Chromatin/Epigenetics
Epigenetics
Epigenetics means above genetics. It determines how much and whether a gene is expressed without changing DNA sequences. Epigenetic regulations include, 1. DNA methylation: the addition of methyl group to DNA, converting cytosine to 5-methylcytosine, mostly at CpG sites; 2. Histone modifications: posttranslational modificationEpigeneticss of histone proteins including acetylation, methylation, ubiquitylation, phosphorylation and sumoylation; 3. miRNAs: non-coding microRNA downregulating gene expression; 4. Prions: infectious proteins viewed as epigenetic agents capable of inducing a phenotype without changing the genome.
Targets for Chromatin/Epigenetics
- Bromodomain(52)
- Aurora Kinase(89)
- DNA Methyltransferase(40)
- HDAC(218)
- Histone Acetyltransferases(67)
- Histone Demethylases(98)
- Histone Methyltransferase(212)
- HIF(101)
- JAK(178)
- MBT Domains(1)
- PARP(128)
- Pim(35)
- Protein Ser/Thr Phosphatases(41)
- RNA Polymerase(8)
- Sirtuin(84)
- Sphingosine Kinase-2(1)
- Polycomb repressive complex(2)
- SUMOylation(3)
- PAD(18)
- Epigenetic Reader Domain(207)
- MicroRNA(13)
- Protein Arginine Deiminase(12)
- Chromodomain(1)
- Citrullination(15)
- DNA/RNA Demethylation(1)
- DNA/RNA Methylation(6)
- Histone Deacetylation(38)
- Histones/Histone Peptides(7)
- PHD Domains(0)
- Tandem Tudor & Tudor-like Domains(1)
- PRMT(2)
Products for Chromatin/Epigenetics
- Cat.No. Product Name Information
-
GC65326
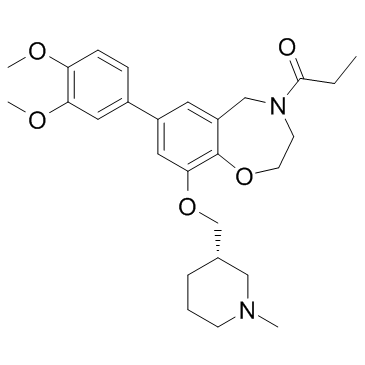
GNE-375
GNE-375 is a potent and highly selective BRD9 inhibitor with an IC50 of 5 nM. GNE-375 shows >100-fold selective for BRD9 over BRD4, TAF1, and CECR2. GNE-375 decreases BRD9 binding to chromatin.
-
GC32081
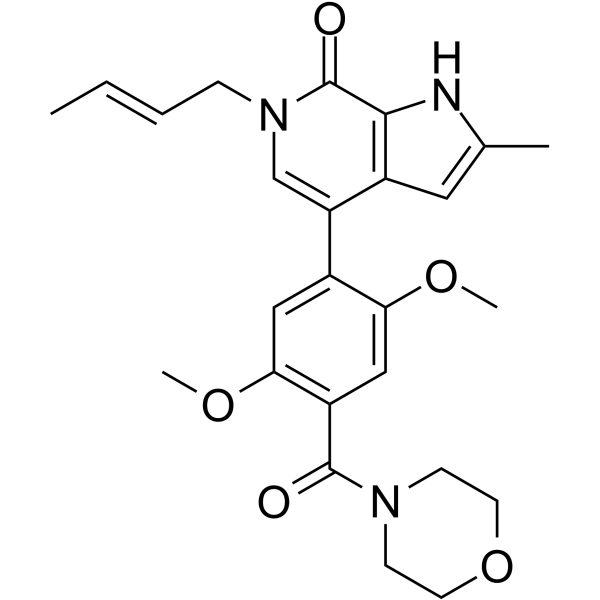
GNE-781
GNE-781 is an orally active, highly potent and selective CBP inhibitor with an IC50 of 0.94 nM in TR-FRET assay. GNE-781 also inhibits BRET and BRD4(1) with IC50s of 6.2 nM and 5100 nM, respectively. GNE-781 displays antitumor activity in an MOLM-16 AML xenograft model.
-
GC33253
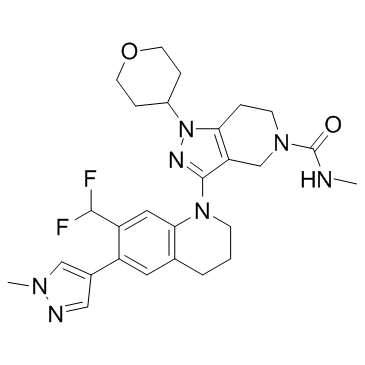
GNE-955
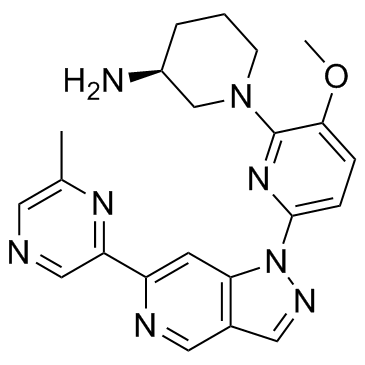
GNE-955 is a potent and orally active pan Pim kinase inhibitor with Kis of 0.018, 0.11, 0.08 nM for Pim1, Pim2, Pim3, respectively.
-
GC40918
Gramicidin A
Gramicidin A is a peptide component of gramicidin, an antibiotic mixture originally isolated from B.
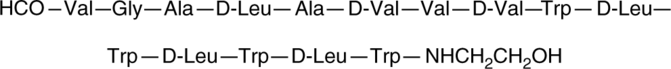
-
GC46152
GS-441524
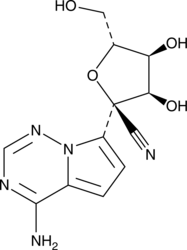
An antiviral nucleoside analog
-
GC33204
GS-626510
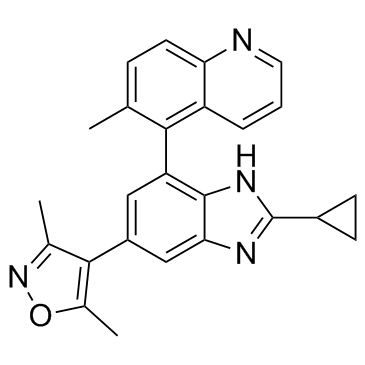
GS-626510 is a potent, and orally active BET family bromodomains inhibitor, with Kd values of 0.59-3.2 nM for BRD2/3/4, with IC50 values of 83 nM and 78 nM foe BD1 and BD2, respectively.
-
GC31731
GSK 4027
A PCAF/GCN5 bromodomain inhibitor
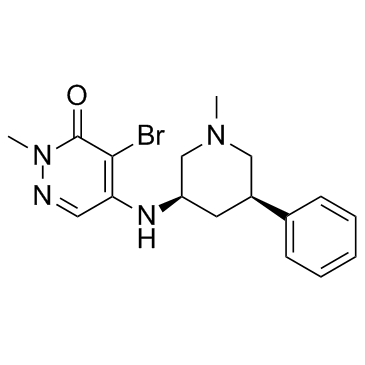
-
GC10524
GSK 525768A
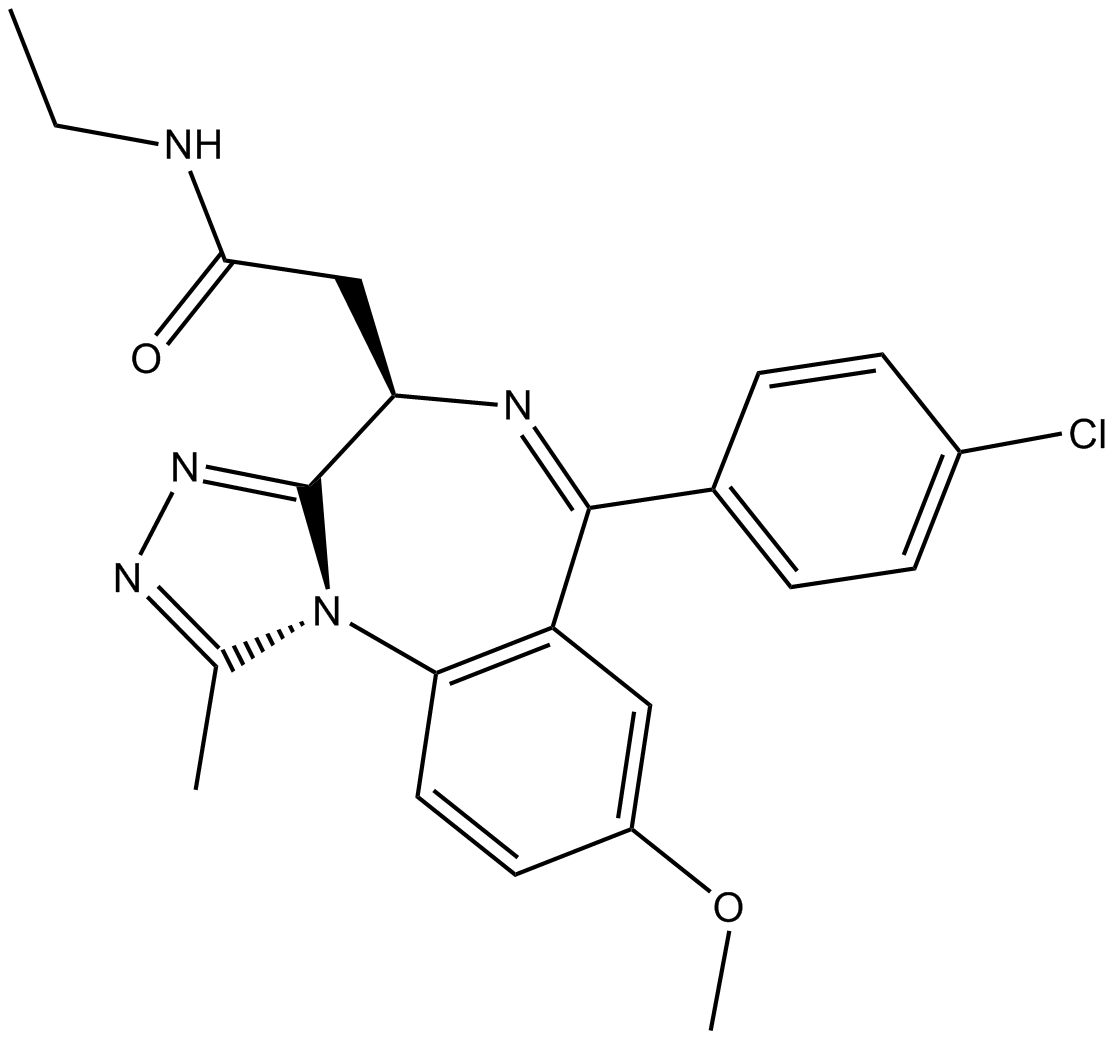
-
GC50697
GSK 591 dihydrochloride
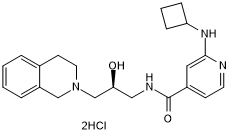
-
GC12440
GSK 5959
BRPF1 bromodomain inhibitor
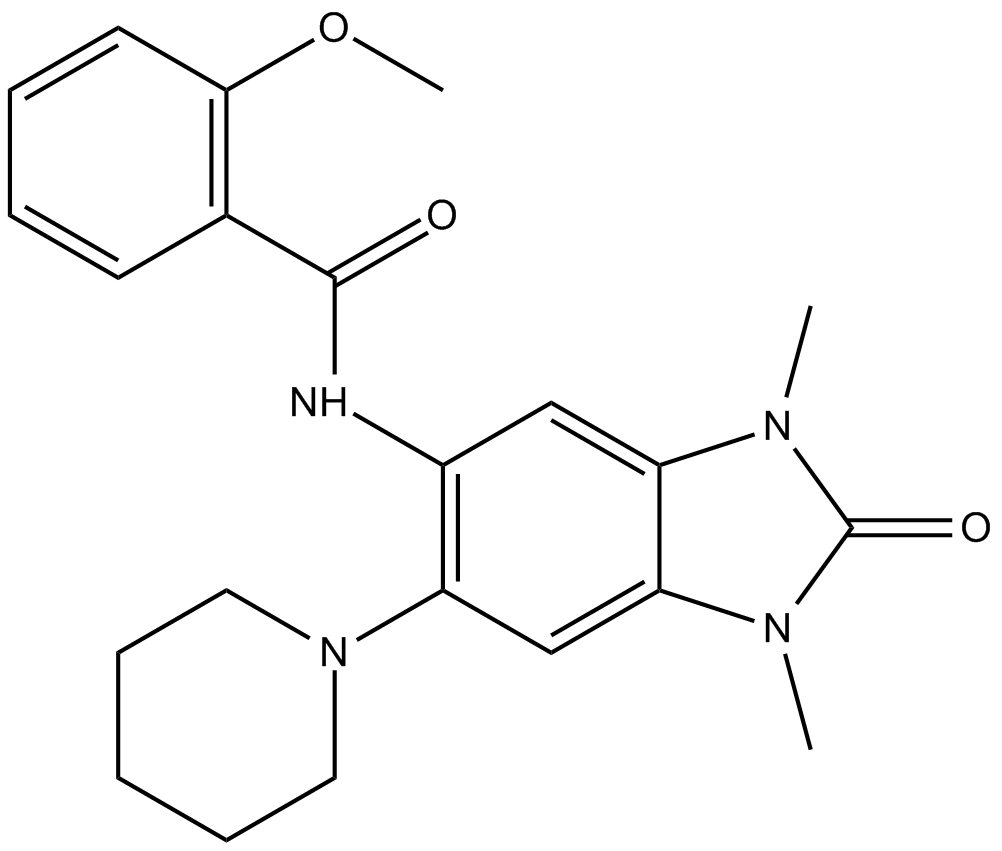
-
GC34314
GSK 690 Hydrochloride
GSK 690 (Hydrochloride) is a reversible inhibitor of lysine specific demethylase 1 (LSD1), with a Kd value of 9 nM and a biochemical IC50 of 37 nM.
-
GC50378
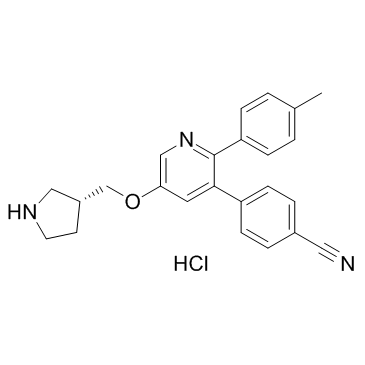
GSK 9311 hydrochloride
Negative control for GSK 6853
-
GC10617
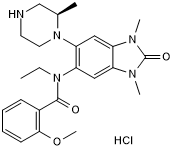
GSK J1
A dual inhibitor of JMJD3 and UTX
-
GC13086
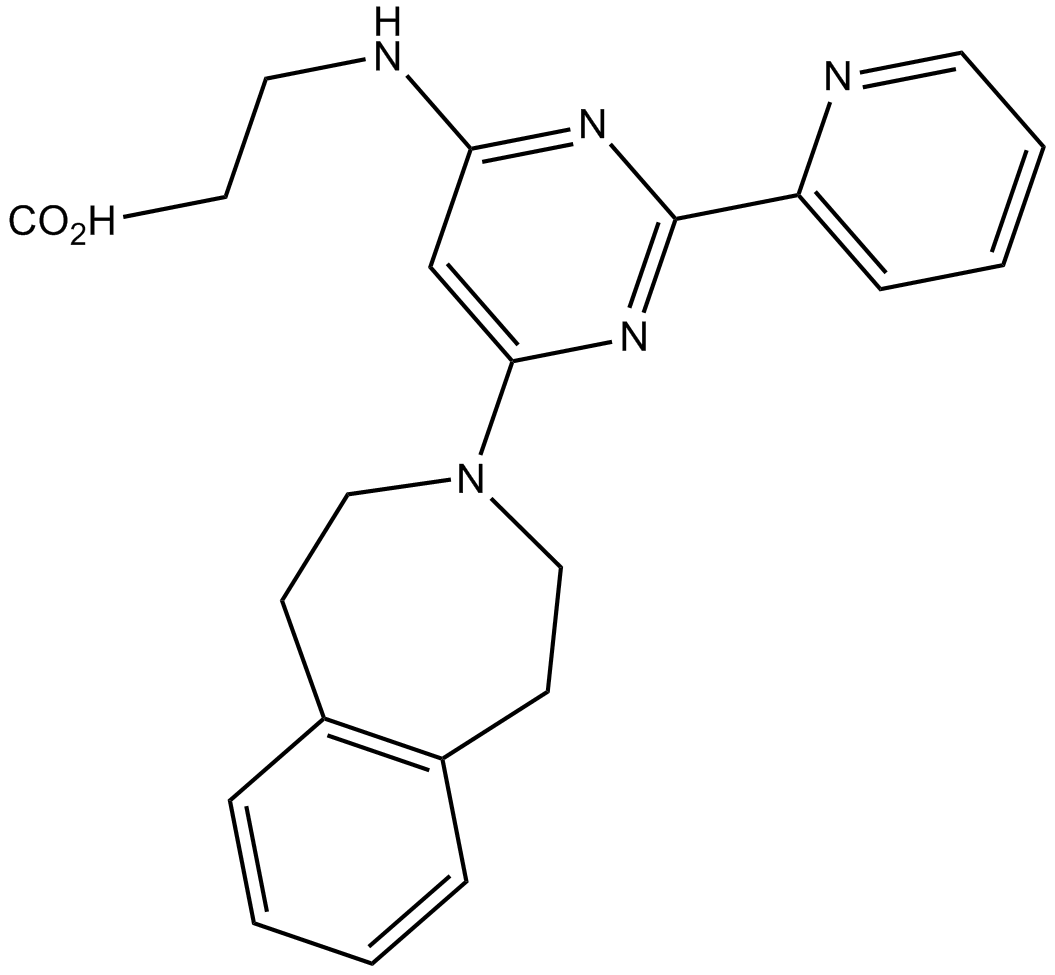
GSK J2
inactive control of GSK J1, JMJD3 (KDM6B) and UTX (KDM6A) inhibitor
-
GC12997
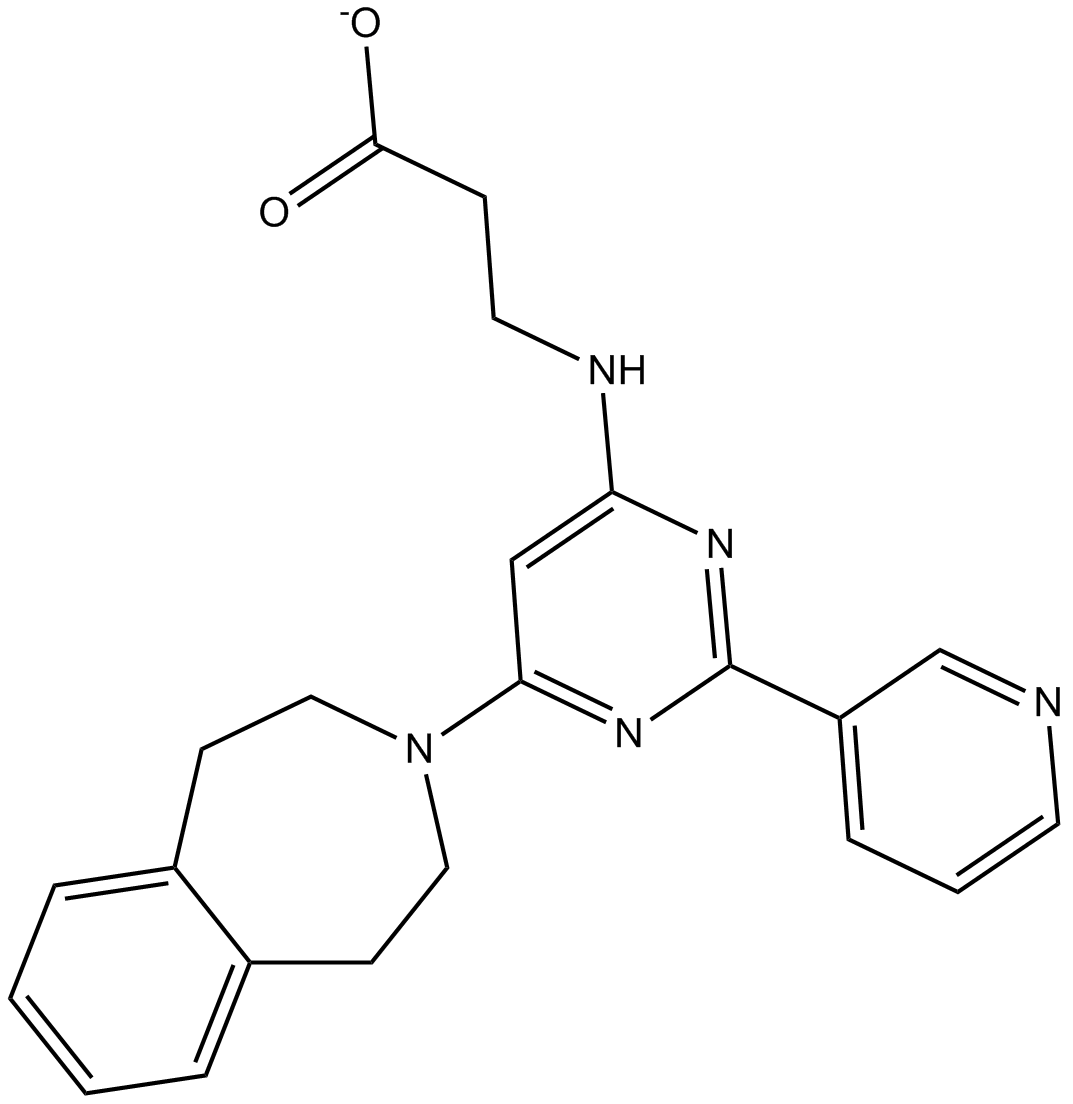
GSK J4 free base
GSK J4 free base is a potent dual inhibitor of H3K27me3/me2-demethylases JMJD3/KDM6B and UTX/KDM6A with IC50s of 8.6 and 6.6 μM, respectively. GSK J4 free base inhibits LPS-induced TNF-α production in human primary macrophages with an IC50 of 9 μM. GSK J4 is a cell permeable prodrug of GSK-J1. GSK J4 free base induces endoplasmic reticulum stress-related apoptosis.
