Tyrosine Kinase
- IGFBR(1)
- Bcr-Abl(73)
- Ack1(4)
- Axl(6)
- ALK(61)
- BMX Kinase(4)
- Broad Spectrum Protein Kinase Inhibitor(11)
- c-FMS(35)
- c-Kit(65)
- c-MET(91)
- c-RET(7)
- CSF-1R(3)
- DDR1/DDR2 Receptor(1)
- EGFR(251)
- EphB4(2)
- FAK(34)
- FGFR(104)
- FLT3(102)
- HER2(19)
- IGF1R(33)
- Insulin Receptor(45)
- IRAK(32)
- ITK(12)
- Lck(2)
- LRRK2(23)
- PDGFR(117)
- PTKs/RTKs(5)
- Pyk2(6)
- ROR(41)
- Spleen Tyrosine Kinase (Syk)(35)
- Src(105)
- Tie-2 (3)
- Trk(37)
- VEGFR(203)
- Kinase(0)
- Discoidin Domain Receptor(15)
- DYRK(26)
- Ephrin Receptor(13)
- ROS(15)
- RET(34)
- TAM Receptor(28)
Products for Tyrosine Kinase
- Cat.No. Product Name Information
-
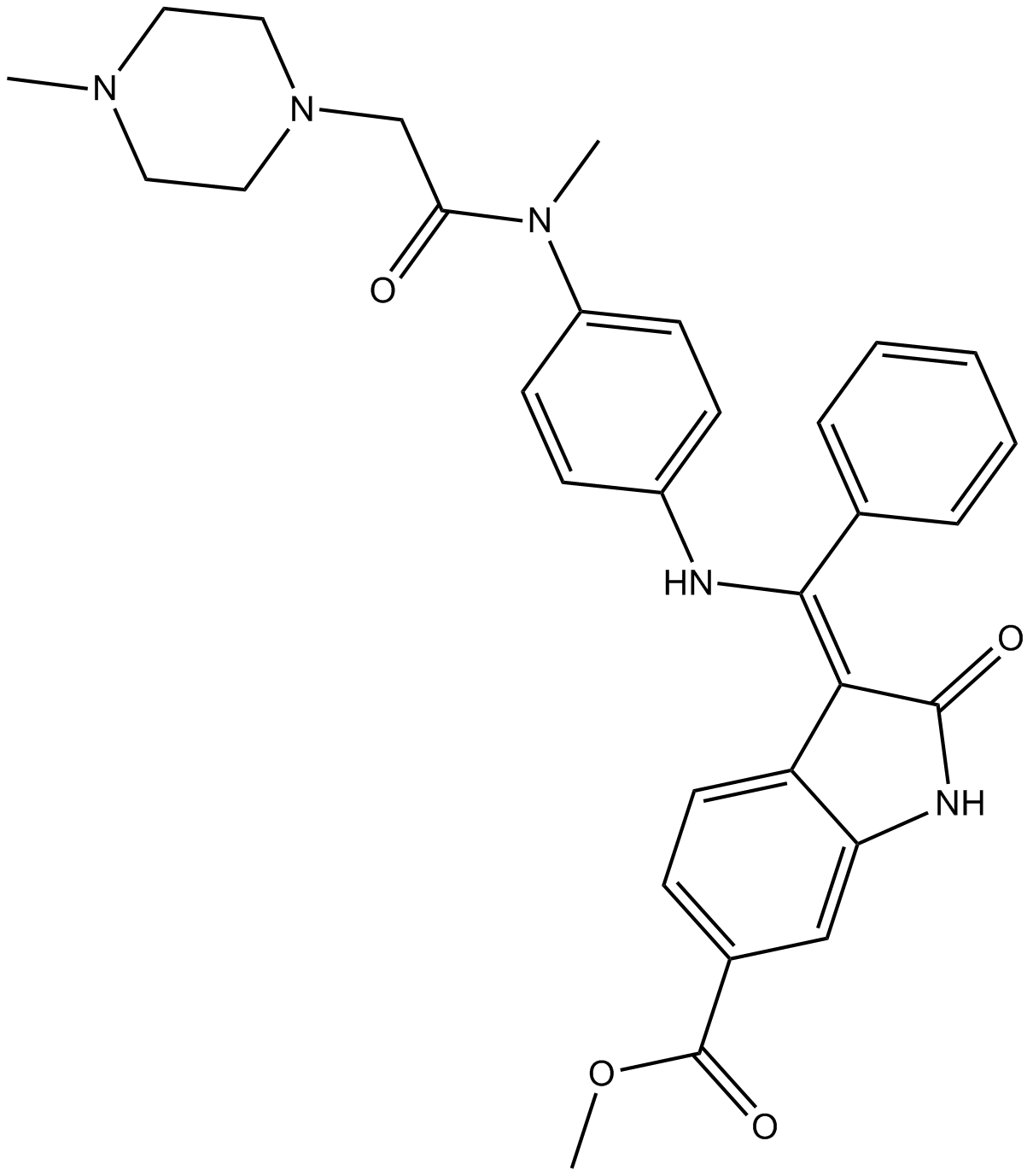
GC67759
Lapatinib-d4
-
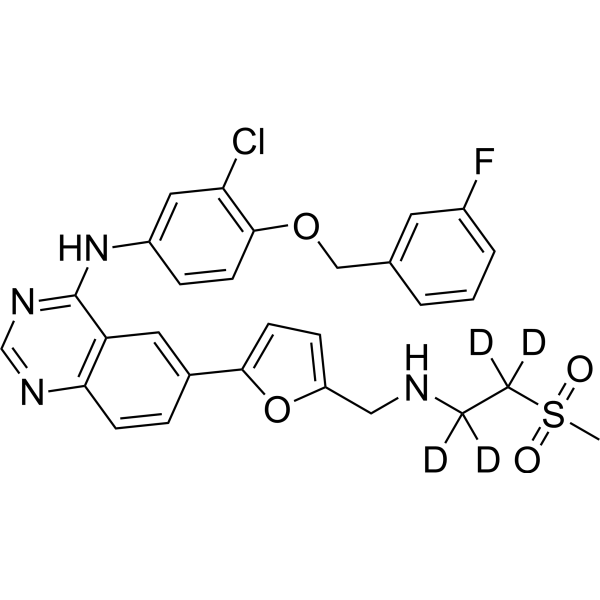
GC16021
Lavendustin A
EGFR tyrosine kinase inhibitor
-
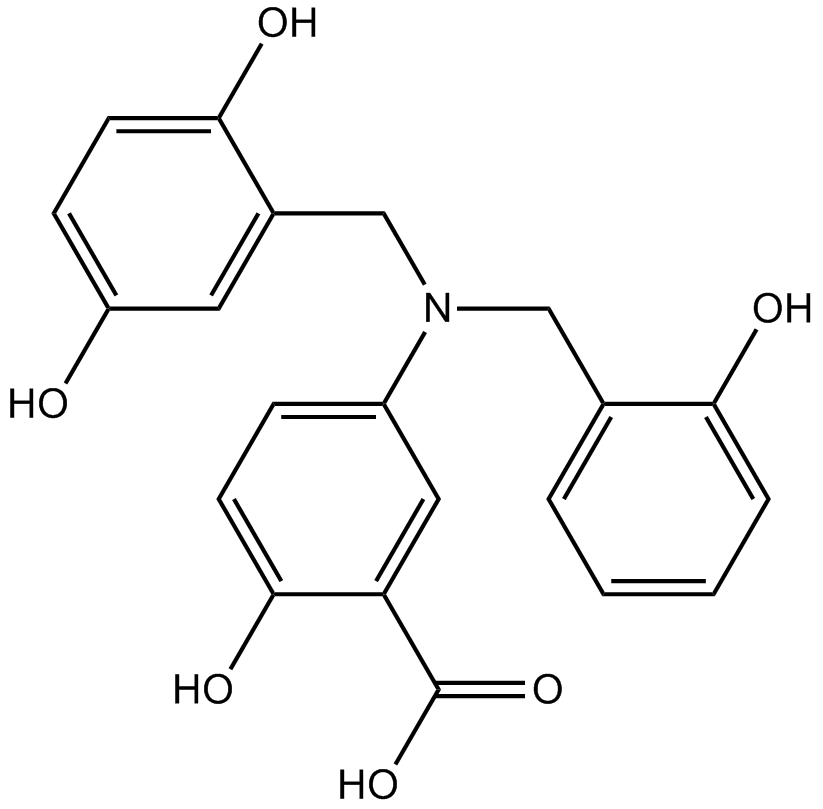
GC19218
Lazertinib
Lazertinib is a potent, highly mutant-selective, blood-brain barrier permeable, orally available and irreversible third-generation EGFR tyrosine kinase inhibitor, and can be used in the research of non-small cell lung cancer.
-
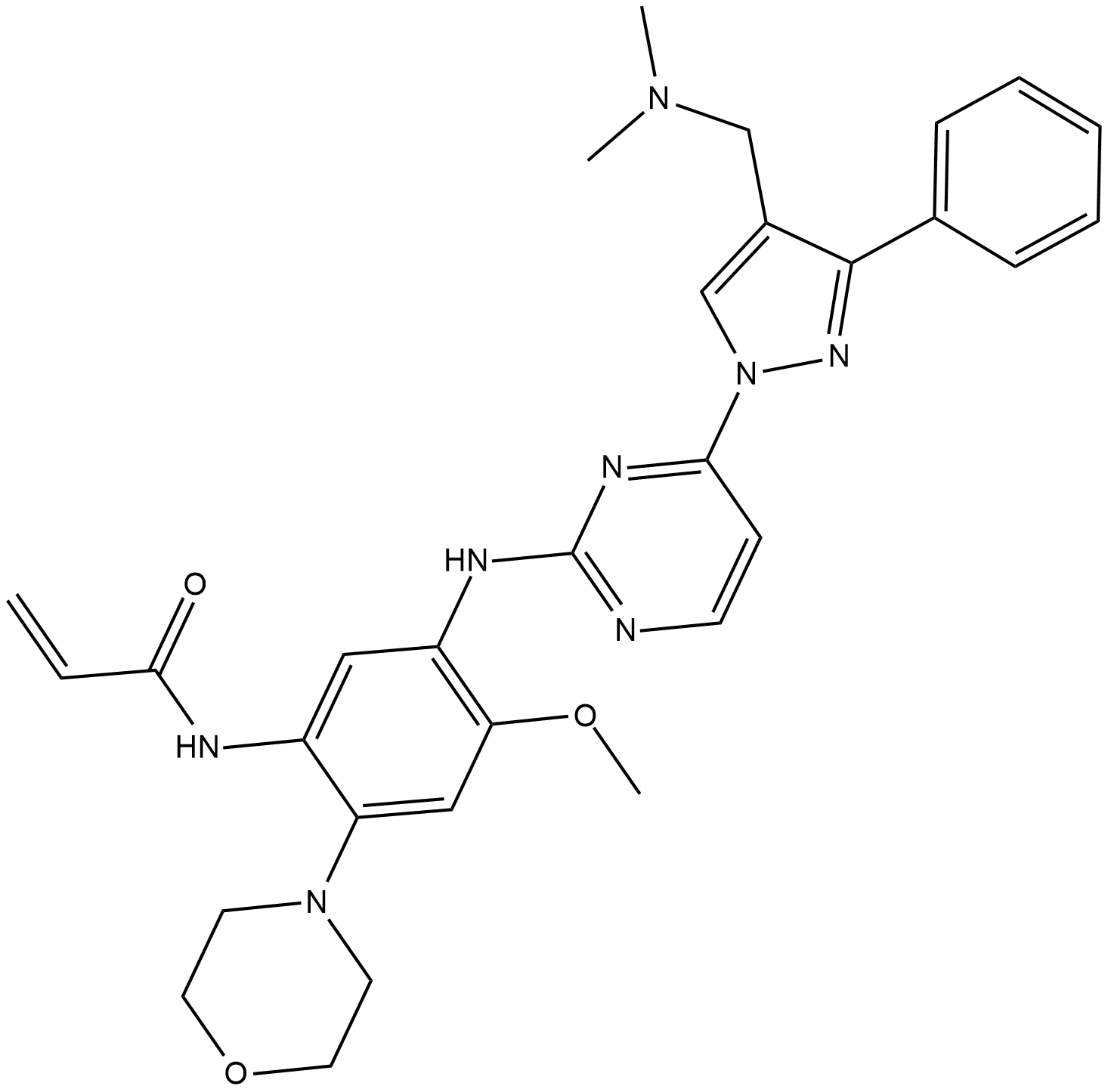
GC13117
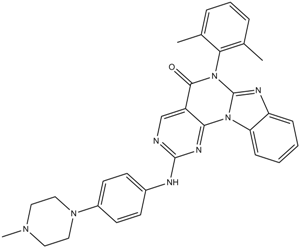
Lck Inhibitor
A selective inhibitor of lymphocyte-specific protein tyrosine kinase
-
GC36430
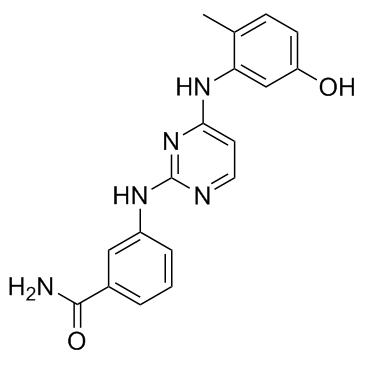
Lck inhibitor 2
Lck inhibitor 2 is a bis-anilinopyrimidine inhibitor of tyrosine kinases including LCK, BTK, LYN, SYK, and TXK. The IC50 values are 13nM, 9nM, 3nM, 26nM and 2nM for Lck, Btk, Lyn, Btk and Txk respectively
-
GC14241
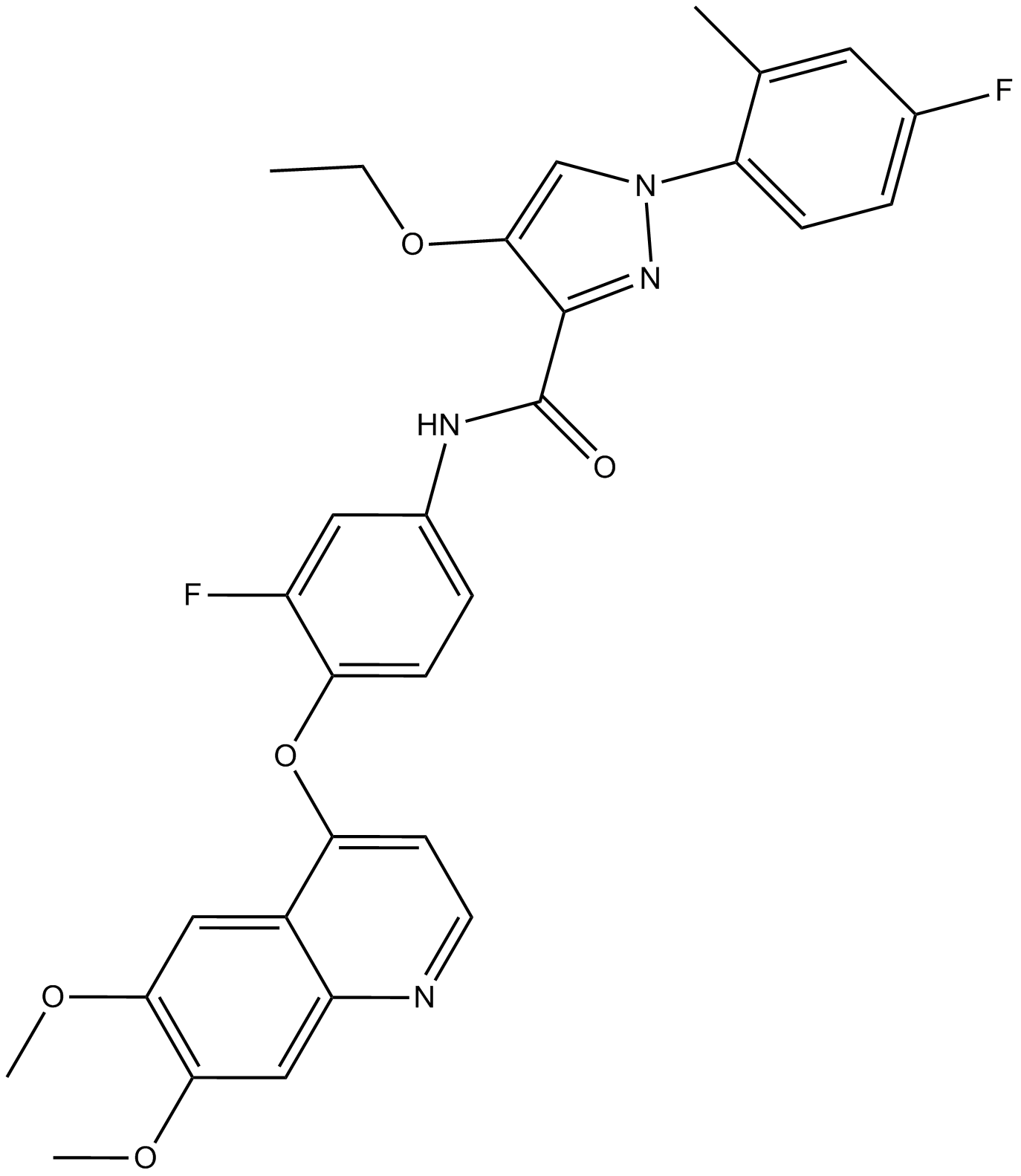
LDC1267
TAM kinase inhibitor,highly selective
-
GC14552
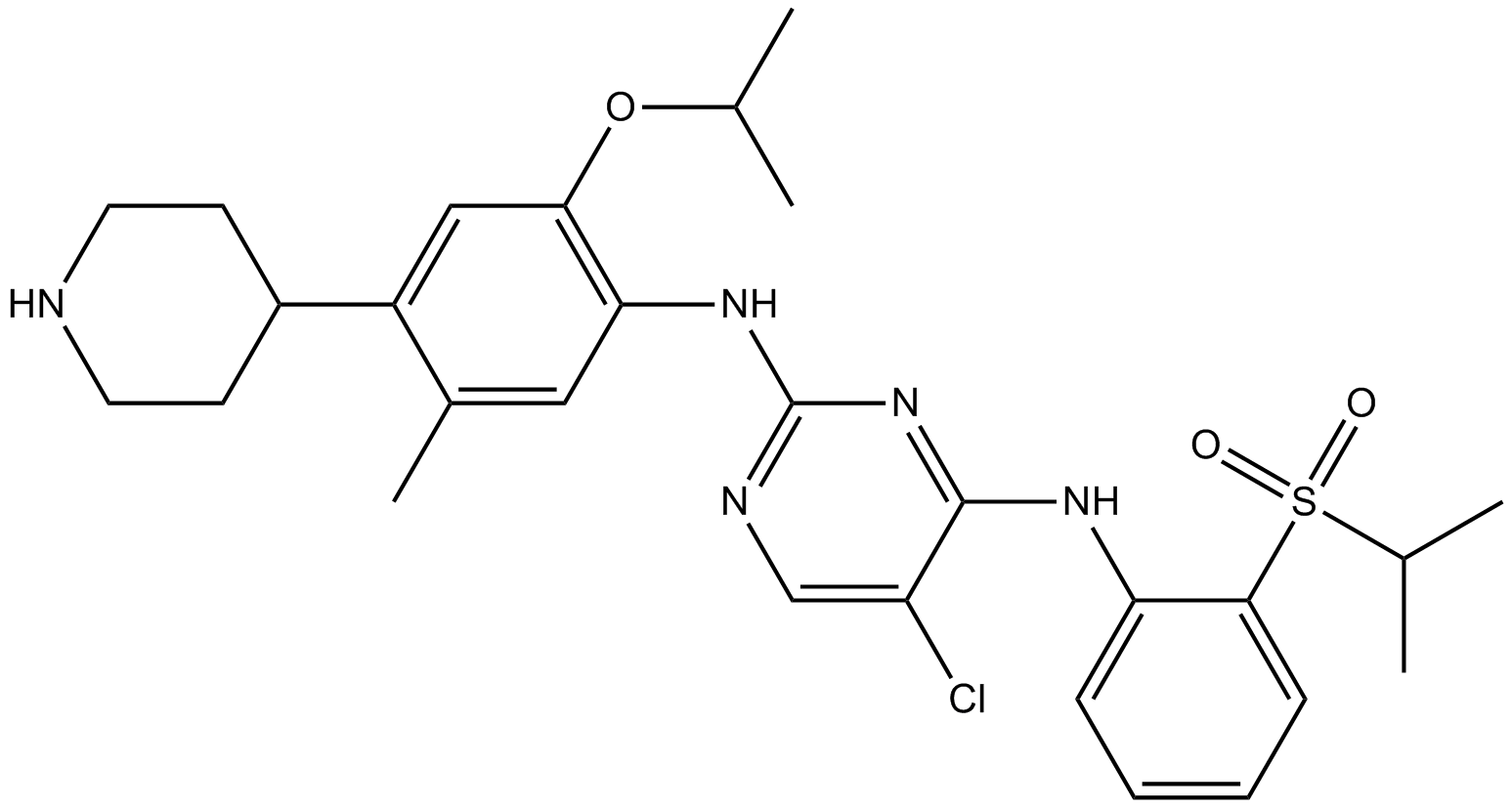
LDK378
LDK378 (LDK378) is a selective, orally bioavailable, and ATP-competitive ALK tyrosine kinase inhibitor with an IC50 of 200 pM.
