Tyrosine Kinase
- IGFBR(1)
- Bcr-Abl(67)
- Ack1(2)
- Axl(6)
- ALK(56)
- BMX Kinase(3)
- Broad Spectrum Protein Kinase Inhibitor(10)
- c-FMS(35)
- c-Kit(56)
- c-MET(84)
- c-RET(0)
- CSF-1R(3)
- DDR1/DDR2 Receptor(1)
- EGFR(235)
- EphB4(1)
- FAK(33)
- FGFR(95)
- FLT3(85)
- HER2(12)
- IGF1R(33)
- Insulin Receptor(44)
- IRAK(28)
- ITK(10)
- Lck(1)
- LRRK2(22)
- PDGFR(104)
- PTKs/RTKs(3)
- Pyk2(6)
- ROR(38)
- Spleen Tyrosine Kinase (Syk)(32)
- Src(97)
- Tie-2 (4)
- Trk(39)
- VEGFR(181)
- Kinase(0)
- Discoidin Domain Receptor(15)
- DYRK(29)
- Ephrin Receptor(12)
- ROS(14)
- RET(29)
- TAM Receptor(29)
Products for Tyrosine Kinase
- Cat.No. Product Name Information
-
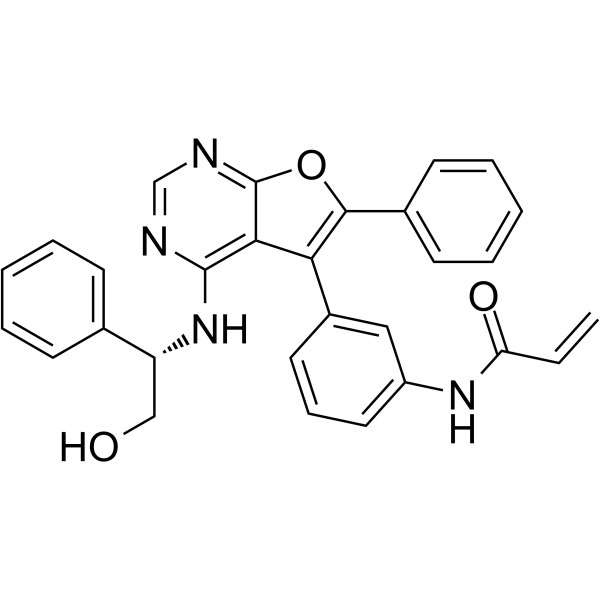
GC12837
Compound 56
A potent EGFR inhibitor
-
GC46123
Comprehensive Kinase Screening Library
For screening of a variety of kinase inhibitors

-
GC35728
Conteltinib
CT-707
Conteltinib (CT-707) is a multi-kinase inhibitor targeting FAK, ALK, and Pyk2. Conteltinib exerts significant inhibitory effect on FAK with an IC50 of 1.6 nM.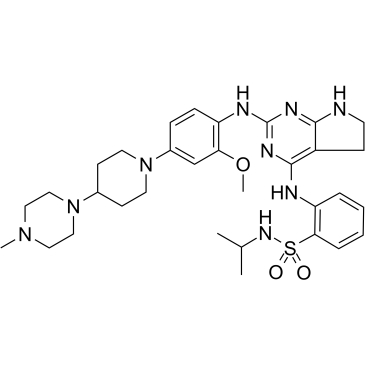
-
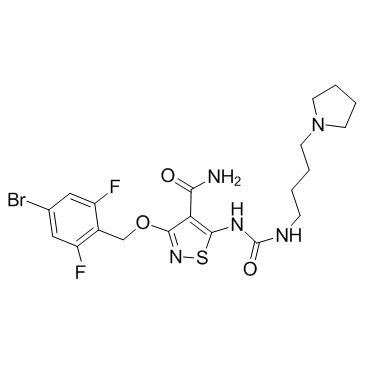
GC33352
CP-547632
A potent inhibitor of VEGFR2 and bFGF
-
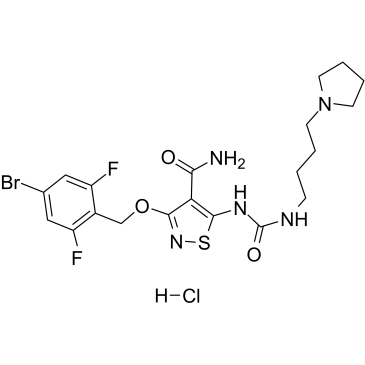
GC38575
CP-547632 hydrochloride
CP-547632 hydrochloride is an orally active, ATP-competitive and potent VEGFR-2 and FGF kinases inhibitor with IC50s of 11 nM and 9 nM, respectively. CP-547632 hydrochloride is selective for VEGFR2 and bFGF over EGFR, PDGFRβ, and related tyrosine kinases (TKs). CP-547632 hydrochloride has antitumor efficacy.
-
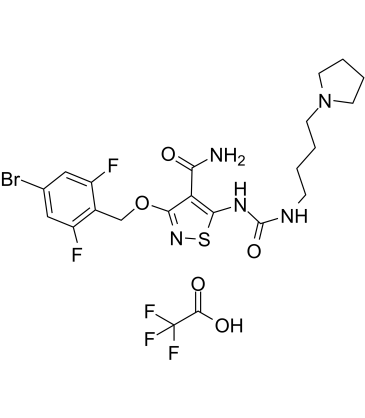
GC60726
CP-547632 TFA
CP-547632 TFA is an orally active, ATP-competitive and potent VEGFR-2 and FGF kinases inhibitor with IC50s of 11 nM and 9 nM, respectively. CP-547632 TFA is selective for VEGFR2 and bFGF over EGFR, PDGFRβ, and related tyrosine kinases (TKs). CP-547632 TFA has antitumor efficacy.
-
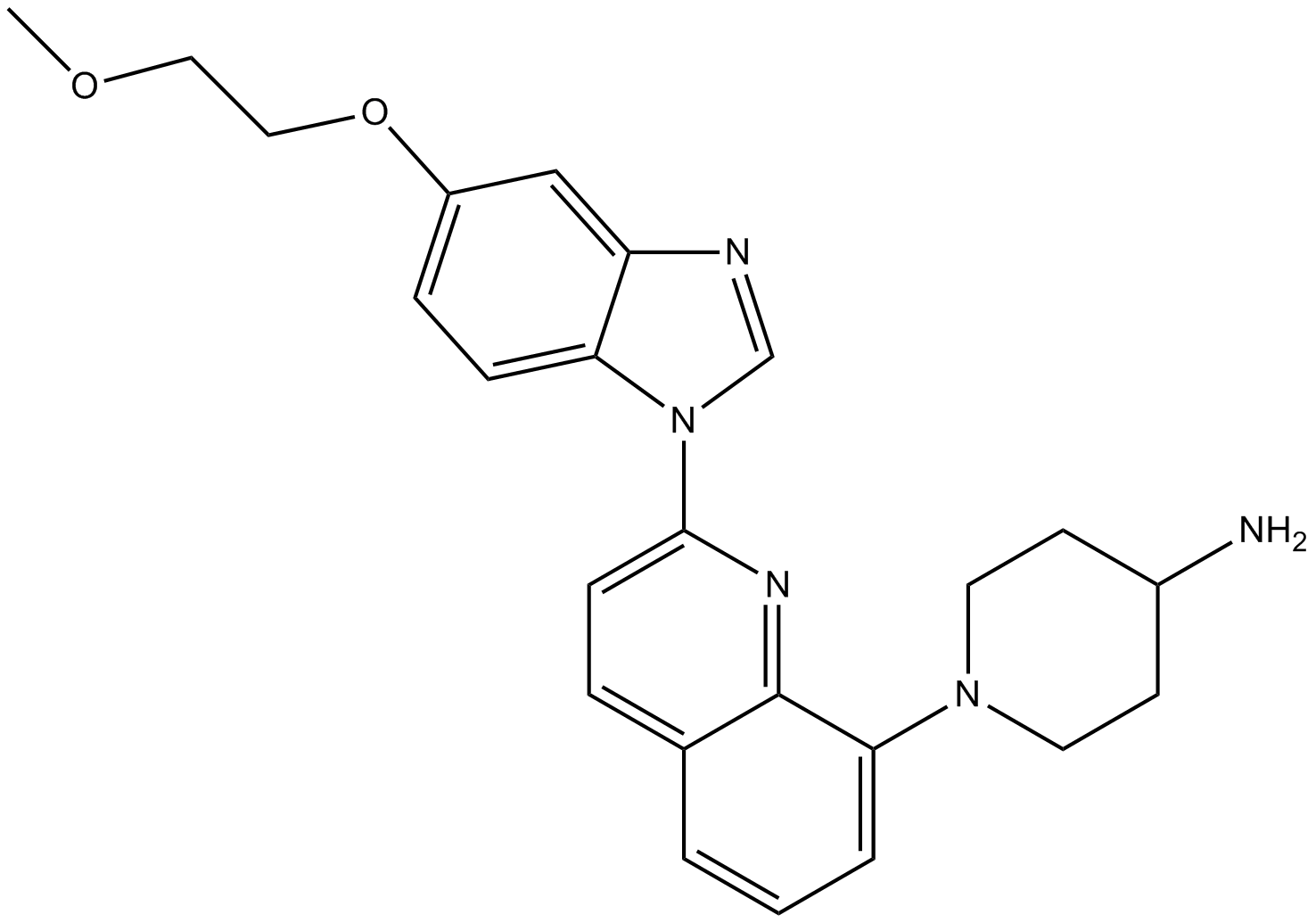
GC12980
CP-673451
PDGFRα/β inhibitor,potent and selective
-
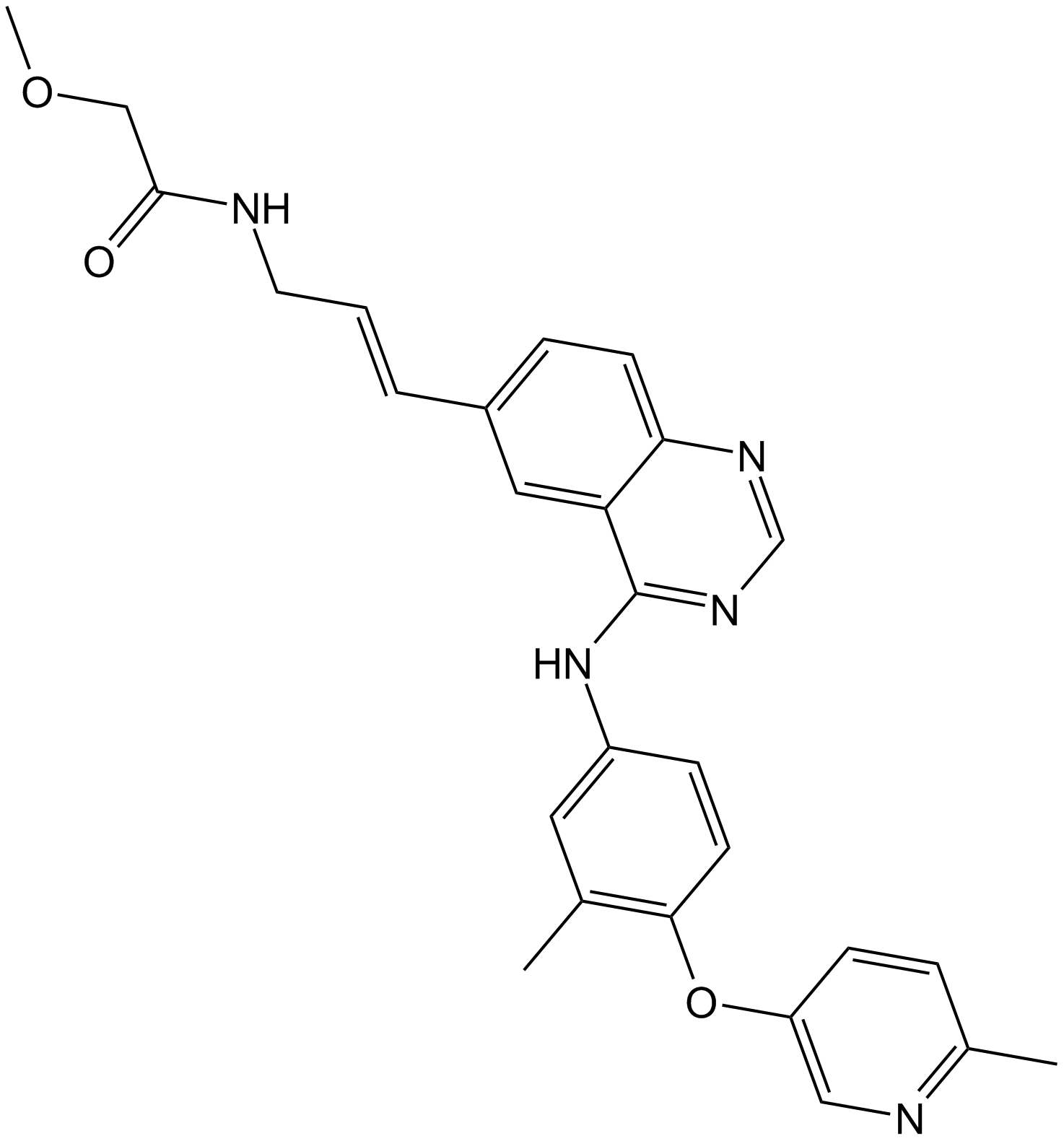
GC13091
CP-724714
HER2 inhibitor,potent and selective
-
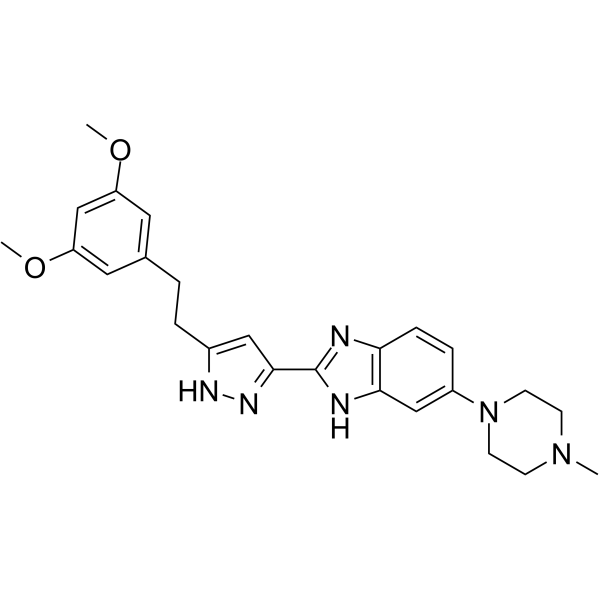
GC62274
CPL304110
CPL304110 is a potent, orally active and selective inhibitor of fibroblast growth factor receptors FGFR (1-3), with IC50 values of 0.75 nM, 0.5 nM, and 3.05 nM for FGFR (1-3), respectively.
-
GC14906
Crenolanib (CP-868596)
CP-868596;CP 868596;CP868596
Crenolanib (CP-868596) is a potent and selective inhibitor of wild-type and mutant isoforms of the class III receptor tyrosine kinases FLT3 and PDGFRα/β with Kds of 0.74 nM and 2.1 nM/3.2 nM, respectively.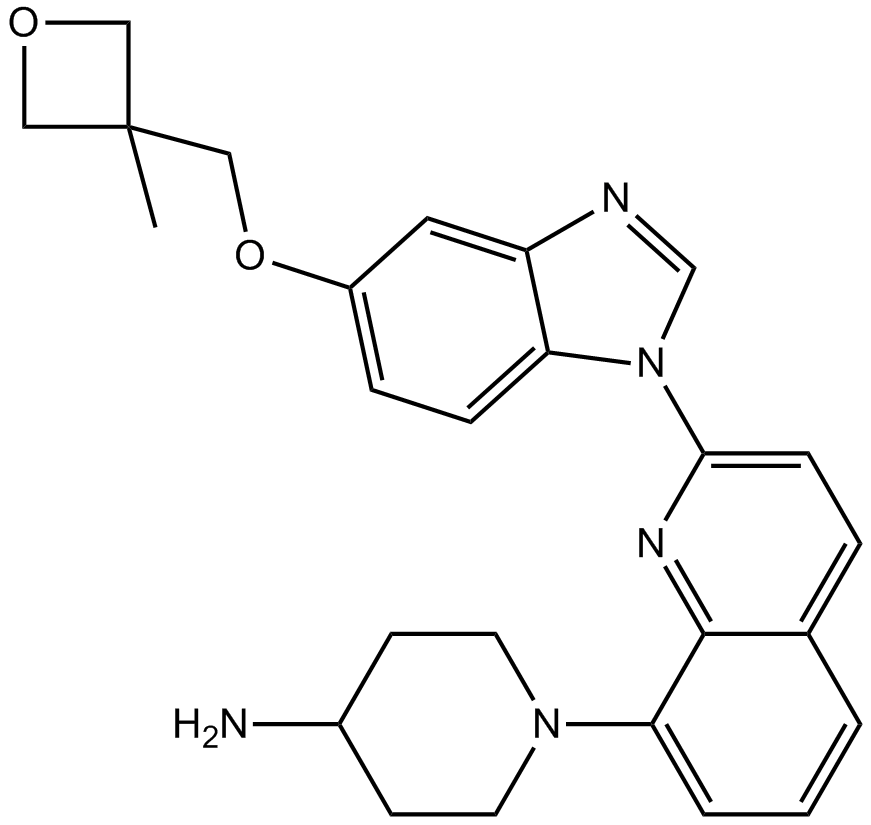
-
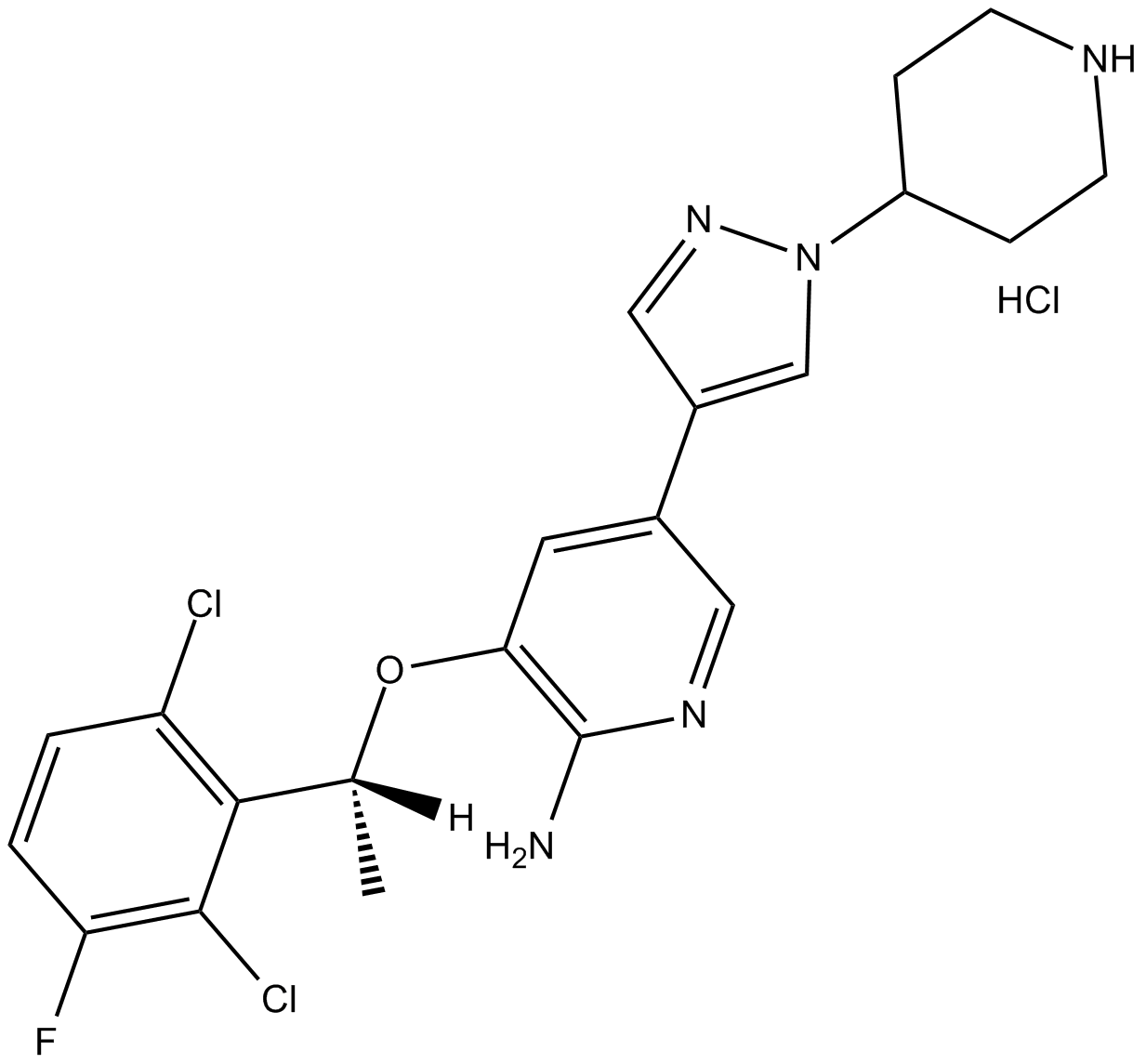
GC12616
Crizotinib hydrochloride
inhibitor of the c-Met kinase and the NPM-ALK
-
GC38412
Crotonoside
2-Hydroxyadenosine, Isoguanine riboside, Isoguanosine, NSC 12161
A guanosine analog with diverse biological activities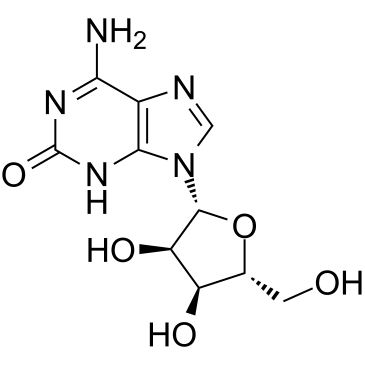
-
GC25313
CS-2660 (JNJ-38158471)
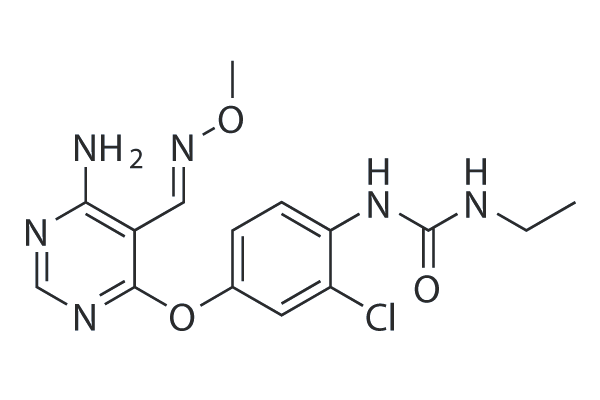
CS-2660 (JNJ-38158471) is a well tolerated, orally available, highly selective VEGFR-2 inhibitor with IC50 of 40 nM. CS-2660 (JNJ-38158471) also inhibits closely related tyrosine kinases such as RET (c-RET) and Kit (c-Kit) with IC50 of 180 nM and 500 nM,while it has no significant activity (>1 microM) against VEGFR-1 and VEGFR-3.
-
GC33241
CSF1R-IN-1
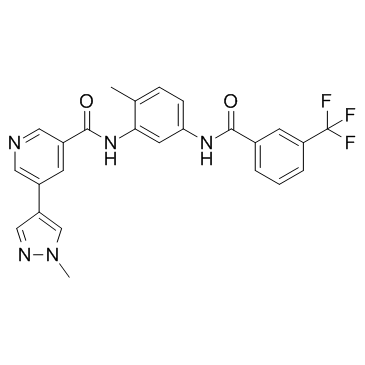
CSF1R-IN-1 is a CSF1R inhibitor with an with an IC50 of 0.5 nM.
-
GC64961
CSF1R-IN-2
CSF1R-IN-2, CSF1R Inhibitor 2, Elzovantinib
CSF1R-IN-2 (compound 5) is an oral-active inhibitor of SRC, MET and c-FMS, with IC50 values of 0.12 nM, 0.14 nM and 0.76 nM for SRC, MET and c-FMS respectively.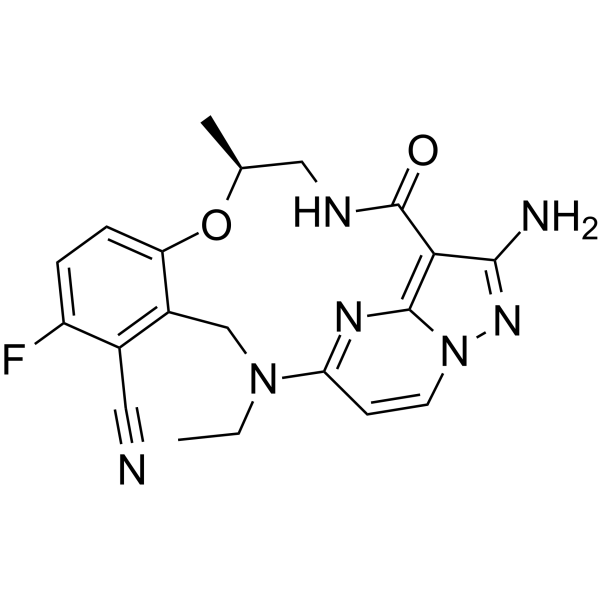
-
GC64805
CSF1R-IN-3
CSF1R-IN-3 (compound 21) is a potent and orally active CSF-1R inhibitor (IC50=2.1 nM). CSF1R-IN-3 is a potent antiproliferative activity against colorectal cancer cells. CSF1R-IN-3 inhibits the progression of colorectal cancer by suppressing the migration of macrophages, reprograming M2-like macrophages to the M1 phenotype, and enhancing the antitumor immunity.