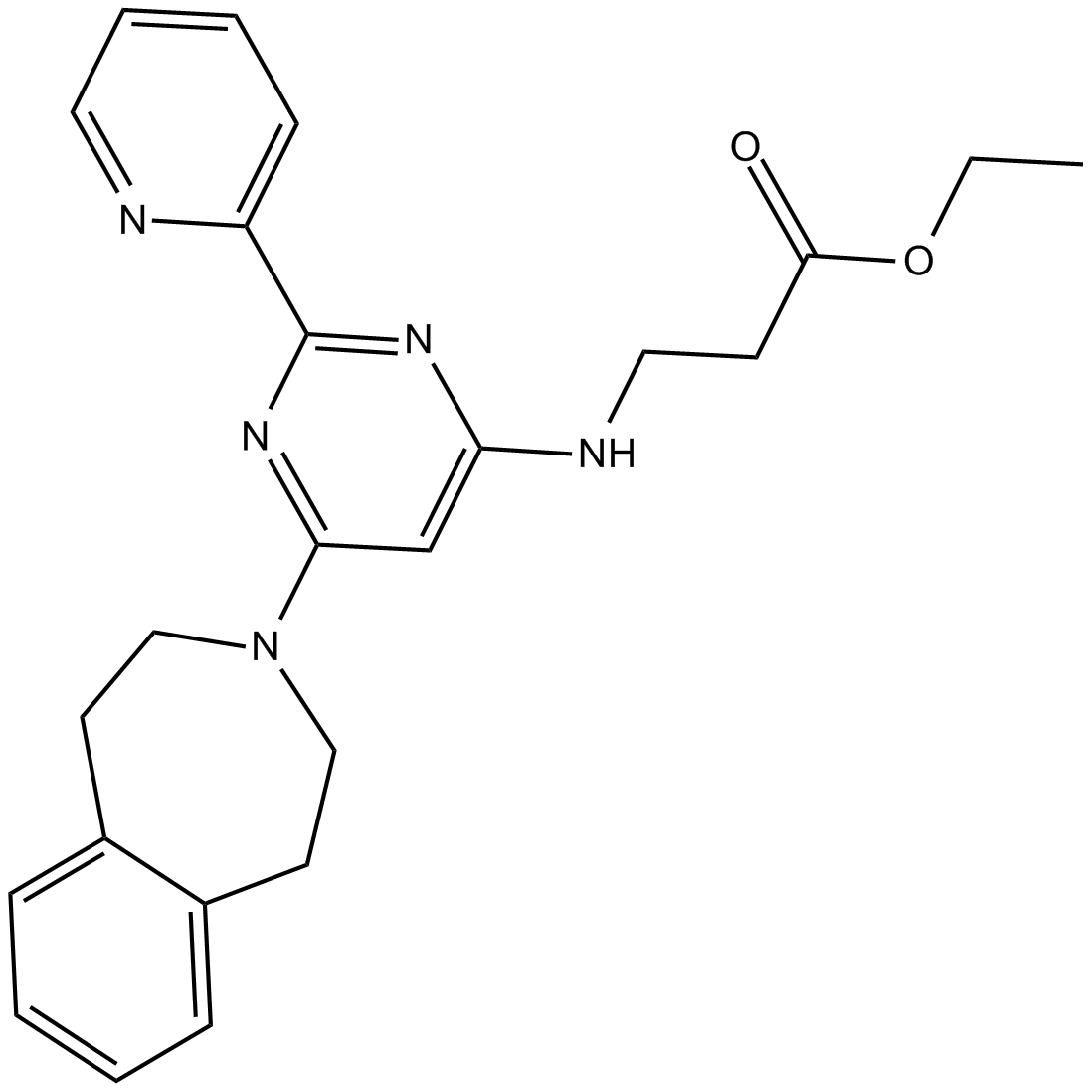
-
GC15497
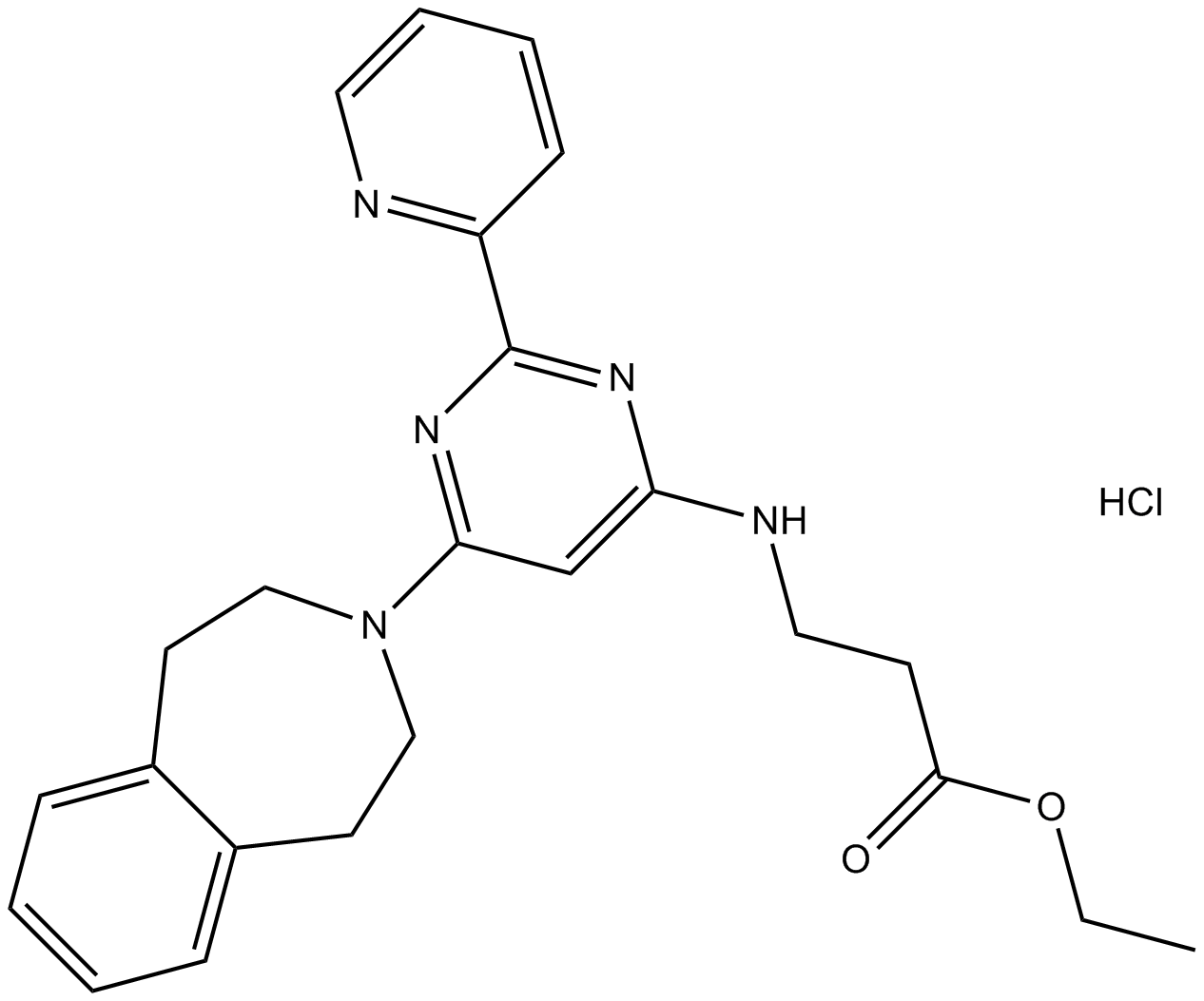
GSK J4 HCl
Prodrug of a selective H3K27 histone demethylase inhibitor
-
GC17118
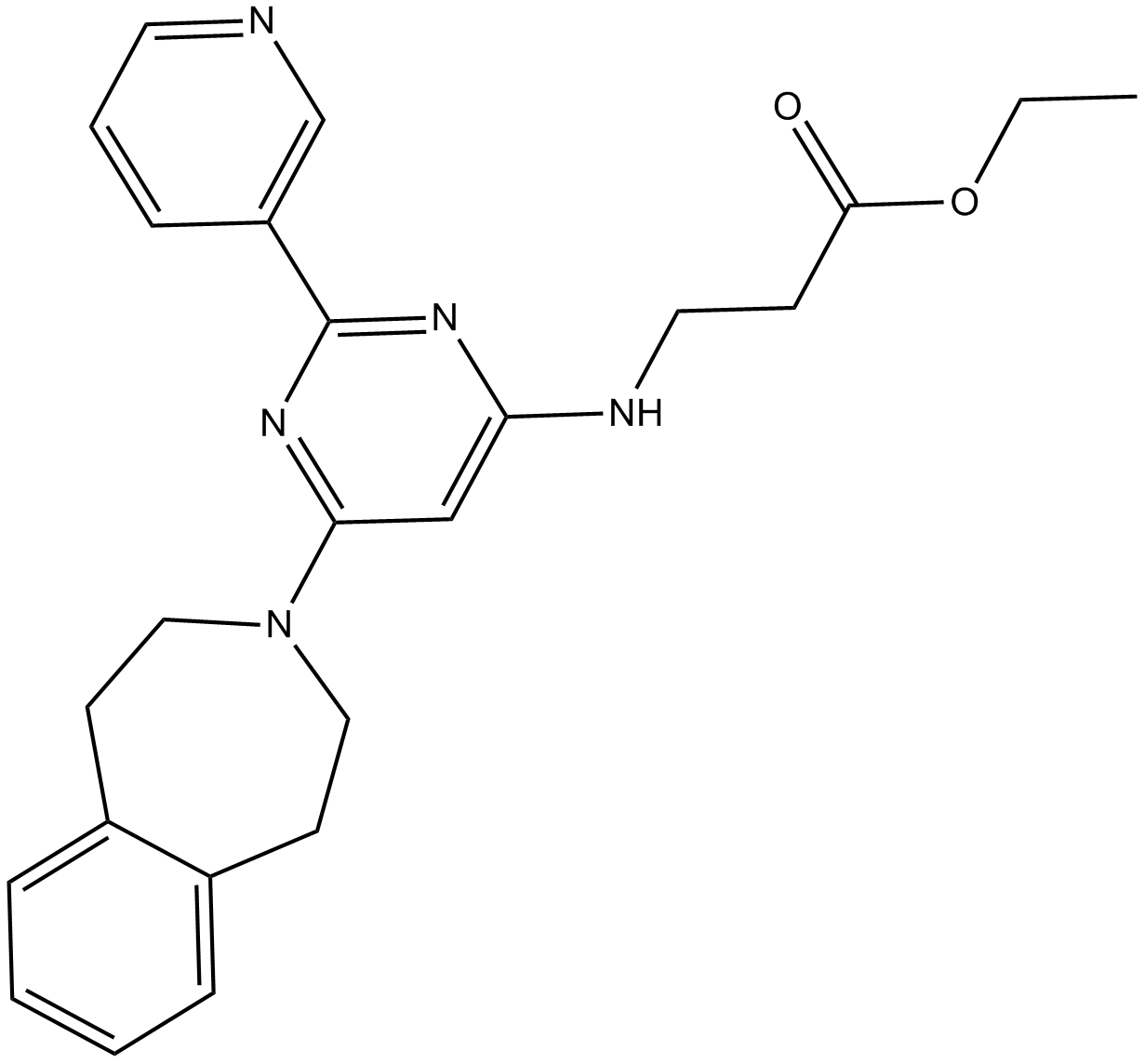
GSK J5
inactive isomer of GSK J4, KDM inhibitor
-
GC63483
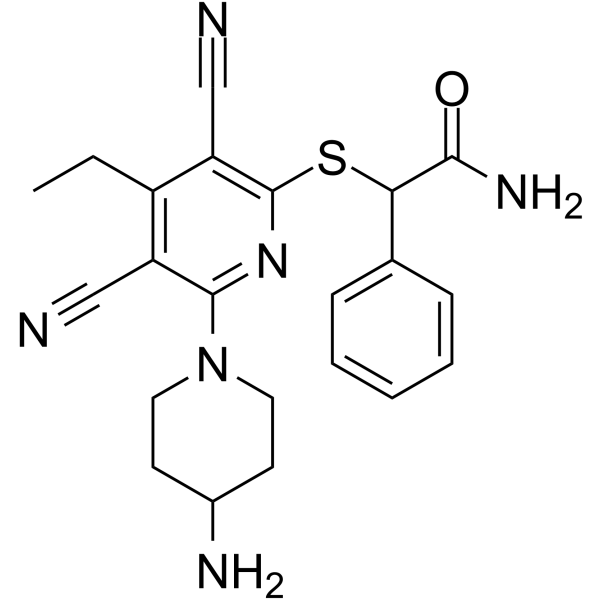
GSK-3685032
GSK-3685032 is a non-time-dependent, noncovalently, first-in-class reversible DNMT1-selective inhibitor, with an IC50 of 0.036 μM. GSK-3685032 induces robust loss of DNA methylation, transcriptional activation, and cancer cell growth inhibition.
-
GC36195
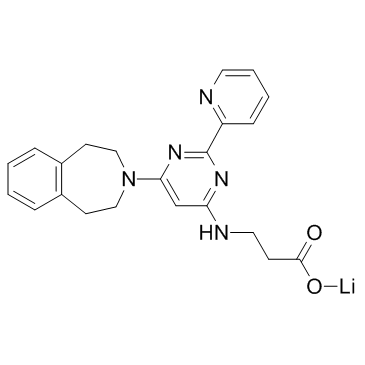
GSK-J1 lithium salt
GSK-J1 lithium salt is a potent inhibitor of H3K27me3/me2-demethylases JMJD3/KDM6B and UTX/KDM6A, with IC50 of 60 nM towards KDM6B.
-
GC43792
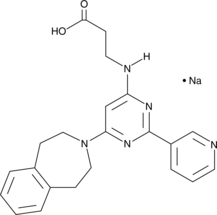
GSK-J2 (sodium salt)
The histone H3 lysine 27 (H3K27) demethylase JMJD3 plays important roles in the transcriptional regulation of cell differentiation, development, the inflammatory response, and cancer.
-
GC43794
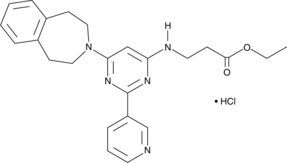
GSK-J5 (hydrochloride)
The histone H3 lysine 27 (H3K27) demethylase JMJD3 plays important roles in the transcriptional regulation of cell differentiation, development, the inflammatory response, and cancer.
-
GC18012
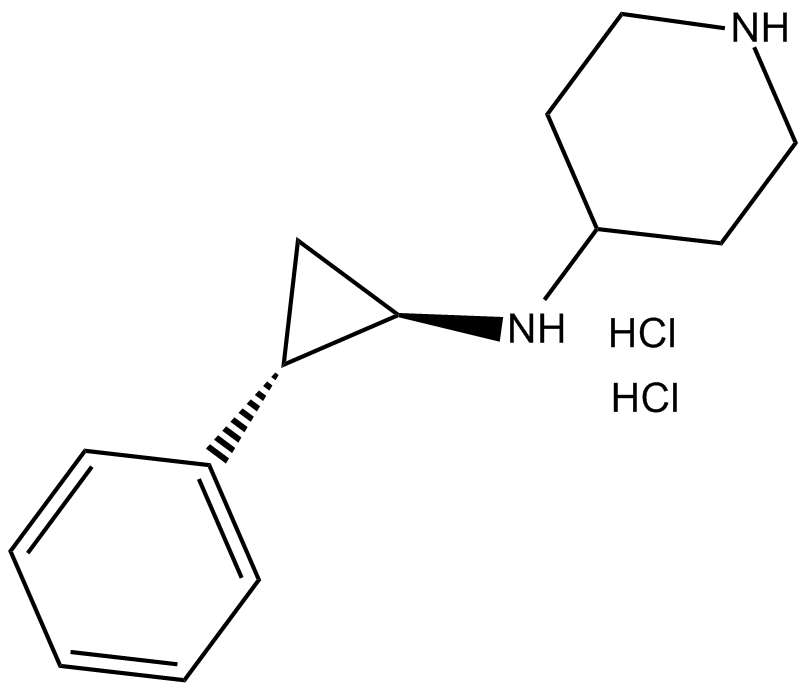
GSK-LSD1 (hydrochloride)
LSD1 inhibitor
-
GC15368
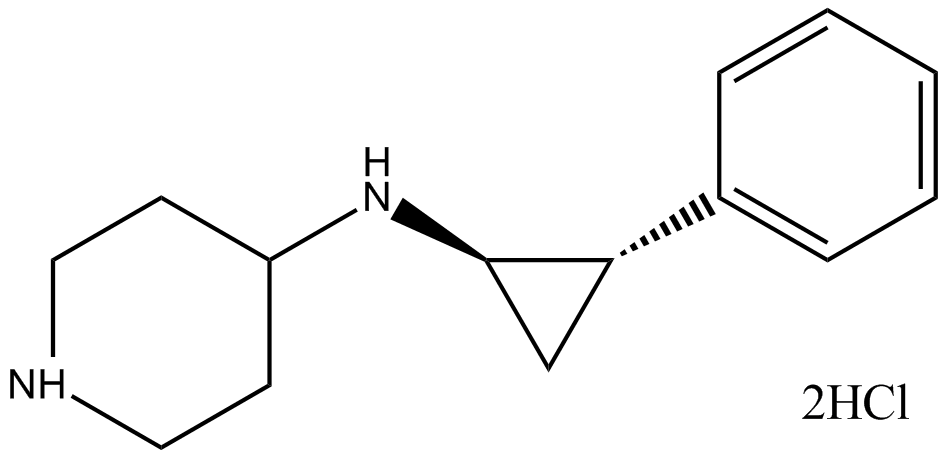
GSK-LSD1 2HCl
irreversible, and selective LSD1 inhibitor
-
GC32764
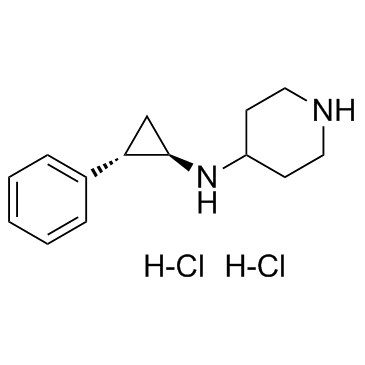
GSK-LSD1 Dihydrochloride
An LSD1 inhibitor
-
GC43787
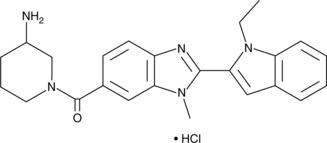
GSK106 (hydrochloride)
GSK106 (hydrochloride) is an inactive control for the selective PAD4 inhibitors, GSK484 and GSK199.