-
GC17452
LDK378 dihydrochloride
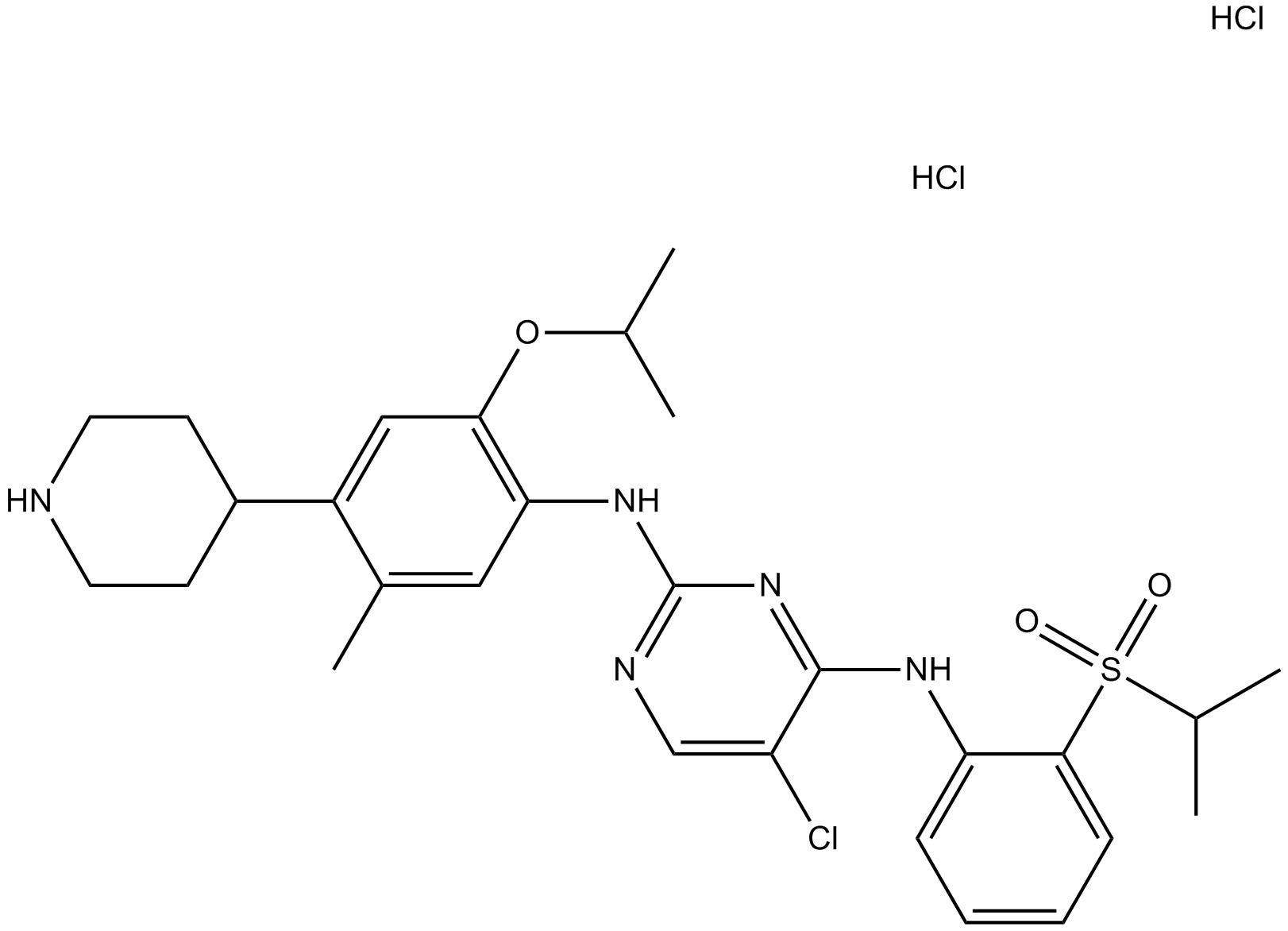
-
GC50327
LDN 193189 dihydrochloride
Potent and selective ALK2 and ALK3 inhibitor; promotes neural induction of hPSCs
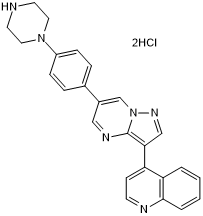
-
GC33086
LDN-192960
LDN-192960 is an inhibitor of Haspin and Dual-specificity Tyrosine-regulated Kinase 2 (DYRK2) with IC50s of 10 nM and 48 nM, respectively.
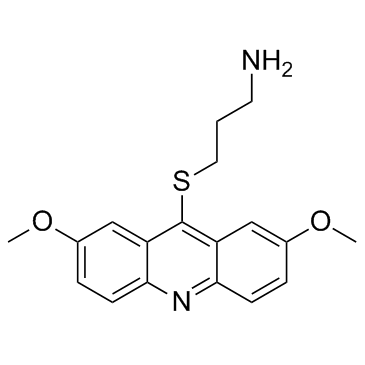
-
GC44047
LDN-192960 (hydrochloride)
LDN-192960 is an inhibitor of haspin protein kinase and dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 (DYRK2) with IC50 values of 10 and 48 nM, respectively.
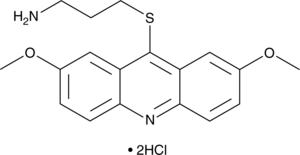
-
GC38403
LDN-192960 hydrochloride
LDN-192960 hydrochloride is an inhibitor of Haspin and Dual-specificity Tyrosine-regulated Kinase 2 (DYRK2) with IC50s of 10 nM and 48 nM, respectively.
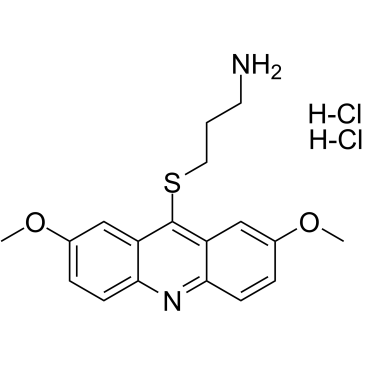
-
GC16580
LDN-193189
ALK inhibitor,potent and selective
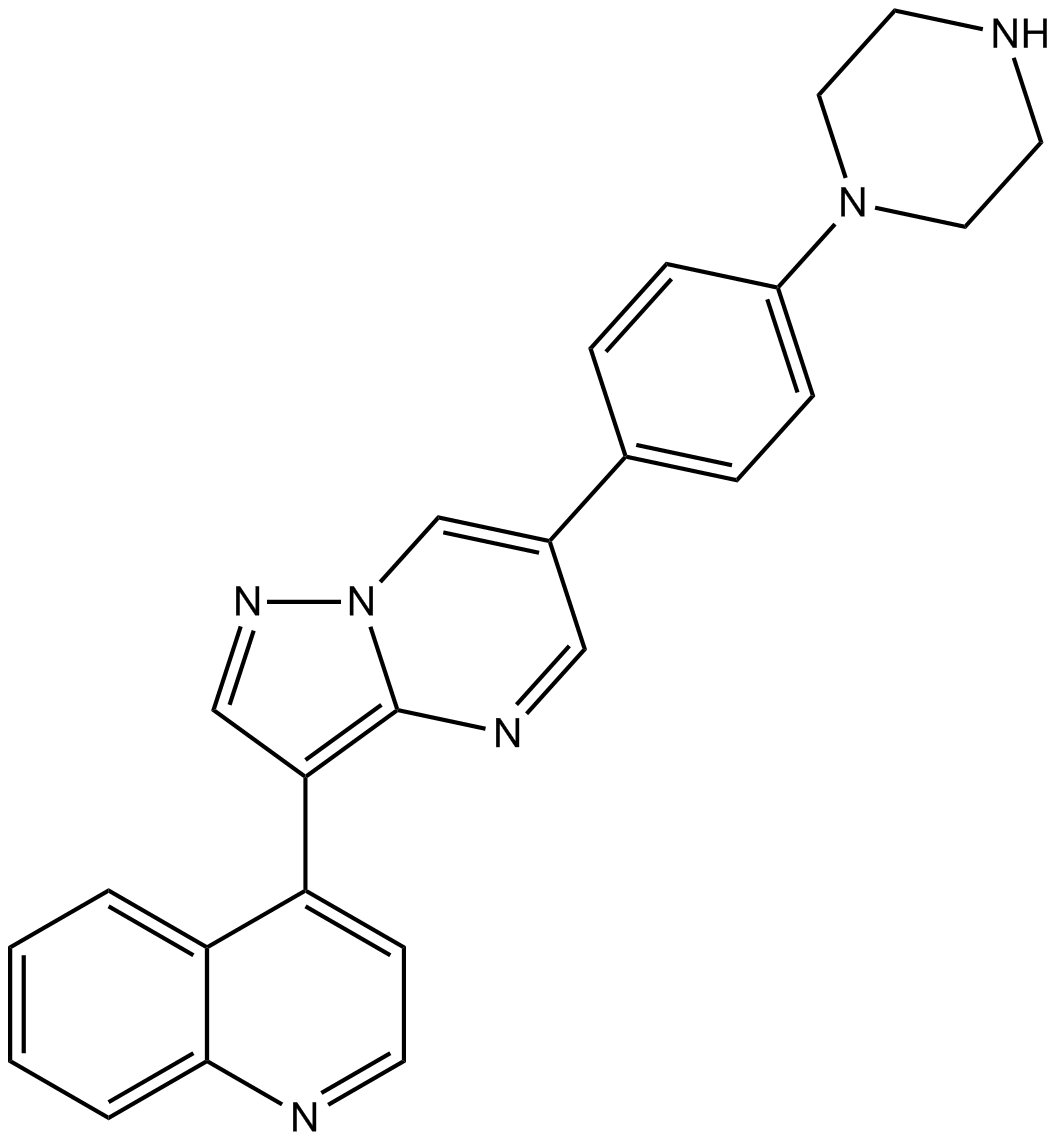
-
GC16798
LDN-211904
LDN-211904 (compound 32) is a potent and selective EphB3 inhibitor with an IC50 of 0.079 μM. LDN-211904 shows good metabolic stability in mouse liver microsomes. LDN-211904 with cetuximab could be effective in inhibiting STAT3-activated CSC stemness and cetuximab resistance in CRC.
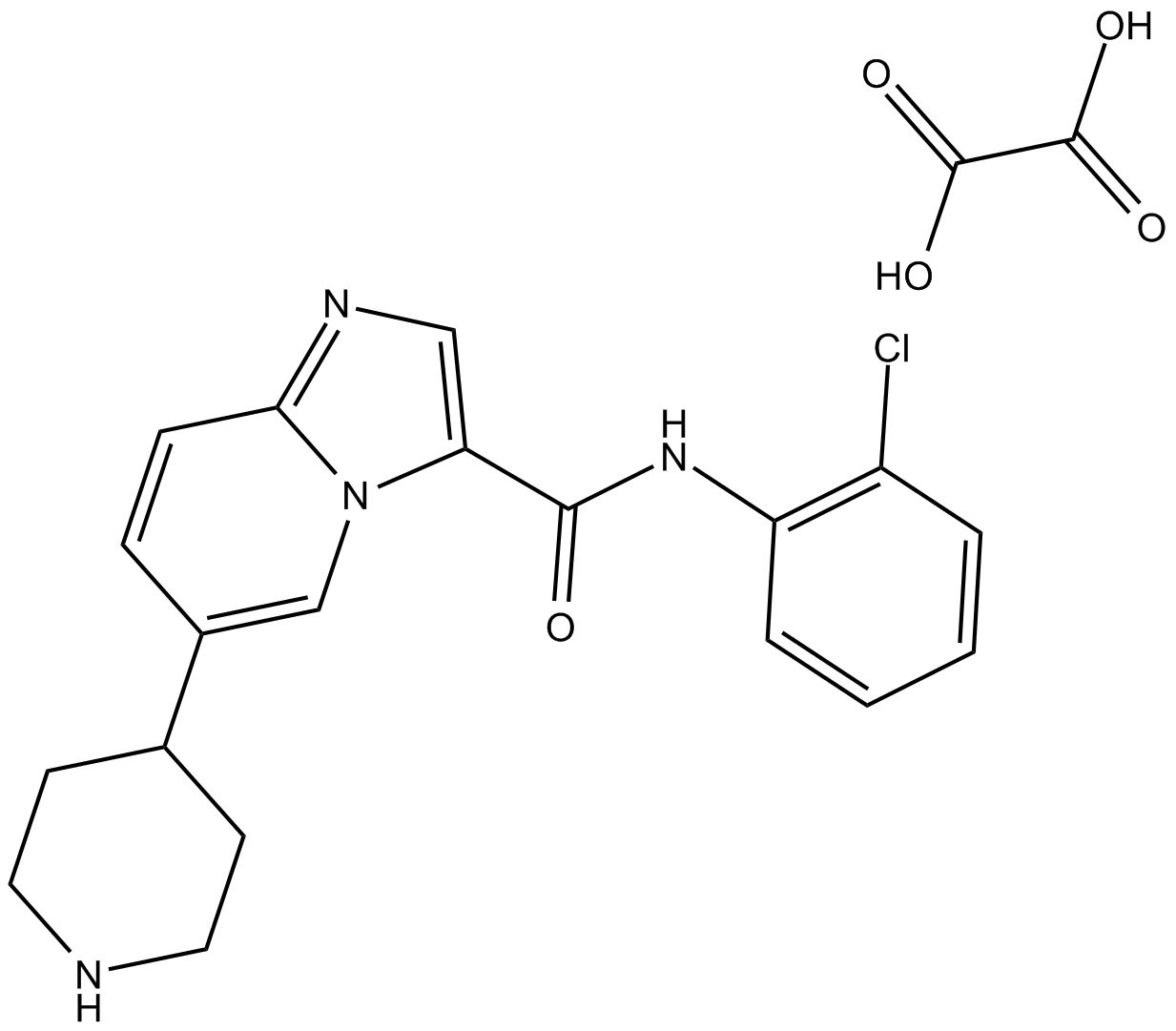
-
GC17035
LDN-212854
BMP receptor inhibitor,potent and selective
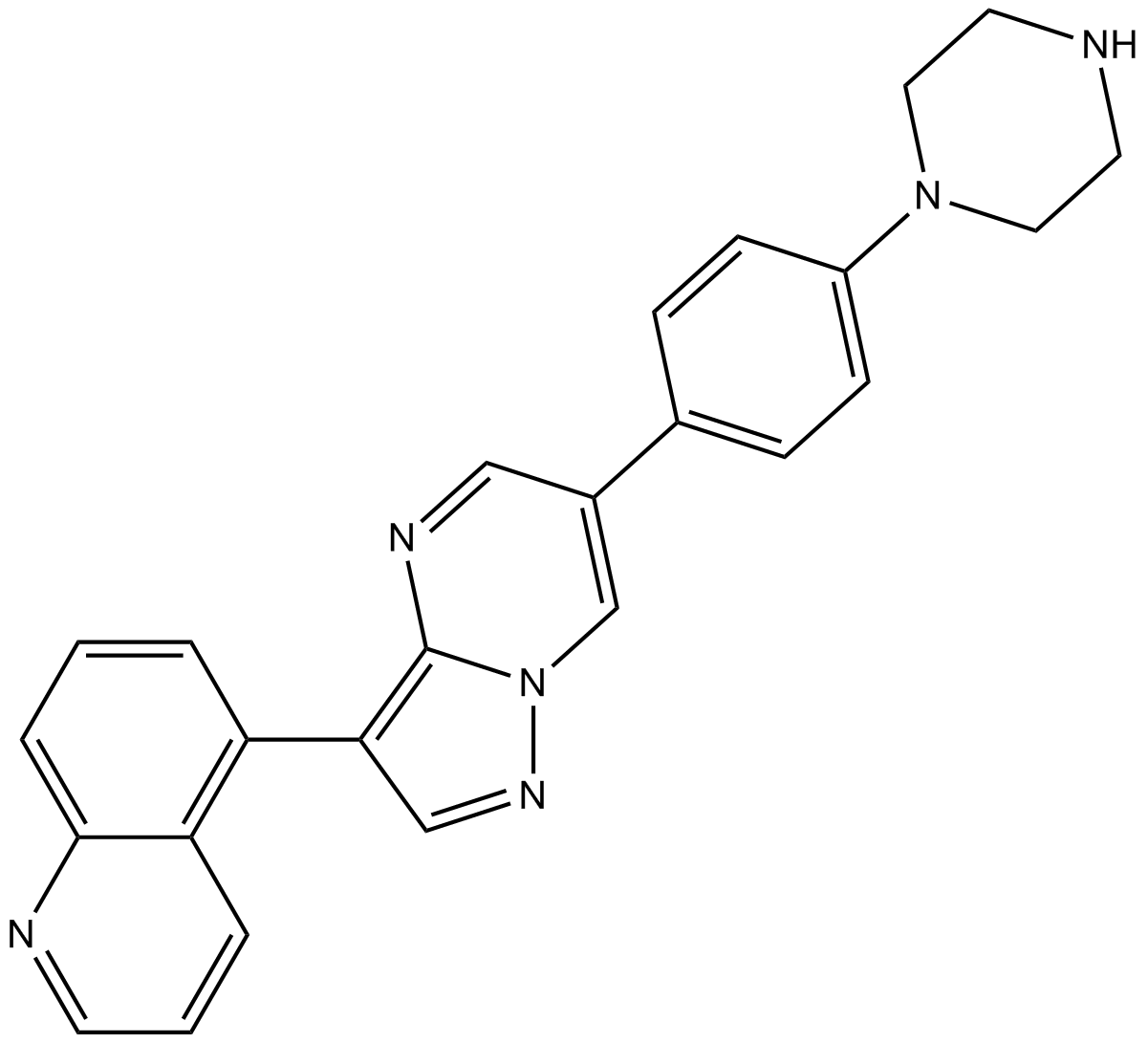
-
GC13225
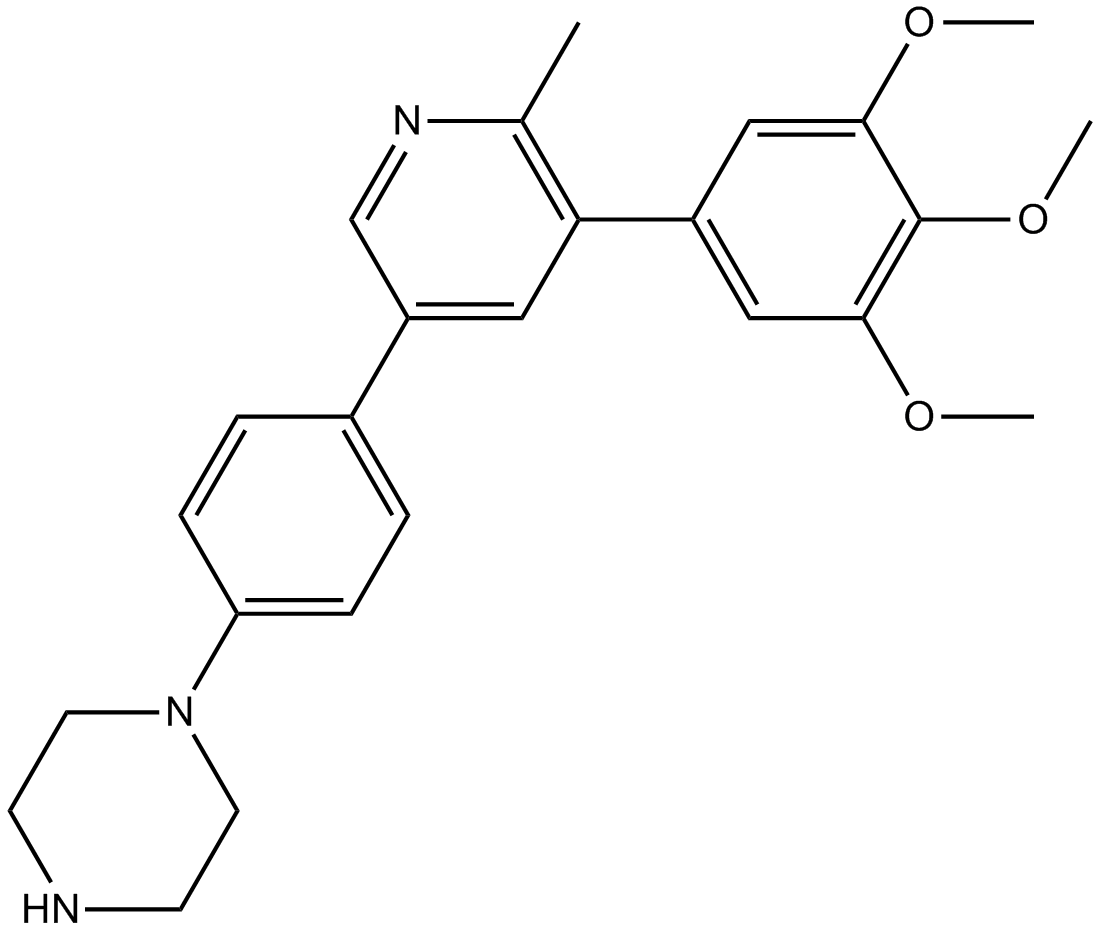
LDN-214117
potent and selective ALK2 inhibitor
-
GC14931
LDN193189 Hydrochloride
An inhibitor of BMP receptors ALK1, ALK2, ALK3, and ALK6

-
GC15454
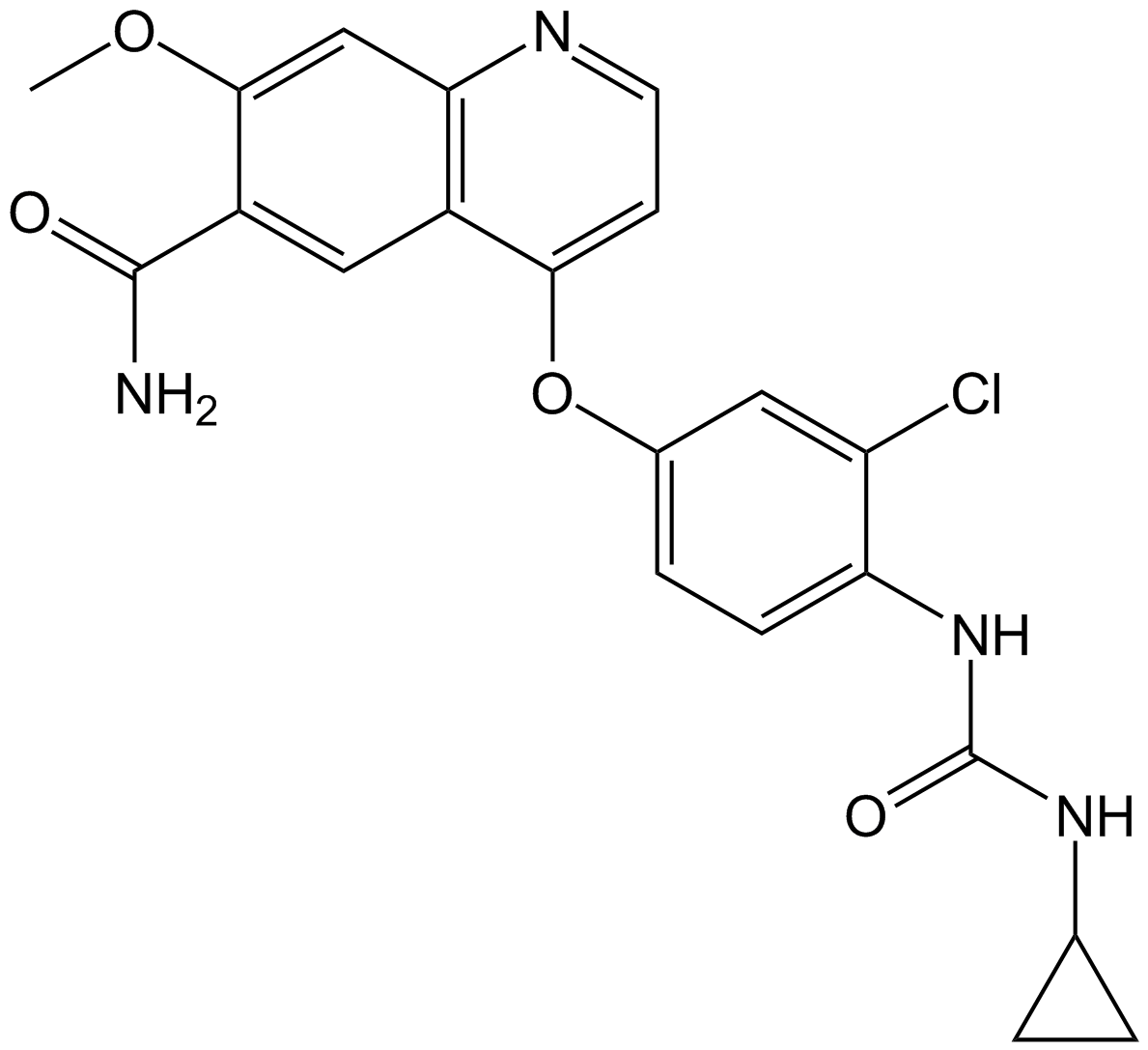
Lenvatinib (E7080)
Lenvatinib (E7080) (E7080) is an oral, multi-targeted tyrosine kinase inhibitor that inhibits VEGFR1-3, FGFR1-4, PDGFR, KIT, and RET, shows potent antitumor activities.