-
GC35753
CT-721
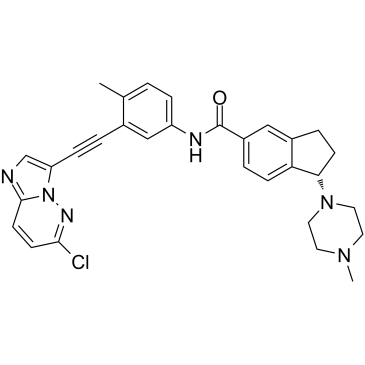
CT-721 is a potent and time-dependent Bcr-Abl kinase inhibitor with an IC50 of 21.3 nM for wild-type Bcr-Abl kinase, and possesses anti-chronic myeloid leukemia (CML) activities.
-
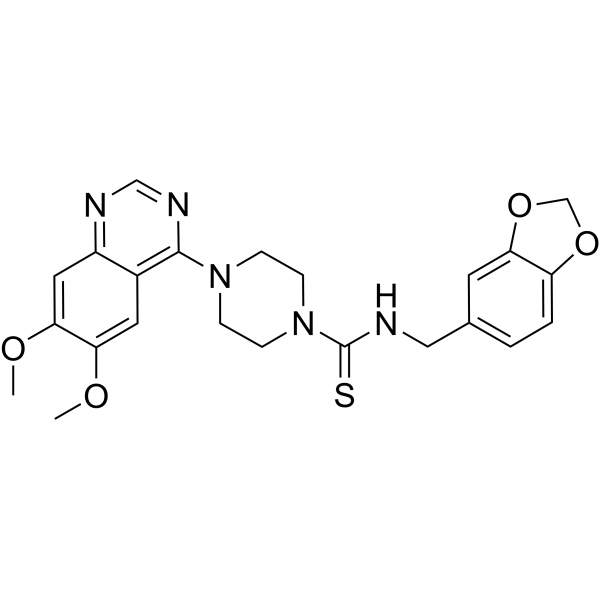
GC67774
CT52923
-
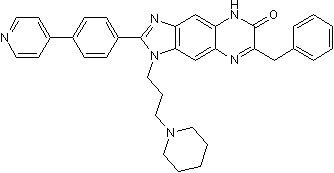
GC50076
CTA 056
ITK inhibitor
-
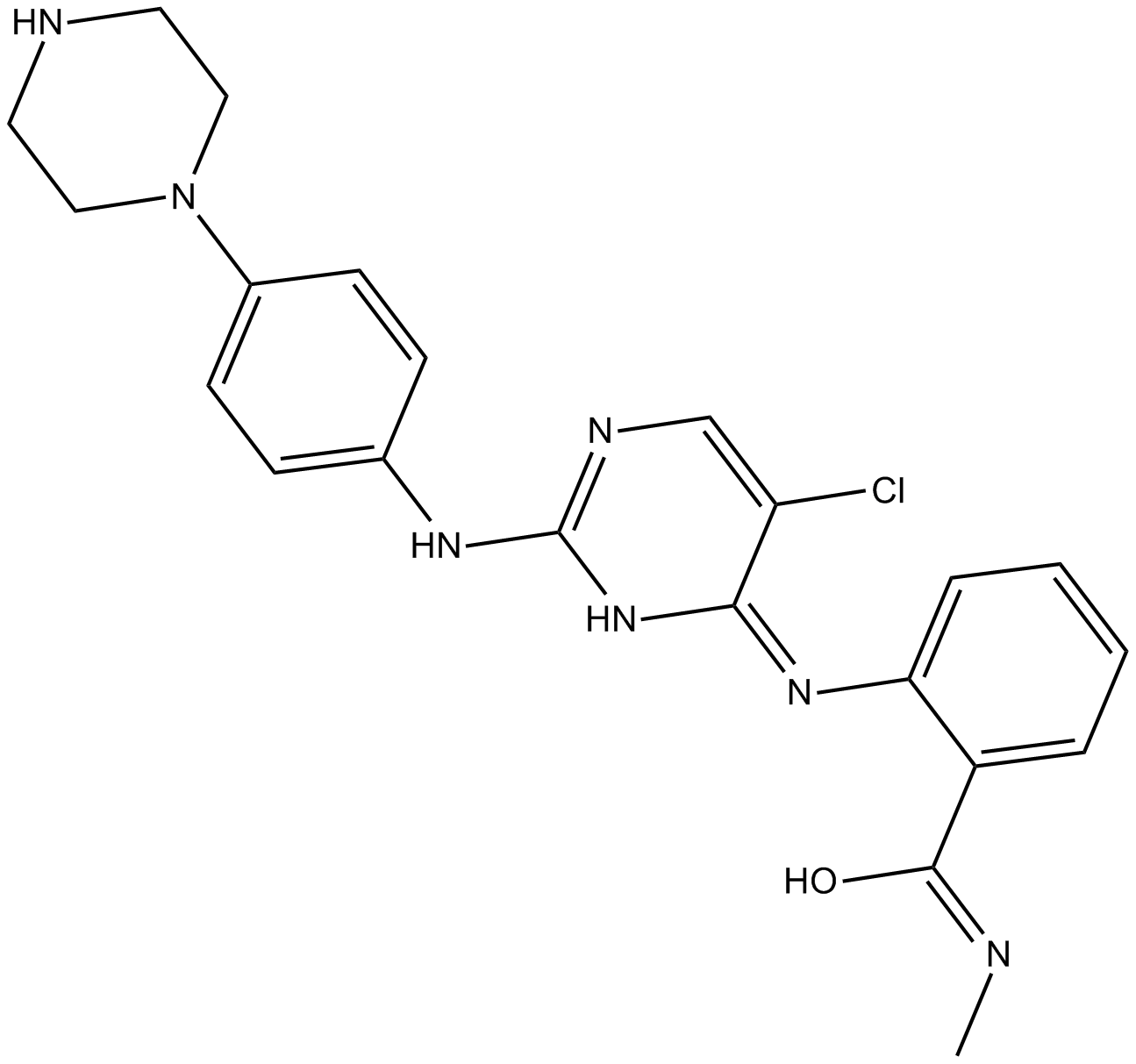
GC17637
CTX0294885
Pan-kinase inhibitor
-
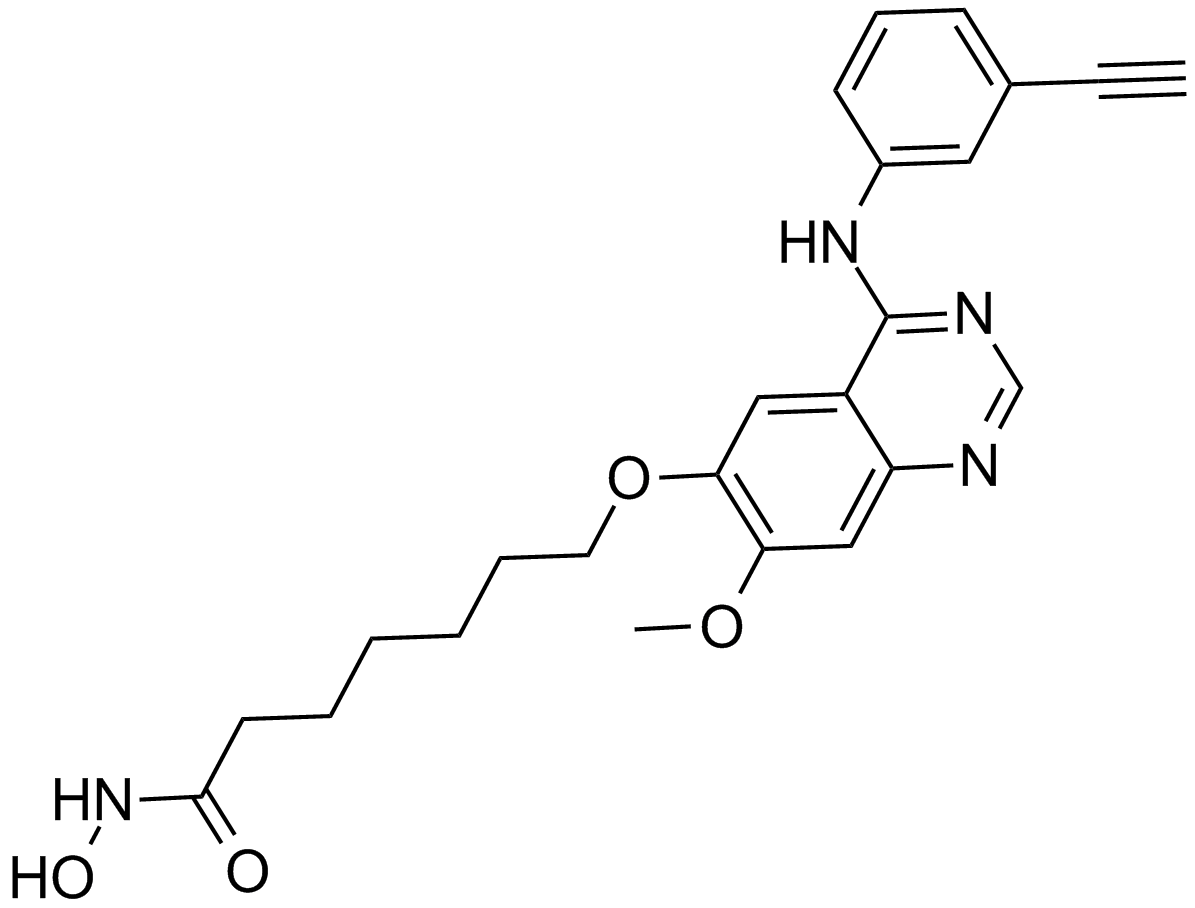
GC16008
CUDC-101
A multi-target inhibitor of HDACs, EGFR, and HER2
-
GC38180
Cyasterone
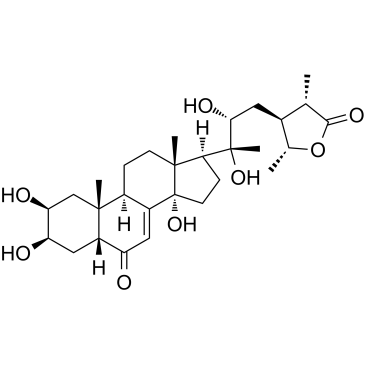
-
GC13780
Cyclotraxin B
TrkB receptor antagonist
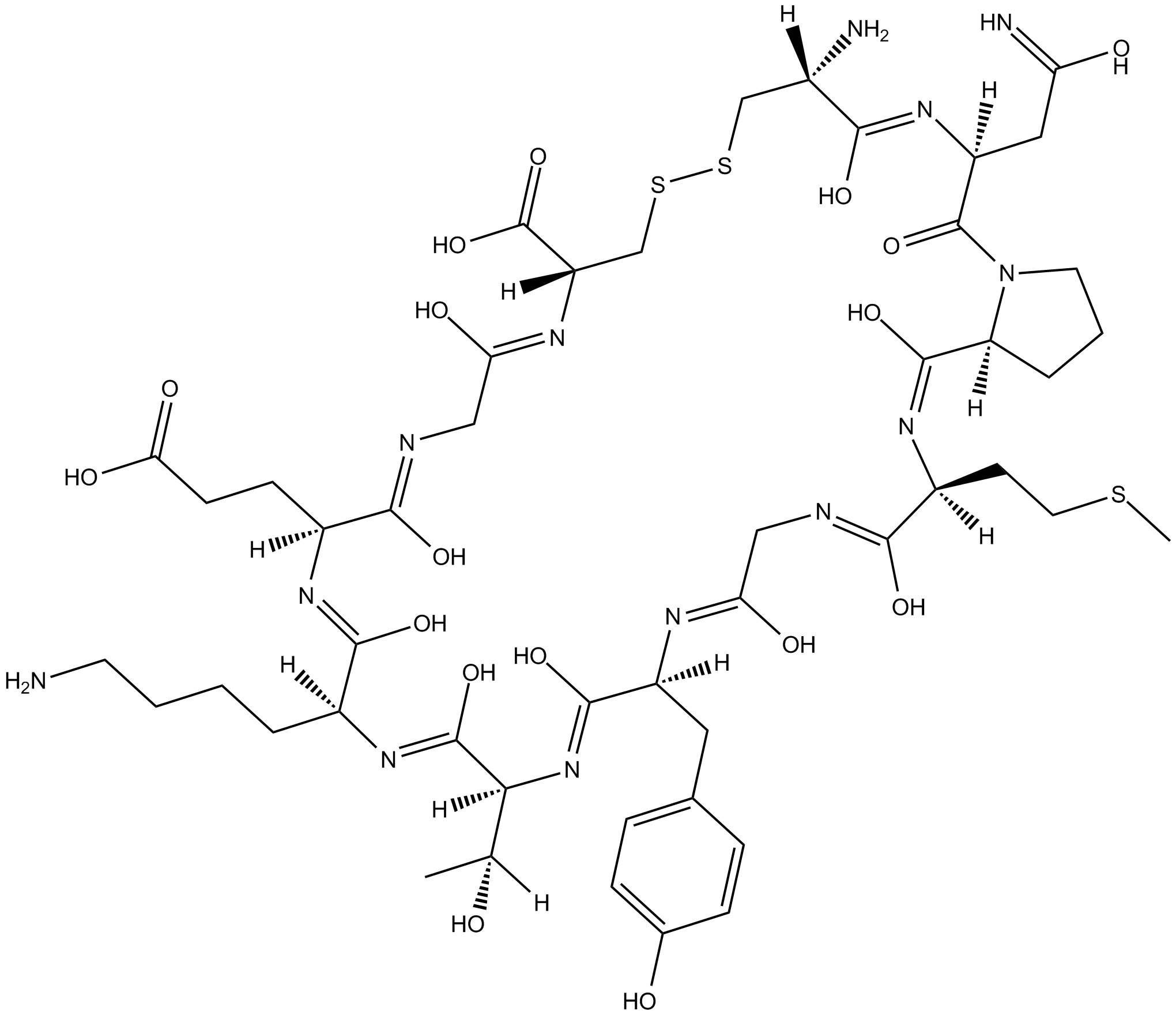
-
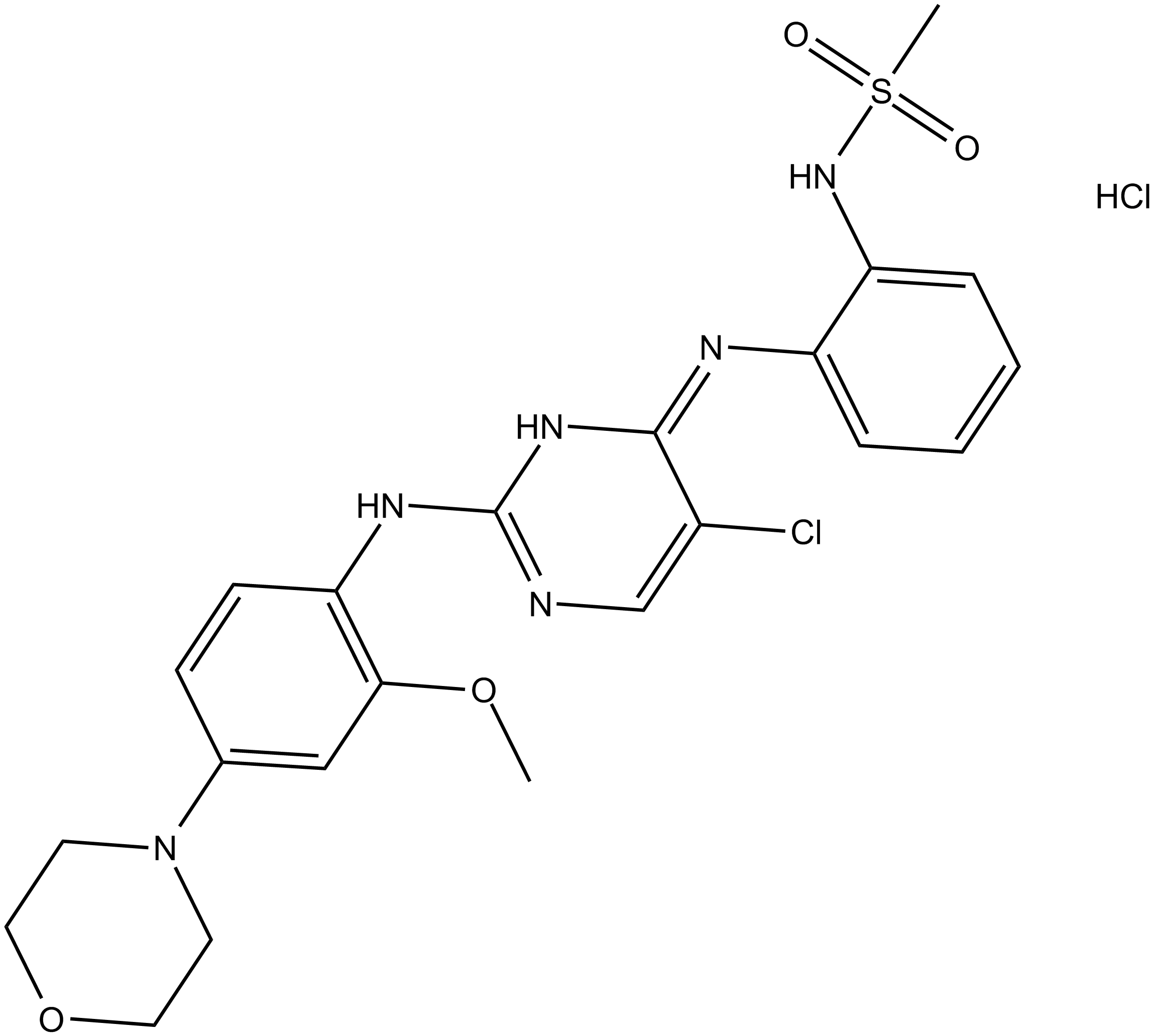
GC16731
CZC 54252 hydrochloride
CZC 54252 hydrochloride is a potent and selective LRRK2 inhibitor with IC50s of 1.28 nM and 1.85 nM for wild-type and G2019S LRRK2, respectively.
-
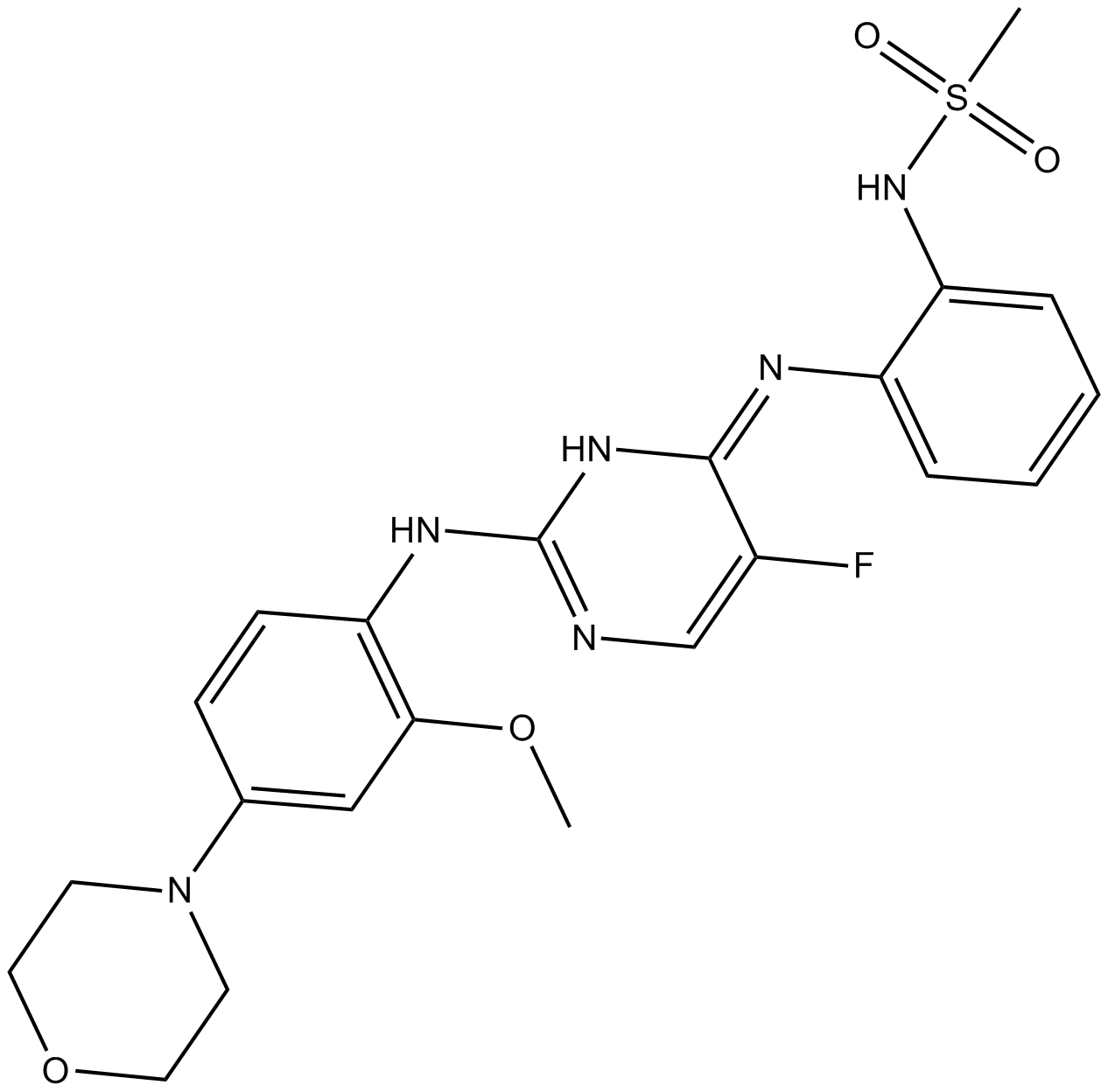
GC16038
CZC-25146
LRRK2 inhibitor
-
GC35792
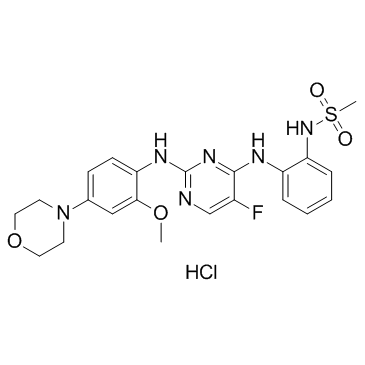
CZC-25146 hydrochloride
CZC-25146 hydrochloride is a potent LRRK2 inhibitor with IC50 values of 4.76 nM and 6.87 nM for wild-type LRRK2 and G2019S LRRK2, respectively.
-
GC33351
CZC-8004 (CZC-00008004)
Dianilinopyrimidine-01
CZC-8004 (CZC-00008004) (CZC-00008004), an aminopyrimidine, is a pan-kinase inhibitor. CZC-8004 (CZC-00008004) can bind a range of tyrosine kinases, including EGFR and VEGFR2 with IC50 values of 650 and 437 nM, respectively.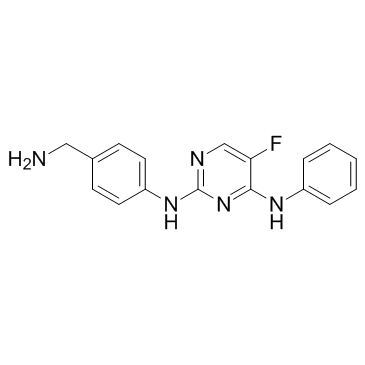
-
GC43433
D-Glucosamine-6-sulfate
GlcN-6S
D-Glucosamine-6-sulfate is a naturally occurring glycosaminoglycan.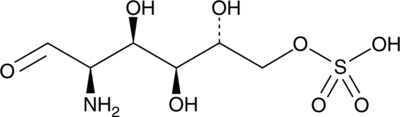
-
GC62712
DA-JC4
DA-JC4 is a dual GLP-1/GIP receptor agonist and can be used for the research of neurological disease and insulin signaling pathways.
-
GC10225
Dacomitinib (PF299804, PF299)
PF-00299804; PF-299804; PF 299804; PF 00299804
Dacomitinib (PF299804, PF299) (PF-00299804) is a specific and irreversible inhibitor of the ERBB family of kinases with IC50s of 6 nM, 45.7 nM and 73.7 nM for EGFR, ERBB2, and ERBB4, respectively.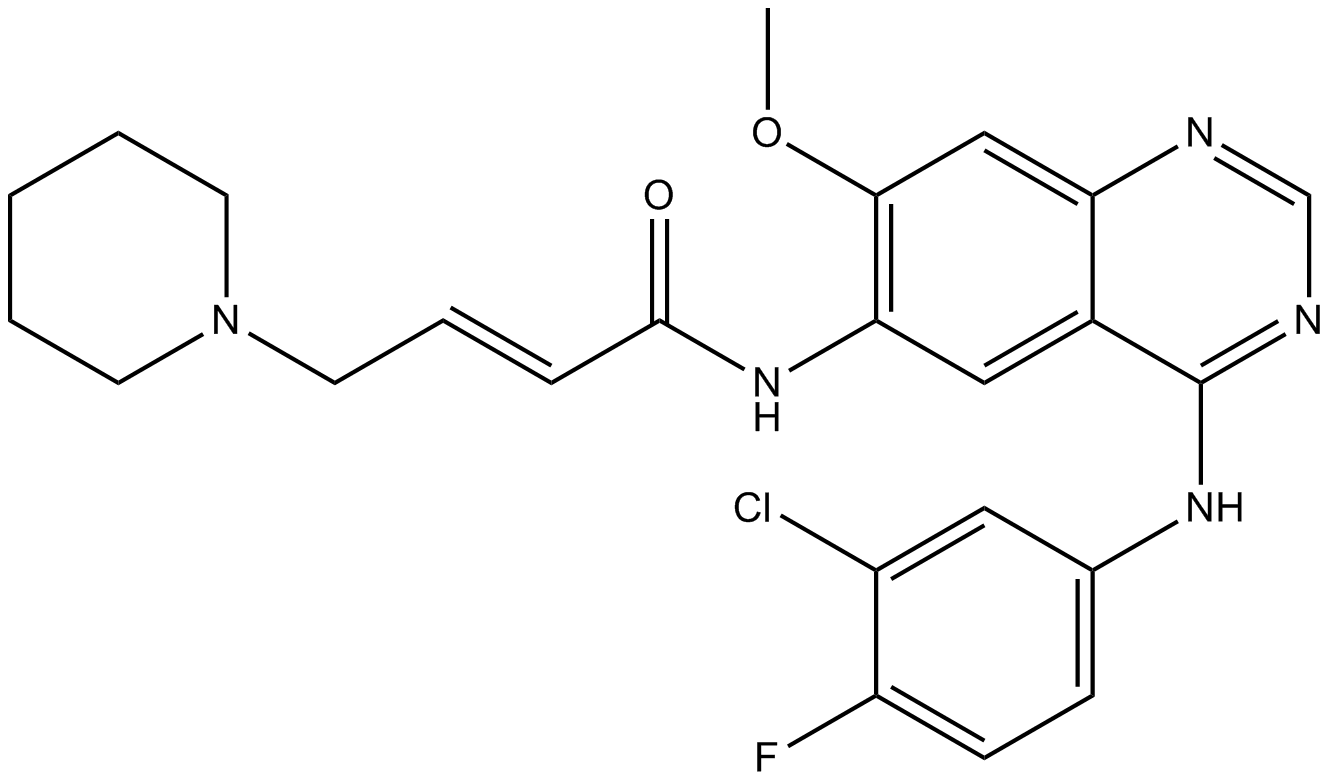
-
GC15211
Damnacanthal
p56lck tyrosine kinase inhibitor
-
GC15217
Danusertib (PHA-739358)
PHA-739358
A pan-Aurora kinase and Abl inhibitor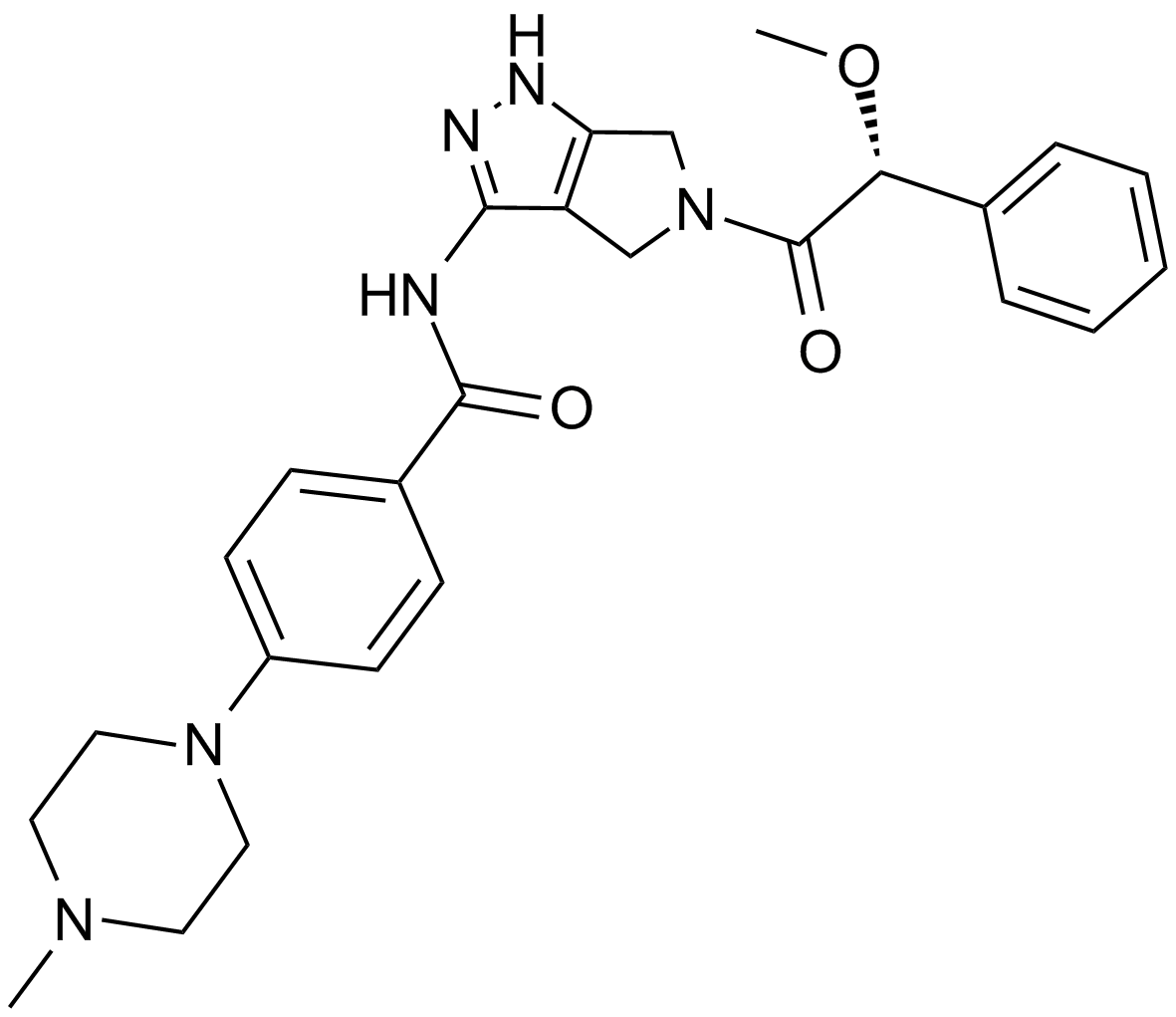
-
GN10336
Daphnetin
7,8-Dihydroxycoumarin, NSC 633563
-
GC15568
Dasatinib (BMS-354825)
BMS 354825, Sprycel
An inhibitor of Abl and Src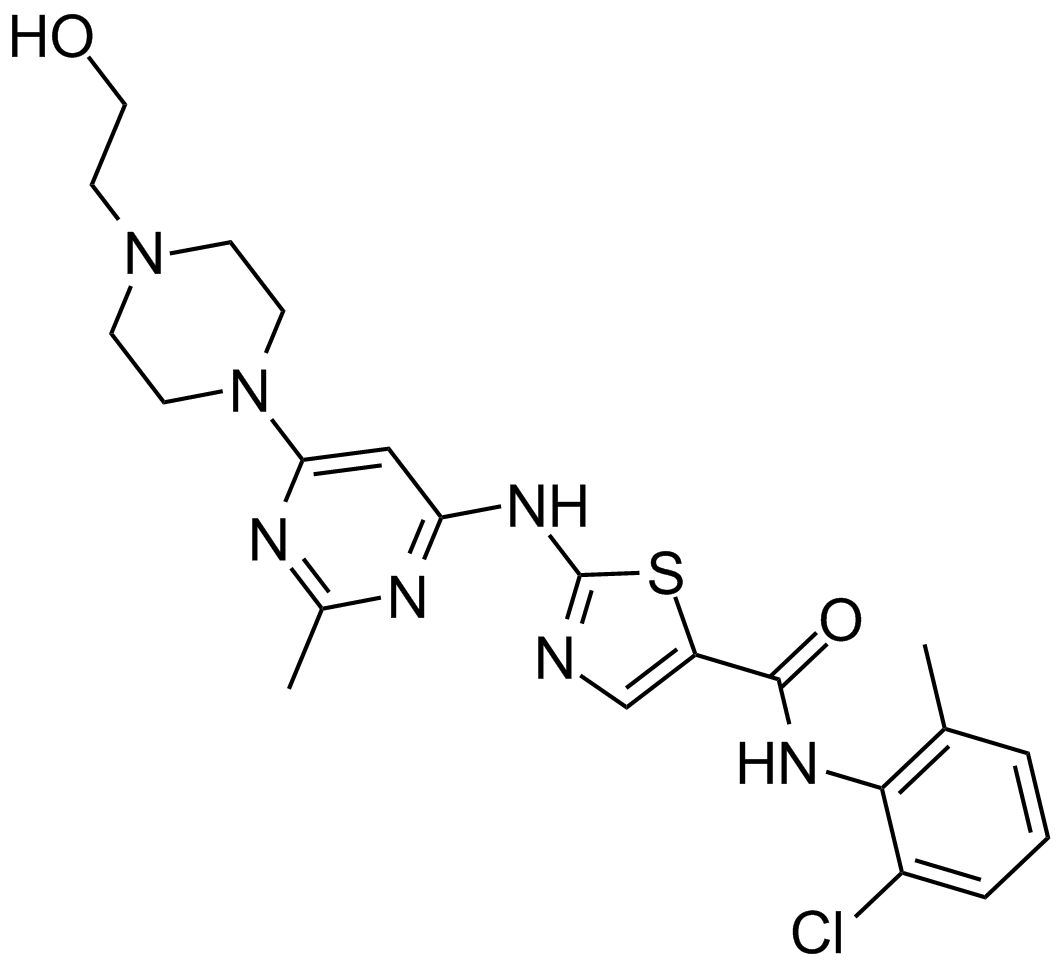
-
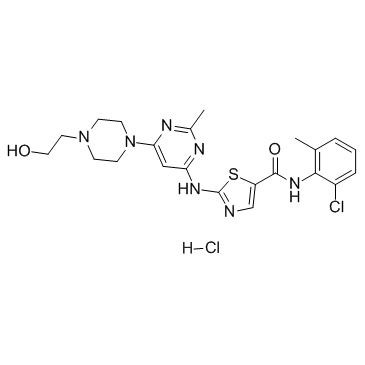
GC35812
Dasatinib hydrochloride
A potent and dual AblWT/Src inhibitor
-
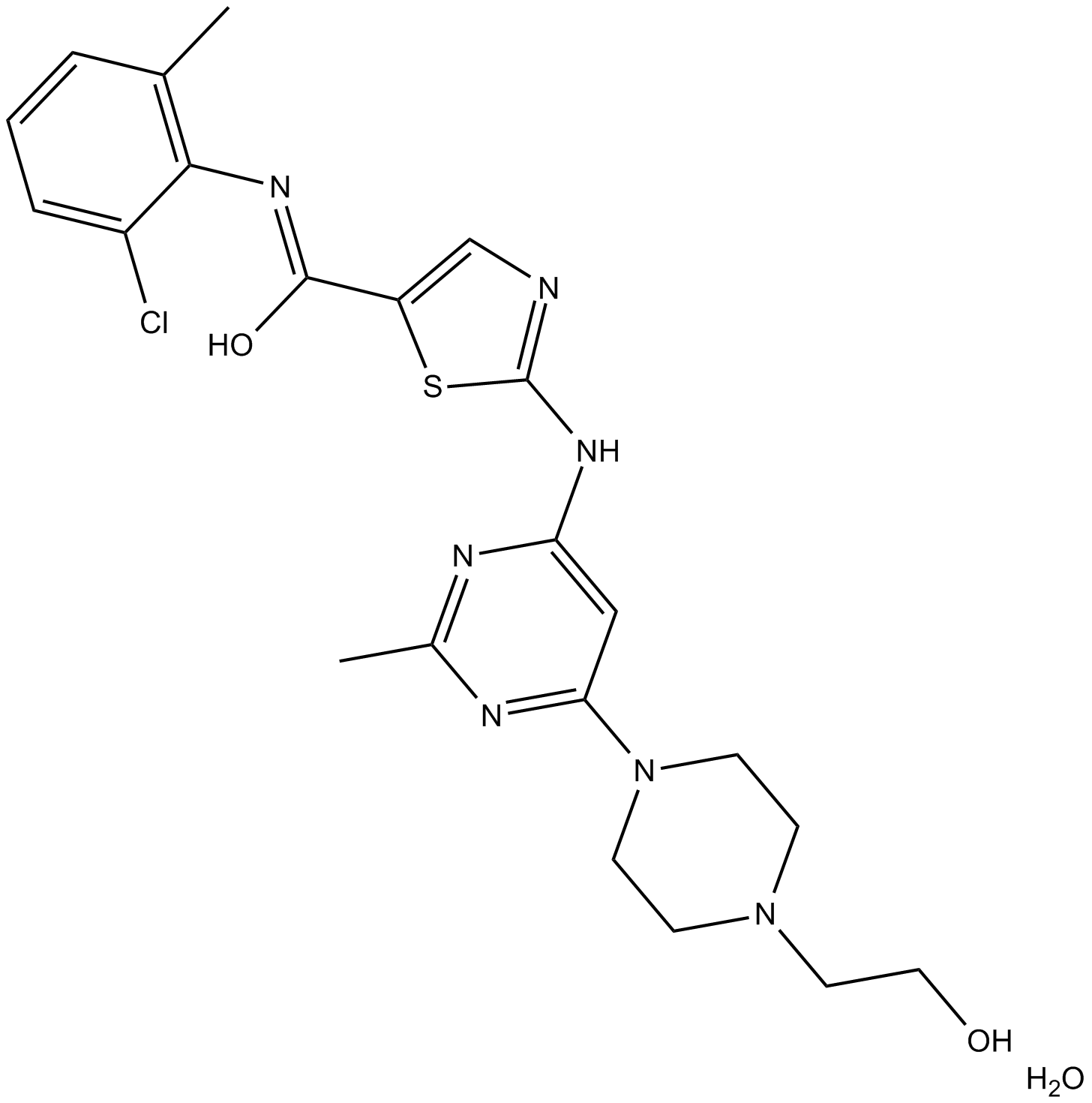
GC15884
Dasatinib Monohydrate
Inhibitor of ABL, SRC, KIT, PDGFR, and other tyrosine kinases.