-
GC10008
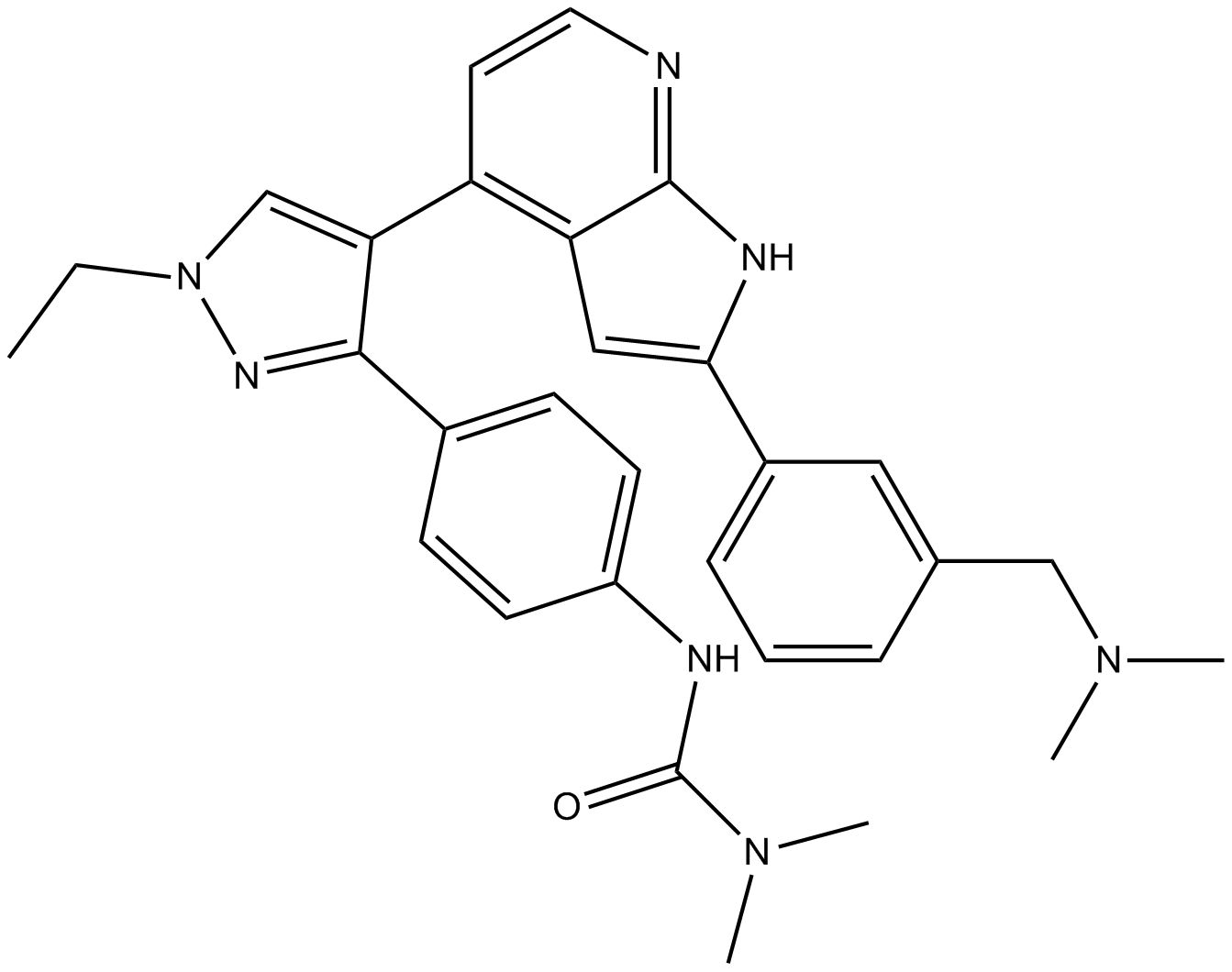
GSK1070916
A potent inhibitor of Aurora B and C kinases
-
GC43788
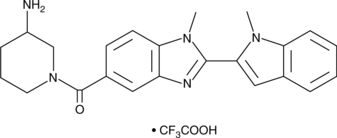
GSK121 (trifluoroacetate salt)
GSK-121 Trifluoroacetates a selective PAD4 inhibitor.
-
GC15783
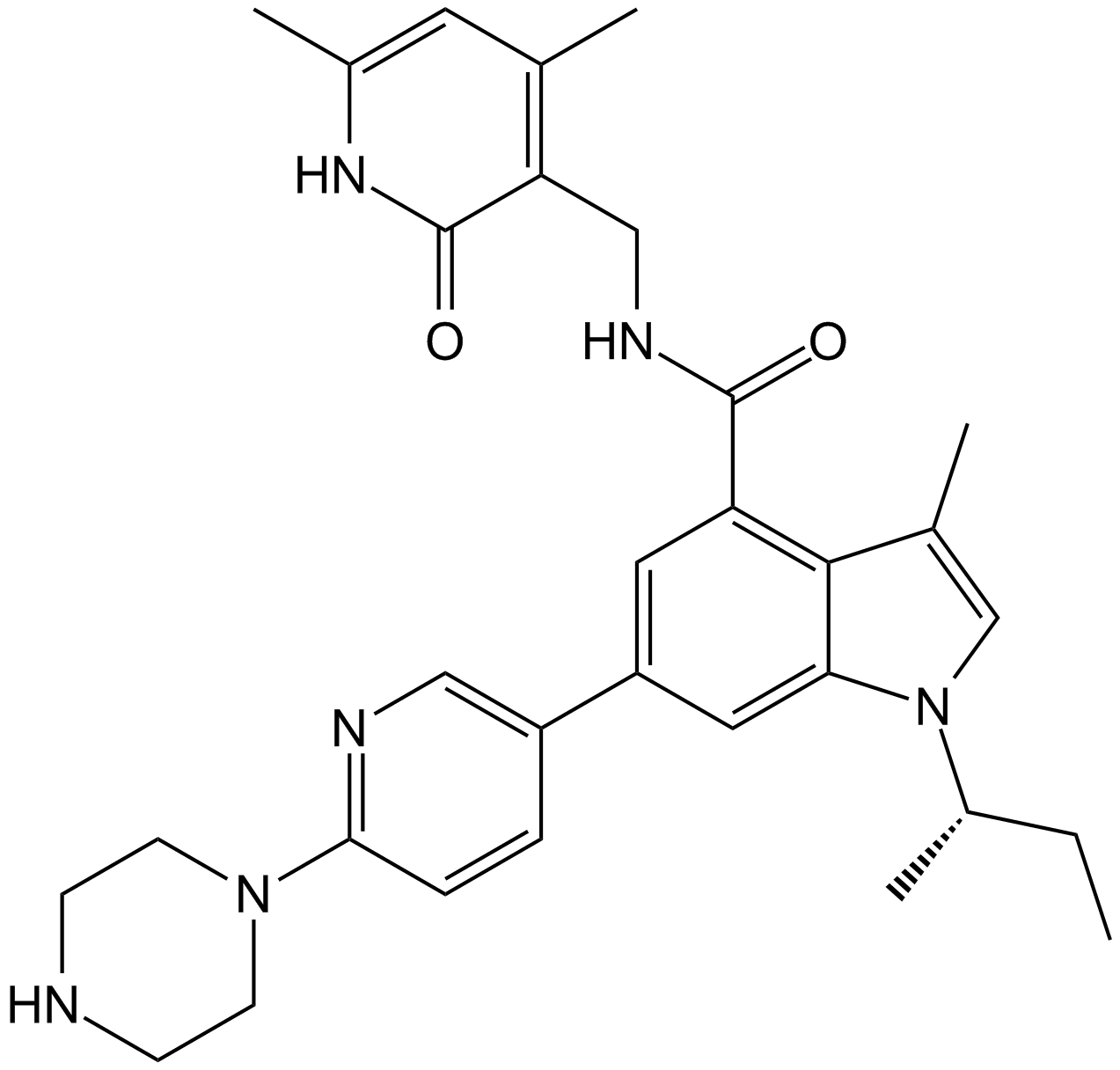
GSK126
A selective EZH2 inhibitor
-
GC14063
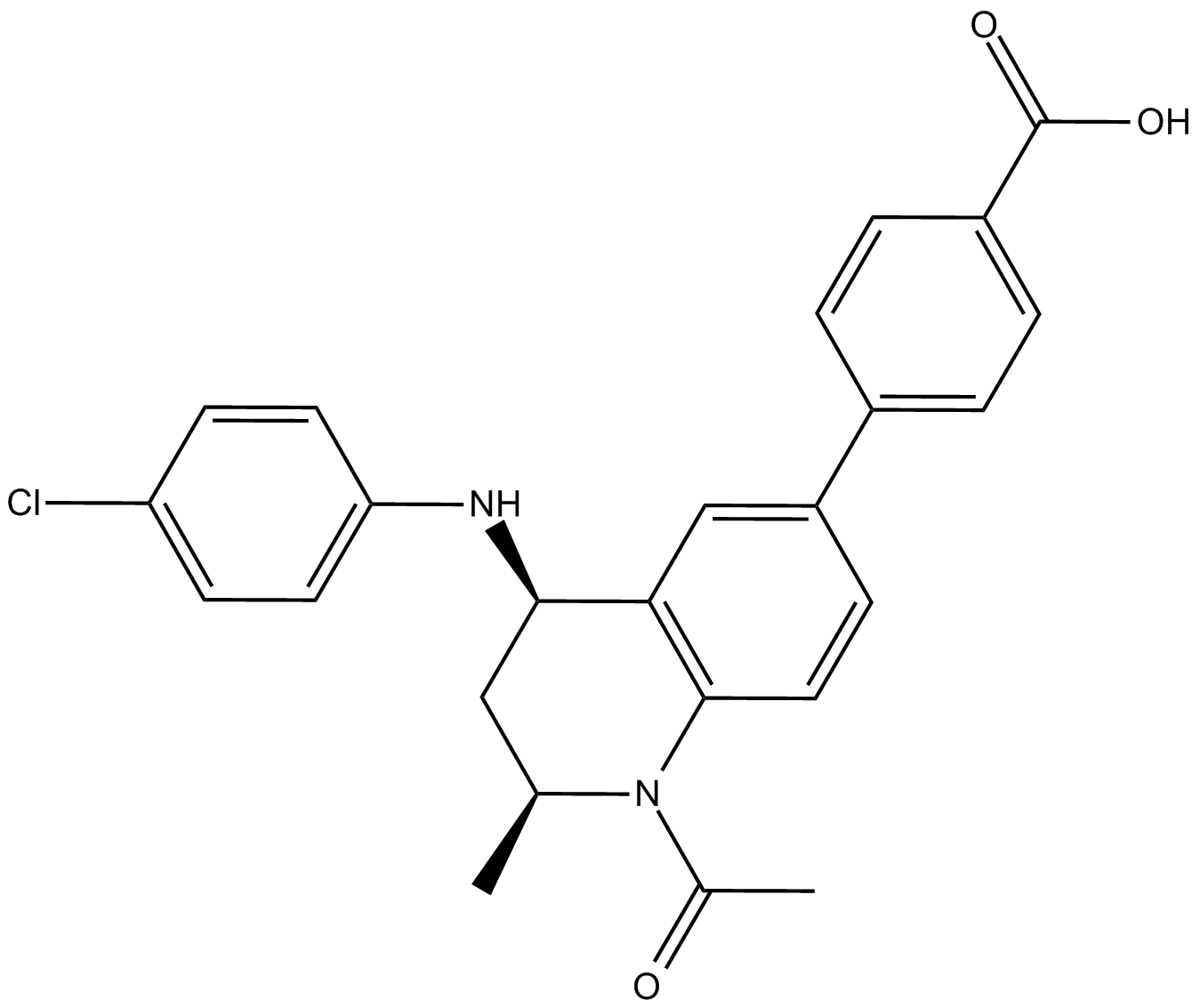
GSK1324726A
BET proteins inhibitor
-
GC43789
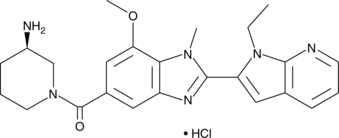
GSK199 (hydrochloride)
GSK199 (hydrochloride) is a reversible and selective PAD4 inhibitor with an IC50 of 200 nM in the absence of calcium.
-
GC15789
GSK2801
inhibitor of BAZ2A and BAZ2B bromodomains
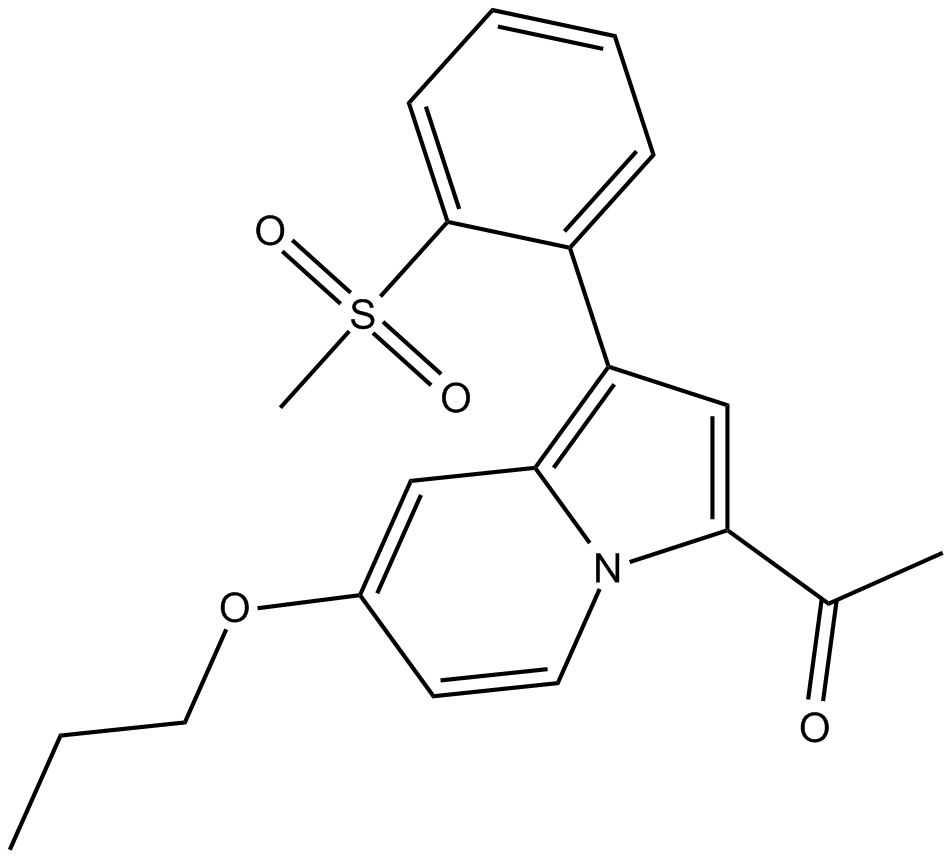
-
GC19181
GSK2807 Trifluoroacetate
GSK2807 Trifluoroacetate is a potent, selective and SAM-competitive inhibitor of SMYD3, with a Ki of 14 nM.