-
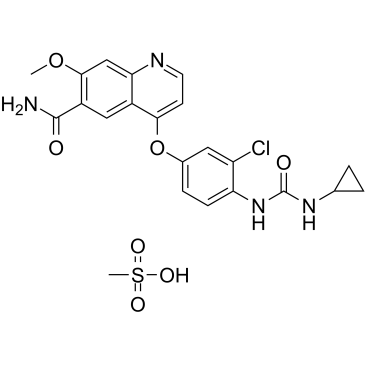
GC36438
Lenvatinib mesylate
An inhibitor of VEGFR2 and VEGFR3
-
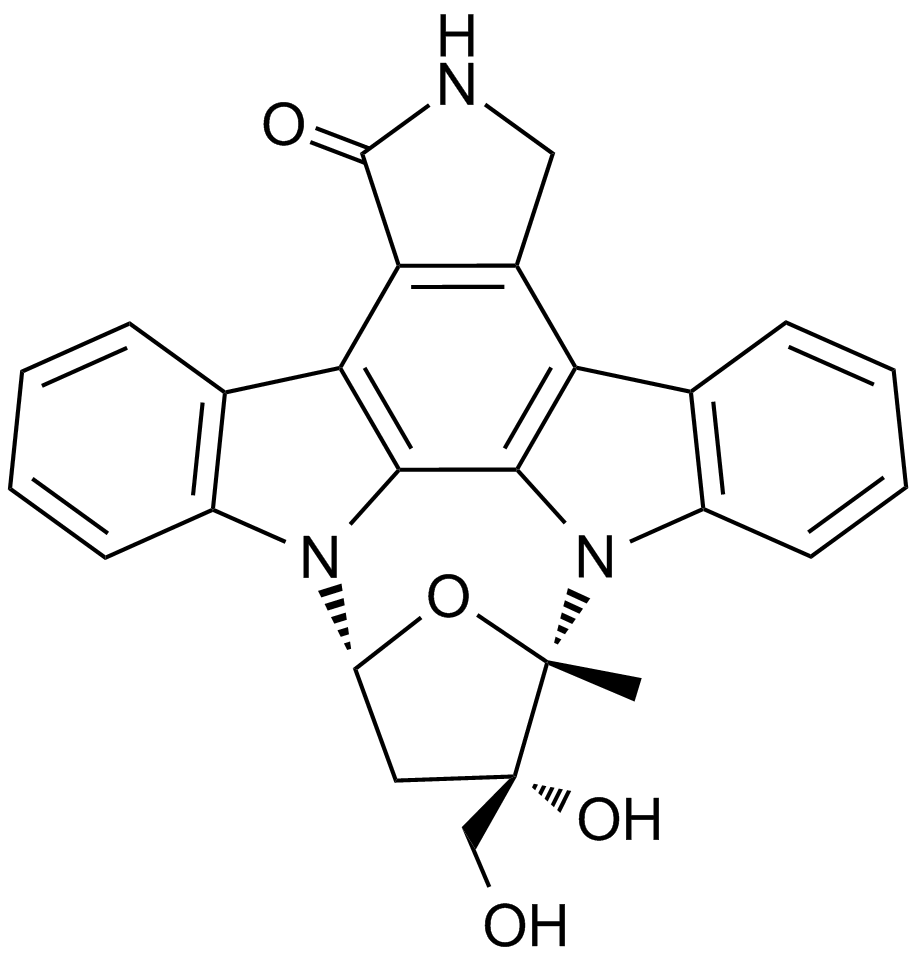
GC17033
Lestaurtinib
A potent JAK2 and PRK1 kinase inhibitor
-
GC17958
Linifanib (ABT-869)
Linifanib (ABT-869) (ABT-869) is a potent and orally active multi-target inhibitor of VEGFR and PDGFR family with IC50s of 4, 3, 66, and 4 nM for KDR, FLT1, PDGFRβ, and FLT3, respectively. Linifanib (ABT-869) shows prominent antitumor activity. Linifanib (ABT-869) has much less activity against unrelated RTKs, soluble tyrosine kinases, or serine/threonine kinases. Linifanib (ABT-869) is a specific miR-10b inhibitor that blocks miR-10b biogenesis.
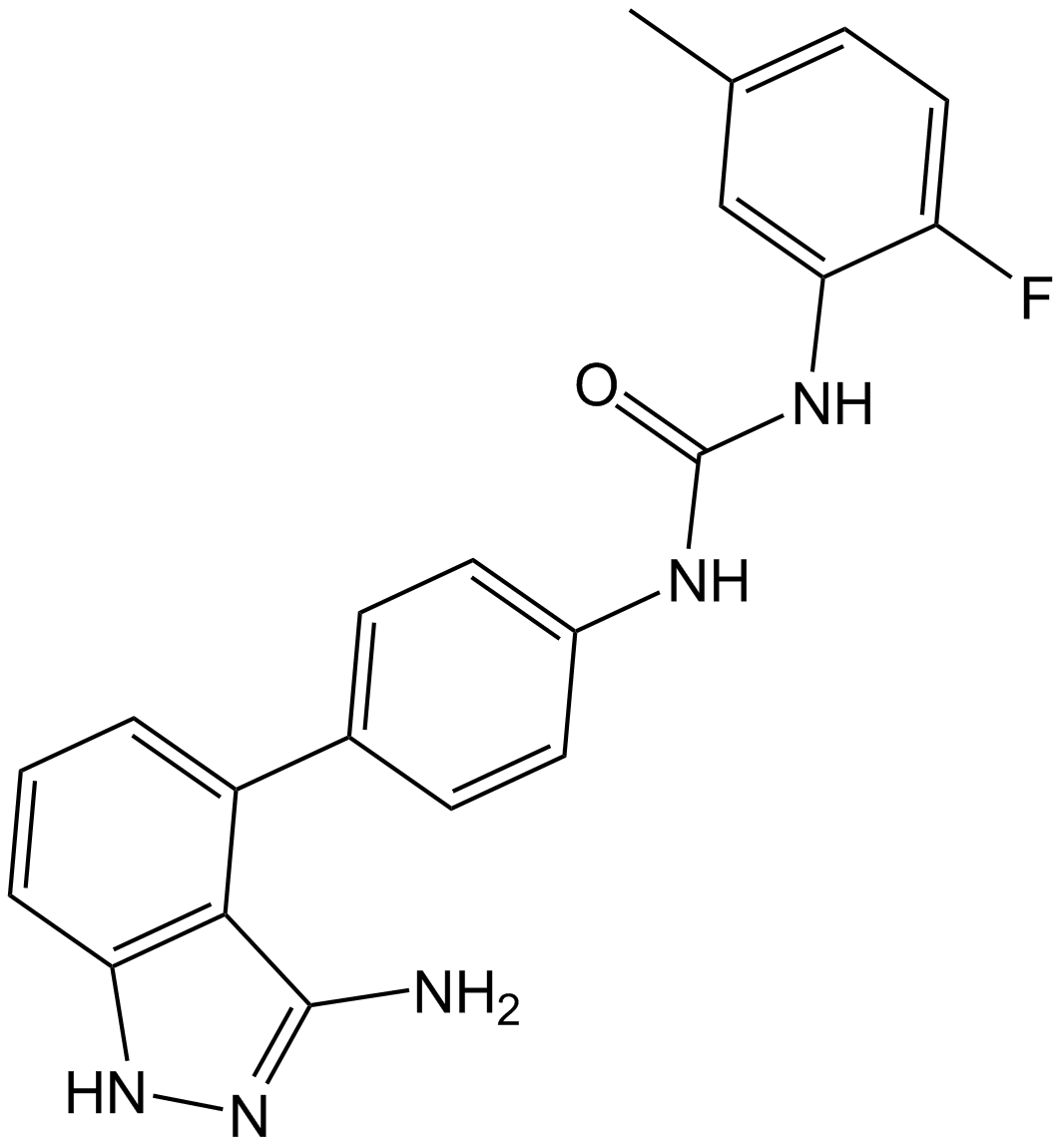
-
GC15749
Linsitinib
IGF1R/IR inhibitor,potent and novel
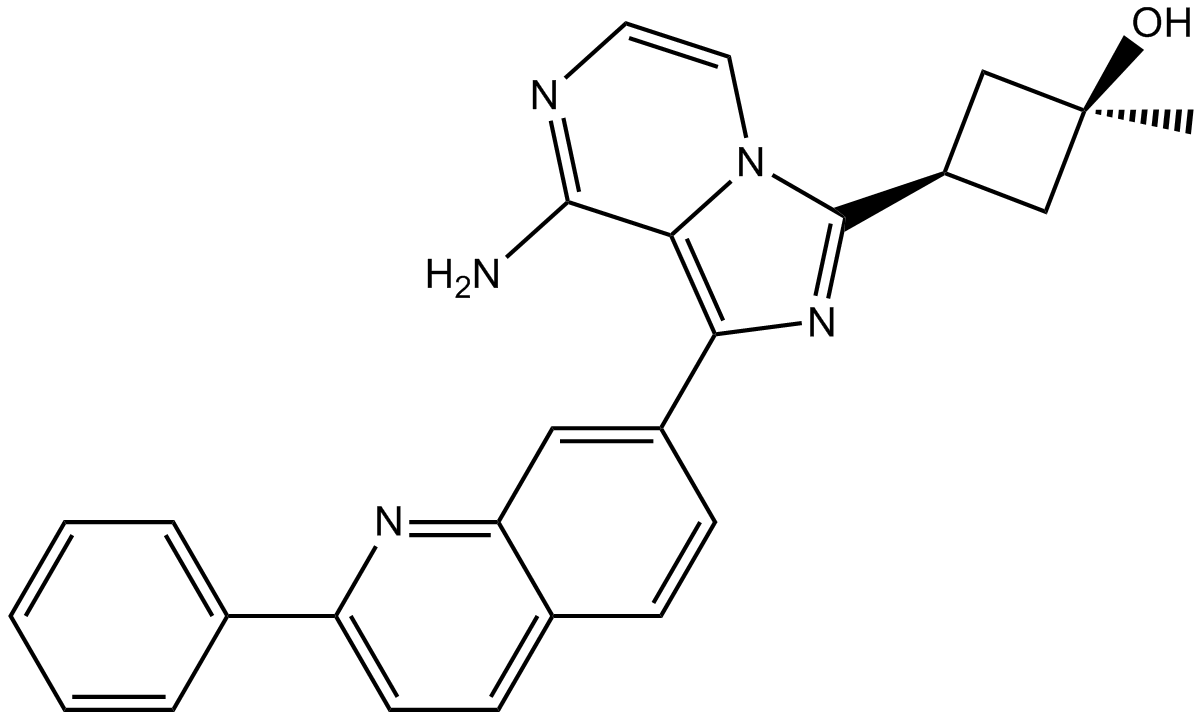
-
GC47570
Lipoxygenin
An inhibitor of 5-LO
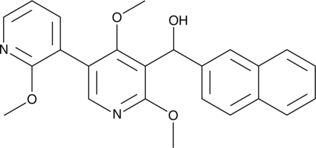
-
GC17835
LM 22A4
tropomyosin-related kinase B (TrkB) agonist
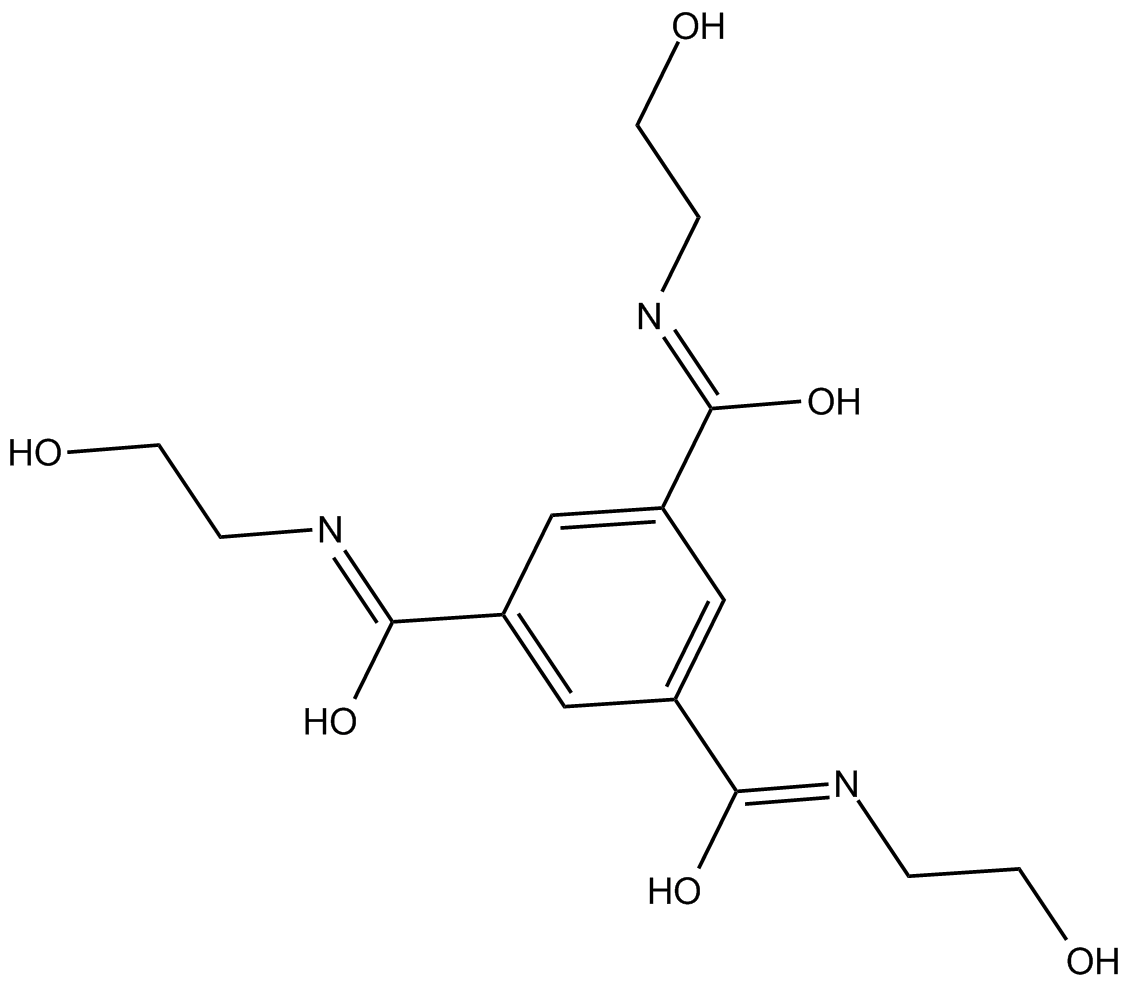
-
GC30770
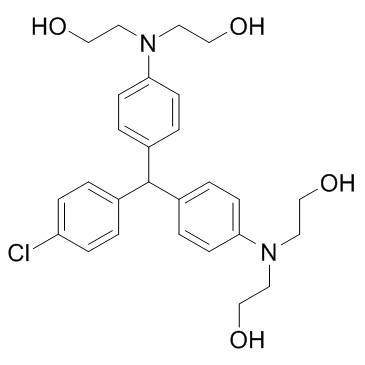
LM22B-10
An activator of TrkB and TrkC
-
GC36425
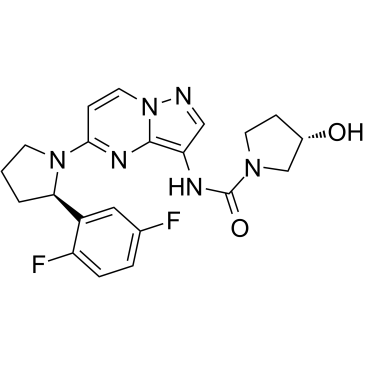
LOXO-101 (Larotrectinib)
LOXO-101 (Larotrectinib) (LOXO-101) is an ATP-competitive oral, selective inhibitor of the tropomyosin-related kinase (TRK) family receptors, with low nanomolar 50% inhibitory concentrations against all three isoforms (TRKA, B, and C).
-
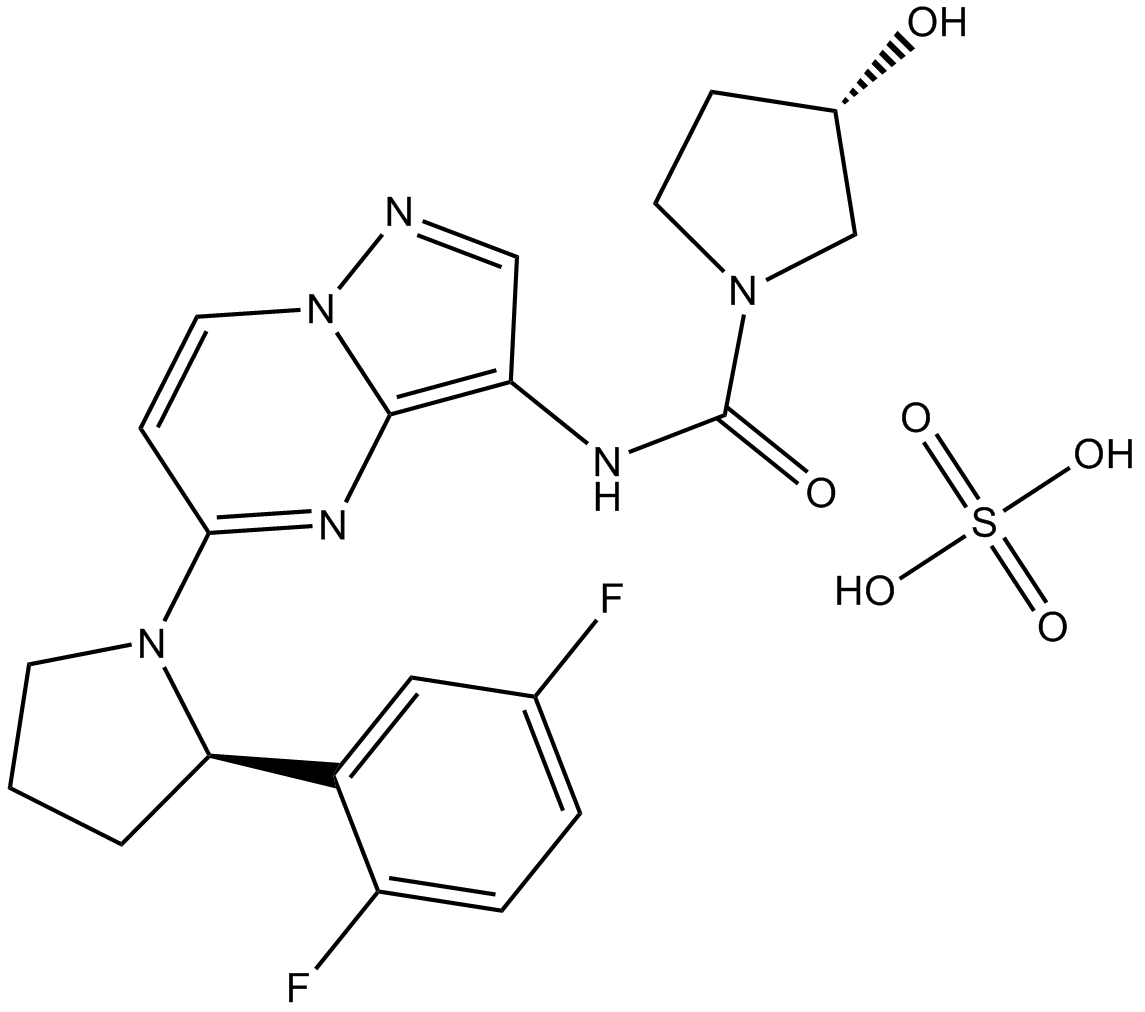
GC15282
LOXO-101 (Larotrectinib) sulfate
LOXO-101 (Larotrectinib) sulfate (LOXO-101 sulfate; ARRY-470 sulfate) is an ATP-competitive oral, selective inhibitor of the tropomyosin-related kinase (TRK) family receptors, with low nanomolar 50% inhibitory concentrations against all three isoforms (TRKA, B, and C).
