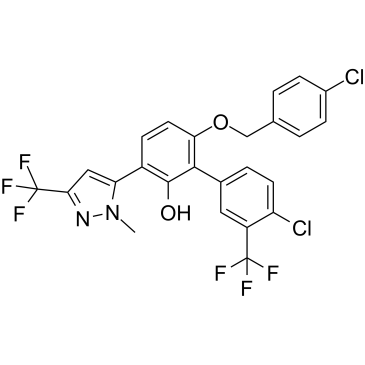
-
GC60259
MYCMI-6
MYCMI-6 (NSC354961) is a potent and selective endogenous MYC:MAX protein interactions inhibitor. MYCMI-6 blocks MYC-driven transcription and binds selectively to the MYC bHLHZip domain with a Kd of 1.6?μM. MYCMI-6 inhibits tumor cell growth in a MYC-dependent manner (IC50<0.5?μM). MYCMI-6 is not cytotoxic to normal human cells. MYCMI-6 induces apoptosis.
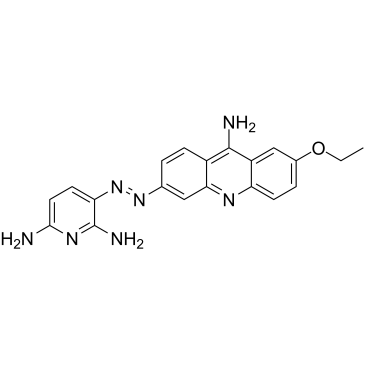
-
GC34094
Mycro 3
Mycro 3 is a potent and selective inhibitor of Myc-associated factor X (MAX) dimerization. Mycro 3 also inhibit DNA binding of c-Myc. Mycro 3 could be used for the research of pancreatic cancer.
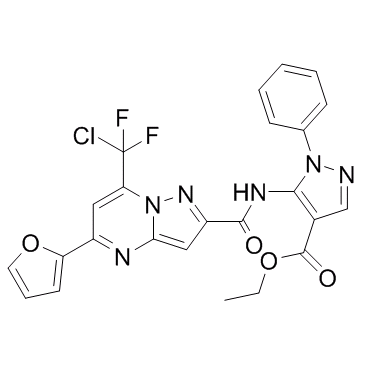
-
GC15773
Myoseverin
microtubule-binding molecule
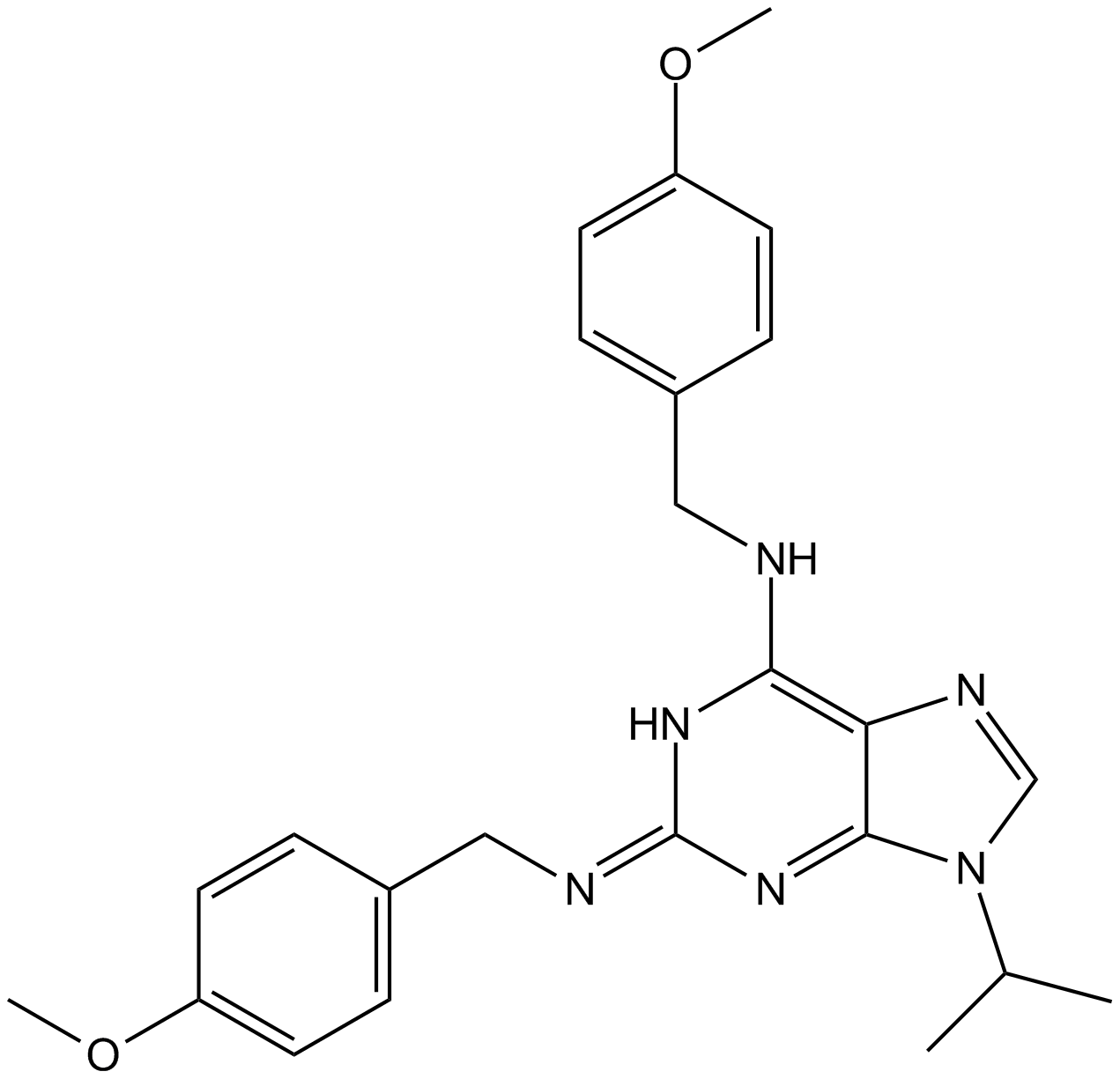
-
GC44260
Myristoyl Coenzyme A (hydrate)