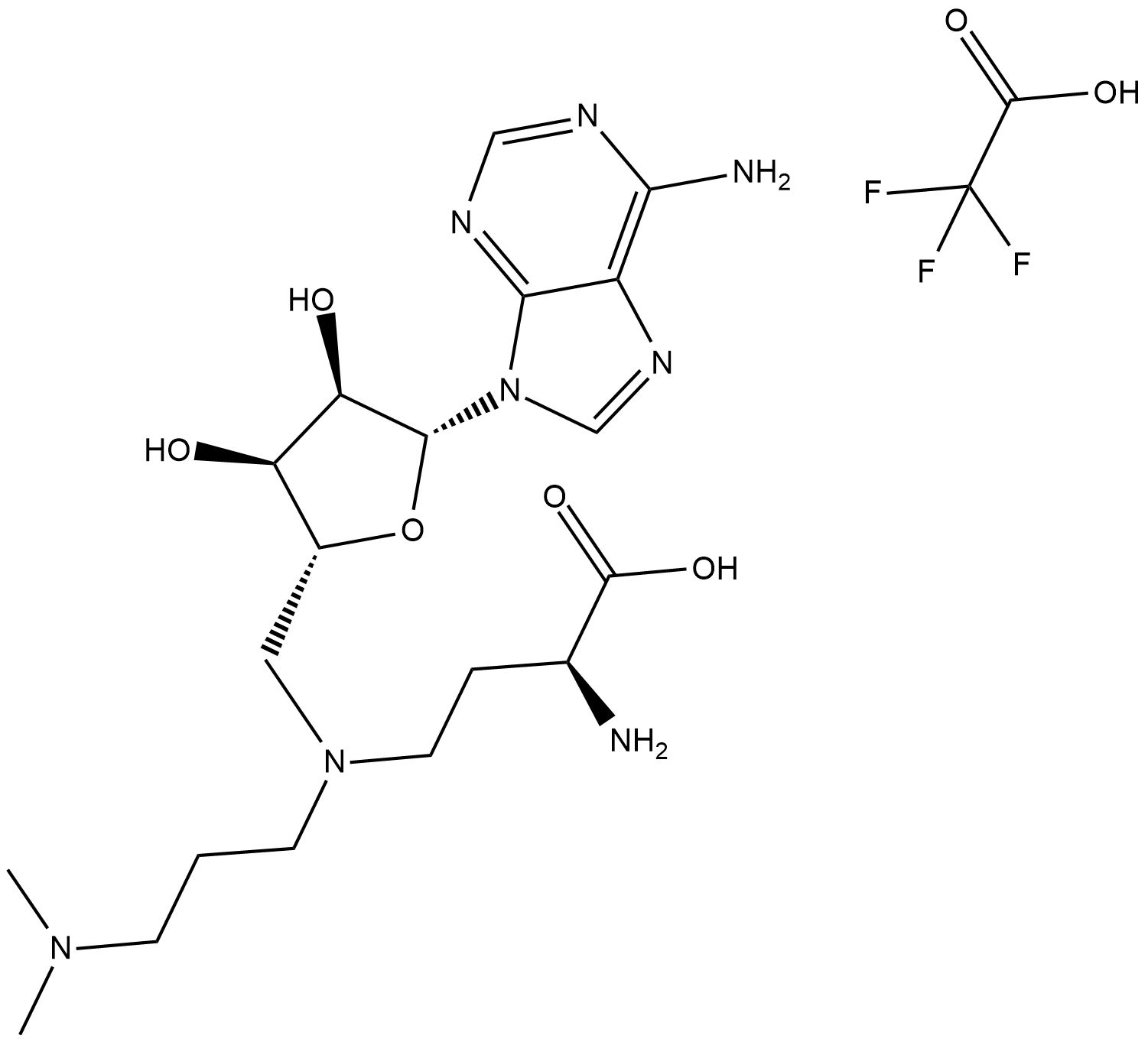
-
GC11578
GSK2879552
Novel and irreversible LSD1 inhibitor
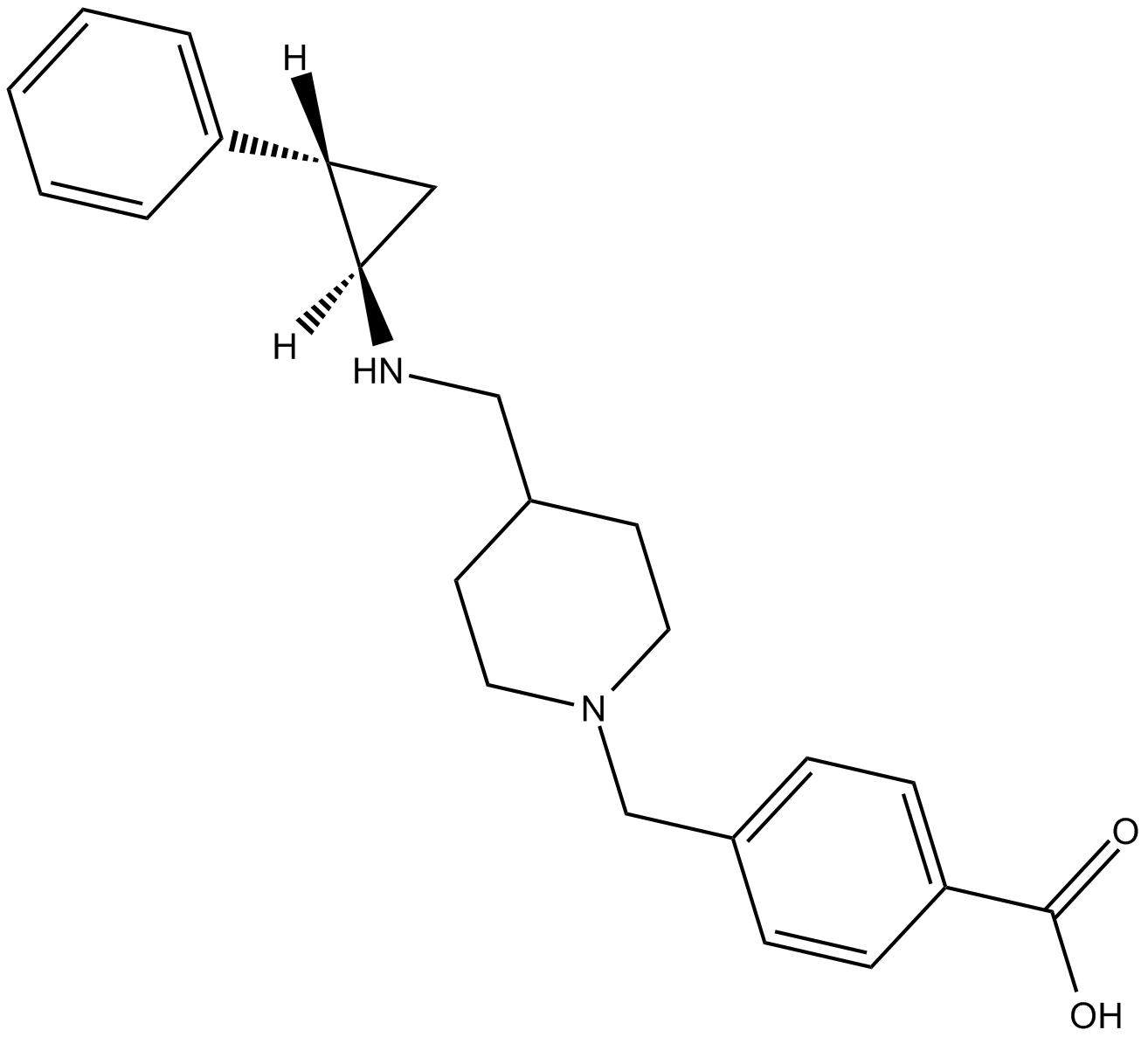
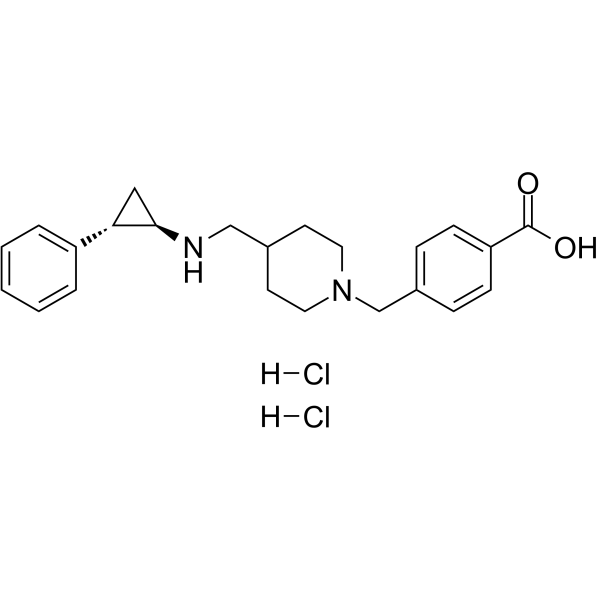
-
GC62719
GSK2879552 dihydrochloride
GSK2879552 dihydrochloride an orally active, selective and irreversible inhibitor of lysine specific demethylase 1 (LSD1/KDM1A), with potential antineoplastic activity.
-
GC18402
GSK3117391
GSK3117391 is a histone deacetylase (HDAC) inhibitor, extracted from patent WO/2008040934 A1.
-
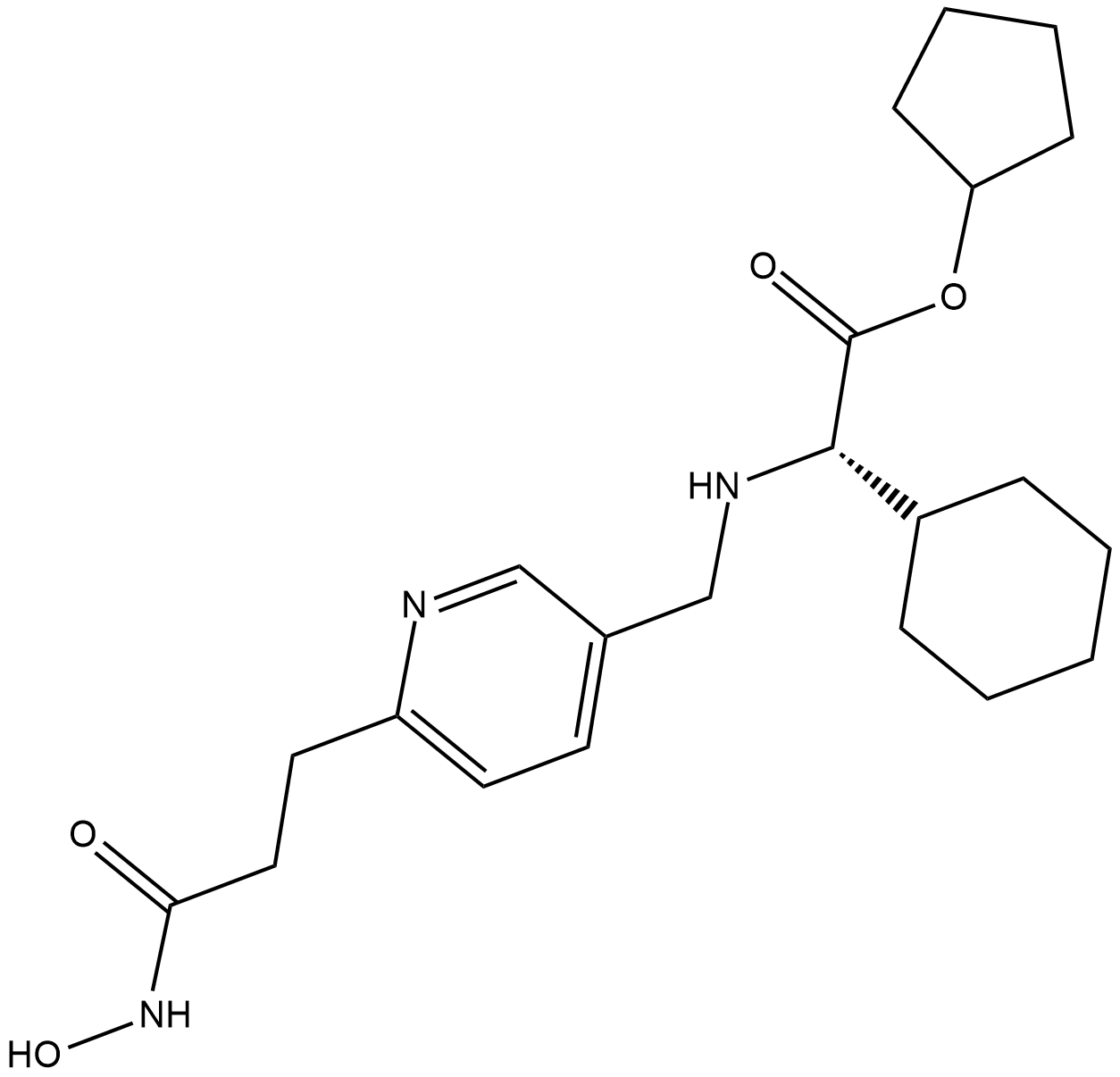
GC32693
GSK3326595 (EPZ015938)
GSK3326595 (EPZ015938) (EPZ015938) is a potent, selective, reversible inhibitor of protein arginine methyltransferase 5 (PRMT5) with an IC50 of 6.2 nM.
-
GC36191
GSK3368715
GSK3368715 (EPZ019997) is an orally active, reversible, and S-adenosyl-L-methionine (SAM) uncompetitive type I protein arginine methyltransferases (PRMTs) inhibitor (IC50=3.1 nM (PRMT1), 48 nM (PRMT3), 1148 nM (PRMT4), 5.7 nM (PRMT6), 1.7 nM (PRMT8)). GSK3368715 (EPZ019997) produces a shift in arginine methylation states, alters exon usage, and has strong anti-cancer activity.
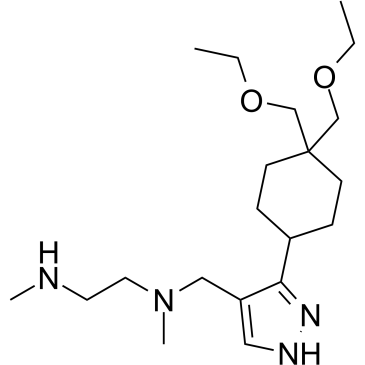
-
GC25483
GSK3368715 (EPZ019997) 3HCl
GSK3368715 is a potent inhibitor of type I protein arginine methyltransferases (PRMT) that inhibits PRMT1, 3, 4, 6 and 8 with Kiapp vaules ranging from 1.5 to 81 nM.
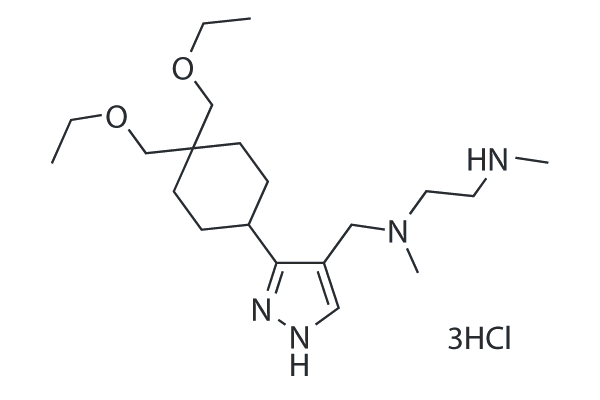
-
GC49398
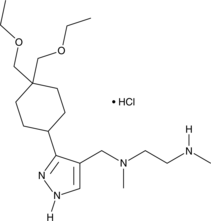
GSK3368715 (hydrochloride)
An inhibitor of type I PRMTs
-
GC36192
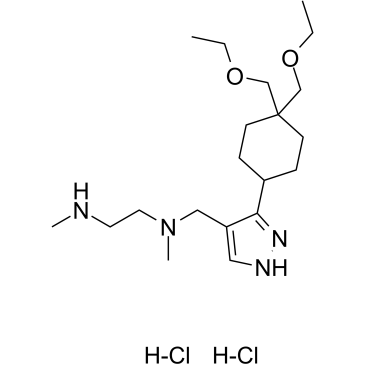
GSK3368715 dihydrochloride
GSK3368715 dihydrochloride (EPZ019997 dihydrochloride) is an orally active, reversible, and S-adenosyl-L-methionine (SAM) uncompetitive type I protein arginine methyltransferases (PRMTs) inhibitor (IC50=3.1 nM (PRMT1), 48 nM (PRMT3), 1148 nM (PRMT4), 5.7 nM (PRMT6), 1.7 nM (PRMT8)). GSK3368715 dihydrochloride (EPZ019997 dihydrochloride) produces a shift in arginine methylation states, alters exon usage, and has strong anti-cancer activity.
-
GC17534
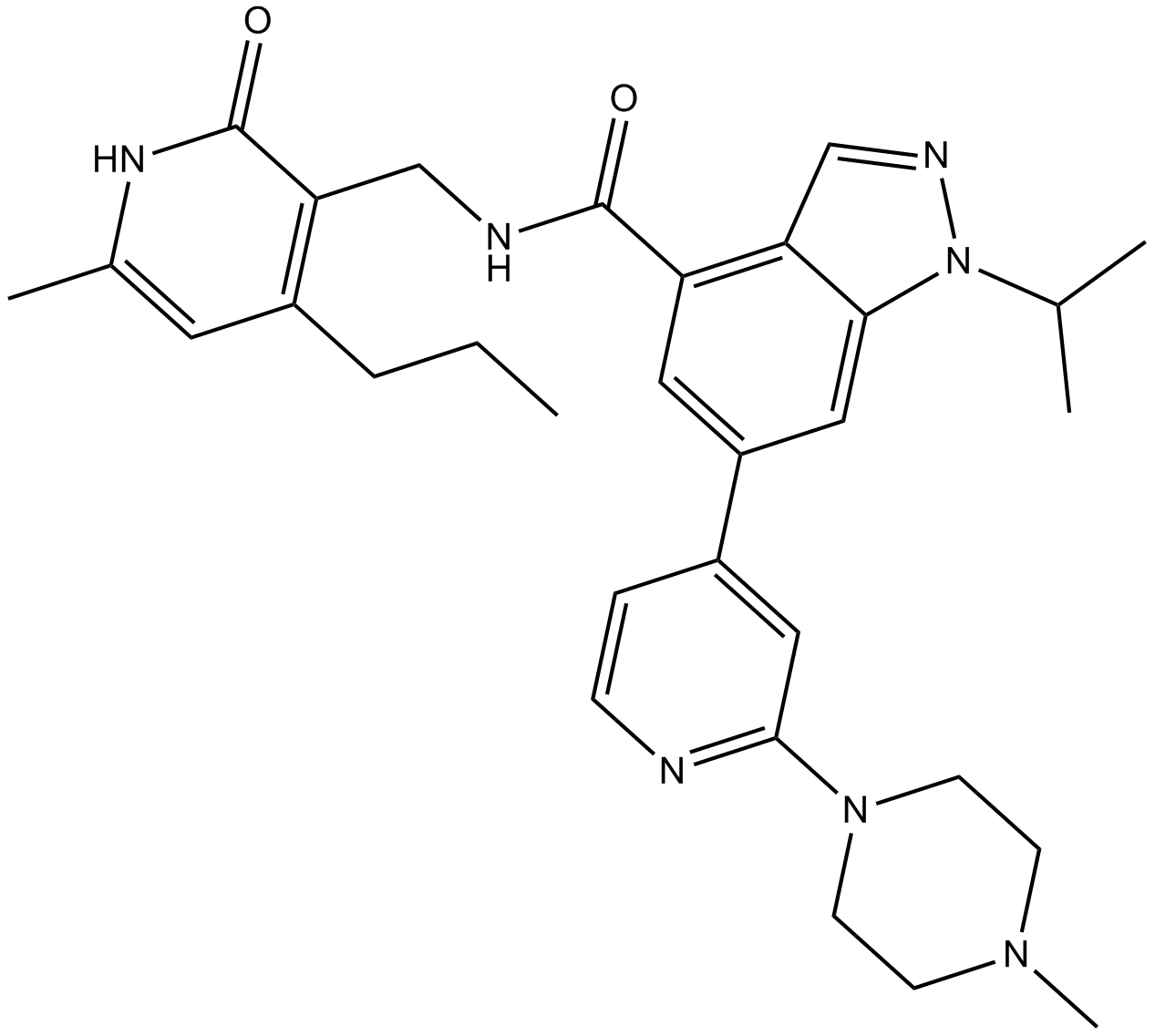
GSK343
A selective, cell-permeable EZH2 inhibitor
-
GC30714
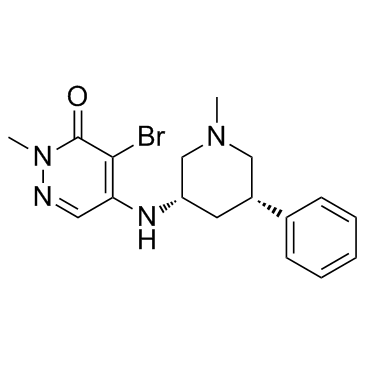
GSK4028
GSK4028 is the enantiomeric negative control of GSK4027, which is a PCAF/GCN5 bromodomain chemical probe, the pIC50 of GSK4028 is 4.9 in a time-resolved fluorescence resonance energy transfer (TR-FRET) assay.
-
GC34604
GSK467
GSK467 is a cell penetrant and selective KDM5B (JARID1B or PLU1) inhibitor with a Ki of 10 nM and an IC50 of 26 nM. GSK467 shows 180-fold selectivity for KDM4C and no measurable inhibitory effects toward KDM6 or other Jumonji family members.