-
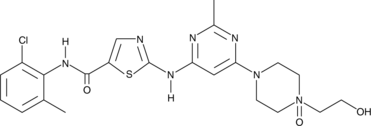
GC45687
Dasatinib N-oxide
A major metabolite of dasatinib
-
GC62569
DBPR112
DBPR112 is an orally active furanopyrimidine-based EGFR inhibitor with IC50s of 15 nM and 48 nM for EGFRWT and EGFRL858R/T790M, respectively. DBPR112 can occupy the ATP-binding site. DBPR112 has significant antitumor efficacy.
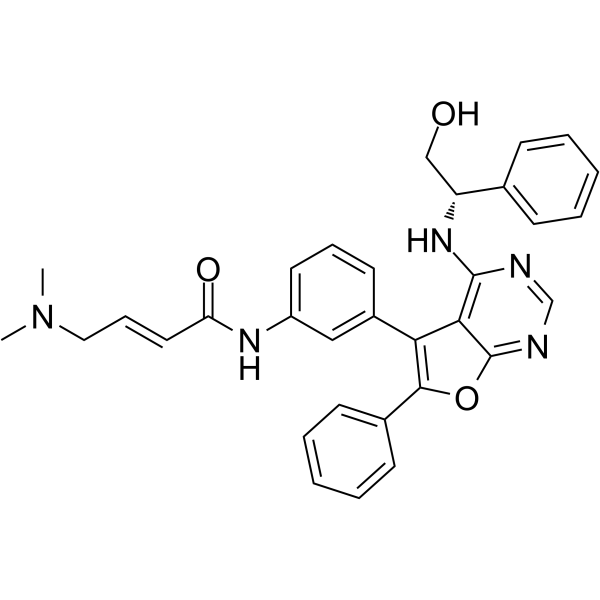
-
GC14007
DCC-2036 (Rebastinib)
DCC-2036
DCC-2036 (Rebastinib) (DCC-2036) is an orally active, non-ATP-competitiveBcr-Abl inhibitor for Abl1WT and Abl1T315I with IC50s of 0.8 nM and 4 nM, respectively. DCC-2036 (Rebastinib) also inhibits SRC, KDR, FLT3, and Tie-2, and has low activity to seen towards c-Kit.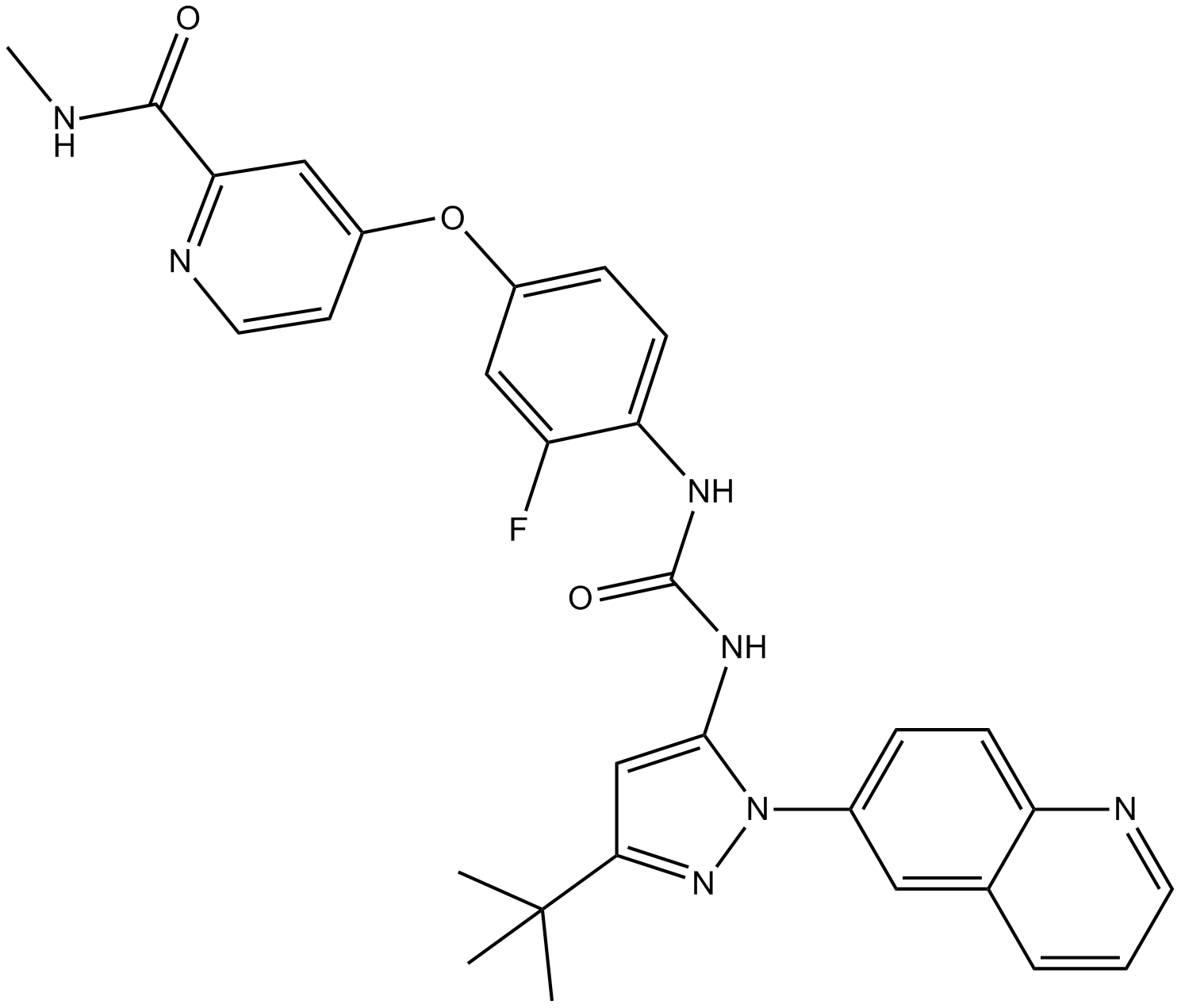
-
GC11171
DCC-2618
DCC2618;DCC 2618
A dual inhibitor of c-Kit and c-MET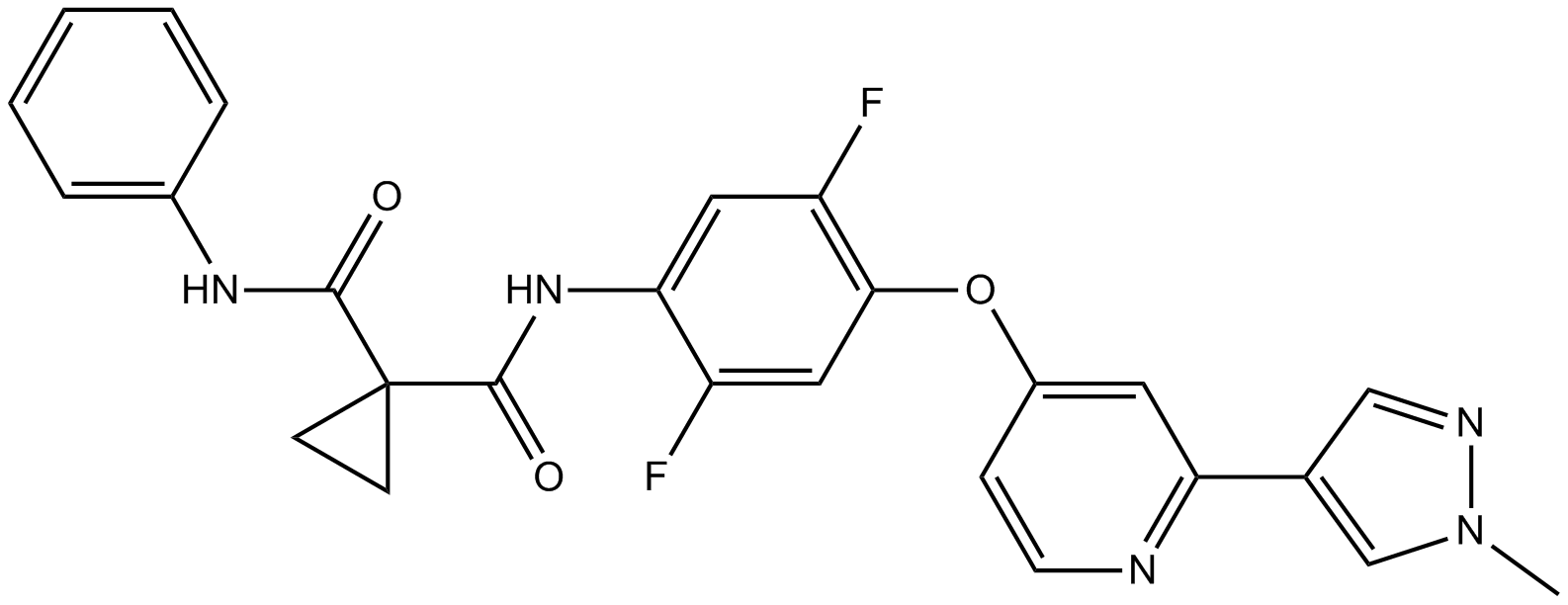
-
GC33361
DDR Inhibitor
DDR Inhibitor is a potent discoidin domain receptor (DDR) inhibitor, with an IC50 of 3.3 nM for DDR2, and shows 53% inhibition on DDR1 at 1.5 nM.
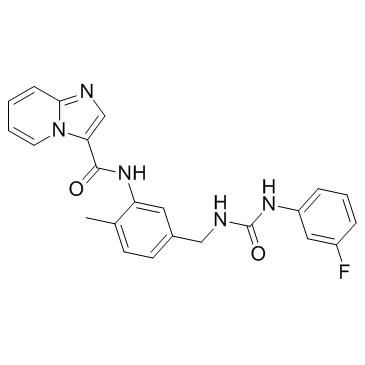
-
GC17365
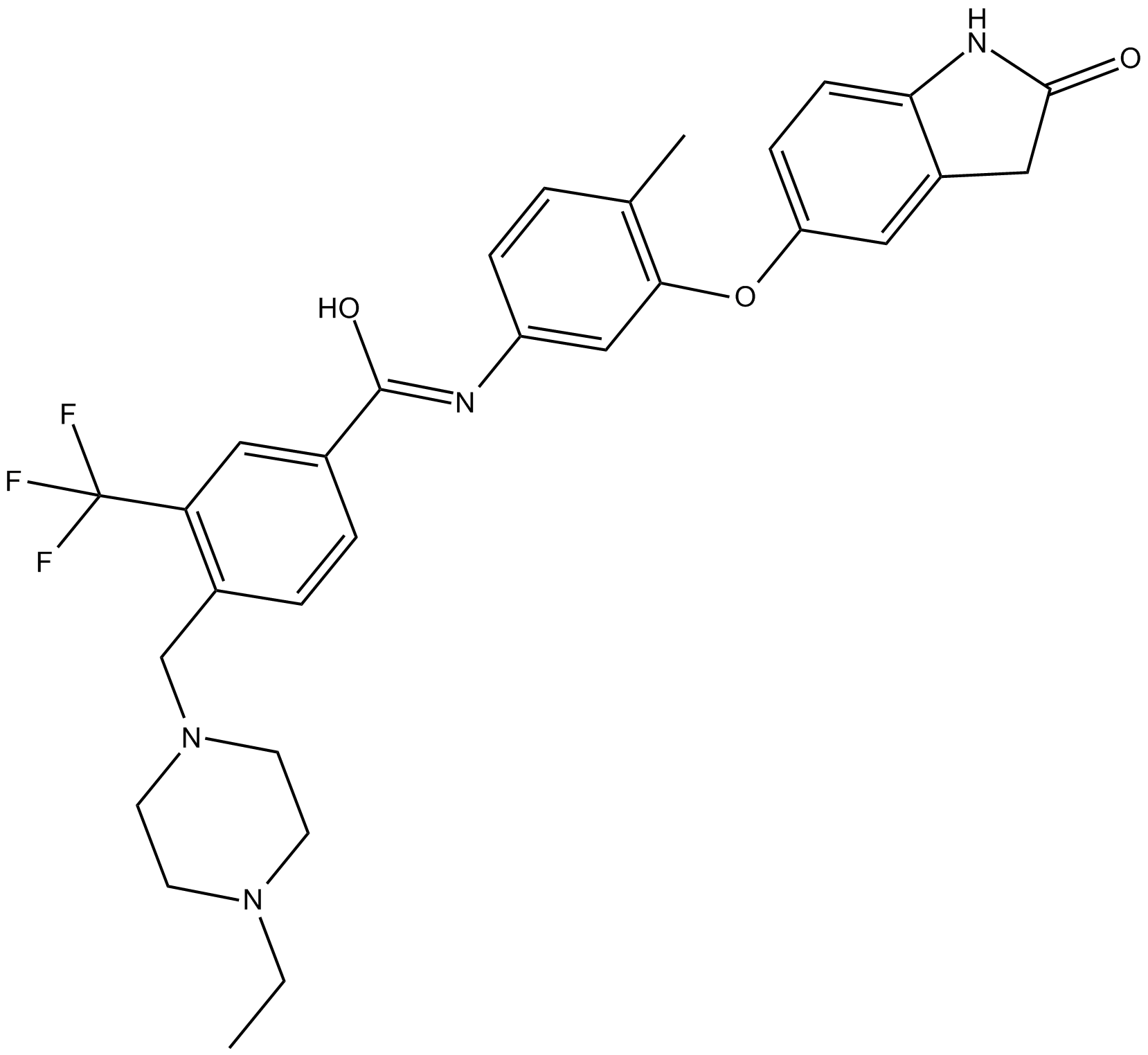
DDR1-IN-1
selective DDR1 receptor tyrosine kinase inhibitor
-
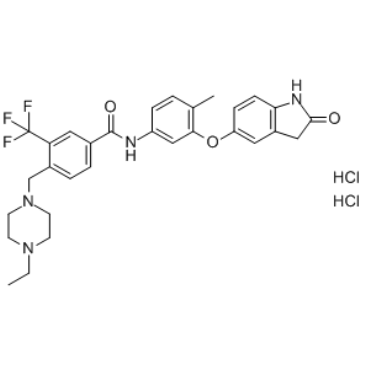
GC35822
DDR1-IN-1 dihydrochloride
DDR1-IN-1 dihydrochloride is a potent and selective DDR1 receptor tyrosine kinase inhibitor with an IC50 of 105 nM; 4-fold less potent for DDR2 (IC50 = 413 nM).
-
GC31724
DDR1-IN-2
7rh
DDR1-IN-2 (DDR1-IN-2) is a potent inhibitor of discoidin domain receptor 1 (DDR1), with an IC50 of 13.1 nM, and also less potently inhibits DDR2, with an IC50 of 203 nM.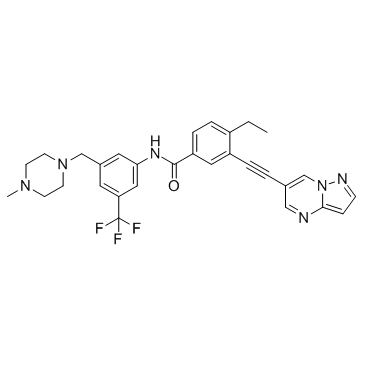
-
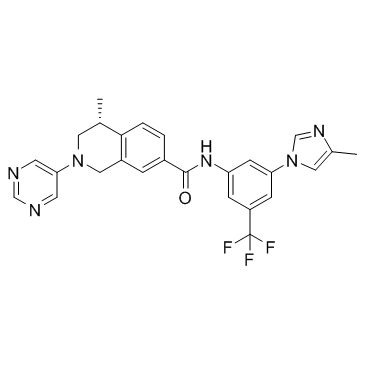
GC33297
DDR1-IN-3
DDR1-IN-3 is a selective Discoidin Domain Receptor 1 (DDR1) inhibitor, with an IC50 value of 9.4 nM. DDR1-IN-3 also inhibits TRK family.
-
GC64268
DDR1-IN-4
DDR1-IN-4 (Compound 2.45) is a selective and potent Discoidin Domain Receptor 1 (DDR1) autophosphorylation inhibitor, with IC50 values of 29 nM and 1.9 μM for DDR1 and DDR2, respectively.
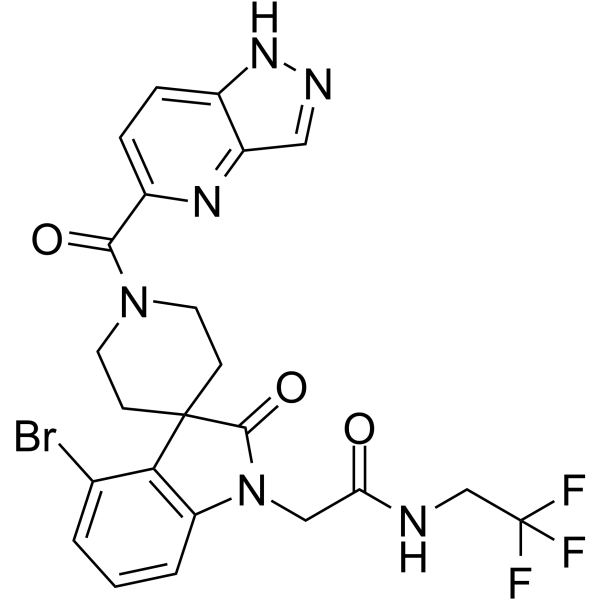
-
GC65378
DDR1-IN-5
DDR1-IN-5 is a selective Discoidin Domain Receptor family, member 1 (DDR1) inhibitor with an IC50 of 7.36 nM. DDR1-IN-5 inhibits auto-phosphorylation DDR1b (Y513) with an IC50 of 4.1 nM. DDR1-IN-5 has anti-cancer activity.
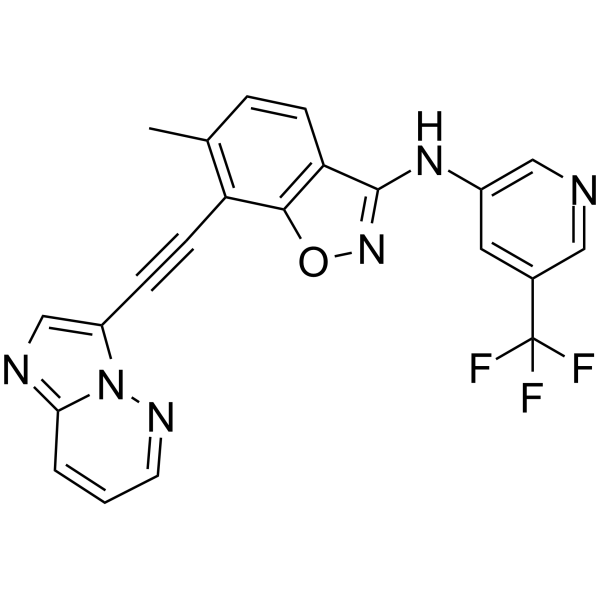
-
GC66476
DDR1-IN-6
DDR1-IN-6 is a selective Discoidin Domain Receptor family, member 1 (DDR1) inhibitor with an IC50 of 9.72 nM. DDR1-IN-6 inhibits auto-phosphorylation DDR1b (Y513) with an IC50 of 9.7 nM. DDR1-IN-6 has anti-cancer activity.
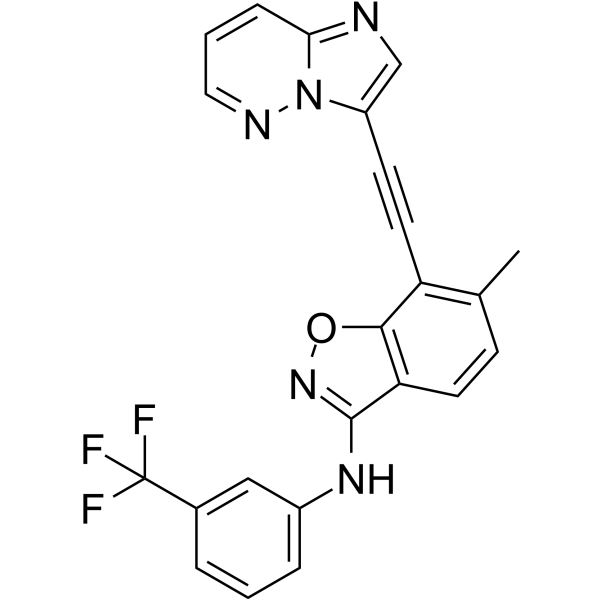
-
GC70345
DDR2-IN-1
DDR2-IN-1 is potent DDR2 inhibitor with an IC50 of 26 nM.

