Tyrosine Kinase
- IGFBR(1)
- Bcr-Abl(73)
- Ack1(4)
- Axl(6)
- ALK(61)
- BMX Kinase(4)
- Broad Spectrum Protein Kinase Inhibitor(11)
- c-FMS(35)
- c-Kit(65)
- c-MET(91)
- c-RET(7)
- CSF-1R(3)
- DDR1/DDR2 Receptor(1)
- EGFR(251)
- EphB4(2)
- FAK(34)
- FGFR(104)
- FLT3(102)
- HER2(19)
- IGF1R(33)
- Insulin Receptor(45)
- IRAK(32)
- ITK(12)
- Lck(2)
- LRRK2(23)
- PDGFR(117)
- PTKs/RTKs(5)
- Pyk2(6)
- ROR(41)
- Spleen Tyrosine Kinase (Syk)(35)
- Src(105)
- Tie-2 (3)
- Trk(37)
- VEGFR(203)
- Kinase(0)
- Discoidin Domain Receptor(15)
- DYRK(26)
- Ephrin Receptor(13)
- ROS(15)
- RET(34)
- TAM Receptor(28)
Products for Tyrosine Kinase
- Cat.No. Product Name Information
-
GC19187
H3B-6527
H3B-6527 (H3 Biomedicine) is a highly selective FGFR4 inhibitor with potent antitumour activity in FGF19 amplified cell lines and mice.
-
GC12461
HA-100 (hydrochloride)
inhibitor of protein kinases (PKs)
-
GC30762
Harmine (Telepathine)
A unique regulator of PPARγ expression
-
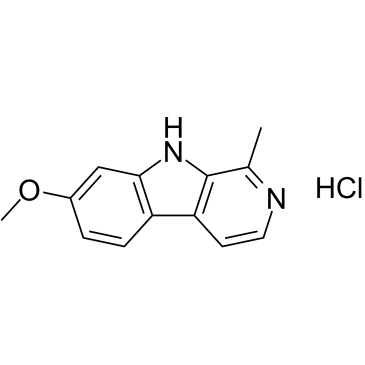
GC38413
Harmine hydrochloride
-
GC15674
HDS 029
HDS 029 (compound 29) is a potent tyrosine kinase inhibitor with IC50s of 0.3, 1.1, 0.5, 2.5, 24 nM for erbB1, erbB2, erbB4, EGF, HER, respectively.
-
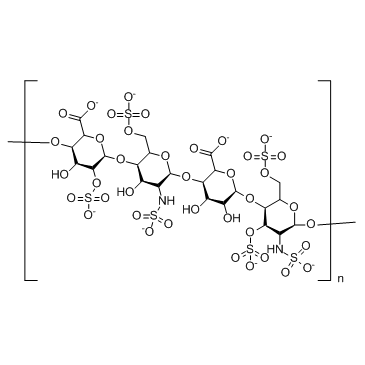
GC30767
Heparan Sulfate
Heparan sulfate (HS) is a complex, polyanionic polysaccharide ubiquitously expressed on cell surfaces and in the extracellular matrix.
-
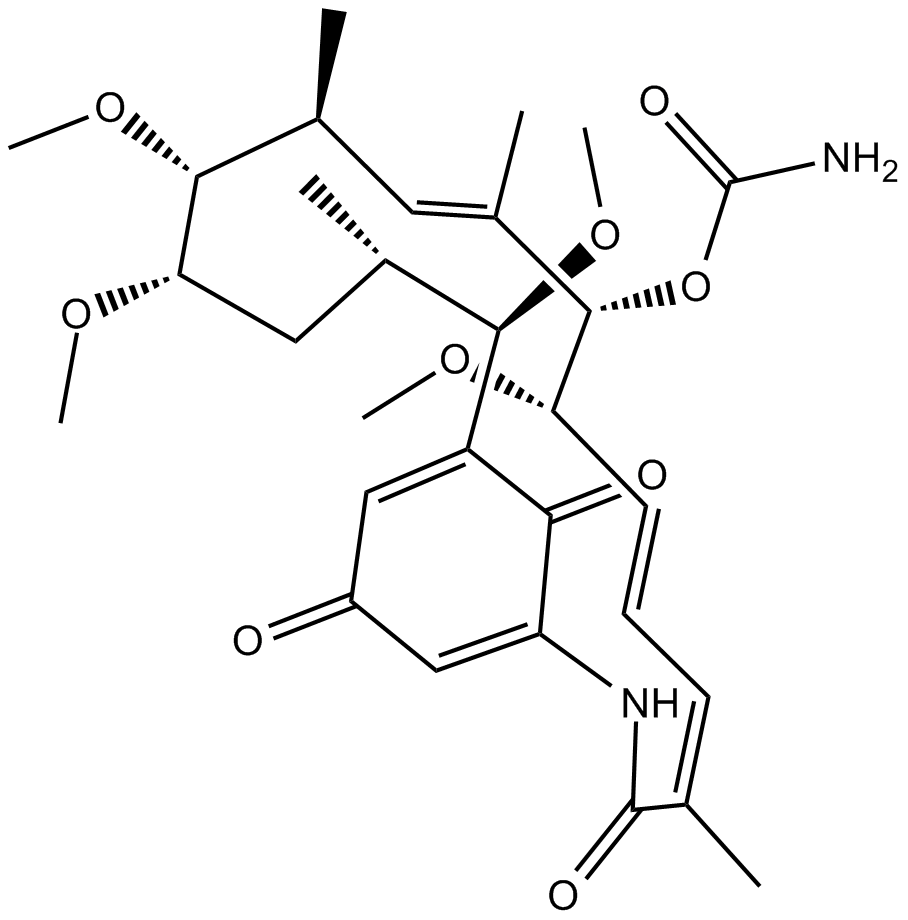
GC15000
Herbimycin A
antibiotic,Src family kinase inhibitor
-
GC13098
HG-10-102-01
LRRK2 inhibitor
-
GC36222
HG-14-10-04
An ALK inhibitor
-
GC50660
HIOC
HIOC is a potent and selective activator of TrkB (tropomyosin related kinase B) receptor.
-
GC65134
HIV-IN-6
HIV-IN-6 is a HIV-Ⅰ viral replication inhibitor by targeting Src family kinases (SFK) that interact with Nef protein of the virus, such as Hck.

-
GC14259
HKI 357
irreversible inhibitor of ErbB2 (HER2) and EGFR
-
GC63507
HM43239
HM43239 is an orally active and selective FLT3 inhibitor with IC50s of 1.1 nM, 1.8 nM and 1.0 nM for FLT3 WT, FLT3 internal tandem duplication (ITD) and FLT3 D835Y kinases, respectively. HM43239 inhibits the kinase activity of FLT3 as a reversible type I inhibitor and modulates p-STAT5, p-ERK, SYK, JAK1/2, and TAK1. HM43239 inhibits the proliferation and induces the apoptosis of leukemic cells.
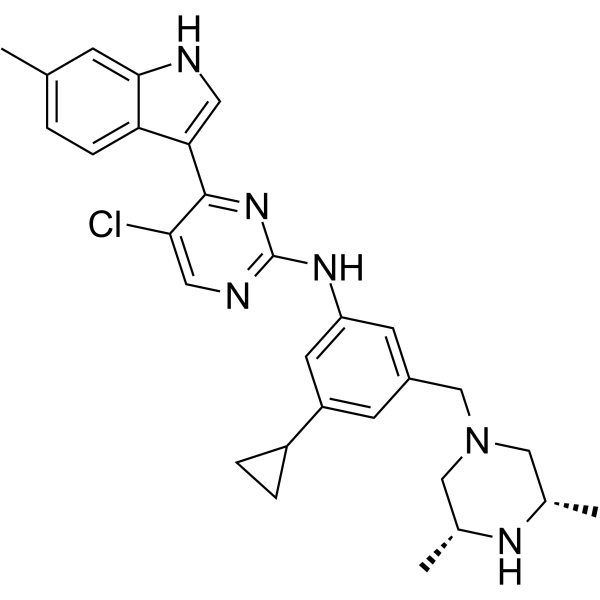
-
GC14654
HNGF6A
increases glucose stimulated insulin secretion and glucose metabolism
-
GC13988
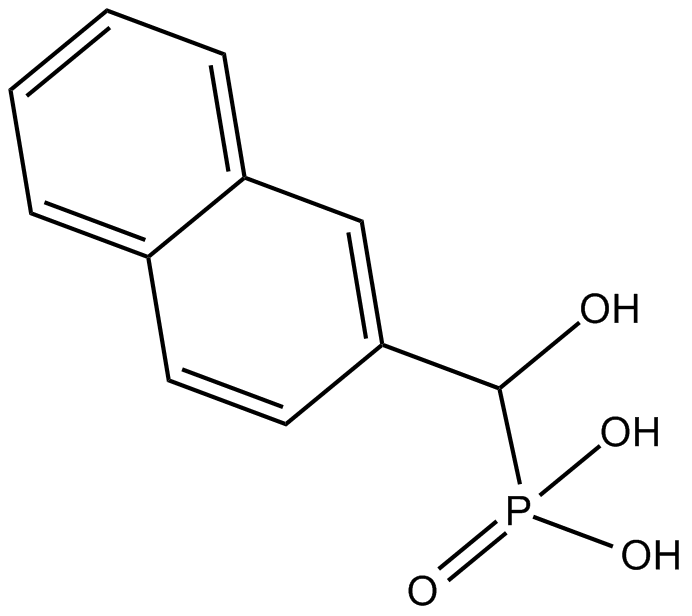
HNMPA
cell impermeable tyrosine kinase inhibitor
-
GC33053
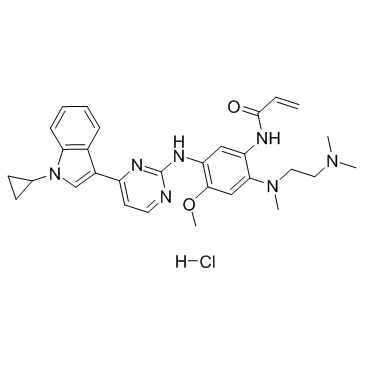
HS-10296 hydrochloride
Almonertinib (HS-10296) hydrochloride is an orally available, irreversible, third-generation EGFR tyrosine kinase inhibitor with high selectivity for EGFR-sensitizing and T790M resistance mutations. HS-10296 hydrochloride shows great inhibitory activity against T790M, T790M/L858R and T790M/Del19 (IC50: 0.37, 0.29 and 0.21 nM, respectively), and is less effective against wild type (3.39 nM). HS-10296 hydrochloride is used for the research of the non-small cell lung cancer.
-
GC68458
HS-243

-
GC65941
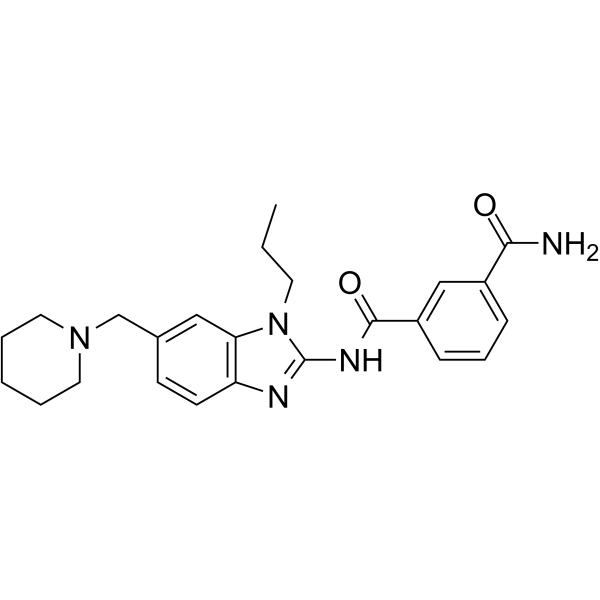
HS-276
HS-276 is an orally active, potent and highly selective TAK1 inhibitor, with a Ki of 2.5 nM. HS-276 shows significant inhibition of TAK1, CLK2, GCK, ULK2, MAP4K5, IRAK1, NUAK, CSNK1G2, CAMKKβ-1, and MLK1, with IC50 values of 8.25, 29, 33, 63, 125, 264, 270, 810, 1280, and 5585 nM, respectively. HS-276 can be used for rheumatoid arthritis (RA) research.
-
GC62429
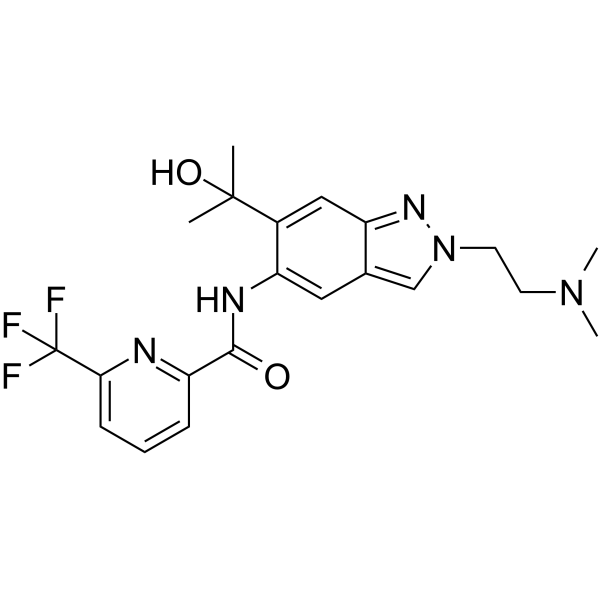
HS271
HS271 is a highly potent, orally active and selective IRAK4 inhibitor, with an IC50 of 7.2 μM.
-
GC32963
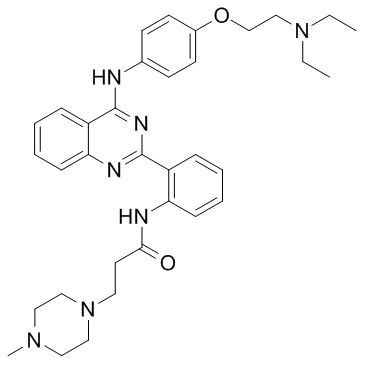
hVEGF-IN-1
hVEGF-IN-1, a quinazoline derivative, could specifically bind to the G-rich sequence in the internal ribosome entry site A (IRES-A) and destabilize the G-quadruplex structure. hVEGF-IN-1 binds to the IRES-A (WT) with a Kd of 0.928 μM in SPR experiments. hVEGF-IN-1 could hinder tumor cells migration and repress tumor growth by decreasing VEGF-A protein expression.
-
GC43885
Hypothemycin
A resorcylic acid lactone polyketide

-
GC63297
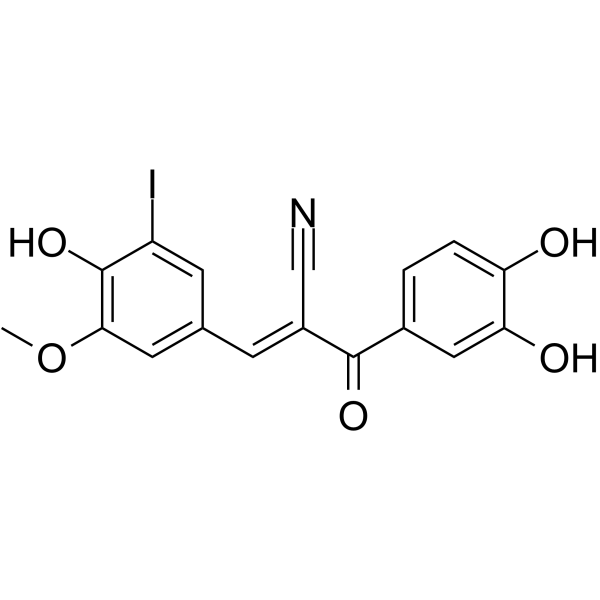
I-OMe-Tyrphostin AG 538
I-OMe-Tyrphostin AG 538 (I-OMe-AG 538) is a specific inhibitor of IGF-1R (insulin-like growth factor-1 receptor tyrosine kinase).