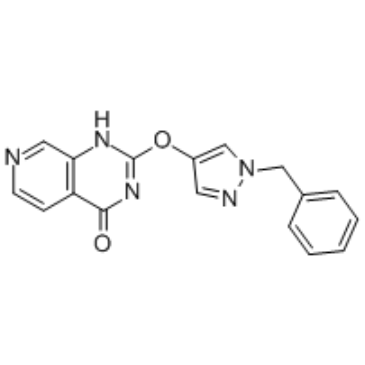
-
GC19184
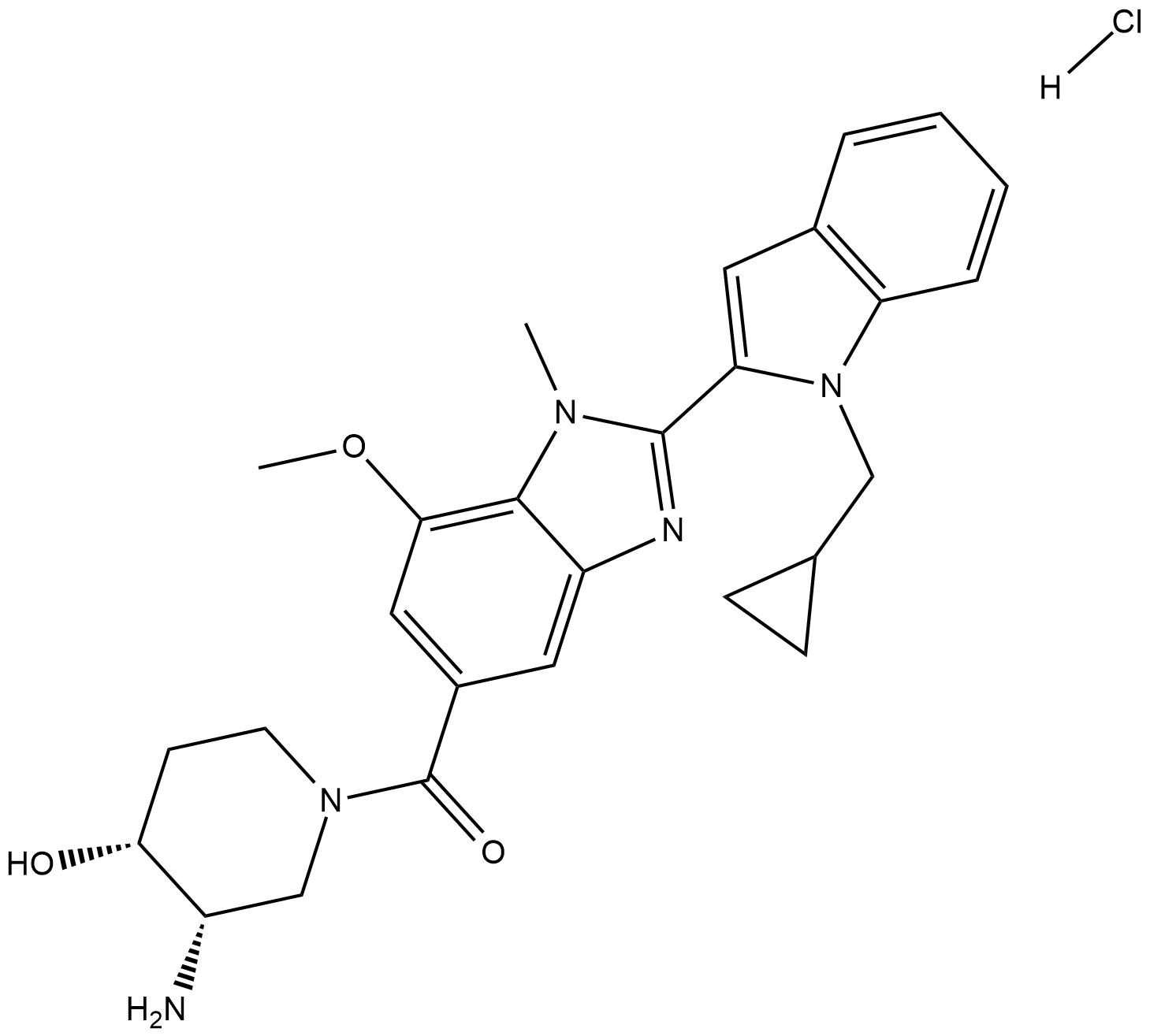
GSK484 hydrochloride
GSK484 is a selective PAD4 inhibitor with an IC50 of 50 nM without calcium.
-
GC11414
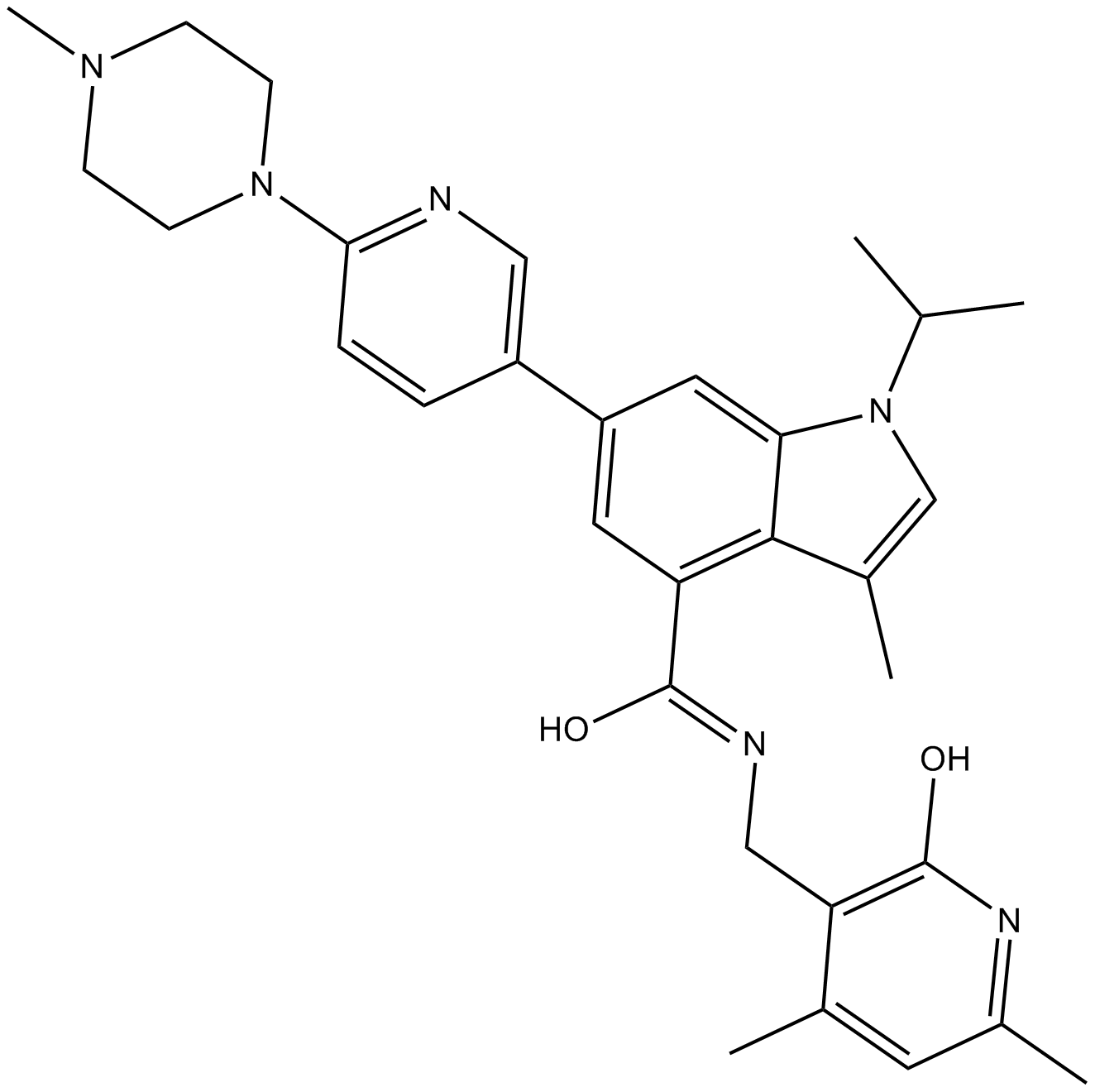
GSK503
EZH2 inhibitor
-
GC14585
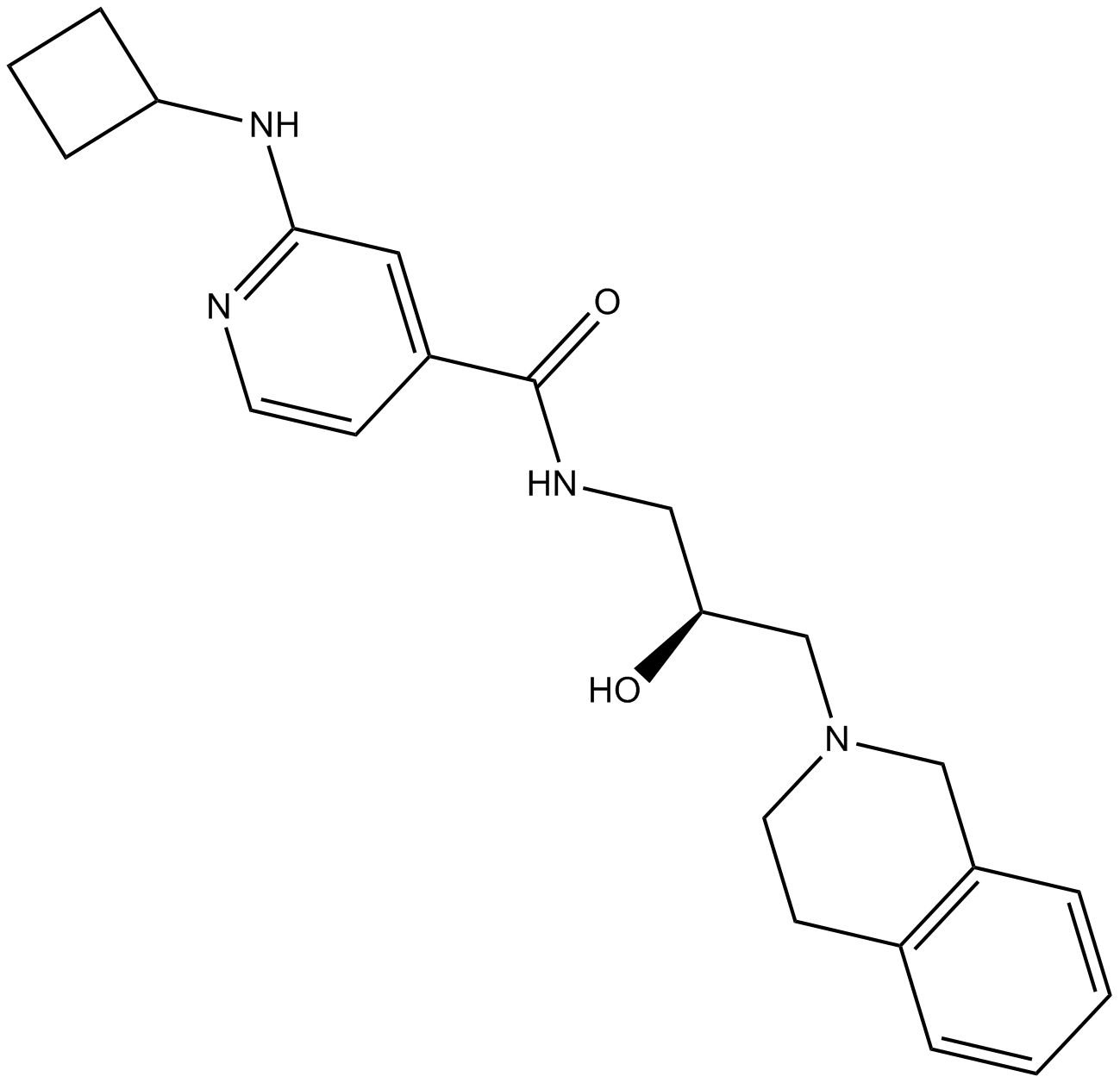
GSK591
PRMT5 inhibitor
-
GC62312
GSK620
GSK620 is a potent and orally active pan-BD2 inhibitor with excellent broad selectivity, developability and in vivo oral pharmacokinetics.
-
GC13025
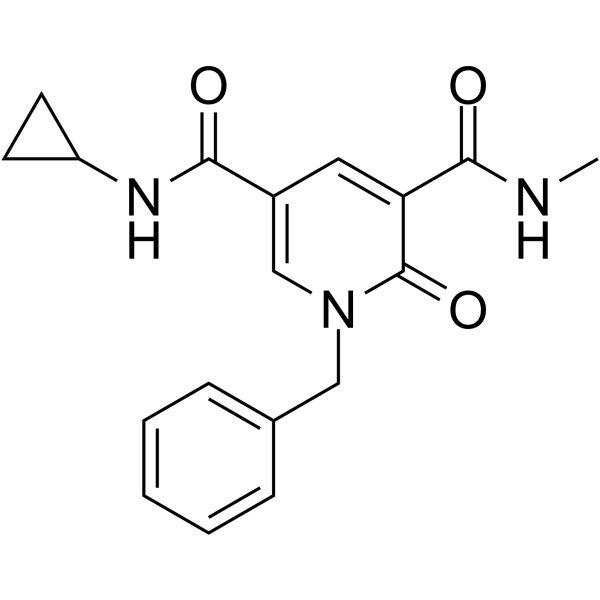
GSK6853
BRPF1 inhibitor
-
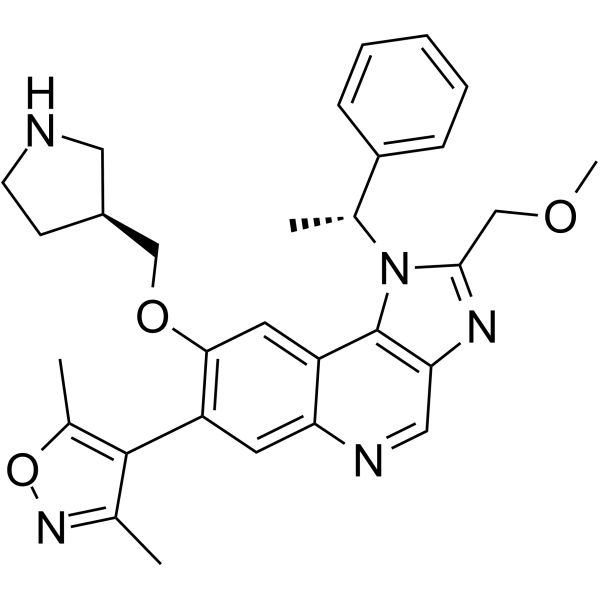
GC62654
GSK778
GSK778 (iBET-BD1) is a potent and selective BD1 bromodomain inhibitor of the BET proteins, with IC50s of 75 nM (BRD2 BD1), 41 nM (BRD3 BD1), 41 nM (BRD4 BD1), and 143 nM (BRDT BD1), respectively.
-
GC38049
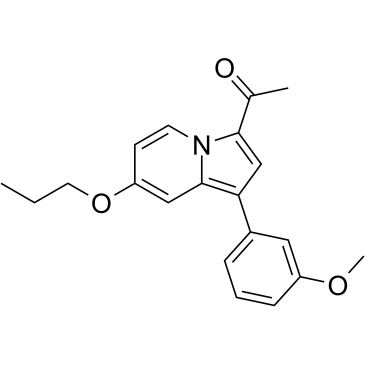
GSK8573
GSK8573 is an inactive control compound for GSK2801 (acetyl-lysine competitive inhibitor of BAZ2A and BAZ2B bromodomains). GSK8573 has binding activity to BRD9 with a Kd value of 1.04 μM and is inactive against BAZ2A/B and other bromodomain familiy. GSK8573 can be used as a structurally related negative control compound in biological experiments.
-
GC60184
GSK8814
GSK8814 is a potent, selective, and ATAD2/2B bromodomain chemical probe and inhibitor, with a binding constant pKd=8.1 and a pKi=8.9 in BROMOscan. GSK8814 binds to ATAD2 and BRD4 BD1 with pIC50s of 7.3 and 4.6, respectively. GSK8814 shows 500-fold selectivity for ATAD2 over BRD4 BD1.
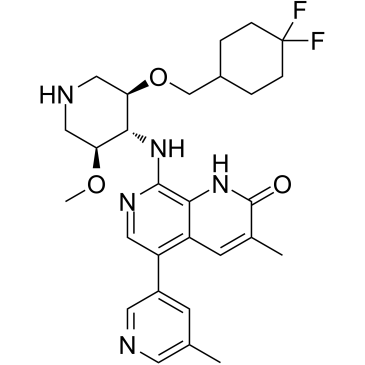
-
GC33324
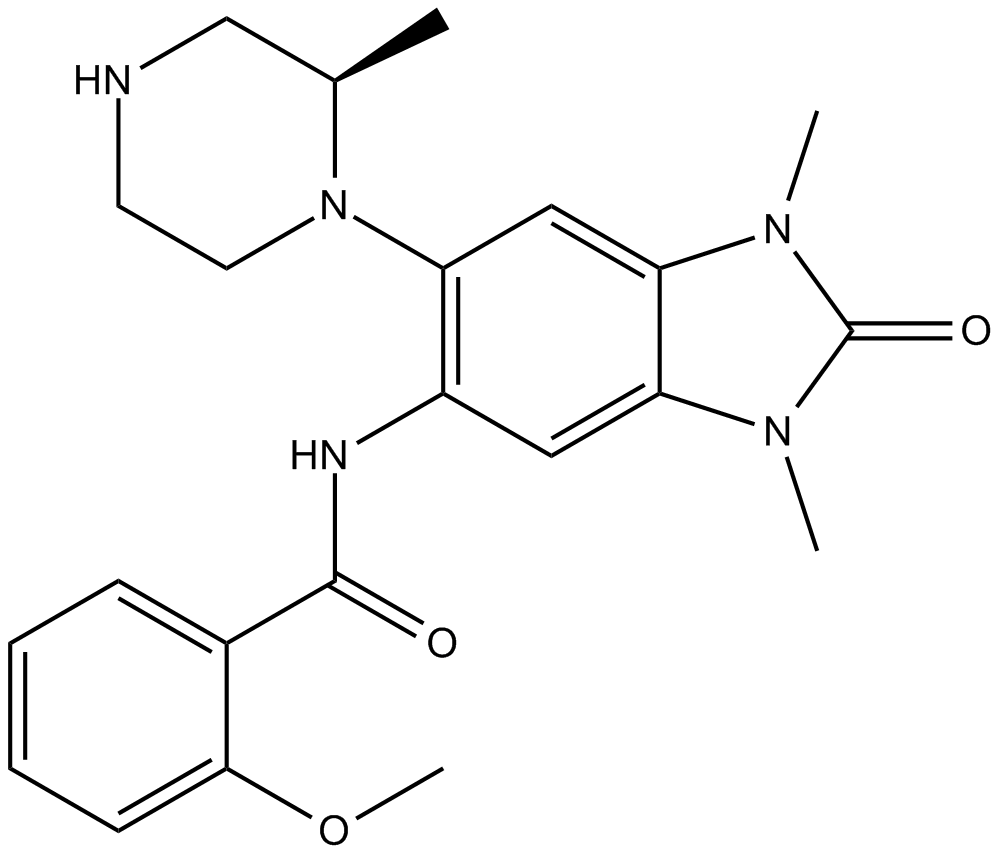
GSK9311
GSK9311, a less active analogue of GSK6853, can be used as a negative control. GSK9311 inhibits BRPF bromodomain with pIC50 values of 6.0 and 4.3 for BRPF1 and BRPF2, respectively.
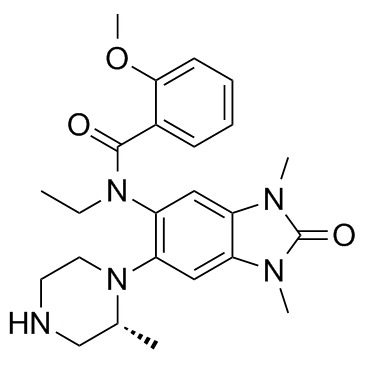
-
GC36196
Guadecitabine sodium
Guadecitabine is a novel hypomethylating dinucleotide of decitabine and deoxyguanosine that is resistant to degradation by cytidine deaminase.
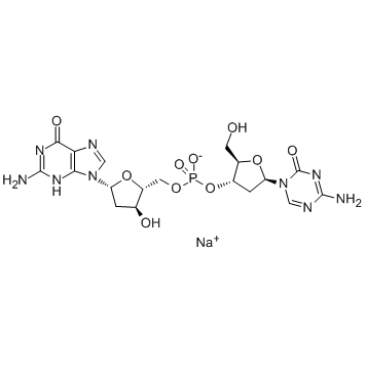
-
GC33041
Gusacitinib (ASN-002)
Gusacitinib (ASN-002) (ASN-002) is an orally active and potent dual inhibitor of spleen tyrosine kinase (SYK) and janus kinase (JAK) with IC50 values of 5-46 nM. Gusacitinib (ASN-002) has anti-cancer activity in both solid and hematological tumor types.
