Cell Cycle/Checkpoint
Cell Cycle
Cells undergo a complex cycle of growth and division that is referred to as the cell cycle. The cell cycle consists of four phases, G1 (GAP 1), S (synthesis), G2 (GAP 2) and M (mitosis). DNA replication occurs during S phase. When cells stop dividing temporarily or indefinitely, they enter a quiescent state called G0.
Targets for Cell Cycle/Checkpoint
- ATM/ATR(26)
- Aurora Kinase(47)
- Cdc42(4)
- Cdc7(4)
- Chk(16)
- c-Myc(20)
- CRM1(8)
- Cyclin-Dependent Kinases(91)
- E1 enzyme(1)
- G-quadruplex(14)
- Haspin(7)
- HMTase(1)
- Kinesin(26)
- Ksp(6)
- Microtubule/Tubulin(243)
- Mps1(15)
- Mitotic(11)
- RAD51(18)
- ROCK(71)
- Rho(13)
- PERK(11)
- PLK(37)
- PTEN(8)
- Wee1(7)
- PAK(21)
- Arp2/3 Complex(8)
- Dynamin(12)
- ECM & Adhesion Molecules(40)
- Cholesterol Metabolism(3)
- Endomembrane System & Vesicular Trafficking(26)
- G1(38)
- G2/M(26)
- G2/S(10)
- Genotoxic Stress(18)
- Inositol Phosphates(18)
- Proteolysis(99)
- Cytoskeleton & Motor Proteins(53)
- Cellular Chaperones(8)
Products for Cell Cycle/Checkpoint
- Cat.No. Product Name Information
-
GC44730
proTAME
proTAME is a cell-permeable prodrug form of N-4-tosyl-L-arginine methyl ester, an inhibitor of the anaphase-promoting complex/cyclosome (APC/C), that is converted to TAME by intracellular esterases.
-
GC44732
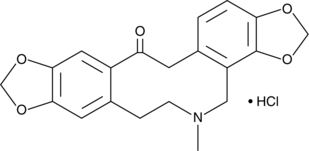
Protopine (hydrochloride)
Protopine is an alkaloid found in Berberidaceae, Ranunculaceae, Rutaceae, Fumariaceae, and Papaveraceae with diverse biological activities.
-
GC49914
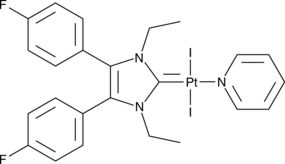
Pt(II)-NHC Complex 2C
An inducer of immunogenic cancer cell death
-
GC69776
PT-262
PT-262 is an effective ROCK inhibitor with an IC50 value of approximately 5 μM. PT-262 induces loss of mitochondrial membrane potential and increases activation of caspase-3 and cell apoptosis. PT-262 inhibits phosphorylation of ERK and CDC2 through a p53-independent pathway. PT-262 blocks cellular cytoskeleton function and cell migration. PT-262 has anti-cancer activity.
-
GC44740
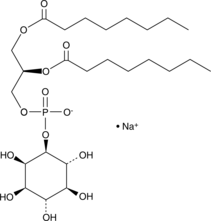
PtdIns-(1,2-dioctanoyl) (sodium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
-
GC44743
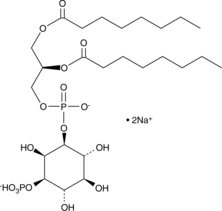
PtdIns-(3)-P1 (1,2-dioctanoyl) (sodium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
-
GC44748
PtdIns-(3,4,5)-P3 (1,2-dihexanoyl) (ammonium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
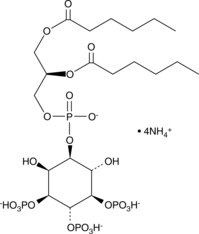
-
GC44750
PtdIns-(3,4,5)-P3 (1,2-dipalmitoyl) (sodium salt)
The phosphatidylinositol phosphates represent a small percentage of total membrane phospholipids.
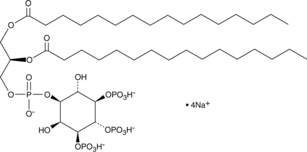
-
GC44753
PtdIns-(3,4,5)-P3-biotin (sodium salt)
The PtdIn phosphates play an important role in the generation and transduction of intracellular signals.
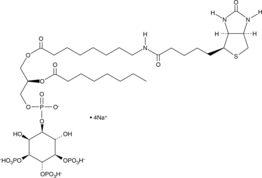
-
GC44759
PtdIns-(4)-P1 (1,2-dioctanoyl) (ammonium salt)
The phosphatidylinositol (PtdIns) phosphates represent a small percentage of total membrane phospholipids.
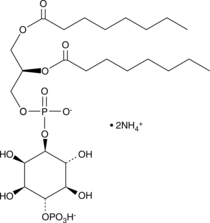
-
GC52484
Purified Ganglioside Mixture (bovine) (ammonium salt)
A mixture of purified bovine gangliosides

-
GC14707
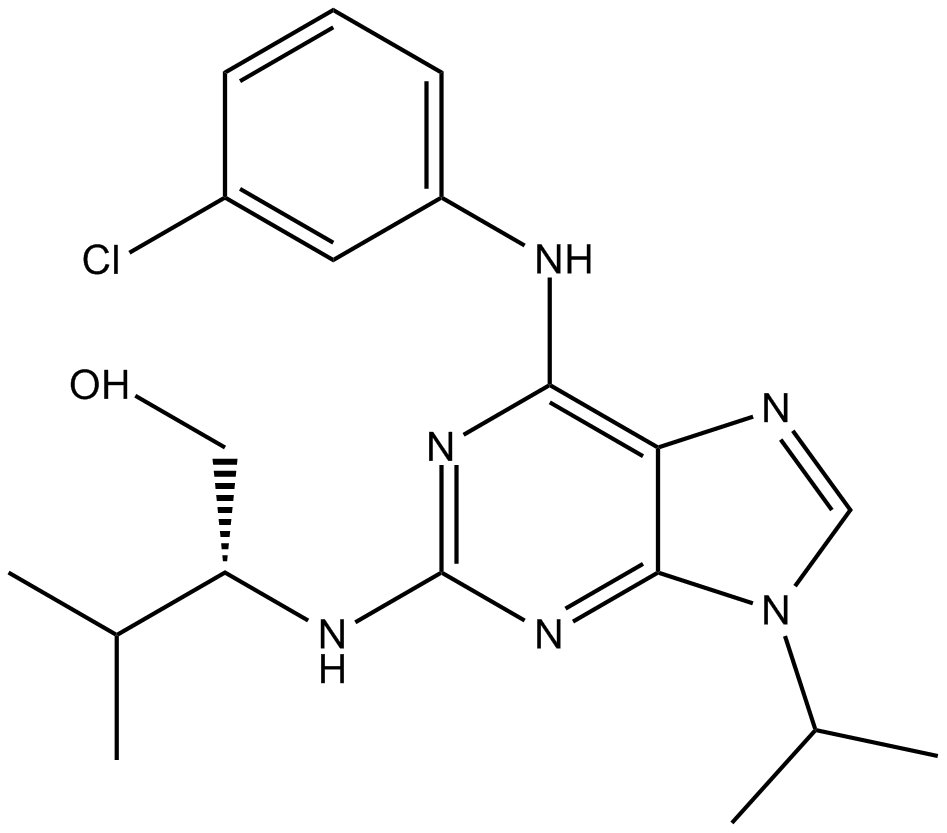
Purvalanol A
potent, and cell-permeable CDK inhibitor
-
GC16268
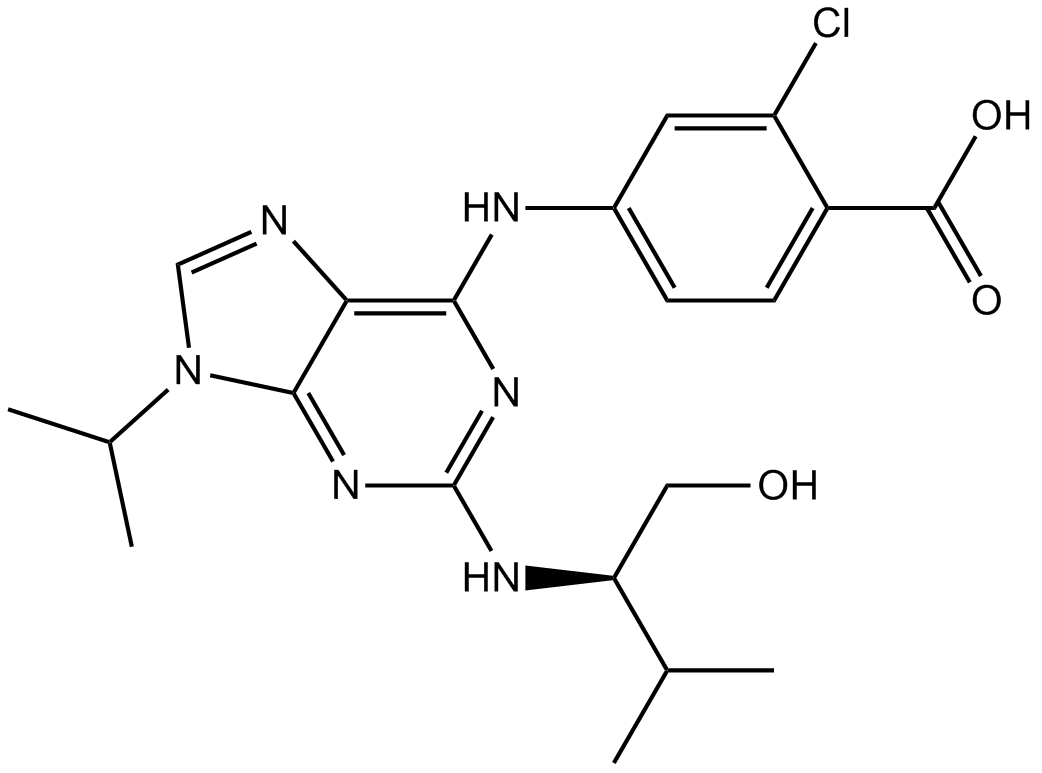
Purvalanol B
CDK1/CDK2/CDK4 inhibitor
-
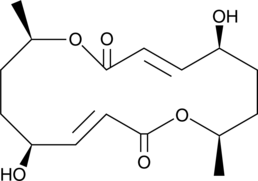
GC45552
Pyrenophorol
-
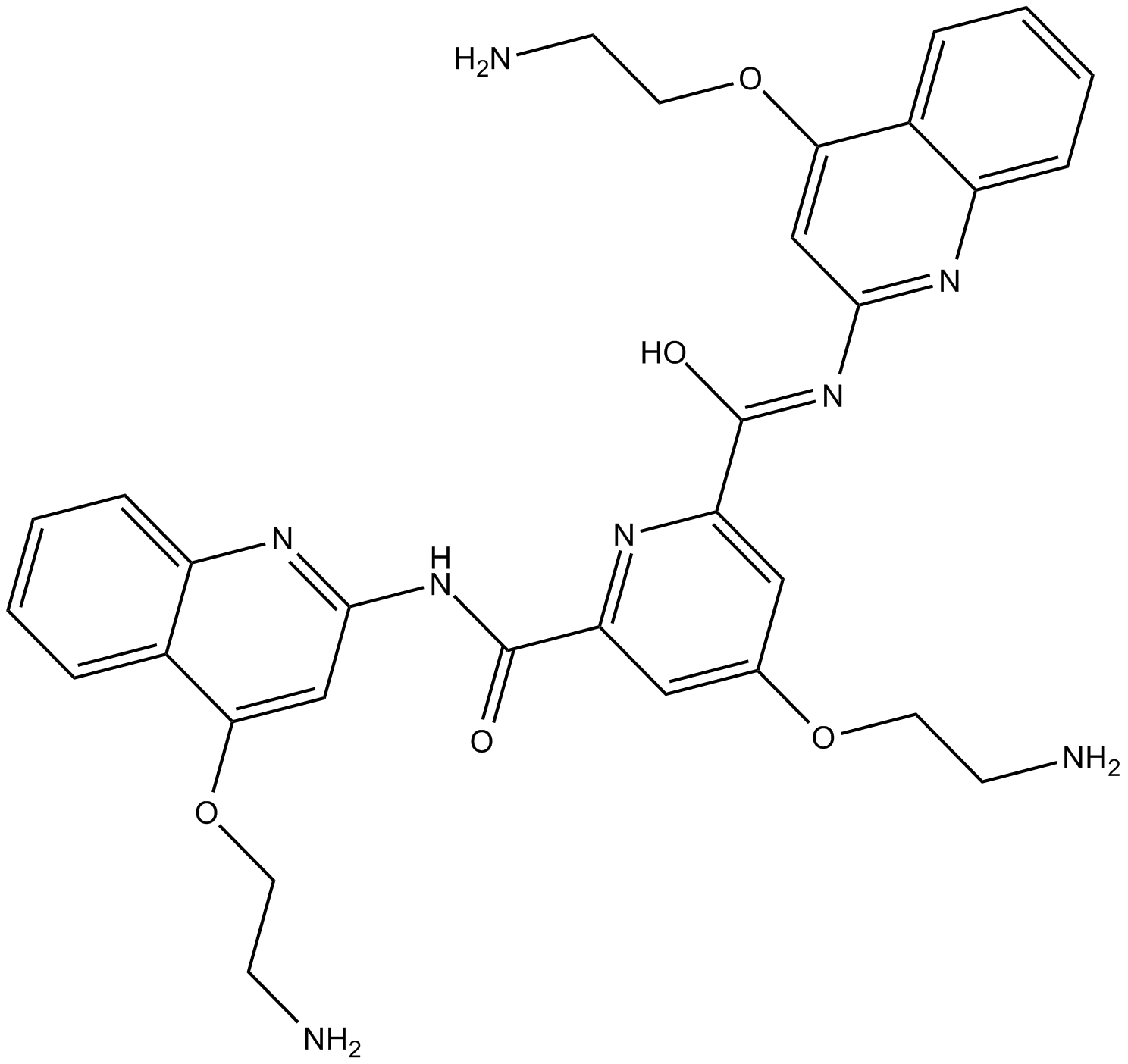
GC15162
Pyridostatin
Pyridostatin is a highly selective G-quadruplex-binding small molecule that was designed to target polymorphic G-quadruplex structures regardless of sequence variability.
-
GC48016
Pyridostatin (trifluoroacetate salt)
Pyridostatin (RR82) TFA is a G-quadruplex DNA stabilizing agent (Kd=490 nM). Pyridostatin (trifluoroacetate salt) promotes growth arrest in human cancer cells by inducing replication- and transcription-dependent DNA damage. Pyridostatin (trifluoroacetate salt) targets the proto-oncogene Src. Pyridostatin (trifluoroacetate salt) reduced SRC protein levels and SRC-dependent cellular motility in human breast cancer cells.