-
GC17982
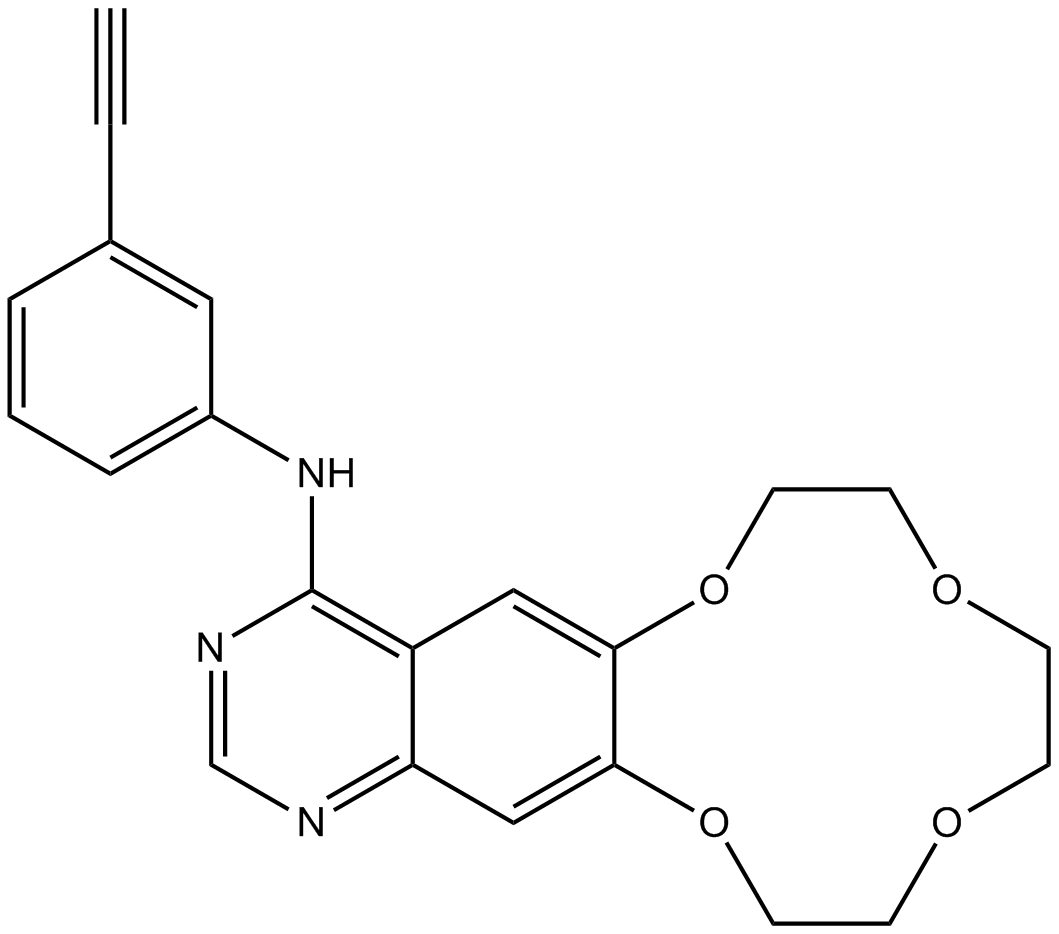
Icotinib
EGFR tyrosine kinase inhibitor
-
GC16244
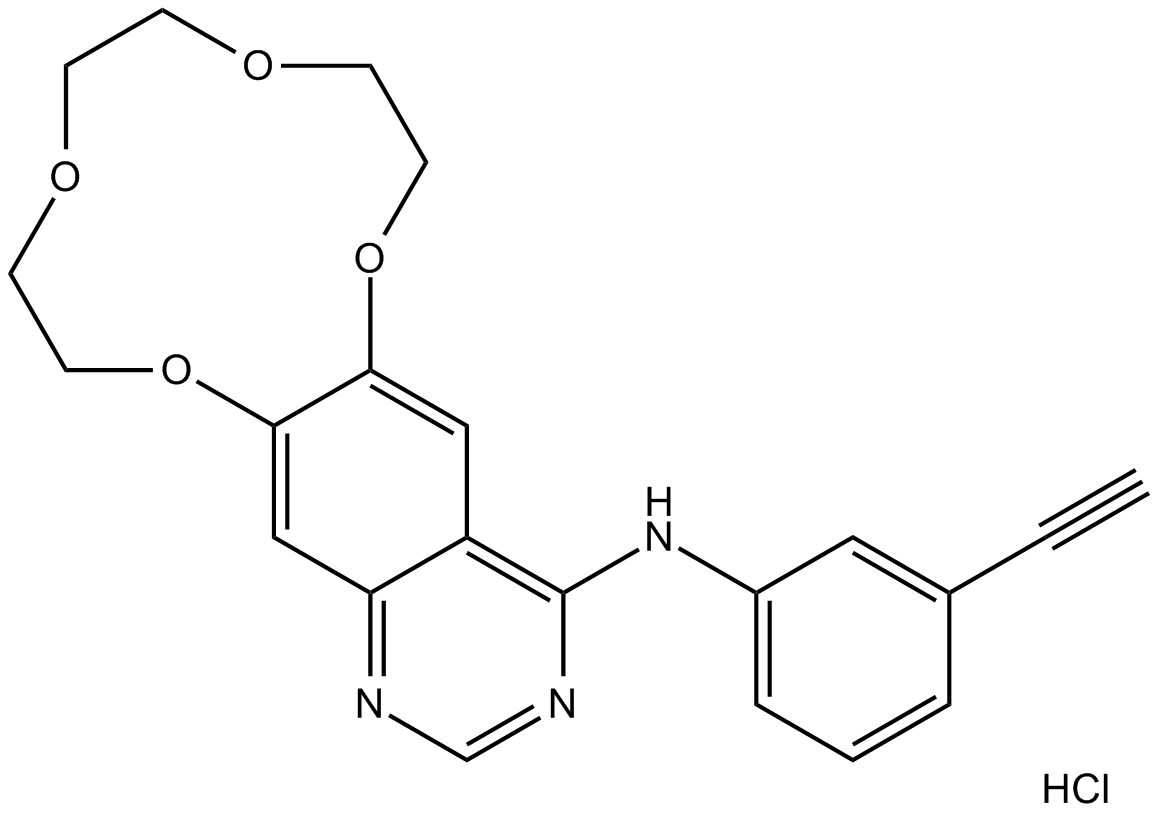
Icotinib Hydrochloride
An EGFR inhibitor
-
GC15647
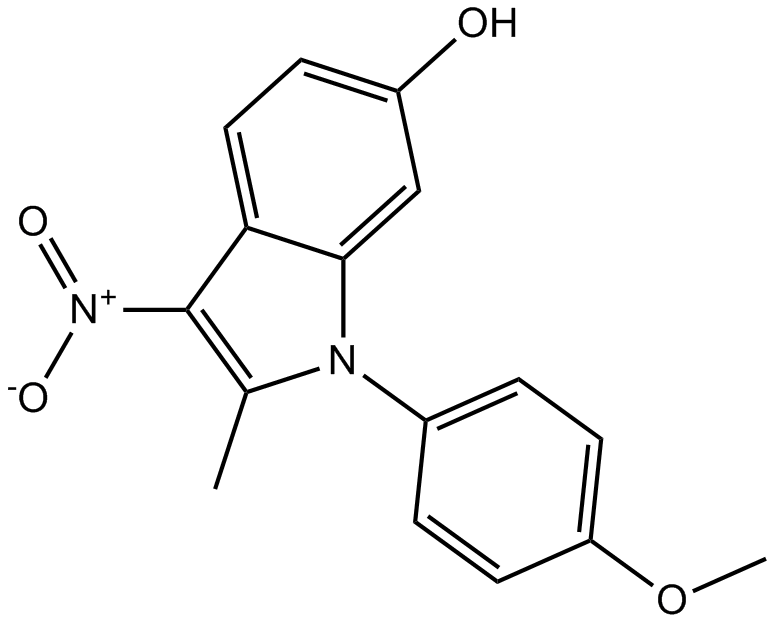
ID-8
DYRK inhibitor
-
GC63606
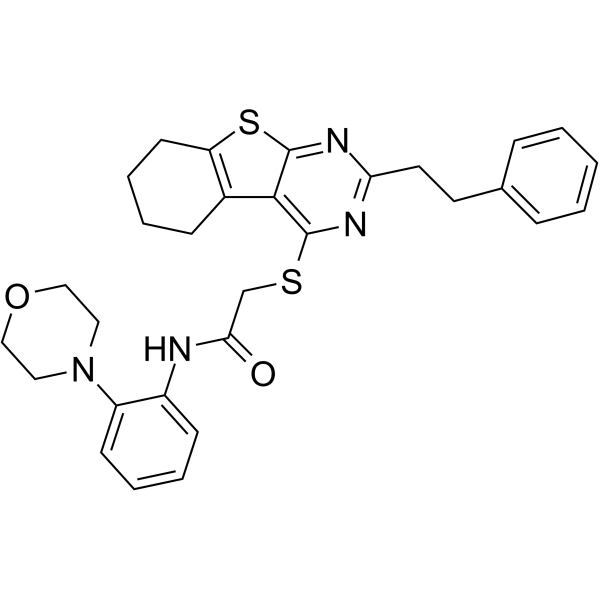
iHCK-37
iHCK-37 (ASN05260065) is a potent and specific Hck inhibitor with a Ki value of 0.22 μM.
-
GC12370
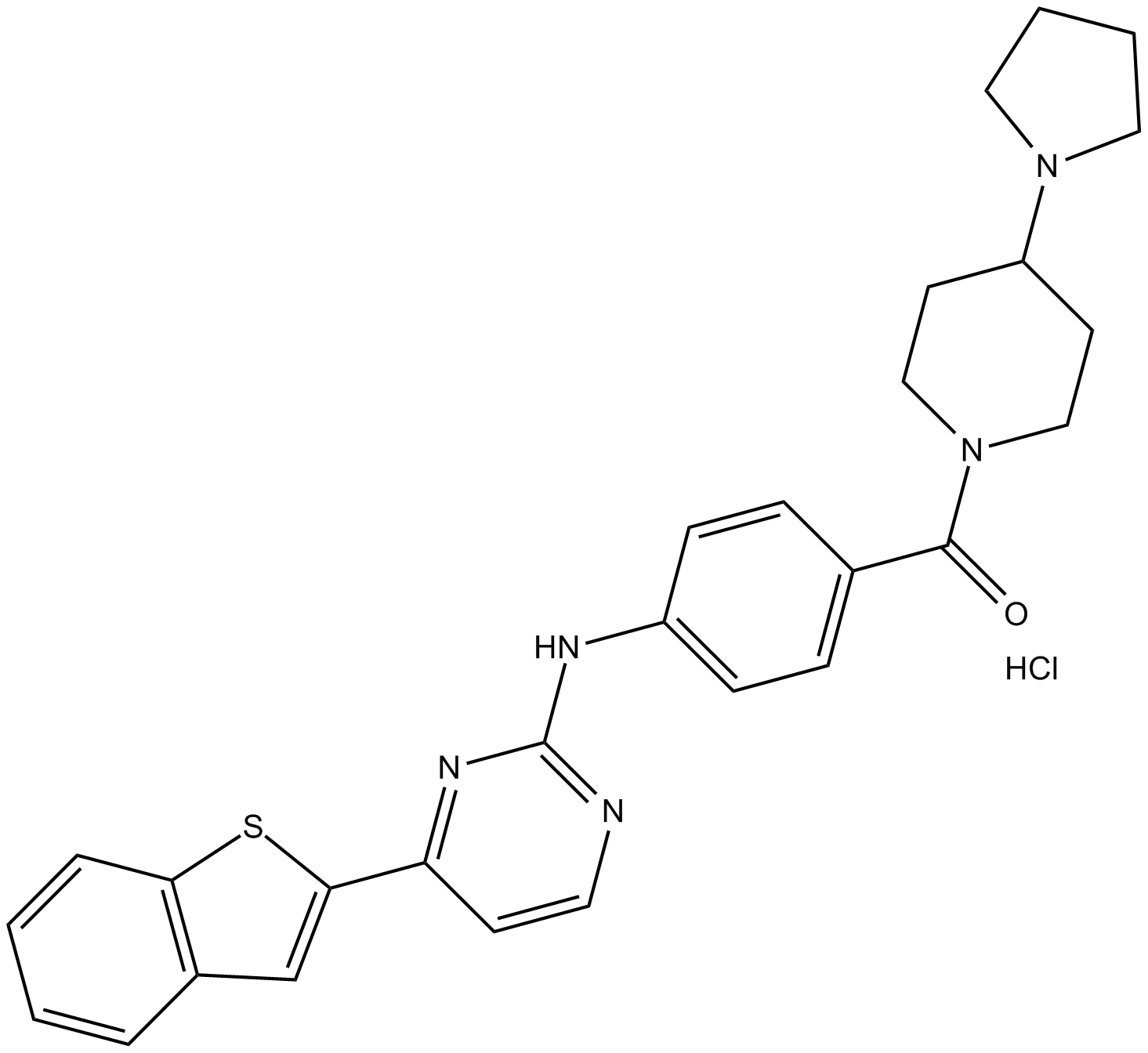
IKK-16 (hydrochloride)
IκB kinases (IKKs) inhibitor
-
GC12180
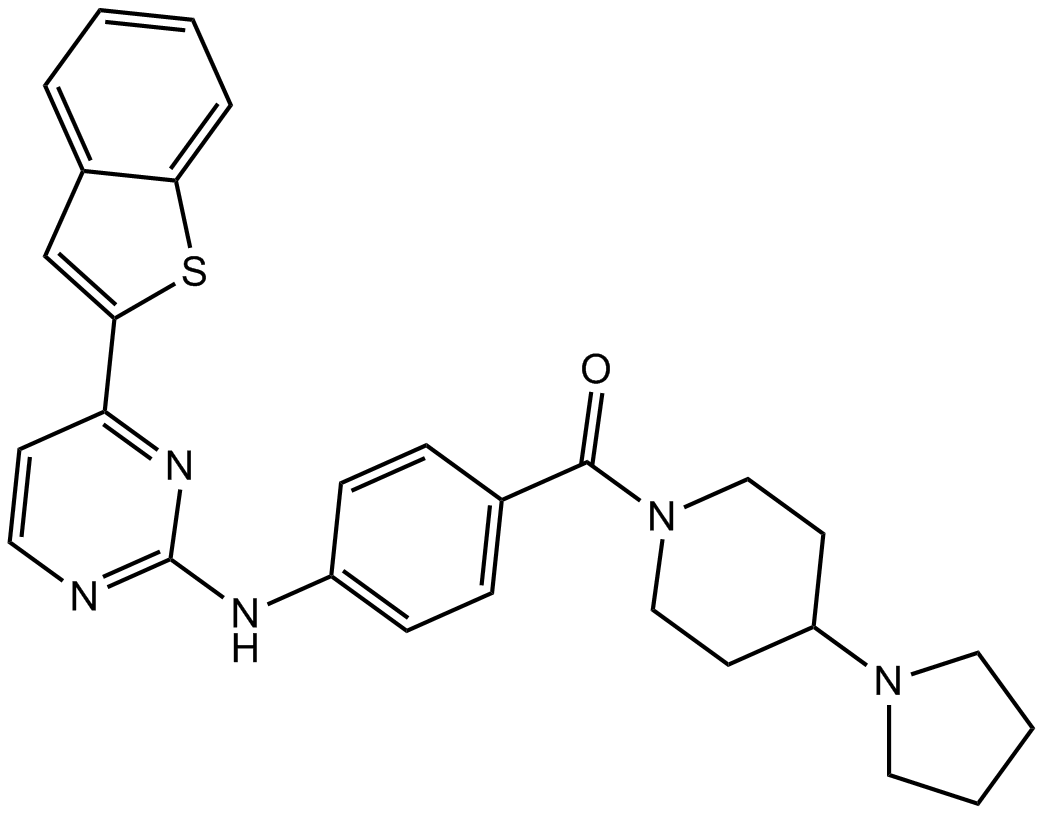
IKK-16 (IKK Inhibitor VII)
IKK-16 (IKK Inhibitor VII) is a selective IκB kinase (IKK) inhibitor for IKK2, IKK complex and IKK1 with IC50s of 40 nM, 70 nM and 200 nM, respectively.