Myristoyl coenzyme A (myristoyl-CoA) is a derivative of CoA that contains the long-chain fatty acid myristic acid.
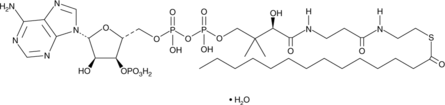
-
GC45958
Myrothecine A
A trichothecene mycotoxin
-
GC49491
N′-Nitrosonornicotine-d4
An internal standard for the quantification of N’-nitrosonornicotine
-
GC49163
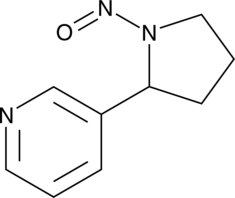
N’-Nitrosonornicotine
An N-nitrosamine and a carcinogen
-
GC45667
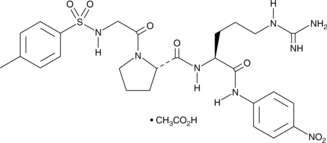
N-(p-Tosyl)-GPR-pNA (acetate)
A colorimetric thrombin substrate
-
GC18445
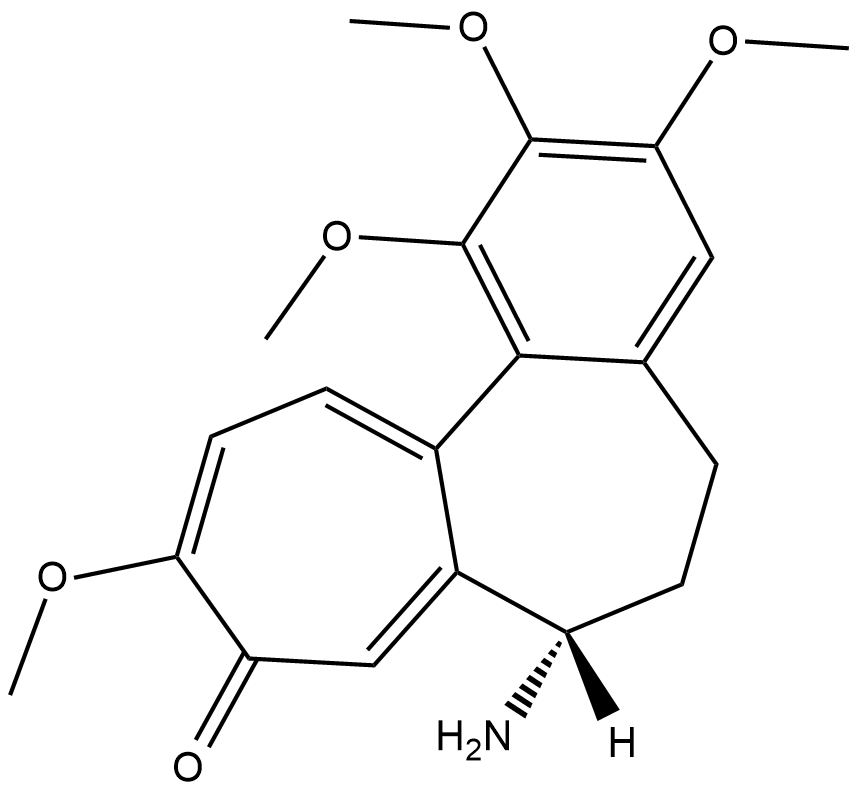
N-Deacetylcolchicine
An inhibitor of microtubule polymerization
-
GC44343
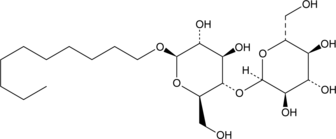
n-Decyl-β-D-maltoside
n-Decyl-β-D-maltoside is a nonionic surfactant that is commonly used to solubilize and stabilize membrane proteins.