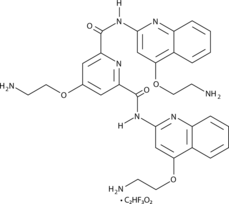
-
GC37043
Pyridostatin hydrochloride
Pyridostatin (RR82) hydrochloride is a G-quadruplex DNA stabilizing agent (Kd=490 nM). Pyridostatin hydrochloride promotes growth arrest in human cancer cells by inducing replication- and transcription-dependent DNA damage. Pyridostatin hydrochloride targets the proto-oncogene Src. Pyridostatin hydrochloride reduced SRC protein levels and SRC-dependent cellular motility in human breast cancer cells.
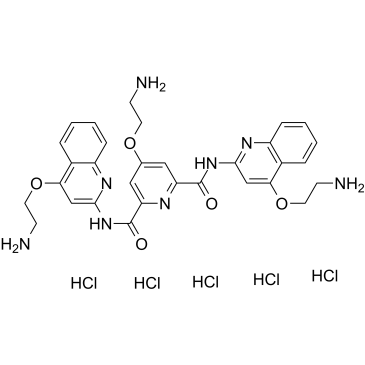
-
GC17801
PYZD-4409
E1 enzyme inhibitor
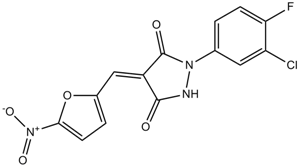
-
GC52057
QN523
An anticancer agent

-
GC45903
Quazinone
A PDE3 inhibitor
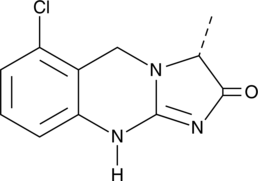
-
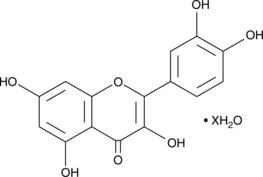
GC48019
Quercetin (hydrate)
A flavonoid with diverse biological activities
-
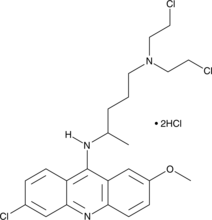
GC46210
Quinacrine mustard (hydrochloride)
Quinacrine mustard (hydrochloride) is a fluorochrome.
-
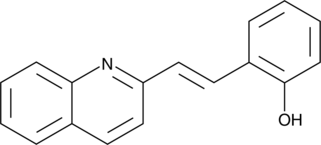
GC44800
Quininib
A CysLT1 and CysLT2 receptor antagonist
-
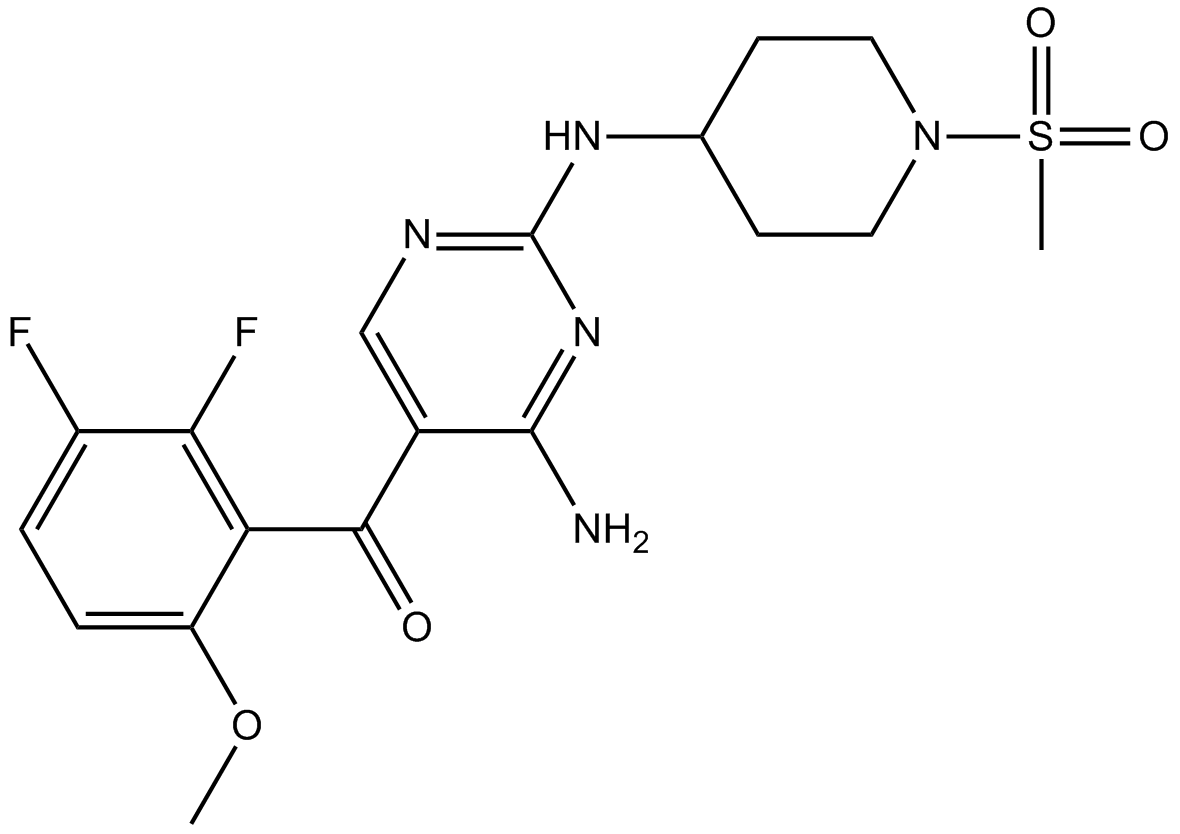
GC17400
R547
CDK1/2/4 inhibitor,ATP-competitive
-
GC49771
rac-Hesperetin-d3
An internal standard for the quantification of hesperetin
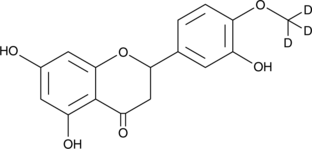
-
GC49108
Racecadotril-d5
An internal standard for the quantification of racecadotril
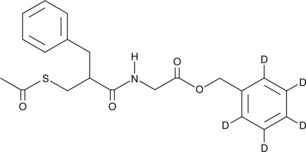
-
GC19306
RAD51 Inhibitor B02
RAD51 Inhibitor B02 (B02) is an inhibitor of human RAD51 with an IC50 of 27.4 uM.

-
GC63165
RAD51-IN-1
RAD51-IN-1, a derivative of ?B02, is a potent inhibitor of RAD51. RAD51-IN-1 can be used for cancer research.
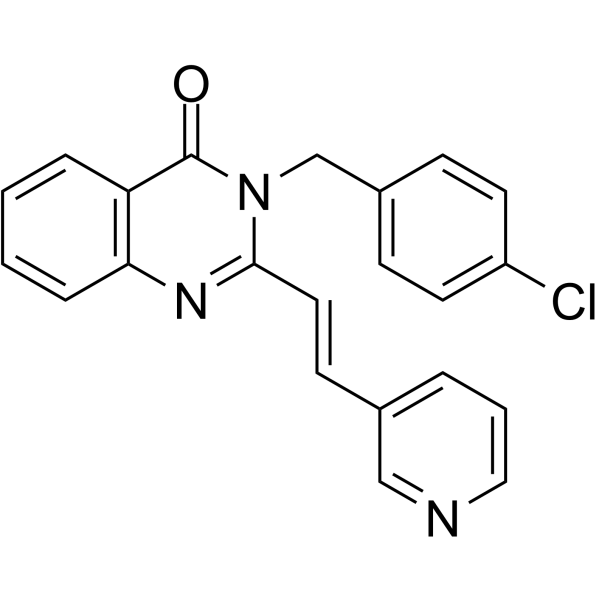
-
GC65383
RAD51-IN-2
RAD51-IN-2 (compound example 67A) is a RAD51 inhibitor extracted from patent WO2019/051465A1.
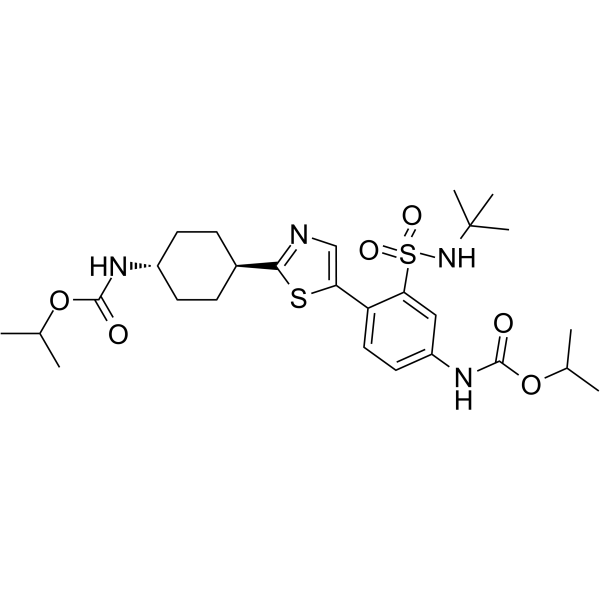
-
GC68416
RAD51-IN-3
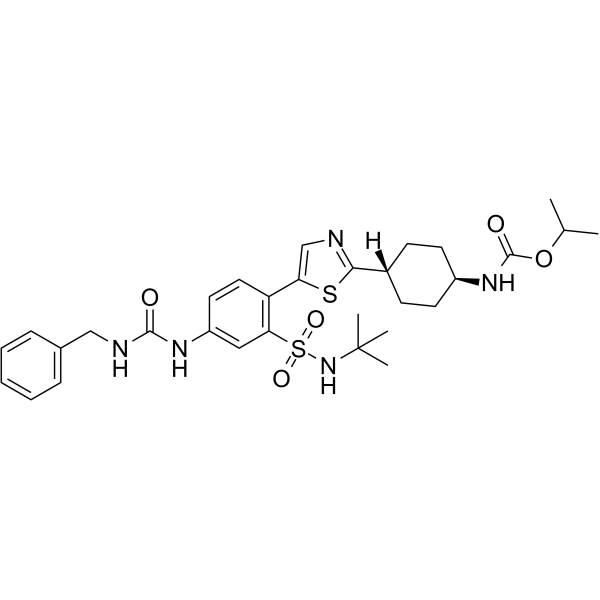
-
GC67888
RAD51-IN-5

-
GC52054
Ranitidine S-oxide
Ranitidine S-oxide is the metabolite of Ranitidine.

-
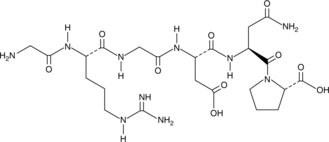
GC52196
RGD Peptide
RGD Peptide acts as an inhibitor of integrin-ligand interactions and plays an important role in cell adhesion, migration, growth, and differentiation.
-
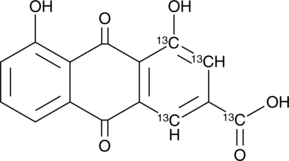
GC45798
Rhein-13C4
An internal standard for the quantification of rhein
-
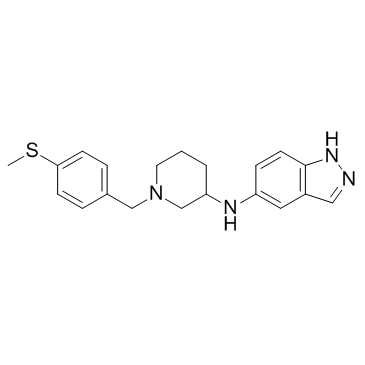
GC33310
Rho-Kinase-IN-1
Rho-Kinase-IN-1 is a Rho kinase (ROCK) inhibitor (Ki values of 30.5 and 3.9 nM for ROCK1 and ROCK2, respectively) extracted from US20090325960A1, compound 1.008.
-
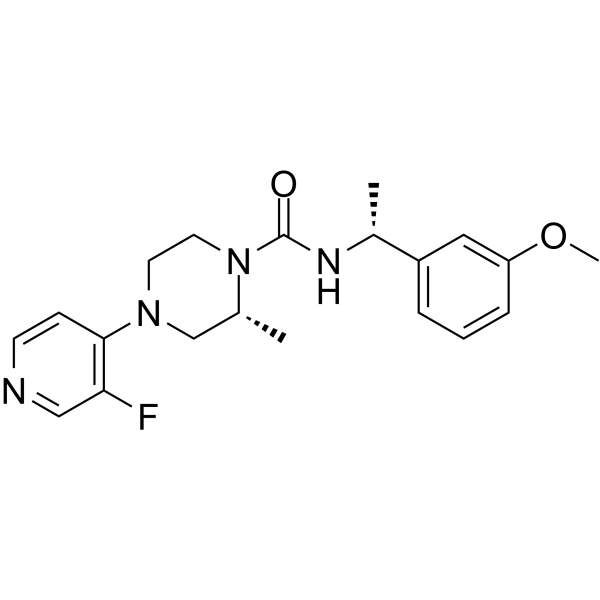
GC66050
Rho-Kinase-IN-2
Rho-Kinase-IN-2 (Compound 23) is an orally active, selective, and central nervous system (CNS)-penetrant Rho Kinase (ROCK) inhibitor (ROCK2 IC50=3 nM). Rho-Kinase-IN-2 can be used in Huntington's research.
-
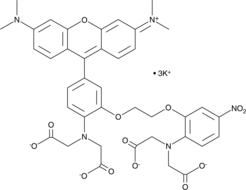
GC44829
Rhod-5N (potassium salt)
Rhod-5N is a low affinity fluorescent calcium probe (Kd = 320 μM).