-
GC34159
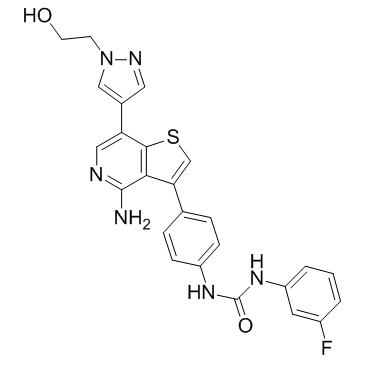
Ilorasertib (ABT-348)
Ilorasertib (ABT-348) (ABT-348) is a potent, orally active and ATP-competitive aurora inhibitor with IC50s of116, 5, 1 nM for aurora A, aurora B, aurora C, respectively. Ilorasertib (ABT-348) also is a potent VEGF, PDGF inhibitor. Ilorasertib (ABT-348) has the potential for the research of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS).
-
GC38519
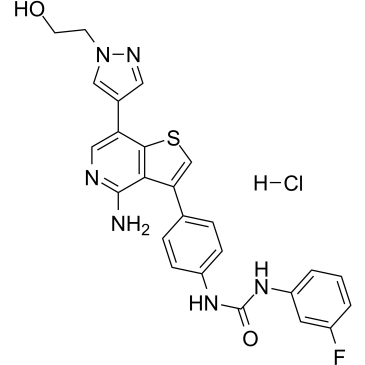
Ilorasertib hydrochloride
Ilorasertib (ABT-348) hydrochloride is a potent, orally active and ATP-competitive aurora inhibitor with IC50s of116, 5, 1 nM for aurora A, aurora B, aurora C, respectively. Ilorasertib hydrochloride also is a potent VEGF, PDGF inhibitor. Ilorasertib hydrochloride has the potential for the research of acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS).
-
GC10314
Imatinib (STI571)
Imatinib (STI571) (STI571) is an orally bioavailable tyrosine kinases inhibitor that selectively inhibits BCR/ABL, v-Abl, PDGFR and c-kit kinase activity. Imatinib (STI571) (STI571) works by binding close to the ATP binding site, locking it in a closed or self-inhibited conformation, therefore inhibiting the enzyme activity of the protein semicompetitively. Imatinib (STI571) also is an inhibitor of SARS-CoV and MERS-CoV.
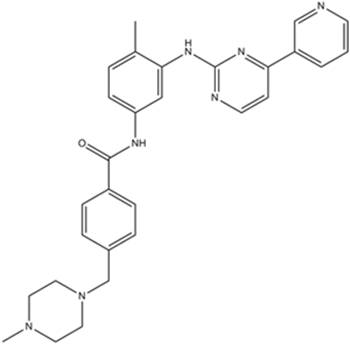
-
GC60930
Imatinib D4
Imatinib D4 (STI571 D4) is a deuterium labeled Imatinib (STI571). Imatinib is an orally bioavailable tyrosine kinases inhibitor that selectively inhibits BCR/ABL, v-Abl, PDGFR and c-kit kinase activity.
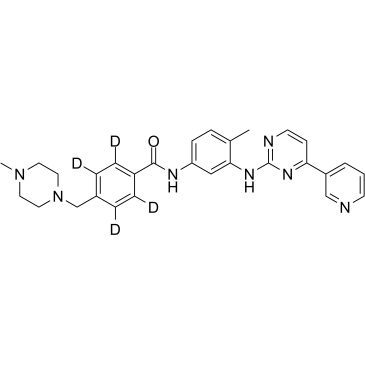
-
GC39612
Imatinib D8
Imatinib D8 (STI571 D8) is a deuterium labeled Imatinib (STI571). Imatinib is an orally bioavailable tyrosine kinases inhibitor that selectively inhibits BCR/ABL, v-Abl, PDGFR and c-kit kinase activity.
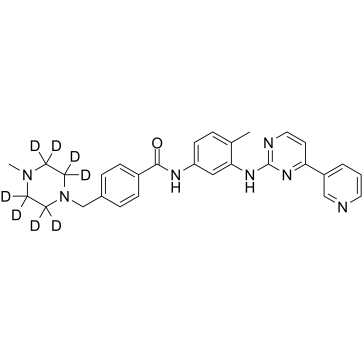
-
GC15263
Imatinib hydrochloride
V-Abl/c-Kit/PDGFR inhibitor
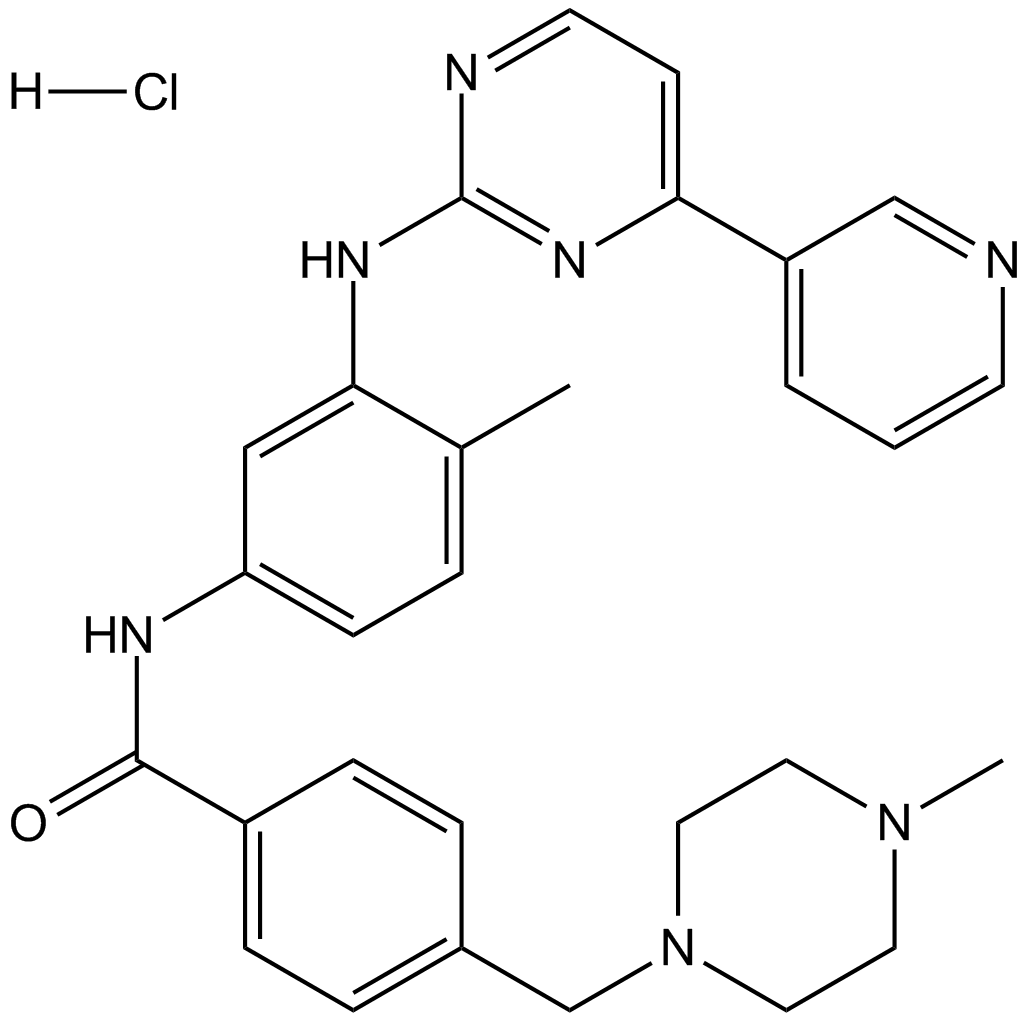
-
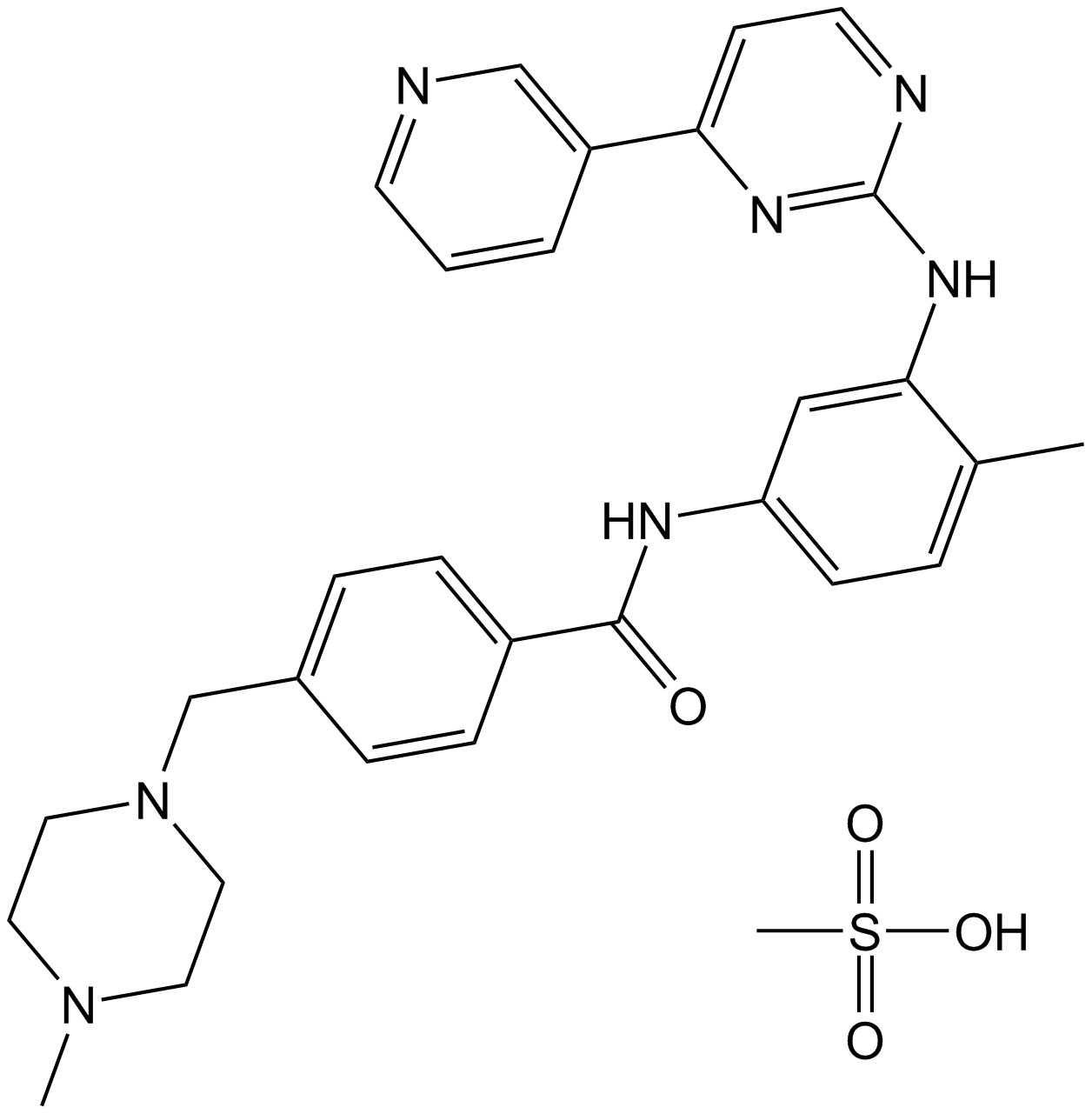
GC11759
Imatinib Mesylate (STI571)
Imatinib Mesylate (STI571) (STI571 Mesylate) is a tyrosine kinases inhibitor that inhibits c-Kit, Bcr-Abl, and PDGFR (IC50=100 nM) tyrosine kinases.
-
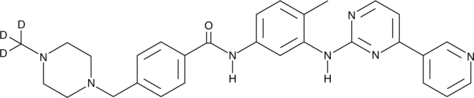
GC47452
Imatinib-d3
Imatinib-d3 (STI571-d3) hydrochloride is the deuterium labeled Imatinib. Imatinib (STI571) is an orally bioavailable tyrosine kinases inhibitor that selectively inhibits BCR/ABL, v-Abl, PDGFR and c-kit kinase activity. Imatinib (STI571) works by binding close to the ATP binding site, locking it in a closed or self-inhibited conformation, therefore inhibiting the enzyme activity of the protein semicompetitively. Imatinib also is an inhibitor of SARS-CoV and MERS-CoV.
-
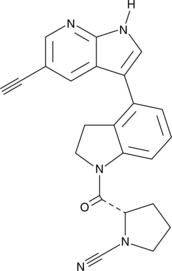
GC48614
IMP-1710
A clickable UCH-L1 inhibitor
-
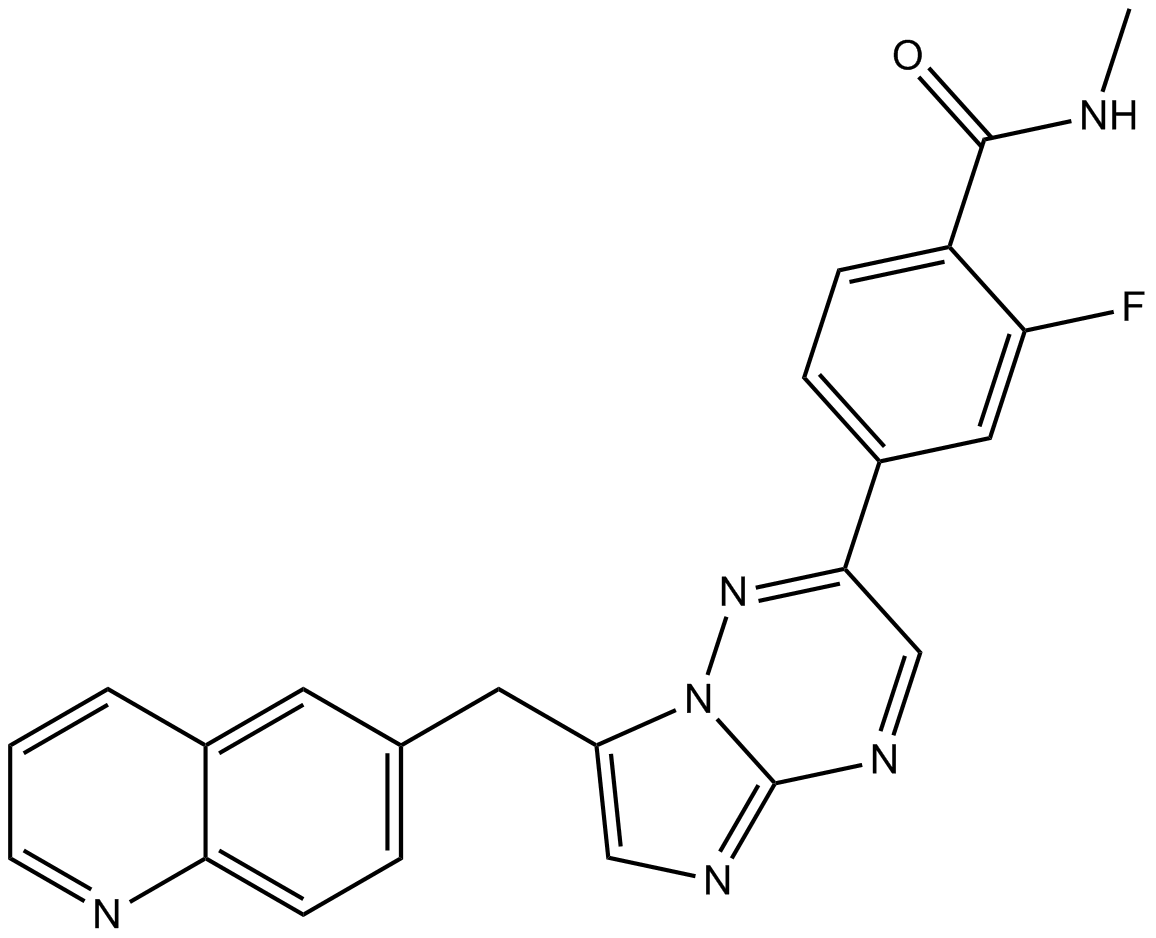
GC17866
INCB28060
INCB28060 (INC280; INCB28060) is a potent, orally active, selective, and ATP competitive c-Met kinase inhibitor (IC50=0.13 nM). INCB28060 can inhibit phosphorylation of c-MET as well as c-MET pathway downstream effectors such as ERK1/2, AKT, FAK, GAB1, and STAT3/5. INCB28060 potently inhibits c-MET-dependent tumor cell proliferation and migration and effectively induces apoptosis. Antitumor activity. INCB28060 is largely metabolized by CYP3A4 and aldehyde oxidase.