-
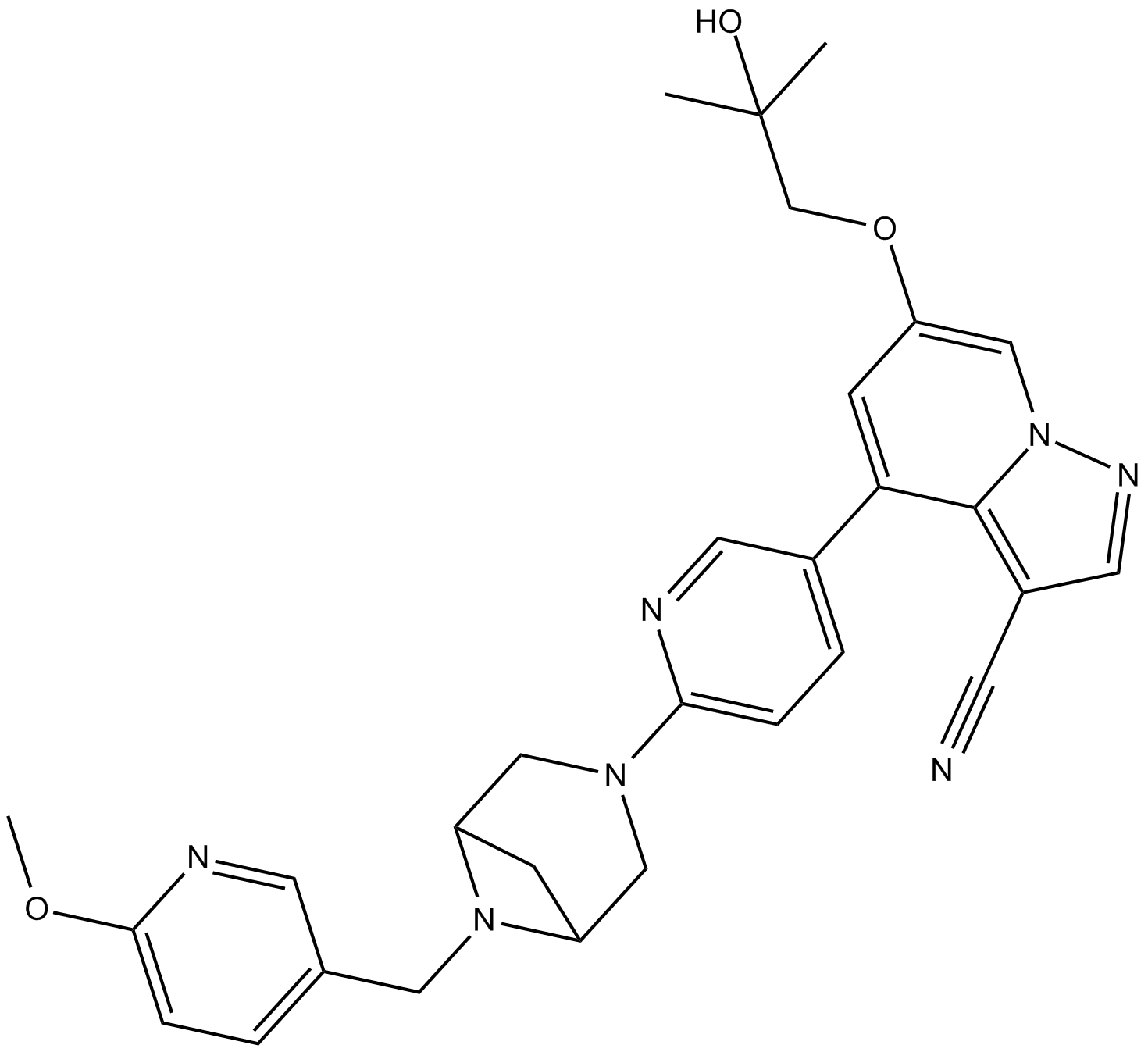
GC19547
LOXO-292
LOXO-292 (LOXO-292) is a potent, selective RET kinase inhibitor with IC50 values of 14.0 nM, 24.1 nM, and 530.7 nM for RET (WT), RET (V804M), and RET (G810R), respectively. LOXO-292 has anticancer activity.
-
GC65390
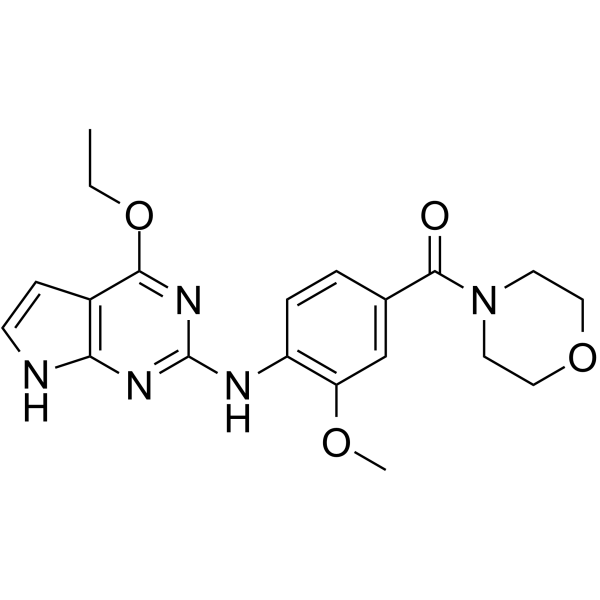
LRRK2 inhibitor 1
LRRK2 inhibitor 1 is a potent, selective and oral LRRK2 inhibitor with an pIC50 of 6.8.
-
GC10809
LRRK2-IN-1
A selective LRRK2 inhibitor

-
GC69410
Lumretuzumab
Lumretuzumab (Anti-Human ERBB3 Recombinant Antibody) is a humanized monoclonal antibody that targets HER3 (ERBB3) and can be used for cancer research.

-
GC13848
LY2784544
Potent inhibitor of JAK2
-
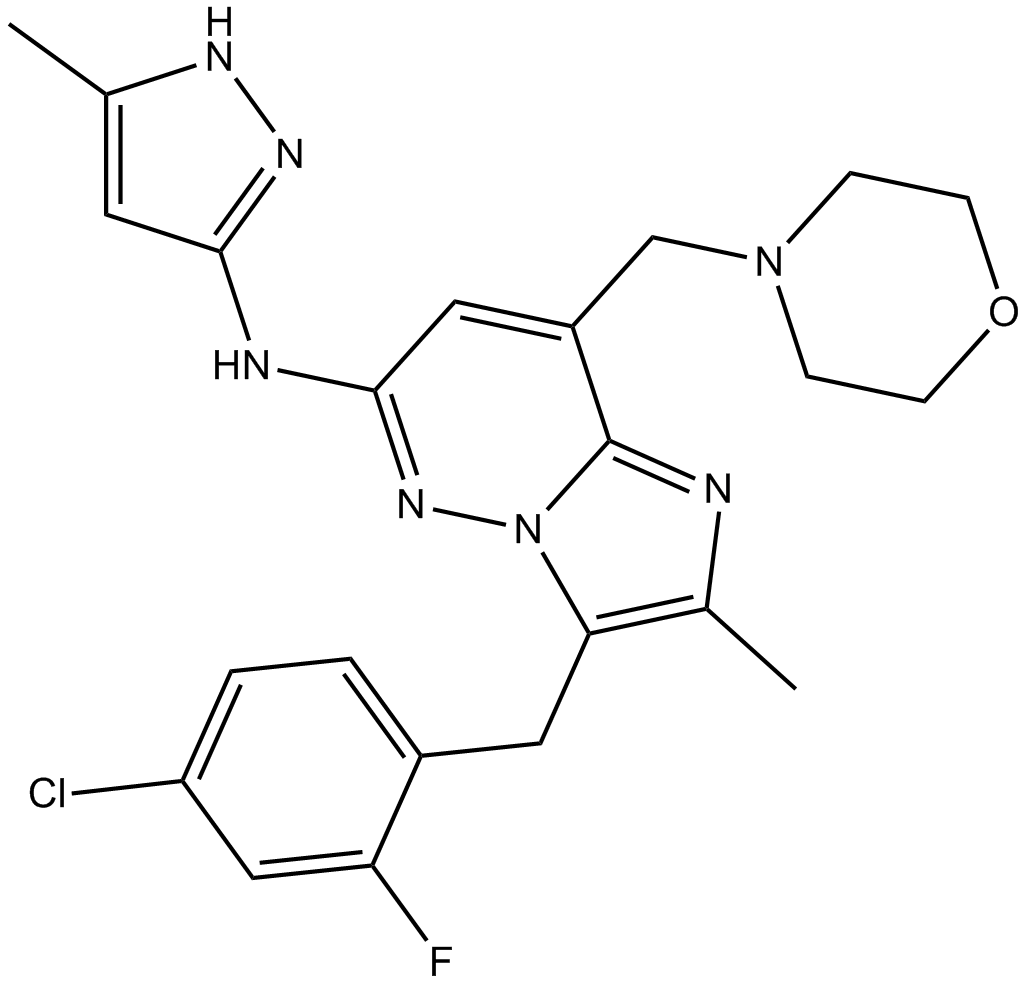
GC11057
LY2801653
A MET kinase inhibitor
-
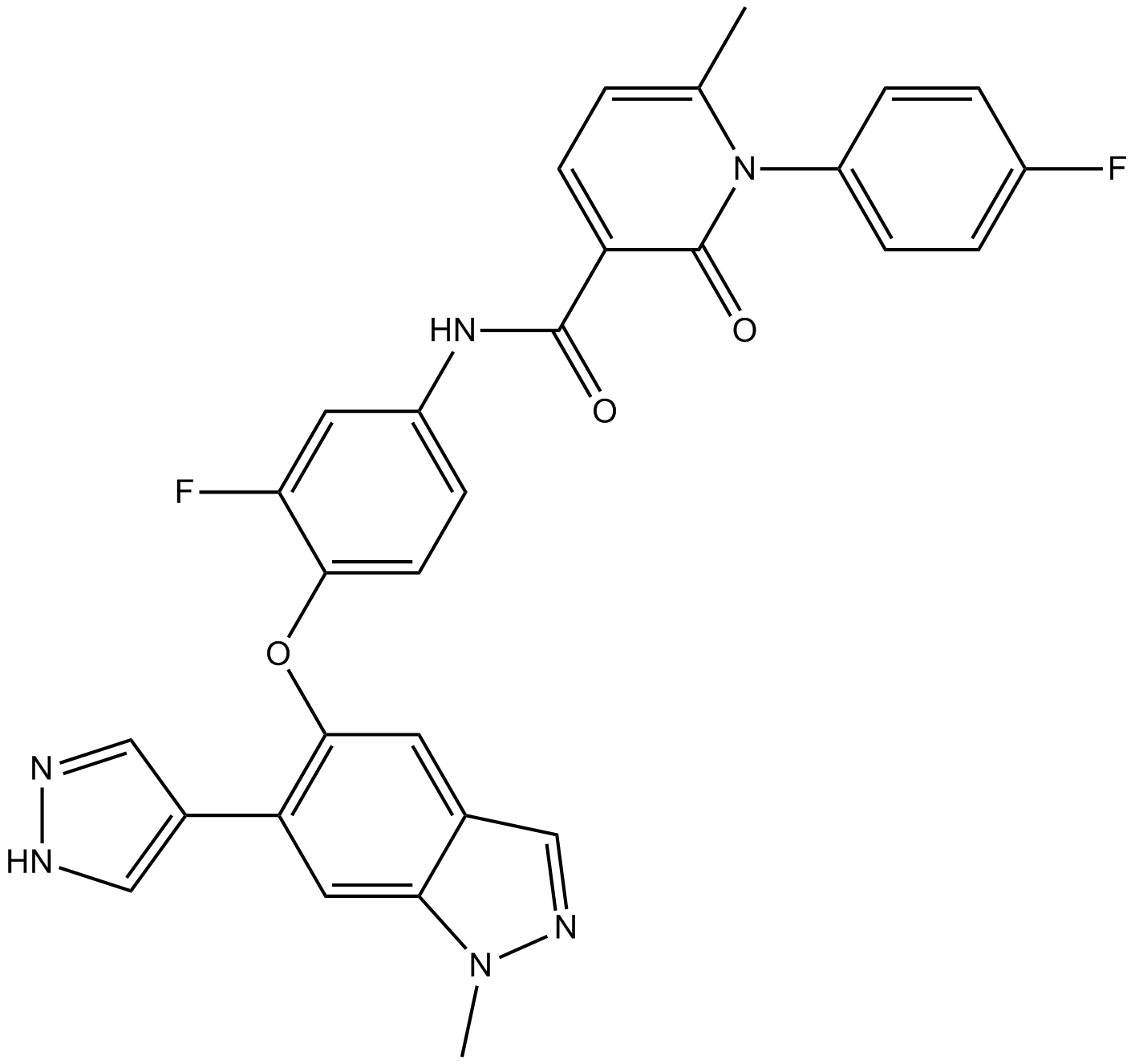
GC13424
LY2874455
A pan-FGFR inhibitor

-
GC40865
LYG-202
LYG-202 is a synthetic flavonoid with anticancer and anti-angiogenic activities.
-
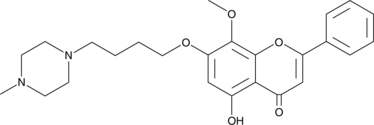
GC62314
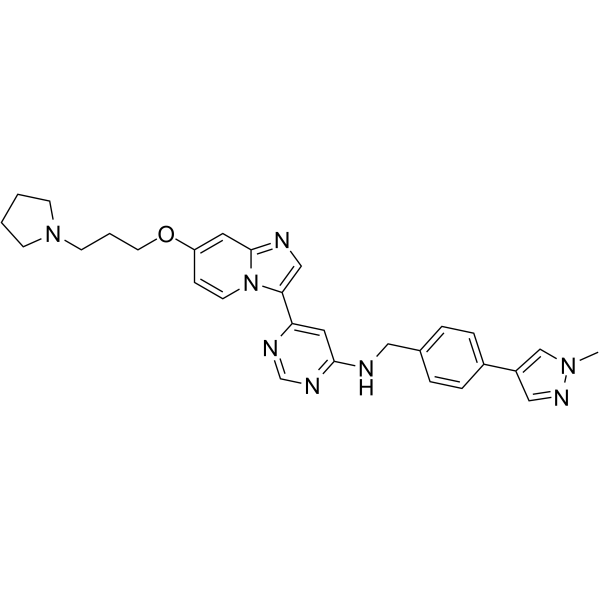
M4205
M4205 is a c-KIT inhibitor, with an IC50 of 10 nM for c-KIT V654A. M4205 has high activity on c-KIT mutations in exon 11, 13, 17.
-
GC68304
Margetuximab

-
GC36546
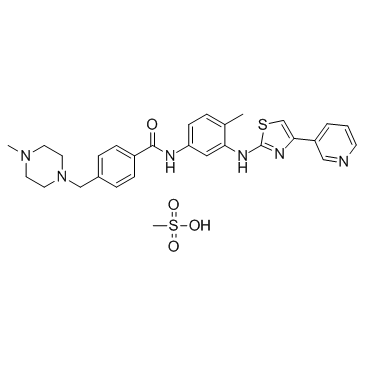
Masitinib mesylate
Masitinib mesylate (AB-1010 mesylate) is a potent, orally bioavailable, and selective inhibitor of c-Kit (IC50=200 nM for human recombinant c-Kit). It also inhibits PDGFRα/β (IC50s=540/800 nM), Lyn (IC50= 510 nM for LynB), Lck, and, to a lesser extent, FGFR3 and FAK. Masitinib mesylate (AB-1010 mesylate) has anti-proliferative, pro-apoptotic activity and low toxicity.
-
GC66349
Mavrilimumab
Mavrilimumab (CAM 3001) is a monoclonal antibody that binds to the α subunit of the granulocyte-macrophage colony stimulating factor (GM-CSF) receptor and blocks intracellular signalling downstream of GM-CSF. GM-CSF might be a mediator of the hyperactive inflammatory response associated with respiratory failure and death.

-
GC65179
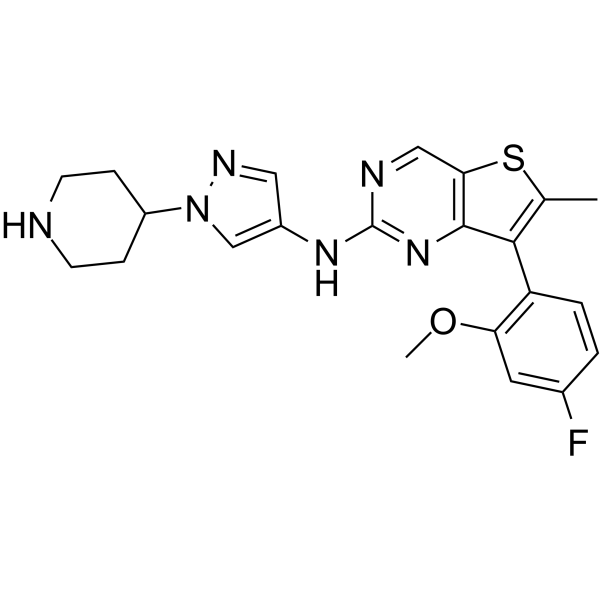
MAX-40279
MAX-40279 is a dual and potent inhibitor of FLT3 kinase and FGFR kinase.
-
GC64583
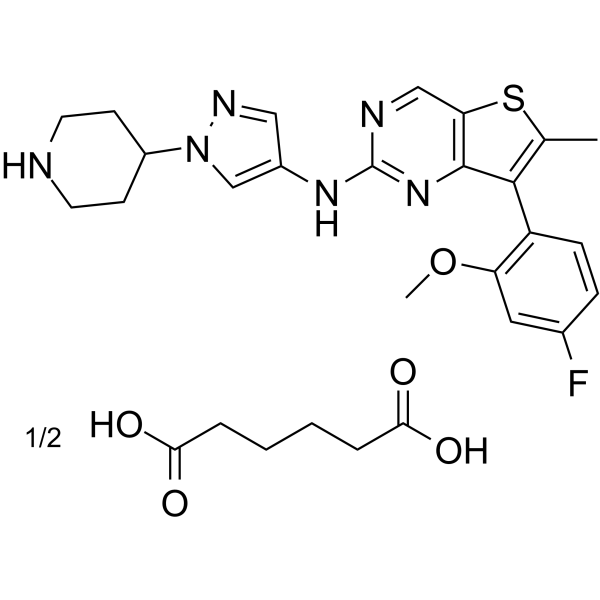
MAX-40279 hemiadipate
MAX-40279 hemiadipate is a dual and potent inhibitor of FLT3 kinase and FGFR kinase.
-
GC64582
MAX-40279 hemifumarate
MAX-40279 hemifumarate is a dual and potent inhibitor of FLT3 kinase and FGFR kinase.
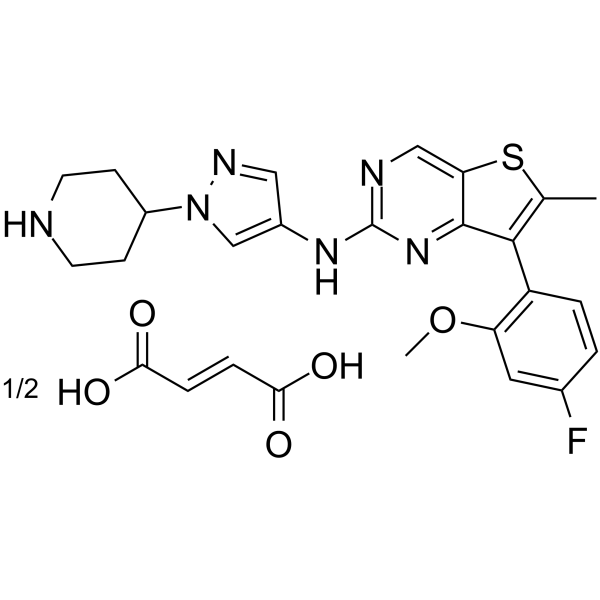
-
GC16483
MAZ51
VEGFR3 antagonist
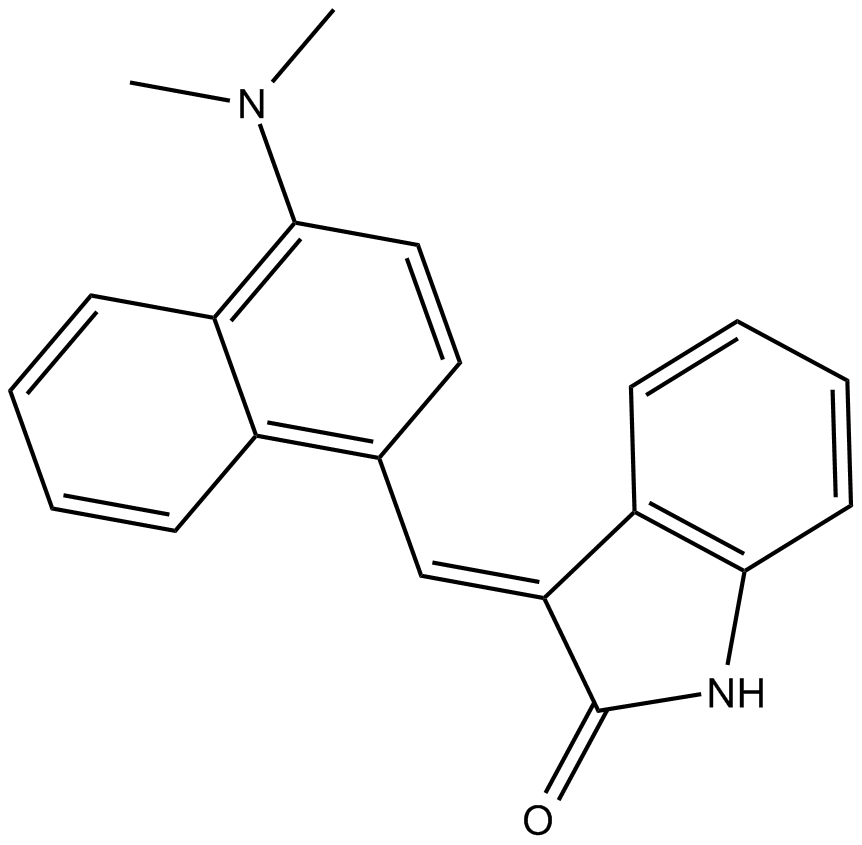
-
GC64710
MC-Val-Cit-PAB-Amide-TLR7 agonist 4
MC-Val-Cit-PAB-Amide-TLR7 agonist 4 (example 15) is a HER2-TLR7 and HER2-TLR8 immune agonist conjugate.