-
GC18140
RI-1
A RAD51 inhibitor

-
GC12752
RI-2
Optimized reversible RAD51 inhibitor
-
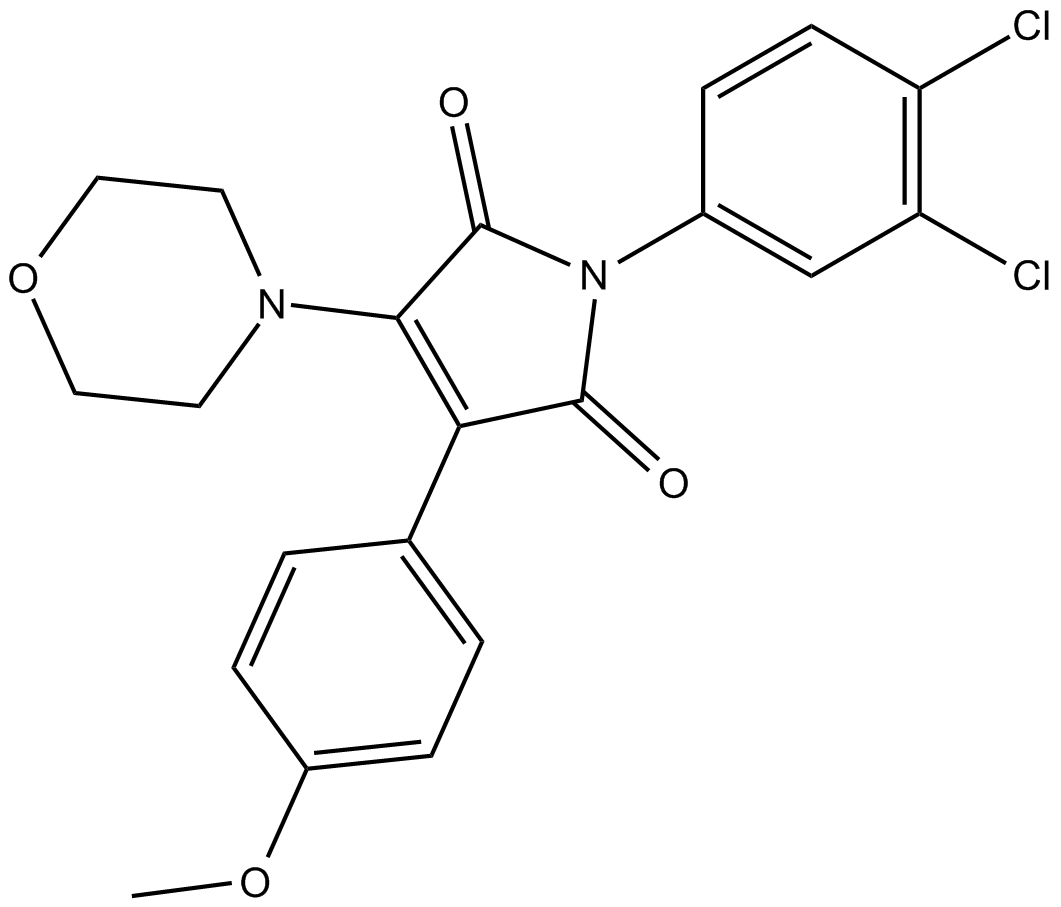
GC12415
Rigosertib
PI3K/PLK1 inhibitor
-
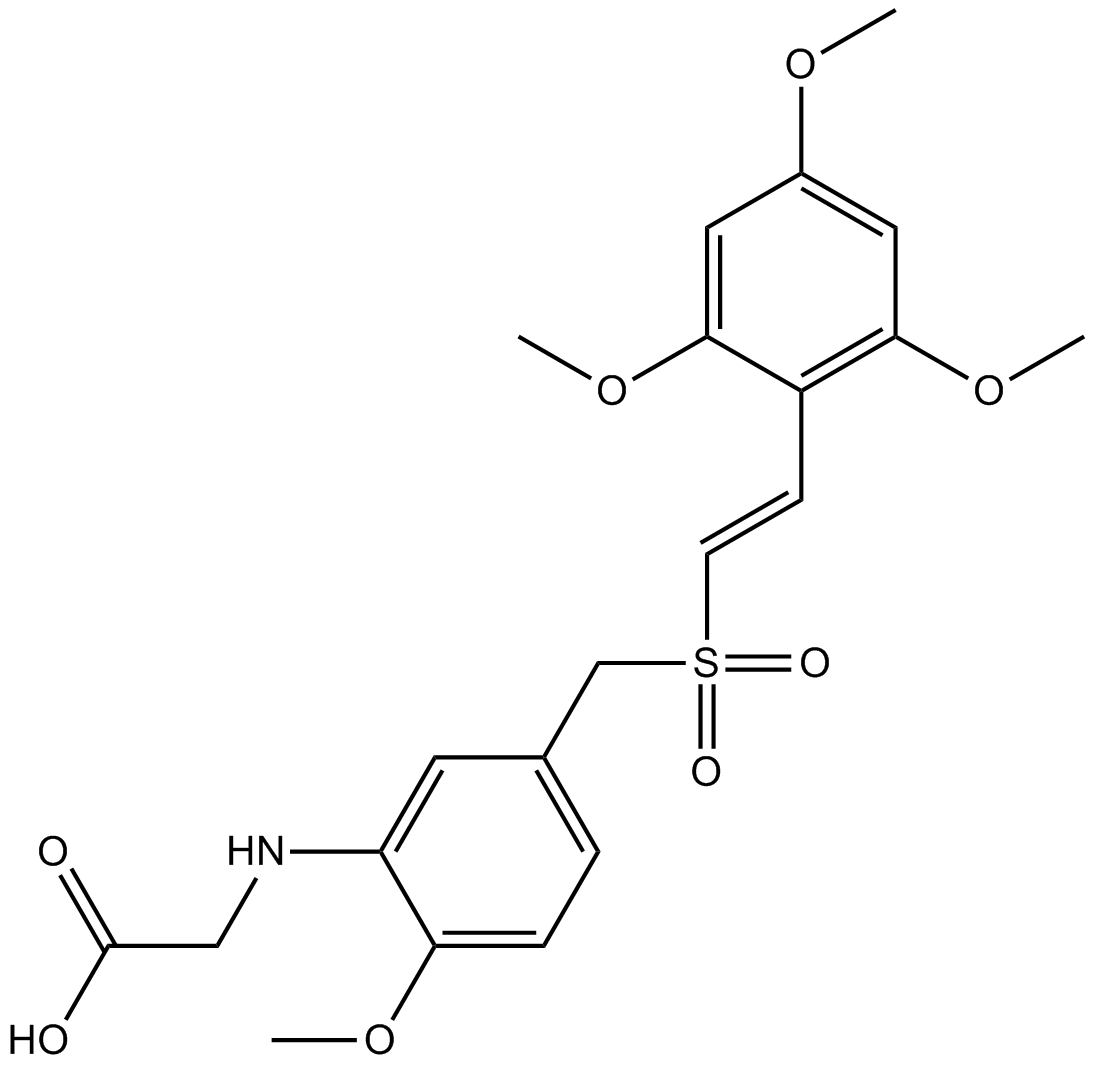
GC14358
Rigosertib (ON-01910,Estybon)
Plk1 inhibitor
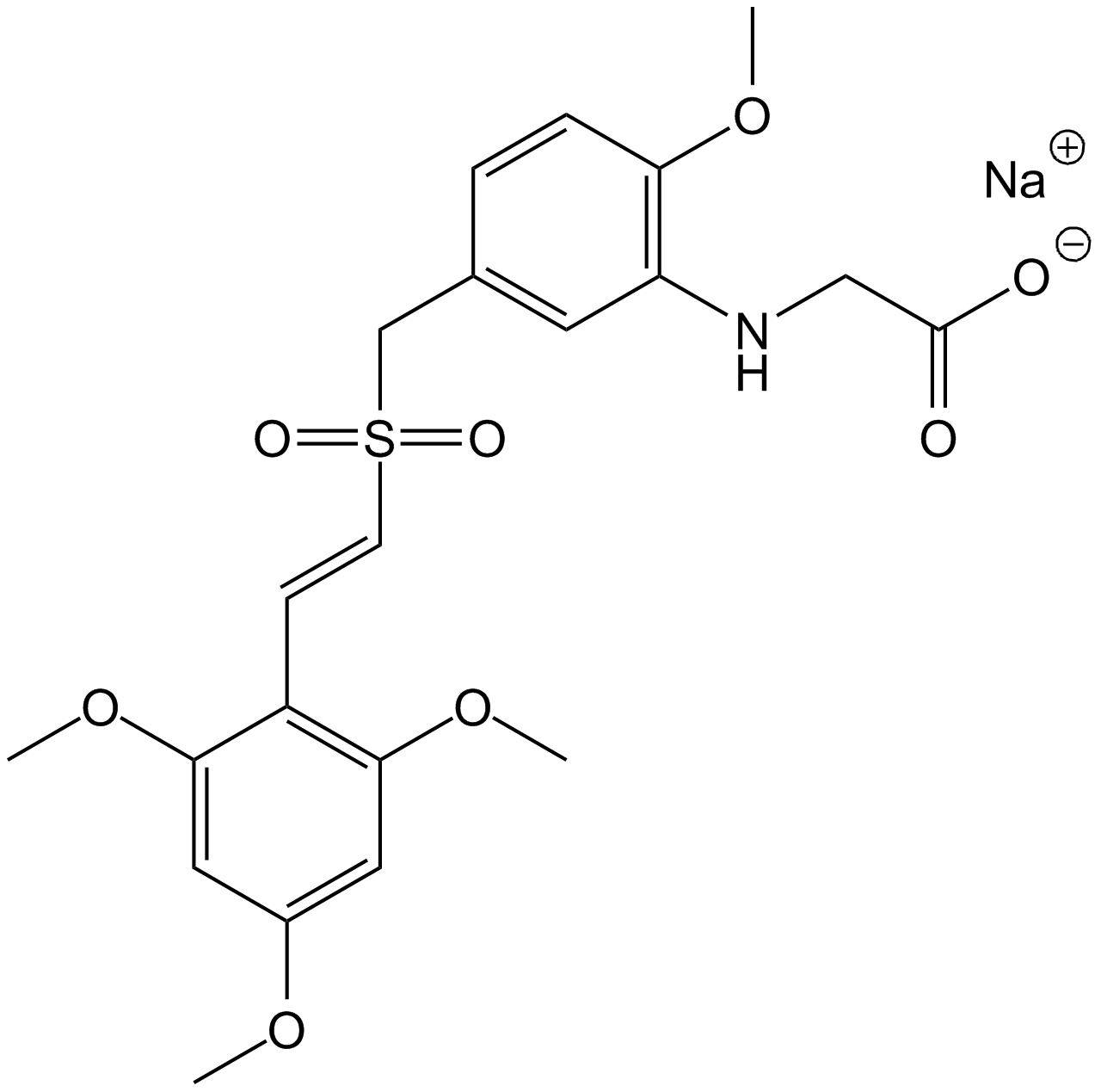
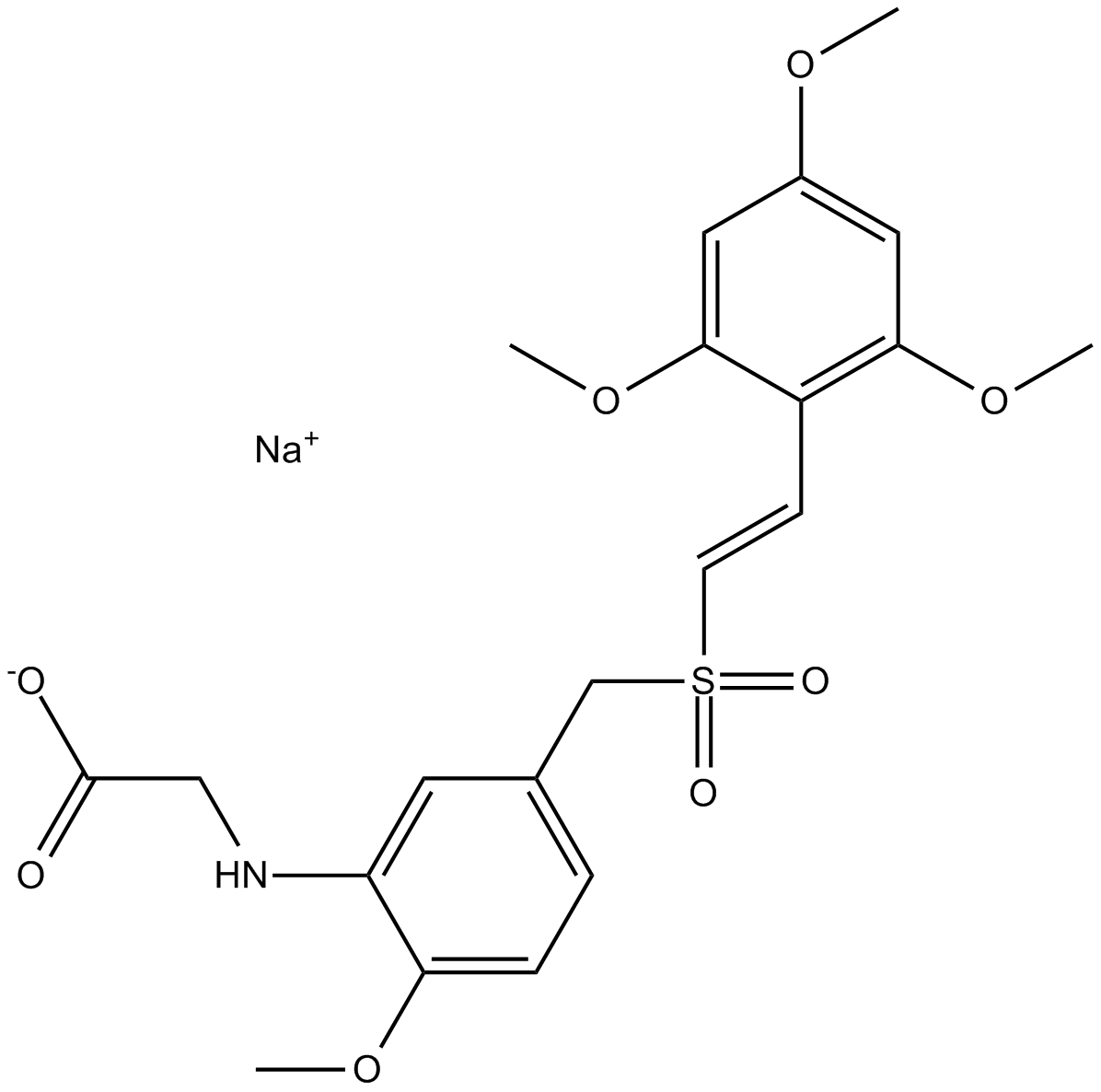
-
GC13580
Rigosertib sodium
Non-ATP-competitive inhibitor of PLK1
-
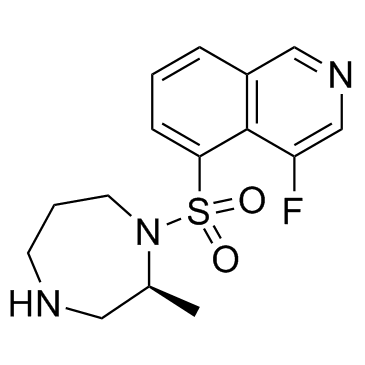
GC37534
Ripasudil free base
Ripasudil free base (K-115 free base) is a specific inhibitor of ROCK, with IC50s of 19 and 51 nM for ROCK2 and ROCK1, respectively.
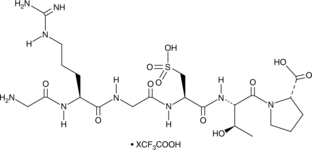
-
GC48401
Risuteganib (trifluoroacetate salt)
An anti-integrin peptide
-
GC44846
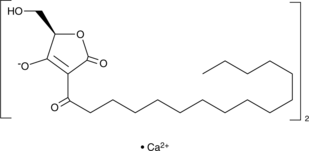
RK-682 (calcium salt)
Protein tyrosine phosphatases (PTPs) remove phosphate from tyrosine residues of cellular proteins.