-
GC36311
Indirubin Derivative E804
Indirubin Derivative E804 is a potent inhibitor of Insulin-like Growth Factor 1 Receptor (IGF1R), with an IC50 of 0.65 μM for IGF1R.
-
GC16126
INDY
Dyrk1A and Dyrk1B inhibitor, selective
-
GC48387
Inostamycin A
A bacterial metabolite with anticancer activity
-
GC52472
Inostamycin A (sodium salt)
A bacterial metabolite with anticancer activity
-
GC18093
Insulin (human) recombinant expressed in yeast
Endogenous insulin receptor agonist
-
GC67998
Insulin degludec

-
GC31526
Insulin levels modulator
Insulin levels modulator could be used to treat diabetes.
-
GC31303
Insulin(cattle) (Insulin from bovine pancreas)
Insulin cattle (Insulin from bovine pancreas) is a two-chain polypeptide hormone produced in vivo in the pancreatic β cells.

-
GC17158
IRAK inhibitor 1
An IRAK4 inhibitor
-
GC12651
IRAK inhibitor 2
-
GC11103
IRAK inhibitor 3
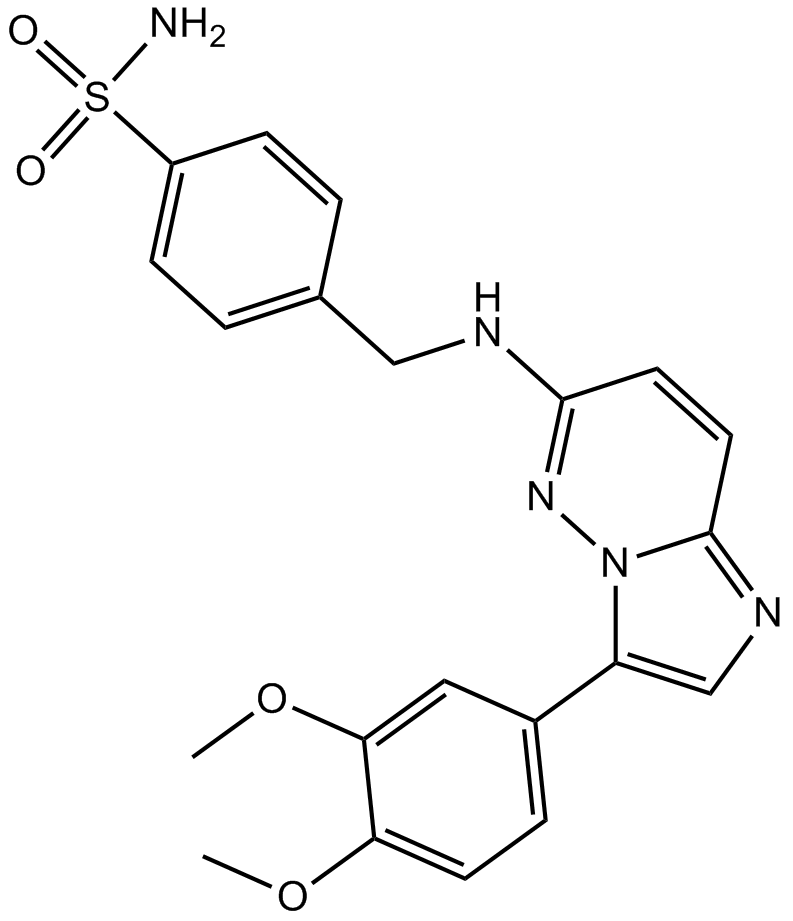
-
GC16264
IRAK inhibitor 4
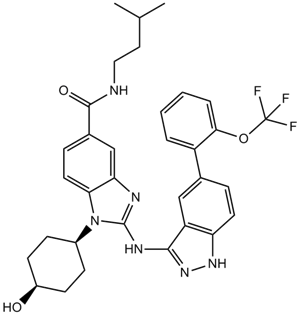
-
GC36328
IRAK inhibitor 4 trans
IRAK inhibitor 4 (trans) is the trans form of IRAK inhibitor 4.
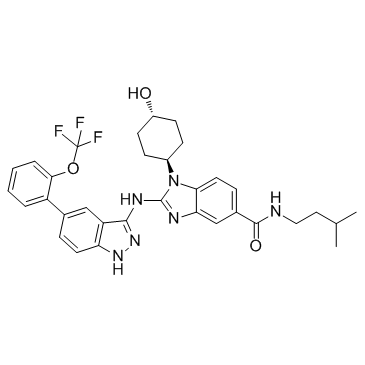
-
GC17371
IRAK inhibitor 6
An IRAK4 inhibitor
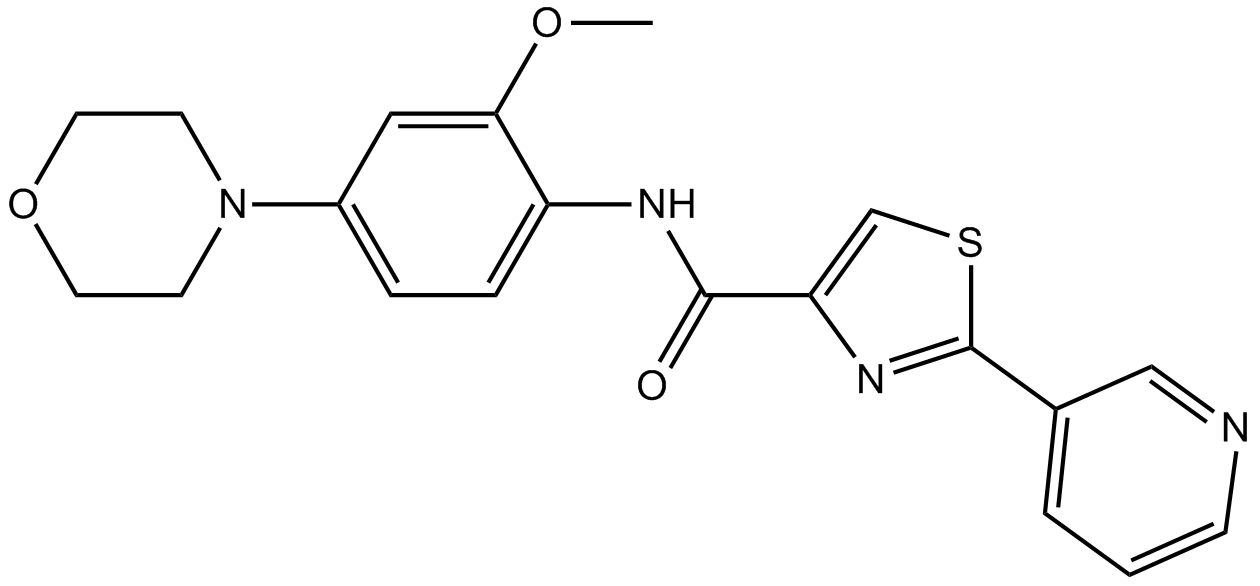
-
GC15999
IRAK-1-4 Inhibitor I
A benzimidazole
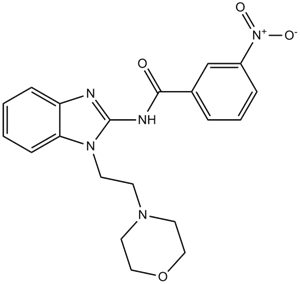
-
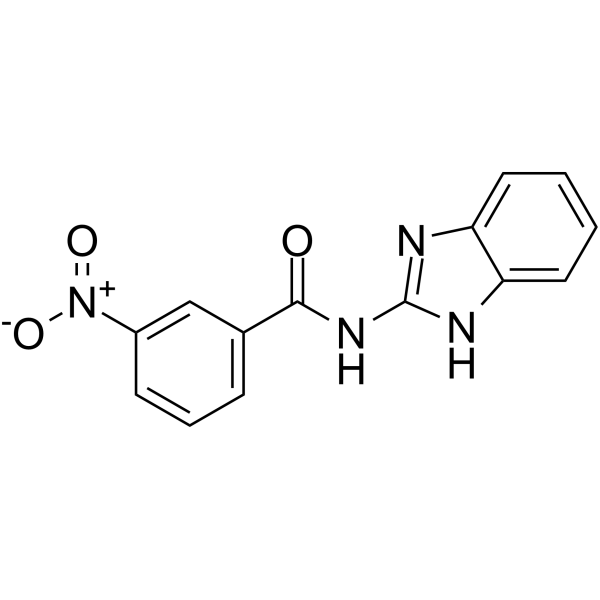
GC62583
IRAK-4 protein kinase inhibitor 2
IRAK-4 protein kinase inhibitor 2 (compound 1) is a potent inhibitor of interleukin-1 (IL-1) receptor-associated kinase-4 (IRAK-4), with an IC50 of 4 μM.
-
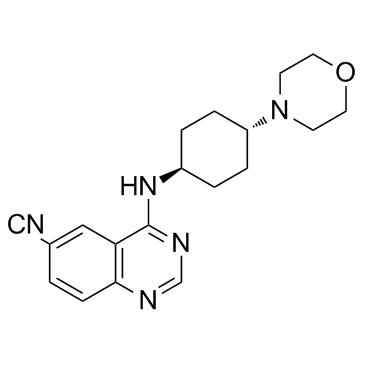
GC31688
IRAK4-IN-1
IRAK4-IN-1 is an interleukin-1 receptor associated kinase 4 (IRAK4) inhibitor with an IC50 of 7 nM.
-
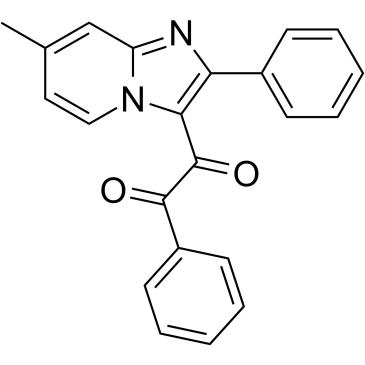
GC38799
IRAK4-IN-4
IRAK4-IN-4 is an interleukin-1 receptor-associated kinase 4 (IRAK4) inhibitor extracted from patent CN107163044A, Compound15, has an IC50 of 2.8 nM.
-
GC61944
IRAK4-IN-6
IRAK4-IN-6 is an orally efficacious and selective IRAK4 inhibitor with an IC50 of 4 nM, and targetes MyD88 L265P mutant diffuse large B cell lymphoma.
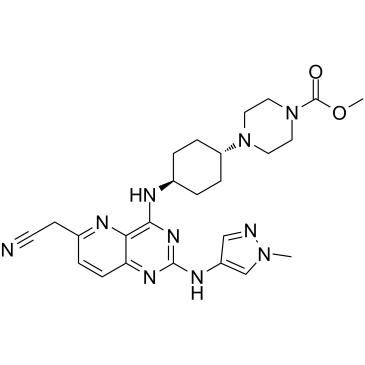
-
GC13961
ISCK03
inhibitor of SCF-mediated c-kit activation
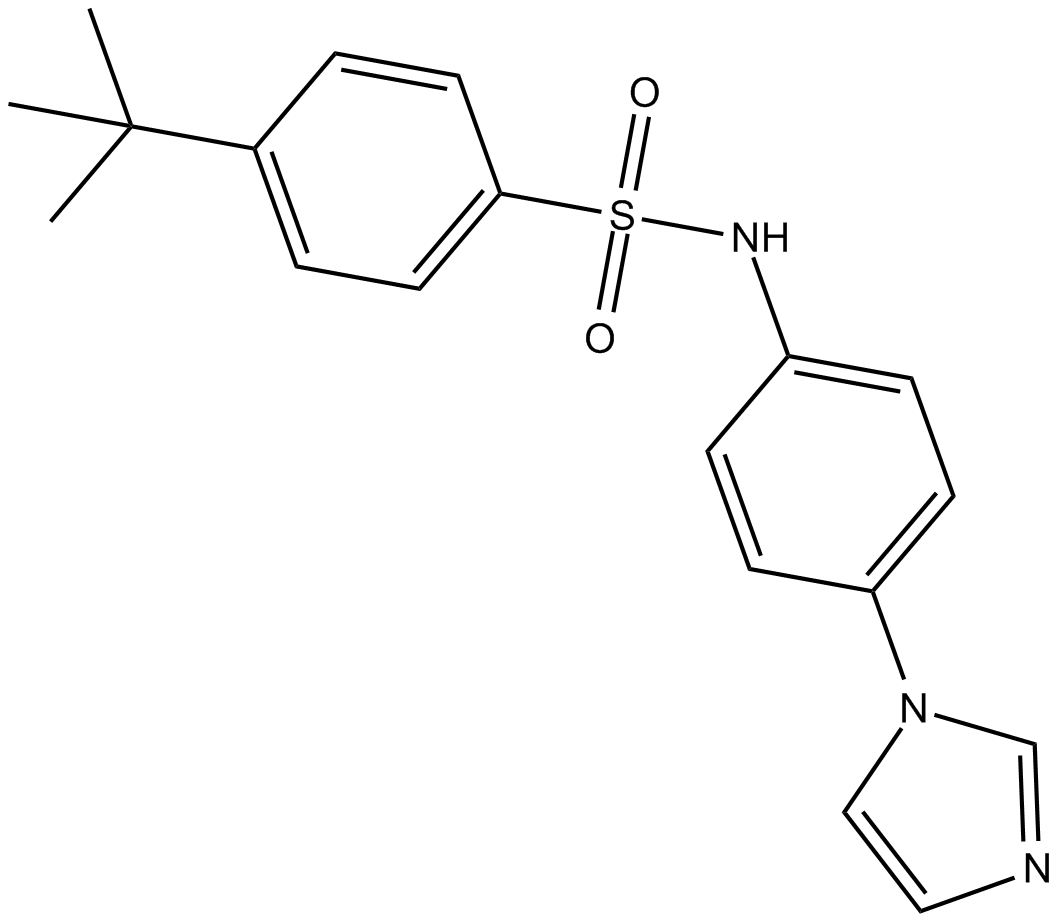
-
GC12167
ITK inhibitor
Potent ITK inhibitor
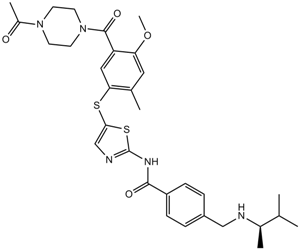
-
GC36354
ITK inhibitor 2
ITK inhibitor 2 is a interleukin-2-inducible T-cell kinase (ITK) inhibitor extracted from patent WO2011065402A1, compound 4, with an IC50 of 2 nM.