-
GC16813
Defactinib
PF-04554878, VS-6063
FAK phosphorylation inhibitor
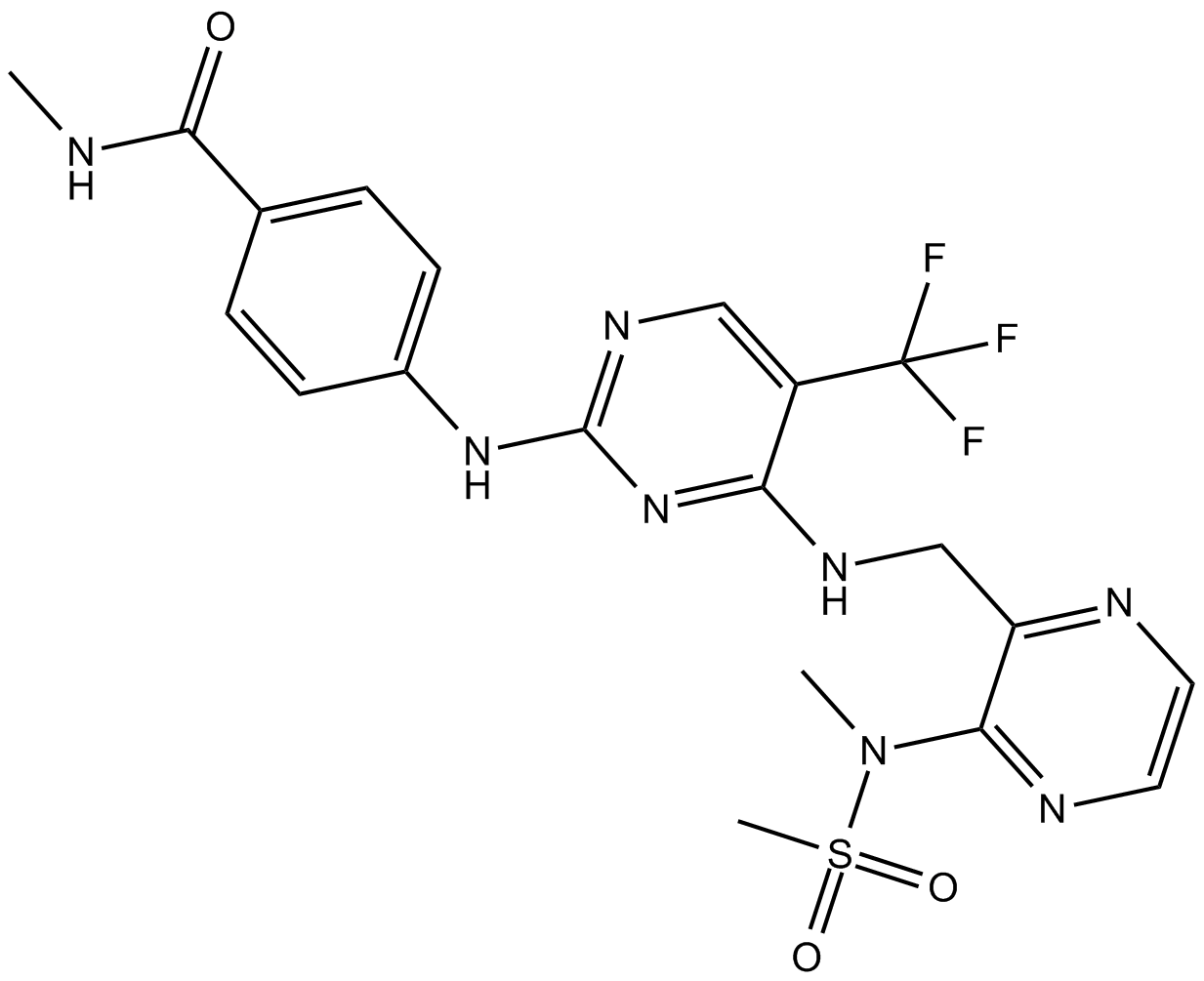
-
GC35827
Defactinib hydrochloride
VS-6063 hydrochloride; PF 04554878 hydrochloride
A novel FAK inhibitor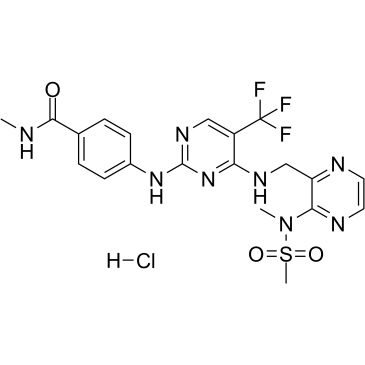
-
GC47182
Dehydrolithocholic Acid
3-keto LCA, 3-keto Lithocholate, 3-keto Lithocholic Acid, 3-KLCA, 3-oxo LCA, 3-oxo Lithocholate, 3-oxo Lithocholic Acid
Dehydrolithocholic Acid (Dehydrolithocholic acid), a bile acid metabolite, inhibits the diferentiation of TH17 cells by directly binding to the key transcription factor RORγt (Kd=1.13 μM).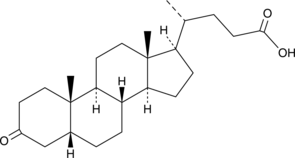
-
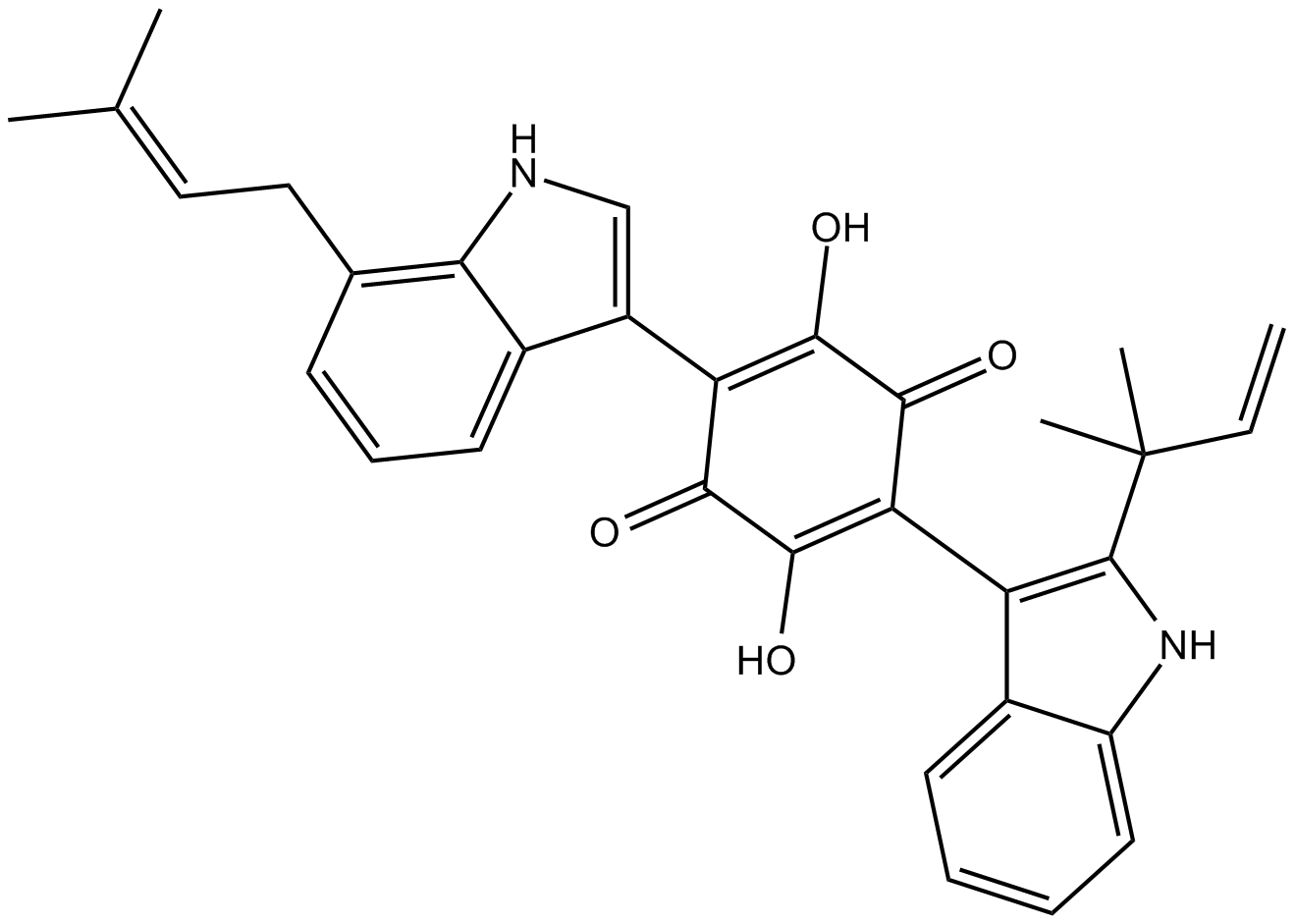
GC14905
Demethylasterriquinone B1
DMAQ B1,L-783,281
An insulin receptor (IR) activator
-
GC19399
Derazantinib
ARQ 087
Derazantinib (ARQ-087) is an ATP competitive tyrosine kinase inhibitor; exhibits potent activity against FGFR1-3 chondrocytes with IC50s of 4.5, 1.8, and 4.5 nM, respectively.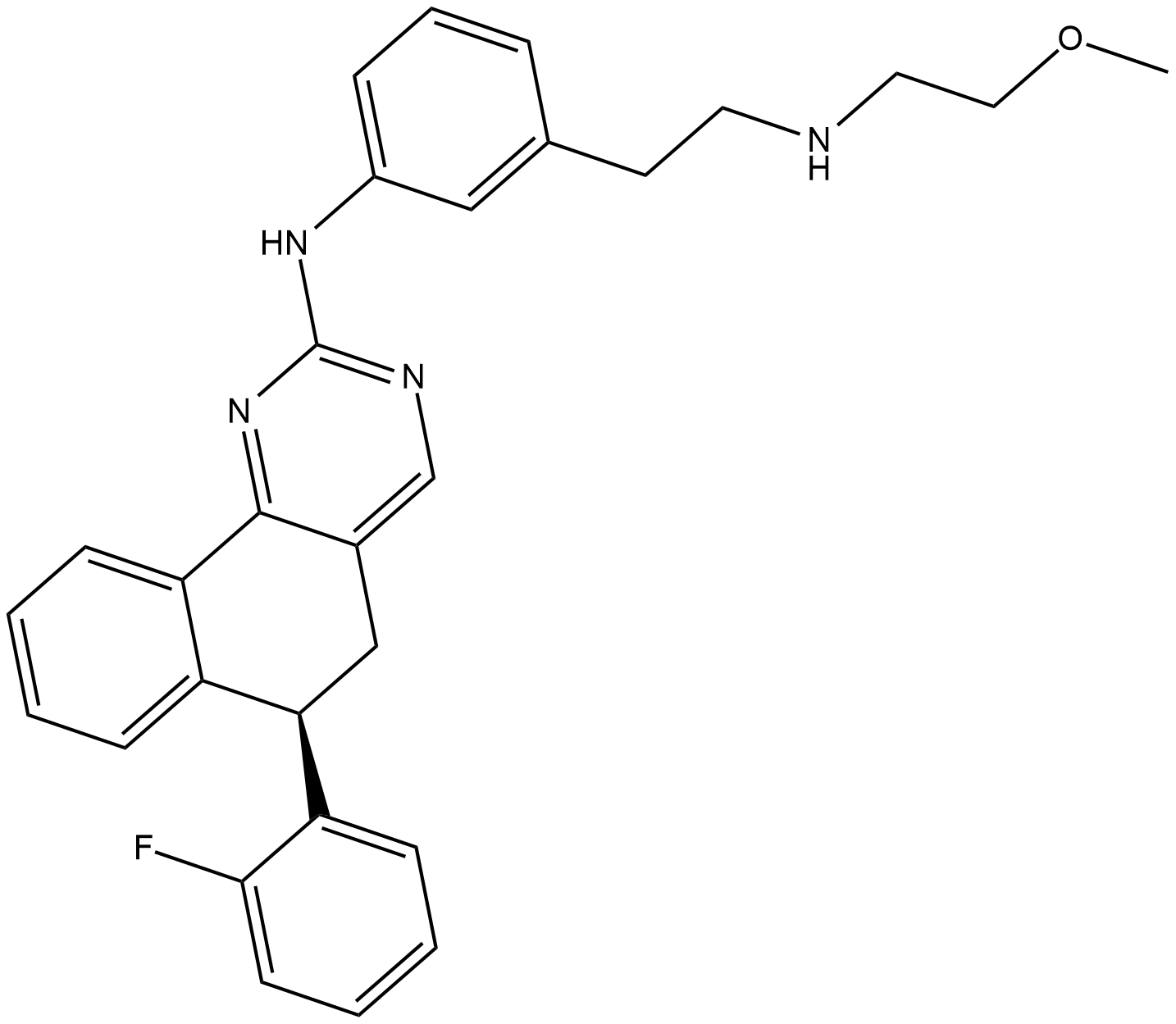
-
GC62107
DGY-06-116
DGY-06-116 is an irreversible covalent, selective Src inhibitor with an IC50 of 3nM. DGY-06-116 inhibits FGFR1 with an IC50 of 8340 nM.
-
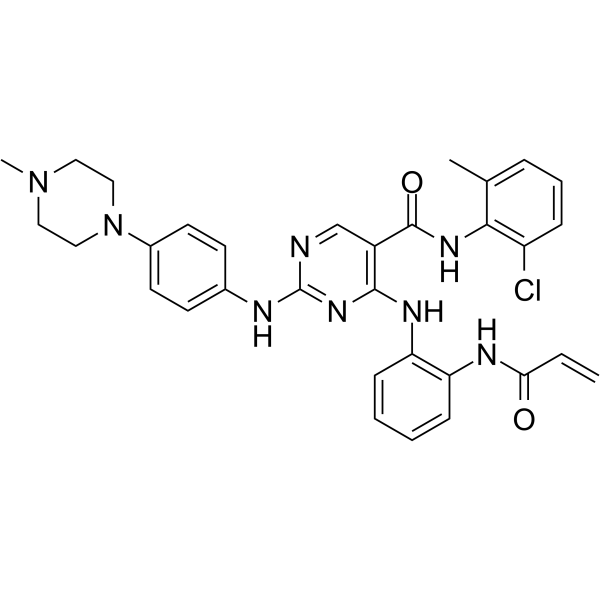
GC30768
Dihexa (PNB-0408)
Dihexa (PNB-0408) (N-hexanoic-Tyr-Ile-(6)-amino hexanoic amide) is an oral active, blood-brain barrier-permeable angiotensin IV analogue.
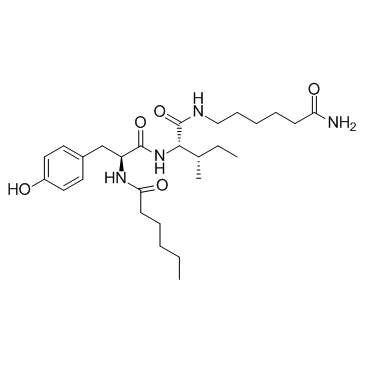
-
GC68986
Disitamab
RC48-0
Disitamab (RC48-0) is a humanized monoclonal antibody that targets HER2. Disitamab can be used to synthesize antibody-drug conjugates (ADCs), such as Disitamab vedotin.