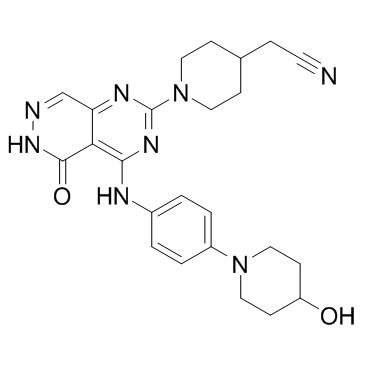
-
GC16611
GW841819X
BET bromodomain inhibitor
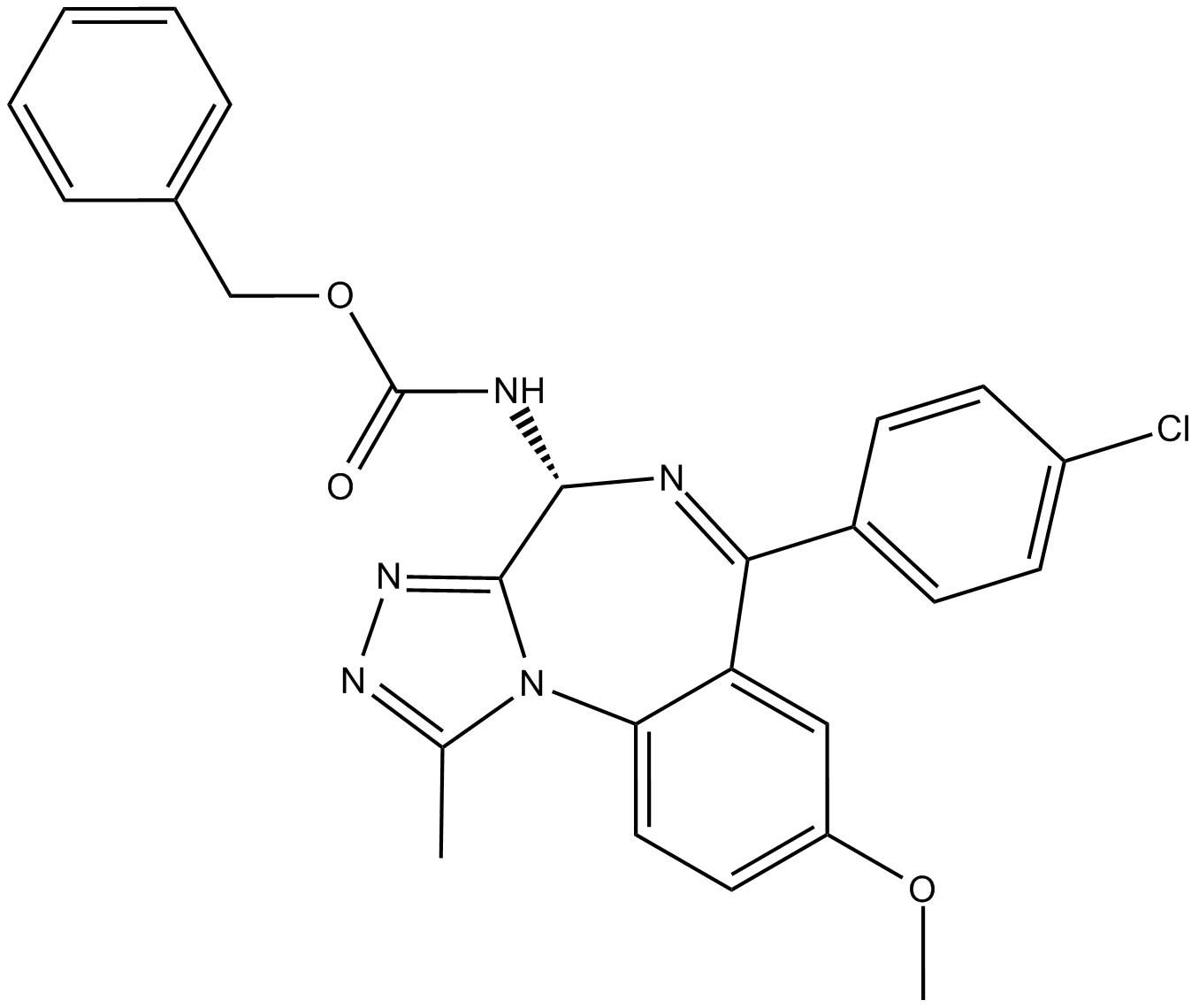
-
GC64115
Gypenoside LI
Gypenoside LI, a gypenoside monomer, possesses anti-tumor activity. Gypenoside LI induces cell apoptosis, cell cycle and migration.
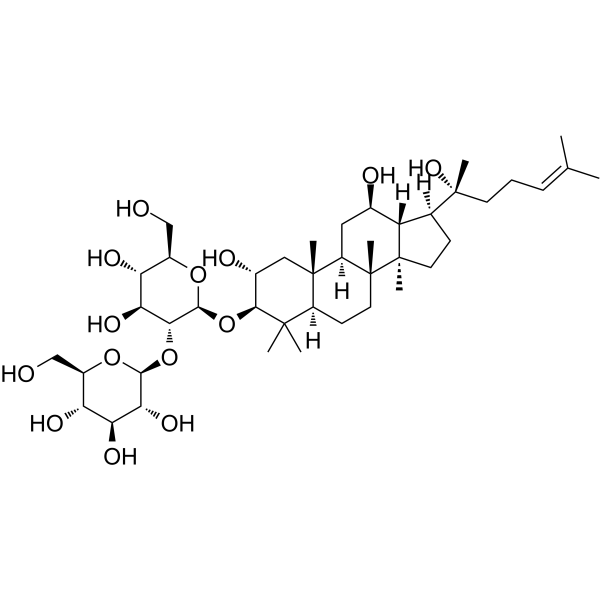
-
GC12009
HAT Inhibitor II
HAT Inhibitor II (compound 2c) is a potent, selective and cell permeable p300 histone acetyltransferase inhibitor, with an IC50 of 5 μM. HAT Inhibitor II shows anti-acetylase activity in mammalian cells.
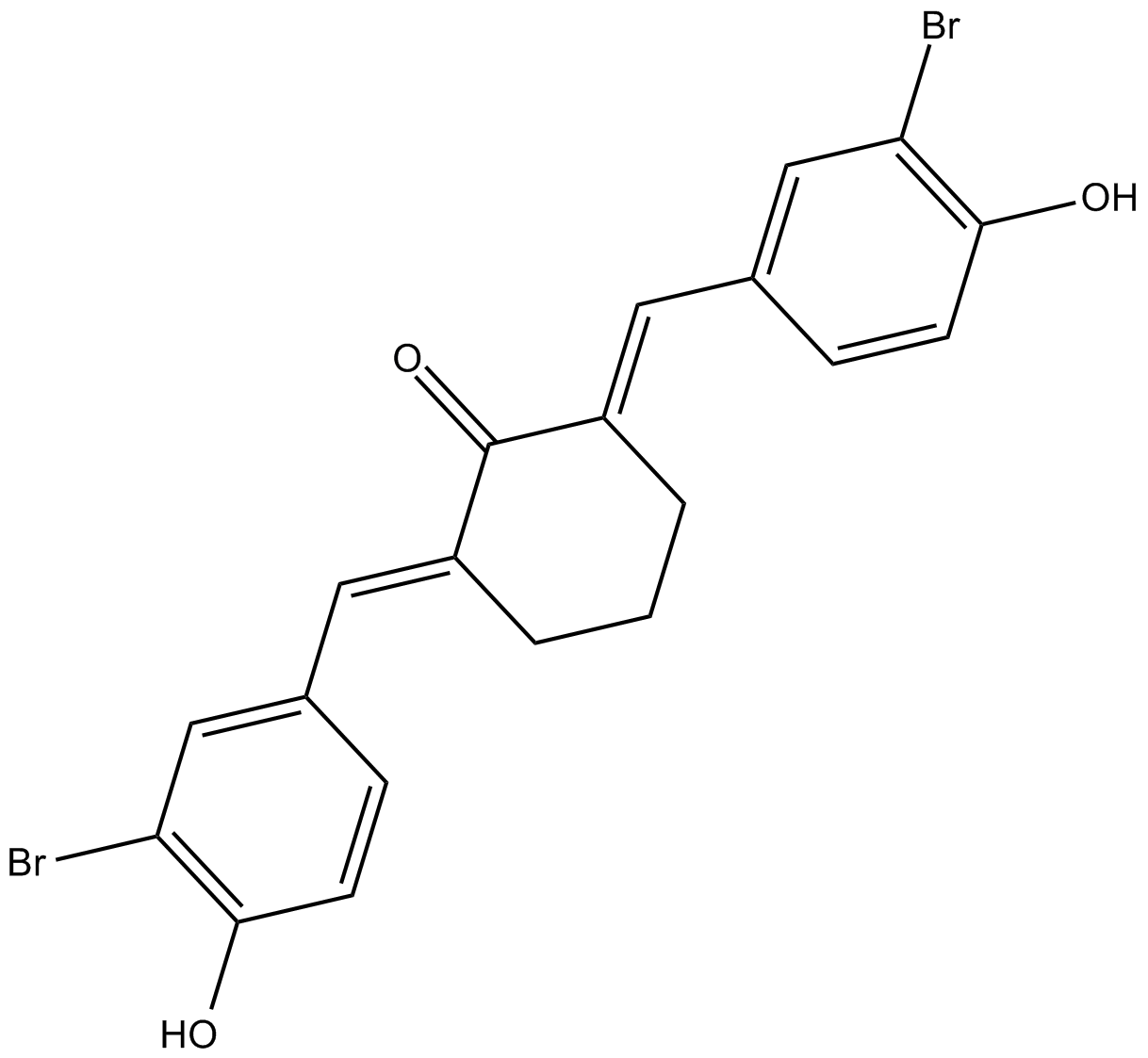
-
GC33422
HAT-IN-1
HAT-IN-1 is an inhibitor of HAT, used in the research of cancer.
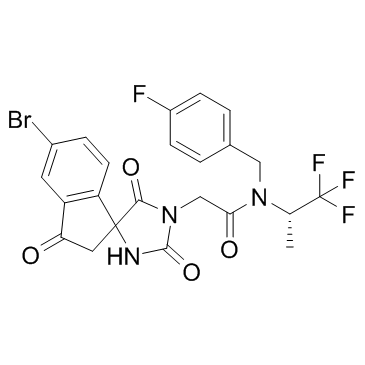
-
GC43806
HC Toxin
HC Toxin is a cell-permeable, reversible inhibitor of histone deacetylases (HDACs) (IC50 = 30 nM).
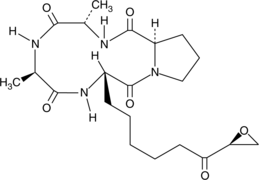
-
GC19190
HDAC-IN-4
HDAC-IN-4 is a potent, selective and orally active class I HDAC inhibitor with IC50s of 63 nM, 570 nM and 550 nM for HDAC1, HDAC2 and HDAC3, respectively. HDAC-IN-4 has no activity against HDAC class II. HDAC-IN-4 has antitumor activity.
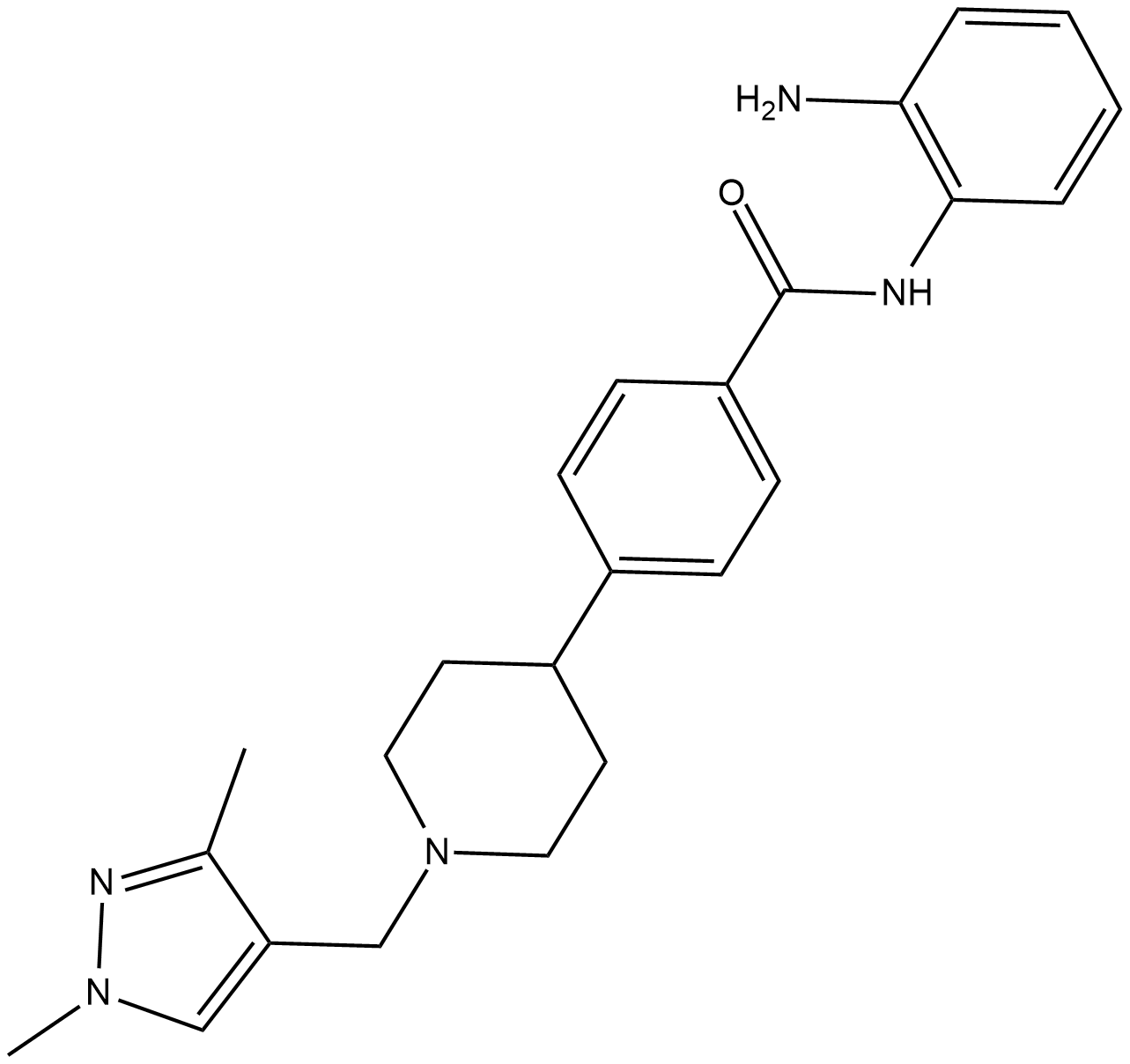
-
GC66052
HDAC-IN-40
HDAC-IN-40 is a potent alkoxyamide-based HDAC inhibitor with Ki values of 60 nM and 30 nM for HDAC2 and HDAC6, respectively. HDAC-IN-40 had antitumor effects.
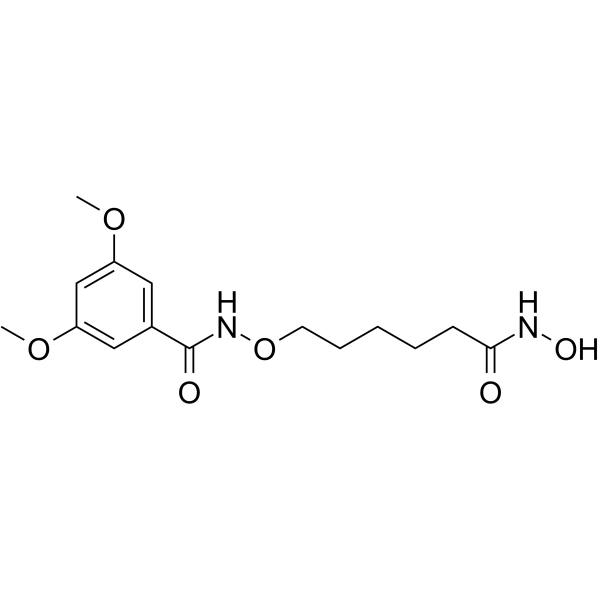
-
GC33395
HDAC-IN-5
HDAC-IN-5 is a histone deacetylase (HDAC) inhibitor.
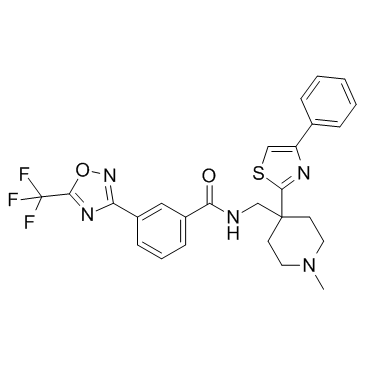
-
GC41495
HDAC3 Inhibitor
HDAC3 Inhibitor (compound 5) is a potent and selective HDAC3 inhibitor, with an IC50 of 5.96 nM.
-
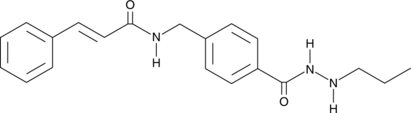
GC68010
HDAC3-IN-T247
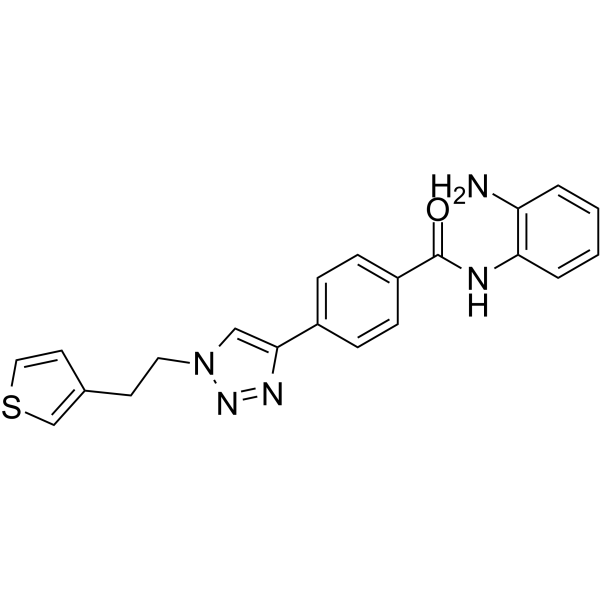
-
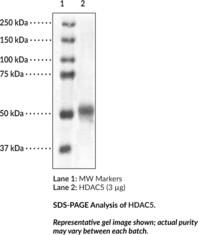
GC49693
HDAC5 (human, recombinant)
Active, pure human recombinant enzyme
-
GC41085
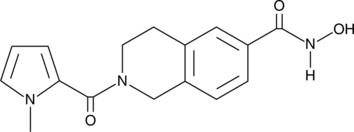
HDAC6 Inhibitor
HDAC6 Inhibitor is a potent and selective HDAC6 inhibitor (IC50=36 nM). HDAC6 Inhibitor weakly inhibits other HDAC isoforms. HDAC6 Inhibitor inhibits acyl-tubulin accumulation in cells with an IC50 value of 210 nM.