-
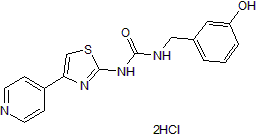
GC50092
RKI 1447 dihydrochloride
Potent and selective ROCK inhibitor; antitumor
-
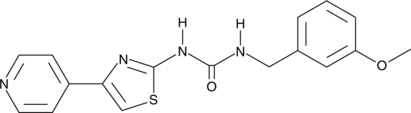
GC44847
RKI-1313
A ROCK1 and ROCK2 inhibitor
-
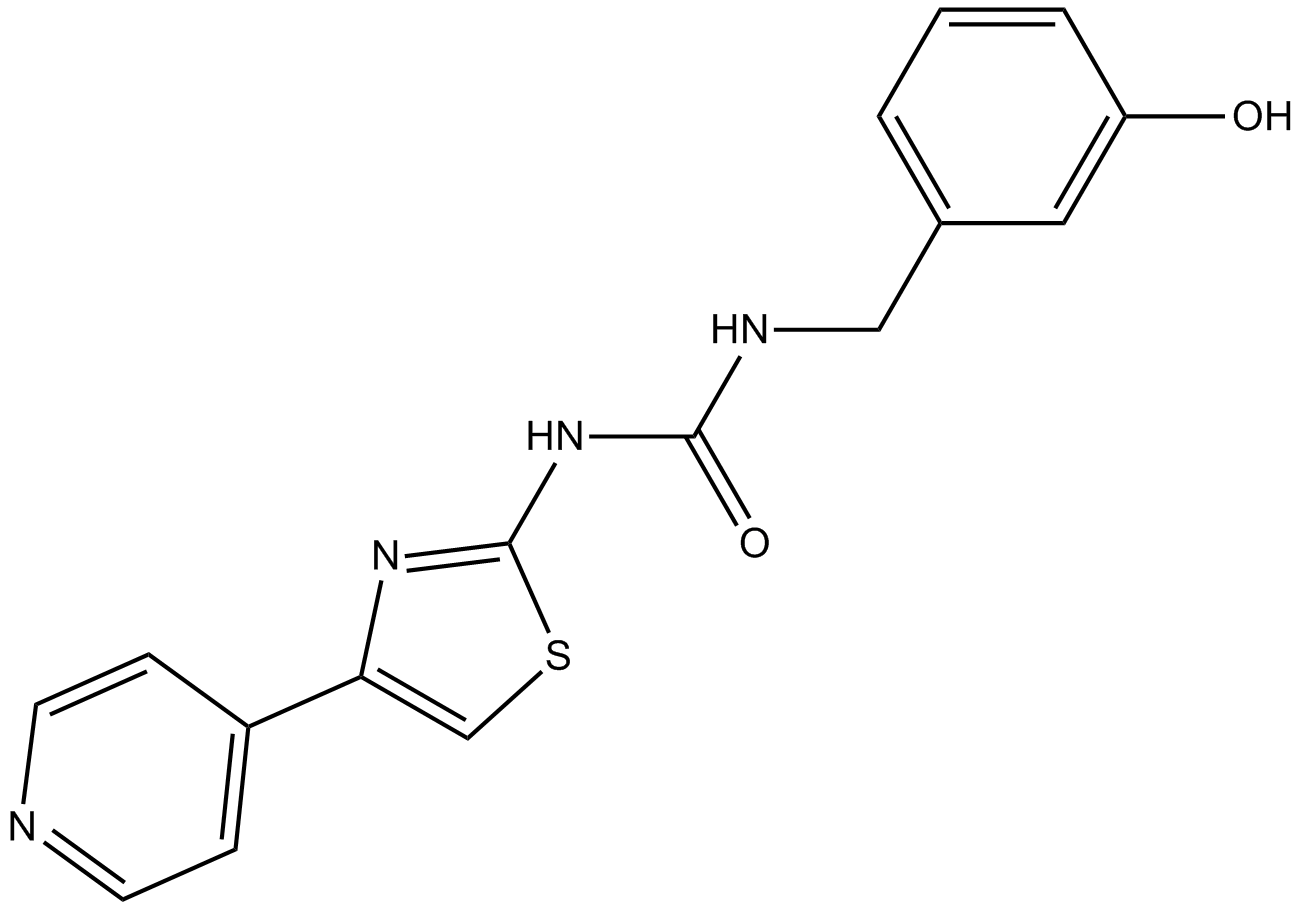
GC15437
RKI-1447
A ROCK1 and ROCK2 inhibitor
-
GC12348
Ro 3306
An ATP-competitive, potent CDK1 inhibitor
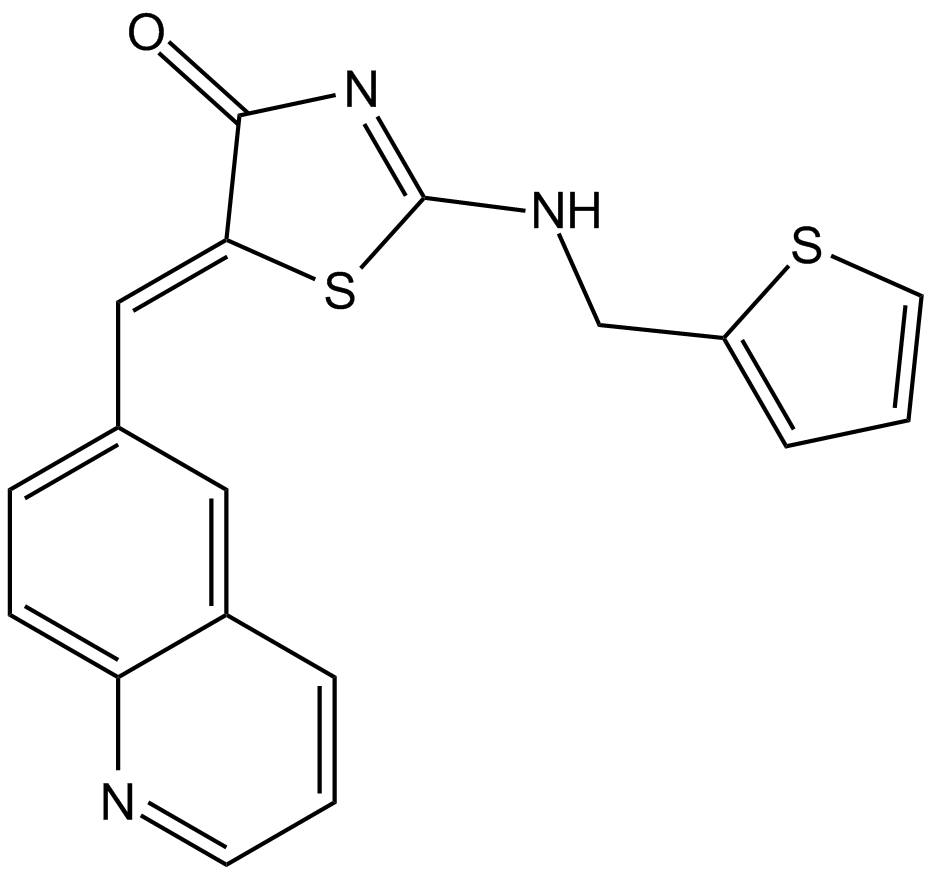
-
GC13903
Ro3280
PLK1 inhibitor,potent and highly selective
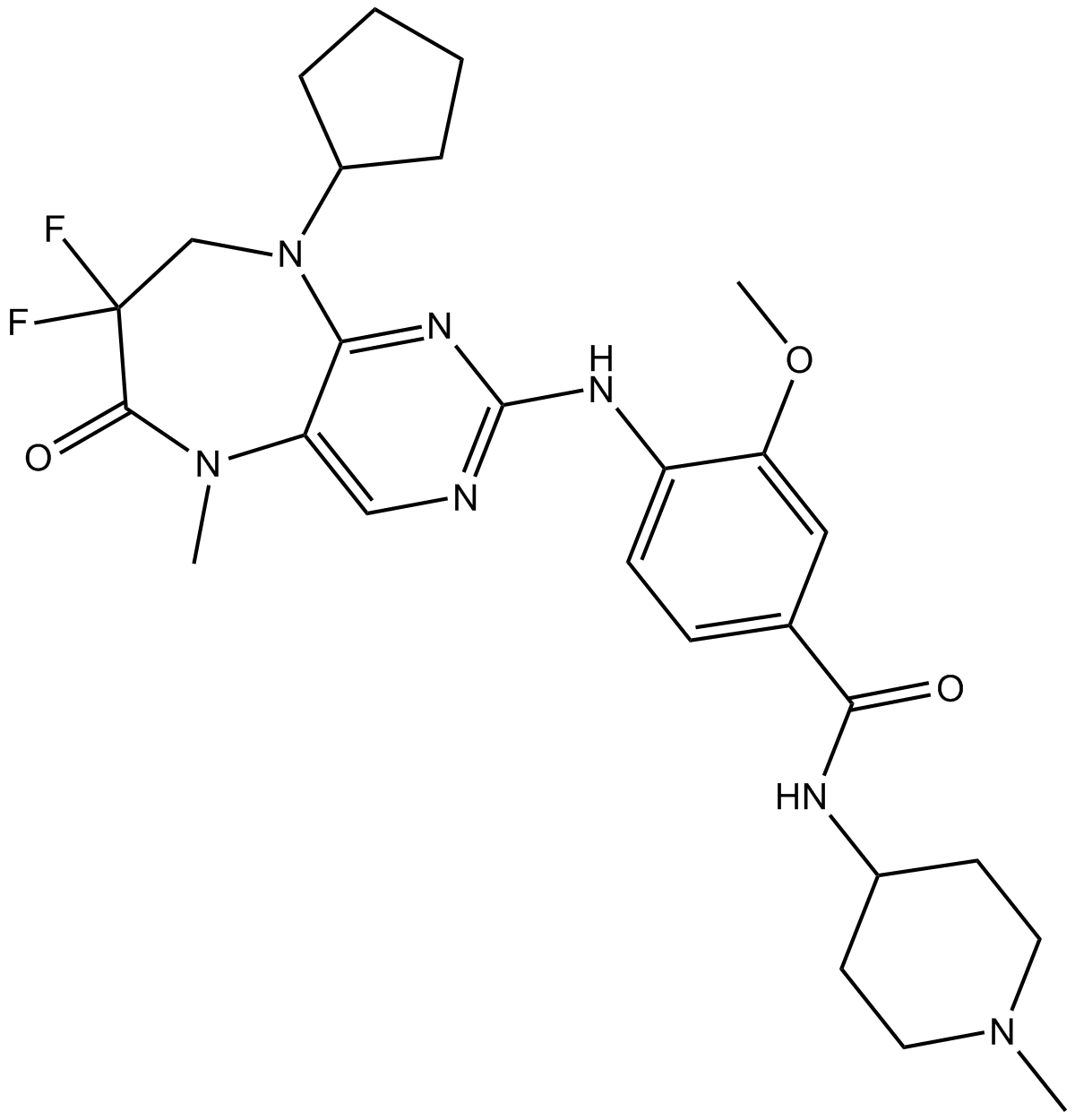
-
GC37551
ROCK inhibitor-2
ROCK inhibitor-2 is a selective dual ROCK1 and ROCK2 inhibitor with IC50s of 17 nM and 2 nM, respectively.
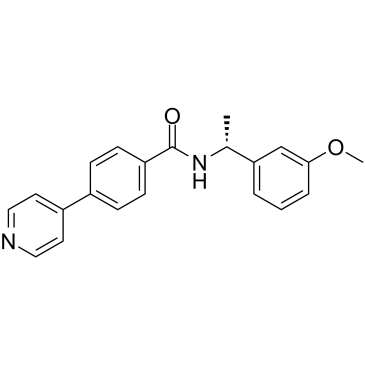
-
GC31952
ROCK-IN-1
ROCK-IN-1 is a potent inhibitor of ROCK, with an IC50 of 1.2 nM for ROCK2.
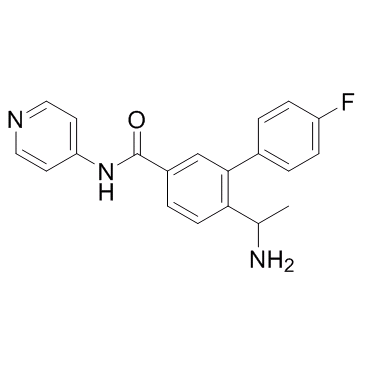
-
GC66338
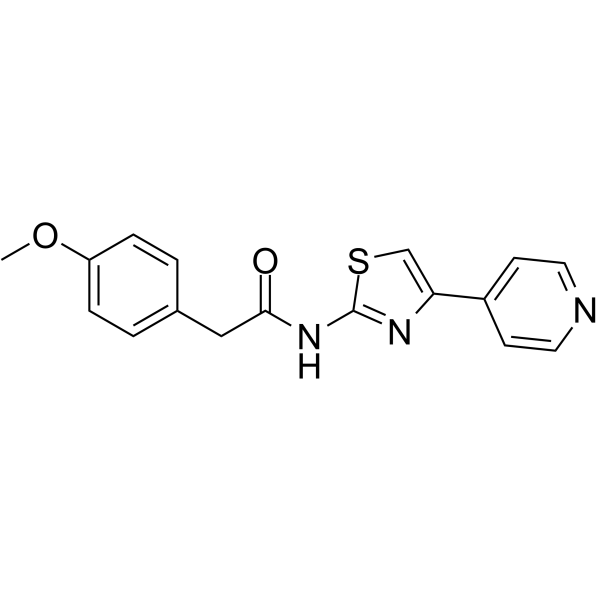
ROCK1-IN-1
ROCK1-IN-1 is a ROCK1 inhibitor with a Ki value of 540 nM. ROCK1-IN-1 can be used for the research of hypertension, glaucoma and erectile dysfunction.
-
GC31883
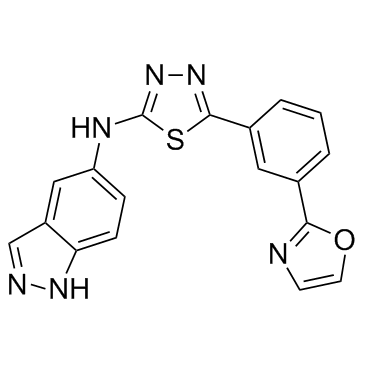
ROCK2-IN-2
ROCK2-IN-2 is a selective ROCK2 inhibitor extracted from patent US20180093978A1, Compound A-30, has an IC50 of <1 μM.
-
GC33169
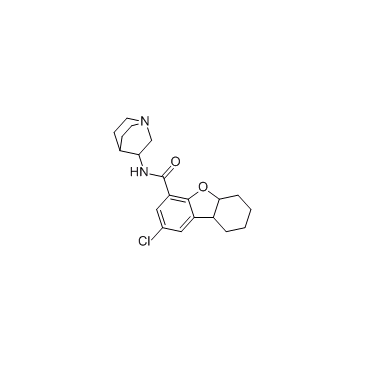
Rosabulin (STA 5312)
Rosabulin (STA 5312) (STA 5312) is a potent and orally active microtubule inhibitor that inhibits microtubule assembly. Rosabulin (STA 5312) has broad-spectrum anti-tumor activity.