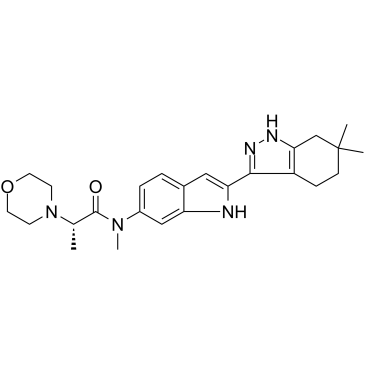
-
GC16869
Ixabepilone
A broad-spectrum anticancer agent
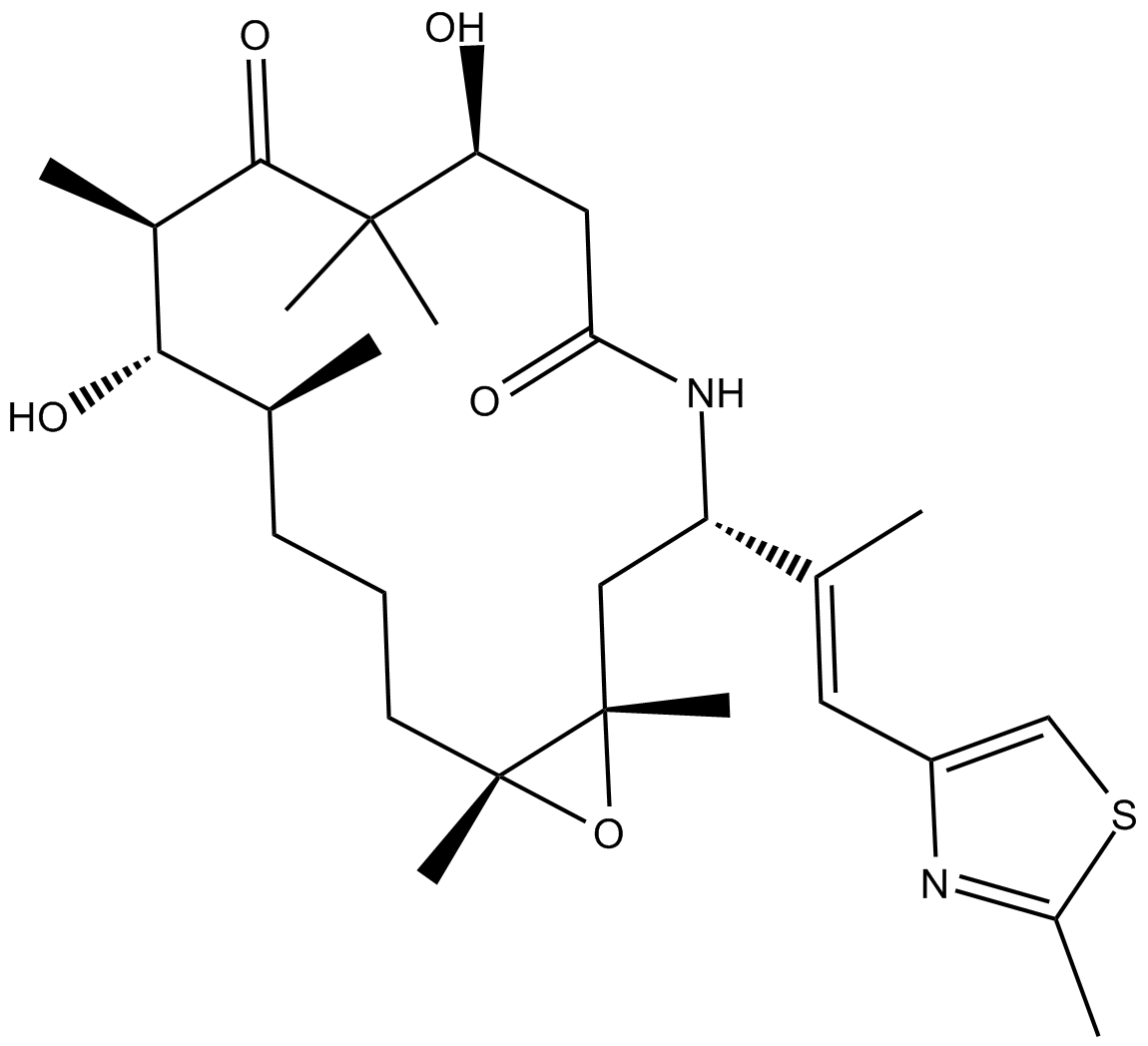
-
GC62500
JAK2-IN-7
JAK2-IN-7 is a selective JAK2 inhibitor with IC50s of 3, 11.7, and 41 nM for JAK2, SET-2, and Ba/F3V617F cells, respectively. JAK2-IN-7 possesses >14-fold selectivity over JAK1, JAK3, FLT3. JAK2-IN-7 stimulates cell cycle arrest in the G0/G1 phase and induces tumor cellapoptosis. Antitumor activities.
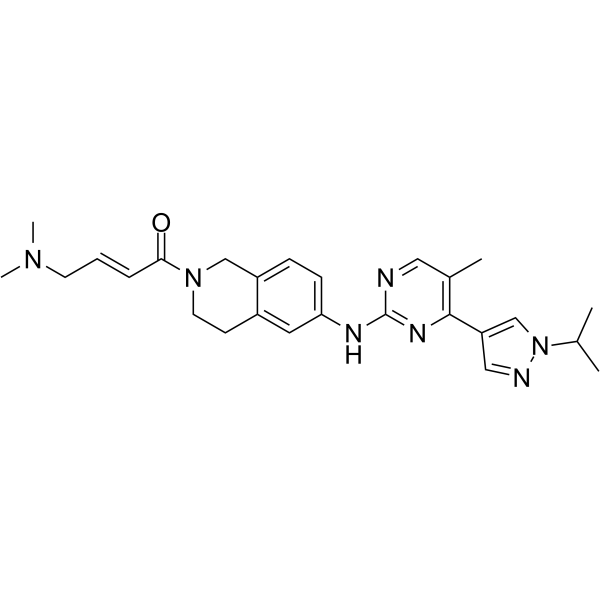
-
GC62665
JAK2/FLT3-IN-1 TFA
JAK2/FLT3-IN-1 (TFA) is a potent and orally active dual JAK2/FLT3 inhibitor with IC50 values of 0.7 nM, 4 nM, 26 nM and 39 nM for JAK2, FLT3, JAK1 and JAK3, respectively. JAK2/FLT3-IN-1 (TFA) has anti-cancer activity.
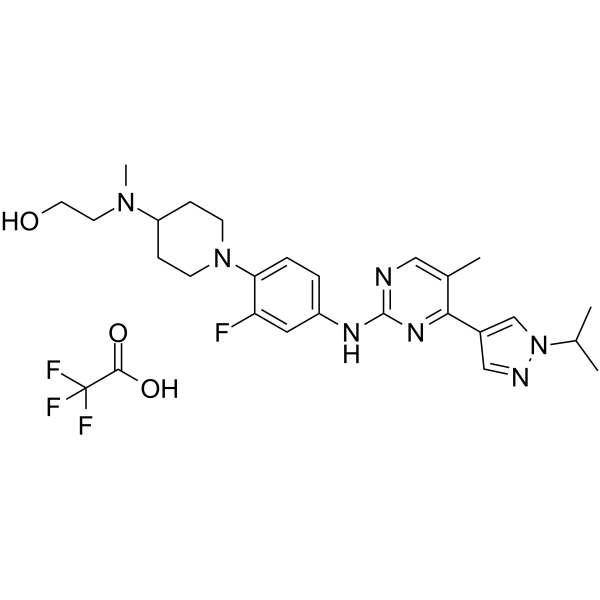
-
GC50706
JBJ-03-142-02
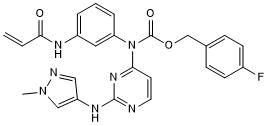
-
GC62632
JBJ-04-125-02
JBJ-04-125-02 is a potent, mutant-selective, allosteric and orally active EGFR inhibitor with an IC50 of 0.26 nM for EGFRL858R/T790M. JBJ-04-125-02 can inhibit cancer cell proliferation and EGFRL858R/T790M/C797S signaling. JBJ-04-125-02 has anti-tumor activities.
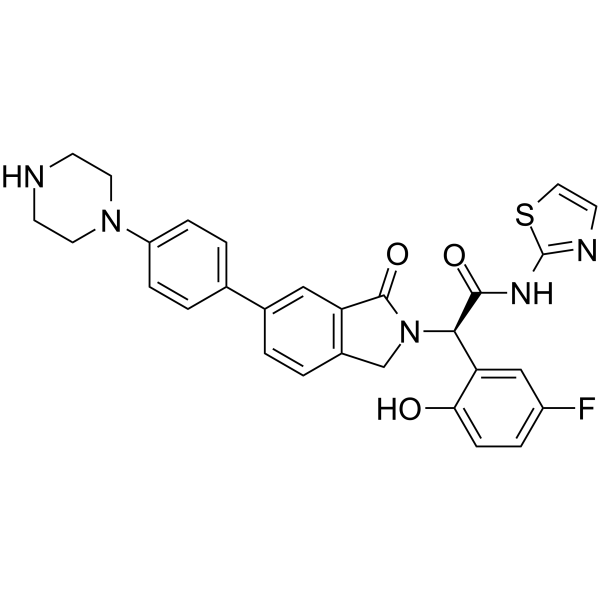
-
GC67690
JBJ-09-063 hydrochloride
JBJ-09-063 hydrochloride is a mutation selective allosteric EGFR inhibitor
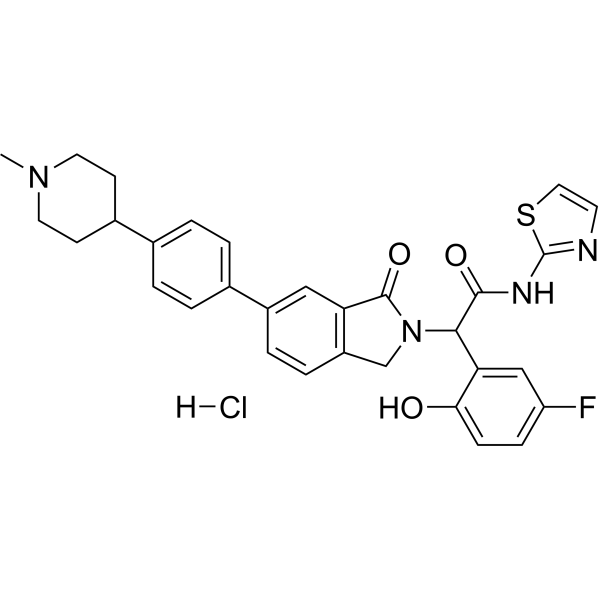
-
GC67860
JBJ-09-063 TFA

-
GC19205
JH-II-127
JH-II-127 is a highly potent, selective, and brain penetrant LRRK2 inhibitor, with IC50 of 6.6 nM, 2.2 nM ,47.7 nM for LRRK2-wild-type, LRRK2-G2019S, LRRK2-A2016T.
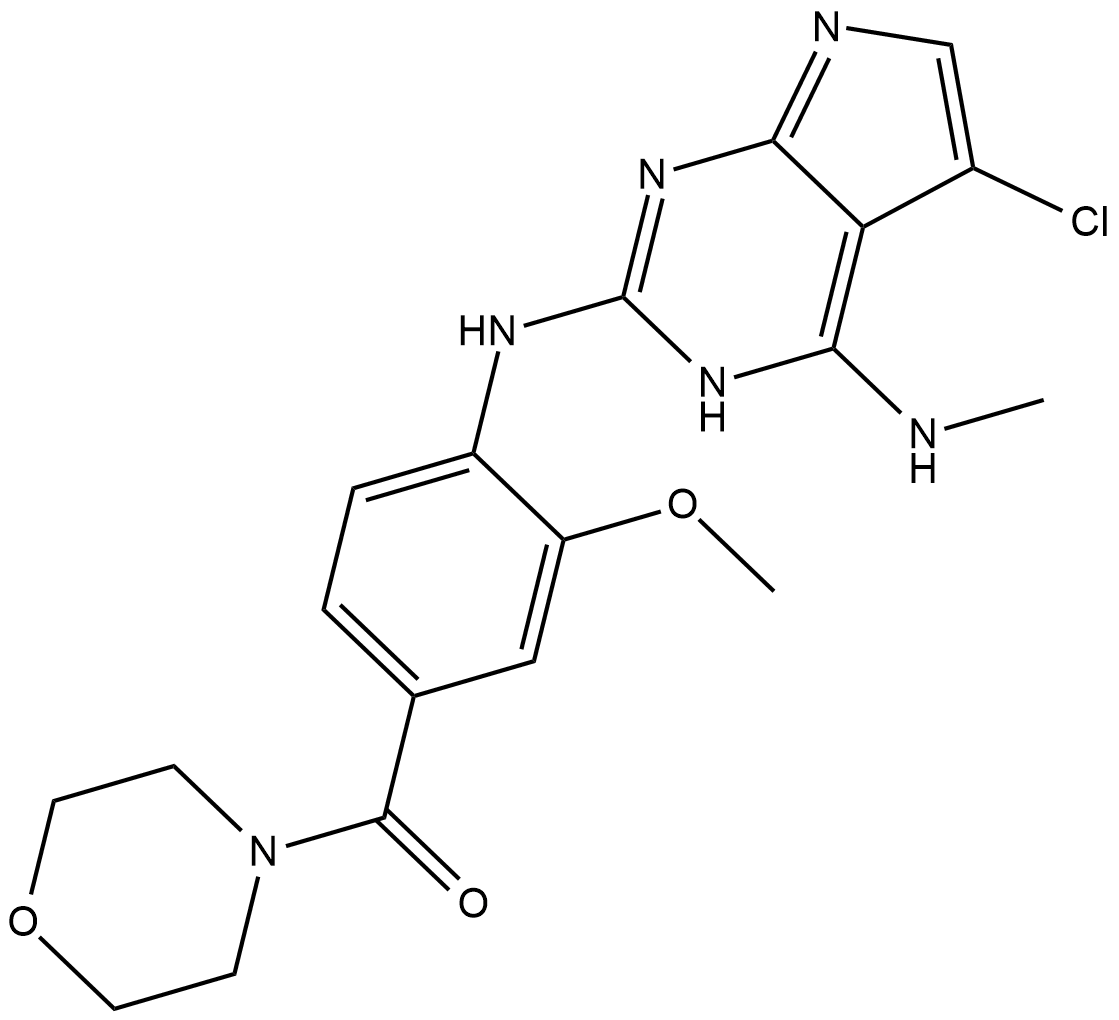
-
GC33062
JH-VIII-157-02
JH-VIII-157-02 is a structural analogue of alectinib, acts as an ALK inhibitor, and shows an IC50 of 2 nM for echinoderm microtubule-associated protein-like 4-ALK (EML4-ALK) G1202R in cells.
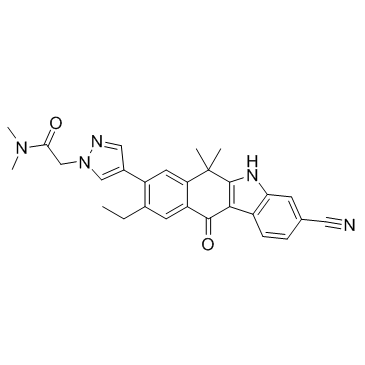
-
GC63925
JH-X-119-01
JH-X-119-01 is a potent and selective interleukin-1 receptor-associated kinases 1 (IRAK1) inhibitor.
-
GC18168
JI-101
An orally active inhibitor
-
GC65436
JND3229
JND3229 is a reversible EGFRC797S inhibitor with IC50 values of 5.8, 6.8 and 30.5 nM for EGFRL858R/T790M/C797S, EGFRWT and EGFRL858R/T790M, respectively. JND3229 has good anti-proliferative activity and can effectively inhibit tumour growth in vivo. JND3229 can be used in cancer research, especially in non-small cell carcinoma.