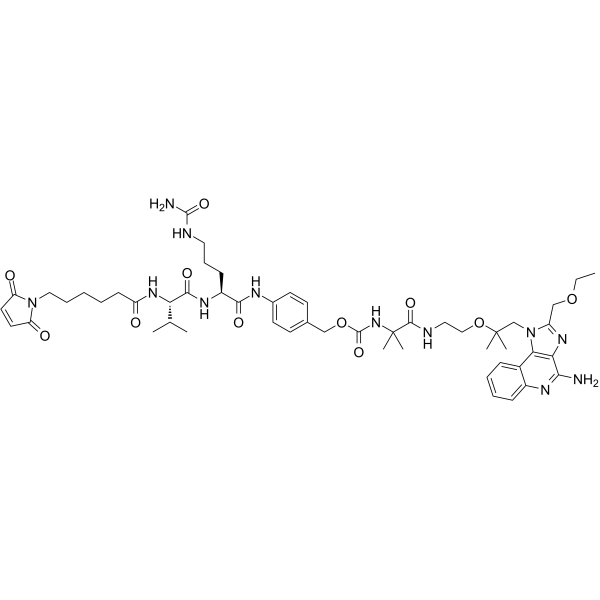
-
GC14951
Meleagrin
antibiotic
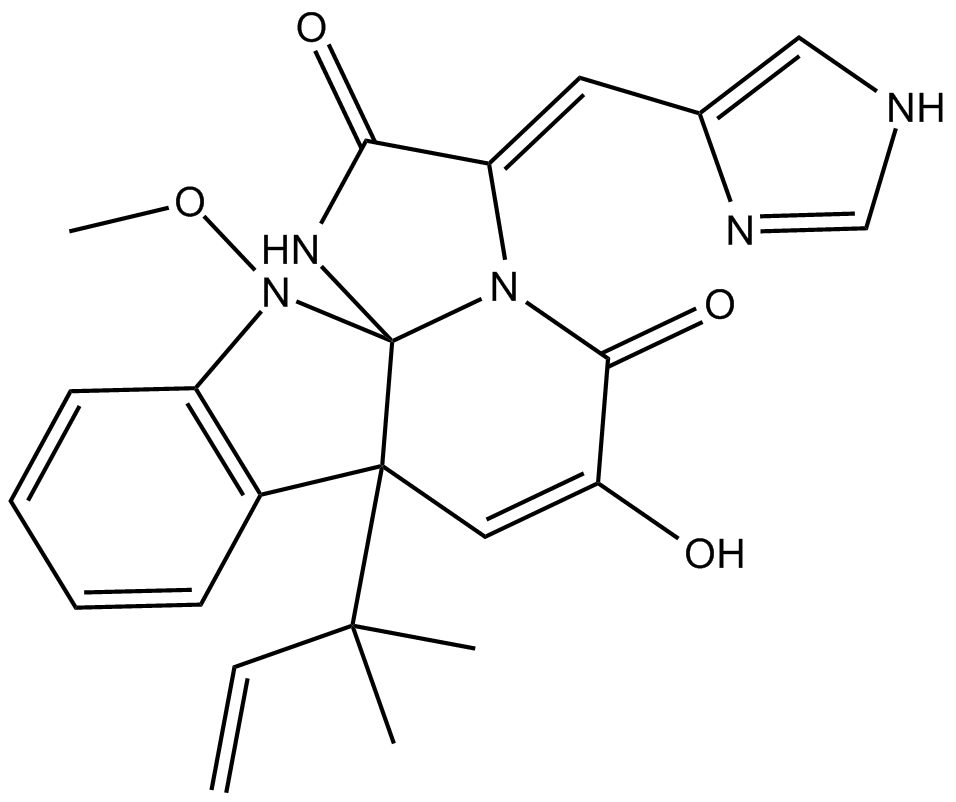
-
GC36585
Merestinib dihydrochloride
Merestinib dihydrochloride (LY2801653 dihydrochloride) is a potent, orally bioavailable c-Met inhibitor (Ki=2 nM) with anti-tumor activities. Merestinib dihydrochloride also has potent activity against MST1R (IC50=11 nM), FLT3 (IC50=7 nM), AXL (IC50=2 nM), MERTK (IC50=10 nM), TEK (IC50=63 nM), ROS1, DDR1/2 (IC50=0.1/7 nM) and MKNK1/2 (IC50=7 nM).
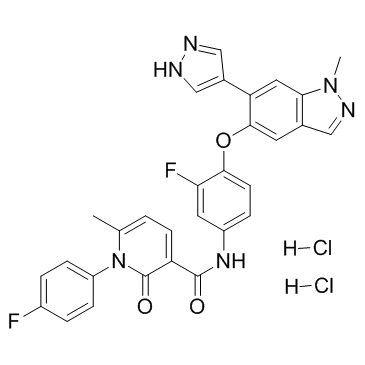
-
GC68018
MET kinase-IN-2
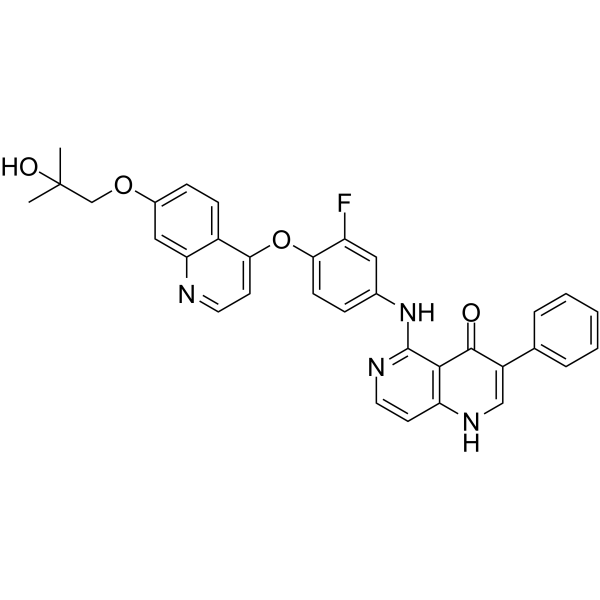
-
GC12069
Methyl 2,5-dihydroxycinnamate
EGF receptor-associated tyrosine kinases inhibitor
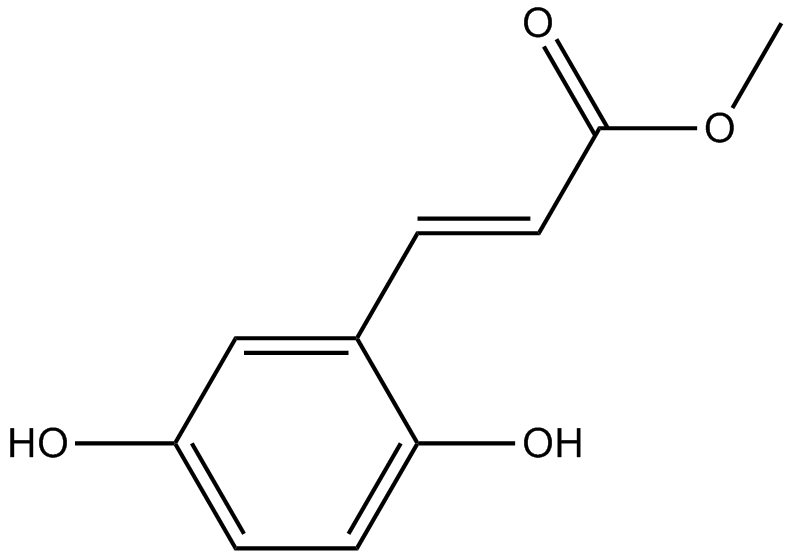
-
GC36596
Methylnissolin
Methylnissolin (Astrapterocarpan), isolated from Astragalus membranaceus, inhibits platelet-derived growth factor (PDGF)-BB-induced cell proliferation with an IC50 of 10 μM.
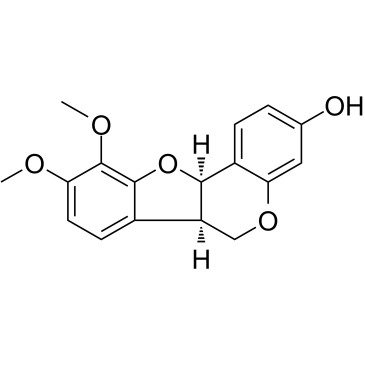
-
GC13598
MGCD-265
MGCD-265 is a potent and oral active inhibitor of c-Met and VEGFR2 tyrosine kinases, with IC50s of 29 nM and 10 nM, respectively. MGCD-265 has significant antitumor activity.
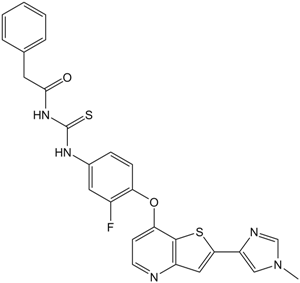
-
GC61516
MID-1
MID-1 is a disruptor of MG53-IRS-1 (Mitsugumin 53-insulin receptor substrate-1) interaction.
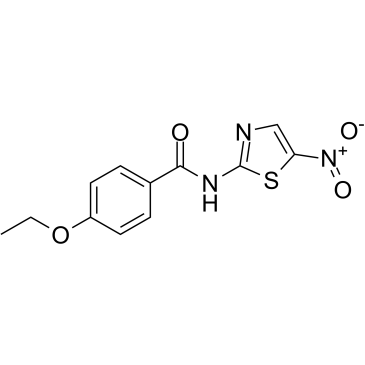
-
GC32819
Mirk-IN-1 (Dyrk1B/A-IN-1)
Mirk-IN-1 (Dyrk1B/A-IN-1) is a potent inhibitor of Dyrk1B(Mirk kianse) and Dyrk1A with IC50 of 68±48 nM and 22±8 nM respectively.
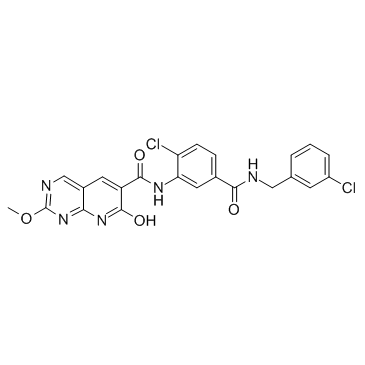
-
GC16337
MK-2461
C-Met (WT/mutants) inhibitor
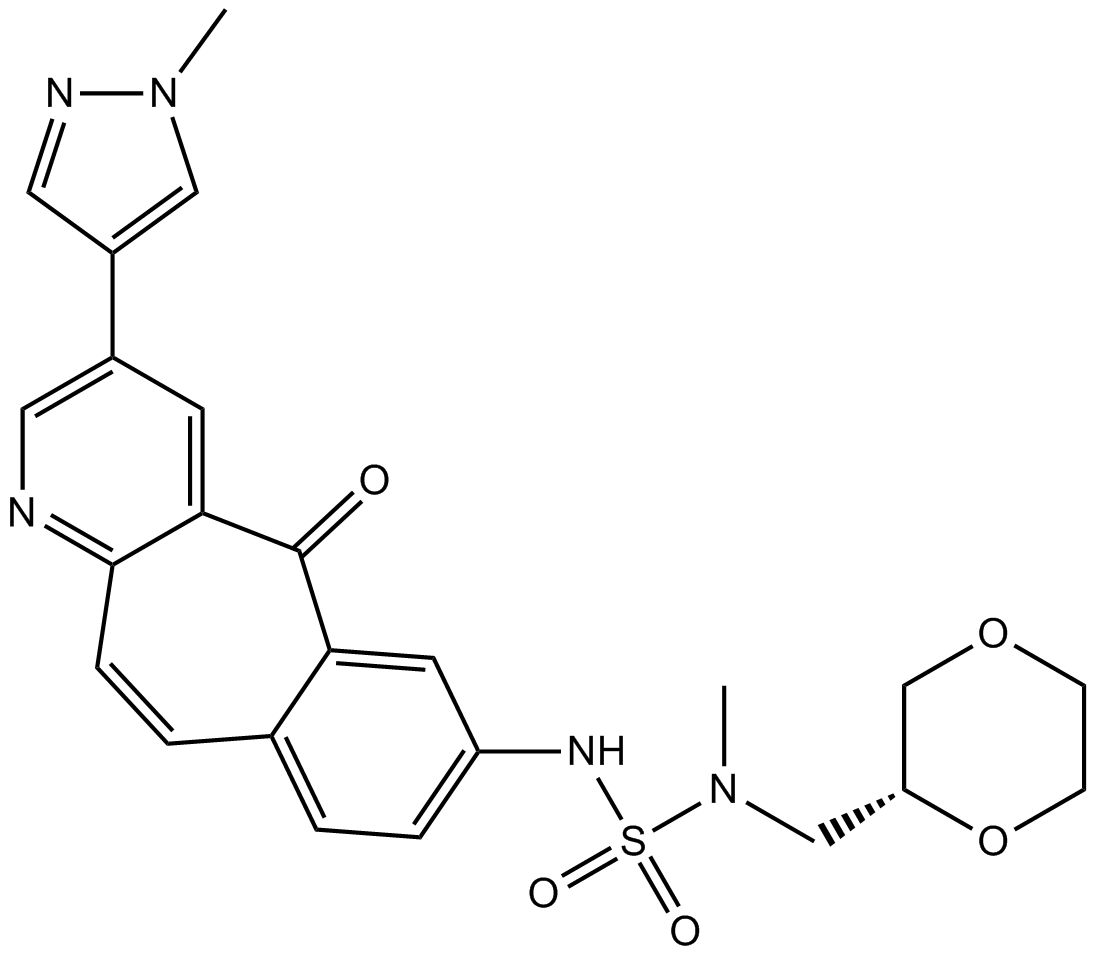
-
GC13140
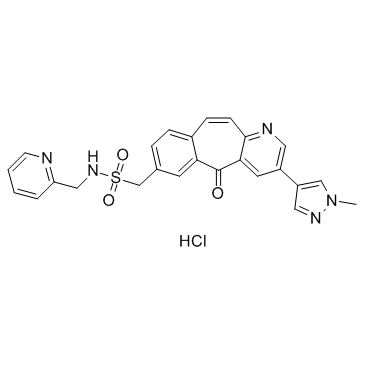
MK-8033
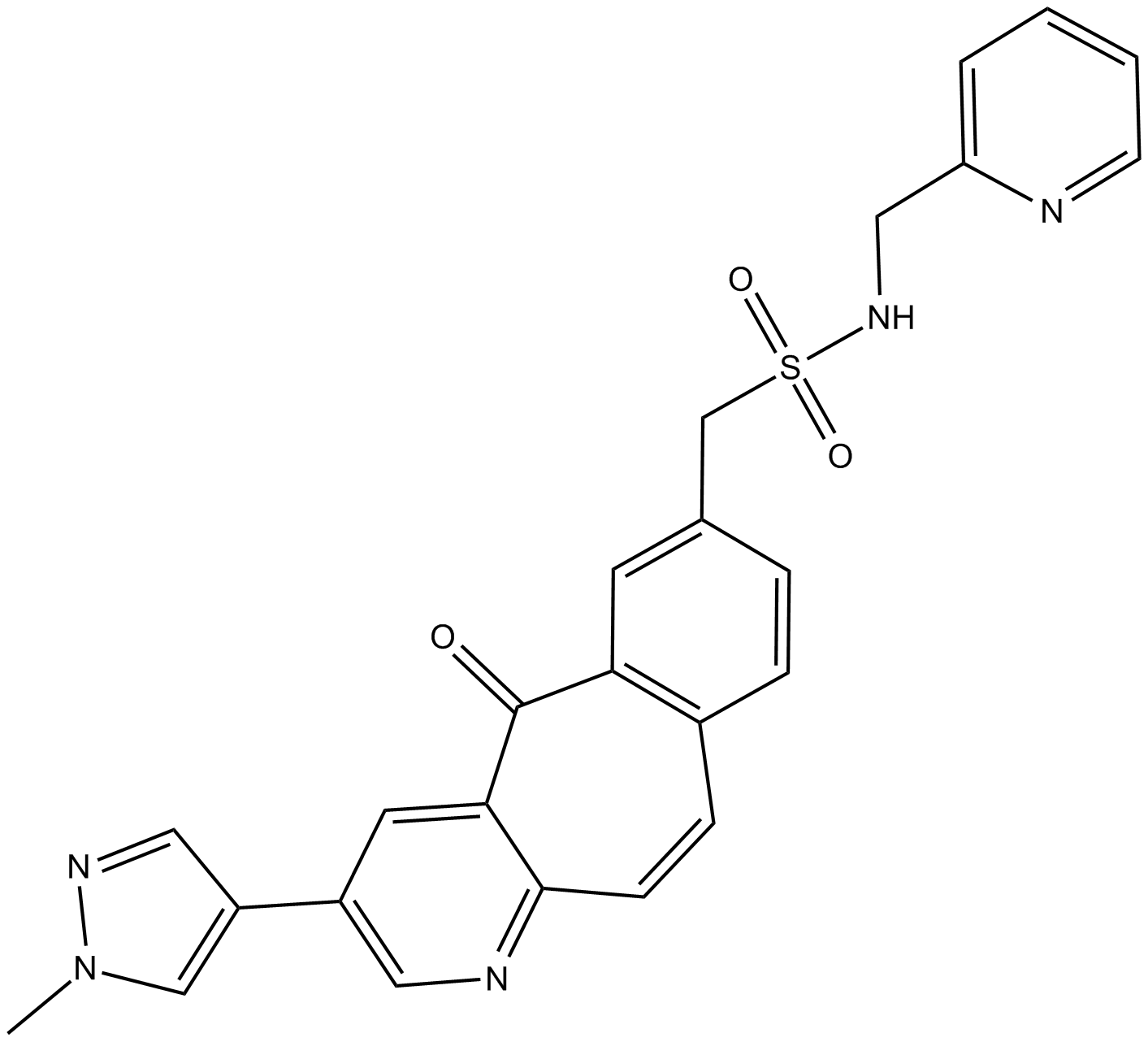
-
GC36625
MK-8033 hydrochloride
MK-8033 hydrochloride is an orally active ATP competitive c-Met/Ron dual inhibitor (IC50s: 1 nM (c-Met),7 nM (Ron)), with preferential binding to the activated kinase conformation. MK-8033 hydrochloride can be used in the research of cancers, such as breast and bladder cancers, non-small cell lung cancers (NSCLCs).
-
GC47687
ML-209
An RORγt antagonist
-
GC17582
ML347
BMP receptor inhibitor,potent and selective
-
GC30769
MLi-2
An LRRK2 inhibitor
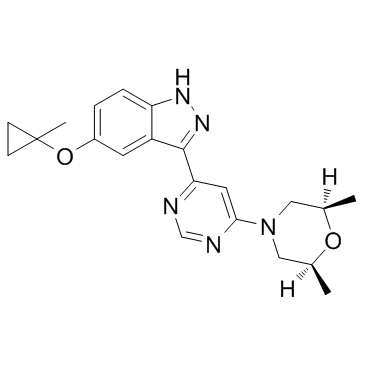
-
GC10775
MLR 1023
MLR 1023 is a potent and selective allosteric activator of Lyn kinase with an EC50 of 63 nM.