-
GC11401
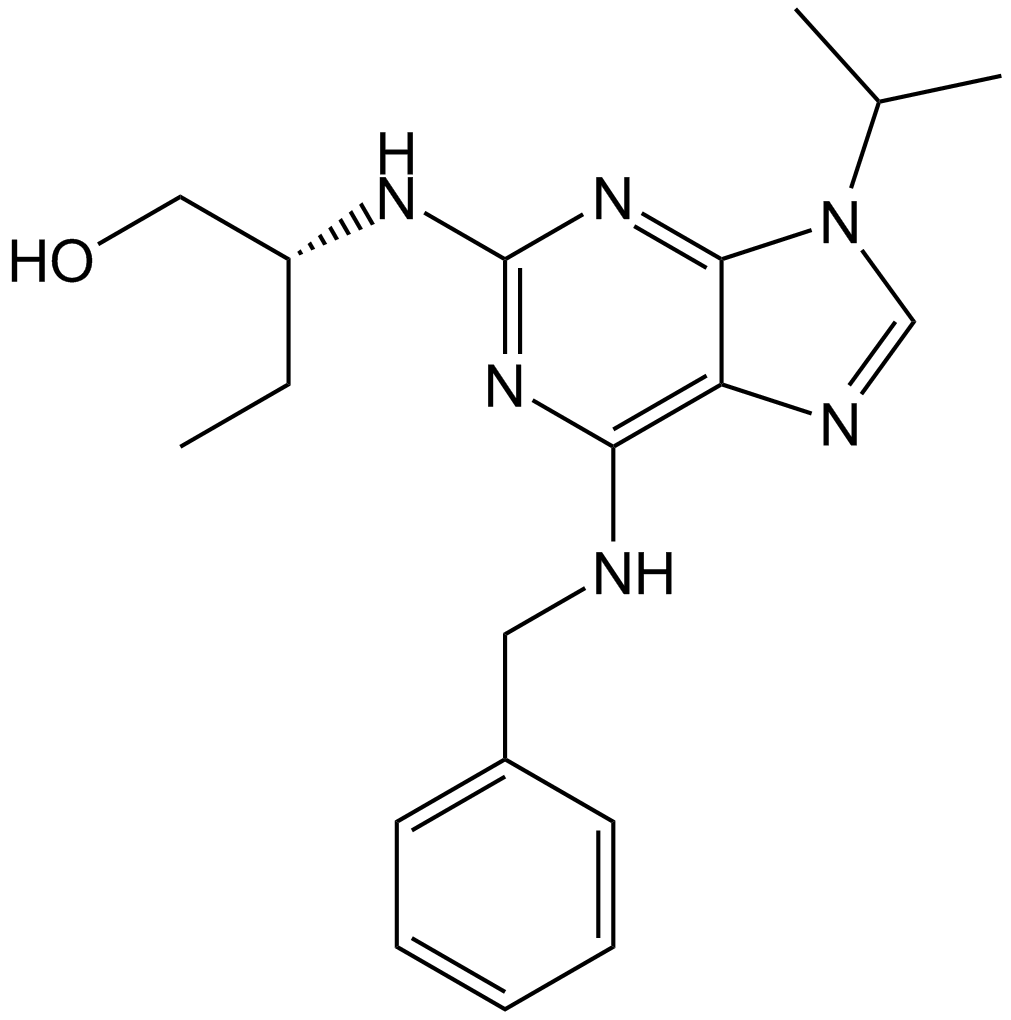
Roscovitine (Seliciclib,CYC202)
Roscovitine (Seliciclib,CYC202) (Roscovitine) is an orally bioavailable and selective CDKs inhibitor with IC50s of 0.2 μM, 0.65 μM, and 0.7 μM for CDK5, Cdc2, and CDK2, respectively.
-
GC64278
RP-3500
RP-3500 (ATR inhibitor 4) is an orally active, selective ATR kinase inhibitor (ATRi) with an IC50 of 1.00 nM in biochemical assays. RP-3500 shows 30-fold selectivity for ATR over mTOR (IC50=120 nM) and >2,000-fold selectivity over ATM, DNA-PK, and PI3Kα kinases. RP-3500 has potent antitumor activity.
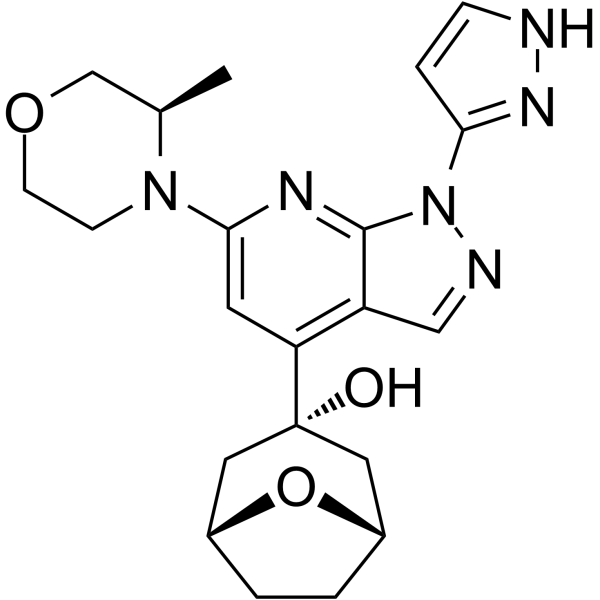
-
GC16265
RS-1
RAD51 activator
-
GC18847
RSM-932A
RSM-932A is a ChoKα inhibitor
-
GC17150
Ryuvidine
SETD8 protein lysine methyltransferase (PKMT) inhibitor
-
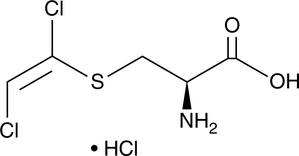
GC49532
S-(1,2-Dichlorovinyl)-Cysteine (hydrochloride)
A nephrotoxin and metabolite of trichloroethylene
-
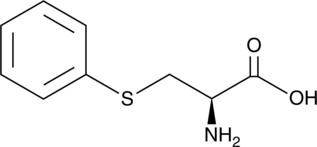
GC46224
S-Phenylcysteine
An adduct
-
GC18852
S14161
D-Cyclins regulate the cell cycle by acting in a complex with cyclin dependent kinases (CDKs) to promote phosphorylation of the retinoblastoma protein and initiate cellular progression from the G1 to the S phase.
-
GC38945
S516
S516 (Compound 22) is an active metabolite of CKD-516 and a potent tubulin polymerization inhibitor with an IC50 of 4.29 μM. S516 has marked antitumor activity.
-
GC49093
Safflower Red
A red pigment with diverse biological activities
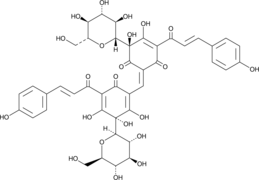
-
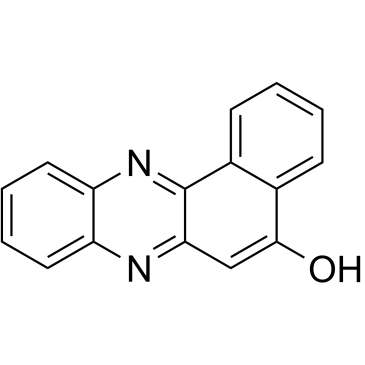
GC38845
sAJM589
sAJM589 is a Myc inhibitor which potently disrupts the Myc-Max heterodimer with an IC50 of 1.8 μM.
-
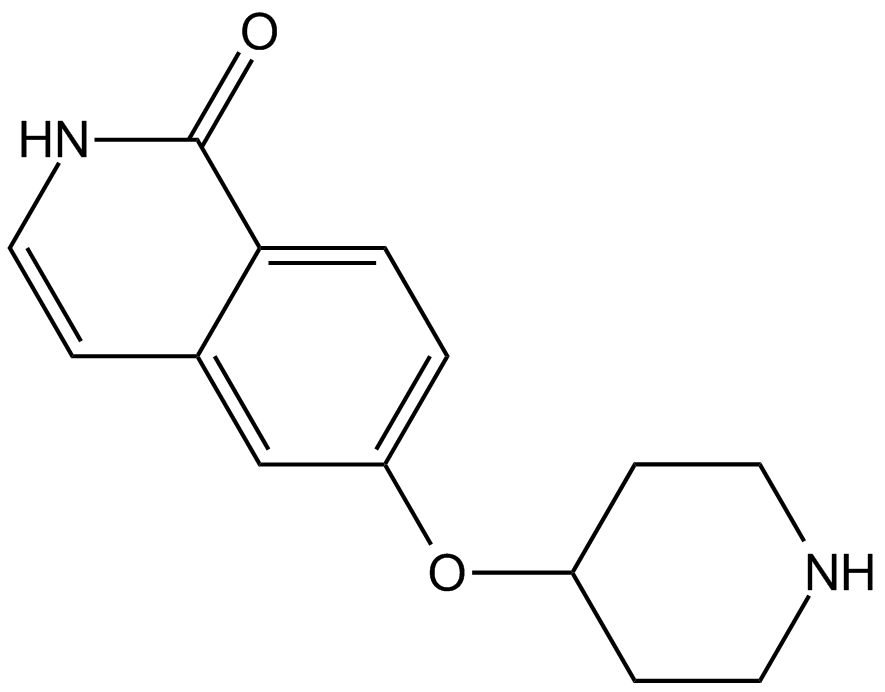
GC13759
SAR407899
ATP-competitive ROCK inhibitor
-
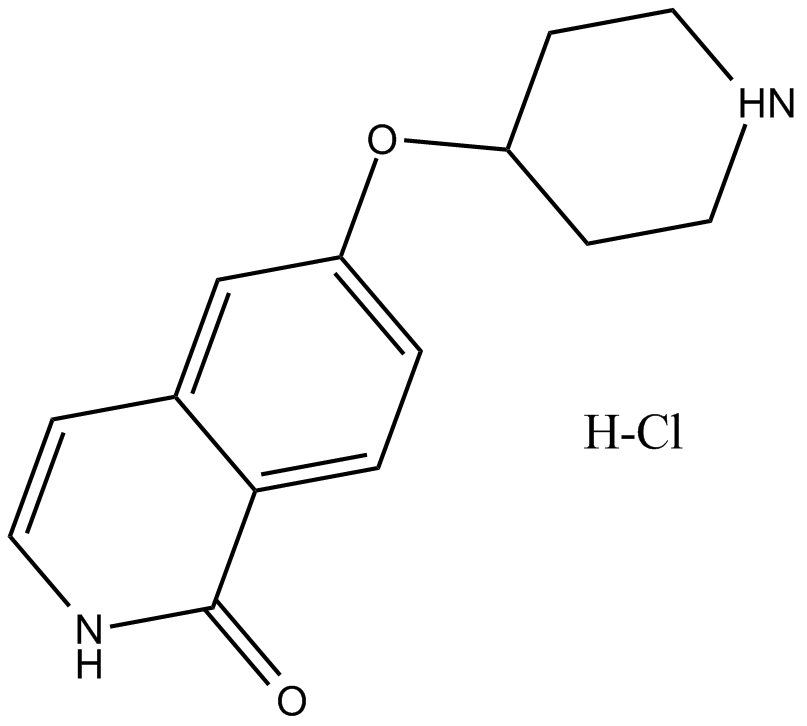
GC16289
SAR407899 hydrochloride
ATP-competitive ROCK inhibitor
-
GC46218
Satratoxin G
A macrocyclic trichothecene mycotoxin
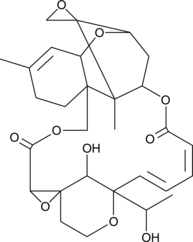
-
GC46219
Satratoxin H
A trichothecene mycotoxin
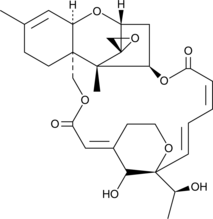
-
GC11022
SB 218078
checkpoint kinase 1 (Chk1) inhibitor
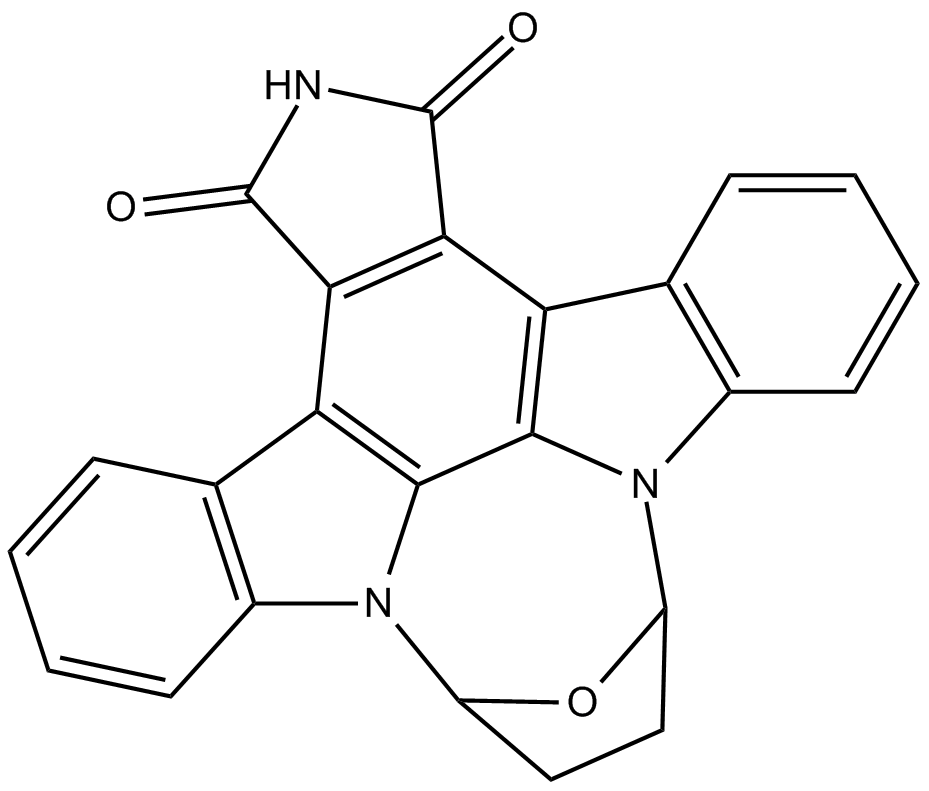
-
GC15170
SB 772077B dihydrochloride
Rho-kinase (ROCK) inhibitor
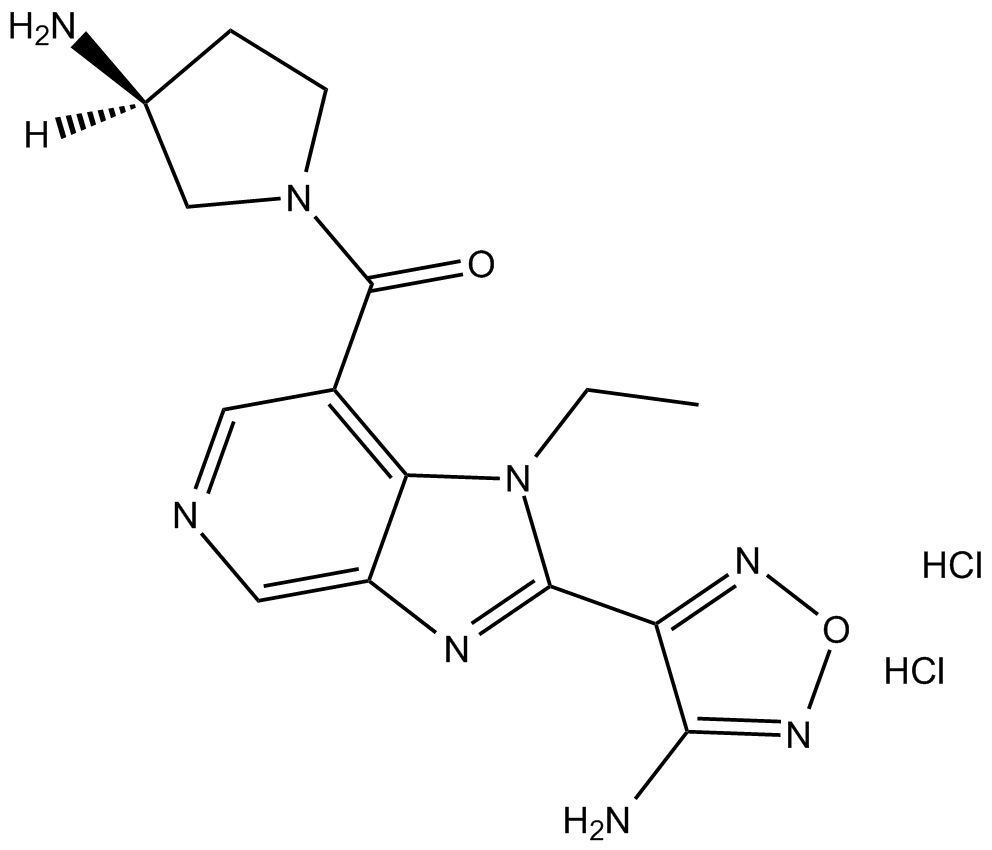
-
GC66437
SB-216
SB-216 is a potent tubulin polymerization inhibitor. SB-216 shows strong antiproliferative potency in a panel of human cancer cell lines, including melanoma, lung cancer, and breast cancer. SB-216 can be used for cancer research.