-
GC52007
N-hydroxylamine Dapsone
An active metabolite of dapsone
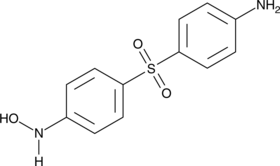
-
GC49351
N-Nitroso Atenolol
A genotoxic derivative of (±)-atenolol
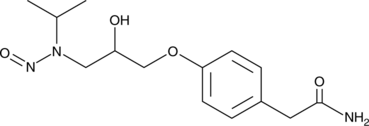
-
GC49394
N-Nitroso Fluoxetine
A derivative of fluoxetine
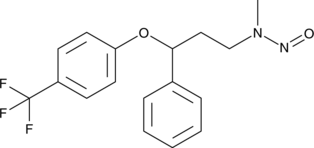
-
GC49353
N-Nitroso Metoprolol
A genotoxic derivative of metoprolol
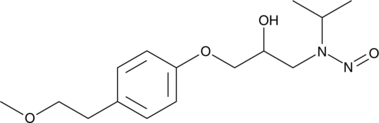
-
GC44430
n-Nonyl-β-D-glucopyranoside
n-Nonyl-β-D-glucopyranoside is an anionic alkylglucoside chiral surfactant that is commonly used for the solubilization and crystallization of biological membrane proteins.
-
GC45833
N-Oleoyl-L-phenylalanine
An N-acyl amide
-
GC47806
N-p-Tosyl-Gly-Pro-Lys-pNA (acetate)
A colorimetric plasmin substrate
-
GC44313
Naphthofluorescein