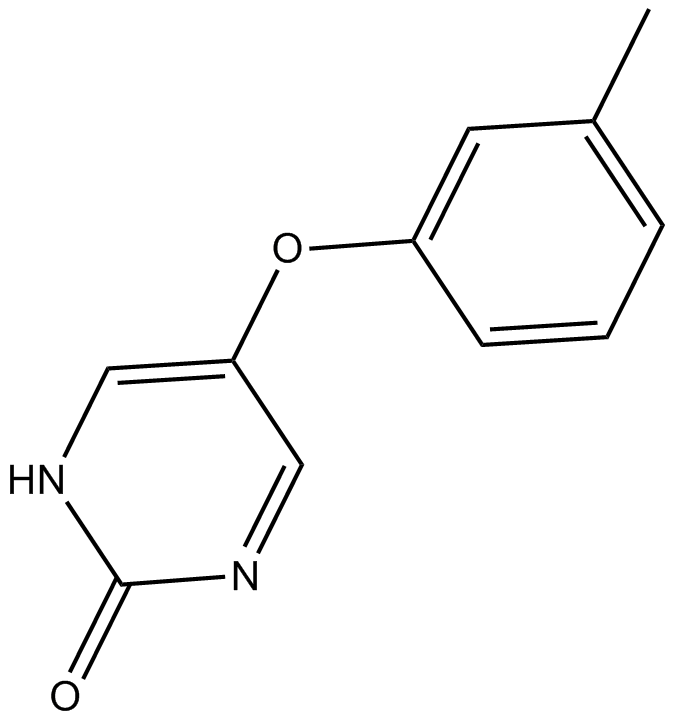
-
GC10048
MNS
Inhibitor of Src/Syk tyrosine kinases
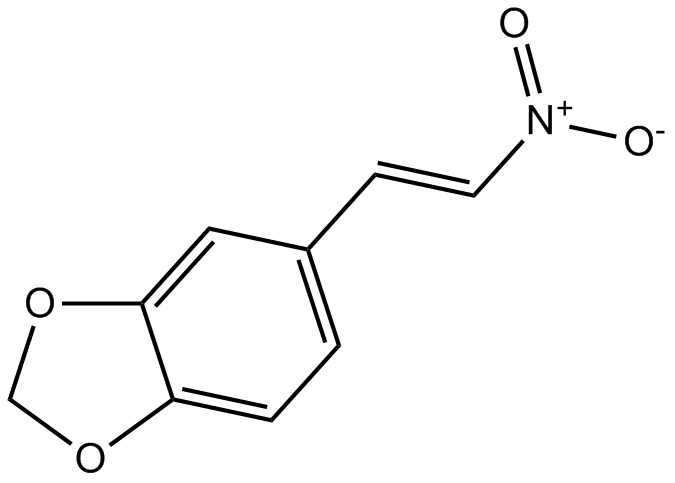
-
GC47697
Mobocertinib
An inhibitor of mutant EGFR and HER2 receptors
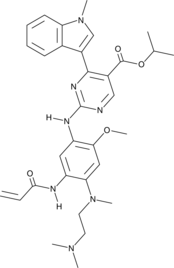
-
GC62160
Mobocertinib succinate
Mobocertinib (TAK-788) succinate is an orally active and irreversible EGFR/HER2 inhibitor. Mobocertinib succinate potently inhibits oncogenic variants containing activating EGFRex20ins mutations with selectivity over wild-type EGFR. Mobocertinib succinate can be used in NSCLC research.
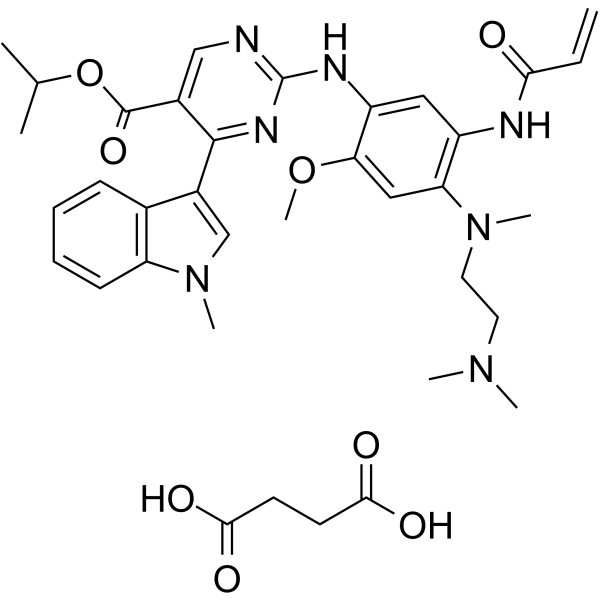
-
GC13012
Motesanib
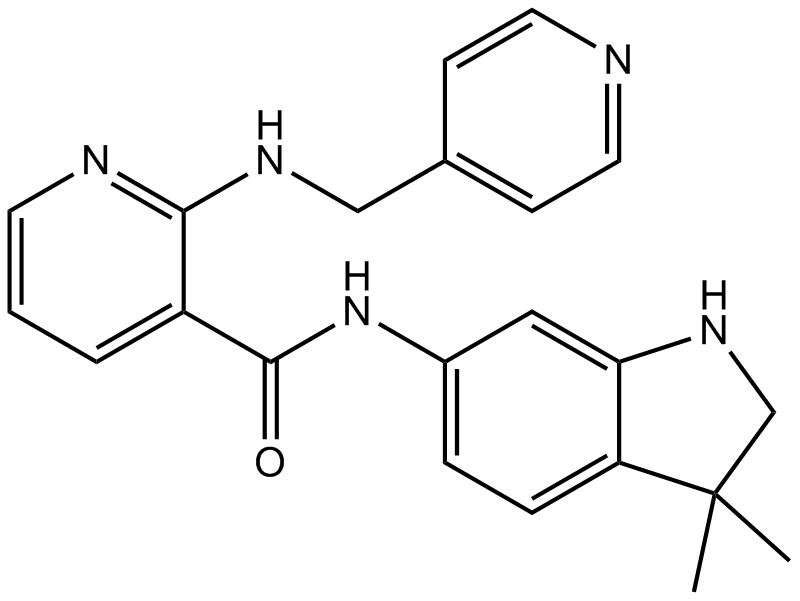
-
GC11336
Motesanib Diphosphate (AMG-706)
Motesanib Diphosphate (AMG-706) (AMG 706 Diphosphate) is a potent ATP-competitive inhibitor of VEGFR1/2/3 with IC50s of 2 nM/3 nM/6 nM, respectively, and has similar activity against Kit, and is approximately 10-fold more selective for VEGFR than PDGFR and Ret.
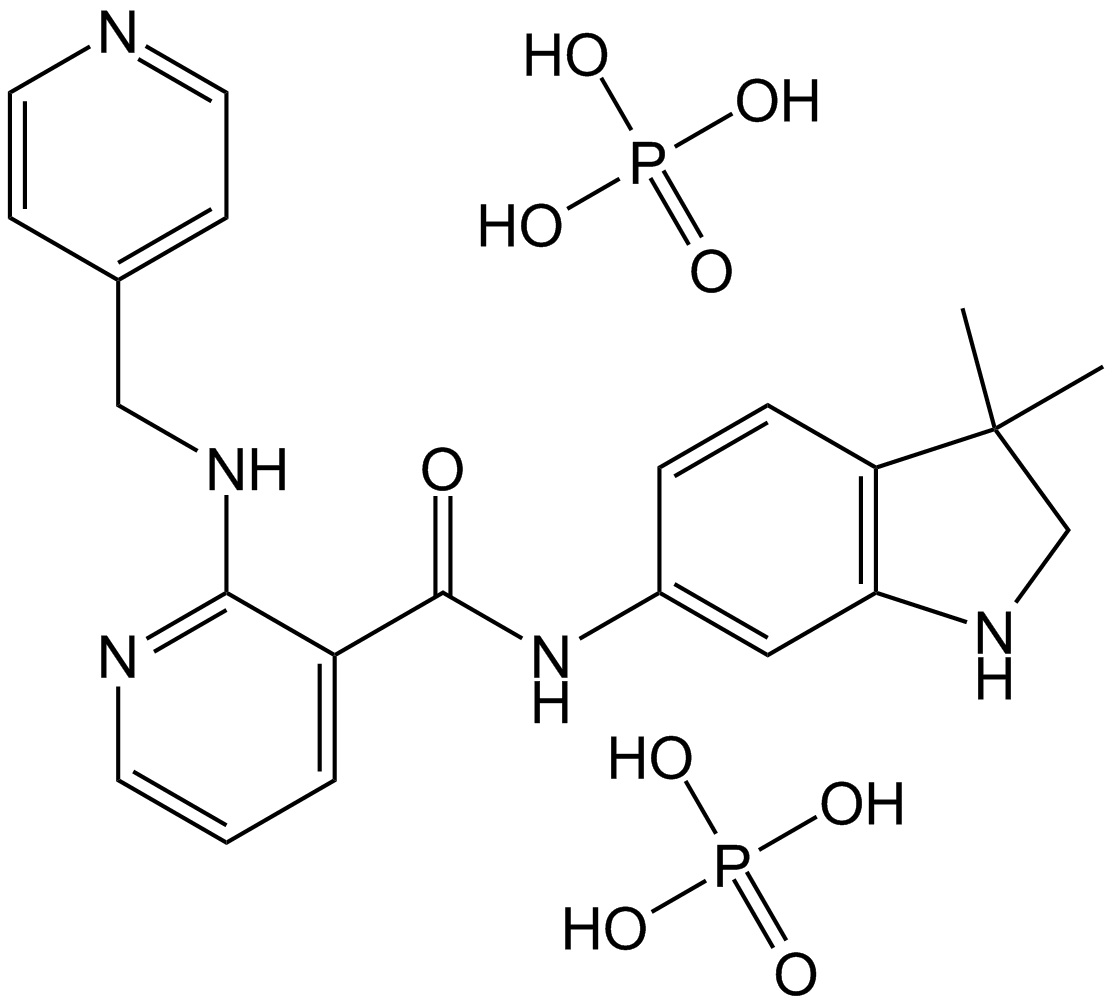
-
GC69496
MRL-871
MRL-871 (compound 3) is an effective inverse agonist of retinoic acid receptor-related orphan receptor gamma t (RORγt), with an IC50 of 264 nM. MRL-871 has a unique imidazole chemical structure and can effectively reduce the generation of IL-17a mRNA in EL4 cells.
-
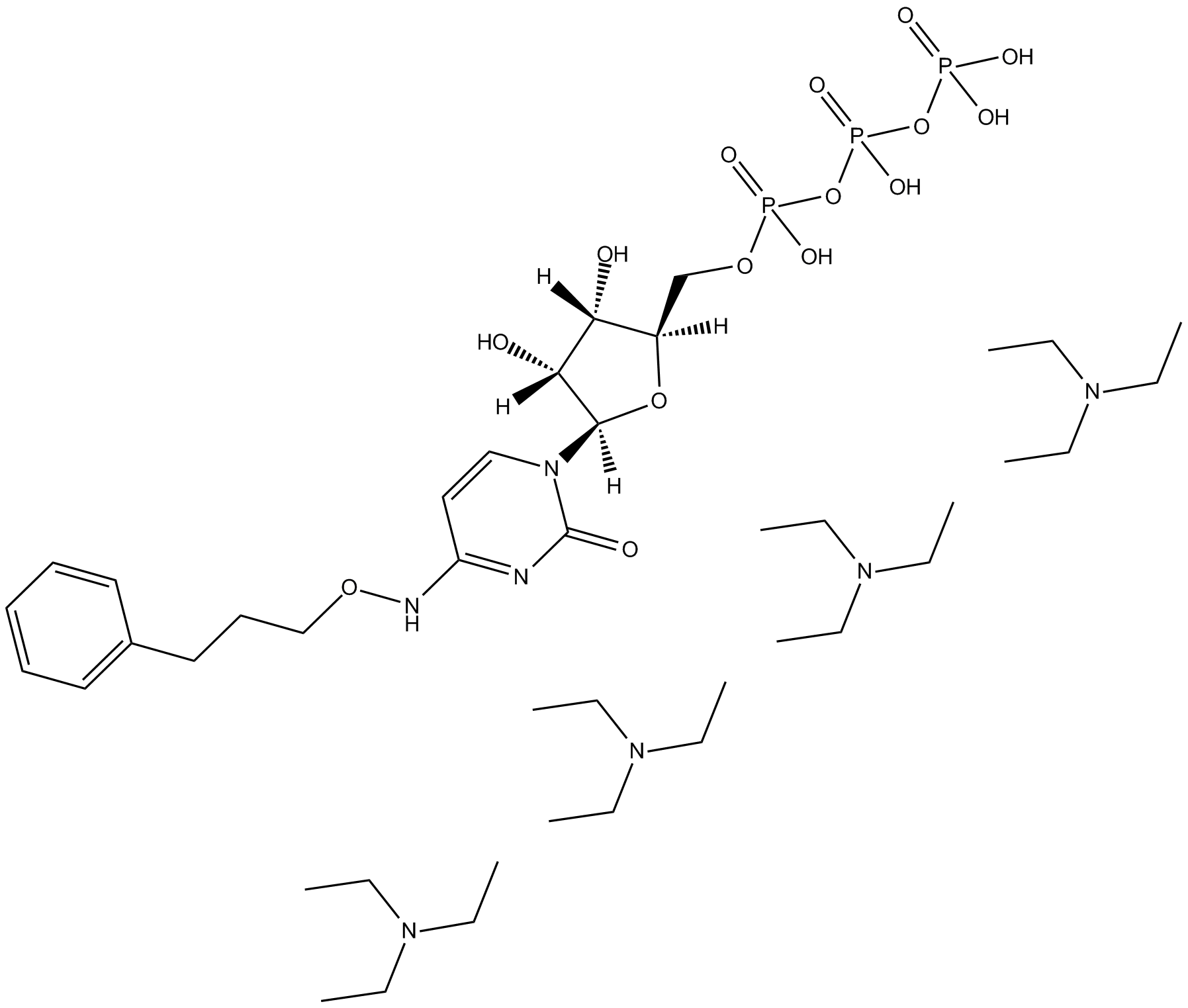
GC13525
MRS 4062 triethylammonium salt
P2Y4 receptor agonist
-
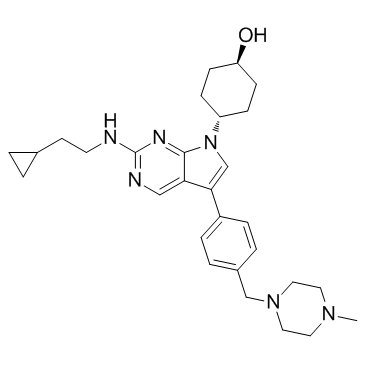
GC32769
MRX-2843 (UNC2371)
MRX-2843 (UNC2371) (UNC2371) is an orally active, ATP-competitive dual MERTK and FLT3 tyrosine kinases inhibitor (TKI) with enzymatic IC50s of 1.3 nM for MERTK and 0.64 nM for FLT3, respectively.
-
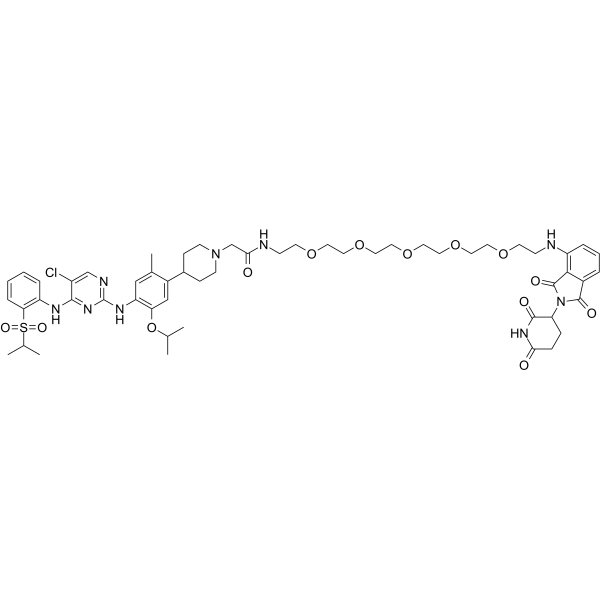
GC65243
MS4077
MS4077 is an anaplastic lymphoma kinase (ALK) PROTAC (degrader) based on Cereblon ligand, with a Kd of 37?nM for binding affinity to ALK.
-
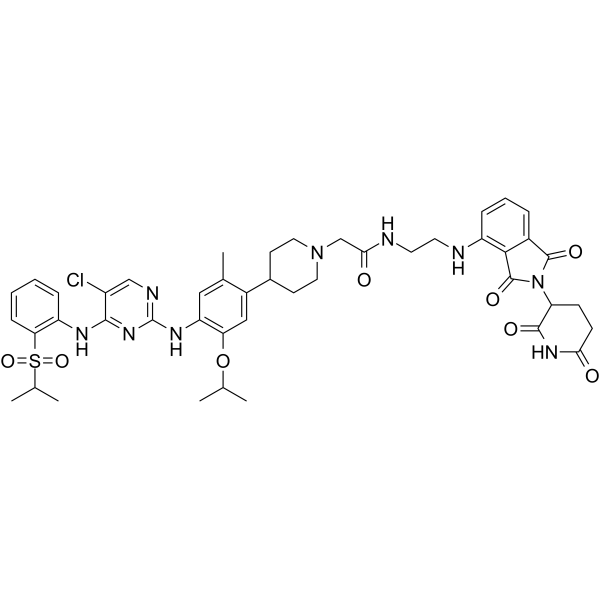
GC64966
MS4078
MS4078 is an anaplastic lymphoma kinase (ALK) PROTAC (degrader) based on Cereblon ligand, with a Kd of 19?nM for binding affinity to ALK.
-
GC15936
MSDC-0160
mTOT-modulating insulin sensitizer
-
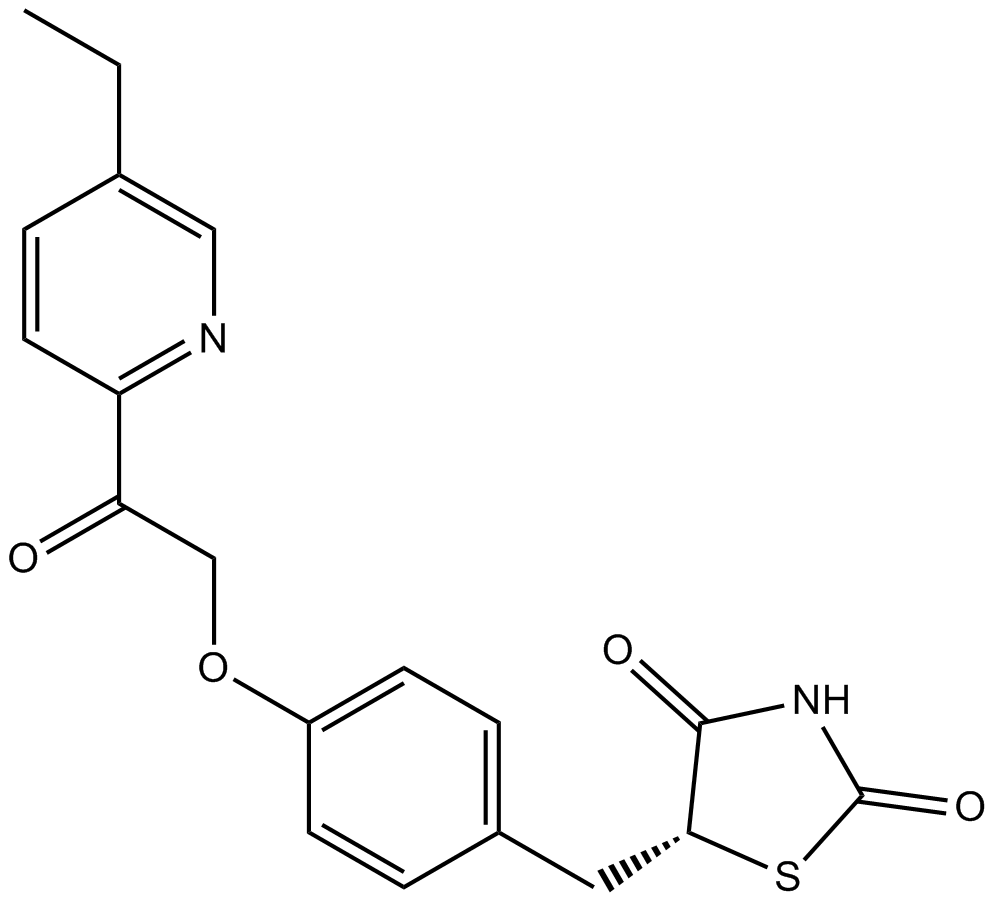
GC64431
MSDC-0602K
MSDC-0602K (Azemiglitazone potassium), a PPARγ-sparing thiazolidinedione (Ps-TZD), binds to PPARγ with the IC50 of 18.25 μM.
-
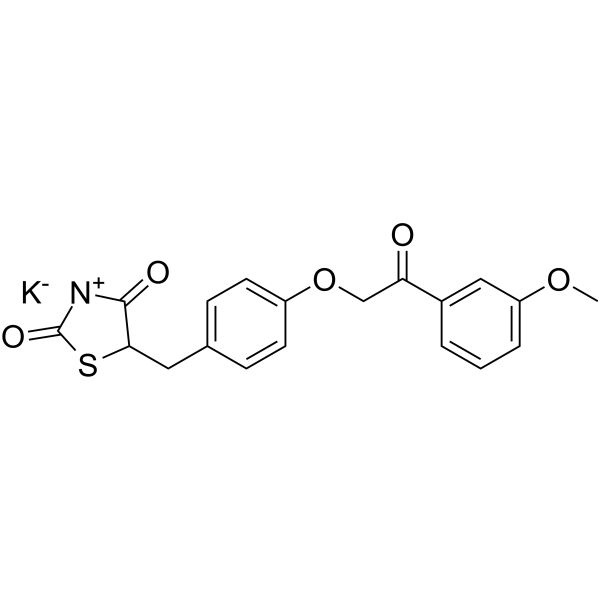
GC19256
MTX-211
MTX-211 is a dual inhibitor of EGFR and PI3K, used for the treatment of cancer and other diseases.