-
GC18030
JNJ 28871063 hydrochloride
ErbB receptor family inhibitor

-
GC38502
JNJ-10198409
A potent PDGF tyrosine kinase inhibitor
-
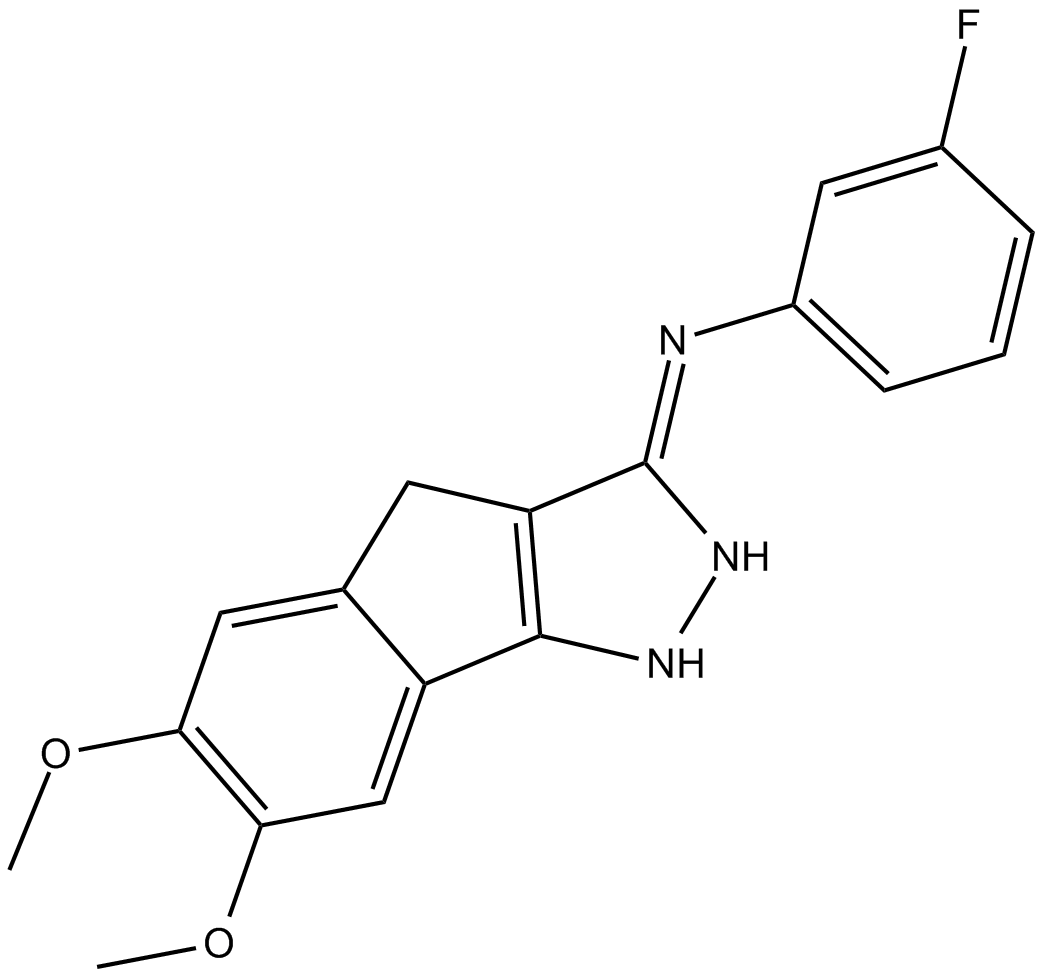
GC14544
JNJ-10198409
inhibitor of platelet-derived growth factor (PDGF-BB) tyrosine kinase
-
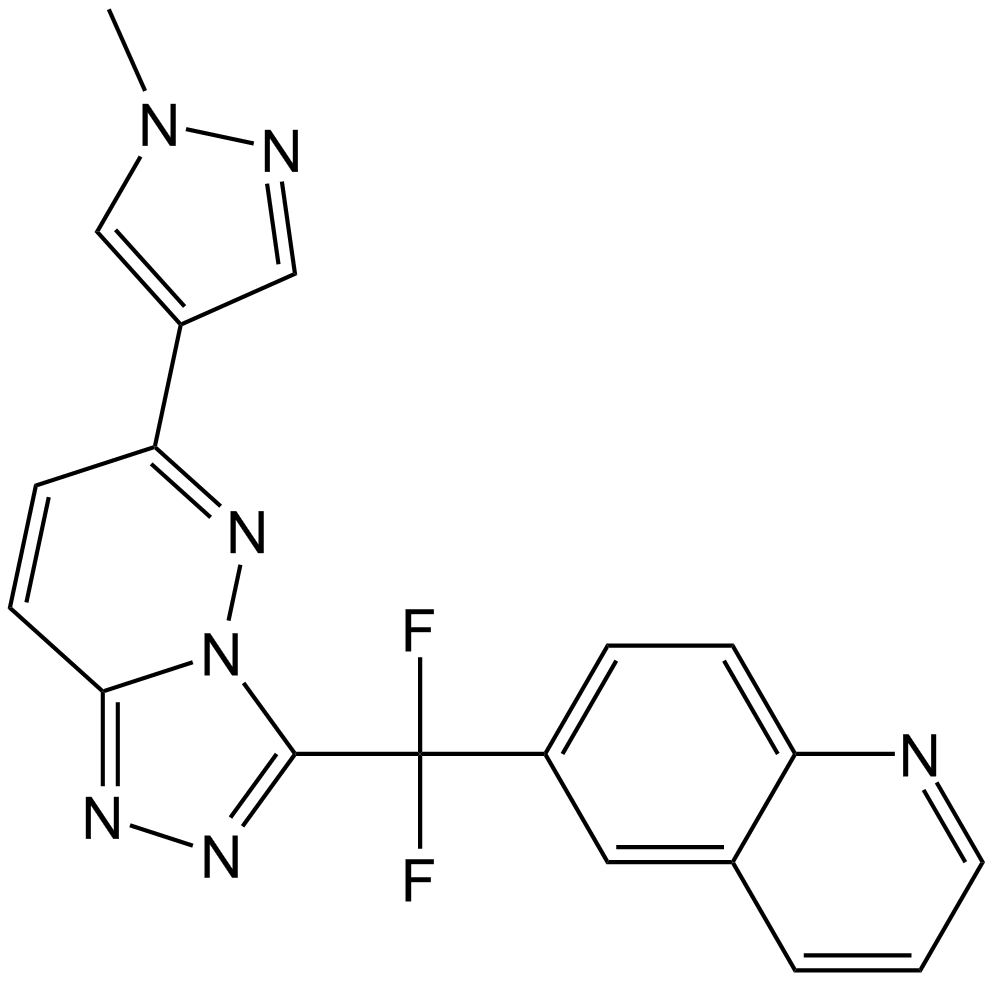
GC12585
JNJ-38877605
C-Met inhibitor,ATP-competitive
-
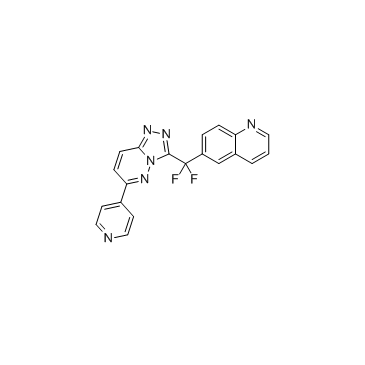
GC33266
JNJ-38877618
JNJ-38877618 is a potent, highly selective, orally bioavailable Met kinase inhibitor with IC50s of 2 and 3 nM for wild type and mutant Met, respectively.
-
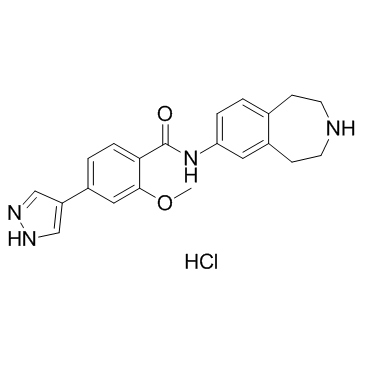
GC32995
JNJ-47117096 hydrochloride (MELK-T1 hydrochloride)
JNJ-47117096 hydrochloride (MELK-T1 hydrochloride) is potent and selective MELK inhibitor, with an IC50 of 23 nM, also effectively inhibits Flt3, with an IC50 of 18 nM.
-
GC11362
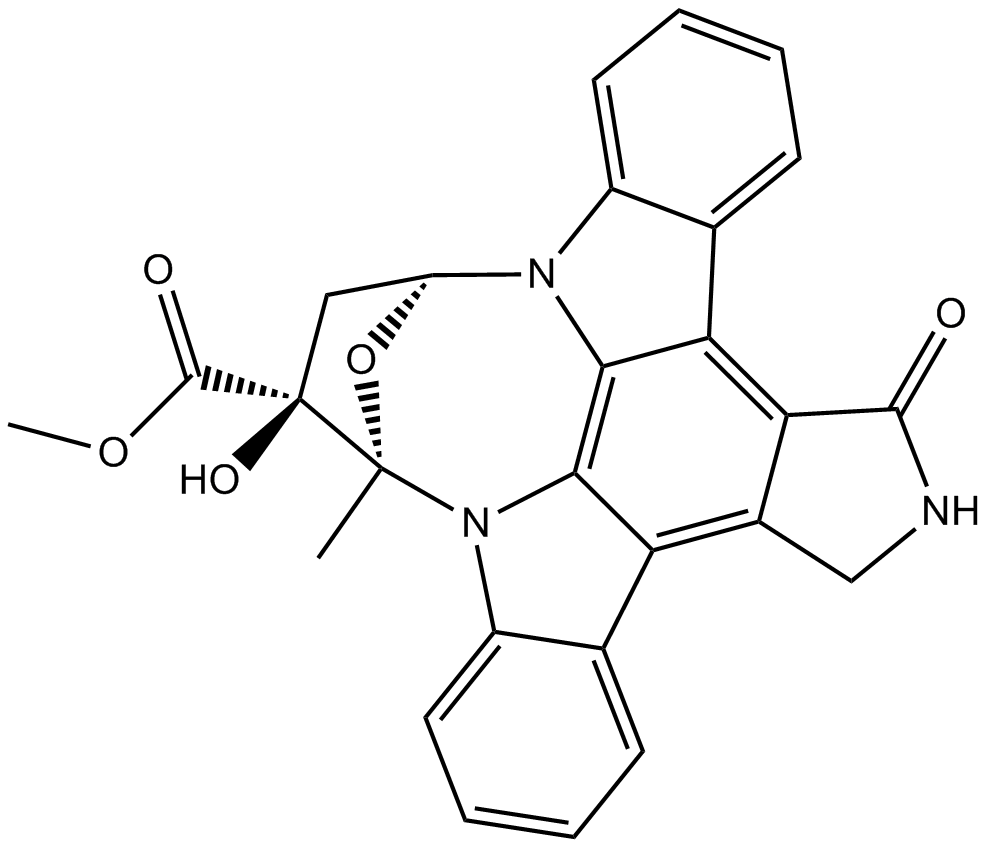
K 252a
A protein kinase inhibitor
-
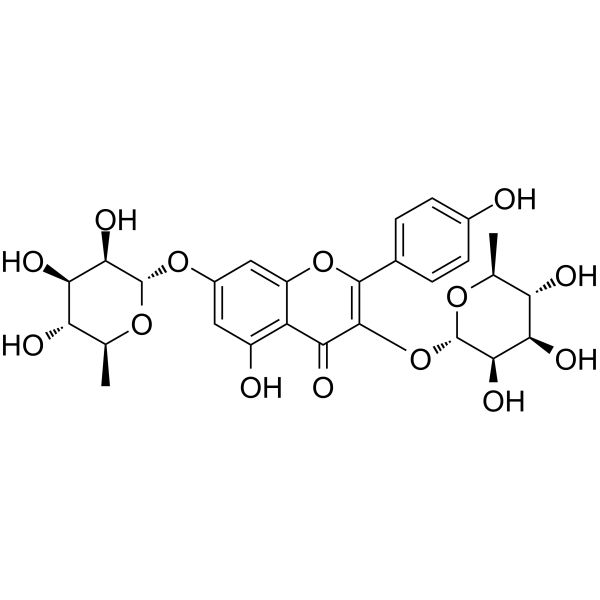
GN10497
Kaempferitrin
-
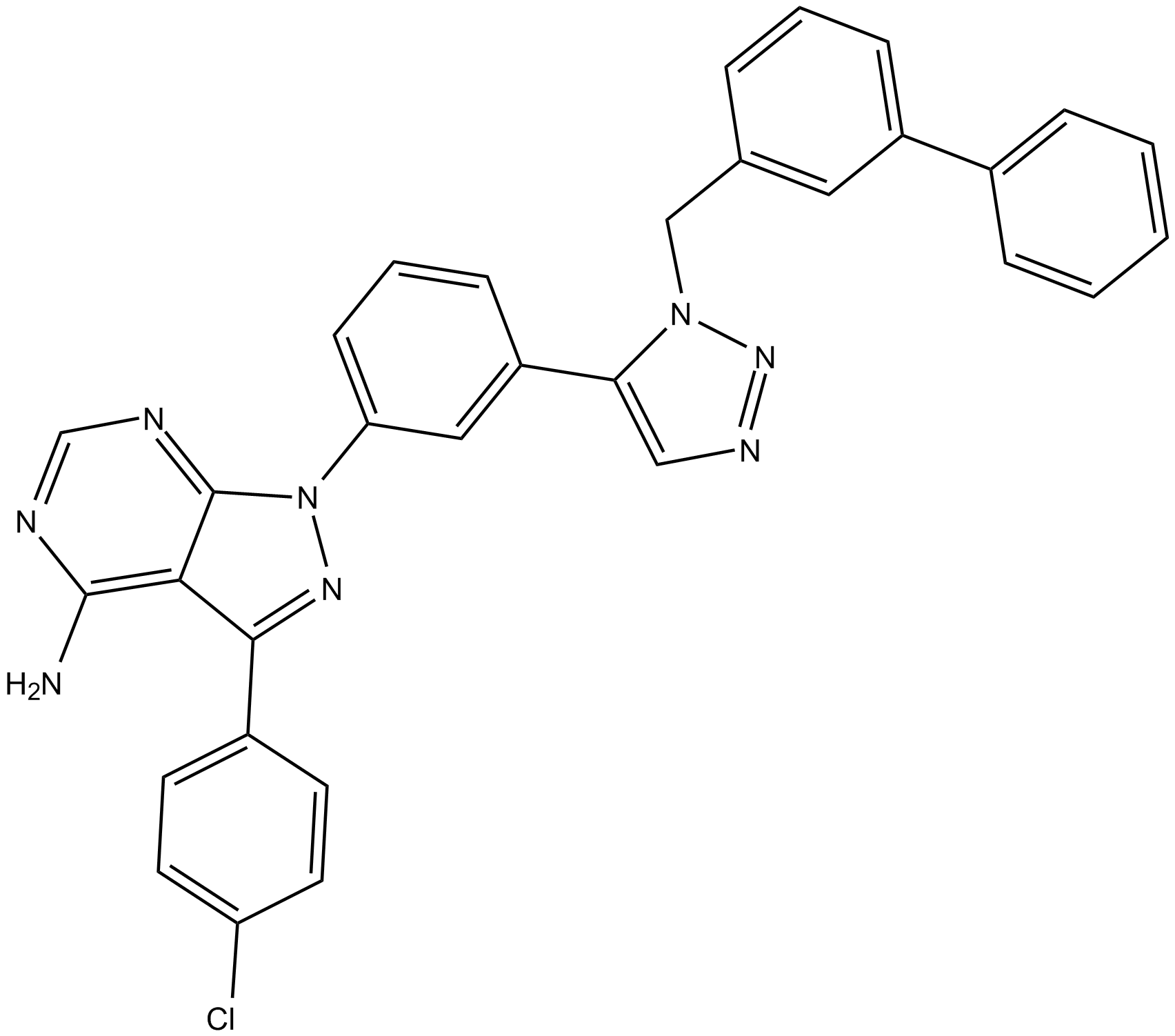
GC17638
KB SRC 4
KB SRC 4 is a potent, and highly selective c-Src inhibitor, with a Ki of 44 nM and a Kd of 86 nM, and shows no inhibition on c-Abl up to 125 μM; KB SRC 4 has antitumor activity.
-
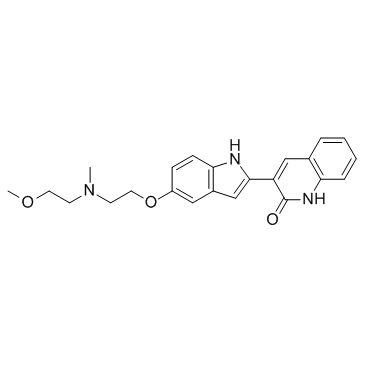
GC32625
KDR-in-4
KDR-in-4 (KDR-in-4) is a potent kinase insert domain-containing receptor (KDR/VEGFR2) inhibitor with an IC50 of 7 nM.
-
GC63943
KH-CB20
KH-CB20, an E/Z mixture, is a potent and selective inhibitor of CLK1 and the closely related isoform CLK4, with an IC50 of 16.5 nM for CLK1.