Furin is a proprotein convertase, converting precursor proteins to functional proteins within the Golgi/trans-Golgi secretory pathway.
-
GC12491
Narciclasine
A plant growth regulator with anticancer activity
-
GC49204
Nefazodone-d6 (hydrochloride)
An internal standard for the quantification of nefazodone
-
GC45523
Nemorosone
-
GC10716
Netarsudil (AR-13324)
ROCK inhibitor
-
GC12515
NMS-1286937
NMS-1286937 is a potent, selective and orally available PLK1 inhibitor, with an IC50 of 2 nM.
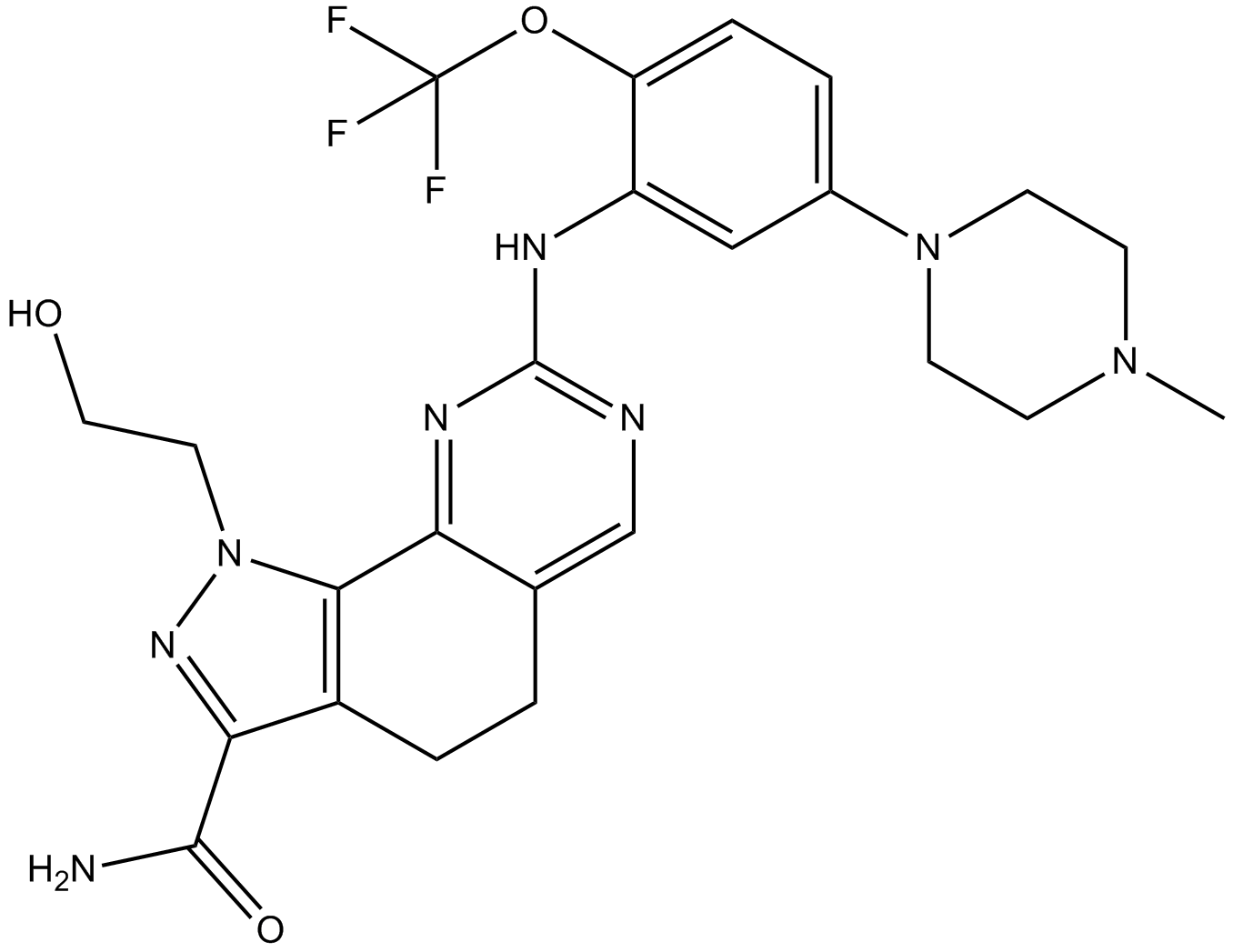
-
GC36752
NMS-P715
An Mps1/TTK inhibitor
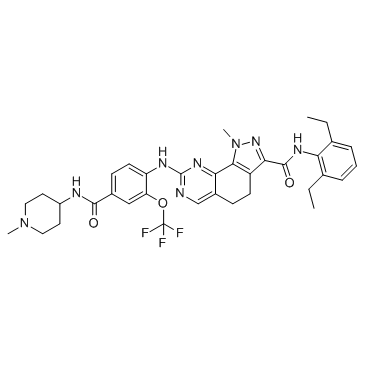
-
GC14075
Nocodazole
A tubulin production inhibitor,anti-neoplastic agent
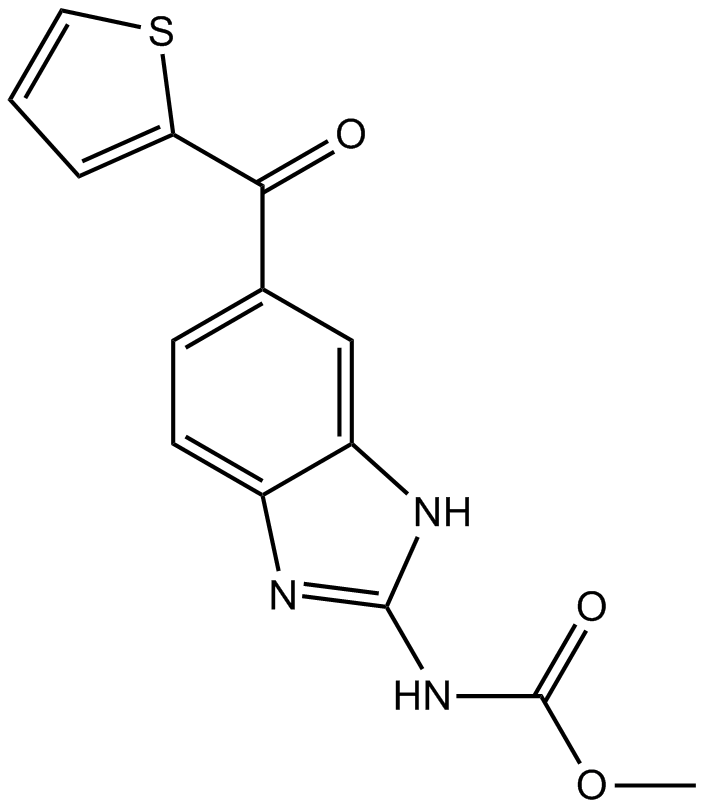
-
GC47803
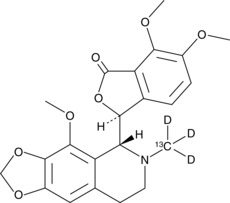
Noscapine-13C-d3
An internal standard for the quantification of noscapine