-
GC64039
Disitamab vedotin
Disitamab vedotin (RC48) is an antibody-drug conjugate (ADC) comprising a monoclonal antibody against human epidermal growth factor receptor 2 (HER2) conjugated via a cleavable linker to the cytotoxic agent Monomethyl auristatin E (MMAE). Disitamab vedotin enhances antitumor immunity.

-
GC14298
DMH-1
BMP Inhibitor II, DorsoMorphin Homolog 1, VU036482
Selective BMP ALK2 receptor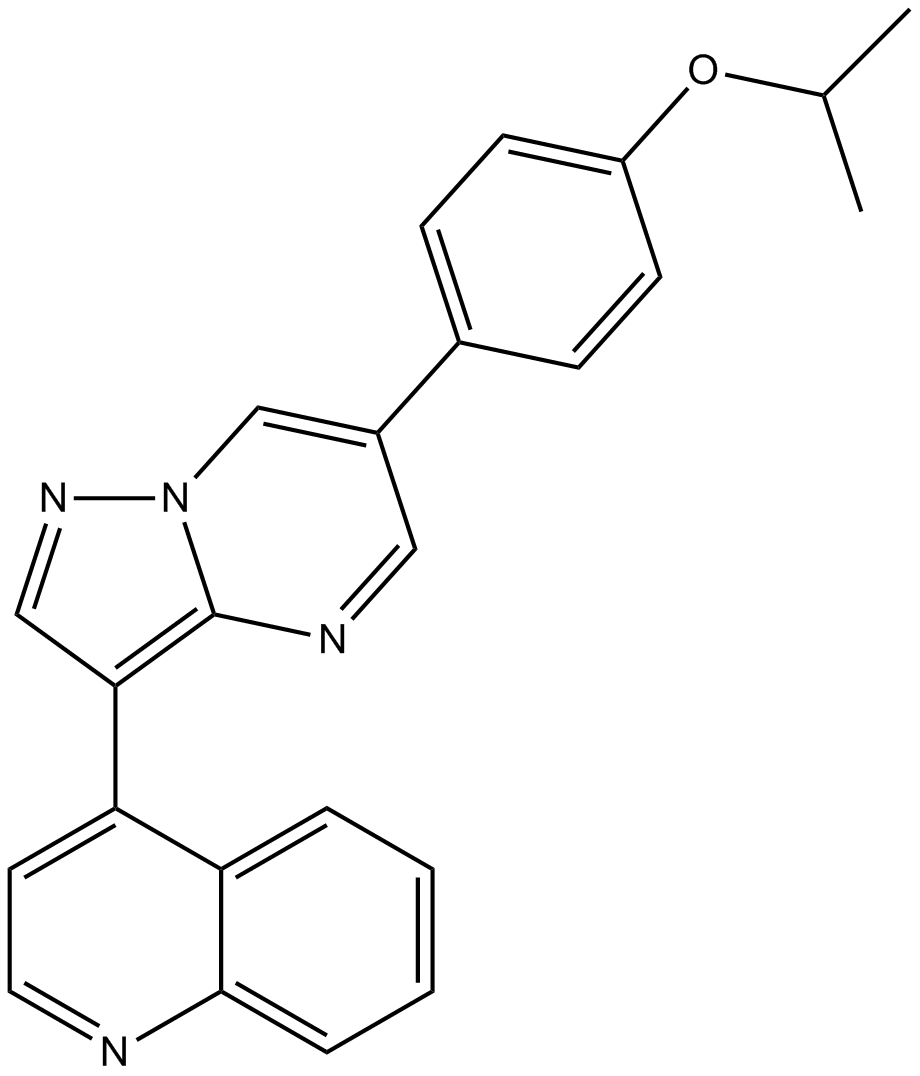
-
GC17098
DMH4
VEGFR-2 inhibitor
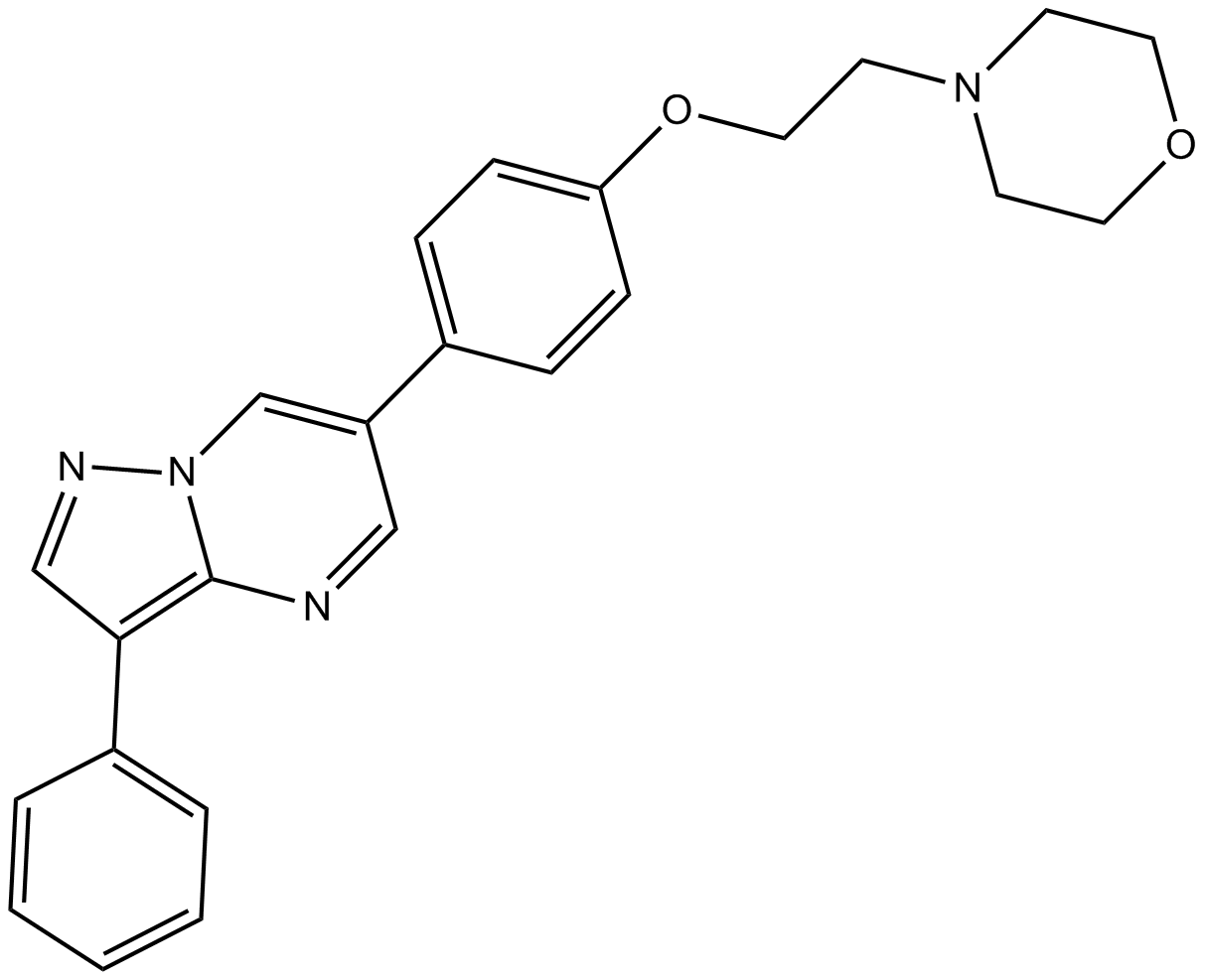
-
GC43503
DMHAPC-Chol
Dimethyl Hydroxyethyl Aminopropane Carbamoyl Cholesterol Iodide
DMHAPC-Chol is a cationic cholesterol.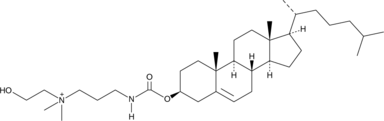
-
GC17024
DMPQ dihydrochloride
PDGFRβ inhibitor
-
GC69000
Dosimertinib-d5 mesylate
Dosimertinib-d5 (mesylate) is an effective orally active EGFR inhibitor. It reduces the expression of p-EGFR and p-ERK proteins. Dosimertinib-d5 (mesylate) exhibits anti-proliferative and anti-tumor activity. Dosimertinib-d5 (mesylate) has potential for research in non-small cell lung cancer (NSCLC).