-
GC33317
HDAC6-IN-1
HDAC6-IN-1 is a potent and selective inhibitor for HDAC6 with an IC50 of 17 nM and shows 25-fold and 200-fold selectivity relative to HDAC1 (IC50=422 nM) and HDAC8 (IC50=3398 nM), respectively.
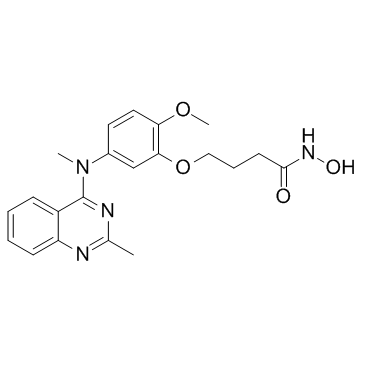
-
GC19189
HDAC8-IN-1
HDAC8-IN-1 is a HDAC8 inhibitor with an IC50 of 27.2 nM.
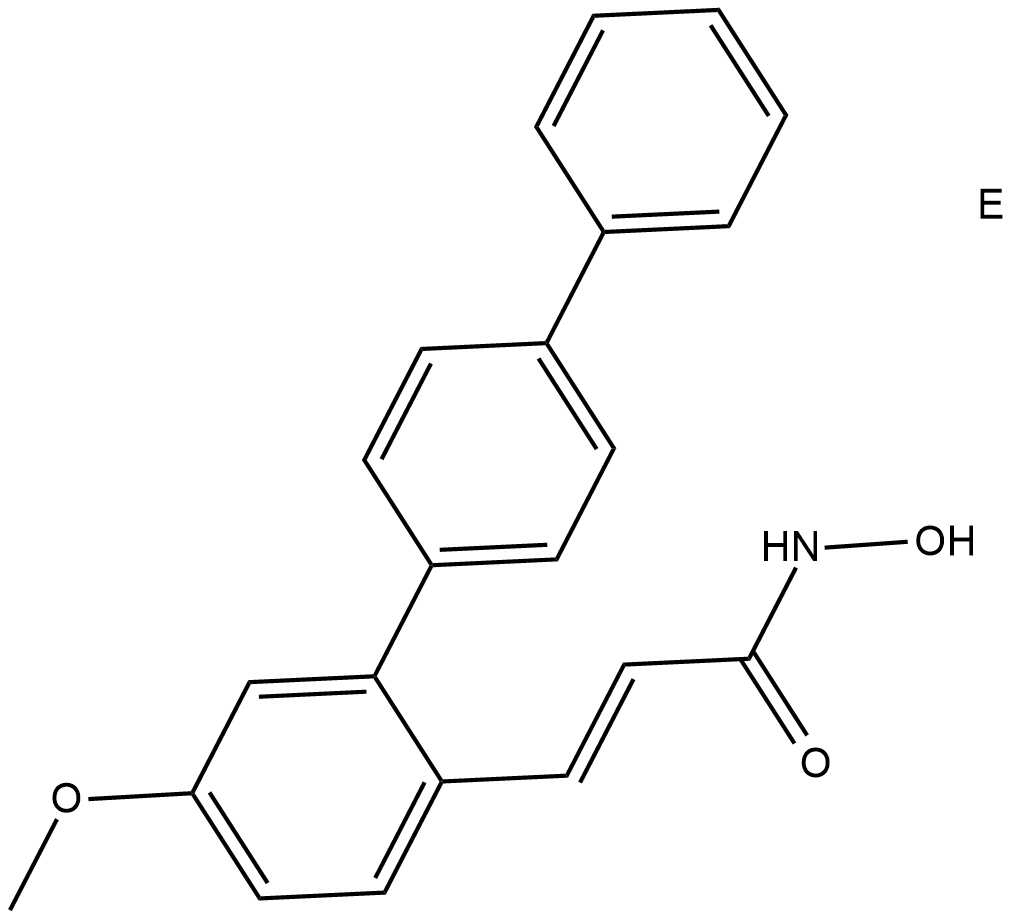
-
GC65460
HDACs/mTOR Inhibitor 1
HDACs/mTOR Inhibitor 1 is a dual Histone Deacetylases (HDACs) and mammalian target of Rapamycin (mTOR) target inhibitor for treating hematologic malignancies, with IC50s of 0.19 nM, 1.8 nM, 1.2 nM and >500 nM for HDAC1, HDAC6, mTOR and PI3Kα, respectively. HDACs/mTOR Inhibitor 1 stimulates cell cycle arrest in G0/G1 phase and induce tumor cell apoptosis with low toxicity in vivo.
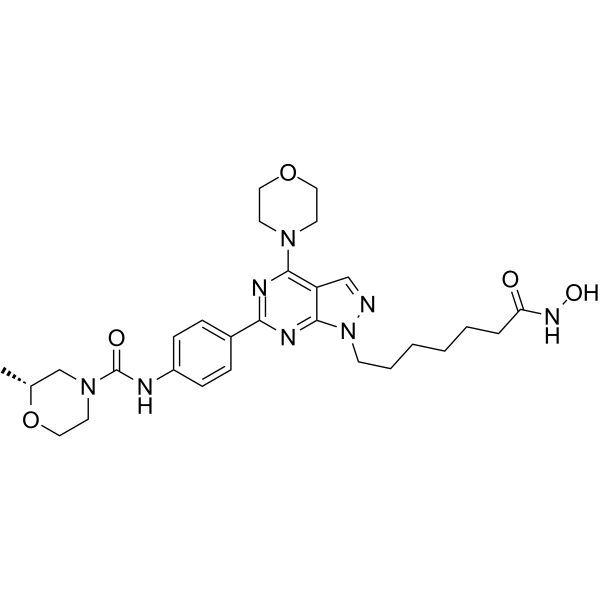
-
GC17196
Hesperadin
A multi-kinase inhibitor
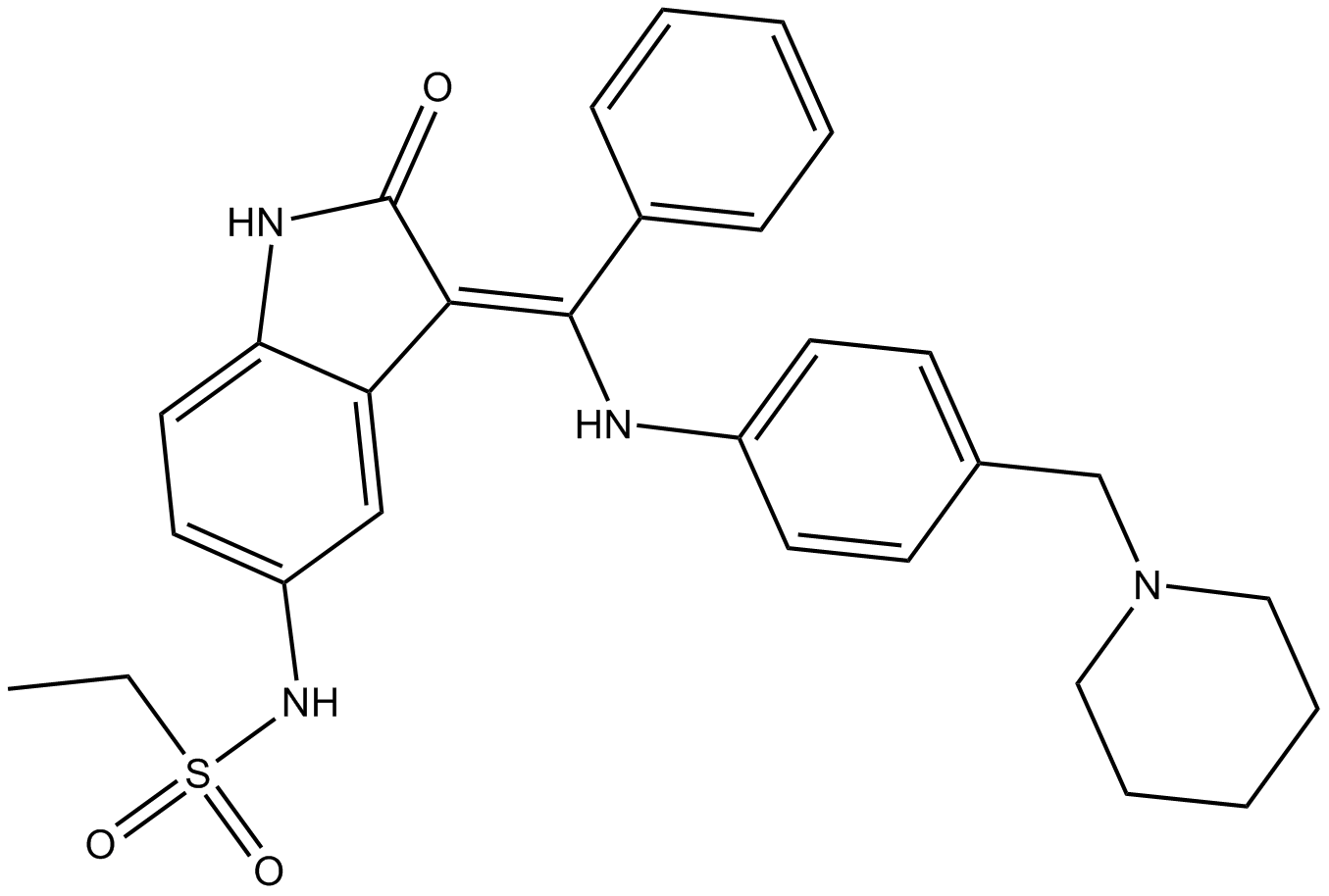
-
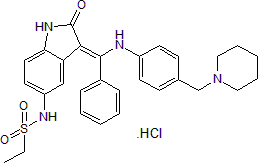
GC50050
Hesperadin hydrochloride
Potent Aurora kinase B inhibitor
-
GC39146
HIF-1 inhibitor-1
An inhibitor of HIF-1 signaling

-
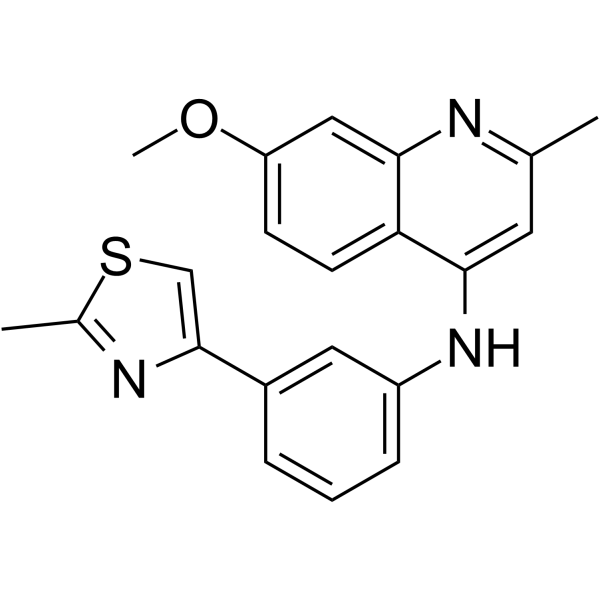
GC66464
HIF-1α-IN-2
HIF-1α-IN-2 is an effective HIF-1α inhibitor with anticancer potencies (IC50s of 28 nM and 15 nM in MDA-MB-231 and MiaPaCa-2 cells, respectively). HIF-1α-IN-2 suppresses HIF-1α expression by blocking transcription and protein translation.
-
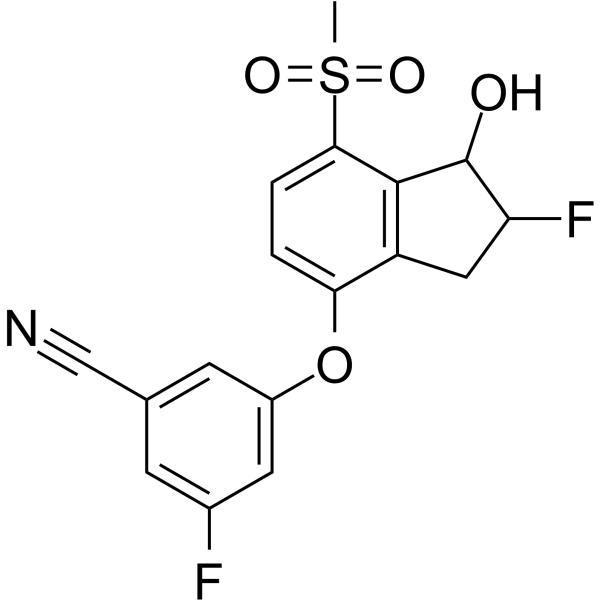
GC62584
HIF-2α-IN-2
HIF-2α-IN-2 is a hypoxia-inducible factors (HIF-2α) inhibitor extracted from patent WO2015035223A1, Compound 232, has an IC50 of 16 nM in scintillation proximity assay (SPA).
-
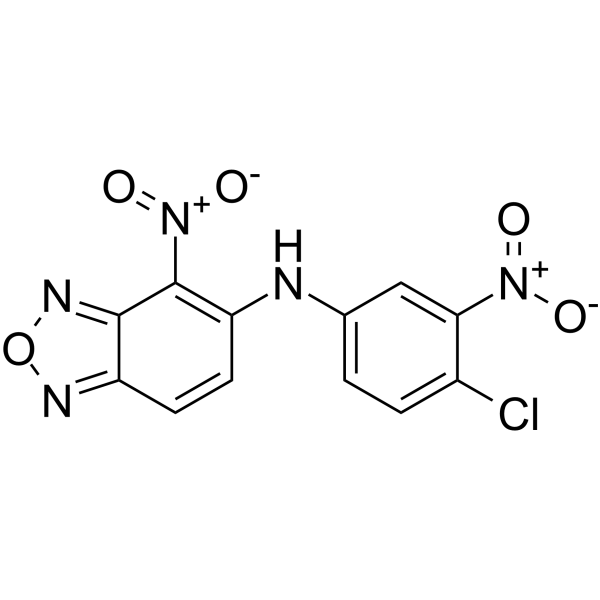
GC62418
HIF-2α-IN-3
HIF-2α-IN-3, an allosteric inhibitor of hypoxia inducible factor-2α (HIF-2α), exhibits an IC50 of 0.4 μM and a KD of 1.1 μM. Anticancer agent.
-
GC63678
HIF-2α-IN-4
HIF-2α-IN-4 is a potent inhibitor of hypoxia inducible factor-2α (HIF-2α) translation, with an IC50 of 5 μM. HIF-2α-IN-4 decreases both constitutive and hypoxia-induced HIF-2α protein expression. HIF-2α-IN-4 links its 5'UTR iron-responsive element to oxygen sensing.
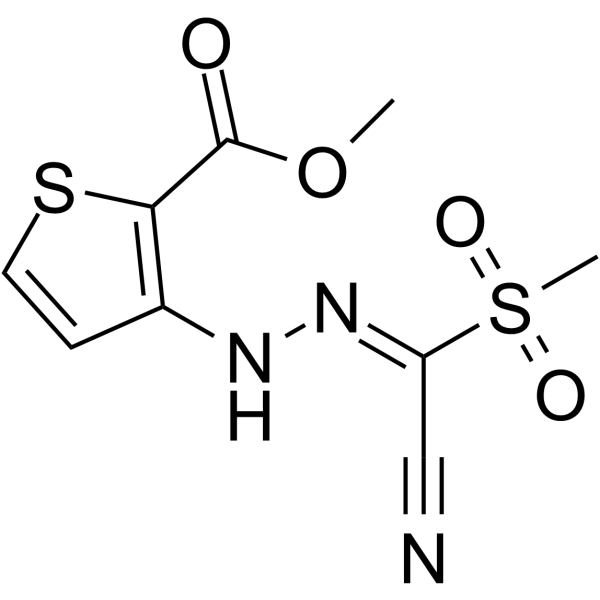
-
GC31358
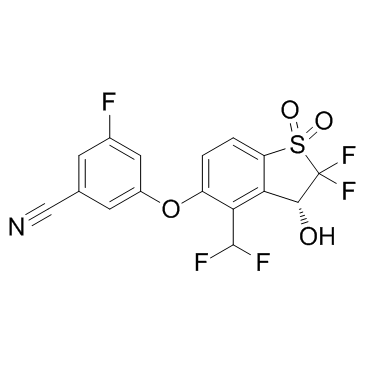
HIF-2α-IN-1
HIF-2α-IN-1 is a HIF-2α inhibitor has an IC50 of less than 500 nM in HIF-2α scintillation proximity assay.
-
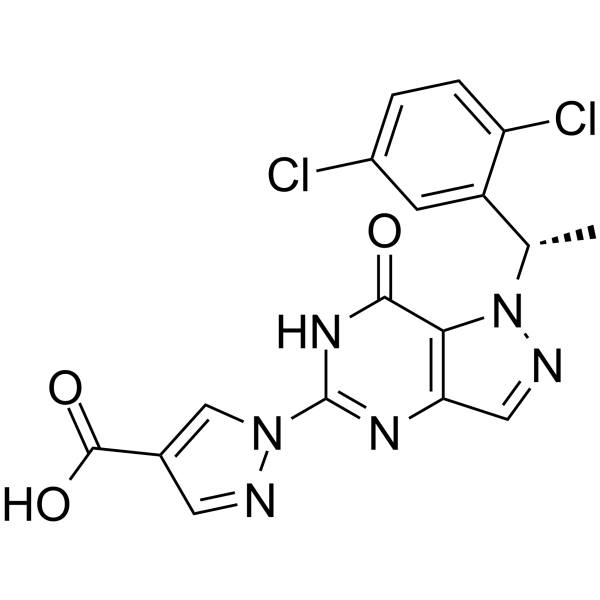
GC67941
HIF-PHD-IN-1
-
GC65570
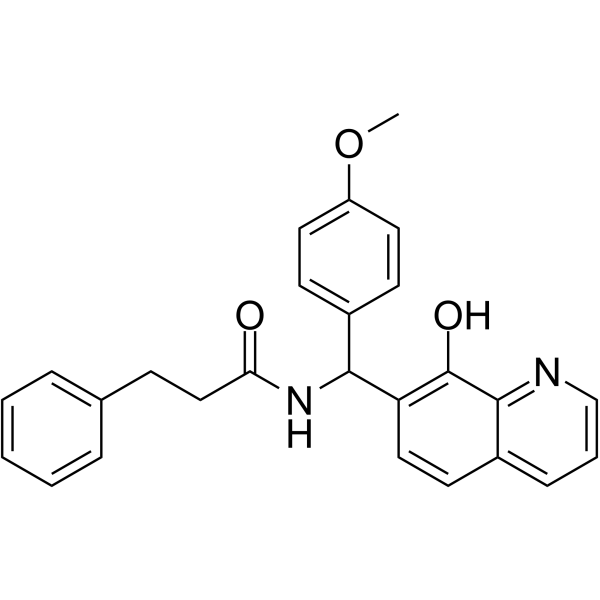
HIF1-IN-3
HIF1-IN-3 (compound F4) is a potent HIF1 inhibitor with an EC50 value of 0.9 μM. HIF1-IN-3 can be used for researching anticancer.