-
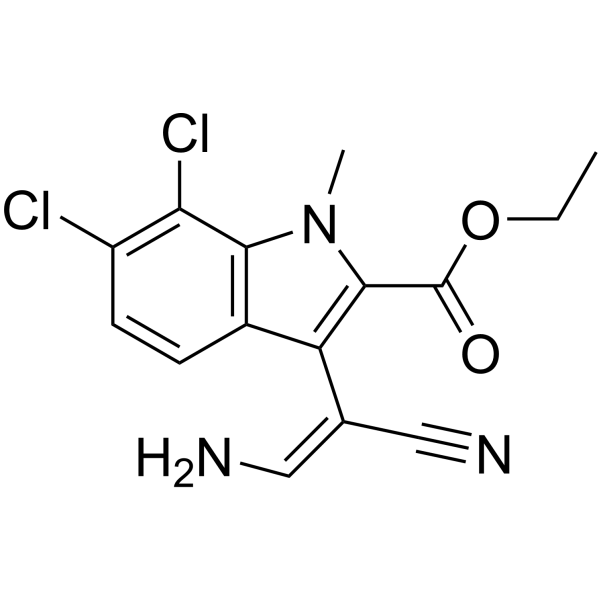
GC13264
Ki20227
A c-Fms kinase inhibitor
-
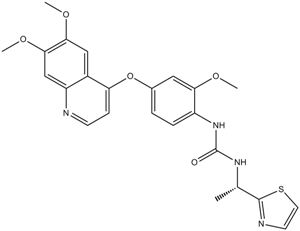
GC11666
Ki8751
VEGFR-2 inhibitor,potent and selective
-
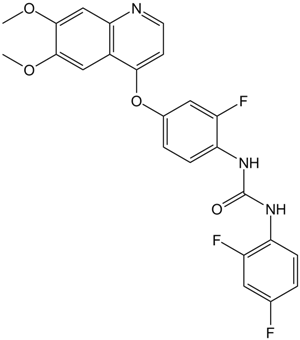
GC13902
KRCA 0008
Ack1 and anaplastic lymphoma kinase (ALK) dual inhibitor
-
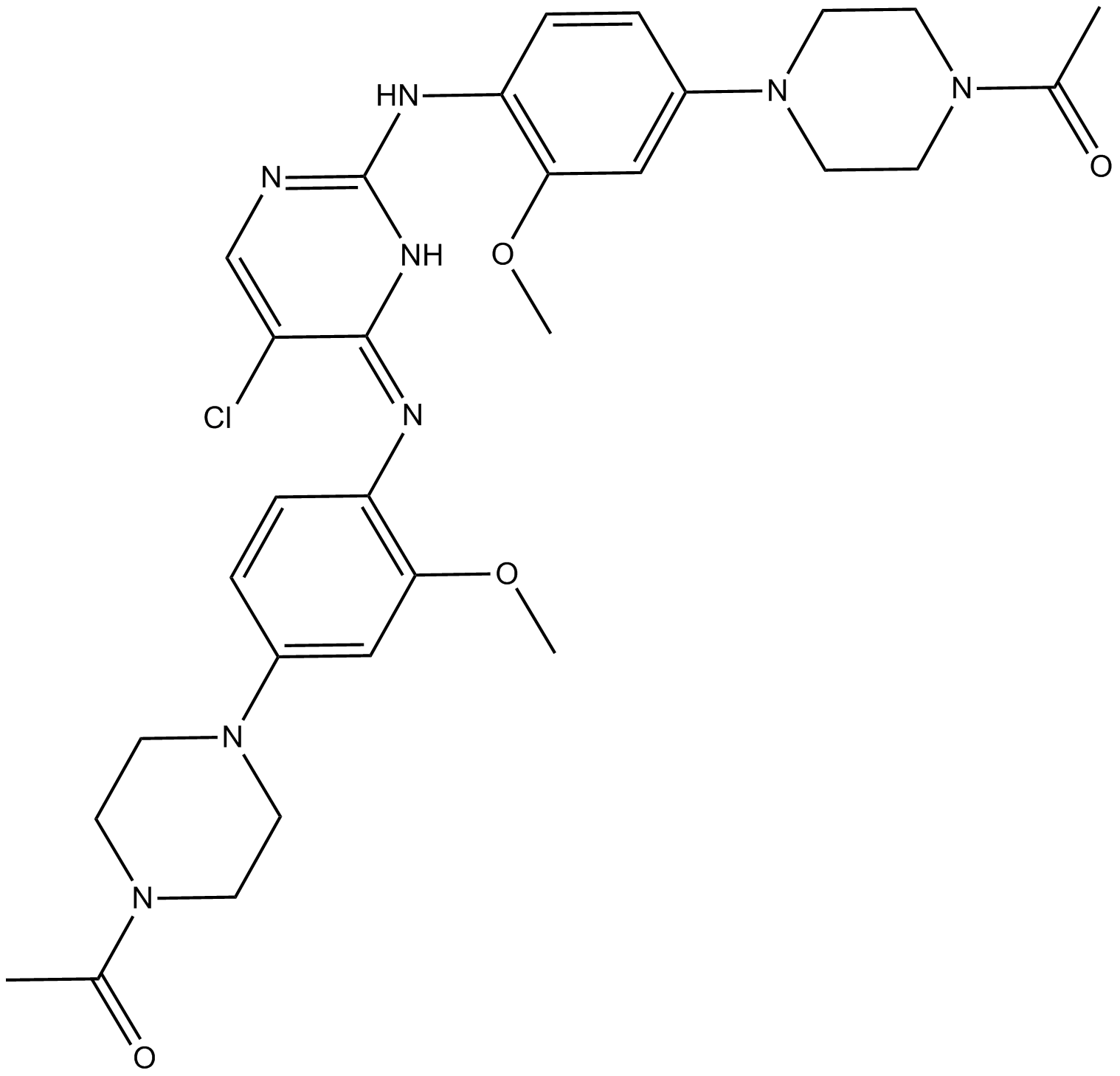
GC12590
KRN 633
VEGFR inhibitor,ATP-competitive
-
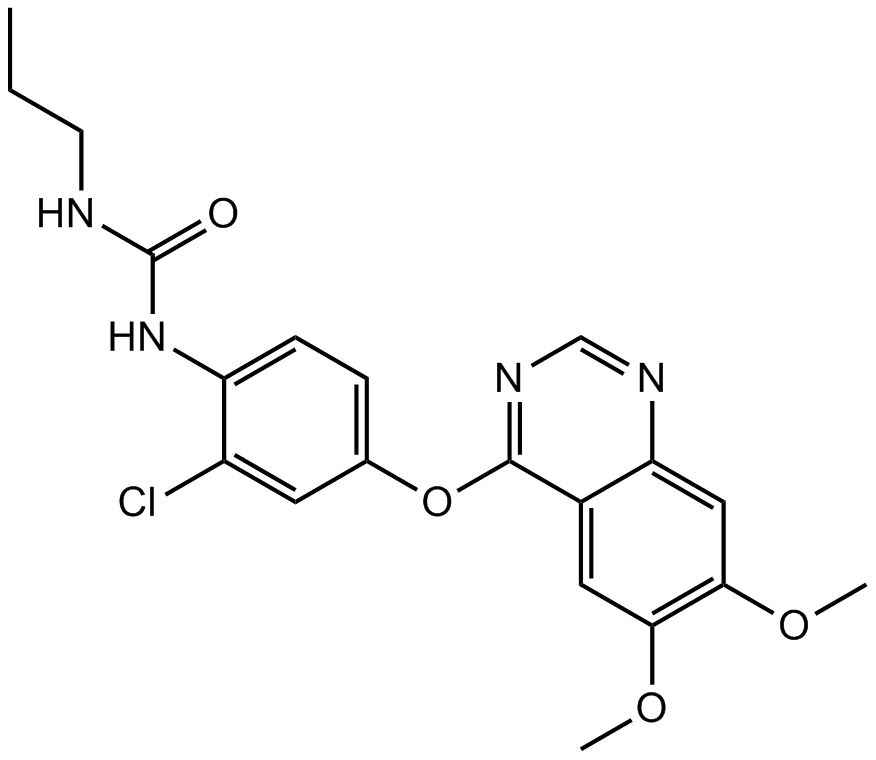
GC10626
KU14R
-
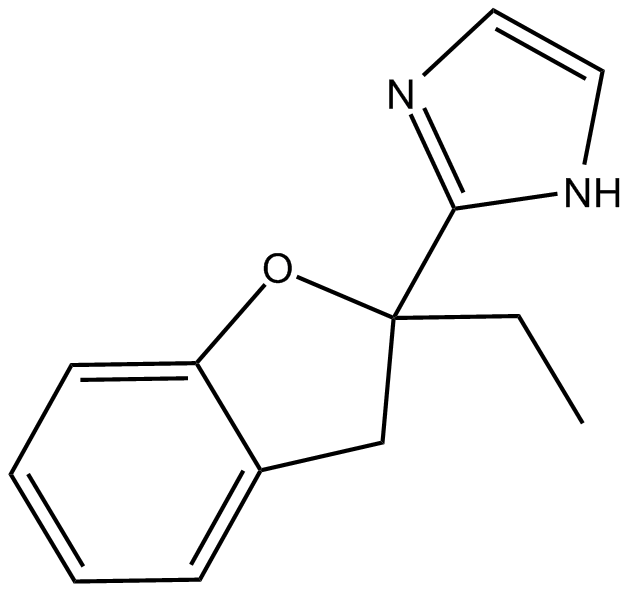
GC14592
KW 2449
A multi-kinase inhibitor
-
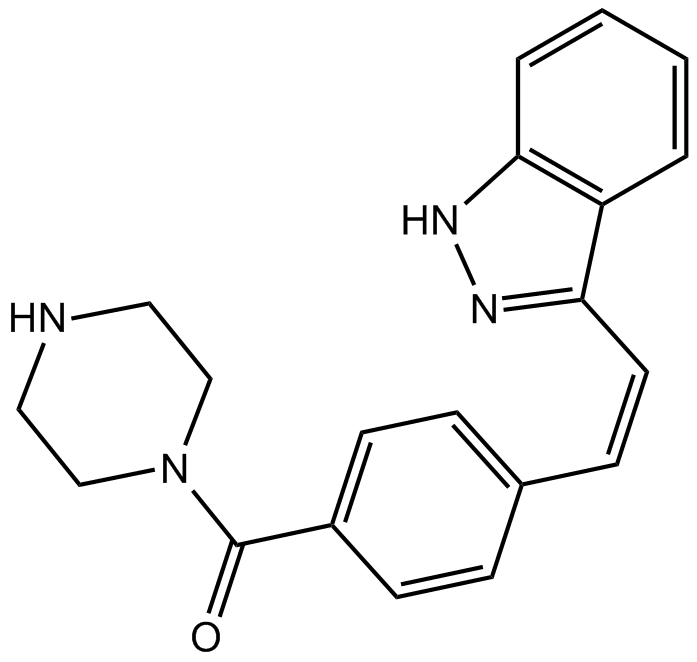
GC10523
KX1-004
Pp60c-src inhibitor
-
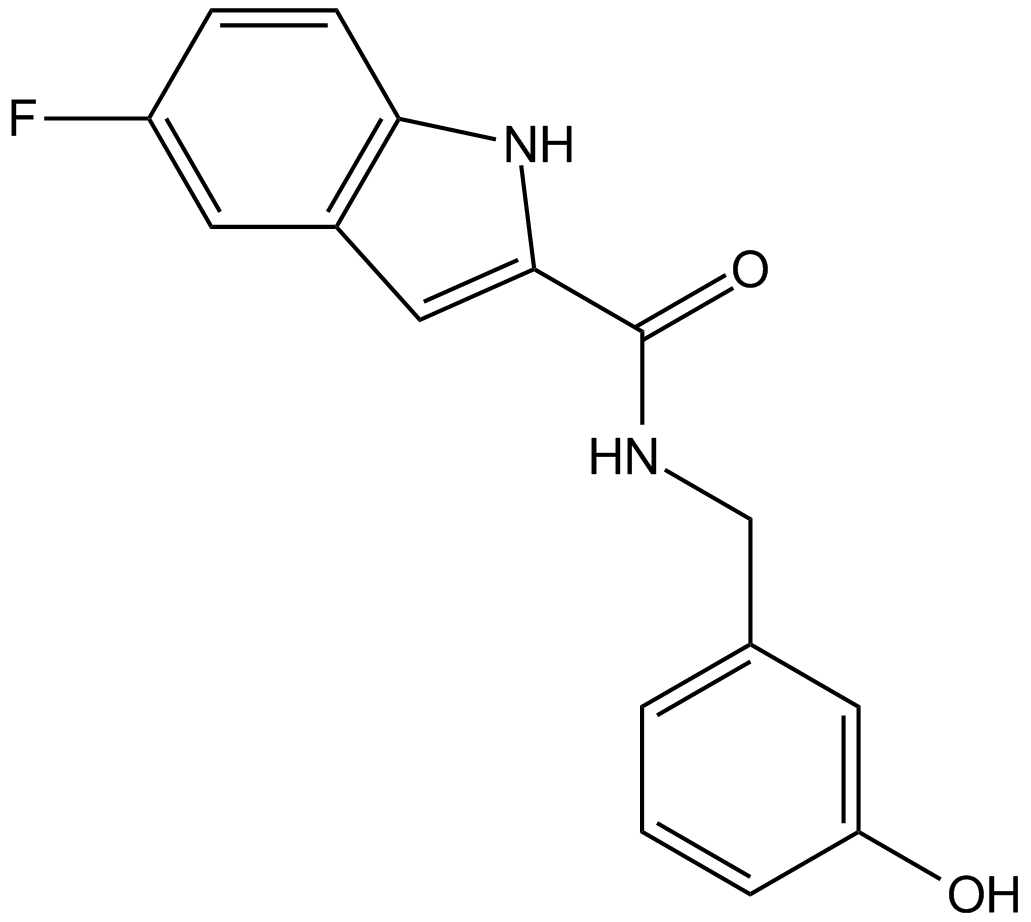
GC14288
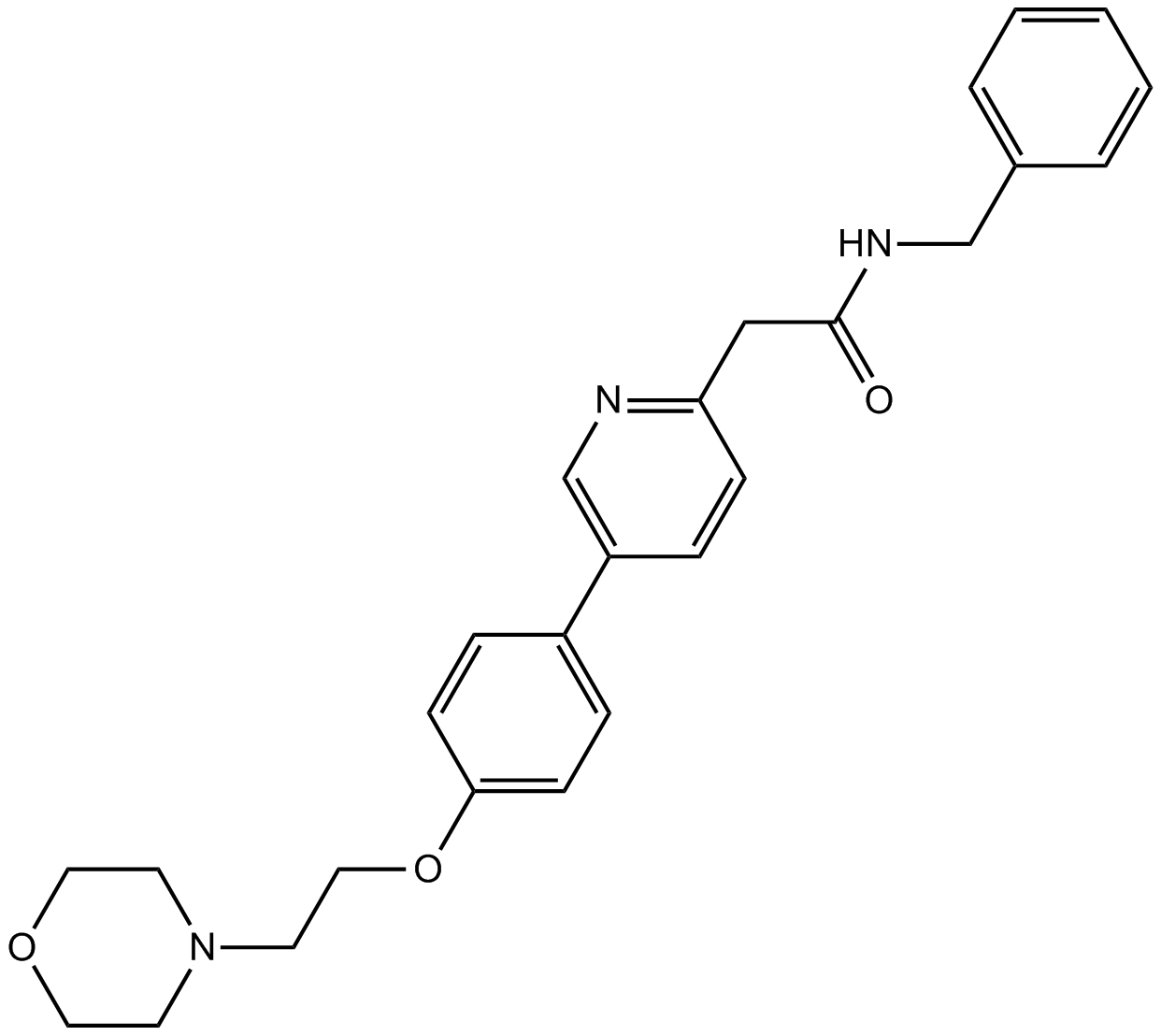
KX2-391
KX2-391 (KX2-391) is an inhibitor of Src that targets the peptide substrate site of Src, with GI50 of 9-60 nM in cancer cell lines.