-
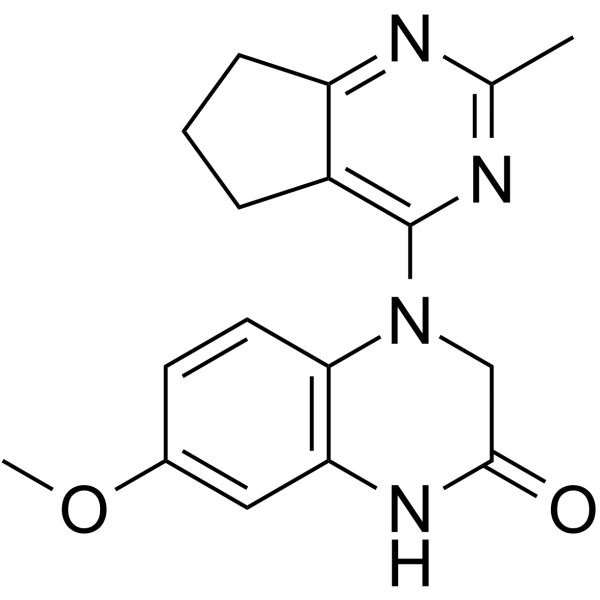
GC12064
SB1317
A multi-kinase inhibitor
-
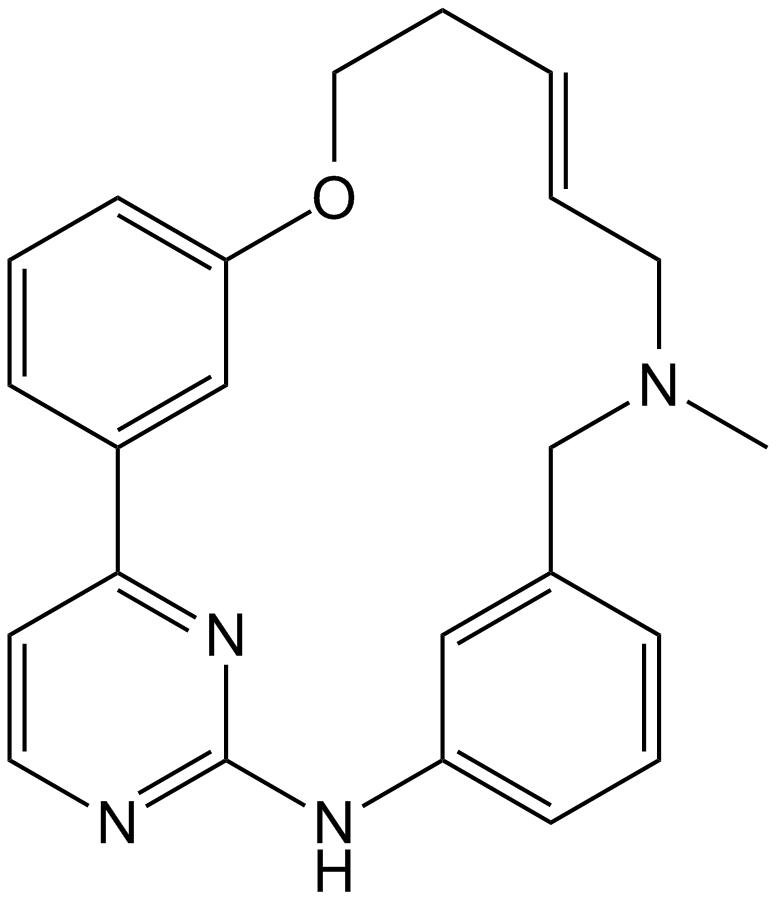
GC14600
SB743921
SB743921 is a potent inhibitor of the mitotic kinesin KSP (Eg5), with a Ki of 0.1 nM.
-
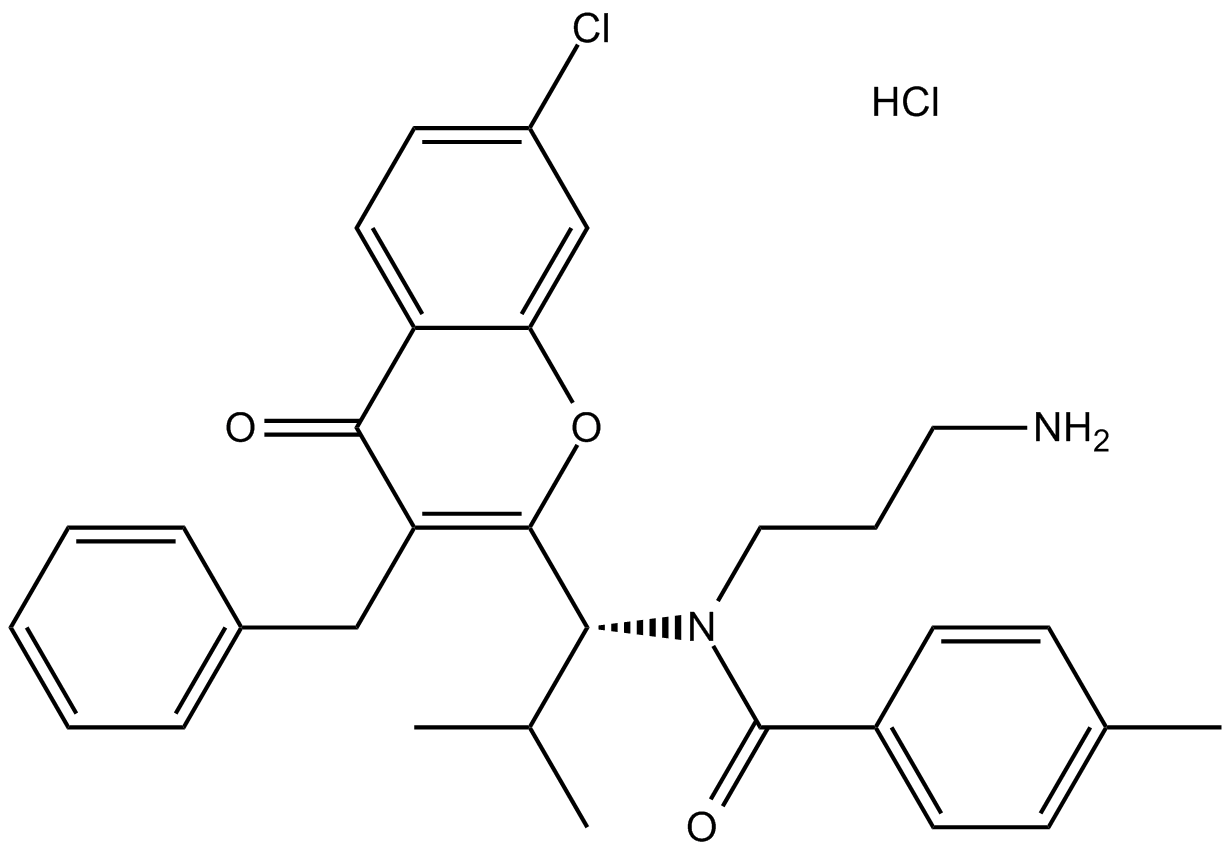
GC14644
SBE 13 HCl
SBE 13 HCl is a potent and selective Plk1 inhibitor, with an IC50 of 200 pM; SBE 13 HCl poorly inhibits Plk2 (IC50>66μM) or Plk3 (IC50=875nM).
-
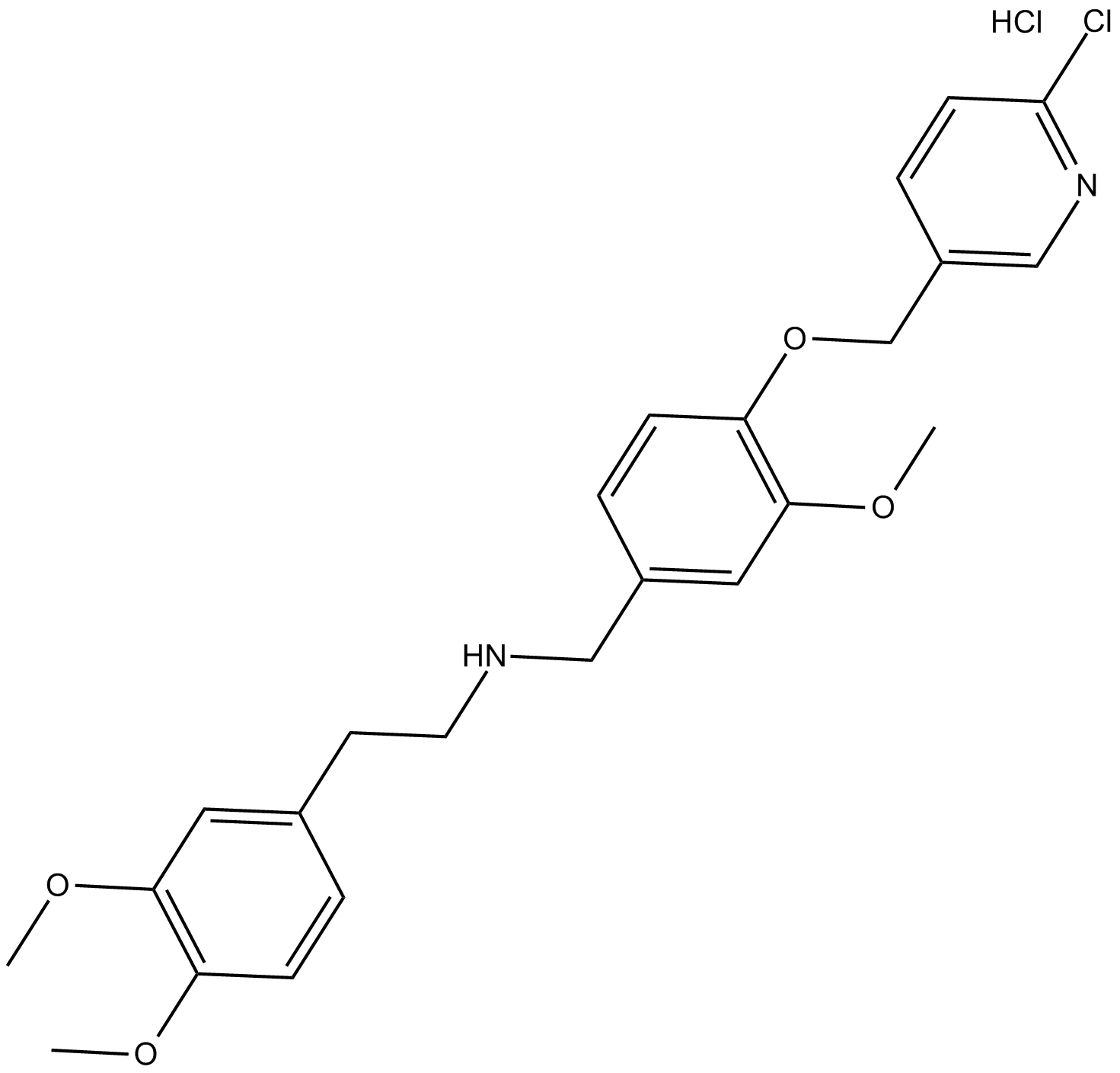
GC37601
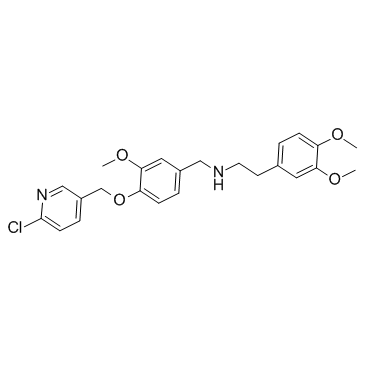
SBE13
SBE13 is a potent and selective Plk1 inhibitor, with an IC50 of 200 pM; SBE13 poorly inhibits Plk2 (IC50>66?μM) or Plk3 (IC50=875?nM).