-
GC11302
Hinokitiol
A tropolone with diverse biological activities

-
GC12359
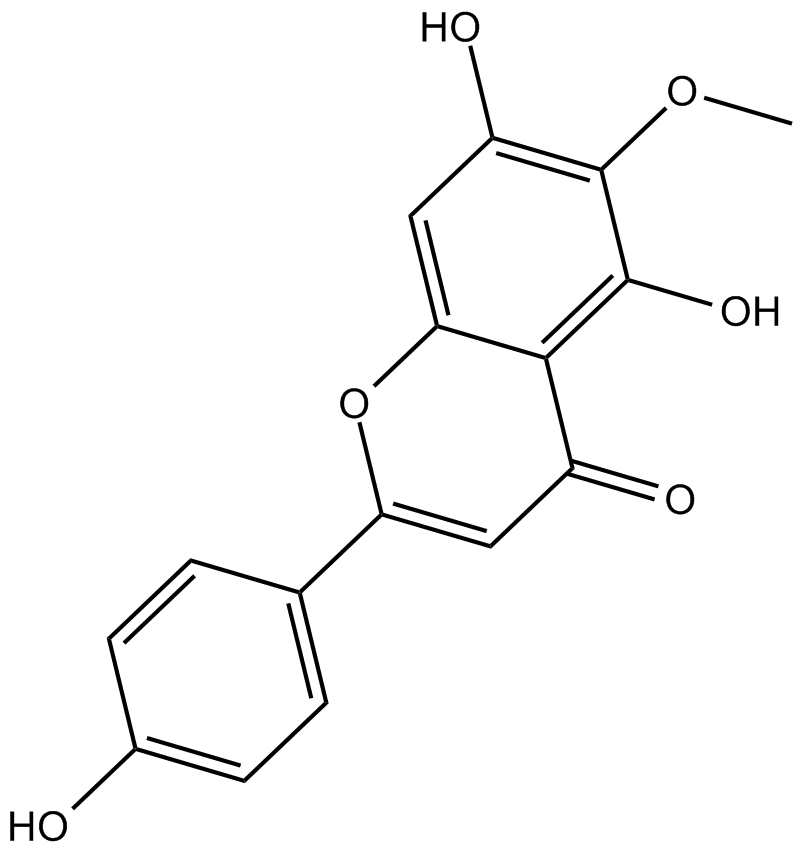
Hispidulin
Partial positive allosteric modulator at the benzodiazepine receptor
-
GC43831
Histone H3 (21-44)-GK-biotin (trifluoroacetate salt)
Histone H3 (21-44)-GK-biotin is a peptide fragment of histone H3 that corresponds to amino acid residues 22-45 of the human histone H3.1 and 3.2 sequences and is biotinylated via a C-terminal GK linker.

-
GC43832
Histone H3 (21-44)-GK-biotin amide (trifluoroacetate salt)
Histone H3 (21-44)-GK-biotin is a peptide fragment of histone H3 that corresponds to amino acid residues 22-45 of the human histone H3.3 sequence and is biotinylated via a C-terminal GK linker.
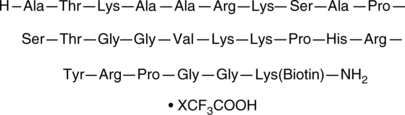
-
GC52479
Histone H3 (Citrullinated R26) (21-44)-GGK-biotin Peptide (trifluoroacetate salt)
A biotinylated peptide fragment of histone H3
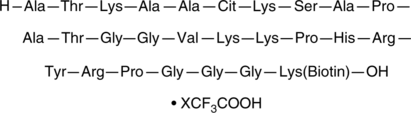
-
GC43846
Histone H3K27Me1 (23-34) (trifluoroacetate salt)
Histone H3K27Me1 (23-34) is a peptide fragment of histone H3 that corresponds to amino acid residues 24-35 of the human histone H3.1 and H3.2 sequences.
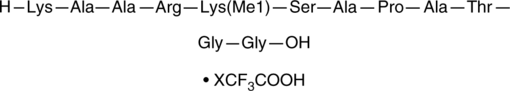
-
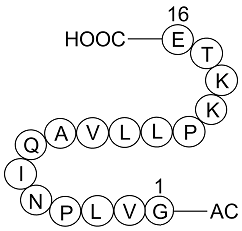
GP10020
Histone-H2A-(107-122)-Ac-OH
Histone-H2A peptide
-
GC33211
HJB97
HJB97 is a high-affinity BET inhibitor with Kis of 0.9 nM (BRD2 BD1), 0.27 nM (BRD2 BD2), 0.18 nM (BRD3 BD1), 0.21 nM (BRD3 BD2), 0.5 nM (BRD4 BD1), 1.0 nM (BRD4 BD2), respectively. HJB97 is employed for the design of potential PROTAC BET degrader and has antitumor activity.
-
GC17023
HLCL-61
HLCL-61 is a first-in-class inhibitor of protein arginine methyltransferase 5 (PRMT5).
-
GC12334
HNHA
HDAC inhibitor
-
GC11574
HPOB
HDAC6 inhibitor, potent and selective
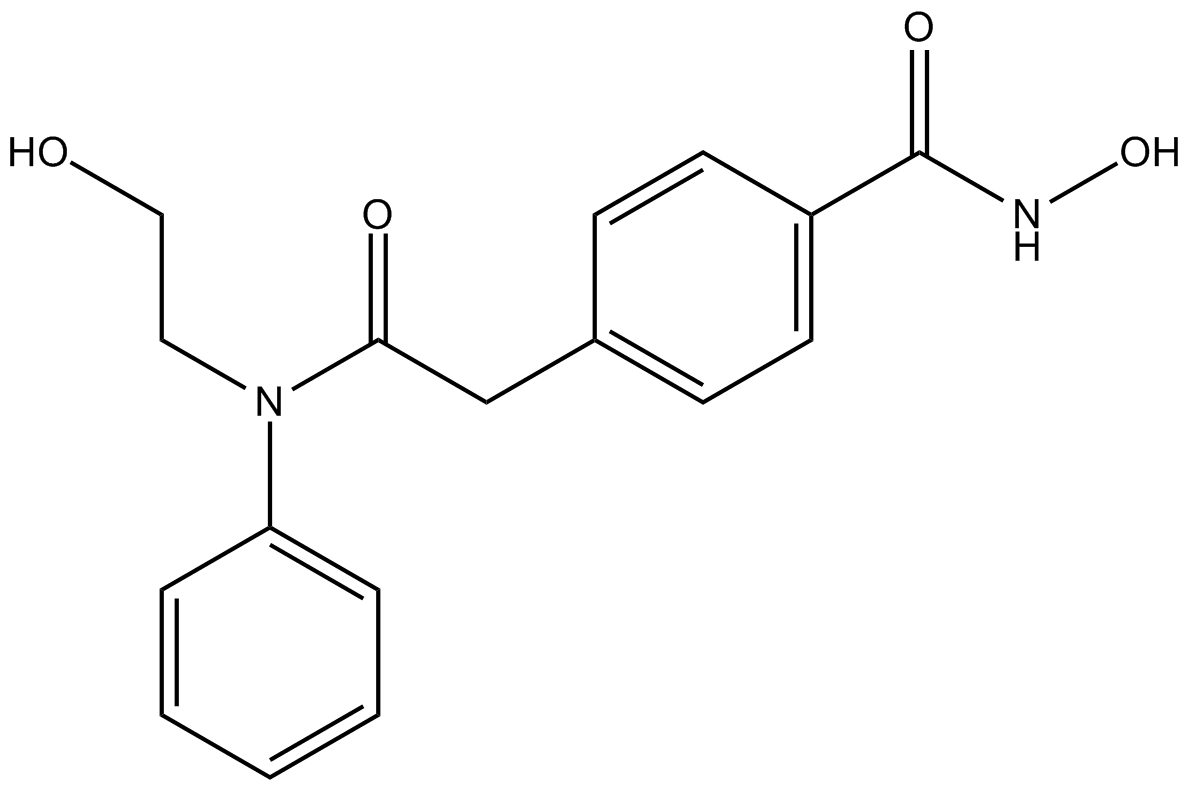
-
GC11767
Hydralazine HCl
Hydralazine HCl is a orally active antihypertensive agent, reduces peripheral resistance directly by relaxing the smooth muscle cell layer in arterial vessel.
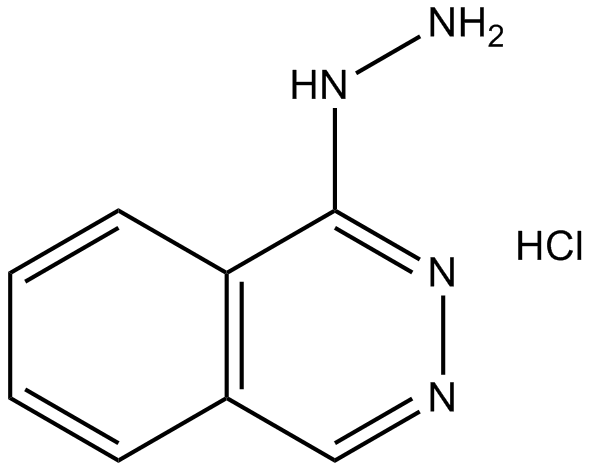
-
GC60919
Hydroxycitric acid tripotassium hydrate
Hydroxycitric acid tripotassium hydrate (Potassium citrate monohydrate) is the major active ingredient of Garcinia cambogia.
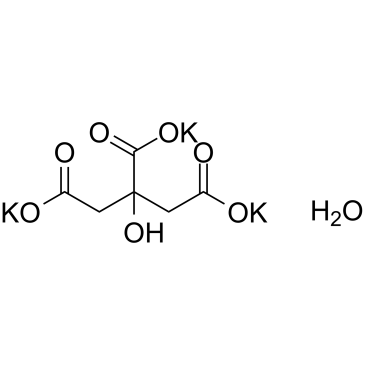
-
GC50070
I-BET 151 dihydrochloride
BET bromodomain inhibitor; also promotes differentiation of hiPSCs into megakaryocytes
-
GC13187
I-BET 151 hydrochloride
BET bromodomain inhibitor
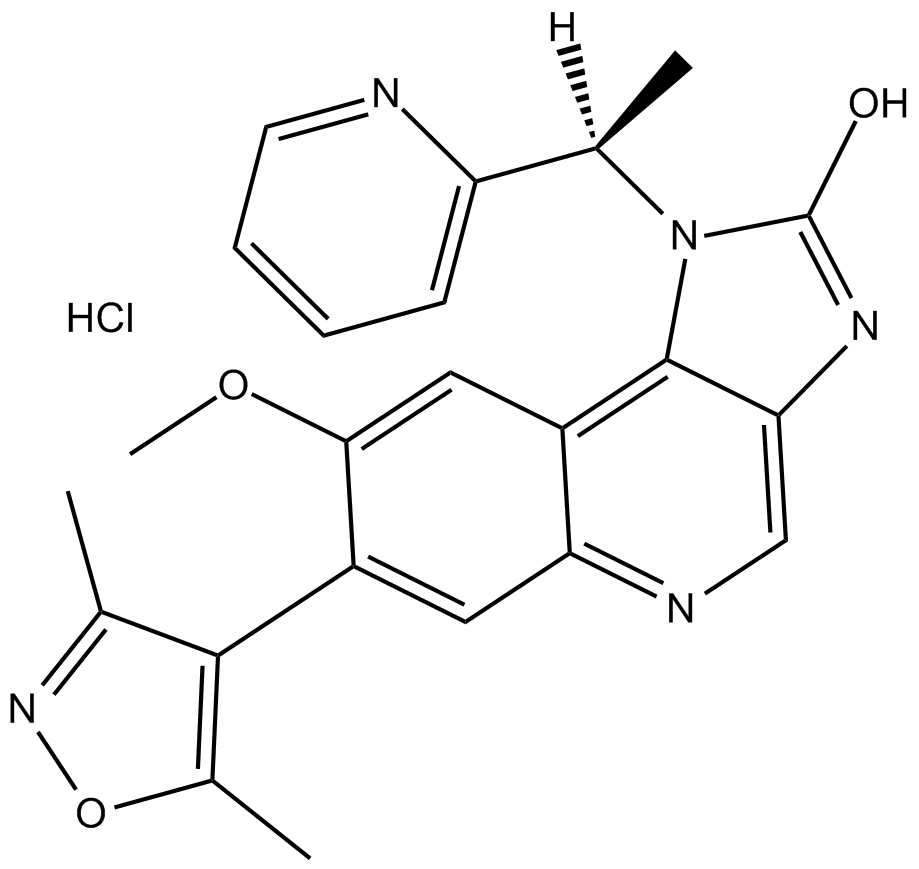
-
GC17073
I-BET-762
I-BET-762 (I-BET762; GSK525762) is a BET bromodomain inhibitor with IC50 of 32.5-42.5 nM.
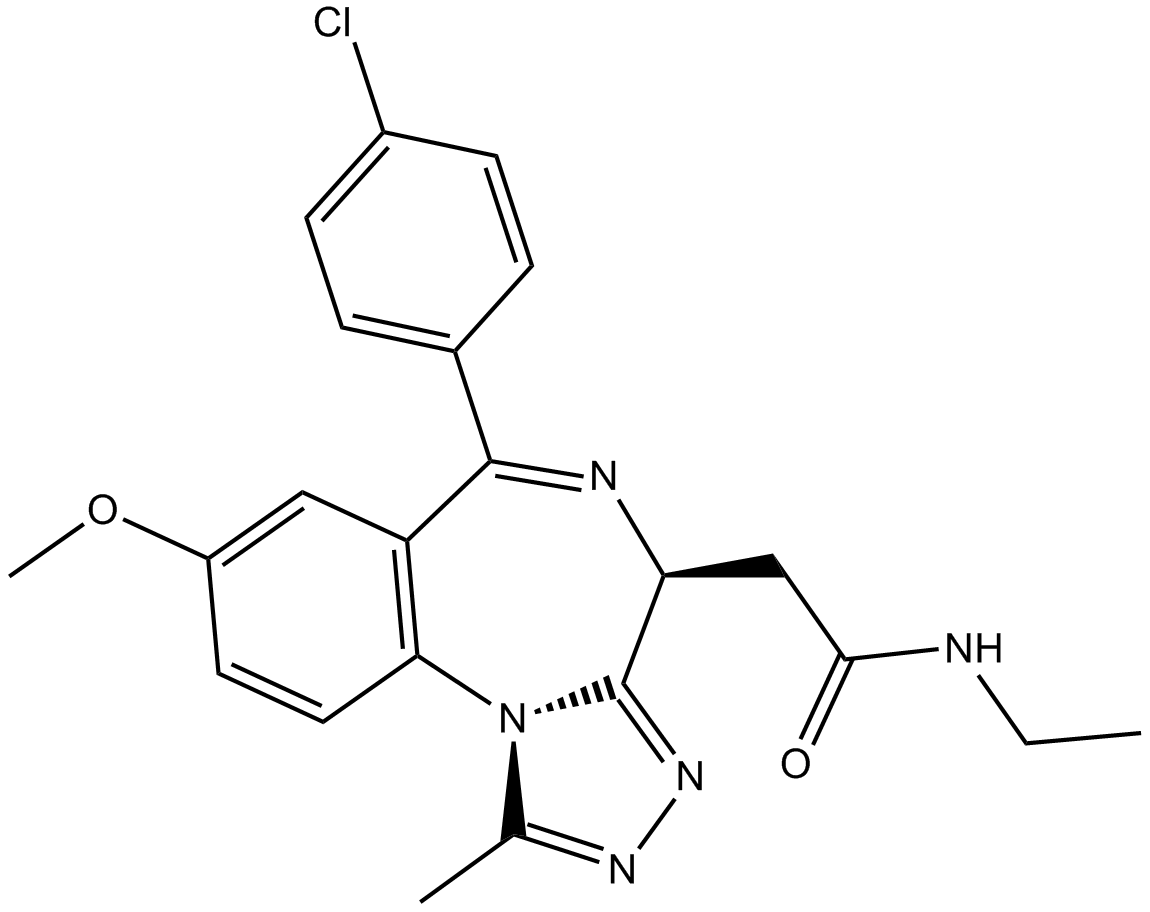
-
GC15747
I-BET151 (GSK1210151A)
I-BET151 (GSK1210151A) (GSK1210151A) is a BET bromodomain inhibitor which inhibits BRD4, BRD2, and BRD3 with pIC50 of 6.1, 6.3, and 6.6, respectively.
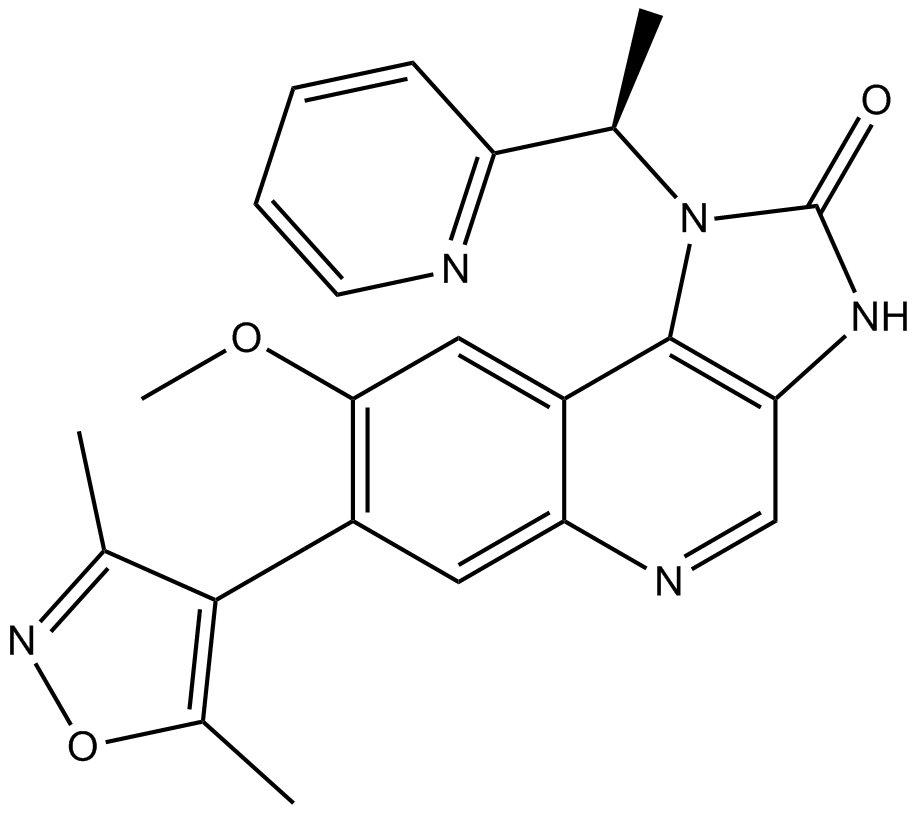
-
GC64297
I-BET567
I-BET567 is a potent and orally active inhibitor of pan-BET candidate with pIC50s of 6.9 and 7.2 for BRD4 BD1 and BD2, respectively.
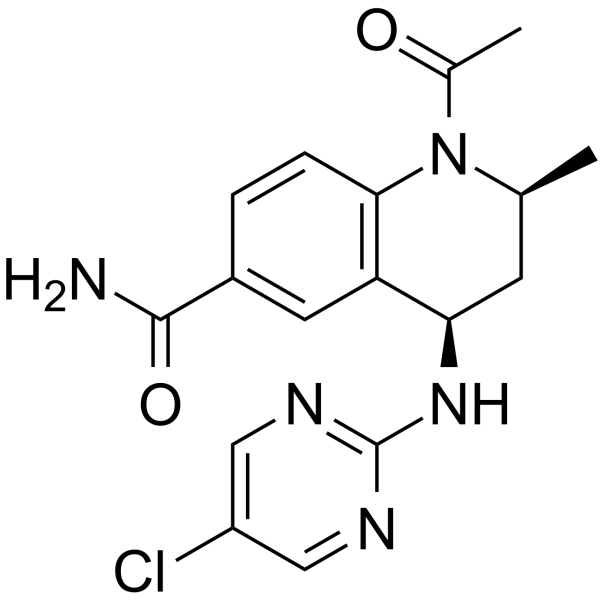
-
GC12588
I-BRD9
BRD9 inhibitor
-
GC17944
I-CBP 112
A p300 and CBP inhibitor
-
GC34078
I-CBP112
I-CBP112 is a specific and potent acetyl-lysine competitive protein-protein interaction inhibitor, that inhibits the CBP/p300 bromodomains, enhances acetylation by p300.