-
GC13547
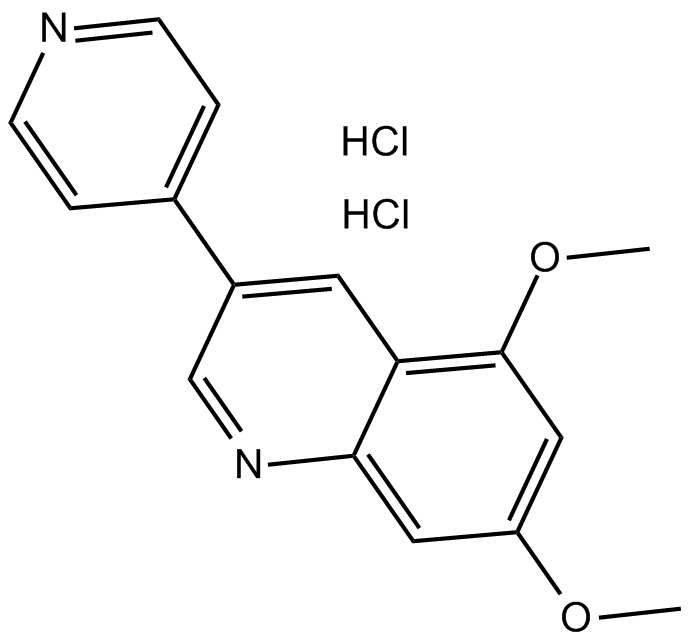
Dovitinib (TKI-258, CHIR-258)
CHIR258, TKI-258
Dovitinib (TKI-258, CHIR-258) (CHIR-258) is an orally active, potent multi-targeted tyrosine kinase (RTK) inhibitor with IC50s of 1, 2, 36, 8/9, 10/13/8, 27/210 nM for FLT3, c-Kit, CSF-1R, FGFR1/FGFR3, VEGFR1/VEGFR2/VEGFR3 and PDGFRα/PDGFRβ, respectively. Dovitinib (TKI-258, CHIR-258) has potent antitumor activity.
-
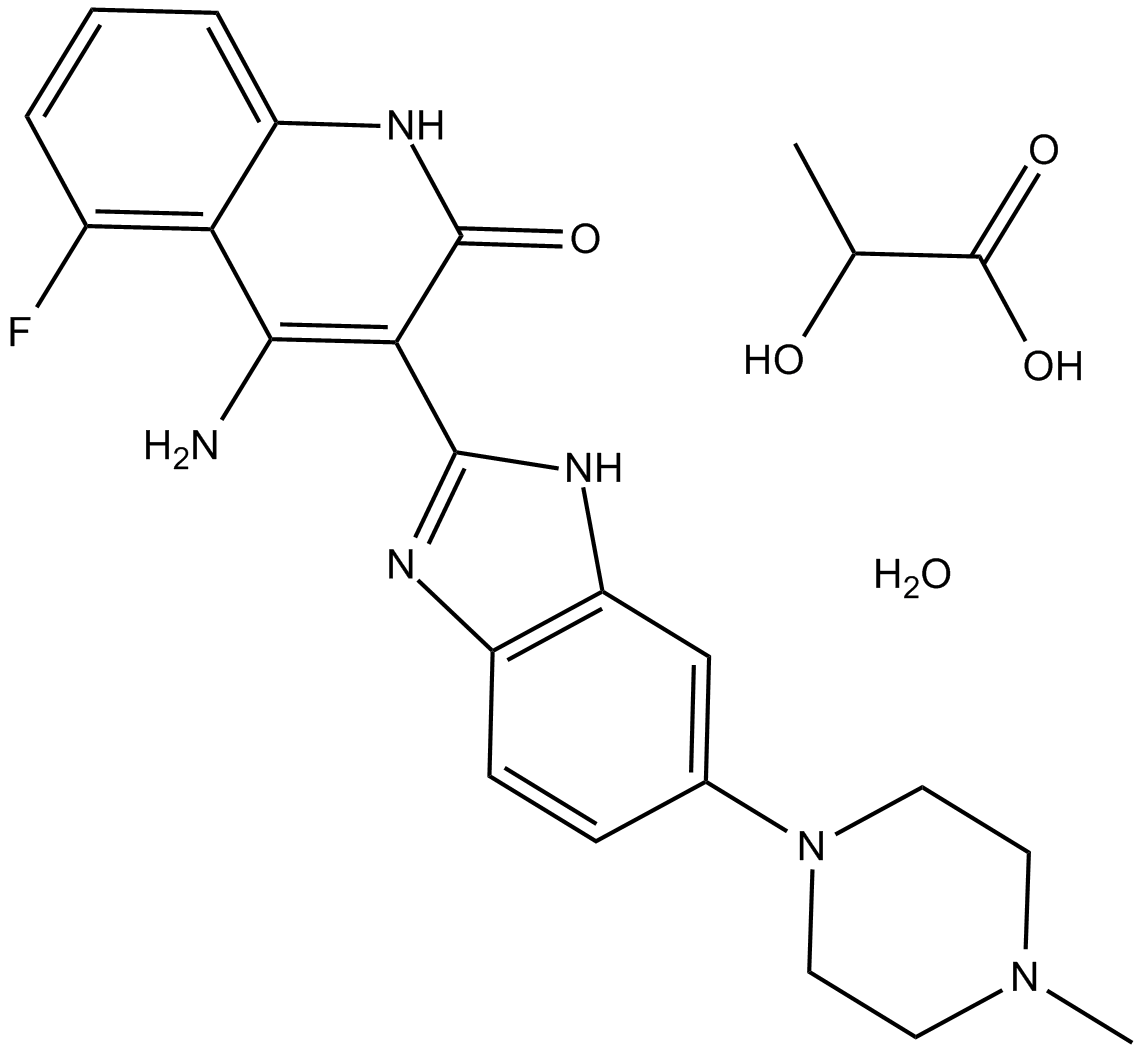
GC10165
Dovitinib (TKI258) Lactate
Dovitinib (TKI258) Lactate (TKI258 lactate hydrate) is a multi-targeted tyrosine kinase inhibitor with IC50s of 1, 2, 8/9, 10/13/8, 27/210 nM for FLT3, c-Kit, FGFR1/3, VEGFR1/2/3 and PDGFRα/β, respectively.
-
GC11372
Dovitinib Dilactic acid
FLT3 inhibitor
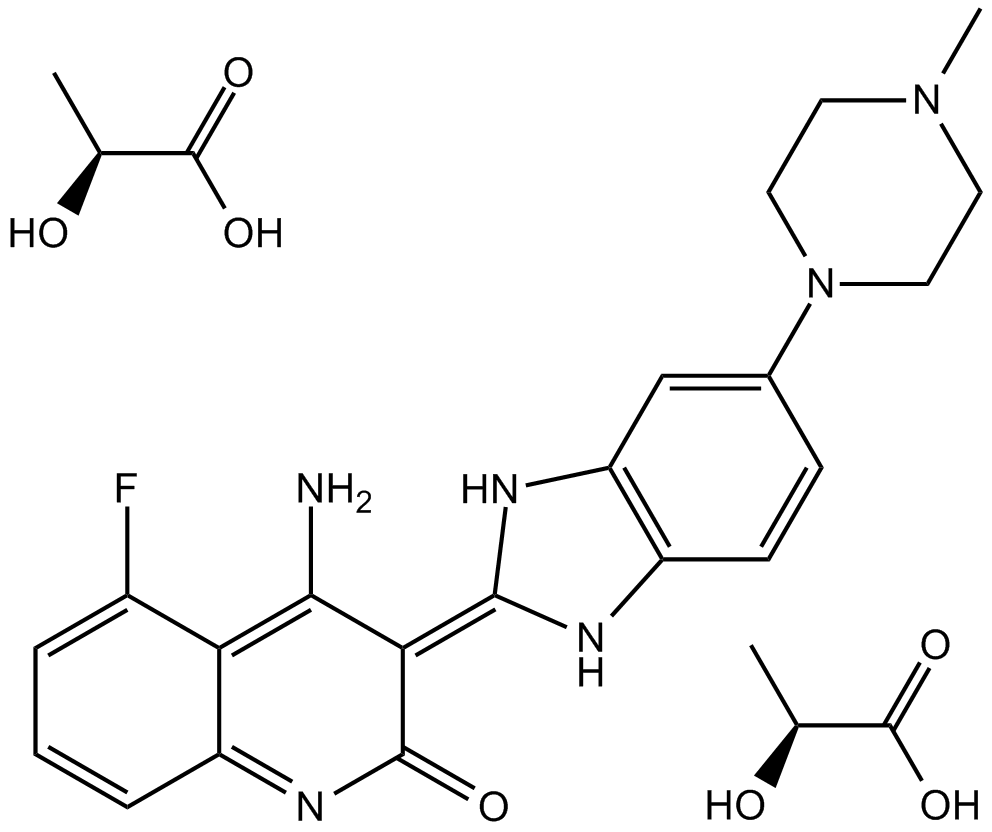
-
GC32760
Dovitinib lactate (CHIR-258 lactate)
Dovitinib lactate (CHIR-258 lactate) (TKI258 lactate) is a multi-targeted tyrosine kinase inhibitor with IC50s of 1, 2, 8/9, 10/13/8, 27/210 nM for FLT3, c-Kit, FGFR1/3, VEGFR1/2/3 and PDGFRα/β, respectively.
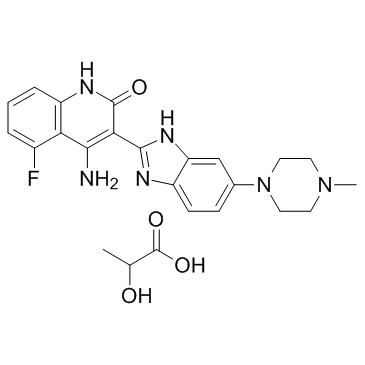
-
GC45767
Dovitinib-d8
An internal standard for the quantification of dovitinib
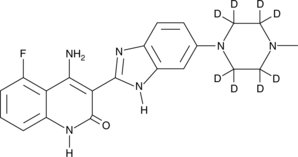
-
GC35897
DPH
A potent cell permeable c-Abl activator
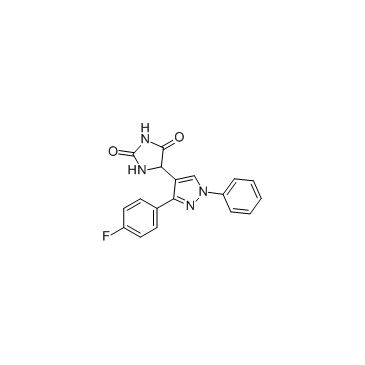
-
GC62597
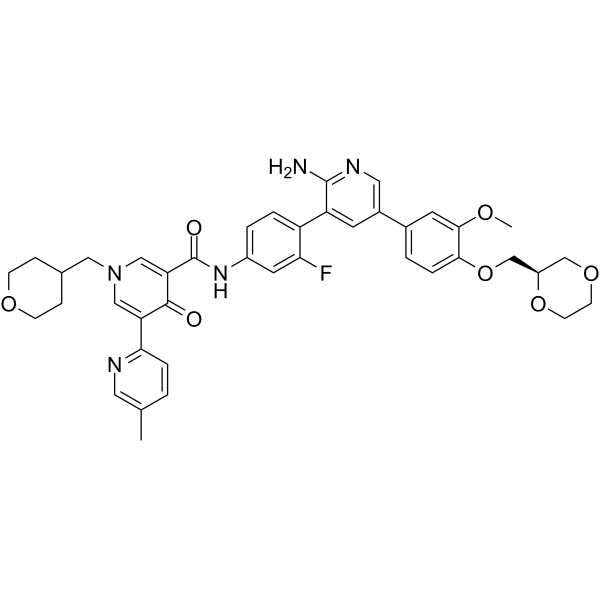
DS-1205b free base
DS-1205b free base is a potent and selective inhibitor of AXL kinase, with an IC50 of 1.3 nM. DS-1205b free base also inhibits MER, MET, and TRKA, with IC50s of 63, 104, and 407 nM, respectively. DS-1205b free base can inhibit cell migration in vitro and tumor growth in vivo.
-
GC72453
Duligotuzumab
Duligotuzumab (MEHD-7945A; RG 7597) is a humanized IgG-κ monoclonal antibody that specifically targets Her3 (ErbB3).