-
GC37606
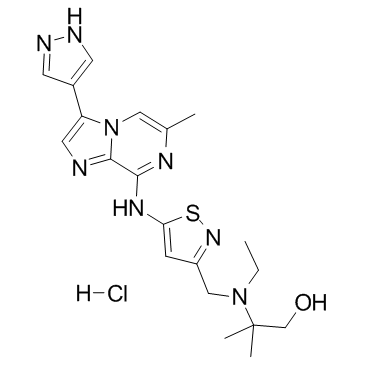
SCH-1473759 hydrochloride
SCH-1473759 hydrochloride is an aurora inhibitor with IC50s of 4 and 13 nM for aurora A and B, respectively.
-
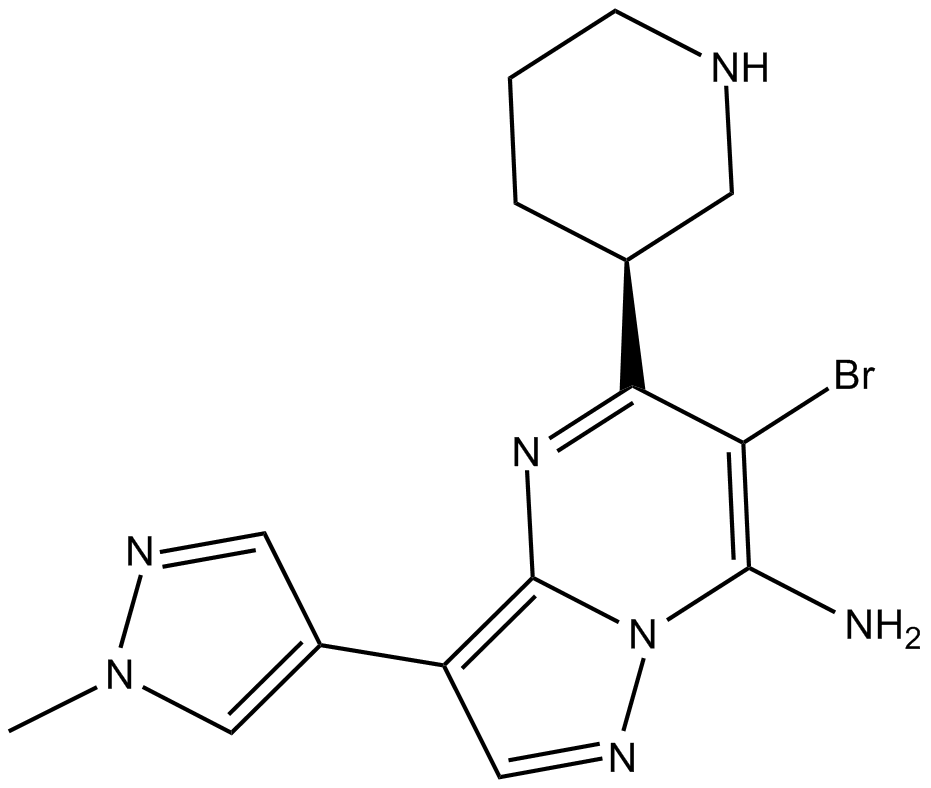
GC17792
SCH900776 S-isomer
-
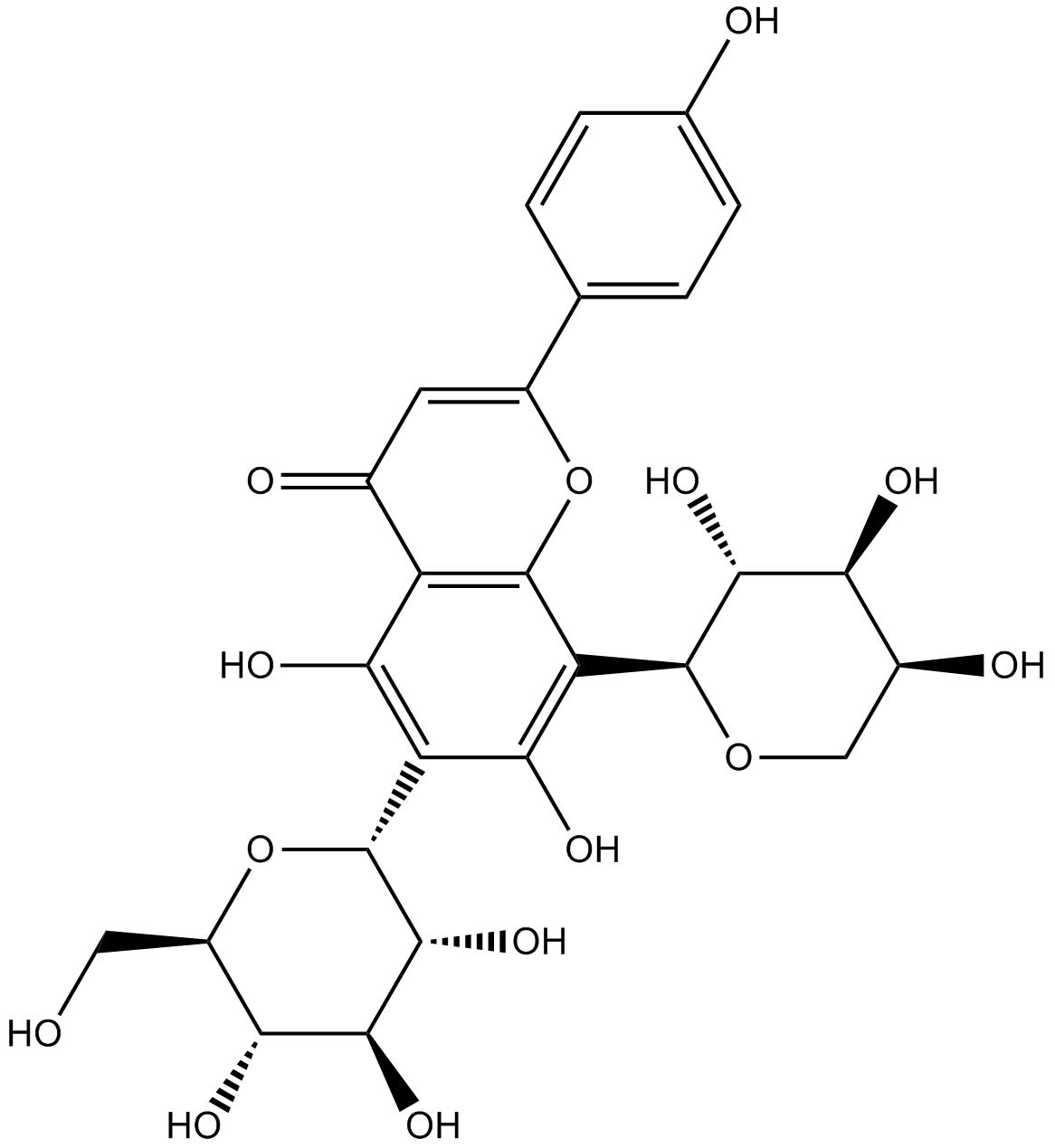
GN10091
Schaftoside
-
GC49674
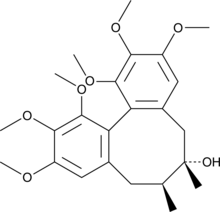
Schizandrin
Schizandrin (Schizandrin), a dibenzocyclooctadiene lignan, is isolated from the fruit of Schisandra chinensis Baill.
-
GC39798
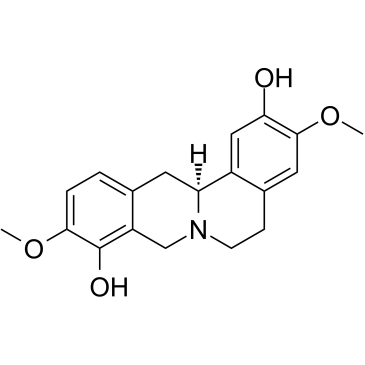
Scoulerine
Scoulerine ((-)-Scoulerine), an isoquinoline alkaloid, is a potent antimitotic compound. Scoulerine is also an inhibitor of BACE1 (?-site amyloid precursor protein cleaving enzyme 1). Scoulerine inhibits proliferation, arrests cell cycle, and induces apoptosis in cancer cells.
-
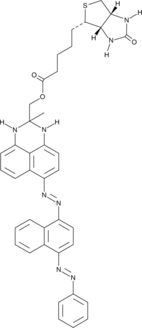
GC49723
SenTraGor™ Cell Senescence Reagent
A cellular senescence detection reagent
-
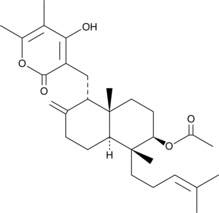
GC48076
Sesquicillin A
A fungal metabolite
-
GC10019
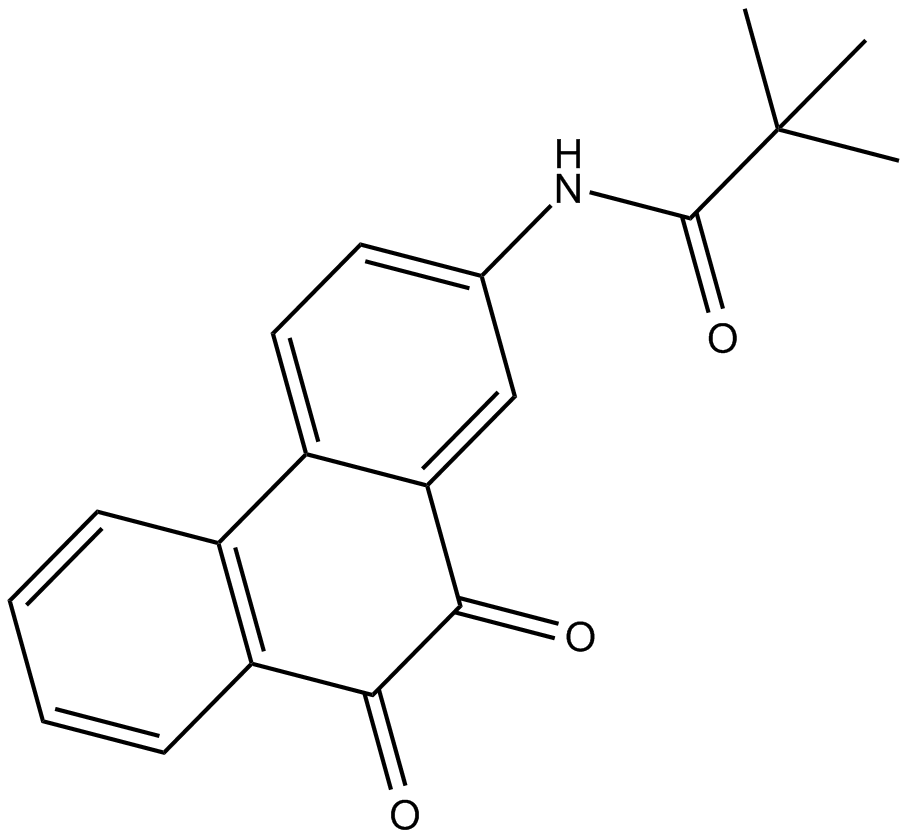
SF1670
PTEN inhibitor, potent and specific
-
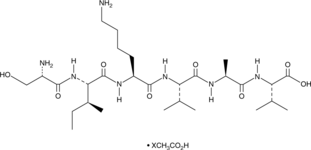
GC49713
SIKVAV (acetate)
A laminin α1-derived peptide
-
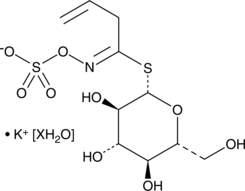
GC49002
Sinigrin (hydrate)
A glucosinolate with diverse biological activities
-
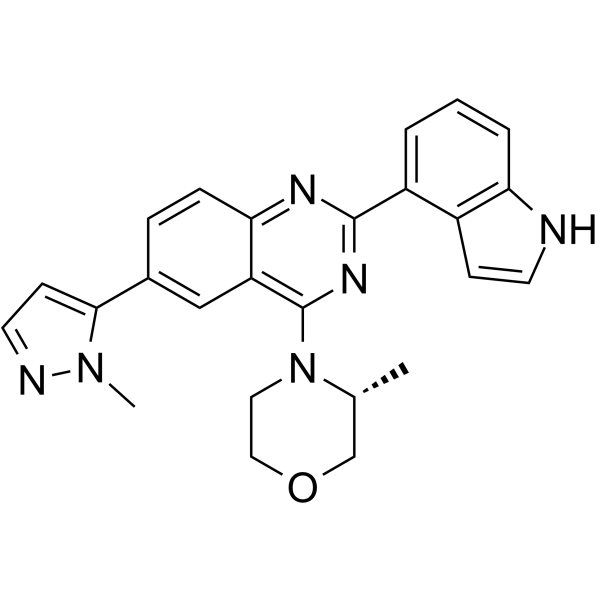
GC64539
SKLB-197
SKLB-197 showed an IC50 value of 0.013 μM against ATR but very weak or no activity against other 402 protein kinases. It displayed potent antitumor activity against ATM-deficent tumors both in vitro and in vivo.