-
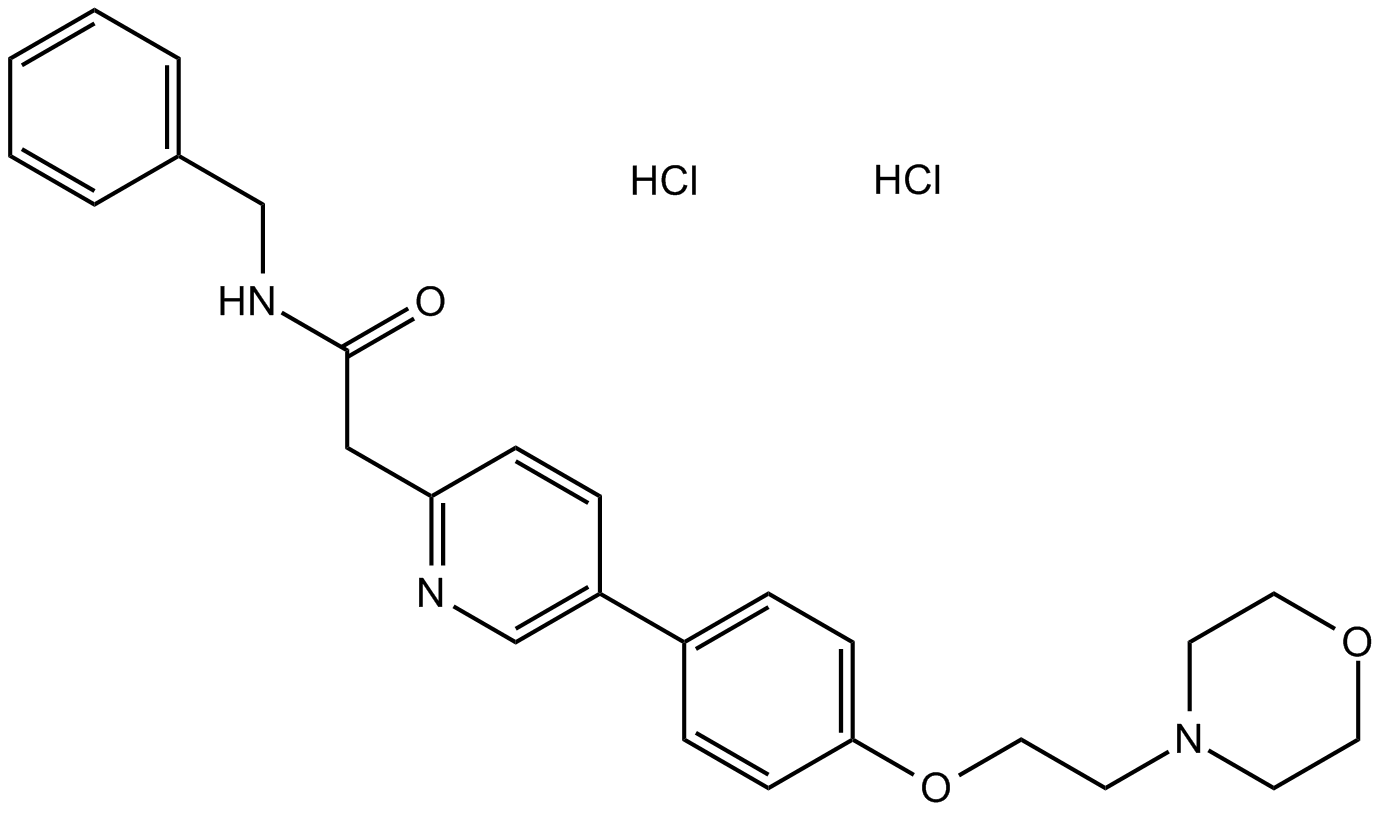
GC10222
KX2-391 dihydrochloride
A Src kinase inhibitor
-
GC50137
KYL
KYL, an antagonistic peptide, selectively targets EphA4 receptor.
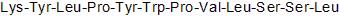
-
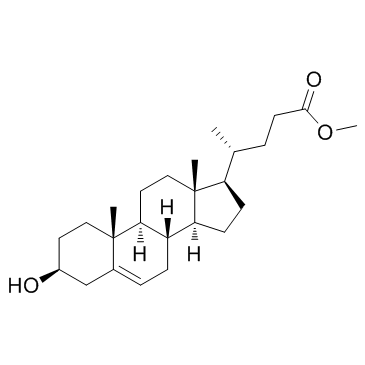
GC32044
L 601920-0 (Methyl-3β-hydroxycholenate)
L 601920-0 (Methyl-3β-hydroxycholenate) is a ROR gamma modulator extracted from patent US20110263046 A1, in figure 2.
-
GC44085
L-Sulforaphene
L-Sulforaphene, isolated from radish seeds, exhibits an ED50 against velvetleaf seedlings approximately 2 x 10-4 M. L-Sulforaphene promotes cancer cells apoptosis and inhibits migration via inhibiting EGFR, p-ERK1/2, NF‐κB and other signals.
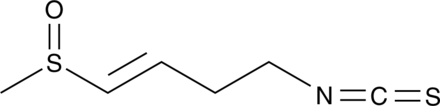
-
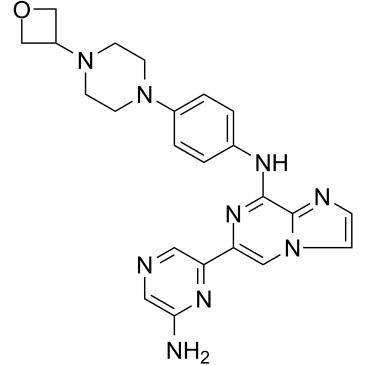
GC36423
Lanraplenib
A Syk inhibitor
-
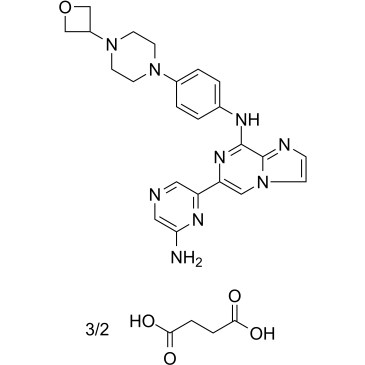
GC38630
Lanraplenib succinate
Lanraplenib succinate (GS-9876 succinate) is a highly selective and orally active SYK inhibitor (IC50=9.5 nM) in development for the treatment of inflammatory diseases.
-
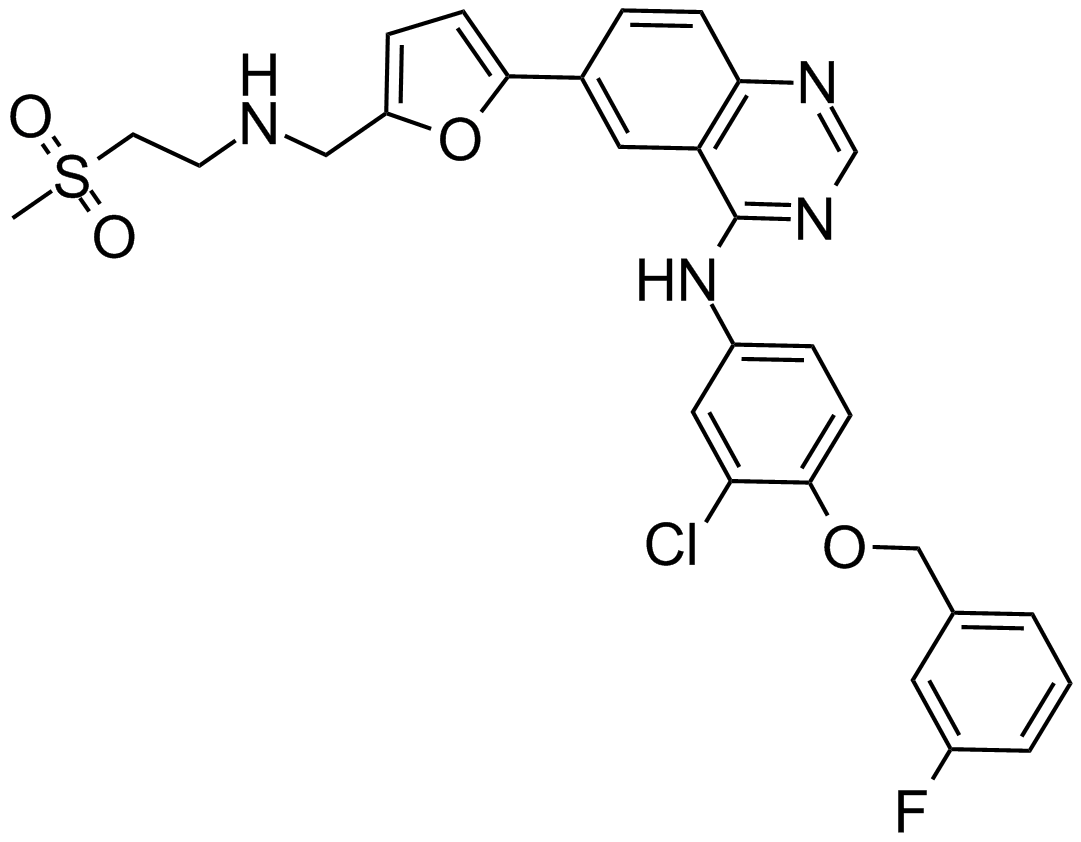
GC13608
Lapatinib
A dual inhibitor of EGFR and ErbB2
-
GC25559
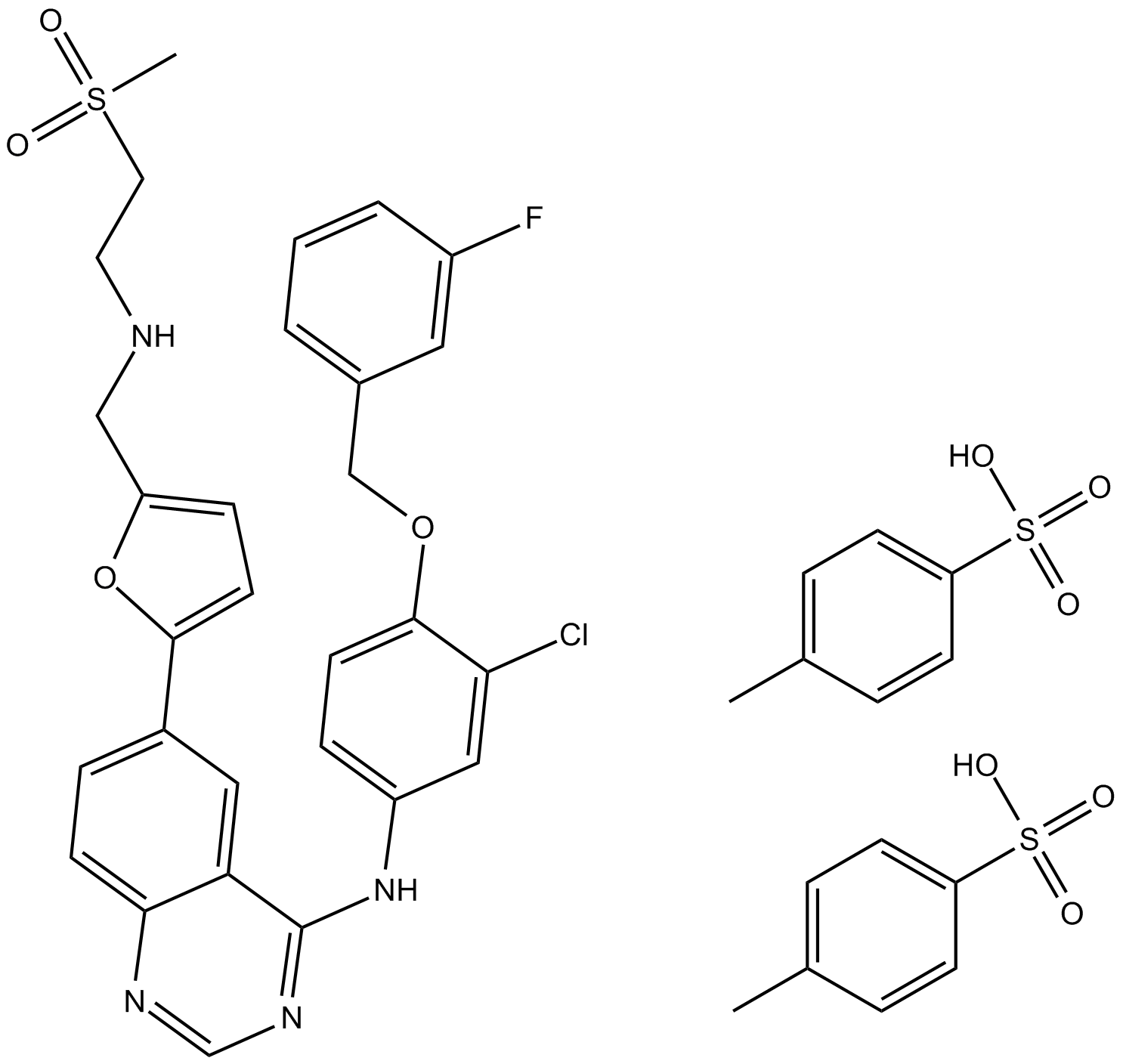
Lapatinib (GW-572016) Ditosylate
Lapatinib (GW-572016) Ditosylate is a potent EGFR and ErbB2 inhibitor with IC50 of 10.8 and 9.2 nM in cell-free assays, respectively.
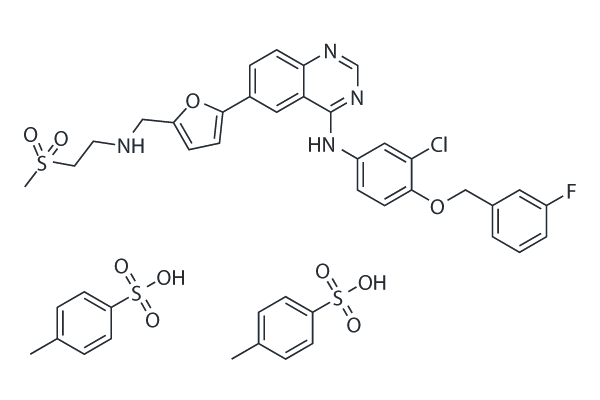
-
GC16593
Lapatinib Ditosylate
Lapatinib Ditosylate is a selective dual inhibitor of ErbB-2 and EGFR with IC50 value against ErbB-2 and EGFR of 9.2 and 10.8 nM in vitro, respectively.