-
GC72454
Dusigitumab
Dusigitumab (MEDI 573) is a human IgG2λ monoclonal antibody that specifically targets IGF2 and IGF1.

-
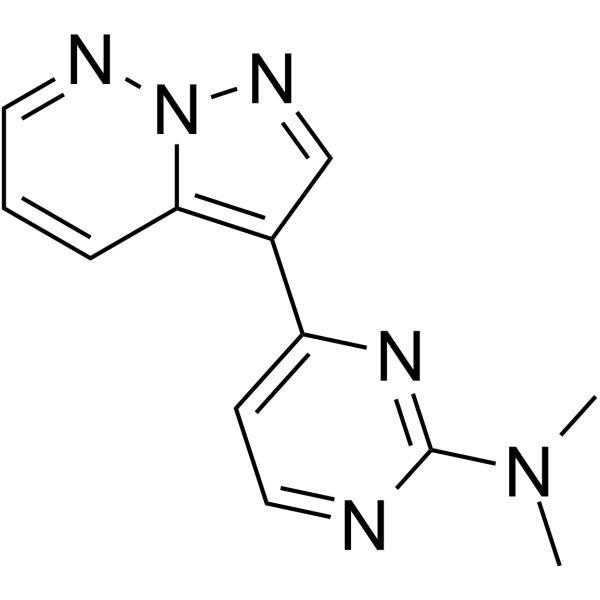
GC62946
DYRK1-IN-1
DYRK1-IN-1 is a highly selective and ligand-efficient DYRK1A inhibitor.
-
GC70985
Dyrk1A-IN-3
Dyrk1A-IN-3 (Compound 8b), a highly selective dual-specificity tyrosine-regulated kinase 1A (DYRK1A)inhibitor, maintains high levels of DYRK1A binding affinity (IC50=76 nM).

-
GC69031
Dyrk1A-IN-5
Dyrk1A-IN-5 (compound 5j) is a potent and selective inhibitor of DYRK1A with an IC50 value of 6 nM. Dyrk1A-IN-5 dose-dependently reduces the phosphorylation of Thr434 in SF3B1, with an IC50 of 0.5 μM. Dyrk1A-IN-5 inhibits the phosphorylation of tau protein at Thr212, with an IC50 of 2.1 μM. Dyrk1A-IN-5 can be used for research on Down syndrome.

-
GC62947
DYRKs-IN-1 hydrochloride
DYRKs-IN-1 hydrochloride is a potent DYRKs (Dual-specificity tyrosine-phosphorylation-regulated kinases) inhibitor with IC50s of 5 nM and 8 nM for DYRK1A and DYRK1B, respectively. DYRKs-IN-1 hydrochloride has antitumor activity.

-
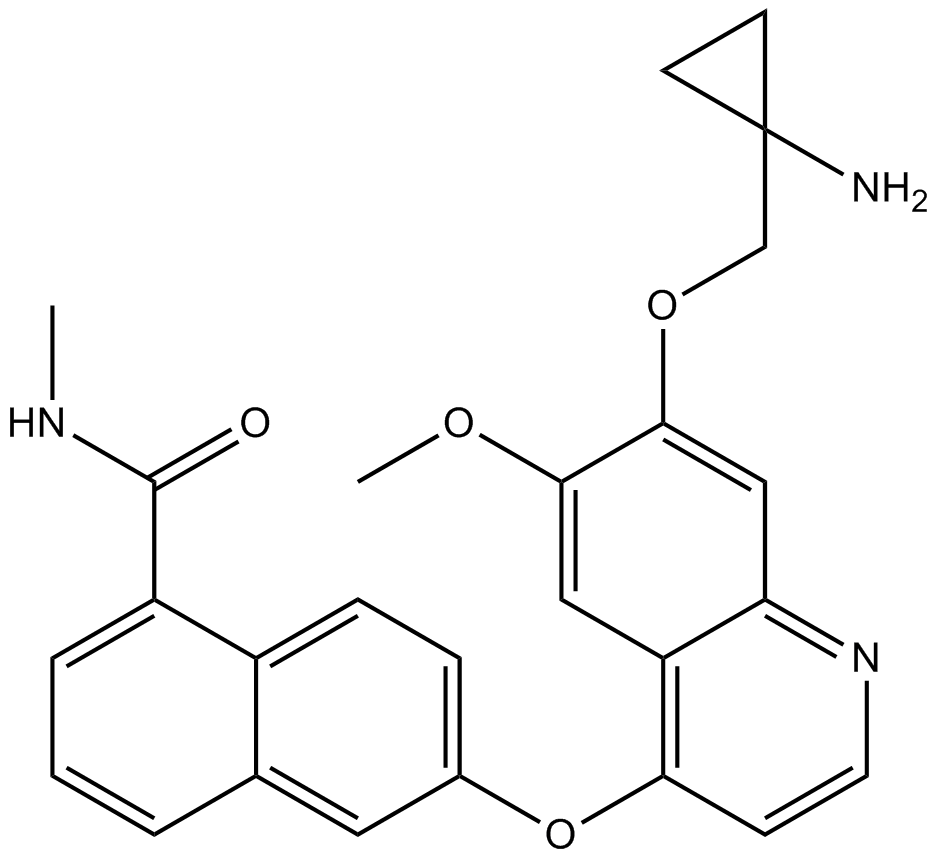
GC12149
E-3810
-
GC69037
E7090
E7090 is an effective selective FGFR inhibitor with oral activity, with IC50 values of 0.71 nM, 0.50 nM, 1.2 nM and 120 nM for FGFR1/FGFR2/FGFR3/FGFR4 respectively.

-
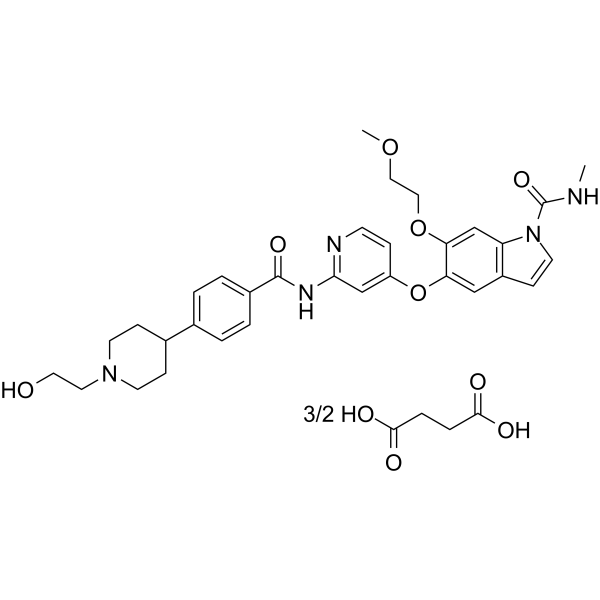
GC62696
E7090 succinate
E7090 succinate is an orally available, selective and potent inhibitor of FGFR1, FGFR2 and FGFR3 tyrosine kinase activities, with IC50 values of 0.71 nM, 0.50 nM, 1.2 nM, and 120 nM for FGFR1/2/3/4, respectively.
-
GC66432
EAI001
EAI001 is a potent, selective mutant epidermal growth factor receptor (EGFR) allosteric inhibitor with an IC50 value of 24 nM for EGFRL858R/T790M. EAI001 can be used for research of cancer.
-
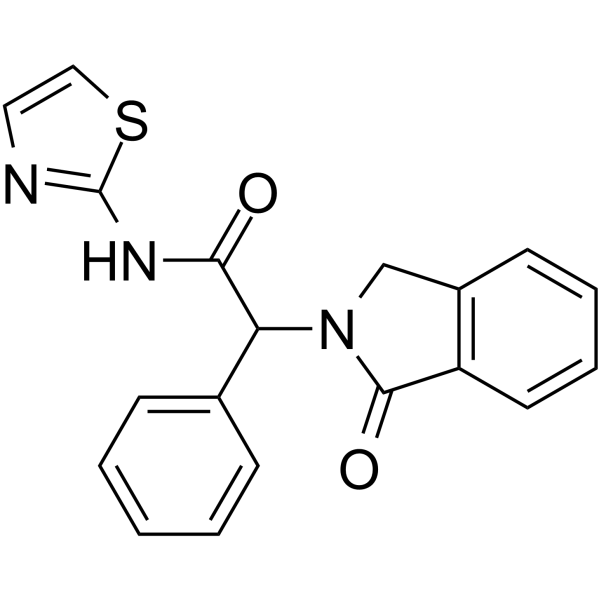
GC12281
EAI045
Inhibitor of L858R/T790M EGFR mutants
-
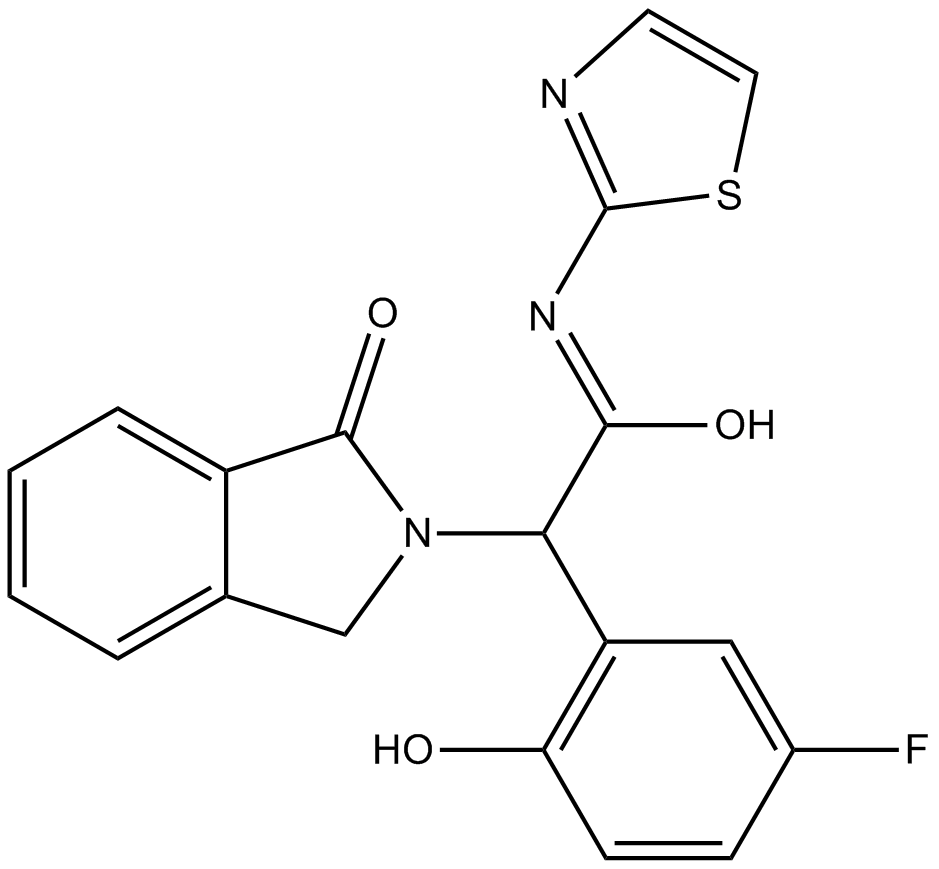
GC64730
EB-42486
EB-42486 is a novel, potent, and highly selective G2019S-LRRK2 inhibitor (IC50 < 0.2 nM).
-
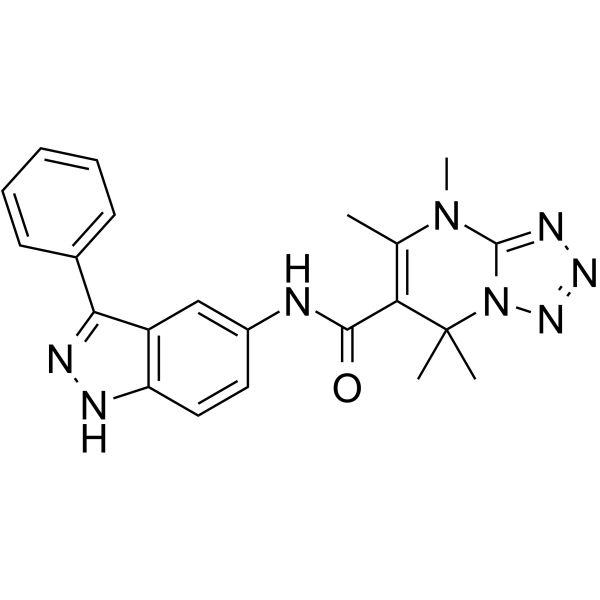
GC32825
eCF506
An inhibitor of Src kinases
-
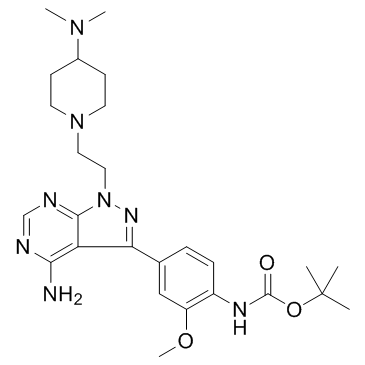
GC67879
Edecesertib
GS-5718
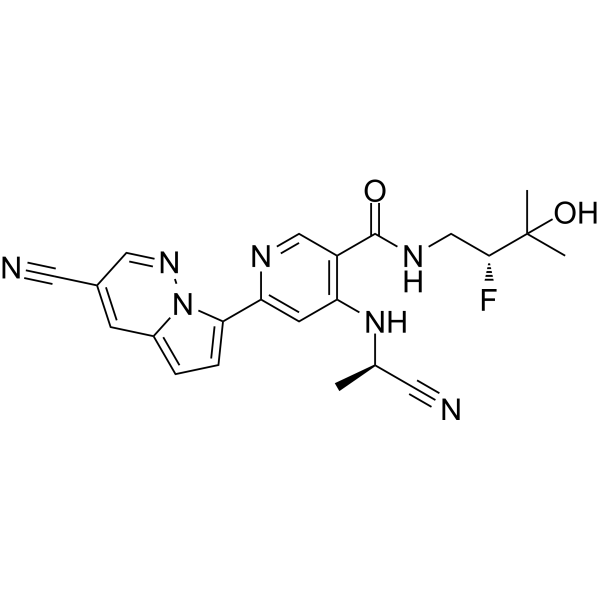
-
GC19418
Edicotinib
JNJ-40346527; JNJ-527
Edicotinib is a selective and orally available colony-stimulating factor-1 (CSF-1) receptor inhibitor, and has entered phase IIA clinical trial to study rheumatoid arthritis (RA) despite disease.
-
GC72468
Efruxifermin
Efruxifermin is an Fc-FGF21 fusion protein (human IgG1 Fc domain linked to a modified human FGF21).