-
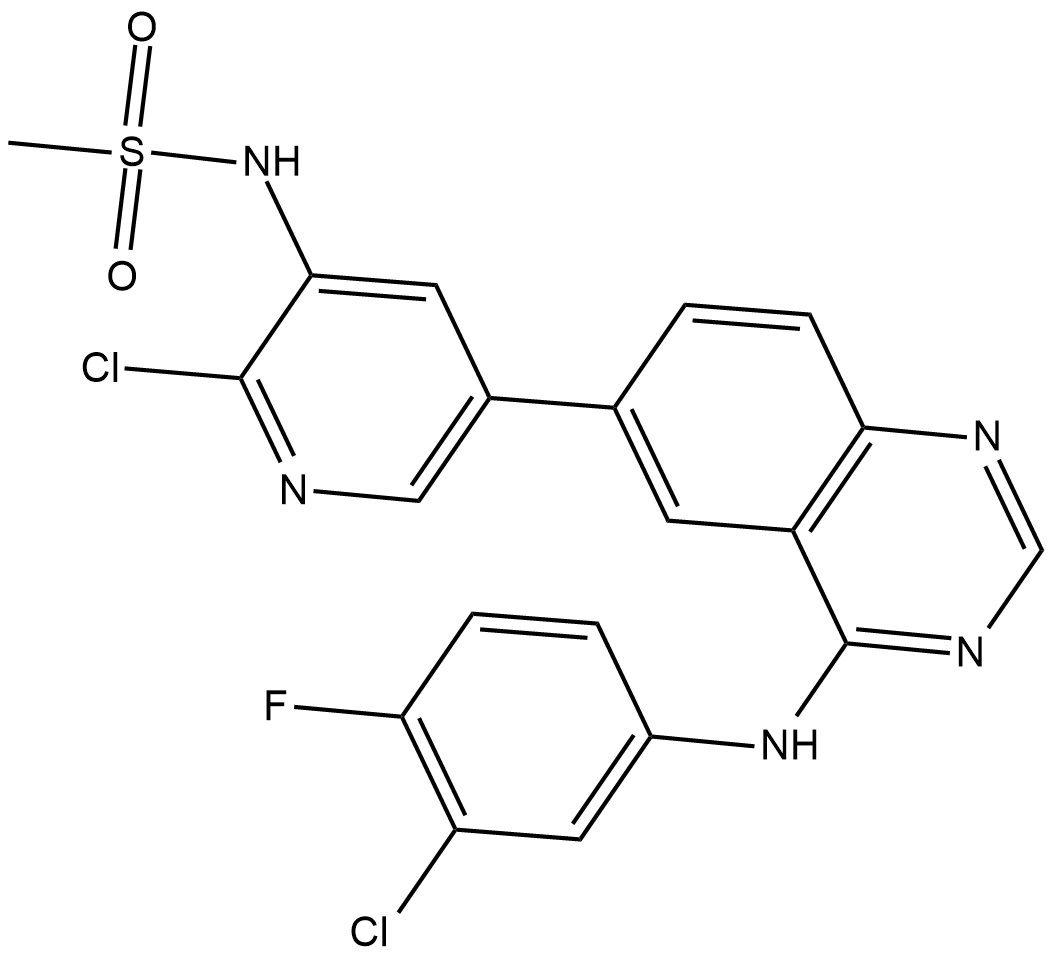
GC10250
Mubritinib (TAK 165)
Mubritinib (TAK 165) (TAK-165) is a potent and selective EGFR2/HER2 inhibitor with an IC50 of 6 nM.
-
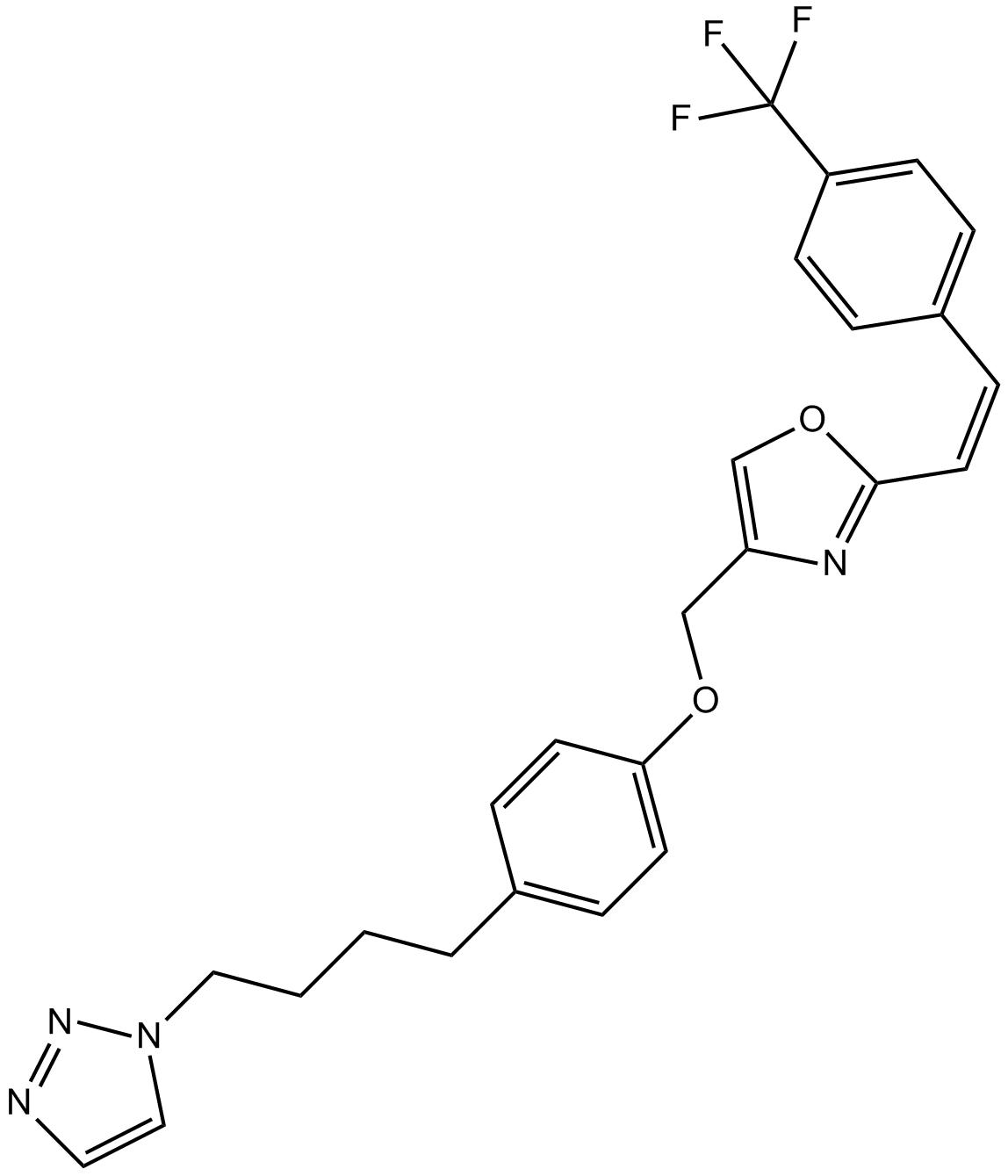
GC11126
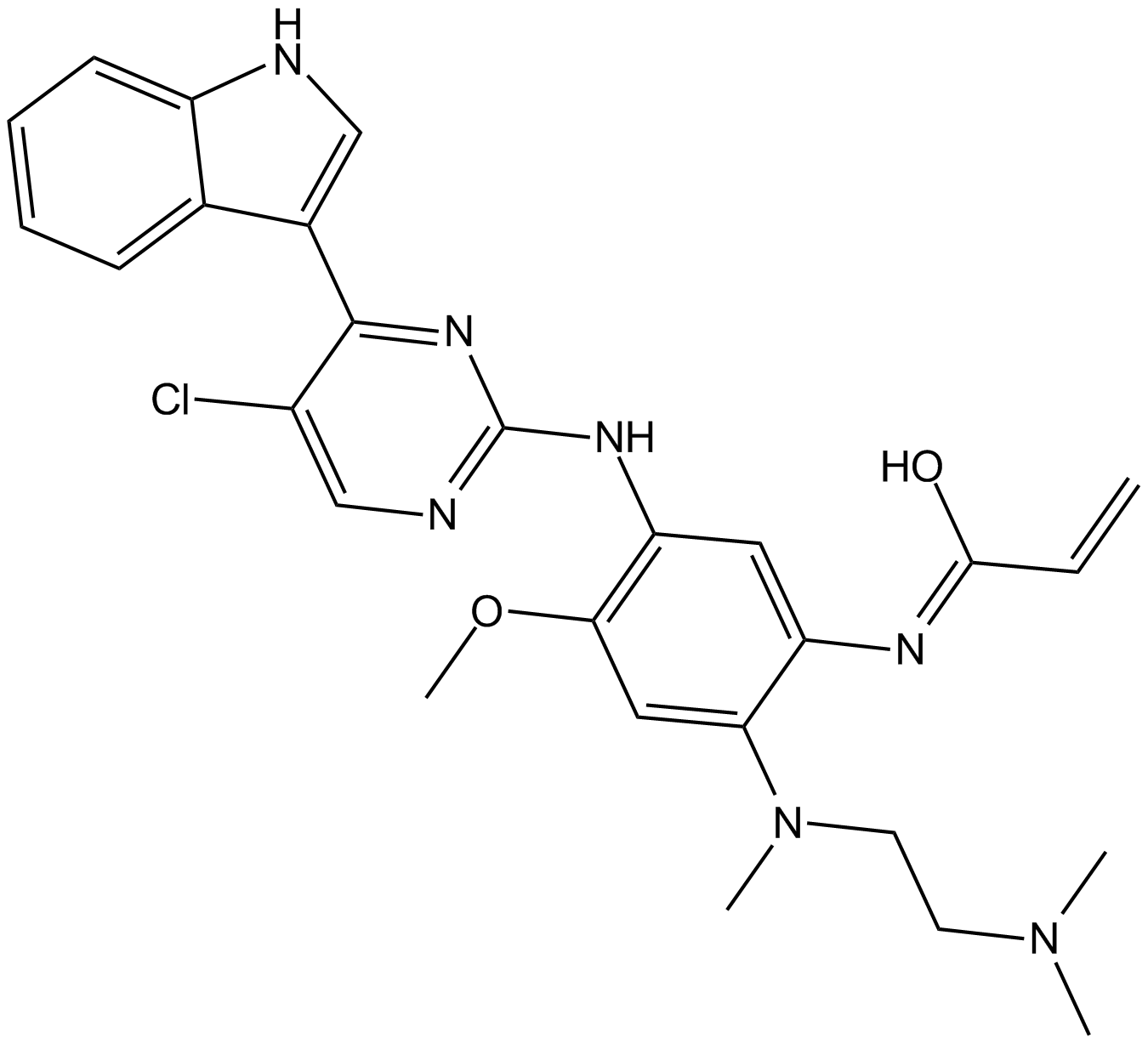
Mutant EGFR inhibitor
Selective Mutated EGFR inhibitor
-
GC36666
Mutated EGFR-IN-1
Mutated EGFR-IN-1 (Osimertinib analog) is a useful intermediate for the inhibitors design for mutated EGFR, such as L858R EGFR, Exonl9 deletion activating mutant and T790M resistance mutant.

-
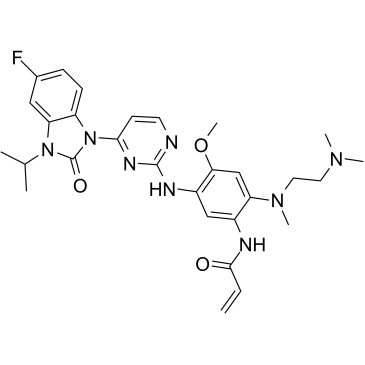
GC36667
Mutated EGFR-IN-2
Mutated EGFR-IN-2 (compound 91) is a mutant-selective EGFR inhibitor extracted from patent WO2017036263A1, which potently inhibits single-mutant EGFR (T790M) and double-mutant EGFR (including L858R/T790M (IC50=<1nM) and ex19del/T790M), and can suppress activity of single gain-of-function mutant EGFR (including L858R and ex19del) as well. Mutated EGFR-IN-2 shows anti-tumor antivity.
-
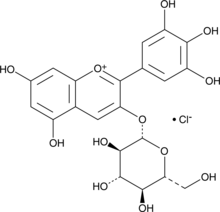
GC44263
Myrtillin
Myrtillin (Delphinidin 3-O-glucoside chloride) is an active anthocyanin found in bilberry extract. Myrtillin induces a pro-apoptotic effect in B cell chronic lymphocytic leukaemia (B CLL). Myrtillin exerts phytoestrogen activity by binding to ERβ, with an IC50 of 9.7 μM. Delphinidin-3-O-glucoside chloride inhibits EGFR with an IC50 of 2.37 μM.
-
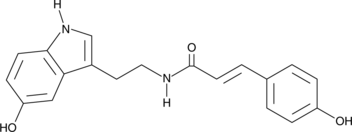
GC40567
N-(p-Coumaroyl) Serotonin
N-(p-Coumaroyl) serotonin is an antioxidative phenolic naturally found in plants, including safflower seed and millet grain.
-
GC11526
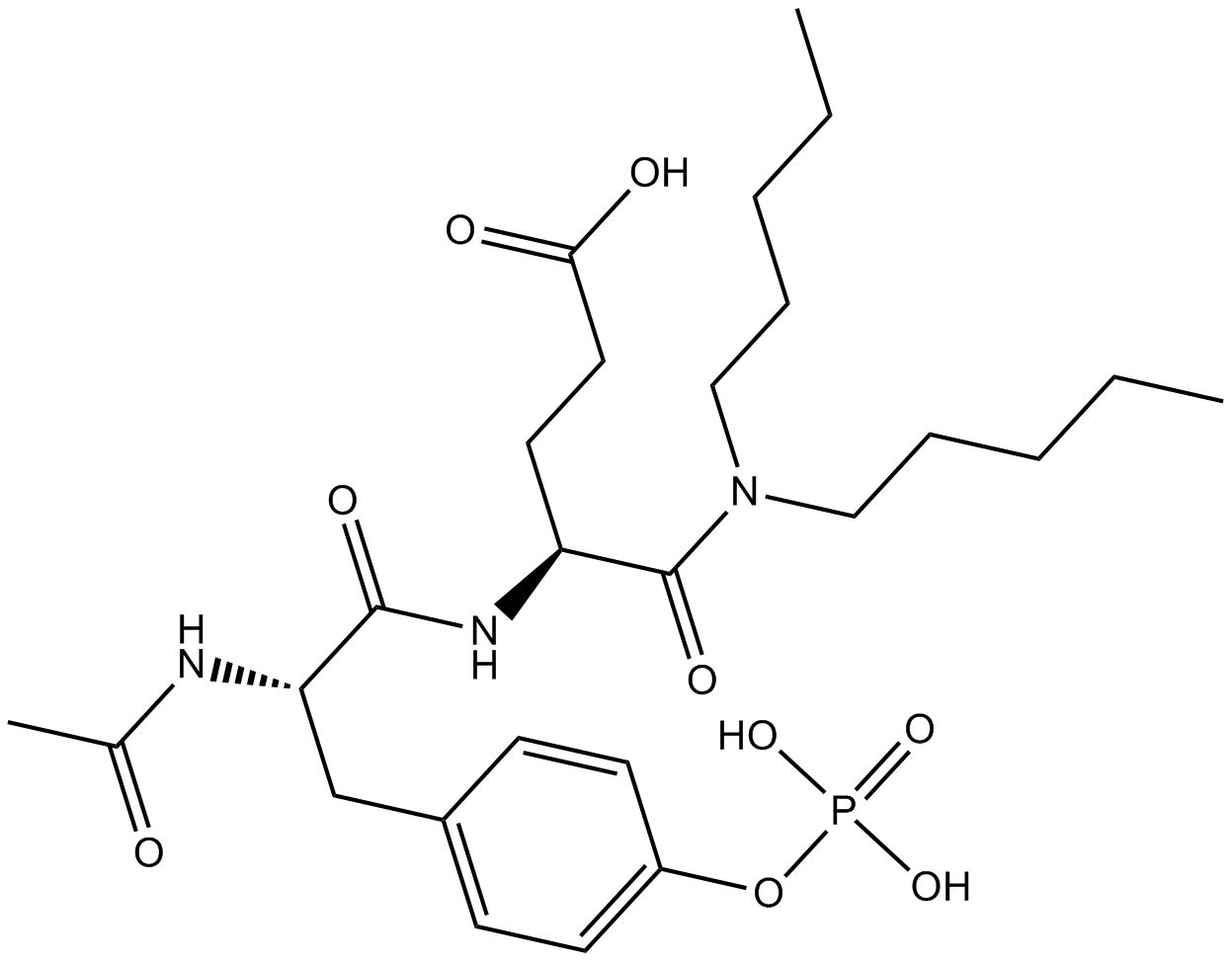
N-Acetyl-O-phosphono-Tyr-Glu Dipentylamide
Phosphopeptide for binding to the src SH2 domain
-
GC12746
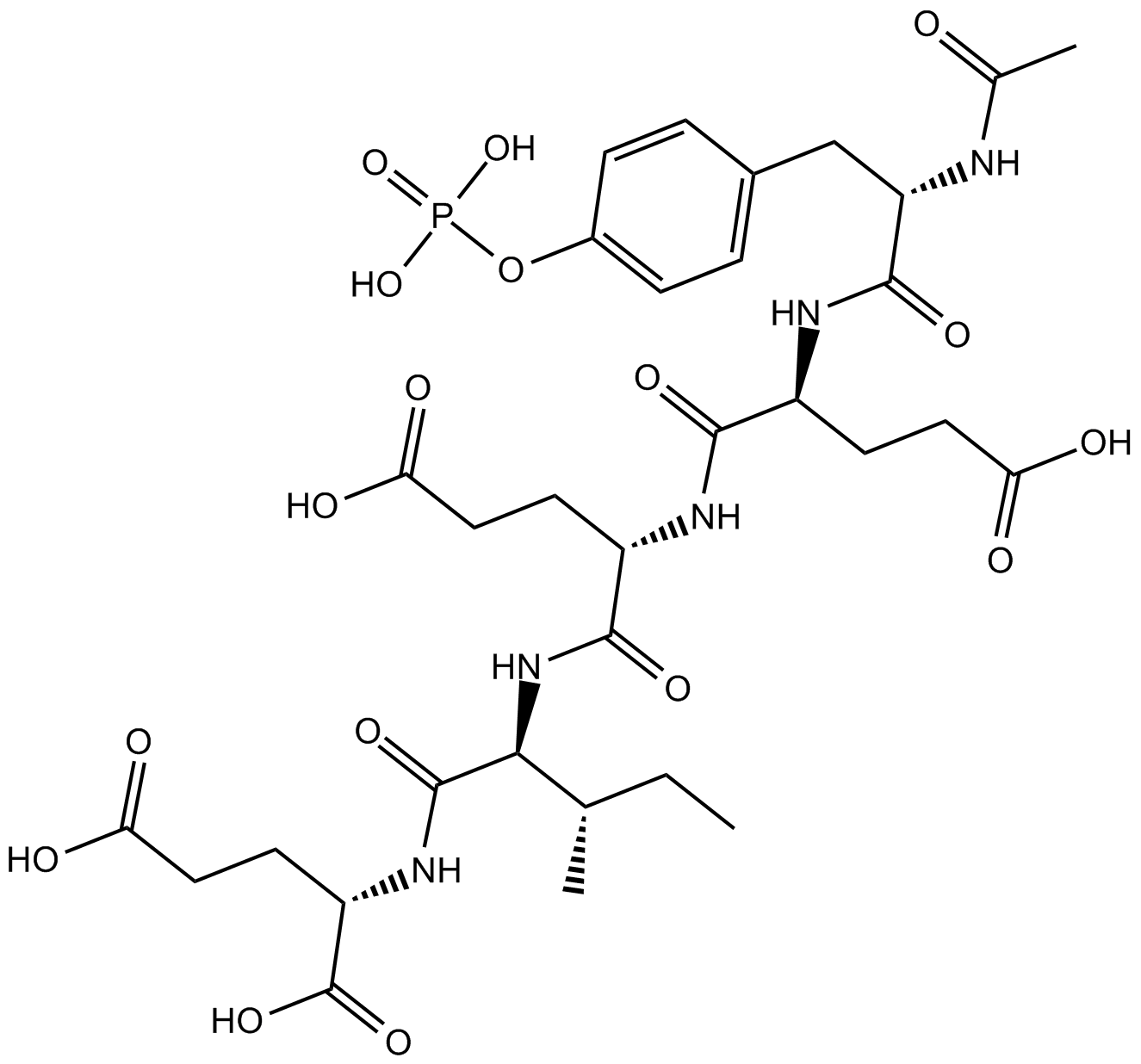
N-Acetyl-O-phosphono-Tyr-Glu-Glu-Ile-Glu
Phosphopeptide ligand for the src SH2 domain
-
GC15365
N-Acetylserotonin
N-Acetylserotonin is a Melatonin precursor, and that it can potently activate TrkB receptor.

-
GC49686
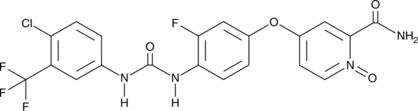
N-desmethyl Regorafenib N-oxide
An active metabolite of regorafenib
-
GC44290
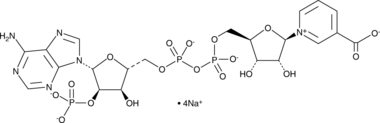
NAADP (sodium salt)
Nicotinic acid adenine dinucleotide phosphate (NAADP) is a secondary messenger that induces calcium mobilization.
-
GC33045
NAMI-A
NAMI-A is a ruthenium-based drug characterised by the selective activity against tumour metastases, inhibits the adhesion and migration.
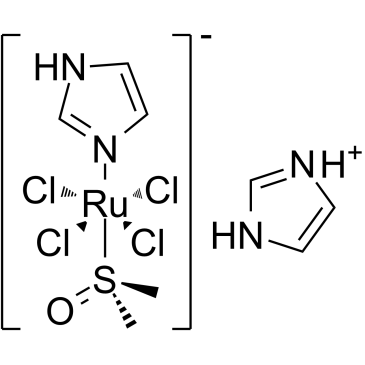
-
GC33022
Naquotinib (ASP8273)
Naquotinib (ASP8273) (ASP8273) is an orally available, mutant-selective and irreversible EGFR inhibitor; with IC50s of 8-33 nM toward EGFR mutants and 230 nM for EGFR.