-
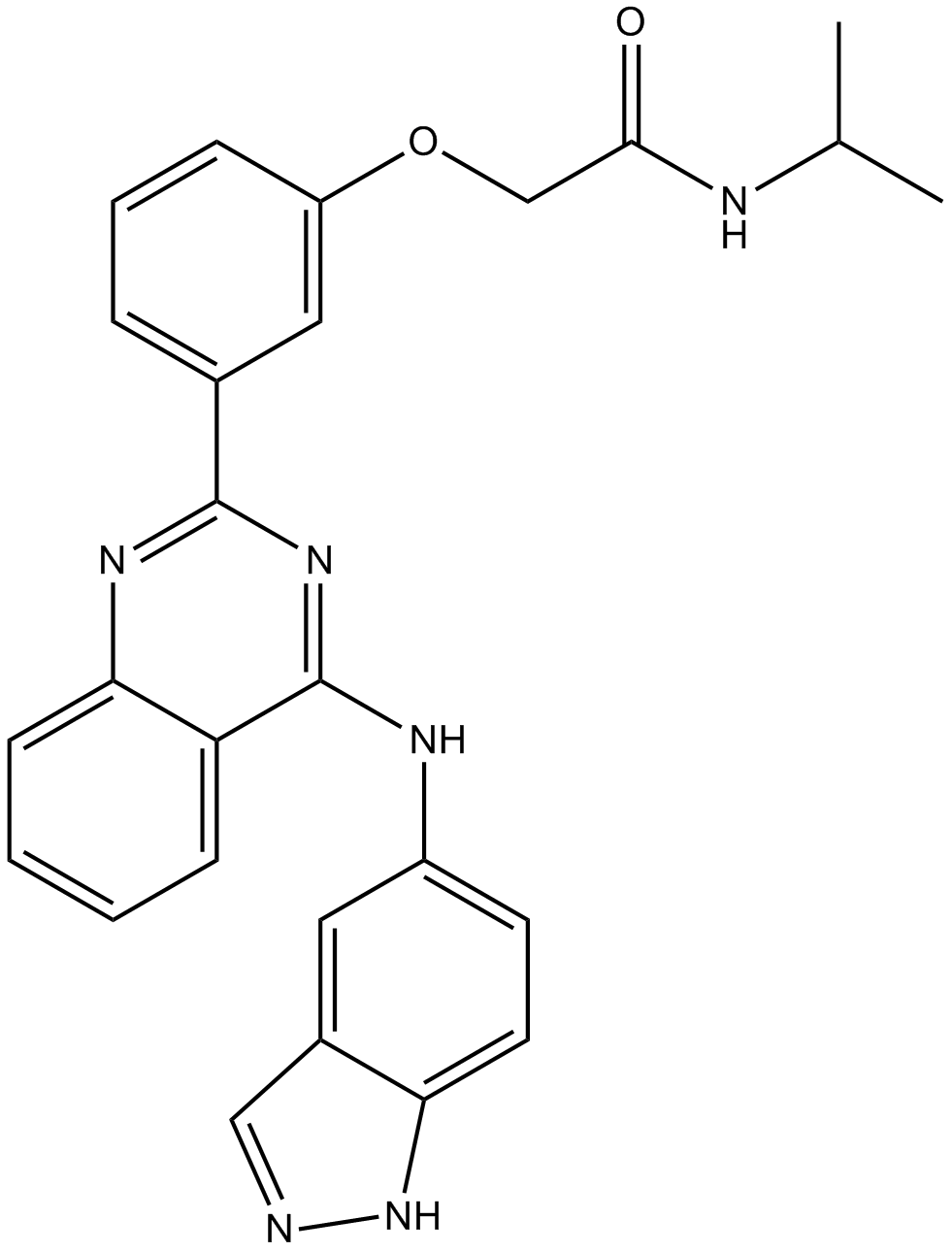
GC12827
SLx-2119
A ROCK2 inhibitor
-
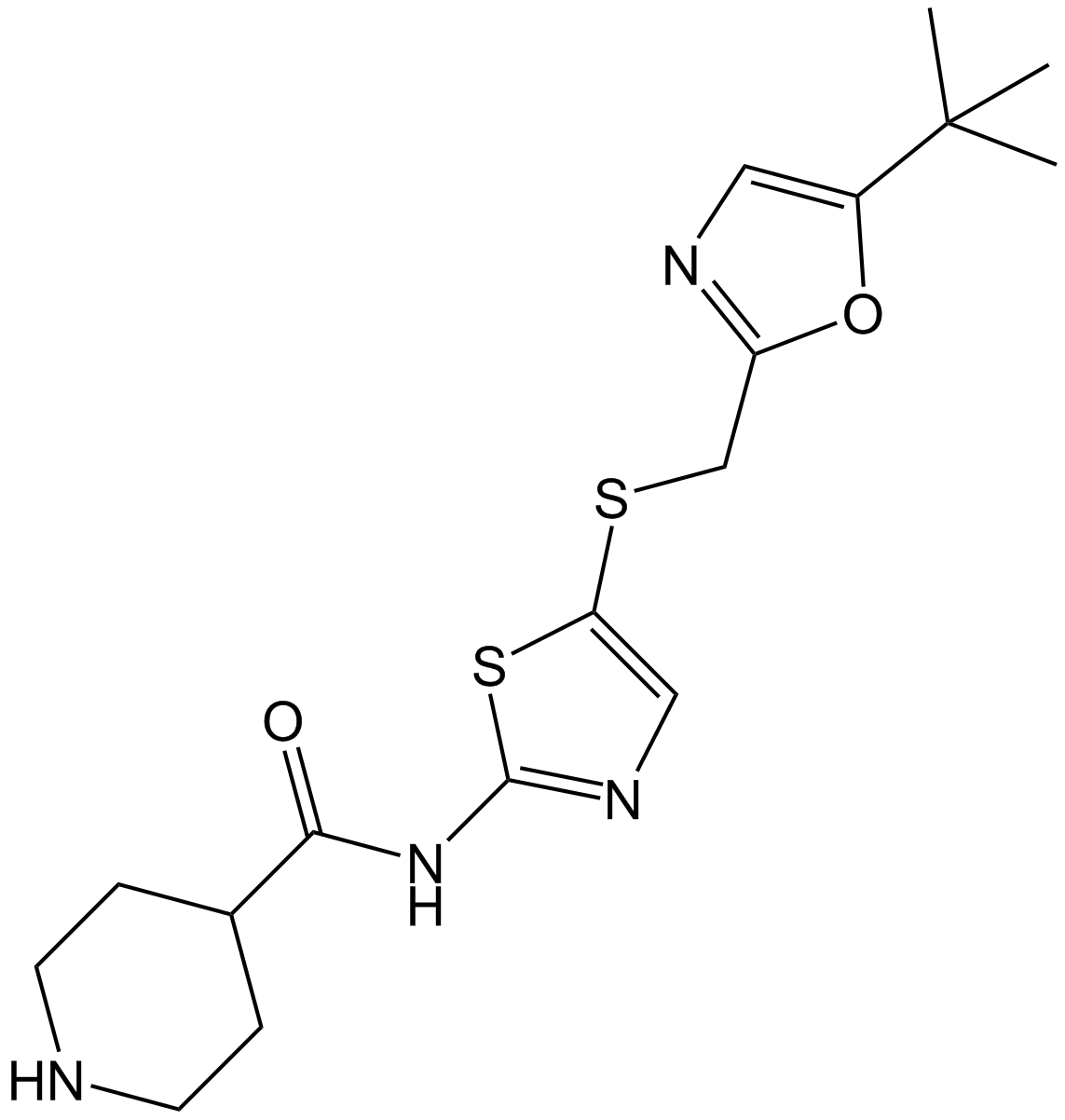
GC11396
SNS-032 (BMS-387032)
SNS-032 (BMS-387032) (BMS-387032) is a potent and selective inhibitor ofCDK2, CDK7, and CDK9 withIC50sof 38 nM, 62 nM and 4 nM, respectively. SNS-032 (BMS-387032) has antitumor effect.
-
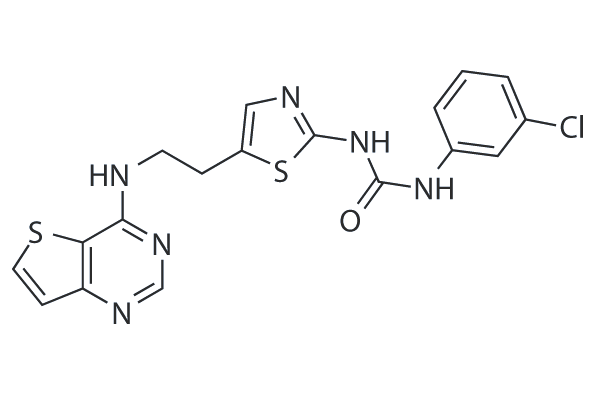
GC25940
SNS-314
SNS-314 is a potent and selective inhibitor of Aurora A, Aurora B and Aurora C with IC50 of 9 nM, 31 nM, and 3 nM, respectively. It is less potent to Trk A/B, Flt4, Fms, Axl, c-Raf and DDR2. Phase 1.
-
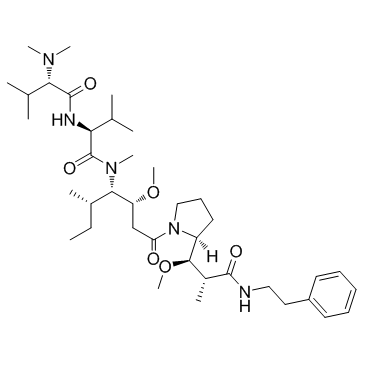
GC32935
Soblidotin (Auristatin PE)
Soblidotin (Auristatin PE) (Auristatin PE) is a novel synthetic Dolastatin 10 derivative and inhibitor of tubulin polymerization.
-
GC48084
Sodium 4-Phenylbutyrate-d11
Phenylbutyrate-d11 (sodium) is deuterium labeled Sodium 4-phenylbutyrate. Sodium 4-phenylbutyrate (4-PBA sodium) is an inhibitor of HDAC and endoplasmic reticulum (ER) stress, used in cancer and infection research.
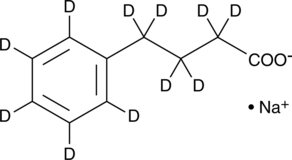
-
GC65887
SOP1812
SOP1812 is a naphthalene diimide (ND) derivative with anti-tumor activity. SOP1812 binds to quadruplex arrangements (G4s), and down-regulates several cancer gene pathways. SOP1812 shows great affinity to hTERT G4 and HuTel21 G4 with KD values of 4.9 and 28.4 nM, respectively. SOP1812 can be used for the research of cancer.
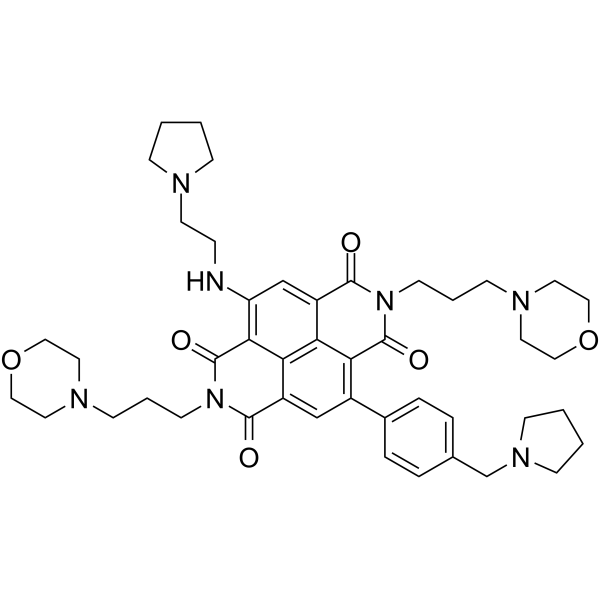
-
GC63351
Sovesudil
Sovesudil (PHP-201) is a potent, ATP-competitive, locally acting Rho kinase (ROCK) inhibitor with IC50s of 3.7 and 2.3 nM for ROCK-I and ROCK-II, respectively.
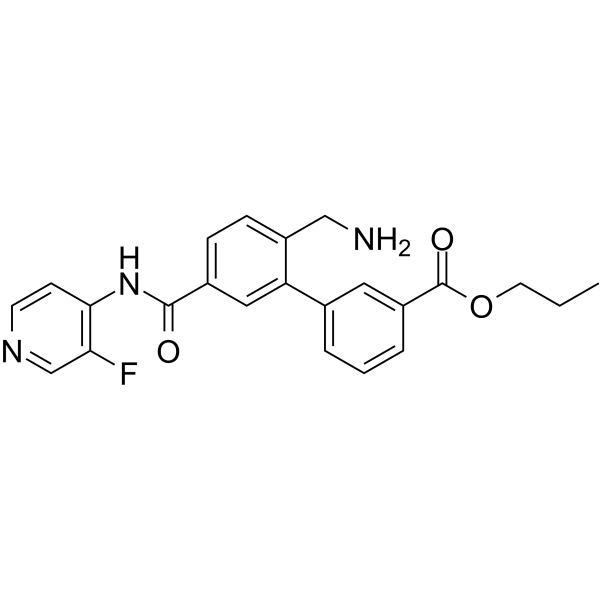
-
GC63716
Sovesudil hydrochloride
Sovesudil (PHP-201) hydrochloride is a potent, ATP-competitive, locally acting Rho kinase (ROCK) inhibitor with IC50s of 3.7 and 2.3 nM for ROCK-I and ROCK-II, respectively.
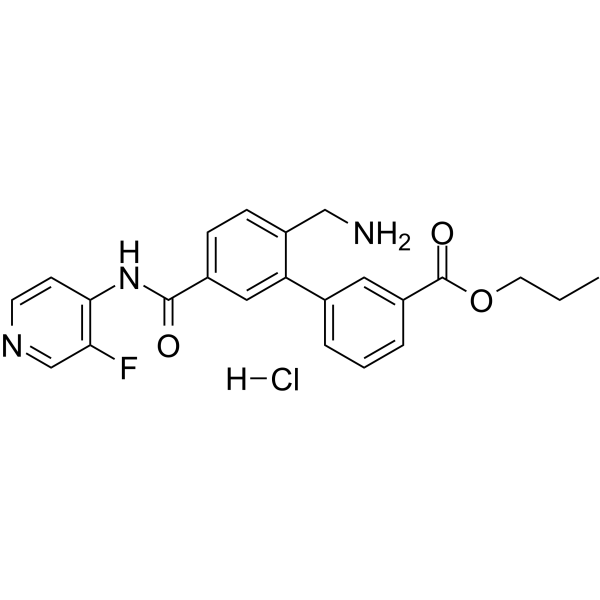
-
GC64019
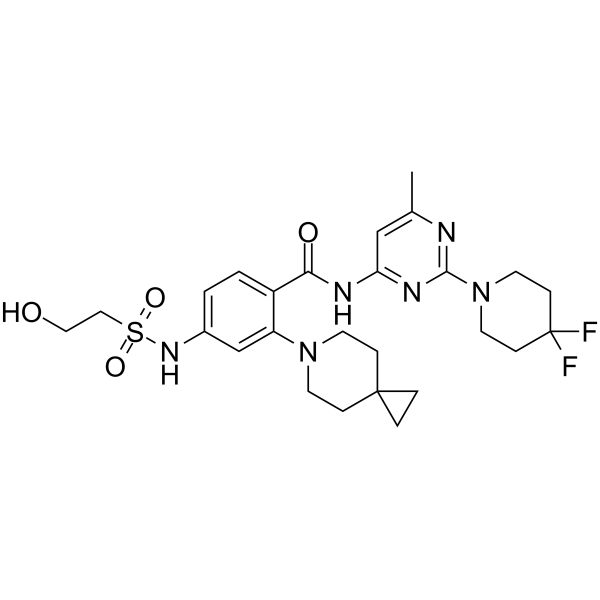
Sovilnesib
Sovilnesib (AMG 650) is a kinesin-like protein KIF18A inhibitor (WO2020132648). Sovilnesib can be used for the research of cancer.
-
GC48092
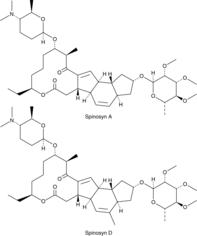
Spinosad
A naturally-occurring insecticide
-
GC41258
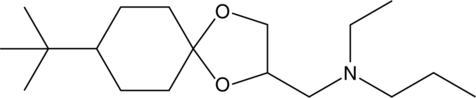
Spiroxamine
Spiroxamine is a tertiary amine fungicide and an inhibitor of δ14 reductase/δ8→δ7 isomerase.
-
GC44946
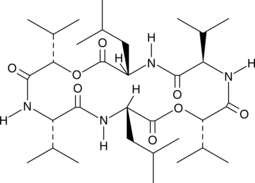
Sporidesmolide III
Sporidesmolide III is a cyclodepsipeptide originally isolated from P.