-
GC16183
EG00229
Nrp1 inhibitor
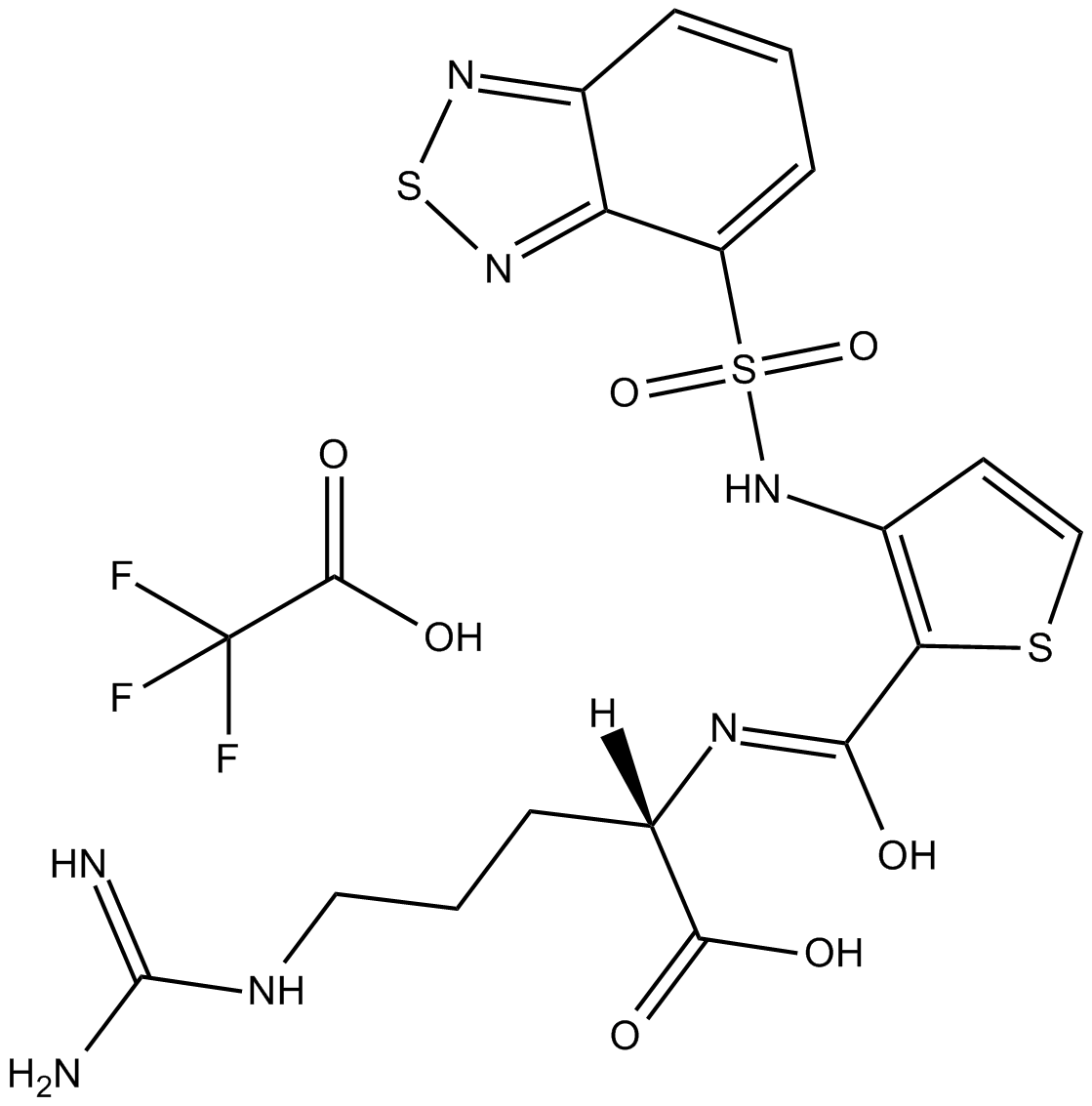
-
GC16321
EGF816
EGF816 (EGF816) is a covalent mutant-selective EGFR inhibitor, with Ki and Kinact of 31 nM and 0.222 min-1 on EGFR(L858R/790M) mutant, respectively.

-
GC11312
EGFR Inhibitor
Epidermal Growth Factor Receptor Inhibitor
EGFR Inhibitor is a 4,6-disubstituted pyrimidine and is a potent, ATP-competitive, irreversible and highly selective EGFR inhibitor with an IC50of 21 nM. EGFR Inhibitor also inhibits mutant EGFRL858R and EGFRL861Q with IC50s of 63 nM and 4 nM, respectively. EGFR Inhibitor displays strong selectivity for EGFR over HER4 (IC50 = 7640 nM) and a panel of 55 other kinases. EGFR Inhibitor induces cells apoptosis and has antitumor activity.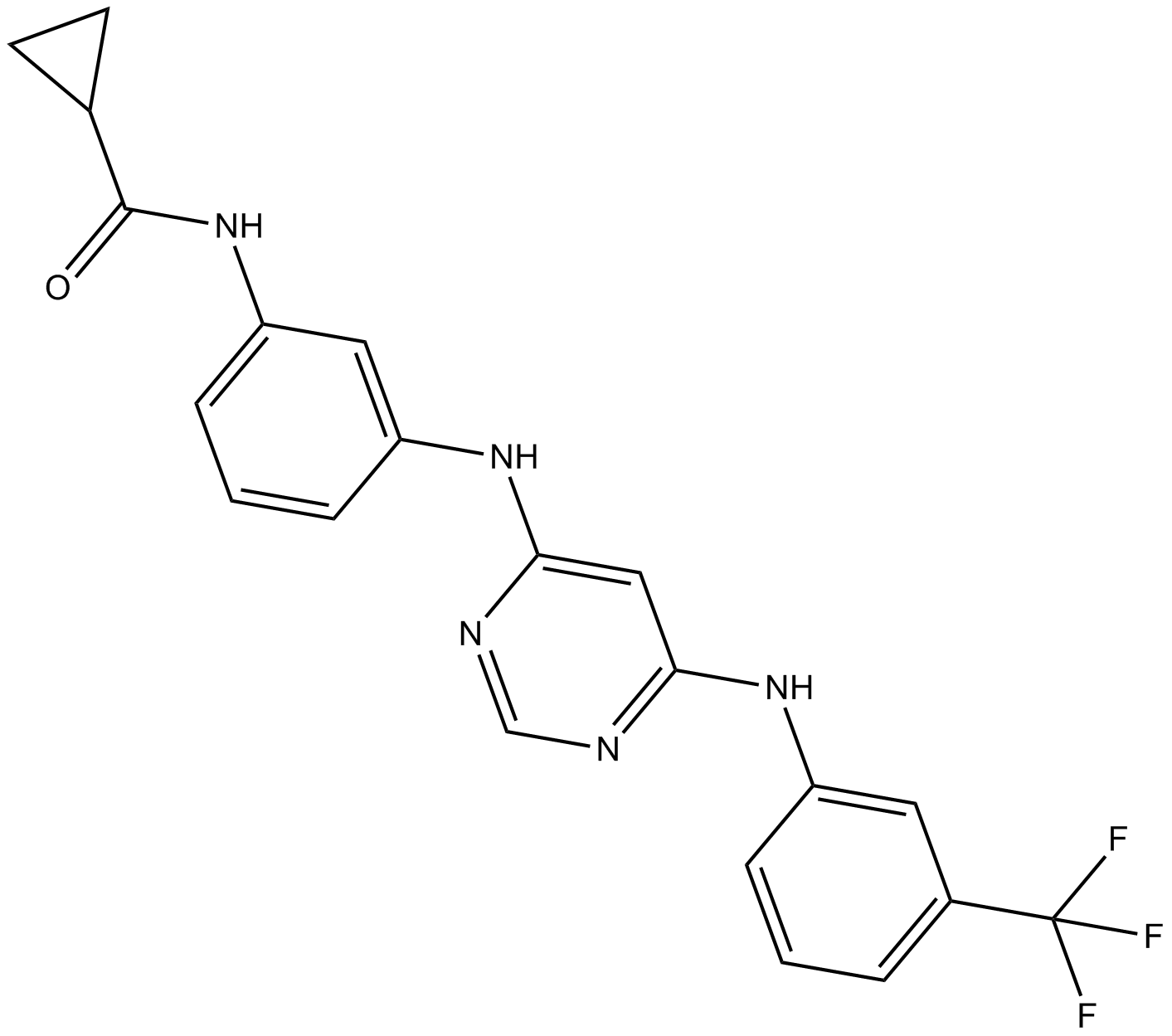
-
GC35965
EGFR mutant-IN-1
EGFR mutant-IN-1, a 5-methylpyrimidopyridone derivative, is a potent and selective EGFRL858R/T790M/C797S mutant inhibitor with an IC50 of 27.5 nM, while being a significantly less potent for EGFRWT (IC50 >1.0 μM).
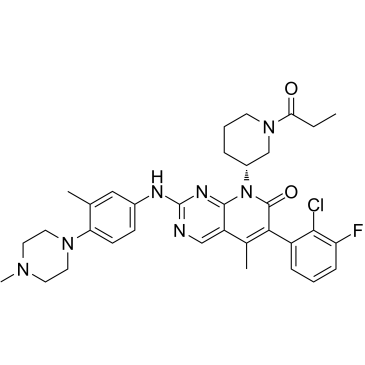
-
GC65572
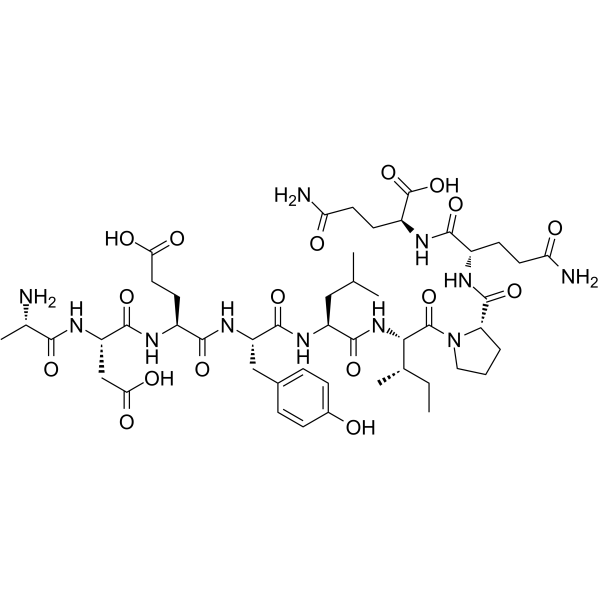
EGFR Protein Tyrosine Kinase Substrate
EGFR Protein Tyrosine Kinase Substrate is a EGFR protein tyrosine kinase substrate.
-
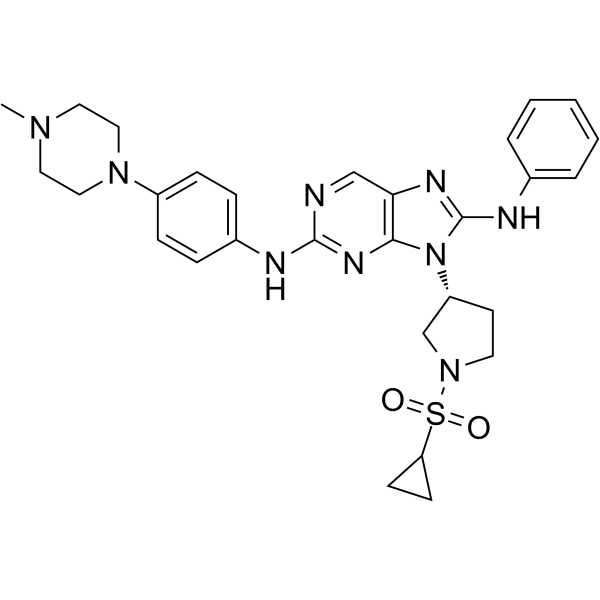
GC62562
EGFR-IN-11
EGFR-IN-11 is a fourth-generation EGFR-tyrosine kinase inhibitor (EGFR-TKI) with an IC50 of 18 nM for triple mutant EGFRL858R/T790M/C797S. EGFR-IN-11 significantly suppresses the EGFR phosphorylation, induce the apoptosis, and arrest cell cycle at G0/G1.
-
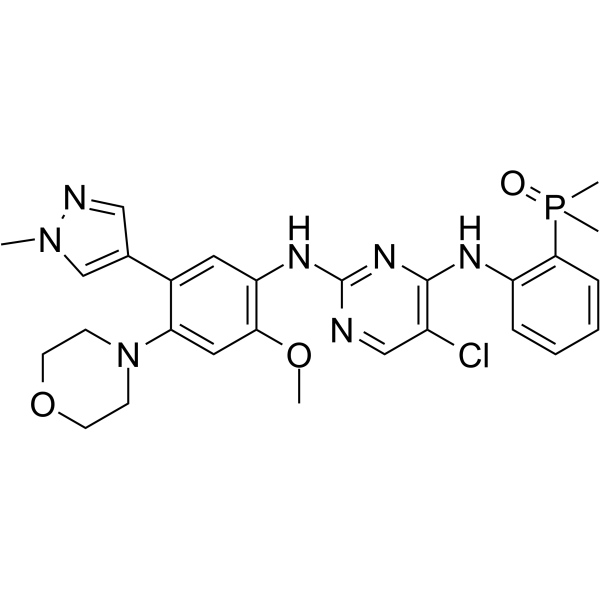
GC64497
EGFR-IN-17
EGFR-IN-17 is a potent and selective inhibitor of the epidermal growth factor receptor ( IC50 0.0002 μM) to overcome C797S-mediated resistance.
-
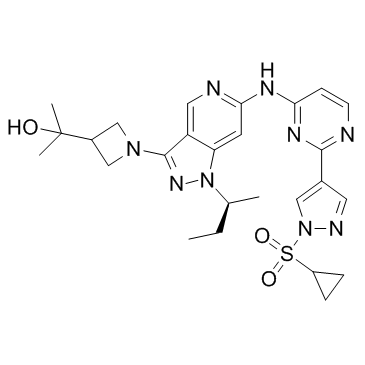
GC33195
EGFR-IN-2
EGFR-IN-2 is a a noncovalent, irreversible, mutant-selective second generation EGFR inhibitor.
-
GC19132
EGFR-IN-3
EGFR-IN-3 is a third-generation EGFR TKI, with GI50 values of 5 nM (EGFR L858R/T790M), 10 nM (EGFR del19) and 689 nM (EGFR WT), respectively. EGFR-IN-3 has the potential for NSCLC research.
-
GC66428
EGFR-IN-5
EGFR-IN-5 is a EGFR inhibitor with IC50s of 10.4, 1.1, 34, 7.2 nM for EGFR, EGFRL858R, EGFRL858R/T790M, and EGFRL858R/T790M/C797S, respectively.
-
GC68440
EGFR-IN-69

-
GC35966
EGFR-IN-7
EGFR-IN-7 is a potent, selective and orally active EGFR kinase inhibitor. EGFR-IN-7 has inhibitory effect for for EGFR (WT) and EGFR (mutant C797S/T790M/L858R) with IC50 values of 7.92 nM and 0.218 nM, respectively. EGFR-IN-7 shows anti-tumor activity. EGFR-IN-7 can be used for the research of various cancers.
-
GC69043
EGFR-IN-70
EGFR-IN-70 (compound 18j) is an effective EGFR inhibitor with IC50 values of 23.6 and 307.5 nM for EGFRLR/TM/CS and EGFRWT, respectively. It has anti-proliferative activity and inhibits the phosphorylation of EGFR. EGFR-IN-70 can be used in cancer research.

-
GC65974
EGFR-IN-9
EGFR-IN-9 (Compound 8) is a potent EGFR kinase inhibitor with IC50s of 7 nM, 28 nM for the wild type EGFR kinase and double mutant EGFR kinase (L858R/T790M). EGFR-IN-9 has antitumor activity.