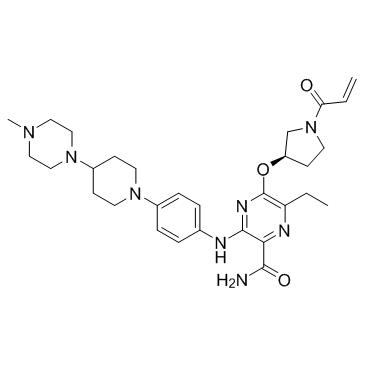
-
GC32836
Naquotinib mesylate (ASP8273)
Naquotinib mesylate (ASP8273) (ASP8273 mesylate) is an orally available, mutant-selective and irreversible EGFR inhibitor; with IC50s of 8-33 nM toward EGFR mutants and 230 nM for EGFR.
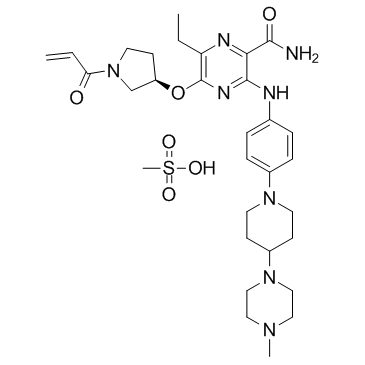
-
GC36699
Nazartinib mesylate
Nazartinib mesylate (EGF816 mesylate) is a novel, covalent mutant-selective EGFR inhibitor, with Ki and Kinact of 31 nM and 0.222 min?1 on EGFR(L858R/790M) mutant, respectively.
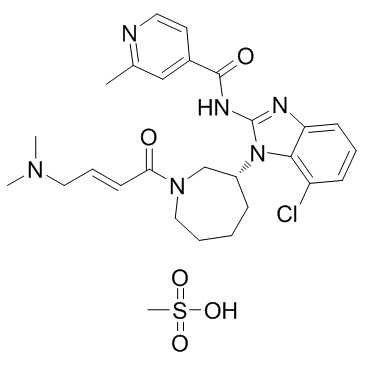
-
GC40623
NBI 31772
NBI 31772 is a nonpeptide ligand that releases insulin-like growth factor-1 (IGF-1) from its binding protein (IGFBP; Kis = 1-24 nM for the six human subtypes of IGFBP).
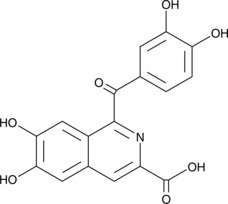
-
GC61111
NBI-31772 hydrate
NBI-31772 hydrate is a potent inhibitor of interaction between insulin-like growth factor (IGF) and IGF-binding proteins (IGFBPs).
-
GC10644
Neoruscogenin
nuclear receptor RORα agonist
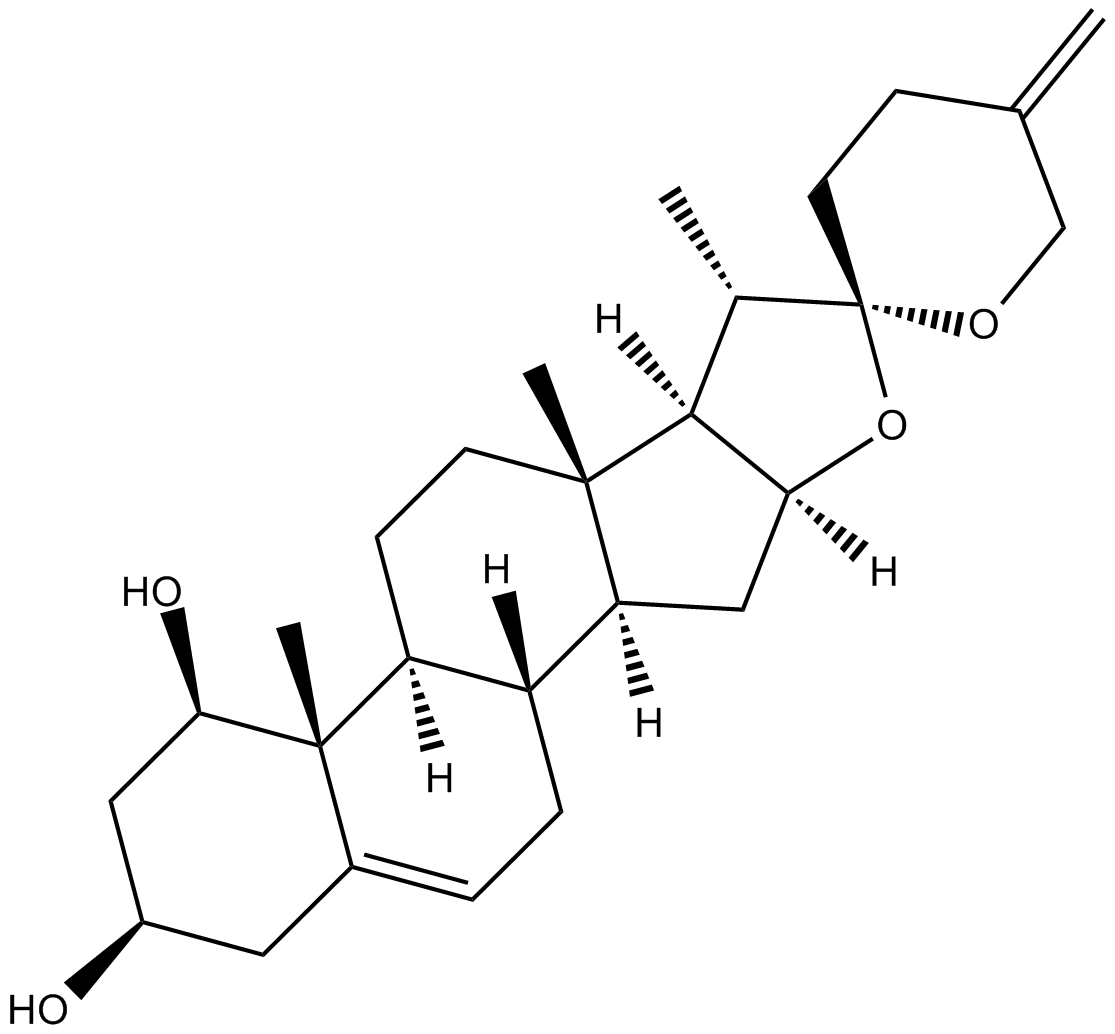
-
GC10362
Neratinib (HKI-272)
Neratinib (HKI-272) (HKI-272) is an orally available, irreversible, highly selective HER2 and EGFR inhibitor with IC50s of 59 nM and 92 nM, respectively.
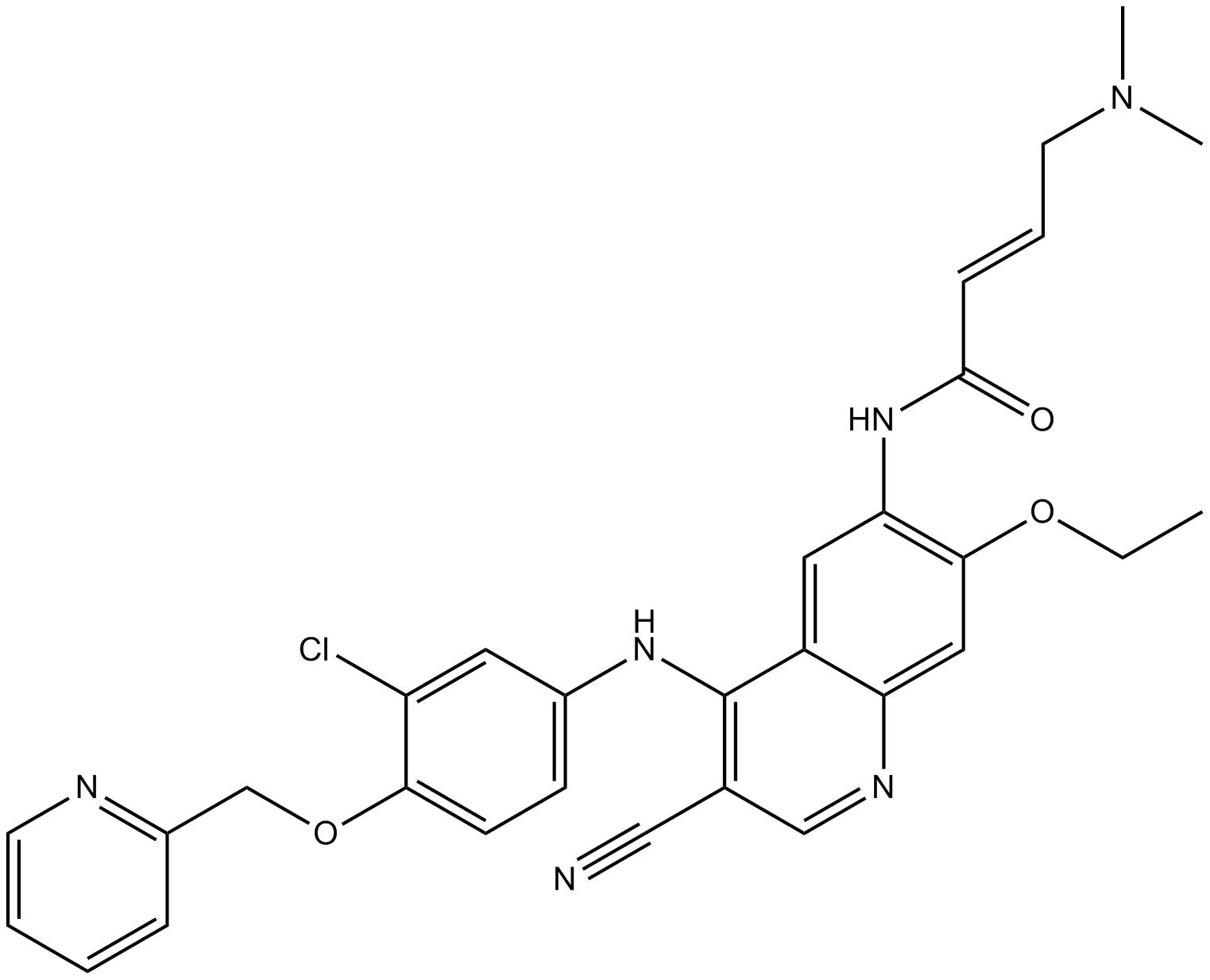
-
GC47771
NG 25 (hydrochloride hydrate)
An inhibitor of MAP4K2 and TAK1
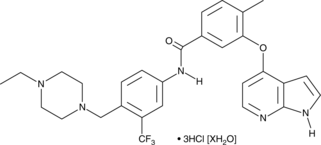
-
GC60270
Nilotinib D6
An internal standard for the quantification of nilotinib
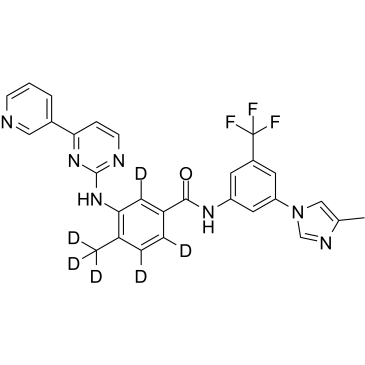
-
GC25669
Nilotinib hydrochloride
Nilotinib hydrochloride (AMN-107) is the hydrochloride salt form of nilotinib, an orally bioavailable Bcr-Abl tyrosine kinase inhibitor with antineoplastic activity.
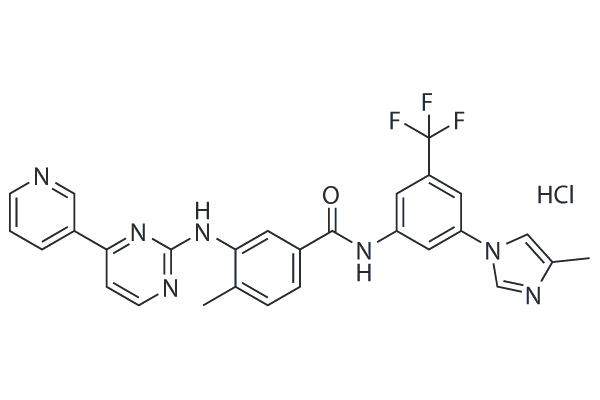
-
GC14237
Nilotinib monohydrochloride monohydrate
A Bcr-Abl inhibitor
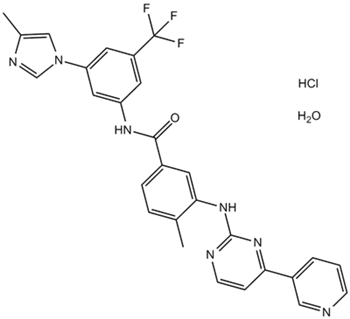
-
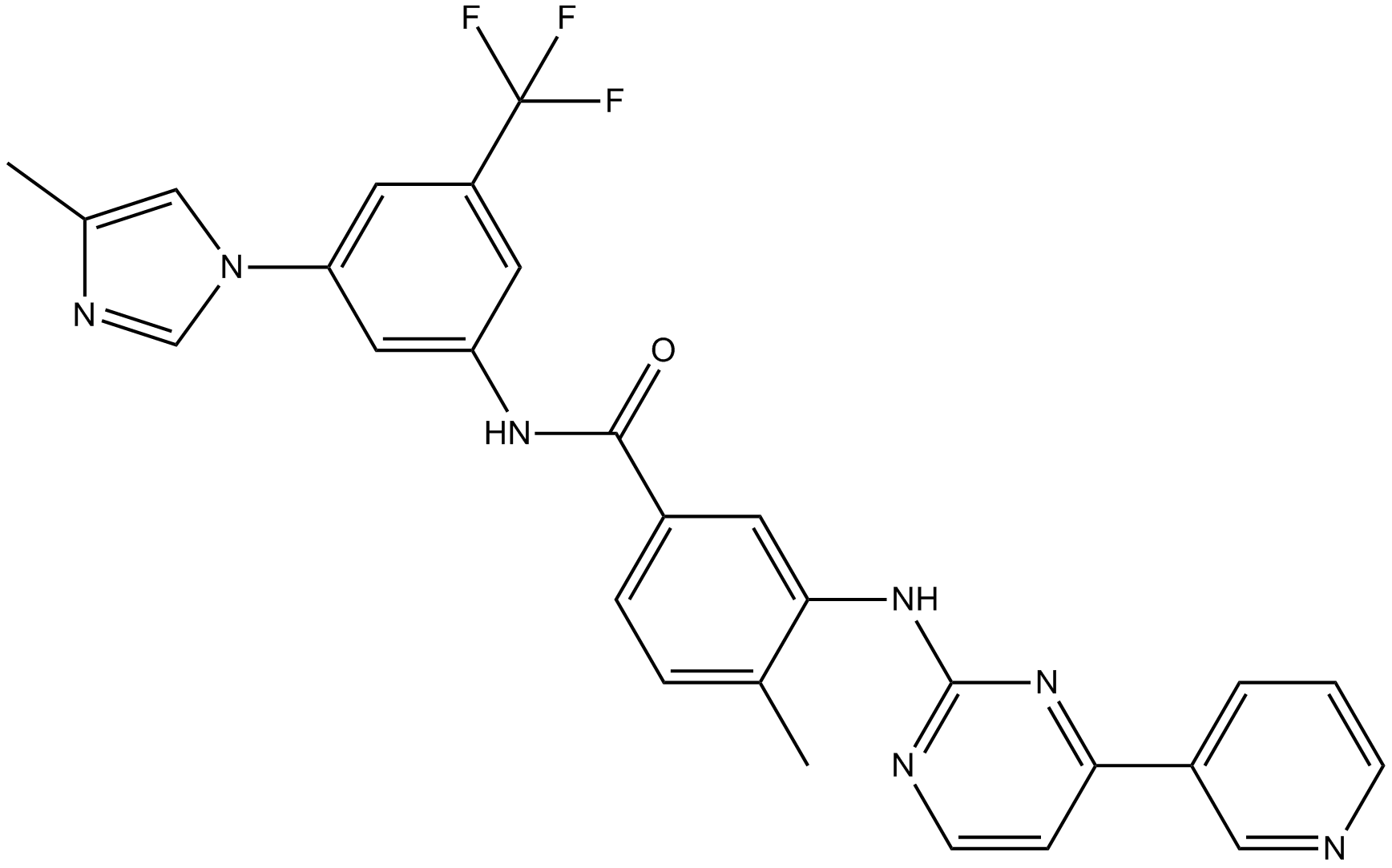
GC14129
Nilotinib(AMN-107)
A Bcr-Abl inhibitor
-
GC62601
Nimotuzumab
Nimotuzumab is a humanized IgG1 monoclonal antibody targeting EGFR with a KD of 0.21 nM. Nimotuzumab is directed against the extracellular domain of the EGFR blocking the binding to its ligands. Nimotuzumab, a strong antitumor drug, is cytolytic on target tumors by its capacity to cause antibody dependent cell mediated cytotoxicity (ADCC) and complement dependent cytotoxicity (CDC).

-
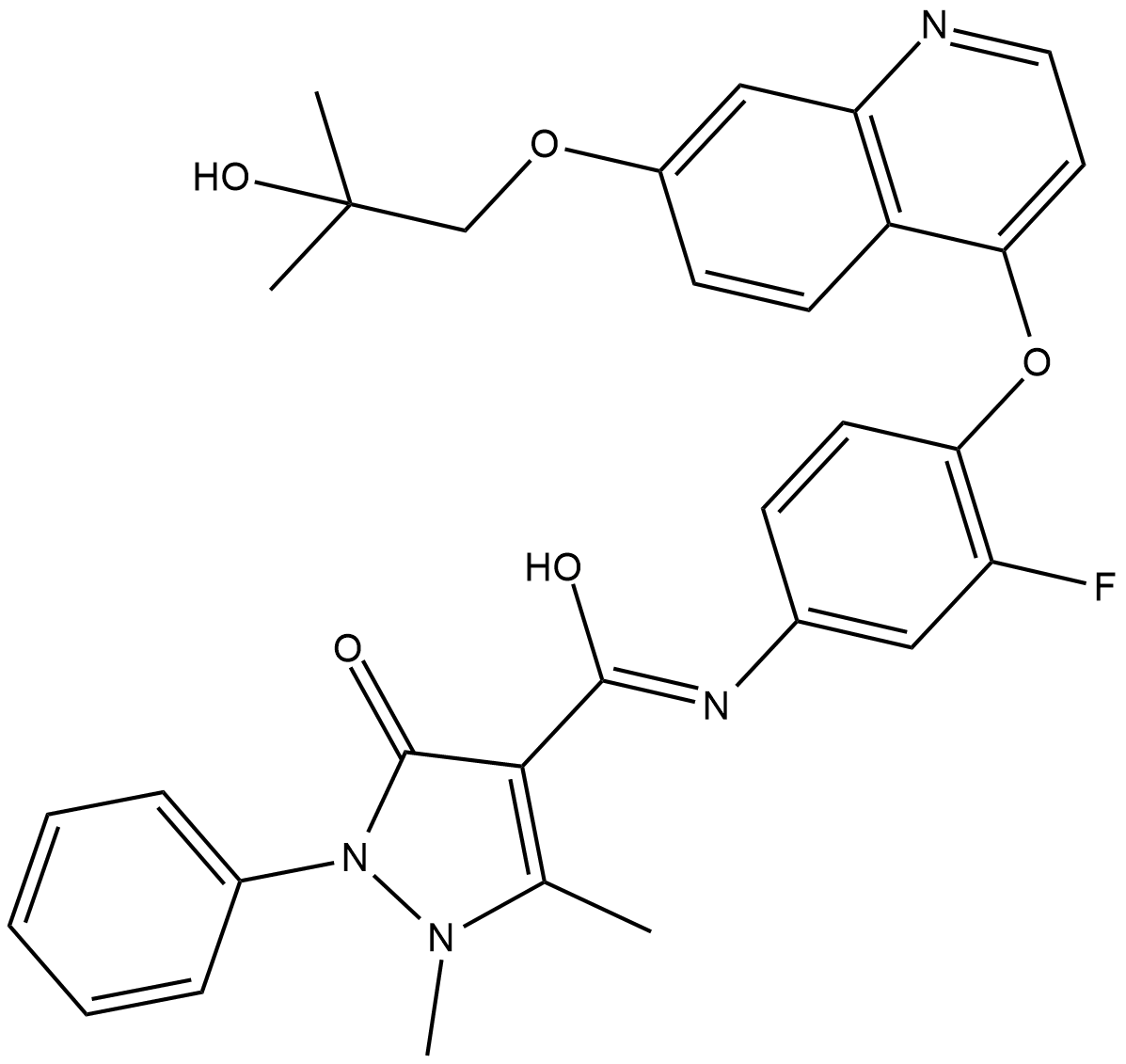
GC18211
Ningetinib
A multi-kinase inhibitor
-
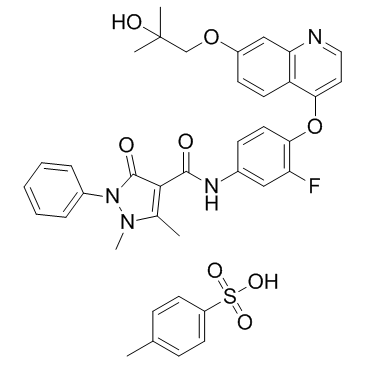
GC36744
Ningetinib Tosylate
Ningetinib Tosylate is a potent, orally bioavailable small molecule tyrosine kinase inhibitor (TKI) with IC50s of 6.7, 1.9 and <1.0 nM for c-Met, VEGFR2 and Axl, respectively.
-
GC11705
Nintedanib (BIBF 1120)
A VEGFR, FGFR, and PDGFR inhibitor